Abstract
During early development, the hindbrain is subdivided into rhombomeres that underlie the organisation of neurons and adjacent craniofacial tissues. A gene regulatory network of signals and transcription factors establish and pattern segments with a distinct anteroposterior identity. Initially, the borders of segmental gene expression are imprecise, but then become sharply defined, and specialised boundary cells form. In this Review, we summarise key aspects of the conserved regulatory cascade that underlies the formation of hindbrain segments. We describe how the pattern is sharpened and stabilised through the dynamic regulation of cell identity, acting in parallel with cell segregation. Finally, we discuss evidence that boundary cells have roles in local patterning, and act as a site of neurogenesis within the hindbrain.
Introduction
During embryonic development, many tissues are regionalised into subdivisions, each with a distinct identity that underlies formation of a specific set of cell types. The boundary of adjacent subdivisions becomes sharp, and in certain tissues forms a signalling centre that regulates local patterning. Important examples of such regionalisation and boundary formation occur in arthropods, and specific tissues in vertebrates, in which repeated segments with distinct anteroposterior (A–P) identity form along the body axis. There has thus been much interest in elucidating the mechanisms that underlie segmentation, A–P specification, and the formation and roles of boundaries (Batlle and Wilkinson, 2012; Dahmann et al., 2011; Kiecker and Lumsden, 2005; Pujades, 2020).
Anatomical studies revealed that segmentation of the hindbrain is central to the organisation of craniofacial tissues. The hindbrain neuroepithelium is transiently subdivided along the A–P axis at an early stage to form a series of rhombomeres that each generate distinct neuronal cell types (Chandrasekhar, 2004; Clarke and Lumsden, 1993; Cordes, 2001; Gilland and Baker, 2005; Kimmel et al., 1988; Lumsden and Keynes, 1989). Rhombomeres become lineage-restricted cellular compartments, with limited cell mixing between neighbouring segments (Calzolari et al., 2014; Fraser et al., 1990; Jimenez-Guri et al., 2010). Concurrently, distinct boundary cells are formed at the interface of hindbrain segments (Guthrie and Lumsden, 1991; Lumsden and Keynes, 1989). The process of segmentation establishes an early ground plan that plays a crucial role in specifying the pattern underlying formation of the neural circuitry associated with the diverse functions of the hindbrain (Briscoe and Wilkinson, 2004; Davenne et al., 1999; Di Bonito et al., 2013; Pasqualetti et al., 2007; Pattyn et al., 2003). Analyses in jawed and jawless vertebrates have revealed that this segmental organisation is a shared trait with origins at the base of vertebrates (Alexander et al., 2009; Lumsden, 2004; Lumsden and Krumlauf, 1996; Moens and Prince, 2002; Parker et al., 2014, 2016). At the molecular level, the formation and A–P identity of hindbrain segments are established by a gene regulatory network (GRN) of cell signalling and transcription factors, which is initiated by graded morphogen signalling along the neuroepithelium. This leads to an initially imprecise pattern of segmental gene expression, which is refined to form segments with a homogeneous regional identity demarcated by sharp borders.
The hindbrain is an excellent model to address questions that have broad relevance in developmental biology; studies of hindbrain segmentation have led to the discovery of mechanisms that also act in many other tissues. In this article, we first give an overview of the functional anatomy of the hindbrain, and then focus on molecular and cellular mechanisms that underlie its patterning along the A–P axis. We present current understanding of the gene regulatory networks that underlie the formation and A-P identity of hindbrain segments. We then discuss how the initially imprecise segmental gene expression is transformed into a sharp pattern, and how this is stabilised despite potential disruption by cell intermingling. Finally, we discuss how distinct boundary cells form at the segment borders, and emerging evidence for the roles that these cells play in hindbrain development.
Functional anatomy of the hindbrain in craniofacial development
The hindbrain is a key coordination centre in the vertebrate central nervous system (CNS) that serves as an important relay hub for control of sensory and motor functions of the head. Cranial somatic and branchiomotor nerves emanating from the hindbrain transmit motor impulses to head muscles, and receive input from cranial sensory organs through their tight association with cranial sensory ganglia (Fig. 1A-C). Through this network of neuronal circuits, the hindbrain relays sensory information from the perception of hearing, touch, taste and balance, and controls facial expressions and jaw, tongue and eye movements. Areas of the hindbrain transfer signals from the spinal cord to higher brain centres and coordinate functions of the autonomic nervous system, such as control of heart rate, respiration, digestion and swallowing. The hindbrain also contains a wide variety of interneurons and relay neurons, including a network of reticulospinal neurons that regulate alertness, sleep, posture, and fine-grained locomotor activities. A series of neuronal circuits are organised in a modular manner in the hindbrain and serve as central pattern generators, producing rhythmic pacemaker-like signals that drive stereotyped behaviours, such as breathing and swallowing (Chatonnet et al., 2002; Fortin et al., 1999; Fortin et al., 1995). Hence, from a functional perspective, the vertebrate hindbrain contains a complex network of dedicated neural circuits that play essential roles in controlling many physiological processes and behaviours. This array of core functions, and the underlying neuroanatomy of their networks, are a common feature of the vertebrate CNS, making the hindbrain one of the most evolutionarily conserved regions of the vertebrate brain (Alexander et al., 2009; Gilland and Baker, 1993, 2005; Kiecker and Lumsden, 2005).
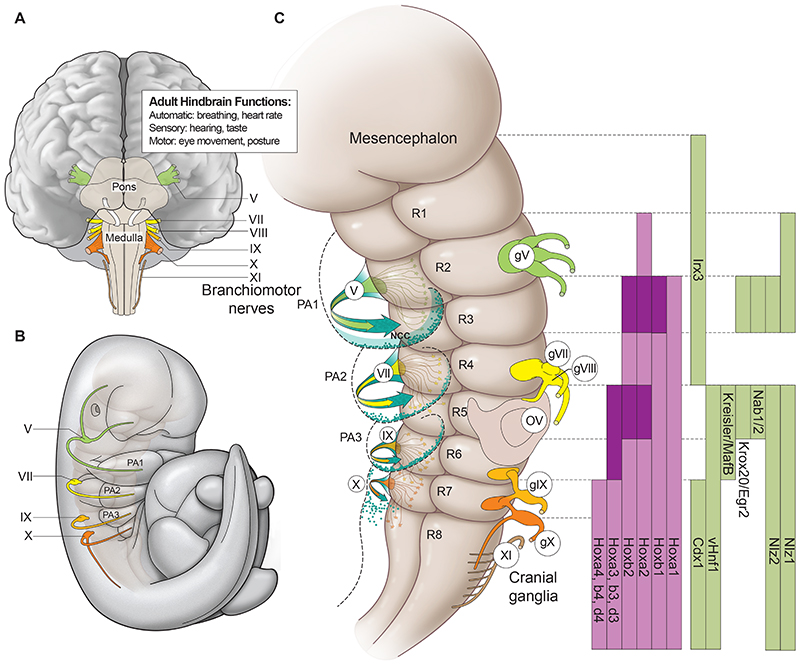
Fig. 1. Anatomy and segmental organisation of the hindbrain.
(A) Diagram of ventral aspect of an adult mouse hindbrain noting the relative positions of branchiomotor nerves and several of its functional roles. (B) Drawing of a lateral view of an E10 mouse embryo showing the paths of branchiomotor nerves and their relationship to the pharyngeal arches (PA). (C) Diagram of a ventral view of a developing hindbrain depicting individual rhombomeres (R) and their relationship with the positions of cranial ganglia (gV-X1), motor nerves (V-X), migrating streams of cranial neural crest cells (NCC) and pharyngeal arches. There is a two-segment periodicity in the segmental organisation, as cranial ganglia, and major streams of migrating neural crest cells (blue arrows and dots) are associated with even-numbered rhombomeres. Each branchiomotor nerve is derived from alternating pairs of even- and odd-numbered segments, exiting to the periphery from the dorsal aspects of only the even-numbered segments. Rhombomere-restricted domains of gene expression of key transcription factors involved in segmentation and segmental patterning are indicated on the right. The gene names correspond to mouse and may vary in other vertebrates. Darker shades of colour (purple, Hox genes; green segmental subdivision genes) in the expression domains indicate higher levels of expression in specific rhombomeres.
Programs of neural differentiation are tightly coupled to the segmental architecture of the hindbrain. For example, a set of projection interneurons display an iterative pattern in every segment (Clarke and Lumsden, 1993; Lumsden, 2004). Neurons that form the trigeminal (V), facial (VII) and glossopharyngeal (IX) branchiomotor nerves arise first in even-numbered rhombomeres r2, r4 and r6. Neurons subsequently differentiate in odd-numbered segments (r3, r5 and r7) and they send axons unidirectionally to the anteriorly adjacent even-numbered rhombomere (Chandrasekhar, 2004; Lumsden and Keynes, 1989). This establishes a two-segment periodicity in the formation and organisation of branchiomotor nerves, whereby each nerve is derived from alternating pairs of even- and odd-numbered segments, exiting to the periphery only from the even-numbered segments (Fig. 1C). In line with this two-segment repeat pattern, the modular neuronal circuits of the GABAergic rhythmic central pattern generators are formed in even-numbered segments (Chatonnet et al., 2002; Fortin et al., 1999; Fortin et al., 1995). This illustrates that spatially and temporally controlled patterns of neurogenesis are coupled to segmentation of the early hindbrain. This relationship leads to the elaboration of a conserved segmental pattern in the organisation of neurons, cranial nerve roots and neuronal connectivity between the hindbrain and peripheral targets, and with other brain centres (Chandrasekhar, 2004; Clarke and Lumsden, 1993; Cordes, 2001; Gilland and Baker, 2005; Kimmel et al., 1988; Lumsden and Keynes, 1989).
Beyond the central nervous system, the hindbrain also makes important contributions to head development and craniofacial patterning through the formation of cranial neural crest cells (cNCC). cNCCs delaminate from the neural epithelium and migrate in discrete streams to populate the pharyngeal arches where their differentiated derivatives form most of the bone and connective tissue of the head (Green et al., 2015; Knecht and Bronner-Fraser, 2002; Le Douarin and Kalcheim, 1999; Santagati and Rijli, 2003; Trainor et al., 2004; Trainor and Krumlauf, 2000b). The formation and patterns of migration of cNCCs are also linked to hindbrain segmentation and its interactions with adjacent tissues (Minoux and Rijli, 2010; Trainor and Krumlauf, 2000b). Major streams of cNCCs emanate from r2, r4 and r6 (Fig. 1C), while smaller numbers of cNCCs migrate from r3 and r5, moving rostrally and caudally to merge with streams from the even-numbered segments (Birgbauer et al., 1995; Couly et al., 1996; Golding et al., 2000; Kontges and Lumsden, 1996; Trainor et al., 2002). This segmental registration between cNCC migration and two-segment periodicity of neuronal differentiation serves to align branchiomotor nerves with their peripheral targets in the pharyngeal arches (Fig. 1C). Thus, while the process of hindbrain segmentation is an early transient state, it establishes a crucial ground plan of regional specification that is progressively elaborated during later development to generate craniofacial structures and neural circuits that underlie functions of the adult hindbrain (Briscoe and Wilkinson, 2004; Di Bonito et al., 2013; Geisen et al., 2008; Pasqualetti et al., 2007).
Gene regulatory networks underlying segmentation and A-P identity
At the molecular level, hindbrain segmentation is coupled to mechanisms that regulate A-P identity in a broad spectrum of animals and tissues (Alexander et al., 2009; Carroll, 1995; Frank and Sela-Donenfeld, 2019; Lowe et al., 2015). An early segmental plan can be visualised through spatially-restricted expression of key developmental genes, which encode transcription factors (TFs) that regulate steps of the segmentation process (Fig. 1C) (Alexander et al., 2009; Lumsden and Krumlauf, 1996; Parker and Krumlauf, 2017; Schneider-Maunoury et al., 1993). For example, the zinc-finger transcription factor Krox20/Egr2 is segmentally-expressed in r3 and r5, and many members of the Hox homeobox gene family are coordinately expressed in nested segmental domains of the hindbrain (Hunt et al., 1991; Murphy et al., 1989; Wilkinson et al., 1989a; Wilkinson et al., 1989b). The segmental patterns of Krox20 and Hox expression in the hindbrain are highly conserved across vertebrates (Godsave et al., 1994; Nieto et al., 1991; Parker et al., 2014, 2019a; Prince et al., 1998).
Because of the conserved segmental organisation of the vertebrate hindbrain, comparative studies between species have facilitated our understanding of GRNs that govern the process of hindbrain segmentation and A-P patterning (Parker et al., 2016; Parker and Krumlauf, 2020). Gene expression, functional perturbation and regulatory analyses in vertebrate models (primarily mouse, chicken, zebrafish and Xenopus) have helped to identify and characterise many of the key genes, signals and regulatory interactions that control the process of segmentation in early hindbrain development (Alexander et al., 2009; Frank and Sela-Donenfeld, 2019; Moens and Prince, 2002; Tumpel et al., 2009). These experimental findings have been integrated to generate a hypothetical hindbrain GRN that depicts the dynamic and progressive steps underlying segmentation and A-P patterning (Fig. 2A) (Parker and Krumlauf, 2020). This provides a useful logistical framework for interpretation of functional studies and evolutionary comparisons between species. The framework of the GRN may be broadly depicted as a series of hierarchical steps associated with cell and developmental processes, each with their own components and layers of regulatory circuits (Fig. 2A). Many of the genes and signals play important roles in multiple steps of the segmentation and patterning process. The components, data and logic used to formulate the structure of the GRN have been previously reviewed in detail (Frank and Sela-Donenfeld, 2019; Moens and Prince, 2002; Parker et al., 2016; Parker and Krumlauf, 2017, 2020; Tumpel et al., 2009)(Fig. 2).
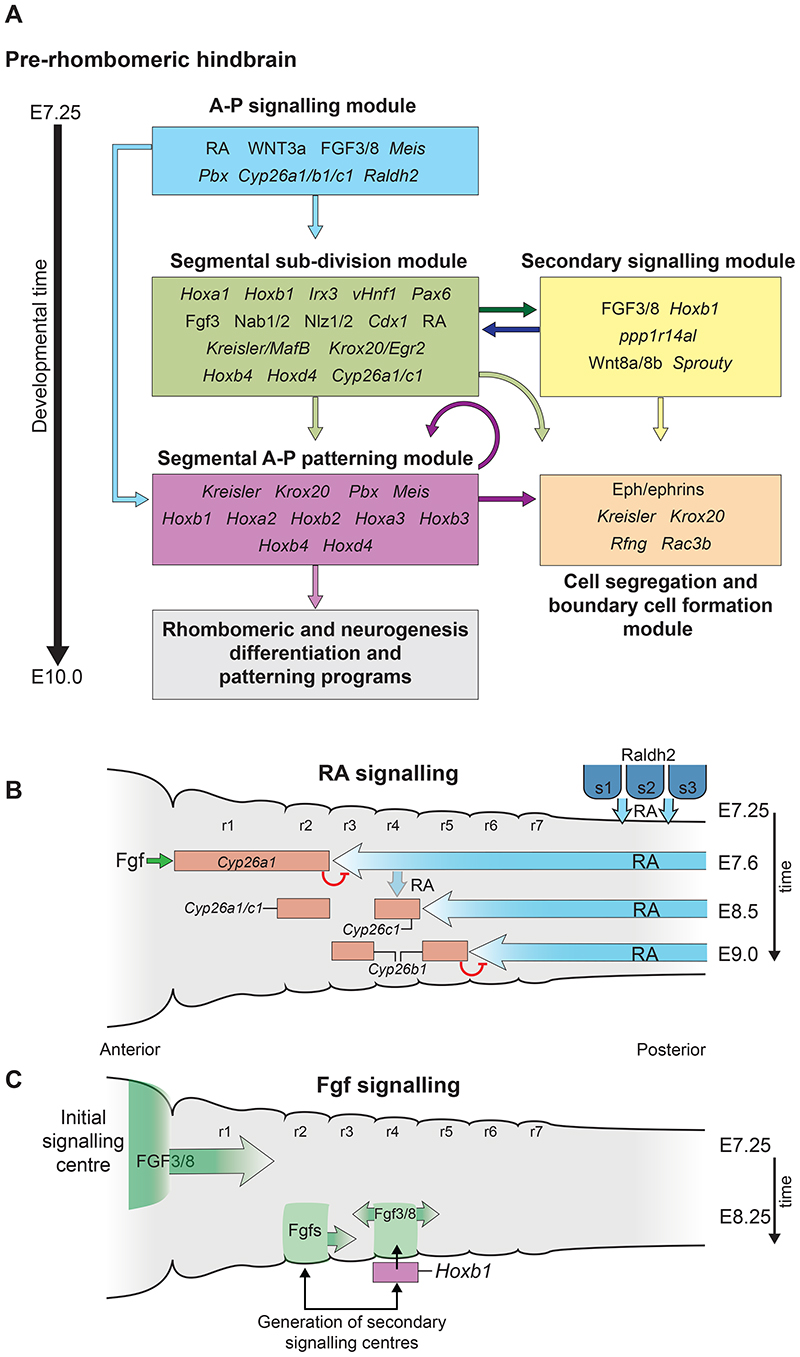
Fig. 2. Hierarchical structure of the gene regulatory network (GRN) governing hindbrain segmentation.
A framework of the GRN model depicted as a progressive series of modules/steps associated with underlying cell and developmental processes in the mouse hindbrain between E7.25 and E10.0. Each module (coloured boxes) has its own layer of regulatory circuits and components, as indicated by the key genes and signals inside each box. The gene names correspond to mouse and may vary in other vertebrates. The arrows indicate the flow of regulatory information between modules. Many genes and signals have fundamental roles in multiple steps of the segmentation and patterning process. Regulatory interactions in the A-P signalling module set up temporally and spatially dynamic domains of signalling. (B) Diagram of shifting RA gradients involved in regulating early events in mouse hindbrain segmentation in stages between E7.5 and E9.0. RA is generated by Raldh2 in somitic mesoderm and diffuses anteriorly in the hindbrain. Fgf signals from the midbrain isthmic organiser induce Cyp26a1 which degrades RA in r1 and r2. This establishes an initial RA signalling domain with an anterior limit at the future r2/3 border at E7.5. Cyp26c1 is expressed in r4 at E7.9, and slightly later (E8.5) Cyp26b1 is induced in r3 and r5. This progressive activation of Cyp26 genes (red rectangles), and degradation of RA, creates shifting gradients of RA (blue arrows) that eventually establish an anterior limit at the r5/6 boundary by E9.0. (C) Diagram of dynamics of initial and secondary Fgf signalling centres. At E7.25 an initial Ffg signalling centre forms at the isthmic organizer in the mid/hindbrain border region and patterns the anterior hindbrain. At E8.5 initiation of Hoxb1 in r4 leads to activation of Fgf3/8 and the formation of a secondary signalling centre in r4 important for patterning the posterior hindbrain. In chick embryos there is evidence for a secondary Fgf signalling centre in r2. Figure adapted from Fig. 2 and 3 of (Parker and Krumlauf, 2020). r, rhombomere; s, somite; RA, retinoic acid.
The first step of the GRN relates to A–P signalling, which is initiated by inputs and cooperative interactions between the fibroblast growth factor (FGF), Wnt and retinoic acid (RA) signalling pathways (Frank and Sela-Donenfeld, 2019). Regulatory interactions in the A–P signalling module create temporally and spatially dynamic domains of signalling (Fig. 2B,C). In mouse embryogenesis at ~E7.25, Wnt3a activates Meis and Pbx genes (Fig. 3A), which induce expression of Fgf3/8 and synthesis of RA by Raldh2 in somitic mesoderm, adjacent to the posterior hindbrain (Fig. 2B). RA spreads anteriorly in the hindbrain, while Fgf and Wnt signals restrict Cyp26a1 to the anterior hindbrain, where it degrades RA in r1 and r2, establishing an initial RA signalling domain with an anterior limit at the future r2/3 border at E7.5. Cyp26c1 is induced in r4 at E7.9, and Cyp26b1 induced in r3 and r5 by E8.5. This progressive activation of Cyp26a1, Cyp26b1 and Cyp26c1 leads to dynamic changes in degradation of RA that eventually establish a gradient with an anterior limit at the r5/6 boundary at E9.0 (Fig. 2B) (Hernandez et al., 2007; Schilling et al., 2012; Sirbu et al., 2005; White et al., 2007b; White and Schilling, 2008).
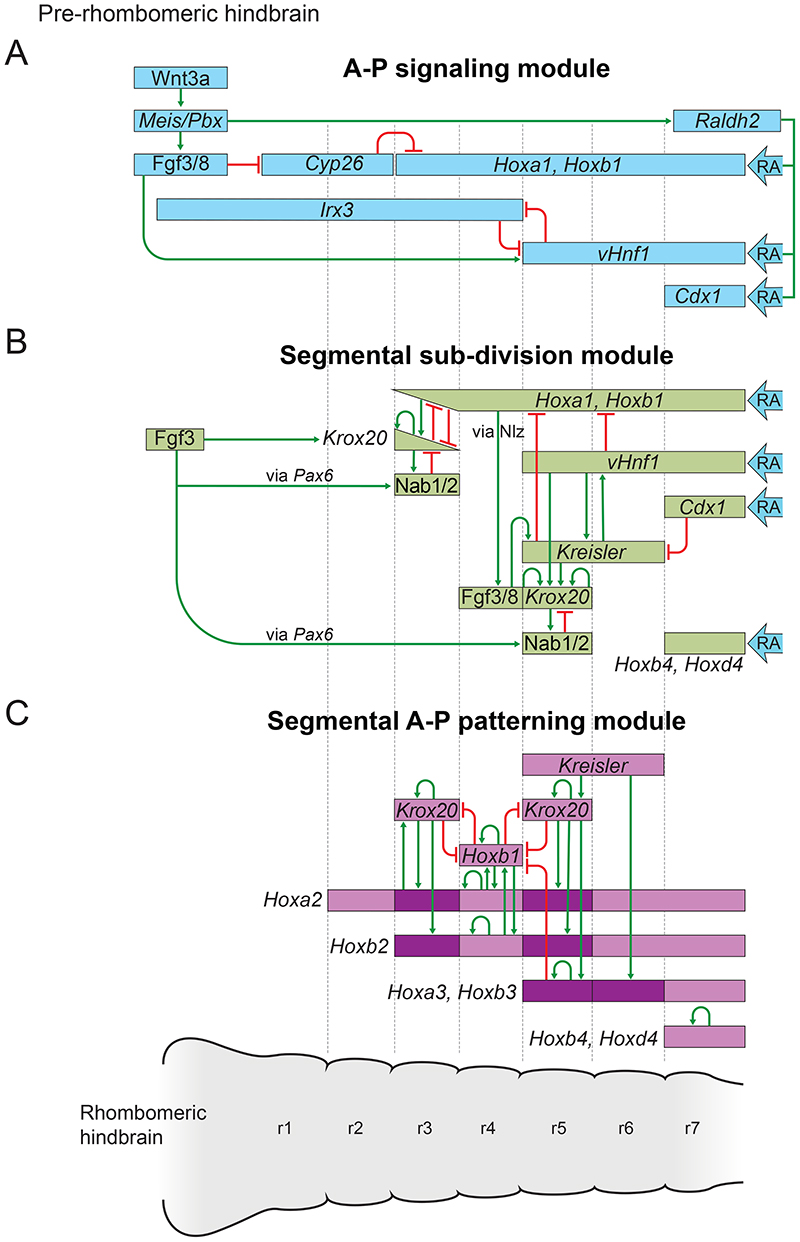
Fig. 3. Regulatory circuits and interactions between genes and signalling components in three modules governing successive steps in the GRN for hindbrain segmentation.
(A) The A–P signalling module initiates the process of segmentation in the pre-rhombomeric hindbrain through combined inputs and cooperative interactions between the fibroblast growth factor (Fgf), Wnt and retinoic acid (RA) signalling pathways. This sets up nested domains of spatially-restricted enzymes, signals and transcription factors (blue rectangles) that then trigger the next step. (B) The segmental sub-division module represents a series of regulatory interactions that establish sharply defined expression domains of segmentation genes (green rectangles) through extensive auto- and cross-regulatory interactions, involving mutual repression and activation. This provides a transcriptional code which sub-divides the hindbrain into rhombomeric compartments. (C) Circuits in the segmental A–P patterning module activate rhombomere-restricted domains of Hox expression (purple rectangles) to specify regional identity of individual segments. Darker shades of colour in the Hox expression domains indicate higher levels of expression in specific rhombomeres. The colours of the domains of expression and activity of genes and signals in each of the three modules correspond to that of the GRN structure in Fig. 2A. In A-C, the interactions depicted within each module are not intended to imply a specific temporal or hierarchical order and represent cumulative interactions associated with each module. Figure adapted from Fig. 4 of (Parker and Krumlauf, 2020). r, rhombomere; RA, retinoic acid.
Collectively, this cascade of events establishes the primary signals and initial nested domains of TF expression in the A–P signalling module (Fig. 3A) that in turn activate a network of spatially-restricted TFs in the segmental sub-division module (Fig. 3B). Through an extensive network of auto- and cross-regulatory interactions, segmentation gene expression domains become progressively refined to generate a pattern of sharply restricted domains of expression. This provides the transcriptional code which divides the hindbrain into rhombomeres (Fig. 2; 3B).
The network of segmentation TFs also provides input into the cell segregation and boundary cell formation module through regulation of Eph receptors (Theil et al., 1998) and set up secondary signalling centres for Fgfs and Wnts in specific rhombomeres, which play an important role in elaborating segment formation (Fig. 2A,C). As rhombomeres begin to form, signalling between them plays a role in regulating segmental identity and cell segregation. The segmentation sub-division module begins to restrict Hoxb1 expression to r4, which leads to expression of Fgf3/8 and the formation of a secondary signalling centre (Fig. 2C; 3B). In zebrafish, r4 serves as an early signalling centre that patterns the posterior hindbrain by regulating the expression of Hoxb1, Krox20 and Kreisler/MafB/valentino (Marin and Charnay, 2000; Maves et al., 2002; Walshe et al., 2002; Wiellette and Sive, 2003, 2004). In combination with vhnf1and FGF signalling, these transcription factors specify individual rhombomere identities in the caudal hindbrain (r4-r7) (Parker and Krumlauf, 2017; Sun and Hopkins, 2001; Wiellette and Sive, 2003). In chick embryos, there is evidence for secondary Fgf signalling centres in r2 and r4 that participate in regulating Krox20-independent EphA4 expression (Cambronero et al., 2020). Collectively, this illustrates the presence of cross-talk and regulatory feedback between modules in the GRN.
In the segmental A–P patterning module, TFs directly regulate segmental expression of Hox genes (Fig. 3C). This generates nested and segment-restricted domains of Hox expression, which form a combinatorial code for specifying distinct A–P identities to each segment. Through the Hox genes, the segmental A–P patterning module also provides input into cell segregation and boundary cell identity, in part through regulation of Eph receptor genes (Prin et al., 2014) (Fig. 2). These progressive steps are dynamic and parts of the regulatory logic may be utilised in multiple steps. For example, the GRN that underlies Hox PG1 gene expression in r4, and Egr2 expression in r3 and r5, are relevant not only for segmentation, but also for the dynamic regulation of cell fate that stabilises the segments in the cell segregation and boundary cell formation module (Fig. 2A).
Conserved network of A-P patterning
Although many of the proposed functional and regulatory interactions have not been independently validated in each jawed vertebrate species used to formulate the GRN model of hindbrain segmentation, it provides a useful regulatory framework for considering hindbrain evolution. Studies in the sea lamprey, a jawless fish, have provided insight into its ancestry at the base of the vertebrate family tree (Shimeld and Donoghue, 2012). Key genes involved in the hindbrain segmental sub-division and A–P segmental patterning modules (Fig. 2A) are present in lamprey and display segment-restricted patterns of expression coupled to hindbrain segmentation and patterning of cNCCs (Jimenez-Guri and Pujades, 2011; Parker et al., 2014, 2019a; Parker et al., 2019b; Smith et al., 2018). Many cis-regulatory elements associated with segmental expression in the GRN of jawed vertebrates can direct analogous rhombomere-restricted domains in the lamprey hindbrain, and conserved cis-regulatory elements that mediate segmental expression have been identified in similar positions in and around lamprey genes (Parker et al., 2014; Parker et al., 2019b). These analyses show that essential aspects of the hindbrain GRN, including upstream regulatory factors and cis-regulatory circuits underlying segmental expression, form part of an ancient regulatory circuit already present in the common ancestor of lamprey and jawed vertebrates. Therefore, hindbrain segmentation appears to be a fundamental innovation that, along with the ability to form neural crest cells, is wired into conserved GRNs for developmental programs governing head development at the base of the vertebrate tree (Parker et al., 2016; Sauka-Spengler et al., 2007).
In light of the high degree of conservation of the hindbrain GRN and its A–P Hox code, it is interesting to consider how diversity in hindbrain and craniofacial development has evolved in vertebrates. One possibility is that diversification between species may have been achieved through differences in downstream targets of TFs in the GRN, impacting elaboration of rhombomeric and cNCC differentiation programs. The genome-wide duplications associated with evolution of vertebrates has generated families of paralogous genes that can partition ancestral activities and evolve new roles. The Hox PG1 genes are a good example, where the expression and function of the three mammalian PG1 genes (Hoxa1, Hoxb1 and Hoxd1) in the hindbrain have been differentially distributed among the paralogues in zebrafish and Xenopus (Frank and Sela-Donenfeld, 2019; Kolm and Sive, 1995; McClintock et al., 2002; Moens and Prince, 2002; Studer et al., 1998; Tvrdik and Capecchi, 2006). Furthermore, analyses of mouse HOX PG1 proteins have uncovered functional divergence between paralogues and found that downstream targets of HOXA1 and HOXB1 are associated with different biological processes, (De Kumar et al., 2017; Singh et al., 2021; Singh et al., 2020). This illustrates how specific functional components and regulatory circuits in the hindbrain GRN can vary between species as a result of diversification in the roles and outputs of paralogous genes.
Another input into diversity between species may arise through differences in roles for genes in the GRN and their targets after hindbrain segmentation is completed. There is evidence for continued roles of these genes in later steps of hindbrain development and we know very little about the GRNs governing these processes (Santagati et al., 2005). In the future, it will be important to explore the nature of similarities and differences in downstream Hox target genes and pathways in different species during head development to extend the hindbrain GRN into stages that govern processes underlying morphological and neuroanatomical diversity.
Since hindbrain segmentation arose during the evolutionary transition to vertebrates, this raises the question of how it became coupled to ancient A-P patterning networks. Comparative regulatory analyses in tunicates (Ciona) and cephalochordates (amphioxus) indicate that the cis-elements responsible for the segmental expression of Hox genes in jawed vertebrates do not appear to be present in these chordates (Manzanares et al., 2000; Natale et al., 2011). Studies in a hemichordate, the acorn worm Saccoglossus kowalevskii, have surprisingly uncovered deep similarities in A-P axis formation and organisation in embryonic stages across deuterostomes. The A-P expression domains of key developmental TFs, including Hox genes, and components of signalling pathways (FGF, Hh and Wnt) are similarly aligned along the bodies of hemichordates and chordates suggesting a deeply conserved axial patterning system (Gerhart et al., 2005; Lowe et al., 2015; Lowe et al., 2003; Pani et al., 2012). Functional studies have shown that Wnt signalling is important for regulating Hox genes in hemichordates (Darras et al., 2018) and also provides multiple inputs into coordinate regulation of mouse HoxA genes during gastrulation and primitive streak formation (Neijts et al., 2017; Neijts et al., 2016; Neijts and Deschamps, 2017). RA-responsive enhancers embedded in mouse Hox clusters are involved in coordinate regulation of multiple genes in each cluster (Ahn et al., 2014; Gould et al., 1997; Nolte et al., 2013; Oosterveen et al., 2003; Qian et al., 2018; Sharpe et al., 1998) and there is experimental evidence for several highly conserved retinoic acid response elements (RAREs) in neural enhancers of amphioxus and mouse Hox clusters (Manzanares et al., 2000; Wada et al., 2006). This suggests that major axial signalling centres evolved long ago in chordate evolution, and an ability to coordinately respond to RA and Wnt signalling may be part of an ancient regulatory mechanism that underlies the generation of nested domains of Hox expression along the A-P axis. Vertebrates may then have co-opted this ancient patterning system and coupled it to hindbrain segmentation through changes in existing control modules and/or the emergence of new cis-regulatory elements.
Generation of sharp and homogeneous hindbrain segments
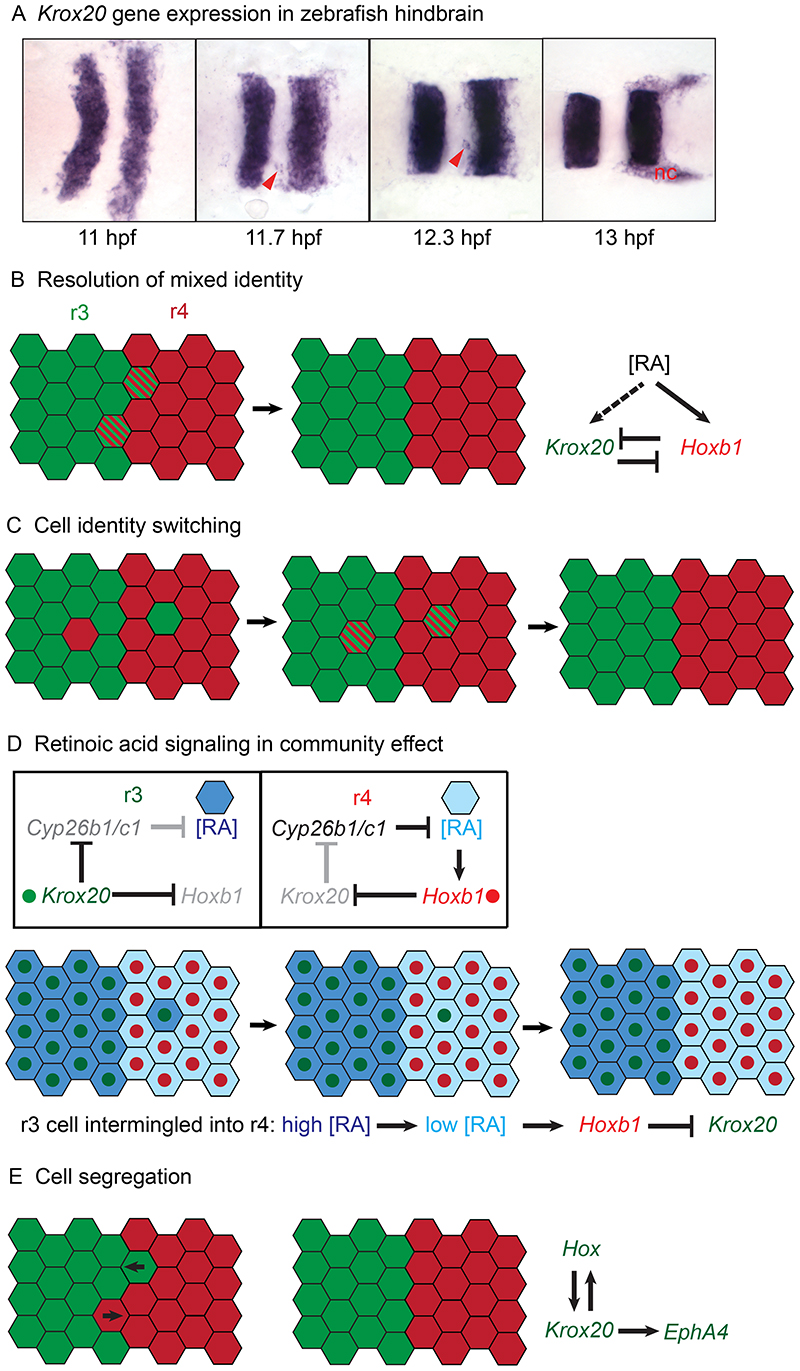
At early stages, the expression domains of genes specifying regional identity are not precise, but are refined to form sharp borders. Studies of hindbrain segmentation have shown that several molecular mechanisms act together to sharpen segment borders. In particular, important insights have come from studies of how complementary and sharp borders of Krox20 and Hoxb1 gene expression are established in r3-r5. At the onset of Krox20 and Hoxb1 upregulation, the borders of their segmental expression are ragged, but once hindbrain boundaries can be seen at the morphological level, the borders have become sharp and straight (Cooke and Moens, 2002; Irving et al., 1996; Murphy et al., 1989; Murphy and Hill, 1991; Prince et al., 1998; Sundin and Eichele, 1990; Wilkinson et al., 1989b). Furthermore, some cells at the borders initially co-express Krox20 and Hoxb1 that specify distinct identities (Zhang et al., 2012). For example, in zebrafish, the fuzzy expression of Krox20 seen at 11 hpf (hours post fertilisation) has become sharpened by 13 hpf (Fig. 4A). The initial imprecision of gene expression domains is likely due to two factors: first, that the generation and interpretation of morphogen gradients that regulate gene expression is not precise; and second, that cell intermingling challenges the formation and maintenance of a sharp border. Studies in chick (Fraser et al., 1990) and zebrafish (Addison et al., 2018) found that some cells intermingle between segments at early stages, due to cell intercalation concomitant with cell proliferation and convergent-extension of the neural epithelium (Kimmel et al., 1994). Subsequently, cell intermingling is restricted across segment borders (Calzolari et al., 2014; Fraser et al., 1990; Jimenez-Guri et al., 2010). This progressive establishment of cell segregation mechanisms during hindbrain segmentation reflects that Eph receptors and ephrins, which restrict intermingling, are regulated downstream of TFs that underlie segmentation. For example, EphA4 is a direct target of Krox20 (Theil et al., 1998) and is repressed by Hoxa3 and Hoxb4 (Prin et al., 2014); EphB4 is regulated downstream of Kreisler (Cooke et al., 2001), and ephrinB2 is regulated by Hoxb4 and Hoxd4 (Prin et al., 2014). Consistent with this, some isolated Krox20-expressing cells are observed in r2+r4+r6 during the period of border sharpening (Cooke and Moens, 2002; Irving et al., 1996) (Fig. 4A). Potentially, such ectopic cells could switch identity to match their neighbours or segregate back to r3 or r5, such that segments maintain a homogeneous identity (Cooke and Moens, 2002; Pasini and Wilkinson, 2002). Recent studies support the idea that both dynamic regulation of cell identity and cell segregation contribute to form a precise segmental pattern.
Fig. 4. Mechanisms of border sharpening in the hindbrain.
(A) Time course of Krox20 gene expression in the zebrafish hindbrain. From 11 to 13 hpf the initial wiggly borders of expression in r3 and r5 become sharp and straight. Some isolated Krox20-expressing cells (red arrowheads) are present in adjacent segments. (B) At early stages, some cells at the borders of r3-r4-r5 co-express Krox20 and Hoxb1, indirectly or directly downstream of retinoic acid (RA) signalling, respectively. The overlapping expression is resolved by mutual repression. (C) Intermingling of cells between segments occurs at early stages, when cell segregation mechanisms are not fully established. Ectopic cells switch identity to match their new neighbours. (D) Cell identity switching is regulated by retinoic acid signalling, illustrated for r3 and r4. In r3, Krox20 reduces expression of Cyp26b1 and Cyp26c1, and consequently the RA level is high. In r4, there is a higher level of Cyp26b1 and Cyp26c1 expression and the RA level is lower. Following intermingling of an r3 cell into r4 its RA level is decreased, leading to upregulation of Hoxb1 and repression of Krox20 expression. (E) At later stages, cell segregation mediated by Eph-ephrin signalling has been established and drives reorganisation of cells to sharpen borders. EphA4 which contributes to cell segregation is directly regulated by Krox20, and thus coupled to Hox gene expression. (A) is adapted from (Addison, 2016). (D) is adapted from (Addison et al., 2018). nc, Krox20 expression in neural crest.
Roles and mechanisms of dynamic regulation of cell identity
Hoxb1 and Krox20 are expressed in, and have key roles in the specification of, r4 and r3+r5, respectively. Segmental expression of these genes is directly (Hoxb1) or indirectly (Krox20) regulated by a gradient of RA in the hindbrain that is established by a counter-gradient of the RA-degrading enzyme, Cyp26a1 (White et al., 2007a). At early stages some cells at the borders of r4 co-express Hoxb1 and Krox20 (Zhang et al., 2012), which is resolved through reciprocal repression (Alexander et al., 2009) (Fig. 4B). Direct visualisation of RA reveals that the gradient is noisy at single-cell resolution (Sosnik et al., 2016), and computer simulations suggest that this noise contributes to the initial overlap of Hoxb1 and Krox20 expression (Zhang et al., 2012). Experiments and simulations support an important role of the RA-binding protein Crabp2a, as well as Cyp26a1, which dampen noise in the regulation of RA target genes (Sosnik et al., 2016). Interestingly, the simulations suggest that an appropriate amount of noise in the level of Hoxb1 and Krox20 expression has a positive role in enabling the transition from overlapping to mutually exclusive expression of these genes (Zhang et al., 2012).
In addition to resolving mixed identity at the border, there is dynamic regulation of segmental gene expression in cells that have intermingled between segments (Fig. 4C). Whereas single cells transplanted between hindbrain segments change identity to match their new location, cells transplanted as a group do not switch identity (Schilling et al., 2001; Trainor and Krumlauf, 2000a). This suggests that there is a community regulation of segmental identity, which in classical models is mediated by positive feedback between TFs and intercellular signalling (Bolouri and Davidson, 2010). Krox20 may participate in such feedback, because its mosaic overexpression induces Krox20 expression in adjacent cells (Giudicelli et al., 2001).
Recent studies have revealed an RA-mediated mechanism that underlies community regulation of Krox20 expression (Fig. 4D). Two Cyp26 family members, Cyp26b1 and Cyp26c1, have dynamic segmental expression in the zebrafish hindbrain that contributes to A-P patterning in parallel with graded Cyp26a1 (Fig. 2B) (Hernandez et al., 2007). Collectively, Cyp26b1 and Cyp26c1 are expressed at a lower level in r3+r5 than in r2+r4+r6, due to their repression downstream of Krox20 (Addison et al., 2018). Since Cyp26 expression influences the level of RA, segmentation genes thus regulate a difference in RA signalling between adjacent segments. Furthermore, high levels of Cyp26 can act as a sink to non-autonomously decrease RA in adjacent cells (Rydeen et al., 2015; Rydeen and Waxman, 2014; White et al., 2007a). Supporting a role for the segmental regulation of RA levels, Cyp26b1 and Cyp26c1 are required for the identity switching of Krox20-expressing cells that have intermingled into adjacent segments (Addison et al., 2018). In r4, the identity switching also requires Hoxb1, which is a direct target of RA signalling (Studer et al., 1994). These findings suggest that coupling of the level of Cyp26b1 and Cyp26c1 expression to segment identity mediates a community effect that switches the identity of Krox20- expressing cells that intermingle into adjacent segments (Fig. 4D). These cells move from a high RA (low Cyp26) to lower RA (higher Cyp26) environment, which by non-autonomously decreasing RA levels leads to upregulation of Hoxb1 and downregulation of Krox20 expression.
These findings suggest a two-step model for the relationship between RA signalling and segmental identity. At early stages, a gradient of Cyp26a1 expression underlies a gradient of RA that regulates segmental identity. Cyp26a1 is RA-inducible and acts in self-enhanced RA degradation that is crucial for gradient formation (White et al., 2007a). Subsequently, Cyp26b1 and Cyp26c1 are expressed in segmental patterns that are downstream of segment identity genes. Consequently, positive feedback can occur in which segment identity regulates the level of RA, which in turn can reinforce segment identity. The findings also provide an explanation of the relationship between cell organisation and regulation of segmental identity. When cells are surrounded by others with a distinct identity they regulate Krox20 expression to match their neighbours, as occurs following mosaic overexpression of Krox20 (Addison et al., 2018; Giudicelli et al., 2001). In contrast, non-autonomous induction of Krox20 expression does not occur once cells have segregated (Addison et al., 2018). Thus identity switching depends upon how many neighbours have the same or different identity, which mechanistically can be explained by the short range of the non-autonomous effect of Cyp26 on RA levels (Rydeen et al., 2015; White et al., 2007a).
Roles and mechanisms of cell segregation
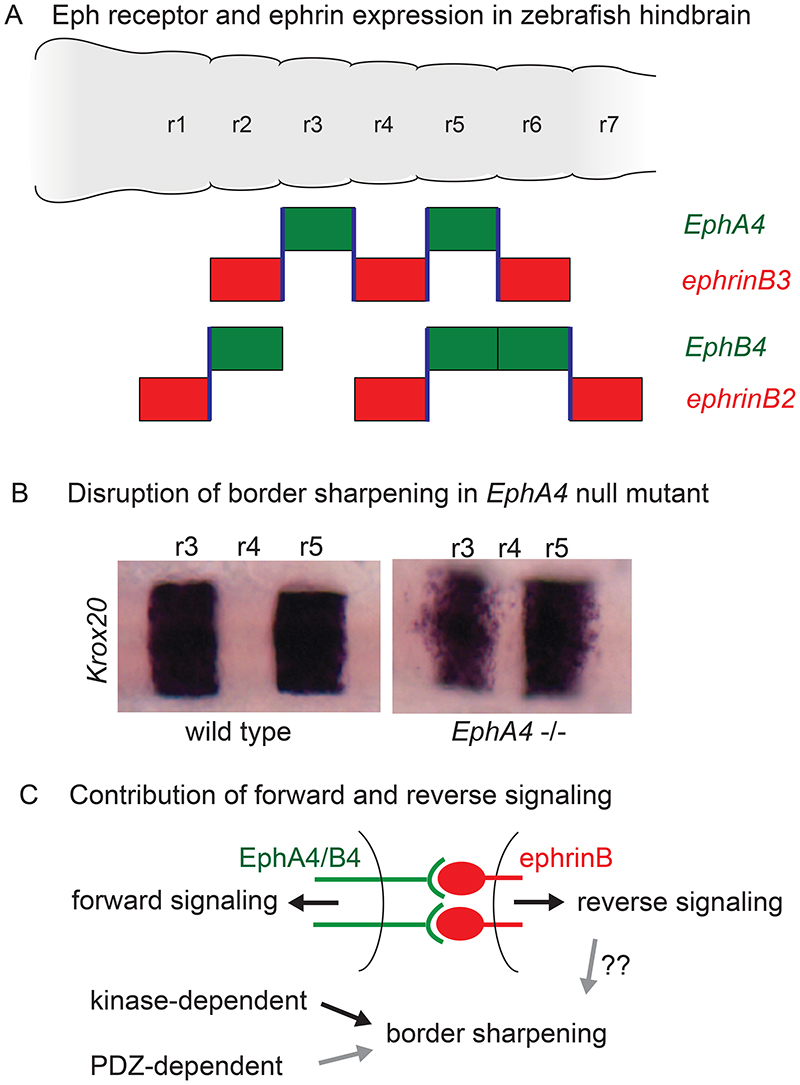
Initial evidence for how cell intermingling is restricted between rhombomeres came from transplantation experiments in chick, which found that it involved cell affinity properties that distinguish r3+r5 from r2+r4+r6 (Guthrie et al., 1993). The identification of segmentally-expressed Eph receptors (Becker et al., 1994; Nieto et al., 1992) led to functional analyses (Calzolari et al., 2014; Cayuso et al., 2019; Cooke et al., 2001; Cooke et al., 2005; Xu et al., 1995; Xu et al., 1999) that revealed key roles in cell segregation that establishes and maintains sharp borders in the hindbrain. Eph receptor tyrosine kinases are clustered and activated upon interacting with membrane-bound ephrins, which also mediate signal transduction, leading to ‘forward’ and ‘reverse’ signalling, respectively (Kania and Klein, 2016; Pasquale, 2008). Eph receptors and ephrins that have a high affinity are expressed in complementary hindbrain segments, and consequently bidirectional signalling occurs at segment borders. For example, in zebrafish this is seen for EphA4 and ephrinB3, and for EphB4 and ephrinB2a (Chan et al., 2001; Cooke et al., 2001; Xu et al., 1995) (Fig. 5A). Mosaic gain or loss of Eph or ephrin function leads to cell segregation within segments (Cooke et al., 2001; Cooke et al., 2005; Kemp et al., 2009; Xu et al., 1999), suggestive of a role in the regulation of cell affinity. Furthermore, EphA4 loss of function in zebrafish (Cayuso et al., 2019; Cooke et al., 2005; Xu et al., 1995) and chick (Sela-Donenfeld et al., 2009) increases cell intermingling and disrupts sharpening of the borders of r3 and r5; only the r4/r5 border remains sharp, which is likely due to functional redundancy with EphB4 (Cooke et al., 2001) (Fig. 5B).
Fig. 5. Eph-ephrin signalling in hindbrain border sharpening.
(A) Depiction of the segmental expression of Eph receptors and ephrins in the zebrafish hindbrain. EphA4 has high affinity for ephrinB2 and ephrinB3, whereas EphB4 binds selectively to ephrinB2. The segmental expression of high-affinity Eph-ephrin pairs is complementary, such that strong activation (blue lines) occurs at segment borders. (B) Krox20 expression in wild type and Epha4 null mutant zebrafish embryos, adapted from (Cayuso et al., 2019). Sharpening is disrupted at r3 and r5 borders, except for the r4/r5 border where there is functional redundancy with EphB4/ephrinB2 as depicted in panel (A). (C) Signalling through Eph receptors occurs through tyrosine phosphorylation following activation of the kinase domain, and also through interaction of PDZ domain proteins with a C-terminal binding motif. Use of truncation and point mutants of EphA4 finds a major input of kinase-dependent forward signalling and minor input of PDZ-dependent signalling in border sharpening. Further studies are needed to determine whether reverse signalling also contributes.
It is now well established that Eph-ephrin signalling is a major player in cell segregation and border formation in many tissues in vertebrates (Batlle and Wilkinson, 2012; Fagotto et al., 2014; Kania and Klein, 2016; Pasquale, 2005). As in the hindbrain, expression of high affinity Eph and ephrin binding partners occurs in complementary domains (Gale et al., 1996), and there is overlapping expression of lower affinity partners (Rohani et al., 2014). Consequently, there is strong activation of Eph and ephrin signalling at the border of the adjacent domains, but also weak activation within each domain. Studies of cell responses and biochemical targets of Eph-ephrin signalling suggest that it can drive cell segregation by decreasing cadherin-mediated adhesion, and/or through increased cell repulsion or cortical tension mediated by actomyosin contraction (Fagotto et al., 2013; O’Neill et al., 2016; Rohani et al., 2011; Rohani et al., 2014; Solanas et al., 2011). The use of quantitative measurements in computer simulations suggest that the principal mechanisms are heterotypic repulsion or cortical tension, which are more efficient than differential adhesion in driving cell segregation (Canty et al., 2017; Taylor et al., 2017). However, cadherin-mediated adhesion has a crucial role in counter-balancing the repulsion or tension response to low level Eph-ephrin signalling that occurs within each tissue or regional domain (Taylor et al., 2017). This latter finding potentially explains the requirement for cadherin function in segregation of cells from different rhombomeres (Wizenmann and Lumsden, 1997).
In the zebrafish hindbrain, increased levels of actomyosin and phosphorylated myosin light chain (pMLC) are detected at rhombomere boundaries from 15 hpf, which for r3 and r5 are dependent upon EphA4 (Calzolari et al., 2014; Cayuso et al., 2019). Furthermore, actomyosin contraction underlies increased tension and the distinctive shape of hindbrain boundary cells (Gutzman and Sive, 2010), and inhibition of myosin function leads to wiggly hindbrain borders (Calzolari et al., 2014). These findings suggest that Eph and/or ephrin activation leads to increased cortical tension required for border sharpness. Since pMLC and increased actomyosin are first detected at segment borders several hours after they have become sharp, increased cortical tension may maintain rather than generate sharpness, but it remains possible that there is a dynamic regulation of tension at early stages that underlies segregation. An important question is whether cell segregation requires forward and/or reverse signalling, but in null Eph or ephrin mutants signalling is disrupted in both directions. To address this problem, deletion and point mutants of EphA4 were generated that disrupt all or specific pathways of forward signalling but leave reverse signalling intact (Cayuso et al., 2019). It was found that border sharpening in the hindbrain requires kinase-dependent forward signalling (Cayuso et al., 2019) (Fig. 5C). A similar picture has come from studies in other tissues, with forward signalling having the dominant role in cell segregation and border sharpening (O’Neill et al., 2016; Rohani et al., 2014). However, studies in a cell culture model suggest that reverse signalling can contribute to sharpening (Wu et al., 2019), but this has yet to be directly tested in vivo.
Interplay of cell identity regulation and cell segregation
The relative contribution of cell identity regulation and cell segregation shifts during the progression of hindbrain border sharpening. Intermingling between segments occurs at early stages, before expression of EphA4 downstream of Krox20 (Theil et al., 1998) has been sufficiently upregulated. Consequently, an early transgenic reporter of Krox20 gene expression detects cell intermingling followed by identity switching (Addison et al., 2018), whereas cells marked by a later Krox20 reporter segregate rather than switch identity (Calzolari et al., 2014). During this progression, hindbrain cells transition from a plastic to a more committed state (Schilling et al., 2001), by which time cell segregation mechanisms have been fully established. Further insights have come from computer simulations that found that cell identity regulation alone, or cell segregation alone, are not able to generate a sharp border, but in combination lead to efficient sharpening (Wang et al., 2017). The simulations suggest that these mechanisms are synergistic as they make distinct contributions to border sharpening: cell segregation is not efficient if the transition zone of intermingled cells is too wide, and cell identity regulation serves to create a narrow transition zone (Wang et al., 2017). Further simulations suggest that cell reorganisation during convergent-extension and identity regulation by Fgf signalling also contribute to border sharpening (Qiu, 2021). Recent studies have found a similar interplay of cell segregation and identity regulation at the midbrain-hindbrain boundary (Kesavan et al., 2020). Some intermingling of cells between midbrain and hindbrain occurs prior to formation of the compartment boundary, and sharp gene expression borders are formed through cell identity switching and cell segregation, which requires Eph-ephrin function (Kesavan et al., 2020). Interestingly, midbrain-hindbrain border sharpening also requires N-cadherin (Kesavan et al., 2020), which as discussed above may be due to an interplay between adhesion and cell responses to Eph-ephrin signalling (Taylor et al., 2017).
Regulation and roles of hindbrain boundary cells
Early studies of hindbrain segmentation revealed that cells at the borders, termed boundary cells, have distinct cellular and molecular properties from cells away from the border (Lumsden and Keynes, 1989). In chick, boundary cells proliferate more slowly and have less interkinetic nuclear migration (Guthrie et al., 1991), and have larger intercellular spaces that are filled with specific extracellular matrix (ECM) components (Heyman et al., 1995; Lumsden and Keynes, 1989; Weisinger et al., 2011). Boundary cells also express a distinct set of genes from non-boundary cells (Cooke et al., 2005; Letelier et al., 2018; Riley et al., 2004; Tambalo et al., 2020; Xu et al., 1995), but with some differences between species, such as Fgf3 in chick (Mahmood et al., 1995) and Rfng in zebrafish (Cheng et al., 2004). Recent studies have started to uncover how boundary cells are formed at segment borders and the roles that they play in hindbrain development (Pujades, 2020).
Boundary cell induction
Ablation and transplantation experiments revealed that boundary cells are induced when odd- and even-numbered segments are juxtaposed (Guthrie and Lumsden, 1991). This correlates with cell segregation that also involves distinct properties of odd versus even segments, but it was argued that boundary cells are not involved in restricting cell intermingling (Guthrie et al., 1993). This receives support from recent studies that have revealed mechanisms of cell segregation and boundary cell formation and how they are linked.
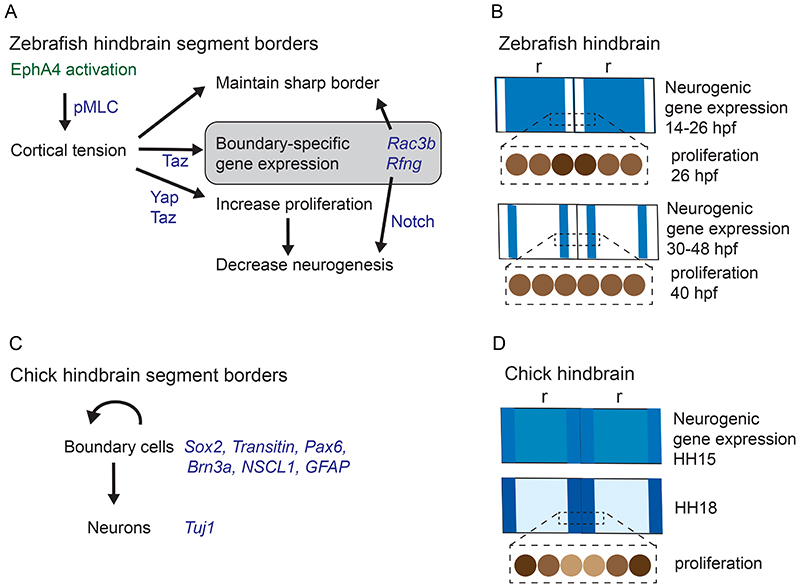
Disruption of EphA4 function leads to loss of boundary cells at the borders of r3 and r5 (Cooke et al., 2005; Xu et al., 1995), suggesting that forward and/or reverse signalling is required for boundary cell induction. Furthermore, there is growing evidence that in some tissues Eph-ephrin signalling is involved in the regulation of cell differentiation (Laussu et al., 2014; Wilkinson, 2014). The use of mutants to dissect EphA4 signalling in zebrafish found that hindbrain boundary cell induction is mainly regulated by kinase-dependent forward signalling, and thus correlates with border sharpening (Cayuso et al., 2019). Expression of Rfng at boundaries is induced by cortical tension generated by myosin activation downstream of EphA4 signalling (Cayuso et al., 2019). Increased cortical tension leads to nuclear translocation of Taz, which in cooperation with Tead1 induces boundary gene expression (Cayuso et al., 2019). There is thus a coupling in which increased cortical tension both maintains border sharpness and induces boundary cells, ensuring that boundary cells form at a sharp interface (Fig. 6A). Boundary cells also express a modulator of the actin cytoskeleton, Rac3b, which is implicated in increasing cortical tension (Letelier et al., 2018). This suggests that in addition to acting directly on actomyosin contraction, Eph receptor signalling maintains border sharpness through transcriptional targets regulated downstream of cortical tension (Fig. 6A). Since EphA4 signalling is also required for boundary cell formation in chick (Sela-Donenfeld et al., 2009), it will be interesting to determine whether this likewise involves the increase in cortical tension that occurs at segment borders (Filas et al., 2012).
Fig. 6. Regulation of hindbrain boundary cells.
(A) Molecular mechanisms of boundary cell regulation in the zebrafish hindbrain. EphA4 activation leads to increased cortical tension at boundaries through myosin light chain phosphorylation (pMLC). Increased tension maintains border sharpness, and through nuclear translocation of Taz leads to upregulation of boundary-specific gene expression. Boundary-specific genes include Rac3b which contributes to border sharpness, and Rfng which inhibits neurogenesis by promoting Notch activation. Cortical tension acts through Yap/Taz activation to increase boundary cell proliferation, thus decreasing neurogenesis. (B) The pattern of neurogenesis in the zebrafish hindbrain is depicted for two adjacent rhombomeres (r) to illustrate the relationship with boundaries. Neurogenic gene expression is widespread but lower at the boundaries from 14-26 hpf. The rate of cell proliferation (light to dark brown shading, low to high rate) at 26 hpf is higher at boundaries than in non-boundary cells. Neurogenic gene expression has become confined to zones adjacent to hindbrain boundaries from 30 hpf onwards. By 40 hpf, there is no longer increased cell proliferation at boundaries. (C) In the chick hindbrain, boundary cells express Sox2 and neurogenic markers, and are self-renewing neural stem cells that can generate neurons. (B) In the chick hindbrain, neurogenic gene expression (blue shading) is widespread at HH15, and has become mainly confined to boundaries by HH18. The rate of cell proliferation is low at the centre of the boundary region and higher adjacent to the non-boundary region.
Role of boundary cells as signalling centres
It is notable that boundary cells express a number of signalling molecules, including members of the Wnt family (Riley et al., 2004), Follistatin family (Connolly et al., 1995; Tambalo et al., 2020; Weisinger et al., 2008) and Fgf3 (Mahmood et al., 1995; Sela-Donenfeld et al., 2009). Fgf3 has been found to act as an autocrine regulator of boundary cells in chick, in which there is also elevated expression of Fgf receptors, ECM components (e.g. HSPG, CSPG) and genes associated with neurogenesis (e.g. Brn3a, NSCL1) at hindbrain boundaries (Weisinger et al., 2011). Fgf3 knockdown abrogates the expression of these ECM and neurogenic genes, although not of Follistatin, suggesting that it is required for specific properties of boundary cells (Weisinger et al., 2011). Thus Fgf3 has stage-specific roles in the hindbrain (Box 1), associated with a switch from segmental to boundary-restricted expression. Interestingly, the downregulation of segmental Fgf3 expression requires signals from boundaries (Sela-Donenfeld et al., 2009), thus ensuring the appropriate spatial restriction of Fgf signalling at late stages.
Box 1. Differences in gene expression and patterning in chick and zebrafish.
Although many aspects of gene regulation and cell organisation are conserved, there are some important differences between species. An example is the spatial regulation of Fgf signalling and its roles in neurogenesis. In all species studied, early segmental Fgf expression (e.g. Fgf3 and Fgf8 in r4) is required for further segmentation of the caudal hindbrain (Hernandez et al., 2004; Maves et al., 2002; Walshe et al., 2002). Following this, the segmental expression is downregulated and, in chick, Fgf3 is upregulated in hindbrain boundaries where it promotes the expression of neurogenic genes and ECM components (Weisinger et al., 2011). Conversely, late Fgf signalling in the zebrafish hindbrain occurs at segment centres (Esain et al., 2010; Gonzalez-Quevedo et al., 2010), where Fgf20-expressing neurons inhibit neurogenesis and are positioned by chemorepulsive signalling from boundaries. What may underlie interspecies differences in the late expression and role of Fgf signalling? A clue comes from the observation that primary reticulospinal neurons are located each at segment centre in zebrafish (Hanneman et al., 1988). These neurons co-localise and may overlap with Fgf20-expressing neurons. Reticulospinal neurons are among the earliest neurons generated and form a neuronal circuit that mediates the escape response that enables aquatic embryos to move away from predators (O’Malley et al., 1996). This is not relevant in amniotes and, as for other neuronal cell types, reticulospinal neurons are not localised to segment centres (Cepeda-Nieto et al., 2005). Thus a requirement for A-P patterning of neurons within segments in zebrafish may have been lost in the evolutionary transition to amniotes, accompanied by changes in the organisation of neurogenesis by Fgf signalling.
The expression of signalling molecules by hindbrain boundaries also suggests potential roles in patterning of cell differentiation within segments, analogous to boundaries in other tissues (Dahmann and Basler, 1999; Kiecker and Lumsden, 2005; Rhinn and Brand, 2001). Direct evidence for a patterning role have come from studies of the spatial regulation of neurogenesis within segments in the zebrafish hindbrain. Expression of proneural genes that initiate neuronal differentiation initially occurs throughout the hindbrain, and later becomes downregulated at boundaries and in the centre of each segment (Cheng et al., 2004; Gonzalez-Quevedo et al., 2010; Riley et al., 2004) (Fig. 6B). Consequently, by 30 hpf neurogenesis has become confined to zones adjacent to hindbrain boundaries. Two mechanisms have been implicated in the decrease in neurogenesis in boundary cells (Fig. 6A). First, Rfng expression is induced downstream of cortical tension and Taz (Cayuso et al., 2019), and this promoter of Notch activation acts to inhibit neuronal differentiation (Cheng et al., 2004). Second, cortical tension acts through Yap/Taz to increase boundary cell proliferation and thus reduce neurogenesis (Voltes et al., 2019). The inhibition of neurogenesis in segment centres is mediated by Fgf20, which is expressed by a subset of neurons in the adjacent mantle zone (Gonzalez-Quevedo et al., 2010). Fgf20-expressing neurons form a cluster in the centre of each segment, which locally inhibit neurogenesis in the adjacent neural epithelium (Gonzalez-Quevedo et al., 2010). Since Fgf signalling is required for gliogenesis in the hindbrain (Esain et al., 2010), Fgf20 may underlie a switch from neuronal to glial cell differentiation at segment centres. The clustering of Fgf20-expressing neurons at segment centres is maintained by chemorepulsion mediated by Semaphorin family members that are expressed by hindbrain boundary cells (Terriente et al., 2012). Thus in zebrafish, boundary cells have a role in the patterning of neurogenesis within segments, albeit through an unconventional mechanism in which they organise Fgf20-expressing neurons that act as a signalling source. It currently remains unclear whether other signals from hindbrain boundary cells mediate direct paracrine regulation of cell differentiation (Amoyel et al., 2005; Gerety and Wilkinson, 2011; Riley et al., 2004).
Role of boundary cells as neural stem cells
Recent studies have found that hindbrain boundary cells are neural stem cells that are a source of neurogenesis, although with differences between species in timing and spatial organisation. In the chick hindbrain, the boundaries become a slowly dividing population of neural stem cells that are the major source of neurogenesis after HH18 (Fig. 6C). This is reflected by expression of markers of neurogenesis which is initially widespread in the hindbrain, and becomes restricted to boundaries by HH18 (Peretz et al., 2016) (Fig. 6D). In addition to expressing proneural genes, boundary cells express Sox2, a key regulator of neural stem cell properties, and can form neurospheres in culture that self-renew and differentiate (Peretz et al., 2016). There is a low rate of cell proliferation at the centre of the boundary cell zone and a higher rate at the outer part of the boundary (Peretz et al., 2016) (Fig. 6D). By analogy with other tissues, the accumulation of ECM at boundaries may contribute to regulation of these properties of boundary cells.
There is a different situation at zebrafish hindbrain boundaries, in which neurogenic gene expression is inhibited downstream of Eph signalling and increased cortical tension. Cortical tension acts through Yap/Taz to increase cell proliferation and upregulate Rfng expression (Cayuso et al., 2019; Voltes et al., 2019), leading to a sharp demarcation between non-neurogenic boundary cells and the adjacent neurogenic zones (Fig. 6B). Subsequently, there is a decrease in cortical tension, leading to a decline in the rate of cell proliferation and shift towards neuronal differentiation of boundary cells after 40 hpf (Voltes et al., 2019).
Although the spatial regulation of cell proliferation and neurogenic gene expression at boundaries seems to differ in chick and zebrafish (Fig. 6B,D), a common feature is that neurogenesis is lower at the segment borders (i.e. the centre of the boundary region) than in flanking regions. The inhibition of neuronal differentiation at the segment borders may serve to maintain a stable cell population that has other functions, such as a signalling source. One interpretation is that the boundary region described in chick is functionally equivalent to the boundary plus flanking neurogenic zones in zebrafish. In both species, boundaries comprise a pool of neural stem cells generating progeny that can move away from the segment border and differentiate. An important difference between chick and zebrafish is that in the latter the boundary cells initially have a higher rather than lower proliferation rate than non-boundary regions. This may reflect that there is a much shorter time interval between segmentation and neurogenesis in zebrafish compared with chick, which requires an early expansion of progenitor cells at boundaries (Voltes et al., 2019).
There is currently less understanding of the relationship between boundaries and neurogenesis in the mouse hindbrain. There is sustained high level expression of Hes1 at boundaries, in contrast to the variable and oscillating expression of this gene away from boundaries (Baek et al., 2006). Since high Hes1 maintains quiescent neural stem cells and inhibits proneural gene expression (Sueda et al., 2019), this suggests that there is decreased neurogenesis at hindbrain boundaries. Consistent with this, Plzf, a transcriptional repressor which inhibits neurogenesis (Sobieszczuk et al., 2010), is expressed at hindbrain boundaries in mouse (Cook et al., 1995). It will be interesting to ascertain whether in mouse, hindbrain boundaries become a source of neurogenic stem cells at later stages, as occurs in chick and zebrafish.
Conclusion
Studies of hindbrain segmentation have given important insights into how mechanisms of cell segregation and cell identity regulation cooperate to generate sharp and homogeneous regional identity. Such cooperation has also been found in recent studies of the midbrain–hindbrain border (Kesavan et al., 2020), and it is likely that similar principles apply to other tissues in which an initial imprecise pattern is sharpened. It will be important to have a deeper understanding of how these mechanisms are embedded in the GRN of hindbrain patterning. With regard to cell segregation, this requires uncovering of how other segmentally-expressed Eph receptors and ephrins are regulated by TFs that underlie segmentation. It will also be important to understand dynamic aspects of segment identity regulation through modelling of the GRN. For example, how do Krox20-expressing cells that intermingle into an adjacent segment switch identity? Such plasticity likely involves indirect responses of the initiator and autoregulatory elements of the Krox20 gene (Labalette et al., 2015) to the lower level of RA that the cell is exposed to in the new environment. Since autoregulation increases and maintains Krox20 expression (Labalette et al., 2015), this potentially contributes to the decrease in cell identity switching at late stages in the hindbrain (Schilling et al., 2001).
Another important area for future work is to understand how the early networks that establish segmental patterning lead to the organisation and coordinated differentiation of neuronal cell types in the hindbrain. Studies have shown that Hox genes have important roles in the trunk in regulating subtype diversity of motor neurons and display multiple functions in diverse neuronal classes to impact neuronal specification and connectivity (Philippidou and Dasen, 2013). In later stages of hindbrain development, Hox genes display dynamic D–V patterns of expression that correlate with the birth of major classes of neurons (Graham et al., 1991), and mutational studies demonstrate that Hox genes play important roles in patterning hindbrain neurons (Arenkiel et al., 2004; Davenne et al., 1999; Gaufo et al., 2003; Gavalas et al., 2003; Pattyn et al., 2003; Philippidou and Dasen, 2013). Existing studies have generated a rich level of knowledge of the molecular mechanisms and cellular processes regulated by the early roles of Hox and other TFs in the GRN of hindbrain patterning. The rapidly emerging array of genomic approaches for investigating small numbers of cells and single cells in developing tissues holds promise for identifying downstream targets and how the TFs they are coupled to control neurogenesis. Single-cell transcriptional profiling in the developing zebrafish hindbrain has begun to unravel the D–V and A–P distribution of neuronal cell types as they differentiate (Tambalo et al., 2020). There are major shifts in the transcriptomes of progenitors and differentiating cells over time that provide molecular insights and novel markers for functional analyses on the regulation and patterning of neural differentiation. A systematic application of this approach in multiple species holds promise for expanding the GRN and unravelling how Hox genes and segmentation regulate neurogenesis programs in the developing hindbrain and have broader roles in circuit formation.
Summary statement.
We discuss how sharp patterns of segments and boundaries are formed during hindbrain development
Acknowledgments
RK is grateful to Hugo Parker and Mark Miller for figure design. We thank members of the Krumlauf and Wilkinson groups for valuable discussions on the topic of this review. The Krumlauf lab is funded by the Stowers Institute for Medical Research (Grant #1001). The Wilkinson lab is supported by the Francis Crick Institute which receives its core funding from Cancer Research UK (FC001217), the UK Medical Research Council (FC001217) and the Wellcome Trust (FC001217).
Contributor Information
Robb Krumlauf, Email: rek@stowers.org.
David G. Wilkinson, Email: david.wilkinson@crick.ac.uk.
References
- Addison M. Investigating the roles of cell identity regulation and Eph/ephrin signalling in early hindbrain segmentation. Ph.D. thesis, University College London; 2016. [Google Scholar]
- Addison M, Xu Q, Cayuso J, Wilkinson DG. Cell identity switching regulated by retinoic acid signaling maintains homogeneous segments in the hindbrain. Dev Cell. 2018;45:606–620. doi: 10.1016/j.devcel.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn Y, Mullan HE, Krumlauf R. Long-range regulation by shared retinoic acid response elements modulates dynamic expression of posterior Hoxb genes in CNS development. Dev Biol. 2014;388:134–144. doi: 10.1016/j.ydbio.2014.01.027. [DOI] [PubMed] [Google Scholar]
- Alexander T, Nolte C, Krumlauf R. Hox genes and segmentation of the hindbrain and axial skeleton. Annu Rev Cell Dev Biol. 2009;25:431–456. doi: 10.1146/annurev.cellbio.042308.113423. [DOI] [PubMed] [Google Scholar]
- Amoyel M, Cheng YC, Jiang YJ, Wilkinson DG. Wnt1 regulates neurogenesis and mediates lateral inhibition of boundary cell specification in the zebrafish hindbrain. Development. 2005;132:775–785. doi: 10.1242/dev.01616. [DOI] [PubMed] [Google Scholar]
- Arenkiel BR, Tvrdik P, Gaufo GO, Capecchi MR. Hoxb1 functions in both motoneurons and in tissues of the periphery to establish and maintain the proper neuronal circuitry. Genes Dev. 2004;18:1539–1552. doi: 10.1101/gad.1207204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek JH, Hatakeyama J, Sakamoto S, Ohtsuka T, Kageyama R. Persistent and high levels of Hes1 expression regulate boundary formation in the developing central nervous system. Development. 2006;133:2467–2476. doi: 10.1242/dev.02403. [DOI] [PubMed] [Google Scholar]
- Batile E, Wilkinson DG. Molecular mechanisms of cell segregation and boundary formation in development and tumorigenesis. Cold Spring Harbor Perspect Biol. 2012;4:a008227. doi: 10.1101/cshperspect.a008227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker N, Seitanidou T, Murphy P, Mattei MG, Topilko P, Nieto MA, Wilkinson DG, Charnay P, Gilardi-Hebenstreit P. Several receptor tyrosine kinase genes of the Eph family are segmentally expressed in the developing hindbrain. Mechanisms of development. 1994;47:3–17. doi: 10.1016/0925-4773(94)90091-4. [DOI] [PubMed] [Google Scholar]
- Birgbauer E, Sechrist J, Bronner-Fraser M, Fraser S. Rhombomeric origin and rostrocaudal reassortment of neural crest cells revealed by intravital microscopy. Development. 1995;121:935–945. doi: 10.1242/dev.121.4.935. [DOI] [PubMed] [Google Scholar]
- Bolouri H, Davidson EH. The gene regulatory network basis of the “community effect,” and analysis of a sea urchin embryo example. Developmental biology. 2010;340:170–178. doi: 10.1016/j.ydbio.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J, Wilkinson DG. Establishing neuronal circuitry: Hox genes make the connection. Genes Dev. 2004;18:1643–1648. doi: 10.1101/gad.1227004. [DOI] [PubMed] [Google Scholar]
- Calzolari S, Terriente J, Pujades C. Cell segregation in the vertebrate hindbrain relies on actomyosin cables located at the interhombomeric boundaries. EMBO J. 2014;33:686–701. doi: 10.1002/embj.201386003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambronero F, Ariza-McNaughton L, Wiedemann LM, Krumlauf R. Inter-rhombomeric interactions reveal roles for fibroblast growth factors signaling in segmental regulation of EphA4 expression. Dev Dyn. 2020;249:354–368. doi: 10.1002/dvdy.101. [DOI] [PubMed] [Google Scholar]
- Canty L, Zarour E, Kashkooli L, Francois P, Fagotto F. Sorting at embryonic boundaries requires high heterotypic interfacial tension. Nature communications. 2017;8:157. doi: 10.1038/s41467-017-00146-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. Homeotic genes and the evolution of arthropods and chordates. Nature. 1995;376:479–485. doi: 10.1038/376479a0. [DOI] [PubMed] [Google Scholar]
- Cayuso J, Xu Q, Addison M, Wilkinson DG. Actomyosin regulation by Eph receptor signaling couples boundary cell formation to border sharpness. eLife. 2019;8 doi: 10.7554/eLife.49696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepeda-Nieto AC, Pfaff SL, Varela-Echavarria A. Homeodomain transcription factors in the development of subsets of hindbrain reticulospinal neurons. Mol Cell Neurosci. 2005;28:30–41. doi: 10.1016/j.mcn.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Chan J, Mably JD, Serluca FC, Chen JN, Goldstein NB, Thomas MC, Cleary JA, Brennan C, Fishman MC, Roberts TM. Morphogenesis of prechordal plate and notochord requires intact Eph/ephrin B signaling. Developmental biology. 2001;234:470–482. doi: 10.1006/dbio.2001.0281. [DOI] [PubMed] [Google Scholar]
- Chandrasekhar A. Turning heads: development of vertebrate branchiomotor neurons. Dev Dyn. 2004;229:143–161. doi: 10.1002/dvdy.10444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatonnet F, Thoby-Brisson M, Abadie V, Dominguez del Toro E, Champagnat J, Fortin G. Early development of respiratory rhythm generation in mouse and chick. Respir Physiol Neurobiol. 2002;131:5–13. doi: 10.1016/s1569-9048(02)00033-2. [DOI] [PubMed] [Google Scholar]
- Cheng YC, Amoyel M, Qiu X, Jiang YJ, Xu Q, Wilkinson DG. Notch activation regulates the segregation and differentiation of rhombomere boundary cells in the zebrafish hindbrain. Dev Cell. 2004;6:539–550. doi: 10.1016/s1534-5807(04)00097-8. [DOI] [PubMed] [Google Scholar]
- Clarke JD, Lumsden A. Segmental repetition of neuronal phenotype sets in the chick embryo hindbrain. Development. 1993;118:151–162. doi: 10.1242/dev.118.1.151. [DOI] [PubMed] [Google Scholar]
- Connolly DJ, Patel K, Seleiro EA, Wilkinson DG, Cooke J. Cloning, sequencing, and expressional analysis of the chick homologue of follistatin. Dev Genet. 1995;17:65–77. doi: 10.1002/dvg.1020170108. [DOI] [PubMed] [Google Scholar]
- Cook M, Gould A, Brand N, Davies J, Strutt P, Shaknovich R, Licht J, Waxman S, Chen Z, Gluecksohn-Waelsch S, et al. Expression of the zinc-finger gene PLZF at rhombomere boundaries in the vertebrate hindbrain. Proc Natl Acad Sci U S A. 1995;92:2249–2253. doi: 10.1073/pnas.92.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke J, Moens C, Roth L, Durbin L, Shiomi K, Brennan C, Kimmel C, Wilson S, Holder N. Eph signalling functions downstream of Val to regulate cell sorting and boundary formation in the caudal hindbrain. Development. 2001;128:571–580. doi: 10.1242/dev.128.4.571. [DOI] [PubMed] [Google Scholar]
- Cooke JE, Kemp HA, Moens CB. EphA4 is required for cell adhesion and rhombomere-boundary formation in the zebrafish. Curr Biol. 2005;15:536–542. doi: 10.1016/j.cub.2005.02.019. [DOI] [PubMed] [Google Scholar]
- Cooke JE, Moens CB. Boundary formation in the hindbrain: Eph only it were simple. Trends Neurosci. 2002;25:260–267. doi: 10.1016/s0166-2236(02)02134-3. [DOI] [PubMed] [Google Scholar]
- Cordes SP. Molecular genetics of cranial nerve development in mouse. Nat Rev Neurosci. 2001;2:611–623. doi: 10.1038/35090039. [DOI] [PubMed] [Google Scholar]
- Couly GF, Grapin-Bottom A, Coltey P, Le Douarin NM. The regeneration of the cephalic neural crest, a problem revisited: the regenerating cells originate from the contralateral or from the anterior and posterior neural folds. Development. 1996;122:3393–3407. doi: 10.1242/dev.122.11.3393. [DOI] [PubMed] [Google Scholar]
- Dahmann C, Basler K. Compartment boundaries: at the edge of development. Trends Genet. 1999;15:320–326. doi: 10.1016/s0168-9525(99)01774-6. [DOI] [PubMed] [Google Scholar]
- Dahmann C, Oates AC, Brand M. Boundary formation and maintenance in tissue development. Nat Rev Genet. 2011;12:43–55. doi: 10.1038/nrg2902. [DOI] [PubMed] [Google Scholar]
- Darras S, Fritzenwanker JH, Uhlinger KR, Farrelly E, Pani AM, Hurley IA, Norris RP, Osovitz M, Terasaki M, Wu M, Aronowicz J, et al. Anteroposterior axis patterning by early canonical Wnt signaling during hemichordate development. PLoS Biol. 2018;16:e2003698. doi: 10.1371/journal.pbio.2003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenne M, Maconochie MK, Neun R, Pattyn A, Chambon P, Krumlauf R, Rijli FM. Hoxa2 and Hoxb2 control dorsoventral patterns of neuronal development in the rostral hindbrain. Neuron. 1999;22:677–691. doi: 10.1016/s0896-6273(00)80728-x. [DOI] [PubMed] [Google Scholar]
- De Kumar B, Parker HJ, Paulson A, Parrish ME, Zeitlinger J, Krumlauf R. Hoxa1 targets signaling pathways during neural differentiation of ES cells and mouse embryogenesis. Dev Biol. 2017;432:151–164. doi: 10.1016/j.ydbio.2017.09.033. [DOI] [PubMed] [Google Scholar]
- Di Bonito M, Narita Y, Avallone B, Sequino L, Mancuso M, Andolfi G, Franze AM, Puelles L, Rijli FM, Studer M. Assembly of the auditory circuitry by a Hox genetic network in the mouse brainstem. PLoS genetics. 2013;9:e1003249. doi: 10.1371/journal.pgen.1003249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esain V, Postlethwait JH, Charnay P, Ghislain J. FGF-receptor signalling controls neural cell diversity in the zebrafish hindbrain by regulating olig2 and sox9. Development. 2010;137:33–42. doi: 10.1242/dev.038026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagotto F, Rohani N, Touret AS, Li R. A molecular base for cell sorting at embryonic boundaries: contact inhibition of cadherin adhesion by ephrin/ Eph-dependent contractility. Dev Cell. 2013;27:72–87. doi: 10.1016/j.devcel.2013.09.004. [DOI] [PubMed] [Google Scholar]
- Fagotto F, Winklbauer R, Rohani N. Ephrin-Eph signaling in embryonic tissue separation. Cell adhesion & migration. 2014;8:308–326. doi: 10.4161/19336918.2014.970028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filas BA, Oltean A, Majidi S, Bayly PV, Beebe DC, Taber LA. Regional differences in actomyosin contraction shape the primary vesicles in the embryonic chicken brain. Phys Biol. 2012;9:066007. doi: 10.1088/1478-3975/9/6/066007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin G, Jungbluth S, Lumsden A, Champagnat J. Segmental specification of GABAergic inhibition during development of hindbrain neural networks. Nat Neurosci. 1999;2:873–877. doi: 10.1038/13172. [DOI] [PubMed] [Google Scholar]
- Fortin G, Kato F, Lumsden A, Champagnat J. Rhythm generation in the segmented hindbrain of chick embryos. J Physiol. 1995;486:735–744. doi: 10.1113/jphysiol.1995.sp020849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D, Sela-Donenfeld D. Hindbrain induction and patterning during early vertebrate development. Cellular and molecular life sciences : CMLS. 2019;76:941–960. doi: 10.1007/s00018-018-2974-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser S, Keynes R, Lumsden A. Segmentation in the chick embryo hindbrain is defined by cell lineage restrictions. Nature. 1990;344:431–435. doi: 10.1038/344431a0. [DOI] [PubMed] [Google Scholar]
- Gale NW, Holland SJ, Valenzuela DM, Flenniken A, Pan L, Ryan TE, Henkemeyer M, Strebhardt K, Hirai H, Wilkinson DG, Pawson T, et al. Eph receptors and ligands comprise two major specificity subclasses and are reciprocally compartmentalized during embryogenesis. Neuron. 1996;17:9–19. doi: 10.1016/s0896-6273(00)80276-7. [DOI] [PubMed] [Google Scholar]
- Gaufo GO, Thomas KR, Capecchi MR. Hox3 genes coordinate mechanisms of genetic suppression and activation in the generation of branchial and somatic motoneurons. Development. 2003;130:5191–5201. doi: 10.1242/dev.00730. [DOI] [PubMed] [Google Scholar]
- Gavalas A, Ruhrberg C, Livet J, Henderson CE, Krumlauf R. Neuronal defects in the hindbrain of Hoxa1, Hoxb1 and Hoxb2 mutants reflect regulatory interactions among these Hox genes. Development. 2003;130:5663–5679. doi: 10.1242/dev.00802. [DOI] [PubMed] [Google Scholar]
- Geisen MJ, Di Meglio T, Pasqualetti M, Ducret S, Brunet JF, Chedotal A, Rijli FM. Hox paralog group 2 genes control the migration of mouse pontine neurons through slit-robo signaling. PLoS Biol. 2008;6:e142. doi: 10.1371/journal.pbio.0060142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerety SS, Wilkinson DG. Morpholino artifacts provide pitfalls and reveal a novel role for pro-apoptotic genes in hindbrain boundary development. Developmental biology. 2011;350:279–289. doi: 10.1016/j.ydbio.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart J, Lowe C, Kirschner M. Hemichordates and the origin of chordates. Curr Opin Genet Dev. 2005;15:461–467. doi: 10.1016/j.gde.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Gilland E, Baker R. Conservation of neuroepithelial and mesodermal segments in the embryonic vertebrate head. Acta anatomica. 1993;148:110–123. doi: 10.1159/000147530. [DOI] [PubMed] [Google Scholar]
- Gilland E, Baker R. Evolutionary patterns of cranial nerve efferent nuclei in vertebrates. Brain Behav Evol. 2005;66:234–254. doi: 10.1159/000088128. [DOI] [PubMed] [Google Scholar]
- Giudicelli F, Taillebourg E, Charnay P, Gilardi-Hebenstreit P. Krox-20 patterns the hindbrain through both cell-autonomous and non cell-autonomous mechanisms. Genes Dev. 2001;15:567–580. doi: 10.1101/gad.189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godsave S, Dekker EJ, Holling T, Pannese M, Boncinelli E, Durston A. Expression patterns of Hoxb genes in the Xenopus embryo suggest roles in anteroposterior specification of the hindbrain and in dorsoventral patterning of the mesoderm. Dev Biol. 1994;166:465–476. doi: 10.1006/dbio.1994.1330. [DOI] [PubMed] [Google Scholar]
- Golding JP, Trainor P, Krumlauf R, Gassmann M. Defects in pathfinding by cranial neural crest cells in mice lacking the Neuregulin receptor ErbB4. Nature cell biology. 2000;2:103–109. doi: 10.1038/35000058. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Quevedo R, Lee Y, Poss KD, Wilkinson DG. Neuronal regulation of the spatial patterning of neurogenesis. Dev Cell. 2010;18:136–147. doi: 10.1016/j.devcel.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould A, Morrison A, Sproat G, White RA, Krumlauf R. Positive cross-regulation and enhancer sharing: two mechanisms for specifying overlapping Hox expression patterns. Genes Dev. 1997;11:900–913. doi: 10.1101/gad.11.7.900. [DOI] [PubMed] [Google Scholar]
- Graham A, Maden M, Krumlauf R. The murine Hox-2 genes display dynamic dorsoventral patterns of expression during central nervous system development. Development. 1991;112:255–264. doi: 10.1242/dev.112.1.255. [DOI] [PubMed] [Google Scholar]
- Green SA, Simoes-Costa M, Bronner ME. Evolution of vertebrates as viewed from the crest. Nature. 2015;520:474–482. doi: 10.1038/nature14436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie S, Butcher M, Lumsden A. Patterns of cell division and interkinetic nuclear migration in the chick embryo hindbrain. Journal of Neurobiology. 1991;22:742–754. doi: 10.1002/neu.480220709. [DOI] [PubMed] [Google Scholar]
- Guthrie S, Lumsden A. Formation and regeneration of rhombomere boundaries in the developing chick hindbrain. Development. 1991;112:221–229. doi: 10.1242/dev.112.1.221. [DOI] [PubMed] [Google Scholar]
- Guthrie S, Prince V, Lumsden A. Selective dispersal of avian rhombomere cells in orthotopic and heterotopic grafts. Development. 1993;118:527–538. doi: 10.1242/dev.118.2.527. [DOI] [PubMed] [Google Scholar]
- Gutzman JH, Sive H. Epithelial relaxation mediated by the myosin phosphatase regulator Mypt1 is required for brain ventricle lumen expansion and hindbrain morphogenesis. Development. 2010;137:795–804. doi: 10.1242/dev.042705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanneman E, Trevarrow B, Metcalfe WK, Kimmel CB, Westerfield M. Segmental pattern of development of the hindbrain and spinal cord of the zebrafish embryo. Development. 1988;103:49–58. doi: 10.1242/dev.103.1.49. [DOI] [PubMed] [Google Scholar]
- Hernandez RE, Putzke AP, Myers JP, Margaretha L, Moens CB. Cyp26 enzymes generate the retinoic acid response pattern necessary for hindbrain development. Development. 2007;134:177–187. doi: 10.1242/dev.02706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez RE, Rikhof HA, Bachmann R, Moens CB. vhnf1 integrates global RA patterning and local FGF signals to direct posterior hindbrain development in zebrafish. Development. 2004;131:4511–4520. doi: 10.1242/dev.01297. [DOI] [PubMed] [Google Scholar]
- Heyman I, Faissner A, Lumsden A. Cell and matrix specialisations of rhombomere boundaries. Dev Dyn. 1995;204:301–315. doi: 10.1002/aja.1002040308. [DOI] [PubMed] [Google Scholar]
- Hunt P, Gulisano M, Cook M, Sham MH, Faiella A, Wilkinson D, Boncinelli E, Krumlauf R. A distinct Hox code for the branchial region of the vertebrate head. Nature. 1991;353:861–864. doi: 10.1038/353861a0. [DOI] [PubMed] [Google Scholar]
- Irving C, Nieto MA, DasGupta R, Charnay P, Wilkinson DG. Progressive spatial restriction of Sek-1 and Krox-20 gene expression during hindbrain segmentation. Developmental biology. 1996;173:26–38. doi: 10.1006/dbio.1996.0004. [DOI] [PubMed] [Google Scholar]
- Jimenez-Guri E, Pujades C. An ancient mechanism of hindbrain patterning has been conserved in vertebrate evolution. Evol Dev. 2011;13:38–46. doi: 10.1111/j.1525-142X.2010.00454.x. [DOI] [PubMed] [Google Scholar]
- Jimenez-Guri E, Udina F, Colas JF, Sharpe J, Padron-Barthe L, Torres M, Pujades C. Clonal analysis in mice underlines the importance of rhombomeric boundaries in cell movement restriction during hindbrain segmentation. PLoS One. 2010;5:e10112. doi: 10.1371/journal.pone.0010112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania A, Klein R. Mechanisms of ephrin-Eph signalling in development, physiology and disease. Nat Rev Mol Cell Biol. 2016;17:240–256. doi: 10.1038/nrm.2015.16. [DOI] [PubMed] [Google Scholar]
- Kemp HA, Cooke JE, Moens CB. EphA4 and EfnB2a maintain rhombomere coherence by independently regulating intercalation of progenitor cells in the zebrafish neural keel. Dev Biol. 2009;327:313–326. doi: 10.1016/j.ydbio.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesavan G, Machate A, Hans S, Brand M. Cell-fate plasticity, adhesion and cell sorting complementarily establish a sharp midbrain-hindbrain boundary. Development. 2020;147 doi: 10.1242/dev.186882. [DOI] [PubMed] [Google Scholar]
- Kiecker C, Lumsden A. Compartments and their boundaries in vertebrate brain development. Nat Rev Neurosci. 2005;6:553–564. doi: 10.1038/nrn1702. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Sepich DS, Trevarrow B. Development of segmentation in zebrafish. Development. 1988;104(Suppl):197–207. doi: 10.1242/dev.104.Supplement.197. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Warga RM, Kane DA. Cell cycles and clonal strings during formation of the zebrafish central nervous system. Development. 1994;120:265–276. doi: 10.1242/dev.120.2.265. [DOI] [PubMed] [Google Scholar]
- Knecht AK, Bronner-Fraser M. Induction of the neural crest: a multigene process. Nat Rev Genet. 2002;3:453–461. doi: 10.1038/nrg819. [DOI] [PubMed] [Google Scholar]
- Kolm PJ, Sive HL. Regulation of the Xenopus labial homeodomain genes, HoxA1 and HoxD1- activation by retinoids and peptide growth factors. Dev Biol. 1995;167:34–49. doi: 10.1006/dbio.1995.1005. [DOI] [PubMed] [Google Scholar]
- Kontges G, Lumsden A. Rhombencephalic neural crest segmentation is preserved throughout craniofacial ontogeny. Development. 1996;122:3229–3242. doi: 10.1242/dev.122.10.3229. [DOI] [PubMed] [Google Scholar]
- Labalette C, Wassef MA, Desmarquet-Trin Dinh C, Bouchoucha YX, Le Men J, Charnay P, Gilardi-Hebenstreit P. Molecular dissection of segment formation in the developing hindbrain. Development. 2015;142:185–195. doi: 10.1242/dev.109652. [DOI] [PubMed] [Google Scholar]
- Laussu J, Khuong A, Gautrais J, Davy A. Beyond boundaries-Eph:ephrin signaling in neurogenesis. Cell adhesion & migration. 2014;8:349–359. doi: 10.4161/19336918.2014.969990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin N, Kalcheim C. The Neural Crest, 2nd ed. Cambridge University Press; Cambridge, UK New York, NY, USA: 1999. [Google Scholar]
- Letelier J, Terriente J, Belzunce I, Voltes A, Undurraga CA, Polvillo R, Devos L, Tena JJ, Maeso I, Retaux S, Gomez-Skarmeta JL, et al. Evolutionary emergence of the rac3b/rfng/sgca regulatory cluster refined mechanisms for hindbrain boundaries formation. Proc Natl Acad Sci U S A. 2018;115:E3731–E3740. doi: 10.1073/pnas.1719885115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe CJ, Clarke DN, Medeiros DM, Rokhsar DS, Gerhart J. The deuterostome context of chordate origins. Nature. 2015;520:456–465. doi: 10.1038/nature14434. [DOI] [PubMed] [Google Scholar]
- Lowe CJ, Wu M, Salic A, Evans L, Lander E, Stange-Thomann N, Gruber CE, Gerhart J, Kirschner M. Anteroposterior patterning in hemichordates and the origins of the chordate nervous system. Cell. 2003;113:853–865. doi: 10.1016/s0092-8674(03)00469-0. [DOI] [PubMed] [Google Scholar]
- Lumsden A. Segmentation and compartition in the early avian hindbrain. Mechanisms of development. 2004;121:1081–1088. doi: 10.1016/j.mod.2004.04.018. [DOI] [PubMed] [Google Scholar]
- Lumsden A, Keynes R. Segmental patterns of neuronal development in the chick hindbrain. Nature. 1989;337:424–428. doi: 10.1038/337424a0. [DOI] [PubMed] [Google Scholar]
- Lumsden A, Krumlauf R. Patterning the vertebrate neuraxis. Science. 1996;274:1109–1115. doi: 10.1126/science.274.5290.1109. [DOI] [PubMed] [Google Scholar]
- Mahmood R, Kiefer P, Guthrie S, Dickson C, Mason I. Multiple roles for FGF-3 during cranial neural development in the chicken. Development. 1995;121:1399–1410. doi: 10.1242/dev.121.5.1399. [DOI] [PubMed] [Google Scholar]
- Manzanares M, Wada H, Itasaki N, Trainor PA, Krumlauf R, Holland PW. Conservation and elaboration of Hox gene regulation during evolution of the vertebrate head. Nature. 2000;408:854–857. doi: 10.1038/35048570. [DOI] [PubMed] [Google Scholar]
- Marin F, Charnay P. Hindbrain patterning: FGFs regulate Krox20 and mafB/kr expression in the otic/preotic region. Development. 2000;127:4925–4935. doi: 10.1242/dev.127.22.4925. [DOI] [PubMed] [Google Scholar]
- Maves L, Jackman W, Kimmel CB. FGF3 and FGF8 mediate a rhombomere 4 signaling activity in the zebrafish hindbrain. Development. 2002;129:3825–3837. doi: 10.1242/dev.129.16.3825. [DOI] [PubMed] [Google Scholar]
- McClintock JM, Kheirbek MA, Prince VE. Knockdown of duplicated zebrafish hoxb1 genes reveals distinct roles in hindbrain patterning and a novel mechanism of duplicate gene retention. Development. 2002;129:2339–2354. doi: 10.1242/dev.129.10.2339. [DOI] [PubMed] [Google Scholar]
- Minoux M, Rijli FM. Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development. Development. 2010;137:2605–2621. doi: 10.1242/dev.040048. [DOI] [PubMed] [Google Scholar]
- Moens CB, Prince VE. Constructing the hindbrain: Insights from the zebrafish. Dev Dyn. 2002;224:1–17. doi: 10.1002/dvdy.10086. [DOI] [PubMed] [Google Scholar]
- Murphy P, Davidson DR, Hill RE. Segment-specific expression of a homeobox-containing gene in the mouse hindbrain. Nature. 1989;341:156–159. doi: 10.1038/341156a0. [DOI] [PubMed] [Google Scholar]
- Murphy P, Hill RE. Expression of the mouse labial-like homeobox-containing genes, Hox 2.9 and Hox 1.6, during segmentation of the hindbrain. Development. 1991;111:61–74. doi: 10.1242/dev.111.1.61. [DOI] [PubMed] [Google Scholar]
- Natale A, Sims C, Chiusano ML, Amoroso A, D’Aniello E, Fucci L, Krumlauf R, Branno M, Locascio A. Evolution of anterior Hox regulatory elements among chordates. BMC evolutionary biology. 2011;11:330. doi: 10.1186/1471-2148-11-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neijts R, Amin S, van Rooijen C, Deschamps J. Cdx is crucial for the timing mechanism driving colinear Hox activation and defines a trunk segment in the Hox cluster topology. Dev Biol. 2017;422:146–154. doi: 10.1016/j.ydbio.2016.12.024. [DOI] [PubMed] [Google Scholar]
- Neijts R, Amin S, van Rooijen C, Tan S, Creyghton MP, de Laat W, Deschamps J. Polarized regulatory landscape and Wnt responsiveness underlie Hox activation in embryos. Genes Dev. 2016;30:1937–1942. doi: 10.1101/gad.285767.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neijts R, Deschamps J. At the base of colinear Hox gene expression: cis-features and trans-factors orchestrating the initial phase of Hox cluster activation. Dev Biol. 2017;428:293–299. doi: 10.1016/j.ydbio.2017.02.009. [DOI] [PubMed] [Google Scholar]
- Nieto MA, Bradley LC, Wilkinson DG. Conserved segmental expression of Krox-20 in the vertebrate hindbrain and its relationship to lineage restriction. Development Supplement. 1991;2:59–62. [PubMed] [Google Scholar]
- Nieto MA, Gilardi-Hebenstreit P, Charnay P, Wilkinson DG. A receptor protein tyrosine kinase implicated in the segmental patterning of the hindbrain and mesoderm. Development. 1992;116:1137–1150. doi: 10.1242/dev.116.4.1137. [DOI] [PubMed] [Google Scholar]
- Nolte C, Jinks T, Wang X, Martinez Pastor MT, Krumlauf R. Shadow enhancers flanking the HoxB cluster direct dynamic Hox expression in early heart and endoderm development. Dev Biol. 2013;383:158–173. doi: 10.1016/j.ydbio.2013.09.016. [DOI] [PubMed] [Google Scholar]
- O’Malley DM, Kao YH, Fetcho JR. Imaging the functional organization of zebrafish hindbrain segments during escape behaviors. Neuron. 1996;17:1145–1155. doi: 10.1016/s0896-6273(00)80246-9. [DOI] [PubMed] [Google Scholar]
- O’Neill AK, Kindberg AA, Niethamer TK, Larson AR, Ho HYH, Greenberg ME, Bush JO. Unidirectional Eph/ephrin signaling creates a cortical actomyosin differential to drive cell segregation. J Cell Biol. 2016;215:217–229. doi: 10.1083/jcb.201604097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterveen T, Niederreither K, Dolle P, Chambon P, Meijlink F, Deschamps J. Retinoids regulate the anterior expression boundaries of 5’ Hoxb genes in posterior hindbrain. EMBO J. 2003;22:262–269. doi: 10.1093/emboj/cdg029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pani AM, Mullarkey EE, Aronowicz J, Assimacopoulos S, Grove EA, Lowe CJ. Ancient deuterostome origins of vertebrate brain signalling centres. Nature. 2012;483:289–294. doi: 10.1038/nature10838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker HJ, Bronner ME, Krumlauf R. A Hox regulatory network of hindbrain segmentation is conserved to the base of vertebrates. Nature. 2014;514:490–493. doi: 10.1038/nature13723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker HJ, Bronner ME, Krumlauf R. The vertebrate Hox gene regulatory network for hindbrain segmentation: Evolution and diversification: Coupling of a Hox gene regulatory network to hindbrain segmentation is an ancient trait originating at the base of vertebrates. BioEssays : news and reviews in molecular, cellular and developmental biology. 2016;38:526–538. doi: 10.1002/bies.201600010. [DOI] [PubMed] [Google Scholar]
- Parker HJ, Bronner ME, Krumlauf R. An atlas of anterior hox gene expression in the embryonic sea lamprey head: Hox-code evolution in vertebrates. Dev Biol. 2019a;453:19–33. doi: 10.1016/j.ydbio.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker HJ, De Kumar B, Green SA, Prummel KD, Hess C, Kaufman CK, Mosimann C, Wiedemann LM, Bronner ME, Krumlauf R. A Hox-TALE regulatory circuit for neural crest patterning is conserved across vertebrates. Nature communications. 2019b;10:1189. doi: 10.1038/s41467-019-09197-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker HJ, Krumlauf R. Segmental arithmetic: summing up the Hox gene regulatory network for hindbrain development in chordates. Wiley interdisciplinary reviews. Developmental biology. 2017;6 doi: 10.1002/wdev.286. [DOI] [PubMed] [Google Scholar]
- Parker HJ, Krumlauf R. A Hox gene regulatory network for hindbrain segmentation. Curr Top Dev Biol. 2020;139:169–203. doi: 10.1016/bs.ctdb.2020.03.001. [DOI] [PubMed] [Google Scholar]
- Pasini A, Wilkinson DG. Stabilizing the regionalisation of the developing vertebrate central nervous system. BioEssays : news and reviews in molecular, cellular and developmental biology. 2002;24:427–438. doi: 10.1002/bies.10085. [DOI] [PubMed] [Google Scholar]
- Pasquale EB. Eph receptor signalling casts a wide net on cell behaviour. Nat Rev Mol Cell Biol. 2005;6:462–475. doi: 10.1038/nrm1662. [DOI] [PubMed] [Google Scholar]
- Pasquale EB. Eph-ephrin bidirectional signaling in physiology and disease. Cell. 2008;133:38–52. doi: 10.1016/j.cell.2008.03.011. [DOI] [PubMed] [Google Scholar]
- Pasqualetti M, Diaz C, Renaud JS, Rijli FM, Glover JC. Fate-mapping the mammalian hindbrain: segmental origins of vestibular projection neurons assessed using rhombomere-specific Hoxa2 enhancer elements in the mouse embryo. J Neurosci. 2007;27:9670–9681. doi: 10.1523/JNEUROSCI.2189-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattyn A, Vallstedt A, Dias JM, Samad OA, Krumlauf R, Rijli FM, Brunet JF, Ericson J. Coordinated temporal and spatial control of motor neuron and serotonergic neuron generation from a common pool of CNS progenitors. Genes and Development. 2003;17:729–737. doi: 10.1101/gad.255803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peretz Y, Eren N, Kohl A, Hen G, Yaniv K, Weisinger K, Cinnamon Y, Sela-Donenfeld D. A new role of hindbrain boundaries as pools of neural stem/progenitor cells regulated by Sox2. Bmc Biol. 2016;14:57. doi: 10.1186/s12915-12016-10277-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippidou P, Dasen JS. Hox genes: choreographers in neural development, architects of circuit organization. Neuron. 2013;80:12–34. doi: 10.1016/j.neuron.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prin F, Serpente P, Itasaki N, Gould AP. Hox proteins drive cell segregation and non-autonomous apical remodelling during hindbrain segmentation. Development. 2014;141:1492–1502. doi: 10.1242/dev.098954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince VE, Moens CB, Kimmel CB, Ho RK. Zebrafish hox genes: expression in the hindbrain region of wild-type and mutants of the segmentation gene, valentino . Development. 1998;125:393–406. doi: 10.1242/dev.125.3.393. [DOI] [PubMed] [Google Scholar]
- Pujades C. The multiple functions of hindbrain boundary cells: Tinkering boundaries? Seminars in cell & developmental biology. 2020;107:179–189. doi: 10.1016/j.semcdb.2020.05.002. [DOI] [PubMed] [Google Scholar]
- Qian P, De Kumar B, He XC, Nolte C, Gogol M, Ahn Y, Chen S, Li Z, Xu H, Perry JM, Hu D, et al. Retinoid-Sensitive Epigenetic Regulation of the Hoxb Cluster Maintains Normal Hematopoiesis and Inhibits Leukemogenesis. Cell stem cell. 2018;22:740–754.:e747. doi: 10.1016/j.stem.2018.04.012. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Fung L, Schilling TF, Nie Q. Multiple morphogens and rapid elongation promote segmental patterning during development. bioRxiv. 2021:2021.04.22.440966. doi: 10.1371/journal.pcbi.1009077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhinn M, Brand M. The midbrain--hindbrain boundary organizer. Curr Opin Neurobiol. 2001;11:34–42. doi: 10.1016/s0959-4388(00)00171-9. [DOI] [PubMed] [Google Scholar]
- Riley BB, Chiang MY, Storch EM, Heck R, Buckles GR, Lekven AC. Rhombomere boundaries are Wnt signaling centers that regulate metameric patterning in the zebrafish hindbrain. Dev Dyn. 2004;231:278–291. doi: 10.1002/dvdy.20133. [DOI] [PubMed] [Google Scholar]
- Rohani N, Canty L, Luu O, Fagotto F, Winklbauer R. EphrinB/EphB Signaling Controls Embryonic Germ Layer Separation by Contact-Induced Cell Detachment. PLoS biology. 2011;9:e1000597. doi: 10.1371/journal.pbio.1000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohani N, Parmeggiani A, Winklbauer R, Fagotto F. Variable combinations of specific ephrin ligand/eph receptor pairs control embryonic tissue separation. PLoS biology. 2014;12:e1001955. doi: 10.1371/journal.pbio.1001955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydeen A, Voisin N, D’Aniello E, Ravisankar P, Devignes CS, Waxman JS. Excessive feedback of Cyp26a1 promotes cell non-autonomous loss of retinoic acid signaling. Developmental biology. 2015;405:47–55. doi: 10.1016/j.ydbio.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydeen AB, Waxman JS. Cyp26 enzymes are required to balance the cardiac and vascular lineages within the anterior lateral plate mesoderm. Development. 2014;141:1638–1648. doi: 10.1242/dev.105874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagati F, Minoux M, Ren SY, Rijli FM. Temporal requirement of Hoxa2 in cranial neural crest skeletal morphogenesis. Development. 2005;132:4927–4936. doi: 10.1242/dev.02078. [DOI] [PubMed] [Google Scholar]
- Santagati F, Rijli FM. Cranial neural crest and the building of the vertebrate head. Nat Rev Neurosci. 2003;4:806–818. doi: 10.1038/nrn1221. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Meulemans D, Jones M, Bronner-Fraser M. Ancient evolutionary origin of the neural crest gene regulatory network. Developmental cell. 2007;13:405–420. doi: 10.1016/j.devcel.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Schilling TF, Nie Q, Lander AD. Dynamics and precision in retinoic acid morphogen gradients. Curr Opin Genet Dev. 2012;22:562–569. doi: 10.1016/j.gde.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling TF, Prince V, Ingham PW. Plasticity in zebrafish hox expression in the hindbrain and cranial neural crest. Developmental biology. 2001;231:201–216. doi: 10.1006/dbio.2000.9997. [DOI] [PubMed] [Google Scholar]
- Schneider-Maunoury S, Topilko P, Seitandou T, Levi G, Cohen-Tannoudji M, Pournin S, Babinet C, Charnay P. Disruption of Krox-20 results in alteration of rhombomeres 3 and 5 in the developing hindbrain. Cell. 1993;75:1199–1214. doi: 10.1016/0092-8674(93)90329-o. [DOI] [PubMed] [Google Scholar]
- Sela-Donenfeld D, Kayam G, Wilkinson DG. Boundary cells regulate a switch in the expression of FGF3 in hindbrain rhombomeres. BMC Dev Biol. 2009;9:16. doi: 10.1186/1471-213X-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe J, Nonchev S, Gould A, Whiting J, Krumlauf R. Selectivity, sharing and competitive interactions in the regulation of Hoxb genes. EMBO J. 1998;17:1788–1798. doi: 10.1093/emboj/17.6.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimeld SM, Donoghue PC. Evolutionary crossroads in developmental biology: cyclostomes (lamprey and hagfish) Development. 2012;139:2091–2099. doi: 10.1242/dev.074716. [DOI] [PubMed] [Google Scholar]
- Singh NP, De Kumar B, Paulson A, Parrish ME, Scott C, Zhang Y, Florens L, Krumlauf R. Genome-Wide Binding Analyses of HOXB1 Revealed a Novel DNA Binding Motif Associated with Gene Repression. Journal of developmental biology. 2021;9 doi: 10.3390/jdb9010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NP, De Kumar B, Paulson A, Parrish ME, Zhang Y, Florens L, Conaway JW, Si K, Krumlauf R. A six-amino-acid motif is a major determinant in functional evolution of HOX1 proteins. Genes Dev. 2020;34:1680–1696. doi: 10.1101/gad.342329.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirbu IO, Gresh L, Barra J, Duester G. Shifting boundaries of retinoic acid activity control hindbrain segmental gene expression. Development. 2005;132:2611–2622. doi: 10.1242/dev.01845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Timoshevskaya N, Ye C, Holt C, Keinath MC, Parker HJ, Cook ME, Hess JE, Narum SR, Lamanna F, Kaessmann H, et al. The sea lamprey germline genome provides insights into programmed genome rearrangement and vertebrate evolution. Nat Genet. 2018;50:270–277. doi: 10.1038/s41588-017-0036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobieszczuk DF, Poliakov A, Xu Q, Wilkinson DG. A feedback loop mediated by degradation of an inhibitor is required to initiate neuronal differentiation. Genes & development. 2010;24:206–218. doi: 10.1101/gad.554510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solanas G, Cortina C, Sevillano M, Batlle E. Cleavage of E-cadherin by ADAM10 mediates epithelial cell sorting downstream of EphB signalling. Nature cell biology. 2011;13:1100–1107. doi: 10.1038/ncb2298. [DOI] [PubMed] [Google Scholar]
- Sosnik J, Zheng L, Rackauckas CV, Digman M, Gratton E, Nie Q, Schilling TF. Noise modulation in retinoic acid signaling sharpens segmental boundaries of gene expression in the embryonic zebrafish hindbrain. eLife. 2016;5:e14034. doi: 10.7554/eLife.14034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer M, Gavalas A, Marshall H, Ariza-McNaughton L, Rijli FM, Chambon P, Krumlauf R. Genetic interactions between Hoxa1 and Hoxb1 reveal new roles in regulation of early hindbrain patterning. Development. 1998;125:1025–1036. doi: 10.1242/dev.125.6.1025. [DOI] [PubMed] [Google Scholar]
- Studer M, Pöpperl H, Marshall H, Kuroiwa A, Krumlauf R. Role of a conserved retinoic acid response element in rhombomere restriction of Hoxb-1. Science. 1994;265:1728–1732. doi: 10.1126/science.7916164. [DOI] [PubMed] [Google Scholar]
- Sueda R, Imayoshi I, Harima Y, Kageyama R. High Hes1 expression and resultant Ascl1 suppression regulate quiescent vs. active neural stem cells in the adult mouse brain. Genes & development. 2019;33:511–523. doi: 10.1101/gad.323196.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Hopkins N. vhnf1, the MODY5 and familial GCKD-associated gene, regulates regional specification of the zebrafish gut, pronephros, and hindbrain. Genes Dev. 2001;15:3217–3229. doi: 10.1101/gad946701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundin OH, Eichele G. A homeo domain protein reveals the metameric nature of the developing chick hindbrain. Genes Dev. 1990;4:1267–1276. doi: 10.1101/gad.4.8.1267. [DOI] [PubMed] [Google Scholar]
- Tambalo M, Mitter R, Wilkinson DG. A single cell transcriptome atlas of the developing zebrafish hindbrain. Development. 2020;147 doi: 10.1242/dev.184143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor HB, Khuong A, Wu Z, Xu Q, Morley R, Gregory L, Poliakov A, Taylor WR, Wilkinson DG. Cell segregation and border sharpening by Eph receptor-ephrin-mediated heterotypic repulsion. J R Soc Interface. 2017;14:20170338. doi: 10.1098/rsif.2017.0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terriente J, Gerety SS, Watanabe-Asaka T, Gonzalez-Quevedo R, Wilkinson DG. Signalling from hindbrain boundaries regulates neuronal clustering that patterns neurogenesis. Development. 2012;139:2978–2987. doi: 10.1242/dev.080135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theil T, Frain M, Gilardi-Hebenstreit P, Flenniken A, Charnay P, Wilkinson DG. Segmental expression of the EphA4 (Sek-1) receptor tyrosine kinase in the hindbrain is under direct transcriptional control of Krox-20. Development. 1998;125:443–452. doi: 10.1242/dev.125.3.443. [DOI] [PubMed] [Google Scholar]
- Trainor P, Krumlauf R. Plasticity in mouse neural crest cells reveals a new patterning role for cranial mesoderm. Nat Cell Biol. 2000a;2:96–102. doi: 10.1038/35000051. [DOI] [PubMed] [Google Scholar]
- Trainor PA, Bronner-Fraser M, Krumlauf R. In: Handbook of Stem Cells. Lanza RP, editor. Elsevier Academic; Boston, MA: 2004. Neural Crest Cells; pp. 219–232. [Google Scholar]
- Trainor PA, Krumlauf R. Patterning the cranial neural crest: Hindbrain segmentation and Hox gene plasticity. Nature reviews Neuroscience. 2000b;1:116–124. doi: 10.1038/35039056. [DOI] [PubMed] [Google Scholar]
- Trainor PA, Sobieszczuk D, Wilkinson D, Krumlauf R. Signalling between the hindbrain and paraxial tissues dictates neural crest migration pathways. Development. 2002;129:433–442. doi: 10.1242/dev.129.2.433. [DOI] [PubMed] [Google Scholar]
- Tumpel S, Wiedemann LM, Krumlauf R. Hox genes and segmentation of the vertebrate hindbrain. Curr Top Dev Biol. 2009;88:103–137. doi: 10.1016/S0070-2153(09)88004-6. [DOI] [PubMed] [Google Scholar]
- Tvrdik P, Capecchi MR. Reversal of hox1 gene subfunctionalization in the mouse. Dev Cell. 2006;11:239–250. doi: 10.1016/j.devcel.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Voltes A, Hevia CF, Engel-Pizcueta C, Dingare C, Calzolari S, Terriente J, Norden C, Lecaudey V, Pujades C. Yap/Taz-TEAD activity links mechanical cues to progenitor cell behavior during zebrafish hindbrain segmentation. Development. 2019;146 doi: 10.1242/dev.176735. [DOI] [PubMed] [Google Scholar]
- Wada H, Escriva H, Zhang S, Laudet V. Conserved RARE localization in amphioxus Hox clusters and implications for Hox code evolution in the vertebrate neural crest. Dev Dyn. 2006;235:1522–1531. doi: 10.1002/dvdy.20730. [DOI] [PubMed] [Google Scholar]
- Walshe J, Maroon H, McGonnell IM, Dickson C, Mason I. Establishment of hindbrain segmental identity requires signaling by FGF3 and FGF8. Curr Biol. 2002;12:1117–1123. doi: 10.1016/s0960-9822(02)00899-0. [DOI] [PubMed] [Google Scholar]
- Wang QX, Holmes WR, Sosnik J, Schilling T, Nie Q. Cell sorting and noise-induced cell plasticity coordinate to sharpen boundaries between gene expression domains. Pios Comput Biol. 2017;13:e1005307. doi: 10.1371/journal.pcbi.1005307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisinger K, Kohl A, Kayam G, Monsonego-Ornan E, Sela-Donenfeld D. Expression of hindbrain boundary markers is regulated by FGF3. Biology Open. 2011;1:67–74. doi: 10.1242/bio.2011032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisinger K, Wilkinson DG, Sela-Donenfeld D. Inhibition of BMPs by follistatin is required for FGF3 expression and segmental patterning of the hindbrain. Developmental biology. 2008;324:213–225. doi: 10.1016/j.ydbio.2008.09.005. [DOI] [PubMed] [Google Scholar]
- White RJ, Nie Q, Lander AD, Schilling TF. Complex regulation of cyp26a1 creates a robust retinoic acid gradient in the zebrafish embryo. PLoS biology. 2007a;5:e304. doi: 10.1371/journal.pbio.0050304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RJ, Nie Q, Lander AD, Schilling TF. Complex regulation of cyp26al creates a robust retinoic acid gradient in the zebrafish embryo. PLoS Biol. 2007b;5:e304. doi: 10.1371/journal.pbio.0050304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RJ, Schilling TF. How degrading: Cyp26s in hindbrain development. Dev Dyn. 2008;237:2775–2790. doi: 10.1002/dvdy.21695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiellette EL, Sive H. vhnfl and Fgf signals synergize to specify rhombomere identity in the zebrafish hindbrain. Development. 2003;130:3821–3829. doi: 10.1242/dev.00572. [DOI] [PubMed] [Google Scholar]
- Wiellette EL, Sive H. Early requirement for fgf8 function during hindbrain pattern formation in zebrafish. Dev Dyn. 2004;229:393–399. doi: 10.1002/dvdy.10464. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG. Regulation of cell differentiation by Eph receptor and ephrin signaling. Cell Adhesion Migration. 2014;8:339–348. doi: 10.4161/19336918.2014.970007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DG, Bhatt S, Chavrier P, Bravo R, Charnay P. Segment-specific expression of a zinc-finger gene in the developing nervous system of the mouse. Nature. 1989a;337:461–464. doi: 10.1038/337461a0. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG, Bhatt S, Cook M, Boncinelli E, Krumlauf R. Segmental expression of Hox-2 homeobox-containing genes in the developing mouse hindbrain. Nature. 1989b;341:405–409. doi: 10.1038/341405a0. [DOI] [PubMed] [Google Scholar]
- Wizenmann A, Lumsden A. Segregation of rhombomeres by differential chemoaffinity. Mol Cell Neurosci. 1997;9:448–459. doi: 10.1006/mcne.1997.0642. [DOI] [PubMed] [Google Scholar]
- Wu Z, Ashlin TG, Xu Q, Wilkinson DG. Role of forward and reverse signaling in Eph receptor and ephrin mediated cell segregation. Exp Cell Res. 2019;381:57–65. doi: 10.1016/j.yexcr.2019.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Alldus G, Holder N, Wilkinson DG. Expression of truncated Sek-1 receptor tyrosine kinase disrupts the segmental restriction of gene expression in the Xenopus and zebrafish hindbrain. Development. 1995;121:4005–4016. doi: 10.1242/dev.121.12.4005. [DOI] [PubMed] [Google Scholar]
- Xu Q, Mellitzer G, Robinson V, Wilkinson DG. In vivo cell sorting in complementary segmental domains mediated by Eph receptors and ephrins. Nature. 1999;399:267–271. doi: 10.1038/20452. [DOI] [PubMed] [Google Scholar]
- Zhang L, Radtke K, Zheng L, Cai AQ, Schilling TF, Nie Q. Noise drives sharpening of gene expression boundaries in the zebrafish hindbrain. Mol Syst Biol. 2012;8:613. doi: 10.1038/msb.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]