Abstract
Objective
Insulin gene (INS) mutations have recently been described as a cause of permanent neonatal diabetes. We aimed to determine the prevalence, genetics and clinical phenotype of INS mutations in large cohorts of patients with neonatal diabetes and permanent diabetes diagnosed in infancy, childhood or adulthood.
Research Design And Methods
The INS gene was sequenced in 285 patients with diabetes diagnosed before 2 years, 296 probands with MODY and 463 patients with young onset T2D (non-obese, diagnosed <45 years). None had a molecular genetic diagnosis of monogenic diabetes.
Results
We identified heterozygous INS mutations in 33/141 probands diagnosed <6 months, 2/86 between 6-12 months and 0/58 between 12-24 months. Three known mutations (A24D, F48C and R89C) account for 46% of cases. There were six novel mutations: H29D, L35P, G84R, C96S, S101C and Y103C. INS mutation carriers were all insulin treated from diagnosis and were diagnosed later than KATP mutation carriers (11 vs 8 weeks, P<0.01). In 279 patients with permanent neonatal diabetes, the frequency of KCNJ11, ABCC8 and INS gene mutations was 31%% 10% and 12%, respectively. A heterozygous R6C mutation co-segregated with diabetes in a MODY family and is probably pathogenic, but the L68M substitution identified in a patient with young onset T2D may be a rare non-functional variant.
Conclusions
We conclude that INS mutations are the second most common cause of permanent neonatal diabetes and a rare cause of MODY. Insulin gene mutation screening is recommended for all diabetic patients diagnosed before one year.
An estimated 1-2% of all diabetes is due to a monogenic etiology. HLA studies show that patients diagnosed with diabetes in the first 6 months of life are very likely to have monogenic neonatal diabetes rather than type 1 diabetes (1; 2). Neonatal diabetes is a rare disorder with an incidence of between 1 in 215,000-500 000 live births (3; 4) with approximately 50% having permanent neonatal diabetes (PNDM). Heterozygous activating mutations in the KCNJ11 and ABCC8 genes which encode the Kir6.2 and SUR1 subunits of the ATP-sensitive potassium (KATP) channel are the commonest causes of PNDM (5–9). A number of other rare genetic aetiologies have been identified (GCK, IPF1, PTF1A, GLIS3, FOXP3, EIF2AK3, GLUT2 and HNF1B). Most show autosomal recessive inheritance and all except for GCK mutations result in additional non-pancreatic features.
We have recently described a new monogenic cause of neonatal diabetes; mutations in the insulin (INS) gene (10). We reported sixteen families with ten different heterozygous missense INS mutations in probands with PNDM. These mutations are predicted to disrupt the folding of the proinsulin molecule and result in misfolded protein or retention of the protein in the endoplasmic reticulum (ER), resulting in ER stress and beta cell apoptosis (11). Disulphide bonds are crucial for proinsulin folding in the ER and sixty percent (6/10) of the mutations either abolish or disrupt disulfide bridge formation within the protein either by substitution of a cysteine residue (eg. C96Y) or creation of an additional cysteine (eg: R89C).
The most common forms of monogenic diabetes are subtypes of maturity-onset diabetes of the young (MODY) due to heterozygous mutations in the transcription factor genes HNF1A/TCF1 or HNF4A, or the GCK gene that encodes the pancreatic glucose sensor, glucokinase (12). HNF1A and HNF4A mutations result in progressive beta-cell dysfunction and diabetes is usually diagnosed before 25 years. GCK mutations cause elevated fasting hyperglycemia from birth, but are often not diagnosed until young adulthood (13). Rare mutations in other transcription factor genes (HNF1B/TCF2, IPF1, NEUROD1) have been reported in MODY but, at least 11% of UK families who fit the clinical criteria for MODY do not have a mutation in any of the known genes (14). The initial report of INS mutations includes the father of a child with neonatal diabetes who was diagnosed with type 2 diabetes at the age of 30 years (10). It is not known if insulin gene mutations can cause MODY.
We now report mutation screening of a large series of patients (n=1044) with permanent diabetes diagnosed during infancy, childhood and adulthood (up to 45 years) in order to determine the prevalence of INS gene mutations. We also studied a series of patients with hyperinsulinism (n=49) where mutations in the KATP channel genes had been excluded by sequencing. This group was investigated to determine whether a similar situation exists for INS as that seen for GCK (15), KCNJ11 and ABCC8 where different mutations result in the opposite phenotypes of diabetes and hyperinsulinism.
Research Design And Methods
Study population
We studied 5 groups of patients with permanent diabetes at the time of referral (Table 1). The permanent neonatal diabetes (PNDM) group consisted of 141 patients with diabetes diagnosed in the first 6 months of life who were on insulin treatment at the time of referral. Mutations in KCNJ11 had been excluded by sequencing in all 141 patients and ABCC8 mutations had been excluded in the 129 cases where there was sufficient DNA. Patients diagnosed outside the first six months were divided into those diagnosed in infancy (between 27 and 52 weeks of life, n=86), and those diagnosed in early childhood (between 53-104 weeks of life n=58). We also screened 296 cases who met minimal diagnostic criteria for MODY (at least two generations affected and at least one subject diagnosed before 25 years of age). Mutations in the HNF1A, HNF4A and GCK genes had been excluded by sequencing in 189, 81 and 56 cases, respectively. Finally we screened 463 non-obese patients with young T2D diagnosed before 45 years of age. Of these, 45% had a diabetic first-degree relative. Mutations in HNF1A, HNF4A and GCK were excluded by sequencing in 180, 76 and 87 cases, respectively.
Table 1. Clinical characteristics of the study groups.
Insulin gene (INS) mutations were investigated in 1044 patients with permanent diabetes diagnosed before 6 months (PNDM, 0-26 weeks), between 6-12 months (infancy, 27-52 weeks), 1-2 years (early childhood, 53-104 weeks), MODY diagnosed <25 years or YT2D diagnosed <45 years, and in 49 patients with hyperinsulinism. The data is shown as the median with the range in parentheses. Data sets for BMI were incomplete (indicated by an asterisk). NA= not available.
| Clinical characteristic |
PNDM | Infancy | Early childhood |
MODY | YT2D | Hyperinsulinism |
|---|---|---|---|---|---|---|
|
Diabetes (Age-at-
diagnosis) |
0-26 weeks | 27-52weeks | 53-104weeks | <25 years | <45 years | |
| n | 141 | 86 | 58 | 296 | 463 | 49 |
| Sex (% Male) | 50% | 58% | 41% | 36% | 48% | 67% |
| Current age (yrs) | 7 (0-69) | 10 (1-51) | 13 (2- 72) | 36 (5-77) | 38 (3-87) | 3 (0.4-21) |
| Age-at-diagnosis | 8 weeks (0-26) |
39 weeks (27-52) |
97 weeks (57-104) |
16 years (3-25) |
29 years (3-46) |
7 weeks (0-728) |
| BMI (kg/m2) | 17 (9-40)* | 19 (13-30)* | 22 (17-25)* | 24 (13-30)* | 23 (15-30) | NA |
|
Number of
patients with an affected first- degree relative |
24 (17%) | 9 (10%) | 11 (19%) | 296 (100%) | 208 (45%) | – |
|
Treatment: INS/
OHA+INS/ OHA/ Diet (% cases) |
100/0/0/0 | 100/0/0/0 | 100/0/0/0 | 49/4/26/21 | 45/2/32/21 | – |
|
KCNJ11/ ABCC8
mutations excluded |
141 / 129 | 86 / 21 | 58 / 2 | 0 / 0 | 0/0 | 49 / 49 |
|
HNF1A / HNF4A /
GCK mutations excluded |
– | – | – | 189/81/56 | 180/76/87 | – |
Patients diagnosed between 0-2 years (n=285) were predominantly recruited in Exeter through the International Society of Paediatric and Adolescent Diabetes (ISPAD) group. These were supplemented by 39 patients from the Barts and Oxford study (BOX) and the British Diabetes Association (BDA) 1972-1981 cohort, as previously described (2). In addition we also screened 49 patients with isolated hyperinsulinism, diagnosed at a median age of 7 weeks (0-728 weeks), where mutations in the KATP channel genes (KCNJ11 or ABCC8) had been excluded by sequence analysis.
Clinical characteristics are presented as median (range) and comparative analysis using a Kruskal Wallis or Mann-Whitney U test. The summary clinical details of these groups are shown in Table 1.
Mutations were tested for co-segregation with diabetes in family members and the INS gene was sequenced in 111 UK Caucasian population controls. Clinical characteristics were obtained from hospital notes with assistance from the referring clinician.
Molecular genetic analysis
Genomic DNA was extracted from peripheral leukocytes using standard procedures. Coding exons 2 and 3 of the INS gene were amplified by the polymerase chain reaction (PCR). Sequence specific primers for each exon (10), were tagged with 5’ M13 tails to allow sequencing to be performed with a “universal” M13 primer. A new primer for the first coding exon was used; INS_Exon2F-TGTAAAACGACGGCCAGTTGGCTGGGCTCGTGAAG and INS_Exon2R CAGGAACACGCTATGACCCCCTTCTGCCCATGCTG.
Single strand sequencing was carried out using standard methods on an ABI 3730 (Applied Biosystems, Warrington, UK). Sequences were compared to the published sequence (NM_000207) using Mutation Surveyor v2.61. Changes in the sequence were checked against published polymorphisms and mutations and for conservation across species. We used a panel of microsatellites for chromosome 20q (16) or chromosome 11p (17) to confirm family relationships for sporadic cases of diabetes where neither parent was shown to carry the mutation found in their child.
Results
Prevalence of mutations in the different patient groups tested
Heterozygous INS mutations were found in 33/141 (23%) probands diagnosed before 6 months, 2/86 (2%) between 6 -12 months and 0/58 (0%) cases diagnosed between 12-24 months. Only 1/296 (0.3%) MODY and 1/463 (0.2%) young T2D probands had possible mutations identified by sequencing. No mutations were found in the 49 patients with hyperinsulinism. The details of the mutations found and the associated clinical characteristics are discussed below.
Molecular genetics
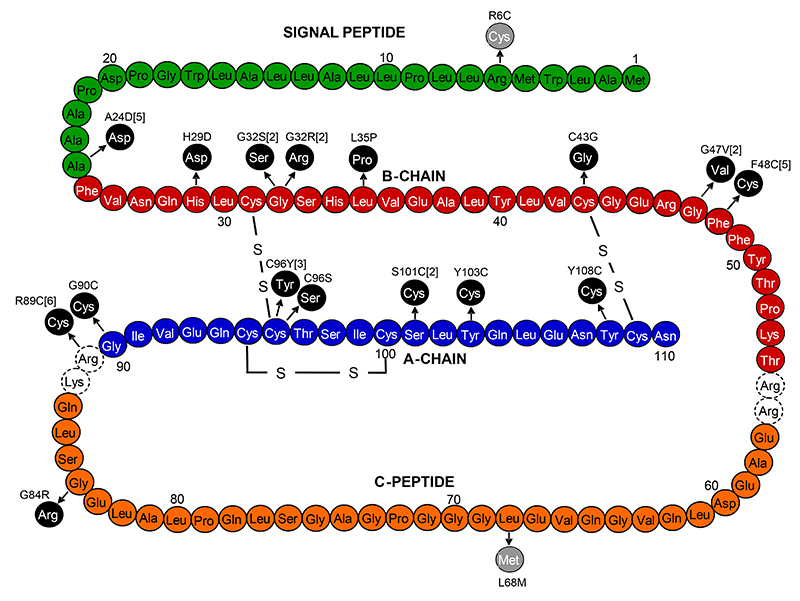
We identified 16 different heterozygous mutations in 35 probands with diabetes diagnosed in the first year of life (see Figure 1). Twelve of the 35 families have been reported previously (10). Ten of the 16 mutations found have previously been described (10): A24D (5 families), G32S(2 families), G32R (2 families), C43G, G47V(2 families), F48C(5 families), R89C (6 families), G90C, C96Y(3 families), Y108C. There are six novel mutations; H29D (c.85C>G, p.His29Asp), L35P (c.104T>C, p.Leu35Pro), G84R (c.250G>A, p.Gly84Arg), C96S (c.287G>C, p.Cys96Ser), S101C (c.302C>G, p.Ser101Cys in two families) and Y103C (c.308A>G, p.Tyr103Cys). These residues are conserved across species (human, mouse, dog, horse, lizard and Xenopus), with the exception of Y103 (F103 in Xenopus), and G84 (R84 in Platypus). These mutations were not seen in 222 UK Caucasian control chromosomes.
FIG. 1. Mutations identified in the preproinsulin molecule.
Black circles represent amino acid changes identified in probands with diabetes diagnosed before 12 months, grey circles represent amino acid changes identified in probands with possible MODY. Where the number of probands with the mutation is greater than one, the total number is indicated in the square brackets.
Three mutations (A24D, F48C and R89C) account for 16/35 (46%) of cases, but only R89C occurs at a CpG dinucleotide where the spontaneous deamination of methylcytosine is frequent. Nine of the 16 mutations either abolish a cysteine residue or create an additional cysteine.
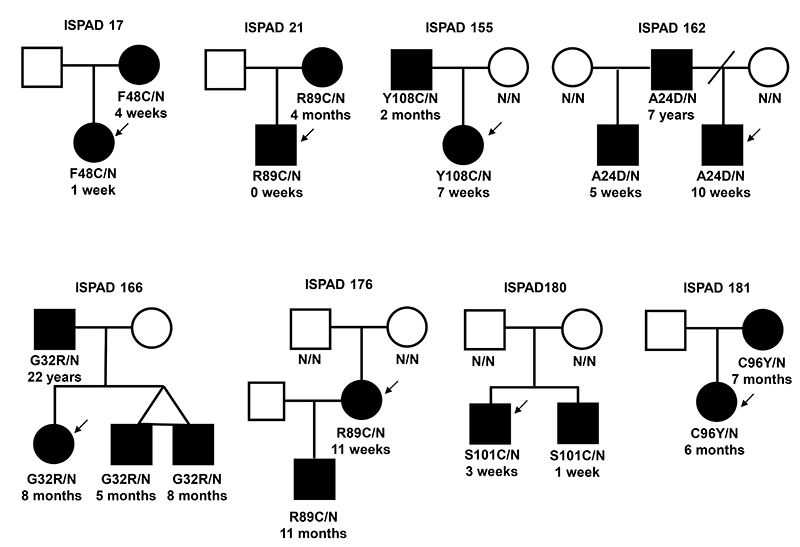
Six families demonstrated autosomal dominant inheritance of diabetes that cosegregated with the INS mutation, including one previously published family ISPAD 155 (10) (Figure 2). An additional 11 family members were shown to have an INS mutation. Six were diagnosed before 6 months, three between 6 and 12 months, one at 7 years and one at 22 years. In 29/35 (83%) of probands an affected child was born to unaffected parents representing probable de novo mutations. In 17 families DNA was available from both unaffected parents and microsatellite analysis established that these were de novo mutations (including eleven previously published families (10)). In family ISPAD180 two affected brothers had the S101C mutation, but the mutation was not detected in leukocyte DNA from either parent (Figure 2). This pattern of inheritance is consistent with germline mosaicism. The clinical features of this family have previously been described (18).
FIG. 2. Partial pedigrees for families with multiple individuals affected with diabetes.
Filled symbols represent patients with diabetes. The genotype is shown underneath each symbol, N/N denotes no mutation identified. Below the genotype is the age of diagnosis of diabetes. An arrow indicates the proband.
INS mutations - Clinical characteristics
Clinical characteristics are provided for all INS mutation carrying probands diagnosed under 1 year and their affected family members (Table 2). The median age at diagnosis of the INS mutation carriers was 11 weeks. Presentation was either with symptomatic hyperglycemia (41%) or diabetic ketoacidosis (59%). All patients were treated with insulin replacement therapy, and most with a full replacement dose (>0.5U/kg/day) (median 0.7 [range 0.3-1.9] U/kg/day). Glycemic control was variable (HbA1c median 7.95% [range 4.6-13.8%]). Autoantibodies, when measured, were not detected in any cases although the two affected siblings from family ISPAD 180 were previously reported with autoantibodies to the exocrine pancreas but not the islet at diagnosis (18).
Table 2. Clinical characteristics of probands diagnosed in infancy and their family members with an INS gene mutation.
The data are shown as the median with range in parentheses. NA= not available.
| Characteristic | Age-at-diagnosis | All subjects | ||
|---|---|---|---|---|
| 0-26 weeks | 27-52 weeks | 1 -45 years | ||
| n | 39 | 5 | 2 | 46 |
| Sex (% Male) | 19/39 (49%) | 2/5 (40%) | 2 (100%) | 23/46 (50%) |
|
Current age
(years) |
10 (0.5-42) | 16 (1-45) | 42 (35-48) | 14 (0.5-48) |
|
Current BMI
(kg/m2) |
17 (11-36) | 18 (16-30) | NA | 17 (11-36) |
| Birth weight (kg) | 2.6 (1.7-3.8) | 3 (2.9-3.9) | 2.7 (2.4-2.9) | 2.7 (1.7-3.9) |
|
Gestational age
(weeks) |
40 (35-42) | 40 (37-42) | 40 | 40 (35-42) |
|
Corrected birth
weight (Centile) |
3 (<1st-87) | 44 (5-83) | 4.5 (<1-8) | 6 (<1st -87) |
| Age-at-diagnosis | 9 weeks (0-26) | 35 weeks (31 -48) | 14.5 years (7-22) | 11 weeks (0-1144) |
| Insulin treatment | 100% | 100% | 100% | 100% |
|
Insulin dose
(U/kg/day) |
0.73 (0.3-1.9) | 0.6 (0.2 -0.7) | 0.45 (0.3-0.6) | 0.7 (0.3-1.9) |
| HbA1c | 7.9 (4.6-13.8) | 7.6 (6.5-9.5) | NA | 7.95 (4.6-13.8) |
|
Antibody status:
Neg/Pos/NA |
18/0/21 | 2/0/3 | 0/0/2 | 19/1/26 |
Birth weights were reduced, consistent with in utero growth retardation due to reduced insulin secretion. The median birth weight was 2.7 kg (range 1.7-3.9 kg), corresponding to the 6th percentile (range <1st -87th). There was no difference in the age at diagnosis for male INS mutation carriers compared to females (median 10 weeks [range 0-1144] for males vs 13 weeks [range 1-35] for females; P=0.9) but birth weight was lower for males (median centile birth weight <1 [range <1-87] for males vs 11th centile [range <1-83] for females; P=0.048).
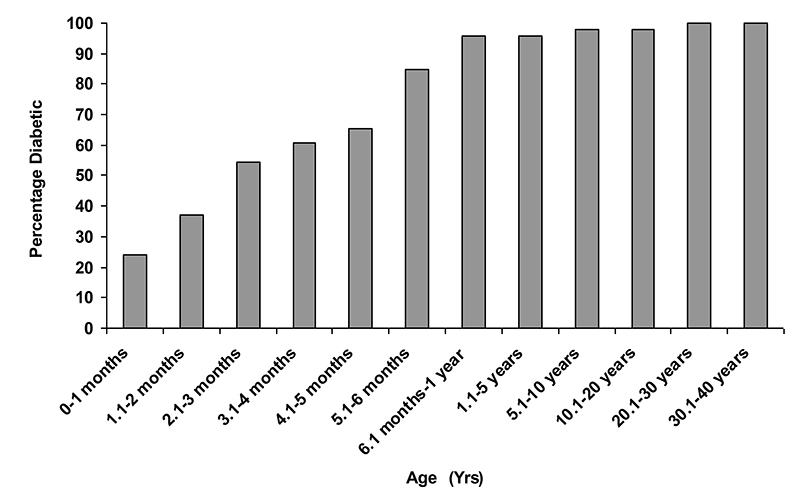
Figure 3 illustrates the penetrance of INS mutations in families where the proband was diagnosed in infancy. The percentage of patients with diabetes increases from 24% in the first month, to 85% at 6 months and 96% at one year.
FIG. 3. Penetrance of INS gene mutations in 46 individuals from 35 families where the proband was diagnosed in the first year of life.
The percentage of patients with an INS mutation and diabetes at any given age is shown.
Comparison of INS and KATP channel mutations
The clinical characteristics of patients with an INS, KCNJ11 or ABCC8 gene mutation were compared (Table 3). No difference in sex, birth weight or gestational age was observed between the three groups but patients with an INS mutation were diagnosed later (median 11 weeks [range 0-1144]) than those with a KATP channel mutation (median 8 weeks [range 0-40], P<0.01).
Table 3. Comparison of the clinical characteristics of INS, KCNJ11 and ABCC8 mutation carriers.
The data are shown as the median with range in parentheses.
| Characteristic | INS | KCNJ11 | ABCC8 | P |
|---|---|---|---|---|
| n | 46 | 100 | 31 | |
| Sex (% Male) | 23/46 (50) | 53/100 (53) | 13/31(42) | 0.6 |
|
Birth weight
(kg) |
2.7 (1.7-3.9) | 2.66 (1.85-3.6) | 2.7 (1.51-4.20) | 0.8 |
|
Gestational
age |
40 (35-42) | 40 (33-42) | 40 (26-40) | 0.2 |
|
Corrected
birth weight (Centile) |
6 (<1st -87) | 7 (<1-91) | 13 (<1-95) | 0.2 |
|
Age-at-
diagnosis (weeks) |
11 (0-1144) | 8 (0-33) | 8 (0-40) | 0.04 |
Frequency of INS mutations in PNDM
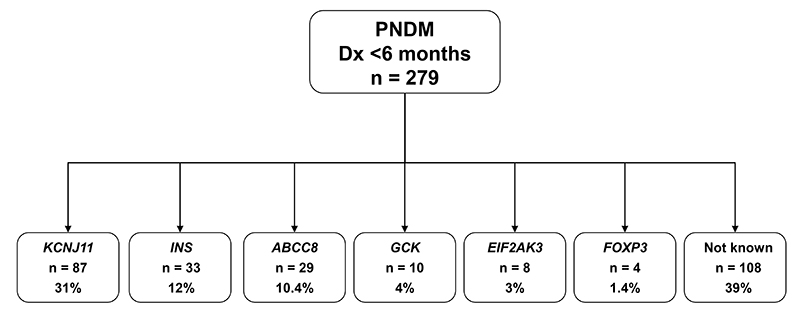
The 33 INS mutation carriers diagnosed before 6 months of age are from a cohort of patients with permanent neonatal diabetes referred to Exeter. This cohort includes 87 patients with KCNJ11 mutations, 29 with ABCC8 mutations and 22 with mutations in the GCK, EIF2AK3 or FOXP3 genes (10, 8 and 4 cases respectively). There are also 108 subjects in whom no mutations were found after sequencing the KCNJ11, ABCC8 and INS genes. We were unable to complete sequencing of these three genes in 21 cases due to insufficient DNA. The prevalence of the different genetic subtypes within the 279 fully tested cohort members is shown in figure 4. Sequence analysis for GCK was performed only for consanguineous pedigrees and EIF2AK3/FOXP3 mutation testing was guided by extra-pancreatic features and/or consanguinity, thus these etiologies may be underestimated.
FIG. 4. Etiology of permanent neonatal diabetes in a cohort of 279 patients diagnosed before 6 months.
The number of probands with mutations identified is shown, together with the percentages for each etiology. All subjects of unknown etiology were sequenced for KCNJ11, ABCC8 and INS.
INS mutations in MODY and young T2D
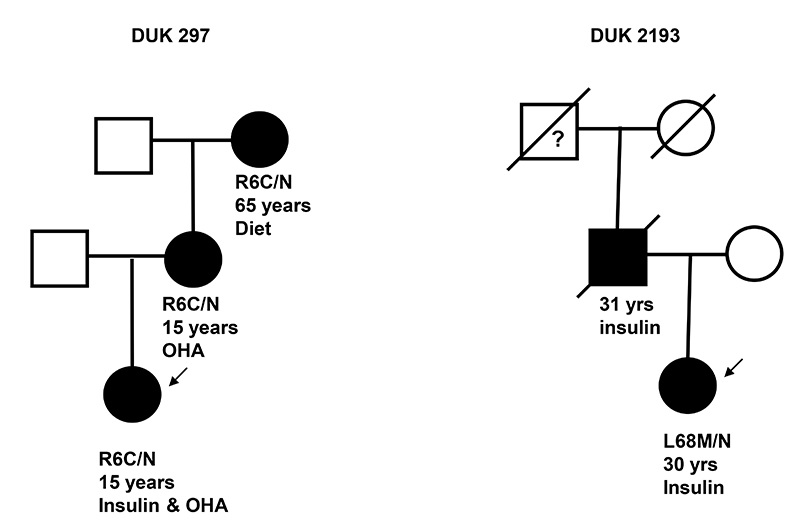
Two different missense mutations were identified in two families; one in a proband with MODY (1/296) and one in a proband with young T2D (1/463). The novel mutation, R6C (c.16C>T, p.Arg6Cys), was found in a UK MODY family and was not present in 222 UK Caucasian control chromosomes. The novel mutation L68M (c.202C>A, p.Leu68Met) was identified in a family of Turkish origin with young T2D and was not seen in 170 Turkish control chromosomes.
The R6C mutation co-segregated with diabetes as it was found in the proband, the proband’s mother and maternal grandmother who were diagnosed with diabetes at 15, 15 and 65 years, respectively. The clinical features in the three affected family members were consistent with a diagnosis of MODY: they were non-obese (BMI 24.1, 26.9 and 29.3 kg/m2) and their diabetes was non-insulin dependent. The proband had 10 years on diet and then oral agents before starting low dose insulin (0.2U/kg/day), her mother was treated with diet for 40 years before starting oral agents at the age of 55 and the maternal grandmother has been on diet treatment for the 8 years since diagnosis. The L68M mutation was found in a proband diagnosed with diabetes at the age of 30 years (BMI 17kg/m2). She was treated with diet for one year, started oral agents but soon required insulin treatment. Her father was diagnosed aged 31 years and was insulin treated. He died from diabetic complications and DNA from her unaffected mother was not available (Figure 5). Mutations in the GCK, HNF1A or 4A genes were excluded in both probands by sequence and dosage analysis.
FIG. 5. Partial pedigrees for novel INS mutations identified in probands with MODY and young T2D.
Filled symbols represent patients with diabetes. The genotype is shown underneath each symbol, N/N denotes no mutation identified. Below the genotype is the age at diagnosis of diabetes and current treatment. An arrow indicates the proband.
The R6 residue is conserved across mammalian species (Rhesus monkey, mouse, dog and horse) and Xenopus, but not conserved in lizard (S6) or platypus (G6). The L68 residue is also conserved in mammals (Rhesus monkey, mouse, dog and horse) but absent from Xenopus, lizard and platypus. The SIFT analysis software (19) predicts that L68M is tolerated but R6C is not. PolyPhen (20) predicts that the L68M variant is benign. Whilst leucine and methionine are both small, hydrophobic amino acids, the R6C substitution creates an additional cysteine residue and the likelihood of disrupted disulfide bridge formation.
In addition to these non-synonymous mutations, we also detected the common intronic SNPs rs689 and rs3842752, a synonymous A12A variant (rs3842744) in 1/759 MODY cases, and a synonymous P21P variant (rs11564720) in 1/759 MODY cases and 1/222 Caucasian controls.
Discussion
We report the largest series of INS gene mutations identified by screening 1044 patients diagnosed with diabetes from birth to 45 years of age. Heterozygous missense mutations were found in 37 probands of whom 35 were diagnosed during infancy (the first 12 months of life) and 33 had permanent neonatal diabetes diagnosed before 6 months of age. INS mutations were common in patients diagnosed with diabetes in the first 6 months of life (23% of cases tested), but far less frequent in patients diagnosed in childhood or early adulthood (two novel mutations; 0.3% of cases). We have also excluded INS mutations as a common cause of congenital hyperinsulinism.
The majority of patients with an INS gene mutation are sporadic cases that result from de novo mutations. Dominant inheritance was observed in 7/35 (20%) families. A similar proportion of KCNJ11 mutations are spontaneous (21), hence permanent neonatal diabetes is primarily a sporadic form of diabetes.
We identified 16 different INS mutations in the 35 patients with permanent diabetes diagnosed in infancy (the first 12 months of life). Ten of these mutations have previously been reported (10), including the three most common mutations A24D, F48C and R89C that account for 16/35 (46%) of cases. Most mutations are located within the A or B chains of insulin and 9 are predicted to interfere with the formation of disulphide bridges between cysteine residues either by replacing a cysteine residue, or introducing an additional cysteine residue. In vitro experimental studies like those described by Liu et al (22) are required to explore the effect of such mutations upon insulin processing and secretion.
Novel INS mutations diagnosed in PNDM
Six novel missense mutations (H29D, L35P, G84R, C96S, S101C, and Y103C) were identified in seven families. Three of these mutations involve cysteine residues within the insulin A chain and are therefore predicted to affect the normal folding of the proinsulin molecule. S101C was identified in two families; in ISPAD 180 the finding that both children are heterozygous for the mutation but neither parent is a carrier suggests that the mutation has arisen de novo in one parent who must be a germline mosaic (see Figure 2). In the second family the mutation was not present in the unaffected father but DNA was not available from the mother.
The novel C96S mutation occurs at the site of a previously reported mutation (C96Y) identified in a patient with PNDM (10) and the Akita mouse model (11). The same amino acid substitution (Cys>Ser) is present at the adjacent residue (C95S) in the Munich mouse model (23). Studies of these mice indicate that mutant proinsulin is trapped and accumulated in the endoplasmic reticulum, leading to induction of the endoplasmic reticulum stress response, inhibition of protein synthesis and ultimately beta-cell death (11; 24–26).
The other three novel mutations, H29D (residue B5), L35P (B11) and G84R, do not involve cysteine residues. The H29D mutation had arisen de novo and affects a residue containing an imidazole ring which packs closely to the C96-C31 inter-disulfide bridge between the A and B chains (27). This mutation has been shown to disrupt the folding and conformation of proinsulin in vitro, decreasing the thermodynamic stability of the protein in the ER (28).
The L35P mutation affects a residue located in the B chain of the insulin molecule. The B chain contains two beta turns at C31-H34 and G44-G47. These turns are highly conserved and guide the folding of the protein to form the inter-disulfide bond with the A chain (29). Residue L35 is highly conserved across all insulins and IGFs, and is located adjacent to beta turn 1. Due to the importance of this region in forming the C96-C31 inter-disulfide bond, we predict that a substitution of this leucine by proline would introduce prohibited steric contacts with C96, resulting in impaired disulfide bond formation.
The pathogenicity of the mutation G84R is less certain. Neither parent is affected with diabetes but DNA samples were not available for testing. G84 is located within the C-peptide, a 31 amino acid linker between the A and B chains of proinsulin that is not highly conserved throughout evolution. The G84 residue is conserved from human to Xenopus, but an arginine (R) residue is present at this position in platypus. The patient with the G84R mutation is Vietnamese and the possibility that this is a rare Asian polymorphism of no clinical significance cannot be excluded. In vitro studies have shown that the proinsulin molecule requires at least 5 linker amino acids (acting as the C peptide) to produce the correct orientation of the molecule for the critical inter-disulfide bond formation between the A and B chains (30) but it is not known which are the crucial amino acids within the C peptide. Functional studies may help to determine the significance of this variant on insulin biosynthesis in vivo.
Novel INS mutations in MODY and young onset T2D
Two novel mutations were identified within the cohort of patients with MODY (R6C) and young onset T2D (L68M). R6C affects a residue within the signal peptide whilst L68 is located within the C-peptide which shows a lower level of evolutionary conservation. The sum of the genetic, evolutionary and structural evidence suggests that while R6C is probably a pathogenic MODY mutation, but L68M is probably a rare, non-functional variant. Functional studies will be required to investigate these mutations further.
Clinical characteristics of patients with INS mutations
All mutation carriers identified through the infancy-onset cohort presented with symptomatic hyperglycemia or diabetic ketoacidosis and were treated with insulin. The majority (85%) of patients with an INS mutation were diagnosed before 6 months of age, 96% had diabetes at one year, but two family members were diagnosed at 7 and 22 years (Figure 3). In contrast none of the three family members with the R6C mutation nor the proband heterozygous for L68M were initially treated with insulin and their age at diagnosis ranged from 15 to 65 years.
Our initial study showed that the age at onset of diabetes can vary within families (10), and in this larger series it has become clear that variable penetrance is common not only within families but also between families with the same mutation. For example, the four affected individuals from family ISPAD 166 with the G32R mutation were diagnosed between 5 months and 22 years, whereas the proband from another family was diagnosed at 4 weeks. The age at diagnosis for A24D mutations carriers ranged from 4 weeks to 7 years. These findings suggest that other genetic or environmental factors may affect penetrance.
Both the mouse models show a more severe phenotype in male than in female mice with more severe diabetic symptoms in the male Akita mice (11) and lower birth weight in the Munich mice (23). Whilst we saw no difference in the age at diagnosis of diabetes, insulin dose or glycemic control, birth weight was lower for male INS mutation carriers compared to females suggesting a more severe insulin secretory defect in utero. The reasons underlying this sex-specific phenotypic variability are not known, either in man or mouse.
No difference in birth weight was observed between patients with INS mutations and those with activating KCNJ11 or ABCC8 mutations. Although patients with INS gene mutations are diagnosed later (median 11 weeks vs 8 weeks, P<0.01), the ranges overlap and hence patients diagnosed within the first 6 months with permanent diabetes require molecular genetic testing to confirm the genetic subtype. We suggest that newly diagnosed patients with neonatal diabetes (aged <6 months) should be referred for testing of the most common forms of TNDM and PNDM to determine the genetic aetiology. This would include analysis for chromosome 6q24 abnormalities as well as KCNJ11 and INS mutations, followed by ABCC8 if these tests are negative.
Prevalence of INS mutations
INS mutations accounted for 23% (33/141) of neonatal diabetes diagnosed with diabetes before 6 months of life when patients with known aetiological mutations were excluded. These patients were selected from the Exeter collection of permanent diabetes diagnosed before 6 months of age. In this series, mutations in the KCNJ11, ABCC8 and INS genes are the cause in 31%, 10% and 12% of cases respectively, while mutations in GCK, EIF2AK3 or FOXP3 genes account for a further 8% of cases. Thus, mutations in known genes account for ~60% of all cases indicating that additional genes for this form of diabetes remain to be identified.
Both KCNJ11 and INS mutations are a rare cause of permanent diabetes diagnosed between 6 and 12 months but INS gene mutations may be more frequent in this group (5 INS mutation carriers identified vs 3 KCNJ11 mutations in our patient cohort).
In summary, we report the largest series of INS mutation carriers. These mutations are the second most common cause of permanent neonatal diabetes but a rare cause of diabetes in childhood or adulthood, including MODY. We recommend that the INS gene should be screened for mutations in all children diagnosed with diabetes in the first year of life.
Acknowledgements
We thank Jon Locke, Piers Fulton, Noaman Hasan and Mindy Kendrick for their technical assistance. Kathleen Gillespie, Paul Lambert, Neil Raymond, Peter Swift, Polly Bingley and Edwin Gale collected the DNA samples and clinical data for the Bart’s Oxford (BOX) study and the BDA 1972-1981 cohort, funded by Diabetes UK and the Wellcome Trust. Mark Strachan and Jill Little assisted with recruitment of the MODY cohort. This work was funded by the European Union (Integrated Project EURODIA LSHM-CT-2006-518153 in the Framework Programme 6 of the European-Community), the Wellcome Trust and funding from the Sir Graham Wilkins studentship to Sarah Flanagan. ATH is a Wellcome Trust Research Leave Fellow. Funding for the work carried out in Chicago was provided by U.S. Public Health Service Grants DK-13914, DK-20595, DK-44752, DK-73541, and DK-77489 and a gift from the Kovler Family Foundation.
Abbreviations
- DUK
Diabetes UK (formerly BDA, British Diabetes Association)
- ER
Endoplasmic Reticulum
- ISPAD
International Society of Paediatric and Adolescent Diabetes
- MODY
Maturity-Onset Diabetes of the Young
- PCR
Polymerase Chain Reaction.
Footnotes
The authors declare no conflict of interest.
Contributor Information
Other members of the Neonatal Diabetes International Collaborative Group::
S. Amemiya, K. Azad, L. Barak, T. Barrett, C. Costigan, D. Darko, S. Diamantopoulos, D. Doyle, M. Densriwiwat, R. P. F. Dullaart, I. Dzivite, J. A. Edge, K. Ekstrom, G. Forsander, D. Gasperikova, V. Hakeem, J. P. Hamilton-Shield, M.L. Hofstra, S-A. Ivarsson, I. Klimes, M. Kocova, O. Kordonouri, A. R. A. Lafferty, S. Likitmaskul, L. Liu, T. Milenkovic, W. Mlynarski, F. Mohsin, A. Noczynska, J. Odrezin, J. Porter, A. Roeleveld, J. Sanchez, M. Schebek, A. Schumacher, D. Segal, J. Stanik, Y. Tomita, and S. Wentworth
References
- 1.Iafusco D, Stazi MA, Cotichini R, Cotellessa M, Martinucci ME, Mazzella M, Cherubini V, Barbetti F, Martinetti M, Cerutti F, Prisco F. Permanent diabetes mellitus in the first year of life. Diabetologia. 2002;45:798–804. doi: 10.1007/s00125-002-0837-2. [DOI] [PubMed] [Google Scholar]
- 2.Edghill EL, Dix RJ, Flanagan SE, Bingley PJ, Hattersley AT, Ellard S, Gillespie KM. HLA genotyping supports a nonautoimmune etiology in patients diagnosed with diabetes under the age of 6 months. Diabetes. 2006;55:1895–1898. doi: 10.2337/db06-0094. [DOI] [PubMed] [Google Scholar]
- 3.Stanik J, Gasperikova D, Paskova M, Barak L, Javorkova J, Jancova E, Ciljakova M, Hlava P, Michalek J, Flanagan SE, Pearson E, et al. Prevalence of permanent neonatal diabetes in Slovakia and successful replacement of insulin with sulfonylurea therapy in KCNJ11 and ABCC8 mutation carriers. J Clin Endocrinol Metab. 2007;92:1276–1282. doi: 10.1210/jc.2006-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Polak M, Shield J. Neonatal and very-early-onset diabetes mellitus. Semin Neonatol. 2004;9:59–65. doi: 10.1016/S1084-2756(03)00064-2. [DOI] [PubMed] [Google Scholar]
- 5.Gloyn AL, Pearson ER, Antcliff JF, Proks P, Bruining GJ, Slingerland AS, Howard N, Srinivasan S, Silva JM, Molnes J, Edghill EL, et al. Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. N Engl J Med. 2004;350:1838–1849. doi: 10.1056/NEJMoa032922. [DOI] [PubMed] [Google Scholar]
- 6.Sagen JV, Raeder H, Hathout E, Shehadeh N, Gudmundsson K, Baevre H, Abuelo D, Phornphutkul C, Molnes J, Bell GI, Gloyn AL, et al. Permanent neonatal diabetes due to mutations in KCNJ11 encoding Kir6.2: patient characteristics and initial response to sulfonylurea therapy. Diabetes. 2004;53:2713–2718. doi: 10.2337/diabetes.53.10.2713. [DOI] [PubMed] [Google Scholar]
- 7.Flanagan SE, Edghill EL, Gloyn AL, Ellard S, Hattersley AT. Mutations in KCNJ11, which encodes Kir6.2, are a common cause of diabetes diagnosed in the first 6 months of life, with the phenotype determined by genotype. Diabetologia. 2006;49:1190–1197. doi: 10.1007/s00125-006-0246-z. [DOI] [PubMed] [Google Scholar]
- 8.Babenko AP, Polak M, Cave H, Busiah K, Czernichow P, Scharfmann R, Bryan J, Aguilar-Bryan L, Vaxillaire M, Froguel P. Activating mutations in the ABCC8 gene in neonatal diabetes mellitus. N Engl J Med. 2006;355:456–466. doi: 10.1056/NEJMoa055068. [DOI] [PubMed] [Google Scholar]
- 9.Ellard S, Flanagan SE, Girard CA, Patch AM, Harries LW, Parrish A, Edghill EL, Mackay DJ, Proks P, Shimomura K, Haberland H, et al. Permanent Neonatal Diabetes Caused by Dominant, Recessive, or Compound Heterozygous SUR1 Mutations with Opposite Functional Effects. Am J Hum Genet. 2007;81:375–382. doi: 10.1086/519174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stoy J, Edghill EL, Flanagan SE, Ye H, Paz VP, Pluzhnikov A, Below JE, Hayes MG, Cox NJ, Lipkind GM, Lipton RB, et al. Insulin gene mutations as a cause of permanent neonatal diabetes. Proc Natl Acad Sci U S A. 2007;104:15040–15044. doi: 10.1073/pnas.0707291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Takeuchi T, Tanaka S, Kubo SK, Kayo T, Lu D, Takata K, Koizumi A, Izumi T. A mutation in the insulin 2 gene induces diabetes with severe pancreatic beta-cell dysfunction in the Mody mouse. Journal of Clinical Investigation. 1999;103:27–37. doi: 10.1172/JCI4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hattersley AT. Molecular genetics goes to the diabetes clinic. Clin Med. 2005;5:476–481. doi: 10.7861/clinmedicine.5-5-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hattersley AT. Maturity-onset diabetes of the young: Clinical heterogeneity explained by genetic hetergeneity. Diabetic Medicine. 1998;15:15–24. doi: 10.1002/(SICI)1096-9136(199801)15:1<15::AID-DIA562>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 14.Hattersley AT, Pearson ER. Minireview: pharmacogenetics and beyond: the interaction of therapeutic response, beta-cell physiology, and genetics in diabetes. Endocrinology. 2006;147:2657–2663. doi: 10.1210/en.2006-0152. [DOI] [PubMed] [Google Scholar]
- 15.Gloyn AL. Glucokinase (GCK) mutations in hyper- and hypoglycemia: Maturity-onset diabetes of the young, permanent neonatal diabetes, and hyperinsulinemia of infancy. Hum Mutat. 2003;22:353–362. doi: 10.1002/humu.10277. [DOI] [PubMed] [Google Scholar]
- 16.Harries LW, Wickham CL, Evans JC, Rule SA, Joyner MV, Ellard S. Analysis of haematopoietic chimaerism by quantitative real-time polymerase chain reaction. Bone Marrow Transplant. 2005;35:283–290. doi: 10.1038/sj.bmt.1704764. [DOI] [PubMed] [Google Scholar]
- 17.Flanagan S E.L. E, A.L. G, K.L. S, S. E, A.T. H. Kir6.2 gene mutations are a common cause of diabetes before 6 months and the presence of neurological features varies with the site of the mutation. Diabet Med. 2005;22:10–28. [Google Scholar]
- 18.Ivarsson SA, Marner B, Lernmark A, Nilsson KO. Nonislet pancreatic autoantibodies in sibship with permanent neonatal insulin-dependent diabetes mellitus. Diabetes. 1988;37:347–350. doi: 10.2337/diab.37.3.347. [DOI] [PubMed] [Google Scholar]
- 19.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sunyaev S, Ramensky V, Bork P. Towards a structural basis of human non-synonymous single nucleotide polymorphisms. Trends Genet. 2000;16:198–200. doi: 10.1016/s0168-9525(00)01988-0. [DOI] [PubMed] [Google Scholar]
- 21.Edghill EL, Gloyn AL, Goriely A, Harries LW, Flanagan SE, Rankin J, Hattersley AT, Ellard S. Origin of de novo KCNJ11 mutations and risk of neonatal diabetes for subsequent siblings. J Clin Endocrinol Metab. 2007;92:1773–1777. doi: 10.1210/jc.2006-2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu M, Hodish I, Rhodes CJ, Arvan P. Proinsulin maturation, misfolding, and proteotoxicity. Proc Natl Acad Sci U S A. 2007;104:15841–15846. doi: 10.1073/pnas.0702697104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herbach N, Rathkolb B, Kemter E, Pichl L, Klaften M, de Angelis MH, Halban PA, Wolf E, Aigner B, Wanke R. Dominant-negative effects of a novel mutated Ins2 allele causes early-onset diabetes and severe beta-cell loss in Munich Ins2C95S mutant mice. Diabetes. 2007;56:1268–1276. doi: 10.2337/db06-0658. [DOI] [PubMed] [Google Scholar]
- 24.Yoshinaga T, Nakatome K, Nozaki J, Naitoh M, Hoseki J, Kubota H, Nagata K, Koizumi A. Proinsulin lacking the A7-B7 disulfide bond, Ins2Akita, tends to aggregate due to the exposed hydrophobic surface. Biol Chem. 2005;386:1077–1085. doi: 10.1515/BC.2005.124. [DOI] [PubMed] [Google Scholar]
- 25.Nozaki J, Kubota H, Yoshida H, Naitoh M, Goji J, Yoshinaga T, Mori K, Koizumi A, Nagata K. The endoplasmic reticulum stress response is stimulated through the continuous activation of transcription factors ATF6 and XBP1 in Ins2+/Akita pancreatic beta cells. Genes Cells. 2004;9:261–270. doi: 10.1111/j.1356-9597.2004.00721.x. [DOI] [PubMed] [Google Scholar]
- 26.Yoshida H. ER stress and diseases. Febs J. 2007;274:630–658. doi: 10.1111/j.1742-4658.2007.05639.x. [DOI] [PubMed] [Google Scholar]
- 27.Baker EN, Blundell TL, Cutfield JF, Cutfield SM, Dodson EJ, Dodson GG, Hodgkin DM, Hubbard RE, Isaacs NW, Reynolds CD. The Structure of 2Zn pig insulin crystals at 1.5A resolution. Philos Trans R Soc Lond Ser. 1988;319:369–456. doi: 10.1098/rstb.1988.0058. [DOI] [PubMed] [Google Scholar]
- 28.Hua QX, Liu M, Hu SQ, Jia W, Arvan P, Weiss MA. A conserved histidine in insulin is required for the foldability of human proinsulin: structure and function of an ALAB5 analog. J Biol Chem. 2006;281:24889–24899. doi: 10.1074/jbc.M602617200. [DOI] [PubMed] [Google Scholar]
- 29.Nakagawa SH, Hua QX, Hu SQ, Jia W, Wang S, Katsoyannis PG, Weiss MA. Chiral mutagenesis of insulin. Contribution of the B20-B23 beta-turn to activity and stability. J Biol Chem. 2006;281:22386–22396. doi: 10.1074/jbc.M603547200. [DOI] [PubMed] [Google Scholar]
- 30.Liu M, Ramos-Castaneda J, Arvan P. Role of the connecting peptide in insulin biosynthesis. J Biol Chem. 2003;278:14798–14805. doi: 10.1074/jbc.M212070200. [DOI] [PubMed] [Google Scholar]