Abstract
Placentas may exhibit chromosomal aberrations that are absent from the fetus1. The basis of this genetic segregation, termed confined placental mosaicism, remains unknown. Here, we investigated the phylogeny of human placental cells reconstructed from somatic mutations, using whole-genome sequencing of 86 bulk placental samples (median weight 28mg) and of 106 microdissections. We found that every bulk placental sample represented a clonal expansion that is genetically distinct, exhibiting a genomic landscape akin to childhood cancer, in terms of mutation burden and mutational imprints. Furthermore, unlike any other normal human tissue studied to date, placental genomes commonly harboured copy number changes. Reconstructing phylogenetic relationships between tissues from the same pregnancy revealed that developmental bottlenecks genetically isolate placental tissues, by separating trophectodermal from inner cell mass derived lineages. Of particular note were cases in which inner cell mass derived and placental lineages fully segregated within a few cell divisions of the zygote. Such early embryonic bottlenecks may enable the normalisation of zygotic aneuploidy. We observed direct evidence for this in a case of mosaic trisomic rescue. Our findings reveal extensive mutagenesis in placental tissues and portray mosaicism as a normal feature of placental development.
Introduction
Dysfunction of the placenta contributes substantially to the global burden of disease2. Amongst its many peculiarities is the occurrence of chromosomal aberrations confined to the placenta which affect 1-2% of pregnancies3. It may be present in either or both components of placental villi, the trophectoderm or the inner cell mass (ICM) derived mesenchyme.
Fetal and placental lineages diverge spatially within the first few days of embryogenesis4. The genetic segregation of confined placental mosaicism (CPM) suggests that developmental bottlenecks exist that genetically isolate individual cells, enabling clonal expansions and mosaicism. These may be physiological genetic bottlenecks underlying normal placental development. Alternatively, genetic segregation may represent pathological perturbation of the normal clonal dynamics of early embryogenesis. For example, it has been suggested that CPM represents a depletion from the ICM of cytogenetically abnormal cells, commonly found in early embryos5.
The clonal dynamics of human embryos cannot be studied prospectively. It is, however, possible to reconstruct embryonic lineage relationships from somatic mutations that have been acquired during cell divisions6–8. Furthermore, these mutations may reveal specific mutagenic processes affecting a tissue9. Here, we studied the somatic genetic architecture of human placentas by whole genome sequencing.
Results
Somatic mutations in bulk placental samples
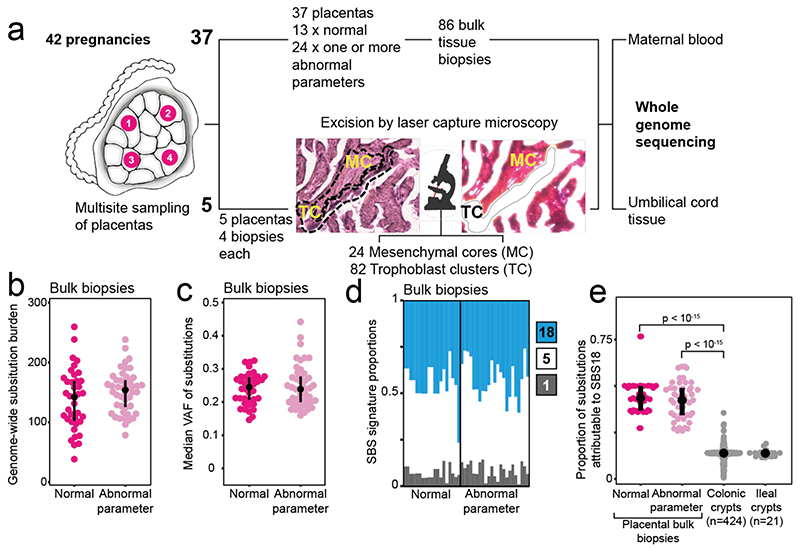
We sequenced the whole genomes of 86 bulk placental samples (median weight 28mg; range 17-79mg) from 37 term placentas along with ICM derived umbilical cord tissue and maternal blood (Fig. 1a), obtained through the Pregnancy Outcome Prediction study, a prospective collection of placental tissue and extensive clinical data10. From each placenta, we studied at least two separate lobules (Supplementary Table 1). We included placentas from normal and complex pregnancies associated with a range of abnormal parameters (Supplementary Tables 2-3). The placental basal plate was trimmed off prior to sampling to eliminate maternal contamination and polyploid extravillous trophoblasts. Each sample was washed in phosphate-buffered saline. Residual contamination was excluded by searching DNA sequences for germline polymorphisms unique to the mother (Supplementary Information). We identified the somatic mutations of each tissue through an extensively validated variant-calling pipeline that provided exquisite sensitivity and high specificity11–14 (Supplementary Tables 4-5). We applied sensitivity corrections to estimates of mutational burdens to adjust for variations in sequence coverage and clonal architecture of samples (Methods).
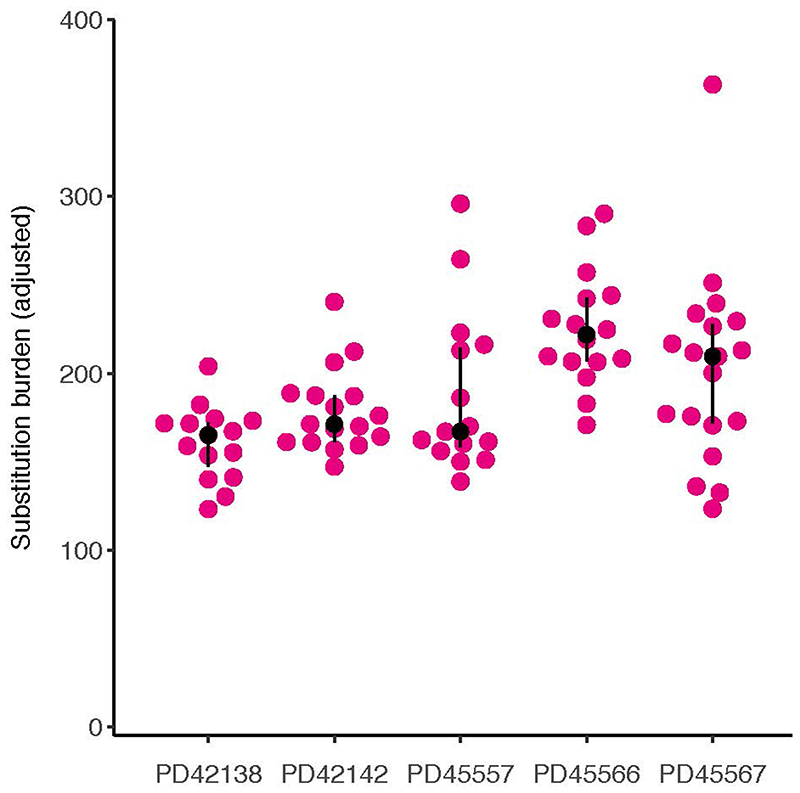
Figure 1. The genomes of bulk placental biopsies.
(a) Workflow detailing experimental design with a photomicrograph demonstrating microdissection of placental tissue. (b) Substitution burden per bulk placental sample, adjusted for coverage and median VAF (Methods). An abnormal pregnancy is defined by the deviation of one or more clinically validated markers from their normal range over the course of pregnancy (Supplementary Tables 2-3). (c) Median VAF of substitutions in each bulk placental sample. (d) SBS signatures in placental biopsies. Each column represents one bulk sample. Colours represent signatures, as per legend. (e) Prevalence of SBS18 mutations in bulk placental biopsies in comparison to human intestinal tissue12, the normal tissue with the highest prevalence of SBS18 reported to date. P-values refer to the two-sided Wilcoxon rank sum test.
We found a high substitution burden within bulk placental samples (Fig. 1b). This was an unusual result because no other bulk normal human tissue studied to date has harboured clones detectable by whole genome sequencing. As they are polyclonal, i.e. comprised of many thousands of distantly related cells (or small clonal groups), the only somatic mutations apparent in polyclonal tissues are typically one to two post-zygotic variants that represent cell divisions of the early embryo6–8. However, in bulk placental samples we found a mean of 145 substitutions (range 38-259). On average, the median variant allele frequency (VAF) of placental mutations within each bulk sample was 0.24 (range 0.15-0.44). Since the proportion of cells carrying a substitution can be estimated by doubling the VAF, this indicated the mutations pervaded ~50% of cells (Fig. 1c). In contrast, umbilical cord samples did not harbour detectable clonal expansions.
Studies of somatic mutations in normal and cancerous human tissues have generated a reference of mutational signatures9. Accordingly, we identified three single base substitution mutational signatures (SBS) in placental tissues: SBS1, SBS5 and SBS18 (Fig. 1d). SBS1 and SBS5 are ubiquitous in human tissues and accumulate throughout life9. SBS18 variants, which may be associated with oxidative stress15, are rare in normal tissues. In bulk placental samples, SBS18 contributed ~43% of substitutions, whereas in normal human colorectal crypts, the normal tissue with the highest prevalence of SBS18 mutations described to date12, SBS18 contributes an average of ~13% of substitutions (Fig. 1e).
Other classes of somatic mutations, small insertions and deletions (indels) and copy number changes (Supplementary Table 1), confirmed the clonality of bulk samples. Of note, 41/86 bulk samples harboured at least one copy number change (gain or loss; median size per unique segment, 73.6 kb), only one of which, a trisomy of chromosome 10, would have been detectable by clinical karyotyping. Within sample size constraints, we did not observe systematic differences in overall mutation burden and spectra between normal and complex pregnancies (Extended Data Fig. 1). The majority of somatic changes in bulk samples from the same placenta - each obtained from separate quadrants, several centimetres apart - were confined to single samples which indicated that the placenta was a patchwork of mosaic, independent genetic units (Extended Data Fig. 2).
Trophoblast clones underpin mosaicism
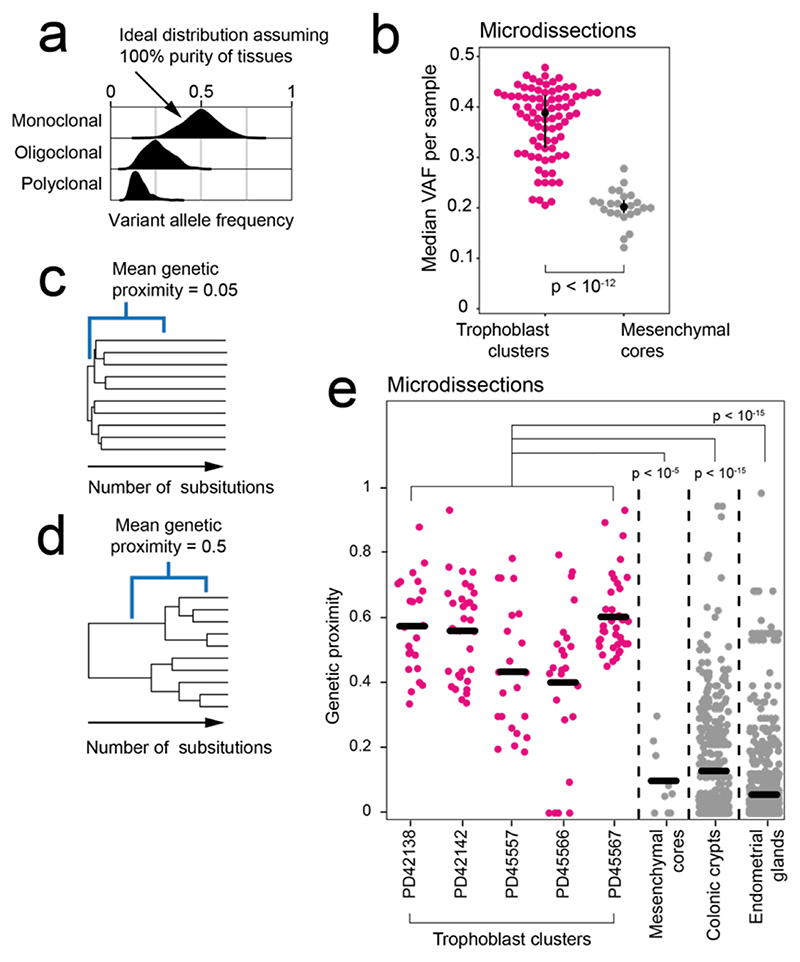
To investigate the cellular origin of the mosaicism in bulk samples, we directly assessed the genomes of the two main elements comprising chorionic villi; the ICM derived fetal mesenchymal cores and the trophoblast (Fig. 1a). Trophoblast microdissections largely represent a single cell type, syncytiotrophoblasts, within the term placenta. Mesenchymal cores consist of a mixture of Hofbauer cells, fibroblasts, smooth muscle and endothelial cells16,17. Using laser capture microscopy, we excised 82 trophoblast clusters and 24 mesenchymal cores from the term placentas of five normal pregnancies, sampling each quadrant at least twice (median 5.5 microdissections, range 2-9, Supplementary Table 1). Following whole-genome sequencing, we called substitutions in each microdissected sample independently and assessed their VAF distribution. If groups of cells were organised as monoclonal patches derived from a single stem cell, their mutations would exhibit a VAF close to 0.5, as seen in single colonic crypts and endometrial glands12,13 (Fig. 2a). Alternatively, if groups of cells were of oligo- or polyclonal origin, their median VAF would be shifted towards zero (Fig. 2a). We found the median VAF of trophoblast clusters and mesenchymal cores significantly differed (0.39 versus 0.20, two-sided Wilcoxon rank sum test, p < 10-12) (Fig. 2b), thus indicating that it was the trophoblast clusters that were monoclonal rather than the mesenchymal cores.
Figure 2. Clonal architecture of microdissected trophoblast clusters and mesenchymal cores.
(a) Theoretical, expected VAF distribution as per different clonal architecture, assuming 100% purity. (b) Comparison of the median substitution VAF between microdissected trophoblast (n=82) and mesenchymal cores (n=24). P-value refers to the two-sided Wilcoxon rank sum test. (c, d) Genetic proximity scores (Methods) were calculated as the fraction of shared mutations between two samples divided by their mean total mutation burden. For example, a mean score of 0.05 conveys little sharing (c), while 0.5 signifies a longer shared development (d). (e). Genetic proximities across trophoblast clusters, mesenchymal cores, colonic crypts12 (n=326) and endometrial glands13 (n=252) from single biopsies. Each dot represents the comparison of two of the same histological unit (e.g., two trophoblast clusters) from the same bulk sample. P-values refer to the two-sided Wilcoxon rank sum test.
We further corroborated this conclusion by studying the genetic relationship between trophoblasts and mesenchymal cores from the same bulk sample. We constructed phylogenetic trees and calculated pairwise genetic proximity scores of microdissections of the two components. This score represented the fraction of total mutations shared between a pair of microdissections. A low score for pairs of trophoblast clusters or mesenchymal cores from the same bulk sample would indicate their precursor cells diverged early in development (Fig. 2c) whilst a high score would suggest a long, shared ancestry (Fig. 2d). This analysis revealed a significant difference in the developmental clonal composition between trophoblast clusters and mesenchymal cores (p < 10-5; two-sided Wilcoxon rank sum test) (Fig. 2e). On average, within each bulk sample, pairs of trophoblast clusters shared 53% of somatic mutations, indicating a long, joint developmental path. In contrast, pairs of mesenchymal cores from the same bulk sample exhibited a mean genetic proximity of 10% and thus a short, shared phylogeny, in line with other ICM-derived tissues, such as colon and endometrium12,13 (Fig. 2e). These observations, in conjunction with monoclonal VAF distributions, suggest that large expansions of single trophoblast progenitors underpin the clonality and confined mosaicism of bulk placental samples.
Cell allocation bias in the early embryo
Our findings indicated that the seeding of a patch of placental tissue represented a genetic bottleneck at which clinically detectable, trophoblast CPM could arise. We now considered whether earlier bottlenecks may exist, amongst the first cell divisions of the embryo. We assessed the distribution of early embryonic lineages across placental and ICM-derived tissues by measuring the VAFs of early embryonic mutations, representing the first cell divisions of the zygote (Fig. 3a,b). These are mutations present in umbilical cord or placenta which, unlike heterozygous germline variants, occur at a variable VAF across tissues.
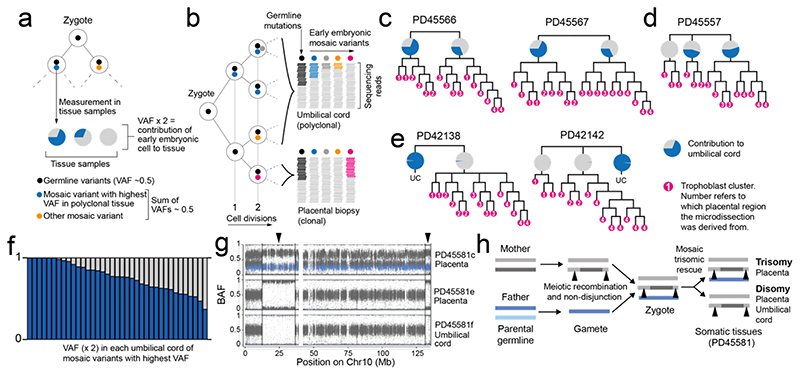
Figure 3. Early embryonic genetic bottlenecks and their relationship to trisomic rescue.
(a) Schematic depicting the detection of the earliest post-zygotic mutations and the estimation of contribution to samples from their VAFs. (b) Hypothetical lineage tree of early embryo showing how measurements of VAF relate to cell divisions. (c) The contribution of the major lineage to the umbilical cord as calculated from the embryonic mutation with the highest VAF. (d) Early trees of trophoblast clusters of PD45566 and PD45567, with the contribution of lineages to the umbilical cord coloured in blue in pie charts. The umbilical cord exhibits an asymmetric contribution of the daughter cells of the zygote. (e) Early cellular contribution in PD45557 shows separation of one placental lineage. (f) In PD42138 and PD42142 the placental and umbilical cord lineages do not share any early embryonic mutations. (g) B-allele frequency (BAF) of germline SNPs on chromosome 10 in PD45581, showing a trisomy in PD45581c (placenta), but a disomy in PD45581e (placenta) and PD45581f (umbilical cord). SNPs absent from mother are coloured in blue. (h) Overview of genomic events in PD45581 and parents leading to the observed mosaic trisomic rescue. The arrowheads highlight areas of two genotypes in PD45581c due to meiotic recombination in the mother.
We directly compared the VAF of early embryonic mutations across bulk and microscopic samples, examining a total of 234 samples from 42 pregnancies. We found three configurations (Fig. 3, c to f). In 19/42 pregnancies, the earliest post-zygotic mutation exhibited an asymmetric VAF across ICM and trophectoderm lineages, without genetically segregated placental samples (Fig. 3c, Extended Data Fig. 3). In about a quarter of pregnancies (12/42), we found that one bulk placental sample did not harbour the early embryonic mutations shared between umbilical cord and the other bulk samples. This indicated that the primordial cell seeding the bulk sample in question segregated in early embryogenesis, thus representing a genetic bottleneck (Fig. 3d, Extended Data Fig. 4). Loss of heterozygosity as an explanation for the absence of early embryonic mutations was excluded (Supplementary Tables 1&5). In the remaining 11/42 pregnancies, the genetic bottleneck generated a complete separation of all placental tissues from umbilical cord samples (Fig. 3e, Extended Data Fig. 5). There were no shared mutations between placental tissues and umbilical cord lineages, consistent with this complete split having occurred at the first cell division of the zygote. Taken together, this data suggests that in about half of placentas, at least one bottleneck exists, which may segregate genomic alterations that arise within the first few cell divisions between placenta and fetal lineages.
Early bottleneck permits trisomic rescue
A striking example of segregating genomic alterations was a pregnancy harbouring trisomy of chromosome 10 in one bulk placental sample, but chromosome 10 disomy elsewhere in the placenta and umbilical cord (Fig. 3g). Analysis of the distribution of parental alleles demonstrated that there were two maternal and one paternal chromosomes in the affected bulk placental sample. Importantly, the two maternal copies were non-identical, generating segments of chromosome 10 with three genotypes in the affected sample. In samples which were disomic for chromosome 10, there were two maternal copies, i.e. uniparental (maternal) disomy (Fig. 3h). Thus, at fertilisation two distinct copies of chromosome 10 had been present in the egg, and resulted in a zygote with trisomy 10. This pattern demonstrates direct evidence for trisomic rescue, i.e. that the trisomy was present in the zygote, but that one cell of the early embryo, which ultimately formed the fetus and some of the placenta, reverted to disomy post-zygotically (Fig. 3h). As the extra chromosome was maternal, and the chromosome lost paternal, the fetus was euploid with uniparental (maternal) disomy. Only a single clonal substitution was detected in the umbilical cord of this pregnancy, indicating that the trisomic rescue had occurred at a genetic bottleneck within the first cell divisions (Extended Data Fig. 5).
Mutational landscape of trophoblasts
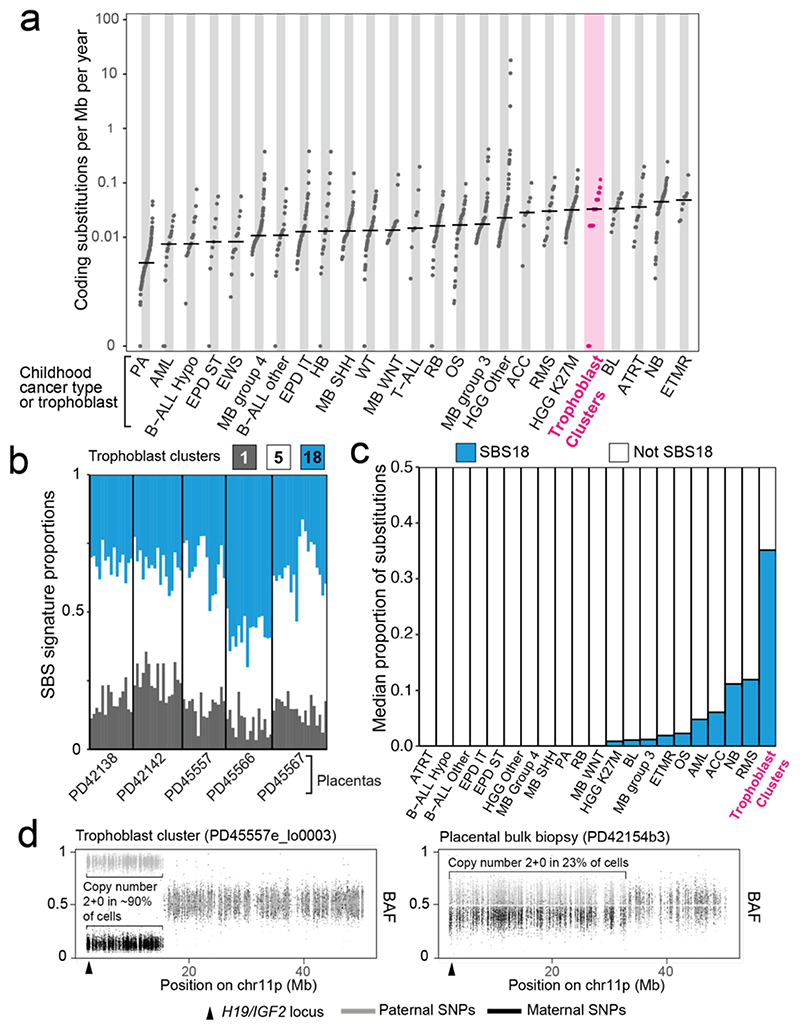
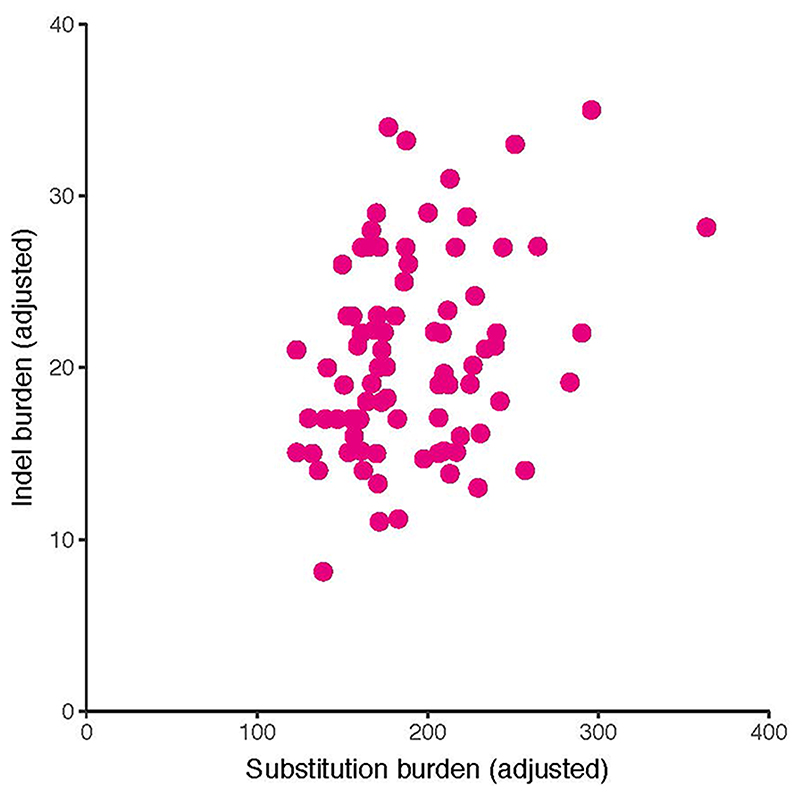
The monoclonal organisation of trophoblast clusters provided the opportunity to examine mutational processes affecting placental tissue in detail. We found an average of 192 variants per trophoblast cluster (Extended Data Fig. 6) with a mutation rate akin to that of childhood cancers, which, like the placenta, are primarily subjected to in utero mutagenesis18 (Fig. 4a). Furthermore, a large proportion of substitutions in each trophoblast sample could be assigned to SBS18 (Fig. 4b and Fig. 4c), exceeding what has been observed in cancer types with the highest relative burden of SBS1818 (Figure 4c). In addition, we found an indel burden proportional to substitutions in each sample and widespread copy number changes (Extended Data Fig. 7, Supplementary Tables 1&5); at least one aberration was detected in 37/82 trophoblast clusters, acquired at an average estimated rate of 0.894 per year. For context, normal colorectal crypts are estimated to accrue ~0.004 structural variants per year12.
Figure 4. The genomes of microdissected trophoblast clusters.
(a) Comparison of the coding substitutions per Mb per year between trophoblast microdissections and paediatric malignancies18. Per year estimates are corrected for gestation. Abbreviations: Pilocytic astrocytoma (PA), acute myeloid leukaemia (AML), hypodiploid B-cell acute lymphoblastic leukaemia (B-ALL Hypo), supratentorial ependymoma (EPD ST), Ewing’s sarcoma (EWS), medulloblastoma group 4 (MB group 4), non-diploid B-cell acute lymphoblastic leukaemia (B-ALL other), infratentorial ependymoma (EPD IT), hepatoblastoma (HB), medulloblastoma SHH subgroup (MB SHH), Wilms tumour (WT), medulloblastoma WNT subgroup (MB WNT), T-cell acute lymphoblastic leukaemia (T-ALL), retinoblastoma (RB), osteosarcoma (OS), medulloblastoma group 3 (MB group 3), high-grade glioma K27wt (HGG Other), adrenocortical carcinoma (ACC), rhabdomyosarcoma (RMS), high-grade glioma K27M (HGG K27M), Burkitt’s lymphoma (BL), atypical teratoid rhabdoid tumour (ATRT), neuroblastoma (NB), embryonal tumours with multilayered rosettes (ETMR). (b) Single base substitution signatures in trophoblast clusters. Each column represents one piece of microdissected tissue. (c) Bar chart showing the median proportion of substitutions attributable to SBS18. Abbreviations as per (a). (d) Partial paternal uniparental disomy of 11p in two samples, represented by the BAF of SNPs across 11p. Grey denotes SNPs contributed by the father and black by the mother.
Annotating somatic placental variants
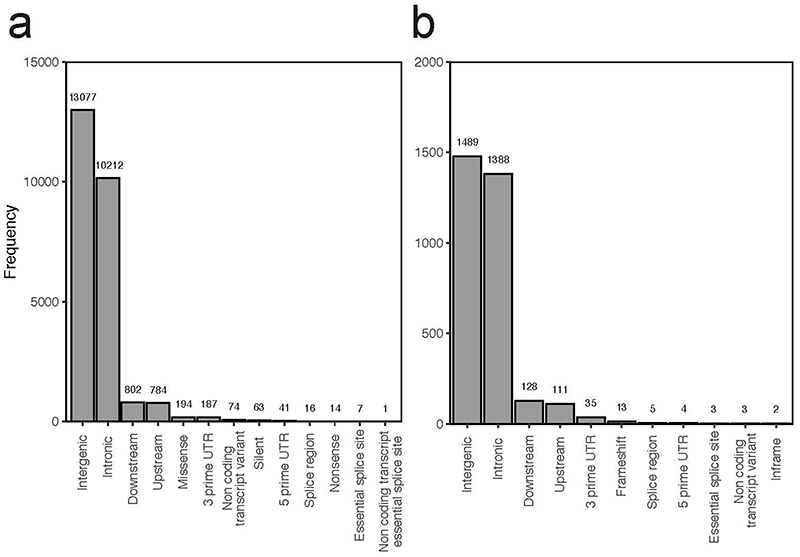
Most somatic variants found in bulk and microscopic trophoblast samples were unlikely to have any sequelae (Extended Data Fig. 8, Supplementary Table 4). The majority of copy number changes (42/80 unique variants) lay within fragile sites (Extended Data Table 4). Interestingly, 2/42 harboured copy number neutral loss of heterozygosity (i.e. paternal uniparental disomy) of chromosome 11p15 (Fig. 4d). Inactivation of this locus in fetal lineages underpins a cancer-predisposing overgrowth syndrome, Beckwith-Wiedemann19. It may also be associated with placental disease, as uniparental disomy of 11p15 has been implicated in driving gestational, trophoblast-derived choriocarcinoma20. However, in both cases here, the pregnancy, the placenta, and histology of the placental sample in question were unremarkable, making the functional significance of these alterations uncertain.
Discussion
Studying the somatic genomes of human placentas, we identified bottlenecks at different developmental stages that confined placental tissues genetically. Most prominently, every bulk placental sample that we examined represented an independent clonal trophoblast unit, suggesting that mosaicism represents the inherent clonal architecture of trophoblastic tissues in human placentas. In some cases, we may have identified the complete genetic separation of fetal and placental lineages, suggesting that placental lineages had passed through genetic bottlenecks preceding the spatial segregation of fetal and placental lineages4. Together these bottlenecks may represent developmental pathways through which cytogenetically abnormal cells phylogenetically and spatially separate, thereby rendering them detectable by genomic assays utilised in the clinical assessment of chorionic villi. Our findings thus provide a plausible mechanism for trophoblast CPM, the prevalence of which would seem to have been substantially underestimated. We expect that, as our understanding of the clonal dynamics of human embryonic lineages grows, we may find additional bottlenecks that account for placental mosaicism affecting mesenchymal lineages too.
The landscape of somatic mutations in placental tissue was an outlier compared to other normal human tissues studied to date. In colon12, endometrium13, esophagus21, liver14 and skin22, clonal fields either represent morphologically discrete, histological units, such as colonic crypts, or clonal expansions associated with oncogenic mutations. In contrast, clonal fields in placental samples were “driverless” developmentally acquired expansions that pervaded areas as large as macroscopic biopsies. As the placenta is a highly arborised structure, it is likely that our bulk samples consisted of villi mostly derived from a single parental branch18. Consequently, the clonality we observed represented single trophoblast clones, spatially fixed from early embryogenesis, expanding over multiple villi. Furthermore, placental tissues exhibited a comparatively high mutation rate, an unusual predominant mutational signature, and – uniquely for a normal human tissue – frequent copy number changes, reminiscent of some types of human tumours.
There may be several reasons for these distinct somatic features. Mutagenesis is likely to broadly differ, quantitatively and qualitatively, between fetal and adult life, as has been seen previously23, reflecting in utero growth demands. Villous trophoblast faces an approximate threefold rise in the local oxygen tension of blood surrounding the villi between eight and twelve week’s gestation24. Finally, it is conceivable that as a temporary, ultimately redundant organ, mechanisms protecting the genome elsewhere do not operate in placental trophoblasts.
Placental genomic alterations may contribute to the pathogenesis of placental dysfunction, which is a key determinant of the “Great Obstetrical Syndromes”, such as preeclampsia, fetal growth restriction and stillbirth2. Previous studies associating CPM with these syndromes have yielded conflicting results3,25–28. From our study and previous work, it is clear that genomic alterations are often not uniformly distributed within placentas, which may explain the variable pregnancy outcomes observed in the presence of mosaicism29–31. Larger scale systematic studies of the genomic architecture of the human placenta in health and disease might establish the role of placental genomic aberrations in driving the placenta-related complications of human pregnancy.
Methods
Ethics statement
All the samples were obtained from the Pregnancy Outcome Prediction (POP) study, a prospective cohort study of nulliparous women attending the Rosie Hospital, Cambridge (UK) for their dating ultrasound scan between January 14, 2008, and July 31, 2012. The study has been previously described in detail11,32. Ethical approval for this study was given by the Cambridgeshire 2 Research Ethics Committee (reference number 07/H0308/163) and all participants provided written informed consent.
Bulk DNA sequencing
DNA was extracted from maternal blood, umbilical cord, and fresh frozen placental biopsies. Short insert (500bp) genomic libraries were constructed, flow cells prepared and 150 base pair paired-end sequencing clusters generated on the Illumina HiSeq X or NovaSeq platform according to Illumina no-PCR library protocols. An overview of samples and sequencing variables, including the average sequence coverage, is shown in Supplementary Table 1.
Laser capture microdissection and low-input DNA sequencing
Tissues were prepared for microdissection and libraries were constructed as described previously12–14 and subsequently submitted for whole-genome sequencing on the Illumina HiSeq X or NovaSeq platform.
DNA sequence alignment
All DNA sequences were aligned to the GRCh37d5 reference genome by the Burrows-Wheeler algorithm (BWA-MEM)33.
Detection of somatic variants
We called all classes of somatic mutations: substitutions (CaVEMan algorithm34, see below), indels (Pindel algorithm35), copy number variation (ASCAT36 and Battenberg12,13 algorithms), and rearrangements (BRASS algorithm12,13). Besides ASCAT and Battenberg, sub-chromosomal copy number variants can also be detected via the breakpoints as predicted by BRASS, providing three independent methods to call copy number variants. The umbilical cord sample functioned as a matched normal sample in variant calling.
Rearrangements were validated by local assembly, as implemented in the BRASS algorithm. To generate a high confidence, final list of structural variants, only rearrangements whose breakpoints were greater than 1,000 base pairs apart, absent in the germline and associated with a copy number change were included in our analysis (see Supplementary Table 5). These copy number changes were validated by visual inspection in the genome browser Jbrowse37; demonstrating changes in sequencing depth and, where heterozygous SNPs were identified between the breakpoints, B allele frequency.
Unmatched substitution calling
Substitutions were called by applying the CaVEMan34 algorithm in an unmatched analysis of each sample against an in silico human reference genome, as applied previously8,38. Beyond the inbuilt post-processing filter of the algorithm, we removed variants affected mapping artefacts associated with BWA-MEM by setting the median alignment score of reads supporting a mutation as greater than or equal to 140 (ASMD>=140) and requiring that fewer than half of the reads were clipped (CLPM=0). We then recounted across samples belonging to the same patient the variant allele frequency of all substitutions with a cut-off for base quality (=25) and read mapping quality (=30). Variants were also filtered out if they were called in a region of consistently low or high depth across all samples from one patient.
To filter out germline variants, we fitted a binomial distribution to the combined read counts of all normal samples from one patient per SNV site, with the total depth as the number of trials, and the total number of reads supporting the variant as number of successes. Germline and somatic variants were differentiated based on a one-sided exact binomial test. For this test, the null hypothesis is that the number of reads supporting the variants across copy number normal samples is drawn from a binomial distribution with p=0.5 (p=0.95 for copy number equal to one), and the alternative hypothesis drawn from a distribution with p<0.5 (or p<0.95). Resulting p-values were corrected for multiple testing with the Benjamini-Hochberg method and a cut-off was set at q < 10-5 to minimize false positives as on average, roughly 40,000 variants were subjected to this statistical test. Variants for which the null hypothesis could be rejected were classified as somatic, otherwise as germline.
Further, remaining artefacts were filtered out by fitting a beta-binomial distribution to the variant counts and total depth for all variants across all samples from one patient. From this set of observations, we quantified the overdispersion parameter (rho). Any variant with an estimated rho smaller than 0.1 was filtered out, as used previously39,40.
Following visual inspection of a subset of these putative variants using Jbrowse37, a small number of substitutions called within the placental biopsies were found to falsely pass at sites of germline indels. To remedy this, substitutions called at the site of an indel were removed.
Phylogeny reconstruction
Phylogenies of microdissected trophoblast clusters were generated from the filtered substitutions using a maximum parsimony algorithm, MPBoot41. Substitutions were mapped onto tree branches using a maximum likelihood approach.
Unmatched indel calling
A similar approach was taken for indel filtering. Variants in each sample were called against the in silico human reference genome using Pindel35. Those that passed and possessed a minimum quality score threshold (>=300) were subject to the same genotyping and fitting of binomial and beta-binomial distributions described above and only variants supported by at least five mutant reads were retained.
Some samples with higher coverage (>50X) retained an inflated number of low VAF indel calls following this filtering approach. Further investigation revealed that most of these excess calls to occur at sites Pindel frequently rejects in other unrelated samples sequenced using the same sequencing platforms, suggesting that they were artefactual in nature. As these samples accounted for the majority of low VAF indels called in the biopsies, indels with a VAF <0.1 in these bulk samples were removed. Again, a subset of called indels were reviewed in Jbrowse37 to check the veracity of the pipeline detailed here.
Exclusion of maternal contamination
To exclude the possibility of any remaining maternal DNA in the placenta to skew results on mutation burden and clonality, we used maternal SNPs to quantify contamination. For each pregnancy, we randomly picked 5,000 rare germline variants (i.e. left in by the common SNP filter in CaVEMan) found in mother but not in umbilical cord. All these variants passed other CaVEMan flags, did not fall in regions of low depth (on average, below 35), and were present at a VAF greater than 0.35 in mother. Their VAFs in all individual placental samples, microdissections and biopsies, are displayed in the Supplementary Information. No sample had a level of support for maternal SNPs that exceeded the expectations for sequencing noise (0.1%), excluding maternal contamination as a plausible origin for any observations made here.
Sensitivity correction of mutation burden
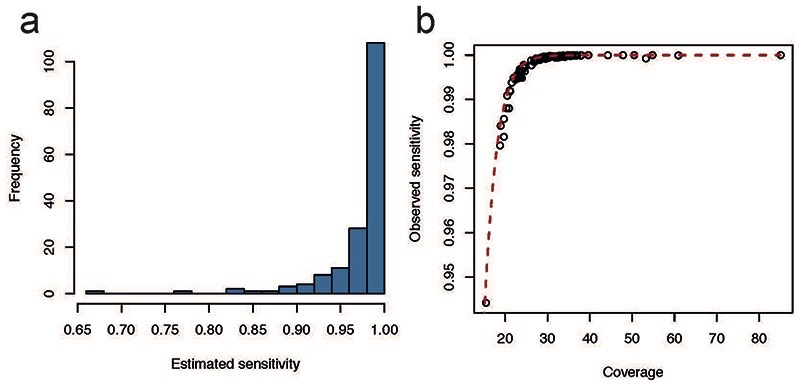
To compensate for the effects of sequencing coverage and low clonality on the final mutation burden per sample, we estimated the sensitivity of variant calling, as done previously39,40. For each sample, we generated an in silico coverage distribution by drawing 100,000 times from a Poisson distribution with the observed median coverage of the sample as its parameter. For each coverage simulation, we calculated the probability of observing at least four mutant reads for SNVs or five for indels (the minimum depth requirement for our CaVEMan and Pindel calls respectively) with the underlying binomial probability given by the observed median VAF of the sample. The average of all these probabilities then represents the sensitivity of variant calling (Extended Data Fig. 9). Final mutation burdens were then obtained by dividing the observed number of mutations by the estimated sensitivity.
To assess the performance of the predicted sensitivity correction, we compared this to the observed sensitivity of germline mutations in our sample set. These germline variants are called when combining all individual samples together, so a subset might not be called in individual placenta samples. Germline variants behave exactly the same way as somatic variants with a true VAF of 0.5. Comparing the fraction of germline variants called in individual samples to the predicted sensitivity based on VAF (0.5) and coverage revealed that our sensitivity correction traces the observed sensitivity curve (Extended Data Fig. 10).
Mutational signature extraction and fitting
To identify possibly undiscovered mutational signatures in human placenta, we ran the hierarchical Dirichlet process (HDP) (https://github.com/nicolaroberts/hdp) on the 96 trinucleotide counts of all microdissected samples, divided into individual branches. To avoid overfitting, branches with fewer than 50 mutations were not included in the signature extraction. HDP was run with individual patients as the hierarchy, in twenty independent chains, for 40,000 iterations, with a burn-in of 20,000.
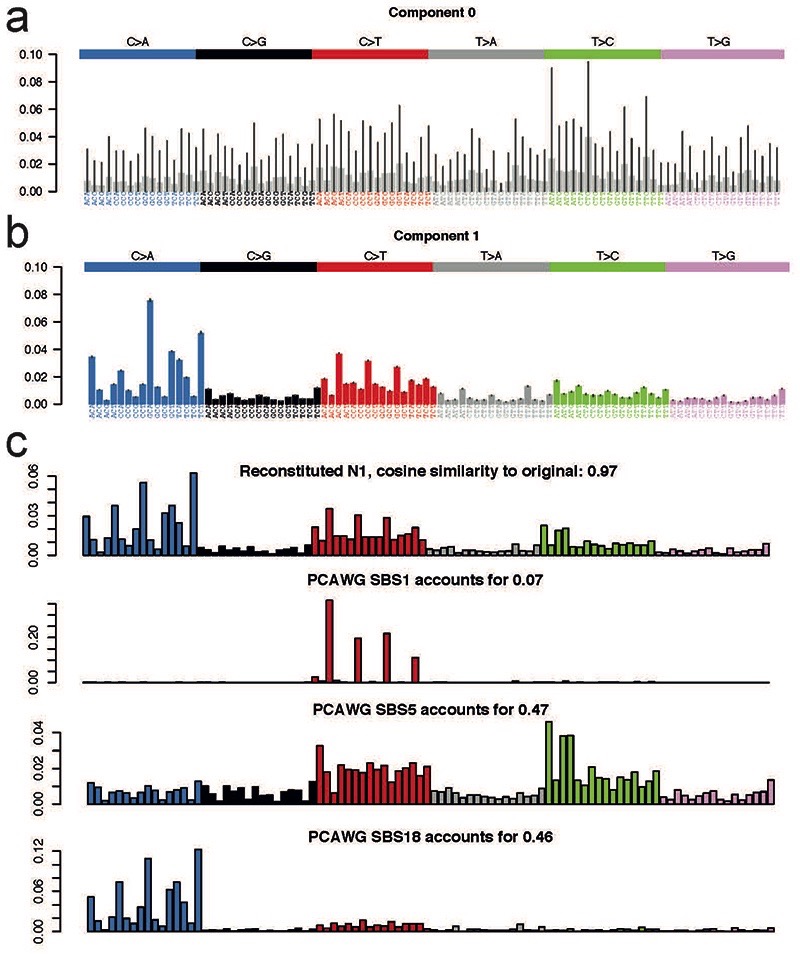
Besides the usual flat noise signature (Component 0) that is usually extracted, only one other signature emerged (Component 1) from the signature extraction. Deconvolution of that signature revealed it could be fully explained by a combination of reference single base substitution (SBS) signatures SBS1, SBS5, and SBS18 (Extended Data Fig. 10), all of which have been previously reported in normal tissues.
Because of the lack of novel signatures in this data set, the remainder of mutational signature analysis was performed by fitting this set of three signatures to trinucleotide counts using the R package deconstructSigs (v1.8.0)42.
Genetic proximity scores
To measure the genetic proximity between any two trophoblast clusters from the same bulk sample, we used the following equation:
Or simply, the fraction of shared mutations between samples i and j divided by their average total mutation burden. The resulting number reflects how much of in utero development was shared between these samples.
However, control data of normal human colon12 and endometrium13 were obtained from adults and their phylogenetic histories will reflect postnatal tissue dynamics as well. To obtain a proxy for the sharedness due to development in utero, we only considered a pair of samples i and j, if they did not split at a mutational time inconsistent with early development. We set this threshold for both colon and endometrium at 100 mutations, a very rough estimate of the maximum burden at birth in these tissues given preliminary studies. Consequently, instead of dividing the number of early shared mutations by the average burden, for adult tissues, these were divided by 100. All samples were subjected to the same variant calling pipeline.
Embryonic mutations
To discover early mutations in the umbilical cord samples, we included these in the unmatched variant calling as described above, either with all bulk placenta samples or microdissections. In the case of the latter, the umbilical cord samples were not included in phylogeny reconstruction due to their polyclonality, but aggregating it with microscopic sampling data allows for effective removal of germline variants due to the high cumulative depth of coverage.
All embryonic variants were visually inspected in Jbrowse37 to exclude any possible remaining sequencing or mapping artefacts.
For the five phylogenies of trophoblast clusters, the contribution of branches to the umbilical cord was measured by the VAF of mutations on these branches. In PD42138 and PD42142, where no variants were shared between the trophoblast phylogeny and the umbilical cord, the earliest mutations were found exclusively in the umbilical cord sample and the mutations with the highest VAF were taken to delineate the major clone, as done for sets of bulk biopsies. In both cases, the VAFs of the earliest mutations reflected a clonal origin for umbilical cord.
For bulk placenta samples and umbilical cord, the asymmetric contribution of the zygote was calculated by converting the highest VAF found in umbilical cord to a contribution (effectively multiplying by two). The alternative lineage was identified using the pigeonhole principle13, i.e. when clustering of the VAFs across placenta and umbilical cord prohibited this lineage from being a sub-clone of the previously identified major clone. In about half of cases (17/37), this yielded an asymmetry in umbilical cord with major and minor lineage also fully accounting for the bulk placental samples (see Extended Data Fig.4). For one case (PD45595), we could not identify any non-artefactual early embryonic mutations in the umbilical cord. This patient is hence omitted from the subsequent analysis concerning the early asymmetries.
In 11 out of 37 cases (Extended Data Fig. 5), one or more of the placental lineages could not be fully explained by the umbilical cord lineages, although the latter exhibited the expected asymmetry. This was established by calculating the 95% confidence intervals around the expected binomial probabilities of both major and minor lineages. If the sum of the higher extremes was less than 0.5 (the expected value to fully account for this lineage), the bulk placental sample was not fully explainable by the umbilical cord lineages.
In the remaining 9 out of 37 cases (Extended Data Fig.6), the umbilical cord showed clonal origins (a major lineage with a VAF around 0.5), which we found to be paired with segregated placental lineages in all cases.
Genotyping germline SNPs on chromosome 10
PD45581c, a bulk placenta bulk sample, exhibited trisomy of chromosome 10, which was absent from PD45581e (placenta) and PD45581f (umbilical cord). This could be the result of either a somatic duplication of chromosome 10 or a trisomy present in the fertilised egg that was post-zygotically reverted to a disomy. These two scenarios can be distinguished from one another by the number of distinct chromosomal alleles: three different chromosomes for a trisomic rescue, two for a somatic duplication. To test this, all SNPs on chromosome 10 reported by the 1000 Genomes project were genotyped across the three samples from the pregnancy, as well as the mother.
Coding substitution rate of trophoblast clusters against paediatric cancers
A recent, large scale, pan-paediatric cancer project provided the data necessary to contrast against the high mutation rate we observe in the trophoblast18. Here, the burden analysis focused on ‘coding mutations’, taken to mean all SNVs and indels that lie within exonic regions. This was adjusted for the callability and expressed per megabase.
To generate comparable results from our data, we used mosdepth (https://github.com/brentp/mosdepth) to estimate the callable length of the autosomal exonic regions. This meant excluding all regions blacklisted during variant calling, such as those with low mappability, and those with insufficient sequencing depth to call substitutions (<4X). Our substitution burden estimates were then adjusted according to what percentage of the total autosomal exonic regions this represented and converted it to a “per megabase” value. To account for the potentially years of additional time the malignant precursor has had to acquire mutations in comparison to the placenta, we divided the coding substitutions per Mb figure by the postpartum age provided (in years) plus 0.75. This would adjust for gestation and so any substitutions gained in the tumour precursor whilst still in utero.
Calculating the burden of SBS18 compared to paediatric malignancies
Using only the tumours that had undergone whole-genome sequencing and SBS signature extraction in the paper listed above18, we simply expressed the SBS18 mutations as a proportion of all SNVs and ranked the median value returned per tumour against the trophoblast clusters.
Chromosome 11p phased B-allele frequency plotting
ASCAT and Battenberg identified two samples, PD45557e_lo0003 and PD42154b3, as having uniparental disomy of part of chromosome 11p. To phase this to a given parent, all SNPs identified by the 1000 Genomes project on chromosome 11p were genotyped for the affected sample, the matched umbilical cord and the maternal blood sample. The SNPs that were homozygous in the mother but heterozygous in the umbilical cord could then be used to phase the loss of heterozygosity in the placental sample as the remaining allele must belong to the father.
Extended Data
Extended Data Figure 1. Differences in substitutions between clinical groups.
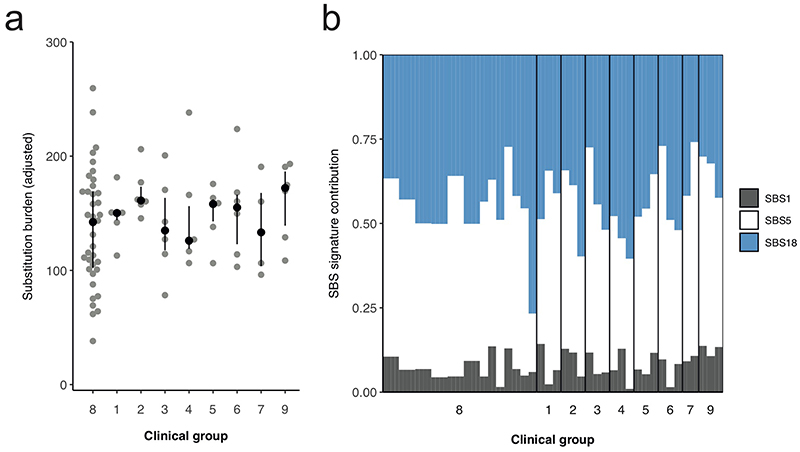
Analysis per clinical group of the absolute substitution burden of each bulk placental sample (a) and their associated mutational signatures (b). The difference in substitution burden between the clinical groups is not significant (Kruskal-Wallis rank sum test, p=0.7438). Each point and bar represent a single bulk placental sample. Clinical groups are defined in Extended Data Tables 2-3.
Extended Data Figure 2. Unique variants in placental biopsies.
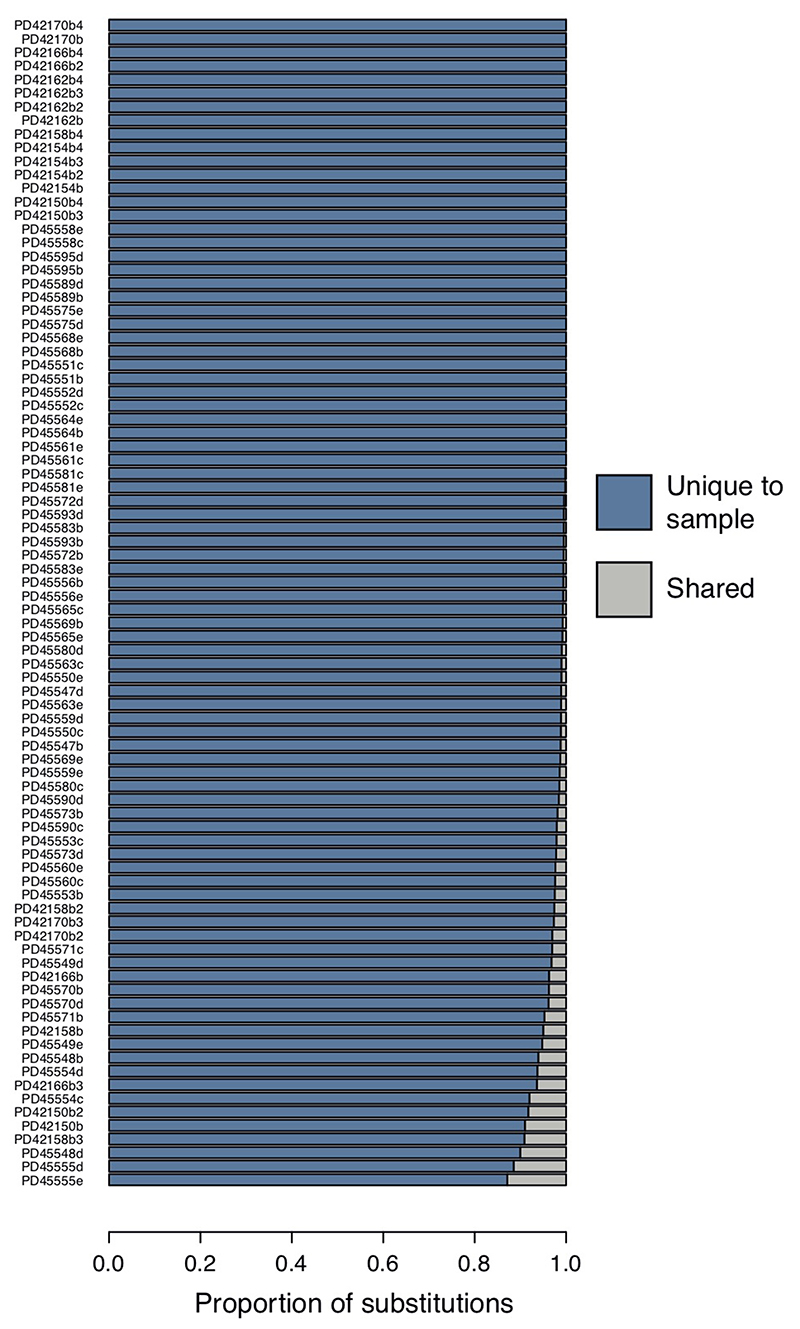
Proportion of variants that are unique to each bulk placental sample (blue), so absent from matched umbilical cord as well as any other bulk placental sample taken from the same case.
Extended Data Figure 3. Asymmetry across trophectoderm and umbilical cord.
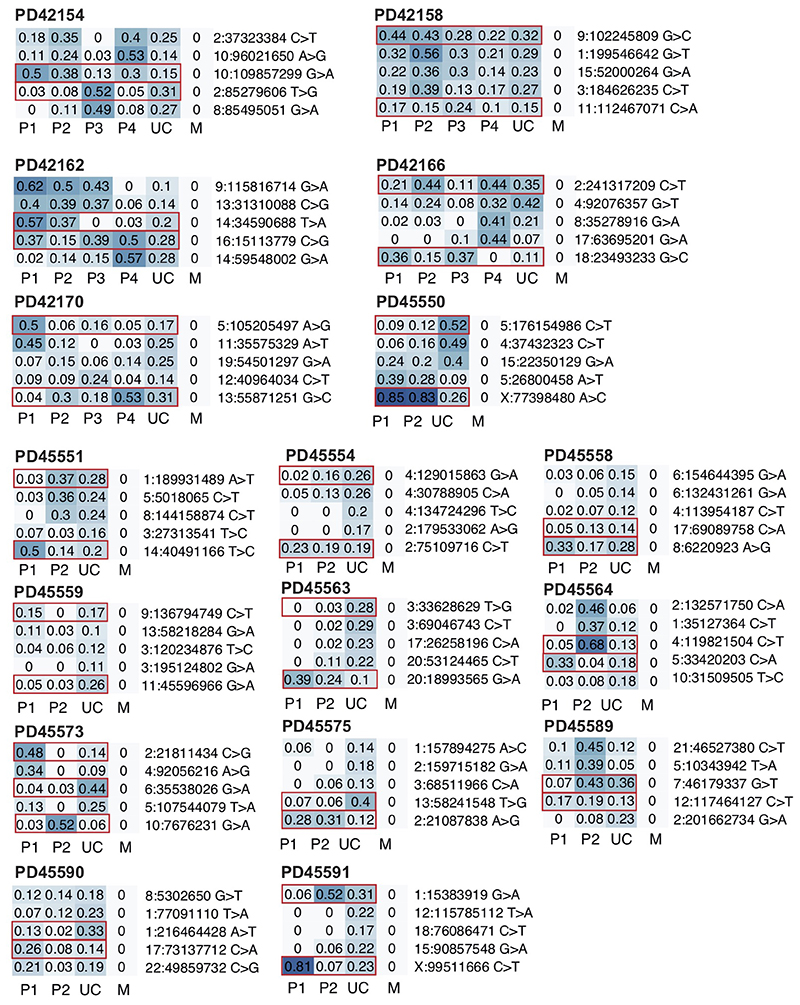
Heatmaps of VAFs of early embryonic mutations with the two earliest lineages contributing both to placenta and umbilical cord. Putative earliest mutations highlighted in red. P, placenta; UC, umbilical cord; M, maternal.
Extended Data Figure 4. Unexplained placental lineages.
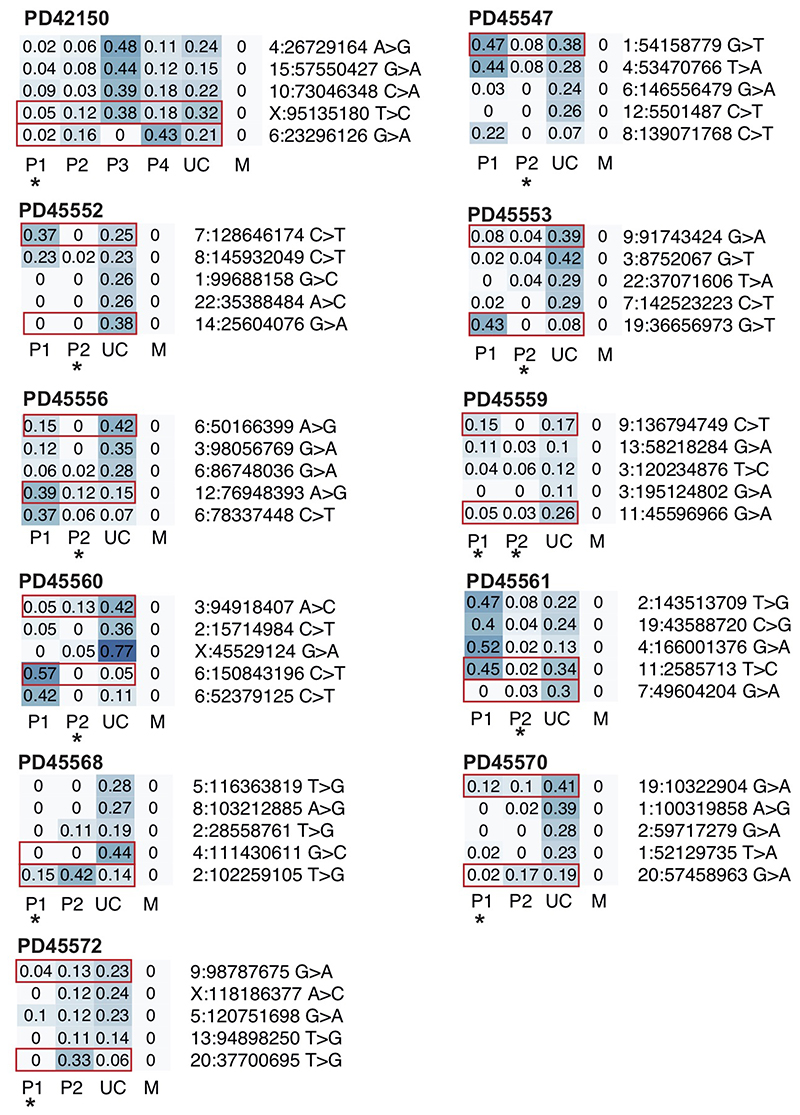
Heatmaps of VAFs of early embryonic mutations with the two earliest lineages contributing umbilical cord. Putative earliest mutations highlighted in red. Asterisk indicates placental lineage is not fully explained by umbilical cord (see Methods). P, placenta; UC, umbilical cord; M, maternal.
Extended Data Figure 5. Full segregation of placental and umbilical cord lineages.
Heatmaps of VAFs of early embryonic mutations with the umbilical cord being derived from one clonal lineage. In all cases, one or more placental lineages do not share any genetic ancestry with umbilical cord and are largely unexplained, as indicated by an asterisk (see Methods). P, placenta; UC, umbilical cord; M, maternal.
Extended Data Figure 6. Substitution burden per individual trophoblast cluster.
Adjusted for coverage and median variant allele frequency.
Extended Data Figure 7. Indels versus substitutions.
Indel burden versus substitution burden per trophoblast cluster. Both are corrected for median VAF and coverage.
Extended Data Figure 8. Impacts of mutations.
Overview of functional consequences of unique SNVs (a) and indels (b) seen in the placental biopsies and trophoblast clusters.
Extended Data Figure 9. Sensitivity of variant calling.
(a) Histogram of the estimated sensitivity for SNV calling in microdissected trophoblast clusters and bulk placenta samples. (b) The observed sensitivity of germline variants across placental samples plotted against the coverage of the sample. The red dashed line indicates our predicted sensitivity, which we would have used as sensitivity correction.
Extended Data Figure 10. Signatures extraction and deconvolution.
Signature extraction by HDP yielded a noise component (a) and one genuine mutational signature (b), which could be convoluted and reconstructed using three reference mutational signatures: SBS1, SBS5 and SBS18 (c).
Supplementary Material
Acknowledgements
We thank Professor Magdalena Zernicka-Goetz, Professor Sir Michael Stratton and Dr Iñigo Martincorena for insightful discussions.
Funding
This experiment was primarily funded by Wellcome (core funding to Wellcome Sanger Institute; personal fellowships to T.H.H.C, T.R.W.O., S.B.). All research at Great Ormond Street Hospital NHS Foundation Trust and UCL Great Ormond Street Institute of Child Health is made possible by the NIHR Great Ormond Street Hospital Biomedical Research Centre. The POP study was supported by the National Institute for Health Research (NIHR) Cambridge Biomedical Research Centre (Women’s Health theme). The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health and Social Care.
Footnotes
Contributions
S.B. designed the experiment. T.H.H.C. performed phylogenetic analyses. T.H.H.C. and T.R.W.O. analysed somatic mutations. T.R.W.O. performed microdissections. R.S., U.S., E.C., R.V.-T., M.H., M.D.Y., and R.R. contributed to experiments or analyses. N.S. provided pathological expertise. P.J.C. contributed to discussions. S.B., T.H.H.C. and T.R.W.O. wrote the manuscript, aided by D.S.C.J. and G.S. D.S.C.J., G.S., and S.B. co-directed this study.
Competing Interests
No competing interests are declared by the authors of this study.
Data Availability
DNA sequencing data are deposited in the European Genome-Phenome Archive (EGA) with accession code EGAD00001006337 (https://ega-archive.org/datasets/EGAD00001006337). Sample information and data on mutation burdens and signatures can be found in Supplementary Table 1. Further clinical information can be found in Supplementary Tables 2 and 3. Somatic mutations and embryonic mutations (bulk samples) can be found in Supplementary Tables 4 and 6, respectively. Calls of structural variants with associated copy number changes can be found in Supplementary Table 5.
Code Availability
Bespoke R scripts used for analysis and visualisation in this study are available online from GitHub (https://github.com/TimCoorens/Placenta).
References
- 1.Kalousek DK, Dill FJ. Chromosomal mosaicism confined to the placenta in human conceptions. Science. 1983;221:665–667. doi: 10.1126/science.6867735. [DOI] [PubMed] [Google Scholar]
- 2.Brosens I, Pijnenborg R, Vercruysse L, Romero R. The “Great Obstetrical Syndromes” are associated with disorders of deep placentation. Am J Obstet Gynecol. 2011;204:193–201. doi: 10.1016/j.ajog.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalousek DK, et al. Confirmation of CVS mosaicism in term placentae and high frequency of intrauterine growth retardation association with confined placental mosaicism. Prenat Diagn. 1991;11:743–750. doi: 10.1002/pd.1970111002. [DOI] [PubMed] [Google Scholar]
- 4.Xenopoulos P, Kang M, Hadjantonakis AK. In: Mouse Development Results and Problems in Cell Differentiation. Kubiak J, editor. Springer; 2012. Cell Lineage Allocation Within the Inner Cell Mass of the Mouse Blastocyst. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolton H, et al. Mouse model of chromosome mosaicism reveals lineage-specific depletion of aneuploid cells and normal developmental potential. Nat Commun. 2016;7:11165. doi: 10.1038/ncomms11165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behjati S, et al. Genome sequencing of normal cells reveals developmental lineages and mutational processes. Nature. 2014;513:422–425. doi: 10.1038/nature13448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ju YS, et al. Somatic mutations reveal asymmetric cellular dynamics in the early human embryo. Nature. 2017;543:714–718. doi: 10.1038/nature21703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coorens THH, et al. Embryonal precursors of Wilms tumor. Science. 2019;366:1247–1251. doi: 10.1126/science.aax1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alexandrov LB, et al. The repertoire of mutational signatures in human cancer. Nature. 2020;578:94–101. doi: 10.1038/s41586-020-1943-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaccioli F, Lager S, Sovio U, Charnock-Jones DS, Smith GCS. The pregnancy outcome prediction (POP) study: Investigating the relationship between serial prenatal ultrasonography, biomarkers, placental phenotype and adverse pregnancy outcomes. Placenta. 2017;59:S17–S25. doi: 10.1016/j.placenta.2016.10.011. [DOI] [Google Scholar]
- 11.The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Campbell PJ, Getz G, et al. Pan-cancer analysis of whole genomes. Nature. 2020;578:82–93. doi: 10.1038/s41586-020-1969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee-Six H, et al. The landscape of somatic mutation in normal colorectal epithelial cells. Nature. 2019;574:532–537. doi: 10.1038/s41586-019-1672-7. [DOI] [PubMed] [Google Scholar]
- 13.Moore L, et al. The mutational landscape of normal human endometrial epithelium. Nature. 2020;580:640–646. doi: 10.1038/s41586-020-2214-z. [DOI] [PubMed] [Google Scholar]
- 14.Brunner SF, et al. Somatic mutations and clonal dynamics in healthy and cirrhotic human liver. Nature. 2019;574:538–542. doi: 10.1038/s41586-019-1670-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poetsch AR. The genomics of oxidative DNA damage, repair, and resulting mutagenesis. Comput Struct Biotechnol J. 2020;18:207–219. doi: 10.1016/j.csbj.2019.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castellucci M, Schepe M, Scheffen I, Celona A, Kaufmann P. The development of the human placental villous tree. Anat Embryol. 1990;181:117–128. doi: 10.1007/BF00198951. [DOI] [PubMed] [Google Scholar]
- 17.Knöfler M, et al. Human placenta and trophoblast development: key molecular mechanisms and model systems. Cell Mol Life Sci. 2019;76:3479–3496. doi: 10.1007/s00018-019-03104-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gröbner SN, et al. The landscape of genomic alterations across childhood cancers. Nature. 2018;555:321–327. doi: 10.1038/nature25480. [DOI] [PubMed] [Google Scholar]
- 19.Choufani S, Shuman C, Weksberg R. Molecular findings in Beckwith-Wiedemann syndrome. Am J Med Genet C Semin Med Genet. 2013;163C:131–140. doi: 10.1002/ajmg.c.31363. [DOI] [PubMed] [Google Scholar]
- 20.Poaty H, et al. Genome-wide high-resolution aCGH analysis of gestational choriocarcinomas. PLoS One. 2012;7:e29426. doi: 10.1371/journal.pone.0029426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martincorena I, et al. Somatic mutant clones colonize the human esophagus with age. Science. 2018;362:911–917. doi: 10.1126/science.aau3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martincorena I, et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015;348:880–886. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuijk E, et al. Early divergence of mutational processes in human fetal tissues. Sci Adv. 2019;5:eaaw1271. doi: 10.1126/sciadv.aaw1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jauniaux E, et al. Onset of maternal arterial blood flow and placental oxidative stress. A possible factor in human early pregnancy failure. Am J Pathol. 2000;157:2111–2122. doi: 10.1016/S0002-9440(10)64849-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amor DJ, et al. Health and developmental outcome of children following prenatal diagnosis of confined placental mosaicism. Prenat Diagn. 2006;26:443–448. doi: 10.1002/pd.1433. [DOI] [PubMed] [Google Scholar]
- 26.Baffero GM, et al. Confined placental mosaicism at chorionic villous sampling: risk factors and pregnancy outcome. Prenat Diagn. 2012;32:1102–1108. doi: 10.1002/pd.3965. [DOI] [PubMed] [Google Scholar]
- 27.Toutain J, Goutte-Gattat D, Horovitz J, Saura R. Confined placental mosaicism revisited: Impact on pregnancy characteristics and outcome. PLoS One. 2018;13:e0195905. doi: 10.1371/journal.pone.0195905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grati FR, et al. Outcomes in pregnancies with a confined placental mosaicism and implications for prenatal screening using cell-free DNA. Genet Med. 2020;22:309–316. doi: 10.1038/s41436-019-0630-y. [DOI] [PubMed] [Google Scholar]
- 29.Henderson KG, et al. Distribution of mosaicism in human placentae. Human genetics. 1996;97:650–654. doi: 10.1007/BF02281877. [DOI] [PubMed] [Google Scholar]
- 30.Peñaherrera MS, et al. Patterns of placental development evaluated by X chromosome inactivation profiling provide a basis to evaluate the origin of epigenetic variation. Human reproduction. 2012;27:1745–1753. doi: 10.1093/humrep/des072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moreira de Mello JC, et al. Random X inactivation and extensive mosaicism in human placenta revealed by analysis of allele-specific gene expression along the X chromosome. PloS one. 2010;5:e10947. doi: 10.1371/journal.pone.0010947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sovio U, White IR, Dacey A, Pasupathy D, Smith GCS. Screening for fetal growth restriction with universal third trimester ultrasonography in nulliparous women in the Pregnancy Outcome Prediction (POP) study: a prospective cohort study. Lancet. 2015;386:2089–2097. doi: 10.1016/S0140-6736(15)00131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones D, et al. cgpCaVEManWrapper: simple execution of CaVEMan in order to detect somatic single nucleotide variants in NGS data. Current protocols in bioinformatics. 2016;56(1):15–10. doi: 10.1002/cpbi.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25(21):2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Loo P, et al. Allele-specific copy number analysis of tumors. Proceedings of the National Academy of Sciences. 2010;107(39):16910–16915. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buels R, et al. JBrowse: a dynamic web platform for genome visualization and analysis. Genome biology. 2016;17(1):66. doi: 10.1186/s13059-016-0924-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coorens THH, et al. Lineage-Independent Tumors in Bilateral Neuroblastoma. N Engl J Med. 2020;383:1860–1865. doi: 10.1056/NEJMoa2000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olafsson S, et al. Somatic evolution in non-neoplastic IBD-affected colon. Cell. 2020;182:672–684. doi: 10.1016/j.cell.2020.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson PS, et al. Elevated somatic mutation burdens in normal human cells due to defective DNA polymerases. bioRxiv. 2020 doi: 10.1038/s41588-021-00930-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoang HT, Vinh LS, Flouri T, Stamatakis A, Von Haeseler A, Minh BQ. MPBoot: fast phylogenetic maximum parsimony tree inference and bootstrap approximation. BMC evolutionary biology. 2018;18(1):1–11. doi: 10.1186/s12862-018-1131-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosenthal R, McGranahan N, Herrero J, Taylors BS, Swanton C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome biology. 2016;17(1):1–11. doi: 10.1186/s13059-016-0893-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequencing data are deposited in the European Genome-Phenome Archive (EGA) with accession code EGAD00001006337 (https://ega-archive.org/datasets/EGAD00001006337). Sample information and data on mutation burdens and signatures can be found in Supplementary Table 1. Further clinical information can be found in Supplementary Tables 2 and 3. Somatic mutations and embryonic mutations (bulk samples) can be found in Supplementary Tables 4 and 6, respectively. Calls of structural variants with associated copy number changes can be found in Supplementary Table 5.
Bespoke R scripts used for analysis and visualisation in this study are available online from GitHub (https://github.com/TimCoorens/Placenta).