Abstract
Autoimmune diseases develop when autoantigens activate previously quiescent self-reactive lymphocytes. Gene-gene interaction between certain human leukocyte antigen (HLA)-class I risk alleles and variants of the endoplasmic reticulum aminopeptidase ERAP1 controls the risk for common immune-mediated diseases including psoriasis, ankylosing spondylitis, and Behçet’s disease. The functional mechanisms underlying this statistical association are unknown. In psoriasis, HLA-C*06:02 mediates an autoimmune response against melanocytes by autoantigen presentation. Using various genetically modified cell lines together with an autoreactive psoriatic TCR in a TCR activation assay, we demonstrate here that in psoriasis ERAP1 generates the causative melanocyte autoantigen through trimming NH2-terminal elongated peptide precursors to the appropriate length for presentation by HLA-C*06:02. An ERAP1 risk haplotype for psoriasis produced the autoantigen much more efficiently and increased HLA-C expression and stimulation of the psoriatic TCR by melanocytes significantly more than a protective haplotype. Compared to the overall HLA-class I molecules, cell surface-expression of HLA-C decreased significantly more upon ERAP1 knockout. The combined upregulation of ERAP1 and HLA-C on melanocytes in psoriasis lesions emphasizes the pathogenic relevance of their interaction in patients. We conclude that in psoriasis pathogenesis the increased generation of an ERAP1-dependent autoantigen by an ERAP1 risk haplotype enhances the likelihood that autoantigen presentation by HLA-C*06:02 will exceed the threshold for activation of potentially autoreactive T cells, thereby triggering CD8+ T-cell mediated autoimmune disease. These data identify ERAP1 function as a central checkpoint and promising therapeutic target in psoriasis and possibly other HLA-class I-associated diseases with a similar genetic predisposition.
Keywords: Autoimmunity, T Cells, T Cell Receptors, MHC, Antigens/Peptides/Epitopes
Free-form key words: Psoriasis, HLA-class I association, ERAP1, HLA-C*06:02
Introduction
Common autoimmune diseases arise from a polygenic predisposition, in which combinations of diverse protective and risk gene variants induce an autoimmune response and ultimately lead to a clinical phenotype (1). The association with certain HLA alleles characterizes virtually any human autoimmune disease. In particular, three of the more than 19,000 HLA-class I alleles currently registered in the IMGT/HLA Database (2) pose a high risk for immune-mediated diseases: HLA-C*06:02 is associated with psoriasis, HLA-B*27 with ankylosing spondylitis, and HLA-B*51 with Behçet’s disease. This is interpreted to indicate that the pathomechanisms in the three diseases are directly related to these HLA-class I alleles. In view of the unexplained autoimmune pathogenesis, the diseases were grouped under the term MHC-I-opathies (3). As another shared feature, genome-wide association studies (GWASs) have revealed that epistasis, i.e. non-additive gene-gene interaction between the HLA-class I risk alleles and certain variants of the endoplasmic reticulum aminopeptidase 1 (ERAP1) controls the risk of contracting these diseases (4–8). The functional interaction between HLA-class I alleles and ERAP1 may thus play a crucial role in the development of autoimmune diseases, the elucidation of which should provide essential insights into disease pathogenesis and enable the development of innovative therapeutic strategies that benefit patients without causing broad immunosuppression.
HLA-class I-molecules and ERAP1 cooperate in antigen processing and presentation. HLA-class I molecules primarily present peptide antigens originating from cytosolic proteins to the TCRs of CD8+ T cells. Thus, CD8+ T cell-mediated immune responses are specifically directed against target cells that express and process these proteins into antigenic peptides (9). The peptide antigens are generated by the multi-catalytic cellular proteasome. It creates mainly NH2-extended peptides with a defined C-terminus that are translocated by the “transporter associated with antigen presentation” (TAP) into the endoplasmic reticulum for loading onto HLA-class I molecules. HLA-class I molecules limit the size of peptide antigens typically to 8 to 10 amino acids, because the peptide binding groove is closed at both sides (10). ERAP1 is a peptidase that may create the appropriate peptide length for HLA-class I binding by NH2-terminal trimming of antigen precursors, but can also destroy HLA-class I ligands by overtrimming (11, 12). Approximately one third of peptides presented by HLA-class I result from processing by ERAP1, while the major part of the HLA-class I peptidome is generated independently from ERAP1 activity (13). Various non-synonymous coding ERAP1 variants modulate the catalytic aminopeptidase activity (14, 15). This may lead to altered peptide repertoires, HLA misfolding and HLA homodimer formation, all of which have been discussed as possible causes of abnormal CD8+ T-cell or natural killer cell activation or autoinflammation (16). However, it is ultimately unresolved how ERAP1 variants, in conjunction with the disease-associated HLA-class I alleles, determine the development of immune-mediated inflammatory diseases.
Psoriasis is a common T-cell mediated autoimmune skin disease. HLA-C*06:02 is the main risk allele (17, 18). Development of psoriasis lesions depends on the epidermal recruitment, activation and clonal expansion of CD8+ T cells (19–21). Through unbiased screening of a Vα3S1/Vβ13S1 TCR of a CD8+ T-cell clone from the pathogenic psoriatic T-cell infiltrate of an HLA-C*06:02-positive donor, we had discovered that HLA-C*06:02 directs an autoimmune CD8+ T-cell response against melanocytes as an underlying pathomechanism of psoriasis (22). We further had identified a peptide from ADAMTS-like protein 5 (ADAMTSL5) as the causative melanocyte autoantigen, which is naturally processed in melanocytes, presented by HLA-C*06:02 and recognized by the Vα3S1/Vβ13S1 TCR. ADAMTSL5 is highly expressed in psoriasis lesions, especially melanocytes (22, 23). ADAMTSL5-specific CD8+ T cells are clearly detectable in psoriasis patients and produce psoriasis key cytokines, IL-17A and IFN-γ, upon antigenic stimulation (22).
These insights now enabled us to investigate the functional mechanisms underlying the statistically defined gene-gene interaction between particular ERAP1 haplotypes and the main HLA-class I risk allele, HLA-C*06:02. For our experiments, we had established a TCR activation assay that reproduces the in vivo psoriatic autoimmune response against melanocytes and ADAMTSL5 as the causative psoriatic autoantigen. It employs the autoreactive HLA-C*06:02-restricted ADAMTSL5-specific Vα3S1/Vβ13S1 TCR expressed in a CD8+ mouse T-hybridoma reporter cell line (22) which indicates TCR stimulation by the induction of super green fluorescent protein (sGFP) under the control of the promoter of nuclear factor of activated T cells (NFAT) (22, 24). Together with HLA-C*06:02, melanocytes as the autoimmune target cells of the Vα3S1/Vβ13S1-TCR, the melanocyte autoantigen ADAMTSL5, and different risk-associated ERAP1 haplotypes, this assay combines the core components of the psoriatic autoimmune response. Such complex experimental conditions of a human autoimmune disease can hardly be simulated in experimental animal models.
Our results demonstrate that ERAP1 elicits melanocyte immunogenicity for the psoriatic autoimmune response by generating the autoantigenic ADAMTSL5 peptide for presentation by HLA-C*06:02. Based on this, different ERAP1 haplotypes control the extent of an autoimmune response against melanocytes and thus probably also the risk of HLA-C*06:02 carriers for psoriasis by different autoantigen yields. Interestingly, cell surface HLA-C expression proved to be much more affected by ERAP activity than that of the overall HLA-class I expression, which comprises HLA-A, HLA-B and HLA-C molecules, resembling the ERAP1-dependence of H-2Ld expression in mice (25). These findings reveal how the interaction between HLA-C*06:02 and ERAP1 drives CD8+ T-cell mediated autoimmune pathogenesis in psoriasis, and they propose a pathomechanism by which ERAP1 risk haplotypes enhance autoantigen presentation to induce CD8+ T-cell autoimmune responses in other HLA-class I-associated diseases as well. This may open new avenues for the treatment of HLA-class I associated autoimmune diseases, which target ERAP1 function.
Materials and Methods
Cell lines and cell culture
Generation of the Vα3S1/Vβ13S1-TCR CD8+ reporter T hybridoma from the αβ-TCR chains of a lesional psoriatic CD8+ T cell clone and its culture conditions have been described (20, 22). WM793 (HLA-A*01:01, HLA-A*29:01, HLA-B*35:01, HLA-B*57:01, HLA-C*04:01, HLA-C*06:02) and WM278 cells (HLA-A*02:01, HLA-A*26:01, HLA-B*13:02, HLA-B*38:01, HLA-C*06:02, HLA-C*12:03) (NCBI Biosample accession: https://www.ncbi.nlm.nih.gov/biosample/?term=SAMN03471796) were originally obtained from the Wistar Institute. They were cultured in TU2% medium containing MCDB153, 20% Leibovitz’s L15, 5 μg/ml Insulin (Sigma-Aldrich), 2% fetal calf serum (FCS) and 1.68 mM CaCl2. WM793 and WM278 cells were tested for DNA short tandem repeat (STR) analysis. HEK293T cells (HLA-A*02:01, HLA-A*03:01, HLA-B*07:02, HLA-B*07:02, HLA-C*07:02, HLA-C*07:02) were maintained in D-MEM medium supplemented with geneticin (500 μg/ml). All cell lines were negative for mycoplasma contamination.
ERAP1 knockout by CRISPR/Cas9 genome editing
Single-guide RNAs (sgRNAs) targeting ERAP1 inserted into the CRISPR/Cas9-encoding px330 plasmid and generation of ERAP1-/- HEK293T cells have been reported (26). WM793 or WM278 cells were transfected with CRISPR plasmid three times and cloned at densities of 0.3 cells/well. Successful gene editing and deletion of protein expression were confirmed by Western blotting (Fig. 1A,B) and by sequencing of targeted sites of genomic DNA.
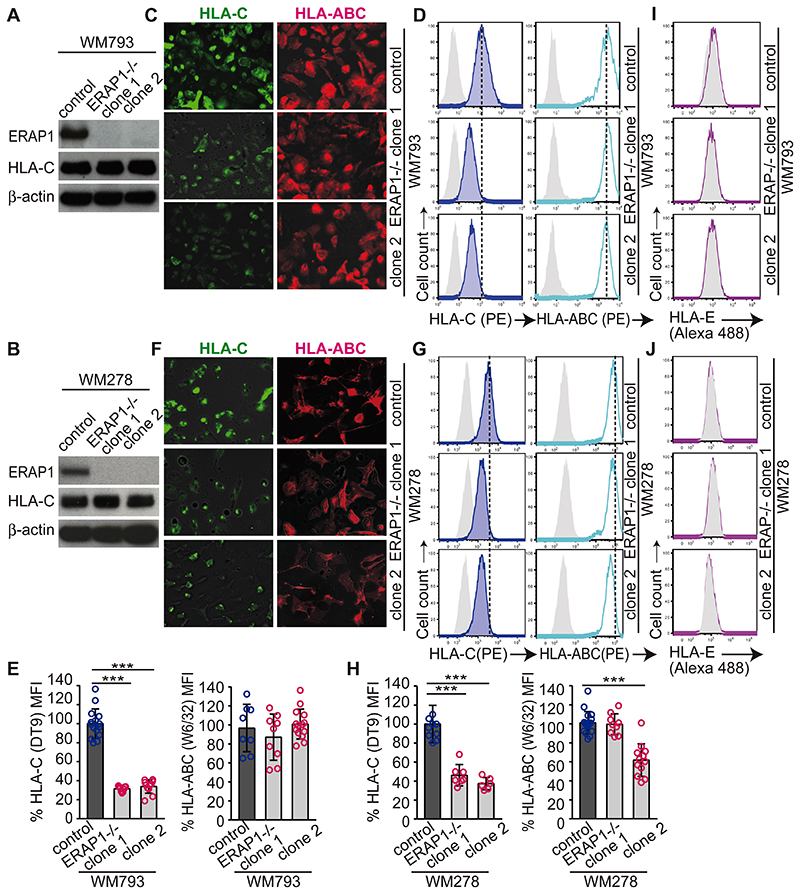
Figure 1. ERAP1 controls cell-surface HLA-C expression.
(A and B) Western blot analyses of ERAP1, HLA-C, and β-actin from parental cell lines (control) or ERAP1-/- WM793 (A) and WM278 (B) clones. (C and F) Representative images of HLA-C or HLA-ABC expression by parental cell lines or ERAP1-/- WM793 (C) and WM278 (F) clones from two independent experiments (original magnification x200). (D and G) Representative flowcytometric panels of HLA-C (DT-9) and HLA-ABC (W6/32) stainings with isotype controls (gray) of parental cell lines or ERAP1-/- WM793 (D) and WM278 (G) clones from technical triplicates of three or more independent experiments. (E and H) Mean fluorescence intensity (MFI) of HLA-C or HLA-ABC staining of parental cell lines and ERAP1-/- WM793 (E) or WM278 (H) clones was assessed by flowcytometry and normalized to means of parental cell-data in each analysis. Data are summarized, compared to parental control cells by non-paired t-test (*** P < 0.005) and shown as mean ± s.e.m. (I and J). Representative flowcytometric analyses of HLA-E with isotype staining (gray) for parental cells or ERAP1-/- WM793 (I), or W278 (J) clones from two independent experiments.
Evaluation of HLA expression
Parental and ERAP1-/- WM793, WM278 and HEK293T cells were seeded at growth-adjusted cell numbers prior to HLA staining in 24 well plates. HEK293T cells were incubated with IFN-γ (1 ng/ml) to induce sufficient HLA-C expression. On the next day, cells at 80-90% density were detached using 0.2% EDTA/PBS, and stained with HLA-C antibody (DT-9, BD Bioscience #566372), HLA-ABC antibody (W6/32, Biolegend #311406), or corresponding isotype IgG (IgG2b, IgG2a Biolegend #401208, #400214), all conjugated with phycoerythrin. Data were acquired by flowcytometry and analysed by FlowJo (TreeStar, 889). Gating strategy is given in Supplemental Fig. 1A. DT-9 antibody is considered the most specific HLA-C antibody, though it also detects HLA-A*23:01, A*80:01, HLA-B*13:01, B*35:01, B*40:06, B*73:01, and HLA-E (27). Only WM793 cells express HLA-B*35:01, which is recognized by DT-9 with low affinity. HLA-E (Abcam #ab11821) and corresponding isotype (BD pharmingen #556648) stainings were detected by goat anti-mouse IgG (H+L) cross-absorbed antibody Alexa Fluor 488 (Invitrogen #A11001).
To stain adherent cells, chamber glass slides were coated with 0.5 mg/ml poly-D-lysine (Sigma Aldrich #P7280) at 4°C for overnight and seeded with parental cell lines or ERAP1-/- WM793 and WM278 cell clones adjusted to yield comparable cell density on analysis. After 2 d culture, cells were reacted with antibodies for HLA-C, HLA-ABC or corresponding isotype control (Biolegend #401216, #400264, #311441, and DT-9, all without azide), and anti-mouse IgG (H+L) Alexa Fluor 488 or 594 secondary antibody (Invitrogen #A11001, #A11005). After washing, the cells were mounted with Fluorescence Mounting Media (DAKO #S3203).
Stimulation of the CD8+ Vα3S1/Vβ13S1-TCR reporter T-hybridoma cell line
Quantitative evaluation of TCR stimulation was validated using TCR cross-linking by increasing concentrations of plate-bound CD3 antibody (eBioscience #14-0032-82) or by stimulation with serially diluted synthetic 9mer ADAMTSL5 peptide presented by stably HLA-C*06:02-transfected COS-7 cell (Supplemental Fig. 2A–C). After 24 h of stimulation, the degree of hybridoma activation was determined with respect to the percentage and mean fluorescence intensity (MFI) of sGFP+ hybridoma cells. The percentage of sGFP+ hybridoma cells showed a more direct association with the degree of TCR ligation than mean fluorescence intensity (MFI) of sGFP and was therefore used for quantification of TCR stimulation in this study. The maximal possible yield of 50% to 70% of sGFP-positive hybridoma cells reflects their maximum activatability also observed in former experiments that had established this technology (24, 28).
To determine hybridoma stimulation by either parental or ERAP1-/- WM793 or WM278 cells, cells were seeded in 48 well plates. WM278 cells required incubation with IFN-γ (1 ng/ml) for TCR stimulation. Vα3S1/Vβ13S1-TCR hybridoma cells were added 24 h later. As in all TCR stimulation experiments, sGFP induction in Vα3S1/Vβ13S1-TCR hybridoma cells was examined after 24 h co-culture by flowcytometry. As negative/positive controls in all stimulation experiments, Vα3S1/Vβ13S1-TCR hybridoma cells were incubated in culture plates either untreated or pre-coated with CD3 antibody (eBioscience 17A2, 2 μg/ml in PBS).
To compare stimulation of the Vα3S1/Vβ13S1-TCR hybridoma by ADAMTSL5 8mer and 9mer, stably HLA-C*06:02-transfected COS-7 cells or IFN-γ-treated (1 ng/ml) WM278 cells were incubated with synthetic ADAMTSL5 peptides (10 μg/ml), and sGFP induction of Vα3S1/Vβ13S1-TCR hybridoma cells was measured after 24 h of coculture (Supplemental Fig. 3B, C). ADAMTSL5 peptides were synthesized with purity greater than 95% by Thermo Fisher.
Plasmid-based transfection to evaluate HLA expression or TCR stimulation
Cloning of ERAP1 haplotypes has been described (26). For expression of short antigenic peptides, forward and reverse oligonucleotides were annealed and ligated into pcDNA3.1D/V5-His-TOPO® vector using the Directional TOPO Expression kit (Invitrogen) as described (22).
To compare ERAP1 Hap2 and Hap10 activity, ERAP1-/- WM793 clones were transfected with pcDNA-ERAP1 Hap2 or Hap10 (100 ng) at 60-80% cell density using FuGENE HD (Promega). 24 h after transfection, HLA expression was evaluated, or Vα3S1/Vβ13S1-TCR hybridoma cells were added and assessed for sGFP induction after 24 h of co-culture by flowcytometry. Parental or ERAP1-/- HEK293T cells were seeded on 48 well plates and co-transfected with pRSV-HLA-C*06:02 (75 ng), plasmid encoded ADAMTSL5 peptides (75 ng) and either pcDNA-ERAP1 Hap2, Hap10, or pcDNA-vector (75 ng) using FuGENE HD. 24 h later, Vα3S1/Vβ13S1-TCR hybridoma cells were added. Staining with CD8-PerCPCy5.5 (Biolegend #344710) differentiated hybridoma cells from ERAP1-/- HEK293T cells. Gating strategy to examine TCR hybridoma stimulation is given in Supplemental Fig. 2A.
RNA isolation and quantification
RNA was isolated from cell lines (RNeasy mini kit, Qiagen). The same amounts of total RNA were reverse transcribed into cDNA with random primers using SuperScript III (Invitrogen) according to the manufacture’s protocol. Quantitative PCR was performed in triplicate using Light Cycler 2.0 (Roche). Porphobilinogen deaminase (PBGD) was used as internal standard for quantifying ADAMTSL5 mRNA.
Analysis of protein expression
Adherent cells were detached using 0.2% EDTA/PBS, washed twice with PBS and lysed in Cold Spring Harbor Buffer: 50 mM Tris, pH 7.4, 0.25 M NaCl, 1 mM EDTA, 0.1% Triton X-100, with phosphatase inhibitors (PhosSTOP, Roche), and protease inhibitors (Complete, Mini, EDTA-free; Roche). Protein concentration was measured using the BCA Protein Assay Kit (Pierce). Protein gel electrophoresis and immunoblotting were performed with same amounts of 5-15 μg of denatured protein lysate by using the Xcell SureLock Mini-Cell system with 4–12% gels in MES SDS buffer and PVDF membranes in each experiment (all from Invitrogen). After blocking (Roche #11921681001), blots were incubated with primary antibodies (R&D Systems # AF2334, 1/2000; Santa Cruz #sc-166088, 1/500; Sigma Aldrich #SAB3500142, 1/1000) overnight at 4°C, washed with 0.1% Tween 20 in PBS and incubated with HRP-conjugated secondary antibodies (Abcam #6885, Cell Signaling #7076S, Abcam #6877, diluted as recommended) for 1 h. Subsequently, blots were washed and visualized by chemoluminescence (GE Healthcare # 89168-782). Protein levels of β-actin served as a control for constant loading and transfer.
In vitro peptide digestion assays
ADAMTSL5 11mer peptide (20 nM) was incubated with purified recombinant ERAP1 Hap2 or Hap10 (2μg) at 37°C in 50 mM Tris-HCl (pH 8.0) for up to 2 h. Reactions were stopped by the addition of 0.5% trifluoroacetic acid. Each sample was analyzed for peptide digestion by RP-HPLC (Shimadzu) on a 2.1 mm × 250 mm C18 column (Vydac) over a gradient of 18-34% acetonitrile and a flow rate of 0.25 ml/min. Peptide peaks were analyzed using LC Solutions software (Shimadzu). Synthetic peptides (20 nM) and buffer only samples were run and analyzed in identical conditions to establish the ADAMTSL5 peptide series retention times and the absence of cross-contamination (Fig. 5A).
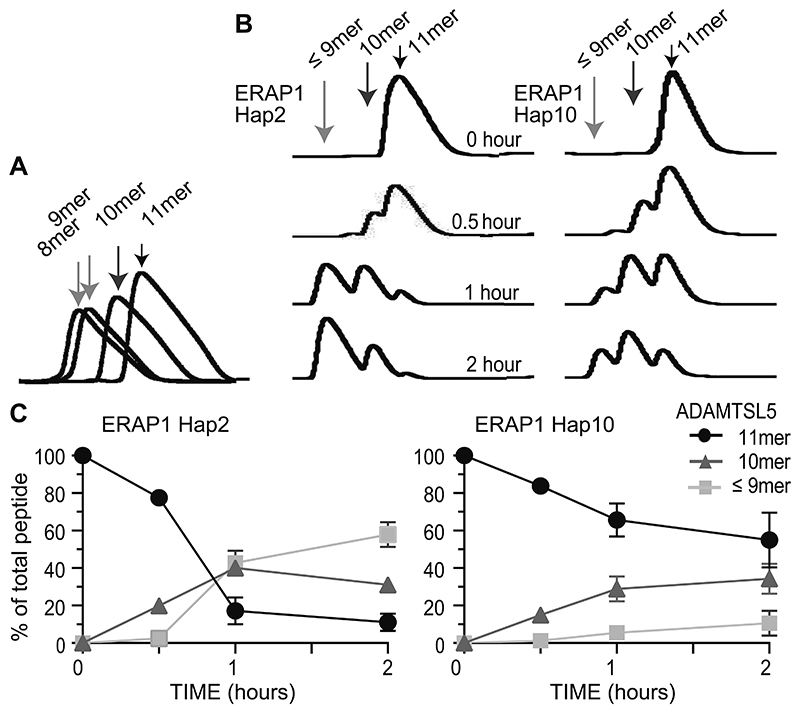
Figure 5. ERAP1 haplotypes differently generate the ERAP1-dependent autoantigenic ADAMTSL5 epitope.
(A) Discrimination of ADAMTSL5 peptides by reversed-phase HPLC. Synthetic peptides (20 nM) and buffer only samples were run and analyzed in identical conditions to establish the ADAMTSL5 peptide series retention times and the absence of cross-contamination. (B) Time-dependent digestion of ADAMTSL5 11mer to 10mer and 9mer/8mer by ERAP1 Hap2 or Hap10 was analyzed by reversed-phase HPLC and is shown by representative plots from three independent experiments. (C) Generation of ADAMTSL5 11mer to the intermediate 10mer and final 9mer/8mer by ERAP1 Hap 2 or Hap10. Data summarize three independent experiments.
Double immunofluorescence staining
Lesional skin biopsies were obtained from patients with clinically and histologically verified chronic plaque psoriasis, normal skin specimens from donors undergoing plastic surgery. Patients and healthy individuals participated voluntarily and gave written informed consent. The study of human material was performed in accordance with the Helsinki Declaration and approved by the Ethics Committee of the Ludwig-Maximilian-University, Munich. Immunofluorescence stainings were performed as previously described with several modifications (22). Heat-induced antigen retrieval used Tris-EDTA buffer for MART1/ERAP1 and citrate buffer (10 mM Citric Acid, 0.05% Tween 20, pH 6.0) for HLA-C/cKit or HLA-C/ERAP1 staining. Slides were incubated with antibodies against ERAP1 (R&D Systems #AF2334, goat, 1/100), MART1 (Leica A103, 1/100), HLA-C (Santa Cruz #sc-166088, 1/100), or cKit (DAKO #A4502, 1/400), and washed with PBS, 0.1% Triton, or with 0.1% saponin-containing PBS for ERAP1 staining. For HLA-C staining, slides were blocked with 5% goat serum at 4 °C overnight. Antibody reactivity was detected using biotinylated anti-goat IgG antibody and streptavidin-Alexa 488, donkey highly cross-adsorbed anti-mouse IgG (H+L) secondary antibody Alexa Fluor 594 (Invitrogen #A21207), Goat anti-mouse IgG (H+L) cross-absorbed antibody Alexa 488 (Invitrogen #A11001), or Goat anti-Rabbit IgG (H+L) highly cross-adsorbed antibody Alexa 594 (Invitrogen #A11037). DAPI counterstained cell nucleoli. MART1 and cKit antibodies marked the same cell population in the epidermis of healthy skin and psoriatic lesions, and ERAP1 reactivity of the goat antibody overlapped with that of the rabbit ERAP1 antibody (Abcam #ab124669), validating the detection methods (Supplemental Fig. 4A).
Epidermal areas of 5 randomly and continuously selected view fields were photodocumented using an Axio Observer microscope (Zeiss, Visitron). Isotype staining with the same concentration of corresponding isotype antibody in each experiment determined background levels and positivity thresholds. Fluorescence intensity was determined using Image J software. Selection gate for whole epidermis or basal layers were defined on DAPI staining and basal layers by one cell-width from basal membrane. MART1+ or cKit+ gate excluded the background staining in keratinized layer and cKit+ cells in the dermis. Selection gate was transferred onto HLA-C or ERAP1 staining, and fluorescence intensity was quantified for each selection (Supplemental Fig. 4B,C). Results represent median values of 5 sequential view fields from each individual.
Statistics
Statistical analyses were performed using GraphPad Prism v.7 (GraphPad Software). Microsoft excel was used to store data. Kruskal-Wallis H-test was used for multiple comparisons and Bonferroni correction was applied. When P-value of Kruskal-Wallis H-test was significant for comparing multiple groups, two group comparison was performed using a parametric t-test for hybridoma experiments or HLA staining, non-parametric Mann-Whitney U-test for unpaired continuous variables, or Wilcoxon signed-rank test for paired clinical samples between two groups. Two tailed P < 0.05 was considered significant. Although some groups were assessed to have non-normal distributions in hybridoma stimulation experiments or HLA staining data (Shapiro-Wilk W Test), significance outcomes based on the results from Mann-Whitney U-test were similar to those from a parametric t-test. In hybridoma stimulation experiments, or HLA staining, group sizes were determined based on the results of preliminary experiments, considering the variation and mean of the samples. In principal, no data were excluded from analyses. Some stimulation experiments were excluded when the positive control samples in the experiments or GFP transfection had failed. Investigators were not blinded for samples. All data are not subjective but based on quantifications.
Results
ERAP1 controls cell surface-expression of HLA-C
To investigate the role of ERAP1 in the immunogenicity of melanocytes, we generated ERAP1-deficient clones from two HLA-C*06:02-positive melanoma cell lines, WM793 and WM278, by CRISPR/Cas9 gene editing (Fig. 1A,B). These cell lines are potent stimulators of the HLA-C*06:02-restricted autoreactive psoriatic Vα3S1/Vβ13S1 TCR and can replace primary melanocytes for analysing melanocyte-specific autoreactivity (22). In our experiments we used each two knockout clones to avoid possible clone-specific effects.
Expression of HLA-class I molecules on the cell surface requires binding of peptides (29). The significance of ERAP1 for cell surface HLA-class I expression by human cells is still debated. ERAP1 knockout in WM793 and WM278 cells reduced HLA-C cell surface expression measured by the reactivity of an HLA-C antibody (DT-9) by >50%, whereas the overall HLA-ABC expression decreased only moderately (Fig. 1C–H). Although the HLA-C antibody also reacts against HLA-E, HLA-E antibody staining revealed only minimal HLA-E expression in these cell lines, which remained essentially unchanged by ERAP1 knockout and thus had no significant influence on the overall result (Fig. 1I,J). Furthermore, DT-9 recognizes HLA-B*35:01 with low affinity (27), which is expressed by WM793 cells. Analysis of WM278 verified that ERAP1 knockout primarily decreased the cell surface expression of HLA-C rather than of HLA-A or HLA-B, as this cell line does not express HLA-A or HLA-B alleles recognized by DT-9. Western blot analysis revealed unchanged total cellular expression of HLA-C upon ERAP1 knockout (Fig. 1A,B), indicating that the lack of ERAP1 activity had not affected the synthesis of HLA-C but caused intracellular HLA-C retention.
To validate these results, we examined the effect of ERAP1 knockout on the HLA-class I expression in another cell line, HEK293T cells (Supplemental Fig. 1B–E). These cells require IFN-γ to induce HLA-C expression. As in WM278 and WM793 cells, HLA-C expression was significantly decreased on the cell surface but not intracellularly in ERAP1-/- HEK293T cells, whereas the overall HLA-ABC cell surface expression as well as HLA-E expression remained largely unaffected. Thus, ERAP1 activity strongly affects HLA-C cell surface expression, presumably by generating self-peptides for presentation.
An ERAP1 risk haplotype for psoriasis is associated with higher HLA-C cell surface expression
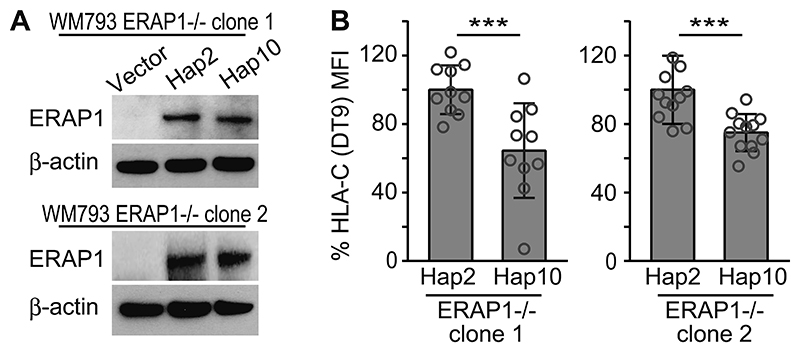
GWASs had revealed that certain ERAP1 SNPs are associated with the risk of psoriasis in carriers of HLA-C*06:02 (6–8). The different coding ERAP1 single nucleotide polymorphisms (SNPs) are not transmitted individually but are inherited in different combinations as sets of DNA variations in the ERAP1 gene that form ten main haplotypes (Hap1-10) (30). Hap2 is a risk haplotype in epistasis with HLA-C*06:02 in psoriasis, while Hap10 protects against the disease. To determine the role of the two ERAP1 haplotypes with opposite effects on psoriasis risk in our experimental system of the psoriatic autoimmune response we restored ERAP1 expression in ERAP1-/- WM793 clones 1 and 2 with Hap2 or Hap10. Western blot showed comparable protein expression of both ERAP1 haplotypes in each clone (Fig. 2A). Reconstitution of the ERAP1-/- WM793 clones with Hap2 induced a significantly higher HLA-C surface expression compared to Hap10 (Fig. 2B). Thus, the psoriasis risk haplotype, Hap2, mediates higher HLA-C expression than Hap10.
Figure 2. Differential effects of ERAP1 haplotypes Hap2 and Hap10 on cell-surface HLA-C expression by ERAP1-/- cell clones.
(A) Western blot analyses of ERAP1 or β-actin from two ERAP1-/- WM793 clones transfected with pcDNA vector control, ERAP1 Hap2 or Hap10. Hap2 is a psoriasis risk haplotype, while Hap10 is protective. (B) ERAP1-/- WM793 clones reconstituted with psoriasis risk-associated ERAP1 haplotypes, Hap2 or Hap10, were assessed for HLA-C antibody (DT-9) reactivity. Mean fluorescence intensity (MFI) was assessed by flowcytometry in technical triplicates in three independent experiments, normalized to mean MFI of Hap2-reconstituted clones in each experiment. Data are summarized, compared by non-paired t-test (*** P < 0.005) and shown as mean ± s.e.m.
ERAP1 controls HLA-C*06:02-restricted melanocyte immunogenicity
HLA-class I expression levels determine target-cell immunogenicity in malignant melanoma and other tumours, and reduced cell membrane HLA expression is thought to contribute to tumour escape from immune recognition (31). To assess the relevance of ERAP1 for the HLA-C*06:02-restricted immunogenicity of melanocytes, we performed stimulation experiments in which we co-cultured WM278 and WM793 cells with the Vα3S1/Vβ13S1 TCR hybridoma. We measured the extent of stimulation by the percentage of hybridoma cells induced to express sGFP, which shows a direct association with the degree of TCR ligation (Supplemental Fig. 2A–C) (32). As observed in former experiments that had established this technology (28), depending on the mode of stimulation it may reach a maximal possible yield of 50% to 70% activatable cells.
In parallel with the reduced HLA-C expression, ERAP1 knockout greatly reduced the ability of WM793 and WM278 cells to stimulate the HLA-C*06:02-restricted Vα3S1/Vβ13S1 TCR, as measured by the percentage of sGFP+ hybridoma cells (Fig. 3A,B). As shown by comparison with untransfected ERAP1-/- cells, reconstitution with the psoriasis risk haplotype Hap2 restored the ability of the ERAP1-/- melanoma cell clones to stimulate the Vα3S1/Vβ13S1 TCR to a significantly greater extent than reconstitution with Hap10 (Fig. 3C,D) at comparable protein expression of both haplotypes (Fig. 2A). Thus, in our in vitro system of the psoriatic autoimmune response, ERAP1 is essential for melanocyte immunogenicity, with the psoriasis risk haplotype, Hap2, conferring higher immunogenicity and cell surface HLA-C expression to melanocytes than the protective haplotype, Hap10.
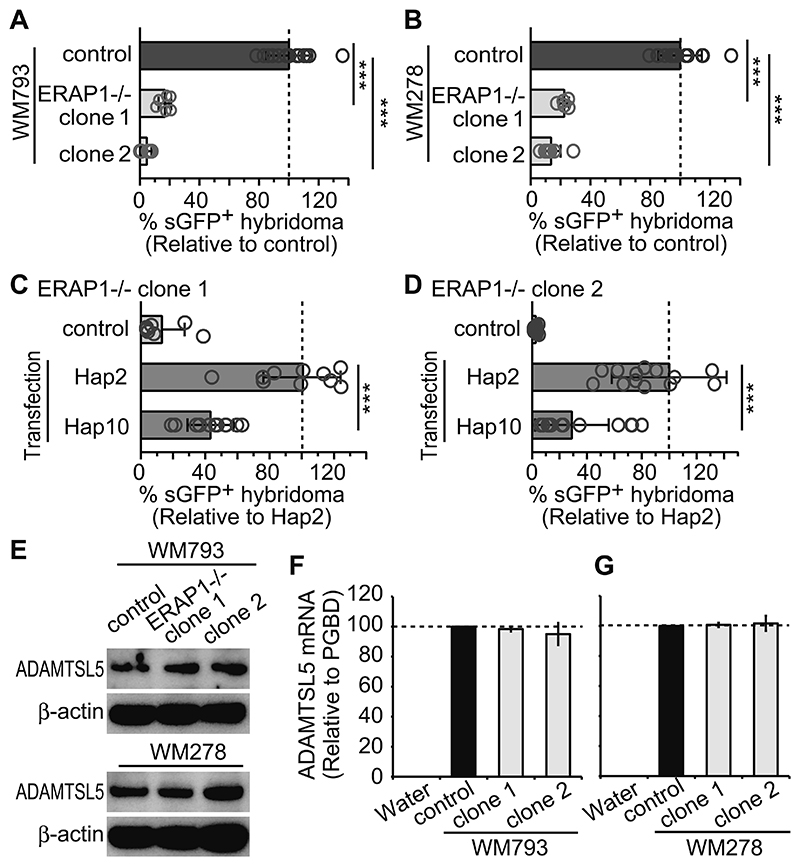
Figure 3. ERAP1 controls melanocyte immunogenicity for the Vα3S1/Vβ13S1 TCR.
(A and B) Vα3S1/Vβ13S1-TCR hybridoma stimulation by co-culture with parental cell lines (control) or ERAP1-/- WM793 (A) or WM278 (B) clones. Percentages of GFP+ hybridoma cells are assessed by flowcytometry and normalized to means of parental cell-data in the same experiment. Data are each summarized from triplicates of three independent experiments and compared to parental control-cell data by non-paired t-test (*** P < 0.005) and shown as mean ± s.e.m. Dashed lines indicate 100% reference value. (C and D) TCR-hybridoma activation by ERAP1-/- WM793 cell clone 1 (C) and clone 2 (D) reconstituted with either ERAP1 Hap2 or Hap10. Stimulation by ERAP1-/- WM793 cells served as control. Percentages of GFP+ hybridoma cells were normalized to mean value induced by Hap2-reconstituted cells in each experiment. Data are summarized from technical triplicates from two independent experiments, and results from Hap2 or Hap10 reconstitution are compared by non-paired t-test and shown as mean ± s.e.m. (E) Western blot analyses of ADAMTSL5 or β-actin from parental cell lines or two ERAP1-/- WM793 and WM278 clones. (F and G) Relative ADAMTSL5 transcript levels in parental cell lines or ERAP1-/- WM793 (F) and WM278 (G) clones were assessed in triplicates by qPCR. Water served as negative control.
Generation of the autoantigenic ADAMTSL5 epitope requires ERAP1 trimming
Stimulation of the Vα3S1/Vβ13S1 TCR by melanocytic cells reflects the autoimmune response against a peptide from the psoriatic autoantigen, ADAMTSL5 (22). Western blot and qPCR analyses showed comparable transcription and expression of ADAMTSL5 in ERAP1-/- WM278 and WM793 clones and in the parental cell lines. (Fig. 3E–G). This rules out that the loss of immunogenicity of ERAP1-/- WM278 and WM793 cells is due to decreased expression of the source protein and suggests instead that the ADAMTSL5 epitope must initially be produced as longer precursors that are subsequently trimmed in the endoplasmic reticulum.
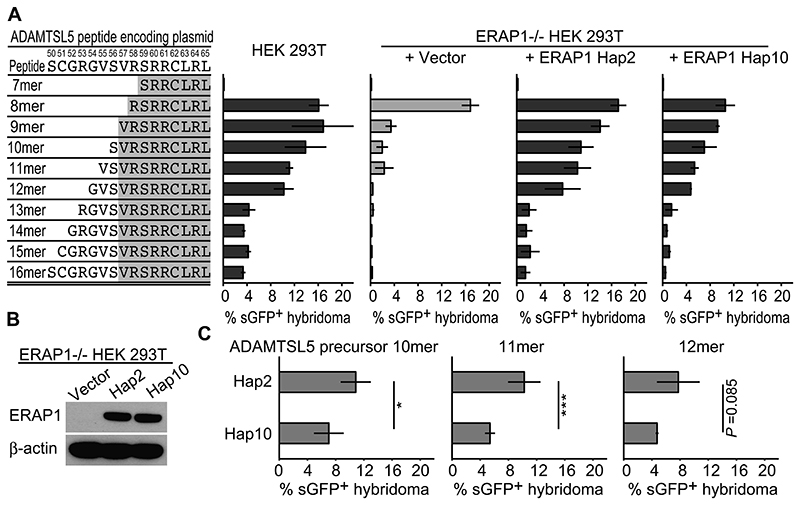
We therefore investigated the role of ERAP1 in the generation of the ADAMTSL5 epitope and cloned elongated ADAMTSL5 precursors or shortened ADAMTSL5 peptides into pcDNA3.1 expression vector. For presentation we co-expressed them together with HLA-C*06:02 in HEK293T cells, which are suitable for investigating the immunogenicity of these self-peptides because they lack endogenous antigenicity for the Vα3S1/Vβ13S1 TCR. Peptide stimulation was measured by coculture-induced sGFP expression in Vα3S1/Vβ13S1-TCR hybridoma cells. Using this experimental approach, recombinant expression of ADAMTSL5 peptides with NH2-terminal extensions up to a length of 12 amino acids in wild-type HEK293T cells induced a substantial activation of the Vα3S1/Vβ13S1 TCR, although the magnitude of TCR activation gradually decreased with increasing peptide length (Fig. 4A). NH2-terminal extensions beyond a length of 12 amino acids strongly reduced the ability of ADAMTSL5 peptides to stimulate the TCR, while C-terminally extended peptides did not ligate the Vα3S1/Vβ13S1 TCR (Supplemental Fig. 3A). This is consistent with findings that the proteasome generates the C-terminus of antigenic peptides (33). Antigenicity was also lost for ADAMTSL5 peptides truncated at the NH2- or C-terminus, as this leads to a loss of anchor residues for HLA-C*06:02 (Fig. 4A, Supplemental Fig. 3A) (34, 35).
Figure 4. Generation of the autoantigenic ADAMTSL5 epitope from NH2-elongated precursor peptides requires ERAP1 trimming.
(A) Vα3S1/Vβ13S1-TCR hybridoma activation by co-culture with the parental cell line (control) or ERAP1-/- HEK293T cells cotransfected with HLA-C*06:02, NH2-terminal extended or shortened plasmid-encoded ADAMTSL5 peptides and either vector control, ERAP Hap2 or Hap10. Percentages of GFP+ hybridoma cells were assessed by flowcytometry. Representative data set is shown as mean ± s.e.m. of technical triplicates from two independent experiments. (B) Western blot analyses of ERAP1 or β-actin in ERAP1-/- HEK293T cells transfected with pcDNA vector control or ERAP1 Hap2 or Hap10. (C) TCR-hybridoma stimulation by ERAP1-/- HEK293T cells co-transfected with NH2-elongated ADAMTSL5 10-12mer peptide-coding plasmids, HLA-C*06:02, and ERAP1 Hap2 or Hap10. Data are summarized from technical triplicates or quadruplicates from two independent experiments, compared by non-paired t-test (* P < 0.05, *** P < 0.005) and shown as mean ± s.e.m.
When presented by ERAP1-/- HEK293T cells, only the plasmid-encoded ADAMTSL5 8mer retained its ability to strongly stimulate the Vα3S1/Vβ13S1 TCR. The ability of ADAMTSL5 peptides 9 to 11 amino acids in length for TCR stimulation was much reduced, while longer peptides completely lost their ability to stimulate the Vα3S1/Vβ13S1 TCR in the absence of ERAP1 (Fig. 4A). Reconstitution of ERAP1-/- HEK293T cells with ERAP1 Hap2 or Hap10 restored the antigenicity of the ADAMTSL5 peptide precursors in a stimulation pattern corresponding to the parental cell lines, with Hap2 producing stronger stimulation by the ADAMTSL5 peptides than Hap10. We thus conclude that ERAP1 is involved in the generation of the autoantigenic ADAMTSL5 epitope from precursors.
The ERAP1 risk haplotype Hap2 generates significantly higher stimulatory activity of ADAMTSL5 precursor peptides than the protective haplotype Hap10
We then directly compared the activity of the two ERAP1 haplotypes with opposite effects on psoriasis risk with respect to their ability to restore the stimulatory capacity of NH2-terminally elongated ADAMTSL5 peptides, focusing on the 12-mer to 10-mer peptides. With similar protein expression (Fig. 4B), reconstitution of ERAP1-/- HEK293T cells with Hap2 mediated significantly higher stimulation of the Vα3S1/Vβ13S1 TCR by the NH2-elongated ADAMTSL5 10mer and 11mer than reconstitution with Hap10. Although we observed stronger stimulatory effects of Hap2 also for the ADAMTSL5 12mer, the difference to Hap10 was not statistically significant (Fig. 4A,C).
These experiments clearly demonstrate that ADAMTSL5 precursor peptides require NH2-terminal trimming by ERAP1 to become immunogenic. Stimulation with the plasmid-encoded peptides presented by the ERAP1-/- cells, however, may not reflect the actual length of the ADAMTSL5 epitope. The plasmid-encoded peptides are produced with an NH2-terminal methionine that should normally be removed by methionine aminopeptidases or ERAP1. Methionine cleavage by the methionine aminopeptidases, however, may be specifically hindered by valine and arginine at the NH2-terminus of the ADAMTSL5 9mer and 8mer (36, 37), so that the plasmid-encoded 9mer may not be adequately presented by HLA-C*06:02. The plasmid-encoded 8mer might instead become a 9mer through the NH2-terminal methionine and thus stimulate the Vα3S1/Vβ13S1 TCR. As synthetic peptides, both the ADAMTSL5 9mer and 8mer activate the Vα3S1/Vβ13S1 TCR, while consistent with the predicted binding affinity for HLA-C*06:02 by NetPanMHC 4.0, the 9mer stimulates the TCR hybridoma more strongly than the 8mer (Supplemental Fig. 3B,C).
In vitro digestion experiments verify that the ERAP1 risk haplotype generates higher amounts of the autoantigenic ADAMTSL5 epitope from precursor peptides
We finally investigated the ability of the ERAP1 haplotypes to trim NH2-elongated ADAMTSL5 precursor peptides by in vitro digestion experiments. We selected the NH2-elongated ADAMTSL5 11mer for this analysis, since the difference in digestion by ERAP1 Hap2 and Hap10 was most pronounced in the co-culture experiments for peptides of this length (Fig. 4C). Direct exposure of synthetic ADAMTSL5 11mer to recombinant ERAP1 haplotypes confirmed that ERAP1 trims NH2-terminally elongated ADAMTSL5 peptides to the length that stimulates the Vα3S1/Vβ13S1 TCR, although HPLC analysis of the digested products was limited in distinguishing 9mer from 8mer (Fig. 5A,B). With repeated NH2-terminal excisions at the same peptide, ERAP1 showed de facto ability for processivity, as formerly suggested by crystal structure analysis (38). The trimming experiments further clearly revealed that Hap2 cleaved the precursor peptides much more efficiently and, in a given time period, produced more of the mature ADAMTSL5 epitope from NH2-elongated precursors than did Hap10 (Fig. 5B,C). After 1 hour, Hap2 had trimmed 89% of the ADAMTSL5 11mer to generate shorter peptides, whereas 69% remained untrimmed by Hap10. Compared to Hap10, the psoriasis risk haplotype, Hap2, thus provides a higher supply of the psoriatic autoantigen from NH2-elongated precursor peptides.
Together, these results reveal that the ADAMTSL5 peptide belongs to the ERAP1-dependent peptidome. The ERAP1 risk haplotype generates more ADAMTSL5 peptide and thus likely increases the immunogenicity of melanocytes by creating a higher density of self-peptide/HLA-C*06:02-complexes on the cell surface of these autoimmune target cells for stimulation of ADAMTSL5-specific CD8+ T cells.
A coordinated upregulation of ERAP1 and HLA-C enhances melanocyte immunogenicity in psoriasis lesions
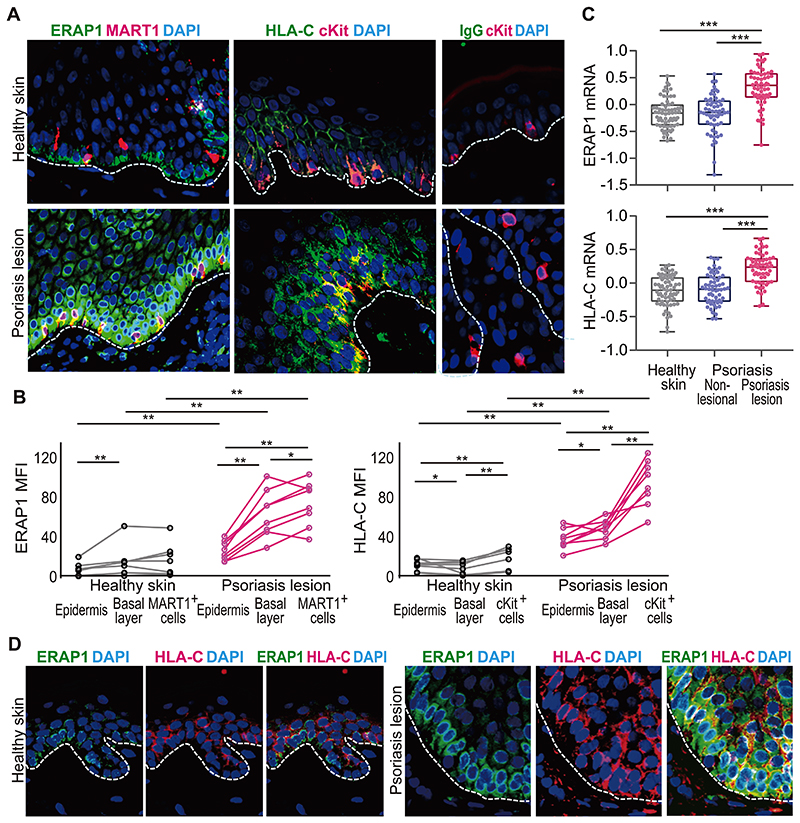
Our results from the in vitro model of the psoriatic autoimmune response reveal a mechanism by which ERAP1 and HLA-C determine the autoimmunogenicity of melanocytes and control the development of an autoimmune response in psoriasis in a mutually dependent manner. To examine the relevance of this mechanism directly in patients, we compared the expression of ERAP1 and HLA-C in healthy skin and psoriasis lesions by immunofluorescence staining. Fluorescence intensity was quantified for the whole epidermis, the basal epidermal cell layer, and specifically for melanocytes, which we distinguished by staining with MART1 (melan-A) or c-kit (CD117) antibodies (Supplemental Fig. 4A).
By immunohistologic staining and analysis of the NCBI GDS4602 data (https://www.ncbi.nlm.nih.gov/sites/GDSbrowser?acc=GDS4602) (39) we found that transcription and protein expression of both, ERAP1 and HLA-C, are dramatically upregulated in psoriasis lesions as compared to healthy skin (Fig. 6A–D). More specifically, we observed that in the epidermis of normal skin, ERAP1 expression was more pronounced in the basal epidermal layer (Fig. 6A,B,D, Supplemental Fig. 4B–D), which consists of germinative keratinocytes and melanocytes. Compared to normal skin, the inflammatory psoriasis lesions showed an overall increase in ERAP1, especially in basal layer cells, where it was most pronounced in melanocytes (Fig. 6A,B,D, Supplemental Fig. 4B–D). This indicates a high enzymatic activity of ERAP1 in psoriatic melanocytes, as it is related to the ERAP1 expression level (40). In healthy skin, melanocytes showed the highest HLA-C expression, while otherwise HLA-C had a mainly suprabasal distribution, as reported (41). In psoriasis lesions, melanocytes exhibited a significant increase in HLA-C (Fig. 6A,B).
Figure 6. Melanocytes in psoriasis lesions highly co-express ERAP1 and HLA-C.
(A), Representative immunofluorescence staining for ERAP1 (green) and MART1 (red), HLA-C (green) and cKit (red), or isotype control staining for mouse IgG and cKit staining from each two independent staining experiments. Overlay of red and green shows as yellow. Dashed lines indicate basal membrane. DAPI stained cell nucleoli (original magnification x200). (B) Mean fluorescence intensity (MFI) of ERAP1 staining in MART1+ cells or of HLA-C staining in epidermal cKit+ cells of healthy skin (n=8) or psoriasis lesions (n=8) are compared by Mann-Whitney U-test for unpaired, and Wilcoxon signed-rank test for paired samples (* P < 0.05, ** P < 0.01, *** P < 0.005). Data summarize each two independent simultaneous stainings of both healthy skin and psoriasis lesions. Each dot represents median values of 5 sequentially selected view fields from each individual. (C) ERAP1 and HLA-C transcripts in whole healthy skin (n=64), non-lesional or lesional psoriasis skin (n=58) were compared by Mann-Whitney U-test for unpaired, and Wilcoxon signed-rank test for paired samples (*** P < 0.005, (39)). (D) Representative double immunofluorescence staining to determine co-localization of ERAP1 (green) and HLA-C (red) in healthy skin and psoriasis lesion (n=3, each). Dashed line indicates basal membrane.
Thus, melanocytes in psoriatic lesions are characterized by a combined upregulation of the proteins encoded by the two most important genes for psoriasis susceptibility, HLA-C and ERAP1 (Fig. 6D). This is suggestive of an enhanced functional interaction of ERAP1 and HLA-C, ultimately leading to higher melanocyte autoimmunogenicity and potentiating the autoimmune CD8+ T-cell response in psoriasis.
Discussion
The results of our study reveal that, in psoriasis, the HLA-C*06:02-restricted autoimmune response against melanocytes is controlled by ERAP1, because the generation of a melanocyte autoantigen for presentation by HLA-C*06:02 is ERAP1-dependent. Our data furthermore show that ERAP1 variants determine the level of melanocyte autoimmunogenicity in psoriasis by generating different amounts of the autoantigenic ADAMTSL5 epitope. This mechanism functionally explains how epistasis between HLA-C*06:02 and ERAP1 variants, which is statistically defined by GWAS, affects the risk for psoriasis. The density of presented antigens on the cell surface crucially determines priming and recall responses of CD8+ T cells (42, 43). The generation of a greater amount of ADAMTSL5 peptides by the ERAP1 risk haplotype should cause a higher density of self-peptide/HLA-C*06:02-complexes on the cells surface of the autoimmune target cells and thus increase the activation of autoreactive CD8+ T cells, as reflected by the direct relationship between antigen concentration and TCR hybridoma activation (Supplemental Fig. 2C). Consequently, different trimming efficacies of different ERAP1 haplotypes for the autoantigen translate directly into different degrees of HLA-class I-restricted immunogenicity of the autoimmune target cells. This may explain why the increased production of the melanocyte autoantigen by the psoriasis risk haplotype, Hap2, mediates a greater risk of psoriasis in HLA-C*06:02 carriers as opposed to Hap10. These findings propose that ERAP1 has a true gatekeeper function for HLA-class I-restricted autoimmune responses, and they define the enzymatic activity of ERAP1 haplotypes as a central checkpoint in the development of psoriasis.
Both, stimulation experiments with NH2-elongated ADAMTSL5 peptides and the ERAP1 digestion data of ADAMTSL5 precursor peptides clearly demonstrate the ability of ERAP1 to successively remove the NH2-terminal residues of ADAMTSL5 precursors, shortening the peptide to a length required for HLA-C*06:02 binding, respectively. Our results thus reveal how the HLA-class I-restricted immunogenicity of the target cells of the psoriatic autoimmune response emerges. The findings from psoriasis propose that the yet unknown (auto)antigens of ankylosing spondylitis and Behçet’s disease, which show strong statistical epistasis between ERAP1 variants and the HLA-class I-risk alleles (4, 5), also belong to the ERAP1-dependent peptidome and become presented by the disease-associated HLA-class I molecules to elicit pathogenic CD8+ T-cell responses. Clonal CD8+ T-cell expansions in tissue lesions of ankylosing spondylitis indicate extensive antigen-driven activation and support a CD8+ T-cell driven autoimmune pathogenesis (44), similar to what we have shown in psoriasis patients (20, 22, 45). In skin lesions of Behcet’s disease, we find that antigen-activated CD8+ T cells appear to drive neutrophil recruitment and vasculitis by a strong IL-17A production (46). Other diseases also involving epistasis between HLA-class I alleles and ERAP1 variants such as bird-shot chorioretinopathy, type I diabetes, multiple sclerosis and Crohn’s disease (reviewed in (47)) may also involve this ERAP1-dependent pathogenic pathway. As such, our experimental work on psoriasis may provide insights into the still controversial role of ERAP1 and HLA-class I risk alleles in other autoimmune diseases.
HLA-class I molecules are transported from the endoplasmic reticulum to the cell surface only if they have bound peptides for presentation. HLA-C cell surface expression is strongly influenced by the diversity of peptides available for HLA-C binding (48). In our experimental system, the cell-surface expression of HLA-C on melanocytes was significantly reduced by ERAP1 knockout compared to the total HLA-class I expression comprising HLA-A, HLA-B and HLA-C. Thus, HLA-C expression in humans resembles the selective ERAP1 dependence of H-2Ld in mice, which decreased by approximately 70% by ERAP1 knockout (25). We therefore conclude that ERAP1 may generate a certain proportion of the immunopeptidome required for HLA-C expression and thus increase the availability of peptides of suitable length and sequence for HLA-C binding. This strongly emphasizes that ERAP1 activity may be of much greater importance for HLA-C- than HLA-A- or HLA-B-restricted immune responses. Furthermore, the different autoantigen densities generated by ERAP1 Hap2 and Hap10 for presentation may not only influence the probability and strength of the autoimmune response, but also have possible effects on the cytokine pattern of the stimulated T cells, as has already been shown for CD4+ T cell-responses (49, 50).
Cell surface-expression of HLA-C molecules is very low compared to HLA-A and HLA-B molecules and requires IFN-γ or other proinflammatory signals for a substantial upregulation (51, 52). Peptides that can bind to HLA-C may therefore be largely sequestered from presentation on the cell surface under immunologically quiet conditions and thus be ignored by T cells. They may, however, become immunogenic under circumstances upregulating HLA-C. By inducing ERAP1, IFN-γ increases the amount of ERAP1-generated self-peptides (53), and by inducing HLA-C (51), IFN-γ promotes their presentation. Up-regulation of HLA-class II molecule expression likely plays an important role in initiating the autoimmune response in HLA-class II-associated autoimmune diseases by enhancing antigen presentation to T cells (54). In a similar fashion, the coordinated upregulation of the two functionally linked molecules by IFN-γ in psoriasis, which is evident from our immunohistologic study of psoriatic skin sections, should enhance the immunogenicity of melanocytes. Our data thus propose that an increase in the density of HLA-class I complexed autoantigens on the cell surface by IFN-γ is an important mechanism amplifying autoreactive CD8+ T-cell activation in psoriasis and presumably also other HLA-class I-associated diseases.
Overall, using melanocytic cells as well-defined target cells and ADAMTSL5 as causative autoantigen of the HLA-C*06:02-restricted autoimmune response in psoriasis, our study unravels essential aspects of the functional mechanisms underlying the statistical epistasis between ERAP1 variants and particular HLA-class I alleles in the risk of HLA-class I-associated autoimmune diseases. Our data thus establish a plausible pathogenic concept for these diseases in which ERAP1 generates the autoantigenic epitopes for presentation by the respective disease-associated HLA-class I molecule to CD8+ T cells. Different ERAP1 haplotypes then control the likelihood and strength of the autoimmune response through generating different amounts of autoantigen for HLA-class I presentation. In this way, ERAP1 determines the HLA-class I restricted immunogenicity of the autoimmune target cells and the subsequent risk of autoimmune CD8+ T-cell activation. Specific generation of certain autoantigens by ERAP1 may thus serve as an essential checkpoint in HLA-class-I restricted CD8+ T-cell mediated autoimmune diseases, suggesting ERAP1 activity as a promising therapeutic target for the downregulation of autoimmune pathology in psoriasis and potentially other diseases with a similar genetic predisposition. Thus, our study uncovers underlying functional mechanisms of the gene-gene interaction between ERAP1 variants and HLA-class I risk alleles identified by GWASs (4–8), providing important novel insights into the pathomechanisms of common human autoimmune diseases.
Supplementary Material
Key points.
ERAP1 generates the autoantigen for presentation by HLA-C*06:02 in psoriasis
ERAP1 haplotypes control the autoimmune response by different autoantigen yields
HLA-C-restricted immune responses may be particularly dependent on ERAP1 function
Acknowledgments
The authors are grateful to Ulla Knaus, Takashi K. Satoh, and Noriaki Sasai for their critical reading. C. Kammerbauer, U. Puchta and A. Yanagida are appreciated for their technical advice. We would like to acknowledge Life Science Editor Helen Pickersgill for editorial assistance.
Abbreviations
- ADAMTSL5
ADAMTS-like protein 5
- ERAP1
endoplasmic reticulum aminopeptidase 1
- GWAS
genome-wide association study
- MFI
mean fluorescence intensity
- TAP
transporter associated with antigen presentation
Footnotes
Competing Interest Statement: The authors declare no competing financial interests.
References
- 1.Theofilopoulos AN, Kono DH, Baccala R. The multiple pathways to autoimmunity. Nat Immunol. 2017;18:716–724. doi: 10.1038/ni.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robinson J, Barker DJ, Georgiou X, Cooper MA, Flicek P, Marsh SGE. IPD-IMGT/HLA Database. Nucl Acids Res. 2020;48:D948–D955. doi: 10.1093/nar/gkz950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGonagle D, Aydin SZ, Gul A, Mahr A, Direskeneli H. ‘MHC-I-opathy’-unified concept for spondyloarthritis and Behcet disease. NatRevRheumatol. 2015;11:731–740. doi: 10.1038/nrrheum.2015.147. [DOI] [PubMed] [Google Scholar]
- 4.Evans DM, Spencer CC, Pointon JJ, Su Z, Harvey D, Kochan G, Oppermann U, Dilthey A, Pirinen M, Stone MA, Appleton L, et al. C. Australo-Anglo-American Spondyloarthritis, and C. Wellcome Trust Case Control. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. NatGenet. 2011;43:761–767. doi: 10.1038/ng.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirino Y, Bertsias G, Ishigatsubo Y, Mizuki N, Seyahi E, Ozyazgan Y, Sacli FS, Erer B, Inoko H, Emrence Z, Cakar A, et al. Genome-wide association analysis identifies new susceptibility loci for Behcet’s disease and epistasis between HLA-B*51 and ERAP1. NatGenet. 2013;45:202–207. doi: 10.1038/ng.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strange A, Capon F, Spencer CC, Knight J, Weale ME, Allen MH, Barton A, Band G, Bellenguez C, Bergboer JG, Blackwell JM, et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet. 2010;42:985–990. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun LD, Cheng H, Wang ZX, Zhang AP, Wang PG, Xu JH, Zhu QX, Zhou HS, Ellinghaus E, Zhang FR, Pu XM, et al. Association analyses identify six new psoriasis susceptibility loci in the Chinese population. Nat Genet. 2010;42:1005–1009. doi: 10.1038/ng.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang H, Jin X, Li Y, Jiang H, Tang X, Yang X, Cheng H, Qiu Y, Chen G, Mei J, Zhou F, et al. A large-scale screen for coding variants predisposing to psoriasis. Nat Genet. 2014;46:45–50. doi: 10.1038/ng.2827. [DOI] [PubMed] [Google Scholar]
- 9.Neefjes J, Jongsma ML, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol. 2011;11:823–836. doi: 10.1038/nri3084. [DOI] [PubMed] [Google Scholar]
- 10.Matsumura M, Fremont DH, Peterson PA, Wilson IA. Emerging principles for the recognition of peptide antigens by MHC class I molecules. Science. 1992;257:927–934. doi: 10.1126/science.1323878. [DOI] [PubMed] [Google Scholar]
- 11.Serwold T, Gonzalez F, Kim J, Jacob R, Shastri N. ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature. 2002;419:480–483. doi: 10.1038/nature01074. [DOI] [PubMed] [Google Scholar]
- 12.York IA, Chang SC, Saric T, Keys JA, Favreau JM, Goldberg AL, Rock KL. The ER aminopeptidase ERAP1 enhances or limits antigen presentation by trimming epitopes to 8-9 residues. Nat Immunol. 2002;3:1177–1184. doi: 10.1038/ni860. [DOI] [PubMed] [Google Scholar]
- 13.Barnea E, Melamed Kadosh D, Haimovich Y, Satumtira N, Dorris ML, Nguyen MT, Hammer RE, Tran TM, Colbert RA, Taurog JD, Admon A. The Human Leukocyte Antigen (HLA)-B27 Peptidome in Vivo, in Spondyloarthritis-susceptible HLA-B27 Transgenic Rats and the Effect of Erap1 Deletion. Mol Cell Proteom: MCP. 2017;16:642–662. doi: 10.1074/mcp.M116.066241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evnouchidou I, Kamal RP, Seregin SS, Goto Y, Tsujimoto M, Hattori A, Voulgari PV, Drosos AA, Amalfitano A, York IA, Stratikos E. Cutting Edge: Coding single nucleotide polymorphisms of endoplasmic reticulum aminopeptidase 1 can affect antigenic peptide generation in vitro by influencing basic enzymatic properties of the enzyme. J Immunol. 2011;186:1909–1913. doi: 10.4049/jimmunol.1003337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reeves E, Edwards CJ, Elliott T, James E. Naturally occurring ERAP1 haplotypes encode functionally distinct alleles with fine substrate specificity. J Immunol. 2013;191:35–43. doi: 10.4049/jimmunol.1300598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alvarez-Navarro C, Lopez de Castro JA. ERAP1 structure, function and pathogenetic role in ankylosing spondylitis and other MHC-associated diseases. Mol Immunol. 2014;57:12–21. doi: 10.1016/j.molimm.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 17.Nair RP, Stuart PE, Nistor I, Hiremagalore R, Chia NV, Jenisch S, Weichenthal M, Abecasis GR, Lim HW, Christophers E, Voorhees JJ, et al. Sequence and haplotype analysis supports HLA-C as the psoriasis susceptibility 1 gene. Am J Hum Genet. 2006;78:827–851. doi: 10.1086/503821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henseler T, Christophers E. Psoriasis of early and late onset: characterization of two types of psoriasis vulgaris. J Am Acad Dermatol. 1985;13:450–456. doi: 10.1016/s0190-9622(85)70188-0. [DOI] [PubMed] [Google Scholar]
- 19.Chang JC, Smith LR, Froning KJ, Schwabe BJ, Laxer JA, Caralli LL, Kurland HH, Karasek MA, Wilkinson DI, Carlo DJ, et al. CD8+ T cells in psoriatic lesions preferentially use T-cell receptor V beta 3 and/or V beta 13.1 genes. Proc Natl Acad Sci USA. 1994;91:9282–9286. doi: 10.1073/pnas.91.20.9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim SM, Bhonsle L, Besgen P, Nickel J, Backes A, Held K, Vollmer S, Dornmair K, Prinz JC. Analysis of the paired TCR alpha- and beta-chains of single human T cells. PloS one. 2012;7:e37338. doi: 10.1371/journal.pone.0037338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DiMeglio P, Villanova F, Navarini AA, Mylonas A, Tosi I, Nestle FO, Conrad C. Targeting CD8(+) T cells prevents psoriasis development. J Allergy Clin Immunol. 2016;138:274–276.:e276. doi: 10.1016/j.jaci.2015.10.046. [DOI] [PubMed] [Google Scholar]
- 22.Arakawa A, Siewert K, Stohr J, Besgen P, Kim SM, Ruhl G, Nickel J, Vollmer S, Thomas P, Krebs S, Pinkert S, et al. Melanocyte antigen triggers autoimmunity in human psoriasis. J Exp Med. 2015;212:2203–2212. doi: 10.1084/jem.20151093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuentes-Duculan J, Bonifacio KM, Hawkes JE, Kunjravia N, Cueto I, Li X, Gonzalez J, Garcet S, Krueger JG. Autoantigens ADAMTSL5 and LL37 are significantly upregulated in active Psoriasis and localized with keratinocytes, dendritic cells and other leukocytes. Exp Dermatol. 2017;26:1075–1082. doi: 10.1111/exd.13378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seitz S, Schneider CK, Malotka J, Nong X, Engel AG, Wekerle H, Hohlfeld R, Dornmair K. Reconstitution of paired T cell receptor alpha- and beta-chains from microdissected single cells of human inflammatory tissues. Proc Natl Acad Sci USA. 2006;103:12057–12062. doi: 10.1073/pnas.0604247103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hammer GE, Gonzalez F, Champsaur M, Cado D, Shastri N. The aminopeptidase ERAAP shapes the peptide repertoire displayed by major histocompatibility complex class I molecules. Nat Immunol. 2006;7:103–112. doi: 10.1038/ni1286. [DOI] [PubMed] [Google Scholar]
- 26.Reeves E, Wood O, Ottensmeier CH, King EV, Thomas GJ, Elliott T, James E. HPV Epitope Processing Differences Correlate with ERAP1 Allotype and Extent of CD8(+) T-cell Tumor Infiltration in OPSCC. Cancer Immunol Res. 2019;7:1202–1213. doi: 10.1158/2326-6066.CIR-18-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas R, Apps R, Qi Y, Gao X, Male V, Ge D, Fellay J, Martin JN, Margolick J, Goedert JJ, Buchbinder S, et al. HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nat Genet. 2009;41:1290–1294. doi: 10.1038/ng.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siewert K, Malotka J, Kawakami N, Wekerle H, Hohlfeld R, Dornmair K. Unbiased identification of target antigens of CD8+ T cells with combinatorial libraries coding for short peptides. Nat Med. 2012;18:824–828. doi: 10.1038/nm.2720. [DOI] [PubMed] [Google Scholar]
- 29.Blum JS, Wearsch PA, Cresswell P. Pathways of antigen processing. Annu Rev Immunol. 2013;31:443–473. doi: 10.1146/annurev-immunol-032712-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ombrello MJ, Kastner DL, Remmers EF. Endoplasmic reticulum-associated amino-peptidase 1 and rheumatic disease: genetics. Curr Opin Rheumatol. 2015;27:349–356. doi: 10.1097/BOR.0000000000000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferrone S, Marincola FM. Loss of HLA class I antigens by melanoma cells: molecular mechanisms, functional significance and clinical relevance. Immunol Today. 1995;16:487–494. doi: 10.1016/0167-5699(95)80033-6. [DOI] [PubMed] [Google Scholar]
- 32.Mann SE, Zhou Z, Landry LG, Anderson AM, Alkanani AK, Fischer J, Peakman M, Mallone R, Campbell K, Michels AW, Nakayama M. Multiplex T Cell Stimulation Assay Utilizing a T Cell Activation Reporter-Based Detection System. Frontiers Immunol. 2020;11:633. doi: 10.3389/fimmu.2020.00633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Craiu A, Akopian T, Goldberg A, Rock KL. Two distinct proteolytic processes in the generation of a major histocompatibility complex class I-presented peptide. Proc Natl Acad Sci USA. 1997:10850–10855. doi: 10.1073/pnas.94.20.10850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DiMarco M, Schuster H, Backert L, Ghosh M, Rammensee HG, Stevanovic S. Unveiling the Peptide Motifs of HLA-C and HLA-G from Naturally Presented Peptides and Generation of Binding Prediction Matrices. J Immunol. 2017;199:2639–2651. doi: 10.4049/jimmunol.1700938. [DOI] [PubMed] [Google Scholar]
- 35.Mobbs JI, Illing PT, Dudek NL, Brooks AG, Baker DG, Purcell AW, Rossjohn J, Vivian JP. The molecular basis for peptide repertoire selection in the human leucocyte antigen (HLA) C*06:02 molecule. J Biol Chem. 2017;292:17203–17215. doi: 10.1074/jbc.M117.806976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frottin F, Martinez A, Peynot P, Mitra S, Holz RC, Giglione C, Meinnel T. The proteomics of N-terminal methionine cleavage. Mol Cell Proteom: MCP. 2006;5:2336–2349. doi: 10.1074/mcp.M600225-MCP200. [DOI] [PubMed] [Google Scholar]
- 37.Wingfield PT. N-Terminal Methionine Processing. Curr Protoc Protein Sci. 2017;88:6.14.11–16.14.13. doi: 10.1002/cpps.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giastas P, Mpakali A, Papakyriakou A, Lelis A, Kokkala P, Neu M, Rowland P, Liddle J, Georgiadis D, Stratikos E. Mechanism for antigenic peptide selection by endoplasmic reticulum aminopeptidase 1. Proc Natl Acad Sci USA. 2019;116:26709–26716. doi: 10.1073/pnas.1912070116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nair RP, Duffin KC, Helms C, Ding J, Stuart PE, Goldgar D, Gudjonsson JE, Li Y, Tejasvi T, Feng BJ, Ruether A, et al. P. Collaborative Association Study. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Costantino F, Talpin A, Evnouchidou I, Kadi A, Leboime A, Bonilla N, Letourneur F, Leturcq T, Ka Z, Garchon HJ, Chiocchia G, et al. ERAP1 Gene Expression Is Influenced by Nonsynonymous Polymorphisms Associated With Predisposition to Spondyloarthritis. Arthritis Rheum. 2015;67:1525–1534. doi: 10.1002/art.39072. [DOI] [PubMed] [Google Scholar]
- 41.Carlen L, Sakuraba K, Stahle M, Sanchez F. HLA-C expression pattern is spatially different between psoriasis and eczema skin lesions. J Invest Dermatol. 2007;127:342–348. doi: 10.1038/sj.jid.5700549. [DOI] [PubMed] [Google Scholar]
- 42.Bullock TN, Mullins DW, Engelhard VH. Antigen density presented by dendritic cells in vivo differentially affects the number and avidity of primary, memory, and recall CD8+ T cells. J Immunol. 2003;170:1822–1829. doi: 10.4049/jimmunol.170.4.1822. [DOI] [PubMed] [Google Scholar]
- 43.Henrickson SE, Mempel TR, Mazo IB, Liu B, Artyomov MN, Zheng H, Peixoto A, Flynn MP, Senman B, Junt T, Wong HC, et al. T cell sensing of antigen dose governs interactive behavior with dendritic cells and sets a threshold for T cell activation. Nat Immunol. 2008;9:282–291. doi: 10.1038/ni1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duchmann R, Lambert C, May E, Hohler T, Marker-Hermann E. CD4+ and CD8+ clonal T cell expansions indicate a role of antigens in ankylosing spondylitis; a study in HLA-B27+ monozygotic twins. Clin Exp Immunol. 2001;123:315–322. doi: 10.1046/j.1365-2249.2001.01440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arakawa A, Vollmer S, Besgen P, Galinski A, Summer B, Kawakami Y, Wollenberg A, Dornmair K, Spannagl M, Ruzicka T, Thomas P, et al. Unopposed IL-36 Activity Promotes Clonal CD4(+) T-Cell Responses with IL-17A Production in Generalized Pustular Psoriasis. J Invest Dermatol. 2018;138:1338–1347. doi: 10.1016/j.jid.2017.12.024. [DOI] [PubMed] [Google Scholar]
- 46.Vural S, Kerl K, Ertop P, Vollmer S, Puchta U, He M, Arakawa Y, Oktem A, Okczu Heper A, Hartmann D, Boyvat A, et al. Lesional activation of Tc17 cells in Behçet’s disease and psoriasis supports HLA-class I-mediated autoimmune responses. Brit J Dermatol. 2021 doi: 10.1111/bjd.20643. in press. [DOI] [PubMed] [Google Scholar]
- 47.Reeves E, James E. The role of polymorphic ERAP1 in autoinflammatory disease. Biosci Rep. 2018;38:BSR20171503. doi: 10.1042/BSR20171503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaur G, Gras S, Mobbs JI, Vivian JP, Cortes A, Barber T, Kuttikkatte SB, Jensen LT, Attfield KE, Dendrou CA, Carrington M, et al. Structural and regulatory diversity shape HLA-C protein expression levels. Nat Comm. 2017;8:15924. doi: 10.1038/ncomms15924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Constant S, Pfeiffer C, Woodard A, Pasqualini T, Bottomly K. Extent of T cell receptor ligation can determine the functional differentiation of naive CD4+ T cells. J Exp Med. 1995;182:1591–1596. doi: 10.1084/jem.182.5.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tao X, Constant S, Jorritsma P, Bottomly K. Strength of TCR signal determines the costimulatory requirements for Th1 and Th2 CD4+ T cell differentiation. J Immunol. 1997;159:5956–5963. [PubMed] [Google Scholar]
- 51.Gobin SJ, van Zutphen M, Woltman AM, van den Elsen PJ. Transactivation of classical and nonclassical HLA class I genes through the IFN-stimulated response element. J Immunol. 1999;163:1428–1434. [PubMed] [Google Scholar]
- 52.Snary D, Barnstable CJ, Bodmer WF, Crumpton MJ. Molecular structure of human histocompatibility antigens: the HLA-C series. Eur J Immunol. 1977;7:580–585. doi: 10.1002/eji.1830070816. [DOI] [PubMed] [Google Scholar]
- 53.Saric T, Chang SC, Hattori A, York IA, Markant S, Rock KL, Tsujimoto M, Goldberg AL. An IFN-gamma-induced aminopeptidase in the ER, ERAP1, trims precursors to MHC class I-presented peptides. Nat Immunol. 2002;3:1169–1176. doi: 10.1038/ni859. [DOI] [PubMed] [Google Scholar]
- 54.Bottazzo GF, Pujol-Borrell R, Hanafusa T, Feldmann M. Role of aberrant HLA-DR expression and antigen presentation in induction of endocrine autoimmunity. Lancet. 1983;2:1115–1119. doi: 10.1016/s0140-6736(83)90629-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.