Abstract
Mutations in the gene encoding the transcription factor CCAAT/enhancer-binding protein alpha (C/EBPα) occur in 10–15% of acute myeloid leukemia (AML). Frameshifts in the CEBPA N-terminus resulting in exclusive expression of a truncated p30 isoform represent the most prevalent type of CEBPA mutations in AML. C/EBPα p30 interacts with the epigenetic machinery, but it is incompletely understood how p30-induced changes cause leukemogenesis. We hypothesized that critical effector genes in CEBPA-mutated AML are dependent on p30-mediated dysregulation of the epigenome. We mapped p30-associated regulatory elements (REs) by ATAC-seq and ChIP-seq in a myeloid progenitor cell model for p30-driven AML that enables inducible RNAi-mediated knockdown of p30. Concomitant p30-dependent changes in gene expression were measured by RNA-seq. Integrative analysis identified 117 p30-dependent REs associated with 33 strongly down-regulated genes upon p30-knockdown. CRISPR/Cas9-mediated mutational disruption of these genes revealed the RNA-binding protein MSI2 as a critical p30-target. MSI2 knockout in p30-driven murine AML cells and in the CEBPA-mutated human AML cell line KO-52 caused proliferation arrest and terminal myeloid differentiation, and delayed leukemia onset in vivo. In summary, this work presents a comprehensive dataset of p30-dependent effects on epigenetic regulation and gene expression and identifies MSI2 as an effector of the C/EBPα p30 oncoprotein.
Introduction
Acute myeloid leukemia (AML) is characterized by aberrant myeloid homeostasis, leading to an increase in myeloid progenitor cells at the expense of mature blood cells [1]. Despite recent advances in therapeutic options, the overall prognosis is poor for most AML patients. Therefore, there is an urgent need for a better understanding of molecular mechanisms of leukemia initiation and maintenance to develop novel therapies.
Ten to fifteen percent of de novo AML patients harbor mutations in the CEBPA gene [2, 3], which encodes the transcription factor (TF) CCAAT/enhancer-binding protein alpha (C/EBPα). C/EBPα directly binds to DNA as a homo- or heterodimer and can regulate gene expression via a C-terminal basic-region leucine zipper domain [4]. Alternative usage of two translation initiation sites leads to the expression of a full-length (42 kDa) and a shorter (30 kDa) isoform of the C/EBPα protein, which are termed p42 and p30, respectively [5]. In AML patients, mutations are found in two hotspot regions of the CEBPA gene: N-terminal frameshift mutations are located between the first and second initiation codons, resulting in selective ablation of p42 expression. C-terminal in-frame mutations affect the structure of the basic region and/or the leucine zipper, leading to loss of DNA binding [3, 6, 7]. It has been reported that most AML patients carry biallelic CEBPA mutations, harboring an N-terminal frameshift together with a C-terminal in-frame mutation, although some datasets prevalently feature patient samples with N-terminal CEBPA mutations [8, 9].
It has been proposed that the leukemia-associated p30 isoform represents a gain-of-function variant [10]. C/EBPα p30 is able to actively change the gene expression pattern of leukemia cells through its ability to bind to chromatin and consequently recruit chromatin modulators, such as histone methyltransferases [7, 11–13]. Indeed, p30 has a strong impact on the transcriptional program of myeloid cells [11, 13, 14]. However, the molecular mechanisms of C/EBPα p30-mediated regulation of gene expression are still incompletely understood.
Regulatory elements (REs) are non-coding regions that can be bound by transcriptional regulators and chromatin modulators in a locus-specific fashion, thereby governing tissue-specific regulation of gene expression. The critical importance of REs in cancer development has repeatedly been demonstrated. For instance, alterations in the genomic landscape can cause the establishment of cancer-specific REs that drive aberrant gene expression and consequently result in oncogenic transformation [15, 16]. In addition, AML oncoproteins can bind to particular enhancer repertoires, thereby recruiting distinct TFs to REs to deregulate gene expression [17, 18]. In fact, it was shown that functional annotation of regulatory regions in cancer is superior to RNA-sequencing data to map cellular lineages during normal hematopoietic development and to classify AML subtypes [19].
Given its potential to interact with various chromatinmodifying enzymes, we hypothesized that C/EBPα p30 plays an important role in the induction and maintenance of global epigenetic changes via the establishment and usage of AML-specific REs. Furthermore, detailed analysis of p30-associated REs could result in the discovery of p30-driven effector genes that are crucial for AML development and maintenance. In this study, we applied a combinatorial functional genomics approach to integrate TF and histone modification ChIP-seq, ATAC-seq, and RNA-seq data from a cellular model of N-terminal CEBPA-mutated AML. Systematic interrogation of the resulting dataset identified 33 genes whose expression was directly driven by the C/EBPα p30 oncoprotein. The functional investigation by CRISPR/Cas9 screening identified the RNA-binding protein (RBP) MUSASHI-2 (MSI2) as a critical downstream effector of p30. AML patients with CEBPA mutations expressed high levels of MSI2, and MSI2 was required for the survival of murine and human AML cells with CEBPA N-terminal or doublemutated genotypes. These data provide a comprehensive evaluation of target genes of the C/EBPα-p30 oncoprotein and establish MSI2 as a potential target in patients with CEBPA-mutated AML.
Materials and methods
CRISPR/Cas9 competition assays
SpCas9-expressing subclones of Cebpap30/p30 and of Cebpap30/C-mut cells were isolated after lentiviral expression of lenti-Cas9-Blast (Addgene #52962). SpCas9-Cebpap30/p30 and SpCas9-Cebpap30/C-mut cells were transduced with sgRNA-containing LentiGuide-Puro-IRES-GFP constructs for single sgRNA expression or pKLV2.2-mU6gRNA5 (SapI)-hU6gRNA5(BbsI)-PGKpuroBFP (Addgene, #72667) for dual sgRNA expression, obtaining transduction efficiencies ranging from 10 to 50%. The dynamics of GFP or iRFP670 levels were monitored over time. Data were normalized to values on day 0 (72 or 96 h after the last infection) and a non-targeting control sgRNA (Ctrl) (for sgRNAs see Supplementary Table S1).
Quantitative real-time PCR analysis
Total RNA was isolated using the RNeasy Mini Kit (Qiagen) and reverse-transcribed into cDNA using the RevertAid first-strand cDNA synthesis Kit (ThermoFisher Scientific). cDNA was amplified on the Bio-Rad CFX96-Real-TimeSystem using SsoAdvanced Universal SYBR green Supermix (Bio-Rad, Hercules, USA). Gene expression levels were calculated by normalizing the values to Gapdh expression (for qPCR primers see Supplemental Table S2).
Chromatin immunoprecipitation (ChIP)
ChIP was performed as previously described [20]. In brief, Cebpap30/p30 cells were crosslinked with 11% formaldehyde (Thermo Fisher Scientific, Waltham, MA, USA), quenched with glycine, and lysed in 10% SDS-buffer. After sonication, cells were incubated overnight with anti-C/EBPα (Santa Cruz, Dallas, Texas, USA, sc-9314) or anti-H3K27ac (Abcam, Cambridge, UK, ab4729) antibodies. Antibody-bound DNA was isolated using G-protein coupled magnetic beads (Dynabeads Protein G, Invitrogen, Camarillo, CA, USA) and decrosslinked. Enrichment of genomic DNA was measured via qRT-PCR.
Statistics
Prism 5.01 software (Graphpad, La Jolla, CA, USA) was used for statistical analyses, and data are shown as mean ± SD. Experiments were performed in duplicates/triplicates and/or repeated at least three times. The individual sample size is reported in figure legends. If not stated differently, the unpaired Student’s t-test was used for p-value determination. Differences in survival were evaluated with the logrank test. The Wilcoxon test was used for statistical analysis of patient data. p < 0.05 was considered statistically significant (*p <0.05, **p <0.01, ***p <0.001, ****p < 0.0001, n.s. = not significant).
Additional materials and methods are described in Supplementary Methods.
Results
C/EBPα p30 knockdown results in global changes in the chromatin landscape
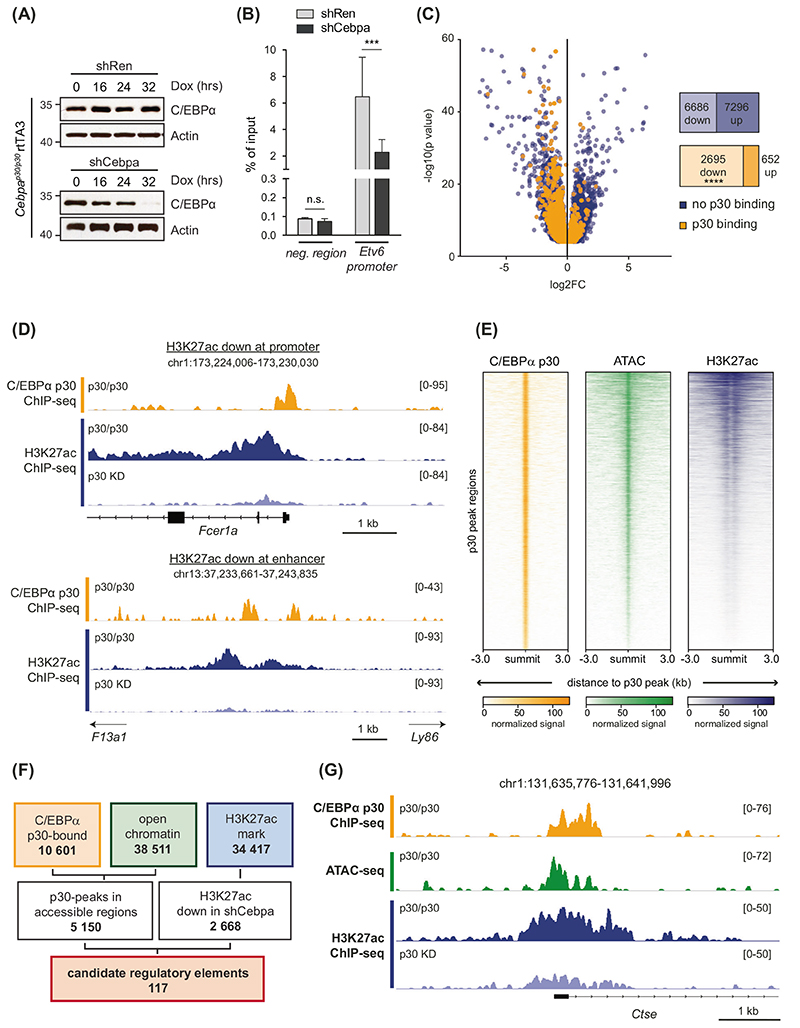
To explore the effects of the C/EBPα p30 isoform on the regulatory landscape in CEBPA-mutated AML, we used a Cebpap30/p30 myeloid progenitor cell line that was derived from a mouse AML model mimicking N-terminal CEBPA mutations [13, 21]. These cells exclusively express the p30 C/EBPα isoform and were classified as myeloid progenitor cells through analysis of global gene expression patterns. The presence of mast cells was excluded by cell surface marker expression and Toluidine blue staining (Supplementary Fig. 1A–D). To identify p30-dependent epigenomic and transcriptomic changes we employed doxycycline (Dox)-inducible RNA interference (RNAi) of Cebpa (Fig. 1A). Despite a slightly lower knockdown efficiency of shCebpa #2, both Cebpa-targeting shRNAs induced similar anti-proliferative effects in Cebpap30/p30 cells (Supplementary Fig. 1E, F), and all subsequent experiments were performed with shCebpa #1 (shCebpa). ChIP using antibodies against histone-3 acetylated at lysine-27 (H3K27ac) was used to characterize the global dynamic regulation of REs including promoters and enhancers in response to p30-expression. ChIP-qPCR analysis revealed that H3K27ac levels at the exemplary promoter of the Etv6 gene were decreased after Dox-induced p30 down-regulation (Fig. 1B). Analysis of ChIP-seq data from C/EBPα p30-proficient (shRen) vs. -deficient (shCebpa) Cebpap30/p30 cells identified 34 417 H3K27ac-marked REs, of which 6 686 (18.6%) were down- and 7 296 (21.1%) were up-regulated upon loss of C/EBPα p30. Integration of these results with data of genomic p30 binding [13] revealed that the majority of REs that also exhibited C/EBPα p30 binding was down-regulated (2 695; 19.6% of regulated REs), while few were up-regulated (652; 4.8%) upon RNAi-induced p30 loss (Fig. 1C). These changes were observed in both promoter and enhancer regions (Fig. 1D). As active gene regulation is associated with open chromatin states, the assay for transposase-accessible chromatin sequencing (ATAC-seq) was used to gain additional information about the chromatin state and activity of REs [22]. Thirty-seven percent of all accessible regions (14 296 of 38 511) were found in the proximity (0–3 kb) of annotated transcriptional start sites (TSS) of genes, indicating active promoters (Supplementary Fig. 1G). A large fraction of ATAC-seq peaks (28%, 10 270 of 38 511) localized to distal areas containing other REs, such as enhancers or insulators (>10 kb from TSS). Global comparison of C/EBPα p30 ChIP-seq data with ATAC-seq and H3K27ac ChIP-seq data in Cebpap30/p30 cells yielded a strong correlation of p30 peak intensity with chromatin accessibility and increased H3K27ac occupancy (Fig. 1E). To identify C/EBPα p30-regulated enhancer and promoter regions that might drive genes, which are crucial for N-terminal CEBPA-mutated AML, we reasoned that such genomic regions 1, are directly bound by p30 2, lie within accessible chromatin, and 3, exhibit reduced H3K27 acetylation upon p30 depletion. This filtering strategy identified 117 candidate REs among promoter and enhancer regions (Fig. 1F, G). In line with their potential involvement in leukemia, functional annotation revealed that these REs were strongly associated with abnormal immune cell function (Supplementary Fig. 1H). In summary, this epigenomic dataset enables detailed studies of the effects of C/EBPα p30 on the epigenomic landscape.
Fig. 1. C/EBPα p30 knockdown results in global changes in the chromatin landscape.
A Western blot analysis of indicated proteins from lysates of Cepbap30/p30 rtTA3 cells expressing Dox-inducible shRNAs against Renilla luciferase (shRen, control) or C/EBPα (shCebpa) at indicated time points after Dox addition. B H3K27ac ChIP-qPCR analysis of indicated regions in Cepbap30/p30 rtTA3 shRen and shCebpa after 48 h of Dox induction, n = 3. neg. region, negative control. C Differential H3K27ac-binding upon Cebpa knockdown. Volcano plot displaying differential H3K27ac-peaks (p <0.05, mean read concentration > 5) in Cebpap30/p30 rtTA3 shCebpa vs. shRen samples. C/EBPα p30-bound peaks are indicated in orange and numbers of differentially regulated regions are given. Statistical analysis was performed using Pearson’s Chi-squared test (X2(1, N = 17 329) = 1 163, p < 0.00001). KD, knockdown. D Representative examples of p30-bound promoters (top) and enhancers (bottom) with down-regulated H3K27ac signals upon p30 knockdown. E Heatmaps showing p30 binding, accessible chromatin (ATAC-seq), and H3K27ac intensities at 10 601 identified p30-bound regions (p < 0.05). F Filtering scheme for the identification of candidate REs. Bold numbers indicate the number of regions identified in each analysis/filtering step. C/EBPα p30-bound regions (p < 0.05) were intersected with open chromatin (p < 10−5), resulting in 5 150 p30 peaks in accessible regions. Combination with down-regulated H3K27ac peaks (fold change < –2; 2 668/34417) resulted in 117 candidate REs. G C/EBPα p30 (orange), ATAC-seq (green) and H3K27ac (blue) ChIP-seq profiles for Cebpap30/p30 shRen cells (p30/p30) and shCebpa cells (p30 KD) at an exemplary candidate RE.
C/EBPα p30 regulates expression of differentiation-associated genes at accessible REs
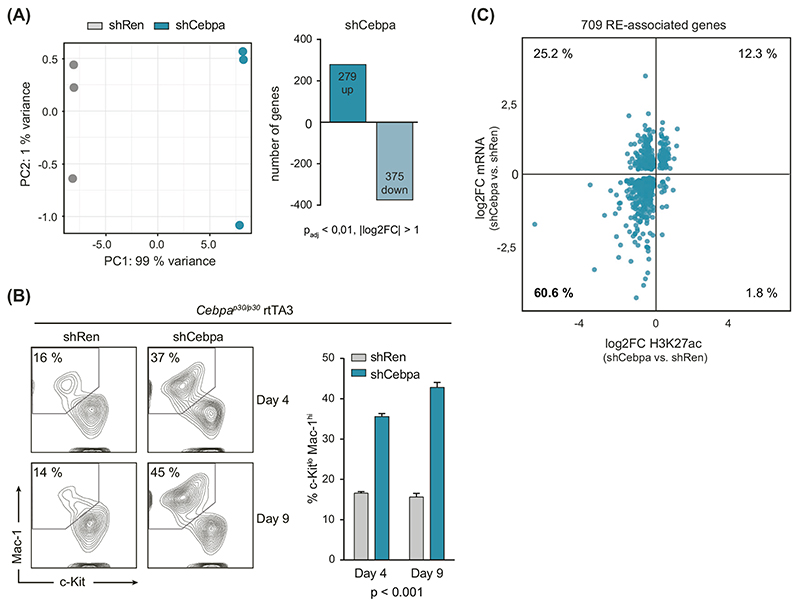
To obtain more information about the direct effects of C/EBPα p30 on target gene expression, we used the Dox-inducible RNAi system for global analysis of gene expression changes upon p30 loss in Cebpap30/p30 cells. RNA-sequencing (RNA-seq) after 48 h of Dox-treatment confirmed successful knockdown of Cebpa (Supplementary Fig. 2A). Global analysis of RNA-seq data identified C/EBPα p30-dependent gene expression programs. p30 knockdown caused the up-regulation of 279 genes, while 375 genes were strongly down-regulated (p < 0.01, fold change > 2; Fig. 2A). Functional annotation of genes that were up-regulated upon loss of C/EBPα p30 using gene ontology revealed an association with terminal myeloid differentiation, as exemplified by the induction of the myeloid genes Egr1, Irf8, Ccl3 and others (Supplementary Fig. 2B, C and Supplementary Table S3). In line with this observation, knockdown of Cebpa in Cebpap30/p30 cells resulted in decreased levels of the progenitor marker c-Kit, while the expression of the myeloid marker Mac-1 was increased (Fig. 2B). Combined analysis of p30-driven REs and their effect on gene expression identified 709 genes that were associated with p30-dependent H3K27 acetylation and whose expression was regulated by p30 (Supplementary Fig. 2D–F). Of note, the majority (60.6%; 430 of 709) of these genes exhibited down-regulated expression together with reduced H3K27ac signals upon RNAi-induced p30 loss (Fig. 2C). These results indicate that C/EBPα p30 occupancy is associated with active gene expression at accessible promoters and enhancers that directly regulate the expression of genes important for the maintenance of the myeloid differentiation block in Cebpap30/p30 cells.
Fig. 2. C/EBPα p30 regulates the expression of differentiation-associated genes in the context of p30-regulated REs.
A Left, principal component analysis (PCA) of transcriptomes from shRen and shCebpa Cebpap30/p30 cells. Right, number of differentially regulated genes between shRen and shCebpa Cebpap30/p30 cells. B Flow cytometric analysis of Mac-1 and c-Kit levels in shRNA-expressing Cepbap30/p30 rtTA3 cells upon Dox addition (left panels) and quantification of c-Kitlt> Mac-1hi population at indicated time points (right panel). The p values for all comparisons of shRen vs. shCebpa are indicated (n = 3). C Scatterplot showing log2FC of H3K27ac (x-axis) and normalized mRNA expression (y-axis) upon loss of p30. The 5 150 C/EBPα p30-bound peaks in accessible regions (Fig. 1F) were intersected with all differential H3K27ac-peaks to generate a list of 1 845 REs, which were associated with 709 differentially expressed genes upon p30 knockdown using GREAT [23]. Each point represents a gene with its associated REs that feature p30-binding and accessible chromatin.
CRISPR/Cas9 screening identifies MSI2 as a critical factor in C/EBPα p30-expressing AML cells
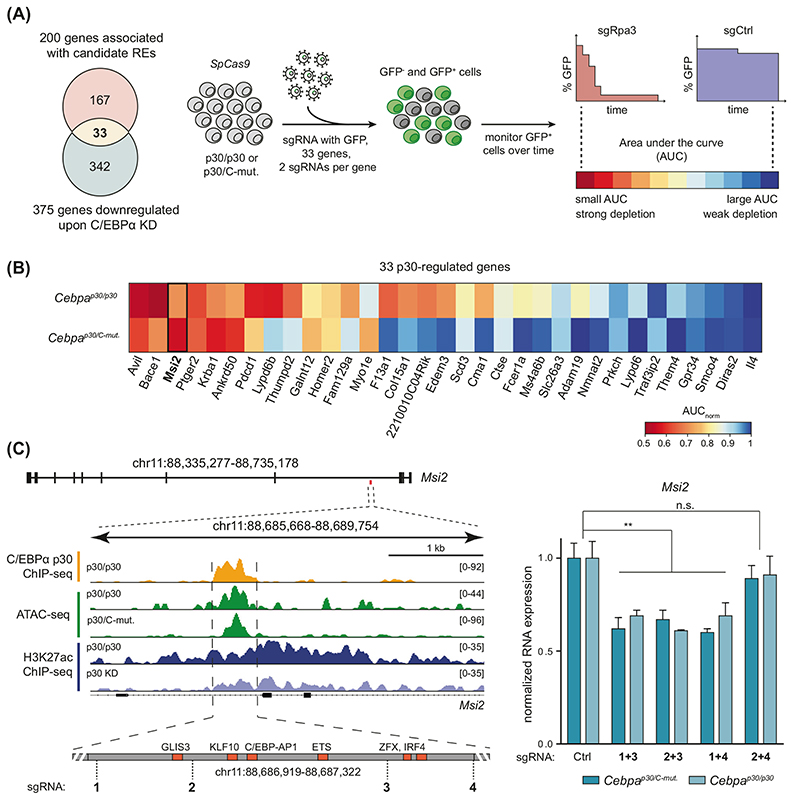
We next reasoned that direct gene targets of C/EBPα p30 are associated with the 117 p30-dependent REs (Fig. 1F) and are down-regulated upon p30 knockdown. 200 genes were predicted to be associated with the 117 p30-dependent candidate REs by the genomic regions enrichment of annotations tool (GREAT) [23]. Of those, 33 genes were strongly down-regulated upon p30-targeted RNAi (p < 0.01, fold change < −2). (Fig. 3A; Supplementary Fig. 2G). This list of genes encodes for proteins with different functions, including proteases (BACE1, CTSE, ADAM19), G-protein coupled receptors (GPR34, PTGER2), and transmembrane receptors involved in the regulation of immune responses (PDCD1, FCER1A).
Fig. 3. CRISPR/Cas9 screening identifies MSI2 as a critical factor in C/EBPα p30-expressing AML cells.
A Left, the overlap of genes associated with candidate REs and genes down-regulated upon p30 knockdown (fold change <–2, p adj<0.01). Right, schematic representation of the CRISPR/Cas9-based competition assay. Each gene is targeted with 2 sgRNAs in SpCas9-expressing Cebpap30/p30 and Cebpap30/C-mut clones. sgRNA-expressing cells (GFP+) are monitored over time and the area under the curve (AUC) is determined for each sgRNA as indicated. KD knockdown, Ctrl negative control, Rpa3 positive control. B Ranked mean AUCs for both sgRNA pairs in each cell line are displayed. AUCnorm, normalized to sgRen. C Left, a schematic overview of the Msi2 enhancer region. Transcription factor binding motifs in the RE (in red) and recognition sites of sgRNAs (dashed lines) are indicated. Right, qRT-PCR analysis of Msi2 in Cebpap30/p30 and Cebpap30/C-mut upon targeting the RE with sgRNA pairs shown in the left schematic, compared to sgRen (ctrl negative control).
We sought to investigate whether the 33 high-confidence p30-target genes identified by our approach are of functional importance for CEBPA-mutated AML. We performed CRISPR/Cas9-mediated mutagenesis in stably SpCas9-expressing clones of myeloid progenitor cells with the genetic background of N-terminally mutated (Cebpap30/p30) or double-mutated (Cebpap30/C-mut) Cebpa (Supplementary Fig. 1A) [24]. A competition assay was used to monitor competing growth kinetics of sgRNA-expressing, mutated GFP-positive (GFP+) cells vs. uninfected GFP-negative (GFP−) SpCas9 cells (Fig. 3A) [13]. While the percentage of GFP+ cells remained stable over time in cells expressing control sgRNAs (sgCtrl), transduced cells were strongly depleted when an essential gene for DNA replication (sgRpa3) was targeted. Similar to Cebpap30/p30 cells [13], the proliferation of Cebpap30/C-mu’ cells was dependent on Cebpa expression, as targeted mutagenesis of Cebpa with three different sgRNAs had strong anti-proliferative effects (Supplementary Fig. 3A, B).
We targeted all 33 candidate p30 target genes with two different sgRNAs (Fig. 3A). The area under the curve (AUC) for each sgRNA in both Cebpa-mutated genotypes (p30/p30 vs. p30/C-mut, resulting in four values per gene) was used to score gene essentiality in this system. Our scoring algorithm revealed Avil, Bace1, and Msi2 as the most essential genes for the proliferation of Cebpa-mutant cells (Fig. 3B, Supplementary Fig. 3C). The Avil gene encodes an actin regulatory protein and Bace1 belongs to the peptidase A1 family of aspartic proteases, which are involved in the formation of amyloid-beta plaques [25, 26]. As neither of these genes has been linked to cancer, we decided to focus on Msi2. The Msi2 gene encodes for the RBP MSI2, which has been implicated in various cancer entities [27–36]. MSI2 expression is frequently dysregulated in leukemia and high MSI2 expression has been linked to poor prognosis in this disease [37]. Msi2 expression was decreased upon C/EBPα p30 knockdown in Cebpap30/p30 cells as shown by quantitative PCR (Supplementary Fig. 3D). Analysis of the MSI2-associated RE revealed p30-binding, accessible chromatin, and loss of H3K27ac upon p30 knockdown. Amongst others, the RE harbored TF binding motifs for members of the ETS family, which are known to interact with C/EBPα. CRISPR/Cas9-mediated mutational disruption of the MSI2-associated RE led to a reduction of Msi2 expression, demonstrating its importance for gene regulation (Fig. 3C). Taken together, our CRISPR/Cas9-mediated mutagenesis survey of candidate p30-driven target genes identified the RBP MSI2 as an essential factor for the proliferation of Cebpa-mutated AML cells.
The RBP MSI2 is critical for Cebpa-mutated AML cells
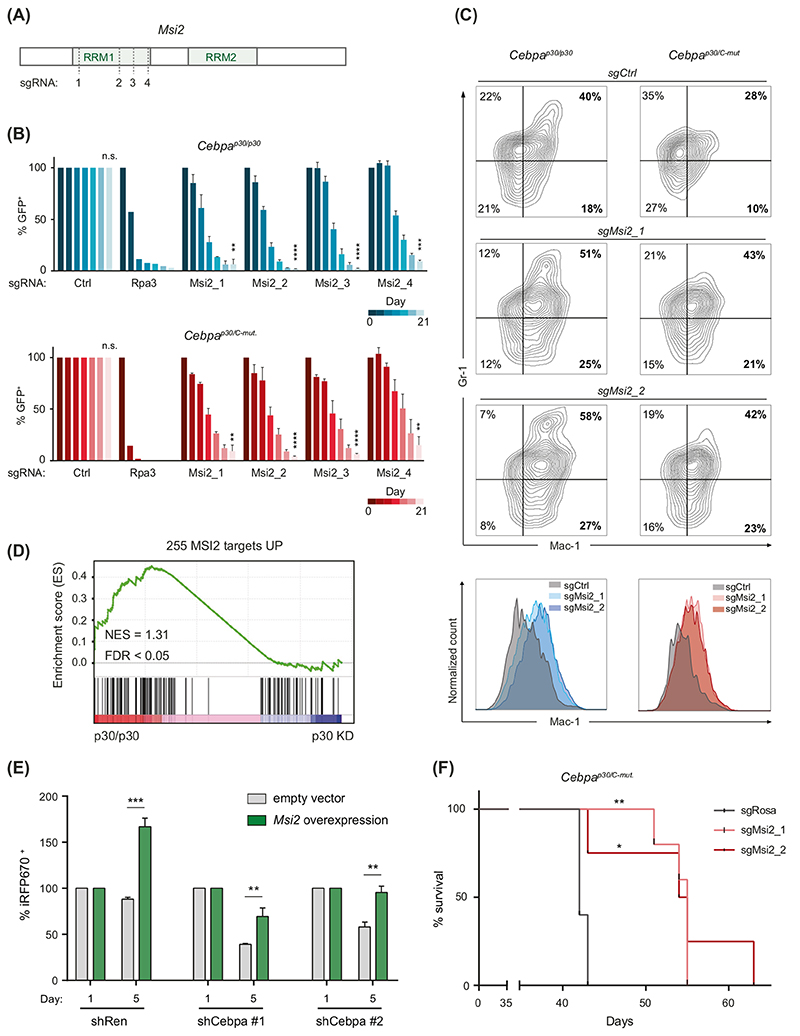
The strong dependency of Cebpa-mutated cells on Msi2 expression was confirmed in independent experiments using four unique sgRNAs targeting the RNA-binding motif 1 (RRM1) of the Msi2 gene (Fig. 4A). All sgRNAs caused strong depletion of transduced cells in independently derived SpCas9-expressing clones of Cebpap30/p30 and Cebpap30/C-mut cells (Fig. 4B). Mutational disruption of Msi2 induced myeloid differentiation in both cell lines, as measured by an increase in the Mac-1 + population (Fig. 4C). Importantly, p30 expression was strongly associated with high expression of 255 previously reported MSI2-target genes (NES = 1.31, p <0.05) (Fig. 4D). Exogenous expression of Msi2 was able to counteract the proliferation arrest induced by Cebpa knockdown, indicating that p30-dependent induction of proliferation is dependent on Msi2 (Fig. 4E, Supplementary Fig. 4A) [38]. Finally, Msi2 knockout in leukemia-initiating cells with biallelic Cebpa mutations delayed leukemia onset in an in vivo transplantation model (Fig. 4F, Supplementary Fig. 4B–D). These data show that MSI2 is a critical effector of the C/EBPα p30-mediated differentiation block and leukemogenesis.
Fig. 4. The RNA-binding protein MSI2 is critical for Cebpa-mutated AML cell proliferation.
A Schematic structure of the MSI2 gene. Recognition sites of sgRNAs are indicated by dashed lines. RRM RNA recognition motif. B Survival of sgRNA-expressing Cebpap30/p30 SpCas9 and Cebpap30/C-mut SpCas9 cells over time normalized to sgRen (Renilla) as a negative control, and day 0. Ctrl negative control, Rpa3 positive control. C Flow cytometric analysis of Mac-1 and Gr-1 levels in sgRNA-expressing SpCas9 Cepbap30/p30 and Cepbap30/C-mut cells. Ctrl negative control. D Gene set enrichment analysis (GSEA) showing global enrichment of MSI2-targets in Cebpap30/p30 shRen (p30/p30) vs. shCebpa (p30 KD) cells. NES normalized enrichment score, FDR false discovery rate. E Viability of Cebpap30/p30 rtTA3 shCebpa (#1 and #2) cells transduced either with empty or Msi2-overexpression vector (coupled to iRFP670), was measured over time. Values were normalized to day 1 posttransduction. F Survival analysis of leukemia-initiating Cebpap30/C-mut . SpCas9 cells transduced with sgRosa (control), sgMsi2_1 and sgMsi2_2 (n ≥ 3). Statistical analysis was performed using the logrank test.
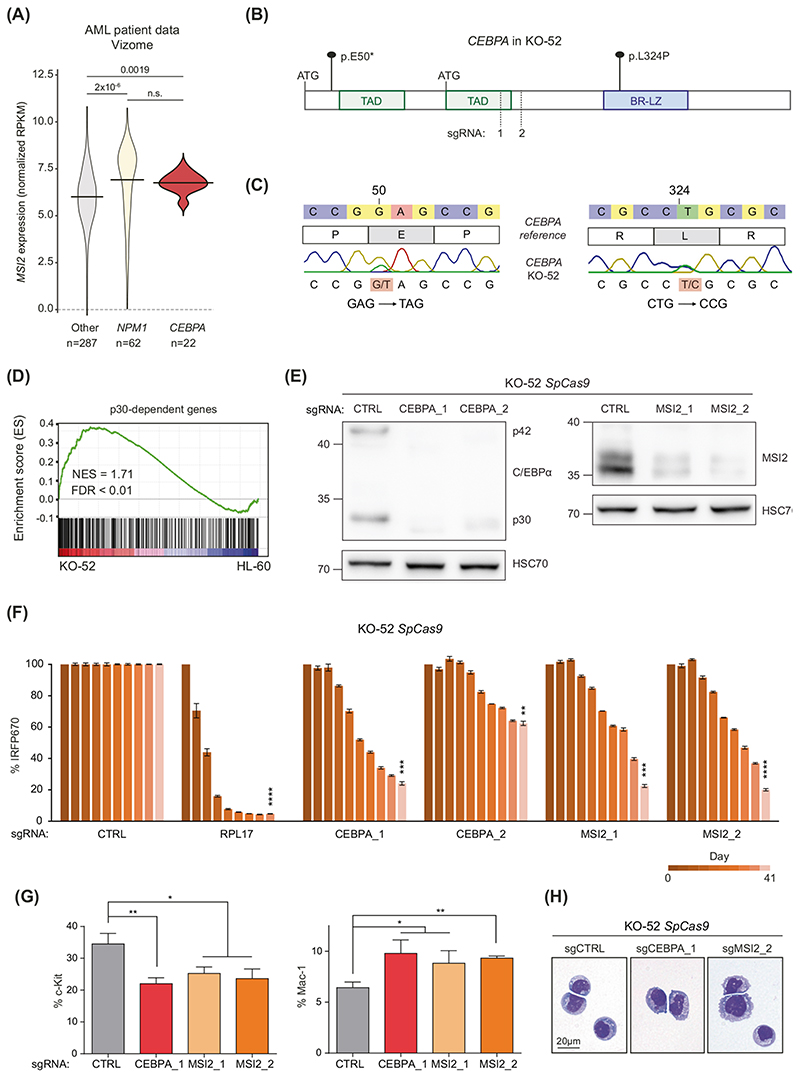
MSI2 is critical for human AML with CEBPA mutations
To assess the human relevance of our findings we first scrutinized publicly available gene expression data in the Vizome database [9]. In addition to high expression in NPM1-mutated AML [39], MSI2 was highly expressed in CEBPA-mutated AML (Fig. 5A). Among AML cell lines, MSI2 expression was highest in the KASUMI-6 line, which serves as a model for C-terminal CEBPA mutations (Supplementary Fig. 5A) [40]. Further interrogation of the DepMap AML cell line panel for additional cell lines with CEBPA mutations revealed that the KO-52 cell line, which originated from a 46-year-old AML patient, harbors both an N-terminal truncating p.E50* and a C-terminal missense p. L324P mutation in CEBPA, which we confirmed by Sanger sequencing (Fig. 5B, C) [41]. KO-52 cells expressed both p42 and p30 C/EBPα isoforms (Supplementary Fig. 5B) and showed a CD34+ c-Kit+ immunophenotype (Supplementary Fig. 5C). The KO-52 transcriptome featured high expression of p30-dependent genes and gene signatures associated with AML with CEBPA mutations, further supporting the use of KO-52 as a model for CEBPA-mutated AML (Fig. 5D, Supplementary Fig. 5D). CRISPR/Cas9-mediated mutagenesis showed that KO-52 cell proliferation was dependent on both C/EBPα and MSI2 expression (Fig. 5B, E, F and Supplementary Fig. 5E, F). Furthermore, loss of C/EBPα or MSI2 led to the induction of myeloid differentiation, as shown by reduced c-Kit expression and increased levels of Mac-1 (Fig. 5G, H, Supplementary Fig. 5G, H). In summary, our data establish the KO-52 cell line as a valuable human cell line model for CEBPA-mutant AML and highlight MSI2 as a critical effector of mutated CEBPA in this system.
Fig. 5. MSI2 is required for the proliferation of human CEBPA-mutated AML cells.
AMSI2 expression in AML patient data obtained from the beatAML dataset [9]. p values were calculated using the Wilcoxon test. RPKM reads per kilobase per million. B Schematic structure of the CEBPA gene. Mutations found in the KO-52 cell line are indicated. Recognition sites of sgRNAs are indicated by dashed lines. TAD transactivation domain, BR-LZ basic-region leucine zipper. C Sanger sequencing analysis of mutated regions in the N- and C-terminus of the human CEBPA gene of the KO-52 cell line. D Gene set enrichment analysis (GSEA) showing global up-regulation of p30-dependent genes in KO-52 cells compared to HL-60 cells. NES normalized enrichment score, FDR false discovery rate. E Western blot analysis of indicated proteins from KO-52 SpCas9 lysates (clone #1) upon targeting of MSI2 or CEBPA with two different sgRNAs, compared to a control guide (CTRL, sgAAVS1). F Survival of sgRNA-expressing KO-52 SpCas9 cells (clone #1) over time, normalized to sgAAVS1 as the negative control, and day 0. G Flow cytometric analysis and quantification of the c-Kit+ (Day 26) and Mac-1+ (Day 40) population of KO-52 SpCas9 cells (clone #1) upon CEBPA or MSI2 knockout. H Histological staining of cytospin preparations of KO-52 SpCas9 (clone #1) cells upon CEBPA or MSI2 knockout.
Discussion
The heterogeneity of AML requires a detailed understanding of the mechanisms and, in particular, the potentially targetable effector proteins of subtype-specific recurrent mutations [42, 43]. The combination of multiple datasets and comprehensive analysis thereof can result in the identification of novel targets. We have created a comprehensive resource for CEBPA-mutated AML featuring ChIP-seq, ATAC-seq and RNA-seq data in myeloid progenitor cells harboring leukemia-associated mutations in CEBPA. Integration of epigenomic and transcriptomic data led to the discovery of MSI2 as a crucial effector in CEBPA-mutated AML. The RNA binding protein MSI2 is required for the proliferation of murine AML cells with Cebpa mutations and Msi2 knockout delayed the onset of Cebpa-driven leukemia in vivo. Consistently, MSI2 is also essential in the human AML cell line KO-52, which we validate as an appropriate human cell line model for CEBPA-mutated AML.
Several studies have demonstrated the importance of the epigenetic landscape and its regulation in the context of multiple malignancies [15–17, 44, 45]. As C/EBPα has been shown to interact with numerous components of the chromatin remodeling machinery [46–48], we investigated the specific role of the AML-associated C/EBPα p30 isoform on the epigenetic landscape in AML. H3K27ac ChIP-seq in p30-proficient vs. p30-deficient cells identified p30-regulated active enhancers and promoters in Cebpap30/p30 cells. Integration of these data with C/EBPα ChIP-seq, ATAC-seq, and RNA-seq datasets resulted in a uniquely comprehensive resource for studies of CEBPA-mutated AML. While this dataset empowers a multitude of orthogonal analyses, we focused on the combinatorial investigation of p30-dependent gene expression changes together with alterations in the corresponding enhancer/promoter regions. In general, genes whose expression was down-regulated upon p30 knockdown also exhibited p30-dependent H3K27ac decoration in their corresponding REs, pointing to direct relationships between gene expression and activity of REs. Cebpa knockdown might differentially impact the kinetics of chromatin composition vs. gene expression. This could be reflected in the higher number of down-regulated H3K27ac regions when compared to down-regulated genes upon p30 knockdown. However, we also found that loss of expression of another group of p30-responsive genes was associated with increased H3K27ac. This scenario indicates that a significant number of genes could be subject to more complex regulatory circuits that might involve multiple enhancers.
The intersection of 200 genes linked to p30-dependent REs with 375 genes down-regulated upon Cebpa knockdown resulted in a list of 33 genes that is likely enriched for direct gene targets of the p30 C/EBPα variant. The remaining 167 genes were not significantly down-regulated after p30 silencing, suggesting that other TFs could maintain their expression independently of p30. The 33 p30-dependent genes encode for proteins with diverse functions, including proteases, G-protein coupled receptors, and proteins involved in the regulation of the immune response. To investigate the functional contribution of these 33 genes to CEBPA-mutated AML we used CRISPR/Cas9-mediated mutagenesis followed by competitive growth assays in both Cebpap30/p30 and Cebpap30/C-mut cells. Both cell lines are strongly dependent on Cebpa expression for survival. The Cebpap30/p30 cell line exclusively expresses the p30 isoform and is thus an ideal model for the study of its specific function. Cebpap30/C-mut cells mimic biallelic CEBPA mutations, where p30 is the only fully functional C/EBPα isoform, while the expressed full-length protein lacks DNA-binding capacity [21, 24]. As the full-length isoform may retain some DNA-independent activity, this could influence the effect of the tested sgRNAs, resulting in non-uniform results over the two cell lines. As we were searching for effectors in both monoallelic and biallelic CEBPA-mutated settings, we analyzed the screening results by investigating the area under the curve over time for tested sgRNAs. We ranked the results by combined mean AUC of all four settings (both cell lines and two sgRNAs per gene) to obtain genes, on which both Cebpap30/p30 and Cebpap30/C-mut cells are dependent. This resulted in the discovery of MSI2 as one of the top hits.
MUSASHI-2 (MSI2) is an RBP that has been implicated in various cancer entities [27–30]. RBPs are required for diverse aspects of mRNA metabolism and can modulate the abundance of central regulatory proteins in stem cells [49]. Any dysbalance of these regulatory and tightly controlled functions through altered expression and/or function of RBPs could therefore pre-dispose cells to cancer [50, 51]. In line with this, a recent CRISPR/Cas9 screen found that RBPs are critically involved in myeloid leukemia [52]. High MSI2 expression has been associated with poor prognosis in hepatocellular cancer, gastric cancer, acute lymphoblastic leukemia, chronic myeloid leukemia, and AML [27, 36, 53–55]. AML with NPM1 mutations and FLT3-ITD were shown to express high levels of MSI2 [37, 53]. We found that MSI2 expression was elevated in CEBPA-mutated patients, supporting the direct association between mutated CEBPA and MSI2. While CEBPA mutations are associated with a good prognosis in AML, high MSI2 expression is associated with a poor outcome. This controversy might be explained by the important functional contribution of additional mutations to overall survival. For instance, mutations in TET2, ASXL1, or GATA2 were reported to frequently co-occur with CEBPA mutations.
In an effort to determine direct MSI2-regulated genes, a recent study identified 255 conserved gene targets of MSI2 in AML [37]. The expression of these top MSI2 targets was highly correlated with p30 expression in Cebpap30/p30 cells, further supporting our findings of MSI2 as a direct effector of oncogenic functions of C/EBPα p30. MSI2 was demonstrated to regulate the self-renewal program of MLL-fusion induced leukemia [56], which is in line with the observation that MLL-AF9 targets are enriched in the transcriptome of leukemia-initiating cells in a Cebpap30/p30 AML mouse model [21].
Therefore, targeting MSI2 could be a promising therapeutic option in CEBPA-mutated AML. The small molecule Ro 08-2750 that was originally designed as a nerve growth factor inhibitor, has been reported to additionally bind and inhibit both MSI1 and MSI2 in murine and human myeloid leukemia cells [28]. MSI2-specific derivatives of this compound could be encouraging for further testing in CEBPA-mutated AML.
Studies of CEBPA-mutated AML have been mainly limited to analyses of murine cellular models, which have led to the discovery of attractive targets [14, 20, 21, 24]. However, the field has lacked appropriate human models for the identification and validation of new targets. Here, we identify and characterize the human cell line KO-52 as a relevant model for CEBPA-mutated AML. KO-52 was established from a 46-year-old AML patient, but it was only recently shown to harbor biallelic CEBPA mutations [57]. We found that KO-52 cell proliferation is C/EBPα-dependent and show that the transcriptome of this cell line features a high expression of gene profiles that were linked to CEBPA-mutated AML. Thus, we propose that this cell line will serve as a valuable human model for further studies of CEBPA-mutated AML.
In summary, this work provides a comprehensive analysis of the epigenetic landscape in Cebpa-mutated cells. This dataset will be useful for further understanding the interaction between epigenetic regulation and transcriptional output in cancer cells. Furthermore, our finding that the oncogenic p30 variant drives the expression of MSI2 highlight this factor as a novel actionable effector in both biand monoallelic CEBPA-mutated AML.
The data reported in this article have been deposited in the Gene Expression Omnibus database (GSE158727).
Supplementary Material
Acknowledgements
We thank the members of the Grebien laboratory for stimulating discussions and T. Weiss, E. Rzepa, and M. Piontek for technical help. We thank D. Berger and G. Stefanzl for skillful technical assistance and P. Valent for providing access to human samples. Next Generation Sequencing was performed at the VBCF NGS Unit (www.viennabiocenter.org/facilities) and at the BSF (https://cemm.at/research/facilities/). This project has received funding from the European Union’s Horizon 2020 research and innovation program (European Research Council grant agreement No 636855 and Marie Sklodowska-Curie grant agreement No 813091). LS is a recipient of the DOC Fellowship of the Austrian Academy of Sciences at the Ludwig Boltzmann Institute for Cancer Research.
Footnotes
Compliance with ethical standards
Conflict of interest The authors declare no competing interest.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015 Sep;373:1136–52. doi: 10.1056/NEJMra1406184. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Wang F, Chen X, Liu W, Fang J, Wang M, et al. Mutation profiling of 16 candidate genes in de novo acute myeloid leukemia patients. Front Med. 2019 May;13:229–37. doi: 10.1007/s11684-018-0616-1. [DOI] [PubMed] [Google Scholar]
- 3.Fasan A, Haferlach C, Alpermann T, Jeromin S, Grossmann V, Eder C, et al. The role of different genetic subtypes of CEBPA mutated AML. Leukemia. 2014;28:794–803. doi: 10.1038/leu.2013.273. [DOI] [PubMed] [Google Scholar]
- 4.Keeshan K, Santilli G, Corradini F, Perrotti D, Calabretta B. Transcription activation function of C/EBPalpha is required for induction of granulocytic differentiation. Blood. 2003 Aug;102:1267–75. doi: 10.1182/blood-2003-02-0477. [DOI] [PubMed] [Google Scholar]
- 5.Lin F-T, MacDougald OA, Diehl AM, Lane MD. A 30-kDa alternative translation product of the CCAAT/enhancer binding protein alpha message: transcriptional activator lacking antimitotic activity. Proc Natl Acad Sci USA. 1993 Oct;90:9606–10. doi: 10.1073/pnas.90.20.9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nerlov C. C/EBPalpha mutations in acute myeloid leukaemias. Nat Rev Cancer. 2004 May;4:394–400. doi: 10.1038/nrc1363. [DOI] [PubMed] [Google Scholar]
- 7.Koschmieder S, Halmos B, Levantini E, Tenen DG. Dysregulation of the C/EBPalpha differentiation pathway in human cancer. J Clin Oncol. 2009 Mar;27:619–28. doi: 10.1200/JCO.2008.17.9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su L, Tan Y, Lin H, Liu X, Yu L, Yang Y, et al. Mutational spectrum of acute myeloid leukemia patients with double CEBPA mutations based on next-generation sequencing and its prognostic significance. Oncotarget. 2018 May;9:24970–9. doi: 10.18632/oncotarget.23873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL, et al. Functional genomic landscape of acute myeloid leukaemia. [Accessed 18 Oct 2018];Nature. 2018 1 doi: 10.1038/s41586-018-0623-z. http://www.nature.com/articles/s41586-018-0623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt L, Heyes E, Grebien F. Gain-of-function effects of N-terminal CEBPA mutations in acute myeloid leukemia. BioEssays. 2020 Feb;42:e1900178. doi: 10.1002/bies.201900178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grebien F, Vedadi M, Getlik M, Giambruno R, Grover A, Avellino R, et al. Pharmacological targeting of the Wdr5-MLL interaction in C/EBPα N-terminal leukemia. Nat Chem Biol. 2015 Aug;11:571–8. doi: 10.1038/nchembio.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pabst T, Mueller BU, Zhang P, Radomska HS, Narravula S, Schnittger S, et al. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (C/EBPalpha), in acute myeloid leukemia. Nat Genet. 2001 Mar;27:263–70. doi: 10.1038/85820. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt L, Heyes E, Scheiblecker L, Eder T, Volpe G, Frampton J, et al. CEBPA-mutated leukemia is sensitive to genetic and pharmacological targeting of the MLL1 complex. Leukemia. 2019 Jul;33:1608–19. doi: 10.1038/s41375-019-0382-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jakobsen JS, Laursen LG, Schuster MB, Pundhir S, Schoof E, Ge Y, et al. Mutant CEBPA directly drives the expression of the targetable tumor-promoting factor CD73 in AML. Sci Adv. 2019 Jul;5 doi: 10.1126/sciadv.aaw4304. eaaw4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gröschel S, Sanders MA, Hoogenboezem R, De Wit E, Bouwman BAM, Erpelinck C, et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in Leukemia. Cell. 2014;157:369–81. doi: 10.1016/j.cell.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 16.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–47. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian Y, Wang G, Hu Q, Xiao X, Chen S. AML1/ETO transactivates c-KIT expression through the long range interaction between promoter and intronic enhancer. J Cell Biochem. 2018;119:3706–15. doi: 10.1002/jcb.26587. [DOI] [PubMed] [Google Scholar]
- 18.Loke J, Assi SA, Imperato MR, Ptasinska A, Cauchy P, Grabovska Y, et al. RUNX1-ETO and RUNX1-EVI1 differentially reprogram the chromatin landscape in t(8;21) and t(3;21) AML. Cell Rep. 2017;19:1654–68. doi: 10.1016/j.celrep.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet. 2016;48:1193–203. doi: 10.1038/ng.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt L, Heyes E, Scheiblecker L, Eder T, Volpe G, Frampton J, et al. CEBPA-mutated leukemia is sensitive to genetic and pharmacological targeting of the MLL1 complex. [Accessed 25 Jun 2019];Leukemia. 2019 Jan;1 doi: 10.1038/s41375-019-0382-3. http://www.nature.com/articles/s41375-019-0382-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirstetter P, Schuster MB, Bereshchenko O, Moore S, Dvinge H, Kurz E, et al. Modeling of C/EBPα mutant acute myeloid leukemia reveals a common expression signature of committed myeloid leukemia-initiating cells. Cancer Cell. 2008 Apr;13:299–310. doi: 10.1016/j.ccr.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 22.Klemm SL, Shipony Z, Greenleaf WJ. Chromatin accessibility and the regulatory epigenome. Nat Rev Genet. 2019 Apr;20:207–20. doi: 10.1038/s41576-018-0089-8. [DOI] [PubMed] [Google Scholar]
- 23.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010 May;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bereshchenko O, Mancini E, Moore S, Bilbao D, Månsson R, Luc S, et al. Hematopoietic stem cell expansion precedes the generation of committed myeloid leukemia-initiating cells in C/EBPα Mutant AML. Cancer Cell. 2009 Nov;16:390–400. doi: 10.1016/j.ccr.2009.09.036. http://www.sciencedirect.com/science/article/pii/S1535610809003444?via%3Dihub. [DOI] [PubMed] [Google Scholar]
- 25.Rao J, Ashraf S, Tan W, Van Der Ven AT, Gee HY, Braun DA, et al. Advillin acts upstream of phospholipase C Є1 in steroid-resistant nephrotic syndrome. J Clin Investig. 2017 Dec;127:4257–69. doi: 10.1172/JCI94138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vassar R. BACE1: The β-secreiase enzyme in Alzheimer’s disease. J Mol Neurosci. 2004;23:105–13. doi: 10.1385/JMN:23:1-2:105. [DOI] [PubMed] [Google Scholar]
- 27.Kaeda J, Ringel F, Oberender C, Mills K, Quintarelli C, Pane F, et al. Up-regulated MSI2 is associated with more aggressive chronic myeloid leukemia. Leuk Lymphoma. 2015;56:2105–13. doi: 10.3109/10428194.2014.981175. [DOI] [PubMed] [Google Scholar]
- 28.Minuesa G, Albanese SK, Xie W, Kazansky Y, Worroll D, Chow A, et al. Small-molecule targeting of MUSASHI RNA-binding activity in acute myeloid leukemia. Nat Commun. 2019;10:2691. doi: 10.1038/s41467-019-10523-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hattori A, McSkimming D, Kannan N, Ito T. RNA binding protein MSI2 positively regulates FLT3 expression in myeloid leukemia. Leuk Res. 2017;54:47–54. doi: 10.1016/j.leukres.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z, Jin H, Mao G, Wu L, Guo Q. Msi2 plays a carcinogenic role in esophageal squamous cell carcinoma via regulation of the Wnt/β-catenin and Hedgehog signaling pathways. Exp Cell Res. 2017;361:170–7. doi: 10.1016/j.yexcr.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Wang R, Bai S, Xiong S, Li Y, Liu M, et al. Musashi2 contributes to the maintenance of CD44v6+ liver cancer stem cells via notch1 signaling pathway. J Exp Clin Cancer Res. 2019 Dec;38:505. doi: 10.1186/s13046-019-1508-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheng W, Shi X, Lin Y, Tang J, Jia C, Cao R, et al. Musashi2 promotes EGF-induced EMT in pancreatic cancer via ZEB1-ERK/MAPK signaling. J Exp Clin Cancer Res. 2020 Jan;39:16. doi: 10.1186/s13046-020-1521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao J, Zhang Y, Liu X-S, Zhu F-M, Xie F, Jiang C-Y, et al. RNA-binding protein Musashi2 stabilizing androgen receptor drives prostate cancer progression. Cancer Sci. 2020 Feb;111:369–82. doi: 10.1111/cas.14280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z-L, Wang C, Liu W, Ai Z-L. Emerging roles of the long non-coding RNA 01296/microRNA-143-3p/MSI2 axis in development of thyroid cancer. Biosci Rep. 2019 Nov;39 doi: 10.1042/BSR20182376. BSR20182376. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Lan L, Xing M, Kashipathy M, Douglas J, Gao P, Battaile K, et al. Crystal and solution structures of human oncoprotein Musashi-2 N-terminal RNA recognition motif 1. Proteins. 2020 Apr;88:573–83. doi: 10.1002/prot.25836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Z, Li J, Shi Y, Li L, Guo X. Increased Musashi 2 expression indicates a poor prognosis and promotes malignant phenotypes in gastric cancer. Oncol Lett. 2019 Mar;17:2599–606. doi: 10.3892/ol.2019.9889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kharas MG, Lengner CJ, Al-Shahrour F, Bullinger L, Ball B, Zaidi S, et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat Med. 2010 Aug;16:903–8. doi: 10.1038/nm.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen DTT, Lu Y, Chu KL, Yang X, Park SM, Choo ZN, et al. HyperTRIBE uncovers increased MUSASHI-2 RNA binding activity and differential regulation in leukemic stem cells. Nat Commun. 2020 Dec;11:1–12. doi: 10.1038/s41467-020-15814-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Byers RJ, Currie T, Tholouli E, Rodig SJ, Kutok JL. MSI2 protein expression predicts unfavorable outcome in acute myeloid leukemia. Blood. 2011 Sep;118:2857–67. doi: 10.1182/blood-2011-04-346767. [DOI] [PubMed] [Google Scholar]
- 40.Asou H, Gombart AF, Takeuchi S, Tanaka H, Tanioka M, Matsui H, et al. Establishment of the acute myeloid leukemia cell line Kasumi-6 from a patient with a dominant-negative mutation in the DNA-binding region of the C/EBPα gene. Genes Chromosom Cancer. 2003 Feb;36:167–74. doi: 10.1002/gcc.10161. [DOI] [PubMed] [Google Scholar]
- 41.DepMap, Broad. DepMap 20Q1 Public. 2020 2020. [Google Scholar]
- 42.De Kouchkovsky I, Abdul-Hay M. Acute myeloid leukemia: a comprehensive review and 2016 update. Blood Cancer J. 2016 Jul;6:e441. doi: 10.1038/bcj.2016.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374:2209–21. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soukup AA, Zheng Y, Mehta C, Wu J, Liu P, Cao M, et al. Single-nucleotide human disease mutation inactivates a blood-regenerative GATA2 enhancer. J Clin Investig. 2019 Mar;129:1180–92. doi: 10.1172/JCI122694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mansour MR, Abraham BJ, Anders L, Berezovskaya A, Gutierrez A, Durbin AD, et al. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science (80-) 2014;346:1373–7. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nerlov C, Ziff EB. CCAAT/enhancer binding protein-alpha amino acid motifs with dual TBP and TFIIB binding ability co-operate to activate transcription in both yeast and mammalian cells. EMBO J. 1995 Sep;14:4318–28. doi: 10.1002/j.1460-2075.1995.tb00106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pedersen TÅ, Kowenz-Leutz E, Leutz A, Nerlov C. Cooperation between C/EBPα TBP/TFIIB and SWI/SNF recruiting domains is required for adipocyte differentiation. Genes Dev. 2001 Dec;15:3208–16. doi: 10.1101/gad.209901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nerlov C, Ziff EB. Three levels of functional interaction determine the activity of CCAAT/enhancer binding protein-α on the serum albumin promoter. Genes Dev. 1994 Feb;8:350–62. doi: 10.1101/gad.8.3.350. [DOI] [PubMed] [Google Scholar]
- 49.Dvinge H, Kim E, Abdel-Wahab O, Bradley RK. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer. 2016;16:413–30. doi: 10.1038/nrc.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang E, Lu SX, Pastore A, Chen X, Imig J, Chun-Wei Lee S, et al. Targeting an RNA-binding protein network in acute myeloid leukemia. Cancer Cell. 2019;35:369–384.e7. doi: 10.1016/j.ccell.2019.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang ZL, Li B, Luo YX, Lin Q, Liu SR, Zhang XQ, et al. Comprehensive genomic characterization of RNA-binding proteins across human cancers. Cell Rep. 2018 Jan;22:286–98. doi: 10.1016/j.celrep.2017.12.035. [DOI] [PubMed] [Google Scholar]
- 52.Bajaj J, Hamilton M, Shima Y, Chambers K, Spinler K, Van Nostrand EL, et al. An in vivo genome-wide CRISPR screen identifies the RNA-binding protein Staufen2 as a key regulator of myeloid leukemia. Nat Cancer. 2020 Apr;1:410–22. doi: 10.1038/s43018-020-0054-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thol F, Winschel C, Sonntag A-K, Damm F, Wagner K, Chaturvedi A, et al. Prognostic significance of expression levels of stem cell regulators MSI2 and NUMB in acute myeloid leukemia. Ann Hematol. 2013 Mar;92:315–23. doi: 10.1007/s00277-012-1637-5. [DOI] [PubMed] [Google Scholar]
- 54.He L, Zhou X, Qu C, Hu L, Tang Y, Zhang Q, et al. Musashi2 predicts poor prognosis and invasion in hepatocellular carcinoma by driving epithelial-mesenchymal transition. J Cell Mol Med. 2014 Jan;18:49–58. doi: 10.1111/jcmm.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aly RM, Ghazy HF. Prognostic significance of MSI2 predicts unfavorable outcome in adult B-acute lymphoblastic leukemia. Int J Lab Hematol. 2015 Apr;37:272–8. doi: 10.1111/ijlh.12284. [DOI] [PubMed] [Google Scholar]
- 56.Park SM, Gönen M, Vu L, Minuesa G, Tivnan P, Barlowe TS, et al. Musashi2 sustains the mixed-lineage leukemia’ driven stem cell regulatory program. J Clin Investig. 2015 Mar;125:1286–98. doi: 10.1172/JCI78440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ghandi M, Huang FW, Jané-Valbuena J, Kryukov GV, Lo CC, McDonald ER, et al. Next-generation characterization of the cancer cell line encyclopedia. Nature. 2019 May;569:503–8. doi: 10.1038/s41586-019-1186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.