Abstract
End-stage liver diseases are an increasing health burden, and liver transplantations are currently the only curative treatment option. Due to a lack of donor livers, alternative treatments are urgently needed. Human liver organoids are very promising for regenerative medicine; however, organoids are currently cultured in Matrigel, which is extracted from the extracellular matrix of the Engelbreth-Holm-Swarm mouse sarcoma. Matrigel is poorly defined, suffers from high batch-to-batch variability and is of xenogeneic origin, which limits the clinical application of organoids. Here, a novel hydrogel based on polyisocyanopeptides (PIC) and laminin-111 is described for human liver organoid cultures. PIC is a synthetic polymer that can form a hydrogel with thermosensitive properties, making it easy to handle and very attractive for clinical applications. Organoids in an optimized PIC hydrogel proliferate at rates comparable to those observed with Matrigel; proliferation rates are stiffness-dependent, with lower stiffnesses being optimal for organoid proliferation. Moreover, organoids can be efficiently differentiated toward a hepatocyte-like phenotype with key liver functions. This proliferation and differentiation potential maintain over at least 14 passages. The results indicate that PIC is very promising for human liver organoid culture and has the potential to be used in a variety of clinical applications including cell therapy and tissue engineering.
Keywords: hepatocyte differentiation, human liver organoids, polyisocyanopeptides, synthetic hydrogels, tissue engineering
1. Introduction
The liver is the largest internal organ in the body and is responsible for crucial metabolic functions. Upon acute damage, the liver has a great regenerative capacity and can restore up to 70% of its mass.[1] However, injury caused by chronic diseases such as viral infections or alcoholic fatty liver disease lead to a gradual decrease in liver function, eventually resulting in end-stage liver disease.[2] For end-stage liver disease, the only curative treatment option is organ transplantation.[3] Unfortunately, the demand for transplantable donor livers far exceeds availability, resulting in approximately 20% of patients dying while on the waitlist.[4] In addition, the experience of waiting for a donor organ is a great burden to the patient and the stress of not knowing if a donor organ will become available in time can negatively impact prognosis.[5] Alternatives for donor livers are thus urgently needed.[6]
Liver organoids hold great promise for regenerative approaches, for example, cell therapy or as cellular building blocks for liver tissue engineering.[7] Organoids are 3D miniature versions of their organ of origin, which contain most, if not all, cell types that are present in vivo.[8] Human liver organoids can be initiated from a variety of cells, including embryonic stem cells,[9] induced pluripotent stem cells (PSCs),[10] multipotent adult tissue-resident stem cells,[11,12] and primary hepatocytes.[13] In this paper, we focus on organoids established from leucine-rich repeat-containing G-protein coupled receptor 5 positive (LGR5+) adult liver stem cells. These organoids can be expanded infinitely in culture, remain genetically stable, and can be differentiated into the hepatocyte[11,12] or cholangiocyte[14] lineages; as such, they reflect important functional and structural aspects of the liver. Transplantation experiments have shown that organoids were able to engraft into the damaged parenchyma of mice and rats, where they became functional hepatocytes, although the repopulation efficiency remained low.[11,12,15] Another benefit is the ability to genetically modify these organoids in vitro using the CRISPR/Cas system, and in theory, enable the correction of relatively simple genetic defects of a patient prior to autologous cell transplantation.[16]
The current gold standard for organoid culture is critically dependent on the use of Matrigel as a 3D matrix. Matrigel is a gelatinous protein mixture extracted from the extracellular matrix (ECM) of the Engelbreth-Holm-Swarm mouse sarcoma, which is propagated in mice.[17] Matrigel is widely used because it is extremely bioactive and therefore supports proliferation of a wide variety of organoid types.[18] Organoids cultured in Matrigel, however, are unsuitable for clinical use due to the murine tumor origin of Matrigel. Another disadvantage of Matrigel is very high (up to 50%) batch-to-batch variations, which may influence reproducibility of organoid experiments.[19]
In this paper, we sought to develop a synthetic, defined and easy-to-handle Matrigel alternative for the expansion and differentiation of human liver organoids. In 2016, Gjorevski et al. published the first landmark paper in the field of synthetic hydrogels for organoid culture.[20] Murine intestinal organoid formation and differentiation was successfully achieved in an enzymatically-crosslinked polyethylene glycol (PEG)-based hydrogel when modified with laminin or Arg-Gly-Asp (RGD) peptides. This group recently published a preprint on bioRxiv, in which they applied the same PEG-based hydrogel for the culture of murine and human liver organoids.[21] Moreover, in 2018, Broguiere et al. published a defined (although not synthetic) hydrogel based on a combination of a thrombin-crosslinked fibrin gel, nanocellulose and laminin-111/entactin complex.[22] The gel was as efficient as Matrigel for the expansion and differentiation of murine intestinal organoids and also seemed to be applicable for the expansion of human organoids from a variety of different organs. However, both PEG and fibrin-based hydrogels must be chemically or enzymatically cross-linked to achieve gelation, which complicates the retrieval of organoids from the hydrogel and may be a disadvantage for certain applications.
In this study, we used hydrogels based on polyisocyanopeptides (PIC), a synthetic nonimmunogenic material, for the expansion and differentiation of human liver organoids.[23] Because PIC-based hydrogels exhibit thermoreversible gelation, they are convenient to work with, provide gentle conditions for organoids during passaging and have a unique range of applications, such as the use of the hydrogel for bioprinting without a necessity for enzymatic or light-induced cross-linking.[24] PIC is a free-flowing liquid below 16 °C;[25] when the temperature rises above 16 °C, the liquid becomes a viscous hydrogel within minutes. This flexible behavior facilitates simple cell recovery protocols that merely rely on changing the temperature. It is also possible to modify the PIC backbone with a variety of desired molecules and bioactive epitopes.[26] Most importantly, PIC is a bioinert material and has already been applied by others in vivo without evoking any adverse immune response.[27] We therefore expect that culturing organoids in a PIC hydrogel will enable their use for organoid-based clinical applications such as cell therapy and tissue engineering approaches.
2. Results and Discussion
2.1. An Optimized PIC Hydrogel Supports Liver Organoid Expansion
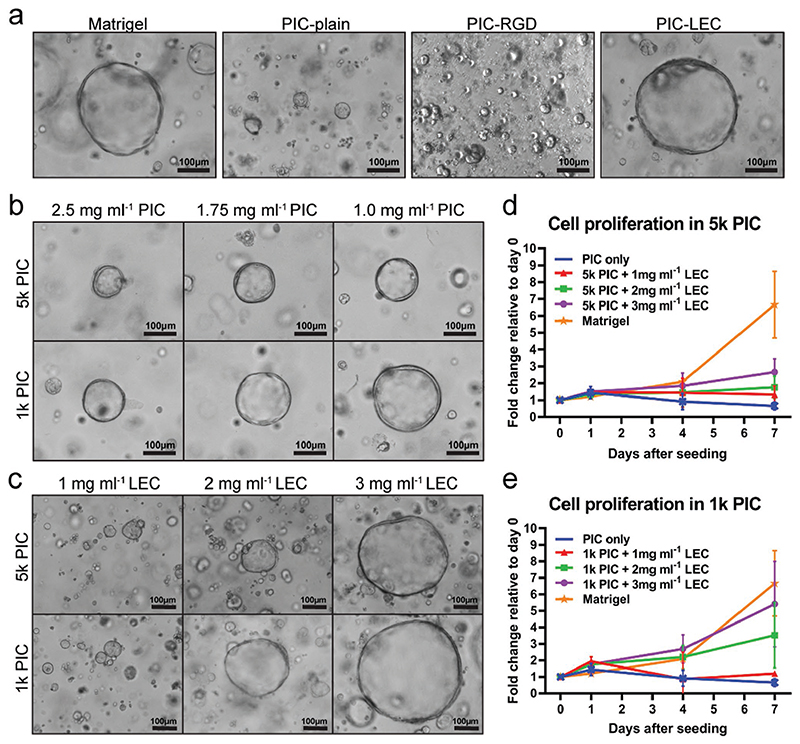
We first developed a PIC hydrogel that supports organoid formation and proliferation. PIC itself does not contain any bioactive components to support cell attachment or induce proliferation. Accordingly, we observed marginal organoid formation, with little to no proliferation when single organoid cells were seeded in plain PIC and cultured for 7 days in organoid expansion medium (EM) (Figure 1a). To stimulate proliferative pathways involved in cell attachment to the ECM, we tested a PIC hydrogel that contained covalently-attached RGD sequences, which can be recognized by cellular integrins.[20] However, the incorporation of RGD motifs in PIC was not sufficient to induce organoid proliferation (Figure 1a). Our findings contrast with the recently published preprint by Sorrentino et al., where the addition of RGD motifs to a PEG hydrogel seemed sufficient for human liver organoid culture; our dissimilar results may be explained by a difference in RGD concentrations. In our commercially purchased hydrogel, RGD peptides are covalently coupled to the PIC at a concentration of ≈0.2 mM, whereas concentrations of 1–2 mM have been used in earlier publications.[21] We continued to add 3 mg mL−1 lamininentactin complex (LEC) to the plain PIC gel, which resulted in efficient organoid formation and proliferation that seemed comparable to the Matrigel controls (Figure 1a). LEC is the main component of Matrigel and has previously been shown to promote formation and proliferation of murine intestinal organoids.[22]
Figure 1. An optimized PIC hydrogel supports liver organoid expansion.
Single organoid cells were seeded at day 0 and cultured in human organoid expansion medium (EM) supplemented with the Rho kinase inhibitor Y-27632 for 14 days. Four different donors were analyzed in independent experiments (N = 4). a) Light microscopy images of organoids at day 7 after single cell seeding. Organoids did not proliferate in PIC-plain and PIC-RGD, but showed a morphology comparable to Matrigel in PIC supplemented with 3 mg mL−1 LEC (PIC-LEC). b) Lower concentrations of PIC improve organoid proliferation. c) Light microscopy pictures reveal a dose-dependent effect of LEC on organoid proliferation. Quantification of organoid proliferation in d) 5k PIC and e) 1k PIC. Organoids from five different donors were cultured in Matrigel or PIC with different concentrations of LEC. Relative cell numbers were determined by an Alamar blue assay every 2-3 days and cell expansion relative to day 0 was calculated. Each dot represents the mean of the four donors with standard deviation.
We then examined the effects of the mechanical properties of PIC hydrogels on liver organoid proliferation. We used PIC at two different molecular weights, 1 kDa PIC (1k PIC) and 5 kDa PIC (5k PIC), and modulated the hydrogels by varying PIC concentrations, resulting in a higher stiffness and lower porosity with increasing concentrations. Light microscopy pictures and Alamar blue assays (ABAs) confirmed that lower PIC concentrations led to a significantly increased organoid proliferation (Figure 1b).
After determining the optimal PIC concentration (1 mg mL−1), we continued to optimize the concentration of LEC in PIC. In both the 1k PIC and 5k PIC hydrogel backbones, we observed a concentration-dependent increase in organoid proliferation (Figure 1c), with highest proliferation rates at 3 mg mL−1 LEC in a 1k PIC hydrogel (Figure 1d,e).
Taken together, a 1k PIC hydrogel at a concentration of 1 mg mL−1 PIC, supplemented with 3 mg mL−1 LEC, provided an environment that optimally supported organoid formation and proliferation with an efficiency comparable to Matrigel.
2.2. Soft PIC Hydrogels Enhance Organoid Expansion
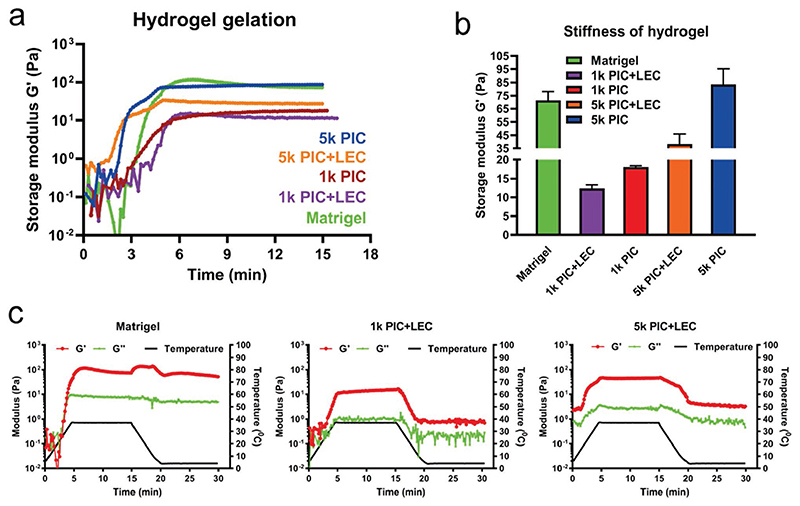
To gain more insights into the different PIC hydrogels, we analyzed their mechanical properties. As the gelation of both Matrigel and PIC hydrogels is temperature-dependent, we placed the different hydrogel premixes on a temperature-controlled plate and followed their gelation in time. The plate was heated from 4 to 37 °C at a speed of +7 °C min−1 and then maintained at a temperature of 37 °C for 10 min. Our analysis of storage modulus G′ showed that for all premixes, gelation started during the heating period and reached a plateau after 10 min at 37 °C (Figure 2a). We observed that the stiffness of the 5k PIC (≈83 Pa) was comparable to that of Matrigel (≈71 Pa), whereas the stiffness of all other PIC hydrogels (with and without LEC) was lower. The 5k PIC hydrogels demonstrated a higher stiffness (≈83 Pa) compared to 1k PIC hydrogels (≈18 Pa) of the same concentration and the addition of LEC resulted in a decreased stiffness for both the 1k (≈12 Pa) and 5k (≈38 Pa) PIC hydrogels (Figure 2a,b), which was most likely caused by sterical interference of PIC network formation by the large laminin molecules. By contrast, increasing LEC concentrations did not significantly affect hydrogel stiffnesses of the same PIC concentration, suggesting that the LEC concentration-dependent organoid proliferation was caused by biological rather than mechanical effects (unpublished data). As such, the softest PIC hydrogel (1k PIC hydrogel at a concentration of 1 mg mL−1 PIC, supplemented with 3 mg mL−1 LEC) best supported liver organoid expansion. This corresponds with our observations that organoids proliferate better in 70% Matrigel (diluted with culture media and thus, softer) compared to 100% Matrigel (unpublished data). Remarkably, Sorrentino et al. observed that, for their PEG hydrogels, a much higher stiffness of 1.3 kPa optimally supported colony formation of human liver organoids, whereas a lower stiffness of 0.3 kPa was much less efficient.[21] The authors, however, did not quantify proliferation of the liver organoids in their various hydrogels.
Figure 2. Soft PIC hydrogels enhance organoid expansion.
Mechanical properties of Matrigel and PIC hydrogels measured at constant frequency (1 rad s−1) and strain (1%). a) Time trace of storage modulus G′ showing hydrogel gelation with the temperature raising from 4 to 37 °C (+7 °C min−1) and holding at 37 °C. For all conditions, the starting modulus at time = 0 min were much lower because all the samples were liquid at 4 °C; the modulus almost reached plateau at time = 6 min when the temperature was held at 37 °C (N = 3). b) Stiffness of hydrogels showing the storage modulus G′ of Matrigel and PIC hydrogels after 10 min incubation at 37 °C (N = 3). c) Time trace of storage modulus G′ and loss modulus G″ of Matrigel and PIC-LEC hydrogels through a heating and cooling cycle. At time = 0 min, G′ and G″ were close to each other in all three conditions showing a liquid state; at time = 5 min, G′ reached a peak higher than G″ after heating from 4 to 37 °C (+7 °C min−1); during time = 5–10 min, all gels remained plateaued when the temperature was held at 37 °C; after time = 15 min, both PIC hydrogels showed a sharp decrease of G′ when the cooling cycle started from 37 to 4 °C (-7 °C min−1), and G′ of both PIC hydrogels dropped to less than 1 Pa after being held at 4 °C for 10 min. However, G′ of Matrigel was still higher than 50 Pa after cooling for 15 min (N = 2).
For serial passaging and long-term expansion of organoids in 3D culture, the thermoreversible character of PIC hydrogels is one of the most significant advantages over other hydrogels. Using a rheometer, we compared the thermoreversible character of PIC hydrogels with Matrigel through a controlled temperature change. We first heated the hydrogel premixes from 4 to 37 °C, allowing them to form a hydrogel. Subsequently, we cooled the hydrogels down to 4 °C at a speed of 7 °C min−1 and then kept the samples at 4 °C for another 10 min to see whether they would become liquid again. Our results show that the optimized PIC-LEC hydrogel promoted a faster gel/sol transition than Matrigel (Figure 2c). After approximately 6 min of heating, the PIC-LEC hydrogels turned into a gel, which immediately turned into solution again upon cooling (Figure 2c). In contrast, Matrigel still had a stiffness of ≈50 Pa after a 15 min cooling period (Figure 2c). This corresponds with our observations that PIC hydrogels are more easily manipulated and thereby easier to handle during organoid passaging, giving them a major advantage over Matrigel.
We also characterized the dynamic properties of PIC hydrogels after gelation, since this may influence organoid shape and size. We investigated the modulus change of the hydrogels in a frequency range from 10 to 0.1 rad s−1. Both Matrigel and PIC hydrogels were stable in the measured frequency range and behaved as a viscoelastic solid (Figure S1, Supporting Information). Interestingly, the addition of LEC did not influence the dynamic properties of the PIC hydrogels, indicating that the dynamic character of the PIC-LEC hydrogels did not play a key role in supporting organoid expansion.
2.3. Organoids in PIC Retain a Stem/Progenitor Phenotype and Are Highly Proliferative
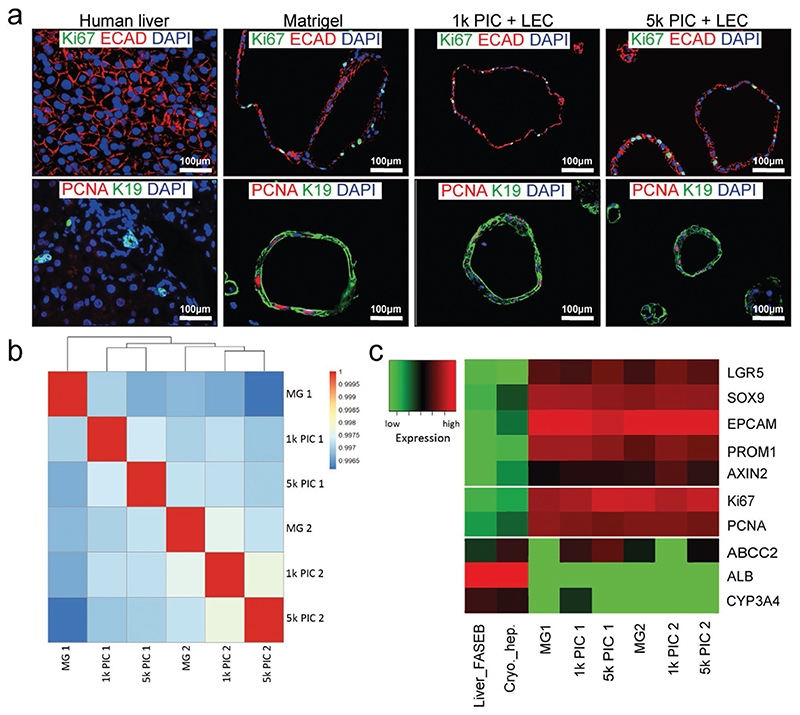
To further characterize liver organoids in the optimized PIC-LEC hydrogel, we seeded single organoid cells in EM conditions and cultured them for 14 days. Single organoid cells seeded in Matrigel were used as a control. Immunofluorescent analysis showed that the cells in all hydrogels retained an epithelial E-cadherin (ECAD)-positive phenotype and were highly proliferative (Figure 3a).
Figure 3. Organoids in PIC retain a stem/progenitor phenotype and are highly proliferative.
Single organoid cells were seeded at day 0 and organoids were expanded in the different hydrogels for 14 days. Three different donors were analyzed in independent experiments (N = 3). a) Immunofluorescent analysis of paraffin-embedded organoids confirmed that the organoids in all gels have an epithelial progenitor phenotype and are highly proliferative. Human liver tissue was used as a control. Whole transcriptome sequencing analysis of organoids from two independent donors at day 7 of expansion in Matrigel, 1k PIC-LEC, and 5k PIC-LEC (N = 2). b) Pearson correlation map of the organoids in different hydrogels. Note that the correlation of cultured organoids is more than 99%. c) Heatmap of markers for stem/progenitor cells, epithelial cells, and mature hepatocytes selected from the mRNA sequencing data.
To further interrogate this proliferative phenotype, we performed mRNA sequencing on organoids from two different donors that were cultured in 1k PIC-LEC, 5k PIC-LEC, or Matrigel for 14 days. A Pearson’s correlation test showed that all hydrogel conditions (Matrigel, 1k PIC, and 5k PIC) displayed a similar correlation coefficient for each donor (Figure 3b). We then analyzed the expression of a few well-known liver stem/progenitor cell markers (LGR5, SOX9, EPCAM, PROM1, AXIN2), proliferation markers (KI67, PCNA), and hepatocyte markers (ABCC2, ALB, CYP3A4), and compared these with gene expression in cryopreserved hepatocytes and human liver tissue. As expected, stem cell markers and proliferation markers were highly expressed in all hydrogel conditions in both donors, whereas hepatocyte markers were low (Figure 3c).
2.4. Organoids in PIC can be Differentiated into Functional Hepatocyte-like Cells
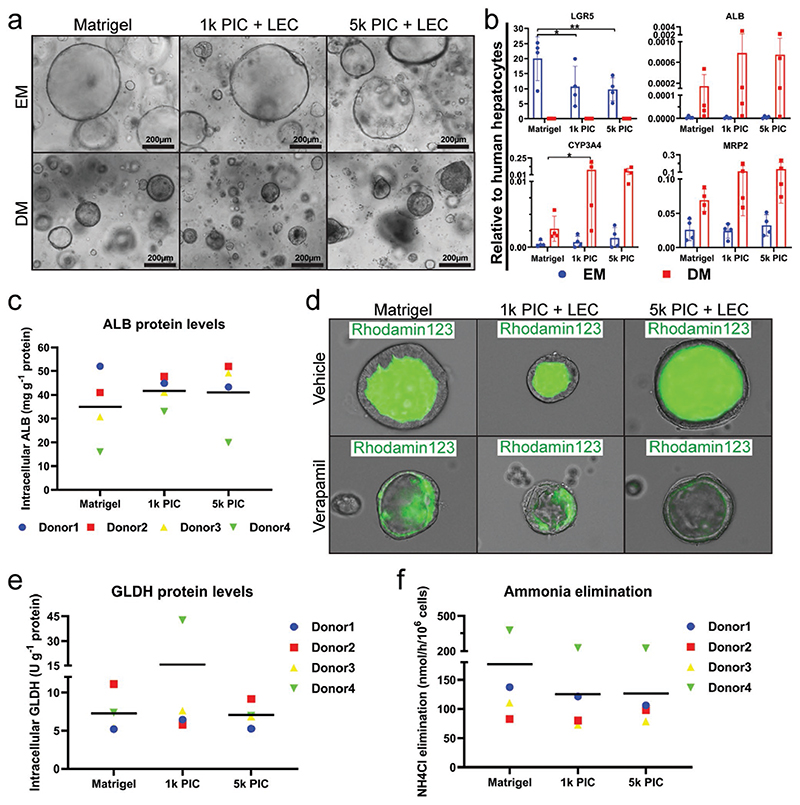
It has previously been shown that human liver organoids can be differentiated toward hepatocyte-like cells when cultured in differentiation medium (DM), which contains N-[(3,5-difluorophenyl)acetyl]-L-alanyl-2-phenyl]glycine-1,1-dimethylethyl ester (DAPT), fibroblast growth factor 19 (FGF19), dexamethasone and bone morphogenetic protein 7 (BMP-7), and is void of inducers of proliferation, such as Rspondin-1 and forskollin (FSK).[12,28] In order to compare the differentiation potential of human liver organoids cultured in PIC-LEC to Matrigel, we differentiated organoids toward hepatocyte-like cells. Light microscopy demonstrated that both Matrigel and PIC-LEC organoids acquired a more dense and dark morphology at day 8 of differentiation compared to expansion conditions (Figure 4a). Analysis of the organoids by quantitative real-time PCR (qRT-PCR) after 8 days of differentiation confirmed that the stem cell marker LGR5 was downregulated in both hydrogels (Figure 4b). In contrast, several hepatocyte markers such as cytochrome p450 3A4 (CYP3A4), albumin (ALB), and multidrug resistance-associated protein 2 (MRP2) were upregulated and reached comparable levels in PIC-LEC and Matrigel (Figure 4b).
Figure 4. Organoids differentiated into functional hepatocyte-like cells in PIC.
Organoids were differentiated in Matrigel, 1k PIC-LEC, or 5k PIC-LEC for 8 days (N = 4). a) Light microscopy images of organoids showing that differentiated organoids became denser and darker compared to EM organoids in all hydrogels. b) Gene expression of stem cell and hepatocyte markers in differentiated organoids from four independent donors. Transcriptional levels of leucine-rich repeat-containing G-protein coupled receptor 5 (LGR5), albumin (ALB), cytochrome p450 3A4 (CYP3A4), and multidrug resistance-associated protein 2 (MRP2) were determined by qRT-PCR and compared to cryopreserved human hepatocytes. Hepatocyte functionality of differentiated organoids in the different hydrogels was assessed. Four different donors were analyzed in independent experiments. d) Rh123 transport was determined as read-out for MDR1 activity. Verapamil was added as an inhibitor of MDR1 function. c) Albumin concentrations and e) GLDH levels in cell lysates were measured after incubation in DM for 24 h. Comparison was normalized to total protein concentrations. Graphs indicate mean ± SD. f) Ammonium elimination from the culture medium was determined as read-out for hepatocyte functionality. *indicates p-value ≤ 0.05.
We also assessed the functionality of the differentiated organoids in both hydrogels with several assays. Important functions of hepatocytes include the production of serum proteins, vectorial uptake, and secretion of several compounds, and urea cycle activity.[29] To determine the production of serum proteins, we quantified the intracellular ALB concentration of the organoids at day 8 of differentiation. We measured comparable concentrations of ALB in PIC-LEC and Matrigel (Figure 4c). To determine vectorial transmembrane transport, we exposed organoids in both hydrogels to rhodamine123 (Rh123), a fluorescent compound that is actively secreted from the apical membrane of hepatocytes by the transporter multidrug resistance gene 1 (MDR1).[30] Organoids in both hydrogels accumulated fluorescence inside their lumens (Figure 4d). To confirm that this accumulation was MDR1-specific, organoids were pretreated with the competitive MDR1 inhibitor verapamil. This resulted in an accumulation of Rh123 in the cytoplasm of the cells, whereas no fluorescence was observed in the lumen of the organoids, confirming the MDR1-specific transport of Rh123. Finally, we tested organoids in both hydrogels for their capacity to eliminate ammonium from the media, a measure for urea cycle activity. Ammonia reacts with α- ketoglutaric acid to form L-glutamate, which is catalyzed by L-glutamate dehydrogenase (GLDH).[31] The intracellular concentration of GLDH was comparable in organoids from both hydrogels (Figure 4e), along with ammonia elimination from the media (Figure 4f).
2.5. Long-Term Expansion of Organoids in PIC
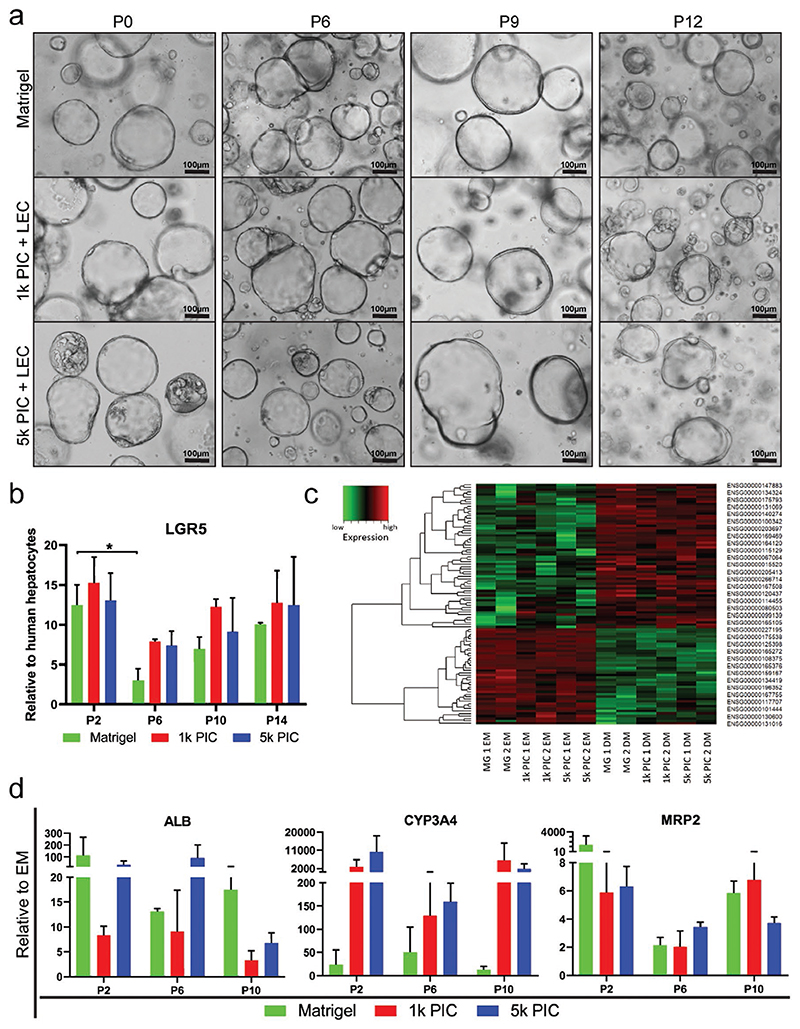
One of the main advantages of organoid cultures is that they can be serially passaged, allowing for seemingly unlimited expansion. This continued expansion is fueled by the LGR5+ stem cells in liver organoid cultures.[32] In order to test if this stem cell phenotype and organoid proliferation capacity can be retained in PIC-LEC over several passages, we cultured human liver organoids from two donors for 14 passages with weekly passaging. Light microscopy images confirmed that the organoids retained their cystic morphology through all passages and remained highly comparable in PIC-LEC and Matrigel (Figure 5a). qRT-PCR showed that the expression of LGR5 in organoids cultured in PIC-LEC or Matrigel was stable over the course of 14 weeks (4 months) (Figure 5b). We then assessed the differentiation potential of organoids that had been cultured in PIC-LEC for 2, 6, and 10 passages (P2, P6, and P10). After those passages in EM, in either PIC-LEC or Matrigel, organoids from two donors were differentiated in DM for 8 days and subsequently analyzed by mRNA sequencing (P6) and qRT-PCR (P2, P6, and P10).
Figure 5. Long-term expansion of organoids in PIC.
Organoids from two different donors were cultured for 12 weeks in Matrigel, 1k PIC-LEC, and 5k PIC-LEC with weekly passaging. a) Light microscopy pictures show that organoids maintained their morphological phenotype during all passages in Matrigel, 1k PIC-LEC, and 5k PIC-LEC. b) qRT-PCR analysis of the stem cell marker LGR5 shows stable expression levels at passage 2, 6, 10, and 14 in all different hydrogels. c) mRNA sequencing on organoids from two independent donors in EM and after differentiation (day 8 in DM). The 100 most significant differentially-expressed genes in DM versus EM are displayed in a heatmap. Note that EM samples from both donors and all three hydrogels show a very similar expression pattern and that all DM samples from both donors and all three different hydrogels are similar. A full list of genes is provided as Table S1, Supporting Information. d) Gene expression of hepatocyte markers in differentiated organoids from two independent donors during long-term culture in Matrigel or PIC hydrogels. Transcriptional levels of hepatocyte markers ALB, CYP3A4, and MRP2 were determined by qRT-PCR and compared to their respective EM controls (N = 2).
At P6, the 100 most significant differentially-expressed genes in DM versus EM displayed an identical pattern in PIC-LEC compared to Matrigel (Figure 5c). Gene ontology analysis[33] of these genes revealed significant enrichment for processes related to liver functions, such as lipid and alcohol metabolism (Tables S1 and S2, Supporting Information). Analysis by qRT-PCR confirmed that the gene expression of several hepatocyte markers in differentiated organoids was upregulated compared to their respective EM controls in all hydrogels and that this upregulation was reproducible at P2, P6, and P10 (Figure 5d), indicating that organoids retain their differentiation potential over several passages in PIC-LEC.
2.6. Human Recombinant Laminin-111 Can Substitute LEC
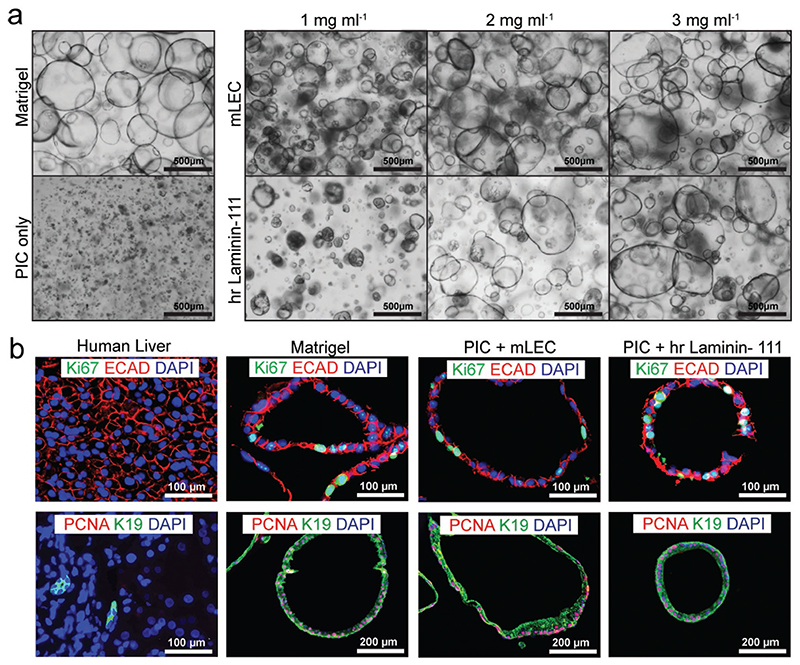
The hydrogel we have developed and extensively characterized for human liver organoid culture contains a chemically well-defined PIC component supplemented by a bioactive LEC, and the combination has several advantages over Matrigel. However, the LEC originates from mouse sarcoma and is thus not xenofree and not suitable for clinical applications. Therefore, we examined the possibility of using human recombinant laminin-111 (hrlaminin-111) as a substitute for LEC. The experiments with LEC showed a concentration-dependent proliferation of organoids in LEC at concentrations of 1, 2, and 3 mg mL−1 (Figure 1c). We prepared PIC hydrogels with the same concentrations of hrlaminin-111 and seeded single organoid cells in those gels. For two of the three analyzed donors, we observed a comparable concentration-dependent proliferation in the PIC-LEC and PIC-hrlaminin-111 (Figure 6a). For the third donor however, proliferation in the PIC-hrlaminin-111 seemed lower compared to PIC-LEC (Figure S2, Supporting Information). To further characterize the phenotype and proliferative potential of organoids cultured in PIC-LEC and PIC-hrlaminin-111, we conducted immunofluorescent stainings for ECAD, keratin 19 (K19), Ki67, and PCNA, which confirmed that the cells in all hydrogels retained an epithelial ECAD-positive phenotype and were highly proliferative (Figure 6b). These data indicate that, although less efficient, it seems possible to replace LEC by hrlaminin-111 and possibly other human recombinant ECM proteins as the bioactive components in PIC, rendering the PIC completely synthetic and applicable for clinical applications such as cell therapy or tissue engineering.
Figure 6. Human recombinant laminin-111 can substitute LEC.
a) Organoids from three different donors were cultured for 7 days in Matrigel, PIC-plain, or 1k PIC supplemented with either LEC or human recombinant Laminin (hrlaminin-111). Light microscopy pictures show a similar morphological phenotype in LEC and hrlaminin-111, with increased organoid size and number at higher laminin concentrations. (N = 3) b) Immunofluorescent analysis of paraffin-embedded organoids confirmed that the organoids in all gels have an epithelial progenitor phenotype and are highly proliferative. Organoids cultured in Matrigel, 5k PIC with 3 mg mL−1 LEC or 5k PIC with 2 mg mL−1 hrlaminin-111 are displayed. Human liver tissue was used as a control (N = 1).
3. Conclusion
In this study, we developed and characterized a novel synthetic hydrogel for the expansion and differentiation of human adult stem cell-derived liver organoids. PIC hydrogel alone was not sufficient to support organoid growth, but PIC mixed with LEC supported organoid formation and proliferation (Figure 1). As no covalent reaction can occur between PIC and LEC in the absence of catalysts, the interaction of PIC and LEC is most probably non-covalent self-assembly, but the exact interaction of PIC and LEC remains to be determined. The optimized PIC-LEC hydrogel supported organoid proliferation at rates that were similar to Matrigel (Figure 1), with lower stiffnesses most favorable for organoid proliferation. We showed that the organoids were highly proliferative in PIC-LEC when cultured in EM (Figure 3) and could be efficiently differentiated toward functional hepatocyte-like cells when cultured in DM (Figure 4). Importantly, the stem cell phenotype and proliferation and differentiation capacity of the organoids could be maintained in PIC-LEC over several passages, enabling their seemingly unlimited expansion and subsequent maturation (Figure 5). Finally, we showed that the LEC in the PIC-LEC gels could be replaced by hrlaminin-111, resulting in a completely synthetic hydrogel for the expansion of human liver organoids. Future studies will have to confirm the suitability of this synthetic hydrogel for differentiation of human liver organoids toward hepatocytes and cholangiocytes. Moreover, hrlaminin-111 might be replaced or complemented by other ECM components to improve the differentiation to either hepatocytes or cholangiocytes.[34] It will also be interesting to replace full-length ECM proteins by peptides derived from laminin-111 and other ECM components. For PEG hydrogels, it has been shown that high concentrations of covalently-linked A55 peptide (GGFLKYTVSYDI) and AG73 peptide (RKRLQVQLSIRT), which are both located on the α-1 laminin chain, supported intestinal organoid proliferation and survival that approached levels in Matrigel.[20]
Together with the recent preprint by Sorrentino et al., this is the first chemically-defined and synthetic hydrogel that supports human liver organoid expansion and differentiation at rates comparable to Matrigel.[21] Both hydrogels, PEG-based and PIC-based, have their own advantages. An enzymatically or chemically crosslinked PEG hydrogel was the first synthetic hydrogel applied for intestinal organoid culture derived from multipotent or PSCs.[20,35] ECM components such as fibronectin, laminin-111, collagen IV, hyaluronic acid, and perlecan were added to the hydrogel, and a minimized PEG-RGD hydrogel was sufficient to support organoid formation and proliferation. This hydrogel system was recently adapted to liver organoids.[21] Proliferation rates of intestinal or liver organoids in the PEG hydrogels were not quantified however, so it remains to be determined how they compare to our PIC hydrogel. In a very elegant approach, the authors also modified the PEG hydrogels to be hydrolytically or protease-degradable, as such mimicking the dynamic character of the in vivo environment.[20,35] Similar modifications may be applied to the PIC hydrogels by applying a copper-free click chemistry as previously published.[36] The most striking difference between the PEG and PIC hydrogels is the mode of gelation. Whereas the chemically- or enzymatically-crosslinked PEG hydrogel is advantageous for applications where a controlled stiffness over a prolonged period of time is necessary, the thermoreversible properties of the PIC hydrogel allow for easier cell recovery during organoid culture and make it advantageous for certain practical purposes such as bioprinting and in vivo cell therapy.[37] PEG-based hydrogels have already been applied as delivery vehicles in vivo, where they supported localized engraftment of human intestinal organoids derived from PSCs in colonic mucosal wounds and enhanced wound closure.[35] In order to facilitate gelation in situ, a custom made-device was applied, in which the hydrogel precursor solution and the crosslinking solution only met in the tubing during injection.[35] PIC hydrogels have also been applied in vivo for wound healing studies and subcutaneous cell transplantations without causing any adverse effects.[26,27] The thermo-responsive properties of the PIC hydrogels made in vivo applications very convenient, and the PIC gelated within 1 min upon contact with the warm skin.[27] These results highlight the great potential for in vivo applications of organoids cultured in PIC. Of note, we recently published a protocol for the large-scale expansion of human liver organoids in suspension culture,[28] and foresee that a combination of this large-scale expansion method and our well-defined synthetic hydrogel will pave the way for clinical applications of human liver organoids in the near future.[38]
4. Experimental Section
Hydrogel Preparation and Characterization
Noviogel (PIC) was purchased from Sopachem (1k-PIC-P, 1k-PIC-RGD, 5k-PIC-P, and 5k-PIC-RGD, followed by catalog numbers: NCN01, NCN03, NCN02, and NCN04, respectively). Stock solutions of 5 mg mL−1 were made by adding 3 mL of advanced DMEM/F12 (AD, Gibco) to each bottle of PIC. To optimize the PIC hydrogels, both 1k and 5k PIC were tested in three concentrations: 1, 1.75, and 2.5 mg mL−1. Where indicated, LEC (Corning) was added to each PIC formulation at concentrations of 1, 2, or 3 mg mL−1 to test the dose effects. When LEC was replaced with human recombinant laminin-111 (hrlaminin-111, Biolamina, LN111-050) the same concentrations were used (1, 2, or 3 mg mL−1).
Working Solutions of Thermoresponsive Materials
PIC, LEC, and Matrigel (Corning, 356 237) were divided into aliquots to reduce frequent freeze-thaw cycles and stored at −20 °C. To make hydrogels, materials were thawed and prepared on ice before gelation. Once plated, hydrogels were allowed to solidify for 15–30 min at 37 °C.
Rheological measurements were conducted on a discovery HR-2 rheometer (TA instruments) to test the mechanical properties of hydrogels made from PIC, PIC-LEC, or Matrigel. A temperature-controlled plate (20 mm parallel plate, aluminium-105381) was used for all measurements. Settings included a geometry diameter of 20 mm and a measuring gap of 200 μm. For hydrogel formation measurements, 150 μL of each hydrogel premix was deposited onto the plate and conditioned for 2 min at 4 °C. The plate was then heated (strain 1%, angular frequency 1.0 rad s−1) from 4 to 37 °C at a speed of 7 °C min−1, and then continuously measured at 37 °C for another 10 min. Following the hydrogel formation step, frequency sweep tests were carried out and data was collected in the frequency range of 10 rad s−1 to 0.1 rad s−1. For thermoreversible characterization, hydrogel formation was carried out as described above. After remaining at 37 °C for 10 min, the plate was cooled from 37 to 4 °C at a rate of 7 °C min−1. When the temperature of the plate reached 4 °C, it was held for another 10 min to observe if the hydrogels could become fluidic again.
Human Liver Organoid Establishment
Liver biopsies were obtained during liver transplantation at the Erasmus Medical Center Rotterdam and in accordance with the ethical standard of the Helsinki Declaration of 1975. Use of the tissue for research purposes was approved by the Medical Ethical Council of the Erasmus Medical Center and informed consent by the liver transplant recipient was given (MEC-2014-060). To establish human liver organoid lines, liver biopsies were cut into small pieces, followed by the enzymatic digestion with type II collagenase (0.125 mg mL−1, Gibco) and dispase (0.125 mg mL−1, Gibco) in DMEM GlutaMAX (Gibco) containing 1% fetal calf serum (FCS, Gibco). The supernatant was collected every hour. Tissue digestion followed by supernatant collection was performed three times. Collected single cells were washed in DMEM GlutaMAX (Gibco) containing 1% FCS (Gibco) and centrifuged for 5 min at 1500 rpm. The cells were resuspended in Matrigel (Corning) at a concentration of ≈400 cells per μL. Cells were seeded in droplets (50 μL) in non-attaching 24-well plates (M9312, Greiner, Merck). EM was added after ≈15 min incubation at 37 °C, 5% CO2.
Organoid Expansion and Differentiation in PIC and Matrigel Droplets
For human liver organoid expansion, previously defined EM was used.[9] EM was based on advanced DMEM/F12 (AD, Gibco) containing 1% GlutaMax (Gibco), HEPES (10 mM, Gibco), and 1% penicillin-streptomycin (Gibco, Dublin, Ireland). EM was supplemented with 10% Rspondin-1 conditioned medium (the Rspo1-Fc-expressing cell line was kindly provided by Calvin J. Kuo), 1% N2 supplement (Invitrogen), 2% B27 supplement without vitamin A (Invitrogen), N-acetylcysteine (1.25 mM, Sigma-Aldrich), nicotinamide (10 mM, Sigma-Aldrich), epidermal growth factor (50 ng mL−1, EGF, Invitrogen), hepatocyte growth factor (25 ng mL−1, HGF, Peprotech), fibroblast growth factor 10 (0.1 μg mL−1, FGF10, Peprotech), recombinant human (Leu15)-gastrin I (GAS, 10 nM, Sigma-Aldrich), FSK (10 μM, FSK), and A83-01 (5 μM, Tocris Bioscience).
During the optimization of PIC hydrogels, single cells were prepared from organoids by trypsinizing with TrypLE Express Enzyme (12604-013, Gibco). Single cells were seeded in droplets (50 μL) at a concentration of 300–1000 cells per μL, in temperature and humidity-balanced 24-well plates (M9312, Greiner, Merck). After seeding, plates were incubated at 37 °C for 15–30 min to facilitate hydrogel gelation and thereafter, EM (500 μL) containing Y-27632 (10 μM, SelleckChem) was added to each well. After three days of culture, Y-27632 (SelleckChem) was no longer used. Medium was refreshed every 2–3 days; organoids were passaged weekly at a 1:3–1:4 split ratio by mechanical dissociation method. All cultures were incubated at 37 °C, 5% CO2.
For liver organoid differentiation, previously-defined differentiation medium (DM) was used. DM was based on advanced DMEM/F12 (AD, Gibco) containing 1% GlutaMax (Gibco), HEPES (10 mM, Gibco), and 1% penicillin-streptomycin (Gibco), and supplemented with 1% N2 supplement (Invitrogen), 2% B27 supplement without vitamin A (Invitrogen), N-acetylcysteine (1.25 mM, Sigma-Aldrich), EGF (50 ng mL−1, Invitrogen), HGF (25 ng mL−1, Peprotech), FGF19 (100 ng mL−1, FGF19, Peprotech), recombinant human GAS (10 nM, Sigma-Aldrich), A83-01 (500 nM, Tocris Bioscience), BMP-7 (25 ng mL−1, Peprotech), 30 μM dexamethasone (Sigma-Aldrich), and DAPT (10 μM, Selleckchem). Prior to switching to DM from EM, liver organoids were primed in EM containing BMP-7 (25 ng mL−1, Peprotech) for three days. Medium was then switched to DM. Medium was refreshed every 2–3 days for 8 days.
Measurement of Cell Proliferation
ABA was used to measure the cell proliferation. For ABA, the stock solution (Dal1100, Life Technologies Europe BV) was diluted 1:10 in DMEM/F12 without phenol red (21 041, Gibco), sterilized through a 0.22 μm filter, and pre-warmed to 37 °C. EM was removed from each well and replaced with pre-warmed ABA solution. Organoids were incubated in ABA solution for 90 min at 37 °C, 5% CO2. After incubation, ABA solution was transferred to a new 24-well plate (Greiner) after which, fresh pre-warmed EM was added to the old plate. For ABA measurement, Fluoroskan Ascent FL (Thermo Fisher Scientific) was used. The wavelength of excitation and emission were 544 and 577 nm, respectively. ABAs were conducted on days 0, 1, 4, and 7 after single-cell seeding. Data analysis was normalized to day 0; results were graphed as fold-changes using GraphPad Prism (GraphPad Software).
Cryopreserved Hepatocyte Culture
LiverPool Cryoplateable Hepatocytes (10-donor, mixed gender) were provided by Bioreclamation IVT. Hepatocytes were cultured in a collagen sandwich according to the manufacturer’s instructions, and supplemented with the recommended InVitroGRO CP media (Bioreclamation IVT). For mRNA sequencing, hepatocytes were harvested after 4 h of sandwich culture.
RNA Isolation and qRT-PCR
The RNeasy Mini Kit (Qiagen, Hilden, Germany) was used to isolate RNA from tissues, hepatocytes, and organoids following the manufacturer’s instructions. RNA quality and quantity was measured with DS-11 spectrophotometer (DeNovix). Complementary DNA (cDNA) was synthesized with the iScript cDNA synthesis kit (Bio-Rad) following the manufacturer’s instructions. qRT-PCR was used to determine relative expression of target genes using validated primers (Table S3, Supporting Information) using the SYBR Green method (Bio-Rad). Normalization was carried out using reference genes Hypoxanthine-guanine phosphoribosyltransferase and ribosomal protein L19.
Rhodamine123 Transport Assay
Liver organoids were differentiated in PIC-LEC or Matrigel droplets for 8 days as previously described. For Rh123 transport assays, organoids were pretreated with DM containing verapamil (10 μM, Sigma-Aldrich) or DMSO for 30 min. Organoids were then removed from PIC-LEC or Matrigel and resuspended in DM containing rh123 (100 μM, Sigma-Aldrich) and incubated at 37 °C for 10 min. Fluorescence was visualized by an EVOS FL cell imaging system (Life Technologies).
Ammonium Elimination Assay
For ammonium elimination assays, liver organoids were differentiated in PIC-LEC or Matrigel droplets for 8 days as previously described. Organoids were incubated with DM supplemented with NH4Cl (2 mM) for 24 h. After 24 h, media samples were harvested and stored at −20 °C. Afterward, Tryple-Express (Gibco) was added to each well and organoids were trypsinized for cell counting. Cell counts were carried out using the TC20 automated cell counter (Bio-Rad). Viable cells were determined using trypan blue exclusion assay. Ammonium concentrations were measured with the urea/ammonia Assay Kit (Megazyme). As a control, DM containing NH4Cl (2 mM) was incubated for 24 h without cells. Ammonia elimination rates were normalized to live cell numbers.
GLDH Expression and Albumin Production
To quantify the intracellular levels of GLDH and ALB, liver organoids were differentiated in PIC-LEC or Matrigel droplets for 8 days, as previously described. Organoids were provided with fresh DM 24 h before being lysed in MilliQ water. GLDH and ALB were measured in the cell lysates using a DxC-600 Beckman chemistry analyzer (Beckman Coulter). Values were normalized to total protein concentrations.
Microscopy and Immunofluorescence Analysis
Imaging of the organoids was performed using an EVOS FL cell imaging system (Life Technologies) and an Olympus BX51 microscope in combination with an Olympus DP73 camera. Detailed information of applied antigen retrieval methods, antibodies, dilutions, and incubation times are listed in Table S4, Supporting Information.
Bright field images were taken to track organoid morphology throughout expansion in different hydrogel formulations. Images were also taken to compare the morphology of organoids in EM and DM.
For immunofluorescent (IF) staining, organoids were fixed with 4% neutral buffered formalin containing 0.1% eosin and incubated at 37 °C overnight. Fixed samples were dehydrated and embedded in paraffin or stored in 70% ethanol at 4 °C for up to 1 month; 4 μm thick paraffin sections were prepared for IF staining. To start the IF staining procedure, the paraffin sections were first heated at 62 °C for 15 min and dewaxed by xylene, followed by rehydration in gradient ethanol concentrations from 100% to 70%. Then, sample sections were incubated in antigen retrieval solution for 30 min at 98 °C. After balancing to room temperature, sample sections were treated with NH4Cl solution (20 mM) for 10 min to reduce background autofluorescence and blocked with 10% goat serum for 1 h to avoid non-specific antibody binding. Next, primary antibodies against ECAD, Ki67, PCNA, and K19 were added to the sections and incubated overnight at 4 °C. After being washed with tris-buffered saline containing Tween 20 three times, sample sections were incubated with secondary antibodies (5 μM), including mouse anti-rabbit Alexa Fluor 488 (Molecular Probes), mouse anti-rabbit Alexa Fluor647 (Molecular Probes), rabbit anti-mouse Alexa Fluor488 (Molecular Probes), and rabbit anti-mouse Alexa Fluor647 (Molecular Probes). Nuclei were stained with DAPI (0.5 μg mL−1, Sigma Aldrich).
Whole Transcriptome Sequencing and Analysis
For whole transcriptome sequencing, RNA was isolated from organoids using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. Library preparation, 75 bp single-end sequencing on Illumina NextSeq500 (Illumina) and mapping raw reads was performed as previously described.[39] The raw files have been uploaded to Gene Expression Omnibus under the accession GSE143223. Data was merged with previously sequenced LiverPool Cryoplateable Hepatocytes (GSE123498)[28] and normalized. Differentially-expressed genes were identified using the DESeq2 package with standard settings.[40] Human liver tissue whole transcriptome sequencing data (Liver_FASEB) was obtained from an online database.[41] Heatmaps were generated using gplots. ToppFun was used for functional enrichment analysis based on functional annotations and protein interactions networks.[33]
Human Recombinant Laminin-111 Condensation
To obtain high-concentration human recombinant laminin-111 (hrlaminin-111), commercial hrlaminin-111 (Biolamina) was condensed with an Amicon Ultra-0.5 centrifugal filter unit (Millipore, Merck) following the manufacturer’s instructions. hrlaminin-111 was concentrated from 100 μg mL−1 to ≈5 mg mL−1, as measured by BCA assay with a Pierce BCA protein assay kit (Thermo Scientific). Afterward, the condensed hrlaminin-111 replaced LEC to make a chemically-defined PIC hydrogel for liver organoid culture.
Statistical Analyses
ABA results (Figure 1d,e) and rheological results (Figure 2) were analyzed in an Excel datasheet and converted into graphs using Graphpad Prism 8. qRT-PCR results (Figures 4b and 5b), ALB secretion (Figure 4c), GLDH expression (Figure 4e), and ammonium elimination (Figure 4f) were analyzed using a Tukey’s multiple comparisons test by two-way ANON multiple comparisons. The p-values are indicated in the respective figures.
Supplementary Material
Supporting Information is available from the Wiley Online Library or from the author.
Acknowledgements
This work was supported by the Dutch Research Council NWO VENI (016.Veni.198.021) to K.S., China Scholarship Council (CSC201808310180) to S.Y., 3 V fonds and Jubileumfonds Veeartsenijkundige Hoogeschool 1921, both named funds of Utrecht University Fund to K.S. The ToC figure was created with BioRender.com.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Dr. Marko Mihajlovic, Department of Pharmaceutics, Utrecht Institute for Pharmaceutical Sciences, Faculty of Science Utrecht University Universiteitsweg 99, Utrecht 3584 CG, The Netherlands; Department of Biomedical Engineering Eindhoven University of Technology Postbus 513, Eindhoven 5600 MB, The Netherlands
Dr. Massimiliano Caiazzo, Department of Pharmaceutics, Utrecht Institute for Pharmaceutical Sciences, Faculty of Science Utrecht University Universiteitsweg 99, Utrecht 3584 CG, The Netherlands; Department of Molecular Medicine and Medical Biotechnology University of Naples ‘Federico II’ Via Pansini 5, Naples 80131, Italy
Prof. Luc J.W. van der Laan, Department of Surgery Erasmus MC-University Medical Center Dr. Molewaterplein 40, Rotterdam 3015 GD, The Netherlands
Louis C. Penning, Department of Clinical Sciences of Companion Animals, Faculty of Veterinary Medicine Utrecht University Uppsalalaan 8, Utrecht 3584 CT, The Netherlands
Dr. Tina Vermonden, Department of Pharmaceutics, Utrecht Institute for Pharmaceutical Sciences, Faculty of Science Utrecht University Universiteitsweg 99, Utrecht 3584 CG, The Netherlands
Dr. Bart Spee, Department of Clinical Sciences of Companion Animals, Faculty of Veterinary Medicine Utrecht University Uppsalalaan 8, Utrecht 3584 CT, The Netherlands
Dr. Kerstin Schneeberger, Department of Clinical Sciences of Companion Animals, Faculty of Veterinary Medicine Utrecht University Uppsalalaan 8, Utrecht 3584 CT, The Netherlands.
Data Availability
The whole transcriptome sequencing datasets generated and analyzed during this study are available in the Gene Expression Omnibus repository under the accession GSE143223. Other datasets generated and analyzed during the current study are available from the corresponding author upon request.
References
- [1].Michalopoulos GK, DeFrances MC. Science. 1997;276:60. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- [2].O’Shea RS, Dasarathy S, McCullough AJ. Hepatology. 2010;51:307. doi: 10.1002/hep.23258. [DOI] [PubMed] [Google Scholar]
- [3].Shaheen NJ, Hansen RA, Morgan DR, Gangarosa LM, Ringel Y, Thiny MT, Russo MW, Sandler RS. Am J Gastroenterol. 2006;101:2128. doi: 10.1111/j.1572-0241.2006.00723.x. [DOI] [PubMed] [Google Scholar]
- [4].Júnior RFM, Salvalaggio P, de Rezende MB, Evangelista AS, Guardia BD, Matielo CEL, Neves DB, Pandullo FL, Felga GEG, Alves JAS, Curvelo LA, et al. Einstein (Sao Paulo) 2015;13:149. doi: 10.1590/S1679-45082015RW3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Tanikella R, Kawut SM, Brown RS, Krowka MJ, Reinen J, Dinasarapu CR, Trotter JF, Roberts KE, Mohd MA, Arnett DK, Fallon MB. Liver Transplant. 2010;16:238. doi: 10.1002/lt.21984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Atala A. J Pediatr Surg. 2012;47:17. doi: 10.1016/j.jpedsurg.2011.10.013. [DOI] [PubMed] [Google Scholar]
- [7].Forbes SJ. Hepatology. 2015;62:1635. doi: 10.1002/hep.27970. [DOI] [PubMed] [Google Scholar]
- [8].Lancaster MA, Knoblich JA. Nat Protoc. 2014;9:2329. doi: 10.1038/nprot.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hay DC, Zhao D, Fletcher J, Hewitt ZA, McLean D, Urruticoechea-Uriguen A, Black JR, Elcombe C, Ross JA, Wolf R, Cui W. Stem Cells. 2008;26:894. doi: 10.1634/stemcells.2007-0718. [DOI] [PubMed] [Google Scholar]
- [10].Sullivan GJ, Hay DC, Park I-H, Fletcher J, Hannoun Z, Payne CM, Dalgetty D, Black JR, Ross JA, Samuel K, Wang G, et al. Hepatology. 2010;51:329. doi: 10.1002/hep.23335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Huch M, Dorrell C, Boj SF, van Es JH, Li VSW, van de Wetering M, Sato T, Hamer K, Sasaki N, Finegold MJ, Haft A, et al. Nature. 2013;494:247. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Huch M, Gehart H, van Boxtel R, Hamer K, Blokzijl F, Verstegen MMA, Ellis E, van Wenum M, Fuchs SA, de Ligt J, van de Wetering M, et al. Cell. 2015;160:299. doi: 10.1016/j.cell.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].a) Zhang K, Zhang L, Liu W, Ma X, Cen J, Sun Z, Wang C, Feng S, Zhang Z, Yue L, Sun L, et al. Cell Stem Cell. 2018;23:806. doi: 10.1016/j.stem.2018.10.018. [DOI] [PubMed] [Google Scholar]; b) Peng WC, Logan CY, Fish M, Anbarchian T, Aguisanda F, Álvarez-Varela A, Wu P, Jin Y, Zhu J, Li B, Grompe M, et al. Cell. 2018;175:1607. doi: 10.1016/j.cell.2018.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Hu H, Gehart H, Artegiani B, LÖpez-Iglesias C, Dekkers F, Basak O, van Es J, de S Lopes SMC, Begthel H, Korving J, van den Born M, et al. Cell. 2018;175:1591. doi: 10.1016/j.cell.2018.11.013. [DOI] [PubMed] [Google Scholar]
- [14].Chen C, Jochems PGM, Salz L, Schneeberger K, Penning LC, van de Graaf SFJ, Beuers U, Clevers H, Geijsen N, Masereeuw R, Spee B. Biofabrication. 2018;10:034103. doi: 10.1088/1758-5090/aac8fd. [DOI] [PubMed] [Google Scholar]
- [15].Kuijk EW, Rasmussen S, Blokzijl F, Huch M, Gehart H, Toonen P, Begthel H, Clevers H, Geurts AM, Cuppen E. Sci Rep. 2016;6:22154. doi: 10.1038/srep22154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].a) Aravalli RN, Steer CJ. Expert Opin Biol Ther. 2016;16:595. doi: 10.1517/14712598.2016.1158808. [DOI] [PubMed] [Google Scholar]; b) Schwank G, Koo B-K, Sasselli V, Dekkers JF, Heo I, Demircan T, Sasaki N, Boymans S, Cuppen E, van der Ent CK, Nieuwenhuis EES, et al. Cell Stem Cell. 2013;13:653. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]; c) Artegiani B, van Voorthuijsen L, Lindeboom RGH, Seinstra D, Heo I, Tapia P, López-Iglesias C, Postrach D, Dayton T, Oka R, Hu H, et al. Cell Stem Cell. 2019;24:927. doi: 10.1016/j.stem.2019.04.017. [DOI] [PubMed] [Google Scholar]
- [17].Kleinman HK, Martin GR. Semin Cancer Biol. 2005;15:378. doi: 10.1016/j.semcancer.2005.05.004. [DOI] [PubMed] [Google Scholar]
- [18].Hughes CS, Postovit LM, Lajoie GA. Proteomics. 2010;10:1886. doi: 10.1002/pmic.200900758. [DOI] [PubMed] [Google Scholar]
- [19].Benton G, Arnaoutova I, George J, Kleinman HK, Koblinski J. Adv Drug Delivery Rev. 2014;79-80:3. doi: 10.1016/j.addr.2014.06.005. [DOI] [PubMed] [Google Scholar]
- [20].Gjorevski N, Sachs N, Manfrin A, Giger S, Bragina ME, Ordóñez-Morán P, Clevers H, Lutolf MP. Nature. 2016;539:560. doi: 10.1038/nature20168. [DOI] [PubMed] [Google Scholar]
- [21].Sorrentino G, Rezakhani S, Yildiz E, Nuciforo S, Heim MH, Lutolf MP, Schoonjans K. bioRxiv. 2019:810275. doi: 10.1038/s41467-020-17161-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Broguiere N, Isenmann L, Hirt C, Ringel T, Placzek S, Cavalli E, Ringnalda F, Villiger L, Züllig R, Lehmann R, Rogler G, et al. Adv Mater. 2018;30:1801621. doi: 10.1002/adma.201801621. [DOI] [PubMed] [Google Scholar]
- [23].Kouwer PHJ, Koepf M, Le Sage VAA, Jaspers M, van Buul AM, Eksteen-Akeroyd ZH, Woltinge T, Schwartz E, Kitto HJ, Hoogenboom R, Picken SJ, et al. Nature. 2013;493:651. doi: 10.1038/nature11839. [DOI] [PubMed] [Google Scholar]
- [24].Ye S, Boeter JWB, Penning LC, Spee B, Schneeberger K. Bioengineering (Basel) 2019;6:59. doi: 10.3390/bioengineering6030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Deshpande SR, Hammink R, Das RK, Nelissen FHT, Blank KG, Rowan AE, Heus HA. Adv Funct Mater. 2016;26:9075. [Google Scholar]
- [26].Zimoch J, Padial JS, Klar AS, Vallmajo-Martin Q, Meuli M, Biedermann T, Wilson CJ, Rowan A, Reichmann E. Acta Biomater. 2018;70:129. doi: 10.1016/j.actbio.2018.01.042. [DOI] [PubMed] [Google Scholar]
- [27].op ‘t Veld RC, van den Boomen OI, Lundvig DMS, Bronkhorst EM, Kouwer PHJ, Jansen JA, Middelkoop E, den Hoff JWV, Rowan AE, Wagener FADTG. Biomaterials. 2018;181:392. doi: 10.1016/j.biomaterials.2018.07.038. [DOI] [PubMed] [Google Scholar]
- [28].Schneeberger K, Sánchez-Romero N, Ye S, van Steenbeek FG, Oosterhoff LA, Palacin IP, Chen C, van Wolferen ME, van Tienderen G, Lieshout R, Colemonts-Vroninks H, et al. Hepatology. 2020 doi: 10.1002/hep.31037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Shatford RA, Nyberg SL, Meier SJ, White JG, Payne WD, Hu W-S, Cerra FB. J Surg Res. 1992;53:549. doi: 10.1016/0022-4804(92)90253-v. [DOI] [PubMed] [Google Scholar]
- [30].Yumoto R, Murakami T, Nakamoto Y, Hasegawa R, Nagai J, Takano M. J Pharmacol Exp Ther. 1999;289:149. [PubMed] [Google Scholar]
- [31].Yelamanchi SD, Jayaram S, Thomas JK, Gundimeda S, Khan AA, Singhal A, Prasad TSK, Pandey A, Somani BL, Gowda H. J Cell Commun Signaling. 2016;10:69. doi: 10.1007/s12079-015-0315-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Aloia L, McKie MA, Vernaz G, Cordero-Espinoza L, Aleksieva N, van den Ameele J, Antonica F, Font-Cunill B, Raven A, Cigliano RA, Belenguer G, et al. Nat Cell Biol. 2019;21:1321. doi: 10.1038/s41556-019-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Chen J, Bardes EE, Aronow BJ, Jegga AG. Nucleic Acids Res. 2009;37:W305. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kourouklis AP, Kaylan KB, Underhill GH. Biomaterials. 2016;99:82. doi: 10.1016/j.biomaterials.2016.05.016. [DOI] [PubMed] [Google Scholar]
- [35].a) Cruz-Acuna R, Quiros M, Farkas AE, Dedhia PH, Huang S, Siuda D, Garcia-Hernandez V, Miller AJ, Spence JR, Nusrat A, Garcia AJ. Nat Cell Biol. 2017;19:1326. doi: 10.1038/ncb3632. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Cruz-Acuna R, Quiros M, Huang S, Siuda D, Spence JR, Nusrat A, Garcia AJ. Nat Protoc. 2018;13:2102. doi: 10.1038/s41596-018-0036-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Mandal S, Eksteen-Akeroyd ZH, Jacobs MJ, Hammink R, Koepf M, Lambeck AJA, van Hest JCM, Wilson CJ, Blank K, Figdor CG, Rowan AE. Chem Sci. 2013;4:4168. [Google Scholar]
- [37].Celikkin N, Padial JS, Costantini M, Hendrikse H, Cohn R, Wilson CJ, Rowan AE, Święszkowski W. Polymers. 2018;10:555. doi: 10.3390/polym10050555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Schneeberger K, Spee B, Costa P, Sachs N, Clevers H, Malda J. Biofabrication. 2017;9:013001. doi: 10.1088/1758-5090/aa6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Oosterhoff LA, Kruitwagen HS, van Wolferen ME, van Balkom BWM, Mokry M, Lansu N, van den Dungen NAM, Penning LC, Spanjersberg TCF, de Graaf JW, Veenendaal T, et al. Front Physiol. 2019;10:101. doi: 10.3389/fphys.2019.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Love MI, Huber W, Anders S. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kampf C, Mardinoglu A, Fagerberg L, Hallstrom BM, Edlund K, Lundberg E, Ponten F, Nielsen J, Uhlen M. FASEB J. 2014;28:2901. doi: 10.1096/fj.14-250555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The whole transcriptome sequencing datasets generated and analyzed during this study are available in the Gene Expression Omnibus repository under the accession GSE143223. Other datasets generated and analyzed during the current study are available from the corresponding author upon request.