Abstract
Cells respond to stress by blocking translation, rewiring metabolism, and forming transient mRNP assemblies called stress granules (SGs). After stress release, re-establishing homeostasis and disassemblying SGs requires ATP-consuming processes. However, the molecular mechanisms whereby cells restore ATP production and disassemble SGs after stress remain poorly understood. Here we show that upon stress, the ATP-producing enzyme Cdc19 forms inactive amyloids, and that their rapid re-solubilization is essential to restore ATP production and disassemble SGs in glucose-containing media. Cdc19 re-solubilization is initiated by the glycolytic metabolite fructose-1,6-bisphosphate (FBP), which directly binds Cdc19 amyloids, allowing Hsp104 and Ssa2 chaperone recruitment and aggregate re-solubilization. FBP then promotes Cdc19 tetramerization, which boosts its activity to further enhance ATP production and SG disassembly. Together, these results describe a molecular mechanism critical for stress recovery, which directly couples cellular metabolism with SG dynamics via regulation of reversible Cdc19 amyloids.
Introduction
To maintain homeostasis, cells need to rapidly sense stress conditions such as heat stress and nutrient starvation, and orchestrate appropriate protective responses, including the formation of stress granules (SGs) [1]. SGs are reversible, membrane-less mRNP organelles that form in the cytoplasm in response to metabolic stress, and are thought to store and protect mRNAs and proteins from stress-induced degradation [1–6]. Formation of persistent SGs due to chronic stress or mutations, and the consequent metabolic changes have been associated with neurodegenerative diseases such as amyotrophic lateral sclerosis and Alzheimer’s disease [7, 8]. Thus, SGs must be tightly controlled and efficiently disassembled. However, despite their patho-physiological importance, the mechanisms regulating SG dynamics remain poorly understood.
Most SG proteins contain intrinsically disordered low-complexity regions (LCRs) that can condense by liquid-liquid phase separation (LLPS) [9, 10]. This process, together with low-affinity RNA-RNA interactions, is thought to underlie SG formation [11, 12]. In contrast, SG disassembly mechanisms are less well understood. ATP plays a crucial role in disassembly, as artificial ATP depletion reduces SG dynamics and results in persistent SGs [3, 9, 13], while adding ATP to stressed cells triggers aggregate disassembly [14]. The ATP-dependent activities of the disaggregase Hsp104 and the Hsp70 chaperones Ssa1 and Ssa2, as well as the kinases Sky1 [15] and ULK1/2 [16] have been implicated in efficient SG disassembly and growth restart after stress [14, 17]. ATP itself was also proposed to act as a biological hydrotrope, modulating cytoplasmic diffusion and further facilitating aggregate re-solubilization [18]. However, ATP reserves are rapidly depleted in stressed cells [14, 19–22], raising the question of how energy production is restored for efficient post-stress SG disassembly.
Yeast cells mainly produce ATP by fermenting glucose through glycolysis [23, 24], in which the conserved enzyme pyruvate kinase (Cdc19) catalyses the ATP-producing and irreversible final step, i.e. the conversion of phosphoenolpyruvate (PEP) and ADP into pyruvate and ATP. Cdc19 activity is modulated by its allosteric regulator fructose-1,6-biphosphate (FBP), which promotes Cdc19 oligomerization into more active tetramers [4, 22, 25, 26]. Moreover, glycolytic flux is altered by trehalose metabolism, which regulates glucose-6-phosphate availability and hexokinase activity [27–29]. We previously showed that Cdc19 reversibly aggregates via its LCR and localizes to SGs in response to glucose starvation and heat stress [4, 30]. Formation and disassembly of Cdc19 aggregates modulates SG dynamics, and ensures stress survival. In particular, failure to timely re-solubilize Cdc19 aggregates results in persistent SGs and no cell cycle restart after stress release. However, how physiological Cdc19 aggregates are re-solubilized and how they regulate SGs disassembly is unclear.
To address these questions, we combined in vitro and in vivo approaches, and exploited a Cdc19 mutant (Cdc19irrev) unable to disassemble Cdc19 aggregates after stress [4]. We found that reversible Cdc19 aggregates assume an enzymatically inactive, amyloid structure in vivo. Their post-stress re-solubilization is necessary to restore ATP production and re-solubilize SGs on fermentable carbon sources. We propose a molecular mechanism in which direct FBP binding to functional Cdc19 aggregates initiates their re-solubilisation and subsequent SG disassembly by promoting recruitment of the molecular chaperones Hsp104 and Ssa2. Together, our results not only provide a mechanism for re-solubilizing functional amyloids in vivo, but also show how metabolism and energy production are coupled to SG dynamics and cell growth.
Results
SG disassembly depends on resolubilization of Cdc19 amyloids
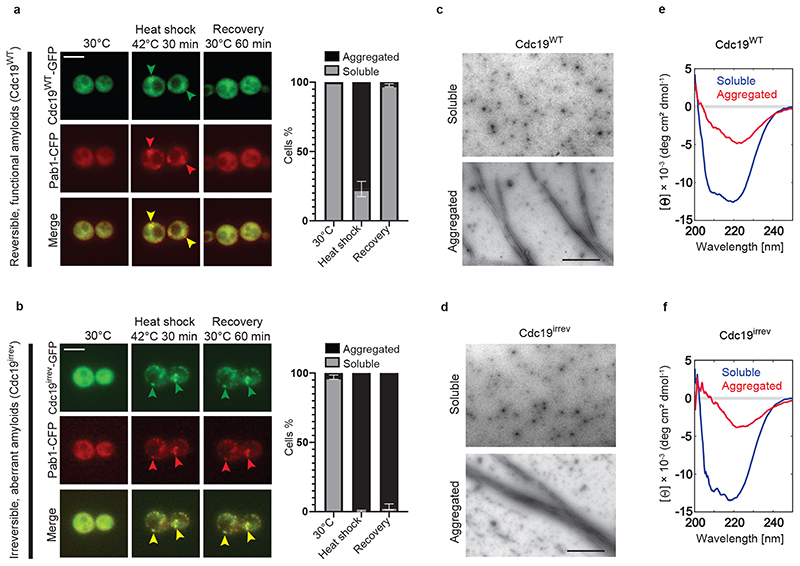
Formation of reversible Cdc19 aggregates upon stress and their timely resolubilization after stress release is required to ensure cell survival [4]. Indeed, cells expressing an irreversibly aggregating mutant of Cdc19 (Cdc19irrev) are unable to restart growth after stress. This Cdc19irrev mutant, called Cdc194D in previous work, bears four phosphomimicking mutations in the aggregation-prone Cdc19 LCR (cdc19-T372D, T376D, S377D, S385D, Extended Data Fig. 1A). Cdc19irrev is active and grows similarly to the wild-type in no stress conditions, but upon stress it forms irreversible aggregates and causes persistent SGs (Fig. 1A and B).
Figure 1. SG disassembly depends on re-solubilization of Cdc19 amyloids.
(A) – (B) Cdc19WT-GFP and Cdc19irrev-GFP cells expressing the SG marker Pab1-CFP were grown at 30 °C, and subjected to heat stress for 30 min at 42 °C before recovery at 30 °C. Representative images taken at the indicated time points are shown. Merged images demonstrate co-localization (arrowheads). Plots indicate mean percentage (%) of cells with Cdc19 aggregates ± S.E.M (n = 3 independent experiments, >30 cells per sample per experiment). Scale bar: 5 μm.
(C) – (D) Cdc19WT or Cdc19irrev proteins purified from E. coli were imaged by negative staining transmission electron microscopy (TEM). Representative images of three independent experiments are shown. Scale bar: 2 μm.
(E) – (F) Cdc19WT or Cdc19irrev proteins purified from E. coli were analyzed by circular dichroism (CD) before (soluble) or after a 10 min heat shock at 42 °C (aggregated). Representative images of three independent experiments are shown. Source data for all graphical representations can be found in Source Data Fig. 1.
To understand how Cdc19 regulates SG disassembly and the mechanisms underlying Cdc19 aggregate re-solubilization, we investigated wild-type and Cdc19irrev aggregate properties and potential differences in vitro and in vivo. We examined biophysical characteristics of Cdc19WT and Cdc19irrev aggregates to understand if irreversibility was caused by an aberrant aggregate structure. Recombinant wild-type and Cdc19irrev proteins were produced from E. coli, and their aggregation was triggered in vitro by heat shock. Both proteins formed needle-like filaments, which were indistinguishable by negative staining transmission electron microscopy (TEM) (Fig. 1C and D). Wild-type and mutant aggregates also revealed similar β-sheet-rich structures by circular dichroism (CD) spectroscopy (Fig. 1E and F), and bound the amyloid dyes Thioflavin T (ThT) and Congo Red (CR) [31] (Extended Data Fig. 1B and C), suggesting that Cdc19WT and Cdc19irrev form similar amyloids in vitro. To test whether Cdc19 aggregates also exhibit amyloid structures in vivo, we heat shocked and lysed yeast cells, and stained both Cdc19WT and Cdc19irrev aggregates with CR. Both cellular Cdc19 aggregates bound the amyloid dye (Extended Data Fig. 1D). Background staining by CR is likely due to its ability to also bind cellular structures other than amyloids (e.g. cell wall) [32]. To further confirm the amyloid nature of Cdc19 aggregates in vivo, we applied limited proteolysis coupled to mass-spectrometry (LiP-MS) (Extended Data Fig. 1E). This technique allows probing subtle conformational changes both in vitro and in complex cell lysates, and offers more precise structural information compared to conventional methods for detecting amyloids [33, 34]. Cdc19 aggregation was induced in vitro by heat shocking purified Cdc19, and in vivo by subjecting cells to starvation stress to achieve full Cdc19 aggregation. LiP-MS analysis revealed that diagnostic Cdc19 LiP-MS peptides characteristic of the amyloid conformation in vitro resemble the structures observed in vivo. Moreover, conformation-sensitive LiP-peptides were mostly similar in wild-type and Cdc19irrev assemblies, suggesting that their different re-solubilization behaviour is not due to radically different structures (Extended Data Fig. 1E). Interestingly, LiP-MS also identified subtle structural alterations in the aggregated catalytic domain of Cdc19. Thus, Cdc19 aggregation and re-solubilization could affect Cdc19 activity and constitute a potential difference between Cdc19WT and Cdc19irrev.
Cdc19 amyloid resolubilization is crucial for ATP production
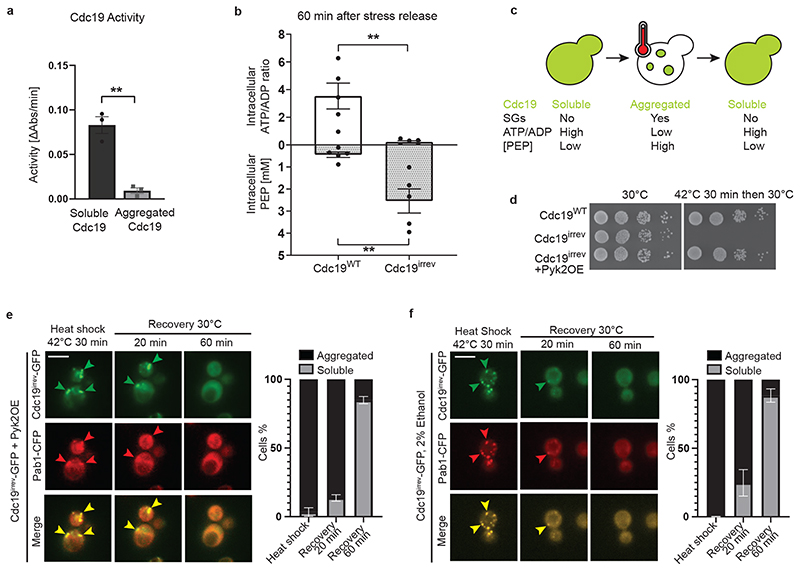
Comparing the enzymatic activity between soluble and aggregated Cdc19 using an established lactate-dehydrogenase-coupled pyruvate kinase activity assay revealed that Cdc19 amyloids are enzymatically inactive (Fig. 2A). Given the large conformational changes required for amyloid fibril formation, the loss of catalytic activity is not surprising. Consistently, cellular energy levels - represented by the ATP/ADP ratio - rapidly dropped in cells exposed to heat stress or glucose starvation, while Cdc19’s substrate PEP accumulated under these conditions [14, 20–22] (Fig. 2C). Thus, to re-activate Cdc19 and resume energy production, Cdc19 amyloids need to be re-solubilized. Since ATP is essential to disassemble SGs, persistent SGs in cdc19irrev cells could be due to impaired ATP production. Indeed, in contrast to wild-type controls, cdc19irrev cells could not metabolize the accumulated PEP and hence failed to restore high ATP levels during stress recovery (Fig. 2B and Extended Data Fig. 1F). We conclude that wild-type and cdc19irrev cells differ primarily in their ability to restore energy production after stress.
Figure 2. SG disassembly depends on Cdc19 re-solubilization and restoration of ATP production.
(A) The activity of purified Cdc19WT before (soluble) or after a 30 min heat shock at 42 °C (aggregated) was measured using a lactate dehydrogenase-coupled activity assay and plotted as the decrease in absorbance at 340 nm per min. Mean ± S.E.M. of n = 3 independent experiments is indicated (two-tailed t-test, P = 0.0018).
(B) cdc19irrev cells are unable to re-activate Cdc19 and resume ATP production after stress release. Intracellular ATP/ADP ratio and phosphoenolpyruvate (PEP) concentrations (mM) were determined in wild-type and cdc19irrev cells, after a 30 min heat shock at 42 °C and recovery at 30 °C for 60 min. Mean ± S.E.M. of n = 5 independent experiments is shown (two-tailed Mann-Whitney test, PATP/ADP = 0.0079, PPEP = 0.0079).
(C) Schematic drawing showing a yeast cell before, during, and after heat stress. Upon stress, Cdc19 (represented in green) forms inactive aggregates, which colocalize with SGs. Energy levels (ATP/ADP ratio) rapidly decrease under these conditions, while Cdc19’s substrate phosphoenolpyruvate (PEP) accumulates. Upon stress release, Cdc19 re-solubilizes, gets re-activated, and consumes PEP to produce ATP, concomitant with SG disassembly.
(D) Serial dilutions of exponentially growing cells of the indicated genotype were spotted on agar plates before or after a 30 min heat shock at 42 °C, and imaged after 3 days at 30 °C (n = 3 independent experiments).
(E) – (F) ATP production using Cdc19-independent pathways allows Cdc19irrev aggregate and SG re-solubilization after stress release. Cells of the indicated genotype were grown at 30 °C with 2% glucose (E) or 2% ethanol (F) as carbon source and subjected to heat stress for 30 min at 42 °C before recovery at 30 °C in the presence of 25 μg/ml cycloheximide to prevent de novo protein synthesis. Representative images are shown as in Fig. 1A. Plots indicate mean percentage (%) of cells with Cdc19 aggregates ± S.E.M (n = 3 independent experiments, >30 cells per sample per experiment). Scale bar: 5 μm. Source data for all graphical representations can be found in Source Data Fig. 2.
To test if increasing ATP levels was sufficient to disassemble SGs, we restored ATP metabolism in heat-stressed cdc19irrev cells by two Cdc19-independent means. First, we artificially overexpressed Pyk2, a pyruvate-kinase isoform expressed at low levels in glucose-rich conditions that like Cdc19 uses PEP to produce ATP [35]. Consistent with our hypothesis, Pyk2 overexpression re-solubilized Cdc19irrev aggregates and SGs, and restored growth of heat-shocked cdc19irrev cells (Fig. 2D and E). Second, we grew cdc19irrev cells with 2% ethanol instead of glucose. Under these conditions, ATP production occurs mainly via oxidative phosphorylation [36]. This also allowed Cdc19irrev aggregate and SG re-solubilization (Fig. 2F). We conclude that under physiological conditions, when yeast cells undergo fermentative growth, re-solubilizing Cdc19 amyloids is critical to resume post-stress ATP production, which is likely required to power ATP-dependent processes critical for SG dissolution and growth restart.
Disassembly of Cdc19 amyloids in vivo is triggered by FBP
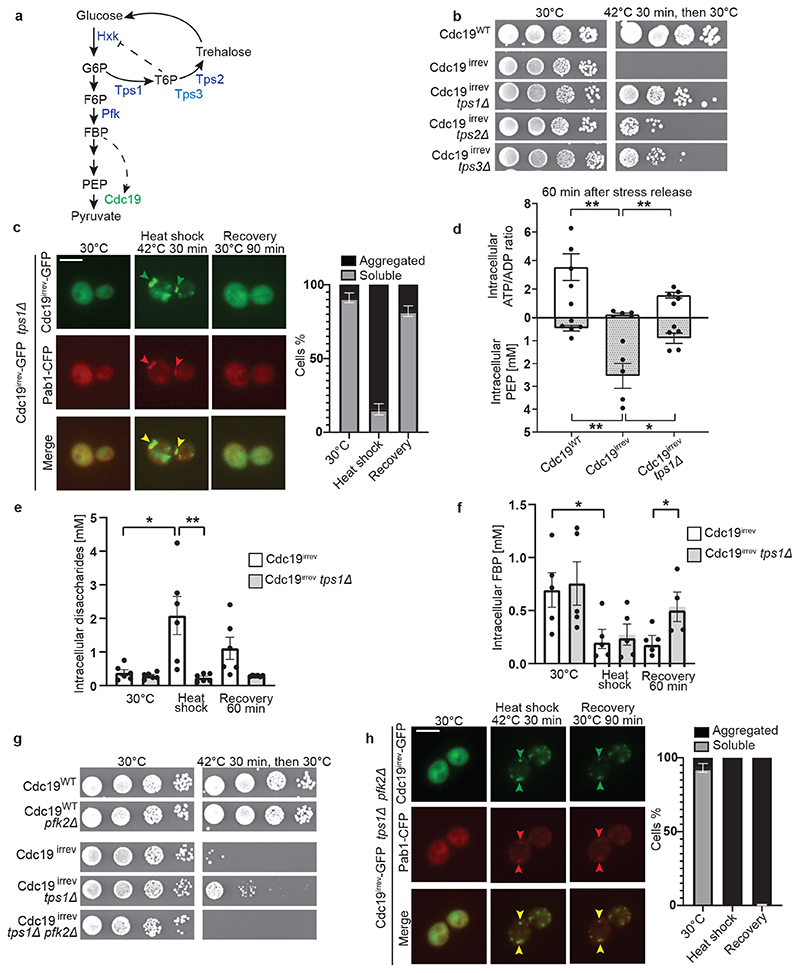
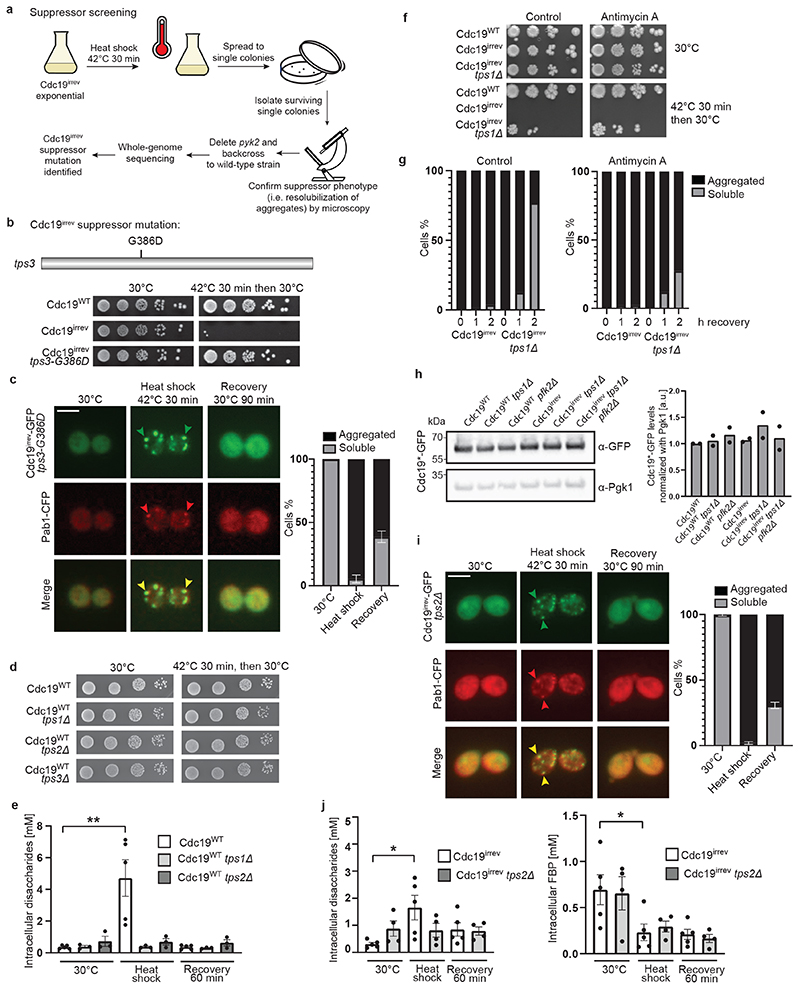
To better understand the molecular mechanisms regulating Cdc19 amyloid disassembly, we isolated rare spontaneous suppressors that restore growth of stressed cdc19irrev cells (Extended Data Fig. 2A). After deleting PYK2 in the isolated mutants to exclude suppression by upregulated Pyk2 activity, we backcrossed them to wild-type cells to confirm Mendelian segregation of the phenotype. Using whole genome sequencing, we identified a single point mutation in Tps3 (tps3-G386D), a protein involved in trehalose biosynthesis, as the likely cause for the Cdc19irrev suppressor phenotype (Extended Data Fig. 2B and C). Trehalose is a disaccharide synthesized in a two-step process (Fig. 3A). Tps1 catalyses the conversion of glucose-6-phosphate (G6P) into trehalose-6-phosphate (T6P), and Tps2 then converts T6P into trehalose. Tps3 acts as cofactor, facilitating trehalose production. Deleting either TPS1, TPS2 or TPS3 to compromise trehalose synthesis (Extended Data Fig. 2E, [37]) did not alter growth compared to wild-type (Extended Data Fig. 2D), but allowed cdc19irrev cells to partially restore growth after stress release (Fig. 3B), suggesting that decreased trehalose metabolism favours Cdc19 re-solubilization. As TPS1 catalyses the first reaction in trehalose synthesis and its deletion resulted in the strongest rescue (Fig. 3B), we focussed on this mutant. Fluorescence microscopy confirmed that growth restart correlated with Cdc19irrev amyloid re-solubilization and SG disassembly in tps1Δ cells (Fig. 3C). Metabolomic data showed that Cdc19 re-solubilization coincided with restored cellular energy levels after stress. Indeed, while cdc19irrev cells failed to increase their ATP/ADP ratio and maintained high intracellular PEP levels after stress, cdc19irrev tps1Δ cells consumed PEP and increased their ATP/ADP ratio and ATP levels (Fig. 3D). As TPS1 deletion could also restore growth and Cdc19irrev aggregate disassembly in the presence of the respiration inhibitor antimycin A, we conclude that restoration of energy metabolism depends mainly on re-activation of glycolysis, while oxidative phosphorylation plays a minor role (Extended Data Fig. 2F and G). Next, we tested how TPS1 deletion mediates this rescue. We reasoned that Tps1 activity decreases glycolytic flux by diverting G6P from glycolysis into trehalose synthesis generating T6P, which inhibits hexokinase (Fig. 3A, [27–29]). Thus, tps1Δ cells contain virtually no trehalose and have enhanced flux through glycolysis, which results in increased levels of the glycolytic intermediate fructose-1,6-bisphosphate (FBP) [29]. FBP levels tightly correlate with glycolytic flux and cell growth, with low FBP reflecting low glycolysis and stressed, quiescent cells, while high FBP levels denoting high glycolytic flux and favourable growth conditions [38, 39]. Interestingly, besides being a “glycolytic flux sensor”, FBP is also a known allosteric regulator of Cdc19 [22]. Thus, increased FBP levels in tps1Δ cells may directly or indirectly play a role in Cdc19 aggregate disassembly. Comparing intracellular trehalose and FBP levels in heat-shocked cdc19irrev and cdc19irrev tps1Δ cells revealed that lack of Tps1 abrogated trehalose production (Fig. 3E). Moreover, similar to glucose starvation [29], heat stress induced a rapid decrease in FBP, but during stress recovery FBP accumulated to significantly higher levels in cdc19irrev tps1Δ cells compared to cdc19irrev controls (Fig. 3F). To genetically test whether increased FBP levels mediate the observed rescue of stressed Cdc19irrev cells by tps1Δ, we impaired FBP production by deleting Pfk2, the main FBP-producing enzyme [29]. In contrast to cdc19irrev tps1Δ, cdc19irrev tps1Δ pfk2Δ mutants were unable to recover from heat stress, while pfk2Δ alone neither significantly reduced growth rates nor impaired survival to stress (Fig. 3G). Moreover, cdc19irrev tps1Δ pfk2Δ cells were also unable to dissolve Cdc19irrev aggregates and SGs (Fig. 3H). Importantly, Cdc19 levels were not altered in any of the mutants (Extended Data Fig. 2H). We conclude that high FBP levels contribute to efficient stress recovery and Cdc19 amyloid and SG disassembly.
Figure 3. Re-solubilization of Cdc19 amyloids and cell cycle restart after stress are regulated by the glycolytic metabolite FBP.
(A) Representation of glycolysis and trehalose metabolism. Abbreviations: glucose-6-phosphate (G6P), fructose-6-phosphate (F6P), fructose-1,6-bisphosphate (FBP), phosphoenolpyruvate (PEP), trehalose-6-phosphate (T6P), hexokinase (Hxk), trehalose synthase (Tps) and phosphofructokinase (Pfk).
(B) Serial dilutions of the indicated strains were spotted on agar plates before or after heat shock (42°C, 30 min) and grown at 30°C for 3 days (n = 3 independent experiments).
(C) cdc19irrev tps1Δ cells expressing Pab1-CFP were heat shocked (42°C, 30 min) and recovered at 30°C. Plots indicate mean percentage (%) of cells with Cdc19 aggregates ± S.E.M (n = 3 independent experiments, >30 cells per sample per experiment). Scale bar: 5 μm.
(D) Intracellular ATP/ADP ratio and phosphoenolpyruvate (PEP) were measured in the indicated strains after heat shock (42°C, 30 min) and 60 min recovery at 30°C. Mean ± S.E.M. is shown (n = 5 independent experiments, two-tailed Mann-Whitney test, cdc19WT–cdc19irrev comparison: PATP/ADP = 0.0079, cdc19irrev-cdc19irrevtps1Δ comparison: PATP/ADP = 0.0079, PPEP = 0.0556).
(E) – (F) Intracellular disaccharides (E) (representing trehalose [46]) and FBP (F) were measured in the indicated strains before, during and after heat shock (42°C, 30 min). Mean ± S.E.M. is shown (n = 6 independent experiments for disaccharides, n = 5 independent experiments for FBP, two-tailed Mann-Whitney test, cdc19irrev 30°C–HS comparison: PDisaccharides = 0.0152, PFBP = 0.0317, cdc19irrev-cdc19irrevtps1Δ HS comparison: P Disaccharides = 0.0022, cdc19irrev-cdc19irrevtps1Δ recovery 60 min comparison: P FBP = 0.0317).
(G) Serial dilutions of the indicated strains were spotted on agar plates before or after heat shock (42°C, 30 min) and imaged after 3 days at 30°C (n = 3 independent experiments).
(H) tps1Δ pfk2Δ cells expressing Pab1-CFP and GFP-tagged Cdc19irrev were heat shocked (42°C, 30 min) and recovered at 30°C. The intensity of the GFP and CFP images before heat shock was adjusted for better visualization. Plot indicates mean percentage (%) of cells with Cdc19 aggregates ± S.E.M (n = 3 independent experiments, >30 cells per sample/experiment). Scale bar: 5 μm. Source data for graphical representations are found in Source Data Fig. 3.
Altered trehalose levels might also influence Cdc19 resolubilization by other means, as trehalose affects SG dynamics by tuning cytoplasmic viscosity [40]. Indeed, deleting TPS2 also rescues the aggregate dissolution defects observed in cdc19irrev cells (Extended Data Fig. 2I), although this effect is weaker compared to tps1Δ cells (Fig. 3B). While both strains are impaired in trehalose synthesis, only deletion of TPS1, but not TPS2, resulted in increased post-stress FBP levels in cdc19irrev cells (Extended Data Fig. 2J). We conclude that FBP-dependent and -independent mechanisms co-operate to achieve efficient Cdc19 amyloid disassembly after stress, but that increasing cytoplasmic viscosity via trehalose breakdown plays a minor role compared to increased FBP levels.
FBP binding regulates Cdc19 amyloid assembly and disassembly
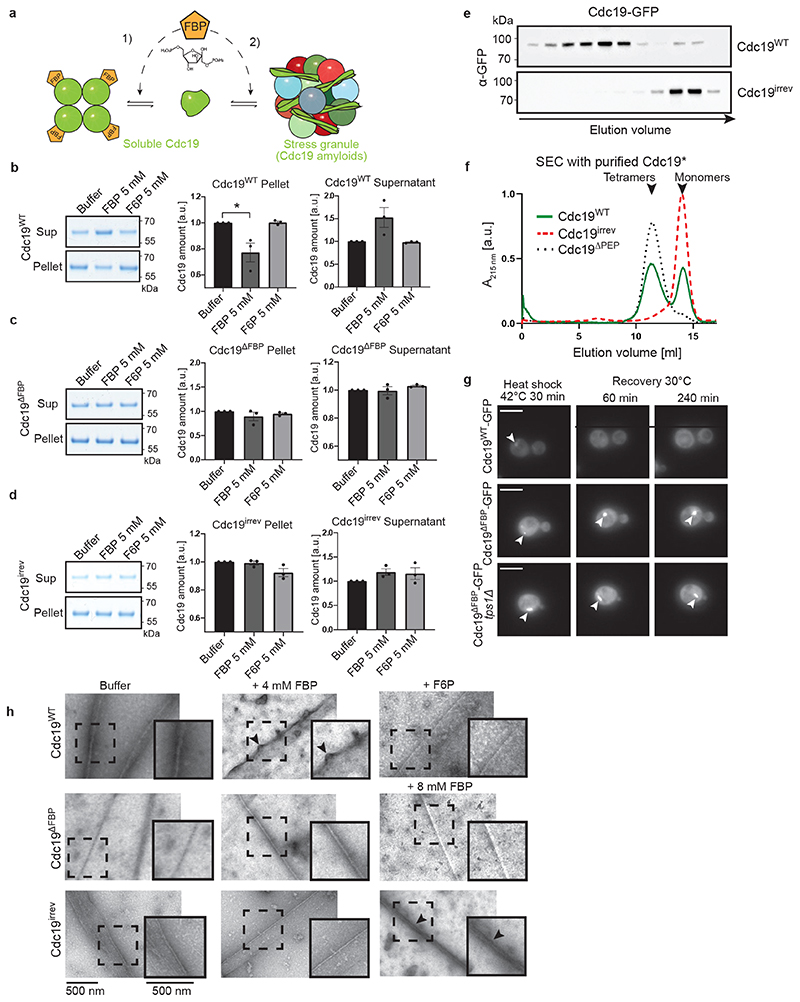
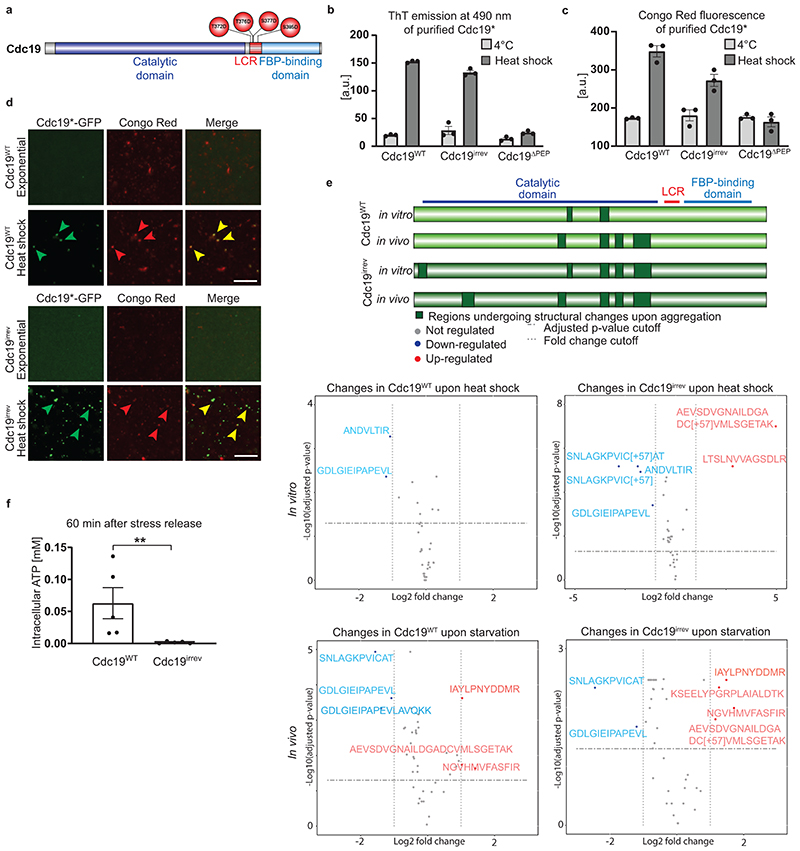
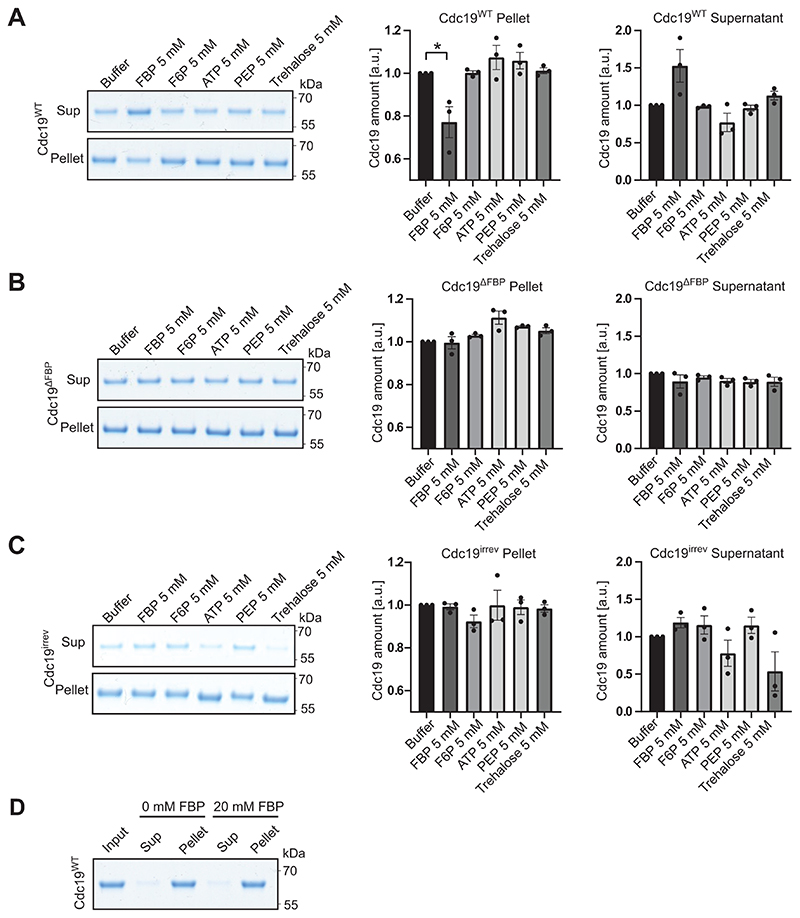
Mechanistically, direct binding of FBP to Cdc19 could reduce aggregation by promoting Cdc19 tetramerization (Fig. 4A, arrow 1), as Cdc19 tetramers are less aggregation-prone than monomers [4]. Alternatively, FBP could directly bind to Cdc19 amyloids and facilitate Cdc19 aggregate and SG disassembly (Fig. 4A, arrow 2). These two action modes are not mutually exclusive, and could co-operate to ensure rapid Cdc19 re-solubilization. A centrifugation assay revealed that incubation of purified Cdc19WT with 5 mM FBP, but not with its structurally-related precursor fructose-6-phosphate (F6P), interfered with aggregate formation (Fig. 4B). Indeed, in the presence of FBP, a significant portion of Cdc19WT remained soluble, with a corresponding decrease in the aggregated pellet fraction. Conversely, addition of 5 mM ATP, PEP or trehalose did not influence Cdc19 aggregation (Extended Data Fig. 3A–C). To confirm that FBP reduces Cdc19 aggregation by direct binding, we measured the aggregation behaviour of a Cdc19 point-mutant (T403E) unable to bind FBP (Cdc19ΔFBP) [41]. FBP addition did not interfere with heat shock-induced Cdc19ΔFBP aggregation (Fig. 4C). Thus, direct FBP binding to Cdc19 monomers partially prevents amyloid formation by promoting the formation of Cdc19 tetramers (Fig. 4A, arrow 1). However, FBP did not affect Cdc19irrev aggregate formation (Fig. 4D), suggesting that this mutant might have a reduced ability to bind FBP and/or to tetramerize. Indeed, size-exclusion chromatography demonstrated that Cdc19irrev is fully monomeric both in vivo and in vitro (Fig. 4E and F). Importantly, these results imply that the FBP-mediated rescue of Cdc19irrev in vivo cannot simply be explained by increased tetramerization (Fig. 4A, arrow 1), but FBP must also promote aggregate disassembly (Fig. 4A, arrow 2). To confirm that direct FBP binding to Cdc19 amyloids is required for their disassembly, we compared aggregation dynamics of Cdc19WT and Cdc19ΔFBP in vivo (Fig. 4G). Interestingly, Cdc19ΔFBP was soluble under favourable growth conditions, but rapidly formed irreversible aggregates upon heat shock that could not be disassembled after stress recovery. In contrast to Cdc19irrev, Cdc19ΔFBP aggregates could not be rescued by increasing FBP levels through TPS1 deletion, implying that direct FBP binding to Cdc19 is essential to disassemble Cdc19 amyloids in vivo.
Figure 4. FBP directly regulates formation and disassembly of reversible Cdc19 amyloids.
(A) Illustration of how FBP binding may regulate Cdc19 amyloid formation and disassembly. FBP (1) depletes aggregation-prone monomers by inducing Cdc19 tetramerization [4], and (2) directly promotes aggregate re-solubilization.
(B) – (D) Purified Cdc19WT (B), Cdc19ΔFBP (C) or Cdc19irrev (D) proteins were mixed with the indicated metabolites and incubated at 30 °C for 14 hours. After centrifugation, the supernatant (Sup) and pellet (Pellet) fractions were analysed by SDS-PAGE and quantified relative to buffer controls (shown as mean ± S.E.M, n = 3 independent experiments, two-tailed t-test, P = 0.0342).
(E) – (F) Cdc19irrev is monomeric in vivo and in vitro. (E) Extracts from cells expressing GFP-tagged Cdc19WT or Cdc19irrev were separated by size-exclusion chromatography (SEC) and selected peak fractions were analysed by Western blot with an α-GFP antibody. Elution volume (ml) and size markers (kDa) are indicated. (F) Recombinant Cdc19WT, Cdc19irrev and for control tetrameric Cdc19ΔPEP mutant proteins were purified and analysed by SEC. Normalized absorbance at 215 nm(arbitrary units; a.u.) was plotted as a function of elution volume (ml). n = 3 independent experiments.
(G) Direct FBP binding to Cdc19 is necessary to re-solubilize Cdc19 amyloids in vivo. Wild-type or tps1Δ cells expressing GFP-tagged Cdc19WT or Cdc19ΔFBP and over-expressing Pyk2 were heat shocked (42 °C, 30 min) and allowed to recover at 30 °C in the presence of 25 μg/ml cycloheximide. Representative GFP-images (three independent experiments) were taken 60 or 240 min after heat shock. Arrowheads indicate Cdc19-GFP aggregates. Scale bars: 5 μm.
(H) Heat shock-induced (42 °C, 10 min) Cdc19WT, Cdc19ΔFBP and Cdc19irrev amyloids were incubated with FBP (4 or 8 mM) or for control with F6P (4 mM) or buffer. Scale bar: 500 nm. Black arrows mark fibrils that became positively stained due to FBP binding. Representative fibrils (dashed squares) were 1.5 x enlarged to better visualize distinct fibril morphologies (inserts). n = 3 independent experiments. Unprocessed original scans of gels and blots and source data for all graphical representations are shown in Source Data Fig. 4.
Next, we examined whether FBP is sufficient to disassemble pre-formed Cdc19 amyloids in vitro. We induced fibril formation of purified Cdc19WT by heat shock, and incubated the obtained amyloids with varying FBP concentrations. SDS-PAGE analysis of the soluble and insoluble fractions after centrifugation revealed no significant Cdc19 re-solubilisation even upon incubation with FBP excess (20 mM), indicating that FBP binding is not sufficient to disassemble Cdc19 amyloids in vitro (Extended Data Fig. 3D). However, FBP bound directly to Cdc19 amyloids and altered their appearance in negative staining TEM. Indeed, incubation of pre-formed Cdc19WT aggregates with FBP (4 mM) resulted in fibrils that were consistently positively stained (i.e. dark colour) (Fig. 4H, +FBP), in contrast to negatively stained fibrils (i.e. white) in absence of FBP. This change from negative (white fibrils) to positive (black fibrils) staining results from acidic FBP molecules bound to fibrils attracting the positively charged uranyl stain. In contrast, incubating pre-formed Cdc19WT amyloids with buffer (Fig. 4H, buffer) or F6P (Fig. 4H, +F6P) did not cause any visible changes, i.e. Cdc19WT amyloids appeared as negatively stained, needle-like fibrils characterized by smooth edges. Cdc19ΔFBP amyloid fibrils incubated with FBP also retained their straight, smooth morphology with negative staining even at high concentations of FBP (8 mM), validating that the observed alterations are induced by direct FBP binding to Cdc19 (Fig. 4H, lower panels). Interestingly, Cdc19irrev showed intermediate signs of fibril curvature, uneven borders and positive staining compared to Cdc19WT and Cdc19ΔFBP, and these changes were apparent only at high FBP concentrations (8 mM), confirming reduced FBP binding by Cdc19irrev. We conclude that although FBP directly binds to Cdc19 fibrils, binding is not sufficient to trigger Cdc19 amyloid re-solubilization in vitro. However, FBP may facilitate morphological changes and/or factor recruitment that promotes efficient amyloid disassembly observed in vivo.
Chaperones contribute to Cdc19 amyloid resolubilization
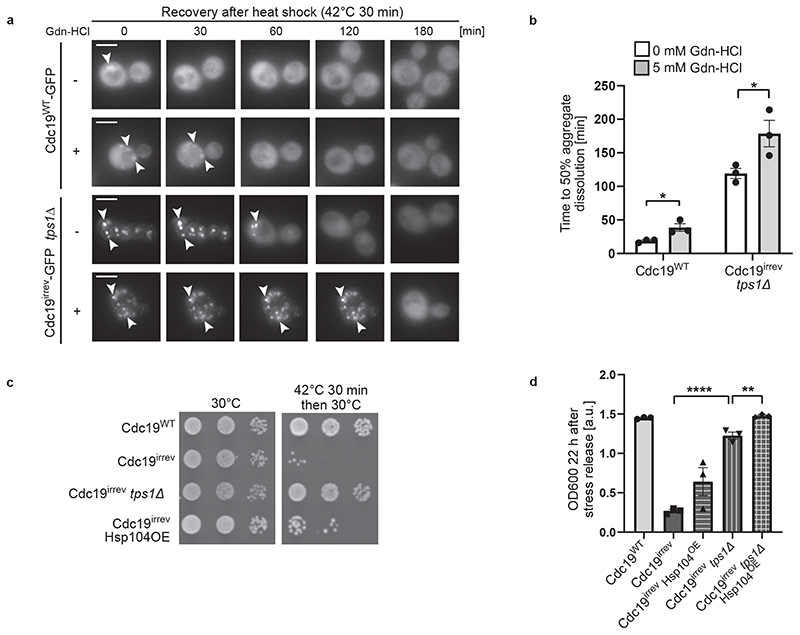
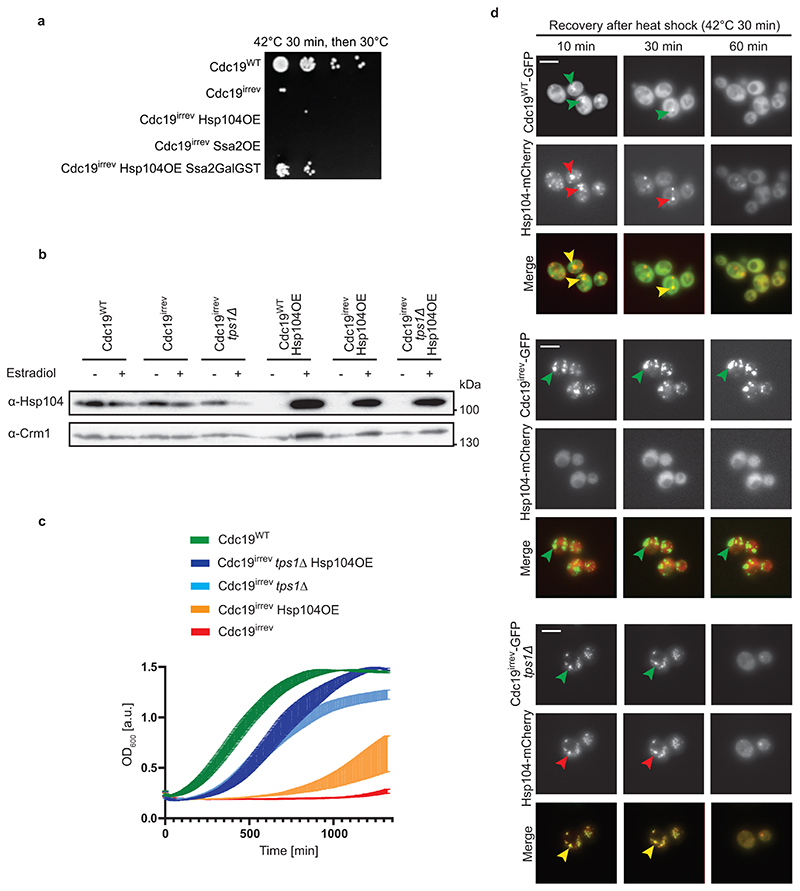
Chaperones such as Hsp104 and Ssa2 have been implicated in re-modelling protein assemblies and SGs, and cells lacking HSP104 or SSA1/2 are slightly delayed in SG disassembly [17, 42, 43]. Thus, we tested if these chaperones are also involved in Cdc19 disaggregation. We observed that Gdn-HCl-mediated inhibition of Hsp104 activity delayed dissolution of both Cdc19WT and Cdc19irrev aggregates in tps1Δ cells (Fig. 5A and B). Conversely, overexpressing Hsp104 was sufficient to partially restore growth of heat-stressed cdc19irrev cells (Fig. 5C), which was further enhanced by simultaneously increasing Ssa2 levels (Extended Data Fig. 4A and B). Moreover, Hsp104 overexpression slightly improved growth of cdc19irrev tps1Δ mutants after heat stress, indicating that increased FBP and chaperone levels have an additive effect (Fig. 5D). Increasing FBP levels by TPS1 deletion allowed rapid growth restart after stress, while Hsp104 overexpression seemed to facilitate growth in a later phase, suggesting that high FBP may be particularly important in the initial steps of Cdc19 amyloid disassembly (Extended Data Fig. 4C). Together, these results imply that FBP binding to Cdc19 initiates aggregate re-solubilization and co-operates with Hsp104 and Ssa2 to disassemble SG in vivo.
Figure 5. Chaperones are involved in Cdc19 amyloid disassembly.
(A) – (B) Inhibition of Hsp104 activity delays Cdc19 and SG re-solubilization. Cells of the indicated genotype were treated with 5 mM Gdn-HCl for 3 hours prior to a 30 min heat shock at 42 °C. (A) Aggregate disassembly after stress release was monitored by fluorescence microscopy. Representative images, arrowheads indicate Cdc19 aggregates (n = 3 independent experiments). Scale bar: 5 μm. (B) Cdc19 aggregate disassembly was quantified by counting the number of foci at different time points after stress. The average time ± S.E.M. to reach a 50% reduction in foci number is displayed (n = 3 independent experiments, >30 cells per sample per experiment, two-tailed t-test, PWT = 0.0268, Pirrev+tps1Δ = 0.048).
(C) – (D) Overexpression of Hsp104 partially rescues growth of cdc19irrev cells after heat shock. (C) Serial dilutions of exponentially growing cells of the indicated genotype were spotted on agar plates before or after a 30 min heat shock of 42 °C, and imaged after 3 days at 30 °C (n = 3 independent experiments). Where indicated, overexpression of Hsp104 was induced by treating cells with 10 mM estradiol for 3 hours. (D) Growth restart was also quantified by measuring cell density (OD600) over time after inoculation of equal cell numbers at 30 °C. Mean cell density 22 hours after stress release ± S.E.M. is shown (n = 3 independent experiments, two-tailed t-test, Pirrev-irrev+tps1Δ = 0.0000413, Pirrev+tps1Δ - irrev+tps1Δ+Hsp104OE = 0.0055). Source data for all graphical representations can be found in Source Data Fig. 5.
FBP binding to Cdc19 amyloids promotes chaperone recruitment
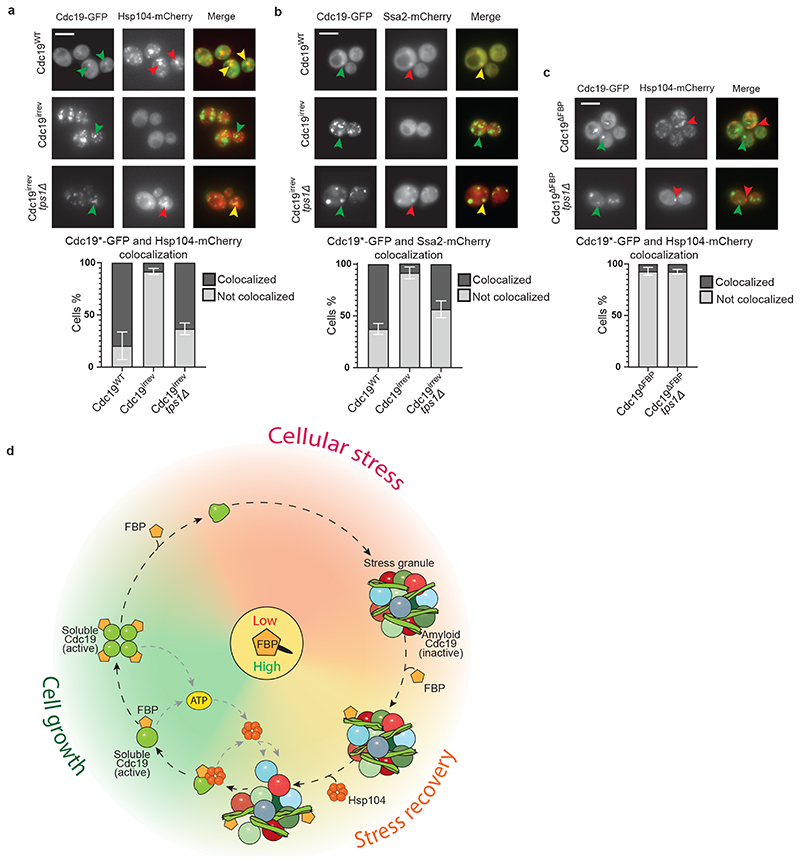
Next, we compared the localization of Cdc19-GFP and Hsp104-mCherry or Ssa2-mCherry in wild-type, cdc19irrev and cdc19irrev tps1Δ cells. Fluorescence microscopy confirmed that Hsp104-mCherry and Ssa2-mCherry co-localized with heat shock-induced Cdc19-GFP aggregates in the majority of wild-type cells (Fig. 6A and B). Surprisingly, however, Hsp104 and Ssa2 were not recruited to irreversible Cdc19irrev aggregates but remained dispersed throughout the cytoplasm. Co-localization was restored upon TPS1 deletion, consistent with the notion that increased FBP levels and FBP binding to Cdc19 allow chaperone recruitment to SGs (Fig. 6A and B) to promote Cdc19 and SG re-solubilization (Extended Data Fig. 4D). To corroborate these results, we examined co-localization of Hsp104-mCherry with GFP-tagged Cdc19ΔFBP. As expected, Cdc19ΔFBP amyloids failed to recruit Hsp104-mCherry, and co-localization could not be restored by TPS1 deletion, confirming that FBP binding to Cdc19 is essential for chaperone recruitment (Fig. 6C). In this background, Hsp104-mCherry showed a punctate pattern by binding to heat-induced aggregates, but these foci did not co-localize with irreversible Cdc19ΔFBP amyloids (Fig. 6C). We thus conclude that FBP binding to Cdc19 aggregates promotes chaperone binding and allows their recruitment to SGs, which is essential for Cdc19 amyloid re-solubilization, restoration of ATP production, and SG disassembly.
Figure 6. FBP binding to Cdc19 amyloids promotes chaperone recruitment triggering Cdc19 re-solubilization and SG disassembly in vivo .
(A) – (C) FBP binding to Cdc19 amyloids promotes chaperone recruitment, which correlates with aggregate disassembly. Wild-type or tps1Δ cells co-expressing mCherry-tagged Hsp104 (A, C) or Ssa2 (B) and either GFP-tagged Cdc19WT, Cdc19irrev or Cdc19ΔFBP were subjected to heat stress for 30 min at 42 °C and imaged by fluorescence microscopy. cdc19ΔFBP cells (C) overexpress Pyk2 to restore ATP production and cell viability. Representative GFP- and mCherry-images are shown, together with the merged image to visualize co-localization of Cdc19 aggregates and the chaperones (n = 3 independent experiments). Scale bar: 5 μm. Plots indicate mean percentage (%) of cells with co-localized Cdc19 and Hsp104 or Ssa2 foci ± S.E.M (n = 3 independent experiments, >300 cells per sample per experiment).
(D) Molecular network coupling cellular metabolism with reversible Cdc19 amyloids and stress granule dynamics. During cell growth, the metabolite FBP is produced through glycolysis and its binding to Cdc19 promotes the formation of active, non-aggregation-prone Cdc19 tetramers. Upon stress, FBP levels rapidly drop, leading to tetramer disassembly and the accumulation of Cdc19 monomers. In this state, Cdc19 exposes its aggregation-prone LCR and is primed to form Cdc19 amyloids, which are inactive and accumulate in SGs. After stress release, FBP levels rise and FBP directly binds to Cdc19 amyloids, thereby facilitating the recruitment of Hsp104 and Ssa2 to the aggregates. These chaperones allow re-solubilisation of Cdc19 amyloids and the release of Cdc19 monomers. Re-solubilised, active Cdc19 in turn uses PEP to produce ATP, which is required to activate chaperones to further solubilize Cdc19 and increase energy production. This positive feedback loop fuels disassembly of Cdc19 amyloids and other SG components. Concomitant breakdown of trehalose liberates glucose moieties for glycolysis and modulates cytoplasmic diffusion. Orchestrated disassembly of SGs then releases several mRNAs and metabolic enzymes, and allows full resumption of cell growth and cell cycle progression. Source data for all graphical representations can be found in Source Data Fig. 6.
Discussion
Cell survival under stress conditions requires rapid and reversible implementation of protective measures, including the timely formation and disassembly of reversible aggregates and SGs. Here, we show that upon stress, the essential ATP-producing enzyme Cdc19 forms proteolysis-resistant reversible amyloids which co-localize with SGs. Aggregation not only protects Cdc19 from stress-induced degradation, but also inactivates the enzyme, leading to a rapid decrease of cellular ATP. Yet, ATP is required for post-stress Cdc19 aggregate and SG disassembly as well as growth restart [14]. We show that Cdc19 amyloid re-solubilization after stress restores cellular energy levels, and that this is essential to disassemble SGs and restart growth. Moreover, our results unravel a multi-step mechanism for Cdc19 amyloid and SG re-solubilization. Indeed, the glycolytic metabolite FBP regulates the assembly state and activity of Cdc19 in two ways (Fig. 6D). First, during exponential growth, FBP binds to soluble Cdc19, promoting the formation of active, non-aggregating Cdc19 tetramers. Upon stress, decreasing FBP levels facilitate the formation of aggregation-prone Cdc19 monomers, which assemble into amyloid structures. Second, upon stress release, glycolysis restarts and increasing FBP levels trigger Cdc19 resolubilization. FBP directly binds to Cdc19 amyloids and promotes efficient recruitment of Hsp104 and Ssa2 chaperones. Indeed, a dedicated algorithm [44] predicts a specific binding site for Hsp70 chaperones co-localizing with the FBP-binding site of Cdc19. Cells expressing a Cdc19ΔFBP mutant, which cannot bind FBP, fail to recruit chaperones and cannot disassemble Cdc19 aggregates after stress release. Moreover, FBP increases Cdc19’s ATP production further facilitating chaperone recruitment and activity. Re-solubilized Cdc19 produces ATP which further fuels Cdc19 amyloid and SG disassembly in a positive feedback loop. While Hsp104 was identified as the critical ATP-consuming enzyme during this process [14], other cellular factors such as RNA/DNA-helicases, protein kinases and AAA-ATPases may also contribute to dissolve SGs and restart cell growth [9]. In summary, FBP functions as a “glycolytic flux sensor” [38, 39] and directly controls Cdc19 amyloid formation and disassembly. The need for ATP production by Cdc19 can be bypassed with non-glycolytic substrates, which are metabolized by oxidative phosphorylation in mitochondria.
Conceptually, our results reveal a mechanism whereby the metabolic and energetic state of the cell is coupled to SG disassembly via regulated formation of Cdc19 amyloids. We uncovered a crucial function of reversible Cdc19 amyloids in SG regulation and cell survival under stress, highlight Cdc19 amyloids as physiological targets of Hsp104 and Ssa2, and elucidate how functional amyloids can be disassembled in a physiological context. As several metabolic enzymes reversibly aggregate during stress [45], similar mechanisms may regulate their activity. Finally, although the interplay between physiological and pathological amyloids remains to be explored, understanding the regulation of physiological amyloids might provide important insights into the pathogenesis of neurodegenerative diseases.
Methods
Media, growth assays, yeast strains and plasmid construction
Yeast strains and plasmids used in this work were obtained using standard molecular biology protocols and are listed in Supplementary Tables S1 and S2, respectively. Unless otherwise stated, yeast cells were grown at 30 °C in synthetic defined (SD) media with 2% glucose (2% glucose, 0.5% NH4-sulfate, 0.17% yeast nitrogen base, and amino acids), as previously described [4]. Where indicated, 2% glucose in SD media was substituted with 2% ethanol. When grown exponentially, Cdc19irrev strains were never allowed to grow above an optical density 600 (OD600) of 0.8 to avoid stress conditions. Heat shock was induced by shifting exponentially growing cells (OD600 0.4-0.6) for 30 min to 42 °C. Where indicated, cycloheximide (Sigma-Aldrich, C-7698) or Antimycin A (Sigma-Aldrich A8674) was added to the medium at 25 μg/ml or 2 μM final concentration (1 μM in SD plates), respectively. Hsp104 activity was inhibited with 5 mM Gdn-HCl (Sigma-Aldrich 50950) for 3 h prior to stress initiation. Growth rates were determined at 30 °C by measuring OD600 in a 96-well plate (Greiner Bio-One) using a plate reader (BioLector m2p-labs or ClarioStar BMG Labtech), or by imaging serial dilution spottings onto appropriate SD plates after 3 days. Protein overexpression from estradiol-inducible promoters was achieved by exposing cells to 10 mM estradiol (Sigma-Aldrich E8875) for 3 hours.
Fluorescence microscopy, image analysis and quantification
Fluorescence microscopy was performed using a Nikon Eclipse Ti-E microscope with MicroManager V 1.4 and NIS-Elements Advanced Research V 5.02 software. For time-lapse experiments, cells were loaded in microfluidic plates (CellASIC ONIX2, Merck Millipore) at 30 °C [4]. Heat shock was induced with the microfluidic plate temperature controller by increasing the temperature to 42 °C for 30 min, before returning it to 30 °C, or by shifting cultures for 30 min in a shaking incubator (Kühner, LT W Lab-Therm) pre-heated at 42 °C. Images were recorded every 10 min, and quantified as described in the figure legends.
Screen for suppressors of irreversible Cdc19 aggregates
Cells expressing Cdc19irrev-GFP were grown in SD-His media to OD600 of 0.6, heat shocked for 30 min at 42 °C in a shaking incubator (Kühner, LT W Lab-Therm), collected by centrifugation and spread on SD-His plates to single colonies. Surviving single colonies were isolated and visually inspected by fluorescence microscopy. PYK2 was deleted by homologous recombination to exclude suppressors that upregulate Pyk2. Suppressors were backcrossed to ensure Mendelian segregation, and suppressor strains with single gene traits were subjected to whole-genome sequencing. Identified mutations were validated by crossing cdc19irrev strains with full-deletions of the mutated gene.
Protein purification
Plasmids expressing wild-type or mutant Cdc19-strep constructs were transformed into E. coli cells (Rosetta). Cells were grown at 37 °C in LB media (1% peptone, 0.5% yeast extract, 0.5% NaCl) with 30 μg/ml chloramphenicol and 100 μg/ml carbenicillin to OD600 0.6. Protein production was induced by adding 0.1 mM IPTG and growing cells at 16 °C overnight. Cells were harvested by centrifugation, resuspended in pre-cooled lysis buffer (100 mM Tris/HCl pH 7.4, 200 mM NaCl, 1 mM MgCl2, 10% glycerol, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride (PMSF), protease inhibitor mix (Roche) and 75 U/ml of Pierce universal nuclease), and processed for lysis by freezer milling (SPEX SamplePrep 6870 Freezer/Mill; five cycles of 2 min cooling and 2 min grinding at setting 15 CPS). Extracts were cleared by centrifugation (4 °C, 30 min, 48000 g), and the supernatant was loaded on a Strep-Tactin Superflow Plus column (Qiagen) at 4 °C following the manufacturer’s instructions. Proteins were eluted using 2.5 mM D-desthiobiotin in purification buffer (100 mM Tris/HCl pH 7.4, 200 mM NaCl, 1 mM MgCl2, 10% glycerol, 1 mM DTT, 1 mM PMSF). Fractions were collected, checked by SDS–PAGE and Coomassie blue staining, and pure fractions were pooled and aliquoted for storage at −80 °C.
Transmission electron microscopy (TEM)
After thawing on ice, Cdc19 samples were cleared by centrifugation (21000 g, 4 °C, 10 min), diluted to 0.2 mg/ml in purification buffer pH 7.4, and the pH was adjusted to 6-7. Subsequently, a non-glow discharged carbon film 300 mesh copper grid (CF300-CU, Electron Microscopy Sciences) was submerged in this Cdc19 dilution and incubated for 10 min at 42 °C. Excess sample was then manually removed with Whatman filter paper. Where indicated, grids with pre-formed Cdc19 aggregates were incubated for 20 min with FBP or F6P dissolved in purification buffer to the indicated concentration, or simply washed twice with purification buffer, and stained with two drops of 2% uranyl acetate. TEM micrographs were acquired with a FEI Morgagni 268 microscope at 100 kV using a CCD 1376 × 1032 pixel camera at different magnifications.
In vitro protein-metabolite precipitation
Purified wild-type or mutant Cdc19 was thawed on ice, cleared by centrifugation (4 °C, 10 min, 21000 g), and diluted to a final protein concentration of 0.5 mg/ml in 100 mM Tris/HCl pH 7.4, 200 mM NaCl, 1 mM MgCl2, 10% glycerol, 1 mM DTT, 1 mM PMSF. Different metabolites (FBP, F6P ATP, PEP, trehalose) were solubilized in the same buffer, added to Cdc19 to the indicated final concentrations, and the protein-metabolite samples were incubated for 14 h at 30 °C. Aggregates were pelleted by centrifugation (4 °C, 10 min, 21000 g), and separated from the supernatant containing soluble Cdc19. Aggregation was quantified by loading pellet and supernatant on a SDS-PAGE gel, followed by Coomassie blue staining and quantification with FIJI (ImageJ V 2.0.0) software.
In vitro aggregate re-solubilization with FBP
Purified wild-type Cdc19 was heat shocked for 10 min at 42 °C and the resulting aggregates were incubated with buffer or 20 mM FBP for 5 h at 4 °C. Aggregates were pelleted by centrifugation (4 °C, 10 min, 21000 g) and Cdc19 re-solubilization in the presence or absence of FBP was quantified by loading supernatant and pellet fractions on a SDS-PAGE gel, followed by Coomassie blue staining.
Metabolite extraction and LC-MS/MS measurements
For metabolite extraction, 1 ml cultures were vacuum-filtered on a 0.45 μm pore size PVDF filter (Millipore) and cell pellets were immediately transferred into 4 ml of −20 °C cold extraction solution [40:40:20 acetonitrile/methanol/water, HPLC-grade solvents] supplemented with 200 ul of 13C internal standard and incubated at −20 °C for 2 h. The extraction solution was centrifuged (15 min, 0 °C, 4000 rpm) and the supernatant containing the metabolites was transferred to fresh tubes. Samples were dried in a speed-vac overnight and stored at −80 °C. Prior to targeted metabolomics measurements, samples were resuspended in 100 μl ddH2O.
Compounds were analyzed using an LC-MS/MS mass spectrometer system consisting of a 1290 Infinity LC (Agilent Technologies) coupled to a 5500 QTRAP triple quadrupole mass spectrometer (AB Sciex) in negative multiple reaction monitoring (MRM) scan mode. Five μl of metabolite extracts were injected on an Agilent PoroShell 120 HILIC-Z column (150 x 2.1 mm, 2.7 μm; Agilent, Santa Clara, CA) using a mobile phase A (water, 10 mM ammonium acetate, 5 μM medronic acid, pH 9) and mobile phase B (90% acetonitrile, 10% water, 10 mM ammonium acetate, 5 μM medronic acid, pH 9) at a constant flow rate of 250 μl/min. Gradient was as follows: 10% A, 2 min: 10% A, 12 min: 40% A, 15 min: 40% A, 16 min: 10% A, 24 min: 10% A. The MRM settings were adapted from Yuan et al. [47]. The raw data were processed and analyzed using a proprietary software in Matlab (Mathworks), the exact choice of software is not essential for reproducing the results.
Intracellular metabolite concentration was calculated based on the measured 12C/13C ratio, assuming a dry weight [48] of 0.53 g*l-1*OD-1 and a cell volume [49] of 0.002 l*g-1. Calibration curves were obtained from dilutions of pure compounds containing same amounts of 13C internal standard.
Protein extraction and Western blotting
Proteins were precipitated using a TCA/acetone-based protocol. Briefly, exponentially growing cells were incubated 10 min on ice after addition of 10% TCA, centrifuged (5 min, 4 °C, 21000 g), and pellets were washed twice with acetone. Precipitated proteins were resuspended using a SDS-based loading buffer and protein levels were quantified by Western blotting with specific antibodies. The following antibodies were used: α-GFP (Roche, 11 814 460 001, 1:3000 dilution), rabbit polyclonal α-Crm1 antibody (kindly provided by Karsten Weis, 1:3000 dilution), α-Pgk1 (Invitrogen, 459250, 1:3000 dilution), HRP-coupled secondary antibody (Biorad, 170-6516, 1:3000 dilution) and α-Hsp104 (Abcam, ab69549, 1:1000 dilution).
Pyruvate kinase activity assay
Pyruvate kinase activity was measured with an enzymatic assay where pyruvate production by Cdc19 is coupled to lactate production by lactate dehydrogenase and the simultaneous conversion of NADH to NAD+ is measured spectrophotometrically at 340 nm. Purified Cdc19 was diluted to 0.2 mg/ml and either kept on ice or heat shocked for 60 min at 42 °C. Soluble or aggregated Cdc19 was diluted in activity buffer (50 mM imidazole pH 7, 100 mM KCl, 25 mM MgCl2, 10 mM ADP, 0.3 mM NADH, 10 U/ml LDH) to a final protein concentration of 2 μg/ml. Reactions were started by adding PEP (final concentration 2 mM), and decrease in absorbance at 340 nm was monitored over time.
Size-exclusion chromatography (SEC)
Purified wild-type or mutant Cdc19 was thawed on ice and cleared by centrifugation (10 min, 21000 g, 4 °C). Protein (0.1 mg) was loaded on a Superdex 200 10/300 GL size-exclusion column (GE Healthcare) connected to an ÄKTA pure (GE Healthcare) at 4 °C. Protein elution was followed by measuring UV absorbance (280 and 215 nm), and collected fractions were analyzed by SDS-PAGE gel and Coomassie blue staining.
Lysates for size-exclusion chromatography were prepared by growing cells expressing wild-type or mutant Cdc19-GFP in SD media to an OD600 of 0.6. Cells were harvested by centrifugation, pellets resuspended in lysis buffer (PBS buffer (pH 7.4), 10% glycerol, 0.5% Tween 20, 20 mM β-glycero-phosphate, 1 mM vanadate, 1 mM NaF, 1 mM PMSF, protease inhibitor mix (Roche)), mixed with glass beads (Sigma-Aldrich, 425-600 μm) and lysed with a FastPrep homogenizer (FastPrep-24 5G, MP Biochemicals). Resulting extracts were cleared by centrifugation (10 min, 21000 g, 4 °C) and filtering (0.2 μm filter, Millipore). Protein concentration of the supernatant was measured by Nanodrop and a total of 4 mg protein was loaded on a Superdex 200 10/300 GL size-exclusion column (GE Healthcare) connected to an ÄKTA pure (GE Healthcare). The column was previously equilibrated in lysis buffer and run according to manufacturer’s instructions. Fractions were collected and analyzed by Western blotting with α-GFP antibodies.
Circular dichroism (CD), Thioflavin T (ThT) and Congo Red (CR) staining in vitro
CD spectra of purified wild-type or mutant Cdc19 (0.2 mg/ml) were measured with a J-710 spectropolarimeter (Jasco) using a quartz cuvette with 0.1 cm path length.
For in vitro measurements with the amyloid dyes, Thioflavin T (ThT, Sigma-Aldrich, T3516) or Congo Red (CR, Sigma-Aldrich, 75768-25MG) were dissolved in purification buffer (100 mM Tris/HCl pH 7.4, 200 mM NaCl, 1 mM MgCl2, 10% glycerol, 1 mM DTT, 1 mM PMSF) to a final concentration of 2.5 mM or 1 mM, respectively, and filtered (0.2 μm, Millipore). Purified wild-type or mutant Cdc19 were thawed on ice, cleared by centrifugation (10 min, 4 °C, 21000 g), diluted to 0.3 mg/ml in purification buffer, and then either heat shocked (10 min, 42 °C) or kept on ice. ThT or CR solution was added (1:10 dilution), and fluorescence intensity was measured in a 384-well plate (Corning Life Sciences) at the indicated temperature using a CLARIOstar plate reader (BMG Labtech). For ThT, excitation was set at 450 nm, and emission spectra were recorded at 490 nm. For CR, fluorescence intensity was measured at 614 nm with excitation at 560 nm.
Congo Red (CR) staining ex vivo
For ex vivo CR staining, cells expressing wild-type or mutant Cdc19-GFP were collected by centrifugation, resuspended in lysis buffer (PBS buffer pH 7.4, 10% glycerol, 0.5% Tween 20, 1 mM PMSF), and lysed at 4 °C with glass beads (Sigma-Aldrich, 425–600 μm) and a FastPrep homogenizer (FastPrep-24 5G, MP Biochemicals). Lysates were cleared by centrifugation (2 min, 2000 g, 4 °C), and total protein concentration was determined by Nanodrop. Glass microscope slides were cleaned with consecutive washes of H2O, 100% ethanol and acetone, before 3 mm-wide stripes of double-sided tape were applied along the length of the glass slides in order to form three channels (approximatively 3 mm-wide). Assembled flow chambers were functionalized by incubation for 20 min with 0.3 mg/ml Avidin DN (Vector Laboratories) in T50 buffer (10 mM Tris pH 8.0 and 50 mM NaCl), blocked for 30 min with 0.1% puronic F-127 (Sigma-Aldrich) in T50 buffer, washed twice with 0.1 mg/ml BSA in T50 buffer, and incubated for 30 min with biotinylated G-protein in lysis buffer. Finally, 10 nM α-GFP antibody in lysis buffer was flushed into the channels, protein extracts were loaded and allowed to bind for 10 min, before washing three times with lysis buffer. Aggregates were stained with 250 μM CR in lysis buffer, excess dye was removed by washing twice with lysis buffer, and analyzed by fluorescence microscopy.
LiP-MS experiments, SRM assays and quantification analysis
For in vivo LiP-MS experiments, cells expressing Cdc19WT-GFP or Cdc19irrev-GFP were grown in SD media and three biological replicates were collected when cells were growing exponentially (OD600 0.6) or following entry into stationary phase (two days). Cells were resuspended in 100 mM HEPES, 150 mM KCl, 1 mM MgCl2, pH 7.6 and disrupted by bead beating in the presence of acid-washed glass beads in three consecutive rounds of 30 s of beating and 4 min of incubation at 4 °C. Protein concentration in the yeast extracts was determined by the bicinchoninic acid assay (BCA Protein Assay Kit, Thermo Scientific). Equal amounts of cell lysates were treated for 3 min with proteinase K (Sigma Aldrich) at a concentration lysate:proteases of 100:1. Protease activity was quenched by heating at 95 °C and adding 5% sodium deoxycholate (Sigma Aldrich) [50]. Equal amounts of cell lysates were treated in parallel with vehicle instead of proteinase K to estimate Cdc19 protein amount variations in the different conditions assayed. Tris(2-carboxyethyl)phosphine-HCl (TCEP-HCl, Thermo Fisher Scientific) was added to a final concentration of 5 mM (35 min, 37 °C) to reduce disulfide bridges, followed by alkylation of free cysteine residues with iodoacetamide (Sigma aldrich) at 20 mM final concentration (30 min, room temperature). Peptide and protein fragments were digested to completion in denaturing conditions with LysC and trypsin proteases. Before adding LysC and trypsin, reaction mixtures were diluted with 0.1 M ammonium bicarbonate (Sigma Aldrich) to reach a sodium deoxycholate concentration lower than 1%. First, LysC digestion was performed at an enzyme to substrate (E/S) ratio of 1:100 for 4 h at 37 °C, then trypsin digestion was carried out at an E/S ratio of 1:100, for 16 h at 37 °C. For LiP experiments with recombinantly purified proteins, aliquots of recombinantly purified Cdc19WT and Cdc19irrev were incubated for 10 min at 25 °C or 42 °C in triplicates to induce Cdc19 structural variations just before the limited proteolysis step.
The development and validation of Selected Reaction Monitoring (SRM) assays to measure Cdc19 levels was guided by a spectral library generated in-house through a LC-MS/MS analysis of the in vivo yeast samples on a QE-plus (Thermo Fisher scientific) mass spectrometer equipped with a nano-electrospray ion source. On-line chromatographic separation of the peptides was achieved with an Easy-nLC 1000 nano liquid chromatography system (Thermo Fisher scientific) equipped with a 40 cm fused-silica column (New Objective), packed in-house with Reprosil Pur C18 Aq, 1.9 μm beads (Dr Maisch). Peptide mixtures (~1 μg) were separated with a linear gradient from 5% to 35% acetonitrile in 120 min. The collected spectra were searched against the S. cerevisiae UniProt reference database with Sorcerer-Sequest (Thermo Electron). Trypsin was set as the digesting protease tolarating two missed cleavages and semi tryptic termini. Cysteine carbamidomethylation (+57.0214 Da) was defined as a fixed modification, and methionine oxidation (+15.99492) as a variable modification. Protein identifications were filtered with a false discovery rate of <1%, calculated on the basis of a target-decoy approach. Proteome discoverer 2.2 (Thermo Fisher scientific) was used to build a consensus spectral library using LiP-MS in vivo samples.
Peptides matching to the spectral library were ranked by intensity using Skyline (v4.1, MacCoss Lab Software, University of Washington), and the 10 most intense transitions consisting of doubly or triply charged precursor ions and singly or doubly charged fragment ions of the y- and b-ion series were experimentally tested in SRM mode to select the most suitable transitions for quantification. Samples were measured on a triple quadrupole/ion-trap mass spectrometer (5500 QTrap, ABSciex) equipped with a nano-electrospray ion source and operated in SRM mode. On-line chromatographic peptide separation was achieved with an Eksigent 1D-plus Nano liquid chromatography system equipped with a 15 cm fused-silica column with 75 μm inner diameter, packed in-house with Reprosil Pur C18 Aq, 5 μm beads (Dr Maisch). The peptide mixtures (~1 μg) were separated with a linear gradient from 5% to 35% acetonitrile in 30 min. SRM analysis was conducted with Q1 and Q3 operated at unit resolution (0.7 m/z half-maximum peak width) with a dwell time of 20 ms and a cycle time <3.0 s.
SRM-MS data was analyzed with Skyline, and for the final SRM assay retention times were scheduled and at least three transitions per precursor ion of Cdc19 were retained. The identity of each SRM peak was confirmed against the spectral library by matching realigned retention times and relative fragment-ion intensities. 199 transitions matching to 44 Cdc19 precursors were selected for Cdc19 peptide quantification purposes. Integration of peak areas was manually inspected and relative quantification was performed for the Cdc19 peptides in the LiP-MS in vitro and in vivo experiments. Statistical significance analysis was performed with MSstats (www.msstats.org). Precursor intensities were normalized using the option “equalize medians” of MSstats. In the LiP-MS in vivo experiments peptide intensities were corrected for variations of Cdc19 protein levels between the exponential and stationary conditions. The normalization factor applied was equivalent to the relative variation of the Cdc19WT or Cdc19irrev proteins between the exponential and stationary conditions.
Statistics and Reproducibility
All data are representative results from at least three independent experiments, unless differently specified in the figure legends. Whenever possible, mean ± S.E.M. and individual data points of individual experiments are shown. GraphPad Prism was used to analyse and plot data. No statistical method was used to predetermine sample size, and the experiments were not randomized. No outlier tests were performed, and no data was excluded from the analysis. The Investigators were not blinded to allocation during experiments and outcome assessment.
Extended Data
Extended Data Fig. 1. Cdc19 aggregates are amyloids both in vitro and in vivo .
(A) Schematic representation of the domains of Cdc19. The four mutated residues located within the LCR in the Cdc19irrev mutant are indicated (red circles).
(B)-(C) Both Cdc19WT and Cdc19irrev form ThT- and Congo Red (CR)-positive aggregates upon heat shock in vitro. Purified Cdc19WT, Cdc19irrev and a non-aggregating Cdc19 mutant as negative control (Cdc19ΔPEP [4]) were incubated with ThT (B) or CR (C) at 4 °C or after heat shock (42 °C, 10 min). Fluorescence emission was measured at 490 nm or 614 nm, respectively. Graphs show mean ± S.E.M. of three independent experiments (two-tailed t-test, ThT: PWT = 0.0000149, Pirrev = 0.0091, CR: PWT = 0.0003, Pirrev = 0.0128).
(D) In vivo-formed Cdc19WT and Cdc19irrev aggregates are CR-positive. Cells expressing GFP-tagged Cdc19WT or Cdc19irrev were harvested when exponentially growing or after heat shock (42 °C, 30 min) and lysed. Cdc19-GFP was immobilized in a GFP-trap microfluidic chamber and stained with CR. GFP and CR signals were detected by fluorescence microscopy, and merged to visualize co-localization. Arrowheads indicate CR-positive Cdc19-GFP aggregates. Images are representative of three independent experiments. Scale bar: 10 μm.
(E) Limited-Proteolysis Mass Spectrometry (LiP-MS) results indicate that Cdc19WT and Cdc19irrev undergo comparable structural transitions upon aggregation in vitro and in vivo. Purified soluble or aggregated (42 °C, 10 min) Cdc19WT or Cdc19irrev, as well as cell extracts obtained from cells expressing Cdc19WT-GFP or Cdc19irrev-GFP harvested during exponential growth or stationary phase (2 days) to induce aggregation were analysed by LiP-MS as described in the Methods (n = 3 independent experiments). Peptides detected in soluble and aggregated Cdc19WT and Cdc19irrev are displayed in volcano plots, and upregulated (red) or downregulated (blue) conformation-sensitive peptides are highlighted. Conformation-specific LiP-MS-peptides detected in vitro and in vivo were mapped to the Cdc19 schematic drawing (green).
(F) Intracellular ATP levels (mM) were determined in the indicated strains after heat shock (42 °C, 30 min) and recovery (30 °C, 60 min). Mean ± S.E.M. of n = 5 independent experiments is shown (two-tailed Mann-Whitney test, P = 0.0079). Source data for all graphical representations are found in Source Data Extended Data Fig. 1.
Extended Data Fig. 2. Genetic screening identifies trehalose metabolism as a regulator of reversible Cdc19 aggregation.
(A) - (D) Screening protocol that identified a single point mutation (G386D) in TPS3 as a suppressor of stress-induced growth arrest of cdc19irrev cells (A). Serial dilutions of the indicated strains were spotted on agar plates before or after heat shock (42 °C, 30 min), and imaged after 3 days at 30 °C (3 independent experiments) (B and D). tps3-G385D cells expressing Cdc19irrev-GFP and Pab1-CFP were heat shocked (42 °C, 30 min) and allowed to recover at 30 °C (C). Plot indicates mean percentage (%) of cells with Cdc19 aggregates ± S.E.M (n = 3 independent experiments, >30 cells per sample/experiment). Scale bar: 5 μm.
(E) Intracellular disaccharides were measured in the indicated strains before, during and after heat shock (42 °C, 30 min), and shown as mean ± S.E.M. (n = 5 independent experiments for wild-type, n = 3 independent experiments for tps1Δ and tps2Δ, two-tailed Mann-Whitney test, P = 0.0079).
(F) and (G) The indicated strains were heat shocked (42 °C, 30 min) and allowed to recover at 30 °C ± antimycin A (1 or 2 μM, respectively). (F) Serial dilutions were spotted on agar plates ± antimycin A and imaged after 3 days at 30 °C (3 independent experiments). (G) Plots indicate mean percentage (%) of cells that re-solubilized Cdc19 from two independent experiments.
(H) Mean Cdc19-GFP levels relative to Pgk1 in the indicated strains are shown from two independent experiments.
(I) tps2Δ cells expressing Cdc19irrev-GFP and Pab1-CFP were heat shocked (42 °C, 30 min) and allowed to recover at 30 °C. Plot indicates mean percentage (%) of cells with Cdc19 aggregates ± S.E.M (n = 3 independent experiments, >30 cells per sample/experiment). Scale bar: 5 μm.
(J) Intracellular disaccharides (mainly trehalose [46]) and FBP were measured in the indicated strains before, during and after heat shock (42 °C, 30 min), and plotted as mean ± S.E.M. (n = 4 independent experiments for cdc19irrev tps2Δ, n = 5 for cdc19irrev, two-tailed Mann-Whitney test, PDisaccharides = 0.0159, PFBP = 0.0317). Source data for all graphical representations and unprocessed Western blots available in Source Data Extended Data Fig. 2.
Extended Data Fig. 3. FBP specifically reduces aggregation of purified Cdc19WT, but not Cdc19ΔFBP and Cdc19irrev mutant proteins.
(A)– (C) Purified wild-type Cdc19 (A), FBP binding-deficient Cdc19ΔFBP mutant (B) or Cdc19irrev mutant (C) proteins were mixed as indicated with 5 mM FBP or for control 5 mM F6P, 5 mM ATP, 5 mM PEP, 5 mM trehalose or buffer and incubated at 30 °C for 14 hours. Cdc19 aggregates were separated from soluble protein by centrifugation and the supernatant (Sup) and pellet (Pellet) fractions were analysed by SDS-PAGE and Coomassie blue staining. Cdc19 amount was quantified in the Pellet and Supernatant by measuring Cdc19 band intensities using ImageJ and normalizing to buffer controls, and is displayed as mean and S.E.M. of three independent experiments (two-tailed t-test, P = 0.0342).
(D) Addition of FBP alone is not sufficient to re-solubilize pre-formed Cdc19WT amyloids in vitro. Purified Cdc19WT (Input) was incubated for 10 min at 42 °C to trigger its aggregation, and the resulting amyloids were incubated with (20 mM) or without (0 mM) FBP for several hours. Cdc19 re-solubilization was then tested by centrifugation and analysis of the resulting supernatant (Sup) and pellet (Pellet) fractions by SDS-PAGE and Coomassie blue staining. The image is representative of three independent experiments. Unprocessed original scans of gels are shown in Source Data Extended Data Fig. 3.
Extended Data Fig. 4. The chaperones Hsp104 and Ssa2 co-operate with FBP to efficiently disassemble Cdc19 amyloids in vivo .
(A) Co-overexpression of Hsp104 and Ssa2 partially restores growth of cdc19irrev cells after heat stress. cdc19WT and cdc19irrev cells overexpressing as indicated Hsp104 or Ssa2, or both together by addition of 10 mM estradiol for 3 hours were treated for 30 min at 42 °C. Serial dilutions were spotted on agar plates and grown at 30 °C for 3 days. The image is representative of three independent experiments.
(B) Hsp104 protein levels were quantified in the indicated strains by Western blot either in the absence (-) or presence (+) of 10 mM estradiol for 3 hours. The image is representative of three independent experiments. Unprocessed original scans of blots are shown in Source data Extended data fig. 4.
(C) Increased FBP levels and Hsp104 co-operate to efficiently restart growth after stress in cdc19irrev cells. Exponentially growing wild-type or tps1Δ cells expressing either Cdc19WT or Cdc19irrev were subjected to a 30 min heat shock at 42 °C. Where indicated, overexpression of Hsp104 was induced by treating cells with 10 mM estradiol for 3 hours. Growth restart after stress release of the indicated strains was quantified by measuring the cell density (OD600) over time after inoculation of equal cell numbers at 30 °C. Mean cell density 22 hours after stress release is shown, with error bars representing S.E.M. of three independent experiments. Note that Hsp104 overexpression and increased FBP levels co-operate to rescue cdc19irrev cells after heat shock.
(D) Wild-type or tps1Δ cells co-expressing mCherry-tagged Hsp104 and either GFP-tagged Cdc19WT or Cdc19irrev mutant were heat stressed for 30 min at 42 °C and imaged by fluorescence microscopy at the times indicated (minutes). Representative GFP- (left row) and mCherry-images (middle row) are shown, together with the merged image (bottom row) to visualize co-localization of Cdc19 aggregates and Hsp104. Images are representative of three independent experiments. Scale bar: 5 μm. Source data for the graphical representation and unprocessed Western blots can be found in Source Data Extended Data Fig. 4.
Supplementary Material
Acknowledgements
We thank C. Boone (University of Toronto) for providing strains for chaperone overexpression; K. Weis (ETH Zürich) for antibodies and yeast strains; C. Kraft (University of Freiburg) for help with yeast cell semi-permeabilization; S.-S. Lee, L. Garbani Marcantini, J. Schleicher, D. M. Szymala and A. Timofiiva for help with microscopy and data analysis; ScopeM and FGCZ for their technical support; A. Smith for critical editing, and P. Arosio, D. Jarosz, A. Sengör and members of the Peter lab for helpful discussions and comments on the manuscript. This work was funded by the Synapsis Foundation, ETH Zürich, and the Swiss National Science Foundation (SNF 200426). In addition, PP received funding from the European Research Council (ERC-CoG), and the EPIC-XS Consortium.
Footnotes
Author Contributions Statement
Conceptualization: GC, RD, MP; Formal analysis: GC, IP; Investigation: GC, CW, VK, MD, AA, IP, HY, SS; Writing – Original Draft Preparation: GC, MP; Writing – Review & Editing: GC, MP, with input from all authors; Visualization: GC, MP; Supervision: MP, PP, US, AD; Funding Acquisition: MP, PP, US, AD.
Competing Interests Statement
The authors declare that they have no competing interests related to this paper.
Data Availability
Mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE [51] partner repository (dataset identifier PXD026060). Numerical source data giving rise to graphical representations (with all independent repeats) and unprocessed images of gels and blots are reported in the Source Data files. Detailed experimental procedures and additional data supporting the findings of this study are available from the corresponding authors upon reasonable request.
References
- 1.Fulda S, et al. Cellular Stress Responses: Cell Survival and Cell Death. International Journal of Cell Biology. 2010;2010:214074. doi: 10.1155/2010/214074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aulas A, et al. Stress-specific differences in assembly and composition of stress granules and related foci. J Cell Sci. 2017;130(5):927–937. doi: 10.1242/jcs.199240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Protter DSW, Parker R. Principles and Properties of Stress Granules. Trends Cell Biol. 2016;26(9):668–679. doi: 10.1016/j.tcb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saad S, et al. Reversible protein aggregation is a protective mechanism to ensure cell cycle restart after stress. Nat Cell Biol. 2017;19(10):1202–1213. doi: 10.1038/ncb3600. [DOI] [PubMed] [Google Scholar]
- 5.Begovich K, et al. Conserved metabolite regulation of stress granule assembly via AdoMet. J Cell Biol. 2020;219(8) doi: 10.1083/jcb.201904141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cabrera M, et al. Chaperone-Facilitated Aggregation of Thermo-Sensitive Proteins Shields Them from Degradation during Heat Stress. Cell Rep. 2020;30(7):2430–2443.:e4. doi: 10.1016/j.celrep.2020.01.077. [DOI] [PubMed] [Google Scholar]
- 7.Wolozin B, Ivanov P. Stress granules and neurodegeneration. Nat Rev Neurosci. 2019;20(11):649–666. doi: 10.1038/s41583-019-0222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao X, Jin X, Liu B. The involvement of stress granules in aging and aging-associated diseases. Aging Cell. 2020;19(4):e13136. doi: 10.1111/acel.13136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jain S, et al. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell. 2016;164(3):487–98. doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Molliex A, et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163(1):123–33. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banani SF, et al. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18(5):285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Treeck B, et al. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc Natl Acad Sci U S A. 2018;115(11):2734–2739. doi: 10.1073/pnas.1800038115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kroschwald S, et al. Different Material States of Pub1 Condensates Define Distinct Modes of Stress Adaptation and Recovery. Cell Rep. 2018;23(11):3327–3339. doi: 10.1016/j.celrep.2018.05.041. [DOI] [PubMed] [Google Scholar]
- 14.Sathyanarayanan U, et al. ATP hydrolysis by yeast Hsp104 determines protein aggregate dissolution and size in vivo. Nat Commun. 2020;11(1):5226. doi: 10.1038/s41467-020-19104-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shattuck JE, et al. The prion-like protein kinase Sky1 is required for efficient stress granule disassembly. Nat Commun. 2019;10(1):3614. doi: 10.1038/s41467-019-11550-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang B, et al. ULK1 and ULK2 Regulate Stress Granule Disassembly Through Phosphorylation and Activation of VCP/p97. Mol Cell. 2019;74(4):742–757.:e8. doi: 10.1016/j.molcel.2019.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walters RW, et al. Differential effects of Ydj1 and Sis1 on Hsp70-mediated clearance of stress granules in Saccharomyces cerevisiae. Rna. 2015;21(9):1660–71. doi: 10.1261/rna.053116.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel A, et al. ATP as a biological hydrotrope. Science. 2017;356(6339):753–756. doi: 10.1126/science.aaf6846. [DOI] [PubMed] [Google Scholar]
- 19.Hofmann S, et al. Translation suppression promotes stress granule formation and cell survival in response to cold shock. Mol Biol Cell. 2012;23(19):3786–800. doi: 10.1091/mbc.E12-04-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weitzel G, Pilatus U, Rensing L. The cytoplasmic pH, ATP content and total protein synthesis rate during heat-shock protein inducing treatments in yeast. Exp Cell Res. 1987;170(1):64–79. doi: 10.1016/0014-4827(87)90117-0. [DOI] [PubMed] [Google Scholar]
- 21.Soini J, et al. Transient increase of ATP as a response to temperature up-shift in Escherichia coli. Microb Cell Fact. 2005;4(1):9. doi: 10.1186/1475-2859-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu YF, et al. Regulation of yeast pyruvate kinase by ultrasensitive allostery independent of phosphorylation. Mol Cell. 2012;48(1):52–62. doi: 10.1016/j.molcel.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fiechter A, Fuhrmann GF, Käppeli O. Regulation of glucose metabolism in growing yeast cells. Adv Microb Physiol. 1981;22:123–83. doi: 10.1016/s0065-2911(08)60327-6. [DOI] [PubMed] [Google Scholar]
- 24.Flores CL, et al. Carbohydrate and energy-yielding metabolism in non-conventional yeasts. FEMS Microbiol Rev. 2000;24(4):507–29. doi: 10.1111/j.1574-6976.2000.tb00553.x. [DOI] [PubMed] [Google Scholar]
- 25.Grignaschi E, et al. A hydrophobic low-complexity region regulates aggregation of the yeast pyruvate kinase Cdc19 into amyloid-like aggregates in vitro. J Biol Chem. 2018;293(29):11424–11432. doi: 10.1074/jbc.RA117.001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anastasiou D, et al. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol. 2012;8(10):839–47. doi: 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blázquez MA, et al. Trehalose-6-phosphate, a new regulator of yeast glycolysis that inhibits hexokinases. FEBS Lett. 1993;329(1–2):51–4. doi: 10.1016/0014-5793(93)80191-v. [DOI] [PubMed] [Google Scholar]
- 28.van Vaeck C, et al. Analysis and modification of trehalose 6-phosphate levels in the yeast Saccharomyces cerevisiae with the use of Bacillus subtilis phosphotrehalase. Biochem J. 2001;353(Pt 1):157–162. [PMC free article] [PubMed] [Google Scholar]
- 29.Peeters K, et al. Fructose-1,6-bisphosphate couples glycolytic flux to activation of Ras. Nat Commun. 2017;8(1):922. doi: 10.1038/s41467-017-01019-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cereghetti G, et al. Reversible, functional amyloids: towards an understanding of their regulation in yeast and humans. Cell Cycle. 2018:1–14. doi: 10.1080/15384101.2018.1480220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Villar-Piqué A, et al. Screening for amyloid aggregation: in-silico, in-vitro and in-vivo detection. Curr Protein Pept Sci. 2014;15(5):477–89. doi: 10.2174/1389203715666140221101038. [DOI] [PubMed] [Google Scholar]
- 32.Linder T. Evaluation of the chitin-binding dye Congo red as a selection agent for the isolation, classification, and enumeration of ascomycete yeasts. Arch Microbiol. 2018;200(4):671–675. doi: 10.1007/s00203-018-1498-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng Y, et al. Global analysis of protein structural changes in complex proteomes. Nat Biotechnol. 2014;32(10):1036–44. doi: 10.1038/nbt.2999. [DOI] [PubMed] [Google Scholar]
- 34.Cappelletti V, et al. Dynamic 3D proteomes reveal protein functional alterations at high resolution in situ. Cell. 2021;184(2):545–559.:e22. doi: 10.1016/j.cell.2020.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boles E, et al. Characterization of a glucose-repressed pyruvate kinase (Pyk2p) in Saccharomyces cerevisiae that is catalytically insensitive to fructose-1,6-bisphosphate. J Bacteriol. 1997;179(9):2987–93. doi: 10.1128/jb.179.9.2987-2993.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orlandi I, et al. Ethanol and acetate acting as carbon/energy sources negatively affect yeast chronological aging. Oxid Med Cell Longev. 2013;2013:802870. doi: 10.1155/2013/802870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bell W, et al. Composition and functional analysis of the Saccharomyces cerevisiae trehalose synthase complex. J Biol Chem. 1998;273(50):33311–9. doi: 10.1074/jbc.273.50.33311. [DOI] [PubMed] [Google Scholar]
- 38.Monteiro F, et al. Measuring glycolytic flux in single yeast cells with an orthogonal synthetic biosensor. Mol Syst Biol. 2019;15(12):e9071. doi: 10.15252/msb.20199071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ji F, et al. Determination of intracellular metabolites concentrations in Escherichia coli under nutrition stress using liquid chromatography-tandem mass spectrometry. Talanta. 2018;189:1–7. doi: 10.1016/j.talanta.2018.06.050. [DOI] [PubMed] [Google Scholar]
- 40.Persson LB, Ambati VS, Brandman O. Cellular Control of Viscosity Counters Changes in Temperature and Energy Availability. Cell. 2020 doi: 10.1016/j.cell.2020.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fenton AW, Blair JB. Kinetic and allosteric consequences of mutations in the subunit and domain interfaces and the allosteric site of yeast pyruvate kinase. Arch Biochem Biophys. 2002;397(1):28–39. doi: 10.1006/abbi.2001.2634. [DOI] [PubMed] [Google Scholar]
- 42.Cherkasov V, et al. Coordination of translational control and protein homeostasis during severe heat stress. Curr Biol. 2013;23(24):2452–62. doi: 10.1016/j.cub.2013.09.058. [DOI] [PubMed] [Google Scholar]
- 43.Mogk A, Bukau B, Kampinga HH. Cellular Handling of Protein Aggregates by Disaggregation Machines. Mol Cell. 2018;69(2):214–226. doi: 10.1016/j.molcel.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 44.Gutierres MBB, Bonorino CBC, Rigo MM. ChaperISM: improved chaperone binding prediction using position-independent scoring matrices. Bioinformatics. 2020;36(3):735–741. doi: 10.1093/bioinformatics/btz670. [DOI] [PubMed] [Google Scholar]
- 45.Narayanaswamy R, et al. Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc Natl Acad Sci U S A. 2009;106(25):10147–52. doi: 10.1073/pnas.0812771106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Plata MR, et al. Determination of carbohydrates present in Saccharomyces cerevisiae using mid-infrared spectroscopy and partial least squares regression. Anal Bioanal Chem. 2013;405(25):8241–50. doi: 10.1007/s00216-013-7239-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan M, et al. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc. 2012;7(5):872–81. doi: 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sonderegger M, Sauer U. Evolutionary engineering of Saccharomyces cerevisiae for anaerobic growth on xylose. Appl Environ Microbiol. 2003;69(4):1990–8. doi: 10.1128/AEM.69.4.1990-1998.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lamprecht I, Schaarschmidt B, Welge G. Microcalorimetric investigation of the metabolism of yeasts. V Influence of ploidy on growth and metabolism. Radiat Environ Biophys. 1976;13(1):57–61. doi: 10.1007/BF01323624. [DOI] [PubMed] [Google Scholar]
- 50.Piazza I, et al. A Map of Protein-Metabolite Interactions Reveals Principles of Chemical Communication. Cell. 2018;172(1-2):358–372.:e23. doi: 10.1016/j.cell.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 51.Perez-Riverol Y, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47(D1):D442–d450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE [51] partner repository (dataset identifier PXD026060). Numerical source data giving rise to graphical representations (with all independent repeats) and unprocessed images of gels and blots are reported in the Source Data files. Detailed experimental procedures and additional data supporting the findings of this study are available from the corresponding authors upon reasonable request.