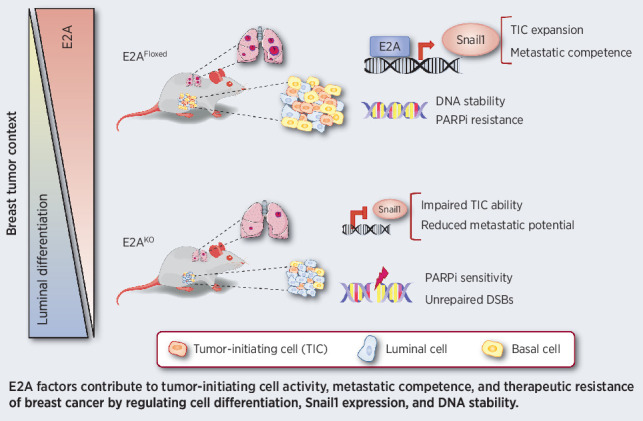
These findings identify key functions of E2A factors in breast cancer cell stemness, metastasis, and drug resistance, supporting a therapeutic vulnerability to targeting E2A proteins in breast cancer.
Abstract
Cancer stem cells (CSC) are considered responsible for tumor initiation, therapeutic resistance, and metastasis. A comprehensive knowledge of the mechanisms governing the acquisition and maintenance of cancer stemness is crucial for the development of new therapeutic approaches in oncology. E2A basic helix–loop–helix (bHLH) transcription factors are associated with epithelial–mesenchymal transition (EMT) and tumor progression, but knowledge of their functional contributions to cancer biology is still limited. Using a combination of in vivo and in vitro analyses in a novel PyMT-E2A conditional knockout mouse model and derived primary tumor cell lines, we report here an essential role of E2A in stemness, metastasis, and therapeutic resistance in breast cancer. Targeted deletion of E2A in the mammary gland impaired tumor-initiating ability and dedifferentiation potential and severely compromised metastatic competence of PyMT-driven mammary tumors. Mechanistic studies in PyMT-derived cell lines indicated that E2A actions are mediated by the upregulation of Snai1 transcription. Importantly, high E2A and SNAIL1 expression occurred in aggressive human basal-like breast carcinomas, highlighting the relevance of the E2A–Snail1 axis in metastatic breast cancer. In addition, E2A factors contributed to the maintenance of genomic integrity and resistance to PARP inhibitors in PyMT and human triple-negative breast cancer cells. Collectively, these results support the potential for E2A transcription factors as novel targets worthy of translational consideration in breast cancer.
Significance:
These findings identify key functions of E2A factors in breast cancer cell stemness, metastasis, and drug resistance, supporting a therapeutic vulnerability to targeting E2A proteins in breast cancer.
Graphical Abstract
Introduction
Metastasis and therapeutic resistance are major challenges in present oncology. Both processes depend on a small subset of tumor cells [cancer stem cells (CSC)] endowed with unlimited self-renewal, high invasive properties, and resistance to standard cancer therapies (1, 2). However, the molecular mechanisms underlying CSC biology and their similarities with those operating in normal stem cells are not completely understood (1, 3). Epithelial–mesenchymal transition (EMT) is a cell-plasticity program that enables epithelial cells to shift toward a mesenchymal-like phenotype with increased migratory and invasive abilities that facilitate metastatic dissemination (4, 5). Induction of EMT relies on several signaling pathways that impinge on a series of EMT-transcription factors (EMT-TF) that are differently expressed in a tissue- and tumor-dependent context (4, 6, 7). EMT has also been associated with the acquisition of stem cell–like properties and therapeutic resistance in several types of carcinomas (8, 9). In particular, Twist and Snail EMT-TFs promote the expression of stem cells markers and confer tumor-initiating activity to normal and transformed mammary epithelial cells (MEC; refs. 10,11,12), indicating a tight interconnection between EMT and the CSC phenotype. Nevertheless, the link of EMT with stemness and metastasis is still under debate (13,14,15). Regarding breast cancer, distinct CSC subpopulations coexist within mammary tumors displaying different capacities to initiate primary tumors [tumor-initiating cells (TIC)] or metastatic foci [metastasis-initiating cells (MIC; ref. 16)]. In this scenario, EMT has been described to promote MIC over TIC activity (16) or to become irrelevant for stemness-enhanced metastatic outgrowth of early-disseminated breast tumor cells (15). Thus, a better understanding of the contribution of EMT to breast CSC activity, together with the characterization of the specific role of distinct EMT-TFs in tumor and metastasis initiation are required.
E2A proteins (E12/E47), members of the basic Helix–Loop–Helix (bHLH) TF family, are encoded by the E2A gene (also known as TCF3) and generated by differential splicing (17). They critically control transcriptional programs that promote commitment and differentiation of the B- and T-cell lineages (18) and participate in the maintenance of the hematopoietic stem cell pool (19), being defects in their expression associated with B- and T-cell leukemias (18, 20). The E47 isoform is a potent E-cadherin repressor and EMT inducer (21), promoting a specific EMT-genetic program and distinct invasive properties compared with Snail1/Snail2 (22, 23). In addition, E47 interacts with lysyl oxidase-like 2 (LOXL2), a key driver of metastatic breast cancer (24), and participates in premetastatic niche formation in syngeneic breast cancer models (25). Although E2A expression associates with poor prognosis in human breast carcinomas (26), the implication of E2A proteins in breast cancer stemness is still unknown.
To evaluate the contribution of specific EMT-TFs, in particular E2A factors, to breast CSC biology, we generated conditional E2A knock-out mice in the context of the MMTV-PyMT model of breast cancer. Using a combination of in vivo and in vitro studies in this mouse model and in primary tumor cells, we here define E2A factors as key regulators of stemness and metastatic competence of breast cancer cells. Mechanistically, E2A proteins favor tumor progression by enhancing Snai1 transcription. Noticeably, E2A and SNAIL1 are coexpressed in human basal-like breast tumors, reinforcing the biological relevance of the epistatic E2A–Snail1 axis. Moreover, E2A participates in the control of genome stability and favors resistance of breast cancer cells to the PARP inhibitor olaparib. Collectively, these data define the role of a novel E2A–Snail1 axis in metastatic breast cancer that can be exploited in future therapeutic strategies.
Materials and Methods
Human tumor samples
A total of 11 infiltrating ductal breast carcinomas (IDBC) were acquired from the MD Anderson Cancer Center Biobank (record number B.0000745, ISCIII National Biobank Record). All tumors were grade 3 and classified with basal-like phenotype following the recommendation described in ref. 27. Patients underwent surgery between 2013 and 2020. The mean patient age at surgery was 67.6 years (range: 41–93 years). Written informed consent was obtained from the patients. This study was performed following standard ethical procedures of the Spanish regulation (Ley de Investigación Orgánica Biomédica, 14 July 2007) and was approved by the ethic committees of the MD Anderson Cancer Center (Madrid, Spain).
Mice
MMTV-PyMT strain was purchased from Jackson laboratory, and MMTV-Cre mice were provided by the CNIO Transgenic Unit. Transgenic mice carrying E2A floxed alelles were kindly provided by Dr. Y Zhuang (Department of Immunology, Duke University Medical Center, Durham, NC; ref. 28). PyMT mice harboring conditional deletion of E2A in the mammary gland were generated by intercrossing MMTV-Cre+/+;E2AFloxed/Floxed females with MMTV-PyMT+/T;MMTV-Cre+/T;E2AFloxed/Floxed males. MMTV-PyMT+/T;MMTV-Cre+/+;E2AFloxed/Floxed littermates females were used as controls. Mice were bred in a mixed genetic background (50% FVB and 50% mix from C57BL/6, CD1 and 129v strains). Experiments were conducted in accordance with protocols approved by the Use Committee for Animal Care from the UAM (Ref# CEI-25–587) and the Comunidad de Madrid (PROEX 122/17). For tumorigenesis studies, females were examined twice a week for tumor development by palpation and, unless otherwise specified, sacrificed at 14 weeks of age. Small pieces were taken from each tumor for genotyping and RNA and protein analyses, and the remnants were included in optimal cutting temperature compound (OCT) and paraffin blocks. For metastasis determination, animals were sacrificed at 16 weeks of age, and lungs were paraffin-embedded. The investigators were not blinded to genotype allocation during experiments and outcome assessment. No randomization method was used as mice were segregated into groups based on genotype.
Genotyping
Primers for conventional PCR-based genotyping of tail genomic DNA are described in Supplementary Table S1.
Isolation and flow cytometry analysis of mammary epithelial cells
MECs were obtained by mechanical dissociation of mammary glands from 5- to 9-week-old virgin females as described previously (29). For flow cytometry, single-cell suspensions were stained with CD45/CD31/CD24/CD49f or CD45/CD31/CD24/CD90 combinations of conjugated antibodies (Supplementary Table S2). Labeled cells were analyzed and sorted on BD FACSCantoII and FACSVantage Flow Cytometers (BD Biosciences), respectively.
Human cell lines
MDA-MB-468 and HCC1937 human breast cancer cell lines were obtained from ATCC and cultured as detailed in Supplementary Data. All experiments were performed within 3–8 passages after thawing. Cells were authenticated by STR profiling and routinely tested for Mycoplasma contamination.
Mammosphere assays
Freshly isolated MECs and stable neoplastic cell lines (NC) were seeded at a density of 5 × 103 cells/well on ultralow-adherence 24-well plates (Corning Inc.) in mammosphere-growing media (see Supplementary Data for details; ref. 30). Every 10 days, mammospheres were dissociated with 0.05% trypsin (Thermo Fisher Scientific) and single-cell suspensions were reseeded and cultured. Mammosphere number was estimated with ImageJ free software (“Cell Counter” plugin; v1.47d).
Transplantation experiments
Sorted CD24+/CD49flow MECs or dissociated first-generation (1stG) mammospheres from NCs were transplanted into the inguinal fat pads of 3-week-old Swiss-Nude females (Charles River Laboratories) cleared of endogenous epithelium as described previously (31). Development of tumors was monitored three times a week by palpation and outgrowths were analyzed 12 weeks after injection. Repopulating unit frequency was calculated with Extreme Limiting Dilution Analysis software (32).
Protein extraction from human tumor samples
Ten sections (10-μm thick) of OCT-embedded human tumors were collected, washed repeatedly with PBS to remove excess preservative, and lysed on Urea-Thiourea-Hepes Lysis Buffer [7 mol/L urea, 2 mol/L thiourea, and 50 mmol/L Hepes pH 7.2], containing 2% N-octylglucoside, 15 mmol/L TCEP-HCl, and 0.1 mg/mL DNAse I, using an Eppendorf pellet pestle. After centrifugation at 13,000 rpm for 20 minutes at 4ºC, supernatants were collected and protein concentration was determined using Pierce 660 nm Protein Assay Kit (Thermo Fisher Scientific).
Metaphase-spread preparation and analysis
Subconfluent cultured cells were treated with 100 ng/mL of colcemid for 2 hours and collected by trypsinization. Cellular pellets were hypotonically swollen in 75 mmol/L KCl for 10 minutes at 37ºC, fixed in an ice-cold fixative solution (75% methanol-25% acetic acid), washed and spread on glass slides. Cells were then steam-treated for 5 seconds, heat-dried, and stained with mounting media containing DAPI (4′,6-diamidino-2-phenylindole). Spreads were imaged using Cytovision GSL 120 scanning system for karyotyping (Leica Microsystems). At least 50 spreads were analyzed in every experiment and chromosome number was determined with ImageJ software (“Cell Counter” plugin).
In silico expression analyses
To assess E2A and SNAI1 mRNA expression in human breast cancer, an in silico study on 499 IDBC were selected from The Cancer Genome Atlas (TCGA) cohort dataset (33). First, E2A microarray expression was log2-transformed (+0.25 to avoid zero misspecifications) and quartile-normalized. SNAI1 expression was then evaluated on tumors previously categorized according to E2A expression as “high” if the value was within the third percentile (top 25% expression from all analyzed samples, n = 125), and as “low” when below the second quartile (bottom 25% expression, n = 125). A Pearson correlation was calculated to estimate the significance between both genes (P < 0.05). Furthermore, tumor samples were subclassified on Normal-Like, luminal A-B, HER2-enriched, and basal-like phenotypes using PAM50 criteria (34), available for each case in the cBioPortal (35). Each breast tumor subtype was also classified as “E2A/SNAI1 high” (top 25% expression, n = 38) and the rest named as “E2A/SNAI1 low” (n = 52) for studying the correlation with breast cancer phenotype. An adjusted for multiple testing using the FDR method with an adjusted P < 0.05 was used. Box and whisker plot analysis were carried out using the GraphPad PRISM 5.0 software (https://www.graphpad.com/scientific-software/prism/). Mutual exclusivity analysis between E2A expression and mutational status of BRCA1/2 was done using cBioPortal criteria in 463 IDBC from TCGA dataset (33). This analysis tests three pairs (E2A expression and BRCA1/2 mutated or not) in the OncoPrint tool. Representation was done, indicating the presence or absence of each track (neither of them, E2A not BRCA1/2, BRCA1/2 not E2A, and both E2A and BRCA1/2). The Log2 OR was used for the estimations of cooccurrence.
Statistics and reproducibility
Unless otherwise stated, numerical data are expressed as mean ± SEM. Sample sizes, number of replicates, and normalization methods are indicated in each figure legend. No statistical methods were used to predetermine sample/group sizes. Statistical analyses were performed as described in each figure legend using GraphPad PRISM 5.0 software. The statistical significance of difference between groups is indicated by asterisks (*, 0.01 < P < 0.05; **, 0.001 < P < 0.01; ***, P < 0.001).
Supplementary materials and methods
Protocols for: mammosphere and human tumor cells culture, generation of immortalized neoplastic cells, viral transductions, whole-mount mammary glands studies, spontaneous and experimental metastasis assays, generation of mSnai1-promoter vectors and promoter activity, invasion, proliferation and cell-cycle analysis, qRT-PCR, histology, immunofluorescence and additional immunologic analyses are described in Supplementary Data. List of secondary antibodies (Supplementary Table S3) and primers used for qRT-PCR analysis (Supplementary Table S4) are also included in Supplementary Data.
Results
Conditional deletion of E2A does not affect mammary gland morphogenesis
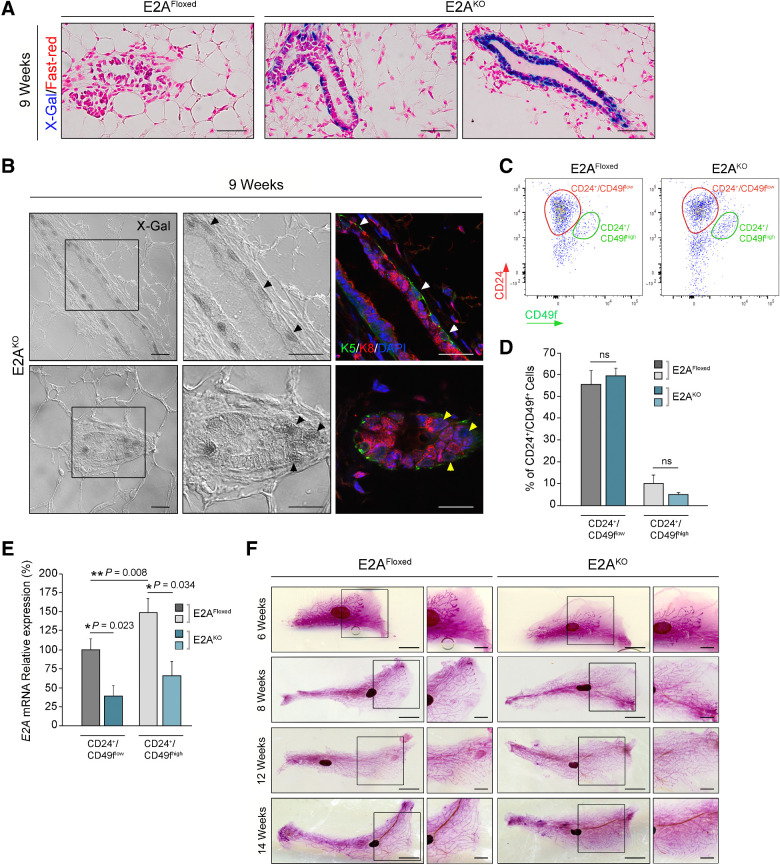
To evaluate whether E2A proteins are required for mammary gland development, we first analyzed the impact of E2A deletion in mammary gland morphogenesis. E2AFloxed/Floxed mice (28) were crossed with MMTV-Cre mice to target Cre-recombinase expression to the mammary epithelium (Supplementary Fig. S1A). E2A deletion was restricted to the mammary glands of MMTV-Cre+/T;E2AFloxed/Floxed mice (E2AKO), while tissue from MMTV-Cre+/+;E2AFloxed/Floxed control littermates (E2AFloxed) was not affected (Supplementary Fig. S1Band S1C). Next, we performed X-Gal staining of the mammary epithelium from 9-week-old females (Fig. 1A and B). Because β-Galactosidase reporter is present in the E2AFloxed allele (28), X-Gal positivity indicates Cre-recombinase expression and active E2A promoter activity in the E2AKO mammary epithelium. Most basal (cytokeratin-5–positive, K5) cells displayed intense X-Gal staining (Fig. 1A and B), while luminal (cytokeratin-8–positive, K8) cells showed X-Gal positivity only in some ducts (Fig. 1A, right; and B, bottom). MECs obtained from 9-week-old females were then sorted using markers of luminal (CD24+/CD49flow) and basal (CD24+/CD49fhigh) epithelial subsets (29). The percentage of CD24+/CD49flow and CD24+/CD49fhigh subpopulations was comparable between E2AFloxed and E2AKO mice (Fig. 1C and D). Consistent with X-Gal staining, qRT-PCR analysis revealed that E2A mRNA levels were notably higher in the basal than in the luminal population of control mammary glands (Fig. 1E). E2A expression was significantly reduced in both populations from E2AKO mammary glands (Fig. 1E), indicating Cre-recombinase activity in both cell layers of the mammary epithelium.
Figure 1.
Mammary gland morphogenesis and MEC populations are not altered in E2AKO mice. A, Sections of whole mounts of virgin mammary glands from 9-week-old E2AFloxed and E2AKO females stained with X-Gal and Fast-red. Images are representative of 3–5 mice per genotype. Scale bar, 100 μm. B, Differential interference contrast (DIC) and confocal microscopy images of mammary glands from 9-week-old E2AKO females (n = 4). X-Gal–positive cells (black arrowheads) were appreciated by DIC microscopy (left and middle panels). K8 (red) and K5 (green) immunofluorescence (right panels) identified luminal and basal populations, respectively (white and yellow arrowheads). Nuclei were counterstained with DAPI (blue). Magnifications correspond to the boxed areas in left panels. Scale bar, 50 μm. C and D, Flow cytometry analysis of LIN− (CD45−/CD31−) single-cell suspensions from 9-week-old E2AFloxed and E2AKO mammary glands for CD24/CD49f surface markers. Biaxial density plot of a representative experiment (C) and quantification (D) of CD24+/CD49flow and CD24+/CD49fhigh cell populations E2AFloxed (n = 4) and E2AKO (n = 5) mammary glands. Data represent the mean ± SEM of five independent experiments. E, qRT-PCR analysis of E2A mRNA levels in CD24+/CD49flow and CD24+/CD49fhigh cells from E2AFloxed and E2AKO mammary glands as in D. Values are expressed relative to those of CD24+/CD49flow cells from E2AFloxed glands. Data represent the mean ± SEM of five independent experiments. F, Whole-mount Carmine-Alum staining of mammary glands from control and E2AKO virgin females at the indicated weeks of age (n = 4–6 mice/group). Magnifications correspond to the boxed areas. Scale bars, 5 mm and 2.5 mm (magnifications). P value was calculated by one-way ANOVA test (D and E). ns, not significant.
We then investigated whether in vivo deletion of E2A alters mammary gland development because E47-mediated regulation of mammary epithelial branching morphogenesis has been described in 3D-organotypic cultures (36). Whole-mount analysis of mammary glands from virgin females at different developmental stages showed no defects in architecture, duct density, or side branching in mammary glands from E2AKO compared with E2AFloxed mice (Fig. 1F). Collectively, these results indicate that specific deletion of E2A in the mammary epithelium does not alter mammary ductal morphogenesis and validate E2AKO transgenic strain as a suitable model to study the role of E2A EMT-TFs in tumorigenesis and metastasis.
Ablation of E2A in the mammary epithelium restrains PyMT-driven tumorigenesis
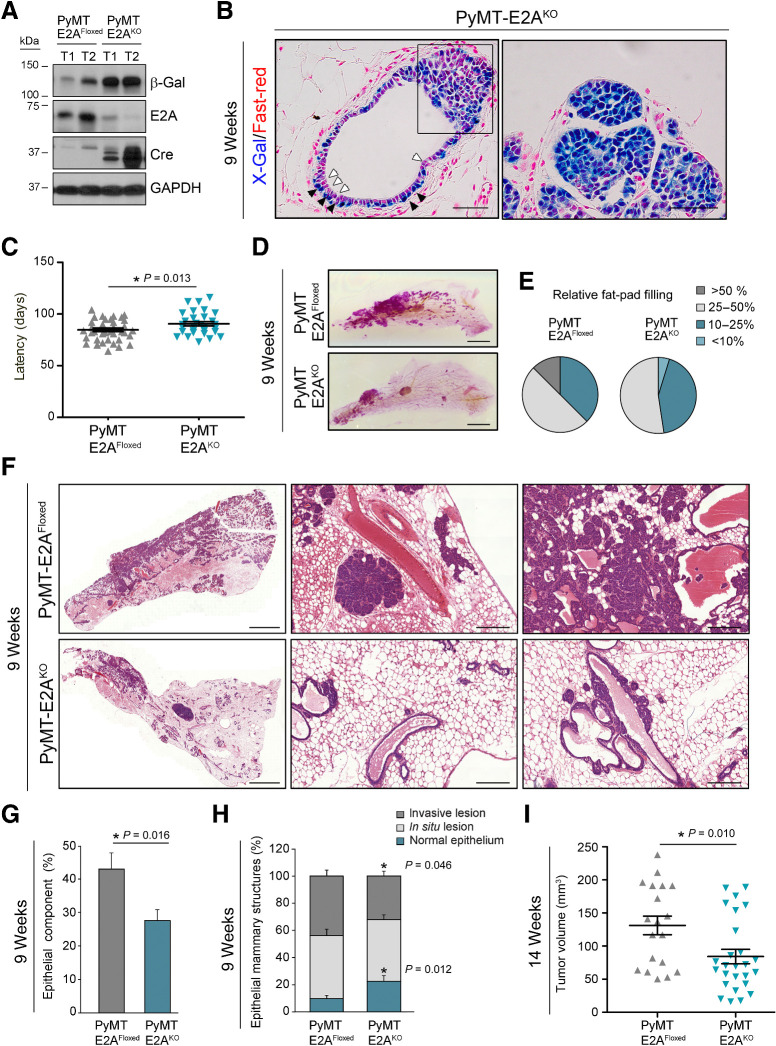
To analyze in vivo the role of endogenous E2A proteins in tumor progression and metastasis, the conditional E2AKO strain was crossed with MMTV-PyMT mice, an established model of metastatic breast cancer (Supplementary Fig. S2A and S2B; ref. 37). Specific deletion of E2A in MMTV-PyMT+/T;MMTV-Cre+/T;E2AFloxed/Floxed mice (PyMT-E2AKO) was accompanied by reduced levels of E2A mRNA and protein within tumor tissue compared with MMTV-PyMT+/T;MMTV-Cre+/+;E2AFloxed/Floxed control cohort (PyMT-E2AFloxed; Fig. 2A; Supplementary Fig. S2C). Of note, PyMT expression was comparable between PyMT-E2AFloxed and PyMT-E2AKO primary lesions (Supplementary Fig. S2D and S2E), demonstrating no interference between the MMTV-promoters driving PyMT and Cre-recombinase expression. Similar to the observations made in E2AKO mice, X-Gal–positive cells preferentially accumulated in the basal layer of PyMT-E2AKO pretumoral mammary epithelium, with luminal cells displaying a weaker staining (Fig. 2B, left). However, in hyperplastic and tumor lesions the majority of epithelial cells were X-Gal–positive, suggesting an increase in E2A expression with tumor progression (Fig. 2B, left (boxed area) and right panel). Indeed, upregulation of E2A mRNA occurs in tumors (14 weeks) compared with mammary glands (9 weeks) from PyMT-E2AFloxed mice (Supplementary Fig. S2F).
Figure 2.
Conditional deletion of E2A in the mammary gland restrains PyMT-driven tumorigenesis. A, Immunoblot analyses of mammary tumors from 14-week-old PyMT-E2AFloxed and PyMT-E2AKO mice. GAPDH detection was used as a loading control. Results are representative of five independent experiments. B, X-Gal/Fast-red staining of PyMT-E2AKO primary tumors from 9-week-old females. Cells from the basal (black arrowheads) and luminal (white arrowheads) compartments are depicted. The boxed area in the left panel comprises one hyperplastic region. Images are representative of four mice. Scale bars, 100 μm. C, Latency to detect palpable tumors in PyMT-E2AFloxed (n = 21) and PyMT-E2AKO (n = 12) mice. Midlines represent the mean value. Error bars, SEM. D, Whole-mount Carmine-Alum staining of mammary glands from 9-week-old PyMT-E2AFloxed and PyMT-E2AKO virgin females. Scale bars, 5 mm. E, Relative fat pad area covered by lesion in control (n = 16) and E2AKO (n = 21) mice, expressed as percentage of the whole fat-pad area. F, Histologic analysis of primary tumors derived from 9-week-old PyMT-E2AFloxed and PyMT-E2AKO mice. Hematoxylin and eosin images are representative of 8–12 mice for each genotype. Scale bars, 2 mm (left) and 250 μm (middle and right panels). G, Quantification of the whole epithelial component present in the hematoxylin and eosin staining of mammary glands from 9-week-old control and E2A-deficient mice. Results are expressed in percentage relative to the whole mammary gland area. Data shown are the mean ± SEM of PyMT-E2AFloxed (n = 12) and PyMT-E2AKO (n = 18) mice. H, Epithelial structures quantified in G were classified into the indicated histopathologic categories and expressed in percentage relative to the whole epithelial area. Data shown are the mean ± SEM. I, Tumor volume of lesions present in 14-week-old PyMT-E2AFloxed (n = 13) and PyMT-E2AKO (n = 26) mice. Midlines represent the mean value. Error bars, SEM. P value was calculated by two-sided unpaired Student t test (C and I) and by two-sided Mann–Whitney U test (G and H).
Compared with the control group, PyMT-E2AKO animals showed a significant delay in tumor latency (Fig. 2C). Likewise, whole mounts of PyMT-E2AKO mammary glands at 9 weeks of age exhibited a marked reduction in the fat pad area occupied by the lesion compared with glands from PyMT-E2AFloxed mice (Fig. 2D–G). According to their multifocal origin (38), PyMT-E2AFloxed and PyMT-E2AKO lesions showed multiple stages of tumor progression (Fig. 2F and H). PyMT-E2AFloxed glands contained only 10% of normal epithelial tissue, while 46% and 44% of the epithelial component corresponded to in situ and invasive carcinomas, respectively. In contrast, PyMT-E2AKO glands presented an increase in normal epithelial structures (23%) and reduced invasive component (32%; Fig. 2F and H). Histologic differences were also found in invasive lesions originated by each cohort. Tubular features were increased in PyMT-E2AKO tumors, while papillary/cystic and solid adenocarcinomas were more prevalent in PyMT-E2AFloxed tumors (Supplementary Fig. S2G). In addition, more than 91% of PyMT-E2AFloxed tumors contained fibrotic areas, while fibrosis decreased to 42% in PyMT-E2AKO tumors (Supplementary Fig. S2H and S2I). These results suggest that at early stages PyMT-E2AKO tumors exhibit a more differentiated phenotype than their corresponding controls. Indeed, expression of the luminal differentiation marker Csn2 (β-casein) was increased, concomitantly with other caseins, in PyMT-E2AKO tumors (Supplementary Fig. S2J).
At 14 weeks of age, tumors from PyMT-E2AKO mice were smaller than those from PyMT-E2AFloxed controls (Fig. 2I), although advanced lesions from both genotypes displayed extensive areas of high-grade carcinomas and little positivity for estrogen (ER), progesterone (PR), and HER2/neu receptors (Supplementary Fig. S2K and S2L), in line with previous reports and a luminal B–like phenotype (38, 39). Collectively, these results indicate that E2A proteins control breast tumor initiation and promote progression to a more dedifferentiated phenotype.
Deletion of E2A impairs self-renewal of PyMT-induced CSCs and favors a luminal differentiation fate
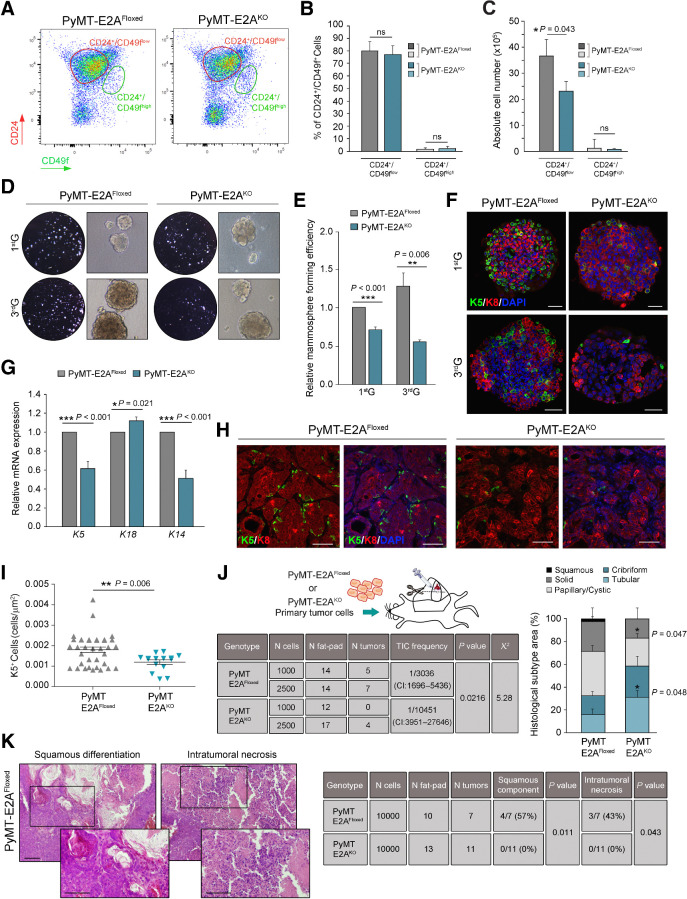
We next investigated whether E2A factors regulate tumor initiation of PyMT-tumor cells by controlling the expansion and activity of CSCs. MECs were recovered from 9-week-old PyMT-E2AFloxed and PyMT-E2AKO mammary glands and profiled using CD24 and CD49f surface markers. Consistent with the luminal-like expression profile of PyMT tumors (39), PyMT-E2AFloxed and PyMT-E2AKO preneoplastic tissues showed a dramatic expansion of the CD24+/CD49flow luminal component, being the CD24+/CD49fhigh basal subpopulation almost undetectable (Fig. 3A and B, compare with Fig. 1C and D). No differences were observed in the percentage of CD24+/CD49flow or CD24+/CD49fhigh cells between PyMT-control and PyMT-E2AKO mice (Fig. 3B), although PyMT-E2AFloxed mammary glands contained a larger absolute number of luminal epithelial cells (Fig. 3C), consistent with histologic analyses of 9-week-old mammary glands (Fig. 2D–G). In contrast, the CD24+/CD90+ subpopulation, reported to exhibit CSC properties in the PyMT model (40), was reduced in PyMT-E2AKO mammary glands (Supplementary Fig. S3A and S3B), suggesting a role for E2A TFs in the regulation of CSC expansion and self-renewal capacity.
Figure 3.
Deletion of E2A impairs self-renewal of PyMT-induced CSCs and maintenance of basal-like features. A–C, Flow cytometry analyses of LIN− (CD45−/CD31−) MECs isolated from PyMT-E2AFloxed (n = 8) or PyMT-E2AKO (n = 9) mammary glands (9 weeks old) for CD24/CD49f surface markers. Biaxial density plot of a representative experiment (A), quantification expressed as percentage (B), and absolute number of cells per mouse (C) of CD24+/CD49flow and CD24+/CD49fhigh populations in mammary glands from both genotypes. Data represent the mean ± SEM of eight independent experiments. D and E, Mammospheres formed by luminal cells isolated from 9-week-old PyMT-E2AFloxed or PyMT-E2AKO mammary glands. Representative microphotographs (D) and relative mammosphere forming efficiency (E) at first (1stG) and third (3rdG) generations. Results in E are expressed relative to those of 1stG of control cells. Data represent the mean ± SEM of six independent experiments. F, Representative confocal microscopy images of mammospheres from PyMT-E2AFloxed and PyMT-E2AKO mammary glands stained for K5 (green) and K8 (red) cytokeratins. Nuclei were counterstained with DAPI (blue). Scale bars, 40 μm. G, qRT-PCR analyses of relative mRNA levels of luminal (K18) and basal (K5 and K14) cytokeratins in 3rdG mammospheres. Results are expressed relative to PyMT-E2AFloxed mammospheres. Data represent the mean ± SEM of six independent experiments. H, Representative confocal microscopy images of K5 (green)/K8 (red) stains in PyMT-E2AFloxed and PyMT-E2AKO tumors (9-week-old mice). Nuclei were counterstained with DAPI (blue). Scale bars, 50 μm. I, Quantification of K5+ cells per tissue area (μm2) in control and E2A-deficient PyMT primary tumors (9-week-old mice). Midlines show the mean and error bars SEM of PyMT-E2AFloxed (n = 9) and PyMT-E2AKO (n = 7) tumors. Two different fields from each lesion were analyzed. J, Schematics, results, and distribution of tumor histologic subtypes of lesions generated in limiting dilution transplantation assays of luminal cells isolated from PyMT-E2AFloxed (n = 3) or PyMT-E2AKO (n = 3) mammary glands (9 weeks old). Left, TIC frequency and associated probability were calculated with ELDA software (CI, 95%). Right, histologic subtype classification of E2AFloxed (n = 12) and E2AKO (n = 4) tumors expressed as percentage of the total lesion area. K, Squamous cell differentiation and necrosis detected in tumors from transplantation assays as depicted in J. Left, representative hematoxylin and eosin images of tumors from transplanted PyMT-E2AFloxed cells. Magnifications correspond to the boxed areas. Scale bars, 50 μm. Right, quantification of both parameters in tumors from E2A-proficient (n = 7) and E2A-deficient (n = 11) cells. P value was calculated by two-sided unpaired Student t test (B, C, E, G, and I), two-sided Mann–Whitney U test (J, right), and by two-sided Fisher exact test (K). ns, not significant.
The effect of E2A on the TIC potential of preneoplastic glands was investigated by mammosphere-formation assays with luminal cells isolated from 9-week-old PyMT-E2AFloxed and PyMT-E2AKO mammary glands. E2A-deficient cells showed decreased mammosphere-forming activity across several generations compared with E2A-proficient cells (Fig. 3D and E). Remarkably, mammospheres from PyMT-E2AFloxed glands contained both luminal K8- and basal K5-expressing cells, while mammospheres from PyMT-E2AKO glands consisted essentially of cells expressing K8 (Fig. 3F). qRT-PCR analysis confirmed decreased expression of basal cytokeratins (K5, K14) in E2A-deficient mammospheres, while K18 expression augmented (Fig. 3G). Mammary basal cells display some EMT-related features (12, 41, 42). Consistent with its higher content in basal cells, PyMT-E2AFloxed mammospheres showed increased expression of mesenchymal markers and EMT-TFs (Supplementary Fig. S3C). Moreover, primary lesions from 9-week-old PyMT-E2AFloxed mice displayed increased number of basal K5-positive cells compared with PyMT-E2AKO tumors (Fig. 3H and I), reinforcing the participation of E2A proteins in the specification or maintenance of the basal cell linage.
The in vivo effect of E2A deletion on TIC capacity was then explored by transplantation experiments of freshly isolated PyMT-tumor cells. Primary PyMT-E2AKO cells showed substantially less tumor-repopulating ability compared with PyMT-E2AFloxed cells, as revealed by reduced tumor incidence at limiting dilution assays (Fig. 3J, left). Moreover, tumors generated from both cell types showed striking changes in their differentiation pattern. Tubular features were more prevalent in tumors generated from PyMT-E2AKO cells, while papillary/cystic and solid adenocarcinomas predominated in those from PyMT-E2AFloxed cells (Fig. 3J, right). Interestingly, 2% of the lesions arising from E2A-proficient cells corresponded to squamous cell carcinomas (SCC), a poorly differentiated histologic subtype more frequently observed in tumors derived from basal- and stem-like cells (43). Additional transplantation assays performed at higher cell density (to minimize differences in TIC frequency) showed that squamous differentiation and necrosis associated with breast SCC (44) were frequent in the lesions from PyMT-E2AFloxed cells, but absent in those from E2AKO cells (Fig. 3K). Besides, sebaceous-like differentiation, present in some tumors originated from basal- and stem-like cells (43), was only detected in some PyMT-E2AFloxed tumors (Supplementary Fig. S3D). Collectively, these results demonstrate that E2A depletion severely compromises self-renewal activity and dedifferentiation potential of PyMT-derived CSC.
E2A proteins are required for lung metastasis of PyMT breast tumors
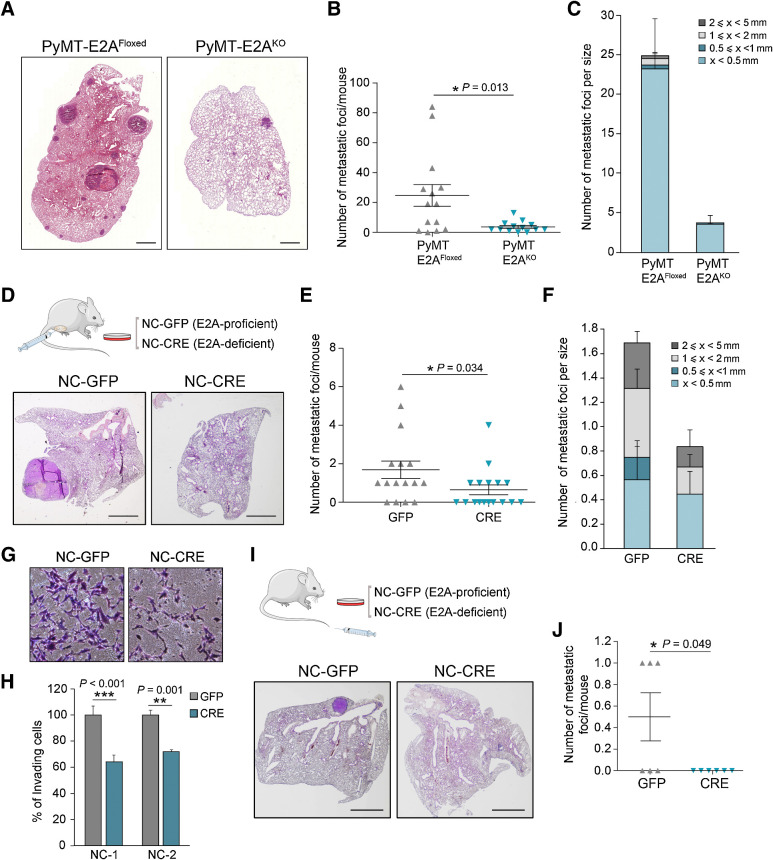
Because CD24+/CD90+ subpopulation has been shown to be specifically enriched in MICs (16, 40), we next analyzed the incidence of pulmonary metastases in 16-week-old PyMT-control and PyMT-E2A–deficient mice. PyMT-E2AKO females displayed a dramatic reduction in the number and size of lung metastatic foci as compared with PyMT-E2A–proficient mice (Fig. 4A–C), indicating a prometastatic action of E2A proteins. To analyze whether this effect is cell-autonomous, we derived two primary neoplastic cell lines (NC-1/NC-2) from independent PyMT-E2AFloxed tumors that were subsequently transduced with adenovirus encoding control GFP or Cre-recombinase. In vitro excision of E2A alleles was confirmed by qRT-PCR and immunoblot analyses (Supplementary Fig. S4A–S4C). E2A-proficient (NC-GFP) and E2A-deficient (NC-CRE) cells exhibited an epithelial-like morphology and showed no differences in their in vitro proliferation rate (Supplementary Fig. S4D and S4E). Spontaneous metastasis assays of PyMT-cells (5 × 105 injected cells/fat pad) showed that control and E2A-deficient NCs produced neoplastic lesions with similar kinetics and proliferation pattern, as confirmed by IHC detection of Ki67 (Supplementary Fig. S4F and S4G). However, pulmonary metastatic burden decreased significantly in mice injected with NC-CRE cells compared with NC-GFP cells (Fig. 4D–F). Notably, some of the mice injected with NC-GFP cells also displayed lymph node metastasis and vascular and lymphatic emboli (Supplementary Fig. S4H), events not detected in mice injected with NC-CRE cells.
Figure 4.
E2A proteins regulate the metastatic potential of PyMT-derived tumor cells. A, Representative hematoxylin and eosin–stained lung sections showing metastatic lesions from 16-week-old PyMT-E2AFloxed and PyMT-E2AKO mice. Scale bar, 1 mm. B and C, Quantification of the number per mouse (B) and number per size category (C) of lung metastatic foci developed by PyMT-E2AFloxed (n = 14) and PyMT-E2AKO (n = 13) mice at 16 weeks of age. Midlines (B) and barplots (C) show the mean values; error bars, SEM. D–F, Hematoxylin and eosin representative images (D), number per mouse (E), and number per size category (F) of lung metastatic foci after orthotopic injection of E2A-proficient (NC-GFP) and E2A-deficient (NC-CRE) cells into the fat pad of immunodeficient mice. Scale bar (D), 2 mm. Midlines (E) and barplots (F) show the mean values; error bars, SEM; n = 16 mice per genotype. G and H, Representative images of invasion assays on Matrigel chambers (G) and their corresponding quantification (H) of NC-GFP or NC-CRE cells. Data are expressed relative to values of NC-GFP cells and represent the mean ± SEM of three independent experiments in two independent PyMT cell lines. I and J, Hematoxylin and eosin representative images (I) and number (J) of lung metastatic foci per mouse after tail vein injection of NC-GFP or NC-CRE cells into immunodeficient mice. Scale bar (I), 2 mm. Midlines (J) represent the mean ± SEM; n = 6 mice per genotype. P value was calculated by two-sided Mann–Whitney U test in B and E and by two-sided unpaired Student t test in H and J. In panels D and I, schematics of the experiments performed are included.
Invasion assays showed reduced invasiveness in NC-CRE cells compared with control counterparts (Fig. 4G and H). Experimental metastasis assays also revealed that E2A deletion resulted in abrogation of lung colonization; half of the mice injected with NC-GFP cells developed lung macrometastasis while none of the NC-CRE–injected animals displayed metastatic foci (Fig. 4I and J). Altogether, these results indicate that E2A proteins are necessary for metastatic competence of PyMT breast tumors, likely influencing metastasis-initiating capacity by cell-autonomous mechanisms.
Snail1 is a transcriptional target of E2A TFs in breast tumors
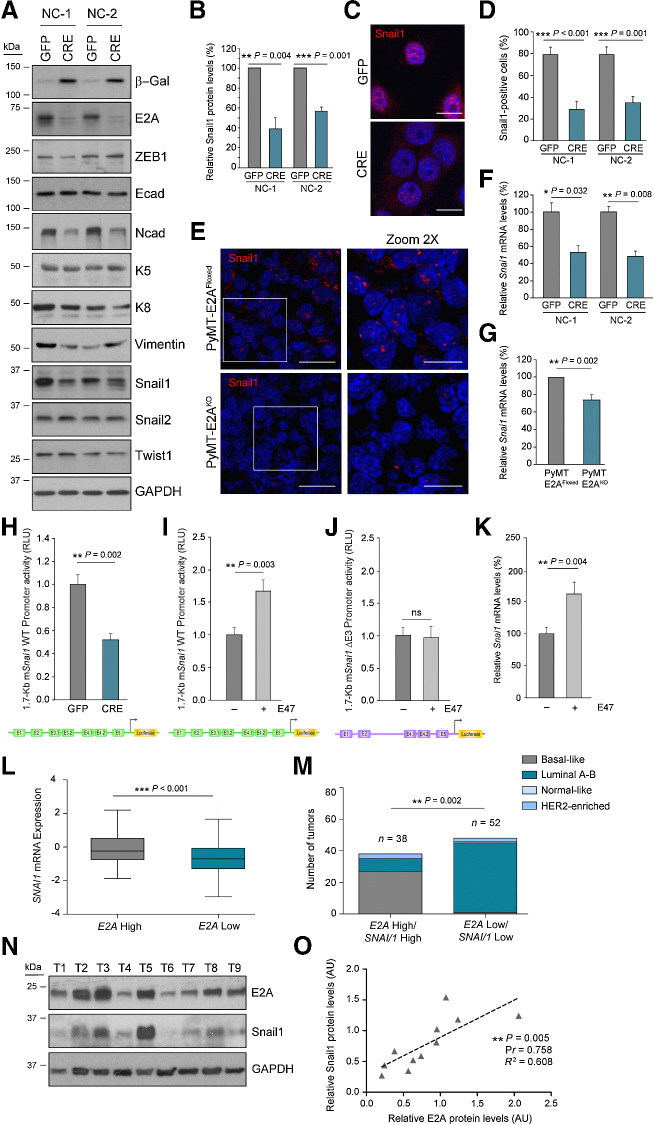
To investigate the molecular mechanisms underlying the ability of E2A to modulate TIC and MIC capacities, we analyzed the expression of different EMT-TFs previously involved in stemness maintenance in breast tumor cells (10,11,12,45,46). Snail1 protein levels decreased in NC-CRE compared with NC-GFP cells, while no significant changes were detected in other EMT-TFs (Snail2, ZEB1, Twist1). No alterations in E-cadherin content or localization was observed, although NC-CRE cells showed reduced N-cadherin expression compared with NC-GFP cells (Fig. 5A and B; Supplementary Fig. S5A and S5B). Immunofluorescence analyses confirmed that NC-CRE cells displayed a marked reduction in nuclear Snail1-positive cells compared with NC-GFP cells (Fig. 5C and D). Similar differences were observed in vivo in neoplastic glands from PyMT-E2AFloxed and PyMT-E2AKO mice (Fig. 5E; Supplementary Fig. S5C and S5D). Decreased Snai1 mRNA levels were also detected in NC-CRE cell lines, as well as in luminal cells from PyMT-E2AKO neoplastic glands, regarding their corresponding controls (Fig. 5F and G), suggesting that E2A TFs could function as transcriptional activators of Snai1 in vitro and in vivo.
Figure 5.
E2A proteins regulate Snail1 expression in breast tumors. A, Representative immunoblots of the indicated EMT markers in two independent PyMT-neoplastic cell lines. GAPDH was used as loading control. B, Quantification of Snail1 protein levels in PyMT cell lines. Data are expressed relative to control NC-GFP cells and represent the mean ± SEM of four independent experiments. C and D, Representative confocal microscopy images (C) and percentage (D) of NC-GFP or NC-CRE cells positive for nuclear Snail1 stain. Nuclei were counterstained with DAPI (blue; C). Scale bar, 10 μm. Results in D correspond to the mean ± SEM of three independent experiments; n = 80–100 cells analyzed per experiment. E, Representative confocal microscopy images of nuclear Snail1 detection in PyMT-E2AFloxed and PyMT-E2AKO tumors (14 weeks old). Magnifications correspond to the boxed areas. Nuclei were counterstained with DAPI (blue). Scale bars, 20 μm and 10 μm (magnification). n = 6 tumors per genotype. F and G, qRT-PCR analyses of relative Snai1 mRNA levels in NC cells (F) and primary luminal cells (G). Data are expressed relative to corresponding controls and represent the mean ± SEM; n = 4 (F) and n = 6 (G) independent cell preparations. H, Relative activity of 1.7-Kb mSnai1 WT promoter in NC-GFP and NC-CRE cells. Data represent the mean ± SEM of four independent experiments. I and J, Relative activity of mSnai1 WT (I) and ΔE3 (J) promoters in NC-GFP cells in the presence or absence of ectopic mE47. Data represent the mean ± SEM of five independent experiments. K, qRT-PCR analysis of endogenous Snai1 mRNA levels in NC-GFP cells in the absence or presence of mE47. Data show the mean ± SEM of three independent experiments. L, Tukey style boxplot showing SNAI1 expression in human IDBC plotted against normalized E2A mRNA levels (first vs. third quartile, n = 125 tumors to each category; TCGA dataset). M, Tumors analyzed in L stratified for normalized E2A and SNAI1 mRNA levels (first vs. third quartile, n = 38 and n = 52, respectively) and classified according to their PAM50 subtype. N, Immunoblot of E2A and SNAIL1 in a representative cohort of human basal-like breast tumors (n = 9). GAPDH was used as a loading control. O, Correlation between E2A and SNAIL1 relative protein levels in human tumors represented in N plus additional samples (n = 11). Correlation between E2A and SNAI1 mRNA (L) and protein expression (O) was done by linear regression analysis and Pearson R coefficient (Pr). Association of high E2A/SNAI1 mRNA expression with PAM50 tumor subtype (M) was calculated by multiple testing using the FDR. Additional statistical analyses were performed by two-sided unpaired Student t test. ns, not significant.
Next, the region of the mouse Snai1 promoter containing 7 E-boxes recognized by E2A and Twist1 factors during palatal fusion (47) was cloned into a reporter vector (Supplementary Fig. S5E and S5F). NC-CRE cells showed a 48% reduction in Snai1-promoter activity compared with NC-GFP cells (Fig. 5H). Besides, transient overexpression of E47 in NC-GFP cultures markedly increased Snai1-promoter activity regarding controls (Fig. 5I) depending on the E-boxes E3.1 and E3.2 (Fig. 5J; Supplementary Fig. S5F). Collectively, these results demonstrate that E47 is a transcriptional activator of Snai1 in PyMT-tumor cells through the E3 region of the Snai1-upstream-promoter. Unlike previous findings in other cellular systems (47), no cooperation between E47 and Twist1 was detected in Snai1-promoter activation in NC-GFP cells (Supplementary Fig. S5G). In line with these results, transient expression of E47 in NC-GFP cells increased endogenous Snai1 mRNA and protein levels compared with controls (Fig. 5K; Supplementary Fig. S5H and S5I).
Remarkably, an in silico study on breast cancer datasets from the TCGA (33, 35) revealed a significant upregulation of SNAI1 mRNA in IDBC displaying high levels of E2A expression (Fig. 5L). Moreover, high coexpression of SNAI1 and E2A was associated with basal-like breast cancer (Fig. 5M), a triple-negative breast cancer (TNBC) subtype, supporting a functional role for the E2A–Snail1 axis in this kind of human breast tumors. Immunoblot analysis of both EMT-TFs in a representative cohort of basal-like breast tumors revealed a direct correlation between E2A and SNAIL1 protein levels (Fig. 5N and O). In addition, depletion of E2A in two human TNBC cell lines, MDA-MB-468 and HCC1937, also decreased SNAIL1 expression (Supplementary Fig. S5J and S5K). Altogether, these results demonstrate a novel action of E47 as a transcriptional activator of Snai1 in PyMT-tumor cells and suggest that E2A-TFs may contribute to human basal-like breast cancer by enhancing SNAI1 transcription.
The E2A–snail1 axis boosts self-renewal and metastatic potential of PyMT tumor cells
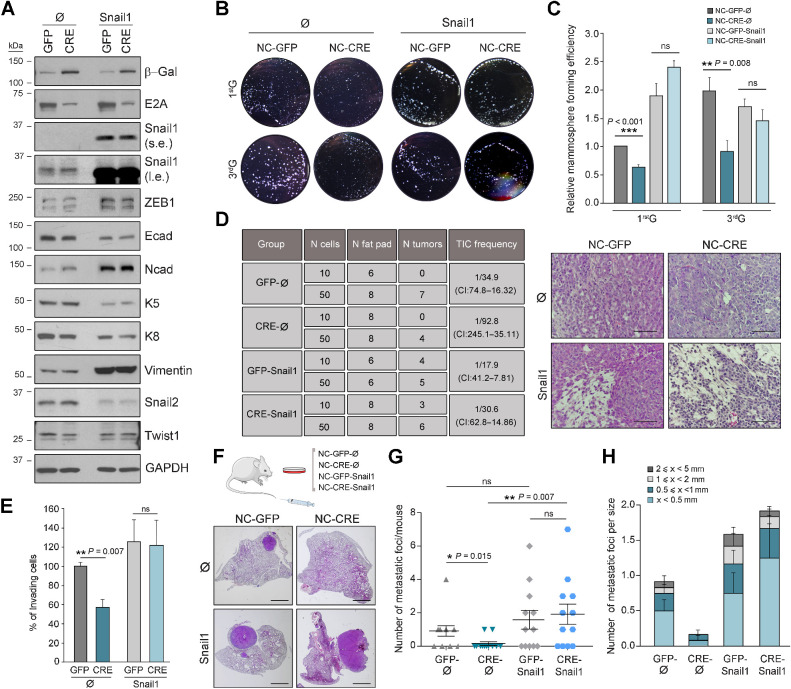
To confirm the functional connection between Snail1 and E2A in breast tumor progression, rescue studies with Snail1 were performed in PyMT-NCs by retroviral transduction (Supplementary Fig. S6A). Increased Snail1 expression was confirmed in Snail1-infected cultures (Snail1) compared with their respective controls (Ø), and to similar levels in NC-CRE and NC-GFP cells (Fig. 6A). Analysis of other EMT-TFs revealed increased expression of ZEB1 and a sharp decrease in Snail2 levels after Snail1 overexpression (Fig. 6A; Supplementary S6B), reproducing the reported switch between Snail2 and Snail1 during PyMT-tumor progression (12). In addition, compared with Ø-controls, Snail1-overexpressing cells showed reduced levels and partial membrane delocalization of E-cadherin and increased expression of N-cadherin and vimentin (Fig. 6A; Supplementary Fig. S6B and S6C). These changes, together with the acquisition of a more elongated morphology (Supplementary Fig. S6D), suggest a transition from an early to a late E/M hybrid phenotype induced by Snail1 overexpression.
Figure 6.
E2A–Snail1 axis favors self-renewal and invasiveness of PyMT-NCs. A, Representative immunoblots of the indicated EMT markers and EMT-TFs in NC cell lines stably expressing Snail1 (GFP-Snail1 and CRE-Snail1) and their corresponding controls (GFP-Ø and CRE-Ø). GAPDH was used as a loading control. (s.e., short exposure; l.e., long exposure). B and C, Mammospheres formed by control and Snail1-overexpressing NC cell lines. Representative microphotographs (B) and relative mammosphere forming efficiency (C) at first (1stG) and third (3rdG) generations of the indicated NC cells. Values in C are expressed relative to those corresponding to 1stG in control GFP-Ø cells. Data represent the mean ± SEM of four independent experiments. D, Limiting dilution transplantation experiments of single-cell suspensions obtained after dissociation of 1stG mammospheres formed by the indicated NC cell lines (n = 3 different cell suspension/condition). TIC frequency and associated probability (left) were calculated with ELDA software (CI, 95%). Representative hematoxylin and eosin images of tumors generated after transplantation of control and Snail1-overexpressing NCs (right). Scale bar, 100 μm. E, Invasion assays on Matrigel chambers of the indicated NC cell lines. Results, expressed in percentage relative to values obtained for NC-GFP-Ø cells, represent the mean ± SEM of three independent experiments. F–H, Schematics of the experimental procedure and hematoxylin and eosin representative lung images (F), number per mouse (G), and number per size category (H) of lung metastatic foci generated by the indicated cell lines upon tail vein injection. Scale bar in F, 2 mm. Midlines in G and barplots in H show the mean values; error bars, SEM; n = 12 mice per condition. P value was calculated by one-way ANOVA test in C, E, and G. ns, not significant.
Mammosphere-forming assays showed that NC-CRE cells gave rise to fewer mammospheres and exhibited lower expression of basal K5 and K14 cytokeratins than NC-GFP cells (Supplementary Fig. S6E–S6H), reproducing the behavior of freshly isolated PyMT-E2AKO luminal cells (Fig. 3D–G). Remarkably, while E2A deletion decreased mammosphere-forming activity of NCs infected with control viruses (Ø), Snail1 overexpression fully restored their mammosphere-forming efficiency (Fig. 6B and C) and their impaired K5 expression (Supplementary Fig. S6I). Moreover, the decreased tumor-repopulating potential of NC-CRE-Ø cells was fully rescued by Snail1 overexpression in transplantation experiments (Fig. 6D, left). All generated tumors corresponded to high-grade invasive carcinomas, but those derived from Snail1-overexpressing cells displayed areas with elongated, less cohesive cells (Fig. 6D, right).
Finally, Snail1 overexpression in NC-CRE cells completely reestablished their invasive abilities to levels similar to those of NC-GFP controls (Fig. 6E) and fully rescued the metastatic potential of E2A-deficient cells (Fig. 6F–H). Of note, NC-CRE-Ø cells gave rise to fewer and smaller lesions than their corresponding NC-GFP-Ø controls (Fig. 6G and H), corroborating results of previous experiments (Fig. 4I and J). Collectively, these data identify Snail1 as a critical E2A target, functionally involved in the regulation of major attributes of breast CSCs such as self-renewal and metastatic potential.
E2A controls genome integrity and therapeutic response of breast cancer cells
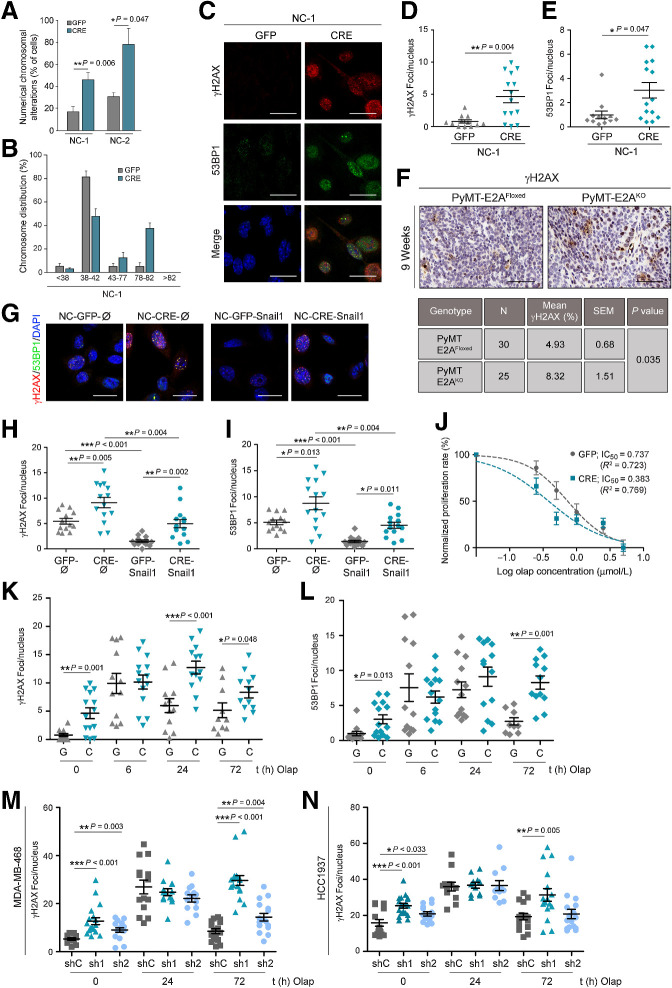
EMT-TFs have been associated with DNA damage (DD), chromosomal instability (CIN), and therapeutic resistance in various types of tumors, including breast cancer (8, 9, 13, 14, 48). More specifically, E2A proteins have been previously related to DD and drug resistance in hematologic malignancies (49, 50), but their contribution to genome integrity and therapeutic resistance in breast cancer remains unexplored. To investigate whether E2A factors regulate genomic stability of PyMT-tumors, we first studied the existence of chromosomal alterations in PyMT-derived cell lines. NC-CRE cells displayed increased numerical chromosomal alterations compared with NC-GFP cultures, being near-tetraploid about 40% of the E2A-deficient cells (Fig. 7A and B; Supplementary Fig. S7A).
Figure 7.
E2A controls genome stability and therapeutic resistance of breast cancer cells. A, Numerical chromosomal alterations of NC-GFP and NC-CRE cell lines determined by metaphase-spread assays. Spreads containing more or less chromosomes than the mean ± 5% were classified as altered. Data, indicated as a percentage, represent the mean ± SEM of two independent experiments. A minimum of 50 spreads were analyzed in every experiment. B, Histogram shows chromosome number distribution in NC-1 cells. Data are representative of two independent experiments. C–E, Representative confocal microscopy images (C) and quantification of the number of γH2AX (D) and 53BP1 (E) foci per nucleus in NC-1-GFP and NC-1-CRE cells. Nuclei were counterstained with DAPI (blue). Scale bar, 20 μm. Results in D and E show the mean ± SEM of three independent experiments. n = 80–100 cells analyzed per experiment. F, Representative images (top) and quantification (bottom) of γH2AX IHC stains in PyMT-E2AFloxed (n = 30) and PyMT-E2AKO (n = 25) tumors from 9-week-old mice. Scale bar, 100 μm. Data are expressed as percentage of positive cells; mean values and SEM are indicated. G–I, Representative confocal microscopy merged images (G) and quantification of γH2AX (H) and 53BP1 (I) foci per nucleus in NC-1 cells stably expressing Snail1 (GFP-Snail1 and CRE-Snail1) and their corresponding controls (GFP-Ø and CRE-Ø). Results show the mean ± SEM of three independent experiments. n = 80–100 cells analyzed per experiment. J, NC-GFP and NC-CRE cells were treated with increasing concentrations of olaparib for 72 hours and their proliferation rate was measured by MTT. Data show the mean ± SEM of six independent experiments. IC50 values were determined after normalization to the results obtained in vehicle-treated cells. K and L, Quantification of γH2AX (K) and 53BP1 (L) foci per nucleus in NC-GFP (G) and NC-CRE (C) cells treated with 1 μmol/L olaparib for the indicated times. Results represent the mean ± SEM of four independent experiments; n = 80–100 cells analyzed per experiment. M and N, Quantification of γH2AX foci per nucleus in MDA-MB-468 (M) and HCC1937 (N) control (shC) and E2A-silenced (sh1 and sh2) cells treated with 5 μmol/L and 50 μmol/L olaparib, respectively, for the indicated times. Results represent the mean ± SEM of three independent experiments. n = 150–200 cells analyzed per experiment. P value was calculated by two-sided unpaired Student t test in A, D, and E; two-sided Fisher exact test in F; and two-way ANOVA test in K–N.
Increased ploidy is associated with spontaneous DD and altered DNA damage response (DDR; refs. 51, 52). Thus, we next examined the DD status in NCs by detecting γH2AX and 53BP1 foci as markers of double-strand breaks (DSB; ref. 53). Increased γH2AX/53BP1 foci numbers were found in NC-CRE cells (Fig. 7C–E; Supplementary Fig. S7B and S7C) and in early primary tumors from PyMT-E2AKO mice (Fig. 7F) compared with their respective controls, suggesting that E2A deletion promotes CIN and the accumulation of DSBs in vitro and in vivo. Because Snail1 has been linked to increased genomic instability and resistance to DNA-damaging agents (13, 48), we next investigated whether this response was attributable to the differential expression of Snail1 between NC-CRE and NC-GFP cells. Noticeably, Snail1 overexpression reduced DSBs compared with control condition (Ø), both in NC-GFP-Snail1 and NC-CRE-Snail1 cells. However, NC-CRE-Snail cells still displayed significantly more γH2AX/53BP1 foci than NC-GFP-Snail1 cultures (Fig. 7G–I), indicating that the accumulation of DSBs in E2A-defficient cells is partially independent of Snail1 expression.
Increased DD, especially unresolved DSBs, is being exploited as a therapeutic weakness in breast cancer and other tumors. In particular, PARP inhibitors (PARPi) are being used for the treatment of breast cancer exhibiting defects on homologous recombination (HR)-mediated DNA repair (54), as well as in recurrent ovarian cancer and other tumor types regardless of their BRCA status or the presence of HR defects (55). Therefore, we investigated whether the PARPi olaparib could differentially affect PyMT-tumor cells depending on E2A expression. As expected, treatment with olaparib diminished the proliferation of NC-GFP cells (Fig. 7J). Interestingly, in these control cells, E2A protein levels increased in response to olaparib treatment along with the activation of the canonical DDR pathways (ATM-Chk2, ATR-Chk1, and p53; Supplementary Fig. S7D and S7E). Abrogation of E2A expression enhanced olaparib cytotoxicity in NC-CRE cells, as indicated by decreased IC50 compared with NC-GFP control (Fig. 7J). Although NC-CRE cells showed a basal increased fraction of cells in G2–M compared with NC-GFP cells (Supplementary Fig. S7F), no differences were detected in the expression levels of cell-cycle markers such as phospho-H3 or p27 upon PARP inhibition (Supplementary Fig. S7E), suggesting that olaparib was not promoting a cell-cycle arrest in E2A-deficient cells. At early time points of olaparib treatment, γH2AX/53BP1 foci increased in PyMT-NCs regardless of their E2A status. However, after 24 hours, γH2AX/53BP1 foci were significantly reduced in NC-GFP cells, while they remained high in NC-CRE cells up to 72 hours (Fig. 7K and L). Accordingly, canonical DDR pathways were similarly activated in NC-GFP and NC-CRE cells at early time-points of olaparib treatment. After 72 hours, most DDR signaling dropped to basal levels in NC-GFP cells, whereas higher levels of phosphorylated Chk1 and p53 remained in NC-CRE cells, together with increased levels of active caspase3 (Supplementary Fig. S7E), in line with enhanced DD and olaparib sensitivity (Fig. 7J–L). This finding suggests that E2A-proficient cells harbor a better competence to repair DNA and that E2A depletion leads to an increase in unresolved DSBs, which ultimately results in apoptosis activation and cleavage of caspase-3. The observed response seems to be specific of PARPi because E2A abrogation slightly increased the sensitivity of NC-CRE cells to paclitaxel, but not as markedly as treatment with olaparib (Supplementary Fig. S7G).
To explore the translational potential of the results obtained in PyMT-derived cells, we next investigated the consequences of E2A interference in the response to PARPi in the TNBC cell lines, MDA-MB-468 and HCC1937, harboring wild-type or mutated BRCA1, respectively (56). Similar to PyMT-NCs, E2A silencing in both cell lines promoted an increase in DNA damage in basal conditions (Supplementary Fig. S7H and S7I). In response to olaparib, E2A-interfered cells were also less capable to repair DNA, as evidenced by a greater number of γH2AX foci than those observed in E2A-proficient MDA-MB-468 and, to a lesser extent, HCC1937 cells after 72 hours of treatment (Fig. 7M and N). In addition, olaparib sensitivity was enhanced to different degrees in E2A-depleted cells (Supplementary Fig. S7J). These results suggest that the loss of E2A in TNBC cell lines favors genomic instability irrespective of their BRCA status. In support of this conclusion, no cooccurrence was found between E2A expression and the mutational status of BRCA1 or BRCA2 in the datasets of human IDBC from the TCGA previously used to identify the association between E2A and SNAI1 expression (Supplementary Fig. S7K).
Altogether, these results indicate that E2A deletion promotes the accumulation of unrepaired DSBs, a situation that favors a greater sensitivity of E2A-deficient cells to PARPi, pointing to E2A factors as promising therapeutic targets in breast cancer.
Discussion
Invasive and stem-like properties are features inherent to some tumor cells, but can also be acquired by certain tumor cell subpopulations in response to appropriate stimuli (1). The impact of EMT to tumor progression and metastasis has been previously explored (4, 5), but the contribution of specific EMT-TFs to tumor biology in certain tumor-contexts still remains elusive. Using the MMTV-PyMT model, we here describe a novel role for E2A proteins (E12/E47 EMT-TFs; ref. 21) in stemness and metastasis of breast cancer. Targeted deletion of E2A impairs the early stages of breast tumor growth and diminishes the self-renewal capacity of PyMT-TICs. This is accompanied by a switch to a luminal cell fate and a more differentiated tumor phenotype, a behavior consistent with the proposal that PyMT-dependent tumor progression is associated with EMT-activation and the acquisition of basal features (12). Considering the key role of E2A proteins in the maintenance of the hematopoietic stem cell pool and differentiation of B- and T-cell lineages (18, 19), it is tempting to speculate that E2A factors may regulate differentiation and self-renewal capacity in the mammary gland. Because in vivo deletion of E2A in the normal mammary gland epithelium does not alter gland morphogenesis, E12/E47 function could be restricted to stem cell/differentiation programs operating in breast CSCs but not in normal MECs. These observations are in line with the hypothesis that normal stem cells and TICs from the same tissue-of-origin exploit different molecular mechanism to activate related signaling pathways, as shown for Snail2 and Snail1 in breast tissue (12, 41). Alternatively, E2A proteins could contribute to the regulation of stem cell–like properties in normal MECs but their absence might be compensated in vivo by other EMT-TFs during mammary homeostasis.
Remarkably, our data from PyMT-E2AKO mice provide compelling evidence for the in vivo prometastatic action of E2A proteins in breast cancer, corroborating previous findings on the contribution of E47 to the premetastatic niche formation (25). The diminished invasive and metastatic potential of E2A-depleted cells is in full agreement with the reduced invasive component of E2A-deficient primary breast tumors and the dramatic decrease in the metastatic burden exhibited by PyMT-E2AKO mice, supporting that E2A factors regulate PyMT-dependent metastatic competence by cell-autonomous mechanisms. In this regard, we herein characterize Snail1 as a transcriptional target of E2A proteins during PyMT-driven tumorigenesis, E47 being able to activate the upstream Snai1-promoter through its E3-boxes. Because Snail1 expression is associated with the acquisition of basal features, dedifferentiation, and increased TIC and MIC activities in various breast cancer models (12, 45, 57), our findings suggest that E2A TFs may regulate malignant breast cancer progression by enhancing Snai1 transcription. Indeed, overexpression of Snail1 in PyMT-E2A–deficient cells fully restored their invasive and metastatic potential, as well as TIC competence concomitant with their switch toward a basal cell fate. Interestingly, PyMT-E2A–proficient and PyMT-E2A–deficient cells maintained E-cadherin expression together with that of several EMT-TFs and mesenchymal markers, which is suggestive of a hybrid E/M phenotype that might confer them high plasticity (5, 8), especially following Snail1 overexpression (10, 11). Accordingly, Snail1-overexpressing PyMT-tumor cells exhibited significant E-cadherin downregulation together with increased expression of the EMT-TF ZEB1 and the mesenchymal markers N-cadherin and vimentin, changes that may reflect their transition from an early to a late E/M phenotype (5). Notably, high E2A and SNAI1 mRNA levels occur in human IDBC and coexpression of E2A and SNAI1 associates with the aggressive basal-like phenotype, a breast tumor subtype compatible with hybrid E/M states (42). Indeed, a directed correlation between E2A and SNAIL1 protein levels was confirmed in a representative cohort of basal-like breast tumors. These observations reinforce the relevance of the E2A–Snail1 axis in human breast cancer biology and support the previous association of E2A expression with poor prognosis in human breast tumors (26). Because transient Snail1 expression seems to be required for efficient PyMT-dependent metastasis (57), we hypothesize that E2A factors might regulate Snail1 expression at early steps of the metastatic cascade, that is, invasion, while other E2A downstream targets could contribute to later stages of metastasis evolution. This reasoning also extends to the acquisition of stem cell–like properties, being plausible that other factors assist Snail1 in E2A-mediated stemness.
Besides the participation of E2A factors in tumor initiation and metastasis, we herein describe their contribution to genomic integrity and therapeutic resistance of breast cancer cells. E2A deletion increases CIN, and favors tetraploidization and the accumulation of DSBs, sensitizing PyMT-E2A–deficient cells to PARPi. Noticeably, E2A depletion also seems to favor a greater sensitivity of TNBC cell lines to olaparib independent of their BRCA1 status, as additionally supported from in silico analyses of E2A expression and BRCA1/2 mutational status of IDBC samples. Although the underlying mechanisms of E2A-mediated drug resistance are presently unknown, it is worth to mention that E2A proteins participate in megakaryocytic polyploidization by regulating Cdc6 transcription (58). Because CDC6 expression has been linked to poor prognosis and resistance to DNA-damaging agents in various types of cancer (59), altered expression of Cdc6 in E2A-deficient cells may contribute to increase genomic instability, eventually favoring olaparib cytotoxicity. However, participation of alternative mechanisms cannot be discarded, such as E2A-dependent regulation of p21/PUMA expression ratio upon p53 activation (60).
In summary, this study demonstrates that E2A EMT-TFs are key players in breast cancer stemness, tumor initiation, and metastasis; characterizes a novel E2A–Snail1 axis participating in mammary gland tumorigenesis and dedifferentiation; and identifies E2A as a vulnerability target to be exploited in novel therapeutic approaches for patients with breast cancer.
Authors' Disclosures
A. Cano reports grants from Spanish Ministry of Economy and Innovation, Spanish Institute of Health Carlos III (Madrid, Spain), and Worldwide Cancer Research during the conduct of the study. No disclosures were reported by the other authors.
Supplementary Material
Acknowledgments
The authors are grateful to Marie-Ange Deugnier (Institut Curie, Paris, France) for advice in transplantation experiments; Eva Díaz (MD Anderson Cancer Center, Madrid, Spain) for her support with the IHC analyses and human sample collection; Alberto Martín (Instituto de Investigación de Enfermedades Raras, ISCIII, Madrid, Spain) for scientific support and advice for the development of experimental protocols; and Guillermo de Cárcer and Bruno Sainz (IIBM, CSIC-UAM) for proving reagents and helpful discussions. The authors also acknowledge members of Amparo Cano's and Gema Moreno-Bueno's laboratories for constructive suggestions, as well as Raquel Arocha (Animal Care Facility, IIBM, CSIC-UAM) and Laura Molero (Flow-Cytometry Facility, SIdI-UAM) for help with the maintenance of mice colonies and FACS analyses, respectively. The authors also thank Genomics and Confocal Microscopy core facilities at IIBM (CSIC-UAM). This work has been supported by grants from the Spanish Ministry of Economy and Innovation (SAF2013–44739-R, SAF2016–76504-R to A. Cano and F. Portillo), PID2019–111052RB-I00 to F. Portillo, PID2019–104644RB-I00 to G. Moreno-Bueno; the Spanish Institute of Health Carlos III (CIBERONC-CB16/12/00295 to A. Cano, G. Moreno-Bueno; all of them partly supported by FEDER funds); the AECC Scientific Foundation 2019 (PROYE19036MOR to G. Moreno-Bueno); the Association International for Cancer Research (AICR; 12–1057 to A. Cano, P.G. Santamaría); and Worldwide Cancer Research (16–0295 to A. Cano, F. Portillo, and P.G. Santamaría). A. Vázquez-Naharro was funded by a PhD contract from SAF2016–76504-R grant; V. Santos by AICR (12–1057) and SAF2016–76504-R grants; P.G. Santamaría by a research contract of the AECC Scientific Foundation, and from Worldwide Cancer Research (16–0295) and SAF2016–76504-R grants.
The costs of publication of this article were defrayed, in part, by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Current address for V. Santos and P.G. Santamaría: Centro Nacional de Investigaciones Oncológicas (CNIO), Madrid, Spain.
Cancer Res 2021;81:4529–44
Authors' Contributions
C. López-Menéndez: Conceptualization, software, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. A. Vázquez-Naharro: Software, validation, investigation, visualization. V. Santos: Validation, investigation, visualization, methodology. P. Dubus: Data curation, formal analysis, validation, visualization, methodology. P.G. Santamaría: Formal analysis, methodology, writing–original draft, writing–review and editing. Á. Martínez-Ramírez: Investigation, visualization, methodology. F. Portillo: Conceptualization, resources, funding acquisition. G. Moreno-Bueno: Data curation, software, formal analysis, funding acquisition, investigation, methodology. M.M. Faraldo: Conceptualization, formal analysis, supervision, investigation, methodology. A. Cano: Conceptualization, resources, formal analysis, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing.
References
- 1. Batlle E, Clevers H. Cancer stem cells revisited. Nat Med 2017;23:1124–34. [DOI] [PubMed] [Google Scholar]
- 2. Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. Cell 2017;168:670–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Celia-Terrassa T, Kang Y. Distinctive properties of metastasis-initiating cells. Genes Dev 2016;30:892–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nieto MA, Huang RY, Jackson RA. TJP Emt: 2016. Cell 2016;166:21–45. [DOI] [PubMed] [Google Scholar]
- 5. Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol 2019;29:212–26. [DOI] [PubMed] [Google Scholar]
- 6. Stemmler MP, Eccles RL, Brabletz S, Brabletz T. Non-redundant functions of EMT transcription factors. Nat Cell Biol 2019;21:102–12. [DOI] [PubMed] [Google Scholar]
- 7. Peinado H, Olmeda D, Snail CA. Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer 2007;7:415–28. [DOI] [PubMed] [Google Scholar]
- 8. Santamaria PG, Moreno-Bueno G, Cano A. Contribution of epithelial plasticity to therapy resistance. J Clin Med 2019;8:676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gupta PB, Pastushenko I, Skibinski A, Blanpain C, Kuperwasser C. Phenotypic plasticity: driver of cancer initiation, progression, and therapy resistance. Cell Stem Cell 2019;24:65–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008;133:704–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morel AP, Lievre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One 2008;3:e2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ye X, Tam WL, Shibue T, Kaygusuz Y, Reinhardt F, Eaton NE, et al. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature 2015;525:256–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 2015;527:525–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 2015;527:472–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Beerling E, Seinstra D, de Wit E, Kester L, van der Velden D, Maynard C, et al. Plasticity between epithelial and mesenchymal states unlinks EMT from metastasis-enhancing stem cell capacity. Cell Rep 2016;14:2281–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fico F, Bousquenaud M, Ruegg C, Santamaria-Martinez A. Breast cancer stem cells with tumor- versus metastasis-initiating capacities are modulated by TGFBR1 inhibition. Stem Cell Reports 2019;13:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol 2000;20:429–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Belle I, Zhuang Y. E proteins in lymphocyte development and lymphoid diseases. Curr Top Dev Biol 2014;110:153–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Semerad CL, Mercer EM, Inlay MA, Weissman IL, Murre C. E2A proteins maintain the hematopoietic stem cell pool and promote the maturation of myelolymphoid and myeloerythroid progenitors. Proc Natl Acad Sci U S A 2009;106:1930–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 2007;446:758–64. [DOI] [PubMed] [Google Scholar]
- 21. Perez-Moreno MA, Locascio A, Rodrigo I, Dhondt G, Portillo F, Nieto MA, et al. A new role for E12/E47 in the repression of E-cadherin expression and epithelial-mesenchymal transitions. J Biol Chem 2001;276:27424–31. [DOI] [PubMed] [Google Scholar]
- 22. Peinado H, Marin F, Cubillo E, Stark HJ, Fusenig N, Nieto MA, et al. Snail and E47 repressors of E-cadherin induce distinct invasive and angiogenic properties in vivo. J Cell Sci 2004;117:2827–39. [DOI] [PubMed] [Google Scholar]
- 23. Moreno-Bueno G, Cubillo E, Sarrio D, Peinado H, Rodriguez-Pinilla SM, Villa S, et al. Genetic profiling of epithelial cells expressing E-cadherin repressors reveals a distinct role for Snail, Slug, and E47 factors in epithelial-mesenchymal transition. Cancer Res 2006;66:9543–56. [DOI] [PubMed] [Google Scholar]
- 24. Salvador F, Martin A, Lopez-Menendez C, Moreno-Bueno G, Santos V, Vazquez-Naharro A, et al. Lysyl oxidase-like protein LOXL2 promotes lung metastasis of breast cancer. Cancer Res 2017;77:5846–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Canesin G, Cuevas EP, Santos V, Lopez-Menendez C, Moreno-Bueno G, Huang Y, et al. Lysyl oxidase-like 2 (LOXL2) and E47 EMT factor: novel partners in E-cadherin repression and early metastasis colonization. Oncogene 2015;34:951–64. [DOI] [PubMed] [Google Scholar]
- 26. Cubillo E, Diaz-Lopez A, Cuevas EP, Moreno-Bueno G, Peinado H, Montes A, et al. E47 and Id1 interplay in epithelial-mesenchymal transition. PLoS One 2013;8:e59948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tang P, Tse GM. Immunohistochemical surrogates for molecular classification of breast carcinoma: a 2015 update. Arch Pathol Lab Med 2016;140:806–14. [DOI] [PubMed] [Google Scholar]
- 28. Pan L, Hanrahan J, Li J, Hale LP, Zhuang Y. An analysis of T cell intrinsic roles of E2A by conditional gene disruption in the thymus. J Immunol 2002;168:3923–32. [DOI] [PubMed] [Google Scholar]
- 29. Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, et al. Purification and unique properties of mammary epithelial stem cells. Nature 2006;439:993–7. [DOI] [PubMed] [Google Scholar]
- 30. Spike BT, Engle DD, Lin JC, Cheung SK, La J, Wahl GM. A mammary stem cell population identified and characterized in late embryogenesis reveals similarities to human breast cancer. Cell Stem Cell 2012;10:183–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Faraldo MM, Glukhova MA, Deugnier MA. The transplantation of mouse mammary epithelial cells into cleared mammary fat pads. Methods Mol Biol 2015;1293:161–72. [DOI] [PubMed] [Google Scholar]
- 32. Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods 2009;347:70–8. [DOI] [PubMed] [Google Scholar]
- 33. Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 2009;27:1160–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee K, Gjorevski N, Boghaert E, Radisky DC, Nelson CM. Snail1, Snail2, and E47 promote mammary epithelial branching morphogenesis. EMBO J 2011;30:2662–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol 1992;12:954–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lin EY, Jones JG, Li P, Zhu L, Whitney KD, Muller WJ, et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol 2003;163:2113–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol 2007;8:R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Malanchi I, Santamaria-Martinez A, Susanto E, Peng H, Lehr HA, Delaloye JF, et al. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature 2011;481:85–9. [DOI] [PubMed] [Google Scholar]
- 41. Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 2012;148:1015–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sarrio D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res 2008;68:989–97. [DOI] [PubMed] [Google Scholar]
- 43. Drobysheva D, Smith BA, McDowell M, Guillen KP, Ekiz HA, Welm BE. Transformation of enriched mammary cell populations with polyomavirus middle T antigen influences tumor subtype and metastatic potential. Breast Cancer Res 2015;17:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kinoshita M, Matsuda Y, Arai T, Soejima Y, Sawabe M, Honma N. Cytological diagnostic clues in poorly differentiated squamous cell carcinomas of the breast: Streaming arrangement, necrotic background, nucleolar enlargement and cannibalism of cancer cells. Cytopathology 2018;29:22–7. [DOI] [PubMed] [Google Scholar]
- 45. Ni T, Li XY, Lu N, An T, Liu ZP, Fu R, et al. Snail1-dependent p53 repression regulates expansion and activity of tumour-initiating cells in breast cancer. Nat Cell Biol 2016;18:1221–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chaffer CL, Marjanovic ND, Lee T, Bell G, Kleer CG, Reinhardt F, et al. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell 2013;154:61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yu W, Zhang Y, Ruest LB, Svoboda KK. Analysis of Snail1 function and regulation by Twist1 in palatal fusion. Front Physiol 2013;4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pyun BJ, Seo HR, Lee HJ, Jin YB, Kim EJ, Kim NH, et al. Mutual regulation between DNA-PKcs and Snail1 leads to increased genomic instability and aggressive tumor characteristics. Cell Death Dis 2013;4:e517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Piao J, Takai S, Kamiya T, Inukai T, Sugita K, Ohyashiki K, et al. Poly ( ADP-ribose) polymerase inhibitors selectively induce cytotoxicity in TCF3-HLF-positive leukemic cells. Cancer Lett 2017;386:131–40. [DOI] [PubMed] [Google Scholar]
- 50. Eldfors S, Kuusanmaki H, Kontro M, Majumder MM, Parsons A, Edgren H, et al. Idelalisib sensitivity and mechanisms of disease progression in relapsed TCF3-PBX1 acute lymphoblastic leukemia. Leukemia 2017;31:51–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Storchova Z, Kuffer C. The consequences of tetraploidy and aneuploidy. J Cell Sci 2008;121:3859–66. [DOI] [PubMed] [Google Scholar]
- 52. Janssen A, van der Burg M, Szuhai K, Kops GJ, Medema RH. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science 2011;333:1895–8. [DOI] [PubMed] [Google Scholar]
- 53. Zimmermann M, de Lange T. 53BP1: pro choice in DNA repair. Trends Cell Biol 2014;24:108–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mateo J, Lord CJ, Serra V, Tutt A, Balmana J, Castroviejo-Bermejo M, et al. A decade of clinical development of PARP inhibitors in perspective. Ann Oncol 2019;30:1437–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ison G, Howie LJ, Amiri-Kordestani L, Zhang L, Tang S, Sridhara R, et al. FDA approval summary: niraparib for the maintenance treatment of patients with recurrent ovarian cancer in response to platinum-based chemotherapy. Clin Cancer Res 2018;24:4066–71. [DOI] [PubMed] [Google Scholar]
- 56. Keung MY, Wu Y, Badar F, Vadgama JV. Response of breast cancer cells to PARP inhibitors is independent of BRCA status. J Clin Med 2020;9:940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tran HD, Luitel K, Kim M, Zhang K, Longmore GD, Tran DD. Transient SNAIL1 expression is necessary for metastatic competence in breast cancer. Cancer Res 2014;74:6330–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vilaboa N, Bermejo R, Martinez P, Bornstein R, Cales C. A novel E2 box-GATA element modulates Cdc6 transcription during human cells polyploidization. Nucleic Acids Res 2004;32:6454–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mahadevappa R, Neves H, Yuen SM, Bai Y, McCrudden CM, Yuen HF, et al. The prognostic significance of Cdc6 and Cdt1 in breast cancer. Sci Rep 2017;7:985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Andrysik Z, Kim J, Tan AC, Espinosa JM. A genetic screen identifies TCF3/E2A and TRIAP1 as pathway-specific regulators of the cellular response to p53 activation. Cell Rep 2013;3:1346–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.