Summary
Childhood tumors that occur synchronously in different anatomical sites usually represent metastatic disease. However, such tumors can be independent neoplasms. We investigated whether cases of bilateral neuroblastoma represented independent tumors in two children with pathogenic germline mutations by genotyping somatic mutations shared between tumors and blood. Our results suggested that in both children, the lineages that had given rise to the tumors had segregated within the first cell divisions of the zygote, without being preceded by a common premalignant clone. In one patient, the tumors had parallel evolution, including distinct second hits in SMARCA4, a putative predisposition gene for neuroblastoma. These findings portray cases of bilateral neuroblastoma as having independent lesions mediated by a germline predisposition. (Funded by Children with Cancer UK and Wellcome.)
TUMORS THAT OCCUR IN MULTIPLE SITES USUALLY REPRESENT METASTATIC disease originating from the same primary tumor. However, in some forms of childhood cancer, it has been proposed that synchronous lesions that are located in separate anatomical regions may represent independent tumors. The only established example of this phenomenon has been shown in children who have pathogenic germline mutations in the gene encoding RB transcriptional corepressor 1 (RB1), a tumor-suppressor retinoblastoma protein that is dysfunctional in several major cancers. Such children are at increased risk for bilateral retinoblastoma, with each tumor harboring a somatic RB1 mutation that is independent of the RB1 mutation in the contralateral tumor.1,2 Other tumor types that may be multifocal include neuroblastoma, in particular “4S” metastatic disease of infants, and bilateral neuroblastoma.3 However, the notion of independent tumor formation has not been substantiated in these diseases. For the clinical treatment of children with multisite neuroblastoma, the distinction between metastatic and multifocal disease is likely to have decisive implications. For example, in patients with bilateral tumors, it may be justifiable to reduce treatment intensity if the tumors were found to be independent, low-risk lesions. By contrast, if such tumors were metastatic, most treatment protocols would stipulate an aggressive, multimodal approach to therapy.
It is possible to establish the phylogenetic relationship between multiple tumors by comparing the somatic mutations that they share. Such analyses have been applied widely to the study of the evolution of human cancers.4 We have recently adapted this approach to enable the reconstruction of the embryologic features of childhood tumors.5 As cells divide and develop, they acquire (post-zygotic) somatic genetic variants that represent embryologic lineages and may be measurable in normal tissues. These variants and lineages render the organism mosaic. Therefore, in principle, it would be possible to relate tumors to embryologic lineages through mosaic variants (Fig. 1), which may unambiguously resolve whether tumors are developmentally related and reveal a precise developmental stage at which tumors formed or segregated. For example, in the case of bilateral Wilms’ tumor, a childhood kidney cancer, we found that tumors originated from a common embryologic precursor that formed after blood and kidney lineages segregated.5 Here, we applied our approach to rare cases of bilateral neuroblastoma in two children to investigate whether these lesions represented independent cancers.
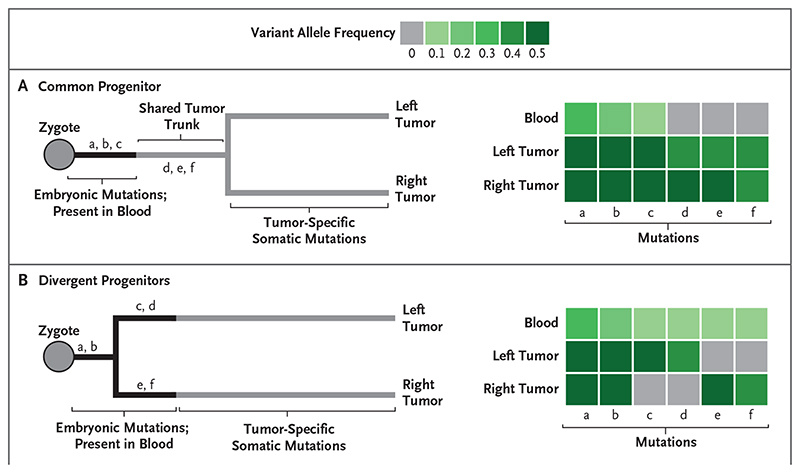
Figure 1. Possible Developmental Origins of Multisite Tumor Formation.
The expected pattern of mutation sharing between tumors and paired normal blood reflects the origin of bilateral tumors. Bilateral tumors can arise from a common progenitor in development, which will be delineated by a shared trunk of mutations that is absent from blood (Panel A). If the tumors diverged early in embryogenesis, one or both lesions will share mutations with blood but not with the other lesion (Panel B). Letters in the phylogenetic trees refer to postzygotic mutations; the variant allele frequency of these mutations is provided in the heat maps.
Case Reports
Patient 1 (case number, PD36812), a previously healthy 4-year-old boy, was referred to us for the evaluation of abdominal masses (Fig. 2A). After performing standard diagnostic and staging investigations that included bilateral biopsies of suprarenal lesions, we derived a diagnosis of bilateral neuroblastoma, with each tumor having poorly differentiated histologic features, no amplification in MYCN (a member of the Myc family of oncogenes), and the deletion of 19p. There was no evidence of distant metastatic disease. We initiated treatment according to the high-risk protocol of Société Internationale d’Oncologie Pédiatrique (SIOP).
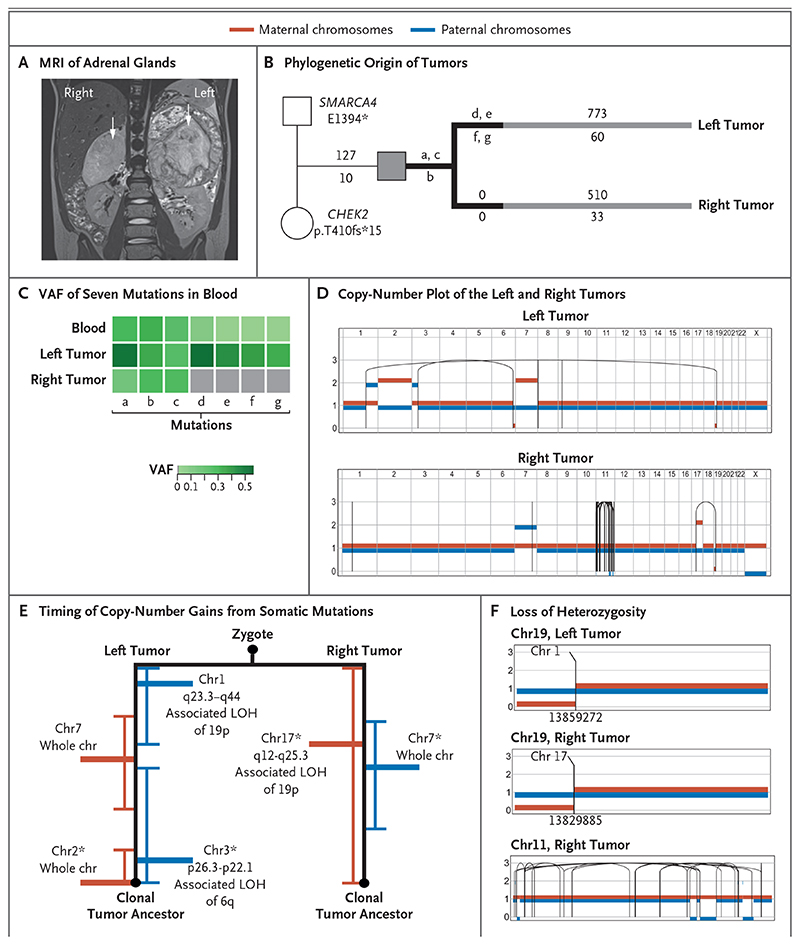
Figure 2. Independent Origin and Convergent Parallel Evolution of Tumors in Patient 1.
Panel A shows coronal magnetic resonance imaging (MRI) of the right and left adrenal glands in Patient 1 (case number, PD36812), with large tumors indicated by white arrows. Panel B shows the phylogenetic origins of the right and left tumors in Patient 1. The patient inherited two pathogenic variants: a truncating missense variant in SMARCA4 from the father and a truncating frameshift CHEK2 variant from the mother. Branches in black depict variants (labeled a through g) in the tumors that were shared with blood. The numbers of substitutions are annotated above the branch, and insertions or deletions (indels) are annotated below the branch. Panel C shows the variant allele frequency (VAF) of the seven mutations present in blood (labeled a through g, as in the phylogenetic tree). Panel D shows a phased copy-number plot of the left and right adrenal tumors. Breakpoints of structural variants are connected by solid lines. In the right tumor, the dense grouping of solid lines on chromosome 11 is the result of chromothripsis. Panel E shows the timing of the acquisition of copy-number gains from somatic mutations before and after duplication. Gains of maternal chromosomes (chr) are indicated in red and gains of paternal chromosomes in blue. Within each branch, asterisks highlight pairs of rearrangements that are unlikely to have occurred at different time points (P>0.05 by the Poisson test). LOH denotes loss of heterozygosity. Panel F shows the different breakpoints that underlie the loss of heterozygosity of chromosome 19 in the left and right tumors. Only the right tumor shows signs of chromothripsis. Translocations that mediate a loss of heterozygosity on chromosome 19p in the left and right tumors have different chromosomal partners and are located in different genomic positions, as indicated by the numbers below the breakpoints.
Patient 2 (case number, PD34954), a 4-year-old girl, was referred to us for the management of bilateral adrenal masses that were shown to be undifferentiated neuroblastoma with wide-spread metastatic disease. In contrast to Patient 1, Patient 2 had a left adrenal tumor that was locally extensively invasive, whereas the right adrenaltumor constituted a relatively small adrenal mass (Fig. 3A). A diagnosis of congenital central hypoventilation syndrome was made in association with a germline mutation in PHOX2B, an alteration that also results in a predisposition to neuroblastoma.6
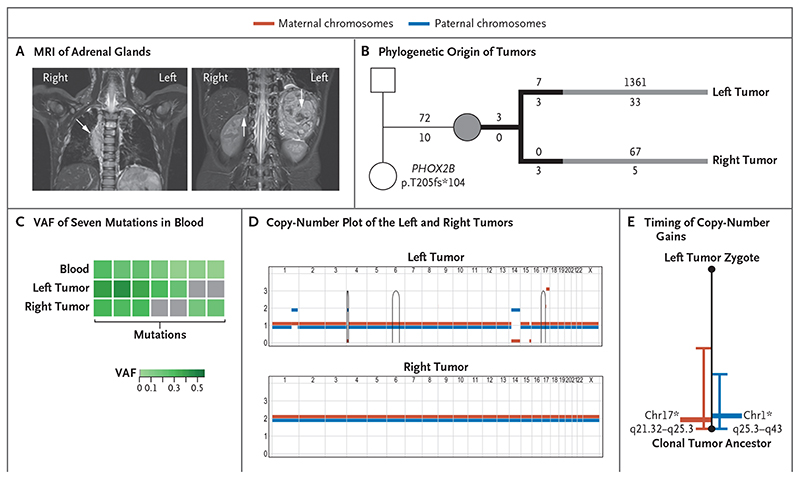
Figure 3. Independent Origin of Bilateral Tumors with Divergent Risk Profiles in Patient 2.
Coronal MRI shows a large left adrenal tumor extending into the right thorax (image at left) and a small right adrenal tumor (with arrows indicating tumors) in Patient 2 (case number, PD34954). Panel B shows the phylogenetic origin of the left and right tumors, with a de novo truncating variant in PHOX2B. Branches in black depict variants shared with blood. The numbers of substitutions are annotated above the branch, and indels are annotated below the branch. Panel C shows the variant allele frequencies of representative variants that are present in blood and that delineate both tumor lineages. A full heat map is provided in Figure S4 in Supplementary Appendix 1. Panel D shows a phased copy-number plot of the left and right tumors. Breakpoints of structural variants are connected by solid lines. Panel E shows the timing of the acquisition of copy-number gains from somatic mutations before and after duplication. Asterisks indicate that there was a lack of evidence that gains occurred at different time points (P>0.05 by the Poisson test).
Genomic Analysis and Results
We performed whole-genome sequencing of DNA derived from each tumor and from peripheral blood samples obtained from each of the children and their parents. Using a validated variantcalling pipeline,5 we determined variants of all classes, which were subdivided into inherited germline, de novo germline, mosaic (embryonic), and somatic variants. From these catalogues, we reconstructed the phylogenetic development of each tumor, as described previously.5 Details about this approach are provided in the Supplementary Methods section and in Figures S1 through S3 in Supplementary Appendix 1; an overview of the two cases is provided in Table S1 in Supplementary Appendix 2, along with details about the two patients in Tables S2 through S5. The two appendixes are available with the full text of this article at NEJM.org.
Patient 1
In Patient 1, we examined inherited and de novo germline variants. We found two bona fide pathogenic mutations, one inherited from each parent. The child carried a paternal truncating variant in SMARCA4, which is a putative predisposition gene for neuroblastoma.7–9 In addition, he carried a maternally inherited truncating mutation in the gene encoding checkpoint kinase 2 (CHEK2), which has recently been found to confer a predisposition to a variety of childhood tumors7–9 (Fig. 2B). Next, we examined mosaic variants that were present in one or both tumors and in blood cells. These variants are nonheterozygous in the blood and can be inferred to have accumulated during the first few cell divisions of the zygote. Three of these variants were present in blood and in both tumors (Fig. 2C). Unexpectedly, four of these variants were present in the left tumor and blood but absent in the right tumor. These variants, which are exclusive to cells of the blood and the left tumor, were not found at chromosomal loci affected by the loss of heterozygosity or copynumber variation at chromosomal loci (Table S5 in Supplementary Appendix 2). The exclusivity of these variants indicated that the tumors had been derived from separate embryologic lineages that segregated within the first few cell divisions before gastrulation. In a separate analysis, we did not find any somatic variants that were shared between the left and right tumors, which was consistent with the previous finding of exclusivity.
After segregation of the two lineages, the tumors had undergone remarkably parallel development (Fig. 2D). Among key somatic changes in each tumor was the gain of chromosome 7, albeit of different parental chromosomes: duplication of the maternal chromosome in the left tumor and duplication of the paternal chromosome in the right. Furthermore, both tumors had a loss of maternal 19p, thus deleting the remaining intact germline copy of SMARCA4. However, different translocations generated the breakpoint on 19p (Fig. 2D, 2E, and 2F). In the right tumor, the loss of 19p was coupled with a gain of 17q, a genomic feature of high-risk neuro-blastoma.6 In addition, on chromosome 11, the right tumor had a high density of intrachromosomal rearrangements associated with copy-number changes, which was consistent with chromothripsis (i.e., multiple chromosomal rearrangements that are thought to occur simultaneously and are commonly found in human cancer). To establish the order of these parallel somatic events, we applied analytic techniques that time the acquisition of copy-number changes relative to that of single-base variants.4 Our findings indicated that these somatic events may have been acquired in the same order on both the left and right sides (Fig. 2E). As has been shown in other cancers,4,10 we identified trisomy 7 as an early acquired event. Among the somatic events unique to each tumor, we did not find any additional driver events.
Patient 2
We performed the same analyses on samples obtained from Patient 2 and her parents, and we again found 3 variants in both tumors and in blood, along with 14 variants that were exclusive to one tumor or the other yet present in blood cells (Fig. 3B and 3C and Fig. S4 in Supplementary Appendix 1). We did not find variants that were common to both tumors in a separate analysis. We concluded that both tumors were derived from independent embryonic lineages that segregated within a few cell divisions after fertilization. In Patient 2, tumor development diverged, leading to a low-risk neuroblastoma in the right adrenal with no additional driver events. By contrast, the tumor in the left adrenal had acquired somatic genetic changes that characterize high-risk tumors, including gain of chromosome 17q (Fig. 3D and 3E), consistent with its characteristics (locally and extensively invasive).
Discussion
Here, we reconstructed the development of two cases of bilateral neuroblastoma from somatic mutations that provide a barcode for embryonic cell divisions and lineages. Our expectation might have been that such tumors stem from a common postzygotic precursor, similar to what has been observed in bilateral Wilms’ tumor.5 In such a case, we would have observed shared mutations between tumors that we did not find in blood. However, we found that such tumors are independent neoplasms that separated early in embryogenesis, thus providing direct phylogenetic proof for the presumption that some multisite neuroblastomas do not represent metastatic disease.
Although our study is limited in scale owing to the rarity of bilateral neuroblastoma, our findings raise questions about multisite tumors in children. In the majority of cases, it is likely that, as commonly assumed, multisite tumors represent metastatic disease. Nevertheless, our observations may lead to some questioning of this assumption in certain clinical scenarios, such as multisite tumors in infants or in children who are predisposed to cancer. One could argue that for a child with independent multisite tumors, treatment should be guided by the lesion associated with the worst prognosis, with a less aggressive treatment regimen in the instance of multiple, independent low-risk tumors.
In our study, Patient 1 had two embryonic lineages that evolved in parallel into high-risk tumors. Although the parallel convergent development of different clones within a tumor is a well-described phenomenon,11 the development of independent synchronous tumors that mirror each other is a remarkable observation. We speculate that each of the germline pathogenic variants sets the stage for tumor formation through a restricted repertoire of additional driver events. Our analytic strategy may provide further insights into the early embryonic development of childhood cancers, especially because whole-genome sequencing in the context of clinical management of cancer is becoming increasingly common.
Supplementary Material
Acknowledgments
Supported by Children with Cancer UK and Wellcome.
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
We thank the children and their families for participating in our research.
Contributor Information
Tim H.H. Coorens, From the Wellcome Sanger Institute, Hinxton
Sarah J. Farndon, Cambridge University Hospitals NHS Foundation Trust
Thomas J. Mitchell, From the Wellcome Sanger Institute, Hinxton; Cambridge University Hospitals NHS Foundation Trust; the Departments of Surgery
Neha Jain, University of Cambridge, Cambridge, and UCL Great Ormond Street Institute of Child Health; Great Ormond Street Hospital for Children NHS Foundation Trust
Sangjin Lee, From the Wellcome Sanger Institute, Hinxton
Michael Hubank, the Royal Marsden NHS Foundation Trust London — all in the United Kingdom
Neil Sebire, University of Cambridge, Cambridge, and UCL Great Ormond Street Institute of Child Health; Great Ormond Street Hospital for Children NHS Foundation Trust
John Anderson, University of Cambridge, Cambridge, and UCL Great Ormond Street Institute of Child Health; Great Ormond Street Hospital for Children NHS Foundation Trust
Sam Behjati, From the Wellcome Sanger Institute, Hinxton; Cambridge University Hospitals NHS Foundation Trust; the Departments of Paediatrics
References
- 1.Knudson AG, Jr, Strong LC. Mutation and cancer: neuroblastoma and pheochromocytoma. Am J Hum Genet. 1972;24:514–32. [PMC free article] [PubMed] [Google Scholar]
- 2.Knudson AG. Hereditary cancer: two hits revisited. J Cancer Res Clin Oncol. 1996;122:135–40. doi: 10.1007/BF01366952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Noesel MM, Hählen K, Hakvoort-Cammel FG, Egeler RM. Neuroblastoma 4S: a heterogeneous disease with variable risk factors and treatment strategies. Cancer. 1997;80:834–43. [PubMed] [Google Scholar]
- 4.Gerstung M, Jolly C, Leshchiner I, et al. The evolutionary history of 2,658 cancers. Nature. 2020;578:122–8. doi: 10.1038/s41586-019-1907-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coorens THH, Treger TD, Al-Saadi R, et al. Embryonal precursors of Wilms tumor. Science. 2019;366:1247–51. doi: 10.1126/science.aax1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rybinski B, Wolinsky T, Brohl A, et al. Multifocal primary neuroblastoma tumor heterogeneity in siblings with co-occurring PHOX2B and NF1 genetic aberrations. Genes Chromosomes Cancer. 2020;59:119–24. doi: 10.1002/gcc.22809. [DOI] [PubMed] [Google Scholar]
- 7.Gröbner SN, Worst BC, Weischenfeldt J, et al. The landscape of genomic alterations across childhood cancers. Nature. 2018;555:321–7. doi: 10.1038/nature25480. [DOI] [PubMed] [Google Scholar]
- 8.Pugh TJ, Morozova O, Attiyeh EF, et al. The genetic landscape of high-risk neuroblastoma. Nat Genet. 2013;45:279–84. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bellini A, Bessoltane-Bentahar N, Bhalshankar J, et al. Study of chromatin remodeling genes implicates SMARCA4 as a putative player in oncogenesis in neuroblastoma. Int J Cancer. 2019;145:2781–91. doi: 10.1002/ijc.32361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heim S, Mandahl N, Jin Y, et al. Trisomy 7 and sex chromosome loss in human brain tissue. Cytogenet Cell Genet. 1989;52:136–8. doi: 10.1159/000132863. [DOI] [PubMed] [Google Scholar]
- 11.Martinez P, Birkbak NJ, Gerlinger M, et al. Parallel evolution of tumour subclones mimics diversity between tumours. J Pathol. 2013;230:356–64. doi: 10.1002/path.4214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.