Abstract
AMPA receptors (AMPARs) mediate the majority of excitatory transmission in the brain, and enable synaptic plasticity that underlies learning1. A diverse array of AMPAR signaling complexes are established by receptor auxiliary subunits, associating in various combinations to modulate trafficking, gating and synaptic strength2. However, their mechanisms of action are poorly understood. Here, we determine cryo-electron microscopy structures of the heteromeric GluA1/2 receptor assembled with both TARP-γ8 and CNIH2, the predominant AMPAR complex in the forebrain, in both resting and active states (at 3.1 and 3.6 Å, respectively). Consequential for gating regulation, two γ8 and two CNIH2 subunits lodge at distinct sites beneath the ligand-binding domains of the receptor, with site-specific lipids shaping each interaction. Activation leads to asymmetry between GluA1 and GluA2 along the ion conduction path, and an outward expansion of the channel triggers counter-rotations of both auxiliary subunit pairs, promoting the active-state conformation. In addition, both γ8 and CNIH2 pivot towards the pore exit on activation, extending their reach for cytoplasmic receptor elements. CNIH2 achieves this through its uniquely extended M2 helix, which has transformed this ER-export factor into a powerful AMPAR modulator, capable of providing hippocampal pyramidal neurons with their integrative synaptic properties.
AMPARs form an array of signaling complexes, specialized for a range of functions - from faithfully decoding high frequency inputs to integration of low-frequency signals in support of synaptic plasticity3. This spectrum results from a mosaic of receptors, assembled from four pore-forming subunits (GluA1-4) and auxiliary proteins that exist in various stoichiometries and exhibit distinct expression patterns in the brain4,5. The predominant subunit, GluA2, dictates ion permeation, and forms heteromers with GluA1 or GluA3 in principal forebrain neurons2,6.
Members of the transmembrane AMPAR regulatory proteins (TARPs-γ2, 3, 4, 5, 7, 8) and cornichon homologue (CNIH2, 3) families are the most widely expressed auxiliary subunits2. TARP-γ8 and CNIH2 are abundant at cortical and hippocampal synapses7,8, and are powerful modulators9,10 that co-purify and synergize to determine AMPAR expression levels, kinetics and pharmacology7,11–13. Both are four-helical bundle transmembrane proteins, yet they have different topologies, resulting in different mechanisms of action. Whilst engaging the receptor transmembrane domain (TMD), TARPs possess an elaborate extracellular beta-sheet structure that contacts the ligand-binding domains (LBDs)14–16. CNIHs are mostly embedded within the membrane17, but have two cytoplasmic loops of unknown structure that contribute to their function as cargo exporters from the endoplasmic reticulum (ER)18. Hetero-auxiliary receptor complexes have yet to be resolved. Therefore, the stoichiometry and distinct modulatory mechanism of each associating protein are poorly understood. To address these questions, we studied a principal AMPAR, the GluA1/2 receptor assembled with TARP-γ8 and CNIH2.
GluA1/2/γ8/CNIH2 resembles native AMPARs
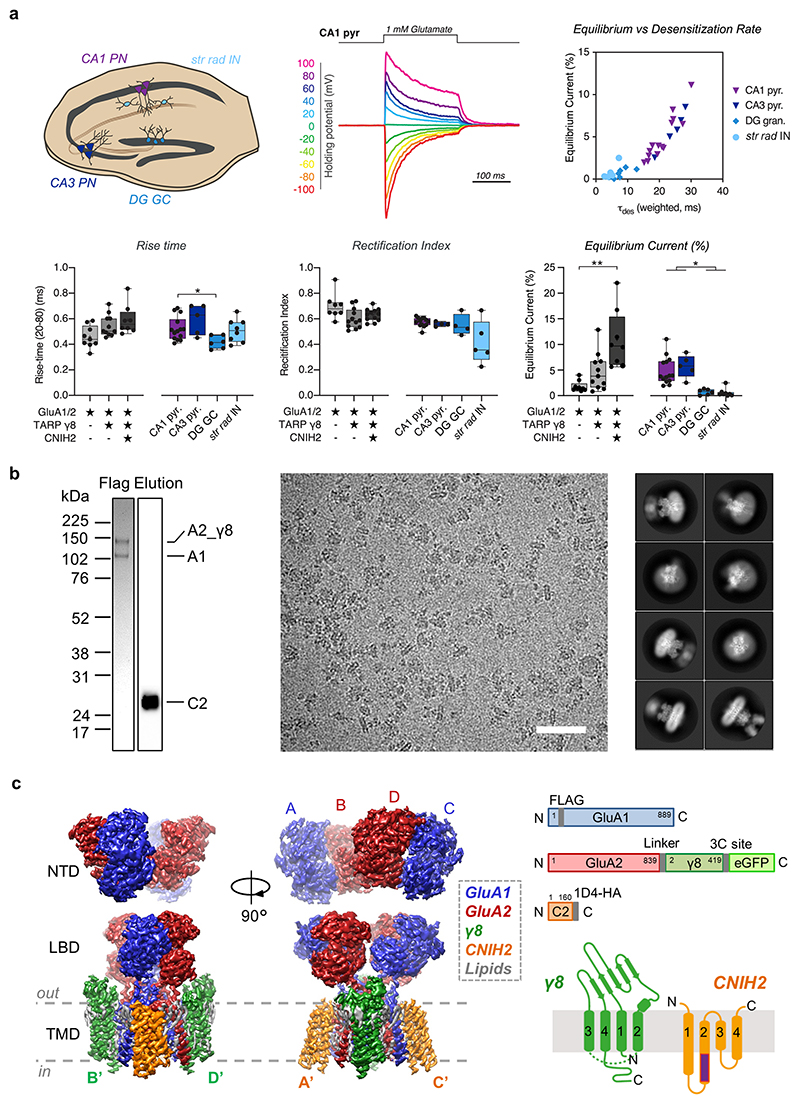
We reconstituted the AMPAR complex and compared its properties to those of hippocampal AMPARs. CNIH2-expressing HEK-Expi293F cells were co-transfected with GluA1 and a GluA2_γ8 (A2_γ8) fusion gene, to ensure the presence of two γ8 subunits15. Although AMPAR stoichiometries with four γ8 molecules have been proposed19, the co-existence of γ8 and CNIH2 predominates in forebrain neurons7,11. In response to rapid glutamate application to excised membrane patches, γ8 slowed desensitization rates about 2-fold compared to GluA1/2 alone, and addition of CNIH2 led to further slowing that was greater than with γ8 alone (Fig. 1a, b)15. CNIH2 also increased the equilibrium current (Extended Data Fig. 1a). Together with previous data13, this confirms incorporation of CNIH2 into a hetero-auxiliary complex.
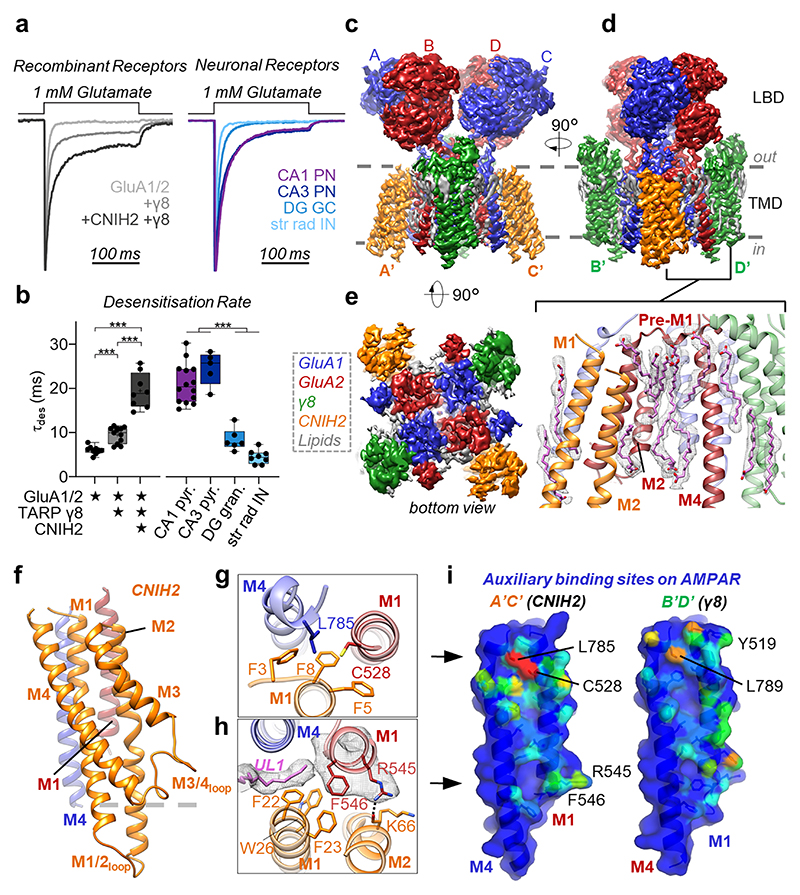
Fig. 1. Physiology and architecture of the GluA1/2_γ8 /CNIH2 complex.
a,b, Auxiliary subunits slow desensitization rates of recombinant receptors (a, left). Native AMPARs show diverse properties (a, right), with A1/2_γ8 +CNIH2 recapitulating CA1 & CA3-like kinetics (b, weighted τdes (ms), mean ± SEM). Recombinant receptors: GluA1/2: 6.01 ± 0.31, n=9; +γ8: 9.29 ± 0.53, n=12; +CNIH2+γ8: 19.71 ± 1.27, n=9. Neuronal receptors: CA1 pyramidal: 21.03 ± 1.19, n=14; CA3 pyramidal 24.51 ± 1.69, n=5; DG granule cell: 8.38 ± 1.01, n=6; CA1 stratum radiatum interneurons: 4.48 ± 0.55, n=8; Welch’s ANOVA tests with Dunnett’s multiple comparisons test - Recombinant: W(2,15.11) = 60.68, p<0.0001; Neurons: W(3,11.60) = 76.28, p<0.0001; see Supplementary Table 1 for details). Boxes represent 25 % to 75 % percentile, whiskers minimum/maximum values and central line median. c-e, Cryo-EM maps, depicting the LBD and TMD domain layers. Core subunits positioned to AC/BD, and auxiliary subunits at A’C’/B’D’ sites are shown. (c) Front view, depicting γ8 at the B’D’ sites. (d) Side view, visualising CNIH2 at A’C’ sites. Inset: lipids concentrating at the TMD, beneath the GluA2 pre-M1 helix. (e) Bottom view, highlighting CNIH2 binding to GluA2 transmembrane helices M1-3 (red), and γ8 to the GluA1 M1-3 helices (blue). f, Model of CNIH2, including the M1/2 and M3/4 cytosolic loops, docking to its binding site (M1GluA2 [red] M4GluA1 [blue]). g, CNIH2 Phe3, -5, -8 slotting into its binding site close to Cys528 (GluA2) and Leu785 (GluA1). h, CNIH2 contacts at the bottom of the binding site, mediated by Phe23 and Lys66; lipid (UL1) penetrates the A’C’ site and interacts with GluA2 Phe546. i, A’C’ (left) and B’D’ (right) site surface representation. M1A2 and M4A1 residues contacted by CNIH2 are coloured depending on the number of atoms contributing to the interaction (red: high, blue: low). Contacts were counted using ‘findNeighbors’ in ProDy38’, with a 4.5 Å cutoff between heavy atoms.
We found close correspondence between responses from the recombinant complex and AMPARs expressed in CA1 and CA3 pyramidal neurons, but not to those from dentate granule cells or CA1 interneurons, where γ8 and other auxiliary subunits prevail (Fig. 1a, b, Extended Data Fig. 1a)20. This demonstrates the diversity of AMPAR complexes, and provides further evidence that CNIH2 contributes to the response properties of pyramidal neurons, with the GluA1/2/γ8/CNIH2 octamer the major assortment in these cells7,11,13. We subjected this native-like complex for structural analysis by cryo-EM.
Trapping active and inactive AMPARs
To resolve gating transitions, the complex was trapped in an inactive state, bound to the competitive antagonist NBQX, and in an active state with the agonist L-glutamate together with the desensitization blocker cyclothiazide (CTZ) (Extended Data Fig. 1b). 3-D reconstructions revealed the three-layered domain architecture, with the extracellular N-terminal domain (NTD) and LBD arranged as dimers of dimers, attached to the ion channel of pseudo four-fold symmetry (Fig. 1c-e, Extended Data Fig. 1c). Focused processing of the LBD-TMD and of individual domain layers resulted in maps ranging from 3.0 Å to 3.7 Å (Extended Data Fig. 2-4, and Extended Data Table 1), permitting us to follow gating transitions at comparable resolutions. Moreover, resting-state particles lacking CNIH2 enabled us to examine the effects of CNIH2-association with the receptor (Extended Data Fig. 2).
Subunit arrangement and stoichiometry
EM-map quality permitted unambiguous assignment of the four core subunits15,21, placing GluA1, marked by unique N-glycans in the NTD (Extended Data Fig. 5a), to the outer (or AC) positions, and GluA2 to the inner (BD) positions (Supplementary Video 1). This arrangement dictates function as the BD subunits exert a greater force on the channel gate than the AC subunits22, with consequences on the entire complex, as we describe below. The core subunits alternate around the pore with their peripheral M1 and M4 transmembrane helices contributing two pairs of non-equivalent binding sites for auxiliary subunits: the A’C’ sites (M1GluA2/M4GluA1) locate beneath the LBD dimers, and are thus spatially more restricted than the B’D’ sites (M1GluA1/M4GluA2), beneath the LBD dimer-of dimers interface (Fig. 1c-e). To reveal the preferred stoichiometry and arrangement of TARP-γ8 association, we determined a structure of a GluA2 homomer fused to γ8. In this receptor, only two γ8 subunits at the B’D’ sites are apparent, while the A’C’ TARPs were displaced (Extended Data Fig. 6). Hence, similar to GSG1l23, another auxiliary subunit with a bulky extracellular region, γ8 disfavors the A’C’ sites, and preferentially associates in a two-TARP stoichiometry. With γ8 binding to the B’D’ site15, CNIH2 occupies the A’C’ positions. This organization determines gating, as each site provides differential access of the auxiliaries to the gating machinery.
Our CNIH2 structural model includes its two sequence-diverse cytoplasmic loops between the M1/2 and the M3/4 helices, which were not resolved in CNIH317 (Extended Data Fig. 5f). Lodging to the A’C’ sites, the CNIH2 M1 and M2 helices project into the cytoplasm (>20 Å) (Fig. 1f), forming a loop with currently unknown function. A comparison of the binding modes for CNIH2 and γ8 shows that both engage the same residues on the receptor, yet their footprints vary, and differences in side chain orientations along the M1 helix are apparent (Fig. 1g-i, Extended Data Fig. 7a). While TARP-γ8 contacts are spread out more evenly, CNIH2 prominently engages two regions: the upper tier of the A’C’ site, centered around GluA2 Cys528, and its base, adjacent to GluA2 Phe546 (Fig. 1i). Three highly conserved N-terminal phenylalanines (Phe3, -5, -8)17 anchor CNIH2 proximal to the gate (Fig. 1g), and these are crucial for modulation, as we show below. At the base, CNIH2 interacts with Arg545 and Phe546 through polar and hydrophobic contacts (Fig. 1h), at a region shaped by lipids.
Lipids shape auxiliary subunit binding
An abundance of lipids ensheath the ion channel (inset in Fig. 1d, Extended Data Fig. 7b). Lipids in the upper leaflet connect the gate-surrounding pre-M1 helices to the M2 pore helices in the lower leaflet. Their location within the channel lateral fenestrations indicate a role in channel modulation. Other lipids, denoted lower-leaflet lipid (LL) 1 and LL2, bridge between auxiliary subunits and the M2 pore helix, thereby extending the reach of these modulating proteins to the conduction path (Extended Data Fig. 7c, d).
The acyl chains of two lipids penetrate the lower part of the CNIH2 binding site: wedging between GluA1 and CNIH2, upper-leaflet lipid (UL) 1 contacts Phe546 (Fig. 1h); immediately beneath, LL3 anchors CNIH2 via its Phe22 and Trp26 side chains to the base of the GluA1 M4 helix (Extended Data Fig. 7d). These acyl chains likely influence CNIH2 binding by extending the hydrophobic network at the A’C’ site. This idea is supported by the GluA2-CNIH3 structure (PDB 6PEQ), where some lipids are also observed17, but LL1 and LL2 are missing. As a result, CNIH3 aligns with the receptor more closely than CNIH2 (Extended Data Fig. 7e). Hence, lipids may regulate CNIH-function.
The GluA2 subunit dominates gating
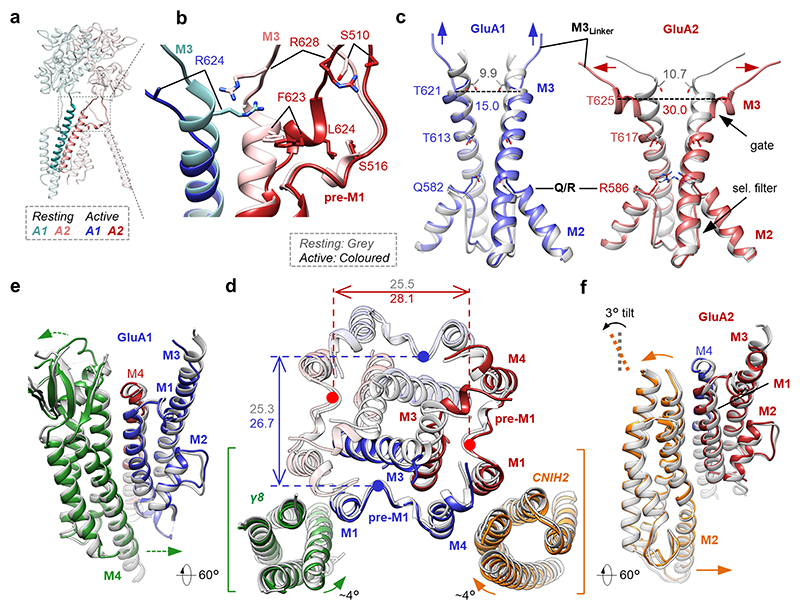
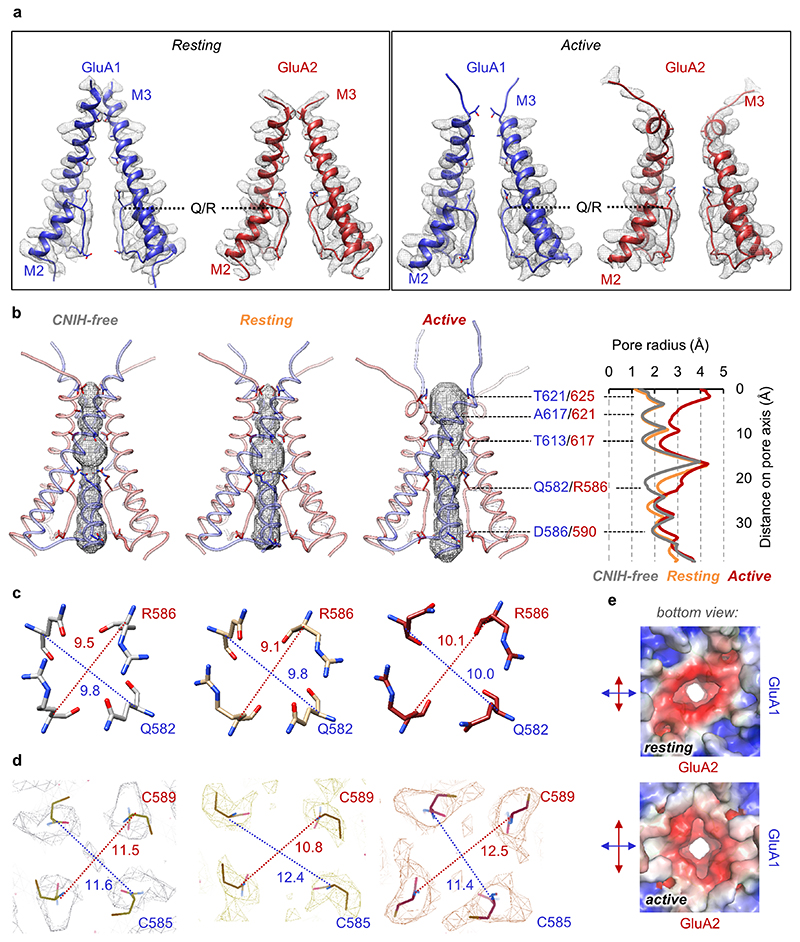
Receptor activation is triggered by glutamate closing the LBD ‘clamshells’24 (Extended Data Fig. 5b-e), leading to separation of the lower lobes within an LBD heterodimer and rearrangement of the LBD-TMD linkers to open the gate14,16. This transition is well resolved. Focusing on the outer gate formed by the M3 transmembrane helices, M3 linker tension ruptures polar contacts between GluA1 Arg624 and GluA2 Arg628, a position determining gating kinetics25, and breaks a stacking interaction between GluA1 Arg624 and Phe623 in GluA2 (Fig. 2a, b). These changes are stabilized by reorganization of the gating linkers and are accompanied by opening of the gate, formed by hydrophobic contacts facing the pore axis along the conserved SYTANLAAFLT motif (Fig. 2b, c), as has been observed for active-state GluA2/γ214,16.
Fig. 2. Gating transitions of the A1/2_γ8/C2 octamer.
a, View onto a GluA1/GluA2 LBD dimer and M3 gating helices (bold) in the resting state. b, Side view onto the M3 gate with open (bold colour) and resting (soft colour) states overlain, depicting rearrangement of the M3 linkers and M3 side chains relative to pre-M1. c, Comparison of GluA1 and GluA2 conduction path; resting state (grey) is superimposed on the active state (colour). Divergence from the closed gate at the top of M3 is more pronounced in GluA2 (Cα distances between opposing T621 (GluA1), and T625 (GluA2) indicated). Also shown is the selectivity filter with the Q/R site. d, Top view onto the closed-(grey) and open-state (colour) models at the level of the M3 gate. The dilation of the gate-surrounding helices is measured between opposing pre-M1 helices. Rotation angles were calculated by averaging angular displacements of all Cα atoms in each auxiliary subunit between the two states relative to center of mass of each auxiliary subunits. e, Side view showing outward expansion of the upper part of GluA1 M1 and M3 and the γ8 β-sheet, accompanied by inward movement of the γ8 M4 helix towards the GluA1 M12 loop. f, CNIH2 undergoes a ~3° pivot (‘tilt’) on activation, moving its M12 loop closer to the pore axis.
Gate dilation is more pronounced between the two opposing GluA2 subunits than between GluA1s, and this continues into the selectivity filter, formed by the M2 pore loops (Fig. 2c). At the Q/R site (Arg586), which determines Ca2+ flux and conductance at the M2-loop apex26,27, activation widens the filter entrance selectively between the GluA2 subunits (Extended Data Fig. 8a-b). This asymmetric dilation, has also been described for active-state GluA2/γ2 (PDB 6DLZ28), and extends down to GluA2 Cys589 in the filter and to the M2 pore helices. The M2 helices separate by > 1 Å in GluA2 but not GluA1, which is apparent at the cytoplasmic face of the pore, a site modulated by polyamines29 (Extended Data Fig. 8c, d, e). We note that CNIH2 association, which increases channel conductance30, does not cause a visible expansion of the selectivity filter, when comparing our resting-states with and without CNIH2 (Extended Data Fig. 8b-d).
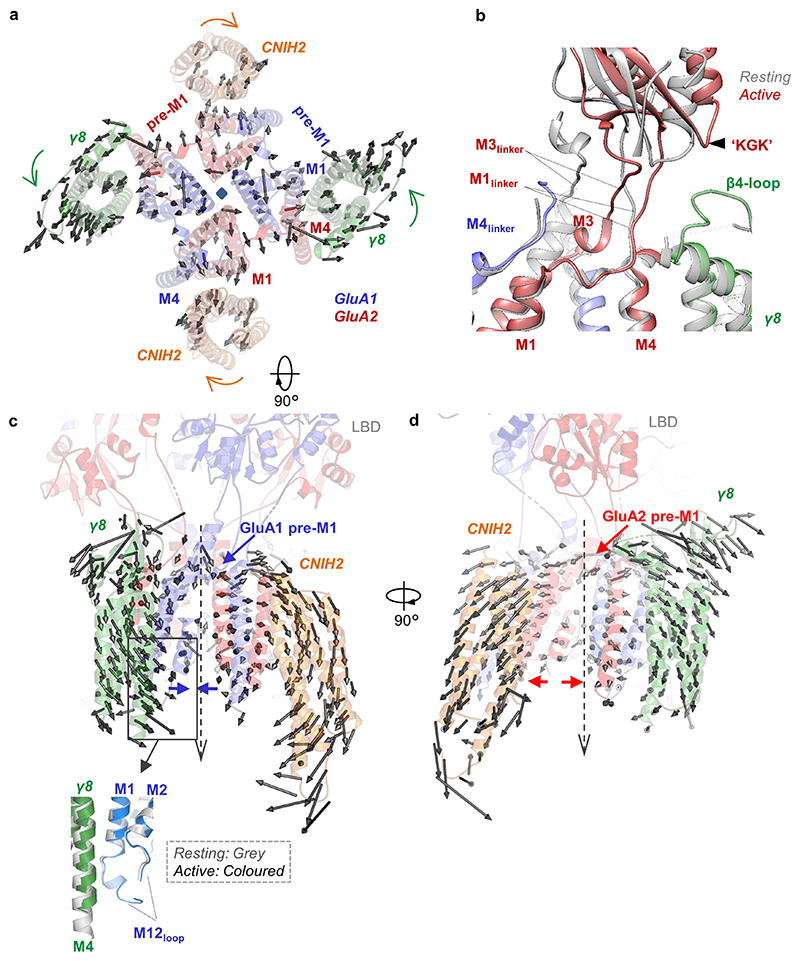
Activation is accompanied by an anti-clockwise twist of the TMD helices, and an expansion in the upper part of the channel surrounding the gate (Extended Data Fig. 9a). This dilation is greater between opposing GluA2 subunits than between GluA1, and includes the channel’s peripheral helices (pre-M1, M1 and M4) and both pairs of auxiliary subunits (Fig. 2d).
Gating transitions in the hetero-octamer
Dictated by its architecture, gating differentially impacts the interactions between core and auxiliary subunits. On activation, the GluA2 M3 gate helix pushes its Phe623 and Leu624 side chains against its pre-M1 helix, triggering channel dilation (Fig. 2b, d, Supplementary Video 2). Pre-M1 couples LBD closure to channel opening, and shapes gating kinetics throughout iGluRs31–33. At the same time, the GluA2 M1 and M3 linkers closely align and approach the TARP extracellular (β4) loop, which is simultaneously contacted by the GluA2 LBD through its ‘KGK’ motif34 (Extended Data Fig. 9b). GluA1 does not experience these large changes, as its M3 linkers are pulled upward (Fig. 2c), a characteristic of AC subunits14,16. As a result, the GluA1 M3 helices minimally contact pre-M1 on activation, which underlies the asymmetric expansion of the channel (Fig. 2d). We also observe newly formed contacts between the GluA1 M4 linkers (at Thr780) with the GluA2 M3 gate on activation, which are not apparent with GluA1 M3. Together, these rearrangements lead to enhanced engagement of GluA2 by CNIH2 and γ8.
Auxiliary subunit counter-rotations
Expansion of the gate transmits to both auxiliary subunit pairs, leading to global structural changes (Fig. 2d): The TARP extracellular beta-sheet segment bends away from the pore axis; its TMD sector undergoes a right-handed twist relative to its center of mass, while CNIH2 turns left-handedly (Fig. 2d-f). As a net result, the GluA2 side of the channel (between the GluA2 M1 and M4 helices) widens on activation, while the GluA1 side (between GluA1 M1 and M4) contracts (Extended Data Fig. 9c, d), contributing to the two-fold symmetry-switch upon opening. The counter-rotations of the two auxiliaries are likely driven by their different topologies (Extended Data Fig. 1c) and is of consequence, as the left-handed twist of CNIH2 follows the rotation of the channel and thereby likely stabilizes an active, open pore conformation. Together, these auxiliary subunits synergise to promote channel activation.
Rotations of the auxiliary subunits are also accompanied by subtle tilts in the vertical plane, bringing them into closer reach of cytoplasmic portions of the receptor (Supplementary Video 3). We observe formation of contacts between the γ8 M4 helix and the start of the GluA1 M1/2 cytoplasmic loop (Fig. 2e, inset in Extended Data Fig. 9c), and tilting of the cytoplasmic part of the CNIH2 M2 helix towards the pore (Fig. 2f). This behaviour is also evident by normal mode analysis (Supplementary Video 4)35,36, where both twisting and pivoting of CNIH2 is apparent in a number of modes. Hence, activation facilitates interaction of both auxiliaries with the AMPAR intracellular portion, which is currently unresolved.
Mechanism of CNIH2 modulation
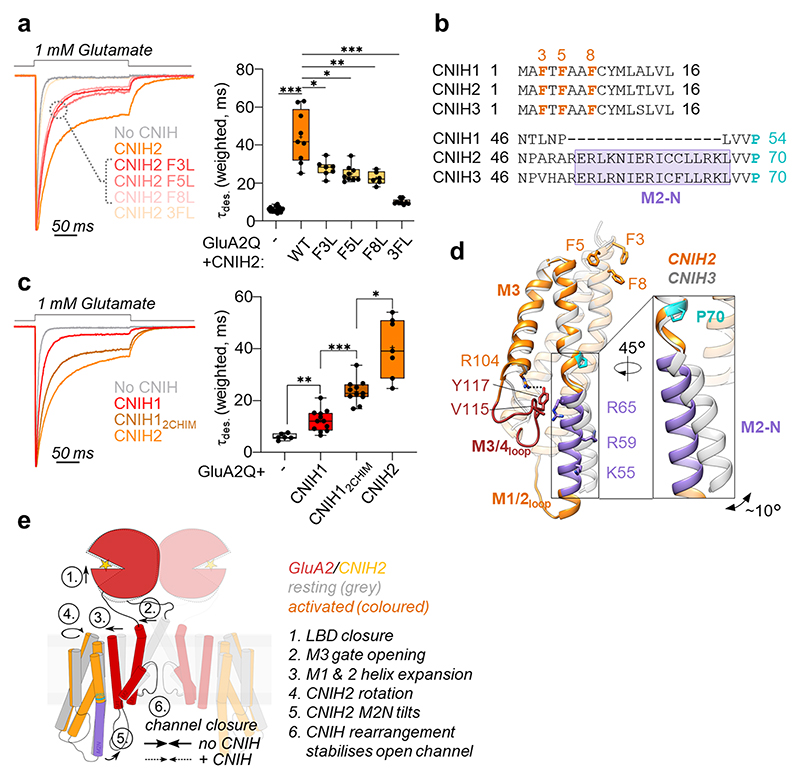
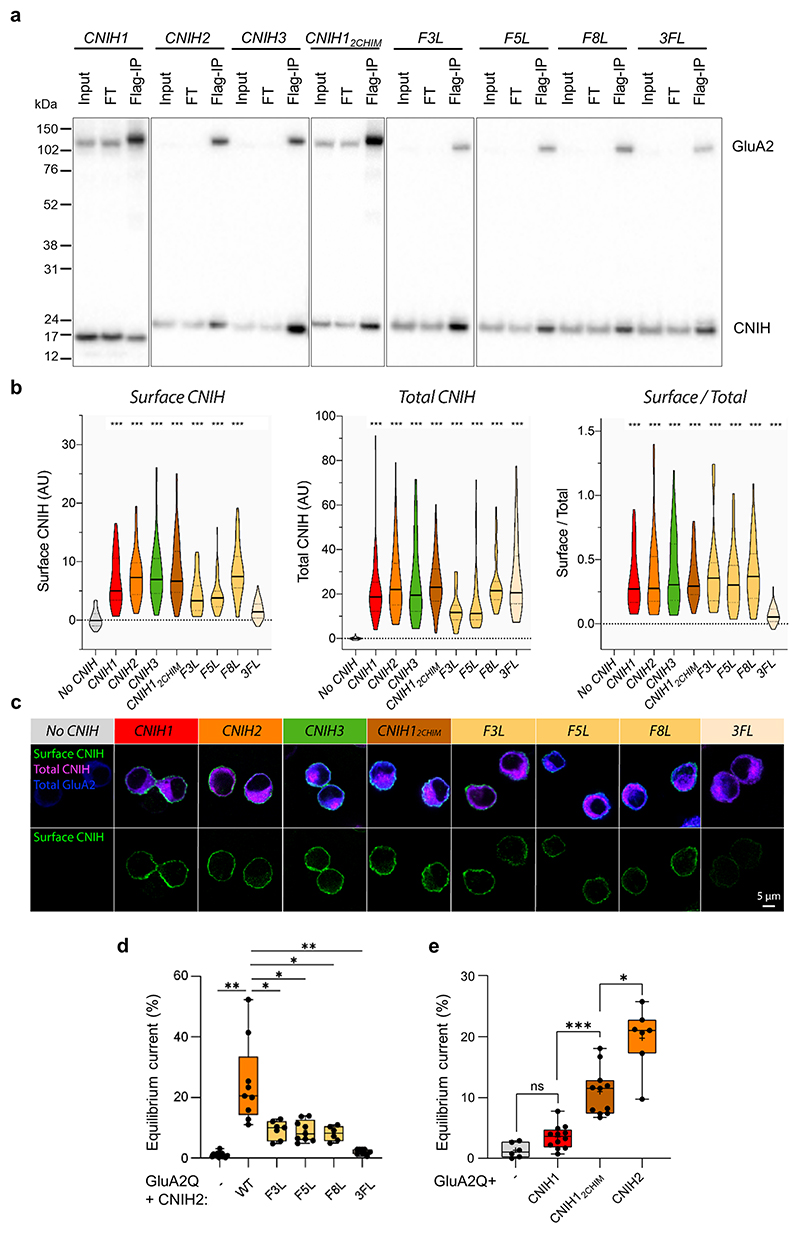
Dictated by their architecture, CNIHs will exert AMPAR modulation exclusively at the TMD or cytoplasmic levels. Three conserved phenylalanines (Phe3, -5, -8) slot CNIH2 (and CNIH317) into the upper tier of the A’C’ sites (Fig. 1g, i). When mutated individually to leucine, all three residues weakened CNIH2’s modulation of GluA2 kinetics, and reduced the equilibrium response, but did not affect complex formation or trafficking (Fig. 3a, Extended Data Fig. 10a-d). Simultaneous mutation of all three residues to leucine reduced trafficking and, consequently, gating modulation (Fig. 3a, Extended Data Fig. 10a-d), highlighting this interaction as an important modulatory site.
Fig. 3. Mechanism of AMPAR modulation by CNIH2.
a, Mutations of three phenylalanines in the CNIH2 N-terminus speed GluA2 desensitization: weighted τdes (ms), mean ± SEM – GluA2 alone: 6.16 ± 0.35, n=15; CNIH2: 44.32 ± 4.58, n=9; F3L: 27.73 ± 1.58, n=7; F5L: 24.85 ± 1.43, n=9; F8L: 22.57 ± 1.40, n=6; 3FL: 10.07 ± 0.46, n=9; Welch’s ANOVA with Dunnett’s multiple comparisons test: W(5,17.83) = 85, p<0.0001; further details in Supplementary Table 2. Boxes represent 25 % to 75 % percentile, whiskers minimum/maximum values and central line median. b, Sequence alignment of mouse CNIH1-3, highlighting the conserved N-terminal phenylalanines and the M2-N region, where CNIH1 lacks 16 residues; Pro70 is shown in cyan. c, The CNIH2 M2-N helix contributes to modulation of GluA2 kinetics, demonstrated by a gain-of-function when transplanted onto CNIH1: weighted τdes (ms), mean ± SEM – GluA2 alone: 6.03 ± 0.47, n=6; CNIH1: 12.22 ± 1.20, n=12; CNIH12chimera: 23.48 ± 1.39, n=11; CNIH2: 40.45 ± 4.32, n=7; Welch’s ANOVA with Dunnett’s multiple comparisons test: W(3,15.30) = 64.09, p<0.0001; further details in Supplementary Table 2. Box plot parameters as in b. d, Superposition of CNIH2 (orange) and CNIH3 (grey; PDB 6PEQ), using the top of their M1 and M2 helices. CNIH2 M2-N (purple) kinks at Pro70 (cyan) and diverges from CNIH3 by ~ 10° (inset); the N-terminal phenylalanines are also indicated. An interaction with the M3/4 loop through Val115 and Arg65 is shown, Arg55 and Arg59 project toward the pore axis. e, Molecular mechanism underlying AMPAR modulation by CNIH2.
Of note, the CNIH2 N-terminus is conserved in CNIH1 (Fig. 3b), which also associates with the receptor and traffics to the cell surface, but modulates gating only weakly (Fig. 3c, Extended Data Fig. 10a-c, e)30,37. CNIH1 lacks an elongated N-terminal segment in its M2 helix (‘M2-N’), which is present in both CNIH2 and CNIH3 (Fig. 3b, d). M2-N is created by a conserved proline (Pro70), that structurally ‘uncouples’ M2-N from the rest of the M2 helix, enabling M2-N tilting towards the pore on activation (Fig. 2f, Supplementary Videos 3, 4). Structural flexibility of M2 is exemplified by its different kink angle in CNIH2 and CNIH3, and by the CNIH2 M3/4 loop, harboring a potential cholesterol binding site (Extended Data Fig. 7f), which straddles M2-N to impact its dynamics (Fig. 3d).
To test the role of M2-N, we inserted this segment from CNIH2 into CNIH1. This chimera strongly gained modulatory activity, slowing GluA2 desensitization and increasing the equilibrium response (Fig. 3c, Extended data Fig.10e), implying a critical role for M2-N interactions with cytoplasmic receptor elements. Given the presence of positively charged side chains projecting from M2-N towards the negatively charged M1/2 receptor loops (Fig. 3d), these two cytosolic elements are candidates to interact and enable this regulation.
Conclusion
Based on these data, we propose the following mechanism (Fig. 3e): Association with CNIH2 stabilises the active-state conformation, through local contacts between Phe3,-5,-8 and gate-surrounding receptor residues. Opening of the M3 gate triggers a left-handed rotation of CNIH2, that promotes the open state by stabilising the expanded states of the A’C’-site helices and M2 loop, with a dilated pore. Full modulation requires additional contacts between M2-N and cytosolic receptor elements, which further maintain the ‘active’ conformation, slowing relaxation of the rotated regions back to the closed conformation. This scheme accounts for residual modulation by CNIH1, which supports the active state locally through its conserved N-terminus, but in lacking M2-N, is unable to maintain the fully active-state conformations.
Dictated by their position in the octameric complex and by topological differences, these auxiliary subunits operate in concert, through their TMD helices and through structurally distinct cytoplasmic regions. However, while TARPs engage the LBDs and the M1&M3 gating linkers via their extracellular portion, CNIH2 & 3 act through their distinct TMD arrangements, and now resolved cytoplasmic loops. These appendages, unique to each auxiliary subunit, determine the rotational behavior of CNIH2 and γ8 to stabilise transient gating conformations. These mechanisms combine to build a relatively slowly deactivating AMPAR complex, providing hippocampal pyramidal neurons with the required synaptic properties to integrate temporally coinciding inputs.
Methods
cDNA constructs
All cDNA constructs were produced using IVA cloning 39. To achieve heteromeric AMPAR expression, GluA1 (rat cDNA sequence, flip isoform) was co-expressed with GluA2 (rat cDNA sequence, flip isoform, R/G edited, Q/R edited), expressed from the pRK5 vector. For TARP-containing complex recordings, GluA2 was expressed in a tandem configuration (denoted GluA2_γ8) with TARP γ8 (rat cDNA sequence) by cloning the TARP γ8 coding sequence (Glu2 - Lys419) at the C-terminus of the GluA2 coding sequence (Val1 to Ser839), in the pRK5 vector, separated by a Gly-Gly-Ser-Gly-Ser-Gly linker sequence. pN1-EGFP (Clontech) was used for visualisation of transfected cells. CNIH2 (rat) was expressed either from a stable cell line (heteromeric AMPAR recordings, see below), or from either a pRK5 or a pBOS vector, with an HA-tag at the extreme C-terminus. A chimera of CNIH1 and CNIH2 (CNIH12CHIM) was made by inserting 16 amino acids from CNIH2 (51-RERLKNIERICCLLRK-66) into CNIH1 at position P50-L51.
Organotypic slice preparation
All procedures were carried out under PPL 70/8135 in accordance with UK Home Office regulations. Experiments conducted in the UK are licensed under the UK Animals (Scientific Procedures) Act of 1986 following local ethical approval.
Organotypic slice cultures were prepared as described previously 40. Briefly, hippocampi from P6-8 C57/Bl6 mice were isolated in high-sucrose Gey’s balanced salt solution containing (in mM): 175 sucrose, 50 NaCl, 2.5 KCl, 0.85 NaH2PO4, 0.66 KH2PO4, 2.7 NaHCO3, 0.28 MgSO4, 2 MgCl2, 0.5 CaCl2 and 25 glucose at pH 7.3. Hippocampi were cut into 300 μm thick slices using a McIlwain tissue chopper and cultured on Millicell cell culture inserts (Millipore Ltd) in equilibrated slice culture medium (37°C/5% CO2). Culture medium contained 78.5% Minimum Essential Medium (MEM), 15% heat-inactivated horse serum, 2% B27 supplement, 2.5% 1 M HEPES, 1.5% 0.2 M GlutaMax supplement, 0.5% 0.05 M ascorbic acid, with additional 1 mM CaCl2 and 1 mM MgSO4 (all from Thermo Fisher Scientific; Waltham, MA). Medium was refreshed every 3-4 days. Recordings were performed at 7-14 days in vitro.
Recombinant cell electrophysiology
For heteromeric AMPAR experiments, suspension HEK-Expi293F™ cells (ThermoFisher Scientific) were cultured in Expi293™ expression media (Gibco) and were transfected using PEI Max with a DNA to PEI ratio at 1:3. GluAl, GluA2 (with or without TARP γ8 tethered via a flexible linker, as described before 15 and EGFP plasmids were transfected at a 2:8:1 stoichiometry to aid heteromeric receptor production, and identification of transfected cells.
For homomeric AMPAR experiments, GluA1 or GluA2Q (pIRES2-mCherry or pIRES2-EGFP) and CNIH2 (pRK5 or pBOS) plasmids were transfected at a 1:2 ratio using Effectene (QIAGEN) into adherent HEK293T cells (ATCC: Cat# CRL-11268, RRID: CVCL_1926, Lot 58483269: identity authenticated by STR analysis, mycoplasma negative). Cells were cultured at 37°C and 5% CO2 in DMEM (Gibco; high glucose, GlutaMAX, pyruvate, Cat#10569010) supplemented with 10% foetal bovine serum (Gibco) and penicillin/streptomycin. Transfected cells were recorded from 36 hours post-transfection. Cells were plated on poly-L-lysine coated glass coverslips approximately 12 hours before recording, to allow outside-out patch recording and 30 μM 2,3-dioxo-6-nitro-1,2,3,4-tetrahydrobenzo[f]quinoxaline-7-sulfonamide (NBQX; Tocris, Cat#1044) was added to media post-transfection to avoid AMPAR-mediated toxicity.
Borosilicate glass electrodes (1.5mm o.d., 0.86mm i.d., Science Products GmbH), pulled with a PC-10 vertical puller (Narishige) with tip resistance of 2 to 6 MΩ, were filled with internal solution containing (in mM) CsF (120), CsCl (10), EGTA (10), HEPES (10), and spermine (0.1), adjusted to pH 7.3 with CsOH. Surface membrane patches were acquired by gentle excision after achieving the whole-cell patch clamp configuration. Patches were voltage clamped at -60 mV, and subject to fast application of 1 mM L-glutamate using two-barrel theta glass tube controlled by a piezoelectric translator (Physik Instrumente or Burleigh), allowing solution exchange in around 200 μs (open tip response). Signals were acquired using the MultiClamp 700B or Axopatch 200B amplifier (Axon Instruments), digitised using a Digidata 1440A interface and recorded with pClamp10 (Molecular Devices). Extracellular solution contained (in mM) NaCl (145), KCl (3), CaCl2 (2), MgCl2 (1), glucose (10), and HEPES (10), adjusted to pH 7.4 using NaOH. 20 μM IEM 1925 dihydrobromide (Tocris,
Cat#4198) was added to the extracellular solution to limit the contribution of homomeric AMPAR complexes to heteromeric recordings.
Current/voltage relationships were recorded for AMPAR responses with holding potentials of -100 mV to +100 mV in 20 mV steps. To ensure heteromeric receptor analysis, patches with a rectification index of less than 0.5 were discarded. Recordings were not corrected for the liquid junction potential. Rectification index was calculated as the receptor current flow at + 40 mV holding potential, divided by that at -60 mV holding potential, with both values relative to the current at 0 mV [RI = -(I40mV - I0mV)/(I-60mV - I0mV)].0
Desensitization entry was determined from the first 150-200 ms after the peak response, which was fitted with a two-exponential function to obtain the (weighted) time constant. ‘Equilibrium’ responses were denoted as the percentage of peak current remaining after 200 ms.
Signal acquisition and data analysis were performed using pClamp10.
Neuronal cell electrophysiology
Cultured organotypic hippocampal slices were submerged in artificial cerebrospinal fluid (aCSF) containing (in mM): 125 NaCl, 2.5 KCl, 1.25 NaH2PO4, 25 NaHCO3, 10 glucose, 1 sodium pyruvate, 4 CaCl2 and 4 MgCl2 at pH 7.3 and saturated with 95% O2/5% CO2. Neuronal surface patches were acquired from the cell soma as detailed above for recombinant cell patches, using borosilicate pipettes containing (in mM): 135 CH3SO3H, 135 CsOH, 4 NaCl, 2 MgCl2, 10 HEPES, 4 Na2-ATP, 0.4 Na-GTP, 0.15 spermine, 0.6 EGTA, 0.1 CaCl2, at pH 7.25. Agonist was applied to neuronal patches using a piezo-driven theta-barrelled perfusion setup, with aCSF containing (in mM): 140 NaCl, 3.5 KCl, 1 MgCl2, 2.5 CaCl2, 10 HEPES, 10 glucose, 1 sodium pyruvate, 2 NaHCO3, at pH 7.3. 100 μM D-APV (Tocris, Cat #0106) was added to the extracellular solution of both barrels to prevent the contribution of NMDA receptor currents. 1 mM L-glutamate was added to one barrel for fast-agonist application (as above). A subset of hippocampal CA1 pyramidal neuron patches were recorded using the same extra- and intracellular solutions as recombinant cell electrophysiology, and no difference in kinetic parameters analysed was observed between recording configurations, therefore these datasets were pooled and presented together.
CA1 and CA3 pyramidal, and dentate gyrus (DG) granule cells were identified by cell morphology and location in tissue. Stratum radiatum interneurons (IN) were selected for analysis as a population of interneuronal cells that can be identified by location, rather than requiring a genetic driver line, and consisted of neurons located between stratum pyramidale and stratum lacunosum-moleculare in the CA1 region of organotypic slices. Cell identity was not confirmed by morphological or other analysis post-hoc, and therefore likely consists of a heterogeneous combination of IN cell types.
Generation of CNIH2-1D4-HA stable cell line
A CNIH2-1D4-HA stable expression cell line was generated using a lentiviral expression system following an established protocol 41. A 1D4 tag followed by an HA tag were included at the extreme C-terminus of rat CNIH2 with a TEV cleavage site, separated by a ‘GGS’ linker sequence. The CNIH2 gene together with the two tags were synthesized and cloned into pHR vector. pHR-CNIH2-ID4-HA was co-transfected with psPAX2 and pMD2.G into HEK293T Lenti-X cells (Takara/Clontech, Cat# 632180). 72 hours after transfection, lentiviral particles were harvested from the media and used to infect HEK-Expi293F cells. 48 hours after infection, fresh Expi293™ expression media (Gibco) was added, cells were surface-labelled with an APC-conjugated anti-HA antibody (Miltenyi Biotec, Cat# 130-098-404, 1:50) and sorted by flow cytometry, collecting HA-positive and DAPI-negative cells. Positive cells were then scaled up for A1/A2_γ8/C2 complex expression.
Expression and purification of A1/A2_γ8/C2 and A2_γ8
For the A1/A2_γ8/C2 heteromeric complex, constructs of FLAG-tagged GluA1(flip) and GluA2(flip)-TARPγ8-eGFP tandem (pRK5) used in this paper are the same as previously reported 15. GluA1 and GluA2-TARPγ8 were co-transfected into the CNIH2-1D4-HA stable expression cell line at a ratio of 1:1. To prevent AMPA-mediated excitotoxicity, AMPAR antagonists ZK200775 (2 nM, Tocris, Cat# 2345) and kynurenic acid (0.1 mM, Sigma, Cat# K335-5G) were added to the culture medium. 36-44 hours post-transfection, cells were harvested and lysed for 3 hours in lysis buffer containing: 25 mM Tris pH 8, 150 mM NaCl, 0.6 % digitonin (w/v) (Sigma, Cat# 300410-5G), 5 μM NBQX, 1 mM PMSF, 1× Protease Inhibitor (Roche, Cat# 05056489001). Insoluble material was then removed by ultracentrifugation (41,000 rpm, 1 hour, rotor 45-50 Ti) and the clarified lysate incubated with anti-GFP beads for 3 hours. After washing with glyco-diosgenin (GDN) (Anatrace, Cat# GDN101) buffer (25 mM Tris pH 8, 150 mM NaCl, 0.02% GDN) the protein was eluted from the beads by digestion with 1 mg/ml 3C protease at 4 °C overnight. Eluted fractions were incubated with FLAG beads (Sigma, Cat# A2220) for 1.5 hours and washed 3 times with GDN buffer. Finally, the complex was eluted using 0.15 mg/ml 3×FLAG peptide (Millipore Cat# F4799) in GDN buffer. Eluted fractions were pooled and concentrated to ~3 mg/ml for cryo-EM grid preparation.
For the A2_γ8 homomeric complex, the same GluA2(flip)-TARPγ8-eGFP tandem construct was used for expression and purification. GluA2-TARPγ8 was transfected into the Expi293™ cell line with the addition of ZK200775 (2 nM) and kynurenic acid (0.1 mM). 36-44 hours post-transfection, cells were harvested, lysed, and purified according to the same procedure as heteromeric receptors (above), until the 3C protease digestion step. After digestion, the eluate was collected and concentrated to < 0.5 ml. Concentrated protein was then loaded onto a Superose6 increase 10/300 GL column (GE Healthcare) with GDN buffer. The peak eluted at around 11 ml was collected and concentrated to ~3 mg/ml for grid preparation.
Cryo-EM grid preparation and data collection
Cryo-EM grids were prepared using a FEI Vitrobot Mark IV. For the resting state A1/A2_γ8/C2 heteromeric complex, protein was incubated with 100 μM NBQX for at least 30 min on ice before freezing. Similarly, 100 μM NBQX was added to the resting sate A2_γ8 homomeric complex. For the active state A1/A2_γ8/C2 heteromeric complex, protein was first incubated with 300 μM cyclothiazide (CTZ, Tocris, Cat# 0713) for at least 30 min on ice and then quickly mixed with 1 M L-glutamate stock solution to a final concentration of 100 mM prior to loading onto the grids. Quantifoil Au 1.2/1.3 grids (300 mesh) were glow-discharged for 30 s at 0.35 mA before use. 3 μl sample was applied to the grids, blotted for 4.5-5 s at 4 °C with 100 % humidity and plunge-frozen in liquid ethane. All cryo-EM data were collected on an FEI Titan Krios operated at 300 kV, equipped with a K3 detector (Gatan) and a GIF Quantum energy filter (slit width 20 eV). Movies at 1.5-2.5 μm underfocus were taken in counting mode with a pixel size of 1.07 Å/pixel. A combined total dose of 50 e/Å2 was applied with each exposure and 50 frames were recorded for each movie. For the A1/A2_γ8/C2 heteromeric complex, two datasets of resting state were collected using SerialEM with 10462 movies in total, and two datasets of active state were collected using EPU2 with 8246 movies in total. For the A2_γ8 homomeric complex, 5733 movies were collected using EPU2.
Cryo-EM data processing and model building
Dose-fractionated image stacks were first motion-corrected using MotionCor2 42. Corrected sums were used for CTF estimation by GCTF 43. All further data processing was performed with RELION 3.1 44. For the A1/A2_γ8/C2 heteromeric complex, automatic particle picking was performed using a Gaussian blob and particles were binned to 4.28 Å/pixel and extracted in a box of 80 pixels. 2 to 3 rounds of 2D classification were carried out to remove particles not showing AMPAR-like features. For the following 3D classification, emd-20332 was used as initial model to further eliminate low-quality particles. Following data clean-up, particles were re-centered, scaled up to 2.14 Å/pixel and re-extracted in a box of 160 pixels. Another 3D classification focused only on the LBD-TMD and C2 symmetry was applied on the re-extracted data set with EMD-20330 as an initial model. After this round of classification, CNIH2 containing and CNIH2-free particles were separated into two datasets with 3D refinement applied on each set. Refined particles were scaled to the original 1.07 Å/pixel size, and refined with C2-symmetry followed by post-processing. Four datasets were processed independently in the beginning, and particles at the same states were joined together before CTF refinement and Bayesian polishing. The final refinement was carried with side-splitter to achieve maps at 3.1 Å (CNIH2 containing) and 3.5 Å (CNIH2 free) resolution in the resting state and 3.6 Å (CNIH2 containing) in the active state (individually based on the standard Fourier shell correlation (FSC) 0.143). To further improve map resolution, we applied masked refinement on TMD or LBD region independently by continuing old run from the last iteration of the global LBD-TMD refinement. Local resolution was also estimated by RELION3.1. To help with model building, we used EMDA (https://www2.mrc-lmb.cam.ac.uk/groups/murshudov/content/emda/emda.html) to generate composite maps of LBD-TMD from the mask refinement. However, the density observed at the CNIH region was weaker than that of the receptor. In order to further enhance the density for model building, symmetry expansion was applied on the aligned particles from TMD reconstruction. A masked classification without alignment was then performed to select CNIH stable particles. When classifying the active state data, a smaller mask and higher T value were required for the focused classification. The following focused refinements improved the density of the CNIH region in both resting and active states. For reconstruction of the NTD in the resting state, after scaling particles back up to 2.14 Å/pixel, 3D classifications against the full-length receptor were performed and particles with a stable NTD signal were selected and subjected to masked refinement with a soft mask placed only on the NTD. Finally, particles were rescaled to original size, CTF refinement and Bayesian polishing were preformed to attain a 3.4 Å map.
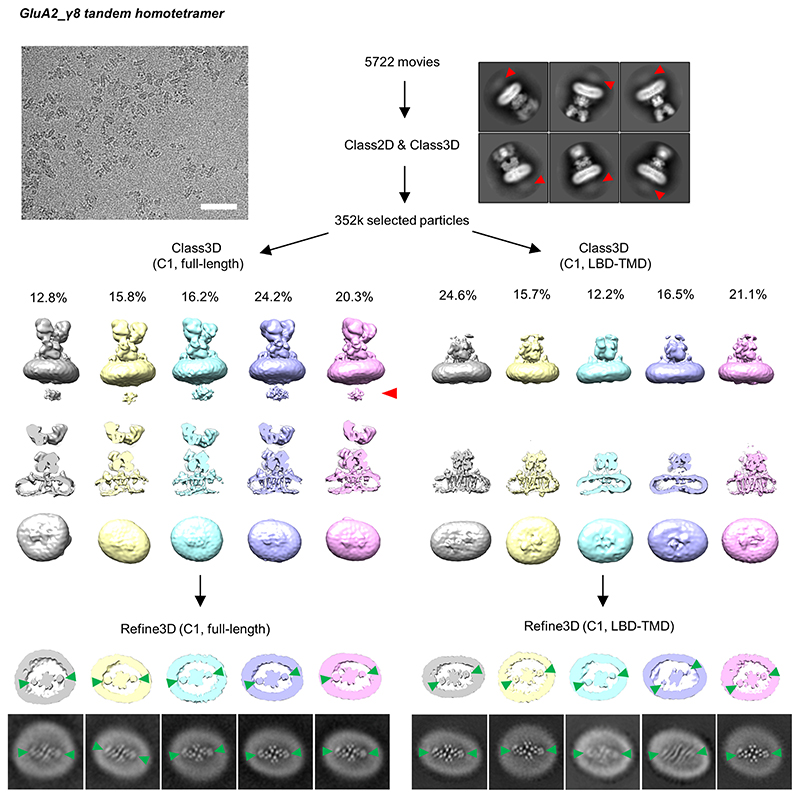
For the A2_γ8 homomeric complex, automatic particle picking was performed using a Gaussian blob and particles were binned to 4.28 Å/pixel and extracted in a box of 88 pixels. Two rounds of 2D classification were performed to remove particles without AMPAR-like features. Selected particles were rescaled to 2.14 Å/pixel for the following 3D classifications. Full-length receptor classification or focused classification on the LBD-TMD region were performed independently. Particles in individual 3D class were then refined separately.
Model building and refinement were performed using Coot 45, REFMAC5 46 and PHENIX 47 real-space refinement. Composite maps of LBD-TMD generated by EMDA were used for general building of A1/A2_γ8 and both states of A1/A2_γ8/C2. Building of the flexible regions in CNIH2 was further supported by EMDA bestmap and focused refinement maps. Initially, the GluA1/A2-γ8 complex (PDB 6QKC and 6QKZ) and CNIH3 in GluA2-CNIH3 complex (PDB 6PEQ) were used as starting points. The individual coordinates were first rigid-body fitted into the map using UCSF chimera (http://www.rbvi.ucsf.edu/chimera) and then automatically refined by REFMAC5. After this, manual refinement through Coot was performed, followed by PHENIX real-space refinement, to further refine the geometry. External restraints generated from LBD crystal structure 3TKD were used to further improve the model quality in the LBD region of the open state. For lipid building, EMDA bestmap was used to enhance the lipid signal in the resting state map. Model validation was performed with MolProbity 48. All graphics figures in the paper were prepared using UCSF Chimera or PyMOL (http://www.pymol.org). Pore radius was calculated using a plugin version of HOLE 49 in Coot.
Normal mode analysis
Normal modes were calculated and visualized using the ANM server 36 and the ProDy API 50 In both cases, the protein was treated as an elastic network model as previously described 35. Briefly, each residue is represented as one bead placed at the Cα position and interactions were treated as uniform harmonic springs within a cutoff distance of 15 Å. This simple representation allows fast analytic calculation of the most energetically favourable modes of motion via normal mode analysis. Calculations were facilitated using the SignDy module within ProDy 51, which enabled easier alignment of the active and resting structures and comparisons of their modes to each other and the transition between them, which were calculated using the correlation cosine overlap between vectors. Movies were created using PyMOL version 1.8.2.0 and Fiji 52.
Flag immunoprecipitation of CNIH homologues and CNIH2 mutants
All CNIH homologues, the CNIH12 chimera and CNIH2 mutants were HA-tagged at the extreme C-terminus and cloned into the pRK5 vector. FLAG-tagged GluA2 was co-transfected alongside CNIH constructs. 10 ml of HEK-Expi293F cells was used for each transfection. 40 hours post-transfection, cells were harvested and resuspended in 1 ml lysis buffer (25 mM Tris pH 8, 150 mM NaCl, 0.6 % digitonin (w/v)). After a 2 hour incubation, cell lysate was centrifuged at 21000 × g for 30 min and the clarified supernatant was taken as the input sample. 10 μl FLAG beads was added to the input and incubated for 1.5 hours. Following incubation, FLAG beads were pelleted and washed 5× with GDN buffer (25 mM Tris pH 8, 150 mM NaCl, 0.02% GDN). To elute samples the beads were then boiled in SDS sample buffer for 5 mins and run on a 4-12% Bis-Tris gel (Invitrogen, Cat# NW04127BOX). Rabbit GluR2 polyclonal antibody (Millipore, Cat# AB1768-l) and rabbit HA antibody (Sigma, Cat #6908) were used to detect the GluA2 and CNIHs signal, respectively.
Immunostaining
Suspension HEK-Expi293F™ cells were transfected with GluA2Q (pRK5) and CNIH (pRK5) at a stoichiometry of 1:1 and settled on poly-L-lysine coated 12 mm glass coverslips (Corning) 36 hours later. Live-labelling of surface CNIH was performed at 48 hours post-transfection by incubation with rabbit anti-HA (Sigma, Cat# H6908; 1:200) for 20 min at room temperature (RT) in Expi293™ expression media (Gibco). Cells were washed 3× in medium and once in PBS before fixation in 4% paraformaldehyde and 4% sucrose for 10 min at RT. Subsequently, cells were washed in PBS before permeabilisation in 0.1% Triton X-100 (Fisher Bioreagents) and blocking in 1% bovine serum albumin (BSA; Fisher Bioreagents) and 10% normal goat serum (NGS; Sigma) in PBS for 30 min. Permeabilised cells were then incubated sequentially in primary and secondary antibodies prepared in 1% BSA and 10% NGS for 2 hours at RT and washed in PBS after each incubation. Total CNIH was detected using mouse anti-HA (BioLegend, Cat# 901501; 1:500) and total GluA2 was detected using guinea pig anti-GluA2 (Synaptic Systems, Cat# 182 105; 1:500), followed by incubation in corresponding secondary antibodies: goat anti-rabbit IgG Alexa Fluor (AF) 568 (Invitrogen, Cat# A-11036; 1:500), goat anti-mouse IgG AF 488 (Invitrogen, Cat# A-11029; 1:500) and goat anti-guinea pig IgG AF 647 (Invitrogen, Cat# A-21450; 1:500). Coverslips were mounted in ProLong Diamond Antifade Mountant (Invitrogen) and left to cure in the dark for 48 hours at RT.
Images were acquired with a Leica TCS SP8 confocal inverted microscope using Leica Application Suite X software. Z-stacks (5x 1 μm steps) of whole cells were acquired using a 63x oil-immersion objective. 6 images were taken per coverslip and repeated from 3 independent preparations. Equivalent laser power settings were applied across all experimental conditions. Z-stacked images were averaged and the fluorescence intensity was measured from individual cells using ImageJ (Fiji) and normalised for cell area and background fluorescence. Brightness and contrast settings were kept consistent across all images.
Extended Data
Extended Data Figure 1. Properties of neuronal and recombinant AMPAR complexes.
a, Electrophysiological properties of neuronal and recombinant AMPAR complexes. Top left: Hippocampus schematic indicating selected cell types. Bottom left: Rise-time of fast-application glutamate responses of recombinant and neuronal AMPAR patches (20-80 % rise time (ms) - Recombinant receptors: GluA1/2: 0.46 ± 0.03, n=9; +γ8: 0.55 ± 0.03, n=11; +γ8+CNIH2: 0.59 ± 0.04, n=8. Neuronal receptors: CA1 pyramidal: 0.52 ± 0.02, n=14; CA3 pyramidal 0.60 ± 0.05, n=5; DG granule cell: 0.42 ± 0.02, n=6; CA1 stratum pyramidale interneurons: 0.51 ± 0.03, n=8; Welch’s ANOVA with Dunnett’s multiple comparison tests – Recombinant: W(2,15.11) = 4.25, p=0.03; Neurons: W(3,12.12) = 5.40, p=0.014) ; further details in Supplementary Table 1. Top middle: Example trace of rectification index (RI) recording from CA1 pyramidal neuron normalized to -100 mV peak amplitude. Bottom middle: Quantified RI from recorded surface patches (Recombinant receptors: GluA1/2: 0.70 ± 0.04, n=8; +γ8: 0.60 ± 0.02, n=12; +γ8+CNIH2: 0.63 ± 0.01, n=12. Neuronal receptors: CA1 pyramidal: 0.58 ± 0.01, n=13; CA3 pyramidal 0.56 ± 0.01, n=4; DG granule cell: 0.55 ± 0.04, n=4; CA1 stratum pyramidale interneurons: 0.42 ± 0.08, n=5; Welch’s ANOVA with Dunnett’s multiple comparisons test – Recombinant: W(2,15.01) = 2.47, p=0.12, Neurons: W(3,7.57) = 1.5, p=0.29) ; further details in Supplementary Table 1. Top right: Strong correlation between equilibrium current and desensitization rate are observed (individual neuronal patches plotted). Bottom right: Equilibrium current for patch responses show auxiliary protein dependent modulation and neuronal heterogeneity (% peak current - Recombinant receptors: GluA1/2: 1.81 ± 0.34, n=9; +γ8: 4.72 ± 1.09, n=11; +γ8+CNIH2: 10.97 ± 2.03, n=8. Neuronal receptors: CA1 pyramidal: 4.86 ± 0.71, n=14; CA3 pyramidal 5.78 ± 1.00, n=5; DG granule cell: 0.75 ± 0.22, n=6; CA1 stratum pyramidale interneurons: 0.59 ± 0.29, n=8; Welch’s ANOVA tests with Dunnett’s multiple comparisons test – Recombinant: W(2,12.09) = 11.93, p=0.002; Neurons: W(3,12.42) = 16.74, p=0.0001); further details in Supplementary Table 1. b, Purification and cryo-EM images of the GluA1/2_γ8/CNIH2 complex. Left: Representative 4-12% Bis-Tris gel stained with coomassie blue, indicating elution of A1/2_γ8/C2 complex from FLAG beads. CNIH2 expression from the same purification is detected by probing for the C-terminal HA tag on western blot. Purification was performed reproducibly (4 times); refer to the Supplementary Figure for uncropped blots. Middle: A representative motion-corrected micrograph of the resting state A1/2_γ8/C2 complex (scale bar, 50 nm) among collected data. Right: Representative 2D class averages of the resting state A1/2_γ8/C2 complex. c, Left: Cryo-EM maps of the full-length AMPAR octamer, depicting the three domain layers, NTD, LBD and TMD, composed of the GluA1 (blue), GluA2 (red) heteromer associated with γ8 (green) and CNIH2 (orange). Right: Schematic of plasmid constructs and secondary protein structure of auxiliary subunits shown alongside.
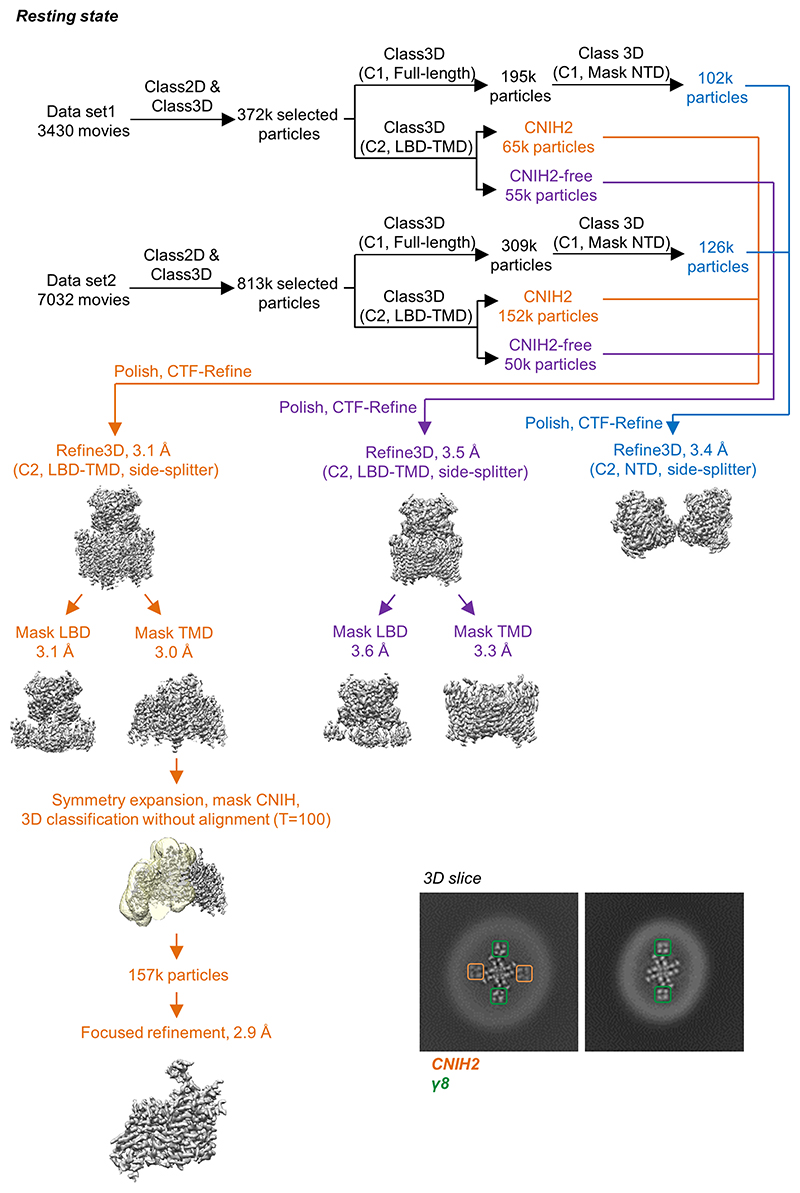
Extended Data Figure 2. Cryo-EM data processing workflow of the resting state A1/2_γ8/C2 complex.
Two datasets were first processed individually to remove particles lacking AMPAR features. Next, classifications focused on the LBD-TMD region were performed to separate CNIH-containing and CNIH-free particles, meanwhile classifications for full-length receptors were conducted to elucidate particles with a stable NTD signal. Subsequently, particles from the two datasets were combined together for refinement. Focused refinements were performed separately on the LBD-TMD gating core and the NTD region. To further improve the resolution, LBD and TMD are refined separately A structure of A1/2_γ8 (lacking CNIH2) was also resolved from the same dataset (containing only γ8 observed in 3D slice). CNIH density was further enhanced by applying first symmetry expansion on aligned particles from the TMD reconstruction, following by focused classification and refinement on only CNIH2 and the surrounding receptor transmembrane helices. Inset: Top view slices of the A1/2_γ8/C2 (left) and A1/2_γ8 (right). 3D maps at the TMD region show signal for transmembrane helices of γ8 (green) and CNIH2 (orange).
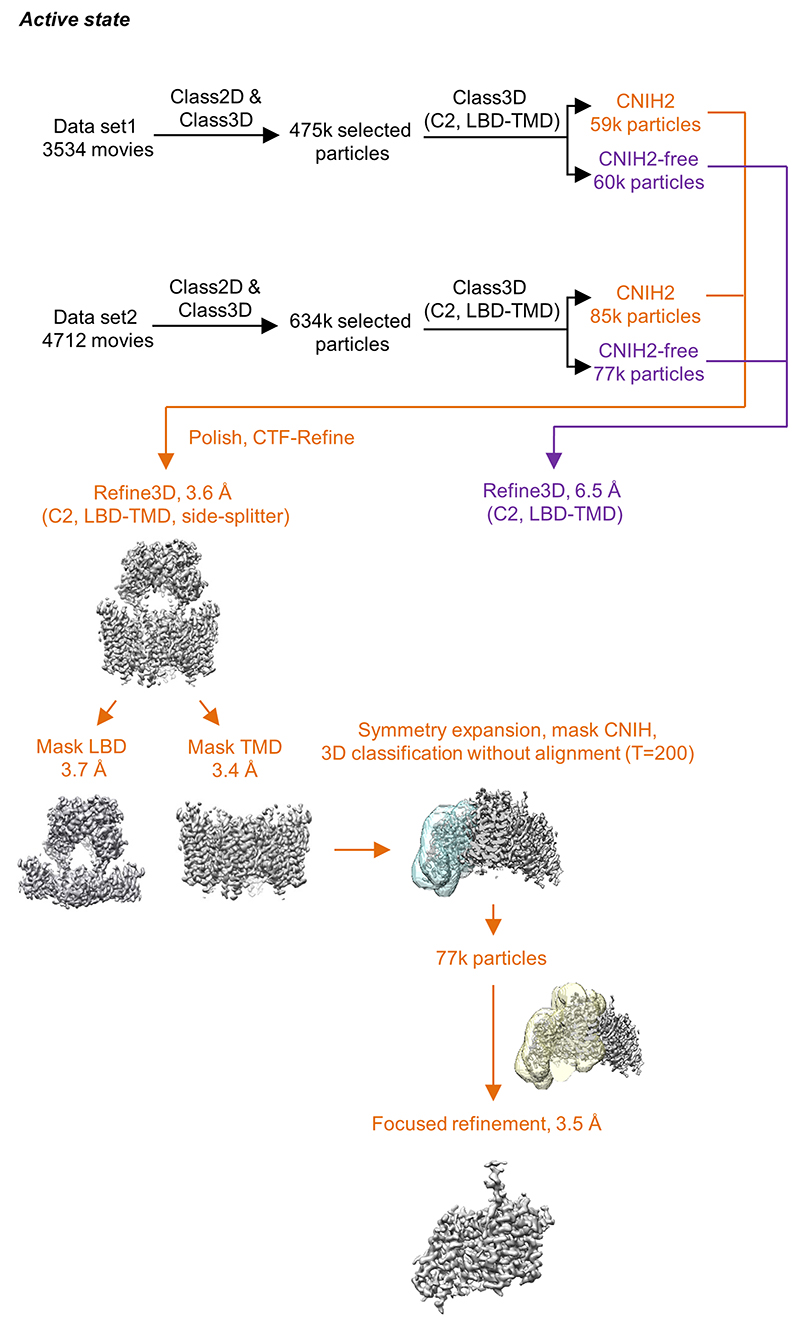
Extended Data Figure 3. Cryo-EM data processing workflow of the active state A1/2_γ8/C2 complex.
The overall data processing procedure for the active state complex is similar to that of resting state complexes. Focused refinement was performed on the LBD-TMD gating core and individual LBD and TMD domain layers of the receptor. CNIH density was further improved by applying first symmetry expansion on aligned particles from the TMD reconstruction, followed by focused classification on CNIH alone, and finally focused refinement on CNIH together with surrounding receptor transmembrane helices. Particles lacking CNIH2 found in these datasets were not of high enough quality to provide a high-resolution structure.
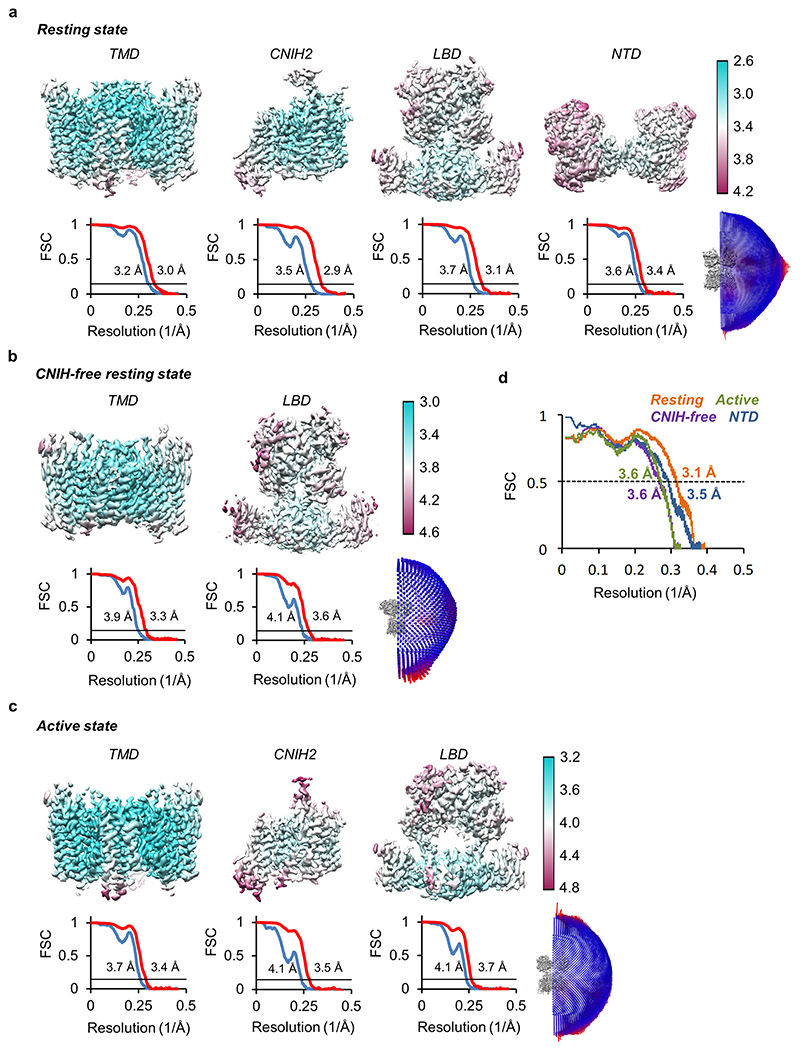
Extended Data Figure 4. Cryo-EM analysis of A1/2_γ8/C2 and A1/2_γ8 complexes.
a, Local resolution and Fourier shell correlation (FSC) of focused refinement maps at the TMD, CNIH2, LBD and NTD. Euler angle distribution of particles for cryo-EM reconstruction of the resting state A1/2_γ8/C2 complex. 3D maps are coloured based on local resolution estimation. Masked (red) or unmasked (blue) FSC of corresponding maps are both shown where FSC=0.143 (black line). b, Local resolution and FSC of focused refinements at the TMD and LBD. Euler angle distribution of particles for cryo-EM reconstruction of the resting state A1/2_γ8 complex. c, Local resolution, FSC of focused refinements at the TMD, CNIH2 and LBD. Euler angle distribution of particles for cryo-EM reconstruction of the active state A1/2_γ8/C2 complex. d, Model to map FSCs of A1/2_γ8/C2 LBD-TMD models in resting and active states, resting state NTD model and resting state A1/2_γ8 LBD-TMD model.
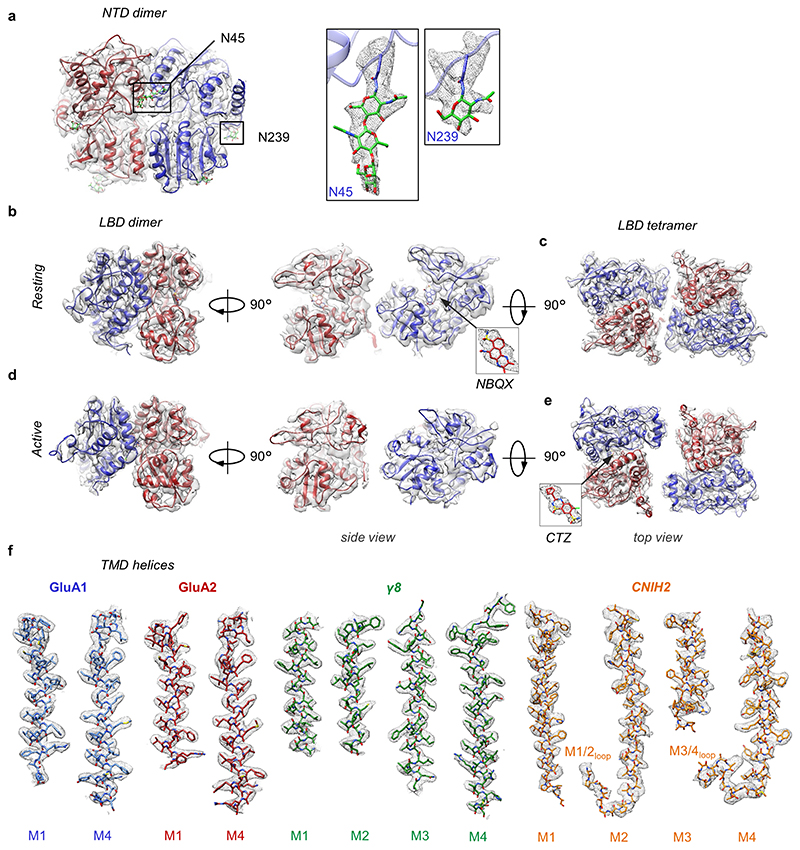
Extended Data Figure 5. Features of A1/2_γ8/C2 NTD and LBD layers and quality of density in the TMD region.
a, Cryo-EM density and model of the resting state GluA1 (blue) and GluA2 (red) NTD dimer. GluA1-specific N-linked glycans are observed at N45 and N239 (green sticks). b, Cryo-EM density and model of A1/2 LBD dimer in the resting state. Density and model for the competitive antagonist NBQX bound to its orthosteric site in the LBD cleft. c, Top view of cryo-EM density and model of A1/2 LBD tetramer in the resting state. d, Cryo-EM density and model of A1/2 LBD dimer in the active state demonstrating a closure of the LBD ‘clamshell’. e, Top view of cryo-EM density and model of A1/2 LBD tetramer in the active state. Density and model of desensitization blocker cyclothiazide (CTZ) bound at the LBD dimer interface are shown in the insert. f, Cryo-EM density and model of transmembrane helices of A1/2_γ8/C2 in the resting state.
Extended Data Figure 6. Cryo-EM data processing workflow of GluA2_γ8 homomeric complex.
Automatic particle picking was first applied on the raw images which had similar features to the A1/A2_γ8/C2 heteromeric complex (scale bar, 50nm). 2D classifications were then performed to remove particles lacking AMPAR features. In several side view 2D class averages, an additional layer of density (marked by a red arrowhead) beneath the micelle can be observed. Next, selected particles were used for separate 3D classifications on the full-length receptor (left panel) or on masked-out LBD-TMD regions (right panels). In each of the two classifications, ~10% of low-quality particles were removed and the remaining AMPAR-shaped class averages are presented (side- and bottom views). An additional layer was observed in the full-length classification (indicated by red arrow). 3D refinement was performed on all classes individually, slices of the TMD region from the refined maps are also shown, with γ8 densities only apparent at the B’D’ sites (indicated by green arrowheads).
Extended Data Figure 7. γ8 and CNIH2 receptor binding sites and their relevant bound lipids.
a, Overlay of the A’C’ and B’D’ binding sites, showing reorientations of five residues along the M1 helices (GluA1 M1, red; GluA2 M1, blue). These changes are likely mediated by γ8 engaging GluA1 M1. b, Strong lipid densities (light blue, density shown in grey mesh) line the cavity between GluA2 M1 and M4. F515, F517 and L518 from GluA2 pre-M1 interact with the lipids from the upper leaflet. Other residues from GluA2 M1, M2 and GluA1 M3 involved in these interactions are shown as stick. c, LL1 binds to the γ8 N224 side chain, connecting TARP-γ8 to the GluA1 M2 pore helix. LL2 bridges between CNIH2 and the GluA2 M1 and M2 helices. d, ‘Open book’ view of the A’C’ binding site, displaying how the UL1, LL2 and LL3 lipids engage the receptor (left) and the CNIH2 M1 and M2 helices (right). Side chains in close proximity to lipids are shown. e, Superposition of CNIHs and their binding peripheral helices from the resting state A1/2_γ8/C2 (orange) and A2_C3 (grey, PDB 6PEQ) complexes. While the upper parts of CNIHs’ M1 and M2 helices are aligned together, the lower part of CNIH2 is kinked away from the receptor relative to CNIH3, this permits the accommodation of three CNIH2 binding-relevant lipids. Distance between W26 (C2) and C811 (A1) in A1/2_γ8/C2 and W26 (C3) and C815 (A2) in A2_C3 are measured. M1 and M4 from A1/A2_γ8/C2 resting state are coloured as in Figure 1. M1 and M4 for A2_C3 (PDB 6PEQ) are coloured in grey. Three CNIH2 binding-relevant lipids LL2, LL3 and UL1 are shown as pink stick. f, A density modeled as cholesterol occupies the pocket between CNIH2 M3 and M4, observed after focused refinement.
Extended Data Figure 8. Features of the A1/2_γ8/C2 and A1/2_γ8 conduction pore.
a, Density of M2/M3 gating regions and their fit against models in the resting and active state. b, Pore dimensions of resting state A1/2_γ8 (left) and the resting (middle) and active (right) state of A1/2_γ8/C2 depicted by space-filling representation (HOLE program) with relevant side chains indicated as sticks. A comparison of pore radius across these three structures indicates a similar diameter of the receptor gate in resting state A1/2_γ8 (grey) and A1/2_γ8/C2 (orange), with a clear expansion observed in the active state A1/2_γ8/C2 (red) complex. Diameter differences at the Q/R site are mainly caused by conformational variations at R586 side chain among these three models. c, Pore dimensions measured between Cα of GluA1 Q582 and GluA2 R586 in resting state A1/2_γ8 (left), A1/2_γ8/C2 (middle) and active state A1/2_γ8/C2 (right). Upon receptor activation, the distance between GluA2 R586 is increased by ~1 Å in A1/2_γ8/C2. d, Distance measured between Cα of GluA1 C585 and GluA2 C589 at A1/2_γ8 resting state (left), A1/2_γ8/C2 resting (middle) and active (right) state. The corresponding EM densities are shown as mesh. Upon receptor activation, the distance between GluA2 C589 also increased by ~1.5 Å in A1/2_γ8/C2. All diameter labels are measured in Å. e, Charge distribution maps of the intracellular face of A1/2_γ8/C2 (red: -5 kBT/e, blue: 5 kBT/e) in the resting (top) and active (bottom) state indicate a dilation of the pore entrance in the direction of GluA2, but not GluA1 during receptor activation.
Extended Data Figure 9. Conformational changes of A1/2_γ8/C2 during receptor activation.
a, Top view superposition along the pore axis of A1/2_γ8/C2 in resting (grey) and active (red) states shows dilation of receptor and rotation of γ8 and C2 during activation. b, Superposition of A1/2_γ8/C2 in resting (grey) and active (coloured) states along the pore axis shows the conformational change of GluA2 M1 and M3 linkers as well as the LBD region upon receptor activation. The GluA2 M3 linker moves towards M1 linker, while the latter approaches γ8 ‘acidic’ β4 loop. The LBD ‘KGK’ motif also moves towards the γ8 ‘acidic’ loop. c, d, Conformational change of γ8 and C2 during receptor activation. Models are aligned along the pore axis. The translation of the Cα atoms from the resting to active state is indicated as arrows for every second residue. Arrows indicate the direction and distance of helical movements; these were determined for all Cα atoms between the two states relative to the COM of a given auxiliary subunit. Auxiliary subunits come together on the GluA1 pre-M1 side (c), but are separated on the GluA2 pre-M1 side (d). Zoomed-in panel (c) indicates a contact between the γ8 M4 helix and the base of the GluA1 M1/2 cytoplasmic loop formed during receptor activation.
Extended Data Figure 10. Flag IP, immunostaining, and electrophysiology of CNIH homologues and CNIH2 mutants in complex with GluA1 or GluA2 homomers.
a, Flag IP of CNIH homologues and CNIH2 mutants in complex with Flag tagged GluA2 homomers. CNIH12CHIM, CNIH 1 and CNIH2 chimera with a fragment of CNIH2 (51-RERLKNIERICCLLRK-66) inserted into CNIH1 between P50 and L51; F3L, CNIH2 F3L; F5L, CNIH2 F5L; F8L, CNIH2 F8L; CNIH2 3FL, mutate all three phenylalanine 3, 5, 8 in CNIH2 to leucine; FT, flow through. IPs were performed reproducibly (3 times); refer to the Supplementary Figure for uncropped blots. b, Surface CNIH fluorescence (left), Total CNIH fluorescence (middle) and Surface/Total ratio (right) for CNIH homologues and CNIH2 mutants in complex with GluA2 (Surface CNIH (AU) - No CNIH: 0.21 ± 0.15, n=80; CNIH1: 6.60 ± 0.63, n=46; CNIH2: 7.54 ± 0.56, n=55; CNIH3: 8.07 ± 0.64, n=61; CNIH12CHIM: 8.56 ± 0.69, n=61; F3L: 4.42 ± 0.80, n=17; F5L: 4.25 ± 0.40, n=50; F8L: 8.36 ± 0.75, n=34; 3FL: 1.67 ± 0.22, n=50; Kruskal-Wallis test: H(8) = 256.3, p<0.0001. Total CNIH (AU) - No CNIH: 0.02 ± 0.06, n=80; CNIH1: 21.5 ± 2.03, n=46; CNIH2: 25.8 ± 2.08, n=55; CNIH3: 25.2 ± 2.19, n=61; CNIH12CHIM: 25.0 ± 1.49, n=61; F3L: 12.7 ± 1.85, n=17; F5L: 16.2 ± 1.97, n=50; F8L: 24.6 ± 2.00, n=34; 3FL: 27.3 ± 2.42, n=50; Kruskal-Wallis test: H(8) = 230.1, p<0.0001. Surface/Total - CNIH1: 0.34 ± 0.03, n=46; CNIH2: 0.39 ± 0.04, n=55; CNIH3: 0.42 ± 0.04, n=61; CNIH12CHIM: 0.36 ± 0.02, n=61; F3L: 0.41 ± 0.08, n=17; F5L: 0.35 ± 0.03, n=50; F8L: 0.39 ± 0.04, n=34; 3FL: 0.07 ± 0.01, n=50; One-sample Wilcoxon test (Median = 0), p<0.0001; further details in Supplementary Table 3. Homologues CNIH1, CNIH2 and CNIH3 show robust surface expression. CNIH2 mutants F3L, F5L and F8L, as well as the CNIH12CHIM chimera, also traffic to the cell surface, whereas 3FL does not. F3L and F5L CNIH2 mutants show decreased total and, consequently, surface expression levels; to ensure the AMPARs in our electrophysiology experiments were still saturated with CNIHs we used a 1:2 AMPAR:CNIH co-transfection ratio. Increasing this ratio further to 1:4 for F3L & F5L did not affect the gating properties, suggesting that the observed change in AMPAR modulation by these mutants is not caused by their lower (surface) expression. c, Representative images showing surface CNIH (green), total CNIH (magenta) and total GluA2 (blue). d, Equilibrium current (Fig. 3a data set): (% peak) – GluA2 alone: 1.03 ± 0.19, n=15; CNIH2 WT: 24.72, ± 4.55, n=9; F3L: 9.25 ± 1.16, n=7; F5L: 8.96 ± 1.16, n=9; F8L: 8.05 ± 1.00, n=6; 3FL: 2.01 ± 0.28, n=9; Welch’s ANOVA with Dunnett’s multiple comparisons test: W(5,17.48) = 27.95, p<0.0001). e, Equilibrium current (Fig. 3c data set): GluA2 alone: 1.33 ± 0.50, n=6; CNIH1: 3.52 ± 0.56, n=12; CNIH12CHIM: 10.93 ± 1.16, n=11; CNIH2: 19.77 ± 1.93, n=7; Welch’s ANOVA with Dunnett’s multiple comparisons test, W(3,15.36) = 40.08, p<0.0001); further details in Supplementary Table 2.
Extended Data Table 1. Cryo-EM data collection, refinement and validation statistics.
| A1/2_γ8/C2 Resting state | A1/2_γ8 Resting state | A1/2_γ8/C2 Active state | ||
|---|---|---|---|---|
| LBD-TMD (EMDB-12805) (PDB 7OCE) | NTD (EMDB-12803) (PDB 7OCC) | LBD-TMD (EMDB-12804) (PDB 7OCD) | LBD-TMD (EMDB-12806) (PDB 7OCF) | |
| Data collection and processing | ||||
| Microscope | FEI Titan Krios | FEI Titan Krios | ||
| Detector | K3 + GIF | K3 + GIF | ||
| Magnification | 81000X | 81000X | ||
| Voltage (kV) | 300 | 300 | ||
| Electron exposure (e-/Å2) | 50 | 50 | ||
| Defocus range (μm) | -1.2 to -2.4 | -1.2 to -2.4 | ||
| Pixel size | 1.07 | 1.07 | ||
| Symmetry imposed | C2 | C2 | ||
| Micrographs | 10462 | 8246 | ||
| Map resolution (Å) | 3.1 | 3.4 | 3.5 | 3.6 |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 |
| Refinement | ||||
| Initial model used (PDB) | 6QKC | 6QKC | 6QKZ | 6QKC |
| Model resolution (Å) | 3.1 | 3.4 | 3.5 | 3.5 |
| FSC threshold | 0.5 | 0.5 | 0.5 | 0.5 |
| Map sharpening B factor (Å2) | -113 | -142 | -115 | -122 |
| Model composition | ||||
| Non-hydrogen atoms | 18092 | 11202 | 13624 | 17092 |
| Protein residues | 2322 | 1480 | 1920 | 2298 |
| Ligands | NBQX: 4 | NAG: 18, BMA: 2 | NBQX: 4 | CTZ: 4 |
| Lipids | 48 | 0 | 6 | 26 |
| B factors (Å2) | ||||
| Protein | 37.44 | 34.10 | 61.09 | 50.60 |
| Ligand | 35.48 | 59.58 | 48.78 | 31.70 |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.006 | 0.010 | 0.007 | 0.007 |
| Bond angles | 0.684 | 0.722 | 0.595 | 0.616 |
| Validation | ||||
| Molprobity score | 1.45 | 1.46 | 1.75 | 1.54 |
| Clashscore | 4.07 | 5.04 | 7.03 | 4.84 |
| Poor rotamers (%) | 0 | 0 | 0 | 0 |
| Ramachandran plot | ||||
| Favoured (%) | 96.22 | 96.87 | 94.73 | 95.79 |
| Allowed (%) | 3.78 | 3.13 | 5.27 | 4.21 |
| Disallowed (%) | 0 | 0 | 0 | 0 |
Supplementary Material
Acknowledgements
We thank the Greger lab, Beatriz Herguedas, James Krieger, and Jan-Niklas Dohrke for comments on the manuscript; James Krieger and Jan-Niklas Dohrke for discussion, James Krieger for help with the normal mode analysis, Bianka Köhegyi for help with EM imaging, Veronica Chang and Kunimichi Suzuki for helping to generate the CNIH2-1D4-HA stable cell line, and Maria Carvalho for assistance at early stages of this project. We are grateful to LMB scientific computing and the EM facility for support, Paul Emsley for help with model building, Takanori Nakane for helpful comments with Relion 3.1, and Rangana Warshamanage for helping with EMDA EM-map processing. We also acknowledge Diamond Light Source for access and support of the Cryo-EM facilities at the UK national electron bio10 imaging centre (eBIC), proposal EM17434, funded by the Wellcome Trust, MRC and BBSRC. This work was supported by grants from the Medical Research Council, as part of United Kingdom Research and Innovation (also known as UK Research and Innovation) [MC_U105174197] and BBSRC (BB/N002113/1) to IHG.
Footnotes
Author contributions:
IHG conceptualized and supervised the study, and wrote the paper with input from JFW. DZ performed protein purification, cryo-EM data collection, data processing and model building. JFW and OC designed and performed electrophysiological experiments. PMM and JFW performed confocal imaging and data analysis.
Competing interests: The authors declare no competing interests.
Data and materials availability
Cryo-EM coordinates are deposited in the Protein Data Bank under the accession codes 7OCA (resting state full-length AMPAR octamer), 7OCC (NTD of the resting state octamer), 7OCD (resting state LBD-TMD with TARP-γ8 only), 7OCE (resting state LBD-TMD of the octamer), 7OCF (active state LBD-TMD of the octamer); the corresponding EM maps are deposited in the EMDB under accession codes EMD-12802, EMD-12803, EMD-12804, EMD-12805, EMD-12806. Source data for Fig. 1 and 3, and Extended Data Fig. 1 and 10 are provided with the paper.
References
- 1.Traynelis SF, et al. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greger IH, Watson JF, Cull-Candy SG. Structural and Functional Architecture of AMPA-Type Glutamate Receptors and Their Auxiliary Proteins. Neuron. 2017;94:713–730. doi: 10.1016/j.neuron.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 3.Jonas P. The Time Course of Signaling at Central Glutamatergic Synapses. News Physiol Sci. 2000;15:83–89. doi: 10.1152/physiologyonline.2000.15.2.83. [DOI] [PubMed] [Google Scholar]
- 4.Schwenk J, et al. High-resolution proteomics unravel architecture and molecular diversity of native AMPA receptor complexes. Neuron. 2012;74:621–633. doi: 10.1016/j.neuron.2012.03.034. [DOI] [PubMed] [Google Scholar]
- 5.Shanks NF, et al. Differences in AMPA and kainate receptor interactomes facilitate identification of AMPA receptor auxiliary subunit GSG1L. Cell Rep. 2012;1:590–598. doi: 10.1016/j.celrep.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu W, et al. Subunit composition of synaptic AMPA receptors revealed by a single-cell genetic approach. Neuron. 2009;62:254–268. doi: 10.1016/j.neuron.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwenk J, et al. Regional diversity and developmental dynamics of the AMPA-receptor proteome in the mammalian brain. Neuron. 2014;84:41–54. doi: 10.1016/j.neuron.2014.08.044. [DOI] [PubMed] [Google Scholar]
- 8.Schwenk J, et al. Functional proteomics identify cornichon proteins as auxiliary subunits of AMPA receptors. Science. 2009;323:1313–1319. doi: 10.1126/science.1167852. [DOI] [PubMed] [Google Scholar]
- 9.Boudkkazi S, Brechet A, Schwenk J, Fakler B. Cornichon2 dictates the time course of excitatory transmission at individual hippocampal synapses. Neuron. 2014;82:848–858. doi: 10.1016/j.neuron.2014.03.031. [DOI] [PubMed] [Google Scholar]
- 10.Rouach N, et al. TARP gamma-8 controls hippocampal AMPA receptor number, distribution and synaptic plasticity. Nat Neurosci. 2005;8:1525–1533. doi: 10.1038/nn1551. [DOI] [PubMed] [Google Scholar]
- 11.Gill MB, et al. Cornichon-2 modulates AMPA receptor-transmembrane AMPA receptor regulatory protein assembly to dictate gating and pharmacology. J Neurosci. 2011;31:6928–6938. doi: 10.1523/JNEUROSCI.6271-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herring BE, et al. Cornichon proteins determine the subunit composition of synaptic AMPA receptors. Neuron. 2013;77:1083–1096. doi: 10.1016/j.neuron.2013.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato AS, et al. Hippocampal AMPA receptor gating controlled by both TARP and cornichon proteins. Neuron. 2010;68:1082–1096. doi: 10.1016/j.neuron.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen S, et al. Activation and Desensitization Mechanism of AMPA Receptor-TARP Complex by Cryo-EM. Cell. 2017;170:1234–1246.:e1214. doi: 10.1016/j.cell.2017.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herguedas B, et al. Architecture of the heteromeric GluA1/2 AMPA receptor in complex with the auxiliary subunit TARP gamma8. Science. 2019;364:eaav9011. doi: 10.1126/science.aav9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Twomey EC, Yelshanskaya MV, Grassucci RA, Frank J, Sobolevsky AI. Channel opening and gating mechanism in AMPA-subtype glutamate receptors. Nature. 2017;549:60–65. doi: 10.1038/nature23479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakagawa T. Structures of the AMPA receptor in complex with its auxiliary subunit cornichon. Science. 2019;366:1259–1263. doi: 10.1126/science.aay2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dancourt J, Barlowe C. Protein sorting receptors in the early secretory pathway. Annu Rev Biochem. 2010;79:777–802. doi: 10.1146/annurev-biochem-061608-091319. [DOI] [PubMed] [Google Scholar]
- 19.Shi Y, Lu W, Milstein AD, Nicoll RA. The stoichiometry of AMPA receptors and TARPs varies by neuronal cell type. Neuron. 2009;62:633–640. doi: 10.1016/j.neuron.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khodosevich K, et al. Coexpressed auxiliary subunits exhibit distinct modulatory profiles on AMPA receptor function. Neuron. 2014;83:601–615. doi: 10.1016/j.neuron.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Y, Chen S, Swensen AC, Qian WJ, Gouaux E. Architecture and subunit arrangement of native AMPA receptors elucidated by cryo-EM. Science. 2019;364:355–362. doi: 10.1126/science.aaw8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Twomey EC, Yelshanskaya MV, Grassucci RA, Frank J, Sobolevsky AI. Structural Bases of Desensitization in AMPA Receptor-Auxiliary Subunit Complexes. Neuron. 2017;94:569–580.:e565. doi: 10.1016/j.neuron.2017.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- 25.Yelshansky MV, Sobolevsky AI, Jatzke C, Wollmuth LP. Block of AMPA receptor desensitization by a point mutation outside the ligand-binding domain. J Neurosci. 2004;24:4728–4736. doi: 10.1523/JNEUROSCI.0757-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burnashev N, Monyer H, Seeburg PH, Sakmann B. Divalent ion permeability of AMPA receptor channels is dominated by the edited form of a single subunit. Neuron. 1992;8:189–198.:0896-6273(92)90120-3 [pii] doi: 10.1016/0896-6273(92)90120-3. [DOI] [PubMed] [Google Scholar]
- 27.Hume RI, Dingledine R, Heinemann SF. Identification of a site in glutamate receptor subunits that controls calcium permeability. Science. 1991;253:1028–1031. doi: 10.1126/science.1653450. [DOI] [PubMed] [Google Scholar]
- 28.Twomey EC, Yelshanskaya MV, Vassilevski AA, Sobolevsky AI. Mechanisms of Channel Block in Calcium-Permeable AMPA Receptors. Neuron. 2018;99:956–968.:e954. doi: 10.1016/j.neuron.2018.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bowie D. Polyamine-mediated channel block of ionotropic glutamate receptors and its regulation by auxiliary proteins. J Biol Chem. 2018;293:18789–18802. doi: 10.1074/jbc.TM118.003794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coombs ID, et al. Cornichons modify channel properties of recombinant and glial AMPA receptors. J Neurosci. 2012;32:9796–9804. doi: 10.1523/JNEUROSCI.0345-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alsaloum M, Kazi R, Gan Q, Amin J, Wollmuth LP. A Molecular Determinant of Subtype-Specific Desensitization in Ionotropic Glutamate Receptors. J Neurosci. 2016;36:2617–2622. doi: 10.1523/JNEUROSCI.2667-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDaniel MJ, et al. NMDA receptor channel gating control by the pre-M1 helix. J Gen Physiol. 2020;152 doi: 10.1085/jgp.201912362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yelshanskaya MV, Mesbahi-Vasey S, Kurnikova MG, Sobolevsky AI. Role of the Ion Channel Extracellular Collar in AMPA Receptor Gating. Sci Rep. 2017;7:1050. doi: 10.1038/s41598-017-01146-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dawe GB, et al. Distinct Structural Pathways Coordinate the Activation of AMPA Receptor-Auxiliary Subunit Complexes. Neuron. 2016;89:1264–1276. doi: 10.1016/j.neuron.2016.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krieger J, Lee JY, Greger IH, Bahar I. Activation and desensitization of ionotropic glutamate receptors by selectively triggering pre-existing motions. Neurosci Lett. 2019;700:22–29. doi: 10.1016/j.neulet.2018.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eyal E, Lum G, Bahar I. The anisotropic network model web server at 2015 (ANM 2.0) Bioinformatics. 2015;31:1487–1489. doi: 10.1093/bioinformatics/btu847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shanks NF, et al. Molecular dissection of the interaction between the AMPA receptor and CNIH-3. J Neurosci. 2014;34:12104–12120. doi: 10.1523/JNEUROSCI.0595-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bakan A, Meireles LM, Bahar I. ProDy: protein dynamics inferred from theory and experiments. Bioinformatics. 2011;27:1575–1577. doi: 10.1093/bioinformatics/btr168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia-Nafria J, Watson JF, Greger IH. IVA cloning: A single-tube universal cloning system exploiting bacterial In Vivo Assembly. Sci Rep. 2016;6:27459. doi: 10.1038/srep27459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stoppini L, Buchs PA, Muller D. A simple method for organotypic cultures of nervous tissue. J Neurosci Methods. 1991;37:173–182. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- 41.Elegheert J, et al. Lentiviral transduction of mammalian cells for fast, scalable and high-level production of soluble and membrane proteins. Nat Protoc. 2018;13:2991–3017. doi: 10.1038/s41596-018-0075-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zivanov J, et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 2018;7 doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liebschner D, et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Williams CJ, et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018;27:293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14:354–360. doi: 10.1016/s0263-7855(97)00009-x. 376. [DOI] [PubMed] [Google Scholar]
- 50.Bakan A, et al. Evol and ProDy for bridging protein sequence evolution and structural dynamics. Bioinformatics. 2014;30:2681–2683. doi: 10.1093/bioinformatics/btu336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang S, Li H, Krieger JM, Bahar I. Shared Signature Dynamics Tempered by Local Fluctuations Enables Fold Adaptability and Specificity. Mol Biol Evol. 2019;36:2053–2068. doi: 10.1093/molbev/msz102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM coordinates are deposited in the Protein Data Bank under the accession codes 7OCA (resting state full-length AMPAR octamer), 7OCC (NTD of the resting state octamer), 7OCD (resting state LBD-TMD with TARP-γ8 only), 7OCE (resting state LBD-TMD of the octamer), 7OCF (active state LBD-TMD of the octamer); the corresponding EM maps are deposited in the EMDB under accession codes EMD-12802, EMD-12803, EMD-12804, EMD-12805, EMD-12806. Source data for Fig. 1 and 3, and Extended Data Fig. 1 and 10 are provided with the paper.