Abstract
The successful in vivo implementation of gene expression modulation strategies relies on effective, non-immunogenic delivery vehicles. Lipid nanoparticles (LNPs) are one of the most advanced, non-viral clinically approved nucleic-acid delivery systems. Yet, LNPs accumulate naturally in liver cells upon intravenous administration and hence there is an urgent need to enhance uptake by other cell types. Here we use a conformation-sensitive targeting strategy to achieve in vivo gene silencing in a selective subset of leukocytes and show potential therapeutic applications in a murine model of colitis. In particular, by targeting the high-affinity (HA) conformation of α4β7 integrin, which is a hallmark of inflammatory, gut-homing leukocytes, we silenced interferon γ in the gut resulting in an improved therapeutic outcome in experimental colitis. The LNPs did not induce adverse immune activation or liver toxicity. These results suggests that our LNP targeting strategy might be applied for selective delivery of payloads to other conformation-sensitive targets.
Keywords: Lipid nanoparticles, RNA therapy, targeting leukocytes, MAdCAM-1, α4β7 integrin, CD45, IFNγ
IBD is a growing problem with rising incidence since the 19th century. Despite several decades of research in both animals and humans, current treatments remain disappointing and do not provide a cure1. Although novel biologics, such as antibodies against TNF-α, revolutionized IBD treatment, not all patients respond and initial responders can lose response over time due to the development of antibody drug antibody (ADA2,3) responses.
To establish a curative solution to IBD, blocking cytokines or receptors with antibodies is most likely insufficient and provides only temporary relief from the symptoms. A curative solution could be achieved by actively modulating gene expression in the aberrantly activated leukocyte population. Altering the expression of specific genes in inflammatory leukocytes and thereby changing their behavior might restore the balance in the intestinal immune response for a longer term. The feasibility of this is supported by several studies that demonstrated modulation of gene expression by, for instance, silencing TNF-α4 or overexpressing IL-105 in a specific subset of cells in experimental colitis. These studies relied on the use of lipid nanoparticles (LNPs) to deliver therapeutic nucleic acids in vivo (Fig. 1a). LNPs have evolved over the years to become one of the most suitable, non-viral methods of delivering nucleic acids in vivo due to their high encapsulation efficiency, low batch-to-batch variation and fusiogenic properties6 7. Since the recent approval of the first-ever RNAi-based drug, Onpattro™ (Patisiran), by the FDA8, the use of LNPs has gained even more momentum. Like most injected particulates, LNPs tend to accumulate in the liver and are thereby limited in their use in other cell types. Recent attempts to generate targeted LNPs that, in addition to accumulating in the liver, are directed to other cells as well4,5,9 generated promising results and opened the door to a new era of targeted delivery of therapeutic nucleic acids.
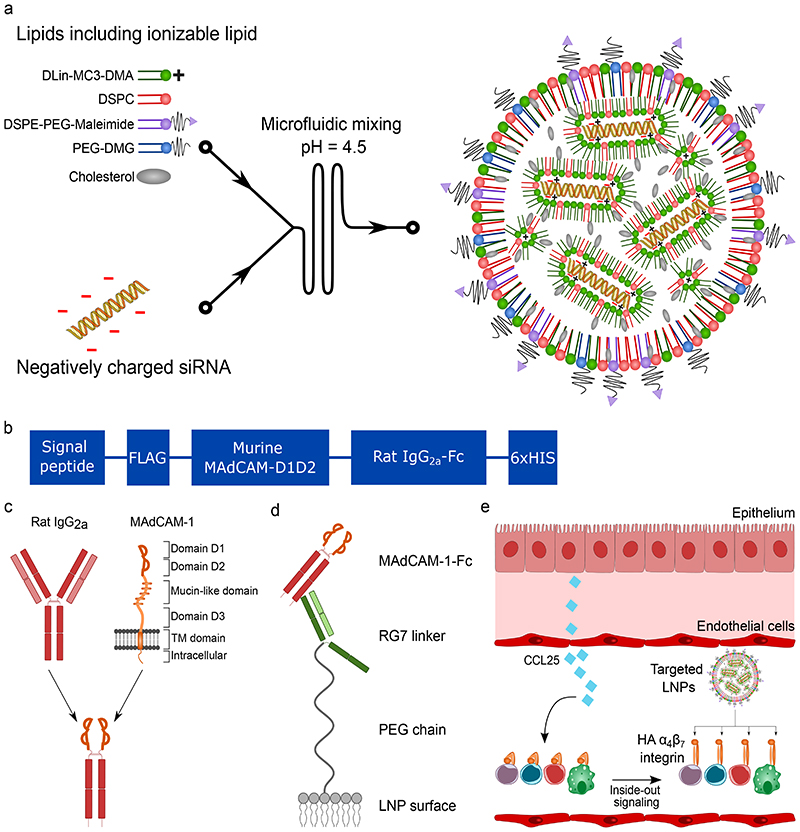
Figure 1. Generation of LNPs to target the high-affinity conformation of integrin α4β7.
a. Illustration of the generation of LNPs using microfluidics. The ionizable lipid facilitates siRNA encapsulation through its positive charge at low pH. b. Overview of the different domains of the MAdCAM-1-Fc fusion protein. c. Overview of the fusion strategy. The different domains of the wild type MAdCAM-1 are shown. Only the integrin binding domains D1 and D2 are used. D1D2 is fused to the hinge of rat IgG2a with a flexible linker. d. Schematic drawing depicting the conjugation strategy of the MAdCAM-1-Fc to the LNPs. The RG7 linker (mAb against rat IgG2a) is chemically conjugated with the LNPs to the maleimide group in the lipid DSPE-PEG-Maleimide. RG7 readily binds the MAdCAM-1-Fc by antibody affinity. e. LNP targeting to HA α4β7 integrin. CCL25 induces the integrin conformational change.
Herein, we report the delivery of siRNAs specifically to activation-sensitive receptors expressed on gut-homing leukocytes in a mouse model of colitis. Our targeting strategy is different from traditional strategies as we employ a targeting moiety that only recognizes a specific protein conformation, namely the high-affinity (HA) conformation of integrin α4β7. Gut-homing leukocytes utilize this pivotal intestinal homing receptor to adhere to the intestinal endothelium. Previously, we exploited this leukocyte integrin and generated lipid nanoparticles that were targeted to the β7 integrin subunit10. Although α4β7 integrin is considered a key protein in homing of leukocytes to the gut during intestinal inflammations, in healthy individuals ~70% of the total intestinal T-cell population and ~35% of circulating CD4+ T-cells express α4β7 integrin11, indicating that merely targeting α4β7 integrin or one of its subunits lacks specificity.
Integrin functionality depends on the conformational state12. Integrins change conformation when stimulated and dramatically increase the affinity for their ligands. Integrin α4β7 has the potential to bind both Vascular Cell Adhesion Molecule 1 (VCAM-1) for homing to peripheral tissues and Mucosal Vascular Addressin Cell Adhesion Molecule 1 (MAdCAM-1) for homing to intestinal tissues, but not simultaneously. Whether integrin α4β7 has affinity for VCAM-1 or MAdCAM-1 depends on the specific stimulus, subsequent signaling and type of conformational change13. As only leukocytes that actively home to the intestinal tissues possess α4β7-integrin in the HA confirmation, we wanted to generate LNPs that can target integrin α4β7 + expressing cells in a conformation-dependent manner (Fig. 1e). This is contrary to commercially available, conformation-insensitive monoclonal antibodies such as natalizumab (anti-α4-integrin) and vedolizumab (anti-α4β7-integrin). The possible dangers associated with insufficient specificity when blocking leukocyte homing is exemplified by natalizumab which was temporally withdrawn from the market for treatment of IBD due to increased risk of opportunistic infections leading to multifocal leukoencephalopathy14.
In this study, we generated a recombinant fusion protein that contains two domains of the intestinal endothelium ligand MAdCAM-1. MAdCAM-1 naturally has an increased affinity to integrin α4β7 in its HA conformation and is therefore an excellent start for the design of the LNP targeting moiety. MAdCAM-1 is a multi-domain protein that is naturally involved in both initial tethering and in firm adhesion of leukocytes to the intestinal endothelium.
To maximize specificity, we only utilized the integrin binding domains, D1 and D2 (Fig. 1b-c). To protect the integrin binding domain while conjugating the protein to the LNPs, we made use of a monoclonal secondary antibody against rat IgG2a, here referred to as RG7, which serves as a linker between the LNPs and the MAdCAM-D1D2 protein. The RG7 linker was chemically conjugated to the LNPs using maleimide/thiol chemistry and the MAdCAM-D1D2 protein was recombinantly fused to the Fc region of rat IgG2a. This way, the RG7 antibody will bind the MAdCAM-1 protein by affinity to the rat IgG2a domain and leave the domains D1 and D2 free for binding to α4β7 integrin (Fig. 1d). This conjugation strategy was compared with two other options: direct conjugation to the DSPE-PEG-maleimide lipid using reduced cysteine residues in the D1D2 protein or by using a previously published conjugation strategy that involves ASSET (Anchored Secondary ScFv Enabling Targeting), a lipidated scFv against rat IgG2a that readily incorporates in the LNPs4. When ASSET is incorporated into the LNPs, it can bind the D1D2-Fc by affinity. For our specific approach, the RG7-mediated conjugation was by far superior over the other two methods (Supplementary Fig. 1). We ensured that this linkage is sufficiently stable in freshly isolated mouse blood plasma for at least 1 hour at 37°C (Supplementary Fig. 2).
Design and production of MAdCAM-1-Fc protein
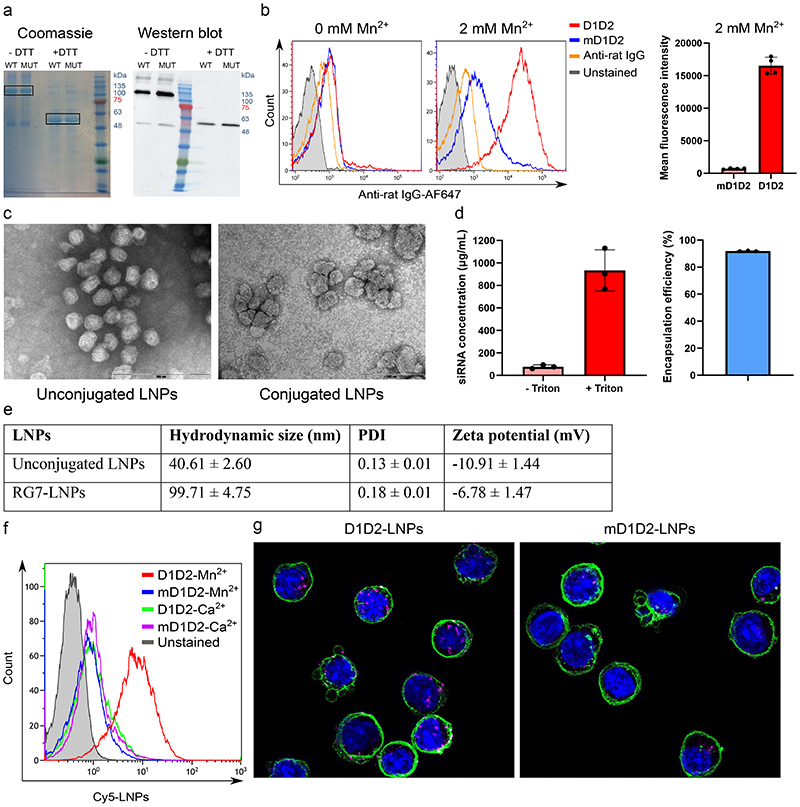
The domains D1 and D2 of murine MAdCAM-1 were fused to the N terminus of the Fc domain of rat IgG2a (including hinge, excluding CH1). A signal peptide for secretion and a FLAG tag were added to the N terminus of the construct and a 6x HIS tag was added at the C terminus for purification purpose. Purity and size (~50 kDa) were confirmed by SDS-PAGE (Fig. 2a).
Figure 2. Characterization of D1D2, LNPs and D1D2-targeted LNPs.
a. SDS PAGE of both the D1D2 (WT) and mD1D2 (MUT) proteins. Dimerization through disulfide bonds is evident when comparing to the lanes without reducing agent. Experiment was repeated 3 times independently. b. In vitro binding of the purified D1D2 protein to TK-1 cells measured by flow cytometry. mD1D2 does not bind the cells while the D1D2 binds only cells with HA integrin α4β7 (after addition of Mn2+). A significant difference between D1D2 and mD1D2 in Mn2+-activated cells was observed (p < 0.0001, n = 4 biologically independent samples, two-sided student’s t-test, data are presented as mean +/- SD). c. Representative transmission electron microscopy images of unconjugated LNPs and LNPs conjugated to RG7. Experiment was repeated 3 times independently. d. siRNA entrapment efficiency assayed by Ribogreen, n = 3 biologically independent samples. Data presented as mean +/- SD. e. Hydrodynamic size, polydispersity index (PDI) and zeta potential of the produced LNPs. f. In vitro binding of Cy5-labeled, D1D2-targeted LNPs to TK1 cells. g. Representative confocal images showing internalization of Cy5-labeled LNPs (magenta) into TK-1 cells. Images were generated by combining 11 frames from Z-stack imaging (0.3 μm/frame).
We also generated a mutated version of the MAdCAM-1-D1D2-Fc that serves as a negative control. This has the D42A mutation that has been reported to severely affect the ability of MAdCAM-1 to bind α4β7 integrin15. This mutation affects the amino acid sequence LDTS, which has been shown to be the critical motif that is actively engaged in integrin binding16. In the rest of this manuscript, we refer with D1D2 to the non-mutated version and with mD1D2 to the mutated version.
After purifying D1D2 and mD1D2, we tested the functionality in vitro by assessing the binding to TK-1 cells using flow cytometry. TK-1 cells have been shown to express high levels of α4β7 integrin10, 17 and are therefore an excellent in vitro model to test the functionality of the recombinant MAdCAM-1 construct. Cells were either treated with Mn2+ to create the HA-α4β7 or with Ca2+ as a LA-α4β7 control. As expected, in the absence of Mn2+, no difference in cell binding between D1D2 and mD1D2 was observed, however, a significant difference (p < 0.0001, n = 4) in binding was visible upon Mn2+ treatment (Fig. 2b).
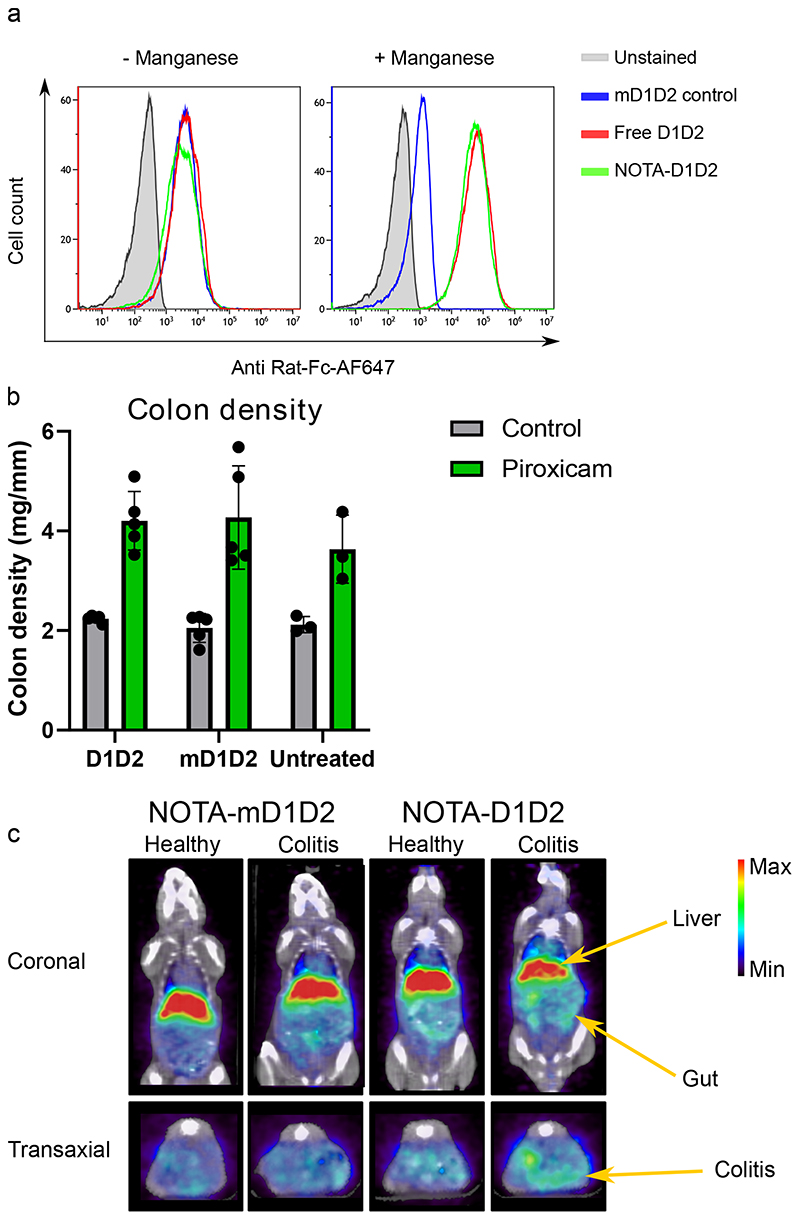
After validating that the D1D2 protein binds exclusively to the HA conformation of integrin α4β7, we used molecular imaging to analyze the biodistribution. We have previously reported that targeting β7 integrin and α4β7 integrin via mAbs facilitate specific imaging of inflammatory leukocytes in DSS colitis (although unspecific accumulation in the liver cannot be excluded)18,19. Here we wanted to test this with the newly developed D1D2 targeting protein and hence we performed a proof-of-concept study on the imaging of inflammatory leukocytes in experimental colitis. We conjugated the D1D2 protein directly to a chelator, NOTA, to enable labeling with the radioisotope 64Cu, this did not affect the protein functionality as validated by binding to TK-1 cells (Fig. 3a).
Figure 3. Molecular imaging of inflammatory leukocytes in experimental colitis using PET/CT and D1D2-NOTA-64Cu.
a. NOTA-conjugation with D1D2 did not affect the protein functionality as was demonstrated with binding to TK-1 cells. b. Treatment with NOTA-conjugated D1D2 or mD1D2 did not significantly affect colitis severity as determined by colon density and analyzed by one-way ANOVA. There was a significant difference (p = 0.0008, n = 10 mice) in colon density between piroxicam-treated animals and controls, confirming active colitis. Data are presented as mean values +/- SD. c. Representative image of PET/CT imaging using D1D2-NOTA-64Cu. Gut uptake was compared to mD1D2-NOTA-64Cu as control.
D1D2-NOTA-64Cu or mD1D2-NOTA-64Cu was injected in both healthy mice and mice with colitis, and 24 h post-injection, the mice were imaged by microPET/CT. Injection of the radiolabeled D1D2 protein did not affect the colitis severity (Fig. 3b). Figure 3c displays a representative image of the biodistribution. The D1D2-NOTA-64Cu shows an increased uptake in the gut of colitic mice. This uptake was significantly correlated to colitis severity (Supplementary Fig. 6) with n = 5 mice/group (p = 0.014). Colon density was used to score colitis severity. This experiment lacked extensive colitis scoring such as histology, but we believe that this still gives a good indication of preferential uptake of D1D2 in the inflamed colons.
Generation of targeted LNPs
Having confirmed the binding of D1D2 to gut-tropic leukocytes, we generated targeted LNPs. To generate uniformly sized LNPs that have a high siRNA encapsulation efficiency and minimal batch-to-batch variation, we used the NanoAssemblr™ microfluidic mixing device20. The ionizable lipid, DLin-MC3-DMA, aids in siRNA encapsulation as previously reported,21 and we used an LNP formulation that we and others previously reported4,5,22.
LNPs had mean diameter of ~ 40 nm, zeta potential of ~ -10mV (Fig. 2e) and encapsulation efficiencies close to 90% (Fig. 2d). The size and uniformity of the particles was confirmed using transmission electron microscopy (Fig. 2c).
The LNPs were conjugated to RG7 using maleimide-thiol chemistry. The targeting protein is bound to the LNP surface through affinity by the RG7 linker.
The biodistribution of the targeted LNPs was analyzed by microPET/CT. We covalently added a TCO group to the DSPE-PEG lipid to enable TCO-tetrazine chemistry to add the chelator (NOTA). However, we did not see increased uptake in target tissue compared with controls (Supplementary Fig. 5). Large macromolecules such as LNPs accumulate in the liver and it is hard to clearly distinguish enhanced uptake in other tissues. Furthermore, tissue-specific targeting is often dependent on passive targeting. Only once the tissue of interest is reached, LNPs can take advantage of the increased affinity of targeting moieties by binding and internalizing to specific cells23. Therefore, it is hard to distinguish tissue distribution between the D1D2-targeted and the mD1D2-targeted LNPs. The targeting is mainly related to enhanced cell-specific uptake rather than the alteration of tissue accumulation by the LNPs.
After preparation, conjugation and purification of the LNPs, the functionality of the D1D2-targeted LNPs was tested. Binding of LNPs to cells (Mn2+ or Ca2+ treated) was tested by flow cytometry using encapsulated Cy5-siRNA that can fluorescently label bound cells. As shown in Fig. 2f, LNP binding increased dramatically when targeted by D1D2 as compared to mD1D2. Furthermore, confocal microscopy indicated internalization of the Cy5-labeled, D1D2-targeted LNPs (Fig. 2g). D1D2-LNP binding was compared to both mD1D2 and anti-α4β7 integrin (mAb clone DATK32) in primary leukocytes. Specific binding of D1D2-LNPs to Mn2+-activated primary cells was evident while DATK32 was conformation insensitive (Supplementary Fig. 3). D1D2-targeted LNPs bound most robustly to CD4+ T-cells from the mLN in a conformation-dependent manner.
CCL25 specifically increases α4β7 integrin’s affinity for MAdCAM-1 by binding to the CCR9 receptor. As expected, CCL25 treatment of cells enhances D1D2-LNP binding compared to CXCL10-treated control cells (CXCL10 increases α4β7 integrin’s affinity for VCAM-1), see Supplementary Fig. 4.
D1D2-LNPs silence CD45 in a conformational-sensitive manner
Before testing therapeutic gene silencing, feasibility was tested with the pan-leukocyte marker CD45. DSS colitis, a common model, did not result in substantial gene silencing, probably because DSS colitis is strongly macrophage driven and T- and B-cell independent24 27. Therefore we explored another animal model: piroxicam-accelerated colitis (PAC) in IL-10KO mice25,26. Although macrophages play an important role in the PAC model as well28, we thought it would be of interest to investigate the effects of targeting a specific subpopulation of CD4 T-cells in the PAC model. Furthermore, IL-10KO mice with active colitis have strongly increased levels of MAdCAM-1 in the gut29, pointing to the importance of MAdCAM-1 in IL-10 deficient mice.
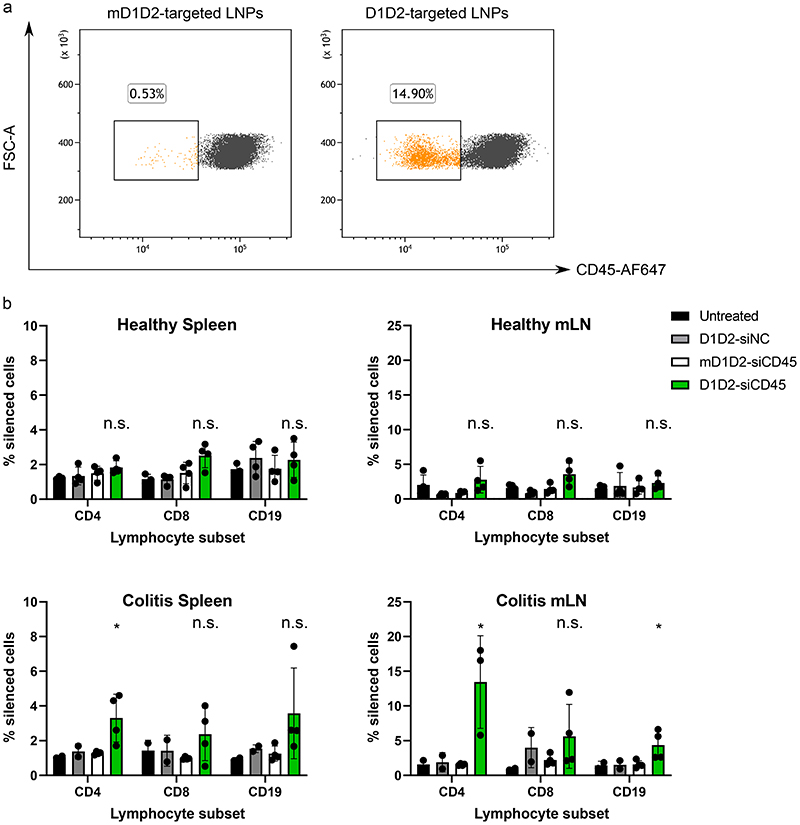
Five days post intravenous injection with siCD45-LNPs, cells from the mesenteric lymph nodes (mLNs) were collected. Cells from the spleen were also collected as a control, peripheral-lymphoid organ. Cells were analyzed for their CD45 expression using flow cytometry (gating strategy: Supplementary Fig. 9). Silencing levels in the spleen were negligible while CD4+ T-cells from mLN were effectively silenced by D1D2-targeted LNPs (Fig. 4a+b). As a negative control, healthy mice were also injected with D1D2-targeted LNPs yielding no significant results in any cell type from any organ (Fig. 4b).
Figure 4. In vivo gene silencing of CD45 using D1D2-targeted LNPs in both healthy mice and mice with colitis.
D1D2-LNPs were either loaded with siRNA against CD45 (siCD45) or with a negative control siRNA (siNC). Untreated mice were used to determine the default CD45 expression levels. a. Strongest gene silencing was seen in the CD4+ T-cell population from the mLN in the piroxicam-accelerated colitis (PAC) model. Observed gene silencing was ~15% as compared to mD1D2 control. b. Healthy mice did not show significant silencing in any cell population while in the PAC model, CD45 was silenced significantly in CD4+ and CD19+ cells. Data are presented as mean values +/- SD, n = 5 mice per group, * p < 0.05, n.s. = not significant (statistics performed with two-sided student’s t-test comparing D1D2 to mD1D2 for each cell subtype).
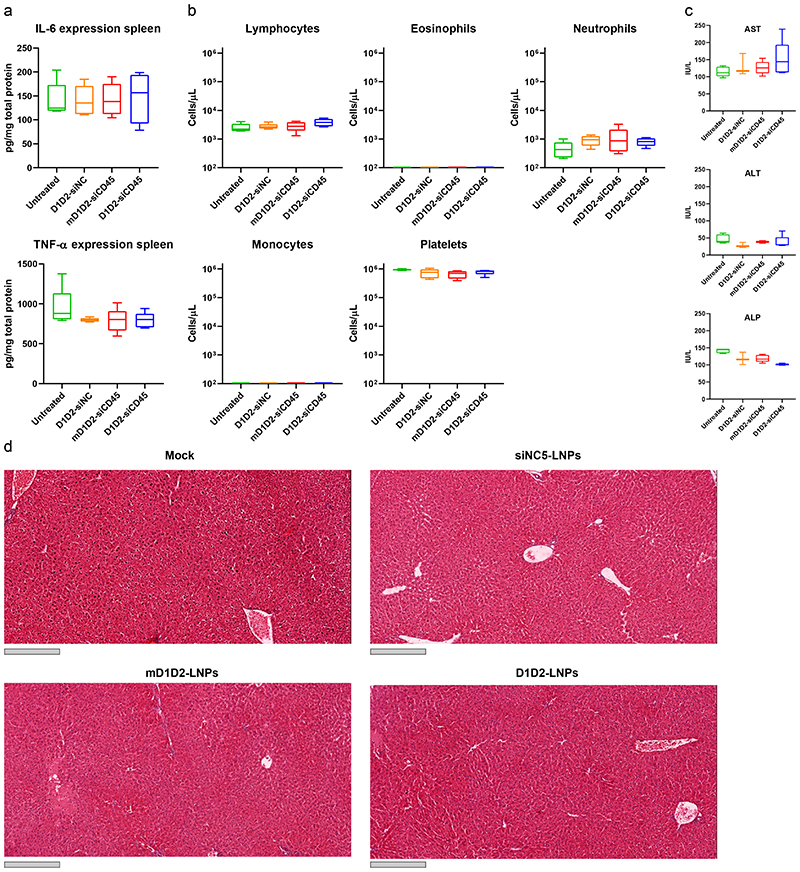
LNPs did not affect the colitis severity as was concluded from colon histology in mice from the different groups (Supplementary Fig. 7). No elevation of the liver enzymes aspartate aminotransferase (AST), alanine transaminase (ALT) and alkaline phosphatase (ALP) was detected (Fig. 5c). In addition, liver histology did not reveal any excessive bleeding or liver damage (Fig. 5d), thus we concluded that injection of the LNPs does not cause any observable liver toxicity. Furthermore, the LNPs did not induce unwanted immune responses as measured by complete blood count (Fig. 5b) and splenic TNF-α and IL-6 expression levels (Fig. 5a). There was no significant difference in counts of the platelets, neutrophils and lymphocytes between the groups and counts of eosinophils and monocytes in the blood were nearly undetectable.
Figure 5. Safety profile of different LNP formulations.
Safety was determined by measuring both liver toxicity and immune activation. a. Splenic expression levels of the pro-inflammatory cytokines TNF-α and IL-6. b. Complete blood count. c. Serum levels of liver enzymes. d. Histology of the liver. Grey scale bar represents 300 μm. The liver was sectioned, and H&E stained. No significant elevation of cytokines, liver enzymes and blood count was detected in the LNP-treated mice compared to the untreated (one-way ANOVA with Dunnett’s test comparing each group to the untreated mice, n = 5). Liver histology did not reveal tissue damage or excessive bleeding in any of the treated groups. Box plots in a-c: minimum = lowest value, maximum = highest value, center = median, bounds of box = interquartile range, lower whisker = lowest value till 25th percentile, upper whisker = 76th percentile till highest value. Experiment was repeated 3 times independently.
Efficacy of siIFNγ-D1D2-LNPs in mice with active colitis
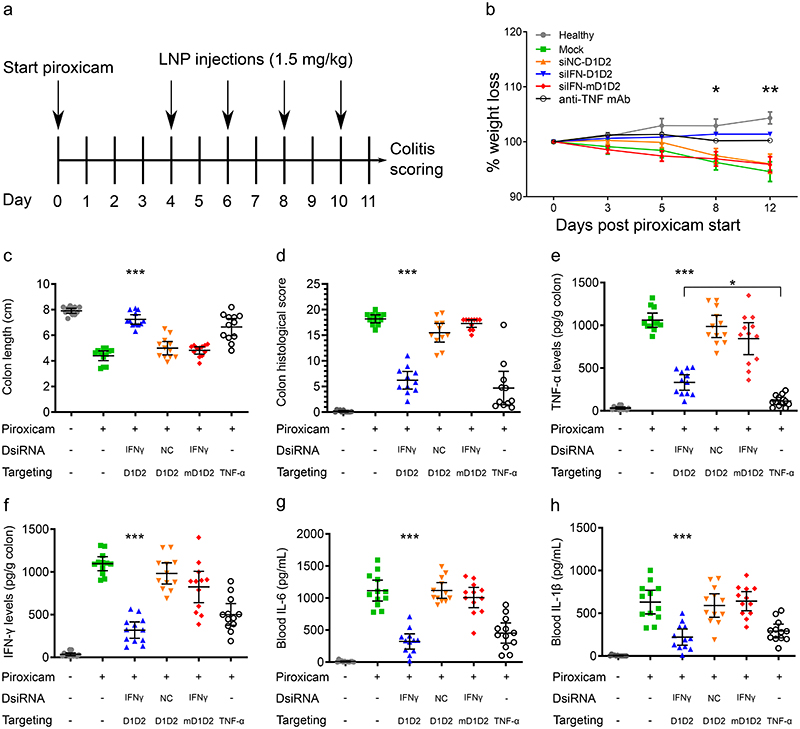
Based on the CD45 silencing results, a therapeutic target gene related to CD4+ T-cell biology was explored for therapeutic efficacy studies. Because IFNγ is secreted by inflammatory Th1 cells and as IFNγ is causatively involved in experimental colitis30, this gene was chosen as a therapeutic target gene. To test the therapeutic efficacy, D1D2-LNPs or mD1D2-LNPs loaded with siIFNγ were injected into mice with colitis 4, 6, 8 and 10 days after initiation of colitis (Fig. 6a). A control mouse group injected with D1D2-LNPs loaded with a siNC was used to correct for possible therapeutic effects unrelated to IFNγ. Mice without piroxicam were used as a healthy control and as positive therapeutic control, a validated mAb against TNF-α was used (administered at days 4, 6, 8 and 10). The antibody against TNF-α was used as a positive control for amelioration of colitis. Blocking of TNF-α with mAbs has been well validated and anti-TNF-α mAbs are currently used in the clinic (e.g. infliximab31). At day 11, mice were sacrificed, and colitis was assessed in all groups (Fig. 6).
Figure 6. Therapeutic gene silencing of IFNγ using D1D2-LNPs in the PAC model.
a. Experimental design. b. Percentage weight change in all treated groups with n = 12 mice/group. D1D2-siIFN showed a significant difference from day 8 onward. Error bars represent the standard error of the mean. In two groups, the error bars are smaller than the symbols and are therefore not displayed. c. Colon length comparison. d. Colon histological score index. e-h. Expression levels of pro-inflammatory cytokines in the colon (e-f) and blood (g-h). Data are represented as scatter dot plots with error bars representing the 95% CI, statistical tests were calculated using one-way ANOVA with Dunnet’s post hoc test, n = 12 mice / group, * p < 0.05, ** p < 0.01, *** p < 0.0001. Statistical significance in the plots indicates a difference in the siIFNγ-D1D2 LNP group compared to the negative control groups (siIFNγ-mD1D2, siNC-D1D2 and untreated mice with colitis). Additionally, plot e indicates a significant difference specifically between the siIFNγ-D1D2 and the mAb-TNF-α groups. In the other plots, significant differences between these 2 groups were absent. Therefore, overall, the therapeutic outcome with D1D2-siIFNγ was comparable to the anti-TNFα mAb.
Starting from day 8, there was a significant difference in weight change in the D1D2-siIFNγ group as compared to the negative control groups (Fig. 6b). Colonic levels of IFNγ decreased dramatically (~2.5-fold) in D1D2-targeted IFNγ-LNPs as compared to the mD1D2 control (Fig. 6f). Colonic IFNγ levels also decreased in mice treated with the anti-TNF-α antibody, likely due to an overall decrease in intestinal inflammation. Because IFNγ affects TNF-α expression32 and because these two cytokines have synergistic effects on NF-ĸB signaling33, we would expect an associated reduction of other pro-inflammatory cytokines by silencing IFNγ. Mice treated with D1D2-siIFNγ-LNPs and mice treated with anti-TNF-α mAb showed a strong decrease in colonic TNF-α levels (Fig. 6e). The colonic TNF-α levels in mice treated with the mAb against TNF-α (positive control) were even lower than the D1D2-LNPs group (p < 0.05). Furthermore, blood IL-6 and IL-1β levels decreased dramatically and equally both in mice treated with D1D2-targeted LNPs and mice treated with anti-TNF-α mAb (Fig. 6g-h). Colon length (colon shortening is an important marker of colonic inflammation) was significantly increased (p < 0.0001) in the D1D2-targeted group as compared to the mD1D2 control (Fig. 6c). A significantly lower colon histological score (p < 0.0001) further supported the improved therapeutic outcome in mice treated with D1D2-targeted IFNγ-LNPs (Fig. 6d). These results were also in accordance with visual examination of photomicrographs of sections of the colon (Supplementary Fig. 8). Taken together, these data demonstrate a strong therapeutic response in mice treated with D1D2-targeted LNPs loaded with siIFNγ while the mutated control (mD1D2) did not lead to a significantly improved therapeutic outcome. This approach opens new potential avenues for conformational sensitive targeting as novel therapeutic modality with reduced adverse effects and increased efficacy.
Conclusions
While novel biologics such as monoclonal antibodies (mAbs) have entered the IBD arena, they lack conformational specificity. Given the chronic nature of inflammatory bowel disease and the lifelong dependence on such medications, non-specific treatments could potentially lead to iatrogenic global immune suppression, sensitizing the patient to opportunistic infections. To overcome this, we report here on a strategy to target specifically gut-homing leukocytes in experimental colitis. We fused the integrin binding domains, D1 and D2, of the natural ligand of α4β7 integrin, MAdCAM-1, to an IgG-Fc and devised a strategy to efficiently conjugate this fusion protein to the surface of lipid nanoparticles.
We silenced genes in an inflammation-dependent manner in lymphocytes (mainly T-cells), cells that are intrinsically hard to transfect. Using siIFNγ we translated this gene knockdown into an improved therapeutic outcome in IL-10KO mice with active colitis.
Taken together, we believe that our results demonstrate the feasibility of conformation-sensitive targeting in general and in IBD in particular. The relationship between conformational state and protein functionality is a commonly seen phenomenon and targeting specific conformations of proteins opens new avenues in LNP targeting strategies.
However, the integrin conformational change that drives leukocyte extravasation is a complex process. The interaction between receptor and ligand in an activation dependent manner is enhanced not only by increased affinity but also by increased avidity. Both processes occur naturally and simultaneously to increase ligand-receptor binding, and it is hard to distinguish between the contributions of each of these two processes34. To gain more insight into these activation-dependent mechanisms, mAbs have been generated that mimic ligand binding in a conformation-sensitive manner. For instance, a mAb against a disulfide-locked HA version of LFA-1 was generated to study the role of affinity enhancement in the adhesion process of lymphocytes34. Furthermore, an activation specific monoclonal antibody that recognizes the HA conformation of the human version of integrin α4β7 was recently developed using glycan wedges that stabilize the HA conformation35. This antibody was generated to study the dynamics of conformational changes and the mechanism of affinity increase.
On a molecular level, much is known about the chemokine-driven, inside-out signaling processes that result in such conformational changes. The conversion of a non-adhesive to an adhesive state of integrins involves extracellular stimuli that activate receptor tyrosine kinases or G protein-coupled receptors, leading to signaling by intracellular integrin binding proteins such as kindlin and talin which in turn induce the conformational change36,37.
In vivo, however, the reality is more complex, and little is known about the dynamics. A recombinant mouse model (C57BL/6-Itgb7tm1Mshi/J) with a disrupted α4β7-integrin domain leading to a permanent HA conformation37, can be used to study the conformational changes in greater detail.
As for this study, we have demonstrated that exploiting the conformational changes of an integrin to enhance cell-specific uptake is feasible and that it translates into therapeutic efficacy in experimental colitis. This is a relatively daunting task as it is unknown what percentage of leukocytes display the HA conformation, at which location in the body and for what period of time. Despite the limited knowledge in these in vivo dynamics, we demonstrated the possibility of using HA-integrin-α4β7-targeted LNPs with a therapeutic payload to produce a therapeutic improvement. In terms of clinical perspective in humans, we envision a novel LNP conjugation strategy without the need for including rat IgG Fc regions (which could render the protein immunogenic). Such conjugation strategies could include a humanized scFv against a small, non-immunogenic peptide tag that can be integrated into the recombinant protein. If this scFv is lipidated similarly to the ASSET (as described earlier), it will facilitate straightforward incorporation into the LNPs. Another option would be to include an Fc region of a human IgG antibody and chemically conjugate this to the LNPs.
This is a first step demonstrating the feasibility of conformational-sensitive targeting of LNPs for potential future clinical applications. While immunologists discover more defined roles of small subsets of leukocytes, the development of more specific drug delivery systems can accordingly deliver therapeutic payloads to more precisely defined cell populations and revolutionize precision nanomedicine.
Material and methods
Antibodies
Rat anti-mouse α4β7 integrin (Clone DATK32, Biolegend)
Rat anti-mouse β7 integrin (Clone FIB504, BioXCell)
Mouse anti-rat IgG2a (Clone RG7/1.30, BioXCell)
Rat IgG2a isotype control (Clone 2A3, BioXCell)
PE-conjugated donkey anti-mouse IgG (Polyclonal, Jackson Immuno Research)
AlexaFluor 647-conjugated goat anti-rat IgG (Polyclonal, Biolegend)
AlexaFluor 647-conjugated mouse anti-human IgG (Polyclonal, Biolegend)
AlexaFluor 647-conjugated rat anti-mouse CD45 (Clone 30-F11, Biolegend)
AlexaFluor 488-conjugated rat anti-mouse CD45 (Clone 30-F11, Biolegend)
BV650-conjugated rat anti-mouse/human CD11b (Clone M1/70, Biolegend)
PE/Cy5-conjugated rat anti-mouse CD19 (Clone 6D5, Biolegend)
BV421-conjugated hamster anti-mouse CD3ε (Clone 145-2C11, Biolegend)
APC/Cy7-conjugated rat anti-mouse CD4 (Clone GK1.5, Biolegend)
AlexaFluor 488-conjugated rat anti-mouse CD8a (Clone 53-6.7, Biolegend)
PE-conjugated rat anti-mouse CD8a (Clone 53-6.7, Biolegend)
PE/Dazzle 594 rat anti-mouse CD25 (Clone PC61, Biolegend)
BV510 armenian hamster anti-mouse CD69 (Clone H1.2F3, Biolegend)
PE-conjugated rat anti-mouse FOXP3 (Clone MF-14, Biolegend)
HRP-conjugated donkey anti-rat IgG (Polyclonal, Jackson Immuno Research)
siRNAs
The siRNA against CD45 and the corresponding negative control siRNA are chemically modified siRNAs and were kindly provided as a gift by Alnylam Pharmaceuticals.
The siRNA against IFNγ and the corresponding negative control siRNA are chemically modified dicer substrates (synthesized by Integrated DNA Technologies).
Mouse CD45 siRNA.
| Sense: | mCmUrGrGmCmUrGrArAmUmUmUmCrArGrArGmCrAdTsdT |
| Antisense: | rUrGrCrUrCrUrGrArArArUrUmCrArGrCmCrArGdTsdT |
Negative control siRNA.
| Sense: | mCmUmUAmCrGmCmUrGrArGmUrAmCmUmUmCrGAdTsdT |
| Antisense: | rUrCrGrArArGmUrArCrUmCrArGrCrGmUrArArGdTsdT |
Mouse IFNγ dicer substrate.
| Sense: | mCmArUmUrCmArUrGrArGrUmArUmUrGmCrCmArArGrUrUrUmGA |
| Antisense: | rUrCmArArArCrUrUmGrGmCrAmArUrArCrUrCrArUrGrAmArUmGmCmA |
Negative control dicer substrate.
| Sense: | mCmArUmArUmUrGrCrGrCrGmUrAmUrAmGrUmCrGrCrGrUrUmAG |
| Antisense: | rCrUmArArCrGrCrGmArCmUrAmUrArCrGrCrGrCrArArUmArUmGmGmU |
m: 2′-OMe-modified nucleotides
r: RNA bases (nucleotides without ‘r’ or ‘m’ represent DNA bases
s: represents a phosphorothioate linkage
Cloning
Sequences of the primers are listed in Supplementary Fig. 10. Murine MAdCAM-1 D1D2 was synthesized as a gBlock gene fragment (Integrated DNA Technologies, USA) and cDNA of rat IgG2a was obtained from an in-house hybridoma clone using RNeasy minkit (Qiagen, Netherlands) and qScript cDNA synthesis kit (Quantabio, USA). The Fc of rat IgG2a was amplified from the obtained cDNA using primer pair F1 and R1, the primers added the required homology sequences for the Gibson assembly to the 5’ ends of the amplicon. Next, the murine MAdCAM-D1D2 and IgG Fc were assembled in the pCMV3-FLAG plasmid using Gibson assembly (New England Biolabs, USA). In a later stage, the entire construct was reassembled in the pcDNA3.4 expression plasmid to improve protein yields using primer pair F2 and R2. The point mutation for the negative control was generated using site-directed mutagenesis, primers F3 and R3 amplified the entire plasmid while generating the point mutation. The resulting PCR product was DpnI digested, purified on a 0.8% agarose gel, phosphorylated using T4 Polynucleotide kinase (New England Biolabs, USA), circularized using T4 DNA Ligase (New England Biolabs, USA) and transformed into chemically competent bacteria. To improve flexibility in the context of the LNPs, the CH1 domain of Rat IgG2a was later removed by PCR amplification of the entire plasmid using primers F4 and R4. See Supplementary Fig. 10 for the sequence of the entire construct.
Cell Culture
Cell lines used: Expi293 (ThermoFisher Scientific), TK-1 (ATCC), HEK293 (ATCC). All cell lines in the laboratory were tested every 2 months for mycoplasma and discarded when positive. Expi293 cells were grown in Expi293 Expression Medium (ThermoFisher Scientific, USA) in disposable Erlenmeyer flasks at 37°C and 8% CO2 on a shaker rotating at 125 rpm. Cells were grown at densities between 0.3x106 – 5x106 cells per mL. At least 3 passages after thawing, cells were transfected with the expression plasmid encoding the MAdCAM-D1D2-Fc using the Expifectamine293 transfection kit (ThermoFisher Scientific, USA). 18 h post transfection, enhancer 1 and enhancer 2 from the Expifectamine293 transfection kit were added to boost recombinant protein expression levels. Five days post transfection the culture medium was harvested for purification of the secreted protein.
TK-1 cells were grown in RPMI1640 medium supplemented with 10% fetal bovine serum, L-glutamine and Pen-Strep-Nystatin (Biological Industries, Israel). Cells were grown at densities between 0.3x106 – 2x106 cells per mL in either T25 or T75 cell culture flasks (Greiner bio-one, Austria).
Protein purification
Conditioned medium was separated from the Expi293 cells by centrifugation at 300 g for five minutes. The supernatant was centrifuged again at 5000 g for 20 min to remove cellular debris. Using the Äkta FPLC protein purification system (GE Healthcare, UK), the protein was purified with a 1 mL Histrap column (GE Healthcare, UK). The sample was adjusted to the composition of the binding buffer (20 mM NaPO4 at pH 7.4 and 500 mM NaCl) and the sample was passed through a 0.2 μm syringe filter (Sartorius, Germany) before loading into the FPLC. Flowrate during binding and elution was 0.5 mL/min and during washes 1 mL/min. The columns were washed with 20 mL of 0.5 M NaOH and 20 mL deionized water before it was equilibrated with 10 mL of binding buffer and loaded with the sample. Protein elution was done using binding buffer supplemented with 0.5 M imidazole. The elution was performed stepwise with incremental increases of ~30 mM imidazole per fraction until the final concentration of 0.5 M was reached. Fractions were loaded on SDS PAGE gel and stained with coomassie to determine which fractions contained the MAdCAM-Fc at sufficient purity. Pooled fractions were buffer exchanged to PBS using PD-10 desalting columns (GE Healthcare, UK). The purified protein was concentrated to >1 mg/mL using Amicon ultra centrifugal filters (EMD Millipore, USA), snap frozen in liquid nitrogen and stored at -80°C.
SDS PAGE gel analysis
Protein sample was diluted in sample buffer with or without the presence of 500 mM DTT to compare between monomers and dimers (presence of the hinge domain should dimerize the D1D2). Samples were loaded on a 10% polyacrylamide gel and either stained with Coomassie (Bio-rad, USA) or transferred to a nitrocellulose membrane using iBlot 2 Dry Blotting System (ThermoFisher Scientific, USA). Membrane was blocked with 5% skim milk in PBS for 2 hours at RT and incubated with anti-rat IgG-HRP. Membrane was washed with PBS + 0.01% Tween20 and developed using SuperSignal West Pico chemiluminescent substrate (ThermoFisher Scientific, USA). Chemiluminescence was measured with the Amersham Imager 600 (GE Healthcare, USA).
In vitro binding
TK-1 cells were activated according to Y. Yang et al.38. Briefly, cells were washed with PBS and resuspended in resuspension buffer (HBSS with 10mM HEPES buffer, 2 mM CaCl2 and 2 mM MgCl2). Non-activated cells were kept on ice while activated cells were resuspended in pre-incubation buffer (HBSS with 10 mM HEPES buffer and 2 mM EDTA), incubated at RT for 30 min. with gentle rotation, washed with PBS and finally resuspended in activation buffer (HBSS with 10 mM HEPES buffer, 2 mM CaCl2 and 2 mM MnCl2). MAdCAM-Fc and the controls were added to both activated and non-activated cells and incubated for 30 min. at 4°C. Cells were subsequently washed and stained with either anti-human IgG or anti-mouse IgG conjugated to AlexaFluor647. Binding of MAdCAM to TK-1 cells was assessed by analyzing the fluorescence of the cells by flow cytometry. Binding of MAdCAM-targeted Cy5-LNPs was detected by flow cytometry directly using the Cy5 fluorescence (without addition of another antibody). Because of a higher background using LNPs, in binding experiments with Cy5-LNPs, the HBSS in the activation buffer was replaced with RPMI + 10% FBS.
Preparation of LNPs
NPs were prepared as previously described 39 by using the Nanoassemblr microfluidic mixer (Precision Nanosystems, Canada). Here, we utilized the current gold standard for LNP production, DLin-MC3-DMA, an ionizable lipid with a pKa of 6.44 that has a positive charge under acidic conditions. LNPs were prepared at pH 4.5 to ensure that DLin-MC3-DMA is ionized and hence siRNA encapsulation is maximized. Lipid mixture (DLin-MC3-DMA, DSPC, Cholesterol, PEG-DMG and DSPE-PEG-Maleimide at 50:10:38:1.5:0.5 molar ratio) in ethanol was mixed with siRNA in acetate buffer, pH 4.5 at a combined flowrate of 2 mL/min. Lipid and siRNA were mixed at a 1:3 volume ratio (1:16 w/w siRNA to lipid). For Cy5-labeled LNPs, 20% Cy5-labeled siRNA was mixed with 80% unlabeled siRNA. The resulting LNPs were dialyzed against PBS for 24 h to remove the ethanol and restore the pH to neutral. The hydrodynamic diameter and Zeta potential of the LNPs were measured by dynamic light scattering using disposable cuvettes in the Malvern Zetasizer (Malvern Instruments, UK).
Determining the siRNA encapsulation efficiency
LNPs were either lysed with Triton X-100 or not and the total amount of siRNA in the sample was measured with the Quant-iT Ribogreen RNA assay kit (ThermoFisher Scientific, USA). After subtracting the blank measurement, the encapsulation efficiency (in percentage) was calculated by
Conjugation of RG7 to LNPs and gel filtration
RG7/1.3 antibody was reduced in PBS supplemented with 1 mM DTT and 5 mM EDTA by incubating 1 h at room temperature. The DTT was subsequently removed by buffer exchange to 5 mM EDTA in PBS using 7K Zeba spin desalting columns (ThermoFisher Scientific, USA). Immediately after buffer exchange, the reduced antibody was added to the LNPs at a ratio of 0.67 mg antibody per mL of LNPs. The mixture was incubated for 2 h at room temperature with gentle shaking followed by overnight incubation at 4 °C. The next day, LNPs were separated from the free antibody using sepharose CL4B beads on a gel filtration column with PBS as the mobile phase. Fractions containing pure LNPs were pooled and concentrated to the initial volume using 100K Amicon centrifugal filters (EMD Millipore, USA). The loss of LNPs during conjugation and gel filtration was estimated by lysing LNPs before and after conjugation with Triton X-100 followed by measuring the amount of released siRNA using Quant-iT Ribogreen RNA assay kit (ThermoFisher Scientific, USA).
Dotblot analysis
Dotblot was performed using Minifold I system 96-well device (GE Healthcare, UK). A nitrocellulose membrane was added on top of two filter papers (Whatman plc, UK) and loaded into the 96 dotblot device. Wells were filled with PBS and vacuum was applied to wet the membrane. Samples (conjugated LNPs, unconjugated LNPs and several different amounts of free antibody) were added to the wells in a 100 μL volume followed by a vacuum to pass the sample through the membrane. Wells were washed with PBS and the device was again applied to a vacuum. Next, the membrane was blocked with 5% skim milk in PBS for 2 h followed by incubation with anti-mouse IgG antibody linked to HRP (diluted in PBS with 1% skim milk) for 1 h at RT. Next, the membrane was washed 3 times with PBST (5 min. per wash) and the samples were detected by adding the SuperSignal West Pico chemiluminescent substrate (ThermoFisher Scientific, USA). Chemiluminescence was measured with the Amersham Imager 600 (GE Healthcare, USA).
Confocal microscopy
Cells were stained with Hoechst (nucleus) and with AlexaFluor 488-conjugated anti-CD45 (cell membrane). Cells were resuspended in PBS and images were taken with a Zeiss confocal microscope. The images were created by merging 11 frames from a Z-stack with 0.3 μm per frame.
Ex vivo binding
Leukocytes were extracted from the spleen and mLN of both healthy C57Bl/6 mice and IL-10KO mice that spontaneously developed colitis. For the mLN, tissue was homogenized and cells were strained through a 70 μm cell strainer. Cells were washed with PBS, centrifuged and the pellet was washed again with PBS and resuspended as a single cell suspension. For the spleen, tissue was homogenized, cells were strained through a 70 μm cell strainer. After a single wash with PBS, red blood cells were lysed with ddH2O for a few seconds followed by addition of 10x HBSS to restore the solution to physiological conditions. Cells were strained a second time to ensure a single-cell suspension. Next, cells were stained with the appropriate cell surface markers (CD4, CD8, CD19, CD11b) and allowed to bind to either D1D2-LNPs or DATK32-LNPs (or its respective controls: mD1D2-LNPs, isotype-LNPs). After a 20-min. incubation at 4 °C, cells were washed and resuspended in FACS buffer. LNP binding for each leukocyte subpopulation was determined by the level of Cy5 as measured by flow cytometry.
Animal experiments
The Tel Aviv Institutional Animal Care and Use Committee approved the animal protocols for all in vivo studies in accordance with current regulations and standards of the Israel Ministry of Health.
In vivo gene silencing
Wild type C57BL/6 and IL-10KO C57BL/6 mice were kept in a specific-pathogen-free animal facility at Tel Aviv University. For DSS colitis model, DSS was added to the drinking water at a concentration of 1.5% for a total of 10 d. Five days after DSS administration, LNPs were injected intravenously and CD45 expression in various organs was assessed 5 d post-injection. For the PAC colitis model, piroxicam was administered to IL-10KO mice in the chow at a concentration of 200 ppm for a total of 11 d. 7 d after administration of piroxicam, LNPs were injected and CD45 expression in various organs was assessed 4 d post-injection. For healthy mice, CD45 expression was assessed 4 d post-injection. After sacrificing the animals, organs were homogenized and single cell suspensions were obtained using 70 μm cell strainers (Corning, USA). Cells were stained with antibodies against CD3 (Brilliant Violet 421), CD4 (APC-Cy7), CD8a (AlexaFluor 488), CD19 (PE-Cy5.5), CD11b (Brilliant Violet 650), F4/80 (Brilliant Violet 605) and CD45 (AlexaFluor 647) and analyzed by flow cytometry.
In vivo safety study
Female, 10-week-old C57Bl/6 mice (Harlan laboratories) were injected with LNPs at a dose of 1.5 mg/kg and sacrificed 24 h later. Blood was collected and analyzed by A.M.L. Israel for complete blood count (Sysmex and Advida-120) and biochemistry (Cobas-6000). Liver samples were used for histology (Histospeck, Israel). Splenic TNF-α and IL-6 levels were measured by DuoSet ELISA kits (R&D Systems, USA).
Therapeutic efficacy studies with siIFNγ
To test in silico optimized siRNA sequences against murine IFNγ, HEK293 cells stably expressing murine IFNγ were generated by transfecting the cells with a pcDNA3 plasmid harboring the murine IFNγ gene. Stably expressing cells were selected with G418. Next, cells were transfected with each of the siRNA sequences and with a control sequence (siNC). 48 h post-transfection, cells were lysed, RNA was extracted and cDNA was generated. Silencing efficiency was determined using qPCR with SYBR green. The sequence that most efficiently silenced IFNγ was used for the subsequent efficacy experiments.
For the efficacy studies, colitis was induced in 9-week-old female C57BL/6 IL-10KO mice by mixing piroxicam (200 ppm) in the food. Freshly prepared LNPs (encapsulated with either siIFNγ or siNC) were conjugated to RG7 and purified using CL4B resin. LNPs were injected intravenously at day 4, 6, 8 and 10, post piroxicam administration. The volume of administered LNPs was calculated with the Ribogreen assay (Thermo Fisher Scientific, USA) for each LNP preparation to ensure a consistent dose of 1.5 mg siRNA per kg body weight. 30 min. prior to injection, LNPs were mixed with the D1D2 or mD1D2 (final protein concentration of 60 μg/mL) and the total volume was completed to 200 μL.
Animals were randomized before piroxicam treatment and the study was performed in a double-blinded fashion. The experiment was performed by a CRO to ensure that the investigator was blinded during group allocation.
Body weight was recorded daily and at day 11 of the experiment the mice were sacrificed. The colon was harvested to measure the length, to analyze colonic cytokine levels and to perform colon histology. Blood samples were collected to measure IL-6 and IL-1β expression levels.
Molecular imaging studies
1,4,7-Triazacyclononane-1,4-bis-acetic acid-7-maleimidoethylacetamide (NOTA-mal: Macrocyclics; Dallas, TX) was conjugated to the immunoprotein using a previously described approach with minor modifications40. Briefly, to 500 μg of protein in 150 μL of phosphate buffer (pH 7.0) was added freshly prepared 2-iminothiolane and then NOTA-mal in phosphate buffer (10% dimethyl sulphoxide) and 2IT such that final concentration ratios were: Ab, 1; NOTA-mal, 20; 2IT, 10. The reaction was mixed by gentle pipetting, briefly centrifuged, and then placed in a 37 °C water bath for 30 min. Unbound chelator was removed using centrifugal filter units (3 kDa MW cut-off; Centricon, Millipore, Billerica MA), and the immunoconjugate was concentrated into phosphate buffer (0.1 M, pH 7.0) and stored in aliquots at -80 °C.
For the radiolabeling, 6 volume equivalents of sodium acetate buffer were added to 178 M Bq (4.78 mCi) 64Cu in 5 μL HCl (0.04 N). Antibody (177 μg in 61 μL phosphate buffer) was then added to 15.8 μL of 64Cu solution (31.7 MBq (858 μCi)). After 30 min incubation at room temperature, the degree of radiolabeling was assessed by thin-layer chromatography (TLC; Whatman No.1 paper eluted with phosphate buffer; 0.1 M, pH 8, 100 mM EDTA) and radiochemical purity was found to be >95%. The radioimmunoconjugate was diluted with saline and sterile filtered (0.2 μm) before injection.
For the imaging studies, 64Cu-labeled antibody (22 μg antibody, 3.12 MBq (84.3 μCi)) was injected into the tail vein. The mice were then anesthetized using isoflurane (1-4% in oxygen) and placed in a Bruker Albira multimodality (PET/CT) small-animal imaging system (Bruker Corporation; Woodbridge CT) for imaging. PET/CT data were collected for 30 min. at 1, 3 and 24 h post-injection (p.i.). At 24 h p.i., the mice were euthanized by CO2 inhalation, and an ex vivo biodistribution analysis was carried out. Tissues were collected and weighed, and the radioactivity was assayed. The large colon was excised, measured and weighed to confirm the presence of colitis in the piroxicam treated group. PET and CT images were registered manually using AMIDE software41. Data from volumes of interest (VOIs) were used to calculate the biodistribution in selected tissues for the small animal PET imaging studies.
Statistical analysis
Data in the bar charts is expressed as mean ± standard deviation unless indicated otherwise. Boxplots center line represents the median, box represents the interquartile range and whiskers represent minimum and maximum values. Statistical analysis was performed in GraphPad Prism. In general, when comparing 2 groups (for instance mD1D2 to D1D2), Student’s t-test was used, and a one-way ANOVA was used when comparing 3 or more groups. A more complex statistical model (2-way interaction mixed model) was used in the molecular imaging section to confirm the correlation between colitis severity and D1D2 uptake, and this is shown in Supplementary Fig. 6.
More specifically: for the CD45 silencing, a two-sided Student’s t-test was performed between the groups mD1D2 and D1D2. For the toxicity study, a one-way ANOVA test was performed to exclude significant differences between any of the groups. For the efficacy study, a one-way ANOVA with Dunnett’s post hoc test was used to demonstrate a significant difference between the D1D2-IFNγ group and the negative control groups (D1D2-NC5, mD1D2-IFNγ and mock-treated). The same post hoc test revealed a significant difference between the groups D1D2-IFNγ and mAb TNF-α only in the colonic TNF-α expression levels (p < 0.05). Group size for the in vivo silencing and toxicity study was 5 mice per group. In the efficacy study, group size was increased to 12 mice per group due to relatively high standard deviations in smaller group sizes. In all figures * for p < 0.05, ** for p < 0.01 and *** for p < 0.0001.
Supplementary Material
Acknowledgments
We thank Dr. Vered Holdengreber for assistance with the TEM analysis, Patrick Johnston for detailed statistical analysis of the molecular imaging part, Dr. Sushmita Chatterjee for help with the confocal microscope and Prof. Steffen Jung for providing the IL-10KO mice. This work was supported by the ERC grant LeukoTheranostics (Award # 647410) to D.P.
Footnotes
Author contributions
N.D. and D.P. conceived the study. N.D., S.R., N.V., J.D. performed the experiments. N.D. J.D., A.P. and D.P. analyzed the data. N.D. and D.P. wrote the manuscript.
Competing interests
D.P. declares financial interests in Quiet Therapeutics and ART Bioscience. None of them relates to this work. The rest of the authors declare no financial interests.
Data availability
All relevant data are available from the authors upon reasonable request.
References
- 1.Elinav E, Peer D. Harnessing nanomedicine for mucosal theranostics-a silver bullet at last? ACS Nano. 2013;7:2883–90. doi: 10.1021/nn400885b. [DOI] [PubMed] [Google Scholar]
- 2.Hanauer SB, et al. Incidence and importance of antibody responses to infliximab after maintenance or episodic treatment in Crohn’s disease. Clin Gastroenterol Hepatol. 2004;2:542–553. doi: 10.1016/s1542-3565(04)00238-1. [DOI] [PubMed] [Google Scholar]
- 3.Baert F, et al. Influence of Immunogenicity on the Long-Term Efficacy of Infliximab in Crohn’s Disease. N Engl J Med. 2003;348:601–608. doi: 10.1056/NEJMoa020888. [DOI] [PubMed] [Google Scholar]
- 4.Kedmi R, et al. A modular platform for targeted RNAi therapeutics. Nat Nanotechnol. 2018;13:214–219. doi: 10.1038/s41565-017-0043-5. [DOI] [PubMed] [Google Scholar]
- 5.Veiga N, et al. Cell specific delivery of modified mRNA expressing therapeutic proteins to leukocytes. Nat Commun. 2018;9 doi: 10.1038/s41467-018-06936-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fenske DB, Chonn A, Cullis PR. Liposomal Nanomedicines: An Emerging Field. Toxicol Pathol. 2008;36:21–29. doi: 10.1177/0192623307310960. [DOI] [PubMed] [Google Scholar]
- 8.Ledford H. Gene silencing technology gets first drug approval after 20-year wait. Nature. 2018;560:291–292. doi: 10.1038/d41586-018-05867-7. [DOI] [PubMed] [Google Scholar]
- 9.Ramishetti S, et al. Systemic Gene Silencing in Primary T Lymphocytes Using Targeted Lipid Nanoparticles. ACS Nano. 2015;9:6706–6716. doi: 10.1021/acsnano.5b02796. [DOI] [PubMed] [Google Scholar]
- 10.Peer D, Park EJ, Morishita Y, Carman CV, Shimaoka M. Systemic leukocyte-directed siRNA delivery revealing cyclin D1 as an anti-inflammatory target. Science. 2008;319:627–30. doi: 10.1126/science.1149859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meenan J, et al. Altered expression of alpha 4 beta 7, a gut homing integrin, by circulating and mucosal T cells in colonic mucosal inflammation. Gut. 1997;40:241–246. doi: 10.1136/gut.40.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu Y, et al. Structural specializations of α4β7, an integrin that mediates rolling adhesion. J Cell Biol. 2012;196:131–46. doi: 10.1083/jcb.201110023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun H, et al. Distinct chemokine signaling regulates integrin ligand specificity to dictate tissue-specific lymphocyte homing. Dev Cell. 2014;30:61–70. doi: 10.1016/j.devcel.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 14.Lichtenstein GR, Hanauer SB, Sandborn WJ. Risk of Biologic Therapy-Associated Progressive Multifocal Leukoencephalopathy: Use of the JC Virus Antibody Assay in the Treatment of Moderate-to-Severe Crohn’s Disease. Gastroenterol Hepatol (N Y) 2012 [PMC free article] [PubMed] [Google Scholar]
- 15.Green N, et al. Mutational analysis of MAdCAM-1/alpha4beta7 interactions reveals significant binding determinants in both the first and second immunuglobulin domains. Cell Adhes Commun. 1999;7:167–81. doi: 10.3109/15419069909010800. [DOI] [PubMed] [Google Scholar]
- 16.Shyjan AM, Bertagnolli M, Kenney CJ, Briskin MJ. Human mucosal addressin cell adhesion molecule-1 (MAdCAM-1) demonstrates structural and functional similarities to the alpha 4 beta 7-integrin binding domains of murine MAdCAM-1, but extreme divergence of mucin-like sequences. J Immunol. 1996;156:2851–7. [PubMed] [Google Scholar]
- 17.Berlin C, et al. α4β7 integrin mediates lymphocyte binding to the mucosal vascular addressin MAdCAM-1. Cell. 1993;74:185–195. doi: 10.1016/0092-8674(93)90305-a. [DOI] [PubMed] [Google Scholar]
- 18.Dearling JLJ, et al. Detection of intestinal inflammation by MicroPET imaging using a 64Cu-labeled anti-β7 integrin antibody. Inflamm Bowel Dis. 2010;16:1458–1466. doi: 10.1002/ibd.21231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dearling JLJ, Daka A, Veiga N, Peer D, Packard AB. Colitis ImmunoPET: Defining Target Cell Populations and Optimizing Pharmacokinetics. Inflamm Bowel Dis. 2016;22:529–538. doi: 10.1097/MIB.0000000000000677. [DOI] [PubMed] [Google Scholar]
- 20.Belliveau NM, et al. Microfluidic Synthesis of Highly Potent Limit-size Lipid Nanoparticles for In Vivo Delivery of siRNA. Mol Ther Nucleic Acids. 2012;1:e37. doi: 10.1038/mtna.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen ZR, et al. Localized RNAi therapeutics of chemoresistant grade IV glioma using hyaluronan-grafted lipid-based nanoparticles. ACS Nano. 2015;9:1581–1591. doi: 10.1021/nn506248s. [DOI] [PubMed] [Google Scholar]
- 22.Veiga N, et al. Leukocyte-specific siRNA delivery revealing IRF8 as a potential anti-inflammatory target. J Control Release. 2019;313:33–41. doi: 10.1016/j.jconrel.2019.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Rosenblum D, Joshi N, Tao W, Karp JM, Peer D. Progress and challenges towards targeted delivery of cancer therapeutics. Nat Commun. 2018;9:1410. doi: 10.1038/s41467-018-03705-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chassaing B, Aitken JD, Malleshappa M, Vijay-Kumar M. Current Protocols in Immunology. Vol. 104. John Wiley & Sons, Inc; 2014. Dextran Sulfate Sodium (DSS)-Induced Colitis in Mice; 15.25.1-15.25.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berg DJ, et al. Rapid development of colitis in NSAID-treated IL-10 deficient mice. Gastroenterology. 2002;123:1527–1542. doi: 10.1053/gast.2002.1231527. [DOI] [PubMed] [Google Scholar]
- 26.Holgersen K, Kvist PH, Markholst H, Kornerup Hansen A, Holm TL. Characterisation of enterocolitis in the piroxicam-accelerated interleukin-10 knock out mouse — A model mimicking inflammatory bowel disease ☆. 2012;8:147–160. doi: 10.1016/j.crohns.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 27.Dieleman LA, et al. Dextran sulfate sodium-induced colitis occurs in severe combined immunodeficient mice. Gastroenterology. 1994;107:1643–1652. doi: 10.1016/0016-5085(94)90803-6. [DOI] [PubMed] [Google Scholar]
- 28.Holgersen K, Kvist PH, Hansen AK, Holm TL. Predictive validity and immune cell involvement in the pathogenesis of piroxicam-accelerated colitis in interleukin-10 knockout mice. Int Immunopharmacol. 2014;21:137–147. doi: 10.1016/j.intimp.2014.04.017. [DOI] [PubMed] [Google Scholar]
- 29.Connor EM, Eppihimer MJ, Morise Z, Granger DN, Grisham MB. Expression of mucosal addressin cell adhesion molecule-1 (MAdCAM-1) in acute and chronic inflammation. doi: 10.1002/jlb.65.3.349. [DOI] [PubMed] [Google Scholar]
- 30.Ito R, et al. Interferon-gamma is causatively involved in experimental inflammatory bowel disease in mice. Clin Exp Immunol. 2006;146:330–8. doi: 10.1111/j.1365-2249.2006.03214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferreiro R, Barreiro-de Acosta M. Infliximab: Pharmacology, Uses and Limitations. Vol. 10. Nova Science Publishers, Inc; 2012. Infliximab in inflammatory bowel disease; pp. 39–74. [Google Scholar]
- 32.Vila-del Sol V, Punzón C, Fresno M. IFN-γ-Induced TNF-α Expression Is Regulated by Interferon Regulatory Factors 1 and 8 in Mouse Macrophages. J Immunol. 2008;181:4461–4470. doi: 10.4049/jimmunol.181.7.4461. [DOI] [PubMed] [Google Scholar]
- 33.Wesemann DR, Benveniste EN. STAT-1α and IFN-γ as Modulators of TNF-α Signaling in Macrophages: Regulation and Functional Implications of the TNF Receptor 1:STAT-1α Complex. J Immunol. 2003;171:5313–5319. doi: 10.4049/jimmunol.171.10.5313. [DOI] [PubMed] [Google Scholar]
- 34.Shimaoka M, et al. AL-57, a ligand-mimetic antibody to integrin LFA-1, reveals chemokine-induced affinity up-regulation in lymphocytes. Proc Natl Acad Sci U S A. 2006;103:13991–13996. doi: 10.1073/pnas.0605716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qi JP, et al. Identification, characterization, and epitope mapping of human monoclonal antibody J19 that specifically recognizes activated integrin α4β7. J Biol Chem. 2012;287:15749–15759. doi: 10.1074/jbc.M112.341263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kinashi T. Intracellular signalling controlling integrin activation in lymphocytes. Nature Reviews Immunology. 2005;5:546–559. doi: 10.1038/nri1646. [DOI] [PubMed] [Google Scholar]
- 37.Eun JP, et al. Aberrant activation of integrin α4β7suppresses lymphocyte migration to the gut. J Clin Invest. 2007;117:2526–2538. doi: 10.1172/JCI31570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.YANG Y, et al. Construction and Adhesive Properties of a Soluble MAdCAM‐1‐Fc Chimera Expressed in a Baculovirus System: Phylogenetic Conservation of Receptor‐Ligand Interaction. Scand J Immunol. 1995;42:235–247. doi: 10.1111/j.1365-3083.1995.tb03650.x. [DOI] [PubMed] [Google Scholar]
- 39.Rungta RL, et al. Lipid Nanoparticle Delivery of siRNA to Silence Neuronal Gene Expression in the Brain. Mol Ther Nucleic Acids. 2013;2:e136. doi: 10.1038/mtna.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCall MJ, Diril H, Meares CF. Simplified Method for Conjugating Macrocyclic Bifunctional Chelating Agents to Antibodies via 2-Iminothiolane. Bioconjugate Chemistry. 1990;1:222–226. doi: 10.1021/bc00003a007. [DOI] [PubMed] [Google Scholar]
- 41.Loening AM, Gambhir SS. AMIDE: A Free Software Tool for Multimodality Medical Image Analysis. Mol Imaging. 2003;2:131–137. doi: 10.1162/15353500200303133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are available from the authors upon reasonable request.