Summary
Innate lymphoid cells (ILCs) have ample roles in tissue homeostasis, inflammation and repair. ILCs are generated early during ontogeny and persist predominantly as tissue-resident cells. How ILCs are maintained and renewed remains unclear. Here, we generated a single cell atlas of lung ILC2s and found that Il18r1+ ILCs comprise circulating and tissue-resident ILC progenitors (ILCP) and effector-cells with heterogenous expression of Tcf7, Zbtb16 and CD103. We revealed a continuous differentiation trajectory and validated the potential of Il18r1+ ILCPs to generate ILC2s. Upon helminth infection, recruited and BM-derived cells generated the entire spectrum of ILC2s in parabiotic and shield chimeric mice, consistent with their potential role in the renewal of tissue ILC2s. Our work identifies local ILCPs and highlights their in situ differentiation and tissue adaptation as a mechanism of ILC maintenance and phenotypic diversification. Our results suggest that local tissue niches, rather than progenitor origin, or the developmental window during ontogeny, dominantly imprint ILC phenotypes in adult tissues.
Introduction
Innate lymphoid cells (ILCs) have been identified in many different tissues where they contribute to immune surveillance, tissue homeostasis and repair, and barrier function. ILCs are found in the bone marrow, in secondary lymphoid organs and, most prominently, in non-lymphoid tissues including mucosal barriers. The size and subset composition of the local pool of ILCs can exhibit dramatic differences between organs. Despite considerable progress in ILC biology, ILC development and their adaptation to heterogeneous tissue environments remain poorly understood. ILCs are believed to seed tissues during embryonic development (Bando et al., 2014), where they expand and differentiate during the first weeks of life (Saluzzo et al., 2017; Schneider et al., 2019). In adult tissues, ILCs persist as tissue-resident cells. Originally established in parabiotic mice (Gasteiger et al., 2015; Huang et al., 2018; Moro et al., 2016; Peng et al., 2013), this concept is supported by analyses of human ILCs in transplanted livers and intestines (Cuff et al., 2016; Weiner et al., 2017). However, only a fraction of ILC2s labelled in the neonatal window persists in adult tissues (Schneider et al., 2019), reinforcing the question of how tissue-resident ILCs are maintained and replenished. Local pools of tissue-ILCs may receive input from distant cellular sources during inflammation, and local changes in ILC subsets and abundance correlate with a number of inflammatory diseases (Mazzurana et al., 2018). Consistently, murine ILC2s can traffic between organs during helminth infection (Gasteiger et al., 2015; Huang et al., 2018). It remains unclear whether recruited cells have a dedicated function, and whether they replenish the local pool of resident cells. Thus, ILCs detected in a given tissue may be generated from different cellular sources and at different times during ontogeny. How the local pools of ILCs are being maintained and renewed is currently unknown. In particular, we lack a detailed understanding of the defined stages of differentiation of tissue ILCs, the phenotype of “immature” ILCs, as well as the sites and tissue niches where maturation occurs.
To address these questions, we generated a comprehensive single-cell atlas of ILC populations in the BM, and neonatal and adult lung at steady-state, as well as during helminth infection in wild-type, parabiotic and shield chimeric mice. We identified immature stages of ILCs, inferred their differentiation trajectories, and validated their developmental potential in vitro and in vivo. We show that subsets of Il18r1+ST2- lung ILC2s express ILC progenitor genes, become proliferative, and gradually mature in the lung. Analysis of Nippostrongylus brasiliensis (Nb) infected parabiotic and shield chimeric mice suggests a contribution of these cells to the renewal of tissue ILC2s upon local inflammation. Furthermore, our study establishes that cells trafficking to the adult lung, particularly from the bone marrow, can give rise to the full spectrum of phenotypes of resident ILC2 subsets that have been generated in distinct waves during ontogeny, implying a dominant role of tissue imprinting on ILC phenotypes. Our work therefore identifies local progenitors as tissue ILCs and highlights tissue adaptation and in situ differentiation as a mechanism of ILC maintenance and phenotypic diversification. In addition, the single-cell RNA-seq datasets (http://murine-ilc-atlas.ie-freiburg.mpg.de/) represent a rich, interactive resource for further exploration of ILC biology.
Results
Heterogeneity of ILC2s in healthy lung tissue
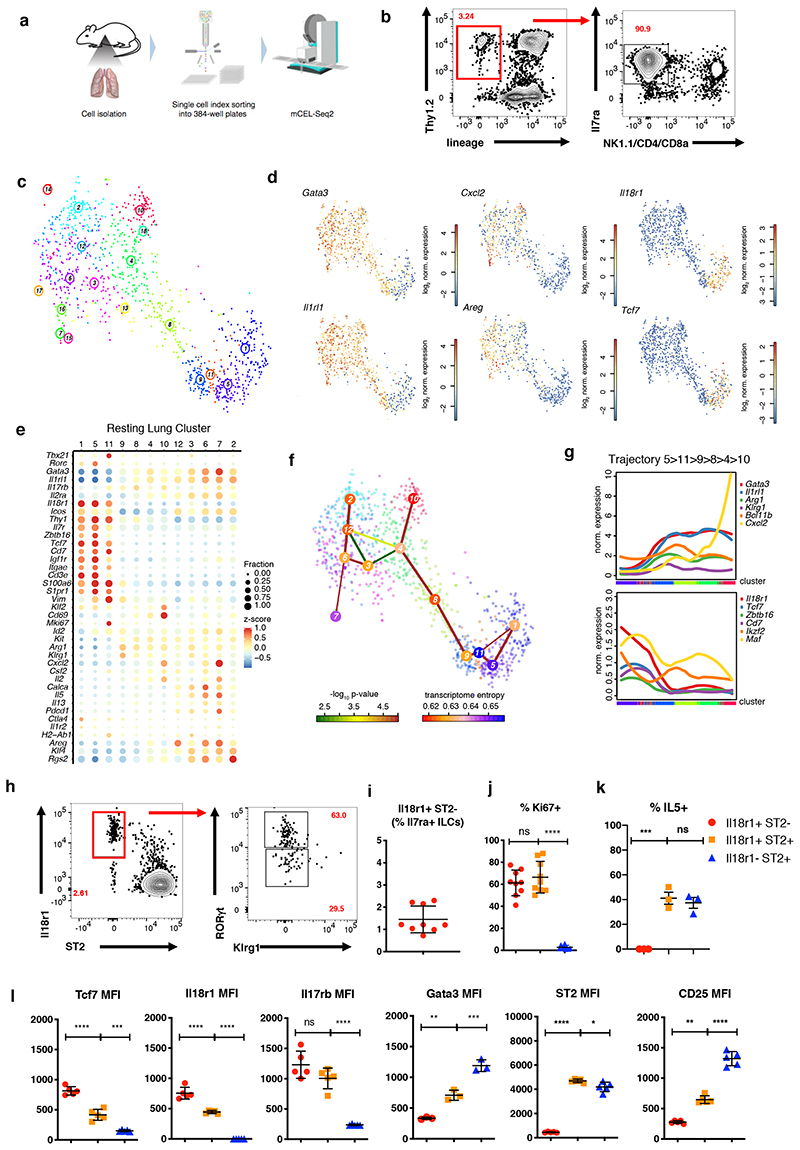
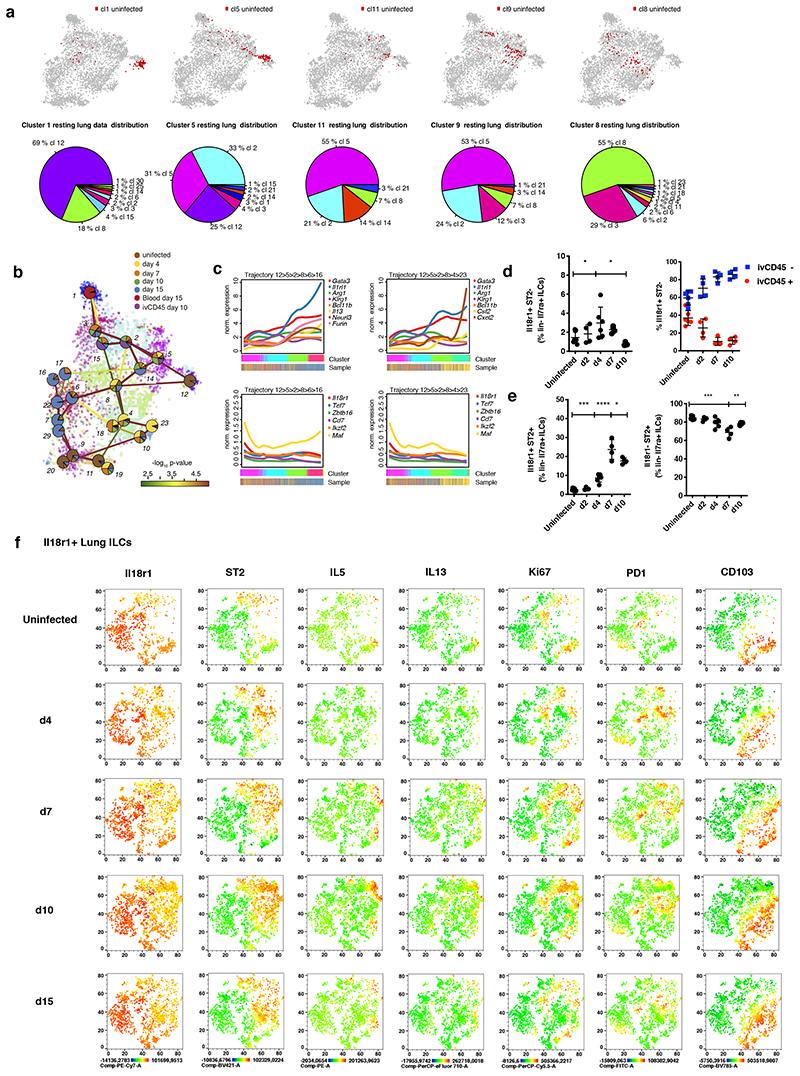
To characterize the full phenotypic spectrum of adult lung ILC2s in an unbiased manner we FACS-purified lineage-negative (Lin-) CD45+I17ra+Thy1+/lowNK1.1- “pan-ILCs” for scRNA-seq, excluding NK cells and ILC1, but not ILC3, which are relatively rare in the lung (Figure 1A, B and S1A). As committed ILC progenitors (ILCPs) and mature tissue ILCs express I17ra (Constantinides et al., 2014; Klose et al., 2014), we expected that this sorting strategy samples the entire differentiation tree of ILC2s including putative early differentiation stages. Clustering cells using RaceID3 (Herman et al., 2018) yielded 12 clusters with >20 cells (Figure 1C). Except for clusters 1, 5 and 11, all cells highly expressed Gata3 and Il1rl1 (Figure 1D, E), which are markers of mature ILC2s in the lung. We detected different effector ILC2 sub-types such as cluster 10, up-regulating the neutrophil recruiting factor Cxcl2, Il2, Klf2 and Cd69, or cluster 12, up-regulating Areg, Calca and Csf2 (Figure 1D, 1E and S1B). In addition, clusters 6 and 7 shared the up-regulation of Cxcl2, Areg, Il13, Il5 and Calca, but were distinct with regard to expression of Calca and Csf2 versus Il2, Cxcl2, and Pdcd1, respectively. Besides these effector-like ILC2s resembling subsets of ST2+ natural ILC2s (nILCs) (Nagashima et al., 2019; Wallrapp et al., 2017) we identified clusters 1, 5 and 11 characterized by low expression of Gata3 and Il1rl1. Only a small fraction of cells in these clusters expressed low levels of ILC2 effector genes such as Cxcl2, Areg and Il13 or Il5. We also detected cells up-regulating Rorc and Tbx21 within cluster 5 and 11, respectively (Figure 1E, S1C), indicating mixed lineage contributions, as previously observed (Wallrapp et al., 2017). Few cells in clusters 1 and 5 expressed T cell markers Cd3e and Trdc, akin to reported subsets of murine and human ILCs (Björklund et al., 2016; Robinette et al., 2015), although a contamination of T cells with downregulated surface CD3ε cannot be ruled out (Figure S1A). Interestingly, cells in clusters 1, 5 and 11 up-regulated Il18r1, Thy1, Cd7, Tcf7. The Notch target Tcf7 is expressed in ILCPs, and CD7 in circulating human ILC precursors (Constantinides et al., 2014; Harly et al., 2019; Ishizuka et al., 2016; Lim et al., 2017). Of note, a subset of cells in clusters 1, 5 and 11 expressed Zbtb16, a marker of committed ILCPs (Ishizuka et al., 2016; Klose et al., 2014; Lim et al., 2017) and few of these cells co-expressed combinations of Rorc, Tbx21 and Il17rb, consistent with a mixed lineage-potential (Figure S1C). In addition, we observed up-regulation of Igfr1 in clusters 1 and 5, a receptor suggested to be critical for recently identified ILC3-progenitors in neonatal lung (Oherle et al., 2020).
Figure 1. Heterogeneity of ILC2s in healthy adult lung tissue and immature Il18r1+ ILCs.
(A) Workflow for lung ILC scRNA-seq library preparation. (B) Representative sorting strategy for Lin- NK1.1-Il7ra+Thy1.2hi/im ‘pan’-ILCs from lung. (C) t-SNE map of single-cell lung ILC transcriptomes clustered with RaceID3. (D) t-SNE maps indicating log2 normalized expression of candidate genes. (E) Candidate gene expression for clusters with at least 20 cells. Color represents z-score mean expression across clusters and dot size represents fraction of cells in the cluster expressing the gene. (F) Lineage inference using StemID2. Node color depicts transcriptome entropy and link color indicates p-value of StemID2 links (P < 0.05, Methods). (G) Pseudo-temporal gene expression profiles (local regression) of representative ILC2 genes (top) and ILC progenitor genes (bottom) along the predicted trajectory. Color bars indicate cluster identity. (H,I), Representative FACS analyses of Lin- Il7ra+ ILCs in adult lung at steady state. Gating (H) and percentage (I) of Il18r1+ST2-RORγt+Klrg1- ILCs. (J-K) Fraction of Ki67+ (J) and Il5+ (K) cells within the indicated lung ILC subsets (ICS after incubation with monensin,RORγt+ ILCs are not excluded). (L) FACS analysis of Lin-Il7ra+ lung ILCs subdivided into Il18r1+ST2-RORγt+Klrg1-, Il18r1+ST2+ and Il18r1-ST2+ ILCs. Depicted is the mean fluorescence intensities (MFI) for Tcf7, Il18r1, Il17rb, Gata3, ST2 and CD25. Data in (H-L) are representative of 2-3 independent experiments (n=4-12 mice). Graphs in (I), (J) and (K) depict data as mean ± SD, (one-way ANOVA Tukey’s multiple comparisons test, *P < 0.05, **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant).
To investigate if these cells represent early differentiation stages of tissue ILCs we analysed the developmental relationship of lung ILC subsets with StemID2, which connects related clusters to construct a lineage tree (Figure 1F). This analysis predicted a differentiation trajectory connecting Il18r1 + Gata3 lo Il1rl1 lo cells (clusters 1, 5, 11), to ILC2s with increasing levels of Il1rl1 and intermediate expression of effector cytokines (clusters 9, 8, 4) and to mature effector clusters (10, 12, 6, 7 and 2) (Figure 1F). Elevated transcriptome entropy for clusters 1 and 5 suggested a more naïve state compared to the other clusters (Grün et al., 2016). After ordering cells along the trajectory starting at cluster 5 with the highest expression of known progenitor marker genes, and extending via clusters 11, 9, 8, and 4 towards cluster 10, pseudo-temporal gene expression profiles were grouped into distinct co-expression modules (Methods, Figure S1D, E). While the Cxcl2-expressing cluster 10 represents one possible endpoint of the trajectory, other mature clusters (2, 6, 7, and 12) correspond to alternative endpoints branching out from cluster 4 (Figure S1F).
Along the trajectory, we observed gradual up-regulation of Gata3, Il1rl1, Klrg1 and Bcl11b, which has been shown to be up-regulated during ILC2 differentiation (Califano et al., 2015; Yu et al., 2015). Il18r1 and Cd7 were down-regulated along with ILC progenitor genes such as Zbtb16, Tcf7, Maf and Ikzf2 (Figure 1G) (Harly et al., 2019; Ishizuka et al., 2016; Seehus et al., 2015). These inferred gene-expression dynamics suggested that mature Gata3hiIl1rl1 hi ILC2s can differentiate from progenitor-like Il18r1 + Gata3 lo Il1rl1 lo ILCs (clusters 1, 5, and 11) in the lung. FACS analysis confirmed that Il18r1+ST2-RORγt-Klrg1- cells, hereafter referred to as Il18r1+ST2- cells, represented about 2% of Lin- Il7ra+ ILCs (Figure 1H, I), and were also detected in lungs of T cell deficient RAG-/- and TCRβδ-/- mice (Figure S1G and data not shown). Consistent with the gene signatures predicted for cluster 11 (Figure 1D), a high fraction of these cells was positive for the proliferation marker Ki67, suggesting their capacity for local expansion or self-renewal (Figure 1J). Together, these observations indicated that Il18r1+ST2- cells comprise immature differentiation stages of ILC2s that are enriched in the lung, where they may undergo local maturation. Consistently, when we analyzed lung ILC2s by dividing them into three subsets that were predicted to map successively along the inferred maturation trajectory (i.e., Il18r1+ST2-, Il18r1+ST2+ and Il18r1-ST2+ cells), we found that cells downregulated Ki67, Il18r1, Il17rb and Tcf7 along this trajectory, while gaining expression of CD25, ST2, Gata3 and Il5 protein, which all characterize mature ILC2s in the lung (Figure 1J- L).
To demonstrate that clustering and trajectory inference were not driven by proliferation associated programs, we removed proliferation associated genes prior to analysis (Methods) and obtained a similar trajectory connecting Il18r1 + Gata3 lo Il1rl1 lo cells to effector-like ILC2s (Figure S1H-K). Together, these observations suggested that Il18r1+ST2- cells comprise early immature differentiation stages of ILC2s that are enriched in the lung, where they may locally mature.
Expression of Il18r1, Tcf7 and Cd7 as well as effector genes Il5, Il13, and Il17a has recently been reported for ILC2s in the skin (Bielecki et al., 2018; Kobayashi et al., 2019; Ricardo-Gonzalez et al., 2018), raising the possibility that Il18r1+ST2- cells comprise heterogeneous ILC subtypes. To better characterize Il18r1 + ILCs across tissues, we integrated our data with lung and skin single-cell transcriptomes from Ricardo-Gonzalez et al. and tested for the presence of discrete progenitor and effector states (Methods, Figure S2A). We found that Il18r1 expressing lung ILCs largely clustered separately from mature skin ILC2s expressing ILC2 effector genes (Figure S2A, B). However, a fraction of skin ILCs clustered together with immature Il18r1 + lung ILCs and up-regulated Igf1r, Tcf7, Zbtb16 and Pdcd1, suggesting an immature progenitor phenotype (Figure S2A-C). It is important to note, that lung and skin-ILCs were sorted as Il5-mRNA-reporter+ cells by Ricardo-Gonzalez et al., and therefore this analysis may underestimate the frequency of progenitor-like cells. Reminiscent of these distinct phenotypes in skin, we detected CD103+Il18r1+ cells in the lung, a fraction of which also expressed cytokines Il5, Il13 and Il17a upon in vitro re-stimulation (Figure S2D, E). These combined analyses reveal a previously unappreciated heterogeneity of Il18r1+ST2- ILC subtypes in skin and lung.
Il18r1+ST2- ILCs in neonatal and adult lung share similarities with Il18r1+ ILC progenitors in the bone marrow
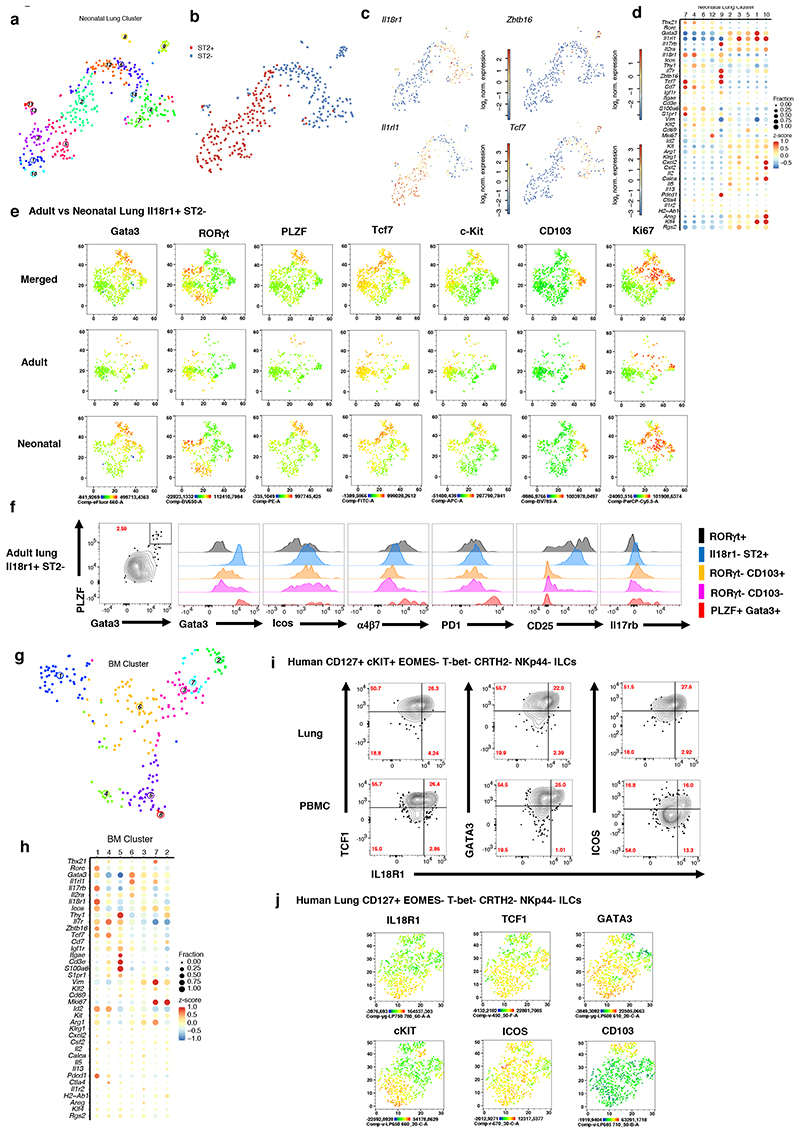
We next asked how the Il18r1+ST2- putative ILC2 progenitors from adult lung compare to known ILC progenitors. ILC2s are generated in different developmental windows during ontogeny, and cells emerging during neonatal life can persist in the adult lung (AL) (Schneider et al., 2019). To test whether corresponding progenitor populations are present in the neonatal lung (NL), we sequenced ST2+ and ST2- ILCs from postnatal day 4 (P4) lung (Figure S3A), and performed clustering analysis (Figure 2A, 2B). Within the ST2- compartment, cluster 9 exhibits a naïve ILC signature akin to AL clusters 1, 5, and 11, with up-regulation of Il18r1, Tcf7, Zbtb16 and Pdcd1, as well as intermediate levels of Gata3 and high expression of Il17rb (Figure 2C, 2D), suggesting predominant ILC2 bias. Cluster 4, 6, 7 and 12 share expression of Il18r1 and Tcf7, and resemble ILC1s or NK cells with expression of Tbx21, Nkg7 and Klrd1 (Figure 2D). FACS analysis confirmed the heterogeneity of Il18r1+ST2- cells and the presence of Il18r1+Tcf7+ cells that expressed Zbtb16 (PLZF), c-Kit, PD1, α4β7, and Ki67 to variable degrees in both NL and AL (Figure 2E, F and S3B-D). These cells shared expression of many genes with Il18r1+Icos+ BM ILCs that we further characterized by scRNA-Seq (Figure 2G, H) and cells that have recently been refined as Id2+ BM ILCPs (Wu et al. (2019). By integrating our data with published BM ILC scRNA-seq data (Walker et al., 2019), we confirmed that BM Il18r1 + Icos + cells contain ILCPs with ILC2 bias (Figure S3E-H). Accordingly, cells of Il18r1 +Icos+ BM cluster 1 up-regulated Tcf7, Zbtb16, Pdcd1 and Il17rb (Figure 2H), reminiscent of NL cluster 9. While Il18r1 + NL clusters lacked expression of Cd3 genes, BM clusters 4 and 5 as well as AL progenitor clusters 1 and 5 up-regulated these genes. In addition, we resolved clusters 2 and 7, up-regulating Mki67 (Figure 2h). These data confirm the recently described phenotypic spectrum of BM ILC progenitors and reveal that corresponding populations with a similar heterogeneity exist in NL and AL (Figure 1, 2A-F). Circulating human ILC progenitors reminiscent of the populations we identified here were described recently (Lim et al., 2017). The IL7R+ c-KIT+ bulk population, which possibly include CRTH2- cells biased toward ILC2 lineage differentiation (Nagasawa et al., 2019) displays the transcriptional profile of IL18R1, TCF7, CD7 and ZBTB16 (Lim et al., 2017). As expression of these genes was not tested previously at the protein level, we analyzed human ILCs in peripheral blood and lungs and identified a population of IL7R+IL18R1+TCF1+ (encoded by the TCF7 gene) cells that expressed GATA3 while lacking markers of mature NK cells, ILC1, ILC2 and ILC3 (i.e., EOMES, T-bet, CRTH2, and NKp44, respectively) (Fig. 2I-K, S3I). Again, similar to cells in the murine lung, we observed graded expression of IL18R1, TCF1, GATA3, c-KIT, ICOS and CD103, reminiscent of the described spectrum of murine Il18r1+ ILCs in neonatal and adult lungs, and BM ILCPs (Walker et al., 2019; Xu et al., 2019).
Figure 2. IL18rl+ST2- ILCs in neonatal and adult lung share similarities with IL18rl+ BM ILCPs.
(A-C), t-SNE maps of neonatal lung ILC single-cell transcriptomes highlighting RaceID3 clusters (A) or sorted ST2- and ST2+ ILCs (B), and log2 normalized expression of representative genes marking putative ILC progenitors (C). (D) Expression of candidate genes within neonatal lung clusters with at least 10 cells. Color represents z-score mean expression across clusters and dot size represents fraction of cells in the cluster expressing the gene. (E) FACS analysis of neonatal (P4) and adult (6-8w) Il18r1+ST2- lung ILCs, represented as t-SNE maps, indicating expression levels for Gata3, RORγt, PLZF, Tcf7, C-kit, CD103 and Ki67. (F) Histograms comparing ILC subsets (PLZF+Gata3+, RORγt+CD103-, RORγt+CD103+, Il18r1-ST2- ILC2s and RORγt+ ILC3) in adult lung for Gata3, Icos, a4p7, PD1, CD25 and Il17rb expression levels. Data are representative of 2-3 independent experiments (n=6-12 mice). (G) t-SNE map of Il18r1+Icos+ BM ILCs highlighting RaceID3 clusters. (H) Expression of candidate genes for Il18r1+Icos+ BM clusters with at least 10 cells. Color represents z-score mean expression across clusters and dot size represents fraction of cells in the cluster expressing the gene. (I-J) FACS analysis of human lung and PB ILCs pre-gated on CD45+Lin- CD127+EOMES-T-bet-CRTH2-NKp44-c-KIT+/- (I) or c-KIT ILCs (J). Figures depict representative staining (I) and t-SNE maps (J) indicating the expression levels of selected markers. Data are representative of 3 independent experiments (n=2-4 samples each).
Together, these observations raise the possibility that we have identified a murine counterpart to ILC progenitors present in human lungs, reinforcing the question about the developmental potential of these cells.
Il18r1+ ILCs comprise immature cells that give rise to Gata3hi ILC2s in the lung
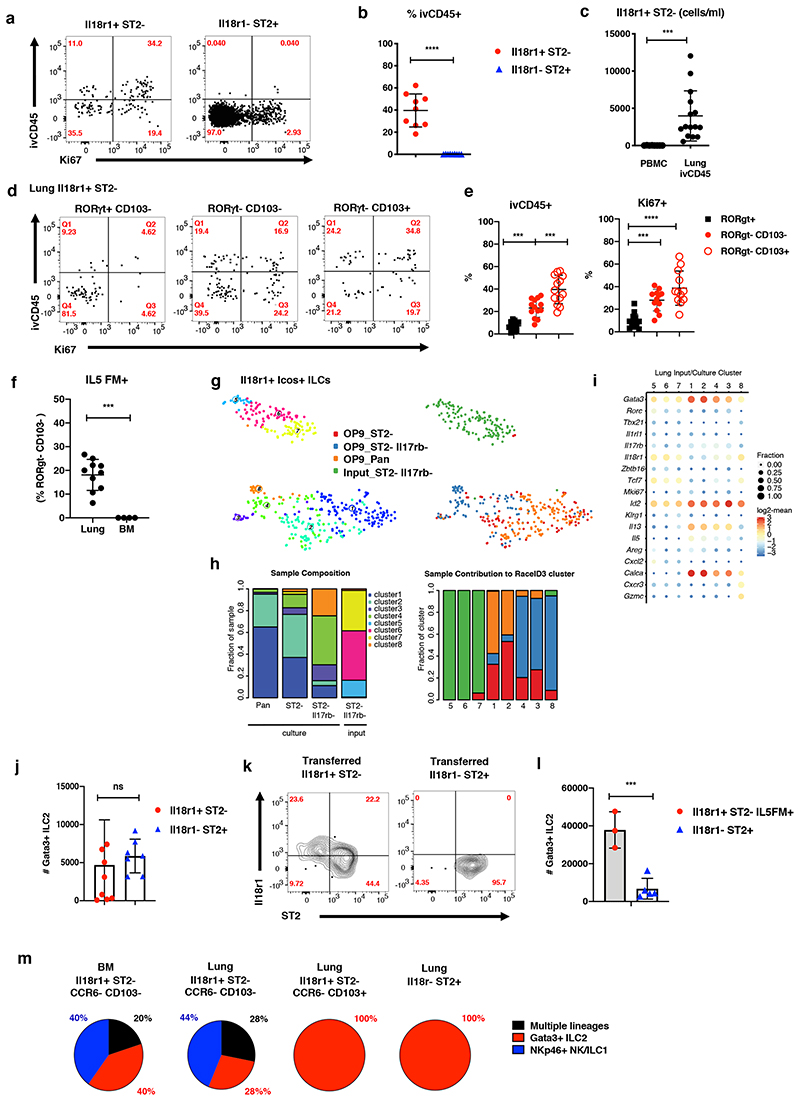
Consistent with the detection of human ILCPs in peripheral blood, we have previously observed rare donor-derived cells among BM ILCPs in parabiotic mice, suggesting that ILCPs, similar to HSCs, can physiologically traffic and systemically disperse at low levels in adult mice (Gasteiger et al., 2015; Wright et al., 2001). Of note, around 30-40% of Il18r1+ST2- cells were labelled with intravenously injected anti-CD45 (ivCD45) (Figure 3A, B), and expressed genes associated with migration such as Vim and S1pr1 (cluster 11, Figure 1E), and therefore could represent cells transiting through the pulmonary circulation. Il18r1+ST2- cells were, however, extremely rare in the peripheral blood but readily detected in the ivCD45+ fraction of the lung (Figure 3C), indicating that they may marginalize and enrich in lung vasculature. Interestingly, while Il18r1+ ILCs comprised a high fraction of ivCD45+ and Ki67+ cells, the majority of them was still ivCD45-, indicating their presence in the lung tissue at steady state (Figure 3D, E). Consistent with their residency in the lung and a minor exchange with circulating cells, all ivCD45-Il18r1+ST2- subtypes were predominantly host-derived in parabiotic mice (Figure S4A). In addition, when comparing cells with an ILCP phenotype (Il18r1+ST2-CD103-RORγt-) in lung versus BM, we found that about 20% fate-mapped positive for Il5 in lungs but not in the BM of Il5-Cre mice, further indicating that they are distinct from BM ILC progenitors “in transit” (Figure 3E). Instead, these data support the idea that ILCPs gained Il5 expression locally and matured towards ILC2s within the lung tissue (Figure 3F).
Figure 3. Il18r1+ ILCs are immature cells that give rise to Gata3hi ILC2s in the lung.
(A-B) FACS analysis of Ki67 expression and intravenous CD45 label (ivCD45) in Il18r1+ST2-RORγt+Klrg1- and Il18r1-ST2+ adult lung ILC2s. Representative gating (A) and percentage of ivCD45+ cells within indicated ILC subsets (B). (C) Number of Lin-Il7ra+Il18r1+ cells per ml blood in PBMCs (Il18r1+ST2- ILCs) and lung (ivCD45+Il18r1+ST2-RORγt+Klrg1- ILCs). Graphs in (B) and (C) depict data as mean ± SD (unpaired t-test, *P < 0.05, **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant). (D-E) Representative gating (D) and percentage of Ki67+ and ivCD45+ cells (E) for indicated Il18r1+ST2- ILC subsets in adult lung. Data are pooled from 3-4 independent experiments (n=4-16 mice). (F) Percentage of Il5Cre fate-mapped (Il5FM+) Il18r1+ST2-RORγt-CD103- ILCs in lung and BM. RORγt staining was used to exclude ILC3. Data are pooled from 2-3 independent experiments (n=4-9 mice). Graphs in (E) and (F) depict data as mean ± SD (one-way ANOVA Tukey’s multiple comparisons test (E) or unpaired t-test (b, c, f), ***P < 0.001; ****P < 0.0001). Data are pooled from 3-4 independent experiments with a total of n = 8-15 mice). (G) t-SNE representation of single-cell transcriptomes of lung Il18r1+Icos+ ILCs before (“input”) and after culture (“output”) highlighting RaceID3 clusters. (H) Cluster composition of samples (left) and sample composition of clusters (right, for legend see (G)). (I) Expression of candidate genes for input and output populations. Color represents log2 mean expression in the respective cluster and dot size indicates fraction of cells expressing the gene in the cluster. (J) Total number of Gata3+ ILC2s after culture of indicated lung ILCs for 15 days in the presence of Il2, Il7, SCF, Il25 and Il33 on OP9-DL1 for 15 days. (K) FACS analysis of Lin-Il7ra+ lung ILCs on d21 post transfer into sublethally irradiated CD45.2+RAG-/-γc-/- mice. Congenically marked Il18r1+ST2- and Il18r1-ST2+ ILCs sorted from lungs of d2 Nb infected mice were cotransferred (2,000 cells each). Data are representative of 2 individual experiments (n=3 mice). (L) Total number of Gata3+ ILC2 after culture of indicated lung ILCs as in (J). Data in (J) and (L), are pooled from 2 independent experiments with n=3-4 repeat wells each. (M) Indicated lung and BM ILC subsets were sorted as single cells onto OP9-DL1 monolayers and cultured for 19 days. Clonal progeny was analyzed by FACS. Pie charts indicate distribution of culture output across positive single cell cultures.
To directly test their cell-intrinsic differentiation potential, we purified Il18r1+Icos+ lung ILCs by FACS for in vitro differentiation assays. In order to deplete cells that may be committed towards the ILC2 lineage, we additionally sorted ST2- and ST2-Il17rb- double negative Il18r1+Icos+ ILCs from adult lungs, and cultured all three populations on OP9 in the presence of Il2, Il7, Il1β, and Il23 (Figure S4B), conditions which were recently used to expand human ILCPs (Lim et al., 2017). After ~21 days we performed scRNA-seq of lineage-negative output from these cultures along with the Il18r1+Icos+ST2-Il17rb- input population. First, we confirmed that the input population co-clustered with the naïve population from the uninfected lung ILC dataset and expressed only very low levels of Il1rl1 and Il17rb (Figure S4C, D). Next, a combined analysis yielded 8 clusters, of which 5 corresponded to cells harvested from the culture (clusters 1, 2, 3, 4, 8) and 3 represent the input population (clusters 5, 6, 7, Figure 3G, H). Clusters 1 and 2 are marked by the expression of Il5 and high levels of Gata3, Il13 and Calca (Figure 3I), and were thus classified as ILC2s. The majority of cells differentiating from Il18r1+Icos+ and Il18r1+Icos+ST2- input populations gave rise to these ILC2s (95 and 77%, respectively), while only ~15% of cells originating from Il18r1+Icos+ST2-Il17rb- cells, were ILC2s in clusters 1 and 2. Instead, these cells predominantly (61%) populated clusters 3 and 4, comprising a mixture of ILC2s and ILC3s with reduced levels of ILC2 specific gene expression. About 25% of cells originating from the Il18r1+Icos+ST2-Il17rb- population gave rise to NK/ILC1 cluster 8 characterized by expression of Cxcr3, Klrk1 and Gzmc. Furthermore, whereas ILC2s largely reduced or lost Il18r1 and Tcf7 expression, cells in cluster 8 maintained high expression of these genes (Figure 3G-I). Of note, only a small fraction of cluster 8 originated from cultured Il18r1+Icos+ST2- cells (Figure 3H), suggesting that depletion of Il17rb expressing cells enriches the fraction of cells with ILC1/NK cell potential. We also assessed the potential of the Il18r1+Icos+ counterparts from the BM in the same conditions. Consistent with lung ILCs, the majority of Il18r1+Icos+ST2- BM ILCs gave rise to ILC2s. In contrast, Il18r1+Icos+ST2-Il17rb- cells from the BM gave rise to only very few Il13 expressing ILC2s (Figure S4E, F). Of interest, the presence of Il18 in these cultures led to the upregulation of ILC2 effector genes (Figure S4H, I), as recently suggested (Ricardo-Gonzalez et al., 2018) but did not change the expansion of Il18r1+ ILC2s in vitro (Fig S4J).
These data suggest that Il18r1 + lung and BM ILCs predominantly differentiated into Gata3 hi Il13 + ILC2. This bias towards ILC2s was evident under culture conditions that were recently shown to expand ILC progenitors (Lim et al., 2017) in the absence of alarmins Il25 and Il33, known to enforce ILC2 development. In order to test the expansion potential under such ILC2-activating conditions we cultured Il18r1+ST2- and Il18r1-ST2+ ILC2s from lung in the presence of Il25 and Il33, and found that both populations generated similar numbers of Gata3hi ILCs as ST2+ cells (Fig 3J). To test the differentiation potential in vivo, we activated the ILC2 compartment through infection with the parasitic helminth Nippostrongylus brasiliensis (Nb), which induces the local expansion of ILC2s. To this end, we sort-purified Il18r1+ST2- and Il18r1-ST2+ ILCs from lungs 2 days (d) post infection (p.i.) and co-transferred cells at a 1:1 ratio into sublethally irradiated RAGγc-/- hosts, as previously established (Ghaedi et al., 2020; Xu et al., 2019). Three weeks after transfer, we found that Il18r1+ST2- cells partly gave rise to Il18r1+ST2-, Il18r1+ST2+ and Il18r1-ST2+ ILC2s, consistent with the idea that these cells can differentiate into mature ILC2s (Figure 3K). Transferred Il18r1-ST2+ ILC2s, in contrast, only generated Il18r1-ST2+ progeny, suggesting that Il18r1+ cells represent early stages on the differentiation trajectory that are not readily generated from more mature cells in lymphopenic recipients.
Notably, Il18r1+ cells are a mixture of Ccr6+RORγt+ ILC3s (Figure S4K) as well as Ccr6-RORγt- immature cells with NK/ILC1 and ILC2 potential, as suggested by the differentiation of these cells in vitro and in vivo, upon transfer (Figure S4 K-M). Therefore, only a fraction of the sorted Il18r1+ subsets will likely represent cells with ILC2 fate. To enrich for these cells, we sorted Il18r1+ST2- Il5FM+ cells for culture in the presence of Il25 and Il33, yielding many more Gata3hi ILC2s than culture of ST2+ cells, highlighting the unique expansion potential of these immature lung ILCs (Figure 3L). Consistently, transfer of Ccr6- cells (i.e., RORγt-depleted and ILCP enriched cells) engrafted better and generated more ILC2s as compared to ST2+ mature ILC2s in vivo (Figure S4M). To compare the clonal developmental potential of lung Il18r1+ST2- subsets and BM ILCPs, we performed single-cell cultures in Il2, Il7, SCF in the presence of Il25 and Il33. Intriguingly, we found that both Il18r1+ST2-CD103-Ccr6- lung and BM ILCPs gave rise to ILC2 or ILC1/NK cells or multiple lineages (Figure 3M). CD103+ and ST2+ cells, in contrast, preferentially generated ILC2s and ILC3s were undetectable under these type-2 polarizing conditions. Together, these data demonstrate that similar to the BM, the lung hosts a spectrum of immature cells with mixed lineage ILC potentials, including cells that are primed for ILC2 fate.
Emergence of ILC2 subset heterogeneity during N. brasiliensis infection
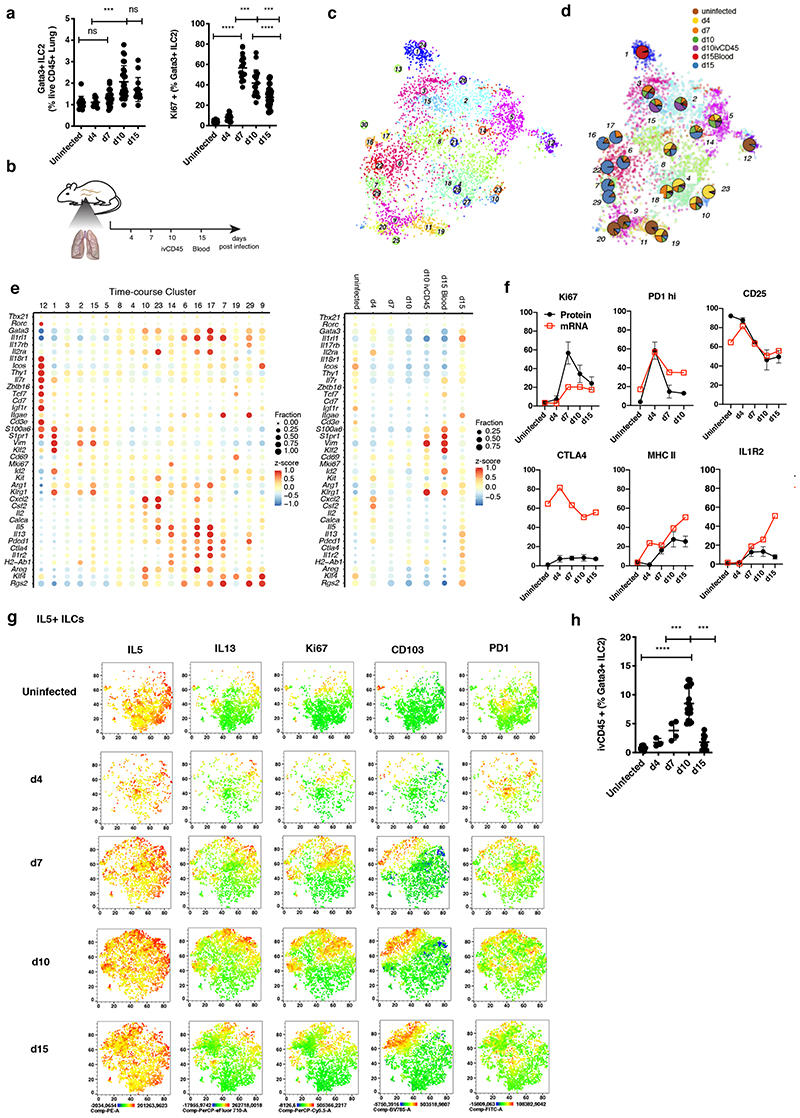
Next, we investigated how the spectrum of phenotypic states of lung ILC2s and their differentiation trajectories change upon immune challenge. Infection with Nb causes local expansion of tissue-resident ILC2s during the acute phase of infection (Figure 4A), and around d15 postinfection (p.i.), in the tissue-repair phase, a fraction of lung ILC2s has emerged from cells recruited through the blood circulation (Gasteiger et al., 2015; Huang et al., 2018). During inflammation, ILCs exit from the small intestine (Huang et al., 2018), the lung (Ricardo-Gonzalez et al., 2020), and the BM (Stier et al., 2018) into the peripheral blood, and, hence, ILCs of different origins can traffic to the lung during infection to extend the local spectrum of phenotypes and functions. In order to dissect the heterogeneity of lung ILC2s upon immune challenge, “pan-ILCs” were sequenced at different time points p.i. along with “pan-ILCs” from the peripheral blood and ivCD45+ ILCs from the lung vasculature (Figure 4B). After combining with the uninfected lung data (Figure 1) and removal of NK cell and macrophage contaminations, we inferred ILC2 sub-types using RaceID3 (Figure 4C). Our analysis highlights the dynamic emergence of ILC heterogeneity and specialization over time (Figure 4D, S5A). For example, cells from the uninfected condition contributed mainly to clusters 9, 11, 12 and 20 and Il18r1 + immature ILCs from the uninfected condition predominantly mapped to cluster 12(Figure 4E, S5B, C). ILC2s derived from d4 p.i. expressed high levels of Calca, Csf2 and Cxcl2 and resided in cluster 4 and 10 together with a subset of ILC2s from the uninfected condition. However, ILC2s became more specialized during infection, exemplified by cluster 23 comprising almost exclusively d4 cells (Figure 4E, S5B), with marked up-regulation of Csf2, Cxcl2, Cxcl1, and Il5, which promote neutrophil and eosinophil recruitment, two myeloid subsets that expand in tissues early during helminth infection (Motran and Cervi, 2018). Similarly, ILC2s isolated at d15 p.i. comprised a variety of different effector subsets, e.g., cluster 14 with high expression of MHCII complex genes Cd74, H2-Ab1 and H2-Aa (Oliphant et al., 2014; Symowski and Voehringer, 2019) and genes associated with T cell regulation such as Il1r2, Ctla4 and Furin, or clusters 16 and 17, expressing high levels of Il5, Il13, Cxcl2 and Areg (Figure 4E, S5B).
Figure 4. Emerging heterogeneity of lung ILC2s during Nippostrongylus brasiliensis infection.
(A) FACS analysis of the fraction of Lin-Gata3+ ILC2s among live CD45+ cells (left) and of Ki67+ cells among Lin-Gata3+ ILC2s (right) in Nb infected lungs. Data are pooled from 3 independent experiments with a total of n=12-16 mice per time point. (B) Sequenced samples at the respective time-points during Nb infection. (C) t-SNE map of combined Nb infection time-course and uninfected lung data (cf. Figure 1) highlighting RaceID3 clusters. (D) Overlay of t-SNE map from (C) with pie charts indicating normalized sample contribution to clusters. (E) Expression of candidate genes for ILC subsets for inferred time-course clusters with at least 20 cells (left) or the respective sample (right). Color represents z-score of mean expression across clusters/samples and dot size represents fraction of cells positive for the gene in the cluster/sample. (F) Time-course FACS analysis of the fraction of ivCD45+ ILC2s in Nb infected lungs. FACS data are pooled from 2-3 independent experiments with n=6-12 mice per time point. (G) Time-course FACS analysis of Il7ra+ ILCs in Nb infected lungs (black) compared with fractions of RNA expressing cells of time-course dataset (red). FACS data are pooled from 2-3 independent experiments with n=6-9 mice per time point. (H) Time-course FACS analysis of Nb infected lungs. Expression of indicated markers represented as t-SNE maps for Il5+ ILCs. ICS after incubation with PMA, ionomycin and monensin.Data are pooled from 5 mice for each time point and merged from equal numbers of Lin-Il7ra+ ILCs. Graphs in (A) and (F) depict data as mean ± SD (one-way ANOVA Tukey’s multiple compari sons test, ***P < 0.001; ****P < 0.0001).
ILCs isolated on d7 and d10 p.i. contributed predominantly to clusters 2, 3, 5, and 15, which up-regulated Vim and, except for cluster 3, down-regulated expression of Il1rl1. Clusters 2 and 15 additionally up-regulated Klrg1 (Figure 4E). We performed ivCD45 labeling to identify ILC2s that circulate through the lung during infection (Huang et al., 2018; Ricardo-Gonzalez et al., 2020) and sequenced ivCD45+ cells on d10 p.i. when their frequency peaked at ~10% of all lung ILC2s (Figure 4F). IvCD45+ ILC2s clustered mainly with d7 and d10 ILCs (cl. 2 and 15) and few ivCD45+ cells also clustered with mature effector ILC2s (e.g. cl. 6, 7 and 23). In contrast, blood ILC2s sorted on d15 p.i., when the wave of tissue-exiting ivCD45+ cells has largely ceded, constitute a separate cluster (cl. 1) with high expression of Vim, Klrg1, Id2, Klf2 and S1pr1, and have only minor contributions to clusters 2 and 3 (Figure 4D, E, S5B). Among them we detected a tiny population of cells expressing Il18r1, Icos, Tcf7 and low levels of Il1rl1, consistent with the idea that Il18r1+ST2- cells may traffic to the lungs where they enrich in the lung vasculature and in the tissue (Figure 3A-C, and S5D, E).
We validated the dynamic regulation of selected markers, including PD1, MHCII, Il1r2, Ctla4, and CD25, on the protein level during the course of infection by flow cytometry, suggesting a close correlation of mRNA and protein levels for most genes (Figure 4G). Consistently, our analyses confirmed the regulated co-expression of Il1r2, PD1 and MHCII during late stages of infection (Figure S5F). Furthermore, we could validate the dynamic emergence of heterogeneous Il5-expressing ILC2s, co-expressing Il13 and/or PD1, resembling cluster 23 at d4 and clusters 16 and 17 at d10/d14 p.i. (Figures 4H).
Differentiation trajectories of lung ILC2s during N. brasiliensis infection
We then investigated the differentiation dynamics of ILC2 subsets during Nb infection. Initially, we highlighted the position of cells from the early part of uninfected lung trajectory (uninfected lung clusters 1, 5, 11, 9, 8 in Figure 1) in the combined dataset (Figure 5A). Cells of the uninfected lung cluster 1 were largely localized to cluster 12, which was dominated by cells from the uninfected condition (Figure S6A). In contrast, the majority of cells from uninfected lung clusters 5, 11 and 9, representing early ILC2 differentiation stages, mixed with cells isolated during infection in clusters 2, 5 and 12 (Figure 5A), raising the question whether these cells may undergo dynamic differentiation and give rise to mature effector ILC2s during Nb infection. We applied StemID2 to infer potential trajectories starting from Il18r1 + immature ILCs in either cluster 5 or 12 and ending in mature ILC2 effector clusters such as cluster 16 or 23 (Figure 5B). A large fraction of immature uninfected lung cells localized to clusters 2 and 8, suggesting that differentiating immature Il18r1 + ILCs transition through these stages. From cluster 8 the trajectory branches out, potentially terminating in early (d4) or late (d15) effector clusters 23 or 16, respectively (Figure 5B). As the link between clusters 12 and 5 was populated well with intermediate single-cell transcriptomes (Figure S6B), we chose cluster 12 as a starting point, traversing cluster 5. Along both trajectories, we detected up-regulation of Arg1, Klrg1, Gata3, Il1rl1, and effector cytokines, as well as down-regulation of ILC progenitor genes (Figure 5C). Of note, we detected cells expressing Il18r1 and Tcf7 also within mature ILC2 compartments, albeit at reduced levels and frequencies compared to the progenitor cluster 12 (Figure 4E, S6C). The cluster composition along the trajectory suggested the presence of alternative starting points. Cluster 2 or 15, marked by a high contribution of potentially infiltrating ivCD45+ cells (Figure 5B), may represent such alternative starting points for the in situ differentiation into mature nILC2 effector cells, consistent with the observation that both recruited and resident cells can contribute to lung ILC2s (Figure S6D) (Gasteiger et al., 2015; Huang et al., 2018).
Figure 5. Differentiation trajectories of lung ILC2s during Nippostrongylus brasiliensis infection.
(A) t-SNE maps showing the distribution of uninfected lung immature clusters (cf. Figure 1) within the combined data set (top) and pie charts of the distributions (bottom) (cf. Figure 4C). (B) StemID2 lineage inference and normalized sample contribution to clusters depicted as pie-charts. Link color indicates p-value of StemID2 links (P < 0.05, Methods). (C) Pseudo-temporal gene expression profiles (local regression) along depicted trajectories. Color bars indicate cluster and sample identity, respectively, of cells ordered along the depicted trajectory by StemID2. (D) Time course analysis for the percentage of Il18r1+ST2-RORγt+Klrg1- ILCs (left) and the comparison of ivCD45 labelled and unlabelled fractions (right). (E) Time course analysis for the percentage of Il18r1+ST2+ (left) and Il18r1-ST2+ (right) ILC2s. Data in (D) and (E) are representative of 2 independent experiments with n=4-6 mice per timepoint. Graphs depict data as mean ± SD (one-way ANOVA Tukey’s multiple comparisons test, *P < 0.05, **P < 0.01; ***P < 0.001; ****P < 0.0001). (F) Time-course FACS analysis of Nb infected lungs. Expression of indicated markers represented as t-SNE maps for Il18r1+ ILCs. ICS after incubation with PMA, ionomycin and monensin. Data are pooled from 5 mice for each time point and merged from equal numbers of Lin-Il7ra+ ILCs.
These observations suggest that lung ILC2s can differentiate from an Il18r1 + Tcf7 + immature population during Nb infection. We found that Il18r1+ST2- cells were maintained during Nb infection while the proportion of ivCD45- cells among them gradually increased, consistent with the local expansion of these cells in the lung tissue (Figure 5D). At the same time, we observed a dramatic increase in highly proliferative Il18r1+ST2+ cells over the course of infection (Figure 5E), which we characterized as differentiation intermediates along an expression gradient of Tcf7 and Gata3 and which were generated in vivo from transfers of Il18r1+ST2- immature cells (Figure 3K). Consistently, we observed the upregulation of ST2, Il5, Il13 and Ki67 within Il18r1+ lung ILC2s over the course of infection (Figure 5F).
Recruited cells contribute to the entire phenotypic spectrum of lung ILC2s during N. brasiliensis infection
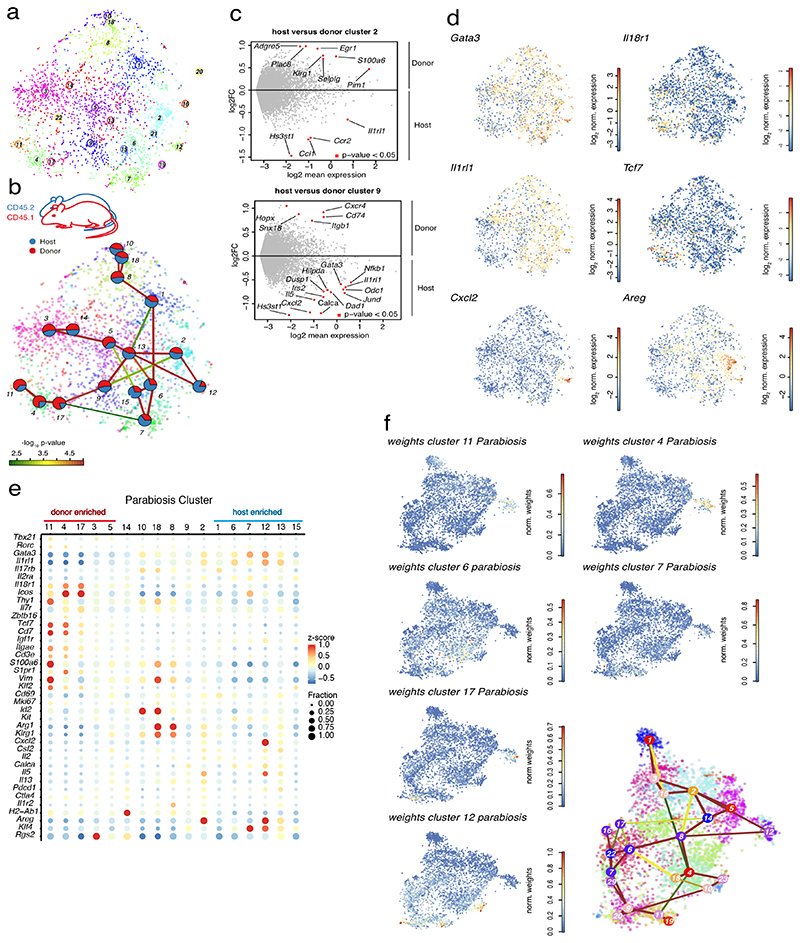
A major open question is how cells recruited during inflammation contribute to the local pool of tissue ILCs, particularly, whether these cells retain specific phenotypes and functions, or adapt to their new tissue-environment. To address this question, we analyzed infiltrating versus resident ILC2s from lungs of Nb infected parabiotic mice. Here, two mice establish a common blood circulation and infiltrated donor-derived cells can be identified through allogeneic markers. Parabionts were infected with Nb, and similar numbers of donor and host ILC2s were sorted for scRNA-seq on d15 p.i,, a time point when donor-derived, i.e., recruited, ILC2s represent about 10-15% of all lung ILC2s (Gasteiger et al., 2015; Huang et al., 2018). After RaceID3 analysis (Figure 6A) clusters were examined for their normalized host and donor contributions. Remarkably, all inferred clusters showed contribution of both host- and donor-derived cells (Figure 6B), with few differentially expressed genes between donor and host cells of the same cluster (Figure 6C). This observation demonstrates that cells recruited into the lung post parabiotic surgery contribute to all subpopulations that can be discriminated by unbiased scRNA-seq analysis at late stages of Nb infection.
Figure 6. Recruited cells contribute to the entire phenotypic spectrum of lung ILC2s during Nippostrongylus brasiliensis infection.
(A) t-SNE map of cells derived from parabiotic mice at d15 p.i. highlighting RaceID3 clusters. (B) StemID2 lineage inference and normalized donor and host contribution to clusters depicted as pie-charts. Link color indicates p-value of StemID2 links (P < 0.05, Methods). (C) Differentially expressed genes (P < 0.05, Methods) between donor and host cells within the depicted clusters. (D) t-SNE maps indicating log2 normalized expression of candidate genes in the parabiosis data. (E) Expression of representative candidate genes for ILC subsets in parabiosis clusters with at least 20 cells. Color represents z-score of mean expression across clusters and dot size represents fraction of cells expressing the gene in the cluster. (F) t-SNE map of parabiosis data showing normalized weights of indicated parabiosis cluster medoids for cells within the Nb timecourse data (cf. Figure 4) as computed by cluster mapping (Methods).
Consistent with the idea of in situ differentiation of recruited cells from immature ILC2s, we found that some clusters were significantly enriched either for donor cells (clusters 3, 4, 5, 11 and 17) or host cells (cluster 1, 6, 7, 12, 13 and 15) (Figure 6B, S7A). While the former exhibited the signature of immature ILC2s with high levels of Il18r1, Tcf7, and Cd7, and low levels of Gata3 and Il1rl1 (clusters 4, 11 and 17), the latter showed expression of Calca and cells of clusters 12 and 13 additionally up-regulated Cxcl2, Areg, and Il5 (Figure 6D, E and S7B). Thus, donor-derived cells are enriched in early differentiation stages. To further test this, we mapped the position of the donor- and host-enriched parabiosis clusters along the inferred trajectory of the time-course data (Figure 5b, Methods). The Il18r1 + donor-dominated clusters 4, 11 and 17 of the parabiosis dataset had indeed the highest similarity to cells of time-course cluster 12, marking one of our inferred origins of ILC2 differentiation during Nb infection. Conversely, clusters enriched for host derived cells predominantly exhibited highest similarities with the terminal clusters of the inferred trajectories from the time-course data (Figure 6F and S7C).
These results demonstrate that donor-derived recruited cells populate all clusters while being enriched in immature Il18r1 +ST2- ILCs, consistent with their in situ differentiation and their potential role in the renewal of tissue ILC2s. Importantly, our analyses reveal that ILCs recruited into the adult lung can generate the entire spectrum of tissue ILC2s based on their single cell transcriptomes.
Bone marrow-derived ILCs give rise to the full phenotypic spectrum of ILC2s in the adult lung
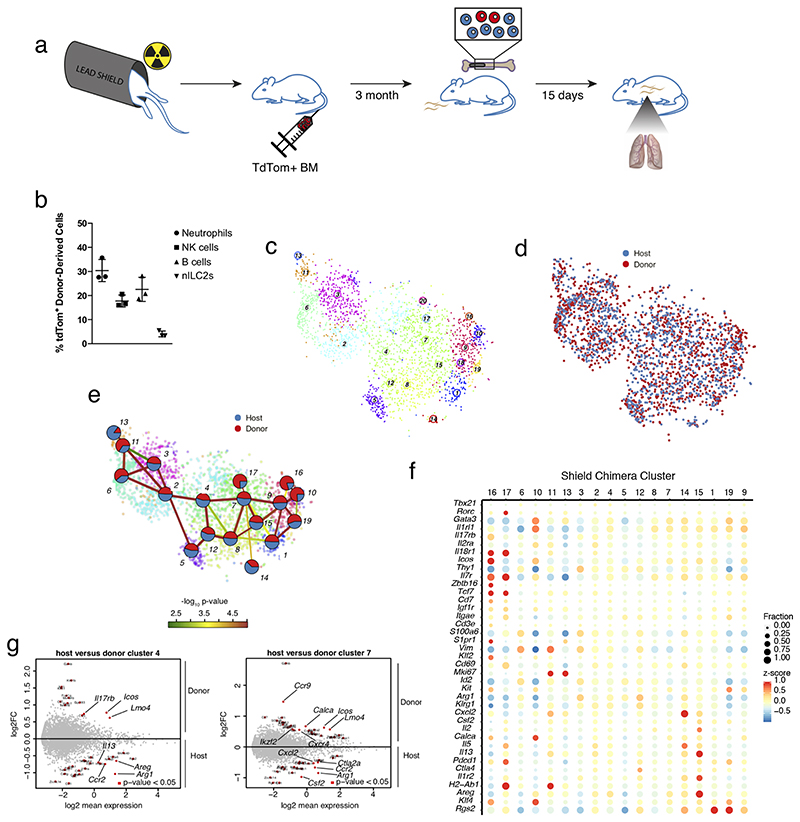
We demonstrated that ILCs entering from the circulation give rise to all subtypes of tissue ILC2s during Nb infection (Figure 6B). However, the infiltrating cells can originate from different sources, including the BM (Constantinides et al., 2014; Klose et al., 2014; Stier et al., 2018), a circulating ILCP (Lim et al., 2017), or cells exiting from inflamed tissues (Huang et al., 2018; Ricardo-Gonzalez et al., 2020). These observations raise the possibility that cells from different origins give rise to limited specialized subsets of ILCs, and only together compose the full spectrum of tissue ILCs. Therefore, we investigated, whether cells originating from a defined source contribute to specialized subsets, or whether they can reconstitute the full phenotypic spectrum of lung ILC2s. To this end we adapted a shield chimera model (Gentek et al., 2018), where partial bone marrow chimerism was established through transfer of bone marrow after local irradiation of the hind legs (Figure 7A). Most of the body, including the lung, was shielded from the irradiation to avoid perturbation of the host tissue and immune cells. This procedure led to partial chimerism of BM-dependent cells of the myeloid and lymphoid lineages (e.g., NK cells), while very few ILC2s were detectable in the lung (Figure 7B). In contrast, ILC2s readily emerged from engrafted BM when shield chimeras were generated during the neonatal time window (data not shown), consistent with the local expansion of lung ILC2s during that time of ontogeny (Huang et al., 2018; Saluzzo et al., 2017; Schneider et al., 2019).
Figure 7. Bone marrow-derived ILCs give rise to the full phenotypic spectrum of ILC2s in the adult lung.
(A) Nb infection of the shield chimera model. (B) Frequency of donor-derived cells from transferred bone marrow (BM). (C-D) t-SNE map of ILCs from shield chimeric mice isolated at d15 p.i. highlighting RaceID3 clusters (C) and donor or host origin for each cell (D). (E) StemID2 lineage inference and normalized donor and host contribution to clusters depicted as pie-charts. Link color indicates p-value of StemID2 links (P < 0.05, Methods). (F) Expression of candidate genes for ILC subsets in the shield chimera clusters with at least 10 cells. Color represents z-score of mean expression in a cluster and dot size represents fraction of cells expressing the gene in the cluster. (G) Differentially expressed genes (P < 0.05, Methods) between donor and host cells within the depicted clusters.
To test the differentiation potential of BM-derived cells in adult mice, adult shield chimeras were infected with Nb, and donor and host ILCs were sequenced on d15 p.i. (Figure 7A). Donor derived cells comprised ~5-8% of lung ILCs (Figure 7C, D). Similar to our observations for the parabiotic mice, all inferred clusters comprised both host and donor derived cells (Figure 7E). Again, we detected donor enriched clusters with immature ILC2 signature up-regulating Il18r1, Tcf7, and Zbtb16 (clusters 16 and 17) (Figure 7E, F and S7D, E). Within-cluster comparison revealed reduced levels of Ccr2, Ctla2a, Cxcl2 and Areg, and higher levels of Lmo4, Icos and Il17rb in donor versus host derived cells (Figure 7G). These small but consistent differences suggest that donor derived cells carry a signature of less mature and migrating cells, consistent with their potential recruitment into the lung during infection. Our analyses do, however, not directly test this recruitment during infection, and donor-derived ILC2s could also differentiate from BM-derived cells that have migrated to tissues before infection. Consistent with both of these possibilities, our data establish that the adult bone marrow is a source of ILC2s, which can traffic to the lung and can generate the full spectrum of ILC2 subsets and differentiation stages in the lung during infection.
Discussion
How ILCs are maintained and regenerated in the adult organism is currently unclear. Here, we provide evidence that differentiation and maturation of ILC2s from immature progenitors occurs within the adult lung. We analyzed the full spectrum of lung ILC2s at single-cell resolution and identified a heterogeneous population of Il18r1+ tissue-associated ILCs, that express Tcf7 and Zbtb16, and low levels of Gata3, ST2 and ILC2 effector molecules. We derived a continuous differentiation trajectory and validated the potential of Il18r1+Tcf7+ cells to differentiate into Gata3hi ILC2s.
Our extensive single-cell transcriptome comparison of ILC populations in the BM, and neonatal and adult lung at steady-state, as well as over the course of helminth infection in wild-type, parabiotic and shield chimeric mice support the idea that core programs of ILC2 differentiation are recapitulated during ontogeny as well as inflammatory challenge.
Intriguingly, recently described systemic ILC progenitors in men exhibit a transcriptional profile reminiscent of Il18r1+ ILCPs in murine lung (Ghaedi et al., 2020; Lim et al., 2017). Here, we confirmed that these cells are present in human lung tissue and can be further characterized by IL18R1 and TCF1 expression. Our findings suggest that Il18r1+ lung ILCPs represent the murine counterpart of human tissue ILCPs and thereby open new avenues to investigate how the local differentiation of tissue ILCs may contribute to inflammatory diseases (Mazzurana et al., 2018).
Il18 is best known as a myeloid derived cytokine that can activate lymphocytes, including ILC2s (Ricardo-Gonzalez et al., 2018). Il18 may, however, have additional functions and Il18r1 should not be considered an exclusive marker of effector cells as it is also expressed by, e.g., BM hematopoietic stem cells (HSCs), common lymphoid progenitors, BM and circulating ILCPs, (Lim et al., 2017; Seillet et al., 2016; Silberstein et al., 2016; Xu et al., 2019), and local production of Il18 regulates the turnover of HSCs in their niche (Silberstein et al., 2016). Future studies should therefore investigate the niches of Il18r1+ ILC progenitors, as well as the potential functions of Il18 in regulating progenitor-like ILCs during homeostasis versus inflammatory challenge.
Il18r1+ ILCs were virtually absent from peripheral blood of uninfected mice, but enriched in the lung vasculature which harbors many leukocytes that marginate from the blood stream, enrich at the endothelium and extravasate upon local inflammation (Anderson et al., 2014; Barletta et al., 2012; Yipp et al., 2017). It is therefore conceivable, that the vasculature may similarly serve as a local reservoir or niche for subsets of Il18r1+ ILCs. A large fraction of Il18r1+ ILCs was nevertheless ivCD45-, confirming their presence in the lung parenchyma. In addition, analysis of parabiotic mice suggested that these cells are resident or exchange only very slowly in the lung-tissue. Importantly, lung-resident and BM ILCPs share core similarities. However, BM ILCPs were negative for Il5 and Il5-fatemap label, while corresponding populations in the lung had intermediate levels of both, consistent with the idea that these cells receive local signals driving their maturation within the tissue. Intriguingly, the lung has been proposed as a reservoir of HSCs (Lefrançais et al., 2017), and may provide niches for the local differentiation of progenitors. Consistent with local differentiation, pulmonary Il18r1+ cells were highly proliferative in uninfected tissue (in striking contrast to BM ILCPs or mature tissue ILCs) and gave rise to Il18r1+ST2-, Il18r1+ST2+ and Il18r1-ST2+ ILCs in recipient lungs upon transfer. Transplanted mature ST2+ cells, however, did not generate Il18r1+ST2- or Il18r1+ST2+ populations, suggesting that they are further downstream of a differentiation cascade, or that they cannot access the niche of immature ILCs in the lung.
Our data raise the possibility that tissue ILCPs may locally self-renew, but also indicate that they can receive input from circulating cells. First, we observed a small but significant donor contribution in the ivCD45+Il18r1+CD103- subset in parabiotic mice. Second, we found that Il18r1+Tcf7+ immature cells are generated from donor BM in shield chimeras. Third, donor-populations in infected parabiotic and shield chimeric mice were enriched with Il18r1+Tcf7+ immature cells. Therefore, tissue ILCPs may receive low-grade influx from systemic counterparts. Increased competitive fitness of these cells may favour their contribution during aging, or when the local tissue niches are reorganized, e.g., upon regeneration of the ILC pool after infection. Such a model may explain why the frequency of donor-derived ILC2s is low early on, but gradually increases several months after parabiotic surgery, or in the reconstitution phase p.i. (Gasteiger et al., 2015; Huang et al., 2018; Ricardo-Gonzalez et al., 2020; Schneider et al., 2019). Despite a burst of ivCD45+ migrating inflammatory cells in the acute phase of helminth infection, these studies consistently showed that the majority of ILC2s were derived from tissue-resident cells around 2 weeks p.i.. Here we provide evidence for the in situ differentiation of ILCs from resident progenitors, highlighting a mechanism for the local, tissue-autonomous maintenance of ILCs in the adult organism. Importantly, we show that tissue ILCPs feature a phenotypic spectrum and mixed-lineage potential similar to the BM ILCP compartment. This heterogeneity may enable local “ILC-poiesis” to flexibly respond and generate the desired output upon distinct immune challenges.
Patients with inflammatory diseases such as psoriasis, hepatic fibrosis, or inflammatory bowel disease have elevated numbers of disease-associated ILCs in the blood. A major open question is how cells recruited during inflammation may contribute to the pool of tissue-resident cells. The extent to which local immune cells that have been generated early during ontogeny can be replenished in the adult organism has also been controversially discussed for macrophages, representing prototypic tissue-resident cells (Ginhoux and Guilliams, 2016; Mass et al., 2016). Here, we show that adult BM cells recruited into the adult lung can generate the full spectrum of lung-resident ILC2 subsets upon helminth infection. Our analysis reveals that ILCs differentiating in the adult organism can locally adapt the phenotypes of ILCs that have been suggested to emerge in distinct waves during ontogeny (Popescu et al., 2019; Schneider et al., 2019). Therefore, local tissue niches, rather than progenitor origin, may dominantly imprint ILC phenotypes. Together, our work highlights the local differentiation of immature ILCs as a mechanism of maintenance, phenotypic diversification and local adaptation of tissue ILCs.
Limitations of Study
One current limitation in the study of ILCPs is the lack of conditional, stage-specific fate-mapping models to delineate the actual contribution of BM or tissue ILCPs during homeostasis or immune challenges in the adult organism. Moreover, we lack examples of conditions in which ILCPs are essential to maintain or regenerate a functional ILC pool. We speculate that ILCPs may have such critical roles upon depletion of ILCs, as documented in mice and men during different types of infection and environmental challenges (Kløverpris et al., 2016; Ng et al., 2018). It will be interesting to test whether local depletion and regeneration of ILC2s occurs during lung diseases as well. We are aware that our work highlights the tissue adaptation and differentiation potential of BM-derived ILCs in adult lungs but does not deconvolute potential contributions of other circulating cells. Future work should address if inflammatory ILC2s are a transient effector population or if (and how) they actually contribute to lung ILC2s once acute infection has resolved. Here, we mapped the heterogeneity of lung ILC2s and highlight multiple subsets that are dynamically emerging during infection, including, e.g., effector subsets expressing different combinations of cytokines and chemokines (Il5/Il2/Cxcl2/Csf2 versus Il5/Il13/Areg), or molecules involved in regulatory functions (Pd1, Ctla4, Il1r2, MHCII) and highly proliferative Il18r1+ and/or CD103+ states. While functional validation of these populations was beyond the scope of this study, we feel that our interactive cell atlas (http://murine-ilc-atlas.ie-freiburg.mpg.de/) provides a rich resource for future studies testing subset-specific functions of ILC2s.
Star Methods
| REAGENT or RESOURCE Antibodies | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| InVivoMAb anti-mouse CD16/CD32 (Clone 2.4G2) | Bioxcell | Cat # BE0307 |
| Fixable Viability Dye eFluor® 780 | eBioscience | Cat # 65-0865-18 |
| Anti-mouse CD3e Biotin (Clone 145-2C11) | BioLegend | Cat# 100304 |
| Anti-mouse CD5 Biotin (Clone 53-7.3) | BioLegend | Cat# 100604 |
| Anti-mouse CD19 Biotin (Clone 6D5) | BioLegend | Cat# 115504 |
| Anti-mouse Gr1 Biotin (Clone RB6-8C5) | BioLegend | Cat# 108404 |
| Anti-mouse F4/80 Biotin (Clone BM8) | BioLegend | Cat# 123106 |
| Anti-mouse CD11b Biotin (Clone M1/70) | BioLegend | Cat# 101204 |
| Anti-mouse CD11c Biotin (Clone N418) | BioLegend | Cat# 117304 |
| Anti-mouse NK1.1 Biotin (Clone PK136) | BioLegend | Cat# 108704 |
| Anti-mouse TCRβ Biotin (Clone H57-597) | BioLegend | Cat# 109204 |
| Anti-mouse TCRγδ Biotin (Clone eBioGL3) | eBioscience | Cat # 13-5711-85 |
| Anti-mouse FceR1 Biotin (Clone MAR-1) | BioLegend | Cat# 134304 |
| Anti-mouse TER-119 Biotin (Clone TER-119) | BioLegend | Cat# 116204 |
| Anti-mouse CD45R/B220 Biotin (Clone RA3-6B2) | BioLegend | Cat# 103204 |
| Anti-mouse Ly6G Biotin (Clone 1A8) | BioLegend | Cat# 127604 |
| Streptavidin V500 | BD Biosciences | Cat# 561419 |
| Anti-mouse Thy1.2 FITC (Clone 30-H12) | BioLegend | Cat# 105306 |
| Anti-mouse Thy1.2 AF786 (Clone 30-H12) | BioLegend | Cat# 105331 |
| Anti-mouse CD127/IL7Rα APC (Clone A7R34) | BioLegend | Cat# 135012 |
| Anti-mouse CD127/IL7Rα BV605 (Clone A7R34) | BioLegend | Cat# 135041 |
| Anti-mouse CD25 PE-Cy7 (Clone PC61) | BioLegend | Cat# 102016 |
| Anti-mouse ICOS PE-Cy7 (Clone C398.4A) | BioLegend | Cat# 313520 |
| Anti-mouse/human KLRG-1 FITC (Clone 2F1/KLRG1) | BioLegend | Cat# 138410 |
| Anti-mouse/human KLRG-1 Percp-Cy5.5 (Clone 2F1/KLRG1) | BioLegend | Cat# 138417 |
| Anti-mouse PD1/CD279 PE-Cy7 (Clone 29F.1A12) | BioLegend | Cat# 135216 |
| Anti-mouse PD1/CD279 FITC (Clone 29F.1A12) | BioLegend | Cat# 135213 |
| Anti-mouse CCR6 PE (29-2L17) | BioLegend | Cat# 129804 |
| Anti-mouse CD103 FITC (2E7) | BioLegend | Cat# 121420 |
| Anti-mouse CD45.1 FITC (Clone A20) | BioLegend | Cat# 110706 |
| Anti-mouse CD45.2 BV711 (Clone 104) | BioLegend | Cat# 109847 |
| Anti-mouse CD45.2 Pacific Blue (Clone 104) | BioLegend | Cat# 109820 |
| Anti-mouse CD45 AF-700 (Clone 30-F11) | BioLegend | Cat# 110724 |
| Anti-mouse CD3 AF-700 (Clone 17A2) | BioLegend | Cat# 100215 |
| Anti-mouse CD3 BV711 (Clone 17A2) | BioLegend | Cat# 100241 |
| Anti-mouse IL-5 PE (Clone TRFK5) | BioLegend | Cat# 504304 |
| Anti-mouse IL-17A AF700 (Clone TC11-18H10.1) | BioLegend | Cat# 506914 |
| Anti-mouse IL13 AF488 (Clone TWAJ) | eBioscience | Cat # 53-7133-82 |
| Anti-mouse GATA-3 eF660 (Clone TWAJ) | eBioscience | Cat # 50-9966-42 |
| Anti-mouse GATA-3 PerCP-eF710 (Clone TWAJ) | eBioscience | Cat # 46-9966-42 |
| Anti-mouse Eomes AF488 (Clone Dan11mag) | eBioscience | Cat # 53-4875-82 |
| Anti-mouse IL25R/IL17RB eF660 (Clone MUNC33) | eBioscience | Cat # 50-7361-82 |
| Anti-mouse IL18Rα PE-Cy7 (Clone P3TUNYA) | eBioscience | Cat # 25-5183-82 |
| Anti-mouse NKp46 / CD335 PercP-eF710 (29A1.4) | eBioscience | Cat # 46-3351-82 |
| Anti-mouse/human TCF1/7 AF488 (Clone C63D9) | Cell Signalling | Cat # 6444S |
| Anti-mouse IL33R/ST2 BV421 (Clone U29-93) | BD Biosciences | Cat# 566309 |
| Anti-mouse RORγt PE (Clone Q31-378) | BD Biosciences | Cat# 562607 |
| Anti-mouse RORγt AF647 (Clone Q31-378) | BD Biosciences | Cat# 562682 |
| Anti-mouse RORγt BV650 (Clone Q31-378) | BD Biosciences | Cat# 564722 |
| Anti-mouse PLZF PE (Clone Q31-378) | BD Biosciences | Cat# 564850 |
| Anti-mouse Ki67 PerCP-Cy5.5 (Clone B56) | BD Biosciences | Cat# 561284 |
| Anti-mouse CTLA-4 PE (Clone UC10-4B9) | BD Biosciences | Cat# 553720 |
| Anti-mouse CD121b PE (Clone 4E2) | BD Biosciences | Cat# 554450 |
| Anti- mouse ICOS (CD278) BUV395 (Clone 398.4A) | BD Biosciences | Cat# 565884 |
| Anti-mouse CD69 BUV805 (Clone H1.2F3) | BD Biosciences | Cat# 741927 |
| Anti-mouse KLRG-1 BUV661 (Clone 2F1) | BD Biosciences | Cat# 741586 |
| Anti-mouse CD279 (PD-1) BUV737 (Clone RMP1-30) | BD Biosciences | Cat# 749306 |
| Anti-mouse CD90.2 BU496 (Clone 30-H12) | BD Biosciences | Cat# 741047 |
| Anti-mouse LPAM-1(a4b7) BV711 (Clone DATK32) | BD Biosciences | Cat# 740701 |
| Anti-mouse CD103 BV786 (Clone M290) | BD Biosciences | Cat# 564322 |
| Anti-mouse CD25 PE-Dazzle 594 (Clone PC61) | BD Biosciences | Cat# 562694 |
| Anti-mouse CD3e PE-Cy5 (Clone 145-2C11) | BD Biosciences | Cat# 553065 |
| Anti-mouse CD45.2 BUV737 (Clone 104) | BD Biosciences | Cat# 612778 |
| Anti-mouse CD45 BUV496 (Clone 30-F11) | BD Biosciences | Cat# 749889 |
| Anti-mouse IL33R/ST2 PE (Clone U29-93) | BD Biosciences | Cat# 566311 |
| Anti-mouse CD90.2 BV605 (Clone 30-H12) | BioLegend | Cat# 140317 |
| Anti-mouse CD45 BV510 (Clone 30-F11) | BioLegend | Cat# 103137 |
| Anti-mouse CD45.2 FITC (Clone 104) | BioLegend | Cat# 109806 |
| Anti-mouse CD45.1 Percp-Cy5.5 (Clone A20) | BioLegend | Cat# 110728 |
| Anti-mouse CD4 PE-Texas Red (Clone RM4-5) | eBioscience | Cat # MCD0417 |
| Anti-mouse CD8a PE-Texas Red (Clone 5H10) | eBioscience | Cat # MCD0817 |
| Anti-mouse I-A/I-E AF-700 (Clone M5/114.15.2) | BioLegend | Cat# 107621 |
| Anti-mouse TCRγδ Biotin (Clone GL3) | BioLegend | Cat# 118103 |
| Anti-mouse Epcam Biotin (Clone G8.8) | BioLegend | Cat# 118203 |
| Anti-mouse CD8α Biotin (Clone 53.67) | BioLegend | Cat# 100704 |
| Anti-mouse CD3 Biotin (Clone 17A2) | BioLegend | Cat# 100243 |
| Anti-mouse CD4 Biotin (Clone GK1.5) | BioLegend | Cat# 100404 |
| Anti-mouse CD45 FITC (Clone 30-F11) | BioLegend | Cat# 103107 |
| Anti-mouse NK1.1 PE-Cy7 (Clone PK136) | BioLegend | Cat# 108713 |
| Anti-mouse CD19 FITC (Clone MB19-1) | BioLegend | Cat# 101505 |
| Anti-mouse CD3e FITC (Clone 145-2C11) | BD Bioscience | Cat# 563061 |
| Anti-mouse CD4 FITC (Clone H1 29.19) | BD Bioscience | Cat# 553651 |
| Anti-mouse CD8a FITC (Clone 53-6.7) | BD Bioscience | Cat# 553031 |
| Anti-mouse Gr1 FITC (Clone R86-8C5) | BD Bioscience | Cat# 553126 |
| Anti-mouse Siglec-H FITC (Clone 551) | BioLegend | Cat# 129603 |
| Anti-mouse B220 FITC (Clone RA3-6B2) | BD Bioscience | Cat#553088 |
| Anti-mouse c-Kit/CD117 BV510 (Clone ACK2) | BioLegend | Cat#135119 |
| Anti-mouse Sca-1 BV650 (Clone D7) | BioLegend | Cat#108143 |
| Anti-mouse Flt3/CD135 PerCP–eFluor 710 (Clone A2F10) | eBioscience | Cat#46-1351-80 |
| Anti-mouse CD27 PE (Clone LG3A10) | BD Bioscience | Cat# 558754 |
| Anti-mouse Thy1.2 AF-700 (Clone 30-H12) | BioLegend | Cat# 105320 |
| Anti-mouse ICOS BV785 (Clone C398.4A) | BioLegend | Cat# 313533 |
| Anti-mouse IL18Rα AF647 (Clone BG/IL18RA) | BioLegend | Cat# 132903 |
| Anti-mouse IL33R/ST2 (ST2) PE-Cy7 (Clone RMST2-2) | eBioscience | Cat#25-9335-82 |
| Streptavidin BV510 | Biolegend | Cat#405233 |
| Anti-mouse IL25R/IL17RB PE (Clone 9B10) | BioLegend | Cat#146305 |
| Anti-mouse CD127/IL7Rα BV421 (Clone A7R34) | BD Biosciences | Cat# 566377 |
| Anti-mouse TCRγδ Antibody APC (Clone GL3) | BioLegend | Cat#118115 |
| Anti-mouse/human CD11b FITC (Clone M1/70) | eBioscience | Cat# 11-0112-82 |
| Anti-mouse TER-119 FITC (Clone TER-119) | eBioscience | Cat# 11-5921-82 |
| Anti-human CD14 APC-eF780 (Clone 61D3) | eBioscience | Cat# 47-0149-42 |
| Anti-human CD19 APC-eF780 (Clone HIB19) | eBioscience | Cat# 47-0199-42 |
| Anti-human CD123 APC-eF780 (Clone 6H6) | eBioscience | Cat# 47-1239-42 |
| Anti-human CD3 APC-eF780 (Clone SK7) | eBioscience | Cat# 47-0038-42 |
| Anti-human FceRIa APC Vio770 (Clone CRA1) | Miltenyi | Cat# 130-105-500 |
| Anti-human CD127 PE (Clone MB15-18C9) | Miltenyi | Cat# 130-113-410 |
| Anti-human CD218(IL18Ra) PEVio770 (Clone H44) | Miltenyi | Cat# 130-101-723 |
| Anti-human ICOS APC (Clone REA192) | Miltenyi | Cat# 130-101-723 |
| Anti-human EOMES FITC (Clone WD1928) | eBioscience | Cat# 11-4877-42 |
| Anti-human CRTH2 PerCP-Cy5.5 (Clone BM16) | BioLegend | Cat# 350116 |
| Anti-mouse/human T-bet BV605 (Clone 4B10) | BioLegend | Cat# 644817 |
| Anti-human CD117(c-KIT) BV650 (Clone 104D2) | BioLegend | Cat# 313222 |
| Anti-human CD103 BV711 (Clone Ber-ACT8) | BioLegend | Cat# 350222 |
| Anti-human CD336(NKP44) Biotin (Clone P44-8) | BioLegend | Cat# 325106 |
| Streptavidin BV785 | BioLegend | Cat# 405249 |
| Experimental Models: Organisms/Strains | ||
| Mouse: wild-type: | C57BL/6, Charles River Laboratories | |
| Mouse: CD45.1/Ly5.1 | B6.SJL-Ptprca Pepcb/BoyJ, Jax mice | JAX stock #006584 |
| Mouse: Ubiquitin tdT (“Ubow”) | (Ghigo et al., 2013) | |
| Mouse: IL5cre x LSLtdTom | (Nussbaum et al, 2013) | |
| Mouse: RAG1 -/- | B6.129S7-Rag1tm1Mom/J | JAX stock #002216 |
| Mouse: RAG2-/- γc-/- | C57BL/6NTac;B10(Cg)-Rag2tm1Fwa Il2rgtm1Wj1, Taconic, 4111-F | |
| Software and Algorithms | ||
| FlowJo v.10.5.0 | FlowJo | https://www.flowjo.com |
| Prism v.6.0. | GraphPad | https://www.graphpad.com |
| R v. 3.5.1 and 3.6.1 | https://www.r-project.org/ | |
Contact For Reagent And Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, D. Grün(gruen@ie-freiburg.mpg.de) and G. Gasteiger (georg.gasteiger@uni-wuerzburg.de)
Experimental Model And Subject Details
Mice
C57BL/6 mice were purchased from Charles River Laboratories. All mice (IL5Cre x LSL-tdTom mice, RAG1-/- mice, RAG2-/- γc-/- mice, etc.) were bred and housed under specific pathogen free conditions at the animal facilities of the University of Würzburg (ZEMM and Institute for Immunology and Virology), Max Planck Institute of Immunobiology and Epigenetics, Memorial Sloan-Kettering Cancer Center and Centre Immunologie Luminy and were used in accordance with institutional guidelines and as approved by the local authorities. For all experiments except for shield chimeras, female mice at 6-8 weeks of age were used.
Method Details
Parabiosis
Congenic CD45.1 and CD45.2 mice were surgically connected in parabiosis as previously described (Gasteiger et al., 2015). After corresponding lateral skin incisions were made from elbow to knee in each mouse, forelimbs and hindlimbs were tied together using nylon suture, and the skin incisions were closed using stainless steel wound clips. After surgery, mice were maintained on a diet supplemented with trimethoprim-sulfamethoxazole for prophylaxis of infection.
Shield chimeras
Shield irradiation was performed as described before (Baratin et al., 2017; Scott et al., 2016). Briefly, mice were placed in a 6mm thick lead cylinder that only exposed their hind legs, irradiated (9Gy) and reconstituted i.v. with BM cells from Ubiquitin tdT (“Ubow”) (Ghigo et al., 2013) donors. Newborn (1-2 days old) mice were transplanted with BM depleted of monoblasts by magnetic depletion of CD11b+ cells. Adult mice were anesthetized with a Ketamine/Xylazine mixture during irradiation and received total BM prepared using standard procedures. In both setups, BM cells were transplanted at a ratio of 1 donor per recipient.
ivCD45 labeling and cell isolation
2ug anti-mouse CD45 AF-700 antibody were injected via retroorbital vein(200ul/mouse) after mice were anesthetized with intraperitoneal injection of ketamine/xylazine. Mice were sacrificed 5 mins after antibody injection. Peripheral blood was taken from the left ventricle and bone marrow cells were prepared using standard procedures. To isolate mononuclear cells from lung, the tissues were physically dissociated using scissors and incubated for 45 minutes in 2ml DMEM (10mM HEPES) digestion buffer (0.25mg/ml Liberase LT, Roche) before being passed through 70-μm strainers. Red blood cells in blood and lung were lysed using ACK lysis buffer.
Helminth Infection
Mice were injected subcutaneously (s.c.) with 350 L3 Nippostrongylus brasiliensis (Nb) larvae cultured as previously described (Symowski and Voehringer, 2019). Briefly, Nb were maintained by passage in rats. These were injected s.c. with 1000 L3 Nb and stool was collected on days 7-9 p.i.. Stool was mixed with water and activated charcoal in petri dishes and cultured at 25°C for up to 4 weeks in a chamber lined with wet paper towels to maintain humidity. L3 Nb were collected as needed using a Baermann apparatus after at least 7 days of culture. L3 larvae were extensively washed with PBS to eliminate culture contaminants before infection.
Murine cell isolation
Lungs were harvested, chopped into small pieces and incubated in digestion buffer DMEM (Gibco 31966021); 0.25 mg/ml Liberase™ TL (Research Grade 05401020001, Roche); 10mM Hepes (Gipco 1563049) for 45 mins at 37°C on Eppendorf rotator at 120rpm. Digestion was stopped with 20ml RPMI 1640 medium Thermo Fisher Scientific; 31870074) added with 50μM 2-mercaptoethonal (AppliChem, A1108). Cells were filtered through 70um cell strainer (Falcon 3522350) and washed and collected in FACS Buffer or 1× MojoSort buffer (BioLegend; 480017). Bone marrow cells from ilia, femora, and tibiae were isolated by crushing the bones in FACS Buffer or 1× MojoSort buffer. Bone marrow cells were filtered (CellTrics, 30 μm; Sysmex) and washed and collected in FACS Buffer or 1× MojoSort buffer.
Human samples and cell isolation
Peripheral blood (PB) leukocytes were isolated from buffy coats of heathy adults provided by the German Red Cross (DRK, Berlin). Lung samples were obtained at the Helios Klinikum, Berlin, from non-microscopically affected surgical excisions of lung tumor patients. All samples were processed within 24 hours. The institutional review boards of the Charité Universitätsmedizin Berlin approved the protocols and all probands gave their written informed consent according to the Declaration of Helsinki. All studies were approved by the Ethics Committee. Human mononuclear cell suspensions from PB and lung were isolated by Ficoll density gradient centrifugation. Lung tissues were thoroughly dissociated in sterile PBS/BSA/2mM EDTA and passed through a 70μm strainer prior to density gradient.
Antibody staining for sorting and flow cytometric analysis
Cell suspensions were prepared as described above and incubated with Fc block (2.4G2, Bioxcell) and Fixable Viability Dye, and were subsequently stained with fluorophore-conjugated antibodies against: CD3ε, CD5, CD19, Gr, F4/80, CD11b, CD11c, NK1.1, TCRβ, TCRγδ, FcεR1, TER-119, CD45R/B220, Ly6G, Thy1.2, CD127/IL7Rα, CD25, IL25R/IL17RB, CD218a/IL18Rα, IL33R/ST2, ICOS, KLRG-1, PD1/CD279, α4β7, C-kit, CD45.1, CD45.2, CD45, CD3, IL-5, IL13, IL17A, NKp46, Eomes, RORγt, Gata3, TCF1/7, PLZF, Ki67, IL1R2, I-A/I-E, CD103 and CTLA-4. Lineage (Lin) positive cells were stained with a cocktail of biotin-labeled antibodies against CD3ε, NK1.1, CD5, Gr-1, CD19, TCRγδ, TCRβ, CD11b, CD11c, F4/80, TER-119, and FcεR1, followed by staining with Streptavidin (V500, BD). For intracellular cytokine staining, cells were incubated in the presence of 1μg/ml monensin (BioLegend) or re-stimulated with 50ng/ml PmA and 500ng/ml Ionomycin in the presence of 1ug/ml monensin with/without 1ug/ml Brefeldin A for 3.5h in indicated experiment. Intracellular staining of CD3ε, IL5, IL13, IL17A Ki67, IL1R2, CTLA-4, PLZF, Tcf7, Eomes, Gata3 and RORγt was performed using Foxp3 / Transcription Factor Staining Buffer Set (eBioscience). To reserve tdTom from IL5Cre fate-mapped mice, cells are first fixed with 2% FA for 1h, and then washed and stained RORγt with Foxp3/Transcription Factor staining buffer. Human cells, were incubated with antibodies against EOMES, CRTH2, TCF1/TCF7, T-bet, NKp44, CD117, CD103, ICOS, CD45, CD127, GATA3 and IL-18R. In addition, Lin was stained using antibodies against CD14, CD19, CD123, CD3, FceRIa. For flow cytometric analysis cells were analyzed on a BD Fortessa, BD Celesta or Cytek Aurora cytometer and analyzed with FlowJo software.
Pan-ILCs were sorted as live Lin- CD45+ Il7ra+ Thy1.2+/lo ILCs for scRNAseq. Additional markers were included for host and donor-derived cells (Parabiosis and Shield chimeras), intravascular cells (ivCD45-labeling) and neonatal lung (ST2).
For transfer and culture experiments, BM and lung ILCs were sorted as live Lin-CD45+ Il18r1+ Icos+ Il7ra+ Thy1.2+ and further divided into subsets based on the expression of ST2 and/or Il17rb for culturing and input for sequencing. Moreover, CD45+ Il7ra+ Thy1.2+ lung ILCs were sorted as Il18r1+ ST2-, Il18r1+ ST2-IL5FM+, Il18r1+ Ccr6+, Il18r1+ Ccr6- and Il18r1- ST2+ subsets for in vitro cultures and transfer experiments as indicated in respective figures. Cells were enriched by negative selection and depletion of Lin+ cells with anti-biotin Microbeads (Miltenyi) or (MojoSort Streptavidin Nanobeads, 480016, Biolegend) according to the manufacturer’s recommendations before sorting.
Staining and enrichment of sorted Il18r1+Icos+ BM ILCs for single-cell sequencing: BM cells were harvested as described above. Hematopoietic progenitors were enriched with the MojoSort mouse hematopoietic progenitor cell isolation kit (BioLegend; 480004) according to the manufacturer’s protocol. After enrichment, 5 x 106 to 7 x 106 cells per 100 μl of 1× MojoSort buffer were stained on ice in the dark for 40 min with following fluorophore-conjugated antibodies against: c-Kit/CD117, Sca-1, Il18r1, Thy1.2, NK1.1, Icos, Flt3/CD135, CD127/Il7rα, CD27 and the lineage markers: CD19, Siglec-H, CD3ε, B220, Gr1, CD74, TER-119 and CD11b/Mac-1. Cells were sorted as live Lin-Il18r1+Icos+ and index information was recorded for the other markers.
Adoptive cell transfer
Lung ILCs were isolated and stained as above. Cells were derived from lungs of congenically marked B6 mice. 1500-2000 cells each were co-transferred into sub-lethally irradiated (3Gy) RAG-/-γc-/- mice. ILCs were analyzed 3 or 6w post transfer as indicated in the Figure legends.
Cell culture of sorted ILCs subsets
Sorted ILCs were seeded onto mitomycin-C-treated OP9 or OP-DL1 feeder cells. Approximately 100 cells were plated in 1 well of a 96-well plate and grown at 5% CO2 in RPMI 1640 medium (complete formulation; Thermo Fisher Scientific; A1049101) supplemented with 10% fetal calf serum (Corning BV; 35-016-CV), 1× penicillin–streptomycin (Gibco; 10378016) and 25 ng/ml IL-2 (BioLegend; 575402), 25ng/ml IL-7 (BioLegend; 577802), complemented with different combinations of the following cytokines at 10ng/ml IL-1β (BioLegend; 575102), IL-23 (BioLegend; 589002), IL25 (R&D, 1399-IL-025/CF), IL33(Biolegend, 580506) and 25ng/ml SCF(Peprotech, 250-03) where indicated. One day after sorting and then every other day, 100 μl of medium was replaced with fresh medium containing cytokines. For clonal analysis of progenitor cell potential, single cells were sorted directly onto OP9-DL1 monolayers and cultured for 19 days when FACS analysis was performed. Cultures were considered to be positive for a cell population if > 20 events appeared within the indicated FACS gates.
Preparation of OP9 feeder cell layer
OP9 feeder cells were grown in 1× MEM Alpha (Gibco; 12561-056) supplemented with 20% fetal calf serum (Corning BV; 35-016-CV), 1× penicillin–streptomycin (Gibco; 10378016). OP9 cells were grown until 80–90% confluent and treated with 10 μg/ml mitomycin C (Sigma-Aldrich; M0503-2MG) for 1.5 h. Cells were washed four times with PBS and detached with trypsin–EDTA (Gibco; 25200056) at 37 °C for 5 min. Cells were resuspended in fresh complete MEM Alpha medium, and cell concentration was adjusted to seed approximately 40,000–50,000 cells in 100 μl of medium per well of a 96-well plate.
Single cell sorting, single-cell RNA amplification and library preparation of ILCs
Single cells were FACS sorted into 384-well plates on a BD AriaIII, BD Influx cell sorter or BD Aria Fusion using single-cell mode and index information was recorded (primary cells) or sorted on BD Aria Fusion using 4-way purity mode (output cultured cells). After sorting, plates were centrifuged for 5 min at 2200g at 4°C, snap-frozen in liquid nitrogen and stored at -80°C until processed. Single cell RNA sequencing was performed using the CEL-Seq2 method (Hashimshony et al., 2016) with modifications as described in (Herman et al., 2018). ILCs were sequenced on a HiSeq 2500 or Hiseq 3000 sequencing system (Illumina paired-end multiplexing run, high output mode) at a depth of ~72,000-~600,000 reads per cell.
Quantification of transcript abundance
Paired end reads were aligned to the transcriptome using bwa (version 0.6.2-r126) with default parameters (Li and Durbin, 2010). The transcriptome contained all gene models based on the mouse ENCODE VM9 release downloaded from the UCSC genome browser comprising 57,207 isoforms with 57,114 isoforms mapping to fully annotated chromosomes (1 to 19, X, Y, M). All isoforms of the same gene were merged to a single gene locus. Furthermore, gene loci overlapped by >75% were merged to larger gene groups. This procedure resulted in 34,111 gene groups. As described in (Wallrapp et al., 2017) the loci for Nmur1 was extended at the 3’ end to position 86,384,000 on the chromosome 1. The right mate of each read pair was mapped to the ensemble of all gene loci and to the set of 92 ERCC spike-ins in sense direction (Baker et al., 2005). Reads mapping to multiple loci were discarded. The left read contained the barcode information: The first six bases corresponded to the unique molecular identifier (UMI), followed by six bases representing the cell specific barcode. The remainder of the left read contained a polyT stretch. The left read was not used for quantification. For each cell barcode, the number of UMIs per transcript was counted and aggregated across all transcripts derived from the same gene locus. Based on binomial statistics, the number of observed UMIs was converted into transcript counts (Grün et al., 2014).
Clustering
The datasets were analyzed using RaceID3 (v0.1.3 and v0.1.6). Rescaling to 3,000 transcripts per cells was used for data normalization (2,000 for the parabiosis dataset due to lower sample quality). Prior to filtering and normalization, ERCCs and mitochondrial genes were excluded and cells expressing >2% of Kcnq1ot1 transcripts, identified as a marker of low-quality cells (Grün et al., 2016) were removed from analysis. RaceID3 was run with the following parameters: mintotal= 3,000, minexpr=5, outminc=5, FSelect=TRUE, probthr=10-3, ccor = 0.65 (probthr=10-4, for the parabiosis and shield chiemera dataset, and ccor=0.4, mintotal= 2000, minexpr=3, outminc=3 for the parabiosis dataset). In addition, we used imputing implemented in RaceID3 with knn=10. For the BM dataset cln=6 was set, since cluster dispersion decreases linearly after 6 clusters. For cultured lung Il18r1+Icos+ ILCs outlier identification was omitted. To remove batch effect and technical variability we initialized CGenes. CGenes and highly correlating genes (Pearson’s correlation > ccor) were removed from clustering, representing a minimal invasive batch effect removal approach. The following sets of genes were initialized as CGenes: ensemble of all Gm, RP, Hsp, Igl, Igh and A4300 genes, Scgb1a1, Jchain, Igkc, Malat1, Xist, Mid1 and Kcnq1ot1. To remove cell-cycle associated variability from the uninfected lung dataset we integrated cell cycle related genes (Buettner et al., 2015) and highly correlating genes(Pearson’s correlation >= 0.4) into CGenes. As a large fraction of neonatal cells expressed proliferation associated genes with a dominant effect on cluster, Mki67, Pcna, Tuba1a/Tuba1b, Top2a, Tubb5 were additionally included into CGenes. We used the internal RaceID batch correction method for the in vitro culture datasets. Briefly, this method identifies a pair of cells across the Il18r1+Icos+ and the Il18r1+Icos+ST2- batches with minimal k nearest neighborhoods distance and infers differentially expressed genes between cells of these two neighborhoods. Thus, Gzmb, Calca and Ucp2 were additionally included in CGenes to remove batch effect associated variability for this dataset. For the parabiosis dataset, Lars2, Mki67 and Pcna were included into CGenes, to remove batch effects or cell cycle associated variability. For contaminant cell removal, cells within clusters with signatures for non-ILCs, NK-cells or low quality cells (Grün et al., 2016) were removed from the dataset after initial clustering and RaceID3 was rerun. The single-cell RNA-seq data of uninfected adult lung comprised (after removal of a cluster of cells highly expressing natural killer (NK) cell markers such as Nkg7, Klrd1 and Klrc2) 1,043 ILC transcriptomes with a median count of 4,470 transcripts and a median of 1,780 different genes per cell. Neonatal lung ILC data encompassed 366 cells with a median of 7,047 transcripts and 2,546 different genes per cell. Il18r1+Icos+ BM cells consisted of 230 transcriptomes with a median transcript count of 6,372 transcripts and a median of 2,609 different genes per cell. Cultured lung and input cells (related to Figure 3G-I) comprised of 436 transcriptomes with a median of 5,035 transcripts and 1,908 different genes per cell. The Nb time-course dataset comprised 3200 ILC transcriptomes(excluding uninfected lung data) with median counts of 6,451 transcripts and 2,147 different genes after removal of NK cell and macrophage contaminants. The parabiosis dataset comprised 2,500 ILC transcriptomes, with a median count of 4,249 transcripts and 1,252 different genes after removal of NK cell and macrophage contaminants. In total, 1,422 cells were derived from host, and 1,078 cells from the enriched donor populations of cells. The shield chimera dataset comprised 2,281 ILC transcriptomes(995 host derived and 1,286 donor derived) with a median count of 5,729 transcripts and 2,047 different genes after removal of macrophage contaminations and low quality cells (Grün et al., 2016).
Cell-cycle Scoring
To evaluate the impact of proliferation signature on clustering we adapted Seurat’s cell cycle scoring. Genes passing filtering of the uninfected lung dataset were defined as universe. S phase and G2/M phase genes proposed by Tirosh and colleagues (Tirosh et al., 2016), which were elements of universe, were utilized. Genes of universe were binned into 24 bins based on the mean expression across cells. For every cell-cycle gene 100 genes were sampled from the respective bin and combined to a control gene set, containing only unique genes. Mean expression across all genes of the control gene set within each cell was subtracted from mean expression across combined S phase and G2/M phase gene sets within each cell. Cells were considered as cycling if this score was >0 and non-cycling if the score was <0. S phase and G2/M phase genes were combined as we detected correlation between genes of the two gene sets. To compare cell-cycle score with and without removal of cell-cycle variability in uninfected lung (see above), universe before removal of cell-cycle variability was used.
Differential expression analysis
Differentially expressed genes between two subgroups of cells were identified similar to a previously published method (Anders and Huber, 2010). First, negative binomial distributions reflecting the gene expression variability within each subgroup were inferred based on the background model for the expected transcript count variability computed by RaceID3. Using these distributions, a p-value for the observed difference in transcript counts between the two subgroups was calculated. Multiple testing correction was performed using the Benjamini-Hochberg method and referred to as “adjusted p-value” (Herman et al., 2018).
Lineage inference, pseudo-temporal ordering and SOM
For derivation of differentiation trajectories the StemID2 algorithm was used. (Grün et al., 2016; Herman et al.,2018). StemID2 was run with the following parameters: cthr=20 (for shield chimera cthr = 10), pdishuf=2000, pthr=0.01 and nmode=TRUE. Pseudo-temporal order was derived based on StemID2 projection coordinates of highest projection of cell to medoid links onto inter-cluster links of the subsequent cluster along the selected trajectories (uninfected lung: 5>11>9>8>4>10, 5>11>9>8>4>12 & time-course: 12>5>2>8>6>16, 12>5>2>8>4>23, 15>2>8>6>16). Self-organizing maps (SOMs) were used to infer models of pseudo-temporal expression profiles using the following parameters: nb=500, alpha=0.5, corthr=0.75, minsom=10.
KEGG/GO pathway enrichment
GO and KEGG enrichment analysis was performed using the clusterProfiler R package (Yu et al., 2012). Genes of SOM inferred gene modules along the respective trajectory were used as input and the ensemble of all genes, excluding ERCCs and mitochondrial genes, were used as universe. In addition the following parameters were used: pAdjustMethod = “BH”, pvalueCutoff = 0.01, qvalueCutoff = 0.05.
Fraction dot plot
For expression color code, log2 mean expression above for 4 or below -4 are replaced by 4 and -4, respectively. Z-scores above 1 and -1 are replaced by 1 and -1, respectively.
Normalisation of pie charts
For pie charts, sample composition of RaceID3 clusters was normalized by dividing the number of cells within a cluster of the respective sample by the total number of cells of the respective sample in the dataset.
Enrichment scores
Enrichment scores in RaceID3 clusters were obtained by dividing the relative frequency of donor or host cells within a cluster by the expected frequency, based on the frequency of donor and host cells in the total population. P-values of enrichments were calculated by a one-sided fisher-test using the R fisher.test function in R. For combined enrichment score and P-value calculation for shield chimera cluster 16 and 17, respective numbers of donor and host cells were aggregated
Dataset integration
Integration of Il18r1+Icos+ BM ILCs with ILCPs and ILC2Ps from Walker et al. (Walker et al., 2019) Initially, RaceID3 was run on the filtered raw counts for cells from cluster 1 and 2 of Walker et al., representing ILCPs and ILC2s, respectively. Cells with expression of Cd302, Sox13, Cd19, Spi1 and Gata1 were considered contaminants and removed prior to further analysis. For data set integration, the normalized counts of the remaining cells from cluster 1 and 2 of Walker et al. for genes shared with our Il18r1+Icos+ BM dataset were used as feature set. For this purpose, Ensembl gene IDs from Walker et al., were matched with their respective gene symbol. If two or more Ensembl IDs were matching the same symbol, their counts were aggregated per cell, if the differences of their aggregated counts across cells was >=1 or averaged per cell if their difference across cells was <1. Dataset integration was performed using Seurat’s standard workflow (Stuart et al. Cell 2019) with nfeatures=3000 as argument for the “FindVariableFeatures” function. Variable features passing RaceID3 filtering (minexpr=5, minnumber=1, CGenes initialization as described above with ccor=0.65) were used for anchoring and integration. The resulting integrated object was subjected to RaceID3 distance matrix calculation and clustering.
Integration of uninfected lung ILCs with lung and skin ILCs from Ricardo-Gonzalez et al.(Ricardo-Gonzalez et al., 2018):
Before integration, cells of our uninfected lung dataset from Figure 1 (after removal of NK cells) and lung and skin ILCs from Ricardo-Gonzalez et al., with less than 2000 transcripts or less than 200 different genes were discarded. Genes which were unique to any of the two datasets were removed and Ensembl gene ID counts matching the same gene symbol were treated as described above. Uninfected lung cells were assigned to batch1, whereas lung and skin cells from Ricardo-Gonzalez et al. were assigned to batch2. Dataset integration was performed using the SCTransform function from the Seurat package with selection of 3000 integration features. Integration features were filtered by removal of CGenes and correlated genes as described before. In addition, cell-cycle genes from Buettner et al.(Buettner et al., 2015) and correlating genes (correlation >= 0.4 across uninfected lung cells) were removed from integration feature set, which was subsequently subjected to anchoring and integration. Clustering was performed using Seurat, using 30 principle components and the resolution factor 1.2 for clustering.
Cluster mapping: Inferring transcriptome similarities between two scRNA-seq datasets based on cluster annotation
The unbiased identification of cells sharing a similar transcriptome in different tissues can be formulated as a classification problem: given a set of classifiers, i.e. cluster medoids in one dataset (e.g., parabiosis data with 22 clusters), we would like to identify non-negative weights reflecting similarities for each single cell in a given query (e.g., a single cell in the time-course data) under the constraint that the weights sum up to one. Hence, if a cell in the query set does not resemble a distinct cluster in the classifier set, it will have uniform weights for all clusters in the classifier set. On the other hand, if a cell resembles a specific cluster, the corresponding weight will be close to one, and all other weights will be close to zero. This optimization problem can be solved by quadratic programming, which was implemented using the solve.QP function from the quadprog R package.
Accession Codes
scRNA-seq data generated in this study are deposited in Gene Expression Omnibus (GEO) under accession code GSE141330.
Supplementary Material
Highlights.
-
-
A single cell atlas of BM and lung ILCs of healthy, infected and parabiotic mice
-
-
Identification of tissue-associated ILC progenitors in neonatal and adult lung
-
-
Cells recruited from BM generate the entire spectrum of ILC2s in infected lungs
-
-
Local cues imprint the phenotypes of ILC2s differentiating in the adult lung
eTOC blurb.
To investigate how ILCs are locally maintained, Zeis et al. generated a single cell atlas of lung ILCs and tracked Il18r1+ progenitor and effector ILC2s. Their work identifies tissue-resident and circulating ILCPs, and highlights in situ differentiation and tissue adaptation as a mechanism of ILC renewal and phenotypic diversification.
Acknowledgments
We thank A. Diefenbach, R. Golub and W. Kastenmüller for critically reading the manuscript, and G. A. Häcker and T. Gogishvilli for their support. This work was supported by grants through the German Research Foundation (DFG) priority program SPP1937 – Innate lymphoid cells to G.G. (GA2129/2-1), D.G. (GR4980/1-1), C.R. (RO3565/4-1) and D.V. (VO944/8-2), through the European Research Council to M.B. (647257-STROMA), G.G. (759176-TissueLymphoContexts) and D.G. (818846-ImmuNiche). G.G. was further supported by the DFG Emmy Noether programme (GA2129/2-1). D.G. was further supported by the DFG (GR4980/3-1, GRK2344 MeInBio), by the DFG under Germany’s Excellence Strategy (CIBSS – EXC-2189 – Project 390939984), and by the Behrens-Weise-Foundation. G.G. and D.G. are supported by the Max Planck Society.
Footnotes
Author Contributions Conceptualization, funding acquisition and project coordination, D.G. and G.G.; scRNA-seq experiments, computational analysis and web interface, P.Z.; FACS analysis and transfers, M.L.; in vitro culture experiments, M.L., P.Z., J.S.H..; parabiosis, X.F., S.E.; shield chimera, R.G.; human ILCs, D.C.H.; experimental support, K.K., N.P., C.F., R.D.L., A.M.K.; Il5 reporter mouse, R.M.L.; Nb culture, C.S.; experimental design, M.L., P.Z., D.G., G.G.; supervision, A.R., D.V., M.B., C.R., D.G., G.G.; paper writing and visualizations, P.Z., M.L., D.G., G.G.; review and editing, all authors.
Declaration of Interests The authors declare no conflict of interests.
References
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11 doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KG, Mayer-Barber K, Sung H, Beura L, James BR, Taylor JJ, Qunaj L, Griffith TS, Vezys V, Barber DL, et al. Intravascular staining for discrimination of vascular and tissue leukocytes. Nat Protoc. 2014;9:209–222. doi: 10.1038/nprot.2014.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SC, Bauer SR, Beyer RP, Brenton JD, Bromley B, Burrill J, Causton H, Conley MP, Elespuru R, Fero M, et al. The external RNA controls consortium: A progress report. Nat Methods. 2005;2:731–734. doi: 10.1038/nmeth1005-731. [DOI] [PubMed] [Google Scholar]
- Bando JK, Liang H-E, Locksley RM. Identification and distribution of developing innate lymphoid cells in the fetal mouse intestine. Nat Immunol. 2014;16:153–160. doi: 10.1038/ni.3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baratin M, Simon L, Jorquera A, Ghigo C, Dembele D, Nowak J, Gentek R, Wienert S, Klauschen F, Malissen B, et al. T Cell Zone Resident Macrophages Silently Dispose of Apoptotic Cells in the Lymph Node. Immunity. 2017;47:349–362.:e5. doi: 10.1016/j.immuni.2017.07.019. [DOI] [PubMed] [Google Scholar]
- Barletta KE, Cagnina RE, Wallace KL, Ramos SI, Mehrad B, Linden J. Leukocyte compartments in the mouse lung: Distinguishing between marginated, interstitial, and alveolar cells in response to injury. J Immunol Methods. 2012;375:100–110. doi: 10.1016/j.jim.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielecki P, Riesenfeld SJ, Kowalczyk MS, Regev A, Flavell RA. Skin inflammation driven by differentiation of quiescent tissue-resident ILCs into a spectrum of pathogenic effectors. BioRxiv. 2018:1–43. [Google Scholar]
- Björklund ÅK, Forkel M, Picelli S, Konya V, Theorell J, Friberg D, Sandberg R, Mjösberg J. resource The heterogeneity of human CD127 + innate lymphoid cells revealed by single-cell RNA sequencing. 2016;17:451–456. doi: 10.1038/ni.3368. [DOI] [PubMed] [Google Scholar]
- Buettner F, Natarajan KN, Casale FP, Proserpio V, Scialdone A, Theis FJ, Teichmann SA, Marioni JC, Stegle O. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechnol. 2015;33:155–160. doi: 10.1038/nbt.3102. [DOI] [PubMed] [Google Scholar]
- Califano D, Cho JJ, Califano D, Cho JJ, Uddin MN, Lorentsen KJ, Yang Q, Bhandoola A. Transcription Factor Bcl11b Controls Identity and Function of Mature Type 2 Innate Lymphoid Cells. Immunity. 2015;43:354–368. doi: 10.1016/j.immuni.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature. 2014;508:397–401. doi: 10.1038/nature13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuff AO, Robertson FP, Stegmann KA, Pallett LJ, Maini MK, Davidson BR, Male V. Eomes hi NK Cells in Human Liver Are Long-Lived and Do Not Recirculate but Can Be Replenished from the Circulation. J Immunol. 2016;197:4283–4291. doi: 10.4049/jimmunol.1601424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger G, Fan X, Dikiy S, Lee SY, Rudensky AY. Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science (80-) 2015;350:981–985. doi: 10.1126/science.aac9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentek R, Hoeffel G, Chen J, Hoeffel G, Bulle MJ, Msallam R, Gautier G. Hemogenic Endothelial Fate Mapping Reveals Dual Developmental Origin of Mast Cells. Immunity. 2018:1160–1171. doi: 10.1016/j.immuni.2018.04.025. [DOI] [PubMed] [Google Scholar]
- Ghaedi M, Shen ZY, Orangi M, Martinez-Gonzalez I, Wei L, Lu X, Das A, Heravi-Moussavi A, Marra MA, Bhandoola A, et al. Single-cell analysis of RORα tracer mouse lung reveals ILC progenitors and effector ILC2 subsets. J Exp Med. 2020;217 doi: 10.1084/jem.20182293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghigo C, Mondor I, Jorquera A, Nowak NJ, Wienert S, Zahner SP, Clausen BE, Luche H, Malissen B, Klauschen F, et al. Multicolor fate mapping of langerhans cell homeostasis. J Exp Med. 2013;210:1657–1664. doi: 10.1084/jem.20130403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Guilliams M. Tissue-Resident Macrophage Ontogeny and Homeostasis. Immunity. 2016;44:439–449. doi: 10.1016/j.immuni.2016.02.024. [DOI] [PubMed] [Google Scholar]
- Grün D, Kester L, van Oudenaarden A. Validation of noise models for single-cell transcriptomics. Nat Methods. 2014;11:637–640. doi: 10.1038/nmeth.2930. [DOI] [PubMed] [Google Scholar]
- Grün D, Muraro MJ, Boisset J-C, Wiebrands K, Lyubimova A, Dharmadhikari G, van den Born M, van Es J, Jansen E, Clevers H, et al. De Novo Prediction of Stem Cell Identity using Single-Cell Transcriptome Data. Cell Stem Cell. 2016;19:266–277. doi: 10.1016/j.stem.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harly C, Kenney D, Ren G, Lai B, Raabe T, Yang Q, Cam MC, Xue H-H, Zhao K, Bhandoola A. The transcription factor TCF-1 enforces commitment to the innate lymphoid cell lineage. Nat Immunol. 2019;20:1150–1160. doi: 10.1038/s41590-019-0445-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimshony T, Senderovich N, Avital G, Klochendler A, de Leeuw Y, Anavy L, Gennert D, Li S, Livak KJ, Rozenblatt-Rosen O, et al. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 2016;17:77. doi: 10.1186/s13059-016-0938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JS, Sagar, Grün D. FateID infers cell fate bias in multipotent progenitors from single-cell RNA-seq data. Nat Methods. 2018;15:379–386. doi: 10.1038/nmeth.4662. [DOI] [PubMed] [Google Scholar]
- Huang Y, Mao K, Chen X, Sun M, Kawabe T, Li W, Usher N, Zhu J, Jr JFU, Paul WE, et al. S1P-dependent interorgan trafficking of group 2 innate lymphoid cells supports host defense. Science (80-) 2018;119:114–119. doi: 10.1126/science.aam5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka IE, Chea S, Gudjonson H, Constantinides MG, Dinner AR, Bendelac A, Golub R. Single-cell analysis defines the divergence between the innate lymphoid cell lineage and lymphoid tissueinducer cell lineage. Nat Immunol. 2016;17:269–276. doi: 10.1038/ni.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose CSN, Flach M, Möhle L, Rogell L, Hoyler T, Ebert K, Fabiunke C, Pfeifer D, Sexl V, Fonseca-Pereira D, et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. 2014;157:340–356. doi: 10.1016/j.cell.2014.03.030. [DOI] [PubMed] [Google Scholar]
- Kløverpris HN, Kazer SW, Mjösberg J, Mabuka JM, Wellmann A, Ndhlovu Z, Yadon MC, Nhamoyebonde S, Muenchhoff M, Simoni Y, et al. Innate Lymphoid Cells Are Depleted Irreversibly during Acute HIV-Infection in the Absence of Viral Suppression. Immunity. 2016;44:391–405. doi: 10.1016/j.immuni.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Voisin B, Kim DY, Kennedy EA, Jo JH, Shih HY, Truong A, Doebel T, Sakamoto K, Cui CY, et al. Homeostatic Control of Sebaceous Glands by Innate Lymphoid Cells Regulates Commensal Bacteria Equilibrium. Cell. 2019;176:982–997.:e16. doi: 10.1016/j.cell.2018.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrançais E, Ortiz-Muñoz G, Caudrillier A, Mallavia B, Liu F, Sayah DM, Thornton EE, Headley MB, David T, Coughlin SR, et al. The lung is a site of platelet biogenesis and a reservoir for haematopoietic progenitors. Nature. 2017;544:105–109. doi: 10.1038/nature21706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim AI, Li Y, Lopez-Lastra S, Stadhouders R, Paul F, Casrouge A, Serafini N, Puel A, Bustamante J, Surace L, et al. Systemic Human ILC Precursors Provide a Substrate for Tissue ILC Differentiation. Cell. 2017;168:1086–1100.:e10. doi: 10.1016/j.cell.2017.02.021. [DOI] [PubMed] [Google Scholar]
- Mass E, Ballesteros I, Farlik M, Halbritter F, Günther P, Crozet L, Jacome-Galarza CE, Händler K, Klughammer J, Kobayashi Y, et al. Specification of tissue-resident macrophages during organogenesis. Science (80-) 2016;353 doi: 10.1126/science.aaf4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzurana L, Rao A, Van Acker A, Mjösberg J. The roles for innate lymphoid cells in the human immune system. Semin Immunopathol. 2018;40:407–419. doi: 10.1007/s00281-018-0688-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro K, Kabata H, Tanabe M, Koga S, Takeno N, Mochizuki M, Fukunaga K, Asano K, Betsuyaku T, Koyasu S. Interferon and IL-27 antagonize the function of group 2 innate lymphoid cells and type 2 innate immune responses. Nat Immunol. 2016;17:76–86. doi: 10.1038/ni.3309. [DOI] [PubMed] [Google Scholar]
- Motran CC, Cervi L. Helminth infections: Recognition and Modulation of the immune Response by innate immune Cells. 2018;9:1–12. doi: 10.3389/fimmu.2018.00664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa M, Heesters BA, Kradolfer CMA, Krabbendam L, Martinez-Gonzalez I, De Bruijn MJW, Golebski K, Hendriks RW, Stadhouders R, Spits H, et al. KLRG1 and NKp46 discriminate subpopulations of human CD117+CRTH2-ILCs biased toward ILC2 or ILC3. J Exp Med. 2019;216:1762–1776. doi: 10.1084/jem.20190490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima H, Mahlakõiv T, Shih H-Y, Davis FP, Meylan F, Huang Y, Harrison OJ, Yao C, Mikami Y, Urban JF, et al. Neuropeptide CGRP Limits Group 2 Innate Lymphoid Cell Responses and Constrains Type 2 Inflammation. Immunity. 2019;51:682–695.:e6. doi: 10.1016/j.immuni.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SS, Souza-Fonseca-Guimaraes F, Rivera FDL, Amante FH, Kumar R, Gao Y, Sheel M, Beattie L, Montes De Oca M, Guillerey C, et al. Rapid loss of group 1 innate lymphoid cells during blood stage Plasmodium infection. Clin Transl Immunol. 2018;7:1–14. doi: 10.1002/cti2.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oherle K, Acker E, Bonfield M, Wang T, Gray J, Lang I, Bridges J, Lewkowich I, Xu Y, Ahlfeld S, et al. Insulin-like Growth Factor 1 Supports a Pulmonary Niche that Promotes Type 3 Innate Lymphoid Cell Development in Newborn Lungs. Immunity. 2020;52:275–294.:e9. doi: 10.1016/j.immuni.2020.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant CJ, Hwang YY, Walker JA, Salimi M, Wong SH, Brewer JM, Englezakis A, Barlow JL, Hams E, Scanlon ST, et al. Article MHCII-Mediated Dialog between Group 2 Innate Lymphoid Cells and CD4 + T Cells Potentiates Type 2 Immunity and Promotes Parasitic Helminth Expulsion. Immunity. 2014;41:283–295. doi: 10.1016/j.immuni.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Jiang X, Chen Y, Sojka DK, Wei H, Gao X, Sun R, Yokoyama WM, Tian Z. Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. J Clin Invest. 2013;123:1444–1456. doi: 10.1172/JCI66381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu D-M, Botting RA, Stephenson E, Green K, Webb S, Jardine L, Calderbank EF, Polanski K, Goh I, Efremova M, et al. Decoding human fetal liver haematopoiesis. Nature. 2019:1–7. doi: 10.1038/s41586-019-1652-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricardo-Gonzalez RR, Van Dyken SJ, Schneider C, Lee J, Nussbaum JC, Liang HE, Vaka D, Eckalbar WL, Molofsky AB, Erle DJ, et al. Tissue signals imprint ILC2 identity with anticipatory function. Nat Immunol. 2018;19:1093–1099. doi: 10.1038/s41590-018-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricardo-Gonzalez RR, Schneider C, Liao C, Lee J, Liang HE, Locksley RM. Tissuespecific pathways extrude activated ILC2s to disseminate type 2 immunity. J Exp Med. 2020;217 doi: 10.1084/jem.20191172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinette ML, Fuchs A, Cortez VS, Lee JS, Wang Y, Durum SK, Gilfillan S, Colonna M, Consortium G. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat Immunolgy. 2015;16:306–318. doi: 10.1038/ni.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saluzzo S, Gorki A-D, Rana BMJ, Martins R, Scanlon S, Starkl P, Lakovits K, Hladik A, Korosec A, Sharif O, et al. First-Breath-Induced Type 2 Pathways Shape the Lung Immune Environment. Cell Rep. 2017;18:1893–1905. doi: 10.1016/j.celrep.2017.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Lee J, Koga S, Ricardo-Gonzalez RR, Nussbaum JC, Smith LK, Villeda SA, Liang H-E, Locksley RM. Tissue-Resident Group 2 Innate Lymphoid Cells Differentiate by Layered Ontogeny and In Situ Perinatal Priming. Immunity. 2019;50:1425–1438.:e5. doi: 10.1016/j.immuni.2019.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott CL, Zheng F, De Baetselier P, Martens L, Saeys Y, De Prijck S, Lippens S, Abels C, Schoonooghe S, Raes G, et al. Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat Commun. 2016;7 doi: 10.1038/ncomms10321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehus CR, Aliahmad P, Torre B, De Iliev ID, Spurka L, Funari VA, Kaye J. Articles The development of innate lymphoid cells requires TOX-dependent generation of a common innate lymphoid cell progenitor. Nat Immunol. 2015;16 doi: 10.1038/ni.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seillet C, Mielke LA, Amann-Zalcenstein DB, Su S, Gao J, Almeida FF, Shi W, Ritchie ME, Naik SH, Huntington ND, et al. Deciphering the Innate Lymphoid Cell Transcriptional Program. Cell Rep. 2016;17:436–447. doi: 10.1016/j.celrep.2016.09.025. [DOI] [PubMed] [Google Scholar]
- Silberstein L, Goncalves KA, Kharchenko PV, Turcotte R, Kfoury Y, Mercier F, Baryawno N, Severe N, Bachand J, Spencer JA, et al. Proximity-Based Differential Single-Cell Analysis of the Niche to Identify Stem/Progenitor Cell Regulators. Cell Stem Cell. 2016;19:530–543. doi: 10.1016/j.stem.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stier MT, Zhang J, Goleniewska K, Cephus JY, Rusznak M, Wu L, Van Kaer L, Zhou B, Newcomb DC, Peebles RS. IL-33 promotes the egress of group 2 innate lymphoid cells from the bone marrow. J Exp Med. 2018;215:263–281. doi: 10.1084/jem.20170449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symowski C, Voehringer D. Th2 cell-derived IL-4/IL-13 promote ILC2 accumulation in the lung by ILC2-intrinsic STAT6 signaling in mice. Eur J Immunol. 2019;49:1421–1432. doi: 10.1002/eji.201948161. [DOI] [PubMed] [Google Scholar]
- Tirosh I, Izar B, Prakadan SM, Wadsworth MH, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science (80-) 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JA, Clark PA, Crisp A, Barlow JL, Szeto A, Ferreira ACF, Rana BMJ, Jolin HE, Rodriguez-Rodriguez N, Sivasubramaniam M, et al. Polychromic Reporter Mice Reveal Unappreciated Innate Lymphoid Cell Progenitor Heterogeneity and Elusive ILC3 Progenitors in Bone Marrow. Immunity. 2019;51:104–118.:e7. doi: 10.1016/j.immuni.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrapp A, Riesenfeld SJ, Burkett PR, Abdulnour RE, Nyman J, Dionne D, Hofree M, Cuoco MS, Rodman C, Farouq D, et al. The neuropeptide NMU amplifies ILC2-driven allergic lung inflammation. Nat Publ Gr. 2017 doi: 10.1038/nature24029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner J, Zuber J, Shonts B, Yang S, Fu J, Martinez M, Farber DL, Kato T, Sykes M. Long-term Persistence of Innate Lymphoid Cells in the Gut after Intestinal Transplantation. Transplantation. 2017;101:2449–2454. doi: 10.1097/TP.0000000000001593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright DE, Wagers AJ, Pathak Gulati A, Johnson FL, Weissman IL. Physiological migration of hematopoietic stem and progenitor cells. Science (80-) 2001;294:1933–1936. doi: 10.1126/science.1064081. [DOI] [PubMed] [Google Scholar]
- Xu W, Cherrier DE, Chea S, Vosshenrich C, Serafini N, Petit M, Liu P, Golub R, Di Santo JP. An Id2RFP-Reporter Mouse Redefines Innate Lymphoid Cell Precursor Potentials. Immunity. 2019;50:1054–1068.:e3. doi: 10.1016/j.immuni.2019.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yipp BG, Kim JH, Lima R, Zbytnuik LD, Petri B, Swanlund N, Ho M, Szeto VG, Tak T, Koenderman L, et al. The lung is a host defense niche for immediate neutrophil-mediated vascular protection. Sci Immunol. 2017;2 doi: 10.1126/sciimmunol.aam8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, He QY. ClusterProfiler: An R package for comparing biological themes among gene clusters. Omi A J Integr Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Wang C, Clare S, Wang J, Lee S, Brandt C, Burke S, Lu L, He D, Jenkins NA, et al. The transcription factor Bcl11b is specifically expressed in group 2 innate lymphoid cells and is essential for their development. J Exp Med. 2015;212:865–874. doi: 10.1084/jem.20142318. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.