Abstract
Despite their simple body plan, stony corals (order Scleractinia, phylum Cnidaria) can produce massive and complex exoskeletal structures in shallow, tropical and subtropical regions of Earth’s oceans. The species-specific macromorphologies of their aragonite skeletons suggest a highly coordinated biomineralization process that is rooted in their genomes, and which has persisted across major climatic shifts over the past 400 + million years. The mechanisms by which stony corals produce their skeletons has been the subject of interest for at least the last 160 years, and the pace of understanding the process has increased dramatically in the past decade since the sequencing of the first coral genome in 2011. In this review, we detail what is known to date about the genetic basis of the stony coral biomineralization process, with a focus on advances in the last several years as well as ways that physical and chemical tools can be combined with genetics, and then propose next steps forward for the coming decade.
Keywords: Skeletal organic matrix, Aragonite, Highly acidic proteins, Proteomic, Genomic
1. Introduction
Biomineralization is a complex process occurring across multiple highly divergent taxa, including some of the most basal metazoan phyla. Scleractinian, or stony, corals (phylum Cnidaria) are some of the earliest metazoans to possess an organized body structure and appear to have evolved the ability to build their skeletons independently of other calcifying cnidarian groups (McFadden, 2021). In colonial corals, radially symmetrical - and frequently genetically identical - polyps of a few mm to ~ 1 cm dwell in individual cup-shaped mineralized corallites and are interconnected by tissue for communication and resource sharing across the colony (Fig. 1, Veron, 2000). The layer of tissue overlying the aragonite exoskeleton of the corallites and coenosteum (skeleton beneath tissue between polyps) contains specialized cells, calicoblasts, of the aboral epithelium. These cells adhere the tissue to new substrate or to existing skeleton and secrete biomolecules, collectively termed the skeletal organic matrix (SOM), to a confined calcifying space where these biomolecules direct the formation of new mineral (review by Drake, 2020). However, as with other biomineralizing taxa, the exact mechanisms of the stony coral biomineralization process remain elusive despite decades of research. Here we argue that genetic tools improved in other systems over the past several years can be combined with physical and chemical analyses to make large strides forward in understanding stony coral biomineralization mechanisms.
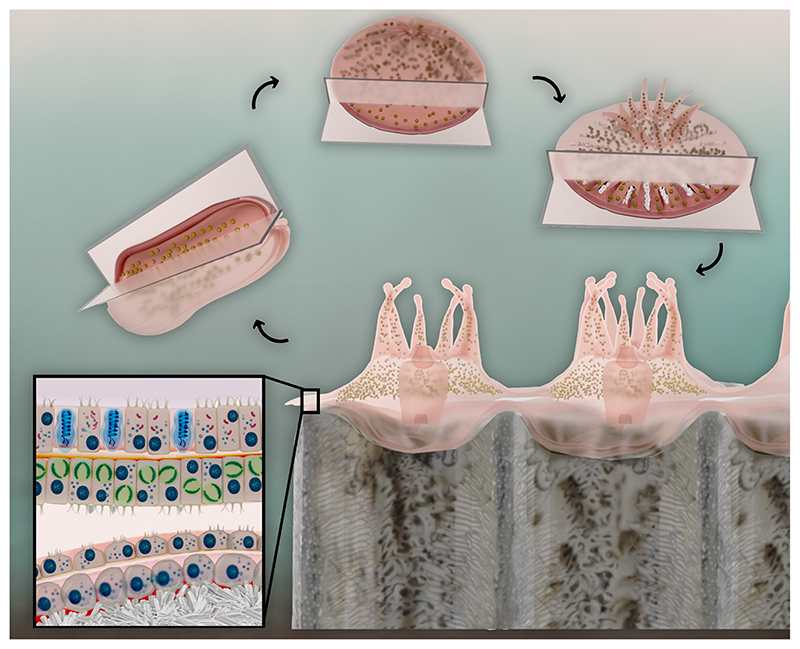
Fig 1.
Pre- (1) and post-metamorphosed (2) stony coral larvae, settled spat (3), and multi-polyp colony cross-section (4) showing the simple body plan and tissue structure containing stinging cells (blue) and endosymbiotic algae (green) in the oral cell layers, and the calcifying cells of the aboral epithelium adjacent to the newly formed skeleton.
Cnidarians have a biphasic life cycle. Egg and sperm packets or fertilized larvae are released from parent polyps to the water column. Larvae do not appear to produce the biomolecular machinery to calcify and lack calicoblasts prior to metamorphosis (Levy, et al., 2021). Mature metamorphosed larvae produce amorphous calcium carbonate and small aragonite crystals inside their bodies (Akiva, 2018) in preparation for rapid commencement of external aragonite calcification upon settling as spat (Fig. 1). Spat and adult polyps exhibit hexagonal symmetry to their tissues that is reflected in patterning of the mineral corallite (Veron, 2000). Since the first coral genome was sequenced 10 years ago (Shinzato, 2011), there have been increasing insights into the biomolecular mechanisms of coral calcification. Studies of the transcriptomic changes across the developmental stages has revealed that genes for known SOM proteins (SOMPs) are up-regulated in mature metamorphosed larvae, spat, and adults, and many are localized to the calicoblasts which are more abundant in spat than adults (Fig. 1, e.g., Levy, et al., 2021). However, these changes have not been associated with proteomic changes of the SOM to date.
Upon settlement, polyps produce new aragonite in the calcifying space between the aboral ectoderm and the substrate by filling the space with calcium, carbonate, and bicarbonate through a combination of transcellular (intracellular contents passed across cell membranes) and paracellular (between cells from the overlying seawater) transport processes (e.g., Venn et al., 2020). The exact contribution of each transport type is presently unknown. While the genetic basis of the paracellular pathway remains to be resolved, several genes have been implicated in the transcellular pathway including calcium transporters and bicarbonate pumps (review by Drake, 2020). Once the chemistry of the calcifying space is established (Sevilgen, 2019), SOM molecules then provide structure and points for mineral nucleation among other functions (review by Drake, 2020), although the visualization of this process through microscopy has not yet been well developed.
Properties of stony coral skeletons and tissue have been studied since at least the 1840s (review by Drake, 2020), revealing universal processes and structures across all stony corals as well as key differences between species. Some of the most outstanding differences across taxa involve the skeleton: overall colony structure, placement of corallites in relation to each other, and the species-specific macromorphology of corallites, all of which have historically been used for species identification (Veron, 2000). Further, efforts to sequence stony coral skeletal proteins have revealed many proteins that appear to be unique at both the family and species levels (Zaquin et al., 2021 and proteomic studies referenced therein). Combined, these findings suggest that there is a genetic basis for inter-specific differences in skeleton formation and understanding these differences may aid in conservation and restoration efforts as corals variably respond to anthropogenic climate change in the coming decades (IPCC, 2021).
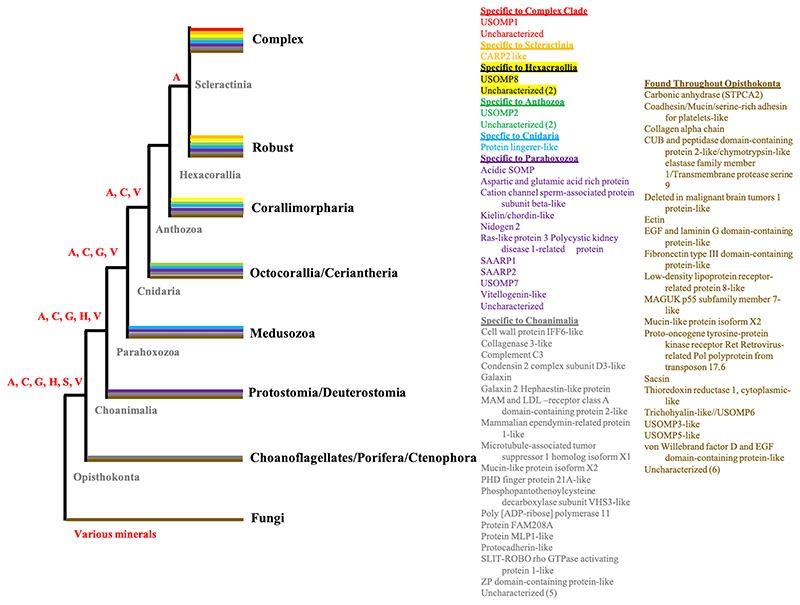
Conversely, there appears to be a conserved genetic basis for some of the shared microstructural aspects of coral skeleton formation. SOM extracted from decalcified coral skeletons of multiple species and incubated in an artificial supersaturated solution biases the mineral polymorph toward aragonite, even when the experimental solution should promote calcite formation (e.g., Sancho-Tomás, 2014). This has also been observed in vivo along with up-regulation of several known biomineralization-related genes (Yuyama and Higuchi, 2019). It is likely that the extracellular SOM biomolecules lead to the reticulate microstructure of the inorganic portion of the mineral (e.g., Mass, 2013). While some of these biomolecules differ between taxa, explorations of the evolutionary history of genes encoding coral SOMPs from divergent coral genera have found that several genes are conserved across Scleractinia and approximately half of SOM genes were co-opted from ancestors shared with other phyla, some of which contain no extant calcifying taxa (Zaquin et al., 2021) (Fig. 2). In addition, recently elucidated interactions between specific coral SOMPs support that they are not randomly arranged in the mineral as contaminants or general mucus proteins (Mummadisetti et al., 2021). The degree to which genetically conserved SOMPs are incorporated into the growing mineral in a variety of other taxa beyond the two coral genera queried at the protein level using species-specific genomes and transcriptomes remains to be shown.
Fig 2.
Conservation of known biomineralization genes from stony corals across the Metazoa with fungi as outgroups, based on Figure 1 and Supplementary Table 2 in (Zaquin et al., 2021). Color coding at branches represents presence of the comparably colored genes in taxa within that branch. Not all listed genes are in every branch of that color. Letters at nodes represent mineral types produced by some of the members in branches derived from those nodes as aragonite (A), calcite (C), vaterite (V), gypsum (G), hydroxyapatite (H), and silica (S). Genes found only in one of the coral taxa with a sequenced skeletal proteome (i.e., appear to be novel to that species) are not represented here.
While great advances have been made in the past fifteen years in our understanding of coral SOMPs, they are not the only biological component of the SOM. Several polysaccharides and lipids have also been detected in SOM (e.g., Takeuchi, 2018; Reggi, 2016). However, the comprehensive identification of many of these biomolecules is complicated as their characterization relies on matching their chromatography or spectroscopy peaks to those of synthetic standards (a relatively small number) and not to predicted sequences or structures based on a genetic database (10 s of thousands of potential matches), as in the case of proteins. Furthermore, they are not directly encoded in the genome and genetic inquiry therefore focuses on the enzymes that construct them. Additionally, modification of these biomolecules as well as proteins is not well-understood, although such modifications as glycosylation, phosphorylation, sulfation, and biomolecule trimming may dramatically alter their functions (review by Beltrao et al., 2013).
2. A path forward
The present state of genetic tools available to coral biologists is ripe for their application to outstanding questions about coral biominerlization mechanisms. One basic question only minimally answered over the past decade is ‘what do the SOMPs do?’. Precipitation experiments in which SOM biomolecule complexes are added to supersaturated solutions are useful in detailing that the SOM affects polymorph type (e.g., Sancho-Tomás, 2014), but the roles of individual proteins in this process are not revealed. Additionally, it is currently unknown how much the SOM extraction process affects their function. Use of recombinant proteins expressed in bacterial cultures has revealed the function of several highly acidic coral SOMPs in precipitating what is likely aragonite from unamended seawater (Mass, 2013) and causing the precipitation of the highly unstable polymorph vaterite in a calcitepromoting super-saturated solution (Gavriel, 2018; Laipnik, 2020). Next steps using recombinant proteins include (1) examining subregions of various highly acidic proteins to see which domains are controlling their calcium carbonate precipitation functions as has been initiated for one protein, CARP3, so far (Gavriel, 2018), (2) comparing precipitation rates between the same protein from different species, (3) and adding the use of tools such as NMR to study at the atomic level how the proteins and peptides interact with calcium (e.g., Gavriel, 2018). Additionally, recent analysis of the evolutionary history of the CARP4-5/SAARP1-2 gene family of highly acidic coral SOMPs has revealed that the ancestral gene was not acidic and evolved through multiple duplication events to have increasingly greater numbers of aspartic acid (Zaquin et al., 2021); therefore, selectively altering protein and peptide sequences, such as in the highly acidic proteins, to test their ancestral state function would be useful. Further, several genes for SOMPs conserved across distantly related stony coral taxa are annotated as ‘uncharacterized’ (Fig. 2); genetic tools will be central in resolving the function of these proteins that remain ambiguous based on BLASTing.
There is recent strong support that key SOMPs structure the calcifying space in a non-random manner (Mummadisetti et al., 2021). Deep learning modeling of the interactions between SOMPs is now a possibility (Sledzieski et al., 2021), and the increasing genetic information from various stony coral species allows inquiry into the differences in this potential structuring capability between species. An open question remains as to how the SOMPs create this structure (Tambutté et al., 2021). Further, the mechanisms by which these and other biomolecules are exported to the calcifying space remain to be resolved. These and other questions can be answered by a combined genetic-proteomic approach.
A very interesting tool that has recently been successfully attempted in corals is genetic modification using the CRISPR/Cas9 technique (Cleves et al., 2018). This gene editing is a further way to study not only the functions of individual SOMPs but also their gene regulatory networks. Obvious initial targets are the highly acidic SOMPs and a well-studied SOM carbonic anhydrase (review by Drake, 2020). Microscopy studies of the calcifying space could incorporate edits to genes for structural SOMPs. Finally, pluripotency (Reyes-Bermudez et al., 2021) and the search for stem cells in stony corals is a current hot topic and gene editing may allow resolution of this interesting area.
As noted above, most research on stony coral SOM has focused on proteins but those are not the only biomolecular component of the SOM (e.g., Takeuchi, 2018; Reggi, 2016). Genome-based inquiry of pathways that produce lipids and polysaccharides may help to overcome some of the technical issues associated with their characterization. Cell-specific transcriptomic studies of genes related to glycosylation, phosphorylation, and sulfation will also allow a more complete understanding of how these modifications affect the biomineralization process. In addition to these downstream post-translational modifications, epi-genetic modifications of DNA upstream of transcription are also being revealed as important to various biological processes in corals (review by Torda, 2017). For instance, DNA methylation of known biomineralization genes was lower than expected for one species tested under pCO2 conditions projected for the end of the present century (Liew, 2018); further study of this as well as other types of epigenetic modifications will be helpful in predicting how stony corals’ biomineralization mechanism will fare in the face of ocean acidification.
3. conclusions
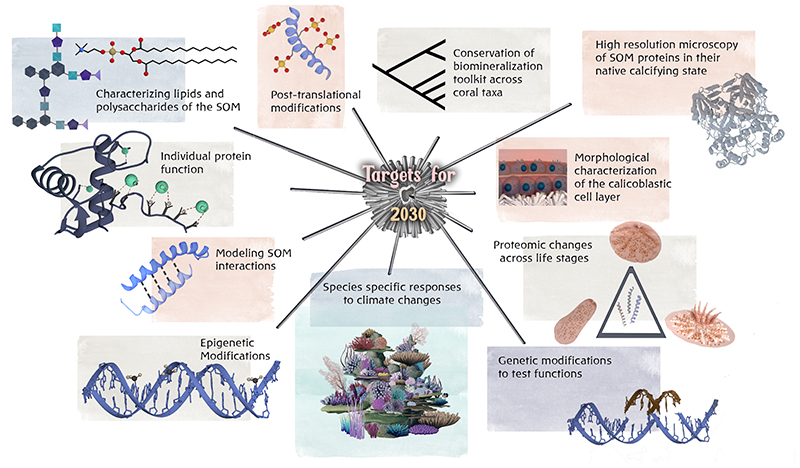
In summary, there have, for the past decade, been many ‘low hanging fruits’ with regards to a better understanding of the genetic basis, and downstream biochemical and mineralogical processes, of the stony coral calcification mechanism. Multiple imaging, physical, chemical, and genomics/transcriptomic tools are primed to help answer the outstanding questions related to the molecular mechanisms of biomineralization in stony corals, some detailed here (Fig. 3), in the next decade. These tools include advanced high resolution microscopy, improved mass spectrometry detection for proteomics, high resolution spectroscopy, various omics strategies, and in-silico analysis. We anticipate that they will soon be combined with genetic tools such as reverse genetics to address the biochemical mechanisms of stony coral biomineralization. While some of these tools are currently in an optimized state that many outstanding questions can be applied to these non-model organisms in the next year or so, others still require some refining for use on these complex organisms.
Fig 3. Proposed targets for the next decade to better understand stony coral biomineralization mechanisms.
Genetic and other tools presently exist to address each of these targets. The length of the line leading to each target is reflective of the ease of answering the major outstanding questions for that target with shorter lines being more easily/quickly achieved.
Acknowledgments
JLD was supported by the Zuckerman STEM Leadership Program. This work has received funding to TM from the European Research Council under the European Union’s Horizon 2020 research and innovation programme (grant agreement number 755876). 3D illustrations were undertaken using the educational license for Autodesk Maya 2020. DNA illustration is by Dietmar Zenker licensed under Creative Commons attribution. We thank an anonymous reviewer for their constructive and thoughtful suggestions to improve the manuscript.
Footnotes
CRediT authorship contribution statement
Jeana L. Drake: Conceptualization, Visualization, Writing - original draft, Writing - review & editing. Neta Varsano: Visualization, Writing - original draft, Writing - review & editing. Tali Mass: Conceptualization, Visualization, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Akiva A, et al. Minerals in the pre-settled coral Stylophora pistillata crystallize via protein and ion changes. Nat Commun. 2018;9:1880. doi: 10.1038/s41467-018-04285-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrao P, Bork P, Krogan NJ, van Noort V. Evolution and functional crosstalk of protein post-translational modifications. Mol Syst Biol. 2013;9:714. doi: 10.1002/msb.201304521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleves PA, Strader ME, Bay LK, Pringle JR, Matz MV. CRISPR/Cas9-mediated genome editing in a reef-building coral. Proc Natl Acad Sci. 2018;115:5235–5240. doi: 10.1073/pnas.1722151115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J, et al. How corals made rocks through the ages. Global Change Biol. 2020;26:31. doi: 10.1111/gcb.14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavriel R, et al. The coral protein CARP3 acts from a disordered mineral surface film to divert aragonite crystallization in favor of Mg-calcite. Adv Funct Mater. 2018;28:1707321 [Google Scholar]
- IPCCMasson-Delmotte V, Zhai P, Pirani A, Connors SL, Péan C, Berger S, Caud N, Chen Y, Goldfarb L, Gomis MI, Huang M, et al., editors. Climate Change 2021: The Physical Science Basis Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge University Press; 2021. In Press. [Google Scholar]
- Laipnik Ra, et al. Coral acid rich protein selects vaterite polymorph in vitro. J Struct Biol. 2020;209:107431. doi: 10.1016/j.jsb.2019.107431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S, et al. A stony coral cell atlas illuminates the molecular and cellular basis of coral symbiosis, calcification, and immunity. Cell. 2021;184:2973–2987.:e2918. doi: 10.1016/j.cell.2021.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew YJ, et al. Epigenome-associated phenotypic acclimatization to ocean acidification in a reef-building coral. Sci Adv. 2018;4:eaar8028. doi: 10.1126/sciadv.aar8028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mass T, et al. Cloning and characterization of four novel coral acid-rich proteins that precipitate carbonates in vitro. Curr Biol. 2013;23:1126–1131. doi: 10.1016/Jcub.2013.05.007. [DOI] [PubMed] [Google Scholar]
- McFadden CS, et al. Phylogenomics, origin, and diversification of anthozoans (phylum Cnidaria) Syst Biol. 2021 doi: 10.1093/sysbio/syaa103. [DOI] [PubMed] [Google Scholar]
- Mummadisetti MP, Drake JL, Falkowski PG. The spatial network of skeletal proteins in a stony coral. J Royal Soc Interface. 2021;18:20200859. doi: 10.1098/rsif.2020.0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggi M, et al. Influence of intra-skeletal coral lipids on calcium carbonate precipitation. CrystEngComm. 2016;18:8829–8833. [Google Scholar]
- Reyes-Bermudez A, Hidaka M, Mikheyev A. Transcription profiling of cultured Acropora digitifera adult cells reveals the existence of ancestral genome regulatory modules underlying pluripotency and cell differentiation in cnidaria. Genome Biol E. 2021;13:evab008. doi: 10.1093/gbe/evab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho-Tomás M, et al. Exploring coral biomineralization in gelling environments by means of a counter diffusion system. CrystEngComm. 2014;16:1257–1267. [Google Scholar]
- Sevilgen DS, et al. Full in vivo characterization of carbonate chemistry at the site of calcification in corals. Sci Adv. 2019;5:eaau7447. doi: 10.1126/sciadv.aau7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzato C, et al. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature. 2011;476:320–323. doi: 10.1038/nature10249. [DOI] [PubMed] [Google Scholar]
- Sledzieski S, Singh R, Cowen L, Berger B. Sequence-based prediction of protein-protein interactions: a structure-aware interpretable deep learning model. bioRxiv. 2021 doi: 10.1016/j.cels.2021.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T, et al. Biochemical characterization of the skeletal matrix of the massive coral, Porites australiensis-The saccharide moieties and their localization. J Struct Biol. 2018;203:219–229. doi: 10.1016/j.jsb.2018.05.011. [DOI] [PubMed] [Google Scholar]
- Tambutté E, Ganot P, Venn AA, Tambutté S. A role for primary cilia in coral calcification? Cell Tissue Res. 2021;383:1093–1102. doi: 10.1007/s00441-020-03343-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torda G, et al. Rapid adaptive responses to climate change in corals. Nature Climate Change. 2017;7:627. [Google Scholar]
- Venn AA, Bernardet C, Chabenat A, Tambutté E, Tambutté S. Paracellular transport to the coral calcifying medium: effects of environmental parameters. J Exp Biol. 2020;223 doi: 10.1242/jeb.227074. [DOI] [PubMed] [Google Scholar]
- Veron JEN. Corals of the World. Sea Challengers. 2000 [Google Scholar]
- Yuyama I, Higuchi T. Differential gene expression in skeletal organic matrix proteins of scleractinian corals associated with mixed aragonite/calcite skeletons under low mMg/Ca conditions. PeerJ. 2019 doi: 10.7717/peerj.7241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaquin T, Malik A, Drake JL, Putnam HM, Mass T. Evolution of protein-mediated biomineralization in scleractinian corals. Front Genet. 2021;12:52. doi: 10.3389/fgene.2021.618517. [DOI] [PMC free article] [PubMed] [Google Scholar]