Abstract
Before squamous cell lung cancer develops, pre-cancerous lesions can be found in the airways. From longitudinal monitoring, we know that only half of such lesions become cancer, whereas a third spontaneously regress. While recent studies have described the presence of an active immune response in high-grade lesions, the mechanisms underpinning clinical regression of pre-cancerous lesions remain unknown. Here, we show that host immune surveillance is strongly implicated in lesion regression. Using bronchoscopic biopsies from human subjects, we find that regressive carcinoma in-situ lesions harbour more infiltrating immune cells than those that progress to cancer. Moreover, molecular profiling of these lesions identifies potential immune escape mechanisms specifically in those that progress to cancer: antigen presentation is impaired by genomic and epigenetic changes, CCL27/CCR10 signalling is upregulated, and the immunomodulator TNFSF9 is downregulated. Changes appear intrinsic to the CIS lesions as the adjacent stroma of progressive and regressive lesions are transcriptomically similar.
Introduction
Before the development of lung squamous cell carcinoma (LUSC), pre-invasive lesions can be observed in the airways. These evolve stepwise, progressing through mild and moderate dysplasia (low-grade lesions) to severe dysplasia and carcinoma in-situ (CIS; high-grade lesions), before the development of invasive cancer(1). In cross-sectional studies, markers of immune sensing and escape have been associated with increasing grade(2). However, longitudinal bronchoscopic surveillance of such lesions has shown that progression of pre-invasive lesions to cancer is not inevitable; only half of high-grade CIS lesions will progress to cancer within two years, whereas a third will spontaneously regress(3). Our previous work defined the genomic, transcriptomic and epigenetic landscape of carefully phenotyped airway CIS lesions(4). Here, we combine these data with immunohistochemistry (IHC), imaging and transcriptomic analysis of adjacent stroma (Table S1; Figure S1) to assess the role of immune surveillance in lesion regression. We identify key immune escape mechanisms enriched in pre-invasive lesions which later progressed to cancer. Understanding these mechanisms may offer new therapeutic strategies to induce regression and prevent the development of invasive disease.
Results
To assess our hypothesis that lesion regression is driven by immune surveillance, we used a deep-learning approach(5) to quantify lymphocytes from hematoxylin and eosin (H&E) stained slides in a large dataset of 112 samples from 62 patients, which contained more infiltrating lymphocytes in regressive lesions than progressive (Figure 1a; p=0.049). We next performed immunohistochemistry (IHC) on 28 progressive and 16 regressive CIS lesions from 29 patients (Figure 1b-c). Regressive lesions showed higher concentrations of intra-lesional CD8+ cytotoxic T-cells (Figure 1a; p=0.055) but no significant difference in CD4+ T helper cells (p=0.26) or FOXP3+ regulatory T cells (p=0.42). We then quantified immune cells in stromal regions adjacent to CIS lesions, but found no significant differences between progressive and regressive lesions for CD8+ (p=0.50), CD4+ (p=0.43) or FOXP3+ (p=0.64) cells. We confirmed these findings in an independent dataset of 19 progressive and 9 regressive samples subjected to multiplex IHC(6,7) (mIHC) using a wider antibody panel of lymphoid biomarkers (Table S2), in which we again observed that regressive lesions had an increased proportion of infiltrating lymphocytes (Figure 2a; p=0.032). Specifically, regressive lesions showed significantly more infiltrating CD3+CD8+ cytotoxic T-cells (p=0.017) but no significant difference in CD3+CD4+ T helper cells (p=0.18), T regulatory cells (p=0.12), B-cells (p=0.12), macrophages (p=0.79) or neutrophils (p=0.53). In the mIHC cohort, the proportion of CD3+CD8+ cells positive for granzyme B and EOMES was similar between progressive and regressive lesions (p=0.63 and p=0.18 respectively) which may indicate that disruption of T-cell infiltration into lesions has a greater impact on their capacity for immune evasion than impairment of cytotoxic function or differentiation. Again, stromal regions in this cohort showed no significant differences between progressive and regressive lesions.
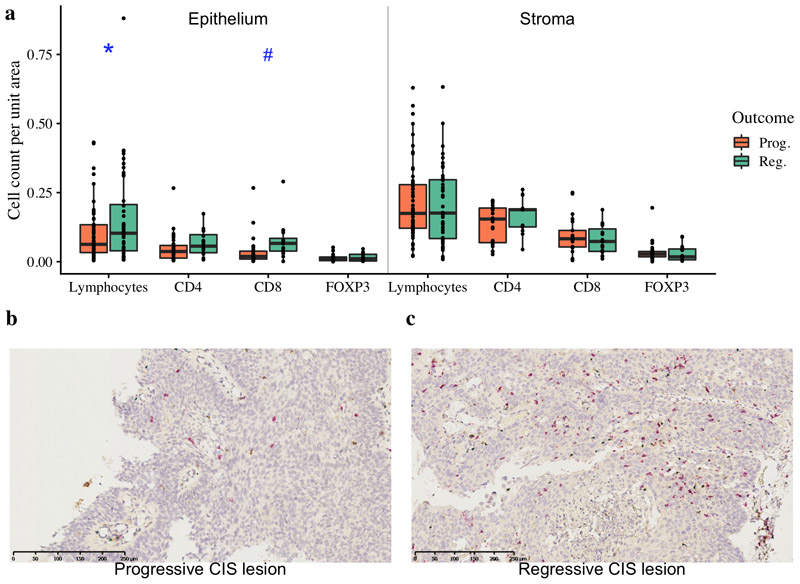
Figure 1. Immune cell infiltration of lung carcinoma-in-situ lesions.
a) Combined quantitative immunohistochemistry data of CD4, CD8 and FOXP3 staining (n=44; 28 progressive, 16 regressive) with total lymphocyte quantification from H&E images (n=112; 68 progressive, 44 regressive) shown. We observe increased lymphocytes (p=0.049) and CD8+ cells (p=0.055) per unit area of epithelium within regressive CIS lesions compared to progressive. Stromal regions adjacent to CIS lesions showed no significant differences in immune cells between progressive and regressive lesions. p-values are calculated using linear mixed effects models to account for samples from the same patient; #p<0.1, *p<0.05. (b-c) Immunohistochemistry images of (b) progressive CIS lesion and (c) regressive CIS lesion with CD4+ T helper cells stained in brown, CD8+ cytotoxic T-cells in red and FOXP3+ T regulatory cells in blue. Immune cells are separately quantified within the CIS lesion and in the surrounding stroma.
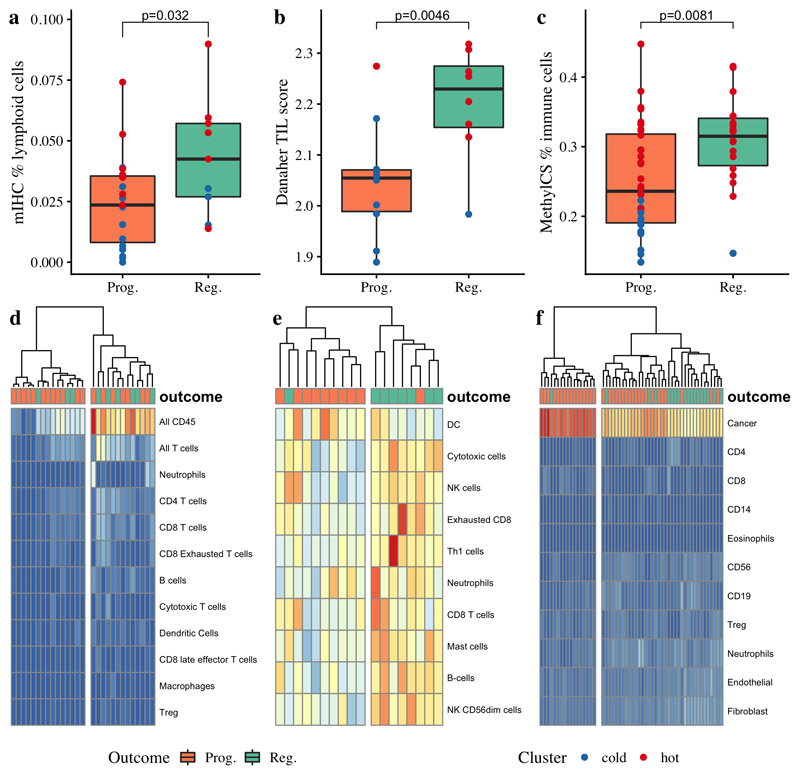
Figure 2. Identification of immune ‘hot’ and ‘cold’ carcinoma in-situ lesions by immune cell clustering.
Regressive lesions harbored significantly more infiltrating lymphocytes as assessed by multiplex immunohistochemistry (a; p=0.032 comparing percentage of all nucleated cells identified as T-cells (CD45+CD3+) or B-cells (CD45+CD3-CD20+) between 19 progressive and 9 regressive lesions). This finding was corroborated by molecular data in partially overlapping datasets; regressive lesions had higher gene-expression derived Tumor Infiltrating Lymphocyte (TIL) scores (b; p=0.0046; n=10 progressive, 8 regressive) and a higher proportion of immune cells as estimated from methylation data using methylCIBERSORT (c; p=0.0081; n=36 progressive, 18 regressive). d) Immune cell quantification from IHC data (n=28) shows an ‘immune cold’ cluster (left) in which most lesions progressed to cancer, and an ‘immune hot’ cluster (right) in which the majority regressed. Similar clustering patterns are seen in deconvoluted gene expression data (e; n=18) and on methylation-derived cell subtypes using methylCIBERSORT (f; n=54). p-values are calculated using mixed effects models to account for samples from the same patient.
For a broader assessment of transcriptomic differences between CIS lesions and their adjacent stroma, we isolated epithelial tissue and paired stroma separately using laser capture microdissection for 10 progressive and 8 regressive CIS lesions. Similarly to IHC data, cell type deconvolution analysis using the Danaher method(8) demonstrated higher infiltrating lymphocytes within regressive lesions (Figure 2b; p=0.0046), as did deconvolution of methylation data from 36 progressive and 18 regressive CIS lesions using methylCIBERSORT(9) (Figure 2c; p=0.0081). Comparing predictions for individual cell types across gene expression and methylation data found an increase in most immune cell types in regressive lesions compared to progressive (Table S3).
Analysis of cytokines classically considered to be pro- or anti-inflammatory within the epithelial compartment (Table S4) demonstrated an increase in pro-inflammatory (p=3.7x10-4) but not anti-inflammatory (p=0.32) response in regressive lesions compared to progressive (Figure S2a-f). IL2, TNF, IL12A and IL23A were all increased in regressive lesions (Figure S3a-b; FDR=0.0081, FDR=0.00051, FDR=0.00078, FDR=0.011 respectively). Only CXCL8 was upregulated in progressive samples compared to regressive (FDR=0.0063); produced by macrophages, the expression of CXCL8 correlated strongly with macrophage quantification from deconvoluted gene expression data (r2=0.62, p=0.007). Taken together, these data are in keeping with a model in which inflammation via pathways including IL-2 and TNF fosters effective immune surveillance, whilst lesion-associated macrophages – similar to tumor-associated macrophages in advanced cancers – have an immunosuppressive effect.
Given the well-known immunosuppressive effects of smoking, we hypothesised that patients who were current smokers were more likely to show reduced immune infiltrate and therefore a higher chance of progression. Smoking status was available for 132 CIS lesions from 59 patients (24 lesions from 13 current smokers; 104 from 43 former smokers; 4 from 3 never smokers; Figure S4a-j). Using a Cochrane-Armitage test to look for a trend from Current to Former to Never smokers, we found a trend towards higher chance of regression (p=0.002) and more infiltrating lymphocytes (p=0.095). This trend is still observed, yet no longer statistically significant, using a bootstrapping method to account for samples from the same patient (p=0.069 for regression; p=0.12 for infiltrating lymphocytes). Interestingly, within the former-smoker group we did not observe increasing lymphocytes or chance of regression with increasing time since quitting smoking, suggesting that the observed differences in outcome are driven by the active process of smoking and its direct effects on the immune response, rather than by chronic processes of airway remodelling and repair(10).
Recent advances have demonstrated heterogeneity of lung cancer immune infiltration, with patients whose tumors have predominantly infiltrated ‘immune hot’ regions having improved survival as compared to those with abundant poorly infiltrated, ‘immune cold’ regions(11,12). Hierarchical clustering of immune cell quantification by mIHC and by deconvolution of both transcriptomic and epigenetic data demonstrated clear clusters of ‘cold’ lesions, almost all of which progressed to cancer (Figure 2d-f). However, we also observed some ‘hot’ progressive lesions, suggesting the presence of other immune evasion mechanisms in these lesions. We therefore sought to address two questions: firstly, could deficits in antigen presentation and immune recruitment in progressive lesions be identified, which could explain the observed ‘cold’ lesions? Secondly, could disordered immune cell function explain the existence of progressive immune ‘hot’ lesions?
The acquisition of mutations that result in clonal neoantigens drives T cell immunoreactivity in cancer(13). We hypothesised that immune-active regressive lesions may contain more neoantigens than progressive lesions, however, this was not supported by whole-genome sequencing data(4) (n=39). Predicted neoantigens correlated very closely with mutational burden (r2=0.94), and progressive lesions have been shown to have significantly higher mutational burden than regressive lesions(4), therefore more neoantigens were identified in progressive than regressive lesions (Figure S5a-b; p=0.088). This remained true when the analysis was limited to clonal neoantigens (Figure S5c; p=0.023) and there was no difference in the proportion of neoantigens that were clonal (Figure S5d; p=0.76). Further, there were no significant differences in binding affinity (p=0.46) or differential agretopicity index(14)(p=0.58) and the ratio of observed to expected neoantigens (“depletion score”(15)) was not significantly different (Figure S5e-h; p=0.94), therefore the putative neoantigens themselves were not qualitatively different in the regressive group. The increased number of neoantigens identified in progressive lesions suggests that immune escape mechanisms must be active in these lesions; indeed, these antigens may act as a selection pressure to promote the development of immune escape(16). Importantly, no overlap in putative tumor neoantigens was observed between different patients suggesting that vaccine-based approaches aiming to prevent progression will most likely need to be designed on a personalised basis.
Given that neoantigens are present in progressive lesions, we assessed the ability of these lesions to present antigens to the immune system. In cancer, genomic alterations have previously been associated with modulation of immune response(17,18). We studied mutations and copy number burden of 62 genes expressed by cancer cells which are involved in the following pathways: antigen presentation by MHC mechanisms, antigen processing and immunomodulation (stimulation and inhibition of T-cell responses) (Figure 3). Mutations and CNAs in these genes were more prevalent in progressive than regressive samples (p=0.003). Four of these genes – B2M, CHUK, KDR and CD80 – had a significantly elevated dN/dS ratio (19) – comparison of the rates of non-synonymous to synonymous mutations – indicating positive selection for acquisition of mutations in these genes. We observe that expression of immunostimulatory genes predominantly positively correlates with infiltrating lymphocytes in CIS, and these genes are mostly downregulated in progressive compared to regressive CIS. Conversely, inhibitory genes predominantly correlate negatively with infiltrating lymphocytes and are upregulated in progressive lesions.
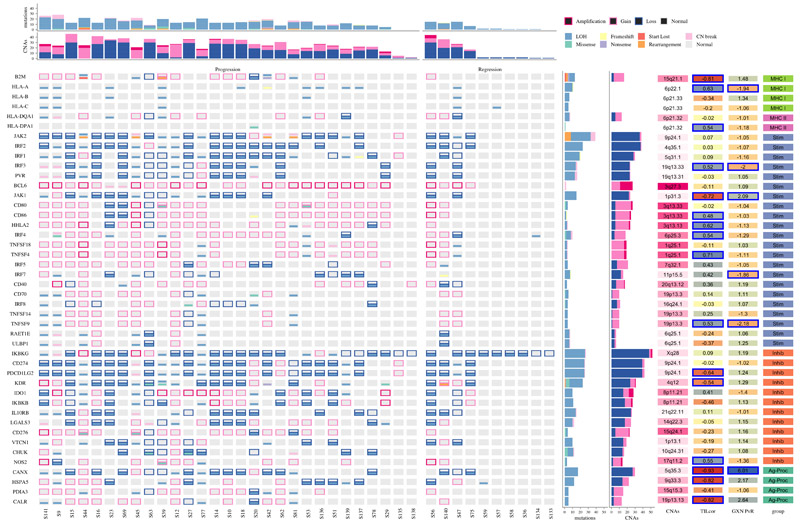
Figure 3. Genomic aberrations affecting immune genes in lung carcinoma in-situ lesions.
The mutational status is shown for 62 genes involved in the immune response, which are expressed by antigen presenting (tumor) cells. Genes are categorized as belonging to the Major Histocompatibility Complex (MHC) class I or II; stimulators (Stim) and inhibitors (Inhib) of the immune response, and genes involved in antigen processing (Ag-Proc). Mutations and copy number aberrations (CNAs) are shown for each of 29 progressive and 10 regressive samples. Loss of heterozygosity (LOH) events are shown as mutations to avoid confusion with copy number loss, relative to ploidy. The GXN PvR column displays the fold-change in expression of each gene between progressive and regressive samples, defined in a partially overlapping set of 18 samples. Significant genes, defined as False Discovery Rate < 0.05, are highlighted in blue. The TILcor column displays the Pearson’s correlation coefficient between the expression of each gene and the gene-expression based tumour infiltrating lymphocyte (TIL) score, derived by the Danaher method. Progressive samples had more mutations (p=0.028) and CNAs (p=0.0038) than regressive in this gene set. dN/dS analysis identified B2M, CHUK, KDR and CD80 as showing evidence of selection.
Loss of heterozygosity (LOH) in the HLA region, which is found in 61% of LUSC patients(20), was identified in 34% of patients with CIS lesions. Interestingly, a similar proportion of LUSC patients (28%) demonstrated clonal HLA LOH(20), suggesting that such clonal events may often occur prior to tumor invasion; future longitudinal studies will be required to confirm this. We did not find a statistically significant difference in the prevalence of HLA LOH between progressive and regressive lesions (p=0.25) although sample numbers were small. Expression of HLA-A was significantly reduced in progressive compared to regressive lesions (p=1.9x10-10).
Additionally, hypermethylation of the HLA region, which is well-described in invasive cancers(21,22), was commonly observed, suggesting that epigenetic HLA silencing may be an important immune escape mechanism in pre-invasive disease. Genome-wide differential methylation analysis between progressive and regressive lesions identified differentially methylated regions (DMRs) including a striking cluster of hypermethylation in chromosome 6 ((4); Figure S6a-b), covering a region containing all of the major HLA genes. This cluster was also identified in analysis of 370 LUSC versus 42 control samples published by the Cancer Genome Atlas(23). Further analysis of TCGA data demonstrate strong evidence for epigenetic silencing of multiple genes in the antigen presentation pathway: mean methylation beta value over the gene is inversely correlated with expression for HLA-A (r2=-0.32, p=2.5x10-10), HLA-B (r2=-0.42, p<2.2x10-16), HLA-C (r2=-0.18, p=3.6x10-4), TAP1 (r2=-0.53, p<2.2x10-16) and B2M (r2=-0.38, p=1.1x10-14). Similar trends were observed in CIS data (Figure S7a-b). The methylation pattern affecting these genes is predominantly promoter hypermethylation (Figure S8).
Demethylating agents have been shown to promote immune activation through improved antigen presentation, immune migration and T cell activity(24–26). These data support the case for moving on-going trials of demethylating agents in combination with immunotherapy from advanced lung cancer into early disease (examples of such trials include NCT01928576 and NCT03220477, registered at https://clinicaltrials.gov/). Additionally, several other cancer-associated pathways are known to be affected by methylation changes(4), therefore the benefits of these drugs may extend beyond immune activation. Nevertheless, we note with caution that some key immune genes demonstrate positive correlations in TCGA data between gene expression and methylation, including the immune co-stimulating ligand TNFSF9 (coding for 4-1BBL) (r2=0.32, p=1.7x10-10) and the MHC class II transcriptional activator CIITA (r2=0.39, p=2.5x10-15) (Figure S7). Further studies will be required to demonstrate that immunological benefits of demethylating agents are not outweighed by effects on these important pathways.
Despite this evidence for impairment of antigen presentation mechanisms in CIS, we do observe ‘immune hot’ CIS lesions which progress to cancer. We therefore next considered functional and microenvironment-related mechanisms to explain how these lesions were able to evade immune predation.
To study microenvironment effects on the immune response, we performed gene expression profiling on laser-captured stromal tissue taken from regions adjacent to CIS lesions. In contrast to data from gastrointestinal pre-invasive lesions(27), no genes were significantly differentially expressed on comparing stromal expression between progressive (n=10) and regressive (n=8) lesions when a FDR of <0.1 was applied. This result holds true with restricted hypothesis testing considering only genes that are related to immunity and inflammation (Figure 4a-b; Table S4).
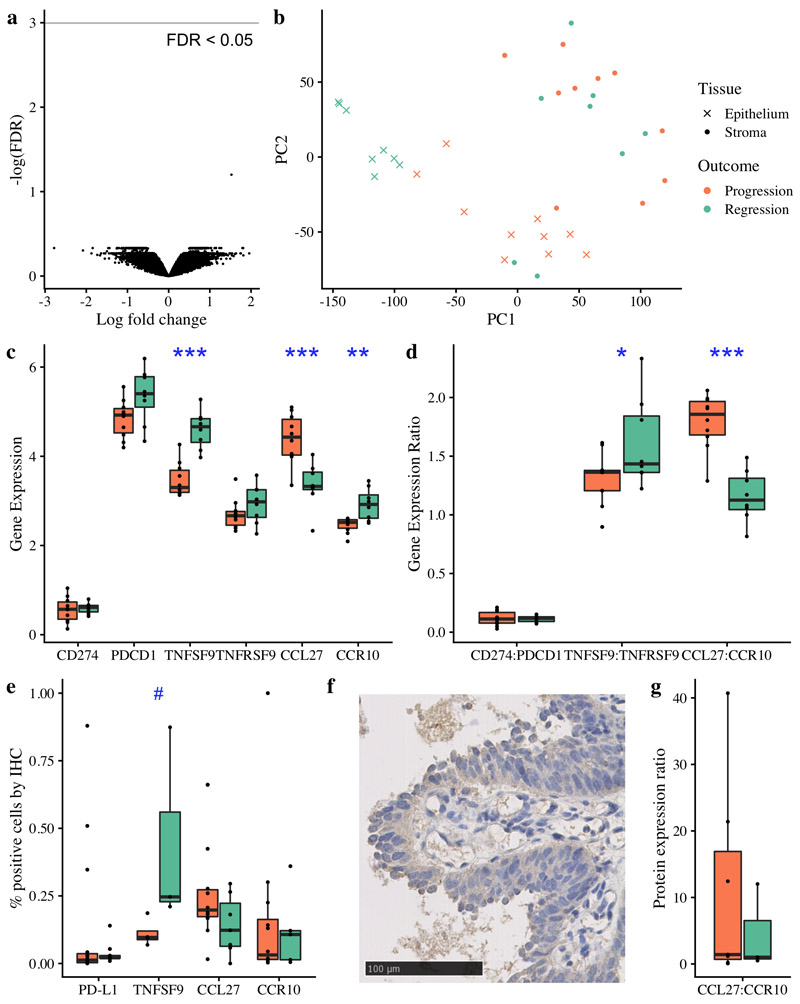
Figure 4. Immune escape mechanisms in CIS beyond antigen presentation.
(a) Volcano plot of gene expression differential analysis of laser-captured stroma comparing progressive (n=10) and regressive (n=8) CIS samples. No genes were significant with FDR < 0.05 following adjustment for multiple testing. (b) Principle component analysis plot of the same 18 CIS samples, showing laser-captured epithelium and matched stroma. (c-d) RNA analysis of immunomodulatory molecules and cytokine:receptor pairs in n=18 CIS samples identified TNFSF9 and CCL27:CCR10 as significantly differentially expressed between progressive and regressive samples (p=0.0000058 and p=0.0000019 respectively). (e) Immunohistochemistry showed that TNFSF9 was similarly differentially expressed at the protein level (p=0.057; n=7 with successful staining). (f) Illustrative immunohistochemistry staining for TNFSF9. CCL27 and CCR10 showed a similar trend at the protein level to the RNA level (e,g); whilst these data did not achieve a significance threshold (g; p=0.49 for CCL27:CCR10 ratio, n=10) we observe several outliers in the progressive group. Analysis of PD-L1 (encoded by CD274) and its receptor PD-1 (encoded by PDCD1) is included due to its relevance in clinical practice; again we do not achieve statistically significant results but do observe three marked outliers with PD-L1 expression >25%, all of which progressed to cancer. All p-values are calculated using linear mixed effects modeling to account for samples from the same patient; ***p < 0.001 **p < 0.01 *p<0.05 #p<0.1. Units for gene expression figures represent normalised microarray intensity values.
Targeting immunomodulatory molecules such as PD-1 now forms part of first-line lung cancer management(28). PD-L1 expression is common in invasive LUSC with estimates of positivity ranging from 34% to 52%, depending on criteria(29). Whilst we did not identify transcriptional upregulation of the PD-L1 gene (CD274; Figure 4c-d), IHC data identified 3 samples with >25% of epithelial cells (PanCK+) also positive for PD-L1 (Figure 4e), all of which progressed to cancer, suggesting that targeting this pathway early in the clinical course may have therapeutic benefit in selected patients.
To investigate the role of immunomodulatory molecules more broadly in pre-invasive immune escape, we performed differential expression analysis between progressive and regressive lesions, focused on 28 known immunomodulatory genes (Table S4). TNFSF9 (4-1BBL, CD137L) was significantly downregulated in progressive lesions (FDR=4.34x10-5; Figure 4c-d) with no corresponding change identified in its receptor TNFRSF9 (FDR=0.6). These findings were corroborated by IHC (Figure 4e-f). TNFSF9 promotes activation of T cells and natural killer (NK) cells(30); in CIS lesions TNFSF9 expression correlates with cytotoxic cell (r2=0.77, p=0.0002) and NK cell infiltration (r2=0.54, p=0.02), as predicted from gene expression data. Agonists of the TNFSF9 receptor have been shown to be clinically efficacious in several cancers(31–33) and these data support their investigation in targeted early lung cancer cohorts. Furthermore, individual lesions showed notably high or low expression of other immunomodulatory genes, raising the possibility that other immunomodulators may be targets for therapy in individual cases (Figure S9).
To identify differences in cytokine responses between progressive and regressive lesions, we calculated the ligand:receptor mRNA expression ratio for 52 known cytokine:receptor pairs(34). Only one, CCL27:CCR10, was significant with FDR < 0.01 (Fold change 1.55, FDR 0.003); progressive samples express more CCL27 (p=2.6x10-6) and less CCR10 (p=0.1x10-4) than regressive (Figure 4c-d). Whilst sample numbers were small, these findings were broadly supported by IHC (Figure 4e-g). CCL27:CCR10 signaling has been associated with immune escape in melanoma through PIK/Akt activation in a mouse model(35); in CIS, CCL27 expression correlates with expression of both PIK3CA (r2=0.61, p=0.008) and AKT1 (r2=0.68, p=0.002) (Figure S10a-b). CCL27 is minimally expressed in both normal lung tissue and invasive squamous cell lung cancer(23,36), suggesting that this effect is specific to early carcinogenesis and therefore warrants further investigation as a target for preventative therapy.
Our previous research highlighted occasional cases of ‘late progressive’ lesions, which met a clinical endpoint of regression (defined by the subsequent biopsy at the same site showing resolution to normal epithelium or low-grade dysplasia) but the index CIS biopsy had the molecular appearance of a progressive lesion, and it indeed subsequently developed cancer months or years later. Clinical review identified 11 lesions across the 53 regressive lesions in our current cohort (20.7%) that at later clinical follow up subsequently progressed to cancer, and hence are termed ‘late progressive’. These included 4 previously published lesions subjected to whole-genome sequencing and/or methylation and shown to display the genomically unstable appearance of progressive lesions, as well as 7 with immunohistochemistry data and 10 with lymphocyte quantification performed from H&E slides (Table S1; Figure S1). Interestingly, based on these data, late progressive lesions appear immunologically similar to regressive lesions, showing increased infiltration with lymphocytes and CD8+ T-cells compared to progressive lesions (Figure S11).
Whilst we acknowledge that sample numbers are small when examining subgroups of regressive lesions in this way, our data support a model in which lesions can be considered on two axes: genomic stability and immune competence. Our previous work predicts that chromosomally unstable lesions will usually progress, implying that they have escaped immune predation. Yet some may regress if they remain immune competent only to later progress, potentially due to their genomic instability making them more likely to evolve immune escape mechanisms during regression, and hence become ‘late progressors’. Of 11 late progressors in this cohort, median time from regressive index biopsy to progression was 3.2 years (range 0.8-4.6 years). This time period represents a change from a point of known immune competence to demonstrated immune escape. Hence, we might estimate that a successful therapeutic strategy to block a particular immune escape mechanism might delay the onset of cancer by around 3 years. Of the remaining 42 regressive samples in this cohort, median follow-up time was 4.73 years (range 0.42-13.5 years), suggesting that genomically ‘stable’ samples are likely to regress and remain regressed long-term. Given their immunological competence, late progressors are included in the regressive cohort when analysing immune escape mechanisms in this study.
Discussion
In summary, we present evidence that immune surveillance may play a critical role in spontaneous regression of pre-cancerous lesions of the airways. Whilst recent cross-sectional studies have greatly furthered our understanding of immune signals prior to cancer invasion, and indeed at earlier disease stages than CIS(2,12), we have for the first time shown an association with lesion regression. Including such outcome data offers insight into the dynamics of immune surveillance and evasion; assuming that lesion regression is driven by immune surveillance – which is likely based on our data – we are able to directly compare preinvasive lesions which are immune competent (regressed) with those that are able to evade immune predation (progressed). Analysis of ‘late progressive’ samples furthers this model by providing estimates of timescales over which immune evasion evolves. Hence we provide a roadmap for manipulation of the immune system as a cancer intervention strategy, by identifying and targeting differences between these two immune states.
To this end, we identify mechanisms of immune escape present before the point of cancer invasion, many of which offer potential therapeutic targets. These data present an opportunity to induce regression and prevent cancer development. Demethylating agents, 4-1BB agonists and CCL27 blockade are therapeutic candidates that warrant further research, as well as targeting the PD-1/PD-L1 axis in highly selected patients. As a result of field carcinogenesis, patients with pre-invasive lesions are at risk of synchronous cancers at other sites, which are likely to be clonally related(4,37) and therefore may benefit from systemic immunomodulatory treatment. The data presented here support a new paradigm of personalised immune-based systemic therapy in early disease.
Methods
Additional methods are provided in a supplementary file accompanying this manuscript.
Ethical approval
All tissue and bronchial brushing samples were obtained under written informed patient consent and were fully anonymized. Study approval was provided by the UCL/UCLH Local Ethics Committee (REC references 06/Q0505/12 and 01/0148). All relevant ethical regulations were followed.
Cohort description and patient characteristics
For over 20 years, patients presenting with pre-invasive lesions, which are precursors of squamous cell lung cancer (LUSC), have been referred to the UCLH Surveillance Study. As previously described(3), patients undergo repeat bronchoscopy every four months, with definitive treatment performed only on detection of invasive cancer. Autofluorescence bronchoscopy is used to ensure the same anatomical site is biopsied at each time point. Gene expression, methylation and whole genome sequencing data of carcinoma in-situ (CIS) samples have been performed on this cohort, and data have been published(4). These data are used in this study.
All patients enrolled in the UCLH Surveillance Study who met a clinical end point of progression or regression were included; by definition they underwent an ‘index’ CIS biopsy followed by a diagnostic cancer biopsy (progression) or a normal/low-grade biopsy (regression) four months later. Index lesions were identified between 1999 and 2017. Cases meeting an end-point of regression underwent clinical review to identify those which subsequently progressed; 11 samples (20.7%) were identified, which are described as ‘late progressors’ in the main text. Of these 11, median time from ‘regressive’ index biopsy to progression was 3.2 years (range 0.8-4.6 years) whilst the remaining 42 samples had a median follow up time of 4.73 years (range 0.42-13.5 years). Whilst we cannot fully exclude that any regressive sample may later develop cancer, the fact that median follow up in the study group was longer than the maximum follow up in the late progression group suggests that late progression in included samples is unlikely.
All samples underwent laser capture microdissection (LCM) to ensure only CIS cells underwent molecular profiling. Methods for sample acquisition, quality control and mutation calling are as previously described, as are full details regarding patient clinical characteristics.
Briefly, gene expression profiling was performed using both Illumina and Affymetrix microarray platforms. Normalisation was performed using proprietary Illumina software and the RMA method of the affy(38) Bioconductor package respectively. This study includes 18 previously unpublished gene expression arrays from stromal tissue. These samples were collected using LCM to identify stromal regions adjacent to 18 already-published CIS samples (corresponding to the 18 samples undergoing Affymetrix microarray profiling described above). These new stromal samples underwent Affymetrix profiling using the exact same methodology as previously described for CIS tissue samples. To avoid issues related to batch effects between platforms, the analyses in this paper utilise only samples profiled on Affymetrix microarrays, which include both CIS and matched stromal samples (see Supplementary Methods and Table S5).
Methylation profiling was performed using the Illumina HumanMethylation450k microarray platform. All data processing was performed using the ChAMP Bioconductor package(39).
Whole genome sequencing data was obtained using the Illumina HiSeq X Ten system. A minimum sequencing depth of 40x was required. BWA-MEM was used to align data to the human genome (NCBI build 37). Unmapped reads and PCR duplicates were removed. Substitutions, insertions-deletions, copy number aberrations and structural rearrangements were called using CaVEMan(40), Pindel(41,42), ASCAT(43) and Brass(44) respectively.
Sample selection for profiling
As previously described, all patients enrolled in the surveillance programme discussed above were considered for this study. For a given CIS lesion under surveillance, when a biopsy from the same site in the lung showed evidence of progression to invasive cancer or regression to normal epithelium or low-grade dysplasia, we defined the preceding CIS biopsy as a progressive or regressive ‘index’ lesion respectively. Due to the small size of bronchoscopic biopsy samples, not all profiling techniques were applied to all samples. Patients with Fresh Frozen (FF) samples underwent whole genome sequencing and/or methylation analysis depending on sample quality. Patients with formalin-fixed paraffin-embedded (FFPE) samples underwent gene expression analysis. Further detail is available in our previous manuscript(4). Additionally, any patient with an available FFPE block underwent image analysis as described below, and all patients with Affymetrix-based gene expression profiling underwent further profiling of laser-captured adjacent stroma.
Statistical Methods
Unless otherwise specified, all analyses were performed in an R statistical environment (v3.5.0; www.r-project.org/) using Bioconductor(45) version 3.7. Code to reproduce a specific statistical test is publicly available at the Github repository above.
Unless otherwise stated, comparisons of means between two independent groups are performed using a two-sided Wilcoxon test. In some cases, multiple samples have been profiled from the same patient, although always from distinct sites within the lung. In such cases we used mixed effects models to compare means between groups, treating the patient ID as a random effect, as implemented in the Bioconductor lme4 library(46), with p-values calculated using the Anova method from the Bioconductor car library (available from https://cran.r-project.org/web/packages/car). Differential expression was performed using the limma(47) Bioconductor package to compare microarray data between two groups. When adjustment for multiple correction is required we quote a False Discovery Rate (FDR) which is calculated using the Benjamini-Hochberg method(48). Cluster analysis and visualization was performed using the pheatmap Bioconductor package (available from https://cran.r-project.org/web/packages/pheatmap/).
Supplementary Material
Statement of Significance.
Immune evasion is a hallmark of cancer. For the first time, this study identifies mechanisms by which pre-cancerous lesions evade immune detection during the earliest stages of carcinogenesis and forms a basis for new therapeutic strategies that treat or prevent early stage lung cancer.
Acknowledgements
We thank all of the patients who participated in this study. We thank P. Rabbitts, A. Banerjee and C. Read for their early development of the study. The results published here are in part based on data generated by a TCGA pilot project established by the National Cancer Institute and National Human Genome Research Institute. Information about TCGA and the investigators and institutions that constitute the TCGA research network can be found at http://cancergenome.nih.gov. R.E.H., N.M., P.J.C., and S.M.J. are supported by Wellcome Trust fellowships. S.M.J. is also supported by the Rosetrees Trust, the Welton Trust, the Garfield Weston Trust, the Stoneygate Trust and UCLH Charitable Foundation, as well as Stand Up to Cancer-LUNGevity-American Lung Association Lung Cancer Interception Dream Team Translational Cancer Research Grant (Grant Number: SU2C-AACR-DT23-17). Stand Up To Cancer is a division of the Entertainment Industry Foundation. The research grant is administered by the American Association for Cancer Research, the scientific partner of SU2C. V.T., C.P., R.E.H. and S.M.J. have been funded by the Roy Castle Lung Cancer Foundation. A.P. and D.C. are funded by Wellcome Trust clinical PhD training fellowships. H.L.-S. is funded by the Wellcome Trust Sanger Institute non-clinical PhD studentship. C.T. was a CRUK Clinician Scientist. This work was partially undertaken at UCLH/UCL, who received a proportion of funding from the Department of Health’s NIHR Biomedical Research Centre’s funding scheme (S.M.J.). R.E.H., D.M., N.M., C.S., and S.M.J. are part of the CRUK Lung Cancer Centre of Excellence. Y.Y. acknowledges funding from Cancer Research UK Career Establishment Award, Breast Cancer, Children’s Cancer and Leukaemia Group, NIH U54 CA217376 and R01 CA185138, CDMRP Breast Cancer Research Program Award, CRUK Brain Cancer Award (TARGET-GBM), European Commission ITN, Wellcome Trust, and The Royal Marsden/ICR National Institute of Health Research Biomedical Research Centre. S.A.Q. is funded by a CRUK Senior Cancer Research Fellowship, a CRUK Biotherapeutic Program Grant, the Cancer Immunotherapy Accelerator Award (CITA-CRUK) and the Rosetrees Trust. LMC acknowledges funding from the National Institutes of Health (1U01 CA224012, U2C CA233280, R01 CA223150, R01, R01 CA226909, R21 HD099367), the Knight Cancer Institue, and the Brenden-Colson Center for Pancreatic Care at OHSU. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Footnotes
Author Contributions
A.P. and V.H.T. contributed equally to this work, as did K.A., S.E.A.R. and T.L.. A.P., V.H.T., N.M. and S.M.J. co-wrote the manuscript. S.M.J., S.A.Q., V.H.T. and A.P. conceived the study design. V.H.T., D.C., F.R.M. and S.A. performed stromal LCM and gene expression profiling experiments. C.P.P. and C.T. performed methylation experiments. H.L-S. and P.J.C. performed genomic experiments. A.A., T.L., J.Y.H., L.K. and T.M. designed and performed IHC experiments. Further quantitative multiplex IHC was performed by C.M., M-L.A-L., W.L., C.B. and L.C.. K.A., S.E.A.R., Y.B.H. and Y.Y. performed cell quantification on H&E and IHC images. S.M.J., P.J.G., B.C. and R.M.T. led the bronchoscopic surveillance programme through which samples were obtained. M.F. and D.M. performed histological review. P.F.D. performed pathological processing. A.P. performed bioinformatic analysis, supported by R.R. and N.M.. R.E.H., K.H.C.G., C.D., A.F., N.M., C.S., C.T., S.A.Q. and N.M. gave advice and reviewed the manuscript. S.M.J. provided overall study oversight.
Conflict of Interest Statement
S.A.Q. and C.S. are co-founders of Achilles Therapeutics. C.S. is a shareholder of Apogen Biotechnologies, Epic Bioscience, GRAIL, and has stock options in Achilles Therapeutics. R.R. and N.M. have stock options in and have consulted for Achilles Therapeutics. L.M.C. is a paid consultant for Cell Signaling Technologies, received reagent and/or research support from Plexxikon Inc., Pharmacyclics, Inc., Acerta Pharma, LLC, Deciphera Pharmaceuticals, LLC, Genentech, Inc., Roche Glycart AG, Syndax Pharmaceuticals Inc., Innate Pharma, and NanoString Technologies, and is a member of the Scientific Advisory Boards of Syndax Pharmaceuticals, Carisma Therapeutics, Zymeworks, Inc, Verseau Therapeutics, Cytomix Therapeutics, Inc., and Kineta Inc.
Data Availability
All raw data used in this study is publicly available. Previously published CIS gene expression and methylation data is stored on GEO under accession number GSE108124; matched stromal gene expression data is stored under accession number GSE133690. Previously published CIS whole genome sequencing data is available from the European Genome Phenome Archive (https://www.ebi.ac.uk/ega/) under accession number EGAD00001003883. Annotated H&E images of all samples used for lymphocyte quantification were deposited to the Image Data Resource (https://idr.openmicroscopy.org) under accession number idr0082.
Code Availability
All code used in our analysis will be made available at http://github.com/ucl-respiratory/cis_immunology on publication. All software dependencies, full version information, and parameters used in our analysis can be found here.
References
- 1.Nicholson AG, Perry LJ, Cury PM, Jackson P, McCormick CM, Corrin B, et al. Reproducibility of the WHO/IASLC grading system for pre-invasive squamous lesions of the bronchus: a study of inter-observer and intra-observer variation. Histopathology. 2001;38:202–8. doi: 10.1046/j.1365-2559.2001.01078.x. [DOI] [PubMed] [Google Scholar]
- 2.Mascaux C, Angelova M, Vasaturo A, Beane J, Hijazi K, Anthoine G, et al. Immune evasion before tumour invasion in early lung squamous carcinogenesis. Nature. 2019;571:570–5. doi: 10.1038/s41586-019-1330-0. [DOI] [PubMed] [Google Scholar]
- 3.George PJ, Banerjee AK, Read CA, O’Sullivan C, Falzon M, Pezzella F, et al. Surveillance for the detection of early lung cancer in patients with bronchial dysplasia. Thorax. 2007;62:43–50. doi: 10.1136/thx.2005.052191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teixeira VH, Pipinikas CP, Pennycuick A, Lee-Six H, Chandrasekharan D, Beane J, et al. Deciphering the genomic, epigenomic, and transcriptomic landscapes of pre-invasive lung cancer lesions. Nat Med. 2019;25:517–25. doi: 10.1038/s41591-018-0323-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.AbdulJabbar K. Geospatial immune variability illuminates differential evolution of lung adenocarcinoma. Nature Medicine (accepted for publication) 2020 doi: 10.1038/s41591-020-0900-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsujikawa T, Kumar S, Borkar RN, Azimi V, Thibault G, Chang YH, et al. Quantitative Multiplex Immunohistochemistry Reveals Myeloid-Inflamed Tumor-Immune Complexity Associated with Poor Prognosis. Cell Reports. 2017;19:203–17. doi: 10.1016/j.celrep.2017.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banik G, Betts CB, Liudahl SM, Sivagnanam S, Kawashima R, Cotechini T, et al. High-dimensional multiplexed immunohistochemical characterization of immune contexture in human cancers. Meth Enzymol. 2020;635:1–20. doi: 10.1016/bs.mie.2019.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danaher P, Warren S, Dennis L, D’Amico L, White A, Disis ML, et al. Gene expression markers of Tumor Infiltrating Leukocytes. J Immunother Cancer. 2017;5:18. doi: 10.1186/s40425-017-0215-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakravarthy A, Furness A, Joshi K, Ghorani E, Ford K, Ward MJ, et al. Pan-cancer deconvolution of tumour composition using DNA methylation. Nature Communications. 2018;9:3220. doi: 10.1038/s41467-018-05570-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshida K, Gowers KHC, Lee-Six H, Chandrasekharan DP, Coorens T, Maughan EF, et al. Tobacco smoking and somatic mutations in human bronchial epithelium. Nature. 2020;578:266–72. doi: 10.1038/s41586-020-1961-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenthal R, Cadieux EL, Salgado R, Bakir MA, Moore DA, Hiley CT, et al. Neoantigen-directed immune escape in lung cancer evolution. Nature. 2019;567:479–85. doi: 10.1038/s41586-019-1032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beane JE, Mazzilli SA, Campbell JD, Duclos G, Krysan K, Moy C, et al. Molecular subtyping reveals immune alterations associated with progression of bronchial premalignant lesions. Nature Communications. 2019;10:1856. doi: 10.1038/s41467-019-09834-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463–9. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghorani E, Rosenthal R, McGranahan N, Reading JL, Lynch M, Peggs KS, et al. Differential binding affinity of mutated peptides for MHC class I is a predictor of survival in advanced lung cancer and melanoma. Ann Oncol. 2018;29:271–9. doi: 10.1093/annonc/mdx687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lakatos E, Williams MJ, Schenck RO, Cross WCH, Househam J, Werner B, et al. Evolutionary dynamics of neoantigens in growing tumours. bioRxiv. 2019:536433. doi: 10.1038/s41588-020-0687-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang T-H, et al. The Immune Landscape of Cancer. Immunity. 2018;48:812–830.:e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wellenstein MD, de Visser KE. Cancer-Cell-Intrinsic Mechanisms Shaping the Tumor Immune Landscape. Immunity. 2018;48:399–416. doi: 10.1016/j.immuni.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 19.Martincorena I, Raine KM, Gerstung M, Dawson KJ, Haase K, Van Loo P, et al. Universal Patterns of Selection in Cancer and Somatic Tissues. Cell. 2017;171:1029–1041.:e21. doi: 10.1016/j.cell.2017.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGranahan N, Rosenthal R, Hiley CT, Rowan AJ, Watkins TBK, Wilson GA, et al. Allele-Specific HLA Loss and Immune Escape in Lung Cancer Evolution. Cell. 2017;171:1259–1271.:e11. doi: 10.1016/j.cell.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gyõrffy B, Bottai G, Fleischer T, Munkácsy G, Budczies J, Paladini L, et al. Aberrant DNA methylation impacts gene expression and prognosis in breast cancer subtypes. International Journal of Cancer. 2016;138:87–97. doi: 10.1002/ijc.29684. [DOI] [PubMed] [Google Scholar]
- 22.Ye Q, Shen Y, Wang X, Yang J, Miao F, Shen C, et al. Hypermethylation of HLA class I gene is associated with HLA class I down-regulation in human gastric cancer. Tissue Antigens. 2010;75:30–9. doi: 10.1111/j.1399-0039.2009.01390.x. [DOI] [PubMed] [Google Scholar]
- 23.Network TCGAR. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Amoozgar Z, Huang J, Saleh MH, Xing D, Orsulic S, et al. Decitabine Enhances Lymphocyte Migration and Function and Synergizes with CTLA-4 Blockade in a Murine Ovarian Cancer Model. Cancer Immunol Res. 2015;3:1030–41. doi: 10.1158/2326-6066.CIR-15-0073. [DOI] [PubMed] [Google Scholar]
- 25.Wang L-X, Mei Z-Y, Zhou J-H, Yao Y-S, Li Y-H, Xu Y-H, et al. Low dose decitabine treatment induces CD80 expression in cancer cells and stimulates tumor specific cytotoxic T lymphocyte responses. PLoS ONE. 2013;8:e62924. doi: 10.1371/journal.pone.0062924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang H, Bueso-Ramos C, DiNardo C, Estecio MR, Davanlou M, Geng Q-R, et al. Expression of PD-L1, PD-L2, PD-1 and CTLA4 in myelodysplastic syndromes is enhanced by treatment with hypomethylating agents. Leukemia. 2014;28:1280–8. doi: 10.1038/leu.2013.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saadi A, Shannon NB, Lao-Sirieix P, O’Donovan M, Walker E, Clemons NJ, et al. Stromal genes discriminate preinvasive from invasive disease, predict outcome, and highlight inflammatory pathways in digestive cancers. PNAS. 2010;107:2177–82. doi: 10.1073/pnas.0909797107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paz-Ares L, Luft A, Vicente D, Tafreshi A, Gümüş M, Mazières J, et al. Pembrolizumab plus Chemotherapy for Squamous Non–Small-Cell Lung Cancer. New England Journal of Medicine. 2018;379:2040–51. doi: 10.1056/NEJMoa1810865. [DOI] [PubMed] [Google Scholar]
- 29.Pawelczyk K, Piotrowska A, Ciesielska U, Jablonska K, Gletzel-Plucinska N, Grzegrzolka J, et al. Role of PD-L1 Expression in Non-Small Cell Lung Cancer and Their Prognostic Significance according to Clinicopathological Factors and Diagnostic Markers. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20040824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qi X, Li F, Wu Y, Cheng C, Han P, Wang J, et al. Optimization of 4-1BB antibody for cancer immunotherapy by balancing agonistic strength with FcyR affinity. Nature Communications. 2019;10:2141. doi: 10.1038/s41467-019-10088-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melero I, Shuford WW, Newby SA, Aruffo A, Ledbetter JA, Hellström KE, et al. Monoclonal antibodies against the 4-1BB T-cell activation molecule eradicate established tumors. Nat Med. 1997;3:682–5. doi: 10.1038/nm0697-682. [DOI] [PubMed] [Google Scholar]
- 32.Bartkowiak T, Curran MA. 4-1BB Agonists: Multi-Potent Potentiators of Tumor Immunity. Front Oncol. 2015;5:117. doi: 10.3389/fonc.2015.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Segal NH, He AR, Doi T, Levy R, Bhatia S, Pishvaian MJ, et al. Phase I Study of Single-Agent Utomilumab (PF-05082566), a 4-1BB/CD137 Agonist, in Patients with Advanced Cancer. Clin Cancer Res. 2018;24:1816–23. doi: 10.1158/1078-0432.CCR-17-1922. [DOI] [PubMed] [Google Scholar]
- 34.Zlotnik A, Yoshie O, Nomiyama H. The chemokine and chemokine receptor superfamilies and their molecular evolution. Genome Biol. 2006;7:243. doi: 10.1186/gb-2006-7-12-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murakami T, Cardones AR, Finkelstein SE, Restifo NP, Klaunberg BA, Nestle FO, et al. Immune Evasion by Murine Melanoma Mediated through CC Chemokine Receptor-10. J Exp Med. 2003;198:1337–47. doi: 10.1084/jem.20030593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–60. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pipinikas CP, Kiropoulos TS, Teixeira VH, Brown JM, Varanou A, Falzon M, et al. Cell migration leads to spatially distinct but clonally related airway cancer precursors. Thorax. 2014;69:548–57. doi: 10.1136/thoraxjnl-2013-204198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–15. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 39.Morris TJ, Butcher LM, Feber A, Teschendorff AE, Chakravarthy AR, Wojdacz TK, et al. ChAMP: 450k Chip Analysis Methylation Pipeline. Bioinformatics. 2014;30:428–30. doi: 10.1093/bioinformatics/btt684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones D, Raine KM, Davies H, Tarpey PS, Butler AP, Teague JW, et al. cgpCaVEManWrapper: Simple Execution of CaVEMan in Order to Detect Somatic Single Nucleotide Variants in NGS Data. Curr Protoc Bioinformatics. 2016;56:15 10 1–15 10 18. doi: 10.1002/cpbi.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–71. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raine KM, Hinton J, Butler AP, Teague JW, Davies H, Tarpey P, et al. cgpPindel: Identifying Somatically Acquired Insertion and Deletion Events from Paired End Sequencing. Curr Protoc Bioinformatics. 2015;52:15 7 1–12. doi: 10.1002/0471250953.bi1507s52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raine KM, Van Loo P, Wedge DC, Jones D, Menzies A, Butler AP, et al. ascatNgs: Identifying Somatically Acquired Copy-Number Alterations from Whole-Genome Sequencing Data. Curr Protoc Bioinformatics. 2016;56:15 9 1–15 9 17. doi: 10.1002/cpbi.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Papaemmanuil E, Rapado I, Li Y, Potter NE, Wedge DC, Tubio J, et al. RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia. Nat Genet. 2014;46:116–25. doi: 10.1038/ng.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nature Methods. 2015;12:115–21. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bates D, Mächler M, Bolker B, Walker S. Fitting Linear Mixed-Effects Models Using lme4. Journal of Statistical Software. 2015;67:1–48. [Google Scholar]
- 47.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Statist. 2001;29:1165–88. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data used in this study is publicly available. Previously published CIS gene expression and methylation data is stored on GEO under accession number GSE108124; matched stromal gene expression data is stored under accession number GSE133690. Previously published CIS whole genome sequencing data is available from the European Genome Phenome Archive (https://www.ebi.ac.uk/ega/) under accession number EGAD00001003883. Annotated H&E images of all samples used for lymphocyte quantification were deposited to the Image Data Resource (https://idr.openmicroscopy.org) under accession number idr0082.
All code used in our analysis will be made available at http://github.com/ucl-respiratory/cis_immunology on publication. All software dependencies, full version information, and parameters used in our analysis can be found here.