Abstract
Current knowledge on environmental distribution and taxon richness of free-living bacteria is mainly based on cultivation-independent investigations employing 16S rRNA gene sequencing methods. Yet, 16S rRNA genes are evolutionarily rather conserved, resulting in limited taxonomic and ecological resolutions provided by this marker. The faster evolving protein-encoding gene priB was used to reveal ecological patterns hidden within a single OTU defined by >99% 16S rRNA sequence similarity. The studied subcluster PnecC of the genus Polynucleobacter represents a ubiquitous group of abundant freshwater bacteria with cosmopolitan distribution, which is very frequently detected by diversity surveys of freshwater systems. Based on genome taxonomy and a large set of genome sequences, a sequence similarity threshold for delineation of species-like taxa could be established. In total, 600 species-like taxa were detected in 99 freshwater habitats scattered across three regions representing a latitudinal range of 3400 km (42°N to 71°N) and a pH gradient of 4.2 to 8.6. Besides the unexpectedly high richness, the increased taxonomic resolution revealed structuring of Polynucleobacter communities by a couple of macroecological trends, which was previously only demonstrated for phylogenetically much broader groups of bacteria. An unexpected pattern was the almost complete compositional separation of Polynucleobacter communities of Ca2+-rich and Ca2+-poor habitats. This compositional pattern strongly resembled the vicariance of plant species on silicate and limestone soils. The presented new cultivation-independent approach opened a window to an incredible, previously unseen diversity, and enables investigations aiming on deeper understanding of how environmental conditions shape bacterial communities and drive evolution of free-living bacteria.
Introduction
Prokaryotes are the most numerous organisms on earth. Our current knowledge on diversity of prokaryotes and composition of prokaryotic communities is mainly based on cultivation-independent investigations employing 16S rRNA gene sequencing methods. The insights obtained by such investigations are, however, limited by the taxonomic resolution of the 16S rRNA gene sequences (Stackebrandt & Ebers, 2006). It was repeatedly shown that prokaryotes affiliated with distinct but related species cannot by discriminated by using their full-length 16S rRNA gene sequences (Jaspers & Overmann, 2004; Stackebrandt & Ebers, 2006; Hahn et al., 2016). In addition to the limited taxonomic resolution of the gene, the majority of the current studies using high-throughput amplicon sequencing of 16S rRNA genes, does not use the entire gene length and thus not the full information content of 16S rRNA gene sequences. Some studies were even based on < 150 bp sequence fragments (García-García et al., 2019) equaling to only about 10% of the entire gene. Furthermore, the taxonomic ranks of the operational taxonomic units (OTUs) typically established in 16S rRNA-sequence-based investigations are unknown. Despite frequently stated in publications and independent of the applied sequence similarity threshold used for OTU demarcation, the established OTUs do not represent species-like taxa. Due to all these limitations, prokaryotic communities remained at the species level black boxes. A few alternative markers and primers were developed for profiling the composition and structure of bacterial communities with higher resolution than that provided by the 16S rRNA gene (Hill et al., 2002; Sánchez et al., 2014; Ogier et al., 2019). Such approaches have to deal with the trade-off of either relying on strongly degenerated primers ensuring a taxonomically broad match of the primers (Hill et al., 2002; Ogier et al., 2019), or to use less degenerated primers, which only match to genes of much more narrow taxonomic groups (Sánchez et al., 2014).
Here we present a method targeting only a phylogenetically narrow (>99% 16S rRNA sequence similarity, OTU99%) but important group of freshwater bacteria. The presented method is not intended to be a replacement of standard 16S rRNA amplicon sequencing but as a supplemental method enabling a species level taxonomic resolution for an important fraction of freshwater bacterioplankton. For the targeted protein-encoding single-copy housekeeping gene (primosomal replication protein N, priB), involved in DNA replication, a sequence similarity threshold could be established, which largely resolves communities of the investigated OTU99% at the species level. The establishment of this threshold is based on large sets of priB gene sequences and corresponding genome sequences obtained from a large culture collection. These two corresponding data sets enabled the search for a priB gene sequence similarity threshold equaling to the 95% genome-wide average nucleotide identity (ANI) threshold used in genome taxonomy and in standard taxonomy of prokaryotes for genome-based species demarcation (Konstantinidis et al., 2006; Jain et al., 2018).
The genus Polynucleobacter can be phylogenetically subdivided in the four subclusters PnecA, PnecB, PnecC and PnecD (Hahn, 2003). In contrast to the ecological diversification at the subcluster level (Jezbera et al., 2012; Newton & McLellan, 2015; Nuy et al., 2020), not much is known about ecological diversification within these subclusters (Jezbera et al., 2011; Hahn et al., 2016). The presented method is targeting only subcluster PnecC (Hahn, 2003), which consists of many described and undescribed species all sharing 16S rRNA sequence similarity values ≥ 99%, thus the whole subcluster PnecC represents a single OTU99%. This subcluster harbors strains of two distinct lifestyles. The evolutionary primary lifestyle is a free-living and planktonic one, while a derived lifestyle is found in strains dwelling as obligate endosymbionts of benthic ciliates (Heckmann & Schmidt, 1987; Boscaro et al., 2013). Strains of the planktonic lifestyle were frequently detected in diversity surveys, which covered together an ecologically broad range of freshwater systems (Bahr et al., 1996; Zwart et al., 2002; Percent et al., 2008; Allen & Cavicchioli, 2017). They represent abundant freshwater bacteria (Jezbera et al., 2012) ubiquitously appearing in the water column of freshwater habitats all over the world (Jezberova et al., 2010; Peixoto et al., 2011; Hahn et al., 2015; Comte et al., 2016). Conversely to the planktonic strains, endosymbiotic strains were never reported to appear with significant abundance in the water column of freshwater systems.
Comparative genomic analyses suggested that species of the genus Polynucleobacter are biologically maintained by intensive intra-specific recombination, which is opposed by inter-specific recombination barriers that separate core gene pools of related species (Hoetzinger & Hahn, 2017). The coherence of the intraspecific gene pool of at least one species is even maintained across populations separated by geographic distances of up to 3000 km, which is indicating a high dispersal potential of the species (Hoetzinger et al., 2021). On the other hand, microdiversification of Polynucleobacter species is influenced by horizontal acquisition of accessory genomic islands that can be transferred among different species (Hoetzinger et al., 2017).
The study presented here was largely designed as an exploratory study to reveal the breadth of the diversity within the targeted OTU99%. We investigated and compared the PnecC diversity of 99 freshwater habitats representing a broad limnological range and scattered across a latitudinal range of 3400 km. This resulted in the detection of an astonishingly high total species richness across the investigated habitats. We tested if the revealed diversity is structured by environmental factors and known macroevolutionary trends. Since the structuring of bacterial communities and evolutionary processes rather take place at the species level and not primarily at the level of 16S rRNA OTUs, the developed method provides a supplementary tool partially increasing the resolution of diversity surveys. In addition, the method is suitable to reveal drivers structuring the community composition and the selective forces of evolutionary adaptation of a model group of planktonic freshwater bacteria.
Materials and Methods
Investigated habitats and sampling
In total, 117 water samples from 102 freshwater habitats were obtained (Suppl. Mat. Table S1 and Fig. S1), however, only results of 114 samples from 99 habitats could be analyzed (see below). This includes eleven habitats sampled two-to three-times. Surface water (0.1-0.5 m depths) samples were taken from the shoreline or from piers if available. Biomasses were collected by filtration onto 0.2 μm Nuclepore membrane filters, preserved by storage in absolute ethanol, and transported in a mobile refrigerator. Water temperature, pH, conductivity and oxygen were measured on location. For determination of concentrations of major ions, water samples were filtered through GF/F filters (Whatman, UK) and measured by ion chromatography (Thermo Scientific DIONEX ICS-1100).
Reference priB sequences of cultured strains
In total, a set of 377 priB (primosomal replication protein N) sequences of cultured Polynucleobacter strains could be established, which represent 254 unique sequences (Suppl. Mat. Table S2). Of those priB sequences, 102 originate from genome sequences. The remaining 275 sequences were obtained by Sanger sequencing of PCR products. The sequenced amplicons were generated by using primers priBausF 5’-CGTCARATGGCTTACATGATC-3’ and priBausR 5’-CAATAACGYTTACGCTTGAAC-3’. The sequenced fragments obtained with this primer pair included the binding sites of the priBinnFd/priBinnRd primer pair (see below), which enabled checking of the binding sites in strains without available genome sequence.
Amplicon sequencing and processing of reads
Genomic DNA was extracted from environmental samples as described previously (Jezbera et al., 2011), and purified using the Wizard DNA clean☐up kit (Promega). The priB gene of Polynucleobacter bacteria (subcluster PnecC) was amplified by using primers priBinnFd 5’-YGGCGTTGAATCATTTMAC-3’ and priBinnRd 5’-TTCCAAACGCCATGRTGATT-3’ (annealing temperature 62°C, 30 cycles, Q5 polymerase (New England Biolabs)). The primers were tagged with Illumina adaptors and sample-specific tags (fusion primers). Amplicons of 117 environmental samples, one technical replicate and two controls consisting of amplicons from one and four cultured strains, respectively, were paired-end sequenced (300 bp) by Illumina MiSeq. Reads were processed by QIIME2 (Bolyen et al., 2019). This included demultiplexing, trimming of adapter and primer sequences, quality filtering, joining of paired reads, exclusion of too long (>288 bp) and too short (<285 bp) sequences (all reference sequences of PnecC strains were in the length range of 285-288 bp), removal of reads with copy number <10 present only in single samples, and rarefication to 25230 reads per sample. Due to too small read numbers, three environmental samples had to be excluded, therefore, only the results from 114 water samples obtained from 99 habitats are presented (Suppl. Mat. Text S1). Reads were clustered into OTUs by employing a 98% sequence similarity threshold (see results). OTUs were taxonomically classified by employing a reference set of priB sequences obtained from Polynucleobacter strains (Suppl. Mat. Table S2). The two groups of OTU98% sharing ≥98% or ≤98% sequence similarity with reference taxa are termed reference operational taxonomic units (refOTUs) and environmental OTUs (eOTUs), respectively.
Data analyses
The OTU table exported from Qiime2 and the environmental data were analyzed using R version 3.6.1 (R Core Team, 2019). The vegan package (Oksanen et al., 2019) was used for most of the performed analyses. Geographic distances between sampled habitats were determined by calculation of great-circle-distance using the haversine method, which assumes a spherical earth, from the R package “geosphere” (Hijmans, 2019). Site-specific climate data were obtained from the WorldClim data set (Fick & Hijmans, 2017) using the DIVA-GIS software (Hijmans et al., 2001). Furthermore, the R packages “ggplot2” (Wickham, 2016) and “maps” (Minka & Deckmyn, 2018) were used.
Bray-Curtis dissimilarities were calculated without prior transformation of the OTU table. Occupancy of OTU was defined and calculated as follows. Because Polynucleobacter species are basically unable to exist across the whole studied environmental pH range of more than four units, occupancy of each OTU was assessed within a specific pH range of two units, i.e. only samples within a pH range of two units around (+1 and -1) the relative-abundance-weighted average pH of samples with detection of the respective OTU were considered. The weighted average pH indicates the pH optimum of the respective OTU. For instance, the relative-abundance-weighted average pH of detections of P. paneuropaeus was 5.9, therefore occupancy refers to the samples of the pH range 4.9 to 6.9. Forty-one of the investigated samples belonged to this pH range and P. paneuropaeus was detected in 31 of them (with >25 reads, i.e. >0.1% of reads per sample), thus showed a pH-specific occupancy of 73%.
Results
Development of PCR primers targeting a protein-encoding gene
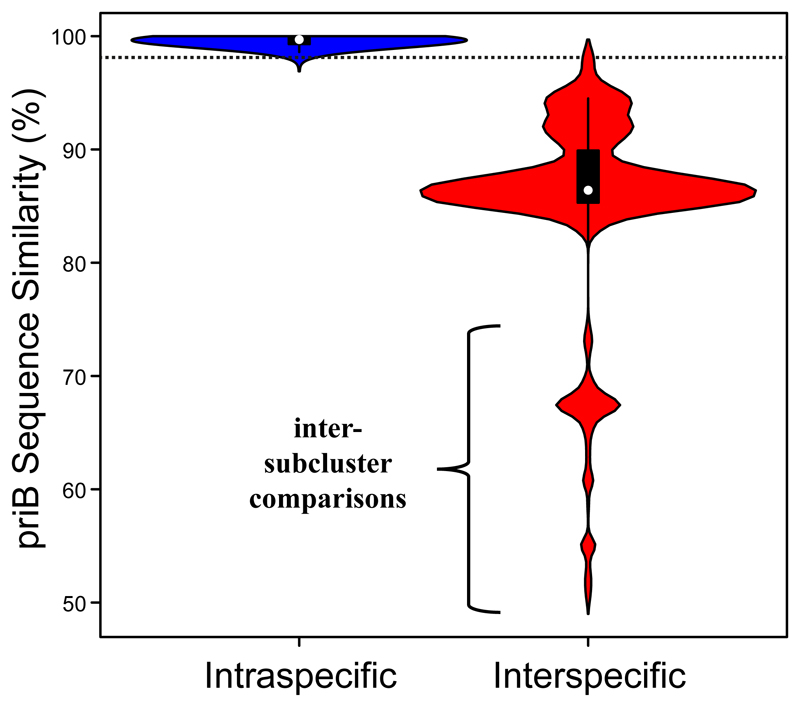
We aimed for development of a primer pair suitable for specific amplification of a protein-encoding gene present in all Polynucleobacter bacteria. The strategy employed for primer development and the faced limitations and results are described in the Suppl. Mat. Text S1. Briefly, a primer pair for amplification of the primosomal replication protein N (priB) gene of Polynucleobacter bacteria affiliated with subcluster PnecC could be developed. Detailed analyses suggested a sequence similarity threshold of 98% for discrimination of species-like OTUs (Fig. 1; Suppl. Mat. Text S1). Discrimination based on this threshold agreed with average nucleotide identity (ANI) based species discrimination (95% identity threshold) for 99.2% of the pairwise comparisons among the 102 strains with available genome sequences (Fig. 1). Species that could not be discriminated by priB similarities of <98% were lumped together to species complexes.
Fig. 1.
Violin plot showing frequencies of priB sequence similarity values for intra- and interspecific (95% ANI threshold) pairwise comparisons of genome-sequenced strains. The dotted horizontal line indicates 98% priB sequence similarity. Sequence similarities of priB genes below 80% represent comparisons of strains affiliated with different Polynucleobacter subclusters (PnecA, PnecB, PnecC and PnecD).
Amplicon sequencing of environmental samples
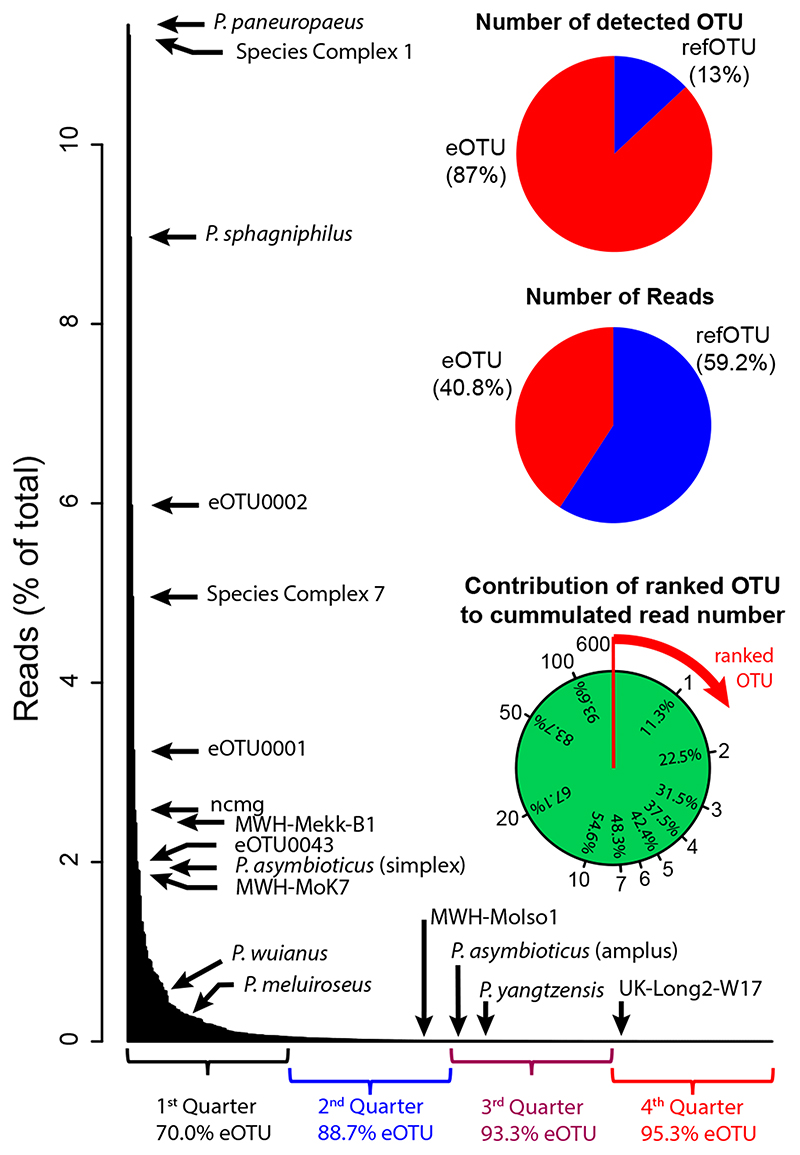
In total, results from 99 freshwater habitats (114 samples) including small ponds, lakes, streams and rivers (Suppl. Mat. Table S1) located in three regions (Lapland, Central Europe, Corsica; Suppl. Mat. Fig. S1) along a European South-North cross-section (42°N to 71°N) were obtained. The selection of habitats aimed for maximizing the covered habitat diversity in order to maximize the insights into diversity of Polynucleobacter taxa. Details on the results of the amplicon sequencing are given in Suppl. Mat. Text S1. Rarefaction analyses suggested that sequencing depth after rarefication (25230 reads) was large enough to completely cover the amplicon sequence variant (ASV) numbers in the respective samples (Suppl. Mat. Fig. S2A). In total, 600 OTUs98% were detected in the samples. Rarefaction analyses of the OTU98% data suggested that the number of investigated samples was not high enough to completely cover the total OTU98% richness in the investigated area (Suppl. Mat. Fig. S2B). The established OTU98% were taxonomically classified by using reference sequences and a threshold of 98% sequence similarity (Fig. 1). The reference sequences represented 108 species-like taxa, seven species complexes, and two taxa representing the same species but separated by priB sequence similarities < 98% (Suppl. Mat. Text S1 and Table S2). Environmental OTU98% classified in one of the reference taxa are called refOTU, while all remaining OTU98% sharing < 98% similarity with reference sequences are called eOTU. Only thirteen percent of the 600 detected OTUs were represented by refOTUs, i.e. by cultivated reference strains (Fig. 2), but this minor fraction of the total number of detected OTUs recruited 59% of the total amount of priB reads. The rank-abundance plot (Fig. 2) sorting the detected taxa according to their relative read abundance shows that only a few OTUs recruited most of the obtained reads. The top ranked OTU (P. paneuropaeus) recruited 11.3% of all reads. The top-seven-ranked OTUs (including two species complexes) recruited together almost half of all reads, while the vast majority of the detected OTUs represented rare taxa. The percentage of eOTUs, i.e. environmental OTUs sharing <98% sequence similarity with all reference strains, increased with decreasing read numbers recruited by the respective OTUs (Fig. 2). While only 30% of the top-ten-ranked OTUs were eOTUs, the first quarter of the ranking contained 70% and the last quarter 95% eOTUs.
Fig. 2.
Frequencies of the detected OTUs98%. The bar plot shows the rank-abundance distribution of the 600 OTUs detected in the 99 investigated habitats. Detections of repeatedly sampled habitats were down-weighted in order to give detections from all habitats the same weight. Individual rank-abundance curves of each investigated sample are shown in Suppl. Mat. Fig. S2C. The pie charts depict shares of refOTUs (representing cultured strains) and eOTUs (sharing <98% sequence similarity with priB sequences of cultured strains). Top pie chart, shares of refOTUs and eOTUs of the total number of detected OTUs98%. Middle, share of reads assigned to refOTUs and eOTUs. Bottom, cumulative contribution of detected OTUs sorted by increasing rank to the total number of reads. For instance, the top-ranked OTU98% (P. paneuropaeus) recruited 11.3% of the total number of reads (habitats weighted equally) and the top-seven ranked OTUs98% recruited in total 48.3% of reads.
Structuring of Polynucleobacter communities by environmental factors
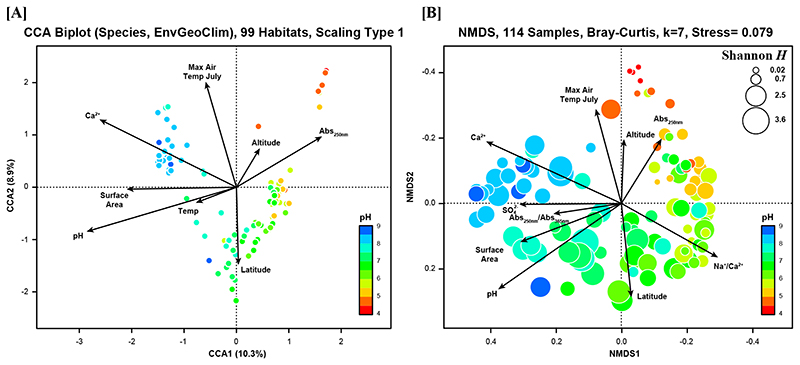
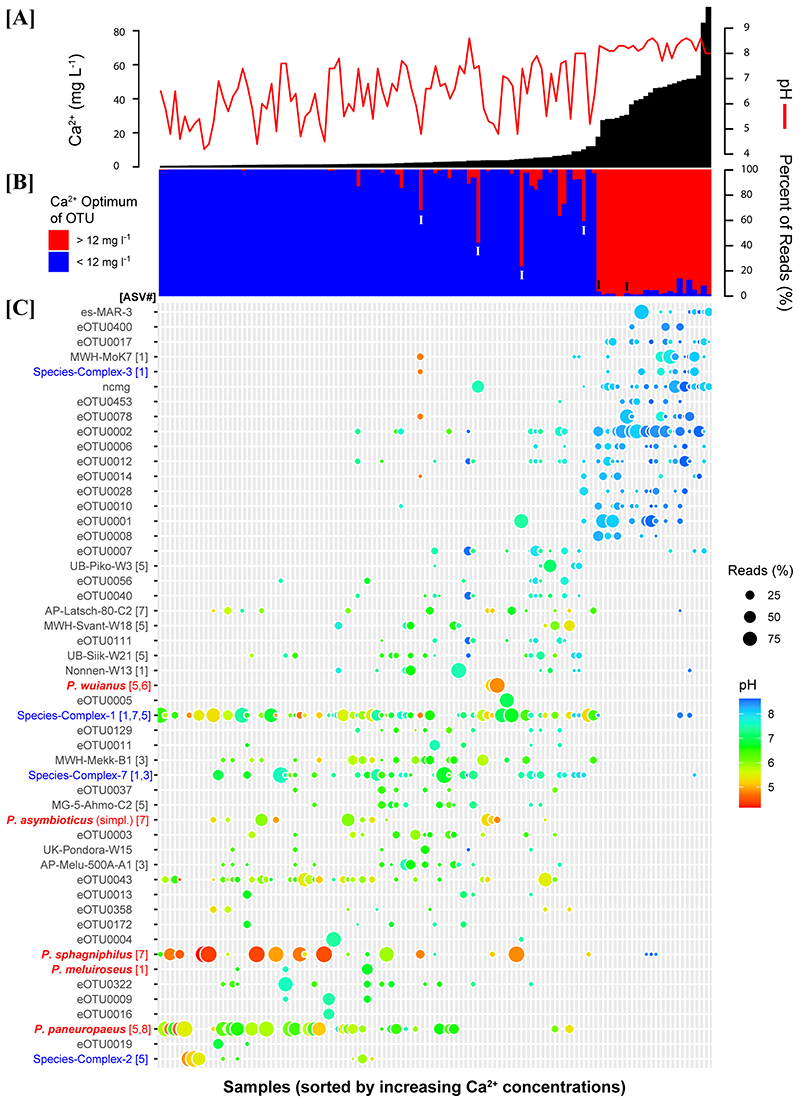
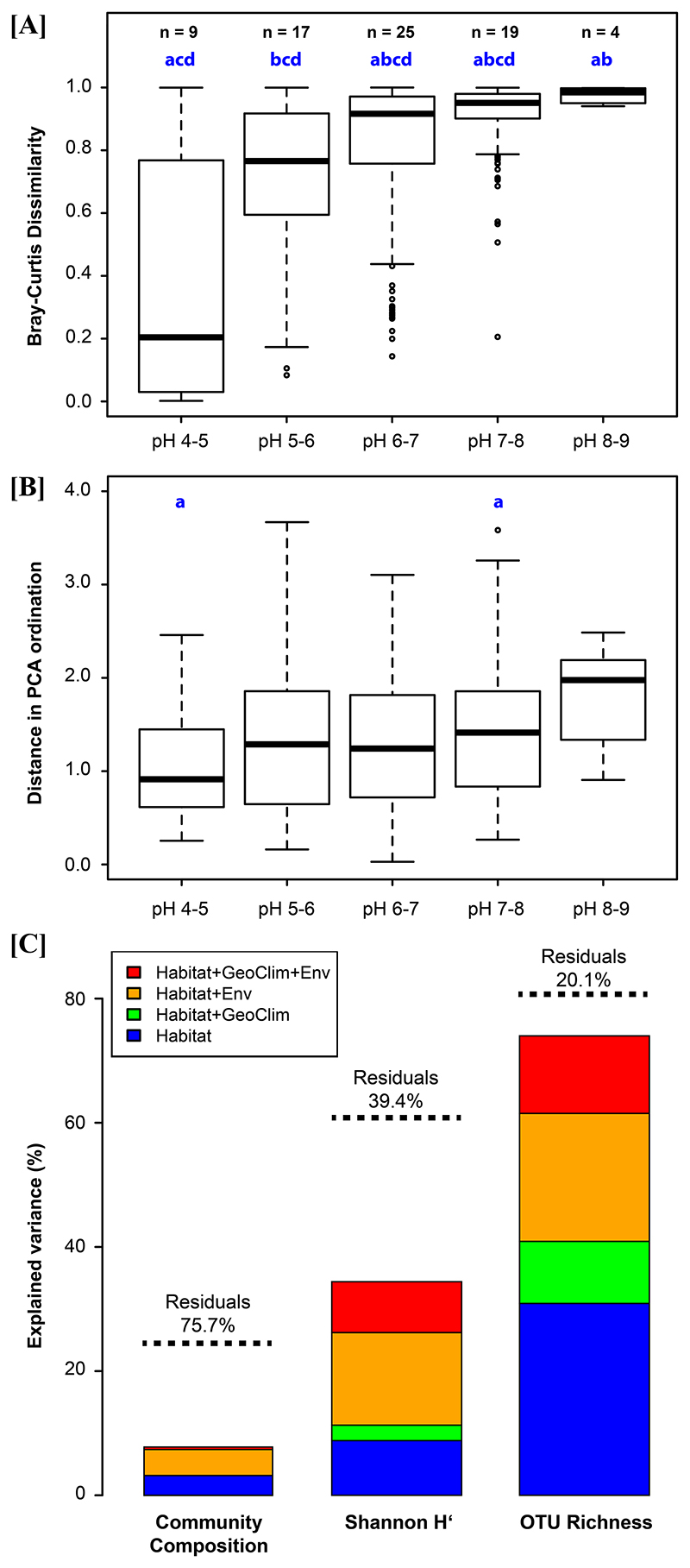
The investigated samples and habitats represent broad ranges of environmental parameters (Suppl. Mat. Table S1), for instance, including a pH range of 4.2 to 8. The composition of the PnecC communities in the investigated samples was strongly changed along the environmental gradients defined by the respective habitats. A constrained gradient analysis by canonical correspondence analysis (CCA) suggested a discontinuous variation in composition mainly corresponding to the concentration of dissolved Ca2+ ions (Fig. 3A). Further analyses of community compositions along the Ca2+ gradient of the investigated samples suggested a sharp breakpoint in community composition at calcium concentrations of about 12 mg Ca2+ l-1 (Fig. 4B). An ANOSIM analysis confirmed significant differences in the composition of communities in samples below and above this concentration (9999 permutations, R=0.6499, p=0.0001). Communities from low Ca2+ habitats were rather diverse and showed continuous changes along environmental gradients (mainly pH), while communities from high Ca2+ environments appeared in the NMDS ordination as a much smaller cluster (Fig. 3B and Suppl. Mat. Fig. S3). A variation partitioning analysis indicated that a set of 15 explanatory variables including physicochemical, geographic and climatic parameters as well as habitat characteristics (Suppl. Mat. Fig. S4) explained together about 25% of variance in community composition across the 114 samples (Fig. 5C). Not much less than half of the explained variance in community composition was explained by the set of physicochemical parameters (Env, 9.7%) alone (Suppl. Mat. Fig. S4), while the habitat characteristics (habitat type and size) explained the variance of composition only marginally. By contrast, variance in OTU richness of samples was much better explained by environmental conditions. About 80% of variation of richness could be explained by the set of environmental variables (Fig. 5C). The largest explained fraction of variance was explained by habitat characteristics (habitat size and type, 31%). The second largest fraction was explained by physicochemical variables (including pH) together with habitat characteristics (21% of variability in OTU richness). The rest of the variance was mainly explained by various combinations of parameters (Suppl. Mat. Fig. S4).
Fig. 3.
(A) Direct gradient analysis by canonical correspondence analysis (CCA) of Bray-Curtis community dissimilarity values and environmental variables (permutation test for the whole CCA model, P = 0.001). In this constrained ordination, dots represent the 99 habitats color-coded by the pH of the respective sample. (B) Indirect gradient analysis by non-metric multidimensional scaling (NMDS) exclusively based on Bray-Curtis dissimilarities of the 114 environmental samples. Each bubble represents a sample color-coded by pH. The diameter of the bubbles is non-linearly scaled by the Shannon index H’ of the respective sample. Both ordinations show environmental variables significantly (P < 0.05) correlated with the ordination models.
Fig. 4.
OTU and community distribution along the Ca2+ gradient of the 114 investigated samples. All three plots show the samples sorted by increasing Ca2+ concentrations. (A) Ca2+ concentrations (black bars) and pH (red line). (B) Community compositions regarding Ca2+ preferences of the OTUs (< or >12 mg Ca2+ l-1) constituting the particular communities. Communities labeled by an “I” represent communities showing an intermediate position in the NMDS ordination (Fig. 3) regarding the Ca2+ vector (Suppl. Mat. Fig. S3F). (C) Detection of the most abundant high and low Ca2+ OTUs. The color code applied to the taxon names indicates described species (red), species complexes (blue), and other OTUs (black). The numbers of the eight 16S rRNA ASVs (compare Fig. 8) are given after the OTU names in squared brackets if known. The colors and the diameters of the bubbles indicate the pH of the samples and the relative read abundances in the respective sample, respectively.
Fig. 5.
(A) Boxplots of pairwise community dissimilarities (Bray-Curtis) within pH classes. Only samples of standing waters with low Ca2+ concentrations (< 12 mg l-1) were considered. The numbers (n) of samples per pH class are given above the bars. Classes with significantly (p < 0.025) different data (Kruskal-Wallis test with Dunn’s post hoc test with Holm correction for multiple comparisons) are labeled with the same blue letter. (B) Boxplots of pairwise comparisons of environmental distance among samples within pH classes. The environmental distance was calculated as Euclidian distance between coordinates of a Principal Component (PCA) ordination of 20 variables. The same set of samples as in the above analysis of Bray-Curtis dissimilarities was used. A test for significant differences between groups was performed as above. (C) Results of variation partitioning analyses on community composition (Bray-Curtis dissimilarity), Shannon H’ and OTU richness (OTUs >1% of priB reads). Three sets of explanatory variables were used. Env, eight environmental variables; GeoClim, five geographic and climatic variables; Habitat, two variables characterizing habitat properties (surface area and type of habitat, i.e., running or standing waters). The stacked bars show only partitions of explained variance including habitat properties as explanatory variables. The total explained variance is indicated by dotted lines.
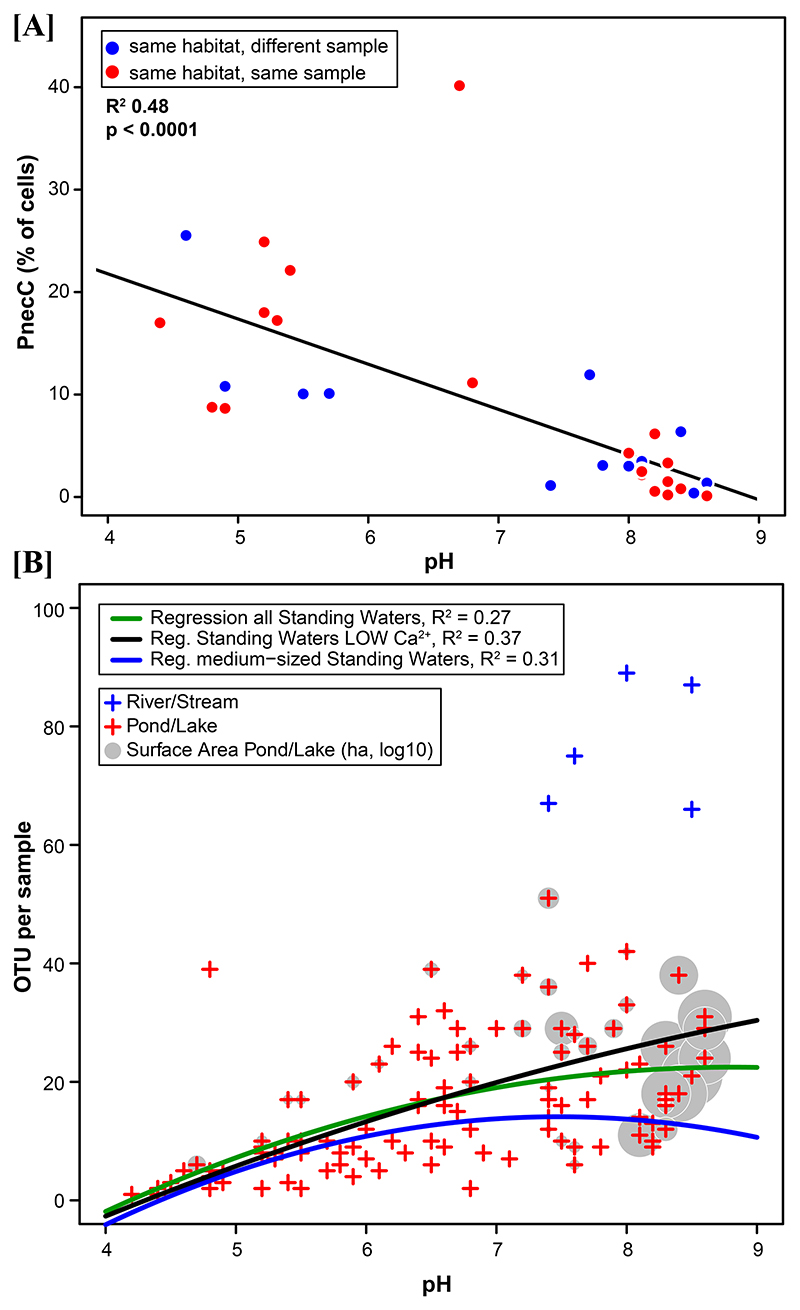
Communities of the most acidic samples tended to be dominated by only a single OTU, i.e., P. sphagniphilus. In the four samples with lowest pH (<4.6) this species represented 99.7 – 99.8% of reads (Fig. 6B). When excluding rare OTUs defined by relative abundances in the samples of <1% (i.e. <1% of reads), these four samples showed an OTU richness of 1.0. By contrast, samples from streams and rivers were characterized by 66 – 89 OTUs when rare OTUs were excluded. In general, both the OTU richness (Fig. 6B) and the size of habitats (Suppl. Mat. Fig. S5) tended to increase with pH values of the habitats. Interestingly, similar z values describing the relationship of habitat size and species (OTU) richness were observed for Polynucleobacter OTU (Suppl. Mat. Fig. S5) and whole bacterioplankton communities of freshwater lakes characterized by using 16S rRNA sequence data (Reche et al., 2005).
Fig. 6.
(A) Relative abundance of PnecC bacteria determined by FISH (data from (Jezbera et al., 2012)). Some of the shown data represents samples included in the priB amplicon sequencing (red dots), other samples were obtained from habitats included in the priB sequencing but taken at other dates (blue dots). (B) Polynomial regressions on relationship between pH and OTU richness. OTU numbers represent only detections >1% of reads per sample. Samples from running waters were completely excluded from the analyses. Regressions were performed on all remaining samples, all remaining samples with low Ca2+ communities, and remaining samples from habitats with mediumsized surface area (0.018 – 0.64 ha). The surface size of all standing water habitats is indicated by grey bubbles (log transformed data).
Occupancy of particular OTUs along the pH gradient
Only eight (including two species complexes) of the 600 detected OTUs represented common taxa with pH-specific occupancy >50%. These 1.5% of all detected OTUs frequently occurred with high average relative abundances and tended to appear with more even relative abundance across the occupied habitats. In contrast to these common taxa, 15.5% of the detected OTUs showed occupancies between 10% and 50% and appeared by average with maximum relative abundances of 15.9% of the reads (range 0.3% to 87%). Interestingly, locally abundant taxa showed occupancy values <10% but rather high relative abundances in a few habitats or samples. Examples for locally abundant taxa are P. wuianus and P. meluiroseus (Suppl. Mat. Fig. S1). The former species was discovered in October 2002 in a small slightly acidic pond designated Pond-1 (Hahn et al., 2005). The priB amplicon sequencing included three samples of that pond, which were taken in August and October 2009, and in June 2010. P. wuianus was detected in all three samples of its type locality with relative abundances ranging from 15% to 86% of the reads. Across the other 111 investigated samples, this species was detected only in six samples from two ponds located 200 meters and 40 km away from Pond-1. Both ponds were sampled three times but P. wuianus was detected only with low relative abundances of 0.004 to 0.135% of the reads. The occupancy of this species was only 3.7% and the average relative abundance was despite the local abundance peak in Pond-1 only 0.56%, which was twenty-times smaller than the relative abundance of the common species P. paneuropaeus. Similarly, P. meluiroseus showed an occupancy of 6.1% with detections in nine samples representing seven habitats but only in three samples this species appeared with relative abundances of >1%. Interestingly, the two highest values of 41 and 8% relative abundance were observed for the lake from which the type strain of the species was isolated (Pitt et al., 2018). In contrast to P. wuianus, the detections of P. meluiroseus were geographically broader scattered (Suppl. Mat. Fig. S1). Obviously, both species are characterized by high local relative abundances, low pH-specific occupancy and high local persistence.
The pH-specific occupancy of OTUs tended to decrease with increasing pH (Suppl. Mat. Fig. S6A). This trend is linked to a trend of increasing community dissimilarities among communities of the same pH class with increasing pH (Fig. 5A). This means that differences in composition among communities present in habitats with similar pH are increasing with pH. This is also obvious in the NMDS ordination where the communities from habitats with similar pH are spread over larger ordination space if pH values of their habitats are higher (Fig. 3B). Interestingly, in the case of high-Ca2+ communities (mainly represented by the pH class 8-9), the link between occupancy of OTUs and community dissimilarities seems to be less strict than in the low Ca2+ communities (Fig. 5A and Suppl. Mat. Fig. S6A).
Biogeography of Polynucleobacter communities
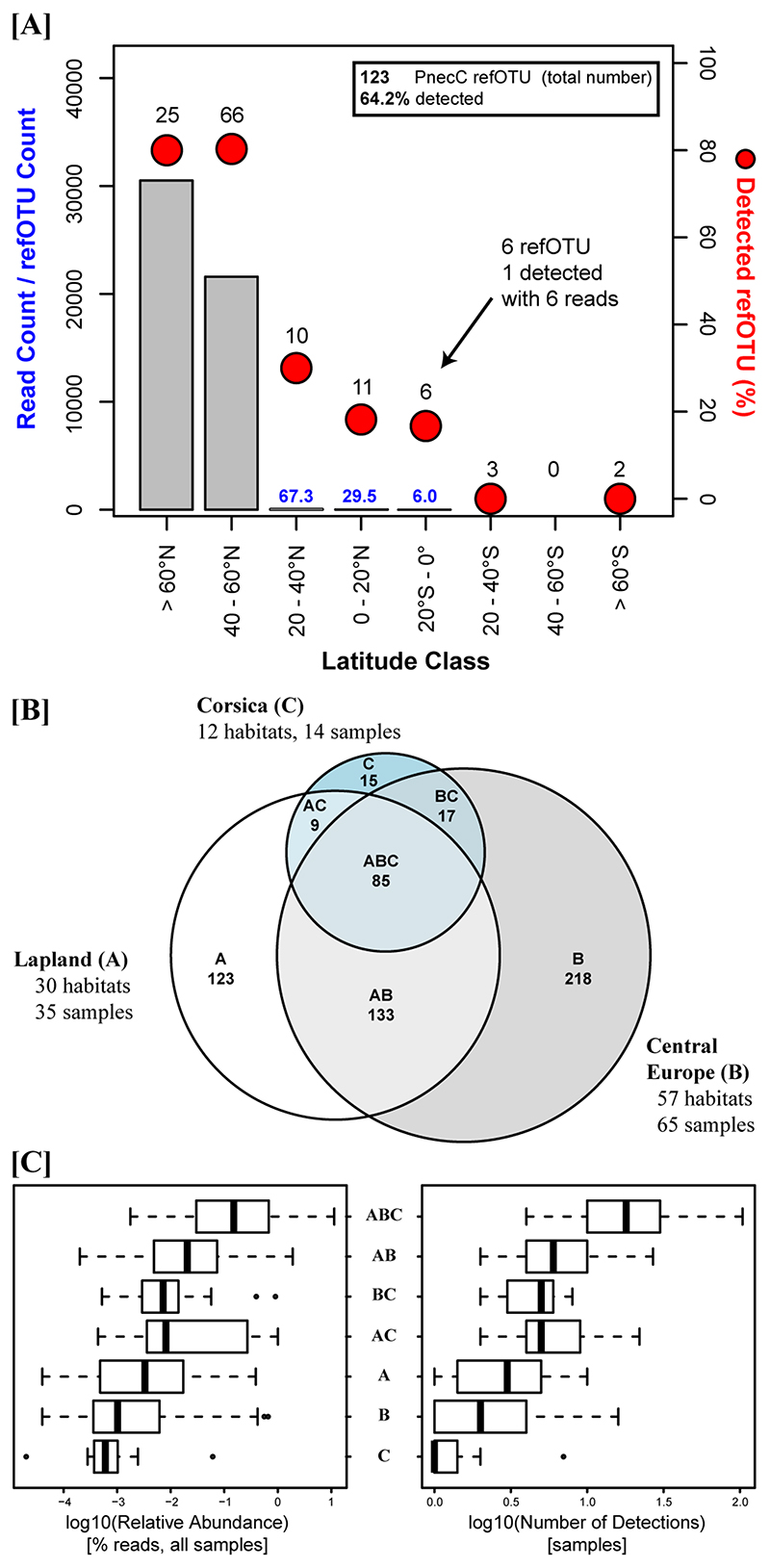
Of the set of 123 reference taxa (Suppl. Mat. Table S2) used for taxonomic classification of the detected OTU98% only 64.2% were detected in the 114 investigated samples, however, the detections differed in relation to the latitudinal origin of the strains representing reference taxa (Fig. 7). We compared the latitudinal origin of the strains representing reference taxa (62°S – 78°N) with the range of geographic origin of the 114 investigated samples (42°N - 71°N). While 80% of the reference taxa represented by strains obtained from sites located at latitudes >40°N were detected, only 19% (0–30% in particular latitude classes) of the reference taxa represented by strains obtained from latitudes <40°N were detected (Fig. 7A). Importantly, all detected reference taxa with strains obtained from latitudes <40°N recruited only very small numbers of reads.
Fig. 7. Biogeography of refOTUs and eOTUs.
(A) Detection of the 123 reference taxa (total number, including undetected) represented by cultured strains obtained from locations of various latitudes. The bars indicate the average number of reads per refOTU assigned to reference taxa grouped according to their respective origin in latitude classes. The numbers above the red dots show the absolute number of reference taxa in a particular latitude class, and the dots indicate the fraction of these reference taxa that was detected in the priB amplicon dataset. For instance, the latitude class 0 - 20°N harbors in total 11 reference taxa of which only 18.2% were detected (2 taxa), with average read numbers of 29.5 reads per refOTU. Note that the latitude range of the habitats investigated by priB amplicon sequencing was 42°N – 71°N. (B) VENN diagram depicting the number of detected OTUs (eOTUs and refOTUs) shared or not shared between the three investigated geographic regions. (C) (Left graph) Boxplot of total relative abundance (in all samples) of detected OTUs grouped in geographic classes according to the VENN diagram shown in (B), and (right graph) boxplot of the number of samples with detections for the same geographic OTU classes. Note the log transformation of the plotted data in both boxplots.
Despite of a latitudinal range spanning almost 30° and a maximum distance between habitats of about 3400 km (Suppl. Mat. Fig. S1), a Mantel test did not suggest that differences in community composition increased with geographic distance between the sampled habitats (Mantel R=-0.01, p=0.66; Suppl. Mat. Table S3). Even when controlling for environmental influences including differences in pH or Ca2+ concentration (partial Mantel tests) no significant correlations between community composition and geographic distance were observed. Different results were obtained when only Polynucleobacter communities from low Ca2+ conditions were considered. Weak but significant correlations (Mantel R <0.13, p < 0.01) were observed with geographic distance, even when controlling for distances in pH, Ca2+ and other chemical parameters. However, when controlling for a broader set of environmental variables (including habitat type and climatic variables), no significant correlation between community composition and geographic distance was observed (Mantel R=0.0514 p=0.0602). By contrast, Mantel tests and partial Mantel tests on correlations between community composition and environmental factors yielded in all cases significant correlations. The highest correlation coefficient (Mantel R=0.53, p=0.0001) was observed for community composition and pH distance between habitats.
Despite of lacking indications for an isolation by distance pattern for the investigated Polynucleobacter communities, geographic structuring was evident. More than 50% of the detected OTUs were exclusively detected in only a single of the three sampled regions (Fig. 7B). Such OTUs only detected in single regions tended to be characterized by low average relative abundances and by detections in only few samples (Fig. 7C).
Predictive power of 16S rRNA sequence ASVs of Polynucleobacter bacteria
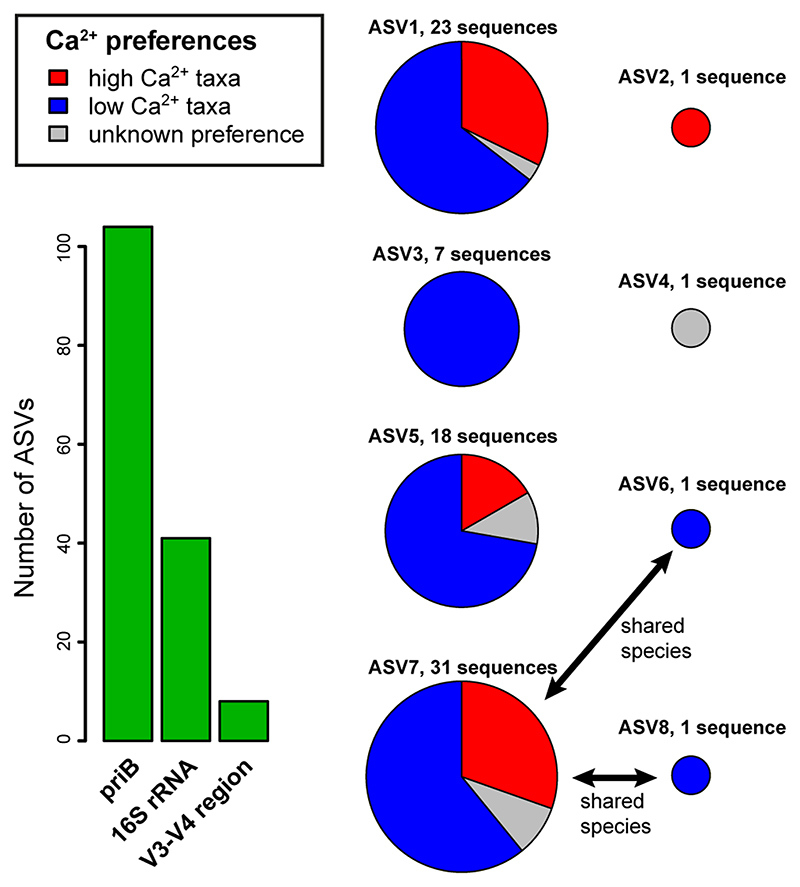
We evaluated if 16S rRNA based ASVs of Polynucleobacter bacteria possess a predictive power regarding environmental preferences of ASVs. A set of 226 strains (Suppl. Mat. Table S2 plus genomes recently published by (Hoetzinger et al., 2021)) affiliated with subcluster PnecC was represented by only eight V3-V4 region ASVs, although these strains represented 80 different species according to genome similarities (95% ANI threshold) (Fig. 8). Six ASVs represented more than one species, respectively, and three of these ASVs consisted of strains with distinct Ca2+ preferences. Thus, bacteria with markedly different environmental preferences (Fig. 4) were lumped together within single 16S rRNA ASVs.
Fig. 8.
Bar chart, theoretical number of ASVs represented by different marker sequences of a set of 226 Polynucleobacter strains all affiliated with subcluster PnecC (excluding endosymbionts) representing 81 different Polynucleobacter species (>95% ANI). Pie charts, Ca2+concentration preferences of priB OTUs represented by the eight V3-V4 region ASVs (16S rRNA gene). Each species is only represented by a single sequence if no intraspecific sequence polymorphism was present. The Ca2+ preferences of the priB OTUs reflect the Ca2+ optima determined through analyses of the investigated environmental samples (Fig. 4). ASVs with unknown preferences were either not detected or were detected with too low read numbers.
Discussion
The limited taxonomic and ecological resolution of the 16S rRNA marker is well known (Jaspers & Overmann, 2004; Stackebrandt & Ebers, 2006; Hahn et al., 2016). An alternative “universal” marker for diversity investigations on bacterial communities is available (Hill et al., 2002) but requires the use of highly degenerated primers strongly substituted with inosine bases. This is potentially biasing comparative compositional analyses of bacterial communities. High degeneration of primers can be avoided if the taxonomic focus of diversity studies is narrowed to genus-like taxa (Sánchez et al., 2014; Pereira et al., 2018).
We developed a new marker and investigated the structure of Polynucleobacter communities along environmental gradients characterized, amongst other parameters, by a pH range of 4.2 to 8.6 and Ca2+ concentrations of 0.1 to 94 mg 1-1. To maximize the studied environmental gradients, a broad variety of freshwater systems was investigated, which range from very small, shallow ponds to large lakes, rivers and streams. The sampled habitats are located in different climate zones and at altitudes ranging from about sea level to more than 2000 meters. The applied method for determination of the community composition provided a resolution largely at the species level, but only covered the multi-species subcluster PnecC of the genus Polynucleobacter. Remarkably, in typical diversity studies based on 16S rRNA amplicon sequences, the targeted Polynucleobacter diversity is represented by only a single OTU99% harboring a rather small number of ASVs (Fig. 8). In 16S rRNA based studies comparing, for instance, acidic and alkaline habitats or systems with low and high Ca2+ concentrations, the OTU99% representing subcluster PnecC of the genus Polynucleobacter is harboring different organisms across the investigated samples. The same OTU99% found in, for instance, samples from acidic and alkaline habitats is comprised by species differing pronouncedly in their respective ecological traits. This heterogenous composition of OTUs potentially results in masking of ecological trends and patterns, and may also blur dispersal and community assembly processes. It is known that the studied group of Polynucleobacter bacteria is not exceptional among free-living bacteria regarding the limited taxonomic and ecological resolution of their 16S rRNA genes (Jaspers & Overmann, 2004; Rodriguez-R et al., 2018; Chevrette et al., 2019).
The priB gene could also be used for biodiversity survey or experimental studies on other genus-like bacterial taxa. However, this would need sufficient knowledge on sequence diversity at the potential primer binding sites (Suppl. Mat. Text S1) and the quality of the taxonomic resolution at the species level would largely depend on a suitable collection of reference genomes enabling a sound search for a species discrimination threshold (Fig. 1).
Enormous but still incompletely covered OTU richness
Our priB-based investigation of 99 freshwater habitats revealed an astonishing total number of 600 species-like Polynucleobacter OTUs. We cannot be sure that the performed read processing and filtering removed all erroneous sequences, however, the strict sequence length filtering and the removal of sequences containing additional stop codons should have helped to exclude many sequences representing PCR artifacts. Especially the search for additional stop codons increased the confidence in the established sequence data, because their appearance in the single-copy, essential house-keeping gene priB clearly indicates erroneous sequences. Importantly, rather few such sequences were found and all of them were present in very low copy numbers (Suppl. Mat. Text S1). We did not perform chimera filtering due to the lack of a suitable priB reference database. However, we used a phylogenetic tree calculated with all reference and eOTU priB sequences to search for eOTUs displaying unusually long branch length. Long branch lengths are expected if two sequences with low similarity contribute larger fractions to a chimeric sequence, however, suspicious long branches were not observed for any eOTU. The increasing percentage of eOTUs towards the rare species end of the rank-abundance distribution (Fig. 2) could indicate erroneous sequences, however, an alternative explanation for the decreasing percentage of refOTUs could be that rare species tend to be underrepresented in collections of cultured strains.
Even if we would assume that 10, 20, or even 30% of the detected 600 OTUs were based on erroneous sequences, an impressive number of detected OTUs would remain. In addition, rarefaction analyses suggested that not even all of the abundant taxa (>10% of priB reads of a particular sample) could be detected by the variety of samples we investigated (Suppl. Mat. Fig. S2B). This indicated that further sampling would increase the detected number of both abundant and rare OTUs. This is not surprising given the observation of taxa with overall low occupancy but locally abundant populations like P. wuianus and P. meluiroseus. In addition, our study could certainly not cover the entire diversity of freshwater systems in the investigated regions. Running water systems, for instance, were only marginally covered and anoxic hypolimnia of lakes known to be rich in Polynucleobacter bacteria (Jezbera et al., 2011; Diao et al., 2017) were completely omitted. The indicated incomplete coverage of OTUs present in the investigated area is further supported by the lack of detection of 20% of the reference taxa originating from this area (Fig. 7A). Consequently, ecologically broader sampling and inclusion of seasonal aspects are both expected to increase the total number of OTUs detected in the investigated regions.
Biogeography of taxa mainly reflected regional differences in ecological conditions
Environmental filtering resulted in a strong biogeographic structuring. For instance, OTUs abundant in limestone areas in Central Europe were not detected in other sampled regions, which lack habitats with high Ca2+ concentrations (Suppl. Mat. Fig. S1), and low pH preferring OTUs were almost absent from the investigated habitats in Scandinavia with mainly circum-neutral pH. On the other hand, hints on biogeographic structuring caused by an isolation by distance mechanism were scarce. Partial Mantel tests controlling for environmental influences did not suggest that dissimilarity of Polynucleobacter communities increased with geographic distance (Suppl. Mat. Table S3). This was in line with a recent study on population structure of P. paneuropaeus along the same South-North range studied here, which suggested a lack of dispersal barriers along this 3400 km latitudinal range (Hoetzinger et al., 2021). The detection of OTUs exclusively found in only one of the three investigated regions (Fig. 7B) is well explained by the abundance-occupancy relationship documented in macroecology (Gaston et al., 2000), which predicts a positive relationship between the abundance of a taxon and its range occupancy (Fig. 7C). However, in the case of Polynucleobacter bacteria it is not known if this relationship simply resulted from undersampling of rare OTUs or if it really reflects restricted biogeographic distributions. Nevertheless, the former explanation seems to be more likely. The rather small fraction of reference taxa originating from lower latitudes and the southern hemisphere detected in the investigated habitats, as well as the very low relative abundance of the detected Sothern taxa (Fig. 7A) confirmed a previously revealed biogeographic pattern (Hahn et al., 2015). Currently, it is still unknown if these pattern result from a distance mechanism (dispersal limitation) or from environmental filtering of Polynucleobacter taxa differing in thermal adaptation (Hahn et al., 2015). In any case, a pronounced further increase in numbers of species-like Polynucleobacter OTUs has to be expected if future cultivation-independent studies would investigate habitats located south of 40°N.
Complex diversity trends along environmental gradients
An uneven distribution of Polynucleobacter subclusters along pH gradients is well known (Jezbera et al., 2012; Nuy et al., 2020), and based on previous investigations, even within subcluster PnecC differences in distribution of species along pH gradients were expected (Jezbera et al., 2011; Hahn et al., 2016). Due to the pronouncedly increased taxonomic resolution provided by the priB marker, much deeper insights into the structuring of PnecC communities by environmental factors were possible. This revealed a couple of diversity trends.
Importantly, the composition of the PnecC communities did not change continuously along all environmental gradients. A previous investigation suggested that the composition of PnecC communities is mainly controlled by pH (Jezbera et al., 2011), however, here we observed that the majority of OTUs preferred either low or high Ca2+ concentrations (Fig. 4). This trend seemed to be at least partially independent of pH, since alkaline habitats with low and high Ca2+ concentrations rarely share their inhabitants. Ca2+ concentrations were tightly correlated with conductivity (R2=0.93, p<0.0001), therefore, it is not known if really Ca2+ concentrations or rather salinity was specifically controlling the composition of the communities. However, coastal habitats with increased NaCl concentrations shared community compositions with low but not with high Ca2+ communities, suggesting that salinity is not the major driver of this distribution. The Ca2+ concentrations of aquatic systems are largely controlled by their geological background. Therefore, high Ca2+ Polynucleobacter communities were restricted to habitats located in limestone areas and characterized by higher Ca2+ concentrations (Suppl. Mat. Fig. S1). But even within limestone areas smaller habitats with low Ca2+ concentrations inhabited by low Ca2+ communities were found. Such habitats are limited to systems influenced by peat bogs or at least influenced by peat moss (Sphagnum spp.) vegetation. Besides Ca2+ concentration, pH had, as expected, a strong influence on the PnecC community composition (Fig. 3), however, OTU composition changed more continuously along the pH gradient.
In botany, it is well known that silicate and limestone soils basically differ in their plant community compositions, at least regarding the non-tree species (Bothe, 2015). These two soil types differ in many variables including pH and CaCO3 content. Due to the manifold factors distinguishing these two soil types, the major drivers of the distinct differences in vegetation composition are unknown (Bothe, 2015). On the other hand, it is well known that in many plant genera pairs of vicarious species evolved, which either dwell on silicate or on limestone soils. Similar vicariances seemed to be given among species of Polynucleobacter subcluster PnecC. Another case of vicariance was previously reported (Schauer et al., 2005) for planktonic freshwater bacteria affiliated with the two related taxa Candidatus Aquirestis calciphila (aka subcluster LD2) and Candidatus Haliscomenobacter calcifugiens (aka subcluster GKS2-217).
In soils, bacterial OTU richness shows a unimodal distribution along pH gradients with a richness peak at about neutral pH and a 3- to 4-fold change of richness across the pH gradient (Fierer & Jackson, 2006; Bickel et al., 2019). In comparison, the increase in PnecC OTU richness with pH was huge (Fig. 6B). The observed maximum increase was about 50-fold for standing waters, and even about 90-fold if running waters were also considered. It is not clear if richness also follows a unimodal trend in PnecC bacteria. Richness is obviously strongly influenced by habitat size and type, as well as several other environmental factors (Fig. 5C). However, if only medium-sized standing waters were considered, a unimodal model of richness along the pH gradient was suggested (Fig. 6B). Probably, a unimodal shape could be clearly confirmed if the alkaline side of the sample distribution could be extended towards higher pH values.
Unexpectedly, OTU richness and relative abundance of PnecC bacteria showed opposing trends along the investigated pH gradient (Fig. 6). It is well known that acidic lakes and ponds tend to possess higher relative abundances of PnecC bacteria ((Jezberova et al., 2010; Jezbera et al., 2012); Fig. 6A). Thus, lower numbers of PnecC OTUs present in acidic habitats contributed larger fractions to total bacterial numbers than PnecC communities in alkaline habitats with manifold higher OTU richness. Obviously, richness and relative abundance are at least partially decoupled across the investigated habitats. This could hint on differences in niche partitioning in acidic and alkaline waters, however, it is unknown which factors are involved in this unexpected diversity pattern.
Along with OTU richness, Shannon diversity increased with pH (Fig. 3B). This increase in diversity was accompanied by an increase of community dissimilarity among communities dwelling in habitats of similar pH (Fig. 3B). This trend was even obvious if habitats with high Ca2+ concentrations were excluded (Fig. 5A). We found no hint on a general increase of environmental diversity between different pH classes along the gradient towards higher pH values (Fig. 5B), however, the measured environmental variables seemed to be only poor predictors of variance of composition of PnecC communities (Fig. 5C). The increase of dissimilarity among communities of the same pH category comes along with a decreasing trend in occupancy of taxa (Suppl. Mat. Fig. S6A). This could be explained by a more stochastic community assembly in habitats with higher pH (Nemergut et al., 2013), combined with a higher number of OTUs able to dwell in systems with higher pH (Fig. 6B). Persistence of taxa with rather low occupancy in particular habitats over periods of more than one year (e.g. P. wuianus and P. meluiroseus) could hint on historical contingencies (Langenheder & Lindström, 2019) combined with potential local adaptation (Kraemer & Boynton, 2017). However, the observed phenomenon could also be linked to unmeasured abiotic environmental variables or to only locally occurring specific biotic interactions (Zhou & Ning, 2017). Time series and broader sets of measured variables are necessary to get insights into mechanisms responsible for this phenomenon.
Obviously, Polynucleobacter species strongly differ in ecophysiological adaptations (e.g. pH, Ca2+-related adaptation) but also in other ecological characteristics like occupancy. The resolution of the priB marker was high enough to reveal these species-specific differences in adaptation and ecological success among Polynucleobacter bacteria, which are undetectable with 16S rRNA sequence-based methods (Fig. 8).
Conclusions
Amplicon sequencing of the priB gene provided an unprecedented insight into the diversity of Polynucleobacter bacteria and structuring of their local communities by environmental factors. The used marker gene revealed patterns and trends invisible to 16S rRNA sequence-based methods. An astonishingly high yet incompletely covered species richness was found in the studied area. The observed high richness could indicate a general huge underestimation of bacterial species richness by 16S rRNA-based methods, if the observed high degree of diversification is also present in other bacterial OTUs99%. Importantly, Polynucleobacter communities showed several patterns well known from macroecological theory, which were previously only observed in phylogenetically much broader microbial taxa and communities (Horner-Devine et al., 2004; Reche et al., 2005; Sogin et al., 2006). This includes species-area and geographic abundance-occupancy relationships, as well as the organization of communities in a few abundant and many rare taxa (rank-abundance curves, (Sogin et al., 2006)). By contrast, the observed opposing trends of abundance and diversity of Polynucleobacter communities along the pH gradient, as well as differences in pH-specific occupancy of taxa along this gradient were unexpected. Obviously, priB amplicon sequencing provides a possibility to study the mechanisms of community assembly in great detail. Furthermore, this method may provide an opportunity to measure the response of some important freshwater bacteria to environmental changes caused by anthropogenic impact (Kraemer et al., 2020) with higher sensitivity than synecological methods based on ribosomal markers. However, detailed studies on the influence of various environmental factors and time series will be needed to better understand the mechanisms structuring Polynucleobacter communities and influencing the occupancy of particular taxa.
Supplementary Material
Acknowledgements
We thank Ulrike Koll and Johanna Schmidt for isolation of strains, DNA isolation, and processing of half of the samples for priB amplicon sequencing, and Johanna Schmidt for determination of major ion concentrations. We thank ‘Le Syndicat Mixte du Parc Naturel Régional de Corse et le gestionnaire des lacs d’altitude sur son territoire’ for the permission to take samples in the Corsica Natural Park (France), and we thank the National Park Hohe Tauern (Austria) and the owners of lakes for sampling permissions.
This research was funded in whole by the Austrian Science Fund (FWF) [project 27160-B22]
Funding information
Austrian Science Fund (FWF) project 27160-B22.
Footnotes
Author contributions
MWH designed research; all authors performed research and analyzed data; MWH wrote the paper and MH and AP commented and edited the draft.
Data Accessibility
Details on the sampled habitats and the measured environmental variable are provided in the Supplemental Information (Table S1 and Fig. S1) The complete set of environmental variables was deposited in the DRYAD repository (https://doi.org/10.5061/dryad.hhmgqnkgq). The nucleotide sequences of priB genes (MT988562-MT989336), genome sequences of Polynucleobacter strains (CP000655; CP007501; CP015017; CP015922; CP023276; CP023277; CP028940-CP028942; CP030085; CP049628; CP049637; CP049645; CP061288; CP061289; CP061291-CP061293; CP061295-CP061300; CP061302; CP061304-CP061306; CP061308-CP061319; JAANGD000000000; JAANHG000000000; JACVOK000000000; JACVOL000000000; JACVOM000000000; JACVON000000000; JACVOO000000000; JACVOP000000000; JACVOQ000000000; JACVOR000000000; JACVOS000000000; JACVOT000000000; JACVOU000000000; JACVOX000000000; JACVOY000000000; JACVOZ000000000; JACVPA000000000; JACVPD000000000; JACVPE000000000; JACVPF000000000; JACVPG000000000; JACVPH000000000; JACVPI000000000; JACVPJ000000000; JACVPM000000000; JACVPN000000000; JACVPP000000000; JACVPQ000000000; JACVPR000000000; JACVPS000000000; JACVPU000000000; JACVPW000000000; JACVPX000000000; JACVPY000000000; JACVPZ000000000; JACVQA000000000; JACVQB000000000; JACVQC000000000; JACVQD000000000; LOJI00000000; LOJJ00000000; LZFI00000000; LZMQ00000000; MPIY00000000; NAIA00000000; NGUO00000000; NGUP00000000; NJGG00000000; NTGB00000000; OANS00000000; PGTX00000000; QMCG00000000), reads obtained by Illumina amplicon sequencing (SRR11117533-SRR11117652), BioProject data (PRJNA607194), and BioSamples data (SAMN02724733; SAMN03430691; SAMN03430798; SAMN04080026; SAMN04086652; SAMN04086667-SAMN04086669; SAMN06014615; SAMN07200920; SAMN08383909; SAMN08383917-SAMN08383921; SAMN14212605-SAMN14212701) associated with this study were deposited in public databases curated by NCBI. Suppl. Mat. Table S2 links reference strains and reference environmental reads to refOTU und eOTU (respectively), accession numbers of priB sequences and genome sequences.
References
- Allen MA, Cavicchioli R. Microbial communities of aquatic environments on Heard Island characterized by pyrotag sequencing and environmental data. Sci Rep. 2017;7:44480. doi: 10.1038/srep44480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahr M, Hobbie JE, Sogin ML. Bacterial diversity in an arctic lake: A freshwater SAR11 cluster. Aquatic Microbial Ecology. 1996;11(3):271–277. doi: 10.3354/ame011271. [DOI] [Google Scholar]
- Bickel S, Chen X, Papritz A, Or D. A hierarchy of environmental covariates control the global biogeography of soil bacterial richness. Sci Rep. 2019;9(1):12129. doi: 10.1038/s41598-019-48571-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Caporaso JG. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology. 2019;37(8):852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscaro V, Felletti M, Vannini C, Ackerman MS, Chain PSG, Malfatti S, et al. Petroni G. Polynucleobacter necessarius a model for genome reduction in both free-living and symbiotic bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(46):18590–18595. doi: 10.1073/pnas.1316687110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothe H. The lime–silicate question. Soil Biology and Biochemistry. 2015;89:172–183. doi: 10.1016/j.soilbio.2015.07.004. [DOI] [Google Scholar]
- Chevrette MG, Carlos-Shanley C, Louie KB, Bowen BP, Northen TR, Currie CR. Taxonomic and Metabolic Incongruence in the Ancient Genus Streptomyces. Frontiers in Microbiology. 2019;10(2170) doi: 10.3389/fmicb.2019.02170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comte J, Monier A, Crevecoeur S, Lovejoy C, Vincent WF. Microbial biogeography of permafrost thaw ponds across the changing northern landscape. Ecography. 2016;39(7):609–618. doi: 10.1111/ecog.01667. [DOI] [Google Scholar]
- Diao M, Sinnige R, Kalbitz K, Huisman J, Muyzer G. Succession of bacterial communities in a seasonally stratified lake with an anoxic and sulfidic hypolimnion. Frontiers in Microbiology. 2017;8(2511) doi: 10.3389/fmicb.2017.02511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fick SE, Hijmans RJ. WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. International Journal of Climatology. 2017;37(12):4302–4315. doi: 10.1002/joc.5086. [DOI] [Google Scholar]
- Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(3):626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-García N, Tamames J, Linz AM, Pedrós-Alió C, Puente-Sánchez F. Microdiversity ensures the maintenance of functional microbial communities under changing environmental conditions. The ISME Journal. 2019;13(12):2969–2983. doi: 10.1038/s41396-019-0487-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaston KJ, Blackburn TM, Greenwood JJD, Gregory RD, Quinn RM, Lawton JH. Abundance–occupancy relationships. Journal of Applied Ecology. 2000;37(s1):39–59. doi: 10.1046/j.1365-2664.2000.00485.x. [DOI] [Google Scholar]
- Hahn MW. Isolation of strains belonging to the cosmopolitan Polynucleobacter necessarius cluster from freshwater habitats located in three climatic zones. Applied and Environmental Microbiology. 2003;69(9):5248–5254. doi: 10.1128/AEM.69.9.5248-5254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Jezberova J, Koll U, Saueressig-Beck T, Schmidt J. Complete ecological isolation and cryptic diversity in Polynucleobacter bacteria not resolved by 16S rRNA gene sequences. The ISME Journal. 2016;10(7):1642–1655. doi: 10.1038/ismej.2015.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Koll U, Jezberova J, Camacho A. Global phylogeography of pelagic Polynucleobacter bacteria: Restricted geographic distribution of subgroups, isolation by distance, and influence of climate. Environmental Microbiology. 2015;17(3):829–840. doi: 10.1111/1462-2920.12532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Pöckl M, Wu QL. Low intraspecific diversity in a Polynucleobacter subcluster population numerically dominating bacterioplankton of a freshwater pond. [yes] Applied and Environmental Microbiology. 2005;71(8):4539–4547. doi: 10.1128/AEM.71.8.4539-4547.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann K, Schmidt HJ. Polynucleobacter necessarius gen. nov., sp. nov., an obligately endosymbiotic bacterium living in the cytoplasm of Euplotes aediculatus . International Journal of Systematic Bacteriology. 1987;37(4):456–457. [Google Scholar]
- Hijmans RJ. Geosphere: Spherical trigonometry. R package version 1.5-10. 2019 https://CRAN.R-project.org/package=geosphere.
- Hijmans RJ, Guarino L, Cruz M, Rojas E. Computer tools for spatial analysis of plant genetic resources data: 1. DIVA-GIS. Plant Genetic Resources Newsletter. 2001;127:15–19. [Google Scholar]
- Hill JE, Seipp RP, Betts M, Hawkins L, Van Kessel AG, Crosby WL, Hemmingsen SM. Extensive profiling of a complex microbial community by high-throughput sequencing. Applied and Environmental Microbiology. 2002;68(6):3055–3066. doi: 10.1128/aem.68.6.3055-3066.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoetzinger M, Hahn MW. Genomic divergence and cohesion in a species of pelagic freshwater bacteria. BMC Genomics. 2017;18(1):794. doi: 10.1186/s12864-017-4199-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoetzinger M, Pitt A, Huemer A, Hahn MW. Continental-scale gene flow prevents allopatric divergence of pelagic freshwater bacteria. Genome Biol Evol. 2021;13(3):evab019. doi: 10.1093/gbe/evab019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoetzinger M, Schmidt J, Jezberová J, Koll U, Hahn MW. Microdiversification of a pelagic Polynucleobacter species is mainly driven by acquisition of genomic islands from a partially interspecific gene pool. Applied and Environmental Microbiology. 2017;83(3):e02266-02216. doi: 10.1128/aem.02266-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner-Devine MC, Lage M, Hughes JB, Bohannan BJ. A taxa-area relationship for bacteria. Nature. 2004;432(7018):750–753. doi: 10.1038/nature03073. [DOI] [PubMed] [Google Scholar]
- Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nature Communications. 2018;9(1):5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspers E, Overmann J. Ecological significance of microdiversity: Identical 16S rRNA gene sequences can be found in bacteria with highly divergent genomes and ecophysiologies. Applied and Environmental Microbiology. 2004;70(8):4831–4839. doi: 10.1128/AEM.70.8.4831-4839.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jezbera J, Jezberova J, Brandt U, Hahn MW. Ubiquity of Polynucleobacter necessarius subspecies asymbioticus results from ecological diversification. Environmental Microbiology. 2011;13(4):922–931. doi: 10.1111/j.1462-2920.2010.02396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jezbera J, Jezberova J, Koll U, Hornak K, Simek K, Hahn MW. Contrasting trends in distribution of four major planktonic betaproteobacterial groups along a pH gradient of epilimnia of 72 freshwater habitats. FEMS Microbiology Ecology. 2012;81(2):467–479. doi: 10.1111/j.1574-6941.2012.01372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jezberova J, Jezbera J, Brandt U, Lindstrom ES, Langenheder S, Hahn MW. Ubiquity of Polynucleobacter necessarius ssp. asymbioticus in lentic freshwater habitats of a heterogenous 2000 km2 area. Environmental Microbiology. 2010;12(3):658–669. doi: 10.1111/j.1462-2920.2009.02106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis KT, Ramette A, Tiedje JM. The bacterial species definition in the genomic era. Philosophical Transactions of the Royal Society B: Biological Sciences. 2006;361(1475):1929–1940. doi: 10.1098/rstb.2006.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer SA, Barbosa da Costa N, Shapiro BJ, Fradette M, Huot Y, Walsh DA. A large-scale assessment of lakes reveals a pervasive signal of land use on bacterial communities. The ISME Journal. 2020 doi: 10.1038/s41396-020-0733-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer SA, Boynton PJ. Evidence for microbial local adaptation in nature. Molecular Ecology. 2017;26(7):1860–1876. doi: 10.1111/mec.13958. [DOI] [PubMed] [Google Scholar]
- Langenheder S, Lindström ES. Factors influencing aquatic and terrestrial bacterial community assembly. Environmental Microbiology Reports. 2019;11(3):306–315. doi: 10.1111/1758-2229.12731. [DOI] [PubMed] [Google Scholar]
- Minka TP, Deckmyn A. Maps: Draw geographical maps. R package version 3.3.0. 2018 https://CRAN.R-project.org/package=maps.
- Nemergut DR, Schmidt SK, Fukami T, O’Neill SP, Bilinski TM, Stanish LF, et al. Ferrenberg S. Patterns and processes of microbial community assembly. Microbiology and molecular biology reviews : MMBR. 2013;77(3):342–356. doi: 10.1128/MMBR.00051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton RJ, McLellan SL. A unique assemblage of cosmopolitan freshwater bacteria and higher community diversity differentiate an urbanized estuary from oligotrophic Lake Michigan. Frontiers in Microbiology. 2015;6:1028. doi: 10.3389/fmicb.2015.01028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuy JK, Hoetzinger M, Hahn MW, Beisser D, Boenigk J. Ecological differentiation in two major freshwater bacterial taxa along environmental gradients. Frontiers in Microbiology. 2020;11(154) doi: 10.3389/fmicb.2020.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogier J-C, Pagès S, Galan M, Barret M, Gaudriault S. rpoB, a promising marker for analyzing the diversity of bacterial communities by amplicon sequencing. BMC Microbiology. 2019;19(1):171. doi: 10.1186/s12866-019-1546-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen FJ, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, et al. Wagner H. vegan: Community Ecology Package. R package version 2.5-6. 2019 https://CRAN.R-project.org/package=vegan.
- Peixoto JC, Leomil L, Souza JV, Peixoto FB, Astolfi-Filho S. Comparison of bacterial communities in the Solimoes and Negro River tributaries of the Amazon River based on small subunit rRNA gene sequences. Genetics and Molecular Research. 2011;10(4):3783–3793. doi: 10.4238/2011.December.8.8. [DOI] [PubMed] [Google Scholar]
- Percent SF, Frischer ME, Vescio PA, Duffy EB, Milano V, McLellan M, et al. Nierzwicki-Bauer SA. Bacterial community structure of acid-impacted lakes: What controls diversity? Applied and Environmental Microbiology. 2008;74(6):1856–1868. doi: 10.1128/aem.01719-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira RPA, Peplies J, Mushi D, Brettar I, Hofle MG. Pseudomonas-Specific NGS Assay Provides Insight Into Abundance and Dynamics of Pseudomonas Species Including P. aeruginosa in a Cooling Tower. Frontiers in Microbiology. 2018;9 doi: 10.3389/fmicb.2018.01958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt A, Schmidt J, Lang E, Whitman WB, Woyke T, Hahn MW. Polynucleobacter meluiroseus sp. nov. a bacterium isolated from a lake located in the mountains of the Mediterranean island of Corsica. International Journal of Systematic and Evolutionary Microbiology. 2018;68(6):1975–1985. doi: 10.1099/ijsem.0.002777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2019. URL https://www.R-project.org/ [Google Scholar]
- Reche I, Pulido-Villena E, Morales-Baquero R, Casamayor EO. Does ecosystem size determine aquatic bacterial richness? Ecology. 2005;86(7):1715–1722. [Google Scholar]
- Rodriguez-R LM, Castro JC, Kyrpides NC, Cole JR, Tiedje JM, Konstantinidis KT. How Much Do rRNA Gene Surveys Underestimate Extant Bacterial Diversity? Applied and Environmental Microbiology. 2018;84(6):e00014-00018. doi: 10.1128/aem.00014-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez D, Matthijs S, Gomila M, Tricot C, Mulet M, García-Valdés E, Lalucat J. rpoD gene pyrosequencing for the assessment of Pseudomonas diversity in a water sample from the Woluwe River. Applied and Environmental Microbiology. 2014;80(15):4738–4744. doi: 10.1128/aem.00412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer M, Kamenik C, Hahn MW. Ecological differentiation within a cosmopolitan group of planktonic freshwater bacteria (SOL cluster, Saprospiraceae, Bacteroidetes) Applied and Environmental Microbiology. 2005;71(10):5900–5907. doi: 10.1128/aem.71.10.5900-5907.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, et al. Herndl GJ. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(32):12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackebrandt E, Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiology Today. 2006;33:152–155. [Google Scholar]
- Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag; New York: 2016. [Google Scholar]
- Zhou J, Ning D. Stochastic community assembly: Does it matter in Microbial Ecology? Microbiology and Molecular Biology Reviews. 2017;81(4):e00002-00017. doi: 10.1128/mmbr.00002-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart G, Crump BC, Agterveld M, Hagen F, Han SK. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquatic Microbial Ecology. 2002;28(2):141–155. doi: 10.3354/ame028141. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.