Abstract
Broad spectrum antiviral platforms that can decrease or inhibit viral infection would alleviate many threats to global public health. Nonetheless, effective technologies of this kind are still not available. Here we describe a programmable icosahedral canvas for the self-assembly of icosahedral shells that have viral trapping and antiviral properties. Programmable triangular building blocks constructed from DNA assemble with high yield into various shell objects with user-defined geometries and apertures. We create shells with molecular masses ranging from 43 to 925 Megadaltons (8 to 180 subunits) and with internal cavity diameters ranging up to 280 nm. The shell interior can be functionalized with virus-specific moieties in a modular fashion. We demonstrate this virus-trap concept by engulfing hepatitis B virus (HBV) core particles and adeno-associated viruses (AAV). We show inhibition of HBV core interactions with surfaces in vitro and demonstrate neutralization of infectious AAV exposed to human cells.
Introduction
For the majority of viral diseases, no effective treatment is available. Broadly applicable antiviral platform technologies do not exist. Here, we propose a platform technology that considers trapping entire virus particles within de novo designed macromolecular shells to inhibit molecular interactions between viruses and host cells (Fig. 1a). We envision shells that can augment and work synergistically with a large variety of virus binding moieties, whether by themselves neutralizing or not, to create an effective antiviral agent.
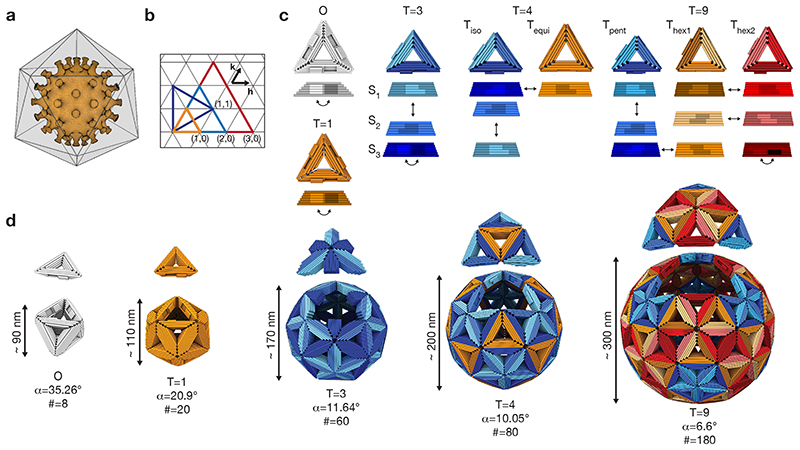
Fig. 1. Design principles.
(a) Icosahedral shell encapsulating a virus capsid. (b) Triangular net representation of icosahedral shells. Each colored triangle represents one of the 20 faces forming an icosahedron. The small triangles represent the triangular building blocks. (h,k) indicates the location of pentamers within a shell. (c) Cylindrical model of DNA-origami triangles assembling into the shells shown in (d). The edges of the triangles are beveled and modified with shape-complementary protrusions (light) and recesses (dark). The arrows indicate shape-complementary combinations. For design details see Supplementary Fig. 37 to 40. (d) Icosahedral shells formed by the triangles shown in (c). For each shell design, one of its 20 icosahedral faces has been displaced (see (b)) to help recognize the icosahedral symmetry. α is the bevel angle of the sides, # the number of DNA-origami triangles building the shell.
To accomplish this function, the shells must on the one hand be large enough to accommodate entire viruses, while also be chemically addressable to allow including virus-specificity conferring moieties on the shell’s interior surface. The extended surface of the shells enables functionalization in a multivalent fashion. Multivalency can support tight binding of a target virus even for individually weakly virus-binding molecules, as exemplified in previous experiments with phage nanoparticles engineered to trivalently bind influenza A hemagglutinin (1), and with star-shaped DNA aptamer clusters that simultaneously target multiple dengue virus envelope proteins (2). With shells that fully cover viruses, an even larger degree of multivalency, and thus stronger binding, can be envisioned. Modular functionalization of the shells with virus binders will enable using the same type of shell platform to target a variety of viruses. Candidate virus binders could be e.g. antibodies, designed proteins (3), nucleic acid aptamers, or polymers such as heparan sulphates (4). In our concept, the shell material, rather than the moieties directly contacting the virus, will mainly prevent access to the viral surface. Therefore, in principle any virus binding molecule could potentially be utilized to convert the shells into an effective virus-neutralizing trap.
Our shell concept requires constructing massive molecular complexes that are adaptable to cover the dimensions of viral pathogens (~ 20 nm to ~ 500 nm (5)), which poses a fundamental nanoengineering challenge. Protein designers have previously succeeded in creating artificial macromolecular cages (7–10). However, the designed protein-cages are much smaller than the vast majority of natural viruses and cannot be easily modified. DNA nanotechnology (11–16) can create discrete objects with structurally well-defined 3D shapes (17, 18), including higher-order objects (19–23) with molecular masses exceeding one Gigadalton (24). However, these previous designs and the underlying concepts yield objects that are either too small, assemble with insufficient yields, do not match the shapes of viruses, or are too flexible or skeletal to be suitable for effectively trapping and occluding entire virus particles.
To build the envisioned virus trap, we created a programmable icosahedral shell “canvas” by adapting symmetry principles known from natural viral capsids. Caspar and Klug elucidated the geometric principles that govern the structure of natural viral capsids in 1962 (25). According to Caspar and Klug theory, which has been expanded recently (26), the number of distinct environments occupied by proteins within an icosahedral capsid is described by its triangulation number (T-number), which can be computed by the arrangement of pentamers and hexamers within an icosahedral capsid (T=h2+hk+k2, Fig. 1b). The total number of proteins required to build a natural capsid is T times sixty. This is because natural protein subunits are, by default, asymmetric and homo-trimerization is minimally required to construct a three-fold symmetric subunit that can assemble into an icosahedral shell with twenty triangular faces. To build larger capsids, viruses use more than one capsid protein or capsid proteins that can adopt different conformations. The structure of natural virus capsids forms the basis for our synthetic programmable icosahedral shell canvasses, which we analogously classify using a T-number.
Shell canvas design principles
To implement the icosahedral canvas concept, we designed pseudo-symmetric triangular subunits (Fig. 1c) based on multi-layer DNA origami concepts (12, 13). Each side of a triangular subunit is the equivalent of one protein subunit of a natural viral capsid. The overall canvas scale and type are controlled by geometric instructions provided by the triangular subunits. These instructions are given by the choice of the length, the topological binding pattern (27), and the bevel angle of each triangular edge. Since in our system each triangular edge represents one protein, the Caspar and Klug triangulation number gives the number of unique triangular edges required to build a particular icosahedral canvas shell. Hence, T=1 and T=3 shells may both be built with a single triangle, with three identical edges for T=1 and three different edges for a T=3 shell (Fig. 1c, 1d, left). A T=4 shell requires two separate triangular subunits, for example, one triangle with three unique edges and another with three identical edges (Fig. 1c, 1d, middle). A T=9 shell requires three different triangles, each having three unique edges (Fig. 1c, 1d, right). The greater the T number, the greater the overall number of triangles per target shell, given by 20T. We used design solutions in which all triangle bevel angles for a particular target shell were the same. While T=9 was the largest canvas we set out to build, we also designed triangular subunits for a smaller octahedral container (“O”) (Fig. 1c, 1d, left).
Subunit and shell canvas assembly
We used iterative design with caDNAno (28) paired with elastic-network-guided molecular dynamics simulations (29) to produce candidate designs. To approximate target bevel angles, we tuned the helical connectivity of the triangle edges in the vertices (Supplementary Fig. 1). These candidate designs were encoded in DNA sequences using the methods of DNA origami (12, 30) and self-assembled in one-pot reaction mixtures (31). Gel-electrophoretic folding quality analysis demanded some design iterations to improve triangular subunit assembly yields (Supplementary Fig. 2). To validate the 3D structures of the designed triangles, we studied all triangle subunits using cryo transmission electron microscopy (cryo-EM) single particle analysis (Fig. 2). The resulting 3D electron maps had resolutions ranging from 13 to 22 Angstroms (Supplementary Fig. 3-10), which allowed us to evaluate the overall 3D shapes, the observed versus desired bevel angles (Supplementary Table 1, deviations within 5°), the correct formation of the binding patterns, and the occurrence of systematic folding defects. For instance, one triangle variant (Thex1) had a defective vertex, which decreased its ability to form lateral edge-to-edge interactions (Supplementary Fig. 11). Based on the cryo-EM data, we refined the design and eliminated the defect.
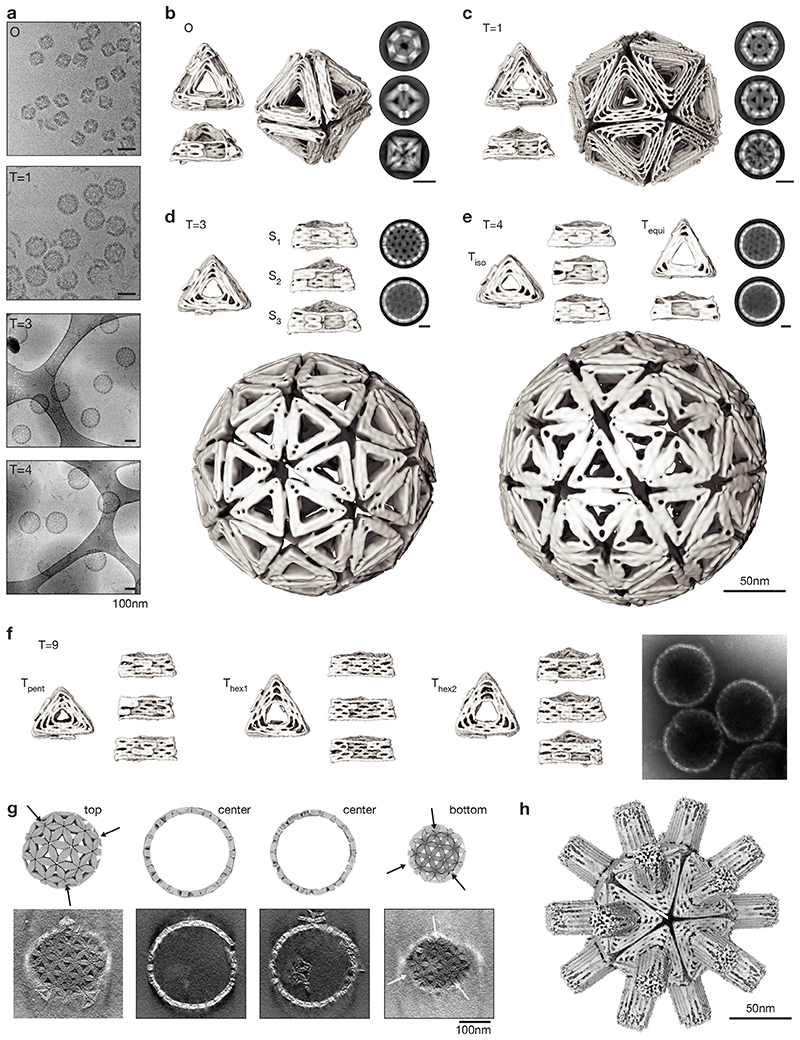
Fig. 2. Structures of shells and of shell subunits.
(a) Cryo-EM micrographs of assembled shells in free-standing ice (O, T=1) and on lacey carbon grids with carbon support (T=3, T=4). (b to e) Cryo-EM reconstructions of shell subunits and fully assembled shells (octahedron to T=4 shells). The two-dimensional class averages show assembled shells from different orientations, scale bars 50nm. See also Supplementary Video 1-4. (f) Cryo-EM reconstructions of the three triangles assembling into a T=9 shell and negatively stained EM micrograph of assembled shells. (g) Comparison of slices through a model T=9 shell to slices of a tomogram calculated from an EM tilt series. The arrows indicate the positions of pentamers within the T=9 shell. See also Supplementary Video 5. (h) Cryo-EM reconstruction of a T=1 shell with a central-cavity blocking DNA “spacer” module. See also Supplementary Video 8.
The triangle variants self-assembled (see methods) successfully into the designed icosahedral shells, as confirmed by direct imaging with cryo-EM (Fig. 2a, Supplementary Fig. 12-16). Inspection of individual particles (Fig. 2a) and of 2D class averages (Fig. 2b-f) revealed particles displaying the designed symmetries. For example, the three symmetry axes of the octahedron (4-fold, 3-fold, 2-fold, Fig. 2b) and T=1 shell (5-fold, 3-fold, 2-fold, Fig. 2c) can be clearly seen. For the higher-T-number shells, the underlying triangular net predicted from the Caspar-and-Klug representation became clearly visible (Fig. 2d-g). We determined 3D EM maps from the image data by imposing the respective symmetry (Fig. 2b-e). The resulting maps had resolutions ranging from 20 to 40 Angstrom. For the octahedron and T=1 shell, 3D maps reconstructed without imposing any a priori symmetry superimposed well with the sibling reconstructed with imposed symmetry (Supplementary Fig. 12,13). We classified and treated cryo-EM maps of shells that lacked one or multiple triangles separately from complete shells to assess quality and yield (Supplementary Fig. 12,13). The largest T=9 shells were imaged using negative stain EM tomography (Fig. 2f,g, Supplementary Fig. 17). Sections through tomograms of assembled T=9 shells show fully closed shells as well as the correct arrangement of pentamers according to the designed T-number (arrows in Fig. 2g and Supplementary Fig. 17).
To elucidate effects of orientational specificity of subunit-subunit interactions, we varied the bevel angle of the T=1 subunits from the ideal geometry (α=20.9°). We designed two additional variants of the T=1 triangle whose bevel angles deviated by +5° or -5° from the icosahedral ideal. The decrease or increase of the bevel angle caused the appearance of larger or smaller, often defective, assemblies in addition to T=1 shells, respectively (Supplementary Fig. 18). Based on these data we conclude that the correct target bevel angle in a T=1 triangle subunit must be matched within a range of +-5°.
As a demonstration for a route for sealing the remaining cavities in the shells, we built a DNA brick having a triangular cross-section roughly corresponding to the dimensions of the triangular cavity in the shell subunits. We anchored the brick via multiple attachment points to the outer surface of a T=1 shell triangle (Supplementary Fig. 19 for design details, Supplementary Fig. 20 for a cryo-EM map). We solved a structure of the spiky T=1 shell using cryo-EM single particle analysis (Fig. 2h, Supplementary Fig. 21). The resulting map overlaps well with those of the unmodified T=1 shell, but the central cavity of the triangle subunits is now blocked by the added brick module (Supplementary Fig. 21). The fact that the cavity-plugging with the DNA brick worked indicates robustness and structural modularity of our shells. The brick may also be considered as a mimic for previously described DNA-based membrane channels (32) or for any other functional module that one wishes to attach to a shell.
Shell yield and stability
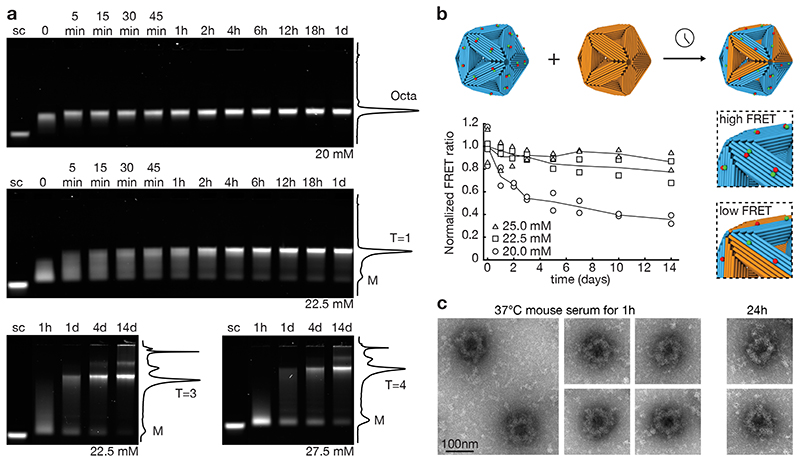
We evaluated practical aspects such as assembly yield and stability in physiological conditions where the system is ultimately expected to be applied. Low-density gel electrophoretic mobility analysis (Fig. 3a, Supplementary Fig. 22) revealed that shell assembly proceeded by disappearance of the triangular monomers, appearance of a smear indicating the presence of oligomeric species, followed by emergence of a dominant high intensity band, corresponding to the fully formed shells. Octahedra and T=1 shells formed within 15 and 60 minutes, respectively, which is sufficiently fast to enable self-assembly of these shells directly during the one-pot triangle-folding reaction (Supplementary Fig. 23). They formed with a final complete shell yield of ~95% and ~70%, respectively. The T=3 and T=4 shells formed with about 40% yield (Fig. 3a). Subunit-exchange experiments with fluorescently labeled subunits revealed that under shell-favoring conditions triangles that are incorporated in closed shells do not exchange with solution (Fig. 3b, Supplementary Fig. 24). Under equilibrium conditions, triangles do exchange (Fig. 3b, Supplementary Fig. 24). To stabilize the shells for application in physiological fluids, we first assembled the shells and then applied UV point welding (33) to create additional internal covalent bonds across the stacking contacts in the triangle subunits. We then coated the shells with a mixture of oligolysine and PEG oligolysine (34). This two-step treatment allowed us to successfully transfer the shells into mouse serum, where the shells remained intact for up to 24 h (Fig. 3c).
Fig. 3. Shell yield and stability.
(a) Laser-scanned fluorescent images of 0.5% agarose gels showing the assembly of octahedra, T=1, T=3 and T=4 shells at 40°C with a monomer concentration of 5 nM at different time points. Solid lines give cross-sectional lane intensity profiles from the 1d (octahedron, T=1) and 14d samples (T=3, T=4). sc: M13-8064 scaffold strand as reference. (b) Triangle exchange experiments. Cyan: FRET-pair labeled T=1 shells. Orange: unlabeled shells. Symbols give FRET signals measured at different incubation times in the presence of the indicated concentrations of Mg2+ for n=2 independent measurements (design details see Supplementary Fig. 44). (c) Negative-staining TEM image of octahedral shells coated with a 1:1 mixture of oligolysine and oligolysine-PEG and incubated for 1 h and 24h in 55% mouse serum at 37°C.
Sculpting on the icosahedral canvas
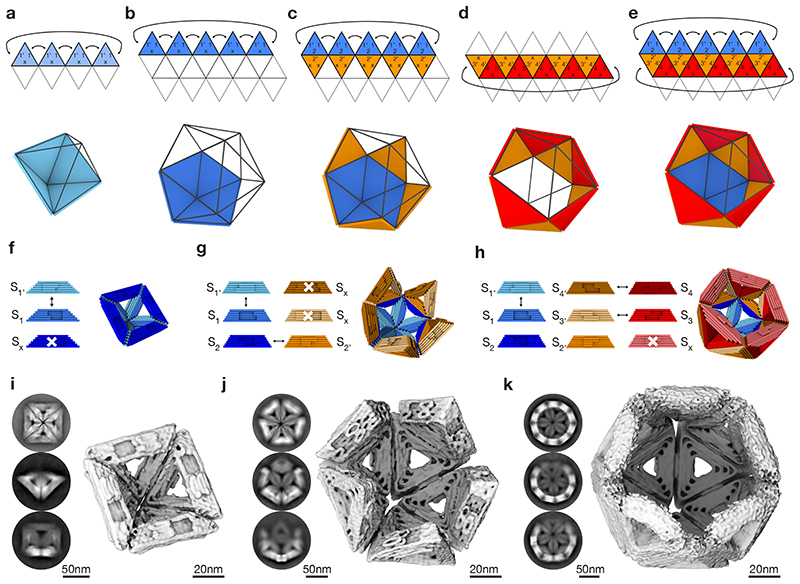
By changing the geometry of the shape-complementary topographic features, the triangular subunits can be programmed to cover only user-defined areas on the icosahedral canvas. To create full shells, only the minimum number of different topographic interaction patterns (“symmetries”) is implemented as discussed above. Introducing additional types of topographic edge-to-edge interactions per triangular subunit allows reducing the symmetry in which the subunit may be integrated in the canvas. Furthermore, the stacking interactions can be modularly activated and de-activated, for example by shortening a strand terminus involved in a stacking contact or by adding unpaired thymidine terminal strand extensions. Together, these features enable sculpting a variety of objects on the icosahedral canvas in a programmable fashion, including full shells, pentagonal vertices, (spherical) half-shells, and shells with virus-sized openings using rational design decisions.
To design such objects, we used the triangular net projection of the chosen icosahedral canvas type as a drawing board (Fig. 4a-e). For example, in order to prepare half instead of full octahedra, complementary lock-and-key interactions of two edges of the triangular subunit are needed and one edge interaction must be deactivated (Fig. 4a). A pentagonal dome can be analogously created based on the T=1 icosahedral canvas (Fig. 4b). Building an icosahedral half shell requires two different triangular subunits, one that forms the pentagonal dome, and another that specifically docks onto the edges of the pentamer (Fig. 4c). A ring-like “sheath” may also be built by two triangles (Fig. 4d). To build a T=1 shell variant with one missing pentagon vertex, three triangular subunit variants with a specific interaction pattern are needed (Fig. 4e). We practically implemented the above discussed design variants using appropriately modified triangular building blocks (Fig. 4f-h, Supplementary Fig. 25). The building blocks self-assembled successfully into the desired higher-order objects based on their icosahedral canvas, which we validated experimentally by determining cryo EM solution structures (Fig. 4i-k, Supplementary Fig. 26-28) and negative stain TEM images (Supplementary Fig. 29).
Fig. 4. Sculpting on an icosahedral canvas.
(a-e) Triangular net projection and schematics of different partial shells: half octahedral shell (a), pentamer (b), half T=1 shell (c), ring (d) and T=1 shell lacking a pentagon vertex (e). (f-h) Cylindrical models of DNA-origami triangles and corresponding partial shells of the half-octahedral shell (f), half T=1 shell (g) and T=1 shell lacking a pentagon vertex (h). The sides of the triangles are modified with protrusions and recesses. The arrows indicate shape-complementary sides. White crosses indicate deactivated interaction sites. For design details see Supplementary Fig. 41, 42. (i-k) Cryo-EM 3D reconstructions of the partial shells shown in (f-h). Insets give typical two-dimensional class averages showing assembled shells from different orientations.
Virus trapping
Viruses can be trapped in, or coordinated by, pre-assembled icosahedral shell segments featuring sufficiently large apertures (Fig. 5a,b, Supplementary Fig. 30). Alternatively, protective shells can be formed directly on the surface of virus particles (Fig. 5c). Both approaches are illustrated in experiments we performed with hepatitis B virus core particles (HBV) (Fig. 5a-c inset, red). To confer specificity to HBV, we conjugated anti-HBc 17H7 (Isotype IgG-2b) to the DNA shells by hybridization of ssDNA-labeled antibodies to a set of anchor points on the triangle subunits (Fig. 5a-c inset, cyan). We did not observe any HBV binding in the absence of HBV antibodies (Supplementary Fig. 31), nor in the presence of antibodies specific for other targets (Supplementary Fig. 31).
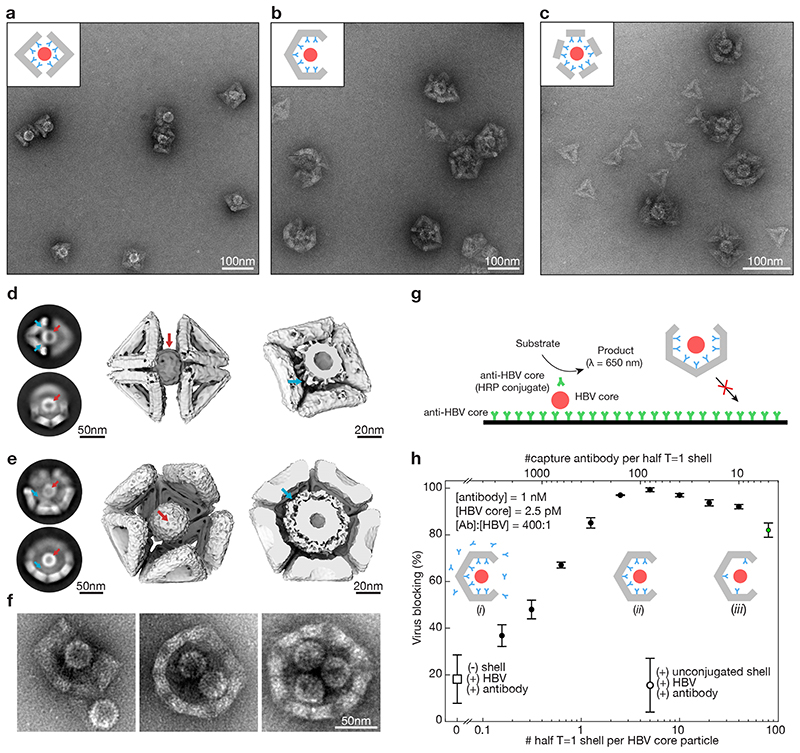
Fig. 5. Trapping of hepatitis B virus (HBV) core particles.
(a) Negative stain TEM images of HBV core particles trapped in octahedral half-shells. Inset: schematic representation of two octahedral half-shells (grey) equipped with antibodies (cyan) with a trapped HBV core particle (red). (b) Same as in (A) with half T=1 shells. (c)) Same as in (A) with monomeric T=1 triangles modified with nine antibodies self-assembled around HBV core particles. (d) Left: Two-dimensional EM class-averages. Middle: Cryo-EM reconstruction of two octahedral half-shells coordinating a trapped hepatitis-B virus particle. See Supplementary Video 6,7. Right: Cut through the cryo-EM map with the HBV core particle trapped. The density around the HBV core particle stems from the antibodies connecting the HBV core particles to the octahedral shell. Red arrows: HBV core particle. Cyan arrows: antibodies connecting the shell to the HBV core particle. (e) Same as in (d) for the T=1 half-shell. The electron density thresholds differ, which makes the HBV core particle look thicker in the T=1 half-shell compared to the half octahedron (right). (f) Negative stain TEM images of T=1 shells with a missing pentagon vertex engulfing up to three HBV core particles. (g) Schematic representation of the in vitro virus blocking ELISA experiment. (h) In vitro virus blocking ELISA experiments. All experiments are done at a ratio of antibody (Ab) to HBV of 400:1. The half-shells have 90 antibody binding sites. Solid filled dots indicate 2.5 pM HBV core particles incubated with pre-assembled mixtures of 1 nM oligonucleotide-conjugated capture antibody and various concentrations of half T=1 shells. Insets: (i) low half-shell concentration for which antibodies saturate the half-shell binding sites and excess antibodies are in solution. (ii) half-shells are saturated with antibody with little antibody remaining in solution. (iii) high half-shell concentration for which antibodies are not saturating all half-shell binding sites. Controls: the open dot represents a mixture of HBV core particles, antibodies and unfunctionalized T=1 half-shells; the open square represents HBV plus antibody without half-shells. The green dot shows a blocking efficiency of about 80% at only a 5:1 ratio of Ab:half-shells. Data is presented as mean ± s.d., n = 3 independent measurements.
We determined 3D cryo EM maps of octahedral and T=1 half shells with trapped HBV core particles (Fig. 5d,e, Supplementary Fig. 32,33). For the half-octahedral variant, the majority of particles consisted of two opposing half octahedra coordinating a single HBV core particle in their middle (Fig. 5d, Supplementary Fig. 32). The micrographs and the cryo EM map also reveal signatures reflecting the antibodies that link the DNA shell to the trapped HBV core particle (Fig. 5d, right). Similar antibody signatures may be found in the image data with the half T=1 shell-HBV complex (Fig. 5e, right). We also trapped HBV core particles in larger T=1 shell variants with a missing pentagon vertex (Fig. 5f) which can accommodate multiple HBV particles in their interior cavities (Fig. 5f, Supplementary Fig. 30).
To test the capacity of our shells to prevent a trapped virus to undergo interactions with surfaces, we performed in vitro virus blocking assays with HBV-binding antibodies immobilized on a solid surface (Fig. 5g, Supplementary Fig. 34). We quantified the extent of HBV core particle binding to the surface via binding of an orthogonal HBV core-specific reporter antibody coupled to horseradish peroxidase (HRP). Residual HBV core particles that are bound to the surface are detected via HRP catalyzed production of a colorimetric signal. In the presence of our virus-engulfing shells (half T=1 shells), virus interactions with the surface were blocked up to 99% (Fig. 5h), thus confirming the interaction-inhibiting capacity of our shells. Control experiments with shells lacking HBV trapping antibody resulted in minimal virus blocking compared to the signal generated by naked HBV core particles that represent baseline 0% virus blocking.
HBV core particles directly incubated with antibodies, but without any shells present, were negligibly blocked from binding the surface. This finding indicates that the antibodies by themselves do not fully passivate the HBV capsid surface even though they were added at 400-fold excess over HBV particles. However, in contrast, when using the shells functionalized with on average as few as five antibodies, we achieved a virus blocking efficiency of greater than 80%. The blocking was nearly complete (up to 99%) when using more than five antibodies in the shells. The data thus shows that the shell-trapping method can be highly effective even when only a handful of physical interactions are formed between the virus surface and surrounding shell. Our data indicates that the shells, and not the antibodies used for holding the virus inside the shell, shield the virus from its exterior by steric occlusion.
Virus neutralization in human cells
We tested the neutralization capacity of the DNA-origami half octahedron shells using adeno-associated virus serotype 2 (AAV2) (35) virions carrying an enhanced green fluorescent protein (eGFP) expression cassette (36) using both microscopy and flow cytometry (Fig. 6a). We stabilized DNA shells with UV point welding and PEG-oligolysine/oligolysine as described above. AAV2 particles were successfully trapped in DNA half shells functionalized with anti-AAV2 antibody in the shell interior, in serum in the presence of bovine serum albumin (BSA) as seen by direct imaging with TEM (Fig. 6b). Since AAV belongs to a completely different virus family than HBV, this data also establishes the modularity of the shell: by swapping out the virus-binding moieties one can trap different types of viruses.
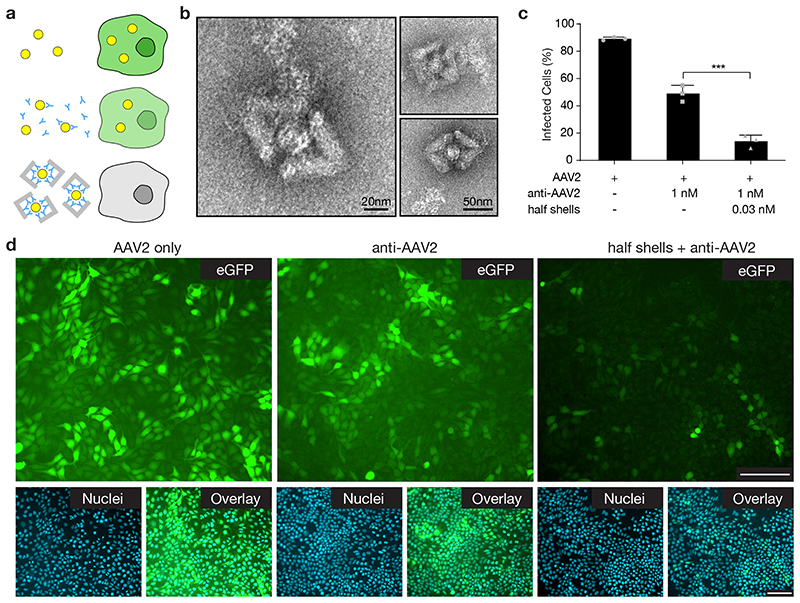
Fig. 6. Neutralization of AAV2 with DNA-origami half shells.
(a) Schematic illustrating that successful infection of HEK293T cells with AAV2 results in the expression of eGFP, while cells exposed to AAV2 captured in DNA half shells do not express eGFP. Yellow circles = AAV2, blue Y = anti-AAV2 IgG antibody, grey angled blocks: DNA half shells. (b) TEM images demonstrating capture of AAV2 virus particles within the DNA-origami half shells. Capture was successful in the presence of BSA. (c) Quantification of infected cells by flow cytometry for the conditions: AAV2 only, anti-AAV2 applied at IC50 concentration (1 nM), and DNA-origami half shells with anti-AAV2 conjugated to the inside. Anti-AAV2 and DNA half shells were preincubated with AAV2, respectively. The half shells were used at an overall identical antibody concentration as the anti-AAV2 only condition, with ~ 36 antibodies per shell, and ~ 7 half shells per virus particle. Data was quantified using flow cytometry, and is presented as mean ± s.d., n = 3 biologically independent experiments. Individual data points are overlayed. One-way ANOVA was performed to test significant inhibition compared to the control, both anti-AAV2 alone and half shell origami + anti-AAV2 demonstrated significant neutralization compared to the AAV2 only control (p ≤ 0.0001). Conjugation of anti-AAV2 to DNA-origami half shells results in significantly greater neutralization capacity than free anti-AAV2 (p = 0.0002). (d) Representative epifluorescent microscopy images demonstrating the expression of eGFP by infected cells. Images are representative from one of n = 3 biologically independent experiments, with similar results each time. For each of the conditions, eGFP expression (green), cell nuclei (blue) and the overlay are given. Scale bars represent 100 μm.
We quantified the efficacy of virus neutralization by determining the dose response curves for DNA half-shells functionalized with on average 36 anti-AAV2 antibodies per half-shell and free anti-AAV2 antibodies as reference (Supplementary Fig. 35). The number of eGFP positive cells served as a readout for infection efficacy using flow cytometry analysis. The DNA half shells neutralized AAV2 with an estimated half maximal inhibitory concentration (IC50) of ~0.3 nM (Supplementary Fig. 35). At our conditions, the IC50 corresponded to approximately 2.5 half-shells per infectious virus particle. The DNA half shells had increased neutralization capacity compared to the activity of the free anti-AAV2 (Fig. 6c,d, Supplementary Fig. 35). This neutralization enhancement is best appreciated in fluorescence microscopy images (Fig. 6d), where few eGFP positive cells remain in the samples with AAV2-trapping DNA half shells, whereas many eGFP positive cells appear in samples exposed to the identical dose of anti-AAV2 antibodies free in solution. This experiment demonstrates that the shells function in physiological conditions with live cells. It also shows that the shells can further augment the already quite potent neutralization capabilities of the anti-AAV2 antibodies. As above with the in vitro HBV blocking experiments in figure 4, the enhanced neutralization suggests that the shells trap viruses in a multivalent fashion and that the shell material additionally contributes as a viral-surface occluding agent.
We also investigated whether the DNA-origami half shells without any conjugated antibody had an effect, and we found a low but non-negligible neutralization activity at the highest origami concentration tested (Supplementary Fig. 35). We suggest this activity arises from electrostatic interactions between the PEG-oligolysine/oligolysine coated DNA-shells, and the AAV2 particles. Finally, we tested if exposure to the DNA half shells had any effect on cell viability and found no significant effect across any of the concentrations used in this study (Supplementary Fig. 35).
Outlook
We envision that trapping viruses in shells can decrease the viral load in acute viral infections by preventing viruses from undergoing host cell interactions. We tested the virus trapping concept successfully with HBV core and AAV2 virus particles. We achieved near complete inactivation by engulfing HBV in a surrounding shell in vitro and could also effectively block AAV from infecting live cells. Due to the modularity of the DNA shells, other virus binders could be used. For example, host receptor domains or peptides known to be targeted by a viral pathogen and DNA / RNA aptamers could be conjugated to the shells. One of our design solutions, the half T=1 shell, featured 90 sites for anchoring virus-binding moieties in the interior cavity. This high level of multivalency will be particularly useful for trapping pathogens for which only low-affinity binders are available. Multiple different antibodies could also be combined to achieve higher specificity against a single target or against a plurality of targets.
Our icosahedral shells consist of DNA, which is durable, available commercially, and easily functionalized and modified. The components needed for our shells can be mass-produced biotechnologically (37). Using DNA-based agents can potentially circumvent neutralization, phagocytosis, and degradation by pathways of the innate and adaptive immune system targeting protein structures. We expect our shells to be largely non-toxic because they do not target any enzymes of the host metabolism as many current antivirals do. However, we cannot exclude that nucleic-acid specific reactions such as activation of pattern-recognition receptors recognizing DNA (38, 39) or induction of DNA-binding antibodies may occur once the shells are applied in vivo. Testing our concept and assessing potentially adverse effects in organisms are important challenges for the future. Beyond the proposed application as virus traps, our programmable icosahedral canvas system also offers opportunities to create antigen-carriers for vaccination, DNA or RNA carriers for gene therapy or gene modification, drug delivery vehicles, and protective storage containers (see Supplementary Fig. 36 for cargo loading examples).
Methods
Self-assembly of shell subunits
All self-assembly experiments were performed in standardized “folding buffers” containing x mM MgCl2 in addition to 5 mM Tris Base, 1 mM EDTA and 5 mM NaCl at pH 8 (FoBx). Single-scaffold-chain DNA-origami objects were self-assembled in one-pot folding reactions containing 50 nM scaffold DNA and 200 nM of each staple strand. The individual scaffolds were produced as described previously (41, 42) (see Supplementary Note for sequence). Folding buffer (FoB20) was used with x = 20 mM MgCl2. All reaction mixtures were subjected to thermal annealing ramps as detailed in Supplementary Table 2 in Tetrad (Bio-Rad) thermal cycling devices. Staple strands were purchased from IDT (Integrated DNA Technologies).
Purification of shell subunits and self-assembly of shells
All shell subunits were purified using gel purification and, if necessary, concentrated with ultrafiltration (Amicon Ultra 500 μl with 100kDa molecular weight cutoff) before self-assembling the subunits into shells. Both procedures were performed as previously described (31) with the following alterations: for gel purification, we used 1.5% agarose gels containing 0.5x TBE and 5.5 mM MgCl2. For ultrafiltration, the same filter was filled with gel-purified sample multiple times (about 2-5 times, ~400 μl every step) in order to increase the concentration of objects that are recovered from the filter. Before putting the filter upside down in a new filter tube, we performed two washing steps with 1xFoB5 (~400 μl) to achieve well-defined buffer conditions for the shell assembly. To assemble the purified (and concentrated) shell subunits into shells we adjusted the subunit and MgCl2 concentrations by adding 1xFoB5 and 1.735 M MgCl2 in suitable amounts. Typical subunit concentrations were in the range of 5 nM and up to 100 nM (for cryo-EM measurements, see Supplementary Table 3). Typical MgCl2 concentrations for shell self-assembly were in the range of 10-40 mM. Shell self-assembly was performed at 40°C. Reaction times were varied depending on the shell type (see Fig. 3a). Both, all shell subunits and assembled shells, can be stored at room temperature for several months.
T=1 shell exterior modification
The T=1 triangle and the triangular brick (Fig. 2h) were dimerized using single stranded DNA sticky ends protruding from the T=1 triangle. The protruding sequences contained three thymidines for flexibility plus 7 base long sequence motifs that were directly complementary to single stranded scaffold domains of the brick (Supplementary Fig. 43B). Dimerization reactions were performed at room temperature overnight using a monomer concentration of 40 nM in the presence of 11 mM MgCl2.
Cargo encapsulation in T=1 shells
Nine staples of the T=1 shell subunits were modified by adding 16 bases on the 5’ ends. These nine modified staples and unmodified T=1 staples are folded with p8064 scaffold to produce T=1 triangles with nine ssDNA “handles” (Supplementary Fig. 36A, left). The 16-base ssDNA handles are located on the shell-inward facing surface of the monomers. 8 of those 9 strands were oriented facing inwards towards the interior of the monomer and consequently may not have been accessible to the cargo. Single-stranded DNA cargo was prepared by attaching staple strands to the p8064 ssDNA circular scaffold with a 16 base-long overhang that was complementary to the handles on the shell subunits. An oligo containing a CY5 dye was also hybridized to the scaffold to enable fluorescence read-out by laser scanning of agarose gels (Supplementary Fig. 36A, middle and Supplementary Fig. 43C). In order to avoid having the unbound staples in cargo solution, which would passivate the monomers, 20 different staples are mixed with the scaffolds in 1:2 ratio. To anneal staples to the circular ssDNA, FOB15 buffer is used with a temperature ramp of 65°C for 15 min, 60°C to 44°C for 1h/1°C. To encapsulate gold nanoparticles, complementary handles of the monomer’s handles are attached to the gold nanoparticles with a diameter of 30 nm (Cytodiagnostics, OligoREADY Gold Nanoparticle Conjugation Kit). A schematic and a negative stain TEM tomogram slice is shown in Supplementary Fig. 36, B and C. To increase the visibility of the encapsulated circular ssDNA in TEM images, gold nanoparticles with a diameter of 20 nm (Cytodiagnostics, OligoREADY Gold Nanoparticle Conjugation Kit) were attached to the circular ssDNA scaffold (schematic and negative stain TEM are shown in Supplementary Fig. 36, B and C last images from the right). T=1 shells with & without cargo were assembled in 1xFoB20 buffer at 40°C for 3 days. Shell subunits were gel purified prior to assembly. Concentration of triangles was 16 nM. Concentration of cargo (of any type) was 0.8 nM.
Half shells and HBV core binding
Nine staples on the inside of the triangles were modified with handles with 26 single-stranded bases at the 5’ ends (seq.: ‘GCAGTAGAGTAGGTAGAGATTAGGCAoligonucleotide’, for design details see Supplementary Figs. 41,42). The triangles were purified and assembled as described above. Oligonucleotides complementary to the handle-sequence and modified with a thiol group at the 3’ end were coupled to the anti-HBc 17H7 (100 μg) using a Sulfo-SMCC (Sulfosuccinimidyl-4-[N-maleimidomethyl]cyclohexane-1-carboxylate) cross-linker. The product was subsequently purified using the proFIRE® from Dynamic Biosensors. The DNA modified antibodies were added to the assembled shells and incubated over night at 25°C. HBV core particles were incubated with the modified shells for 1-4 hours at 25°C. To assemble T=1 triangles around HBV core particles, the modified antibodies were added to single triangles. These triangles were then incubated with HBV core particles at a MgCl2 concentration of 19 mM for one day.
Shell oligolysine stabilization
The complete octahedral shells were assembled at 35 mM MgCl2 and UV cross-linked as described in (33) for 1h at 310 nm wavelength using the Asahi Spectra Xenon Light source 300W MAX-303. The shells were incubated in a 0.6:1 ratio of N:P with a mixture of K10 oligolysine and K10-PEG5K oligolysine (1:1) for 1h at room temperature as similarly described in (34). The octahedra were incubated in 55% mouse serum for 1h and 24h at 37°C. To allow imaging with negative stain the samples were diluted with PBS to a final mouse serum concentration of 5%, immediately before application to the negative stain grids.
The partial shells used for virus neutralization experiments in vivo were assembled at 60 mM MgCl2 and UV cross-linked as described in (33) for 30 min using the Asahi Spectra Xenon Light source 300W MAX-303. We introduced three-bases long sticky overhangs at every stacking contact and added one thymidine at the ends of both oligonucleotides (see Supplementary Fig. 45) to covalently crosslink the triangular subunits. The sticky overhangs were necessary to compensate for the decrease in blunt-end stacking induced by addition of the thymidines for UV point welding. The shells were incubated in a 0.6:1 ratio N:P with a mixture of K10 oligolysine and K10-PEG5K oligolysine (1:1) for 1h at room temperature as similarly described in (34). The DNA modified antibodies were added to the assembled shells and incubated over night at room temperature.
Gel electrophoresis
The size distribution of folding reactions or shell assemblies was investigated using agarose gel electrophoresis. For solutions including only shell subunits, we used 1.5% agarose gels containing 0.5xTBE Buffer (22.25 mM Tris Base, 22.25 mM Boric Acid, 0.5 mM EDTA) and 5.5 mM MgCl2. For solutions including oligomeric assemblies such as shells, an agarose concentration of 0.5% was used. The gel electrophoresis was performed in 0.5xTBE buffers supplemented with the same MgCl2 concentration as the solutions in which the shells were incubated in. For MgCl2 concentration larger than 15 mM, a surrounding ice-water bath was used for cooling the gel. The gel electrophoresis was performed for 1.5 to 2 hours at 90 V bias voltage. For gels with MgCl2 concentrations larger than 15 mM, we exchanged the buffer after 45 min. The agarose gels were then scanned with a Typhoon FLA 9500 laser scanner (GE Healthcare) with a pixel size of 50 μm/pix.
Negative-staining TEM
Samples were incubated on glow-discharged collodion-supported carbon-coated Cu400 TEM grids (in-house production) for 30 to 120 s depending on structure and MgCl2 concentration. The grids were stained with 2% aqueous uranyl formate solution containing 25 mM sodium hydroxide. Imaging was performed with magnifications between 10000x to 42000x. T=3 triangles were imaged on a Phillips CM100 equipped with a AMT 4Mpx CCD camera. All other negative staining data was acquired with SerialEM at a FEI Tecnai T12 microscope operated at 120 kV with a Tietz TEMCAM-F416 camera. TEM micrographs were high-pass filtered to remove long-range staining gradients and the contrast was auto-leveled (Adobe Photoshop CS6). To obtain detailed information on individual particles and investigate successful encapsulation negative stain EM tomography was used as a visualization technique. The grids were prepared as described above, and the tilt series acquired with magnifications between 15000x and 30000x using the FEI Tecnai 120. The stage was tilted from -50° to 50° and micrographs were acquired in 2° increments.
All tilt series were subsequently processed with Etomo (IMOD) (43) to acquire tomograms. The micrographs were aligned to each other by calculating a cross correlation of the consecutive tilt series images. The tomogram is subsequently generated using a filtered back-projection. The Gaussian-Filter used a cutoff between 0.25 and 0.5 and a fall-off of 0.035.
Cryo electron microscopy
The DNA origami concentrations used for preparing the cryo-EM grids are summarized in Supplementary Table 3. Samples with concentrations higher than 100 nM were applied to glow-discharged C-flat 1.2/1.3 or 2/1 thick grids (Protochip). Samples containing shells with less than 30 nM monomer concentrations were incubated on glow-discharged grids with an ultrathin carbon film supported by a lacey carbon film on a 400-mesh copper grid (Ted Pella). The concentration of all single triangles was increased above 500 nM with PEG precipitation (31). 1 ml of folding reaction (~50 nM monomer concentration) was mixed with 1 ml of PEG, centrifuged at 21k rcf for 25 min and re-suspended in 50 to 100 μl 1xFoB5. The DNA-origami triangles used for assembling the shells were all gel purified and concentrated with ultrafiltration as described above before increasing the MgCl2 concentration. Plunge freezing in liquid ethane was performed with a FEI Vitrobot Mark V with a blot time of 1.5 to 2 s, a blot force of -1 and a drain time of 0 s at 22°C and 100% humidity. The samples with less than 100 nM monomer concentrations were incubated on the support layer for 60 to 90 s before blotting. All cryo-EM images were acquired with a Cs-corrected Titan Krios G2 electron microscope (Thermo Fisher) operated at 300 kV and equipped with a Falcon III 4k direct electron detector (Thermo Fisher). We used EPU 1.2 up to 2.6 (Thermo Fisher Scientific) for automated single particle acquisition. See Supplementary Table 3 for microscope settings for all individual datasets. The defocus for all acquisitions was set to -2 μm. The image processing was done at first in RELION-2 (44) and then later in RELION-3 (45). The recorded movies were subjected to MotionCor2 (46) for movie alignment and CTFFIND4.1 (47) for CTF estimation. After reference-free 2D classification the best 2D class averages, as judged by visual inspection, were selected for further processing. A subset of these particles was used to calculate an initial model. After one to two rounds of 3D classification, the classes showing the most features or completely assembled shells were selected for 3D auto-refinement and post-processing. For the corresponding shells octahedral (O) or icosahedral (I1) symmetry was used for the last two steps. All post-processed maps are deposited in the Electron Microscopy Data Bank (EMDB) (see Supplementary Table 3).
In vitro virus blocking ELISA
Various concentrations of assembled half-T1 shells were incubated overnight at room temperature with 2 nM oligonucleotide-conjugated capture antibody (anti-HBc 17H7, Isotype IgG-2b) in FoB30-T (FoB30 + 0.05% Tween-20). The next day the pre-incubated mixtures were added to 5 pM HBV core particles and incubated overnight at room temperature, yielding 1 nM capture antibody, 2.5 pM HBV core particle and 0-200 pM half-T=1 shells. A flat-bottom transparent 96 well microplate (Nunc MaxiSorp) was treated overnight at 4 °C with 100 μl/well anti-IFA-HepBcore (1 μg/ml in PBS). After washing 4 times with 200 μl/well PBS-T (PBS + 0.05% Tween-20) the well surface was blocked by incubating with 200 μl/well 5% bovine serum albumin in PBS for 2 hours at room temperature. After washing 4 times with 200 μl/well FoB30-T, 90 μl of the pre-incubated samples were added to the wells and incubated for 2 hours at room temperature, followed by washing and subsequent incubation for 1 hour with 100 μl/well horseradish peroxidase conjugated detection antibody (anti-HepBcore-HRP in FoB30-T). After washing with FoB30-T, 100 μl/well HRP substrate (3,3’,5,5’-Tetramethylbenzidine, lifetechnologies) was added and product formation was monitored in time by measuring the absorbance at 650 nm with a 60 s interval in a platereader pre-equilibrated to 30 °C (CLARIOstar, BMG labtech). HRP activity was calculated by fitting linear regression slopes to the linear regime of the kinetic data (typically the first 5 minutes). Virus blocking efficiency was calculated relative to a control of HBV core particles only and blank measurements where no HBV core particle was present during all the incubation and washing steps. All experiments were performed in triplicates. Antibodies used for the ELISA were kindly provided by Centro De Ingenieria Genetica y Biotecnologia de sancti spiritus in Cuba.
Helium Ion Microscopy (HIM)
Imaging was performed with negative-stained TEM grids coated with a 5 nm layer of AuPd using a Quorum Q150T sputter coater in ORION Nanofab (Zeiss). We used an acceleration voltage of 30 kV and a beam current of 0.3 to 0.4 pA. The images were acquired in scanning mode with an Everhart-Thornley 2k detector.
Production of HBV core particles
Hepatitis B virus core particles of genotype D (subtype ayw2) were produced recombinantly in E. coli K802 and BL21 cells (purchased from the Latvian Biomedical Research and Study Centre, Riga, Latvia). Briefly, particles were obtained by sonication and clarification from bacterial protein extracts and purified by ammonium sulphate precipitation and subsequent anion exchange and size exclusion chromatography as described (48). Final preparations were constantly kept at 4 °C in the dark in conventional PBS (including 0.05% NaN3, 1 mM DTT).
Production of Anti-HBc
Anti-HBV core antibody (anti-HBc) 17H7 (Isotype IgG-2b) was produced by the Monoclonal Antibody Core Facility at Helmholtz Zentrum München in Munich (HMGU). Briefly, mouse HBc-recognizing B cells were generated by common hybridoma technology. The mice were challenged with the peptide NLEDPASRDLVVC (aa 75-86 of HBV core). Mouse hybridoma clones were selected and secreted antibodies were analyzed by immune staining and precipitation of HBcAg and ELISA for native antigen recognition and by Western Blot analysis for detection of denatured antigen. Final 17H7 preparations were purified via standard affinity chromatography using a protein A/G column and concentrated to 0.8 mg/mL (5.33 μM) of protein and kept in conventional PBS (137 mM NaCl, 10 mM Phosphate, 2.7 mM KCl, pH 7.4) at 4 °C in the dark.
Cell culture and neutralization assays
HEK293T (human embryonic kidney cell line, DSMZ) cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco, cat. no. 31966047) with 10% heat-inactivated Fetal Bovine Serum (FBS, Sigma-Aldrich, cat. no. F9665). Cells were cultured routinely in a humidified incubator at 37 °C with 5% CO2. AAV2 carrying eGFP (Biocat, cat. no. AA002-GVO-GC) were utilized for transduction experiments, where the concentration of infectious particles was determined by titration as per the manufactures protocol. Briefly, cells were seeded in 24-well plates at 80,000 cells mL-1 16–24 h prior to transduction, and harvested 72 h after transduction for quantification of transduction efficiency by flow cytometry. Samples were acquired and analyzed using Attune™ NxT Flow Cytometer and software (Thermofisher) respectively. 20,000 single cell events, gated on side scatter area vs height were recorded for analysis. eGFP was excited by 488 nm laser, and emission was measured with a 530/30 nm bandpass filter. Untreated cells were used as a negative control. Concentration of infectious particles was determined to be 1.23 × 109 IFU mL-1. Total number of AAV2 virus particles was measured by ELISA as per manufacture’s protocol (Progen, cat. No. PRATV), and determined to be 2.24 × 1012 VP mL-1.
For neutralization experiments, cells were cultured as above. 48-well plates were coated with poly-L-lysine (Sigma Aldrich cat. no. P2636, 0.1 mg mL-1, 10 min r.t. incubation) and then washed 2× with H20 and then PBS. HEK293T cells were seeded at 80,000 cells mL-1 16–24 h prior to transduction. Stock solutions were prepared for the overnight binding of conjugated anti-AAV2 to half shell origamis. Binding occurred in the presence of 0.1 mg mL-1 bovine serum albumin (BSA). Similarly, conjugated anti-AAV2, and half shells without antibody were also prepared in an identical manner.
The next day, the half shells were coated with PEG-oligolysine/oligolysine by r.t. incubation for 2 h. Next, each of the different titration conditions were prepared and diluted to a total of 33.5 μL per condition with PBS. 4 μL of diluted AAV-2 sample (1/100, in PBS) was added and mixed, and samples were left to incubate (2 h, r.t.). Cells were washed with PBS and 62.5 μL of DMEM with 2% FBS was added to each well. Mixtures (37.5 μL) were then added dropwise to each well. Cells were incubated for 2 h, before 100 μL of DMEM with 18% FBS and 1× antibiotic/antimycotic was added. The cells were incubated for a further 22 h, before media was removed, cells were washed with 1? PBS, and 250 μL of DMEM with 10% FBS and 1× antibiotic/antimycotic was added. At 48 h post-transduction, the cells were trypsinized and prepared for flow cytometry. Transduction efficiency was quantified by flow cytometry as above, representative gates are given in Supplementary Fig. 46. Statistical analyses were performed with Graphpad Prism (GraphPad Software Inc.).
For epifluorescence imaging, the procedure was identical as above, with the exception that the cells were seeded in 8-chambered well slides (Nunc™ Lab-Tek™, Thermofisher). After the total 48 h timepoint, cells were washed with 1× PBS and then fixed with 2% paraformaldehyde (PFA). Cells were washed again (1× PBS), and cell nuclei were stained (Hoescht 3342, diluted in PBS, 5 min, r.t.). Cells were washed with PBS, and the samples were mounted using Fluoromount-G aqueous mounting media. Samples were imaged using a Tikon Eclipse Ti2-E inverted microscope, using a 10× objective. Images were collected using NIS-Elements AR software, and processed using ImageJ.
Viability Assay
Cytotoxicity was quantified by cell viability following 24 or 48 h incubation of cells with the half-shell mixtures. HEK293T cells were seeded in poly-L-lysine treated 96-well plates at 80,000 cells mL-1. Cells were allowed to settle overnight; media was removed and cells exposed to half shell mixtures in an identical procedure to the neutralisation assays. Cells were incubated with half shell solutions for a further 24 h or 48 h, and alamarBlue reagent (Invitrogen, 10 μL per well) was added. Plates were mixed and incubated for 4 h before being read on plate reader (CLARIOstar). Absorbance readings at 570 nm and 600 nm were taken, as per manufacturer’s protocol. Measured values were normalized to control wells, which were treated identically, but received PBS containing no origami structures. All conditions were measured at least in triplicate.
Statistics and Reproducibility
All cryo-EM micrographs, negative stain TEM images and epifluorescent microscopy images presented in the manuscript and the supplementary information are exemplary micrographs and show representative images of many acquired micrographs. All negative stain TEM images, epifluorescent microscopy images and agarose gel electrophoresis experiments were repeated independently multiple times and reproduced reliably the same results.
Supplementary Material
Acknowledgements
We thank B. Kick for help with the antibody-DNA conjugation and for scaffold preparation. We also thank the Monoclonal Antibody Facility of Helmholtz Zentrum München (head: Regina Feederle) for help with generating and providing anti-HBc 17H7. This work was supported by a European Research Council Consolidator Grant to H.D. (GA no. 724261), the Deutsche Forschungsgemeinschaft through grants provided within the Gottfried-Wilhelm-Leibniz Program and the SFB863 TPA9 (to H.D.), the TRR179 (TP14 to U.P.), the German Ministry for Education and Research (BMBF) through StabVacB and DZIF project 05.806/907 (U.P.), the European Commission FET Open Grant VIROFIGHT (GA no. 899619) (H.D., U.P.), the Netherlands Organization for Scientific Research (NWO, Rubicon program, project no. 019.182EN.037 to W.E.), the USA National Science Foundation through the Brandeis University Materials Research Science and Engineering Center NSF DMR-1420382 and NSF DMR-2011486 (S.F. and M.F.H), Award Number R01GM108021 from the National Institute Of General Medical Sciences (M.F.H), and the Alexander von Humboldt Foundation (Humboldt Research Fellowship to J.A.K).
Footnotes
Author contributions
H.D. designed the research. S.F. co-designed icosahedral shell self-assembly studies (Figs. 1, 2). C.S. performed shell subunit design, shell assembly and all structural studies (Figs. 1-6). E.W. performed shell modification and stabilization, and HBV virus-binding inhibition experiments (Figs. 4-6), supported by A.L.. W.E. performed subunit exchange and HBV virus-binding inhibition experiments (Figs. 4, 5). J.K. performed the cell culture AAV neutralization experiments (Fig. 6). K.S. performed auxiliary shell subunit geometry alteration experiments (Supplementary Fig. 18). F.K., F.W., U.P. contributed HBV samples, and generated and provided anti-HBc (Fig. 5). S.A. performed cargo encapsulation (Supplementary Fig. 36). M.H. contributed to shell design choices.
Competing interests
A provisional patent has been filed by TUM (#PCT/EP2021/054307).
Data availability
The data supporting the findings of this study are available within the paper and its supplementary information files, and are available from the corresponding author upon reasonable request. Cryo-EM data of this study have been deposited in the Electron Microscopy Data Bank (EMDB) with accession codes: EMD-12007, EMD-12008, EMD-12009, EMD-12010, EMD-12011, EMD-12012, EMD-12013, EMD-12014, EMD-12015, EMD-12016, EMD-12019, EMD-12020, EMD-12021, EMD-12022, EMD-12023, EMD-12024, EMD-12044, EMD-12045, EMD-12046, EMD-12049
References
- 1.Lauster D, et al. Phage capsid nanoparticles with defined ligand arrangement block influenza virus entry. Nature nanotechnology. 2020;15:373–379. doi: 10.1038/s41565-020-0660-2. [DOI] [PubMed] [Google Scholar]
- 2.Kwon PS, et al. Designer DNA architecture offers precise and multivalent spatial pattern-recognition for viral sensing and inhibition. Nature chemistry. 2020;12:26–35. doi: 10.1038/s41557-019-0369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao L, et al. De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science. 2020 doi: 10.1126/science.abd9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cagno V, Tseligka ED, Jones ST, Tapparel C. Heparan Sulfate Proteoglycans and Viral Attachment: True Receptors or Adaptation Bias? Viruses. 2019;11 doi: 10.3390/v11070596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Legendre M, et al. Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:4274–4279. doi: 10.1073/pnas.1320670111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crick FH, Watson JD. Structure of small viruses. Nature. 1956;177:473–475. doi: 10.1038/177473a0. [DOI] [PubMed] [Google Scholar]
- 7.Bale JB, et al. Accurate design of megadalton-scale two-component icosahedral protein complexes. Science. 2016;353:389–394. doi: 10.1126/science.aaf8818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.King NP, et al. Accurate design of co-assembling multi-component protein nanomaterials. Nature. 2014;510:103–108. doi: 10.1038/nature13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lai YT, et al. Structure of a designed protein cage that self-assembles into a highly porous cube. Nature chemistry. 2014;6:1065–1071. doi: 10.1038/nchem.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butterfield GL, et al. Evolution of a designed protein assembly encapsulating its own RNA genome. Nature. 2017;552:415–420. doi: 10.1038/nature25157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 12.Douglas SM, et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature. 2009;459:414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castro CE, et al. A primer to scaffolded DNA origami. Nature methods. 2011;8:221–229. doi: 10.1038/nmeth.1570. [DOI] [PubMed] [Google Scholar]
- 14.Veneziano R, et al. Designer nanoscale DNA assemblies programmed from the top down. Science. 2016;352:1534. doi: 10.1126/science.aaf4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benson E, et al. DNA rendering of polyhedral meshes at the nanoscale. Nature. 2015;523:441–444. doi: 10.1038/nature14586. [DOI] [PubMed] [Google Scholar]
- 16.Dunn KE, et al. Guiding the folding pathway of DNA origami. Nature. 2015;525:82–86. doi: 10.1038/nature14860. [DOI] [PubMed] [Google Scholar]
- 17.Bai XC, Martin TG, Scheres SH, Dietz H. Cryo-EM structure of a 3D DNA-origami object. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:20012–20017. doi: 10.1073/pnas.1215713109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Funke JJ, Dietz H. Placing molecules with Bohr radius resolution using DNA origami. Nature nanotechnology. 2016;11:47–52. doi: 10.1038/nnano.2015.240. [DOI] [PubMed] [Google Scholar]
- 19.Iinuma R, et al. Polyhedra self-assembled from DNA tripods and characterized with 3D DNA-PAINT. Science. 2014;344:65–69. doi: 10.1126/science.1250944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jungmann R, et al. DNA origami-based nanoribbons: assembly, length distribution, and twist. Nanotechnology. 2011;22:275301. doi: 10.1088/0957-4484/22/27/275301. [DOI] [PubMed] [Google Scholar]
- 21.Liu W, Zhong H, Wang R, Seeman NC. Crystalline two-dimensional DNA-origami arrays. Angewandte Chemie. 2011;50:264–267. doi: 10.1002/anie.201005911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki Y, Endo M, Sugiyama H. Lipid-bilayer-assisted two-dimensional self-assembly of DNA origami nanostructures. Nature communications. 2015;6:8052. doi: 10.1038/ncomms9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ke Y, et al. DNA brick crystals with prescribed depths. Nature chemistry. 2014;6:994–1002. doi: 10.1038/nchem.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wagenbauer KF, Sigl C, Dietz H. Gigadalton-scale shape-programmable DNA assemblies. Nature. 2017;552:78–83. doi: 10.1038/nature24651. [DOI] [PubMed] [Google Scholar]
- 25.Caspar DL, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harbor symposia on quantitative biology. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 26.Twarock R, Luque A. Structural puzzles in virology solved with an overarching icosahedral design principle. Nature communications. 2019;10:4414. doi: 10.1038/s41467-019-12367-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerling T, Wagenbauer KF, Neuner AM, Dietz H. Dynamic DNA devices and assemblies formed by shape-complementary, non-base pairing 3D components. Science. 2015;347:1446–1452. doi: 10.1126/science.aaa5372. [DOI] [PubMed] [Google Scholar]
- 28.Douglas SM, et al. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic acids research. 2009;37:5001–5006. doi: 10.1093/nar/gkp436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maffeo C, Yoo J, Aksimentiev A. De novo reconstruction of DNA origami structures through atomistic molecular dynamics simulation. Nucleic acids research. 2016;44:3013–3019. doi: 10.1093/nar/gkw155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rothemund PW. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 31.Wagenbauer KF, et al. How we make DNA origami. Chembiochem : a European journal of chemical biology. 2017 doi: 10.1002/cbic.201700377. [DOI] [PubMed] [Google Scholar]
- 32.Langecker M, et al. Synthetic lipid membrane channels formed by designed DNA nanostructures. Science. 2012;338:932–936. doi: 10.1126/science.1225624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerling T, Kube M, Kick B, Dietz H. Sequence-programmable covalent bonding of designed DNA assemblies. Sci Adv. 2018;4:eaau1157. doi: 10.1126/sciadv.aau1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ponnuswamy N, et al. Oligolysine-based coating protects DNA nanostructures from low-salt denaturation and nuclease degradation. Nature communications. 2017;8:15654. doi: 10.1038/ncomms15654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang D, Tai PWL, Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat Rev Drug Discov. 2019;18:358–378. doi: 10.1038/s41573-019-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo P, et al. Rapid AAV-Neutralizing Antibody Determination with a Cell-Binding Assay. Mol Ther Methods Clin Dev. 2019;13:40–46. doi: 10.1016/j.omtm.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Praetorius F, et al. Biotechnological mass production of DNA origami. Nature. 2017;552:84–87. doi: 10.1038/nature24650. [DOI] [PubMed] [Google Scholar]
- 38.Ohto U, et al. Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9. Nature. 2015;520:702–705. doi: 10.1038/nature14138. [DOI] [PubMed] [Google Scholar]
- 39.Andreeva L, et al. cGAS senses long and HMGB/TFAM-bound U-turn DNA by forming protein-DNA ladders. Nature. 2017;549:394–398. doi: 10.1038/nature23890. [DOI] [PubMed] [Google Scholar]
- 40.Naso MF, Tomkowicz B, Perry WL, 3rd, Strohl WR. Adeno-Associated Virus (AAV) as a Vector for Gene Therapy. BioDrugs. 2017;31:317–334. doi: 10.1007/s40259-017-0234-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Engelhardt FAS, et al. Custom-Size, Functional, and Durable DNA Origami with Design-Specific Scaffolds. ACS nano. 2019 doi: 10.1021/acsnano.9b01025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kick B, Praetorius F, Dietz H, Weuster-Botz D. Efficient Production of Single-Stranded Phage DNA as Scaffolds for DNA Origami. Nano letters. 2015 doi: 10.1021/acs.nanolett.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. Journal of structural biology. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 44.Kimanius D, Forsberg BO, Scheres SH, Lindahl E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife. 2016;5 doi: 10.7554/eLife.18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zivanov J, et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 2018;7 doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nature methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. Journal of structural biology. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sominskaya I, et al. A VLP library of C-terminally truncated Hepatitis B core proteins: correlation of RNA encapsidation with a Th1/Th2 switch in the immune responses of mice. PloS one. 2013;8:e75938. doi: 10.1371/journal.pone.0075938. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available within the paper and its supplementary information files, and are available from the corresponding author upon reasonable request. Cryo-EM data of this study have been deposited in the Electron Microscopy Data Bank (EMDB) with accession codes: EMD-12007, EMD-12008, EMD-12009, EMD-12010, EMD-12011, EMD-12012, EMD-12013, EMD-12014, EMD-12015, EMD-12016, EMD-12019, EMD-12020, EMD-12021, EMD-12022, EMD-12023, EMD-12024, EMD-12044, EMD-12045, EMD-12046, EMD-12049