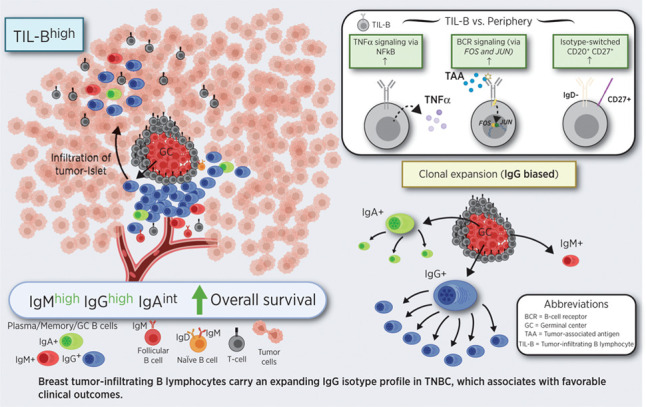
Tumor-infiltrating B lymphocytes assemble in clusters, undergoing B-cell receptor–driven activation, proliferation, and isotype switching. Clonally expanded, IgG isotype-biased humoral immunity associates with favorable prognosis primarily in triple-negative breast cancers.
Abstract
In breast cancer, humoral immune responses may contribute to clinical outcomes, especially in more immunogenic subtypes. Here, we investigated B lymphocyte subsets, immunoglobulin expression, and clonal features in breast tumors, focusing on aggressive triple-negative breast cancers (TNBC). In samples from patients with TNBC and healthy volunteers, circulating and tumor-infiltrating B lymphocytes (TIL-B) were evaluated. CD20+CD27+IgD− isotype-switched B lymphocytes were increased in tumors, compared with matched blood. TIL-B frequently formed stromal clusters with T lymphocytes and engaged in bidirectional functional cross-talk, consistent with gene signatures associated with lymphoid assembly, costimulation, cytokine–cytokine receptor interactions, cytotoxic T-cell activation, and T-cell–dependent B-cell activation. TIL-B–upregulated B-cell receptor (BCR) pathway molecules FOS and JUN, germinal center chemokine regulator RGS1, activation marker CD69, and TNFα signal transduction via NFκB, suggesting BCR–immune complex formation. Expression of genes associated with B lymphocyte recruitment and lymphoid assembly, including CXCL13, CXCR4, and DC-LAMP, was elevated in TNBC compared with other subtypes and normal breast. TIL-B–rich tumors showed expansion of IgG but not IgA isotypes, and IgG isotype switching positively associated with survival outcomes in TNBC. Clonal expansion was biased toward IgG, showing expansive clonal families with specific variable region gene combinations and narrow repertoires. Stronger positive selection pressure was present in the complementarity determining regions of IgG compared with their clonally related IgA in tumor samples. Overall, class-switched B lymphocyte lineage traits were conspicuous in TNBC, associated with improved clinical outcomes, and conferred IgG-biased, clonally expanded, and likely antigen-driven humoral responses.
Significance:
Tumor-infiltrating B lymphocytes assemble in clusters, undergoing B-cell receptor–driven activation, proliferation, and isotype switching. Clonally expanded, IgG isotype-biased humoral immunity associates with favorable prognosis primarily in triple-negative breast cancers.
Graphical Abstract
Introduction
Initiation of effective adaptive immunity may contribute to tumor growth restriction through specific antigen-directed responses. The T lymphocyte component of antitumor immunity has received significant attention (1). In contrast, B lymphocytes, especially the memory and isotype-switched B lymphocyte compartments, and their expressed antibody profiles remain only partially elucidated. Emerging findings suggest that aspects of humoral immune responses may correlate with improved clinical outcomes via B lymphocyte tumor-infiltration and expression of antibodies in lesions or in the circulation (2, 3). These could differ across tumor types, potentially offering opportunities for stratification and for guiding therapy options.
Breast cancer is one of the most frequently diagnosed malignancies, divided into biological, and differential therapy-associated subtypes based on estrogen receptor (ER), progesterone receptor (PR), and HER2 expression, with specific prognostic and predictive biomarker implications. Triple-negative breast cancers (TNBC) do not express any of these markers and demonstrate the least-favorable prognosis due to both an aggressive phenotype and limited targeted therapies (4). Although breast cancer has not traditionally been regarded as a typical immunogenic malignancy, emerging studies report the presence and potential clinical significance of tumor-infiltrating immune cells for clinical outcomes (5). Paradoxically, despite an overall poor prognosis of patients with TNBC, immune infiltration is more pronounced compared with other breast cancer types. Consistent with an immunogenic tumor microenvironment (TME), some patients with TNBC may benefit from anti-programmed death-ligand 1 (PD-L1) and anti–PD-1 immunotherapy with atezolizumab in combination with chemotherapy (6). TNBCs are characterized by immunologically variable and compartmentalized tumors with structural features in the tumor–immune interphase and large variability across individuals, mandating the need for patient stratification for therapy selection (7). The most thoroughly studied effector cells within the breast cancer setting are CD8+ cytotoxic T lymphocytes and natural killer (NK) cells (8). However, tumor-infiltrating B lymphocytes (TIL-B) aggregating within tertiary lymphoid structures (TLS; ref. 9) may have an antigen-educated phenotype (10) and autoantibodies are thought to trigger tumor cell clearance (11). TIL-Bs might also serve as antigen-presenting cells to promote antitumor Th responses (12). Therefore, it is increasingly recognized that humoral immune responses may be important contributors to breast cancer outcome, especially in more immunogenic TNBCs.
The interaction between the immune system and malignant cells, therefore, constitutes a major focus of current translational and clinical investigation (13). Recent studies have provided evidence of TIL-Bs and tumor-reactive immunoglobulin (Ig) in several solid tumors, including in breast cancer, and TIL-Bs have been reported to respond to B-cell receptor stimulation and produce Igs ex vivo (14–16).
Here, in peripheral blood and cancer lesions of patients with breast cancer, specifically in individuals with TNBC, we performed flow cytofluorimetric, transcriptomic, immunofluorescence, single-cell RNA-sequencing (scRNA-seq), and long-read Ig repertoire studies to evaluate isotype-switched and memory B lymphocyte subsets, Ig isotype distribution, and clonal expansion profiles.
Materials and Methods
Clinical sample collection and cohort descriptions
A collection of internal and external cohorts of healthy volunteer (HV) and patient samples, including the unique accession numbers, are summarized in Supplementary Table S1 and Supplementary Materials and Methods. All internal King's College London (KCL) samples were collected with informed written consent, in accordance with the Helsinki Declaration [study design was approved by the Guy's Research Ethics Committee (REC No. 07/H0804/131), Guy's and St. Thomas' NHS Foundation Trust]. Peripheral blood mononuclear cells were isolated using Ficoll–Paque PLUS density gradient centrifugation, and single cells from breast tissues were isolated as described previously in Supplementary Materials and Methods.
Gene expression profiling of lymphocyte and lymphoid assembly markers
Gene expression (GEx) levels were analyzed from internal Guy's Hospital and The Cancer Genome Atlas (TCGA) Breast cohorts (KCL and TCGA GEx cohorts), and compared between PAM50 (basal-like, HER2, luminal A, luminal B, and normal-like) and TNBC subtypes (basal-like 1/2, immunomodulatory, mesenchymal, mesenchymal stem-like, and luminal androgen receptor) according to classification described previously in Brasó-Maristany and colleagues (17). A B lymphocyte metagene signature was analyzed from published NanoString data of primary and metastatic breast cancers (GSE102818; ref. 18). Kaplan–Meier (KM) plotter tool was used to generate survival plots (19). CIBERSORT was applied to evaluate naïve B, plasma, and memory B cells identified among 22 immune subsets in the KCL GEx cohort (20). Univariate Cox proportional hazards regression models were used to investigate the prognostic importance of these immune subsets in high and low TIL-infiltrated tumors (semiquantitative TIL classification was performed by a trained histopathologist using tissue microarrays; Supplementary Materials and Methods). B lymphocyte function–associated gene sets were identified from gene ontology (GO) database (Supplementary Materials and Methods). A lymphoid assembly–associated gene signature was compiled from a set of known markers (21).
scRNA-seq analysis
Analyses were performed on a published scRNA-seq dataset (GSE114725; ref. 22; single-cell cohort) using R package Seurat (23). Dimensionality reduction was performed using Uniform Manifold Approximation and Projection (UMAP) and cells were clustered using the Louvain algorithm (23). Ig isotypes were detected based upon heavy-chain gene expression. Differentially expressed genes identified by Seurat were used to perform gene set enrichment analysis (GSEA) using the fgsea package. Gene sets were obtained from Broad Institute Molecular Signature Database using R package msigdbr (24). CellPhoneDB v2.0 was used to analyze B-cell–T-cell interactions (25).
Immunohistochemical/immunofluorescence evaluations of TIL-B distribution and surface Ig expression
Three sections per tissue sample were stained with fluorescently labeled antibodies conferring three panels: TIL classification (DAPI/CD20/CD3/PanCK), naïve B lymphocyte identification (DAPI/CD20/IgD), and Ig isotype expression (DAPI/CD27/IgG/IgA/IgM). Antibodies used are detailed in Supplementary Table S2. TIL-B structural features were evaluated (Supplementary Materials and Methods) following TIL working group guidelines (26), guided by trained pathologists. Olympus VS120-S5 and Nikon TE 2000-U microscopes were used for imaging.
Long-read Ig repertoire analysis
Ig repertoire analysis was performed from cDNA synthesized from breast tissues with the 5′ RACE template switch method (Supplementary Materials and Methods). Full-length Ig cDNAs were PCR-amplified with primers containing unique molecular barcodes. Purified DNA samples were sequenced using PacBio Single Molecule, Real-Time (SMRT) Sequencing platform (27). Redundant sequences with identical molecular barcodes were removed. Ig genes and complementarity determining region (CDR)3 sequences were determined using IMGT/HighV-QUEST (28). Relatedness among sequences were estimated using BRepertoire webserver (29) “Clonotype clustering” function, after partitioning all CDR3 DNA sequences by the sample and the V gene family used (30). Selection pressure analysis was performed using R package shazam (31).
Data availability
The R code used to analyze scRNA-seq data from GSE114725 (22) can be accessed from https://codeocean.com/capsule/7488142. The R code to analyze BCR Repertoire data collected from breast tissues can be accessed at https://codeocean.com/capsule/4153741.
Statistical analyses
GraphPad Prism and R were used for statistical analyses of paired and unpaired datasets. Data are presented as mean ± SEM. P values reported as: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 and all tests were two-sided.
Further details can be found in Supplementary Materials and Methods.
Results
Collapse in circulating memory B lymphocytes contrasts with amplification of intratumoral class-switched memory B lymphocyte compartment
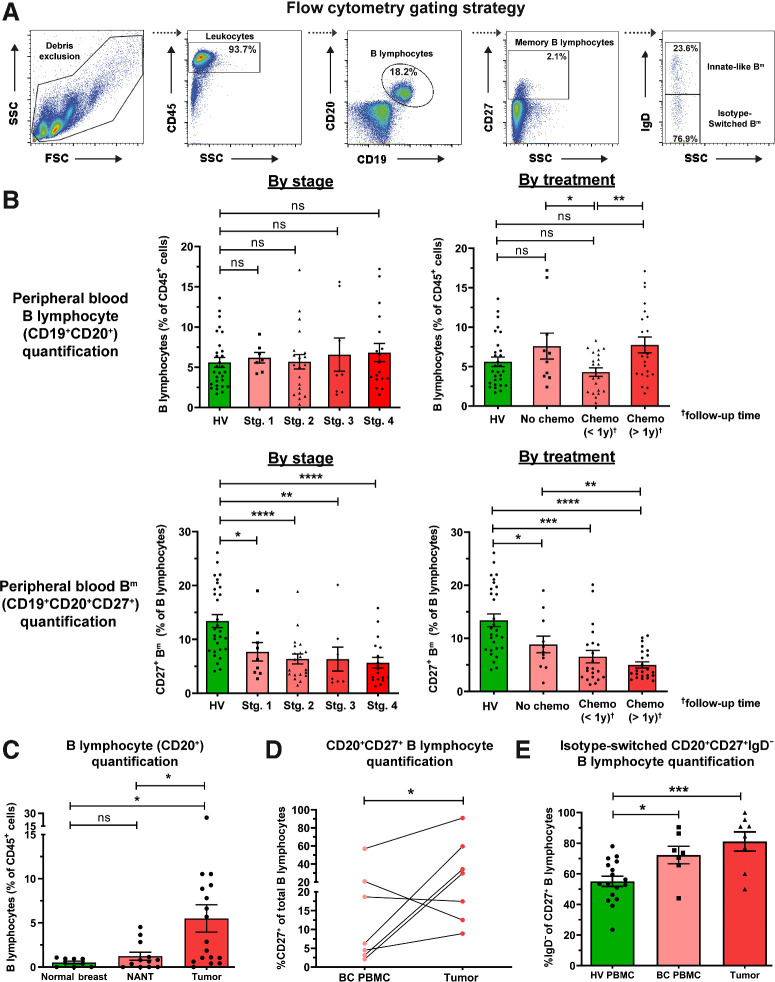
We compared B lymphocyte subsets between breast cancer patient blood and tumors (KCL flow cohort; see Supplementary Table S1 for information of all cohorts used). We quantified percentages of B lymphocytes (CD19+CD20+) and memory (CD19+CD20+CD27+) B lymphocytes (Bm) in patients (N = 55) and healthy subjects (N = 48) by flow cytometry (Fig. 1A and B). Consistent with a report in melanoma (32), we identified a fall in peripheral Bm in patients compared with HVs. This was independent of disease stage or chemotherapy treatment status. We observed significantly lower proportions of B lymphocytes (of CD45+ cells) in the circulation of patients with recent chemotherapy compared with those without treatment (Fig. 1B), alongside reduced serum Ig titers in chemotherapy-treated individuals (Supplementary Fig. S1A and S1B, KCL Luminex cohort). Baseline serum Ig isotype titers in patients were not predictive of breast cancer–specific death [Supplementary Fig. S2, Apolipoprotein Mortality Risk (AMORIS) cohort; ref. 33].
Figure 1.
Flow cytometric analyses reveal reduced circulating CD20+CD27+ memory and amplification of tumor-infiltrating CD20+CD27+IgD− class-switched subsets among B lymphocytes. A, Gating strategy for identification of B lymphocytes and memory (Bm) lymphocytes derived from peripheral blood mononuclear cell (example patient peripheral blood mononuclear cell shown). B, Quantification of total circulating B cells (top) and Bm cells (bottom) as percentage of CD45+ cells in HV (N = 48) and patient (N = 55) peripheral blood (KCL flow cohort; Supplementary Table S1 for patient information), stratified according to stage and treatment status. C, Quantification of B lymphocytes (CD20+) from single cell suspensions of normal breast (N = 9), NANT (N = 12), and cancer tissue (N = 17) samples. D, Quantification of matched patient circulating- and tumor-infiltrating CD20+CD27+ B cells (matched samples of 7 patients). E, Quantification of CD20+CD27+IgD− B cells in HV (N = 17), patient peripheral blood (N = 7), and cancers (N = 8) of total CD20+CD27+ B cells. Statistical significance was determined using the Student t test. ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
In contrast, using tumor single-cell suspensions (N = 17), we identified a higher B lymphocyte infiltration compared with non-adjacent, nontumor (NANT; N = 12) and normal breast (N = 9) tissues (Fig. 1C). In matched blood and tumor samples (N = 7), tumors contained higher CD20+CD27+ B-cell proportions (Fig. 1D). Both peripheral and intratumoral CD20+CD27+ B-cell populations showed a bias toward the loss of IgD expression (N = 32; Fig. 1E). Additional quantitative single B lymphocyte gene expression analyses (single-cell cohort) also confirmed larger proportions of CD20+CD27+ and CD20+CD27+IgD− B lymphocytes in tumors compared with blood (Supplementary Fig. S3A and S3B). Furthermore, isotype-switched TIL-B populations comprise memory, germinal center (GC) B cells and plasma cells in non-TNBC and TNBC [immunohistochemical/immunofluorescence (IHC/IF), KCL IHC cohort and Single cell cohort; Supplementary Fig. S3A–S3E].
These findings reveal a reduced peripheral Bm population in patients compared with HVs, independently of disease stage or treatment, in parallel with an enriched tumor-infiltrating class-switched B-cell compartment in the TME compared with matched patient blood.
Elevated TIL-B signatures and assembly within stromal clusters in TNBC
We asked whether TNBC demonstrate increased immune cell infiltration, particularly concentrating on the B-cell population, compared with normal breast or other breast cancer types. Quantitative IHC/IF evaluations (Bart's IHC cohort) and GEx analyses (KCL and TCGA GEx cohorts) revealed an expansion of CD20+ TIL-B within TNBC lesions compared with normal tissues (Fig. 2A), and compared with non-TNBC carcinomas, expression of B lymphocyte (CD20 and BCR complex CD79A) and T lymphocyte (CD3D and CD3G) markers were elevated in TNBC. Within different TNBC subtypes, these markers were elevated especially in the Lehmann's immunomodulatory (IM) molecular TNBC subtype, known to be enriched in core immune signal transduction pathways and cytokine signaling (N = 122; Fig. 2A; Supplementary Fig. S4A and S4B; ref. 34). TIL-B density and CD20 gene expression positively correlated with tumor-infiltrating T lymphocyte (TIL-T) density and CD3 gene expression, respectively (Fig. 2B). The expression of a B lymphocyte metagene signature (NanoString cohort) was significantly higher in primary cancers compared with patient-matched metastatic sites (N = 31 vs. 17), suggesting a diminished immune TME with advanced disease (Fig. 2C).
Figure 2.
B lymphocyte infiltration and its positive prognostic value in TNBC. A, TIL-B quantification by IHC within normal breasts and TNBC (N = 15 each, Bart's IHC cohort), and by GEx (normal breast) vs. TNBC (N = 10 vs. 131, KCL GEx cohort); non-TNBC vs. TNBC (N = 515 vs. 123, TCGA GEx cohort); TNBC subtypes [mesenchymal (M), luminal androgen receptor (LAR), basal-like 1 and 2 (BL1 and 2), and immunomodulatory (IM; N = 122, KCL GEx cohort]. The Mann–Whitney test was used for statistical significance. B, TNBC TIL-B correlation with TIL-T by IHC (r = 0.72, Bart's IHC cohort) and by GEx (r = 0.73, KCL GEx cohort). Linear regression analysis was used to calculate correlation coefficients (r) and P values. C, B lymphocyte metagene NanoString GEx data comparing B lymphocytes in primary tumors with metastatic sites (N = 31 vs. 17; NanoString cohort). D, High (above median) TIL-B densities by IHC were associated with better overall survival in TNBC (N = 15; Bart's IHC cohort), and in the basal-like subtype by high CD20 GEx (N = 241; KM plotter cohort; log rank test used to assess statistical significance). E, Kaplan–Meier survival curves display DMFS for naïve B cells, plasma cells, and memory B cells in TNBC (KCL GEx cohort) using CIBERSORT (20). Data were divided into four groups based on B lymphocyte subset and TIL levels stratified by semiquantitative TIL classification. Statistical significance was assessed using univariate Cox proportional hazards regression models. F, Representative IHC/IF images (Bart's IHC cohort) depicting nucleated cells (DAPI), epithelial cells (PanCK), B lymphocytes (CD20), and T lymphocytes (CD3) within TNBC TIL-Blow and TIL-Bhigh lesions. Scale bar, 50 μm. G, Representative TNBC (TIL-Bhigh) images highlighting numerous lymphoid aggregates (within white dash lines) consisting of B lymphocytes assembled adjacent to a T lymphocyte zone. Brown dash lines indicate carcinoma edge. Scale bar, 2 mm. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Both quantitative IHC/IF (N = 15) and KM-plotter survival analysis (N = 241) demonstrated that TIL-B in primary tumors were significantly associated with more favorable overall survival, most prominently in basal-like/TNBC (Fig. 2D; Supplementary Fig. S4C). Additional KM analyses using CIBERSORT (from KCL GEx cohort, see Supplementary Materials and Methods; ref. 20) investigated the associations between distant metastasis-free survival (DMFS) and naïve, plasma, and memory B-cell phenotypes in TNBC (N = 89). In tumors with high TIL infiltrates (classified by using tissue microarrays), memory B cells were associated with a more-favorable DMFS, whereas a negative prognostic value was shown for naïve B cells in this cohort (Fig. 2E). IHC/IF staining analyses (Bart's IHC cohort) confirmed the heterogeneity of TIL-T and TIL-B (Fig. 2F), with evidence of expansive cluster formation [demarcated in white (>30 TIL-B and >30 TIL-T aggregated), typically of a B lymphocyte assembly adjacent to a T lymphocyte zone; Fig. 2G]. This highlighted close B–T lymphocyte interactions within the TME.
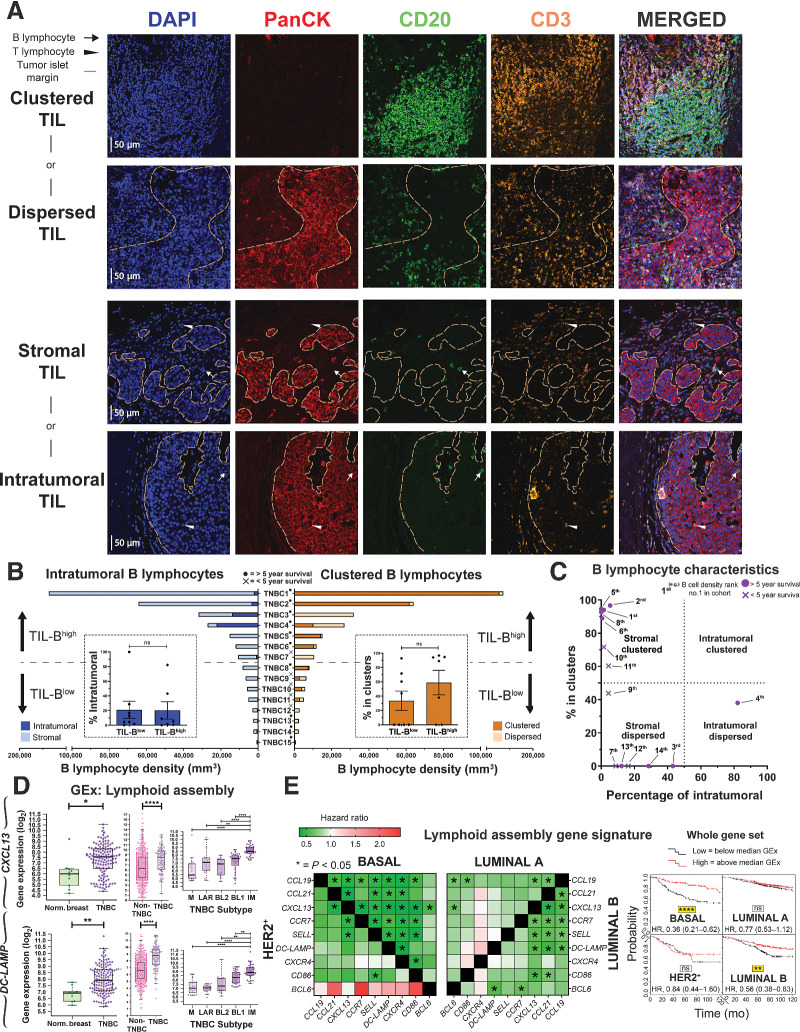
Because we found B–T lymphocyte clusters in a large proportion (9 of 15) of TNBCs (Bart's IHC cohort), we next evaluated TIL-B spatial and structural characteristics. We characterized TIL-B as either stromal or intratumoral, according to penetration within PanCK+ tumor islets. Cells were further categorized as forming either clusters or dispersions, according to the extent of aggregation along with CD3+ TIL-T (Fig. 3A). Tumors were split into TIL-Bhigh and TIL-Blow groups, as defined by median TIL-B density. B lymphocytes typically formed within stromal clusters irrespective of overall TIL-B density (Fig. 3B). Five individuals featuring highly clustered TIL-B characteristics showed a >5 year survival post-surgery (Fig. 3C). Consistent with the B–T lymphocyte cluster formation observed, we found elevated expression of B lymphocyte recruitment and lymphoid assembly marker genes (CXCL13, CXCR4, and DC-LAMP) in TNBC compared with both non-TNBC and normal breast, and within the TNBC cohort the highest expression of these genes was detected in IM tumors (Fig. 3D; Supplementary Fig. S4D, KCL and TCGA GEx cohorts). Higher expression of these signatures were also associated with significantly improved odds of overall survival (10-year follow-up) in basal-like/TNBC (Fig. 3E, KM plotter cohort).
Figure 3.
Occurrence of B lymphocytes in stromal clusters. A, Representative IHC/IF images (Bart's IHC cohort) highlighting key TIL characteristics: clustered TIL versus dispersed TIL and stromal TIL (outside tumor nests) versus intratumoral TIL (within tumor nests). Scale bar, 50 μm. B, Quantitative assessment of TIL-B spatial and structural characteristics within TNBC (N = 15; left) intratumoral versus stromal; clustered versus dispersed (right). Patients ranked according to CD20+ TIL-B density (TNBC1 = highest), and overall survival data are indicated. Inset, patient samples split at median density into high and low TIL-B groups and the percentage of intratumoral (left) or clustered (right) B lymphocytes were analyzed. Statistical significance was determined using the Student t test. C, Characterization of TNBC TIL-B profile as stromal clustered, intratumoral clustered, stromal dispersed, or intratumoral dispersed. Overall survival data are indicated. D, GEx data for lymphoid assembly marker genes CXCL13 and DC-LAMP: normal breast versus TNBC (N = 10 vs. 131, KCL GEx cohort; left); non-TNBC vs. TNBC (N = 515 vs. 123, TCGA GEx cohort; middle); TNBC subtypes (N = 122, KCL GEx cohort; right). E, Survival analysis in KM Plotter of determined ER−HER2−/basal surrogate, HER2+, luminal A, and luminal B subtype KM plotter surrogate subgroups (KM plotter cohort; ref. 19). These indicate that expression of lymphoid cell assembly genes carries positive prognostic value in TNBC/basal-like and luminal B subtypes. Individual genes were evaluated in combination with each other gene (left) and gene set as a whole (right). ns, nonsignificant; *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
Therefore, substantial TIL-B densities in TNBC typically form clusters along with T lymphocytes. TIL-B marker (CD20 and CD79A) and GEx features conferring B lymphocyte recruitment and lymphoid assembly were elevated compared with non-TNBC and normal breast tissues, and associated with more favorable survival in basal-like/TNBC subtype.
TIL-Bs are activated via the B-cell receptor
We sought direct evidence of active roles for B lymphocytes in the TME, using a previously published dataset (single-cell cohort). We applied dimensionality reduction (UMAP) to single-cell B lymphocyte populations and revealed distinct CD20+ and CD27+ TIL-B populations compared with those in the circulation (blood (N = 1,476 B cells) and tumors (N = 1,021 B cells) of eight patients; Fig. 4A). Downstream differential expression gene (DEG) analysis identified several upregulated genes in TIL-B: FOS and JUN, molecules downstream of the BCR complex pathway (35); RGS1, germinal center B lymphocyte regulator of chemokine receptor signaling (36); and the lymphocyte activation marker CD69, triggered through cross-linking of surface Ig (Fig. 4B; ref. 37). Hallmark enrichment analysis (38) revealed significantly enhanced expression among TIL-B for genes controlling TNFα signaling via NFκB, hypoxia, and UV response pathways, in comparison with circulating B lymphocytes (Fig. 4C). Several genes, known to positively regulate B lymphocyte activation, proliferation, and differentiation functions were upregulated in TNBC compared with normal breast (Supplementary Fig. S5, KCL and TCGA GEx cohorts). Survival analysis of these gene signatures revealed positive associations with overall survival (10-year follow up), most pronounced in basal-like/TNBC (Fig. 4D, KM plotter cohort). We next evaluated B-cell–T-cell interactions in the TME (single-cell cohort) using CellPhoneDB (25). We identified cell communication pathways associated with lymphoid assembly (CXCL12, CCL19, CCL21, and CXCL13), cytokine signaling (IL4, IL6, IL13, IL15, IL17, and IFNγ), costimulation (CD28 and ICOS) and immune activation (CD40 and CD226). These findings support a bidirectional functional cross-talk between tumor-infiltrating B and T lymphocytes (Fig. 4E).
Figure 4.
scRNA-seq analysis reveals BCR-driven TIL-B activatory signatures, and B lymphocyte functional trait gene markers predict positive survival outcome. A, UMAP visualization according to global GEx of single B lymphocytes pooled from the peripheral blood (1,476 cells) and tumors (1,021 cells) of eight patients (single-cell cohort), colored by relative normalized gene expression levels for CD20 and CD27. B, The detection of differentially expressed genes [DEG; on cells originally annotated by Azizi and colleagues (22) as B cells] demonstrates elevated expression of FOS, JUN, RGS1, and CD69, indicated by fold change (FC; determined using Wilcoxon rank sum test). C, Gene set enrichment analysis of TIL-B relative to circulating B lymphocytes using hallmark gene sets. Red, positive normalized enrichment scores (hallmark expression enhanced in TIL-B). D, Survival analysis in KM Plotter of determined ER−HER2−/basal surrogate, HER2+, luminal A, and luminal B subtype KM plotter surrogate subgroups (KM plotter cohort; ref. 19) for expression of gene signatures positively regulating key B lymphocyte properties (activation, proliferation, and differentiation). Representative genes listed for B lymphocyte proliferation (44 total in set). Signatures from all three functions carry positive prognostic value in the basal-like cancer subtype. Individual genes were evaluated in combination with each other gene (left) and gene set as a whole (right). E, CellPhoneDB (25) was applied to analyze B-cell–T-cell interactions (single-cell cohort). After FDR (FDR < 0.001) correction, communication pathways identified included lymphoid assembly, cytokine signaling, costimulation, T-cell–dependent B-cell activation, and cytotoxic T lymphocyte (CTL) activation. Circle sizes indicate P value, whereas color-coding represents the average expression level of interacting molecule 1 in cluster 1 and interacting molecule 2 in cluster 2. ****, P < 0.0001.
These findings indicate the presence of distinct B lymphocyte populations between patient blood and cancers and evidence of TIL-B antigen–Ig complexing, BCR pathway stimulation, and evidence of B-cell–T-cell cross-talk. Key functional attributes for the initiation of B lymphocyte responses are associated with improved patient outcomes, especially in TNBC.
IgG+ B lymphocyte densities are elevated in tumors, and IgG isotype switching predicts positive survival outcomes in TNBC
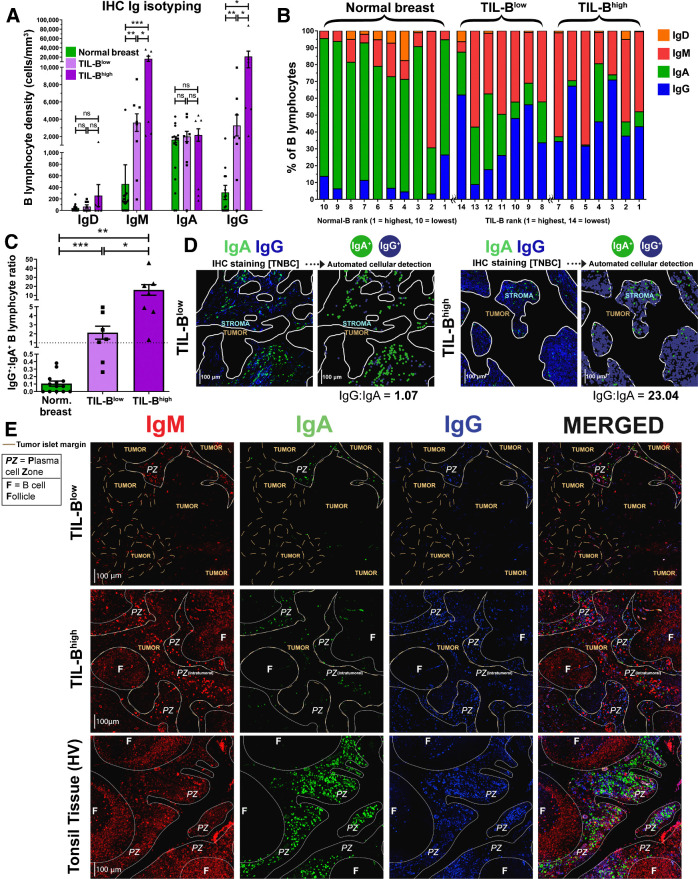
We studied B lymphocyte densities according to Ig isotype expression in primary breast cancers by quantitative IHC/IF evaluations of IgD, IgM, IgA, and IgG isotype-expressing B lymphocytes [Bart's IHC cohort; normal breast tissue (N = 10), TNBC (N = 14)]. Naïve (IgD+) B lymphocytes were found at low densities in normal breast, TIL-Blow and TIL-Bhigh tumors (Supplementary Fig. S6A). Higher IgG+ and IgM+ B lymphocyte densities were found in TIL-Bhigh and, to a lesser extent, in TIL-Blow cancers compared with normal breast (Fig. 5A and B). In contrast, IgA+ B lymphocyte densities were consistent among tissue compartments, highlighting an overall bias toward increased IgG+:IgA+ B lymphocyte ratio within cancers compared with normal breast, and within TIL-Bhigh compared with TIL-Blow cancers (Fig. 5C and D).
Figure 5.
Quantitative fluorescence IHC reveals elevated IgG+:IgA+ ratio within high TIL-B tumors, implicating expansion of IgG+ B lymphocytes within TNBC tumor microenvironment. A, Comparison of surface immunoglobulin-expressing B lymphocyte density in normal breast and TNBC with low TIL-B density (below median CD20+) and high TIL-B density (above median CD20+). B, Quantitative IHC analysis profiling the proportions of B lymphocytes present within the microenvironment of normal breast tissue (N = 10) and TNBC (N = 14) expressing each Ig isotype. C, Enumeration of IgG+:IgA+ B lymphocyte ratios. D, Right, example images illustrating IgG+ and IgA+ B lymphocytes in a typical TIL-B low individual and a TIL-B high individual. Automated cellular detection identifies IgA (green) and IgG (blue) B lymphocytes. Scale bar, 100 μm. E, Representative images depicting typical Ig isotype expression among B lymphocytes: IgM+ (red), IgA+ (green), and IgG+ (blue). Images from Bart's IHC cohort. Brown dash lines indicate margin of the cancer. White lines separate distinct regions of B lymphocyte compartments (PZ, plasma cell zone; F, B lymphocyte follicle). Scale bar, 100 μm. Statistical significance was determined using the Student t-test. ns, nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
IHC/IF showed the dominating IgA+ B lymphocyte profile present in normal breast is contained within normal lobules and their periphery (Supplementary Fig. S6B). Although TIL-Blow cancers lacked large B lymphocyte follicles and contained small IgM/IgA/IgG plasma cell zones, TIL-Bhigh cancers typically contained denser IgM/IgA/IgG plasma cell zones surrounding expansive IgM+ follicular B lymphocyte clusters with defined germinal centers (Fig. 5E). The resulting B lymphocyte assembly in these TIL-Bhigh tumors shares some structural similarities with those in tonsil tissues. IgG and IgA expression were accompanied with CD27 upregulation, validating the differentiated, isotype-switched status of these cells (Supplementary Fig. S7A–S7C). We further confirmed that intratumoral IgG+ cells comprised both CD20+ as well as CD138+ (plasma) cell populations, the latter being low or negative for CD20 expression (Supplementary Figs. S7D and S3C).
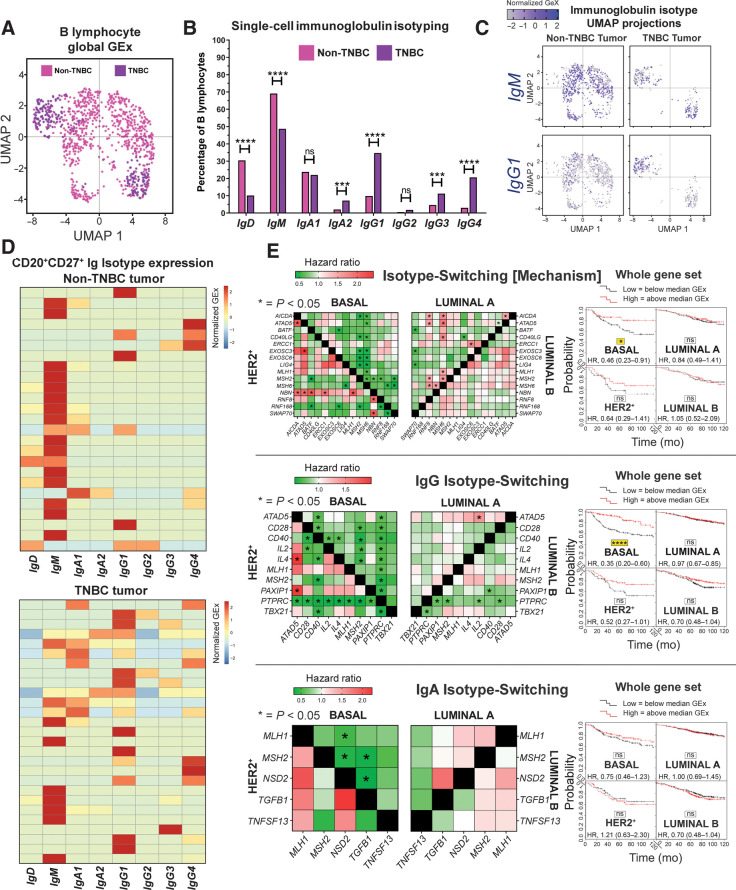
Because TNBCs feature high levels of IgG-expressing B lymphocytes, we studied Ig isotypes (Ig heavy chain) using a published scRNA-seq dataset (single-cell cohort; N = 1,021 TIL-B across eight patients). UMAP applied to single-cell B lymphocyte populations revealed distinct TIL-B populations in TNBC compared with non-TNBC samples. This analysis also revealed enhanced isotype-switching to IgG1, IgG3, IgG4 and IgA2 subclasses in TNBC (Fig. 6A and B). UMAP confirmed differential isotype-expressing B lymphocyte compartments in TNBC compared with non-TNBC for IgM and IgG1 based on clustering (Fig. 6C). Heatmaps of single CD20+CD27+ B lymphocyte IgCH expression showed a propensity toward non-switched IgM transcripts in blood in contrast with high levels of isotype-switched IgG and IgA B lymphocytes in tumor samples (Supplementary Fig. S8), and higher levels of isotype-switching in TNBC compared with non-TNBC tumor samples (Fig. 6D). These support an active tumor-resident humoral response, different to the equivalent response in the circulation, and likely driven by inflammatory and possibly antigenic signals that may support Ig class-switch recombination in the breast cancer microenvironment, especially in TNBC.
Figure 6.
Single-cell RNA-seq data analysis reveals favor of IgG isotypes in TNBC, while isotype-switching gene markers predict positive survival outcome. A, UMAP visualization of B lymphocyte populations in non-TNBC versus TNBC (single-cell cohort). B, Percentage of each Ig isotype based upon raw data of Ig heavy-chain (Student t test). C, UMAP visualization for IgM and IgG1 isotypes colored by relative normalized gene expression levels (N = 1,021 cells). D, IgCH switch transcripts of single B lymphocytes (CD19+CD27+/CD20+CD27+/CD22+CD27+ single cells) were analyzed in non-TNBC and TNBC tissues and demonstrated more Ig isotype-switching events in the TNBC samples. E, Survival analysis in KM Plotter of determined ER−HER2−/basal surrogate, HER2+, luminal A, and luminal B subtype KM plotter surrogate subgroups (KM plotter cohort; ref. 19) for expression of gene signatures conferring isotype switching and those positively regulating isotype switching (IgG and IgA isotype switching). Signatures from all three functions carry positive prognostic value in basal-like cancer. Individual genes were evaluated in combination with each other gene (left) and the gene set as a whole (right). ns, nonsignificant; ***, P < 0.001; ****, P < 0.0001.
Survival analysis of isotype-switching gene signatures revealed a positive association with 10-year overall survival for IgG, but not for IgA, isotype switching, most pronounced in basal-like/TNBC (Fig. 6E, KM plotter cohort). Moreover, as expected, several genes involved in the mechanism of and/or positively regulating isotype switching were upregulated in TNBC compared with normal breast (Supplementary Fig. S9, KCL and TCGA GEx cohorts).
These findings indicate a shift in favor of IgG+ B lymphocytes in TIL-Bhigh TNBC and point to IgG isotype switching as a contributor to the overall positive role of B lymphocyte responses to breast cancers.
IgG-biased, clonally expanded, and Ig repertoires in breast cancer
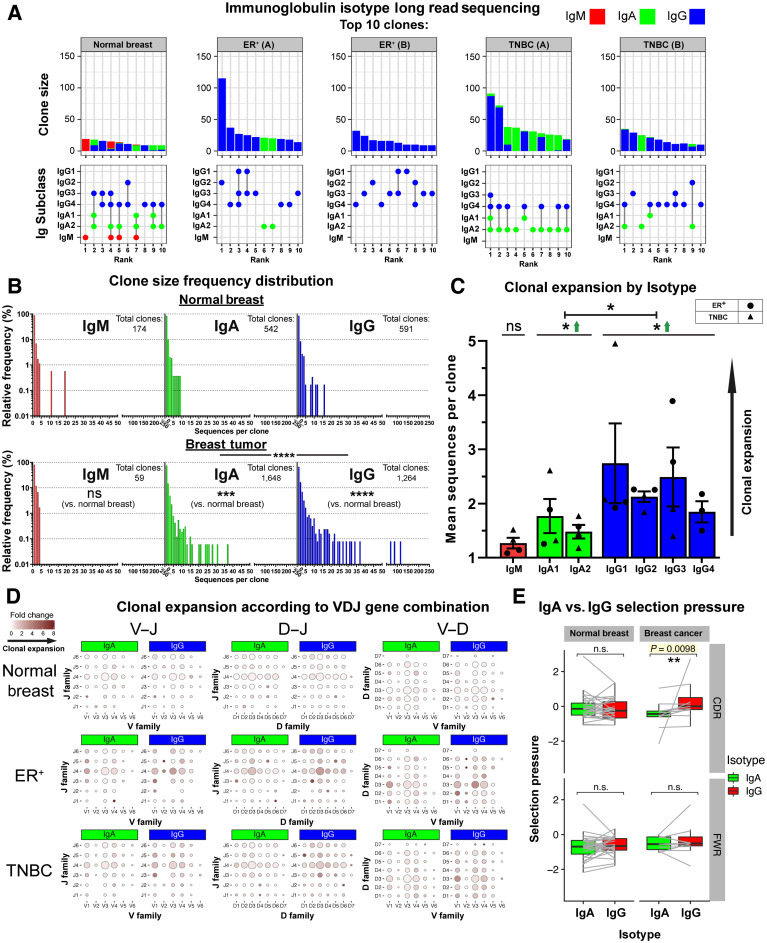
Using long-read sequencing, we next generated a dataset of Ig heavy-chain repertoires (N = 7,670) from two TNBCs, two ER+ cancers, and one normal breast tissue (KCL sequencing cohort) and performed sequence clustering analyses to define clonotypes and compare the distributions of Ig isotypes following B lymphocyte clonal expansion. We observed consistently larger clonal family sizes within the 10 largest clones of cancers compared with normal breast (Fig. 7A, top). B lymphocyte sequences from ER+ cancers featured a heterogeneous IgG subclass expansion with few IgA clones. In contrast, TNBCs showed clones with coexisting IgG1 and IgA1 subclasses, suggested intraclone isotype switching (Fig. 7A, bottom). Kolmogorov–Smirnov analyses revealed that IgG and IgA clonal family frequency distributions were significantly different between carcinomas and normal breast (Fig. 7B). Accordingly, IgG and IgA were clonally expanded within cancers (Supplementary Fig. S10A and S10B), and on average IgG+ B lymphocytes belonged to larger clonal families than IgA+ cells (Fig. 7C). These data point to an inherent bias toward the preferential clonal expansion of IgG isotypes within breast cancers. When we compared variable region gene usage, we found specific V(D)J genes whose combined usage was overrepresented in clonally expanded IgG and IgA sequences in cancers. Specific combinatorial gene usages appeared to expand in TNBC but not ER+ lesions, and vice versa, suggestive of clonally restricted, likely antigen-focused, Ig repertoires (Fig. 7D; Supplementary Fig. S10C). Furthermore, Ig light chain repertoire transcripts from single CD20+CD27+ B lymphocytes from two matched blood and tumor samples (single-cell cohort, Supplementary Fig. S10D) showed similarities in kappa and lambda chain V genes (IGKV, IGLV) and J genes (IGKJ, IGLJ), pointing to potential common clonal origins. Furthermore, we observed a stronger positive selection pressure in the CDRs of IgG compared with their clonally related IgA in tumor samples, whereas such a relationship was absent in the framework region (FWR), and in BCRs from normal breast (Fig. 7E). This hints at different aspects of B lymphocyte responses where IgA expression may act as an early response and IgG may confer higher affinity antigen-driven responses.
Figure 7.
B lymphocyte repertoire analyses of immunoglobulin isotype switching and clonal expansion in breast cancers. A, A total of 7,670 immunoglobulin heavy-chain sequences were analyzed (KCL sequencing cohort). Top 10 clones determined by B lymphocyte repertoire long read data analyses. Clonotypes were estimated via clustering CDR3 sequences. Top, bars depict sizes of clones and their breakdown by isotypes. Bottom, isotypes present in each clone are indicated by dots. Vertical lines signify co-occurrence of isotypes in the same clone. B, Clone size frequency distribution of IgM/IgA/IgG sequences in normal breast (1,771 sequences) and breast cancer (5,899 sequences). Kolmogorov–Smirnov analysis highlights significant differences in clone size frequency distributions. C, Mean sequences per clone of IgM/IgA/IgG isotypes. IgG and, to a lesser extent, IgA isotypes are clonally expanded, whereas on average, IgG isotypes have significantly larger clone sizes than IgA. D, Comparisons of IgA and IgG variable usage of V–J, D–J, and V–D genes extracted from normal breast, ER+ cancer, and TNBC. For each gene usage combination, dot size is proportional to the frequency before clonal expansion. Dot colors correspond to fold change in the number of sequences following clonal expansion, indicating the preference of B lymphocytes with that specific VDJ combination to be clonally expanded. E, Selection pressure in clonally related IgA and IgG. Clonally related sequences are represented as paired observations (gray lines), and selection pressure was considered separately for the complementarity determining regions and framework regions. Sequences are grouped into normal breast and breast cancer (containing two TNBC and two ER+ samples). Paired Wilcoxon tests were conducted, and P values were corrected (Benjamini–Hochberg) for multiple comparisons. ns, nonsignificant; *, P < 0.05; **, P < 0.01.
Large clonal families, isotype switching within clonally expanded TIL-B, and a bias for IgG subclasses accumulating mutations on specific variable region gene combinations, together suggest a mature humoral immune response driven toward specific antigenic stimuli in breast cancer. Further understanding of these features may reveal therapeutic targets, prognostic biomarkers, and patient subpopulations upon whom to focus adaptive immune response-enhancing therapies.
Discussion
Clinical outcomes in patients with cancer may be influenced by the initiation of effective antitumoral adaptive T lymphocyte responses, but these are likely to be significantly more effective when launched in combination with humoral immunity, including induction of isotype-switched B lymphocytes and secretion of antibodies. We undertook flow cytometric, IHC/IF, bulk GEx, scRNA-seq, and long-read Ig sequencing analyses to investigate activated, memory and isotype-switched B lymphocytes in breast cancers, including the more-aggressive and more-immunogenic TNBC subtypes. Consistent with previous reports (39, 40), the TIL-B compartment in tumor stroma is typically organized into clusters of lymphocytes, including isotype-switched memory, GC B cells, and plasma cells. Enhanced isotype-switched B lymphocytes, with an IgG-isotype bias may be part of the humoral clonal expansion mechanism, especially pronounced in TIL-Bhigh TNBC. The accompanied narrow mature Ig variable region repertoires and enhanced BCR signaling in TIL-B strongly signify antigen-driven responses. This dynamic humoral immune profile is especially associated with immunogenic TNBC and indicative of more favorable patient outcomes.
We found a depleted memory CD19+CD20+CD27+ B lymphocyte repertoire in patients' peripheral blood regardless of stage or treatment history for their carcinoma. Moreover, this effect appears to be exacerbated in patients with breast cancer who have undergone chemotherapy, possibly owing to the depletion of B lymphocytes during treatment, which may irreversibly diminish long-lived memory populations. Accordingly, patients receiving chemotherapy have lower total serum Ig titers, indicative of a depleted circulating antibody-secreting B lymphocyte population. In contrast, our flow cytometric analyses revealed an upregulated CD20+CD27+IgD− B-cell compartment among TIL-B. In agreement, single-cell transcriptomic analyses point to a bias toward non-switched IgM transcripts in blood memory CD20+CD27+ B lymphocytes and higher levels of isotype-switched IgG and IgA CD20+CD27+ B lymphocytes in tumor samples. Consistent with an immunogenic signature in TNBCs, we found higher levels of isotype switching shown in single CD20+CD27+ B lymphocyte transcripts in TNBC samples compared with non-TNBC tumors. These may be driven by a combination of antigen exposure and inflammation in the breast cancer microenvironment that promotes Ig class-switch recombination.
The TIL-B population is largely assembled in clusters (>30 TIL-B and >30 TIL-T aggregated) and positively associates with overall survival. Although evident across breast cancer types, transcriptomic analysis suggests that TIL-B infiltration is more pronounced in TNBC compared with non-TNBC. TNBC, especially the IM molecular subtype, featured elevated expression of B lymphocyte recruitment and lymphoid cell assembly (CXCL13, CXCR4, DC-LAMP) genes, compared with normal breast. Enhanced local expression of CXCL13 in arthritic synovial fluids can draw circulating B lymphocytes to inflammation sites (41) and B lymphocytes may traffic from the blood towards lymphoid tissues via CXCR4 stimulation (42). The evident expression of these signals within the TME may recruit B lymphocytes, including Bm, from the periphery to cancer lesions. Consistent with this, we report reduced circulating and enhanced intratumoral CD20+CD27+ B-cell compartments in patients. Such chemoattractant signals may also promote local B lymphocyte assembly into B–T clusters, in line with our observations of close proximity and strong correlation between tumor-infiltrating B and T lymphocytes. Although the B–T clusters we describe are not entirely equivalent to TLSs identified in routine histology (43), within these clusters, B–T lymphocytic cross-talk can lead to B lymphocyte activation, Ig isotype-switching and local clonal expansion (39, 44).
Alongside IHC evidence of spatial B–T association, an active and functional B lymphocyte compartment is also indicated by BCR signaling and isotype switching. Analyses of scRNA-seq data demonstrated that TIL-B are phenotypically distinct to those in blood, featuring upregulated BCR complex pathway molecules FOS and JUN, germinal center chemokine regulator RGS1, lymphocyte activation marker CD69 and TNFα signaling via NFκB. These implicate active BCR engagement by immune complexes. In concordance, Ig repertoire analyses revealed IgG-skewed clonal family expansion with clonally restricted Ig variable regions. We detected significantly larger IgG and to a lesser extent IgA, clones with narrow variable region repertoires in cancers compared with normal breast, indicative of antigen-focused clonal expansion. Together, these suggest a dynamic expanded IgG-biased humoral response focused toward a small repertoire of antigenic stimuli in the TME the greater understanding of which has the potential to inform therapeutic and biomarker strategies in patients.
Several studies reported that TIL-B carry positive prognostic value (5, 26), whereas others found no significant effect (45), or even poorer survival with CD138+ plasma cell infiltrates (46). However, the extent to which B lymphocyte activity may correlate with prognosis has not been addressed. In our TNBC cohort, we report positive prognostic associations of memory, but not of naïve or plasma, B lymphocyte infiltration in high-TIL tumors. This may point to positive contributions of memory B lymphocytes as part of the humoral response in immunogenic breast cancers and especially in TNBC. In concordance, our findings indicate that genes that positively regulate B lymphocyte functions, particularly those involved in activation, proliferation, differentiation, and isotype switching, and especially genes associated with isotype switching to IgG, may carry positive prognostic value. Furthermore, evidence of a bidirectional functional cross-talk between B and T cells reveals expression and interaction of gene pairs associated with lymphoid assembly, costimulation, cytokine–cytokine receptor interactions, cytotoxic T-cell activation, and T-cell–dependent B-cell activation. These interactions between B and T cells may have functional relevance in driving B-cell stimulation and maturation. Positive associations with prognosis may stem from the observed cross-talk between B and T lymphocytes within TLS (47), where antigen presentation and antibody affinity maturation may occur. B lymphocyte–mediated T-helper lymphocyte activation may contribute to immunotherapy response in TNBC with high mutation burden (48) and local antigen presentation could amplify tumor antigen-specific immune responses (12). Our findings also provide support for the involvement of TIL-B in tumor immune surveillance through secretion of cytokines such as TNFα, which may promote differentiation of Th1 cells and polarize immune effector cells towards classically activated phenotypes (49). Tumor-associated B lymphocytes may, therefore, receive, trigger, and respond to significant T lymphocyte–mediated innate and antigenic signals, including BCR–immune complex formation and costimulation. These may induce Ig class-switch recombination and affinity maturation, especially in TNBC.
We identified an IgG-dominated clonally expanded B lymphocyte response in those breast cancers that were highly infiltrated by immune cells, and positive associations between IgG class regulator signatures and patient outcome, especially in TNBC. The Ig isotypes produced in the TME may be critical for containing tumor growth. Antibodies can directly block cancer cell signaling, and if expressed of the IgG class, and specifically IgG1, they engender immune-mediated clearance of cancer cells via complement activation and engagement of Fc receptor–expressing monocytes, macrophages, and NK cells (50). In cancer lesions, we observed a higher mutation load in the CDRs of IgG compared with their clonally related IgA, absent in the FWR, and in BCRs from normal breast. This stronger positive selection pressure on IgG+ B lymphocytes may represent different aspects of humoral immunity, likely arising from a common B-cell precursor, with IgA+ and IgG+ B lymphocytes driven to generate highly specific but divergent BCRs. Our work suggests significant implications regarding the effectiveness of antitumor B lymphocyte responses within highly infiltrated cancers, where a higher IgG+:IgA+ ratio among the expansive B lymphocyte infiltrate could engender potent antitumor responses through the increased relative proportion of IgG isotypes, featuring increased capacity to trigger antibody-dependent cellular cytotoxicity by NK cells and macrophages. Future therapeutic interventions may facilitate or take advantage of IgG+ memory and plasma cell infiltrates, and thus influence the balance in favor of immune-activatory Ig isotypes in tumors.
Collectively, our findings indicate a highly activated B lymphocyte compartment in breast cancers. Intratumoral B lymphocytes are spatially associated with T lymphocytes within large stromal clusters, isotype-switch, expand into large clonal families featuring a bias toward IgG subclasses, and carry specific variable region gene combinations with narrow repertoires, all suggesting targeted humoral responses to specific antigenic stimuli. Clonally restricted IgG-expressing B cells may include in situ generated germinal center and activated memory B cells and terminally differentiated plasma cells. Together, these may contribute to humoral immune responses in breast cancer, likely more prominent in TNBC. These expansive and clonally skewed Ig repertoires, and in particular those switched to IgG isotypes, may be associated with more-favorable patient survival. Although found across breast cancer types, these dynamic B lymphocyte traits are highly prominent in TNBC. Despite the usually poor prognosis and aggressive nature of TNBC, this analysis implicates considerable biological and associated prognostic heterogeneity that extends to the TME. Expansive and active B lymphocyte cancer infiltrates, in a proportion of individuals, provide a degree of antitumor activity that may, in combination with other immune responses, confer a survival benefit and may be exploited with immunotherapies to aid in tumor clearance. Elucidating the microenvironmental conditions required to initiate, sustain, and enhance these beneficial antitumor responses may be key to developing novel treatments.
Authors' Disclosures
D.K. Dunn-Walters reports grants from MRC during the conduct of the study. A.N.J. Tutt reports grants from Breast Cancer Now Charity and CRUK during the conduct of the study as well as reports personal fees from Inbiomotion, CRUK, MD Anderson, and reports other support from Tesaro/GlaxoSmithKline, AstraZeneca, AstraZeneca, Merck KGAA, as well as personal fees from Pfizer, Vertex, Artios, Prime Oncology, and reports other support from Medivation, Myriad Genetics, and reports personal fees from Gilead, other support from AstraZeneca, and grants from AstraZeneca outside the submitted work; as well as a patent for AstraZeneca with royalties paid to the Institute of Cancer Research with royalties paid from AstraZeneca. S.N. Karagiannis reports grants from Breast Cancer Now, Cancer Research UK, Medical Research Council, and National Institute for Health Research during the conduct of the study as well as reports grants from Epsilogen Ltd. outside the submitted work; and patents on novel antibodies for cancer therapy. No disclosures were reported by the other authors.
Acknowledgments
The authors acknowledge support by Breast Cancer Now (147; KCL-BCN-Q3); the Cancer Research UK King's Health Partners Center at King's College London (C604/A25135); Cancer Research UK (C30122/A11527; C30122/A15774); the Medical Research Council (MR/L023091/1); CR UK//NIHR in England/DoH for Scotland, Wales and Northern Ireland Experimental Cancer Medicine Center (C10355/A15587). The research was supported by the National Institute for Health Research Biomedical Research Center based at Guy's and St Thomas' NHS Foundation Trust and King's College London (IS-BRC-1215-20006). The authors are solely responsible for study design, data collection, analysis, decision to publish, and preparation of the manuscript. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. The authors acknowledge the Breast Cancer Now Tissue Bank in collecting and making available samples used in the generation of this publication. They acknowledge the Biomedical Research Center Immune Monitoring Core Facility at Guy's and St Thomas' NHS Foundation Trust and the Nikon Imaging Center at Kings College London for assistance.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Contributions
R.J. Harris: Conceptualization, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft. A. Cheung: Conceptualization, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. J.C. Ng: Data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. R. Laddach: Data curation, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. A.M. Chenoweth: Data curation, investigation, writing–review and editing. S. Crescioli: Conceptualization, writing–review and editing. M. Fittall: Conceptualization, data curation, validation, investigation, methodology. D. Dominguez Rodriguez: Data curation, validation, investigation. J. Roberts: Data curation, formal analysis, methodology. D. Levi: Data curation, formal analysis. F. Liu: Formal analysis, supervision. E. Alberts: Data curation, formal analysis, visualization. J. Quist: Data curation, formal analysis. A. Santaolalla: Methodology. S.E. Pinder: Supervision, writing–review and editing. C. Gillett: Supervision. N. Hammar: Supervision. S. Irshad: Resources. M. Van Hemelrijck: Supervision, writing–review and editing. D.K. Dunn-Walters: Resources, supervision. F. Fraternali: Supervision, investigation, methodology. J.F. Spicer: Supervision. K.E. Lacy: Supervision. S. Tsoka: Supervision. A. Grigoriadis: Conceptualization, resources, supervision, funding acquisition, writing–review and editing. A.N.J. Tutt: Conceptualization, resources, supervision, funding acquisition, writing–review and editing. S.N. Karagiannis: Conceptualization, resources, supervision, funding acquisition, writing–original draft, writing–review and editing.
References
- 1. Erdag G, Schaefer JT, Smolkin ME, Deacon DH, Shea SM, Dengel LT, et al. Immunotype and immunohistologic characteristics of tumor-infiltrating immune cells are associated with clinical outcome in metastatic melanoma. Cancer Res 2012;72:1070–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ladanyi A, Kiss J, Mohos A, Somlai B, Liszkay G, Gilde K, et al. Prognostic impact of B-cell density in cutaneous melanoma. Cancer Immunol Immunother 2011;60:1729–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Garaud S, Zayakin P, Buisseret L, Rulle U, Silina K, de Wind A, et al. Antigen specificity and clinical significance of IgG and IgA autoantibodies produced in situ by tumor-infiltrating B cells in breast cancer. Front Immunol 2018;9:2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lehmann BD, Pietenpol JA, Tan AR. Triple-negative breast cancer: molecular subtypes and new targets for therapy. Am Soc Clin Oncol Educ Book 2015:e31–9. DOI: 10.14694/EdBook_AM.2015.35.e31. [DOI] [PubMed] [Google Scholar]
- 5. Loi S, Michiels S, Salgado R, Sirtaine N, Jose V, Fumagalli D, et al. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: results from the FinHER trial. Ann Oncol 2014;25:1544–50. [DOI] [PubMed] [Google Scholar]
- 6. Schmid P, Rugo HS, Adams S, Schneeweiss A, Barrios CH, Iwata H, et al. Atezolizumab plus nab-paclitaxel as first-line treatment for unresectable, locally advanced or metastatic triple-negative breast cancer (IMpassion130): updated efficacy results from a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2020;21:44–59. [DOI] [PubMed] [Google Scholar]
- 7. Keren L, Bosse M, Marquez D, Angoshtari R, Jain S, Varma S, et al. A structured tumor-immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. Cell 2018;174:1373–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ramakrishnan R, Assudani D, Nagaraj S, Hunter T, Cho HI, Antonia S, et al. Chemotherapy enhances tumor cell susceptibility to CTL-mediated killing during cancer immunotherapy in mice. J Clin Invest 2010;120:1111–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Seow DYB, Yeong JPS, Lim JX, Chia N, Lim JCT, Ong CCH, et al. Tertiary lymphoid structures and associated plasma cells play an important role in the biology of triple-negative breast cancers. Breast Cancer Res Treat 2020;180:369–77. [DOI] [PubMed] [Google Scholar]
- 10. Singh M, Al-Eryani G, Carswell S, Ferguson JM, Blackburn J, Barton K, et al. High-throughput targeted long-read single cell sequencing reveals the clonal and transcriptional landscape of lymphocytes. Nat Commun 2019;10:3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Scott AM, Wolchok JD, Old LJ. Antibody therapy of cancer. Nat Rev Cancer 2012;12:278–87. [DOI] [PubMed] [Google Scholar]
- 12. Rossetti RAM, Lorenzi NPC, Yokochi K, Rosa M, Benevides L, Margarido PFR, et al. B lymphocytes can be activated to act as antigen presenting cells to promote anti-tumor responses. PLoS ONE 2018;13:e0199034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science 2015;348:69–74. [DOI] [PubMed] [Google Scholar]
- 14. Hu X, Zhang J, Wang J, Fu J, Li T, Zheng X, et al. Landscape of B cell immunity and related immune evasion in human cancers. Nat Genet 2019;51:560–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Helmink BA, Reddy SM, Gao J, Zhang S, Basar R, Thakur R, et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature 2020;577:549–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Garaud S, Buisseret L, Solinas C, Gu-Trantien C, de Wind A, Van den Eynden G, et al. Tumor infiltrating B cells signal functional humoral immune responses in breast cancer. JCI Insight 2019;5:e129641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Braso-Maristany F, Filosto S, Catchpole S, Marlow R, Quist J, Francesch-Domenech E, et al. PIM1 kinase regulates cell death, tumor growth and chemotherapy response in triple-negative breast cancer. Nat Med 2016;22:1303–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Szekely B, Bossuyt V, Li X, Wali VB, Patwardhan GA, Frederick C, et al. Immunological differences between primary and metastatic breast cancer. Ann Oncol 2018;29:2232–9. [DOI] [PubMed] [Google Scholar]
- 19. Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 2010;123:725–31. [DOI] [PubMed] [Google Scholar]
- 20. Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dieu-Nosjean MC, Goc J, Giraldo NA, Sautes-Fridman C, Fridman WH. Tertiary lymphoid structures in cancer and beyond. Trends Immunol 2014;35:571–80. [DOI] [PubMed] [Google Scholar]
- 22. Azizi E, Carr AJ, Plitas G, Cornish AE, Konopacki C, Prabhakaran S, et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell 2018;174:1293–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36:411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc 2020;15:1484–506. [DOI] [PubMed] [Google Scholar]
- 26. Salgado R, Denkert C, Demaria S, Sirtaine N, Klauschen F, Pruneri G, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol 2015;26:259–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu YC, Kipling D, Dunn-Walters D. Assessment of B-cell repertoire in humans. Methods Mol Biol 2015;1343:199–218. [DOI] [PubMed] [Google Scholar]
- 28. Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res 2008;36:W503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Margreitter C, Lu HC, Townsend C, Stewart A, Dunn-Walters DK, Fraternali F. BRepertoire: a user-friendly web server for analysing antibody repertoire data. Nucleic Acids Res 2018;46:W264–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Townsend CL, Laffy JM, Wu YB, Silva O'Hare J, Martin V, Kipling D, et al. Significant differences in physicochemical properties of human immunoglobulin kappa and lambda CDR3 regions. Front Immunol 2016;7:388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yaari G, Uduman M, Kleinstein SH. Quantifying selection in high-throughput immunoglobulin sequencing datasets. Nucleic Acids Res 2012;40:e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Carpenter EL, Mick R, Rech AJ, Beatty GL, Colligon TA, Rosenfeld MR, et al. Collapse of the CD27+ B-cell compartment associated with systemic plasmacytosis in patients with advanced melanoma and other cancers. Clin Cancer Res 2009;15:4277–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Walldius G, Malmstrom H, Jungner I, de Faire U, Lambe M, Van Hemelrijck M, et al. Cohort profile: the AMORIS cohort. Int J Epidemiol 2017;46:1103–i. [DOI] [PubMed] [Google Scholar]
- 34. Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011;121:2750–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yin Q, Wang X, McBride J, Fewell C, Flemington E. B-cell receptor activation induces BIC/miR-155 expression through a conserved AP-1 element. J Biol Chem 2008;283:2654–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moratz C, Kang VH, Druey KM, Shi CS, Scheschonka A, Murphy PM, et al. Regulator of G protein signaling 1 (RGS1) markedly impairs Gi alpha signaling responses of B lymphocytes. J Immunol 2000;164:1829–38. [DOI] [PubMed] [Google Scholar]
- 37. D'Arena G, Musto P, Nunziata G, Cascavilla N, Savino L, Pistolese G. CD69 expression in B-cell chronic lymphocytic leukemia: a new prognostic marker? Haematologica 2001;86:995–6. [PubMed] [Google Scholar]
- 38. Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maletzki C, Jahnke A, Ostwald C, Klar E, Prall F, Linnebacher M. Ex-vivo clonally expanded B lymphocytes infiltrating colorectal carcinoma are of mature immunophenotype and produce functional IgG. PLoS ONE 2012;7:e32639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Silina K, Rulle U, Kalnina Z, Line A. Manipulation of tumour-infiltrating B cells and tertiary lymphoid structures: a novel anti-cancer treatment avenue? Cancer Immunol Immunother 2014;63:643–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Armas-Gonzalez E, Dominguez-Luis MJ, Diaz-Martin A, Arce-Franco M, Castro-Hernandez J, Danelon G, et al. Role of CXCL13 and CCL20 in the recruitment of B cells to inflammatory foci in chronic arthritis. Arthritis Res Ther 2018;20:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Okada T, Ngo VN, Ekland EH, Forster R, Lipp M, Littman DR, et al. Chemokine requirements for B-cell entry to lymph nodes and Peyer's patches. J Exp Med 2002;196:65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pavoni E, Monteriu G, Santapaola D, Petronzelli F, Anastasi AM, Pelliccia A, et al. Tumor-infiltrating B lymphocytes as an efficient source of highly specific immunoglobulins recognizing tumor cells. BMC Biotechnol 2007;7:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Buisseret L, Desmedt C, Garaud S, Fornili M, Wang X, Van den Eyden G, et al. Reliability of tumor-infiltrating lymphocyte and tertiary lymphoid structure assessment in human breast cancer. Mod Pathol 2017;30:1204–12. [DOI] [PubMed] [Google Scholar]
- 45. Song IH, Heo SH, Bang WS, Park HS, Park IA, Kim YA, et al. Predictive value of tertiary lymphoid structures assessed by high endothelial venule counts in the neoadjuvant setting of triple-negative breast cancer. Cancer Res Treat 2017;49:399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mohammed ZM, Going JJ, Edwards J, Elsberger B, McMillan DC. The relationship between lymphocyte subsets and clinico-pathological determinants of survival in patients with primary operable invasive ductal breast cancer. Br J Cancer 2013;109:1676–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Garnelo M, Tan A, Her Z, Yeong J, Lim CJ, Chen J, et al. Interaction between tumour-infiltrating B cells and T cells controls the progression of hepatocellular carcinoma. Gut 2017;66:342–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hollern DP, Xu N, Thennavan A, Glodowski C, Garcia-Recio S, Mott KR, et al. B cells and T follicular helper cells mediate response to checkpoint inhibitors in high mutation burden mouse models of breast cancer. Cell 2019;179:1191–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lund FE. Cytokine-producing B lymphocytes-key regulators of immunity. Curr Opin Immunol 2008;20:332–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cheung A, Opzoomer J, Ilieva KM, Gazinska P, Hoffmann RM, Mirza H, et al. Anti-folate receptor alpha-directed antibody therapies restrict the growth of triple-negative breast cancer. Clin Cancer Res 2018;24:5098–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The R code used to analyze scRNA-seq data from GSE114725 (22) can be accessed from https://codeocean.com/capsule/7488142. The R code to analyze BCR Repertoire data collected from breast tissues can be accessed at https://codeocean.com/capsule/4153741.



![Figure 2. B lymphocyte infiltration and its positive prognostic value in TNBC. A, TIL-B quantification by IHC within normal breasts and TNBC (N = 15 each, Bart's IHC cohort), and by GEx (normal breast) vs. TNBC (N = 10 vs. 131, KCL GEx cohort); non-TNBC vs. TNBC (N = 515 vs. 123, TCGA GEx cohort); TNBC subtypes [mesenchymal (M), luminal androgen receptor (LAR), basal-like 1 and 2 (BL1 and 2), and immunomodulatory (IM; N = 122, KCL GEx cohort]. The Mann–Whitney test was used for statistical significance. B, TNBC TIL-B correlation with TIL-T by IHC (r = 0.72, Bart's IHC cohort) and by GEx (r = 0.73, KCL GEx cohort). Linear regression analysis was used to calculate correlation coefficients (r) and P values. C, B lymphocyte metagene NanoString GEx data comparing B lymphocytes in primary tumors with metastatic sites (N = 31 vs. 17; NanoString cohort). D, High (above median) TIL-B densities by IHC were associated with better overall survival in TNBC (N = 15; Bart's IHC cohort), and in the basal-like subtype by high CD20 GEx (N = 241; KM plotter cohort; log rank test used to assess statistical significance). E, Kaplan–Meier survival curves display DMFS for naïve B cells, plasma cells, and memory B cells in TNBC (KCL GEx cohort) using CIBERSORT (20). Data were divided into four groups based on B lymphocyte subset and TIL levels stratified by semiquantitative TIL classification. Statistical significance was assessed using univariate Cox proportional hazards regression models. F, Representative IHC/IF images (Bart's IHC cohort) depicting nucleated cells (DAPI), epithelial cells (PanCK), B lymphocytes (CD20), and T lymphocytes (CD3) within TNBC TIL-Blow and TIL-Bhigh lesions. Scale bar, 50 μm. G, Representative TNBC (TIL-Bhigh) images highlighting numerous lymphoid aggregates (within white dash lines) consisting of B lymphocytes assembled adjacent to a T lymphocyte zone. Brown dash lines indicate carcinoma edge. Scale bar, 2 mm. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/a589/9398116/79435c19899c/4290fig2.jpg)
![Figure 4. scRNA-seq analysis reveals BCR-driven TIL-B activatory signatures, and B lymphocyte functional trait gene markers predict positive survival outcome. A, UMAP visualization according to global GEx of single B lymphocytes pooled from the peripheral blood (1,476 cells) and tumors (1,021 cells) of eight patients (single-cell cohort), colored by relative normalized gene expression levels for CD20 and CD27. B, The detection of differentially expressed genes [DEG; on cells originally annotated by Azizi and colleagues (22) as B cells] demonstrates elevated expression of FOS, JUN, RGS1, and CD69, indicated by fold change (FC; determined using Wilcoxon rank sum test). C, Gene set enrichment analysis of TIL-B relative to circulating B lymphocytes using hallmark gene sets. Red, positive normalized enrichment scores (hallmark expression enhanced in TIL-B). D, Survival analysis in KM Plotter of determined ER−HER2−/basal surrogate, HER2+, luminal A, and luminal B subtype KM plotter surrogate subgroups (KM plotter cohort; ref. 19) for expression of gene signatures positively regulating key B lymphocyte properties (activation, proliferation, and differentiation). Representative genes listed for B lymphocyte proliferation (44 total in set). Signatures from all three functions carry positive prognostic value in the basal-like cancer subtype. Individual genes were evaluated in combination with each other gene (left) and gene set as a whole (right). E, CellPhoneDB (25) was applied to analyze B-cell–T-cell interactions (single-cell cohort). After FDR (FDR < 0.001) correction, communication pathways identified included lymphoid assembly, cytokine signaling, costimulation, T-cell–dependent B-cell activation, and cytotoxic T lymphocyte (CTL) activation. Circle sizes indicate P value, whereas color-coding represents the average expression level of interacting molecule 1 in cluster 1 and interacting molecule 2 in cluster 2. ****, P < 0.0001.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/a589/9398116/9d36b38b731e/4290fig4.jpg)