Abstract
Mitochondrial genome (mtDNA) variation in common diseases has been under-explored, partly due to a lack of genotype calling and quality control procedures. Developing an at-scale workflow for mtDNA variant analyses, we show correlations between nuclear and mitochondrial genomic structures within sub-populations of Great Britain and establish a UK Biobank reference atlas of mtDNA-phenotype associations. A total of 260 mtDNA-phenotype associations were novel (P<1x10-5) including, rs2853822/m.8655C>T (MT-ATP6) with type 2 diabetes, rs878966690/m.13117A>G (MT-ND5) with multiple sclerosis, six mtDNA associations with adult height, 24 with two liver biomarkers and 16 with parameters of renal function. Rare variant gene-based tests implicated Complex I genes modulating mean corpuscular volume and mean corpuscular hemoglobin. Seven traits had both rare and common mtDNA associations where rare variants tended to have larger effects than common variants. Our work illustrates the value of studying mtDNA variants in common complex diseases and lays foundations for future large-scale mtDNA association studies.
The 16,569bp human mitochondrial genome has a compact genomic organization, with ~95% of the sequence encoding 13 proteins, 22 transfer RNAs and 2 ribosomal RNAs that are essential for oxidative phosphorylation (OXPHOS) and production of cellular energy in the form of adenosine triphosphate1 (ATP). Being maternally inherited2, mtDNA undergoes negligible population level inter-molecular recombination3. As humans migrated out of Africa and populated the globe, they have acquired mitochondrial (mt) single nucleotide variants (mtSNVs), which define geographical region-specific macro-haplogroups (related haplotypes)4,5. Many mtSNVs either directly affect mitochondrial function, or are in linkage disequilibrium with variants known to influence mitochondrial metabolism6, and have been associated with common complex diseases including type 2 diabetes7 (T2D), cardiomyopathy and neurodegenerative disorders8,9.
Initial mtDNA association studies in complex traits were under-powered and yielded conflicting findings which rarely replicated10,11. The inclusion of 265 mtDNA variants on the Affymetrix genotyping arrays used in UK Biobank (UKBB)12, coupled with deep phenotypic data collected on half a million participants, provides an opportunity to address these issues and perform robust mtDNA genome-wide association studies (mtGWAS). Here, we establish a workflow for high quality variant calling and imputation and evaluate their role in 877 complex traits, providing a comprehensive atlas of mtSNV associations with diseases and endophenotypes in the UKBB population.
Results
Two hundred and sixty-five mtSNVs were genotyped in 488,377 UKBB participants (Supplementary Table 1). In the absence of standard procedures for quality control (QC) and imputation of mtSNVs from genotyping arrays, we adapted existing algorithms13,14 (Fig. 1, Methods), to generate a QC set of 719 mtSNVs in 483,626 pan-ancestry individuals, referred to as the “Full Set” (Supplementary Tables 2-4). Next, we defined a set of European (EUR) unrelated individuals (based on the kinship coefficient, removing up to 3rd degree relatives; N=358,916; Fig. 1, Supplementary Note, Methods). We excluded mtSNVs with minor allele count (MAC)<10 or imputation INFO score<0.7 (Fig. 1), resulting in 473 mtSNVs for association analyses in the unrelated EUR set (Supplementary Tables 5-6). In accordance with our power calculations (Supplementary Fig. 1), we restricted the analyses of binary traits to 416 mtSNVs with MAF>0.0001.
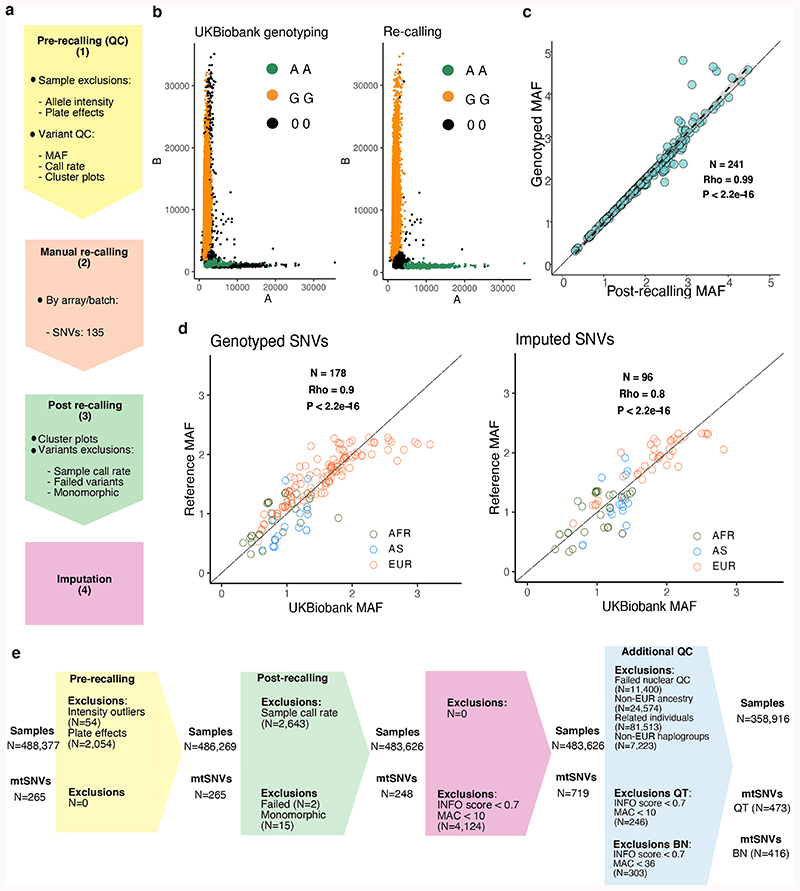
Figure 1. Mitochondrial genome PheWAS workflow.
(a) Quality control (QC) workflow: the steps taken to assure genotype quality are listed. The stages were as follows: (1) pre-recalling QC, (2) manual re-calling, (3) post-re-calling QC, and (4) imputation of mtSNVs not genotyped on the arrays. (b) Examples of probe intensities cluster plots for a mtSNV (m.14869G>A) pre- and post-recalling genotyped in the “Full set” (N = 483,626 participants); color legend corresponds to genotype assignment with black dots indicate missing genotypes. (c) Scatterplot showing correlation of -log10 MAFs of the 241 recalled mtSNVs compared to UKBB genotyped mtSNVs. The long dashed lines indicate y=x and the short dashed lines the linear regression fit. The grey shaded area represents the 95% confidence interval of the regression fit. Spearman’s correlation, two-sided P-value (P = 1.8x10-205) and rho are provided. (d) Scatterplots showing correlation of -log10 MAFs of the genotyped mtSNVs post-recalling (left plot) and the imputed variants (right plot) in UKBB mtSNVs compared to GenBank mtSNVs (MAC≥30). Spearman’s correlation, two-sided P-value (P = 8.6x10-65 for genotyped SNVs; P = 1.8x10-26 for imputed SNVs) and rho are provided. Color coding represents the population each mtSNV is tagging (green = African, blue = Asian, orange = European population). The UKBB individuals with nuclear-mitochondrial matched African (AFR, N=2012 participants), Asian (AS, N=888 participants) and European (EUR, N=358,916, unrelated participants) ancestries were compared to corresponding GenBank genomes (EUR, N=6,593, AFR, N = 704, AS, N = 3,587). (e) CONSORT-like diagram showing the breakdown of people and mtSNVs excluded at each step of the study. Colors correspond to the following steps: light yellow = pre-calling, peach = manual re-calling, light green = post re-calling, pink = imputation. INFO = IMPUTE2 score; MAC = minor allele count; BT = binary trait; QT = quantitative trait.
Calling and imputation of mtSNV genotypes
Genotype calling algorithms trained to detect three nuclear genotype clusters cannot reliably be employed for mtSNVs where two clusters overwhelmingly predominate (Supplementary Fig. 2). To address this we developed a four-stage genotype QC procedure (Methods, Supplementary Figs. 3-6): (1) pre-recalling QC, (2) manual re-calling, (3) post-re-calling QC, and (4) imputation of mtSNVs not genotyped on the array (Fig. 1, Supplementary Note). In stage 1 we excluded probe intensity outliers (Supplementary Figs. 3-4) and identified mtSNVs to re-call in stage 2. We re-called 135 mtSNVs (50.9%) in stage 2 and, in stage 3, 248 high quality mtSNVs passed genotype QC (Supplementary Tables 2-3, Supplementary Fig. 5), increasing the mean per sample call rate from 0.820 to 0.997 (Fig. 1, Supplementary Table 2). In stage 4, we imputed 719 additional mtSNVs in the full UKBB set using 5,271 biallelic (homoplasmic) mtSNVs from a combined reference panel of 17,815 European, African and Asian mtDNA complete genomes (Fig. 1, Supplementary Fig. 5, Methods). Our genotype QC workflow tripled the number of variants available for subsequent analyses (Fig. 1, Supplementary Fig. 5) and quadrupled the number of rare mtSNVs (MAF≤0.01; N=553). Our estimates of MAFs for re-called variants in the UKBB EUR set showed high correlation (average Spearman’s rho=0.84), with the allele frequencies from the EUR ancestry components of three reference datasets (GenBank, 1000 Genomes and the Wellcome Trust Case Control Consortium cohort; Methods, Fig. 1, Supplementary Table 7). Furthermore, the mtSNV imputation facilitated by the re-called variants together improved mtDNA haplogroup calling accuracy (median Haplogrep 2 overall rank=0.73, compared to 0.65 for genotyped variants pre-QC) (Supplementary Fig. 6, Supplementary Tables 8-9).
mtDNA & nuclear genetic structure is correlated
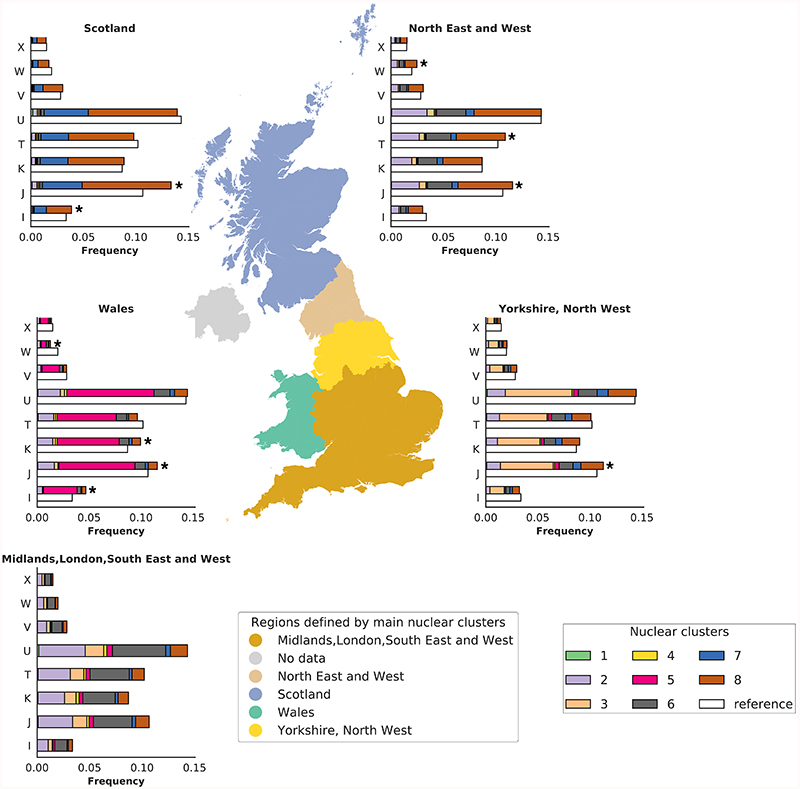
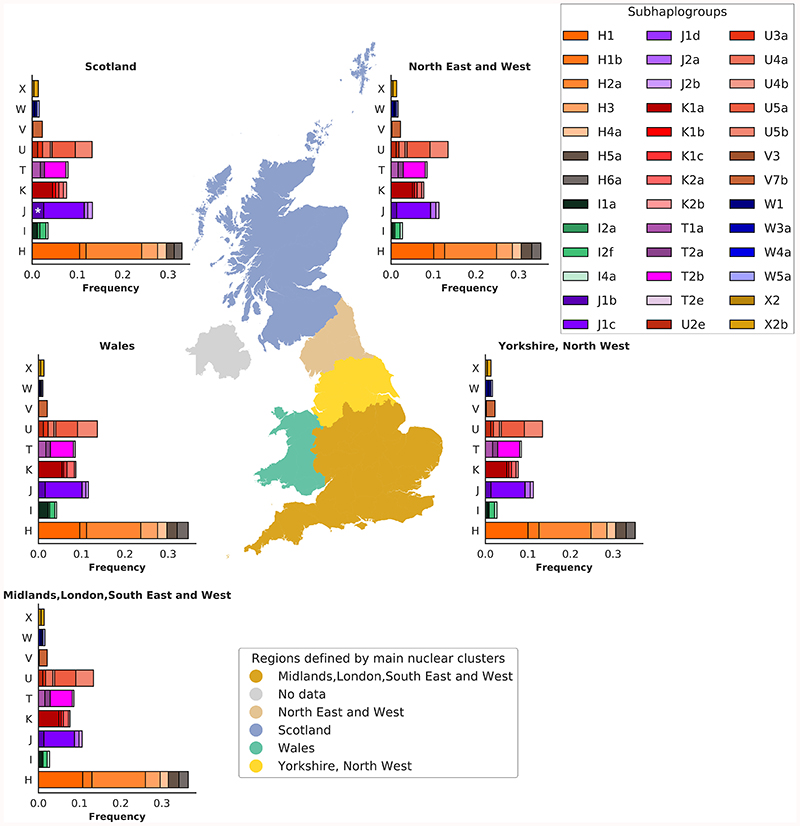
In contrast to the structure of the nuclear genome in Great Britain (GB), less is known about the structure of mtDNA variation, and given the uniparental mtDNA inheritance, the two genomes are thought to be uncorrelated15. In the unrelated EUR set of UKBB (358,916 participants) we observed differences in frequencies of ~2% for macro-haplogroups J, W, and I, in three areas, Scotland, Northumberland/Tyne and Wear and Wales, compared to London (P=7x10-16, 5x10-8, 1x10-15 for J, W and I, respectively; Supplementary Tables 10-11). Amongst the sub-haplogroups with >100 carriers i.e. where we had statistical power to discern differences, we observed over-representation of one sub-haplogroup, J1b in Scotland (P=3.5x10-8; Extended Data Fig. 1).
We observed that 332 mtSNVs were associated (P<5x10-5) with at least one of the first 10 nucPCs (Extended Data Fig. 2). We also observed an association between macro-haplogroups I, J and K and nucPCs, specifically with clusters of individuals of Scottish, Northumbrian or Welsh ancestry (Fig. 2, Supplementary Fig. 7, Supplementary Tables 12-13). We next tested whether the observed correlation of the macro-haplogroups with the nucPCs could be attributed to specific sub-haplogroups. The major sub-haplogroups (I1a, I2a, I2f, J1b, J1c, K1a, K2a, K2b) were correlated with nucPCs reflecting the findings with the macro-haplogroups and H5a and H1b were additionally correlated. Sub-haplogroup distributions were comparable to those in the WTCCC (Supplementary Table 9).
Figure 2. Distribution of the eight nuclear genome clusters and mtDNA haplogroups across Great Britain.
The European unrelated individuals with birth coordinates (N=327,665 participants) were clustered based on the first 10 nucPCs, resulting in eight nuclear clusters. The map of Great Britain and Northern Ireland is colored according to the five regions identified by the most common clusters or combination of clusters in each region: 1) Scotland; 2) North of England (North East and West); 3) North of England (Yorkshire and the Humber, North West of England); 4) South of England (Midlands, London, South East and West of England); 5) Wales. No data was available for Northern Ireland as participants were not recruited to UKBB from Northern Ireland. The stacked bar charts represent the frequency of unrelated individuals in each of the eight identified nuclear clusters across eight European macro-haplogroups in each region (X, W, V, U, T, K, J, I). The macro-haplogroup H (the most common among European macro-haplogroups) was used as baseline in the multinomial regression analysis and has been omitted. The white bars indicate the frequencies of individuals in the region used as reference to compare macro-haplogroups distribution, i.e. the area corresponding to South of England. * denotes macro-haplogroups that are distributed differently (likelihood ratio test, two-sided, P<5x10-5) between South of England and the rest of the country. Colors in the ‘Nuclear clusters’ box refer to haplogroup frequency barcharts, while colors in the ‘Regions defined by the main nuclear clusters’ are used to mark the five regions of the country. The map was plot using the GeoPandas package (https://geopandas.org/) under python 2.7.
Our evaluation of both mtDNA principal components (mtPCs, Extended Data Fig. 2, 3, Supplementary Fig. 8, Supplementary Note) and nucPCs showed that adjusting for the latter was sufficient to account for any major mtDNA geographical allele frequency difference because adding mtPCs did not result in a reduction in inflation factors, nor did we observe an increase in trait variance explained by the mtDNA variants.
mtSNV phenome-wide association study
Next, we performed both single-variant and gene-based PheWAS in the UKBB EUR set. UKBB is a general population cohort and so is underpowered for many diseases with the exception of the most prevalent such as coronary artery disease, type 2 diabetes, hypertension12. For conditions with less than 500 cases, our study lacked statistical power (Supplementary Fig. 1), we therefore restricted analysis of binary traits to 767 conditions derived from ICD-10 and self-report (Methods; Supplementary Tables 14, 15). For the same reason, we also excluded mtSNVs with MAF≤0.0001. We analyzed 126 quantitative traits including anthropometric and blood cell traits, serum and urinary biomarkers (Methods; Supplementary Tables 15-16). We excluded trait outliers and inverse normal transformed quantitative traits as appropriate (Supplementary Table 16). In total, we explored 378,696 mtSNV-trait associations including both binary and quantitative traits and up to 473 mtSNVs. We found 88 mtSNVs were associated with one or more of 94 binary traits (Supplementary Table 17) at mitochondrial genome-wide significance (Bonferroni adjusted P-value<5x10-5=0.05/1000 independent haplotypes; Methods), and 66 mtSNVs were associated with one or more of 27 quantitative traits (Tables 1-4, Figures 3-4, Supplementary Table 18). Two hundred and sixty mtSNV-trait associations were novel, and all associations that reached the mtGWAS P-value threshold had a FDR<5.6% (Supplementary Table 17-18). We have used “lead” variant throughout the manuscript to describe the variant with the smallest association P-value for a given trait.
Table 1. New trait-mtSNV associations defined using UKBB ICD10 and self-report codes.
| Chapter | Trait | Definition | Locus | rsID, position (EA, EAF) |
AA change | OR | 95% CI | P (FDR) | Haplogroup |
|---|---|---|---|---|---|---|---|---|---|
| I. Infectious and parasitic diseases | Other specified bacterial agents* | ICD10: B968 | CO1 | rs879058417; 6528 (T,0.006)G | L209L | 1.36 | 1.18-1.57 | 3.1x10-5 (0.04) | W5; K1a1b1f |
| II. Neoplasm | Skin of other and unspecified parts of face | ICD10: C443 | ND3 | rs41487950; 10084 (C,0.007)I | I9T | 1.36 | 1.17-1.57 | 4.7x10-5 (0.05) | homoplastic |
| Descending colon | ICD10: D124 | ND5 | rs1556424100;12397(G,0.002)G | T21A | 1.92 | 1.41-2.64 | 4.4x10-5 (0.05) | homoplastic | |
| CYB | rs200336777; 15812 (A,0.008)G | V356M | 1.45 | 1.22-1.72 | 3.2x10-5 (0.03) | ||||
| Type 2 diabetes | ICD10, ICD9, #20002, #20003 | ATP6 | rs2853822; 8655 (T,0.001)Ga | I43I | 1.48 | 1.23-1.78 | 3.9x10-5 (0.05) | ancestral variant common to L | |
| ND5 | rs878966690; 13117(G, 0.005)G | I261V | 1.65 | 1.30-2.11 | 4.2x10-5 (0.05) | K1a3a | |||
| Lesion of plantar nerve | ICD10: G576 | CO3 | rs41482146; 9667 (G, 0.01)Ga | N154S | 1.49 | 1.24-1.80 | 3.2x10-5 (0.04) | U5a1b; J1b2a | |
| VII. Eye and adnexa | Ptosis of eyelid | ICD10: H02.4 | CYB | rs193302994; 15452 (A, 0.21)G | L236I | 1.15 | 1.07-1.23 | 4.8x10-5 (0.05) | J; T |
| IX. Circulatory system | Abdominal aortic aneurysm | ICD10: I71.4 | DLOOP | rs369669319; 207 (A, 0.04)I | - | 1.38 | 1.18-1.61 | 4.6x10-5 (0.04) | homoplastic |
| TT | rs193303002; 15927 (A,0.009)I | 1.70 | 1.35-2.15 | 8.0x10-6 (0.02) | |||||
| CYB | rs193302994; 15452 (A, 0.21)G | L236I | 1.22 | 1.11-1.34 | 3.7x10-5 (0.05) | ||||
| Joint disorder | #20002: 1295 | ND2 | rs1556422875; 4592 (C, 0.003)I | I41I | 1.91 | 1.44-2.53 | 8.2x10-6 (0.02) | H2a5a1a; U5a1h | |
| Bladder problem (not cancer) | #20002: 1201 | DLOOP | rs147029798; 16126 (C, 0.21)I | - | 1.12 | 1.06-1.17 | 1.3x10-5 (0.02) | homoplastic | |
| Abnormal findings on diagnostic imaging of other parts of digestive tract | ICD10: R93.3 | ND3 | rs41487950; 10084 (C, 0.007)I | I9T | 1.68 | 1.32-2.14 | 3.1x10-5 (0.04) | homoplastic |
Summary of the single-variant mtDNA PheWas associations identified with P<5x10-5 in UKBB (in up to 358,618 participants). Each variant had at least 10 cases carrying the effect-allele. Chapter=ICD-10 chapters. Definition: ICD-10 or non-cancer illness self-reported diseases codes. Locus=mtDNA encoded gene; rsID=SNP id as of dbSNP 153; position=mtDNA nucleotide position on rCRS (NC_012920); EA=effect allele; EAF=effect allele frequency; AA change = amino acid change. OR=odds ratio; 95% CI=95% confidence interval; P=P-value for the corresponding EA. FDR=False Discovery Rate calculated with Benjamini-Hochberg procedure. Haplogroup=Haplogroup(s) defined by the EA, according to Phylotree (build 17); SNVs tagging more than two European haplogroups are reported as “homoplastic”. G=genotyped; I=imputed; M=mixed, i.e. genotyped one array only and imputed on the other; Ga=mtSNVs genotyped on one array only (i.e. either the UKBB array or the UKBL array) and not imputed on the other array (or excluded because of low INFO score on the other array).
Table 4. New mtSNV-blood cell trait associations.
| Trait | Locus | rsID; position (EA, EAF) | AA change | beta | se | P (FDR) | Haplogroup |
|---|---|---|---|---|---|---|---|
| MPV | CO3 | rs2853493; 11467 (G, 0.21)G | L236L | -0.0147 | 0.002 | 1.5x10-11 (1.7x10-7) | U; K |
| PCT | RNR1 | rs267606617; 1555 (G, 0.002)G | - | -0.0829 | 0.002 | 7.1x10-6 (0.01) | na |
| CO3 | rs2853825; 9477 (A, 0.09)M | V91I | -0.0206 | 0.003 | 1.8x10-11 (2x10-7) | U5 | |
| CYB | rs41518645; 15257 (A, 0.02)G | D171N | -0.034 | 0.0061 | 3.1x10-8 (1x10-4) | J2; K1b1a | |
| RBC# | CO3 | rs2853493; 11467 (G, 0.21)G | L236L | -0.011 | 0.002 | 2.5x10-7 (0.001) | U; K |
| ND4 | rs199476112; 11778 (A, 0.0004)G | R340H | -0.3306 | 0.05 | 4.0x10-13 (6x10-9) | T3; X2p1 | |
| CYB | rs41518645; 15257 (A, 0.02)G | D171N | -0.0285 | 0.006 | 2.10x10-6 (0.005) | J2; K1b1a | |
| MCV | RNR1 | rs267606617; 1555 (G, 0.002)G | - | 0.1427 | 0.02 | 3.4x10-15 (8x10-11) | na |
| CO3 | rs2853493; 11467 (G, 0.21)G | L236L | 0.017 | 0.002 | 1.8x10-15 (5x10-11) | U; K | |
| ND4 | rs2853495; 11719 (G, 0.48)M | G320G | -0.0156 | 0.002 | 1.2x10-18 (9 x10-14) | H; V | |
| rs199476112; 11778 (A, 0.0004)G | R340H | 0.3114 | 0.05 | 7.6x10-12 (9 x10-8) | T3; X2p1 | ||
| ND5 | rs28359178; 13708 (A, 0.13)G | A458T | 0.017 | 0.003 | 6.8x10-11 (6 x10-7) | homoplastic | |
| HCT | ND4 | rs199476112; 11778 (A, 0.0004)G | R340H | -0.1863 | 0.05 | 4.4x10-5 (0.05) | T3; X2p1 |
| MCH | RNR1 | rs267606617; 1555 (G, 0.002)G | - | 0.1399 | 0.02 | 1.2x10-14 (2x10-10) | na |
| CO3 | rs2853493; 11467 (G, 0.21)G | L236L | 0.0185 | 0.002 | 5.4x10-18 (5x10-13) | U; K | |
| ND4 | rs2853495; 11719 (G, 0.48)M | G320G | -0.0162 | 0.002 | 4.5x10-20 (2x10-14) | H; V | |
| rs199476112; 11778 (A, 0.0004)G | R340H | 0.3077 | 0.05 | 1.3x10-11 (2x10-7) | T3; X2p1 | ||
| ND5 | rs28359178; 13708 (A, 0.13)G | A458T | 0.0152 | 0.003 | 5.5x10-9 (4x10-5) | homoplastic | |
| MCHC | RNR2 | rs2854128; 2706 (A, 0.44)G | - | -0.0072 | 0.002 | 4.7x10-5 (0.05) | H |
| RDW | TL2 | rs2853498; 12308 (G, 0.23)M | - | -0.0138 | 0.002 | 3.5x10-11 (3x10-7) | U; K |
| ND5 | rs2853499; 12372 (A, 0.23)G | L12L | -0.0139 | 0.002 | 3.0x10-11 (3x10-7) | U; K | |
| EO# | CYB | rs41518645; 15257 (A, 0.02)G | D171N | -0.0316 | 0.006 | 1.7x10-7 (7x10-4) | J2; K1b1a |
| GRAN# | CO1 | rs41413745; 6734 (A, 0.01)G | M277M | 0.0349 | 0.008 | 4.3x10-5 (0.05) | homoplastic |
| GRAN% MYELOID | DLOOP | rs62581312;150 (T, 0.09)G | - | -0.0128 | 0.003 | 3.3x10-5 (0.04) | homoplastic |
| LYMPH# | TL2 | rs2853498; 12308 (G, 0.23)M | - | -0.01 | 0.002 | 1.5x10-6 (0.004) | U; K |
| WBC# | CO3 | rs2853493; 11467 (G, 0.21)G | L236L | -0.0103 | 0.002 | 1.4x10-6 (0.004) | U; K |
| MONO% | CO2 | rs41534044; 7768 (G, 0.04)G | M61M | 0.0184 | 0.004 | 2.7x10-5 (0.04) | U5b |
| EO% | CYB | rs41518645; 15257 (A, 0.02)G | D171N | -0.0258 | 0.006 | 2.0x10-5 (0.03) | J2; K1b1a |
Summary of the 28 lead mtSNVs-blood cell traits associations (P < 5X10-5) and associated mtSNVs identified as independent signals by FINEMAP (P < 5X10-5 in single-mtSNVs GWAS analysis) in UKBB (in up to 345,714 individuals). Each cell contains the P-value for association between the mtSNV and the listed trait. Traits are as follows: MPV = mean platelet volume; PCT = plateletcrit; RBC# = red blood cell count; MCV = mean corpuscular volume; HCT = hematocrit; MCH = mean corpuscular hemoglobin; MCHC = mean corpuscular hemoglobin concentration; RDW = red blood cell width; EO# = eosinophil count; GRAN# = granulocyte count; GRAN%MYELOID = % of granulocytes in the myeloid fraction; LYMPH# = lymphocyte count; WBC# = white blood cell count; MONO% = % of monocytes; EO% = % of eosinophils. Locus = mtDNA gene/locus; position = mtDNA nucleotide position on rCRS (NC_012920.1); rsID = SNP id as of dbSNP 153; EA = effect allele; EAF = effect allele frequency; Haplogroups = haplogroups defined by the EA, according to Phylotree (build 17). beta= effect size of the association. se = standard error. FDR = False Discovery Rate calculated with Benjamini-Hochberg procedure; mtSNVs tagging more than two European haplogroups are reported as “homoplastic”; “na” values indicate alleles that were not observed in Phylotree (build 17). G=genotyped; I=imputed; M=mixed, i.e. genotyped on one array only and imputed on the other; Ga=genotyped on one array only but not imputed or excluded because of low INFO score on the other array. P-values for the lead mtSNV association for a given trait are in bold.
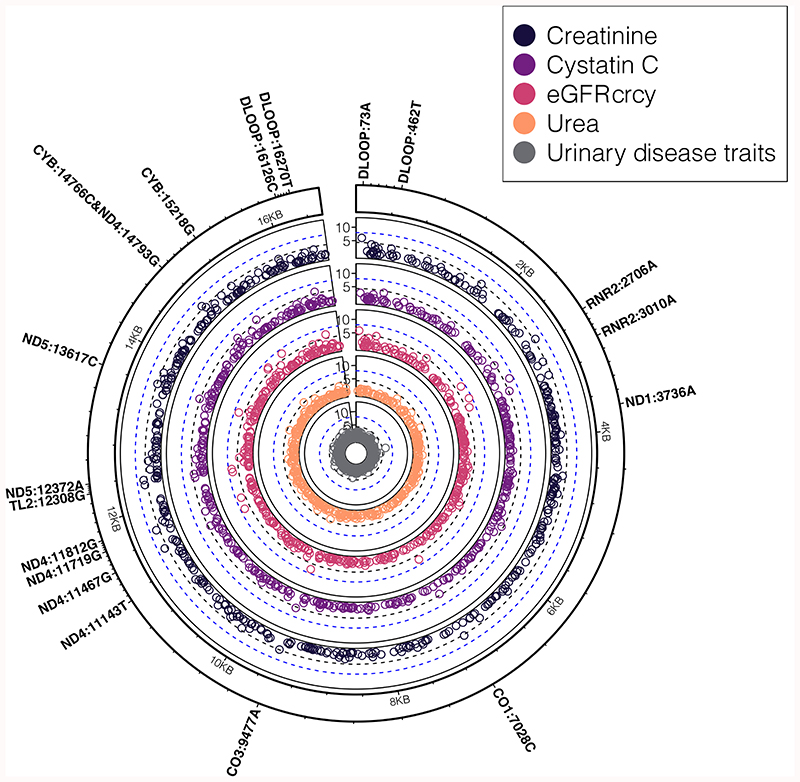
Figure 3. mtSNV associations with kidney related traits.
Concentric circular Manhattan plots showing a summary of associations (two-sided, P<5x10-5) between mtSNVs and traits related to kidney function: creatinine (N=341,440 participants), cystatin C (N=341,197 participants), estimated glomerular filtration rate (eGFR) calculated using both creatinine and cystatin C (crcy, N=342,007 participants), urea (N=341,276 participants), kidney related disease traits (calculus of the kidney [ICD10:N20.0] (N=279,179 participants); polyuria [ICD10:R35], (N=279,179 participants); urinary tract infection/kidney infection [#20002:1196] (N = 271,331 participants); bladder problem (not cancer) [#20002:1201] (N=271,331 participants)). The first four traits are part of (or derived from) the serum biomarker kidney function panel. The black dashed line denotes the mitochondrial genome multiple testing adjusted significance threshold (two-sided, P=5x10-5) while the blue dashed line denotes the nuclear GWAS significance threshold (two-sided, P=5x10-8). mtSNVs passing the mitochondrial genome multiple testing threshold are annotated by their locus, position and effect allele.
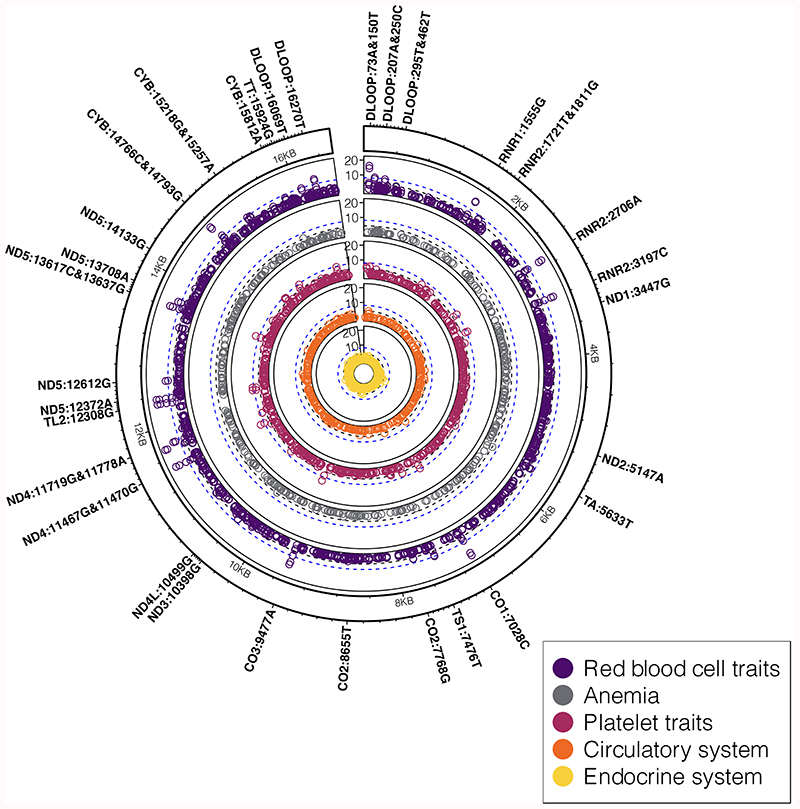
Figure 4. PheWAS association results for blood cell and cardiometabolic traits.
Circular concentric Manhattan plots showing a summary of associations (two-sided, P<5x10-5) between mtSNVs and traits related to cardio-metabolic health and endocrine traits, including red blood cell and platelet traits (quantitative traits, in up to N=325,670 participants), iron deficiency anemia (N=279,179 participants), and associated binary traits belonging to circulatory (N=279,179 participants) and endocrine systems (N=322,038 participants) according to ICD-10 codes (Table 1). The black dashed line denotes the mitochondrial genome multiple testing adjusted significance threshold (two-sided, P=5x10-5) while the blue dashed line denotes the nuclear genome significance threshold (two-sided, P=5x10-8). mtSNVs passing the mitochondrial genome multiple testing adjusted significance threshold are annotated by their locus, position and effect allele.
We performed fine-mapping and found for eight traits (mean platelet volume (MPV), plateletcrit (PCT), red blood cell count (RBC#), mean corpuscular volume (MCV), mean corpuscular haemoglobin (MCH) creatinine, estimated glomerular filtration rate (eGFR) from creatinine (eGFRcr) and height, associations with mtSNVs were due to multiple different mtSNVs using FINEMAP (Methods, Supplementary Table 19).
Finally, we tested for haplogroup specific effects. Of the 22 binary traits tested, we found no evidence of haplogroup specific effects for 18, meaning the lead variant (or variants in LD with it not genotyped or imputed in our study) captured the association and making it unlikely that additional haplogroup-specific variants were responsible for the phenotype association. For four (ptosis of the eye, abdominal aortic aneurysm, volvulus and bladder problem), either the haplogroups or the lead variants model the association equally well (P>0.001; Methods; Supplementary Table 20). Of the 24 continuous traits tested, four were associated with haplogroups in addition to mtSNVs (cystatin C, cystatin C derived eGFR, MCH and WBC#; Supplementary Table 20).
The observed associations were robust to sensitivity analyses assessing the analysis models used (Supplementary Information, Supplementary Tables 21-22). There was no significant evidence of association between mtDNA variants and age (P>1x10-5; minimum P=0.003 for association; minimum P=0.002 for interaction; Supplementary Tables 21-24, Supplementary Fig. 9). We found one variant (rs2853516/m.3316G>A; MT-ND1: p.Ala4Thr) was less common in men than women (MAFmen=0.0018 vs. MAFwomen=0.0025; P=5x10-5; Supplementary Tables 21-24). We observed an enrichment of associations with non-synonymous mtSNVs for binary traits (N=14; P=6x10-5) but not for quantitative traits (N=17; P=0.65) (P<0.017=0.05/3; Methods).
New mtSNV multiple sclerosis associations
We identified three mtSNVs associated with multiple sclerosis (Table 1), including two novel mtSNV associations tagging the K1a3 haplogroup (m.7559A>G in MT-TD, OR[95% CI]=2.06[1.59–2.67], P=5.0x10-8 and non-synonymous m.13117A>G in MT-ND5; OR[95%CI]=1.65[1.30–2.11], P=4.2x10-5), and replicating a previously reported association with rs2853826/m.10398A>G (P=4.3x10-5; Supplementary Note). We found that mtSNVs rather than the macro-haplogroups under-pinned the association (P=0.15; Supplementary Table 20) resolving previous inconsistencies16–18.
Rare L-clade mtSNV is associated with type 2 diabetes
We found a novel association between rs2853822/m.8655C>T (MAF=0.001) in MT-ATP6 with T2D (OR[95%CI]=1.48[1.23–1.78], P=3.9x10-5; Table 1 and Fig. 4). The m.8655T is an ancestral L-lineage allele common in Africa (MAF=0.41)19 and rare in Europeans (MAF=0.006)19, consistent with a recurrent mutation in Europeans (homoplasy, Supplementary Note). As rs2853822/m.8655C>T is synonymous, it is unlikely to be the causal variant but likely tags the effect of a mtDNA variant not captured in our study. L clade variants have been associated with T2D previously, including rs28693675/m.16189T>C in African Americans20 and in white British7, but this variant was not available in UKBB. We found no additional effect of the haplogroups on T2D risk in addition to rs2853822/m.8655C>T (P=0.58; Supplementary Table 20).
mtSNVs associated with decreased height
Five mtSNVs tagging macro-haplogroup J, and a rare non-coding variant, rs267606617/m.1555A>G (MAF=0.002; MT-RNR1) were associated with reduced height (Table 2, Supplementary Table 18). Our fine-mapping analyses showed that rs28359172/m.12612A>G (synonymous in MT-ND5, MAF=0.11) was independently associated with height (Supplementary Table 19) and this macro-haplogroup J-tagging variant was not correlated with the rare variant, rs267606617 (R2=0.001). The common variant (rs28359172/m.12612A>G) was associated with a ~0.8mm reduction in height, while the rare variant, rs267606617/m.1555A>G, which has previously been associated with non-syndromic deafness21–23, was associated with ~4.3mm reduction in height (Table 2). The effect size of these associations are comparable to the top 10% of effects reported for the nuclear genome24. Short stature is a well-recognized feature of inherited mitochondrial diseases25. In some patients this has a neuroendocrine basis, but this is not the case in most, where impaired cartilage-mediated growth has been implicated25,26. Given that the known differences in OXPHOS and ATP synthesis are linked to different mtDNA haplogroups27,28, the observed decrease in height might be linked to a less efficient ATP synthesis in individuals with haplogroup J28 which can have an impact on poor growth and short stature, as previously described in patients with mitochondrial dysfunction25. The lack of effect of mtDNA haplogroups after including the two height-associated mtSNVs in the model (P=0.04; Methods; Supplementary Table 20), implies that other mtSNVs not directly genotyped or imputed in this study are likely to account for the association with synonymous variant rs28359172/m.12612A>G.
Table 2. mtSNV associations with height, airways function and longevity.
| Trait | Locus | rsID; position (EA, EAF) | AA change | beta | se | P (FDR) | Haplogroup |
|---|---|---|---|---|---|---|---|
| ND5 | rs28359172; 12612 (G, 0.11)G | V92V | -0.010 | 0.003 | 2.0x10-6 (0.005) | J; K1a4c1 | |
| Airway function (FeV1/FVC) | ND4 | rs3088053; 11812 (G, 0.0001)G | L351L | -0.001 | 0.0003 | 1.9x10-5 (0.03) | T2 |
| DLOOP | rs2853513; 16223 (T, 0.07)Ga | - | 0.020 | 0.003 | 1.1x10-5 (0.02) | homoplastic |
Summary of the single-variant mitochondrial PheWas associations identified with height (N=358,045, participants), airways function (N=266,818, participants) and longevity (up to 348,257 participants) traits, found at discovery P<5x10-5. Z score: when both parents’ ages were modelled together, they were first standardised within sex prior to the analysis. Locus = mtDNA encoded gene. rsID = SNP id as of dbSNP 153. Position = mtDNA nucleotide position on rCRS (NC_012920). EA = effect allele. EAF=effect allele frequency, calculated on the set of samples with non-missing genotype/covariates. AA change = amino acid change. Beta = effect size of the association. se = standard error. P = P-value for association of the EA with the listed trait. FDR = False Discovery Rate calculated with Benjamini-Hochberg procedure. Haplogroup = Haplogroup(s) defined by the EA, according to Phylotree (build 17); SNVs tagging more than two European haplogroups are reported as “homoplastic”; “na” values indicate alleles that were not observed in Phylotree (build 17). FeV1 = Forced expiratory Volume in 1 sec; FVC = Forced Volume Capacity. G = genotyped; M = mixed, i.e. genotyped one array only and imputed on the other; Ga = mtSNVs genotyped on one array only (i.e. either the UKBB array or the UKBL array) and not imputed on the other array (or excluded because of low INFO score on the other array).
mtSNV associations with longevity
We used parental age as a proxy for longevity (Table 2) and found two variants, rs3021089/m.8251G>A (MAF=0.06; synonymous, MT-CO2) and rs2853513/m.16223C>T (MAF=0.07; DLOOP), associated with >30 day increase in attained maternal age (P<1x10-5) but not with paternal age (Table 2). These associations persisted when longevity was treated as a binary variable (maternal age ≥90years versus mothers <90yrs; OR=1.05, P<1x10-5). The same effect was seen after excluding living mothers (β=0.02 standard deviations (SD)), excluding censoring bias. Of the two mtSNVs previously associated with longevity29,30,31, only rs62581312/m.150C>T was available in UKBB (MAF=0.09) but we did not replicate published findings (P=0.16) (and rs28625645/m.489T>C was absent in our data set, R2<0.2 with the available variants). Longevity has been associated with macro-haplogroup J previously30,32–36, however, we found the association unconvincing in UKBB (P=0.05). Our analyses showed there were no haplogroup specific effects on longevity (P=0.13; Supplementary Table 20).
New mtSNV liver biomarker associations
We tested all eight liver function biomarkers available in UKBB (Supplementary Table 16). We identified 23 mtSNVs associated with aspartate aminotransferase (AST) and nine mtSNVs with alanine aminotransferase (ALT, P<5x10-5; Supplementary Tables 18-19). The lead AST-associated variant, rs193302927/m.10238T>C (MAF=0.03; MT-ND3), was associated with an increase of ~0.2U/L in AST and a missense variant rs193302980/m.14766T>C (MAF=0.49) in MT-CYB was associated with an ~0.1U/L reduction in ALT (Table 3, Supplementary Table 18). Notably, mtDNA variants have been associated with AST in the Japanese Biobank15, but these were tagging the B4f haplogroup, which is absent in Europeans. Our haplogroup-specific analyses showed that either the haplogroups or the non-synonymous variant, rs193302980/m.14766 mtSNV could model the association with ALT, while for AST there was no effect of the haplogroups in addition to the synonymous variant rs193302927/m.10238T>C (Supplementary Table 20) suggesting a variant not captured in this study may account for the AST association.
Table 3. New mtSNV associations with serum biomarkers.
| ALT | CYB | rs193302980; 14766 (C, 0.49)M | T7I | -0.008 | 0.002 | 2.8x10-7 (0.001) | homoplastic |
| AST | ND3 | rs193302927; 10238 (C, 0.03)G | I60I | 0.03 | 0.005 | 1.3x10-14 (2x10-10) | I |
| Creatinine | DLOOP | rs869183622; 73 (A, 0.45)G | - | 0.007 | 0.001 | 8.9x10-7 (0.003) | HV |
| Cystatin C | RNR2 | rs3928306; 3010 (A, 0.26)G | - | 0.009 | 0.002 | 2.9x10-7 (0.001) | H1; J1 |
| eGFRcr | DLOOP | rs869183622; 73 (A, 0.45)G | - | -0.007 | 0.001 | 9.6x10-7 (0.003) | HV |
| eGFRcy | RNR2 | rs3928306; 3010 (A, 0.26)G | - | -0.008 | 0.001 | 2.9x10-7 (0.001) | H1; J1 |
| rs3928306; 3010 (A, 0.26)G | - | -0.008 | 0.001 | 1.2x10-7 (0.0005) | H1; J1 | ||
| Urea | CYB | rs2853504; 14793 (G, 0.05)Ga | H16R | -0.015 | 0.004 | 3.9x10-5 (0.05) | U5a |
Summary of the 11 lead mtSNVs-serum biomarker associations (P < 5X10-5) and associated mtSNVs identified as independent signals by FINEMAP (P < 5X10-5 in single-mtSNVs GWAS analysis) in UKBB (in up to 358,640 individuals). Each cell contains the P-value for association between the mtSNV and the listed trait. Locus = mtDNA gene/locus; position = mtDNA nucleotide position on rCRS (NC_012920); rsID = SNP id as of dbSNP 153; EA = effect allele; EAF = effect allele frequency. beta = effect size of the association. se = standard error. FDR = False Discovery Rate calculated with Benjamini-Hochberg procedure; Haplogroups = haplogroups defined by the EA, according to Phylotree (build 17); mtSNVs tagging more than two European haplogroups are reported as “homoplastic”. G = genotyped; I = imputed; M=mixed, i.e. genotyped one array only and imputed on the other; Ga=mtSNVs genotyped on one array only (i.e. either the UKBB array or the UKBL array) and not imputed on the other array (or excluded because of low INFO score on the other array). AST = aspartate aminotransferase; ALT = alanine aminotransferase; eGFRcr = eGFR estimated using creatinine; eGFRcy = eGFR estimated using cystatin C; eGFRcrcy = eGFR estimated using both creatinine and cystatin C. P-values for the lead mt-SNV association for a given trait are in bold.
New mtSNV renal biomarker associations
Creatinine, cystatin C, urea and estimated glomerular filtration rate (eGFR) were associated with one or more of 16 mtSNVs (Fig. 3, Table 3, Supplementary Tables 18-19). We observed distinct mtSNV-creatinine and mtSNV-cystatin C associations (lead mtSNVs rs869183622/m.73A>G MAF=0.45 (allele A in UKBB); and rs3928306/m.3010G>A, MAF=0.26, MT-RNR2 respectively) which were broadly reflected by the creatinine and cystatin C derived eGFR associations (Table 3, Methods). rs2853504/m.14793A>G (MAF=0.05) was associated with reduced urea (Table 3) and cystatin C (Supplementary Table 18) and a lower frequency of polyuria (OR [95%CI] = 0.83[0.76-0.91], P=4.6x10-5; Table 1). The creatinine associations were tagged by rs869183622/m.73 (Table 3) and rs28359172/m.12612 (MAF=0.11; Supplementary Table 19), while the cystatin C association was tagged by the homoplastic variant, rs3928306/m.3010G>A (Table 3), which confers increased mitochondrial sensitivity to the antibiotic linezolid37. None of the individuals in the EUR set reported taking linezolid (or linezolid-containing drugs) (Supplementary Table 22). rs201513497/m.3736G>A (MAF=0.001), rs1556423898/m.11143C>T (MAF=0.002) and rs147029798/m.16126T>C (MAF=0.21) were associated (P<1x10-5) with calculus of kidney, bladder problems and urinary tract infections, respectively (Fig. 3, Table 1) and rs879066842/m.13500T>C (Supplementary Table 18), a rare (MAF=0.003) synonymous variant, (MT-ND5) was associated with a ~2.4mM/L decrease in urine potassium (P=2.5x10-5). Associations between mtSNVs and creatinine and eGFR were also observed in the Japanese Biobank15 but with variants absent in Europeans. We observed an effect of haplogroup U with cystatin C and the cystatin C derived eGFR (eGFRcy) in addition to rs3928306/m.3010G>A (Supplementary Table 20). However, the second most significantly associated variants in both cases (m.16270C>T in cystatin C with MAF=0.09 and m.15218A>G with MAF=0.04 in eGFRcy, Supplementary Table 18) tagged the U haplogroup.
New mtSNV associations with blood cell traits
We identified 44 mtSNVs associated with at least one of 15 blood cell traits (P<5x10-5; Fig. 4, Table 4, Supplementary Table 18-19). Many of the blood cell trait-mtSNV associations were shared across traits e.g. 29 were associated with both mean corpuscular hemoglobin and mean corpuscular volume (Supplementary Table 18). Our fine-mapping analyses identified multiple distinct mtSNV associations for five traits: red blood cell number, mean corpuscular haemoglobin, mean corpuscular volume, plateletcrit, mean platelet volume (Table 4, Supplementary Table 19). We found two mutations (rs199476112, rs267606617) known to cause mitochondrial diseases that were associated with mean corpuscular volume and mean corpuscular haemoglobin (Table 4, Supplementary Table 19). rs199476112/m.11778G>A (MAF=0.0004; MT-ND4) is found in 95% of Europeans with Leber’s hereditary optic atrophy (LHON)38,39, while rs267606617/m.1555A>G (MAF=0.002) in MT-RNR2 is associated with non-syndromic deafness21,22,23 (in addition to our observed association with height). In both cases, the effect sizes were subtle (βmax = 0.3 SD). Interestingly, we found the mean corpuscular volume and mean corpuscular haemoglobin-associated variants, rs200336777/m.15812G>A (MAF=0.008) and rs878944253/m.5633C>T (MAF=0.008; Supplementary Table 18), were also associated with anaemia (Table 1). We identified an effect of haplogroup I in addition to the nonsynonymous mtSNV associations with mean corpuscular haemoglobin (P=4x10-4; Supplementary Table 20).
Rare variant and gene-based tests
The mtSNV-trait associations we identified included 13 rare mtSNVs (MAF < 0.01). Across the seven traits (height, multiple sclerosis, AST, PCT, RBC#, MCV, MCH) that had both rare (MAF ≤ 0.01) and common (MAF > 0.05) mtSNV associations, the rare variants tended to have larger effect sizes than common variants. For example, for multiple sclerosis p.Thr114Ala (MAF=0.21) in MT-ND3 had OR=1.15, compared to the rare variants with OR=1.65-2.06 for p.Ile261Val in MT-ND5 (MAF=0.005) and mt.7559 in MT-TD (MAF=0.005) (Table 1). While for AST, common mtSNVs had β = [-0.007-0.015] SD and rare variants had β = [0.03-0.04] SD (Supplementary Table 18). This difference in effect size could reflect statistical power (Supplementary Fig. 1), but we note that the study was well-powered to identify common mtSNVs with large effects for these phenotypes, but failed to do so.
Gene-based tests identified novel associations between blood cell traits and seven individual genes or clusters of respiratory chain complex genes (P < 0.001 = 0.05/46 aggregate tests; Methods): MCH with MT-ND2 (P = 3.7x10-4), MT-ND3 (P = 6.0x10-4), MT-ND genes coding for respiratory chain Complex I (CI, P = 6.0x10-6), and the DLOOP (P = 1.8x10-4); MCV with Complex I (P = 7.6x10-6) and the DLOOP (P = 4.9x10-5); RBC# with Complex I (P = 2.7x10-4, Supplementary Table 25). A further 85 gene/complex-trait associations involving 31 traits were identified but these associations were driven by a single mtSNV (Supplementary Table 25).
Pathogenic mtSNVs occur on specific haplogroups
Our analysis also included six pathogenic mtSNVs causing mitochondrial diseases (non-syndromic deafness: rs267606617/m.1555A>G with MAF=0.002; LHON: rs28616230/m.4171A>G with MAF=0.0006, rs199476112/m.11778G>A with MAF=0.0003, rs199476104/m.14484T>C with MAF=0.0008; Mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes (MELAS) and Leigh syndrome: rs199476122/m.3697G>A with MAF=0.0002; myopathy: rs3879064421/m.14674T>C with MAF=0.00003)40 (Supplementary Note, Supplementary Fig. 10, Supplementary Table 5), which typically have an incomplete clinical penetrance and are associated with specific mtDNA haplogroups41. For example, patients with maternally inherited blindness due to mtDNA mutations causing LHON, overwhelmingly belong to macro-haplogroup J42, and patients with deafness due to the rs267606617/m.1555A>G variant preferentially belong to macro-haplogroup H43. Although well established, these haplogroup associations are based on ascertaining individuals who have the disease. It is therefore not clear whether the haplogroup influences the clinical penetrance of the mtDNA mutation, whether the haplogroup predisposes to the mtDNA mutation, or whether founder effects contribute to the associations.
We compared the carrier frequencies for the six mtSNVs in the EUR set, using macro-haplogroup H as reference (Supplementary Table 25). rs199476112/m.11778G>A occurred equally across all nine European macro-haplogroups, but the odds of carrying the rs199476104/m.14484T>C on J macro-haplogroup were lower (OR [95% CI]=0.31 [0.18-0.55], P=6.4x10-5), while on the U macro-haplogroup, were higher (OR [95% CI]=1.58 [1.38-1.80], P=1.8x10-11). Similarly, the odds of carrying the rs267606617/m.1555A>G on J and X were higher (OR [95% CI]=1.95 [1.79-2.13] and OR [95% CI]=3.29 [2.91-3.71], respectively; P<1.0x10-5 in both cases) and slightly lower on T (OR [95% CI]= 0.68 [0.58-0.78]; P=1.3x10-4). This adds to the evidence that haplogroup J influences the clinical penetrance of LHON, and excludes a single founder effect for each pathogenic mutation (Supplementary Fig. 9, Supplementary Table 26). We did not analyse heteroplasmic mtDNA mutations because of unreliable allele calling.
Discussion
We have developed a workflow for quality control, imputation that analysis of mtDNA genotypes that results in a set of variants of similarly high quality to the nuclear-encoded variants and will facilitate future mtDNA analyses. When applied to UKBB, the workflow has provided a comprehensive reference dataset of mtDNA variant-trait associations to date, highlighting 260 novel mtDNA-phenotype associations (P<1x10-5). In addition, the gene-based tests identified novel associations between red blood cell traits and MT-ND genes coding for respiratory chain Complex I.
Mitochondrial dysfunction has been observed in several of the diseases that were associated with mtSNVs in our analyses, such as multiple sclerosis44, T2D45, and abdominal aortic aneurysm46. Similarly, some of the associations are with conditions that are complications of mitochondrial or mitochondrial-related diseases e.g. volvulus in a MELAS patient47, eyelid ptosis in chronic progressive ophthalmoplegia48, and bladder problems in multiple sclerosis49. Interestingly, a recent study reported a link between mitochondrial dysfunction and pneumothorax consistent with the association we observe50. We replicate some previously reported disease associations and discovered many more, despite limited statistical power due to UKBB being a general population cohort and the resulting low prevalence of many diseases.
Sixteen, predominantly homoplastic and macro-haplogroup tagging variants (e.g. rs28359172/m.12612A>G), showed pleotropic effects across blood cell traits, liver biomarkers, and renal biomarkers (Supplementary Table 18). Four of the mtSNVs associated with liver biomarkers showed an opposite direction to the effect on creatinine, and sixteen mtSNVs associated with RBC# showed opposite directions of effect for platelet parameters.
The associations we observed between individual mtSNVs and specific traits could either be directly due to the variants themselves (as for non-synonymous or RNA variants) or reflect ‘tagging’ of the real functional variants that reside within the same mtDNA haplogroup or sub-haplogroup, and the lack of whole mtDNA genome sequencing data in UKBB currently precludes in-depth fine-mapping. However, our findings are supported by related clinical observations. For example height and eGFR are known to be affected in severe inherited mtDNA diseases25,51, which is consistent with the idea that ‘extreme phenotypes’ caused by pathogenic mutations and milder quantitative phenotype seen in the general population constitute a spectrum of genetic effects arising from the mitochondrial genome52. Mitochondria also play key roles in the liver and the kidney. Liver mitochondria are a central site for integration of metabolic processes (including the urea cycle) and for exchange of metabolic intermediates, which is linked to oxidative phosphorylation53,54. For example, AST and ALT, the classical liver function biomarkers, are mitochondrial enzymes that play a central role in amino acid metabolism and in the replenishment of intermediates of the tricarboxylic acid cycle within mitochondria55,56,57, and their synthesis is energy dependent. The kidneys have the second highest mitochondrial content and oxygen consumption after the heart58. Renal mitochondria power the active transport needed for ion reabsorptions and play a role in nutrient sensing58. Most liver and kidney pathologies are characterized by mitochondrial dysfunction59,53,60,61,62 and variants in a variety of nuclear-encoded mitochondrial genes have been linked to both mitochondrial and common complex diseases affecting the liver63 and kidneys64,65.
Several mechanisms can explain the mtDNA associations we have observed. For some time it has been known that common polymorphic mtDNA variants, in vertebrate and invertebrate model systems, regulate respiratory chain complex function66–68, membrane potential27, ATP levels69,70 and calcium uptake71. However, even subtle bioenergetic effects can have marked down-stream consequences on cell mtDNA content, intramitochondrial transcription72–74, and cell growth27. Moreover, given emerging evidence that mitochondria act as metabolic hubs controlling diverse cellular processes75, it is plausible that mtDNA variants also modulate canonical cellular signaling pathways76,77 in a tissue specific manner78, explaining why the same mtDNA variants are associated with different phenotypes16, and sometimes with opposing effects. The experimental dissection of these mechanisms is a key next step in the interpretation of our findings.
Lastly, this is the most comprehensive study to explore the population structure of mtDNA in Great Britain to date. We showed that mtDNA structure is reflective of nuclear genetic ancestry and that macro-haplogroups J, K, I and W were more common in the Welsh, Northumbrians and the Scottish, probably reflecting known admixture from the Celts and Vikings (which is also apparent from analyses of the nuclear genome79). The reasons for the associations are not clear, but given recent evidence that the nuclear genome appears to shape mtDNA evolution80–82, it is tempting to speculate that maintaining nuclear-mitochondrial compatibility is driving this at a population level. However, we cannot exclude an ascertainment bias given the homogeneity of the European UKBB subset. We note our study has limitations, including the known inaccuracy of self-reported phenotypes, and a single time-point for the cross-sectional data collection. However, the quantitative phenotype data provides novel insights and forms the baseline for future prospective investigation.
In conclusion, understanding mitochondrial genetic architecture and the interaction between the nuclear and mitochondrial genomes will be important for reducing the burden of cardio-metabolic and neurodegenerative diseases, among others. Our current findings establish the key role played by mtDNA variants in many quantitative human traits, and confirm their contribution to common disease risk. The atlas of UKBB mtSNV-trait associations provided here lays a firm foundation for future studies at the whole mitochondrial genome level.
Methods
Study populations
UK Biobank (UKBB)
The UKBB study is a prospective population study (N=502,682, age range: 40-69, predominantly western-Europeans) of UK residents recruited at 22 assessment centers across the UK between 2006 and 201083. Participants attended a center, where they were deeply phenotyped, including various physical measurements, extensive health and lifestyle questionnaires, and biological samples. DNA was extracted from buffy coat at UK Biocenter (Stockport, UK) using a Promega Maxwell® 16 Blood DNA Purification Kit (AS1010). Samples with sufficient DNA concentration and purity (as measured by 260/280 ratio) were aliquoted and 50 μL were shipped for genotyping at Affymetrix (Santa Clara, Ca, USA), where the majority of participants were genotyped using the UKBB Affymetrix Axiom array (UKBB). This is a customized genotyping array comprising 845,485 probesets for the assay of 820,967 SNVs and short insertions/deletions, including 243 mtSNVs.
A subset of the UK Biobank individuals (N=50,008, males to females ratio: 1:1; white European ancestry, either never smokers or ‘heavy smokers’ (mean 35 pack years) with lung function measurements), were selected to investigate the genetic determinants of smoking behavior, lung function and chronic obstructive pulmonary disorder (COPD) as part of the UKBB Lung Exome Variant Evaluation (UK BiLEVE) study84. As the UK BiLEVE participants are a subset of the UKBB study, DNA extraction, aliquoting and shipment procedures were the same as the rest of the UKBB but UK BiLEVE participants were genotyped using the UK BiLEVE Affymetrix Axiom array (UKBL), which has>95% content overlap with the UKBB array, including 161 mtSNVs in common between the two arrays and 22 unique UKBL mtSNVs. A detailed description of the sample- and variant-based QC procedures85 and genetic principal components estimation we performed using FLASHPCA2 and the genotyped nuclear variants is available in Supplementary Note. UK Biobank has approval from the North West Multi-centre Research Ethics Committee (MREC).
Design of a bespoke workflow for quality control, re-calling and imputation of mtDNA variants
This section summarizes the workflow for quality control, re-calling and imputation of mtSNVs we designed. Further, in depth description of the workflow is available in Supplementary Note. The workflow uses the standard genotype call, intensity and array manifest files provided after genotyping, as well as publicly available whole mitochondrial genomes for imputation purposes.
Stage1. Pre-re-calling QC procedures: Within each genotyping array, we determined the per mtSNV and per sample call rates. We selected for re-calling mtSNVs (Fig.1) with call rate <0.990 (N = 7) and mtSNVs with lower call rates than in the 150,000 UKBB release (N = 8). Further, we excluded 54 samples who were outliers in terms of mean intensities over all SNVs, irrespective of batch, as well as 2,054 samples, because of plate effects (Supplementary Fig. 3 and 4). Next, in the remaining individuals, variants with discordant MAF (> 3%), compared to at least one of the reference data sets (GenBank, 1000 Genomes, WTCCC), were selected for re-calling, only if they also showed suboptimal clustering. Finally, cluster plots for each variant were produced and visually inspected and variants were selected for re-calling on bases of those plots (Supplementary Note, Supplementary Table 1). This resulted in 135 poorly called mtSNVs, out of 265 mtSNVs genotyped in UKBB.
Stages 2 and 3. Re-calling procedures and post-recalling QC: Re-calling of mtSNVs, either per array (N=53, 39.3%) or per batch (N=66, (48.9%) was done using a bespoke validated R script (https://github.com/clody23/UKBiobank_mtPheWas/tree/master/scripts/recalling). Variant cluster plots (per array and per batch) were generated and visually inspected and mtSNVs (N=2, 1.5%) that showed overlapping clusters to an extent where re-calling was not possible and monomorphic variants (N=15, 10.7%) were excluded, resulting in a set of 248 (Supplementary Tables 1-2, Supplementary Fig. 5). Finally, 2,643 samples with low call rate within each array (call rate≤0.97) were excluded.
Stage4. Imputation (using IMPUTE2): Prior to attempting imputation, we explored whether imputation of mtDNA variants was at all possible and tested haploid and diploid settings (Supplementary Note). We found no differences between the two settings and that, with 248 mtSNVs in common between the reference and imputation sets, mtSNVs could be imputed with concordance>90% for both the major and the minor alleles when the following cut-offs were implemented: INFO score≥0.7 and mac≥10 (Supplementary Fig. 6).
We then imputed the UKBB dataset (Supplementary Note, Extended Data Fig. 2) against the 17,815 GenBank complete genomes and 5,271 biallelic SNVs. Imputation was performed separately on each array (N=49,945 for UKBL and N=438,377 for UKBB).
Haplogroup prediction
We predicted participants’ haplogroups using the command line version of the Haplogrep2 v2.1.1 software86 on 483,626 UKBB individuals that passed QC (Supplementary Table 8, Supplementary Fig. 6). Prediction were successful for all the 483,626 individuals (Full Set) when using the genotyped SNVs before QC (N=265), for 462,366 individuals using only post-recalling SNVs (N=248) and for all 483,626 individuals (Full Set) using post-recalling and post-imputed SNVs (N=768). For imputed SNVs, we selected those with INFO score ≥ 0.7 in at least one of the two arrays and mac≥1 (N=520), then imputed genotypes were set to hard calls with gtool (https://www.well.ox.ac.uk/~cfreeman/software/gwas/gtool.html) conversion to ped, setting a probability threshold≥0.9. Haplogroup predictions were based on the rCRS-oriented Phylotree build 1787 human phylogeny using VCF files as input. We also performed Haplogrep2 performed on 17,815 GenBank complete genomes, 2,419 1000 Genomes and 763 WTCCC individuals, using only the subset of homoplasmic variants identified by the MToolBox variant calling88 (Supplementary Table 9, Supplementary Fig. 7).
To compare UKBB haplogroup predictions obtained with the three different reference mtSNV sets we used Haplogrep 2 overall rank score, a value ranging from 0.5 to 1 that reflects the ratio of observed and expected haplogroup-defining mtSNVs for that sample (Supplementary Note).
mtDNA variants annotation
We annotated the high-quality set of 473 minor alleles (including 248 genotyped and 225 imputed SNVs) identified in the UKBB EUR set using a programmatic API query of the HmtVar resource19. These included mitochondrial locus, variant type, amino acid change (for coding variants only), disease annotations in Clinvar89 and MITOMAP40 databases and several pathogenicity predictions and conservation scores (Supplementary Note, Supplementary Table 5, Supplementary Figure 11). We considered pathogenic variants with indication of association with diseases in both Clinvar and MITOMAP databases. We further selected variants with confirmed MITOMAP pathogenic status based on the list provided as of April 2018 (https://www.mitomap.org/foswiki/bin/view/MITOMAP/ConfirmedMutations). We used the Phylotree build 17 human phylogeny87 to retrieve the list of haplogroups tagged by minor alleles, and further integrated using the MITOMAP list of top level haplogroup markers (https://www.mitomap.org/foswiki/bin/view/MITOMAP/HaplogroupMarkers).
Modelling the geographic distribution of mitochondrial variation in Great Britain
To assess the distribution of mitochondrial variation in Great Britain as well as the relationship between the population structures of the two genomes, we first tested whether there was an association between mtSNVs and nucPCs (Extended Data Fig. 2). Then we calculated mtPCs using either all genotyped mtSNVs or genotyped mtSNVs with MAF>0.01. mtPCs were calculated with (R2<0.2) or without pruning over the Full Set or the EUR Set and these were compared to similarly calculated mtPCs in the 3 reference data sets (Extended Data Fig. 3, Supplementary Fig. 8). The correlation between the nucPCs and mtPCs in UKBB was assessed (Extended Data Fig. 2).
Next we added information on where participants were born (east-ness and north-ness of birth postcode) and attempted to predict those using mtSNVs (Supplementary Table 13). Based on the postcodes we assigned people to level 2 territory units of the Nomenclature of Territorial Units for Statistics (NUTS2) and assessed the distribution of macro-haplogroups across those units and whether any differences in frequencies were adjusted for by adding the first 10 nucPCs as covariates. Finally, in order to understand the overlaps between the nuclear genome population structure and macro-haplogroups we performed k-means clustering using the nucPCs and compared the distribution of these clusters and of the macro-haplogroups within the different NUTS2 territory units (Supplementary Tables 11-12, Supplementary Fig. 7). Further, in depth description of the analyses is available in Supplementary Note.
Trait selection
To avoid under-powered analyses, we tested 767 categorical traits that had at least 500 cases each and MAF>0.0001 (Supplementary Note, Supplementary Figure 1, Supplementary Table 14). Traits with low heritability, with less objective measurements or that were not fully available at the time we performed our analysis (e.g. socio-economic, lifestyle traits, MRI phenotypes) were not analyzed. Analyzed traits included International Classification of Diseases version-10 (ICD-10, N=528) codes, non-cancer illness self-reported diseases (#20002, N=166), health and medical history records (codes under category 100036, N=12) and other remaining traits (N=56), including combined categories (Supplementary Table 15). We tested 15 ICD-10 chapters and diseases of the digestive system (chapter XI), musculoskeletal system and connective tissue (chapter XIII) and of the genitourinary system (chapter XIV) represented the biggest portion of traits tested (altogether ~45% of all traits).
We also tested 126 quantitative traits, primarily from amongst the metabolic and cardiovascular endophenotypes, including anthropometric, major lipids, blood pressure, blood cell traits and serum biomarkers amongst others (Supplementary Note, Supplementary Table 16). We did not analyse the 1000s of continuous traits that were socio-economic/lifestyle related e.g. economic status, education, smoking, alcohol consumption, coffee drinking as these are more subjective measures and tend to be less heritable.
Determining a multiple testing threshold for mtSNVs
As opposed to nuclear genome, there is no conventional mitochondrial multiple testing threshold. One could adopt a Bonferroni threshold over the total number of bases in the mt-genome (16,569, P<3x10-6). Such a threshold is overly conservative as it does not recognize the known correlations between multiple variants which reflects the phylogenetic structure of mtDNA inheritance, nor the presence of known mutational cold-spots. Previous candidate-variant studies corrected for number of variants tested, failing to account for correlation between variants, while studies focusing on haplogroups mostly used P<0.0590,91. In a recent study, Kraja et al.92 performed a permutation analysis in one of the cohorts used in their meta-analysis and concluded that 49 SNVs represented the number of independent genetic effects. If we were to adopt similar strategies we would arrive at P=0.05/473=1x10-4 (European Set) or P=0.05/220=2x10-4 after linkage disequilibrium (LD) pruning (R2<0.2). Both strategies do not take into account the actual number of independent variants one could test if whole-genome data was available and may be too liberal. Here we propose a Bonferroni threshold of P=5x10-5=0.05/1000. This threshold reflects the number of distinct mtDNA haplotypes in the whole of UKBB (N=1,141) (Supplementary Table 8) and is also equivalent to the number of independent variants (N=1,036) with MAC≥10 at R2<0.2 within the GenBank reference panel. Additionally, we have calculated the Benjamini-Hochberg false discovery rate (FDR) across all the PheWas associations to contextualise the reported P-values for the association tests (N=378,696; Supplementary Table 17-18).
Statistical analyses
Unless stated otherwise, we performed all the statistical analyses using mtSNV allele dosages, calculated using QCTOOL version 2 (https://www.well.ox.ac.uk/~gav/qctool_v2/).
PheWAS
For binary traits we performed a two-sided Wald test for association between each trait and the mtSNVs was performed using RVTESTS93, adjusting for the effects of sex, array and the first 10 nucPCs. For quantitative traits we performed a (two-tailed) Score test and adjusted for traits specific covariates (Supplementary Table 16). Additional analyses regarding the relationship between age, sex and mtSNVs are outlined in Supplementary Notes and the effects of additional covariates used in the sensitivity analysis are presented in Supplementary Table 21-22). We considered significant association at two-sided P<5x10-5 (see above). Associations where none of the leading mtSNVs were in high LD (R2 >0.8) with previously associated variants we classified as novel. We calculated enrichment of specific types of mtSNVs by applying a two-tailed Z-score test to compare the proportions of non-synonymous, synonymous and non-coding variants within the leading associated-mtSNVs over a background set (binary traits-associated mtSNVs N = 416, quantitative traits-associated mtSNVs N = 473). Notably, the background distribution of mtSNVs did not change from that observed in the GenBank reference set of variants. All our associations analyses were performed using mtSNV allele dosages.
Statistical model assumptions
Analyses of binary traits in the context of extreme case/control imbalance using logistic regression can be anti-conservative94. Therefore, we used both the score and Wald tests when testing mtSNVs for association with binary traits. For the 29 that were significant (P<5x10-5, MAC in cases ≥ 10) we further calculated the likelihood ratio test to ensure robustness. We report the Wald test results as they were the most conservative. The score and Wald test are implemented in RVTEST and STATA 14.2 (https://www.stata.com/stata14/) was used for the likelihood ratio test. Finally, due to the modest number of variants tested, the correlation between variants and high identity by descent (median ratio of shared mtSNVs [MAF>0.01, R2 <0.2] between pairs of individuals=0.92[min=0.68] and 0.90[min=0.71] in GenBank and 1000 Genomes, respectively), we simulated inflation factor (lambda) distributions for the quantitative traits to test if they were in an acceptable range (Supplementary Note, Supplementary Fig. 11).
Interaction analysis: We explored whether there was any evidence for age-by-mtSNV or sex-by-mtSNV interactions, by adding interaction terms to the regression models (STATA 14.2, likelihood ratio test).
Drugs affecting mitochondrial function: We tested whether the observed associations with quantitative traits were independent of the effects of drugs known to affect mitochondrial function such as antibiotics, drugs with heart or liver mitotoxicity, or metformin37,95,96 (Supplementary Note).
Nuclear variants: To explore whether the mtSNV associations were independent of the effects of nuclear variants, we calculated, whenever possible, a nuclear polygenic score (nPGS) for each trait and modeled that together with the standard set of covariates described above. The nPGS was calculated for variants (P< 5x10-8) as follows:
Either conditionally independent or LD-pruned variants (R2 <0.2, PLINK 1.974) were used for nPGS calculations. The β estimates were derived from the following studies: Astle et al, 2016 for blood cell traits97; Sinnot-Armstrong et al, 2019 for serum biomarkers98.
Statistical fine-mapping
We used Bayesian stochastic search approach (using FINEMAP v1.399) on the summary statistics from the single variant analyses to finemap loci with multiple mtSNVs associations. Here we used default priors set --corr-config and --corr-group to 0.7. Correlation between the mtSNVs needed for the fine-mapping was calculated using LDSTORE100 and individuals from both UKBB arrays. We considered evidence for multiple signals present if log10 Bayes factor in favour of one or more underlying variants was > 2.
We also performed sensitivity analyses using stepwise bidirectional regression on the individual level data (cut-off for inclusion was set at the mtGWAS threshold: two-sided P<5x10-5; STATA version 14; https://www.stata.com/stata14/); Sensitivity analyses using conditional regression gave broadly consistent results, with no trait having completely different mtSNVs selected by the two methods. Where inconsistencies occurred, these were usually because the conditional regression included an additional variant, or included variants correlated with (but not the same as) those from FINEMAP (Methods, Supplementary Table 19).
Haplogroup analysis
For all associated traits we tested the distinct contributions of haplogroups and independently associated mtSNVs as defined by FINEMAP. We did this by comparing the model fit (by likelihood ratio test) for the following three models:
Macro-haplogroups were modelled as a factorial variable with haplogroup H as reference for all models.
Haplotype analysis
We tested associations between known pathogenic mutations and haplotype backgrounds using multinomial logistic regression (multinom function of the nnet R package v.7.3-12; https://www.rdocumentation.org/packages/nnet/versions/7.3-12/topics/multinom). We treated the nine European macro-haplogroups as outcome and the pathogenic variants as predictors, using the H haplogroup as reference and adjusting for the effect of array, sex, 10 nucPCs and NUTS2 defined J and W geographical parameters (Supplementary Note).
Gene level analysis
Variants with MAF≤0.02 were selected for gene-based testing. Score tests and covariance matrices for the mtDNA variants were generated for each phenotype using RVTEST93 with the meta-analysis command (--meta). We preformed gene level analyses using the SKAT method (rareMETALS.range function) in the rareMETALs R package101 and two-sided gene-level P-values. Mitochondrial coding and non-coding annotated features of rCRS, downloaded from HmtVar19 were selected for inclusion in the gene-based tests (Supplementary Table 5). We considered 46 features to group the mtSNVs as follows: 37 genes, 3 complexes (I,IV,V), all tRNAs together as a single feature, all rRNAs together as a single feature, the whole of DLOOP and its two hypervariable regions separately (HVSI and HVSII) and all mtSNVs in non-coding regions as a single feature.
Extended Data
Extended Data Fig. 1. Distribution of mitochondrial sub-haplogroups across Great Britain.
The European unrelated individuals with birth coordinates (N=327,665) were clustered based on the first 10 nucPCs, resulting in eight nuclear clusters. The map of Great Britain is colored according to the five regions identified by the most common clusters or combination of clusters in each region: 1) Scotland; 2) North of England (North East and West); 3) North of England (Yorkshire and the Humber, North West of England); 4) South of England (Midlands, London, South East and West of England); 5) Wales. No data were available for Northern Ireland. The stacked bar charts represent the frequency of unrelated individuals in each mitochondrial sub-haplogroups, in the five regions identified by the most common nuclear clusters or combination of nuclear clusters. The star indicates an over-representation (likelihood ratio test, two-sided P<5x10-5) of J1b sub-haplogroup in Scotland compared to the Midlands, London, South East and West region.
Extended Data Fig. 2. Relationship between population structure in the nuclear and mitochondrial genomes.
The figure shows (a) circular Manhattan plots of the association between the first 10 nucPCs and mtSNVs. For each mtSNV, the association was tested using a linear regression model: Y~ β1 x X1 + β2 x X2 + β3 x X3 + β4 x X4 + β5 x X5 where Y is a vector containing the values of a nucPC, X1 is a vector of mtSNV dosages and X2-X5 are vectors containing covariate values (age, age squared, sex, and array) and β1-5 represent the effect of each variable on the mean of Y. Wald test two-sided P-values are presented. The nucPCs are ordered from PC1 to PC10 from outside to in and black dots represent (Wald test, two-sided) P <5x10-5; (b) 3D plots of the first three mtPCs; and (c) the relationship between the first three nuclear principal components (nucPCs, nucPC1 - left, nucPC2 - middle, nucPC3 - right) and the first two mitochondrial principal components (mtPCs). The latter were calculated using mtSNVs with MAF>0.01 and R2<0.2. The mtPCs in (a) and (b) were calculated using the following sets of genotyped mtSNV: (from left to right) all mtSNVs; mtSNVs with MAF>0.01 only; and mtSNVs with MAF>0.01 after LD-pruning at R2<0.2. N=the number of mtSNVs included in a given analysis. In (b) and (c) individuals are coloured according to macro-haplogroup carrier status.
Extended Data Fig. 3. Principal components analysis of the European set of UK Biobank participants in comparison to European participants in GenBank, 1000 genomes and WTCCC.
Plots of the first three mitochondrial principal components (mtPCs) for individuals in: (a) the European set of UK Biobank (N=358,916), (b) GenBank reference set used for imputation (N=6,593), (c) 1000 Genomes individuals (N=498) and (d) WTCCC controls (N=747). For each of the three data sets, plots on the left-hand side show mtPCs calculated using pruned SNVs (R2<0.2 for UK Biobank and R2<0.1 for GenBank, 1000 Genomes and WTCCC) while the plots on the right were generated without LD-pruning. Individuals are colored according to macro-haplogroup carrier status. mtPCs were calculated using genotyped SNVs (MAF>0.01).
Supplementary Material
Acknowledgements
We are grateful to: G. Hudson and H. Griffin for discussions and the preliminary exploratory work that proceeded this study; P. Surendran and T. Jiang for assistance with the genotype calling scripts; W. Astle from the University of Cambridge for providing blood cell trait phenotypes and summary statistics. The BHF Cardiovascular Epidemiology Unit is supported by the UK Medical Research Council [MR/L003120/1], British Heart Foundation [RG/13/13/30194], and UK National Institute for Health Research Cambridge Biomedical Research Centre. P. Chinnery is a Wellcome Trust Principal Research Fellow (212219/Z/18/Z), and a UK NIHR Senior Investigator, who receives support from the Medical Research Council Mitochondrial Biology Unit (MC_UU_00015/9), the Medical Research Council (MRC) International Centre for Genomic Medicine in Neuromuscular Disease, the Evelyn Trust, and the National Institute for Health Research (NIHR) Biomedical Research Centre based at Cambridge University Hospitals NHS Foundation Trust and the University of Cambridge. J. Howson is funded by the British Heart Foundation (RG/13/13/30194; RG/18/13/33946) and the National Institute for Health Research [Cambridge Biomedical Research Centre at the Cambridge University Hospitals NHS Foundation Trust] [*]. E. Yonova-Doing is funded by Isaac Newton Trust/Wellcome Trust ISSF/University of Cambridge. A. Gomez-Duran is funded by the National Institute for Health Research (NIHR - 146281) Cambridge Biomedical Research Centre. This research was conducted using the UKBB Resource under Application Numbers: 20480, 7439 and 18794.
Footnotes
Author Contributions
EY-D has performed analyses and drafted the manuscript. CC has performed analyses and drafted the manuscript. KS selected binary traits for analysis and filtering. AG-D selected binary traits for analysis and filtering. WW performed GenBank data retrieval and initial QC. SK performed initial QC of UK Biobank data. PFC and JMMH drafted the manuscript and supervised the work. All authors approved the final version of the manuscript.
Conflicts of interest
JMMH and EYD become full time employees of Novo Nordisk during the drafting of the manuscript. The remaining authors declare no conflicts of interest.
Data availability
Full summary statistics are provided at: https://app.box.com/s/vu9ewgd9sv7ga3ua0lsk0cql444xm3ht. and at Zenodo: https://doi.org/10.5281/zenodo.4609973
We have used data from the following publicly available databases: www.mitomap.org; https://www.ncbi.nlm.nih.gov/clinvar/; https://www.internationalgenome.org/.
Code availability
Code used to process UKBB data and source data used to generate the main figures is available at: https://github.com/clody23/UKBiobank_mtPheWas.
Source data for Figures 1, 2, 3 and 4 are available from: https://github.com/clody23/UKBiobank_mtPheWas/tree/master/source_files/Figure1, https://github.com/clody23/UKBiobank_mtPheWas/tree/master/source_files/Figure2, https://github.com/clody23/UKBiobank_mtPheWas/tree/master/source_files/Figure3_and_4
Source data for Extended Data Figures 1, 2 and 3 are available from: https://github.com/clody23/UKBiobank_mtPheWas/tree/master/source_files/EDF1,
https://github.com/clody23/UKBiobank_mtPheWas/tree/master/source_files/EDF2,
https://github.com/clody23/UKBiobank_mtPheWas/tree/master/source_files/EDF3
References
- 1.Saraste M. Oxidative phosphorylation at the fin de siècle. Science. 1999;283:1488–1493. doi: 10.1126/science.283.5407.1488. [DOI] [PubMed] [Google Scholar]
- 2.Giles RE, Blanc H, Cann HM, Wallace DC. Maternal inheritance of human mitochondrial DNA. Proceedings of the National Academy of Sciences; 1980. pp. 6715–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elson JL, et al. Analysis of European mtDNAs for recombination. Am J Hum Genet. 2001;68:145–153. doi: 10.1086/316938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wallace DC. Mitochondrial DNA sequence variation in human evolution and disease. Proc Natl Acad Sci USA. 1994;91:8739–8746. doi: 10.1073/pnas.91.19.8739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wallace DC, Brown MD, Lott MT. Mitochondrial DNA variation in human evolution and disease. Gene. 1999;238:211–230. doi: 10.1016/s0378-1119(99)00295-4. [DOI] [PubMed] [Google Scholar]
- 6.Elson JL, Majamaa K, Howell N, Chinnery PF. Associating mitochondrial DNA variation with complex traits. Am J Hum Genet. 2007;80:378–382. doi: 10.1086/511652. author reply 382-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poulton J, et al. Type 2 diabetes is associated with a common mitochondrial variant: evidence from a population-based case-control study. Hum Mol Genet. 2002;11:1581–1583. doi: 10.1093/hmg/11.13.1581. [DOI] [PubMed] [Google Scholar]
- 8.Wallace DC, Chalkia D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb Perspect Biol. 2013;5:a021220. doi: 10.1101/cshperspect.a021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keogh MJ, Chinnery PF. Mitochondrial DNA mutations in neurodegeneration. Biochim Biophys Acta. 2015;1847:1401–1411. doi: 10.1016/j.bbabio.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 10.Herrnstadt C, Howell N. An evolutionary perspective on pathogenic mtDNA mutations: haplogroup associations of clinical disorders. Mitochondrion. 2004;4:791–798. doi: 10.1016/j.mito.2004.07.041. [DOI] [PubMed] [Google Scholar]
- 11.Samuels DC, Carothers AD, Horton R, Chinnery PF. The power to detect disease associations with mitochondrial DNA haplogroups. Am J Hum Genet. 2006;78:713–720. doi: 10.1086/502682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bycroft C, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laurie CC, et al. Quality control and quality assurance in genotypic data for genome-wide association studies. Genet Epidemiol. 2010;34:591–602. doi: 10.1002/gepi.20516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao S, et al. Strategies for processing and quality control of Illumina genotyping arrays. Brief Bioinform. 2017;19:765–775. doi: 10.1093/bib/bbx012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamamoto K, et al. Genetic and phenotypic landscape of the mitochondrial genome in the Japanese population. Commun Biol. 2020;3:104. doi: 10.1038/s42003-020-0812-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hudson G, Gomez-Duran A, Wilson IJ, Chinnery PF. Recent mitochondrial DNA mutations increase the risk of developing common late-onset human diseases. PLoS Genet. 2014;10:e1004369. doi: 10.1371/journal.pgen.1004369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozin MS, et al. Variants of Mitochondrial Genome and Risk of Multiple Sclerosis Development in Russians. Acta Naturae. 2018;10:79–86. [PMC free article] [PubMed] [Google Scholar]
- 18.Tranah GJ, et al. Mitochondrial DNA sequence variation in multiple sclerosis. Neurology. 2015;85:325–330. doi: 10.1212/WNL.0000000000001744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Preste R, Vitale O, Clima R, Gasparre G, Attimonelli M. HmtVar: a new resource for human mitochondrial variations and pathogenicity data. Nucleic Acids Res. 2019;47:D1202–D1210. doi: 10.1093/nar/gky1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell SL, et al. Investigating the relationship between mitochondrial genetic variation and cardiovascular-related traits to develop a framework for mitochondrial phenome-wide association studies. BioData Min. 2014;7:6. doi: 10.1186/1756-0381-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.el-Schahawi M, et al. Two large Spanish pedigrees with nonsyndromic sensorineural deafness and the mtDNA mutation at nt 1555 in the 12s rRNA gene: evidence of heteroplasmy. Neurology. 1997;48:453–456. doi: 10.1212/wnl.48.2.453. [DOI] [PubMed] [Google Scholar]
- 22.Casano RA, et al. Hearing loss due to the mitochondrial A1555G mutation in Italian families. Am J Med Genet. 1998;79:388–391. [PubMed] [Google Scholar]
- 23.Bravo O, Ballana E, Estivill X. Cochlear alterations in deaf and unaffected subjects carrying the deafness-associated A1555G mutation in the mitochondrial 12S rRNA gene. Biochem Biophys Res Commun. 2006;344:511–516. doi: 10.1016/j.bbrc.2006.03.143. [DOI] [PubMed] [Google Scholar]
- 24.Yengo L, et al. Meta-analysis of genome-wide association studies for height and body mass index in ∼700000 individuals of European ancestry. Hum Mol Genet. 2018;27:3641–3649. doi: 10.1093/hmg/ddy271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boal RL, et al. Height as a Clinical Biomarker of Disease Burden in Adult Mitochondrial Disease. J Clin Endocrinol Metab. 2019;104:2057–2066. doi: 10.1210/jc.2018-00957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holzer T, et al. Respiratory chain inactivation links cartilage-mediated growth retardation to mitochondrial diseases. J Cell Biol. 2019;218:1853–1870. doi: 10.1083/jcb.201809056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gómez-Durán A, et al. Oxidative phosphorylation differences between mitochondrial DNA haplogroups modify the risk of Leber’s hereditary optic neuropathy. Biochim Biophys Acta. 2012;1822:1216–1222. doi: 10.1016/j.bbadis.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 28.Gómez-Durán A, et al. Unmasking the causes of multifactorial disorders: OXPHOS differences between mitochondrial haplogroups. Hum Mol Genet. 2010;19:3343–3353. doi: 10.1093/hmg/ddq246. [DOI] [PubMed] [Google Scholar]
- 29.Chen A, Raule N, Chomyn A, Attardi G. Decreased reactive oxygen species production in cells with mitochondrial haplogroups associated with longevity. PLoS ONE. 2012;7:e46473. doi: 10.1371/journal.pone.0046473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niemi A-K, et al. A combination of three common inherited mitochondrial DNA polymorphisms promotes longevity in Finnish and Japanese subjects. Eur J Hum Genet. 2005;13:166–170. doi: 10.1038/sj.ejhg.5201308. [DOI] [PubMed] [Google Scholar]
- 31.Zhang J, et al. Strikingly higher frequency in centenarians and twins of mtDNA mutation causing remodeling of replication origin in leukocytes. Proc Natl Acad Sci USA. 2003;100:1116–1121. doi: 10.1073/pnas.242719399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niemi A-K, et al. Mitochondrial DNA polymorphisms associated with longevity in a Finnish population. Hum Genet. 2003;112:29–33. doi: 10.1007/s00439-002-0843-y. [DOI] [PubMed] [Google Scholar]
- 33.Santoro A, et al. Mitochondrial DNA involvement in human longevity. Biochim Biophys Acta. 2006;1757:1388–1399. doi: 10.1016/j.bbabio.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 34.Dato S, et al. Association of the mitochondrial DNA haplogroup J with longevity is population specific. Eur J Hum Genet. 2004;12:1080–1082. doi: 10.1038/sj.ejhg.5201278. [DOI] [PubMed] [Google Scholar]
- 35.De Benedictis G, et al. Mitochondrial DNA inherited variants are associated with successful aging and longevity in humans. FASEB J. 1999;13:1532–1536. doi: 10.1096/fasebj.13.12.1532. [DOI] [PubMed] [Google Scholar]
- 36.Rose G, et al. Paradoxes in longevity: sequence analysis of mtDNA haplogroup J in centenarians. Eur J Hum Genet. 2001;9:701–707. doi: 10.1038/sj.ejhg.5200703. [DOI] [PubMed] [Google Scholar]
- 37.Pacheu-Grau D, et al. Mitochondrial antibiograms in personalized medicine. Hum Mol Genet. 2013;22:1132–1139. doi: 10.1093/hmg/dds517. [DOI] [PubMed] [Google Scholar]
- 38.Jurkute N, Yu-Wai-Man P. Leber hereditary optic neuropathy: bridging the translational gap. Curr Opin Ophthalmol. 2017;28:403–409. doi: 10.1097/ICU.0000000000000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu-Wai-Man P, Turnbull DM, Chinnery PF. Leber hereditary optic neuropathy. J Med Genet. 2002;39:162–169. doi: 10.1136/jmg.39.3.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kogelnik AM, Lott MT, Brown MD, Navathe SB, Wallace DC. MITOMAP: a human mitochondrial genome database. Nucleic Acids Res. 1996;24:177–179. doi: 10.1093/nar/24.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chinnery PF, Gomez-Duran A. Oldies but Goldies mtDNA Population Variants and Neurodegenerative Diseases. Front Neurosci. 2018;12:682. doi: 10.3389/fnins.2018.00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elliott HR, Samuels DC, Eden JA, Relton CL, Chinnery PF. Pathogenic Mitochondrial DNA Mutations Are Common in the General Population. Am J Hum Genet. 2008;83:254–260. doi: 10.1016/j.ajhg.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Achilli A, et al. The molecular dissection of mtDNA haplogroup H confirms that the Franco-Cantabrian glacial refuge was a major source for the European gene pool. Am J Hum Genet. 2004;75:910–918. doi: 10.1086/425590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patergnani S, et al. Mitochondria in Multiple Sclerosis: Molecular Mechanisms of Pathogenesis. Int Rev Cell Mol Biol. 2017;328:49–103. doi: 10.1016/bs.ircmb.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 45.Achilli A, et al. Mitochondrial DNA backgrounds might modulate diabetes complications rather than T2DM as a whole. PLoS ONE. 2011;6:e21029. doi: 10.1371/journal.pone.0021029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Navas-Madroñal M, et al. Enhanced endoplasmic reticulum and mitochondrial stress in abdominal aortic aneurysm. Clin Sci. 2019;133:1421–1438. doi: 10.1042/CS20190399. [DOI] [PubMed] [Google Scholar]
- 47.Hallac A, Keshava HB, Morris-Stiff G, Ibrahim S. Sigmoid volvulus in a patient with mitochondrial encephalomyopathy, lactic acidosis and stroke-like episodes (MELAS): a rare occurrence. BMJ Case Rep. 2016;2016 doi: 10.1136/bcr-2015-213718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu-Wai-Man P, Newman NJ. Inherited eye-related disorders due to mitochondrial dysfunction. Hum Mol Genet. 2017;26:R12–R20. doi: 10.1093/hmg/ddx182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Compston A, Coles A. Multiple sclerosis. Lancet. 2002;359:1221–1231. doi: 10.1016/S0140-6736(02)08220-X. [DOI] [PubMed] [Google Scholar]
- 50.Carstens P-O, et al. X-linked myotubular myopathy and recurrent spontaneous pneumothorax: A new phenotype? Neurol Genet. 2019;5:e327. doi: 10.1212/NXG.0000000000000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martín-Hernández E, et al. Renal pathology in children with mitochondrial diseases. Pediatr Nephrol. 2005;20:1299–1305. doi: 10.1007/s00467-005-1948-z. [DOI] [PubMed] [Google Scholar]
- 52.Stewart JB, Chinnery PF. The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat Rev Genet. 2015;16:530–542. doi: 10.1038/nrg3966. [DOI] [PubMed] [Google Scholar]
- 53.Degli Esposti D, et al. Mitochondrial roles and cytoprotection in chronic liver injury. Biochem Res Int. 2012;2012:387626. doi: 10.1155/2012/387626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Houten SM, Wanders RJA. A general introduction to the biochemistry of mitochondrial fatty acid β-oxidation. J Inherit Metab Dis. 2010;33:469–477. doi: 10.1007/s10545-010-9061-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Owen OE, Kalhan SC, Hanson RW. The key role of anaplerosis and cataplerosis for citric acid cycle function. J Biol Chem. 2002;277:30409–30412. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- 56.Pesi R, Balestri F, Ipata PL. Metabolic interaction between urea cycle and citric acid cycle shunt: A guided approach. Biochem Mol Biol Educ. 2018;46:182–185. doi: 10.1002/bmb.21099. [DOI] [PubMed] [Google Scholar]
- 57.Martínez-Reyes I, Chandel NS. Mitochondrial TCA cycle metabolites control physiology and disease. Nat Commun. 2020;11:102. doi: 10.1038/s41467-019-13668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bhargava P, Schnellmann RG. Mitochondrial energetics in the kidney. Nat Rev Nephrol. 2017;13:629–646. doi: 10.1038/nrneph.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Connor TM, et al. Mutations in mitochondrial DNA causing tubulointerstitial kidney disease. PLoS Genet. 2017;13:e1006620. doi: 10.1371/journal.pgen.1006620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Galvan DL, Green NH, Danesh FR. The hallmarks of mitochondrial dysfunction in chronic kidney disease. Kidney Int. 2017;92:1051–1057. doi: 10.1016/j.kint.2017.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hunt NJ, Kang SWS, Lockwood GP, Le Couteur DG, Cogger VC. Hallmarks of Aging in the Liver. Comput Struct Biotechnol J. 2019;17:1151–1161. doi: 10.1016/j.csbj.2019.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mansouri A, Gattolliat C-H, Asselah T. Mitochondrial Dysfunction and Signaling in Chronic Liver Diseases. Gastroenterology. 2018;155:629–647. doi: 10.1053/j.gastro.2018.06.083. [DOI] [PubMed] [Google Scholar]
- 63.Lee WS, Sokol RJ. Liver disease in mitochondrial disorders. Semin Liver Dis. 2007;27:259–273. doi: 10.1055/s-2007-985071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.O’Toole JF. Renal manifestations of genetic mitochondrial disease. Int J Nephrol Renovasc Dis. 2014;7:57–67. doi: 10.2147/IJNRD.S37887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eirin A, Lerman A, Lerman LO. The Emerging Role of Mitochondrial Targeting in Kidney Disease. Handb Exp Pharmacol. 2017;240:229–250. doi: 10.1007/164_2016_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moreno-Loshuertos R, et al. Differences in reactive oxygen species production explain the phenotypes associated with common mouse mitochondrial DNA variants. Nat Genet. 2006;38:1261–1268. doi: 10.1038/ng1897. [DOI] [PubMed] [Google Scholar]
- 67.Correa CC, Aw WC, Melvin RG, Pichaud N, Ballard JWO. Mitochondrial DNA variants influence mitochondrial bioenergetics in Drosophila melanogaster. Mitochondrion. 2012;12:459–464. doi: 10.1016/j.mito.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 68.Ji F, et al. Mitochondrial DNA variant associated with Leber hereditary optic neuropathy and high-altitude Tibetans. Proc Natl Acad Sci USA. 2012;109:7391–7396. doi: 10.1073/pnas.1202484109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bellizzi D, D’Aquila P, Giordano M, Montesanto A, Passarino G. Global DNA methylation levels are modulated by mitochondrial DNA variants. Epigenomics. 2012;4:17–27. doi: 10.2217/epi.11.109. [DOI] [PubMed] [Google Scholar]
- 70.Fernández-Moreno M, et al. Mitochondrial DNA haplogroups influence the risk of incident knee osteoarthritis in OAI and CHECK cohorts. A meta-analysis and functional study. Ann Rheum Dis. 2017;76:1114–1122. doi: 10.1136/annrheumdis-2016-210131. [DOI] [PubMed] [Google Scholar]
- 71.Kazuno A, et al. Identification of mitochondrial DNA polymorphisms that alter mitochondrial matrix pH and intracellular calcium dynamics. PLoS Genet. 2006;2:e128. doi: 10.1371/journal.pgen.0020128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Suissa S, et al. Ancient mtDNA genetic variants modulate mtDNA transcription and replication. PLoS Genet. 2009;5:e1000474. doi: 10.1371/journal.pgen.1000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Salminen TS, et al. Mitochondrial genotype modulates mtDNA copy number and organismal phenotype in Drosophila. Mitochondrion. 2017;34:75–83. doi: 10.1016/j.mito.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 74.Picard M, et al. Progressive increase in mtDNA 3243A>G heteroplasmy causes abrupt transcriptional reprogramming. Proc Natl Acad Sci USA. 2014;111:E4033-4042. doi: 10.1073/pnas.1414028111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mottis A, Herzig S, Auwerx J. Mitocellular communication: Shaping health and disease. Science. 2019;366:827–832. doi: 10.1126/science.aax3768. [DOI] [PubMed] [Google Scholar]
- 76.Fang H, et al. mtDNA Haplogroup N9a Increases the Risk of Type 2 Diabetes by Altering Mitochondrial Function and Intracellular Mitochondrial Signals. Diabetes. 2018;67:1441–1453. doi: 10.2337/db17-0974. [DOI] [PubMed] [Google Scholar]
- 77.D’Aquila P, Rose G, Panno ML, Passarino G, Bellizzi D. SIRT3 gene expression: a link between inherited mitochondrial DNA variants and oxidative stress. Gene. 2012;497:323–329. doi: 10.1016/j.gene.2012.01.042. [DOI] [PubMed] [Google Scholar]
- 78.Dunbar DR, Moonie PA, Jacobs HT, Holt IJ. Different cellular backgrounds confer a marked advantage to either mutant or wild-type mitochondrial genomes. Proc Natl Acad Sci USA. 1995;92:6562–6566. doi: 10.1073/pnas.92.14.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Leslie S, et al. The fine-scale genetic structure of the British population. Nature. 2015;519:309–314. doi: 10.1038/nature14230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wei W, et al. Germline selection shapes human mitochondrial DNA diversity. Science. 2019;364 doi: 10.1126/science.aau6520. [DOI] [PubMed] [Google Scholar]
- 81.Latorre-Pellicer A, et al. Regulation of Mother-to-Offspring Transmission of mtDNA Heteroplasmy. Cell Metabolism. 2019;30:1120–1130.:e5. doi: 10.1016/j.cmet.2019.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Latorre-Pellicer A, et al. Mitochondrial and nuclear DNA matching shapes metabolism and healthy ageing. Nature. 2016;535:561–565. doi: 10.1038/nature18618. a genetic association study in UK Biobank. Lancet Respir Med 3, 769-781 (2015) [DOI] [PubMed] [Google Scholar]
- 83.Sudlow C, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wain LV, et al. Novel insights into the genetics of smoking behaviour, lung function, and chronic obstructive pulmonary disease (UK BiLEVE): a genetic association study in UK Biobank. Lancet Respir Med. 2015;3:769–781. doi: 10.1016/S2213-2600(15)00283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Surendran P, et al. Discovery of rare variants associated with blood pressure regulation through meta-analysis of 1.3 million individuals. Nat Genet. 2020;52:1314–1332. doi: 10.1038/s41588-020-00713-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Weissensteiner H, et al. HaploGrep 2: mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. 2016;44:W58–63. doi: 10.1093/nar/gkw233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30:E386-394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 88.Calabrese C, et al. MToolBox: a highly automated pipeline for heteroplasmy annotation and prioritization analysis of human mitochondrial variants in high-throughput sequencing. Bioinformatics. 2014;30:3115–3117. doi: 10.1093/bioinformatics/btu483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Landrum MJ, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980-985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chalkia D, et al. Association Between Mitochondrial DNA Haplogroup Variation and Autism Spectrum Disorders. JAMA Psychiatry. 2017;74:1161–1168. doi: 10.1001/jamapsychiatry.2017.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hudson G, et al. Two-stage association study and meta-analysis of mitochondrial DNA variants in Parkinson disease. Neurology. 2013;80:2042–2048. doi: 10.1212/WNL.0b013e318294b434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kraja AT, et al. Associations of Mitochondrial and Nuclear Mitochondrial Variants and Genes with Seven Metabolic Traits. Am J Hum Genet. 2019;104:112–138. doi: 10.1016/j.ajhg.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhan X, Hu Y, Li B, Abecasis GR, Liu DJ. RVTESTS: an efficient and comprehensive tool for rare variant association analysis using sequence data. Bioinformatics. 2016;32:1423–1426. doi: 10.1093/bioinformatics/btw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ma C, Blackwell T, Boehnke M, Scott LJ, GoT2D investigators Recommended joint and meta-analysis strategies for case-control association testing of single low-count variants. Genet Epidemiol. 2013;37:539–550. doi: 10.1002/gepi.21742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Meyer JN, Hartman JH, Mello DF. Mitochondrial Toxicity. Toxicol Sci. 2018;162:15–23. doi: 10.1093/toxsci/kfy008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vial G, Detaille D, Guigas B. Role of Mitochondria in the Mechanism(s) of Action of Metformin. Front Endocrinol (Lausanne) 2019;10:294. doi: 10.3389/fendo.2019.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Astle WJ, et al. The Allelic Landscape of Human Blood Cell Trait Variation and Links to Common Complex Disease. Cell. 2016;167:1415–1429.:e19. doi: 10.1016/j.cell.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sinnott-Armstrong N, et al. Genetics of 38 blood and urine biomarkers in the UK Biobank. 2019 doi: 10.1101/660506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Benner C, et al. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics. 2016;32:1493–1501. doi: 10.1093/bioinformatics/btw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Benner C, et al. Prospects of Fine-Mapping Trait-Associated Genomic Regions by Using Summary Statistics from Genome-wide Association Studies. Am J Hum Genet. 2017;101:539–551. doi: 10.1016/j.ajhg.2017.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Feng S, Liu D, Zhan X, Wing MK, Abecasis GR. RAREMETAL: fast and powerful meta-analysis for rare variants. Bioinformatics. 2014;30:2828–2829. doi: 10.1093/bioinformatics/btu367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Full summary statistics are provided at: https://app.box.com/s/vu9ewgd9sv7ga3ua0lsk0cql444xm3ht. and at Zenodo: https://doi.org/10.5281/zenodo.4609973
We have used data from the following publicly available databases: www.mitomap.org; https://www.ncbi.nlm.nih.gov/clinvar/; https://www.internationalgenome.org/.
Code used to process UKBB data and source data used to generate the main figures is available at: https://github.com/clody23/UKBiobank_mtPheWas.
Source data for Figures 1, 2, 3 and 4 are available from: https://github.com/clody23/UKBiobank_mtPheWas/tree/master/source_files/Figure1, https://github.com/clody23/UKBiobank_mtPheWas/tree/master/source_files/Figure2, https://github.com/clody23/UKBiobank_mtPheWas/tree/master/source_files/Figure3_and_4
Source data for Extended Data Figures 1, 2 and 3 are available from: https://github.com/clody23/UKBiobank_mtPheWas/tree/master/source_files/EDF1,
https://github.com/clody23/UKBiobank_mtPheWas/tree/master/source_files/EDF2,
https://github.com/clody23/UKBiobank_mtPheWas/tree/master/source_files/EDF3