Leukemia cells evade immune control by engaging the AXL receptor tyrosine kinase in innate immune cells; targeting AXL lifts the innate immune barriers, triggers lymphocyte-mediated leukemic clearance, and elicits response to adaptive checkpoint therapy.
Abstract
Acute leukemias are systemic malignancies associated with a dire outcome. Because of low immunogenicity, leukemias display a remarkable ability to evade immune control and are often resistant to checkpoint blockade. Here, we discover that leukemia cells actively establish a suppressive environment to prevent immune attacks by co-opting a signaling axis that skews macrophages toward a tumor-promoting tissue repair phenotype, namely the GAS6/AXL axis. Using aggressive leukemia models, we demonstrate that ablation of the AXL receptor specifically in macrophages, or its ligand GAS6 in the environment, stimulates antileukemic immunity and elicits effective and lasting natural killer cell– and T cell–dependent immune response against naïve and treatment-resistant leukemia. Remarkably, AXL deficiency in macrophages also enables PD-1 checkpoint blockade in PD-1–refractory leukemias. Finally, we provide proof-of-concept that a clinical-grade AXL inhibitor can be used in combination with standard-of-care therapy to cure established leukemia, regardless of AXL expression in malignant cells.
Significance:
Alternatively primed myeloid cells predict negative outcome in leukemia. By demonstrating that leukemia cells actively evade immune control by engaging AXL receptor tyrosine kinase in macrophages and promoting their alternative priming, we identified a target which blockade, using a clinical-grade inhibitor, is vital to unleashing the therapeutic potential of myeloid-centered immunotherapy.
This article is highlighted in the In This Issue feature, p. 2659
INTRODUCTION
Acute leukemia is a heterogenous group of devastating and rapidly progressing blood cancers that have a dismal outcome. Despite therapeutic progress, acute leukemia remains the leading cause of cancer-related death in children and is an appalling clinical challenge in particular in adults and elderly, whose overall survival (OS) remains below 50% (1, 2). Similar to solid cancers, immune evasion is a hallmark of acute leukemia (3). Several studies have identified leukemia-intrinsic mechanisms that promote immune escape, including loss of HLA molecules (4–6), expression of inhibitory ligands that dampen T-cell response (7), as well as downregulation of ligands that activate cytotoxic lymphocytes, such as natural killer (NK) cells (8). Moreover, the disseminated nature of acute leukemias, their rapid disease course, as well as their notoriously low mutational load (9) represent specific features that likely limit the initial engagement of antileukemic immunity (3, 10). This is exemplified by the recent finding that disseminated acute myeloid leukemia (AML) cells fail to induce host type I IFN response and effective antileukemic immunity (11). Besides these leukemia-intrinsic features, the extrinsic environment is also believed to heavily contribute to immune evasion, but the underlying molecular mechanisms remain largely unknown in hematologic malignancies. Therefore, identifying the pathways that impose a suppressive environment is critical to close this gap in knowledge and inform the development of more effective therapies.
Tumor-associated myeloid cells significantly affect tumor progression through a plethora of mechanisms including dampening protective adaptive immunity (12). In B-cell acute lymphoblastic leukemia (B-ALL), a recent single-cell RNA study revealed that monocyte abundance, and in particular nonclassical monocytes, is predictive of patient survival (13). Likewise, a high number of CD68+CD163+ M2-like macrophages or CD206 immune-suppressive myeloid cells are associated with poor outcome in adult T-cell leukemia and AML, respectively (14, 15). These studies suggest that, similar to their counterpart found in solid tumors, leukemia-associated myeloid cells likely contribute to disease progression; however, the underlying molecular mechanisms remain undefined.
Under physiologic conditions, a well-known immune regulatory mechanism that is primarily active in myeloid cells is driven by the TYRO3, AXL, and MERTK (collectively termed TAM) receptor tyrosine kinases (RTK). These RTKs are differentially activated by their ligands GAS6 and PROS1, with GAS6 showing highest affinity for AXL. When engaged in innate immune cells, namely macrophages and dendritic cells (DC), TAM RTKs drive the acquisition of a noninflammatory phenotype that promotes tissue repair and resolution of inflammation (16–18). In cancer, AXL overexpression is frequently associated with poor prognosis, in both solid and hematologic malignancies (19). This tumor-promoting function of AXL is primarily attributed to its tumor cell–intrinsic ability to promote proliferation, epithelial-to-mesenchymal transition, survival, and resistance to cancer therapy, including in hematologic malignancies (20–29). In addition, tumor-intrinsic AXL expression can exert immune-suppressive functions by suppression of MHC-I expression, induction of PD-L1 expression, and altered expression of cytokines and chemokines that promote recruitment of myeloid cells (30–36). Notably, although AXL is expressed in immune cells within the tumor microenvironment, its potential tumor immune-modulatory function in tumor-associated immune cells per se remains largely unexplored.
Here we demonstrate that acute leukemia cells establish a self-reinforcing immune suppressive microenvironment by co-opting a host-derived mechanism, driven by the GAS6/AXL axis in macrophages, to dampen innate immunity and limit protective inflammation. Combining different mouse models and clinical-grade pharmacologic inhibitors, we show that targeting AXL specifically in macrophages promotes antileukemic immunity and elicits susceptibility to PD-1 blockade. When further combined with the standard-of-care treatment, GAS6/AXL blockade leads to unprecedented cure rates in high-risk B-ALL in mice, including those resistant to BCR–ABL1 inhibition.
RESULTS
GAS6 Is Induced in the Bone Marrow Microenvironment of Patients with Hematologic Malignancies and Its Expression Correlates with Poor Outcome
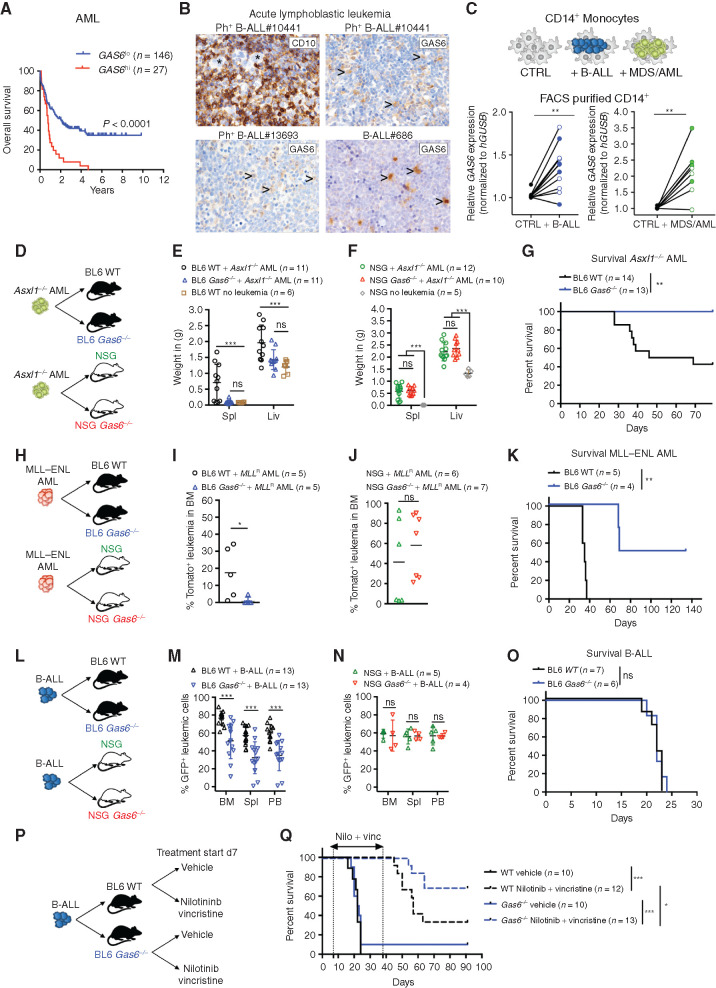
Using publicly available data sets, we found that high GAS6 expression correlates with poor outcome in AML, the most frequent form of acute leukemia in adults, as well as B-cell lymphoma (Fig. 1A; Supplementary Fig. S1A). Consistently, high GAS6 expression was previously associated with high-risk adult patients with de novo AML (28). To extend these in silico findings to other hematologic malignancies and pinpoint the cellular source of GAS6, we used IHC to evaluate GAS6 expression in situ, in bone marrow trephine biopsies from patients with B-ALL and T-cell acute lymphoblastic leukemia (T-ALL), at diagnosis (Supplementary Table S1). In B-ALL, GAS6 was undetectable in malignant blasts marked by CD10 expression (Fig. 1B, top left) that often constitute more than 95% of the cells in diagnostic biopsies, and readily produced by stromal cells, megakaryocytes, and hematopoietic cells with typical myeloid morphology (Fig. 1B; Supplementary Fig. S1B). Using an ex vivo coculture system, we show that both Philadelphia chromosome positive (Ph+) B-ALL and myeloid leukemia cells [myelodysplastic syndrome (MDS) and AML] instructively enhance GAS6 expression in human monocytes (CD14+) isolated from healthy donors (Fig. 1C). Overall, these human data indicate that leukemic cells enhance GAS6 expression in the microenvironment in a spectrum of hematologic malignancies and, most importantly, that GAS6 expression correlates with poor outcome.
Figure 1.
Leukemia-induced GAS6 contributes to immune evasion and leukemic progression. A, Prognostic value of GAS6 expression in AML (TCGA LAML, n = 173). Data was generated using the KaplanScan mode from the R2 Genomics Analysis and Visualization Platform (http://r2.amc.nl). Survival analysis by log-rank (Mantel–Cox) test. B, IHC of GAS6 on bone marrow trephine biopsies from representative patients with Ph+ (BCR–ABL1+) B-ALL at diagnosis. CD10 marks B-ALL blasts (upper left). Arrowhead marks myeloid cells. C, CD14+ peripheral blood monocytes from healthy donors were cultured with leukemia cells from either patients with Ph+ B-ALL (left, CD14+ from 11 donors cultured with 2 Ph+ B-ALL) or patients with myeloid diseases (right, CD14+ from 8 donors cultured with 1 AML and 1 higher-risk MDS). GAS6 mRNA levels were then determined by real-time PCR. Each data point represents a mean value obtained from two technical replicates, after normalization to a reference gene, GUSB. **, P < 0.01, paired two-tailed Student t test. Characteristics of all patients and healthy donors are described in Supplementary Table S1. D–F, Wild-type (WT) and Gas6−/− C57BL/6 or NSG (NSG Gas6−/−, line#697-31) mice were challenged with Asxl1−/− leukemia cells (105). Weight of spleens and livers on day 19 days post–leukemia challenge are displayed. NSG (n = 5) and C57BL/6 (n = 6) mice without leukemia are used as reference. ns, not significant. ***, P < 0.001, paired two-tailed Student t test. G, Kaplan–Meier survival analysis of WT and Gas6−/− C57BL/6 challenged with Asxl1−/− leukemia cells (105). **, P < 0.01, log-rank (Mantel–Cox) test. H–J, WT and Gas6−/− C57BL/6 or NSG (NSG Gas6−/−, line#697-31) mice were challenged with MLL–ENL AML cells (105). BM aspiration was performed after 22 days to determine leukemic burden (Tomato+). ns, not significant. *, P < 0.05, paired two-tailed Student t test. K, Kaplan–Meier survival analysis of WT and Gas6−/− C57BL/6 challenged with MLL–ENL AML cells (105). **, P < 0.01, log-rank (Mantel–Cox) test. L–N, WT and Gas6−/− C57BL/6 or NSG (NSG Gas6−/−, line#697-29) mice were challenged with B-ALL cells (103) and analyzed two weeks post–leukemia injection for leukemic burden (B220+GFP+) in bone marrow (BM), spleen (Spl), and peripheral blood (PB). This experiment was repeated with a different NSG Gas6−/− mouse line (line#697-31) with similar results (Supplementary Fig. S3E). ns, not significant. ***, P < 0.001, paired two-tailed Student t test. O, Kaplan–Meier survival analysis of WT and Gas6−/− C57BL/6 challenged with B-ALL cells (103). ns, not significant, log-rank (Mantel–Cox) test. P and Q, Kaplan–Meier survival analysis of WT and Gas6−/− C57BL/6 challenged with B-ALL cells (103) and subjected to either vehicle or nilotinib plus vincristine treatment combination. Treatment was initiated on day 7 and terminated on day 39. Data representative of at least two independent experiments. *, P < 0.05; ***, P < 0.001, log-rank (Mantel–Cox) test.
Induction of Successful Antileukemic Immunity by Gas6 Ablation in the Host Environment
To functionally test the role of GAS6 in possibly abetting leukemia progression, three syngeneic leukemia models were used in this study. In the myeloid models, disease was initiated by either the loss of Asxl1 (ref. 37; Supplementary Fig. S1C–S1H; Supplementary Methods) or the expression of the MLL–ENL fusion oncogene coupled to a tomato reporter (ref. 38; Supplementary Fig. S1I). For lymphoblastic leukemia, we used a highly aggressive B-ALL model driven by the expression of the BCR–ABL1 fusion oncogene coupled to GFP, in an Arf-null genetic background (Supplementary Fig. S1J–S1L). This secondary genetic lesion is frequently found in patients with high-risk B-ALL bearing the Philadelphia chromosome (the chromosomal translocation encoding the BCR–ABL1 oncoprotein) and is associated with an inferior outcome (39). Leukemic burden was defined by the percentage of B220dimGFP+ and CD11bdim tomato+ cells in the B-ALL and MLL–ENL models, respectively. The ASXL1 model was characterized by massive hepatosplenomegaly and myeloblast infiltration; hence, spleen and liver weight were often used as a surrogate for disease burden (Supplementary Fig. S1F and S1G). Notably, all three disease models show undetectable leukemia-intrinsic expression of Axl and Mertk, but do express Tyro3, albeit at variable levels (Supplementary Fig. S2A–S2J), and therefore could possibly derive cell-intrinsic benefit from GAS6 in the microenvironment, as previously reported by others (20–25, 27, 28, 40). In line with the human data, we show increased GAS6 protein expression in leukemia-associated Iba1+ myeloid cells (Supplementary Fig. S2K). Several cytokines are proposed to induce Gas6 expression, including TSG6, IL4, M-CSF, and IL10 (22, 41). We found Il10 to be readily upregulated by B-ALL blasts upon in vivo transplantation (Supplementary Fig. S2L). Moreover, IL10 blockade using a neutralizing antibody in vivo significantly blunts GAS6 induction and dramatically reduces the positive correlation seen between the percentage of GFP+ leukemic blasts and GAS6-expressing IBA1+ myeloid cells (Supplementary Fig. S2M–S2O). These data demonstrate that IL10, at least in part, contributes to GAS6 induction in leukemia-associated macrophages.
To functionally distinguish between the possibilities that GAS6 stimulates leukemic growth by activating oncogenic TAM receptor signaling in the blasts versus whether GAS6 functions via negatively regulating the antileukemic immune response, we transplanted established leukemia from three different models to wild-type (WT) and newly generated Gas6-deficient (Gas6−/−) hosts in C57BL/6 immune-competent or NOD.Cg-Prkdcscid IL2rgtmWjl/Sz (NSG) severely immune-compromised backgrounds (Fig. 1D–O; Supplementary Fig. S3A–S3D). In all three models, leukemic burden was significantly reduced in immune-competent (Fig. 1E, I, and M) but not immune-compromised (Fig. 1F, J, N; Supplementary Fig. S3E) Gas6-deficient animals. The reduction in leukemic burden observed in immune-competent Gas6-deficient animals translated into significantly prolonged survivals in the myeloid disease models (Fig. 1G and K), but not in the aggressive B-ALL model (Fig. 1O). Together, these data demonstrate that the functional relevance of GAS6 in leukemia goes well beyond its previously recognized role as a cell-intrinsic growth-promoting factor and relies on its ability to effectively suppress the immune response against leukemia.
GAS6 Deficiency Synergizes with Standard-of-Care Therapy to Enable a Powerful and Durable Antileukemic Immune Response against BCR–ABL1-Positive B-ALL
In the clinical setting, the standard of care for patients with Ph+ B-ALL combines a tyrosine kinase inhibitor (TKI) targeting the ABL1 kinase and intensive chemotherapy (including vincristine) followed by allogeneic hematopoietic cell transplantation (HCT) for clinically fit adult patients. Because efferocytic clearance of apoptotic cells by TAM receptors expressing phagocytes promotes the resolution of inflammation by both avoiding secondary inflammatory cell death and altering phagocyte priming (42), we speculated that induction of apoptotic cell death may synergize with a Gas6-deficient environment to promote antitumor immunity. Leukemia-challenged immune-competent WT and Gas6−/− mice were therefore subjected to a second-generation TKI targeting the BCR–ABL1 oncogene (nilotinib) and vincristine treatment (Fig. 1P), both of which induce effective apoptotic cell death. While vehicle-treated animals rapidly succumbed to leukemia with a median survival of 20 days, regardless of genotype (Fig. 1Q), 70% (9/13) of nilotinib plus vincristine–treated Gas6−/− mice remained leukemia free and achieved long-term disease-free survival (DFS; Fig. 1Q; Supplementary Fig. S4A). In stark contrast, only 30% (4/12) of WT mice showed durable responses (Fig. 1Q; Supplementary Fig. S4A), a result that is well in line with the 5-year OS observed in patients (43). In addition, bone marrow cells from long-lived Gas6−/− mice failed to transfer disease to secondary immune-compromised NSG hosts (Supplementary Fig. S4B), thereby demonstrating leukemia eradication in the primary hosts.
To further model the clinical situation often encountered with elderly and frail patients with Ph+ B-ALL who cannot tolerate intensive chemotherapy, we also evaluated the impact of treatment with nilotinib alone. Although nilotinib-treated WT mice showed a significantly prolonged survival, they ultimately succumbed to full-blown leukemia (Supplementary Fig. S4C), hence recapitulating the relapses seen in patients treated with TKI alone (44). In stark contrast, 30% (3/10) of nilotinib-treated Gas6−/− mice showed durable responses and remained leukemia free (Supplementary Fig. S4C). Bone marrow cells from long-lived nilotinib-treated Gas6−/− mice also failed to propagate leukemia upon transfer to secondary NSG recipients, indicating effective cure of the primary host (Supplementary Fig. S4D). Importantly, when immune-compromised NSG mice bearing the same leukemia cells were subjected to the same treatment, the synergistic effects between Gas6 deficiency and nilotinib were abrogated (Supplementary Fig. S4E). Together, these data unequivocally show that blockade of GAS6-mediated immune suppression effectively synergizes with standard-of-care regimens to achieve eradication of leukemic stem cells (i.e., cells with leukemia propagating activity) in B-ALL, thereby leading to long-term disease-free survival. This new strategy could considerably enhance the effectiveness of TKI treatment in frail patients who are urgently in need for alternative therapeutic approaches.
AXL-Expressing Leukemia-Associated Macrophages Contribute to Immune Suppression
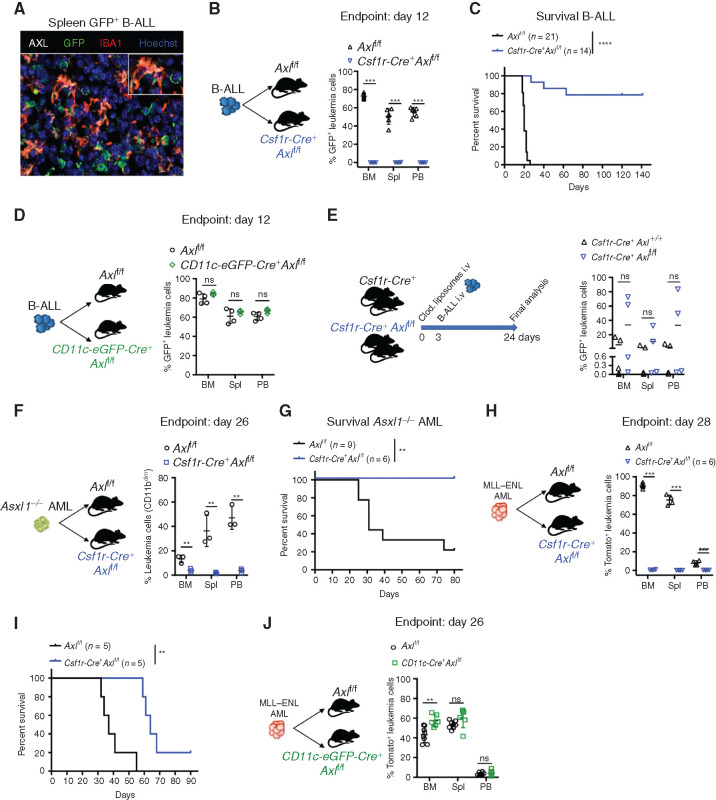
AXL, the TAM RTK with the highest affinity for GAS6, is readily expressed in leukemia-associated myeloid cells, in particular IBA1+ macrophages (Fig. 2A). High Axl expression in mononuclear phagocytes, namely monocytes, macrophages, and dendritic cells (45) was also seen using the Immunological Genome project (https://www.immgen.org) (Supplementary Fig. S5A). We next speculated that GAS6 expression could promote leukemic progression by signaling through AXL in immune cells and tested whether selective Axl deletion in leukemia-associated myeloid cells (both macrophages and DCs) enhances antileukemic immunity using mice with floxed alleles of Axl plus a Cre recombinase driven by the Csf1r promoter (Csf1r-Cre+ Axlf/f), as described previously (ref. 42; Supplementary Fig. S5B). When challenged with B-ALL, control mice (Axlf/f) exhibited full-blown leukemia, while Csf1r-Cre+ Axlf/f mice showed no sign of disease as evidenced by macroscopic analysis, flow cytometry, and IHC (Fig. 2B; Supplementary Fig. S5C–S5E). Remarkably, this translated into long-term DFS of >140 days in 80% of the animals (Fig. 2C). Long-term survivors had no detectable GFP+ leukemia cells in the bone marrow, indicating effective leukemia clearance (Supplementary Fig. S5F). Potential Csf1r-Cre toxicity was carefully excluded as both Axlf/+ and Csf1r-Cre+ Axlf/+ mice had full-blown leukemia after challenge with B-ALL (Supplementary Fig. S5G-H). Notably, myeloid-specific Axl deletion has more prominent antileukemic effects than constitutive Gas6 loss in B-ALL (compare Fig. 1M and O with Fig. 2B and C). We speculate that this might be mitigated by the increased expression of other TAM receptor ligands, such as Pros1, which is significantly induced in macrophages in response to B-ALL, but not AML (Supplementary Fig. S5I). Similar to GAS6, PROS1 promotes the resolution of inflammation by phagocytes (16), and tumor-derived PROS1 has been proposed to limit antitumor immune response in the B16F10 melanoma model (46).
Figure 2.
Selective Axl ablation in macrophages confers effective protection against leukemia. A, Representative immune fluorescence showing AXL expression (white) in IBA1+ leukemia-associated macrophages (red) in the spleen of a B-ALL leukemia–bearing mouse. B, Leukemic burden (% of GFP+ B220+) in bone marrow (BM), spleen (Spl), and peripheral blood (PB) of Axlf/f (n = 6) and Csf1r-Cre+ Axlf/f (n = 4) animals 12 days after challenge with 103 B-ALL cells. ***, P < 0.001, unpaired two-tailed Student t test. Experiment is representative of at least three experiments. C, Kaplan–Meier survival analysis of control Axlf/f and Csf1r-Cre+ Axlf/f animals challenged with 103 Ph+ B-ALL. Data are from two independent experiments. Similar results obtained in a third experiment using a different primary B-ALL. ****, P < 0.0001, log-rank (Mantel–Cox) test. D, Leukemic burden (% of GFP+ B220+) in bone marrow, spleen, and peripheral blood of Axlf/f (n = 4) and CD11c-Cre+ (CD11c-eGFP-Cre+ Axlf/f, n = 3) mice 12 days after challenge with 103 B-ALL cells. ns, not significant, unpaired two-tailed Student t test. Experiment is representative of two independent experiments. E,Csf1r-Cre+ control mice (n = 5) and Csf1r-Cre+ Axlf/f mice (n = 4) received 1 injection of clodronate liposomes (250 μL i.v./mouse) 3 days before challenge with 103 B-ALL cells. Three weeks later, when the first mouse was terminally ill, all mice were sacrificed and leukemic burden evaluated in bone marrow, spleen, and peripheral blood. ns, not significant, unpaired two-tailed Student t test. F,Axlf/f (n = 3) and Csf1r-Cre+ Axlf/f (n = 3) animals were challenged with 5 × 105Asxl1−/− AML cells. At day 26, leukemic burden (CD11bdimB220dim) in bone marrow, spleen, and peripheral blood is depicted. **, P < 0.01, unpaired two-tailed Student t test. G, Kaplan–Meier survival analysis of control Axlf/f (n = 9) and Csf1r-Cre+ Axlf/f (n = 6) animals challenged with 105Asxl1−/− AML as in F. **, P < 0.01, log-rank (Mantel–Cox) test. H,Axlf/f (n = 4) and Csf1r-Cre+ Axlf/f (n = 4) animals were challenged with 105MLL–ENL AML cells. At day 28, leukemic burden (% tomato+ CD11b+) in bone marrow, spleen, and peripheral blood is depicted. ***, P < 0.001, unpaired two-tailed Student t test. I, Kaplan–Meier survival analysis of control Axlf/f and Csf1r-Cre+ Axlf/f animals challenged with 105 MLL–ENL AML. These mice are also depicted in Fig. 4I. **, P < 0.01, log-rank (Mantel–Cox) test. J,Axlf/f (n = 8) and CD11c-Cre+ (CD11c-eGFP-Cre+ Axlf/f, n = 6) mice were challenged with 105MLL–ENL AML. On day 26, leukemic burden (% tomato+ CD11b+) in bone marrow, spleen, and peripheral blood is depicted. ns, not significant, unpaired two-tailed Student t test.
Because of its physiologic expression in DCs and the recent finding that AXL partially marks a subpopulation of immunoregulatory DCs that restrain DC immunostimulatory function, termed mregDCs (47), we also explored the impact of selective Axl deletion in DCs, using the CD11c-eGFP-Cre line, as reported previously (Supplementary Fig. S5B; ref. 48). We found that Axl deletion in DCs alone is not sufficient to elicit antileukemic immunity (Fig. 2D), while depletion of macrophages using clodronate liposomes in Csf1r-Cre+ Axlf/f mice prior to leukemia challenge abolished antileukemic immunity (Fig. 2E; Supplementary Fig. S5J). Importantly, the antileukemic effects and prolongation of survival conferred by Axl deletion in Csf1r-expressing cells were also recapitulated in the myeloid leukemia models (Fig. 2F–I). Likewise, AML burden remained unaffected when Axl was specifically ablated in DCs (Fig. 2J), further confirming the finding from the B-ALL model. Notably, however, Axl deficiency did not lead to cure in the MLL–ENL model, possibly reflecting a difference in the downstream immune response to these different types of leukemia. Collectively, these data point to AXL as a bona fide innate immune checkpoint in leukemia-associated macrophages.
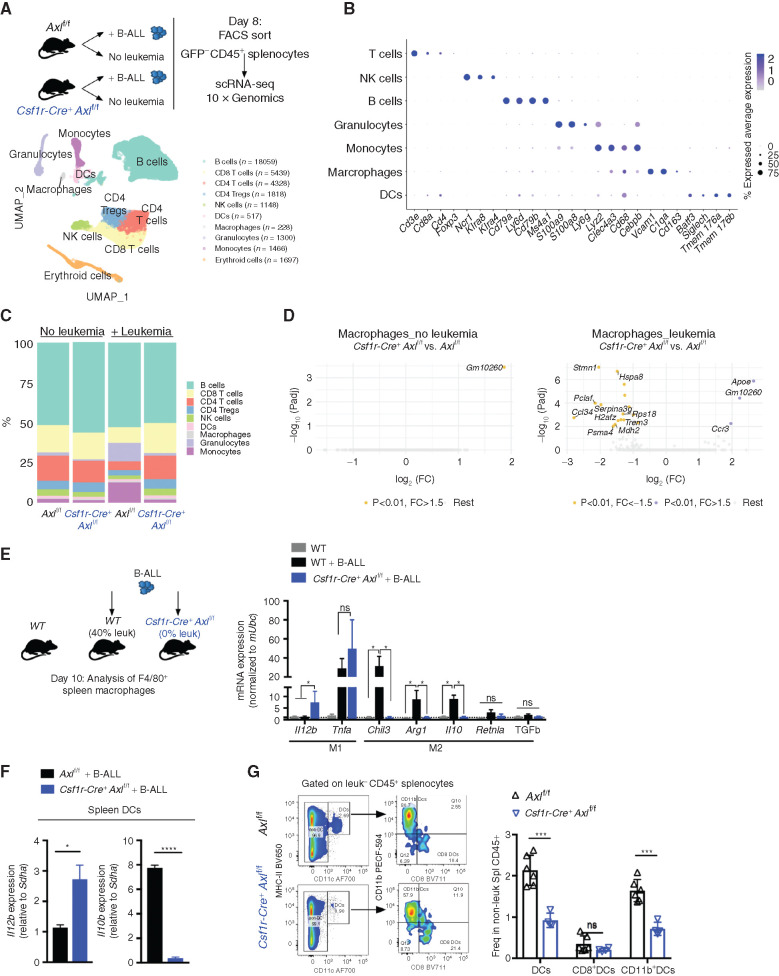
AXL Ablation in Leukemia-Associated Macrophages Prevents the Establishment of an Immune-Suppressive Microenvironment
To further characterize the immune changes associated with leukemia and hampered by Axl ablation in macrophages, we FACS purified nonleukemic CD45+ spleen leukocytes from Axlf/f control and Csf1r-Cre+Axlf/f animals that were either challenged or not with B-ALL, and subsequently subjected them to single-cell RNA sequencing (scRNA-seq) using the 10X Genomics platform. A total of 36,000 cells with a median number of 1,529 quantified genes/cell were analyzed. Downstream analysis (detailed in the Methods) identified distinct clusters that were subsequently classified into major hematopoietic cell types (Fig. 3A). To confirm the validity of the inferred cell types, we identified conserved gene markers for each cluster and evaluated their expression across all identified clusters (Fig. 3B; Supplementary Fig. S6A). In response to leukemia, Axlf/f animals showed a marked expansion of the monocyte and granulocyte clusters with a massive drop in lymphoid cells, likely reflecting the accumulation of immune-suppressive myeloid populations (Fig. 3C) including monocytes, which have recently been linked to poor outcome in both adult and childhood BCR–ABL1+ B-ALL (13). These immune-suppressive changes, including the accumulation of immune-suppressive CD11b+Ly6G−Ly6Chi myeloid-derived suppressor cells (MDSC), were abrogated upon Axl ablation in Csf1r-expressing cells (Fig. 3C) and verified to occur in both the spleen and bone marrow, using flow cytometry (Supplementary Fig. S6B and S6C). Differential gene expression (DGE) analysis further revealed that leukemia-challenged Csf1r-Cre+Axlf/f macrophages display reduced expression of proliferation-associated genes (Pclaf, H2afz, Hspa8) and drastic decrease in Stathmin expression (Stmn1), encoding a microtubule-binding protein whose downregulation is required for classical priming of macrophages (49), as well as Ccl24, a chemokine gene that is highly expressed in M2-polarized macrophages (ref. 50; Fig. 3D). Targeted expression analysis of key polarization genes additionally showed that, in response to leukemia challenge, Axl-deficient macrophages exhibit an enhanced expression of immune-stimulatory cytokines (Il12, Tnfα; refs. 51, 52) and blunted expression of genes associated with tissue repair and immune-suppressive functions (Retnla, Chil3, Il10, Arg1, and Tgfb; ref. 53; Fig. 3E), which further support the single-cell transcriptomic results. Of note, detection of these genes in the scRNA-seq results was hampered by the limited number of recovered macrophages and DCs, as well as the sequencing depth achieved with the 10X Genomics platform. Besides macrophages, DCs from Csf1rCre+Axlf/f mice also displayed increased expression of the immune-stimulatory cytokine Il12, while higher expression of Il10 was prominent in leukemia-challenged Axlf/f control animals (Fig. 3F) and correlated with increased abundance of CD11b+CD11c+ tolerogenic cDC2 in these mice (Fig. 3G; ref. 54). Ultimately, this prominent shift in myeloid priming resulted in reduced frequency of suppressive Foxp3+ T regulatory cells (Supplementary Fig. S6D and S6E), higher ratio of CD8 to CD4 T cells (Supplementary Fig. S6F–S6H), and higher frequency of NK cells (Supplementary Fig. S6I and S6J), reflecting the potent antileukemic immune response observed in Csf1rCre+Axlf/f animals.
Figure 3.
Axl-deficient macrophages prevent the establishment of an immune suppressive environment in response to leukemia. A, Nonleukemic (GFP−) spleen leukocytes were FACS purified from control Axlf/f mice and Csf1r-Cre+ Axlf/f mice that were either naïve (Axlf/f n = 2; Csf1r-Cre+ Axlf/f n = 2) or challenged with 103 B-ALL cells (n = 2 Axlf/f + B-ALL; n = 2 Csf1r-Cre+ Axlf/f+ B-ALL) for 8 days and subjected to 10X Genomics scRNA-seq. Data clustering, UMAP visualization of 36,000 individual cells (pooled from all conditions) followed by marker-based cell type annotation identified 10 broad immune subsets across all profiled single cells. B, Dot plot of selected cluster-specific marker genes. C, Relative abundance of identified cell types across conditions. D, Volcano plots showing the DEG (Padj < 0.01 and fold change >1.5) in macrophages comparing Axlf/f and Csf1r-Cre+ Axlf/f under steady-state conditions (left) and upon leukemia challenge (right), with the significant genes (max 10) annotated. E, Real-time PCR expression data in F4/80+ spleen macrophages purified using magnetic beads from naïve WT mice (n = 4) and mice transplanted with 103 B-ALL (WT n = 4; Csf1r-Cre+ Axlf/f n = 4). Data are normalized to a reference gene, Ubc, and are mean ± SEM. *, P <0.05, unpaired two-tailed Student t test. F, Real-time PCR expression data in dendritic cells (DC) isolated by flow cytometry as CD45+GFP−MHC-II+CD11c+ from the spleen of B-ALL challenged Axlf/f (n = 4) and Csf1r-Cre+ Axlf/f mice (n = 3). Data are normalized to a reference gene, Sdha, and are mean ± SEM. *, P < 0.05; ****, P < 0.0001, unpaired two-tailed Student t test. G, Representative gating and flow cytometry based quantification of total classical dendritic cells (DCs: CD45+GFP−MHC-II+CD11c+) as well as subsets: cDC1 (CD8+DCs: MHC-II+CD11c+CD8+CD11b−) and cDC2 (CD11b+DCs: MHC-II+CD11c+CD8−CD11b+) within nonleukemic splenocytes (GFP−CD45+) from B-ALL challenged Axlf/f (n = 6) and Csf1r-Cre+ Axlf/f (n = 4) mice. ns, not significant; ***, P < 0.001, unpaired two-tailed Student t test.
In a recent study, systemic blockade of MERTK, another TAM receptor family member, was shown to enhance antitumor immunity in the MC38 model of colorectal cancer by engaging the cGAS–STING pathway in phagocytes (55). In our leukemia model, we found STING expression in immune cells to be dispensable for the leukemia rejection phenotype imposed by AXL blockade (Supplementary Fig. S7A and S7B) while myeloid selective ablation of the suppressor of cytokine signaling 3 (SOCS3; Supplementary Fig. S7C), an AXL downstream target that actively impairs type I IFN response and proinflammatory cytokine signaling, readily recapitulates the protective effects associated with myeloid selective Axl ablation in all three leukemia models (Supplementary Fig. S7D–S7G). This hints toward a potentially essential role of pro-inflammatory and immune-stimulatory cytokines in this process and argues that, although the outcomes of blocking MERTK in the MC38 model, or AXL in leukemia, are similar, the underlying downstream molecular mechanisms are likely distinct.
Axl Ablation in Leukemia-Associated Macrophages Unleashes an Effective NK Cell– and T Cell–Mediated Killing of Leukemia Cells
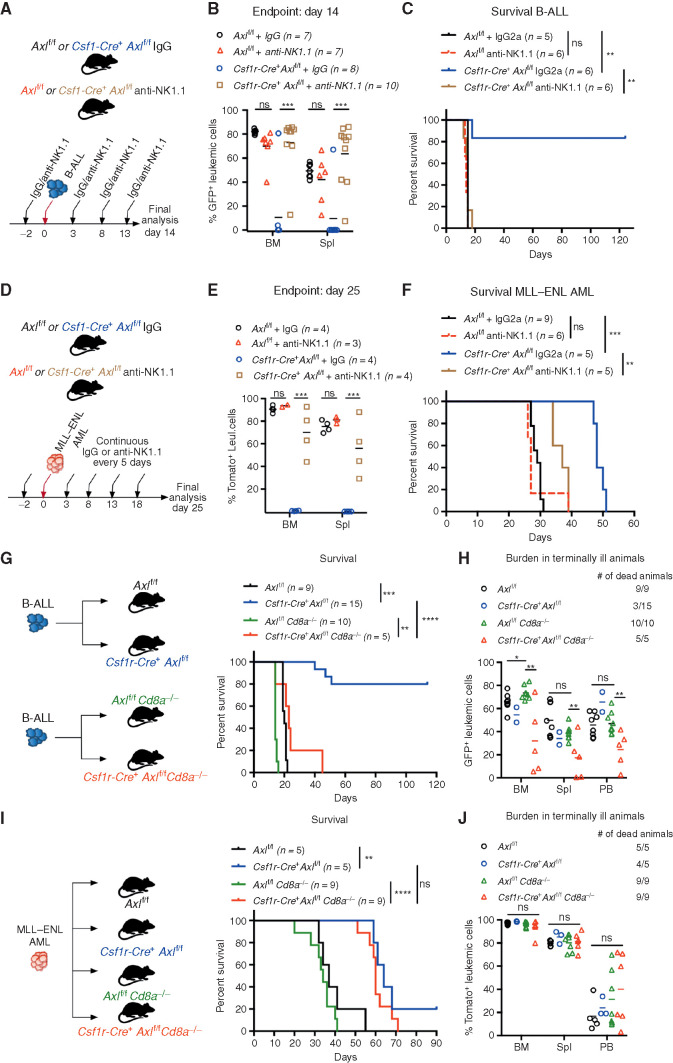
Because the activity of NK cells is tightly modulated by macrophages and greatly enhanced by IL12, we speculated that NKs might be involved in the leukemia clearance phenotype. Intraperitoneal administration of an anti-NK1.1 antibody every 5 days, starting 2 days before leukemia transplantation, achieved effective and continuous depletion of NK cells (Supplementary Fig. S7H and S7I). In B-ALL (Fig. 4A–C) and AML (Fig. 4D–F) challenged mice, both NK depleted and nondepleted Axlf/f control animals showed comparable leukemic burden and succumbed to disease with similar latencies, suggesting that NK cells are functionally impaired. In Csf1r-Cre+ Axlf/f mice, however, NK depletion abrogated antileukemic immunity and resulted in an accelerated disease course in both leukemia types, while IgG-treated counterparts remained largely protected against B-ALL (Fig. 4C) and maintained a prolonged survival in AML (Fig. 4F). This demonstrates that Axl ablation in phagocytes is sufficient to elicit a powerful NK-cell response that is essential for leukemic clearance.
Figure 4.
Axl-deficient macrophages trigger a robust NK-cell and T-cell immune response that suppresses leukemia. A and B,Axlf/f and Csf1r-Cre+ Axlf/f mice were challenged with 103 B-ALL cells and treated with either an anti-NK1.1 antibody or a mouse IgG2a isotype control (50 μg/mouse) every 5 days as indicated. Leukemic burden (% GFP+) in the bone marrow and spleen on day 14 is depicted (B). ns, not significant. ***, P < 0.001, unpaired two-tailed Student t test. C, Kaplan–Meier survival analysis of mice of the indicated genotypes challenged with 103 B-ALL cells and treated as in A. Treatments stopped once all anti-NK1.1–treated mice were dead. ns, not significant. **, P < 0.01, log-rank (Mantel–Cox) test. D and E, Same as in A and B using 105MLL–ENL AML cells. Leukemic burden (% Tomato+) on day 25 is depicted. ns, not significant. **, P < 0.01; ***, P < 0.001, unpaired two-tailed Student t test. F, Kaplan–Meier survival analysis of mice of the indicated genotypes challenged with 105MLL–ENL AML cells and treated as in D. Treatments stopped once all anti-NK1.1–treated mice were dead. ns, not significant. **, P < 0.01; ***, P < 0.001, log-rank (Mantel–Cox) test. G, Kaplan-Meier survival analysis of mice of the indicated genotypes challenged with 103 B-ALL cells. Data are pooled from two independent experiments as indicated in the scheme. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, log-rank (Mantel–Cox) test. H, Leukemic burden (% GFP+) in all terminally ill animals that could be analyzed from G. Note that burden from animals found dead cannot be depicted. ns, not significant. *, P < 0.05; **, P < 0.01, unpaired two-tailed Student t test. I, Kaplan–Meier survival curve of mice of the indicated genotypes challenged with 105 MLL–ENL cells. Survival of the reference groups (Axlf/f and Csf1r-Cre+ Axlf/f) is also depicted in Fig. 2I. ns, not significant. **, P < 0.01; ***, P < 0.001, log-rank (Mantel–Cox) test. J, Leukemic burden (% tomato+) in all terminally ill animals that could be analyzed from I. Note that burden from animals found dead cannot be depicted. ns, not significant, unpaired two-tailed Student t test.
To further interrogate the functional relevance of T cells as additional downstream effectors, we generated compound mice that lack CD8 T cells as well as Axl expression in phagocytes (Csf1r-Cre+ Axlf/f Cd8a−/− and Axlf/f Cd8a−/−) and challenged them with B-ALL or AML. In B-ALL, CD8 deficiency abrogated the survival advantage observed in Csf1r-Cre+ Axlf/f compared with Axlf/f control mice (Fig. 4G and H). These results were recapitulated by depletion of CD8+ T cells using an antibody approach (Supplementary Fig. S7J and S7K). Notably, the lower leukemic burden observed in Csf1r-Cre+ Axlf/f Cd8a−/− animals may reflect the productive engagement of NK cells that are unaffected in this model. In the MLL–ENL AML model, the survival benefit conferred by Axl-deficient phagocytes was maintained in CD8-deficient animals (Fig. 4I and J). Together, these data reveal that productive engagement of NK cells is a shared downstream mechanism by which Axl-deficient macrophages elicit antileukemic immunity, while the engagement of CD8 T cells appears to be model dependent.
Axl Ablation in Macrophages Elicits Susceptibility to PD-1 Checkpoint Blockade in PD-1 Refractory Treatment-Naïve B-ALL
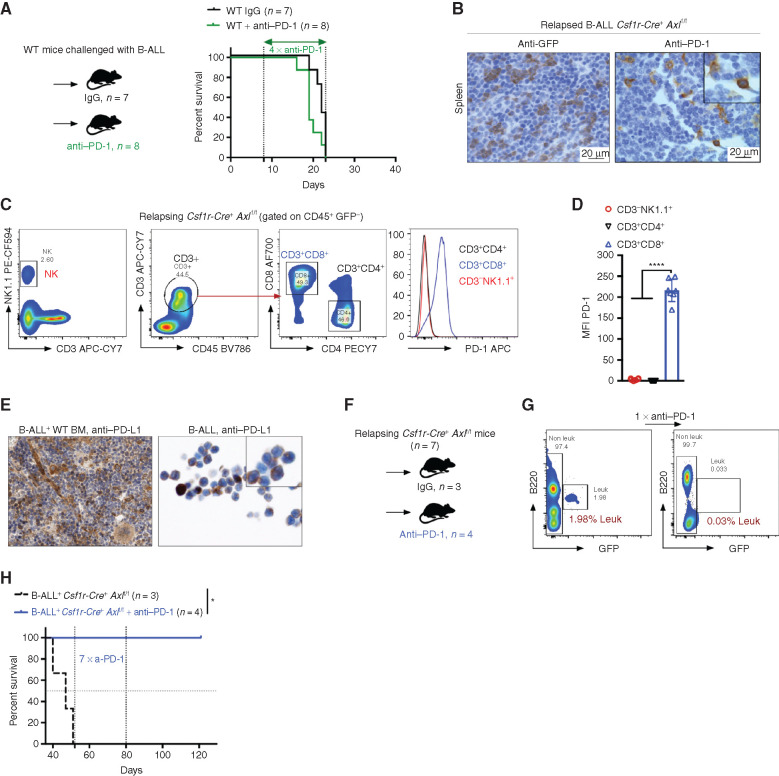
The fact that the enhanced antileukemic immunity observed in B-ALL challenged Csf1r-Cre+ Axlf/f mice depends, at least in part, on T cells prompted us to test whether interfering with this axis could also sensitize PD-1 refractory B-ALL (Fig. 5A) to PD-1 checkpoint blockade therapy. To address this issue, we took advantage of the limited fraction (10%–20%) of Csf1r-Cre+Axlf/f mice that eventually escape immune control and develop leukemia in the B-ALL model with a significantly delayed latency of >40 days (Fig. 2C). In situ IHC analysis of bone marrow and spleen sections revealed prominent PD-1 expression in these mice (Fig. 5B), which was largely restricted to the CD8+ T-cell subset, as demonstrated by flow cytometry (Fig. 5C and D). On the other hand, PD-1 ligand (PD-L1) is readily expressed by stromal and immune cells in the bone marrow as well as leukemic cells themselves (Fig. 5E). PD-1 blockade in Csf1r-Cre+ Axlf/f mice in early-stage relapse (up to 2% leukemic burden in peripheral blood) led to rapid leukemic clearance (Fig. 5F and G) and long-term DFS of more than 120 days (Fig. 5H). Hence, our data demonstrate that AXL blockade in phagocytes not only triggers a potent and lasting immune response against BCR–ABL1+ B-ALL, but also elicits susceptibility to checkpoint blockade in case of disease recurrence.
Figure 5.
Axl deficient macrophages trigger antileukemic immunity and elicit PD-1 checkpoint blockade. A, Kaplan–Meier survival analysis of WT mice challenged with 103 B-ALL cells and treated with either anti–PD-1 (n = 8) or isotype control (n = 7). B, GFP+ blasts (left) and PD-1+ cells (right) by IHC in the spleen of Csf1r-Cre+ Axlf/f mice that succumbed to B-ALL with a delayed latency of >40 days (Mice depicted in Fig. 2C). C and D,Csf1r-Cre+ Axlf/f from three independent experiments were followed by weekly bleeding to identify mice with late disease recurrence (n = 7). Flow cytometry data depicting PD-1 expression in peripheral blood lymphocytes (CD4 and CD8 T cells, NK cells) and corresponding PD-1 mean fluorescence intensity (MFI) from Csf1r-Cre+ Axlf/f mice showing signs of relapse (detectable GFP+ cells, representative data in G). ****, P < 0.0001, unpaired two-tailed Student t test. E, PD-1 ligand (PD-L1) expression by IHC in bone marrow cells with both stromal and hematopoietic morphology (left), as well as on cytospined B-ALL cells (right). F and G,Csf1r-Cre+ Axlf/f mice with late disease recurrence (n = 7, depicted in C and D) were either left untreated (n = 3) or subjected to 7 cycles of anti–PD-1 treatment (n = 4; 200 μg/mouse every 4 days). Representative FACS plot depicting leukemic burden (GFP+ B220dim) in the peripheral blood of the same mouse before and after one shot of anti–PD-1 treatment. H, Kaplan–Meier survival analysis of mice from F. *, P < 0.05, log-rank (Mantel–Cox) test.
AXL Inhibition in Nonmalignant Cellular Components of the Leukemic Microenvironment Unleashes Potent Antileukemic Immunity In Vivo and Synergizes with Standard-of-Care Therapy to Eradicate Leukemia
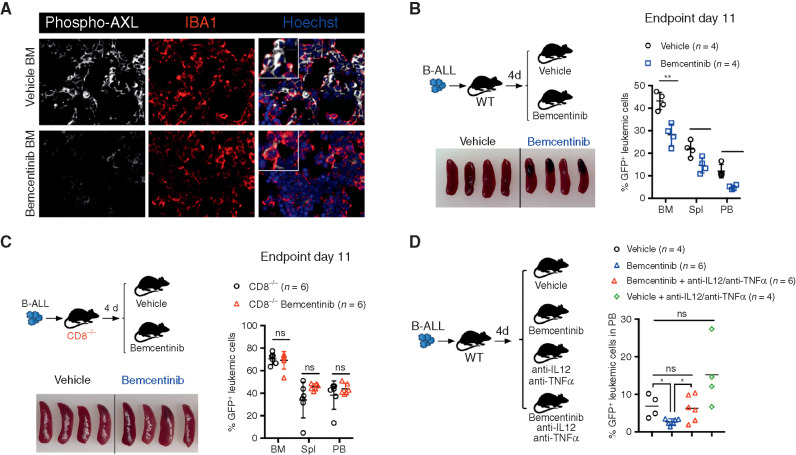
To determine the translational relevance of these findings, we pharmacologically interfered with AXL in vivo, using bemcentinib (also known as R428 or BGB324; ref. 56), an orally available and selective inhibitor for AXL currently undergoing clinical evaluation in cancers in which AXL expression, in tumor cells, is thought to contribute to disease pathogenesis (e.g., NCT02488408; NCT02424617; NCT02922777). In this study, by using leukemia models that do not express AXL protein (Supplementary Fig. S2J), we primarily used bemcentinib to evaluate its potential immune-modulatory effects on the nonmalignant components of the leukemic microenvironment that express AXL, namely myeloid cells (Fig. 2A). To model an intervention trial in which patients would exhibit low leukemic burden, such as those with measurable minimal residual disease (MRD+) after induction therapy, bemcentinib treatment was initiated several days post–leukemia challenge when leukemic cells were readily detected in the bone marrow and administered 7 days a week, twice daily at a dose of 50 mg/kg body weight. This resulted in reduced AXL phosphorylation in IBA1-expressing macrophages (Fig. 6A) and led to a significant reduction in leukemic burden across all analyzed organs in the highly aggressive B-ALL model (Fig. 6B). As anticipated, in this model the antileukemic effects were dependent on the engagement of CD8 T cells, as demonstrated by the lack of therapeutic efficacy of bemcentinib in CD8-deficient mice (Fig. 6C). In line with our observations in Axl-deleted macrophages, bemcentinib significantly and consistently enhanced the proinflammatory priming of macrophages as evidenced by increased expression of Il12 and Tnfα in response to LPS and IFNγ ex vivo (Supplementary Fig. S8A). Consequently, we show that the therapeutic effects of bemcentinib are curtailed upon IL12 and TNFα blockade in vivo (Fig. 6D), indicating that these proinflammatory cytokines indeed contribute to the overall immune-stimulatory effects observed upon AXL inhibition in vivo.
Figure 6.
Bemcentinib, a clinical-grade AXL inhibitor, triggers effective antileukemic immunity in B-ALL that depends on IL12, TNFα, and engagement of CD8 T cells. A, Representative phospho-AXL (white) expression and IBA1+ leukemia-associated macrophages (red) by immune fluorescence in frozen bone sections from vehicle and bemcentinib-treated leukemia-bearing mice depicted in B, at final analysis. B, Leukemic burden (% GFP+ B220dim in bone marrow, spleen, and peripheral blood) and spleen pictures of WT mice challenged with 103 B-ALL cells and treated twice daily with either vehicle or bemcentinib at 50 mg/kg. Treatment was initiated on day 4 post–leukemia injection and mice were analyzed on day 11. *, P < 0.05; **, P < 0.01, two-tailed Student t test. C, Same as B, using CD8-deficient mice. ns, not significant, two-tailed Student t test. D, Day 10 leukemic burden (% GFP+ B220dim in peripheral blood) in WT mice challenged with 103 B-ALL cells treated as in B, in the presence or absence of blocking antibodies against IL12 (300 μg/mouse) and TNFα (400 μg/mouse). Blocking antibodies for IL12 and TNFα were administered daily starting from day 4 post–leukemia challenge. Each dot represents an individual mouse and mean value is depicted. ns, not significant, *, P < 0.05, unpaired two-tailed Student t test.
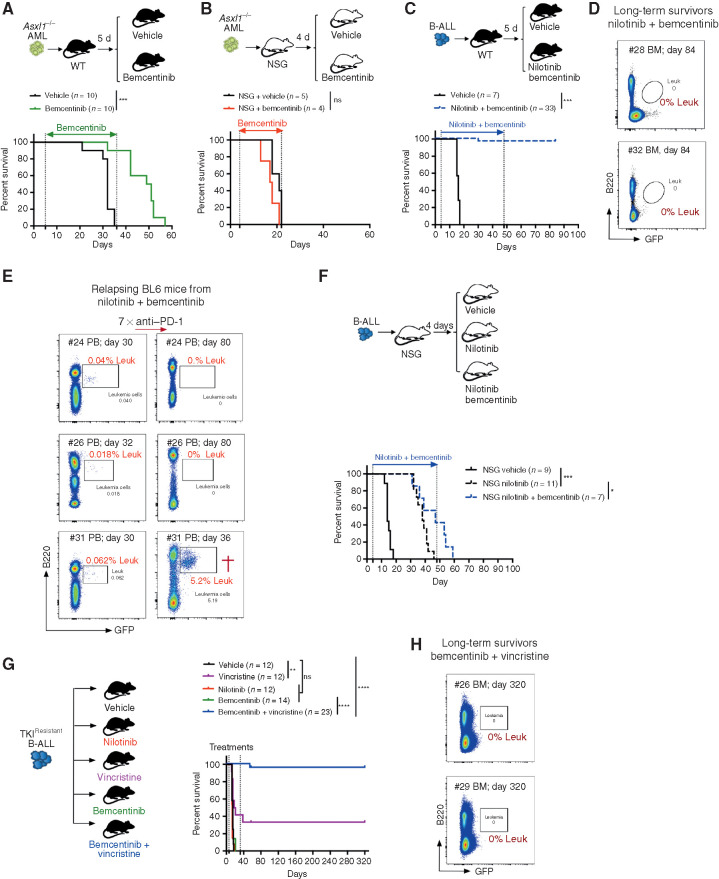
To further evaluate whether AXL inhibition may improve OS, leukemia-bearing mice were first treated with bemcentinib as a single agent, on an intermittent schedule of 5 days on, 2 days off for the indicated duration. Under these conditions, bemcentinib led to a significant increase in OS in the Asxl1−/− model (Fig. 7A). These effects were lost in immune-compromised NSG mice, in line with bemcentinib's predicted effects on AXL-positive immune cells (Fig. 7B). Of note, terminally ill mice exhibited the same disease phenotype regardless of treatment status (Supplementary Fig. S8B). In the high-risk B-ALL model, single-agent treatment with bemcentinib (Supplementary Fig. S8C) or nilotinib (Supplementary Fig. S8D) significantly extended survival, but all animals eventually succumbed to bona fide B-ALL within 50 days, despite continuous drug treatment. Remarkably, however, combination treatment with nilotinib and bemcentinib exhibited a prominent synergistic effect that led to complete remission and disease eradication in more than 90% (30/33) of the animals, with no sign of drug toxicity (Fig. 7C). Notably, although combination treatment was stopped on day 48, all mice remained leukemia-free, as demonstrated by the analysis of their bone marrow using flow cytometry (Fig. 7D), while vehicle-treated mice succumbed to bona fide leukemia within 15 to 20 days. In this experiment, weekly monitoring revealed that 3 of 33 mice (9%) treated with nilotinib plus bemcentinib showed signs of disease recurrence around day 30, at which point these mice were subjected to anti–PD-1 treatment. Similar to our observation in relapsing Csf1r-Cre+ Axlf/f mice (Fig. 5), checkpoint blockade led to disease clearance in 2 of the 3 relapsing mice (Fig. 7E). In addition, when NSG mice bearing the same leukemia were subjected to combined treatment with nilotinib and bemcentinib, the latter failed to drastically potentiate nilotinib effects, as demonstrated by the fact that 100% of the mice succumbed to leukemia with a median survival that was comparable to nilotinib only treated animals (Fig. 7F).
Figure 7.
Systemic AXL inhibition induces potent antileukemic immunity and eliminates leukemic blasts in AXL-negative leukemias. A and B, Kaplan–Meier survival curves of C57BL/6 WT mice (A) or NSG mice (B) challenged with 5.105Asxl1−/− AML cells and treated with either vehicle or bemcentinib (50 mg/kg, twice daily). ns, not significant. ***, P < 0.001, log-rank (Mantel–Cox) test. C, Kaplan–Meier survival analysis of C57BL/6 WT mice challenged with 103 B-ALL cells and treated with either vehicle (n = 7) or nilotinib (80 mg/kg, once a day) plus bemcentinib (50 mg/kg, twice daily; n = 33) for a total of 44 days. Data are pooled from two independent experiments. ***, P < 0.001, log-rank (Mantel–Cox) test. D, Representative FACS plots depicting absence of GFP+ B220dim leukemic cells in the bone marrow of long-term survivors from C. E, Mice from C were followed by weekly bleeding. Three of 33 mice (#24, #26, and #31) showed GFP+ cells indicative of disease recurrence and were subjected to anti–PD-1 treatment as indicated (7 × 200 μg/mL every fourth day). Mouse #31 succumbed to full-blown leukemia on day 36, while #24 and #26 remained leukemia-free. F, Kaplan–Meier survival analysis of NSG mice challenged with 103 B-ALL cells and treated with either vehicle, nilotinib, or nilotinib plus bemcentinib for a total of 44 days as in C. ns, not significant, log-rank (Mantel–Cox) test. G, WT mice were injected with 103 TKIR B-ALL cells. After 5 days, mice were randomly attributed to the indicated vehicle or treatment groups and their survival depicted using a Kaplan–Meier analysis. Data are pooled from two independent experiments. ns, not significant, **, P < 0.01; ****, P < 0.0001, log-rank (Mantel–Cox) test. H, Representative FACS plots depicting absence of GFP+ B220dim leukemic cells in the bone marrow of bemcentinib + vincristine–treated long-term survivors from G. In all experiments, treatments were initiated and stopped on the days indicated by dotted lines.
AXL Inhibition Synergizes with Chemotherapy to Eradicate Leukemia and Promote Disease-Free Survival in TKI-Resistant BCR–ABL1+ B-ALL
Because treatment resistance is a major source of relapse and a leading cause of acute leukemia–related deaths, we explored the therapeutic efficacy of AXL inhibition in the context of B-ALL resistant to BCR–ABL1 inhibition. B-ALL cells were exposed to increasing doses of nilotinib ex vivo to obtain a nilotinib-resistant subclone (referred to as TKIR) that was proven to be equally resistant to nilotinib treatment in vivo as shown by the lack of survival advantage (Fig. 7G). This is in stark contrast to the increased survival of nilotinib-treated animals challenged with the parental (nilotinib sensitive) B-ALL cells (Supplementary Fig. S8D). Bemcentinib alone did not significantly prolong survival in this TKIR B-ALL model, while chemotherapy using vincristine showed some efficacy, with 4 of 12 mice (33.3%) achieving long-term DFS (Fig. 7G). Most importantly, addition of the AXL inhibitor to vincristine showed remarkable synergistic effects, with 22 of 23 mice (95.6%) achieving leukemic clearance and long-term DFS of over 180 days (Fig. 7G and H). In addition, secondary transplantation of bone marrow cells from these long-term survivors failed to propagate disease to secondary NSG recipients, thereby indicating effective eradication of leukemia in the treated mice (Supplementary Fig. S8E and S8F). Notably, TKIR blasts remained AXL-negative, thereby excluding a potential leukemia-intrinsic effect of AXL inhibition in this setting (Supplementary Fig. S8G). Together, these data unambiguously demonstrate that systemic AXL inhibition using a clinical-grade inhibitor shows significant immune-dependent therapeutic efficacy that further synergizes with standard-of-care treatment to promote leukemic clearance, prolong OS, and even lead to cure in leukemia-bearing mice, including in a TKIR setting. In addition, our findings confirm that, similar to Axl ablation, pharmacologic inhibition of AXL also elicits susceptibility to PD-1 checkpoint blockade in case of disease recurrence. Collectively this highlights the broad clinical applicability of this new immunotherapeutic modality that could constitute a lifesaving alternative strategy for patients who develop therapy-resistant disease.
DISCUSSION
In this study, we show that leukemic cells engage a pathway that physiologically enables the noninflammatory clearance of apoptotic cells by phagocytes (18), namely the GAS6–AXL axis, to usurp macrophages to evade immune control and convert the environment into a highly immune-suppressive milieu that reinforces leukemic expansion. In addition, we comprehensively demonstrate that ablating AXL, specifically in leukemia-associated macrophages, or its high-affinity ligand GAS6 in the host environment, prevents the establishment of a suppressive immune architecture and converts leukemia cells with notoriously low mutational load (9) into potent immune-stimulatory triggers. This is further supported by the fact that combining GAS6–AXL blockade with treatment regimens that increase apoptosis of leukemia cells (e.g., TKI, chemotherapy) leads to enhanced antileukemic effects. Of utmost clinical importance is the demonstration that these unprecedented antileukemic effects can be effectively recapitulated, in vivo, by subjecting mice bearing AXL-negative leukemias to a selective clinical-grade AXL inhibitor (56) that can remarkably synergize with standard-of-care therapy, such as chemotherapy to potentiate antileukemic immunity. In highly aggressive Ph+ B-ALL, this approach effectively eradicates leukemia-propagating stem cells, including in TKI-resistant models, an outcome that is unprecedented with current therapies. In addition, we carefully demonstrate that with both the genetic and pharmacologic approach the observed therapeutic effects are strictly immune-dependent, as efficacy can be shown in leukemia-bearing immune-competent but not immune-deficient animals.
Interestingly, a recent study showed that higher expression of GAS6 correlates with adverse effects in patients with AML who underwent HCT (57). The fact that HCT, the curative potential of which primarily relies on the induction of potent graft-versus-leukemia (GvL) effect, cannot overcome the harmful effects imposed by high GAS6 expression further supports our conclusion that, in vivo, GAS6–AXL axis primarily promotes leukemic progression by its suppressive effects in leukemia-associated immune cells. These findings are of high clinical significance, as failure to achieve long-term survival in patients is primarily due to a high rate of relapse and ability of treatment-resistant leukemic stem cells to escape immune control (3). Consequently, AXL inhibition may represent a promising postremission strategy after HCT to boost the donor immune system to eradicate residual malignant cells and thus prevent relapse. Moreover, because AXL inhibition has remarkable immune-sensitizing effects when combined with reduced intensity single-agent chemotherapy regimen (Fig. 7G), a combination treatment could empower the patient's own immune system and provide hope for cure to adult and frail patients who cannot be exposed to intensive chemotherapy or HCT, including those with measurable MRD+ after induction therapy.
Mechanistically, we found that AXL blockade in leukemia-associated macrophages triggers productive inflammation by skewing their priming toward a leukemia-suppressive phenotype. This prevents the accumulation of MDSCs and stimulates the acquisition of immune-stimulatory features in DCs and production of key cytokines such as IL12 and TNFα that we demonstrate to be essential for the potent antileukemic immunity observed upon AXL inhibition. In line with our finding, myeloid cells engineered to express high levels of IL12 have recently been shown to reverse immune suppression and activate antitumor immunity in preclinical models of metastasis (58). In addition, we show that rewiring of the myeloid compartment in our setting kick-starts the immunity cycle (59) and results in major changes in downstream effector cells, including productive engagement of NK cells, suppression of T regulatory cells, as well as potent CD8 response, the latter being most prominently seen in Ph+ B-ALL. Notably, in Ph+ B-ALL, our approach leads to complete eradication of leukemia-propagating stem cells and elicits susceptibility to PD-1 checkpoint blockade upon relapse, an outcome that is unprecedented in this highly aggressive disease model. We speculate that the differential engagement of CD8+ T cells, observed in B-ALL versus MLL–ENL AML, may reflect the contribution of leukemia-intrinsic determinants, such as the immunogenic potential of specific genetic alterations.
To the best of our knowledge, this study is the first to demonstrate that AXL blockade in leukemia-associated myeloid cells triggers effective and durable antileukemic immunity, in particular in highly aggressive acute leukemia subtypes, such as Ph+ B-ALL. Because TAMs are key components of the tumor microenvironment and potent drivers of immune suppression, our study goes well beyond the existing cancer literature that only provides rationale for AXL targeting in AXL-positive tumors and warrants the clinical evaluation of AXL targeting strategies in other cancer types, including those with AXL-negative tumor cells. Furthermore, while tumors evolve under selective pressure of therapies and rapidly acquire resistance, which may limit the long-term benefits associated with tumor-intrinsic AXL inhibition, its targeting in nonmalignant tumor-associated immune cells may result in lasting efficacy.
Because of its selectivity toward AXL, bemcentinib triggers robust antileukemic immunity without inducing the autoimmune manifestations that are reported in mice with deletion of all three TAM receptors (60). Bemcentinib has so far showed favorable safety data in three phase II clinical trials in AML (NCT02488408; NCT02424617; NCT02922777), with interim reports providing evidence of TCR repertoire diversification in some patients (61), thereby hinting toward a potential immune-modulatory effect. In light of our work, it would be important to extend such studies to other hematologic malignancies such as Ph+ B-ALL, and more specifically to evaluate the potential tumor-extrinsic immune-modulatory function of AXL-targeting compounds, including bemcentinib and other compounds such as gilteritinib, a dual FLT3/AXL inhibitor that has shown efficacy in FLT3-mutated relapsed or refractory AML (62).
By demonstrating the key immune-suppressive function of the GAS6–AXL axis in leukemia-associated macrophages, this work provides a conceptual advance in our understanding of the molecular mechanisms underlying immune suppression in leukemia and can effectively be translated into a treatment strategy that not only empowers the patient's own immune system to fight leukemia but can also be harnessed, in specific contexts, to overcome the major issue of primary resistance to PD-1 checkpoint blockade. We believe that our work as a whole paves the way for the design of new combinatorial therapeutic strategies that can enhance the effectiveness of standard and immune-based therapies, while limiting treatment-associated toxicity, in aggregate to significantly improve the outcome of patients with leukemia. Moreover, because AXL blockade demonstrates efficacy in AXL-negative tumors, this work has far-reaching clinical implications, as it extends the potential clinical benefit of AXL inhibition to a wider population of patients with cancer. Within the hematologic malignancy field, our study also stands as an important report demonstrating that effective rewiring of alternatively primed macrophages toward a proinflammatory fate is sufficient to “lift the barriers” toward potent antitumor immunity, kick-start the immunity cycle, and even elicit susceptibility to PD-1 checkpoint blockade in highly aggressive PD-1–refractory leukemia. As such, our work puts AXL on the list of promising cancer therapeutic targets that could improve efficacy of current therapeutic strategies by virtue of stimulating the innate immune system.
METHODS
Animal Studies
MxCre Asxl1f/f mice were a kind gift from Dr. Omar Abdel-Wahab (37) and used to generate a transplantable Asxl1−/− AML model after inducible ablation of Asxl1 in aged animals. All experiments were carried out using cells from a diseased primary mouse that displayed expansion of immature CD11bdimB220dim blasts that transferred leukemia to nonirradiated secondary recipients. Details about the generation of both BCR–ABL1+ B-ALL and the serially transplantable Asxl1−/− AML models are described in Supplementary Methods and Supplementary Fig. S1. Immune-competent C57BL/6N Gas6 knockout mice (Gas6tm1.1(KOMP)Vlcg) were obtained from the Knock Out Mouse Project (KOMP) Repository. A scheme (Supplementary Fig. S3A) and detailed description of the knockout allele are available at http://www.mousephenotype.org/data/alleles/MGI:95660/tm1.1(KOMP)Vlcg. Absence of Gas6 was validated by real-time PCR and ELISA (Supplementary Fig. S3B and S3D). For Gas6−/− mice, C57BL/6N mice were used as controls (Jackson Laboratories; line #005304). In all other experiments with immune-competent mice, control wild-type mice were from the C57BL/6J sub-strain (Jackson laboratories; line #000664). For bone marrow transplantation experiments, B6.SJL-Ptprca Pepcb/BoyJ (CD45.1) were used as recipients (Jax line #002014). Immune-deficient Gas6 knockout mice were generated by inactivation of the Gas6 gene using CRISPR/Cas9 editing in NOD.Cg-Prkdcscid IL2rgtmWjl/Sz (NSG) zygotes using a workflow that was recently described by our group and detailed in Supplementary Methods and Supplementary Fig. S3C (63). NSG mice were obtained from the Jackson Laboratories (line #005557). The Csf1r-Cre Axlf/f and CD11c-eGFP-Cre Axlf/f mice were obtained from Carla Rothlin and Sourav Ghosh (Yale University, New Haven, CT) and described previously (42, 48). Cd8a−/− mice (Jax line #002665), Sting−/− mice (Jax line # 025805), and Socs3f/f mice (Jax line #010944) were obtained from the Jackson Laboratories. Mice were bred and maintained at the animal facility of the Institute for Tumor Biology and Experimental Therapy (Frankfurt am Main, Germany) in accordance with regulatory guidelines. All experiments were approved under protocols G50/15, F123/1034, and F123/2003.
Data Reporting
No statistical methods were used to predefine sample size. In drug treatment experiments, mice were randomized to different treatment groups. Monitoring of mice was done with blinding.
Cell Lines
The MS-5 cell line was acquired from the DSMZ-German Collection of Microorganisms and Cell Cultures. The PlatE cell line (64) was a gift from Dr. Jacques Ghysdael, and used to generate retroviral stocks expressing BCR–ABL1 as described previously (65). Primary leukemia lines and cell lines were routinely checked for Mycoplasma using the Venor GeM OneStep Mycoplasma PCR Kit (Minerva Biolabs).
Leukemia Transplantation Experiments
For primary BCR–ABL1+ B-ALL generation (Supplementary Fig. S1J), transduced cells were transplanted in lethally irradiated C57BL/6J mice (9 Gy). For all experiments, leukemia cells from primary mice were transplanted in nonirradiated secondary recipients to maintain the integrity of the microenvironment. The number of cells injected is indicated in each figure legend. Generation of BCR–ABL1+ B-ALL and a serially transplantable Asxl1−/− AML model are described in more detail in Supplementary Methods and Supplementary Fig. S1. The MLL–ENL leukemia model has been described previously (38, 66). For this model, transplantation of 105 leukemic cells (tomato+) cells was used to carry out all described experiments.
Generation of Bone Marrow Chimeras
WT B6.SJL-Ptprca Pepcb/BoyJ (CD45.1; Jax line #002014) mice were lethally irradiated (9 Gy) and subsequently reconstituted by intravenous injection of 1.5 × 106 whole bone marrow cells isolated from either WT C57BL/6J (CD45.2) mice or Sting −/− (CD45.2) mice. Hematopoietic reconstitution was verified by bleeding and flow analysis before mice were used in experiments (Supplementary Fig. S7A).
Peripheral Blood Analysis
Blood was collected by bleeding from the vena facialis using an EDTA-containing microvette (Microvette 200 K3E, SARSTEDT).
Isolation of Human Monocytes and Coculture Experiments
Peripheral blood was obtained from healthy adults, and mononuclear cells isolated by Ficoll density gradient centrifugation using Ficoll-Paque Plus (1,077 g/mL; GE Healthcare). Monocytes were isolated using MACS Human CD14 microbeads (#130-050-201; Miltenyi Biotec) according to the manufacturer's instructions. Purity was confirmed by FACS to be >95%. Cocultures of monocytes and Cell Trace Violet (Invitrogen, #C34557)–labeled leukemia cells were carried out in StemSpan serum-free medium (Stemcell Technologies) supplemented with 1% penicillin/streptomycin and glutamine for 24 hours in 12-well plates. CD45+CD14+ monocytes were subsequently purified using flow cytometry before RNA was isolated using the PicoPure RNA Isolation Kit (Applied Biosystems, #KIT0204).
RNA Isolation and Real-Time PCR
Spleen-derived macrophages were obtained using ultrapure mouse anti-F4/80 microbeads (#130-110-443, Miltenyi Biotec) as per the manufacturer's instructions. Purified cells were verified to be at least 95% CD45+CD11b+F4/80+. Assessment of Axl excision in the Csf1r-Cre+ Axlf/f mice has been evaluated as previously described (42). RNA was isolated using the PicoPure RNA Isolation Kit (Applied Biosystems, #KIT0204) according to the manufacturer's instructions and converted into cDNA using the SuperScript VILO cDNA Synthesis Kit (Thermo Fischer Scientific, #11754050). cDNA was diluted 1:4 before usage. Real-time PCR for assessment of Axl excision was carried out using Axl primers described in ref. 42 and expression was normalized to Sdha. Primers were purchased from Sigma-Aldrich and sequences provided in Supplementary Methods. ABI Power SYBR Green Master Mix (#4368702, Thermo Fisher Scientific) was used. For all other real-time PCR experiments, Taqman-based real-time PCR assays using the TaqMan Gene Expression Master Mix (Thermo Fisher Scientific, #4369016) and Taqman probes listed in Supplementary Methods, all purchased from Thermo Fisher Scientific, were used. Reactions were all performed on Viia7 system (Thermo Fisher Scientific).
Analysis of Publicly Available Datasets
RSEM-normalized RNA-sequencing expression data of 173 primary AML samples from The Cancer Genome Atlas (TCGA-LAML; ref. 67) and corresponding clinical data were downloaded using the cBioPortal for Cancer Genomics (https://www.cbioportal.org). The B-cell lymphoma dataset (GSE4475, n = 159) was retrieved from The SurvExpress database (68). Both datasets were imported into the R2 Genomics Analysis and Visualization Platform (http://r2.amc.nl) and subjected to a KaplanScan analysis to stratify patients according to GAS6 expression using the “scan” mode to define the best expression cutoff. Results were exported and plotted using GraphPad Prism 7 software and survival analysis performed using the log-rank (Mantel–Cox) test.
Flow Cytometry
Cells were prepared as single cell suspension and blocked with CD16/32 Fc Block (BD Biosciences, #553441) then subjected to multicolor panel staining. Staining was performed for 45 minutes on ice, in the dark. Antibodies and secondary reagents were titrated to determine optimal concentrations. CompBeads (BD Biosciences) were used for single-color compensation to create multicolor compensation matrices. The mouse antibodies used in this study were as follows: anti-CD45 BV786 (BD Biosciences, #564225), anti-CD45.1 FITC (eBioscience, #11-0453-85), anti-CD45.2 PE (eBioscience, #12-0454-83), anti-Gr1 PE-Cy7 (BD Biosciences, #552985), anti-CD3 APC-CY7 (BioLegend, #100222), anti-CD8a AF700 (BD Biosciences, #564986) or anti-CD8 PECY7 (eBioscience, #25-0081-82), anti-CD4 PECY7 (eBioscience, #25-0041-82), anti-B220 APC (BD Biosciences, #553092) or anti-B220 BV711 (BD Biosciences, #563892), anti-NK1.1 PECF594 (BD Biosciences, #562864), anti-CD11b FITC (BD Biosciences, #553310) or anti-CD11b PECF594 (BD Biosciences, #562317), anti-CD11c AF700 (BD Biosciences, #560583), anti-F4/80 PE (BD Biosciences, #565410), anti-MHC-II BV650 (BD Biosciences, #563415), anti-Annexin V-APC (BD Biosciences, #550475) and anti-FoxP3 PE (eBioscience, #12-5773-82), anti-Axl APC (eBioscience, #17-1084-82), anti-DX5 APC (eBioscience, #17-5971-81). The human antibodies used in this study were as follows: hCD45 PE (BD Bioscience; catalog 555483), hCD14 APC-CY7 (BD Biosciences; catalog no. 557831). For Foxp3 staining, following cell surface staining cells were fixed and permeabilized using the Cytofix/Cytoperm plus (BD Biosciences, #555028) according to the manufacturer instructions, and Foxp3 staining was performed overnight at 4°C in the dark using an antibody dilution of 1:200. Propidium iodide (catalog no. 25535-16-4, Sigma-Aldrich) or the AF700 fixable viability dye (BD Biosciences; catalog no. 564997) was used for live- and dead-cell discrimination. Gating strategies are depicted in the main figures and Supplementary Figures. FACS data were acquired on a BD LSRFortessa (BD Biosciences). FACS was done using a FACSAria Fusion (BD Biosciences). BD FACS Diva software version 8.0.1 was used for data collection. FlowJo version 10.4.2 was used for data analysis. Postsort purity was >95% and determined by reanalysis of sorted cells.
scRNA-seq
Viable nonleukemic (GFP−) leukocytes were FACS sorted from the spleen of n = 8 mice (no leukemia Axlf/f, n = 2; no leukemia Csf1r-Cre+ Axlf/f, n = 2; B-ALL Axlf/f, n = 2; B-ALL Csf1r-Cre+ Axlf/f, n = 2) and subjected to scRNA-seq following a standard 10 × Genomics workflow. Libraries were generated using the Chromium Next GEM Single Cell 3′ v3.1 kit. cDNA QC and quantification was measured using Bioanalyzer High Sensitivity DNA chip (Agilent) and Qubit dsDNA High Sensitivity Kit (Thermo Fisher Scientific). The sequencing was performed on NextSeq500 platform (Illumina) with a sequencing depth of at least 20,000 reads per cell using the NextSeq500/550 high output kit v2.5 (75 cycles; Illumina; catalog no. 20024906). Two runs were conducted, thus generating 2 libraries, using 1 mouse from each condition. 10X Genomics demultiplexed sequencing reads were obtained using cellranger mkfastq (version 3.1.0) from 10X Genomics and used to align the reads to the mouse genome (refdata-cellranger-mm10-3.0.0). The data from all samples were loaded in R (R version 3.6.2) and processed using the Seurat package (version 3.2.0; ref. 69). Cells with at least 1,000 UMIs per cell and less than 20% mitochondrial gene content were retained for analysis. To increase our analytic power, data from all mice, regardless of genotype or disease status, were initially combined into a single set leading to a total cell number of 36,000 cells. Merged dataset was normalized for sequencing depth per cell and log-transformed using a scaling factor of 10,000. The most variable genes in the dataset were identified and the top 2,000 were used for dimensionality reduction using Uniform Manifold Approximation and Projection (UMAP) dimension reduction technique (70) followed by density-based clustering using the Seurat tool (69). The top differentially expressed genes per cluster were used to identify cell types. To evaluate the differences between samples, differential expression analysis was performed using the MAST test using the 10X run number as latent variable (71).
Administration of Drugs
Nilotinib (catalog no. A8232) was purchased from APExBIO and administered once daily by oral gavage at a dose of 80 mg/kg (72). Bemcentininb (BGB324; CAS Nr-1037624-75-1) was kindly provided by BerGenBio, ASA and administered twice daily by oral gavage at a dose of 50 mg/kg as previously reported (35). Vincristine sulfate was purchased from APExBIO (catalog no. A1765) and administered by intraperitoneal injection once a week at a dose of 0.5 mg/kg for 2 weeks. The vehicle used for both bemcentinib and nilotinib was 0.5 % (W/W) Methyl Cellulose 400cp (#M0262, Sigma-Aldrich)/0.1 % (W/W) Tween 80 (#P4780, Sigma-Aldrich) in water. Vincristine sulfate was prepared in PBS. Nilotinib and bemcentinib were administered on 5 days on/2 days off schedule, unless otherwise indicated in text or figure legends. Nilotinib and bemcentinib were prepared fresh every day.
ELISA
Peripheral blood serum was isolated using Microvette 500 Z-Gel (#20.1344, SARSTEDT). Samples were then analyzed using the Mouse Gas6 DuoSet ELISA (DY986: R&D Systems), according to the manufacturer's instructions. Results were plotted using GraphPad Prism 7 software.
Generation of Bone Marrow–Derived Macrophages and Polarization Experiments
Bone marrow cells from 2 femurs were cultured for 6 days in 30 mL size teflon bags (#PL30, PermaLife, Origen) in DMEM (#21969-035, Gibco, Life Technologies) complemented with 10% FBS (#10270106, Gibco, Thermo Fisher Scientific), 1% l-Glutamine (#25030-024, Gibco, Thermo Fisher Scientific), 1% HEPES 1 mol/L (#H0887, Sigma-Aldrich), 1% penicillin–streptomycin (#15140-122, Gibco, Thermo Fisher Scientific), and 10 ng/mL mouse M-CSF (#14-8983-80, Thermo Fisher Scientific; M0). Media were exchanged every 2 days. For polarization experiments, cells were seeded in Nunc Multidishes with UpCell Surface (#174899, Thermo Fisher Scientific) with 10 ng/mL mouse M-CSF for M0 or 10 ng/mL mouse IFNγ (#315-05, PeproTech) plus 10 ng/mL LPS (#L4391, Sigma) for M1. Polarization was done for 24 hours in the presence or absence of 0.5 μmol/L of bemcentinib (BergenBio, ASA).
Administration of Antibodies and Liposome Suspension
Clodronate liposomes were purchased from Liposoma Research and delivered intravenously at a dose of 250 μL/mouse as indicated in figure legends. All depletion antibodies were purchased from Hölzel Diagnostika and administered via intraperitoneal injection.
CD8 Depletion
InVivoMab anti-mouse CD8alpha (BE0061) and InVivoMab rat IgG2b isotype control (BE0090) were administered at a dose of 50 μg/mouse.
NK Depletion
InVivoMab anti-mouse NK1.1 (BE0036) and InVivoMab mouse IgG2a isotype control (BE0085) were diluted in the InVivoPure pH7.0 dilution buffer (Hölzel Diagnostika; IP0070) and administered at a dose of 50 μg/mouse.
PD-1 Blockade
InVivoMab anti-mouse PD-1 (BE0146) and InVivoMab Rat IgG2a isotype control (BE0089) were administered at a dose of 200 μg/mouse every 4 days. Anti–PD-1 and Rat IgG2a isotype control antibodies were diluted in the InVivoPure pH 7.0 and pH 6.5 dilution buffers, respectively, (Hölzel Diagnostika; IP0070) as recommended by the manufacturer.
IL10, IL12, and TNFa-Blocking antibodies
InVivoMab anti-mouse IL12p40 (BE0051) and anti-mouse TNFα (BE0058) were administered via intraperitoneal injection at a dose of 300 and 400 μg/mouse/day, respectively, for the whole duration of the experiment, as indicated in Fig. 6D. IL10-neutralizing antibody (BE0049) or rat IgG1 isotype control (BE0088) were administered daily at 300 μg/mouse as described I Supplementary Fig. S2M.
Histologic Analyses
Tissue samples (spleen, liver, brain, and femur) were fixed in ROTI Histofix 4% (#P087.3, Carl Roth), dehydrated, and embedded in paraffin. Bones were decalcified in 0.5 mol/L EDTA PH7.4 (#ED2SS, Sigma-Aldrich) at 4°C under constant agitation, for 1–2 weeks, prior to embedding. Paraffin sections (3 μm) were subjected to either hematoxylin and eosin staining or immunohistochemistry staining on a Leica Bond-Max using the detection systems Bond Polymer Refine Detection (Leica) for anti-rabbit and anti-mouse antibodies or Bond Intense R Detection (Leica) for anti-goat antibodies. Primary antibodies were diluted with the Bond Primary Antibody Diluent (Leica). Anti-human GAS6 (1:100, rabbit polyclonal, HPA008275; Sigma-Aldrich), anti-human CD10 (1:100, ORG-8941, monoclonal mouse clone 56C6, Novocastra); anti-mouse PD1 (1:200, goat polyclonal, R&D Systems, AF1021), anti-human PDL1 (1:200; rabbit monoclonal, Cell Signaling Technology, 13684) were used. Antigen retrieval was performed within the detection system using a citrate or EDTA (CD10) buffer solution. Anti-GFP (1:1,000, polyclonal goat, ab6673; Abcam). Antigen retrieval and a secondary antibody staining were performed within the detection system using an EDTA buffer solution and a biotinylated rabbit anti-Goat IgG Antibody (BA-5000; Vector Laboratories), respectively. Myeloperoxidase (MPO) staining was performed using the ready to use Rabbit Polyclonal Antibody MPO Ab-1, #RB-373-R7, Thermo Scientific). Slides were examined with a Zeiss Axio Imager 2 microscope and pictures were taken using the AxioVision SE64 Rel.4.9 software. The analysis depicted in Supplementary Fig. S2M was carried out using QuPath v0.2.3 digital pathology software (73).
Tissue Preparation, Immunofluorescence Staining, and Microscopy
Tissues were fixed in ROTI Histofix 4% (#P087.3, Carl Roth) for 24 hours, then washed in PBS for an additional 24 hours before transfer into 30% sucrose (#9097.1, Carl Roth) in PBS until full equilibration. Tissues were then embedded in OCT (#600001, Weckert Labortechnik) and 5-μm tissue sections were cut at a cryostat (Leica). For immunofluorescence staining, frozen tissue sections were thawed, dried for 1 hour at room temperature and rehydrated for 30 minutes in PBS. Subsequently, tissue sections were blocked in 3% BSA (#BSA-50, BiomoL) + 0.1% Triton-X100 (# X100, Sigma Aldrich) in PBS for 1 hour at room temperature, followed by incubation with primary antibodies in 1% BSA overnight at 4°C in a humidified chamber. Primary antibodies were used at the following dilutions: anti-AXL (#AF854, R&D Systems; 1:50); anti-phospho AXL (#AF2228, R&D Systems; 1:50); anti-GAS6 (#AF986, R&D Systems; 1:50); anti-GFP (#ab13970, Abcam; 1:500); anti-Iba1 (rabbit; #019-19741, Wako Chemicals; 1:500); anti-Iba1 (Goat; #ab48004, Abcam; 1:200). Fluorophore-conjugated secondary antibodies were used at a dilution of 1:500 in 1% BSA in PBS for 1 hour at room temperature. Hoechst 33342 (#H3570, Thermo Fisher Scientific) was used to counterstain nuclei at a dilution of 1:2,500 prior to covering tissues with Fluoromount Aqueous Mounting Medium (#F4680, Sigma Aldrich) and cover slides (#631-0158, VWR). Immunofluorescence was visualized with a Yokogawa CQ1 confocal microscope using a 40x objective.
Human Samples and Ethical Compliance
All human specimens were obtained after written informed consent in compliance with the institutional review board at the Faculty of Medicine of the Technical University of Munich (ethics vote number 538/16) and the university hospital Carl Gustav Carus (ethics vote number EK49022018). Paraffin-embedded bone marrow trephine biopsies were retrieved from archived diagnostic samples (patient information is presented in Supplementary Table S1). The medical charts of all patients were reviewed by a physician to confirm the diagnosis and clinical data. Evaluation of GAS6 staining on bone marrow biopsies was carried out by a trained pathologist. Pseudonymized use of healthy donor buffy coat preparations from whole blood donations was approved by the Ethics Committee of Goethe University of Medicine, under ethics vote 329/10.
Quantification and Statistical Analysis
All statistical analyses were carried out using version 7 of the GraphPad Prism software. Kaplan–Meier survival curves with two-sided log-rank Mantel–Cox analysis was used to evaluate the difference in survival in vivo. Comparison of leukemic burdens and target expression levels were carried out using two-sided Student t tests.
Data and Code Availability
All raw sequencing data have been deposited in the European Nucleotide Archive (https://www.ebi.ac.uk/ena/) under the accession number PRJEB43830. Further information and requests for resources and reagents may be directed to and will be fulfilled by the corresponding author, Hind Medyouf (hind.medyouf@medyouflab.com). All unique/stable reagents generated in this study are available from the corresponding author with a completed Materials Transfer Agreement.
Authors' Disclosures
I. Tirado-Gonzalez reports non-financial support from Bergenbio and other support from European Research Council during the conduct of the study. A. Descot reports non-financial support from Bergenbio and other support from European Research Council during the conduct of the study. D. Soetopo reports non-financial support from Bergenbio during the conduct of the study. A. Schäffer reports other support from DFG during the conduct of the study. I. Kur reports other support from German Cancer Consortium during the conduct of the study. E. Czlonka reports other support from Deutsche José Carreras Leukämie-Stiftung e.V. during the conduct of the study. Dr Schäfer reports other support from European Research Council during the conduct of the study. A. Kubasch reports personal fees from Novartis, personal fees from Celgene, and personal fees from Janssen outside the submitted work. O. Abdel-Wahab has served as a consultant for H3B Biomedicine, Foundation Medicine Inc, Merck, Prelude Therapeutics, and Janssen, and is on the Scientific Advisory Board of Envisagenics Inc., Pfizer Boulder, and AIChemy Inc.; O. Abdel-Wahab has received prior research funding from Loxo Oncology and H3 Biomedicine unrelated to the current manuscript. U. Platzbecker reports grants and personal fees from BMS, Novartis, grants from Jazz, BergenBio, and grants from Amgen outside the submitted work. C.V. Rothlin reports personal fees from Surface Oncology and grants from Mirati Therapeutics outside the submitted work. H. Medyouf reports non-financial support from BergenBio, Inc, other support from Hessen State Ministry for Higher Education, Research, and the Arts, grants from European Research Council, and other support from EMBO during the conduct of the study; grants and personal fees from BergenBio, Inc; and grants from Janssen outside the submitted work. No disclosures were reported by the other authors.
Supplementary Material
Supplementary Methods
Supplementary Figures and Figure legends
Acknowledgments
We thank Diana Passaro and Owen Williams for the MLL–ENL tomato+ leukemia cells, Jan Rehwinkel for the Sting-deficient bone marrow cells, Lisa Sevenich for the Socs3-floxed mice, Sophia Thevissen for assistance with animal monitoring and processing, Birgitta E. Michels for help with the analysis of the Immunological Genome Project data, Boris Brill and Margit Wagenblast for assistance with animal applications, Stefan Stein, Annette Trzmiel, and Malte Paulsen for assistance with cell sorting, and Nathalie Groen for assistance with scRNA-seq data analysis. We are thankful to Florent Ginhoux and Florian R. Greten for helpful discussions and feedback and to BerGenBio for providing bemcentinib for this study. The Klinikum rechts der Isar/Technical University Munich (MTBIO) and the Klinikum Carl Gustav Carus MK1/Technical University Dresden biobanks are acknowledged for support with collection of human bone marrow biopsies. This work was supported by the European Research Council (ERC-Stg-639795, to H. Medyouf), the EMBO Young Investigator program (#4027, to H. Medyouf), the LOEWE Center Frankfurt Cancer Institute (FCI) funded by the Hessen State Ministry for Higher Education, Research and the Arts (to H. Medyouf), the German Research Council (DFG ME4214/2-1 within SPP 2084, to H. Medyouf) and the German Cancer Consortium (DKTK) Joint Funding (CHOICE consortium; to H. Medyouf, U. Platzbecker, K.S. Götze). E. Czlonka was supported by a fellowship from the German José Carreras Leukemia Foundation (DJCLS 04FN/2018). K.S. Götze is supported by the German Research Council (DFG, SFB 1243 project A09) and the German Jose Carreras Leukemia Foundation (DJCLS, 14 R/2018).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors' Contributions
I. Tirado-Gonzalez: Investigation, writing–review and editing. A. Descot: Investigation, writing–review and editing. D. Soetopo: Investigation, writing–review and editing. A. Nevmerzhitskaya: Investigation. A. Schäffer: Investigation. I. Kur: Investigation. E. Czlonka: Investigation. C. Wachtel: Investigation. I. Tsoukala: Investigation. L. Muller: Investigation, methodology. A.-L. Schäfer: Investigation. M. Weitmann: Investigation. P. Dinse: Methodology. E. Alberto: Resources. M.C. Buck: Investigation. J.J. Landry: Investigation, visualization, methodology. B. Baying: Investigation, methodology. J. Slotta-Huspenina: Resources, investigation. J. Roesler: Investigation. P.N. Harter: Supervision, investigation. A. Kubasch: Resources. J. Meinel: Resources. E. Elwakeel: Investigation. E. Strack: Investigation. C. Tran Quang: Resources, investigation. O. Abdel-Wahab: Resources. M. Schmitz: Supervision, methodology. A. Weigert: Supervision, investigation, writing–review and editing. T. Schmid: Investigation, methodology, writing–review and editing. U. Platzbecker: Resources. V. Benes: Supervision, investigation, methodology. J. Ghysdael: Resources, writing–review and editing. H. Bonig: Resources. K.S. Götze: Resources, investigation. C.V. Rothlin: Resources, writing–review and editing. S. Ghosh: Resources, writing–review and editing. H. Medyouf: Conceptualization, data curation, supervision, funding acquisition, investigation, writing–original draft, project administration, writing–review and editing.
References
- 1. Bassan R, Bourquin JP, DeAngelo DJ, Chiaretti S. New approaches to the management of adult acute lymphoblastic leukemia. J Clin Oncol 2018:JCO2017773648. [DOI] [PubMed] [Google Scholar]
- 2. Dohner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med 2015;373:1136–52. [DOI] [PubMed] [Google Scholar]
- 3. Witkowski MT, Lasry A, Carroll WL, Aifantis I. Immune-based therapies in acute leukemia. Trends Cancer 2019;5:604–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Christopher MJ, Petti AA, Rettig MP, Miller CA, Chendamarai E, Duncavage EJet al. Immune escape of relapsed AML cells after allogeneic transplantation. N Engl J Med 2018;379:2330–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Toffalori C, Zito L, Gambacorta V, Riba M, Oliveira G, Bucci Get al. Immune signature drives leukemia escape and relapse after hematopoietic cell transplantation. Nat Med 2019;25:603–11. [DOI] [PubMed] [Google Scholar]
- 6. Vago L, Perna SK, Zanussi M, Mazzi B, Barlassina C, Stanghellini MTet al. Loss of mismatched HLA in leukemia after stem-cell transplantation. N Engl J Med 2009;361:478–88. [DOI] [PubMed] [Google Scholar]
- 7. Giannopoulos K. Targeting immune signaling checkpoints in acute myeloid leukemia. J Clin Med 2019;8:236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Paczulla AM, Rothfelder K, Raffel S, Konantz M, Steinbacher J, Wang Het al. Absence of NKG2D ligands defines leukaemia stem cells and mediates their immune evasion. Nature 2019;572:254–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AVet al. Signatures of mutational processes in human cancer. Nature 2013;500:415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Curran EK, Godfrey J, Kline J. Mechanisms of immune tolerance in leukemia and lymphoma. Trends Immunol 2017;38:513–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Curran E, Chen X, Corrales L, Kline DE, Dubensky TW Jr, Duttagupta Pet al. STING pathway activation stimulates potent immunity against acute myeloid leukemia. Cell Rep 2016;15:2357–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Engblom C, Pfirschke C, Pittet MJ. The role of myeloid cells in cancer therapies. Nat Rev Cancer 2016;16:447–62. [DOI] [PubMed] [Google Scholar]
- 13. Witkowski MT, Dolgalev I, Evensen NA, Ma C, Chambers T, Roberts KGet al. Extensive remodeling of the immune microenvironment in B cell acute lymphoblastic leukemia. Cancer Cell 2020;37:867–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Komohara Y, Niino D, Saito Y, Ohnishi K, Horlad H, Ohshima Ket al. Clinical significance of CD163(+) tumor-associated macrophages in patients with adult T-cell leukemia/lymphoma. Cancer Sci 2013;104:945–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van Galen P, Hovestadt V, Wadsworth Ii MH, Hughes TK, Griffin GK, Battaglia Set al. Single-cell RNA-seq reveals AML hierarchies relevant to disease progression and immunity. Cell 2019;176:1265–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lumbroso D, Soboh S, Maimon A, Schif-Zuck S, Ariel A, Burstyn-Cohen T. Macrophage-derived protein S facilitates apoptotic polymorphonuclear cell clearance by resolution phase macrophages and supports their reprogramming. Front Immunol 2018;9:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bosurgi L, Hughes LD, Rothlin CV, Ghosh S. Death begets a new beginning. Immunol Rev 2017;280:8–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rothlin CV, Carrera-Silva EA, Bosurgi L, Ghosh S. TAM receptor signaling in immune homeostasis. Annu Rev Immunol 2015;33:355–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gay CM, Balaji K, Byers LA. Giving AXL the axe: targeting AXL in human malignancy. Br J Cancer 2017;116:415–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Graham DK, DeRyckere D, Davies KD, Earp HS. The TAM family: phosphatidylserine sensing receptor tyrosine kinases gone awry in cancer. Nat Rev Cancer 2014;14:769–85. [DOI] [PubMed] [Google Scholar]
- 21. Ben-Batalla I, Erdmann R, Jorgensen H, Mitchell R, Ernst T, von Amsberg Get al. Axl blockade by BGB324 inhibits BCR-ABL tyrosine kinase inhibitor-sensitive and -resistant chronic myeloid leukemia. Clin Cancer Res 2017;23:2289–300. [DOI] [PubMed] [Google Scholar]
- 22. Ben-Batalla I, Schultze A, Wroblewski M, Erdmann R, Heuser M, Waizenegger JSet al. Axl, a prognostic and therapeutic target in acute myeloid leukemia mediates paracrine crosstalk of leukemia cells with bone marrow stroma. Blood 2013;122:2443–52. [DOI] [PubMed] [Google Scholar]
- 23. Brandao LN, Winges A, Christoph S, Sather S, Migdall-Wilson J, Schlegel Jet al. Inhibition of MerTK increases chemosensitivity and decreases oncogenic potential in T-cell acute lymphoblastic leukemia. Blood Cancer J 2013;3:e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jin Y, Nie D, Li J, Du X, Lu Y, Li Yet al. Gas6/AXL signaling regulates self-renewal of chronic myelogenous leukemia stem cells by stabilizing beta-catenin. Clin Cancer Res 2017;23:2842–55. [DOI] [PubMed] [Google Scholar]
- 25. Lee-Sherick AB, Eisenman KM, Sather S, McGranahan A, Armistead PM, McGary CSet al. Aberrant Mer receptor tyrosine kinase expression contributes to leukemogenesis in acute myeloid leukemia. Oncogene 2013;32:5359–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Park IK, Mundy-Bosse B, Whitman SP, Zhang X, Warner SL, Bearss DJet al. Receptor tyrosine kinase Axl is required for resistance of leukemic cells to FLT3-targeted therapy in acute myeloid leukemia. Leukemia 2015;29:2382–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sinha S, Boysen J, Nelson M, Warner SL, Bearss D, Kay NEet al. Axl activates fibroblast growth factor receptor pathway to potentiate survival signals in B-cell chronic lymphocytic leukemia cells. Leukemia 2016;30:1431–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Whitman SP, Kohlschmidt J, Maharry K, Volinia S, Mrozek K, Nicolet Det al. GAS6 expression identifies high-risk adult AML patients: potential implications for therapy. Leukemia 2014;28:1252–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Niu X, Rothe K, Chen M, Grasedieck S, Li R, Nam SEet al. Targeting AXL kinase sensitizes leukemic stem and progenitor cells to venetoclax treatment in acute myeloid leukemia. Blood 2021;137:3641–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Aguilera TA, Rafat M, Castellini L, Shehade H, Kariolis MS, Hui ABet al. Reprogramming the immunological microenvironment through radiation and targeting Axl. Nat Commun 2016;7:13898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tsukita Y, Fujino N, Miyauchi E, Saito R, Fujishima F, Itakura Ket al. Axl kinase drives immune checkpoint and chemokine signalling pathways in lung adenocarcinomas. Mol Cancer 2019;18:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Terry S, Abdou A, Engelsen AST, Buart S, Dessen P, Corgnac Set al. AXL targeting overcomes human lung cancer cell resistance to NK- and CTL-mediated cytotoxicity. Cancer Immunol Res 2019;7:1789–802. [DOI] [PubMed] [Google Scholar]
- 33. Skinner HD, Giri U, Yang LP, Kumar M, Liu Y, Story MDet al. Integrative analysis identifies a novel AXL-PI3 kinase-PD-L1 signaling axis associated with radiation resistance in head and neck cancer. Clin Cancer Res 2017;23:2713–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sadahiro H, Kang KD, Gibson JT, Minata M, Yu H, Shi Jet al. Activation of the receptor tyrosine kinase AXL regulates the immune microenvironment in glioblastoma. Cancer Res 2018;78:3002–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ludwig KF, Du W, Sorrelle NB, Wnuk-Lipinska K, Topalovski M, Toombs JEet al. Small-molecule inhibition of Axl targets tumor immune suppression and enhances chemotherapy in pancreatic cancer. Cancer Res 2018;78:246–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan Set al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 2016;165:35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Abdel-Wahab O, Gao J, Adli M, Dey A, Trimarchi T, Chung YRet al. Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. J Exp Med 2013;210:2641–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Horton SJ, Walf-Vorderwulbecke V, Chatters SJ, Sebire NJ, de Boer J, Williams O. Acute myeloid leukemia induced by MLL-ENL is cured by oncogene ablation despite acquisition of complex genetic abnormalities. Blood 2009;113:4922–9. [DOI] [PubMed] [Google Scholar]
- 39. Pfeifer H, Raum K, Markovic S, Nowak V, Fey S, Oblander Jet al. Genomic CDKN2A/2B deletions in adult Ph(+) ALL are adverse despite allogeneic stem cell transplantation. Blood 2018;131:1464–75. [DOI] [PubMed] [Google Scholar]
- 40. Waizenegger JS, Ben-Batalla I, Weinhold N, Meissner T, Wroblewski M, Janning Met al. Role of growth arrest-specific gene 6-Mer axis in multiple myeloma. Leukemia 2015;29:696–704. [DOI] [PubMed] [Google Scholar]
- 41. Nepal S, Tiruppathi C, Tsukasaki Y, Farahany J, Mittal M, Rehman Jet al. STAT6 induces expression of Gas6 in macrophages to clear apoptotic neutrophils and resolve inflammation. Proc Natl Acad Sci U S A 2019;116:16513–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bosurgi L, Cao YG, Cabeza-Cabrerizo M, Tucci A, Hughes LD, Kong Yet al. Macrophage function in tissue repair and remodeling requires IL-4 or IL-13 with apoptotic cells. Science 2017;356:1072–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gokbuget N. Treatment of older patients with acute lymphoblastic leukemia. Hematology Am Soc Hematol Educ Program 2016;2016:573–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vignetti M, Fazi P, Cimino G, Martinelli G, Di Raimondo F, Ferrara Fet al. Imatinib plus steroids induces complete remissions and prolonged survival in elderly Philadelphia chromosome-positive patients with acute lymphoblastic leukemia without additional chemotherapy: results of the Gruppo Italiano Malattie Ematologiche dell'Adulto (GIMEMA) LAL0201-B protocol. Blood 2007;109:3676–8. [DOI] [PubMed] [Google Scholar]
- 45. Heng TS, Painter MW, Immunological Genome Project Consortium. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol 2008;9:1091–4. [DOI] [PubMed] [Google Scholar]
- 46. Ubil E, Caskey L, Holtzhausen A, Hunter D, Story C, Earp HS. Tumor-secreted Pros1 inhibits macrophage M1 polarization to reduce antitumor immune response. J Clin Invest 2018;128:2356–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maier B, Leader AM, Chen ST, Tung N, Chang C, LeBerichel Jet al. A conserved dendritic-cell regulatory program limits antitumour immunity. Nature 2020;580:257–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schmid ET, Pang IK, Carrera Silva EA, Bosurgi L, Miner JJ, Diamond MSet al. AXL receptor tyrosine kinase is required for T cell priming and antiviral immunity. Elife 2016;5:e12414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xu K, Harrison RE. Down-regulation of stathmin is required for the phenotypic changes and classical activation of macrophages. J Biol Chem 2015;290:19245–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol 2004;25:677–86. [DOI] [PubMed] [Google Scholar]
- 51. Balkwill F. Tumour necrosis factor and cancer. Nat Rev Cancer 2009;9:361–71. [DOI] [PubMed] [Google Scholar]
- 52. Tugues S, Burkhard SH, Ohs I, Vrohlings M, Nussbaum K, Vom Berg Jet al. New insights into IL-12-mediated tumor suppression. Cell Death Differ 2015;22:237–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Murray PJ. Macrophage polarization. Annu Rev Physiol 2017;79:541–66. [DOI] [PubMed] [Google Scholar]
- 54. Merad M, Sathe P, Helft J, Miller J, Mortha A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu Rev Immunol 2013;31:563–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhou Y, Fei M, Zhang G, Liang WC, Lin W, Wu Yet al. Blockade of the phagocytic receptor MerTK on tumor-associated macrophages enhances P2X7R-dependent STING activation by tumor-derived cGAMP. Immunity 2020;52:357–73. [DOI] [PubMed] [Google Scholar]
- 56. Holland SJ, Pan A, Franci C, Hu Y, Chang B, Li Wet al. R428, a selective small molecule inhibitor of Axl kinase, blocks tumor spread and prolongs survival in models of metastatic breast cancer. Cancer Res 2010;70:1544–54. [DOI] [PubMed] [Google Scholar]
- 57. Yang X, Shi J, Zhang X, Zhang G, Zhang J, Yang Set al. ssExpression level of GAS6-mRNA influences the prognosis of acute myeloid leukemia patients with allogeneic hematopoietic stem cell transplantation. Biosci Rep 2019;39:BSR20190389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kaczanowska S, Beury DW, Gopalan V, Tycko AK, Qin H, Clements MEet al. Genetically engineered myeloid cells rebalance the core immune suppression program in metastasis. Cell 2021;184:2033–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity 2013;39:1–10. [DOI] [PubMed] [Google Scholar]
- 60. Lu Q, Lemke G. Homeostatic regulation of the immune system by receptor tyrosine kinases of the Tyro 3 family. Science 2001;293:306–11. [DOI] [PubMed] [Google Scholar]
- 61. Loges S, Heuser M, Chromik J, Vigil CE, Paschka P, Ben-Batalla Iet al. Final analysis of the dose escalation, expansion and biomarker correlations in the Ph I/II trial BGBC003 with the selective Oral AXL inhibitor bemcentinib (BGB324) in relapsed/refractory AML and MDS. Blood 2018;132:2672. [Google Scholar]
- 62. Perl AE, Martinelli G, Cortes JE, Neubauer A, Berman E, Paolini Set al. Gilteritinib or chemotherapy for relapsed or refractory FLT3-mutated AML. N Engl J Med 2019;381:1728–40. [DOI] [PubMed] [Google Scholar]
- 63. Tirado-Gonzalez I, Czlonka E, Nevmerzhitskaya A, Soetopo D, Bergonzani E, Mahmoud Aet al. CRISPR/Cas9-edited NSG mice as PDX models of human leukemia to address the role of niche-derived SPARC. Leukemia 2018;32:1049–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Morita S, Kojima T, Kitamura T. Plat-E: an efficient and sSupplementary table system for transient packaging of retroviruses. Gene Ther 2000;7:1063–6. [DOI] [PubMed] [Google Scholar]
- 65. Medyouf H, Alcalde H, Berthier C, Guillemin MC, dos Santos NR, Janin Aet al. Targeting calcineurin activation as a therapeutic strategy for T-cell acute lymphoblastic leukemia. Nat Med 2007;13:736–41. [DOI] [PubMed] [Google Scholar]
- 66. Passaro D, Di Tullio A, Abarrategi A, Rouault-Pierre K, Foster K, Ariza-McNaughton Let al. Increased vascular permeability in the bone marrow microenvironment contributes to disease progression and drug response in acute myeloid leukemia. Cancer Cell 2017;32:324–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cancer Genome Atlas Research Network, Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJet al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013;368:2059–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Aguirre-Gamboa R, Gomez-Rueda H, Martinez-Ledesma E, Martinez-Torteya A, Chacolla-Huaringa R, Rodriguez-Barrientos Aet al. SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS One 2013;8:e74250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM IIIet al. Comprehensive integration of single-cell data. Cell 2019;177:1888–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Diaz-Papkovich A, Anderson-Trocme L, Ben-Eghan C, Gravel S. UMAP reveals cryptic population structure and phenotype heterogeneity in large genomic cohorts. PLoS Genet 2019;15:e1008432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AKet al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol 2015;16:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wolf A, Couttet P, Dong M, Grenet O, Heron M, Junker Uet al. Preclinical evaluation of potential nilotinib cardiotoxicity. Leuk Res 2011;35:631–7. [DOI] [PubMed] [Google Scholar]
- 73. Bankhead P, Loughrey MB, Fernandez JA, Dombrowski Y, McArt DG, Dunne PDet al. QuPath: open source software for digital pathology image analysis. Sci Rep 2017;7:16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods
Supplementary Figures and Figure legends
Data Availability Statement
All raw sequencing data have been deposited in the European Nucleotide Archive (https://www.ebi.ac.uk/ena/) under the accession number PRJEB43830. Further information and requests for resources and reagents may be directed to and will be fulfilled by the corresponding author, Hind Medyouf (hind.medyouf@medyouflab.com). All unique/stable reagents generated in this study are available from the corresponding author with a completed Materials Transfer Agreement.