Summary
Regulatory sequences or erroneous incorporations during DNA transcription cause RNA polymerase backtracking, and inactivation in all kingdoms of life. Reactivation requires RNA transcript cleavage. Essential transcription factors (GreA/GreB, or TFIIS) accelerate this reaction. We report four cryo-EM reconstructions of Escherichia coli RNA polymerase representing the entire reaction pathway: A backtracked complex (i); a backtracked complex with GreB before (ii), and after (iii) RNA cleavage; and a reactivated, substrate bound complex with GreB before RNA extension (iv).
Compared with eukaryotes, the backtracked RNA adopts a different conformation. RNA polymerase conformational changes cause distinct GreB states: i) a fully engaged GreB before cleavage; ii) a disengaged GreB after cleavage; and iii) a dislodged, loosely bound GreB removed from the active site to allow RNA extension. These reconstructions give insights on the catalytic mechanism and dynamics of RNA cleavage and extension, and suggest how GreB targets backtracked complexes without interfering with canonical transcription.
Keywords: Transcription, RNA polymerase structure, transcriptional arrest, backtracking, GreB, transcriptional rescue, Cryo-EM
Introduction
Transcription of DNA into RNA is the first step in gene expression. DNA-dependent RNA polymerase (RNAP), a universally conserved protein enzyme, carries out transcription with high accuracy (subunit composition α 2 ββ′ω). RNAP uses one DNA strand as template to synthesize the complementary RNA. The RNA first forms a 9 to 10 base pair long RNA-DNA hybrid before it is guided into the RNA exit channel. The 3′-end of the RNA transcript occupies the product-site (P-site, also called i-site, register -1) in the enzyme active site at the beginning of the nucleotide addition cycle. To extend the RNA, a nucleoside triphosphate (rNTP) enters the active site and base pairs with the template DNA in the acceptor-site (A-site, also called i+1-site, register +1). The 3′-OH of the RNA attacks the correctly paired rNTP substrate to extend the transcript by one base. Forward translocation of RNAP by one base pair relative to the DNA concludes the nucleotide addition cycle and repositions the RNA 3′-end in the P-site (Gnatt et al., 2001; Vassylyev et al., 2007)(Figure 1A, top).
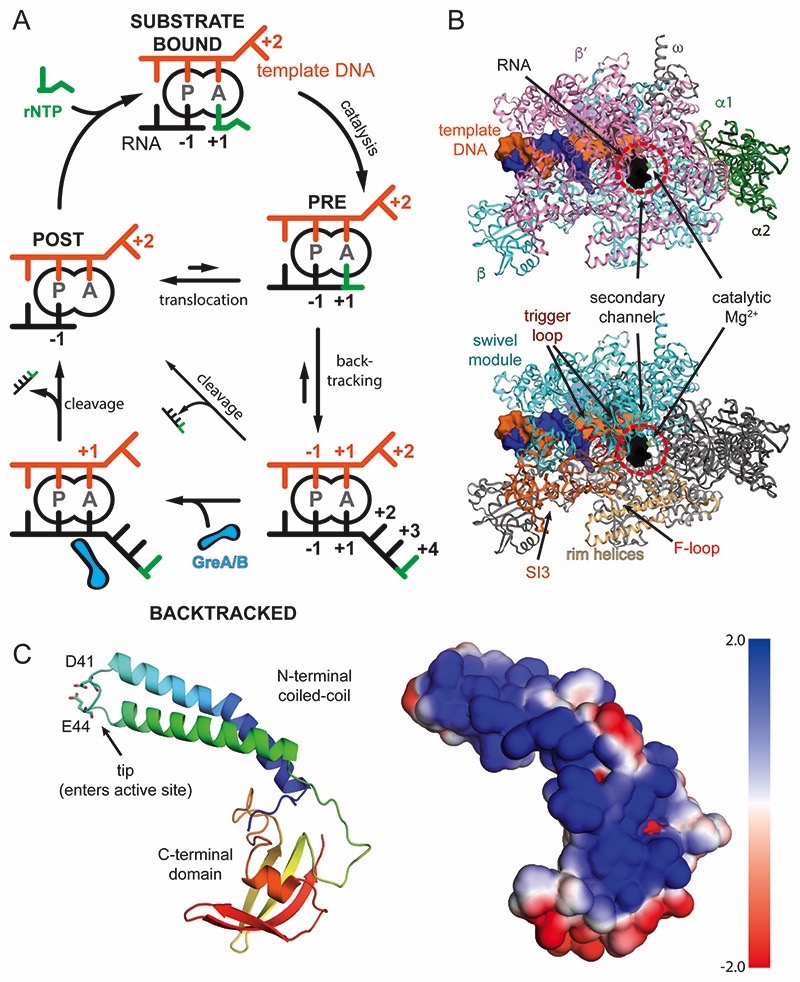
Figure 1. Schematic of transcription, RNAP, and GreB.
(A) Nucleotide addition and backtracking: In the post-translocated state (POST), the RNA 3′ end occupies the P-site, while the A-site holds the +1 template DNA, which dictates the next rNTP substrate to bind (green, top). Catalysis extends the RNA giving rise to the pre-translocated state (PRE). RNAP translocates relative to DNA by one base pair and concludes the cycle. Misincorporations, weak RNA-DNA hybrids or pause signals can cause backtracking and extrusion of the RNA from the active site (bottom, right). Reactivation requires forward translocation (slow, upward arrow), intrinsic RNA cleavage (slow, diagonal arrow), or Gre-factor assisted cleavage (bottom, left). (B) RNAP structure: View into the secondary channel with the five subunits, DNA, and RNA indicated (top). RNAP modules, which will be referred to throughout the manuscript are indicated (bottom) (C) GreB contains two domains. The N-terminal coiled-coil inserts its tip with two conserved acidic residues into the active site through the secondary channel. The C-terminal domain interacts with the RNAP surface. The electrostatic surface potential of GreB shows one side is highly positively charged and predicted to interact with backtracked RNA (right).
Misincorporations in the growing RNA transcript, regulatory DNA sequences, weak RNA-DNA hybrids, or abortive transcription initiation on the other hand can, despite their distinct nature, induce RNAP to reverse translocate relative to DNA for various distances (Artsimovitch and Landick, 2000; Komissarova and Kashlev, 1997; Lerner et al., 2016; Nudler et al., 1997). This process, termed backtracking, inactivates the transcription machinery because the RNA 3′-end is no longer aligned with the active site. As a result, it cannot be extended, and instead is extruded through a pore in RNAP called the secondary channel (termed pore and funnel in eukaryotic RNAP II) (Cheung and Cramer, 2011; Sekine et al., 2015; Wang et al., 2009). Backtracking can result in long-lived arrested transcription complexes, which require reactivation (Figures 1A and 1B).
Backtracking and subsequent reactivation are important processes involved in proof-reading (Erie et al., 1993). Transcriptional pausing, a temporary halt of the nucleotide addition cycle, is a prominent example for mechanisms involved in regulating gene expression in all kingdoms of life (Jonkers and Lis, 2015; Landick, 2006). Transcriptional pauses can be stabilized and prolonged by RNAP backtracking (Adelman et al., 2005; Landick, 2006; Nechaev et al., 2010). Subsequent forward translocation allows pause escape but this is often too slow. Instead, cleavage of the RNA is required. While the RNAP active site can catalyse endonucleolytic RNA hydrolysis to produce a shortened transcript whose 3′-end is newly engaged with the enzyme active site, this reaction is slow under physiological conditions (Figures 1A and S1A-S1D) (Borukhov et al., 1992; Orlova et al., 1995; Shaevitz et al., 2003; Surratt et al., 1991). A set of elongation factors, including bacterial GreA and GreB, and eukaryotic TFIIS, stimulate the intrinsic RNA cleavage activity of RNAP (Borukhov et al., 1993; Reines et al., 1992). While Gre factors share structure (Stebbins et al., 1995; Vassylyeva et al., 2007) and sequence homology with a variety of secondary channel binding proteins in bacteria (e.g. E. coli DksA, Rnk, and Thermus thermophilus GfhI), the eukaryotic counterpart (TFIIS) is homologous to Gre factors in function but not structure.
Gre factors consist of two domains. The N-terminal α-helical coiled-coil domain (NTD) is about 45Å long and inserts its tip with several conserved residues into the RNAP active site (Opalka et al., 2003; Sekine et al., 2015). This includes conserved acidic residues (E. coli GreB D41 and E44), which were proposed to complement the active site and coordinate a magnesium ion involved in activating the nucleophile (Figure 1C) (Sosunova et al., 2003). Likewise, TFIIS in eukaryotes also contains conserved acidic residues, which were predicted to have a similar role. Mutation of these acidic residues in Gre/TFIIS leads to impaired function (Jeon et al., 1994; Opalka et al., 2003). The globular C-terminal domain (CTD) consists of anti-parallel β-sheets flanked by a small α-helix and is connected through a flexible linker to the NTD. With the exception of cyanobacteria (Imashimizu et al., 2011), all species appear to have at least one cleavage factor (Borukhov et al., 1993; Hausner et al., 2000; Izban and Luse, 1992). Some bacterial species such as E. coli, have two: GreA and GreB. GreA and GreB exhibit different RNA cleavage preferences: GreA-induced cleavage shortens the RNA 3′-end by two to three nucleotides, whereas GreB can cleave fragments longer than three nucleotides (Borukhov et al., 1993). These preferences suggested that GreA targets complexes backtracked by one or two nucleotides, such as during erroneous rNTP incorporation, while GreB targets complexes with longer backtracked RNA (Borukhov et al., 1993; Shaevitz et al., 2003).
Previous efforts to gain a structural understanding of this fundamental process employed crystallographic studies on prokaryotic and eukaryotic RNAP. Short RNA and DNA oligonucleotides forming tailed templates were used along with inactive or chimeric factors (Cheung and Cramer, 2011; Sekine et al., 2015; Wang et al., 2009). For bacterial RNAP, no structures are available that resolved the backtracked RNA in presence of Gre factors. Using single particle electron cryo-microscopy (cryo-EM) on functional RNAP complexes, we obtained reconstructions unaffected by crystal packing forces. The resulting atomic models cover the entire reaction pathway: i) an arrested, backtracked complex, ii) a GreB bound complex before RNA cleavage, iii) a GreB bound complex after cleavage, and iv) a reactivated complex with a bound rNTP substrate and GreB prior to RNA extension. The structures provide new details on the mechanism of intrinsic and transcription factor-assisted RNA cleavage. In addition, the structures provide a more complete picture of the dynamics of RNAP before, during, and after arrest and reactivation.
Results
A backtracked RNA polymerase elongation complex
Short DNA and RNA oligonucleotides were annealed to form a 10 base pair DNA bubble with complementary RNA hybridised to one strand. The RNA contained three mismatched bases at the 3′-end (Figure S1A). This transcription bubble mimic was mixed with purified E. coli RNAP to directly assemble a functional elongation complex (EC) backtracked by three bases (backtracked complex). We monitored RNA cleavage in absence of additional factors. As expected, RNAP slowly cleaves off four nucleotides from the mismatched 3′-end (Figure S1D). To prepare a homogeneous sample for structural analysis, we used an RNA with two phosphorothioate modifications (Figure S1B). These modifications do not affect RNA conformation in a double helix (Smith and Nikonowicz, 2000). However, they sufficiently decrease the intrinsic cleavage rate of RNAP to provide time for EM grid preparation (Tetone et al., 2017). Most importantly, the modifications affected the cleavage rate but not the cleavage position (Methods and Figure S1D).
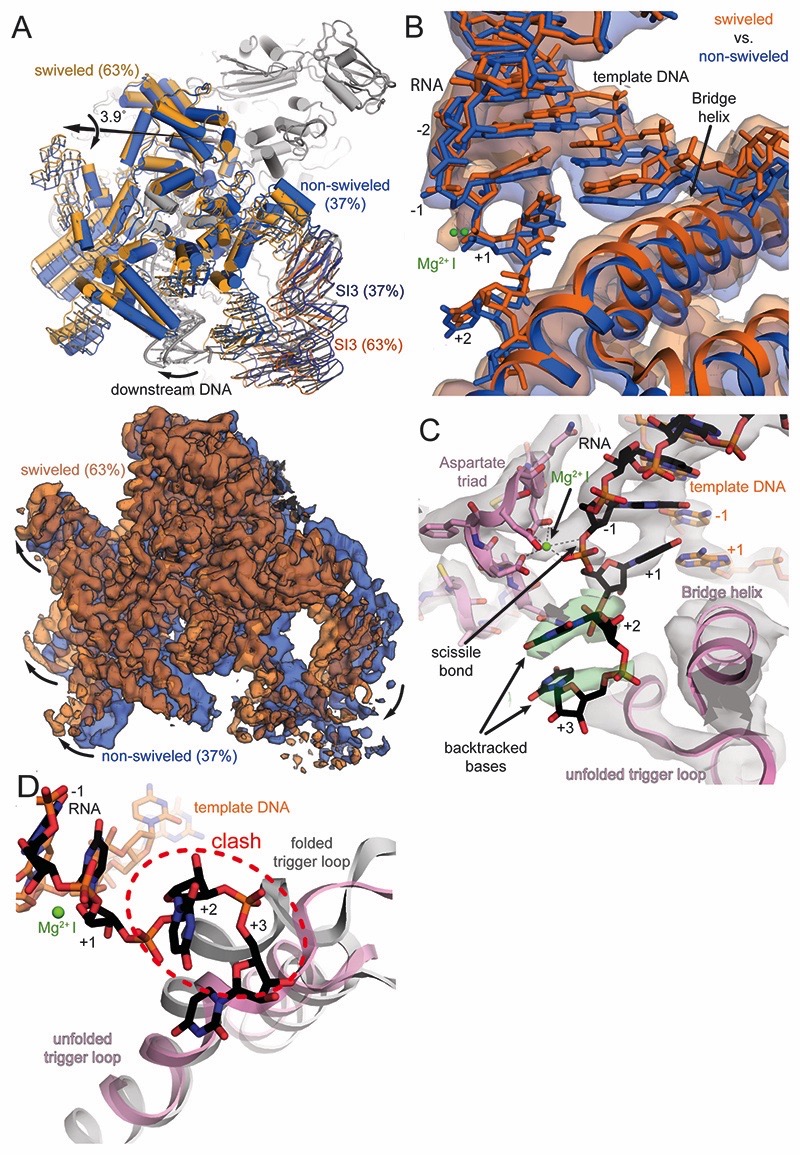
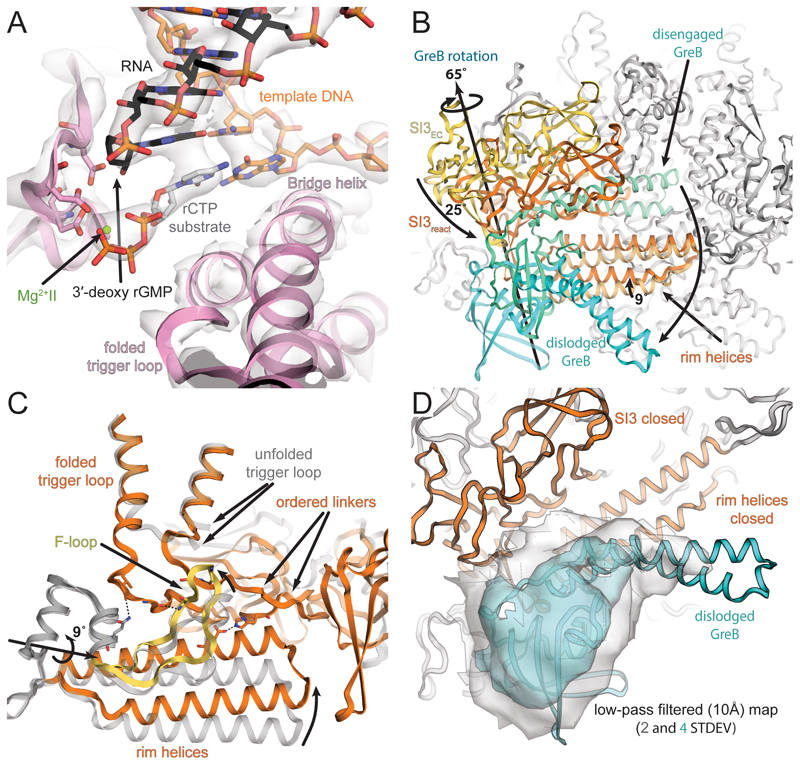
We obtained a 3D reconstruction of the backtracked RNAP complex and assessment of conformational heterogeneity by 3D classification indicated the presence of two populations (Figures 2A and S2, Table S2). The clamp and shelf domain form the swivel module (residues β1244-1342, β′1-499, and β′800-1407, Figure 1B). In paused RNAP complexes, this module has been observed to rotate relative to the core module (2 α-N-terminal domains, β514-828, β1071-1235, and β′504-771, Figure 1B, Table S1) in a plane approximately parallel to the DNA upstream and downstream duplex (Guo et al., 2018; Kang et al., 2018a; 2017). The reconstruction for about one third of the particles refined to a nominal resolution of 3.7Å and the swivel module adopts a conformation less swivelled (~ -1.2˚, Tables S1 and S2, Movie S1) compared to a canonical EC (Kang et al., 2017). The reconstruction for the remaining two thirds of particles refined to a nominal resolution of 3.4Å and the swivel module was rotated ~2.9˚ relative to an EC (Figure 2A, Tables S1 and S2, Movie S1). This is the same direction as observed for a paused EC (Guo et al., 2018; Kang et al., 2018a). Thus, the swivel module can rotate by up to 4˚ in the backtracked state. Importantly, the downstream DNA duplex moves along, which affects the downstream end of the RNA-DNA hybrid because the template DNA shifts relative to the RNA by about 2Å (Figure 2B). We will focus mostly on the swiveled conformation because it comprises the larger fraction of particles and refined to higher resolution (see also Discussion).
Figure 2. RNAP and active site conformation of backtracked complex.
(A) The swivel module (clamp and shelf domain of RNAP, shades of blue and orange) can rotate relative to the core module by almost 4˚ in the backtracked complex. The trigger loop insertion domain (SI3, orange and blue) and downstream DNA move along with the swivel module. Superposition of EM maps from the two 3D classes (blue 37%, orange 63%) confirms swiveling. (B) Downstream DNA movement affects the downstream end of the RNA-DNA hybrid. The template DNA shifts around 2Å between the two conformations and affects the RNA in A- (+1) and P-site (-1). The two positions are reminiscent of an oscillation around the position observed in an EC (Kang et al., 2017). (C) Cryo-EM density (grey and green transparent surface) for the active site reveals the backtracked RNA at lower contour level (black sticks and green surface), the BH, unfolded TL, and Aspartate triad (pink), the template DNA (orange) and MgI (green). (D) Superposition of a substrate bound EC shows the steric clash between the folded trigger loop (grey) with the backtracked RNA (black) in the present reconstruction.
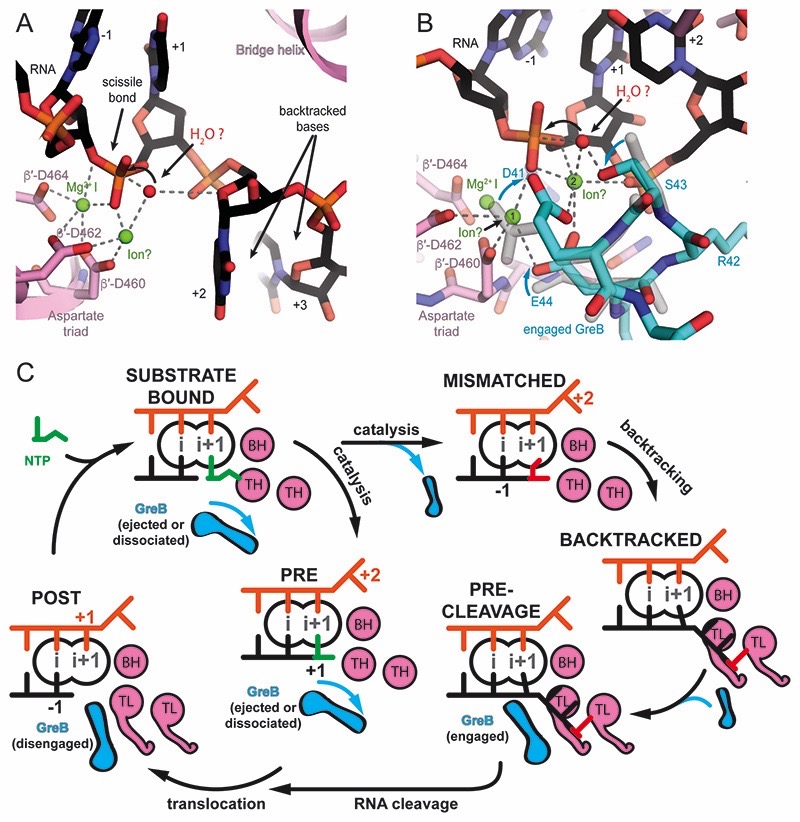
The electron density in the active site allows to discriminate purines from pyrimidines, which confirmed the nucleic acid register and was consistent with scaffold design and the number of cleaved nucleotides (Figure S1D). In the A-site (register +1), the RNA forms a complementary base pair with the corresponding template DNA (rUMP-dAMP). The base pair in the A-site exhibits a slightly distorted geometry with a larger buckle (~27˚) compared with a standard Watson-Crick base pair. A Magnesium ion (MgI) involved in catalysis and bound to a conserved triad of aspartates (E. coli β′ D460, D462 and D464) in the active site is visible and is also coordinated by the RNA base in the A-site (Figure 2B and 2C). The first backtracked RNA nucleotide rUMP (register +2) adopts a new conformation not observed before (Figure 2C). It is buried in a previously predicted pocket called the B-site (Sosunova et al., 2013), at the interface between the β and β’ subunits. The base forms polar interactions with basic residues β R678 and R1106. The backbone of the RNA between rUMP +1 and rUMP +2 is bent over ~125˚ out of the path of the hybrid helix. The second backtracked residue, rUMP +3, contacts the trigger loop (TL) residue Q929. The TL, a mobile element of the RNAP active site, folds into trigger helices (TH) upon substrate binding. This contact has also been observed in the backtracked eukaryotic RNAP II (Cheung and Cramer, 2011). The third backtracked base, rUMP +4, was not resolved, likely because it is disordered. However, the extrapolated RNA trajectory is consistent with crosslinking experiments (Markovtsov et al., 1996).
In the present reconstruction, two out of three resolved backtracked RNA bases block TL to TH folding (Figure 2D). The folded TH are thus not required for intrinsic cleavage of RNA backtracked by more than one base. Consistent with this observation, deletion of the TL had no influence on the rate of intrinsic RNA cleavage backtracked by two nucleotides (Zhang et al., 2010).
A GreB bound backtracked complex before RNA cleavage
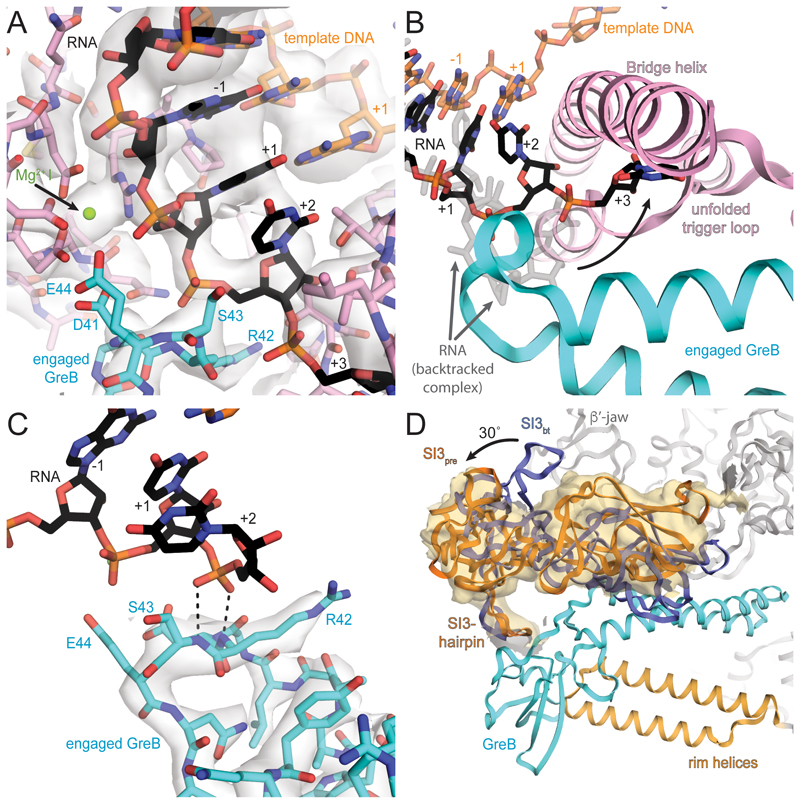
To gain insight into the role of cleavage factors, we assembled the same backtracked EC with an excess of GreB. GreB accelerated cleavage of unmodified and modified RNA at the same position compared to RNAP alone (Figure S1D). However, the reaction for phosphorothioate modified RNA was slow enough to prepare EM samples before significant cleavage occurred (Figures S1B and S1D). Thus, we could capture an active, backtracked RNAP EC with a functional GreB bound to the secondary channel prior to RNA cleavage (pre-cleavage complex). It was refined to a nominal resolution of 3.7Å and adopts the non-swiveled conformation (Figures 3A and S3, Tables S1 and S2, Movie S1).
Figure 3. Active site of pre-cleavage complex and GreB interaction with RNAP.
(A) Cryo-EM density (light grey, transparent surface) for the active site reveals density for the RNA (black) and template DNA (orange), the Aspartate triad and MgI (pink and green respectively) and GreB (cyan). (B) Comparing the backtracked complex (RNA transparent grey) with the pre-cleavage complex (RNA black), shows GreB shifts the backtracked portion of the RNA towards the BH and unfolded TL (pink). (C) A different view of the active site shows well-resolved density (grey transparent surface) for the tip of GreB (cyan). The backtracked RNA base in position +2 (black), stabilizes the backbone of GreB and helps to orient the catalytic residues. (D) GreB (cyan) interacts through the CTD with the secondary channel rim helices (light orange) and the trigger loop insertion SI3 (orange, SI3pre), which rotates almost 30° relative to the orientation in the backtracked state (blue, SI3bt). A low-pass filtered map confirms SI3 rotation (orange surface).
Two backtracked RNA bases are resolved but the tip of GreB pushed them closer to the RNAP bridge helix (BH) and TL compared to the backtracked complex (Figure 3B). Similarly to the backtracked complex the base pair in the A-site (rUMP-dAMP, register +1) is not planar with a more pronounced buckle and propeller twist than the remaining base pairs in the RNA-DNA hybrid. The first backtracked nucleotide (rUMP, register +2) forms different contacts with the BH residue T790. The second backtracked nucleotide (rUMP, register +3) is more disordered but appears to adopt a new position with polar contacts to BH residue β′-K789 and GreB residue Q49 and R56. The third backtracked residue (rUMP, register +4) was not resolved. These new contacts observed in the presence of GreB stabilize the RNA, resulting in lower temperature factors and increased density at higher contour levels. The backbone at the tip of the GreB NTD is well resolved and complements the RNAP active site. We call this the engaged state of GreB.
Although side chain density is weak, the backbone conformation orients the two essential and conserved acidic residues in the tip of GreB (D41 and E44) towards the active site. The side chain of R42, which is conserved in GreB but not in GreA, contacts the TL residues β′-T928 and β′-Q929. In addition, the phosphate of the first backtracked base hydrogen bonds with the backbone amides of GreB R42 and S43, stabilizing this backbone conformation (Figure 3C). Together with the BH, the TL and GreB form a narrow cleft that defines the path of RNA backtracked by more than one base. Furthermore, a positively charged surface of GreB provides a route for backtracked RNA to the surface of RNAP (Figure 1C). This surface has previously been suggested to determine the preference of GreB for longer backtracked RNA and longer cleavage products in contrast to GreA (Kulish et al., 2000). The conserved acidic residues at the tip of GreB, D41 and E44, were suggested to coordinate a second magnesium ion (MgII) in the active site (Opalka et al., 2003; Sosunov et al., 2003). However, while density for MgI was apparent, density for MgII was too weak to reliably model it. MgII occupancy must be low (likely the result of the phosphorothioate modification), which is consistent with the decreased cleavage rate compared to unmodified RNA.
In E. coli, a ~200-residue domain (sequence insertion 3, SI3) is inserted into the TL via two flexible linkers (Artsimovitch et al., 2003). The GreB NTD is inserted into the secondary channel, and restricts the available space for the SI3 linkers. This improved the density for the linkers, allowing us to model them. The GreB CTD interacts with the secondary channel rim-helices, and with SI3. The interactions with the rim-helices are mostly hydrophobic, consistent with mutation studies (Vassylyeva et al., 2007) (Figure 3D). A hairpin loop in SI3 (β′1052-1056) contacts the GreB CTD with a high degree of shape complementarity. The latter interaction explains why SI3 deletions or antibody binding interfered with GreB activity (Zakharova et al., 1998; Zhang et al., 2010). While the overall RNAP conformation is similar to the non-swivelled, backtracked population (Table S1), the interaction with GreB and the restriction of the SI3 linker flexibility, causes a rotation of SI3 compared to a canonical EC of about 30˚ around an axis close to the distal end of the linkers (Figure 3D, Movie S1). The rotation of SI3 causes a change in the interaction surface with the β′ jaw domain (residues β′1149-1214) consistent with chemical probing experiments (Laptenko et al., 2003).
Analogous to the backtracked complex, the engaged GreB is incompatible with TH folding. This is consistent with observations that TL to TH transition is dispensable for GreB-assisted cleavage (Kulish et al., 2000; Zhang et al., 2010). Therefore, while TH folding is required for RNA extension and pyrophosphorolysis (the direct reversal of RNA extension), it is not required for hydrolysis of RNA backtracked by more than one base.
Post cleavage complex
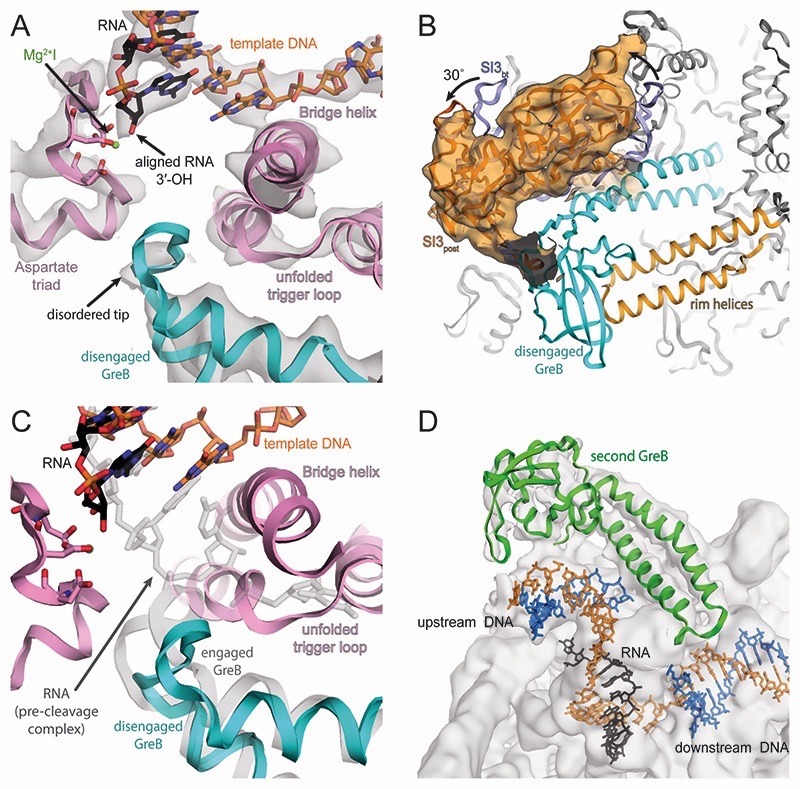
Cleavage of the scissile phosphodiester bond releases a short RNA oligonucleotide from the 3′-end. This results in a post-translocated EC with the newly generated RNA 3′-OH aligned in the active site (post-cleavage complex, Figure 4A). To understand the consequence of RNA cleavage on the conformation of GreB and RNAP, we assembled a post-cleavage complex using an unmodified RNA forming a fully complementary 9 base pair long RNA-DNA hybrid (Figure S1E) and added GreB in excess. The reconstruction refined to 3.9Å nominal resolution (Figures S4 and Table S2). RNAP adopts the swiveled conformation (∼4˚ rotation relative to non-swiveled backtracked population, ∼3˚ rotation relative to EC, Table S1) (Kang et al., 2017). SI3 adopts a similar orientation as in the pre-cleavage complex (Figure 4B and Movie S1). The RNA 3′-OH is aligned in the active centre, coordinates MgI and occupies the P-site of RNAP (Figure 4A). Consistent with the high affinity, GreB is still bound to the secondary channel, and the complex is stable enough for size exclusion chromatography (Figure S1H). In contrast to the pre-cleavage complex, the tip of the GreB NTD is more disordered and the backbone adopts a different conformation (Figure 4C). We call this the disengaged state of GreB. GreB R42 no longer interacts with the TL. In addition, because there is no backtracked RNA, the backbone amides of R42 and S43 have no phosphate to interact with, explaining the increased disorder. Unexpectedly, 3D classification of the data resulted in 2 classes (Figure S4B). Both contained disengaged GreB in the secondary channel. However, a subset of particles (62%) had apparently bound a second GreB, close to the upstream DNA (Figure 4D). We suspect this interaction is driven by non-specific electrostatic interactions between the positively charged GreB NTD and the upstream DNA and may not play any physiological role. In agreement, quantitative E. coli proteomics and quantitative Western blots suggest different degrees of molar excess of RNAP over GreB during various growth phases (Rutherford et al., 2007; Schmidt et al., 2016). We conclude that RNAP unlikely binds more than one GreB in vivo.
Figure 4. Active site of post-cleavage complex and additional GreB binding.
(A) Density for the active site (grey transparent surface) reveals the RNA in a post-translocated register (black), the template DNA (orange) as well as RNAP components (pink). Density for MgI (green) is weak but visible at lower contour level. The tip of GreB (cyan) is more disordered than in the pre-cleavage complex. (B) Similar to the pre-cleavage complex, a low-pass filtered map confirms SI3 rotates relative to the backtracked state as a result of GreB binding. (C) Superposition of pre- (grey transparent) and post-cleavage (colored) complex indicates that GreB disengages from the active site as a result of RNA cleavage. (D) 3D classification indicates a population of RNAP that bound a second copy of GreB (green) close to the upstream DNA, which becomes apparent at low contour level.
Reactivation of the GreB bound transcription complex
How does RNAP get reactivated after GreB-induced cleavage? To shed light on this aspect, we reconstituted a post-translocated RNAP complex using an RNA lacking the 3′-OH (Figure S1F). Addition of GreB as well as the next cognate substrate (rCTP) allowed us to reconstitute a substrate bound, reactivated RNAP EC trapped prior to RNA extension. We reasoned that this complex could potentially still bind GreB in an alternative conformation. We obtained a reconstruction that refined to 3.6Å (reactivated complex) (Figures S5 and Table S2). The electron density for the bound substrate in the A-site was readily discernible (Figure 5A). This complex resembles a canonical substrate bound EC prior to RNA extension. However, SI3 and the secondary channel rim helices moved to close the secondary channel, a striking difference to the post-translocated complex (Figure 5B and Movie S1). Substrate binding induces TH folding (Vassylyev et al., 2007), which shortens the linkers connecting the TH to SI3 (Movie S1). As a result, SI3 approaches the downstream DNA in a depression formed by the β′ jaw and β domain II. The distal lobe of SI3 moved more than 20Å from its position in the pre- and post-cleavage complexes. In this state, RNAP favours the non-swiveled conformation. The secondary channel rim helices rotate by 9˚ around an axis at their base, moving their tip into a concave surface of SI3. Similar changes have been observed for pre-translocated initiation complexes but in the present reconstruction the rotation of the rim helices is more pronounced (Liu et al., 2016; Zuo and Steitz, 2015). The adjacent F loop (FL) moves along and approaches the TH. FL residues β′-A748 and β′-G752 contact SI3 linker residues β′-G939, and β′-T1131 through backbone interactions and likely stabilize the folded TH conformation (Figure 5C). This would be consistent with reports suggesting FL involvement in promoting TL to TH transitions. (Miropolskaya et al., 2014). The folded TH and the restricted secondary channel are incompatible with bound GreB. As a consequence, no density for GreB was initially visible in the secondary channel.
Figure 5. Active site of reactivated, substrate bound complex and RNAP conformational changes.
(A) Two thirds of particles, showed density (grey transparent surface) for the substrate and MgII in the active site. The substrate base pairs to the template DNA (orange). The TL (pink) is folded but density for MgI is absent presumably because the RNA (black) lacks a 3′-OH. (B) TL folding repositions SI3, which approaches the secondary channel (compare SI3EC vs. SI3react). Likewise, the rim helices approach SI3 and close the secondary channel for GreB access. (C) Movement of the F-loop along with the rim helices establishes new contacts between F-loop and SI3 linkers, which may stabilize the folded TH. (D) Weak density for GreB was apparent on the surface of RNAP, suggesting it can maintain loose interactions with RNAP. Shown here is a 10 Å low-pass filtered map at two contour levels.
However, 3D classification suggested the presence of two populations (Figure S5B): i) about 36% of the particles were lacking density for the bound substrate, are thus in a post-translocated state, and GreB bound to the secondary channel. This subset appears identical to the post-cleavage complex; ii) about 64% of the particles formed a sub-population with rCTP bound in the A-site. In this case, weak density for a loosely bound GreB was visible on the surface of RNAP. GreB appears to maintain its interaction with SI3, but the NTD moved out of the active site and points away from RNAP (Figure 5D). We call this the dislodged state of GreB. Further sub-classification (not shown) indicated that dislodged GreB is flexibly bound, explaining the low resolution. Recent single molecule experiments performed on transcribing RNAP suggested a rapid exchange of GreB on timescales comparable to nucleotide addition (Tetone et al., 2017). In the present reconstruction, no structural requirement for complete GreB dissociation is evident. However, we suspect the dislodged state reflects an intermediate binding state, which we were able to observe because no productive RNA extension occurs and because we used a high concentration of GreB. Excess of RNAP over GreB and competition with other secondary channel binding proteins in vivo supports a model in which GreB can rapidly screen ECs (Schmidt et al., 2016; Tetone et al., 2017).
Discussion
The present reconstructions visualize intermediates covering the entire reaction pathway of RNAP backtracking, GreB-assisted cleavage, and transcription reactivation. For the first time, we obtained high-resolution structures of active intermediates using wild-type protein, without mutation of catalytically important residues or incomplete nucleic acid scaffolds. This allowed us to model the state before cleavage including two backtracked RNA bases with and without GreB. It allows us to speculate about the endonuclease reaction and propose a comprehensive model of how GreB facilitates RNA cleavage and re-activates transcription complexes without interfering with canonical RNA extension (Figures 6A-6C). Finally, we gain insights into the dynamic nature of RNAP by comparing conformations before and after substrate binding to the active site.
Figure 6. Models for RNA cleavage and the role of GreB during transcription elongation.
(A) In the backtracked complex, the nucleophile (red sphere) can be placed in line with the scissile bond. The O4′ of the first backtracked base (+2, black) may help to coordinate the nucleophile. MgII (modelled), could be coordinated by β′-D460, and β′-D462, but would be 3Å away from the attacking nucleophile. Small changes in the RNA backbone conformation may allow to adopt optimal geometry for the nucleophilic attack. (B) In the pre-cleavage complex, the nucleophile (red sphere) can be placed in line with the scissile bond. MgII can be modeled to be coordinated by β′-D460, β′-D462, the phosphate of the base in the A-site (+1) as well as GreB D41 and E44. However, this position (position 1) is 5Å away from the nucleophile. A change in the RNA backbone conformation may allow direct coordination of the nucleophile by MgII. Alternatively, a hypothetical third ion (modelled in position 2) as observed in DNA polymerase could activate the nucleophile. S43 in GreB might help orient the nucleophile consistent with a subtle decrease in cleavage rates upon mutation to Alanine. (C) Model for GreB’s role in transcription elongation: During a canonical elongation cycle (left), GreB cannot access the active site of substrate-bound and pre-translocated complexes because of the folded TH and secondary channel closure (not shown). As a result of erroneous incorporations or pause inducing DNA sequences, RNAP can backtrack and extrude the RNA 3′-end from the active site (right). RNA backtracking by 2 or more bases results in an unfolded TL and allows GreB to access the active site. GreB accelerates RNA cleavage and gives rise to a post-translocated EC, which can resume transcription (bottom).
Although we lack mechanistic details on how backtracking occurs, existing biochemical evidence suggests the thermodynamic stability of the RNA-DNA hybrid at a given position is a major contributor. Thus, weak RNA-DNA hybrids or mismatches are expected to favor backtracking (Nudler et al., 1997; Tadigotla et al., 2006). The extent of backtracking, and the sequence of the backtracked RNA are different in available structures (Cheung and Cramer, 2011; Sekine et al., 2015; Wang et al., 2009). As a result, the precise conformation of the backtracked RNA also differs in those structures and from our reconstructions. In agreement with this notion, we had to test about 15 RNAs varying in sequence and length of their backtracked portion. Some of them produced several cleavage products, and for others the backtracked portion could not be resolved in the reconstruction (data not shown).
In yeast PolII, the first backtracked base is oriented towards the BH (Cheung and Cramer, 2011; Wang et al., 2009), while in Thermus thermophilus the backtracked base is almost perpendicular to this orientation pointing into the secondary channel. In the present reconstruction, the first backtracked base (register +2, Figure 2C) is in a third orientation: opposite to the yeast counterpart, with the Watson-Crick edge towards the beta subunit (e.g. β-R678 and R1106). This alternative orientation is consistent with a model that has been proposed for E. coli based on mutational studies (Sosunova et al., 2013). The predominant conformation may depend on the sequence context, the number of backtracked nucleotides, and the species under investigation. This would agree with species-specific features of the reaction. For example, endopyrophosphorolysis is efficient in mammalian RNAP but not in E. coli (Rudd et al., 1994; Sosunov et al., 2003; Sosunova et al., 2013). Interestingly the first backtracked base occupies the proposed binding site for pyrophosphate, and thus provides a structural explanation for this observation (Freudenthal et al., 2013; Sosunova et al., 2013).
The conformation we observe allows speculation about some of the key players involved in intrinsic endonucleolysis. The nucleophile (water), needs to be in line with the leaving group (the rNMP occupying the P-site) for the SN2 reaction. Interestingly, the O4′ of the first backtracked base could help to position the nucleophile in agreement with earlier proposals (Sosunova et al., 2013). Estimates for the dissociation constant for a second Magnesium ion (MgII) in the endonucleolytic reaction vary but seem to be at least 10mM (Sosunova et al., 2003; Zhang et al., 2010). This is higher than the intracellular magnesium concentration (1-3 mM) (Dann et al., 2007; Grubbs, 2002). Thus MgII is bound to a potentially small fraction of complexes at any given time likely explaining the slow reaction rates. Nevertheless, we can model MgII for swiveled and non-swiveled populations so it is coordinated by D460 and D462 of the Aspartate triad as well as a non-bridging oxygen from the phosphodiester linkage. In this position it would be about 3Å, slightly too far, from the nucleophile (Figure 6A). Based on this model, a small change in the RNA backbone is sufficient to reach optimal geometry for cleavage.
Here we provide evidence that intrinsic, endonucleolytic cleavage of RNA backtracked by more than one nucleotide does not require folded TH. Biochemical studies and available crystal structures are consistent with this (Cheung and Cramer, 2011; Wang et al., 2009; Zhang et al., 2010). For RNAP backtracked by one nucleotide, conflicting models have been put forward. One set of biochemical and mutational studies suggest a role of the TL in an endonucleolytic cleavage reaction that releases a 2 nucleotide long RNA (Turtola et al., 2018; Yuzenkova and Zenkin, 2010). Structural studies of bacterial and eukaryotic RNAP indicate the TL is unfolded for RNAP backtracked by one nucleotide and residues proposed to be involved in acid-base catalysis (e.g. E. coli β’-H936, Yeast Rpb1 H1085) are more than 25Å away from the scissile bond (Sekine et al., 2015; Wang et al., 2009). More recent biochemical studies suggested a role of the TL as a positional rather than acid-base catalyst, helping to stabilize RNA backtracked by one nucleotide (Mishanina et al., 2017). Future studies employing cryo-EM and extensive 3D classification on complexes backtracked by one nucleotide may shed light on this state.
RNA backtracked by at least two nucleotides blocks TL folding (Figures 2C and 2D). This keeps the secondary channel open so GreB can access the active site. The backtracked RNA binds to the positively charged surface of GreB, and likely stabilizes GreB binding. This is consistent with Fe-mediated hydroxyl radical cleavage assays, which indicated GreB binds strongest to backtracked complexes (Furman et al., 2013). Importantly, the phosphate of the first backtracked nucleotide (+2) stabilizes the coiled-coil tip of the engaged GreB. This orients the conserved acidic residues D41 and E44, which are involved in cleavage catalysis, to face the active site (Figure 3C). Those residues can coordinate a magnesium ion involved in catalysis and thus accelerate the cleavage of the RNA 3′-end. However, despite the use of the wild-type protein, we did not see strong density for MgII. The phosphorothioate modifications in the RNA have likely lowered the affinity for magnesium (Pecoraro et al., 1984). Nevertheless, because we see the backtracked RNA at sufficient resolution for the first time, we can infer the nucleophile position for an SN2 reaction and model the likely position of some key players. Sosunova et al. proposed β′-D460 and β′-D462 of the Aspartate triad, a non-bridging oxygen of the scissile phosphate and GreB D41 and E44 to coordinate MgII, which in turn coordinates and activates the nucleophile (Sosunova et al., 2003). While these contacts are consistent with our structure and weak density appears in locally sharpened maps in this region (Jakobi et al., 2017), MgII in this position would be too far away to activate the nucleophile directly (∼5Å, ion in position 1 in Figure 6B). We envision two possibilities: Either, the RNA backbone conformation changes for the nucleophilic attack to reorient the scissile phosphodiester bond and allow direct coordination of the nucleophile by MgII bound in position 1 (Figure 6B). Alternatively, a third ion (for example in position 2 of Figure 6B), as observed for pyrophosphorolysis in DNA polymerase activates the nucleophile and the role of MgII is to stabilize and activate the RNA substrate (Freudenthal et al., 2013; Perera et al., 2015). A high-resolution structure of a transition state analogue would provide valuable information to answer this question.
According to our reconstruction a serine conserved in GreB (S43), but replaced by Lysine in GreA, in the coiled-coil tip, could help position the nucleophile. Consistent with this proposal, the S43A mutation reduced cleavage rates slightly (Figure S6A).
According to our model, the main factor contributing to the higher affinity of MgII in a GreB complex as opposed to a backtracked complex is additional negatively charged side chains provided by GreB, which could coordinate MgII. Furthermore, the first backtracked RNA residue (register +2) interacts with GreB. As a result, both GreB and the RNA substrate are more ordered. To summarize, the mechanism we propose for endonucleolytic cleavage of RNA backtracked by at least two nucleotides and GreB assisted cleavage are similar because the same scissile bond needs to be cleaved. The main differences are additional coordination partners for a potential magnesium ion involved in catalysis and a higher degree of order for the cleavage substrate in presence of GreB.
After cleavage, the RNAP is in a post-translocated register. No cognate rNTP substrate is bound and the TL is unfolded called the open state (Kettenberger et al., 2004). Gre factors can bind a post-translocated EC. This temporarily blocks substrate binding and traps the TL in a different state called the locked state (Cheung and Cramer, 2011) explaining observations of GreB decreasing transcription rates (Roghanian et al., 2011; Tetone et al., 2017). It is also exploited by transcription inhibitors, like Thermus thermophilus GfhI, which binds the secondary channel and blocks rNTP access and TH folding (Tagami et al., 2010). How does the transcription machinery minimize GreB interference with canonical elongation? Binding of a cognate substrate induces TH folding (Vassylyev et al., 2007; Wang et al., 2006), giving rise to the closed state (Vassylyev et al., 2007; Wang et al., 2006). Structural and biochemical evidence indicates that a pre-translocated complex (i.e. after catalysis but prior to translocation) also contains folded TH (Vassylyev et al., 2007; Yuzenkova et al., 2013; Zuo and Steitz, 2015). Steric clashes between the TH and the tip of Gre factors prevent access to the active site. This is likely universally true for secondary channel binding proteins, which insert into the active site. In addition, species that contain an SI3 domain, such as E. coli, are able to prevent Gre factor binding by re-positioning of SI3 and movement of the secondary channel rim helices that close the secondary channel. Thus, Gre factors are unable to induce cleavage of correctly incorporated substrates. According to single molecule studies, GreB has high on- and off-rates and presumably rapidly dissociates from the secondary channel, but it may keep the contact to the SI3 domain to allow rebinding in absence of a competing nucleotide (Tetone et al., 2017).
3D classification indicates changes in the equilibria between the swivelled and non-swivelled state of RNAP in the course of backtracking and subsequent reactivation (Figures S6B and S6C). The state of the active site and binding of transcription factors to RNAP may favour one over the other conformation (Kang et al., 2018b). Importantly, swivelling is distinct from the ratcheting movement observed for thermophilic RNAP. Here, the clamp domain rotates around an axis at approximately 45˚ relative to the swivelling axis opening the main nucleic acid binding channel (Figure S6D) (Sekine et al., 2015; Tagami et al., 2010).
It appears that conditions, which suppress productive elongation, favour swiveling and vice versa. In particular, it has been proposed that the swiveled conformation is incompatible with folded TH because of potential steric clashes between the repositioned SI3 domain and RNAP β-domain II (β143-448) (Kang et al., 2018a). The backtracked complex prefers the swiveled state. For the pre-cleavage and post-cleavage complexes the equilibrium is shifted to non-swiveled and swiveled respectively. Finally, for the reactivated complex containing rCTP, the non-swiveled state dominates. However, we were surprised to find only two thirds of the particles had rCTP bound given a concentration of 2mM during complex formation and substrate affinities in the range of 100 μM (Erie et al., 1992). This likely reflects the equilibrium of rCTP- and GreB-binding to an active site lacking an RNA 3’-OH and MgI under the conditions used for sample preparation. However, in vivo competition of GreB and rNTP substrates for the active site is probably minimized by the substoichiometric amounts of GreB compared to RNAP (Schmidt et al., 2016). In addition, the equilibrium of swiveled and non-swiveled RNAP may also contribute to the partitioning into rCTP versus GreB bound populations. Although further studies will be required, it is tempting to speculate whether the equilibrium between the swiveled and non-swiveled conformation explains widespread observations of two kinetic species in transcription elongation.
Star Methods
Key Resources Table.
| Reagent or Resource | Source | Identifier |
|---|---|---|
| Bacterial and Virus Strains | ||
| Escherichia coli LACR II rna-, rnb- (E. coli LOBSTR RNase I and II knock-out strain) | (Andersen et al., 2013); and to be published | |
| Chemicals, Peptides, and Recombinant Proteins | ||
| pVS11_His10_HRV3C_Ec_RNAP (E. coli RNAP co-expression plasmid for α-, β-, C-terminally His10- tagged β-, and ω-subunits) | (Twist et al., 2011) | |
| pACYC_Duet1_rpoZ (E. coli RNAP ω-subunit expression plasmid) | (Twist et al., 2011) | |
| pSKB2_His6_HRV3C_Ec_GreB (E. coli GreB with cleavable N-terminal His6-tag | This work | |
| Deposited Data | ||
| E. coli Backtracked complex (non-swiveled) | This work | PDB ID: VVVV |
| Backtracked complex density map (non-swiveled) | This work | EMD-VVVV |
| E. coli Backtracked complex (swiveled) | This work | PDB ID: WWWW |
| Backtracked complex density map (swiveled) | This work | EMD-WWWW |
| E. coli Pre-cleavage complex | This work | PDB ID: XXXX |
| Pre-cleavage complex density map | This work | EMD-XXXX |
| E. coli Post-cleavage complex | This work | PDB ID: YYYY |
| Post-cleavage complex density map | This work | EMD-YYYY |
| E. coli Substrate-bound complex | This work | PDB ID: ZZZZ |
| Substrate-bound complex density map | This work | EMD-ZZZZ |
| Experimental Models: Organisms/Strains | ||
| Escherichia coli | ||
| Oligonucleotides | ||
| Template DNA (used in the backtracked and pre-cleavage complexes): 3’- CGTGTAGTGGGTAAGGTCCGCTACACGTACCGAT CGACG-5’ |
This work | |
| Non-template DNA (used in the backtracked and pre-cleavage complexes): 5’- GCACATCACCCATTCAGAAGCTAAGGCATGGCTA GCTGC-3’ |
This work | |
| RNA (used in the backtracked and pre-cleavage complexes): 5’-UCAGGCGAU**UUUU-3’ |
This work | |
| Template DNA (used in the post-cleavage and substrate-bound complexes): 3’- CCAGTCATGCAGGGCTACACACGACCTTCTCTAA GTCTC-5’ |
This work | |
| Non-template DNA (used in the post-cleavage and substrate-bound complexes): 5’- GGTCAGTACGTCCCGTCGATCTTCGGAAGAGATT CAGAG-3’ |
This work | |
| RNA (used in the post-cleavage complex): 5’- UCAGGCGAUGUGUG -3’ |
This work | |
| RNA (used in the substrate-bound complex): 5’-UCAGGCGAUGUGUG** -3’ |
This work | |
| Software and Algorithms | ||
| Relion v2.1 | (Scheres, 2012) | https://www2.mrc-lmb.cam.ac.uk/relion/index.php/Main_Page |
| CryoSPARC v0.6.5 | (Punjani et al., 2017) | https://cryosparc.com |
| Motioncor2 | (Zheng et al., 2016) | http://msg.ucsf.edu/em/software/motioncor2.html |
| Gctf | (Zhang, 2016) | https://www.mrc-lmb.cam.ac.uk/kzhang/Gctf/ |
| Gautomatch | https://www.mrc-lmb.cam.ac.uk/kzhang/Gautomatch/ | |
| EMAN v2.2 | (Tang et al., 2007) | http://blake.bcm.edu/emanwiki/EMAN2 |
| CCP4 suite | (Winn et al., 2011) | http://www.ccp4.ac.uk/ |
| Phenix suite | (Adams et al., 2010) | https://www.phenix-online.org/ |
| COOT v0.8.3 | (Emsley and Cowtan, 2004) | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| PyMOL v1.6 | (Schrodinger, LLC, 2015) | https://pymol.org/2/ |
| UCSF Chimera v1.11.2 | (Pettersen et al., 2004) | https://www.cgl.ucsf.edu/chimera/download.html |
| LocScale density scaling (sharpening) procedure | (Jakobi et al., 2017) | https://git.embl.de/jakobi/LocScale |
indicates phosphorothioate modification
indicates terminal 3’-deoxyguanosine-5’-monophosphate (3’-dGMP)
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Albert Weixlbaumer (albert.weixlbaumer@igbmc.fr).
Methods Details
Purification of RNAP and GreB
E. coli RNA polymerase with a C-terminally His10-tagged β’-subunit was prepared from E. coli LACRII cells as described previously (Guo et al., 2018). Briefly, 6 liters of LB culture containing (100 μg/ml Ampicillin and 34 μg/ml Chloramphenicol) were induced at an OD600 of 0.7 with 0.5 mM IPTG for 2 hours at 37°C. Cells were harvested by centrifugation, resuspended in 5 volumes of lysis buffer (50 mM Tris-HCl pH 8.0, 5% glycerol, 1 mM EDTA, 10 mM DTT, 0.1 mM PMSF, 1mM benzamidine, 10 μM ZnCl2) and DNase I (~20 μg/10g cell) and EDTA-free protease inhibitor cocktail was added (Sigma-Aldrich cOmplete, 1 tablet/50ml), and sonicated. The lysate was clarified by centrifugation at 40,000 g for 30 minutes followed by polyethyleneimine fractionation and ammonium sulfate precipitation (Vassylyeva et al., 2002). The precipitate was resuspended in IMAC buffer (20 mM Tris-HCl, pH 8.0, 1 M NaCl, 5% glycerol, 5 mM β-mercaptoethanol, 0.1 mM PMSF, 1 mM Benzamidine, 10 μM ZnCl2), applied to a 20 ml Ni-IMAC Sepharose HP column (GE Healthcare), and eluted using IMAC buffer plus 250 mM imidazole. Peak fractions were pooled and dialyzed overnight in the presence of His-tagged HRV3C (PreScission) protease into dialysis buffer (20 mM Tris-HCl, pH 8.0, 1 M NaCl, 5% glycerol, 5 mM β-mercaptoethanol, 10 μM ZnCl2). Cleaved RNAP was loaded on a subtractive Ni-IMAC column, isolated from the flow-through, and dialyzed into Bio-Rex buffer (10 mM Tris-HCl, pH 8.0, 5% glycerol, 0.1 mM EDTA, 0.1 M NaCl, 1 mM DTT, 0.1 mM PMSF, 1 mM Benzamidine, 10 μM ZnCl2). RNAP was then loaded on a 50 mL Bio-Rex 70 column (Bio-Rad) and eluted into Bio-Rex buffer plus 1 M NaCl. The peak was concentrated and further purified by gel filtration using a HiLoad Superdex 200 PG 26/600 column (GE Healthcare) equilibrated with GF buffer (10 mM HEPES, pH 8.0, 0.5 M KCl, 1% Glycerol, 2 mM DTT, 0.1 mM PMSF, 1 mM benzamidine, 10 μM ZnCl2, 1 mM MgCl2). The final protein was concentrated to ~80mg/ml, dialyzed into cryo-EM buffer (10 mM HEPES, pH 8.0, 150 mM KOAc, 2 mM DTT, 10 μM ZnCl2, 5 mM Mg(OAc)2) and aliquots were flash frozen and stored at −80°C.
E. coli GreB with N-terminal His6-tag was overexpressed in E. coli LACRII strain by inducing 6 liters of LB culture containing (50 μg/ml Kanamycin) at an OD600 of 0.7 with 1 mM IPTG for 3 hours at 37°C. Cells were harvested by centrifugation, resuspended in 5 volumes of lysis buffer (40 mM Tris-HCl pH 8.0, 1 M NaCl, 5% glycerol, 10 mM EDTA, 1 mM DTT, 0.1 mM PMSF, 1mM benzamidine) containing EDTA-free protease inhibitor cocktail (Sigma-Aldrich cOmplete, 1 tablet/50ml), and lysed by sonication. The lysate was clarified by centrifugation at 40,000 g for 30 minutes. Polyethyleneimine (0.3% v/v) was added to the cleared lysate followed by centrifugation at 10,000 g for 15 minutes and the supernatant was further purified by addition of 0.8 M (NH4)2SO4 and centrifugation at 10,000 g for 25 minutes. The supernatant was dialyzed against IMAC binding buffer (40 mM Tris-HCl pH 8.0, 0.6 M NaCl, 0.8 M (NH4)2SO4, 5mM imidazole, 2 mM β-mercaptoethanol, 0.1 mM PMSF, 1mM benzamidine) and was loaded on a 5 mL Ni-IMAC Sepharose HP column (GE Healthcare) equilibrated with the binding buffer, and eluted using a step gradient into IMAC binding buffer containing 250 mM imidazole (20 mM imidazole wash for 5 column volumes (CVs), 40 mM imidazole wash for 5CVs, gradient starting from 40 mM to 250 mM imidazole over 20 CVs, target peak starts to elute at 50mM). Peak fractions were pooled and dialyzed overnight in the presence of His-tagged HRV3C protease into cleavage buffer (40 mM Tris-HCl, pH 8.0, 0.6 M NaCl, 0.8 M (NH4)2SO4, 2 mM β-mercaptoethanol). Cleaved GreB was reloaded on the Ni-IMAC column and was isolated from the flow-through. GreB was then applied to a 10 ml Butyl-Sepharose HP column (GE Healthcare) equilibrated with HIC buffer (40 mM Tris-HCl, pH 8.0, 0.6 M NaCl, 0.8 M (NH4)2SO4, 0.1 mM EDTA, 1 mM DTT, 0.1 mM PMSF, 1mM benzamidine) and eluted using a gradient over 10 CVs into HIC buffer without (NH4)2SO4, supplemented with 5% glycerol. The final protein was concentrated to 60 mg/ml, dialyzed against storage buffer (40 mM Tris-HCl, pH 8.0, 1 M NaCl, 10% glycerol, 0.1 mM EDTA, 1 mM DTT), and aliquots were flash frozen and stored at −80°C.
DNA (TriLink) and RNA (Dharmacon) oligonucleotides were chemically synthesized and gel purified by the manufacturer. RNA was deprotected following the protocols provided by the manufacturer. Both DNA and RNA were dissolved in RNase free water and aliquots were stored at −80°C.
In vitro cleavage assays
To test protein activity, cleavage assays were conducted using reconstituted nucleic acid scaffolds (template DNA, tDNA; non-template DNA, ntDNA, and RNA). The RNA was first 5′-labeled with [γ-32P] ATP using T4 polynucleotide kinase (NEB). Nucleic acid scaffolds were annealed in reconstitution buffer (10 mM Tris-HCl, pH 8.0, 40 mM KCl, 5 mM MgCl2), by mixing a 2-fold molar excess of ntDNA and tDNA over RNA (5 μM), incubation for 2 min at 95°C, and slowly cooling to room temperature in a water bath. RNA cleavage kinetics were measured at 37°C by mixing 0.5 μM scaffold, 1 μM RNAP and 5 μM GreB (if required) in cryo-EM buffer in the presence of 20 μg/ml BSA and 8 mM CHAPSO. Aliquots were taken at predetermined time points and quenched with an equal volume of loading buffer (8 M urea, 20 mM EDTA pH 8, 5 mM Tris-HCl pH 7.5, 0.5% bromophenol blue, and 0.5% xylene cyanol). Cleavage reactions were chased by adding 1 mM ATP, CTP, UTP and GTP and analyzed using denaturing polyacrylamide gels. For data analysis, gels were exposed to storage phosphor screens and quantified using a Typhoon PhosphorImager and ImageQuant software (GE Healthcare).
Cryo-EM sample preparation
Functional complexes were prepared right before grid freezing by mixing E. coli RNAP (20 μM), nucleic acid scaffold, and wild type GreB (if required) in cryo-EM buffer using the following molar ratios: 1:1.5 (backtracked complex), 1:1.5:8 (post-cleavage complex), 1:2:5 (pre-cleavage complex), 1:1.5:4 (substrate bound complex), respectively.
To prepare the backtracked complex, RNAP and nucleic acid scaffold (containing phosphorothioate RNA) were incubated for 3 minutes at 37°C prior to grid freezing. For the pre-cleavage complex, RNAP and nucleic acid scaffold (containing phosphorothioate RNA) were mixed together first, incubated for 3 minutes at 37°C followed by addition of GreB and then centrifuged at 14,000 rpm for one minute at 25°C, to pellet down potential aggregates. The post-cleavage complex was assembled by mixing RNAP, nucleic acid scaffold, and GreB and incubating the mixture for 3 minutes at 37°C followed by centrifugation at 14,000 rpm for one minute at 25°C. Finally, to prepare the substrate bound complex, RNAP and nucleic acid scaffold (containing 3’-deoxy RNA) were incubated for 3 minutes at 37°C in presence of 2 mM CTP followed by adding GreB and further incubation for an additional minute at 37°C followed by centrifugation at 14,000 rpm for one minute at 25°C. Immediately before freezing, 8 mM CHAPSO was added to the complexes. C-flat holey carbon copper grids (CF-2/2 400 mesh, Protochips, Morrisville, NC, USA) were used for the pre-cleavage, post-cleavage, and substrate bound complexes. Quantifoil holey carbon copper grids (R2/2 300 mesh, Quantifoil Micro Tools, Großlöbichau, Germany) were used for the backtracked complex. C-flat grids were glow-discharged for 25 seconds using an ELMO glow discharge system (Cordouan Tech.), while Quantifoil grids were glow-discharged for one minute using a NanoClean model 1070 (Fishione Instruments) with a gas mixture of 20% oxygen and 80% argon. 4 μL of the sample was applied to the grid, blotted and plunge-frozen in liquid ethane using a Vitrobot Mark IV (FEI) at 95% chamber humidity and 10°C.
Cryo-EM data acquisition
Movies of the four different complexes were recorded on four different Titan Krios (FEI) microscopes, at an acceleration voltage of 300 kV, defocus range −0.8 to −3.2 μm, and 40 frames per movie. The microscopes were each equipped with a K2 Summit camera (Gatan, Inc., Pleasanton, CA) and operated either in counting mode (backtracked complex) or super-resolution counting mode (pre-cleavage, post-cleavage, and substrate bound complex). The detectors were placed at the end of a GIF Quantum energy filter (Gatan, Inc.), operated in zero-energy-loss mode with a slit width of 20 eV. The backtracked complex was collected at the ESRF, Grenoble, France (4279 movies) with a pixel size of 1.067 Å/pixel, an exposure rate of 4.1 e- /Å2/s, and total dose of ~49 e-/Å2. The pre-cleavage complex was collected at the IGBMC, Illkirch, France (6946 movies) with a super resolution pixel size of 0.55 Å/pixel, an exposure rate of 5.3 e-/Å2/s, and total dose of 51 e-/Å2. The post-cleavage complex was collected at the Biozentrum, University of Basel, Switzerland (3960 movies) with a super resolution pixel size of 0.52 Å/pixel, an exposure rate of 4.9 e- /Å2/s, and total dose of 59 e-/Å2. The substrate bound complex was collected at the EMBL, Heidelberg, Germany (5700 movies) with a super resolution pixel size of 0.52 Å/pixel, an exposure rate of 2.3 e-/Å2/s, and total dose of ~46 e-/Å2.
Image processing
Dose fractionated frames were aligned and summed using Motioncor2 (Zheng et al., 2016). The contrast transfer function (CTF) of each micrograph was estimated with Gctf (Zhang, 2016). Initially, ~3000 particles were manually picked with EMAN (Tang et al., 2007) and subjected to 2D classification with Relion (Scheres, 2012). 7 representative 2D class averages were selected as templates for automatic picking by Gautomatch. Particles were extracted in Relion and further processed using CryoSPARC (Punjani et al., 2017). The best class averages resulting from 2D classification were used for a reference-free ab-initio 3D reconstruction and classification. After visual inspection, the selected map was further refined and used as a reference for the multi-refine procedure.
For the backtracked complex, 268851 particles were automatically picked. After removing bad particles by 2D classification and multi-refine, 162979 particles were remaining. 3D classification resulted in class 1 (60080 particles) and class 2 (102899 particles) that were refined to 3.7 Å and 3.4 Å, respectively. The two classes differ in the orientation of the swivel module relative to the core module of RNAP.
For the pre-cleavage complex, 758120 particles were automatically picked. After removing bad particles by 2D classification and multi-refine, 574584 particles were remaining. 3D classification did not produce any noticeable difference in the resulted maps. Hence, particles were further refined to 3.7 Å final map.
In case of the post-cleavage complex, 270554 particles were automatically picked. After removing bad particles by 2D classification and multi-refine, 121680 particles were remaining. 3D classification resulted in class 1 (76069 particles) and class 2 (45611 particles) that were refined to 4 Å and 4.3 Å, respectively. Unlike class 2, we observed density for a second GreB near the upstream DNA in class 1. Particles from the two classes were merged and used for homogeneous refinement, which yielded a map at 3.9 Å that was used for model building.
For the substrate bound complex, 380141 particles were initially picked. After removing bad particles by 2D classification and multi-refine, 170328 particles were remaining. 3D classification generated 3 main classes. Class 1 (63204 particles) lacked the substrate in the active site and was refined to 4 Å. Class 2 (69718 particles) and class 3 (37406 particles) showed discernible density for the substrate in the active site, were refined to 3.7 Å and 4.2 Å, respectively, and showed a different conformation for the dislodged GreB. Particle images from class 2 and 3 were merged and used for homogeneous refinement yielding a map at 3.6 Å.
Model building
We constructed an initial model of the different complexes by combining the cryo-EM structure of E. coli RNAP (PDB: 6ALH) and the crystal structure of E. coli GreB (PDB: 2P4V), if required. Model placement into the EM maps was performed with UCSF Chimera (Pettersen et al., 2004). Model building and manual modification was done in Coot (Emsley and Cowtan, 2004) followed by several rounds of real space refinement using Phenix (Adams et al., 2010). The RNA-DNA hybrid and downstream DNA duplex were built de novo in Coot. The resulting models were used for several rounds of real-space refinement using secondary structure restraints and geometry optimization in Phenix against density maps sharpened by applying a B-factor that was estimated using automated procedures (Punjani et al., 2017). Models with refined B-factors were then used for a final round of building and refinement against maps locally sharpened using LocScale (Jakobi et al., 2017). In case of the substrate bound complex, a custom dictionary entry for the RNA 3’-end (3’-deoxyguanosine) and CTP substrate were generated in JLigand (Lebedev et al., 2012) and added to the RNA chain in Coot.
Supplementary Material
Highlights.
Four cryo-EM structures of RNAP cover backtracking, cleavage and reactivation
Backtracking allows active site access for GreB
GreB gets ejected from active site upon substrate binding
Substrate binding induces global conformational change in RNAP
Acknowledgements
We thank Lubomir Kovacik and Mohamed Chami for help with data collection at the University of Basel, Switzerland. We thank Wim Hagen and Felix Weis for help with data collection at the EMBL in Heidelberg, Germany and we acknowledge the support and use of resources of Instruct-ERIC. We acknowledge the European Synchrotron Radiation Facility for provision of microscope time on CM01 and we thank Eaazhisai Kandiah and Gregory Effantin for their assistance. We thank Seth A. Darst and Robert Landick for insightful discussions and comments on the manuscript. The authors were supported by the French Infrastructure for Integrated Structural Biology (FRISBI ANR-10-INBS-05, Instruct-ERIC, and grant ANR-10-LABX-0030-INRT, a French State fund managed by the Agence Nationale de la Recherche under the program Investissements d’Avenir ANR-10-IDEX-0002-02). The work was supported by PhD fellowships from Région Alsace, Inserm and La Fondation pour la recherche médicale to M.A. and the ERC starting grant TRANSREG (679734) to A.W.
Footnotes
Author Contributions
Conceptualization, A.W. and M.A.; Sample preparation, M.A., C.SA., and M.T.; Data collection and processing, M.A., C.SA., X.G., J.O., G.P., C.C.; Investigation and analysis, M.A., A.W.; Writing, M.A., A.W.; Supervision, A.W.
Declaration of Interests
The authors declare no competing interests.
Data and Software Availability
The accession numbers for the five cryo-EM reconstructions (non-swiveled backtracked complex, swiveled backtracked complex, pre-cleavage complex, post-cleavage complex, reactivated complex) reported in this paper are … Fitted models were deposited in the PDB under accession numbers VVVV (all atom model for non-swiveled backtracked complex), WWWW (all atom model for swiveled backtracked complex), XXXX (all atom model for pre-cleavage complex), YYYY (all atom model for post-cleavage complex), ZZZZ (all atom model for reactivated complex).
References
- Adelman K, Marr MT, Werner J, Saunders A, Ni Z, Andrulis ED, Lis JT. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol Cell. 2005;17:103–112. doi: 10.1016/j.molcel.2004.11.028. [DOI] [PubMed] [Google Scholar]
- Artsimovitch I, Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc Natl Acad Sci USA. 2000;97:7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I, Svetlov V, Murakami KS, Landick R. Co-overexpression of Escherichia coli RNA polymerase subunits allows isolation and analysis of mutant enzymes lacking lineage-specific sequence insertions. J Biol Chem. 2003;278:12344–12355. doi: 10.1074/jbc.M211214200. [DOI] [PubMed] [Google Scholar]
- Borukhov S, Sagitov V, Goldfarb A. Transcript cleavage factors from E coli. Cell. 1993;72:459–466. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- Cheung ACM, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471:–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- Dann CE, Wakeman CA, Sieling CL, Baker SC, Irnov I, Winkler WC. Structure and mechanism of a metal-sensing regulatory RNA. Cell. 2007;130:878–892. doi: 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- Erie DA, Hajiseyedjavadi O, Young MC, von Hippel PH. Multiple RNA polymerase conformations and GreA: control of the fidelity of transcription. Science. 1993;262:867–873. doi: 10.1126/science.8235608. [DOI] [PubMed] [Google Scholar]
- Erie DA, Yager TD, von Hippel PH. The single-nucleotide addition cycle in transcription: a biophysical and biochemical perspective. Annual Review of Biophysics and Biomolecular Structure. 1992;21:379–415. doi: 10.1146/annurev.bb.21.060192.002115. [DOI] [PubMed] [Google Scholar]
- Freudenthal BD, Beard WA, Shock DD, Wilson SH. Observing a DNA polymerase choose right from wrong. Cell. 2013;154:157–168. doi: 10.1016/j.cell.2013.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman R, Tsodikov OV, Wolf YI, Artsimovitch I. An insertion in the catalytic trigger loop gates the secondary channel of RNA polymerase. J Mol Biol. 2013;425:82–93. doi: 10.1016/j.jmb.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- Grubbs RD. Intracellular magnesium and magnesium buffering. Biometals. 2002;15:251–259. doi: 10.1023/a:1016026831789. [DOI] [PubMed] [Google Scholar]
- Guo X, Myasnikov AG, Chen J, Crucifix C, Papai G, Takacs M, Schultz P, Weixlbaumer A. Structural Basis for NusA Stabilized Transcriptional Pausing. Mol Cell. 2018;69:816–827.:e4. doi: 10.1016/j.molcel.2018.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausner W, Lange U, Musfeldt M. Transcription factor S, a cleavage induction factor of the archaeal RNA polymerase. J Biol Chem. 2000;275:12393–12399. doi: 10.1074/jbc.275.17.12393. [DOI] [PubMed] [Google Scholar]
- Imashimizu M, Tanaka K, Shimamoto N. Comparative Study of Cyanobacterial and E.coli RNA Polymerases: Misincorporation, Abortive Transcription, and Dependence on Divalent Cations. Genet Res Int. 2011;2011:572689. doi: 10.4061/2011/572689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izban MG, Luse DS. The RNA polymerase II ternary complex cleaves the nascent transcript in a 3“---5” direction in the presence of elongation factor SII. 1992;6:1342–1356. doi: 10.1101/gad.6.7.1342. [DOI] [PubMed] [Google Scholar]
- Jakobi AJ, Wilmanns M, Sachse C. Model-based local density sharpening of cryo-EM maps. Elife. 2017;6:e27131. doi: 10.7554/eLife.27131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon C, Yoon H, Agarwal K. The transcription factor TFIIS zinc ribbon dipeptide Asp-Glu is critical for stimulation of elongation and RNA cleavage by RNA polymerase II. Proc Natl Acad Sci USA. 1994;91:9106–9110. doi: 10.1073/pnas.91.19.9106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015;16:167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Mishanina TV, Bellecourt MJ, Mooney RA, Darst SA, Landick R. RNA Polymerase Accommodates a Pause RNA Hairpin by Global Conformational Rearrangements that Prolong Pausing. Mol Cell. 2018a;69:802–815.:e1. doi: 10.1016/j.molcel.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Mooney RA, Nedialkov Y, Saba J, Mishanina TV, Artsimovitch I, Landick R, Darst SA. Structural Basis for Transcript Elongation Control by NusG Family Universal Regulators. Cell. 2018b doi: 10.1016/j.cell.2018.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Olinares PDB, Chen J, Campbell EA, Mustaev A, Chait BT, Gottesman ME, Darst SA. Structural basis of transcription arrest by coliphage HK022 nun in an Escherichia coli RNA polymerase elongation complex. Elife. 2017;6 doi: 10.7554/eLife.25478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenberger H, Armache K-J, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- Komissarova N, Kashlev M. Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3’ end of the RNA intact and extruded. Proc Natl Acad Sci USA. 1997;94:1755–1760. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulish D, Lee J, Lomakin I, Nowicka B, Das A, Darst S, Normet K, Borukhov S. The functional role of basic patch, a structural element of Escherichia coli transcript cleavage factors GreA and GreB. J Biol Chem. 2000;275:12789–12798. doi: 10.1074/jbc.275.17.12789. [DOI] [PubMed] [Google Scholar]
- Landick R. The regulatory roles and mechanism of transcriptional pausing. Biochem Soc Trans. 2006;34:1062–1066. doi: 10.1042/BST0341062. [DOI] [PubMed] [Google Scholar]
- Laptenko O, Lee J, Lomakin I, Borukhov S. Transcript cleavage factors GreA and GreB act as transient catalytic components of RNA polymerase. Embo J. 2003;22:6322–6334. doi: 10.1093/emboj/cdg610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner E, Chung S, Allen BL, Wang S, Lee J, Lu SW, Grimaud LW, Ingargiola A, Michalet X, Alhadid Y, et al. Backtracked and paused transcription initiation intermediate of Escherichia coli RNA polymerase. Proc Natl Acad Sci USA. 2016;113:E6562–E6571. doi: 10.1073/pnas.1605038113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Zuo Y, Steitz TA. Structures of E.coli σS-transcription initiation complexes provide new insights into polymerase mechanism. Proc Natl Acad Sci USA. 2016;113:4051–4056. doi: 10.1073/pnas.1520555113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovtsov V, Mustaev A, Goldfarb A. Protein-RNA interactions in the active center of transcription elongation complex. Proc Natl Acad Sci USA. 1996;93:3221–3226. doi: 10.1073/pnas.93.8.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miropolskaya N, Esyunina D, Klimasauskas S, Nikiforov V, Artsimovitch I, Kulbachinskiy A. Interplay between the trigger loop and the F loop during RNA polymerase catalysis. Nucleic Acids Res. 2014;42:544–552. doi: 10.1093/nar/gkt877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishanina TV, Palo MZ, Nayak D, Mooney RA, Landick R. Trigger loop of RNA polymerase is a positional, not acid-base, catalyst for both transcription and proofreading. Proc Natl Acad Sci USA. 2017;114:E5103–E5112. doi: 10.1073/pnas.1702383114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 2010;327:335–338. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- Opalka N, Chlenov M, Chacon P, Rice WJ, Wriggers W, Darst SA. Structure and function of the transcription elongation factor GreB bound to bacterial RNA polymerase. Cell. 2003;114:335–345. doi: 10.1016/s0092-8674(03)00600-7. [DOI] [PubMed] [Google Scholar]
- Pecoraro VL, Hermes JD, Cleland WW. Stability constants of Mg2+ and Cd2+ complexes of adenine nucleotides and thionucleotides and rate constants for formation and dissociation of MgATP and MgADP. Biochemistry. 1984;23:5262–5271. doi: 10.1021/bi00317a026. [DOI] [PubMed] [Google Scholar]
- Perera L, Freudenthal BD, Beard WA, Shock DD, Pedersen LG, Wilson SH. Requirement for transient metal ions revealed through computational analysis for DNA polymerase going in reverse. Proc Natl Acad Sci USA. 2015;112:E5228–E5236. doi: 10.1073/pnas.1511207112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reines D, Ghanouni P, Li QQ, Mote J. The RNA polymerase II elongation complex. Factor-dependent transcription elongation involves nascent RNA cleavage. J Biol Chem. 1992;267:15516–15522. [PMC free article] [PubMed] [Google Scholar]
- Roghanian M, Yuzenkova Y, Zenkin N. Controlled interplay between trigger loop and Gre factor in the RNA polymerase active centre. Nucleic Acids Res. 2011;39:4352–4359. doi: 10.1093/nar/gkq1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd MD, Izban MG, Luse DS. The active site of RNA polymerase II participates in transcript cleavage within arrested ternary complexes. Proc Natl Acad Sci USA. 1994;91:8057–8061. doi: 10.1073/pnas.91.17.8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford ST, Lemke JJ, Vrentas CE, Gaal T, Ross W, Gourse RL. Effects of DksA, GreA, and GreB on transcription initiation: insights into the mechanisms of factors that bind in the secondary channel of RNA polymerase. J Mol Biol. 2007;366:1243–1257. doi: 10.1016/j.jmb.2006.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Kochanowski K, Vedelaar S, Ahrné E, Volkmer B, Callipo L, Knoops K, Bauer M, Aebersold R, Heinemann M. The quantitative and condition-dependent Escherichia coli proteome. Nat Biotechnol. 2016;34:104–110. doi: 10.1038/nbt.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine S-I, Murayama Y, Svetlov V, Nudler E, Yokoyama S. The Ratcheted and Ratchetable Structural States of RNA Polymerase Underlie Multiple Transcriptional Functions. Mol Cell. 2015 doi: 10.1016/j.molcel.2014.12.014. [DOI] [PubMed] [Google Scholar]
- Shaevitz JW, Abbondanzieri EA, Landick R, Block SM. Backtracking by single RNA polymerase molecules observed at near-base-pair resolution. Nature. 2003;426:684–687. doi: 10.1038/nature02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Nikonowicz EP. Phosphorothioate substitution can substantially alter RNA conformation. Biochemistry. 2000;39:5642–5652. doi: 10.1021/bi992712b. [DOI] [PubMed] [Google Scholar]
- Sosunov V, Sosunova E, Mustaev A, Bass I, Nikiforov V, Goldfarb A. Unified two-metal mechanism of RNA synthesis and degradation by RNA polymerase. Embo J. 2003;22:2234–2244. doi: 10.1093/emboj/cdg193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosunova E, Sosunov V, Epshtein V, Nikiforov V, Mustaev A. Control of Transcriptional Fidelity by Active Center Tuning as Derived from RNA Polymerase Endonuclease Reaction. J Biol Chem. 2013;288:6688–6703. doi: 10.1074/jbc.M112.424002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosunova E, Sosunov V, Kozlov M, Nikiforov V, Goldfarb A, Mustaev A. Donation of catalytic residues to RNA polymerase active center by transcription factor Gre. Proc Natl Acad Sci USA. 2003;100:15469–15474. doi: 10.1073/pnas.2536698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins CE, Borukhov S, Orlova M, Polyakov A, Goldfarb A, Darst SA. Crystal structure of the GreA transcript cleavage factor from Escherichia coli. Nature. 1995;373:636–640. doi: 10.1038/373636a0. [DOI] [PubMed] [Google Scholar]
- Tadigotla VR, Maoilé O, idigh D, Sengupta AM, Epshtein V, Ebright RH, Nudler E, Ruckenstein AE. Thermodynamic and kinetic modeling of transcriptional pausing. Proc Natl Acad Sci USA. 2006;103:4439–4444. doi: 10.1073/pnas.0600508103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami S, Sekine S-I, Kumarevel T, Hino N, Murayama Y, Kamegamori S, Yamamoto M, Sakamoto K, Yokoyama S. Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein. Nature. 2010;468:978–982. doi: 10.1038/nature09573. [DOI] [PubMed] [Google Scholar]
- Tetone LE, Friedman LJ, Osborne ML, Ravi H, Kyzer S, Stumper SK, Mooney RA, Landick R, Gelles J. Dynamics of GreB-RNA polymerase interaction allow a proofreading accessory protein to patrol for transcription complexes needing rescue. Proc Natl Acad Sci USA. 2017 doi: 10.1073/pnas.1616525114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turtola M, Mäkinen JJ, Belogurov GA. Active site closure stabilizes the backtracked state of RNA polymerase. Nucleic Acids Res. 2018 doi: 10.1093/nar/gky883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- Vassylyeva MN, Svetlov V, Dearborn AD, Klyuyev S, Artsimovitch I, Vassylyev DG. The carboxy-terminal coiled-coil of the RNA polymerase beta’-subunit is the main binding site for Gre factors. EMBO Rep. 2007;8:1038–1043. doi: 10.1038/sj.embor.7401079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Bushnell DA, Huang X, Westover KD, Levitt M, Kornberg RD. Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution. Science. 2009;324:1203–1206. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzenkova Y, Zenkin N. Central role of the RNA polymerase trigger loop in intrinsic RNA hydrolysis. Proc Natl Acad Sci USA. 2010;107:10878–10883. doi: 10.1073/pnas.0914424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzenkova Y, Roghanian M, Bochkareva A, Zenkin N. Tagetitoxin inhibits transcription by stabilizing pre-translocated state of the elongation complex. Nucleic Acids Res. 2013;41:9257–9265. doi: 10.1093/nar/gkt708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharova N, Bass I, Arsenieva E, Nikiforov V, Severinov K. Mutations in and monoclonal antibody binding to evolutionary hypervariable region of Escherichia coli RNA polymerase beta’ subunit inhibit transcript cleavage and transcript elongation. J Biol Chem. 1998;273:24912–24920. doi: 10.1074/jbc.273.38.24912. [DOI] [PubMed] [Google Scholar]
- Zhang J, Palangat M, Landick R. Role of the RNA polymerase trigger loop in catalysis and pausing. Nat Struct Mol Biol. 2010;17:99–104. doi: 10.1038/nsmb.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y, Steitz TA. Crystal structures of the E.coli transcription initiation complexes with a complete bubble. Mol Cell. 2015;58:534–540. doi: 10.1016/j.molcel.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen KR, Leksa NC, Schwartz TU. Optimized E.coli expression strain LOBSTR eliminates common contaminants from His-tag purification. Proteins. 2013;81:1857–1861. doi: 10.1002/prot.24364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Guo X, Myasnikov AG, Chen J, Crucifix C, Papai G, Takacs M, Schultz P, Weixlbaumer A. Structural Basis for NusA Stabilized Transcriptional Pausing. Molecular Cell. 2018;69:816–827.:e814. doi: 10.1016/j.molcel.2018.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobi AJ, Wilmanns M, Sachse C. Model-based local density sharpening of cryo-EM maps. eLife. 2017;6 doi: 10.7554/eLife.27131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedev AA, Young P, Isupov MN, Moroz OV, Vagin AA, Murshudov GN. JLigand: a graphical tool for the CCP4 template-restraint library. Acta Crystallographica Section D: Biological Crystallography. 2012;68:431–440. doi: 10.1107/S090744491200251X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nature Methods. 2017;14:290. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Scheres SHW. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. Journal of Structural Biology. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Twist KA, Husnain SI, Franke JD, Jain D, Campbell EA, Nickels BE, Thomas MS, Darst SA, Westblade LF. A novel method for the production of in vivo-assembled, recombinant Escherichia coli RNA polymerase lacking the alpha C-terminal domain. Protein science : a publication of the Protein Society. 2011;20:986–995. doi: 10.1002/pro.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassylyeva MN, Lee J, Sekine S-i, Laptenko O, Kuramitsu S, Shibata T, Inoue Y, Borukhov S, Vassylyev DG, Yokoyama S. Purification, crystallization and initial crystallographic analysis of RNA polymerase holoenzyme from Thermus thermophilus. Acta Crystallographica Section D. 2002;58:1497–1500. doi: 10.1107/S0907444902011770. [DOI] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta Crystallographica Section D. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Palovcak E, Armache J-P, Cheng Y, Agard D. Anisotropic Correction of Beam-induced Motion for Improved Single-particle Electron Cryo-microscopy. bioRxiv. 2016 doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession numbers for the five cryo-EM reconstructions (non-swiveled backtracked complex, swiveled backtracked complex, pre-cleavage complex, post-cleavage complex, reactivated complex) reported in this paper are … Fitted models were deposited in the PDB under accession numbers VVVV (all atom model for non-swiveled backtracked complex), WWWW (all atom model for swiveled backtracked complex), XXXX (all atom model for pre-cleavage complex), YYYY (all atom model for post-cleavage complex), ZZZZ (all atom model for reactivated complex).