Abstract
Membrane potential reflects the difference between cytoplasmic and apoplastic electrical potentials and is essential for cellular operation. The application of the phytohormone auxin (IAA) causes instantaneous membrane depolarization in various cell types1–6, making depolarization a hallmark of IAA-induced rapid responses. In root hairs, depolarization requires functional IAA transport and TIR1/AFB signalling5 but its physiological significance is not understood. Specifically in roots, auxin triggers rapid growth inhibition7–9 (RGI); a process required for gravitropic bending. RGI is initiated by the TIR1/AFB co-receptors, AFB1 paralogue playing a crucial role10,11. The nature of the underlying rapid signalling is unknown, as well as the molecular machinery executing it. Even though the growth and depolarization responses to auxin show remarkable similarities, the significance of membrane depolarization for root growth inhibition and gravitropism is unclear. Here, by combining the DISBAC2(3) voltage sensor with microfluidics and vertical-stage microscopy, we show that rapid auxin-induced membrane depolarization tightly correlates with rapid RGI. Rapid depolarization and RGI require the AFB1 auxin co-receptor. Finally, AFB1 is essential for the rapid formation of membrane depolarization gradient across the gravistimulated root. These results clarify the role of AFB1 as the central receptor for rapid auxin responses.
Keywords: AUX1, cvxIAA, DISBAC2(3), Gravitropism, PIN2, Vertical stage microscopy
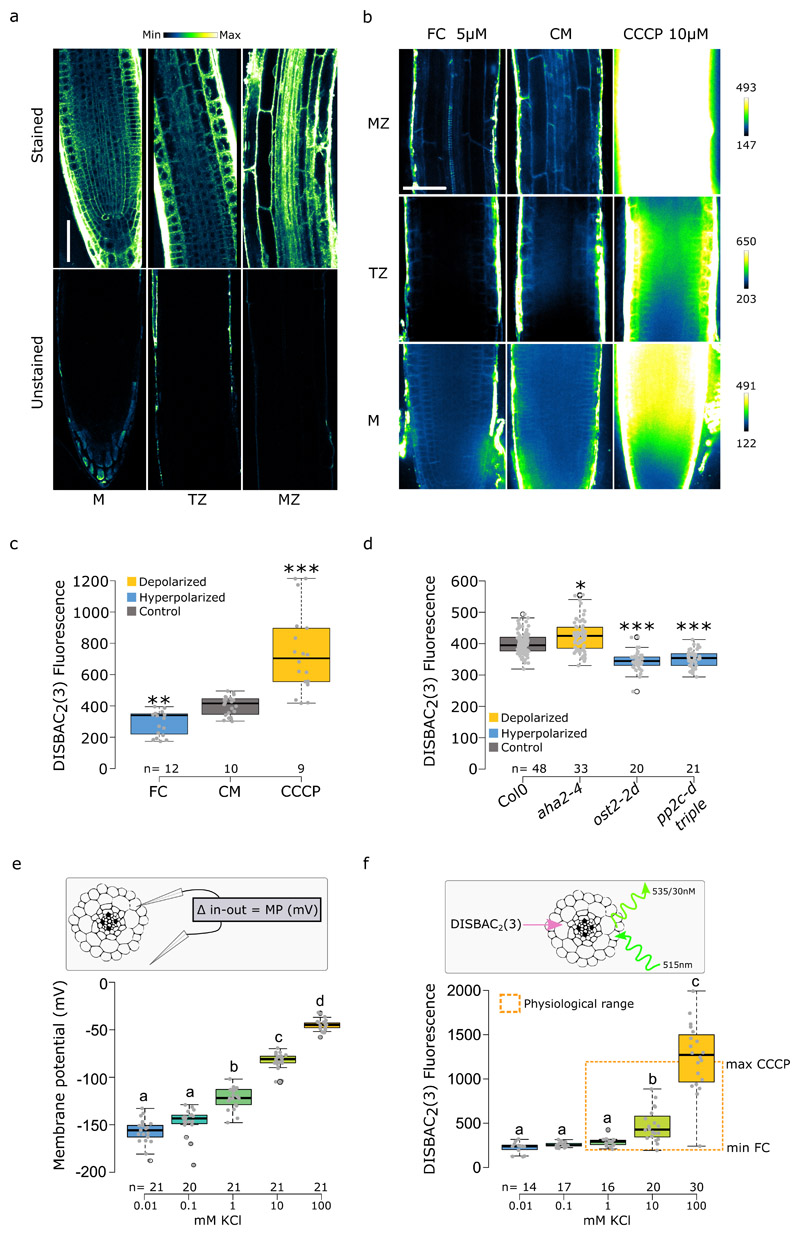
As the traditional membrane potential measurement by impaling cells with microelectrodes is incompatible with undisturbed root growth, we tested the membrane potential fluorescent dye DISBAC2(3)12 to optimize simultaneous detection of growth and membrane potential in Arabidopsis thaliana roots. The voltage-sensor visibly stained all root tissues in seedlings observed on a horizontal stage microscope with a multi-photon excitation (Fig.1a), with a stronger signal in the epidermis of the meristematic and elongation zones as well as a nonspecific signal in the dead lateral root cap cells. To follow root elongation and gravitropic response, we imaged growing roots using a vertical-stage13 spinning disk microscope. Spinning disk microscopy is characteristic by high temporal resolution and low phototoxicity, features ideal for imaging of rapid responses of roots to auxin. However, it is less optimal for imaging of deep tissues, therefore the inner tissues appeared less stained by the voltage sensor (Fig.1b). DISBAC2(3) staining was not toxic as judged by the growth of the primary root (Fig.S1a,b). To verify that we could report changes in membrane potential with the voltage sensor we first, quantified the effects of two known membrane polarization disruptors on the fluorescence at the interface between inner epidermis/outer cortex cells. Application of fusicoccin (FC), an activator of the plasma membrane proton pump14, decreased the DISBAC2(3) fluorescence compared to the control medium (CM) indicating hyperpolarization15,16 (Fig.1b,c). On the other hand, Carbonyl cyanide m-chlorophenyl hydrazone (CCCP), an uncoupler that dissipates the proton gradient17, significantly increased the fluorescence reporting a depolarized state (Fig. 1b,c). Secondly, to test whether the sensor can detect more subtle changes, we genetically disrupted the resting membrane potential using mutants affected in the expression or activity of the plasma membrane AHA H+-ATPase proton pumps. The aha2-4 mutant lacks one of the two major AHA isoforms, the ost2-2d harbors a mutation in AHA1 which renders it constitutively active18, and, similarly, the absence of PP2C-D2,5,6 phosphatases in the pp2c-d triple mutant leads to an increased activity of the proton pumps19. The DISBAC2(3) staining revealed that aha2-4 mutant roots were slightly depolarized compared to Col0 (Fig.1d), while the constitutive activation of the proton pumps in ost2-2d and pp2c-d triple mutants led to a significant hyperpolarization (Fig.1d) similar to the FC treatment (Fig1b,c). Next, to gain insight into the physiological detection range of DISBAC2(3), we compared direct membrane potential measurements (using an impaled microprobe, Fig.1e) with the fluorescence measurements in response to a KCl gradient (Fig.1f, S1c). Both methods recorded similar profiles. However, DISBAC2(3) displayed lower sensitivity in the detection of deep hyperpolarized states, with no detectable differences between 0.01mM, 0.1mM and 1mM KCl. Nevertheless, the range of significant fluorescence detection was covering the fluorescence observed in the control medium, FC and CCCP treatments (Fig.1b,c) and the membrane potential values in resting state or treatment from various plant species recovered from the literature (around -120mV, Supplemental table I). Finally, we compared the membrane potential of seedlings transferred to ½ MS +/- sucrose measured by both methods (Fig.S1d,e). Both methods reported a similar significant hyperpolarization of the root when sucrose was added to the medium. This finding is in accordance with the literature, as adding 1% sucrose to the ½ MS made the medium more hypertonic due to sucrose osmotic properties. Hypertonic solution with other sugars (mannitol and sorbitol) were shown to hyperpolarize Arabidopsis thaliana roots20.
Fig. 1. DISBAC2(3) reports membrane potential in Arabidopsis thaliana roots.
(a) DISBAC2(3) stains all tissues of the meristem (M) transition zone (TZ) and mature zone (MZ). Unstained roots shown on the bottom. Roots were imaged using the multi-photon excitation in photon-counting mode (lookup table on top, scale bar = 50μm). (b,c) Effect of 5μM FC and 10μM CCCP compared to control medium (CM) on DISBAC2(3) fluorescence after 20 minutes of treatment; (b) representative images (lookup table for each zone on the right, scale bar = 25μm) and (c) fluorescence quantification. (d) Comparison of Col0 DISBAC2(3) basal fluorescence to the basal fluorescence of mutants affected in the activity of the AHA H+-ATPases: aha2-4, ost2-2d and pp2c-d triple (2,5,6). (e) Comparison of impaling probe membrane potential measurements (in mV) and f) DISBAC2(3) fluorescence (arbitrary unit) in response to a KCl gradient. Orange dash box represents DISBAC2(3) maximum fluorescence values of CCCP treatment and minimum value of FC treatment, indicating that DISBAC2(3) detection range covers the physiological range of membrane potential. n= is indicated on figures. (a) Representative microscopy pictures of two independent experiments. Statistical analysis for (c,d) were conducted with a two-sided non-parametric multiple comparison with Dunnett contrast and logit approximation. ***: p-value<0.0005, **: p-value<0.005, *: p-value<0.05. For (e,f) statistics were conducted with Tukey contrast. Letters indicates the significantly different statistical group with a p-value<0.05 minimum.
Taken together, these results indicated that, despite its non-linear responses and reduced sensitivity in the 0.01 to 1mM KCl range, DISBAC2(3) can reliably report membrane potential variation under majority of physiologically relevant conditions, in both the basic (buffered KCl) or complex media (½ MS +/- sucrose). DISBAC2(3) can, then, be used to non-invasively monitor spatiotemporal kinetics of membrane potential changes. It should be kept in mind, however, that as the fluorescence intensity depends on variables such as staining efficiency, laser intensity, or microscope detector sensitivity, all these parameters should be “locked” (set identical) for any comparative analysis, and the method should be used to determine relative values of membrane potential in relation to an internal control (such as control imaged alongside with mutants or the pre-treatment fluorescence level).
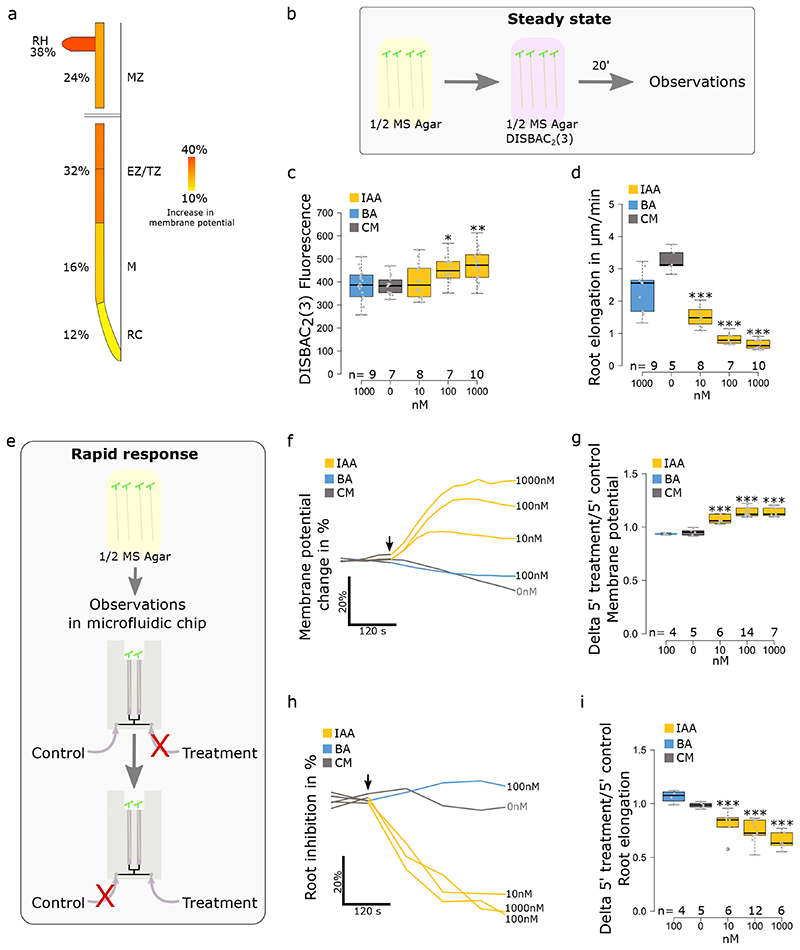
Auxin triggers root growth inhibition within ~30 seconds and this rapid and reversible response lasts for tens of minutes10. Therefore, we measured the steady state growth and membrane potential response (>20 min after IAA application) by imaging seedlings in microscopy chambers containing agar medium with the respective treatments. To capture the initial phases of the response (<5 minutes), we imaged the seedlings in closable microfluidic chips (Fig.S2a,b). Applied at 100nM, the native auxin IAA triggered a spatially heterogeneous depolarization in Arabidopsis thaliana roots (Fig.2a, Sup. movie 1). After 5 minutes of treatment, root hairs and epidermal cells in the transition/elongation zones showed the strongest depolarization. For this reason and given that this zone is a hotspot of root growth regulation21, we decided to focus on membrane potential responses in the cells of the transition zone for further experiments. To analyze whether membrane depolarization is proportional to RGI, we analyzed root response to a gradient of IAA (0, 10, 100, 1000nM; Fig.2b–d and S2c). In steady state (Fig.2b), a significant depolarization was only detected at high IAA concentrations (100 and 1000nM; Fig.2c and S2c) while growth was strongly inhibited by 10nM IAA (Fig.2d). We further focused on the initial stages of the response (Fig.2e). Application of 10 nM IAA (or higher) triggered an instantaneous (<30s) membrane depolarization and the response amplitude was positively correlated with IAA concentration (Fig.2f,g). It is worth noting that in microfluidic experiments, the switch from control-to-control condition triggers a small decrease in fluorescence, which we attribute to sticking of the dye to the microfluidic tubing in the non-flowing inlet. In parallel, RGI began instantly (<30s), but the amplitude was similar between concentrations after 5 minutes (Fig.2h,i). Benzoic acid did not elicit membrane depolarization (Fig.2c,f,g) and did not inhibit growth (Fig.2h,i), apart from a partial inhibition at a high concentration (Fig.2d). These results show that physiological IAA levels elicit membrane depolarization and growth inhibition with a similar temporal dynamics and concentration dependence. Neither growth inhibition nor membrane depolarization were caused by the weak acid properties of IAA. The membrane depolarization was only maintained over time at higher concentrations of IAA (100, 1000nM).
Fig. 2. Auxin induces depolarization and root growth inhibition in Arabidopsis thaliana root tip.
(a) Root epidermis membrane potential response to 100nM IAA in the root cap (RC), meristematic zone (M), transition/elongations zone (TZ and EZ) and mature zone (MZ). Percentages represent the delta of DISBAC2(3) fluorescence between control media and 5 minutes of treatment. n= 5 individual roots. (b) Schematic of the steady state response measurements. Seedlings were transferred to a ½ MS medium containing DISBAC2(3) and stained for 20 minutes before imaging. (c-d) Effects of an IAA concentration gradient on (c) steady state root tip membrane potential and (d) primary root elongation (μm/minute). (e) Schematic of the rapid response measurements. Seedlings were transferred to a microfluidic chip filled with ½ MS medium containing DISBAC2(3) and stained for 20 minutes before imaging. During imaging, the medium is switched from control to treatment. (f) Rapid membrane potential response average change (in %) over 5 minutes of treatment and (g) boxplot of the ratio of 5 minutes treatment over 5 minutes control media. h) rapid root elongation response to an auxin gradient, average change (in %) over 5 minutes of treatment and (i) boxplot of delta of 5 minutes treatment over 5 minutes control media. Application of treatments is indicated by a black arrow. For (f) and (h), standard errors were not added to simplify reading, instead the data dispersion is represented in (g) and (h). n= is indicated on figures. Statistical analysis for (c,d,g,i) were conducted with a two-sided non-parametric multiple comparison with Dunnett contrast and logit approximation. ***: p-value<0.0005, **: p-value<0.005, *: p-value<0.05.
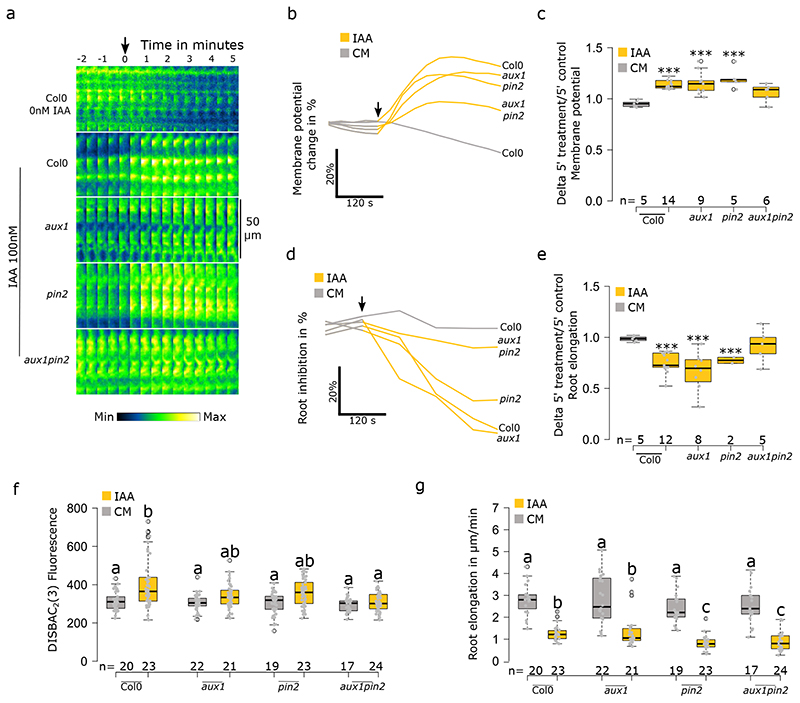
RGI requires intracellular accumulation of auxin, but not active transport across the plasma membrane per se 10. On the other hand, previous studies reported the importance of AUX1-mediated influx and PIN2-mediated efflux in the auxin-induced membrane depolarization in root hairs5,6. To investigate this contradiction, we genetically perturbed auxin influx, efflux or both in epidermal cells using aux1, pin2 and aux1pin2 mutants, respectively. Firstly, in rapid response to 10nM IAA, disruption of active IAA influx in aux1 resulted in no depolarization (Fig.S3a–c) correlated with the absence of rapid RGI over 5 minutes of treatment10 (Fig.S3d,e). On the other hand, the pin2 mutant impaired in IAA efflux, showed no difference compared to control response in both rapid depolarization (Fig.S3a-c) and rapid RGI (Fig.S3d,e). The simultaneous disruption of IAA influx and efflux in aux1pin2 resulted in a similar response to aux1 with no measurable rapid responses (Fig.S3a–e). Secondly, to investigate if these results were a consequence of an impaired transport or the lack of the transporters themselves, we saturated transport across the PM with 100nM IAA. In these conditions, aux1 and pin2 displayed statistically significant depolarizations, similarly to Col0, while only aux1pin2 displayed a depolarization which was not significant after 5 minutes of treatment (Fig.3a-c). These responses fully correlated with rapid RGI (Fig.3d,e). It is worth highlighting that aux1 took slightly longer to reach its maximum depolarization. Thirdly, in the steady state experiment with 1000nM IAA, we observed statistically non-significant depolarization tendencies in all three mutants, while the control roots were significantly depolarized (Fig.3f). On the other hand, root growth was inhibited in all three mutants and the presence of the pin2 mutation caused a significant hypersensitivity to IAA (Fig.3g). Together, these results showed that AUX1-mediated IAA influx and PIN2-mediated efflux contributed to the maintenance of membrane depolarization, possibly by a constitutive cycling of IAA in and out of cells and the associated proton fluxes. In our experiments, the PIN2-mediated efflux was not required for rapid membrane depolarization, a result contradicting previous studies conducted with 10μM IAA in root hairs showing that both aux1 and pin2 mutants are impaired in the membrane potential response5,6. To put these observations in perspective, these measurements were obtained from trichoblasts which might react differently than primary root epidermis. Moreover, in pin2, the length and density of root hairs are diminished22 and thus, could be impaired in auxin response. Finally, at physiological IAA concentrations, AUX1-mediated auxin influx is required for rapid membrane depolarization. Nevertheless, depolarization can be achieved even in the aux1 mutant with high IAA concentrations, at which IAA diffuses into cells. This shows that the depolarization response is not caused by the process of AUX1-mediated IAA influx, but by the presence of auxin in the cell. Accordingly, it was shown that the intracellular injection of IAA into Arabidopsis epidermal cells can trigger calcium fluxes5. Thus, these data point towards the activity of an intracellular IAA receptor.
Fig. 3. Auxin transport through PIN2 and AUX1 is not essential for rapid membrane depolarization.
(a-e) Rapid membrane potential response to 100nM IAA in Col0, aux1, pin2, and aux1pin2. (a) Representative images (median measured values) of the membrane potential reporter in time. (b) average change (in %) over 5 minutes of treatment and (c) delta of 5 minutes treatment over 5 minutes control media. (d) Root elongation rapid response over 10 minutes of treatment; average change (in %) over 5 minutes of treatment and (e) delta of 5 minutes treatment over 5 minutes control media. (f) Effect of 1000nM IAA on Col0, aux1, pin2 and aux1pin2 on the steady state response of root tip membrane potential and (g) primary root elongation. Steady state corresponds to the fluorescence of roots after 20 minutes of treatment in agar medium and root elongation measured over 20 minutes. Rapid response corresponds to roots treated in microfluidics and measured every 30 seconds. Application of treatments is indicated by a black arrow. For (b) and (d), standard errors were not added to simplify reading, instead the data dispersion is represented in (c and e. In (a) the lookup table is indicated on the bottom. n= is indicated on figures. Statistical analysis for (c,e) were conducted with a two-sided non-parametric multiple comparison with Dunnett contrast and logit approximation. ***: p-value<0.0005, **: p-value<0.005, *: p-value<0.05. For (f,g) statistics were conducted with a two-sided non-parametric multiple comparison with Tukey contrast and logit approximation. Letters indicates the significantly different statistical group with a p-value<0.05 minimum.
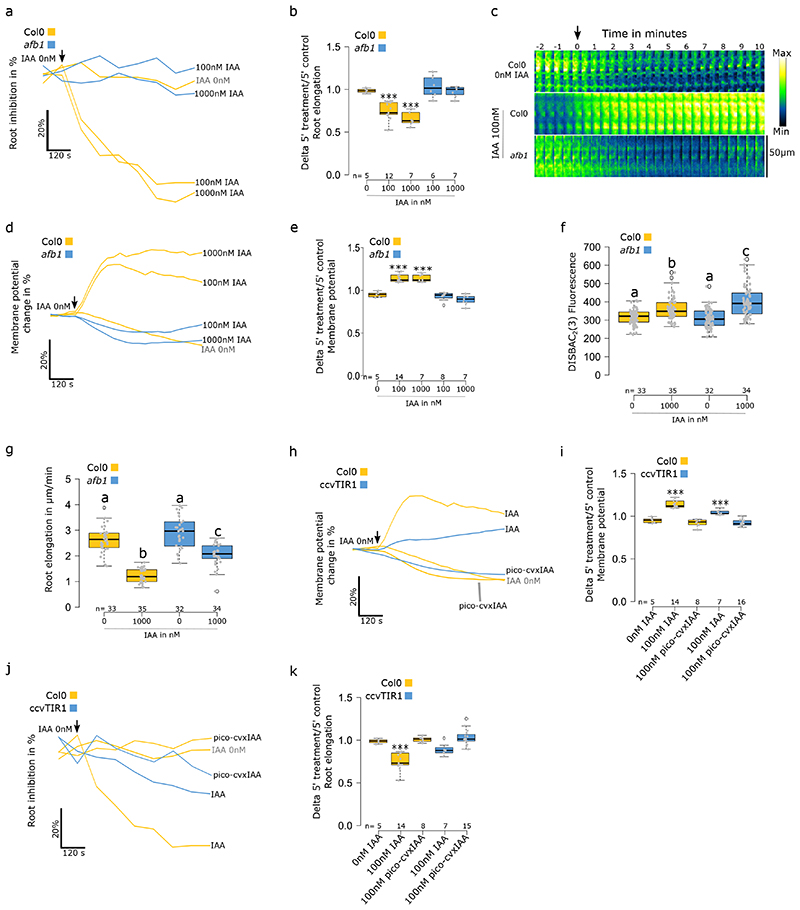
The Arabidopsis thaliana TIR1/AFB auxin coreceptor, encoded by six paralogous genes TIR1 and AFB1-523, has been independently reported as being involved in both rapid RGI and membrane depolarization induced by auxin. The tir1–1/afb2–1/afb3–1 mutant showed a reduced rapid RGI9,10 and decreased depolarization in trichoblast cells5. Recently, the so-far enigmatic AFB1 paralogue was revealed to play an important role in the rapid root growth response to auxin11. We confirmed the lack of rapid root inhibition response to auxin in afb1-3 (Fig.4a). At the same time, the afb1-3 mutant showed a total lack of immediate membrane depolarization response to auxin, even when high IAA concentration was applied (Fig.4c-e). Interestingly, after 5 minutes of treatment, afb1-3 cells initiated a trend for a mild depolarization (Fig.4c,d). In the steady state, afb1-3 showed a slightly stronger depolarization in response to IAA (Fig.4f) while the root growth was inhibited, albeit less than Col0 (Fig.4g). This indicated a delay in the mutant’s response to auxin and highlights the crucial role for AFB1 in the earliest auxin response.
Fig. 4. AFB1 triggers rapid membrane depolarization in response to IAA.
(a-g) Effect of IAA on Col0 and afb1-3. (a) Rapid root elongation response, average change (in %) over 10 minutes of treatment and (b) delta of 5 minutes treatment over 5 minutes control media. (c-e) Membrane potential rapid response changes over 10 minutes of treatment. (c) Representative images of the membrane potential reporter in time (median measured values), (d) average change (in %) over 10 minutes of treatment and (e) delta of 5 minutes treatment over 5 minutes control media. (f) Steady state response of root tip membrane potential and (g) primary root elongation. (h-k) Effect of IAA and pico-cvxIAA on Col0 and ccvTIR1 roots. (h) Rapid membrane potential response; average change (in %) over 10 minutes of treatment and (i) delta of 5 minutes treatment over 5 minutes control media. (j) Root elongation rapid response over 10 minutes of treatment; average change (in %) over 5 minutes of treatment and (k) delta of 5 minutes treatment over 5 minutes control media. Steady state corresponds to the fluorescence of roots after 20 minutes of treatment in agar medium and root elongation measured over 20 minutes. Rapid response corresponds to roots treated in microfluidics and measured every 30 seconds. Application of treatments is indicated by a black arrow. For (a), (c), (f) and h, standard errors were not added to simplify reading, instead the data dispersion is represented in (b), (e), (i) and (k). (c) Fluorescence intensity lookup table is indicated on the right. n= is indicated on figures. Statistical analysis for (b,e,I,k) were conducted with a two-sided non-parametric multiple comparison with Dunnett contrast and logit approximation. ***: p-value<0.0005, **: p-value<0.005, *: p-value<0.05. For (f,g) statistics were conducted with a two-sided non-parametric multiple comparison with Tukey contrast and logit approximation. Letters indicates the significantly different statistical group with a p-value<0.05 minimum.
Further, we used the orthogonal ccvTIR1-cvxIAA receptor-auxin pair24, in which all the native TIR1/AFB paralogues are shunted and only the synthetic ccvTIR1 receptor is active when synthetic cvxIAA is applied. The cvxIAA was shown to rapidly inhibit growth of ccvTIR1 roots. However, a several-minute delay in the growth response was recorded when compared to the IAA - wild type root combination10. We used the pico-cvxIAA synthetic auxin that is active in lower doses compared to cvxIAA25. In ccvTIR1 roots, pico-cvxIAA did not trigger an observable rapid depolarization (Fig.4h,i). In the observation window (0 to 10 minutes), pico-cvxIAA-treated roots initiated a trend to inhibit root growth around 7 minutes of treatment (Fig.4j,k). This trend was significant after 15 minutes. (Fig.S4a). In the steady state, the 1000nM pico-cvxIAA hyperpolarized Col0’s root membranes while ccvTIR1 was not significantly affected (Fig.S4b). Col0 was unaffected by both 100 and 1000nM of pico-cvxIAA, while ccvTIR1 showed a strong RGI (Fig.S4c). From the perspective of the cvxIAA, the ccvTIR1 line effectively mimics a quintuple afb1/2/3/4/5 mutant and these results therefore mostly fit the afb1-3 results.
Finally, as the ccvTIR1 receptor is expressed in the tir1/afb2 mutant background, we analyzed the response of this line to IAA to address to what extent the TIR1 and AFB2 are necessary for the rapid membrane depolarization. We observed an instantaneous but weaker and less dynamic IAA-induced rapid depolarization (Fig.4h,i) and impaired rapid RGI (Fig.4j,k) compared to control. In the steady state, no significant depolarization was observed in response to high IAA concentration (Fig.S4b). However, with this treatment, the mutant displayed similar RGI as Col0 (Fig.S4c).
Altogether, these results clarify the role of the TIR1/AFB family of auxin receptors in auxin-triggered membrane depolarization: The initial instantaneous depolarization and rapid RGI strictly depend on AFB1. When AFB1 is missing, as in the afb1-3 mutant or in the pico-cvxIAA -ccvTIR1 combination, the depolarization response is delayed or absent. The ccvTIR1 receptor itself is unable to trigger the full rapid auxin response that involves membrane depolarization, which explains the delay in root growth inhibition observed before10. Together with the tight correlation of auxin-induced depolarization with rapid RGI, the lack of depolarization in the afb1-3 mutant suggest that AFB1 initiates membrane depolarization that is crucial for the rapid RGI and likely other rapid auxin responses as well. It is intriguing to speculate that this prominent role correlates with the cytoplasmic localization of AFB1 that was demonstrated recently11.
At the same time, the other paralogues such as TIR1 and AFB2 clearly participate in the rapid response, as shown by the decreased intensity of depolarization in the mutant tir1/afb2 background, as well as by the data in root hairs5,6. The steady state depolarization was not always fully correlated with the mutant’s growth response, similarly to perturbations in auxin transport, indicating that the maintenance of the depolarization is a very sensitive readout of genetic perturbation. Accordingly, the inability of the cvxIAA-ccvTIR1 pair to trigger a significant long-term membrane depolarization might be caused by the lack of cvxIAA transport by the auxin carriers.
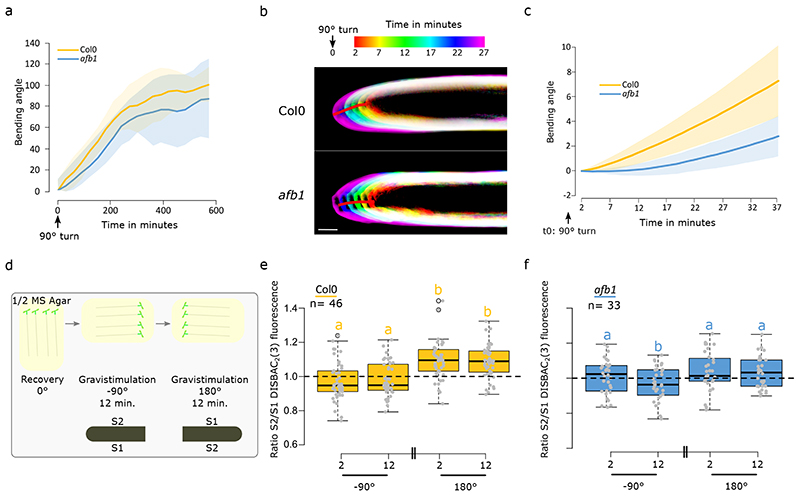
We showed that the AFB1 receptor is essential for the rapid initiation of IAA-induced depolarization and rapid RGI. It was previously shown that despite the defects in auxin response, the afb1-3 single mutant has a normal gravitropic response11. In a low-resolution gravitropic experiment, we obtained similar results, where the afb1-3 gravitropism was not significantly different from the behavior of the control (Fig.5a). AFB1 is essential for the rapid auxin responses and afb1-3 is mostly impaired in the earliest stages of auxin response. Therefore, we optimized high spatiotemporal resolution imaging of early root gravitropism (Fig.S5a) to quantify the root bending angle using an unbiased semi-automated image analysis workflow (Supplementary methods). This approach revealed that the control plants initiated gravitropic bending within 2-4 minutes after the gravitropic stimulus, while the afb1-3 roots showed an approximately 10-minute delay (Fig.5b,c, Supplemental movie 2 and 3). These results spectacularly match up with the defects in membrane depolarization observed during external application of IAA, as well as with the growth inhibition impairment observed in the afb1-3 mutant. Intriguingly, the afb1-3 mutant phenotype resembles the behavior of the roots lacking the calcium channel CNGC1426. On the other hand, the swiftness of the gravitropic response in the control roots confirms that rapid responses to auxin are indeed happening and are relevant for plant cell responses to the internal auxin fluxes.
Fig. 5. AFB1-induced growth inhibition drives the early stages of root gravitropic response.
(a) Mean +/- SD bending (SD represented as shaded areas) angle during 550 minutes after 90° gravistimulation of Col0 and afb1-3. Images were taken every 30 minutes. n= [22-25] individual roots. (b-c) High spatio-temporal resolution imaging of Col0 and afb1-3 response to gravistimulation. b) Temporal color code images of representative Col0 and afb1-3 roots. (c) Quantification of the mean +/- SD (SD represented as shaded areas) root tip angles. n= [8-12] individual roots. ‘d) Schematic representing the gravitropic experiment: After 25 minutes of recovery, chamber was turned 90° clockwise. After 12 minutes of imaging, the chamber was rotated 180° anticlockwise for 12 more minutes of imaging. ’e) Quantification of Col0 transition zone membrane potential ratio S2/S1 after 90° clockwise rotation and S2/S1 after 180° anticlockwise rotation. f) Quantification of afb1-3 transition zone membrane potential ratio S2/S1 after 90° clockwise rotation and S2/S1 after 180° anticlockwise rotation. Scale bar = 50μm. For (e,f) statistics were conducted with a two-sided non-parametric multiple comparison with Tukey contrast and logit approximation. Letters indicates the significantly different statistical group with a p-value<0.05 minimum.
During gravitropic bending, the lower root side accumulates IAA, leading to differential root elongation between the lower and upper side resulting in root bending. The impaling probe technique showed that the lower epidermis of gravitropically bending maize roots was depolarized while the upper side showed hyperpolarization; in comparison to the situation during vertical growth27,28. To test the significance of auxin induced depolarization for gravitropism, we imaged membrane potential during the gravitropic response. The staining with DISBAC2(3) did not interfere with the gravitropic response when grown on the agar surface (Fig.S5b). However, when the stained roots were grown between the agar and the imaging cover glass, a required condition to observe fluorescence, the staining slowed down the gravitropic response (Fig.S5c). To avoid artifacts in our measurements, we restricted our observation of membrane potential response to the first 12 minutes of the gravitropic response.
We did not observe any gradient in membrane potential of the two sides of the root prior to gravistimulation. A 90° gravistimulation of Col-0 roots (Fig.S5d) led to a tendency of higher depolarization of the lower root side, but this difference was not significant (Fig. S5e). This was probably caused by the fact that the changes in auxin fluxes that occur during gravistimulation are not as sudden as the external auxin application and therefore the responses are close to the detection limit of the DISBAC2(3) membrane potential visualization. We therefore performed an experiment where we first gravistimulated the roots by a 90° (clockwise) rotation and then by 180° (anticlockwise) rotation (Fig.5d). We expected that this treatment will lead to more abrupt changes in auxin concentrations at the respective sides of the gravistimulated roots. The first 90° gravistimulation again produced a non-significant tendency of a more depolarized lower root side (Fig.5e). However, the subsequent 180° stimulation triggered a significant increase in the membrane potential gradient between the two sides of the root which was caused either by depolarization in the lower side and/or by hyperpolarization of the upper side (Fig.5e). As for afb1-3, no significant difference or tendency was observed between polarization profiles obtained before stimulation (vertical) and 90° orientation (Fig.S5f) or after a 180° gravistimulation (Fig.5f). These results confirmed that AFB1 is essential for the auxin-induced depolarization.
The membrane potential changes during gravistimulation in Col0 and afb1-3 strongly resemble the results obtained with external application of IAA. Furthermore, the polarization changes we observed during gravistimulation are in accordance with the literature27,28. Altogether, these results demonstrate the physiological relevance of AFB1-dependent membrane depolarization by auxin.
We optimized and established a method to dynamically visualize membrane potential in vivo in Arabidopsis thaliana roots by combining the DISBAC2(3) fluorescent probe with microfluidics and vertical stage microscopy. This allowed us to show that auxin-induced membrane depolarization tightly correlates with rapid RGI and that the cells of the transition zone/early elongation zone are the most responsive to auxin. Further, we demonstrated that auxin cycling in and out of the cells through AUX1 influx and PIN2 efflux per se is not essential for membrane depolarization and rapid RGI but facilitates these responses. Instead, the rapid membrane depolarization by auxin strictly depends on the AFB1 auxin receptor, while the other TIR1/AFB paralogues contribute to this response. The lack of membrane depolarization in the afb1-3 mutant explains the lack of the immediate root growth inhibition. Finally, we provide evidence that AFB1 is required for the early stage of gravitropic root response. We propose that AFB1-dependent membrane depolarization is the earliest auxin response that initiates the growth inhibition and thus the gravitropic bending of the root. Taken together, these results mark a major step in understanding the physiological significance of membrane depolarization for the gravitropic response of the root and clarify the role of AFB1 as the receptor central for rapid auxin responses, adding another piece to the puzzle in understanding the biology of the phytohormone auxin.
Material and methods
Plant material and growth conditions
Wild-type Arabidopsis thaliana ecotype Columbia (Col0) and the following transgenic lines were used in this study: aha2-4 (SALK082786), ost2-2d 18, pp2c-d triple 19 (pp2c-d2 WsDsLOX493G12, pp2c-d5 GABI_330E08, pp2c-d6 SAIL_171H03), aux1 (SALK_020355), pin2 (NASC_N16706), aux1pin2 (SALK_020355/SALK_091142), ccvTIR124 (tir1-afb2) and afb1-3 29 (SALK_070172). Genotypes were verified by PCR-genotyping using the following primers. SALK LB1.3 (ATTTTGCCGATTTCGGAAC), aux1-R (AGCTGCGCATCTAACCAAGT) aux1-L primers (GTTTCACACCTTCCGCCTAA), pin2-R (AAGCACCAAAGACTATAACTA) and PIN2 L primers (CAACGCGAAGAATGCTATGA), afb1-3-RP (GCAACAGCTTCAAGACCTTTG) and afb1-3-LP (AACGGAAGACTAGGAAGCGAG). ccvTIR1-R was verified by its ability to react to pico-cvxIAA. pin2 single mutant has a single nucleotide (G to A) insertion which was verified by comparison of Col0 and pin2 sequencing of the PCR product obtained with the primers: PIN2-R (AAGCACCAAAGACTATAACTA) and PIN2-F (CAACGCGAAGAATGCTATGA).
Seeds were surface sterilized by chlorine gas for 2 hours30. Seeds were sown on 1% (w/v) agar (Duchefa) with ½ Murashige and Skoog (MS, Duchefa, 1 % (w/v) sucrose, adjusted to pH 5.8 with KOH 1M, and stratified for 2 days at 4°C. Seedlings were grown vertically in a growth chamber with 23°C by day (16h), 18°C by night (8h), 60% humidity, light intensity of 120 μmol photons m-2 s-1.
Pharmacological treatments
Treatments were prepared using the following chemicals: 3-indoleacetic acid (IAA, 10mM stock in 96% ethanol, Sigma Aldrich), N-1-naphthylphthalamic acid (NPA, 10mM stock in DMSO, Sigma Aldrich), Carbonyl cyanide m-chlorophenyl hydrazone (CCCP, 10mM stock in DMSO, Sigma Aldrich), Fusicoccin (FC, 1mM stock in 96% ethanol, Sigma Aldrich), 5-Adamantyl-IAA (pico-cvxIAA, 10mM stock in DMSO, TCI Chemicals).
Microfluidic chip description and manufacturing
Microfluidics experiments were conducted using a closable single-layer PDMS silicone chip (Fig.S1g). The chip contained two inlet channels with dimensions of 200 × 50 μm (w×h) and two channels accommodating the growing roots of 1000 × 100 × 20000 μm (w×h×l). To facilitate the sealing of the PDMS and the coverslip, the PDMS resin (Sylgard 184, Dow Corning, USA) was prepared by mixing 15:1 parts of PDMS base:curing agent, resulting in a stickier surface. The master for PDMS casting was made from a two-component polyurethane resin (PUR, F32, Axson, Czech Republic) that consists of Polyol (main base) and Isocyanate (curing agent). These were mixed in a 1:1 volumetric ratio and cured for 15 minutes. To prepare the master, 4 ml of resin was poured on a plexiglass plate with microfluidic structures identical to those in the final chip. These structures were sculptured by micromachining using a micro-milling machine (GV21, GRAVOS, Czechia).
Microfluidic experiments
Five-day-old seedlings were transferred to the chip (Fig.S2a) and enclosed by careful placement of a pre-cleaned microscopic glass coverslip (117μm thick) on the PDMS cast. The closed chip was then placed in a home-made plexiglass holder with screws ensuring tight sealing between the PDMS layer and the coverslip (Fig.S2b). This setup was then placed on the vertical microscope stage for 20 minutes for seedlings to recover before imaging. A constant flow of 3 μL/minute was maintained using a piezo electric pressure controller (OBI1, Elveflow, France) coupled with micro-flow sensors (MFS2, Elveflow, France) and the dedicated Elveflow software ESI (v3.04.1) to control both recording and the flow/pressure feedback. To switch between control and treatment solutions, we set the desired solution flow to 3 +/- 0.01μL/minute and the other to 0 +/-0.01μL/min.
Membrane potential recording
Conventional 1M KCl-filled Ag/AgCl microelectrodes with a tip diameter of 0.5μm were used31 (Shabala et al., 2005). The electrodes were connected to the MIFE amplifier (University of Tasmania, Hobart, Australia) and voltage reading recorded with the CHART software (see32 for details). For Fig.1e, seedlings were immobilized in 0.5 g/L MES solution containing various amounts of KCl (ranging from 10μM to 100mM). After 30 min of exposure, electrodes were impaled into epidermal cells in the mature root zone, and membrane potential values recorded for at least 20 sec. At least 5 individual plants were measured for each treatment, with 4 to 6 cells impaled in each (n = [20-30]).
In another set of experiments (Fig.S1d), we have tested whether the presence of sucrose or other ions in the MS media affects MP. Seedlings were immobilized in the liquid ½ strength MS solution (pH 5.8, buffered with 0.5 g/L MES, +/-1% (w/v). Concentration of K+ in the media was 12mM (measured by flame spectrophotometry). The measurements were obtain as described above.
For membrane potential measurement using DISBAC2(3) fluorescence for the KCl gradient, 5-day old seedlings were transferred to 0.7% agarose (VWR Life Science) and MES 0.5g/L adjusted to pH 5.8, containing 15μM DISBAC2(3) (Sigma-Aldrich) and increasing concentrations of KCl (ranging from 10μM to 100mM). Seedlings and staining media were then placed into a custom 3D printed chambered cover glass (24 x 60mm) and treated for 20 minutes before imaging.
For membrane potential measurement using DISBAC2(3) fluorescence, steady state experiments were conducted using 5-day old seedlings transferred to ½ MS, 1% (w/v) sucrose in 1% agar (Duchefa) and 15μM DISBAC2(3) (Sigma-Aldrich). Seedlings and staining/treatment media were then placed into a custom 3D printed chambered cover glass (24 x 60mm) and treated for 20 minutes before imaging every 10 minutes for 40 minutes. Control media contained the same amount of the chemical solvent added to the treatment mediums (½ MS, DMSO or ethanol 96%). For microfluidics experiments, both control and treatment solutions were prepared by adding 1% (w/v) sucrose and then 20μM DISBAC2(3) to a pre-solution, ensuring that both mediums contained the same amount of sucrose and dye. This pre-solution was then split in two and chemical(s) were added to the treatment solution. The same volume of chemical solvent was added to the control solution (½ MS, DMSO or ethanol 96%). Two seedlings were transferred to the two channels pre-filled with control solution. After closing the chip, the channels were constantly flushed with control solution and placed on the vertical microscopy stage for 20 minutes before imaging. Both seedlings were then imaged every 30 seconds. We recorded 6 minutes of control and 12 minutes of treatment (except specified otherwise).
Analysis of the gravitropic response
The low-resolution analysis of gravitropic response was performed using a vertically placed flatbed scanner (Perfection V600, Epson) with the Epson Scanner software v3.9.2.1US. 5-day old seedlings were transferred to plates containing the desired media and let to recover in a growth chamber. After an hour, plates were turned 90° and imaged every 30 minutes.
For high-spatiotemporal analysis of gravitropism, 4-day old seedlings were placed onto a thin layer of ½ MS medium placed inside a custom 3D printed chambered cover glass (24 x 50mm). The seedlings were let to recover vertically for at least 30 minutes before gravistimulation. In this setup, the roots were growing unobstructed on the surface of the agar and the imaging was performed through the cover glass and the agar (Fig.S5a). Three roots of control and mutant plants were imaged at the same time every 1 minute for 40 minutes.
Microscopic imaging
Imaging in Fig.1a was performed using a horizontal stage Carl Zeiss Axio Observer.Z1 with LSM 880 confocal module via a C-Apochromat 40x/1.2 objective. The dye was excited using a pulsed MP Laser Chameleon Ultra II tuned to 980 nm with the intensity of 3%. The GaAsP detector was operated in the photon counting mode and the exposition (pixel dwell time) was set to 8.92μs.
Imaging was performed using a vertical stage13 Zeiss Axio Observer 7 coupled to a Yokogawa CSU-W1-T2 spinning disk unit with 50 μm pinholes and equipped with a VS-HOM1000 excitation light homogenizer (Visitron Systems). Images were acquired using the VisiView software (Visitron Systems, v4.4.0.14) and Zen Blue (Zeiss, v2.5) for Fig. 5b,c. We used the Zeiss Plan-Apochromat 20x/0.8 and Plan-Apochromat 10x/0.45 objectives. DISBAC2(3) was excited with a 515nm laser and the emission was filtered by a 535/30nm band pass filter. Signal was detected using a PRIME-95B Back-Illuminated sCMOS Camera (1200 x 1200 px; Photometrics) or Orca Flash 4.0 V3 (2048 x 2048 px; Hamamatsu) for Fig. 5b,c.
Image analysis
All microscopy image analysis were conducted using the software ImageJ Fiji33. DISBAC2(3) fluorescence was measured using the segmented line selection tool (20 pixels width). Transition zone was defined as starting at the first epidermis cell (from the quiescent center) that doubled in length compared to the previous one and ending 6 cells shootward (approximately 100μm). The selection line was placed at the intersection of epidermis and cortex cells. Fluorescence was measured in both sides of the root.
Root growth rate was measured with the FiJi plugin Correct 3D drift34 by stabilizing the drift of the root tip. From the drift exported file, distance between root tip positions in consecutive frames was calculated using the formula to calculate distance between 2 sets of coordinates in space.
Root bending angle in the low-resolution scanner experiments was measured after transforming the roots into a string of pixels. The angle between the root tip pixel and the 10th pixel before the root tip was then calculated for every time point (https://sourceforge.net/projects/lbopsis/).
Root bending during high-resolution microscopy experiments was measured using the method described in Supplementary methods (https://sourceforge.net/projects/gravifast/). In brief, to produce homogeneous measurements and unbiased analysis, measurements of root growth and root bending in scanner and microscope experiments were semi-automated to limit human-picture interpretation using ImageJ macros and R scripts (R Core Team, R Foundation for Statistical Computing, Austria, R Cran v3.5.3 and R Studio v1.1.463). The following R packages were used: LearnGeom35, spdep36, stringr(v1.4.0), dplyr(v1.0.2), REdaS37, reshape238, matrixstats(v0.57.0).
Graphics and statistical analysis
Graphics and statistical analysis were performed using the R software. The box plot represents median, 1st and 3rd quartile; the whiskers extend to data points < 1.5 interquartile range away from the 1st or 3rd quartile; all data points are shown as grey dots. As our samples were mostly composed of less than 40 individuals and were thus unlikely to follow a normal distribution and respect the equality of variances, two-sided non-parametric comparison tests were used with 95% confidence using the nparcomp R package39. To compare two sets of samples we used a two-sided non-parametric Student test (npar.t.test function). To compare several samples, we used two-sided non-parametric multiple contrast tests (mctp function). We adjusted the type of comparison by using either a Dunnett (every sample compared to a control) or Tukey (every sample between them) contrast method according to the statistical analysis desired. Data obtained at different time points (paired) during gravitropic experiment in Fig.5 and S5 were analyzed with pairwise Student tests with Bonferroni correction (pairwise.t.test function) after verification that all sample were following a normal distribution and that the equality of variances was respected.
Every steady state experiment was conducted two to four times using different seedlings sown on different days. All the conditions presented in one boxplot were imaged and measured at the same time. Small physiological stresses, differences in incubation time and/or pipetting and/or plant age led to variability in DISBAC2(3) basal staining from one experiment to another. To compare several experiments in one statistical analysis and graphic, data were normalized. Fluorescence values obtained in agar experiments were normalized to an arbitrary value of 400 by dividing 400 by the mean of the control value (Col0 control condition) and multiplying every single fluorescence value by this ratio for one experiment.
For microfluidics experiments, the results correspond to a minimum of 4 individual chips containing two seedlings. The measurements were conducted using seedlings sown on at least 2 different days. As microfluidics experiments are independent to each other (control and treatment for one genotype in one experiment) we took the liberty to reuse, on different graphics and analysis, the data collected with Col0 and the following treatment: control medium (CM) to CM, CM to IAA 10nM, CM to IAA 100nM. Both fluorescence and root elongation data presented have been normalized to reflect changes in percentages. The normalization for one individual root was performed by dividing each value for each time point by the mean of 5 minutes of control measurements (every 30 seconds, otherwise specified). This ratio was then multiplied by 100 to obtain percentages. The ratio shown in boxplot figures represents the mean of 5 minutes of treatment measurements (every 30 seconds, otherwise specified) by the mean of 5 minutes (except specified otherwise) of control measurements.
Supplementary Material
Acknowledgements
This work was supported by the European Research Council (Grant No. 803048), Charles University Primus (Grant No. PRIMUS/19/SCI/09), and in the initial stages by the Czech Science Foundation (GAČR Grant No. 18-10116Y). Sergey Shabala acknowledged support from the Australian Research Council and China National Distinguished Expert Project (WQ20174400441).
The authors thank Eva Medvecká for technical support, Mark Estelle, Michael Prigge and William Gray for Arabidopsis seeds, Jiří Friml for the initial aux1pin2 cross, Jack Merrin for guidance in microfluidics.
Footnotes
Author contributions
NBCS and MF conceived the project, performed the experimental work, analyzed, and interpreted the data. PY and SS conceived the impaling electrode membrane potential experiment and PY measured and analyzed the data. DK, ZS and MF conceived the microfluidics chip design and DK fabricated and optimized it. NBCS, MF and SS wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Data availability
All raw images used in this study are available on Zenodo (https://doi.org/10.5281/zenodo.4922659). All the measured datasets are available as an excel file in supplementary materials.
Code availability
All the script used in this study are available at https://sourceforge.net/projects/lbopsis/ and https://sourceforge.net/projects/gravifast/.
References
- 1.Goering H, Stahlberg R. Depolarization of transmembrane potential of corn and wheat coleoptiles under reduced water potential and after IAA application. Plant and Cell Physiology. 1979 doi: 10.1093/oxfordjournalspcp.a075851. [DOI] [Google Scholar]
- 2.Bates GW, Goldsmith MH. Rapid response of the plasma-membrane potential in oat coleoptiles to auxin and other weak acids. Planta. 1983;159:231–237. doi: 10.1007/BF00397530. [DOI] [PubMed] [Google Scholar]
- 3.Tretyn A, Wagner G, Felle HH. Signal Transduction in Sinapis alba Root Hairs: Auxins as External Messengers. Journal of Plant Physiology. 1991;139:187–193. [Google Scholar]
- 4.Felle H, Peters W, Palme K. The electrical response of maize to auxins. Biochimica et Biophysica Acta (BBA) - Biomembranes. 1991;1064:199–204. doi: 10.1016/0005-2736(91)90302-o. [DOI] [PubMed] [Google Scholar]
- 5.Dindas J, et al. AUX1-mediated root hair auxin influx governs SCFTIR1/AFB-type Ca2+ signaling. Nature Communications. 2018;9 doi: 10.1038/s41467-018-03582-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paponov IA, et al. Auxin-Induced Plasma Membrane Depolarization Is Regulated by Auxin Transport and Not by AUXIN BINDING PROTEIN1. Frontiers in Plant Science. 2019;9 doi: 10.3389/fpls.2018.01953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans ML, Mulkey TJ, Vesper MJ. Auxin action on proton influx in corn roots and its correlation with growth. p. 3. [DOI] [PubMed]
- 8.Monshausen GB, Miller ND, Murphy AS, Gilroy S. Dynamics of auxin-dependent Ca2+ and pH signaling in root growth revealed by integrating high-resolution imaging with automated computer vision-based analysis: Calcium and auxin signaling. The Plant Journal. 2011;65:309–318. doi: 10.1111/j.1365-313X.2010.04423.x. [DOI] [PubMed] [Google Scholar]
- 9.Scheitz K, Lüthen H, Schenck D. Rapid auxin-induced root growth inhibition requires the TIR and AFB auxin receptors. Planta. 2013;238:1171–1176. doi: 10.1007/s00425-013-1941-x. [DOI] [PubMed] [Google Scholar]
- 10.Fendrych M, et al. Rapid and reversible root growth inhibition by TIR1 auxin signalling. Nature Plants. 2018;4:453–459. doi: 10.1038/s41477-018-0190-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prigge MJ, et al. Genetic analysis of the Arabidopsis TIR1/AFB auxin receptors reveals both overlapping and specialized functions. eLife. 2020;9:e54740. doi: 10.7554/eLife.54740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Renier M, et al. Use of a Membrane Potential-Sensitive Probe to Assess Biological Expression of the Cystic Fibrosis Transmembrane Conductance Regulator. Human Gene Therapy. 1995;6:1275–1283. doi: 10.1089/hum.1995.6.10-1275. [DOI] [PubMed] [Google Scholar]
- 13.von Wangenheim D, et al. Live tracking of moving samples in confocal microscopy for vertically grown roots. eLife. 2017;6 doi: 10.7554/eLife.26792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baunsgaard L, et al. The 14-3-3 proteins associate with the plant plasma membrane H(+)-ATPase to generate a fusicoccin binding complex and a fusicoccin responsive system. Plant J. 1998;13:661–671. doi: 10.1046/j.1365-313x.1998.00083.x. [DOI] [PubMed] [Google Scholar]
- 15.Senn AP, Goldsmith MHM. Regulation of Electrogenic Proton Pumping by Auxin and Fusicoccin as Related to the Growth of Avena Coleoptiles. PLANT PHYSIOLOGY. 1988;88:131–138. doi: 10.1104/pp.88.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ullrich CI, Novacky AJ. Extra- and Intracellular pH and Membrane Potential Changes Induced by K+, Cl−, H2PO4 −, and NO3 −Uptake and Fusicoccin in Root Hairs of Limnobium stoloniferum . Plant Physiol. 1990;94:1561–1567. doi: 10.1104/pp.94.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Labady A, Thomas D, Shvetsova T, Volkov AG. Plant bioelectrochemistry: effects of CCCP on electrical signaling in soybean. Bioelectrochemistry. 2002;57:47–53. doi: 10.1016/s1567-5394(01)00175-x. [DOI] [PubMed] [Google Scholar]
- 18.Merlot S, et al. Constitutive activation of a plasma membrane H+-ATPase prevents abscisic acid-mediated stomatal closure. EMBO J. 2007;26:3216–3226. doi: 10.1038/sj.emboj.7601750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren H, Park MY, Spartz AK, Wong JH, Gray WM. A subset of plasma membrane-localized PP2C.D phosphatases negatively regulate SAUR-mediated cell expansion in Arabidopsis. PLoS Genet. 2018;14:e1007455. doi: 10.1371/journal.pgen.1007455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shabala SN, Lew RR. Turgor Regulation in Osmotically Stressed Arabidopsis Epidermal Root Cells Direct Support for the Role of Inorganic Ion Uptake as Revealed by Concurrent Flux and Cell Turgor Measurements. Plant Physiol. 2002;129:290–299. doi: 10.1104/pp.020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verbelen J-P, Cnodder TD, Le J, Vissenberg K, Baluška F. The Root Apex of Arabidopsis thaliana Consists of Four Distinct Zones of Growth Activities: Meristematic Zone, Transition Zone, Fast Elongation Zone and Growth Terminating Zone. Plant Signaling & Behavior. 2006;1:296–304. doi: 10.4161/psb.1.6.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rigas S, et al. Root gravitropism and root hair development constitute coupled developmental responses regulated by auxin homeostasis in the Arabidopsis root apex. New Phytol. 2013;197:1130–1141. doi: 10.1111/nph.12092. [DOI] [PubMed] [Google Scholar]
- 23.Dharmasiri N, et al. Plant Development Is Regulated by a Family of Auxin Receptor F Box Proteins. Developmental Cell. 2005;9:109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 24.Uchida N, et al. Chemical hijacking of auxin signaling with an engineered auxin-TIR1 pair. Nat Chem Biol. 2018;14:299–305. doi: 10.1038/nchembio.2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamada R, et al. A Super Strong Engineered Auxin-TIR1 Pair. Plant Cell Physiol. 2018;59:1538–1544. doi: 10.1093/pcp/pcy127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shih H-W, De Pew CL, Miller ND, Monshausen GB. The Cyclic Nucleotide-Gated Channel CNGC14 Regulates Root Gravitropism in Arabidopsis thaliana. Curr Biol. 2015;25:3119–3125. doi: 10.1016/j.cub.2015.10.025. [DOI] [PubMed] [Google Scholar]
- 27.Behrens HM, Gradmann D, Sievers A. Membrane-potential responses following gravistimulation in roots of Lepidium sativum L. Planta. 1985;163:463–472. doi: 10.1007/BF00392703. [DOI] [PubMed] [Google Scholar]
- 28.Evans ML, Ishikawa H. Responses of Arabidopsis roots to auxin studied with high temporal resolution: Comparison of wild type and auxin-response mutants. p. 8.
- 29.Savaldi-Goldstein S, et al. New auxin analogs with growth-promoting effects in intact plants reveal a chemical strategy to improve hormone delivery. Proceedings of the National Academy of Sciences. 2008;105:15190–15195. doi: 10.1073/pnas.0806324105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindsey BE, Rivero L, Calhoun CS, Grotewold E, Brkljacic J. Standardized Method for High-throughput Sterilization of Arabidopsis Seeds. JoVE. 2017:56587. doi: 10.3791/56587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shabala L, Cuin TA, Newman IA, Shabala S. Salinity-induced ion flux patterns from the excised roots of Arabidopsis sos mutants. Planta. 2005;222:1041–1050. doi: 10.1007/s00425-005-0074-2. [DOI] [PubMed] [Google Scholar]
- 32.Shabala S, et al. Extracellular Ca2+ Ameliorates NaCl-Induced K+ Loss from Arabidopsis Root and Leaf Cells by Controlling Plasma Membrane K+-Permeable Channels. Plant Physiol. 2006;141:1653–1665. doi: 10.1104/pp.106.082388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parslow A, Cardona A, Bryson-Richardson RJ. Sample Drift Correction Following 4D Confocal Time-lapse Imaging. JoVE. 2014:51086. doi: 10.3791/51086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Briz-Redón Á, Serrano-Aroca Á. Novel pedagogical tool for simultaneous learning of plane geometry and R programming. RIO. 2018;4:e25485 [Google Scholar]
- 36.Bivand RS, Wong DWS. Comparing implementations of global and local indicators of spatial association. TEST. 2018;27:716–748. [Google Scholar]
- 37.Hatzinger R, Hornik K, Nagel H, Maier MJ. R: Einführung durch angewandte Statistik. Pearson; 2014. [Google Scholar]
- 38.Wickham H. Reshaping Data with the reshape Package. J Stat Soft. 2007;21 [Google Scholar]
- 39.Konietschke F, Placzek M, Schaarschmidt F, Hothorn LA. nparcomp: An R Software Package for Nonparametric Multiple Comparisons and Simultaneous Confidence Intervals. Journal of Statistical Software. 2015;64 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw images used in this study are available on Zenodo (https://doi.org/10.5281/zenodo.4922659). All the measured datasets are available as an excel file in supplementary materials.
All the script used in this study are available at https://sourceforge.net/projects/lbopsis/ and https://sourceforge.net/projects/gravifast/.