Abstract
Many natural and designed proteins are only marginally stable limiting their usefulness in research and applications. Recently, we described an automated structure and sequence-based design method, called PROSS, for optimizing protein stability and heterologous expression levels that has since been validated on dozens of proteins. Here, we introduce improvements to the method, workflow and presentation, including more accurate sequence analysis, error handling and automated analysis of the quality of the sequence alignment that is used in design calculations. PROSS2 is freely available for academic use at https://pross.weizmann.ac.il.
Introduction
The marginal stability of many natural and engineered proteins is a major bottleneck in using proteins in basic and applied research (Goldenzweig and Fleishman, 2018). In many cases, proteins require specific hosts to enable their functional expression and they may exhibit low production yields and low tolerance to heat or other insults, complicating experimental protocols and raising production costs. A variety of experimental, semi-rational, and computational approaches have been developed to identify mutants that improve protein stability, but these typically require iterative and laborious experimental screening (Romero and Arnold, 2009; Magliery, 2015).
To address this general problem, we recently described an automated sequence and structure-based design method, called PROSS, which optimizes the energy of the native state subject to constraints that are inferred from a multiple sequence alignment of homologs (Goldenzweig et al., 2016). PROSS has enabled us and many other labs to dramatically improve protein stability and heterologous expression levels including in proteins that defied state-of-the-art experimental and computational optimization methods, such as large human enzymes, potential therapeutics and vaccine immunogens from HIV and malaria (Georgoulia et al., 2020; Lambert et al., 2020; Malladi et al., 2020; Tullman et al., 2020; Warszawski et al., 2020; Trudeau et al., 2018; Campeotto et al., 2017; Goldsmith et al., 2017; Brazzolotto et al., 2017).
Methods
Based on the constructive feedback of many users, we now present improvements to the method’s workflow, output, and presentation:
(1) Disabling design in active-site positions. Previously, we identified interacting positions by their distance from the ligand, without taking into account the positions’ orientation. This simplification led to the undesirable restriction of large segments of the protein, particularly in small proteins. Now, positions are selected according to their distance from the ligand (8 and 5 Å for small molecules and protein chains, respectively) and their directionality (the ligand must be within 90° of the position’s Cα-Cβ vector), allowing more positions to be designed. (2) The original PROSS method suggested seven designs, typically comprising an increasing number of mutations relative to one another. We have found that some use-cases require a smaller number of mutations and others a larger one. PROSS2 adds two additional designs, one more conservative and one more permissive than those provided previously. (3) PROSS2 enables two all-atom Rosetta energy functions, talaris (O’Meara et al., 2015), which was used in the previous server, and the newer Rosetta energy function 2015 (ref2015) (Park et al., 2016). Both energy functions are dominated by van der Waals interactions, solvation and electrostatics, and the latter improves the treatment of solvation and electrostatic interactions. (4) In the original PROSS algorithm, the sequence alignment in loop regions excluded sequences that exhibited insertions or deletions (indels) relative to the query as indels may alter the local backbone structure (Netzer et al., 2018). We now segment all secondary structure elements in the query and eliminate from the alignment sequences that exhibit indels relative to the query in loops, α helices, or β sheets. (5) Based on typical user queries and errors, the PROSS2 web interface provides detailed warnings and error messages, including possible solutions. (6) The new results page provides detailed information on the sequence analysis, including a table of all of the designed mutations, a sequence viewer denoting the amino acid identities at each position, and the depth of the sequence alignment that underlies the analysis (Figure 1). The designed-mutations table provides a detailed view for comparing designs. The results page includes warnings based on the depth and coverage of the multiple sequence alignment relative to the query, flagging designed mutations that may require close user inspection. A help page for the results can be found at https://pross.weizmann.ac.il/pross-results/. (7) PROSS2 uses the NGL viewer (Rose et al., 2016; Rose and Hildebrand, 2015) to present the designs and enables the rendering even of large proteins. (8) The downloadable results now include the files needed to run the last step of Rosetta combinatorial design on the user’s local computer to provide manual control over this step.
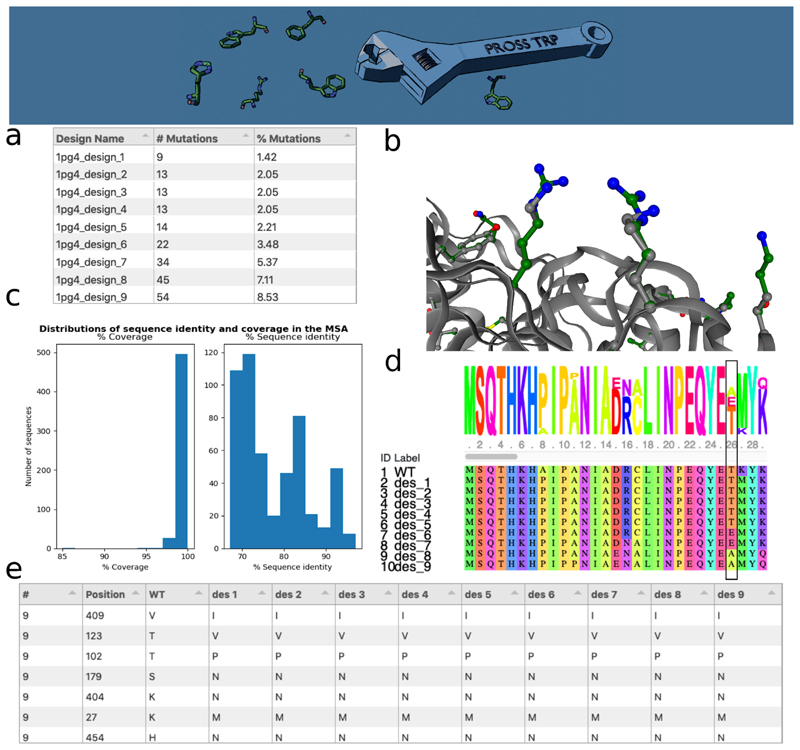
Figure 1. Features from the PROSS2 results page.
(a) A tabular summary of designs. (b) An interactive NGL rendering of the designed mutations (green sticks and balls) relative to the query (gray). (c) Histograms depicting the quality of the multiple sequence alignment according to their sequence diversity and coverage relative to the query sequence. (d) Sequence view comparing all designs to the query. (e) Part of a table reporting the identities at all mutated positions across the designs.
Acknowledgments
We thank users from around the world for their feedback and support of the PROSS web server. We also thank Shiran Barber-Zucker for valuable feedback on the new server, Ziv Avizemer, Dina Listov and Olga Khersonsky for testing PROSS2, and Jaime Prilusky and Rotem Barzilay for excellent technical support. Research was funded by the European Research Council (815379), the Israel Science Foundation (1844), the Israel Ministry of Science (128625), the Milner Foundation and a charitable donation from Sam Switzer and family.
References
- Brazzolotto X, et al. Bacterial Expression of Human Butyrylcholinesterase as a Tool for Nerve Agent Bioscavengers Development. Molecules. 2017:22. doi: 10.3390/molecules22111828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campeotto I, et al. One-step design of a stable variant of the malaria invasion protein RH5 for use as a vaccine immunogen. Proc Natl Acad Sci U S A. 2017;114:998–1002. doi: 10.1073/pnas.1616903114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgoulia PS, et al. Deciphering the molecular mechanism of FLT3 resistance mutations. FEBS J. 2020 doi: 10.1111/febs.15209. [DOI] [PubMed] [Google Scholar]
- Goldenzweig A, et al. Automated Structure-and Sequence-Based Design of Proteins for High Bacterial Expression and Stability. Mol Cell. 2016;63:1–10. doi: 10.1016/j.molcel.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenzweig A, Fleishman SJ. Principles of Protein Stability and Their Application in Computational Design. Annu Rev Biochem. 2018;87:105–129. doi: 10.1146/annurev-biochem-062917-012102. [DOI] [PubMed] [Google Scholar]
- Goldsmith M, et al. Overcoming an optimization plateau in the directed evolution of highly efficient nerve agent bioscavengers. Protein Eng Des Sel. 2017;30:333–345. doi: 10.1093/protein/gzx003. [DOI] [PubMed] [Google Scholar]
- Lambert AR, et al. Optimization of Protein Thermostability and Exploitation of Recognition Behavior to Engineer Altered Protein-DNA Recognition. Structure. 2020;28:760–775.e8. doi: 10.1016/j.str.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magliery TJ. Protein stability: computation, sequence statistics, and new experimental methods. Curr Opin Struct Biol. 2015;33:161–168. doi: 10.1016/j.sbi.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malladi SK, et al. One-step sequence and structure-guided optimization of HIV-1 envelope gp140. Current Research in Structural Biology. 2020;2:45–55. doi: 10.1016/j.crstbi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netzer R, et al. Ultrahigh specificity in a network of computationally designed protein-interaction pairs. Nat. Commun. 2018;9:5286. doi: 10.1038/s41467-018-07722-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Meara MJ, et al. Combined covalent-electrostatic model of hydrogen bonding improves structure prediction with Rosetta. J Chem Theory Comput. 2015;11:609–622. doi: 10.1021/ct500864r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, et al. Simultaneous Optimization of Biomolecular Energy Functions on Features from Small Molecules and Macromolecules. J Chem Theory Comput. 2016;12:6201–6212. doi: 10.1021/acs.jctc.6b00819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero PA, Arnold FH. Exploring protein fitness landscapes by directed evolution. Nat Rev Mol Cell Biol. 2009;10:866–876. doi: 10.1038/nrm2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose AS, et al. Web-based molecular graphics for large complexes. Proceedings of the 21st International Conference on Web3D Technology, Web3D ’16. Association for Computing Machinery; New York, NY, USA. 2016. pp. 185–186. [Google Scholar]
- Rose AS, Hildebrand PW. NGL Viewer: a web application for molecular visualization. Nucleic Acids Res. 2015;43:W576–9. doi: 10.1093/nar/gkv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudeau DL, et al. Design and in vitro realization of carbon-conserving photorespiration. Proceedings of the National Academy of Sciences. 2018;115:E11455–E11464. doi: 10.1073/pnas.1812605115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullman J, et al. A ClpS-based N-terminal amino acid binding reagent with improved thermostability and selectivity. Biochem Eng J. 2020;154 107438. [Google Scholar]
- Warszawski S, et al. Design of a basigin-mimicking inhibitor targeting the malaria invasion protein RH5. Proteins. 2020;88:187–195. doi: 10.1002/prot.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]