Abstract
The classical Arp2/3-mediated dendritic network defines the cytoskeleton at the leading edge of crawling cells, and it is generally assumed that Arp2/3-mediated actin polymerization generates the force necessary to extend lamellipods. Our previous work suggested that successful lamellipod extension required not only free barbed ends for actin polymerization but also a proper ultrastructural organization of the cytoskeleton. To further explore the structural role of the Arp2/3 complex-mediated network in lamellipod morphology and function, we performed a detailed analysis of the ultrastructure of the Arp2/3-mediated networks, using the WA domains of Scar and WASp to generate mislocalised Arp2/3 networks in vivo, and to reconstruct de novo Arp2/3-mediated actin nucleation and polymerization on extracted cytoskeletons. We present here evidence that spatially unrestricted Arp2/3-mediated networks are intrinsically 3 dimensional and multilayered by nature and, as such, cannot sustain significant polarized extension. Furthermore, such networks polymerize only at preferred locations in extracted cells, corresponding to pre-existing Arp2/3 networks, suggesting that the specific molecular organization of the actin cytoskeleton, in terms of structure and/or biochemical composition, dictates the location of Arp2/3 complex-mediated actin polymerization. We propose that successful lamellipod extension depends not only on localized actin polymerization mediated through local signalling, but also on spatial restriction of the Arp2/3 complex-mediated nucleation of actin polymerization, both in terms of location within the cell and ultrastructural organization of the resulting network.
Keywords: actin, Arp2/3 complex, lamellipod, cytoskeleton ultrastructure
Introduction
Lamellipod extension is an essential step in cell motility and chemotaxis, and is driven by localized actin polymerization at the leading edge (Ridley et al, 2003; Pollard and Borisy, 2003). While the Arp2/3 complex is acknowledged as one of the main players in the establishment and progression of lamellipods (Pollard and Borisy, 2003; Bailly et al, 1999; Svitkina et al, 1999), recent evidence has shown that control of the ultrastructural filament organization at the leading edge is an essential determinant of protrusion shape, velocity and stability (Gupton et al, 2005; Mejillano et al, 2004; Desmarais et al, 2004; Bear et al, 2002). In particular, a range of actin-associated molecules, including capping protein and proteins of the ENA/VASP family, can control both the length and the organization of the actin filaments downstream of the Arp2/3 complex nucleation activity, thus modulating protrusion velocity and structure (Mejillano et al, 2004; Bear et al, 2002). Furthermore, cell protrusion and migration can also be achieved efficiently in the absence of a characteristic Arp2/3-rich lamellipod (Gupton et al, 2005). Hence, while studies with purified protein systems have provided “proof of principle” that Arp2/3-mediated nucleation of actin polymerization was sufficient to generate a protrusive force and displace significant structures such as polystyrene beads or bacteria (Bernheim-Groswasser et al, 2002; Weisner et al 2003), they can only provide partial insights into the molecular organization of protrusions in cells. Furthermore, such models do not address fundamental differences between the motility assay in a soluble protein mix and the periphery of the cell where the protrusion will occur within a pre-existing network of polymerized actin in a space restricted by the membrane. Indeed, to date, no one has been able to reconstruct a protruding lamellipod from a pre-existing cytoskeleton using pure proteins.
We have previously demonstrated that EGF-mediated lamellipod extension is dependent on the synergistic activities of cofilin and the Arp2/3 complex to generate a transient localized increase in nucleation sites for actin polymerization at the tip of lamellipods (Bailly et al., 1999; Chan et al., 2000; Zebda et al, 2000; Bailly et al, 2001; Desmarais et al, 2004). Further evidence suggested that while the generation of free barbed ends is necessary for lamellipod extension, it is not sufficient, and proper geometry of the actin network at the leading edge is required for successful protrusion (Desmarais et al, 2004). We have attempted here to use this well characterized model to further explore the mechanism of lamellipod extension through Arp2/3 complex-mediated actin polymerization. One of the main families of activators of the Arp2/3 complex in vivo is the Scar/WASp family of proteins (Welch and Mullins, 2002; Millard et al, 2004). Both WASp and Scar/WAVE contain a C-terminal region (WA), which binds to the Arp2/3 complex and monomeric actin, and activates the Arp2/3 complex to nucleate branched actin filaments (Machesky et al., 1999, Blanchoin et al., 2000). Overexpression of the Scar/WAVE1-WA domain in fibroblasts abolished the ability to form membrane ruffles induced by growth factor stimulation (Machesky and Insall, 1998), and microinjection of the WASp-WA domain into macrophages resulted in an increase in filamentous actin and podosome disruption (Linder et al., 1999 and 2000). We used here the WA domains of Scar and WASp to perform a deeper study of the structural and functional role of the Arp2/3 complex in lamellipod extension, by using the purified peptides to both analyze Arp2/3 complex-mediated polymerization in the cells and recreate Arp2/3-mediated polymerization of actin networks on isolated cell skeletons. We present evidence that spatially unrestricted WA-activated Arp2/3-mediated networks are intrinsically 3 dimensional and multilayered by nature, and as such cannot sustain significant polarized extension. Furthermore, such network polymerize only at preferred locations in extracted cells, suggesting that the specific molecular organization of the preexisting actin cytoskeleton, in terms of structure and/or biochemical composition, dictates the location of Arp2/3 complex-mediated actin polymerization. Successful lamellipod extension will then depend on further spatial restriction of the Arp2/3 complex-mediated nucleation and polymerization.
Results
WA-domain microinjection causes a reorganization of the F-actin cytoskeleton that displaces the Arp2/3 complex from the leading edge
Our previous work using antibodies altering Arp2/3 complex function in the cells suggested that maintaining the proper cytoskeleton structure at the leading edge was an essential component of successful lamellipod extension (Bailly et al., 2001, Desmarais et al., 2004). To further explore the relationship between structure and function of the Arp2/3 complex at the leading edge, we used another means of altering the structure of Arp2/3 networks without directly altering Arp2/3 function. To this end, we microinjected into the cells purified peptides encompassing the C-terminal (WA) Arp2/3- and actin-binding domains of Scar and WASp (Machesky and Insall, 1998). Both peptides are potent activators of Arp2/3-mediated nucleation of actin polymerization in vitro (data not shown), and are thought to sequester the Arp2/3 complex in vivo (Machesky and Insall, 1998; Machesky et al., 1999). MTLn3 cells were injected with purified Scar and WASp C-terminal fragments (WA domains) at needle concentration of 130 μM, resulting in an intracellular concentration of approximately 10 μM. The cell morphology and cytoskeleton were analyzed 10, 30 and 60 minutes after microinjection using phalloidin staining. Following microinjection of either WA-domain, the cells retracted within 30 minutes, and maintained a significantly smaller surface area for at least an hour after injection (Figure 1, A and B). This was accompanied by a pronounced remodelling of the filamentous actin network, marked by the rapid appearance of a dense perinuclear cloud of F-actin. This was followed after 30 min by the emergence of unusually marked F-actin rich focal contact-like structures, and a thinning or complete disappearance of the stress fibres (Fig. 1A), particularly after Scar-WA microinjection (Figure 1A, g, k). To determine if the remodelling of the F-actin network in the cells after microinjection led to a net increase in the amount of F-actin in the cells, we analyzed the kinetics of accumulation of F-actin in cells injected with Scar-WA and WASp-WA, as compared with control cells injected with dextran and cells left uninjected. There was a significant 20 % increase in the total F-actin content of the WA-domain injected cells compared to the controls within the first 10 min after injection, which subsided within 30 minutes (Figure 1C). Conversely, the level of G-actin, as measured using fluorescently labelled Dnase I (Cramer et al., 2000), was reduced by approximately 20% in cells 30 min after microinjection with Scar-WA domain as compared to control cells (data not shown).
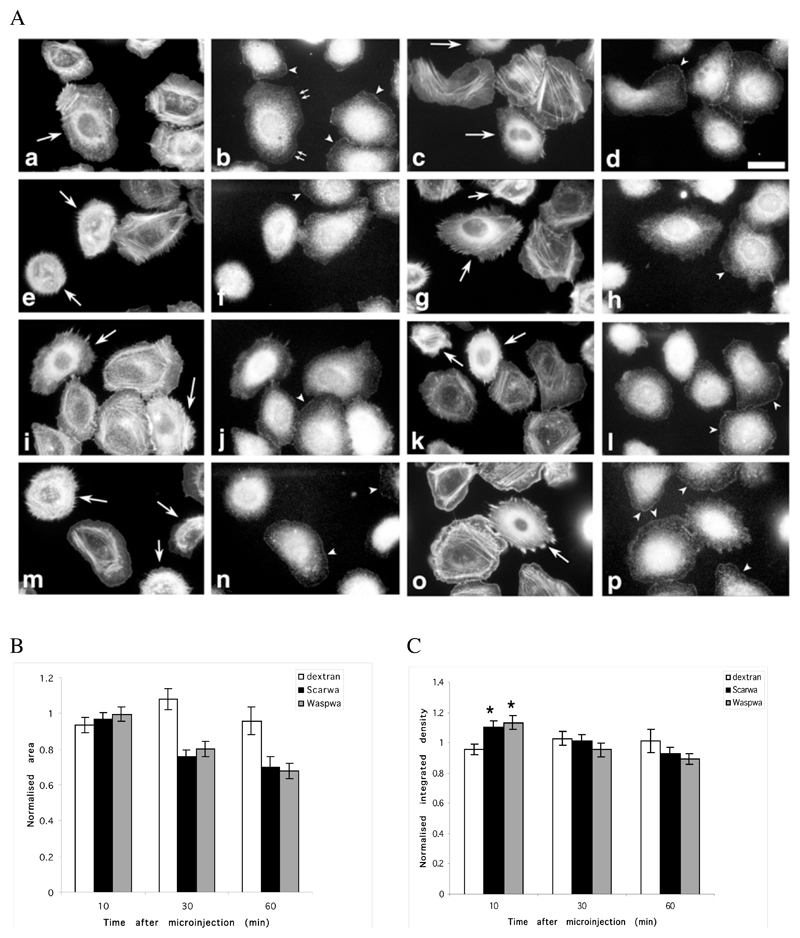
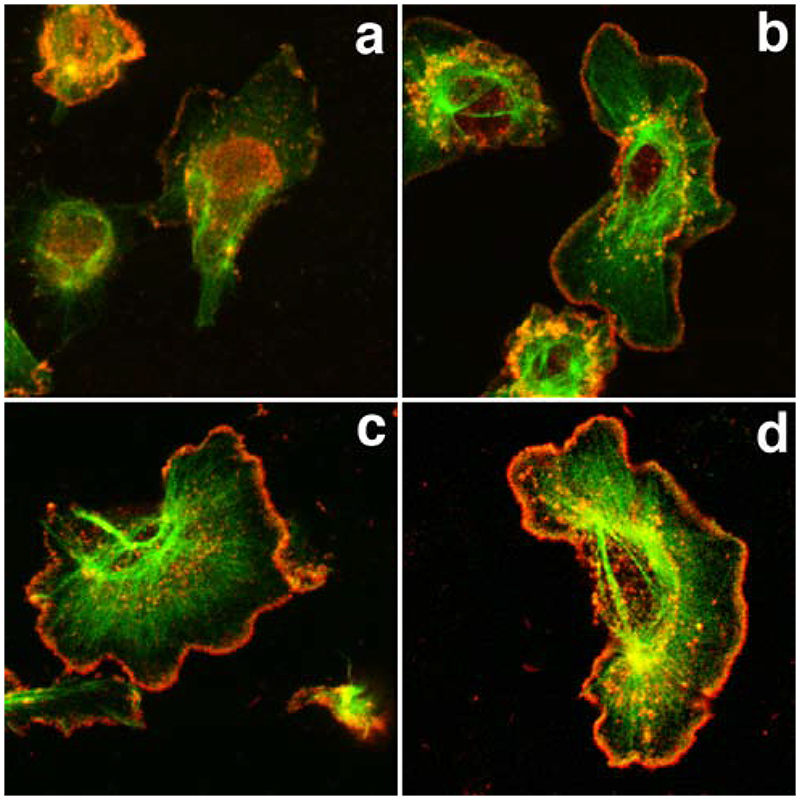
Figure 1.
Microinjection of WA domains causes a rapid reorganization of the actin cytoskeleton and alters Arp2/3 location in resting and stimulated cells. MTLn3 cells were microinjected with a WASP-WA or Scar-WA peptide/dextran mix or with dextran alone, and fixed and stained for F-actin (phalloidin) and Arp2/3 (p34 antibodies) at different times after injection (a-d, 10 min; e-h, 30 min; i-l, 60 min after injection), or after stimulation with 5 nM EGF for 3 min 30 min after injection (m-p). A. F-actin (1 and 3rd column) and Arp2/3 (2nd and 4th column) distribution patterns: cells microinjected with the WA domains (WASP-WA peptide, left 2 columns; Scar-WA peptide, right 2 columns) accumulate F-actin in perinuclear region, while Arp2/3 is depleted from leading edges, even in stimulated cells. Arrows, injected cells; arrowheads, Arp2/3 complex at the leading edge; small double arrows, Arp2/3 complex at the leading edge in an injected cell where actin has already accumulated in the perinuclear region. Scale bar: 20μm. B. Quantification of cell area: the cell area was normalized to the area of control non-injected cells. WA-injected cells display significantly smaller area (P<0.001) than control cells. C. Quantification of total F-actin in microinjected cells: the integrated density value for the phalloidin fluorescence was used as a measure of total F-actin content in the cells, normalized to the levels in control non injected cells. An average of 30 cells (13-54) was measured for each time point. Both WASP-WA and Scar-WA injected cells display a significant increase in F-actin content 10 minutes after injection (* P<0.001 and P<0.005 respectively for Scar-WA and WASP-WA).
We then investigated how microinjection of the WA domains was altering the localization of the Arp2/3 complex and its recruitment to the leading edge after stimulation (Bailly et al., 1999 and 2001; Desmarais et al., 2004). In resting MTLn3 cells, the distribution of the Arp2/3 complex is largely cytoplasmic, with a clear portion located at the leading edge (Bailly et al., 2001 and Figure 1A, arrowheads on control cells). Dextran injection did not affect its localization (data not shown). WA domain microinjection displaced the Arp2/3 complex from the leading edge and concentrated it within the actin rich perinuclear region (Figure 1A). Using the WA domain of WASp whose alterations to the actin cytoskeleton occur slightly slower, we could show that the displacement of the Arp2/3 complex was not actually concurrent to the initial changes in actin, but happened after the F-actin had accumulated significantly within the perinuclear region. Indeed, 10 min after injection of WASp-WA domain, some cells still had Arp2/3 complex localized at the periphery while having already developed a significant increase in perinuclear F-actin (Figure 1A b, small arrows). Furthermore, while control cells showed an increase in Arp2/3 recruitment to the leading edge after EGF stimulation as previously shown (Bailly et al., 2001, Desmarais et al., 2004; arrowheads in Figure 1A n and p), cells microinjected with WA domains fail to show any relocalisation of the Arp2/3 complex after stimulation.
WA domain injection blocks EGF-stimulated lamellipod extension but does not prevent an increase in nucleation activity in response to EGF
Since the WA domain-injected cells displayed an altered actin cytoskeleton and Arp2/3 mislocalisation, we tested whether these cells could still extend lamellipods in response to stimulation by EGF (Bailly et al., 1999). Stimulation with EGF triggers a characteristic rapid circumferential lamellipod extension in control and dextran-injected MTLn3 cells (Figure 2, and Bailly et al., 1999, Chan et al., 2000, Desmarais et al., 2004). On the other hand, cells injected with the WA domain of Scar or WASp failed to extend significant lamellipods or show any other form of protrusive activity upon EGF stimulation (Figure 2A, quantified in B).
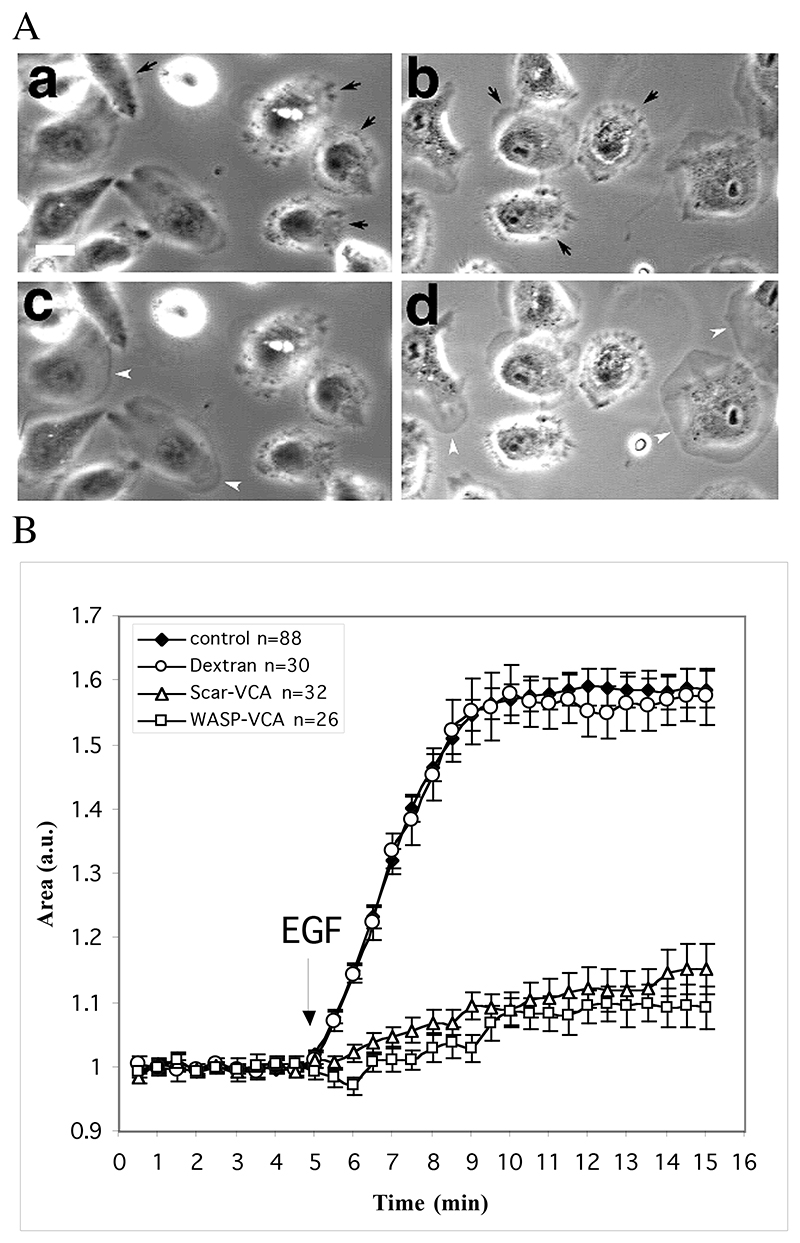
Figure 2.
Microinjection of the WA domains inhibits EGF stimulated lamellipod extension. MTln3 cells were starved for 3 hours and microinjected with WA-domains 30 minutes prior to stimulation with 5 nM EGF. Phase contrast images were recorded every minutes using an OpenLab driven CCD camera and the resulting movies were processed for cell area measurements as described in Material and Methods A. Phase contrast images of injected cells before (a,b) and after (c,d) stimulation with EGF for 3 min. EGF stimulation triggers broad lamellipod extension in control cells (white arrow heads) but not in WASp-WA (a,c)- or Scar-WA (b,d)-domain injected cells (black arrows). Bar =20μm. B. Quantification of area change as a readout of EGF stimulated lamellipod extension. Arrow indicates time point when EGF was added.
We have shown previously that actin polymerization at the leading edge is essential for EGF-stimulated lamellipod extension, where Arp2/3 complex and cofilin appear to act in synergy to create localized new nucleation sites for actin polymerization (Chan et al 1998, Chan et al 2000, Bailly et al 2001, DesMarais et al., 2004). In particular, blocking cofilin or Arp2/3 complex activity abolished for the former, and greatly diminished for the latter, EGF-mediated barbed end generation, and prevented the cells from extending lamellipods after stimulation (Chan et al, 2000; Zebda et al 2001, Desmarais et al 2004). We thus investigated if the incapacity of the peptide-injected cells to extend lamellipods after EFG stimulation was the result of a failure to generate free barbed ends. We analyzed the level and distribution of free barbed ends in cells injected with the WA domain of Scar, as it was the most efficient peptide in affecting the cytoskeleton. The barbed ends were localized by incorporation of biotin-labelled actin monomers in permeabilised cells using our standard assay (Bailly et al., 1999). In unstimulated cells, free barbed ends are typically localized at the edge of lamellipods, the tips of stress fibres and in a diffuse fashion in the perinuclear area (Figure 3Aa, 5f and Bailly et al, 1999). EGF stimulation causes a major increase in barbed ends at the leading edge around the whole cell periphery, which peaks with 50-60 sec after stimulation (Figure 3A b and c; and Bailly et al., 1999; Chan et al, 2000, Desmarais et al., 2004). Scar-WA microinjection caused major disturbances to both the number and the localization of the barbed ends. Unlike in the control cells, the nucleation activity in Scar-WA microinjected cells covered most of the cell area, with a particularly strong presence in the perinuclear region and marked focal contacts (Fig. 3Ad). While the overall amount of barbed ends in cells 30 min after injection is similar to that of control cells (Figure 3C), the mean intensity at the edge of the cells is significantly higher than in the control cells (Figure 3B), presumably because of the multiple bright focal contacts localized at the edge (Figure 3Bd). Furthermore, Scar-WA microinjected cells responded to EGF stimulation better and faster than control cells, reaching a peak number of barbed ends 30 seconds after stimulation as opposed to the characteristic 60 sec of the control cells (Figure 3B and C). In addition, the overall level of barbed ends 30 and 60 sec after stimulation was significantly higher in microinjected cells as compared to control cells (Figure 3C). In control experiments, cells microinjected with control dextran showed no disturbance of barbed end generation or distribution (data not shown).
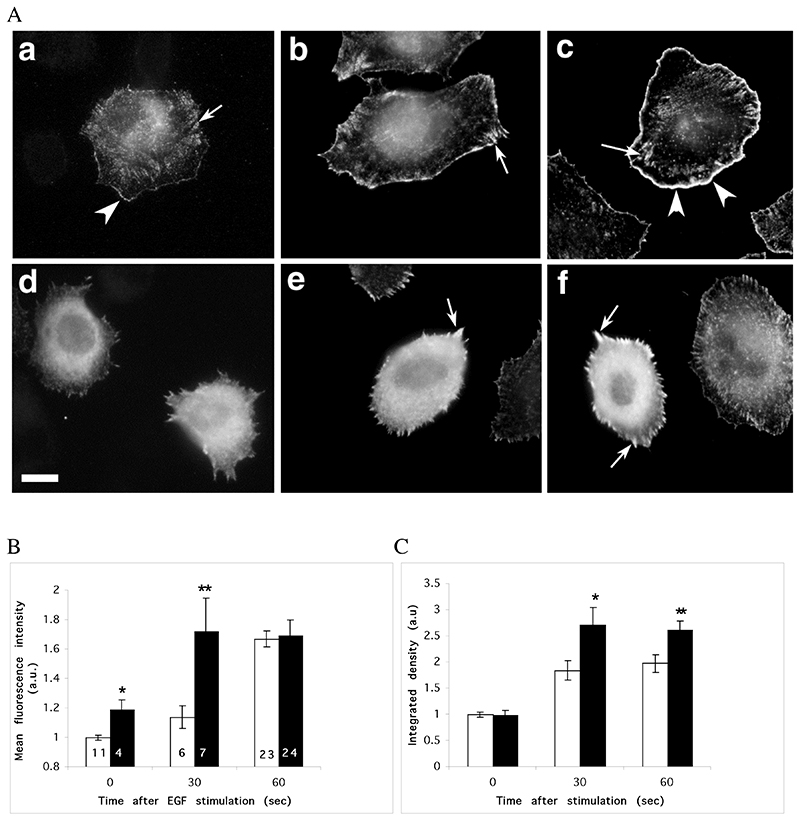
Figure 3.
Microinjection of Scar-WA domain triggers a complete reorganisation of barbed ends in resting cells, but does not prevent a further increase in barbed ends following EGF stimulation. MTLn3 cells were starved for 3 hours and microinjected with Scar-WA domain 30 minutes prior to barbed end labelling. Barbed ends were visualised by permeabilising the cells in presence of 0.45 uM labelled monomeric actin, and quantified as detailed in Material and Methods. A. Barbed ends staining before (a, d) and 30 sec (b, e) or 1 min (c, f) after EGF stimulation. In resting cells (a), barbed ends are localised at the edge of existing lamellipods (arrowheads), in focal contacts (arrows) and in a perinuclear diffuse pattern. Stimulation with EGF triggers a large increase in barbed ends specifically at the leading edge (arrowheads). Cells injected with Scar-WA domain display a diffuse and strong cytosplamic barbed end pattern (d), with an enrichment at focal contacts (arrows) after stimulation but not at the leading edge (e, f). Scale bar =10μm. B, C. Quantification of barbed ends at the leading edge (B) and total barbed ends in the cells (C). The barbed ends were measured locally as the mean fluorescence within 1.1 um at the leading edge (B), or globally as the integrated density of the fluorescence over the total cell area (C). Both numbers were normalised to control uninjected cells within the Scar-WA unstimulated sample (EGF 0). White bars, control cells; black bars, Scar-WA injected cells. The numbers in bar represents the number of cells analysed. Stars: significantly different from control (*, P<0.05; **, P<0.01).
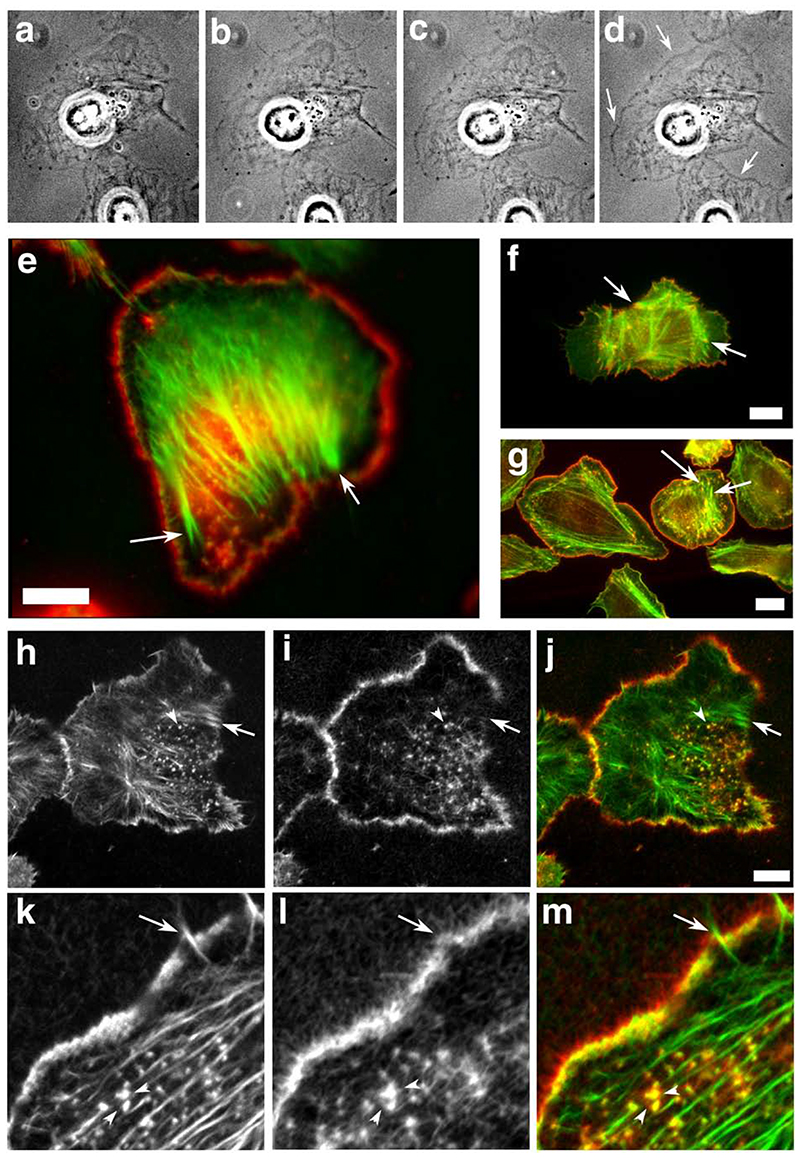
Figure 5.
Scar-WA-activated, -Arp2/3-mediated actin polymerisation in permeabilised resting cells using purified proteins results in a leading edge nucleation pattern similar to that of EGF-stimulated cells. Resting MTln3 cells were membrane-extracted and the resulting cytoskeletons were incubated with an Arp2/3 polymerisation mix (2 uM biotinylated actin, 5 nM Arp2/3, 20 nM Scar-WA, 0.2 uM cofilin) for 5-10 minutes. The samples were imaged live using fluorescence contrast (a-d), and/or fixed and processed for light (e) or confocal (h-m) microscopy. WA domain-activated Arp2/3 complex preferentially induces actin polymerisation at the leading edge in permeabilized cells in a pattern similar to barbed end generation after EGF stimulation. a-d: time course of the actin mix polymerisation in permeabilised cells as visualised by phase contrast microscopy. Actin accumulation around the cell circumference is visible as a denser grey outline of the cell (arrows). a, 40 sec after the beginning of membrane extraction, immediately before polymerisation mix addition; b-d, 1, 3 and 8 min after mix addition. e, h-m: analysis of the distribution of the de-novo Arp2/3 mediated network (green and h, k: pre-existing filaments as identified by phalloidin staining; red and i, l, newly polymerised network as identified using Cy3 coupled anti-biotin antibodies). k-m shows a detail of the leading edge of a cell after polymerisation of the mix. For comparison, light microscopy images of standard nucleation activity using monomeric actin polymerisation (standard barbed end distribution as in Figure 3; green, F-actin, red, newly polymerised actin) is shown for resting (f) and EGF-stimulated cells (g). Arrows point to focal contacts that show standard nucleation activity in both resting and stimulated cells, but are devoid of newly polymerised Arp2/3 mediated network. Arrowheads points at actin-rich dots inside the cell which show both standard actin nucleation and Arp2/3-mediated nucleation activity. Bars, 10 um.
WA-domain microinjection disturbs the ultrastructural organization of the cytoskeleton at the edge of the cells
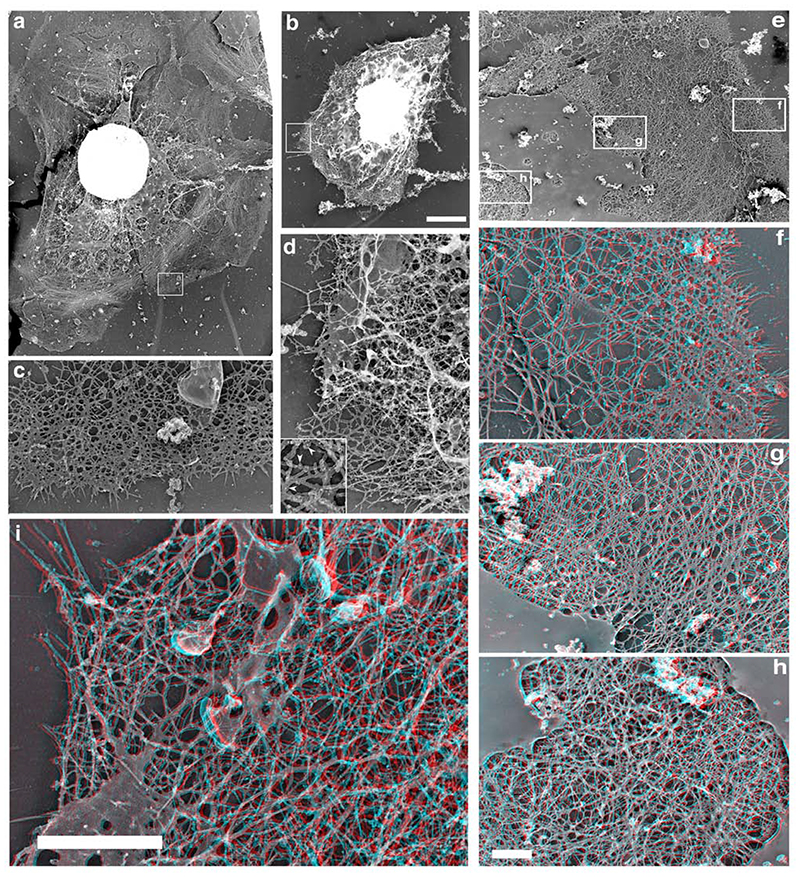
The results above showed that the inability of Scar-WA microinjected cells to extend lamellipods was not due to a failure to generate new barbed ends, suggesting that the WA domain did not interfere in a major way in the pathway mediating barbed end generation after EGF stimulation. However, cells microinjected with Scar-WA domain still were unable to organize any protrusive structure, or to localize the newly generated barbed ends specifically at the cell periphery. Our previous work suggested that disturbances in the ultrastructural organization of the cytoskeleton at the leading edge were sufficient to prevent lamellipod extension, despite the cells having a significant increase in nucleation activity after EGF stimulation (Desmarais et al., 2004). We thus investigated whether the cytoskeletal organization in these cells could explain their failure to respond to EGF, using our rapid freeze/freeze dry/metal shadowing technique, which allows a high-resolution 3-dimensional analysis of the actin network (Bailly et al., 1999, Desmarais et al., 2004). In a resting cell, the leading edge is usually comprised of a characteristic dendritic network spanning over 1-2 microns which is backed by a zone of loosely organized less dense network (Figure 4 a, c, e, f). The density of the filament network further inside then gradually increases, mostly by building different layers in depth, as shown by the 3D view (Figure 4 f-h). As the perinuclear area is reached, the actin network is extremely dense and clearly multilayered (Figure 4h). In contrast, despite being quite dense as well, the dendritic network at the leading edge tends to be very 2 dimensional, with mostly one layer of filaments in a 2D network parallel to the substrate and a few individual filaments pointing upwards (Figure 4f). Scar-WA injected cells displayed a complete reorganization of the actin network (Figure 4 b, d, i). These cells failed to display any typical dendritic network at the periphery (Figure 4d), but showed a quite homogeneous dense network of actin filaments (Figure 4 d, i), whose multi-layered arrangement resembles closely that normally found in the perinuclear region of resting uninjected cells (compare i and h).
Figure 4.
Scar microinjection alters both the structure and the depth of the actin network at the leading edge. Resting MTLn3 cells were microinjected with Scar-WA domain and processed for rapid freeze/freeze dry and rotary shadowing to generate metal replicas. 3 dimensional red/blue views of the cytoskeleton were reconstructed from stereo pairs (f-i; proper viewing of these images require red/blue 3D glasses). a, c, e-h: control cells; b, d, i: Scar-WA injected cells. Control resting cells display broad lamellipods (a) with a typical dendritic-like network at the leading edge (c). The leading edge network is mostly a 2 dimensional array of filaments with a few filaments pointing upwards (e, f). After a relatively less dense of actin filaments, the network further inside becomes profoundly denser, mostly by building different layers in depth (g). The perinuclear actin network is extremely dense and multilayered (h). Scar-WA injected cells display a complete reorganisation of the actin network at the periphery (b, d, i). These cells fail to display any typical dendritic network at the periphery (d), but show a poorly organised dense network of actin, which is a multilayered 3D structure,, (i) similar to that normally found in the perinuclear region of resting cells (d). Inset, enlargement showing the 5 nm gold label on the filaments that allowed for identification of the microinjected cells (see Material and methods). Bars: a-d: bar= 2 um (a, b) and 0.5 um (c, d); e-h: bar= 2um (e) and 0.5 um (f-h); i: bar =1 um
In situ Arp2/3-mediated actin polymerization recapitulates EGF-mediated specific barbed end generation pattern
Since a) microinjected WA peptides induce a dense actin network in MTLn3 cells, b) WA-activated Arp2/3 nucleation of actin polymerization generates typical branched filaments in vitro (Bailly et al, 2001; Ichetovkin et al, 2002), and c) Arp2/3 function is essential for barbed end generation at the leading edge and lamellipod extension (Bailly et al, 2001; Desmarais et al, 2004), we then attempted to see if WA-activated-Arp2/3-mediated actin polymerization would be sufficient to construct a fully functional (extending) lamellipodial structure from a pre-existing cell cytoskeleton using purified proteins. Resting MTLn3 cells were permeabilised with Triton to extract the plasma membrane and cytosolic proteins, and the remaining cytoskeletons were exposed to a standard polymerization cocktail containing Arp2/3 complex, Scar-WA domain, cofilin and G-actin (biotin-labelled) in a polymerization-competent buffer. We took advantage of the pH sensitivity of cofilin, which binds actin but does not sever filaments at pH under 7, to use it as a marker to allow us to see separately the old pre-existing filaments and the newly polymerized ones, and to stabilize the newly polymerized network. At pH 6.7 (our buffer conditions), the newly formed biotin-labelled filaments are thus refractory to phalloidin binding since they are mostly covered with cofilin. This allowed us to visualize clearly any overlap between the 2 compartments. Under these conditions, live observations of the permeabilised cells showed the progressive appearance of a dense peripheral actin accumulation (visible as a discrete grey line outlining the cell contour, Figure 5A, a-d), within 5 minutes after adding the polymerization mix. However, no significant net advance of the leading edge could be measured (data not shown). Immunofluorescence of fixed samples revealed that the newly polymerized actin (in red, Figure 5) incorporated almost exclusively at the leading edge all over the cell periphery in resting cells, surprisingly mimicking the barbed end distribution after EGF stimulation in intact cells (compare Fig 5 e to Fig 5g). The Arp2/3 polymerization mix did not incorporate at the tips of the stress fibres (Figure 5e, h-j, arrows) or filopods (Fig 5, k-m), but it did incorporate into F-actin-rich dots inside the cell (Figure 5, h-m, arrowheads). Further experiments confirmed that while cofilin was dispensable in this assay as intended (data not shown), the presence of an activated Arp2/3 complex was necessary to obtain the specific circumferential de novo polymerization pattern. In the absence of Arp2/3 complex in the polymerization mix (actin only), very few cells were left on the coverslip after 7 min of incubation, and amongst the few cells left, only 24% showed significant (covering at least ¼ of the cell perimeter) incorporation of labelled actin and 16% presented the typical circumferential pattern as shown in Figure 5. On the other hand, when the polymerization mix included activated Arp2/3 complex (actin plus Arp2/3 complex plus VCA), the number of cells preserved on the coverslip more than doubled and 61% of the cells showed a significant polymerization at the edge, while 48% presented the full circular pattern.
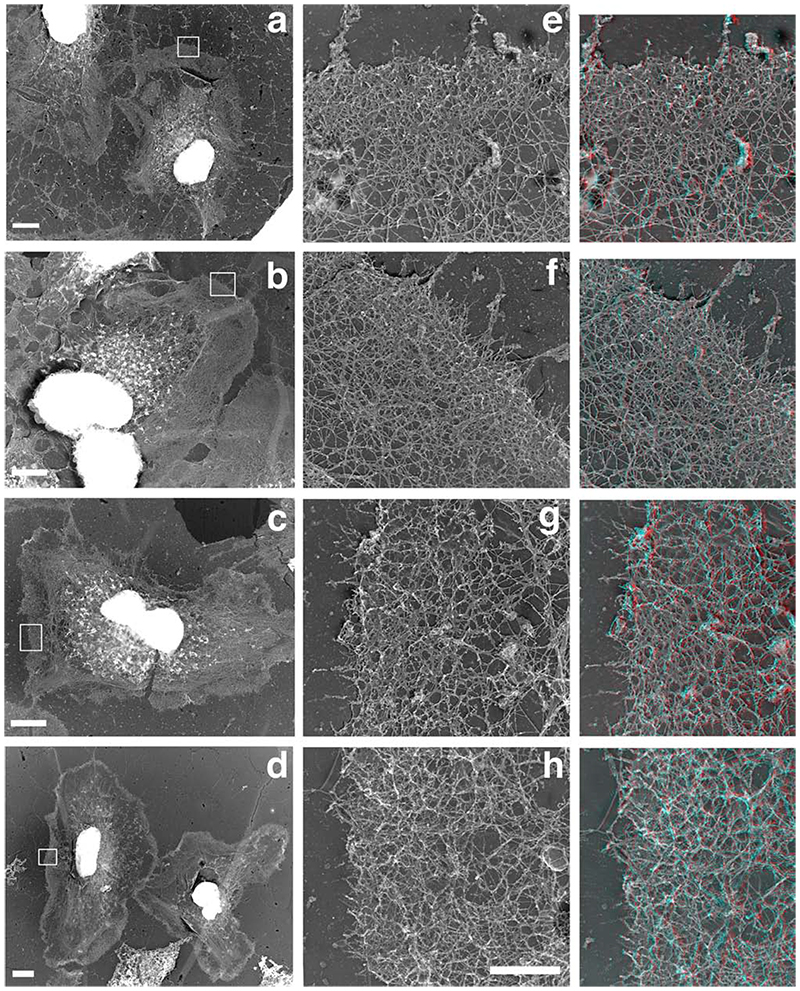
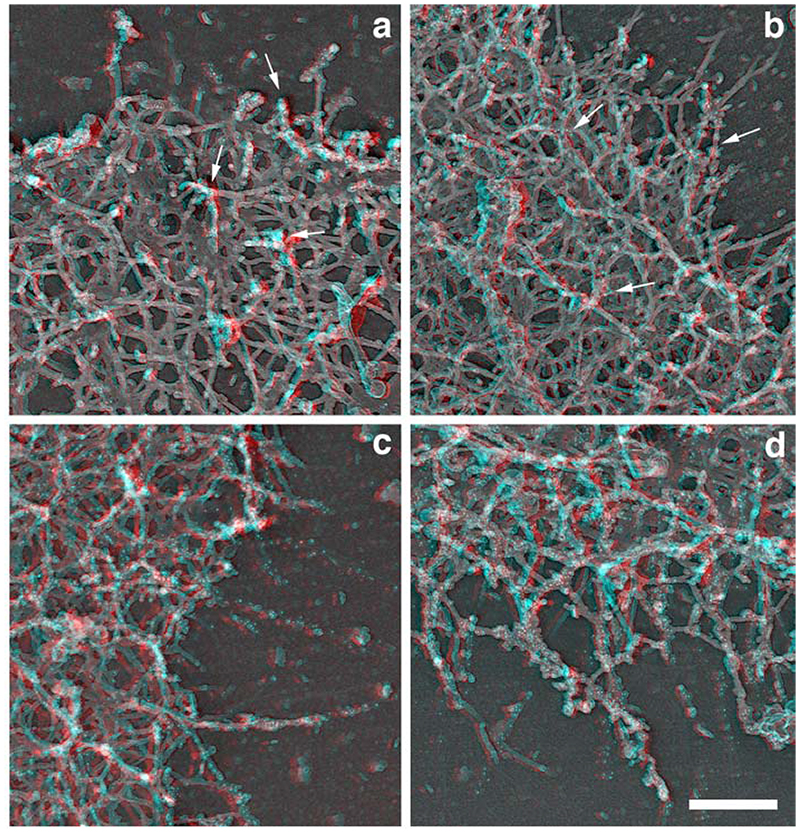
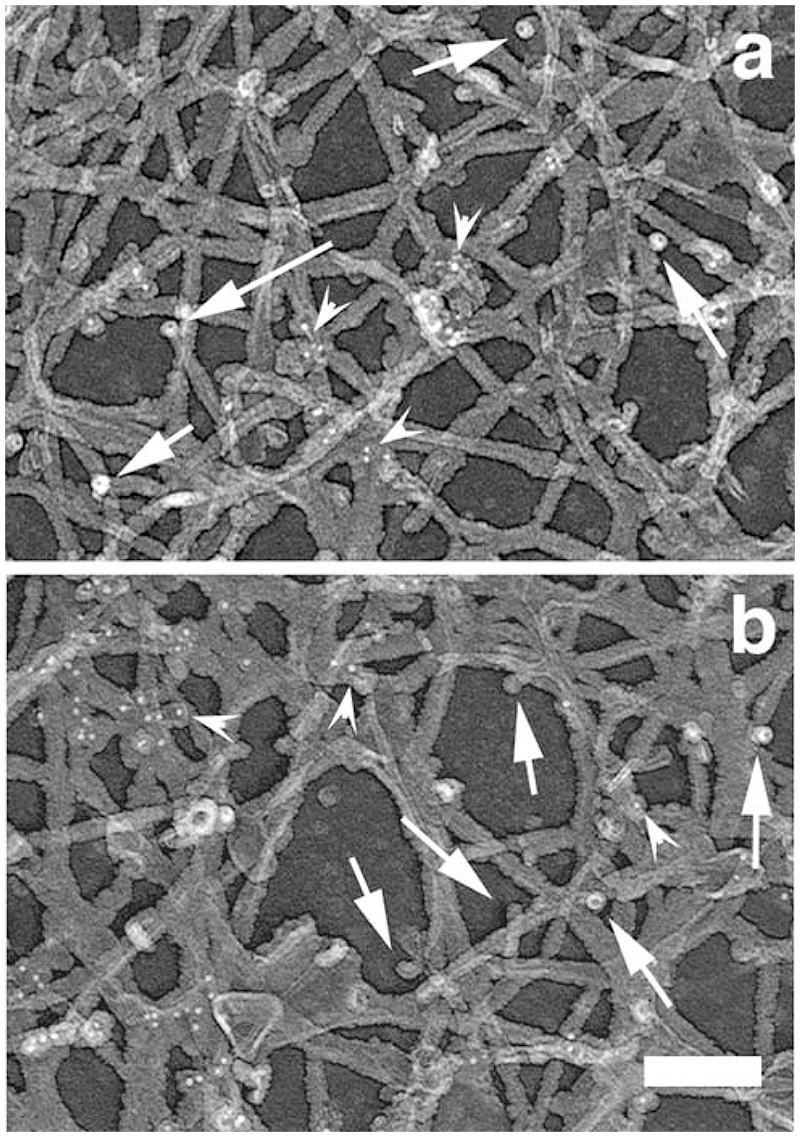
We then analyzed the spatial and temporal organization of this newly polymerized network in greater detail using electron microscopy. The polymerization mix containing actin only or actin and activated Arp2/3 was incubated for 1 or 5 min and the cells were then processed for replica microscopy. As expected from the light microscopy studies, the “actin only” samples contained few intact spread cells, while a much higher proportion of the cells in the “actin + activated Arp2/3 “ samples were nicely spread with extended leading edges (data not shown). A significant proportion of the cells were partially extracted, especially in the “actin only” samples, as would be expected from the drastic permeabilisation of the samples (Cramer, 1999) and as predicted by the light microscopy assay. However, a clear zone of dense actin network was visible in most cells all around the cell circumference after 5 min polymerization in the “actin + activated Arp2/3” samples (Figure 6d), which was absent from the “actin alone” sample (Fig6c). The leading edges of the cells appeared denser in the “actin + activated Arp2/3” samples (Fig6 f,h) compared to the actin alone samples (Figure 6 e,g), both after 1 and 5 min of incubation. Examination at a higher magnification revealed that this higher density was due to the 3 dimensional growth of a newly polymerized network on top of the pre-existing network. This newly polymerized network was more developed and complex in the “actin plus activated Arp2/3” samples compared to the “actin alone” samples, with more branched filaments (Figure 6, 3D right panels, and Figure 7). The labelled network clearly formed on top of the pre-existing network, by growing essentially in the vertical direction from the pre-existing unlabeled filaments. The pre-existing network was seen as a largely unlabeled 2D layer beneath the labelled network, with even a significant proportion of unlabeled, free-end outward pointing filaments at the edge (Figure 7b). The newly polymerized networks observed in the “actin only” samples were of a similar structure, but they were less complex with shorter actin filaments and less branches (Figure 7 a and c). Despite the Arp2/3-mediated network being much denser after 5 minutes polymerization, it appeared still largely restricted to the leading edge of the cells, as in the 1 min samples. Examination of the actin filaments far beneath the edge (2-3 um inside the cell) revealed that the labelled actin incorporation was minimal, even after 5 min polymerization (Figure 8, arrowheads), and that these networks still contained a significant proportion of unlabeled free-end filaments (Figure 8, arrows), confirming that the newly polymerized network does not just incorporate onto the ends of free-end filaments but builds on the whole of the pre-existing network at the leading edge.
Figure 6.
The Arp2/3-mediated in situ polymerisation is concentrated as a 3 dimensional network in a circumferential pattern at the edge of the cells. A polymerisation mix containing actin (“actin only”, a, c) or actin plus Arp2/3 plus VCA (“actin + activated Arp2/3”, b, d) was incubated on extracted cytoskeleton for 1 (a, b) or 5 (c, d) min before the cells were fixed and processed for replica microscopy. Right column: Red/blue 3D version of images e-h. Note that the “actin + activated Arp2/3” samples present leading edges with a much denser actin network, which is largely the result of a strong 3 dimensional growth of a newly formed network on top of the pre-existing one (see 3D montages on the right). Bars: a-d, 5um; e-h, 1 um.
Figure 7.
Structural organization of the de novo polymerised network in permeabilized cells. A polymerisation mix containing actin (“actin only”, a, c) or actin plus Arp2/3 plus VCA (“actin + activated Arp2/3”, b, d) was incubated on extracted cytoskeleton for 1 (a, b) or 5 (c, d) min before the cells were fixed and processed for replica microscopy. Newly polymerised labelled actin filaments (arrows pointing to gold particles) can be seen emerging from a largely unlabeled network, especially after a short time polymerisation (a, b), and with a greater density in the “actin + activated Arp2/3” sample (b). After 5 min polymerisation, a very dense 3 dimensional highly branched and strongly labelled network has been built at the edge of the cells, particularly in the “actin + activated Arp2/3” sample (d). Bar, 250 um.
Figure 8. The Arp2/3-mediated in situ polymerisation is minimal further back from the edge even after 5 min.
A polymerisation mix containing actin plus Arp2/3 plus VCA (“actin + activated Arp2/3”) was incubated on extracted cytoskeleton for 1 (a) or 5 (c) min before the cells were fixed and processed for replica microscopy. Arrowheads pointing at gold particles illustrate the minimal incorporation of exogenous actin, even after 5 min polymerisation (b). Arrows point at typical unlabelled free-end filaments. Bar, 100 nm.
In situ Arp2/3-mediated actin polymerization occurs on the pre-existing Arp2/3 rich network at the leading edge rather than at barbed end location
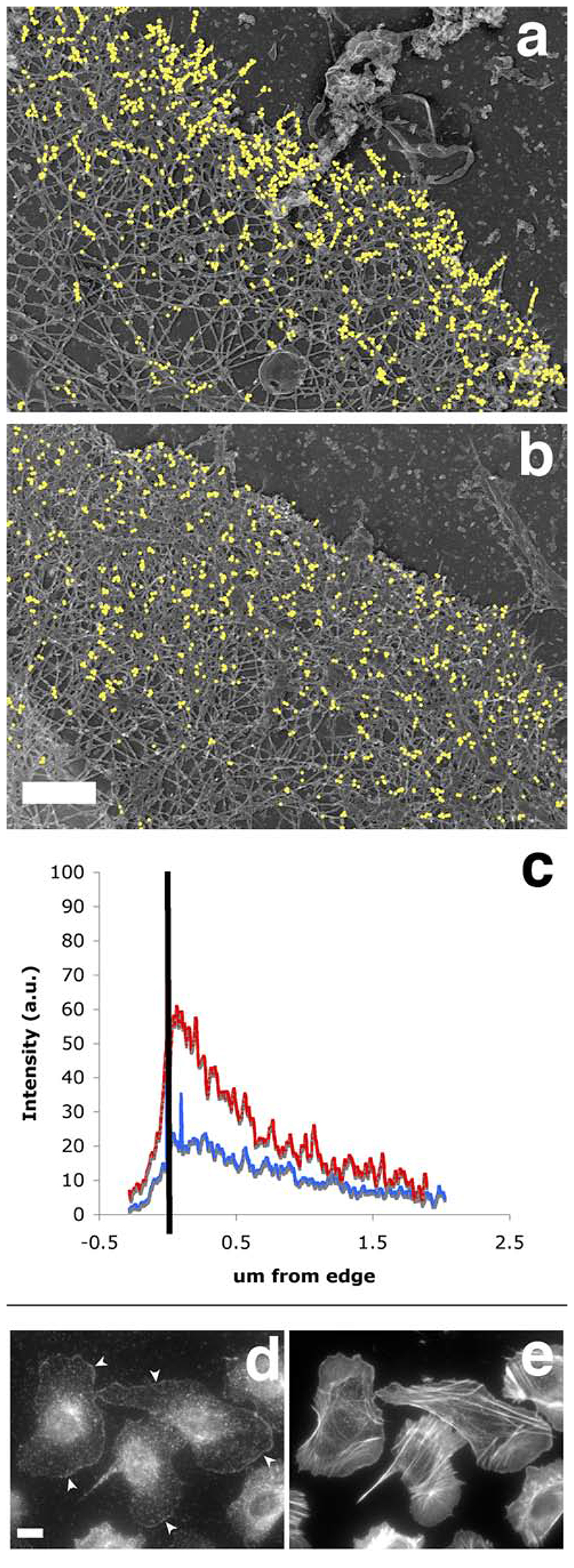
The pattern of Arp2/3 mediated polymerization did not match the barbed end distribution in unstimulated cells, and we observed a lot of unlabeled free-end filaments beneath the newly polymerized network at the leading edge. However, we couldn’t rule out the possibility that the polymerization that we observed was mostly the result of actin polymerization on free barbed ends, as our in situ polymerization assay conditions (full membrane extraction with triton before a polymerization step) were quite different from the standard barbed end localization assay (one step saponin permeabilisation and incubation). We thus performed our in situ Arp2/3 mediated polymerization assay in parallel with a modified version of the barbed end localization assay using a 1 min triton extraction in standard buffer C instead of saponin, as previously used for the detection of barbed ends at the electron microscopy level (Bailly et al., 1999). Both assays were performed on resting starved cells as well as cells stimulated with EGF for one minute, corresponding to the peak of free barbed end appearance at the leading edge (Bailly et al, 1999). As expected from previous work (data not shown), the pattern of barbed ends obtained was typical for these cells, i.e. the incorporation was limited to the edges of pre-existing lamellipods in unstimulated cells, and covered the whole cell circumference with greater intensity in the stimulated cells (Figure 9a, b). The only noticeable difference compared to the saponin protocol was an absence of incorporation at the tip of the stress fibres, suggesting that the corresponding focal contacts might be disassembled during the strong triton extraction. The Arp2/3-medaited polymerization samples done in parallel showed the typical circumferential polymerization pattern both in unstimulated and stimulated cells (Figure 9c,d), but the incorporation was clearly more intense at the leading edge for cells stimulated with EGF (Figure 9d and data not shown). As both the barbed ends and the Arp2/3 complex amount are increased at the leading edge after EGF stimulation for 1 min (Bailly et al, 1999; Desmarais et al, 2004), but their distribution is clearly distinct at the ultrastructural level (Bailly et al, 1999), we turned to high resolution electron microscopy to analyze the distribution of the newly polymerized Arp2/3 – mediated network with reference to the leading edge. The samples were analyzed after only 1 min polymerisation to obtain greater spatial resolution, and compared with the distribution of labelled actin incorporation at the leading edge in “actin only” samples. Consistent with our previous work (Bailly et al, 1999), the distribution of incorporated labelled actin at the leading edge in “actin only” samples was concentrated right beneath the edge, with a small peak within 400 nm of the membrane position, reflecting the distribution of free barbed ends (Figure 10, a and c). However, the distribution of labelled incorporated actin in the “actin plus activated Arp2/3” samples was flatter and broader, matching precisely the distribution of the Arp2/3 complex at the leading edge as previously published (Figure 10b, and c; Bailly et al, 1999). Incidentally, further localization of the Arp2/3 complex in unstimulated cells after triton extraction revealed a clear circumferential pattern, similar to the in situ Arp2/3 mediated polymerisation pattern that we observed (Figure 10, d and e).
Figure 9.
The Arp2/3-mediated in situ polymerisation pattern partially matches the barbed end pattern both spatially and temporally. MTln3 cells, starved (a,c) or stimulated with EGF for 1 min (b,d) were permeabilised with triton and stained for barbed end distribution (a,b) or Arp2/3-mediated in situ polymerisation (c,d). The barbed end distribution was assessed after 1 min incorporation of biotin labelled actin in standard polymerisation-enabling buffer, while the Arp2/3-mediated in situ polymerisation pattern was revealed after 5 min incubation of an actin/Arp2/3/VCA polymerisation mix. The Arp2/3-mediated in situ polymerisation pattern was similar in resting and stimulated cells and matched the barbed end distribution in stimulated cells. Exogenous actin accumulation around the cell circumference also appeared increased in stimulated cells as opposed to starved cells.
Figure 10.
The Arp2/3-mediated in situ polymerisation pattern is distinct from the free barbed end distribution, and closely matches the distribution of the cytoskeletal-bound fraction of the ar2/3 complex. A polymerisation mix containing actin (“actin only”, a, c) or actin plus Arp2/3 plus VCA (“actin + activated Arp2/3”, b, c) was incubated on extracted cytoskeleton for 1 min before the cells were fixed and processed for replica microscopy. Photos of the leading edges were taken at high resolution and these were processed to analyse the distribution of incorporated labelled actin as described in Material and Methods. a and b show the exogenous actin incorporation in representative leading edges from an “actin only “ sample and an “actin+ activated Arp2/3” sample respectively, where gold particles have been digitally enhanced for easier visualisation (bar, 0.5 um). c: average distribution of gold particles at the leading edge of the cells. Black bar represents the position of the leading edge, with the area on the left being outside the cell. Red, average distribution of exogenous actin in 6 “actin only “ samples; blue, average distribution of exogenous actin in 16 “actin+ activated Arp2/3” samples. d: the cytoskeleton–bound pool of Arp2/3 complex is mostly at the leading edge, mimicking the nucleation pattern in cells. Resting MTLn3 cells were extracted for 1 minute with 0.1% of Triton in cytoskeletal buffer containing 1 ug/ml phallacidin, and fixed and stained for F-actin using labelled phalloidin (right) and for Arp2/3 complex using anti-p34 antibodies (left). Detergent extraction of live cells eliminates most of the Arp2/3 complex in cytoplasm whilst a large portion of the Arp2/3 complex at the leading edge remains intact (arrowheads). Bar. 10 um.
Discussion
Actin polymerization at the leading edge of the lamellipod plays an essential role in cell motility, and the Arp2/3 complex has been proposed as a major player in this process (Pollard and Borisy, 2003). However, while its functional role in nucleating the polymerization of actin filaments has been extensively explored in vitro, much less is known about the relationship between this nucleation function and its ability to form specific and localized structured actin networks in cells. We have previously shown that both localized generation of free barbed ends and proper ultrastructural organization of the actin network are necessary for successful lamellipod extension (Desmarais et al., 2004). Particularly, by blocking the branching and nucleation activity of Arp2/3 complex in vivo using specific function-blocking antibodies, we showed that the specific organization of the network with filaments abutting the membrane at or around an optimal angle of 35° was necessary for lamellipod extension (Bailly et al., 2001; Desmarais et al., 2004). To further explore the relationship between the structural and functional role of the Arp2/3 complex, we have used here microinjection of the WA-domain of Scar and WASp to perturb the function of the Arp2/3 complex function in the cell by generating a sudden unlocalised burst of nucleation activity, and we have reconstructed WA- activated Arp2/3-mediated actin polymerization on extracted cytoskeletons.
As expected from previous work (Machesky and Insall, 1998, Hufner et al., 2001; Linder et al, 1999), microinjection of the WA domain of Scar and WASp proteins caused a rapid and transient increase in filamentous actin, which was accompanied by a dramatic reorganization of the polymerized actin network. The most striking effect was an accumulation of filamentous actin in the perinuclear region, with subsequent changes in stress fibre and focal contact patterns. Interestingly, while the initial increase in polymerized actin subsided rapidly (within 15 minutes), the resulting changes in the cytoskeleton organization persisted for much longer (over hours). This and our findings that the Arp2/3 complex within the leading edge network seems rather resistant to extraction suggests that Arp2/3-mediated networks may be quite stable in vivo, as opposed to their previously described instability in vitro using purified protein preparations (Le Clainche et al., 2003). Similarly, the perinuclear localization of the WA-mediated actin polymerization in the microinjected cells is consistent with this region harbouring most of the cytosolic free Arp2/3, while the cytoskeleton-bound Arp2/3 pool at the leading edge is only displaced when the cytoplasmic pool has been exhausted through extensive polymerization. An increased stability of the Arp2/3 networks could indeed explain the intriguing abundance of cytosolic Arp2/3 in cells, as a large pool of free protein would be then required for the rapid nucleation and polymerization activity that follows growth factor activation for example (the Arp2/3 incorporated in pre-existing networks being only slowly released as these networks remodel).
While we were using the microinjection of the WA domain as a mean to disturb the function of the Arp2/3 complex, we actually observed that the microinjected cells were even more efficient than the control cells in generating barbed ends after EGF stimulation, despite having similar background levels of barbed ends before stimulation. Similar results have been obtained in a neutrophil system, where loading of the Scar-WA domain did not affect the cells’ response to chemokine (Anderson et al, 2003). It is unlikely that this is due to a failure of Scar-WA to activate all of the Arp2/3 complex because we could show that the Arp2/3 complex is largely removed from the leading edge in our cells and locked in a multilayered complex network throughout the cells. Furthermore, cells transiently transfected with an expression vector containing the Scar-WA domain, presumably yielding maximum amounts of cytoplasmic Scar-WA, showed a phenotype similar to the microinjected cells (data not shown). Rather, we favour the idea that the presence of an increased Arp2/3 network in the cells is indirectly responsible for an increased number of barbed ends after stimulation. Since the Arp2/3 complex is already locked within a fully formed cytoplasmic network, it is also unlikely that Arp2/3 nucleation activity is responsible for the increase in barbed ends. However, it would be highly consistent with a direct role for cofilin not only in generating the barbed ends as suggested before (Chan et al, 2001; Desmarais et al, 2004; Ghosh et al, 2005), but also in generating them preferentially on Arp2/3 mediated networks. Indeed, recent work using caged cofilin showed that, even after global activation of cofilin over the whole cell area, the barbed end pattern observed after stimulation with EGF is still restricted to leading edge in the typical circumferential pattern (Ghosh et al, 2005), mimicking the pattern of pre-existing Arp2/3 networks as we describe here.
Rapid actin polymerization at the leading edge upon EGF stimulation requires the localized generation of free barbed ends (Bailly et al., 1999 and 2001), which is the result of a synergistic cooperation between cofilin and the Arp2/3 complex (DesMarais et al., 2004, Ichetovkin et al., 2002). However, confirming our previous work, we have shown here that the mere generation of new barbed ends is not sufficient to trigger lamellipod extension. Despite being more efficient at generating barbed ends after EGF stimulation, the WA-injected cells are incapable of building proper lamellipods before or after stimulation. We show here that is likely due to the fact that not only do the free barbed ends need to be localized, but also the growth of the Arp2/3-mediated network itself needs to be spatially restricted. Our analysis of Arp2/3-mediated networks, either in intact cells injected with WA-peptide or in de novo Arp2/3 networks polymerized on extracted cytoskeletons, showed that the WA-Arp2/3-mediated network is a multilayered 3D structure, with filaments pointing in every direction. This is in accord with recent theoretical insights into the structure of the lamellipod, predicting that unrestricted Arp2/3–mediated branching activity would generate a 3 dimensional multilayered structure because of the variation in the spatial orientation of the branching following the inherent rotation of the actin filament (Atilgan et al, 2005). The preferred lamellipod thickness, necessary to maintain extension, is only achieved if the filaments grow essentially in 2D, which corresponds to the filament organization that we observed in control lamellipods. The inability of the in vitro generated networks to restrict growth in the 2D plane would explain their failure to extend as proper lamellipodial structures (Atligan et al, 2005). How exactly the cells restrict the filament growth within the 2D plane is still controversial. It has been proposed that a competition between capping and branching favours the filaments oriented towards the membrane in the classical ±35° direction (Maly and Borisy, 2001). It is interesting to note here that even in the absence of capping protein, the Arp2/3-mediated networks we observed were essentially composed of short interconnected filaments. This suggests that at least the control of the filament size, which is necessary to maintain efficient protrusion, does not systematically require capping protein. Other work suggested that the spatial restriction of the growth was mostly reliant on biophysical and mechanical constraints within the membrane environment (Atilgan et al, 2005; Bernheim-groswasser et al, 2002). In both arguments however, the Arp2/3 complex must be localized at the leading edge for the network to extend, and it is likely that restriction in the spatial orientation of the Arp2/3 complex is essential for proper orientational pattern of the network (Atilgan et al, 2005). Our observations on the de novo generated networks on permeabilised cells indeed shows that the mere re-creation of a localized Arp2/3 network at the edge of the cell is not sufficient to promote a protrusion, suggesting that the presence of the membrane might indeed be essential to restrict polymerization within a 2D plane and preserve preferred lamellipod thickness. Altogether, this data concurs with recent evidence suggesting that, although the Arp2/3 complex is an important factor in determining the morphology of actin-based cellular structures (Svitkina and Borisy, 1999; Vignjevic et al, 2003), proper geometry of the actin cytoskeleton in cells is essential for lamellipod protrusion and cell motility (Atilgan et al, 2005, Plastino et al, 2004; Mejillano et al, 2004; Vignjevic et al., 2003; Bear et al, 2002).
It is generally assumed that barbed end generation and filament growth is restricted at the leading edge because of local signalling. While this is most certainly the case for polarized lamellipod extension during chemotaxis, where receptor engagement and subsequent signalling are localized in response to the gradient (Ridley et al, 2003; Bailly et al., 2000; Servant et al, 2000), it is less clear how global stimulation with growth factors generates localized protrusive activity and peripheral ruffling as commonly observed (Bailly et al, 1998; Machesky and Insall, 1998). Indeed, EGF receptor distribution on MTLn3 cells appears quite homogeneous, with no concentration at the edges of lamellipods, even after stimulation (Bailly et al., 2000). And yet, a homogenous up shift in EGF, which would then presumably bind all over the cell surface, activates actin polymerisation only locally at the extreme perimeter of the cell (Bailly et al., 1998, 1999). We provide here evidence that this specific pattern of polymerization can be recreated on extracted cytoskeletons by WA-activated- Arp2/3-mediated nucleation of actin polymerization using pure proteins. We show that this pattern could be partially influenced by the presence of barbed ends, which is consistent with previous works showing that the Arp2/3 complex preferred positions at or near the barbed ends to nucleate polymerization (Pantaloni et al, 2000; Falet et al, 2000). However, our high-resolution analysis showed that the pattern is more likely to be determined by the distribution of the cytoskeleton-bound Arp2/3 complex. This suggests that the de novo Arp2/3-mediated polymerization occurs preferentially on pre-existing Arp2/3-rich actin networks. One likely explanation is that these networks are preferred for polymerization because they are tropomyosin free (Desmarais et al, 2002 and data not shown). The tropomyosin distribution can potentially dictate the network structure by inhibiting F-actin binding in specific areas, thus assigning a lamellipod structure only to areas where absence of tropomyosin on the filaments allows the binding of the lamellipod signature proteins such as Arp2/3 (Gupton S et al, 2005; Desmarais et al, 2002). Alternatively, Arp2/3 networks might be preferred because of their net-like easily accessible structure, as opposed to tightly bundled filaments in stress fibres (Machesky et al., 1999). Clearly more work will be needed to discriminate between these two possibilities but it is interesting to notice as discussed above that the increase in Arp2/3-mediated networks in cells microinjected with the WA-domains was accompanied by an increased number of barbed ends after EGF stimulation as compared to control.
Interestingly, when the intrinsic cellular pool of Arp2/3 was activated by microinjection of the WA peptides, we did not observe a specific polymerization at the leading edge. However, it should be noted that the expected cellular concentrations of G-actin, and WA peptides and Arp2/3 in these conditions are respectively about 30 and 500 times higher than the concentrations we used in the polymerization mix, thus predicting a much faster nucleation and polymerization. Furthermore, diffusion in the cytosol is expected to be considerably slowed down compared to the solution of pure proteins, making it more likely that the polymerization would occur extremely rapidly within the perinuclear cytosol where the Arp2/3 complex is more abundant.
The results presented here, combined with our previous work, confirm that if the generation of free barbed ends is a prerequisite for lamellipod extension, it is clearly not sufficient for protrusion. We also show that, as predicted from the molecular mechanism of branching, unrestricted Arp2/3 –mediated nucleation and polymerization of actin generates multilayered 3 dimensional networks, whose growth doesn’t generate a lamellipodial-like net protrusion. In addition, in the presence of a pre-existing cytoskeleton, the Arp2/3 –mediated polymerization is restricted to specific areas within the cell, potentially corresponding to pre-existing Arp2/3-rich networks. Altogether, this suggest that successful lamellipod extension is an extremely well organized process governed by the molecular composition and structural organization of the cytoskeleton at the leading edge of the cell. This process appears to be regulated at multiple levels involving localized barbed end generation, restriction of the Arp2/3-mediated actin polymerization to defined subcellular compartments, and confinement of the growth of the Arp2/3 complex –mediated network mostly within a 2-dimensional plane. This raises the intriguing possibility that cell protrusive activity and shape is governed at multiple levels in the cell, not only by local activation of specific biochemical pathways, but also by the molecular composition and ultrastructural organization of the actin cytoskeleton. Such a hypothesis would actually be consistent with recent observations in MTLn3 cells that the in vivo pattern of activation of N-WASP (Lorenz et al., 2004), a direct activator of Arp2/3, is much broader than the pattern of nucleation sites previously described in detail in these cells (Bailly et al., 1999), suggesting that nucleation of actin polymerization is further restricted to a specific area by a mechanism other than local activation of a nucleation-promoting factor. Clearly further work will be needed to test this idea but as new and more powerful tools are developed to study the biology of the cell, the challenge will be to integrate all the data obtained using in vitro models into the actual 3 dimensional cell structure to gain novel insights into cell function.
Materials and methods
Proteins and reagents
Actin and Arp2/3 complex were purified from rabbit muscle acetone powder and human platelets (Bailly et al, 2001). Recombinant cofilin was prepared as previously described (Zebda et al, 2000). Human WASp-WA and Scar1-WA domains (Machesky et al, 1998) were expressed in bacteria and purified as GST-fusion proteins. The GST tag was removed by thrombin (Sigma) digestion, and the WA domains were further purified by Mono-Q and Superdex-75 filtration (Amersham). Fluorescently- labelled phalloidins, Alexa 488 labelled Dnase I, lysine fixable FITC- and Rhodamine-labeled dextran (MW 10,000 were from Molecular Probes. Biotin-labelled actin and bovine brain Arp2/3 complex were purchased from Cytoskeleton Ltd.
Cells and antibodies
Rat MTLn3 cells were maintained and stimulated with EGF as previously described (Bailly et al. 1998, Bailly et al., 1999). Anti-p34 antibody was raised against a peptide containing amino acids 286-298 of the human p34 subunit (Bailly et al., 2001). Cy3-labeled anti-biotin antibodies were from Sigma-Aldrich. FITC and TRITC- labelled secondary antibodies were from Jackson Laboratories. 5nm-gold coupled anti-biotin antibodies were from British BioCell.
Microinjection
Cells were grown on Mattek dishes as previously described (Bailly et al, 2001), and placed in L15 medium (Life Technologies) with 0.35% BSA for 1–2 h before injection. Microinjection was conducted using an Eppendorf semi-automated microinjection system using needles pulled on a Sutter p87 micropipette puller. WA domains at a concentration of 136 μM in PBS (phosphate buffer saline) were mixed with FITC- or Rhodamine labelled dextran (0.8 mg/ml final concentration) for identification of the injected cells. Cells were usually allowed to recover for 30 min before further manipulations.
Immunofluorescence
F-actin and p34 staining were performed as described (Bailly et al., 1998; 1999, 2001). G-actin staining using fluorescently–labelled DNAse was performed as described in Cramer et al., 2002. Samples were examined on a Zeiss Axiovert 100M using a Plan Apochromat 63X oil immersion objective (NA 1.4), and images were taken using a CCD camera coupled to an OpenLab-driven acquisition system (Improvision), or on a Zeiss Axiovert S100 TV using a comparable 63X objective, and coupled to a Biorad Radiance 2000 confocal microscope for image acquisition. To identify the fraction of the Arp2/3 complex bound to the cytoskeleton, the cells were extracted for 1 minute in a Triton containing-buffer (0.1% Triton X100 in cytoskeletal buffer with 1 ug/ml phallacidin) prior to fixation and processing for standard immunofluorescence. For figure purposes, all images were processed in Adobe Photoshop to enhance contrast using identical settings for matching group of images.
Barbed end localization
The standard barbed end assay was performed as previously described (Chan et al., 1998, Bailly et al., 1999) by permeabilizing the cells for 1 minute in a saponin-containing polymerization-enabling buffer containing 0.45 μM of biotin-labelled G-actin. The cells were then fixed and barbed ends were detected using Cy3-conjugated mouse anti-biotin antibodies. For barbed end localization after triton extraction, the protocol originally described for barbed end localization in electron microscopy samples was followed (Bailly et al, 1999), except that the samples were first washed once briefly with 0.25% triton in Buffer C/1%BSA before adding the triton/BSA/rhodamin actin mix for 1 min.
Lamellipod Extension Assay
Lamellipod extension after EGF stimulation was quantified as a net increase in the cell area (Bailly et al, 1998, 1999). MTLn3 cells were plated on Mattek dishes, starved for 3 hours in L15 medium containing 0.35% BSA prior to stimulation with 5 nM EFG, and microinjected as described above. Cells were placed in an environmental chamber at 37°C on a Zeiss Axiovert 100M and one image under fluorescence was taken to identify the microinjected cells. Phase contrast images were taken every minute using a CCD camera coupled to an OpenLab-driven acquisition system (Improvision). Resulting image stacks were loaded onto the NIH Image software where the cells’ contours were manually traced, and the resulting cell areas calculated. Relative area increase was calculated by dividing the cell area at each time point by the average cell area measured in the few minutes prior to stimulation.
Fluorescence Quantification
Full range fluorescence digital images of total F-actin and barbed ends were captured as described above using identical settings for matching sets of data. The images were converted linearly in NIH Image. The mean pixel intensity per cell and the cell area were determined, as described previously (Bailly et al., 1999), and the resulting integrated density value per cell was calculated (area x mean pixel intensity) as a relative measure of the total F-actin or barbed end content in the cell. Intensities values were normalized against that of control uninjected cells within the same field to overcome the variability between experiments and imaging fields. Barbed ends at the leading edge were calculated as the relative mean fluorescence of a cell perimeter within 1.1 um at the leading edge using a previously described NIH macros (Bailly et al 2001, Desmarais et al, 2004).
Electron microscopy
For metal replica microscopy, MTLn3 cells were grown on customized rectangular small glass coverslips for 24 hours, and microinjected with a mix of Scar-WA domain and biotinylated G-actin at a concentration of 2.5 mg/ml to serve as a marker to identify microinjected cells at the end of the procedure (Desmarais et al, 2004), and left to recover for 30 minutes. The cells were then permeabilized and immunostained using anti-biotin antibodies as previously described (Bailly et al, 1999; Desmarais et al, 2004). After post-fixation in with 1% glutaraldehyde and 5 uM phallacidin for 15 min, and the samples were transferred to cytoskeletal buffer containing 5 uM phallacidin (Bailly et al, 1999). To prepare samples for rotary shadow, the coverslips were transferred to distilled water with 5 uM phallacidin, washed twice for 5 minutes each with distilled water containing 0.1 uM phallacidin, and transferred to distilled water. The coverslips were then plunge-frozen in a liquid-nitrogen-cooled 1:4 mixture of isopentane:propane. The samples were transferred to the specimen mount of a freeze fracture unit (Balzers BAF400D) and rotary shadowed at a 45° angle with 1.2-1.3 nm tantalum-tungsten or 1.5 nm platinum, followed by respectively 2.5 or 5 nm carbon at 90°. The replicas were separated from the glass coverslips with 8% hydrofluoric acid, washed into distilled water, and picked up onto the surface of formvar-coated copper grids. Samples were examined using a JEOL 1010 transmission electron microscope at 80 kV. Negatives were scanned and processed for increased contrast in Photoshop. To obtain 3D rendering of the samples, ± 10 degree tilt stereo pair images were generated, merged and converted to a red/blue 3D image in Photoshop, so that sample depth can be viewed using standard red/blue 3D glasses. For the analysis of the distribution of incorporated labelled actin at the leading edge, the negatives representing leading edges at high resolution were rotated in Adobe Photoshop so as to place all of them in a left/right orientation (with the outside of the cell on the left and the inside on the right) with the leading edge perpendicular to this orientation. The leading edge was marked manually on the modified negatives as a vertical line and the gold particles representing the exogenous actin incorporation were marked in colour using the pen tool (see Figure 10a,b). The particle distribution as well as the leading edge mark were then extracted from the original negative and processed in Image J using the plot profile function, generating a profile of the actin incorporation with a reference to the leading edge. The profiles of individual leading edges from different cells were then averaged in Excel with reference to the position of the leading edge for each of them.
In situ actin polymerization using a purified protein mix
Resting cells were permeabilized with 0.25% Triton in cytoskeletal preservation buffer for 45 sec (Bailly et al, 1999), and following a rapid wash, the remaining cytoskeletons were exposed to an actin polymerisation cocktail of purified proteins (20 nM Scar-WA, 5 nM Arp2/3 complex bovine or human, 0.2 uM cofilin and 2 uM biotin-actin in ISAP buffer pH 6.7; Ichetovkin et al, 2002). Live imaging was performed on a Zeiss Axiovert 100M using a Plan Apochromat 63X oil immersion objective (NA 1.4), and images were taken every 30 seconds using a CCD camera coupled to an OpenLab-driven acquisition system (Improvision). Alternatively, after 5-10 min polymerisation at room temperature, the samples were fixed and stained with FITC-Phalloidin and Cy3-labelled anti-biotin for light microscopy. For further high resolution analysis and comparison with barbed end distribution, cofilin was omitted from the Arp2/3 polymerisation mix, and the polymerisation was allowed to proceed for only 1or 5 min, after which the cells were processed for light microscopy or metal replica electron microscopy as described above.
Acknowledgements
The authors wish to thank Dr Laura Machesky for the Scar and WASp-WA constructs, Drs Vera Desmarais and John Condeelis for sharing unpublished data and for the GFP-Beta-actin construct, Michael Cammer for advice on image quantitation, and Dr Alpa Ahir for help in the preparation of the anti-p34 antibody. This work was supported by grants from the Wellcome Trust, the University College London Central Research Funds and the Special Trustees of Moorfields Eye Hospital (MB).
References
- Anderson SI, Behrendt B, Machesky LM, Insall RH, Nash GB. Linked regulation of motility and integrin function in activated migrating neutrophils revealed by interference in remodelling of the cytoskeleton. Cell Motil Cytoskeleton. 2003;54:135–46. doi: 10.1002/cm.10091. [DOI] [PubMed] [Google Scholar]
- Atilgan E, Wirtz D, Sun S. Morphology of the Lamellipodium and Organization of Actin Filaments at the Leading Edge of Crawling Cells. Biophys J. 2005;89:3589–602. doi: 10.1529/biophysj.105.065383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly M, Ichetovkin I, Grant W, Zebda N, Machesky LM, Segall JE, Condeelis J. The F-actin side binding activity of the Arp2/3 complex is essential for actin nucleation and lamellipod extension. Curr Biol. 2001;11:620–625. doi: 10.1016/s0960-9822(01)00152-x. [DOI] [PubMed] [Google Scholar]
- Bailly M, Macaluso F, Cammer M, Chan A, Segall JE, Condeelis JS. Relationship between Arp2/3 complex and the barbed ends of actin filaments at the leading edge of carcinoma cells after epidermal growth factor stimulation. J Cell Biol. 1999;145:331–345. doi: 10.1083/jcb.145.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly M, Wyckoff J, Bouzahzah B, Hammerman R, Sylvestre V, Cammer M, Pestell R, Segall JE. Epidermal growth factor receptor distribution during chemotactic responses. Mol Biol Cell. 2000;11:3873–3883. doi: 10.1091/mbc.11.11.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly M, Yan L, Whitesides GM, Condeelis JS, Segall JE. Regulation of protrusion shape and adhesion to the substratum during chemotactic responses of mammalian carcinoma cells. Exp Cell Res. 1998;241:285–299. doi: 10.1006/excr.1998.4031. [DOI] [PubMed] [Google Scholar]
- Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, Maly IV, Chaga OY, Cooper JA, Borisy GG, Gertler FB. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 2002;109:509–21. doi: 10.1016/s0092-8674(02)00731-6. [DOI] [PubMed] [Google Scholar]
- Bernheim-Groswasser A, Wiesner S, Golsteyn RM, Carlier MF, Sykes C. The dynamics of actin-based motility depend on surface parameters. Nature. 2002;417:308–11. doi: 10.1038/417308a. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD, Mullins RD. Interactions of ADF/cofilin, Arp2/3 complex, capping protein and profilin in remodeling of branched actin filament networks. Curr Biol. 2000;10:1273–1282. doi: 10.1016/s0960-9822(00)00749-1. [DOI] [PubMed] [Google Scholar]
- Chan AY, Bailly M, Zebda N, Segall JE, Condeelis JS. Role of cofilin in epidermal growth factor-stimulated actin polymerization and lamellipod protrusion. J Cell Biol. 2000;148:531–542. doi: 10.1083/jcb.148.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AY, Raft S, Bailly M, Wyckoff JB, Segall JE, Condeelis JS. EGF stimulates an increase in actin nucleation and filament number at the leading edge of the lamellipod in mammary adenocarcinoma cells. J Cell Sci. 1998;111(Pt 2):199–211. doi: 10.1242/jcs.111.2.199. [DOI] [PubMed] [Google Scholar]
- Cramer LP. Role of actin-filament disassembly in lamellipodium protrusion in motile cells revealed using the drug jasplakinolide. Curr Biol. 1999;9:1095–105. doi: 10.1016/s0960-9822(99)80478-3. [DOI] [PubMed] [Google Scholar]
- Cramer LP, Briggs LJ, Dawe HR. Use of fluorescently labelled deoxyribonuclease I to spatially measure G-actin levels in migrating and non-migrating cells. Cell Motil Cytoskeleton. 2002;51:27–38. doi: 10.1002/cm.10013. [DOI] [PubMed] [Google Scholar]
- DesMarais V, Ichetovkin I, Condeelis J, Hitchcock-DeGregori SE. Spatial regulation of actin dynamics: a tropomyosin-free, actin-rich compartment at the leading edge. J Cell Sci. 2002;115:4649–4660. doi: 10.1242/jcs.00147. [DOI] [PubMed] [Google Scholar]
- DesMarais V, Macaluso F, Condeelis J, Bailly M. Synergistic interaction between the Arp2/3 complex and cofilin drives stimulated lamellipod extension. J Cell Sci. 2004;117:3499–3510. doi: 10.1242/jcs.01211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falet H, Hoffmeister KM, Neujahr R, Italiano JE, Jr, Stossel TP, Southwick FS, Hartwig JH. Importance of free actin filament barbed ends for Arp2/3 complex function in platelets and fibroblasts. Proc Natl Acad Sci U S A. 2002;99:16782–7. doi: 10.1073/pnas.222652499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M, Song X, Mouneimne G, Sidani M, Lawrence DS, Condeelis JS. Cofilin promotes actin polymerization and defines the direction of cell motility. Science. 2004;304:743–6. doi: 10.1126/science.1094561. [DOI] [PubMed] [Google Scholar]
- Gupton SL, Anderson KL, Kole TP, Fischer RS, Ponti A, Hitchcock-DeGregori SE, Danuser G, Fowler VM, Wirtz D, Hanein D, Waterman-Storer CM. Cell migration without a lamellipodium: translation of actin dynamics into cell movement mediated by tropomyosin. J Cell Biol. 2005;168:619–31. doi: 10.1083/jcb.200406063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufner K, Higgs HN, Pollard TD, Jacobi C, Aepfelbacher M, Linder S. The verprolin-like central (vc) region of Wiskott-Aldrich syndrome protein induces Arp2/3 complex-dependent actin nucleation. J Biol Chem. 2001;276:35761–35767. doi: 10.1074/jbc.M106520200. [DOI] [PubMed] [Google Scholar]
- Ichetovkin I, Grant W, Condeelis J. Cofilin produces newly polymerized actin filaments that are preferred for dendritic nucleation by the Arp2/3 complex. Curr Biol. 2002;12:79–84. doi: 10.1016/s0960-9822(01)00629-7. [DOI] [PubMed] [Google Scholar]
- Le Clainche C, Pantaloni D, Carlier MF. ATP hydrolysis on actin-related protein 2/3 complex causes debranching of dendritic actin arrays. Proc Natl Acad Sci U S A. 2003;100:6337–6342. doi: 10.1073/pnas.1130513100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder S, Higgs H, Hufner K, Schwarz K, Pannicke U, Aepfelbacher M. The polarization defect of Wiskott-Aldrich syndrome macrophages is linked to dislocalization of the Arp2/3 complex. J Immunol. 2000;165:221–225. doi: 10.4049/jimmunol.165.1.221. [DOI] [PubMed] [Google Scholar]
- Linder S, Nelson D, Weiss M, Aepfelbacher M. Wiskott-Aldrich syndrome protein regulates podosomes in primary human macrophages. Proc Natl Acad Sci U S A. 1999;96:9648–9653. doi: 10.1073/pnas.96.17.9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M, Yamaguchi H, Wang Y, Singer RH, Condeelis J. Imaging sites of N-wasp activity in lamellipodia and invadopodia of carcinoma cells. Curr Biol. 2004;14:697–703. doi: 10.1016/j.cub.2004.04.008. [DOI] [PubMed] [Google Scholar]
- Machesky LM, Insall RH. Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr Biol. 1998;8:1347–1356. doi: 10.1016/s0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
- Machesky LM, Mullins RD, Higgs HN, Kaiser DA, Blanchoin L, May RC, Hall ME, Pollard TD. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc Natl Acad Sci U S A. 1999;96:3739–3744. doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maly IV, Borisy GG. Self-organization of a propulsive actin network as an evolutionary process. Proc Natl Acad Sci U S A. 2001;98:11324–9. doi: 10.1073/pnas.181338798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard TH, Sharp SJ, Machesky LM. Signalling to actin assembly via the WASP (Wiskott-Aldrich syndrome protein)-family proteins and the Arp2/3 complex. Biochem J. 2004;380:1–17. doi: 10.1042/BJ20040176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejillano MR, Kojima S, Applewhite DA, Gertler FB, Svitkina TM, Borisy GG. Lamellipodial versus filopodial mode of the actin nanomachinery: pivotal role of the filament barbed end. Cell. 2004;118:363–73. doi: 10.1016/j.cell.2004.07.019. [DOI] [PubMed] [Google Scholar]
- Pantaloni D, Boujemaa R, Didry D, Gounon P, Carlier MF. The Arp2/3 complex branches filament barbed ends: functional antagonism with capping proteins. Nat Cell Biol. 2000;2:385–91. doi: 10.1038/35017011. [DOI] [PubMed] [Google Scholar]
- Plastino J, Olivier S, Sykes C. Actin Filaments Align into Hollow Comets for Rapid VASP-Mediated Propulsion. Curr Biol. 2004;14:1766–71. doi: 10.1016/j.cub.2004.09.054. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–9. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- Servant G, Weiner OD, Herzmark P, Balla T, Sedat JW, Bourne HR. Polarization of chemoattractant receptor signaling during neutrophil chemotaxis. Science. 2000;287:1037–40. doi: 10.1126/science.287.5455.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina TM, Borisy GG. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignjevic D, Yarar DM, Welch D, Peloquin J, Svitkina T, Borisy GG. Formation of filopodia-like bundles in vitro from a dendritic network. J Cell Biol. 2003;160:951–962. doi: 10.1083/jcb.200208059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch MD, Mullins RD. Cellular control of actin nucleation. Annu Rev Cell Dev Biol. 2002;18:247–288. doi: 10.1146/annurev.cellbio.18.040202.112133. [DOI] [PubMed] [Google Scholar]
- Wiesner S, Helfer E, Didry D, Ducouret G, Lafuma F, Carlier MF, Pantaloni D. A biomimetic motility assay provides insight into the mechanism of actin-based motility. J Cell Biol. 2003;160:387–98. doi: 10.1083/jcb.200207148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebda N, Bernard O, Bailly M, Welti S, Lawrence DS, Condeelis JS. Phosphorylation of ADF/cofilin abolishes EGF-induced actin nucleation at the leading edge and subsequent lamellipod extension. J Cell Biol. 2000;151:1119–1128. doi: 10.1083/jcb.151.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]