Abstract
The right timing of animal physiology and behavior ensures the stability of populations and ecosystems. In order to predict anthropogenic impacts on these timings, more insight is needed into the interplay between environment and molecular timing mechanisms. This is particularly true in marine environments.
Using high-resolution, long-term daylight measurements from a habitat of the marine annelid Platynereis dumerilii, we find that temporal changes in UVA/deep violet intensities, more than longer wavelengths, can provide annual time information, which differs from annual changes in photoperiod. We developed experimental setups that resemble natural daylight illumination conditions, and automated, quantifiable behavioral tracking. Experimental reduction of UVA/deep violet light (app. 370-430nm) under long photoperiod (LD16:8) significantly decreases locomotor activities, comparable to the decrease caused by short photoperiod (8:16). In contrast, altering UVA/deep violet light intensities does not cause differences in locomotor levels under short photoperiod. This modulation of locomotion by UVA/deep violet light under long photoperiod requires c-opsin1, an UVA/deep violet-sensor employing Gi-signalling. C-opsin1 also regulates the levels of rate-limiting enzymes for monogenic amine synthesis and of several neurohormones, including PDF, Vasotocin (Vasopressin/Oxytocin) and NPY-1.
Our analyses indicate a complex inteplay between UVA intensities and photoperiod as indicators of annual time.
Since its beginning in the marine environment, life has been exposed to the rhythmic changes generated by the impact of the sun and the moon. Organisms have adapted to these changes and today, many marine organisms precisely time their physiology and behavior to specific months of the year, specific days during the month and specific hours during those days. This timing is not just impressive, like illustrated in the precisely timed mass spawning events of species ranging from corals to worms to fishes 1–3, but crucial for successful reproductive synchronization 4 and a major ecological aspect of food web architecture in an environment that covers more than 71% of the earth’s surface 5–7. It is also of relevance for freshwater and terrestrial ecology 8. The importance of timing for food webs has been especially well documented for seasonal interactions 5–7. Evidence accumulates that the on-going climate change desynchronizes intra- and interspecies interactions and thereby endangers ecosystem stability 4,9. A prominent example are seasonal phytoplankton blooms, which tend to occur earlier, while the behavioral and physiological rhythms of higher trophic levels change less. This results in timing mismatches in food webs with possibly severe consequences for species reproduction 10–13.
In order to understand the differential effects that anthropogenic impacts can have on the timing of different species 9, we need knowledge about key environmental parameters, and the underlying molecular mechanisms interpreting these environmental cues and driving the timing mechanisms in animals 13. Significant mechanistic insight exists for understanding daily and seasonal timing in the terrestrial animal model species, especially Drosophila and mouse, but also for birds, hamsters and sheep (e.g. reviewed in 14–19). However, the molecular and cellular foundations of the mechanisms underlying timed processes in the marine environment, despite their ecological importance, are still little understood. This has multiple reasons. First, marine environments are highly complex and many geophysical factors, such as light or temperature, do not only differ with latitude and time, but also depend on depth, currents and local water properties, like absorption coefficients, stratification and changes thereof. Second, the existing experimental analyses are predominantly descriptive, mainly because of the lack of marine animal models exhibiting diverse rhythms in the lab and being amenable to functional investigations.
Platynereis dumerilii is a marine bristle worm, whose physiology and behavior are governed by an interplay of multiple rhythms at its natural habitat, as well as in the lab 20–23. This likely cosmopolitan annelid worm 24 can serve as a representative for many marine invertebrates 24, while also being amenable to the functional manipulations and experimental analyses methods 25 required to unravel the mechanisms underlying marine timing.
Here we combine high resolution light measurements from the natural habitat of Platynereis at two different depths (10m and 4m) and newly established illumination systems and automated behavioral tracking for the laboratory with genetic functional manipulation, peptidomics, proteomics and transcript quantifications, as well as photoreceptor signaling analyses to dissect the impact of seasonal light changes on animals. We show that UVA/ deep violet light (app. 370-430nm) at environmentally relevant intensity impacts the hormonal and behavioral status of the marine bristle worm via the UVA-sensitive photoreceptor c-opsin1. Thus, differences in UVA/deep violet light intensities might provide information on annual time to marine animals in addition to photoperiod. While our data provide significant new insight, it is clear that no laboratory setting will mimic the complexity of natural environments. It remains a challenge for the future to unravel how UVA/deep violet light signals via c-opsin1 integrate with the full complexity of other natural signals and their changes over time. This includes the intricacy of the natural light spectrum, but also variations in temperature, currents, oxygen content, symbiotic interactions and predation pressure. Yet, in order to understand (and predict) the responses to complex mixtures of environmental cues, understanding the mechanisms underlying individual responses are essential.
Results
UVA/deep violet light changes between seasons in the natural Platynereis habitat
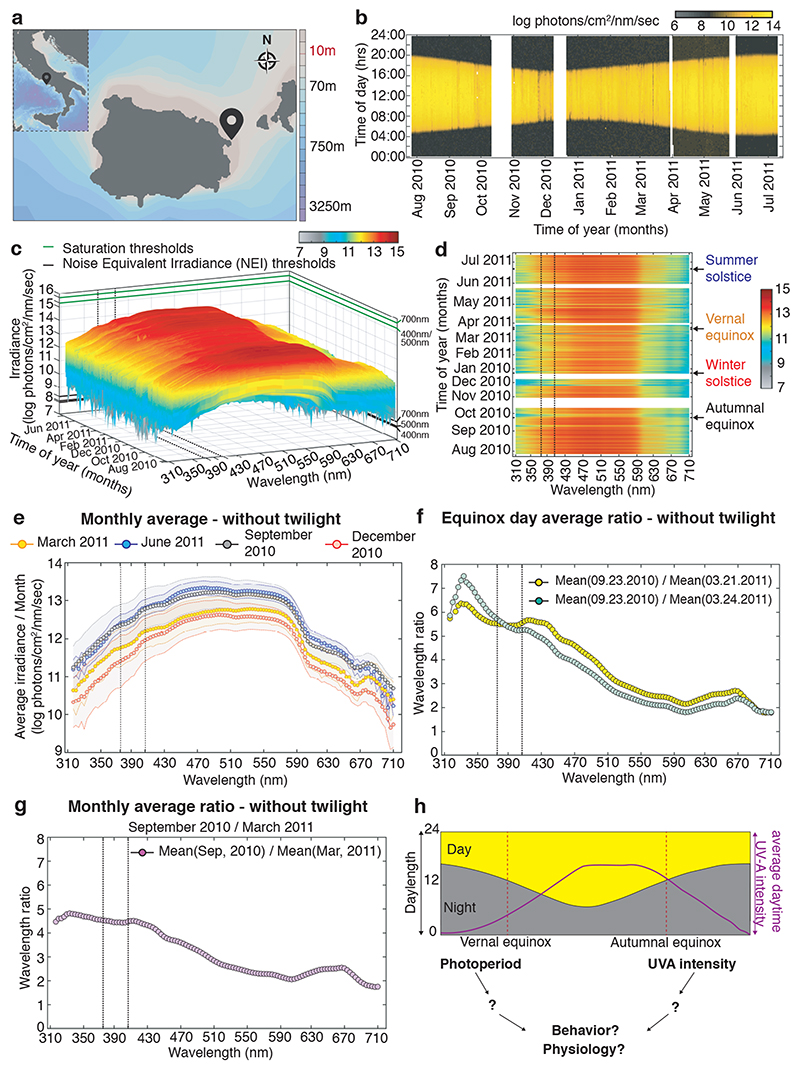
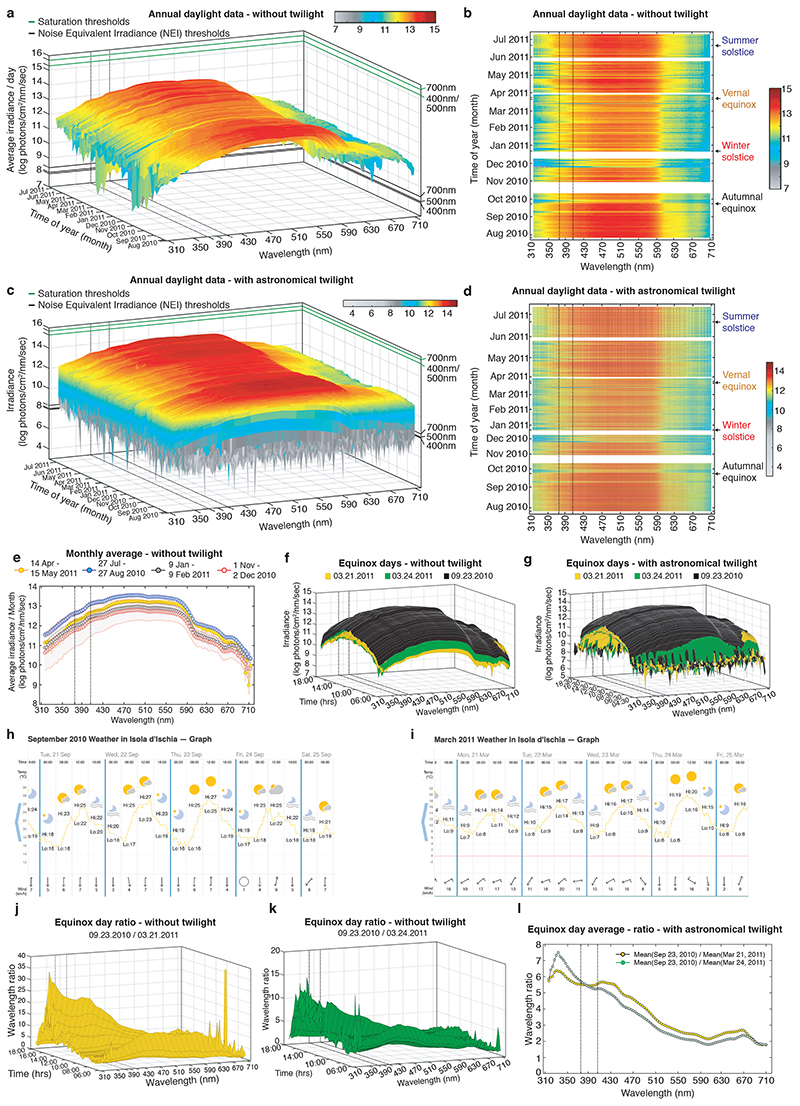
In order to obtain a better understanding how light can impact on the physiology and behavior of marine animals within their complex light environment displaying various rhythms of different period lengths, we performed detailed light measurements. We selected a marine habitat that is inhabited by a marine bristle worm, Platynereis dumerilii, which can also be functionally studied in the laboratory. Platynereis has a particularly well documented natural occurrence in Mediterranean and Atlantic coastal habitats across a range of depths from 1m-5m, but also at 10m and below 26–28. We deposited two highly sensitive Ramses hyperspectral radiometers, calibrated by TriOS GmbH prior to their usage following NIST standards over the entire measurement range, near Ischia island, Italy (Fig.1a), the geographic origin of our laboratory culture. The presence of Platynereis spec. at the measurement sites (10m and 4m water depth) was confirmed by diving. We measured and analyzed the light spectrum from 310nm-710nm in 3nm, 15min intervals over almost an entire year between July 27, 2010 – July 10, 2011 at 10m depth and for overlapping time from May, 31st 2011 - July, 10th 2011 at 4m. For technical reasons we could not start the measurements at 4m earlier and continue longer at the same location. We compared several examples of our daylight dataset against measured or calculated underwater spectra previously reported in the literature (Extended Data Fig.1). This benchmarking confirmed that our measurements were within the expected spectral intensity distributions. We also verified that all daytime measurements were distant to the saturation and noise equivalent intensities (NEI) (Fig.1c, Suppl.Data 1, Extended Data Fig.1c,f,i,l). We next analyzed the longer dataset (10m) for light intensity across the spectrum and time (Fig.1b-d, Extended Data Fig. 2a-d, Supplementary Data 1-3). When analyzing photoperiod (Fig.1b) vs. irradiance levels (Fig.1c,d; Extended Data Fig. 2a-d), we noticed that these two read-outs exhibit different dynamics across the year. While day length changes symmetrically relative to the longest day of the year (Fig.1b, https://www.timeanddate.com/sun/italy/naples), the daylight spectral intensities have a shifted distribution (Fig.1c-e: compare December/January and March vs. September and June/July, Supplementary Data 1-3; Extended Data Fig.2a-d). This shifted distribution relative to photoperiod can be well demonstrated for individual days. We selected the spring vs. fall equinox days as an example (Extended Data Fig. 2f,g). We build the ratios between the same wavelengths of the September equinox day (September, 23rd, Extended Data Fig.2f,g,j,k) and two different days of March, March 21st (the exact spring equinox day, Fig.1f: yellow line, Extended Data Fig.2f,g,j) and March 24th (daytime 8mins longer, Fig.1f: blue line, Extended Data Fig.2f,g,k, Supplementary Data 6-8). We analyzed September/March ratios from two different March days to test for the effects of few clouds (for corresponding weather data see Extended Data Fig.2h,i), which overall caused only minor changes. The least difference between the different cloud conditions was present for ratios between 370-420nm. Intensity differences between the September and March equinoxes were observable across the day (albeit slightly stronger during the afternoon, Extended Data Fig.2f,g,j,k, Supplementary Data 6-8) and independent of twilight (compare Fig. 1f and Extended Data Fig.2l, Extended Data Figs. 2f,g).
Figure 1. Light intensity ratios provide seasonal information different to photoperiod.
(a) Location of RAMSES hyperspectral radiometers installed at the Platynereis natural habitat at 10m (40°43’50.6”N 13°58’02.9”E) and 4m (40°43’56”N 13°57’44”E) depths for light data collection: GPS symbol. (b) Photoperiod (at 10m) as determined by averaging all wavelengths per timepoint. White bars: times when sensor was out of the water for cleaning. (c) Daylight spectrum (10m, raw data) across the year, sunrise to sunset, without twilight times. For data plotted as average across day or including twilight: see Extended Data Fig.2a,c; for 3D-rotational graphs: Supp.Data 1-3. Green lines: saturation thresholds, black lines: noise equivalent intensity (NEI) thresholds for exemplary wavelengths. (d) 2-D pcolor plot of (c). For data plotted as average across day or including twilight: see Extended Data Fig.2b,d. (e) Daytime monthly irradiance averages from dataset without twilight for March 2011 (yellow), June 2011 (blue), September 2010 (black) and December 2010 (red). (f) Wavelength ratios of equinox days averaged across day. (g) Monthly wavelength average ratio of September/ March. For additional ratios (days, including twilight, different calculation of averages) see Extended Data Fig.2e-g,j-l, 3D-plots: Supp.Data 7-8. (h) Hypothesis of an integrative light detection model further tested in the following experiments.
Black dotted lines: range of strong c-opsin1 activation. Original measurements were corrected for daylight saving timeshifts. Maps created with Ocean Data View (v. 5.1.7).
The presence of shifted irradiance levels relative to photoperiod was not restricted to the equinox times, but more generally noticeable for time spans surrounding equinox, and between the equinoxes and summer solstice (Fig. 1g, Extended Data Fig.2e), but less or absent for the short photoperiods surrounding winter solstice (Extended Data Fig.2e). The differences were particularly noticeable in the UVA/ deep violet range (315-430nm, Fig. 1e-g, Extended Data Fig.2e,j-l). This allows for the possibility that the differences in UV/deep violet light intensity provide additional information about the time of year, especially discriminating times of equal photoperiod (Fig.1h). Interestingly, while the UVA/deep violet light intensity differs from photoperiod information, its timing is similar to the changes in water temperature across the year in the Bay of Naples, which is higher in fall than in spring 29.
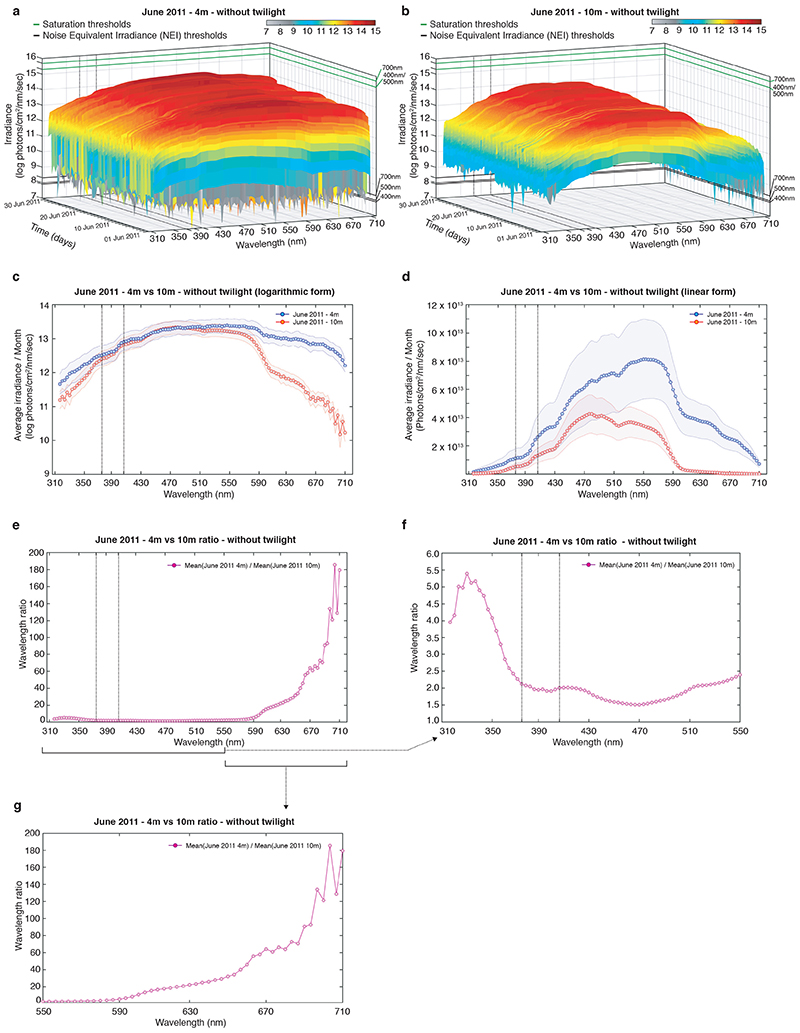
As spectral distribution also changes across water depth, we wondered how much of the UVA signal would be altered by the water column. We thus analyzed 4m vs. 10m data recorded at identical time periods and in spatial proximity (Fig.1a, Extended Data Fig.3a,b, Supplementary Data 4,5). These data show that wavelengths above 590nm diminish more strongly with depth than shorter wavelengths (including UVA) (Extended Data Fig. 3c-g), in line with other underwater measurements and calculations (Extended Data Fig.1). This suggests that seasonal changes in UVA intensity can occur across depth (at least in the natural habitat range of Platynereis dumerilii).
So far, the biggest focus for seasonal light reception has been on day length and it is clear that changes in day length have important instructive meanings for seasonal animals 30. However, it should be noted that any work carried out under standard laboratory white light does not typically possess wavelengths smaller than or equaling 400nm 31. Thus, effects of UVA/deep violet light on animals can be easily overlooked in laboratory settings.
Given the seasonal information potential and the fact that many animals from flies to humans possess light receptors that can respond to UVA/deep violet light 32–35, we next wondered if UVA light could contribute to seasonal responses in physiology and/or behavior in marine animals.
Differences in UVA light regulate locomotor behavior
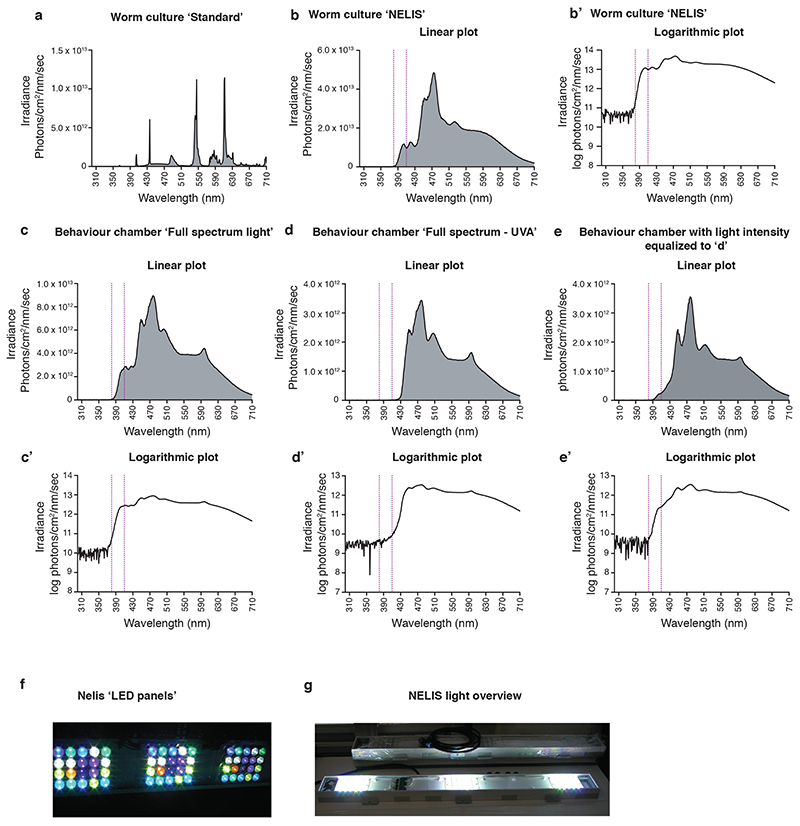
We next quested for evidence from the literature if Platynereis might displays seasonal changes in its physiology or behavior in its natural habitat. We then aimed to test if aspects of those changes could be mimicked by differences in UVA/deep violet light intensities. UVA is defined as wavelength from 315 to 400nm (ISO 21348 Definitions of Solar Irradiance Spectral Categories). However, the borders of the different subsections of the electromagnetic spectrum are historical and do not correspond to a fixed physical property 36,37. For experiments under lab conditions, we focused our attention on UVA/deep-violet light in the range of 370-430nm, but the “Nelis sunlight source” used emits light across the entire UVA spectrum (see below and Materials and Methods section for further details).
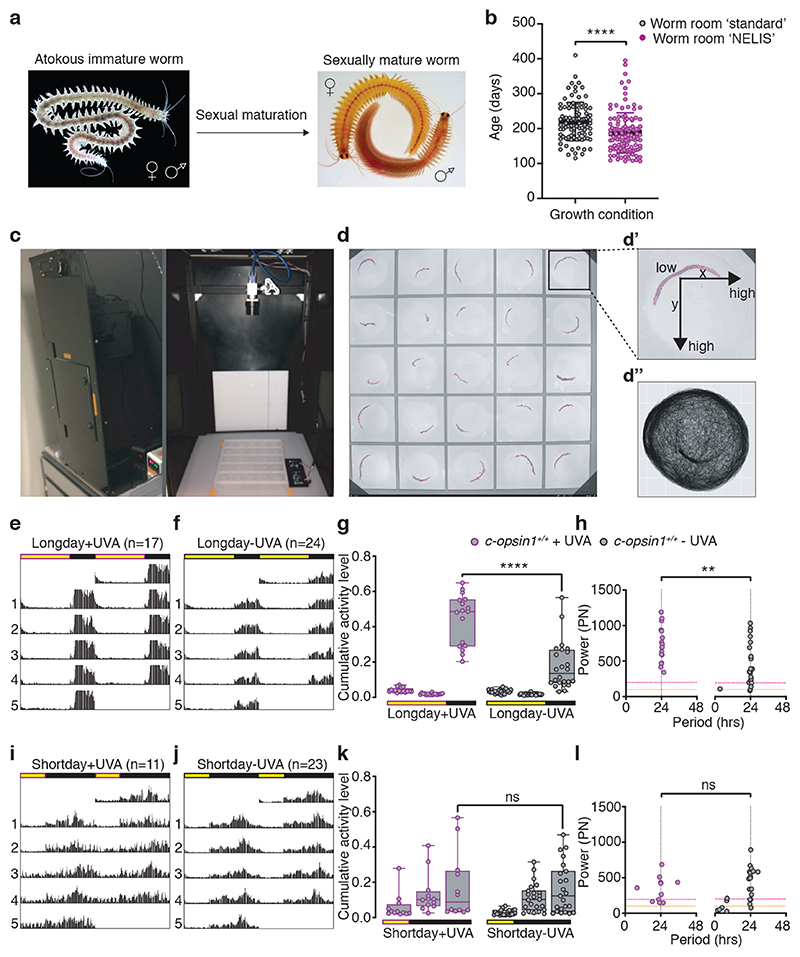
Detailed catch data of swarming mature worms, as well as recruitment analyses of Platynereis dumerilii in the Gulf of Naples suggest that the worms exhibit a seasonal regulation of their reproduction 1,20,21,38. We thus analyzed the timing of maturation between Platynereis subcultures grown under significant intensities versus minimal intensities of UVA light (Extended Data Fig.4a,b,b’). Wildtype worms exhibited a significantly faster maturation in conditions including UVA light compared to those with minimal UVA (Fig.2a,b).
Figure 2. UVA/deep violet light affects locomotor behavior in Platynereis dumerilii.
(a) sexual maturation in Platynereis dumerilii. (b) Platynereis wildtype worms grown under spectral conditions mimicking sunlight (“NELIS” culture, Extended Data Fig.4b,b’) mature significantly faster than sibling worms reared under wormroom ‘standard’-white light (Extended Data Fig.4a), Mann-Whitney U test. Worm density and feeding was as similar as possible for both conditions. Worms were scored mature when exhibiting the mature appearance and exhibited the characteristic “nuptial dance” behavior. (c) Platynereis behavioral recording chamber (51cm x 51cm x 101cm) with broad spectrum lighting system, IR-camera and IR-light. Real time light intensity and temperature were recorded during experiment by light/temperature detector positioned next to the behavioral grid. (d) Behavioral grid with worms tracked by ‘Motif’, an automated worm tracking software developed to detect individual experimental worms (magenta contour outline). (d’) The individual distance moved is calculated from the mid-point of the detected shape in relation to the xy-pixel coordinates in subsequent frames. (d”) Exemplary movement plot of an immature worm recorded and tracked for 10 days. (e,f) Double plotted average actograms of c-opsin1+/+ worms under long day including UVA (e: n=17) and long day filter-reduced UVA (f: n=24). (g) c-opsin1+/+ worms recorded in (f) show significantly decrease locomotor activity compared to (e) (One-way ANOVA, Sidak’s multiple comparison test) and (h) a significant decrease in power (PN) and rhythmicity (Lomb-Scargle periodogram test, Mann-Whitney-Wilcoxon test). Rhythmic strength thresholds were manually annotated: PN≥200 – rhythmic (above magenta dotted line), PN≤100 – arrhythmic (below orange dotted line) and PN>100<200 – weakly rhythmic. (i,j) Double plotted average actograms of c-opsin1+/+ worms under short day including UVA (i, n=11) and short day filter-reduced UVA (j, n=23). (k,l) c-opsin1+/+ worms recorded in (i,j) showed no difference in locomotor activity level (k) and rhythmicity (l). *p<0.05, ** p<0.01, *** p<0.001. Full statistical analyses: Supp.Fig. 7. Locomotor data of individual worms: Supp.Figs 2a,4a,5a,6a. Individual worm rhythm period and power were determined via Lomb-Scargle periodograms using ActogramJ.
However, while this result was interesting, we next thought of an alternative assay under better controlled light conditions that can also be faster analyzed than maturation timing. There were three main reasons for developing such an alternative assay. First, the lights in the worm culture rooms still included residual amounts of UVA light (Extended Data Fig.4a). Second, light intensity varied depending on wormbox position and was technically difficult to change evenly across the spectrum. Third, indirect effects, e.g. on algal growth (i.e. food) were difficult to exclude.
Changes in behavioral activity patterns across seasons have been documented for a variety of species. Strong examples of such changes are long distance migrations or diapause/hibernation 39–42, but are also present as subtler differences 43–46, including diel locomotor activity in the close nereidid relative Nereis virens 47,48. We thus investigated Platynereis locomotion over the day/night cycle under different UVA light conditions. To enable this, we further advanced and improved our ‘worm behavioral box’ (Fig.2c, 22). We installed an illumination that closely resembled the underwater sunlight spectrum, including UVA, (Extended Data Fig.4c-e’,f,g) and also developed a set-up and computer program that allowed fully automated underwater worm tracking (Fig.2d-d”). Using this set-up in combination with a spectral filter that specifically reduced light below 430nm (Extended Data Fig.4c-e’), allowed us to directly assess the effects of UVA/deep violet light and different photoperiods on the worms’ locomotor behavior.
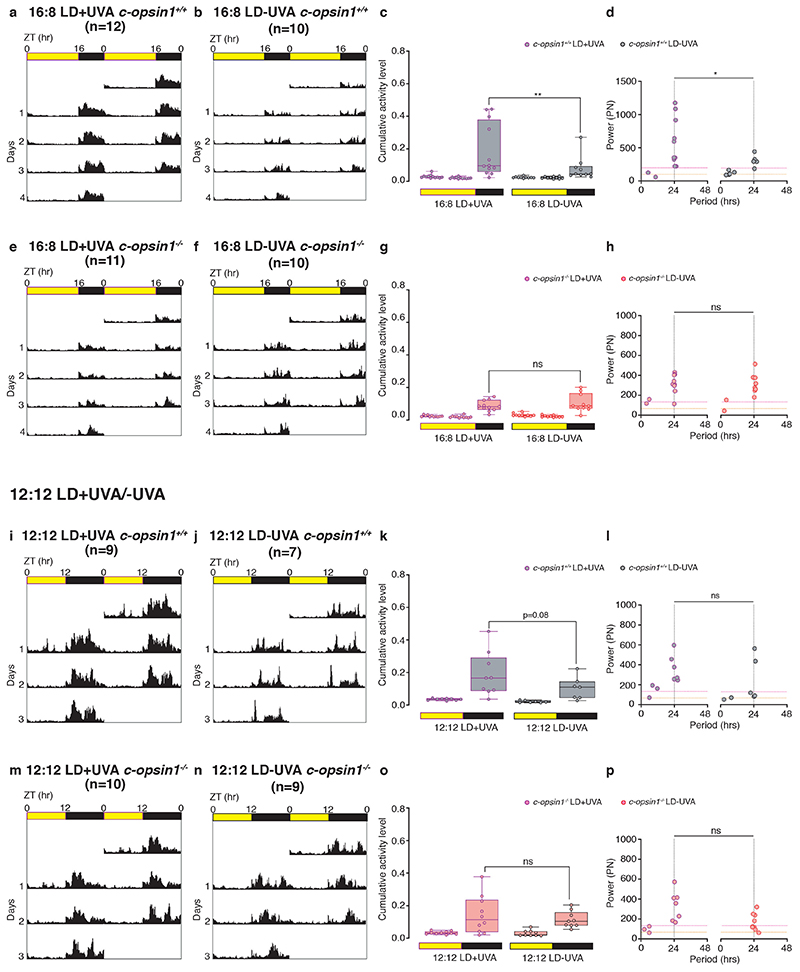
The presence of intense UVA/deep violet light (+UVA condition) led to significant changes in the locomotor behavior of the worms. Animals exposed to white light including UVA/deep violet under long day (LD 16h:8h) moved significantly more than worms under the same photoperiod (LD 16h:8h) exposed to white light with filter-reduced UVA/deep violet light (– UVA condition, Fig.2e-g, Supp.Figs.1a,b,2a, 3a, 4a, 7). While period length remained similarly stable under both conditions, the strength of the rhythmic activity, i.e. its power in the Lomb-Scargle analysis, was significantly lower under filter-reduced UVA/deep violet light (Fig.2h, Suppl.Tables S5,6,9). We also compared these data to locomotor behavior under short day (LD 8h:16h) under the same white light spectrum with intense and filter-reduced intensities of UVA/deep violet light (Supp.Figs 1c,d). Locomotor distance of wildtype worms decreased under short day, white light, including intense UVA/deep violet light conditions (+UVA) to levels observed under long day with filter-reduced UVA/ deep violet light (–UVA, Fig.2i-k, compare to 2e-g). Reduction of UVA/ deep violet light (–UVA) by filter under short day conditions had no additional effects on locomotor activity (Fig.2i-k, SuppFigs.5a,6a, full statistics SuppFig.7), suggesting that the seasonal UVA/ deep violet light intensity differences are most relevant outside the short day times, consistent with the natural light situation (Extended Data Figure 2e). Daily period length of individual worms was generally more variable under short day than long day conditions (Fig.2h,l; Suppl.Tables S5-9).
It should be noted that Platynereis worms are nocturnal, i.e. the majority of their locomotion occurs during the night. This indicates that the differences in locomotor behavior in the absence versus presence of UVA/ deep violet light (under long day conditions) are not an immediate response to the light itself, which only occurs during the daytime, but likely rather manifest themselves in molecular and/or cellular changes that modify the worms’ behavior over time.
Platynereis c-opsin1 is a UVA/ deep violet light sensor signaling via Gi/o
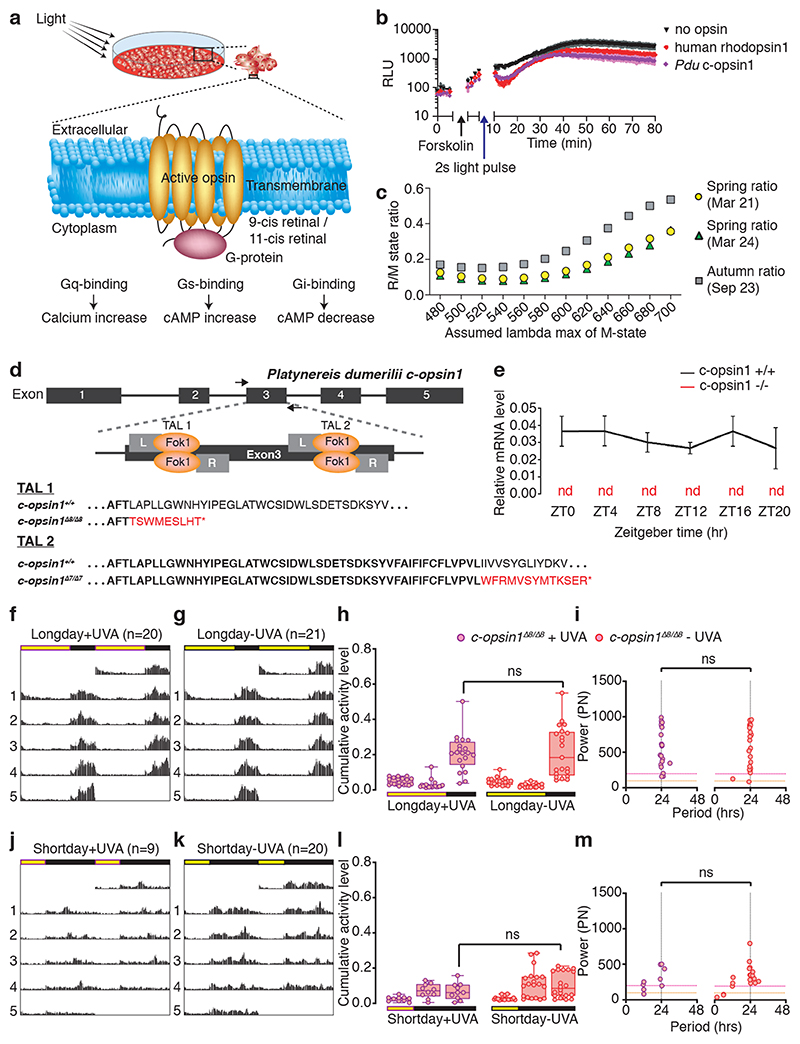
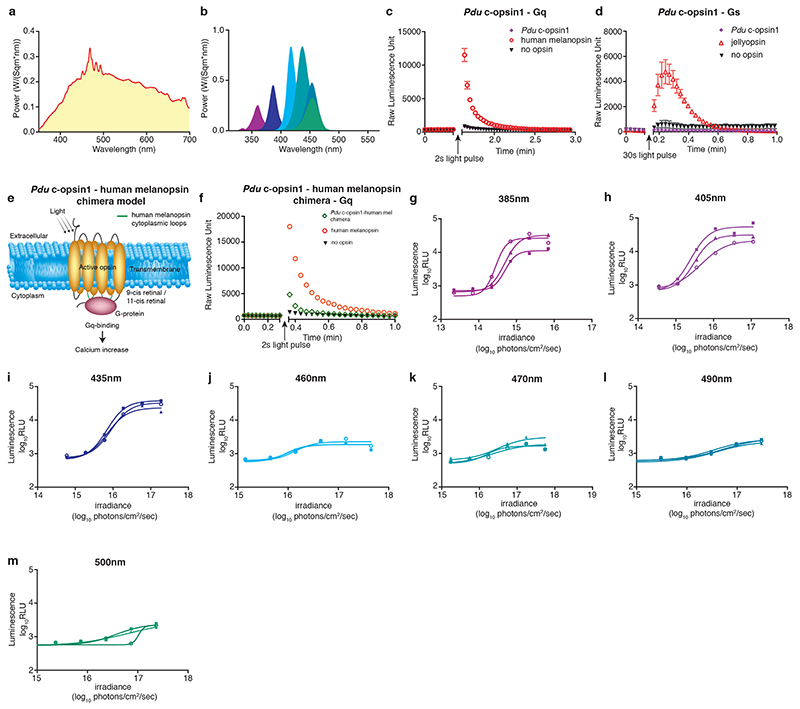
Previous work on the absorbance spectrum of Platynereis ciliary Opsin1, expressed in the medial brain of the worm 49, showed that this Opsin is activated by UVA/ deep violet light 50,51, bistable, i.e. the activated form can be reverted to the dark form by visible (non-UVA) light and likely coupled to the Gi-signaling pathway 50. We first confirmed more directly that Pdu-c-opsin1 signals via Gi-signaling (Fig.3a, b, Extended Data Fig.5a), using a previously established tissue culture assay that tests for decrease in cAMP upon light exposure, indicative of Gi-signaling 52,53. Using an equivalent test for cAMP and Ca2+ increase, indicative for Gs and Gq-signaling 52,53, we also found that c-opsin1 does not signal via Gs or Gq (Extended Data Fig.5c,d). Given that it is technically challenging to determine an action spectrum with an Opsin coupled to an inhibitory G-protein cascade, we generated a c-opsin chimera in which cytoplasmic loops of Pdu-c-opsin1 were replaced by those of human melanopsin (Extended Data Fig.5e, for details see methods). Such chimera change the G-protein signaling cascade of the Opsin, but should leave the activating Λmax unaltered 54–56. The Pdu-c-opsin1-human-Melanopsin chimera was expressed in tissue culture cells and tested for its response to different wavelengths (Extended Data Fig.5b,f-m). By this we confirmed that also in a cellular context (action spectrum) cOpsin1 responds strongest to UVA/ deep violet light.
Figure 3. Pdu-c-opsin1 mediates UVA/deep violet light input via Gi-signaling and regulates locomotor activity.
(a) Simplified model of phototransduction machinery with different downstream second-messenger signaling systems and relevant readouts used in the HEK293-based second messenger assays 52,53 shown in 3b and Extended Data Fig.5c,d). (b) Luminescent assay identified Gi-protein as signaling partner for Pdu c-opsin1. black arrow: forskolin addition (to increase cAMP levels), blue arrow: light pulse, purple diamond: Pdu-c-opsin1, red circle: human rhodopsin1 (positive control), black triangle: reporter construct. See Extended Data Fig.5c,d for Gs and Gq-signaling tests. (c) Ratios between Pdu-c-opsin1 rhodopsin state (R-state) total power value (λmax ≅ 380nm) and metarhodopsin state (M-state) total power value (testing M-state λmax ≅ for 480-700nm; in 20nm intervals) for autumn vs. spring equinox days. The total power value of each state is calculated as the absorbance efficiency of the photoreceptor across the measured averaged equinox day spectra assuming the indicated λmax of R- and M-states. (d) Pdu-c-opsin1 genomic locus with the two independent TALEN target sites in exon 3, resulting in two independent mutations: Δ8bp and Δ7bp, both resulting in early frameshifts and stop codons (red). (e) c-opsin1 mRNA was absent in qPCRs of Pdu c-opsin1Δ8/Δ8 worms, compared to sibling controls. (f,g) Double plotted average actogram of c-opsin1Δ8/Δ8 worms under long day including UVA (f: n=20) and long day filter-reduced UVA (g: n=21). (h) Statistical analysis of (f,g) shows lack of locomotor activity level difference observed for wildtype siblings under different UVA conditions (One-way ANOVA, Sidak’s multiple comparison test). (i) Rhythmicity and power analyses of f,g (Lomb-Scargle, Mann-Whitney-Wilcoxon test). (j,k) Double average actogram plot of c-opsin1Δ8/Δ8 worms under short day including UVA (j: n=9) and short day filter-reduced UVA (k: n=20). (l) Statistical analysis of j,k shows no activity level difference under different UVA conditions (One-way ANOVA, Sidak’s multiple comparison test), full statistical analysis: Supplementary Fig.7. (m) Rhythmicity and power analyses of j,k (Lomb-Scargle, Mann-Whitney-Wilcoxon test). *p<0.05, ** p<0.01, *** p<0.001. Locomotor data of individual worms: Supp.Figs 2c,4b,5b,6b, see Extended Data Fig.6,7, Supp.Figs 2,3 for analyses including heterozygous and transheterozygous worms. Individual worm rhythm period and power were determined via Lomb-Scargle periodograms using ActogramJ.
The previously unraveled bistability of c-opsin1 implies that this photoreceptor might detect ratios of UVA (R-state) and longer wavelength light (M-state). We thus wondered, if the spectral difference for instance present in spring versus fall would also be present, if both detection states of c-opsin1 are taken into account. Again, we used the days around equinox as representative examples for spring versus autumn. While the maximally activating wavelength of the R-state of c-opsin1 is relatively well-defined (λmax 380nm), the maximally effective wavelength on the M-state is not well defined (i.e. >480nm) 50. We hence calculated the R/M ratios for spring vs. fall with a fixed R-state λmax at 380nm and tested different M-state λmax. In all cases was the R/M state ratio for the autumn equinox higher than for the spring equinox (Fig.3c). Modest clouding had again no effect on the outcome (Fig.3c green versus yellow points; weather data Extended Data Fig.2i). These analyses suggest that also in case c-opsin1 would function as a wavelength ratio detector, the differences in the annual spectral distribution could be detected by it.
Based on its UVA/ deep violet light sensitivity, c-opsin1 has been implicated in depths measurements of the freely swimming (pelagic) Platynereis larvae 50,51. However, the pelagic larval stage during which such a depth gauge sensor will be relevant lasts only few days during the several months long life of Platynereis dumerilii. Already at one week of age, the worms will have settled, start building tubes and switch to a benthic life style 24. The majorly benthic life-style and the fact that the worms are nocturnal 22,23 (Fig.2e-l) is difficult to reconcile with a function of c-opsin1 as a gauge sensor beyond its short larval stage. Furthermore, while complete data on annual reproduction do not exist, one detailed study spanning June-December suggest that the highest level of reproduction occurs in July/August 1. Thus, enhanced locomotion (and physiological adaptations) at the long photoperiods between summer solstice and autumnal equinox, could have beneficial effects on the developing young worms and help to prepare them for short photoperiod times. To therefore explore the role of this light receptor in benthic worms, we tested, if c-opsin1 mutants display behavioral or physiological aspects reminiscent of those observed under the lack of UVA/ deep violet light.
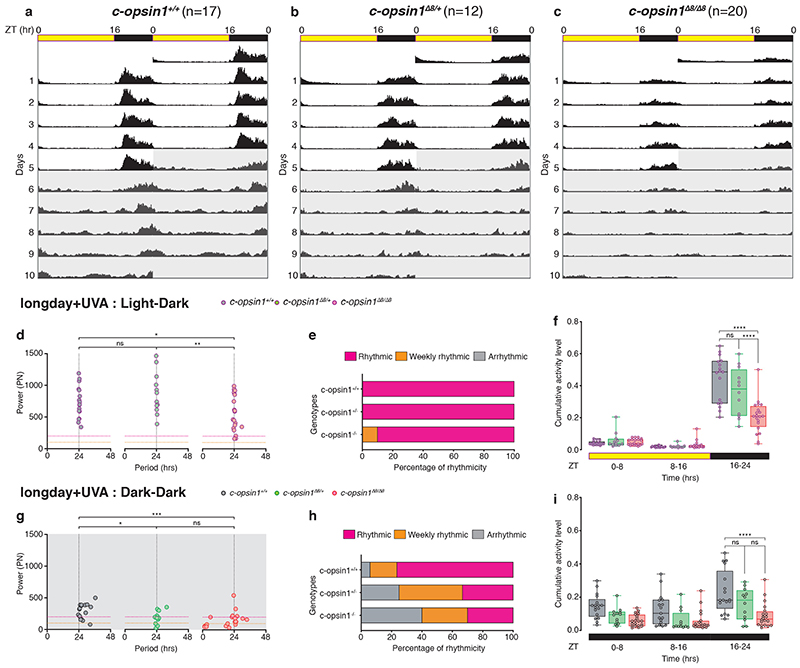
Platynereis c-opsin1 mutants exhibit lowered locomotor activity levels
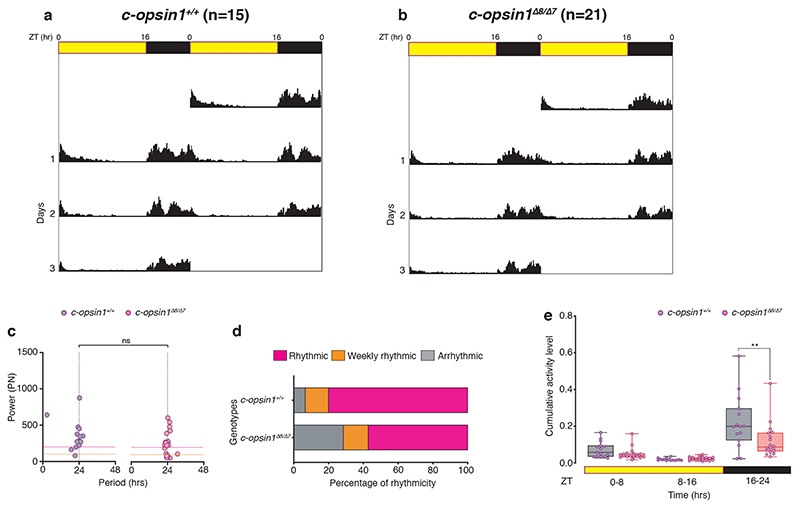
We used TALEN-technology to generate two independent alleles of c-opsin1. The two alleles Δ8 (previously published 51) and a new 7bps deletion mutant (Δ7) generated with two independent TALEN pairs at two separate sites of exon 3 led to premature stop-codons in the N-terminal region of the protein (Fig.3d). qPCR revealed that c-opsin1 transcripts were not detectable in mutant worms compared to wildtype sibling controls (Fig.3e), suggesting a complete loss-of-function phenotype. We next tested the locomotor behavior of c-opsin1-/- worms, using the behavioral assay outlined above. Indeed, c-opsin1 mutant worms showed a significant decrease in locomotor activity compared to wildtype (long day, +UVA condition), for both c-opsin1 Δ8/Δ8 (Extended Data Fig.6, 8a,c,e,g, Supp.Fig.2) and c-opsin1 Δ7/Δ8 (Extended Data Fig.7, Supp.Fig.3). In addition, mutant worms exhibited a weaker 24hrs rhythmicity under LD and DD (Extended Data Figs.6d,e,g,h, 7c,d, 8d,h). We then compared the locomotor activity levels of c-opsin1 Δ8/Δ8 under long day conditions with intense and filter-reduced UVA/ deep violet light levels. c-opsin1 Δ8/Δ8 worms did not show the decrease in locomotor activity visible for their sibling wildtype worms under the corresponding conditions, instead their locomotor activity was constantly at the low level observed under long day, filter-reduced UVA/ deep violet light (–UVA) conditions (compare Fig.2e-g, 3f-h, Extended Data Fig.8a-c and e-g Supp.Figs.2,4,8,9).
Short day conditions lead to an additional activity decrease in mutants (Fig.3h,l, Supp.Figs 5,6: hrs 16-24 c-ops1 Δ8/Δ8 in LD vs SD +UVA: Sidak’s multiple comparison test: p<0.0001, hrs 16-24 c-ops1 Δ8/Δ8 in LD vs SD–UVA: Sidak’s multiple comparison test: p<0.0001) suggesting that photoperiod detection is independent of c-opsin1. As for sibling wildtype worms, daily period length of individual worms was generally more variable and rhythmicity had a lower power under short day conditions (Fig.3i, m, Supp.Tables S5-9).
Finally, given the differential effects of UVA/ deep violet light intensity depending on photoperiod, we tested worm locomotion also under the intermediate photoperiod LD 12:12. The effect of intense UVA/ deep violet light (+UVA condition) versus filter-reduced UVA/ deep violet light (–UVA condition) on locomotion was intermediate to the results obtained for the two different UVA/ deep violet light conditions under long day versus short day photoperiods for wildtype (Extended Data Figure 8i-l, Supplementary Figs.7, 9). Wildtype worms exhibited a trend to less locomotion under –UVA close to statistical significance (p=0.08, n=9 worms +UVA and 7 worms -UVA). For comparison, under LD16:8 the difference is at p< 0.0001 (initial dataset with n=20 worms +UVA and 21 worms -UVA) and p=0.0056 (new repetition dataset with about half the number of worms), while under LD 8:16 p=0.9863 (Supplementary Fig. 7). In contrast to wildtype, there was no trend to less locomotion under LD 12:12 -UVA versus +UVA condition in c-opsin1-/- worms (Extended Data Figure 8m-p, p= 0.5743, n=10 and 9 worms, respectively, Supplementary Figs. 7, 9), again consistent with the suggestion that the effects of the +UVA condition on wildtype are mediated by c-opsin1.
Platynereis c-opsin1 mutants have lowered levels of key enzymes and hormones regulating physiology and behavior
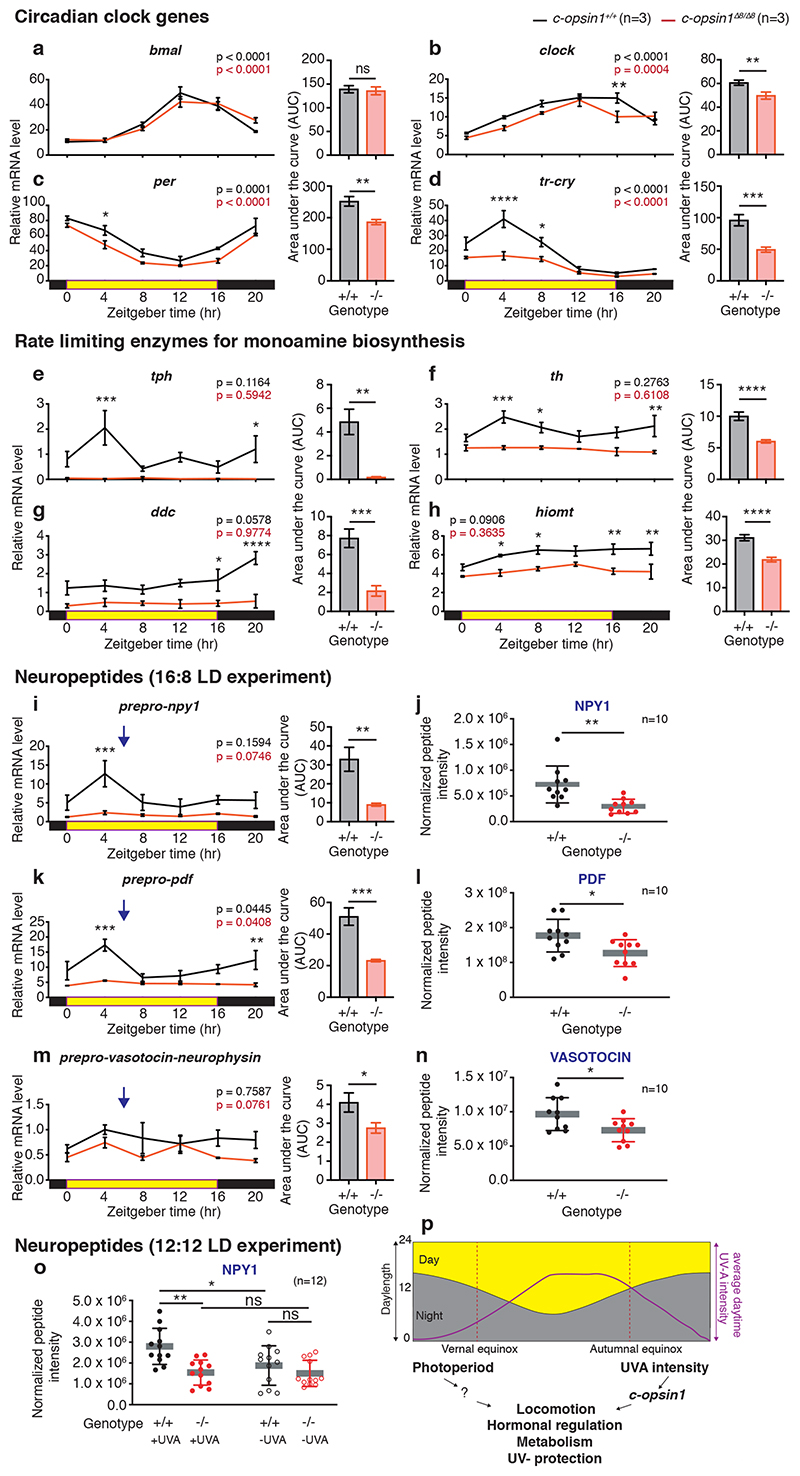
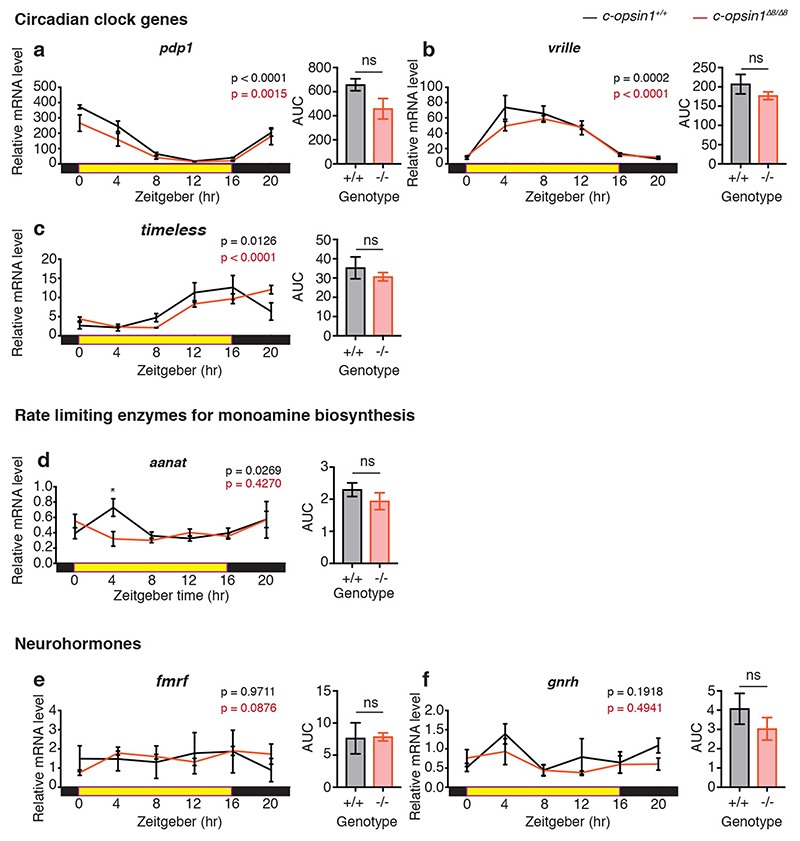
In order to gain a first understanding how c-opsin1 might lead to the observed behavioral alterations, we tested for differences in the transcript levels of key neurotransmitter enzymes, as well as core circadian clock genes. We previously established that the products of the genes Pdu-per, Pdu-tr-cry, Pdu-bmal, Pdu-clock can function as circadian repressor/activators, respectively 22. We analyzed their temporal expression profiles in c-opsin1 wildtype versus c-opsin1Δ8/Δ8 animals. Phase and period of bmal, per, clock, pdp1, vrille and tim appeared unchanged in mutants, albeit a small significant reduction in overall levels was present in per and clock (Fig4.a-c, Extended Data Fig. 9a-c). tr-cry exhibited stronger changes, but was still significantly rhythmic (Fig.4d). In contrast to these rather minor changes in core circadian clock gene levels, we observed strong reductions in the transcript levels of several key neurotransmitter enzymes critically required for the synthesis of biogenic amines (Fig4e-h). Furthermore, the transcripts of the preprohormones neuropeptide Y (npy-1), pigment dispersing factor (pdf) and vasotocin- neurophysin (vtn) were strongly decreased (Fig.4i,k,m). We confirmed this reduction of peptide hormones using targeted nano-LC-MS/MS for the corresponding mature peptides (Fig.4j,l,n, Supp.Table S11).
Figure 4. Loss of Pdu-c-opsin1 affects brain hormone synthesis.
(a-h, i,k,m) Diel profiles of immature head mRNA levels after 5 days of entrainment under long day including UVA n=3, 5heads/BR, data shown as mean±S.E.M. Black: c-opsin1+/+, red: c-opsin1Δ8/Δ8. Tested gene indicated above each graph. Statistics: One-way ANOVA with Sidak’s multiple comparison test. Unpaired student’s t-test with Welch’s correction was used to test for changes in overall transcript levels (AUC). (j,l,n) Targeted LC-MS/MS analyses of mature neuropeptides (j) NPY-1, (l) PDF, (n) VASOTOCIN sampled under long day+UVA condition. The sampling timepoint (blue arrows: i, k, m) was chosen 2hours after the maximally detected wildtype transcript levels to account for translational and peptide processing delays. n=10 BRs/genotype. 3heads/BR. Details: Supplementary Table S11. (o) Targeted LC-MS/MS analyses of NPY1 under LD12:12. 12BRs/ genotype. 3heads/BR; Statistics: Unpaired student’s t-test with Welch’s correction and One-way ANOVA with sidak’s multiple correction for peptide quantification. Details: Supplementary Table S12. *p<0.05, ** p<0.01, *** p<0.001, n.s.- non significant (p) Integrative light detection model in which photoperiod and UVA light provide differential information about seasonal time via c-opsin1.
c-opsin1+/+ : black, c-opsin1Δ8/Δ8 : red; abbreviations: BR: biological replicate, tph: tryptophan hydroxylase, th: tyrosine hydroxylase, ddc: dopa decarboxylase, hiomt: acetylserotonin O-methyltransferase.
Based on these transcriptomic and peptidomic changes, we suggest that UVA/ deep violet light via c-opsin1 impacts on multiple neurohormone and transmitter systems, which jointly leads to the observed alteration in locomotor behavior. These strong changes are specific given that transcript levels for the pre-pro-hormones fmrf and gnrh-like1 were unaffected (Extended Data Fig.9e,f) and only affected at ZT4 for the enzyme aanat (Extended Data Fig.9d).
Our natural light measurements suggest that the UVA/ deep violet light signaling via c-opsin1 might provide particularly relevant information to discriminate between equal photoperiods close to equinox and long day photoperiods equally distant before and after summer solstice. While our molecular analyses so far only analyzed the effects of c-opsin1 mediated UVA/ deep violet light input under long day (LD 16:8), we next ask, if there are also differences under a photoperiod of LD 12:12. We performed targeted nano-LC-MS/MS on NPY-1, PDF and VTN under LD12:12, comparing conditions that just differed in the intensity of UVA/ deep violet light (+UVA and – UVA conditions) and compared c-opsin1 wildtype vs. c-opsin1Δ8/Δ8 animals (SuppFig1E,F). While PDF and VTN amounts were highly variable and thus did not show statistical significant differences (data not shown), we identified clear effects on NPY-1. Both the filter-reduced levels of UVA/ deep violet light (– UVA), as well as the c-opsin1 mutation in the presence of high UVA/ deep violet light levels (+UVA) showed significantly lowered neurohormone concentrations (Fig.4o, Supp.Table S12).
These data provide strong evidence that UVA/ deep violet light levels, in a c-opsin1-dependent manner, can drive the animals’ hormonal status in a specific direction when photoperiod is at least LD 12:12.
At present we do not have a full explanation why the c-opsin1 mutation has stronger effects on some neurohormones at LD 16:8, compared to LD 12:12. These data might point in the direction of an interplay between photoperiod and UVA signaling. In combination with spectral intensity differences, specific photoperiods will enhance or diminish the effects of UVA/ deep violet light - signaling via c-opsin1. One hint further supporting this interpretation comes from the results that the same c-opsin1 mediated UVA/ deep violet light signal in the locomotion behavioral analyses also causes different effects depending on photoperiod (Fig.2I-L, 3J-M, Extended Data Figure 8, Supplementary Fig.7).
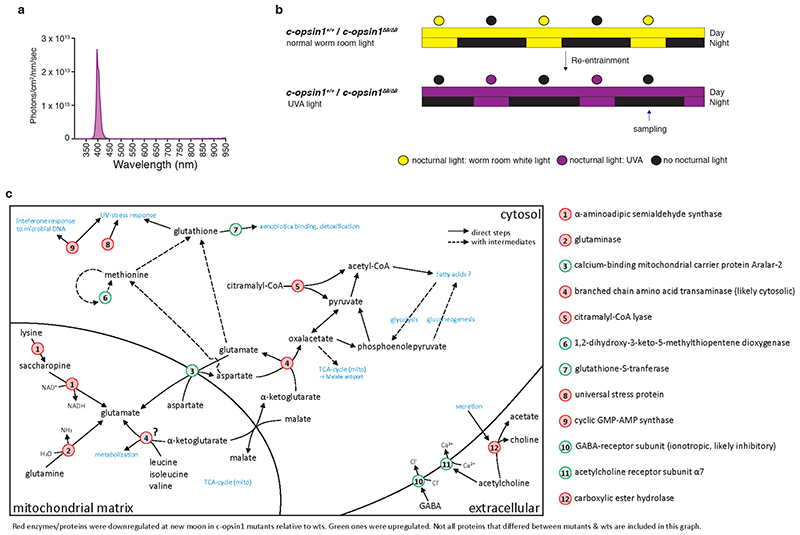
Complementing the candidate qPCR and LC-MS/MS approach above, we performed untargeted quantitative mass spectrometry on c-opsin1 Δ8/Δ8 mutant versus sibling wildtype worms grown under UVA conditions (LD 16:8; Extended Data Fig.10a,b). We found three groups of regulated proteins (Extended Data Fig.10c, Supp.Tables S13-15).
-
1.)
Proteins impacting neurotransmission: Several enzymes impacting glutamate levels are downregulated in the c-opsin1 mutant (e.g. Glutaminase), while orthologs of the Glycine Receptor alpha2 subunit and the Acetylcholine Receptor alpha7 are upregulated, as is the synaptic serine protease Neurotrypsin, which has been implicated in synaptic restructuring and corresponding changes in nervous system activity 57,58. These changes further support the notion that UVA light via c-opsin1 mediates significant molecular changes in nervous system function.
-
2.)
Enzymes involved stress responses: UV- stress responding enzymes were downregulated in c-opsin1 mutants consistent with the worms sensing less energy-rich light. This was also apparent for a sulfotransferase-domain containing protein, which can be induced by irradiation 59. while Glutathione-S-transferase, an enzyme involved in detoxification and oxidative stress response 60, was upregulated in c-opsin1 mutants, as was Thioredoxin domain-containing protein 5. These changes further indicate that c-opsin1 can also convey information about potential UVA stress to the worms.
-
3.)
Enzymes involved in metabolic processes: In addition to several enzymes linked to the synthesis and transport of glutamate (Extended Data Fig.10c, Supp.Tables S13-15), we identified two enzymes (Citramalal-CoA lyase and cytosolic branched chain amino acid transaminase), which were downregulated in the mutant and whose activities impact the levels of available pyruvate. Pyruvate is a key metabolic intermediate, for instance important for the cells’ energy supply.
In summary, these changes in metabolic enzymes, combined with the reduction of NPY1 suggest adaptive metabolic changes to low UVA conditions in addition to locomotor activity changes.
Evidence from multiple techniques provides the consistent picture that UVA light, at naturally occurring levels, is sensed by c-opsin1 leading to multiple changes in the worm’s brain, likely optimizing the animals’ behavior, physiology and irradiance protection in addition to photoperiod information (Fig.4p).
Discussion
Here we show that UVA/ deep violet light intensities change across the year in a different manner than photoperiod, but similar to water temperature. The marine annelid Platynereis dumerilii can sense differences in UVA light by the c-opsin1 photoreceptor. Changes in UVA/ deep violet light intensity significantly impact on the worm’s behavior, as do loss-of-function mutations in c-opsin1. Correspondingly, c-opsin1 mutants exhibit multiple, significant changes in key neurotransmitter enzymes, hormones and receptors, likely resulting in altered locomotor behavior and physiology and by this helping the worms to adjust to the appropriate environmental conditions. Our analyses of the effects of UVA/ deep violet light on worm neuropeptides and locomotion under different photoperiods suggests that photoperiod and UVA/ deep violet light intensity jointly modulate worm physiology and behavior. They might more specifically suggests that the effect of UVA/ deep violet light will be stronger the longer the photoperiod is. However, this specific hypothesis remains to be tested, as well as the effects of other ecological important parameters on worm physiology and behavior.
Nevertheless our findings have at least three larger implications.
First, so far research on light as a signal to regulate seasonal biology has basically exclusively focused on photoperiod and temperature as well-established signals, which either act directly or indirectly by the entrainment of endogenous circannual timers 30,61. The nature of the photoreceptor(s) mediating the photoperiodic input has been a matter of debates and is incompletely resolved. Two main proposals exist for non-mammalian vertebrates, endowing either VA-opsins or Opn5-type opsins with this role (reviewed in 62). Interestingly, ranging from fish to humans, vertebrate Opn5-type opsins have very similar properties to Pdu-c-opsin1, in their bistability (activated by UVA- absorption peak at 380nm-; recovery by longer wavelength light) and Gi-signaling 32–34. They were proposed to function as seasonal light receptors in birds, based on site of expression and light-dependent responses in brain slices 63. However, VA(L)-opsins, show a better match with the determined action spectrum relevant for the avian photoperiod response 64,65.
Our work might help to solve this dispute, suggesting that UVA-sensing opsins may contribute to the detection of seasonal UVA-differences, but not photoperiod. The exact nature of such seasonal UVA differences is likely dependent on the habitat. The use of seasonal UVA-light and - more generally -spectral information could provide fitness benefits for animals in temperate and polar marine habitats to fine-tune their annual timing. A major regular seasonal phenomenon in the oceans are phytoplankton blooms, which will cause spectral alterations of underwater light conditions 66,67, and are themselves often important food sources. Furthermore, in polar regions, the amount of sea ice and snow will also affect the intensity of UVA (and overall light spectrum). The combination of different, partly interlinked, environmental variables make the exact spectral changes across the year difficult to predict, but in any case, UVA levels would provide valuable additional temporal information on regular annual changes of animal habitats. Phenomena like phytoplankton blooms or sea ice cover are cyclic, but not as precise in time as photoperiod. Thus, having additional information on their exact occurrence will help animals to fine tune their physiology and behavior, and- where existent- annual timers. Such an ability of temporal fine-tuning could make animals also more robust against climate change, which has resulted in temporal shifts of boreal and polar phytoplankton blooms 11,12. Unlike photoperiod-based seasonality, UVA-based seasonal adjustments have the potential to detect and respond to these shifts, enabling species to adapt their phenology and avoid seasonal mismatch situations.
In polar habitats, which are characterized by prolonged periods of constant light (midnight sun) or darkness (polar night), UV-cues could further allow for seasonal entrainment in the absence of a clearly defined photoperiod. An UV-effect during the polar night is very unlikely due to the general lack of light. However, UVA-changes during midnight sun could support seasonal, but also diel rhythmicity. A diel change in light intensity during midnight sun does persist. Several reports of animals maintaining diel rhythms during this time indicate the potential for seasonal entrainment via clock-based photoperiod-sensing 68–72, while other animal populations appear to loose diel rhythmicity, but could still achieve seasonal timing via UV-sensing 73–77. It is noteworthy, that diel light intensity changes during midnight sun and very long photoperiods are especially pronounced in the UV-range 77,78. Furthermore, UV-changes alone (under otherwise constant illumination) are sufficient to entrain diel rhythmicity in bumblebees 79. UVA-sensing could thus help to maintain diel rhythmicity and photoperiod detection under extreme photic environments condition under which diel changes in longer wavelengths and overall light intensities can be weak.
Given the widespread occurrence of UVA/ deep violet light -sensitive opsins in the animal kingdom, it will therefore be highly interesting to test if UVA/ deep violet light might mediate similar changes in neurohormones and transmitters in other taxa, including key ecologically relevant marine invertebrates, but also terrestrial vertebrates.
Second, the implication of UVA/ deep violet light in (marine) timing processes also raises a new aspect of possible vulnerability. While UVA/ deep violet light detection might enhance temporal fine-tuning in marine environments, any pollution altering turbidity, and hence light spectrum in the water, could disrupt it. This aspect can now be put to further tests in more direct ecological studies and models. Light pollution on the other hand will likely have little or only indirect effect on UVA-controlled processes as anthropogenic light sources typically do not include UV-wavelengths.
Third, an aspect of evolutionary relevance is the location and molecular fingerprint of the Pdu-c-opsin1-positive cells. It was previously shown that Pdu-c-opsin1 is expressed in large ciliated cells in the medial forebrain of the worm 49,51. Based on their morphological position and molecular fingerprint [i.e. the presence of core circadian clock genes 22,49,80, opsin-based phototransduction cascade genes and hiomt 80 (the rate-limiting enzyme for melatonin biosynthesis in vertebrates 81)], these cells have been suggested to be part of an evolutionary ancestral timing region 49. In general, our data support such an ancient “brain timing nucleus”. More specifically they hint, however, at the possibility that the ancestral timing mechanisms did not only include photoperiod detection, but also short wavelength intensity as a predictor of seasonal change. This work also leaves several open questions, such as the original function of Opn-5-type opsins and the evolutionary relationship of ciliary-type opsins to photoperiod detection.
Finally, while our data provide a novel and important link between natural UVA/ deep violet light changes, behavior and metabolism, more work under truly natural conditions will be required to work out the exact interplay of different environmental factors. Work under laboratory conditions is highly useful to obtain first insights, because it allows to vary one environmental stimulus precisely and in a defined direction. This permits the check of specific hypotheses, like the tests for differential responses to strong versus filter-reduced intensities of UVA/ deep violet light. Under true environmental conditions different environmental factors co-vary, making causal conclusions about one specific environmental factor more difficult to impossible. It is clear that these approaches in combination can be highly powerful to comprehend how animals will respond to the up-coming climate change.
Materials and Methods
Worm culture
Platynereis dumerilii were grown as previously described 82, with the following modification: Light spectra- see Extended Data Fig.4. All cultures involving UVA light were grown in light-tight shelves. “Full moon” for the entrainment of the worm’s circalunar clock was given for 8 nights (except during 2018: 6 nights). Worms were initially raised under worm room conditions and then shifted to the respective experimental light and photoperiod regimes (Supplementary Fig.1). Therefore, the worms are very likely not in the same seasonal physiological state as they would be in nature, an aspect that should be considered for data interpretation and future experiments.
Natural light measurements and analyses
Measurements were recorded every 15min between July, 27th 2010 and July, 10th 2011 at 10m depth and between May, 31st 2011 - July, 10th 2011 at 4m depth. Natural light measurements were acquired by using two RAMSES-ACC-VIS hyperspectral radiometers (TriOS GmbH) for UV to IR spectral range. Both instruments were tied to a cement plate and deployed on a sand flat on the east coast of the island of Ischia, in the Gulf of Naples (Italy) at about 10m (40°43’50.6”N - 13°58’02.9”E) and 4m (40°43’56”N 13°57’44”E) depth, respectively. The locations were near Posidonia oceanica meadows, a natural habitat of Platynereis. Data were retrieved and instrument sensors cleaned every 2-3 months. The radiometers provided individual values of light intensity over 255 individual wavelengths between 305 nm and 1150 nm, the wavelength range for which they had been calibrated directly before usage by the company Trios. As sensor sensitivity drops significantly below 310nm and above 710nm (Suppl.Fig 10), we concentrated our analyses on the wavelengths range between 310-710nm. Following the manufacturer’s instructions and associated software (msda_xe v8.9), the background data and electronic offset of the instrument were subtracted from the raw data, and the spectrum’s values were calibrated for underwater conditions. The final calibrated raw data for light and temperature were deposited in Supp.Tables S1 and S2.
Next, the acquired raw measurements in mW/m2/nm/sec were converted to corresponding number of photons/cm2/nm/sec or, to enable comparisons with previous published works, also converted to uW/cm2/nm/sec, uMolphotons/m2/nm/sec, photons/m2/um/sec and log photons/m2/nm/sec (Extended Data Fig.1, Supp.Tables S3,S4).
To plot the photoperiod changes the annual light data in photon units were analyzed by averaging across all wavelengths for each measured time point.
Idtλ – Light intensity measured at specific day (d) at specific time (t) between wavelengths (λ)
di = Individual days (July 27, 2010 – July 10, 2011)
ti = Individual time points (00:00 – 23:45)
The daylight values between sunrise to sunset/no twilight (referred to as NT dataset) and values between astronomical dawn to astronomical dusk (referred to as AT dataset) were extracted based on the daylight and nightlight timings for the Ischia geographic location as mentioned in the validated astronomical websites (NOAA solar calculator and timeanddate.com). The daylight data for both NT and AT datasets were plotted as surface and pcolor plot in log photon scale using MATLAB. The average of the measurements across the day for the entire NT dataset was calculated for surface and pcolor plots.
To look at the annual variation of daylight spectral intensities, the monthly average for each month from NT dataset (September 2010, December 2010, March 2011 and June 2011), was calculated and plotted as 2D line graph using shadedErrorBar MATLAB function. The shaded error bar in the monthly average plot indicates the intensity variation between individual average day values over month. Further, the monthly average ratio was calculated between autumn versus spring (September/March).
where dm = average daylight of particular month (dm2 – September 2010; dm1 – March 2011) and DtiNT = daylight from individual timepoint from NT dataset.
To investigate the spectral variation over time between equinox days, the ratio was calculated in two ways, 1. Ratio of individual wavelengths from each timepoint measured over day between equinox days, 2. Ratio of individual wavelengths between equinox day average.
- Equinox day average ratio for NT and AT dataset
where d1 = equinox day1 (March 21 / March 24), d2 = equinox day2 (September 23), DtiNT = daylight from individual timepoint from NT dataset and DtiAT = daylight from individual timepoint from AT dataset.
All above calculations inspecting spectral variation between equinox days was also calculated for astronomical twilight dataset mentioned in Extended Data Fig.2.
To understand light intensity variation between different water depths, the radiometer was also placed at 4m depth. Raw measurements were treated as described above. For differences in intensity, we calculated the monthly average from June 2011 4m and June 2011 10m NT dataset. The shaded error bar in the monthly average plot indicates the variation of photon values between individual day averages over month. Additionally, the ratios of individual wavelengths between 4m and 10m June 2011 monthly average were calculated.
R/M state ratio calculation
The c-opsin1 photo-absorbance values across wavelengths were calculated for rhodopsin (R-state) and metarhodopsin (M-states) based on the photopigment absorbance spectra of Govardovskii 83 and the formula modeled on the primary visual processes of insect photoreceptors 84. R-state λmax: 380nm; M-state λmax: 480nm-700nm in 20nm intervals. The calculated photo-absorbance values for R and M-state were normalized to their respective maximum absorbance measurement. The normalized absorbance values were multiplied with the average photon count (310-710nm) across March 21th or March 24th (for R/M spring) and September 23 (for R/M autumn). The resultant power values were summed to calculate the ‘total power’. The final R/M ratio was calculated as R-state-total power / M-state-total power.
Artificial sunlight emulation (NELIS devices)
Artificial sunlight, with a spectrum from around 5m below water surface was realized using NELIS (Natural Environment Light Intensity System) SL-5M LED panels. The panels were supplied with a selection of 20 different LEDs using Cree XT-E Series, Luxeon Rebel and Rebel plus, SemiLeds C3535U and Everlight Shwo 1W LEDs. Different Bin selections were used ranging from 390 nm to „amber“ and “cool white” and were driven by different currents to reach the desired spectrum. The drivers were built using non-switching, continuous constant current sources.
An ORBIT controller was used to continuously calculate sun intensities according to sun elevation above horizon at the selected time and date by a computing program calculates the sun’s right ascension and declination 85 for every minute. These are then transferred via geocentric coordinates to topocentric coordinates to determine the sun’s elevation above horizon and therefore its intensity. Refraction, twilight and weather phenomena (e.g. clouds) are not taken into consideration. Our light source was built based on the underwater light measurements and is hence an improvement over the “normal” indoor illumination usually used in lab experiments. However, it is important to note that it still does not fully match the sunlight spectrum. It is likely that an even further optimized light source might reveal additional biological effects.
The UVAR filter was purchased from Pixelteq, Salvo Technologies, USA: https://opticalfiltershop.com/shop/specialty-filters/uv-ar-filter-438nm/, reducing the intensity of wavelengths that cause strong c-opsin1 activation by at least 102.
Worm cultures and light system were placed in a shelf covered with black acrylic to prevent room light from disturbing the experiment. Air for cooling the lighting device was taken from outside the shelf. The outlet was placed outside the shelf to minimize temperature fluctuations.
Platynereis c-opsin1 G-protein signaling and action spectra determination
Construction of expression vector
The open reading frame sequence of Platynereis c-opsin1 containing kozak sequence at the N-terminus were cloned to pcDNA3.1 expression vector by replacing its stop codon with 1D4 tag sequence from bovine rhodopsin at the c-terminal region. Additionally, Platynereis c-opsin1-melanopsin chimera was generated by replacing the second and third cytoplasmic loop of c-opsin1 sequence with human melanopsin sequence respectively. The cloning was performed using Gibson assembly protocol (Gibson Assembly Cloning Kit, NEB). Primers used for cloning Pdu c-opsin1 expression vector and chimeras: Supplementary Table S10. All constructs were sequence confirmed and endotoxin-free plasmid DNA were prepared for transfection procedure (Endofree Plasmid Maxi Kit, Qiagen).
Target opsin and reporter transfection
HEK293 cells with 80% confluence was cultured and maintained in Dulbecco’s Modified Eagle’s Medium (DMEM), 10% Fetal Calf Serum (FCS, Sigma) with penicillin/streptomycin at 37°c in 5% CO2 atmosphere. These cells were further washed with PBS, trypsinized, neutralized and concentration of 6x104 cells were plated per well in a 96 well solid white plate (Greiner) 24 hours before transfection. Platynereis c-opsin1 expressing plasmid and reporter (based on luminescent second messenger assay) at 200ng concentration each were transiently co-transfected using 0.2ul Lipofectamine 2000 (Invitrogen) prior to the assay. Following 6hrs of incubation, media was then replaced with L15 medium without phenol red (Invitrogen), 10% FCS, 300ng/ml tetracycline, 10uM 9-cis retinal (Sigma) and incubated for 16hrs at 37C. All these above steps after initial transfection were carried out under dim red light.
Luminescent second messenger assay: G-protein signaling
Opsin coupling to Gα q GPCR was accessed by the aequorin reporter 52,53. The assay used similar transfection procedure as mentioned above. Cells transfected with pcDNA5/FRT/TO mtAeq along with Platynereis c-opsin1 were incubated with 10uM Coelenterazine-h in the dark for 2hrs at room temperature. Raw Luminescence Unit (RLU) was measured from a single well with a resolution of 0.5s and cycle of 2s on a Fluostar optima plate reader (BMG Labtech, Germany). After baseline recording, the well was exposed to 2sec light pulse using arc lamp while all the other wells were protected with light-tight black sheet. The time between the light exposure of the well and the recording of next RLU measurement was at least 3s.
Changes in cyclic nucleotide level serve as an indication for opsin binding to Gα s (increase in cAMP level) or Gi/o subunit of GPCR (decrease in cAMP level). To evaluate Pdu c-opsin1 binding to Gα s or Gi/o, cells co-transfected with Pdu c-opsin1 and pcDNA5/FRT/TO Glo22F were incubated with 0.1M Luciferin in the dark for 2hrs at room temperature. RLU was recorded with 1s resolution and cycle of 30s using plate reader as baseline. To screen for Gα s binding, the well was exposed to 30sec light pulse using arc lamp while all the other wells were protected with light-tight black sheet. For Gi/o binding tests, cells were treated with 2uM Forskolin (Sigma-Aldrich) to artificially increase the cAMP level, remeasured, after few cycles, the well was exposed to 2sec light pulse and remeasure.
Spectral sensitivity measurement of Platynereis c-opsin1
To investigate the spectral sensitivity of Platynereis c-opsin1, cells transfected with c-opsin1-melanopsin chimera were exposed to individual monochromatic wavelength of different intensities using pE-4000 CoolLED light source with neutral density (ND) filters and accessed for changes in second messenger level by measuring luminescence signal and tested for different wavelengths: 385, 405, 435, 460, 470, 490, 500nm with decreasing intensity (ND0, ND0.5, ND1, ND1.5, ND2.0, ND2.5, ND3.0, ND3.5) to calculate the irradiance dose response curve. Cells cultured in 96-well plates were exposed to single light exposure on each well with the appearance of different irradiances randomly assigned to wells across the plate. A 96-well plate with clear base containing similar volume of media/well was used to measure the irradiance of light at each wavelength using a spectrophotometer (SpectroCAL MKII, Cambridge Instruments). Each experiment was done with at least 3BRs.
c-opsin1 mutant generation and genotyping
The c-opsin1 genomic region was amplified to screen for sequence polymorphisms in different Platynereis inbred lines (PIN, VIO, FL and ORA strain) 51,86. The in silico-based TALEN prediction for Platynereis c-opsin1 in non-polymorphic exon region was performed using TAL Effector Nucleotide Targeter 2.0 webserver; https://tale-nt.cac.cornell.edu as per customized design conditions mentioned in reference 51. Two pairs of final predicted c-opsin1 TALENs (TAL1 and TAL2) were constructed using Golden Gate assembly protocol (Golden Gate TAL Effector Kit 2.0, Addgene #1000000024) 87. The assembled TALEN pairs were cloned into heterodimeric FokI expression plasmids (pCS2TAL3-DD: left array and pCS2TAL3-RR: right array) and sequence-verified using TAL_F1 and TAL_R2 primers 88. Each array of c-opsin1 TALEN mRNA was generated by linearizing the final cloned plasmid using NotI restriction enzyme and invitro transcribed using mMESSAGE mMACHINE Sp6 kit (AM1340, Life Technologies). c-opsin1 TALEN mRNA of TAL1/TAL2 at concentration of 200 ng/ul/array was microinjected into Platynereis zygotes. TALEN-induced mutations were screened as described 86 using primers cops1_TAL_R1/cops1_TAL_L2 followed by restriction digest by the enzyme BanI for TAL1 and MluC1 for TAL2.
c-opsin1 knockout adult or mature worms were genotyped by isolating genomic DNA from tail piece or whole worm using hotshot method 89 or Nucleospin Tissue kit (#740952, Machery-Nagel) respectively. The DNA was amplified by using specific primers (cops1_del8m_F/cops1_com_R for TAL1 and cops1_del7_Fm/cops1_com_R for TAL2) and the mutations were verified by band size difference compared to WT band in agarose gel and sequencing.
Worm behavioral analyses
Videos were recorded using a Motif video recording system (Loopbio Gmbh, Austria). Images with a resolution of 2048x2048 pixels were acquired with a frame rate of 15fps (frames per second), along with environmental data such as temperature and humidity measured at 1Hz, with both saved to imgstore format (https://github.com/loopbio/imgstore) to allow precise timekeeping and storage of both data for 10 day recordings.
For detecting the position of each worm the video was cropped with a mask to represent single wells. Each well contained one worm. The position of the freely moving worm was tracked by performing a sequence of image processing operations. Detection is based on thresholding brightness with subsequent morphological operations for smoothing (opening-closing mechanism). A contour was fitted to each resulting detection and the enclosed area was calculated. Normally only one detection remained, if its area was larger than a defined minimum it was considered a positive detection of the worm and the two-dimensional coordinates (xy-pixel coordinates) of its center of mass was saved. In the case of multiple detections per well, the largest contour was considered the worm and others discarded.
Additional data processing was applied on top of the xy-pixel coordinates. The euclidean distance between subsequent detections of the worm was calculated. Data smoothing with a rolling average with a window length of 60 frames was applied on the distance. For down-sampling, the time series was grouped by non-overlapping, equally long-time windows, called bins. A window size for each bin of 1 minute (60 sec equals to 900 datapoints) was chosen and the mean for each window was computed. Data analysis was done on the down-sampled data.
Untargeted Mass-Spectrometry (Proteomics)
Platynereis c-opsin1+/+ and c-opsin1Δ8/Δ8 immature worms were grown at long day (LD 16:8) under monochromatic UVA-light (Extended Data Fig.10a,b) for three consecutive lunar months. After entrainment, the heads (n=3BR; 20heads/BR) were sampled on new moon phase at ZT4, ZT19 and ZT22 by anesthetizing the worm with 1:1 7.5% MgCl2:Natural Seawater (NSW) solution. The sampled heads were collected in 2ml tubes containing metal beads on ice, snap-frozen in liquid nitrogen and stored at -80°C until further use.
For tissue lysis, worm heads were resuspended in sample buffer (7.5 M urea, 1.5 M thiourea, 4 % CHAPS. 0.05 % SDS, 100 mM DTT) and sonicated. After centrifugation, protein concentrations were assessed by applying a Bradford assay (Bio-Rad-Laboratories, Vienna, Austria). Protein samples were subjected to a filter-assisted proteolytic digestion with a modified version of the FASP protocol (Wisniewski et al. 2009, PMID: 19377485). In short, 20 μg protein were loaded onto a pre-wetted MWCO filter (Pall Austria Filter GmbH, Vienna, Austria) with a pore size of 3 kD, followed by reduction of disulfide bonds with dithiothreitol (DTT), alkylation with iodoacetamide (IAA) and washing steps with 50 mM ammonium bicarbonate buffer. Digestion of proteins was achieved by applying Trypsin/Lys-C with Mass Spec Grade quality (Promega, Mannheim, Germany) for an overnight digest followed by the further addition of fresh enzyme and an additional incubation for four hours. Resulting peptides were eluted through the filter by centrifugation, and a clean-up step was performed using C-18 spin columns (Pierce, Thermo Fisher Scientific, Austria).
LC-MS/MS analysis:
For LC-MS/MS analyses, samples were reconstituted in 5 μl 30 % formic acid (FA), supplemented with four synthetic peptide standards for internal quality control, and diluted with 40 μl mobile phase A (97.9 % H2O, 2 % ACN, 0.1 % FA). Of this solution, 5 μl were injected into a Dionex Ultimate 3000 nano LC-system coupled to a Q Exactive orbitrap mass spectrometer equipped with a nanospray ion source (Thermo Fisher Scientific, Austria). All samples were analyzed as technical replicates. As a pre-concentration step, peptides were loaded on a 2 cm x 75 μm C18 Pepmap100 pre-column (Thermo Fisher Scientific, Austria) at a flow rate of 10 μl/min using mobile phase A. Elution from the pre-column to a 50 cm x 75 μm Pepmap100 analytical column (Thermo Fisher Scientific, Austria) and subsequent separation was achieved at a flow rate of 300 nl/min using a gradient of 8 % to 40 % mobile phase B (79.9 % ACN, 2 % H2O, 0.1 % FA) over 235 min with a total chromatographic run time of 280 min. For mass spectrometric detection, MS scans were performed in the range from m/z 400-1400 at a resolution of 70000 (at m/z =200). MS/MS scans of the eight most abundant ions were achieved through HCD fragmentation at 30 % normalized collision energy and analyzed in the orbitrap at a resolution of 17500 (at m/z =200).
Proteome data analysis:
The MaxQuant software (version 1.6.0.1), including the Andromeda search engine, was used for data analysis (Cox and Mann, 2008, PMID: 19029910). For positive protein identification, as a minimum two peptides, at least one of them being unique, had to be detected. Trypsin/P was specified in the digestion mode. Peptide mass tolerance was set to 50 and 25 ppm for the first and the main search, respectively. The false discovery rate (FDR) was set to 0.01 both on peptide and protein level. Peptides were mapped against a reference proteome set established before 82. Carbamidomethylation was set as fixed modification, methionine oxidation and N-terminal acetylation as variable modifications. Each peptide was allowed to have a maximum of two missed cleavages and two modifications. “Match between runs” was enabled and the alignment and match time window set to 10 and 1 min, respectively. The analysis of the quantitative protein abundance has been performed using the Perseus software platform (Tyanova et al. 2016, PMID: 27348712), and the FDR (according to the Benjamini-Hochberg procedure) has been set at 0.2. Only proteins detected in at least four biological replicates have been considered in the analysis.
Data analyzed using the Perseus computational platform 90 were processed additionally and independently. Arithmetic means were calculated from technical duplicates and then filtered with all treatments (combinations of genotype and diel time point) being excluded where no protein was detected in >1 of the 3 replicates. Diel time points were then pooled and genotypes were compared via 2-sided t-tests. p-values were corrected for multiple testing using the “Permutation-based FDR” option (250 randomizations). Significant hits were annotated via BLAST against a recently published Platynereis proteome 82.
Targeted Mass Spectrometry (Peptidomics)
Platynereis heads (3heads/BR; n=10BRs for LD16:8; n=12BRs for LD12:12) were sampled from both c-opsin1+/+ and c-opsin1Δ8/Δ8 worms after 5 days of entrainment to respective photoperiod under broad spectrum white light containing UVA wavelength. LD 12:12 experiment entrained by white light without UVA (n=12BRs) was achieved by installing UVAR filters. The heads were first rinsed in natural sea water (NSW) and collected in 200ul of ice-cold acidified methanol (methanol/water/acetic acid – 90/9/1 – v/v/v) to extract peptides 91. The collected heads were homogenized by sonicating three times for 30sec on ice and the supernatant was collected in a fresh collection tube. This procedure was repeated twice by adding 200ul of ice-cold acidified methanol to the remaining pellet each time for all samples to dissociate remaining pellets and the resultant supernatants in each step were collected in same collection tube resulting in about 600ul of total volume.
Synthetic PDF, NPY-1, NPY-4, GnRH-1 and Vasotocin peptides were dissolved in 50% acetonitrile at a final concentration of 5 mM and mixed in a 40:10:2:2:1 ratio to obtain similar signal strength in the mass spectrometer, as determined by a scouting run. To avoid disulfide bond formation 1 uL (10 pmol) of the peptide mixture was dissolved in 45 uL ammonium bicarbonate (ABC), reduced with 10 mM dithiothreitol (DTT) for 30 min at room temperature, alkylated with 20 mM iodoacetamide (IAA) for 30 min at room temperature in the dark and the remaining IAA was quenched by adding 5 mM DTT.
The extracts were concentrated by vacuum centrifugation, freeze-dried and resuspended in 8 M urea, 50 mM ABC. Reduction and alkylation of disulfide bonds was performed as described for the synthetic peptides. Samples were diluted with 50 mM ABC to 4 M urea and peptides were digested with 500 ng Lys-C (Wako) at 25°C for 4 hours. After stopping the digestion by addition of 0.5% trifluoroacetic acid (TFA), the peptides were desalted using C18 stagetips prior to LC-MS 92.
Peptides were separated on an Ultimate 3000 RSLC nano-flow chromatography system (Thermo-Fisher), using a pre-column for sample loading (Acclaim PepMap C18, 2 cm × 0.1 mm, 5 μm, Thermo-Fisher), and a C18 analytical column (Acclaim PepMap C18, 50 cm × 0.75 mm, 2 μm, Thermo-Fisher), applying a segmented linear gradient from 2% to 45% solvent B (80% acetonitrile, 0.1% formic acid; solvent A 0.1% formic acid) at a flow rate of 230 nL/min over 60 minutes. Eluting peptides were analyzed on a Q Exactive HF-X Orbitrap mass spectrometer (Thermo-Fisher), which was coupled to the column with a customized nano-spray EASY-Spray ion-source (Thermo-Fisher) using coated emitter tips (New Objective). For data-dependent acquisition (DDA) of the synthetic peptides we operated the instrument with following MS parameters: survey scan with 60k resolution, AGC 3E6, 60 ms IT, over a range of 300 to 1500 m/z, top 8 ions selected for MS2 scans with 30k resolution, AGC 1E5, 250 ms IT, isolation window of 1.6 m/z, and NCE of 28%. For parallel reaction monitoring (PRM) data acquisition of c-opsin1+/+ and c-opsin1Δ8/Δ8 samples we operated the instrument with the following MS parameters: survey scan with 60k resolution, AGC 1E6, 50 ms IT, over a range of 400 to 1300 m/z, PRM scan with 60k resolution, AGC 2E5, 500 ms IT, isolation window of 0.7 m/z with 0.2 m/z offset, and NCE of 27%.
PRM assays were set up based on a DDA measurement of PDF, NPY-1, NPY-4, GnRH-1 and Vasotocin synthetic peptides, with Lys-C protease specificity and amidated C-termini. The acquired spectra were used to prepare a spectral library in Proteome Discoverer (version 2.3) using MS Amanda 93 and Percolator 94 with the following search parameters: database of target peptides and common contaminants, 10 ppm MS1 and 20 ppm MS2 mass tolerance, carbamidomethylation of Cys and amidated peptide c-termini as fixed and oxidation of Met as variable modifications, 5% FDR on PSM level. PRM assay generation was performed using Skyline software (version: daily 19.0.9.149) 95. Samples were spiked with 100 fmol Pierce Peptide Retention Time Calibration Mixture (PRTC, Thermo-Fisher) to monitor the chromatographic and nano-spray stability across the PRM measurements of all samples. A wild-type sample spiked with the synthetic peptides served as positive control and retention time reference. The samples were measured in a randomized order. Peptide Retention Time Calibration Mixture (PRTC, Thermo Fisher Scientific) was spiked in all samples to monitor chromatographic and MS performance, and their precursor intensities were extracted using Skyline. PRTC peptide intensities indicated that the instrument performance was stable across all runs.
Data analysis, manual validation of all peptides and their transitions (based on retention time, relative ion intensities, and mass accuracy) was performed in Skyline. All peptides were quantified across all samples, except GnRH-1, which was not consistently detected and therefore removed from further analysis. The most intense non-interfering transitions were selected and their peak areas were summed up for each target (total peak area). Intensities of unmodified and oxidized (Met) peptide species were summed up for quantification of the neuropeptides. To correct for varying peptide amounts across replicates and instrument stability over the measurements, the peptide intensities were normalized based on the relative total MS1 ion current, which was extracted using the MaxQuant software (version 1.6.0.16) 96. MaxQuant was run with default settings and the msScans table was used to extract the total ion current for each sample. After normalization, c-opsin1+/+ and c-opsin1Δ8/Δ8 intensity means, standard deviations and a p-value (t-test) were calculated for each of the neuropeptides. The targeted mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via Panorama Public 97 with the identifier PXD014682.
RNA expression analysis
Sample preparation
Platynereis c-opsin1+/+ and c-opsin1Δ8/Δ8 immature worms were grown under long day photoperiod (LD 16:8) under broad spectrum white light containing UVA for 5 days. Subsequently the heads (n=3BR; 5heads/BR) were sampled at ZT0, ZT4, ZT8, ZT12, ZT16 and ZT20 from worms immobilized with 1:1 7.5% MgCl2:NSW. The sampled heads were collected in 2ml tubes containing metal beads on ice, snap-frozen in liquid nitrogen and stored at -80°C until further use. Total RNA was extracted from frozen head samples using Direct-zol RNA miniprep kit (R2052, Zymo Research).
cDNA synthesis and qRT-PCR analyses
cDNA was generated from total RNA (0.8ug) using Quantitect Reverse Transcription kit (Qiagen, Cat#205310). RT-PCR analyses were performed using LightCycler 480 (Roche) in 10ul final volume, using 1ul of cDNA template. qPCR primers: 22 and also new primers were designed (Universal Probe Library assay design tool, Roche, Supp.Table S10). Analyses were performed as previously described 22.
Statistics
Individual worm rhythm period and power were determined via Lomb-Scargle periodograms using ActogramJ. All other statistical analyses were performed using GraphPad prism software version 8.2.0 and R package (http://www.R-project.org). Behavioral activity level between different time-bins of c-opsin1+/+ and c-opsin1-KO were calculated using one-way ANOVA with Sidak’s multiple comparison test. Overall power strength in rhythmicity was determined by Mann-Whitney-Wilcoxon test. Statistical significance between c-opsin1+/+ and c-opsin1-KO for specific ZTs were accessed by one-way ANOVA with Sidak’s multiple comparison test. Unpaired student’s t-test with Welch’s correction was used for changes in overall transcript levels and peptide quantification. One-way ANOVA with Sidak’s multiple correction test was used to compare between LD 12:12 +UVA vs -UVA experimental conditions. Proteomic data was analyzed using Perseus software, version 1.613 90 or MaxQuant software, version 1.6.0.16 96. Differentially regulated protein was calculated using two-sided student’s t-test (permutation-based FDR correction). GO-term analysis of proteome data was performed using GOstats package (v2.46.0) as mentioned in the reference 82. Significance levels of p-values and adjusted p-values are indicated by *p<0.05, ** p<0.01, *** p<0.001 and **** p<0.0001.
Extended Data
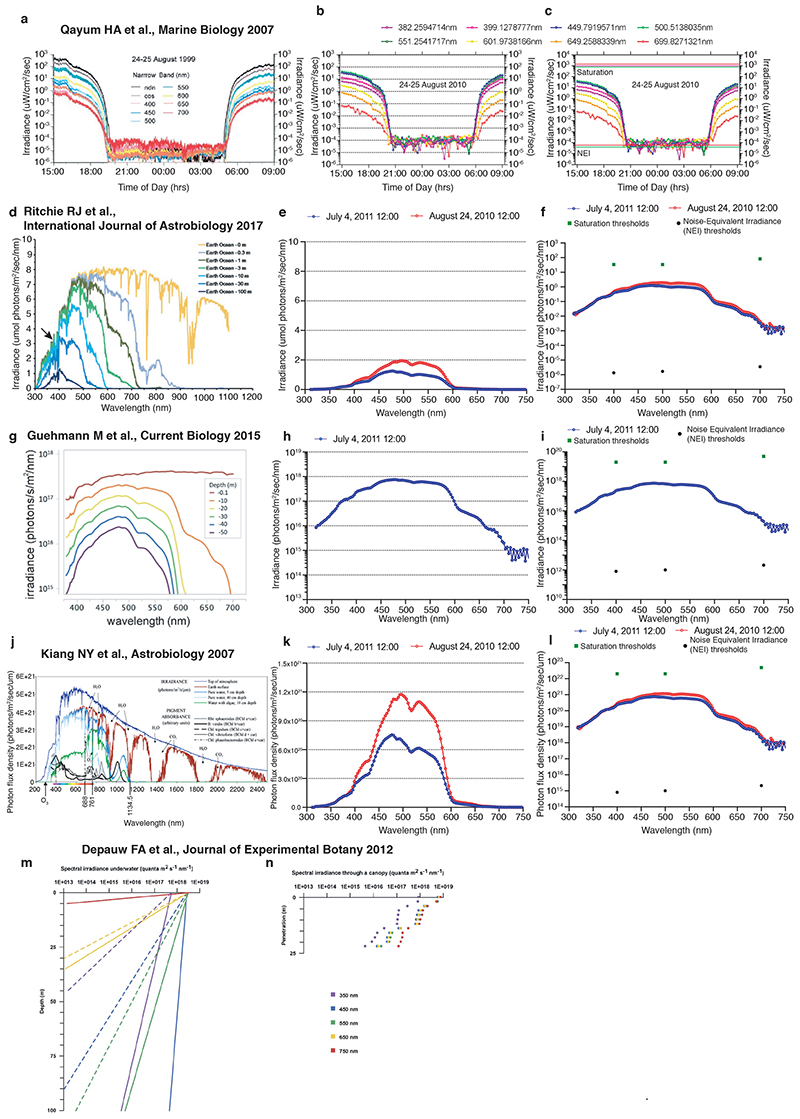
Extended Data Fig. 1. Benchmarking of daylight intensity measurements at 10m with published measurements and calculations.
The units from our data are in each case converted and plotted corresponding to the units and type of plot used in the respective compared publication. (a) Nightlight data measured for individual wavelengths between August 24-25, 1999 98. (b,c) Analysis from (a) performed on natural light data measurement for August 24-25 2010. (c): saturation and noise-equivalent irradiance thresholds are indicated 400nm (pink line), 500nm (green line) and 700nm (red line). (d) Calculations of light irradiance at different ocean depths 99. Black arrow points at UVA spectral range clearly present at 10m water depth and below. (e,f) Our measurements from July 4, 2011 and August 24, 2010 at 12:00 noon for comparison. (g) Irradiance data calculated for different water depths based on in situ measurements of the attenuation coefficients in coastal waters in Corsica using a PhotoreSearch PR-670 spectrophotometer in a custom UW housing on July 4th, 2010, under bright sun at noon 100. (h,i) Our measurements from July 4, 2011 at 12:00 noon timepoint. (j) Irradiance in atmosphere and in water at different depths 101. (k,l) Representative daylight measurements from July 4, 2011 and August 24, 2010 at 12:00 noon timepoint from our 10m measurement set for comparsion. (m,n) Average spectral irradiance and individual wavelength penetration under different ocean depths 102. (f,i,l) Examplary saturation and noise equivalent irradiance (NEI) levels of the RAMSES hyperspectral radiometer indicated as dots.
Extended Data Fig. 2. Daytime spectral irradiance and ratios with and without twilight (10m depth).
(a) Daylight average per day across the year between sunrise to sunset without twilight times for each wavelength. For 3D-rotational graph: Supplementary Data 2. (b) 2-D pcolor plot of (a). (c) Daylight spectrum across the year for each wavelength between astronomical dawn to astronomical dusk (raw data). For 3D-rotational graph: Supplementary Data 3. (d) 2-D pcolor plot of (c). (e) Daytime monthly irradiance averages comparing periods of equal photoperiods. Long day photoperiod example: 14 April – 15 May 2011 (yellow) and 27 July – 27 August 2010 (blue), short day photoperiod example: 9 January – 9 February 2011 (black) and 1 November – 2 December 2010 (red). (f, g) Equinox day spectra without twilight (f) and between astronomical twilight (g). For 3D-plot: Supp.Data 6 and 9. (h,i) Screenshot of weather data for spring and autumnal equinox days: https://www.timeanddate.com/weather. (j, k) 3D-surface plot of equinox day ratio - September 23, 2010/March 21, 2011 (j) and September 23, 2010/March 24, 2011 (k). For 3D-rotational graph: Supplementary Data 7 and 8. (l) Ratios of wavelengths averaged across the day for fall and spring equinox days including twilight times. Black dotted lines: range of strong c-opsin1 activation. Data were corrected for daylight saving time.
Extended Data Fig. 3. Irradiance differences between 4m and 10m water depth.
(a,b) Daylight spectra without twilight of 4m (a) and 10m (b) water depth during June 2011. For 3D-rotational graph: Supplementary data 4 and 5. (c,d) Monthly average 2D plot of (a,b) in logarithmic (c) and in linear scale (d). Blue: 4m, Red: 10m. (e) Wavelength ratios between monthly average of June 2011-4m/June 2011-10m. (f,g) Zoom-in for specific wavelengths ranges from plot (d) for better visualization. Black dotted lines: range of strong c-opsin1 activation. Data were corrected for daylight saving time shifts.
Extended Data Fig. 4. Light spectra and intensity data for experimental light sources.
(a-e) Light intensity and spectra measured using an ILT950 spectrometer (International Light Technologies Inc, Peabody, USA) for (a) standard worm culture illumination, (b,b’) worm culture using ‘NELIS’ in linear plot (b) and logarithmic plot (b’), (c,c’) Behavioural chamber with ‘Nelis’ white light with UVA as linear plot (c) and logarithmic plot (c’). (d,d’) Behavioural chamber with ‘Nelis’ white light and a filter reducing light below 430nm (UVAR filter, Pixelteq, Salvo Technologies, USA) as linear plot (d) and logarithmic plot (d’), (e,e’) Behavioural chamber with ‘Nelis’ white light matching the intensities of the spectrum >430nm from (d,d’) as linear (e) and logarithmic plot (e’). Purple dotted line in b-e indicates the c-opsin1 high activation range. (f,g) Picture details of ‘Nelis’ light source.
Extended Data Fig. 5. G-protein signaling analyses, irradiance dose response curves and spectral sensitivity analyses of Platynereis c-opsin1.
(a) Broad spectrum white light from Arc lamp used for G-protein selectivity assay. (b) Wavelength spectra from CoolLED light source used for spectral characterization of Platynereis c-opsin1. (c) Cells transfected with Platynereis c-opsin1 showed no increase in calcium concentration after 2s white light pulse in calcium bioluminescence assay testing for Gαq binding (purple diamonds: Pdu-c-opsin1, red open circles: human melanopsin, black inverted triangles: no opsin, black arrow: 2s white light pulse). (d) No luminescence increase after 30s white light pulse indicates that Platynereis c-opsin1 does not signal via Gαs (Pdu c-opsin1, Purple diamond; Jellyopsin, red open triangle; no opsin, black inverted triangle; 30s white light pulse, black arrow). (e) Schematic diagram of Platynereis c-opsin1 chimera with second and third intracellular loop regions replaced by corresponding human melanopsin loop region and downstream signaling. (f) Platynereis c-opsin1-human melanopsin chimera depicted in (e) shows a clear response in the calcium luminescence assay, indicative of Gαq-signaling. (g-m) Irradiance dose response curve of Platynereis c-opsin1-melanopsin chimera at 7 different wavelengths.
Extended Data Fig. 6. Platynereis c-opsin1Δ8/Δ8 allele exhibit lowered locomotor activity under longday, including UVA during LD and DD.
(a-c) Double plotted average actogram plot of c-opsin1+/+ (a: n=17), c-opsin1Δ8/+ (b: n=12) and c-opsin1Δ8/Δ8 worms (c: n=20) under longday, including strong UVA under Light-Dark (LD, days 1-5) and Dark-Dark condition (DD, days 6-10, grey shaded). (d) Significant difference in rhythmicity power (PN) exist for c-opsin1Δ8/+ vs c-opsin1Δ8/Δ8 and c-opsin1+/+ vs c-opsin1Δ8/Δ8, but not for c-opsin1+/+ vs c-opsin1Δ8/+ (Mann-Whitney-Wilcoxon test). (e) Percentage of rhythmicity calculated for all genotypes under LD condition (c-opsin1+/+: 100%R, c-opsin1Δ8/+: 100%R and c-opsin1Δ8/ Δ8: 90%R + 10%WR). (f) c-opsin1Δ8/Δ8 worms showed significant decrease in nocturnal locomotor activity compared to c-opsin1+/+ and c-opsin1Δ8/+ (One-way ANOVA with Sidak’s multiple comparison test). (g) Under DD free-running conditions, significant differences in rhythmicity power (PN) exist for c-opsin1+/+ vs c-opsin1Δ8/Δ8 and c-opsin1+/+ vs c-opsin1Δ8/+, but no difference for c-opsin1Δ8/+ vs c-opsin1Δ8/Δ8 (Mann-Whitney-Wilcoxon test). (h) Percentage of rhythmicity calculated for all genotypes under DD condition (c-opsin1+/+: 76.48%R+17.64%WR+5.88%AR, c-opsin1Δ8/+: 33.33%R+41.67%WR+25%AR and c-opsin1Δ8/Δ8: 30%R+30%WR+40%AR). (i) c-opsin1Δ8/Δ8 worms recorded under DD condition showed significant decrease in nocturnal locomotor activity compared to c-opsin1+/+ and c-opsin1Δ8/+ (One-way ANOVA with Sidak’s multiple comparison test). *p<0.05, ** p<0.01, *** p<0.001. For individual actograms see SuppFig.2.
Extended Data Fig. 7. Platynereis c-opsin1Δ8/Δ7 transheterozygous worms exhibit lowered locomotor activity under longday, including UVA conditions.
(a-b) Average, double-plotted actogram of c-opsin1+/+ (a: n=15), c-opsin1Δ8/Δ7 (b: n=21). 3 days of LD. (c) No difference in power (PN) was observed between c-opsin1+/+ and c-opsin1Δ8/Δ7 (Mann-Whitney-Wilcoxon test). (d) Percentage of rhythmicity calculated for all genotypes under LD condition (c-opsin1+/+: 80%R+13.33WR+6.67AR; c-opsin1Δ8/Δ7: 57.14%R+14.29%WR+28.57AR). (e) c-opsin1Δ8/Δ7 worms showed a significant decrease in nocturnal locomotor activity compared to c-opsin1+/+ (One-way ANOVA with sidak’s multiple comparison test). *p<0.05, ** p<0.01, *** p<0.001. For individual actograms see SupplFig.3.
Extended Data Fig. 8. Locomotion under long day (LD 16:8) and intermediate photoperiod (LD 12:12) with full and filter-reduced UVA.
Locomotor behaviour of Platynereis c-opsin1Δ8/Δ8 mutant and its corresponding wt siblings. (a,b) Double plotted average actograms of c-opsin1+/+ worms under long day Nelis white light with intense UVA (+UVA) (a: n=12) and with filter-reduced UVA (-UVA) (b: n=10). (c) c-opsin1+/+ worms under – UVA conditions showed a significantly decrease locomotor activity compared to worms under +UVA conditions and (d) significant decrease in power (PN) and rhythmicity. (e,f) Double plotted average actograms of c-opsin1Δ8/Δ8 worms under LD16:8 +UVA (e, n=11) and -UVA (f, n=10). (g,h) c-opsin1Δ8/ Δ8 worms recorded in (e,f) showed no difference in locomotor activity level (g) and rhythmicity (h). (i,j) Double plotted average actograms of c-opsin1+/+ worms under LD 12:12 +UVA (i: n=9) and -UVA (j: n=7). (k,l) c-opsin1+/+ worms recorded in (i,j) with a difference in locomotor activity close to statistical significance (k), and no difference in rhythmicity (l). (m,n) Double plotted average actograms of c-opsin1Δ8/Δ8 worms under LD 12:12 +UVA (m: n=10) and -UVA (n: n=9). (o,p) c-opsin1Δ8/Δ8 worms recorded in (m,n) showed no difference trend in locomotor activity level (o) and rhythmicity (p). For all statistical comparisons and p values: Suppl.Fig7 Statistics: locomotor activity: One-way ANOVA, Sidak’s multiple comparison test, period, power and rhythmicity: Individual worm rhythmicity and power were determined via Lomb-Scargle periodograms using ActogramJ. The averages of multiple worms were tested by Mann-Whitney-Wilcoxon test. *p<0.05, ** p<0.01, *** p<0.001. Locomotor data of individual worms: Supp.Figs 8, 9.
Extended Data Fig. 9. Head transcript level analyses for additional candidate genes in c-opsin1Δ/8Δ8 and corresponding wildtypes.
Genes as indicated in each panel. The p-value for differences across time was determined by one-way ANOVA. One-way ANOVA with Sidak’s multiple comparison test was used for differences at specific timepoints. Differences between overall transcript levels (measured as AUC) were tested for by Unpaired student’s t-test with Welch’s correction. n.s.- non significant Data displayed as mean+/- S.E.M., n=3BR (5 heads/BR).
Extended Data Fig. 10. Overview of untargeted proteomics experiment under UVA condition.
(a) UVA light used for untargeted proteomics experiment on c-opsin1+/+ and c-opsin1Δ8/Δ8 worms. (b) Sampling scheme. (c) Cellular and pathway model representing differentially regulated protein candidates. For primary data see: Supplementary Tables S13-S15. N=3BRs (20heads/BR), The significance was calculated by two-sided students t-test with adjusted p-value 0.05 (Permutation based FDR correction). Only proteins with least 2 peptides (at least one of it unique) were included in analyses.
Supplementary Material
Acknowledgements
We thank the members of the Tessmar-Raible and Raible groups for discussions. Andrej Belokurov and Margaryta Borysova for excellent worm care at the MFPL aquatic facility, Monika Waldherr, Lukas Orel and Netsaneh Getachew for excellent technical assistance and Natascha Hartl for technical assistance with PRM assays. Targeted proteomics experiments were performed using the Vienna BioCenter Core Facilities (VBCF) instrument pool.
Funding
K.T-R. received funding for this research from the European Research Council under the European Community’s Seventh Framework Programme (FP7/2007–2013) ERC Grant Agreement 337011 and the Horizon 2020 Programme ERC Grant Agreement 819952, the research platform ‘Rhythms of Life’ of the University of Vienna, the Austrian Science Fund (FWF, http://www.fwf.ac.at/en/): START award (#AY0041321), research project grant (#P28970), SFB grant (#SFB F78) and the HFSP (http://www.hfsp.org/) research grant (#RGY0082/2010).
None of the funding bodies was involved in the design of the study, the collection, analysis, and interpretation of data or in writing the manuscript.
Footnotes
Competing interest statement: Max Hofbauer is the CEO of loopbio GmbH, a company developing commercial animal behavioral tracking solutions. C.M. is the CEO of Marine Breeding Systems GmbH, a company developing commercial illumination systems for aquaculture.
All other authors declare no conflict of interest.
Author contributions
V.B.V.R.: experimental work, project planning, data analysis and interpretation, design of figures/data representation, N.S.H.: data analyses, detailed input to/writing of manuscript’s ecological parts, E.A.: natural light and temperature measurements, data analyses, B.P.: light measurements, helped with the design of the automated worm locomotor analysis set-up and NELIS, T.G and M.H..: targeted proteomics data acquisition and analyses, E.G. and R.J.L.: experimental work, data analyses, experimental plans for c-Opsin action spectrum, signalling and Opsin R/M state ratio calculations, M.H.: design of the automated worm locomotor analysis set-up, C.M.: design of the NELIS illumination set-ups, A.B. and C.G.: untargeted proteomics data acquisition and analyses, M.R.d’A: natural light and temperature measurements, detailed feed-back on ecological perspectives, M.C.B: natural light and temperature measurements, K.T-R.: project planning, experimental concepts, data discussion, analyses and interpretation, manuscript writing
Data availability statement
All data generated or analysed during this study are included in this published article and its supplementary information/data files.
The targeted mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via Panorama Public 97 with the identifier PXD014682. The light and temperature measurements, as well as the untargeted proteomics raw data have been deposited at DRYAD: DOI https://doi.org/10.5061/dryad.73n5tb2vv.
References
- 1.Korringa P. Relations between the moon and periodicity in the breeding of marine animals. Ecological Monographs. 1947;17:347–381. [Google Scholar]
- 2.Numata H, Helm B. Annual, lunar, and tidal clocks : patterns and mechanisms of nature’s enigmatic rhythms. Springer; 2014. [Google Scholar]
- 3.Tessmar-Raible K, Raible F, Arboleda E. Another place, another timer: Marine species and the rhythms of life. Bioessays. 2011;33:165–172. doi: 10.1002/bies.201000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shlesinger T, Loya Y. Breakdown in spawning synchrony: A silent threat to coral persistence. Science. 2019;365:1002–1007. doi: 10.1126/science.aax0110. [DOI] [PubMed] [Google Scholar]
- 5.Humphries MM, Studd EK, Menzies AK, Boutin S. To Everything There Is a Season: Summer-to-Winter Food Webs and the Functional Traits of Keystone Species. Integr Comp Biol. 2017;57:961–976. doi: 10.1093/icb/icx119. [DOI] [PubMed] [Google Scholar]
- 6.Burthe S, et al. Phenological trends and trophic mismatch across multiple levels of a North Sea pelagic food web. Marine Ecology Progress Series. 2012;454:119. doi: 10.3354/meps09520. [DOI] [Google Scholar]
- 7.Thackeray SJ, et al. Trophic level asynchrony in rates of phenological change for marine, freshwater and terrestrial environments. Global Change Biology. 2010;16:3304–3313. doi: 10.1111/j.1365-2486.2010.02165.x. [DOI] [Google Scholar]
- 8.Monecke S, et al. Circannual Phase Response Curves to Short and Long Photoperiod in the European Hamster. J Biol Rhythms. 2009;24:413–426. doi: 10.1177/0748730409344502. [DOI] [PubMed] [Google Scholar]
- 9.Thackeray SJ, et al. Phenological sensitivity to climate across taxa and trophic levels. Nature. 2016;535:241–U294. doi: 10.1038/nature18608. [DOI] [PubMed] [Google Scholar]
- 10.Beaugrand G, Brander KM, Lindley JA, Souissi S, Reid PC. Plankton effect on cod recruitment in the North Sea. Nature. 2003;426:661–664. doi: 10.1038/nature02164. [DOI] [PubMed] [Google Scholar]
- 11.Edwards M, Richardson AJ. Impact of climate change on marine pelagic phenology and trophic mismatch. Nature. 2004;430:881–884. doi: 10.1038/nature02808. [DOI] [PubMed] [Google Scholar]
- 12.Soreide JE, Leu E, Berge J, Graeve M, Falk-Petersen S. Timing of blooms, algal food quality and Calanus glacialis reproduction and growth in a changing Arctic. Global Change Biology. 2010;16:3154–3163. doi: 10.1111/J1365-2486.2010.02175.x. [DOI] [Google Scholar]
- 13.Häfker NS, Tessmar-Raible K. Rhythms of behavior: are the times changin’? Curr Opin Neurobiol. 2020;60:55–66. doi: 10.1016/Jconb.2019.10.005. [DOI] [PubMed] [Google Scholar]
- 14.Schiesari L, Kyriacou CP, Costa R. The hormonal and circadian basis for insect photoperiodic timing. FEBS Lett. 2011;585:1450–1460. doi: 10.1016/Jfebslet.2011.02.026. [DOI] [PubMed] [Google Scholar]
- 15.Collins BH, Rosato E, Kyriacou CP. Seasonal behavior in Drosophila melanogaster requires the photoreceptors, the circadian clock, and phospholipase C. Proc Natl Acad Sci U S A. 2004;101:1945–1950. doi: 10.1073/pnas.0308240100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tataroglu O, Emery P. Studying circadian rhythms in Drosophila melanogaster. Methods. 2014;68:140–150. doi: 10.1016/Jymeth.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hegazi S, et al. A Symphony of Signals: Intercellular and Intracellular Signaling Mechanisms Underlying Circadian Timekeeping in Mice and Flies. Int J Mol Sci. 2019;20:2363. doi: 10.3390/ijms20092363. ARTN. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hastings MH, Maywood ES, Brancaccio M. The Mammalian Circadian Timing System and the Suprachiasmatic Nucleus as Its Pacemaker. Biology-Basel. 2019;8:13. doi: 10.3390/biology8010013. ARTN. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dardente H, Wood S, Ebling F, de Miera CS. An integrative view of mammalian seasonal neuroendocrinology. J Neuroendocrinol. 2019;31:e12729. doi: 10.1111/jne.12729. UNSP. [DOI] [PubMed] [Google Scholar]
- 20.Ranzi S. Ricerche sulla biologia sessuale degli Anellidi. Pubbl Staz Zool Napoli. 1931;11:271–292. [Google Scholar]
- 21.Ranzi S. Maturita sessuale degli Anellidi e fasi lunari. Boll Soc Ital Biol Sperim. 1931;6:18. [Google Scholar]
- 22.Zantke J, et al. Circadian and Circalunar Clock Interactions in a Marine Annelid. Cell Reports. 2013;5:99–113. doi: 10.1016/Jcelrep.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zantke J, Oberlerchner H, Tessmar-Raible K. In: Annual, Lunar and Tidal Clocks: Patterns and Mechanisms of Nature’s Enigmatic Rhythms. Numata Hideharu, Helm Barbara., editors. Springer; Japan: 2015. [Google Scholar]
- 24.Fischer A, Dorresteijn AWC. The polychaete Platynereis dumerilii (Annelida): a laboratory animal with spiralian cleavage, lifelong segment proliferation and a mixed benthic/pelagic life cycle. Bioessays. 2004;3:314–325. doi: 10.1002/bies.10409. [DOI] [PubMed] [Google Scholar]
- 25.Zantke J, Bannister S, Veedin Rajan VB, Raible F, Tessmar-Raible K. Genetic and Genomic Tools for the marine annelid Platynereis dumerilii . Genetics. 2014;197:9–31. doi: 10.1534/genetics.112.148254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gambi MC, Lorenti M, Russo GF, Scipione MB, Zupo V. Depth and Seasonal Distribution of Some Groups of the Vagile Fauna of the Posidonia oceanica Leaf Stratum: Structural and Trophic Analyses. Marine Ecology. 1992;13:17–39. doi: 10.1111/J1439-0485.1992.tb00337.x. [DOI] [Google Scholar]
- 27.Somaschini A, et al. Characterization and cartography of some Mediterranean soft-bottom benthic communities (Ligurian Sea, Italy) Scientia Marina. 1998;62:27–36. doi: 10.3989/scimar1998.62n1-227. [DOI] [Google Scholar]
- 28.Galparsoro I, et al. In: Seafloor Geomorphology as Benthic Habitat. Harris Peter T, Baker Elaine K., editors. Elsevier; 2012. pp. 493–507. [Google Scholar]
- 29.Ribera d’ Alcalà M, et al. Seasonal patterns in plankton communities in a pluriannual time series at a coastal Mediterranean site (Gulf of Naples): an attempt to discern recurrences and trends. Scientia Marina. 2004;68:65–83. doi: 10.3989/scimar2004.68s165. [DOI] [Google Scholar]
- 30.Hut RA, Paolucci S, Dor R, Kyriacou CP, Daan S. Latitudinal clines: an evolutionary view on biological rhythms. Proc Biol Sci. 2013;280:20130433. doi: 10.1098/rspb.2013.0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dekens MP, Foulkes NS, Tessmar-Raible K. Instrument design and protocol for the study of light controlled processes in aquatic organisms, and its application to examine the effect of infrared light on zebrafish. PLoS ONE. 2017;12:e0172038. doi: 10.1371/journalpone.0172038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kojima D, et al. UV-sensitive photoreceptor protein OPN5 in humans and mice. PLoS ONE. 2011;6:e26388. doi: 10.1371/journalpone.0026388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato K, et al. Two UV-Sensitive Photoreceptor Proteins, Opn5m and Opn5m2 in Ray-Finned Fish with Distinct Molecular Properties and Broad Distribution in the Retina and Brain. PLoS ONE. 2016;11:e0155339. doi: 10.1371/journalpone.0155339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamashita T, et al. Opn5 is a UV-sensitive bistable pigment that couples with Gi subtype of G protein. Proc Natl Acad Sci U S A. 2010;107:22084–22089. doi: 10.1073/pnas.1012498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ni JD, Baik LS, Holmes TC, Montell C. A rhodopsin in the brain functions in circadian photoentrainment in Drosophila. Nature. 2017;545:340–344. doi: 10.1038/nature22325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zwinkels J. 2015. pp. 1–8.
- 37.Maverakis E, et al. Light, including ultraviolet. J Autoimmun. 2010;34:J247–J257. doi: 10.1016/Jjaut2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ricevuto E, Kroeker KJ, Ferrigno F, Micheli F, Gambi MC. Spatio-temporal variability of polychaete colonization at volcanic CO2 vents indicates high tolerance to ocean acidification. Marine Biology. 2014;161:2909–2919. doi: 10.1007/s00227-014-2555-y. [DOI] [Google Scholar]
- 39.Chapman JW, Reynolds DR, Wilson K. Long-range seasonal migration in insects: mechanisms, evolutionary drivers and ecological consequences. Ecology Letters. 2015;18:287–302. doi: 10.1111/ele.12407. [DOI] [PubMed] [Google Scholar]
- 40.Staples JF. Metabolic Flexibility: Hibernation, Torpor, and Estivation. Comprehensive Physiology. 2016;6:737–771. doi: 10.1002/cphy.c140064. [DOI] [PubMed] [Google Scholar]
- 41.Putman N. Marine migrations. Curr Biol. 2018;28:R972–R976. doi: 10.1016/Jcub.2018.07.036. [DOI] [PubMed] [Google Scholar]
- 42.Alerstam T, Backman J. Ecology of animal migration. Curr Biol. 2018;28:R968–R972. doi: 10.1016/Jcub.2018.04.043. [DOI] [PubMed] [Google Scholar]
- 43.Yokota T, Oishi T. Seasonal Change in the Locomotor-Activity Rhythm of the Medaka, Oryzias-Latipes. Int J Biometeorol. 1992;36:39–44. doi: 10.1007/Bf01208733. [DOI] [PubMed] [Google Scholar]
- 44.Foa A, et al. Seasonal-Changes of Locomotor-Activity Patterns in Ruin Lizards Podarcis-Sicula .1. Endogenous Control by the Circadian System. Behav Ecol Sociobiol. 1994;34:267–274. [Google Scholar]
- 45.Sztarker J, Tomsic D. Neuronal correlates of the visually elicited escape response of the crab Chasmagnathus upon seasonal variations, stimuli changes and perceptual alterations. Journal of Comparative Physiology a-Neuroethology Sensory Neural and Behavioral Physiology. 2008;194:587–596. doi: 10.1007/s00359-008-0333-3. [DOI] [PubMed] [Google Scholar]
- 46.Varpe O. Life History Adaptations to Seasonality. Integrative and Comparative Biology. 2017;57:943–960. doi: 10.1093/icb/icx123. [DOI] [PubMed] [Google Scholar]
- 47.Last KS, Olive PJW, Edwards AJ. An actographic study of diel activity in the semelparous polychaete Nereis (Neanthes) virens Sars in relation to the annual cycle of growth and reproduction. Invertebrate Reproduction & Development. 1999;35:141–145. doi: 10.1080/07924259.1999.9652377. [DOI] [Google Scholar]
- 48.Last KS, Olive PJ. Interaction between photoperiod and an endogenous seasonal factor in influencing the diel locomotor activity of the benthic polychaete Nereis virens . The Biological Bulletin. 2004;206:103–112. doi: 10.2307/1543541. [DOI] [PubMed] [Google Scholar]
- 49.Arendt D, Tessmar-Raible K, Snyman H, Dorresteijn AW, Wittbrodt J. Ciliary photoreceptors with a vertebrate-type opsin in an invertebrate brain. Science. 2004;306:869–871. doi: 10.1126/science.1099955. [DOI] [PubMed] [Google Scholar]
- 50.Tsukamoto H, Chen IS, Kubo Y, Furutani Y. A ciliary opsin in the brain of a marine annelid zooplankton is ultraviolet-sensitive, and the sensitivity is tuned by a single amino acid residue. J Biol Chem. 2017;292:12971–12980. doi: 10.1074/jbcM117.793539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Veraszto C, et al. Ciliary and rhabdomeric photoreceptor-cell circuits form a spectral depth gauge in marine zooplankton. Elife. 2018;7:e36440. doi: 10.7554/eLife.36440. ARTN. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bailes HJ, Lucas RJ. Human melanopsin forms a pigment maximally sensitive to blue light (lambdamax approximately 479 nm) supporting activation of G(q/11) and G(i/o) signalling cascades. Proc Biol Sci. 2013;280:20122987. doi: 10.1098/rspb.2012.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bailes HJ, Zhuang LY, Lucas RJ. Reproducible and sustained regulation of Galphas signalling using a metazoan opsin as an optogenetic tool. PLoS ONE. 2012;7:e30774. doi: 10.1371/journalpone.0030774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sugihara T, Nagata T, Mason B, Koyanagi M, Terakita A. Absorption Characteristics of Vertebrate Non-Visual Opsin, Opn3. PLoS ONE. 2016;11:e0161215. doi: 10.1371/journalpone.0161215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ballister ER, Rodgers J, Martial F, Lucas RJ. A live cell assay of GPCR coupling allows identification of optogenetic tools for controlling Go and Gi signaling. BMC Biol. 2018;16:10. doi: 10.1186/s12915-017-0475-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tichy AM, Gerrard EJ, Sexton PM, Janovjak H. Light-activated chimeric GPCRs: limitations and opportunities. Curr Opin Struct Biol. 2019;57:196–203. doi: 10.1016/Jsbi2019.05.006. [DOI] [PubMed] [Google Scholar]
- 57.Almenar-Queralt A, et al. Presenilins regulate neurotrypsin gene expression and neurotrypsin-dependent agrin cleavage via cyclic AMP response element-binding protein (CREB) modulation. J Biol Chem. 2013;288:35222–35236. doi: 10.1074/jbcM113.513705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hidaka C, Kashio T, Uchigaki D, Mitsui S. Vulnerability or resilience of motopsin knockout mice to maternal separation stress depending on adulthood behaviors. Neuropsychiatr Dis Treat. 2018;14:2255–2268. doi: 10.2147/NDtS170281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakajima T. Roles of Sulfur Metabolism and Rhodanese in Detoxification and Anti-Oxidative Stress Functions in the Liver: Responses to Radiation Exposure. Med Sci Monit. 2015;21:1721–1725. doi: 10.12659/MSM.893234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu J, Holmgren A. The thioredoxin antioxidant system. Free Radic Biol Med. 2014;66:75–87. doi: 10.1016/Jfreeradbiomed2013.07.036. [DOI] [PubMed] [Google Scholar]
- 61.Lincoln GA, Clarke IJ, Hut RA, Hazlerigg DG. Characterizing a mammalian circannual pacemaker. Science. 2006;314:1941–1944. doi: 10.1126/science.1132009. [DOI] [PubMed] [Google Scholar]
- 62.Hankins MW, Davies WIL, Foster RG. In: Evolution of Visual and Non-visual Pigments. Hunt David M, Hankins Mark W, Collin Shaun P, Marshall N Justin., editors. Springer US; 2014. pp. 65–103. [Google Scholar]
- 63.Nakane Y, Shimmura T, Abe H, Yoshimura T. Intrinsic photosensitivity of a deep brain photoreceptor. Curr Biol. 2014;24:R596–597. doi: 10.1016/Jcub.2014.05.038. [DOI] [PubMed] [Google Scholar]
- 64.Halford S, et al. VA opsin-based photoreceptors in the hypothalamus of birds. Curr Biol. 2009;19:1396–1402. doi: 10.1016/Jcub.2009.06.066. [DOI] [PubMed] [Google Scholar]
- 65.Hunt DM, Hankins MW, Collin SP, Marshall NJ. Evolution of visual and non-visual pigments. Springer; 2014. [Google Scholar]
- 66.Kiang NY, Siefert J, Govindjee, Blankenship RE. Spectral signatures of photosynthesis. i Review of Earth organisms. Astrobiology. 2007;7:222–251. doi: 10.1089/ast2006.0105. [DOI] [PubMed] [Google Scholar]
- 67.Bracher AU, Tilzer MM. Underwater light field and phytoplankton absorbance in different surface water masses of the Atlantic sector of the Southern Ocean. Polar Biology. 2001;24:687–696. doi: 10.1007/s003000100269. [DOI] [Google Scholar]
- 68.Reierth E, Van’t Hof TJ, Stokkan KA. Seasonal and daily variations in plasma melatonin in the high-arctic Svalbard ptarmigan (Lagopus mutus hyperboreus) J Biol Rhythms. 1999;14:314–319. doi: 10.1177/074873099129000731. [DOI] [PubMed] [Google Scholar]
- 69.Arnold W, et al. Circadian rhythmicity persists through the Polar night and midnight sun in Svalbard reindeer. Scientific Reports. 2018;8:14466. doi: 10.1038/s41598-018-32778-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Häfker N, Teschke M, Hüppe L, Meyer B. Calanus finmarchicus diel and seasonal rhythmicity in relation to endogenous timing under extreme polar photoperiods. Marine Ecology Progress Series. 2018;603 doi: 10.3354/meps12696. [DOI] [Google Scholar]
- 71.Hüppe L, et al. Evidence for oscillating circadian clock genes in the copepod Calanus finmarchicus during the summer solstice in the high Arctic. Biol Lett. 2020;16:20200257. doi: 10.1098/rsbl2020.0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ashley NT, et al. Revealing a circadian clock in captive arctic-breeding songbirds, lapland longspurs (Calcarius lapponicus), under constant illumination. J Biol Rhythms. 2014;29:456–469. doi: 10.1177/0748730414552323. [DOI] [PubMed] [Google Scholar]
- 73.Reierth E, Stokkan K-A. Activity rhythm in High Arctic Svalbard ptarmigan (Lagopus mutus hyperboreus) Can J Zool. 1998;76:2031–2039. doi: 10.1139/z98-173. [DOI] [Google Scholar]
- 74.Lu W, Meng QJ, Tyler NJ, Stokkan KA, Loudon AS. A circadian clock is not required in an arctic mammal. Curr Biol. 2010;20:533–537. doi: 10.1016/Jcub.2010.01.042. [DOI] [PubMed] [Google Scholar]
- 75.Wallace M, et al. Comparison of zooplankton vertical migration in an ice-free and a seasonally ice-covered Arctic fjord: An insight into the influence of sea ice cover on zooplankton behavior. Limnology and oceanography. 2010;55:831–845. doi: 10.4319/lo.2009.55.2.0831. [DOI] [Google Scholar]
- 76.Kobelkova A, et al. Continuous activity and no cycling of clock genes in the Antarctic midge during the polar summer. J Insect Physiol. 2015;81 doi: 10.1016/Jjinsphys.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 77.Stelzer RJ, Chittka L. Bumblebee foraging rhythms under the midnight sun measured with radiofrequency identification. BMC Biol. 2010;8:93. doi: 10.1186/1741-7007-8-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nordtug T, Thor BM. Diurnal variations in natural light conditions at summer time in arctic and subarctic areas in relation to light detection in insects. Ecography. 1988;11:2020–2209. [Google Scholar]
- 79.Chittka L, Stelzer RJ, Stanewsky R. Daily changes in ultraviolet light levels can synchronize the circadian clock of bumblebees Bombus terrestris . Chronobiol Int. 2013;30:434–442. doi: 10.3109/07420528.2012.741168. [DOI] [PubMed] [Google Scholar]
- 80.Tosches MA, Bucher D, Vopalensky P, Arendt D. Melatonin Signaling Controls Circadian Swimming Behavior in Marine Zooplankton. Cell. 2014;159:46–57. doi: 10.1016/Jcell2014.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sugden D, Cena V, Klein DC. Hydroxyindole O-Methyltransferase. Methods Enzymol. 1987;142:590–596. doi: 10.1016/s0076-6879(87)42070-3. [DOI] [PubMed] [Google Scholar]
- 82.Schenk S, et al. Combined transcriptome and proteome profiling reveals specific molecular brain signatures for sex, maturation and circalunar clock phase. Elife. 2019;8 doi: 10.7554/eLife.41556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Govardovskii VI, Fyhrquist N, Reuter T, Kuzmin DG, Donner K. In search of the visual pigment template. Vis Neurosci. 2000;17:509–528. doi: 10.1017/s0952523800174036. [DOI] [PubMed] [Google Scholar]
- 84.Stavenga DG, Oberwinkler J, Postma M. In: Handbook of Biological Physics. Stavenga DG, DeGrip WJ, Pugh EN, editors. Vol. 3. North-Holland: 2000. pp. 527–574. [Google Scholar]
- 85.Duffett-Smith P, Zwart J. Practical Astronomy with your Calculator or Spreadsheet. 4 edn. Cambridge University Press; 2011. [Google Scholar]
- 86.Bannister S, et al. TALE Nucleases mediate efficient, heritable genome modifications in the marine annelid Platynereis dumerilii . Genetics. 2014;197:19–31. doi: 10.1534/genetics112.148254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cermak T, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dahlem TJ, et al. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 2012;8:e1002861. doi: 10.1371/journalpgen1002861. PGENETICS-D-12-00762 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Meeker ND, Hutchinson SA, Ho L, Trede NS. Method for isolation of PCR-ready genomic DNA from zebrafish tissues. Biotechniques. 2007;43:610, 612, 614. doi: 10.2144/000112619. [DOI] [PubMed] [Google Scholar]
- 90.Tyanova S, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 91.Caers J, et al. Peptidomics of Neuropeptidergic Tissues of the Tsetse Fly Glossina morsitans morsitans. J Am Soc Mass Spectrom. 2015;26:2024–2038. doi: 10.1007/s13361-015-1248-1. [DOI] [PubMed] [Google Scholar]
- 92.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–1906. doi: 10.1038/nprot2007.261. [DOI] [PubMed] [Google Scholar]
- 93.Dorfer V, et al. MS Amanda, a universal identification algorithm optimized for high accuracy tandem mass spectra. J Proteome Res. 2014;13:3679–3684. doi: 10.1021/pr500202e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Käll L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 95.MacLean B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 2016;11:2301–2319. doi: 10.1038/nprot2016.136. [DOI] [PubMed] [Google Scholar]
- 97.Sharma V, et al. Panorama Public: A Public Repository for Quantitative Data Sets Processed in Skyline. Mol Cell Proteomics. 2018;17:1239–1244. doi: 10.1074/mcp.RA117.000543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Qayum HA, Klimley AP, Newton R, Richert JE. Broad-band versus narrow-band irradiance for estimating latitude by archival tags. Marine Biology. 2007;151:467–481. doi: 10.1007/s00227-006-0514-y. [DOI] [Google Scholar]
- 99.Ritchie RJ, Larkum AWD, Ribas I. Could photosynthesis function on Proxima Centauri b? International Journal of Astrobiology. 2018;17:147–176. doi: 10.1017/S1473550417000167. [DOI] [Google Scholar]
- 100.Guehmann M, et al. Spectral Tuning of Phototaxis by a Go-Opsin in the Rhabdomeric Eyes of Platynereis. Curr Biol. 2015;25:2265–2271. doi: 10.1016/Jcub.2015.07.017. [DOI] [PubMed] [Google Scholar]
- 101.Kiang NY, Siefert J, Govindjee, Blankenship RE. Spectral signatures of photosynthesis i Review of Earth organisms. Astrobiology. 2007;7:222–251. doi: 10.1089/ast2006.0105. [DOI] [PubMed] [Google Scholar]
- 102.Depauw FA, Rogato A, Ribera d’Alcalá M, Falciatore A. Exploring the molecular basis of responses to light in marine diatoms. Journal of Experimental Botany. 2012;63:1575–1591. doi: 10.1093/jxb/ers005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information/data files.
The targeted mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via Panorama Public 97 with the identifier PXD014682. The light and temperature measurements, as well as the untargeted proteomics raw data have been deposited at DRYAD: DOI https://doi.org/10.5061/dryad.73n5tb2vv.