Abstract
Almost all haplotype-based variant callers were designed specifically for detecting common germline variation in diploid populations, and give suboptimal results in other scenarios. Here we present Octopus, a variant caller that uses a polymorphic Bayesian genotyping model capable of modeling sequencing data from a range of experimental designs within a unified haplotype-aware framework. Octopus combines sequencing reads and prior information to phase-called genotypes of arbitrary ploidy, including those with somatic mutations. We show that Octopus accurately calls germline variants in individuals, including single nucleotide variants, indels and small complex replacements such as microinversions. Using a synthetic tumor data set derived from clean sequencing data from a sample with known germline haplotypes and observed mutations in a large cohort of tumor samples, we show that Octopus is more sensitive to low-frequency somatic variation, yet calls considerably fewer false positives than other methods. Octopus also outputs realigned evidence BAM files to aid validation and interpretation.
Haplotype-based approaches have emerged as the method of choice for calling germline variants because these methods are robust to alignment errors from read mappers and have better signal-to-noise characteristics than positional approaches1–7. However, existing haplotype-based variant callers have several limitations. First, existing tools are suboptimal for many problems as most implement models that assume either diploidy1–3 or constant copy number4–6, and assume that samples are selected from an idealized population of unrelated individuals. Such models are appropriate for calling germline variants in small cohorts but provide a poorer fit to data generated in other experimental designs, such as studies involving samples with known relatedness such as paired tumors, single cells and parent-offspring trios, or in pooled tumor and bacterial sequencing where samples are often heterogeneous. These limitations cause researchers to implement custom pipelines that may integrate various callers and involve post hoc filtering and interpretation8–16. Second, existing haplotype-based methods suffer from windowing artifacts as variants are evaluated in independent nonoverlapping regions. This can lead to false calls in complex regions where reads support variants that fall outside the region being evaluated. Third, existing methods do not make a clear distinction between the haplotype sequence supported by the read data and the mutation events that gave rise to it. This makes it challenging to assign appropriate prior probabilities to these haplotype sequences, because different sets of mutations can have very different biological plausibility, despite giving rise to the same haplotype sequence. Fourth, haplotype-based methods, by design, are able to physically phase variants, but existing tools are limited to phasing diploid genotypes, and none report potentially clinically relevant17 phase information for somatic mutations with respect to germline variants or other somatic mutations.
To meet the growing demand for variant calling in experimental designs other than diploid population sequencing, we designed an algorithm that can accommodate distinct genotype models within a unified haplotype-aware framework. We took inspiration from particle filtering18 and developed a haplotype inference procedure that typically produces longer haplotypes than other methods, reducing the chance of windowing artifacts and improving the signal-to-noise ratio, resulting in more accurate variant calls. Furthermore, our method can propose and compare haplotypes composed of distinct sets of mutation events that nevertheless result in identical sequences, allowing us to consider the biological plausibility of mutations. We propose a probabilistic phasing algorithm that leverages both prior and read information, and can phase genotypes with arbitrary ploidy, including those that contain somatic mutations.
We present an implementation of our algorithm, Octopus, written in C++. We show that Octopus is more accurate than specialized state-of-the-art tools on several common experimental designs: germline calling in individuals, somatic variant calling in tumors with and without paired normal samples and de novo calling in parent-offspring trios. Octopus is freely available under the MIT license at https://github.com/luntergroup/octopus.
Results
A unified variant calling algorithm
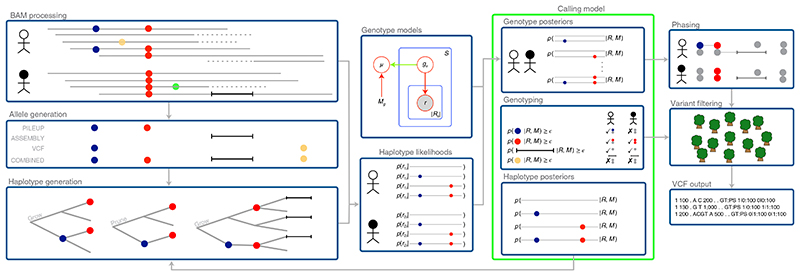
Octopus accepts sequencing data in the BAM and CRAM formats, and performs internal preprocessing, including PCR duplicate removal and adapter masking. Candidate variants are identified from the reads using a combination of local reassembly and pileup inspection with repeat awareness. In addition, variants from existing variant call format (VCF) files may also be considered as candidates. Haplotypes are then constructed exhaustively using a tree data structure (with nodes representing alleles and root-to-tip paths representing haplotypes) that is dynamically pruned, extended and collapsed based on partial read evidence (Fig. 1). Calls are made once there is sufficient evidence that haplotypes represented in the haplotype tree explain all surrounding reads sufficiently well. Haplotype likelihoods are computed for each read and haplotype using a hidden Markov model with context-aware single nucleotide variant (SNV) and indel penalties. These likelihoods are the input to a polymorphic genotype calling model, the form of which depends on the experiment that generated the sequencing data (Table 1). Although calling models are responsible for calling variants, genotypes and any other model-specific inferences, each must be able to compute posterior distributions over haplotypes and genotypes, which are used for updating the haplotype tree and for phasing, respectively. Variants are phased by evaluating the entropy of the computed genotype posterior distribution. Variant calls are then filtered, either with hard filters or a random forest classifier. Octopus optionally creates realigned evidence BAMs after calling by assigning and realigning reads to called haplotypes.
Fig. 1. overview of the unified haplotype-based algorithm, showing joint calling of two samples with the population calling model.
Two SNVs (blue and red) are detected from read pileups, a deletion from local reassembly, and a third SNV (yellow) from input VCF. The first two SNVs are added to the haplotype tree, which then contains four haplotypes. After computing likelihoods for read-haplotype pairs, the haplotype posterior distribution computed by the calling model is used to prune the haplotype tree by removing one haplotype (containing just the blue SNV). Next, the haplotype tree is extended with the deletion, and the process repeats. The polymorphic calling model is shown in the green box. Only the population genotype model (Methods) is shown in plate notation. Calling models also compute any model-specific inferences, such as de novo or somatic classification.
Table 1. Description of Bayesian calling models.
| Model name | Class | Description |
|---|---|---|
| Individuala | Germline | Calls germline variation in an individual with known ploidy. Haplotypes are expected to be observed at a frequency proportional to their copy number. |
| Population | Germline | Jointly calls germline variation in two or more samples with known ploidy but unknown relationship. Uses a joint genotype prior that can improve power to detect variation compared with individual calling by sharing information between samples. |
| Trioa | Germline De novo | Jointly calls inherited and de novo germline variation in a diploid parent-offspring trio. By explicitly modeling inheritance patterns and de novo mutations, the model has higher power compared with typical joint calling. Allosome calling is supported. |
| Cancera | Germline Somatic | Jointly calls germline and somatic variation in paired tumor-normal or tumor-only samples. The number of somatic haplotypes and their frequency are inferred from the data and used to call variants. Multiple tumors from the same individual may be called jointly. |
| Polyclone | Germline | Calls variation in an unknown mixture of haploid clones. Such samples often arise in bacterial or viral sequencing data where multiple clones may form due to sample contamination, mixed infection or in-host evolution. The number of haplotypes and their frequency are inferred from the data. |
Evaluated in this article.
Germline variants in individuals
To assess diploid germline calling accuracy, we called variants in three well-characterized Genome in a Bottle (GIAB)19 samples: HG001 (NA12878), HG002 (NA24385) and HG005 (NA24631), as well as the synthetic-diploid (SynDip) sample CHM1-CHM13 (ref. 20) that includes a validation set compiled using an approach orthogonal to that used for the GIAB truth sets. To account for different sequencing conditions, we tested several whole-genome replicates of the GIAB samples, including PCR-free, PCR-amplified and 10X Genomics Chromium prepared library designs (Supplementary Note 1). We also evaluated a whole-exome library, resulting in 13 tests in total. All raw data were downloaded from publicly available sources. BWA-MEM21 was used to map raw FASTQ files to the human reference sequence (GRCh38 for SynDip, GRCh37 for other tests). We compared Octopus to GATK4 (ref. 6), DeepVariant3, Strelka2 (ref. 2), FreeBayes5 and Platypus1. We ran each caller according to the authors’ recommended settings (Supplementary Note 2). To filter variants, we trained Octopus’ random forest classifier on several independent datasets (Supplementary Note 1). Default filters were used for DeepVariant and Strelka2. For Freebayes and Platypus, we tried recommended hard filters, but found that this deteriorated performance as quantified by the F measure (the harmonic mean of precision and recall), so we did not apply filters other than those based on variant or genotype quality (QUAL and GQ). Similarly, we found that using variant quality score recalibration filtering for GATK4—as recommended by the best practice guidelines—degraded performance, so we only used QUAL for filtering GATK4 calls. All calls were evaluated with RTG Tools vcfeval22.
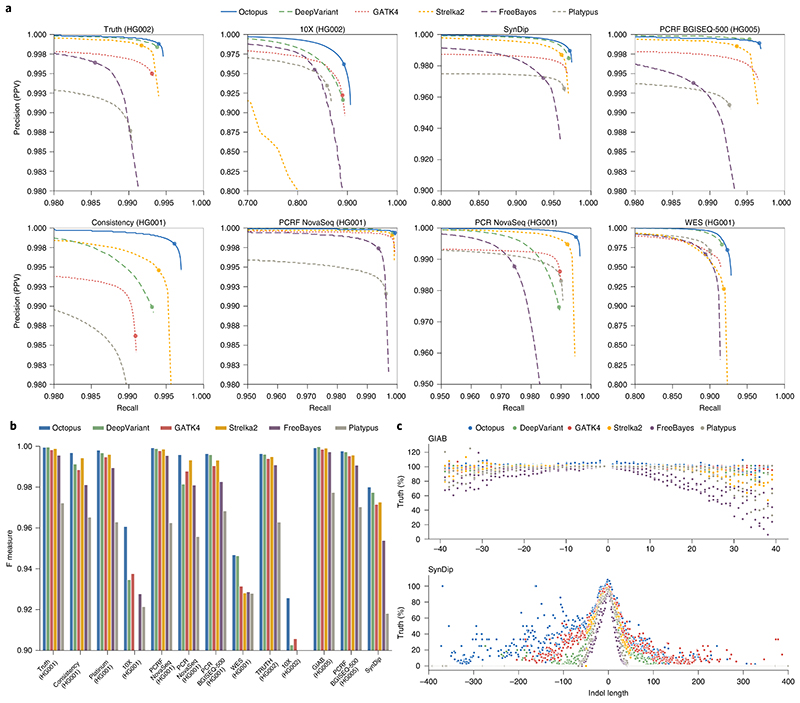
Octopus had the highest F measure in 11 tests while DeepVariant had the highest F measure in the remaining two tests (Fig. 2a,b and Supplementary Table 1). Performance differences were marginal (<1,000 false call difference) between Octopus and DeepVariant in four tests while Octopus substantially outperformed all other callers in nine tests, including all PCR-positive whole-genome sequence (WGS) libraries. Octopus showed the largest F measure improvement on the two 10X samples that have lower coverage and shorter read lengths than the other WGS samples (Supplementary Note 1), in addition to barcoded library preparation. It is unclear why Strelka2 does not perform well on these data. The next largest improvement was on the ‘SynDip’ test, which uses a truth set containing a considerably larger fraction of the genome than the other GIAB-based tests. We found that DeepVariant outperforms Octopus on the HG002 Precision FDA Truth test when compared against the GIAB v.3.3.2 truth set, but the opposite is true on the latest v.4.1 truth set where the high confidence regions cover a greater fraction of the genome. Taken together, these results suggest that Octopus is better able to call ‘difficult’ regions in the genome. Unfortunately, the v.4.1 truth sets for HG001 and HG005 are not yet available. There were also notable improvements on the ‘PCR NovaSeq’, ‘Consistency’ and ‘PCR BGISEQ-500’ tests, which in addition to being PCR-positive, also have the lower coverage compared with the remaining WGS tests. Overall, the average (geometric mean) F measure for Octopus on all 13 tests was 0.984, compared with 0.978 for DeepVariant, 0.976 for GATK4, 0.969 for FreeBayes, 0.962 for Strelka2 and 0.948 for Platypus.
Fig. 2. Germline variant calling accuracy.
Comparison of Octopus with other methods on 13 datasets: Precision FDA Truth HG001, Precision FDA Consistency HG001, Platinum Genomes HG001, PCR NovaSeq HG001, PCRF NovaSeq HG001, PCR BGISEQ-500 HG001, 10X HG001, WES HG001, Precision FDA Truth HG002, 10X HG002, GIAB HG005, PCRF BGISEQ-500 HG005 and SynDip. The average sequencing depths of these datasets are approximately 50×, 40×, 50×, 29×, 41×, 31×, 34×, 30×, 50×, 25×, 50×, 42× and 45×, respectively. SynDip and PCRF NovaSeq are mapped to GRCh38, all other datasets are mapped to GRCh37. All comparisons to the GIAB (latest versions v.3.3.2 for HG001 and HG005, v.4.1 for HG002) and CHM1-CHM13 (v.0.5) truth sets were performed using RTG Tools vcfeval (v.3.11). a, Precision-recall curves showing accuracy on 8 = 13 tests. Scoring metrics used to generate curves were RFGQ (Octopus), GQ (DeepVariant), QUAL (GATK4), GQX (Strelka2), GQ (FreeBayes) and QUAL (Platypus). The dots show typical PASS thresholds: three for Octopus, DeepVariant and Strelka2; 20 for GATK4, FreeBayes and Platypus. b, F measures at PASS thresholds for each test set. c, Proportions of true indels called in comparison to the number in the truth set by indel length. Positive lengths are insertion and negative lengths are deletions. Top, GIAB HiSeq tests (Precision FDA Truth Challenge HG001 and HG002, and GIAB HG005). Bottom, SynDip. The SynDip validation set has a larger range of indel sizes than the GIAB validation sets.
Contrary to common practice, we did not stratify our evaluation into SNVs and indels, because the true mutations that results in a haplotype are generally uncertain. For example, a sequence change of …AAACCC… to …AACCCC… could be explained either by a single SNV, or two homopolymer indels. We found that the representation used for the ground truth is biased toward the tools used to derive it. To demonstrate this, we compared the proportion of indels classified as true on the basis of haplotype matches, with the number of indels in the respective truth sets for the two Precision FDA Truth tests and GIAB HG005 tests. We observed that Octopus had over 2% more true indels by this classification than than the total number of indels in the validation sets, while all other callers called 0.5% or fewer ‘true’ indels than in the validation set (Fig. 2c), despite there being less than 0.1% difference in overall sensitivity between Octopus and DeepVariant on these tests. This apparent contradiction is due to Octopus having made indel calls in regions where other tools—and the validation sets—call SNVs that result in the same haplotype sequence. We observed similar behavior in the SynDip test for indel lengths up to 5 bp (Fig. 2c). Finally, we found evidence that complex replacements such as microinversions may be commonly misrepresented in terms of SNVs and indels (Supplementary Note 3).
Runtimes for Octopus were similar to DeepVariant, both of which were slower than Platypus, Strelka2 and FreeBayes. GATK4 ran slowest as the recommended pipeline does not make use of all available CPU cores. Memory usage was higher for Octopus than for the other tools (Supplementary Table 2); however, we note that Octopus allows users to control the size of internal buffers for the reference genome (default 500 Mb) and read data (default 6 Gb) that are intended to improve disk-access patterns.
De novo mutations in parent-offspring trios
Random germline de novo mutations resulting from imperfections in the DNA replication process during meiosis provide the necessary genetic variation for evolution, and are causative of several Mendelian and polygenic diseases23–25. The number of de novo mutations per genome duplication event is estimated to average around 70 per meiosis in humans26. However, there is uncertainty in this estimate because accurate calling of de novo mutations remains challenging, particularly for indels12–15,26.
To assess de novo calling performance, we ran Octopus using the trio calling model on whole-genome data from a previously studied parent–offspring trio from the WGS500 project1. We selected these data as the libraries were prepared directly from blood rather than from cell lines. We compared calls made by all other tools using recommended trio calling pipelines where documented (Supplementary Note 2). In addition to the 63 de novo mutations in this sample that were previously validated by Sanger sequencing1, we manually identified a further 40 mutations by inspecting unfiltered de novo calls made by three or more callers, as well as all passing de novo calls from Octopus and GATK4, using realigned BAMs that both GATK4 and Octopus are able to generate (Supplementary Fig. 1).
Only Octopus called a plausible number of de novo mutations; all other callers called more false de novo mutations than true ones (Table 2). DeepVariant and Strelka2, despite being the two most accurate germline callers after Octopus, called considerably more false positive de novo mutations than Octopus, demonstrating that strong germline calling performance does not guarantee accurate de novo calls. While the performance of the other callers could likely be improved with additional filtering, it is not always obvious how this is best achieved. For example, filtering DeepVariant calls by GQ or QUAL resulted in complete loss of sensitivity before the number of false positives fell below 100.
Table 2. De novo mutations called in a WGS500 trio.
| True positive SNVs | True positive indels | False negative SNVs | False negative indels | False positive | |
|---|---|---|---|---|---|
| Octopus | 73 | 10 | 6 | 14 | 6 |
| DeepVariant | 30 | 4 | 49 | 20 | 9,287 |
| Strelka2 | 77 | 10 | 2 | 14 | 270 |
| GATK4 | 46 | 9 | 33 | 15 | 144 |
| FreeBayes | 78 | 11 | 1 | 13 | 3,999 |
| Platypus | 75 | 9 | 4 | 49 | 159 |
Synthetic tumors
Comprehensive evaluation of somatic mutation calls is challenging because there is no gold standard reference material to compare with, and different tumor types have distinct mutation profiles27. Although calls may be manually validated to obtain an estimate of the false positive rate, it is not straightforward to estimate sensitivity as the ground truth remains uncertain. Although attempts have been made to accurately characterize somatic mutation profiles in real tumors by manual inspection28, this process is limited by the sensitivity of existing tools and is too time consuming to perform across a range of tumor types. An alternative strategy is to mix reads from unrelated individuals to create virtual tumors2,29. However, this approach is unlikely to yield data with realistic mutation profiles, error profiles and haplotype structure. A third approach is to spike mutations directly into raw sequencing reads from normal samples, which was the approach taken by the ICGC-TCGA-DREAM challenge30. However, this approach will also result in unrealistic haplotypes and inconsistent mutation spike-in positions, unless the germline haplotypes are known, which is unfortunately not the case for the ICGC-TCGA-DREAM challenge data.
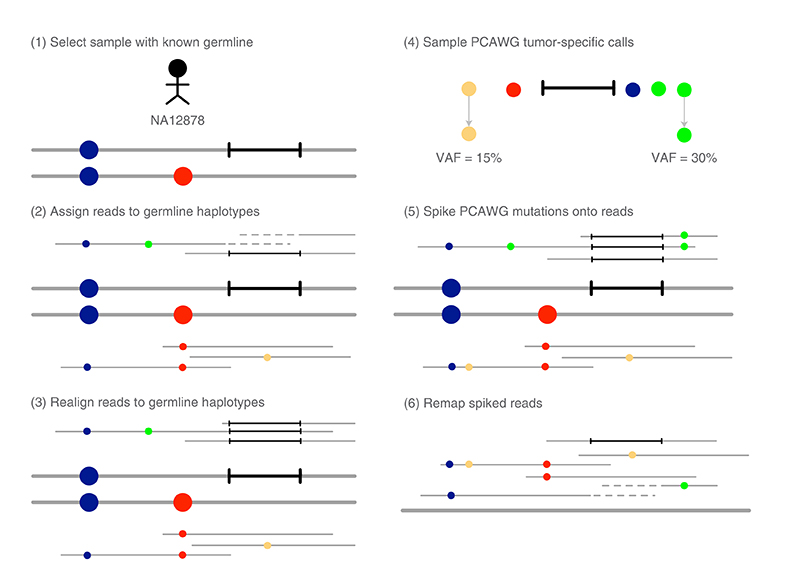
We designed an unbiased and comprehensive somatic mutation calling performance test by improving the method used by the ICGC-TCGA-DREAM challenge to ensure that synthetic tumors would have realistic mutation profiles, error profiles and local haplotype structure (Fig. 3). We created two synthetic tumors by applying this method to reads from GIAB’s NA24631 and NA12878 high-coverage Illumina data (Supplementary Note 4). The first (NA24631) was derived from pancreatic cancer (PACA) mutations using a mutation rate of ten per megabase (60,110 mutations) and spike-in frequencies uniformly sampled between 0.5% and 50%, the second (NA12878) was derived from breast cancer (BRCA) mutations using a mutation rate of one per megabase (5,895 mutations) and spike-in frequencies uniformly sampled between 1 and 20% (Supplementary Fig. 2). We used uniform spike-in frequencies, rather than simulating subclonal architecture, so that we could more thoroughly assess sensitivity across a range of variant allele frequencies (VAFs). We withheld a fraction of reads from spike-in for use as a control sample resulting in average depths of 265× for the NA24631/PACA tumor, 90× NA24631 normal, 220× NA12878/BRCA tumor and 75× NA12878 normal.
Fig. 3. overview of synthetic-tumor creation.
We used germline sequence data from a sample for which high-quality germline haplotypes are available (for example, NA12878), and assigned and realigned reads to these haplotypes (Methods). This ensures that mutations are spiked onto consistent germline haplotypes and minimizes spike-in errors due to indels. We used spike-in mutations from tumor-specific whole-genome somatic mutation calls from the PCAWG consortium37 to ensure realistic somatic mutation profiles. Mutations were spiked in using a modified version of BAMSurgeon30 (Methods). Reads were merged and remapped before variant calling to remove all realignment information.
Somatic mutations in paired tumor-normal samples
We evaluated the accuracy of Octopus at calling somatic mutations in typical WGS tumor-normal paired experiments by calling variants in the PACA and BRCA synthetic tumors downsampled to 60×, with the matched normals downsampled to 30×. Calls were compared to Mutect2 (ref. 29), Strelka2 (ref. 2), LoFreq31, Lancet32 and VarDict33. We trained Octopus’s random forest classifier on chromosome X of the BRCA data and chromosomes 11–22 of the PACA data, which we removed from the test sets. However, we noted that Octopus was comparatively less reliant on filtering than other methods (Supplementary Fig. 3). Recommended filters were used for other methods (Supplementary Note 2).
During the course of evaluation, we discovered a small number of mutations that were not in the truth sets but appeared real. This is not surprising since these data are derived from cell lines. To discount such cases, we identified calls not in the truth set but called by at least three of the six callers tested and ignored these calls during evaluation (BRCA, 843; PACA, 788).
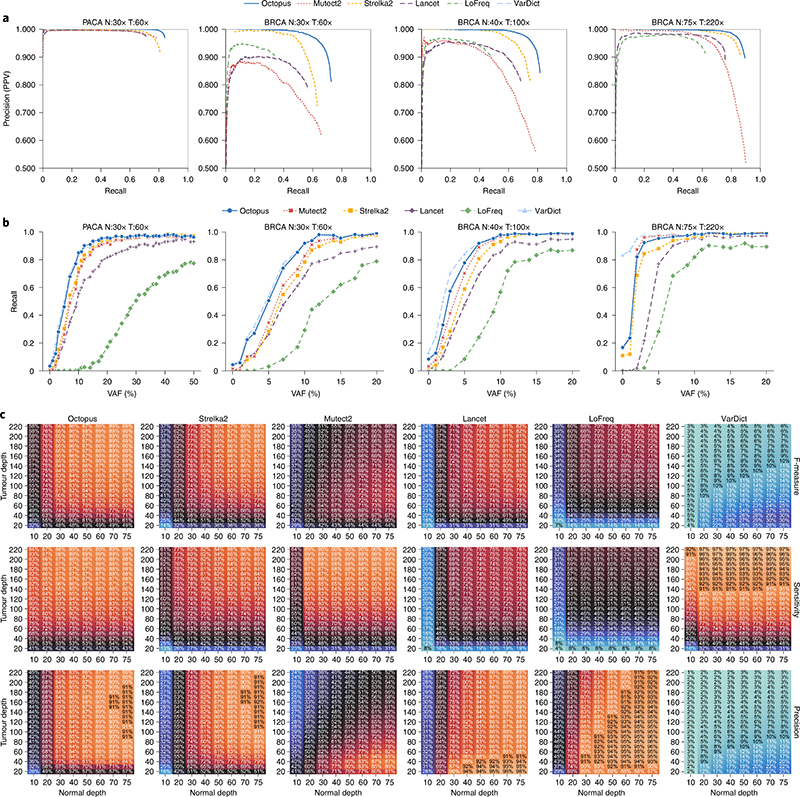
There was a clear trade-off between recall (sensitivity) and precision (positive predictive value) between callers (Fig. 4a). VarDict had the highest recall on both tests but also had the lowest precision and LoFreq had the highest precision but the lowest sensitivity. Lancet had moderate precision and recall compared to other methods. Mutect2 showed higher sensitivity but lower precision and F measure than Strelka2 on the BRCA test while the opposite was true for the PACA test. Octopus had the second highest sensitivity behind VarDict on both tests but only LoFreq and Lancet (on the PACA test) had higher precision. The number of false positives called by each caller is similar in both tests suggesting that each caller had unique biases, although it is possible that some of these false positive calls are genuine cell-line artifacts. Overall, Octopus had a substantially higher F measure than all other methods on both tests (Supplementary Table 3).
Fig. 4. Somatic mutation calling accuracy with a paired normal sample.
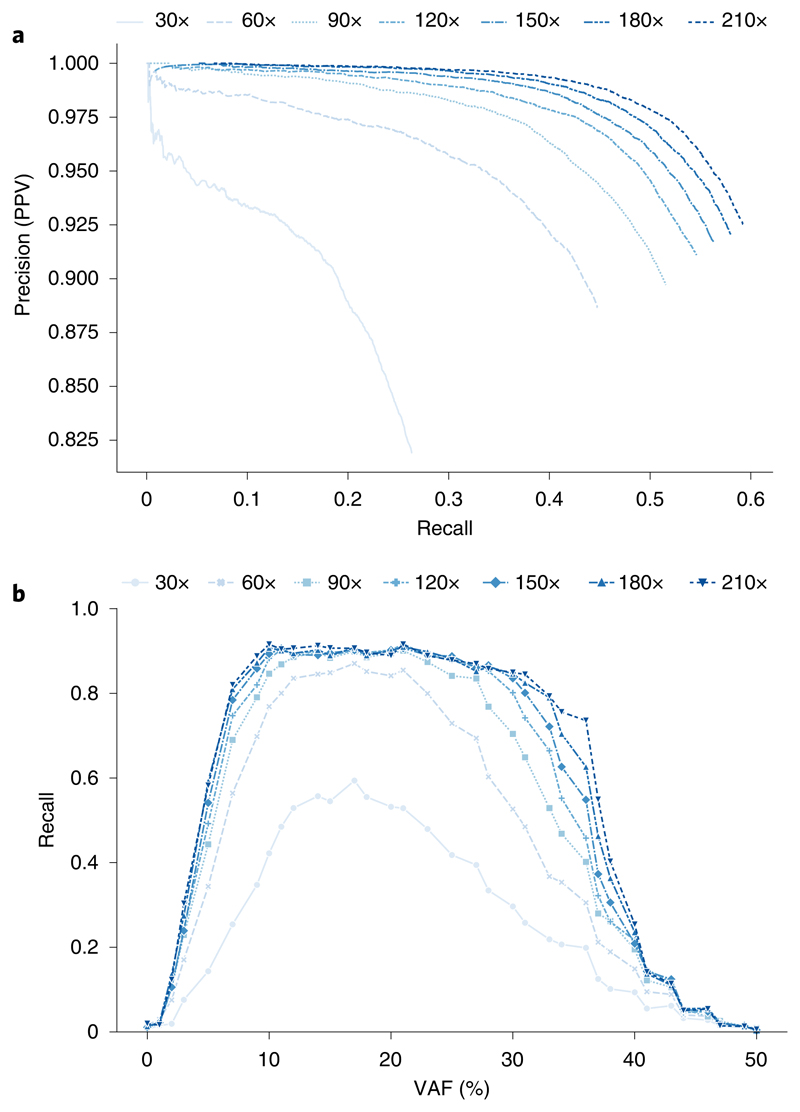
a, Precision-recall curves. Scoring metrics used to generate curves were RFGQ ALL (Octopus), TLOD (Mutect2), SomaticEVS (Strelka2), QUAL (Lancet), QUAL (LoFreq) and SSF (VarDict). Only PASS calls are used. VarDict is not visible as it is outside the axis limits due to low precision. b, Recalls for each VAF using PASS variants. Points show true spike-in VAFs. All comparisons to the synthetic-tumor truth sets were performed using RTG Tools vcfeval. c, Heatmaps showing performance (F measure, recall and precision) on all BRCA test depth combinations.
Octopus also shows considerably better precision-recall trade-off than other caller, most notably at higher recalls. For all recall values, Octopus has higher precision than the other callers. The call filter threshold for Octopus is set reasonably low by default (RFGQ_ALL 3) to achieve high sensitivity; however, increasing this to 5 reduces the number of false positives by approximately 2/3 in both tests, while only reducing the number of true positives by roughly 5% (Fig. 4a). Octopus also showed better quality score calibration consistency across the VAF spectrum (Supplementary Fig. 4).
Most of the differences in recall, particularly between the best performing tools (Octopus, Strelka2, Mutect2, Lancet), were due to low-frequency mutations (Fig. 4b). Sensitivity for mutations below 3% is poor for all callers, although Octopus was still over twice as sensitive than Mutect2 and Strelka2 in this range. At 60× sequencing depth, a 3% VAF corresponds to an expectation of fewer than two observations. However, Octopus had considerably better sensitivity for mutations with VAFs between 4–10% (3–5 expected observations at 60×) and had only slightly worse recall than VarDict, which represents an approximate upper bound on sensitivity.
To assess the influence of sequencing depth in the normal and tumor on calling accuracy, we performed a series of downsampling experiments on the NA12878/BRCA tumor (Fig. 4c and Supplementary Fig. 5). Higher tumor depths increase sensitivity, particularly for low-frequency variants, while higher normal depth improves precision. The sensitivity of Octopus, Mutect2, LoFreq and VarDict showed less dependence on the normal depth than Strelka2 and Lancet, although Mutect2 and LoFreq had substantially decreased sensitivity with a normal depth of ten. Oddly, we found that precision for all variant callers decreases with increased tumor depth for a given normal depth beyond a certain tumor depth, although this may be influenced by undetected cell-line artifacts. Overall, Octopus had the highest F measure on every test, where the greatest improvement compared with other callers was on tests with lower normal and tumor depths.
Runtimes for Octopus were average compared to the other methods (Supplementary Table 2); these were shorter than Lancet, which runs slowly, but greater than Strelka2, which runs very fast. We note, however, that Octopus calls germline variants jointly with somatic variant, and germline variants are often required as part of a tumor analysis and would otherwise be called using the normal sample independently.
Mutations in tumor-only samples
Most somatic detection tools require a paired normal sample2,31,32, but paired control material is not always available. We tested Octopus’s ability to call and classify mutations in tumor-only data by calling variants in the PACA synthetic tumor without providing the paired normal sample. We performed tests on several downsampled subsets of the full data to access the impact of average sequencing depth on performance.
Octopus’ somatic calling accuracy was worse compared with the paired tests. The number of false positive calls for the 60×-tumor-only test was higher than the paired T60×:N30× test (1,945 versus 961). Increased sequencing depth did not notably decrease in the false discovery rate, suggesting that some of these false calls may be unscreened cell-line artifacts. Sensitivity was affected more strongly: 0.45 for the 60×-tumor-only test compared with 0.84 for the paired T60×:N30× test. We also found good quality score calibration for all tested depths (Fig. 5a).
Fig. 5. Somatic mutation calling accuracy in synthetic PACA tumors without a paired normal sample for various sequencing depths.
a, Precision-recall curves. RFGQ ALL was used to generate the curves.
b, Recalls for each VAF. Classified somatic calls were compared to the truth sets with RTG Tools vcfeval.
Most of the reduction in sensitivity compared with the paired tests can be attributed to VAFs approaching 50% (Fig. 5b)—the expectation for heterozygous germline variants. If somatic classification is ignored then sensitivity for somatic mutations on the 60× test increases to 0.78 (Supplementary Table 4), indicating that most false negative somatic variants are actually called but missclassified as germline variants. Depending on the application, classification may or may not be important; it may be sufficient to know whether the variant is present or not.
We observed that the germline/somatic VAF ‘decision boundary’ automatically adjusts depending on sequencing depth due to Octopus’ Bayesian approach to classification that does not use require preset frequency thresholds. We also found that some true positive somatic mutations with high VAFs were correctly classified due to being phased with nearby germline variants.
Phasing somatic mutations
In some situations, such as when compound heterozygous mutations are suspected, it is clinically relevant to be able to determine the germline haplotype affected by a mutation34. Furthermore, phasing information is informative of tumor clonal architecture. To the best of our knowledge, no existing caller is able to phase somatic mutations, either with germline variants or other somatic mutations.
Since we designed the synthetic tumors used to benchmark somatic mutation calling so that individual reads would respect haplotype structure (but not necessarily read pairs), we know the local phase of all somatic mutations. We investigated Octopus’ ability to phase somatic mutations by evaluating how well phasing information was recovered in the paired synthetic PACA tumor test. We found that of the 52,373 somatic mutations that Octopus calls, 13,584 (26%) were phased with one or more heterozygous germline variant. Of these, the maximum and mean phase set length was 1,359 and 222 nucleotides, respectively (Supplementary Fig 6). Furthermore, 141 (0.3%) were phased with at least one other somatic mutation, and 116 (0.2% overall) of these were also phased with a heterozygous germline variant. We found that approximately 97, 98 and 99% of reported somatic-germline phasings with phase quality ≥5, ≥10 and ≥20 (Phred scaled), respectively, were correct, indicating that the phase quality score is well calibrated.
Discussion
We have shown that Octopus is more accurate than state-of-the-art variant callers on several germline samples using two independent validation sets. Performance differences were most evident on samples with 10X and PCR-positive library design, arguably the most challenging tests because of lower read depths and a higher rate of sequencing artifacts than in the other samples. Octopus also had best performance on the SynDip test, indicating that Octopus is better able to call complex regions in the genome. These results are likely due to Octopus’s use of longer haplotypes, effective error modeling and more realistic mutation priors.
Our analysis of germline indels indicate that Octopus is able to call a wider range of indels than other methods. Octopus calls considerably more true short (<15 bp) indels than other methods, and even more than are represented in the truth sets. One explanation for this is that existing methods systematically miscall a large number of indels as SNVs in tandem repeat regions leading to under-representation in the truth sets. Although both representations result in the same haplotype sequence, the distinction could have relevant clinical consequences, such as for mutation signature profiling27 or microsatellite instability analysis35,36. Moreover, around half of our manually curated de novo calls occur in microsatellites. Such sites are known to have higher mutation rates than average but are almost always ignored in de novo mutation studies because such regions are also difficult to call accurately. Our results indicate that Octopus is sufficiently specific for these mutations to be considered. We also found that in the SynDip test, Octopus and GATK4 call considerably higher proportions of true large indels (>50 bp) than other callers, further supporting Octopus’ sensitivity for indels.
High-throughput sequencing is widely used throughout the genomics community, yet the most powerful variant calling methods are optimized for human germline population data. A key advantage of Octopus is its polymorphic calling model, allowing it to optimize performance for other experimental designs as well, including for calling somatic variants from tumor samples.
Sensitivity to a wide range of VAFs is crucial for fully characterizing the mutation profiles of tumor genomes; however, our results suggest that existing somatic callers have poor sensitivity for variation occurring below 10% frequency at typical sequencing depths—unless a large number of false positives are accepted. Octopus shows near optimal sensitivity across all VAFs tested while remaining highly precise. Furthermore, Octopus also showed better precision-recall trade-off than other methods, showing that call sets can be refined on the basis of a single score. Octopus was more robust to changes in sequencing depth than other methods, both in the tumor and normal samples. Notably, for other methods to match the accuracy of Octopus on a typical 30× normal 60× tumor experiment, they required 50% more data overall (for example, 50× normal 90× tumor).
While a matched normal sample is always preferable, our tumor-only analysis indicates that Octopus provides reasonable somatic calling accuracy, particularly for depths over 100×, when one is not available. Our results also indicate that some level of normal contamination may help improve classification of clonal somatic variants in tumor-only samples as the largest fraction of missclassified somatic mutations are found at allele frequencies approaching the germline heterozygous expectation.
Our analysis of somatic mutation phasing indicates that Octopus could be used to detect cases of bi-allelic loss-of-function mutations in tumors, and provide information on tumor clonal architecture—beyond the information already provided by VAF inference. Our phasing method works equally well for SNVs and indels and the algorithm only depends on genotype posterior distributions.
Octopus has a number of usability advantages over existing tools. For example, reads do not require preprocessing as this is done internally, simplifying workflows and eliminating the need for intermediate BAM files. As an example, we required over 20 commands to call de novo mutations using the GATK4 pipeline compared with a single command for Octopus. Furthermore, multithreading is built in, and disk access is optimized for fewer long accesses rather than many short accesses, improving I/O throughput. Octopus is capable of producing realigned ‘evidence’ BAMs, including for somatically mutated haplotypes. We hope that clinicians in particular will find this feature useful by aiding variant and phase call validation and interpretation.
Overall, Octopus is highly accurate on several relevant experimental designs, demonstrating the advantage of our unified haplotype-based algorithm. As new technologies and experimental designs emerge, the flexibility of our method will allow us to rapidly incorporate new calling models that take full advantage of the information present in the data generated by each experiment.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests and statements of data and code availability are available at https://doi.org/10.1038/s41587-021-00861-3.
Methods
Read preprocessing
Input reads are scanned in random subregions of the input region set to estimate basic statistics such as average depth and read lengths. These statistics are used to determine subregions of the input regions to buffer input reads, so that the memory occupied by read data is below a user-defined limit. If multiple threads are requested, then the buffer limit is shared evenly between threads. Input read files can contain multiple samples but must have associated read group information.
Transformations
Read transformations adjust the data contained in a read observation without removing the read. Most of the transformations recalibrate base qualities in certain ways. By default, the only read transformations are to mask (set the base quality to zero) all bases considered to overlap sequencing adapters and mask fragments of reads that overlap other fragments of the same read template.
Filtering
Read filters remove reads that are likely problematic. Read filtering is applied after read transformations. Reads are removed if any of the given filtering predicates fails. By default, reads are filtered if they: (1) have malformed CIGAR strings, (2) are unmapped, (3) have mapping quality below 20, (4) are marked as quality control fails, (5) are identified as being duplicates by Octopus, (6) are marked as duplicates, (7) are secondary or supplementary alignments and/or (8) have fewer than 20 bases with a base quality of 20 or greater.
Downsampling
Read downsampling removes reads to satisfy user-specified depth criteria. Sample read sets are downsampled independently. First, regions that have depth above a certain threshold are detected. Using input alignments, the number of bases that need to be removed is calculated for each position. Reads are then removed iteratively by first selecting a position to downsample with probability proportional to the required depth reduction at each position, and then selecting a read overlapping that position with uniform probability. By default, regions with a depth above 1,000 are downsampled to reach an average read depth of 500.
Candidate allele discovery
Candidate alleles are generated jointly for all samples by taking the union of candidates generated from a set of orthogonal methods (generators). Users can choose which generators to use to optimize accuracy and runtime. The read-backed generators can be tuned to increase sensitivity for low-frequency variation.
Pileups
This uses the read mapping and alignment information present in the input BAM files and proposes candidates based on mismatches present in these alignments. Alleles are only proposed if the observation of a particular candidate satisfies an inclusion predicate, which primarily depends on the observation frequency, observed base qualities and observed read strands.
Local reassembly
The assembler discards read alignment information (but keeps mapping location) and builds a k-mer based assembly graph (that is, de Bruijn graph) at regions considered likely to contain variation. Once the graph is constructed, paths with low observation k-mer counts and cycles are pruned. Candidate alleles are extracted by enumerating the highest scoring nonreference bubbles, where the score is determined by the sum of the k-mer counts, weighted by strand bias. The nonreference path of the bubble is aligned to the reference to form candidate alleles. Complex variation such as microinversions are proposed when bubble alignments cannot be trivially decomposed into SNVs and indels.
Repeat realignment
This identifies common patterns of misalignments in tandem repeat regions that result in runs of regularly spaced SNV mismatches and proposes relevant indel candidates.
Input VCF
This reads a set of user-specified VCFs and extracts all alleles present in the input regions.
Candidate haplotype generation
Haplotype tree construction
Haplotypes are exhaustively constructed from all candidate allele combinations. This approach differs from other methods that construct haplotypes directly from read observations4,5. The primary advantage of our method is that haplotypes with no direct read support are proposed, so the length of haplotypes is not limited by read length. However, since the number of haplotypes is exponential in the number of alleles, this approach is usually only feasible for very short haplotypes. To allow construction of long haplotypes, we use a graph data structure, called a haplotype tree, where tree nodes are alleles and branches are haplotypes. The key property of the haplotype tree is that individual branches (haplotypes) can be removed or extended, which allows us to limit the haplotypes in the tree to only the most likely ones given partial data.
Controlling tree growth and active regions
The size of the tree (number of haplotypes) is controlled by a user-defined parameter (haplotype limit, default 200). The tree is grown by adding alleles sequentially in position order until the size of the tree reaches the haplotype limit. Growth rate is also controlled by checking whether reads overlapping alleles at the frontier of the tree overlap with alleles that are to be added next. Alleles that overlap with the tree frontier are always added, which can cause the tree to exceed the haplotype limit. In this case, the tree is either pruned (Filtering) or some alleles (usually large deletions) are identified to be temporarily removed from the active set, and are added again later once the tree has been sufficiently pruned. The alleles in the tree are called active. Alleles that are active but have already been evaluated by the calling model are called indicators. Periodically, indicator alleles from the root of the tree are dropped to allow room for new active alleles. Only once alleles are dropped from the tree are they viable to be called. The frequency at which this is done is user controlled, and may be turned off completely to give nonoverlapping active regions. The default behavior is to drop indicator alleles based on the current tree size, and by checking reads overlapping indicator alleles and new active alleles.
Deduplication
It is possible that duplicate haplotypes (that is, identical sequence) exist in the haplotype tree since candidate alleles are exhaustively combined. Duplicate haplotypes will have identical likelihoods as the probability of generating a read from a haplotype is only a function of the sequence itself. However, duplicate haplotypes may not have equal posterior probability as the prior probability of a haplotype segregating depends on the alleles that compose the haplotypes. Since the posterior of duplicate haplotypes is only dependent on the prior, we just keep the duplicate with the greatest prior probability.
Filtering
There are two haplotype filtering stages: prior and post to genotype inference. The latter is always preferable as it is possible to compute a marginal posterior probability for each haplotype segregating in the samples, which integrates all available information. However, it is sometimes necessary to reduce the number of haplotypes considered by the genotype model as the haplotype tree can exceed the provided haplotype limit. We considered a number of alternatives and found that likelihood-based statistics are most effective. In particular, we rank haplotypes by the number of reads assigned to each haplotype calculated by maximum likelihood.
Haplotype likelihood calculation
Remapping
The first step of the likelihood calculation is to remap reads to candidate haplotypes. This is necessary because the likelihood model requires that reads already be reasonably well placed, and the mapping position provided by the read mapper may not be accurate with respect to certain haplotypes (for example, when indels are present).
We use a simple k-mer-based mapper to find putative mapping locations. Briefly, the k-mer (k is hard coded) starting at each read and haplotype base are calculated. For each k-mer in the read we then check if the k-mer exists in the haplotype and, if so, calculate which position in the haplotype the read would start assuming perfect alignment between the read and haplotype up until the k-mer (that is, offset by the k-mer position in the read). After doing this for all k-mers in the read, we find positions in the haplotype that have a high number of putative read starts and emit these mapping positions.
Error models
The pair-hidden Markov model (pHMM, described below) for calculating read likelihoods is parameterized by a sequencing error model with indel gap open and gap extension penalties and, optionally, SNV mismatch caps. The current implementation uses a constant gap extension penalty. The penalties are set according to local repeat context, up to some maximum repeat period (currently trinucleotide repeats). There are currently two sequence error models: the default, which is intended for typical Illumina HiSeq 2500 quality data and one intended for platforms with higher error rates, such as the Illumina HiSeq X Ten. We did not use any automated inference procedure to arrive at the parameters for these two models, but set them based on experience and observation.
pHMM
Haplotype likelihoods are obtained using a pHMM that computes the approximate Viterbi probability of the read given the haplotype. We use the Viterbi probability rather than the forward probability since the Viterbi probability is considerably cheaper to compute in log space, and in practice the difference between using the two probabilities is small. The simplest pHMM implementation has positional gap open penalties that are a parameter to the model. There is a second version of the pHMM that also has SNV mismatch caps: a vector of nucleotides and maximum base mismatch penalties, one for each position in the haplotype sequence, that limit the penalty of a mismatch aligned to that position where the mismatching read base is the nucleotide indicated in the vector at that position. The intention of this is to model a common error mode in sequencing data in repetitive regions such as homopolymers, where a single base on the edge of the repeat ‘falls over’ to the leading base of the repeat.
The pHMM is a performance bottleneck in Octopus and it therefore uses a highly optimized banded SIMD implementation. Being banded, the likelihood calculation only explores parts of alignment space that results in indel errors smaller than the band size (currently eight). For short Illumina quality reads, this limitation is almost never an issue as indel errors greater than this are extremely rare. For longer, noisier reads, the band size would need to be increased, but this is currently hard coded through the width of the SIMD register used by the implementation (currently SSE2).
Inactive flank scoring
If there are read observations that partially overlap the current set of active alleles, but also support inactive alleles, then the likelihood for a true haplotype could be lower than a false one only because the false one better supports a true haplotype that has yet to be considered. This, in short, is the ‘windowing’ problem that all haplotype methods must address. Octopus’ solution is to only commit to calling candidate alleles once there is reasonable confidence that the reads supporting the alleles have had likelihoods evaluated on all the true alleles they support. However, the problem still remains that we must evaluate the likelihood function with a haplotype that is correct in the active region, but potentially incorrect outside this region (sequences outside the active region are always padded with reference). Our solution is to ‘discount’ any reductions to the likelihood that arise from mismatches outside the active region. We do this by retracing the Viterbi path and subtract terms from the log likelihood that are due to mismatches outside the active region.
Mapping qualities
Mapping quality is a statistic that reflects the trustworthiness of a read’s mapping location, r m. Formally, it has been defined as the posterior probability the read alignment is wrong: where denotes all genomic positions and typically for In Octopus, we are not as concerned with the alignment of an input read as with the mapping location, since all reads are realigned internally. If the read is incorrectly mapped to a degree that Octopus’ remapping step cannot place the read correctly, then the read is not informative of the true haplotype and should be ignored. To account for mismapped reads, we optionally factor mapping quality into the read likelihood calculation using the formula
where r mq is the read’s mapping quality in Phreds and q is some (possibly unnormalized) probability function. Currently, we use q(r) = 1.
Mutation models
Indel mutation model
Local gap open and extension probabilities are modeled for germline, de novo and somatic mutations with a single indel mutation model. The model takes as input a base rate parameter that is scaled according to the local repeat composition of the sequence using the model in Montgomery et al.38. Gap extension probabilities are assigned based on repeat composition and the current gap length. The extension model encourages the inclusion of whole repeat periods by assigning high probability to extensions of incomplete repeat periods, as indels in tandem repeats almost always occur in whole periods.
Coalescent mutation model
The coalescent mutation model, assigns probabilities to sets of haplotypes assumed to be sampled randomly from an idealized population. For a given set of haplotypes h we first calculate k 1 and k 2, the number of unique segregating SNV and indel alleles observed in h. Both k 1 and k 2 are calculated by comparing the alleles composing haplotypes to those in the reference haplotype.
The model has two parameters: the SNV heterozygosity, θ 1, set depending on user input and the indel heterozygosity, θ 2, set according to a user-supplied base indel heterozygosity and the reference sequence, by taking the maximum indel gap open heterozygosity for all segregating indels in h according to the indel model referred to above, and scaling θ 2 by this value. Probability is then assigned to h by extending the distribution for the number of segregating sites under the coalescent model39 with two heterozygosities:
where
We note that a limitation of this model is that only a single indel heterozygosity value is used for all positions in the observed haplotypes. While this assumption is unrealistic, it is rarely detrimental since the most relevant aspect of the model is to assign high probability to indels in repeat regions. The likelihood of proposing some other spurious indel in a region outside the repeat region is small given the haplotype lengths normally considered.
De novo and somatic mutation models
The de novo mutation model is intended to assign probabilities to de novo mutation occurring on a single haplotype during a single DNA replication. For haplotypes h 1 and h 2, the model assigns probabilities p(h 2|h 1) according to the indel mutation model and a SNV mutation rate that are parameters to the model. The somatic mutation model assigns probabilities to somatic mutations occurring on a single haplotype over some time period and is identical to the de novo mutation model.
Genotype prior models
Genotype prior models are used to assign prior probability to arrangements of genotypes, g = (h 1, …,hm) for ploidy m. There are two types of genotype prior model: single and joint. Single genotype prior models assign probability to single genotypes, joint genotype prior models assign probability to a combination of genotypes, for arbitary s (representing, for example, a family trio or individuals from a population). In some cases, such as the coalescent genotype prior model, the latter is simply a particular instance of the first. We report the unnormalized versions of each genotype prior model since normalization is trivial, and is always performed as part of the genotype posterior calculation.
Uniform genotype prior model
The uniform genotype prior model, is the simplest genotype prior model. We have
for the single case and the joint case, respectively.
Hardy-Weinberg equilibrium (HWE) genotype prior model
The HWE genotype prior model, MHWE, models HWE for a single individual. The model is parameterized by a set of known haplotypes h = {h 1, …, hk}, and their frequencies, f 1, …,f k The haplotype frequencies may be set explicitly or calculated with empirical Bayes. The HWE prior is a multinomial distribution:
where oi(g) is the number of haplotype i occurrences in genotype g and m = |g| is the ploidy.
Coalescent-HWE genotype prior model
The coalescent-HWE genotype prior model, is suitable for modeling genotypes randomly sampled from an idealized population; it is the default germline prior model for all calling models when the sample relationship is unknown. There are two components to this model: a segregation model that assigns probability to the pattern of observed alleles in the genotype(s) and a frequency model that assigns probability to the frequency each haplotype is observed. In particular, the segregation model is just the coalescent mutation model and the frequency model is a Hardy–Weinberg model parameterized with empirical Bayes. We then have
where h = {h ∈ g : g ∈ g} are the haplotypes segregating in the population.
Trio genotype prior model
The trio genotype prior model, assigns probabilities to triplets of genotypes that originate from parent-offspring trios. This model encapsulates two elements of uncertainty: inheritance patterns, and parental haplotype modification due to de novo mutations. The model uses a coalescent-HWE genotype prior model or uniform prior model to assign probability to parental genotypes and the de novo mutation model to model modifications of parental haplotypes. Letting g m, g p and g o denote the maternal, paternal and offspring genotypes, respectively, the model calculates
The form of the latter term, is dependent on meiosis and fertilization in the species under consideration. We consider only the mammalian case.
In the autosomal (that is, all diploid) case, we have
(writing for brevity and g o1, g o2 indicate the offsprings haplotypes), reflecting the uncertainty in parental origin of the offspring haplotypes, and where (where s = mor p, and i = 0 or 1) models uncertainty of which parental haplotype is inherited. is the probability the haplotype g oi is inherited by the offspring given that the haplotype gsj is the one provided by the parent s for fertilization; it models de novo mutations.
For the female offspring X chromosome case we have the same form for as the autosomal case, but
For the male offspring X chromosome case we have
Finally, in the male offspring Y chromosome case, we simply have
Cancer genotype prior model
The cancer genotype prior model, is used to assign probability to cancer genotypes, that is, a pair of regular genotypes, g cancer = (g germ, g som), where g germ is the germline and g som is acquired somatically. The model must explain both the germline and the somatic genotypes. No assumptions of either germline or somatic genotype ploidy are made.
The model is the composition of two separate models: a germline prior model, (for example, the coalescent-HWE model) and a conditional somatic model, We then have (omitting models for brevity)
The second term, models the pattern of somatic haplotypes. In the simplest case, when |g som| = 1 (that is, there is a single somatic haplotype), then we have
where is the probability of observing the somatic haplotype given the germline haplotype suffers some mutational event assigned by the somatic mutation model.
More interesting is when (that is, there are multiple somatic haplotypes). In principle, we must consider that any of the somatic haplotypes could have originated from either the germline or any other somatic haplotype and we should consider possible tumor phylogenies. We have not implemented such a model in Octopus; instead, we assume independence between somatic haplotypes:
Genotype posterior models
Genotype posterior models combine genotype prior models with genotype likelihood models.
Population genotype model
All samples have known ploidy and copy number, so the likelihood function of reads R given genotype g is
where |g| is the ploidy and |R| is the number of reads. The joint genotype posterior for S samples is therefore given by
where the genotype prior model, is either the uniform or HWE-coalescent prior. Unfortunately, the number of genotype combinations g grows exponentially in the number of samples S, so we cannot evaluate the full posterior distribution in general. Therefore, other than for trivial cases, we first approximate the sample marginal genotype posterior distribution under the HWE model (without mutations) and use these marginal probabilities to select K genotype combinations g 1, …, gK (K is user-defined) to evaluate under the full joint genotype model. The approximate posterior marginals are computed with expectation maximization.
Individual genotype model
The individual model is simply a case of the population model without the initial approximation step. This model is always fully evaluated.
Trio genotype model
The trio genotype model has the same likelihood function as the population model, but assigns prior probabilities to genotype combinations with the trio genotype prior model. As for the population model, this model is intractable in general, so we only evaluate the posterior partially. Briefly, we evaluate approximate marginal probabilities for each sample under independence, and use these likelihoods to first combine parental genotypes and then formulate a list of trio genotypes by combining parental combinations with offspring genotypes.
Subclone genotype model
Unlike the other genotype posterior models, this model does not assume known copy number—or mixture frequency—of sample genotypes. The model assumes all read observations originate from the same underlying genotype, but may have been observed at different mixture frequencies. The mixture frequencies for each sample therefore becomes a latent variable that we infer. We use Dirichlet distributions to assign probability to mixture frequencies, so different mixture priors may be specified for each sample by controlling the concentration parameter of the respective Dirichlet distribution. The joint posterior distribution is
where α are prior concentration parameters, ϕ are mixture frequencies and is the genotype prior model. We cannot compute this model exactly—or even partially—due to the integral over mixture frequencies. We therefore compute approximate posteriors for each latent variable using variational Bayes. Namely, we assume the posterior distribution has factorization
where Z s is a latent binary indicator matrix specifying read-component assignments. The associated probabilities, q(Z snk), are often called responsibilities—the responsibility haplotype k assumes for read n in sample s. With this factorization, the posterior distribution for each latent variable is conjugate with the prior and we infer Dirichlet posterior distributions. An important feature of the variational Bayes approximation is that we can easily calculate a lower bound for the data likelihood—the model evidence.
Calling models
Each calling model is responsible for calling variants given candidate alleles, haplotypes and haplotype likelihoods. Although calling models are free to choose which latent variables and genotype models to use, all calling models must be able to infer posterior distributions over candidate haplotypes (for filtering) and genotypes (for phasing).
Individual
The individual calling model uses the individual genotype model for genotype inference. Haplotype posteriors are computed by marginalizing over the genotype posterior distribution:
where the sum extends over genotypes g that contain h at least once. To call variants, we first calculate the posterior probability of all candidate nonreference alleles by marginalizing over the genotype posterior distribution:
where the sum extends over genotypes g with at least one haplotype that contains variant a. We then select alleles with posterior probability above some user-specified threshold.
Next, we identify the genotype with the greatest posterior probability (that is, the MAP genotype). All selected alleles that appear on the MAP genotype are called, where the variant quality is determined by the marginalized allele posterior computed previously.
For each called allele, we then call genotypes at the loci of those alleles. In particular, for each allele we identify the alleles present in the called genotype at the loci of the allele, and once again marginalize over the genotype posterior distribution to compute the posterior probability for that local genotype. This is used for the genotype quality score.
Population
Inference for the population model is similar to the individual and variant calls are made based on sample marginal posteriors.
Trio
The trio calling model first infers a joint genotype posterior distribution p(g m,g p,g o|R) with the trio genotype posterior distribution. We then infer sample marginal genotype posteriors for each sample by marginalizing over the joint posterior distribution. Haplotype posteriors are calculated by integrating over the marginal posterior:
To calculate the probability an allele segregates in the trio, we also integrate over the joint genotype posterior distribution:
Similarly, we calculate the posterior probability an allele is de novo in the child:
Finally, we find the MAP genotype combination using the joint genotype posterior distribution, and only call variants that are included in this trio.
Cancer
The cancer calling model is intended to detect somatic mutations in a set of tumor samples from a single individual. The set of samples may also include a sample that is not expected to contain somatic mutations—a normal or control sample. We attempt to model three data characteristics that result from tumor biology and experimental protocol:
-
(1)
There are no somatic mutational events, reads are generated from a clean germline.
-
(2)
Copy number changes have occurred, but no somatic mutations.
-
(3)
Somatic mutations have occurred and possibly copy number changes.
Each of these three cases is modeled by fitting a unique genotype posterior model:
-
(1)
The individual model with any germline genotype prior model (all read observations are merged): :
-
(2)
The subclone model with a germline genotype prior model (for example, coalescent-HWE):
-
(3)
The subclone model with the cancer genotype prior model:
The posterior probability for each model is calculated using Bayes theorem:
where is the evidence for model x.
For the somatic case, we must also infer the number of segregating somatic haplotypes. To do this we start by assuming a single somatic haplotype, and incrementally add more while the evidence for the model is the greatest observed so far, up to a user-defined limit.
For inference, we must marginalize over models. For example, we calculate posteriors for germline genotypes by marginalization:
where (that is, marginalize over cancer genotypes that contain the matching germline component). The posterior probability of an allele a segregating in the germline is then
Germline candidates are called if the posterior is above a user-defined threshold, and if the candidate is present in the called germline genotype. If the allele is not called in the germline then it is added to a list of candidate somatic alleles.
To calculate the posterior probability an allele a is somatic, we marginalize over conditional on the somatic mutation frequency being above a user-defined threshold τ.
First, we calculate the posterior mass for credible somatic frequencies. Supposing that we inferred a model with K somatic haplotypes, we assign probability to each somatic haplotype k = 1…K if it occurs as a frequency above τ:
where the equality holds since the posterior distribution for ϕs is Dirichlet. The overall credible somatic mass λs is then calculated with
Finally, we set We then calculate the posterior probability that an allele is a somatic mutation by marginalization:
If this probability is greater than some user-defined threshold, we call the allele somatic.
For both called germline and somatic mutations, we also calculate the probability that the variant segregates regardless of classification, by marginalizing over all three models.
Polyclone
The polyclone calling model is similar to the cancer calling model without the third somatic mutation model. It compares the individual genotype model with haploid genotypes to the subclone genotype model where the number of haplotypes is determined iteratively by comparing model evidences. The genotype prior model in both cases is either the uniform or the coalescent-HWE model (depending on user choice).
Probabilistic phasing
Although each caller implements different genotype models, each is required to infer the marginal posterior probability of genotypes for each sample. This posterior distribution is used to infer physical phasing of called variant sites. Direct read data are not required as all information available from the reads, in addition to any prior information, is already contained in the posterior distribution. The advantage of this approach is most evident when calling trios as the genotype prior can be strongly informative about phase due to identity by decent. The method applies to genotypes of arbitrary zygosity and is therefore applicable to nondiploid samples.
Samples are phased independently and the sample marginal genotype posterior distribution is used for phasing. The input to the algorithm is a set of haplotypes genotypes and variation sites (that is, genomic regions) . The algorithm is as follows: (1) an undirected weighted phase graph is created, where nodes are nonoverlapping variation sites and edge weights are phase qualities between the two sites (detailed below). Only edges with phase qualities above some user provided minimum are added to the graph. (2) The maximum clique problem is solved for the phase graph. (3) The maximum cliques are used to find phase sets consisting of variation sites. Each variation site may only belong to one phase set and the variation sites in a phase set must be a subset of a clique in the phase graph. In some cases, a site will exist in multiple cliques (for example, confident homozygous sites). Our approach is therefore to first build phase sets from sites with a unique clique and then assign the other sites to these phase sets by closest genomic distance. (4) Phase qualities for each phase set are calculated by computing the geometric mean of phase qualities for each pair of sites in the phase set.
The idea behind the phase quality is that the uncertainty in variant phasing between two nonoverlapping sites should depend only on the uncertainty in the arrangement of alleles at the sites, rather than the alleles themselves; phase quality should not depend on genotype errors. To achieve this, the full genotype space is partitioned into nonoverlapping sets where all the genotypes in the subset share the same set of alleles at both sites. For the genotypes in each partition, only variation at the two input sites are of interest, so the partition is ’collapsed’ into a new subset where each genotype is mapped onto the genotype in containing only the same arrangement of alleles at the two sites. The uncertainty in the phase of this subset of genotypes is then determined by evaluating how ‘good’ the maximum a posterior (that is, the probability mass of the MAP genotype) genotype of this remapped partition is. More formally, for any pair of nonoverlapping variation sites the phase quality of the two sites is defined
where is the set of alleles at v; hv denotes the allele at site v in haplotype h and denotes the power set.
where we use Iverson bracket notation. Given the map such that where hvi,vj denotes the haplotype formed by copying alleles at sites vi and vj in h, then
Variant filtering
As with any model, there are some error modes that are not well captured by Octopus’ calling models that can lead to false inferences. For example, Octopus assumes read sequencing and mapping errors are independent, which is not true in general. To identify false calls due to model error, we developed classifiers to filter Octopus’s raw calls using statistics, called measures in Octopus, that may be derived directly from the input read data.
Hard filtering
Hard filters are Boolean expressions where the terms of the expression are comparison operations. Currently, only or binary operations are permitted; if any of the individual operations is true, then the call is filtered. Different filter expressions can be specified for germline variant, de novo, somatic and homozygous reference calls.
Random forest filtering
Octopus uses the Ranger library40 for random forest classification. Different random forests may be used for germline and somatic calls.
BAM realignments
Octopus is capable of producing realigned BAM files that provide further evidence of a call’s reliability. These BAM files are especially helpful in cases where there are complex indel variants and the input alignments are notably different from the alignments supported by the called haplotypes. The realignment process for each read is:
-
(1)
Identify the called haplotype where the read originated from.
-
(2)
Align the haplotype to the reference sequence.
-
(3)
Align the read to the called generating haplotype (mismatches due to sequencing or calling errors).
-
(4)
Compute the composition of the two alignments to obtain a single alignment to the reference.
For the first step, the called haplotype with the maximum likelihood of generating the read is used. If there are more than one called haplotypes that have equal likelihood, then we label the read ambiguous. For realignment, ambiguous reads are assigned randomly to one of the equally well supported haplotypes. The second step is trivial in Octopus since haplotypes are defined explicitly by alleles that are reported in the VCF output. The third step is computed using the Viterbi alignment found from calculating the maximum likelihood in the first step.
Since a hard choice of generating haplotype is made in the first step, we can report this information by generating separate BAM files for each called haplotype and another for ambiguous reads.
Synthetic tumors
To generate synthetic-tumor BAM files we followed a similar approach to Ewing et al.30 with the following differences. First, we obtained unmapped reads from the GIAB HiSeq 2500 high-coverage datasets (roughly 300× total each). From the full high-coverage set, we extracted two nonoverlapping subsets each, one for the normal and one for the tumor (Supplementary Note 4). To maintain realistic sequencing conditions, we ensured that reads from same library and sequenced on the same lane were kept together.
For each of the two neo-synthetic-tumor BAMs, we then performed a haplotype-based read assignment and realignment using Octopus’ BAM realignment feature. This results in three BAM files: two containing reads that were assigned to a called germline haplotype and another containing ambiguous reads. The purpose of the assignment step is to ensure spike-in mutations fall on the same germline haplotype. The realignment step is to ensure consistency of spike-in location. Neither of these are guaranteed by the method described in Ewing et al. due to limitations with BAMSurgeon. In summary, this procedure results in three ‘cleaned’ BAM subsets, each of which should be haploid and contain few alignment errors.
We then simulated two sets of somatic mutations by sampling putative real somatic mutations called by the pan-cancer analysis of whole genomes (PCAWG) consortium. In particular, we uniformly sampled PCAWG calls from pancreatic tumors and from breast tumor, to achieve mutation densities close to 10 and 1 Mb-1 for the pancreatic and breast sets, respectively. These densities were chosen from the upper expected range for each tumor type27. For each sampled mutation, we uniformly assigned a spike-in VAF from 0.005 to 0.5 (from 51 equally spaced bins) for PACA mutations, and 0.01 to 0.2 (from 20 equally spaced bins) for BRCA mutations. Although this frequency distribution is unlikely to be biologically realistic, we used this approach to evaluate the performance of each algorithm across a range of VAFs and, to the best of our knowledge, none of the evaluated methods consider genome-wide subclonal tumor architecture when calling variants. Each sampled mutation was finally assigned to a random germline haplotype.
We used a slightly modified version of BAMSurgeon (https://github.com/dancooke/bamsurgeon) to spike in mutations into each putative haploid neo-tumor BAM. The main modification that we made to BAMSurgeon was to ensure any ‘pad’ sequence used for deletion spike-ins came from the originating germline haplotype, rather than the reference sequence. We also needed to make minor modifications to handle our split BAM files, which may not contain proper read pairs. The spike-in VAF used for the BAM containing ambiguous reads, which should contain reads from both parental haplotypes, was always half the chosen spike-in VAF.
Finally, we created paired raw synthetic-tumor FASTQ files by merging and extracting reads from the three spiked haploid BAM files. We emphasize that this final step removes all alignment and phasing information.
Supplementary Material
Additional information
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41587-021-00861-3.
Acknowledgements
This work was supported by The Wellcome Trust Genomic Medicine and Statistics PhD Program (grant nos. 203735/Z/16/Z to D.P.C.). The computational aspects of this research were supported by the Wellcome Trust Core Award grant number 203141/Z/16/Z and the NIHR Oxford BRC. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health.
Footnotes
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author contributions
D.P.C. and G.L. designed the algorithm and wrote the manuscript. D.P.C. implemented the algorithm and performed the evaluation. D.C.W. provided data for the synthetic tumors and critically reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Peer review information Nature Biotechnology thanks Federico Abascal and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Reprints and permissions information is available at www.nature.com/reprints.
Data availability
All germline data used in this manuscript are publicly available from GIAB, Precision FDA and ENA. Links are provided in Supplementary Note 1. Trio data from the WGS500 project are available from the European Nucleotide Archive under accession no. PRJEB9151 (samples AW_SC_4654, AW_SC_4655 and AW_SC_4659). The synthetic-tumor data have been deposited in the Sequence Read Archive under BioProject accession no. PRJNA694520. The corresponding truth sets have been deposited to figshare (https://doi.org/10.6084/m9.figshare.13902212).
Code availability
Octopus source code and documentation is freely available under the MIT licence from https://github.com/luntergroup/octopus. Custom code used for data analysis is available from https://github.com/luntergroup/octopus-paper.
References
- 1.Rimmer A, et al. Integrating mapping-, assembly-and haplotype-based approaches for calling variants in clinical sequencing applications. Nat Genet. 2014;46:912–918. doi: 10.1038/ng.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim S, et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat Methods. 2018;15:591–594. doi: 10.1038/s41592-018-0051-x. [DOI] [PubMed] [Google Scholar]
- 3.Poplin R, et al. A universal SNP and small-indel variant caller using deep neural networks. Nat Biotechnol. 2018;36:983–987. doi: 10.1038/nbt.4235. [DOI] [PubMed] [Google Scholar]
- 4.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. 2012 Preprint at https://arxiv.org/abs/1207.3907. [Google Scholar]
- 6.Poplin R, et al. Scaling accurate genetic variant discovery to tens of thousands of samples. Preprint at bioRxiv. 2017 doi: 10.1101/201178. [DOI] [Google Scholar]
- 7.Lo Y, et al. Comparing variant calling algorithms for target-exon sequencing in a large sample. BMC Bioinf. 2015;16:75. doi: 10.1186/s12859-015-0489-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nik-Zainal S, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayward NK, et al. Whole-genome landscapes of major melanoma subtypes. Nature. 2017;545:175–180. doi: 10.1038/nature22071. [DOI] [PubMed] [Google Scholar]
- 10.Northcott PA, et al. The whole-genome landscape of medulloblastoma subtypes. Nature. 2017;547:311–317. doi: 10.1038/nature22973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waddell N, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Besenbacher S, et al. Multi-nucleotide de novo mutations in humans. PLoS Genet. 2016;12:e1006315. doi: 10.1371/journal.pgen.1006315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jonsson H, et al. Parental influence on human germline de novo mutations in 1,548 trios from Iceland. Nature. 2017;549:519–522. doi: 10.1038/nature24018. [DOI] [PubMed] [Google Scholar]
- 14.Deciphering Developmental Disorders, S. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldmann JM, et al. Germline de novo mutation clusters arise during oocyte aging in genomic regions with high double-strand-break incidence. Nat Genet. 2018;50:487–492. doi: 10.1038/s41588-018-0071-6. [DOI] [PubMed] [Google Scholar]
- 16.Walker TM, et al. Whole-genome sequencing for prediction of mycobacterium tuberculosis drug susceptibility and resistance: a retrospective cohort study. Lancet Infect Dis. 2015;15:1193–1202. doi: 10.1016/S1473-3099(15)00062-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tewhey R, Bansal V, Torkamani A, Topol EJ, Schork NJ. The importance of phase information for human genomics. Nat Rev Genet. 2011;12:215–223. doi: 10.1038/nrg2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doucet A, Johansen AM. A tutorial on particle filtering and smoothing: fifteen years later. In Handbook of Nonlinear Filtering. 2009;12:656–704. [Google Scholar]
- 19.Zook JM, et al. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat Biotechnol. 2014;32:246–251. doi: 10.1038/nbt.2835. [DOI] [PubMed] [Google Scholar]
- 20.Li H, et al. A synthetic-diploid benchmark for accurate variant-calling evaluation. Nat Methods. 2018;15:595–597. doi: 10.1038/s41592-018-0054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. 2013 Preprint at https://arxiv.org/abs/1303.3997. [Google Scholar]
- 22.Cleary JG, et al. Comparing variant call files for performance benchmarking of next-generation sequencing variant calling pipelines. Preprint at. 2015 doi: 10.1101/023754v2. [DOI] [Google Scholar]
- 23.Veltman JA, Brunner HG. De novo mutations in human genetic disease. Nat Rev Genet. 2012;13:565–575. doi: 10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- 24.Xu B, et al. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat Genet. 2012;44:1365–1369. doi: 10.1038/ng.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilissen C, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–347. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- 26.Kong A, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alioto TS, et al. A comprehensive assessment of somatic mutation detection in cancer using whole-genome sequencing. Nat Commun. 2015;6:10001. doi: 10.1038/ncomms10001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ewing AD, et al. Combining tumor genome simulation with crowdsourcing to benchmark somatic single-nucleotide-variant detection. Nat Methods. 2015;12:623–630. doi: 10.1038/nmeth.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilm A, et al. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012;40:11189–11201. doi: 10.1093/nar/gks918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narzisi G, et al. Genome-wide somatic variant calling using localized colored De Bruijn graphs. Commun Biol. 2018;1:20. doi: 10.1038/s42003-018-0023-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lai Z, et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 2016;44:e108. doi: 10.1093/nar/gkw227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Decker B, et al. Biallelic BRCA2 mutations shape the somatic mutational landscape of aggressive prostate tumors. Am J Hum Genet. 2016;98:818–829. doi: 10.1016/j.ajhg.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hause RJ, Pritchard CC, Shendure J, Salipante SJ. Classification and characterization of microsatellite instability across 18 cancer types. Nat Med. 2016;22:1342–1350. doi: 10.1038/nm.4191. [DOI] [PubMed] [Google Scholar]
- 36.Maruvka YE, et al. Analysis of somatic microsatellite indels identifies driver events in human tumors. Nat Biotechnol. 2017;35:951–959. doi: 10.1038/nbt.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature. 2020;578:82–93. doi: 10.1038/s41586-020-1969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montgomery SB, et al. The origin, evolution, and functional impact of short insertion-deletion variants identified in 179 human genomes. Genome Res. 2013;23:749–761. doi: 10.1101/gr.148718.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu YX. Probability of a segregating pattern in a sample of DNA sequences. Theor Popul Biol. 1998;54:1–10. doi: 10.1006/tpbi.1997.1359. [DOI] [PubMed] [Google Scholar]
- 40.Wright MN, Ziegler A. Ranger: a fast implementation of random forests for high dimensional data in C++ and R. J Stat Softw. 2017;77:1–17. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All germline data used in this manuscript are publicly available from GIAB, Precision FDA and ENA. Links are provided in Supplementary Note 1. Trio data from the WGS500 project are available from the European Nucleotide Archive under accession no. PRJEB9151 (samples AW_SC_4654, AW_SC_4655 and AW_SC_4659). The synthetic-tumor data have been deposited in the Sequence Read Archive under BioProject accession no. PRJNA694520. The corresponding truth sets have been deposited to figshare (https://doi.org/10.6084/m9.figshare.13902212).
Octopus source code and documentation is freely available under the MIT licence from https://github.com/luntergroup/octopus. Custom code used for data analysis is available from https://github.com/luntergroup/octopus-paper.