Abstract
Creating in vitro models of diseases of the pancreatic ductal compartment requires a comprehensive understanding of the developmental trajectories of pancreas-specific cell types. Here, we report the single-cell characterization of the differentiation of pancreatic duct-like organoids (PDLOs) from human induced pluripotent stem cells (hiPSCs) on a microwell chip that facilitates the uniform aggregation and chemical induction of hiPSC-derived pancreatic progenitors. Via time-resolved single-cell transcriptional profiling and immunofluorescence imaging of the forming PDLOs, we identified differentiation routes from pancreas progenitors through ductal intermediates to two types of mature duct-like cell and a few non-ductal cell types. PDLO subpopulations expressed either mucins or the cystic fibrosis transmembrane conductance regulator, and resembled human adult duct cells. We also used the chip to uncover ductal markers relevant to pancreatic carcinogenesis, and to establish PDLO co-cultures with stellate cells, which allowed for the study of epithelial–mesenchymal signalling. The PDLO microsystem could be used to establish patient-specific pancreatic-duct models.
Pancreatic ductal cells are organized in tubular ductal networks. Ductal cells secrete a bicarbonate-rich alkaline aqueous solution to transport zymogens produced by acinar cells1. Dysfunction of ductal cells can affect fluid composition in cystic fibrosis, leading to a subsequent deterioration of the entire organ1,2. Dysplastic events within the ductal epithelium can lead to pancreatic ductal adenocarcinoma (PDAC)3–5, one of the most lethal cancer types6. Shortage of healthy and early disease-affected primary ductal material hampers the discovery of biomarkers for diagnostics and drug development. To overcome the limitation imposed by the donor material shortage, lumen-forming pancreatic organoid cultures have been derived from resected healthy and diseased pancreata7,8. Pancreatic organoids preserve the cellular heterogeneity of the human pancreas and can be used to establish functional tests or discover biomarkers for PDAC in vitro 9,10. Pancreatic cancer-derived organoids: (i) exhibit an undefined genetic background, (ii) are generated from a tumour origin, (iii) mimic the end stage of cancer, and (iv) are unsuitable for biomarker discovery in the earliest stages of pancreatic dysplasia11. Adult human pancreatic organoids12,13 are challenging to establish and to culture in an untransformed state and moreover do not provide access to developmental intermediates. Lineage-committed pancreatic ductal cells generated from human pluripotent stem cells (hPSCs) could be an alternative source of pancreatic organoids to overcome these obstacles14,15.
The engineering and translation of in vitro ductal disease models require a full mechanistic understanding of the in vitro ductal differentiation landscape, cell type composition, and functionality at the ductal stage. Unfortunately, there is a dearth of knowledge on human embryonic duct development. Previous studies in rodents revealed that all pancreatic cell lineages evolve from a common pancreatic endoderm-derived cell type, named pancreatic progenitor cell16. Pancreatic progenitors organized in the pancreatic bud undergo tip/trunk patterning. While the acinar cells evolve from the tip domain, ductal cells together with subsequently delaminating endocrine cell types evolve from presumably bipotent trunk cells17,18. Time-resolved immunostaining of developing human embryos suggest a similar mechanism of human pancreatic cell type development19. Recently, we and others developed a differentiation protocol to guide hPSCs into pancreatic duct-like organoids (PDLOs)20,21. Single-cell RNA sequencing (scRNA-seq) has become the method of choice for studying cell differentiation in organoids or tissue development to reveal cell heterogeneity, quantify cell types, resolve molecular mechanisms leading to cell lineage bifurcations and their subsequent transitions22. While scRNA-seq of primary human pancreatic tissue is well-established23–28, a time-resolved single-cell transcriptional roadmap of developmental trajectories from hPSCs toward ducts does not exist. Organoid formation and stem cell differentiation are non-linear deterministic systems; thus, slight variations in initial conditions can influence the outcome of the process. Therefore, controlling the initial organoid size and shape in microwells is thought to improve the reproducibility of differentiation29,30. Microwells are the most simplistic chip technologies for assisting the formation of organoids and their culture. Further, microenvironmental signals of confined organoids and the organoids themselves are directly accessible for downstream analysis from the open microwells. Thus, a microwell chip design offers simple cell sample collection for scRNA-seq, proteome, and/or secretome analyses during ductal differentiation of hPSCs.
In this study, we designed a microwell chip to generate defined 3D aggregates of human induced pluripotent stem cells (hiPSCs) derived pancreatic progenitors, and subsequently to induce their differentiation toward PDLOs. Time-resolved scRNA-seq combined with cleared immunofluorescence imaging provided a deep understanding of in vitro ductal cell type differentiation. By defining the emergent cell types at each stage of differentiation based on their gene expression profiles and organoid structures, we provide a precise cell-by-cell description of the in vitro differentiation trajectory. Transcriptional data of PDLOs were complemented by their proteome and secretome, allowing the identification and validation of prognostic cancer markers. Thus, we show the applicability of hiPSC-derived PDLOs on a microwell chip for future ductal disease modelling.
Results
A microwell chip to form 3D aggregates from hiPSC-derived pancreatic progenitors
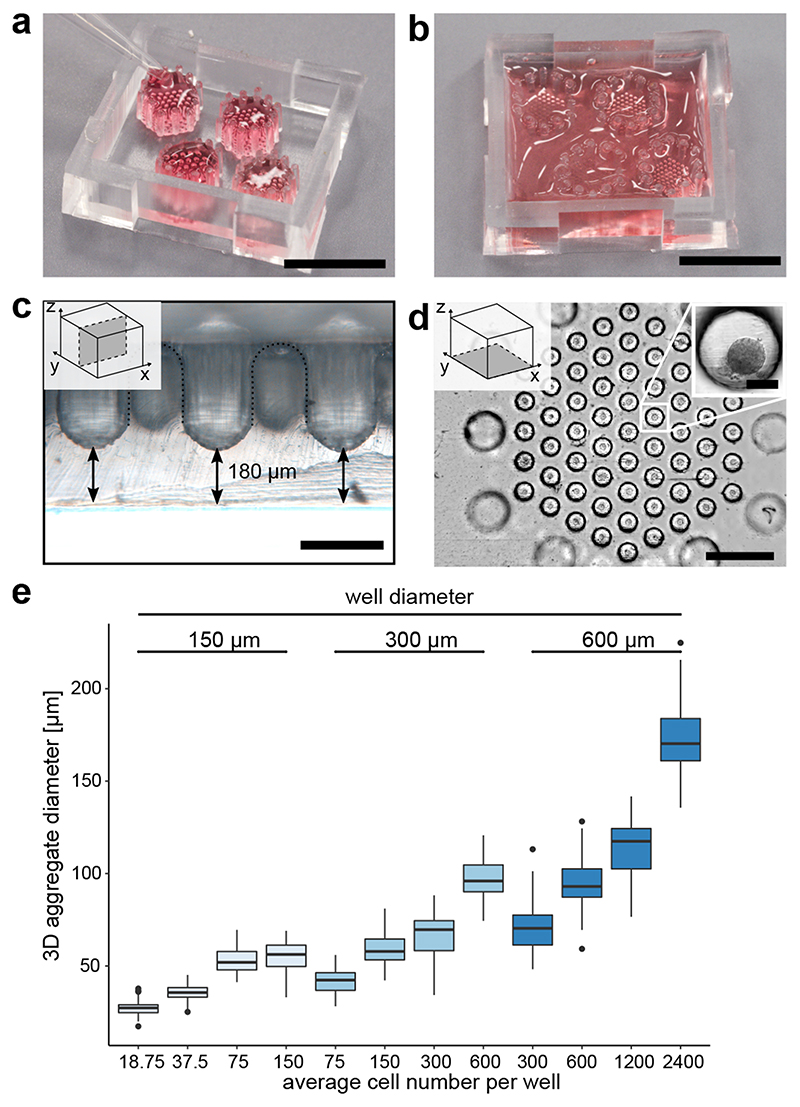
To allow engineering and long-term culture of duct-like organoids from hiPSC-derived pancreatic progenitors, we designed a microwell chip technology (Fig. 1a-d). The microwell chip was produced by soft-lithography with polydimethylsiloxane (PDMS), where the casting moulds were prepared using 3D stereolithography printing (Supplementary Fig. 1a). Each microwell chip contained four hexagonal arrays with cone-shaped wells, and 12 round-shaped pillars surrounded each array. The function of the pillars was to retain an aqueous drop (20–40 μL) above the array by surface tension (Fig. 1a,b). A 180-μm-thin PDMS bottom layer allowed high-resolution imaging of confined organoids (Fig. 1c). Together with roundings between wells, the pillars enabled an efficient and homogenous cell seeding process without blind spots. Details of the production method and workflow for cell seeding and culturing are given in Supplementary Fig. 1b. First, we validated the stem cell viability within differently sized microwells (150, 300, and 600 μm). Next, we systematically altered the initial cell number and well size to optimize the formation of hiPSC-derived aggregates of pancreatic progenitor. Pancreatic progenitor aggregates formed on the microwell chip within 4 h with uniform size according to variable well diameters and cell numbers (Fig. 1d,e). A low number of cells (<50 cells/well) compromised the aggregation step in the microwells, while the generation of cell aggregates with diameters over ~250 μm is known to impair nutrient supply31. For the subsequent ductal differentiation, we selected an initial pancreatic-progenitor-aggregate size of 96 μm on average (600 cells in 300 μm diameter wells).
Fig. 1. Microwell chips for generating and culturing 3D cell aggregates.
a, Image of the microwell chip with four microwell arrays (top-down view). Each hexagonal array is surrounded by 12 pillars to keep fluid volume above the microwell. Cells were seeded in 20-40 μL. Scale bar is 1 cm. b, After cell seeding, the entire microwell chip was filled up to 800 μL media for long-term cell culture. Scale bar is 1 cm. c, Cross-sectional view of the microwell chip bottom. Scale bar is 300 μm. d, Bottom view on one microwell array. Scale bar is 1 mm. Inset: Higher magnification of a pancreatic-progenitor-derived 3D aggregate formed from 600 cells. Scale bar is 100 μm. e, Aggregate size in dependence on the cell number and well diameter after 24 hours of seeding pancreatic progenitors, including at least 58 pancreatic-progenitor-derived 3D aggregates from three different microwell arrays. Boxplots display the median with the first and third quartile, whiskers denote the 1.5x interquartile range and outliers are marked as dots.
Ductal differentiation on the microwell chip
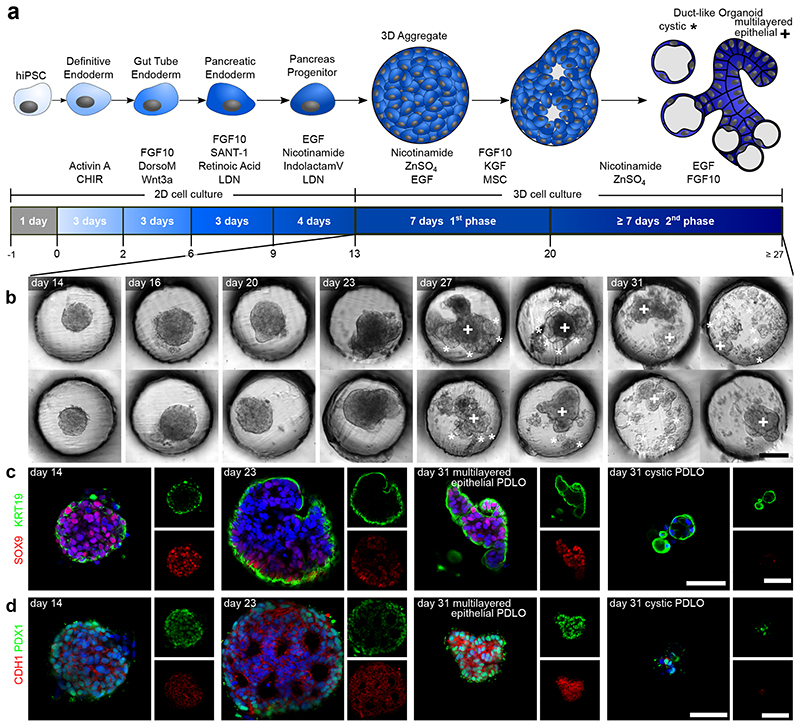
We differentiated 3D pancreatic-progenitor aggregates toward PDLOs using the microwell chip. Pancreatic progenitors generated in monolayer culture formed aggregates on the chip, followed by a two-step differentiation process with respective growth factors (Fig. 2a). Subsequently, pancreatic-progenitor aggregates underwent large morphological changes within the microwells, as indicated in the representative bright-field images (Fig. 2b; Supplementary Fig. 3a). Within the first ductal induction phase, the uniform round-shaped pancreatic-progenitor aggregate structure was broken up by newly formed multi-layered epithelial protrusions (Fig. 2b). In the second ductal differentiation phase, multi-layered epithelium organoids reduced their number of layers and cystic organoids were segregated from the outer layer of the PDLOs. Notably, a few cystic PDLO structures remained connected to the multi-layered epithelial PDLOs at day 31 of differentiation. The morphological transformation of the PDLOs is visualized in Supplementary Movie 1. High-resolution immunofluorescence images of cleared organoids confirmed cellular re-organization toward the end of phase 1 of ductal differentiation (Fig. 2c,d). Epithelial nature and pancreatic ductal identity of the cells at the final stage were confirmed by the upregulation of E-cadherin (CDH1), cytokeratin 19 (KRT19), aquaporin 5 (AQP5), mucin 1 (MUC1) carbonic anhydrase II (CA2), cytokeratin 7 (KRT7), claudin 1 (CLDN1), and cystic fibrosis transmembrane conductance regulator (CFTR) (Fig. 2c,d; Supplementary Fig. 3b). Expression of ductal markers, which were already detected at the pancreatic-progenitor stage, such as cytokeratin 8 (KRT8), SRY-box transcription factor 9 (SOX9), hepatocyte nuclear factor 1 homeobox B (HNF1B), and pancreatic and duodenal homeobox 1 (PDX1) were maintained in PDLOs (Fig. 2c,d; Supplementary Fig. 3b,c). Differently, the progenitor marker homeobox protein NKX-6.1 (NKX6-1), which becomes restricted to endocrine cells during pancreatic development, was downregulated on protein level (Supplementary Fig. 3c). Stemness marker SRY-box 2 (SOX2) and octamer-binding transcription factor 4 (OCT4) were absent (Supplementary Fig. 3d), as were non-ductal pancreas markers, except for few endocrine cells located at the periphery of PDLOs (Supplementary Fig. 3e). In agreement with the marker panel, a forskolin-induced swelling assay confirmed pancreatic ductal functionality (Supplementary Fig. 4). To demonstrate that the microwell-chip-derived-PDLOs are lineage-committed, we transplanted PDLOs differentiated until day 27 orthotopically into the pancreas of immunocompromised mice (Fig. 3a). PDLO engraftments formed tubular duct-like tissue after 8 weeks and homogenously expressed epithelial ductal markers such as SOX9, KRT19, AQP5, and CDH1 and were negative for endocrine cell types (Fig. 3b,c).
Fig. 2. PDLO differentiation in the microwell chip.
a, Schematic of cell differentiation from hiPSCs to PDLOs. b, Representative bright-field images of 3D pancreatic progenitors aggregates after 14, 16, 20, 23, 27, and 31 days of differentiation (DOD). Morphological changes of organoids within the first differentiation phase between 14 to 20 DOD, and formation of two morphologically distinct PDLO types within the second differentiation phase. Multi-layered epithelial PDLOs are marked with a cross (+) and cystic types with a star (*), respectively. See Supplementary Fig. 3a for overview images. c and d, Fluorescence images of the 3D aggregates along the time course of differentiation with co-stained ductal protein markers SOX9, KRT19, PDX1, and CDH1, respectively. A comprehensive set of progenitor and ductal markers is depicted in Supplementary Fig. 3b-e. The nucleus was counterstained with DAPI. Scale bar denotes 50 μm.
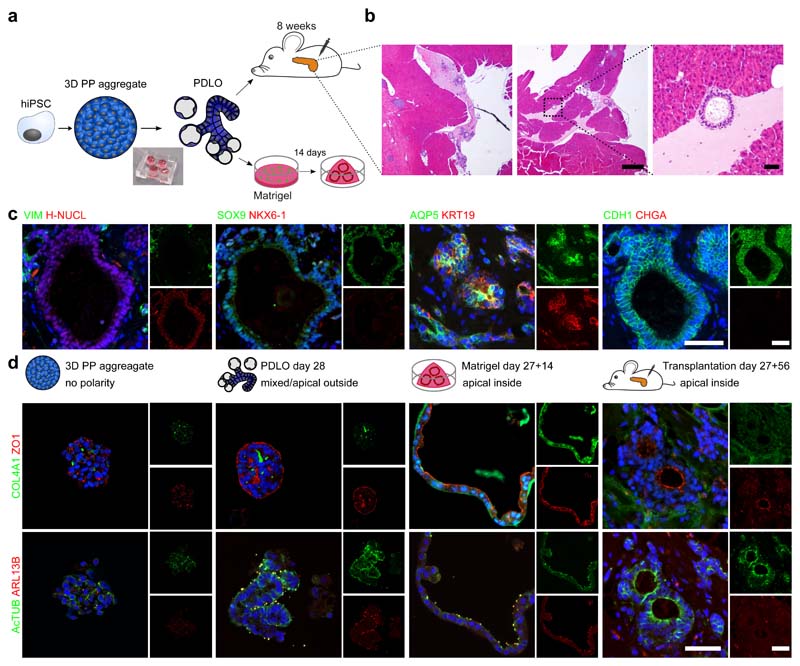
Fig. 3. Apical-out polarity of the microwell-chip-derived PDLOs switched upon orthotopic transplantation or embedding into Matrigel.
a, Schematic of the Matrigel and orthotopic transplantation experiment. PDLOs were transplanted on day 27 and mice were sacrificed after 8 weeks. b, Overview haematoxylin-eosin (HE) staining and magnification of the engraftment site depicted by the dashed square (n=2 mice). Scale bar: 500 μm for overviews, 50 μm for magnification. c, PDLOs formed human epithelial duct-like tissue in vivo. (H-NUCL: human-specific nucleoli). Scale bar denotes 50 μm. d, IF images for the apical markers ZO1 and AcTUB and basal markers COL4A1 and ARL13B on 3D pancreatic-progenitor aggregates, PDLOs, Matrigel PDLOs and transplanted PDLOs. Complementary images are depicted in Supplementary Fig. 5. The nuclei were counterstained with DAPI. Scale bar denotes 50 μm.
PDLOs exhibited a predominant apical-out polarity, although apical markers tight junction protein-1 (ZO1), and cilia marker acetylated tubulin (acTUB), and ADP-ribosylation factor-like protein 13B (ARL13B) were also partly visible on the opposing membrane side, indicating very small lumen within the organoid (Fig. 3d, Supplementary Fig. 5). One explanation of the observed PDLO apical-out polarity could be the lack of extracellular matrix (ECM) deposition in suspension culture format32–34. We hypothesize that providing a basement membrane, as mimicked by Matrigel, to the microwell-chip-derived PDLOs could facilitate the formation of an epithelium with a strictly apical-inside organization. Indeed, upon transfer of microwell chip-derived PDLOs into a 3D Matrigel culture or after transplantation an apical-out to apical-in polarity switch was observed (Fig. 3d).
Single-cell characterization of duct-like organoids
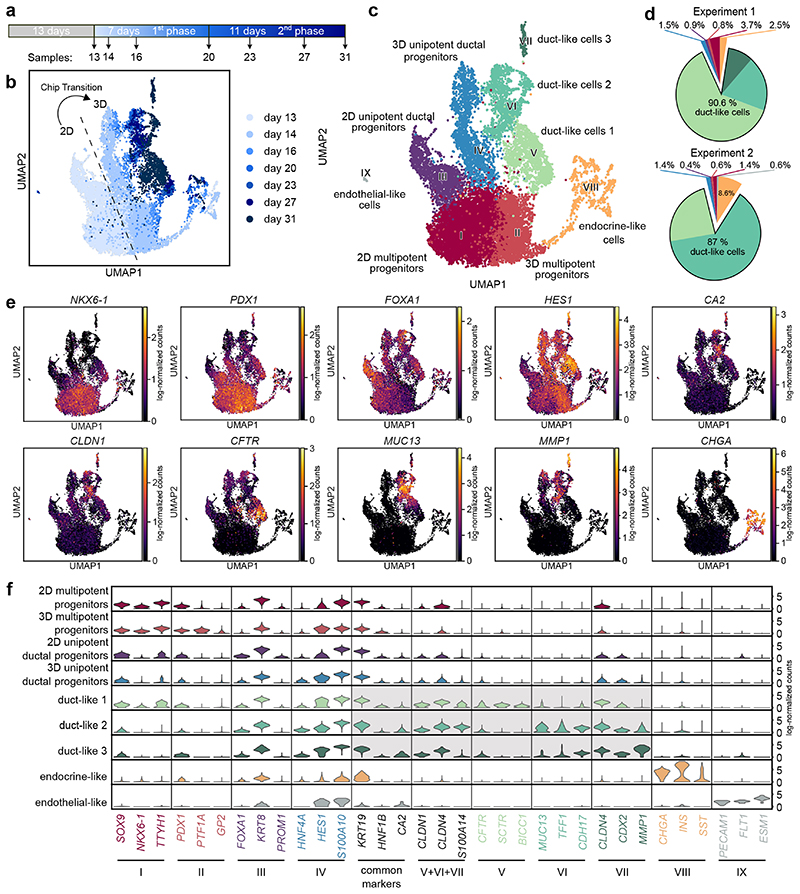
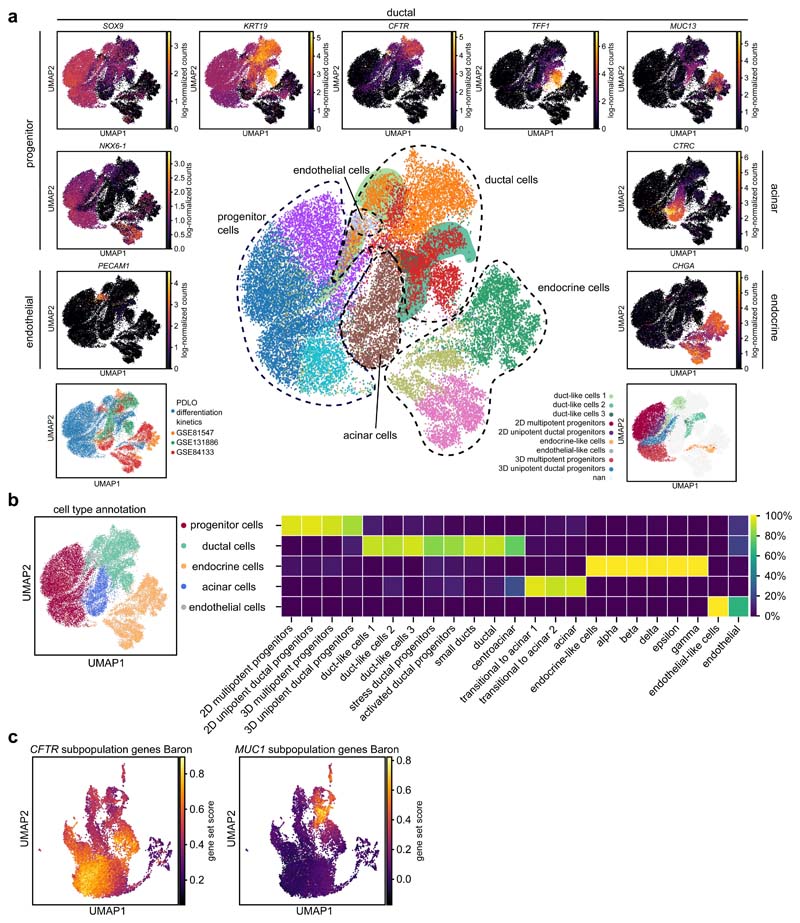
To reconstruct ductal cell type development in our microwell-chip model and to define time-resolved cell composition according to the transcriptional identity, we performed scRNA-seq analysis of 14,811 cells (Fig. 4a; Supplementary Fig. 6a). Louvain clustering and scRNA-seq data analysis with the two-dimensional data reduction algorithm Uniform Manifold Approximation and Projection (UMAP) identified nine cell clusters (Fig. 4b,c). With the progression of the growth factor-induced differentiation process, the recorded single-cell transcriptomes changed substantially indicated by the time-dependent emergence of the distinct cell clusters (Fig. 4b). All cell clusters could be assigned to cell types by matching known pancreatic developmental genes to the differentially expressed genes (DEG) in the respective cluster including three duct-like clusters (Fig. 4c–e; Supplementary Data 1). To test the robustness of the differentiation approach on the single-cell level, we sequenced the end stages of two independent experiments. In both cases, approximately 90% of pancreatic progenitors developed into cells with a transcriptomic profile of one of the duct-like clusters (Fig. 4d). Violin plots highlight a selection of DEGs specific for each cluster (Fig. 4f). The major cell populations identified included four presumptive progenitor cell types (clusters I–IV), three types of duct-like cells (clusters V–VII), a small endocrine-like cell population (cluster VIII), and a subset of non-pancreatic cells, namely endothelial-like cells (cluster IX). Intriguingly, our starting pancreatic-progenitor-cell population, which was generated in a 2D cell culture, contained cells with different transcriptional profiles suggestive of multipotent and unipotent ductal progenitor cells. In both cell clusters, common progenitor markers were expressed, including PDX1, HNF1B, and SOX9 (Fig. 4e,f; Supplementary Fig. 6c). Cluster III, hereafter named unipotent ductal progenitors (UDP), showed high expression of KRT8 and low expression of NKX6-1, suggesting that these cells committed to a ductal-primed fate at the end of the 2D cell differentiation at day 1335,36. Upon aggregation of the 2D cultured progenitors on microwell chips, both corresponding 3D progenitor clusters were transcriptionally re-identified, however, a subset of pancreatic-progenitor markers showed specific alteration patterns (Fig. 4 b,f): 3D UDPs (cluster IV) upregulated the markers hes family bHLH transcription factor 1 (HES1) and S100 calcium binding protein A10 (S100A10), whereas cluster II showed an increased expression of glycoprotein 2 (GP2) and pancreas associated transcription factor 1a (PTF1A), which were specific in this subpopulation (Fig. 4f; Supplementary Fig. 6c). PTF1A and NKX6-1 were only co-expressed in such cluster II, suggestive of multipotent progenitors (MPP), which can give rise to acinar, endocrine, and ductal cells in mice37,38. With the progression of differentiation, three duct-like cell clusters emerged, characterized by high KRT19, SOX9, and intermediate CA2 expression; the latter being a hallmark enzyme of mature pancreatic ducts (Fig. 4f; Supplementary Fig. 6c)39. Differential gene expression of the combined three duct-like clusters revealed significant upregulation of ductal epithelial markers such as CLDN1 and S100A14. While duct-like 1 cells specifically expressed HCO3 - secretion-related proteins such as CFTR, duct-like 2 cells were enriched for mucin-related genes, e.g., MUC13 and trefoil factor 1 (TFF1). The duct-like 3 cluster contained only 134 cells and showed similarities to duct-like 2 cluster with significantly higher expression of CLDN4 and MMP1. We also resolved a small fraction of endothelial cells and pancreatic endocrine cells. Doubling the initial cell number of the pancreatic-progenitor aggregate did not influence the outcome of the ductal differentiation (Supplementary Fig. 6b).
Fig. 4. scRNA-seq identifies cellular heterogeneity along the differentiation from pancreatic progenitors to PDLOs.
a, Schematic representing the sampling time during the ductal differentiation. b, UMAP plot of the single-cell transcriptomes recorded along the time course of ductal differentiation. The light to dark blue colour denotes the time points of sampling. c, UMAP cell cluster representation of the scRNA-seq data. Nine unique cell clusters were identified during the ductal differentiation. d, Cell type distribution at the end-point of two independent ductal differentiation experiments. e, Expression patterns of selected cell marker genes used for pancreatic cluster assignments. Further marker genes are depicted in Supplementary Fig. 7a. f, Violin plot shows mRNA expression levels of differentially expressed genes specific to one of the nine cell clusters and commonly used pancreatic ductal markers as indicated below the graph.
Ductal cell subpopulations in PDLOs
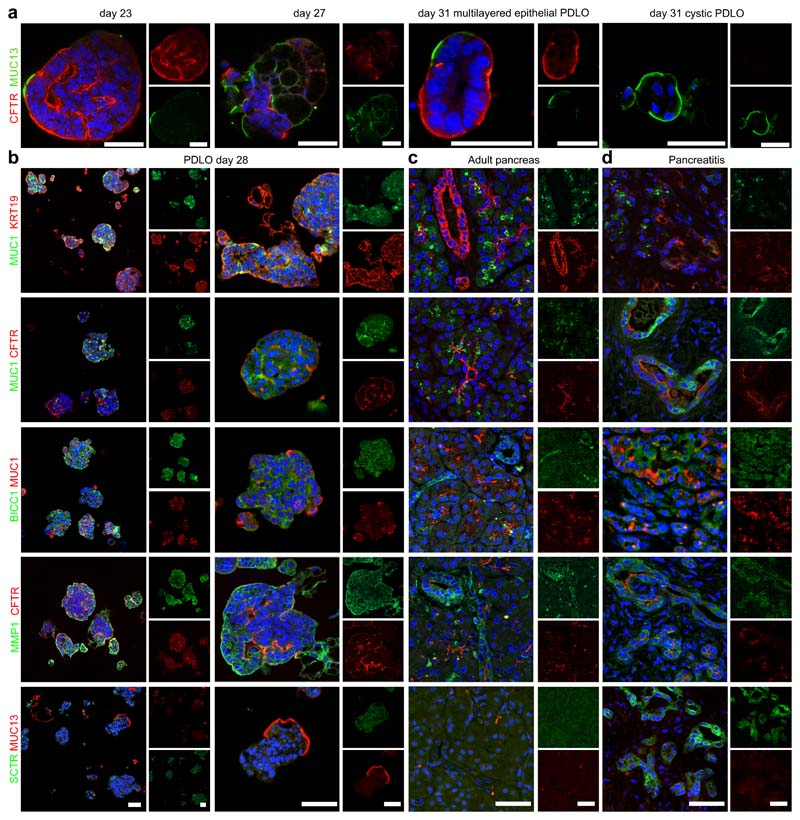
To validate duct-like cell types identified in single-cell transcriptomics on protein level, IF stainings for respective cluster markers were performed on microwell-chip-derived PDLOs. For this, CFTR and MUC13 were stained in PDLOs differentiated on our microwell chip to day 23, 27, and 31. While CFTR was only expressed in cells of multi-layered epithelial PDLOs, MUC13 was expressed in a different subset of cells at the outer side of multi-layered epithelial and in all cystic PDLOs (Fig. 5a,b). A differential expression pattern of a mucin-rich (MUC1+) and a CFTR+ ductal subtype within the pancreas has been previously reported 24. While MUC1 transcripts were not detected in the PDLO scRNA-seq data, MUC1 protein expression was readily found by immunostaining and label-free mass spectrometry on bulk PDLOs (Fig. 5b; Supplementary Fig. 7a; Supplementary Data 4). Next, we performed combinatorial staining of PDLOs for further duct-like subcluster DEGs. Indeed, we found distinct expression patterns and fluorescence intensities across multi-layered epithelial PDLOs for MUC1, CFTR, BICC1, MMP1, and TFF1 (Fig. 5b; Supplementary Fig. 7a), for instance, BICC1 was broadly detected but often appeared weaker in MUC1 positive cells. In larger PDLOs, MMP1 was localized to the peripheral layers while CFTR also appeared in luminal structures inside the organoid (Fig. 5b; Supplementary Fig. 7a). Overall, protein expression of the scRNA-seq based duct-like subtype markers were not mutually exclusive, which is in agreement with the single-cell transcriptomic data (Fig. 4e,f).
Fig. 5. Ductal subcluster-specific genes located within PLDOs and primary pancreas tissue on the protein level.
a, PDLOs after 23, 27, and 31 DOD were stained for duct-like cluster 1 marker CFTR, and duct-like cluster 2 marker MUC13, respectively. b, PDLOs differentiated until day 28 stained for cell subtype markers identified by the scRNA- seq analysis. c,d, The same markers are used to locate the in-vitro-generated duct-like cell subtypes in primary human pancreas tissue, either (c) healthy or (d) pancreatitis tissue. Images are complemented by Supplementary Fig. 7. Scale bar denotes 50 μm.
To translate these spatial expression patterns from microwell-chip-derived PDLOs to primary human pancreatic tissue, the cluster-specific markers illustrated above were stained in both healthy pancreata and chronic pancreatitis specimen (Fig. 5c,d, Supplementary Fig. 7b,c). MUC1 was restricted to acinar structures, centroacinar cells, and connected intercalated ducts, the latter also expressing CFTR (Fig. 5c; first and second row; Supplementary Fig. 7b). KRT19 was additionally stained confirming ductal identity. Dependent on the size of the branching ducts, the marker pattern changed pointing to a transition of different co-expressions. For example, CFTR expression decreased in intralobular ducts and was hardly found in larger ducts, while BICC1 became more prominent (Fig. 5c; third row). In addition, BICC1 was rarely detected in cells in direct proximity to the MUC1 positive cells. On the other hand, MMP1 was basically absent in intercalated ducts but showed a variable staining intensity within larger ductal structures (Fig. 5c; fourth row; Supplementary Fig. 7b). MUC13, TFF1, and SCTR could not be detected in healthy ducts (Fig. 5c; fifth row; Supplementary Fig. 7b). Immunostainings of chronic pancreatitis tissue further confirmed duct specificity with differential expression patterns of the described markers and revealed SCTR expression in metaplastic ductal epithelium (Fig. 5d; Supplementary Fig. 7c). Taken together, we could demonstrate spatially changing expression patterns of the ductal cell type markers MUC1, CFTR, BICC1, MMP1, TFF1, and SCTR on protein level in PDLOs and in human primary tissue. Herein, expression states are likely more dynamic and complex than initial transcriptomic subgrouping had suggested.
Trajectory of the in vitro ductal development
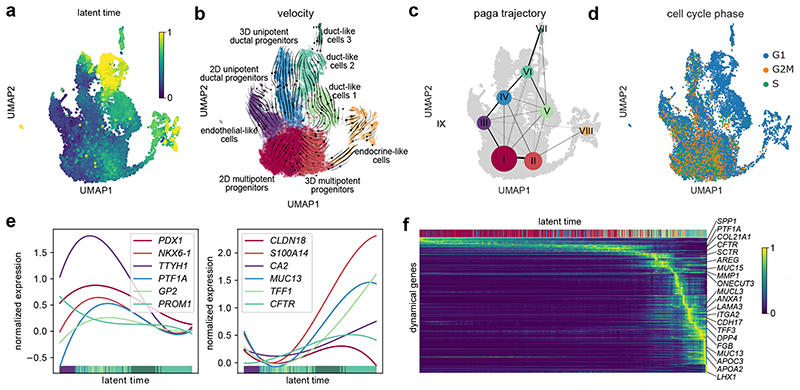
To resolve time-dependent relationships during PDLO differentiation, we performed dynamic RNA velocity analysis40,41. First, we calculated a latent time based on the balance of spliced and unspliced RNA transcripts within the single-cell transcriptomes (Fig. 6a). Indeed, the theoretical latent time matched true chronological differentiation times (compare to Fig. 4a). The corresponding RNA velocity streamlines indicate two differentiation routes from pancreatic progenitors toward duct-like cells: (i) duct-like 1 cells evolved from MPPs, and (ii) duct-like 2 cells mainly evolved from UDPs, which were already present at the pancreatic-progenitor stage (Fig. 6b). Velocity streamlines also directed from duct-like 1 to duct-like 2 cells, indicating a relevant degree of plasticity as reported previously in the pancreas42. Few endocrine cells emerged from the MPP cell cluster. Consistent with the velocity analysis, partition-based graph abstraction (PAGA) analysis showed a connectivity (edges) between clusters (dots) along the second ductal differentiation route (Fig. 6c). Evaluation of cell cycle states showed that maturation of duct-like cells was accompanied by a gradually decreasing fraction of cells in G2 and S phase (Fig. 6d). Subsequently, we plotted the changes in the expression of common stage-specific markers along the latent time to trace ductal differentiation. In line with mouse development data, pancreatic-progenitor markers including GP2, tweety family member 1 (TTYH1), PDX1, and PTF1A decreased over time. Conversely, ductal markers such as S100A14, CFTR, TFF1, and CA2 were upregulated (Fig. 6e). Concordance between transcriptional dynamics of MPP markers and genes essential during mitosis (topoisomerase 2 (TOP2) and cyclin B2 (CCNB2)) was observed, all decreasing during the differentiation process (Fig. 6f). The top 300 dynamic genes are listed in Supplementary Data 2. Upon ductal cell fate commitment, genes characteristic for pancreatic secretion (CFTR and SCTR) were transiently induced, accompanied by gradual increase in genes involved in mineral absorption (metallothionein 1E (MT1E)). Further, MUC13 was upregulated, showing a similar temporal expression profile as genes associated with lipid transport/metabolism (apoliprotein B (APOB)) and genes less studied in the pancreatic duct such as TFF3 or macrophage stimulating 1 (MST1), the latter being critical for maintaining an exocrine differentiation status and tissue integrity43. Since we also detected dynamical expression of several ECM-related genes, we analysed the scRNA-seq data in more detail regarding changes of ECM components during PDLO maturation. Duct-like cells increased the expression of laminin α3 and α5 subunits (Supplementary Fig. 8a). At the same time, corresponding laminin-binding integrins were expressed, supporting ECM formation along the differentiation trajectory. Concomitantly, duct-like cells downregulated the expression of basal matrix collagens accompanied by upregulation of collagenases MMP1/MMP10 (Supplementary Fig. 8b). Pancreatic tubulogenesis requires the basement membrane laminin-1 and an α6-containing integrin receptor for proper initiation, provided by the pancreatic mesenchyme during physiological development32–34. Accordingly, progenitor and duct-like cluster 1 cells expressed α1 laminin, duct-like cells 2 and 3 expressed α3, α5 laminin (Supplementary Fig. 8a). Of note, α4 laminin44 and α5 integrin, which are central for β-cell formation45, were neither expressed in PDLOs nor in the progenitor state. In contrast, αVβ5 integrins were upregulated in duct-like cells (Supplementary Fig. 8b) in agreement with previous findings46. It can be concluded that PDLOs are likely to secrete soluble ECM and corresponding binding proteins as seen under in vivo conditions. Within the open microwell culture format, however, the proteins are most likely resolved in the media which would explain the polarity switch of the microwell-chip-derived PDLOs upon either in vivo transplantation or 3D Matrigel culture (Fig. 3d). In addition to the ECM genes, we integrated a set of signalling pathways related to the applied growth factor stimuli and found that EGF/FGF10 mediated MAPK/ERK, and ErbB signalling peaked in the duct-like clusters 1 and 2. Compatibly, charting gene signatures representative for processes occurring in mature ducts such as pancreatic secretion or mucin type O-glycan biosynthesis finally fostered our generation of duct-like cells on our microwell chip (Supplementary Fig. 9a-d).
Fig. 6. Recovery of transcriptome dynamics predicts differentiation paths for duct-like cell types.
a, UMAP cluster plot coloured with the latent time calculated based on dynamic RNA analysis. b, Velocity and c, PAGA maps reveal the cellular state progressions during ductal differentiation. Streamlines in b indicate the direction of cellular state changes in the velocity map, and weighted lines in c within the graph network indicate higher cell cluster relations. d, UMAP plot highlighting the cell cycle state of differentiating cells. e, Gene expression profiles of progenitor or duct characteristic marker genes along the latent time, respectively. f, Top 300 dynamical genes sorted accordingly to their likelihood score and cells arranged along latent time.
CFTR+-/mucin+-subpopulations in primary duct tissue
One demand for the microwell-chip-derived PDLOs is that the duct-like cell types closely reflect human tissue; thus, we integrated three scRNA-seq data sets from primary human pancreas tissue23,24,26 into our PDLO differentiation kinetics (Fig. 7a). Within the re-clustered combined data set, duct-like clusters from PDLOs mapped with primary duct cells. Ductal progenitors from the in vitro differentiation trajectory clustered separately, as well as endocrine, acinar, and endothelial cells. A comparison of the initial cell type assignments with the cluster location in the combined data set further substantiated the integration approach (Fig. 7b). Previous reference data from primary ducts described two ductal cell types24, i.e. one with MUC1/TFF1 and a second with CFTR as the cluster marker gene. Highlighting CFTR and TFF1 within the combined data set confirmed the presence of ductal cell subtypes in the PDLOs (Fig. 7a, Supplementary Data 3). In fact, PDLO cells positive for CFTR clustered with the primary CFTR duct cell type, and PDLO cells positive for MUC13/TFF1 clustered with primary MUC1/TFF1 duct cells. Data integration can lead to an overinterpretation of cell type similarities. To confirm our ductal cell subpopulation analysis without data integration, we re-clustered the largest primary human ductal single-cell data set (Supplementary Fig. 10a,b) and searched for our cluster markers21 (Supplementary Fig. 10c-e). Indeed, the duct-like 1 cluster markers CFTR, BICC1 and SCTR were expressed in cell clusters separated from the duct-like 2 and 3 cluster markers MMP1 and TFF1 (Supplementary Fig. 10d,e). In addition, we calculated a gene expression enrichment score of the top 100 DEGs of the primary CFTR + and MUC1 + duct subpopulations from Baron et al.22 across the PDLO differentiation kinetics. Indeed, CFTR + and MUC13 + PDLO cells correlated again with the corresponding primary ductal subpopulations (Fig. 7c). In summary, both primary ductal cell subtypes could be found with a similar scRNA-seq expression pattern in vitro on the microwell-chip technology.
Fig. 7. Duct-like cells of the PDLOs clustered with primary ductal cells and resembled CFTR+-/mucin+-subpopulations.
a, Integration of three primary pancreas scRNA-seq data sets23,24,26 (bottom left) into 10 Louvain clusters (middle plot, duct-like PDLO cells are highlighted by areas in their cluster colour). UMAP expression plots display cell type-specific marker gene expression (surrounding graphs). Bottom right illustrates the location of our PDLO differentiation kinetics. Analysis of the CFTR+-/mucin+- subpopulations in the isolated scRNA-seq data set of Qadir, et al.26 can be found in Supplementary Fig. 10. b, Annotated cell types with their percentile distribution of the original clusters. c, Enriched expression of the marker genes for the CFTR high/MUC1 low and MUC1 high/CFTR low subpopulations defined in Baron, et al.24 during PDLO differentiation.
Application of the microwell chip
The microwell chip can be applied for various applications. One example is to investigate cell-cell communication between pancreatic ducts and various kinds of stromal cells. For this, four fluidic separable hexagonal arrays on the microwell chip were exploited to establish a cross contamination-free co-culture between PDLOs and human pancreatic stellate cells (HPaSteC) (Supplementary Fig. 11a). The latter resemble quiescent stromal cells in the pancreas capable to convert during inflammation, injury, and cancer development via auto- and paracrine signals to a metabolically active state, then serving as a central player in the pathogenesis of pancreatic disease47. Quantitative proteome analysis of both PDLOs and HPaSteCs separated co-cultured cells from their individually cultured counterparts (Supplementary Fig. 11b). None of the high abundance proteins of the individual cultured HPaSteCs and only 2% of the PDLOs were found in the upregulated protein set of the co-cultured PDLOs and HPaSteCs, which suggest neglectable cross-contamination on the microwell chip (Supplementary Fig. 11c). Enrichment for similar gene ontology (GO) terms indicated reciprocal signalling between both cell types. Pathways involved in energy metabolism and cellular signalling were enriched in co-cultures compared to single cultures (Supplementary Fig. 11c). Alongside, an in silico constructed protein network in co-cultured PDLOs and HPaSteCs resembled a mitogenic pattern in support of metabolic activation47 (Supplementary Fig. 11d).
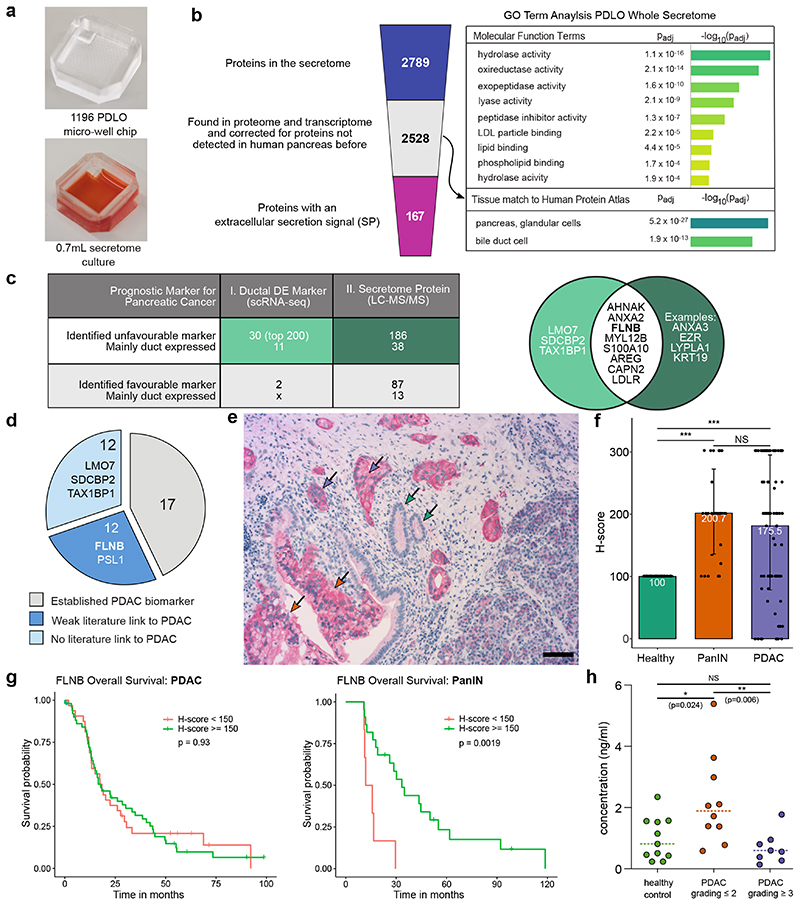
Another microwell chip application is the discovery of prognosticators and/or early-stage pancreatic cancer biomarker. Secretomes derived from either wild type or genetically modified pancreatic ducts could be a promising resource and the open-access of microwells allows their determination from a hydrogel-free environment of the PDLOs by collecting the supernatant. To obtain sufficient protein quantity we made use of label-free mass spectrometry (LS-MS/MS) and upscaled the microwell chip to 1,196 PDLOs (Fig. 8a). 2,528 secreted proteins with high confidence level (Supplementary Data 4), retrieved over a time interval of 8 h, were identified by mass spectrometry, from which 167 contained a signal peptide for active secretion into the extracellular space48. GO term analysis of the PDLO secretome showed an enrichment of enzymes with hydrolytic and lipid binding function (Fig. 8b). Scoring the filtered secretome against all human tissue types revealed significant enrichment for the term “pancreas, glandular cells” (Fig. 8b). Finally, we used the secretome and scRNA-seq data to identify potential prognostic markers of PDAC leveraged by our microwell technology. Within the top 200 DEGs of the duct-like clusters and the 2,528 proteins of the filtered PDLO secretome, 30 and 186 unfavourable prognostic markers for pancreatic cancer49 were found, respectively (Fig. 8c). Eleven of the transcriptomic proteins and 38 of the markers identified in the secretome were predominantly expressed in pancreatic duct cells.
Fig. 8. Potential PDAC biomarkers within the secretome and transcriptome of PDLOs.
a, Microwell chip used for the determination of the PDLO secretome by LC-MS/MS of three biological replicates, including one, two and five technical replicates. b, Left: Filter process applied to the secretome data. Right: GO terms enriched within the filtered PDLO secretome with relevance for ductal function. c, Prognostic pancreatic cancer markers within the top 200 DEGs of the combined duct-like clusters from the scRNA-seq analysis and within the PDLO secretome. The Venn diagram highlights the overlap between the revealed unfavourable diagnostic markers for pancreatic cancer from the scRNA-seq and secretome analysis. Pancreatic duct expression was assessed by examination of Human Protein Atlas staining63. d, Literature survey for the set of unfavourable diagnostic markers from table c. e, IHC staining of FLNB within PDAC tissue. Purple arrows indicate PDAC, orange arrows PanINs, and green arrows healthy ducts. Scale bar denotes 50 μm. Complementary images are shown in Supplementary Fig. 12. f, Mean FLNB H-score in heathy, (n=86), PanIN (n=28) and PDAC (n=84) tissue. Error bars denote standard deviation. The Mann-Whitney U test was applied to calculate statistical significance. Significance levels are indicated as follows: *: p-value<0.05; **: p-value< 0.01; ***: p-value<0.001. g, Overall survival curve for patients with high and low FLNB H-score. High FLNB H-scores in PanINs correlated with a favourable prognosis for patients. P-value was calculated with the log-rank test. h, Median FLNB blood concentration in a healthy control group (n=11) and patients with PDAC grade ≤2 (n=10) and ≥3 (n=7). The Mann-Whitney U test was used to calculate statistical significance. Significance levels are the same as in f.
Filamin b expression in human pancreatic cancer cohorts
One unfavourable PDAC marker in the overlapping protein set was filamin b (FLNB), which has been identified in a secretome screen from pancreatic cancer cell lines without validation in patients50 (Fig. 8d). FLNB exerts tissue- and context-dependent functions in distinct cancers, whereas both gain and loss of function have been shown to foster cancerous properties51,52. To investigate FLNB expression during pancreatic carcinogenesis53, we assessed FLNB protein expression in an independent cohort of resected PDACs54–56 by IHC (Fig. 8e). Normal pancreatic ductal epithelium and some acinar glands were faintly FLNB positive at their luminal surface. As expected, microwell-chip-derived PDLOs were also FLNB positive (Fig. 8e; Supplementary Fig. 12a,b). By contrast, PDAC strongly expressed FLNB in the cytoplasm and on the entire cell surface in concordance with the loss of polarity in the carcinoma cells (Fig. 8e; Supplementary Fig. 12b). IHC observation was specified using the semiquantitative H-score method followed by correlation with clinical data57. Comparing normal ductal epithelium with corresponding cancer tissue revealed significantly higher H-scores in PDAC (Fig. 8f), while H-scores did not correlate with any clinical parameter including survival (Fig. 8g). Pancreatic intraepithelial neoplasia (PanIN) represent the most relevant PDAC precursor lesions and can be frequently found adjacent to PDAC and their presence is of prognostic relevance58–60. Interestingly, H-scores were also elevated in PanINs in comparison to normal ducts (Fig. 8f). FLNB expression in PanIN lesions significantly correlated with higher survival of patients (mOS; p=0.0019) (Fig. 8g; Supplementary Fig. 12c). A gradual increase from normal to preneoplastic lesions has been described e.g. for the epigenetic silencer enhancer of zeste homolog 2 (EZH2), albeit in established cancers higher expression levels ascribed better prognosis61. As alternative splicing can lead to shorter FLNB isoforms being strongly associated with EMT gene signatures in basal-like breast cancer patient samples, distinct forms might also be present in pancreatic cancer precursor lesions ascribing distinct biological outcome62. To finally probe FLNB feasibility as a liquid biopsy-based biomarker, we consulted an independent cohort of human metastatic PDAC patients and measured FLNB levels in peripheral blood (PB). When comparing FLNB levels in PB, no obvious difference in metastatic PDAC patients and healthy volunteers was observed, however, metastatic samples clustered into two FLNB groups. To dissect this clustering in more detail, we correlated clinical and histological characteristics of the PDAC patients with individual FLNB levels in PB. Intriguingly, differentiated tumours (≤G2) had significantly higher FLNB PB levels when compared to less differentiated tumours (≥G3) or healthy donors, the latter two having more or less similar levels (Fig. 8h). We conclude that FLNB might be a suitable blood biomarker for differentiated PDACs and therefore could complement biomarker panels detecting early PDAC formation or discriminating differentiated and dedifferentiated PDAC.
Discussion
We have developed a microwell chip to engineer pancreatic duct-like organoids from hiPSCs, and charted their cellular heterogeneity during the differentiation trajectory. The advances of the microwell chip are (i) the low consumption of cells and materials, (ii) defined and homogeneous size of generated 3D aggregates, (iii) the possibility for long-term 3D cell culture, (iv) sample retrieval for downstream analysis with minimal perturbation, and (v) the possibility to establish co-cultures. We challenged the application potential of this microwell chip and comprehensively analysed the secretome/proteome to identify potential prognostic and diagnostic PDAC biomarkers.
Microwell culture is a low-cost producible cell culture technology offering uniformity for the aggregation process and flexibility in design for multiple downstream applications. The microwell chip implements 3D pancreatic-progenitor aggregates to generate two morphologically distinct PDLO types: a multi-layered epithelial and a cystic PDLO type. Pancreatic organoids generated from healthy or cancerous human pancreata grown in hydrogels show cystic morphologies7,12,64, with an apical-in polarity. In contrast, microwell-chip-derived PDLOs exhibited predominantly but not entirely apical-out polarity. After engraftment in mice, they formed ductal structures exhibiting their apical side to the lumen. The same switch to apical-in polarity was found when PDLOs were transferred from the microwell chip to an embedding laminin-rich hydrogel culture as mimicked by Matrigel. This argues that ECM components are also involved in the induction of ductal epithelial polarity. The inversion of apical-in to apical-out cell polarity upon transferring organoids from hydrogel to suspension cultures has been previously described for enteroids65. The dominant apical-out conformation of PDLOs enhances constant exposure to external stimuli, allows easy access to the secretome of cells, and makes the apical membrane accessible for various studies, such as the study of membrane barrier functions or pathogen infections. Nevertheless, swelling in cystic PDLOs upon stimulation with forskolin suggests a relevant degree of luminal CFTR activity at the inner membrane of PDLOs.
PDLO development in microwells also facilitates the stepwise study of synchronous human duct development. Time-resolved single-cell transcriptomics characterized the entire cell populations evolving from the pancreatic progenitor to duct-like cell stage. Interestingly, the duct-like cells differentiated on microwells clustered together with three primary duct cell references of published scRNA-seq data sets and could be stained in primary human pancreas tissue. Furthermore, counterparts of the previously identified ductal cell subtypes in the adult pancreas with mucosal restitution (MUC1) and HCO3 - secretion (CFTR) transcriptomic profile24 were identified within the in vitro generated PDLOs. Besides ductal heterogeneity at terminal differentiation stages, emerging intermediates revealed important details about ductal cell type commitment. Hitherto, it has been suggested that endocrine precursors delaminate from a common trunk domain arising at E12.5 in mice, which further undergoes tubular morphogenesis to subsequently form the ductal network38. Although the timing of particular marker expression is slightly different, detection of similar markers suggests the presence of a trunk and tip domain during human pancreas development19,38. An unexpected finding of our analysis was the lack of a trunk domain subcluster, at least based on the in vitro marker gene analysis. Velocity analysis suggests that the microwell-derived duct cell types generated from distinct temporarily co-existing progenitor populations harboured distinct potency. The few endocrine cells present in PDLOs evolved from the 3D MPPs, which were positive for classical tripotent pancreatic-progenitor labels, including GP2 and PTF1A, contradicting a trunk-like state18. We did not detect a PTF1A-/NKX6.1+-cell cluster in close proximity to endocrine and ductal cells that would represent an intermediate population giving rise to those two lineages. Possible explanations for the absence of such a trunk domain as described in the mouse development18 could be an early ductal priming of PDLOs or insufficient scRNA-seq sampling time intervals. In contrast, MUC13 positive ductal cells evolved from UDPs, being present at day 13, and to a smaller extent, from the CFTR positive duct cell type. We cannot exclude that the applied chemical induction protocol does not entirely resemble the in vivo development of the pancreas, and that artificial aberrations of the trunk model are induced. Differences between in vitro and in vivo progenitors have been observed during endocrine development, which also did not report a distinct trunk-like stage66–68. From in vivo mouse scRNA-seq data, additional developmental routes of pancreatic progenitors have been postulated69, and the development of a ductal subpopulation from Pdx1+ Ptf1a- pancreatic progenitor cells has been directly demonstrated in vivo by lineage tracing experiments70,71. Thus, it appears that the heterogeneity of pancreatic progenitors is larger than expected. Only single-cell data of primary human ductal embryonic development, which we currently lack, could further refine the in vitro PDLO engineering approach. Our finding that CFTR+/mucin+ duct-like cell types in the PDLOs resemble primary adult human duct cells indicates that PDLOs can be applicable for modelling diseases of the pancreatic ductal compartment, especially pancreatic cancer.
Currently, no efficient screening approach for early tumour detection exists. Personalized treatment of pancreatic cancer is still in its infancy, and predictive biomarkers are largely missing. Thus, secreted biomarkers from early neoplastic ducts could overcome this lingering unsolved obstacle. In this regard, the microwell chip can be a central interface tool for analytical methodologies such as mass spectrometry. PDLOs expressing oncogenic driver genes and probed onto our microwell chip could serve as a hub to dissect oncogene-specific secretomes. Our data provide proof-of-concept by determining the secretome of the genetically unaltered PDLOs and implementation of a systematic downstream biomarker classification. Herein, we investigate potential diagnostic and prognostic pancreatic cancer markers against a comprehensive PDAC biobank.
Methods
Microwell chips
All microwell chips used within this study consisted of four hexagonal microwell arrays surrounded by 12 pillars. Each pillar had a diameter of 0.6 mm and a height of 4 mm. The three different microwell chips had a well diameter of 150 μm, 300 μm, and 600 μm with 251, 61, and 19 wells per array, respectively. The well diameter to well depth ratio on each microwell chip was 1:1.5. For visualization, the 3D print model of the 600 μm microwell chip is displayed in Fig. 1a.
Melds of the PDMS microwell chips were printed using the Freeprint® mould (Detax) resin in a SLA 3D printer (Asiga PICO2 HD 27) with a slice thickness of 10 μm. After washing the prints twice with isopropanol for 10 min, moulds were incubated at 80°C for 1 h. Post-curing of the parts was achieved with an Otoflash G171 (NK-Optik) unit by exposing each side of the mild to 2000 flashes under nitrogen environment. To ensure complete evaporation of isopropanol, moulds were incubated at 80°C overnight.
PDMS microwell chips were produced by standard soft-lithography. In short, 3D printed moulds were precoated with 0.1% hydroxypropylmethylcellulose (Fluka Analytical) dissolved in 0.2 M sodium phosphate (Sigma) (pH 3, adjusted with 0.1 M citric acid (Sigma) for 10 min) based on Gitlin, et al.72. The 3D printed moulds were washed with deionized water and under nitrogen atmosphere. 1:10 PDMS (SYLGARD™ 184 Silicone Elastomer Kit, Dow Chemical Company) was cast and degassed in a vacuum chamber. A glass substrate was placed onto the microwell chip spacer structures and the PDMS was cured for 1.5 h at 80°C.
Cell seeding onto the microwell duct-on-chip technology
Before cell seeding, microwell chips were coated with 10% Pluronic F-127 (Sigma) overnight and sterilized for 30 min using 254 nm UV light (NK-Optik). On the next day, microwell chips were washed twice with Dulbecco’s Phosphate Buffered Saline (PBS) (Gibco) and once with DMEM:F12 (Gibco). For ductal differentiation on microwell chips, pancreatic progenitors were washed with PBS, then incubated for 3-8 min with TrypLE Select (Gibco) at 37°C for detachment. pancreatic progenitors were centrifuged at 200 g for 5 min and then seeded in 35 μL PDLO medium per array. The surface tension on top of the arrays allowed an equal distribution of the cell-suspension. After cell settling at 37°C for 45 min, additional 660 μL PDLO medium was carefully added to the side of the microwell chip. For preliminary determination of ideal cell numbers for organoid formation, hiPSCs were seeded on the microwell chip in mTeSR1 medium, supplemented with 10 μM ROCK inhibitor Y-27632 (abcam) during the first 24h
Culture of hiPSCs
Generation and culturing of the hiPSC line has been described previously14. Briefly, hiPSCs were cultured on hESC Matrigel-precoated plates according to manufacturer’s recommendations (Corning) in mTeSR1 medium (Stemcell Technologies) at 5% CO2, 5% O2, and 37°C with daily medium change. The general scientific use of the cells was approved by the local ethics committee at Ulm University (reference no. 68/11-UBB/bal.). The exact isolation method, culture and pancreatic differentiation of hiPSCs, and the study of such hiPSC derivatives was approved by the local ethics committee at Ulm University (reference no. 159/19) under informed consent of donors.
HiPSC differentiation to PDLOs
HiPSCs were differentiated into pancreatic progenitors in a monolayer culture based on a fusion protocol from two previously published studies14,73. In brief, 2.5x 105 hiPSCs were seeded in mTeSR1 with 10 μM Rock inhibitor per well of a 24-well plate, precoated with growth factor reduced (GFR) Matrigel (1:18 in DMEM:F12, Corning). For the first 6 days, cells were differentiated in the backbone media BE1: MCDB131 (Invitrogen) supplemented with 1% Glutamax (Gibco), 0.8 g/L glucose (Sigma), 1.174 g/L sodium bicarbonate (Sigma), and 5 g/L fatty acid free (FAF) Bovine Serum Albumin (BSA) (Proliant). While 100 ng/mL Activin A (PeproTech) and 2 μM GSK3β-inhibitor (CHIR99021) (Axon MedChem) were added for 24 h, the medium contained 100 ng/mL Activin A and 5 ng/mL bFGF (Novoprotein) for the following two days. From day 3 until day 6 the BE1 media was complemented with 50 ng/mL FGF10 (R&D), 0.75 μM Dorsomorphin (Sigma), and 3 ng/mL mouse Wnt3a (PeproTech). The subsequent backbone media BE3 was composed of MCDB131, 1% Glutamax, 3.3 g/L glucose, 1,754 g/L sodium bicarbonate, 20 g/L FAF-BSA, and 0.5% insulin-transferrin-selenium-X (Gibco). From day 6 until day 9 50 ng/mL FGF10, 200 nM LDN-193189, 0.25 μM SANT-1, 2 μM retinoic acid, and 0.044 g/L L-ascorbic acid (all Sigma) were added. From day 9 until day 13, BE3 media was supplemented with 200 nM LDN-193189, 100 ng/mL EGF (R&D), 10 mM nicotinamide (Sigma), 330 nM Indolactam V (StemCell Technologies), and 0.044 g/L L-ascorbic acid. The media was prepared freshly and changed every day. At day 3 (definitive endoderm), day 9 (pancreatic endoderm), and day 13 (pancreas progenitor stage) differentiation efficiencies were analysed by flow cytometry. Pancreas progenitors were only used when at least 70% of the cells were PDX1 and NKX6-1 double-positive.
For differentiation from pancreatic progenitors to PDLOs, cells were transferred on the microwell chip, facilitating a 3D cell culture. For the first seven days on the microwell chip, BE3 medium was supplemented with 1% Penicillin/Streptomycin (Sigma), 10 mM nicotinamide, 10 μM ZnSO4 (Sigma), 50 ng/mL EGF, 50 ng/mL FGF10, 50 ng/mL KGF (PeproTech), 50 nM MSC2530818 (Selleckchem), and 0.044 g/L L-ascorbic acid (PDLO medium phase I). 10 μM Rock inhibitor was additionally added for the transfer of pancreatic progenitors on the microwell chip. From day 20 on, cells were differentiated in BE3 containing 1% Penicillin/Streptomycin, 10 mM nicotinamide, 10 μM ZnSO4, 50 ng/mL EGF, 50 n/mL FGF10, and 0.044 g/L L-ascorbic acid (PDLO medium phase II). The media was changed twice a week. The complete differentiation scheme from hiPSCs to PDLOs is displayed in Fig. 2a.
Flow cytometry
Differentiation efficiencies were controlled on day 3, 9, and 13. The percentage of definitive endoderm cells was assessed by staining the surface marker CXCR4 and c-KIT on day 3. Pancreatic endoderm and pancreas progenitor cells were formaldehyde fixed (4% PFA) (Thermo Fisher Scientific) on day 9 and 13 for intracellular staining of PDX1, and PDX1/NKX6-1, respectively. Details of the staining procedure have been previously described14.
Live Imaging
For live cell imaging, PDLOs were imaged from day 24 to 31 of differentiation every 2 hours with a Zeiss Axio Observer Z1 microscope. The PDLOs on microwell chip were kept in cell culturing conditions with a stage top incubator (Tokai Hit).
Matrigel culture
For studying niche-dependent polarity changes, microwell-chip-derived PDLOs were harvested and transferred to a Matrigel 3D culture. After pipetting PBS directly on top of the microwells, PDLO cells at day 27 were washed in BE3 medium and PDLOs from ¼ array were resuspended in 50 μL GFR-Matrigel. Domes of 50 μL were seeded on 24-well plate wells and after solidification for 10 min at 37 °C, PDLO medium phase II supplemented with 10 μM Y-27632 was added. After 14 days of culture with medium change (PDLO medium phase II) every 2-3 days, PDLOs were fixated with 4% PFA 10 %Sucrose for histological analysis and processed as described in later sections.
Mouse model
NOD scid gamma (NSG) mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ strain (Charles River); RRID:BCBC_4142) were used for xenotransplantation of PDLOs into the pancreas with permission of the “Regierungspräsidium Tübingen” (TVA-1406). All animal work was performed under ethical and animal protection regulations of the German animal protection law. Husbandry was performed in standardized hygiene barrier rooms with reduced pathogen microorganism burden. Female 10-week-old mice were used for xenotransplantation experiments. Housing was performed in groups of two to four mice per cage.
Orthotopic transplantation of PDLOs
Microwell-chip-derived PDLOs at day 27 were harvested and singularized. After pipetting PBS directly on top of the microwells, cells were washed two more times with PBS and singularized with Accutase (Sigma) at 37°C for 30min. The enzymatic reaction was neutralized with 1% BSA (Proliant) 1% P/S (Thermo) in DMEM/F12 (Gibco), cells were washed in BE3 medium and resuspended in PDLO medium phase II supplemented with 20 μM Y-27632 and GFR-Matrigel in a 1:1 ratio. Aliquots with cell/Matrigel mixture were kept on ice until transplantation. Mice were pain mediated starting three days before transplantation by addition of 1 mg/mL Tramadol (Grünenthal) to the drinking water. To improve the take-rate when transplanting low numbers of PDLO cells, an acute pancreatitis was induced using caerulein (Sigma). 10 μL of a 5 μg/mL Caerulin 0.9% NaCl solution was injected intraperitoneally every hour six times prior to transplantation. After anesthesia with isoflurane a small cutaneous midline incision was followed by a small incision into the peritoneum. A volume of 50 μL with 100,000 cells was injected per mouse directly into the pancreatic tail. Carefully, pancreas and spleen were repositioned in the abdomen before the peritoneum was closed by medical sewing using 5-0 polyglactin coated vicryl suture (Ethicon). Surgical staples were used for closing the skin and removed one week after transplantation, when also Tramadol treatment was stopped. Mice were sacrificed after eight weeks and pancreata were collected, fixated with 4% PFA at 4°C o/n, and processed for histological analysis. All animal experiments were performed in compliance with the institutional guidelines, under ethical and animal protection regulations of Ulm University.
Fast acrylamide free tissue clearing and immunofluorescence staining
To allow imaging of whole organoids on microwell chips, the organoids were cleared with a protocol based on fast acrylamide free tissue clearing (FACT)74. Therefore, organoids were washed once with PBS and then PFA fixed for three days at 4°C. Washing with PBS was followed by incubation at 37°C for 1-3 h with 8% ultra-pure SDS (Invitrogen) in PBS for actual clearing. After washing the organoids twice with PBS and PBS-T (0.01% Tween 20 (Roth)), the organoids were blocked and permeabilized with 1% BSA (Roche), 22.52 mg Glycin (Roth), and 0.1% Tween 20 for 1 h. The primary antibody was diluted in 1% BSA and 0.1% Tween 20 and samples were stained for 3 days at 4°C. After washing twice with TBS-T (Duolink), organoids were incubated with the secondary antibody diluted in 1% BSA overnight at 4°C. The organoids were washed twice with TBS (Duolink) and incubated with 1 μg/mL DAPI (Sigma) for 15 min. Subsequently, the organoids were washed twice with TBS-T and post-fixated for 30 min with 4% PFA. During confocal imaging (Zeiss Axio Observer LSM 880) the organoids were kept on microwell chips in X-CLARITY Mounting Solution (Logos Biosystems).
Immunofluorescence (IF/ICC) staining was performed using the following primary antibodies: 1:200 PDX1 (R&D AF2419), 1:200 CDH1 (Cell Signaling 3195), 1:100 KRT19 (Dako M08889), 1:400 and 1:500 SOX9 (Cell Signaling 82630 and Millipore AB5535), 1:800 CFTR (Cell Signaling 78335), 1:500 MUC13 (BioLegend 363902), 1:200 ZO1 (Invitrogen MA3-39100-A555), 1:500 COL4A1 (Abcam ab6586) and 1:200 CLDN1 (R&D MAB4618). The following secondary antibodies were used: 1:500 anti-rabbit Alexa488 (Invitrogen A21206), 1:2000 anti-mouse Alexa488 (Invitrogen A21202), 1:1000 anti-rabbit atto550 (Sigma Aldrich 43328), 1:800 anti-goat Alexa488 (Invitrogen A11055), and 1:800 anti-rat Alexa488 (Invitrogen A21208).
Paraffin embedding of PDLOs
PDLO cultures were harvested with PBS as described in the Matrigel culture section. Harvesting PDLOs was followed by fixation of PDLOs in 4% PFA with 100 mM sucrose. After incubation at 4°C o/n, PFA was removed carefully and PDLOs were washed twice with PBS. Samples were pre-embedded in 2% agarose (Sigma) and further processed according to standard histology procedures. After serial dehydration, PDLOs were embedded in paraffin and sectioned at 4 μm. Tissue slices were mounted on SuperFrost Ultra Plus microscope slides (Thermo). Pancreatic tissue from transplantation experiments were fixated, embedded, and sectioned as described above without pre-embedding in 2% agarose.
Histological standard techniques
Histological staining including Hematoxylin and Eosin (H&E) staining were performed according to standard protocols. Paraffin sections of PDLOs or pancreatic tissue were rehydrated in ethanol series followed by either heat-mediated or enzymatic antigen retrieval, depending on the antibody (Supplement Data 6). Commercial Tris buffer (pH 9) or Citrate buffer (pH 6, both Vector Laboratories) were used for heat mediated antigen retrieval in a steamer, while a self-made Citrate buffer (pH 6, 1.9 g/l citric acid; Sigma) was used in the pressure cooker. To continue with immunofluorescence-paraffin (IF-p) staining, tissue permeabilization was performed with 0.5% Triton X-100/PBS (PBS-T) for 30 min at RT. After washing twice, primary antibodies diluted in Antibody Diluent (Zytomed) were added to the slides, which were then incubated o/n at 4°C in a wet chamber. After washing three times with PBS-T for 5 min, slides were stained with Alexa Fluor secondary antibodies (Thermo) and 500 ng/mL DAPI diluted in Antibody Diluent for 90 min at RT in the dark. Slides where washed three times with PBS-T and finally with dH2O before sections were mounted with Fluoromount-G (SouthernBiotech).
IHC staining on paraffin tissue sections
Immunohistochemistry (IHC-p) staining of FLNB was performed on a comprehensive human PDAC patient cohort consisting of 86 available tissues from resected PDACs75–77 on a fully automated OMNIS staining device (Dako) using Envision FLEX HRP Magenta high pH kit (Dako, GV900) according to manufacturer’s recommendations. Briefly, paraffin slides were incubated with primary FLNB (rabbit, 1:50, Merck, HPA004886) antibody for 30 min at RT in a wet chamber, secondary reagent for 10 min, polymer reagent for 20 min and chromogen for 5 min for colour development. Washing steps were performed as described in manufacturer’s recommendations. Nuclei were counterstained with 20% haematoxylin (Dako, GC808).
The images were graded according to PDAC and PanIN cells. The intensity of FLNB was denoted from 0 to 3 (0 negative, 3 strongly positive). The percentage of cells with positive FLNB cells within cancerous structures was graded from 1 to 5 (1 denotes 20% and 5 100% of FLNB positive cells). All healthy ductal cells were slightly positive and rated with 5 (number of positive cells) and 1 (intensity).
The H-score78,79 was calculated by multiplication of intensity scores (intensity of staining) with percentage scores (number of positive cells) in PDAC, PanIN, and healthy cells. For the Kaplan-Meier plots, the maximal H-Score per lesion was used for each patient. The patients with non-tumour related death were treated as alive. For the bar plot in Fig. 8e a Mann-Whitney-U Test was used to calculate statistical significance.
Forskolin swelling assay
For the swelling assay, the organoids were transferred to a bright field imaging microwell chip, which was composed of microwells with a diameter of 300 μm and a flat bottom to allow improved bright field imaging. Cells were stimulated at day 28 with either 20 μM forskolin (Abcam) and 100 μM 3-Isobutyl-1-methylxanthine (Sigma) (forskolin treatment) or 1:1000 DMSO (control) in DMEM:F12 medium. Live cell imaging was performed on a Zeiss Axio Observer Z1 microscope for 18 h after treatment. Images of 16 different positions were taken for each sample every 20 min.
Pancreatic stellate cells
Human pancreatic stellate cells (HPaSteCs), that were isolated from a chronic pancreatitis resection and immortalized by SV40 large T antigen and the catalytic subunit of hTERT80, were kindly provided by Prof. Matthias Löhr (Karolinska Institute). Cells were cultured in DMEM supplemented with 10% FCS and 1% Penicillin/Streptomycin and split using 0.05% Trypsin-EDTA (Sigma) twice a week in a 1:6 ratio.
PDLO/Stellate co-culture
PDLOs and HPaSteCs were seeded on a fluidic hexagonal microwell chip, facilitating paracrine signalling between PDLOs and HPaSteCs in the same microwell chip without direct cellular contact. For this, 150 cells were seeded per well following the proeceedure describe above (schematic see Supplementary Fig. 10a). The PDLOs were derived on the upscaled microwell chip (Fig. 8a), harvested at day 31, and transferred onto two arrays of the microwell chip (Fig. 1). The microwell chip was filled with phase II ductal media and co-cultured for 3 days without media change. The 3D HPaSteC aggregates and PDLOs were harvested at day 34. Therefore, the media was removed, and PBS drops of 40 μL were put on top of each arrays. Utilizing surface tension, the HPaSteC aggregates and PDLOs were harvested without cross-contamination. They were washed three times with PBS with an incubation time of 10 min in between. The 3D HPaSteCs aggregates and PDLOs were centrifuged with 200 g for 5 min within the washing steps. The dry pellet was frozen and kept at -80 °C upon sample preparation for proteomic measurements.
PDLO Secretome – Sample preparation for mass spectrometry
For determining the secretome of PDLOs, an upscaled microwell chip was manufactured. The microwell chip contained 1196 microwells with a well diameter of 400 μm, a well depth of 600 μm, and 600 cells were seeded per well (Fig. 8a). At day 28 of differentiation, PDLOs were washed three times with blank DMEM:F12 medium. 10 min incubation time between the washing steps ensured settling of eventually washed out PDLOs. Then, 700 μL blank DMEM:F12 was added and the supernatant was taken after 8 h for subsequent analysis by mass spectrometry. For the parallel PDLO proteome determination, PDLOs were harvested at the end of the experiment from the microwell chips and washed twice with ice-cold PBS and then lysed for 15 min on ice in 200 μL RIPA buffer (Thermo Fisher Scientific). For disruption of the DNA, the samples were additionally sonicated and then incubated for another 15 min on ice. Protein lysates were centrifuged at 13,000 g for 5 min at 4°C and the supernatant was collected. Each 10 μg lysate and 20 μg supernatant were subjected to tryptic digest applying a modified filter aided sample preparation (FASP) procedure as described81. Peptides were collected by centrifugation (10 min at 14,000 g) and acidified with 0.5% trifluoroacetic acid (TFA) and stored at -20°C.
Mass spectrometric measurements
LC-MSMS analysis was performed in data-dependent acquisition (DDA) mode. MS data were acquired on a Q-Exactive HF-X mass spectrometer (Thermo Scientific) each online coupled to a nano-RSLC (Ultimate 3000 RSLC; Dionex). Tryptic peptides were automatically loaded on a C18 trap column (300 μm inner diameter (ID) × 5 mm, Acclaim PepMap100 C18, 5 μm, 100 Å, LC Packings) at 30 μL/min flow rate. For chromatography, a C18 reversed phase analytical column (nanoEase MZ HSS T3 Column, 100 Å, 1.8 μm, 75 μm x 250 mm, Waters) at 250 nL/min flow rate in a 95-min non-linear acetonitrile gradient from 3 to 40% in 0.1% formic acid was used. The high-resolution (60,000 full width at half-maximum) MS spectrum was acquired with a mass range from 300 to 1500 m/z with automatic gain control target set to 3x 106 and a maximum of 30 ms injection time. From the MS pre-scan, the 15 most abundant peptide ions were selected for fragmentation (MSMS) if at least doubly charged, with a dynamic exclusion of 30 s. MSMS spectra were recorded at 15,000 resolution with automatic gain control target set to 5x 102 and a maximum of 50 ms injection time. The normalized collision energy was 28, and the spectra were recorded in profile mode.
Data Processing – Protein Identification
Proteome Discoverer 2.4 software (Thermo Fisher Scientific; version 2.4.1.15) was used for peptide and protein identification via a database search (Sequest HT search engine) against Swissprot human data base (Release 2020_02, 20349 sequences), considering full tryptic specificity, allowing for one missed tryptic cleavage sites, precursor mass tolerance 10 ppm, fragment mass tolerance 0.02 Da. Carbamidomethylation of Cys was set as a static modification. Dynamic modifications included deamidation of Asn, Gln, oxidation of Met; and a combination of Met loss with acetylation on protein N-terminus. Percolator was used for validating peptide spectrum matches and peptides, accepting only the top-scoring hit for each spectrum, and satisfying the cut-off values for FDR <1%, and posterior error probability <0.01. The final list of proteins complied with the strict parsimony principle.
Data processing – Label-free quantification
Proteins were quantified based on abundance values for unique peptides. For this, abundance values were first normalized to the total amount of peptides in each sample to account for sample loading errors. The protein abundances were calculated summing up the abundance values for admissible peptides. In the following only proteins with more than 1 unique peptide hit were used for downstream analysis.
The secretome data was filtered as follows: First proteins were filtered, which were not detected in the parallel recorded proteome or scRNA transcriptome datasets of the duct-like clusters. Secondly, proteins which were never detected before in the pancreas were filtered out. Therefore, we used the protein list “not detected proteins in the pancreas” of the Human Protein Atlas project63. To determine the fraction of proteins which were actively secreted we matched the PDLO secretome against the refined human secretome48. Here, only proteins with the label blood secretion and extracellular space were used.
Protein abundance changes within the co-culture experiment were calculated by normalizing the proteomes to the mean abundance levels. The PC were calculated on the normalized abundances. In the following the log2 fold changes of the proteins were calculated between individual and co-culture. Proteins, which were upregulated in co-culture experiments compared to the respective separately cultured PDLOs or HPaSteCs were further subjected to overrepresentation analyses. Enrichment of upregulated proteins against gene sets in common databases (Gene Ontology, KEGG, Reactome) was tested using g:Profiler [version: e100_eg47_p14_7733820]82 browser tool, and EnrichR83,84 allowed expansion to an EnrichR collective database comprising transcription factor protein-protein interaction networks. The co-culture signalling networks were generated by the X2Kweb85.
Patient material
Archival samples of FFPE material and clinical data from clinical reports originate from the previously published ULM cohort which included 122 patients with resected PDAC55,56. Data collection was done retrospectively and included cases from 1997 to 2008. The study was approved by the local ethics committee at Ulm University (reference no. #67, 105/98, 211/2002 and 268/2008).
Blood plasma of patients with metastasising PDAC was provided by the biobank of Ulm University Hospital. A group of healthy subjects was used as controls. The study was approved by the local ethics committee at Ulm University (reference no. 159/19). Written informed consent of all patients was given for material extraction and scientific use.
Enzyme-linked immunosorbent assay
Levels of Human Filamin B in PDAC patient serum were analysed using the FLNB ELISA kit (MyBioSource, MBS731914) according to manufacturer’s guidelines. Before assaying, plasma was centrifuged at 1000 x g for 15 min at 4 °C and the undiluted supernatant was added in duplicates to the ELISA plate. Absorbance at 450 nm was measured with Tecan Infinite M200 Pro plate reader. Concentration of the samples was interpolated from the standard curve that was determined within the same experiment run. The results and patient characteristics can be found in Supplementary Data 5. Statistical significance between the control patients and PDAC patients with grading 2 and 3 was calculated with t-test (Fig. 8h). One patient with a tumour grading of 2-3 was excluded for the calculation of statistical significance.
Image analysis
IF, IHC and bright-field images were cropped, rotated, aligned, enlarged (with black background to unify the picture size), and brightness and contrast edited with ImageJ. The measurements of the organoid diameters for the comparison of different cell numbers and well diameters were performed in ImageJ. In Fig. 1e at least 58 3D pancreatic-progenitor aggregates from three different microwell-chip arrays were measured for each condition. In Supplementary Fig. 2b 3D hiPSC aggregates from four technical replicates were measured over three days. One-sided students t-test was used to analyse size changes in R. Normal distribution was confirmed by the Shapiro-Wilk test. For the image analysis and editing ImageJ version 1.52p was used86.
Sample preparation for scRNA-seq
While pancreatic progenitors at day 13 were harvested with TrypLE Select as described above, organoids on the microwell chip were harvested at day 14, 16, 20, 23, 27 (samples from experiment 2: 600 cells, 300 μm microwell diameter), and 31 (samples from experiment 1: 300 cells, 300 μm microwell diameter and 600 cells, 600 μm microwell diameter) by washing the microwell chip three times with PBS. For the dissociation of PDLOs into single cells, organoids were incubated in Accutase for 30-45 min at 37°C. In experiment 2, single cells were cryo-preserved in DMEM with 10% heat-inactivated FBS (Thermo Fisher Scientific) and 10% DMSO based on a previously described scRNA-seq sample preparation protocol87. For sequencing, cryo-preserved cells were thawed in DMEM:F12 and then live-dead filtered like described in the 10x Genomics protocol CG000093. Cells from Experiment 1 (Fig. 2a) were not cryo-preserved and not filtered, but directly processed for actual scRNA-seq measurements. A RNA library was generated using Chromium Single Cell 3’ library and gel bead kit v3.1 (10x Genomics). The amplified cDNA library was sequenced on a NovaSeq 6000 S2 flow cell from Illumina. The sequenced cell numbers can be found in Supplementary Fig. 6b.
ScRNA-seq data pre-processing
Raw sequencing data files were demultiplexed, aligned (reference genome hg38_ensrel96), filtered, barcodes and UMIs counted, and subjected to a quality filter with CellRanger (10xGenomics). The pre-processing and downstream analysis were performed with the package ‘Scanpy API’88 in python with default parameters, if not stated differently. First, dead or stressed cells, identified by a percentage of mitochondrial genes higher than 15%, were filtered out. Next, cells with less than 1,200 or more than 104 expressed genes and genes expressed in less than 3 cells were excluded. Afterwards, the datasets of different days and experiments were concatenated, normalized to 104 gene counts per cell and log-transformed. Batch effects were corrected using ComBat. Further on, the top 4,000 highly variable genes were used for the downstream analysis. As discussed by Luecken and Theis 89, we corrected for the total gene counts, percentage of mitochondrial genes, and the cell cycle distribution of S, G2 and M phase to investigate differentiation-dependent changes on the transcriptome level.
Dimensionality reduction, clustering, and cell type annotation
The single-cell neighbourhood graph was computed with the first 50 principal components and 10 nearest neighbours and the cells were clustered with the Louvain algorithm90 at a resolution of 0.4. For visualization, the dimensionality of the data was reduced using Uniform Manifold Approximation and Projection (UMAP)91. For cell type annotation, 300 DEGs for each of the nine Louvain clusters were calculated by ranking the clusters against all remaining cells with the t-test method (Supplementary Data 1). The clusters were then annotated based on known marker genes.
RNA velocity through dynamical modelling
To investigate developmental trajectories, we analysed the RNA velocity by recovering directed dynamic gene information through splicing kinetics. Informations like clustering and UMAP coordinates were retrieved from the Scanpy analysis. The preprocessing and downstream analysis were performed with scVelo40 using default parameters. Splice variants and cells were filtered, normalized, and logarithmized with the function scv.pp.filter_and_normalize (parameters: min_cells=3, min_counts=200, min_shared_counts=20, n_top_genes=500). In a next step, the moments, based on the connectivities, were calculated with 30 PCAs and 30 neighbours. After recovering the dynamics, the latent time was calculated with a root cell from day 13. Based on this latent time, the velocity was calculated as a dynamical model40.
For the poly fit from Fig. 5e, we excluded the endocrine and the endothelial cell clusters, as well as all cells from the progenitor clusters with a latent time higher than 0.5. This mainly excluded the progenitor cells present at late time points. Afterwards, the cells were sorted by their latent time and the gene expression was fitted to a 3rd degree polynomial, following the code published by Bastidas-Ponce, et al.92.
Enriched gene expression of gene sets
The gene enrichment UMAP plots from Supplementary Fig. 9 and Fig. 8c were generated using the scanpy command sc.tl.score_genes. The score function subtracts the average expression of a set of genes with a reference gene set expression, randomly sampled from the whole gene pool. The gene lists for the pathway analysis were downloaded with the R package KEGGREST.
Integration of primary pancreas datasets
For the integration of the primary pancreas, three human datasets were used. While GSE8413324 (human samples GSM2230757, GSM2230759 and GSM2230760) and GSE8154723 mainly focused on endocrine cells, GSE13188626 described more ductal cell types. Before the datasets were concatenated, the datasets were preprocessed, clustered, and aligned as described for the PDLO scRNA-seq data. For calculating the neighbourhood graph, 32 PCAs (based on an elbow plot) and 20 nearest neighbours were considered. For integration and correction of the datasets, we applied bbknn to the datasets (neighbors_within_batch=5, n_pcs=32, trim=0, copy=True) and then reclustered the cells with the Louvain algorithm90 at a resolution of 1.3.
Reclustering of the GSE13188626 dataset
In order to further investigate our identified duct-like cell cluster markers, the GSE13188626 dataset was re-clustered similar to our main analysis. Cells with less than 800 different genes and more than 15% of mitochondrial counts were filtered out. For the re-clustering a Louvain algorithm with a resolution of 0.06 was applied, and 2000 variable genes and 27 PCs were taken into account.
Software specifications
The scRNA-seq alignment was run in CellRanger version 3.0.1 and the analyses were run in python 3.7.4 with Scanpy API version 1.4.4 or 1.5.1, anndata version 0.6.22 or 0.7.4, umap version 0.3.10, numpy version 1.17.4, scipy version 1.5.1, pandas version 0.25.3 or 1.0.5, scikit-learn version 0.22, statsmodels version 0.10.1, python-igraph version 0.7.1, louvain version 0.6.1, scvelo version 0.1.26 development, matplotlib version 3.2.1, seaborn version 0.9.0, loompy version 3.0.6, XlsxWriter version 1.2.6, bbknn version 1.3.6 and scrublet version 0.2.1.
The plots from Fig. 1, 8, Supplementary Fig. 2 and 12 were generated in RStudio with R version 3.6.0 with the R packages readxl version 1.3.1, ggplot2 version 3.3.0, dplyr version 1.0.4, survminer version 0.4.8, ggpubr version 0.4.0, reshape2 version 1.4.4, survival version 3.1-12 and ggsignif version 0.6.0. Dot plots in Fig. 8e and bar graphs in Supplementary Fig. 12b were generated using GraphPad Prism version 8.4.3. PCs (Supplementary Fig.11b) were calculated with stats version 3.6.0 and plotted with factoextra version 1.0.7.
Supplementary Material
Supplementary information is available for this paper at https://doi.org/10.1038/s41551-01X-XXXX-X.
Acknowledgements
This work is supported by the Helmholtz Pioneer Campus, the German Ministry of Education and Research (INDIMED-PancChip), the Baden-Württemberg Stiftung (Project “ExPoChip”), ERC (Consolidator Grant Number 772646), Deutsche Forschungsgemeinschaft (DFG) “Sachbeihilfe” KL 2544/7-1, GRK 2254/1 and 2, “Heisenberg-Programm” KL 2544/6-1, German Cancer Aid (Grant 111879), Else Kröner-Fresenius-Stiftung (supporting A.K. with an Excellence grant and M.H. with a First Application grant), Bausteinprogramm of Ulm University (granted to M.H.). We thank NK Optik for instrumental support. We thank Prof. Matthias Löhr (Karolinska Institute) for providing the human pancreatic stellate cells. We thank Thomas Walzthöni for bioinformatics support provided at the Bioinformatics Core Facility, Institute of Computational Biology, Helmholtz Zentrum München.
Footnotes
Author contributions
S.Wiedenmann, M.B., M.H., A.K. and M.Meier designed the study. S.Wiedenmann designed and produced the microwell chips, based on work from M.Moussus. S.Wiedenmann, M.B., J.M., and M.H. executed the biological experiments. S.Wiedenmann, M.B., M.H. and T.G. did the imaging and image analysis. S.Wiedenmann, M.S. and H.L. performed the scRNA-seq processing and S.Wiedenmann did the analysis. C.v.T. and S.H. did the mass spectrometric measurements and data processing. S.Weissinger and P.M. stained and evaluated the FLNB patient cohort. L.S. and T.S. took the serum samples of the PDAC patient cohort, M.H. performed the ELISA. S.Wiedenmann, M.B. and M.Meier analysed the MS/MS and FLNB screening results. M.H., A.K and M.Meier received the funding and supervised the study. The manuscript was written by S.Wiedenmann, M.B., M.H., A.K. and M.Meier. All authors corrected and approved the paper.
Competing interests
The authors declare no competing interests.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Peer review information Nature Biomedical Engineering thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Reprints and permissions information is available at www.nature.com/reprints.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Data availability
The main data supporting the results in this study are available within the paper and its Supplementary Information. Raw data, read counts and the analysed datasets from scRNA-seq can be accessed from the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE162547) repository via the accession code GSE162547. Mass-spectrometry data have been deposited on the PRIDE database (https://www.ebi.ac.uk/pride/archive/projects/PXD024461) and can be accessed via the identifier PXD024461.
Code availability
The codes for scRNA-seq analysis are available on Zenodo at https://doi.org/10.5281/zenodo.4738625.
References
- 1.Lee MG, Ohana E, Park HW, Yang D, Muallem S. Molecular mechanism of pancreatic and salivary gland fluid and HCO3 secretion. Physiol Rev. 2012;92:39–74. doi: 10.1152/physrev.00011.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hohwieler M, et al. Stem cell-derived organoids to model gastrointestinal facets of cystic fibrosis. United European Gastroenterol J. 2017;5:609–624. doi: 10.1177/2050640616670565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferreira RMM, et al. Duct- and Acinar-Derived Pancreatic Ductal Adenocarcinomas Show Distinct Tumor Progression and Marker Expression. Cell Rep. 2017;21:966–978. doi: 10.1016/j.celrep.2017.09.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey JM, et al. p53 mutations cooperate with oncogenic Kras to promote adenocarcinoma from pancreatic ductal cells. Oncogene. 2016;35:4282–4288. doi: 10.1038/onc.2015.441. [DOI] [PubMed] [Google Scholar]
- 5.Lee AYL, et al. Cell of origin affects tumour development and phenotype in pancreatic ductal adenocarcinoma. Gut. 2018 doi: 10.1136/gutjnl-2017-314426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 7.Boj SF, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huch M, et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 2013;32:2708–2721. doi: 10.1038/emboj.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuveson D, Clevers H. Cancer modeling meets human organoid technology. Science. 2019;364:952–955. doi: 10.1126/science.aaw6985. [DOI] [PubMed] [Google Scholar]
- 10.Frappart PO, et al. Pancreatic cancer-derived organoids - a disease modeling tool to predict drug response. United European Gastroenterol J. 2020:2050640620905183. doi: 10.1177/2050640620905183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tiriac H, et al. Organoid Profiling Identifies Common Responders to Chemotherapy in Pancreatic Cancer. Cancer Discov. 2018;8:1112–1129. doi: 10.1158/2159-8290.CD-18-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Georgakopoulos N, et al. Long-term expansion, genomic stability and in vivo safety of adult human pancreas organoids. BMC Dev Biol. 2020;20:4. doi: 10.1186/s12861-020-0209-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boj SF, et al. Model organoids provide new research opportunities for ductal pancreatic cancer. Molecular & cellular oncology. 2016;3:e1014757. doi: 10.1080/23723556.2015.1014757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hohwieler M, et al. Human pluripotent stem cell-derived acinar/ductal organoids generate human pancreas upon orthotopic transplantation and allow disease modelling. Gut. 2017;66:473–486. doi: 10.1136/gutjnl-2016-312423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang L, et al. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat Med. 2015 doi: 10.1038/nm.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Q, et al. A multipotent progenitor domain guides pancreatic organogenesis. Dev Cell. 2007;13:103–114. doi: 10.1016/j.devcel.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 17.Villani V, et al. SOX9+/PTF1A+ Cells Define the Tip Progenitor Cells of the Human Fetal Pancreas of the Second Trimester. Stem Cells Transl Med. 2019;8:1249–1264. doi: 10.1002/sctm.19-0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaffer AE, Freude KK, Nelson SB, Sander M. Nkx6 transcription factors and Ptf1a function as antagonistic lineage determinants in multipotent pancreatic progenitors. Dev Cell. 2010;18:1022–1029. doi: 10.1016/j.devcel.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jennings RE, et al. Development of the human pancreas from foregut to endocrine commitment. Diabetes. 2013;62:3514–3522. doi: 10.2337/db12-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Breunig M, et al. Modelling Plasticity and Dysplasia of Pancreatic Ductal Organoids Derived from Human Pluripotent Stem Cells. Cell Stem Cell. 2021 doi: 10.1016/j.stem.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang L, et al. Commitment and oncogene-induced plasticity of human stem cell-derived pancreatic acinar and ductal organoids. Cell Stem Cell. 2021 doi: 10.1016/j.stem.2021.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaum N, et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562:367–372. doi: 10.1038/s41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Enge M, et al. Single-Cell Analysis of Human Pancreas Reveals Transcriptional Signatures of Aging and Somatic Mutation Patterns. Cell. 2017;171:321–330.:e314. doi: 10.1016/j.cell.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baron M, et al. A Single-Cell Transcriptomic Map of the Human and Mouse Pancreas Reveals Inter- and Intra-cell Population Structure. Cell Syst. 2016;3:346–360.:e344. doi: 10.1016/j.cels.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muraro MJ, et al. A Single-Cell Transcriptome Atlas of the Human Pancreas. Cell Syst. 2016;3:385–394.:e383. doi: 10.1016/j.cels.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qadir MMF, et al. Single-cell resolution analysis of the human pancreatic ductal progenitor cell niche. Proc Natl Acad Sci U S A. 2020;117:10876–10887. doi: 10.1073/pnas.1918314117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Segerstolpe Å, et al. Single-Cell Transcriptome Profiling of Human Pancreatic Islets in Health and Type 2 Diabetes. Cell Metab. 2016;24:593–607. doi: 10.1016/j.cmet.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang YJ, et al. Single-Cell Transcriptomics of the Human Endocrine Pancreas. Diabetes. 2016;65:3028–3038. doi: 10.2337/db16-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hwang YS, et al. Microwell-mediated control of embryoid body size regulates embryonic stem cell fate via differential expression of WNT5a and WNT11. Proc National Acad Sci. 2009;106:16978–16983. doi: 10.1073/pnas.0905550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brandenberg N, et al. High-throughput automated organoid culture via stem-cell aggregation in microcavity arrays. Nature Biomedical Engineering. 2020 doi: 10.1038/s41551-020-0565-2. [DOI] [PubMed] [Google Scholar]
- 31.Freyer JP. Role of necrosis in regulating the growth saturation of multicellular spheroids. Cancer Res. 1988;48:2432–2439. [PubMed] [Google Scholar]
- 32.Crisera CA, et al. Expression and role of laminin-1 in mouse pancreatic organogenesis. Diabetes. 2000;49:936–944. doi: 10.2337/diabetes.49.6.936. [DOI] [PubMed] [Google Scholar]
- 33.Miner JH, Yurchenco PD. Laminin functions in tissue morphogenesis. Annu Rev Cell Dev Biol. 2004;20:255–284. doi: 10.1146/annurev.cellbio.20.010403.094555. [DOI] [PubMed] [Google Scholar]
- 34.Jiang FX, Cram DS, DeAizpurua HJ, Harrison LC. Laminin-1 promotes differentiation of fetal mouse pancreatic beta-cells. Diabetes. 1999;48:722–730. doi: 10.2337/diabetes.48.4.722. [DOI] [PubMed] [Google Scholar]
- 35.Kopp JL, et al. Progenitor cell domains in the developing and adult pancreas. Cell Cycle. 2011;10:1921–1927. doi: 10.4161/cc.10.12.16010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kopp JL, et al. Sox9+ ductal cells are multipotent progenitors throughout development but do not produce new endocrine cells in the normal or injured adult pancreas. Development. 2011;138:653–665. doi: 10.1242/dev.056499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan FC, Wright C. Pancreas organogenesis: from bud to plexus to gland. Dev Dyn. 2011;240:530–565. doi: 10.1002/dvdy.22584. [DOI] [PubMed] [Google Scholar]
- 38.Nair G, Hebrok M. Islet formation in mice and men: lessons for the generation of functional insulin-producing beta-cells from human pluripotent stem cells. Curr Opin Genet Dev. 2015;32:171–180. doi: 10.1016/j.gde.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inada A, Nienaber C, Fonseca S, Bonner-Weir S. Timing and expression pattern of carbonic anhydrase II in pancreas. Dev Dyn. 2006;235:1571–1577. doi: 10.1002/dvdy.20754. [DOI] [PubMed] [Google Scholar]
- 40.Bergen V, Lange M, Peidli S, Wolf FA, Theis FJ. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat Biotechnol. 2020 doi: 10.1038/s41587-020-0591-3. [DOI] [PubMed] [Google Scholar]
- 41.La Manno G, et al. RNA velocity of single cells. Nature. 2018;560:494–498. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puri S, Folias AE, Hebrok M. Plasticity and dedifferentiation within the pancreas: development, homeostasis, and disease. Cell Stem Cell. 2015;16:18–31. doi: 10.1016/j.stem.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.George NM, Day CE, Boerner BP, Johnson RL, Sarvetnick NE. Hippo signaling regulates pancreas development through inactivation of Yap. Mol Cell Biol. 2012;32:5116–5128. doi: 10.1128/mcb.01034-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qu H, et al. Laminin 411 acts as a potent inducer of umbilical cord mesenchymal stem cell differentiation into insulin-producing cells. J Transl Med. 2014;12:135. doi: 10.1186/1479-5876-12-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mamidi A, et al. Mechanosignalling via integrins directs fate decisions of pancreatic progenitors. Nature. 2018;564:114–118. doi: 10.1038/s41586-018-0762-2. [DOI] [PubMed] [Google Scholar]
- 46.Cirulli V, et al. Expression and function of alpha(v)beta(3) and alpha(v)beta(5) integrins in the developing pancreas: roles in the adhesion and migration of putative endocrine progenitor cells. J Cell Biol. 2000;150:1445–1460. doi: 10.1083/jcb.150.6.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Erkan M, et al. StellaTUM: current consensus and discussion on pancreatic stellate cell research. Gut. 2012;61:172–178. doi: 10.1136/gutjnl-2011-301220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uhlén M, et al. The human secretome. Sci Signal. 2019;12:eaaz0274. doi: 10.1126/scisignal.aaz0274. [DOI] [PubMed] [Google Scholar]
- 49.Uhlen M, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357:eaan2507. doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 50.Zubair H, et al. Proteomic Analysis of MYB-Regulated Secretome Identifies Functional Pathways and Biomarkers: Potential Pathobiological and Clinical Implications. J Proteome Res. 2020;19:794–804. doi: 10.1021/acs.jproteome.9b00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bandaru S, et al. Targeting filamin B induces tumor growth and metastasis via enhanced activity of matrix metalloproteinase-9 and secretion of VEGF-A. Oncogenesis. 2014;3:e119. doi: 10.1038/oncsis.2014.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iguchi Y, et al. Filamin B Enhances the Invasiveness of Cancer Cells into 3D Collagen Matrices. Cell Struct Funct. 2015;40:61–67. doi: 10.1247/csf.15001. [DOI] [PubMed] [Google Scholar]
- 53.Surcel A, et al. Targeting Mechanoresponsive Proteins in Pancreatic Cancer: 4-Hydroxyacetophenone Blocks Dissemination and Invasion by Activating MYH14. Cancer research. 2019;79:4665–4678. doi: 10.1158/0008-5472.CAN-18-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arnold F, et al. RINT1 regulates SUMOylation and the DNA damage response to preserve cellular homeostasis in pancreatic cancer. Cancer Res. 2021 doi: 10.1158/0008-5472.CAN-20-2633. [DOI] [PubMed] [Google Scholar]
- 55.Feld FM, et al. GOT1/AST1 expression status as a prognostic biomarker in pancreatic ductal adenocarcinoma. Oncotarget. 2015;6:4516–4526. doi: 10.18632/oncotarget.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmid SJ, et al. Absence of FLICE-inhibitory protein is a novel independent prognostic marker for very short survival in pancreatic ductal adenocarcinoma. Pancreas. 2013;42:1114–1119. doi: 10.1097/MPA.0b013e31829655ed. [DOI] [PubMed] [Google Scholar]
- 57.Hirsch FR, et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol. 2003;21:3798–3807. doi: 10.1200/JCO.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 58.Kleger A, Perkhofer L, Seufferlein T. Smarter drugs emerging in pancreatic cancer therapy. Ann Oncol. 2014;25:1260–1270. doi: 10.1093/annonc/mdu013. [DOI] [PubMed] [Google Scholar]
- 59.Reichert M, Blume K, Kleger A, Hartmann D, von Figura G. Developmental Pathways Direct Pancreatic Cancer Initiation from Its Cellular Origin. Stem Cells Int. 2016;2016:9298535. doi: 10.1155/2016/9298535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hassid BG, et al. Absence of pancreatic intraepithelial neoplasia predicts poor survival after resection of pancreatic cancer. Pancreas. 2014;43:1073–1077. doi: 10.1097/MPA.0000000000000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bremer SCB, et al. Enhancer of Zeste Homolog 2 in Colorectal Cancer Development and Progression. Digestion. 2019:1–9. doi: 10.1159/000504093. [DOI] [PubMed] [Google Scholar]
- 62.Li J, et al. An alternative splicing switch in FLNB promotes the mesenchymal cell state in human breast cancer. eLife. 2018;7 doi: 10.7554/eLife.37184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uhlén M, et al. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 64.Driehuis E, et al. Pancreatic cancer organoids recapitulate disease and allow personalized drug screening. Proc Natl Acad Sci U S A. 2019 doi: 10.1073/pnas.1911273116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Co JY, et al. Controlling Epithelial Polarity: A Human Enteroid Model for Host-Pathogen Interactions. Cell Rep. 2019;26:2509–2520.:e2504. doi: 10.1016/j.celrep.2019.01.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Veres A, et al. Charting cellular identity during human in vitro beta-cell differentiation. Nature. 2019;569:368–373. doi: 10.1038/s41586-019-1168-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krentz NAJ, et al. Single-Cell Transcriptome Profiling of Mouse and hESC-Derived Pancreatic Progenitors. Stem Cell Reports. 2018;11:1551–1564. doi: 10.1016/j.stemcr.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Petersen MBK, et al. Single-Cell Gene Expression Analysis of a Human ESC Model of Pancreatic Endocrine Development Reveals Different Paths to β-Cell Differentiation. Stem Cell Reports. 2017;9:1246–1261. doi: 10.1016/j.stemcr.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang J, Yuan R, Zhu X, Ao P. Adaptive Landscape Shaped by Core Endogenous Network Coordinates Complex Early Progenitor Fate Commitments in Embryonic Pancreas. Sci Rep. 2020;10:1112. doi: 10.1038/s41598-020-57903-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen C, et al. Evidence of a developmental origin for β-cell heterogeneity using a dual lineage-tracing technology. Development. 2019;146 doi: 10.1242/dev.164913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Larsen HL, et al. Stochastic priming and spatial cues orchestrate heterogeneous clonal contribution to mouse pancreas organogenesis. Nat Commun. 2017;8:605. doi: 10.1038/s41467-017-00258-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gitlin L, Schulze P, Belder D. Rapid replication of master structures by double casting with PDMS. Lab Chip. 2009;9:3000–3002. doi: 10.1039/b904684d. [DOI] [PubMed] [Google Scholar]
- 73.Nostro MC, et al. Efficient generation of NKX6-1+ pancreatic progenitors from multiple human pluripotent stem cell lines. Stem Cell Reports. 2015;4:591–604. doi: 10.1016/j.stemcr.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mohammad Rezazadeh F, et al. Fast free of acrylamide clearing tissue (FACT) for clearing, immunolabelling and three-dimensional imaging of partridge tissues. Microsc Res Tech. 2018;81:1374–1382. doi: 10.1002/jemt.23078. [DOI] [PubMed] [Google Scholar]
- 75.Arnold F, et al. RINT1 regulates SUMOylation and the DNA damage response to preserve cellular homeostasis in pancreatic cancer. Cancer Res. 2021 doi: 10.1158/0008-5472.Can-20-2633. [DOI] [PubMed] [Google Scholar]
- 76.Schmid SJ, et al. Absence of FLICE-inhibitory protein is a novel independent prognostic marker for very short survival in pancreatic ductal adenocarcinoma. Pancreas. 2013;42:1114–1119. doi: 10.1097/MPA.0b013e31829655ed. [DOI] [PubMed] [Google Scholar]
- 77.Feld FM, et al. GOT1/AST1 expression status as a prognostic biomarker in pancreatic ductal adenocarcinoma. Oncotarget. 2015;6:4516–4526. doi: 10.18632/oncotarget.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hirsch FR, et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol. 2003;21:3798–3807. doi: 10.1200/jco.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 79.John T, Liu G, Tsao MS. Overview of molecular testing in non-small-cell lung cancer: mutational analysis, gene copy number, protein expression and other biomarkers of EGFR for the prediction of response to tyrosine kinase inhibitors. Oncogene. 2009;28(Suppl 1):S14–23. doi: 10.1038/onc.2009.197. [DOI] [PubMed] [Google Scholar]
- 80.Jesnowski R, et al. Immortalization of pancreatic stellate cells as an in vitro model of pancreatic fibrosis: deactivation is induced by matrigel and N-acetylcysteine. Lab Invest. 2005;85:1276–1291. doi: 10.1038/labinvest.3700329. [DOI] [PubMed] [Google Scholar]
- 81.Livnat-Levanon N, et al. Reversible 26S proteasome disassembly upon mitochondrial stress. Cell Rep. 2014;7:1371–1380. doi: 10.1016/j.celrep.2014.04.030. [DOI] [PubMed] [Google Scholar]
- 82.Raudvere U, et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47:W191–w198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen EY, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kuleshov MV, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen EY, et al. Expression2Kinases: mRNA profiling linked to multiple upstream regulatory layers. Bioinformatics. 2012;28:105–111. doi: 10.1093/bioinformatics/btr625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Guillaumet-Adkins A, et al. Single-cell transcriptome conservation in cryopreserved cells and tissues. Genome Biol. 2017;18:45. doi: 10.1186/s13059-017-1171-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Luecken MD, Theis FJ. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol Syst Biol. 2019;15:e8746. doi: 10.15252/msb.20188746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. Journal of Statistical Mechanics: Theory and Experiment. 2008;2008:P10008. doi: 10.1088/1742-5468/2008/10/p10008. [DOI] [Google Scholar]
- 91.McInnes L, Healy J, Melville J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. ArXiv. 2018 e-prints 1802.03426. [Google Scholar]
- 92.Bastidas-Ponce A, et al. Comprehensive single cell mRNA profiling reveals a detailed roadmap for pancreatic endocrinogenesis. Development. 2019;146 doi: 10.1242/dev.173849. [DOI] [PubMed] [Google Scholar]
- 93.Clarke DJB, et al. eXpression2Kinases (X2K) Web: linking expression signatures to upstream cell signaling networks. Nucleic Acids Res. 2018;46:W171–W179. doi: 10.1093/nar/gky458. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The main data supporting the results in this study are available within the paper and its Supplementary Information. Raw data, read counts and the analysed datasets from scRNA-seq can be accessed from the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE162547) repository via the accession code GSE162547. Mass-spectrometry data have been deposited on the PRIDE database (https://www.ebi.ac.uk/pride/archive/projects/PXD024461) and can be accessed via the identifier PXD024461.
The codes for scRNA-seq analysis are available on Zenodo at https://doi.org/10.5281/zenodo.4738625.