Introductory Paragraph
Although deep learning algorithms show increasing promise for disease diagnosis, their use with rapid diagnostic tests performed in the field has not been extensively tested. Here, we use deep learning to classify images of rapid HIV tests acquired in rural South Africa. Using newly developed image capture protocols with the Samsung SM-P585 tablet, 60 fieldworkers routinely collected images of HIV lateral-flow tests. From a library of 11,374 images, deep learning algorithms were trained to classify tests as positive or negative. A pilot field study of the algorithms deployed as a mobile application demonstrated high levels of sensitivity (97.8%) and specificity (100%), compared to traditional visual interpretation by humans -experienced nurses and newly trained community health worker staff - and reduced the number of false positives and false negatives. Our findings lay the foundations for a new paradigm of deep learning-enabled diagnostics in low- and middle-income countries, termed REASSURED diagnostics1, for Real-time connectivity, Ease of specimen collection, Affordable, Sensitive, Specific, User-friendly, Rapid, Equipment-free, and Deliverable. Such diagnostics have the potential to provide a platform for workforce training, quality assurance, decision support, and mobile connectivity to inform disease control strategies, strengthen healthcare system efficiency, and improve patient outcomes and outbreak management of emerging infections.
Keywords: Rapid diagnostic test, deep learning, HIV
Rapid diagnostic tests (RDTs) save lives by informing case management, treatment, screening, disease control and elimination programmes1. Lateral flow tests are among the most common RDTs and hundreds of millions of these tests are performed worldwide each year. They have the potential to support near person testing and decentralised management of a range of clinically important diseases (including malaria, HIV, syphilis, tuberculosis, influenza and non-communicable diseases2), making it convenient for the end-user and more affordable for health systems3. RDT also present some issues, namely: errors in performing the test and interpreting the result4,5, quality control, and lack of electronic data capture records of the test and results within health systems and surveillance. Many of these would be overcome with the ‘R’ in REASSURED -the new criteria for an ideal test to reflect the importance of digital connectivity, coined by Peeling and coworkers1. The ‘R’ stands for ‘real-time connectivity’ using mobile phone connected RDTs. To date there have been few peer reviewed studies or evaluations of the effectiveness of connected lateral flow tests at scale in populations in need in low- and middle-income countries.
Recent studies that compare the human interpretation of a HIV RDT to various gold standards, such as Western Blot6–9, Enzyme Immunoassay7,9–11, standardised test panels12 or different HIV RDTs13–15, have highlighted the common issue of subjective interpretation of the test result, which can lead to incorrect diagnosis. User error (especially in the case of weak reactive lines) and inadequate supervision of testers were identified as prime factors for misinterpretation16. In a study of differently experienced users interpreting results of HIV RDTs by looking at pictures of tests17, the accuracy of interpretation varied between 80% and 97%. This highlights the importance of experience in reading the test, as well as the subjectivity involved in reading a weak test line. Evidence also suggests that some fieldworkers struggle to interpret RDTs because of colour blindness or short-sightedness.18 Another study used photographs of HIV RDTs to quantify the subtle difference in tests with faint lines declared as True- or False-positive by a panel of human users19. While these were small-scale studies (N = 148 and 8, respectively), both highlighted the potential for photographs to improve quality control and decision-making.
Deep learning algorithms, harnessing advances in large data sets and processing power, have recently shown the ability to exceed human performance in a plethora of visual tasks, including cell-based diagnostics20, interpreting dermatology21, ophthalmology22 and radiography images23, playing strategic games24, and in clinical medicine when used alongside appropriate guidelines25,26. While some studies are emerging looking at applying deep learning to the interpretation of RDT27,28, little is known about the ability of machine learning models to analyse field-acquired diagnostic test data, with concerns about the potential uniformity of images (e.g. focus, tilt), harsh environmental factors such as lighting, and the variety of test types. In addition, there is a general lack of large real-world datasets available to successfully train deep learning classifiers, particularly from low- and middle-income countries. Recent advances in consumer electronic devices and deep learning, have the potential to improve RDT quality assurance, staff training and connectivity, eventually supporting self-testing, such as HIV-self-testing, which has been shown to be cost-effective29, to appeal to young people30 and help reduce anxiety31.
Mobile health (mHealth) approaches, which marry RDTs with widely available mobile phones, take advantage of inbuilt sensors (e.g. cameras) found in the phones, battery life, processing power, screens to display results, and connectivity to send results to health databases. A recent field study has shown high levels of acceptability for a device sending HIV RDT results to online data bases in real-time32. An array of approaches have been piloted at small scales (N ≤ 283) and have shown good performance. However, most require a physical attachment, such as a dongle (92-100% sensitivity, 97-100% specificity)33, a cradle34, or a portable reader (97-98% sensitivity)35, which increases cost and complexity, and typically rely on simple image analysis software.
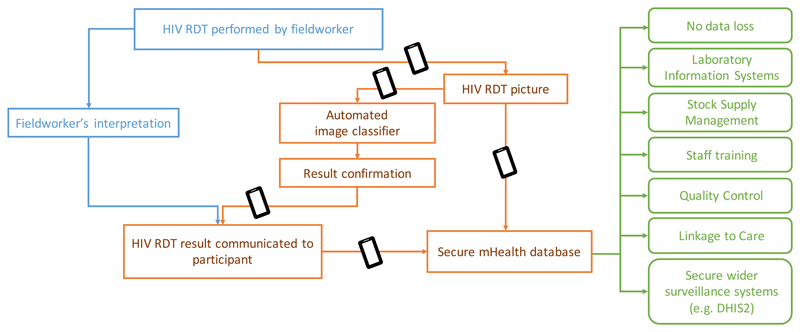
We explore the potential of deep learning algorithms to classify field-based RDT images as either positive or negative, focusing on HIV as an exemplar, and piloting at scale in population ‘test beds’ in KwaZulu-Natal, typical of semi-rural settings in Sub-Saharan Africa. Figure 1 shows the concept of our deep learning-enabled REASSURED diagnostic system to capture and interpret RDT results. Our approach first involved building a large image library of field-acquired test images as training data set, optimising algorithms for high sensitivity and specificity, and then to deploy our classifier in a pilot study to assess its performance compared to traditional visual interpretation with a range of end users with varying levels of training.
Figure 1. Infographic to illustrate the benefits of data capture to support field decisions.
In blue, the current workflow used by fieldworkers. In orange, our proposed mHealth system of automated RDT classifier plus data capture and transmission to a secure mHealth database. In green, the benefits arising from deploying the proposed system. The black rectangle represents a tablet or smartphone.
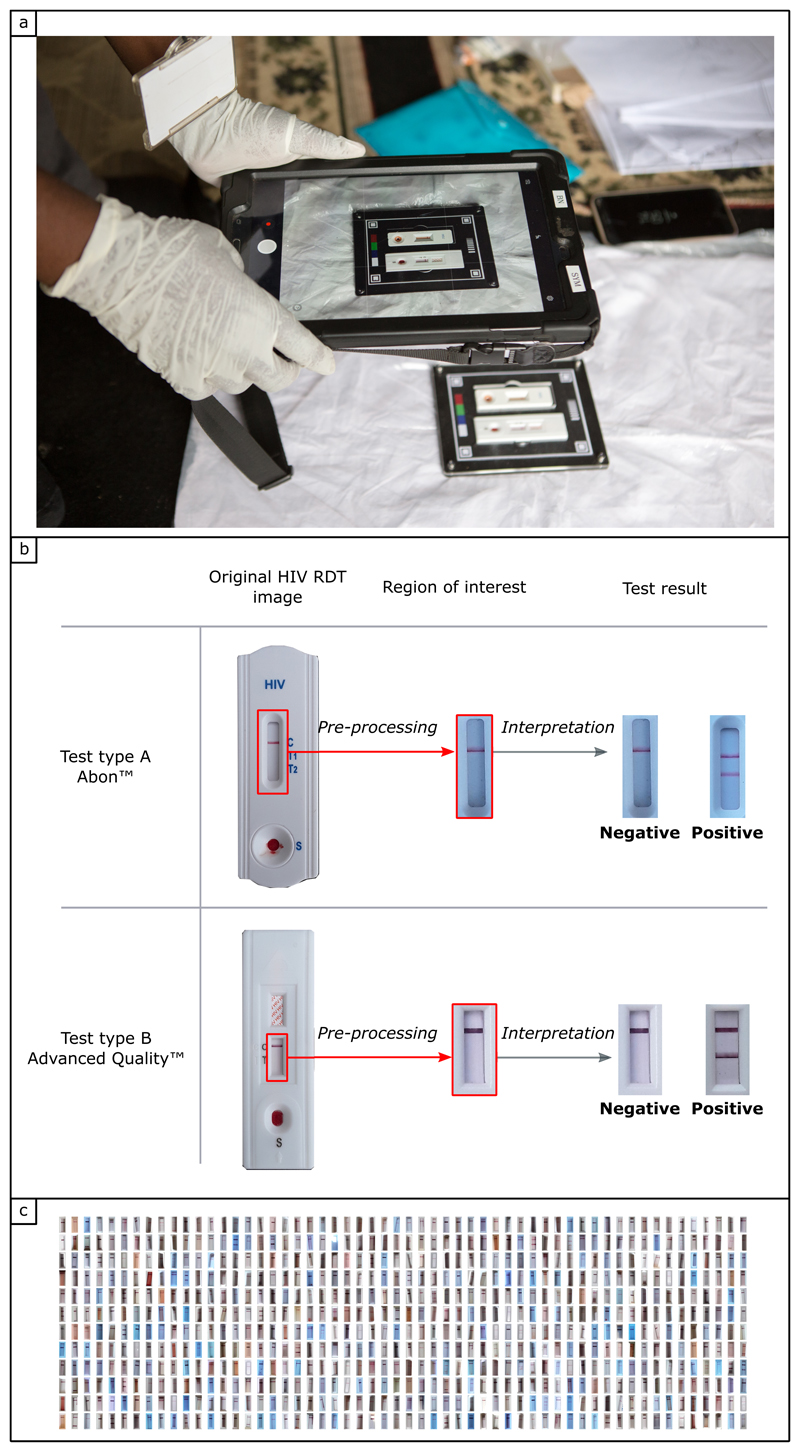
Our standard image collection protocol (Figure 2a) and library are described in the Methods section. In brief, 11,374 photographs of HIV RDT were captured by over 60 fieldworkers using Samsung tablets (SM-P585, 8Megapixel camera, f1/9, with autofocus capability). Embedding routine image collection into staff workflows was acceptable and feasible, and participant consent rate was 96%. We optimised our mHealth system for the two different HIV RDTs used in the study as part of routine household population surveillance. At first glance these RDTs appear similar but have different features and number of test lines. To reduce the number of variables, we cropped the images around the region of interest (ROI) (Figure 2b). Figure 2c shows a snapshot of the very diverse real-world field conditions where the images were captured (indoors, outdoors, in the shade and in direct sunlight).
Figure 2. Standardisation of image capture, image pre-processing and training library.
a) Fieldworker capturing a photograph of two HIV RDTs at the time of interpretation, in the field in rural South Africa (photo credit: Africa Health Research Institute). The two HIV RDTs are fitted in a plastic tray designed to standardise image capture and facilitate image pre-processing. b) Interpretation process, starting from the original picture of HIV RDTs used during the study, pre-processing to select the region of interest (ROI), then interpretation of the test result. If two lines (control + test) are present on the paper strip at the time of interpretation, the test result is positive. Note: for the ABON HIV RDT, one or two different test lines can appear (T1 and T2) depending on the type of HIV infection (HIV-1 and HIV-2, respectively). The test result is positive regardless of which test line is present, or if both test lines are present on the paer strip at the time of interpretation. If only the top line (control) is present, the test is negative. If no control line can be seen, the test is deemed invalid. c) Snapshot of the image library of HIV RDTs collected in the field in rural South Africa (162 randomly selected images out of 11374), illustrating the diversity of the colour, background and brightness.
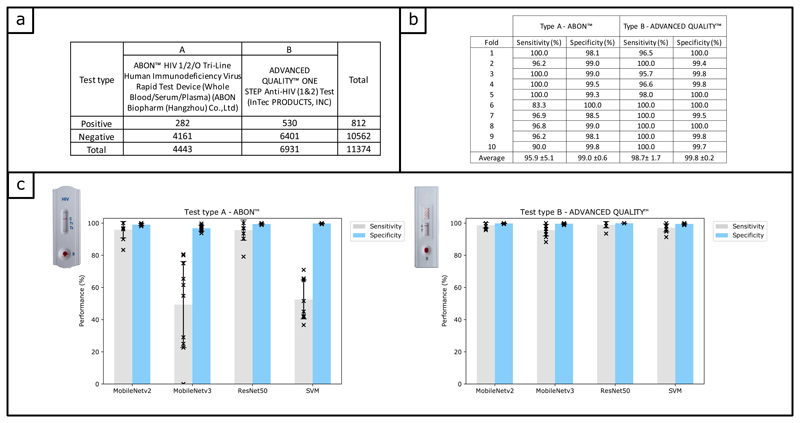
Each image was labelled (see Online Methods) according to the test result. Figure 3a details the number of images used to train classifiers to automatically read the result of HIV RDT images. The training process is described in the Online Methods section. In order to test the reproducibility of the process, we performed a 10-fold cross validation. As can be seen in Figure 3b, the average sensitivity (95.9% ±5.1 for type A, 98.7% ±1.7 for type B) and specificity (99.0% ±0.6 for type A, 99.8% ±0.2 for type B) achieved across the 10 folds was high and consistent for both types of HIV RDT. We therefore used all the available data to train a final classifier for each type of test, which were used in our field study. We investigated different common classification methods being used for clinical diagnostic (Support Vector Machine36 (SVM) and Convolutional Neural Networks (CNN)) including 3 different CNN architectures (ResNet5037, MobileNetV238,39 and MobileNetV340), and found MobileNetV2 was the most appropriate for our task, as can be seen in Figure 3c.
Figure 3. Algorithm training and performance.
a) Table showing the number of images in the training library, divided in two labels categories (‘positive’ and ‘negative’) as well as two sub-categories corresponding to the test type. b) Table to summarise the training process using cross-validation, with a training set of N = 3998 (test type A) and N = 6221 (test type B). The sensitivity and specificity were obtained using a hold-out testing dataset of N = 445 (test type A) and N = 693 (test type B). c) Barplots showing the average performance (sensitivity and specificity) of 4 classification methods trained on our dataset, using cross validation (the error bars represent the standard devidation from the mean). The three CNN pretrained on the ImageNet dataset (ResNet50, MobileNetV2 and MobileNetV3) were retrained and tested using our dataset. The SVM was trained using features extracted by Histogram of Oriented Gradients. All four classifiers were trained using the same training set described in panel b). The sensitivity and specificity were obtained using the hold-out testing dataset described in panel b).
We then conducted a field pilot study in rural South Africa to assess the performance of our mHealth system compared to visual interpretation with a range of end-users with varying levels of training (see Online Methods). Five participants (2 nurses, 3 newly trained community healthworkers) were each asked to give their interpretation of 40 HIV RDTs and to acquire a photograph of the RDT via the app. All five participants (100%) were able to use our mHealth system without training, demonstrating its feasibility and acceptability. The photographs were then evaluated by an expert RDT interpreter, followed by our deep learning algorithms on a secure server. The results were not fed back to the study participants to avoid confirmation bias. The performance results can be seen in Figure 4.
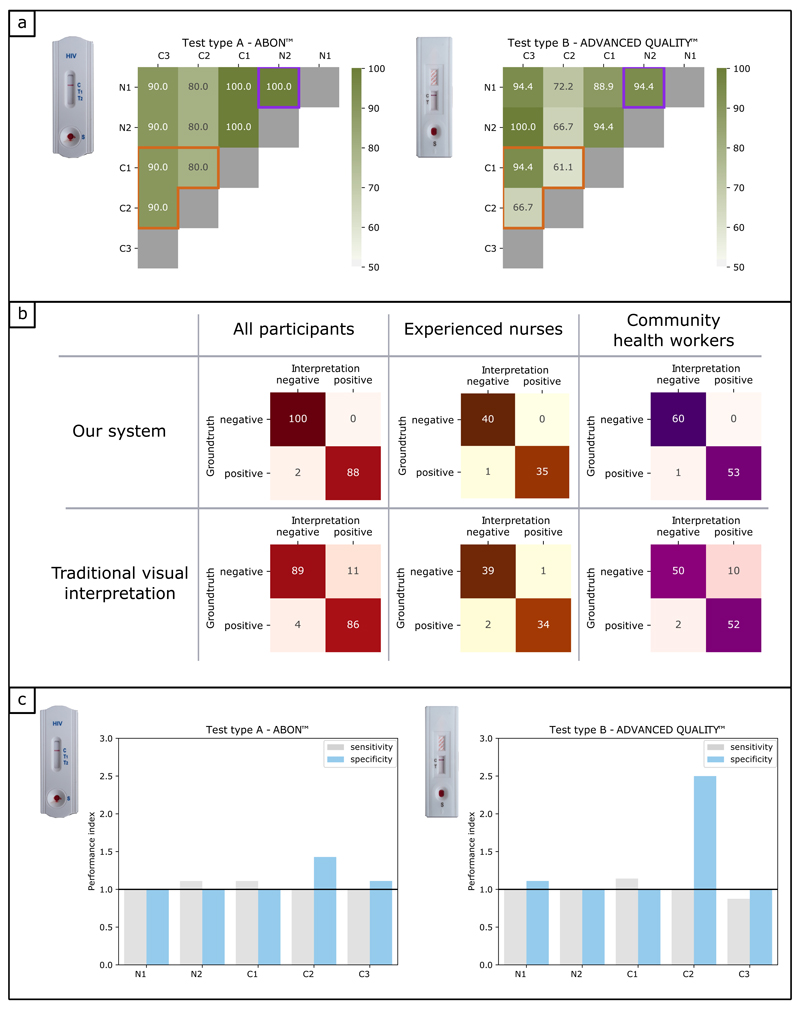
Figure 4. Performance evaluation of our mHealth system compared to traditional visual interpretation, field pilot study.
a) Graphics showing the agreement (%) between pairs of study participants, when asked to interpret HIV RDTs results using traditional visual interpretation. Participants are divided between experienced nurses (N1, N2) and community health workers (C1, C2, C3). For each pair of participants, the number of HIV RDTs was N = 38. The observations are separated according to the two types of HIV RDTs used in the study. The purple square on both graphics highlights the agreement between the two experienced nurses, while the orange polygon highlights the agreement between the three pairs of community health workers. b) Confusion matrices showing the number of True Negative, False Positive, False Negative and True Positive results, when comparing the interpretation of our mHealth system (top row) and traditional visual interpretation (bottom row) to the groundtruth. Red matrices on the left include the results for all study participants, which are broken down into experienced nurses (orange matrices) and community health workers (purple matrices). c) Barplots showing the performance index for individual participants. Participants are divided between experienced nurses (N1, N2) and Community health workers (C1, C2, C3). The performance index is the ratio of the performance of our mHealth system over that of traditional visual interpretation. A performance index greater (or equal) to one indicates our mHealth system performed better than (or as well as) traditional visual interpretation. The observations are separated according to the two types of HIV RDTs used in the study.
When comparing the traditional visual interpretation of the RDTs, we observed varied levels of agreement between participants, (61-100%) as can be seen in Figure 4a. As expected, agreement between nurses (N1 & N2: 100% and 94.4% agreement for test types A and B respectively) was greater than between newly trained community health workers (C1, C2 & C3: 80-90% and 61.1-94.4% for test types A and B, respectively). Test type B showed the lower level of agreement. The low level of agreement between participants, and variability due to the type of HIV RDT, were of concern and highlighted the need for a more objective and consistent method to interpret HIV RDTs in the field. The confusion matrices in Figure 4b, demonstrate our mHealth system reduced the number of errors in reading RDTs. The number of False Positive results from our mHealth system was found to be significantly lower than for the traditional visual interpretation (0 compared to 11 – the largest variation being observed for community health workers, 10), which translates as an improvement in specificity from 89% to 100%, and an improvement in Positive Predictive Value from 88.7% to 100%. Similarly, the number of False Negative results was just two in our mHealth system, compared to four in traditional visual interpretation, which translates as an improvement in sensitivity from 95.6% to 97.8%, and an improvement in Negative Predictive Value from 95.7% to 98%. We plotted the ratio of our mHealth system performance to the participant performance, both for sensitivity and specificity (Figure 4c). All participants had a sensitivity index equal or greater than one for test type A; four out of five participants (N1, N2, C1, C2) also did for test type B, demonstrating our mHealth system was better than those participants at reading positive test results. Our system was also more reliable at reading negative tests, as all participants had a specificity index equal or greater than one for both types of HIV RDTs.
We acknowledge the following limitations of our study. Firstly, our pilot study involved a relatively small number of participants (five), although we note this is comparable to other similar pilot studies reported in the field. In future, larger evaluation studies and clinical trials are needed to assess the performance of the system, involving participants with a broader range of demographics including age, gender and different levels of digital literacy, as well as more expert readers. In addition, future studies would benefit from including an invalid test classifier and different mobile phone types with varying camera specifications. The images were analysed on a secure server, however, future analysis could be on-device overcoming the need to upload images. We are also currently investigating a picture segmentation approach using deep learning for the next iteration of the smartphone application.
To conclude, we demonstrated the potential of deep learning to accurately classify RDT images, with an overall performance of 98.9% accuracy, significantly higher than traditional visual interpretation of study partipants (92.1%), which are comparable with reports of 80-97% accuracy17. Given that over 100 million HIV tests are performed annually, even a small improvement in quality assurance could impact the lives of millions of people by reducing the risk of false positives and negatives. To the best of our knowledge our real-world image library is the first of its kind at this scale and we demonstrate that deep learning models can be deployed in mobile devices in the field, without the need for cradles, dongles or other attachments. It lays the foundation for deep learning enabled REASSURED diagnostics, demonstrating that RDTs linked to a mobile device could standardise capture and interpretation of test results for decision-makers, reducing interpretation and transcription errors and workforce training. Our findings are based on HIV testing decision support for fieldworkers, nurses and community health workers, but in future could be applicable to decision support for self-testing. We focused on HIV as an exemplar, but the capacity of the classifier to adapt to two different test types suggests that it is amenable to a large range of RDTs spanning communicable and non-communicable diseases. This platform could be utilised for workforce training, quality assurance, decision support, and mobile connectivity to inform disease control strategies, strengthen healthcare systems efficiency, and improve patient outcomes, and outbreak management. The ideal connected system would link to connected RDTs to laboratory systems, whereby remote monitoring of RDT functionality and utilisation could also allow health programmes to optimise testing deployment and supply management to deliver the Sustainable Development Goals and ensure no one is left behind. The real-time alerting capability of connected RDTs could also support public health outbreak management, by mapping ‘hotspots’ for epidemics including COVID-19 to protect populations.
Methods
Ethics
Ethical approval for the demographic surveillance study was granted by the Biomedical Research Ethics Committee of the University of KwaZulu-Natal, South Africa, Reference Number BE435/17. Separate informed consent is required for the main household survey, for the HIV sero-survey, the HIV point of care test and the photographs of the HIV test.
Ethical approval for the collection of human blood samples used in the pilot study was granted by the Biomedical Research Ethics Committee of the University of KwaZulu-Natal, South Africa, Reference Number BFCJ 11/18.
Recruitment of participants to AHRI Population Implementation Platform for the image library
Eligible participants are all individuals age 15 years and older resident within the geographic boundaries of the AHRI population intervention programme surveillance area (Cohort profile: Africa Centre demographic information system (ACDIS) and population-based HIV survey. International journal of epidemiology. 2007 Nov 12;37(5):956-62.). Individuals who have died or outmigrated prior to the surveillance visit are no longer eligible. There are three contact attempts by the fieldworker team and a further three contact attempts by a tracking team before the individual is considered to be uncontactable. All individuals in the study gave informed consent. Specifically, all contacted eligible individuals who gave informed consent for this study were offered a rapid HIV test if they were not currently on anti-retroviral therapy. For children under the age of 18, written consent for Rapid HIV testing was obtained for the parent or guardian and assent from the participant.
HIV RDT Image library collection
The original RDT images library was collected in rural South Africa by a team of 60 fieldworkers (between 2017 and 2019). AHRI fieldworkers survey a population of 170,000 people in rural KwaZulu-Natal. Participants were visited at their home, those giving informed consent were tested for HIV using a combination of two HIV RDTs, and upon further consent, a picture of their two HIV RDTs was captured by the fieldworker on a tablet at the time of interpretation. Both HIV RDTs were used as part of routine demographic surveillance in Africa Health Research Institute. The test type continued to change during this study following recommendations by the South African government, exemplifying the need for robust systems to read multiple test formats.
While the two HIV RDTs used in this study have their own instructions for use (see manufacturer’s instructions), they all generally follow the same principle of collecting a drop of blood from the participant’s fingertip, delivering that drop of blood to the sample pad and using a drop of chase buffer to help the blood sample flow through the length of the paper strip. The result (a combination of one or two lines appearing on the paper strip) is then read out after a period of 10 to 40 min, depending on the type of HIV RDT used.
In order to least disturb the fieldworker’s workflow, a plastic tray designed to hold both HIV RDT was given to each fieldworker. A picture of the tray can be seen in Figure 2a. This ensured the fieldworkers only had to capture one picture per participant. The tasks of separating the two HIV RDT and isolating the ROI used to train the classifier were conducted down the line as part of data pre-processing.
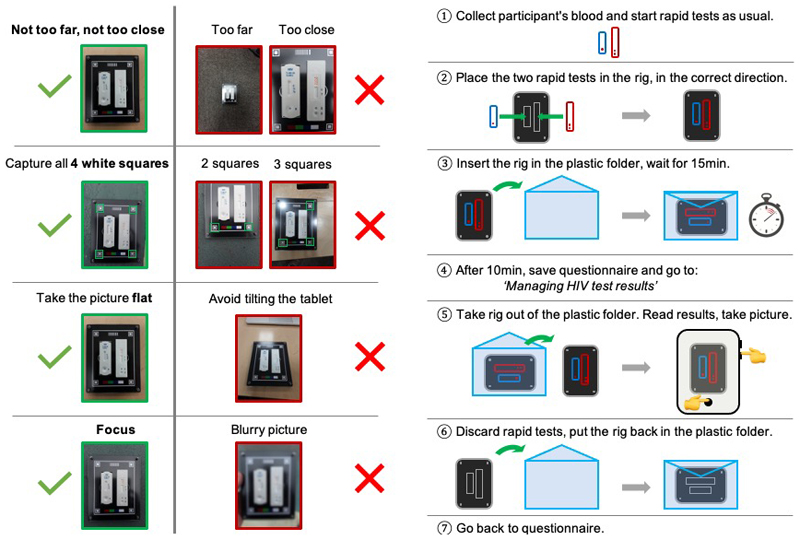
A standard operating procedure (SOP) on how to capture the image was co-created and optimised with the team of fieldworkers. A copy of the SOP can be found in the Extended Data section (Extended Data Figure 1). The SOP was designed to minimise the impact of environmental factors, as well as to ensure a standard way of capturing the pictures. All fieldworkers attended a two-day initial training programme during which the objectives of the data collection and design of the plastic tray were clearly explained, and each fieldworker was personally trained and given feedback on how to capture valid photographs. A training protocol was also established, in order to ensure newly enrolled fieldworkers who did not attend the initial training session could also be trained to capture pictures for the project. Finally, picture quality assessment sessions were conducted in order to give the fieldworkers team feedback, and to ensure most pictures were of high enough quality to be used for training the classifier.
All pictures were captured using Samsung tablets (SM-P585, 8MPixels camera, f1/9, with autofocus capability) using the native Android camera application, stored on the device until the end of the day when they were transferred to a secure database at AHRI. Our mHealth system only allows one picture per test and per participant to be saved to the tablet and uploaded to the AHRI database. After anonymisation (including stripping geo-coordinates from the picture EXIF data), batches of 2000-3000 pictures were securely transferred to UCL team members on a quarterly basis, and stored securely in a ‘Data Safe Haven’ managed by the university.
Both the feasibility (93%) and acceptability (98%) of the system used to capture the HIV RDTs pictures were high, according to a survey taken by the fieldworkers involved in the study.
For the purpose of this study, an initial batch of 11, 374 images were used. As only very few invalid results were obtained from the field, it was decided, for the purpose of this proof of concept study, to focus on training the classifier to distinguish between positive and negative results. In order to optimise this task, the ROI around each HIV RDT was isolated and used to train the classifier.
Image labelling
All pre-processed images were labelled by a group of three RDT experts (99.2% agreement with fieldworkers labelling). Labelling is the process of sorting the images into categories, which are then used to train the classifier. The categories chosen here correspond to the possibilities for the HIV RDT result, i.e. ‘positive’ and ‘negative’. We recognise that a third outcome, ‘invalid’, is also possible and needs to be considered when using the system to provide a confident diagnostic. However, the absence of invalid test results in our library of images collected by fieldworkers did not allow us to train the classifier on this third category in this study. We therefore focused the training on the two main categories (‘positive’ and ‘negative’), and are exploring other ways to incorporate the ‘invalid’ outcome in our mHealth system. This could mean either using data augmentation techniques on the low numbers of invalid test results images, or adding a pre-processing step to detect the presence of a control line on the image before deciding to feed it (or not, in case the control line is absent) to the classifier.
Training library
The labelled images were divided into two sub-categories corresponding to the HIV RDT type. The two types of tests in our library are:
Type A: ABON™ HIV 1/2/O Tri-Line Human Immunodeficiency Virus Rapid Test Device (Whole Blood/Serum/Plasma) (ABON Biopharm (Hangzhou) Co., Ltd)
Type B: ADVANCED QUALITY™ ONE STEP Anti-HIV (1&2) Test (InTec PRODUCTS, INC)
While there are two tests per patient, herein in this study we treat each test individually since the tests are from different manufacturers and therefore could respond differently to the same blood sample. The collection system design also guaranteed that there was never more than one image of a given test per participant.
Image normalisation
Before being used for training, each image was resized to the dimensions of the input layer then standardised. Standardisation of the data was performed using equation (1) below, where xs is the standardised pixel value, xo the original pixel value, μ and σ are the mean and standard deviation of all pixels in the image, respectively.
| Equation (1) |
Cross-validation
Each dataset (one for each type of HIV RDT) was randomly divided into 10 equal folds. Using the leave-one-out method, 10 classifiers were trained using nine folds as the training set (further randomly divided into 80% training and 20% validation). To account for imbalanced datasets (roughly 13:1 negative:positive ratio), we forced every batch during training to contain 50% positive images and 50% negative images using random sampling. Each model was then optimised by creating a ROC curve using the validation set. This yielded an optimal threshold which was used to evaluate the model performance model on the testing set (remaining 10th fold). The deployment models were obtained by retraining using all the available data, for each type of HIV RDT. All training and evaluation were conducted using the scikit-learn and Tensorflow libraries in Python.
Comparison with established classification methods
The SVM was trained using pre-processed features extracted using Histogram of Oriented Gradients (HOG), with Principal Component Analysis used to filter out less significant features. The three CNN (ResNet50, MobileNetV2 and MobileNetV3) were pre-trained using the ImageNet dataset, and re-trained using our dataset. For all four methods, training and evaluation was conducted using the scikit-learn and Tensorflow libraries in Python.
Android application
We developed a smartphone/tablet Android application designed for end-users to capture a picture of their HIV RDT, at the time of reading the test result. Together with end users, we optimised the design so as to maximise the simplicity of the process, in order to make our mHealth system accessible to end users with a broad range of digital literacy. All that is required from the end user is to roughly align a semi-transparent template of the HIV RDT with their HIV RDT and press a button to capture a picture. Cropping around the ROI was then performed automatically in the background (using the pixel coordinates of the template overlay), as was the process of sending the ROI to our classifier and receiving our mHealth system’s result. For the purpose of this pilot study, participants were not made aware of our mHealth system’s interpretation of the test results, so as to avoid bias for their own interpretation. Screenshots of the application can be found in the Extended Data section (Extended Data Figure 2).
Field pilot study protocol
The Android application was deployed in a field pilot study in KwaZulu Natal, South Africa. Five participants were randomly selected from the staff at AHRI – two experienced nurses and three community healthworkers. 40 HIV RDT (20 of type A, 20 of type B) were performed following manufacter’s guidelines using discarded anonymised human blood samples (10 positive, 10 negative according to ELISA). For each of the 40 HIV RDTs, each participant was asked to record their visual interpretation of the test result, then use our mHealth system on a tablet to capture a photograph of the HIV RDT. The system consisted of our Android app (described above), installed on a single Samsung SM-P585 tablet, identical to the ones used by fieldworkers for data collection. Participants were not shown the automated interpretation of the test result provided by our mHealth system in order to avoid confirmation bias. The field pilot study took place at the AHRI rural site at the heart of the community (Mtubatuba, KwaZulu-Natal), under lighting conditions identical to the ones the mHealth system is intended to be used. A short (10 minutes) demonstration on how to use the smartphone application was given to all participants, who were then left on their own to proceed with the task of reading the HIV RDTs and capturing pictures.
Field pilot study data analysis
The data analysis consisted of the comparison of three datasets:
-
i)
Traditional visual interpretation by study participants
-
ii)
Independent expert interpretation of the images captured by study participants
-
iii)
Automated machine learning interpretation by our classifier
Traditional visual interpretaiton was recorded on the tablet by each study participant immediately after being shown the HIV RDTs. Only two of the 40 HIV RDTs (corresponding to 10 images out of 200) had to be discarded from the analysis, as one participant took a photograph of the wrong HIV RDTs and it was therefore not possible to compare interpretation results across all five participants.
An independent RDT expert subsequently visually interpreted all 190 HIV RDTs images. The independent RDT expert had significant experience conducting performance evaluations of lateral flow rapid tests for ocular and genital Chlamydia trachomatis in The Phillippines, The Gambia and Senegal. The visual interpretation occurred 1-5 hours after sample addition. The independent expert certified that none of the HIV RDT results had changed during this time frame.
The automated machine learning interpretation by our classifiers occurred on our secured server. The results were compared to traditional visual interpretation and the independent RDT expert, shown in the confusion matrices in Figure 4, then analysed using the performance indicators described below.
Performance indicators
The four indicators of performance investigated were sensitivity, specificity, positive predictive value (PPV) and negative predicitve value (NPV). For each image, the classifier produces an outcome that belongs to either of the four categories: True Positive (TP), True Negative (TN), False Positive (FP) and False Negative (FN). Whether the outcome is True or False depends on the comparison with the gold standard chosen.
The sensitivity is the ability of the classifier to correctly detect a positive result, by measuring the ratio , while the specificity is the ratio and translates the ability of the classifier to correctly detect a negative result. The PPV is the ratio , the NPV is the ratio . They indicate the proportion of positive and negative results (respecitvely) by a diagnostic test that are true positves and true negatives (respectively).
Extended Data
Extended Data Figure 1.
Extended Data Figure 2.
Acknowledgements
We thank the community of the uMkhanyakude district and the study participants, as well as the AHRI team of fieldworkers and their supervisors. We thank A. Koza, Z. Thabethe, T. Madini, N. Okesola and S. Msane for their help with the pilot study; D. Gareta and J. Dreyer for IT support; V. Lampos and I. J. Cox for useful discussions; and E. Manning and J. McHugh for their help with editing and project management. This research was funded by the m-Africa Medical Research Council GCRF Global Infections Foundation Award (no. MR/P024378/1, to C.H., D.P., K.H., M.S., R.A.M. and V.T.) and is part of the EDCTP2 program supported by the European Union, i-sense Engineering and Physical Sciences Research Council Interdisciplinary Research Collaboration (EPSRC IRC) in Early Warning Sensing Systems for Infectious Disease (no. EP/K031953/1, to R.A.M., V.T., D.P., S.M., S.G., N.A. and M.S.), the i-sense: EPSRC IRC in Agile Early Warning Sensing Systems for Infectious Diseases and Antimicrobial Resistance (no. EP/R00529X/1, to R.A.M., V.T., D.P., S.G., N.A. and S.M.) and supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre (R.A.M. and S.M.). We thank the m-Africa and i-sense investigators and advisory boards. The AHRI is supported by core funding from the Wellcome Trust (core grant no. 082384/Z/07/Z, to T.S., D.P. and K.H.).
Footnotes
Author Contribution Statement
VT and RAM wrote the manuscript with input from co-authors; VT, CH, TMn, ND and TMh collected the field data; VT and SM developed the machine learning models with contributions from VC, KS SG and RAM; VT, NA and JB were involved in manual data pre-processing; KH oversaw the data collection and data management; TS and MS provided access to anonymised blood samples used in the pilot study. RAM, VT, MS, KH and DP conceived the overall project, designed the study and secured the funding. RAM was the PI with overall responsibility for the i-sense EPSRC IRC and the m-Africa programmes. She was the supervisor of the Research Associates (VT, SM and NA) and students (VC, KS and JB) involved in this study.
Competing Interests Statement
The authors declare no competing interests.
Data availability
The datasets generated during and/or analysed during the current study are available from the AHRI data repository:
Herbst, K., & McKendry, R. (2019). m-Africa: Building mobile phone-connected diagnostics and online care pathways for optimal delivery of population HIV testing, prevention and care in decentralised settings (Version 1) [Data set]. Africa Health Research Institute (AHRI). https://doi.org/10.23664/AHRI.M-AFRICA.2019.V1
Code availability
Custom code used in this study is available on the public repository: https://xip.uclb.com/product/classify_ai
References
- 1.Land KJ, Boeras DI, Chen X-S, Ramsay AR, Peeling RW. REASSURED diagnostics to inform disease control strategies, strengthen health systems and improve patient outcomes. Nat Microbiol. 2019;4:46–54. doi: 10.1038/s41564-018-0295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Second WHO Model List of Essential In Vitro Diagnostics. 2019 [Google Scholar]
- 3.Peeling RW. Diagnostics in a digital age: an opportunity to strengthen health systems and improve health outcomes. 2015 doi: 10.1093/inthealth/ihv062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghani AC, Burgess DH, Reynolds A, Rousseau C. Expanding the role of diagnostic and prognostic tools for infectious diseases in resource-poor settings. Nature. 2015;528:S50–S52. doi: 10.1038/nature16038. [DOI] [PubMed] [Google Scholar]
- 5.Figueroa C, et al. Reliability of HIV rapid diagnostic tests for self-testing compared with testing by health-care workers: a systematic review and meta-analysis. Lancet HIV. 2018;5:e277–e290. doi: 10.1016/S2352-3018(18)30044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klarkowski DB, et al. The evaluation of a rapid in situ HIV confirmation test in a programme with a high failure rate of the WHO HIV two-test diagnostic algorithm. PLoS One. 2009;4:e4351. doi: 10.1371/journal.pone.0004351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gray RH, et al. Limitations of rapid HIV-1 tests during screening for trials in Uganda: diagnostic test accuracy study. BMJ. 2007;335:188. doi: 10.1136/bmj.39210.582801.BE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin EG, Salaru G, Paul SM, Cadoff EM. Use of a rapid HIV testing algorithm to improve linkage to care. J Clin Virol. 2011;52:S11–S15. doi: 10.1016/j.jcv.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 9.Cham F, et al. The World Health Organization African region external quality assessment scheme for anti-HIV serology. Afr J Lab Med. 2012;1:39. doi: 10.4102/ajlm.v1i1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galiwango RM, et al. Evaluation of current rapid HIV test algorithms in Rakai, Uganda. J Virol Methods. 2013;192:25–7. doi: 10.1016/j.jviromet.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Louis FJ, et al. Evaluation of an external quality assessment program for HIV testing in Haiti, 2006-2011. Am J Clin Pathol. 2013;140:867–71. doi: 10.1309/AJCPYWX49IZSQKFS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peck RB, et al. What Should the Ideal HIV Self-Test Look Like? A Usability Study of Test Prototypes in Unsupervised HIV Self-Testing in Kenya, Malawi, and South Africa. AIDS Behav. 2014;18:422–432. doi: 10.1007/s10461-014-0818-8. [DOI] [PubMed] [Google Scholar]
- 13.Baveewo S, et al. Potential for false positive HIV test results with the serial rapid HIV testing algorithm. BMC ReS Notes. 2012;5:154. doi: 10.1186/1756-0500-5-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crucitti T, Taylor D, Beelaert G, Fransen K, Van Damme L. Performance of a Rapid and Simple HIV Testing Algorithm in a Multicenter Phase III Microbicide Clinical Trial. Clin Vaccine Immunol. 2011;18:1480. doi: 10.1128/CVI.05069-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tegbaru B, et al. Assessment of the implementation of HIV-rapid test kits at different levels of health institutions in Ethiopia. Ethiop Med J. 2007;45:293–9. [PubMed] [Google Scholar]
- 16.Johnson CC, et al. To err is human, to correct is public health: a systematic review examining poor quality testing and misdiagnosis of HIV status. J Int AIDS Soc. 2017;20:21755. doi: 10.7448/IAS.20.7.21755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Learmonth KM, et al. Assessing proficiency of interpretation of rapid human immunodeficiency virus assays in nonlaboratory settings: ensuring quality of testing. J Clin Microbiol. 2008;46:1692–7. doi: 10.1128/JCM.01761-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.García pJ, et al. Rapid Syphilis Tests as Catalysts for Health Systems Strengthening: A Case Study from Peru. PLoS One. 2013;8:e66905. doi: 10.1371/journal.pone.0066905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sacks R, Omodele-Lucien A, Whitbread N, Muir D, Smith A. Rapid HIV testing using Determine™ HIV 1/2 antibody tests: is there a difference between the visual appearance of true- and false-positive tests? Int J STD AIDS. 2012;23:644–646. doi: 10.1258/ijsa.2012.011422. [DOI] [PubMed] [Google Scholar]
- 20.Doan M, Carpenter AE. Leveraging machine vision in cell-based diagnostics to do more with less. Nat Mater. 2019;18:414–418. doi: 10.1038/s41563-019-0339-y. [DOI] [PubMed] [Google Scholar]
- 21.Esteva A, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Fauw J, et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat Med. 2018;24:1342–1350. doi: 10.1038/s41591-018-0107-6. [DOI] [PubMed] [Google Scholar]
- 23.Xu Y, et al. Deep Learning Predicts Lung Cancer Treatment Response from Serial Medical Imaging. Clin Cancer Res. 2019;25:3266–3275. doi: 10.1158/1078-0432.CCR-18-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silver D, et al. A general reinforcement learning algorithm that masters chess, shogi, and Go through self-play. Science (80) 2018;362:1140–1144. doi: 10.1126/science.aar6404. [DOI] [PubMed] [Google Scholar]
- 25.Ascent of machine learning in medicine. Nat Mater. 2019;18:407. doi: 10.1038/s41563-019-0360-1. [DOI] [PubMed] [Google Scholar]
- 26.Ching T, et al. Opportunities and obstacles for deep learning in biology and medicine. bioRxiv. 2018:142760. doi: 10.1101/142760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng N, Wang Z, Zhang H, Liu W, Alsaadi FE. Deep Belief Networks for Quantitative Analysis of a Gold Immunochromatographic Strip. Cognit Comput. 2016;8:684–692. [Google Scholar]
- 28.Carrio A, Sampedro C, Sanchez-Lopez JL, Pimienta M, Campoy P. Automated low-cost smartphone-based lateral flow saliva test reader for drugs-of-abuse detection. Sensors (Switzerland) 2015;15:29569–29593. doi: 10.3390/s151129569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neuman M, et al. The effectiveness and cost-effectiveness of community-based lay distribution of HIV self-tests in increasing uptake of HIV testing among adults in rural Malawi and rural and peri-urban Zambia: protocol for STAR (self-testing for Africa) cluster randomized evaluations. BMC Public Health. 2018;18:1234. doi: 10.1186/s12889-018-6120-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aicken CRH, et al. Young people’s perceptions of smartphone-enabled self-testing and online care for sexually transmitted infections: qualitative interview study. BMC Public Health. 2016;16:974. doi: 10.1186/s12889-016-3648-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Witzel TC, Weatherburn P, Rodger AJ, Bourne AH, Burns FM. Risk, reassurance and routine: a qualitative study of narrative understandings of the potential for HIV self-testing among men who have sex with men in England. BMC Public Health. 2017;17:491. doi: 10.1186/s12889-017-4370-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nsabimana AP, et al. Bringing Real-Time Geospatial Precision to HIV Surveillance Through Smartphones: Feasibility Study. JMIR public Heal Surveill. 2018;4:e11203. doi: 10.2196/11203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laksanasopin T, et al. A smartphone dongle for diagnosis of infectious diseases at the point of care. Sci Transl Med. 2015;7:273re1. doi: 10.1126/scitranslmed.aaa0056. [DOI] [PubMed] [Google Scholar]
- 34.Mudanyali O, et al. Integrated rapid-diagnostic-test reader platform on a cellphone. Lab Chip. 2012;12:2678. doi: 10.1039/c2lc40235a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allan-Blitz L-T, et al. Field evaluation of a smartphone-based electronic reader of rapid dual HIV and syphilis point-of-care immunoassays. Sex Transm Infect. 2018;94:589–593. doi: 10.1136/sextrans-2017-053511. [DOI] [PubMed] [Google Scholar]
- 36.S F, et al. Immunochromatographic Diagnostic Test Analysis Using Google Glass. ACS Nano. 2014;8 doi: 10.1021/nn500614k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guan Q, et al. Diagnose like a Radiologist: Attention Guided Convolutional Neural Network for Thorax Disease Classification. 2018 [Google Scholar]
- 38.Sandler M, Howard A, Zhu M, Zhmoginov A, Chen L-C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. 2018 [Google Scholar]
- 39.Chaturvedi SS, Gupta K, Prasad PS. Skin Lesion Analyser: An Efficient Seven-Way Multi-class Skin Cancer Classification Using MobileNet. Springer; Singapore: 2021. pp. 165–176. [DOI] [Google Scholar]
- 40.Howard A, et al. Searching for MobileNetV3. 2019 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the AHRI data repository:
Herbst, K., & McKendry, R. (2019). m-Africa: Building mobile phone-connected diagnostics and online care pathways for optimal delivery of population HIV testing, prevention and care in decentralised settings (Version 1) [Data set]. Africa Health Research Institute (AHRI). https://doi.org/10.23664/AHRI.M-AFRICA.2019.V1
Custom code used in this study is available on the public repository: https://xip.uclb.com/product/classify_ai