Abstract
Gag – the main structural protein of HIV-1 – is recruited to the plasma membrane for virus assembly by its matrix (MA) domain. Gag is subsequently cleaved into its component domains, causing structural maturation to repurpose the virion for cell entry. We determined the structure and arrangement of MA within immature and mature HIV-1 by cryo-electron tomography. We found that MA rearranges between two different hexameric lattices upon maturation. In mature HIV-1, a lipid extends out of the membrane to bind a pocket in MA. Our data suggest that proteolytic maturation of HIV-1 not only achieves assembly of the viral capsid surrounding the genome, but extends to repurpose the membrane-bound MA lattice for an entry or post-entry function, and causes partial removal of up to 2,500 lipids from the viral membrane.
Assembly and budding of HIV-1 is initiated at the plasma membrane (PM) and driven primarily by a 55-kDa viral polyprotein named Gag. Gag consists of an N-terminal matrix (MA) domain, responsible for recruitment to the PM, the capsid (CA) domain, which induces Gag self-assembly through protein-protein interactions, the nucleocapsid (NC) domain, which recruits the viral RNA genome to the assembly site, as well as some small peptide domains (1, 2). Proteinprotein, protein-lipid and protein-RNA interactions lead to clustering of Gag at the assembly site, membrane bending, and subsequent release of the membrane-enveloped immature HIV-1 particle. Concomitant with, or shortly after release, the viral protease (PR) cleaves Gag at multiple positions leading to a dramatic structural rearrangement to repurpose the virus particle for entry into a target cell (3, 4). Maturation results in condensation of a ribonucleoprotein complex (RNP) from NC and RNA, which becomes surrounded by the cone-shaped capsid made of CA, while mature MA is thought to remain associated with the viral membrane. Rearrangement of the HIV-1 envelope (Env) glycoproteins on the particle surface is presumed to render the virion fusogenic (2, 3, 5, 6).
The structure of the heterologously-expressed, 17-kDa MA protein has been determined, revealing a small folded domain composed of five alpha helices and one 310 helix between helices 2 and 3 (7, 8). MA crystallizes as a trimer (8), and multimerizes on artificial membrane monolayers into a hexameric lattice of trimers with holes or gaps at the six-fold symmetric positions in the lattice (9). These holes have been suggested to provide binding sites for the C-terminal tail of HIV-1 Env and promote Env incorporation into assembling virions (10, 11).
Membrane recruitment of Gag by MA is mediated by an N-terminal myristate moiety as well as by a highly basic region (HBR, residues 17-31) (12, 13). The N-terminal myristate is thought to be in equilibrium between sequestration in a pocket in the MA domain, and an exposed conformation (14) – the so-called myristoyl switch. Myristate exposure can be promoted by trimerization of MA (14). Additionally, PI(4,5)P2 in the PM has been shown to be required for correct targeting of Gag (15, 16) and for stably retaining Gag at the PM (17). Early NMR studies suggested that insertion of the 2` acyl chain of PI(4,5)P2 in an extended-lipid conformation into a pocket on the side of MA may promote exposure of the myristate and its insertion into the lipid bilayer (18). However, this mode of PI(4,5)P2 binding, in which the 2` acyl chain would have to be pulled out of the bilayer, is no longer widely considered relevant for PM binding. Instead, MA-membrane interactions are believed to involve the PI(4,5)P2 headgroup and the membranefacing HBR (15, 17, 19, 20). MA interacts with nucleic acids in the cytosol, in particular tRNA, through the HBR (21–23). These observations support a model in which exchange of nucleic acid for the PI(4,5)P2 head group promotes myristate exposure and stabilization of PM binding during virus assembly (21, 23, 24).
The known roles of MA are performed during virus assembly, and no function for MA during entry or post-entry stages is currently established. Models for how MA functions during assembly are limited by the current lack of structural information on MA within virions. It is unclear what conformation and arrangement MA adopts in situ, and whether MA undergoes any structural changes in the process of virus maturation. Here, we set out to determine the structure and arrangement of MA within immature and mature virus particles.
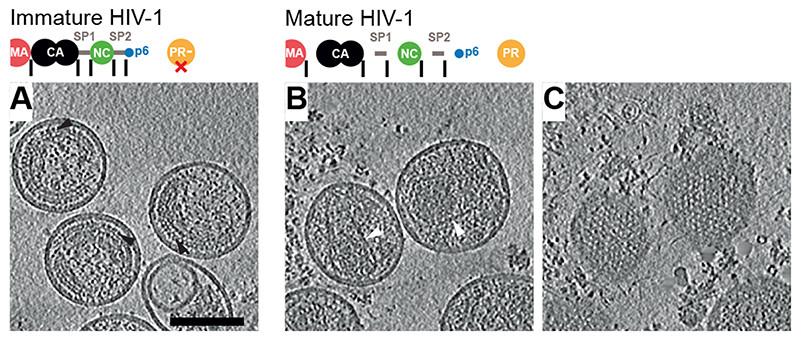
Purified mature particles were generated by transfection of a non-infectious HIV-1 proviral derivative that expresses all viral proteins except for the virulence factor Nef (pcHIV plasmid generating cHIV particles) (25), and immature cHIV particles were generated by transfection of pcHIV carrying an active site mutation in PR (pcHIV PR-). Particles were imaged without chemical fixation using cryo-electron tomography (cryo-ET). Visual inspection of tomograms revealed the expected features: a striated CA layer in immature virus particles, and conical CA cores in mature virus particles (Fig. 1A-B). In some regions underlying the lipid envelope of mature virions, we observed regular lattice-like features (Fig. 1C).
Fig. 1. Tomograms of immature and mature HIV-1 particles.
(A) Top: Schematic of the Gag protein domain architecture with cleavage sites marked as vertical black lines. The D25N mutation (red X) inactivates the viral PR. Bottom: Computational slices through a representative tomogram of immature HIV-1 (cHIV PR-) particles. Density is black. The striated Gag layer is distinctly observed in immature particles (black arrowheads). (B) As in (A) for cleaved, mature HIV-1 (cHIV) particles. Distinct conical CA cores are observed (white arrowheads). (C) Slices grazing the inner surface of the membrane at the top of the particles in (B) revealing a regular MA lattice. Gold fiducials are removed from the images. Scale bar: 100 nm.
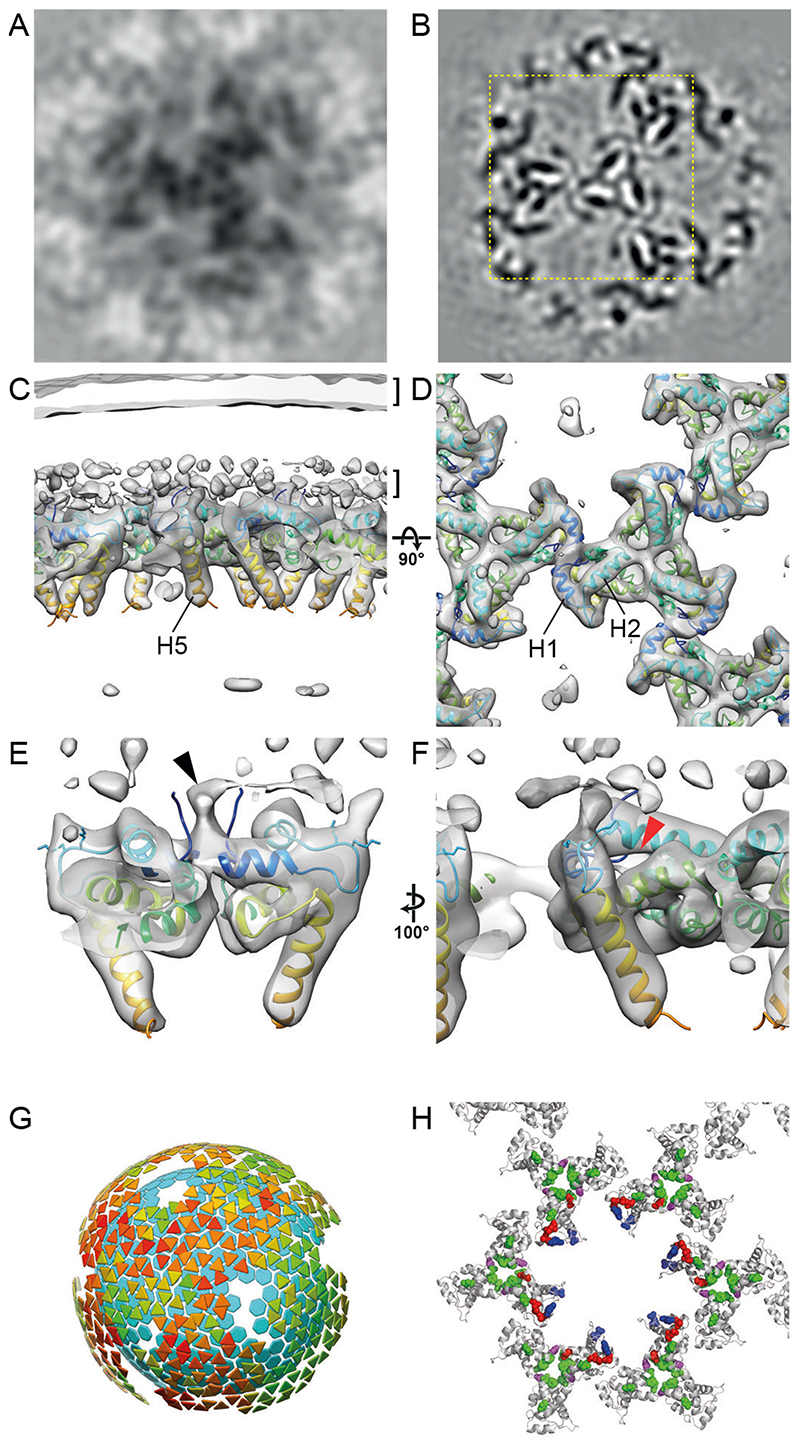
We subjected the region underlying the viral membrane in immature cHIV particles to reference-free subtomogram averaging. This analysis revealed a hexameric lattice of MA trimers with a hexamer-hexamer spacing of 9.8 nm containing large holes at the six-fold axes (Fig. 2A), reminiscent of the lattices observed for membrane-associated purified MA in vitro (9). In order to improve the spatial resolution of the immature MA structure, we analysed a cryo-ET dataset of fixed, immature, complete HIV-1NL4-3 particles (26). From this dataset, we obtained the same MA lattice structure at 7.2 A resolution (Fig. 2B-F, Fig. S1). The poorly-ordered and sparse MA lattice covered only small patches of the inner membrane leaflet, while in other areas MA did not appear to form a lattice, suggesting heterogeneity in MA packing (Fig. 2G, Fig. S2). Despite this heterogeneity, we could clearly resolve alpha-helices (Fig. 2C-F). The MA trimer structure determined by crystallography (PDBID: 1HIW (8)) could be fitted into the density as a rigid body. The MA domain of Gag is therefore present as a trimer within immature HIV-1 particles (Fig. 2D, Fig. S3A).
Fig. 2. The immature HIV-1 matrix structure.
(A, B) Slices through reconstructions of the immature HIV-1 MA lattice in cHIV PR- (A) and HIV-1NL4-3 PR- (B). The boxed region in (B) is shown at a higher magnification in (D). Density is black. (C, D) Isosurface views of the cryo-ET reconstruction (grey) for immature HIV-1NL4-3 PR- MA lattice cut perpendicular to the membrane (C) or viewed from the top towards the virus center (D). The two layers of density corresponding to the lipid headgroup layers are indicated by brackets. The structure of monomeric MA determined by NMR (PDBID: 2H3Q (18); colored blue to red from N to C terminus) was fitted as a rigid body into the density. Helices 1, 2 and 5 are marked. (E) As in (C), enlarged and cut to reveal density corresponding to the N-terminal residues (black arrowhead). (F) As in (E) but rotated. The red arrowhead indicates the unoccupied PI(4,5)P2 binding pocket. (G) Lattice map derived from subtomogram averaging for the immature HIV-1NL4-3 PR- MA lattice. Lattice maps illustrate positions and orientations of MA trimers as triangles colored on a scale from red (lower cross correlation to average structure) to green (higher cross correlation to average structure). The positions of CA hexamers are indicated as cyan hexagons to illustrate the relationship between MA and CA layers. (H) Immature MA lattice is shown as grey ribbons and residues whose mutation has been reported to modulate Env incorporation are shown as colored spheres. Mutations at residues L12, L30 and L74 (red), E16 and E98 (blue), and T69 (purple) impair Env incorporation (11, 28, 30–33). Mutations in V34, F43, Q62 and S66 (green) can rescue Env incorporation defects (11, 28, 31, 43), except those caused by mutations at T69 (purple) (28). E16 and E98 (blue) face the hole in the MA lattice in the immature virion.
MA helices 1 and 2 lie approximately parallel to the membrane and form a surface rich in charged and hydrophobic residues that attach MA to the inner membrane leaflet (Fig. 2C, F). MA trimers are packed together to form a lattice by a novel dimeric interface where the N-terminal residues and the N-terminus of helix 1 interact with themselves and the 310 helix (Fig. 2D, E). We did not observe any density that could correspond to lipid bound in the described PI(4,5)P2 binding site on the side of MA next to the HBR (18) (Fig. 2F). Its absence is consistent with current models in which interactions between the head group of PI(4,5)P2 and the membrane-proximal face of MA function as the major cellular determinant of efficient MA-PM targeting (17, 19). The N-terminal region of MA (residues 1-11) that changes conformation during the myristoyl switch (14, 27), is directly involved in the trimer-trimer interactions (Fig. S3B). At lower isosurface thresholds, a connection to the membrane appears in the vicinity of the N-terminus of helix 1 (Fig. 2E), while no notable density was observed in the myristoyl pocket (Fig. S3C). The conformational equilibrium is therefore shifted towards the exposed, membrane-inserted conformation in the immature virion, consistent with the myristate moiety becoming exposed upon MA oligomerization or membrane recruitment.
The C-terminal helix 5 of MA extends towards the centre of the virus particle and is separated from the underlying CA layer by a disordered stretch containing the MA/CA cleavage site (Fig. 2C). CA is therefore not resolved when MA is aligned, and equivalently MA is not resolved when CA is aligned (Fig. S4). The position of MA is thus constrained but not fixed relative to CA.
A number of studies have implied that holes in a hypothetical MA lattice may present a binding site for the cytoplasmic tail of Env, and mutations affecting MA trimerization have been shown to affect Env incorporation (9, 10, 28). Here we found that MA does form a lattice containing holes. We did not observe density for the Env tail in the holes, consistent with the low Env copy number per particle (29). A number of mutations in MA have been reported to result in defects in Env incorporation (10, 11, 28, 30–33) (Fig. 2H, red, purple and blue spheres). Many of the mutated residues (red and purple spheres) are not exposed towards the central hole, but are rather located at sites close to the N-terminal residues of MA or the intra- and inter-trimer interfaces, consistent with their phenotype in MA trimerization (Fig. 2H). These mutations are thus likely to modulate MA trimerization or lattice formation, and/or the myristoyl switch, but would not be presumed to directly control Env cytoplasmic tail binding within the holes. This is consistent with the model that correct MA oligomerization is required for Env incorporation (11, 34).
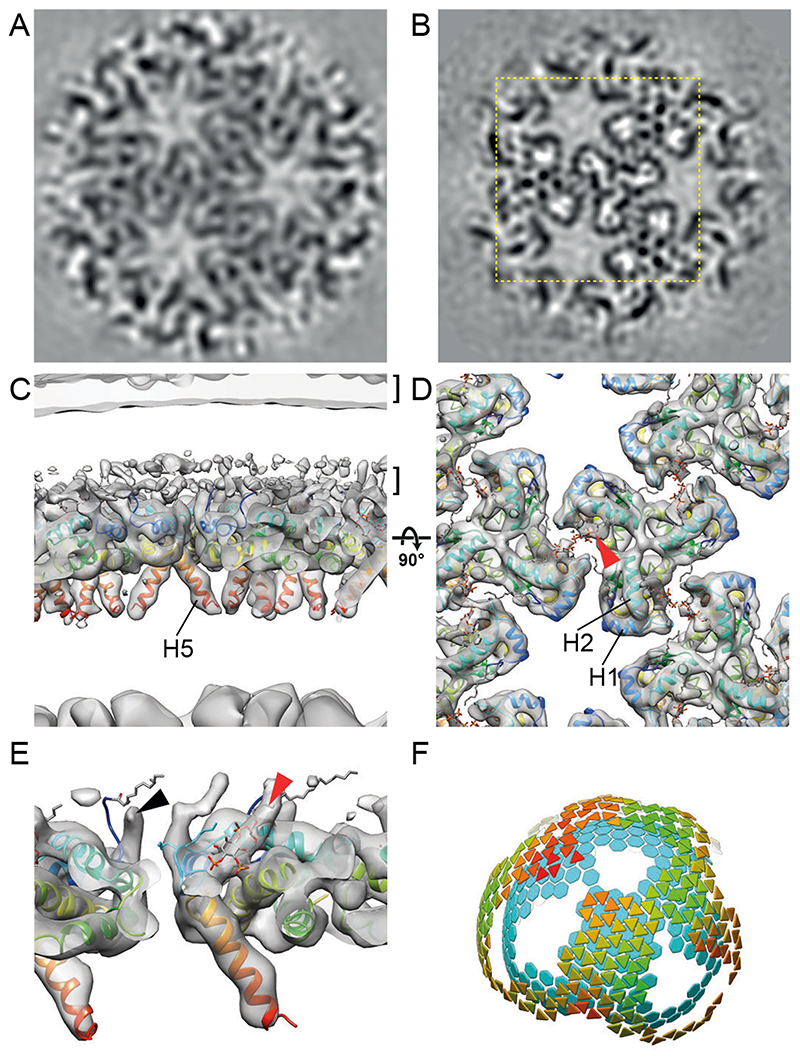
We next subjected the region underlying the membrane in mature cHIV particles to reference-free subtomogram averaging. This analysis revealed large patches of a hexagonal lattice of MA trimers with a hexamer-hexamer spacing of ~8.8 nm. These dimensions are similar to the 9 nm repeating MA lattice observed in anomalous, multi-cored, membranous particles (35). We resolved the structure of the lattice to a resolution of 9.5 Å (Fig. 3A). The mature lattice, like the immature lattice, is formed from MA trimers arranged in a hexagonal lattice, though it has a higher degree of regularity than the immature MA lattice (Fig. S2). The arrangement of MA within the immature and mature MA lattices is, however, strikingly different (compare Figs. 2 and 3) – the MA lattice undergoes structural maturation to form a new, different hexagonal lattice in the mature virion.
Fig. 3. The mature HIV-1 matrix structure.
(A, B) Slices through reconstructions of the mature HIV-1 MA lattice in cHIV (A) and cHIV MA-SP1 (B). The boxed region in (B) is shown at a higher magnification in (D). (C, D) An isosurface view of the cryo-ET reconstruction for mature cHIV MA-SP1 MA lattice fitted with the structure of monomeric MA, cut perpendicular to the membrane (C) or viewed from the top towards the virus center (D). The two layers of density corresponding to the lipid headgroup layers are indicated by brackets. The structure of monomeric MA determined by NMR (PDBID: 2H3Q (18); colored blue to red from N to C terminus) was fitted as a rigid body into the density. Helices 1, 2 and 5 are marked. Density is observed in the PI(4,5)P2 binding site (red arrowhead), and the structure of PI(4,5)P2 as resolved bound to MA by NMR (PDBID: 2H3V (18)), is shown as a stick model. (E) As in (C), enlarged and cut to reveal density corresponding to the N-terminal residues (black arrowhead), and PI(4,5)P2 (red arrowhead). (F) As in Fig. 2G, lattice map for the mature cHIV MA-SP1 MA lattice and the underlying CA lattice.
We had previously determined the structure of the CA layer within cHIV derivatives in which either the PR cleavage site between MA and CA was blocked by mutation (HIV-1 MA-CA) or in which the cleavage site between CA and SP1 was blocked in addition (HIV-1 MA-SP1) (36–38). CA and the CA lattice remain structurally immature in MA-CA and MA-SP1 particles. We now analysed the MA layer in these datasets, and found that the MA lattice in HIV-1 MA-CA and MA-SP1 corresponds to that seen in mature particles, despite the absence of cleavage between MA and CA (Fig. 3B, Fig. S5). Cleavages downstream of SP1 are therefore sufficient for MA lattice maturation, which can occur without CA lattice maturation. It is unlikely that proteolytic cleavage between CA and NC could directly trigger MA maturation through a structural signal passed through CA without inducing any detectable structural change in CA. We therefore suggest that maturation of MA must be triggered “in trans” by another effector in the viroplasm or in the viral membrane. Further experiments will be required to elucidate the mechanism of MA maturation. Immature virions are fusion incompetent (5), and cleavages downstream of CA are sufficient to overcome this defect (39). These observations suggest that structural maturation of MA, rather than of CA, correlates with HIV-1 becoming fusogenic, potentially by allowing Env to redistribute on the virus surface (6).
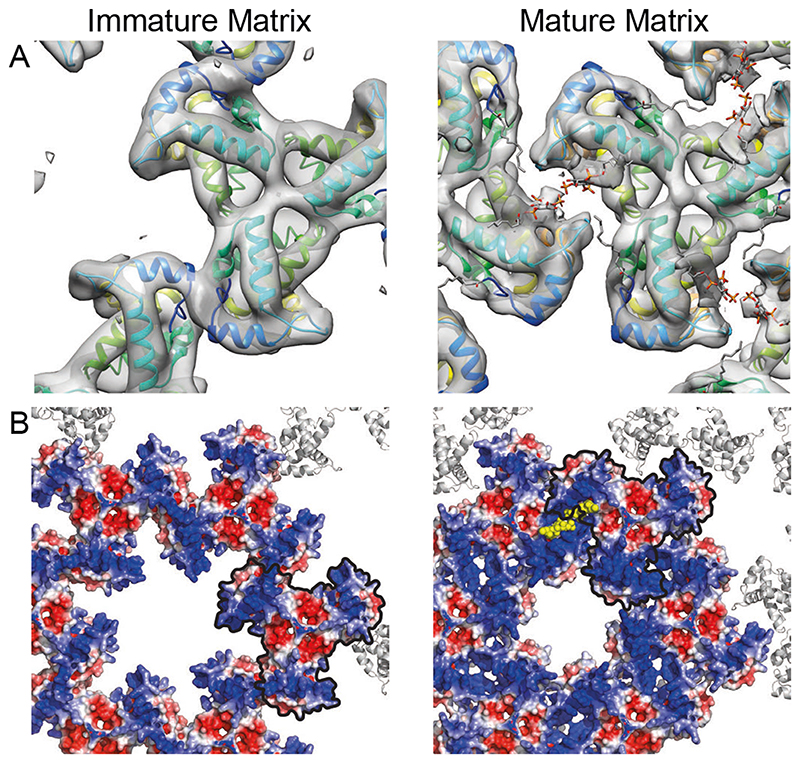
We obtained a higher-resolution, 7.0 Å structure of the mature MA lattice from MA-SP1 particles, and – as for the immature MA lattice – the crystallographic trimer could be fitted into the density as a rigid body (Fig. 3B-E, Fig. S1). Comparison of the immature and mature MA structures revealed important differences (Fig. 4). In the immature lattice the HBR faces the holes at the six-fold axes, which are therefore surrounded by a basic surface (Fig. 4B). In contrast, in the mature virus, MA presents a largely neutral surface towards the holes (Fig. 4B). This change could direct structural changes in the cytoplasmic tails of Env trimers that may alter their distribution and fusogenicity. In the mature virus, basic residues in the HBR loop face acidic residues in the N-terminus of helix 4 (E72) and the 310 helix (E51) of the adjacent MA monomer to form the dimeric interface that links trimers together (Fig. 3D, Fig. 4A, Fig. S3B). The PI(4,5)P2 binding pocket on the side of MA, adjacent to the HBR (18), is positioned right at the centre of the dimer interface. Unlike in the immature virus, where the PI(4,5)P2 binding pocket is empty, it contains a density in the mature virus. Our structure does not resolve the headgroup or acyl chain of the bound lipid, but the density is consistent with the previously-described extended-lipid conformation of bound PI(4,5)P2 in which the 2’ acyl chain is removed from the bilayer, while the 1 ` acyl chain extends upwards towards the bilayer (PDBID: 2H3Q and 2H3V (18)) (Fig. 3D, E, Fig. S6). We consider it reasonable to assign the observed density to PI(4,5)P2. The N-terminus of helix 1 prominently protrudes up into the PM, indicating that the myristate moiety predominantly retains an exposed, membrane-inserted conformation in the mature MA lattice.
Fig. 4. Comparison of the immature and mature HIV-1 MA lattices.
(A) Immature and mature MA lattices are shown with one trimer aligned. In the immature lattice, contact with neighbouring trimers is mediated by N-terminal regions and the PI(4,5)P2 binding site is empty. In the mature lattice, contact with neighbouring trimers is mediated by the region surrounding the occupied PI(4,5)P2 binding site. (B) Electrostatic surface potential maps of the hexameric lattice of immature and mature MA trimers. The red (-5 kT/e) and blue (+5 kT/e) colours represent negatively and positively charged electric potentials. PI(4,5)P2 is shown in yellow. The negatively-charged lipid headgroups are surrounded by positively-charged residues. One MA trimer aligned as in (A) is outlined in black. The potential of the surface of MA facing the holes at the hexamer positions in the lattice changes from positive to neutral/negative during maturation. The hole becomes smaller.
To biochemically determine a potential interaction of MA with PI(4,5)P2 in the immature or mature lattice, we prepared immature and mature particles from cells incorporating a functionalized PI(4,5)P2 derivative (f-PI(4,5)P2) with a diazirine group in the 1 ’ acyl chain, which can be photo-crosslinked to nearby proteins (Fig. S7A). We found that, upon photoactivation, f-PI(4,5)P2 was efficiently crosslinked to Gag within immature PR- particles, but not to MA within mature particles (Fig. S7B). This observation confirms direct interaction of the MA domain of the Gag polyprotein with PI(4,5)P2, reveals that proteolytic cleavage of Gag alters the relative positions of MA and PI(4,5)P2, and provides direct biochemical evidence for a change in MA-lipid interactions upon HIV-1 maturation.
Taken together, our results (summarized in Fig. S8) revealed that MA – similar to CA and NC – undergoes dramatic structural maturation to form very different lattices in immature and mature HIV-1 (Movie S1). Immature virions contain MA trimers packed together through their N-termini into a poorly-ordered hexagonal lattice containing basic-charged holes and disordered regions, which could accommodate the cytoplasmic tails of Env. The N-terminal myristate is predominantly membrane-inserted in the immature and mature particle. The immature MA structure provides a framework for understanding the established roles of MA in virus assembly and Env incorporation. Mature virions, in contrast, contain MA trimers packed together via their HBRs, resulting in a well-ordered hexagonal lattice with neutral holes. The maturation of MA is reminiscent of that of CA, which matures between two different hexameric protein lattices with two different functional roles (3). Upon maturation, both the protein arrangement and the lipid-binding properties of MA change. The lipid-binding pocket on the side of MA is occupied in the mature structure, containing density consistent with the previously-described extended-lipid conformation of PI(4,5)P2 (Fig. S6B) in which one acyl chain is removed from the membrane (18). The resolution of our density does not allow us to distinguish PI(4,5)P2 from other lipids, but its position is such that any lipid positioned in this density is pulled outwards from the lipid bilayer. The virion contains approximately three PI(4,5)P2 molecules per Gag (40) – we assume that the remaining PI(4,5)P2 molecules may interact with the membrane proximal surface of MA via their headgroups as in the immature virus. Our data therefore imply that maturation of Gag results in the partial removal of up to ~2,500 of the ~150,000 lipids in the inner leaflet of the viral membrane (40). The energy barrier to the removal of acyl chains from the bilayer may be overcome by binding of PI(4,5)P2 to MA and stabilization of the MA lattice.
In conclusion, our data suggest that the structural maturation of Gag not only condenses the RNP and achieves assembly of a viral capsid core surrounding the genome, but also rearranges the membrane-bound MA layer and modulates the lipid bilayer itself. How these changes alter the properties of the virus remains to be elucidated, but we speculate that they impact not only MA and Env function, but also the physical properties of the viral envelope, possibly correlating with the observed reduction in stiffness of the virus particle (41, 42). The unexpected presence of a different well-defined MA structure in mature virions suggests that MA performs new roles in the mature virion or after entry into the target cell. We speculate that PI(4,5)P2-stabilized MA lattices may remain membrane associated after virion fusion and have a post-entry function, perhaps acting as signalling platforms to prepare the target cell for early HIV-1 replication.
Supplementary Material
One Sentence Summary.
The structure of HIV-1 MA, determined in immature and mature virions, reveals maturation of MA and the lipid membrane.
Acknowledgements
We thank Wim Hagen, Dustin Morado, Aaron Tan, Florian Schur, and Simone Mattei for technical assistance, Leo James and Petr Chlanda for helpful discussions. This study made use of electron microscopes at EMBL and the MRC-LMB EM Facility, as well as high-performance computing resources set-up and maintained by EMBL IT Services and LMB Scientific Computing; we thank the staff who maintain those resources. JAGB has a secondary affiliation as director at the Max Planck Institute of Biochemistry.
Funding
Funding was provided to JAGB by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (ERC-CoG-648432 MEMBRANEFUSION), the Medical Research Council (MC_UP_1201/16) and the European Molecular Biology Laboratory; to HGK by the Deutsche Forschungsgemeinschaft (112927078 – TRR 83 and 240245660 – SFB1129 project 5); to BM by the Deutsche Forschungsgemeinschaft (MU885-1 and 240245660 – SFB1129 project 6); to CS by the NIH (R01GM127631) and the Deutsche Forschungsgemeinschaft (278001972 – TRR186 project Z1).
Footnotes
Author contributions:
KQ, BM, HGK and JAGB conceived the project. KQ, ZK, VZ, BM, HGK and JAGB designed experiments. VZ, MAÖ and BG purified virus particles. KQ and ZK performed electron microscopy. KQ and ZK performed computational image processing. RM and CS provided the f-PI(4,5)P2 derivative. FM established and VZ performed crosslinking experiments. KQ and JAGB wrote the original draft with assistance from BM and HGK which was edited and reviewed by all authors. KQ and VZ prepared figures. BM, HGK and JAGB obtained funding, provided supervision and managed the project.
Competing interests:
Authors declare no competing interests.
Data and materials availability
All reasonable requests for materials will be fulfilled under a material transfer agreement with the providing institution. Representative tomograms are deposited in the Electron Microscopy Data Bank (EMDB) under accession codes EMD-4020, EMD-13079, EMD-13085 and EMD-13086; the immature and mature lattice structures are deposited under accession codes EMD-13087 and EMD-13088. The associated molecular models are deposited in the protein data bank (PDB) under accession 7OVQ and 7OVR.
References and Notes
- 1.Sundquist WI, Kräusslich H-G. HIV-1 assembly, budding, and maturation. Cold Spring Harbor Perspectives in Medicine. 2012;2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freed EO. HIV-1 assembly, release and maturation. Nature Reviews Microbiology. 2015;13:484–496. doi: 10.1038/nrmicro3490. 2015 13:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mattei S, Schur FK, Briggs JAG. Retrovirus maturation — an extraordinary structural transformation. Current Opinion in Virology. 2016;18:27–35. doi: 10.1016/j.coviro.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Pornillos O, Ganser-Pornillos BK. Maturation of retroviruses. Current Opinion in Virology. 2019;36:47–55. doi: 10.1016/j.coviro.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murakami T, Ablan S, Freed EO, Tanaka Y. Regulation of human immunodeficiency virus type 1 Env-mediated membrane fusion by viral protease activity. Journal of Virology. 2004;78:1026–1031. doi: 10.1128/JVI.78.2.1026-1031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chojnacki J, et al. Maturation-Dependent HIV-1 Surface Protein Redistribution Revealed by Fluorescence Nanoscopy. Science. 2012;338:524–528. doi: 10.1126/science.1226359. [DOI] [PubMed] [Google Scholar]
- 7.Massiah MA, et al. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. Journal of Molecular Biology. 1994;244:198–223. doi: 10.1006/jmbi.1994.1719. [DOI] [PubMed] [Google Scholar]
- 8.Hill CP, Worthylake D, Bancroft DP, Christensen AM, Sundquist WI. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alfadhli A, Barklis RL, Barklis E. HIV-1 matrix organizes as a hexamer of trimers on membranes containing phosphatidylinositol-(4,5)-bisphosphate. Virology. 2009;387:466–472. doi: 10.1016/j.virol.2009.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tedbury PR, Freed EO. The role of matrix in HIV-1 envelope glycoprotein incorporation. Trends in Microbiology. 2014;22:372–378. doi: 10.1016/j.tim.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tedbury PR, et al. HIV-1 Matrix Trimerization-Impaired Mutants Are Rescued by Matrix Substitutions That Enhance Envelope Glycoprotein Incorporation. Journal of Virology. 2019;94 doi: 10.1128/JVI.01526-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bryant M, Ratner L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. PNAS. 1990;87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou W, Parent LJ, Wills JW, Resh MD. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. Journal of Virology. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang C, et al. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. PNAS. 2004;101:517–522. doi: 10.1073/pnas.0305665101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ono A, Ablan SD, Lockett SJ, Nagashima K, Freed EO. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. PNAS. 2004;101:14889–14894. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chukkapalli V, Hogue IB, Boyko V, Hu W-S, Ono A. Interaction between the human immunodeficiency virus type 1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphosphate is essential for efficient gag membrane binding. Journal of Virology. 2008;82:2405–2417. doi: 10.1128/JVI.01614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mücksch F, Laketa V, Müller B, Schultz C, Kräusslich H-G. Synchronized HIV assembly by tunable PIP2 changes reveals PIP2 requirement for stable Gag anchoring. eLife. 2017;6:284. doi: 10.7554/eLife.25287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saad JS, et al. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. PNAS. 2006;103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mercredi PY, et al. Structural and Molecular Determinants of Membrane Binding by the HIV-1 Matrix Protein. Journal of Molecular Biology. 2016;428:1637–1655. doi: 10.1016/j.jmb.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murphy RE, Saad JS. The Interplay between HIV-1 Gag Binding to the Plasma Membrane and Env Incorporation. Viruses. 2020;12:548. doi: 10.3390/v12050548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chukkapalli V, Oh SJ, Ono A. Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. PNAS. 2010;107:1600–1605. doi: 10.1073/pnas.0908661107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alfadhli A, et al. HIV-1 matrix protein binding to RNA. Journal of Molecular Biology. 2011;410:653–666. doi: 10.1016/j.jmb.2011.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaines CR, et al. HIV-1 Matrix Protein Interactions with tRNA: Implications for Membrane Targeting. Journal of Molecular Biology. 2018;430:2113–2127. doi: 10.1016/j.jmb.2018.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alfadhli A, Still A, Barklis E. Analysis of Human Immunodeficiency Virus Type 1 Matrix Binding to Membranes and Nucleic Acids. Journal of Virology. 2009;83:12196–12203. doi: 10.1128/JVI.01197-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lampe M, et al. Double-labelled HIV-1 particles for study of virus-cell interaction. Virology. 2007;360:92–104. doi: 10.1016/j.virol.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 26.Schur FKM, et al. An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science. 2016;353:506–508. doi: 10.1126/science.aaf9620. [DOI] [PubMed] [Google Scholar]
- 27.Saad JS, et al. Point mutations in the HIV-1 matrix protein turn off the myristyl switch. Journal of Molecular Biology. 2007;366:574–585. doi: 10.1016/j.jmb.2006.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tedbury PR, Ablan SD, Freed EO. Global rescue of defects in HIV-1 envelope glycoprotein incorporation: implications for matrix structure. PLoS Pathog. 2013;9:e1003739. doi: 10.1371/journal.ppat.1003739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chertova E, et al. Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), Is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. Journal of Virology. 2002;76:5315–5325. doi: 10.1128/JVI.76.11.5315-5325.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freed EO, Martin MA. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. Journal of Virology. 1995;69:1984–1989. doi: 10.1128/jvi.69.3.1984-1989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freed EO, Martin MA. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. Journal of Virology. 1996;70:341–351. doi: 10.1128/jvi.70.1.341-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joshi A, Ablan SD, Soheilian F, Nagashima K, Freed EO. Evidence that productive human immunodeficiency virus type 1 assembly can occur in an intracellular compartment. Journal of Virology. 2009;83:5375–5387. doi: 10.1128/JVI.00109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brandano L, Stevenson M. A Highly Conserved Residue in the C-Terminal Helix of HIV-1 Matrix Is Required for Envelope Incorporation into Virus Particles. Journal of Virology. 2012;86:2347–2359. doi: 10.1128/JVI.06047-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tedbury PR, Novikova M, Ablan SD, Freed EO. Biochemical evidence of a role for matrix trimerization in HIV-1 envelope glycoprotein incorporation. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E182–E190. doi: 10.1073/pnas.1516618113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frank GA, et al. Maturation of the HIV-1 core by a non-diffusional phase transition. Nature Communications. 2015;6:5854. doi: 10.1038/ncomms6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiegers K, et al. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. Journal of Virology. 1998;72:2846–2854. doi: 10.1128/jvi.72.4.2846-2854.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Marco A, et al. Structural Analysis of HIV-1 Maturation Using Cryo-Electron Tomography. PLoSPathog. 2010;6:e1001215–9. doi: 10.1371/journal.ppat.1001215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mattei S, et al. High-resolution structures of HIV-1 Gag cleavage mutants determine structural switch for virus maturation. Proceedings of the National Academy of Sciences of the United States of America. 2018;115:E9401–E9410. doi: 10.1073/pnas.1811237115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wyma DJ, et al. Coupling of human immunodeficiency virus type 1 fusion to virion maturation: a novel role of the gp41 cytoplasmic tail. Journal of Virology. 2004;78:3429–3435. doi: 10.1128/JVI.78.7.3429-3435.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mücksch F, et al. Quantification of phosphoinositides reveals strong enrichment of PIP2 in HIV-1 compared to producer cell membranes. Sci Rep. 2019;9:17661–13. doi: 10.1038/s41598-019-53939-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kol N, et al. A stiffness switch in human immunodeficiency virus. Biophys J. 2007;92:17771783. doi: 10.1529/biophysj.106.093914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pang H-B, et al. Virion stiffness regulates immature HIV-1 entry. Retrovirology 2006 3:1. 2013;10:4–11. doi: 10.1186/1742-4690-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ono A, Huang M, Freed EO. Characterization of human immunodeficiency virus type 1 matrix revertants: effects on virus assembly, Gag processing, and Env incorporation into virions. Journal of Virology. 1997;71:4409–4418. doi: 10.1128/jvi.71.6.4409-4418.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adachi A, et al. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. Journal of Virology. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Konvalinka J, et al. An active-site mutation in the human immunodeficiency virus type 1 proteinase (PR) causes reduced PR activity and loss of PR-mediated cytotoxicity without apparent effect on virus maturation and infectivity. Journal of Virology. 1995;69:7180–7186. doi: 10.1128/jvi.69.11.7180-7186.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mattei S, et al. Induced maturation of human immunodeficiency virus. Journal of Virology. 2014;88:13722–13731. doi: 10.1128/JVI.02271-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Welker R, Hohenberg H, Tessmer U, Huckhagel C, Krausslich HG. Biochemical and structural analysis of isolated mature cores of human immunodeficiency virus type 1. Journal of Virology. 2000;74:1168–1177. doi: 10.1128/jvi.74.3.1168-1177.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. Journal of Structural Biology. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 49.Hagen WJH, Wan W, Briggs JAG. Implementation of a cryo-electron tomography tilt-scheme optimized for high resolution subtomogram averaging. Journal of Structural Biology. 2017;197:191–198. doi: 10.1016/j.jsb.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kremer JR, Mastronarde DN, McIntosh JR. Computer Visualization of Three Dimensional Image Data Using IMOD. Journal of Structural Biology. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 51.Grant T, Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. eLife. 2015;4:e01963. doi: 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. Journal of Structural Biology. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiong Q, Morphew MK, Schwartz CL, Hoenger AH, Mastronarde DN. CTF determination and correction for low dose tomographic tilt series. Journal of Structural Biology. 2009;168:378–387. doi: 10.1016/j.jsb.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turonova B, Schur FKM, Wan W, Briggs JAG. Efficient 3D-CTF correction for cryo-electron tomography using NovaCTF improves subtomogram averaging resolution to 3.4A. Journal of Structural Biology. 2017;199:187–195. doi: 10.1016/j.jsb.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nickell S, et al. TOM software toolbox: acquisition and analysis for electron tomography. Journal of Structural Biology. 2005;149:227–234. doi: 10.1016/j.jsb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 56.Förster F, Medalia O, Baumeister W, Fass D. Retrovirus envelope protein complex structure in situ studied by cryo-electron tomography. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:4729–4734. doi: 10.1073/pnas.0409178102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 58.Zivanov J, et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018;7:163. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Berg S, et al. ilastik: interactive machine learning for (bio)image analysis. Nature Methods 2013 10:6. 2019;16:1226–1232. doi: 10.1038/s41592-019-0582-9. [DOI] [PubMed] [Google Scholar]
- 60.Heumann JM, Hoenger A, Mastronarde DN. Clustering and variance maps for cryoelectron tomography using wedge-masked differences. Journal of Structural Biology. 2011;175:288–299. doi: 10.1016/j.jsb.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tan A, Pak AJ, Morado DR, Voth GA, Briggs JAG. Immature HIV-1 assembles from Gag dimers leaving partial hexamers at lattice edges as potential substrates for proteolytic maturation. Proceedings of the National Academy of Sciences of the United States of America. 2021;118 doi: 10.1073/pnas.2020054118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qu K, et al. Structure and architecture of immature and mature murine leukemia virus capsids. Proceedings of the National Academy of Sciences of the United States of America. 2018;115:E11751–E11760. doi: 10.1073/pnas.1811580115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jurrus E, et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018;27:112–128. doi: 10.1002/pro.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vlach J, Saad JS. Trio engagement via plasma membrane phospholipids and the myristoyl moiety governs HIV-1 matrix binding to bilayers. PNAS. 2013;110:3525–3530. doi: 10.1073/pnas.1216655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heller H, Schaefer M, Schulten K. Molecular dynamics simulation of a bilayer of 200 lipids in the gel and in the liquid crystal phases. J Phys Chem. 1993;97:8343–8360. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All reasonable requests for materials will be fulfilled under a material transfer agreement with the providing institution. Representative tomograms are deposited in the Electron Microscopy Data Bank (EMDB) under accession codes EMD-4020, EMD-13079, EMD-13085 and EMD-13086; the immature and mature lattice structures are deposited under accession codes EMD-13087 and EMD-13088. The associated molecular models are deposited in the protein data bank (PDB) under accession 7OVQ and 7OVR.