Abstract
The functional activity and differentiation potential of cells is determined by their interaction with surrounding cells. Approaches that allow unbiased characterization of cell states while at the same time providing spatial information are of major value to assess this environmental influence. However, most current techniques are hampered by a trade-off between spatial resolution and cell profiling depth. Here, we developed a photocage-based technology that allows isolation and in-depth analysis of live cells from regions of interest in complex ex vivo systems, including primary human tissues. The use of a highly sensitive 4-nitrophenyl(benzofuran)-cage coupled to a set of nanobodies allowed high-resolution photo-uncaging of different cell types in areas of interest. Single cell RNA-sequencing of spatially defined CD8+ T cells was used to exemplify the feasibility of identifying location-dependent cell states. The technology described here provides a valuable tool for analysis of spatially defined cells in diverse biological systems, including clinical samples.
Introduction
Methods for the in-depth characterization of individual cells, such as single cell transcriptomics and proteomics, form an essential approach for our understanding of cellular function in human tissues. While these technologies are well-suited to describe the diversity of physiological and pathophysiological cell states, information on the spatial localization of the analyzed cells is lost upon tissue dissociation. Knowledge on the spatial context of individual cells is, however, critical to understand how locoregional differences in environmental signals (e.g. through cell-cell interactions or soluble mediators) impact cellular state and cell differentiation. For example, within human cancers, immune cells are found both at peritumoral and intratumoral sites. In addition, intratumoral immune cells may be further subdivided into, for instance, cells located within tumor cell nests and tertiary lymphoid organs. However, the relationship between cell state and any of these different cell locations is poorly understood1. Classical methods that are used to simultaneously assess the localization and phenotypic properties of individual cells, such as immunohistochemistry and confocal microscopy, are limited by the number of parameters that can be analyzed. While next generation imaging techniques, such as imaging mass cytometry2, multi ion beam imaging by time of flight (MIBI-TOF)3 and co-detection by indexing (CODEX)4, allow the analysis of multiple markers on tissue slides, these technologies still require upfront decisions on the genes or proteins that are assessed or lack the resolution offered by the former techniques.
The combination of the spatial resolution from microscopy approaches with the unbiased profiling capacity of single cell techniques has the potential to allow the ab initio analysis of the relationship between cell state and cellular localization. Work in transgenic mouse models has demonstrated that the localized switching of photoactivatable proteins, such as paGFP, Dronpa, or Dendra, allows the analysis of single cell transcriptomes in regions of interest with high spatial resolution5. However, approaches based on the genetic encoding of photo-sensitive proteins are not applicable to primary human tissues. To address this need, a number of methods, including Slide-seq6 and related spatial transcriptomics approaches7–12, have been developed that can be used to couple transcriptome data and spatial information from cells in human tissues. In slide-based spatial transcriptomics approaches, mRNA molecules from tissue slices are transferred to barcode-labeled surfaces6–8, thereby providing the possibility to perform an unbiased analysis of transcriptional activity at defined sites, but with the caveat that the gene expression patterns obtained are often averages from multiple cells. The specific labeling of spatially defined cells in live tissues followed by single cell analysis of those cells upon dissociation on the other hand does not suffer from this limitation. Recent work demonstrated how local uncaging of DNA barcodes that are protected by photolabile 6-nitropiperonyloxylmethyl (NPOM) groups can be used to specifically mark cells in areas of interest in tissue sections13. Here, we developed a distinct technology with a unique set of features that exploits a 4-nitrophenyl(benzofuran) (NPBF) caged protein tag to uniformly label cells in situ in live, intact human tissue. The use of the NPBF group allows highly-efficient uncaging under mild conditions (i.e. using low-intensity violet light), resulting in minimal phototoxicity. In addition, the implementation of a signal switch system, in which uncaging leads to the simultaneous loss of a first signal and gain of a second signal, allows superior separation of uncaged and caged cells. The single cell analysis of regions of interest (SCARI) technology that we describe can be used to specifically isolate cell populations from defined regions with high spatial resolution, and we exemplify the value of this approach through single cell mRNA sequencing of human T cells at defined sites.
Results
Conceptual approach to achieve localized cell marking
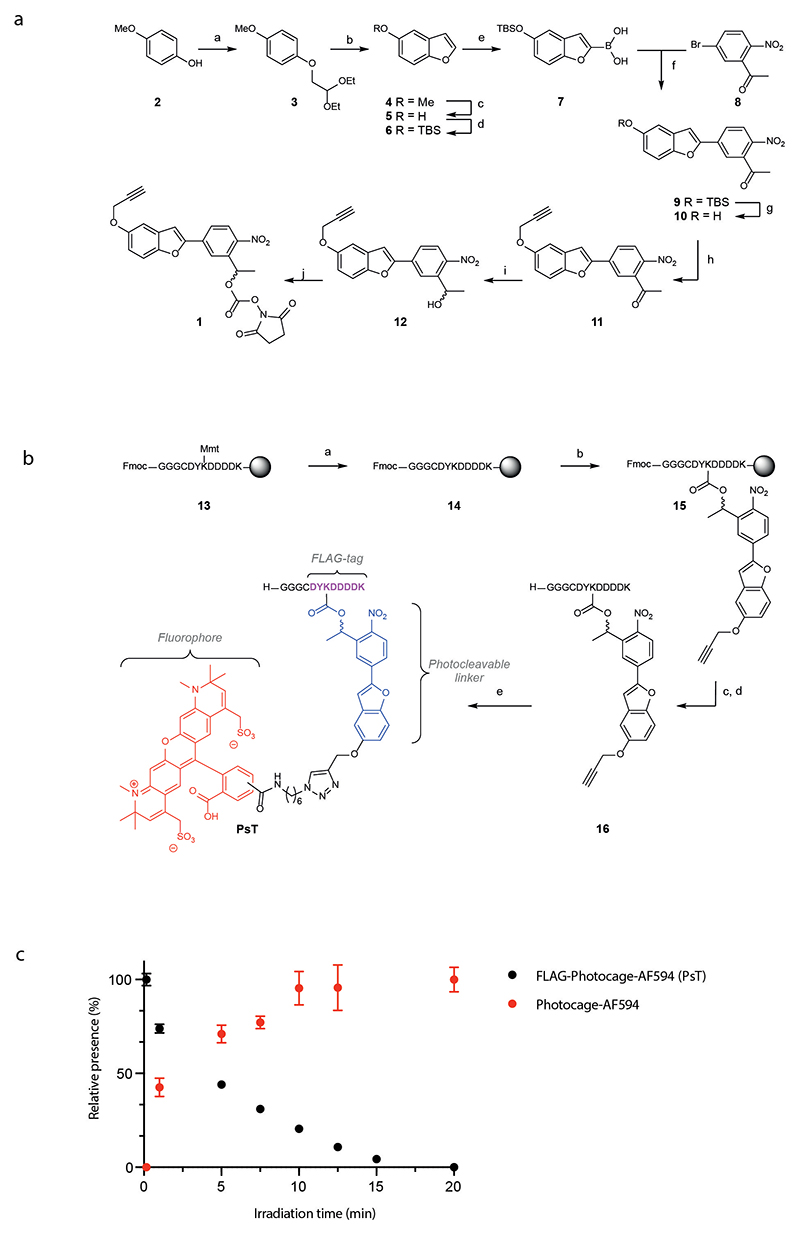
To label cells at specific tissue sites with minimal phototoxicity, we set out to create a photo-sensitive molecule in which a detectable group was shielded by a photolabile protecting group (PPG). The most commonly used PPGs for photo-uncaging in biological systems are the o-nitrobenzyl based chromophores (e.g. 6-nitroveratryloxycarbonyl (NVOC) and 6-nitropiperonyloxymethyl (NPOM)) that have, amongst others, been used to study T-cell activation kinetics14, the liberation of pro-drugs15–18 and variation in immune cell states13. These chromophores, however, have a low quantum yield (ϕu NVOC = 0.0013, ϕu NPOM = 0.0075) and therefore require either long light exposure or high intensity light to remove the photolabile group, potentially resulting in light-induced cellular damage19. In addition, in case of NVOC, photo-uncaging is accompanied by the release of toxic benzaldehyde byproducts20,21. The recently reported 4-nitrophenyl(benzofuran) (NPBF) chromophore has a superior uncaging efficiency (ϕu NPBF = 0.09)22 and, while less information is available with respect to toxicity of the released byproducts upon light exposure, this makes it an attractive starting point for the design of a photo-uncaging system. To achieve an optimal distinction between uncaged and caged cells, we designed an approach in which removal of the NPBF chromophore simultaneously leads to the loss of a first detectable (fluorescent) signal and gain of a second signal. To accomplish this, we developed a photo-sensitive tag (PsT) containing a FLAG-peptide (DYKDDDDK) that is protected by an Alexa Fluor 594 (AF594)-conjugated NPBF-protecting group (Fig. 1, first panel). We envisioned that this photocage would interfere with the binding of αFLAG antibodies, and uncaging of the FLAG-tag could thus be used to simultaneously release the AF594 dye and create a novel antibody binding site. To this end, a bifunctional NPBF photocleavable linker bearing an N-hydroxysuccinimide (NHS) carbonate on one end and an alkyne handle on the other end was developed to allow one to orthogonally install the photocage on the FLAG-epitope and click the alkyne handle of the photocage with an azide functionalized fluorophore23. To be able to label specific cell types, the caged FLAG-tag was subsequently coupled to cell lineage-specific nanobodies through sortase-based reactions24. To this purpose, the canonical FLAG-octapeptide sequence (DYKDDDDK) was extended with three N-terminal glycines25, to allow conjugation to LPXTG-modified nanobodies. In addition, a cysteine was placed in between the N-terminal GGG and FLAG-tag sequence to prevent aspartimide formation26, resulting in the final sequence GGGCDYKDDDDK. Depending on whether labeling of nanobodies is performed under reducing or oxidizing conditions, the presence of the cysteine residue yields either a single copy or two copies of the (caged) FLAG-epitope per nanobody (see below for further characterization). The FLAG-epitope contains two lysine residues that are suitable for installing the photocage. Prior work has demonstrated that the C-terminal lysine does not significantly influence antibody binding, and is thus not suitable for the intended epitope deprotection strategy27. In contrast, it has been demonstrated that the N-terminal DYKD sequence of the FLAG-peptide can be used in immunological detection procedures28, making it plausible that modification of the side chain amine of this lysine residue (Lys7) would abolish antigen recognition for at least some αFLAG antibodies.
Figure 1. Schematic approach for single cell analysis of regions of interest.
Schematic representation of the experimental approach for spatial analysis of cells. Nanobodies are labeled with the photo-sensitive tag (PsT) that consists of a FLAG-tag that is caged by a NPBF-photo-sensitive group coupled to an Alexa Fluor 594 (AF594) fluorochrome. Synthesis of the PsT is shown in Extended Data Fig. 1B. Cells or tissues are stained with PsT-labeled nanobody, and PsT-stained cells in areas of interest are uncaged using 405nm violet light. After cell harvest or tissue dissociation, single cell suspensions are stained with an αFLAG antibody that only binds to the uncaged FLAG-tag. Cells from the area of interest can after uncaging thus be identified on the basis of both their low AF594 signal and high αFLAG signal. Isolation of cells based on both properties (i.e. AF594low αFLAGhigh cells as cells within the area of interest, and AF594high αFLAGlow cells as cells outside the area of interest) results in an optimal separation between the uncaged and caged cell populations.
Synthesis of the photo-sensitive tag
The bifunctional NPBF photocage 1, was synthesized essentially as reported previously (Extended Data Fig. 1A, 2-12, final product 1)23. The modified FLAG-tag sequence GGGCDYKDDDDK was synthesized using acid labile 4-methoxytrityl (Mmt) as the protecting group for the amine on Lys7, to allow orthogonal deprotection of this side chain. An optimized method for Fmoc-based microwave-assisted solid-phase peptide synthesis (SPPS), which prevents racemization and aspartimide formation, was applied to obtain the core oligopeptide 13 (Extended Data Fig. 1B) 29. Mmt was removed under mild conditions (1% TFA), and the sidechain amine of Lys7 of 14 was reacted with the NHS carbonate of 1 to yield photocaged peptide 15 on-resin. After cleavage from the resin and deprotection of all other amino acids, the photocaged peptide 16 was purified by reverse phase HPLC, followed by copper catalysed alkyne-azide cycloaddition with AF594 azide, yielding the PsT (Fig. 1, first panel, and Extended Data Fig. 1B). Analysis of the uncaging efficiency of the PsT using 420nm light demonstrated a t1/2 of less than 5 minutes (Extended Data Fig. 1C).
Characterization of photo-sensitive αCD8 antibody reagents
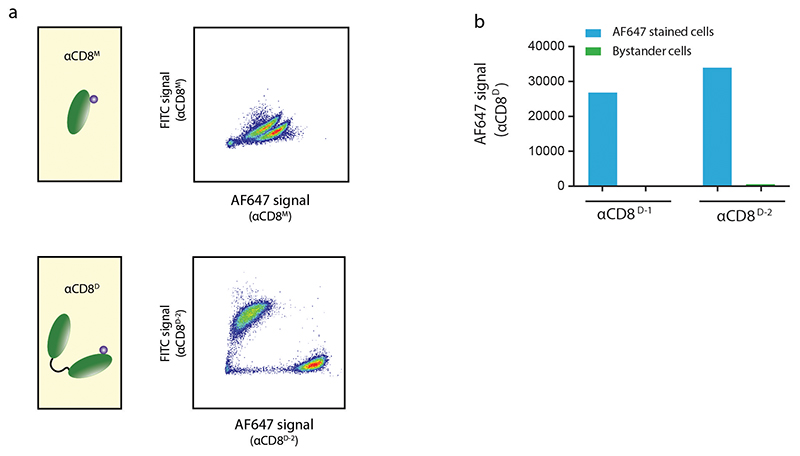
To determine the feasibility of using PsT-labeled antibodies to selectively mark cells at defined tissue sites (Fig. 1), we generated fluorochrome labeled camelid heavy chain-only fragments (nanobodies) specific for the human T cell marker CD8. Nanobodies display superior tissue penetration capacity as compared to regular antibodies due to their small size (~15kDa versus ~150kD)30, a property that may be particularly useful for in vivo or ex vivo staining of intact tissues with dense cellular and extracellular structures. To probe the effect of avidity on the selectivity and stability of cell marking, we first designed monomeric and dimeric fluorochrome-labeled αCD8 nanobodies (αCD8M or αCD8D respectively). Subsequently, the stability of cell labeling when using either monomeric or dimeric αCD8 nanobodies was determined by staining two separate human CD8+ T cell populations with αCD8 nanobodies coupled to distinct fluorochromes and subsequent mixing. Following mixing of the differentially labeled cell populations, a rapid exchange of monomeric αCD8 nanobodies was observed. In contrast, for two different dimeric αCD8 nanobody clones tested (αCD8D-1 and αCD8D-2), highly stable cell binding was observed (Extended Data Fig. 2A and B), a property essential for the intended localized cell marking.
We subsequently generated αCD8D-1 nanobodies labeled with the photo-sensitive tag (Extended Data Fig. 3A) and determined whether the NPBF cage prevents antibody binding to the FLAG-tag. To this purpose, primary human CD8+ T cells were stained with FLAG-tag αCD8D-1 nanobodies that either contain or lack the NPBF cage (αCD8D-1-PsT and αCD8D-1-T, respectively), and accessibility of the FLAG-epitope was probed using a set of different αFLAG antibodies. Notably, while certain αFLAG antibodies were insensitive to the NPBF cage (Extended Data Fig. 3B), binding of the D6W5B antibody was reduced to background levels upon caging of the lysine sidechain (Extended Data Fig. 3C).
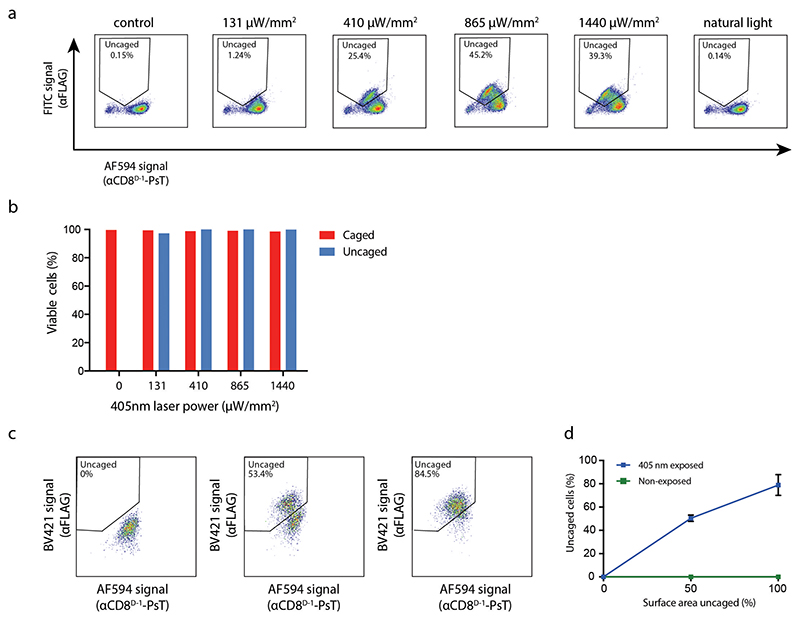
To understand whether photo-uncaging could be used to both remove the cage, and thereby the AF594 signal, and gain an αFLAG signal under conditions with limited toxicity, PsT-labeled cells were photo-uncaged by exposure to 405nm laser light, and then stained with αFLAG antibody. Flow cytometric analyses of the resulting cell populations demonstrated that uncaging both led to the intended loss in AF594 signal and gain in αFLAG antibody binding (Fig. 2A–B, Extended Data Fig. 4). Comparison of the uncaging efficiency of nanobodies that contained one or two copies of the PsT (Extended Data Fig. 5A) showed equally efficient separation between uncaged (AF594low αFLAGhigh) and caged (AF594high αFLAGlow) cells for both molecules (Extended Data Fig. 5B). Notably, while uncaging was already maximally effective at 865 μW/mm2 of violet light-exposure, cell viability remained unaffected (> 95%) up to 1440 μW/mm2 (Fig. 2C and Extended Data Fig. 6A–B). To explore the specificity of uncaging, we uncaged increasing surface areas of microwells containing CD8+ T cells in a heterogeneous population of peripheral blood mononuclear cells (PBMCs). Importantly, the fraction uncaged surface area was tightly correlated with the fraction uncaged CD8+ T cells, as measured by flow cytometry (Extended Data Fig. 6C). Of note, mixing of cell samples that did or did not contain an uncaged CD8+ T cell population showed no detectable αCD8D-1-PsT exchange between cells, confirming the stable binding of αCD8D-1-PsT throughout the sample processing pipeline (Extended Data Fig. 6D). To explore the resolution of our method, we uncaged areas with decreasing dimensions (Fig. 2D). CD8+ T cells from areas as small as 3x103μm2 (58x58μm), corresponding to 30-60 cells, could be clearly and reproducibly distinguished from background (Fig. 2E–F). Finally, the flexibility of the method was demonstrated by the successful uncaging of cell populations labeled with two additional PsT-linked nanobodies, specific for either the CD47 immune checkpoint expressed by myeloid cells, or the HLA class I molecules that are expressed by all nucleated cells (Fig. 2G).
Figure 2. Uncaging of PsT-labeled primary human CD8+ T cells.
A). Left: Confocal image of CD8+ T cells stained with αCD8D-1-PsT and αCD8D-2-FITC shown in magenta and green respectively. Overlapping signals are shown in white. Area in box with solid line is shown at higher magnification in the bottom. Scale bars represent 100μm (top) and 10μm (bottom). Right: Flow cytometry analysis of αCD8D-1-PsT labeled CD8+ T cells after αFLAG staining. Representative data from 3 independent experiments.
B) Left: Confocal image of CD8+ T cells as in A. Box with dotted line indicates area of uncaging by 405nm light exposure. Area in box with solid line is shown at higher magnification in the bottom. Scale bars represent 100μm (top) and 10μm (bottom). Right: Flow cytometry analysis of CD8+ T cells following uncaging and subsequent staining with αFLAG antibody. Representative data from 3 independent experiments.
C) Viability of uncaged and caged CD8+ T cells from partially uncaged samples (as in Fig. 2B), as measured by flow cytometric analysis of IR-dye live-dead staining. Data of 3 independent experiments (n=2 technical replicates for one experiment), plot shows mean + SD.
D) Representative microscopy images of uncaged areas with decreasing dimensions (ranging from 216x103μm2 (465x465μm) to 0.8x103μm2 (29x29μm)) containing CD8+ T cells stained with αCD8D-1-PsT, shown in magenta. Data representative of 3 independent experiments.
E) Cell recovery upon uncaging of areas of the indicated size. Single data points represent the number of uncaged αCD8D-1-PsT cells recovered in flow cytometry analysis (n=1-3 technical replicates per condition). Data representative of 3 independent experiments, plot shows mean +/-SD.
F) Quantification of true uncaging events over background by flow cytometry, after mixing cells from exposed and non-exposed samples. Bars represent fraction of cells derived from exposed (i.e. truly uncaged, blue) or non-exposed samples (i.e. background, green) within the uncaged (AF594low αFLAGhigh) population. Total cell numbers (mean across replicates) within the uncaged gate are depicted on top (n=1-3 technical replicates). Data representative of 3 independent experiments.
G) Flow cytometry analysis showing uncaging of BA/F3 cells and human T cells stained with CD47 (αCD47D-PsT) or HLA-class-I (αHLA-ID-PsT) nanobodies, respectively. Data obtained from a single experiment (n=2 technical replicates).
Local uncaging in human tumor tissue and cell systems
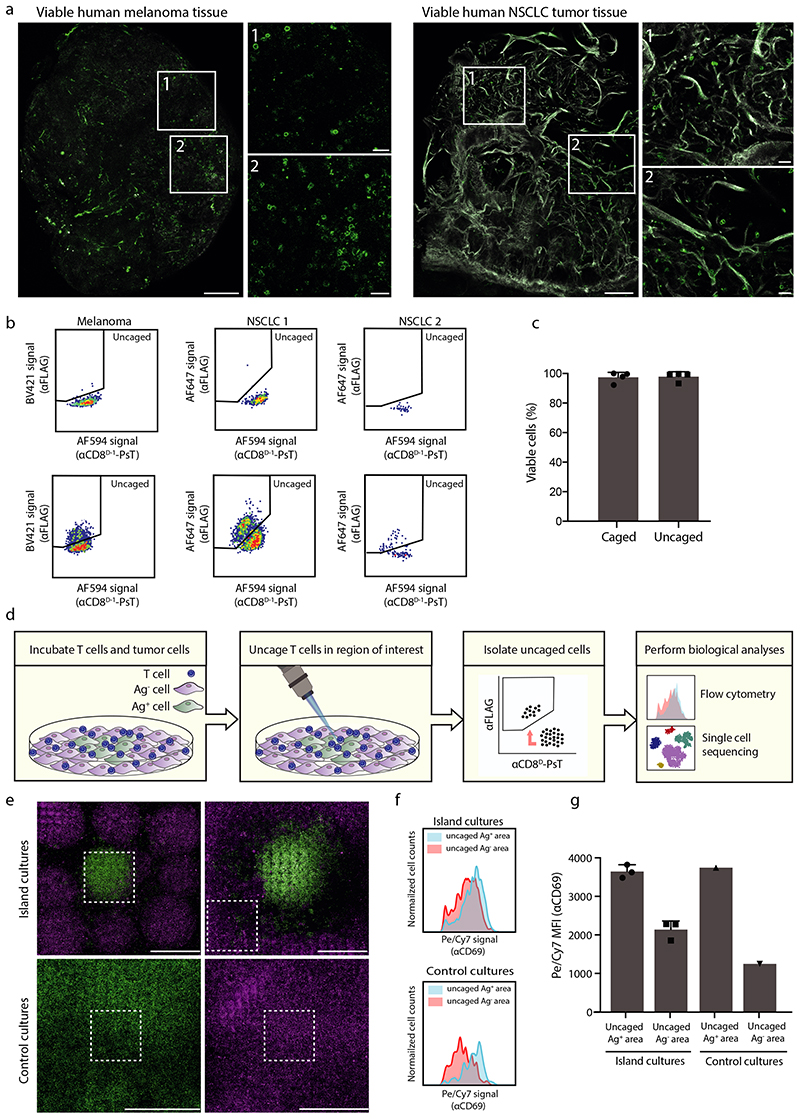
To determine the feasibility of local uncaging in more complex biological structures, we tested the efficiency of staining and uncaging of CD8+ T cells in viable human melanoma and non-squamous cell lung cancer (NSCLC) tissue. CD8+ T cells present within viable human tumor material were readily detected upon staining with αCD8D-1-PsT and αCD8D-2-FITC (Fig. 3A). Furthermore, uncaging of the αCD8D-1-PsT in areas of melanoma and NSCLC tumors resulted in a discrete population of AF594low and αFLAGhigh cells that was not observed in non-exposed tumor tissue (Fig. 3B), while viability of these CD8+ T cells remained unaffected (Fig. 3C).
Figure 3. Uncaging of CD8+ T cells in primary human tumor tissue and cell culture systems.
A) Confocal images of CD8+ T cells stained with αCD8D-1-PsT and αCD8D-2-FITC in viable human melanoma and non-small cell lung carcinoma (NSCLC) tumor tissue. CD8+ T cells are depicted in green, collagen is depicted in white. Enlarged areas are indicated with white boxes. Scale bars represent 100μm (overview) and 10μm (enlarged images). Images are representative of 3 (melanoma) and 1 (NSCLC) experiments.
B) Uncaging of αCD8D-1-PsT labeled CD8+ T cells in human tumor tissue. Indicated samples were left unexposed (top) or locally exposed to 405nm light (bottom).
C) Viability of CD8+ T cells from human tumor tissue. Viability of CD8+ T cells, measured by flow cytometric analysis of IR-dye stain, from locally uncaged regions (AF594low αFLAGhigh) is compared to viability of CD8+ T cells from non-exposed areas (AF594highαFLAGlow) (n=4 technical replicates per group). Representative data from 2 experiments are presented as mean + SD.
D) Approach to validate the ability to identify local differences in cell state.
E) Confocal images of cocultures of Ag-specific CD8+ T cells with tumor-cell islands. Top: island cultures containing Ag+ tumor cell areas (green) surrounded by Ag− tumor cell areas (magenta). Bottom: control cultures that only contain Ag+ tumor cells (bottom, left) or Ag− tumor cells (bottom, right). White boxes indicate regions uncaged by 405nm light exposure. Scale bars represent 1mm. Images are representative of 2 experiments.
F) CD69 expression on αCD8D-1-PsT and αCD8D-2-FITC stained CD8+ T cells that were uncaged in Ag+ or Ag- tumor cell islands. Top: Histograms showing CD69 expression on uncaged CD8+ T cells from Ag+ (blue) or Ag− (red) tumor islands. Bottom: Histogram of CD69 expression on uncaged CD8+ T cells from control cultures that only contained Ag+ tumor cells (blue) or Ag− cells (red).
G) Quantification of αCD69 fluorescence intensity on αCD8D-1-PsT labeled CD8+ T cells uncaged in Ag+ or Ag− regions (as shown in Fig. 3F), analyzed by flow cytometry (for islands cultures n=3 and for control cultures n=1 technical replicates per group). Representative data from 2 independent experiments are presented as mean values + SD.
To subsequently understand whether local uncaging could be used to identify location-dependent differences in cell states, we developed an in vitro cell system in which specific differences in cell states are induced in a controlled setting (Fig. 3D). To this purpose, adjacent islands of tumor cells that either lacked or expressed the HLA class I-restricted CDK4R>L neoantigen (Katushka-positive Ag− regions and GFP-positive Ag+ regions, respectively) were generated. Subsequently, αCD8D-1-PsT labeled CD8+ T cells specific for the CDK4R>L neoantigen were added to such cultures, with the expectation that T cell activation would be induced in Ag+ but not in Ag− areas. Following 4 hours of co-culture, CD8+ T cells in either Ag+ or in Ag− areas were uncaged and then isolated by cell sorting (Fig. 3E). As a control, uncaged CD8+ T cells were isolated from separate control cultures that either only contained Ag+ tumor cells or Ag− tumor cells. To test for spatial differences in T cell activation, expression of the T cell activation marker CD69 was compared on uncaged T cells that were either derived from Ag+ tumor cell areas or from Ag− tumor cell areas. Consistent with expectations, AF594low αFLAGhigh CD8+ T cells isolated from cultures in which uncaging was limited to Ag+ tumor cell areas displayed a substantial increase in CD69 expression relative to AF594low αFLAGhigh CD8+ T cells from cultures in which uncaging was limited to Ag− tumor cell areas (Fig. 3F–G).
Single cell analysis of spatially defined CD8+ T cells
We subsequently analyzed the transcriptomes of CD8+ T cells isolated from Ag+ and Ag− regions by massive parallel single-cell mRNA sequencing (MARS sequencing)31. To determine whether local uncaging could be used to reveal location-dependent transcriptional differences, two parallel approaches were used. First, in a cell-centric approach, cell states were identified using cells from all conditions, and enrichment of specific transcriptional states in uncaged (AF594low αFLAGhigh) cells from either Ag+ or Ag− areas was determined. Second, in gene set-centric approach, gene modules were defined based on the most variable genes in the full dataset, and differential expression of such gene modules between uncaged cells from Ag+ and Ag− areas was subsequently analyzed.
To test for location-dependent differences in cell states, T cells from all conditions were partitioned into groups of cells (“metacells”) with similar gene expression patterns, using the MetaCell algorithm (Supplementary Data 1)32. This partitioning revealed a large group of T cells that unanimously expressed T cell activation markers such as GZMB and CRTAM (T-act), and a second large group of T cells that lacked expression of these marker genes (T-non-act, Fig. 4A and Extended Data Fig. 7A–B, further characterization below). Notably, comparison of cell states of uncaged and caged cells in the control conditions (i.e. that only contained Ag+ tumor cells or only contained Ag− tumor cells) demonstrated that the uncaging procedure did not influence cell states (Extended Data Fig. 7C). Furthermore, uncaging did not induce detectable expression of stress-related genes (Fig. 4B), demonstrating that the PsT uncaging method allows for in depth analysis of viable cells with unperturbed cell-intrinsic gene expression patterns.
Figure 4. Single cell transcriptomics of spatially defined CD8+ T cells.
A) 2D projection of transcriptional relatedness of 9,237 cells on which single cell mRNA sequencing was performed and that passed filtering. Individual cells are depicted as dots. Main cell groups are annotated by color, with T-act1 and T-act2 (red) showing an activated cell state and T-non-act1-3 (blue) showing a non-activated state. Tumor cells are depicted in green.
B) Stress gene expression signature in caged and uncaged CD8+ T cells derived from tumor island cultures. Fraction of UMIs from stress-related transcripts (Supplementary Data 4) of total UMIs per cell in uncaged or caged cells are depicted. Box plot shows the median, 25th and 75th percentile. Whiskers depict 1.5xIQR. Two-tailed Mann-Whitney U test was performed (ns indicates p=0.0586). Data are representative of 2 independent experiments.
C) Quantification of T cell states of uncaged CD8+ T cells from Ag+ and Ag− areas. Plots depict 2D projections of uncaged cells from Ag+ (left) or Ag− (right) areas, with cells from other conditions depicted in grey. Cells from uncaged areas are colored by their cell states as defined in Fig. 4A and Fig. S7A–B. Bar plots show quantification of cell states that are present in the uncaged (AF594low αFLAGhigh) CD8+ T cell populations from samples in which either Ag+ or Ag− areas were uncaged (630 and 615 cells, respectively). Tumor cells are excluded from bar plots. Two-tailed Chi-square test was performed (**** indicates p<1*10-15). Data are representative of 2 independent experiments.
D) Quantification of cell states of CD8+ T cells showing different levels of uncaging. CD8+ T cells from Ag+ areas that show varying degrees of uncaging, based on the AF594 and αFLAG signals, were divided into 5 bins containing 127 cells each (shown in alternating green and black), based on their distance from the cut-off line that distinguishes uncaged from caged cells (left panel). Bin 1 contains cells furthest from the cutoff and bin 5 contains cells closest to the cut-off. Tumor cells were excluded from analysis. Data are representative of 2 independent experiments.
Subsequent comparison of cell states of uncaged CD8+ T cells derived from Ag+ or Ag− areas (Extended Data Fig. 7D) revealed that the T-act state was highly enriched in Ag+ areas (77%), while T-non-act cells showed an increased abundance in Ag− areas (82%, Fig. 4C and Extended Data Fig. 7E). Similarly, at the subgroup level (T-act1, T-act2 and T-non-act1–3, Extended Data Fig. 7B), a substantial enrichment of activated CD8+ T cell states was observed in Ag+ areas, while all three non-activated CD8+ T cell populations showed enrichment in the Ag− region (Fig. 4C). To determine whether the uncaged cell population was homogeneous, or whether cell pools with a lower level of uncaging (i.e. cells with intermediate AF594 and αFLAG signal) showed an increased contamination with adjacent cells (with e.g. partially uncaged cells showing a non-activated cell state in samples in which uncaging was aimed at Ag+ areas), we divided the uncaged T cells from Ag+ areas into bins based on their level of uncaging (bin 1 containing ‘highly uncaged’ cells to bin 5 containing ‘lowly uncaged’ cells). Notably, enrichment in activated T cell states was consistently observed across bins (Fig. 4D), demonstrating the efficient separation of cells located in different areas.
To assess whether variability in the data at the gene level could be mapped to the location of cells (i.e. in Ag+ or Ag− areas), we next selected the top 30 genes with the highest variance throughout the entire dataset (Fig. 5A–B and Supplementary Data 2). This list contained a considerable number of genes encoding soluble mediators, such as IFNG, CCL4, and CXCL8, factors that are known for their role in inflammation33–35. Furthermore, a substantial part (67%) of the top 30 most variable genes showed increased expression in the T-act CD8+ T cell population (Fig. 5B). We then established a gene module containing genes with an expression pattern that was strongly correlated to that of IFNG, the most variable gene in the dataset (Extended Data Fig. 7F). As a control, gene correlations to alternative anchor genes CCL4 and CXCL8 resulted in very similar gene lists (Extended Data Fig. 7G). Notably, CD8+ T cells that were uncaged in Ag+ regions showed increased expression of the IFNG module as compared to caged cells (i.e. cells from Ag− areas) from the same tumor island culture (Fig. 5C). Likewise, expression of the IFNG module was significantly higher in AF594low αFLAGhigh CD8+ T cells that were derived from uncaged Ag+ regions, as compared to AF594low αFLAGhigh CD8+ T cells derived from uncaged Ag− regions (Fig. 5D). Differential gene analysis between uncaged cells from Ag+ areas and uncaged cells from Ag− areas confirmed the enrichment of soluble mediator genes as well as activation marker genes in the former cell population (Extended Data Fig. 7H–I). These data demonstrate that location-dependent transcriptional differences can be readily revealed at the gene level. Differences in cell states could also be identified in more complex systems, containing many distinct adjacent cell populations, as shown by analysis of T cells co-cultured with a large number of intermingled areas of Ag+ and Ag− tumor cells (Extended Data Fig. 8A–B). Finally, we explored the possibility to perform multiplexed analysis of cells residing in distinct regions of the same cell culture. To this end, we set up a strategy involving subsequent rounds of uncaging and in situ labeling with αFLAG antibodies conjugated to distinct fluorochromes, thereby providing cells that are uncaged in different areas with distinct marks (Extended Data Fig. 9A–B). Analysis of the resulting cell pools revealed that cell populations uncaged in subsequent rounds, and hence derived from separate areas, could be readily distinguished (Extended Data Fig. 9C), enabling the direct comparison of transcriptional profiles of cells residing in different areas of the same tissue.
Figure 5. Detection of diverse activation programs in locally activated CD8+ T cells.
A) Bar plots showing the expression levels (transcripts / 1,000 UMIs) of six of the top variable genes throughout the dataset (i.e. all cells from all experiments) across all metacells. For each metacell, the mean number of UMIs across cells is depicted. Bars are colored by the cell state annotations as used in Fig. 4A.
B) Heatmap depicting the log fold change in expression relative to the median expression for the top 30 most variable genes in the dataset across all metacells.
C) Expression of the IFNG module per cell in a sample in which αCD8D-1-PsT labeled CD8+ T cells in Ag+ areas were uncaged. Color gradient depicts the fraction of UMIs from the IFNG module of total UMI count per cell divided by the median fraction of IFNG module-related UMIs over all cells. Values were transformed for visualization purposes and are projected on the index plot of the sample in which the Ag+ area was uncaged (same index plot as in Extended Data Fig. 7D, left panel). Note that expression of the IFNG module is more profound in AF594low αFLAGhigh CD8+ T cells relative to AF594high αFLAGlow CD8+ T cells from the same sample. Data are representative of 2 independent experiments.
D) Expression of the IFNG module in spatially defined CD8+ T cells. Fraction of UMIs from the IFNG module of total UMI count per cell are depicted for uncaged (AF594low αFLAGhigh) CD8+ T cells from samples in which uncaging was either restricted to Ag+ areas or Ag− areas. Box plot shows the median, 25th and 75th percentile. Whiskers depict 1.5xIQR and circles depict outlier cells. Two-tailed Mann-Whitney U test was performed (**** indicates p<1*10−15). Data are representative of 2 independent experiments.
Discussion
The activation and differentiation state of immune cells and other cells is critically dependent on their interaction with environmental signals. For example, recent data demonstrate that differences in the genetic make-up of distinct areas within individual human tumors coincide with variability in immune infiltrate of these areas36. In other words, within human tumors, the local microenvironment is shaped by cellular interactions, either involving direct cell-cell contact or soluble mediators. To dissect how cell states are influenced by the local microenvironment, we developed a small-protein based photo-uncaging technology that allows the selective isolation of cells from regions of interest in human primary tissue and that enables analysis of single cell transcriptomes without detectable induction of stress signatures or cell toxicity. The photo-uncaging techniques as described in the present work and in the recent work presenting the Zip-Seq technology13 share two main advantages as compared to slide-based transcriptomics technologies6–12. First, as these approaches allow isolation of viable cells from defined sites, downstream analyses are not restricted to transcriptional or epigenetic profiling, but can also include analysis of functional properties, such as T cell antigen specificity. Second, while rare cell types and cell states may be missed using grid-based approaches that do not provide full separation between neighboring cells6–8, the current approaches do provide information that is unambiguously derived from single cell units. Of note, photo-uncaging methods do require tissue dissociation for the analysis of cells. While such tissue dissociation is a common step in single cell transcriptomics, this may for instance preclude the use of photo-uncaging methods in settings in which only fixed tissue is available.
While our method and the Zip-Seq method13 share a similar strategy to analyze cells in their environmental context, both methods exhibit unique strengths. While our photo-sensitive tag can be used for the uncaging of multiple areas in the same tissue using sequential rounds of uncaging (Extended Data Fig. 9A–C), the use of caged DNA barcodes likely allows a higher degree of multiplexing. On the other hand, the current data establish that SCARI can effectively be used to achieve the isolation of specifically uncaged cells from tissue structures as small as 30-60 cells, whereas such data are not available for Zip-Seq. Furthermore, uncaging of the NPBF chromophore as used in SCARI can be performed using low-intensity violet light, rather than the possibly phototoxic ultraviolet light that is generally used to uncage photolabile groups37. This makes SCARI of particular interest for analysis of sensitive cell types and tissues. Finally, the sensitivity of the NPBF cage to two-photon excitation22 and the simultaneous gain and loss of signals that is achieved upon uncaging will both be helpful to allow selective marking and isolation of cells located more deeply in live tissues. While we have here focused on site-specific uncaging of CD8+ T cells, exploiting nanobodies that stably bind to other cell markers, including HLA class I, is also feasible. Using such a pan-cell marker makes it possible to identify cell types that reside in or around specific tumor structures, such as high endothelial venules or tertiary lymphoid structures38. Collectively, technologies such as SCARI should contribute to a further understanding of the relationship between cellular location and cell state in human tissues.
Methods
Human material
Human tumor tissue was obtained either following opt-out procedure or upon prior informed consent, in accordance with national guidelines and after approval by the local medical ethical committee (institutional review board, IRB) of The Netherlands Cancer Institute. Tumor tissue was collected from surgical specimens after macroscopic examination of the tissue by a pathologist. Tumor tissue was dissected into fragments of 1-2mm3 and frozen in 90% fetal calf serum (FCS, Sigma) and 10% dimethyl sulfoxide (DMSO, Sigma). Peripheral blood mononuclear cells (PBMCs) were isolated from blood of healthy donors (Sanquin) using standard Ficoll (GE Healthcare) gradient centrifugation separation. PBMCs were stored in liquid nitrogen in 90% FCS and 10% DMSO until further use.
Production of recombinant nanobodies in Escherichia coli
Monomeric and dimeric variants of two human αCD8 nanobody clones (αCD8M, αCD8D-1, αCD8D-2) and dimeric variants of αHLA class I (specific for the β2M subunit of HLA class I, αHLA-ID) and mouse αCD47 (αCD47D) nanobody clones were generated as follows: Escherichia coli WK6 cells were transformed with the pHEN6 expression vector (for production of the αCD8M nanobody) and Escherichia coli BL21 cells were transformed with the pET22b expression vector (for production of the αCD8D, αHLA-ID and αCD47D nanobodies) encoding the relevant nanobody sequence, followed by an LPETGG-6xH sequence. αCD8D nanobodies are formed by coupling two monomeric αCD8 domains with a flexible GC-rich linker. Protein production was induced with IPTG (Thermo Fisher Scientific) and recombinant proteins were isolated from the periplasmic fraction using Ni-NTA beads (Qiagen). Following washing and subsequent elution with 50 mM Tris (pH 8), 150 mM NaCl, 500 mM imidazole, samples were purified by gel filtration chromatography on a Phenomenex Biosep SEC-S3000 column in phosphate-buffered saline (PBS) and material was concentrated using an Amicon 10 kDa MWCO filtration unit (Millipore). Nanobodies were stored at −80 °C until further use.
Labeling of nanobodies with photo-sensitive tag
Maleimide dyes (maleimide-AF647, maleimide-FITC) were coupled to GGGC peptide by incubation of 1 mg (~20 μg/ml) of the fluorescent maleimide with 175-200 μM GGGC peptide for a minimum of 2 hours at room temperature in 10-12.5 mM NaHCO3. Subsequently, conjugates were purified by reverse phase HPLC on a C18 column (Waters) and identity of the obtained material was confirmed by mass spectrometry. Resulting molecules, and the PsT that also contains a GGGC motif, were coupled to the indicated nanobodies by sortase reactions. In brief, 2.5 μM purified nanobody-LPETGG-6xH protein was incubated with either 40 μM GGGC-dye or 40 μM PsT and 0.4 μM hepta-(7M) mutant sortase for 2 hours at 4 °C in 50 mM Tris (pH 8) and 150 mM NaCl. Hepta-(7M) mutant sortase was produced in-house, as described39. Unreacted nanobodies and sortase were removed by adsorption onto Ni-NTA agarose beads (Qiagen). Subsequently, the suspension was added on top of a 100 kDa cut-off filter to remove Ni-NTA agarose beads, and flow-through (containing labeled nanobody and unconjugated GGGC-dye or PsT) was further purified and concentrated. Unconjugated GGGC-dye or PsT was removed using an Amicon 10 kDa MWCO filtration unit (Millipore), and the material was further purified using a zeba spin column (Thermo Fisher Scientific). To create nanobodies that contain a monomeric PsT, PsT-labeled nanobodies were incubated with 5mM DTT for 30 minutes at room temperature, followed by addition of 8mM iodoacetamide for 30 minutes at room temperature. Reduced nanobody-PsTs were purified using zeba spin column and concentrated using an Amicon 10 kDa MWCO filtration unit. Labeled nanobodies were stored in PBS at −20°C. Protein concentrations were determined by spectrophotometry and individual batches of labeled nanobodies were titrated for optimal usage (final concentrations ranging from 5-10 μg/ml). Purity of unlabeled and PsT-labeled nanobodies were assessed using a NuPAGE 4-12% Bis Tris gel (Thermo Fisher Scientific) and InstantBlue Protein Stain (Novus Biologicals). Protein Stain and fluorescence signals were measured with a Typhoon FLA 9500 laser scanner.
Cell lines and tissue preparations
CD8+ T cells were isolated from PBMCs using the CD8+ T cell Isolation Kit (Miltenyi Biotec). PBMCs and CD8+ T cells were cultured in RPMI (Gibco) supplemented with 10% human serum (HS, Sigma), penicillin (100 U/ml, Roche), streptomycin (100 μg/ml, Roche) and recombinant hIL-2 (60 IU/ml, Novartis). BA/F3 cells (kindly provided by J. Leusen, UMC Utrecht, The Netherlands40) were cultured in RPMI supplemented with 10% FCS, penicillin (100 U/ml), streptomycin (100 μg/ml) and 0.2 ng/mL mouse IL-3 (Immunotools). OVCAR5 cells (kindly provided by F. Scheeren, The Netherlands Cancer Institute, The Netherlands) were cultured in IMDM medium (Gibco) supplemented with 10% FCS, penicillin (100 μg/ml), streptomycin (100 μg/ml) and GlutaMax (1×, Gibco). Viable human tumor tissue pieces of ~1-2mm3 were thawed in prewarmed DMEM (Gibco) supplemented with 10% FCS, penicillin (100 U/ml), streptomycin (100 μg/ml), sodium pyruvate (1 mM, Sigma), MEM non-essential amino acids (1x, Sigma) and GlutaMax (1x). Tumor tissue was subsequently washed three times by thoroughly submerging and shaking the tissue pieces in fresh prewarmed medium.
Viral transduction of tumor cells and T cells
CDK4R>L + GFP+ OVCAR5 cells (Ag+ tumor cells), Katushka+ OVCAR5 cells (Ag− tumor cells) and IFNgR−/− OVCAR5 cells (see below) were generated as described previously41. CDK4R>L-specific CD8+ T cells were generated41,42 and expanded43 as described previously. Upon recognition of Ag+ tumor cells, CDK4R>L-specific CD8+ T cells are activated.
Analysis of nanobody binding stability
CD8+ T cells were stained with either αCD8M, αCD8D-1 or αCD8D-2 (FITC or AF647 labeled), as indicated in Extended Data Fig. 2A–B, in PBS supplemented with 0.5% bovine serum albumin (BSA, Sigma) and EDTA (2 mM, Life Technologies) for 30 minutes at 4 °C. After three washes with PBS containing 0.5% BSA and EDTA, stained CD8+ T cells were mixed as indicated, followed by a 30 minutes incubation at 37 °C in RPMI with penicillin, streptomycin and 10% HS. Afterwards, cells were resuspended in PBS with 0.5% BSA and EDTA (2 mM) and analyzed by flow cytometry.
Staining of cells and tissues prior to uncaging
Where indicated, human CD8+ T cells, PBMCs and tumor tissue were stained with αCD8D-1-PsT or αHLA-ID-PsT and αCD8D-2 (FITC or AF647, serving as a stable signal to identify cells after loss of the AF594-positive cage) in PBS supplemented with 0.5% BSA and EDTA (2 mM) for 30 minutes at 4 °C while gently shaking. Where mixing of cells from exposed and non-exposed samples is indicated, exposed cells were additionally stained with αCD3-FITC (SK7, BD Biosciences, 1:30) and non-exposed cells were stained with either αCD3-BV711 (UCHT1, BD Biosciences, 1:100) or αCD3-APC (SK7, BD Biosciences, 1:30) to allow distinction between cells from exposed and non-exposed samples. BA/F3 cells were stained with mouse αCD47D-PsT and αCD44-AF647 antibody (IM7, Biolegend, 1:50, serving as stable marker) in PBS with 0.5% BSA and EDTA for 30 minutes at 4 °C. Cells and tumor tissue were washed and taken up in PBS supplemented with 0.5% BSA and EDTA for confocal microscopy. For staining of OVCAR5 cells for multiplex experiments, see below. Where indicated, the reduced version of PsT-labeled nanobodies, containing a monomeric PsT, was used.
Tumor cell – T cell cultures
3-5 days before coculture, tumor cells were plated in small droplets (±5,000 cells per 5ul droplet) on polymer or glass bottom 8 well μ-slides (Ibidi) in IMDM medium supplemented with 8% FCS, penicillin (100 U/ml), streptomycin (100 μg/ml) and GlutaMax (1×). Ag+ GFP+ and Ag− Katushka+ tumor cells were plated as indicated per experiment. Following tumor cell adherence, remaining non-adherent cells were removed by washing and cells were cultured in IMDM with 8% FCS, penicillin, streptomycin and GlutaMax. Subsequently, αCD8D-1-PsT and αCD8D-2-FITC stained CDK4R>L TCR+ CD8+ T cells were added, and cells were cultured for 4 hours at 37 °C in the climate chamber of a Leica SP8 Confocal system (Leica Microsystems) microscope, as discussed below.
Confocal microscopy imaging and local uncaging
All images were acquired using an inverted Leica SP8 Confocal system equipped with 4 tunable hybrid detectors, visible lasers (405 nm Argon, DPSS 561 nm, and HeNe 633 nm), and an Insight X3 multi-photon laser (Spectra Physics). All images were collected at 12 bit and acquired with a 25x water immersion objective with a free working distance of 2.40 mm (HC FLUOTAR L 25x/0.95 W VISIR 0.17). Fluorophores were excited as follows: FITC and GFP at 488 nm, AF594 and Katushka at 561 nm, and AF647 at 633 nm. FITC and GFP signals were collected between 510-590 nm, AF594 signal was collected between 610-650 nm, Katushka signal was collected between 620-720 nm, and AF647 signal was collected between 680-750 nm. CD8+ T cells, PBMCs and BA/F3 cells were seeded in a Micro-Insert 4 well u-Dish (Ibidi), and placed onto the microscope with a climate chamber adjusted to 37 °C. Similarly, multiplexing cultures and tumor cell – T cell cocultures were imaged and incubated in 8 well μ-slides in the climate chamber at 37 °C. Overview scans of the entire well were acquired. Tumor tissue was placed in between two cover slips (Duran), and was kept ice-cold using custom-made cool packs during image acquisition. Both overview scans and three-dimensional tile scans of the entire tumor fragments (with 1μm Z-step size) were acquired. Note that in tumor tissues, the αCD8D-2-FITC signal is predominantly detected and therefore, while also stained with αCD8D-1-PsT (AF594), CD8+ T cells are depicted in green in microscopy images.
To uncage the αCD8D-1-PsT, αβ2MD-PsT and αCD47D-PsT in defined areas, a population of cells was selected by drawing a region of interest (ROI). For each defined ROI, a Z-stack was made with step sizes of 1μm. Unless indicated otherwise, uncaging was performed using the 405nm laser line at 15% power (equivalent to 865μW/mm2), 600Hz, 25x magnification, and 1024x1024 pixels, with a pixel dwell time of 600 nanoseconds. For the experiments testing the resolution of SCARI (Fig. 2D–F), zoom (ranging from 1x to 16x) and number of pixels (ranging from 1024x1024 to 64x64 pixels per uncaging field) were adjusted in order to keep laser exposure comparable between the conditions.
Harvest and dissociation of cells and tumor tissue
CD8+ T cells, PBMCs and BA/F3 cells were harvested through resuspension. Adherent cells were trypsinized with PBS supplemented with trypsin-EDTA (1x, Thermo Fisher Scientific) at 37 °C and harvested cell fractions were pooled for subsequent staining steps in case of tumor cell-T cell cocultures. Tumor tissue was dissociated by incubation with collagenase IV (1 mg/ml, Sigma-Aldrich) and pulmozyme (12.5 μg/ml, Roche) in RPMI for 20 minutes at 37°C. After dissociation, tumor cell suspensions were filtered through a 35 μm cell strainer (Falcon tube with cell strainer cap, Corning) and washed with cold PBS supplemented with 0.5% BSA and EDTA (2 mM).
αFLAG staining of cell suspensions after uncaging
Where indicated, PBMCs, BA/F3 cells, CD8+ T cells, dissociated tumor tissue, and T cell-tumor cell suspensions were stained in cold PBS with 0.5% BSA and EDTA (2 mM) for 20-30 minutes at 4°C with the following: live-dead fixable near-IR dead cell stain (IR-dye, Thermo Fisher Scientific), αCD3-BV711 antibody, and αFLAG-AF647 antibody (D6W5B, Cell Signaling Technology, 1:50), polyclonal αFLAG-AF647 (Cell Signaling Technology, 1:200), αFLAG-BV421 (L5, Biolegend, 1:50), or primary unlabeled αFLAG antibody (D6W5B, Cell Signaling Technology, 1:800) followed by secondary αRabbit-IgG-BV421 antibody (BD Biosciences, 1:200). In tumor – T cell coculture experiments, cells were also stained with αCD69-PeCy7 (H57-597, Biolegend, 1:100). Following staining, cells were washed three times and resuspended in cold PBS with 0.5% BSA and EDTA (2 mM) for flow cytometry.
Multiplexed uncaging of tumor islands
Tumor islands containing either GFP+ OVCAR5 cells or IFNγR−/− OVCAR5 cells (in order to distinguish the two cell populations in flow cytometry based on GFP signal and IFNγR signal) were generated as discussed above. Plated GFP+ and IFNγR−/− OVCAR5 cells were stained with αHLA-ID-PsT and αHLA-A2-FITC (BB7.2, BD Biosciences, 1:50, serving as stable membrane marker) in IMDM with 8% FCS, penicillin, streptomycin and GlutaMax for 30 minutes at 37 °C. After the cells were washed with IMDM with 8% FCS, penicillin, streptomycin and GlutaMax, cells were covered in PBS supplemented with 8% FCS for confocal microscopy. In a first uncaging round, cells located in a GFP+ tumor island were uncaged following the uncaging procedure described above, and cells were subsequently stained with αFLAG-AF647 (D6W5B) (αFLAG1) for 30 minutes in the climate chamber of the Leica SP8 Confocal system at 37 °C. Cells were then washed with IMDM with 8% FCS, penicillin, streptomycin and GlutaMax and covered in PBS with 8% FCS. Next, in a second uncaging round, cells located in an IFNγR−/− cell island were uncaged, followed by trypsinization of cells with PBS supplemented with trypsin-EDTA for 4 minutes at 37 °C. After trypsinization, tumor cells were stained with αIFNγR1 antibody (GIR-208; eBioscience, 1:50, to distinguish GFP+ IFNγR-proficient cells from membrane-stained IFNγR−/− cells) and primary unlabeled αFLAG antibody (D6W5B) (αFLAG2) followed by secondary αRabbit-IgG-BV421 antibody (BD Biosciences) and IR-dye. Following staining, cells were washed three times and resuspended in cold PBS with 0.5% BSA and EDTA (2 mM), and analyzed by flow cytometry.
Single cell sorting of CD8+ T cells
CD8+ T cells were single cell sorted based on the following gating strategy. Forward and sideward scatter were used to exclude doublets and to distinguish CD8+ T cells from tumor cells. Viable CD8+ T cells were identified by expression of CD3, CD8 (as reflected by staining with αCD8D-1-PsT and αCD8D-2-FITC), and low IR-dye signal. In addition, CD69 expression was measured. For each sample, uncaged CD8+ T cells (AF594low αFLAGhigh) and total CD8+ T cells were sorted using index sorting into 384-well plates containing 2 μl of lysis solution with barcoded poly(T) reverse-transcription (RT) primers (IDT, Li et al.44) per well. Four wells were left empty in each 384 well plate to be used as background controls in single cell sequencing. Following cell sorting, plates were briefly centrifuged, snap frozen on dry ice, and stored at −80 °C.
Single cell library preparation
Single cell libraries were prepared as described previously using the Massively Parallel Single-Cell RNA-seq method (MARS-seq)31. In brief, upon single cell sorting and cell lysis in 384 well capture plates, mRNA was barcoded and converted into cDNA. cDNA was pooled using an automated pipeline and the pooled sample was linearly amplified by T7 in vitro transcription. Resulting RNA was fragmented and converted into a sequencing-ready library by tagging the samples with pool barcodes and Illumina sequences during ligation, reverse transcription, and PCR. For each pool of cells, both library quality and library concentration were assessed.
MARS seq data processing
Sequencing of scRNA-seq libraries that were pooled at equimolar concentration was done on a NextSeq 500 (Illumina) with a median sequencing depth of ~40,000 reads per cell. Sequences were mapped to the human genome (hg19), demultiplexed and filtered as described in Jaitin et al.31, with the modifications reported in Li et al.44.
Metacell modeling and analysis
For modeling of scRNAseq data, we used the MetaCell package version 3.4132, using a similar strategy as described in Li et al.44. In brief, sets of mitochondrial genes, immunoglobulin genes, ribosomal protein genes and long noncoding RNA genes (Supplementary Data 3) were removed. Cells with less than 500 UMIs were filtered out, as well as cells with a fraction of mitochondrial gene expression that exceeded 0.6. Feature genes with a Tvm=0.08 and 100 total UMIs minimal were selected. Gene features that were associated with lateral processes, such as cell cycle, type I IFN response, or stress (adapted from Li et al.44, Supplementary Data 4) were excluded from metacell formation.
Metacell generation was performed on 9,237 cells using 444 genes that passed the filtering steps. K=100 and 500 bootstrap iteration steps were done and heterogeneous metacells were split. The metacell confusion matrix was used to annotate groups of metacells that showed similar expression profiles. Three main cell groups (activated CD8+ T cells, non-activated CD8+ T cells, and tumor cells) were classified based on the expression of marker genes. Tumor cells were excluded from further analysis where indicated. Supervised analysis of cell states was performed as described in the main text.
Cell state analysis of uncaged cells
To annotate single cells as caged or uncaged, mean fluorescence intensity values of the αCD8D-1-PsT and αFLAG signals per cell were used and defined as above cut-off (AF594low αFLAGhigh; uncaged) or below cut-off (AF594high αFLAGlow; caged). To analyze the states of CD8+ T cells with different levels of uncaging, the total uncaged population was divided into five bins containing equal numbers of cells (before exclusion of tumor cells), based on their distance from the cut-off line between the uncaged and caged populations, with bin 1 containing cells with the highest level of uncaging, and bin 5 containing cells that were closest to the cut-off. For subsequent analysis of T cell states per bin, tumor cells were excluded.
The most variable genes within the dataset were defined based on their variance over all cells divided by the mean. To generate gene modules that were associated with the expression of the most variable genes, we identified the top 30 genes that correlated to one of the indicated anchor genes, IFNG, CCL4 and CXCL8, using a linear correlation of the log fold change of the expression value of a gene in each metacell over the median expression value over all metacells. Genes that were part of the cell cycle, type I IFN response, or stress modules were excluded from this analysis. The expression of gene signatures (“signature score”) for both the stress gene module and the IFNG gene module was plotted as the fraction of signature-related UMIs of total UMIs per cell.
Software
For acquisition of microscopic images in this study, LASX 3.5.5 was used. Flow cytometry data acquisition and analysis were done in FACS Diva 7 and FlowJo 10.6.2. Statistical analyses were performed in Prism (GraphPad) 8.0.0. Analysis of single cell RNA sequencing data was done using R 3.6.2 and Rstudio 1.2.1335.
Extended Data
Extended Data Fig. 1. Synthesis of bis functionalized NPBF based photocleavable linker and the photo-sensitive tag (PsT).
A) Reagents and conditions for synthesis of the NPBF cage: a) bromoacetaldehyde diethyl acetal, KOH, NMP, 70 °C, 15 hours, 90%; b) polyphosphoric acid, toluene, 111 °C, 16 hours, 33%; c) BBr3, DCM, 1 hour at - 78 °C then 1 hour at room temperature, 89%; d) TBDMSCl, imidazole, DMF, 1 hour, room temperature, 70%; e) (i) n-BuLi, THF, -78 °C, 1 hour, (ii) triisopropyl borate, 30 minutes at -78 °C then 30 minutes at room temperature; f) Pd(PPh3)4, K2CO3, THF/H2O, 75 °C, 18 hours, 75%; g) HF-pyridine, THF, room temperature, 1 hour, 77%; h) K2CO3, propargyl bromide, room temperature, 5 hours, 86%; i) NaBH4, 1,4-dioxane/methanol, room temperature, 1 hour, 0 °C to room temperature, 88%; j) N,N’-disuccinimidyl carbonate, Et3N, DMF, room temperature, 4 hours, 79%.
B) Reagents and conditions for synthesis of the PsT: a) 1% TFA, DCM, room temperature, 30 minutes (2x); b) 1, DiPEA, DMF, room temperature, 18 hours; C) 20% piperidine, DMF, room temperature, 2 minutes (3x), d) TFA, TIS, H2O 95/2.5/2.5, room temperature, 3 hours; e) CuSO4, sodium ascorbate, THPTA, AF594-N3, H2O, t-BuOH room temperature, 2 hours.
C) Uncaging efficiency of the PsT over time. 10μM PsT (in water) was uncaged with 420nm light using a 1W LED-lamp for the indicated times and reaction products were measured using LC-MS analysis. Data points represent the mean +/- SD of technical triplicates (n=3) from a single experiment.
Extended Data Fig. 2. Generation of stable dimeric αCD8 nanobodies.
A) Binding stability of αCD8 nanobodies to CD8+ T cells. Top panel: a first population of CD8+ T cells was stained with FITC labeled monomeric nanobody (αCD8M-FITc) and a second population was stained with αCD8M-AF647. Bottom panel: a first population of CD8+ T cells was stained with FITC labeled dimeric nanobody containing recognition domains that are identical to the αCD8M (αCD8D-2-FITc) and a second population was stained with αCD8D-2-AF647. Subsequently, two cell populations labeled with either monomeric or dimeric αCD8 reagents were mixed, incubated at 37oC, and exchange of αCD8 nanobodies was measured by flow cytometry. Quantification of this αCD8D-2 clone and of a second nanobody clone αCD8D-1 is depicted in B.
B) Data depict AF647 fluorescence signal of two CD8+ T cell populations that were either stained with the indicated αCD8D-AF647 or with the corresponding αCD8D-FITC, and that were mixed and subsequently incubated at 37 °C. Note the lack of substantial AF647 signal on the FITC-labeled bystander pool. Data are representative of 2 independent experiments.
Extended Data Fig. 3. αCD8-PsT nanobody generation and recognition by αFLAG antibodies clones.
A) Coomassie staining of sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel showing the indicated nanobodies before and after labeling with the PsT, FITC or AF647. Band marked with the open triangle represents the PsT-labeled αCD8D-1 and band marked with the closed triangle represents a hydrolysis product, that has previously been described in C-terminal protein labeling45. Data obtained from a single experiment.
B) Binding of αFLAG antibodies to the FLAg-tag with or without the NPBF cage (αCD8D-1-PsT and αCD8D-1-T respectively). Cells labeled with αCD8D-1-PsT (left panel) or αCD8D-1-T (right panel) were stained with polyclonal αFLAG antibodies or αFLAG clone L5. Data obtained from a single experiment.
C) Specific binding of αFLAG antibody clone D6W5B to the uncaged FLAg-tag. Cells stained with αCD8D-1 nanobody conjugated to either the FLAg-tag without cage (αCD8D-1-T, upper panel) or the FLAg-tag containing the NPBF cage (αCD8D-1-PsT, bottom panel) were stained with αFLAG antibody clone D6W5B. Note that appreciable AF488 signal is only observed when the uncaged FLAg-tag is used. Representative data from 3 independent experiments are depicted.
Extended Data Fig. 4. Flow cytometry gating strategy to distinguish uncaged and caged CD8+ T cells.
Data depict gating strategy used for flow cytometry analysis as well as for index sorting of uncaged CD8+ T cells. Viable single CD8+ T cells are selected based on double staining with αCD8D-1-PsT and αCD8D-2-FITC (serving as a stable signal to identify cells after loss of the AF594-positive cage). Uncaged cells are distinguished from caged cells based on their elevated αFLAG signal and reduced αCD8D-1-PsT signal (AF594low αFLAGhigh).
Extended Data Fig. 5. Characterization of nanobodies labeled with monomeric or dimeric PsT.
A) SDS-PAGE analysis of nanobody-PsT generated under non-reducing or reducing conditions. Left panel: Coomassie stained SDS-PAGE gel showing the reduced (left) or non-reduced (right) versions of PsT-labeled αCD8D-1, αCD47D and αHLA-ID nanobodies. Right panel: AF594 fluorescence signal of the same gel. Note the increased size of PsT-nanobodies when coupled under non-reducing conditions as compared to PsT-nanobodies generated under reducing condition, indicating the presence of PsT dimers under non-reducing conditions (see also LC-MS and NMR spectra in Supplementary Information). Lower band in both conditions represents the non-fluorescent hydrolysis product that is formed during sortase reactions45. Data obtained from a single experiment.
B) Flow cytometric analysis of the uncaging of CD8+ T cells labeled with αCD8D-1-PsT nanobodies generated under reducing or non-reducing conditions (n=2 technical replicates). Data were obtained from a single experiment.
Extended Data Fig. 6. Quantification of uncaging efficiency.
A) Flow cytometric analysis of αCD8D-1-PsT-labeled CD8+ T cells upon exposure to the indicated laser intensities or 1 hour to natural light.
B) Viability of uncaged (AF594low αFLAGhigh) or caged (AF594high αFLAGlow) CD8+ T cells from samples exposed to the indicated laser intensities, as measured by flow cytometric analysis following IR-dye live-dead staining. Data shown are obtained from a single experiment.
C) Correlation between uncaged surface area and the observed fraction uncaged αCD8D-1-PsT labeled CD8+ T cells in peripheral blood mononuclear cells (PBMCs). 0%, 50%, or 100% of the surface area of wells containing αCD8D-1-PsT labeled cells was uncaged, and samples were analyzed by flow cytometry.
D) Quantification of the correlation between uncaged surface area and fraction uncaged CD8+ T cells as measured with flow cytometry, as shown in C. After uncaging of the indicated surface areas, cells were mixed at a 1:1 ratio with PBMCs from non-exposed samples. Cells from exposed and non-exposed samples were distinguished by labeling samples with different αCD3 antibodies before mixing. Blue line represents the fraction of uncaged (AF594low αFLAGhigh) cells after local exposure with 405nm light. Green line represents the fraction of AF594low αFLAGhigh cells within the second sample that was not exposed to 405nm (i.e. not uncaged). Note that αCD8D-1-PsT binding is stable throughout the experimental pipeline, as demonstrated by the absence of AF594low αFLAGhigh cells in the non-exposed sample. Line graphs show mean of technical triplicates from one experiment. Data are presented as mean values +/- SD.
Extended Data Fig. 7. Single cell mRNA sequencing of locally activated CD8+ T cells.
A) 2D projection of the 9,237 cells as shown in Fig. 4A divided into 99 metacells. Single cells are depicted as dots. Metacells are uniquely numbered and depicted as circles. Lines indicate relatedness between metacells.
B) Confusion matrix of all metacells. Clusters indicate groups of related metacells and are defined as main cell groups as shown by the color code below the confusion matrix that corresponds to the color code used in Fig. 4A and Extended data Fig. 7A.
C) Quantification of T cell states of uncaged (AF594low αFLAGhigh) CD8+ T cell populations from control samples that only contained either Ag+ or Ag- tumor cells, and in which an area was uncaged that is comparable to the areas uncaged in the tumor island conditions. Tumor cells are excluded from the analysis. Data are representative of 2 independent experiments.
D) αFLAG and αCD8D-1-PsT signals, as acquired during index sorting. αFLAG and αCD8D-1-PsT signals are depicted for those cells in the gene expression dataset that are derived from tumor island cultures in which either CD8+ T cells in an Ag+ area (left panel) or in an Ag- area (middle panel) were uncaged, or from the control condition (right panel) in which no cells were uncaged. Line depicts the cut-off between caged and uncaged CD8+ T cells.
E) Quantification of T cell states for all technical replicates of uncaged (AF594low αFLAGhigh) CD8+ T cells from antigen positive (Ag+, 193-230 cells) and antigen negative (Ag-, 186-232 cells) tumor cell areas. The three replicates jointly underlie the data in Fig. 4C. Tumor cells are excluded from the analysis. Representative data of 2 independent experiments.
F) Bar plots depicting the top 30 genes of which expression correlates most strongly with IFNG expression, together defining the IFNG module.
G) Bar plots depicting the top 30 genes of which expression correlates most strongly with the two alternative anchor genes CCL4 (left) and CXCL8 (right).
H) Gene expression enrichment in uncaged (AF594low αFLAGhigh) CD8+ T cells from Ag+ versus Ag- areas. Bar plot depicts the top 30 genes showing increased expression in uncaged CD8+ T cells from an Ag+ area as compared to uncaged CD8+ T cells from an Ag- area. Gene enrichments are based on data from a single experiment.
I) Correlation of variable genes between uncaged CD8+ T cells from Ag+ versus Ag- areas in independent experiments. Scatterplot shows the gene enrichment scores in AF594low αFLAGhigh CD8+ T cells from samples in which Ag+ versus Ag- areas were uncaged for 2 independent experiments.
Extended Data Fig. 8. Cell states from CD8+ T cells residing in intermingled tumor areas.
A) Confocal image of coculture of Ag-specific CD8+ T cells with tumor cell islands in which Ag+ tumor cell areas (green) and Ag- tumor cell areas (magenta) are intermingled. Scale bar represents 2mm. Representative of 2 independent experiments.
B) Quantification of T cell states in uncaged αCD8D-1-PsT labeled CD8+ T cells from Ag+ and Ag- areas in a mixed tumor culture as shown in A. Bar plots show the quantification of T cell states as defined in Fig. 4A of uncaged CD8+ T cell populations from samples in which either Ag+ or Ag- areas were uncaged (252 and 236 cells, respectively). Tumor cells are excluded from the analysis. Two-tailed Chi-square test was performed (**** indicates p<1*10-15). Data shown were obtained in a single experiment.
Extended Data Fig. 9. Multiplexed uncaging of cells in different areas.
A) Schematic representation of multiplexed analysis of cells in adjacent areas.
B) Representative confocal image of OVCAR5 tumor cell culture consisting of adjacent islands of GFP+ cells (population 1) and GFP- cells stained for expression of a cell membrane marker (population 2). Scale bar represents 250μm. Enlarged areas are indicated by the boxes. Tumor cells in a first island (area A, marked with the dashed white line), containing cells from population 1, were uncaged in a first uncaging round and in situ stained with a first fluorescently labeled αFLAG antibody (αFLAG1). Subsequently, tumor cells in a second island in the same culture (area B, marked by the dashed yellow line), containing cells from population 2, were uncaged in a second round of uncaging. After staining with a second fluorescent αFLAG antibody (αFLAG2), flow cytometric analysis was performed. Images are representative of 2 independent experiments.
C) Bar plot shows the fraction of population 1 (GFP+) and population 2 (membrane-stained) cells within the AF594low and αFLAG1-high (i.e. uncaged in round 1) population, or within the AF594low and αFLAG2-high (i.e. uncaged in round 2) population (n=3 technical replicates). Data are representative of 2 independent experiments.
Supplementary Material
Acknowledgements
Plasmid sequences for αCD8 nanobodies were kindly provided by 121Bio with support of M. Gostissa and G. Grotenbreg. We thank H. Ploegh for providing the sortase expression vector. We thank M. Marqvorsen, M. de Weert, M. de Bruijn, K. Bresser, D. Philips, D. Elatmioui, P. Hekking, N. Meeuwenoord, H. van den Elst, B. Florea, L. Bornes and staff of the NKI Flow Cytometry facility for technical support and input, and members of the van Kasteren, van Rheenen, and Schumacher laboratories for discussions. This work was supported by ERC AdG SENSIT (742259) to T.N.S., and ERC StG Crosstag (639005) and ERC Cog KineTic (865175) to S.I.v.K.
Footnotes
Contributions
A.M.v.d.L., M.E.H., L.R., M.J.v.d.G., S.I.v.K., and T.N.S. conceived the project and contributed to experimental design. A.M.v.d.L., M.E.H., C.L.G.J.S. and M.T., designed, performed and analyzed biological experiments. C.L.G.J.S. and L.Y.Y.C. performed microscopy experiments. L.R., M.T., M.J.v.d.G., and S.v.K. designed, synthesized and validated the photocaged compounds. L.R. and C.L.G.J.S. equally contributed to the execution of the experiments. H.L. performed single cell sequencing and E.D. performed sequence alignments. A.M.v.d.L. performed computational analysis. H.L., A.B., Y.L. and A.T. gave input on the computational analysis. D.S.T. was responsible for human tumor sample acquisition. A.M.v.d.L., M.E.H., L.R., S.I.v.K., and T.N.S. wrote the manuscript with input of all other co-authors. J.v.R., I.A., S.I.v.K., and T.N.S. supervised the project.
Competing interests
The authors declare no competing interests.
Data availability
The processed single cell RNA sequencing data are deposited in the NCBI Gene Expression Omnibus (GEO), accession number GSE175813. Source data are provided with this paper.
Code availability
The code reproducing the analyses in this study is available as supplementary file to the NCBI GEO accession number GSE175813.
References
- 1.Thommen DS, Schumacher TN. T Cell Dysfunction in Cancer. Cancer Cell. 2018;33:547–562. doi: 10.1016/j.ccell.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giesen C, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods. 2014;11:417–422. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- 3.Keren L, et al. MIBI-TOF: A multiplexed imaging platform relates cellular phenotypes and tissue structure. Sci Adv. 2019;5 doi: 10.1126/sciadv.aax5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goltsev Y, et al. Deep Profiling of Mouse Splenic Architecture with CODEX Multiplexed Imaging. Cell. 2018;174:968–981.:e15. doi: 10.1016/j.cell.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Medaglia C, et al. Spatial reconstruction of immune niches by combining photoactivatable reporters and scRNA-seq. Science. 2017;358:1622–1626. doi: 10.1126/science.aao4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodriques SG, et al. Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science. 2019;363:1463–1467. doi: 10.1126/science.aaw1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moncada R, et al. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat Biotechnol. 2020;38:333–342. doi: 10.1038/s41587-019-0392-8. [DOI] [PubMed] [Google Scholar]
- 8.Vickovic S, et al. High-definition spatial transcriptomics for in situ tissue profiling. Nat Methods. 2019;16:987–990. doi: 10.1038/s41592-019-0548-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thrane K, Eriksson H, Maaskola J, Hansson J, Lundeberg J. Spatially resolved transcriptomics enables dissection of genetic heterogeneity in stage III cutaneous malignant melanoma. Cancer Res. 2018;78:5970–5979. doi: 10.1158/0008-5472.CAN-18-0747. [DOI] [PubMed] [Google Scholar]
- 10.Ståhl PL, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353:78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- 11.Berglund E, et al. Spatial maps of prostate cancer transcriptomes reveal an unexplored landscape of heterogeneity. Nat Commun. 2018;9 doi: 10.1038/s41467-018-04724-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maniatis S, et al. Spatiotemporal dynamics of molecular pathology in amyotrophic lateral sclerosis. Science. 2019;364:89–93. doi: 10.1126/science.aav9776. [DOI] [PubMed] [Google Scholar]
- 13.Hu KH, et al. ZipSeq: barcoding for real-time mapping of single cell transcriptomes. Nat Methods. 2020;17:833–843. doi: 10.1038/s41592-020-0880-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanchez E, Huse M. Spatial and temporal control of T cell activation using a photoactivatable agonist. J Vis Exp. 2018;2018 doi: 10.3791/56655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayer G, Hechel A. Biologically active molecules with a ‘light switch’. Angewandte Chemie - International Edition. 2006;45:4900–4921. doi: 10.1002/anie.200600387. [DOI] [PubMed] [Google Scholar]
- 16.Klán P, et al. Photoremovable protecting groups in chemistry and biology: Reaction mechanisms and efficacy. Chemical Reviews. 2013;113:119–191. doi: 10.1021/cr300177k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu H, Li J, Wu D, Qiu Z, Zhang Y. Chemistry and biological applications of photo-labile organic molecules. Chem Soc Rev. 2010;39:464–473. doi: 10.1039/b901255a. [DOI] [PubMed] [Google Scholar]
- 18.Lee HM, Larson DR, Lawrence DS. Illuminating the chemistry of life: Design, synthesis, and applications of ‘caged’ and related photoresponsive compounds. ACS Chemical Biology. 2009;4:409–427. doi: 10.1021/cb900036s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Icha J, Weber M, Waters JC, Norden C. Phototoxicity in live fluorescence microscopy, and how to avoid it. BioEssays. 2017;39 doi: 10.1002/bies.201700003. [DOI] [PubMed] [Google Scholar]
- 20.Lusic H, Deiters A. A new photocaging group for aromatic N-heterocycles. Synthesis (Stuttg) 2006:2147–2150. doi: 10.1055/s-2006-942424. [DOI] [Google Scholar]
- 21.Young DD, Deiters A. Photochemical hammerhead ribozyme activation. Bioorganic Med Chem Lett. 2006;16:2658–2661. doi: 10.1016/j.bmcl.2006.02.034. [DOI] [PubMed] [Google Scholar]
- 22.Komori N, et al. Design and synthesis of a new chromophore, 2-(4-nitrophenyl)benzofuran, for two-photon uncaging using near-IR light. Chem Commun. 2016;52:331–334. doi: 10.1039/c5cc07664a. [DOI] [PubMed] [Google Scholar]
- 23.Maurits E, et al. Immunoproteasome Inhibitor-Doxorubicin Conjugates Target Multiple Myeloma Cells and Release Doxorubicin upon Low-Dose Photon Irradiation. J Am Chem Soc. 2020;142:7250–7253. doi: 10.1021/jacs.9b11969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guimaraes CP, et al. Site-specific C-terminal and internal loop labeling of proteins using sortase-mediated reactions. Nat Protoc. 2013;8:1787–1799. doi: 10.1038/nprot.2013.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dorr BM, Ham HO, An C, Chaikof EL, Liu DR. Reprogramming the specificity of sortase enzymes. Proc Natl Acad Sci U S A. 2014;111:13343–13348. doi: 10.1073/pnas.1411179111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Capasso S, Mazzarella L, Sica F, Zagari A, Salvadori S. Spontaneous cyclization of the aspartic acid side chain to the succinimide derivative. J Chem Soc Chem Commun. 1992:919–921. doi: 10.1039/C39920000919. [DOI] [Google Scholar]
- 27.Johnson KY, Liu L, Vincent TS. Minimal FLAG sequence useful in the functional epitope tagging of H-ras. Biotechniques. 2002;32:1270–1280. doi: 10.2144/02326st02. [DOI] [PubMed] [Google Scholar]
- 28.Einhauer A, Jungbauer A. The FLAGTM peptide, a versatile fusion tag for the purification of recombinant proteins. Journal of Biochemical and Biophysical Methods. 2001;49:455–465. doi: 10.1016/s0165-022x(01)00213-5. [DOI] [PubMed] [Google Scholar]
- 29.Vanier GS. Microwave-assisted solid-phase peptide synthesis based on the fmoc protecting group strategy (CEM) Methods Mol Biol. 2013;1047:235–249. doi: 10.1007/978-1-62703-544-6_17. [DOI] [PubMed] [Google Scholar]
- 30.Bannas P, et al. Molecular imaging of tumors with nanobodies and antibodies: Timing and dosage are crucial factors for improved in vivo detection. Contrast Media Mol Imaging. 2015;10:367–378. doi: 10.1002/cmmi.1637. [DOI] [PubMed] [Google Scholar]
- 31.Jaitin DA, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baran Y, et al. MetaCell: Analysis of single-cell RNA-seq data using K-nn graph partitions. Genome Biol. 2019;20 doi: 10.1186/s13059-019-1812-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ayers M, et al. IFN-Γ-related mRNA profile predicts clinical response to PD-1 blockade. J CliN Invest. 2017;127:2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russo RC, Garcia CC, Teixeira MM, Amaral FA. The CXCL8/IL-8 chemokine family and its receptors in inflammatory diseases. Expert Review of Clinical Immunology. 2014;10:593–619. doi: 10.1586/1744666X.2014.894886. [DOI] [PubMed] [Google Scholar]
- 35.Repeke CE, Garlet TP, Fonseca AC, Silveira EM, Garlet GP. Ccl4. in Encyclopedia of Signaling Molecules. 2018:805–809. doi: 10.1007/978-3-319-67199-4_10. [DOI] [Google Scholar]
- 36.Ghorani E, et al. The T cell differentiation landscape is shaped by tumour mutations in lung cancer. Nat Cancer. 2020;1:546–561. doi: 10.1038/s43018-020-0066-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van De Graaff MJ, et al. Conditionally Controlling Human TLR2 Activity via Trans-Cyclooctene Caged Ligands. Bioconjug Chem. 2020;31:1685–1692. doi: 10.1021/acs.bioconjchem.0c00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dieu-Nosjean MC, Goc J, Giraldo NA, Sautès-Fridman C, Fridman WH. Tertiary lymphoid structures in cancer and beyond. Trends in Immunology. 2014;35:571–580. doi: 10.1016/j.it.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 39.Dijkgraaf FE, et al. Tissue patrol by resident memory CD8+ T cells in human skin. Nat Immunol. 2019;20:756–764. doi: 10.1038/s41590-019-0404-3. [DOI] [PubMed] [Google Scholar]
- 40.Bracke M, Lammers JWJ, Coffer PJ, Koenderman L. Cytokine-induced inside-out activation of FcαR (CD89) is mediated by a single serine residue (S263) in the intracellular domain of the receptor. Blood. 2001;97:3478–3483. doi: 10.1182/blood.v97.11.3478. [DOI] [PubMed] [Google Scholar]
- 41.Hoekstra ME, et al. Long-distance modulation of bystander tumor cells by CD8+ T-cell-secreted IFN-Γ. Nat Cancer. 2020;1:291–301. doi: 10.1038/s43018-020-0036-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Rooij N, et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J CliN Oncol. 2013;31 doi: 10.1200/JCO.2012.47.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Linnemann C, et al. High-throughput identification of antigen-specific TCRs by TCR gene capture. Nat Med. 2013;19:1534–1541. doi: 10.1038/nm.3359. [DOI] [PubMed] [Google Scholar]
- 44.Li H, et al. Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma. Cell. 2019;176:775–789.:e18. doi: 10.1016/j.cell.2018.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Antos JM, et al. Site-Specific Protein Labeling via Sortase-Mediated Transpeptidation. Curr Protoc protein Sci. 2017;89:15.3.1–15.3.19. doi: 10.1002/cpps.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The processed single cell RNA sequencing data are deposited in the NCBI Gene Expression Omnibus (GEO), accession number GSE175813. Source data are provided with this paper.
The code reproducing the analyses in this study is available as supplementary file to the NCBI GEO accession number GSE175813.