Introductory paragraph
Macrophages exhibit a spectrum of activation states ranging from classical to alternative activation1. Alternatively, activated macrophages are involved in diverse pathophysiological processes such as confining tissue parasites2, improving insulin sensitivity3 or promoting an immune tolerant microenvironment that facilitates tumour growth and metastasis4. Recently, the role of metabolism regulating macrophage function has come into focus as both the classical and alternative activation programmes require specific regulated metabolic reprogramming5. While most of the studies regarding immunometabolism have focussed on the catabolic pathways activated to provide energy, little is known about the anabolic pathways mediating macrophage alternative activation. In this study, we show that the anabolic transcription factor sterol regulatory element binding protein 1 (SREBP1) is activated in response to the canonical Th2 cytokine interleukin 4 (IL-4) to trigger the de novo lipogenesis (DNL) programme, as a necessary step for macrophage alternative activation. Mechanistically, DNL consumes NADPH, partitioning it away from cellular antioxidant defences and raising ROS levels. ROS serves as a second messenger, signalling sufficient DNL, and promoting macrophage alternative activation. The pathophysiological relevance of this mechanism is validated by showing that SREBP1/DNL is essential for macrophage alternative activation in vivo in a helminth infection model.
We analysed RNAseq from human macrophages (GSE117040)6 and identified SREBP1 as the most upregulated transcription factor in M(IL-4) compared to M(LPS) macrophages (Fig. 1a). In murine bone marrow-derived macrophages (BMMФ), SREBP1 was among the most activated transcription factors in response to IL-4 (Fig. 1b). We confirmed SREBP1 activation based on levels of the cleaved active protein (Fig. 1c), and the upregulation of SREBP target genes upon IL-4 stimulation (Fig. 1d).
Figure 1. SREBP1 activation is a feature of alternatively activated macrophages.
a, Transcriptional regulator analysis of RNA sequencing comparing LPS and IL-4 treated human macrophages (GSE117040). Transcriptional regulators enriched in LPS or IL-4 are represented in red or blue, respectively.
b, Transcriptional regulator analysis of RNA sequencing comparing control (CTR) and IL-4 treated bone marrow-derived macrophages (BMMФ). Repressed and activated transcriptional regulators in M(IL-4) BMMФ are respectively presented in red and blue. Data of n=6 biological replicates from 2 independent experiments per group.
c, Protein expression of SREBP1 in BMMФ in response to IL-4. The upper band represents full-length (FL) SREBP1 (~125kDa, immature) and the lower band cleaved SREBP1 (~68kDa, mature). Western-blot representative picture of n=3 biological replicates. ß-actin has been used as loading control for the western blot analysis. Quantification of the ratio between cleaved SREBP1 and ß-actin is presented in the panel below as mean ± SEM.
d, Gene expression profile of SREBP target genes of as defined by Horton et al. (22) from RNA sequencing comparing CTR and IL-4-treated BMMФ of n=6 biological replicates per group. Repressed and activated genes in M(IL-4) BMMФ are respectively presented in red and blue.
e, mRNA expression over the best keeper (BK) of lipopolysaccharide (LPS,100ng/mL, 24h), Dexamethasone (DEX, 100nM, 24h) or IL-4 (10ng/mL, 24h) treated BMMФ of n=4 biological replicates.
f, SREBP1 target gene induction in response to IL-4 of LysM+/+ SCAPflox/flox (WT) and LysMCre/+ SCAPflox/flox (SCAP-KO) BMMФ. mRNA expression over BK of n=4 (Scap) or n=8 biological replicates from 2 independent experiments.
g, Mice were injected with IL-4c or PBS on day 0. After 2 days, the peritoneal cavity was washed, and the peritoneal exudate cells were sorted by magnetic associated cell sorting (MACS) using F4/80 positive selection.
h, Gene expression of SREBP1 target genes in macrophages harvested from 4 mice per group injected with IL-4c or PBS. Data of the mRNA expression over BK presented as mean ± SEM of n=4 mice per group.
Statistical analysis of the RNAseq data is detailed in the methods section. Otherwise, data has been analysed by a one-way ANOVA followed by a Dunnett’s multiple comparisons test (Fig. 1c) or a two-tailed Student’s t-test (Fig. 1h).
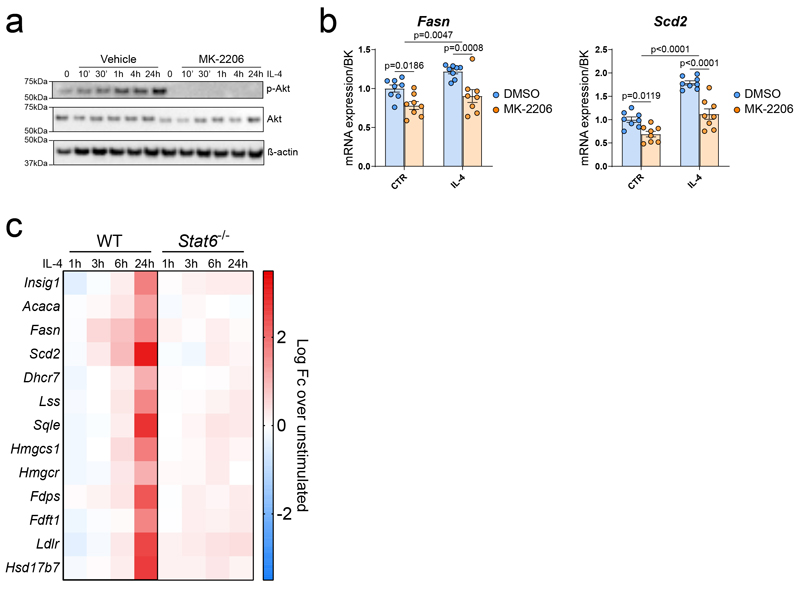
Downstream of IL-4, inhibition of AKT phosphorylation reduced the induction of SREBP1 target genes (Extended data Fig.1a and b). Using a publicly available dataset (GSE106706)7, we observed that induction of SREBP1 target is also Stat6-dependent (Extended data Fig. 1c). Altogether, these data demonstrate that SREBP1 activation in alternatively activated macrophages requires both major IL-4 signalling pathways7,8.
SIRT1 and AMPK reduce SREBP1 activity in other models9,10. While not affecting Fasn expression, SIRT1 activation (SIRT1 activator II) slightly increased the expression of Scd2, without affecting M(IL-4) activation (Extended data Fig. 2a). SIRT1 inhibition (EX-527) did not affect the expression of the SREBP1 target genes Fasn and Scd2, but reduced M(IL-4) activation (Extended data Fig. 2b), likely in a SREBP1-independent fashion. Inhibition of AMPK activation using Compound C had no effect on Fasn and Scd2 expression but induced the expression of the alternative activation marker Retnla while reducing Mgl2 expression (Extended data Fig. 2c). AMPK activation, using AICAR, only reduced the expression of the SREBP1 target genes Fasn and Scd2 at high concentration. As previously described11,12, AICAR reduced M(IL-4) activation (Extended data Fig. 2d). Altogether, these data show that AMPK and SIRT1 do not regulate SREBP1 activation in M(IL-4) cells.
To address the specificity of IL-4 activating SREBP1, we stimulated BMMФ with two other macrophage polarising agents, LPS and dexamethasone1. Both LPS and dexamethasone failed to upregulate SREBP1 target genes (Fig. 1e), demonstrating the specificity of IL-4 to activate SREBP1 in macrophages. To confirm that the induction of the SREBP target genes required SREBP activation, we used LysMCre/+ x SREBP cleavage-activating protein (SCAP)fl/fl (MФ-SCAP-KO) mice and cells to disable SREBP genetically13. As hypothesised, the induction of SREBP1 target genes in response to IL-4 was reduced in SCAP-KO macrophages (Fig. 1f). Furthermore, pharmacological inhibition of SREBP activation (using 25-hydroxycholesterol (25HC), an inhibitor of SCAP transport14), or deletion of SREBP1c, blocked IL-4-induced expression of Fasn and Scd1 (Extended data Fig. 2e and f). Next, we confirmed IL-4 activated SREBP in vivo using IL-4 complex (IL-4c) (Fig. 1g), which increased the expression of SREBP1 target genes (Fig. 1h) in peritoneal macrophages. Altogether, these data demonstrate that IL-4 is an activator of SREBP1 in macrophages.
While IL-4 strongly activated SREBP1, whether SREBP1 was required for alternative macrophage activation remained unknown. When stimulated with IL-4, BMMФs lacking SCAP had impaired alternative activation compared to WT (Fig. 2a and b and Extended data 3a), however the inhibition of pro-inflammatory markers was not affected (Extended data Fig. 3b), indicating that the phenotype was not secondary to a failure to inhibit classical activation. Gene enrichment analysis confirmed the inhibition of alternative activation in SCAP-KO BMMФ compared to controls (Fig. 2c). The dependency of M(IL-4) polarisation on SREBP activation was further confirmed pharmacologically using 25HC (Extended data Fig. 3c-e). Both SREBP1 and 2 are inhibited by 25HC and by SCAP deletion13, so we next determined the importance of the isoforms. Deletion of SREBP1c was sufficient to prevent IL-4 induced alternative activation (Extended data Fig. 3f). We further confirmed that SREBP1 but not SREBP2 was required for macrophage alternative activation in RAW264.7 cells (Extended data Fig. 3g).
Figure 2. Inhibition of SREBP1 activation impairs macrophages alternative activation and immune response to helminths.
a, Alternative activation assessed by the co-expression of RELMα and CD206 by flow cytometry of WT and SCAP-KO BMMФ in response to IL-4. Quantification of the number of M(IL-4) macrophages is presented as mean ± SEM in b. Data of n=8 biological replicates from 2 independent experiments.
c, Gene enrichment analysis of the pathways associated with response to IL-4 and innate immune response of the interaction effect of IL-4 in WT and SCAP-KO BMMФ. Data of n=6 biological replicates from 2 independent experiments per group.
d, Experimental design for N. brasiliensis infection.
e, Macrophage number in the lungs of naïve or 5- and 7-days post N. brasiliensis inoculation WT and MФ-SCAP-KO mice. Data is expressed as mean ± SEM.
f, Percentage of alternatively activated alveolar macrophages assessed by the co-expression of RELMα and CD206 in the lungs of naïve or 5-and 7-days post N. brasiliensis inoculation WT and MФ-SCAP-KO. The number of alternatively activated alveolar macrophages is presented in g. Data is expressed as mean ± SEM.
h, Red blood cells counts in the bronchoalveolar lavage of naïve or 5 and 7-days post N. brasiliensis inoculation WT and SCAP-KO mice. Data is expressed as mean ± SEM.
i, Representative pictures of haematoxylin and eosin staining of lung section from four WT and four MФ-SCAP-KO mice 5-days post N. brasiliensis inoculation. Black arrows indicate alveolar proteinaceous debris.
j, Histological score of alveolar proteinaceous debris naïve or 5 and 7-days post N. brasiliensis inoculation WT and MФ-SCAP-KO mice. Data is expressed as mean ± SEM.
k, Adult worms count in the gut of naïve or 5 and 7-days post N. brasiliensis inoculation WT and MФ-SCAP-KO mice. Data is expressed as mean ± SEM.
For N. brasiliensis infection experiment, data of n=6 (WT D0), n=7 (KO D0), n=11 (WT D5) and n=12 (WT D7 and KO D5 and D7) mice from 2 independent experiments is presented.
Data was analysed using a two-way ANOVA followed by Sidak post-hoc test for comparison between genotypes in control or IL-4 treated cells or at different days post-N. brasiliensis inoculation.
We next assessed the functional relevance of SREBP1 activation in alternative macrophage activation using a pathophysiological infection model, the parasitic helminth Nippostrongylus brasiliensis. Infection with N. brasiliensis triggers a type-2 immune response characterised by ILC2s, Th2 cells, eosinophils and alternatively activated macrophages in the lungs2,15,16. We infected WT or MФ-SCAP-KO mice with N. brasiliensis and assessed the lungs five- and seven-days post-inoculation (Fig. 2d). As expected, N. brasiliensis infection led to the recruitment of eosinophils, macrophages and neutrophils in the lungs (Fig. 2e and Extended data Fig. 4a and b). Strikingly, MФ-SCAP-KO mice showed both decreased alveolar macrophage number and alternative activation (Fig. 2e-g and Extended data Fig. 4c), while interstitial macrophages were not affected (Extended data Fig. 4d-f). By contrast, the levels of neutrophils17 and eosinophils in the lungs were not altered (Extended data Fig. 4a and b). We confirmed by histology that the neutrophil score in both alveolar and interstitial spaces was unchanged between genotypes (Extended data Fig. 4g and h). Alternatively activated alveolar macrophages are essential in limiting tissue damage, notably in the lungs of N. brasilensis infected mice2,16,18,19. MФ-SCAP-KO mice had increased red blood cell count in the bronchoalveolar lavage and a trend toward increased alveolar proteinaceous debris compared to controls (Fig. 2h-j). Notably, the alveolar proteinaceous debris score inversely correlated with alveolar macrophage number 5-day post-inoculation (Extended data Fig. 4i). MФ-SCAP-KO mice also displayed a high number of adult worms in their gut compared to controls at day 5 (Fig. 2k). The worm phenotype could be secondary to the reduced alveolar macrophages number (Fig. 2e-g), as alveolar macrophages kill N. brasiliensis larvae in the lung, limiting their arrival in the gut20. Overall, these data demonstrate the pathophysiological relevance of SREBP1 in macrophages.
Our data indicated that SREBP1 was required for macrophage alternative activation, but the underlying mechanism remained unknown. We investigated how the loss of SREBP1 prevented alternative polarisation. SREBP1 is a well-established regulator of de novo lipid synthesis21. Accordingly, RNAseq gene enrichment analysis confirmed that pathways associated with DNL were upregulated in response to IL-4 and blunted in SCAP-KO macrophages (Extended data Fig. 5a and b). Fatty acid synthase (FASN) protein level was strongly induced by IL-4 in vivo and in vitro in WT but not SCAP-KO cells (Fig. 3a and b). Lipid biosynthesis was significantly induced in IL-4 stimulated macrophages (Fig. 3c) and SCAP-KO cells had reduced lipid synthesis, showing that a loss of SREBP1 activity blunted IL-4-induced DNL (Fig. 3d). While IL-4 induces lipogenesis, LPS-stimulated macrophages only displayed a transient increase in DNL 6h post-stimulation (Extended data Fig. 5c), showing that sustained lipogenesis is a specific feature of M(IL-4) cells. We next determined the synthesis rate of palmitate. Under control conditions, BMMФ synthesised 0.23 nmol.h-1 per 106 cells (1.5% per hour) of palmitate, compared to 0.48 nmol.h-1 per 106 cells (2.8% per hour) in IL-4 treated cells (Fig. 3e and Extended data Fig. 5d). This result confirmed the strong upregulation of DNL by IL-4 and demonstrated that fatty acid synthesis is a significant anabolic process in macrophages.
Figure 3. Limiting SREBP1-induced lipid synthesis reduces macrophages alternative activation without altering macrophages lipid composition.
a, In vivo induction of FASN protein expression in MACS-sorted peritoneal macrophages (F4/80+) from mice injected with IL-4c or PBS. ß-actin was used as loading control.
b, Protein expression of FASN in WT and SCAP-KO BMMФ in response to IL-4. Western-blot representative picture of n=4 biological replicates. ß-actin has been used as loading control for the western blot analysis.
c, Lipid synthesis rate in control (CTR) or IL-4-treated BMMФ. The data represents the incorporation of radiolabelled 14C-acetate in the lipid fraction as mean ± SEM of n=4 biological replicates per group.
d, Lipid synthesis of WT and SCAP-KO BMMФ in response to IL-4. The data represents the incorporation of radiolabelled 14C-acetate in the lipid fraction as mean ± SEM of n=4 biological replicates.
e, Palmitate synthesis rate in response to IL-4. The data are presented as mean ± SEM of n=4 biological replicates. control or IL-4 stimulated BMMФ
f, FAME composition in order of increasing chain length and desaturation of control or IL-4 stimulated BMMФ. Data is presented as mean ± SEM of n=4 biological replicates.
g, Total, essential and non-essential fatty acid content of control or IL-4 stimulated BMMФ. Data is presented as mean ± SEM of n=4 biological replicates.
h, Alternative activation assessed by flow cytometry using the co-expression of RELMα and CD301of BMMФ in response to IL-4 and pre-treated with increasing doses of C75. The quantification of the number of alternatively activated macrophages of n=4 biological replicates is presented as mean ± SEM in i.
j, Alternative activation assessed by flow cytometry using the co-expression of RELMα and CD301of DMSO or C75 (10 μM)-treated BMMФ in response to IL-4 and Palmitate (PA, 10μM) or Oleate (OA, 10μM). The quantification of the number of alternatively activated macrophages of n=4 biological replicates is presented as mean ± SEM in k.
Data has been analysed using a two-tailed Student’s t-test (Fig. 3c and e) or a 2-way ANOVA followed by a Sidak (Fig. 3d, f, and g), Dunnett (Fig. 3i) or Tukey (Fig. 3k) post-hoc test.
In line with an accumulation of DNL products, M(IL-4) cells showed an increased level of palmitic (C16:0), palmitoleic (C16:1n7), oleic (C18:1n9) and vaccenic (C18:1n7) acids (Fig. 3f). Overall, IL-4 increased the fatty acid content of the cells by 8.3 nmol per 106 cells (Fig. 3g) while palmitic acid uptake remained unchanged (Extended data Fig. 5e). Conversely, SCAP-KO cells exhibited a reduction in DNL-derived fatty acids compared to WT cells (Extended data Fig. 5f and g), which was not compensated for by increased palmitate uptake (Extended data Fig. 5h).
To study the role of DNL on M(IL-4) activation, we used two selective FASN inhibitors, C75 and Cerulenin. We showed that both inhibitors dose-dependently decreased IL-4-induced lipid synthesis (Extended data Fig. 6a) and prevented macrophage alternative activation (Fig. 3h and i and Extended data Fig. 6b-e). However, as for SCAP-KO macrophages, FASN inhibition did not alter the capacity of IL-4 to reduce the expression of the pro-inflammatory cytokines Tnf and Il1b (Extended data Fig. 6f).
Since SREBPs also regulate cholesterol biosynthesis13, we investigated whether this process was also required for macrophages alternative activation. Inhibition of HMG-CoA reductase with Simvastatin did not alter macrophage alternative activation, despite a compensatory induction of SREBP2 target genes (Extended data Fig. 6g and h). In response to LPS, cholesterol supplementation rescues inflammatory responses in fatty acid synthase deficient macrophages22. However, in our model, cholesterol supplementation did not restore the defective alternative activation of SCAP-KO or C75-treated macrophages (Extended data Fig. 7). Given that FASN inhibition with C75 also alters acetoacetate production23, we complemented C75-treated macrophages with HMG-CoA to provide substrates to the mevalonate pathway. However, HMG-CoA supplementation failed to rescue the defective alternative activation of C75-treated cells (Extended data Fig. 6i), further demonstrating that cholesterol biosynthesis and the mevalonate pathway are dispensable for macrophage alternative activation.
We then attempted to rescue DNL defect by supplementing C75-treated or SCAP-KO cells with fatty acids. We used either 10 μM or 50 μM of palmitic (PA) or oleic acid (OA) that correspond to addition of either 20 or 100 nmol per 106 cells for 24h. Neither PA nor OA significantly reduced Fasn expression at these doses, but OA reduced Scd2 expression (Extended data Fig. 7). However, neither palmitate nor oleate supplementation restored the alternative activation defects caused by SCAP-KO or FASN inhibition (Fig. 3j and k and Extended data Fig. 7), indicating that the lipids produced by DNL were not the signal regulating alternative activation.
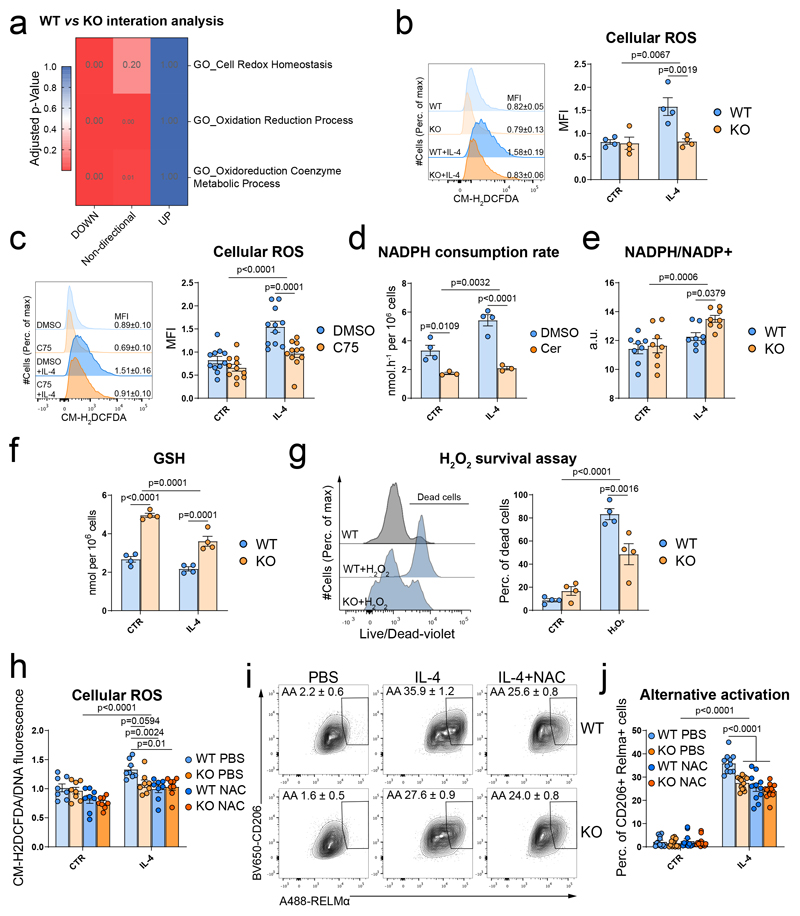
We then investigated the mechanisms by which macrophages sense DNL. We noted that pathways associated with redox homeostasis and oxidoreductive processes were upregulated in response to IL-4 in WT cells (Extended data Fig. 8a) and failed to be induced by IL-4 in SCAP-KO BMMФ (Fig. 4a). Given that reactive oxygen species (ROS) would be a plausible secondary messenger for macrophage alternative activation24–28, we assessed ROS levels in WT and SCAP-KO cells and showed that IL-4 induced ROS in WT but not in SCAP-KO BMMФ (Fig. 4b). Similarly, blocking SREBP activation using 25HC treatment also reduced ROS levels upon IL-4 stimulation in BMMФ (Extended data Fig. 8b). The induction of cellular ROS levels by IL-4 was also blunted when FASN was inhibited with C75 or Cerulenin, indicating that the loss of FASN activity mediated the ROS depletion observed when SREBP1 was inactivated (Fig. 4c and Extended data Fig. 8c).
Figure 4. SREBP1-dependent lipid synthesis increases NADPH utilisation to reduce antioxidant defences and permit macrophage alternative activation.
a, Gene enrichment analysis of the pathways associated with oxidoreduction processes of the interaction effect of IL-4 in WT and SCAP-KO BMMФ. Data of n=6 biological replicates from 2 independent experiments per group.
b, Reactive oxygen species (ROS) levels in WT and SCAP-KO BMMФ in CTR or IL-4 treated cells. ROS were quantified by the median fluorescence intensity (MFI) of CM-H2DCFDA by flow cytometry. Data presented as mean ± SEM of n=4 biological replicates.
c, Reactive oxygen species (ROS) levels in DMSO or C75-treated (10μM) BMMФ in CTR or IL-4 treated cells. ROS were quantified by the MFI of CM-H2DCFDA by flow cytometry. Data presented as mean ± SEM of n=11 biological replicates from 3 independent experiments.
d, NADPH consumption rate by palmitate synthesis in BMMФ treated with DMSO or Cerulenin (1μg/mL) in response to IL-4. Data presented as mean ± SEM from n=4 (DMSO) and n=3 (Cerulenin) biological replicates.
e, NADPH/NADP+ ratio in WT and SCAP-KO BMMФ in response to IL-4. Data presented as mean ± SEM from n=8 biological replicate from 2 independent experiments.
f, Reduced glutathione (GSH) levels in WT and SCAP-KO BMMФ in response to IL-4. Data presented as mean ± SEM of n=4 biological replicates.
g, H2O2-induced cell death (100 μM, 24h) challenge in WT and SCAP-KO BMMФ determined by flow cytometry using a live/dead dye. The quantification of the dead cells percentage is presented in the right panel. Data presented as mean ± SEM of n=4 biological replicates.
h, Cellular ROS levels in CTR or IL-4-stimulated WT and SCAP-KO BMMФ pre-treated with N-acetyl cysteine (NAC). ROS were quantified by the fluorescence ratio of CM-H2DCFDA over DNA (Hoechst). Data presented as mean ± SEM of n=8 biological replicates from 2 independent experiments.
i, Alternative activation (AA) assessed by the co-expression of RELMα and CD206 by flow cytometry of WT and SCAP-KO BMMФ in response to IL-4 and NAC. The quantification of the number of M(IL-4) macrophages as mean ± SEM of n=11 (WT) and n=13 (KO) biological replicates from 3 independent experiments is presented in j.
Statistical analysis of the RNAseq data is detailed in the methods section.
Data was analysed using a two-way ANOVA followed by Sidak post-hoc test (Fig. 4b-j).
We next determined the source of ROS in IL-4 macrophages. Inhibition of canonical ROS producers, such as NADPH oxidases, nitric oxide synthase and xanthine oxidase did not affect ROS levels in M(IL-4) cells, but reduced LPS-induced ROS (Extended data Fig. 8d). Conversely, we confirmed the findings of previous studies that reported increased mitochondrial ROS production in response to IL-4 (Extended data Fig. 8e)27,29.
Previous studies suggested that fatty acid oxidation (FAO) and OXPHOS are required for macrophage alternative activation30,31, and that OXPHOS is a potential source of ROS. However, in agreement with more recent studies showing that FAO and OXPHOS are dispensable for macrophage alternative activation32–36, we did not observe an increase in oxygen consumption or FAO in response to IL-4 (Extended data Fig. 9g-h). In line with this observation, activation of FAO at the expense of DNL using the AMPK activator AICAR (Extended data Fig. 9i and j), inhibited macrophage alternative activation (Extended data Fig. 2d). Also, while SCAP-KO cells displayed a reduced FAO capacity, inhibition of FASN with C75 or Cerulenin did not affect FAO (Extended data Fig. 9k and i), demonstrating that DNL was not required for FAO in response to IL-4. Moreover, inhibition of FAO with Etomoxir did not affect ROS levels, even when FASN was inhibited (Extended data Fig. 8m). Finally, inhibition of DNL in SCAP-KO or C75-treated BMMФ did not affect mitochondrial ROS levels (Extended data Fig. 8n and o). Altogether, these results demonstrated that DNL did not regulate ROS at the level of the mitochondria.
Seeking an alternative explanation for the reduced ROS levels observed when FASN activity was decreased, we focused on the bioenergetic requirements of DNL. FASN uses NADPH, a cofactor also required for the replenishment of the glutathione antioxidant defences. To determine if DNL consumed sufficient NADPH in macrophages to dysregulate the glutathione antioxidant system, we used 3-2H,glucose to measure NADPH consumption by palmitic acid synthesis37.
Palmitate synthesis consumed ~3.3 nmol.h-1 of NADPH per 106 cells under basal conditions, rising to ~5.4 nmol.h-1 of NADPH per 106 cells in alternatively activated macrophages and NADPH consumption was reduced by FASN inhibition (Fig. 4d). Mimicking the reduced NADPH consumption due to DNL inhibition, SCAP-KO BMMФ had an increased NADPH/NADP+ ratio following IL-4 treatment relative to controls (Fig. 4e). In support of the concept that fatty acid synthesis and antioxidant defences compete for NADPH in alternatively activated macrophages, reduced glutathione (GSH) levels were decreased following IL-4 stimulation and increased in both SCAP-KO and C75-treated cells (Fig. 4f and Extended data Fig. 8p). Importantly, GSH levels and the NADPH consumption rate due to palmitate synthesis were both within the 2-4 nmol per 106 cells range. As the half-life of GSH in macrophages has been reported to be 1.9 hours38, consumption of NADPH by DNL could plausibly regulate GSH levels. Finally, SCAP-deficient macrophages were protected against H2O2-induced cell death (Fig. 4g), further indicating they possessed increased antioxidant defences. Altogether, our results demonstrated that, in macrophages, IL4-induced DNL could consume enough NADPH to impact on antioxidant defences, thus increasing ROS levels.
We confirmed the relevance of IL-4-induced ROS for alternatively activated macrophages by treating BMMФ with N-Acetyl cysteine (NAC) to artificially replenish the GSH pool, independently of NADPH levels. NAC treatment blunted the accumulation of ROS following IL-4 stimulation (Extended data Fig. 9a) and impaired the M(IL-4) polarisation of the BMMФ (Extended data Fig. 9b-d). As SCAP-KO BMMФ already had increased antioxidant defences due to their lack of DNL, treating them with NAC did not reduce ROS levels (Fig. 4h) or alternative activation any further (Fig. 4i and j) in M(IL-4). Overall, our data support a mechanism whereby DNL increases ROS levels to enable M(IL-4) activation.
This study has identified the SREBP1-DNL anabolic network as a major downstream effector of IL-4 signalling in macrophages (Extended data Fig. 10) and the pathophysiological relevance of SREBP1 activation in macrophages for the immune response in vivo using a parasite infection model. The activation of SREBP1 in response to IL-4 is conserved in mouse and human macrophages, suggesting targeting this pathway may have therapeutic value39. Our results provide an additional rationale for the efficacy of fatty acid synthesis inhibitors in the treatment of cancer40, where systemic inhibition of SREBP1 improves response to PD-L1 inhibition, potentially through reduced macrophage alternative activation41.
Previous studies emphasised the catabolic role of lipid metabolism in alternatively activated macrophages, with fatty acids being used as a fuel for ATP production through mitochondrial OXPHOS30,31. However, recent studies have challenged this concept by showing that both FAO and OXPHOS are dispensable for macrophage alternative activation32–36. Much of the confusion surrounding the requirement for FAO in macrophage alternative activation comes from the use of Etomoxir, a CPT1a inhibitor. While Etomoxir is known to impair FAO, many studies used high doses that cause off-target effects32. Recently, Divakaruni et al. showed that Etomoxir depletes macrophage-CoA levels, blocking macrophage alternative activation33. This result is consistent with our data, as CoA depletion prevents DNL42.
One of our most striking observations was that the lipid biosynthetic pathway in macrophages is exceptionally active. We quantified that in a resting macrophage, ~1.5% of the total palmitate is de novo synthesised in an hour, or ~36% per day. This agrees with a previous report that fractional synthesis rates of palmitate are nearly 60% over 48h13. The importance of SREBP and DNL in macrophages has been previously demonstrated in LPS- or IFNβ-activated macrophages13,22,23,43–46. Accordingly, both SREBP1a44 and SREBP246,47 bind the promoter of IL1b to promote its transcription in response to LPS. SREBP1 is also required for the resolution of inflammation and efficient phagocytosis post-LPS activation43,45. Furthermore, fatty acid synthesis is needed for classical activation to ensure the correct membrane composition for TLR4 activation22,23. However, the role of fatty acid synthesis in classical activation post-TLR4 signalling remains unclear. Our data show a dramatic reduction in DNL 24h post-LPS stimulation, indicating that lipid accumulation in pro-inflammatory macrophages is likely to come from uptake/or reduced FAO48 rather than increased fatty acid synthesis49.
In M(IL-4) cells we show the high rate of lipid synthesis in macrophages consumes substantial quantities of NADPH, limiting its availability for antioxidant defences and lead to ROS accumulation. ROS acts as a necessary second messenger linking SREBP1 activation to macrophage alternative activation. While ROS is already established as a regulator of macrophage alternative polarisation24–28, a limitation of our study is the absence of any rescue of alternative activation due to inhibition of DNL by pharmacological ROS-inducing agents. This result highlights the complexity of oxidative and reductive stresses and potentially indicates that ROS production’s specific source may be significant, and the link between DNL and ROS production, notably in vivo, requires further investigation.
While the importance of ROS levels have been established, the role of the actual lipids synthesised in response to IL-4 remains to be elucidated. Of potential relevance, while in vitro IL-4 does not alter macrophage number, IL-4 is a potent driver of macrophage proliferation in vivo 50, a process that requires the biosynthesis of new lipids to supply the formation of new cell membranes. Therefore, it could be argued that DNL could support new membrane synthesis required for IL-4-induced macrophage proliferation50. In this context, ROS may serve as a proxy of enough anabolic activity to engage in a demanding proliferative alternative activation. Notably, the lower number of alveolar macrophages in our helminth infection model supports this hypothesis. In support of the concept that anabolism and macrophage alternative activation are intrinsically linked, inhibition of the non-oxidative arm of the PPP, which is required for nucleotide synthesis, also impairs macrophage alternative activation11. Moreover, in vitro inhibition of Myc, a major transcription factor governing cellular proliferation also impedes macrophage alternative activation51. Altogether, these studies indicate the relevance of anabolic pathways for proliferation ability and alternative activation in macrophages. Further studies should address the interdependence of these pathways in disease models involving macrophage alternative activation and proliferation, such as cancer and parasitic infections.
Methods
Animal models
All animal work was carried out in the Disease Model Core or MRC-LMB Ares unit.
Animals were housed in a temperature-controlled room (22°C) with a 12-h light/dark cycle with 55% relative humidity (DMC) and 45-65% relative humidity (MRC-LMB Ares). Food and water were available ad libitum. This research has been regulated under the Animals (Scientific Procedures) Act 1986 Amendment Regulations 2012 following ethical review by the University of Cambridge Animal Welfare and Ethical Review Body (AWERB) and Medical Research Council (MRC) ARES animal facility, under pathogen-free conditions and housed according to UK Home Office guidelines.
For macrophage-specific gene deletion, a mouse model expressing Cre recombinase from bacteriophage P1 under LysM promoter (LysM-Cre). LysM-Cre mouse was generated by replacing a single allele of LysM gene with Cre recombinase coding sequence as described 52 and was gifted to us on a mixed C57Bl/6J, 129/Sv background by Dr. Suzanne Jackowski. LysM-Cre line was backcrossed to C57Bl6/J background using Marker-Assisted Accelerated Backcrossing (MAX-BAX, Charles River, UK) technology until SNP genotyping confirmed >99% background purity.
Macrophage specific SCAP ablation model was generated by crossing the B6;129-Scaptm1Mbjg/J obtain from Jackson laboratories (004162) with the LysM-Cre mouse model. SCAP macrophage-specific knockout mice were produced by crossing LysM+/+ with LysMCre/+ animals on a floxed/floxed background, yielding a 1:1 Mendelian ratio of control (floxed/floxed LysM+/+) to knockout (floxed/floxed LysMCre/+) offspring.
SREBP1c-KO mice were previously described53.
Studies were conducted in 2-4 months old males using littermate controls. Details of the mouse models are provided in Supplementary Table 2.
Bone-marrow derived macrophages cell culture
6-16 weeks old male C57/Bl6J, obtained from Charles River, LysM+/+ or LysMCre/+ SCAPfl/fl, or SREBP1c knockout mice53 were culled and the bone-marrow was flushed from the femur and tibia in RPMI1640. Total bone-marrow cells were passed into a 100 μm cell strainer and counted using Countess II automated cell counter (Thermofisher). Cells were spun (400g, 5 minutes), resuspended in BMMФ culture medium (RPMI1640 supplemented with 20% of L929-conditioned cell medium, 10% heat-inactivated foetal bovine serum (HI-FBS), and 1% penicillin and streptomycin). Total bone-marrow cells were seeded in 10 cm non-culture treated plates (Falcon) at a density of 5x106 cells per plate per 10 ml of macrophage differentiation medium and cultured for 7 days at 37 °C in 5% CO2. On day 5 of differentiation, medium was removed and replace with 10 ml of fresh BMMФ culture medium. On day 7, BMMФ were detached using ice-cold PBS-EDTA 1mM, spun (400xg, 5 minutes) and resuspended in fresh BMMФ culture medium. Differentiated BMMФ were counted using Countess II automated cell counter and cell concentration adjusted to 5x105 cells/ml. Immediately after, cells were plated for experiments at the following densities: 100μl/well of 96-well plate, 500 μl/well of 24-well plate, 1 ml/well of 12-well plate, 2 ml/well of 6-well plate and 10 ml per 10 cm plate. Cells were incubated for 16-24 h after plating before conducting experiments.
Macrophage purity of the culture was routinely tested by the expression of CD11b and F4/80 by flow cytometry. 95-99% of the cells express high levels of CD11b and F4/80 after 7 days of differentiation.
To make L929-conditioned medium, L929 cells (CCL-1, ATCC) were seeded in DMEM supplemented with 10% heat-inactivated FBS, 100 U/ml penicillin-streptomycin and 2 mM L-glutamine (Sigma) at a density of 250,000 cells per 50 ml of medium per T175 tissue culture flask. Medium was harvested after 1 week of culture, and then 50 mL of fresh DMEM supplemented with 10% heat-inactivated FBS, 100 U/ml penicillin-streptomycin and 2 mM L-glutamine was added onto cells and harvested 1 week later. Batches obtained after the first and second weeks of culture were mixed at a 1:1 ratio, aliquoted and stored at -20 °C.
Bone-marrow derived macrophages stimulation
BMMФ were stimulated either with IL-4 (10 ng/ml, AF-214-14, Peprotech), lipopolysaccharide (100 ng/ml, Sigma) or dexamethasone (100nM, D4902, Sigma) to respectively achieve M(IL-4), M(LPS) or M(GC) polarization state. In some experiments, BMMФ were treated with C75 (Sigma), Cerulenin (Sigma), Etomoxir (Sigma), pan-AKT inhibitor MK-2206 (1 mM, Cayman Chemical), 25-hydroxycholesterol (10 μM, Cayman chemical), N-acetyl cysteine (10mM, Sigma), Diphenyleneiodonium chloride (DPI, 5 μM, Sigma), L-NG-Nitroarginine methyl ester (L-NAME, 1mM, Cayman Chemical), Allopurinol (100 μM, Sigma), hydrogen peroxide (Sigma), SIRT1 activator II (Sigma), EX-527 (Sigma), Compound C (Sigma), AICAR (Abcam), HMGCoA (Sigma), water-soluble cholesterol (Cholesterol–methyl-β-cyclodextrin, Sigma) or Simvastatin (Sigma).
For fatty acid treatment, palmitic or oleic acid (Cayman) were solubilized at 100mM in absolute ethanol at 60°C. Fatty acid were conjugated at the desired concentration using a sonicator water bath into the macrophage culture medium to avoid endotoxin contamination from BSA54.
RAW264.7 macrophage cell culture and transfection
RAW264.7 cells (TIB-71, ATCC) were cultured in DMEM (4.5g/L glucose) supplemented with 10% HI-FBS, 100 U/ml penicillin-streptomycin and 2 mM L-glutamine (Sigma). For knockdown experiment, 100,000 cells per well were seeded in a 24-well plate. The next day cells were transfected using Lipofectamine RNAiMAX (Invitrogen, ThermoFisher) according to manufacturer instructions. ON-TARGET plus siRNA against SREBP1 or SREBP2 or CTR were obtained from Dharmacon (Horizon). 24h post transfection, RAW264.7 cells were stimulated for 24h with 10ng/mL IL-4.
Nippostrongylus brasiliensis infection
N. brasiliensis was maintained by passage through Wistar strains rats. LysM+/+ or LysMCre/+ SCAPfl/fl mice were inoculated subcutaneously with 400 viable third-stage N. brasiliensis larvae and euthanized 5- and 7-days post-inoculation. Tissues were collected for analysis and small intestine worm burdens were enumerated using a dissection microscope.
IL-4 complex administration
Animal-free recombinant murine IL-4 (AF-214-14, Peprotech) was suspended at a concentration of 500 μg/ml and mixed with anti-mouse IL-4 antibody (BioXcell clone 11b11) at a molar ratio of 2:1 (weight 1:5) for 1–2 minutes at room temperature to complex IL-4 with the antibody. The complex (IL-4c) was suspended in PBS to a concentration of 25μg/ml IL-4 and 125 μg/ml of 11b11. Each mouse was injected intraperitoneally with 200μl of IL-4c (5 μg IL-4 and 25 μg 11b11) on day 0 and peritoneal exudate cells (PECs) were collected on day 2.
Isolation of cells from mice
Lungs were minced with scissors and digested for 30 minutes at 4 °C in RPMI1640 with 2.5 mg/ml collagenase I and 0.25 mg/ml DNase I. The digest was sequentially passed and mashed through a 70 μm cell strainer and the strainer washed with FACS buffer. The cell suspension was filtered with a 50 μm cell strainer and red blood cells were lysed. Cells were counted using Countess automated cell counter (Invitrogen) and further process for multicolour flow cytometry. Cells were kept on ice until use or analysis.
PBS or IL-4c injected mice were killed by cervical dislocation, peritoneal exudate cells (PECs) were harvested by lavage of the peritoneal cavity with sterile PBS. Total peritoneal exudate cells were counted after red blood cell lysis (Biolegend) using Countess II automated cell counter (Invitrogen) and further process for magnetic-activated cell sorting (MACS). Cells were kept on ice until use or analysis.
Flow cytometry staining and analysis
For BMMФ, cells were seeded in non-tissue culture treated plate and detached using ice-cold PBS-EDTA 1mM. After collection, lung digest or PECs or BMMФ were kept in FACS buffer (PBS, 1mM EDTA, 3% HI-FBS) on ice. Non-specific binding was blocked with 5 μg/ml anti-CD16/32 (Clone 93, Biolegend) for 20 minutes at 4 °C. Cell surfaces were then stained with anti-CD45 (30-F11, BD biosciences), anti-CD11b (M1/70, BD biosciences), anti-Siglec-F (E50-2240, BD biosciences), anti-F4/80 (BM8, Biolegend), anti-CD301 (LOM-14), anti-CD206 (C068C2, Biolegend), anti-CD11c (N418, Biolegend), anti-Ly6G (1A8, Biolegend) and Live/Dead dye (Invitrogen). For intracellular staining, cells were fixed and permeabilized using the FoxP3 staining kit (eBioscience) and stained for 1 h at 4 °C with rabbit anti-RELMα antibody (500-P214, PeproTech). For the BMMФ experiments, we excluded debris on FSC-A/SSC-A plot, then doublets and clumps on an FSC-A/H plot and dead cells using a live/dead staining; macrophages were then identified as CD11b+ F4/80+ cells. The gating strategy for the lung immune cells was as follow. First, we excluded debris on FSC-A/SSC-A plot, then doublets and clumps on an FSC-A/H plot and dead cells using a live/dead staining. Leukocytes were identified as positive for CD4555. Lung tissue is composed of two macrophages subsets, alveolar and interstitial56. In order to gate alveolar macrophages, we initially selected CD11c + CD11b low/- cell57–59 and further identified them as F4/80 low/-57,60 and SiglecF +57,61. Alternatively activated alveolar macrophages were defined as CD206 + Relma +. Positivity for Relmα was defined using fluorescence minus one (FMO) control and positivity for CD206 was defined using eosinophil expression of CD206 as negative control, since eosinophils are known to be CD206 -57. Neutrophils were identified as CD11b + Ly6G+57. Eosinophils were then identified as Ly6G- CD11b + SiglecF + and interstitial macrophages as Ly6G- SiglecF - CD11b + F4/80 low/-57. Alternatively activated interstitial macrophages were defined as Relmα + using FMO control. Gating strategy for the lungs is presented in the Supplementary Information file.
Data were acquired on a LSR Fortessa (BD Biosciences) using FACS Diva software and analysed with TreeStar FlowJo (Version V10.6.1). A detailed list of the antibodies used for this study is presented in Supplementary Table 3.
Lung histology
After collection of BAL, lungs were excised and fixed in formalin. Subsequently, lungs were embedded in paraffin blocks and sectioned. Serial 4-μM sections were obtained from FFPE blocks and extra-coated with paraffin to preserve tissue integrity. Sections were dried overnight at 37°C, dewaxed with xylene and 100% ethanol, and washed in running water for a minimum of 4 minutes. The sections were stained with standard haematoxylin and eosin (H&E) as previously described62.
For each mouse, a blinded histopathologist scored 20 randomly chosen fields for the presence of neutrophils in the alveolar and interstitial space and the presence of proteinaceous debris according to the recommendation of the American Thoracic Society63. The scoring data is expressed as the sum of the score of the 20 fields.
Magnetic-activated cell sorting
PECs were resuspended in MACS buffer (PBS, 2mM EDTA (sterile), 0.5% Bovine Serum Albumin) and incubated with biotin-conjugated anti-F4/80 antibody (Clone REA126, Miltenyi Biotech, 130-101-893) for 10 minutes at 4 °C. Then, PECs were incubated with Streptavidin Microbeads (Miltenyi Biotech, 130-048-101) for 15 minutes at 4 °C. Finally, the F4/80+ fraction was isolated using MACS LS columns according to manufacturer instructions (Miltenyi Biotech). Samples were further processed for immuno-blot or gene expression analysis as described below.
De novo lipid synthesis assay
BMMФ seeded in a non-tissue culture treated 24-well plate at a density of 250,000 cells per well and stimulated as describe in the figure legends. Then, incorporation of 14C-sodium acetate into lipids was assessed over a 2 hours period. After medium removal, cells were detached using PBS-EDTA 1mM, pelleted and snap frozen in dry ice. The lipid fraction was extracted using Folch’s extraction method and extracted lipid were mixed with Opti-Fluor scintillation liquid, and radioactivity was measured using liquid scintillation counter.
Fatty acid oxidation assay
BMMФ were seeded in a 24-well plate at a density of 250,000 cells per well and stimulated as defined in the figure legends. 24h after IL-4 stimulation, cells were washed with PBS and incubated in the presence of 0.5 mL of fatty acid oxidation (FAO) medium (RPMI containing 12.5 mM HEPES, 1 mM L-carnitine, 10% HI-FBS and 0.2 μCi of 1-14C-Oleate (NEC317250UC, Perkin Elmer)). Plates were then immediately sealed with parafilm M and placed at 37 °C for 3 h. Meanwhile, CO2 traps were prepared by adding 200 μl of concentrated HCl into the 1.5 ml Eppendorf tubes containing paper discs, wetted with 20 μl of 1 M NaOH, in the inner side of their lids. Once the FAO reaction finished, 400 μl of medium from BMMФ were transferred into the CO2 traps, lids were immediately closed and tubes were incubated for 1 h at room temperature, allowing CO2 to escape the medium and react with NaOH in the paper disc. Paper discs were then transferred to scintillation vials containing 5 ml of Hionic-Fluor scintillation liquid and radioactivity was measured using liquid scintillation counter.
Lipid extraction
One million cells were seeded in non-tissue culture plates and detached with ice-cold PBS-EDTA. Cell suspensions were transferred into glass vials, counted, spun and the supernatant discarded. The cell pellets were snap-frozen in dry ice prior lipid extraction. Total lipids from cells were extracted using the protein precipitation method64. Glass pipettes and vials were used throughout the procedure in order to avoid plastic-bound lipid contamination. Cell pellets were resuspended in 1 ml of HPLC-grade chloroform:methanol 2:1 v/v mixture. Deuterated tridecanoic acid (DLM-1392-PK, Cambridge Isotopes) was included in extraction mixture as internal standard. Samples were homogenised by vortexing for 15 s. 400 μl of HPLC-grade acetone was added to each sample before vortexing for 2 min and centrifuging at 4000 g for 30 min. The single layer supernatant was pipetted into separate glass vial being careful not to break up the solid pellet at the bottom of the tube. Collected lipid fractions were dried under nitrogen stream.
Quantitative analysis of fatty acid methyl esters (FAMEs)
In order to derive FFAs and esterified fatty acids from complex lipids into FAMEs, 750 μl of HPLC-grade chloroform: methanol 1:1 v/v solution was added to previously dried lipids in 7 ml glass vials. 90 μl of 13% boron trifluoride in methanol (134821, Sigma) was then added into each vial. Vials were sealed and incubated in an oven at 80°C for 90 min in order to hydrolyse fatty acid-glycerol and fatty acid-cholesterol ester bonds and form FAMEs. Samples were allowed to cool, and 1 ml of HPLC-grade n-Hexane and 500 μl of HPLC-grade water were added. Samples were briefly vortexed and centrifuged at 2000. The upper organic layer was transferred into 2 ml gas chromatography glass vials and dried under nitrogen stream.
Gas chromatography-mass spectrometry was performed with Agilent 7890B gas chromatography system linked to Agilent 5977A mass spectrometer, using AS3000 auto sampler and data was acquired using MassHunter Workstation Software. A TR-FAME column (length: 30 m, inter diameter: 0.25 mm, film size: 0.25 μm, 260M142P, Thermofisher Scientific) was used with helium as carrier gas. Inlet temperature was set at 230°C. Dried FAME samples were re-suspended in 200 μl HPLC-grade n-Hexane. 1 μl of this solution was injected for analysis. The oven programme used for separation was as follows: 100°C hold for 2 min, ramp at 25°C/min to 150°C, ramp at 2.5°C/min to 162°C and hold for 3.8 min, ramp at 4.5°C/min to 173°C and hold for 5 min, ramp at 5°C/min to 210°C, ramp at 40°C/min to 230°C and hold for 0.5 min. Carrier gas flow was set to constant 1.5 ml/min. If the height of any FAME peaks exceeded 108 units, sample was re-injected with 10:1 – 100:1 split ratio. Identification of FAME peaks was based on retention time and made by comparison with those in external standards (Food industry FAME mix, 35077, Restek).
Peak integration and quantification was performed using MassHunter Workstation Quantitative Analysis software (version B.07.00, Agilent). Specific high-abundance ions from total ion chromatogram were chosen to calculate each fatty acid peak.
Mass isotopomer distribution analysis
NADPH consumption rate
One million cells were seeded in non-tissue culture plates and stimulated as described above. 4h prior harvesting, the medium was replaced with glucose-free RPMI supplemented with 11.1 mM 3-2H,glucose (GLC-034, Omicron), 10% HI-FBS, 20% L929-conditionned medium and 1% P/S. Cells were detached with ice-cold PBS-EDTA and lipids extracted and derivatized to FAMEs as detailed above.
Mass isotopomer distribution analysis (MIDA) was performed on palmitate. In order to perform MIDA we extracted the M+0 –M+5 ions of palmitate methyl esters (m/z 270-275). From these we calculated the fractional concentration of each ion. All equations below use fractional concentrations.
We determined a theoretical distribution for a newly synthesised molecule that was dependent on the precursor labelling pool p, whereby p was the fraction of deuterated NADPH. Firstly we calculated the isotopomer pattern of palmitate caused by the presence of additional 2H atoms in the molecule. The number of available sites in palmitate that a deuterium could be incorporated via NADPH (N) was assumed to be 14 based on previous publications65,66. Ions dependent on deuterium incorporation where called M’. For each ion M’0-M’5 the following equation gave the expected relative abundance:
The pattern of labelling based on deuterium was then corrected for the presence of naturally occurring oxygen and carbon isotopes using the following equations for ions M0n-M5n, where Mxn stood for the ion of a newly synthesised palmitate molecule:
M’ = labelling due to deuterium
M = natural isotopic labelling
These equations provided us with the ion pattern for a newly synthesised palmitate M0n-M5n
To determine the relative contribution of newly synthesised palmitate and existing palmitate within the cell we set the following equations, where Mxn is newly synthesised palmitate, Mx was endogenous pre-existing palmitate and Mxobs was the observed Mx0 fractional concentration. M0obs’ was the calculated fraction of M0 based on combining newly synthesised and pre-existing palmitate as follows:
We then calculated M0obs-M0obs’ through to M5obs-M5obs’ and calculated the sum of squares for these 6 equations.
We used the GRG Non-Linear Engine of the Solver function of excel to minimise the sum of squares of M0obs-M0obs’+…M5obs-M5obs’ by permitting the values of p and f to be adjusted. This enabled us to calculate the fractional synthesis rate for palmitate.
NADPH consumption rate was determined using the quantification of palmitate quantity within the cell and the fraction synthesis rate (f) determined as detailed above using 3-2H,glucose tracing and the number of NADPH molecules required to make one molecule of palmitate (14):
Palmitate synthesis and uptake rate using deuterated water and stably-labelled palmitate
One million cells were seeded in non-tissue culture plates and stimulated as described above. 2 h prior harvesting, the medium was replaced with BMMФ culture medium supplemented with 8% deuterated water (151882, Sigma) and 10 μM U13C-palmitate (Cambridge Isotope Laboratories, CLM-409-PK). The cells were moved into an incubator filled with 8% deuterated water in order to maintain the concentration of deuterated water in the wells at 8%. Cells were detached with ice-cold PBS-EDTA and lipids extracted and derivatized as FAME as detailed above.
We determined a theoretical distribution for a newly synthesised molecule that was dependent on the precursor labelling pool p, hereby fixed at 8%. Firstly, we calculated the isotopomer pattern of palmitate caused by the presence of additional 2H atoms in the molecule. The number of available sites in palmitate that a deuterium could be incorporated is defined as N. Ions dependent on deuterium incorporation were called M’. For each ion M’0-M’5 the following equation gave the expected relative abundance:
The pattern of labelling based on deuterium was then corrected for the presence of naturally occurring oxygen and carbon isotopes using the following equations for ions M0n-M5n, where Mxn stood for the ion of a newly synthesised palmitate molecule:
M’ = labelling due to deuterium
M = natural isotopic labelling
These equations provided us with the ion pattern for a newly synthesised palmitate M0n-M5n.
To determine the relative contribution of newly synthesised palmitate and existing palmitate within the cell we set the following equations, where M0n is newly synthesised palmitate, M0 was endogenous pre-existing palmitate and M0obs was the observed M0 fractional concentration of the ion measured using the mass spectrometer. M0obs’ was the calculated fraction of M0 based on combining newly synthesised and pre-existing palmitate as follows:
We then calculated M0obs-M0obs’ through to M5obs-M5obs’ and calculated the sum of squares for these 6 equations.
We used the GRG Non-Linear Engine of the Solver function of excel to minimise the sum of squares of M0obs-M0obs’+…M5obs-M5obs’ by changing the values of N and f.
While in vivo, the N for palmitate is established to be 22, there is considerable evidence that it varies in vitro. Given p was known and experimentally fixed at 8%, we therefore determined N by a trial and error and selected an N that gave a p of 8%. This enabled us to calculate the fractional synthesis rate for palmitate and the N number.
Rate of synthesis of palmitate from de novo lipogenesis was determined using the palmitate quantity within the cell and the fraction synthesis rate (f) determined as detailed above:
We divided by 2 as we labelled for 2 hours.
Exogenous palmitate uptake was measured using the sum of the integration of M+16 peaks of palmitate, palmitoleate, stearate, oleate and vaccinate and expressed as nmol/hr/106 cells.
Oxygen-consumption rate measurement
BMMФ were plated in XF-96 cell culture plates at a density of 50,000 cells per well, treated or not with C75 (10 μM) and polarized into M(IL-4) macrophages with IL-4 (10ng/mL) for 24 hours. Cells were washed and the medium change for the XF assay medium (unbuffered DMEM pH 7.4 supplemented with 2 mM L-glutamine with or without 10mM glucose), then incubated for 1h in a non-CO2 incubator, following manufacturer instructions. Real-time measurement of oxygen-consumption rate (OCR) was performed using an XF-96 Extracellular Flux Analyser (Seahorse Bioscience). To assess mitochondrial respiration, three to four consecutive measurements were obtained under basal conditions and after the sequential addition of 1 μM oligomycin, to inhibit mitochondrial ATP synthase; 1.5 μM FCCP (fluoro-carbonyl cyanide phenylhydrazone), a protonophore that uncouples ATP synthesis from oxygen consumption by the electron-transport chain; and 100 nM rotenone plus 1 μM antimycin A, which inhibit the electron transport chain. All drugs were purchased from Sigma.
Reactive oxygen species measurement
For the measurement of intracellular and mitochondrial reactive oxygen species (ROS) production, BMMФ were seeded at 50,000 cells per well in a black 96-wells plate and polarized and/or treated as described above. BMMФ were incubated in RPMI1640 at 37 °C in 5% CO2 with 10 μM CM-H2DCFDA (Thermofisher Scientific), as cytosolic ROS indicator, for 15 or 30 minutes respectively. Cells were counterstained with Hoechst 33342 (Sigma), as read-out of DNA concentration, and a non-stained well was used to correct for autofluorescence signal for each condition. After washes, fluorescence intensity at determined using a Tecan Infinite M1000 Pro Plate reader at excitation/emission spectra of 490/520nm (CM-H2DCFDA) and 350/488nm (Hoechst 33342). Alternatively, reactive oxygen species were measured by flow cytometry. Cells were seeded in non-tissue culture treated 24-well plate at a density of 250,000 cells per well and stimulated as mentioned in the figure legends. Cells were detached with ice-cold PBS-EDTA 1mM and pelleted in 5mL round-bottom tubes. Cells were resuspended in RPMI1640 containing 10 μM CM-H2DCFDA (Thermofisher Scientific) and Live/dead dye (Invitrogen) and stained for 30 minutes in the dark at 37 °C in 5% CO2. Cells were then spun, washed in ice-cold FACS buffer and resuspended in FACS buffer. Cells were kept on ice in the dark and the fluorescence immediately acquired on a LSR Fortessa (BD Biosciences) using FACS Diva software and data were analysed with TreeStar FlowJo (Version V10.6.1).
Gene expression analysis
Whole cell RNA was extracted using QIAShredder and RNeasy mini kit (Invitrogen) according to manufacturer’s procedures. RNA was eluted in 30 μl RNA-free water.
Complementary DNA (cDNA) was generated using Promega reagents in a 20 μl reaction as follows: 250 ng RNA was added to 1 x M-MLV reverse transcriptase master mix (M351A) with 2.5 mM MgCl2 (A351B), 1.25 mM dNTPs (U151B), and 5 μg/mL random hexamers (C118A), and denatured at 65°C for 5 min before being transferred directly to ice in order to prevent the reassembly of the secondary structures of RNA. After the addition of 1 μL of M-MLV reverse transcriptase (M170b), the reaction was incubated at 37°C for 1 h for cDNA synthesis and 95°C for 5 min for enzyme denaturation. cDNA was diluted 50-fold in RNAse-free water and stored at -20°C.
qRT-PCR was performed in a 13 μL reaction with 5 μl of diluted cDNA, 6.5 μl of 2x TaqMan or SYBR Green reagent (Applied Biosystems), 1.3 μl of 3 mM forward and reverse primer mix (including 1.5 mM of probe for TaqMan reactions) and 0.2 μl of RNAse-free water according to the default manufacturer’s protocol (Applied Biosystems). Primer sequences are described in Table S3. Reactions were run in duplicate for each sample and quantified using the ABI Prism 7900 sequence detection system (Applied Biosystems). Duplicates were checked for reproducibility, and then averaged; ‘no reverse transcriptase’ controls were included to check for genomic DNA contamination, and ‘no template’ controls were included to check for the formation of primer dimers. Product specificity was determined using a dissociation curve for SYBR green reactions. A standard curve generated from a pool of all cDNA samples was used for quantification. The expression of genes of interest was normalized using BestKeeper (BK) method to the geometric average of 3 housekeeping genes (18s, 36b4 and Tbp), and data was expressed as arbitrary units or normalised to the average of control group. Primer list is presented in Supplementary Table 4.
Library preparation and RNA-seq
1 μg of total RNA was quality checked (RIN >7) using an Agilent Bioanalyser 2100 system and then used to construct barcoded sequencing libraries with Illumina’s TruSeq Stranded mRNA Library Prep Kit following manufacturer’s instruction. All the libraries where then multiplexed and sequenced on one lane of Illumina HiSeq 4000 at SE50 at CRUK Cambridge Institute Genomics Core Facility.
RNAseq data analysis
Sequence reads were mapped onto the GRCm38 genome using TopHat v2.0.11 and then genes were counted using htseq-count V0.6.1p1. Statistical analysis of the RNAseq data was performed in R (build 3.4.1 Single Candle). Ensembl IDs were converted to gene symbol using the biomaRt “mmusculus_gene_ensembl” dataset67. Duplicated genes counts were summed and genes that had an average number of counts of less than five were filtered out. Counts were log2 normalised using Variance Normalisation Stabilisation (vsn library, 68) for each of the two batches individually, then batch effect was corrected using combat (sva library, 69). PCA was performed with the prcomp function and cross correlation was done using Pearson correlation and complete clustering method on Euclidian distances with pheatmap library. Differential expression analysis was performed using limma library70. Contrast were made to compare the effect of IL4 compared to control (contrast: IL4 WT – control WT), the effect of KO compared to WT (contrast: control KO – control WT) and the specific effect of the KO on the IL-4 response (contrast: IL-4 KO – IL-4 WT + control WT – control KO).
Enrichment analysis was performed using a consensus of multiple enrichment methods with PIANO71 (all available methods in PIANO were used) with a geneset collection comprising msigdb c2 canonical pathways, GO biological processes terms and chemical and genetic perturbations (cgp). Pathway analysis results of the different contrasts are available in the Supplementary dataset section. Results are expressed as adjusted p-values for “distinct”, “mixed” (up or down-regulated) and “non-directional”. “Distinct” refers to an enrichment accounting for the overall deregulations of all measured genes in a pathway with respect to all measured genes in the experiment. A mixed-up enrichment corresponds to an enrichment of only significantly up-regulated genes in a pathway with respect to only up-regulated genes in the experiment, and vice-versa for mixed-down. In the figures, only the “distinct” and “non-directional” results are presented. The pathway analyses are presented as Supplementary Table.
The transcriptional regulators prediction analysis was performed using the upstream regulator function of Ingenuity Pathway Analysis. Analysis was performed to predict for “transcription regulator” and “ligand-dependent nuclear receptor”. The data presented in Fig. 1a has been produced with a cut-off of FDR<0.001 and the human macrophages using the GSE117040 dataset72 with a cut-off of FDR < 0.05 and |log2FC(M1/M2)| > 1.0. Plots were made using ggplot2 library.
Immunoblot analysis
Cells were washed in ice-cold PBS and lysed in RIPA buffer (50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 0.5% sodium deoxycholate, 1% NP-40, pH 7.4) supplemented with cOmplete™, Mini, EDTA-free Protease Inhibitor Cocktail (000000004693159001, Sigma) and Roche PhosSTOP™ (000000004906845001). Protein concentration was quantified by DC protein assay (5000111, Biorad) and adjusted in RIPA buffer. Lysates were diluted in NuPAGE™ LDS sample buffer (NP0007, Thermofisher Scientific) containing 2.5% 2-mercaptoethanol and boiled at 95°C for 5 min. For the detection of SREBP1, the samples were not boiled but incubated at 37°C for 1h prior loading into the gel. 10-25 μg of protein was then separated by electrophoresis using NuPAGE™ SDS-polyacrylamide gels (Thermofisher Scientific) and transferred to nitrocellulose membranes using the iBlot® Dry Blotting System (Thermofisher Scientific). Membranes were blocked for 1 h in 3% BSA in Tris-buffered saline containing 0.05% Tween (TBS-T) at room temperature. Membranes were incubated overnight with the following primary antibodies: mouse anti-SREBP1 (ab3259, clone 2A4, Abcam), rabbit anti-fatty acid synthase (3180, clone C20G5, Cell Signaling), mouse anti-β-actin (3700, clone 8H10D10, Cell Signaling), rabbit anti-phospho-AKT Ser473 (4060, clone D9E, Cell Signaling), rabbit anti-AKT (9272, Cell Signaling). Bound primary antibodies were detected using peroxidase-coupled secondary anti-rabbit (7074, Cell signalling) or anti-mouse (7076, Cell Signaling) and enhanced chemiluminescence (WBLUF0500, Millipore). Images were acquired using the ChemiDoc MP System (Bio-Rad), and bands were quantified using Fiji software (https://imagej.net/software/fiji/). The expression of proteins was normalised to a housekeeping protein (β-actin), and the phosphorylation status was determined by normalising to a respective total protein. All protein quantification data is expressed as arbitrary units. A detailed list of the antibodies used for this study is presented in Extended data Table 1.
NADPH assay
NADPH concentration was determined using the NADP/NADPH-Glo Assay (Promega), following manufacturer instructions. NADP+ levels were determined from the standard curve.
Glutathione assay
Reduced glutathione concentration was determined using the GSH-Glo assay (V6911, Promega) according to manufacturer instructions.
Statistical analysis
RNAseq statistical analysis is detailed in the RNAseq data analysis section. Otherwise, statistical significance was determined using two-tailed Student’s t-test, one-way or two-way analysis of variance (ANOVA) followed by appropriate post-hoc test as indicated in the figure legends. For the ANOVA, the result of the post-hoc test was only considered if the genotype/treatment effect was significant. P<0.05 was considered as statistically significant and all the data were analysed using GraphPad Prism (version 9.1.0).
Extended Data
Extended Data Fig. 1.
a, Protein expression of phosphorylated (Ser473) and total Akt in Vehicle (DMSO) or MK-2206-treated BMMФ in response to IL-4. Representative picture of n=4 biological replicates.
b, mRNA expression over BK of the SREBP1 target genes Fasn and Scd2 AKT in Vehicle (DMSO) or MK-2206-treated BMMФ in response to IL-4. Data are presented as the mean ± SEM of n=8 biological replicates from 2 independent experiments.
c, Gene expression analysis of the SREBP target genes in WT and Stat6-/- BMMФ. Data analysed from the publicly available dataset (GSE106706)59 with FDR<0.05 and Fc>2.
Data has been analysed using a 2-way ANOVA followed by Sidak post-hoc test.
Extended Data Fig. 2.
a-d, mRNA expression of the SREBP1 target genes Fasn and Scd2 and of the macrophages alternative activation markers Retnla and Mgl2 in BMMФ treated with SIRT1 activator II (a) or SIRT1 inhibitor (EX-527, b) or the AMPK inhibitor Compound C (c) or the AMPK activator AICAR (d) in response to IL-4. Data are presented as the mean ± SEM of n=4 biological replicates per group.
e and f, mRNA expression over BK of the SREBP1 target genes in 25-hydroxycholesterol-treated (25HC, n=8 biological replicates from 2 independent experiments) (e) or SREBP-1c KO (n=3 biological replicates) (f) BMMФ in response to IL-4.
Data has been analysed using a 2-way ANOVA followed by a Dunnett (a-d) or Sidak (e and f) post-hoc test.
Extended Data Fig. 3.
a and b, mRNA expression of Retnla and Mgl1 (a) and Tnf and Il1b (b) in WT and SCAP-KO BMMФ, 4h and 24h post IL-4 stimulation. mRNA expression over BK as mean ± SEM of n=8 biological replicates from 2 independent experiments.
c, Alternative activation of Vehicle (EtOH) or 25-hydroxycholesterol (25HC)-treated BMMФ in response to IL-4. Alternative activation was assessed by the expression of RELMα and CD206 by flow cytometry. The quantification of the number of alternatively activated macrophages is presented as mean ± SEM in d. Data of n=4 biological replicates per group.
e, Expression of the macrophage alternative activation markers Mrc1, Mgl1 Retlna and Arg1 in Vehicle (EtOH) or 25-hydroxycholesterol (25HC)-treated BMMФ in response to IL-4. mRNA expression over BK as mean ± SEM of n=4 biological replicates per group.
f, Expression of the macrophage alternative activation markers of Mrc1, Arg1 and Mgl1 in WT and SREBP1c-KO BMMФ in response to IL-4. mRNA expression over BK of n=3 biological replicates.
g, mRNA expression over BK of RAW264.7 macrophages transfected with siRNA against SREBP1, SREBP2 or both in response to IL-4. Data is expressed as mean ± SEM of n=6 different experiments.
Data has been analysed using a 2-way ANOVA followed by Sidak (a, b, d-f) or Tukey (g) post-hoc test.
Extended Data Fig. 4.
a, Neutrophil number presented as mean ± SEM in the lungs of naïve or 5- and 7-days post N. brasiliensis inoculation in WT and SCAP-KO mice.
b, Eosinophil number presented as mean ± SEM in the lungs of naïve or 5- and 7-days post N. brasiliensis inoculation in WT and SCAP-KO mice.
c, Percentage of alternatively activated alveolar macrophages in the lungs of naïve 5- and 7-days post N. brasiliensis inoculation in WT and SCAP-KO mice within the alveolar macrophage population. Alternative polarization was quantified by the expression of RELMα and CD206 by flow cytometry.
d-f, Interstitial macrophages number (d), percentage of alternative activation (e) and number of alternatively activated interstitial macrophages (f) presented as mean ± SEM in the lungs of naïve or 5- and 7-days post N. brasiliensis inoculation in WT and SCAP-KO mice.
g and h, Alveolar (g) and interstitial (h) neutrophil histological score in the lungs of naïve or 5- and 7-days post N. brasiliensis inoculation in WT and SCAP-KO mice.
i, Correlation between the alveolar proteinaceous debris score and the number of alveolar macrophages 5 days post N. brasiliensis inoculation in WT and SCAP-KO mice.
Pooled data as mean ± SEM n=6-12 mice per group from 2 independent experiments.
Data was analyzed using a two-way ANOVA followed by Sidak post-hoc test for comparison between genotypes at different days of post inoculation (a-h) or linear regression modelling (i).
Extended Data Fig. 5.
a and b, Gene enrichment analysis of the pathways associated with de novo lipogenesis and SREBP activation from RNA sequencing comparing CTR and IL-4-treated BMMФ or of the interaction effect of IL-4 in WT and SCAP-KO BMMФ. Data from n=6 biological replicates per group.
c, Lipid synthesis rate in Lipopolysaccharide (LPS) or IL-4-treated BMMФ. The data represents the incorporation of radiolabelled 14C-acetate in the lipid fraction as mean ± SEM of n=4 biological replicates per group.
d, Proportion of newly synthesized palmitate per hour in control or IL-4 stimulated BMMФ. The data are presented as mean ± SEM of n=4 biological replicates.
e, Exogenous palmitate uptake rate of control or IL-4 stimulated BMMФ. The data are presented as mean ± SEM of n=4 biological replicates.
f, FAME composition in order of increasing chain length and desaturation of WT and SCAP-KO BMMФ in response to IL-4. Data is presented as mean ± SEM of n=4 biological replicates.
g, Total, essential and non-essential fatty acid content of WT and SCAP-KO BMMФ in response to IL-4. Data is presented as mean ± SEM of n=4 biological replicates.
h, Exogenous palmitate uptake rate of WT and SCAP-KO BMMФ in response to IL-4. The data are presented as mean ± SEM of n=4 biological replicates.
Statistical analysis of the RNAseq data is detailed in the methods section.
Data has been analysed using a 2-way ANOVA followed by a Dunnett (c) or Sidak post-hoc test (f-h) or a two-tailed Student’s t-test (d and e).
Extended Data Fig. 6.
a, Lipid synthesis in BMMФ pre-treated with increasing doses of the FASN inhibitors C75 and cerulenin (Cer) 30 minutes prior 24h IL-4 stimulation. The data represents the incorporation of radiolabelled 14C-acetate in the lipid fraction as mean ± SEM of n=7 biological replicates from 2 independent experiments.
b, Expression of the macrophage alternative activation markers Mrc1, Mgl2, Arg1, Retlna and Il4i1 in Vehicle (DMSO) or C75 (10μM)-treated BMMФ in response to IL-4. mRNA expression over BK as mean ± SEM of n=4 biological replicates per group.
c, Alternative activation of BMMФ in response to IL-4 and pre-treated with increasing doses of Cerulenin (Cer). Alternative activation was assessed by flow cytometry using the co-expression of RELMα and CD301. The quantification of the number of M(IL-4) macrophages as mean ± SEM of n=4 biological replicates is presented in d.
e, Expression of the macrophage alternative activation markers Retlna and Mgl1 in Vehicle (DMSO) or cerulenin (2.5μg/mL)-treated BMMФ in response to IL-4. mRNA expression over BK as mean ± SEM of n=4 biological replicates per group.
f, Expression of the inflammatory cytokine Tnf and Il1b in Vehicle (DMSO) or C75 (10μM)-treated BMMФ in response to IL-4. mRNA expression over BK as mean ± SEM of n=4 biological replicates per group.
g and h, Expression of the SREBP2-target genes (g) and macrophage activation markers Tnf, Retlna and Mgl2 (h) in Vehicle (DMSO) or Simvastatin (10μM)-treated BMMФ in response to IL-4. mRNA expression over BK as mean ± SEM of n=4 biological replicates per group.
i, mRNA expression of the macrophage activation markers Retlna and Mgl2 in Vehicle (DMSO) or C75 (10μM)-treated BMMФ in response to IL-4 supplemented or not with HMG-CoA (1mM). mRNA expression over BK as mean ± SEM of n=4 biological replicates per group.
Data has been analysed using a 2-way ANOVA followed by a Dunnett (a and d) or Sidak post-hoc test (e-h) or one-way ANOVA followed by Tukey post-hoc test (i).
Extended Data Fig. 7.
mRNA expression of the SREBP1 target genes Fasn and Scd2 in C75-treated (a) or SCAP-KO (c) macrophages and of the macrophages alternative activation markers in C75-treated (b) or SCAP-KO (d) macrophages in response to IL-4 and/or palmitic acid (PA, 10 or 50 μM), oleic acid (OA, 10 or 50 μM) or water-soluble cholesterol (50 μM). mRNA expression of BK of n=4 (C75) or n=3 (SCAP-KO) biological replicates.
Data has been analysed using a 2-way ANOVA followed by a Tukey post-hoc test.
Extended Data Fig. 8.
a, Gene enrichment analysis of the pathways associated with redox homeostasis and response to oxidative stress in IL-4 treated BMMФ. Data from n=6 biological replicates per group. Data of n=6 biological replicates from 2 independent experiments per group.
b, ROS accumulation in Vehicle (EtOH) or 25-hydroxycholesterol (25HC)-treated BMMФ in response to IL-4. ROS were quantified by the fluorescence ratio of CM-H2DCFDA over DNA (Hoechst). Data as mean ± SEM of n=7 biological replicates from 2 independent experiments.
c, ROS accumulation in Vehicle (DMSO) or cerulenin (1μg/mL)-treated BMMФ in response to IL-4. ROS were quantified by the median fluorescence intesnity of CM-H2DCFDA by flow cytometry. Data as mean ± SEM of n=4 biological replicates.
d, Reactive oxygen species (ROS) levels in BMMФ pre-treated with DPI (NADPH oxidase inhibitor), Allopurinol (xanthine oxidase inhibitor) or L-NAME (nitric oxide synthase inhibitor) prior 24h stimulation with IL-4 or LPS. ROS were quantified by the fluorescence ratio of CM-H2DCFDA over DNA (Hoechst). Data are presented as mean ± SEM of n=4 biological replicates.
e, Mitochondrial ROS production in response to IL-4. Mitochondrial ROS levels were determined by the fluorescence ratio of MitoSox over DNA (Hoechst). Data are presented as mean ± SEM of n=8 biological replicates from 2 independent experiments.
f, Time-course of fatty acid oxidation in response to IL-4 (10ng/mL). Data of n=4 biological replicates.
g and h, Oxygen consumption rate (OCR) in WT and SCAP-KO BMMФ (g) or in Vehicle (DMSO) or C75 (10 μM)-treated BMMФ (h) in control or IL-4 stimulated macrophages. OCR was monitored using an XF-96 Extracellular Flux Analyzer following the sequential treatments with oligomycin (oligo), FCCP and rotenone/antimycin (R/A). Data are presented as mean ± SEM of n=4 (WT vs KO) and n=8 from 2 independent experiments (DMSO vs C75) biological replicates per group.
i, Fatty acid oxidation in IL-4 treated BMMФ in response to the AMPK activator AICAR (100 or 500 μM). Data of n=4 biological replicates.
j, Lipid synthesis in BMMФ treated with increasing doses of the AMPK activator AICAR in response to IL-4 (10ng/mL, 24h). Data of n=4 biological replicates.
k, Fatty acid oxidation assay of SCAP-KO macrophages in response to IL-4 (10ng/mL, 24h). Etomoxir (40 μM) was used as a negative control for FAO. Data of n=4 biological replicates.
l, Fatty acid oxidation assay in control or IL-4 stimulated macrophages treated or not with C75 (10 μM) or Cerulenin (1μg/mL) for 24h. Data of n=4 biological replicates.
m, ROS levels in C75 (10 μM) and/or Etomoxir (ETO, 40 μM)-treated BMMФ in response to IL-4. ROS were quantified by the fluorescence ratio of CM-H2DCFDA over DNA (Hoechst). Data of n=4 biological replicates.
n and o, Mitochondrial ROS production in WT and SCAP-KO BMMФ (n=12 biological replicates from 3 independent experiments) (n) or in Vehicle (DMSO) or C75 (10 μM)-treated (n=12 biological replicates from 3 independent experiments) BMMФ (o) in M(IL-4) macrophages. Mitochondrial ROS levels were determined by the fluorescence ratio of MitoSox over DNA (Hoechst).
p, Reduced glutathione (GSH) levels in C75 (10 μM)-treated BMMФ in response to IL-4. Data presented as mean ± SEM of n=4 biological replicates.
Data has been analysed using a 2-way ANOVA followed by a Sidak (b, c, k, l and n-p) or Dunnett (d and j) or Tukey (g and h) post-hoc test or a two-tailed Student’s t-test (e) or a one-way ANOVA followed by Dunnett post-hoc test (f and i).
Extended Data Fig. 9.
a, ROS levels in N-acetyl cysteine (NAC, 10 mM)-treated BMMФ in response to IL-4. ROS were quantified by the fluorescence ratio of CM-H2DCFDA over DNA (Hoechst). Data as mean ± SEM of n=8 biological replicates from 2 independent experiments.
b and c, Alternative activation of NAC-treated BMMФ in response to IL-4. Alternative activation was assessed by the expression of RELMα and CD206 by flow cytometry. The quantification of the number of M(IL-4) macrophages is presented as mean ± SEM in c. Data of n=8 biological replicates from 2 independent experiments.
d, mRNA expression over BK of Arg1, Mgl1 and Retnla in NAC-treated BMMФ in response to IL-4. Data as mean ± SEM of n=4 biological replicates.
Data was analysed using a two-way ANOVA followed by Sidak post-hoc test.
Extended Data Fig. 10. Schematic representation of the mechanism by which DNL is activated and sensed in alternatively activated macrophages.
Supplementary Material
Acknowledgments
LysM-Cre mice were a kind gift from Dr. Susan Jackowski. We thank Daniel Hart, Sarah Grocott, Charley Beresford, Jade Bacon, Laura McKinven, Eerika Rasijeff and Agnes Lukasik and James Warner from the Histology core for their excellent technical assistance. This research was supported by the Cambridge NIHR BRC Cell Phenotyping Hub. In particular, we wish to thank Esther Perez for her advice and support in flow cytometry. We thank Dr. Michael Murphy and Hiran Prag from the MRC Mitochondrial Biology Unit who kindly helped us with the Seahorse experiments. We thank Christian Frezza, Laura Tronci and Efterpi Nikitopoulou from the MRC cancer unit for their help in stable isotope tracer experimental design. We thank the IMS Genomics and Transcriptomics core for the RNA sequencing analysis.
This work was supported by the British Heart Foundation (RG/18/7/33636), the Medical Research Council (MRC) (MC_UU_00014/2) and the FP7 MITIN (223450). K.P. was a recipient of a fellowship from the Wellcome Trust. A.N.J.M and E.J are supported by the Wellcome Trust (100963/Z/13/Z) and the MRC (U105178805). J.L. is a recipient fellowship of the British Heart Foundation. A.D. was a Marie-Curie Early-Stage Researcher supported by the European Union’s Horizon 2020 research and innovation program (675585 Marie-Curie ITN “SymBioSys”) to J.S-R. A.K. is supported by the Wellcome Trust (106260/Z/14/Z) and ERC award (648889). P.F. is supported by the Science Foundation Ireland (10/IN.1/B3004). The IMS Genomics and Transcriptomics and Histology cores (B.M-A, B.Y.H.L and M.M.) are funded by the UK MRC Metabolic Disease Unit (MRC_MC_UU_12012/5), a Wellcome Trust Strategic Award (100574/Z/12/Z). The Disease Model Core is part of the MRC Metabolic Diseases Unit [MRC_MC_UU_12012/5] and Wellcome Trust Strategic Award [100574/Z/12/Z].
Footnotes
Author contributions
G.B. conceived the study, designed, performed and analysed the results of experiments; S.V. provided advice on design and discussion and analysed the lipidomic experiments; K.P. significantly helped with experiments; A.D. performed the bioinformatics pathway analysis; H.E.J. performed the helminths infection and collected the tissues; A-C.G. helped with the bioinformatics analysis; B.M.A scored the lungs histology; B.L. analysed the RNAseq; M.M. performed the library preparation; J.L. and M.D. provided technical assistance; S.C. provide expertise in the lipid synthesis assay; A.K. provide expertise on the manuscript; P.G.F. and A.N.J.M. provided expertise and designed of the helminth infection study; A.V-P supervised the entire study; and G.B., S.V. and A.V-P. prepared the manuscript. All authors contributed to and approved the final manuscript.
Declaration of interests
K.P. is currently employed by AstraZenaca. J.S-R. has received funding from GSK and Sanofi and consultant fees from Travere Therapeutics. All the other authors declare no competing interests.
Data and materials availability
All the data presented in this manuscript is available upon reasonable request to the corresponding authors. Transfer of materials requires materials transfer agreements. The RNA sequencing dataset is deposited in GEO under accession number GSE179066.
References
- 1.Murray PJ, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen F, et al. An essential role for TH2-type responses in limiting acute tissue damage during experimental helminth infection. Nat Med. 2012;18:260–266. doi: 10.1038/nm.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chawla A, Nguyen KD, Goh YP. Macrophage-mediated inflammation in metabolic disease. Nat Rev Immunol. 2011;11:738–749. doi: 10.1038/nri3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 5.O’Neill LA, Pearce EJ. Immunometabolism governs dendritic cell and macrophage function. J Exp Med. 2016;213:15–23. doi: 10.1084/jem.20151570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerrick KY, et al. Transcriptional profiling identifies novel regulators of macrophage polarization. PLoS One. 2018;13:e0208602. doi: 10.1371/journal.pone.0208602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Czimmerer Z, et al. The Transcription Factor STAT6 Mediates Direct Repression of Inflammatory Enhancers and Limits Activation of Alternatively Polarized Macrophages. Immunity. 2018;48:75–90.:e76. doi: 10.1016/j.immuni.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byles V, et al. The TSC-mTOR pathway regulates macrophage polarization. Nat Commun. 2013;4:2834. doi: 10.1038/ncomms3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, et al. AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab. 2011;13:376–388. doi: 10.1016/j.cmet.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walker AK, et al. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev. 2010;24:1403–1417. doi: 10.1101/gad.1901210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haschemi A, et al. The sedoheptulose kinase CARKL directs macrophage polarization through control of glucose metabolism. Cell Metab. 2012;15:813–826. doi: 10.1016/j.cmet.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shay AE, et al. IL-4 up-regulates cyclooxygenase-1 expression in macrophages. J Biol Chem. 2017;292:14544–14555. doi: 10.1074/jbc.M117.785014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.York AG, et al. Limiting Cholesterol Biosynthetic Flux Spontaneously Engages Type I IFN Signaling. Cell. 2015;163:1716–1729. doi: 10.1016/j.cell.2015.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun LP, Seemann J, Goldstein JL, Brown MS. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Insig renders sorting signal in Scap inaccessible to COPII proteins. Proc Natl Acad Sci U S A. 2007;104:6519–6526. doi: 10.1073/pnas.0700907104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neill DR, et al. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature. 2010;464:1367–1370. doi: 10.1038/nature08900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allen JE, Sutherland TE. Host protective roles of type 2 immunity: parasite killing and tissue repair, flip sides of the same coin. Semin Immunol. 2014;26:329–340. doi: 10.1016/j.smim.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen C, Wu D, Zhang L, Zhao Y, Guo L. Comparative phosphoproteomics studies of macrophage response to bacterial virulence effectors. J Proteomics. 2012;77:251–261. doi: 10.1016/j.jprot.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 18.Hastings RH, Folkesson HG, Matthay MA. Mechanisms of alveolar protein clearance in the intact lung. Am J Physiol Lung Cell Mol Physiol. 2004;286:L679–689. doi: 10.1152/ajplung.00205.2003. [DOI] [PubMed] [Google Scholar]
- 19.Berthiaume Y, Albertine KH, Grady M, Fick G, Matthay MA. Protein clearance from the air spaces and lungs of unanesthetized sheep over 144 h. J Appl Physiol (1985) 1989;67:1887–1897. doi: 10.1152/jappl.1989.67.5.1887. [DOI] [PubMed] [Google Scholar]
- 20.Chen F, et al. Monocyte-derived alveolar macrophages mediate resistance to migrating helminths through depletion of arginine availability. bioRxiv. 2020:2020.2010.2002.322388. doi: 10.1101/2020.10.02.322388. [DOI] [Google Scholar]
- 21.Shimano H, Sato R. SREBP-regulated lipid metabolism: convergent physiology - divergent pathophysiology. Nat Rev Endocrinol. 2017;13:710–730. doi: 10.1038/nrendo.2017.91. [DOI] [PubMed] [Google Scholar]
- 22.Wei X, et al. Fatty acid synthesis configures the plasma membrane for inflammation in diabetes. Nature. 2016;539:294–298. doi: 10.1038/nature20117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carroll RG, et al. An unexpected link between fatty acid synthase and cholesterol synthesis in proinflammatory macrophage activation. J Biol Chem. 2018;293:5509–5521. doi: 10.1074/jbc.RA118.001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griess B, Mir S, Datta K, Teoh-Fitzgerald M. Scavenging reactive oxygen species selectively inhibits M2 macrophage polarization and their pro-tumorigenic function in part, via Stat3 suppression. Free Radic Biol Med. 2020;147:48–60. doi: 10.1016/j.freeradbiomed.2019.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kapoor N, et al. Transcription factors STAT6 and KLF4 implement macrophage polarization via the dual catalytic powers of MCPIP. J Immunol. 2015;194:6011–6023. doi: 10.4049/jimmunol.1402797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He C, et al. Mitochondrial Cu,Zn-superoxide dismutase mediates pulmonary fibrosis by augmenting H2O2 generation. J Biol Chem. 2011;286:15597–15607. doi: 10.1074/jbc.M110.187377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He C, Ryan AJ, Murthy S, Carter AB. Accelerated development of pulmonary fibrosis via Cu,Zn-superoxide dismutase-induced alternative activation of macrophages. J Biol Chem. 2013;288:20745–20757. doi: 10.1074/jbc.M112.410720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, et al. ROS play a critical role in the differentiation of alternatively activated macrophages and the occurrence of tumor-associated macrophages. Cell Res. 2013;23:898–914. doi: 10.1038/cr.2013.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanin DE, et al. Mitochondrial Membrane Potential Regulates Nuclear Gene Expression in Macrophages Exposed to Prostaglandin E2. Immunity. 2018;49:1021–1033.:e1026. doi: 10.1016/j.immuni.2018.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang SC, et al. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat Immunol. 2014;15:846–855. doi: 10.1038/ni.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vats D, et al. Oxidative metabolism and PGC-1beta attenuate macrophage-mediated inflammation. Cell Metab. 2006;4:13–24. doi: 10.1016/j.cmet.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van den Bossche J, van der Windt GJW. Fatty Acid Oxidation in Macrophages and T Cells: Time for Reassessment? Cell Metab. 2018;28:538–540. doi: 10.1016/j.cmet.2018.09.018. [DOI] [PubMed] [Google Scholar]
- 33.Divakaruni AS, et al. Etomoxir Inhibits Macrophage Polarization by Disrupting CoA Homeostasis. Cell Metab. 2018;28:490–503.:e497. doi: 10.1016/j.cmet.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nomura M, et al. Fatty acid oxidation in macrophage polarization. Nat Immunol. 2016;17:216–217. doi: 10.1038/ni.3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Namgaladze D, Brune B. Fatty acid oxidation is dispensable for human macrophage IL-4-induced polarization. Biochim Biophys Acta. 2014;1841:1329–1335. doi: 10.1016/j.bbalip.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 36.Wang F, et al. Glycolytic Stimulation Is Not a Requirement for M2 Macrophage Differentiation. Cell Metab. 2018;28:463–475.:e464. doi: 10.1016/j.cmet.2018.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis CA, et al. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol Cell. 2014;55:253–263. doi: 10.1016/j.molcel.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rouzer CA, Scott WA, Griffith OW, Hamill AL, Cohn ZA. Glutathione metabolism in resting and phagocytizing peritoneal macrophages. J Biol Chem. 1982;257:2002–2008. [PubMed] [Google Scholar]
- 39.Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 40.Svensson T, et al. Coping strategies and cancer incidence and mortality: The Japan Public Health Center-based prospective study. Cancer Epidemiol. 2016;40:126–133. doi: 10.1016/j.canep.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 41.Liu C, et al. Treg Cells Promote the SREBP1-Dependent Metabolic Fitness of Tumor-Promoting Macrophages via Repression of CD8(+) T Cell-Derived Interferon-gamma. Immunity. 2019;51:381–397.:e386. doi: 10.1016/j.immuni.2019.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Agius L, Meredith EJ, Sherratt HS. Stereospecificity of the inhibition by etomoxir of fatty acid and cholesterol synthesis in isolated rat hepatocytes. Biochem Pharmacol. 1991;42:1717–1720. doi: 10.1016/0006-2952(91)90507-2. [DOI] [PubMed] [Google Scholar]
- 43.Oishi Y, et al. SREBP1 Contributes to Resolution of Pro-inflammatory TLR4 Signaling by Reprogramming Fatty Acid Metabolism. Cell Metab. 2017;25:412–427. doi: 10.1016/j.cmet.2016.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Im SS, et al. Linking lipid metabolism to the innate immune response in macrophages through sterol regulatory element binding protein-1a. Cell Metab. 2011;13:540–549. doi: 10.1016/j.cmet.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee JH, et al. SREBP-1a-stimulated lipid synthesis is required for macrophage phagocytosis downstream of TLR4-directed mTORC1. Proc Natl Acad Sci U S A. 2018;115:E12228–E12234. doi: 10.1073/pnas.1813458115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reboldi A, et al. Inflammation. 25-Hydroxycholesterol suppresses interleukin-1-driven inflammation downstream of type I interferon. Science. 2014;345:679–684. doi: 10.1126/science.1254790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo C, et al. Cholesterol Homeostatic Regulator SCAP-SREBP2 Integrates NLRP3 Inflammasome Activation and Cholesterol Biosynthetic Signaling in Macrophages. Immunity. 2018;49:842–856.:e847. doi: 10.1016/j.immuni.2018.08.021. [DOI] [PubMed] [Google Scholar]
- 48.Feingold KR, et al. Mechanisms of triglyceride accumulation in activated macrophages. J Leukoc Biol. 2012;92:829–839. doi: 10.1189/jlb.1111537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosas-Ballina M, Guan XL, Schmidt A, Bumann D. Classical Activation of Macrophages Leads to Lipid Droplet Formation Without de novo Fatty Acid Synthesis. Front Immunol. 2020;11:131. doi: 10.3389/fimmu.2020.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jenkins SJ, et al. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science. 2011;332:1284–1288. doi: 10.1126/science.1204351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pello OM, et al. Role of c-MYC in alternative activation of human macrophages and tumor-associated macrophage biology. Blood. 2012;119:411–421. doi: 10.1182/blood-2011-02-339911. [DOI] [PubMed] [Google Scholar]
- 52.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 53.Tan CY, et al. Brown Adipose Tissue Thermogenic Capacity Is Regulated by Elovl6. Cell Rep. 2015;13:2039–2047. doi: 10.1016/j.celrep.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim SK, Oh E, Yun M, Lee SB, Chae GT. Palmitate induces cisternal ER expansion via the activation of XBP-1/CCTalpha-mediated phospholipid accumulation in RAW 264.7 cells. Lipids Health Dis. 2015;14:73. doi: 10.1186/s12944-015-0077-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakano A, Harada T, Morikawa S, Kato Y. Expression of leukocyte common antigen (CD45) on various human leukemia/lymphoma cell lines. Acta Pathol Jpn. 1990;40:107–115. doi: 10.1111/j.1440-1827.1990.tb01549.x. [DOI] [PubMed] [Google Scholar]
- 56.Kopf M, Schneider C, Nobs SP. The development and function of lung-resident macrophages and dendritic cells. Nat Immunol. 2015;16:36–44. doi: 10.1038/ni.3052. [DOI] [PubMed] [Google Scholar]
- 57.Misharin AV, Morales-Nebreda L, Mutlu GM, Budinger GR, Perlman H. Flow cytometric analysis of macrophages and dendritic cell subsets in the mouse lung. Am J Respir Cell Mol Biol. 2013;49:503–510. doi: 10.1165/rcmb.2013-0086MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guth AM, et al. Lung environment determines unique phenotype of alveolar macrophages. Am J Physiol Lung Cell Mol Physiol. 2009;296:L936–946. doi: 10.1152/ajplung.90625.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hussell T, Bell TJ. Alveolar macrophages: plasticity in a tissue-specific context. Nat Rev Immunol. 2014;14:81–93. doi: 10.1038/nri3600. [DOI] [PubMed] [Google Scholar]
- 60.Austyn JM, Gordon S. F4/80, a monoclonal antibody directed specifically against the mouse macrophage. Eur J Immunol. 1981;11:805–815. doi: 10.1002/eji.1830111013. [DOI] [PubMed] [Google Scholar]
- 61.Feng YH, Mao H. Expression and preliminary functional analysis of Siglec-F on mouse macrophages. J Zhejiang Univ Sci B. 2012;13:386–394. doi: 10.1631/jzus.B1100218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Azzu V, et al. Suppression of insulin-induced gene 1 (INSIG1) function promotes hepatic lipid remodelling and restrains NASH progression. Mol Metab. 2021:101210. doi: 10.1016/j.molmet.2021.101210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matute-Bello G, et al. An official American Thoracic Society workshop report: features and measurements of experimental acute lung injury in animals. Am J Respir Cell Mol Biol. 2011;44:725–738. doi: 10.1165/rcmb.2009-0210ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jenkins B, Ronis M, Koulman A. LC-MS Lipidomics: Exploiting a Simple High-Throughput Method for the Comprehensive Extraction of Lipids in a Ruminant Fat Dose-Response Study. Metabolites. 2020;10 doi: 10.3390/metabo10070296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen L, et al. NADPH production by the oxidative pentose-phosphate pathway supports folate metabolism. Nat Metab. 2019;1:404–415. [PMC free article] [PubMed] [Google Scholar]
- 66.Lewis CA, et al. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol Cell. 2014;55:253–263. doi: 10.1016/j.molcel.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(Suppl 1):S96–104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 69.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 70.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Varemo L, Nielsen J, Nookaew I. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. 2013;41:4378–4391. doi: 10.1093/nar/gkt111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gerrick KY, et al. Transcriptional profiling identifies novel regulators of macrophage polarization. PLoS One. 2018;13:e0208602. doi: 10.1371/journal.pone.0208602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data presented in this manuscript is available upon reasonable request to the corresponding authors. Transfer of materials requires materials transfer agreements. The RNA sequencing dataset is deposited in GEO under accession number GSE179066.