Abstract
HUWE1 is a universal quality-control E3 ligase that marks diverse client proteins for proteasomal degradation. Although the giant HECT enzyme is an essential component of the ubiquitin-proteasome system closely linked with severe human diseases, its molecular mechanism is little understood. Here, we present the crystal structure of Nematocida HUWE1, revealing how a single E3 enzyme has specificity for a multitude of unrelated substrates. The protein adopts a remarkable snake-like structure where the C-terminal HECT domain heads an extended alpha solenoid body that coils in on itself and houses various protein-protein interaction modules. Our integrative structural analysis shows that this ring structure is highly dynamic, enabling the flexible HECT domain to reach protein targets presented by the various acceptor sites. Together, our data demonstrate how HUWE1 is regulated by its unique structure, adapting a promiscuous E3 ligase to selectively target unassembled orphan proteins.
Introduction
E3 ligases regulate diverse cellular processes by conjugating ubiquitin, a small protein post-translational modifier, to lysines of target proteins1. Not only do E3s have the critical role of regulating which proteins are ubiquitinated and when, but they also team up with specific partner E2s to determine whether a single ubiquitin or different types of polyubiquitin chain are attached, where the linkage type determines the function of the mark2. Many E3 ligases attach K48-linked chains to target proteins for degradation, either as a form of regulation or for general protein quality control. Other linkage types and monoubiquitination have a range of functions, for example in DNA repair, the immune system and protein trafficking. Thus to understand a vast swathe of biology, we need to understand the specificity and mechanisms of E3 ligases.
There are over 600 E3 ligases in humans, with some of the better understood ubiquitination enzymes belonging to the RING family, including SCF ligases3 and the anaphase promoting complex4. These are often large modular complexes with different subunits to specify the target and regulate activity. In contrast, ubiquitin ligases of the HECT family usually act as a single polypeptide chain, where the catalytic HECT domain is at the very C-terminus and the rest of the protein has a regulatory and substrate recognition role5,6. The HECT E3 ligases are often very large, ranging from 700 amino acids to giant enzymes of more than 3000 residues. Owing to their large size, HECT enzymes are less well characterized than their RING counterparts with regards to molecular mechanism, target selection and regulation7. Indeed, there is no structure of any full-length HECT E3 ligase meaning that a critically important part of our biological understanding is missing.
One of the largest human HECT E3 ligases is HUWE1 (4374 residues, 482 kDa), an essential protein which regulates a myriad of biological processes through ubiquitination8 (Fig. 1a). The giant E3 initially gained the spotlight as a specific regulator of many key cellular factors, although the exact ubiquitin chain-types and downstream effects are not always clear. The targeted substrates include apoptosis factors like Mcl-19, Miz110 and p5311, proliferation and developmental factors like c-Myc12, Ascl-113, FOXP314 and Dvl15, and DNA replication and repair proteins like Cdc616, Pol β17 and BRCA118. Moreover, it is increasingly clear that HUWE1 also has a general protein quality control function, whereby it corrects proteome imbalances by acting in the unassembled soluble protein degradation pathway (USPD,19). Here, the E3 enzyme targets ‘orphan’ proteins, those missing their complex partners, for proteasomal degradation, particularly the unassembled subunits of macromolecular assemblies, like ribosomes20 or nucleosomes21,22. Such proteome imbalances are likely to occur in cancer cells23, most notably in aneuploid cells24. Considering that many HUWE1 targets are involved in cancer-related processes and that HUWE1 itself is frequently mutated in cancers25, the giant HECT enzyme is a highly attractive target for therapeutic intervention26. Additionally, mutation of HUWE1 is associated with severe X-linked intellectual disability27, further highlighting the urgent need to unravel the function of this enigmatic E3 ligase at a molecular level.
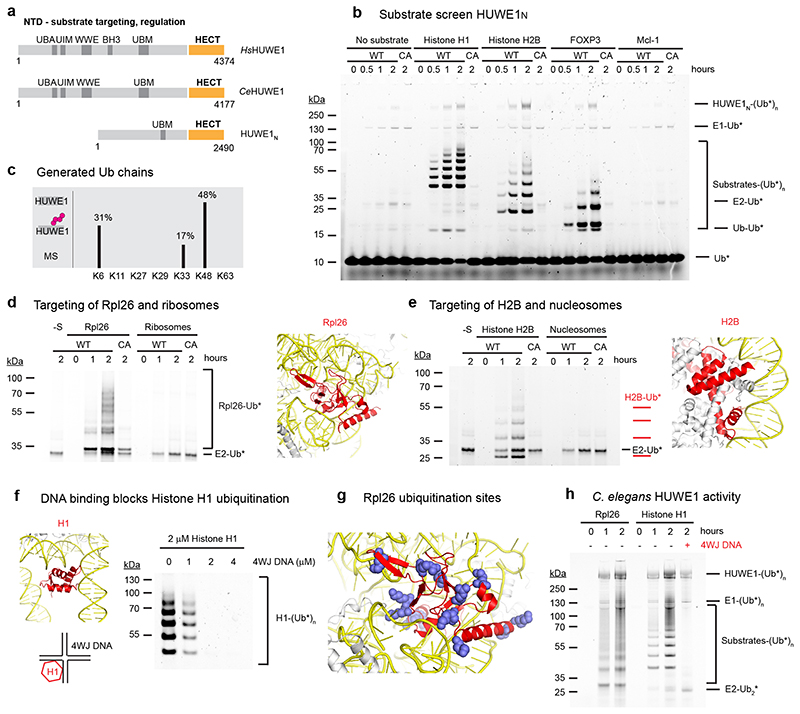
Figure 1. HUWE1N targets orphan proteins for degradation.
(a) Cartoon depicting the domain organization and functional roles of HUWE1. (b) HUWE1N substrate screen. E1-E2-E3 ubiquitination assays were performed using 4 μM of the indicated substrates and either wild-type HUWE1N or the inactive HUWE1N-CA variant. The ubiquitinated products were visualized by using ubiquitin labeled with the DyLight488 fluorophore. (c) Linkage specificity of HUWE1N as seen in autoubiquitination. Ubiquitin chains with the indicated linkage types were detected by mass spectrometry. The ratio of ubiquitinated to total detected peptides for each linkage type is shown as a percentage (d-e) Ubiquitination of an orphan ribosomal subunit and an orphan nucleosome subunit, respectively. Assays were performed with 500 nM substrate and only show the region of the gel containing ubiquitinated substrates, which were visualized with Ub-DyLight800 in (c) and Ub-DyLight488 in (d). The position of Rpl26 (colored red) within the ribosome (PDB 6T59) and histone H2B (colored red) within the nucleosome (PDB 3AFA) are displayed with nucleic acids colored yellow. (f) Histone H1 ubiquitination is blocked by 4WJ DNA. Assays were performed for 120 minutes. (g) An Rpl26 ubiquitination reaction was analyzed by mass spectrometry. Detected ubiquitinated lysines are indicated in the context of the ribosome (PDB 6T59). (h) Ubiquitination of orphan proteins by CeHUWE1, using 2 μM substrate and 4 μM 4WJ DNA in the last lane, and visualized with Ub-DyLight800.
It is puzzling how one polypeptide can regulate so many distinct targets, ranging from specific proliferation factors to theoretically any misassembled complex subunit. The huge E3 portion preceding the HECT domain is likely to hold the answer to this, but is currently largely uncharacterized except for a few known protein-interaction modules, such as a poly-ADP-ribose interaction WWE domain (PDB 6MIW), various ubiquitin binding domains (PDB 2MUL, 2EKK) and an Mcl-1 interacting BH3 motif (PDB 5C6H). In order to understand the promiscuous specificity and the regulation of HUWE1, we used the compact protein from Nematocida as a model system (HUWE1N, 2490 residues, 287 kDa), reconstituted the full-length protein in functional form and determined its crystal structure. The full-length structure of HUWE1 reveals the bipartite organization of the giant E3 ubiquitin ligase, which is composed of a catalytic HECT domain and a large, ring-shaped scaffold defining substrate specificity and E3 activity. Our data suggest that an evolutionarily conserved function of HUWE1 is as a quality control enzyme for misassembled complexes, and we show which structural features of HUWE1 are required for targeting substrates and regulating the HECT domain.
Results
Nematocida HUWE1N as model system
Due to its large size, human HUWE1 is a challenging protein for biochemical and structural analysis. Thus, most in vitro studies so far are limited to its isolated C-terminal HECT domain, a small fraction of the full protein (40 kDa from 482 kDa). To obtain well-behaving full-length HUWE1 we searched for a more compact variant. The smaller 374 kDa budding yeast orthologue, Tom1, has been shown to have similar functions as HUWE1 in marking DNA replication factors, histone proteins and unassembled ribosomal proteins for proteasomal degradation16,20,22. We found an even shorter fungal orthologue, HUWE1N (287 kDa), from the parasite Nematocida sp. ERTm5. Full-length HUWE1N could be produced in large amounts and in a functional form in insect cells, enabling a detailed structure-function analysis.
To demonstrate that HUWE1N is a bona fide orthologue of human HUWE1, we tested its activity on a panel of known in vivo substrates (Fig. 1b). In the presence of UbcH5b, HUWE1N strongly ubiquitinated the transcription factor FOXP3, histone H1 and histone H2B, generating ladders of ubiquitinated species in each case. No activity was seen against Mcl-1, consistent with the lack of the BH3 domain implicated in Mcl-1 binding9 (Fig. 1a). A HUWE1N-CA variant with the catalytic cysteine mutated was inactive, confirming the activity we observed was due to specific E3 activity (Fig. 1b). Autoubiquitination by HUWE1N as well as free di-ubiquitin generation were rather weak, but these activities were stimulated by addition of substrate, hinting that the catalytic activity of HUWE1N is regulated by substrate binding (Fig. 1b). To determine the types of ubiquitin chains generated by HUWE1N we performed a mass spectrometry analysis of the ubiquitinated products (Fig. 1c). These data reveal the predominance of K48, K6 and K33-linked chains, consistent with previous reports for human HUWE128,29, and suggesting that ubiquitination by HUWE1N has a degradative function.
The best substrates that we identified in a screen of reported in vivo targets were positively charged proteins with an isoelectric point (pI) > 9, and missing their complex partners. To test whether HUWE1N recognizes the orphan nature of these proteins, we first compared ubiquitination of the isolated ribosomal protein Rpl2620 with intact ribosomes. At the same molar concentration of substrate, we observed clear ubiquitination of the orphan subunit, but no ubiquitination of proteins integrated into the ribosome (Fig. 1d). Similarly, histone H2B was readily ubiquitinated but intact nucleosomes were not (Fig. 1e). We next directly tested whether HUWE1N recognizes surfaces normally blocked by protein or nucleic acid interactions using the simple system of histone H1, which is integrated into the nucleosome only through protein-DNA interactions (Fig. 1f). Four-way junction (4WJ) DNA mimics this interaction with sub-nanomolar affinity30. We titrated this into the reaction and observed complete loss of histone H1 ubiquitination by HUWE1N at a 1:1 ratio of histone H1 and 4WJ DNA (Fig. 1f). Thus, HUWE1N recognises portions of histone H1 which are normally shielded by DNA. To provide further evidence of this targeting mechanism, we mapped the ubiquitination sites of Rpl26 by mass spectrometry (Fig. 1g). These data reveal that many of the modified lysines would be occluded within the context of the intact ribosome and are thus only accessible for the dissociated Rpl26. Finally, we tested whether these substrate recognition principles are conserved in metazoan HUWE1. In these experiments, we used the C. elegans orthologue (CeHUWE1), which could be functionally produced using insect cell expression, as a model system. CeHUWE1 also strongly ubiquitinated Rpl26 and histone H1, and the ubiquitination of histone H1 was also blocked by 4WJ DNA (Fig. 1h). In conclusion, our data show that the compact Nematocida HUWE1N is an excellent model for full-length metazoan HUWE1, and that one evolutionary conserved function of this giant enzyme is to mark unassembled orphan proteins for degradation.
Determining the crystal structure of HUWE1N
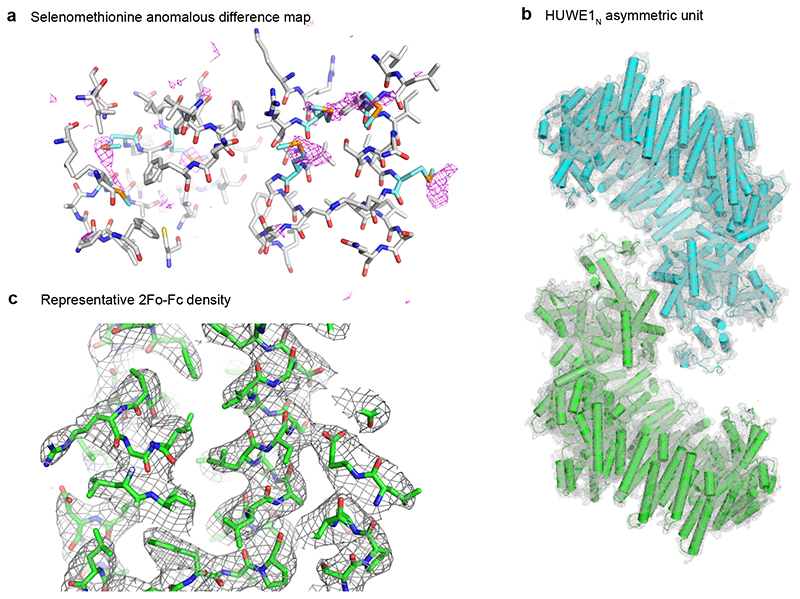
To reveal how the structurally uncharacterized 2100 amino acid N-terminal region of HUWE1N contribute to E3 function and regulation, we crystallized full-length Huwe1N-CA and obtained diffraction data to 3.05 Å resolution (Supplementary Table 1). We were able to partially solve the phase problem by molecular replacement using the human HUWE1 HECT domain as search model. To build the backbone of the large N-terminal region, we used an experimental density map that we obtained by collecting anomalous diffraction data from crystals soaked with Ta6Br14 clusters or grown with selenomethionine substituted protein. Guided by the numerous Se sites identified in the anomalous map (Extended Data Fig. 1a), and contacts predicted by the RaptorX server31, we were able to build an atomic model of the two HUWE1N proteins present in the asymmetric unit of the HUWE1N crystals (Extended Data Fig. 1b). We could model almost the entire polypeptide chain, except two prolonged intrinsically disordered regions (IDR: residues 1442-1521 and 1576-1670), the UBM domain (1671-1764) and several flexible loops connecting helical repeats (4-13, 199-207, 1210-1239, 1828-1850 and 2483-2490). Ultimately, the crystal structure was refined to a final Rfree of 23.3% and Molprobity score of 2.3532. The final electron density was of excellent quality, revealing side-chain conformations for most residues (Extended Data Fig. 1c, Supplementary Video 1).
HUWE1 is a ring forming HECT ubiquitin ligase
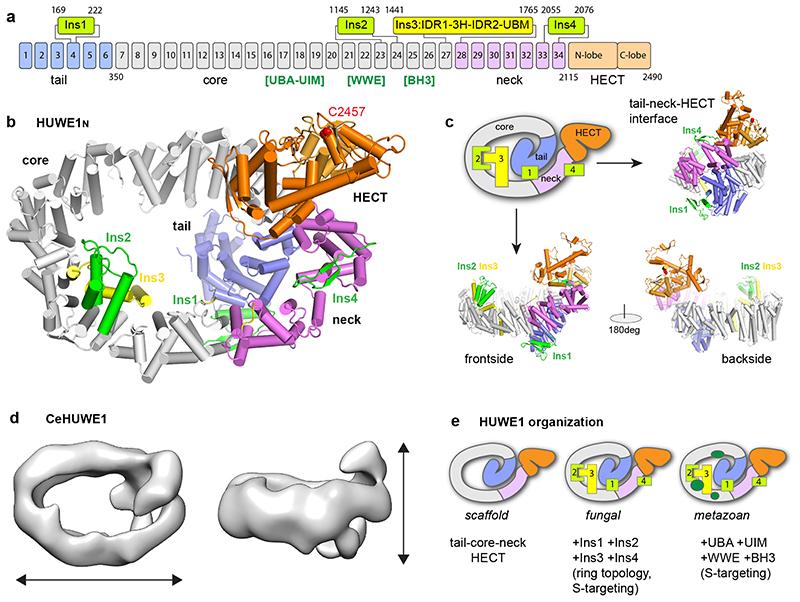
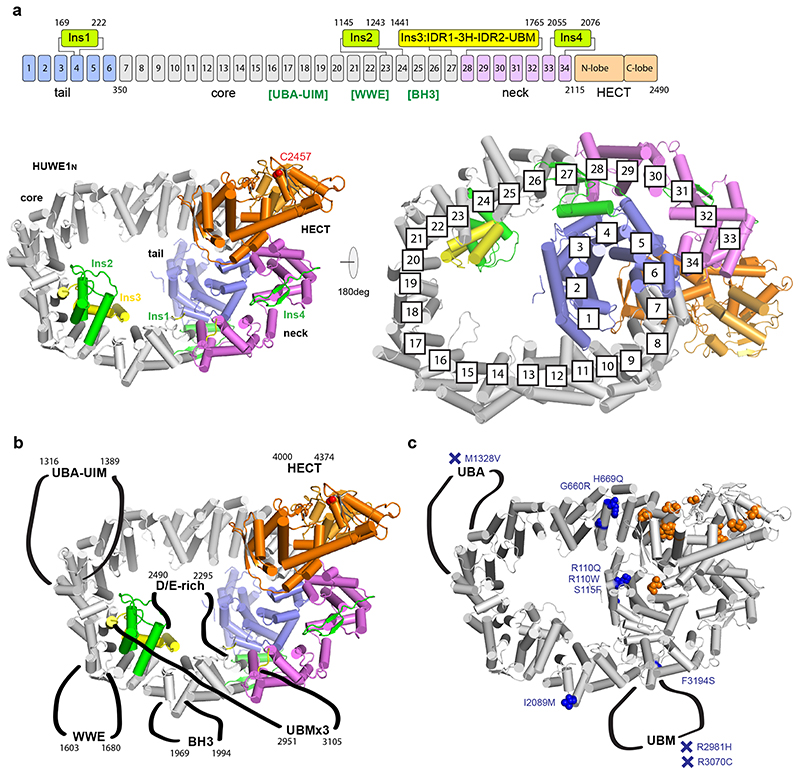
In our crystal structure, HUWE1N adopts a snake-like scaffold composed of an N-terminal tail (residues 1-349), an elongated body (350-1441), a neck region (1765-2114) and the head - the HECT domain (2115-2490) (Fig. 2a). The tail, body and neck, preceding the HECT domain, together form an α-solenoid structure, composed of 34 Armadillo-like (ARM) repeats (Extended Data Fig. 2a). This coils in on itself to form a giant ellipsoidal ring, measuring approximately 140 Å by 90 Å, and an inner diameter of about 60 Å (Fig. 2bc). Rather than being a simple repeat structure, the α-solenoid is distorted and decorated by four insertions (Ins1-4) (Fig. 2a-c). The HECT is connected to the solenoid through the deformed final helix of repeat ARM34, which packs loosely on helix α1 of the HECT domain.
Figure 2. Overall architecture of the giant E3 ligase HUWE1N.
(a) Domain architecture of HUWE1N. ARM repeats 1-34 are numbered, with the four insertions indicated. The position of human HUWE1 insertions, absent in HUWE1N, are shown in square brackets. (b-c) Crystal structure of HUWE1N. The structure is shown in cartoon representation from four different views, using the same color code as in a (catalytic Cys in red). A schematic cartoon illustrates the snake-like organization of the E3 ligase. (d) Negative stain EM analysis of CeHUWE1. The obtained EM density is shown from two viewpoints with approximate dimensions indicated. HECT and tail domains are labelled based on homology to HUWE1N. (e) Organization and increasing complexity of HUWE1.
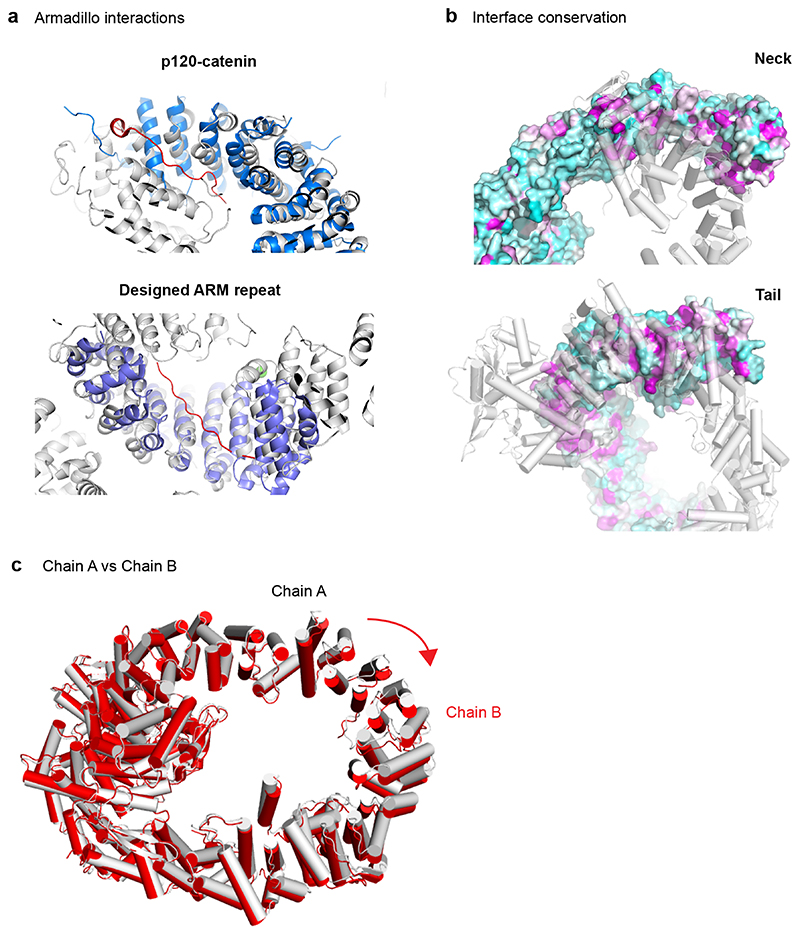
Considering the large difference in sequence length, we also tested if metazoan HUWE1 has a similar architecture. For this, we performed negative stain EM analysis on the 4177 amino acid CeHUWE1. The resulting low-resolution 3D model showed high similarity to Huwe1N in both its dimensions and overall shape (Fig. 2d). Likewise, the recently reported cryo-EM structure of human HUWE1 shows a ring-shaped structure with similar diameter and domain architecture33. We thus propose that the core structure of HUWE1 is conserved and that the additional 1700-2000 residues of metazoan orthologues constitute flexible appendices. The insertions include several previously characterised domains of human HUWE1 which are not present in Huwe1N: the ubiquitin association domain (UBA) and the ubiquitin interaction motif (UIM), a polyADPribose-interacting WWE domain and an Mcl-1 interacting BH3 motif. Considering that these features would be positioned around the solenoid body of HUWE1 (Fig. 2e, Extended Data Fig. 2b), an appealing mechanism emerges for how HUWE1 may function: the ring could act as a substrate binding platform decorated by different docking sites, which could feed proteins to the HECT domain. Additionally, ARM repeat proteins frequently bind peptides within their interior34,35 (Extended Data Fig. 3a), suggesting that the HUWE1 solenoid itself could act as a generic interaction surface. In support of this model, reported intellectual disability mutations are not just located on the HECT domain, but also on the solenoid and the inserted domains (Extended Data Fig. 2c).
Functional insertions within the HUWE1N ring
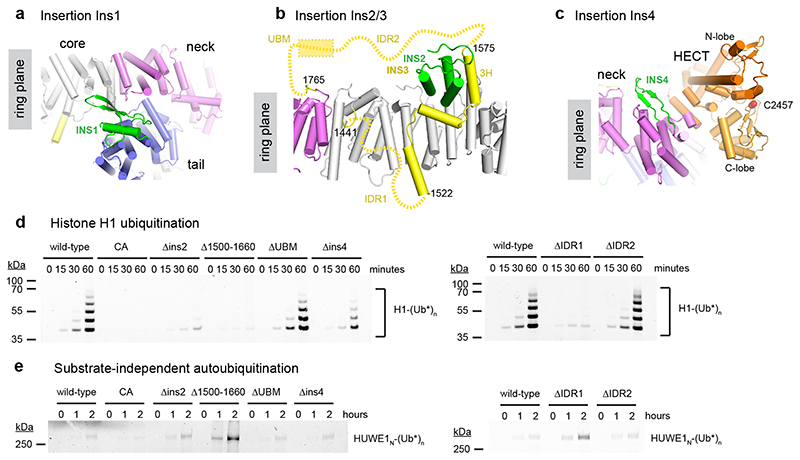
The four insertions within the solenoid form unusual structures which link together different parts of HUWE1N (Fig. 2b, 3a-c). Insertion 1 (Ins1) causes a sharp bend of the solenoid structure. It consists of a helix and flexible β-hairpin-like loops between ARM4 and ARM5 (Fig. 3a) and forms part of the interface between the tail and neck. Ins2 is a three-helix bundle between ARM23 and ARM24 resting on top of the solenoid on the opposite side of the ring relative to the HECT domain (Fig. 3b). Curiously, Ins3 (residues 1442-1764), positioned between ARM27 and ARM28, backtracks three ARM repeats, such that three linearly arranged helices (Ins3-3H) can interact closely with Ins2 and the ARM repeats beneath, yielding a composite portion of the solenoid backbone (Fig. 3b). Ins3-3H is sandwiched between two conserved highly acidic unstructured regions. Ins3-IDR1 runs from 1442-1521, contains 41% Glu/Asp residues and has a predicted pI of 2.7, while Ins3-IDR2 is between 1576-1670 and contains 45% Glu/Asp contributing to a pI of 3.6. At the C-terminal end of Ins3 lies the unresolved ubiquitin binding UBM domain (Ins3-UBM, 1671-1764). The “empty” ring-like structure as observed in the electron density map may be misleading. The unusual topology of Ins3 (Fig. 3b), crossing the plane of the solenoid in a defined manner, implies that the ring is filled with the disordered IDR1 and IDR2 segments generating a 173-residue mesh of negatively charged residues in the center of the HUWE1 solenoid. The final insertion, Ins4 (residues 2055-2076), forms a β-hairpin between ARM33 and ARM34, protruding towards the HECT domain (Fig. 3c).
Figure 3. Functional insertions within the HUWE1N ring.
(abc) Molecular details of insertions Ins1, Ins2, Ins3 and Ins4 within the alpha-solenoid. (d) Histone H1 ubiquitination assay of the HUWE1N insertion deletion variants. Ubiquitination of 4 μM histone H1 was visualized by fluorescence with Ub-DyLight488. (e) Autoubiquitination activity of the HUWE1N variants in the absence of a substrate. Only the region of the gel with HUWE1N-Ub* band is shown.
To determine whether these inserted features contribute to the specificity or regulation of the HECT domain, we generated four deletion variants to remove the inserts without affecting the Armadillo backbone – HUWE1N-ΔIns2 (1146-1243 replaced by GSSG), HUWE1N-ΔIns4 (2057-2070 replaced by GSSG), HUWE1N-Δ1500-1660 (removing ins3-3H, and parts of the two IDRs) and HUWE1N-ΔUBM (1671-1750 replaced by GGSGG). We first tested the variants for specific substrate ubiquitination using histone H1, as well as for presumably unspecific autoubiquitination (Fig. 3d). The Ins4 and UBM deletions had only minor effects on ubiquitination in these assays. In contrast, both the HUWE1N-Δ1500-1660 and the HUWE1N-ΔIns2 variants showed much lower histone H1 ubiquitination activity than the wild-type enzyme. This was not due to a decrease in the catalytic activity of these variants, because their substrate-independent autoubiquitination activity was not reduced. In fact, this activity was much stronger for HUWE1N-Δ1500-1660 than for the wild-type E3 (Fig. 3e). Together, these data suggest that the topologically unusual interaction between Ins2 and Ins3 is crucial for both the specific recognition of substrates and for inhibiting the basal autoubiquitination activity of the enzyme. This could be either because the Ins2-Ins3 interaction maintains the integrity of the ring structure or because it correctly positions the acidic IDR1/IDR2 regions. We next replaced either IDR1 (1442-1522) or IDR2 (1610-1670) with an 18-residue glycine-serine linker to probe the importance of these regions without disrupting the ins2-ins3 interaction. While IDR2 deletion showed no effect, the ΔIDR1 variant displayed greatly reduced histone H1 ubiquitination and increased autoubiquitination (Fig. 3de). Thus, IDR1 plays an important role in orphan substrate recognition and HECT regulation.
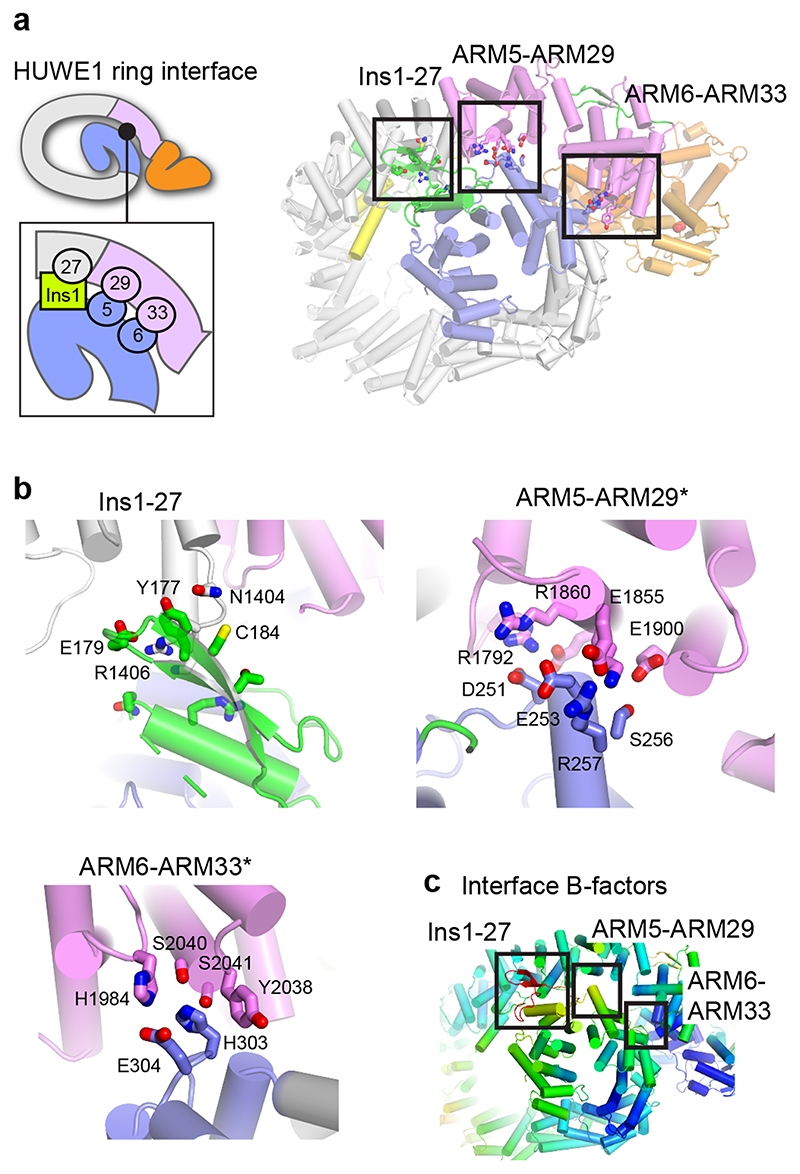
Dynamic interfaces stabilize the HUWE1N ring
Unlike most α-solenoid proteins, Huwe1N coils tightly enough to form a closed ring with three non-contiguous contacts between the N-terminal tail and C-terminal neck portions (Fig. 2b, 4a): The Ins1-ARM27, ARM5-ARM29 and ARM6-ARM33 interfaces are small (total surface area of 654 Å2), contain few specific interactions, and involve loop regions with elevated thermal motion factors (B-factors), suggesting that the interfaces are rather dynamic and flexible (Fig. 4bc). The small Ins1-ARM27 interface mostly consists of van der Waals contacts that occur between aliphatic side chains of various residues, organized around Y177 (Fig. 4b). However, this site may also comprise additional regions not well resolved in our structure, such as residues 1828-1850. The ARM5-ARM29 interface is larger and established by electrostatic interactions with potential salt bridges between R257 and E1855 plus D251 and R1792, as well as putative hydrogen bonding interactions between S252 and E1863 plus S256 and E1900 (Fig. 4b). The most clearly resolved interaction is between ARM6 and ARM33 and involves H303 hydrogen bonding with S2040, stacking interactions with Y2038 and a probable salt bridge between E304 and H1984 (Fig. 4b).
Figure 4. Dynamic interfaces stabilize the HUWE1N ring.
(a) Overview of the interface between the tail and neck regions of HUWE1N. A cartoon schematic illustrates the three main interacting surfaces, highlighted in the right panel. (b) Detailed view of the molecular interactions at each of the three interfaces, which close the HUWE1N ring. (c) HUWE1N colored by B-factor, from low (blue) to high (red), with the three interfaces highlighted. The average B-factor of the HUWE1N crystal structure is 105 Å2.
To dissect the function of these interfaces, which are highly conserved, (Extended Data Fig. 3b), we designed site-specific mutations to disrupt them. However, both the variants we attempted to purify, H303A/E304A and D251R, were unstable HUWE1N proteins prone to precipitation. This suggests that closure of the ring through these interfaces is necessary for stabilizing the HUWE1N structure.
Dynamics of substrate-binding ring and attached HECT domain
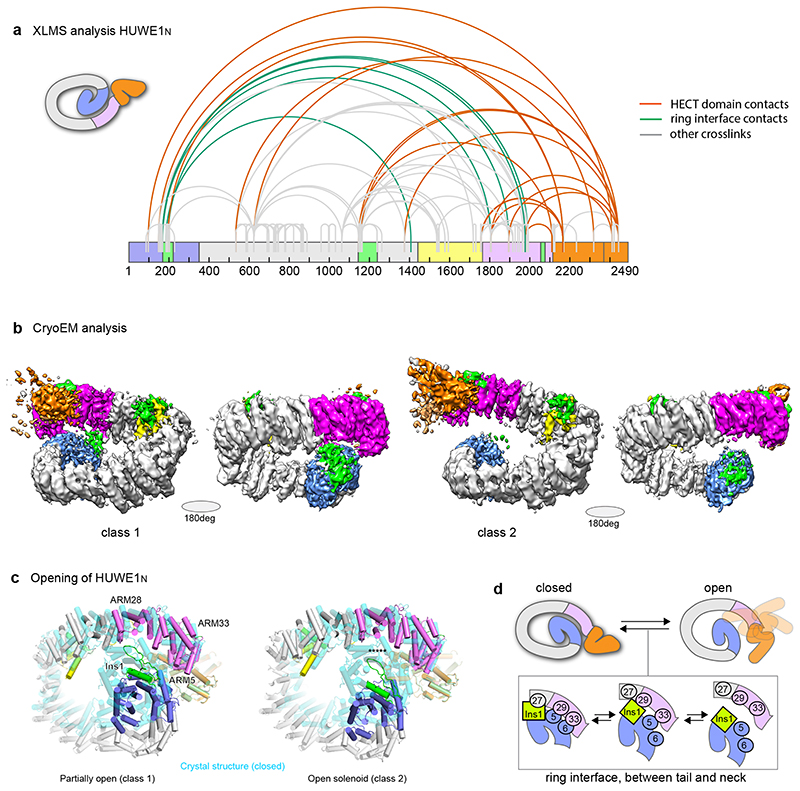
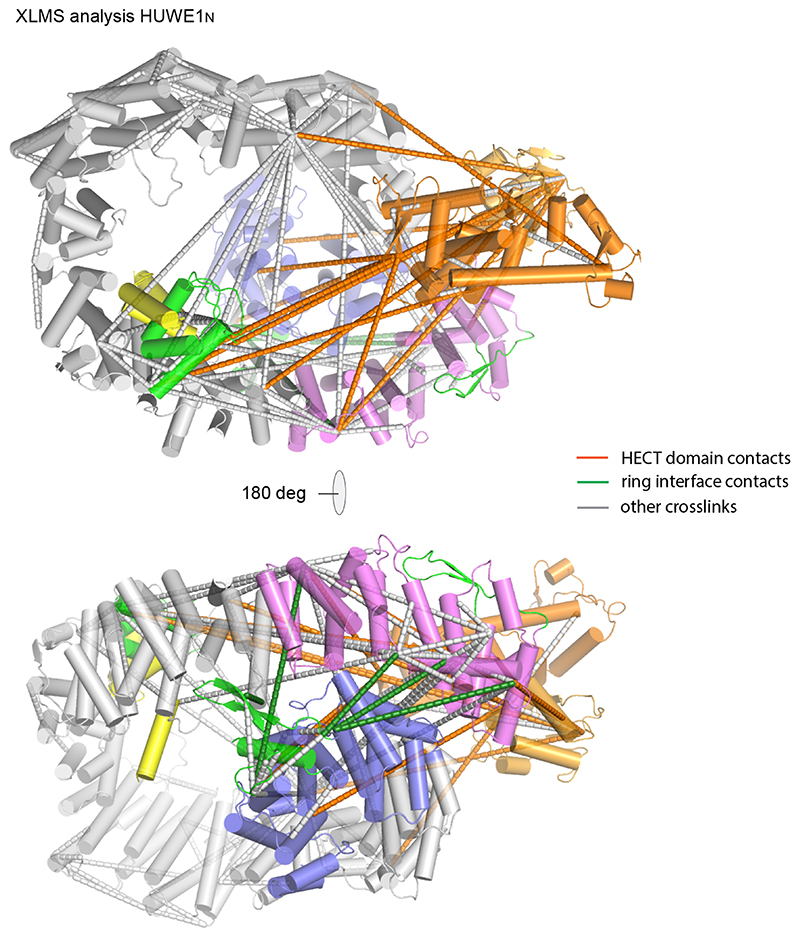
For the HECT domain to ubiquitinate substrates bound to different parts of the ring, a high degree of flexibility would be required. Indeed, comparison of the two copies of HUWE1N in the asymmetric unit of the protein crystals shows a certain breathability of the ARM repeats (Extended Data Fig. 3c). Therefore, we sought to determine how this structure behaves in solution. Cross-linking mass spectrometry of HUWE1N resulted in several cross-links between the N-terminus and C-terminus of the protein (Fig. 5a, Extended Data Fig. 4), showing that the ring-like structure is the prevalent conformation in solution. Additionally, cross-links were detected between Ins2 and Ins3-3H, confirming the composite topology of this region. Notably, the C-lobe of the HECT domain, housing the catalytic cysteine, formed cross-links with sites throughout the solenoid ring (Fig. 5a, Extended Data Fig. 4). This suggests that the HECT domain has sufficient flexibility to reach substrates bound to any part of the solenoid.
Figure 5. Solution behavior of HUWE1N.
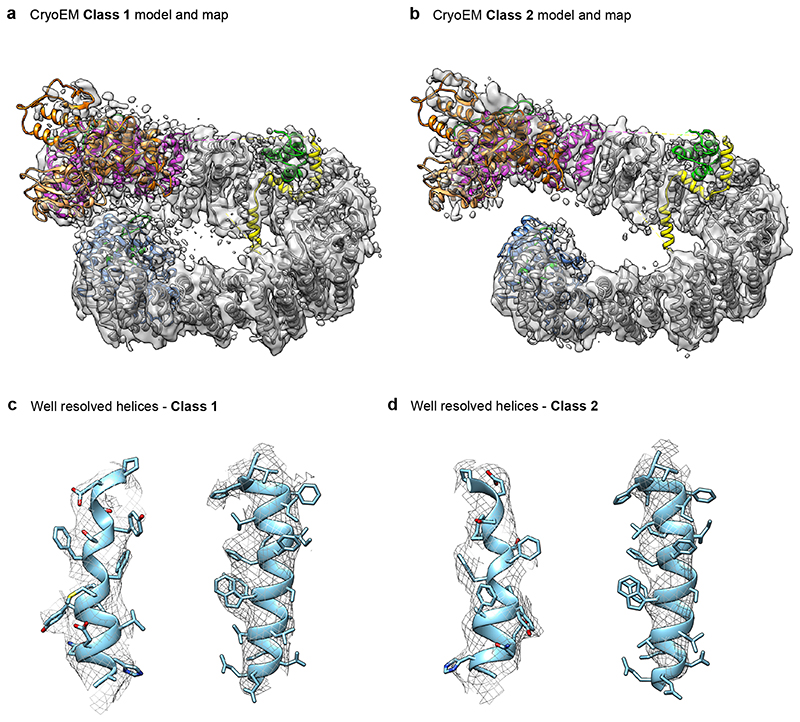
(a) XL-MS analysis of HUWE1N. The highest confidence detected cross-links are displayed on the primary structure of HUWE1N. Cross-links between the neck and tail, supporting the closed ring structure, are shown in green, while cross-links between the HECT domain and the solenoid ring are shown in orange and all other cross-links in gray. (b) Sharpened electron density maps of the two CryoEM classes in two different views showing different degrees of ring opening. Maps are colored according to the schematic in Fig. 2a. (c) Breaking and shifting the ring-closure interfaces. The neck and tail contacts are compared between the crystal structure (cyan) and two cryo-EM classes (domain-based color code). The differences are summarized in a schematic cartoon (d).
To probe the conformational flexibility of HUWE1N at higher resolution we turned to cryo-EM. Classification of the particles revealed that the E3 ligase is highly dynamic in its ring structure and adopts a continuum of coiled conformations ranging from closed to fully open (Supplementary Figure 1). We refined two classes at 5.3 Å (Class 1) and 6.4 Å (Class 2) resolution (Fig. 5b, Supplementary Table 2), allowing us to unambiguously dock the secondary structure of our model (Extended Data Fig. 5). Compared to the crystal structure, the cryo-EM structures exhibited a striking relaxation over the entire length of the solenoid, leading to breaks and shifts in the interfaces between the neck and tail (Fig. 5cd). In the slightly opened Class 1, Ins1 still stabilizes the ring-like structure but now its interaction has been shifted from ARM27 to ARM28. Further along, ARM5 now interacts with ARM33 instead of ARM29, while ARM6 has lost all interactions. In contrast, in Class 2, the solenoid is opened and shows no visible contacts between tail and neck. Together, our structural data demonstrate a high level of plasticity and dynamicity in the intramolecular contacts which maintain the overall architecture of the ring but allow for structural flexibility. The adaptable ring structure is enabled by the small size of the interfaces, the lack of hydrophobic interactions and the flexibility of involved motifs (Fig. 4). This dynamicity could be critical to allow large-scale movements of the HECT domain and to properly position the various substrate binding sites. In fact, the inherent en-bloc flexibility of the HECT domain was clearly seen in the EM densities, as the domain was less well-resolved than the rest of the structure (Supplementary Figure 1, Supplementary Video 2, 3).
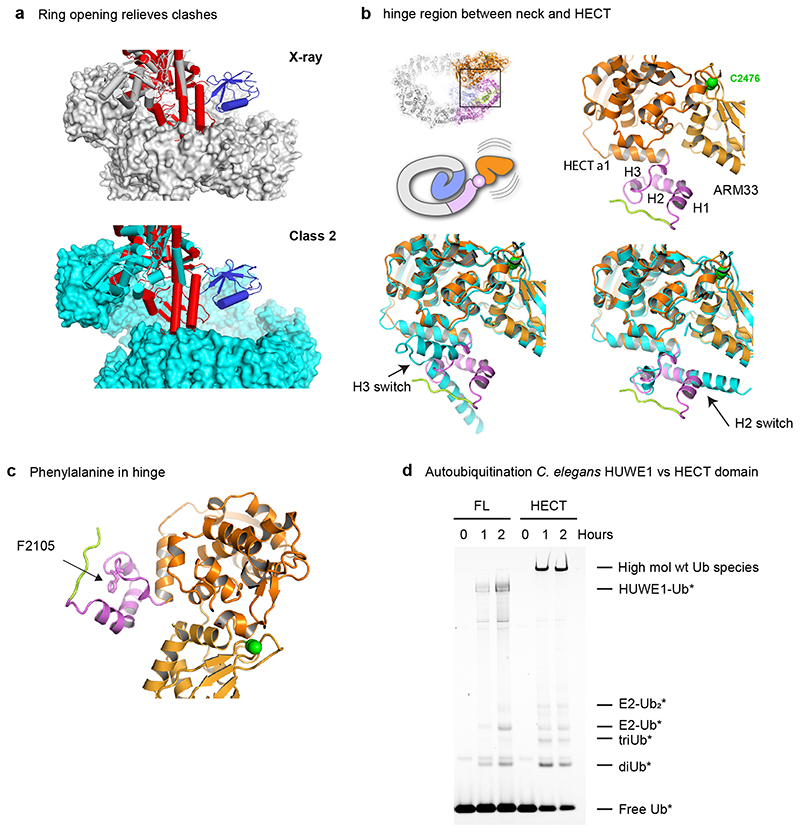
The HECT domain is inhibited in full-length HUWE1N
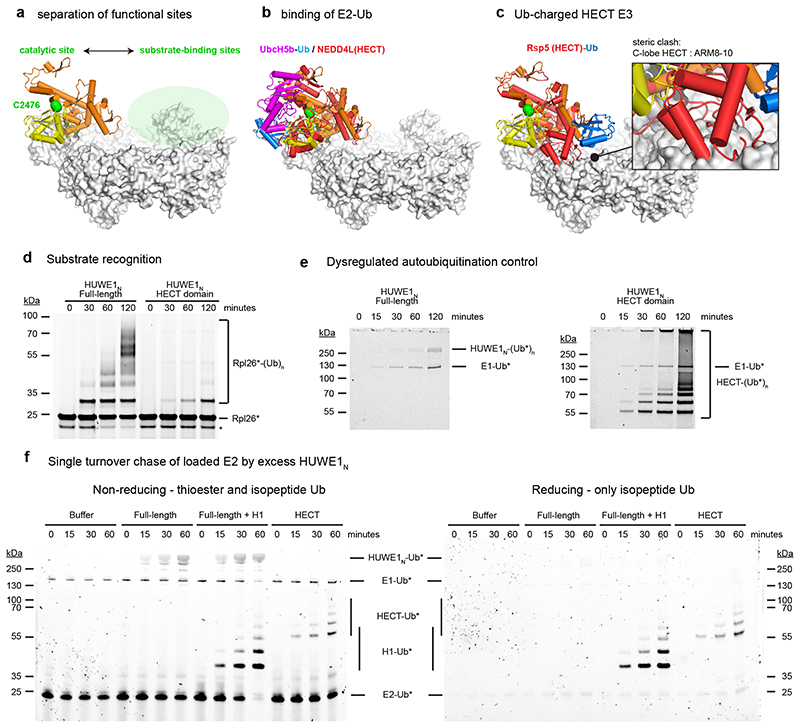
The E3 active site of HUWE1 is located on the face of the HECT domain that is opposite to the substrate-capturing solenoid ring (Fig. 6a). Accordingly, in the first step of the ligation reaction, the E2-Ub complex can bind and transfer Ub to the HECT catalytic cysteine without interference from the solenoid ring, as seen when superimposing the structure of a HECT/E2-Ub complex (Fig. 6b)36. Accessing substrates bound to the ring would require a 180° rotation of the HECT C-lobe. In fact, such a movement is expected in the second step of the reaction, as seen for a trapped HECT-Ub/substrate complex5. However, when this state is overlaid on the HUWE1N crystal structure, there is a steric clash between the predicted position of the C-lobe and the solenoid body of HUWE1N (Fig. 6c). We thus presume that the crystallized HUWE1N represents the latent E3 state, in which the solenoid ring presents a steric block preventing substrate ubiquitination. When we instead overlay the HECT-Ub/substrate complex on our open Cryo-EM Class 2, these clashes are relieved (Extended Data Fig. 6a), suggesting that a conformational rearrangement of the ring could be linked to HECT domain regulation. Another interesting comparison is with the previous structure of the N-terminally extended HECT domain of human HUWE137. The ‘pointer’ and ‘thumb’ helices equivalent to ARM34 in our structure have remarkably different structural organisation, which also differs in the two copies present in the reported crystal form (Extended Data Fig. 6b). Given its structural plasticity and immediate proximity to the HECT domain, ARM34 could serve as a molecular hinge controlling the orientation of the catalytic domain. Moreover, it was shown that mutation of F3892A, a central hinge residue equivalent to Huwe1N F2105, causes a strong increase in activity in vivo 37. A closer look at our full-length structure reveals such a mutation could greatly increase the flexibility of the hinge region, thus bypassing steric inhibition imposed by the ring (Extended Data Fig. 6c).
Figure 6. Regulation of the HUWE1N HECT domain.
(a) The HUWE1N active site is spatially separated from the substrate binding ring. Catalytic cysteine and center of substrate binding ring are highlighted in green. (b) Binding of E2-Ub to the HECT domain is not impeded in the closed state of the full-length enzyme. The HUWE1N HECT domain (N-lobe, orange; C-lobe, yellow) is superimposed with the crystal structure of the NEDD4L HECT domain (red) in complex with Ub (blue)-charged UbcH5b (magenta) (PDB 3JVZ) (c) Steric clashes between the HUWE1N solenoid and a ubiquitin-charged HECT domain. The structure of the Rsp5 HECT domain (in red), charged with Ub (in blue) and in complex with a peptide substrate (PDB 4LCD), is superimposed on the HUWE1N HECT domain. Resultant clashes between the HECT C-lobe and ARM8-10 are highlighted in the zoomed-in drawing. (d) The isolated HUWE1N HECT domain shows weak substrate ubiquitination. Ubiquitination of DyLight488-labelled Rpl26 was visualized by in-gel fluorescence. (e) Dysregulation of the isolated HECT domain. Autoubiquitination in the absence of a substrate was followed by using DyLight488-labelled ubiquitin. The autoubiquitinated products are indicated. (f) Single-turnover chase experiments of 1 μM E2 loaded with DyLight488-labelled ubiquitin by 2 μM HUWE1N (Full-length and HECT), with or without 2 μM histone H1.
To understand how the HECT is regulated in the context of full-length HUWE1N, we tested the activity of the isolated HECT domain (residues 2100-2490). For this construct, we observed very low ubiquitination activity on the Rpl26 substrate (Fig. 6d), which, in contrast, is strongly ubiquitinated by the full-length protein. This is consistent with the idea that the solenoid ring, and its insertions, are critical for substrate targeting. Moreover, the isolated HECT domain autoubiquitinated itself much more efficiently than the full-length enzyme, resulting in extended poly-ubiquitin chains (Fig. 6e). This suggests that, in the full-length protein, the solenoid inhibits the non-specific activity of the HECT domain, both in terms of substrate recruitment and substrate poly-ubiquitination. We observed similar behaviour for the metazoan CeHUWE1, where the isolated HECT domain rapidly depleted ubiquitin to generate high molecular weight ubiquitinated species (Extended Data Fig. 6d), pointing to a common auto-regulation mechanism.
To dissect how the non-specific activity of the HECT is inhibited, and how the full-length protein is activated by substrates (Fig. 1b), we used single turnover assays following discharge of ubiquitin-loaded E2 onto excess HUWE1N (Fig. 6f). Consistent with a previous report for the human HUWE1 HECT domain38, the discharge activity was slow and not complete within an hour. The reaction resulted in accumulation of thioester-loaded HUWE1N (Fig. 6f), as indicated by the sensitivity of the attached ubiquitin to chemical reduction (Fig. 6f). In the presence of the histone H1 substrate, the rate of discharge increased markedly, resulting in complete loss of the E2-Ub signal within an hour (Fig. 6f). This suggests substrate-induced activation occurs at the thioester transfer step. In contrast, when we used the isolated HECT domain, the rate of discharge remained very slow (Fig. 6f). However, here the ubiquitinated HECT species were insensitive to reduction pointing to isopeptide-linked, rather than thioester-linked ubiquitin species (Fig. 6f). Together, these findings indicate that in the absence of the solenoid the thioester-loaded HECT is rapidly discharged by autoubiquitination. Thus, our data highlight the complex regulation of this promiscuous E3 ligase at various steps of the ubiquitination reaction.
Discussion
The giant E3 ligase HUWE1 adopts an intricately coiled structure comprising a ring-shaped, solenoid body serving as substrate binding platform and an attached HECT domain that ubiquitinates bound proteins within its radius of manoeuvre. Aside from the characterized docking sites for poly-ADP-ribose, ubiquitin and Mcl-1 (Fig. 2e), the ring-shaped solenoid likely harbours further recognition modules, for example ligand binding sites within its ARM repeats. Intriguingly, our data show that the IDR1 region, a segment of the Ins3 appendix, constitutes an Asp/Glu-rich molecular mesh in the centre of the solenoid, used to recognize histone H1 and presumably other positively charged proteins (Fig. 3). The architecture of HUWE1, combining a multivalent docking platform with a flexible catalytic head, presents an alternative E3 strategy compared to the prominent class of SCF RING ligases3. These enzymes use a modular setup where promiscuity is achieved through exchangeable F-box protein adaptor modules to target different substrates. The advantage of using a large platform with multiple substrate docking sites, like HUWE1, is that it can act as a general-purpose E3 enzyme ready for all cell states. If additional regulation is required, one attractive possibility is that the individual docking sites may be post-translationally modified to favour or disfavour ubiquitination of certain substrates. Such regulated substrate targeting has been reported for Atoh1 (a transcription factor involved in cerebellar development) and the Wnt pathway component Dvl, both of which are ubiquitinated by HUWE1 in a phospho-dependent manner15,39.
How could the HECT domain reach all these substrates? Our integrative structural biology data show that the overall HUWE1 architecture is highly dynamic and flexible. The ring can adopt an entire range of conformations, opened to different extents, with this plasticity supported by the use of a multitude of small interfaces with low specificity. These large-scale movements are then likely to be combined with flexibility in orientation of the HECT domain relative to the neck. As compared to SCF complexes, where the catalytic RING domain is connected by a flexible peptide linker to a rigid scaffold40, the HUWE1 architecture offers an alternative solution to ubiquitinate a variety of substrate proteins in a reaction sphere. In HUWE1, the HECT domain is enabled to scan a series of docking sites arranged in a defined radius along the ring-shaped scaffold.
Our data suggest that the huge N-terminal ring of HUWE1N is not only responsible for substrate specificity but also regulates the activity of the HECT domain. One possible way this could be achieved is that the large conformational changes required during the catalytic cycle are blocked by the N-terminal region of the solenoid (Fig. 6c). Alternatively, there could be a direct allosteric effect on the catalytic HECT domain, promoted by the intricate connection between ARM34 and the HECT α1 helix. Interestingly, a previous study showed that removing the α1 helix increases the rate of thioester transfer to HUWE138, which is similar to the effect we observe for our substrate-induced activation (Fig. 6f). Auto-inhibition of a HECT by its N-terminal region has also been reported for WWP26, and may represent a general strategy for this class of ubiquitination enzymes. In addition to linking substrate binding with E3 activation, the observed ring structure offers a way to regulate the E3 activity of HUWE1 in trans. By modulating the transition of open and closed states, regulatory proteins could adjust the activity of HUWE1 to the needs of the cell.
In conclusion, the unique architecture of HUWE1 explains its remarkable ability to function as a promiscuous E3 ligase, targeting a broad range of unassembled orphan proteins as well as diverse regulatory proteins. Of note, further giant ubiquitin ligases like UBR4, HECTD1 and Mycbp2 exhibit similar non-catalytic scaffolds, being composed of extended ARM repeats interrupted by diverse protein-protein interaction motifs. Thus, HUWE1 can be a paradigm for understanding how full-length E3 ligases target their substrates and adapt their activities within the complex ubiquitin-proteasome system. With the Nematocida model system, we now can explore the molecular details of how HUWE1 is regulated. These studies will provide the framework to rationally develop HUWE1 directed therapeutics, including small-molecule degraders that could exploit the promiscuity of an essential E3 ligase to induce proteasomal degradation of diverse neo-substrates.
Methods
Protein expression and purification
Expression of HUWE1N in insect cells
The HUWE1 genes from Nematocida sp. ERTm5 and C. elegans were codon optimized for expression in insect cells and synthesized in, respectively, three and five BsaI-flanked fragments. Fragments were scarlessly assembled via the BsaI-GoldenGate reaction into a pGBdest vector from the GoldenBac system41 together with a 3C-cleavable N-terminal hexahistidine tag, a Polyhedrin promoter and a SV40 terminator. The HUWE1N-CA mutation (C2457A), HUWE1N-IDR1 (1442-1522 replaced by GGSx6) and HUWE1N-IDR2 (1610-1670 replaced by GGSx6) were generated by PCR-based site-directed mutagenesis prior to assembly.
Source plasmids containing the target constructs were transformed into DH10EMBacY cells. Blue-white screening was used to isolate colonies containing recombinant baculoviral shuttle vectors (bacmids). Bacmid DNA was extracted by alkaline lysis and isopropanol precipitation. Bacmids were then transfected into adherent Spodoptera frugiperda (Sf9) insect cells in 6-well plates, using Fugene HD transfection reagent (Promega). Successful transfection was tracked by monitoring fluorescence of YFP, encoded by the bacmid backbone. All Sf9 insect cell culture works were performed using ESF921 serum-free growth medium (Expression Systems) without antibiotic supplementation. Following virus amplification in Sf9 suspension insect cell culture, recombinant HUWE1N were expressed in Trichoplusia ni High-Five insect cells (Thermo Fisher) infected at a density of 2×106ml−1 with the appropriate virus and grown at 21°C and 120 r.p.m. using Insect Xpress Protein-free Insect Cell Medium (Lonza) supplemented with GlutaMAX (GIBCO) and Pen/Strep Amphotericin B (Lonza). Cells were harvested 4 days after transfection by centrifugation at 4000 x g for 20 minutes, and pellets were flash-frozen in liquid nitrogen and stored at –80 °C. For generation of selenomethionine-substituted protein Hi5 cells were grown in ESF 921 Delta Series medium (expressionsystems). Afterwards cells were centrifuged and media exchanged to ESF 921 Delta Series Methionine Deficient medium (expressionsystems). Cell suspension (2x106 cells per liter) was infected with baculovirus, and swayed for 8h at 27°C. Then 100 mg/L Seleno-methionine were supplemented and cultures swayed for 3 days at 21°C before harvesting.
Purification of HUWE1N
Purification procedures were carried out at 4°C, using ÄKTA pure 25 (GE Healthcare) instruments. Frozen cell pellet from 2 L of expression culture was rapidly thawed and resuspended in 150 mL of Buffer A (50 mM HEPES, 300 mM NaCl, 0.5 mM TCEP, 20mM imidazole, pH 7.5) with addition of four tablets of Complete EDTA-free Protease Inhibitor (Roche) and 50 μL Benzonase (IMP Molecular Biology Service), followed by centrifugation for 30 min at 40000 × g.. After centrifugation, the soluble fraction was applied on a 5 mL HisTrap HP column (GE Healthcare) pre-equilibrated with Buffer A. The column was washed with 10 column volumes (CV) with Buffer A, followed by 7 CV of 9% Buffer B (50 mM HEPES, 300 mM NaCl, 0.5 mM TCEP, 300 mM imidazole, pH 7.5) and 3 CV of 12% Buffer B, after which HUWE1N was eluted in 100% Buffer B. Fractions containing HUWE1N were identified by SDS-PAGE and dialyzed for 12 hours against Buffer C (50 mM HEPES, 150 mM NaCl 0.5 mM TCEP, pH 7.5). After the dialysis protein was loaded onto a 6 mL Resource Q anion exchange column (GE Healthcare) equilibrated with Buffer C. The column was washed for 3 CV with Buffer C, and then the protein was eluted with a linear 0–100% gradient for 30 CV against Buffer D (50 mM HEPES, 1000 mM NaCl, 0.5 mM TCEP, pH 7.5). Pooled HUWE1N fractions were concentrated to a final volume of 5 mL using a Vivaspin 6 centrifugal concentrator (100 kDa cutoff, Sigma-Aldrich) and applied to a Superose 6 Prep Grade HiLoad column (GE Healthcare) equilibrated with Buffer E (10 mM Na2(HPO4), 50 mM NaCl, 0.5 mM TCEP, pH 7.2).
For purification of HUWE1N-CA for crystallization, the purification tag was cleaved with 3C PreScission Protease (IMP Molecular Biology Service) after the first HisTrap step. Protein sample was again applied on a 5 mL HisTrap HP column pre-equilibrated with Buffer A, untagged protein was collected from the flow-through and loaded on 6 mL Resource Q anion exchange column as described, followed by final step of size-exclusion chromatography. When HUWE1N and variants were purified for assays, the reverse HisTrap step was performed in Buffer C, after which the protein was concentrated and flash frozen. CeHUWE1 purification followed this method, but with an additional SEC step.
The HUWE1N HECT domain was cloned into a pET28 vector with a PreScission cleavable N-terminal His6 tag, and expressed in BL21 cells with IPTG induction at 18°C. Cells were harvested by sonication and purified using a 5 mL HisTrap (GE Healthcare) in buffer containing 50 mM HEPES, 500 mM NaCl, pH 7.4, before size-exclusion chromatography using a HiLoad 16/60 Superdex 200 column (GE Healthcare) equilibrated in 50 mM Tris-HCl, 200 mM NaCl, pH 7.4.
Ribosome preparation
HEK293T cells in DMEM medium were incubated with 10 ng/mL cycloheximide and lysed with 1% Triton X-100 and 1% sodium deoxycholate. The cleared lysate was applied to 6 x 2 mL 30% Sucrose Cushion (10mM Tris pH 7.5, 100 mM NaCl, 10mM MgCl2) and centrifuged at 116 000 x g at 4°C for 5 hours. The pellets were washed with 50mM Tris, 10mM NaCl, 10mM MgCl2, 100 mM NH4Cl pH 8 and resuspended in 30mM Tris, 100 mM NaCl, 10 mM MgCl2. Insoluble contaminants were removed by centrifugation. The soluble material was then applied to a 12 mL 15-30% sucrose gradient in 20mM Tris pH 7.5, 100mM NaCl, 10mM MgCl2. Gradients were centrifuged for 100 min at 4°C and 100 000 x g and fractions containing ribosomes were pooled and concentrated.
Reagents for ubiquitination assays
Pure human Histone H1, Histone H2B and native nucleosomes were purchased from Sigma-Aldrich. 4WJ DNA was generated by mixing equimolar oligonucleotide substrates (Sigma-Aldrich): 5’GAATTCAGCACGAGTCCTAACGCCAGATCT, 5’AGATCTGGCGTTAGGTGATACCGATGCATC, 5’GATGCATCGGTATCAGGCTTACGACTAGTG, and 5’CACTAGTCGTAAGCCACTCGTGCTGAATTC at 100 μM in buffer containing 10 mM Tris-HCl, 100 mM NaCl, pH 8.0, incubating at 95°C for two minutes and then slowly cooling.
Human Mcl-1, amino acids 2-330 was produced as a GST fusion protein in E. coli, using strain BL21(DE3) AI. Cells were grown in TB media, induced with 0.2% arabinose, and grown overnight at 22°C. Cells were lysed by sonication in 50 mM Tris buffer at pH 7.5, containing 200 mM NaCl and 5 mM DTT. After centrifugation to remove cell debris, the protein was bound to glutathione agarose and was eluted in the same buffer containing 25 mM reduced glutathione. The GST tag was removed by overnight cleavage with TEV protease followed by reverse affinity after desalting into buffer without glutathione. Untagged Mcl-1 was further purified by separation on a MonoQ ion exchange column, in 50 mM Tris buffer at pH 7.8 containing 1 mM TCEP, using a gradient from 50 mM to 1 M NaCl over 5 column volumes. Mcl-1 eluted at 220 mM NaCl, was concentrated to 13 mg/mL and stored at -80°C.
The forkhead domain of human FoxP3, amino acids 336 to 419, was expressed as a His-tagged protein in E. coli strain BL21(DE3) in autoinduction media at 18 degrees C. Cells were lysed by sonication in 25 mM Tris buffer at pH 7.8 containing 200 mM NaCl, 1 mM TCEP, 50 mM arginine, 0.5% glycerol, and 0.02% CHAPS. After centrifugation to remove cell debris, the protein was bound to Nickel beads, the beads were washed with 20 mM Tris buffer at pH 8 containing 200 mM NaCl, 1 mM TCEP, 50 mM arginine, 0.5% glycerol, and eluted in the same buffer plus 500 mM imidazole. The protein was incubated overnight with TEV protease to remove the His tag and simultaneously dialyzed into 20 mM Tris buffer at pH 8.0 containing 200 mM NaCl and 1 mM TCEP. After dialysis the protein was concentrated and further purified over a Superdex 75 size exclusion column equilibrated in 25 mM Tris buffer at pH 7.5 containing 150 mM NaCl and 1 mM TCEP. The protein was concentrated to 4.7 mg/mL and stored at -80°C.
Recombinant human UBA1 was provided by the Vienna BioCenter core facilities. Ubiquitin was expressed in BL21(DE3) cells using autoinduction media. Cells were lysed by sonication in 50 mM ammonium acetate pH 4.5, and the cleared lysate was subjected to heat treatment at 70°C to remove impurities. Ubiquitin was then further purified with a SP Sepharose Fast Flow ion-exchange column (GE Healthcare) followed by SEC using a Superdex 75 Increase column (GE Healthcare).
The Rpl-26 and Let-70 (UbcH5b) genes from C. elegans were generated as double-stranded DNA fragments (IDT) and cloned into a modified pET29b expression vector (containing an N-terminal His6-tag and TEV cleavage site) using NEB HiFi assembly (New England Biolabs). The C. elegans HECT domain (3793-4180) vector was constructed similarly. Expression was performed by IPTG induction in E. coli BL21 (DE3) cells at 20°C overnight. Cell pellets were resuspended in lysis buffer (50 mM Tris, 150 mM NaCl, 10 mM imidazole, Complete EDTA-free protease inhibitor cocktail at pH 7.5) before sonication to induce cell lysis. Cell lysates were clarified through centrifugation at 18,500 g for 20 minutes at 4°C. Clarified lysates were then incubated with Ni-NTA resin (Qiagen) for 1 hour at 4°C with mild agitation. Ni-NTA resin was washed before elution with 150 mM imidazole. Proteins were further purified by SEC using a Superdex 75 16/600 column (GE Healthcare) into 50 mM Tris, 150 mM NaCl, 0.5 mM TCEP at pH 7.5. Rpl-26 or ubiquitin bearing a single N-terminal cysteine residue were buffer exchanged into degassed labelling buffer (50 mM Tris, 150 mM NaCl at pH 7), before addition of a 2:1 molar excess of maleimide fluorophore (Dylight488 or Dylight800, Thermo Fischer Scientific). Reactions were flushed with argon and incubated in the dark for 2 hours at room temperature. Reactions were stopped by desalting to remove excess fluorophore using Centri Pure P2 columns.
Ubiquitination assays
Ubiquitination reactions were performed in buffer containing 50 mM HEPES pH 7.5, 150 mM NaCl, 5 mM MgCl2 and 0.05 mM TCEP. Reactions contained 0.5 μM Uba1, 2.5 μM CeUbcH5b, 10 μM ubiquitin and 0.5 μM HUWE1N. Substrates were usually added at 4 μM concentration. For fluorescent visualization of ubiquitinated products, reactions were spiked with 0.5 μM Ub-Dylight488. For the comparative reactions of Rpl26 and ribosomes, or histone H2B and nucleosomes, only 0.5 μM substrate was used, and 2 μM labelled ubiquitin (Ub-DyLight488 for the nucleosomes, Ub-DyLight800 for the ribosomes) to increase sensitivity. To compare the ubiquitination of Rpl26 by the HECT domain and full-length HUWE1N, labelled substrate, Rpl26-DyLight488, was employed to avoid overlapping autoubiquitination bands. All reactions were initiated by addition of 5 mM ATP and incubated at 30°C for the indicated time. Samples were quenched by addition of 2x reducing SDS gel loading buffer and run on 4-12% NuPAGE Bis-Tris gradient gels (Invitrogen) in MES running buffer. Fluorescence imaging was performed using a ChemiDoc MP system (Bio-Rad), and then gels were coomassie stained to visualize total protein content. CeHUWE1 reactions used the same method, but with 2 μM substrate and 0.5 μM Ub-Dylight800. For single turnover experiments, 0.5 μM Uba1, 8 μM CeUbcH5b, 10 μM ubiquitin and 0.5 μM ubiquitin-DyLight488 were incubated at 37°C in the usual reaction buffer but with 3 mM MgCl2 and 3 mM ATP. The reactions were then quenched with 20 mM EDTA on ice and diluted 1 in 8 (final E2 concentration of 1 μM) in reactions containing 2 μM HUWE1N or HECT domain with or without 2 μM histone H1, in the normal reaction buffer but without MgCl2, and then incubated at 30°C for the indicated time points before quenching with either reducing or non-reducing 2x SDS gel loading buffer. Technical replicates of all biochemical assays were performed.
Crystallization and tantalum bromide soaking of HUWE1N-CA
HUWE1N-CA crystals were obtained using the sitting drop vapor diffusion method at 277K. 1.5 μL protein solution (5.1 mg/mL) was mixed with 0.75 μL reservoir solution containing 10.4% (w/v) PEG 4000, 100 mM MgCl2, 100mM HEPES pH 6.6 and 6 mM DTT. The crystals were cryoprotected with 23% ethylene glycolbefore flash freezing. Crystals of selenomethionine-substituted HUWE1N-CA were obtained in the same condition with a protein concentration of 5.6 mg/mL.
To obtain a heavy-atom derivative for experimental phasing, crystals were soaked with hexatantalum tetradecabromide from the Tantalum Cluster Derivatization kit (Jena Biosciences). 1.8 μL of mother liquor supplemented with hexatantalum tetradecabromide were mixed with drops containing crystals and incubated for 1 hour before crystals were flash-frozen in liquid nitrogen.
HUWE1N-CA structure solution and refinement
Diffraction data was collected at X06SA at the Swiss Light Source at a wavelength of 1 Å. The data were autoprocessed in P1 using autoPROC42, implementing XDS43, the STARANISO server44 and Aimless45 for data integration, scaling and merging. Two copies of the human HUWE1 HECT domain (PDB 3G1N) were placed by molecular replacement using PHASER46, however did not allow to build the huge N-terminal part into the electron density map. For experimental phasing, we used anomalous diffraction data from Ta6Br14 soaked crystals. Tantalum sites were localized and refined with PHENIX AutoSol47. The initial model, which comprised the HECT domain and a poly-alanine scaffold of the ARM solenoid, was manually built. Further rounds of modeling and refinement were performed with Coot48 and BUSTER49 using NCS restraints and TLS. The sequence was placed with the aid of an anomalous map calculated from the SeMet dataset to 5 Å resolution using BUSTER and co-evolutionary contact prediction by the RaptorX sever31. Specifically, for small canonical ARM repeat stretches, the predicted RaptorX model was docked as an initial structure. For the rest of the protein, predicted contacts were used alongside bulky residue density and anomalous SeMet signal to initially assign the register of helices that were next to helices of known sequence. Additionally, the regions outside of the solenoid, i.e. Ins1-4, could be readily identified due to their distinct contact pattern, preventing large errors in initial sequence assignment. The final structure had Rwork/Rfree values of 20.24/23.30% and 0.21% Ramachandran outliers and 94.7% favored. Chain A was used for all structural figures, made with PyMOL.
Negative stain EM of CeHUWE1
Copper/palladium grids (Agar Scientific) were covered with a 4 nm carbon-layer, performed in-house. Glow-discharge of grids was performed using a BAL-TEC sputter coater CD005 for 120 seconds. CeHUWE1 was diluted to 0.02 mg/ml in 10 mM HEPES, 100 mM NaCl pH 7.5 and applied to the grid for 60 seconds prior to staining with 2% uranyl acetate. A 200kV FEI Tecnai G2 20 (T20) transmission electron microscope was used to image CeHUWE1 grids at a 50,000x magnification (pixel size 2.211 Å). The micrographs obtained were processed using Relion 3.150. Initial number of particles following auto-picking was ~65,000, reduced to ~14,000 at the final 2D classification, and ~9,000 for the final 3D model.
Cryo-EM sample preparation and data collection
4 μL of HUWE1N at 0.8 mg/mL were applied to a Quantifoil R2/2 Cu 200 mesh grid, freshly glow-discharged at 20 mA for 60 s in a SCD 005 Sputter Coater (BAL-TEC). After 1 s pre-blotting time, the sample was blotted for 2 s with a relative humidity of 70% and a temperature of 4°C in an EM GP freeze plunger (Leica Microsystems). Cryo-EM grids were screened using a Glacios equipped with a Falcon3EC camera (Thermo Fisher Scientific). The selected grid was recorded on a Titan Krios electron microscope (ThermoFisher) equipped with a K3 camera (Gatan) in counting mode at the Institute of Science and Technology Austria. The dataset was recorded both untilted and at 25° stage tilt. The microscope was operated in nanoprobe mode with a beam diameter of 1200 nm at a nominal magnification of 105 000x, resulting in a calibrated pixel size of 0.86 Å per pixel. Images were collected in super-resolution. Four images were taken per hole from different non-overlapping areas using image beam shift. The total electron dose of 60 electrons per Å2 was fractionated into 40 frames. A total number of 2580 movies were acquired without tilt, and 1958 movies were acquired at tilt. A defocus range from −2 to −3.5 μm was used.
Cryo-EM image processing and model refinement
Processing was performed in Relion 3.1.050. Movie frames were motion-corrected, dose-weighted and super-resolution images were binned two-fold using MotionCor251. Tilted and non-tilted micrographs were merged afterwards (4538 micrographs). Micrographs with strong drift were discarded. The Contrast transfer function (CTF) parameters were determined using CTFFIND4.1.1352. Only the micrographs in which Thon rings extended to 5 Å in the untilted or to 7 Å in tilted dataset were included in the further processing steps. This yielded 1847 good micrographs for the non-tilted and 919 for the tilted part of the dataset. Particles were picked using CrYOLO53. As a first step, particles from 8 micrographs were picked manually in order to train the neural network. After the training, particles were picked using the threshold of 0.3 and extracted at 2 x pixel size. Subsequently, several iterations of 2D classification were performed, using subsets with maximum 200000 particles. After this procedure, the best class averages were used to generate an initial 3D model. Afterwards 3D classification of all picked particles with a 3D auto-refined map as initial reference, filtered to 30 Å. This resulted in two good classes. CTF parameters and per-particle trajectories were then refined in an iterative manner, one round of CTF refinement was followed by Bayesian polishing and another CTF refinement. This led to a refined HUWE1N map of 6.49 Å resolution. Particles were further 3D classified without searches and with increased T value to remove damaged particles and sort the heterogeneity. This resulted in two classes, corresponding to a closed (Class 1) and open (Class 2) state, with an overall resolution, as judged by an FSC of 0.143, of 5.3 Å and 6.4 Å respectively. Heterogeneity analysis was performed with CryoDRGN54.
For modelling, Class 1 and Class 2 were sharpened with a B factors of -137 Å2 and -178 Å2 respectively. Our HUWE1N crystal structure was docked onto the maps using UCSF Chimera55. The structure was then manually modeled in Coot48 mostly through rigid body fitting of sections of the structure. Real space refinement was performed using Phenix56.
Mass spectrometry analysis
To determine ubiquitination linkage types, a HUWE1N autoubiquitination reaction was performed overnight and separated by SDS-PAGE. To determine Rpl26 ubiquitination sites, the reaction was performed for two hours. The ubiquitinated products were excised and then digested with trypsin. NanoLC-MS analysis was performed using an UltiMate 3000 RSLC nano system (Thermo Fisher Scientific) coupled to a Q Exactive HF-X mass spectrometer (Thermo Fisher Scientific), equipped with a Proxeon nanospray source (Thermo Fisher Scientific). Peptides were loaded onto a PepMap C18 trap column (Thermo Fisher Scientific) at a flow rate of 25 μL min-1 using 0.1% TFA as mobile phase. After 10 min, the trap column was switched in line with a PepMap C18 analytical column (Thermo Fisher Scientific). Peptides were eluted using a binary 2 hour gradient, starting with 98% A (water/formic acid, 99.9/0.1, v/v) and 2% B (water/acetonitrile/formic acid, 19.92/80/0.08, v/v/v), increasing to 35% B over 180 min, to 90% B over 5 min before decreasing back to 98% A and 2% B.
The mass spectrometer was operated in data-dependent mode, using a full scan (m/z range 350-1600) followed by 15 MS/MS scans of the 15 most abundant ions. MS/MS spectra were acquired using normalized collision energy of 26, 28, 30, isolation width of 1.0 m/z, resolution of 30,000 and the target value was set to 50, 000. Precursor ions of charge state +3 to +7 were fragmented.
For peptide identification, the RAW-files were loaded into Proteome Discoverer (version 2.3.0.523, Thermo Scientific). All MS/MS spectra were searched using MSAmanda v2.0.0.984957. The peptide mass tolerance was set to ±5 ppm and the fragment mass tolerance to 15ppm. The maximal number of missed cleavages was set to 2, using tryptic enzymatic specificity. The result was filtered to 1% FDR on protein level in Thermo Proteome Discoverer. This sub-database was then searched again for all standard amino acid modifications. Peptide areas were quantified using in-house-developed tool apQuant58.
Crosslinking for XL-MS analysis was performed with 0.06 mM DSS (Creative Molecule) for 30 min at 20°C, and quenched with ammonium bicarbonate. The sample was carboxymethylated in urea and then trypsin-digested overnight. Trypsin was deactivated with 0.4% TFA, before loading on Sep-Pak cartridges (Fischer Chemicals) equilibrated in 5% acetonitrile, 0.1% formic acid, and eluted with 50% acetonitrile. SEC in 30% acetonitrile, 0.1% (v/v) TFA was performed using a Superdex 30 Increase 3.2/300 column (GE Healthcare) to enrich for crosslinked peptides. NanoLS-MS analysis was performed as above, but with a 3 hour HPLC gradient.
Identified crosslinks were then filtered to 0.1% FDR on link level and visualized with XiView59. Only one self-link was observed.
Statistics and Reproducibility
The experiments shown in Figures Fig. 1b, Fig. 1d-e, Fi.g 1f, Fig. 1h, Fig. 3d-e, Fig. 6d-f and Extended Data Fig. 6d were independently performed twice and showed the same result. There was no instance where we failed to reproduce these experiments and so no data were excluded.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Figure 1.
Extended Data Figure 2.
Extended Data Figure 3.
Extended Data Figure 4.
Extended Data Figure 5.
Extended Data Figure 6.
Supplementary Material
Acknowledgements
We thank all members of the Clausen group for remarks on the manuscript and discussions, and the Mass Spectrometry and ProTech services from the Vienna BioCenter Core Facilities for their support. We are grateful to the scientific staff at the beamline X10SA, Paul Scherrer Institute (Villigen, Switzerland), as well as the beamline P11, DESY (Hamburg, Germany) for support in crystallographic data collection. We acknowledge the following cryo-EM facilities for their access and support: CEITEC MU of CIISB, Instruct-CZ Centre (proposal LM2018127), UK’s national Electron Bio-imaging Centre (Diamond Lightsource, proposal EM BI25222), EM facility at the Institute of Science and Technology (IST) Austria, and the EM facility at the Vienna BioCenter Core Facilities (VBCF). We also acknowledge members of the Protein Science lab at Boehringer Ingelheim for support with expression and purification of HUWE1N, and Victor-Valentin Hodirnau for help with the cryo-EM analysis. The project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (AdG 694978) and Marie Skłodowska-Curie Grant Agreement Nr. 847548, an FFG Headquarter Grant (No 852936) and the Austrian Science Fund (FWF, SFB F 79). P.M. and O.A.P are members of the Boehringer Ingelheim Discovery Research global post-doc program. The IMP is supported by Boehringer Ingelheim.
Footnotes
Author contributions statement
D.B.G. and T.C. designed experiments; L.D., O.A.P., D.B.G., R.K., P.M., R.G., V.F., D.K., A.L., J.N. prepared expression constructs, purified HUWE1 and performed biochemical assays; O.A.P., I.G., J.A., D.H. performed the cryo-EM analysis, A.V., R.I. the XL-MS analysis, A.S. the bioinformatic analysis, D.B.G., T.C., A.M., G.B., P.S.B., J.B., B.W., G.F., D.K. the crystallographic analysis; D.B.G. built the molecular models; D.B.G. and T.C. coordinated the research project and prepared the manuscript with inputs from all authors.
Competing interests statement
G.B., J.B., P.S.-B., G.F., B.W. And D.K. are currently employees of Boehringer Ingelheim.
Data availability
Coordinates of the HUWE1N crystal structure have been deposited at the Protein Data Bank (PDBe) under accession code 7BII. Cryo-EM maps and atomic coordinates have been deposited in the EMDB with accession codes EMD-12318 and EMD-12319 and PDB under 7NH1 and 7NH3. Source data for all non-structural figures is provided with this paper.
References
- 1.Ciechanover A, Orian A, Schwartz AL. Ubiquitin-mediated proteolysis: biological regulation via destruction. Bioessays. 2000;22:442–51. doi: 10.1002/(SICI)1521-1878(200005)22:5<442::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 2.Hochstrasser M. Lingering mysteries of ubiquitin-chain assembly. Cell. 2006;124:27–34. doi: 10.1016/j.cell.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 3.Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–51. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- 4.Alfieri C, Zhang S, Barford D. Visualizing the complex functions and mechanisms of the anaphase promoting complex/cyclosome (APC/C) Open Biol. 2017;7 doi: 10.1098/rsob.170204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamadurai HB, et al. Mechanism of ubiquitin ligation and lysine prioritization by a HECT E3. Elife. 2013;2:e00828. doi: 10.7554/eLife.00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Z, et al. A Tunable Brake for HECT Ubiquitin Ligases. Mol Cell. 2017;66:345–357.:e6. doi: 10.1016/j.molcel.2017.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lorenz S. Structural mechanisms of HECT-type ubiquitin ligases. Biol Chem. 2018;399:127–145. doi: 10.1515/hsz-2017-0184. [DOI] [PubMed] [Google Scholar]
- 8.Kao SH, Wu HT, Wu KJ. Ubiquitination by HUWE1 in tumorigenesis and beyond. J Biomed Sci. 2018;25:67. doi: 10.1186/s12929-018-0470-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong Q, Gao W, Du F, Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 2005;121:1085–95. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 10.Inoue S, et al. Mule/Huwe1/Arf-BP1 suppresses Ras-driven tumorigenesis by preventing c-Myc/Miz1-mediated down-regulation of p21 and p15. Genes Dev. 2013;27:1101–14. doi: 10.1101/gad.214577.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen D, et al. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071–83. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 12.Adhikary S, et al. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell. 2005;123:409–21. doi: 10.1016/j.cell.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 13.Urbán N, et al. Return to quiescence of mouse neural stem cells by degradation of a proactivation protein. Science. 2016;353:292–5. doi: 10.1126/science.aaf4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hao Z, et al. K48-linked KLF4 ubiquitination by E3 ligase Mule controls T-cell proliferation and cell cycle progression. Nat Commun. 2017;8:14003. doi: 10.1038/ncomms14003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Groot REA, et al. Huwe1-Mediated Ubiquitylation of Dishevelled Defines a Negative Feedback Loop in the Wnt Signaling Pathway. Science Signalling. 2014;7:ra26. doi: 10.1126/scisignal.2004985. [DOI] [PubMed] [Google Scholar]
- 16.Hall JR, et al. Cdc6 stability is regulated by the Huwe1 ubiquitin ligase after DNA damage. Mol Biol Cell. 2007;18:3340–50. doi: 10.1091/mbc.E07-02-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parsons JL, et al. Ubiquitin ligase ARF-BP1/Mule modulates base excision repair. EMBO J. 2009;28:3207–15. doi: 10.1038/emboj.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, et al. HUWE1 interacts with BRCA1 and promotes its degradation in the ubiquitin-proteasome pathway. Biochem Biophys Res Commun. 2014;444:549–54. doi: 10.1016/j.bbrc.2014.01.075. [DOI] [PubMed] [Google Scholar]
- 19.Xu Y, Anderson DE, Ye Y. The HECT domain ubiquitin ligase HUWE1 targets unassembled soluble proteins for degradation. Cell Discov. 2016;2:16040. doi: 10.1038/celldisc.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sung MK, et al. A conserved quality-control pathway that mediates degradation of unassembled ribosomal proteins. Elife. 2016;5 doi: 10.7554/eLife.19105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Z, Oughtred R, Wing SS. Characterization of E3Histone, a novel testis ubiquitin protein ligase which ubiquitinates histones. Mol Cell Biol. 2005;25:2819–31. doi: 10.1128/MCB.25.7.2819-2831.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh RK, Kabbaj MH, Paik J, Gunjan A. Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat Cell Biol. 2009;11:925–33. doi: 10.1038/ncb1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deshaies RJ. Proteotoxic crisis, the ubiquitin-proteasome system, and cancer therapy. BMC Biol. 2014;12:94. doi: 10.1186/s12915-014-0094-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oromendia AB, Amon A. Aneuploidy: implications for protein homeostasis and disease. Dis Model Mech. 2014;7:15–20. doi: 10.1242/dmm.013391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myant KB, et al. HUWE1 is a critical colonic tumour suppressor gene that prevents MYC signalling, DNA damage accumulation and tumour initiation. EMBO Mol Med. 2017;9:181–197. doi: 10.15252/emmm.201606684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peter S, et al. Tumor cell-specific inhibition of MYC function using small molecule inhibitors of the HUWE1 ubiquitin ligase. EMBO Mol Med. 2014;6:1525–41. doi: 10.15252/emmm.201403927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Froyen G, et al. Submicroscopic duplications of the hydroxysteroid dehydrogenase HSD17B10 and the E3 ubiquitin ligase HUWE1 are associated with mental retardation. Am J Hum Genet. 2008;82:432–43. doi: 10.1016/j.ajhg.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jäckl M, et al. β-Sheet Augmentation Is a Conserved Mechanism of Priming HECT E3 Ligases for Ubiquitin Ligation. J Mol Biol. 2018;430:3218–3233. doi: 10.1016/j.jmb.2018.06.044. [DOI] [PubMed] [Google Scholar]
- 29.Michel MA, Swatek KN, Hospenthal MK, Komander D. Ubiquitin Linkage-Specific Affimers Reveal Insights into K6-Linked Ubiquitin Signaling. Mol Cell. 2017;68:233–246.:e5. doi: 10.1016/j.molcel.2017.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White AE, Hieb AR, Luger K. A quantitative investigation of linker histone interactions with nucleosomes and chromatin. Sci Rep. 2016;6:19122. doi: 10.1038/srep19122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S, Sun S, Li Z, Zhang R, Xu J. Accurate De Novo Prediction of Protein Contact Map by Ultra-Deep Learning Model. PLoS Comput Biol. 2017;13:e1005324. doi: 10.1371/journal.pcbi.1005324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hunkeler M, et al. Modular HUWE1 architecture serves as hub for degradation of cell-fate decision factors. bioRxiv. 2020:2020.08.19.257352 [Google Scholar]
- 34.Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- 35.Huber AH, Weis WI. The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin. Cell. 2001;105:391–402. doi: 10.1016/s0092-8674(01)00330-0. [DOI] [PubMed] [Google Scholar]
- 36.Kamadurai HB, et al. Insights into ubiquitin transfer cascades from a structure of a UbcH5B approximately ubiquitin-HECT(NEDD4L) complex. Mol Cell. 2009;36:1095–102. doi: 10.1016/j.molcel.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sander B, Xu W, Eilers M, Popov N, Lorenz S. A conformational switch regulates the ubiquitin ligase HUWE1. Elife. 2017;6 doi: 10.7554/eLife.21036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pandya RK, Partridge JR, Love KR, Schwartz TU, Ploegh HL. A structural element within the HUWE1 HECT domain modulates self-ubiquitination and substrate ubiquitination activities. J Biol Chem. 2010;285:5664–73. doi: 10.1074/jbc.M109.051805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Forget A, et al. Shh signaling protects Atoh1 from degradation mediated by the E3 ubiquitin ligase Huwe1 in neural precursors. Dev Cell. 2014;29:649–61. doi: 10.1016/j.devcel.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 40.Baek K, et al. NEDD8 nucleates a multivalent cullin-RING-UBE2D ubiquitin ligation assembly. Nature. 2020;578:461–466. doi: 10.1038/s41586-020-2000-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neuhold J, et al. GoldenBac: a simple, highly efficient, and widely applicable system for construction of multi-gene expression vectors for use with the baculovirus expression vector system. BMC Biotechnol. 2020;20:26. doi: 10.1186/s12896-020-00616-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vonrhein C, et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr D Biol Crystallogr. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tickle IJ, Flensburg C, Keller P, Paciorek W, Sharff A, Vonrhein C, Bricogne G. STARANISO. Global Phasing Ltd; Cambridge, United Kingdom: 2018. [Google Scholar]
- 45.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–14. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Terwilliger TC, et al. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr D Biol Crystallogr. 2009;65:582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smart OS, et al. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr D Biol Crystallogr. 2012;68:368–80. doi: 10.1107/S0907444911056058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zivanov J, et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 2018;7 doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–21. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wagner T, et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun Biol. 2019;2:218. doi: 10.1038/s42003-019-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhong ED, Bepler T, Berger B, Davis JH. CryoDRGN: reconstruction of heterogeneous cryo-EM structures using neural networks. Nat Methods. 2021;18:176–185. doi: 10.1038/s41592-020-01049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 56.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dorfer V, et al. MS Amanda, a universal identification algorithm optimized for high accuracy tandem mass spectra. J Proteome Res. 2014;13:3679–84. doi: 10.1021/pr500202e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Doblmann J, et al. apQuant: Accurate Label-Free Quantification by Quality Filtering. J Proteome Res. 2019;18:535–541. doi: 10.1021/acs.jproteome.8b00113. [DOI] [PubMed] [Google Scholar]
- 59.Graham M, Combe C, Kolbowski L, Rappsilber J. xiView: A common platform for the downstream analysis of Crosslinking Mass Spectrometry data. bioRxiv. 2019:561829 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates of the HUWE1N crystal structure have been deposited at the Protein Data Bank (PDBe) under accession code 7BII. Cryo-EM maps and atomic coordinates have been deposited in the EMDB with accession codes EMD-12318 and EMD-12319 and PDB under 7NH1 and 7NH3. Source data for all non-structural figures is provided with this paper.