A subset of pediatric AML cases harbors more somatic mutations in their genomes compared to normal blood progenitors. This subset displays expression profiles that resemble more committed progenitors and associates with better patient survival.
Abstract
Acquisition of oncogenic mutations with age is believed to be rate limiting for carcinogenesis. However, the incidence of leukemia in children is higher than in young adults. Here we compare somatic mutations across pediatric acute myeloid leukemia (pAML) patient-matched leukemic blasts and hematopoietic stem and progenitor cells (HSPC), as well as HSPCs from age-matched healthy donors. HSPCs in the leukemic bone marrow have limited genetic relatedness and share few somatic mutations with the cell of origin of the malignant blasts, suggesting polyclonal hematopoiesis in patients with pAML. Compared with normal HSPCs, a subset of pAML cases harbored more somatic mutations and a distinct composition of mutational process signatures. We hypothesize that these cases might have arisen from a more committed progenitor. This subset had better outcomes than pAML cases with mutation burden comparable with age-matched healthy HSPCs. Our study provides insights into the etiology and patient stratification of pAML.
Significance:
Genome-wide analysis of pAML and patient-matched HSPCs provides new insights into the etiology of the disease and shows the clinical potential of these analyses for patient stratification.
Introduction
Somatic mutations gradually accumulate throughout life, which is thought to underlie the increased incidence of cancer with age (1). The more mutations a cell has, the higher the chance that one of these is an oncogenic mutation that can drive cancer development. However, some cancers, such as leukemia and brain tumors, show a relatively high incidence in young children (2, 3) even though their young cells are less damaged by age (4,5,6). Indeed, pediatric cancers display fewer cancer driver mutations compared with adult cancers (7, 8). Therefore, the etiology of pediatric cancers is likely to differ from cancer in the elderly.
In this study, we focused on acute myeloid leukemia (AML), a cancer that occurs in both children and adults. Leukemia is the most common form of childhood cancer, and AML constitutes about 15% to 20% of all childhood leukemias. Although the outcome of children with all types of leukemia has improved significantly over the past decades (9, 10), for pediatric AML (pAML) a therapeutic plateau of approximately 70% overall survival has been reached with current therapies (11). Relapse rates for pAML remain high at 25% to 30%, and this is associated with poor outcome (12). In addition, childhood cancer survivors suffer from late effects of the cancer and treatment, although the molecular causes for this remain unclear.
The molecular heterogeneity of pAML has been studied extensively during the last few years, and various driver genes and structural variants (SV) have been identified (7, 8, 13, 14). Both adult and pediatric AML are thought to arise from hematopoietic stem or progenitor cells (HSPC) that have acquired oncogenic mutations (15, 16). The landscape of somatic driver mutations in AML varies considerably with age (13). Pediatric patients have more somatic SVs compared with adult AML patients, while DNMT3A and TP53 mutations, the most important drivers in adult AML, are virtually absent in pAML. Thus, the driving potential of specific oncogenic mutations seems to critically depend on the timing of acquisition (i.e., prenatal, early childhood, late adulthood). Indeed, KMT2A (also known as MLL) fusions are more common in infant AML, while t(8;21) translocations resulting in the fusion gene RUNX1–RUNXT1 are more common later during childhood (13).
Cancers are formed by evolutionary processes acting in normal tissues (17). Characterization of mutational landscapes in normal cells has advanced our understanding of these processes as well as provided insight into tumorigenesis (4, 5, 13). Here, we compared mutation accumulation in HSPCs and leukemic cells of children suffering from pAML as well as with HSPCs from healthy individuals. We found that the normal HSPCs in leukemic bone marrow are unaffected in terms of somatic mutation numbers, mutation spectra, and clonal composition as compared with healthy individuals. The number of clonal mutations in a subset of pAML cases was increased as compared with normal HSPCs, which was caused by oxidative stress–induced mutagenesis and correlated with a more differentiated leukemic cell-of-origin phenotype and better patient survival. Our work provides insight into the processes that shape pAML as well as the consequences of the disease on blood.
Results
Establishing a Baseline for Mutation Accumulation in Normal Blood during Human Life
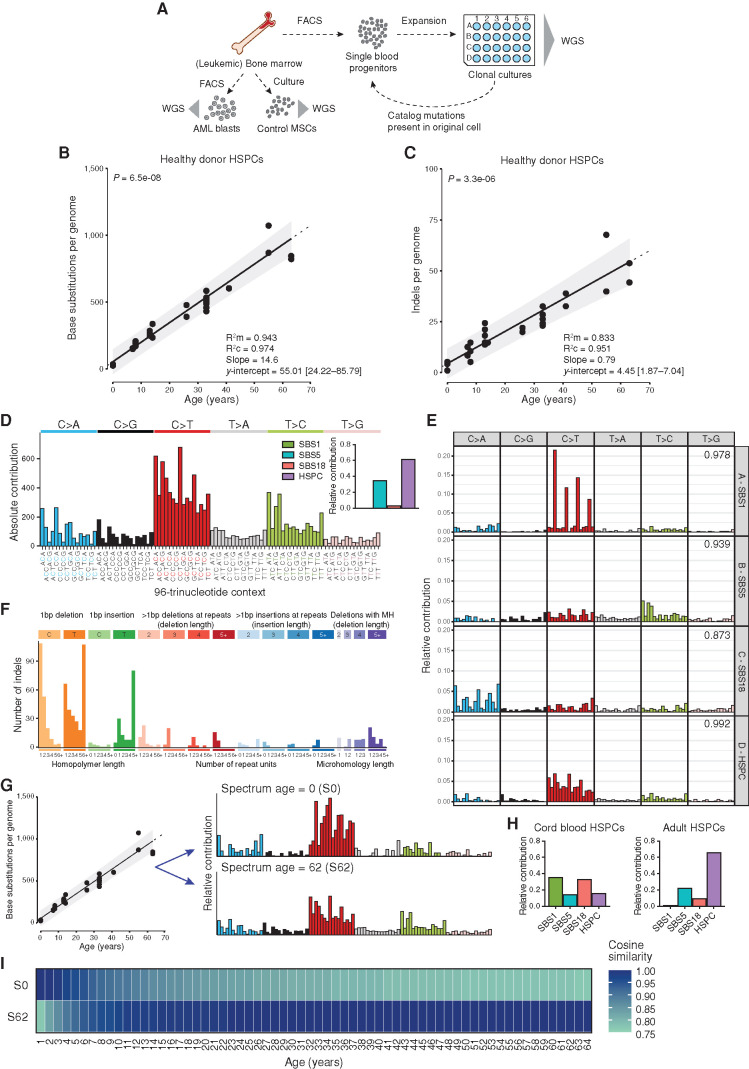
We have previously reported using whole-genome sequencing (WGS) of individual HSPCs that mutations accumulate in a linear fashion in blood of healthy adults (5). To allow direct comparison with normal HSPCs and AML blasts of children with leukemia, here we extend these data to also include the pediatric age range. For this, we included 11 additional genomes of 4 healthy children who donated bone marrow in our institute for an allogeneic hematopoietic stem cell transplantation (HSCT) for their affected sibling. These children were unrelated to any of the patients with AML in this study. We used multiparameter flow cytometry to sort single HSPCs, which were subsequently clonally expanded to obtain sufficient DNA for WGS analysis (Fig. 1A). This procedure allowed us to catalog all the somatic mutations present in the original stem cell that accumulated during the life of the cell. Somatic mutations in the HSPCs displayed a variant allele frequency (VAF) cluster around 0.5, indicating that these mutations were shared by all cells in the culture and therefore present in the expanded parental stem cell (Supplementary Fig. S1A). A smaller, second VAF peak could sometimes be observed around 0.2, which likely represents subclonal mutations that accumulated after the first cell division in vitro and are not shared by all cells in the culture (Supplementary Fig. S1B). These in vitro accumulated mutations are discarded for further downstream analyses based on the low VAF (Supplementary Fig. S1; Methods). When combined with our previous study (5), the final dataset was comprised of 34 HSPCs of 11 healthy donors, ranging from 0 to 63 years of age. In total, we identified 13,662 base substitutions and 760 small insertions and deletions (indels). We did not observe nonsynonymous or truncating mutations in cancer-driving genes associated with hematologic neoplasms (Supplementary Table S1; ref. 18). A positive correlation (P < 0.05; t test linear mixed model) between the number of base substitutions in HSPCs and age of the donors was observed (Fig. 1B), showing an accumulation of 14.6 base substitutions per year of life. Only a limited number of mutations [55.01; 95% confidence intervals (CI) are 24.2–85.8] were acquired before birth. Similarly, the number of indels correlated (P < 0.05; t test linear mixed model) with donor age (Fig. 1C), showing an accumulation of 0.79 indels per year throughout life. Only a few (4.45; 95% CI, 1.9–7.0) indels are acquired prenatally.
Figure 1.
Healthy baseline of mutation accumulation in HSPCs. A, Schematic overview of experimental setup to catalog somatic mutations in single human blood progenitors derived from healthy bone marrow or from leukemic bone marrow at diagnosis. AML blasts were FACS sorted in bulk from the leukemic bone marrow. B and C, Correlation of the number of base substitutions (B) or indels (C) accumulated per genome with age of the independent donors. Each dot represents data from a clonally expanded HSPC from bone marrow of healthy children and adults or from cord blood. Two to ten HSPC clones were analyzed per individual. P value of the age effect in the linear mixed model is indicated above the plot (two-tailed t test). The sample size is 11 healthy donors with a total of 34 clones sequenced. Linear mixed model was performed on all clones using “age” as a fixed effect and “(1 + age | Donor)” as random effects. Dotted line indicates the extrapolation of this correlation. The 95% probability interval of the linear mixed model is depicted in gray. Marginal R2 (R2m), condition R2 (R2c), slope, and y-intercept (with 95% CI) of the models are depicted on the right. D, Total 96-trinucleotide spectrum for all mutations in HSPC clones from healthy individuals. Inset depicts the relative contribution of four mutational signatures to each spectrum. Mutational signatures: HSPC, a specific pattern predominantly found in healthy adult HSPCs; SBS1, spontaneous deamination of methylated cytosines; SBS5, unknown etiology; SBS18, oxidative stress–induced mutagenesis. E, Ninety-six–trinucleotide spectra for the four extracted signatures after de novo signature extraction (Methods). Cosine similarities to Catalogue of Somatic Mutations in Cancer (COSMIC) signatures are indicated in the top right corner, and corresponding names to COSMIC signatures are depicted. F, Total indel spectrum for all mutations in HSPC clones from healthy individuals. MH, microhomology. G, Schematic representation of the application of the reconstructed 96-trinucleotide spectrum based on the slopes estimated by the linear mixed model in B for each trinucleotide. For any desired age, the expected 96-trinucleotide spectrum can be depicted. Ninety-six–trinucleotide spectra of the reconstructed baseline model at age 0 (S0) and age 62 (S62) are depicted. H, The relative contribution of four mutational signatures to each spectrum of all cord blood HSPC clones (left) or all adult HSPC clones (right). I, The expected 96-trinucleotide spectrum at age 0 and age 62 (S0 and S62) was compared with the expected 96-trinucleotide spectrum at any age between 1 and 64 years. Spectra were based on the reconstructed model and cosine similarities are depicted.
Different mutational processes often generate different combinations of mutation types, termed mutational signatures (19). The mutation spectra of healthy HSPCs could be explained by two mutational signatures, namely the “HSPC” signature, previously identified as a specific pattern predominantly found in healthy adult HSPCs (5, 20, 21), and single base substitution (SBS) signature 5, for which the underlying process is still unknown (Fig. 1D and E). The majority of the indels were 1 bp deletions of a C or T and 1 bp insertions of a T (Fig. 1F), which has been attributed to polymerase slippage during replication of the replicated DNA strand (22). Using the mutations that we observed in healthy individuals, we generated a mathematical model that can predict the absolute base substitution load and spectrum at any given age (Fig. 1G), which we can use to determine any additive mutational load in pAML. Our model predicts that the types of mutations that accumulate early in life vary from those in adult life (Fig. 1G–I). As expected, the adult HPSCs mainly show contribution of the HSPC signature, while the cord blood–derived HSPCs show different signature contributions (Fig. 1H). This finding suggests that distinct mutational processes are active during development as compared to mature hematopoiesis, which is in line with previous reports (5, 23).
HSPCs in the Blood System of Patients with pAML
We obtained bone marrow biopsies of pAML patients of various cytogenetic subgroups at diagnosis and before treatment initiation. For each patient, we isolated bulk AML blasts and clonally expanded normal HSPCs that were obtained from the same bone marrow (Fig. 1A; Supplementary Fig. S2A and S2B). The clonal outgrowth of HSPCs obtained from leukemic bone marrow was comparable to those isolated from healthy bone marrow except for reduced outgrowth in the cytogenetic subgroup characterized by t(8;21) (Supplementary Fig. S2C). Because the number of sorted HSPCs was similar for all samples (Supplementary Table S2), the reduced clonal outgrowth of HSPCs from t(8;21) patients with AML might indicate a functional impairment of the HSPC compartment in these patients. To investigate this, we analyzed a cohort of 237 patients with pAML for neutrophil recovery time after chemotherapy as a measure of HSPC function (Supplementary Fig. S2D and S2E). However, no difference in neutrophil recovery time between pAML subtypes could be observed (Supplementary Fig. S2F and S2G). Nonleukemic stem cells expressing the t(8;21) fusion gene–specific transcript have been described before (24). Thus, our data might indicate that the t(8;21) translocation may be present in some of the sorted HSPCs, but that these fail to grow further ex vivo.
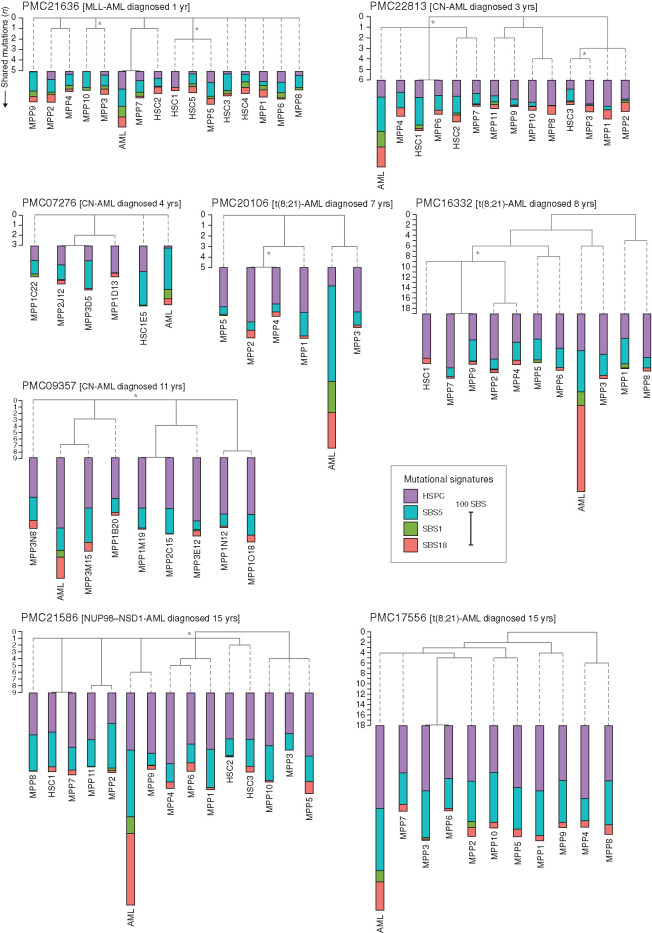
Of nine patients with AML (referred to as “PMC” patients, as they were treated in the Princess Máxima Center), we subjected DNA of bulk sorted AML blasts and matching cultured mesenchymal stromal cells (MSC) to WGS analysis (Fig. 1A). WGS data of MSCs were used to filter out germline variants, allowing us to obtain catalogs of somatic mutations in pAML. In addition, for 8 out of 9 patients, we subjected DNA of 5 to 15 clonally expanded HSPCs per patient to WGS analysis (80 HSPCs in total). For the leukemic blasts, we only considered somatic mutations that were clonally present in the pAML genomes, as these were present in the most recent common ancestor (MRCA) of the malignant blasts and likely represent the leukemic cell of origin (Supplementary Figs. S1B and S2B; ref. 25). In total, we identified 14,174 base substitutions and 878 indels in the HSPCs and 3,661 base substitutions and 383 indels in the AML blasts. Independent validations using molecular inversion probes (MIP) for a subset of the somatic mutations revealed an overall true-positive discovery rate of 97.7% (Supplementary Fig. S3A). We explored the phylogenetic relations between each of the HSPC clones and AML blasts by assessing mutations that are shared between the different cells of the same patient. We observed a limited number of shared (passenger) mutations between HSPC clones and the matching AML (Fig. 2). This finding indicates that although the leukemic blast percentage in pAML can be as high as 77% in this study (Supplementary Table S2), the HSPCs in the leukemic bone marrow are limited in their genetic relatedness, suggesting a polyclonal hematopoietic system in children with AML, similarly as previously observed in healthy adults and fetuses (5, 21). This is different from what has been observed in adult AML and other blood cancers, where clonal hematopoiesis often precedes cancer development and genetic drivers can be detected years before cancer onset in healthy HSPCs (26, 27). The genetic relatedness, reflected by more shared mutations, of the HSPC clones tends to be higher in the older patients (≥7 years) compared with the younger patients (≤4 years; Fig. 2), possibly reflecting the maturation of the blood system during childhood, with more dominant HSPC clones becoming apparent. The pAML MRCA shares very few (zero to seven) somatic mutations with any of the analyzed HSPC clones in all eight patients. To further confirm our results, we genotyped 43% of the pAML mutations in 64 additional HSPCs of one patient (PMC22813) using targeted sequencing (see Methods). Only two of the assessed HSPCs showed subclonal evidence of a passenger somatic mutation with the clonal AML blast mutations (Supplementary Table S3). In addition, for patient PMC21636, DNA from 92 additional HSPCs was analyzed for the presence of the main genetic driver using MLL fusion–specific primers, and all HSPCs tested negative (Supplementary Fig. S3B). Thus, we did not identify large preleukemic clones in the HSPCs analyzed in this study. Together, these data further support our observation that blood lineages within the leukemic bone marrow display limited genetic relatedness and separate early during development.
Figure 2.
Genetic relatedness between AML and patient-derived HSPCs. Phylogenetic trees of n = 8 pAML cases representing different subtypes and ages at diagnosis. Five to fifteen HSPC clones as well as bulk AML blasts were analyzed by WGS for each patient. Each branch represents an individual sequencing sample: either a hematopoietic stem cell (HSC) clone, a multipotent progenitor (MPP) clone, or AML blasts (indicated as AML). Shared branches represent those mutations, both base substitutions and indels, present across all downstream descendent clones; number of shared mutations are depicted on the left axes. Dashed gray line indicates no additional shared mutations. The absolute contribution of each mutational signature to the 96-trinucleotide spectrum is also depicted. Lengths are proportional to total somatic SBS counts as shown by the scale bar of 100 mutations. Age at diagnosis and pAML subtype are indicated above each tree. Asterisk (*) indicates branch with mutations subclonally present in the patient-matched MSC sample. CN, cytogenetically normal.
Mutation Accumulation in Normal HSPCs Is Unaltered in the Leukemic Niche
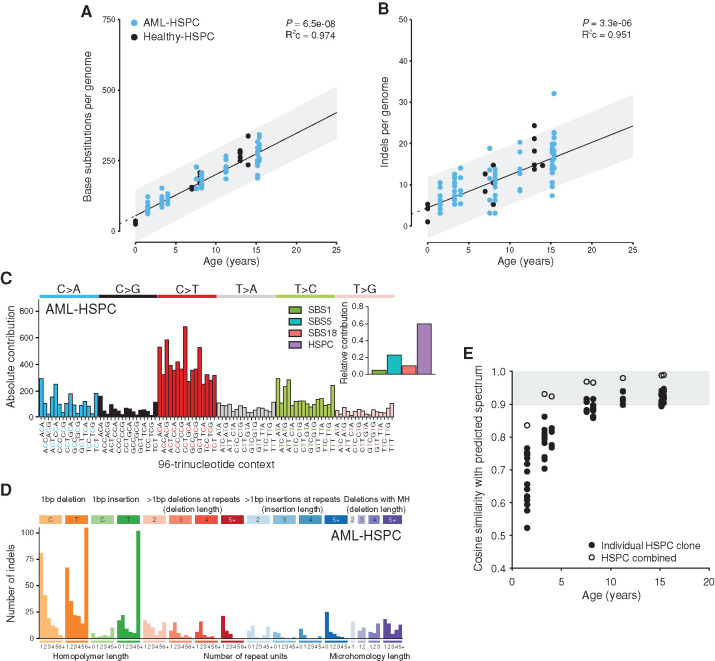
To further study the effect of leukemia on normal HSPCs in the leukemic bone marrow, we compared the genomes of these HSPCs to our healthy baseline (Supplementary Table S4). The number of base substitutions and indels in these HSPCs was similar as observed in healthy blood (Fig. 3A and B). In line with this, the base substitution spectrum and signature contributions of the HSPCs isolated from leukemic bone marrow were similar to those of healthy individuals (cosine similarity = 0.99; Fig. 3C). The indel spectrum of HSPCs from pAML bone marrow was also very similar to that of healthy individuals (cosine similarity = 0.949; Fig. 3D). These observations indicate that HSPCs in leukemic bone marrow have not been exposed to additional mutational process activity. Indeed, the mutation spectra in each HSPC clone as well as of all HSPC clones per pAML patient combined were similar as predicted by our baseline model, which is based on lifelong mutation accumulation in blood (Fig. 3E); however, for the youngest patient, the cosine similarities are lower due to lower numbers of mutations per HSPC clone. Of note, no SVs or copy-number changes were observed in any of the assessed HSPCs. These data indicate that the genomes of normal HSPCs in the leukemic niche are unaffected.
Figure 3.
Mutation accumulation in normal HSPCs from the leukemic niche. A and B, Correlation of the number of base substitutions (A) or indels (B) accumulated per genome with age of the assessed patients. Each dot represents data from a clonally expanded HSPC from bone marrow of patients with pAML (AML-HSPC) and healthy individuals (Healthy-HSPC). Two to fifteen HSPC clones were analyzed per individual. Healthy-HSPC data points are the same as in Fig. 1B and C. P value of the age effect in the linear mixed model of healthy donors (two-tailed t test) and the condition R2 (R2c) of the model are depicted. C, Total 96-trinucleotide spectrum for all mutations in HSPC clones from patients with pAML. Inset depicts the relative contribution of four mutational signatures to each spectrum. D, Total indel spectrum for mutations in HSPC clones from patients with pAML. MH, microhomology. E, Cosine similarity between actual and expected 96-trinucleotide spectrum of AML-HSPC, correlated with patient age. Spectra of individual HSPC clones and the spectrum of all HSPCs combined from one patient were compared to the expected spectrum. Gray indicates a cosine similarity >0.9.
Processes Underlying the Increased Mutation Load in pAML
Most pAML samples showed a higher number of somatic mutations compared with their patient-matched HSPCs (Fig. 2; Supplementary Fig. S4A and S4B). Importantly, we are comparing the clonal mutations of bulk AML cells, i.e., mutations that are shared in all AML cells and represent the MRCA that existed in the past, to those of single clonally expanded HSPCs at the time of diagnosis. Therefore, the difference in somatic mutation load might be even greater, as the MRCA arose before the time of diagnosis at a younger age of the patient. To identify the processes that underlie the increased mutation load in pAML, we analyzed mutation spectra and underlying signatures. The mutation spectra and the mutational signature contributions of the pAML samples were markedly different than those of patient-derived normal HSPCs (Fig. 2; Supplementary Fig. S4C). The pAML genomes of some patients showed a pronounced C>T at NCG profile, while others were characterized by a predominant contribution of C>A mutations (Supplementary Fig. S4C). We mainly observed a higher contribution of SBS1 and SBS18 to the spectra of pAML as compared to patient-matched normal HSPCs (Fig. 2). SBS1 is believed to reflect the spontaneous deamination of methylated cytosines into thymines (28) and likely reflects a cell cycle–dependent mutational clock (4, 5, 29). HSPCs are thought to proliferate extensively during fetal development, whereas postnatally, the majority of HSPCs become quiescent (30). The higher SBS1 contribution could indicate that these pAML cases may be derived from an HSPC “arrested” in this high proliferative, developmental state. SBS18 is thought to reflect oxidative stress–induced mutagenesis (31, 32). It has previously been shown that RUNX1–RUNX1T1, the fusion protein resulting from the t(8;21) translocation, downregulates the expression of the base excision repair gene OGG1, which recognizes and excises oxidized guanines (33). However, not all t(8;21) AML showed strong SBS18 contribution, and we also observed SBS18 in other pAML subtypes, suggesting that downregulation of OGG1 is probably not the only mechanism for increased SBS18 mutations. Indeed, these SBS18 mutations are clonally present in the MRCA of pAML, indicating that these mutations might have occurred before the outgrowth of the MRCA of the leukemia. The increased mutation load in pAML could not be attributed to the HSPC signature; rather, more SBS1, SBS5, and/or SBS18 were observed. Finally, the HSPC signature becomes more pronounced in the healthy HSPC of patients with ≥7 years of age, in line with our model that “mature hematopoiesis,” based on mutation spectra, occurs around 5 years of age (Fig. 1H and I). Overall, these data show that different mutagenic processes might contribute to pAML development, which are absent during healthy postnatal hematopoiesis.
Increased Mutation Load in pAML Compared with Normal HSPCs
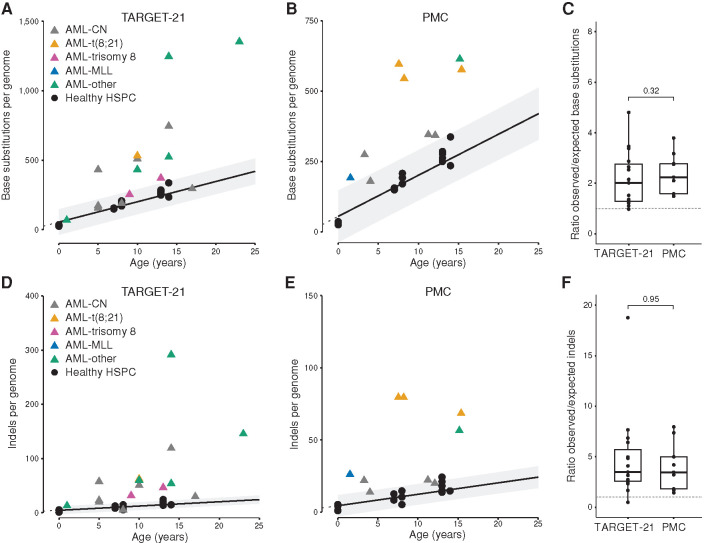
Next, we combined our pAML genome dataset (n = 9) with samples of 15 patients from the Therapeutically Applicable Research to Generate Effective Treatments (TARGET; 13) initiative. We only included 15 patients with a blast percentage ≥80% to be able to call all clonal leukemic mutations with high accuracy (referred to as “TARGET-21” patients; Supplementary Table S5; Methods). In this extended dataset (n = 24), 17 pAML genomes had an increased clonal somatic mutation load compared with healthy HSPCs irrespective of pAML subtype (Fig. 4A and B), and this corresponded to an increased number of indels per genome (Fig. 4D and E). On average, pAML had 2.25-fold more base substitutions and 4.53-fold more indels over the healthy baseline (Fig. 4C and F). In contrast, adult AML has a similar number and types of somatic mutations with age-matched HSPCs (5, 6), indicating that age-related mutation accumulation in normal blood can explain mutation loads in adult leukemia. Thus, these data could indicate that at the moment of pAML leukemogenesis, the leukemic cell of origin of this subset has a higher number of somatic mutations compared with age-matched healthy HSPC.
Figure 4.
Somatic mutations in pAML. A, Correlation of the number of base substitutions accumulated per genome with age of the assessed patients. Data from 15 patients with pAML of the TARGET initiative (TARGET-21) are depicted (13). Each data point represents a single HSPC clone (circles) or bulk AML (triangles). CN, cytogenetically normal. B, Correlation of the number of base substitutions accumulated per genome with age of the assessed patients. Data of nine patients with pAML of the PMC are depicted. Legend is the same as in A. C, Ratio between the observed and expected number of base substitutions per genome. Expected number is extrapolated from the linear mixed model in Fig. 1B. P value indicates a nonsignificant difference between patients with AML of the two institutes (Mann–Whitney test). D, Correlation of the number of indels accumulated per genome with age of the assessed TARGET-21 patients. E, Correlation of the number of indels accumulated per genome with age of the assessed PMC patients. Legend is the same as in D. F, Ratio between the observed and expected number of indels per genome. Expected number is extrapolated from the linear mixed model in Fig. 1C. P value indicates a nonsignificant difference between AML patients of the two institutes (Mann–Whitney test).
Higher Somatic Mutation Load in pAML Correlates with Better Patient Survival
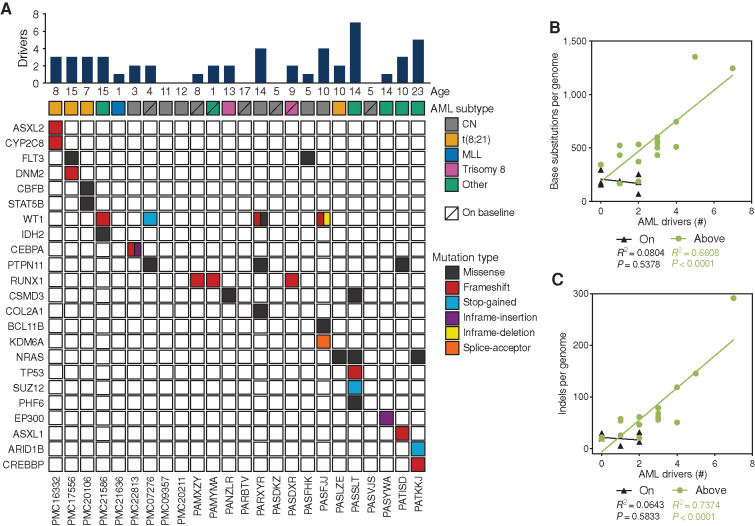
Although the majority of pAML displayed an increased overall mutation load compared with the healthy baseline, 7 of 24 pAML showed a similar number of somatic base substitutions as healthy age-matched HSPCs (Fig. 4A and B). To test whether the enhanced mutation load is a consequence of selection dynamics, we calculated the number of cancer- driving events per patient (Supplementary Table S6). In line with previous studies, only a limited number (on average 1.5 per patient) of the mutations in pAML are in cancer driver genes (Fig. 5A; ref. 13). Although identifying novel drivers of pAML was not the aim of this study, we observed a KMT3A/SORBS2 fusion gene in one patient, which has been described in only one case before (34). The average number of genetic driver events is significantly increased in pAML cases with an increased mutational load as compared to cases that are similar to the healthy baseline (Supplementary Fig. S5A). In fact, the number of genetic driver events in the “above” baseline pAML cases correlated with the total number of mutations, while no correlation could be observed for the “on” baseline pAML cases (Fig. 5B and C). This observation could suggest that for the initiation of “above” baseline AML, mutation accumulation in the leukemic cell of origin could have been rate limiting, since cells with a higher overall mutation burden have more chance of harboring an oncogenic hit (1). In contrast, the genesis of “on” baseline pAML does not necessarily require enhanced mutagenesis, similarly as previously reported for adult AML (5). To study whether these two patient groups have different clinical outcome, the event-free and overall survival of patients was assessed. We found that patients with pAML whose mutational load is higher than the healthy baseline tend to have better survival than patients whose pAML falls on the healthy baseline (Supplementary Fig. S5B and S5C).
Figure 5.
AML driver mutations. A, Overview of the identified clonal AML driver mutations. Bar graph (top) indicates total number of cancer-driving events identified, including known AML driving mutations, fusion genes, trisomy, or loss-of-heterozygosity events. Details can be found in Supplementary Table S6. CN, cytogenetically normal. B and C, Correlation between number of base substitutions (B) or indels (C) per genome and AML drivers in 24 patients with pAML (TARGET-21 and PMC cohort combined) divided in “above” and “on” healthy baseline groups. Linear regression analysis indicates a significant correlation between the number of AML drivers and the number of base substitutions for the “above” baseline group.
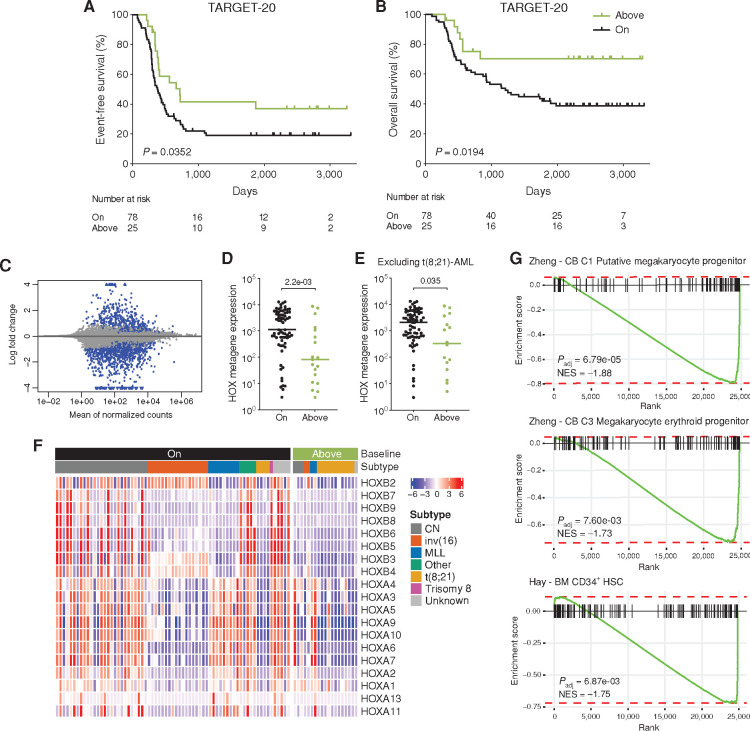
To validate these findings, we analyzed an additional, separate cohort of pAML patient samples with available Complete Genomics (CGI) somatic mutation data (TARGET-20; ref. 7). After filtering out low purity and low sequencing quality samples, the second cohort of pAML comprised 103 patients (see Methods). Twenty-five patients with pAML (24.3%) had an increased clonal somatic mutation load compared with the healthy baseline (Supplementary Fig. S5D), confirming our observation that a subset of pAML has increased somatic mutations. Patients classified as “above” baseline had a significantly higher event-free and overall survival compared with patients with a mutation load comparable with healthy subjects (Fig. 6A and B). Of note, the number of “above” baseline patients in the TARGET-20 cohort is lower as compared with the initial cohort of 24 patients. This difference can be attributed to variation in patient selection criteria between the two cohorts. TARGET-20 is a cohort that is enriched for patients with relapsed pAML, while the TARGET-21 cohort is enriched for patients who showed failure of induction treatment (7, 13). Therefore, these cohorts are enriched for patients with poor outcome and more “on” baseline mutation load. In contrast, the patients we assessed from our institute were randomly selected. Interestingly, 48% of the pAML samples “above” the healthy baseline were of the t(8;21) subtype compared with 5% of the samples “on” the healthy baseline (Supplementary Fig. S6A). After correcting for multiple testing, including pAML subtype, the “above” baseline group still had a significantly decreased HR for both event-free and overall survival (Supplementary Fig. S6B). As the t(8;21) pAML subtype was enriched in the “above” baseline group and this subtype is associated with a more favorable prognosis (12), we reanalyzed the survival data without the patients with t(8;21) pAML and again found a significantly higher event-free and overall survival of the “above” baseline pAML (Supplementary Fig. S6C and S6D), although the effect is less pronounced. Together, these findings show that increased somatic mutation load might be a predictor of better pAML patient survival, which cannot be attributed solely to the t(8;21) subtype.
Figure 6.
Survival and gene expression analysis of “above” and “on” baseline pAML. A and B, Kaplan–Meier survival curves for event-free (A) and overall (B) survival of 103 TARGET-20 patients with pAML classified as “above” or “on” healthy baseline. “Above” baseline indicates that the number of base substitutions in AML is above the 95% prediction interval of the linear mixed model in Fig. 1B. P value indicates a significant difference between above and on baseline patients (log-rank Mantel–Cox test). C, Differential gene expression of 88 TARGET-20 pAML patients classified as “above” or “on” healthy baseline. “Above” baseline pAML was compared with “on” baseline as a reference. Blue indicates significantly upregulated (log2 fold change > 0.585) or downregulated (log2 fold change < –0.585) genes (padj < 0.05). D, Mean normalized expression of 19 HOX genes, depicted in F, for each pAML sample. P value indicates a significant difference between above and on baseline patients (Mann–Whitney test). E, Same as D but excluding t(8;21) pAML (TARGET-20 and TARGET-21 combined). F, Heatmap depicting the expression of 19 HOX genes in 88 TARGET-20 pAML, expression mean is normalized to 0. Legends indicate “above” or “on” baseline and AML subtype. Samples were ordered by pAML subtype. CN, cytogenetically normal. G, Gene-set enrichment plots of the indicated gene sets using log fold change shrinkage data of the 88 TARGET-20 patients with pAML. The normalized enrichment score (NES) corrects for multiple testing. A negative NES indicates genes enriched in “on” baseline pAML, as these were used as reference.
Normal Mutation Burden in pAML Correlates with an Early Progenitor Phenotype
One explanation for the differences in mutation burden and associated prognosis, is that a more committed (more differentiated) progenitor might be the leukemic cell of origin of the pAML with a higher mutation load than the healthy baseline. To test this, we first attempted to train a classifier using a machine learning approach to distinguish hematopoietic stem cells (HSC) from multipotent progenitor cells (MPP) based on our own and publicly available (21) genome-wide mutation data. We constructed a dataset containing 70 HSCs and 70 MPPs of healthy adult donors profiled with WGS and used as variable features the relative contribution of the 96-trinucleotide changes, genomic locations, and the total mutation count. The random forest model trained on these data resulted in an out-of-bag error of 40.71% (Supplementary Fig. S6E), indicating that based on these genomic features, HSCs and MPPs cannot be distinguished. This analysis suggested that HSCs and MPPs have similar mutation burdens and mutational profiles (Fig. 2; refs. 5, 21). Next, we built a model to directly separate pAML “above” and “on” the healthy baseline using genomic features based on the ratio between the observed (measured) and expected (healthy baseline) number of mutations (Methods). We used 75% of the available TARGET patients (n = 90) to train a linear regression model and validated it on the remaining 25% and the PMC patients (n = 37). We found a significant correlation between the predicted ratio and actual ratio of mutations to the healthy baseline (Pearson correlation = 0.73, P = 2e-07; Supplementary Fig. S6F), suggesting there are different mutational processes active in the pAML samples “above” the healthy baseline. The final regression model included the relative contribution of T>A mutations and the cosine similarity to a variety of mutational signatures as features (Supplementary Fig. S6F). While the ratio to the healthy baseline can be predicted to a certain extent using these genomic features, the processes behind them are not easily interpretable. Thus, we looked further into gene expression data to investigate possible processes underlying the two different pAML patient groups we defined in this study.
We used the RNA-sequencing data available for most of the TARGET-20 patients (n = 88). Differential gene expression analyses indicated that many HOX genes were among the most downregulated genes when comparing “above” with “on” baseline pAML (Fig. 6C; Supplementary Table S7). The average HOX metagene expression (comprising 19 HOX genes) was higher in pAML that fall “on” the healthy baseline (Fig. 6D), and this does not seem to directly correlate with pAML subtype, as the analysis yields similar results after excluding t(8;21) pAML (Fig. 6E and F). HOX genes are a family of homeodomain-containing transcription factors that are highly expressed in the most primitive HSCs and progenitors (35), suggesting that these leukemias are arrested in a more primitive stem cell/progenitor state in contrast to pAML cases that have a higher mutation burden as compared with the healthy baseline. To further validate this, we performed gene-set enrichment analysis (GSEA) on the RNA-sequencing data of the TARGET-20 patients using cell-type signature gene sets. We observed a significant enrichment in the “on” baseline group for gene sets of cord blood putative megakaryocyte progenitor, cord blood megakaryocyte progenitor, and bone marrow CD34+ HSC signatures (Fig. 6G, negative enrichment by comparing “above” with “on” baseline; Supplementary Table S8; refs. 36, 37). Similar results were obtained for the 15 TARGET-21 patients included in our initial cohort (Supplementary Fig. S6G–S6J). These data support the idea that pAML with a comparable mutation burden as normal hematopoietic cells most likely originate from early progenitor cells, such as HSCs, which are often more megakaryocyte biased (38). In addition, these findings therefore suggest that t(8;21) AML, as the main subtype of “above” baseline pAML, likely arises in a more committed progenitor with a relatively higher mutation load. Similar findings were obtained for other subtypes in the “above” baseline pAML cases, indicating that this is not a feature unique to t(8;21) pAML. Taken together, our data indicate that pAML with an increased mutation load compared to healthy HSPCs might originate from a more committed progenitor cell, and this correlates with better event-free and overall survival of these patients.
SBS1 and SBS18 Significantly Contribute to the Increased Somatic Mutation Load of pAML
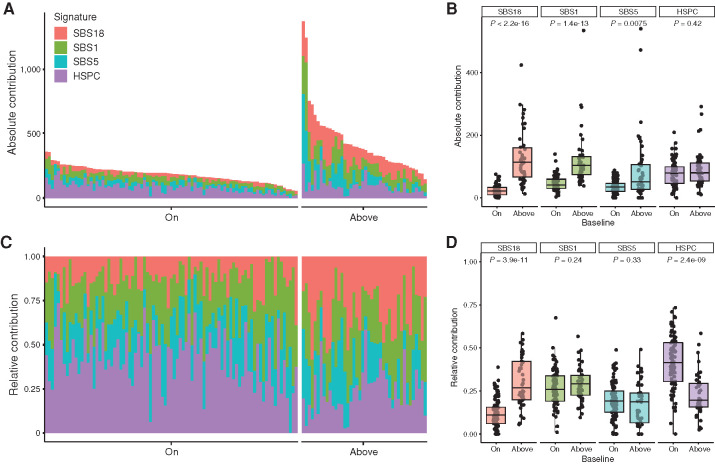
We combined the two cohorts of this study into one final cohort of 127 patients with pAML (Supplementary Table S9) and analyzed the somatic clonal mutation patterns by refitting them to the four mutational signatures SBS1, SBS5, SBS18, and HSPC. An absolute increase in SBS1 was observed in the “above” baseline pAML (Fig. 7A and B), suggesting that the leukemia MRCA has undergone increased proliferation. However, we predominantly observed an increased contribution of SBS18 in pAML with an increased mutation burden as compared with the healthy baseline. Interestingly, high SBS18 contribution correlates with low expression of MEIS1 in “above” baseline pAML (Supplementary Fig. S6K), a transcription factor involved in limiting oxidative stress in HSCs (39). The absolute number of HSPC mutations was similar in both groups, but the relative contribution of the HSPC signature was higher in the “on” baseline pAML (Fig. 7C and D). In conclusion, the increased somatic mutation load of pAML is caused by a higher number of SBS1 and SBS18 mutations, indicating more replication- and oxidative stress–related mutagenesis.
Figure 7.
Mutational signatures in pAML. A, Absolute contribution of the four extracted mutational signatures to the somatic mutation spectrum. Each bar represents a pAML sample, and n = 127 samples are included (PMC, TARGET-20, and TARGET-21). Samples are ordered on the basis of their total absolute mutation count and split per “on” or “above” baseline group. B, Bar graph of data depicted in A. Each dot is a pAML sample. P value indicates significant differences between “on” and “above” baseline patients (Wilcoxon Mann–Whitney test) for SBS18, SBS1, and SBS5. C, Relative contribution of four extracted mutational signatures to the somatic mutation spectrum. D, Bar graph of data depicted in C. P value indicates significant differences between “on” and “above” baseline patients (Wilcoxon Mann–Whitney test) for SBS18 and HSPC signature.
Discussion
Here, we studied the mutational landscapes of HSPCs in the leukemic bone marrow of pAML patients. We show that a subset of pAML has an increased mutation burden compared with age-matched normal HSPCs, which could be explained by increased exposure to replication- and oxidative stress–related mutagenesis during the etiology of the disease. Patients whose AML has this increased mutation burden have a better event-free and overall survival.
During development, a substantial expansion of the HSPCs occurs in the fetal liver (30), whereas in adults, HSPCs are mostly quiescent and reside in the bone marrow. In line with this, fetal and umbilical cord blood–derived HSPCs display a predominant contribution of SBS1, which is thought to be a cell cycle–dependent mutational clock (4, 5, 23, 29). Several pAML genomes also showed a predominant contribution of SBS1, which would suggest that the MRCA of the leukemia has had an extended proliferative history as compared to age-matched HSPCs. The majority of these pAML were categorized as “above” baseline pAML by having a higher mutation load. These pAML often had a more differentiated phenotype and significantly more genetic driver events. These observations fit with a model in which more differentiated progenitors require more genetic driver events to transform into a leukemic cell of origin. Cells with an elevated mutation frequency have an increased chance of harboring an oncogenic hit (1). Indeed, we observe a significant correlation between genome-wide mutation burden and numbers of drivers, suggesting that the “above” baseline AML mutation accumulation in the preleukemic cells was rate limiting for the initiation of disease. Of note, in this study, we used the clonal mutations of bulk AML blasts, which are the mutations that are shared by all blasts and thus represent the MRCA and likely the cell of origin of the leukemia.
Besides SBS1, the only other process that could explain the increased mutation loads in a subset of pAML was oxidative stress–associated mutagenesis by reactive oxygen species (ROS), as reflected by SBS18 (31, 32). The source of the ROS is most likely endogenous, because the normal HSPCs of the same bone marrow do not show enhanced SBS18. Alternatively, the leukemic cell of origin might have been more sensitive yet received the same exposure to oxidative stress as the other HSPCs (33). ROS might be produced as a result of replication stress (40), which could be caused by the high cell division rate during fetal development. ROS can in turn induce direct DNA damage or indirect via interference with the replication machinery that might lead to chromosome instability (41). Standard-of-care chemotherapeutics for pAML, such as cytarabine and anthracyclines, have been described to result in increased intracellular ROS production (42, 43). The presence of SBS18 in pAML might also indicate that these cells are more sensitive to ROS and thus to chemotherapeutics inducing ROS, possibly explaining the better survival of the “above” baseline pAML group.
The t(8;21) pAML subtype was enriched in the “above” baseline group, and this subtype normally has a favorable prognosis (12). However, the inv(16) subtype, which forms the core-binding factor AML group together with t(8;21), is also associated with a favorable prognosis yet was found more frequently in the “on” baseline group with poorer survival. Nonetheless, the limited number of inv(16) pAML cases in this cohort prevents strong conclusions. We correlated the “on” baseline group to a more stem cell/early progenitor state using RNA expression data and propose that leukemogenesis in this group occurs in a noncommitted progenitor. This idea may explain the similar mutation load and profiles as compared to healthy HPSCs. Our data are in line with observations that the cell of origin in which the leukemogenesis occurs influences AML gene expression and drug response, as was shown with murine cells transduced with MLL–AF9 (44). In this study, HSC-derived AML were more resistant to chemotherapy than more committed progenitor-derived AML, and the HSC-derived AML gene expression correlated with poorer prognosis in MLL-AML (44).
Together, our study provides new insights into the etiology of pAML and underscores the clinical potential of WGS in pediatric leukemia for patient stratification.
Methods
See Supplementary Table S10 for key resources.
Patient Samples and Clinical Data
Bone marrow mononuclear cells from healthy children were collected in the context of donation for allogeneic HSCT. These children had no known genetic predisposition toward cancer and were unrelated to the patients with AML analyzed in this study. Residual bone marrow mononuclear cells, left over after diagnostic testing of graft viability, were viably frozen in the HSCT Biobank of the UMC Utrecht. Use of this material was approved by the Biobank Committee of the UMC Utrecht (study TCBIO18–231) and by the Medical Ethical Committee Utrecht (study 19–243). Written informed consent was provided by all children and/or their legal guardians. Bone marrow mononuclear cells at diagnosis of nine patients with pAML were obtained from the Biobank of the Princess Máxima Center (study PMCLAB2018–007). Data on subtype and neutrophil time after chemotherapy of pAML were also obtained from the Biobank of the Princess Máxima Center (study PMCLAB2019–051). This study included 237 primary pAML cases diagnosed in the Netherlands between 2005 and 2019. Both studies were approved by the Biobank and Data Access Committee. Patient written informed consents were obtained by the Princess Máxima Center.
FACS and Clonal HSPC Cultures
Bone marrow mononuclear cells were stained for FACS after thawing using the following surface markers for each cell population (5): HSC, CD34+CD38−CD45RA−CD90+Lin−CD11c−CD16−, and MPP, CD34+CD38−CD45RA−CD90−Lin−CD11c−CD16−. AML blasts were defined on the basis of diagnostic immunophenotyping data, and AML blasts of all patients were CD33+ (Supplementary Table S2). Different cell populations were purified on a SH800S Cell Sorter (Sony). A representative example of sorted populations is shown in Supplementary Fig. S2A. Flow cytometry data were analyzed using FlowJo software. After sorting a bulk AML blast population into a collection tube, single-cell HSCs and MPPs were index sorted into flat-bottom, 384-well plates containing 75 μL HSPC culture medium. HSPC culture medium consisted of StemSpan SFEM medium supplemented with SCF (100 ng/mL), FLT3-L (100 ng/mL), TPO (50 ng/mL), IL-6 (20 ng/mL), and IL-3 (10 ng/mL), and cells were cultured at 37°C, 5% CO2, for 4 to 5 weeks before collection. Polyclonal mesenchymal stromal cell (MSC) cultures were established from a fraction of bone marrow cells by plating cells in 12-well culture dishes in Advanced DMEM/F-12 medium (Gibco) supplemented with 10% FBS. MSCs were kept in culture for 2 weeks, and medium was replaced every other day to remove nonadherent cells.
FACS Antibodies
All antibodies were obtained from BioLegend. Antibodies used for AML blast and HSPC populations were as follows: CD34-BV421 (clone 561, 343609, 1:20), CD38-PE (clone HIT2, 303505, 1:50), CD45RA-PerCP/Cy5.5 (clone HI100, 304121, 1:20), CD90-APC (clone 5E10, 328113, 1:200), Lineage (CD3/CD14/CD19/CD20/CD56)-FITC (clones UCHT1, HCD14, HIB19, HCD56, 348701, 1:20), CD11c-FITC (clone 3.9, 301603, 1:20), CD16-FITC (clone 3G8, 302005, 1:100), and CD33-PE/Cy7 (clone WM53, 303433, 1:100).
WGS and Read Alignment
DNA libraries for Illumina sequencing were generated by using standard protocols (Illumina) from 50 to 150 ng of genomic DNA isolated from the clonally expanded HSPCs using QIAamp DNA Micro Kit (QIAGEN) according to manufacturers' instructions. For the MSC and AML blast samples, 100 to 300 ng of genomic DNA was used as input. All samples were sequenced (2 × 150 bp) by using Illumina HiSeq X Ten or NovaSeq 6000 sequencers to 30× base coverage (pAML, MSCs, 26 HSPC clones) or to 15× base coverage (29 HSPC clones). WGS data were mapped against human reference genome GRCh38 by using Burrows-Wheeler Aligner v0.7.5a mapping tool (45) with settings “bwa mem -c 100 -M”. Sequence reads were marked for duplicates by using Sambamba v0.6.8 markdup. Full pipeline description and settings are also available at https://github.com/UMCUGenetics/IAP.
Mutation Calling and Filtering
Raw variants were multisample called by using the GATK HaplotypeCaller v3.8–1-0 (46) and GATK-Queue v3.8–1-0 with default settings and additional option “EMIT_ALL_CONFIDENT_SITES.” The quality of variant and reference positions was evaluated by using GATK VariantFiltration v3.8–1-0 with options -snpFilterName SNP_LowQualityDepth -snpFilterExpression “QD < 2.0” -snpFilterNameSNP_MappingQuality -snpFilterExpression “MQ < 40.0” -snpFilterName SNP_StrandBias -snpFilterExpression “FS > 60.0” -snpFilterName SNP_HaplotypeScoreHigh -snpFilterExpression “HaplotypeScore > 13.0” -snpFilterName SNP_MQRankSumLow -snpFilterExpression “MQRankSum < -12.5” -snpFilterName SNP_ReadPosRankSumLow -snpFilterExpression “ReadPosRankSum < -8.0” -snpFilterName SNP_HardToValidate -snpFilterExpression “MQ0 > = 4 && ((MQ0/(1.0 * DP)) > 0.1)” -snpFilterName SNP_LowCoverage -snpFilterExpression “DP < 5” -snpFilterName SNP_VeryLowQual -snpFilterExpression “QUAL < 30” -snpFilterName SNP_LowQual -snpFilterExpression “QUAL > = 30.0 && QUAL < 50.0 “ -snpFilterName SNP_SOR -snpFilterExpression “SOR > 4.0” -cluster 3 -window 10 -indelType INDEL -indelType MIXED -indelFilterName INDEL_LowQualityDepth -indelFilterExpression “QD < 2.0” -indelFilterName INDEL_StrandBias -indelFilterExpression “FS > 200.0” -indelFilterName INDEL_ReadPosRankSumLow -indelFilterExpression “ReadPosRankSum < -20.0” -indelFilterName INDEL_HardToValidate -indelFilterExpression “MQ0 > = 4 && ((MQ0/(1.0 * DP)) > 0.1)” -indelFilterName INDEL_LowCoverage -indelFilterExpression “DP < 5” -indelFilterName INDEL_VeryLowQual -indelFilterExpression “QUAL < 30.0” -indelFilterName INDEL_LowQual -indelFilterExpression “QUAL > = 30.0 && QUAL < 50.0” -indelFilterName INDEL_SOR -indelFilterExpression “SOR > 10.0.” To obtain high-quality somatic mutation catalogs, we applied postprocessing filters as described previously (4). Briefly, we considered variants at autosomal chromosomes without any evidence from a paired control sample (MSCs isolated from the same bone marrow); passed by VariantFiltration with a GATK phred-scaled quality score R 100; a base coverage of at least 5× in the clonal and paired control sample; mapping quality (MQ) of 60; no overlap with single nucleotide polymorphisms (SNP) in the Single Nucleotide Polymorphism Database v146; and absence of the variant in a panel of unmatched normal human genomes (BED-file available upon request). We additionally filtered base substitutions with a GATK genotype score (GQ) lower than 99 or 10 in clonal or paired control sample, respectively. For indels, we filtered variants with a GQ score lower than 99 in both clonal and paired control sample (4, 47). We used Bayesian Dirichlet modeling to check the clonality of the clones as described previously (23, 25). Mutations with a VAF ≥0.3 were assumed to be clonal. For TARGET AML, the VAF threshold was adjusted according to the estimated purity based on the VAF plot. The script is available at https://github.com/ToolsVanBox/SMuRF. For downsampling of the 30× coverage sequenced HSPC clones of PMC16332 to 15× coverage, 50% of the mapped reads were randomly selected using “samtools (v1.0) view -s 0.5.” After this, mutation calling and filtering were performed as described above.
Construction of Phylogenetic Trees
For the construction of phylogenetic trees, an initial phylogenetic tree was constructed by cataloging somatic base substitutions and indels that were shared between two or more HSPC clones and completely absent in at least one other HSPC clone (or the patient-matched AML). To exclude germline variants, variants that were clonally present in the MSC control samples were filtered out, but variants subclonally present in MSC control that passed all quality checks were included in the phylogenetic tree, as they represent mutations that were acquired early during embryonic development. All of these shared mutations were manually inspected using Integrative Genomics Viewer (IGV) and false-positive calls were excluded from the final tree. The number of unique and shared mutations as well as the relationship between the clones deduced from the shared mutations were visualized using UpSetR package in R (48).
Healthy Baseline
The number of single-nucleotide variants (SNV) or indels reported are normalized for the length of CALLABLE loci reported by GATK (v3.8–1-0) CallableLoci. For the slope estimation, the linear mixed model was used to take donor dependency into account and the P values are indicated in the figures using lme4 packege (49) in R with “age” as fixed effect and “(1 + age | Donor)” as random effects. The 95% CI was calculated using ggpredict package in R (50). The statistical significance was computed using ggsignif package in R (https://github.com/const-ae/ggsignif). The R2 values were calculated by using the r.squaredGLMM in R (https://cran.r-project.org/src/contrib/MuMIn_1.43.17.tar.gz).
TARGET Patient Selection
We obtained WGS data of bone marrow mononuclear cells and ex vivo expanded MSCs from 31 patients with pAML at diagnosis from the TARGET initiative, termed TARGET-21 patients (13). For one patient, the pipeline failed. All remaining 30 patients were included, except when two of three criteria were below limits: (i) karyotype plots indicate sample is not pure, (ii) blast percentage in bone marrow <80%, and (iii) purity estimate based on VAF <80%. For several patients, criterion 1 was uninformative, because these patients had karyotypically normal AML; for two of these patients, the purity estimate was below the threshold, and, therefore, these patients were excluded. The remaining 18 patients were further analyzed, including SV analysis. Three patients had abnormal SVs, with SVs on each chromosome, while karyotypes were normal, for which we could not find an explanation in the SV calling analysis. Therefore, these three patients were excluded too. The final cohort consisted of 15 TARGET-21 pAML patients (Supplementary Table S5).
In addition, we analyzed somatic mutation calls of 197 patients with pAML that were whole genome sequenced using CGI as part of the TARGET-20 cohort (7), a separate cohort of pAML patients. Mutations with a VAF >0.3 were assumed to be clonal and included in the analyses. pAML samples with an estimated purity ≥80% and at least 40 clonal mutations were included in our analysis, leading to 103 pAML samples in our second TARGET-20 pAML cohort.
Driver Analysis
We applied the following filtering to obtain potential driver variants to the IAP output: passed by VariantFiltration with high or moderate expected effect on the gene reported by SnpEff annotation with a GATK phred-scaled quality score R 60; a base coverage of at least 10× in the clonal and paired control sample; MQ of 30; no overlap with SNPs in the Single Nucleotide Polymorphism Database v146; and located in commonly identified driver genes. We additionally filtered base substitutions with a GATK genotype score (GQ) lower than 10 for homozygous SNV in sample and its paired control, 20 and 10 for heterozygous SNV in sample and its paired control, or 60 for homozygous and heterozygous indels in sample, respectively. Large SVs were detected using grids-purple-linx pipeline (https://github.com/hartwigmedical/gridss-purple-linx) with grids v2.7.2, amber v3.2, cobalt v1.7, purple v2.34, linx v0.69–6, and circos v0.69–9. Chromosomal copy-number alteration was detected using Control-FREEC v11.4 as a part of IAP pipeline (https://github.com/UMCUGenetics/IAP). To calculate the total number of cancer-driving events, we combined the SNV and indel drivers with large SVs, including fusion genes, trisomy, or loss of heterozygosity (Supplementary Table S6). The majority of SNV and indel drivers were in known adult or pediatric hematologic cancer driver genes (75%; refs. 13, 18).
Mutational Profile and Signature Analysis
First, the pooled HSPC profiles from healthy donors and patients with AML were compared, and cosine similarity was calculated for SNVs and indels. On the basis of healthy HSPC data, the linear mixed model was used to estimate the annual accumulation for the 96-mutation type to model the expected mutational profile for every age. Cosine similarity was calculated to indicate how similar our prediction and experimental data were. Then, the AML and its matching HSPC profiles were compared with the cosine similarity. De novo mutational signature extraction was performed using the data reported here, in combination with genome-wide mutation data of healthy stem cells of human small intestine, colon, and liver (4) as described previously (5). For this, we applied nonnegative matrix factorization using an in-house developed R package “MutationalPatterns” (51). Four signatures were extracted on the basis of the residual sum of squares (RSS) plot, as the inflection point between the input matrix and its estimate is at rank 4. The four de novo extracted signatures were compared to the COSMIC v3 signatures (22) and based on a cosine similarity being >0.85 were identified as SBS1, 5, 18 and “HSPC” (Fig. 1D). These four COSMIC signatures were subsequently used to refit and reconstruct the original AML blast and HSPC spectra, which allowed us to obtain their relative contributions. MutationalPatterns was used for the SNV signature analysis with our newly developed functions for indel profile analysis (https://github.com/FreekManders/MutationalPatterns).
MIP Analysis of SNVs
AML blast-specific MIPs of PMC22813 were designed as described previously (52, 53). The genomic regions of interest were captured using 15 to 50 ng DNA of 89 HSPC clones. MIP reads were mapped to the human reference genome (GRCh38) using BWA-mem algorithm with -M option (45). The depth at every targeted position was determined using sambamba v0.6.8 depth, excluding positions without a coverage and any positions with log(coverage) lower than 95% CI. Out of 87 positions, or variants, which passed the coverage cutoff, 85 of them were confirmed in AML blast DNA, resulting in a 97.7% true-positive rate (Supplementary Fig. S3A). Median coverage over 87 positions was determined in 89 HSPC clones and any HSPC with a coverage <100 reads was excluded. Sixty-four HSPCs passed the coverage cutoff, and none of them shared any of the unique AML blast mutations (Supplementary Table S3).
PCR for MLL Fusion
PCR for the MLL fusion of PMC21636 was performed using GoTaq DNA polymerase (Promega) according to manufacturer's protocol with 1 ng of DNA as input per sample. A “touchdown” PCR protocol was used with 15 cycles of (i) 30 seconds 92°C, (ii) 30 seconds 65°C with a decrement of 0.2°C per cycle, and (iii) 60 seconds 72°C followed by 30 cycles of 30 seconds 92°C, 30 seconds 58°C, and 60 seconds 72°C. Primers used were as follows: MLL-fusion specific (F) fw 5′-GGAACATGGACATTCCTTTGA-3′ and rv 5′-GCAGCAGTTATTTTTGGACTCA-3′; wild-type MLL (WT) fw 5′-TCCTGGGGTACAAAGAAGCA-3′ and rv 5′-CACAGGAGGATTGTGAAGCA-3′.
Random Forest and Regression Model
For the random forest model, 70 HSCs and 70 MPPs were randomly sampled from the healthy baseline HSC and MPP clones as well as from another study using bone marrow–derived HSC and progenitor clones to prevent sampling bias (5, 21). The relative contribution of the 96 mutation types and the total mutation count were used as features. This total mutation count was normalized to the donor age using the log2 value of the total mutation count divided by the mean number of mutations in all cells of this donor. In addition, the genome was binned in nine bins based on replication timing, and relative mutation burden per bin was used as feature. The random forest model, consisting of 1,000 trees and with mtry = 10, was then trained using the R package randomForest. For the regression model, the ratio between the observed number of somatic mutations in pAML and the expected number based on the healthy baseline was calculated (range 0.45–4.81) and used as label. The cosine similarity to mutational signatures and the relative contribution of seven mutation types (C>A, C>G, C>T, C>T at CpG, T>A, T>C, T>G) were used as features. The 118 TARGET patients were divided in four equal parts. The last 25% of the data, combined with six PMC patients, was used as hold out data to validate the model (n = 37). The remaining 75% of the data was used as training data. The training data (n = 87) was split in five equal folds, and backward feature selection was performed in a linear regression model to determine the optimal number of features, using R package “caret.” The best model was the one resulting in the lowest Root Mean Square Error (RMSE). We trained a final model with the selected features on all training data and validated this model on the hold out data.
RNA-Sequencing Data Analysis
For 15 TARGET-21 and 88 TARGET-20 patients, RNA-sequencing data were available of the pAML at diagnosis, which were analyzed as separate cohorts. Differential gene expression (DE) analysis was performed using the DESeq2 package v1.30.1 in R (54), and significant DE was determined with a padj < 0.05. HOX metagene expression was calculated per sample as the average normalized gene expression of the 19 HOX genes depicted in Fig. 6F. The ComplexHeatmap package v2.6.2 in R (55) was used to visualize the HOX gene heatmap; data were scaled to mean 0. For GSEA, log fold change shrinkage was applied to the data using the “ashr” method v2.2–47 (56). Subsequently, the fgsea package v1.16.0 was used to perform GSEA with the cell-type signature gene sets (C8) of MSigDB in R (57, 58).
Statistical Analysis
Unpaired two-tailed Student t test was used for direct comparisons of under the assumption of normal distributions, while Mann–Whitney tests were performed for data where a normal distribution was not likely. The statistical parameters are described in the individual figure legends and the related Methods section. For the survival analyses, event-free survival and overall survival were estimated with the Kaplan–Meier method and compared using log-rank tests. Statistical analyses were performed using GraphPad Prism 9 and R studio. AP value less than 0.05 was considered statistically significant.
Resource, Data, and Code Availability
Resource Availability.
Further information and requests for resources should be directed to and will be fulfilled by Ruben van Boxtel (R.vanBoxtel@prinsesmaximacentrum.nl).
Data Availability.
The accession numbers for the WGS data reported in this article are EGAS00001004593 and database of Genotypes and Phenotypes (dbGaP) phs000218 for the TARGET initiative AML data.
Code Availability.
Mutation calling and filtering pipelines are available at https://github.com/UMCUGenetics/IAP and https://github.com/ToolsVanBox/SMuRF, new functions for mutational patterns at https://github.com/FreekManders/MutationalPatterns, and MIP analysis script at https://github.com/ToolsVanBox/smMIPfil. The other scripts are available on request.
Authors' Disclosures
R. van Boxtel reports grants from Dutch Cancer Society (KWF) during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
A.M. Brandsma: Conceptualization, formal analysis, investigation, visualization, writing–original draft, project administration, writing–review and editing. E.J.M. Bertrums: Resources, investigation. M.J. van Roosmalen: Software, formal analysis. D.A. Hofman: Formal analysis. R. Oka: Formal analysis. M. Verheul: Investigation. F. Manders: Software, formal analysis. J. Ubels: Software, formal analysis. M.E. Belderbos: Resources, investigation. R. van Boxtel: Conceptualization, supervision, writing–original draft, project administration, writing–review and editing.
Supplementary Material
Acknowledgments
This study was financially supported by two grants from the Dutch Cancer Society (KWF Research Project 12682 and 11307) to R. van Boxtel. The authors thank the Hartwig Medical Foundation (Amsterdam, the Netherlands) for facilitating low-input WGS. The results published here are in part based upon data generated by the TARGET initiative managed by the NCI. The data used for this analysis are available at the dbGaP (accession number: phs000218). Information about TARGET can be found at http://ocg.cancer.gov/programs/target.
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
Blood Cancer Discov 2021;2:484–99
References
- 1.Tomasetti C, Vogelstein B.Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 2015;347:78–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Magalhães JP.How ageing processes influence cancer. Nat Rev Cancer 2013;13:357–65. [DOI] [PubMed] [Google Scholar]
- 3.Rozhok AI, Salstrom JL, DeGregori J.Stochastic modeling reveals an evolutionary mechanism underlying elevated rates of childhood leukemia. Proc Natl Acad Sci U S A 2016;113:1050–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blokzijl F, de Ligt J, Jager M, Sasselli V, Roerink S, Sasaki N, et al. Tissue-specific mutation accumulation in human adult stem cells during life. Nature 2016;538:260–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osorio FG, Rosendahl Huber A, Oka R, Verheul M, Patel SH, Hasaart K, et al. Somatic mutations reveal lineage relationships and age-related mutagenesis in human hematopoiesis. Cell Rep 2018;25:2308–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell 2012;150:264–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma X, Liu Y, Liu Y, Alexandrov LB, Edmonson MN, Gawad C, et al. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature 2018;555:371–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gröbner SN, Worst BC, Weischenfeldt J, Buchhalter I, Kleinheinz K, Rudneva VA, et al. The landscape of genomic alterations across childhood cancers. Nature 2018;555:321–7. [DOI] [PubMed] [Google Scholar]
- 9.Klein K, de Haas V, Kaspers GJL.Clinical challenges in de novo pediatric acute myeloid leukemia. Expert Rev Anticancer Ther 2018;18:277–93. [DOI] [PubMed] [Google Scholar]
- 10.Pui CH, Nichols KE, Yang JJ.Somatic and germline genomics in paediatric acute lymphoblastic leukaemia. Nat Rev Clin Oncol 2019;16:227–40. [DOI] [PubMed] [Google Scholar]
- 11.de Rooij JDE, Zwaan CM, van den Heuvel-Eibrink M.Pediatric AML: from biology to clinical management. J Clin Med 2015;4:127–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zwaan CM, Kolb EA, Reinhardt D, Abrahamsson J, Adachi S, Aplenc R, et al. Collaborative efforts driving progress in pediatric acute myeloid leukemia. J Clin Oncol 2015;33:2949–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolouri H, Farrar JE, Triche T, Ries RE, Lim EL, Alonzo TA, et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat Med 2018;24:103–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farrar JE, Schuback HL, Ries RE, Wai D, Hampton OA, Trevino LR, et al. Genomic profiling of pediatric acute myeloid leukemia reveals a changing mutational landscape from disease diagnosis to relapse. Cancer Res 2016;76:2197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature 2014;506:328–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greaves M.A causal mechanism for childhood acute lymphoblastic leukaemia. Nat Rev Cancer 2018;18:471–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liggett LA, DeGregori J.Changing mutational and adaptive landscapes and the genesis of cancer. Biochim Biophys Acta Rev Cancer 2017;1867:84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ju YS, Martincorena I, Gerstung M, Petljak M, Alexandrov LB, Rahbari R, et al. Somatic mutations reveal asymmetric cellular dynamics in the early human embryo. Nature 2017;543:714–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helleday T, Eshtad S, Nik-Zainal S.Mechanisms underlying mutational signatures in human cancers. Nat Rev Genet 2014;15:585–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maura F, Degasperi A, Nadeu F, Leongamornlert D, Davies H, Moore L, et al. A practical guide for mutational signature analysis in hematological malignancies. Nat Commun 2019;10:2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee-Six H, Øbro NF, Shepherd MS, Grossmann S, Dawson K, Belmonte M, et al. Population dynamics of normal human blood inferred from somatic mutations. Nature 2018;561:473–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, et al. The repertoire of mutational signatures in human cancer. Nature 2020;578:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hasaart KAL, Manders F, van der Hoorn ML, Verheul M, Poplonski T, Kuijk E, et al. Mutation accumulation and developmental lineages in normal and Down syndrome human fetal haematopoiesis. Sci Rep 2020;10:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyamoto T, Weissman IL, Akashi K.AML1/ETO-expressing nonleukemic stem cells in acute myelogenous leukemia with 8;21 chromosomal translocation. Proc Natl Acad Sci U S A 2000;97:7521–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, Greenman CD, Lau KW, et al. The life history of 21 breast cancers. Cell 2012;149:994–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desai P, Hassane D, Roboz GJ.Clonal hematopoiesis and risk of acute myeloid leukemia. Best Pract Res Clin Haematol 2019;32:177–85. [DOI] [PubMed] [Google Scholar]
- 27.Williams N, Lee J, Moore L, Baxter EJ, Hewinson J, Dawson KJ, et al. Phylogenetic reconstruction of myeloproliferative neoplasm reveals very early origins and lifelong evolution. BioRxiv 2020.11.09.374710 [Preprint]. 2020. Available from: 10.1101/2020.11.09.374710. [DOI]
- 28.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alexandrov LB, Jones PH, Wedge DC, Sale JE, Campbell PJ, Nik-Zainal S, et al. Clock-like mutational processes in human somatic cells. Nat Genet 2015;47:1402–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bowie MB, McKnight KD, Kent DG, McCaffrey L, Hoodless PA, Eaves CJ.Hematopoietic stem cells proliferate until after birth and show a reversible phase-specific engraftment defect. J Clin Invest 2006;116:2808–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pilati C, Shinde J, Alexandrov LB, Assié G, André T, Hélias-Rodzewicz Z, et al. Mutational signature analysis identifies MUTYH deficiency in colorectal cancers and adrenocortical carcinomas. J Pathol 2017;242:10–5. [DOI] [PubMed] [Google Scholar]
- 32.Kucab JE, Zou X, Morganella S, Joel M, Nanda AS, Nagy E, et al. A compendium of mutational signatures of environmental agents. Cell 2019;177:821–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forster VJ, Nahari MH, Martinez-Soria N, Bradburn AK, Ptasinska A, Assi SA, et al. The leukemia-associated RUNX1/ETO oncoprotein confers a mutator phenotype. Leukemia 2016;30:250–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pession A, Lo Nigro L, Montemurro L, Serravalle S, Fazzina R, Izzi G, et al. ArgBP2, encoding a negative regulator of ABL, is fused to MLL in a case of infant M5 acute myeloid leukemia involving 4q35 and 11q23 [10]. Leukemia 2006;20:1310–3. [DOI] [PubMed] [Google Scholar]
- 35.Alharbi RA, Pettengell R, Pandha HS, Morgan R.The role of HOX genes in normal hematopoiesis and acute leukemia. Leukemia 2013;27:1000–8. [DOI] [PubMed] [Google Scholar]
- 36.Zheng S, Papalexi E, Butler A, Stephenson W, Satija R.Molecular transitions in early progenitors during human cord blood hematopoiesis. Mol Syst Biol 2018;14:e8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hay SB, Ferchen K, Chetal K, Grimes HL, Salomonis N.The Human Cell Atlas bone marrow single-cell interactive web portal. Exp Hematol 2018;68:51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishikii H, Kurita N, Chiba S.The road map for megakaryopoietic lineage from hematopoietic stem/progenitor cells. Stem Cells Transl Med 2017;6:1661–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Unnisa Z, Clark JP, Roychoudhury J, Thomas E, Tessarollo L, Copeland NG, et al. Meis1 preserves hematopoietic stem cells in mice by limiting oxidative stress. Blood 2012;120:4973–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marchetti MA, Weinberger M, Murakami Y, Burhans WC, Huberman JA.Production of reactive oxygen species in response to replication stress and inappropriate mitosis in fission yeast. J Cell Sci 2006;119:124–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coluzzi E, Leone S, Sgura A.Oxidative stress induces telomere dysfunction and senescence by replication fork arrest. Cells 2019;8:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iacobini M, Menichelli A, Palumbo G, Multari G, Werner B, Del Principe D.Involvement of oxygen radicals in cytarabine-induced apoptosis in human polymorphonuclear cells. Biochem Pharmacol 2001;61:1033–40. [DOI] [PubMed] [Google Scholar]
- 43.Doroshow JH.Prevention of doxorubicin-induced killing of MCF-7 human breast cancer cells by oxygen radical scavengers and iron chelating agents. Biochem Biophys Res Commun 1986;135:330–5. [DOI] [PubMed] [Google Scholar]
- 44.Krivtsov AV, Figueroa ME, Sinha AU, Stubbs MC, Feng Z, Valk PJM, et al. Cell of origin determines clinically relevant subtypes of MLL-rearranged AML. Leukemia 2013;27:852–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, Durbin R.Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26:589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Depristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jager M, Blokzijl F, Sasselli V, Boymans S, Janssen R, Besselink N, et al. Measuring mutation accumulation in single human adult stem cells by whole-genome sequencing of organoid cultures. Nat Protoc 2018;13:59–78. [DOI] [PubMed] [Google Scholar]
- 48.Conway JR, Lex A, Gehlenborg N.UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 2017;33:2938–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bates D, Mächler M, Bolker BM, Walker SC.Fitting linear mixed-effects models using lme4. J Stat Softw 2015;67:1–48. [Google Scholar]
- 50.Lüdecke D.ggeffects: tidy data frames of marginal effects from regression models. J Open Source Softw 2018;3:772. [Google Scholar]
- 51.Blokzijl F, Janssen R, van Boxtel R, Cuppen E.MutationalPatterns: comprehensive genome-wide analysis of mutational processes. Genome Med 2018;10:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hiatt JB, Pritchard CC, Salipante SJ, O'Roak BJ, Shendure J.Single molecule molecular inversion probes for targeted, high-accuracy detection of low-frequency variation. Genome Res 2013;23:843–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu J, Antić Ž, van Reijmersdal SV, Hoischen A, Sonneveld E, Waanders E, et al. Accurate detection of low-level mosaic mutations in pediatric acute lymphoblastic leukemia using single molecule tagging and deep-sequencing. Leuk Lymphoma 2018;59:1690–9. [DOI] [PubMed] [Google Scholar]
- 54.Love MI, Huber W, Anders S.Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gu Z, Eils R, Schlesner M.Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016;32:2847–9. [DOI] [PubMed] [Google Scholar]
- 56.Stephens M.False discovery rates: a new deal. Biostatistics 2016;18:kxw041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov MN, Sergushichev A.Fast gene set enrichment analysis. BioRxiv 060012 [Preprint]. 2021. Available from: 10.1101/060012. [DOI]
- 58.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession numbers for the WGS data reported in this article are EGAS00001004593 and database of Genotypes and Phenotypes (dbGaP) phs000218 for the TARGET initiative AML data.