Abstract
Kidney fibrosis is the hallmark of chronic kidney disease progression, however, currently no antifibrotic therapies exist. This is largely because the origin, functional heterogeneity and regulation of scar-forming cells during human kidney fibrosis remains poorly understood. Here, using single cell RNA-seq, we profiled the transcriptomes of proximal tubule and non-proximal tubule cells in healthy and fibrotic human kidneys to map the entire human kidney in an unbiased approach. This enabled mapping of all matrix-producing cells at high resolution, revealing distinct subpopulations of pericytes and fibroblasts as the major cellular sources of scar forming myofibroblasts during human kidney fibrosis. We used genetic fate-tracing, time-course single cell RNA-seq and ATAC-seq experiments in mice, and spatial transcriptomics in human kidney fibrosis to functionally interrogate these findings, shedding new light on the origin, heterogeneity and differentiation of human kidney myofibroblasts and their fibroblast and pericyte precursors at unprecedented resolution. Finally, we used this strategy to facilitate target discovery, identifying Nkd2 as a myofibroblast-specific target in human kidney fibrosis.
Chronic kidney disease (CKD) affects more than 10% of the world population. The final common pathway of kidney injury is fibrosis and its extent is inextricably linked to clinical outcomes.1,2 No approved therapies exist and the cellular origin, functional heterogeneity and regulation of scar-producing cells in the human kidney continues to be debated.1,2 Using single cell (sc)RNASeq, we profiled ~135,000 human and mouse kidney cells during homeostasis and fibrosis, allowing the dissection of heterogeneity of extracellular matrix (ECM)-producing cells at high resolution. We identified multiple subpopulations of mesenchymal cells as major contributors to human kidney fibrosis, whereas injured tubular epithelia, endothelium and monocytes only exhibited minor ECM expression. Genetic fate-tracing and time-course scRNA-seq and ATAC-seq experiments in mice, and spatial transcriptomics in human kidney fibrosis, validated these findings, shedding new light on the origin and regulation of human kidney myofibroblasts. This approach also identified novel candidate therapeutic targets, such as the myofibroblast specific naked cuticle homolog 2 (Nkd2).
Single cell atlas of human chronic kidney disease
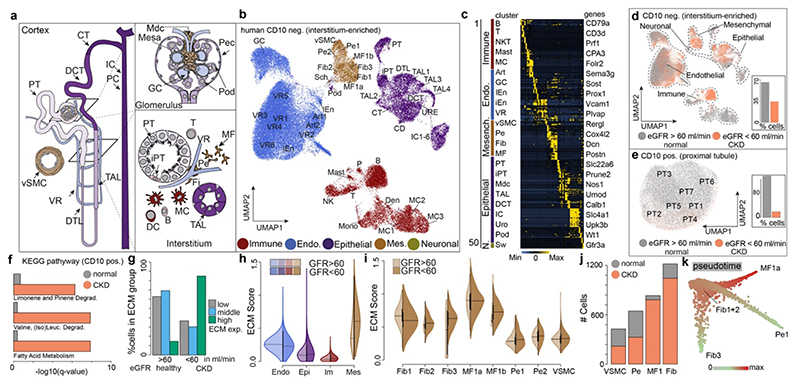
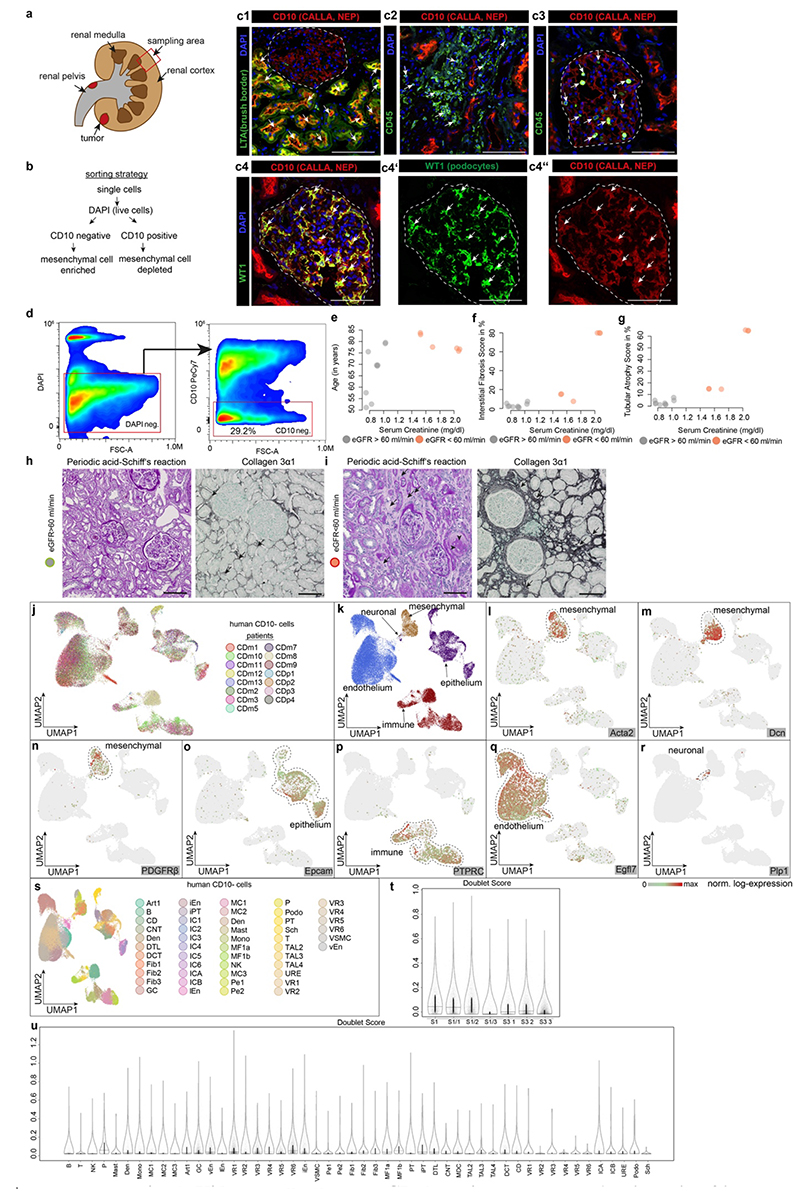
To understand which resident human renal cell types secrete ECM during homeostasis and CKD, we generated a single cell map of kidneys with a focus on the tubulointerstitium. Over 80% of renal cortical cells are proximal tubule epithelial cells (PT) and thus dominated previous single cell maps, masking other populations.3 We therefore sorted for viable, non PT (CD10-) and CD10+ PT to map the entire kidney (Extended Data Fig. 1a-b). While CD10 is also expressed by other cell types, this strategy allows an enrichment/depletion of PT. Both CD10+ and CD10- fractions from 13 patients with CKD due to hypertensive nephrosclerosis (n=7; estimated Glomerular Filtration Rate, eGFR>60 and n=6; eGFR<60) were subjected to scRNAseq (Extended Data Fig. 1a-i and Table 1). We profiled 53,672 CD10- cells from 11 patients, (n=7 eGFR>60; n=4 eGFR<60, Table 1). To integrate the data across patients, we employed an unsupervised graph-based clustering method and identified 50 different CD10- cell clusters represented in both eGFR groups (Fig. 1a-d). Our strategy allowed us to appreciate the heterogeneity of the renal interstitium including identification of rare cell types such as Schwann cells (Figs. 1a-d, Extended Data Figs. 1j-u, 2a-d).
Figure 1. Single cell atlas of human chronic kidney disease (CKD).
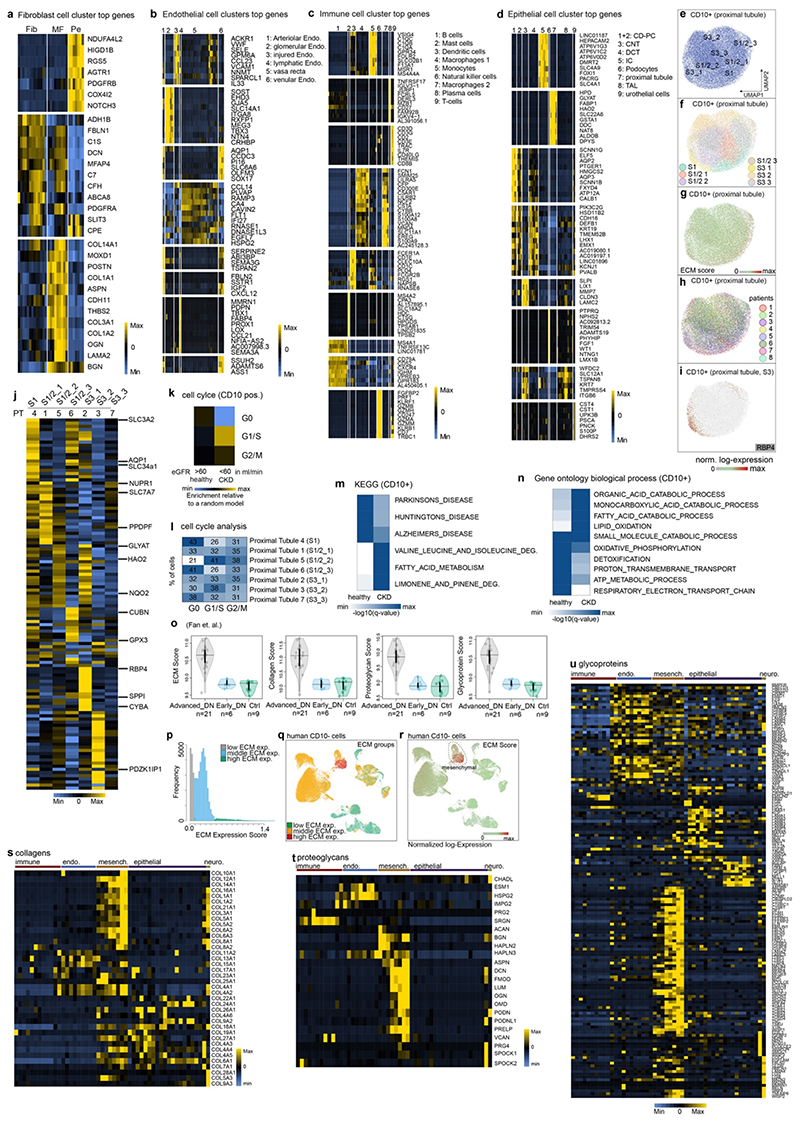
a. Scheme of the kidney b. UMAP embedding of 51,849 CD10- single cells from 15 human kidneys. Labels refer to 50 clusters identified, see Supplementary File 1. c. Scaled gene expression of the top 10 specific genes in each cluster (see Supplementary File 2 for detailed information). Each column is the average expression of all cells in a cluster. d. Stratification of cells by estimated glomerular filtration rate (eGFR). e. UMAP embedding of 31,875 CD10+ single cells stratified by eGFR f. KEGG pathway enrichment for CD10+ cells. g. CD10- clustering by ECM (extracellular matrix) score stratified by eGFR (see Extended Data Figure 2p). h. ECM score stratified by cell type and eGFR, Mesenchymal (p ~0), Immune (p ~0), Epithelial (p ~0), Endothelial (p ~0) i. Single cell ECM score for mesenchymal cells, stratified by major cell types and by eGFR. P-value of differences in eGFR categories: Fib1 (0.00015), Fib2 (1), Fib3 (0.54), MF1a (1), MF1b (0.59), Pe1 (0.096), Pe2 (1), SMC (0.162). (h.-i.) Bonferroni corrected p-values based on two-sided t-test. j. Number of cells per mesenchymal cell type and clinical parameter. Hypergeometric test, adjusted p value for fibroblast and myofibroblast = ~0 - for pericyte and vascular smooth muscle cells ~ 1. k. Diffusion mapping of mesenchymal cells, pseudotime indicates cell ordering along putative differentiation processes. For details on statistics and reproducibility, please see Methods.
Next, 33,690 CD10+ PT cells were profiled (5 patients with eGFR>60 and 3 eGFR<60) and arranged into 7 clusters (Fig. 1e, Extended Data Fig. 2e-j). Cell-cycle analysis indicated increased cycling in CKD likely reflecting epithelial repair (Extended Data Fig. 2k-l). KEGG pathways and Gene Ontology (GO) terms in CD10+ cells suggested increased fatty acid metabolism and dysregulated metabolism in CKD (Fig. 1f, Extended Data Fig. 2m-n). Dysregulated fatty acid metabolism has been shown to cause tubular dedifferentiation and fibrosis.4
Origin of extracellular matrix in human chronic kidney disease
To identify cell types contributing to ECM production in kidney fibrosis, we established a single cell ECM expression score that included collagens, glycoproteins and proteoglycans5 and confirmed an increased score in published CKD data6 (Extended Data Fig. 2o-u). ECM scores demonstrated a clear shift towards high ECM expressing cells in CKD (Fig. 1g). Mesenchymal cells exhibited the highest ECM expression and this increased further in CKD (Fig 1h-i, Extended Data Fig. 2q-u). All fibroblasts and myofibroblasts, expanded in CKD (Fig. 1j). While historically Acta2 was used as a myofibroblast marker, we defined myofibroblasts as cells that express most ECM genes. To assess putative myofibroblast differentiation processes we generated a Uniform Manifold Approximation and Projection (UMAP) embedding of (myo)fibroblasts and pericytes (Extended Data Fig 3a-c). This embedding agreed with our unsupervised graph clustering (Fig. 1b), highlighting the heterogeneity of the renal mesenchyme. Myofibroblasts were identified as periostin (Postn) expressing cells (Extended Data Fig. 3b). Diffusion mapping of high ECM expressing mesenchymal cells suggested that myofibroblasts arise from pericytes and fibroblasts (Fig. 1k, Extended Data Fig. 3d).
Minor upregulation of ECM genes occurred in epithelial cells (Fig. 1h), suggestive of a minor contribution of the long debated epithelial mesenchymal transition (EMT).1,7,8 Injured PT showed the highest expression of ECM genes among CD10- epithelium with various expressed genes and GO terms suggesting de-differentiation (Extended Data Fig. 3e-j). In CD10+ PT ECM expression increased slightly in CKD (Extended data Fig. 3k-n). Injured cells were defined by expression of Sox9, CD24 and CD133 for PT and Vcam1 and Ackr1 for endothelium.9–11
Thus, the vast majority of ECM in human kidney fibrosis originates from mesenchymal cells, with a minor contribution from de-differentiated PT.
Distinct pericyte and fibroblast subpopulations are the major source of myofibroblasts in human kidney fibrosis
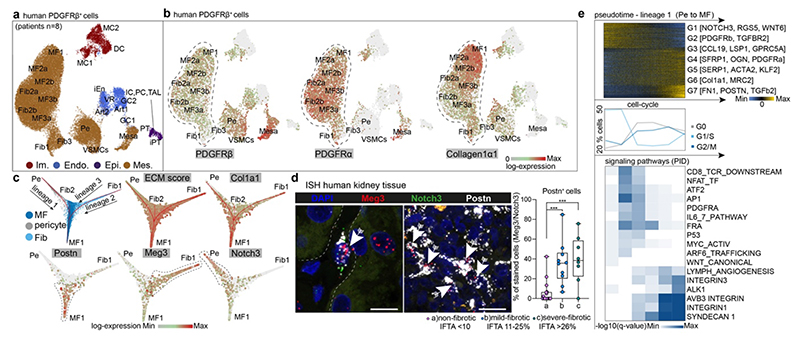
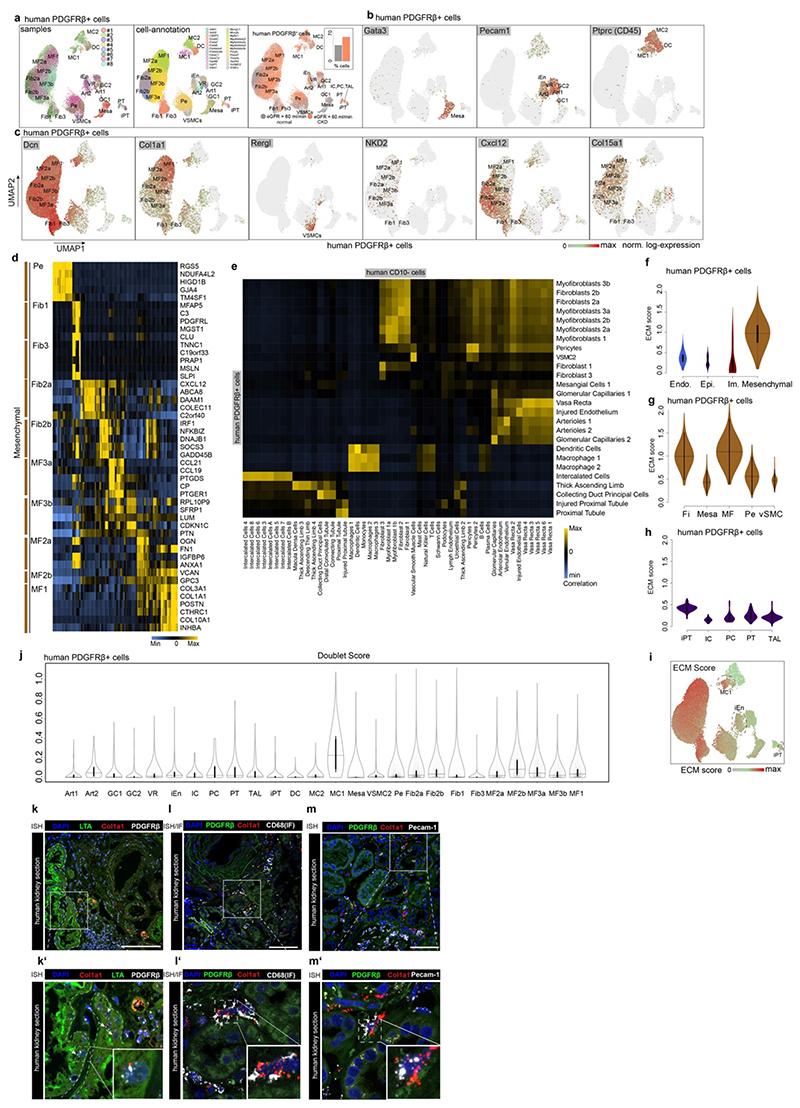
Our CD10- scRNA-seq data identified the majority of Col1a1 expressing cells as PDGFRb+ (Extended data Fig. 3o). Unsupervised clustering of 37,380 PDGFRb+ cells sorted from human kidneys (n=4; eGFR>60 and n=4; eGFR<60; Extended Data Table 1) identified mesenchymal populations and some epithelial, endothelial and immune cells, which were annotated by correlation with the CD10- populations (Fig. 2a-b, Extended Data Fig. 4a-e). ECM gene expression again dominated in pericyte, fibroblast and myofibroblast clusters (Extended Data Fig. 4f-i). Some macrophage/monocyte, endothelial and injured epithelial populations also expressed collagen1a1 and PDGFRb, but at much lower levels (Fig. 2a-b, Extended Data Fig. 4f-i). Doublet-likelihood scores were low for endothelial and injured epithelial cells, however, slightly increased in macrophages (Extended Data Fig. 4j). We verified Col1a1 mRNA expression in these cells by in situ hybridisation (ISH, Extended Data Fig. 4k-m). These data partially explain the controversy regarding the contributions of non-mesenchymal lineages to fibrosis,1,12 since we indeed observed minor ECM gene expression in these cells, whilst the majority of ECM is mesenchymal cell-derived.
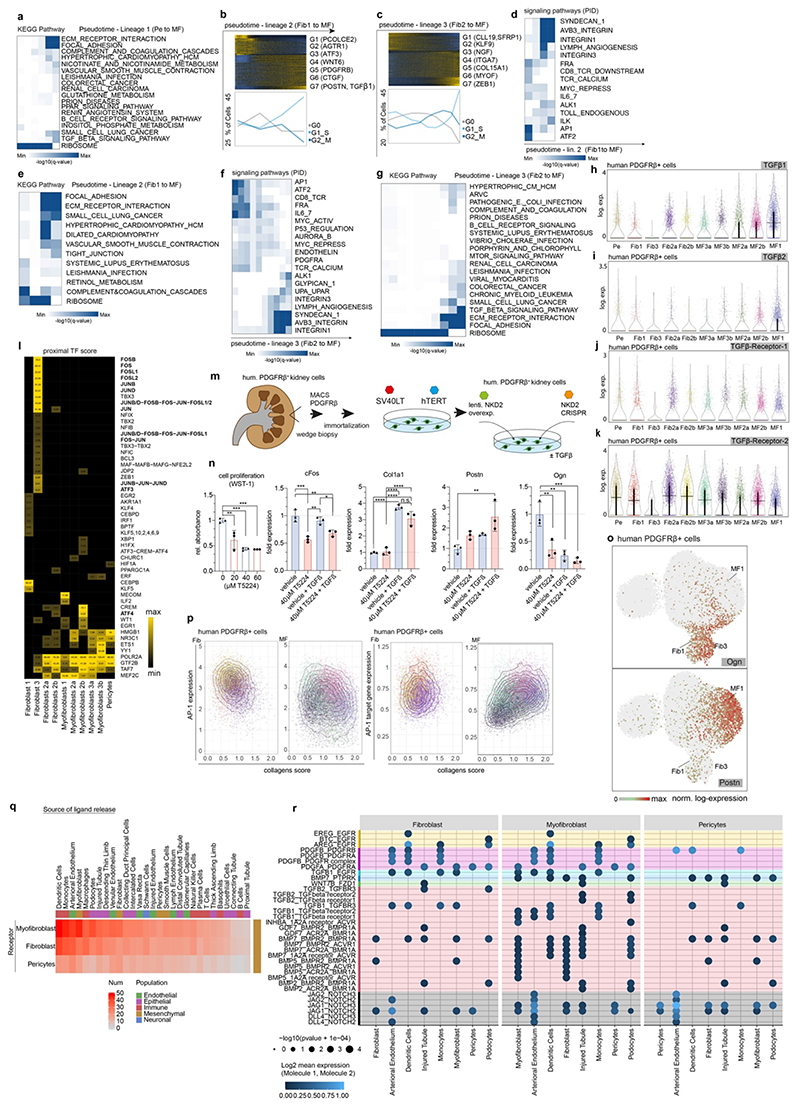
Figure 2. Origin of myofibroblasts in the human kidney.
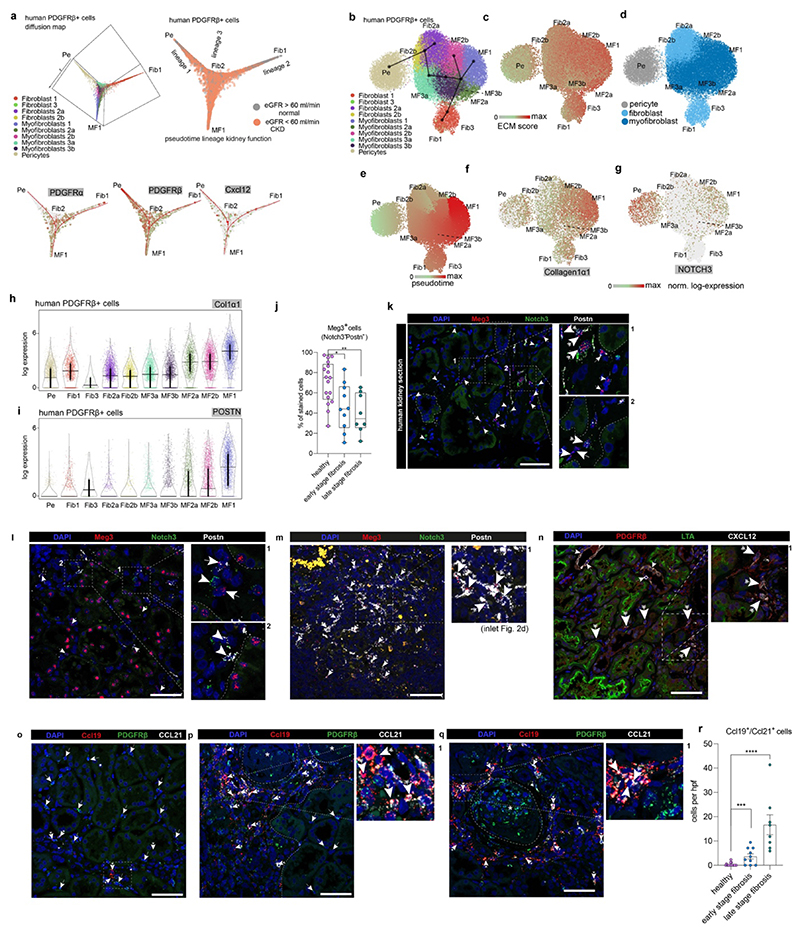
a. UMAP embedding of 37,800 Pdgfrb+ single cells from 8 human kidneys. Labels refer to identified cell-types by unsupervised clustering (see Supplementary File 1). b. Expression of selected genes on the embedding from a. c. Diffusion Map embedding of Pdgfrb+ fibroblasts, myofibroblasts and pericytes (n=23,883) and the expression of selected genes on the same embedding. Red lines correspond to the three lineage trajectories (L1, L2 and L3) predicted by Slingshot given the Diffusion Map coordinates and the clusters depicted in Extended Data Figure 5b. d. Representative images of RNA-in-situ hybridization for Meg3, Notch3, Postn in 35 human kidneys (Patient Data in Extended Data Table 2, IFTA=interstitial fibrosis, tubular atrophy score). n=17 (a), 10 (b) and 8(c); ***p < 0.001 by 1-way ANOVA followed by Bonferroni’s correction. Tukey box whisker plot. Scale bar left 10 μm, right 25 μm. e. Top: Gene expression dynamics along pseudotime for Lineage 1 (see c., see Methods). Middle: Cell cycle stage as percent of each 2000 cells along pseudotime. Bottom: PID Signaling pathway enrichment along pseudotime. For details on statistics and reproducibility, please see Methods.
Pseudotime trajectory and diffusion map analysis of major ECM expressing cells from the Pdgfrb+ populations indicated three major sources of myofibroblasts in human kidneys: 1) Notch3+/RGS5+/Pdgfra- pericytes, 2) Meg3+/Pdgfra+ fibroblasts and 3) Colec11+/Cxcl12+ fibroblasts (Fig. 2c, Extended Data Fig. 5a). Diffusion mapping places non-CKD cells within low ECM-expressing pericyte and fibroblast populations, indicating a differentiation trajectory from low-ECM, non-CKD pericytes and fibroblasts to high-ECM CKD myofibroblasts (Figure 2c, Extended Data Figure 5a-i). We verified this directionality and also the main lineages of the diffusion map, consisting of Notch3+ pericytes (lineage 1) and Meg3+ fibroblasts (lineage 2) using ISH in human kidneys (Fig 2d, Extended Data Fig. 5j-m). We observed a potential intermediate stage of Notch3/Meg3/Postn co-expressing cells possibly representing differentiating cells in the center of the diffusion map (Fig 2d, Extended Data Fig. 5k-m).
Distinct spatial tissue locations could be identified for myofibroblast 1 (Postn+), which increased in fibrosis and for myofibroblast 3 (Ccl19+/Ccl21+) which were enriched around glomeruli (Extended Data Fig. 5n-r).
The gene expression program of pericyte-to-myofibroblast differentiation (Lineage 1) demonstrated cell cycle changes, consistent with differentiation and expansion (Fig. 2e). Ordering their pathway enrichment along pseudotime yielded early canonical Wnt and activator protein-1 (AP1), intermediate ATF2, Pdgfra and late integrin, ECM receptor interaction and TGFb signaling among other pathways (Fig. 2e bottom, Extended Data Fig. 6a).
Cell cycle cessation also characterized fibroblast-to-myofibroblast differentiation, followed by increased proliferation (lineages 2 and 3, Extended Data Fig. 6b-c) with early AP1, and inflammatory pathways, followed by integrin and ECM interaction pathways (Extended Data Fig. 6d-g).
Late TGFb signaling was prevalent in the analysis of lineage 1 and 3 (Fig. 2e, Extended Data Fig. 6a,g). Comparing ligand and receptor expression within this pathway suggested a mechanism whereby myofibroblasts promote differentiation of fibroblasts and pericytes by TGFb signaling (Extended Data Fig. 6h-k).
Many of the above pathways are known regulators of fibrosis, including integrins13 and AP1 signaling.14 To further understand transcriptional regulation of mesenchymal populations, we performed transcription factor DNA sequence motif enrichment analysis in promoters and distal regions of marker genes. This highlighted a potential key regulatory role of AP-1 (Jun/Fos) in fibroblast to myofibroblast differentiation (Extended Data Fig. 6l). To functionally validate this, we generated a human Pdgfrb+ kidney cell line (Extended Data Fig. 6m). Inhibition of AP1 significantly decreased proliferation and osteoglycin (Ogn) expression, whilst Postn expression was increased, suggesting myofibroblast differentiation (Extended Data Fig. 6n). In the human Pdgfrb data, Ogn marked fibroblast 1/3 while Postn marked myofibroblasts 1 (Extended Data Fig. 6o). Consistent with this, AP1 expression negatively correlated with average collagen expression while expression of putative AP1 target genes positively correlated with average collagen expression (Extended Data Figure 6p), possibly indicating a repressor role of AP1. However, the role of AP1 is likely multifunctional and it may have additional roles that could also promote fibrosis.
We next studied which cells signal towards the key ECM expressing cells (Extended Data Fig. 6q). Lowest signaling came from healthy PT, while injured PT were among the top signaling partners, suggesting tubule-interstitial signaling as a hallmark of fibrosis15 (Extended Data Fig. 6q). This interaction involves Notch, TGFb, Wnt and PDGFa signaling (Extended Data Fig. 6r).
Dual-positive PDGFRa+/PDGFRb+ mesenchymal cells represent the majority of ECM-expressing cells in kidney fibrosis
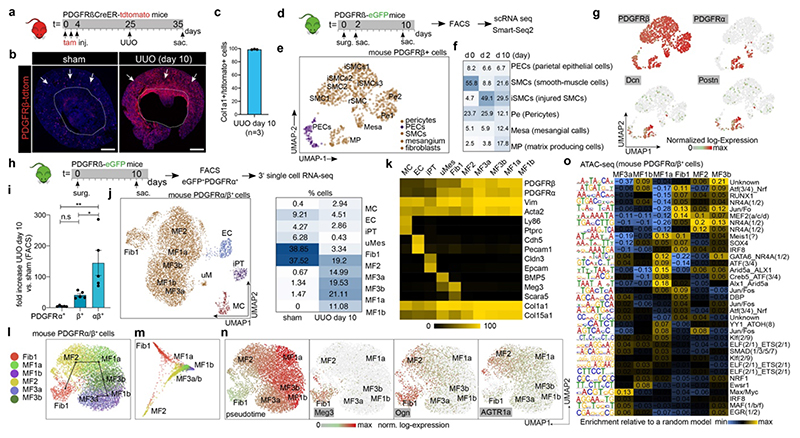
In genetic fate tracing kidney fibrosis experiments of PdgfrbCreER-tdTomato mice ISH and immunostaining confirmed that virtually all myofibroblasts are Pdgfrb lineage derived (Fig. 3a-c; Extended Data Fig. 7a-c). A Smart-Seq2 time-course study in Pdgfrb-eGFP mice demonstrated that smooth muscle cell and pericyte abundance decreased following UUO, whereas mesangial cells and Col1a1+/Pdgfra+ matrix producing cells increased (Fig. 3d-f, Extended Data Fig. 7d-e). Similar to the human kidney, the major ECM-expressing cell population exhibited Pdgfra/Pdgfrb and Postn expression (Fig. 3g, Extended Data Fig. 7e-g). Other cells showed significantly lower ECM expression than the Pdgfra/Pdgfrb population (Extended Data Fig. 7f-g).
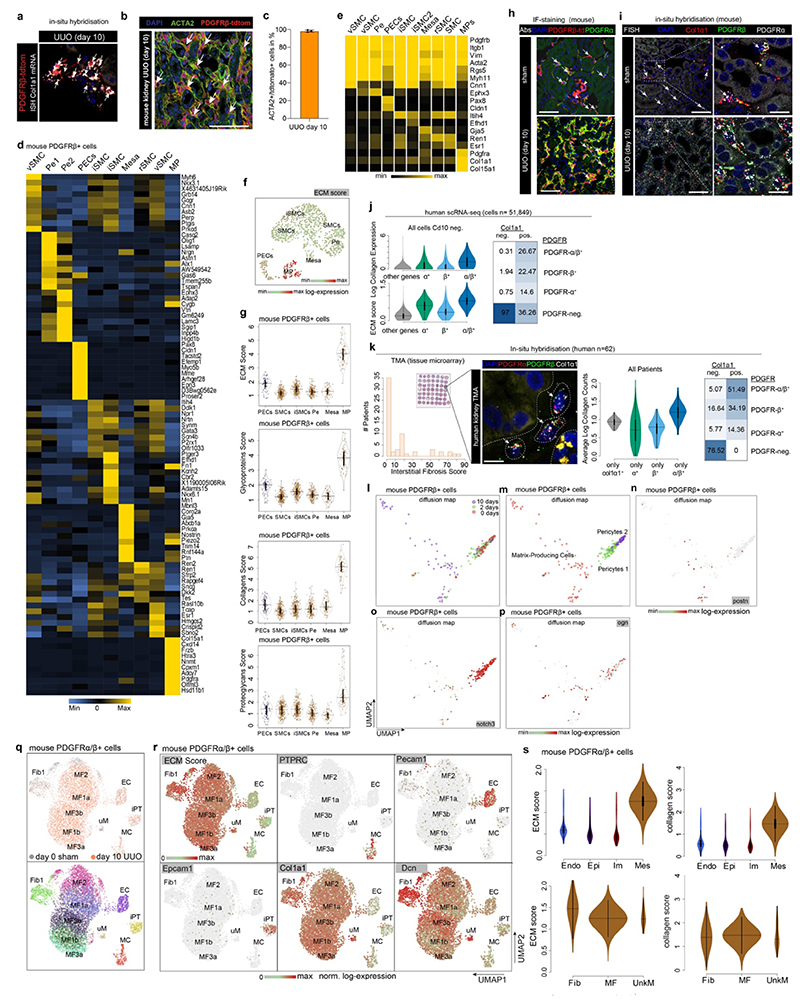
Figure 3. Origin of myofibroblasts in mice.
a. Fate tracing experiment design b. Col1a1 in-situ hybridization in a PdgfrbCreER;tdTomato kidney. Scale bar 1000 μm. c. Percentage of Col1a1-mRNA expressing cells that co-express tdTomato at day10 after (unilateral ureteral obstruction, UUO, n = 3, shown mean). d. Time-course UUO experiment design. e. UMAP embedding of the mouse Pdgfrb+ cells. Labels refer to a cell-types identified. f. Percent of cells per cell type and time-point. g. Expression of selected genes on the UMAP embedding from e. h. Scheme of the PDGFRa/PDGFRb isolation UUO experiment. i. Quantification of Pdgfra+/Pdgfrb+ cells by flow cytometry (n=5 per group). *p<0.05; **p<0.01 by one-way ANOVA with post-hoc Bonferroni correction. Data shown as mean ± s.e.m. j. Left: UMAP embedding of the Pdgfra+/Pdgfrb+ cells Right: Percent of cells per cluster. k. Expression of selected genes in each of the cell clusters from j. n., o. UMAP and diffusion map embedding of fibroblasts and myofibroblasts. p. Computational cell ordering (pseudotime) and expression of selected genes on the embedding in n. q. Enrichment of transcription factor motif occurrence in fibroblasts and myofibroblasts. TF motifs were identified from Pdgfra+/Pdgfrb+ day 10 UUO ATAC-Seq data (see Methods). For details on statistics and reproducibility, please see Methods.
Immunostaining and ISH in mice confirmed double positivity for Pdgfra+ and tdTomato in Col1a1- expressing cells confirming Pdgfra/Pdgfrb expressing cells as the major ECM source (Extended Data Fig. 7h-i). This was confirmed via multiplex ISH in a cohort of 62 patients (Extended Data Fig. 7j-k). Diffusion map embedding of matrix producing cells and pericytes also agreed with our human Pdgfrb data, and suggested that pericytes (Pdgfrb+, Pdgfra-, Notch3+) are one origin of the major ECM-producing cells (Pdgfrb+, Pdgfra+, Col1a1+, Postn+) (Extended Data Fig. 7l-p).
Combined, our data demonstrate that Pdgfra+/Pdgfrb+ dual-positive mesenchymal cells, including all fibroblast and myofibroblast populations but not non-activated Pdgfra-/Pdgfrb+ pericytes (i.e. low ECM expressing pericytes), represent the majority of ECM expressing cells.
Pdgfra+/Pdgfrb+ cells consist of different fibroblast cell states
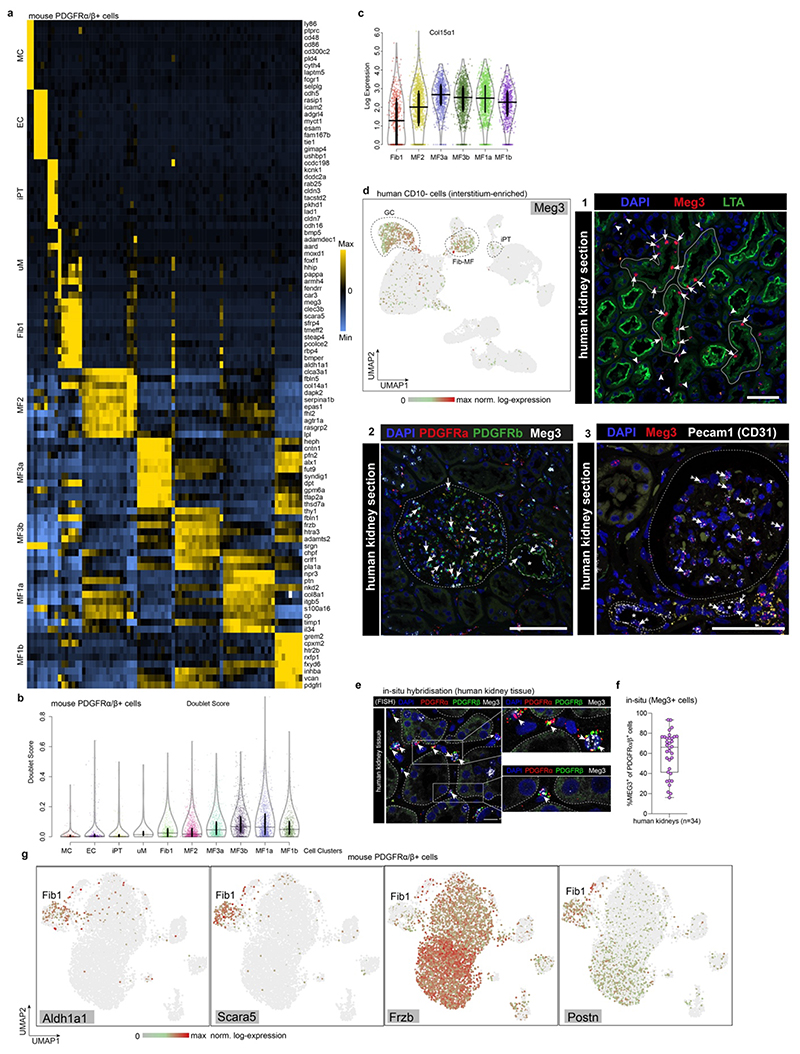
We next generated scRNA-Seq data from 7,245 Pdgfra+/Pdgfrb+ cells in mouse kidney fibrosis experiments (Fig. 3h). These cells expanded ~140-fold after injury and UMAP embedding revealed four major, distinct populations corresponding to mesenchyme, epithelial, endothelial and immune cells (Fig. 3i-k, Extended Data Fig. 7q-r), all of which have been described as origins of kidney fibrosis.1,12,16 We did not detect undifferentiated pericytes in this data, since pericytes are Pdgfra- in humans and mice (Fig. 2c, Extended Data 7e). Non-mesenchymal cells expressed markedly lower ECM and collagen levels than mesenchymal cells (Fig. 3k, Extended Data Fig. 7r-s, 8a), supporting our human data that non-mesenchymal cells contribute little to scarring (Fig. 1,2). Doublet scores were low in these clusters (Extended Data Fig. 8b).
Unsupervised clustering revealed two key classes within mesenchymal cells in this dataset: (1) fibroblast 1 marked by Scara5 and Meg3 expression and (2) myofibroblasts consisting of various myofibroblast subpopulations (Fig. 3j-k, Extended Data Fig. 8a). In our human data, myofibroblasts 1 correspond to terminally differentiated myofibroblasts with the highest ECM expression preceded in differentiation pseudotime by myofibroblast 2 (Ogn+), while fibroblasts 1 appeared as a “progenitor” non-activated fibroblast population (Fig. 2c). Fibroblast 1 cells differed from myofibroblasts in the Pdgfra+/Pdgfrb+ data by three major features: (1) Col15a1, a murine myofibroblast-specific collagen (Extended Data Fig. 7e), was expressed at lower levels in fibroblasts 1 as compared to myofibroblasts (Extended Data Fig. 8c); (2) although Meg3 was expressed in some other cells (Extended Data Fig. 8e), it was confined to fibroblasts 1 within the mesenchyme (Fig. 3k) as validated by ISH in human (Extended Data 8 d-f); (3) Fibroblast 1 are Scara5+ but Frzb- (Extended Data Fig. 8g), again demonstrating that they are distinct from myofibroblasts.
Having established fibroblasts 1 as a distinct population, we generated UMAP and diffusion map embeddings and performed pseudotime analyses of all Pdgfra+/Pdgfrb+ mesenchymal cells to gain insight into their lineage relationships (Fig. 3l-n). This analysis suggested fibroblast 1 (Meg3+, Scara5+) and myofibroblast 2 (Col14a1+, Ogn+) as early states, myofibroblast 3a as an intermediate state, and myofibroblast 1a (Nrp3+, Nkd2+), 1b (Grem2+) and 3b (Frzb+) as terminal states (Fig. 3l-n). Thus, fibroblasts 1 and myofibroblasts 2 are the major source of myofibroblasts in mouse kidney fibrosis. Myofibroblasts 2 (Ogn+/Col14a1+) might exist in healthy mouse kidneys or may arise as an intermediate state via pericyte to myofibroblast differentiation (Fig. 2c, human data). Angiotensin receptor 1 (Agtr1a) expression in these cells points towards a pericyte origin (Fig. 3o).
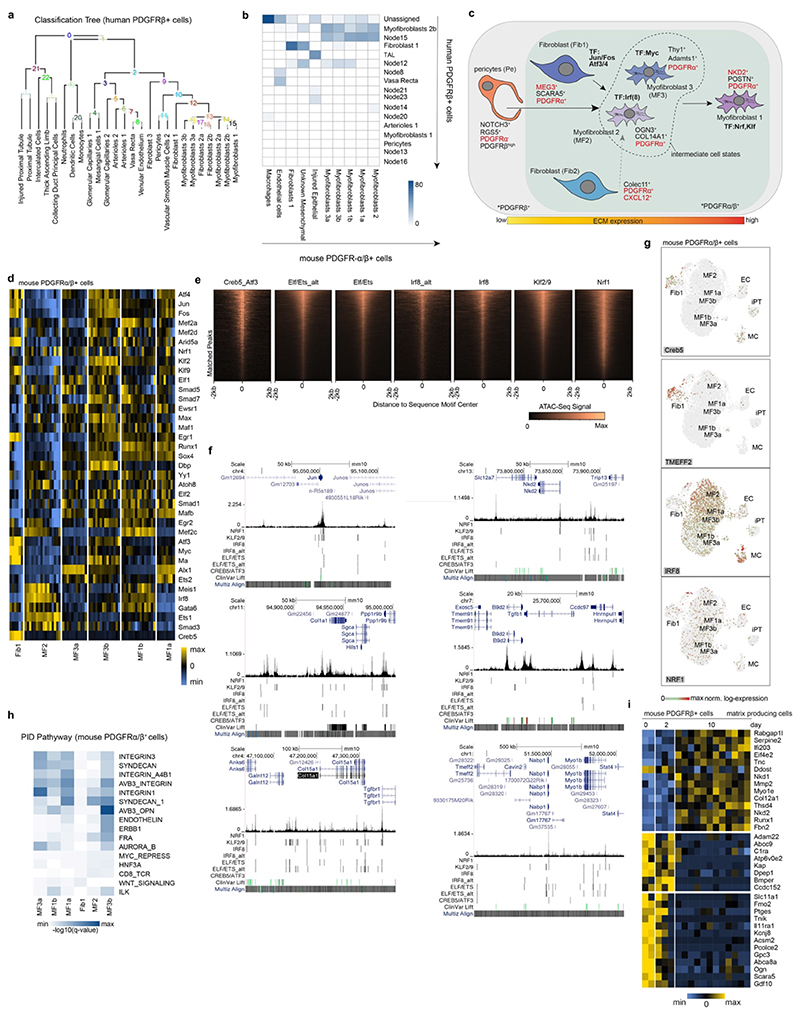
Supervised classification of the mouse Pdgfra+/Pdgfrb+ single cell data based on our human Pdgfrb+ cells confirmed the distinctness of fibroblasts 1 and myofibroblasts in both species (Extended Data Fig. 9a-b).
Our data suggest a model in which Pdgfrb+/Pdgfra+/Postn+ high-ECM expressing myofibroblasts (here termed myofibroblast 1) arise from Pdgfrb+/Pdgfra-/Notch3+ pericytes, Pdgfrb+/Pdgfra+/Scara5+ fibroblasts (fibroblasts 1) and Pdgfrb+/Pdgfra+/Cxcl12+ fibroblasts (fibroblasts 2) (Extended Data Fig. 9c). Pericytes differentiate potentially through an intermediate ECM-expressing Pdgfrb+/Pdgfra+/Ogn+/Col14a1+ (myofibroblasts 2) state into myofibroblasts 1 (Extended Data Fig. 9c)
Distinct fibroblast and myofibroblast cell states are distinguished by specific transcription factor regulatory programs
Next we asked whether the above fibroblast and myofibroblast cell states represent distinct cell types with distinct gene regulatory profiles.17 We generated bulk ATAC-Seq18 data from Pdgfra+/Pdgfrb+ mouse kidneys after UUO and deconvoluted the open chromatin region (OCR) signatures based on OCR proximity to marker genes identified in the scRNA-Seq clusters (Fig. 3q). Fibroblasts 1 and myofibroblasts 2 were distinct from each other and from other myofibroblasts. Myofibroblasts 1a were distinct from myofibroblasts 1b and featured enrichment of ATF factors. Myofibroblasts 2 and 3b showed enrichment of the orphan receptor Nrf4a1, previously reported as a regulator of TGFb signaling and fibrosis.19 Fibroblasts 1 showed enrichment of AP-1 (Jun/Fos) motifs (Fig. 3q), in line with the human data (Extended Data Fig. 6l). RNA expression of these ATAC-Seq selected factors (Extended Data Fig. 9d-g) confirmed the sequence motif enrichment (Fig. 3q), highlighting divergent transcriptional regulation in these populations.
Congruent with our ATAC-Seq data, signaling pathway analysis based on our scRNA-Seq data indicated that fibroblasts 1 and myofibroblasts are distinct populations with different regulatory programs (Extended Data Fig. 9h).
Nkd2 is required for collagen expression in human kidney Pdgfrb+ cells and is a potential therapeutic target in kidney fibrosis
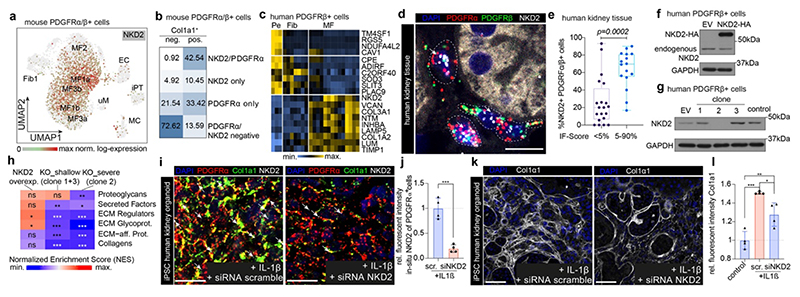
We analysed our data to identify potential therapeutic targets for kidney fibrosis. Nkd2 is specifically expressed in mouse Pdgfra+/Pdgfrb+ terminally differentiated myofibroblasts (Fig. 4a, Extended Data Fig. 9i) and Nkd2/Pdgfra dual positive cells constituted >40% of all Col1a1+ cells (Fig. 4b). In human PdgfrRb+ cells, Nkd2 marks high ECM myofibroblasts, its expression correlates positively with Postn and ECM and negatively with genes associated with pericytes and fibroblasts (Fig. 4c and Extended data Fig. 10a-b). Nkd2+ myofibroblasts exhibited increased TGFb, Wnt and TNFa pathway activity compared to Nkd2- cells (Extended Data Fig. 10c). Multiplex ISH in 36 patients confirmed that a subpopulation of Pdgfra+/Pdgfrb+ cells expresses Nkd2 and expands in fibrosis (Fig. 4d-e).
Figure 4. Nkd2 as therapeutic target.
a. Expression of Nkd2 visualized on the UMAP embedding from Figure 3j. b. Percent of Col1a1+/- cells in mouse Pdgfra+/Pdgfrb+ cells (Figure 3j, stratified by Pdgfra and Nkd2 expression). c. Scaled gene expression of Nkd2 correlating or anti-correlating genes in human Pdgfrb+ cells (Figure 2). d.-e. RNA in-situ hybridization (ISH) of PDGFRa, PDGFRb and NKD2 in human kidneys and quantification of triple positive cells (n=36, Patient data in Extended Data Table 2). n=20 and 16. Two-tailed Mann-Whitney test. Tukey box whisker plot. IF-score = interstitial fibrosis score. Scale bar 10μm. f.-g. Representative Western blots of Nkd2 overexpression and KO cells. For gel source data, see Extended Data Fig. 10e. h. GSEA (Gene set enrichment analysis) of ECM genes in Nkd2-perturbed PDGFRb- kidney cells. n=3 each. *P < 0.05, **p< 0.01, and ***p < 0.001 as determined by FGSEA-multilevel method after adjusting p-values for multiple testing correction (Benjamini & Hochberg). i. ISH of Pdgfra, Pdgfrb and Nkd2 in human iPSC derived kidney organoids. j. Quantification of Nkd2 RNA expression in organoids. n=4 each. Two-tailed unpaired t-test. k.-l. Immunofluorescence staining and quantification of Col1a1 in organoids. n=4 each. *P < 0.05, **p< 0.01, and ***p < 0.001 by 1-way ANOVA followed by Bonferroni’s correction. Scale bar in i+k 50 μm. Data shown as mean±SD. For details on statistics and reproducibility, please see Methods.
Nkd2 is a Wnt pathway and TNFa modulator.20,21 To study the role of Nkd2 in kidney fibrosis, we used our human Pdgfrb+ data to predict a gene regulatory network focused on genes correlated with Nkd2, using the GRNboost2 framework22 (Extended Data Fig. 10d-f). This analysis suggests regulation of Nkd2 by Etv1 and Nkd2 affecting paracrine signaling through Lamp5 (Extended data Fig. 10f-g).
Lentiviral overexpression of Nkd2 in our human Pdgfrb cell line induced expression of ECM molecules in response to TGFb while knockout of Nkd2 markedly reduced col1a1, fibronectin and ACTA2 expression in the presence or absence of TGFb (Fig. 4f-g, Extended Data Fig. 10h-j). RNA-seq from cells overexpressing Nkd2 demonstrated upregulated ECM regulators and glycoproteins, whereas Nkd2 knockout cells exhibited loss of ECM regulators, glycoproteins and collagens (Fig. 4h). Pathway and GO analysis placed Nkd2 in ECM expression programs and suggested interplay with AP1 and integrin signaling (Extended Data Fig. 10k-l). We further observed strong changes in the expression of Wnt receptors and ligands following Nkd2 knockout (Extended Data Fig. 10m).
To validate Nkd2 as a therapeutic target, we generated induced pluripotent stem cell (iPSC) derived kidney organoids containing all major compartments of human kidney (Extended Data Fig. 10n-p). IL1b can induce fibrosis in iPSC derived kidney organoids23 and siRNA mediated knockdown of Nkd2 inhibited IL1b-induced Col1a1 expression (Fig. 4i-l). Thus, Nkd2 marks myofibroblasts in kidney fibrosis, is required for collagen expression, and represents a potential therapeutic target. However, since these organoids do not contain immune cells additional in vivo data will be required to fully verify this finding.
Discussion
Myofibroblasts represent the major source of ECM during kidney fibrosis, but their cellular origin was controversial.1,7 Single cell RNA sequencing allows the dissection of cellular heterogeneity of complex tissues and disease processes, generating novel insights into disease mechanisms at unprecedented resolution.11,24,25
Genetic fate tracing data in mice and histology analyses of human tissue have suggested epithelial, endothelial, hematopoietic cells and resident mesenchymal cells to contribute to fibrosis.1 Here we provide a comprehensive cell atlas of human and mouse kidney fibrosis demonstrating that the majority of scar tissue originates from Pdgfra+/Pdgfrb+ dual-positive fibroblasts and myofibroblasts. In both man and mice these myofibroblasts predominantly derive from pericytes and fibroblasts. Our scRNAseq strategy pointed to novel disease mechanisms and potential therapeutic targets, such as myofibroblast-expressed Nkd2. While Nkd2 has been reported as a Wnt inhibitor our data indicates that it may act also as an activator as some aspects of Wnt signaling.
Our work highlights the intricate cell differentiation mechanisms involved in fibrosis and provides a resource for future clinical research in kidney disease.
Methods
Ethics
The local ethics committee of the University Hospital RWTH Aachen approved all human tissue protocols (EK-016/17). Kidney tissue was collected from the Urology Department of the Hospital Eschweiler from patients undergoing (partial) nephrectomy due to kidney cancer. All patients provided informed consent and the study was performed in accordance with the Declaration of Helsinki.
Human tissue Processing
The tissue was snap-frozen on dry-ice or placed in prechilled in University of Wisconsin solution (#BTLBUW, Bridge to Life Ltd., Columbia, U.S.). Tissues were sliced into approximately 0.5-1mm3 pieces and transferred to a C-tube (Miltenyi Biotec) and processed on a gentle-MACS (Miltenyi Biotec) using the program spleen 4. The tissue was digested for 30 min at 37°C with agitation at 300 RPM in a digestion solution containing 25μg/ml Liberase TL (Roche) and 50μg/ml DNase (Sigma) in RPMI (Gibco). Following incubation, samples were processed again on a gentle-MACS (Miltenyi Biotec) using the same program. The resulting suspension was passed through a 70μm cells strainer (Falcon), washed with 45 ml cold PBS and centrifuged for 5 minutes at 500 g at 4°C. Live, single cells were enriched by FACS-sorting and gating on DAPI negative cells with further enrichment of epithelial cells by CD10 staining or PDGFRß staining for fibroblasts.
Mice
PDGFRßCreERt2 (i.e. B6-Cg-Gt(Pdgfrß-cre/ERT2)6096Rha/J, JAX Stock #029684) and Rosa26tdTomato (i.e. B6-Cg-Gt(ROSA)26Sorttm(CAG-tdTomato)Hze/J JAX Stock #007909) were purchased from Jackson Laboratories (Bar Harbor, ME, USA). Pdgfrb-BAC-eGFP reporter mice were developed by N. Heintz (The Rockefeller University) for the GENSAT project. UUO was performed as previously described using male and female mice.26 Animal experiment protocols were approved by the LANUV-NRW, Germany and by the UK Home Office Regulations. For Smart-Seq2, PDGFRbeGFP male mice were used born within 10 days of each other, and between 9 and 11 weeks old at the time of surgery. For inducible fate tracing PDGFRbCreER;tdTomato mice (8 weeks of age) received tamoxifen (10mg p.o.) 3 times via gavage followed by a washout period of 21 days and then subjected to UUO surgery or sham (as above) and sacrificed at 10 days after surgery. Mice were housed two to five animals per cage with a 12-h light–dark cycle (lights on from 0700 to 1900 h) at sustained temperature (20 °C±0.5°C) and humidity (~50%±10%) with ad libitum access to food and water.
Single cell isolation in mouse
Euthanized mice were perfused via the left heart with 20 ml NaCl 0.9% to remove blood residues from the vasculature.To isolate single kidney cells, a combination of enzymatic and mechanical disruption was used as described above for human single cell isolation. Overall the viability was over 80% using this method.
FACS
Cells were labeled with the following antibodies: anti-CD10 human (clone HI10a, biolegend, 1:100), anti-PDGFRb mouse (clone PR7212, R&D, 1:100), anti-PDGFRalpha mouse (clone APA5, biolegend, 1:100)), anti-CD45 mouse (clone 30_F11). Isolated cells were resuspended in 1% PBS-FBS on ice at a final concentration of 1x107 cells/ml. Cells were pre-incubated with Fc-Block (TruStainFx human, TruStainFx mouse Clone 91, biolegend) and then incubated with the above antibodies for 30 minutes on ice protected from light diluted 1:100 in 2% FBS/PBS. For human anti-PDGFRb staining goat anti-mouse Dyelight 405 (poly24091, biolegend, 1:100) was used as a secondary antibody. All compensation was performed at the time of acquisition using single color staining and negative staining and fluorescence minus one controls. The cells were sorted in the semi-purity mode targeting an efficiency of >80% with the SONY SH800 sorter (Sony Biotechnology; 100 um nozzle sorting chip Sony). For plate based sorting for SMART-Seq, cell sorting was performed on a FACS Aria II machine (Becton Dickinson, Basel, Switzerland) using BD FACSDiva software. FACS data analysis was performed using FlowJo.
Single cell assays incl. Smart-Seq2 and 10X Genomics 3’ sc-RNA-Seq (V2 and V3)
For Smart-Seq2 single cells were processed by SciLifeLab – Eukaryotic Single cell Genomic Facility (Karolinska Institute). Before shipping single cells were sorted into wells of a 384-well plate containing pre-prepared lysis buffer. The single cell solution of cells and primary human kidney cells were run in parallel on a Chromium Single Cell Chip kit and libraries were performed using Chromium Single Cell 3’ library kit V2 and i7 Multiplex kit (PN-120236, PN-120237, PN-120262, 10x Genomics) according to the manufacturer’s protocol.
Human kidney fibrosis evaluation
PAS stained sections of the kidneys were analyzed and scored in a blinded fashion. The extent of interstitial fibrosis and tubular atrophy were assessed as two separate parameters as % of affected cortical area. For collagen I and III immunohistochemistry [Collagen I (Southern Biotech) Cat No. 1310-01; Collagen III (Southern Biotech) Cat No. 1330-01], sections of formalin-fixed and paraffin-embedded renal tissues were processed for indirect immunoperoxidase staining as previously described26. Using a whole slide scanner (NanoZoomer HT, Hamamatsu Photonics, Hamamatsu, Japan), fully digitalized images of immunohistochemically stained slides were further processed and analyzed using the viewing software NDP.view (Hamamatsu Photonics, Hamamatsu, Japan) and ImageJ (National Institutes of Health, Bethesda, MD). The percentage of positively stained area was analysed in the kidney cortex in blinded fashion.
Antibodies and immunofluorescence stainings
Kidney tissues were fixed in 4% formalin for 2 hours at RT and frozen in OCT after dehydration in 30% sucrose overnight. Using 5-10 μm cryosections, slides were blocked in 5% donkey serum followed by 1-hour incubation of the primary antibody, washing 3 times for 5 minutes in PBS and subsequent incubation of the secondary antibodies for 45 minutes. Following DAPI (4′,6′-′diamidino-2-phenylindole) staining (Roche, 1:10.000) the slides were mounted with ProLong Gold (Invitrogen, #P10144). The following antibodies were used: anti-mouse PDGFRa (AF1062, 1:100, R&D), anti-CD10 human (clone HI10a, 1:100, biolegend), anti-HNF4a (clone C11F12, 1:100, Cell Signalling), anti-Pan-Cytokeratin TypeI/II (Invitrogen, Ref. MA1-82041), anti-Dach1 (Sigma, HPA012672, 1:100), anti-Col1a1 (Abcam, ab34710, 1:100), anti-ERG (abcam, ab92513, 1:100), anti-CXCL12/SDF-1 (R&D, MAB350, 1:100), AF488 donkey anti goat (1:200, Jackson Immuno Research), AF647 donkey anti-rabbit (1:200, Jackson Immuno Research)
Confocal imaging
Images were acquired using a Nikon A1R confocal microscope using 40X and 60X objectives (Nikon). Raw imaging data was processed using Nikon Software, ImageJ, Adobe Photoshop and Adobe Illustrator.
Human kidney tissue microarray
Paraffin-embedded, formalin-fixed kidney specimens from 98 non-tumorous human kidney samples of the Eschweiler/Aachen biobank were selected based on a previously performed PAS staining. Areas were randomly selected per sample and one 2-mm core was taken from each kidney sample using the TMArrayer™ (Pathology Devices, Beecher Instruments, Westminster, USA). Each core was arrayed into a recipient block in a 2mm-spaced grid covering approximately 2.5 square cm, and 5-micron thick sections were cut and processed using standard histological techniques.
RNA in-situ hybridization
In situ hybridization was performed using formalin-fixed paraffin embedded tissue samples and the RNAScope Multiplex Detection KIT V2 (RNAScope, #323100) following the manufacturer’s protocol with minor modifications. The antigen retrieval was performed for 22 min at 96°C instead of 15 min at 99°C in a water bath. 3-5 drops of pretreatment 1 solution were incubated at RT for 10 minutes after performing antigen retrieval. The washing steps were performed 5 minutes three times. The following probes were used for the RNAscope assay: Hs-PDGFRß #548991-C1, Hs-PDGFRa #604481-C3, Hs-Col1a1 #401891, Hs-COL1A1 #401891-C2, Hs-MEG3 #400821, Hs-NKD2 #581951-C2 (targeting 236-1694 of NM_033120.3), Hs-Postn #409181-C2 and 409181-C3, Hs-Pecam1 #487381-C2, Hs-Ccl19 #474361-C3, Hs-Ccl21 #474371-C2, Hs-Notch3 #558991-C2, Mm-Col1a1 #319371, Mm-PDGFRa #480661-C2, Mm-PDGFRb #411381-C3.
Image Quantification - ISH image analysis
Systematic random sampling was applied to subsample at least 3 representative tubulo-interstitial areas per image. Next, every fluorescent dot (transcript) was manually annotated using the cell counting tool from Fiji (Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany). Single nuclei were then isolated using an in-house made tool (https://gitlab.com/mklaus/segment_cells_register_marker) based on watershed (limits: 0.1-0.4) to identify neighbouring nuclei, edge detection for incomplete objects and object size selection (limits: 12-180 μm2). The total number of individual dots was then retried for every isolated nucleus. Dots located outside of nuclei were not included in this analysisFor Meg3 and NKD2 analysis of PDGFRa/b cells images were analyzed using QuPath after segmenting the nuclei and counting cells based on >1 pos. spot per imaging channel. For Col1a1-IF quantification or NKD2-ISH quantification images were split in RGB channels and the integrated fluorescent density was determined per image using ImageJ.
Quantitative RT-PCR
Cell pellets were harvested and washed with PBS followed by RNA extraction according to the manufacturer's instructions using the RNeasy Mini Kit (qiagen). 200 ng total RNA was reverse transcribed with High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) and qRT-PCR was carried out further as previously described26 Data were analyzed using the 2-CT method. The primers used are listed in Extended Data Table 3.
Generation of a human PDGFRb+ cell-line
PDGFRb+ cells were isolated from healthy human kidney cortex of a nephrectomy specimen (71 years old male patient) by generating a single cell suspension (as above). For the isolation the cells were stained in two steps using a specific PDGFRb antibody (R&D # MAB1263 antibody, dilution 1:100) followed by Anti-Mouse IgG1-MicroBeads solution (Miltenyi, #130-047-102). Following MACS cells were cultured in DMEM media (Thermo Fisher # 31885) for 14 days and immortalized using SV40LT and HTERT. Retroviral particles were produced by transient transfection of HEK293T cells using TransIT-LT (Mirus). Two types of amphotropic particles were generated by co-transfection of plasmids pBABE-puro-SV40-LT (Addgene #13970) or xlox-dNGFR-TERT (Addgene #69805) in combination with a packaging plasmid pUMVC (Addgene #8449) and a pseudotyping plasmid pMD2.G (Addgene #12259). Retroviral particles were 100x concentrated using Retro-X concentrator (Clontech) 48hrs post-transfection. Cell transduction was performed by incubating the target cells with serial dilutions of the retroviral supernatants (1:1 mix of concentrated particles containing SV40-LT or rather hTERT) for 48hrs. Subsequently the infected PDGFRb+ cells were selected with 2 μg/ml puromycin at 72 h after transfection for 7 d.
Culturing human induced pluripotent stem cell (iPSC) derived kidney organoids
Human iPSC-15 clone 0001 was received from the Stem Cell Facility of the Radboud University Center, Nijmegen, The Netherlands. Human iPSCs were grown on Geltrex-coated plates using E8 medium (Life Technologies). Upon 70-80% confluency, iPSCs were detached using 0.5 mM EDTA and cell aggregates were reseeded by splitting 1:3. Human iPSC were differentiated using a modified protocol based on Takasato et al. (Nature, 2015) and seeded at a density of 18,000 cells per cm2 on geltrex-coated plates (Greiner). Differentiation towards intermediate mesoderm was initiated using CHIR99021 (6 μM, Tocris) in E6 medium (Life Technologies) for 3 and 5 days, followed by FGF 9 (200 ng/ml, RD systems) and heparin (1 μg/ml, Sigma Aldrich) supplementation in E6 up to day 7. After 7 days of differentiation, cell aggregates (300,000 cells per organoid, mixture of 3 and 5 day CHIR-differentiated cells) were cultured on Costar Transwell inserts to stimulate self-organizing nephrogenesis using E6 differentiation medium. On day 7+18 the kidney organoids were used for siRNA knockdown experiments as described below.
siRNA knockdown of NKD2 in human iPSC-derived kidney organoids
NKD2 siRNA knockdown was carried out according to the manufacturer protocol (DharmaFECT transfection reagent and NKD2-specific smartpool siRNA, both Horizon Discovery). The transfection master mix and scrambled controls were prepared in Essential 6 medium (Gibco) and added to the organoids. After an initial incubation of 24 h, the transfection master mixes were refreshed and IL-1β (Sigma-Aldrich) was added at a concentration of 100 ng/ml to induce fibrosis. The IL-1β exposure together with refreshing the transfection master mix was repeated every 24h for two upcoming days. 96h post transfection initiation, the organoids were harvested and processed for paraffin sectioning. Fluorescence in-situ hybridisation (FISH) and immunofluorescence staining was performed as described above.
TGFb- treatment experiments
TGFb (100-21-10UG, Peprotech) at a concentration of 10 ng/ml in PBS was added to 75% confluent PDGFRb cells for 24 hours after 24 hours serum starvation with 0.5% FCS containing medium. For inhibitor experiments with T-5224 the inhibitor (or vehicle) was added to the culture wells 1 hour before adding TGFb. All experiments were performed in triplicates.
AP-1 inhibitor treatment
T-5224 (c-Fos/AP-1inhibitor, Cayman Chemicals, #22904) was dissolved in DMSO and stored at - 80°C. DMSO was always added in the same proportions to control wells.
Cell proliferation (WST-1 assay)
WST-1 assay with PDGFRb-cells was performed in 96-wells as recommended by the manufacturer (Roche Applied Science). In brief, 1 × 10^4 PDGFRb cells were seeded into each well of 96-well plates and the cells were treated with T-5224 or vehicle (DMSO) with the indicated concentrations in triplicates. Cells were incubated with WST-1 reagent for 2h before harvesting at the indicated time points. Both 450 nm and 650 nm (as a reference) absorbance were measured.
sgRNA:CRISPR-Cas9 vector construction, virus production and transduction
The NKD2-specific guide RNA (forward 5′-CACCGACTCCAGTGCGATGTCTCGG -3′; reverse 5′-AAACCCGAGACATCGCACTGGAGTC -3′) were cloned into pL-CRISPR.EFS.GFP (Addgene #57818) using BsmBI restriction digestion. Lentiviral particles were produced by transient co transfection of HEK293T cells with lentiviral transfer plasmid, packaging plasmid psPAX2 (Addgene #12260) and VSVG packaging plasmid pMD2.G (Addgene #12259) using TransIT-LT (Mirus). Viral supernatants were collected 48-72 hours after transfection, clarified by centrifugation, supplemented with 10% FCS and Polybrene (Sigma-Aldrich, final concentration of 8μg/ml) and 0.45μm filtered (Millipore; SLHP033RS). Cell transduction was performed by incubating the PDGFRß cells with viral supernatants for 48hrs. eGFP expressing cells were single cell sorted into 96-well plates. Expanded colonies were assessed for mutations with mismatch detection assay: gDNA spanning the CRISPR target site was PCR amplified and analyzed by T7EI digest (T7 Endonuclease, NEB M0302S). To determine specific mutation events on both alleles within the clones grown, the PCR product was subcloned into the pCR™ 4Blunt-TOPO vector (Thermo Scientific K287520). Minimum 6 colonies per CRISPR-clone were grown and sent for sanger sequencing (Clone C2: 30 colonies have been sequenced).
Western blot
Cell lysates were prepared by RIPA buffer with protease inhibitor cocktail (Roche). The protein concentrations of the lysates were quantified using BCA assay (#23225, Pierce, ThermoScientific). The protein lysates were heated for 5 min at 95°C in 4x SDS sample loading buffer (BioRad) and loaded into 10% SDS-Page gels. Afterwards samples were transferred onto PVDF membranes and the blots were probed with primary antibody in 5% Blotto (Thermo Fisher): (1:3000 rabbit anti-human NKD2 polyclonal antibody, Invitrogen PA5-61979) for 2 hours, followed by incubation with secondary antibody for 1 hour after washing (1:5000 horseradish-peroxidase -HRP-conjugated anti rabbit, Vector Laboratories) and developed using Pierce™ ECL Western Blotting Substrate A and B. Mouse monoclonal anti-GAPDH antibody (NovusBiologicals NB300-320; 1:1000) followed by HRP conjugated anti-mouse secondary antibody (Vector laboratories) was used as a loading control.
Lentiviral overexpression of Nkd2
The human cDNA of NKD2 was PCR amplified using the primer sequences 5’-atggggaaactgcagtcgaag-3’ and 5’ ctaggacgggtggaagtggt-3’. Restriction sites and N-terminal 1xHA-Tag have been introduced into the PCR product using the primer 5’-cactcgaggccaccatgtacccatacgatgttccagattacgctgggaaactgcagtcgaag -3’ and 5’-acggaattcctaggacgggtggaagtg-3’. Subsequently, the PCR product was digested with XhoI and EcoRI and cloned into pMIG (pMIG was a gift from William Hahn (Addgene plasmid # 9044; http://n2t.net/addgene:9044 ; RRID:Addgene_9044). Retroviral particles were produced by transient transfection in combination with packaging plasmid pUMVC (pUMVC was a gift from Bob Weinberg (Addgene plasmid # 8449)) and pseudotyping plasmid pMD2.G (pMD2.G was a gift from Didier Trono (Addgene plasmid # 12259 ; http://n2t.net/addgene:12259 ; RRID:Addgene_12259)) using TransIT-LT (Mirus). Viral supernatants were collected 48-72 hours after transfection, clarified by centrifugation, supplemented with 10% FCS and Polybrene (Sigma-Aldrich, final concentration of 8μg/ml) and 0.45μm filtered (Millipore; SLHP033RS). Cell transduction was performed by incubating the PDGFß cells with viral supernatants for 48hrs. eGFP expressing cells were single cell sorted.
Bulk RNA sequencing
RNA was extracted according to the manufactureŕs instructions using the RNeasy Mini Kit (QIAGEN). For rRNA-depleted RNA-seq using 1 and 10 ng of diluted total RNA, sequencing libraries were prepared with KAPA RNA HyperPrep Kit with RiboErase (Kapa Biosystems) according to the manufacturer’s protocol.
ATAC-seq preparation
PDGFRa/b+ cells were FACS sorted from freshly isolated UUO kidneys as described above, washed twice with cold PBS and centrifuged at 500g for 5 minutes. Cell pellets were lysed in 50μl ice-cold lysis buffer (10mM Tris-HCl, pH7.5; 10mM NaCl, 3mM MgCl2, 0.08% NP40 substitute [74385, Sigma], 0.01% Digitonin [G9441, Promega]), and immediately centrifuged at 500g for 9 minutes. Pellets were resuspended in 50μl of a transposase reaction mix as previously described27. Transposed DNA was amplified by PCR using NEBNext 2x Master mix (M0541S; New England Biolabs) with custom Nextera PCR primers. The first PCR was performed with 50μl volume and 6 cycles using NEBNext 2x Master mix and 1.25μM custom primers; the second RT-PCR was performed with 15μl volume for 20 cycles using 5μl (10%) of the pre-amplified mixture plus 0.125μM primers to determine the number of additional cycles needed as described previously27. The amplified DNA library was purified using MinElute PCR Purification kit (28004, Qiagen) and eluted in 20μl of 10 mM Tris-HCl (pH 8) for subsequent sequencing.
Smart-Seq2 Data Processing
The initial single-cell transcriptomic data was processed at the Eukaryotic Single-Cell Genomics Facility at the Science for Life Laboratory in Stockholm, Sweden. Obtained reads were mapped to the mm10 build of the mouse genome (concatenated with transcripts for eGFP and the ERCC spike-in set) to yield a count for each endogenous gene, spike-in, and eGFP transcript per cell. Ribosomal RNA genes, ribosomal proteins and ribosomal pseudo-genes were filtered out. We noticed that cells that did not feature any alignments assigned to either eGFP or PDGFRb clustered into a single cluster after unsupervised cell clustering (see below). Therefore, we opted to remove those cells, and performed all analysis and clustering without considering those cells (17 cells).
10x single cell RNA-Seq Data Processing
Fastq files were processed using Alevin28 and Salmon (Alevin parameters -l ISR, Salmon version 0.13.1)29, using Gencode v29 human transcriptome and Gencode vM20 mouse transcriptome as reference transcriptomes30. Alevin’s expected Cells parameter was set according to thrice the number of cells estimated according to the knee-method applied to the read counts per cell barcodes distribution. Therefore, UMI count matrix produced by Alevin produced a large number of putative cells which we could filter later (see next paragraph).
10x scRNA-Seq Cell Filtering
We moved ribosomal RNA genes (0-1% on average of detected RNA content per cell) and mitochondrially-encoded genes (0-80% on average of detected RNA content per cell) from the main gene expression matrix. Mitochondrially-encoded genes were removed to avoid introducing unwanted variation between cells that might be solely dependent on changes in mitochondrial content31. log10(total UMI counts per cell) distribution from the count matrix produced by Alevin (see above) typically showed a bimodal distribution, therefore log10(total UMI counts per cell) were clustered into two clusters using mclust R package v5.4.3 setting modelNames to “E” 32. Cells that belong to the cluster with the higher counts were kept. Then cells were filtered based on mitochondrial RNA content and bias toward highly expressed genes as follows: (1) cells were clustered into two clusters using a bivariate Gaussian mixture with two components learned on log10(total UMI counts per cell) and percent of mitochondrial UMI per cell. Clustering was performed using the R package Mclust setting modelNames to “EII”. Cells falling into the cluster with higher mitochondrial content cells were excluded. This filtering step was followed only for libraries which showed a clear bimodal distribution of mitochondrial content (only three 10x libraries in this study) (2) The total number of UMIs per cell should correlate with the total number of unique detected genes. Cells that do not follow this relationship (outliers) were filtered by clustering nuclei using a bivariate Gaussian mixture model on log10(total UMI counts) and log10 total unique detected genes using the mclust R package setting modelNames to “VEV”,”VEE”. (3) Cells whose percent of total counts in the top 500 genes represented more than 5 times absolute median deviation for all cells were removed. (4) Finally, to exclude cells comprised mainly of ribosomal proteins and pseudo-genes, we removed cells whose percent of ribosomal protein and pseudo-gene expression represented more than 5 absolute median deviations of all other cells. Mitochondrial-based filtering was not performed for CD10+ libraries since libraries from proximal tubule epithelial cells are expected to result in a high number of mitochondrial reads. Note that not all filtering steps were performed for all libraries as this depends on each library’s quality and UMI-cell-gene distribution. The script for quality control, cell filtering is available here: https://github.com/mahmoudibrahim/KidneyMap/blob/master/templates/process_scRNA.r
Human 10x Single Cell Data Integration Strategy
Upon initial analysis of our data, we noted several points: (1) Cell types are not guaranteed to be equally represented across patients and across conditions (healthy or CKD). This is because the cell types captured in any single 10x Chromium run are determined by random sampling of cells. (2) Both healthy and CKD patient samples consist of cells in healthy and disease states, since this categorization is based on clinical parameters and not on molecular data or a controlled in vitro experiment. We would expect mainly a change in proportion of healthy and disease cell states between healthy and diseased patient samples. (3) Samples from different patients were processed and prepared on different days as dictated by the surgery schedule at the Eschweiler hospital. Therefore, potential technical (batch) effects could not be controlled on the experimental side. (4) The ability to discover highly resolved cell clusters in under-represented cell types might be affected by class imbalance since certain cell types may be significantly more abundant than others, and the size of the dataset (number of cells) which affects clustering results using unsupervised modularity-based graph clustering algorithms33.
Our experimental strategy involved obtaining separate libraries from CD10+ and CD10- cell fractions (see Main Text), which was designed to mitigate class imbalance on the level of cell type capturing frequency by the 10x Chromium protocol. To further mitigate the points discussed above we aimed to (1) cluster the data on a local level while keeping global information on the relation between cell types intact and (2) to correct for potential technical (batch) differences between samples while retaining important differences, such as different cell types or different states of cell types due to disease. To do so, we followed a strategy comprised of the following steps:
Step One: After quality control and cell filtering (see above), cells in each 10x library were clustered separately and each cell cluster was assigned to one of 6 major cell types: CD10+ epithelial, CD10- epithelial, Immune, Endothelial, Mesenchymal and Neuronal cells.
Step Two: For each one of the 6 major cell types, cells from all 10x libraries were integrated together. Variability between cells due to technical reasons was corrected and cells were clustered using unsupervised graph clustering. This process resulted in 6 separate endothelial, CD10+ epithelial, CD10- epithelial, mesenchymal, immune and neuronal maps. Each map composed of cells from multiple 10x libraries.
Step Three: We integrated 3 single cell maps for: (1) CD10+ cells (proximal tubule / Figure 1), (2) CD10- cells (proximal tubule-depleted / Figure 1) and (3) PDGFRb+ cells (mesenchymal / Figure 2), by combining single cell expression (UMI counts) and clustering information from all main cell type individual maps of each data set from Step Two. All plots in the manuscript are thereafter reproducible from those 3 integrated maps.
This approach accomplished local clustering and technical variability removal, and allowed for high resolution discovery of cell states regardless of highly variable cell cluster sizes. The smallest cluster consisted of 24 cells, while the largest cluster consisted of 5355 cells. Relative to “a high-level clustering followed by sub-clustering” approach, our approach produces highly resolved clusters in a data-driven unbiased manner, while avoiding the question of which clusters to subcluster altogether. We note that Zeisel et al. followed a somewhat similar data integration approach34.
Details, Step One
Cell clustering
After cell filtering and quality control (see above), we used marker genes compiled from Lake et al.35, Clark et al.36 and BioGPS37 (for neuronal genes) as a priori defined highly variable gene list. Two lists were constructed for human and mouse based on gene symbol conversion according to the biomaRt database38,39. We followed a graph clustering approach to determine cell clusters, similar to that of Seurat40 and inspired initially by Xu et al. 2015, Bioinformatics and Macosko et al. Cell 2015, among others The clustering approach consisted of dimensionality reduction of the normalized expression matrix (restricted to the highly variable gene list) using Singular Value Decomposition as a first step. The left singular vectors are Eigengenes that describe gene expression programs across single cells 41. The top n left singular vectors were selected based on the knee of the singular values curve, and used to construct a k-nearest neighbor graph, where average k per cell was defined as the square root of the number of cells. The function nn2 from the R package RANN was first used to define the k-nearest neighbors (https://CRAN.R-project.org/package=RANN) and the final graph was constructed based on the top n nearest neighbors by similarity where n=k*number_of_cells. Cells were clustered on the graph using the Infomap graph clustering method 42 as implemented in the iGraph R package (https://igraph.org). Infomap is a state-of-the-art graph community detection method which we selected for this step as we noticed it tends to produce higher resolved clusters than other graph clustering methods. At this step, we also calculated a single cell doublet score for all cells using the doublet score function in the Scran R package which implements the doublet score method from Dahlin et al. 201843. This score is aggregated per cluster and reported for each integrated map (see Extended Data Figures 4i nad 8e), but not used to exclude cells.
Assigning cell clusters to 5 major types
We obtained a ranking for each gene in each cluster according to whether it is unique to a cluster and also highly expressed in this cluster using the function sortGenes in the genesorteR R package 44, setting binarizeMethod to “naive”. We intersected the top 50 genes in each cluster with the a priori highly variable gene list (see above) and used this intersection to determine which major cell type (epithelial, endothelial, immune, neuronal, mesenchymal) the cell cluster belongs to.
Scripts and meta-data
The a priori putative variable gene list is provided here: https://github.com/mahmoudibrahim/KidneyMap/blob/master/assets/public/all_markers_Human_MMI_Apr2020.txt and https://github.com/mahmoudibrahim/KidneyMap/blob/master/assets/public/all_markers_Moue_MMI_Apr2020.txt. The script for quality control, cell filtering, clustering and cell type assignment is provided here: https://github.com/mahmoudibrahim/KidneyMap/blob/master/templates/process_scRNA.r
Details, Step Two
Combining Data
We combined all cells belonging to each major cell type from all samples and patients (all 10x libraries) as well as their clustering information obtained via graph clustering in Step One. Then for each major cell type the following steps were followed:
Data Integration and Iterative Clustering
We have previously observed that marker genes or differentially expressed genes identified after cell clustering can often differ from those used as a feature set input to the clustering procedure44. It is also generally established that clustering results will vary depending on the input feature set. Therefore we followed an iterative clustering approach that cyclicly refines the variable gene set that is input to the clustering procedure, the technical effect mitigation parameters and the cell cluster assignments. In detail, the algorithm consists of the following steps: (a) given the clustering obtained from Step One we define highly variable features based on gene specificity ranking per cluster using the sortGenes function in the genesorteR R package setting binarizeMethod to “naive” (see above). We use the combined set of the top 500 genes in each cluster as highly variable genes. (b) remove technical effects using the mutual nearest neighbor (MNN) method 45 as implemented in the fastMNN function of the batchelor R package45,31, setting the number of dimensions to 30 and auto.order to TRUE. This method removes technical differences while retaining differences due to cell types and returns reduced dimensions directly. (c) Cluster the cells based on the reduced dimensions returned by fastMNN. The clustering approach is similar to that followed for clustering in Step One except that we use the Louvain algorithm, a widely used algorithm for community detection on Graphs and for single cell clustering 46. To control the resolution at which the clustering occurs, we define the average number of k nearest neighbors used to construct the graph as r. squareroot(n) and vary r between 1 and 0.01. We select the r that returns the most informative clustering as determined using the getClassAUC function from the genesorteR R package 44. This function defines clustering quality by an internal evaluation procedure, and expresses clustering quality as a function of the specificity of the marker genes in each cell cluster. The number of nearest neighbors that produces the clustering with highest average class AUC is selected. (d) Raw gene expression counts (UMI counts) are normalized using the deconvolution strategy for scaling normalization 47 as implemented in the computeSumFactors function in the Scran R package 31, setting the clusters argument to the cluster labels obtained from (c). We repeat steps a-d until there is no longer any appreciable increase in agreement in cell cluster assignments between consecutive iteration, quantified by the slope of change of the adjusted rand index48. We noticed that this algorithm results in a progressive increase in the rand index (between cluster assignments in the i-th iteration and those in the i-1-iteration) and increase in class AUC value measured by genesorteR’s getClassAUC function. Typically no more than 3 iterations are needed. An approach to refine the variable gene list and cell clustering was proposed in Zeisel et al.34 and in Yang et al.49.
Custer Quality Control
We then determine low quality cell clusters as those with no differentially expressed genes at a p-value cutoff of 0.05, as determined by the getPValues function from genesorteR R package44, or those whose differentially expressed genes are dominated by ribosomal proteins or genes typically known as house-keeping genes (such as B2M, GAPDH). ,We also controlled for potential doublet clusters based on marker gene expression. For example, if a cell cluster expresses both Epcam (epithelial marker) and Ptprc (CD45, immune marker) at high levels simultaneously, we assume it may represent an epithelial cell / immune cell doublet. This is a similar approach to the one deployed in a previous study (Karaiskos et al., Science, 2017). We repeated the clustering procedure again after this cell removal.
Scripts and meta-data
Scripts for data integration and clustering is provided here: https://github.com/mahmoudibrahim/KidneyMap/blob/master/templates/clusterCells.r
Details, Step Three
Integrated Maps
Integrated maps were generated by combining the clustering results (Step Two), patient or mouse meta-data and cell expression (UMI count) information as detailed below. For the whole kidney CD10+/- data, we generated two maps accordingly. The CD10- map contained all epithelial, immune, endothelial, mesenchymal and neuronal cells, while the CD10+ combined all epithelial CD10+ sorted cells. PDGFRb+ data was analyzed separately from CD10+/- data. We generated one integrated map comprising all cells from all PDGFRb+ libraries.
Cluster Merging and Filtering
We first removed genes that were detected in less than 0.1% of all cells (ie. at least in 1 out of every 1000 cells) given the full integrated map, and used the remaining genes to produce gene specificity ranking per cell cluster using the sortGenes function from the genesorteR R package setting binarizeMethod to “naive”. Clusters which shared more than 80 out of the top 100 specific genes were merged. We have experimented with different ways to merge similar clusters, and this was our choice as a conservative method that tended to maintain different cell states and merge only very highly similar clusters. Despite our efforts to remove low quality droplets during cell filtering and low quality clusters in Step Two, we still noticed the possibility of observing low quality clusters given the entire integrated map. Therefore, having merged the cell clusters, we checked cell clusters for differential expression using the getPValues function in the genesorteR R package setting numPerm to 20 and removed cell clusters with no differentially expressed genes. Those were consistently low quality cells with lower transcript capture rate overall. For the PDGFRb+ data, we also removed cell clusters where PDGFRb was detected in less than 1 median absolute deviation of its expression in all cell clusters (calculated cutoff was: 4% of cells in the cluster); those were immune and epithelial cell clusters. After removing those cell clusters, we reformed an expression matrix containing all possible genes and performed gene filtering again (see above). We normalized gene expression over the full integrated map using the computeSumFactor function from the Scran R package 31 using the clustering information from Step Two.
Scripts and meta-data
Scripts for combining data into full integrated maps and producing all subsequent plots in the manuscript are available here: https://github.com/mahmoudibrahim/KidneyMap/tree/master/make_intergrated_maps. Details for various analyses are described below.
Overall, this approach was biologically informed, and allowed us to correct for potential technical effects during cell clustering such that almost all cell clusters contained cells from more than one patient/library, while preserving interesting differences between patients such as diseased cell states (for example injured Proximal tubule cells), differences in (myo)fibroblast states and differences in ECM expression.
Mouse 10x Single Cell Data Integration Strategy
Mouse 10x data were analyzed and integrated in the same way as described for human data. The script used to produce the integrated map is available here: https://raw.githubusercontent.com/mahmoudibrahim/KidneyMap/master/make_intergrated_maps/mouse_PDGFRABpositive.r
Mouse Smart-Seq2 Single Cell Data Integration Strategy
Since single cell plate sorting was performed such that cells from all three timepoints were equally represented in all plates, no further batch effect mitigation was performed during the analysis. Variable genes were determined using the Scran R package decomposeVar function, after running the trendVar function on the ERCC transcripts31. Genes with an FDR value < 0.01 and biological variance component > 1 were kept as highly variable genes. Using those variable genes we followed the same clustering approach as described for the 10x Chromium data, but we ran only 2 clustering iteration and did not vary the number of nearest neighbours. Script used for analysis of mouse Smart-Seq2 data is available here: https://github.com/mahmoudibrahim/KidneyMap/blob/master/make_intergrated_maps/mouse_PDGFRBpositive.r.
Cluster Annotation
A gene ranking per cluster was produced using the sortGenes function in the genesorteR R package44 setting binarizeMethod to “adaptiveMedian” (Smart-Seq2 Data) or to “naive” (10x Data). We then annotated our highly resolved cell clusters manually based on prior knowledge and information from literature. We refer to this annotation as “Level 3 Annotation” in supplementary files. There were 50 such clusters in CD10- data, 7 clusters in CD10+ data, 26 clusters in PDGFRb+ human data, 10 clusters in mouse Smart-Seq2 data and 10 clusters in mouse PDGFRa+/b+ data. At that highly-resolved level (level 3), a cell cluster can either represent a bona fide cell type or a different cell state. Thus, we also grouped those highly-resolved cell clusters into canonical cell types based on our annotation. This resulted in 29 cell types in CD10- map, 1 cell type in CD10+ map, 16 cell types in PDGFRb+ map, 5 cell types in mouse PDGFRa+/b+ map and 6 cell types in Smart-Seq2 mouse PDGFRb+ map. We refer to this cell grouping as “Level 2 Annotation” in Supplementary Files. We then further annotated the cell clusters as either epithelial, endothelial, mesenchymal, immune or neuronal for plot and figure annotation in order to enable easier data interpretation.
UMAPs and Diffusion Maps
Integrated full-map UMAP50 projections (Figure 1, 2, 3, 4, 5) were generated via the UMAP Python package (https://github.com/lmcinnes/umap) on the reduced corrected dimensions returned from fastMNN setting min_dist to 0.6 and the number of neighbours to square root the number of cells. Local UMAP projections (Figure 1, Figure 4 and Extended Data Fig. 5) were produced setting min_dist to 1, as those parameters tend to produce more geometrically accurate embeddings (see https://umap-learn.readthedocs.io/en/latest/). Diffusion Maps were produced using the Destiny R package (https://github.com/theislab/destiny) also using the reduced dimensions returned from fastMNN as input and setting the number of neighbours to square root the number of cells.
We tested various randomization seeds for UMAP and Diffusion Map and various Diffusion Map distance metrics (as recommended in the Destiny R package manual) and confirmed that no qualitative difference occurs in the resulting single cell projections.
Lineage Trees/Trajectories and Pseudotime
The Slingshot R package51 was used for lineage tree inference and pseudotime cell ordering inference based on the UMAP/Diffusion Map projection. The cell clustering (Step Two from integration strategy, see above) was used as input cell clusters. Start and end clusters were chosen based on reasonable expectation given our prior knowledge as discussed and recommended in Street et al.51 (for example, myofibroblast is the end cluster in a pericyte/fibroblast/myofibroblast map).
Gene Dynamics along Pseudotime
Genes whose expression varied with cell ordering were defined as those whose normalized expression correlated with cell ordering as quantified by the spearman correlation coefficient at a Bonferroni-Hochberg corrected p-value cutoff of 0.001. Gene clusters and expression heatmaps (for example, Fig. 2f-top) were produced by ordering cells along the pseudotime predicted by SlingShot and using the genesorteR function plotMarkerHeat. This function clusters genes using the k-means algorithm, and we set the plot and clustering to average every 10 cells along pseudotime. Pathway enrichment and cell cycle analyses were calculated by grouping every 2000 cells along pseudotime.
Pathway Enrichment and Gene Ontology Analysis
For the single-cell data, we used KEGG pathway and PID pathway data downloaded in November 2019 from MSigDB 352,53 as “.gmt” files. Pathway enrichment analysis was performed using the clusterProfiler R package54 using the top 100 genes for each cell cluster/group as defined by the sortGenes function from the genesorteR package. The enricher function was used setting minGSSize to 10 and maxGSize to 200. The top 5 terms by q-value for each cell cluster/group were plotted as heatmaps of -log10(q-value). Gene Ontology Biological Process55 analysis was performed on the top 200 genes via the same method. The enricher function was used setting minGSSize to 100 and maxGSize to 500. To compare pathway activity between NKD2+ and NKD2- mesenchymal cells, we used PROGENy to estimate the activity of 14 pathways in a single-cell basis 56,57, using the top 500 most responsive genes from the model as it is recommended from a benchmark study 57.
Cell Cycle Analysis
Cell cycle analysis was done following the method used in Macosko et al.58 and explained in the tutorial by Po-Yuan Tung (https://jdblischak.github.io/singleCellSeq/analysis/cell-cycle.html, date: 06-07-2015), using normalized gene expression as input and setting the gene correlation value to 0.1. We used cell cycle gene sets provided in from Yang et al.59. To quantify enrichment/depletion of single cell cycle assignments (Figure 1g), we plot the log2 fold-change of those frequencies relative to the average frequency obtained by randomizing the true frequency matrix 1000 times while keeping row and column sums constant. Randomization was performed using the R package Vegan (https://CRAN.R-project.org/package=vegan). Positive numbers indicate enrichment relative to what would be expected by chance, negative numbers indicate depletion.
ECM and Collagen Score
The expression of core matrisome genes provided in Naba et al.5 were summarized based on normalized gene expression data using the same method used for cell cycle analysis. Also see Extended Data Figure S4.
Gene Expression Heatmaps
Scaled gene expression heatmaps such as those in Figure 2d were produced using the plotMarkerHeat and plotTopMarkerHeat functions in the genesorteR R package44. The fraction of expressed cells heatmaps such as Figure 3d were produced using plotBinaryHeat function from the genesorteR R package. Heatmaps showing log2-fold-changes and enrichments of features such as Figure 5j,k were produced using ComplexHeatmap R package (v. 2.4.2)60.
ATAC-Seq Analysis
Illumina Tn5 adapter sequences were trimmed from ATAC-Seq reads using bbduk command from BBmap suite (version 38.32, settings: trimq=18, k=20, mink=5, hdist=2, hdist2=0)61. STAR (version2.7.0e) was used to map ATAC-Seq reads to the mm10 genome assembly retaining only uniquely mapped pairs (settings: alignEndsType EndToEnd, alignIntronMax 1, alignMatesGapMax 2000, alignEndsProtrude 100 ConcordantPair, outFilterMultimapNmax 1, outFilterScoreMinOverLread 0.9, outFilterMatchNminOverLread 0.9) 62. Picard’s MarkDuplicates command (version 2.18.27) was used to remove sequence duplicates (settings: remove_duplicates=TRUE, http://broadinstitute.github.io/picard/). Non-concordant read pairs were then removed from the BAM file using Samtools (version 1.3.1)63. bedtools (version 2.17.0) was used to convert BAM files to BED files and to extend each read to 15bp upstream and 22bp downstream from the read 5’-end in a stranded manner 64, in order to account for steric hindrance of Tn5-DNA contacts 65. JAMM (version 1.0.7rev5) was used to identify open regions from the final BED files keeping the two replicates separate, retaining peaks that were at least 50bp in width in the all list for further analysis (parameters: -r peak, -f 38,38, -e auto, -b 100)66. ATAC-Seq signal bigwig files were produced using JAMM SignalGenerator pipeline (settings: -f 38,38 -n depth).
To deconvolute ATAC-Seq signal from bulk ATAC-Seq data according to scRNA-Seq clustering, we followed the following strategy. To deconvolute the ATAC-Seq signal three main steps in the data analysis were taken: 1) each open chromatin peak (where TFs are expected to bind DNA) was first assigned to a specific gene. 2) these genes were ranked per scRNA-Seq cluster (Fib, MF1/2 etc) depending on their expression in the single-cell RNA-Seq dataset. 3) The top 2000 ATAC peaks were used to identify enriched transcription factor motif sequences.
In more detail, each open chromatin ATAC-Seq peak was assigned to a gene according to its closest annotated transcription start site using the bedtools closest function, setting 100kb as the maximum possible assignment distance. ATAC-Seq peak ranking per scRNA-Seq cluster was obtained by ranking the peaks according to the ranking of their assigned gene in the single cell RNA-Seq cluster. The top 2000 ATAC-Seq peaks for each scRNA-Seq cluster were selected and XXmotif 67 was used for de novo motif finding for each scRNA-Seq cluster open chromatin regions separately (settings: -- revcomp --merge-motif-threshold MEDIUM). We kept only motifs whose occurrence was more than 5%, as defined by XXmotif, for further analysis. Motif occurrence from all motifs from all 4 scRNA-Seq clusters were quantified using FIMO 68 with default parameters (MEME version 5.0.1) in the peaks assigned to the top 200 genes in each single cell RNA-Seq cluster. This produced a frequency matrix of motif occurrence in scRNA-Seq clusters. To quantify enrichment/depletion of motif occurrence in scRNA-Seq clusters we plot the log2 fold-change of those frequencies relative to the average frequency obtained by randomizing the true frequency matrix 1000 times while keeping row and column sums constant. Randomization was performed using the R package Vegan (https://CRAN.R-project.org/package=vegan). Positive numbers indicate enrichment relative to what would be expected by chance, negative numbers indicate depletion (see Main Figure 4k). We selected Irf8, Nrf,Creb5/Atf3, Elf/Ets and Klf for further investigation. We plotted the signal from all peaks that contained those motifs using DeepTools version 3.3.1 69, using the bigwig file generated by JAMM as input (see above, Figure S11). We visualized the same bigwig file and motif occurrence in the Integrative Genomics Viewer 70 (version 2.4.10, Figure S11).
Other Visualization / Analysis
Heatmaps that do not quantify gene expression were produced using the heatmap2 function in the gplots R package (https://CRAN.R-project.org/package=gplots). Violin plots were produced using the vioplot R package (https://CRAN.R-project.org/package=vioplot).
Quantification and Statistical Analysis used outside of the single cell sequencing data
Data are presented as mean±SEM if not specified otherwise in the legends. Comparison of two groups was performed using unpaired t-test. For multiple group comparison one-way ANOVA with Bonferroni’s multiple comparison test was applied or two-way ANOVA with Sidak’s multiple comparisons test. Statistical analyses were performed using GraphPad Prism 8 (GraphPad Software Inc., San Diego, CA). A p-value of less than 0.05 was considered significant.
Gene Regulatory Network Analysis
Gene expression was l1-scaled per gene and the pearson correlation coefficient was calculated between Nkd2 and all other genes along pericyte, fibroblast and myofibroblast single cells. The top 100 correlating and top 100 anti-correlating genes were selected for pathway enrichment analysis. Further the expression of those 200 genes along single cells was used as input to GRNboost2+ python package to predict putative regulatory links between genes. The output network was filtered by removing connections with strength <= 10. The resulting network was plotted as an undirected network (since regulators are not known beforehand) using ggraph package (https://cran.r-proiect.org/web/packages/ggraph/index.html) and clustered into 4 modules using the Louvain algorithm as implemented in the igraph package.
Transcription Factor Predictions from Single Cell Data
To obtain transcription factor scores in distal and proximal regions, we used the top 200 marker genes for fibroblast, pericyte and myofibroblast cell clusters as input gene lists to RCisTarget71. We followed the RCisTarget Vignette to perform the analysis with default parameters (available https://bioconductor.org/packages/release/bioc/vignettes/RcisTarget/inst/doc/RcisTarget.html).
To quantify AP1 expression, we used all Jun and Fos genes as a geneset and applied the same method to obtain an AP1 score as we did for ECM score. To quantify AP1 activity (defined as the expression of putative target genes72,73, we defined AP1 target genes according to the Dorothea regulon database57,74 and applied the same method as ECM score to obtain a single cell AP1 activity score.
Mouse Supervised Cell Classification
We classified single cells in the mouse PDGFRa+b+ dataset using the human PDGFRb+ dataset as a reference using the CHETAH algorithm with default parameters75. Human gene symbols were converted to mouse gene symbols using the biomaRt database39.
CellphoneDB Analysis
CellPhoneDB (v.2.1.1) was used to estimate cell-cell interactions among the cell types found in the human CD10- fraction using the version 2.0.0 of the database76, and the normalized gene expression as input, with default parameters (10% of cells expressing the ligand/receptor). Interactions with p-value < 0.05 were considered significant. We consider only ligand-receptor interactions based on the annotation from the database, for which only and at least one partner of the interacting pair was a receptor, thus discarding receptor-receptor and other interactions without a clear receptor. Ligand-receptor interactions from pathways involved in kidney fibrosis were selected using the membership from KEGG database for Hedgehog, Notch, TGFb and WNT signaling, and REACTOME database for EGFR signaling from MSigDB 352,53, and manual curation for PDGF signaling.
Bulk RNA-Seq Data Analysis
Gene expression was quantified on the transcript level using Salmon v1.1.0, with the --validatMappings and --gcBias parameters switched on, to the human Gencode v29 transcriptome. Transcript level counts were aggregated to gene level counts using the import in tximport R package, setting countsFromAbundance to “lengthScaledTPM”77. Limma R package (v.3.44.1) was used to test for differential gene expression between Nkd2-perturbed human kidney PDGFRb+ as compared to their control using the empirical Bayes method after voom transformation78. We found that two out of the three clones of CRISPR-Cas9 NKD2 Knock-Out group together in the principal component analysis and exhibited a shallow phenotype, while the third clone grouped independently and presented a more severe phenotype. Thus, we grouped the two first clone knock-outs, to have two independent Knock-Out conditions for the statistical contrasts. Differentially expressed genes were ranked by the moderated t-statistic from the statistical test for pathway and gene ontology analysis. P-values were adjusted for multiple testing using Benjamini & Hochberg method. Genes and pathways with FDR < 0.05 were considered significant.
For pathway and gene ontology analysis, we also used clusterProfiler R package with KEGG and PID pathways using genes with adjusted p-value less than 0.01 in the Nkd2-perturbed cells as compared to the control and absolute log fold-change higher than 1 for knockout comparison (higher than 0 for over-expression comparison) with a maximum of 200 genes, ranked by the adjusted p-value. We used GSEA-preranked to test for an enrichment of ECM genes in the phenotypes using fgsea R package (v.1.14.0)79, with MatrisomeDB gene set collection5.
Statistics and reproducibility
Data are presented as mean±SEM if not specified otherwise in the legends. Unless otherwise stated, statistical significance was assessed by a two-tailed Student’s t-test or one ANOVA with Bonferroni’s multiple comparison with P value < 0.05 being considered statistically significant. Statistical analyses were performed using GraphPad Prism 8 (GraphPad Software Inc., San Diego, CA) or as decribed in the Methods above. Results are presented in dot plots, with dots representing individual values, violin plots (horizontal line indicates the median, the box indicates the span of the 25% to the 75% percentiles, whiskers extend to maximum 1.5x this interquartile range) and Tukey box-whisker-plots (horizontal line indicates the median, the box indicates the span of the 25% to the 75% percentiles, whiskers extend to max. and min. values). The number of samples for each group was chosen on the basis of the expected levels of variation and consistency. The depicted RNAscope, immunofluorescence micrographs and western blot micrographs are representative. All studies were performed at least twice, and all repeats were successful.
Extended Data
Extended Data Fig. 1. Kidneys cell atlas and CD10 sorting strategy.
a. A schematic of human nephrectomy kidneys. Kidney samples were sampled from the tumor-free kidney cortex distant from the tumor region. b. A schematic of the whole kidney sorting strategy. Single cell 10x Genomics RNA-Seq libraries were prepared from CD10 negative, living (DAPI-) and CD10 positive, living (DAPI-) cells separately. CD10 negative cells are enriched for mesenchymal cells. c. Immunofluorescence staining of CD10, LTA, CD45 and WT1. CD10 expression labels proximal tubule epithelial cells. d. Representative flow cytometric plots from the sorting and gating strategy described in c enriching for CD10- cells. e. Relationship between serum creatinine and age in patients included in the scRNA-seq experiments of Fig. 1. f. Relationship between serum creatinine and degree of interstitial fibrosis scored by a blinded nephropathologist for the same patients in e. g. Relationship between serum creatinine and tubular atrophy as scored by a blinded nephropathologist for the same patients in e and f. h. Representative images of PAS stained (left) and Collagen 3 immunostained kidneys of patients with eGFR>60 ml/min/1.73 m2 body surface area. i. Same as h but patient with eGFR<60 ml/min/1.73 m2 body surface area. j. Each patient visualized in the UMAP of Fig. 1b. k. The main 5 cell types found in the CD10- fraction, illustrated on the same UMAP embedding from Figure 1b. l-r. Expression of select marker genes visualized on the same UMAP embedding. s. Each cell state/type visualized in the UMAP of Fig.1b. t. Doublet Score (see Methods) for human CD10+ cells. u. Doublet Score (see Methods) for human CD10- cells. Scale bars: in c1-c3 50 μm, in c4-c6 30 μm, in h-i 75 μm.
Extended Data Fig. 2. Expression of cell type markers and ECM score.
a. Scaled gene expression of marker genes in mesenchymal cell clusters of the CD10- data depicted in Fig. 1b-e. Each 100 cells are averaged in one column. b. Same as a. but endothelial clusters c. Same as a. but immune clusters. d. Same as a. but epithelial clusters. e. The 7 cell clusters found in the CD10+ fraction cells visualized on the UMAP embedding from Fig. 1e. f. UMAP embedding of e. with colors representing the cell types/states. g. Scaled ECM score on UMAP from e. h. 8 patients visualized on UMAP embedding as e. i. Expression of RBP4 visualized on UMAP embedding from e. j. Scaled gene expression of the top 20 genes by specificity of each of the 7 cell clusters discovered in the CD10+ data depicted in e-h. k. Log fold change of cell cycle stage assignment frequencies in healthy and CKD epithelial cells relative to permuted frequencies. Positive numbers represent enrichment, negative numbers represent depletion. l. Percentage of cells per cell cluster in each cell cycle phase as predicted from gene expression data. m-n. KEGG and GEO Process terms enriched in cells belonging to healthy or CKD patients, according to differentially expressed genes between healthy and diseased patients (see Fig. 1f). Note Fatty Acid Catabolic Process and Lipid Oxidation consistent with KEGG pathway enrichment results o. ECM, collagen, proteoglycan and glycoprotein score of human diabetic kidney dataset (Fan Y. et. al. Diabetes 2019). Advanced DN (diabetic nephropathy) n=21, early DN n=6, control n=9. p. Distribution of single cell ECM scores for all cells in the CD10- cell fraction, colors indicate cell groups obtained by unsupervised mixture model clustering of ECM scores. q. ECM groups visualized on the UMAP of Fig. 1b. r. The same ECM scores as in p. but scaled and visualized on the UMAP embedding from Fig. 1b. s.-u. Scaled expression of gene groups summarized in the ECM score including collagens (r), glycoproteins (s) and proteoglycans (t). All 50 cell clusters are shown, all cells from each cluster are averaged in one column.
Extended Data Fig. 3. Kidney mesenchymal cells and proximal tubules.
a. UMAP embedding of Fibroblast/Pericyte/Myofibroblast cells from 13 human kidneys (n=2,689). Colors represent the cell types. Lines refer to a lineage trajectory predicted by slingshot (see Methods). b. Expression of selected genes on the embedding of a. c. Gene Ontology Biological Process analysis for Pericyte (Pe), Myofibroblast (MF), Fibroblast cell clusters (Fib) and vascular smooth muscle cells (VSMCs) based on the top marker genes for each cluster (CD10- data, see Methods). d. ECM score and scaled expression of select genes visualized on the Mesenchymal cell Diffusion Map embedding of Figure 1o. e.-h. The distribution of ECM score, collagen score, glycoprotein score and proteoglycan scores stratified by epithelial cell clusters in the CD10- -cell fraction. i. Scaled expression of select genes in proximal tubules and injured proximal tubule cell clusters. Each 100 cells are averaged in one column. j. Gene Ontology Biological Process analysis based on differential expression between proximal tubules and injured proximal tubules. k.-n. The distribution of ECM score, collagen score, glycoprotein score and proteoglycan scores for epithelial cells (CD10+ cell fraction) o. Percentage of cells expressing PDGFRb and Col1a1 in each main cell niche. Neuronal Schwann cells were excluded since they are represented by a small number of cells.
Extended Data Fig. 4. PDGFRb+ cell enrichment.
a. Patient samples (n=8) visualized on the UMAP from Figure 2a. Different cell clusters are indicated by different colors. Stratification of single cells according to patient clinical parameters (CKD=chronic kidney disease, eGFR=estimated glomerular filtration rate). b-c. Expression of select genes on the same UMAP embedding from a. d. Scaled gene expression of the top 10 genes in each cell type/state cluster. Gene ranking per cell cluster was determined by genesorteR. e. Correlation between cell clusters identified in CD10- data (Figure 1, columns) and PDGFRb+ data (Figure 2, rows). f. ECM score stratified by 4 main cell types in PDGFRb+ data. g. ECM score stratified by main mesenchymal cell types. h. ECM score stratified by 5 epithelial cells clusters. i. ECM score visualized on the UMAP embedding from a. j. Doublet Score (see Methods) for human PDGFRb+ cells. k. Representative image of combined immunofluorescent and multiplex RNA in-situ hybridization of LTA (proximal tubular marker), Col1a1 and PDGFRb+. Note Col1a1 and PDGFRb expression in LTA+ tubular cells (j’ arrows). l. Representative image of combined immunofluorescent and multiplex RNA in-situ hybridization of CD68 (macrophage marker), Col1a1 and PDGFRb. m. Representative image of multiplex RNA in-situ hybridization of Pecam1, Col1a1 and PDGFRb. Scale bars k-m 50 μm.
Extended Data Fig. 5. Lineage trajectories and spatial localization.
a. The mesenchymal cell clusters in Figure 2 here indicated on the Diffusion Map embedding from Figure 2c (left) and stratified by eGFR class (right) and the expression of selected genes on the same embedding. b. UMAP embedding of mesenchymal cell populations from Fig. 2a. Colors represent the cell types/states shown in Fig 2a. c. ECM score visualized on the UMAP in b. UMAP embedding indicates distinct and separate pericyte and fibroblast origins for myofibroblasts, consistent with Diffusion Map embedding of the same cells (Figure 2e) d. 3 main pericyte and (myo-) fibroblast cell types indicated on the same UMAP embedding. e. Pseudotime as predicted by the Slingshot algorithm on the same UMAP embedding from b. f.-g. Col1a1 and Notch3 expression on the UMAP embedding from b. h-i. Violin plots across mesenchymal cells types of Col1a1 and Postn of human PDGFRb+ dataset in Figure 2. j. Quantification of Meg- (Notch3/Postn-) cells in human kidneys (n=35) (see patient data Extended data table 2). n=17 (healthy), 10 (early) and 8(late); *p<0.05, **p < 0.01 by 1-way ANOVA followed by Bonferroni’s correction. Tukey box whisker plot. k-m. Representative image of multiplex RNA in-situ hybridization of Meg3, Notch3 and Postn. Note that triple positive cells (arrow with tails) or double positive cells (Notch3+Postn+, l magnification 2 arrow heads) can be detected in the kidney interstitium. n. Immunofluorescence staining of Cxcl12(SDF-1). Note expression in the kidney interstitium in PDGFRb+ cells (arrow with tails) and LTA- tubular cells (arrows). o.-q. Representative image of multiplex RNA in-situ hybridization of Ccl19, Ccl21 and PDGFRb. r. Quantification of Ccl19/Ccl21+ cells in human kidneys (n=35) (see Patient data Extended data table 2). n=17 (healthy), 10 (early) and 8(late); ***P < 0.001, ****P <0.0001 by 1-way ANOVA followed by Bonferroni’ post-hoc test. Tukey box whisker plot. For details on statistics and reproducibility, please see Methods.
Extended Data Fig. 6. Mesenchymal pathway activity and role of AP-1.
a. KEGG pathway enrichment analysis along pseudotime for lineage 1 (see Figure 2c.) b. Top: Gene expression dynamics along pseudotime for lineage 2 (Fibroblasts to Myofibroblasts, see Figure 2c.). Cells (in columns) were ordered along pseudotime and genes (in rows) that correlate with pseudotime were selected and plotted along pseudotime (see Methods). Each 10 cells were averaged in one column. Genes were grouped signifying their pseudotime expression pattern. Selected example genes for each group are indicated. See Supplementary File 3 for gene cluster assignments. Bottom: Cell cycle stage along pseudotime as percent of each 500 cells along pseudotime. c. Same as in b. but for lineage 3 cells (see Figure 2c) d. PID signaling pathway enrichment analysis along pseudotime for lineage 2 cells ordered along pseudotime as in b. e. KEGG pathway enrichment analysis along pseudotime for lineage 2. f. Same as in d. but for lineage 3 cells (see Figure 2c). g. Same as in e. but for lineage 3 cells. h.-k. Violin plots across mesenchymal cells types of selected genes of the human PDGFRb+ dataset in Figure 2. l. TF scores for proximal promoter regions (l) and distal regions (m) obtained by TF sequence motif enrichment analysis for top marker genes for the mesenchymal cell clusters of the human PDGFRb+ dataset (see Methods). Note enrichment of Fos and Jun motifs in promoters of fibroblast marker genes. m. Schematic of human kidney PDGFRb+ cell generation and immortalization. n. Cell proliferation (WST-1) and expression of cFos, Col1a1, Postn and Ogn by RNA qPCR after AP-1 inhibitor treatment (T-5224) and/or TGFb treatment of immortalized human PDGFRb kidney cells. n=3 per group. *P < 0.05, **P<0.01, ***P < 0.001, ****P <0.0001 by 1-way ANOVA followed by Bonferroni’ post-hoc test. Mean± S.D. o. Expression of Ogn (Fib1+3) and Postn (MF1) visualized on the same UMAP embedding from Extended data Fig. 5b. p. AP-1 average TF expression (left) and average expression of putative AP-1-regulated genes (right) against Collagen scores stratified by fibroblast and myofibroblast cells. Interestingly, the expression of AP-1 anti-correlates with collagen score but the expression of its target genes positively correlates with collagen score, potentially pointing towards an inhibitory role for AP-1. q. The number of statistically significant receptor-ligand interactions between mesenchymal cells and all other cell types (CD10- fraction, Figure 1) according to CellphoneDB Analysis. Dendritic cells, monocytes, myofibroblasts, podocytes, arteriolar endothelial cells and injured tubules as major sources of signaling ligands to pericytes fibroblasts and myofibroblasts. r. Dot plot for significant ligand-receptor interactions from the selected signaling pathways EGFR, PDGF, WNT, TGFb, Notch and Hedgehog for pericytes, fibroblast and myofibroblasts. Interacting ligand-receptor and cell types are shown by pairs. The first cell type of the interacting pair expresses the ligand and the second cell type expresses the receptor (i.e. first and second proteins from the interaction, respectively). Ligand-receptor interactions are grouped by signaling pathways. Yellow: EGFR, pink: PDGF, green: WNT, red: TGFb, black: Notch, blue (light or dark): mixed of TGFb and EGFR. None of the hedgehog interactions were significant. For details on statistics and reproducibility, please see Methods.
Extended Data Fig. 7. Origin of myofibroblast in murine kidney.
a. Representative image of Col1a1 in-situ hybridization in a PdgfrbCreER;tdTomato kidney after UUO surgery. Scale bar 10 μm b-c. Quantification of aSMA+ cells in PDGFRbtdtom+ kidneys from UUO day 10. n=3. Mean± SD. d. Scaled expression of the top 10 genes by specificity in each cell cluster depicted in Figure 3e. All cells from each cell cluster are averaged in one column. e. Expression of select genes in all 10 cell clusters from Figure 3e. f. ECM score visualized on the same UMAP embedding from Figure 3e. g. Distribution of ECM score, collagen score, glycoprotein score and proteoglycan score per cell cluster. h. Immunofluorescence (IF) staining in sham and UUO (day 10) mouse kidney showing Pdgfra expression in a subset of PDGFRbCreER;tdTomato positive cells (arrows). i. RNA in-situ hybridization showing colocalization of Col1a1 expression in PDGFRa/PDGFRb double-positive cells. Col1a1/PDFGRa/PDFRb triple-positive cells (arrows) occur solely in the kidney interstitium. j. Left: Col1a1 expression and ECM score in CD10 negative cells (Figure 1b) stratified according to PDGFRa and PDGFRb expression. Right: Percent of Col1a1 positive and negative cells in the same data, stratified in the same way. Col1a1 negative cells occur mostly in PDGFRa/b double-negative cells while Col1a1 positive cells occur predominantly in PDGFRa/b double-positive cells (n=51,849). Group comparisons: (other genes) vs. (a/b): p~0, (a-) vs. (a/b): p~0, (b) vs. (a/b): p~0, (other genes) vs. (a): p~0, (b) vs. (a): p~0, (other genes) vs. (b):p~0. Bonferroni corrected p-values based on a two-sided Wilcoxon rank sum test. k. Distribution of IF/TA-Score over 62 patients and representative image of a trichrome stained human kidney tissue microarray (TMA) stained by multiplex RNA in-situ hybridization using PDGFRa, PDGFRb and Col1a1 probes with nuclear counterstain (DAPI) of 62 kidneys (patient data in Extended Data Table 2) (left), average scaled Col1a1 expression in the in-situ hybridization data stratified by PDGFRa/PDGFRb detection in the same data (middle) and percent of Col1a1 positive and negative cells in the same data stratified in the same way (right). Group comparison: (a/b) vs. (col1α1): p~0, (a/b) vs. (b): p~0, (a/-) vs. (a): p~0. Bonferroni corrected p-values based on a two-sided Wilcoxon rank sum test. l-p. A Diffusion Map embedding of pericytes and matrix producing cells with annotation of the different time points in m, cell cluster annotation in n and scaled expression of selected genes in o-q. q. The surgery type per cell (sham versus UUO) visualized on the same UMAP embedding from Figure 4c (top), or with colors representing the cell types/states (bottom). r. Expression of select genes on the same UMAP embedding from 3j. s. ECM and collagens score distribution for the 4 major cell types (top) and for mesenchymal clusters (bottom). Scale bars h+j 50 μm, in k 10 μm. For details on statistics and reproducibility, please see Methods.
Extended Data Fig. 8. PDGFRa+/PDGFRb+ cells in kidney fibrosis.
a. Scaled expression of the top 10 genes by specificity in each of the mesenchymal cell clusters depicted in Figure 3d. Each 100 cells are averaged in one column. b. Cell doublet score (see Methods) of mouse PDGFRa/b+ dataset per cell cluster. c. A violin plot of Col15a1 expression per cell cluster. Only mesenchymal cells are shown. Bonferroni corrected p-values based on a two-sided Wilcoxon rank sum test in Supplemental File 4. d. A UMAP embedding of Meg3 as in Figure 2a and multiplex in-situ staining of Meg3 on human kidney tissue. Scale bars d1 30 μm, d2+3 40 μm e. Representative image of multiplex RNA in-situ hybridization for PDGFRa, PDGFRb and Meg3 in n=34 human kidneys (Patient Data in Extended Data Table 2). Meg3 colocalizes with PDGFRa and PDGFRb. Scale bar 10 μm f. Percent of Meg3-cells out of PDGFRa/b double-positive cells, quantified from RNA in-situ hybridization. n=34. Tukey box-whisker plot. g. Expression of select genes on the same UMAP embedding from Figure 3j. For details on statistics and reproducibility, please see Methods.
Extended Data Fig. 9. Correlation of human and mouse populations and distinct gene-regulatory programs of the mesenchyme.
a. Classification tree of human PDGFRb dataset derived by the CHETAH algorithm based on single cell expression and clustering information. b. Supervised classification of mouse PDGFRa+/b+ cells using human PDGFRb+ cells as a reference (see classification tree in a.). Heatmap displays percentage of mouse PDGFRa+/b+ cells in each mouse cell cluster. Fibroblasts 1 in mice are largely classified as Fibroblasts 1 according to human data. Mouse myofibroblasts are classified as Node 15 and myofibroblasts 2b in humans indicating variability between mouse and human with myofibroblast states. c. Schematic of proposed cellular origin of fibrosis. d. Scaled gene expression of transcription factors discovered by ATAC-Seq (see Figure 3q) in six fibroblasts and myofibroblast cell populations. e. ATAC-Seq signal for motif matches inside open chromatin regions for five selected transcription factors. f. Genome browser snapshots for select genes. ATAC-Seq signal and motif matches in open chromatin regions are shown. Multiz Align is conservation scores between mouse and human, ClinVar lift is clinical variants lift to mouse genome coordinates. Nrf, Irf8, Elf/Ets and Klf motifs are located in promoter and enhancer open chromatin regions of myofibroblast associated genes such as Col1a1, Col15a1, Tgfb and Nkd2. Creb5_Atf3 is found in genes associated to Fib1. cluster, such as Tmeff2. g. Expression of some of the genes investigated in g-i. Visualized on the same UMAP embedding from Figure 4c. i. Scaled expression of genes that correlate or anti-correlate with injury time across matrix producing cells (mouse PDGFRb+ data). Note the expression of Ogn, Scara5 and Pcolce2 is largely specific to day0-day2 cells while the expression Nkd2, Fbn2 and Nkd1 is specific is increased in day 10 after UUO. h. Signaling pathway enrichment in the same mesenchymal cell clusters depicted in Figure 3q.
Extended Data Fig. 10. NKD2 is a potential target in kidney fibrosis.
a-b. Gene ontology Biological Process terms for genes that correlate or anti-correlate with Nkd2+ expression across single cells in pericytes fibroblasts and myofibroblasts in mouse PDGFRa+/b+ data (a) and human PDGFRb+ data (b). Genes correlated with Nkd2+ expression are related to ECM expression, integrin signaling and focal adhesion. c. Pathway activity as estimated by the PROGENy algorithm in NKD2+ vs. NKD2- cells from the human PDGFRb+ dataset. p>0.05 n.s., *p<0.05, **p<0.01, ***p<0.001, p values were adjusted for multiple testing using Benjamin/Hochberg method (FDR) (c). d. Scaled gene expression of top 100 genes whose expression is correlated or anti-correlated with Nkd2 expression across single cells in human PDGFRb+ data (see also b.) e. Gene regulatory network predicted based on the expression of cells and genes depicted in l. using the GRNBoost2+ algorithm. Connection between genes indicate putative direct or indirect regulatory interactions. Colors indicate clustering of the gene regulatory network using the Louvain algorithm and highlights the regulatory network of ECM expression (module 2, Nkd2+) and fibroblast and pericyte maintenance (module 4 and 3) f. Module 2 from l. Depicted separately, connections of Nkd2 are highlighted in red. g. Expression of genes highlighted in e. and f. including Etv1 transcription factor and Lamp5 which are both directly connected to Nkd2 in e. and f. h. Expression of Col1a1, Fibronectin (Fn) and Acta2 (aSMA) by qPCR after Nkd2 over-expression in human immortalized PDGFRb+ cells treated with transforming growth factor beta (TGFb) or vehicle (PBS). n=3 per group. 1-way ANOVA followed by Bonferroni’ post-hoc test. Data represent the mean ± SD. i. Expression of NKD2 by RNA qPCR in NKD2 KO cells. ****P <0.0001 by 1-way ANOVA followed by Bonferroni’ post-hoc test. Data represent the mean ± SD. j. Expression of Col1a1, Fibronectin (Fn) and Acta2 by RNA qPCR after Nkd2 knock-out in the same clones depicted in h. n=3 per group. #p<0.05, ##p<0.01, ###p<0.001, ####p<0.0001 (vs. control NTG); ****p <0.0001 (vs. TGFb NTG) by 2-way ANOVA followed by Sidak’s post-hoc test. Data represent mean ± SD. k. PID signaling pathways enriched in PDGFRb+ NKD2-KO clones and overexpression (up indicates up-regulated genes in indicated condition, and down indicates down regulated genes). l. Gene ontology Biological Process terms enriched in PDGFRb+ NKD2-KO clones (up indicates up-regulated genes in KO condition, and down indicates down regulated genes). m. Scaled gene expression of WNT pathway receptors and ligands in Nkd2-perturbed human kidney PDGFRb+ cells.*p< 0.05, **p< 0.01, and ***p < 0.001 as determined by the empirical Bayes from the test for differential expression after adjusting p-values for multiple testing correction (Benjamini & Hochberg) n. Representative image of multiplex RNA in-situ hybridization of PDGFRa, PDGFRb and NKD2 in human iPSC derived kidney organoids. o. Immunofluorescence stainings of human iPSC derived kidney organoids (day 7+18). LTA and HNF4a mark proximal tubular like-cells. pan-CK (Cytokeratin) marks epithelial-like cells. ERG (ETS regulated-gene) marks endothelial-like cells. Dach1 and Nephs1 mark podocyte-like cells. Col1a1 marks fibroblast/myofibroblasts. p. Immunofluorescence stainings of Col1a1 in IL1b treated kidney organoids. Scale bar in n, o, p=50 μm. For details on statistics and reproducibility, please see Methods.
Acknowledgements
Funding
This work was supported by grants of the German Research Foundation (DFG: SFBTRR57, P 25 and 30; SFBTRR219 C01 and C05, CRU344, P1 and SCHN 1188/5-1), by a Grant of the European Research Council (ERC-StG 677448), a Grant of the State of North Rhine-Westphalia (Return to NRW) and a Grant of the Interdisciplinary Centre for Clinical Research (IZKF) within the faculty of Medicine at the RWTH Aachen University (O3-11) all to RK and by a START grant of the RWTH Aachen University to MMI (691906/2018-1) and a grant of the Germany Society of Internal Medicine (DGIM) to CK. PB received funding from the DFG (SFB/TRR57, SFB/TRR219, BO3755/3-1, BO3755/6-1) and the German Ministry of Education and Research (BMBF: STOP-FSGS-01GM1901A). This work was also supported by the BMBF eMed Consortia Fibromap (to VGP, RK and RKS), iPRIME-EKFS to MK, and DFG (CRC/1192 to TBH and VGP). J.R.S. was supported by a Wellcome Trust PhD fellowship (ref. 104366/Z/14/Z). N.C.H. is supported by a Wellcome Trust Senior Research Fellowship in Clinical Science (ref. 219542/Z/19/Z), Medical Research Council, Chan Zuckerberg Initiative Seed Network Grant, and British Heart Foundation grants (RM/17/3/33381; RE/18/5/34216). We thank the Eukaryotic Single-Cell Genomics facility at SciLifeLab Stockholm for generating Smart-Seq2 single cell sequencing libraries.
Footnotes
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Author contributions
CK, MMI and RK designed the study and interpreted the data. MMI designed the data analysis plan. CK, MMI and RK wrote the manuscript and organized the figures. NCH, RKS, and JF edited the manuscript and advised on data interpretation. CK, JK and JS organized patient tissue collection. JK and JS consented the patients. JRS, RD, JWRK and NCH designed, performed and analysed the data from the SSeq2 experiments. CK and SM carried out all other mouse experiments with assistance from MH, XZ and YX. NKab. established the PDGFRb cell line. SZ, XZ and CK carried out the knock-out and overexpression studies. PB provided mice and scored the human samples blinded. CK carried out all other single cell and imaging experiments with assistance from NKae and XZ. RMH and EMJB validated and sequenced the single cell libraries. VGP, MK, LG, THB and MMI analyzed the in-situ hybridization data. JJ and KR performed the organoid experiments. JP and JSR carried out cell-cell communication analysis and bulk cell line RNA-Seq. MMI carried out all single cell and high-throughput data analysis. CK, NCH, MMI and RK initiated the study. All authors read and approved the final manuscript.
Competing interest
The authors have no competing interests.
Data availability
Processed data for all human and mouse RNA-Seq and ATAC-Seq libraries produced in this study are available at the Zenodo data archive (https://zenodo.org/record/4059315, DOI: 10.5281/zenodo.4059315). Processed and raw mouse data are available via the Gene Expression Omnibus at the following links: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE145173 for Mouse PDGFRab scRNA-Seq and ATAC-Seq), https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE144528 for Mouse PDGFRb Smart-Seq.).
Code availability
Custom scripts used in single cell and bulk RNA-seq data analysis are available at: https://github.com/mahmoudibrahim/KidneyMap. Scripts used for imaging in-situ hybridization data quantification are available at: https://gitlab.com/mklaus/segment_cells_register_marker.
References
- 1.Duffield JS. Cellular and molecular mechanisms in kidney fibrosis. J Clin Invest. 2014;124:2299–2306. doi: 10.1172/JCI72267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falke LL, Gholizadeh S, Goldschmeding R, Kok RJ, Nguyen TQ. Diverse origins of the myofibroblast—implications for kidney fibrosis. Nat Rev Nephrol. 2015;11:233–244. doi: 10.1038/nrneph.2014.246. [DOI] [PubMed] [Google Scholar]
- 3.Young MD, et al. Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science. 2018;361:594–599. doi: 10.1126/science.aat1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang HM, et al. Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat Med. 2015;21:37–46. doi: 10.1038/nm.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naba A, et al. The extracellular matrix: Tools and insights for the ‘omics’ era. Matrix Biol. 2016;49:10–24. doi: 10.1016/j.matbio.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan Y, et al. Comparison of Kidney Transcriptomic Profiles of Early and Advanced Diabetic Nephropathy Reveals Potential New Mechanisms for Disease Progression. Diabetes. 2019;68:2301–2314. doi: 10.2337/db19-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kriz W, Kaissling B, Le Hir M. Epithelial-mesenchymal transition (EMT) in kidney fibrosis: fact or fantasy? J Clin Invest. 2011;121:468–474. doi: 10.1172/JCI44595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang S, Susztak K. Epithelial Plasticity versus EMT in Kidney Fibrosis. Trends in molecular medicine. 2016;22:4–6. doi: 10.1016/j.molmed.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elices MJ, et al. VCAM-1 on activated endothelium interacts with the leukocyte integrin VLA-4 at a site distinct from the VLA-4/fibronectin binding site. Cell. 1990;60:577–584. doi: 10.1016/0092-8674(90)90661-w. [DOI] [PubMed] [Google Scholar]
- 10.Kang HM, et al. Sox9-Positive Progenitor Cells Play a Key Role in Renal Tubule Epithelial Regeneration in Mice. Cell Rep. 2016;14:861–871. doi: 10.1016/j.celrep.2015.12.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramachandran P, et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature. 2019;575:512–518. doi: 10.1038/s41586-019-1631-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y-Y, et al. Macrophage-to-Myofibroblast Transition Contributes to Interstitial Fibrosis in Chronic Renal Allograft Injury. J Am Soc Nephrol. 2017;28:2053–2067. doi: 10.1681/ASN.2016050573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henderson NC, et al. Targeting of αv integrin identifies a core molecular pathway that regulates fibrosis in several organs. Nat Med. 2013;19:1617–1624. doi: 10.1038/nm.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wernig G, et al. Unifying mechanism for different fibrotic diseases. Proc Natl Acad Sci U S A. 2017;114:4757–4762. doi: 10.1073/pnas.1621375114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Venkatachalam MA, Weinberg JM, Kriz W, Bidani AK. Failed Tubule Recovery, AKI-CKD Transition, and Kidney Disease Progression. J Am Soc Nephrol. 2015;26:1765–1776. doi: 10.1681/ASN.2015010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kramann R, et al. Parabiosis and single-cell RNA sequencing reveal a limited contribution of monocytes to myofibroblasts in kidney fibrosis. JCI Insight. 2018;3 doi: 10.1172/jci.insight.99561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerstein MB, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palumbo-Zerr K, et al. Orphan nuclear receptor NR4A1 regulates transforming growth factor-β signaling and fibrosis. Nat Med. 2015;21:150–158. doi: 10.1038/nm.3777. [DOI] [PubMed] [Google Scholar]
- 20.Zhao S, et al. NKD2, a negative regulator of Wnt signaling, suppresses tumor growth and metastasis in osteosarcoma. Oncogene. 2015;34:5069–5079. doi: 10.1038/onc.2014.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li C, et al. Myristoylated Naked2 escorts transforming growth factor α to the basolateral plasma membrane of polarized epithelial cells. Proc Natl Acad Sci U S A. 2004;101:5571–5576. doi: 10.1073/pnas.0401294101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moerman T, et al. GRNBoost2 and Arboreto: efficient and scalable inference of gene regulatory networks. Bioinformatics. 2019;35:2159–2161. doi: 10.1093/bioinformatics/bty916. [DOI] [PubMed] [Google Scholar]
- 23.Lemos DR, et al. Interleukin-1β Activates a MYC-Dependent Metabolic Switch in Kidney Stromal Cells Necessary for Progressive Tubulointerstitial Fibrosis. J Am Soc Nephrol. 2018;29:1690–1705. doi: 10.1681/ASN.2017121283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsukui T, et al. Collagen-producing lung cell atlas identifies multiple subsets with distinct localization and relevance to fibrosis. Nat Commun. 2020;11:1920. doi: 10.1038/s41467-020-15647-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adams TS, et al. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Science Advances. 2020;6:eaba1983. doi: 10.1126/sciadv.aba1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kramann R, et al. Perivascular Gli1+ progenitors are key contributors to injury-induced organ fibrosis. Cell Stem Cell. 2015;16:51–66. doi: 10.1016/j.stem.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corces MR, et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods. 2017;14:959–962. doi: 10.1038/nmeth.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Srivastava A, Malik L, Smith T, Sudbery I, Patro R. Alevin efficiently estimates accurate gene abundances from dscRNA-seq data. Genome Biol. 2019;20:65. doi: 10.1186/s13059-019-1670-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frankish A, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47:D766–D773. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lun ATL, McCarthy DJ, Marioni JC. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res. 2016;5:2122. doi: 10.12688/f1000research.9501.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scrucca L, Fop M, Murphy TB, Raftery AE. mclust 5: Clustering, Classification and Density Estimation Using Gaussian Finite Mixture Models. R J. 2016;8:289–317. [PMC free article] [PubMed] [Google Scholar]
- 33.Fortunato S, Barthélemy M. Resolution limit in community detection. Proc Natl Acad Sci U S A. 2007;104:36–41. doi: 10.1073/pnas.0605965104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeisel A, et al. Molecular Architecture of the Mouse Nervous System. Cell. 2018;174:999–1014.:e22. doi: 10.1016/j.cell.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lake BB, et al. A single-nucleus RNA-sequencing pipeline to decipher the molecular anatomy and pathophysiology of human kidneys. Nat Commun. 2019;10:2832. doi: 10.1038/s41467-019-10861-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clark JZ, et al. Representation and relative abundance of cell-type selective markers in whole-kidney RNA-Seq data. Kidney Int. 2019;95:787–796. doi: 10.1016/j.kint.2018.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu C, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Durinck S, et al. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- 39.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stuart T, et al. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.:e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alter O, Brown PO, Botstein D. Singular value decomposition for genome-wide expression data processing and modeling. Proc Natl Acad Sci U S A. 2000;97:10101–10106. doi: 10.1073/pnas.97.18.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosvall M, Bergstrom CT. Maps of random walks on complex networks reveal community structure. Proc Natl Acad Sci U S A. 2008;105:1118–1123. doi: 10.1073/pnas.0706851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dahlin JS, et al. A single-cell hematopoietic landscape resolves 8 lineage trajectories and defects in Kit mutant mice. Blood. 2018;131:e1–e11. doi: 10.1182/blood-2017-12-821413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ibrahim MM, Kramann R. genesorteR: Feature Ranking in Clustered Single Cell Data. bioRxiv. 2019:676379. doi: 10.1101/676379. [DOI] [Google Scholar]
- 45.Haghverdi L, Lun ATL, Morgan MD, Marioni JC. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol. 2018;36:421–427. doi: 10.1038/nbt.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. J Stat Mech. 2008;2008:P10008 [Google Scholar]
- 47.Lun ATL, Bach K, Marioni JC. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 2016;17:75. doi: 10.1186/s13059-016-0947-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rand WM. Objective Criteria for the Evaluation of Clustering Methods. J Am Stat Assoc. 1971;66:846–850. [Google Scholar]
- 49.Yang L, Liu J, Lu Q, Riggs AD, Wu X. SAIC: an iterative clustering approach for analysis of single cell RNA-seq data. BMC Genomics. 2017;18:689. doi: 10.1186/s12864-017-4019-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McInnes L, Healy J, Melville J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv [statML] 2018 [Google Scholar]
- 51.Street K, et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics. 2018;19:477. doi: 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liberzon A, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schubert M, et al. Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat Commun. 2018;9:20. doi: 10.1038/s41467-017-02391-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holland CH, et al. Robustness and applicability of transcription factor and pathway analysis tools on single-cell RNA-seq data. Genome Biol. 2020;21:36. doi: 10.1186/s13059-020-1949-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang J, et al. Single cell transcriptomics reveals unanticipated features of early hematopoietic precursors. Nucleic Acids Res. 2017;45:1281–1296. doi: 10.1093/nar/gkw1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 61.Bushnell B. BBMap: a fast, accurate, splice-aware aligner. 2014 https://www.osti.gov/biblio/1241166.
- 62.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Quinlan AR. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr Protoc Bioinformatics. 2014;47:11.12.1–34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Adey A, et al. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 2010;11:R119. doi: 10.1186/gb-2010-11-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ibrahim MM, Lacadie SA, Ohler U. JAMM: a peak finder for joint analysis of NGS replicates. Bioinformatics. 2015;31:48–55. doi: 10.1093/bioinformatics/btu568. [DOI] [PubMed] [Google Scholar]
- 67.Luehr S, Hartmann H, Söding J. The XXmotif web server for eXhaustive, weight matriX-based motif discovery in nucleotide sequences. Nucleic Acids Res. 2012;40:W104–9. doi: 10.1093/nar/gks602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ramírez F, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–5. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aibar S, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schacht T, Oswald M, Eils R, Eichmüller SB, König R. Estimating the activity of transcription factors by the effect on their target genes. Bioinformatics. 2014;30:i401–7. doi: 10.1093/bioinformatics/btu446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alvarez MJ, et al. Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat Genet. 2016;48:838–847. doi: 10.1038/ng.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Garcia-Alonso L, Holland CH, Ibrahim MM, Turei D, Saez-Rodriguez J. Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res. 2019;29:1363–1375. doi: 10.1101/gr.240663.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.de Kanter JK, Lijnzaad P, Candelli T, Margaritis T, Holstege FCP. CHETAH: a selective, hierarchical cell type identification method for single-cell RNA sequencing. Nucleic Acids Res. 2019;47:e95. doi: 10.1093/nar/gkz543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat Protoc. 2020;15:1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 77.Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2015;4:1521. doi: 10.12688/f1000research.7563.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Korotkevich G, Sukhov V, Sergushichev A. Fast gene set enrichment analysis. Bioinformatics. 2016:471. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Processed data for all human and mouse RNA-Seq and ATAC-Seq libraries produced in this study are available at the Zenodo data archive (https://zenodo.org/record/4059315, DOI: 10.5281/zenodo.4059315). Processed and raw mouse data are available via the Gene Expression Omnibus at the following links: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE145173 for Mouse PDGFRab scRNA-Seq and ATAC-Seq), https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE144528 for Mouse PDGFRb Smart-Seq.).
Custom scripts used in single cell and bulk RNA-seq data analysis are available at: https://github.com/mahmoudibrahim/KidneyMap. Scripts used for imaging in-situ hybridization data quantification are available at: https://gitlab.com/mklaus/segment_cells_register_marker.