Abstract
Background
Autophagy is the major intracellular degradation route in mammalian cells. Systemic ablation of core autophagy-related (ATG) genes in mice leads to embryonic or perinatal lethality, and conditional models show neurodegeneration. Impaired autophagy has been associated with a range of complex human diseases, yet congenital autophagy disorders are rare.
Methods
We performed a genetic, clinical, and neuroimaging analysis involving five families. Mechanistic investigations were conducted with the use of patient-derived fibroblasts, skeletal muscle-biopsy specimens, mouse embryonic fibroblasts, and yeast.
Results
We found deleterious, recessive variants in human ATG7, a core autophagy-related gene encoding a protein that is indispensable to classical degradative autophagy. Twelve patients from five families with distinct ATG7 variants had complex neuro-developmental disorders with brain, muscle, and endocrine involvement. Patients had abnormalities of the cerebellum and corpus callosum and various degrees of facial dysmorphism. These patients have survived with impaired autophagic flux arising from a diminishment or absence of ATG7 protein. Although autophagic sequestration was markedly reduced, evidence of basal autophagy was readily identified in fibroblasts and skeletal muscle with loss of ATG7. Complementation of different model systems by deleterious ATG7 variants resulted in poor or absent autophagic function as compared with the reintroduction of wild-type ATG7.
Conclusions
We identified several patients with a neurodevelopmental disorder who have survived with a severe loss or complete absence of ATG7, an essential effector enzyme for autophagy without a known functional paralogue. (Funded by the Wellcome Centre for Mitochondrial Research and others.)
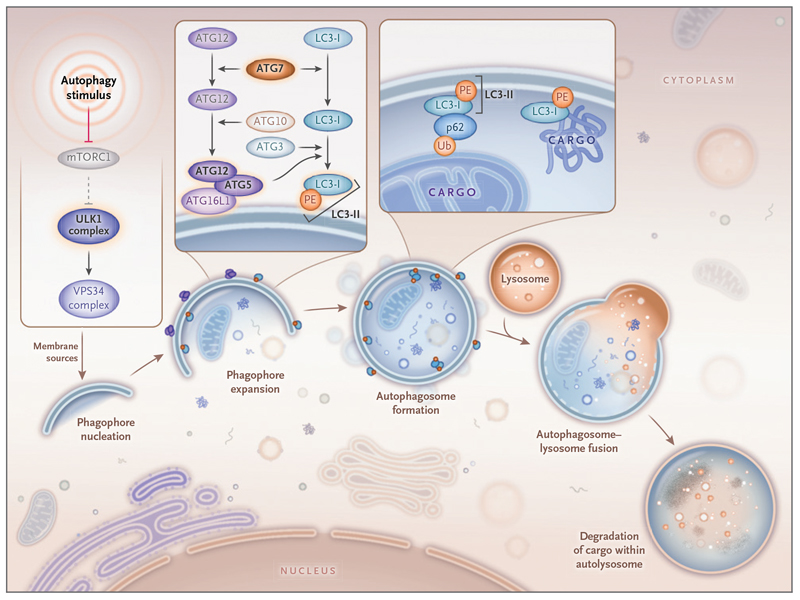
Macroautophagy (hereafter “autophagy”) serves to protect the cell from cytotoxicity through degradation of toxic protein aggregates, pathogens, and damaged organelles. It also sustains homeostasis by recycling essential metabolites.1 Autophagy involves the formation of a transient double membrane-bound autophagosome that encapsulates and delivers cytoplasmic cargo to acidic subcompartments of the endolysosomal system for degradation by hydrolysis.2–5 A core set of autophagy-related (ATG) genes orchestrates the fundamental stages of canonical autophagy (Fig. 1).1
Figure 1. Classical Degradative Autophagy.
The Unc-51-like kinase 1 (ULK1) complex receives and integrates various upstream signals to execute the autophagic response. This leads to the emergence of a transient, double-membrane-bound structure known as a phagophore, which expands through the addition of ATG5-ATG12 conjugates and ATG8 proteins (e.g., LC3-II) to become an autophagosome. ATG5-ATG12 and LC3-II are generated through autophagy conjugation systems, which are driven by ATG7. The autophagosome sequesters cytoplasmic cargo recruited by LC3-II, which interacts directly with cargo or indirectly through autophagy receptors, including p62, as shown in the magnified image. The mature autophagosome then fuses with acidic subcompartments of the endolysosomal system, which includes resident hydrolytic enzymes that degrade the encapsulated cargo. The abbreviation mTORC1 denotes mechanistic target of rapamycin complex 1, PE phosphatidylethanolamine, Ub ubiquitin, and VPS34 class III phosphatidylinositol 3-kinase.
Approximately 20 core ATG genes are conserved across eukaryotes and are critical to the process of canonical autophagy, yet only 4 of these genes (ATG5,6,7 WDR45,8,9 WDR45B,10 and WIPI2 11) are implicated in mendelian disease.4,5 The identification of cohorts of patients with autophagy deficiencies provides an opportunity to investigate the systemic role of core autophagy-related proteins. An understanding of the clinical and functional profiles in such patients could provide further insights into the etiologic links between aberrant autophagy and the many complex disease states it is predicted to underpin, from neurodegeneration to cancer.5,12,13 Indeed, access to biologic specimens from patients with autophagy deficiencies might also accelerate the development of autophagy-augmenting therapeutics.
The Unc-51-like kinase 1 (ULK1) complex integrates multiple upstream signals and transmits them to initiate canonical autophagy, which depends on ubiquitin-like conjugation systems to drive autophagosome biogenesis (Fig. 1).1,14 ATG7 encodes an E1-like enzyme that activates ATG12 before its conjugation to ATG5, promoting expansion of the preautophagosomal phagophore (Fig. 1).15 ATG7 also facilitates lipidation of the protein LC3-I with phosphatidylethanolamine to generate LC3-II, which is found on the inner and outer autophagosomal membranes and recruits cytoplasmic cargo to the autophagosome either directly or through selective adaptor proteins (Fig. 1).16 Studies in mice have shown the physiological significance of endogenous ATG7; Atg7-null mice die within 24 hours after birth.17,18 Thus, autophagy is regarded as an essential process in mammals. The subsequent characterization of mouse models has revealed the profound importance of Atg7 in nerve and muscle, wherein tissue-specific Atg7 ablation leads to ataxia and myopathy, respectively.19–21 Classically, the loss of mammalian ATG7 is regarded as rendering cells and tissues “autophagy-deficient.”17,22,23
We report the discovery of five unrelated families with recessive ATG7 variants that were both deleterious (i.e., predicted to reduce fitness, as determined by cross-species comparison of protein sequences) and damaging (i.e., shown through biochemical assays to interfere with the function of the protein). Affected family members had a neurodevelopmental syndrome that was distinguished by cerebellar hypoplasia, a thin posterior corpus callosum, ataxia, developmental delay, musculoskeletal abnormalities, and facial dysmorphism.
Methods
Patients
We conducted a study involving 12 patients with ataxia and developmental delay from five families. Details of the clinical findings are provided in Table S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org. Written informed consent was obtained from all persons in the study (or from their parents or guardians) in accordance with the Declaration of Helsinki protocols, and our experimental protocols were approved by local institutional review boards. The authors vouch for the accuracy and completeness of the data presented in this report.
Genetic Analyses
Exome sequencing was performed with DNA from each family. Data were filtered, and computer modeling tools were used to predict variant pathogenicity. Full details are provided in the Methods section of the Supplementary Appendix.
Histochemical Analyses
For routine microscopic evaluation of the skeletal muscle-biopsy specimen from Patient 2, we stained 10-μm cryosections with hematoxylin and eosin, periodic acid-Schiff (PAS), diastase-PAS, Sudan black, and Gomori trichrome. Enzyme histochemical analyses included measurement of esterase, phosphorylase, acid phosphatase, ATPase activity at a pH of 4.3 and a pH of 4.6, cytochrome c oxidase (COX), succinate dehydrogenase (SDH), and sequential COX-SDH and NADH tetrazolium reductase activity.24 Immunohistochemical analysis (7-μm sections) with antibodies against p62, neonatal myosin heavy chain, slow myosin heavy chain, fast myosin heavy chain, and HLA-A, -B, and -C was also performed.
Cell Studies
Primary or immortalized dermal fibroblasts from patients and controls were cultured. For functional studies, autophagy was induced with AZD8055 (1 μmol per liter) or by means of acute serum and amino acid withdrawal performed with the use of Earle’s balanced salt solution in the presence or absence of a late-stage autophagy blocker (chloroquine [60 μmol per liter] or bafilomycin A1 [100 nmol per liter]) for the indicated times. Detailed descriptions of the methods used for immunoblotting, immunofluorescence, long-lived protein degradation, and electron microscopy are provided in the Supplementary Appendix.
Protein Modeling
Three-dimensional models of homology between the wild-type and mutated ATG7 C-terminal domain were built from the crystal structure of the yeast Atg7-Atg3 (crystal structure 4GSL in the Protein Data Bank) and Atg7-Atg8 (crystal structure 3VH3 in the Protein Data Bank) complexes with the use of Modeller software, version 10.1 (https://salilab.org/modeller/), with standard parameters.
Yeast Studies
Yeast strains were grown at 28°C in a synthetic medium consisting of 2% glucose and a 0.67% yeast nitrogen base without amino acids. A synthetic medium consisting of 2% glucose and a 0.17% yeast nitrogen base without ammonium sulfate was used as a starvation medium for the autophagy assays. Detailed methods are provided in the Supplementary Appendix.
Statistical Analysis
Data are provided as means and standard errors or standard deviations, as indicated. Ordinary one-way analysis of variance and Sidak’s multiple-comparison test were used to assess differences between samples in the autophagic sequestration assay.
Results
Families with Deleterious ATG7 Variants
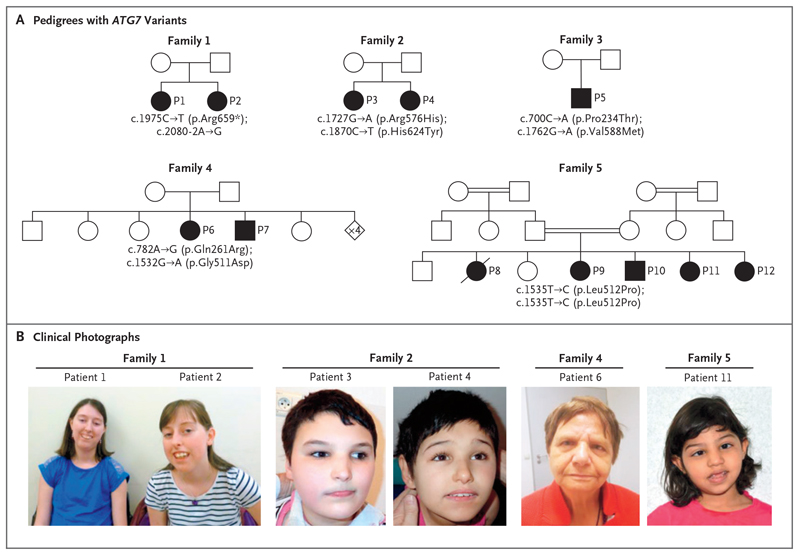
Two female siblings in Family 1 — Patient 1 (28 years old) and Patient 2 (18 years old) — have mild-to-moderate learning difficulties and ataxia, tremor, and proximal muscle weakness (Fig. 2A). They also have bilateral sensorineural hearing loss, as well as eye abnormalities including optic atrophy, chronic progressive external ophthalmoplegia, and ptosis. Patient 2 also has early-onset cataracts. Both siblings have facial dysmorphism comprising a high arched palate; gum hypertrophy; a long, narrow face; and retrognathia (Fig. 2B). Neuroimaging in Patient 1 identified moderate cerebellar hypoplasia and a thin posterior corpus callosum (see Fig. S1, which shows findings on magnetic resonance imaging [MRI] in one or more of the patients from each family in this study). Exome sequencing revealed recessively inherited loss-of-function ATG7 variants in Family 1: c.1975C→T (p.Arg659*) and c.2080-2A→G (RefSeq accession number, NM_006395.2). RNA-sequencing studies showed that ATG7 expression was reduced and that the c.2080-2A→G splice-site variant causes replacement of canonical exon 19 with intron 18, leading to premature termination (Figs. S2 and S3).
Figure 2. Identification of Five Families with Ataxia and Developmental Delay and Deleterious, Biallelic ATG7 Variants.
The pedigrees shown in Panel A represent families with ATG7 variants. Circles denote female family members, squares male family members, diamonds family members of unknown sex or undisclosed gender, and shaded symbols affected family members. Double horizontal lines between two symbols indicate a consanguineous relationship, and a diagonal line through a symbol indicates a deceased family member. P1 through P12 denote Patients 1 through 12. Clinical photographs are shown in Panel B: Patient 1 (at 27 years of age) and Patient 2 (at 17 years of age) from Family 1, Patient 3 (at 18 years of age) and Patient 4 (at 15 years of age) from Family 2, Patient 6 (at 71 years of age) from Family 4, and Patient 11 (at 5 years of age) from Family 5.
Family 2 also has two female siblings. Patient 3 (18 years old) and Patient 4 (15 years old) are wheelchair-bound as a result of spastic paraplegia and severe developmental delay (Fig. 2A). They presented with congenital encephalopathy, axial hypotonia, truncal ataxia, and facial dysmorphism (Fig. 2B). Patient 3 has tonic-clonic seizures with recurrent episodes of status epilepticus. Both sisters have retinopathy, and Patient 4 has optic atrophy. Complex brain abnormalities were identified through neuroimaging and included severe cerebellar hypoplasia, a thin posterior corpus callosum, and bilateral optochiasmatic atrophy. Electroencephalographic abnormalities were also observed. Exome sequencing identified biallelic missense ATG7 variants — c.1727G→A (p.Arg576His) and c.1870C→T (p.His624Tyr) — affecting amino acid residues that are highly conserved (see Fig. S4, which shows the amino acid change or changes in each family in the study as compared with the sequences in several different species).
In Family 3, Patient 5 is a 34-year-old wheelchair-bound man who presented with moderate developmental delay and congenital ataxia (Fig. 2A). This patient has facial dysmorphism comprising a high arched palate and smooth philtrum. MRI revealed cerebellar hypoplasia and a thin posterior corpus callosum. He also has hypogonadotropic hypogonadism, gynecomastia, and hypertrophic cardiomyopathy. Patient 5 has biallelic missense ATG7 variants affecting highly conserved residues: c.700C→A (p.Pro234Thr) and c.1762G→A (p.Val588Met).
Family 4 includes Patient 6 (a 71-year-old woman) and Patient 7 (a 68-year-old man) (Fig. 2A). These siblings have mild-to-moderate intellectual disability with ataxia and tremor (Patient 6) or dyskinesia (Patient 7). Neuroimaging revealed cerebellar hypoplasia and a thin posterior corpus callosum in both siblings, who also have facial dysmorphism and psychiatric involvement: Patient 6 has schizophrenic psychosis, and Patient 7 displays aggression and self-mutilating behavior (Fig. 2B). Patient 6 also has late-onset dementia and an acoustic neuroma that is visible in the brain stem on neuroimaging. Exome sequencing identified recessive missense ATG7 variants in Family 4: c.782A→G (p.Gln261Arg) is predicted to alter two exonic splicing enhancer sites that lead to skipping of coding exon 8, which was confirmed by reverse-transcriptase polymerase chain reaction and Sanger sequencing (Fig. S5), and c.1532G→A (p.Gly511Asp) affects a highly conserved residue.
Five of the seven siblings in Family 5 (Patients 8 through 12) are affected (Fig. 2A). Patient 8, a female infant, presented at 6 weeks of age with strabismus; at that time, there were also concerns regarding her vision. An ophthalmic examination revealed optic atrophy. Subsequently, at the age of 3 months, seizures developed, and she had severe global neurodevelopmental delay. Cranial MRI revealed diffuse brain atrophy, and the patient died at 2 years of age. Three of her siblings (Patients 9, 10, and 12) also presented in infancy with strabismus and visual impairment and were noted to have optic atrophy, whereas Patient 11 (Fig. 2B) was initially identified because of congenital microcephaly; optic atrophy was identified later in the course of her disease. The clinical course for all the surviving siblings has been similar, with mild-to-moderate intellectual disability, motor and language developmental delay, ataxia, and tremor being common features. In addition, axial hypotonia and spastic diplegia developed in Patients 9 and 10. The oldest surviving sibling is now 16 years of age. Patients 9 and 12 also underwent cranial MRI examinations, which showed diffuse brain volume loss and, in the case of Patient 12, a thin posterior corpus callosum. Exome sequencing identified a homozygous ATG7 variant — c.1535T→C (p.Leu512Pro) — in Patients 9 through 12. Patient 8 did not undergo sequencing.
Collaboration among research centers in the United Kingdom, France, Switzerland, Germany, and Saudi Arabia was facilitated by the online tool GeneMatcher (http://genematcher.org).25
Myopathy in ATG7-Deficient Skeletal Muscle
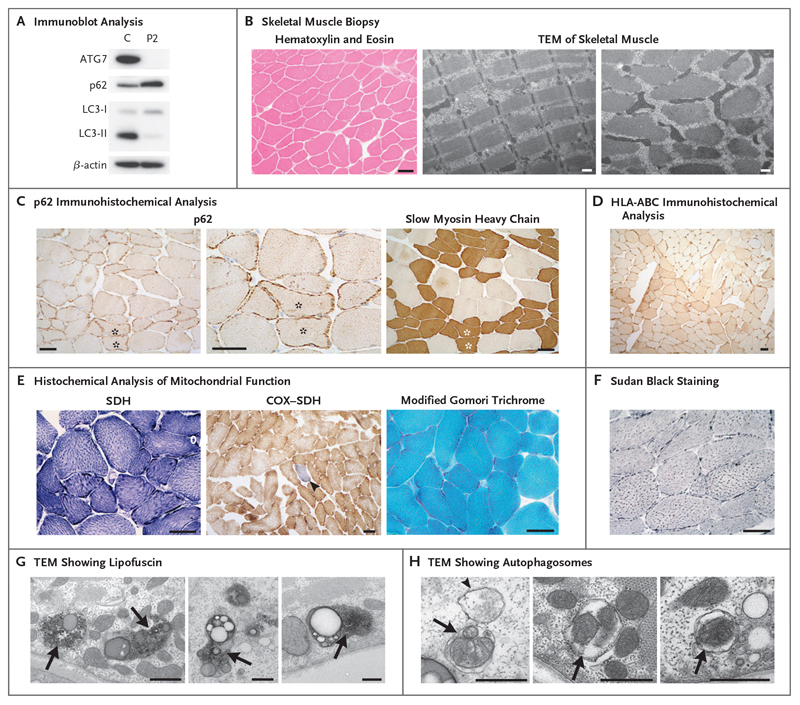
Immunoblotting of a skeletal muscle-biopsy specimen and cultured myoblasts from Patient 2 indicated that ATG7 protein was undetectable (Fig. 3A). ATG7 is required for the lipidation of LC3-I, which is needed in order to generate LC3-II, the levels of which were severely diminished in myoblasts from this patient (Fig. 3A). Increased levels of p62 (also known as SQSTM1), a classical autophagy receptor that recruits cytoplasmic cargo to autophagosomes through interaction with LC3-II and is subsequently degraded after autolysosome formation, were noted in myoblasts from Patient 2 (Fig. 3A). Overall, the muscle-biopsy specimen showed mild myopathic changes. No vacuoles or internalized nuclei were present, and transmission electron micrographs showed normal organization of muscle fibers (Fig. 3B).
Figure 3. Mild Myopathic Changes in ATG7-Deficient Human Skeletal Muscle.
Panel A shows immunoblot analysis of steady-state levels of autophagy proteins ATG7, p62, and LC3 in myoblasts from Patient 2 relative to a control (C) cell line; results reflect one analysis each. Panel B shows hematoxylin and eosin staining and transmission electron microscopy (TEM) indicated that no vacuoles or internalized nuclei were present and that the organization of skeletal muscle fibers was normal. Panel C shows immunohistochemical analysis with an antibody reactive to p62, which showed abnormal aggregation of p62 within the subsarcolemmal region of muscle. An accentuation of these accumulations within slow myosin heavy chain-positive (type I) muscle fibers can be seen in serial sections (highlighted with asterisks). Panel D shows immunohistochemical analysis with antibodies against the inflammatory markers HLA-A, -B, and -C (HLA-ABC). Panel E shows histochemical reactions for mitochondrial succinate dehydrogenase (SDH) and sequential cytochrome c oxidase (COX)-SDH activity; the blue fiber (arrowhead) is COX-deficient. Modified Gomori trichrome staining showed no evidence of abnormal mitochondrial subsarcolemmal accumulation. Panel F shows Sudan black (neutral lipid) staining, which was slightly accentuated. Panels G and H show transmission electron micrographs in which an accumulation of lipofuscin (Panel G, arrows) and the presence of autophagosomes engulfing mitochondria (Panel H, arrows) and general cytoplasmic constituents (Panel H, arrowhead) can be seen. Scale bars indicate 50 μm (immunohistochemical analyses in Panels B through F) and 500 nm (TEM in Panels B, G, and H).
In support of the findings from immunoblotting, immunohistochemical analysis revealed abnormal p62 accumulation in the subsarcolemmal zone (Fig. 3C). Structures that were p62-positive were more apparent in type I fibers than in type II fibers, a finding highlighted by slow myosin heavy chain reactivity (Fig. 3C). Moreover, type I fibers appeared to be smaller than type II fibers. Increased reactivity to major histocompatibility complex HLA-A, -B, and -C across muscle fibers suggested an up-regulated immune response within the muscle from Patient 2 (Fig. 3D). Histochemical analysis of mitochondrial function within muscle was also undertaken and revealed accentuated subsarcolemmal accumulation of SDH activity and focal COX-deficient fibers (Fig. 3E). No ragged-red fibers were identified with modified Gomori trichrome staining, and Sudan black staining revealed mild lipid accumulation (Fig. 3E and 3F). An abnormal accumulation of lipofuscin — an age-related, lipid-containing pigment granule — was readily identified on transmission electron microscopy. Autophagosomes were also present, including some engulfing mitochondria (Fig. 3G and 3H). Autophagic structures were also readily observed in primary fibroblasts from Patient 1 (the sibling of Patient 2) under basal conditions after treatment with the lysosomal inhibitor bafilomycin A1 (Fig. S6).
Functional Autophagy Studies in Cultured Fibroblasts
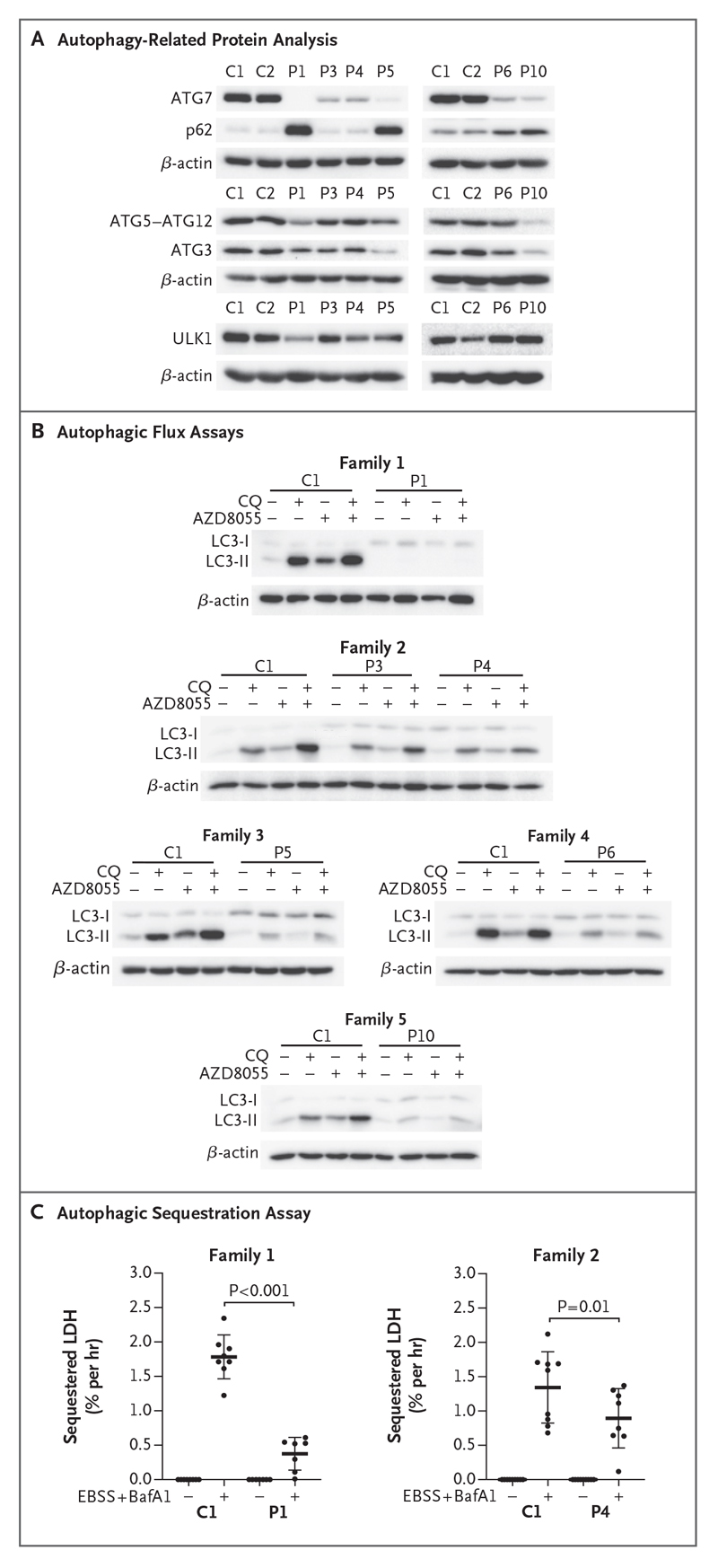
Immunoblot analysis of primary patient-derived fibroblasts cultured under basal conditions showed that ATG7 protein was undetectable in Patient 1 and was present at severely diminished levels in Patients 3 through 6 and 10 (Fig. 4A). Steady-state levels of other autophagy-related proteins, including ATG3, ATG5-ATG12 conjugates, and ULK1, were affected to various degrees in these patients (Fig. 4A). These deficits led to accumulation of p62 in Patients 1, 5, 6, and 10 under basal conditions (Fig. 4A). We immortalized fibroblasts from Patients 1, 3, and 4 and Controls 1 and 2. These cells recapitulated the aforementioned biochemical phenotypes, and an accumulation of large p62 puncta in fibroblasts from Patient 1 was observed on immunofluorescence, a finding consistent with the immunoblotting findings (Fig. S7).
Figure 4. Defective ATG7-Dependent Autophagy in Patients with ATG7 Variants.
Panel A shows a representative Western blot analysis from duplicate or triplicate assessment of autophagy-related proteins in primary fibroblasts cultured in basal conditions and immunoblotted against ATG7, p62, ATG5-ATG12, ATG3, and ULK1. β-Actin was used as a loading control. Panel B shows autophagic flux analyzed by Western blotting after treatment of immortalized or primary fibroblasts with (+) or without (-) chloroquine (CQ) (60 μmol per liter) and in the presence or absence of AZD8055 (1 μmol per liter) for 2 hours before immunoblot detection of LC3 and β-actin (loading control). Western blots are representative of triplicate experiments, except for those in the analysis of fibroblasts from Patient 5, which were completed in duplicate. Panel C shows estimated autophagic cargo sequestration, which was assessed as sequestered lactate dehydrogenase (LDH) in the presence or absence of autophagy induction (starvation with Earl’s balanced salt solution [EBSS]) and late-stage blockade (bafilomycin A1 [BafA1, 100 nmol per liter]) for 3 hours in primary dermal fibroblasts derived from Control 1 and Patients 1 and 4. This assay was performed in triplicate. Adjusted P values are based on ordinary one-way analysis of variance and Sidak’s multiple-comparison test. Horizontal bars indicate mean values, and I bars indicate standard deviations.
The rate of autophagic flux can be inferred by monitoring levels of LC3-II under different conditions.26 Patients’ fibroblasts were incubated with or without the mechanistic target of rapamycin (mTOR) kinase inhibitor AZD8055 to induce autophagy, as well as in the presence or absence of appropriate inhibitors (chloroquine or bafilomycin A1) that block autophagic flux. In fibroblasts from Patient 1, LC3-II was nearly absent even after concomitant autophagy induction and blockade, and flux was diminished in fibroblasts from Patients 3 through 6 and 10 (Fig. 4B). Complementing these approaches, a quantitative assay was also used to directly measure nonselective autophagic sequestration of endogenous cargo in control and patient fibroblasts.27,28 Bulk autophagic sequestration activity was significantly reduced in primary fibroblasts from Patients 1 and 4, which confirmed that autophagic flux was defective in these patients with ATG7 deficiency (Fig. 4C).
Computer Modeling
Next, we sought to understand the effect of missense variants on ATG7 structure. Homology models of the ATG7 homodimer predicted that Arg576His (Family 2), His624Tyr (Family 2), Val588Met (Family 3), Gly511Asp (Family 4), and Leu512Pro (Family 5) would prevent optimal folding of ATG7 protein and interfere with homodimerization (Fig. S8). Western blotting with nonreducing gel electrophoresis supported these predictions, suggesting a greater amount of ATG7 monomer relative to ATG7 dimer in fibroblasts from Patients 3 through 5 than in fibroblasts from controls (Fig. S8).
Complementation Studies
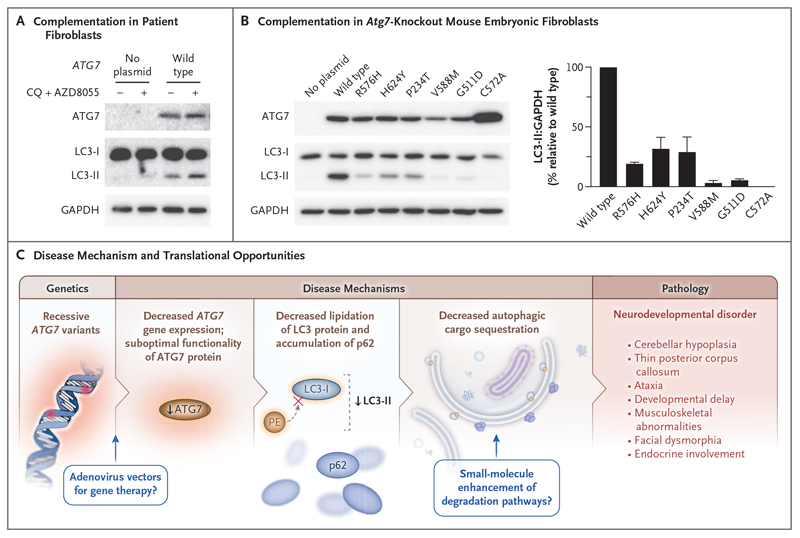
To authenticate the effect of the ATG7 missense variants from Families 2, 3, and 4 on defective autophagy with greater mechanistic precision, we performed functional complementation experiments. In human cells, complementation of wild-type ATG7 in fibroblasts from Patient 1 led to successful LC3 lipidation (Fig. 5A). Next, we transiently introduced plasmids encoding human wild-type, mutant, and catalytic-null (Cys572Ala) ATG7 into Atg7-knockout mouse embryonic fibro-blasts before autophagy induction and blockade of LC3-II degradation through treatment with AZD8055 and chloroquine. Atg7-knockout mouse embryonic fibroblasts expressing patient-associated ATG7 variants did not have recovery of LC3 lipidation to levels associated with expression of wild-type ATG7, thus consolidating the damaging nature of the variants studied (Fig. 5B). Because autophagy is an evolutionarily conserved process and the ATG7 protein sequence is maintained across species, we also used complementation studies to assess autophagy in Saccharomyces cerevisiae expressing homologous atg7 mutations (Fig. S9). The yeast Pho8Δ60 assay,29 which quantitatively measures the vacuolar activity of a modified version of the alkaline phosphatase Pho8 (Pho8Δ60) that is delivered to the vacuole only by bulk autophagy, supported our previous findings by showing attenuated autophagy after starvation in yeast expressing mutated atg7 (Fig. S9). Analogous investigations in which the yeast green fluorescent protein-atg8 assay was used were generally supportive, but experimental data showed variability (Fig. S9).30
Figure 5. Complementation Studies with Fibroblasts and a Summary of Findings.
Panel A shows the results of transient introduction of plasmids encoding wild-type ATG7 into immortalized fibroblasts from Patient 1 for 24 hours. Cells were then treated with or without CQ (60 μmol per liter) and AZD8055 (1 μmol per liter) for 2 hours before immunoblot detection of ATG7, LC3, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH, loading control). The Western blots shown are representative of triplicate experiments. Panel B shows the results of transient introduction of plasmids encoding wild-type, catalytic-null (C572A), and missense ATG7 variants into Atg7-knockout mouse embryonic fibroblasts for 24 hours. Cells were then treated with CQ (60 μmol per liter) and AZD8055 (1 μmol per liter) for 2 hours. Immunoblotting results are representative of three independent experiments with antibodies against ATG7 and LC3. GAPDH was used as a loading control. The graph in Panel B shows the results of a densitometric analysis of the ratio of LC3 to GAPDH, normalized to wild-type values. The bars represent means, and T bars indicate standard deviations. Panel C shows a summary of our findings.
Discussion
Our work shows the clinical significance of dysfunctional autophagy in humans. We identified five unrelated families with deleterious, recessive variants in human ATG7, a gene encoding an essential effector enzyme for canonical autophagy.
Defective autophagy underlies the profound neurologic and developmental impairments in our patients. In Family 1, the absence of ATG7 protein impairs LC3 lipidation and causes an accumulation of p62 in patient-derived fibroblasts and skeletal muscle. The observed low levels of ATG7 steady-state protein and diminished autophagic flux in affected members of Families 2 through 5 are consistent with impaired ATG7 homodimerization caused by the missense variants in these families. Moreover, we observed p62 accumulation in the fibroblasts from Families 3, 4, and 5 and further confirmed functional deficiencies caused by these missense variants through complementation in mouse and yeast systems.
Our comprehensive clinical investigation of these patients consolidates the critical importance of basal autophagy in human neural and musculoskeletal integrity. Many of the clinical features observed in these patients are recapitulated in the conditional Atg7-knockout mouse models, including brain abnormalities19,20 and hypotonia and muscle weakness.21 All the patients for whom MRI findings were available had cerebellar hypoplasia and corpus callosum abnormalities, although in some patients the effect on brain development was more diffuse. In contrast, to date, no liver abnormalities have been reported in these patients. ATG7 may play a role in human endocrine development: patients from Family 2 had late-onset or no puberty, and Patient 5 presented with hypogonadotropic hypogonadism and gynecomastia. Some patients also presented with distinctive facial dysmorphism; other clinical features included hypertrophic cardiomyopathy (Family 3), ocular abnormalities (Families 1, 2, and 5), and deafness (Family 1).
Our investigations indicate that patients with this condition can approach population life expectancy (affected members of Family 4 have reached 68 and 71 years of age) despite severe attenuation of autophagic flux. Moreover, assessment of Family 1 supports the idea that, in rare circumstances, biallelic loss-of-function variants in a core autophagy gene without a functional paralogue are compatible with human life. It is important to note that loss-of-function variants have been reported in core autophagy genes WDR45 and WDR45B, homologues of yeast atg18, but only simultaneous knockout of these two genes causes perinatal lethality in mice, which suggests a degree of functional redundancy.31
Our data present a conundrum: affected members of Family 2 had a greater disease burden than those in the other families despite having the mildest impairment in autophagy. Nevertheless, we have provided strong evidence of pathogenicity. In support of our findings, studies involving patient fibroblasts, mouse embryonic fibroblasts, and yeast have shown the functional deficiencies caused by the p.Arg576His and p.His624Tyr variants. Although it is estimated that 5% of patients who undergo exome sequencing might have more than one genetic diagnosis,32 and there is evidence of secondary-variant genetic burdens in cohorts of patients with a given disease,33 our exome and RNA sequencing did not elucidate any potential disease-modifying variants.
Despite the identification of basal autophagic structures through transmission electron microscopy in muscle and fibroblasts from Family 1, our converging data indicate that bulk autophagy is impaired. The origin of autophagosomes that are generated in the absence of core autophagy conjugation-system proteins is not known. In the absence of mouse Atg7 or Atg5, the trans-Golgi network is proposed to generate autophagosomes by a process termed alternative autophagy.34 It has also been shown that canonical autophagy can still proceed in the absence of ATG proteins, albeit at a reduced rate as a result of impaired inner autophagosomal membrane degradation,35 and there is evidence that ATG8 proteins (there are six known human ATG8 orthologues) mediate autophagosome-lysosome fusion but not autophagosome formation.36 The delineation of these mechanisms, in addition to autophagy-independent turnover pathways, will be critical to understanding intracellular degradation in human health and disease.37
Our data suggest that impaired autophagy resulting from biallelic deleterious ATG7 variants is a cause of neurodevelopmental disorders involving neurologic, muscular, and endocrine hypofunction. These findings strengthen our understanding of autophagy in human disease and expand the spectrum of clinical phenotypes and genetic loci associated with congenital autophagy-deficient syndromes. Given that the perinatal lethality of Atg5-null mice can be avoided through selective restoration of autophagy in the nervous system,38 it is tempting to speculate that analogous neural restoration (perhaps through small-molecule or gene-therapy approaches) may prove to be a vital therapeutic strategy for this series of patients and other persons with diseases driven by impaired autophagy (Fig. 5C).
Supplementary Material
Acknowledgments
Supported by grants from the Wellcome Centre for Mitochondrial Research (203105/Z/16/Z), the Medical Research Council International Centre for Genomic Medicine in Neuromuscular Disease (MR/S005021/1), the Mitochondrial Disease Patient Cohort (United Kingdom) (G0800674), the Lily Foundation, and the NHS Specialised Commissioners, who fund the “Rare Mitochondrial Disorders of Adults and Children” Service in Newcastle Upon Tyne (to Drs. McFarland and Taylor); Ph.D. funding from the Barbour Foundation and an EMBO Short Term Fellowship (to Dr. Collier); the Academy of Finland, Novo Nordisk Foundation, Sigrid Juselius Foundation, Finnish Cardiovascular Foundation, and University of Helsinki (to Dr. McWilliams); grants from the French National Agency for Research (ANR-16-CE16-0025-04) and the “Association Française contre les Myopathies” (AFM-MITOSCREEN, project 17122) (to Ms. Piron-Prunier and Dr. Delahodde); the European Union Horizon 2020 research and innovation program under the frame of ERA-NET Cofund action 643578 network PREPARE (BMBF 01GM1607, to Drs. Synofzik, Koenig, and Zuchner); a grant (779257 “Solve-RD,” to Ms. Reich and Dr. Synofzik) from the European Union Horizon 2020 program; a grant (320030_179547, to Dr. Rauch) from the Swiss National Science Foundation; and a Medical Research Council UK Clinician Scientist Fellowship (MR/N008324/1, to Dr. Ryten).
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
We thank Ross Laws and Tracey Davey at Newcastle University EM Research Services and BioImaging for assistance with microscopy; David Baux for the development of bioinformatics tools; Pascal Joset for his technical help; Nikolai Engedal (University of Oslo) for his help and advice with cargo sequestration assays; and Sara Aguti, Haiyan Zhou, Francesco Muntoni, Joanna Poulton, and Robert Pitceathly for providing the patient-derived fibroblast cell lines that were used as RNA-sequencing controls.
We dedicate this article to the memory of Agnès Delahodde.
Contributor Information
Jack J. Collier, Wellcome Centre for Mitochondrial Research, London, United Kingdom; Translational and Clinical Research Institute, London, United Kingdom
Claire Guissart, (Institut Universitaire de Recherche Clinique and Laboratoire de Génétique Moléculaire, University of Montpellier and Centre Hospitalier Universitaire (CHU) de Montpellier, France)
Monika Oláhová, Wellcome Centre for Mitochondrial Research, London, United Kingdom; Translational and Clinical Research Institute, London, United Kingdom
Souphatta Sasorith, (Institut Universitaire de Recherche Clinique and Laboratoire de Génétique Moléculaire, University of Montpellier and Centre Hospitalier Universitaire (CHU) de Montpellier, France)
Florence Piron-Prunier, Montpellier, and the Institute for Integrative Biology of the Cell (I2BC), Université Paris-Saclay, Alternative Energies and Atomic Energy Commission (CEA), CNRS Gif-sur-Yvette, France
Fumi Suomi, Translational Stem Cell Biology and Metabolism Program, Research Programs Unit, and the Department of Anatomy, Faculty of Medicine, University of Helsinki, Helsinki
David Zhang, Newcastle University, Newcastle Upon Tyne, and the Institute of Child Health, Department of Molecular Neuroscience, University College London Institute of Neurology, London, United Kingdom
Nuria Martinez-Lopez, Wellcome Centre for Mitochondrial Research, London, United Kingdom; Radiation Oncology, Albert Einstein College of Medicine, New York
Nicolas Leboucq, Departments of Neuroradiology, France
Tuomo M. Polvikoski, Translational and Clinical Research Institute, London, United Kingdom
Pierre Meyer, Pediatric Neurology, France; Reference Center for Neuromuscular Diseases Atlantic–Occitania–Caribbean (AOC), France, CHU de Montpellier, and Laboratoire de Physiologie et Médecine Expérimentale du Coeur et des Muscles (PhyMedExp), INSERM, CNRS, University of Montpellier, France
Lise Larrieu, (Institut Universitaire de Recherche Clinique and Laboratoire de Génétique Moléculaire, University of Montpellier and Centre Hospitalier Universitaire (CHU) de Montpellier, France)
Andrew M. Schaefer, Wellcome Centre for Mitochondrial Research, London, United Kingdom; Translational and Clinical Research Institute, London, United Kingdom; NHS Highly Specialised Service for Rare Mitochondrial Disorders of Adults and Children, London, United Kingdom
Hessa S. Alsaif, Departments of Genetics
Suad Alyamani, Neuroscience
Stephan Zuchner, King Faisal Specialist Hospital and Research Center, Riyadh, Saudi Arabia; and the Dr. John T. Macdonald Foundation, Department of Human Genetics, and John P. Hussman Institute for Human Genomics, Miller School of Medicine, University of Miami, Miami
Inês A. Barbosa, Division of Genetics and Molecular Medicine, Guy’s Hospital, King’s College London School of Medicine, London, United Kingdom
Charu Deshpande, Clinical Genetics Unit, Guy’s and St. Thomas’ NHS Foundation Trust, London, United Kingdom
Angela Pyle, Wellcome Centre for Mitochondrial Research, London, United Kingdom; Translational and Clinical Research Institute, London, United Kingdom
Anita Rauch, Institute of Medical Genetics, University of Zurich, Zurich, Switzerland
Matthis Synofzik, Hertie Institute for Clinical Brain Research and Center of Neurology, and the German Center for Neurodegenerative Diseases, University of Tübingen, Tübingen, Germany
Fowzan S. Alkuraya, Departments of Genetics
François Rivier, Pediatric Neurology, France; Reference Center for Neuromuscular Diseases Atlantic–Occitania–Caribbean (AOC), France; CHU de Montpellier, and Laboratoire de Physiologie et Médecine Expérimentale du Coeur et des Muscles (PhyMedExp), INSERM, CNRS, University of Montpellier, France
Mina Ryten, Newcastle University, Newcastle Upon Tyne, and the Institute of Child Health, Department of Molecular Neuroscience, University College London Institute of Neurology, London, United Kingdom
Robert McFarland, Wellcome Centre for Mitochondrial Research, London, United Kingdom; Translational and Clinical Research Institute, London, United Kingdom; NHS Highly Specialised Service for Rare Mitochondrial Disorders of Adults and Children, London, United Kingdom
Agnès Delahodde, Montpellier, and the Institute for Integrative Biology of the Cell (I2BC), Université Paris-Saclay, Alternative Energies and Atomic Energy Commission (CEA), CNRS Gif-sur-Yvette, France
Thomas G. McWilliams, Translational Stem Cell Biology and Metabolism Program, Research Programs Unit, and the Department of Anatomy, Faculty of Medicine, University of Helsinki, Helsinki
Michel Koenig, (Institut Universitaire de Recherche Clinique and Laboratoire de Génétique Moléculaire, University of Montpellier and Centre Hospitalier Universitaire (CHU) de Montpellier, France)
Robert W. Taylor, Wellcome Centre for Mitochondrial Research, London, United Kingdom; Translational and Clinical Research Institute, London, United Kingdom; NHS Highly Specialised Service for Rare Mitochondrial Disorders of Adults and Children, London, United Kingdom
References
- 1.Galluzzi L, Baehrecke EH, Ballabio A, et al. Molecular definitions of autophagy and related processes. EMBO J. 2017;36:1811–36. doi: 10.15252/embj.201796697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu L, Chen Y, Tooze SA. Autophagy pathway: cellular and molecular mechanisms. Autophagy. 2018;14:207–15. doi: 10.1080/15548627.2017.1378838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018;19:349–64. doi: 10.1038/s41580-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 4.Mizushima N. A brief history of autophagy from cell biology to physiology and disease. Nat Cell Biol. 2018;20:521–17. doi: 10.1038/s41556-018-0092-5. [DOI] [PubMed] [Google Scholar]
- 5.Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176:11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yapici Z, Eraksoy M. Non-progressive congenital ataxia with cerebellar hypoplasia in three families. Acta Paediatr. 2005;94:248–53. doi: 10.1111/j.1651-2227.2005.tb01902.x. [DOI] [PubMed] [Google Scholar]
- 7.Kim M, Sandford E, Gatica D, et al. Mutation in ATG5 reduces autophagy and leads to ataxia with developmental delay. Elife. 2016;5:e12245. doi: 10.7554/eLife.12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saitsu H, Nishimura T, Muramatsu K, et al. De novo mutations in the autophagy gene WDR45 cause static encephalopathy of childhood with neurodegeneration in adulthood. Nat Genet. 2013;45(4):445–449.:449e1. doi: 10.1038/ng.2562. [DOI] [PubMed] [Google Scholar]
- 9.Haack TB, Hogarth P, Kruer MC, et al. Exome sequencing reveals de novo WDR45 mutations causing a phenotypically distinct, X-linked dominant form of NBIA. Am J Hum Genet. 2012;91:1144–9. doi: 10.1016/j.ajhg.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suleiman J, Allingham-Hawkins D, Hashem M, Shamseldin HE, Alkuraya FS, El-Hattab AW. WDR45B-related intellectual disability, spastic quadriplegia, epilepsy, and cerebral hypoplasia: a consistent neurodevelopmental syndrome. Clin Genet. 2018;93:360–4. doi: 10.1111/cge.13054. [DOI] [PubMed] [Google Scholar]
- 11.Jelani M, Dooley HC, Gubas A, et al. A mutation in the major autophagy gene, WIPI2, associated with global developmental abnormalities. Brain. 2019;142:1242–54. doi: 10.1093/brain/awz075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mizushima N, Levine B. Autophagy in human diseases. N Engl J Med. 2020;383:1564–76. doi: 10.1056/NEJMra2022774. [DOI] [PubMed] [Google Scholar]
- 13.Suomi F, McWilliams TG. Autophagy in the mammalian nervous system: a primer for neuroscientists. Neuronal Signal. 2019;3(3):NS20180134. doi: 10.1042/NS20180134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zachari M, Ganley IG. The mammalian ULK1 complex and autophagy initiation. Essays Biochem. 2017;61:585–96. doi: 10.1042/EBC20170021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mizushima N, Noda T, Yoshimori T, et al. A protein conjugation system essential for autophagy. Nature. 1998;395:395–8. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 16.Ichimura Y, Kirisako T, Takao T, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–92. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 17.Komatsu M, Waguri S, Ueno T, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–34. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiong J. Atg7 in development and disease: panacea or Pandora’s Box? Protein Cell. 2015;6:722–34. doi: 10.1007/s13238-015-0195-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komatsu M, Wang QJ, Holstein GR, et al. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci U S A. 2007;104:14489–94. doi: 10.1073/pnas.0701311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komatsu M, Waguri S, Chiba T, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–4. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 21.Masiero E, Agatea L, Mammucari C, et al. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10:507–15. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 22.Takamura A, Komatsu M, Hara T, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komatsu M, Waguri S, Koike M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–63. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 24.Old SL, Johnson MA. Methods of microphotometric assay of succinate dehydrogenase and cytochrome c oxidase activities for use on human skeletal muscle. Histochem J. 1989;21:545–55. doi: 10.1007/BF01753355. [DOI] [PubMed] [Google Scholar]
- 25.Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36:928–30. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klionsky DJ, Abdelmohsen K, Abe A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luhr M, Szalai P, Engedal N. The lactate dehydrogenase sequestration assay — a simple and reliable method to determine bulk autophagic sequestration activity in mammalian cells. J Vis Exp. 2018;137:57971. doi: 10.3791/57971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luhr M, Szalai P, Sætre F, Gerner L, Seglen PO, Engedal N. A simple cargo sequestration assay for quantitative measurement of nonselective autophagy in cultured cells. Methods Enzymol. 2017;587:351–64. doi: 10.1016/bs.mie.2016.09.064. [DOI] [PubMed] [Google Scholar]
- 29.Noda T, Klionsky DJ. The quantitative Pho8Delta60 assay of nonspecific autophagy. Methods Enzymol. 2008;451:33–42. doi: 10.1016/S0076-6879(08)03203-5. [DOI] [PubMed] [Google Scholar]
- 30.Kainz K, Tadic J, Zimmermann A, et al. Methods to assess autophagy and chronological aging in yeast. Methods Enzymol. 2017;588:367–94. doi: 10.1016/bs.mie.2016.09.086. [DOI] [PubMed] [Google Scholar]
- 31.Ji C, Zhao H, Li D, et al. Role of Wdr45b in maintaining neural autophagy and cognitive function. Autophagy. 2020;16:615–25. doi: 10.1080/15548627.2019.1632621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Posey JE, Harel T, Liu P, et al. Resolution of disease phenotypes resulting from multilocus genomic variation. N Engl J Med. 2017;376:21–31. doi: 10.1056/NEJMoa1516767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kousi M, Söylemez O, Ozanturk A, et al. Evidence for secondary-variant genetic burden and non-random distribution across biological modules in a recessive ciliopathy. Nat Genet. 2020;52:1145–50. doi: 10.1038/s41588-020-0707-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishida Y, Arakawa S, Fujitani K, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–8. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 35.Tsuboyama K, Koyama-Honda I, Sakamaki Y, Koike M, Morishita H, Mizushima N. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science. 2016;354:1036–41. doi: 10.1126/science.aaf6136. [DOI] [PubMed] [Google Scholar]
- 36.Nguyen TN, Padman BS, Usher J, Oorschot V, Ramm G, Lazarou M. Atg8 family LC3/GABARAP proteins are crucial for autophagosome-lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. J Cell Biol. 2016;215:857–74. doi: 10.1083/jcb.201607039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizushima N. The ATG conjugation systems in autophagy. Curr Opin Cell Biol. 2020;63:1–10. doi: 10.1016/j.ceb.2019.12.001. [DOI] [PubMed] [Google Scholar]
- 38.Yoshii SR, Kuma A, Akashi T, et al. Systemic analysis of Atg5-null mice rescued from neonatal lethality by transgenic ATG5 expression in neurons. Dev Cell. 2016;39:116–30. doi: 10.1016/j.devcel.2016.09.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.