Abstract
Grapevine leafroll disease (GLD) is a globally spreading viral infection that causes major economic losses by reducing crop yield, plant longevity and berry quality, with no effective treatment. Grapevine leafroll associated virus-3 (GLRaV-3) is the most severe and prevalent GLD strain. Here, we evaluated the ability of RNA interference (RNAi), a non-GMO gene-silencing pathway, to treat GLRaV-3 in infected Cabernet Sauvignon grapevines.
We synthesized lipid-modified polyethylenimine (lmPEI) as a carrier for long double-stranded RNA (dsRNA, 250-bp-long) that targets RNA polymerase and coat protein genes that are conserved in the GLRaV-3 genome. Self-assembled dsRNA-lmPEI particles, 220 nm in diameter, displayed inner ordered domains spaced 7.3±2 nm from one another, correlating to lmPEI wrapping spirally around the dsRNA. The particles effectively protected RNA from degradation by ribonucleases, and Europium-loaded particles applied to grapevine leaves were detected as far as 60-cm from the foliar application point. In three field experiments, a single dose of foliar administration knocked down GLRaV-3 titer, and multiple doses of the treatment kept the viral titer at baseline and triggered recovery of the vine and berries.
This study demonstrates RNAi as a promising platform for treating viral diseases in agriculture.
Keywords: viral disease, nanotechnology, siRNA, RNAi, agriculture
1. Introduction
The wine industry in both ancient and modern times depends greatly on healthy vines and fine grapes.[1] In recent decades, viral grapevine leafroll disease (GLD) poses a major economic threat to wine production by reducing crop yield and hampering grape quality including cluster size, pH, sugar level and color.[2] Among eleven viruses associated with GLD, grapevine leafroll associated virus 3 (GLRaV-3) is the most prevalent and severe strain inducing robust symptoms by decreasing plant vigor and longevity.[3] In New Zealand, for example, GLRaV-3 delayed the ripening of Sauvignon Blanc berries and reduced their acidity.[4] In Benton Harbor, Michigan, yield per vine along with soluble solids content declined in infected Cabernet Franc vines.[5]
Genetically, GLRaV-3 belongs to the Ampelovirus genus and consists of helical, positive sense single stranded RNA (ssRNA) genome approximately 18.5 kb in size.[6] The virus has 12 open reading frames coding for replication and structure-related proteins such as RNA dependent RNA polymerase (RdRp) and coat protein (CP), respectively, among other essential proteins.[7] Current approaches for dealing with GLD include uprooting and incinerating infected vines to curb virus transmission and progression.[8] Thus, new technological approaches are warranted in order to mitigate GLD’s negative impact.
RNA interference (RNAi) is a gene regulation mechanism also known as post-transcriptional gene silencing (PTGS).[9] This mechanism is sequence-specific due to its dependency on double stranded RNA (dsRNA) precursors to trigger gene silencing,[10] although ssRNA precursors were shown to share the same RNAi pathway but result in lower efficiency.[11] In plants, RNase III-like enzyme, dicer-like protein, processes dsRNA into 21-24 bp short interfering RNA (siRNA) which guide transcript recognition and degradation downstream.[12] Unlike mammalian cells, dicer-like protein preferably processes long dsRNA sequences to yield multiple siRNAs which transfer between plant cells via plasmodesmata.[13] In order to utilize RNAi, dsRNA needs to enter the cell cytoplasm. RNA particles, complexed with lipids and polymers, have been used for triggering RNAi in medicine and aquaculture.[14] Direct foliar application of naked dsRNA may result in RNA degradation prior to penetrating the cell. In addition, standard delivery methods (e.g. Agrobacterium and DNA vectors) have limitations of their own such as off-target effects, while formulation can extend the silencing duration and specificity.[15]
Here, we evaluate a delivery platform for long dsRNA based on lipid-modified polyethylenimine (lmPEI) for efficient systemic silencing of GLRaV-3 in grapevines. We assessed the ability to use RNAi for treating viral infections in grapevines. dsRNA-lmPEI particles synthesis is rapid and scalable resulting in stable particles in ambient conditions. In addition, dsRNA-lmPEI particles are shown to protect the RNA payload from ribonuclease activity and trigger GLRaV-3 viral knockdown in three consecutive field experiments following foliar administration. Altogether, this study opens a frontier for using RNAi to treat viral infections in grapevines.
2. Results and Discussion
2.1. Preparation and characterization of dsRNA-lmPEI particles
We synthesized lmPEI as a RNA carrier and tested its effect on GLRaV-3 titer in grapevines. Briefly, 14-carbon lipid was conjugated to branched PEI in a 3:1 (epoxide tail:PEI head group) molar ratio to form lmPEI. Next, 250 bp dsRNA was complexed with lmPEI under acidic conditions (pH=5.2) to establish electrostatic interactions between the negatively charged dsRNA and cationic lmPEI, to formulate dsRNA-lmPEI particles, as illustrated in Figure 1A. To target GLRaV-3’s ability to replicate and assemble, we chose to knockdown RNA-dependent RNA polymerase and coat protein genes using two conserved sequences (Figure S1 and S2). The sequence design excluded unintended off-targets within the GLRaV-3 and wine grapevine (e.g. Vitis vinifera) genomes. The dsRNA length was optimized to trigger the dicer-like protein at different locations along the sequence in order to generate multiple siRNAs and increase knockdown probability.[14, 16]
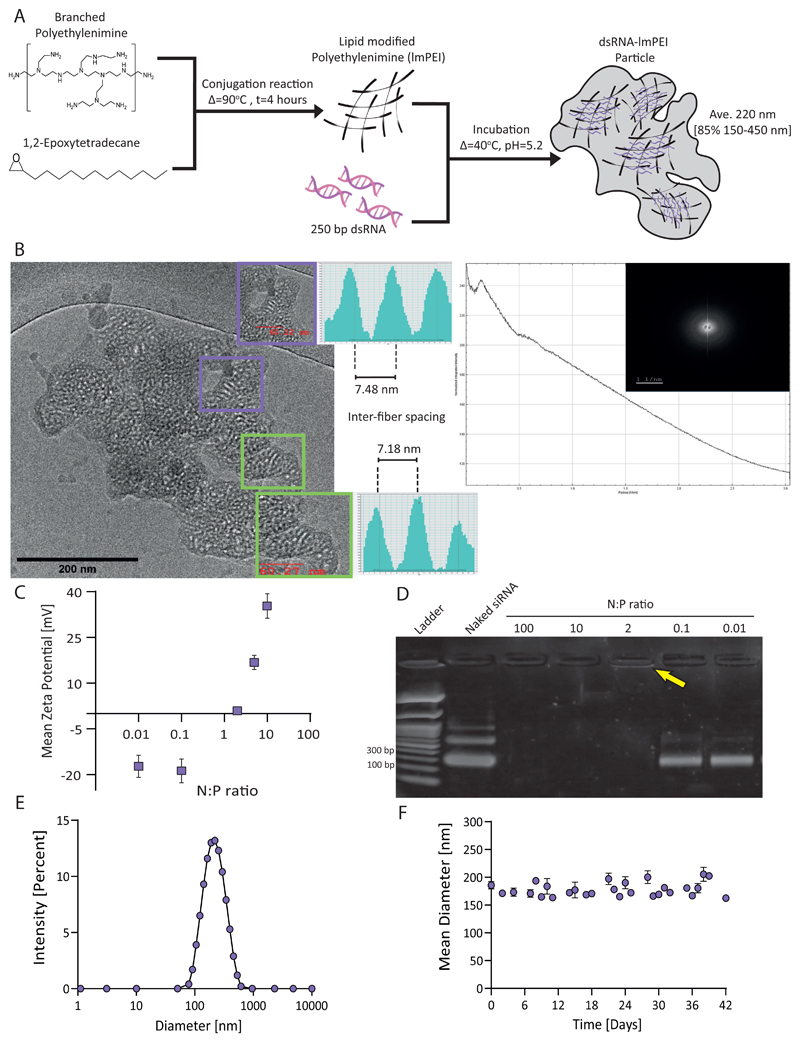
Figure 1. dsRNA-lmPEI particles.
Illustration of particle synthesis including both lipidated tail conjugate and particle formulation (A). cryoTEM image presenting dsRNA-lmPEI particle with inner ordered domains and its relevant fast fourier transformation (FFT) (B). Enhanced insets show gray values profile measurements indicating inter-fiber spacing (7.3±2 nm) from one dsRNA center of mass to another. We examined particles with different N:P ratios according to their Zeta potential (N=3, C) and long dsRNA binding capacity, analyzed by gel electrophoresis (2% agarose, 100V, 35 minutes, D). At low N:P ratios (0.01 and 0.1), bands signify unbound dsRNA matching to naked dsRNA. The yellow arrow highlights band corresponding to complexed dsRNA (0.91±0.08 mV) indicating successful complexation. Particles were tested for their size distribution using dynamic light scattering, peaking at 220 ± 7.75 nm (E). Particles’ stability was coherent for over 40 days at room temperature, 25mM sodium acetate, pH=5.2, with no significant change in size measured (F).
To be effective, the dsRNA-lmPEI particle must bind, protect and release the RNA at the target site.[17] Since binding and release rely on lmPEI electrostatic affinity to dsRNA, they can be controlled through the N:P ratio. In our study, the N:P ratio is defined as the molar ratio between positively charged amine groups present in lmPEI and negatively charged phosphate groups present on the dsRNA backbone. Increasing the N:P ratio elevated the particle’s surface charge (Figure 1C), while below an N:P ratio of 2 (i.e. 0.01 and 0.1) particles were not formed, indicating insufficient lmPEI to bind dsRNA, Figure 1D. Therefore, we conducted our next experiments using N:P=2 particles which bind dsRNA, carrying a weak positive charge (0.91 ± 0.08 mV). The encapsulation efficiency of dsRNA was 92% (Figure S3) and the particle size averaged 220 nm (Figure 1E) with 85% of particles ranging 150-450 nm. The dsRNA-lmPEI particles were imaged using cryo-TEM. Low and high contrast patterns indicate fibrillar high contrast aggregated structure. Some domains exhibited local order within the particle. The high contrast features correspond to sp2 carbons of the π-stacked system representing dsRNA and the low contrast represents sp3 hybridized atoms of the lmPEI, respectively.[18] Relevant radial integration of fast fourier transform (FFT) of the imaged particle was employed in the investigation of the inner structure of the particles (Figure 1B). Previous studies have shown that DNA/PEI complexation is highly kinetic. This is due to electrostatic forces, the main driving force for binding, being affected by the percentage of protonated groups within PEI.[19] Inter-fiber spacing between one dsRNA center of mass to another was 7.3 ± 2 nm, as observed from the FFT and from the gray values profile measurements (see insets in Figure 1B; additional measurements are found in Figure S4). This spacing seems to be due to the lipid tails presence within the particle and their protrusion and possible interaction with other? lmPEI molecules attached to other parts of the dsRNA fiber. The lipophilic character of the alkylic chains of the lmPEI also contribute to the aggregate formation in the aquatic environment as observed in the cryo-TEM image and from partially energy minimized molecular mechanics model (Figure S5). Similar to the proposed model by Ziebarth and colleagues,[20] our findings may also show a possible model of lmPEI wrapping around dsRNA in a spiral manner. Finally, we tested the particle size as a measure of stability and did not notice significant changes over a period of 40 days (Figure 1F).
2.2. Particle biodistribution in the leaf and vine
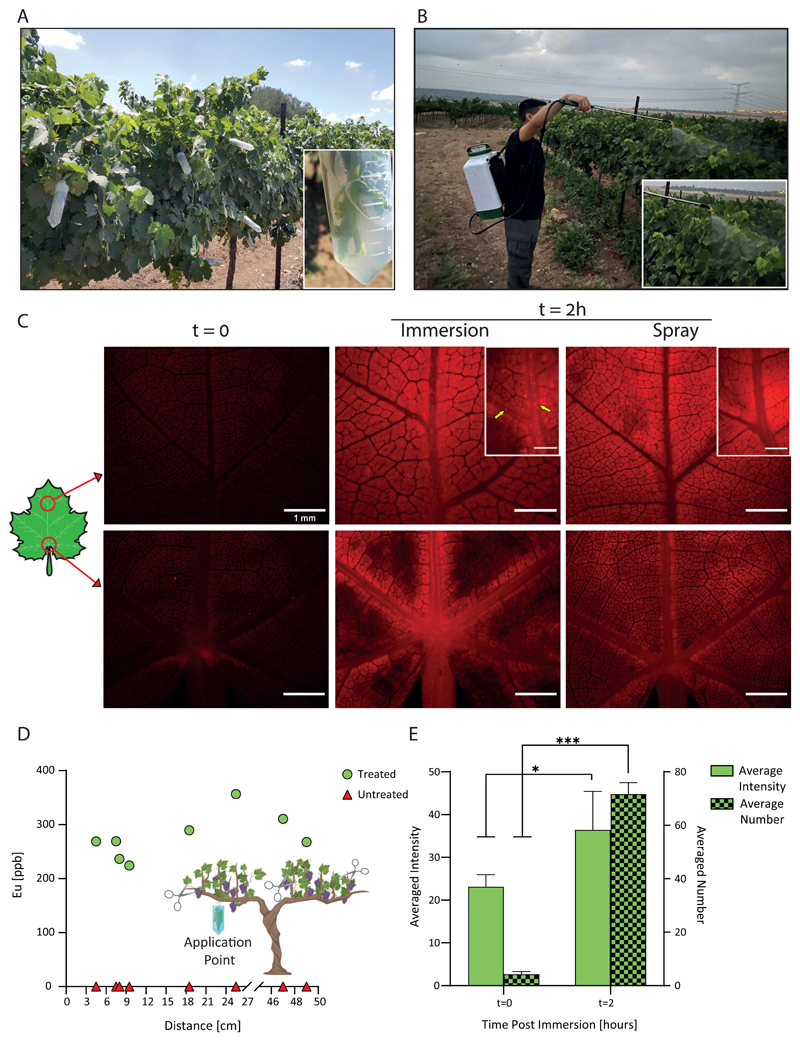
We evaluated the ability of dsRNA-lmPEI particles to be taken up and distribute within vines. The lmPEI carrier was covalently labeled with a Cyanine 5 (Cy5) and then complexed with dsRNA. Treatment groups (N=5) received five different treatments: free Cy5 or Cy5 labeled particles, each given by spray and immersion methods, and 25 mM sodium acetate buffer (control). The immersed leaves’ petioles were embedded within 600 μl of treatment solution while sprayed leaves were treated with 10 mL of relevant infiltrate to cover both their ventral and dorsal sides. The accumulation of particles was imaged and quantified 2 hours after the treatment, and six different locations were used to record the full plant biodistribution profile. Basal autofluorescence was recorded at the initial time and accumulation was seen in both spray and immersion administrations after the 2-hour treatment (Figure 2C, control is presented in Figure S6). Particle accumulation was visualized within both the primary and secondary veins of a leaf when its petiole was immersed within labeled particles, in contrast to sprayed treatment where fluorescence was dispersed throughout the leaf and no particle accumulation was seen within the veins. Moreover, the signal in the immersed leaves after 2 hours significantly increased as demonstrated by quantification of both the average intensity and the number of particles (P<0.05 and P<0.001, respectively Figure 2E). These results suggest particles can penetrate through the stomata after spraying as well as enter a leaf’s veins following immersion, ultimately distributing within the vine.
Figure 2. Particle biodistribution.
A representation of the field experiments’ administration methods: shoot immersion (year 2018, A) and canopy spraying (years 2019 and 2020, B). Distribution of Cy5-labeled particles within vine leaves after a 2-hour administration via spray or immersion (scale bar - 1 mm, C). In contrast to basal auto-fluorescence at the initial state, particle accumulation is observed within leaf veins after 2 hours as emphasized by yellow arrows in the upper middle inset (scale bar - 500 μm). Particles distribute up to 60 cm from the leaf submersion point after a 72-hour treatment with Europium-loaded particles, as quantified by elemental analysis using ICP-OES (D). Particles’ average intensity and number increase before and after a 2-hour immersion with fluorescent particles (N=5, 1.5-fold and 16.5-fold, respectively, E). Results are shown as mean ± SD. Two-tailed paired t-test was used for the statistical analysis of E. *p<0.05, ***p<0.001.
To quantify the biodistribution in the entire plant, nanoparticles were loaded with Europium (EuCl3) and administered to leaves by submersion. Leaves located at different distances were analyzed using elemental analysis by ICP-OES (Figure 2D). Nanoparticles were detected up to 60 cm from the application point, indicating that the nanoparticles penetrated through the leaf stoma and distributed along vine shoots.
2.3. dsRNA stability and release
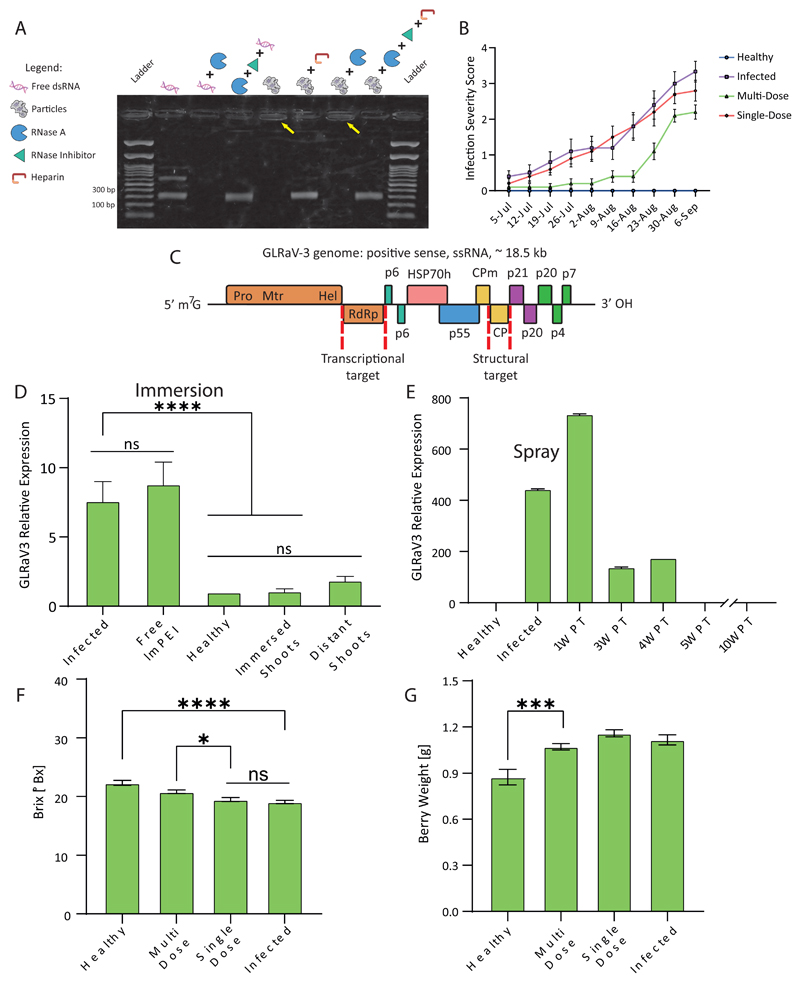
For particles to provide RNAi-mediated knockdown, dsRNA needs to avoid degradation until it reaches the target site where it can be released safely. We tested the ability of the lmPEI carrier to protect dsRNA from RNase-A (ribonuclease) degradation and simulated cargo release by using Heparin. Heparin is a highly negatively-charged molecule that competes with dsRNA for electrostatic interactions, releasing the dsRNA from the lmPEI complex (Figure S7).[21] We assessed the protection capacity of the complex against RNase-A before and after releasing the dsRNA from the particles, and demonstrated that complexation with lmPEI protected the RNA from degradation (Figure 3A). The protection was similar for RdRp and CP sequences, suggesting that the particle protection against nuclease is independent of the RNA sequence (Figure 3C).
Figure 3. Particle efficacy.
Gel image implies that the lmPEI carrier can protect dsRNA from RNase A activity (2% agarose, 100 mV, 35 minutes, A). When naked, RNA sequence is degraded by RNase A (third well to the left) as opposed to maintaining its integrity when complexed with lmPEI in particle form (yellow arrows). Moreover, dsRNA release from complexes is identical both with and without RNase A activity and inhibition (second and fourth well to the right). Infection severity scoring throughout 2020 field experiment shows delayed GLD symptomes in multi-dose treated vines in comparison to single-dose (B). An illustration of grapevine leafroll associated virus 3 open reading frames with transcriptional and structural RNAi targets (red dashed lines; C). Compared to untreated infected vines, GLRaV3 expression is reduced after a single administration of particles in 2018 field experiment. Knockdown effect is apparent within shoot distant from immersion treatment point (D). Respectively, change in administration method to canopy spraying managed to retain virus down-regulation three weeks post-treatment (PT, E). Grape quality parameters values of Brix (F) and berry weight (G) acquired after 2020 harvest indicate multiple treatments are preferable over a single treatment in recovering treated vines’ berries and suggests multiple administrations may be needed to fully recover fruit quality. Results are shown as mean ± SD. Ordinary one-way ANOVA tests were used for statistical analysis of D, F and G. ns – not significant, *p<0.05, ***p<0.001, ****p<0.0001.
2.4. Field experiments – antiviral activity of lmPEI-dsRNA particles
To test the antiviral activity of the dsRNA-lmPEI particles, the ability to knockdown GLRaV-3 in infected vines was evaluated. For this, three field experiments were conducted in vineyards located in the Judean foothills in central Israel during the summer months (June-September) of years 2018, 2019 and 2020. Experiments took place in a Cabernet Sauvignon plot (31°50’17”N, 34°53’57”E, 140 meters above sea level) grafted upon Ruggeri rootstock planted in soil mainly composed of clay, sand, and silt.[22] Vines were randomly divided into treatment groups by creating spaced blocks which were evenly spread across each row. To follow GLD symptoms throughout the experiments, an infection severity assessment table (Table S3) was designed, and half of each treatment group was scored once a week. At the end of each experiment, the shoots were pruned, and RT-PCR analysis was performed to assess GLRaV-3 titer within phloem tissue. Additionally, upon harvest, the berries were tested for grape quality parameters. Two different administration methods were applied as a single dose in 2018 and 2019 while multi-dose treatment was examined in 2020. In 2018 (N=28), to allow infiltrate uptake, leaves were brushed with particle solution and shoots were cut and immersed into treatment infiltrate for 24 hours (Figure 2A). In 2019, vines (N=47) were treated mainly by canopy spraying (Figure 2B) and only a few by shoot immersion to serve as a control. In 2020, vines (N=80) were treated only by canopy spraying either with a single or multi-dose (five treatments). In all consecutive years, there was a significant difference in GLRaV-3 titer between healthy and infected groups as well as between infected and particle-treated groups (P<0.0001; Figure 3D, S8 and S9). These results imply that the dsRNA-lmPEI particles penetrated and distributed within the vine, inducing viral knockdown.[23] Moreover, gene knockdown RT-PCR levels show that three weeks after a single-dose administration the virus titer decreased (Figure 3E). In contrast to virus expression, GLD symptoms improved only when the vine was treated multiple times throughout the growing season (Figure 3B). The berries’ Brix and weight values were measured after harvest. Brix values indicate a single dose was not sufficient to cause recovery in infected berries, whereas a multi-dose treatment significantly improved sugar level within the infected fruits bringing it nearer to healthy berries’ values (P<0.05, Figure 3F). This effect was measured to a lower extent in berry weight, suggesting repeated treatment is superior in improving the viral symptoms. Additional parameters such as pH, total acid, tannin index, color density and softness ratio were examined at different time points after veraison (Figure S10-S14). As ripening progressed, pH levels increased, acidity levels decreased and there was an overall tendency for tannin index to elevate, demonstrating that dsRNA-lmPEI administration did not harm grape quality parameters of the treated vines. Therefore, this suggests that a single dose application of dsRNA-lmPEI is sufficient to reduce the viral titer, but multiple applications are needed to achieve full recovery of fruit quality.
3. Conclusion
GLD infects agricultural vineyards around the world.[6] Our findings show that dsRNA-lmPEI particles carry, protect and distribute within the grapevine’s transportation system, thereby constituting a potent delivery system for long dsRNA. Furthermore, this study demonstrates that dsRNA-lmPEI particles act as a biological, non-GMO contender to treat GLRaV-3 infections in vines via foliar application. These findings may also be leveraged to treat other viral infections in food crops.
4. Experimental Section
4.1. Modified branched Polyethylenimine synthesis
Branched Polyethylenimine (Mw=800 gr/mol, Sigma-Aldrich - Merck) was conjugated with 1,2-Epoxytetradecane (Tokyo chemical industry co.) to formulate 14 carbon lipid-conjugated branched PEI. Conjugation was conducted through an epoxide ring opening reaction, maintaining a 3:1 molar ratio (Epoxide:bPEI) mixture. Briefly, 4.75 gr of bPEI were dissolved into 150 mL of pure ethanol and heated (55°C) to reach homogenous solution. Next, pre-calculated epoxide volume was added to the solution while keeping vigorous mixing. Mixture was incubated for 4 hours maintaining constant heating (90°C) and mixing (600 rpm) using thermocouple. After reaction was completed, verification of successful reaction was performed by loading 2 μl of 20-fold diluted reaction product in ethanol on a silica-coated thin-layer chromatography plate (Biotage) with Chloroform:n-Hexane 1:1 v/v as the mobile phase.
4.2. dsRNA-lmPEI complexation
Complexation based on electrostatic interaction between dsRNA and lmPEI was conducted by applying ethanol injection technique. N:P ratio is defined as the ratio between positively charged amine groups and negatively charged phosphate groups and plays an important role in complexation calculations. In general, pre-heated dsRNA and lmPEI were added to 25 mM sodium acetate buffer (pH=5.2) in 1:1 v/v ratio, keeping a 2:1 N:P molar ratio. lmPEI was injected using a pipettor while vigorous mixing was achieved by vortex. Subsequently, mixture was incubated in Eppendorf shaker for 20 minutes at 40°C and 1000 rpm. Particles’ characteristics such as size distribution, stability and mean diameter and charge were further determined at room temperature by dynamic light scattering using Nano ZSP (Malvern, United Kingdom).
4.3. dsRNA retention and release
Evaluation of dsRNA complexation and its release was assessed using Heparin release assay. Heparin is a strong negatively charged molecule that can compete with dsRNA for electrostatic interactions, thus releasing it from particles. Following particles formation, 1 μl of diluted Heparin sodium salt (0.14 M) from porcine intestinal mucosa (Sigma-Aldrich) was added to approximately 250 ng of complexed dsRNA and incubated for 20 minutes at 35°C. Subsequently, 2% gel agarose (Hy-Labs) in TAE (X1) with Ethidium Bromide (Hy-labs) was visualized under UV light after 35-minute run at 100 V.
4.4. RNase assay
Particles ability to protect dsRNA from degradation was examined using RNase A (Thermo-Scientific, Cat. EN0531) and RiboLock RNase Inhibitor (Thermo-Scientific, Cat. EO0381). Briefly, 20 μl of dsRNA-lmPEI particles (approximately 700 ng dsRNA) were incubated with 2 ng RNase A for 2 hours at 37°C following enzyme inactivation through incubation with RiboLock RNase Inhibitor (20 units per 70 ng of complexed dsRNA) for 1 hour at 37°C. Next, solution was incubated with diluted Heparin for another 20 minutes at 37°C to release dsRNA from particles. Final product was applied with adequate controls to a 2% agarose gel electrophoresis for 35 minutes at 100 V.
4.5. cryo-TEM imaging and Fast Fourier Transformation analysis
Cryogenic transmission electron microscopy (cryo-TEM) imaging was performed by Technion Center for Electron Microscopy of Soft Matter (TCEMSM) on a Thermo-Fisher Talos F200C, FEG-equipped high resolution-TEM, operated at 200kV. Specimens were transferred into a Gatan 626.6 cryo-holder and equilibrated below -170 °C. Micrographs were recorded by a Thermo-Fisher Falcon III direct detector camera, at a 4k × 4k resolution. Specimens were examined at TEM nanoprobe mode using volta phase plates for contrast enhancement. Imaging was performed at a low dose mode of work to minimize the exposure of the imaged area to electrons. Images were acquired using the TEM Imaging and Acquisition (TIA) software. Inter-fiber spacing was deduced by performing radial integration on FFT of the relevant obtained images. Integration was done using FIJI software plugin by Paul Baggethun, 2009 version.
4.6. Cy5-lmPEI labeling
To visualize particles under microscopy instrumentation, amine-reactive red emitting fluorescent dye Cyanine5 NHS ester (Ex/Em:646/662nm, Abcam) was conjugated to lmPEI prior dsRNA complexation through carbodiimide reaction using NHS as a coupling reagent. Following epoxide ring opening reaction described previously Cy5 NHS ester and lmPEI were reacted in 1:4 (Cy5:NH2) molar ratio and incubated for 90 minutes at 37°C, 600 rpm to yield Cy5-lmPEI. One step size exclusion procedure was held using G-10 Sephadex beads (Sigma Aldrich) to isolate desired product from raw reactants and other by products. Each step of the process was verified using TLC with Chloroform:Methanol 1:1 v/v as the mobile phase.
4.7. Fluorescent Microscopy
To investigate particles uptake by vine leaves following spray and immersion administration routes, Cy5-labeled dsRNA-lmPEI were synthesized and complexed as described above. In addition, free Cy5 was treated the same as labeled particles to be administered as control. Vine leaves were divided into five treatment groups (N=5 per group) as follows: 25mM sodium acetate buffer (pH-5.2), sprayed particles, spray control, immersion particles and immersion control. Immersion leaves’ petioles were embedded into 600 μl of infiltrate treatment whereas sprayed leaves were sprayed with treatment to reach seepage (~10 mL). Each group was imaged after 2 hours and six different locations were chosen within each leaf to best average total image signal. Samples were exposed for 400 ms and images were obtained using Olympus SZX16 fluorescent binocular equipped with DP72 CCD camera combined with x1.6 0.3 NA objective lens and Olympus mcherry filter (Excitation: 542-582, Emission: 603-678).
4.8. Nanoparticle biodistribution
To prove NP distribution within vines, a rare earth metal, EuCl3 (12-20 ppm) was encapsulated within 95 ± 25 nm sized liposomes and vine leaves were embedded into infiltrate solution for 72-hour period. Leaves were taken from treated and untreated vines at different distances from application point and dehydrated in an oven (BIFA Electro-therm MS8 multi stage laboratory furnace, Middlesex, UK) for 2 hours at 105°C. Dry matter was weighted and cremated for 5 hours at 550°C. Ash samples were dissolved in 1% HNO3, collected into 10 mL tubes, filtered (0.45 μm filter) and analyzed for Europium presence by ICP-OES apparatus (Agilent), using pre-prepared calibration curve obtained with Eu ICP standard (Sigma Aldrich).
4.9. Field experiments
Field trials (2018, 2019 and 2020) were conducted at ‘Bravdo’ vineyard located near Carmi Yossef, Israel. Vines showed various symptoms consistent with GLRaV3 infection, from small red dots on leaves to leaves that are completely red and curled inwards. At first experiment (June-September 2018), total of 28 vines were selected and divided into seven treatment groups (N=4) as follows: 1) Untreated healthy vines; 2) Untreated infected vines; 3) Infected vines treated with 25mM Sodium acetate buffer; 4) Infected vines treated with lmPEI solution; 5) Infected vines treated with naked RdRp sequence; 6) Infected vines treated with RdRp-lmPEI particles; 7) Infected vines treated with both RdRp-lmPEI and CP-lmPEI particles. To improve infiltrate’s uptake, two administration methods were employed - selected leaves were brushed with 5 mL of treatment solution and trimmed shoots were embedded within 20 mL of treatment solution for 24-hour period. Shoots and berries were taken for further analysis 6, 10 and 21 days and 8 and 9 weeks post treatment, respectively. At following year (June-September 2019), administration methods used were canopy spraying and shoot immersion. Total of 47 vines were distributed to construct the following treatment groups (N=10, except group number 4 where N=7): 1) Untreated healthy vines; 2) Untreated infected vines; 3) Healthy vines sprayed with lmPEI solution; 4) Infected vines’ trimmed shoots immersed within both RdRp-lmPEI and CP-lmPEI particles; 5) Infected vines sprayed with both RdRp-lmPEI and CP-lmPEI particles. Shoots were sampled at harvest whereas berry collection took place 3,5 and 8 weeks after veraison. In the 2020 field experiment (June-September), a manual back-sprayer was used to spray each vine with 400 mL of the RdRp-lmPEI and CP-lmPEI particle solution to reach seepage. Spraying took place every two weeks half an hour before sunrise. Total of 80 vines were divided into four treatment groups (N=20 per group): 1) Untreated healthy vines; 2) Untreated infected vines; 3) Single dose treated infected vines; 4) Multi-dose treated infected vines (sprayed five times). Shoots were sampled before applying the first treatment, at the middle of the experiment and at harvest. Berry collection took place at harvest and once a week after veraison from ten constant vines in each treatment for an on-site Brix measurement. Six berries were samples from three different locations on the same cluster. In addition, to follow GLD symptoms throughout the experiment and further evaluate GLRaV-3 effect on leaves phenotype, ten constant vines from each treatment group were visually tested and scored in a scale of 0-4 according to their severity symptoms by a pre-determined table (Table S3).
4.10. Plant material, RNA extraction and cDNA production
Random shoots were pruned, leaves were disposed and periderm was peeled off using a scalpel exposing inner tissues. Next, xylem was peeled out retaining the green phloem immediately frozen in liquid nitrogen and eventually transferred to – 80°C. 500 mg of frozen phloem was ground to a powder in liquid nitrogen using mortar and pestle. Total RNA was extracted using a Cetyltrimethyl Ammonium Bromide (CTAB)-based extraction protocol.[24] Another approach used for RNA extraction was using industrial SpectrumTM Plant Total RNA Kit (Sigma-Aldrich) following the manufactures’ instructions. After RNA was extracted from the shoots, RNA purity and quantity were evaluated using Nanodrop spectrophotometer; RNA integrity was assessed using gel-electrophoresis. Lastly, total cDNA was collected using Maxima First Strand (Thermo Fisher Ltd) and qScript® (QuantaBio) cDNA Synthesis Kit for RT-qPCR following the manufacturer’s protocol.
4.11. GLRaV identification
cDNA was amplified using specific primers (Table S1) for various GLRaV strains by polymerase chain reaction (PCR). 2X PCRBIO Taq Mix Red (PCRBIOSYSTEMS) PCR kit was used. PCR products were sent to HyLabs IL Ltd for Sanger sequencing, without further purification.
4.12. Viral titer assessment
Quantitative real-time PCR (qRT-PCR) was performed using Power SYBR™ Green PCR Master Mix (Thermo Fisher Ltd, Applied Biosystems™) and qPCR BIO SyGreen Mix (PCRBIOSYSTEMS) with cycling conditions implemented according to manufacturer’s instructions in Rotor-Gene™ 6000 (Corbett Research Ltd), qTOWER3 (Analytik Jena AG) and QuantStudio1 (Applied Biosystems™) real-time PCR thermal cyclers. Before operating qRT-PCR, specific primers were evaluated for their amplification efficiencies (Table S2) as well as confirmed with standard controls of non-template control minus reverse transcriptase control. Moreover, primers specificity was tested any analyzing dissociation curve ranging from 60°C to 95°C. GLRaV-3 Relative expression was calculated based on 2-ΔΔCt method.[25]
4.13. Grape quality parameters
Berries were tested for Brix (sugar) levels, weight, pH levels, color density, tannin index and softness ratio. Each vine’s berries were crushed, and the mixture was filtered using a strainer to measure brix. For acidity measurement, 10 mL of the same mixture were taken and tittered using 0.1N NaOH until pH reached 8.15. The volume of NaOH needed was used to calculated total acid content as follows: 0.75*NaOH volume. Tartaric acid correction was done to mimic wine’s natural acidity level. To calculate averaged berry weight, fresh batch of berries were weighed, counting the number of berries needed to reach 132 grams. Pure ethanol was added to constitute 12% by weight/volume and the berries were crushed. After 48 hours precipitation in 4°C mixture was centrifuged at 1500 rpm for 10 minutes. To assess optical density (OD), mixture was diluted 25-fold and OD was measured using a spectrophotometer at wavelength of 520 and 420 nm. Furthermore, color density was calculated as follow: (25∗OD420nm) + (25∗OD520nm). To evaluate the tannin index (total phenols), mixture was diluted 100-fold, then measured using spectrophotometer at wavelength of 280 nm. Tannin index was calculated as follows: (100∗OD280nm). Lastly, softness ratio was calculated as 10*(color density/tannin index).
During harvest of summer 2019 and 2020, the total yield of the grapes harvested from each vine was calculated.
4.14. Statistical analysis and image acquisition
Data is presented as mean values and error bars indicate SD. Statistical studies such as two-tailed paired t-test in Figure 2 and multiple comparison of one-way ANOVA in Figure 3 were conducted using Prism software version 9.0. Gel and binocular images were processed using Fiji software. Particles were identified using Imaris 9.1.2 software (Oxford, Bitplane) spots module to detect spots as spherical objects with 0.2 μm diameter and quality threshold value above 3.
Supplementary Material
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Acknowledgements
This project has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement No 680242-ERC-[Next-Generation Personalized Diagnostic Nanotechnologies for Predicting Response to Cancer Medicine]
The authors also acknowledge the support of the Technion Integrated Cancer Center (TICC), the Russell Berrie Nanotechnology Institute, the Lorry I. Lokey Interdisciplinary Center for Life Sciences & Engineering, The Israel Ministry of Economy for a Kamin Grant (52752, 69230); the Israel Ministry of Science Technology and Space – Office of the Chief Scientist (3-11878); Israel Innovation Authority for Nofar Grant (67967), the Israel Science Foundation (1778/13, 1421/17); the Israel Ministry of Science & Technology (3-16963); the Israel Cancer Association (2015-0116); Leventhal 2020 COVID19 Research Fund (ATS #11947), the German-Israeli Foundation for Scientific Research and Development for a GIF Young grant (I-2328-1139.10/2012); the European Union FP-7 IRG Program for a Career Integration Grant (908049); the Phospholipid Research Center Grant (ASC-2018-062/1-1); the Louis family Cancer Research Fund, a Mallat Family Foundation Grant; The Unger Family Fund; a Carrie Rosenblatt Cancer Research Fund, A. Schroeder acknowledges Alon and Taub Fellowships. The authors also acknowledge Dr. Nitsan Dahan for his help during the microscopy analysis. The help of Dr. Ruth Jalfon in editing this manuscript is greatly appreciated. Images in this paper were created with BioRender.com.
References
- [1].Bisson LF, Waterhouse AL, Ebeler SE, Walker MA, Lapsley JT. Nature. 2002;418:696. doi: 10.1038/nature01018. [DOI] [PubMed] [Google Scholar]
- [2].a) Alabi OJ, Casassa LF, Gutha LR, Larsen RC, Henick-Kling T, Harbertson JF, Naidu RA. PLoS One. 2016;11:e0149666. doi: 10.1371/journal.pone.0149666. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Atallah SS, Gomez MI, Fuchs MF, Martinson TE. American Journal of Enology and Viticulture. 2011;63:73. [Google Scholar]
- [3].Naidu RA, Maree HJ, Burger JT. Annu Rev Phytopathol. 2015;53:613. doi: 10.1146/annurev-phyto-102313-045946. [DOI] [PubMed] [Google Scholar]
- [4].Montero R, Mundy D, Albright A, Grose C, Trought MC, Cohen D, Chooi KM, MacDiarmid R, Flexas J, Bota J. Food Chem. 2016;197(Pt B):1177. doi: 10.1016/j.foodchem.2015.11.086. [DOI] [PubMed] [Google Scholar]
- [5].Endeshaw ST, Sabbatini P, Romanazzi G, Schilder AC, Neri D. Scientia Horticulturae. 2014;170:228. [Google Scholar]
- [6].Maree HJ, Almeida RP, Bester R, Chooi KM, Cohen D, Dolja VV, Fuchs MF, Golino DA, Jooste AE, Martelli GP, Naidu RA, et al. Front Microbiol. 2013;4:82. doi: 10.3389/fmicb.2013.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Martelli GP, Agranovsky AA, Bar-Joseph M, Boscia D, Candresse T, Coutts RHA, Dolja VV, Hu JS, Jelkmann W, Karasev AV, Martin RR, et al. Virus Taxonomy. 2012 [Google Scholar]
- [8].a) Almeida R, Daane K, Bell V, Blaisdell GK, Cooper M, Herrbach E, Pietersen G. Frontiers in Microbiology. 2013;4 doi: 10.3389/fmicb.2013.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Naidu R, Rowhani A, Fuchs M, Golino D, et al. Plant Dis. 2014;1172;98 doi: 10.1094/PDIS-08-13-0880-FE. [DOI] [PubMed] [Google Scholar]; c) Maliogka VI, Martelli GP, Fuchs M, Katis NI. Adv Virus Res. 2015;91:175. doi: 10.1016/bs.aivir.2014.11.002. [DOI] [PubMed] [Google Scholar]
- [9].Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Nature. 1998;391:806. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- [10].Baulcombe D. Nature. 2004;431:356. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- [11].a) Holen T, Amarzguioui M, Babaie E, Prydz H. Nucleic Acids Research. 2003;2401;31 doi: 10.1093/nar/gkg338. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Martinez J, Patkaniowska A, Urlaub H, Lührmann R, Tuschl T. Cell. 2002;110:563. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]; c) Haringsma HJ, Li JJ, et al. Nucleic Acids Research. 2012;40:4125. doi: 10.1093/nar/gkr1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Meister G, Tuschl T. Nature. 2004;431:343. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- [13].a) Brosnan CA, Voinnet O. Current Opinion in Plant Biology. 2011;14:580. doi: 10.1016/j.pbi.2011.07.011. [DOI] [PubMed] [Google Scholar]; b) Dunoyer P, Schott G, Himber C, Meyer D, Takeda A, Carrington JC, Voinnet O. Science. 2010;328:912. doi: 10.1126/science.1185880. [DOI] [PubMed] [Google Scholar]; c) Duan C-G, Wang C-H, et al. Silence. 2012;3:5. doi: 10.1186/1758-907X-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Robinson KE, Worrall EA, Mitter N. Journal of Plant Biochemistry and Biotechnology. 2014;23:231. [Google Scholar]
- [14].Ufaz S, Balter A, Tzror C, Einbender S, Koshet O, Shainsky-Roitman J, Yaari Z, Schroeder A. Molecular Systems Design & Engineering. 2018;3:38. [Google Scholar]
- [15].a) Silva AT, Nguyen A, Ye C, Verchot J, Moon JH. BMC Plant Biology. 2010;10:291. doi: 10.1186/1471-2229-10-291. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Burch-Smith TM, Anderson JC, Martin GB, Dinesh-Kumar SP. Plant J. 2004;39:734. doi: 10.1111/j.1365-313X.2004.02158.x. [DOI] [PubMed] [Google Scholar]; c) Gan D, Zhang J, et al. Plant Cell Reports. 2010;29:1261. doi: 10.1007/s00299-010-0911-z. [DOI] [PubMed] [Google Scholar]
- [16].Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS. RNA. 2006;12:1179. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].a) Whitehead KA, Langer R, Anderson DG. Nat Rev Drug Discov. 2009;8:129. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kesharwani P, Gajbhiye V, Jain NK. Biomaterials. 2012;33:7138. doi: 10.1016/j.biomaterials.2012.06.068. [DOI] [PubMed] [Google Scholar]
- [18].a) Mishra AK, Weissman H, Krieg E, Votaw KA, McCullagh M, Rybtchinski B, Lewis FD. Chemistry – A European Journal. 2017;23:10328. doi: 10.1002/chem.201700752. [DOI] [PubMed] [Google Scholar]; b) Neelakandan PP, Pan Z, Hariharan M, Zheng Y, et al. Journal of the American Chemical Society. 2010;132:15808. doi: 10.1021/ja1076525. [DOI] [PubMed] [Google Scholar]
- [19].Sun C, Tang T, Uludağ H, Cuervo Javier E. Biophysical Journal. 2011;100:2754. doi: 10.1016/j.bpj.2011.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ziebarth J, Wang Y. Biophysical Journal. 2009;97:1971. doi: 10.1016/j.bpj.2009.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bertschinger M, Backliwal G, Schertenleib A, Jordan M, Hacker DL, Wurm FM. Journal of Controlled Release. 2006;116:96. doi: 10.1016/j.jconrel.2006.09.006. [DOI] [PubMed] [Google Scholar]
- [22].MacDonald SL, Staid M, Staid M, Cooper ML. Computers and Electronics in Agriculture. 2016;130:109. [Google Scholar]
- [23].a) Teszlák P, Kocsis M, Scarpellini A, Jakab G, Kőrosi L. Photosynthetica. 2018;56:1378. [Google Scholar]; b) Valletta A, Chronopoulou L, Palocci C, Baldan B, Donati L, Pasqua G. Journal of Nanoparticle Research. 2014;16:2744. [Google Scholar]; c) et al. Plant Cell Reports. 2017;36:1917. doi: 10.1007/s00299-017-2206-0. [DOI] [PubMed] [Google Scholar]
- [24].Blanco-Ulate B, Vincenti E, Powell A, Cantu D. Frontiers in Plant Science. 2013;4 doi: 10.3389/fpls.2013.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Livak KJ, Schmittgen TD. Methods. 2001;25:402. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- [26].a) Bester R, Pepler PT, Burger JT, Maree HJ. J Virol Methods. 2014;210:67. doi: 10.1016/j.jviromet.2014.09.022. [DOI] [PubMed] [Google Scholar]; b) Bruisson S, Lebel S, Walter B, Prevotat L, Seddas S, Schellenbaum P. Journal of Virological Methods. 2017;240:73. doi: 10.1016/j.jviromet.2016.12.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.