Abstract
Innate Lymphoid Cells (ILCs) are guardians of mucosal immunity, yet the transcriptional networks that support their function remain poorly understood. We employed inducible combinatorial deletion of key transcription factors (TFs) required for ILC development (RORγt, RORα and T-bet) to determine their necessity in maintaining ILC3 identity and function. Both RORγt and RORα were required to preserve optimum effector functions, however RORα was sufficient to support robust IL-22 production among the LTi-like ILC3 subset, but not NCR+ ILC3s. LTi-like ILC3s persisted with only selective loss of phenotype and effector functions even after the loss of both TFs. In contrast, continued RORγt expression was essential to restrain transcriptional networks associated with type 1 immunity within NCR+ ILC3s, which co-express T-bet. Full differentiation to an ILC1-like population required the additional loss of RORα. Together, these data demonstrate how TF networks integrate within mature ILCs post-development to sustain effector functions, imprint phenotype and restrict alternative differentiation programs.
Keywords: Innate lymphoid cells, Transcription factors, Cytokines, Intestine, Retinoic acid orphan receptors, single cell RNA sequencing
Introduction
Innate lymphoid cells (ILCs) are integral to the maintenance of tissue homeostasis1, functioning to interpret a wealth of environmental cues and translate these signals into appropriate responses2. This role is orchestrated through production of effector cytokine profiles once considered limited to CD4 helper T cell (Th) subsets3. Group 1 ILCs (ILC1s) express IFNγ and T-bet, group 2 ILCs (ILC2s) produce IL-4, IL-5, IL-9, IL-13 and express high levels of GATA3, while group 3 ILCs (ILC3s) are characterized by production of IL-22 and IL-17 and RORγt expression4. However, unlike T cells, ILCs do not require the provision of polarising signals or antigen in order to express subset-associated TFs during development, nor to acquire core effector functions5. Nonetheless, ILCs exhibit extensive plasticity and heterogeneity, even within subgroups that express the same “master” TFs6,7. Crucially, our previous findings indicate that developmentally required “master” TFs may in fact be dispensable in mature ILCs8. Dysregulated ILC TF expression, function and activity have been implicated in human inflammatory conditions such as inflammatory bowel disease, allergic asthma and atopic dermatitis9–11, as well as impaired anti-tumour immunity12. Thus, understanding the regulation of ILC functions post-development offers the potential to rectify impaired tissue homeostasis.
In this study, we utilized inducible transgenic mice to determine the requirements for continued expression of core ILC TFs in shaping ILC3 function and identity. We show that continued expression of both RORγt and RORα are required to preserve optimum effector functions, while also uncovering unique functions and subset-specific requirements for these related TFs. Notably, RORα was sufficient to support robust IL-22 production amongst the LTi-like ILC3 subset, and these cells could persist with only selective loss of phenotype and effector functions after the loss of both TFs. In contrast, in NCR+ ILC3s - where T-bet is co-expressed - RORγt is critical to restrain transcriptional networks associated with type 1 immunity. However, full conversion to an ILC1-like population requires the additional loss of RORα - suggesting ROR TFs act to limit T-bet driven transdifferentiation of NCR-expressing ILCs. Thus, our data provides in vivo demonstration of how these transcriptional networks interact to regulate function and phenotype of mature ILC3 subsets, revealing a complex interplay between, and differential needs for, canonical ILC-associated TFs post-development.
Results
ILC3 subsets have different requirements for RORγt and RORα
Our previous findings indicated that while RORγt expression is critical for ILC3 development13, core ILC3 functions - such as IL-22 production - could be maintained in adult, tissue-resident cells for several weeks in the absence of this “master” TF8. These findings suggested that effector functions could either be licensed by RORγt during development – after which the TF is no longer required – or that there are additional TFs that can compensate for the loss of RORγt. A potential candidate for maintaining ILC3 functions in these circumstances is the related TF RORα, which was recently revealed to be expressed by all ILCs14–16, rather than predominantly by ILC2s as initially reported17. To test this in ILCs in vivo – while circumnavigating developmental effects - we utilized Id2 creERT2 mice crossed with Rorc f/f or Rorc f/f × Rora f/f mice to enable inducible deletion of these TFs in adult tissue-resident ILCs. The resulting id2 lΔRORγt and Id2 iΔRORγt/RORα models were additionally crossed to Rosa26 flSTOP-tdRFP mice18 to facilitate identification of cre-recombinase expressing ILCs via tandem-dimer red fluorescent protein (tdRFP) expression. A dosing regimen to assess the impact of prolonged loss (>1 month) of TF expression was devised (Fig. 1a) and we focused our analysis upon the small intestine lamina propria (SILP) given the abundance of ILCs within this tissue (Fig. 1b).
Figure 1. RORα and RORγt combine to preserve functions in NCR+, but not LTi-like, ILC3 subsets.
To investigate the requirements for continued expression of RORγt and RORα in supporting ILC3 functions, mouse models enabling the inducible deletion of RORγt alone (Id2 iΔRORγt) or in combination with RORα (Id2 iΔRORγt/RORα) were established, with expression of tdRFP used to reveal activity of cre-recombinase. ILCs were isolated from SILP in all experiments. Cytokine expression was assessed after ex vivo stimulation with a cocktail of cytokines (IL-1β, IL-2, IL-6, IL-23) then PMA/Ionomycin/BFA. a Schematic outlining experimental design. b Gating strategy for identification of tdRFP+ ILCs from SILP. c Expression of IL-17A versus IL-22 by tdRFP+ ILCs. The percentage (d) and total number (e) of tdRFP+ ILCs expressing IL-22 (top) and IL-17A (bottom) (Id2 creERT2: n=17; Id2 iΔRORγt: n=15; Id2 iΔRORγt/RORα: n=14; from 7 independent experiments). f Expression of IL-17A and IL-22 versus RORγt by tdRFP+ ILCs. g Analysis of CCR6, c-Kit and NKp46 expression by tdRFP+ ILC3s from the SILP of Id2 creERT2 mice. h Representative expression of IL-17A and IL-22 by tdRFP+ KLRG1- ILCs split into the LTi-like, NCR+ and DN ILC3 subsets. i Total number of tdRFP+ ILCs isolated from SILP of Id2 creERT2, Id2 iΔRORγt and Id2 iΔRORγt/RORα mice (Id2 creERT2: n=17; Id2 iΔRORγt: n=16; Id2 iΔRORγt/RORα: n=15; from 8 independent experiments). Each data point on graphs is a mouse, horizontal bars denote the median. Statistical significance was tested using an ordinary one-way ANOVA with Tukey’s test for multiple comparisons, ns = not significant, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. Exact P values are provided in the source data.
To investigate the consequences of TF loss on ILC3 functions, SILP cells were stimulated ex vivo to potently activate ILC3s and promote type 3 cytokine production19. Consistent with our previous study8 and despite the prolonged absence of RORγt expression, approximately 75% of tdRFP+ ILC from id2 lΔRORγt mice produced IL-22. In contrast there was a more marked decrease in the proportion and total number of ILC3s that produced IL-17A (Fig. 1c-e). Moreover, while deletion of both RORγt and RORα completely ablated IL-17A production, it surprisingly resulted in only a 50% decrease in the proportion and number of tdRFP+ ILCs that produced IL-22 (Fig. 1c-e), suggesting a proportion of ILCs retain the capacity to produce IL-22 despite efficient deletion of RORγt and RORα in these cells (Fig. 1f).
To further dissect these discrepancies, we hypothesized that the partial reduction in IL-22 expression could be due to preferential changes within a specific ILC3 subset. The SILP contains diverse ILC3 subsets20,21, which can be differentiated through multiple surface markers including CCR6, c-Kit and NKp46 expression6. Analysis of the SILP of tamoxifen-fed Id2 creERT2 controls confirmed that within the tdRFP+ RORγt+ population, CCR6+ NKp46- (LTi-like ILC3s; which further express c-Kit), CCR6- NKp46+ (NCR+ ILC3s) and CCR6- NKp46- ‘double negative’ (DN) ILC3s were all present (Fig. 1g). Subset specific analysis of cytokine expression revealed that IL-22 expression by NCR+ ILC3s was heavily reduced following loss of RORγt and RORα expression, while LTi-like and DN subsets largely retained their ability to produce IL-22 (Fig. 1h). It was notable, however, that the total numbers of tdRFP+ ILCs between the mouse strains remained comparable (Fig. 1i), suggesting cellular differentiation or reduced function, rather than the loss of ILC populations after TF-deletion.
To further validate ILC3 subset-specific TF requirements, we used an Il17a cre × Rosa26 flSTOP-tdRFP model22 to generate Il17a ΔRORγt and Il17a ΔRORγt/RORα mice. Here, Il17a fate-mapping marked a proportion of the LTi-like and DN ILC3s, but not NCR+ ILC3s (Extended Data Fig. 1a-g). No significant difference in the number of IL-22+ tdRFP+ ILC3s was observed between Il17a ΔRORγt, Il17a ΔRORγt/RORα and controls, while the number of IL-17A+ ILCs was reduced approximately 4fold (Extended Data Fig. 1h,i). Collectively, these data indicated that NCR+ ILC3s were highly dependent upon RORγt and RORα to maintain IL-22 production, while in contrast mature LTi-like and DN ILC3 subsets retain the ability to produce IL-22 even in the combined absence of both of these TFs, as opposed to IL-17A.
TF-deletion drives subset-specific transcriptomic changes
Our data indicated ILC3 subset-specific requirements for the TFs RORγt and RORα in maintaining core effector functions. ILC3 subsets differ in their phenotype and functions6 and co-operative TFs can support ILC3 subset cellular identity and function in part through repression of other differentiation programs. In the case of NCR+ ILC3s RORγt is known to be co-expressed alongside the canonical type 1 TF T-bet23. However, the impact of concomitant deletion of these TFs - either constitutively or inducibly - on ILC3 heterogeneity, phenotype or function has never been assessed. To specifically test this, we further generated Id2 iΔRORγt/Tbet mice, and confirmed efficient deletion of all TFs targeted across these different mouse models (Extended Data Fig. 2a,b). In the absence of reagents to assess RORα protein, we confirmed efficient deletion of the LoxP flanked exon of Rora, alongside a similar analysis of Rorc expression as a control (Extended Data Fig. 2c-e). Conversely, we detected expression of transcripts of the non-LoxP flanked exons of both Rorc and Rora, albeit at reduced frequency compared to controls, reflecting continued transcription across the deleted exons to produce a non-functional truncated mRNA, as expected (Extended Data Fig. 2d,e).
To interrogate the impact of single and combinatorial TF deletion in the intestinal ILC family we employed droplet-based single cell RNA-sequencing (scRNA-seq). Unbiased clustering of total Id2+ (tdRFP+) ILCs from all samples revealed both overlapping and distinct clustering of ILCs by donor genotype (Fig. 2a-e). Further analysis revealed 11 distinct clusters across all samples, with marked enrichment for certain clusters by genotype (Fig. 2b). All cells exhibited high expression of core ILC genes including Id2 and Il7r (Fig. 2c), while expression of known subset-defining genes (e.g. Klrg1, Ccr6 and Ncr1) enabled broad assignment of individual clusters to the previously defined major ILC groups and subsets (Fig. 2c, d). Broadly all 11 clusters could be designated into five ‘superclusters’: ILC2, LTi-like ILC3, NCR+ ILC3, ex-ILC3/ILC1 based upon shared expression of known ILC subset genes, as well as an “unknown” or dysregulated phenotype (Fig. 2b-d, Extended Data Fig. 3a,b). Moreover, marked transcriptional heterogeneity was induced by TF deletion and evident amongst ILC superclusters – most notably within the LTi-like ILC3 and ex-ILC3/ILC1-like superclusters (Fig. 2b,d,e). Amongst the 11 clusters present across all four genotypes, we identified a single cluster of ILC2s (cluster 1; expressing Gata3, Klrg1, Il4, Areg, Ly6a), while clusters 2-4 exhibited gene signatures associated with LTi-like ILC3s (expressing Rorc, Ccr6, Cd4, Nrp1, H2-Aa), cluster 5 was identified as NCR+ ILC3s (expressing Rorc, Ncr1, Tbx21), clusters 6-9 as the ex-ILC3s/ILC1 supercluster (expressing Ncr1, Klrb1c, Tbx21, Il12rb2, Ifng) and two further clusters with a dysregulated transcriptional signature, of unknown identity (clusters 10 and 11) (Fig. 2c,f, Extended Data Fig. 3b). As detailed above, Rorc transcripts driven by non-floxed exons were also detected within our scRNA-seq data across multiple clusters (Extended Data Fig. 2c-e), however we further confirmed the efficient deletion of LoxP-flanked exons (exons 3-6) by mapping of the scRNA-seq reads (Extended Data Fig. 2f). Thus, as previously reported for comparable TF deletion studies24, non-functional (floxed) mRNA was detected in our scRNA-seq data in the absence of functional protein.
Figure 2. NCR+ ILC3s undergo extensive transcriptomic changes upon TF deletion.
To better understand the transcriptional changes and cellular heterogeneity amongst ILC3 subsets upon TF deletion, scRNA-seq was used to compare control (Id2 creERT2) ILCs with ILCs after inducible deletion of RORγt alone (Id2 iΔRORγt) or in combination with RORα (Id2 iΔRORγt/RORα) or T-bet (Id2 iΔRORγt/Tbet). a t-SNE plot showing ILCs derived from all samples by unsupervised clustering analysis (n = 17,347 cells of tdRFP+ ILCs isolated from 3 pooled SILP preps derived from Id2 creERT2, Id2 iΔRORγt, Id2 iΔRORγt/RORα and Id2 iΔRORγt/Tbet mice). b t-SNE plots showing a total of 11 ILC clusters across the 4 samples: Id2 creERT2 (n = 4194 cells), Id2 iΔRORγt (n = 5006 cells), Id2 iΔRORγt/RORα(n = 3921 cells) and Id2 iΔRORγt/Tbet (n = 4226 cells). c t-SNE plots of log-normalised counts (ALRA-imputed values) demonstrating expression of canonical ILC and ILC subset signature genes. d t-SNE plot with inferred ILC subset identity across all sequenced cells. e Relative enrichment of individual ILC clusters (z-scores) across genotypes. f Curated heatmap of highly differentially expressed genes amongst ILC clusters across all cells sequenced (z-scores). Heatmap shows significantly enriched genes expressed on average ≥0.5 fold higher within the specific cluster. Data representative of one independent experiment, cells sequenced within each experimental group derived from a pool of n=3 mice.
To interrogate the breadth of the TF network that might underlie the transcriptomic changes observed, we employed SCENIC analysis25 on our scRNA-seq data to identify predicted TF-associated gene signatures (regulons25,26). This identified 245 TF-associated regulons (each defined by >20 genes with predicted TF binding motifs) within ILCs derived from all four genotypes and notably, alternative clustering of ILCs by predicted regulon activity independently recapitulated the superclusters predicted by gene signature-based clustering methods (Extended Data Fig. 4a,b) and revealed extended TF networks associated with specific ILC subsets and superclusters (Extended Data Fig. 4c,d). Regulon analysis accurately predicted robust and discrete signatures of type 1, 2 and 3 immunity associated with GATA3, RORγt and T-bet despite these genes being otherwise expressed more broadly amongst multiple clusters (Extended Data Fig. 4e). This was particularly notable for Gata3 which has been previously shown to be expressed by all ILC subsets27, but which has the highest expression of its regulon associated predominantly with ILC2. Similarly, we observed that predicted T-bet regulon activity was only associated with a minor cluster of cells exhibiting an ex-ILC3/ILC1-like phenotype, despite the fact that this TF was more widely expressed by other cells, including NCR+ ILC3s. Notably, while RORγt regulon activity was broadly predicted amongst ILC3 subsets as expected, it was also predicted within a cluster of unknown cells (cluster 11), provoking the possibility that - despite the absence of the TF protein – a gene signature typical of RORγt and type 3 immunity may be maintained in ILC clusters emerging following TF perturbation.
To assess the epigenetic landscape of ILCs following deletion of core subset-associated TFs we performed an assay for transposase-accessible chromatin using sequencing (ATAC-seq). As ILC2s were largely unperturbed by TF deletion we assessed the chromatin landscape of FACS-isolated tdRFP+ KLRG1- ILCs. Focusing the analysis on differences within the Id2 iΔRORγt, Id2 iΔRORγt/RORα and Id2 iΔRORγ/Tbet samples (with the control samples plotted as a reference), we identified 2306 differentially represented peaks, the vast majority of which were within enhancer sequences (Extended data Fig. 5a-d)28. Five distinct patterns describing changes in the chromatin across the TF-deleting samples were observed, which we termed Groups A to E. The progressive loss (Group A) or gain (Group E) of peaks from the control (Id2 creERT2) through the Id2 lΔRORγt, Id2 lΔRORγt/RORα samples accounted for the vast majority (>80%) of the changes observed (Extended Data Fig. 5d). Consistent with our initial characterization of cytokine expression, open chromatin overlying ROR response element (RORE) motifs in the Il17a/Il17f loci were partially reduced and then totally lost in the Id2 iΔRORγt and Id2 iΔRORγt/RORαsamples respectively (Extended Data Fig. 5e). Interestingly, chromatin accessibility close to the Il22 gene was maintained, although partially reduced in Id2 iΔRORγt/RORα samples, however no predicted RORE motifs were detected (Extended Data Fig. 5e), suggesting a possible role for other TFs29,30 influenced by RORγt and RORα through indirect regulation.
Altered metabolic gene signatures following TF deletion
While a single LTi-like ILC3 population (cluster 2) was identified in control Id2 creERT2 animals, deletion of RORγt resulted in the marked emergence of two further LTi-like ILC3 clusters (clusters 3 and 4) the latter of which was particularly enriched upon deletion of both RORγt and RORα (Fig. 2b, e, f). Differential gene analysis across the LTi-like ILC3 clusters identified reduced expression of effector molecules and surface markers including Ccr6, Nrp1, Kit, H2-Ab1, ll22 in Id2 iΔRORγt and Id2 iΔRORγt/RORα samples (Extended Data Fig. 6a). However, as with IL-22 production, this translated to only a graded loss of protein expression for markers such as NRP1 and c-Kit, which remained above that detected on NCR+ ILC3s (Extended Data Fig. 6b). In contrast, expression of CCR6, like IL-17A, was reduced in the absence of RORγt and completely lost in the absence of both RORγt and RORα (Extended Data Fig. 6b). Expression of MHCII31 was similarly reduced in ILCs upon compound deletion of both TFs (Extended Data Fig. 6c).
To further investigate the transcriptional changes that occur in LTi-like ILC3s, we performed an unbiased comparison of clusters 2-4, which revealed 5 core modules of differentially expressed genes (DEGs) (Extended Data Fig. 6d, clusters I-V). These included several genes known to be involved in metabolic functions, in particular genes associated with polyamine synthesis (Arg1, Odc1, Sat1) and lipid storage (Dgat1) were reduced following loss of RORγt and/or compound loss of RORγt and RORα expression (Extended Data Fig. 6e). Reduced ARG1 expression was confirmed by flow cytometry (Extended Data Fig. 6f,g). Interestingly, while transcriptional changes within ILC2s were not sufficient to drive unique cell clusters following TF deletion, analysis of DEGs within ILC2s from control and Id2 iΔRORγt/RORα samples revealed a number of similar transcriptional changes that included reduced expression of Arg1 (Extended Data Fig. 7). Combined, these data suggest RORα acts to regulate optimal Arg1 and metabolic gene expression amongst ILCs, potentially contributing to the regulation of ILC metabolism32,33.
Loss of RORγt and RORα forces differentiation of NCR+ ILCs
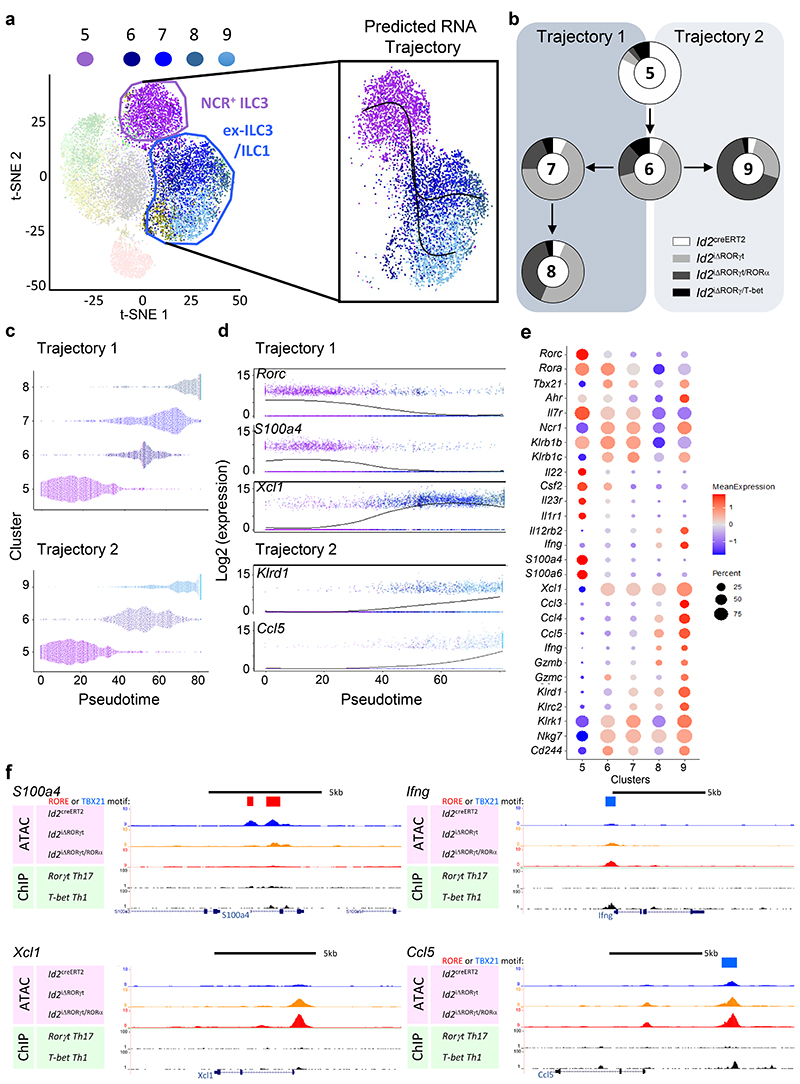
In contrast to LTi-like ILC3s, NCR+ ILC3s appeared highly sensitive to TF deletion with loss of RORγt/RORα resulting in the enrichment and expansion of a markedly heterogeneous population of cells (cluster 6-9), which exhibited gene signatures consistent with an ex-ILC3 or ILC1 phenotype (Fig. 2e,f, Extended Data Fig. 3b). To investigate the relationship between these clusters, we employed a trajectory analysis rooted in cluster 5 (Fig. 3a). This revealed two trajectories branching from cluster 6 either through clusters 7-8 (Trajectory 1), or to cluster 9 (Trajectory 2, Fig. 3a). Clusters 6-8 were dominated by ILCs from Id2 iΔRORγt mice, while the majority of the cells within cluster 9 arose from Id2 iΔRORγt/RORα mice (Fig. 3b). In both trajectories there was the rapid loss of the ILC3 program, characterized by the loss of Rorc, Il22, Il23r, Il1r1 and S100a4/a6 and graded expression of genes associated with ILC1 phenotype and type 1 immunity, including the chemokine Xcl1 (Fig. 3c-e). Changes to S100a4 and Xcl1 expression were further validated by qPCR (Extended Data Fig. 8a,b). Trajectory 2 was additionally defined by acquisition of a more robust ILC1-like phenotype, evidenced by highest expression of genes associated with type 1 effector mechanisms including Ifng, Ccl3, Ccl4, Ccl5 and Gzmb/c (Fig. 3c-e). Moreover, we noted increased expression of a wide range of natural cytotoxicity receptors (NCR) and NK cell-associated surface proteins across both trajectories (Fig. 3e).
Figure 3. Loss of both RORγt and RORα expression within NCR+ ILC3s drives full differentiation to mature ILC1-like phenotype.
To further understand how loss of RORγt and RORα expression affected the NCR+ ILC3 subset specifically, we sought to order the differentiation of these cells after TF deletion. a Trajectory analysis of clusters 5-9 using Slingshot. b Composition of clusters 5-9 based upon the proportion of cells contributed by the different samples: Id2 creERT2, Id2 iΔRORγt, Id2 iΔRORγt/RORα and Id2iΔRORγt/Tbet. c Emergence of clusters in Trajectories 1 and 2 over pseudotime. d Changes in expression of Rorc, S100a4, Xcl1 (Trajectory 1) and Klrd1, Ccl5 (Trajectory 2) over pseudotime. e Mean expression values of key genes associated with ILC3 and ILC1 cells. f UCSC genome browser display of mouse S100a4, Ifng, Xcl1 and Ccl5 loci with average traces of Id2 creERT2 (blue), Id2 iΔRORγt (yellow) and Id2 iΔRORγt/RORα(red) mice, alongside RORγt (49) and T-bet (50) ChIP-seq data, with RORE and TBX21 motifs marked.
These transcriptional changes were mirrored by the chromatin landscape, with S100a4 and Xcl1 highlighting contrasting changes in open chromatin regions (Fig. 3f). Many of the genes upregulated through clusters 6-9, including Ifng, Ccl3, Ccl4, Ccl5, Gzma, Klrc1, Klrd1 (Fig. 3f, Extended Data Fig. 8d) had progressively enhanced chromatin accessibility peaks through the Id2 iΔRORγt and Id2 iΔRORγt/RORα samples overlapping TBX21 motifs. Indeed, while RORγt-associated regulon activity was predicted within the NCR+ ILC3 cluster (cluster 5) and subsequently lost in clusters 6-9, T-bet-associated regulon activity was only predicted within cluster 8 and particularly cluster 9 (Extended Data Fig. 4c-e), despite Tbx21 transcripts throughout clusters 5-9 – suggesting transcriptional changes culminate in a terminal ILC1-like state. Thus, loss of RORγt expression initiates a graded differentiation towards an ILC1-like phenotype and in the additional absence of RORα, the T-bet-orchestrated type 1 immunity program is fully realized.
Transcriptional signature of mature ILC1s
To determine whether the spectrum of ex-ILC3/ILC1-like populations evident at the transcriptomic level existed at the cellular level, we utilized signatures identified in our scRNA-seq to validate and define cells with transitional phenotypes as well as a terminal/mature ILC1-like phenotype. Deletion of RORγt, alone or in combination with RORα, resulted in a striking increase in NKp46+ NK1.1hI cells (Extended data 8c), and coincided with enhanced expression of multiple transcripts further validated at the protein level, including CD94 (Klrd1), NKG2A (Klrc1), CD244 (Cd244), NKp46 (Ncr1) and NK1.1 (Klrb1) (Extended Data Fig. 8c). Indeed, NCR+ ILC3s were distinguishable from these ex-ILC3/ILC1-like cells34 based on lower expression of NK1.1 and very limited production of IFNγ after ex vivo restimulation (Fig. 4a). Based on these gene signatures, NKp46+ NK1.1hi IFNγ+ cells could be further segregated based upon co-expression of CD94 and NKG2A/C/E (Fig. 4b). Notably, CCL5 expression - associated with a terminal type 1 state - was restricted to a CD94+ NKG2A/C/E+ subpopulation amongst NKp46+ NK1.1hi IFNγ+ cells - while XCL1 was more widely expressed across the continuum of NCR+ ILCs (Fig. 4b) - consistent with the transcriptomic analysis. To further validate the spectrum of states, we performed t-SNE analysis of flow cytometry data amongst tdRFP+ ILCs from Id2 creERT2, Id2 iΔRORγt and Id2 iΔRORγt/RORαmice (Fig. 4c). This approach independently revealed the heterogeneity and changes in abundance observed in our scRNA-seq data (Fig. 4d), most notably expression of CD94 and NKG2A/C/E were clearly limited to a subset of the NK1.1+ NKp46+ IFNγ+ T-bet+ cells, found to be most abundant within the Id2 iΔRORγt/RORα mice (Fig. 4d). However, in contrast to recent findings in humans35, transition of NCR+ ILCs from an ILC3 to ILC1-like state did not correlate with expression of Aiolos in mice (Extended Data Fig. 8e), which surprisingly exhibited dichotomous expression between LTi-like ILC3s and the remaining helper ILC family members (Extended Data Fig. 8f,g). Collectively these data indicate loss of both RORγt and RORα expression results in expansion of the most functionally mature ILC1-like population and outline a distinctive phenotype to aid identification of these cells in other contexts.
Figure 4. Ex-ILC3s and ILC1s exhibit comparable spectrum of phenotypes within the SILP.
The transcriptomic changes induced upon deletion of RORγt and RORα in NCR+ ILC3s were assessed at the protein level using flow cytometry. a Representative expression of NKp46, NK1.1 and IFNγ by tdRFP+ KLRG1- ILCs from SILP of Id2 creERT2 and Id2 iΔRORγt/RORα mice after ex vivo stimulation with IL-12, IL-18 and PMA/Ionomycin/BFA. b Representative expression of CCL5 and XCL1 by tdRFP+ ILC subsets identified in ‘a’, gating on NKp46+ NK1.1hi IFNγ+ populations except where compared to DN/LTi-like ILC3s. c t-SNE plots showing tdRFP+ ILCs derived from SILP of Id2 creERT2 (turquoise), Id2 iΔRORγt (pink) and Id2 iΔRORγt/RORα(green) mice after ex vivo stimulation as shown in ‘a’ and ‘b’, by unsupervised clustering analysis (performed with the native platforms in FlowJo). d Relative expression of KLRG1, NKp46, NK1.1, NKG2A/C/E, CD94, IFNγ, CCL5, XCL1 and T-bet by tdRFP+ ILCs within clusters identified in ‘c’. To determine whether ex-ILC3s and ILC1s in the SILP exhibit comparable phenotypes, RORγt expression was fate-mapped using Rorc cre x Rosa26 tdRFP mice, enabling a definitive comparison of ex-ILC3s and bona fide ILC1s within the SILP. e Representative expression of NKp46, NK1.1 and IFNγ by tdRFP+ RORγt+ ILC3s (blue), tdRFP+ RORγt- ex-ILC3s (red) and tdRFP- RORγt- ILC1s (yellow) from SILP after ex vivo stimulation with IL-12, IL-18 and PMA/Ionomycin/BFA. f Representative expression of CD94 and NKG2A/C/E by NKp46+ NK1.1lo IFNγ+ ILC3, and NKp46+ NK1.1hi IFNγ+ ex-ILC3 and ILC1 populations. g Representative expression of CCL5 and XCL1 by populations identified in ‘f’. h Percentage of ILCs identified in ‘f’ expressing CCL5 or XCL1 (n=7 per cell type, data are representative of 3 independent experiments). Each data point on graphs is a mouse, horizontal bars denote the median.
While bona fide ILC1s have been identified in the liver36–38, a separate ILC1 lineage in the intestinal tract remains less clearly defined, particularly given ex-NCR+ ILC3s can take on an ILC1-like phenotype39. We investigated whether the phenotypic markers identified after inducible TF deletion were sufficient to distinguish ex-ILC3s with an ILC1-like phenotype from bona fide ILC1s emerging through a distinct developmental lineage. Using Rorc cre x Rosa26 tdRFP mice to fate-map ex-ILC3s as tdRFP+, we compared ex-ILC3s with NCR+ ILC3s (NKp46+ NK1.1- tdRFP+ RORγt+) and presumed bona fide ILC1s (NKp46+ NK1.1+ tdRFP- RORγt-) (Fig. 4e,f). Amongst NKp46+ NK1.1hi IFNγ+ cells, both RORγt fate-mapped ex-ILC3s and (non-fate-mapped) ILC1s exhibited a sub-population co-expressing CD94 and NKG2A/C/E, in marked contrast to NCR+ ILC3s with de facto RORγt protein. All ex-ILC3 populations and ILC1s expressed XCL1, whereas CCL5 expression was absent in NCR+ ILC3s and conversely enriched amongst CD94+ NKG2A/C/E+ cells, regardless of their origins, although non fate-mapped CD94+ NKG2A/C/E+ ILC1s expressed the highest level of CCL5 overall (Fig. 4g,h). Combined with our T-bet associated regulon analysis (Extended data Fig. 4e), these data suggest that a terminal ILC1 phenotype emerges following compound TF deletion that can be identified using a combination of markers identified above, although both ex-ILC3s and bona fide ILC1s can contribute to this cellular pool.
T-bet and RORα determine IL-22 expression amongst ILC3s
The most striking effect of inducibly deleting RORγt expression in mature NCR+ ILC3s was the radical shift in gene regulatory networks to those associated with type 1 immunity. Thus, we hypothesized that a key role of RORγt expression within NCR+ ILC3s was to check differentiation to an ex-ILC3/ILC1 fate through antagonism of the T-bet program. Further analysis of the id2 iΔRORγt/Tbet mice revealed that the population of NKp46+ RORγt- ILCs that was dramatically expanded upon deletion of RORγt alone, was almost entirely absent, replaced by the greatly increased proportion of NKp46- c-Kit- (DN) ILCs (Fig. 5a). This DN population (amongst the Id2 iΔRORγt/Tbet ILCs) lacked expression of NK1.1, NKG2A/C/E and CD49a, all of which were expressed by the enlarged NCR+ ILC population in the Id2 iΔRORγt mice (Fig. 5b,c). Thus, the combined absence of T-bet and RORγt failed to generate cells with an ILC1-like phenotype, instead resulting in a population of ILCs of unknown function. However, similarities in the chromatin landscape and regulon activity of ILCs from Id2 iΔRORγt/Tbet mice (cluster 11) with control ILC3s, suggested the maintenance of type 3 functions amongst ex-NCR+ ILCs in the absence of both RORγt and T-bet (Extended Data Fig. 5d, Extended Data Fig. 4c-e).
Figure 5. RORα expression is sufficient to preserve IL-23 mediated expression of IL-22 by non LTi-like ILC3s in the absence of RORγt and T-bet.
To investigate whether ILC3 effector functions were preserved in non-LTi-like ILC3 subsets in the absence of T-bet mediated differentiation towards an ILC1 state, ILC3 populations from SILP of Id2 creERT2 control, Id2 iΔRORγt and Id2 iΔRORγt/Tbet mice were assessed. a Representative flow cytometry plots showing NKp46 vs c-Kit expression by tdRFP+ KLRG1- ILCs, with further gating on ILC3 subsets and analysis of T-bet expression. b Representative histograms showing expression of NK1.1, NKG2A/C/E and CD49a by NCR+ ILC3s (blue) from Id2 creERT2 and Id2 iΔRORγt mice versus DN ILC3s (purple) from Id2 iΔRORγt/Tbet mice, compared with LTi-like ILC3s (grey). c Enumeration of expression of NK1.1 (MFI), NKG2A/C/E (%) and CD49a (MFI) (Id2 creERT2: n=4; Id2 iΔRORγt: n=3; Id2 iΔRORγt/T-bet: n=3; from one representative experiment). d Relative expression of key cytokine receptors across clusters 5 (Id2 creERT2), 6-8 (Id2 iΔRORγt) and 11 (Id2 iΔRORγt/Tbet). e UCSC genome browser display of mouse Il23r//l12rb2 loci with average traces of Id2 creERT2 (blue), Id2 iΔRORγt (yellow), Id2 iΔRORγt/RORα (red) and Id2 iΔRORγt/Tbet (green) mice, alongside RORγt (49) and T-bet (50) ChIP-seq data, with RORE and TBX21 motifs marked. f Representative expression of IL-22, IFNγ and XCL1 by c-Kit- (red) and c-Kit+ (grey) tdRFP+ KLRG1- RORγt+ ILCs from Id2 creERT2 mice and RORγt- ILCs from Id2 iΔRORγt and Id2 iΔRORγt/Tbet mice, after ex vivo stimulation with either IL-23 or IL-12 and IL-18. g Proportion of IL-22, IFNγ and XCL1 expressing ILCs from ‘f’ (Top graph Id2 creERT2: n=14; Id2 iΔRORγt: n=13; Id2 iΔRORγt/T-bet: n=12. Middle and bottom graphs n=4 for each genotype. Data representative of 2 independent experiments). h Representative expression of IFNγ versus IL-22 by tdRFP+ KLRG1- c-Kit-RORγt+, RORγt- and RORγt- T-bet- ILCs from Id2 creERT2, Id2 iΔRORγt/Tbet and Id2 iΔRORγt/Tbet/RORα mice, respectively, after stimulation with IL-23. i Proportion of tdRFP+ KLRG1- c-Kit- ILCs expressing IL-22 and MFI of IL-22 expression from ‘h’ (n=4 for each genotype, data representative of one experiment). Each data point on graphs is a mouse, horizontal bars denote the median. Statistical significance was tested using an ordinary one-way ANOVA with Tukey’s test for multiple comparisons. ns = not significant, * P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. Exact P values are provided in the source data.
To determine the functional capacity of these cells, we defined the relative cytokine receptor profile of cluster 11 at the transcriptional level. The loss of Il23r expression and upregulation of Il12rb2 in clusters 6-8 (Id2 iΔRORγt mice), as NCR+ ILC3 transition to ex-ILC3/ILC1, was completely blocked and type 3 cytokine receptors including Il23r and Il1r1 were restored (Fig. 5d), supported by changes to the chromatin landscape (Fig. 5e). To determine the ability of these cells to respond to type 3 and type 1 signals, we compared cytokine production after ex vivo restimulation with only recombinant IL-23 or IL-12/IL-18. NCR+ ILC3s from control mice produced IL-22 in response to IL-23, but neither IFNγ nor XCL1 in response to IL-12/IL-18 (Fig. 5f,g). Conversely, while the expanded population of NKp46+ ex-ILC3s/ILC1s from Id2 iΔRORγt mice was unable to produce IL-22 in response to IL-23, production of IFNγ and XCL1 was readily detected in response to the type 1 associated cytokines. In comparison, the LTi-like ILC3 population remained unchanged in its functions. Importantly, the NCR- c-Kit- ILC population dominant in Id2 iΔRORγt/Tbet mice produced IL-22 in response to IL-23, but lacked ability to make IFNγ or XCL1, suggesting that in the absence of both RORγt and T-bet, ILCs formerly of the NCR+ ILC lineage exhibit core effector functions associated with an ILC3 phenotype. To investigate the functional impacts of targeting RORγt and T-bet in ILCs, we used Citrobacter rodentium infection to test the impact upon ILC-associated protective mechanisms in the intestinal barrier. Comparing Id2 iΔRORγt/Tbet mice with ‘cre-negative’ littermate controls, we observed no differences in the initial colonisation or clearance of C. rodentium, levels of anti-microbial peptides nor histological changes (Extended Data Fig. 9).
To finally test whether continued RORα expression within the ex-NCR+ / c-Kit- ILC population - representative of cluster 11 (Fig. 2) - was sufficient to support ILC3 effector functions, we additionally generated Id2 iΔRORγt/Tbet/RORα mice. Ex vivo stimulation with IL-23 failed to induce IL-22 expression by c-Kit- ILCs from Id2 iΔRORγt/Tbet/RORα mice in contrast to controls, indicating that continued RORα expression was required to sustain IL-23R expression and IL-22 production in response to type 3 signals (Fig. 5h,i). Collectively these data demonstrate the crucial role of continued RORα expression in supporting ILC3 functions, acting in concert with RORγt although the latter is pivotal for preventing expression of a type 1 immunity program and differentiation towards an ILC1-state through inhibiting the actions of T-bet.
Discussion
Here we determined how co-operative and antagonistic transcriptional networks interact to dictate intestinal ILC effector functions post-development. Through inducible targeting of either RORγt alone or in combination with RORα, we discovered both critical shared roles and distinct contributions of these TFs in maintaining ILC3 subset phenotypes and effector cytokine production. These findings further develop our initial observation that some ILC3 functions were maintained after inducible deletion of RORγt8, and demonstrate that this can be only partly explained by the co-expression of RORα and the ability of this TF to sustain ILC3 functions. The impact of RORγt deletion in adult ILC3s was subset-specific, largely due to the differential expression of further master TF programs - namely T-bet - within these populations. ILC plasticity was first identified as the natural transition of NCR+ T-bet+ ILC3s to an ex-ILC3 or ILC1-like phenotype associated with the progressive loss of RORγt expression39,40. Our data demonstrate that sustained RORγt expression is critical to prevent the differentiation of ILCs that co-express T-bet towards an ex-ILC3 phenotype. However, differentiation of ex-ILC3s to a fully mature type 1 effector state - indistinguishable from bona fide intestinal ILC1s - required deletion of both RORγt and RORα. Thus collectively, our data highlight the extent to which ILC effector function and fate is dictated by the fine balance of TF programs within these cells and the role this plays post-development.
Our study reveals that intestinal ILCs require continued expression of neither RORγt nor RORα for survival, in contrast to Th17 cells8. Thus, ILC persistence and aspects of phenotype and effector functions in vivo can be separated from the programs that dictate their development and initial acquisition of subset-specific gene programs. However, both RORγt and RORα clearly contribute to sustaining the transcriptional networks associated with ILC3 function and phenotype, including Il17a/f, Ccr6 and Il23r, presumably through their shared recognition of RORE motifs. A notable exception to this was IL-22, which was surprisingly still produced by LTi-like cells from Id2 iΔRORγt/RORα mice upon ex vivo stimulation, suggesting distinct regulation independent of either TF. Additionally, our data implicate RORα as a potential regulator of metabolic fitness in ILCs. We observed that a number of genes, including Arg1, exhibited diminished expression in both ILC2s and LTi-like ILC3s in the absence of RORα, and Arginase-1 has previously been demonstrated to act as a crucial regulator of ILC metabolism, proliferation and effector function32. Given that all ILC populations express Rora 14 our data suggest a potential role for this TF in regulating conserved modules of genes across ILC groups, an exciting area for future research.
Within the NCR+ ILC3s that additionally express T-bet, deletion of Rorc expression resulted in the differentiation towards a type 1 immunity program, recapitulating the transition of ILC3 to an ex-ILC3 phenotype previously described39. Analysis of Id2 iΔRORγt and Id2 iΔRORγt/RORα mice suggested that RORα restrained full conversion to an ILC1-like state defined by the highest expression of type 1 effector cytokines, chemokines and granzymes. Given the extensive transdifferentiation of the NCR+ ILC3 subset in Id2 iΔRORγt mice, we hypothesized that programs able to sustain type 3 immunity might be masked by the dominance of T-bet activity. Using Id2 iΔRORγt/Tbet mice to test this, we discovered that deletion of both RORγt and T-bet blocked differentiation to an ex-ILC3 phenotype, while surprisingly preserving IL-23-dependent expression of IL-22. These data indicate that continued RORγt expression is necessary to restrain the type 1 immunity program orchestrated by T-bet, but in the absence of both TFs, RORα can sustain type 3 immune functions. In this regard, the cells within cluster 11 resembled the poorly described ‘DN’ ILC3 subset, which are currently defined only by the absence of CCR6 and NKp46 protein expression39. It was notable that our unbiased scRNA-seq analysis of small intestinal ILCs failed to identify a separate cluster of DN ILC3s despite their identification within the tdRFP+ population of the Id2 creERT2 mice by flow cytometry. These cells may be transcriptionally indistinguishable from NCR+ ILC3s due to their shared expression of T-bet and ability to upregulate NKp46 – as previously reported39 - thus lacking a sufficiently differential gene signature to drive separate clustering. As such it is attractive to hypothesize that the type 3 ILC cluster that emerges in the absence of both Rorc and Tbx21 is analogous to DN ILC3s, but clustered separately as a consequence of the loss of T-bet. Further studies are required to identify surface markers than can positively identify this progenitor-like DN ILC3 population and help define the role(s) of this subset.
We were able to identify and subsequently validate a novel functional signature of mature ILC1-like cells defined by co-expression of multiple specific NCR (i.e. NK1.1, CD94, NKG2A/C/E), granzymes and chemokines including Xcl1 and Ccl5. DC recruitment and activation via XCL1 and CCL5 has been attributed to NK and CD8 T cell populations41,42, however, ex-ILC3s and ILC1s may perform a similar role within some tissues and their role in orchestrating and modulating the wider immune response requires further investigation. Indeed, using the expression signatures defined by TF deletion alongside fate-mapping of Rorc expression, our data further indicates that small intestinal ex-ILC3s and bona fide ILC1s converge upon the same functional and transcriptional state, regardless of ontogeny. Together our findings provide novel combinations of TFs, surface markers and effector molecules that will aid future studies aimed at dissecting the origins and functional biology of ILC family members.
In summary, our study has enabled the first detailed assessment of how the TFs RORγt, RORα and T-bet combine to regulate ILC3 function and determine the balance of type 3 and type 1 immune programmes in vivo. The importance of TF interplay in ILC function reported here is echoed in findings from Romagnani and colleagues, who discovered that embryonic lymph node (LN) development was rescued in the absence of RORγt, when T-bet was additionally deleted (Stehle et al., in press43). Here, RORα was sufficient and necessary to support embryonic LTi cell functions in the absence of RORγt, but only having removed the T-bet-mediated pull to a distinct differentiation program. Such division of labour between antagonizing T-bet expression versus sustaining expression of type 3 immune genes enables finer control of cell function and likely reflects the benefit of expressing multiple ROR TFs44. Collectively, these data further highlight the importance of considering both co-operative and antagonistic TFs as we seek to define how ILCs retain their identity and core functions post-development and thus maintain tissue homeostasis.
Methods
Experimental design
Experimental groups of mice were not randomized. Data collection and analysis were not performed blind to the conditions of the experiments.
Mice
All mice used were C57BL/6 background and bred at the University of Birmingham. Mice were age and sex matched whenever possible, both males and females were used experimentally. Littermates were used when ‘cre-negative’ controls were appropriate. Strains used were Id2 creERT2 (JAX stock #016222)45, Il17a cre 46, Rosa26 tdRFP 47, Rorc f/f (JAX stock # 008771)48, Rora f/f 49, Tbx21 f/f 50, Rorc cre 51. All animal experiments were conducted in accordance of Home Office Guidelines, authorized under PPL P53927F5E granted to D. Withers after ethical review by the Animal Welfare and Ethical Review Body at the University of Birmingham, UK. Mice were housed at 21°C +/− 2°C, 55% humidity (+/- 10%) with 12 hrs light dark/ cycle in 7-7 IVC caging with environmental enrichment of plastic houses plus paper bedding. Tamoxifen was administered either ad libitum in the diet or by oral gavage (20 mg/mL, 3-5 doses of 200 μL given within 1 wk). Mice were aged between 6 and 12 wks of age at the start of the experiment. Where tamoxifen diet was used, mice were aged between 4-6 wks of age at the start of the regimen to ensure that the diet was tolerated.
Small intestine digestion protocol
The small intestine (SI) was dissected from below the stomach and above the caecum then placed in a Petri dish containing Hank’s Balanced Salt Solution (HBSS) (Sigma-Aldrich) 2% Foetal Bovine Serum (FBS) (Sigma-Aldrich). The fat and Peyer’s patches were removed before the SI was cut longitudinally and the contents washed out. The tissue was cut into small pieces, placed in HBSS 2% FBS and shaken vigorously, before being filtered through nitex mesh. The SI underwent a series of incubations in various digestion media; it was placed in a specific digestion medium, shaken vigorously for 20 seconds, incubated and shaken at 37°C for 20 min (HBSS/EDTA wash) or 15 min (collagenase digestion). This is followed by a washing process where the SI was filtered, resuspended in HBSS and vigorously shaken for a further 20 seconds before being filtered again. This process was carried out with the following digestion media: HBSS 2.5mM ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich) (20 min) twice and pre-warmed culture medium containing 1 mg/mL collagenase VIII (Sigma-Aldrich) (15 min). The SI was filtered through 100 μm and 70 μm cell strainers before being centrifuged and resuspended in an appropriate amount of DPBS supplemented with 2% FBS and 2.5mM EDTA (when cells were stained straightaway) or culture medium (when cells were cultured and stimulated in vitro before staining). On very rare occasions, digestion of the small intestine resulted in heavily reduced cell yields due to excessive cell death and these samples were then excluded from further analysis.
In vitro cell culture
Cells were cultured for 5 hrs in RPMI 10% FBS, penicillin/streptomycin, L-glutamine (Gibco™ ThermoFisher Scientific) with combinations of 20 ng/mL recombinant mouse IL-1β, IL-2, IL-6 (all obtained from Peprotech) and IL-23 (R&D Systems), where indicated. After 2 hrs, 50 ng/mL Phorbol 12-myristate 13-acetate (PMA), 1.5 μM ionomycin (IONO) and 10 μg/mL Brefeldin A (all obtained from Sigma-Aldrich) were added. In other experiments, cells were cultured for 5 hrs either without cytokine, 20-50 ng/mL IL-23, or 20-50 ng/mL recombinant mouse IL-12 (Peprotech) and IL-18 (R&D Systems), Brefeldin A (BFA) was added for the last 3 hrs of culture as well as PMA/IONO in some experiments.
Flow cytometry
Dead cells were excluded using LIVE/DEAD™ Fixable Near-IR Dead Cell Stain Kit (Invitrogen™ ThermoFisher Scientific) according to the manufacturer’s instructions. Surface staining was performed at 4°C for 30 to 40 min with antibodies diluted in DPBS supplemented with 2% FBS and 2.5mM EDTA. Intracellular staining was done using Cytofix/Cytoperm Plus (BD Biosciences) for cell fixation to preserve tdRFP signal and the Foxp3 / Transcription Factor Staining Buffer Set (eBioscience™ ThermoFisher Scientific) for permeabilization, both according to manufacturer’s instructions. Antibodies for intracellular staining were incubated overnight at room temperature in 1X permeabilization buffer. A complete list of antibodies used for flow cytometry in this study are provided in Supplementary Table 1. Lineage gating was performed by excluding cells stained with antibodies against B220, CD3, CD5, CD11b and CD11c labelled with the same fluorophore. Addition of Spherotech Accucount blank particles was used to calculate total cell numbers. Flow cytometric data was collected using a BD LSRFortessa™ X-20 analyzer (BD Biosciences) using FACSDiva 8.0.2 software (BD), with data subsequently analyzed with FlowJo (version 10) software (Tree Star).
Preparation of ATAC-seq samples
To isolate ILCs for ATAC-seq, magnetic enrichment and then FACS was employed. SILP cell suspensions were first stained with FITC-conjugated anti-CD45.2 antibodies, then incubated with anti-FITC microbeads (Miltenyi Biotec) and enriched through an LS column (Miltenyi Biotec) on a QuadroMACS™ Separator (Miltenyi Biotec), according to the manufacturer’s instructions. Cells in the retained fraction (CD45.2+ cells) were recovered and then stained with anti-CD45.2-FITC, anti-CD127-BV421, anti-CD3-PE-Cy7, anti-CD5-PE-Cy7, anti-CD11b-PE-Cy7, anti-CD11c-PE-Cy7, anti-B220-PE-Cy7 and anti-KLRG1-PerCP-eFluor710 antibodies. CD45.2+ Lin(CD3, CD5, CD11b, CD11c, B220)-CD127+ tdRFP+ KLRG1-/lo ILCs were eventually sorted on a BD FACSAria™ Fusion flow cytometer (BD Biosciences) using FACSDiva 8.0.2 software (BD). Each sample was obtained by first pooling SILP cell suspensions from 2 or 3 mice and enriched as described above, resulting in between 50,000 to 90,000 CD45.2+ Lin- CD127+ tdRFP+ KLRG1-/lo ILCs being sorted. Their DNA was first transposed using the Nextera® DNA Library Prep Kit (Illumina), with 0.5% digitonin, at 37°C for 10 min, with gentle shaking. Libraries were prepared according to the protocol published in the literature52. Libraries were sequenced using a NextSeq 500/550 High Output Kit v2.5 (Illumina) using the NextSeq500 platform (Illumina).
RNA purification and reverse transcription
RNA was extracted from sorted cells or tissue fragments with RNeasy® Mini Kit (Qiagen), with 1% β-mercaptoethanol (Sigma-Aldrich) Buffer RLT, according to the manufacturer’s instructions. Tissue fragments were stored in RNAlater (Invitrogen™ ThermoFisher Scientific) after collection and disrupted for 2 min, 30Hz, using a TissueLyser II (Qiagen). RNA was reverse-transcribed with SuperScript™ III Reverse Transcriptase (Invitrogen™ ThermoFisher Scientific), according to the manufacturer’s instructions.
Quantitative Real-time PCR
cDNA was amplified with SensiFAST™ SYBR® Hi-Rox Kit (Bioline Meridian Bioscience), according to the manufacturer’s instructions. Amplification reactions and detections were performed on ABI PRISM 7900HT Sequence Detection System (Applied Biosystems™ ThermoFisher Scientific): 50°C 2 min, 95°C 10 min, [95°C 15 s; 60°C 1 min] x 40. Primers used are described in Supplementary Table 2 (primers purchased from Sigma-Aldrich were designed using Primer-BLAST). Each sample was run as technical triplicates. Levels of mRNA were normalized to β-actin and data were analyzed in Microsoft Excel by calculating the relative expression of the target to β-actin in each sample= 2ΔCt (with ΔCt = Ctβ-actin – Cttarget).
Single-cell isolation, library construction and sequencing
ILCs were isolated by FACS as described above, but without prior magnetic enrichment. Gene expression libraries were prepared from single cells using the Chromium Controller and Single Cell 3’ Reagent Kits v2 (10x Genomics, Inc. Pleasanton, USA) according to the manufacturer’s protocol (CG00052 Rev B). The resulting sequencing libraries comprised standard Illumina paired-end constructs flanked with P5 and P7 sequences. The 16 bp 10x Barcode and 10 bp UMI were encoded in Read 1, while Read 2 was used to sequence the cDNA fragment. Sample index sequences were incorporated as the i7 index read. Paired-end sequencing (26:98) was performed on the Illumina NextSeq500 platform using NextSeq 500/550 High Output v2.5 (150 Cycles) reagents. The .bcl sequence data were processed for QC purposes using bcl2fastq software (v. 2.20.0.422) and the resulting .fastq files assessed using FastQC (v. 0.11.3), FastqScreen (v. 0.9.2) and FastqStrand (v. 0.0.5) prior to pre-processing with the CellRanger pipeline53.
Data pre-processing, filtering and quality control of scRNA-seq data
The sequence files generated from the sequencer were processed using 10x Genomics custom pipeline Cell Ranger v3.0.1. Three parameters were used for cell quality evaluation: the number of UMIs per cell barcode (library size), the number of genes per cell barcode and the proportion of UMIs that are mapped to mitochondrial genes. Cells with lower UMI counts than three Median Absolute Deviation (MAD) for the first two metrices and cells having higher proportion of reads mapped to mitochondrial genes with a cutoff of three MADs were filtered out. After this initial filtering, 17,347 cells (4194 sample A, 5006 sample B, 3921 sample C and 4226 sample D) out of 20,576 cells remained for downstream analysis.
We used the cyclone method to calculate for each cell the score of G1 phase and G2M phase. Cyclone uses a pre-build model that is trained on a known cell-cycle dataset where the sign of difference in expression between a pair of genes was used to mark the cell-cycle stages. The cell cycle phase of each cell is identified based on the observed sign for each marker pair of each of the phases.
Genes with an average UMI count below 0.009 were filtered out54. After this filtering 10,070 genes were left for downstream analysis. In order to account for the various sequencing depth of each cell, we normalized the raw counts using the deconvolution-based method55.
Visualization & Clustering of scRNA-seq data
Highly Variable Genes (HVGs) were identified and used to reduce the dimensions of the dataset using PCA. The dimension of dataset were further reduced to 2D using t-SNE and UMAP, where 1 to 14 components of the PCA were given as input. The cells were grouped into their putative clusters using the dynamic tree cut method. Minor contaminating clusters identified as expressing gene signatures associated with B cells and monocytes and were removed from the analysis prior to final clustering, which identified 11 clusters. We used the edgeR package to identify marker genes56, which were then used to annotate the cell types of a cluster. As the dataset had multiple samples and cluster, we formulated different combinations of samples and clusters and identified marker genes for these combinations using sSeq package [6].
Pseudotime analysis of scRNA-seq data
For pseudotime analysis, we applied Slingshot57 to cluster 5-9 from ILC samples isolated from Id2 creERT2, Id2 iΔRORγt and Id2 iΔRORγt/RORα mice. We applied an additional gene filtering where a gene is retained if it is expressed in at least 10 cells with a minimum UMI count of 3. After selection of cells and genes that are robustly expressed, data was re-normalized using pool based normalization. Slingshot was then applied with setting the parameter start.clus to 1. We then used SwitchDE to identify genes that have a switch-like differential expression across the pseudotime.
SCENIC Transcription factor analysis
A single-cell transcription factor (TF) gene regulatory network was inferred using PySCENIC (v0.9.19)58, the python implementation of SCENIC59. First, using normalized log-transformed counts for cells which passed the QC steps described above, gene regulatory networks (regulons) were inferred based on co-expression modules using the GRNBoost2 algorithm (https://github.com/tmoerman/arboreto). Next, the number of regulons were pruned to retain only those regulons whose target genes contain matching transcription factor DNA binding motif within their regulatory regions. This was achieved using the mouse mm9 version 9 (mc9nr) cisTARGET databases (https://resources.aertslab.org/cistarget/). Finally, regulon activity was quantified by calculating an enrichment score for the target genes within each regulon using AUCell. To identify regulatory networks specific for each cluster/supercluster we used the pySCENIC regulon specificity score (RSS) function which makes use of Jensen-Shannon Divergence60. Regulon activity was determined to be ON/OFF in each individual cell using the pySCENIC binarization function. A 2D t-SNE embedding of this binarized regulon activity was then generated using the t-SNE R package. Heatmaps of the mean of the sum of binary regulon activity for all cells within each cluster/supercluster were produced using the pheatmap R package. For this we excluded those regulons which we deemed to be constitutively expressed (ON in at least 60% of cells) or rarely expressed (ON in less than 4% of cells).
ATAC-seq data analysis
ATAC-seq data was quality and adapter trimmed using Trim Galore v0.6.5 using default parameters. It was mapped to the mouse GRCm38 genome assembly using bowtie2 v2.3.5.1 with default parameters. Alignments with a MAPQ score of < 20 were discarded. Peaks were called using MACS2 v2.1.2 with no input sequences, using options --nomodel --extsize 200 --shift -100 --broad. A merged peak set was generated by combining and merging overlapping peaks from all of the different samples. For all visualisations, peaks were quantitated with log2 transformed overlapping read counts and normalised using size factor normalisation. Initial differentially enriched regions were identified by feeding raw overlap counts into DESeq2 using the Likelihood Ratio Test across all sample groups with a threshold of FDR < 0.0.5. The reduced peak hit list repeated the DESeq2 analysis but using only the three knockout sample groups. Clustered differentially accessible regions were generated by applying correlation clustering to all replicates of all conditions and cutting the resulting tree at an R value of >=0.6. Clustered with fewer than 10 peaks in were discarded.
Genomic locations of peaks were assessed using the annotations from Ensembl Release 95. Promoters were taken to be +/- 1kbp from the start of mRNA features. An initial comparison of all the samples revealed 10,910 significant changes in areas of open chromatin, dominated by differences between the Id2 iΔRORγt, Id2 iΔRORγt/RORα and Id2iΔRORγ/T-bet mice versus control ILCs from Id2 creERT2 mice. A refined analysis focused on differences within the Id2 iΔRORγt, Id2 iΔRORγt/RORα and Id2 iΔRORγ/T-bet samples (with the control samples plotted as a reference) identified 2306 differentially represented peaks, the vast majority of which were likely within Enhancer sequences. Regions close to a gene were within 20kbp of a gene feature. Intergenic features were more than 20kbp from a gene feature. Relevant transcription factors were identified by analysing the sequences under each peak cluster using AME v5.1.0 using the 2018 non-redundant JASPAR vertebrate database. Counts of individual motif occurrences were extracted from the JASPAR hit data at http://expdata.cmmt.ubc.ca/JASPAR/downloads/UCSC_tracks/2018/mm10/ which were filtered to retain only sites with a confidence value of >=925.
Citrobacter rodentium infection
Nalidixic-acid-resistant C. rodentium ICC16961 was pre-cultured overnight, 37°C, 200 to 250 rpm, in 15 mL LB broth + 50μg/mL nalidixic acid (Sigma-Aldrich) seeded with an aliquot from a frozen stock in LB broth containing 15% glycerol (Sigma-Aldrich). Pre-cultures were then centrifuged and bacterial pellets resuspended in 1.5 mL sterile DPBS to produce the inoculum. Each mouse was orally gavaged with 200 μL of freshly prepared inoculum (which invariably contained 2-3x109 CFUs). The number of 15-mL pre-cultures was scaled accordingly to the size of the cohort to infect. Quantification of faecal CFU was determined by homogenization in sterile DPBS (10 μL per mg of faeces) and serial dilutions on LB agar + 50μg/mL nalidixic acid.
Histological sections
Proximal colons were fixed with 10% formalin and embedded in paraffin, 5-μm sections were cut and stained with hematoxylin and eosin (H&E) and Periodic acid-Schiff (PAS).
Statistical analysis
Details of the statistical tests applied to datasets shown in figures can be found in the corresponding figure legends. No statistical methods were used to predetermine sample sizes but our sample sizes are similar to those reported in previous publications62. Statistical analyses were performed using GraphPad Prism software version 8. Normal distribution was tested using Anderson-Darling, D’Agostino & Pearson, Shapiro-Wilk and Kolmogorov-Smirnov tests. Normally distributed data were analyzed with unpaired Student’s t tests or ordinary one-way ANOVA followed by Tukey’s multiple comparison tests. Non-normally distributed data were analyzed with Kruskal-Wallis tests followed by Dunn’s tests for multiple comparisons. In the entire study, two data points were identified as ‘Significant outliers’, as calculated by the Grubbs’ test, so these two values were removed from the analysis.
Extended Data
Extended Data Fig. 1. ILC3 subset-specific deletion of TFs reveals that LTi-like cells express normal levels of IL-22 in the absence of both RORγt and RORα.
Il-17a cre mice were used to enable ILC3 subset specific targeting of TF-deletion. a Gating strategy for identification of tdRFP+ ILCs from SILP of Il17a cre mice. b Expression of RORγt versus NKp46 by tdRFP- and tdRFP+ ILCs from SILP of Il17a cre mice. c Proportion of ILCs in the SILP of Il17a cre mice that express tdRFP at different ages (wks) post birth. d Representative CCR6, CD4, c-Kit, NK1.1 and NKp46 expression by tdRFP+ ILCs from SILP of Il17a cre mice. e Proportion of tdRFP+ ILCs from SILP of Il17a cre mice expressing molecules assessed in ‘d’. f Representative expression of RORγt versus CD127 by tdRFP- and tdRFP+ ILCs from SILP of Il17a cre, Il17a ΔRORγt and Il17a ΔRORγt/RORα mice. g Percentage of tdRFP- and tdRFP+ ILCs expressing RORγt (Il17a cre: n=12; Il17a ΔRORγt: n=11; from 4 independent experiments). h Representative expression of RORγt versus IL-17A and IL-22 by tdRFP+ ILCs from SILP of Il17a cre, Il17a ΔRORγt and Il17a ΔRORγt/RORα mice. i Total number of tdRFP+ ILCs from SILP of Il17a cre, Il17a ΔRORγt and Il17a ΔRORγt/RORα mice expressing IL-17A (top) and IL-22 (bottom) (Il17a cre: n=19; Il17a ΔRORγt: n=14; Il17a ΔRORγt/RORα: n=19; from 6 independent experiments). Each data point on graphs is a mouse, horizontal bars denote the median. Statistical significance in ‘g’ was tested using unpaired two-tailed Student’s t tests, and in ‘i’ an ordinary one-way ANOVA with Tukey’s test for multiple comparisons. ns = not significant, **P ≤ 0.01, ****P ≤ 0.0001. Exact P values are provided in the source data.
Extended Data Fig. 2. Efficient inducible deletion of targeted transcription factors in Id2 creERT2, Id2 iΔRORγt, Id2 iΔRORγt/RORα and Id2 iΔRORγt/Tbet mice.
a Representative histograms showing expression of RORγt (upper row) and T-bet (lower row) by tdRFP+KLRG1- ILCs from SILP of Id2 creERT2, Id2 iΔRORγt, Id2 iΔRORγt/RORα and Id2 iΔRORγt/Tbet mice, dotted line denotes positive/negative staining. b Proportion of tdRFP+KLRG1- ILCs expressing RORγt and T-bet in Id2 creERT2 (n=20), Id2 iΔRORγt (n=13), Id2 iΔRORγt/RORα (n=8) and Id2 iΔRORγt/Tbet (n=17) mice (Data are representative of 7 independent experiments). c Schematics of Rorc and Rora cDNA showing exons, the location of LoxP sites and the qPCR primer annealing sites, floxed exons removed by cre-mediated deletion are in light grey. Relative expression of transcripts for the exon3-exon4, exon7-exon8 and exon8-exon9 junctions of the Rorc locus (d) and for the exon3-exon4, exon7-exon8 and exon9-exon10 junctions of the Rora locus (e) as depicted in ‘c’, determined by qPCR analysis of cDNA prepared from tdRFP+ ILCs sorted from SILP of Id2 creERT2 (n=5), Id2 iΔRORγt (n=7), Id2 iΔRORγt/RORα (n=4) and Id2 iΔRORγt/Tbet (n=2) mice (Data are representative of 3 independent experiments). f Mapping of the reads detected by scRNA-seq along the Rorc locus in tdRFP+ ILCs from SILP of Id2 creERT2, Id2 iΔRORγt, Id2 iΔRORγt/RORα and Id2 iΔRORγt/Tbet mice. The localisation of LoxP-flanked exons is depicted by a grey box. Each data point on graphs is a mouse, horizontal bars in ‘b’ denote the median, bars in ‘d’ and ‘e’ denote the mean, with error bars denoting the SEM. Statistical significance in ‘b’ was tested using an ordinary one-way ANOVA with Dunnett’s test for multiple comparisons. ****P ≤ 0.0001.
Extended Data Fig. 3. Expression of key genes associated with ILC superclusters.
a t-SNE plot showing ILC superclusters within all samples (Id2 creERT2, Id2 iΔRORγt, Id2 iΔRORγt/RORα and Id2 iΔRORγt/Tbet mice). b Gene expression (ALRA-imputed values) of core ILC genes unique and shared amongst ILC superclusters.
Extended Data Fig. 4. Binary regulon activity within ILC clusters.
a t-SNE plots generated from transcriptome (normalised and log-transformed counts) and b binary regulon activity (On/Off) generated via SCENIC analysis of cells within the 4 samples. c Heatmap of the mean binary regulon activity for all cells within each of the 11 transcriptomically-defined clusters. Regulons were filtered to exclude those which were rarely (< 4%) or constitutively (> 60%) active across all cells in the dataset. d Regulon specificity score for each of the 5 superclusters generated from regulon enrichment scores in each cell calculated using AUCell step of the SCENIC pipeline. Top 8 most specific regulons in each supercluster, relative to the entire dataset, are highlighted in red. e Gata3, Rorc and Tbx21 mRNA expression (ALRA-imputed values, top) and binary regulon activity (bottom).
Extended Data Fig. 5. RORγt and RORα combine to regulate the chromatin landscape of ILC3s.
To further investigate how transcriptomic changes after TF deletion might be influenced by alterations to the chromatin landscape, FACS-isolated tdRFP+ KLRG1- ILCs from Id2 creERT2, Id2 iΔRORγt, Id2 iΔRORγt/RORα and Id2iΔRORγt/Tbet mice were assessed by ATAC-seq. a Principal component analysis of all samples using merged MACS2 broad peaks quantitated with size factor normalized reads per million (RPM). b Heat map showing all initial comparison of 10,910 significant changes in areas of open chromatin. c Primary genomic location of peaks and relative peak abundance around transcription start sites. d Heatmap of differentially enriched peaks between all knockouts (n = 2306) based on median centered size factor normalized log2RPM values, also showing the abundance of ROR response elements (RORE) and TBX21 motifs. Groups A to E refer to the distinct patterns of changes to open chromatin across the mouse models. e UCSC genome browser display of mouse Id2, Il17a/Il17f and Il22 loci with average traces from ILCs from Id2 creERT2 (blue), Id2 iΔRORγt (yellow), Id2 iΔRORγt/RORα (red) and Id2 iΔRORγt/Tbet (green) mice, alongside RORγt (49) and T-bet (50) ChIP-seq data, with RORE and TBX21 motifs marked.
Extended Data Fig. 6. Altered metabolism profile in LTi-like ILC3s lacking RORγt and RORα.
a Relative expression (z-score) of key genes associated with LTi-like ILC3 identification and function in clusters 2-4 (LTi-like supercluster) versus clusters 5 (NCR+ ILC3) and 9 (ILC1-like cells enriched in Id2 iΔRORγt/RORα mice). b Representative expression of CCR6, c-Kit and NRP1 by NCR- ILC3s, identified as tdRFP+ KLRG1- NKp46- cells from SILP of Id2 creERT2, Id2 iΔRORγt and Id2 iΔRORγt/RORα mice, grey shaded histograms show expression by NCR+ ILC3s as a control. c Representative contour plots showing expression of MHCII by NCR- ILC3s from the SILP of Id2 creERT2, Id2 iΔRORγt and Id2 iΔRORγt/RORα mice. d Heatmap (z-score) of differentially expressed genes identified via a three way comparison of LTi-like ILC3 cells (clusters 2-4 combined) derived from Id2 creERT2, Id2 iΔRORγt and Id2 iΔRORγt/RORα mice and selected on the basis of a mean expression ≥0.1 fpkm, False Discovery Rate ≤0.001 and Fold Change in cluster average Z-score > +/− 0.4. e Relative expression (z-score) of selected metabolism associated genes in total LTi-like ILC3 cells (cluster 2-4 combined) derived from Id2 creERT2, Id2 iΔRORγt and Id2 iΔRORγt/RORα mice. f Representative protein expression of ARG1 in tdRFP+ LTi-like ILC3s from SILP of Id2 creERT2, Id2 iΔRORγt and Id2 iΔRORγt/RORα mice. g Enumeration of % ARG1+ LTi-like ILCs and of the mean fluorescence intensity (MFI) of ARG1 expression assessed in ‘f’ (Id2 cre: n=3; Id2 iΔRORγt: n=3; Id2 iΔRORγt/RORα: n=2. Data from one representative experiment out of two independent experiments). Each data point on the graph is a mouse, horizontal bars denote the median. Statistical significance was tested using an ordinary one-way ANOVA with Tukey’s test for multiple comparisons. ns = not significant, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. Exact P values are provided in the source data.
Extended Data Fig. 7. Continued RORα expression maintains optimum Arg1 expression by ILC2s.
a Differentially expressed genes amongst ILC2 clusters in Id2 creERT2 and Id2 iΔRORγt/RORα. Log fold change ≥1.0, mean expression ≥0.1 fpkm, False Discovery Rate ≤0.02. Core ILC2 genes (no significant change) included for comparison. b Violin plots showing Arg1 expression (normalised and log-transformed counts) within the ILC2 cluster identified in Id2 creERT2, Id2 iΔRORγt, Id2 iΔRORγt/RORα and Id2 iΔRORγt/Tbet samples. c Representative protein expression of ARG1 in tdRFP+ ILC2s (KLRG1+ ILCs) from SILP of control Id2 creERT2 and Id2 iΔRORγt/RORα mice. d Percentage of ARG1+ ILC2s (tdRFP+ KLRG1+ ILCs) and MFI of ARG1 expression by ARG1+ ILC2s isolated from the SILP of control Id2 creERT2 and Id2 iΔRORγt/RORα mice (Id2 creERT2: n=5; Id2 iΔRORγt/RORα: n=3). Each data point on graphs is a mouse, horizontal bars denote the median. Statistical significance was tested using unpaired two-tailed Student’s t tests, *P ≤ 0.05. Exact P values are provided in the source data.
Extended Data Fig. 8. Validation of transcriptomic changes induced by deletion of RORγt and RORα.
Relative expression of S100a4 (a) and Xcl1 (b) transcripts determined by qPCR analysis of cDNA prepared from tdRFP+ ILCs sorted from SILP of Id2 creERT2 (n=5), Id2 iΔRORγt (n=7), Id2 iΔRORγt/RORα (n=4) and Id2 iΔRORγt/Tbet (n=2) mice (Data are representative of 3 independent experiments). c Representative expression of CD94, NKG2, CD244 and NKp46, versus NK1.1, by tdRFP+ ILCs from SILP of Id2 creERT2, Id2 iΔRORγt, Id2 iΔRORγt/RORα and Id2 iΔRORγt/Tbet mice. d UCSC genome browser display of mouse Klrc1, Klrd1, Gzma and Ccl3/4 loci with average traces from ILCs from Id2 creERT2 (blue), Id2 iΔRORγt (yellow), Id2 iΔRORγt/RORα (red) and Id2 iΔRORγt/Tbet (green) mice, alongside RORγt (49) and T-bet (50) ChIP-seq data, with RORE and TBX21 motifs marked. e Violin plots of Ikzf1 and Ikzf3 expression across clusters 1-11. f Representative expression of Aiolos by ILC2s, NCR+ ILC3s, ILC1s/ex-ILC3s, DN ILC3s and LTi-like ILC3s using flow cytometry. g Percentage of ILC2s, NCR+ ILC3s, ILC1s/ex-ILC3s, DN ILC3s and LTi-like ILC3s expressing Aiolos (n=4 for each ILC group/subset, data are representative of two independent experiments). Each data point on graphs is a mouse, horizontal bars in ‘a’ and ‘b’ denote the mean, with error bars denoting the SEM; horizontal bars in ‘g’ denote the median. Statistical significance in ‘g’ was tested using an ordinary one-way ANOVA with Tukey’s test for multiple comparisons. ns = not significant, **P ≤ 0.01, ****P ≤ 0.0001. Exact P values are provided in the source data.
Extended Data Fig. 9. Id2 İΔRORγt/Tbet mice are able to control and clear Citrobacter rodentium infection.
To functionally test whether protective intestinal mechanisms were maintained in Id2 iΔRORγt/Tbet mice, infection with Citrobacter rodentium was used. a C. rodentium counts expressed as CFU/g feces over the time course of infection in Id2 iΔRORγt (n=4) and Id2 iΔRORγt/Tbet (n=5 or 6 depending on the time point) mice versus littermate ‘no cre’ controls (n=8; to ensure comparable microbiome). b Relative expression of RegIIIb and RegIIIg in Id2 iΔRORγt and Id2 iΔRORγt/Tbet mice versus littermate ‘no cre’ controls at 21 dpi with C. rodentium (no cre control: n=8; Id2 iΔRORγt: n=4; Id2 iΔRORγt/Tbet: n=6). c Histological analysis of colon isolated from Id2 creERT2, Id2 iΔRORγt and Id2 iΔRORγt/Tbet mice at 15 dpi with C. rodentium (n=3 for each strain, one representative picture from one representative animal per strain). Each data point on graphs is a mouse, bars denote the mean with error bars denoting the SEM. Statistical significance was tested using a one-sided Kruskal-Wallis test with a Dunn’s test for multiple comparisons. *P ≤ 0.05. Exact P values are provided in the source data.
Supplementary Material
Acknowledgments
We thank A. Ptasinska (Institute of Cancer and Genomic Sciences, University of Birmingham), for advice and help with ATAC-seq experiments, Genomics Birmingham, the genomic and sequencing facility of the University of Birmingham, and the University of Birmingham Flow Cytometry Platform. We thank G. Howell and the University of Manchester flow cytometry core, and A. Hayes and C. Morrisroe in the University of Manchester Genomic Technologies core facility for their help with single-cell RNA sequencing. We thank G. Frankel (Imperial College, London), for the provision of C. rodentium ICC169. We thank the following for their kind provision of mice: D. Littman (Howard Hughes Medical Institute, New York, USA; Rorc cre mice), A. McKenzie (MRC-LMB, Cambridge, UK; Rora f/f mice), B. Stockinger (The Francis Crick Institute, London, UK; Il17a cre mice), S. Reiner (Columbia University, New York, USA; Tbx21 f/f mice), J. Fehling (Ulm University, Germany; Rosa26 tdRFP mice). We thank C. Stehle and C. Romagnani (Charité University Medicine Berlin, Germany) for critical discussion of data and the manuscript. This work was supported by a Senior Research Fellowship from the Wellcome Trust to DRW (110199/Z/15/Z) and by a Royal Society and Wellcome Trust Sir Henry Dale Fellowship (105644/Z/14/Z) and a Lister Institute of Preventative Medicine Prize to MRH. This work was also supported by a Wellcome Trust ISSF funding award (105610/Z/14/Z) to MRH. CF was supported by a Wellcome Investigator award (106898/A/15/Z) to J. E. Allen.
Abbreviations
- DEG
differentially expressed gene
- ILC
Innate lymphoid cell
- SILP
small intestine lamina propria
- ROR
retinoic acid orphan receptor
- RORE
ROR response element
- T-bet
T-box expressed in T cells
- tdRFP
tandem-dimer red fluorescent protein
- TF
transcription factor
- Th
helper T cell
Footnotes
Author Contributions statement
RF designed and performed experiments, analyzed data and contributed to writing the manuscript, CF analyzed sequencing data and performed in silico experiments, CW, SLB and JS performed and assisted with experiments, STHN, SMB and SA analyzed sequencing data and provided extensive bioinformatics support, MRH designed and performed experiments, analyzed data and wrote the manuscript, DRW designed experiments, analyzed data and wrote the manuscript.
Competing Interests statement
The authors declare no competing interests.
Data Availability
Data availability statement
scRNA-seq data are available in the ArrayExpress database (accession numbers E-MTAB-9795), ATAC-seq data are available in the GEO data base (accession number GSE163149). Further information and requests for resources should be directed to and will be fulfilled by the lead contacts D.R.W. (d.withers@bham.ac.uk) and M.R.H (matthew.hepworth@manchester.ac.uk). Source data are provided with this paper.
scRNA-seq data: “Understanding transcriptional identity in Innate Lymphoid Cells (ILC)” has been assigned ArrayExpress accession E-MTAB-9795.
ATAC-seq data: The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus63 and are accessible through GEO Series accession number GSE163149 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE163149).
Code Availability
The custom code used did not differ significantly from the intended implementation that we referenced and didn’t contain any new math, the custom R code was all for visualisation was used in this study.
References
- 1.Artis D, Spits H. The biology of innate lymphoid cells. Nature. 2015;517:293–301. doi: 10.1038/nature14189. [DOI] [PubMed] [Google Scholar]
- 2.Withers DR, Hepworth MR. Group 3 Innate Lymphoid Cells: Communications Hubs of the Intestinal Immune System. Front Immunol. 2017;8:1298. doi: 10.3389/fimmu.2017.01298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walker JA, Barlow JL, McKenzie AN. Innate lymphoid cells--how did we miss them? Nat Rev Immunol. 2013;13:75–87. doi: 10.1038/nri3349. [DOI] [PubMed] [Google Scholar]
- 4.Spits H, et al. Innate lymphoid cells--a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13:145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- 5.Shih HY, et al. Developmental Acquisition of Regulomes Underlies Innate Lymphoid Cell Functionality. Cell. 2016;165:1120–1133. doi: 10.1016/j.cell.2016.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melo-Gonzalez F, Hepworth MR. Functional and phenotypic heterogeneity of group 3 innate lymphoid cells. Immunology. 2017;150:265–275. doi: 10.1111/imm.12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colonna M. Innate Lymphoid Cells: Diversity, Plasticity, and Unique Functions in Immunity. Immunity. 2018;48:1104–1117. doi: 10.1016/j.immuni.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Withers DR, et al. Transient inhibition of ROR-gammat therapeutically limits intestinal inflammation by reducing TH17 cells and preserving group 3 innate lymphoid cells. Nat Med. 2016;22:319–323. doi: 10.1038/nm.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mjosberg JM, et al. Human IL-25 and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat Immunol. 2011;12:1055–1062. doi: 10.1038/ni.2104. [DOI] [PubMed] [Google Scholar]
- 10.Geremia A, et al. IL-23-responsive innate lymphoid cells are increased in inflammatory bowel disease. J Exp Med. 2011;208:1127–1133. doi: 10.1084/jem.20101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vivier E, et al. Innate Lymphoid Cells: 10 Years On. Cell. 2018;174:1054–1066. doi: 10.1016/j.cell.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 12.Gao Y, et al. Tumor immunoevasion by the conversion of effector NK cells into type 1 innate lymphoid cells. Nat Immunol. 2017;18:1004–1015. doi: 10.1038/ni.3800. [DOI] [PubMed] [Google Scholar]
- 13.Eberl G, et al. An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat Immunol. 2004;5:64–73. doi: 10.1038/ni1022. [DOI] [PubMed] [Google Scholar]
- 14.Walker JA, et al. Polychromic Reporter Mice Reveal Unappreciated Innate Lymphoid Cell Progenitor Heterogeneity and Elusive ILC3 Progenitors in Bone Marrow. Immunity. 2019;51:104–118.:e107. doi: 10.1016/j.immuni.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lo BC, et al. The orphan nuclear receptor RORalpha and group 3 innate lymphoid cells drive fibrosis in a mouse model of Crohn’s disease. Sci Immunol. 2016;1:eaaf8864. doi: 10.1126/sciimmunol.aaf8864. [DOI] [PubMed] [Google Scholar]
- 16.Lo BC, et al. The Transcription Factor RORalpha Preserves ILC3 Lineage Identity and Function during Chronic Intestinal Infection. J Immunol. 2019;203:3209–3215. doi: 10.4049/jimmunol.1900781. [DOI] [PubMed] [Google Scholar]
- 17.Halim TY, et al. Retinoic-acid-receptor-related orphan nuclear receptor alpha is required for natural helper cell development and allergic inflammation. Immunity. 2012;37:463–474. doi: 10.1016/j.immuni.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 18.Luche H, Weber O, Nageswara Rao T, Blum C, Fehling HJ. Faithful activation of an extra-bright red fluorescent protein in “knock-in” Cre-reporter mice ideally suited for lineage tracing studies. Eur J Immunol. 2007;37:43–53. doi: 10.1002/eji.200636745. [DOI] [PubMed] [Google Scholar]
- 19.Cave Edward John., M.D., F.R.C.P Br Med J. 1934;1:359. [Google Scholar]
- 20.Satoh-Takayama N, et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29:958–970. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 21.Sawa S, et al. Lineage relationship analysis of RORgammat+ innate lymphoid cells. Science. 2010;330:665–669. doi: 10.1126/science.1194597. [DOI] [PubMed] [Google Scholar]
- 22.Hirota K, et al. Fate mapping of IL-17-producing T cells in inflammatory responses. Nat Immunol. 2011;12:255–263. doi: 10.1038/ni.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lazarevic V, et al. T-bet represses T(H)17 differentiation by preventing Runx1-mediated activation of the gene encoding RORgammat. Nat Immunol. 2011;12:96–104. doi: 10.1038/ni.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scott CL, et al. The Transcription Factor ZEB2 Is Required to Maintain the Tissue-Specific Identities of Macrophages. Immunity. 2018;49:312–325.:e315. doi: 10.1016/j.immuni.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aibar S, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van de Sande B, et al. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat Protoc. 2020;15:2247–2276. doi: 10.1038/s41596-020-0336-2. [DOI] [PubMed] [Google Scholar]
- 27.Yagi R, et al. The Transcription Factor GATA3 Is Critical for the Development of All IL-7Ralpha-Expressing Innate Lymphoid Cells. Immunity. 2014;40:378388. doi: 10.1016/j.immuni.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koues OI, et al. Distinct Gene Regulatory Pathways for Human Innate versus Adaptive Lymphoid Cells. Cell. 2016;165:1134–1146. doi: 10.1016/j.cell.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Veldhoen M, et al. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- 30.Quintana FJ, et al. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;453:65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]
- 31.Hepworth MR, et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 2013;498:113–117. doi: 10.1038/nature12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monticelli LA, et al. Arginase 1 is an innate lymphoid-cell-intrinsic metabolic checkpoint controlling type 2 inflammation. Nat Immunol. 2016;17:656–665. doi: 10.1038/ni.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karagiannis F, et al. Lipid-Droplet Formation Drives Pathogenic Group 2 Innate Lymphoid Cells in Airway Inflammation. Immunity. 2020;52:885. doi: 10.1016/j.immuni.2020.04.021. [DOI] [PubMed] [Google Scholar]
- 34.Sanos SL, et al. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat Immunol. 2009;10:83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cella M, et al. Subsets of ILC3-ILC1-like cells generate a diversity spectrum of innate lymphoid cells in human mucosal tissues. Nat Immunol. 2019;20:980–991. doi: 10.1038/s41590-019-0425-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng H, et al. Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. J Clin Invest. 2013;123:1444–1456. doi: 10.1172/JCI66381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sojka DK, et al. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. Elife. 2014;3:e01659. doi: 10.7554/eLife.01659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bai L, et al. Liver type 1 innate lymphoid cells develop locally via an interferon-gamma-dependent loop. Science. 2021;371 doi: 10.1126/science.aba4177. [DOI] [PubMed] [Google Scholar]
- 39.Klose CS, et al. A T-bet gradient controls the fate and function of CCR6-RORgammat+ innate lymphoid cells. Nature. 2013;494:261–265. doi: 10.1038/nature11813. [DOI] [PubMed] [Google Scholar]
- 40.Vonarbourg C, et al. Regulated expression of nuclear receptor RORgammat confers distinct functional fates to NK cell receptor-expressing RORgammat(+) innate lymphocytes. Immunity. 2010;33:736–751. doi: 10.1016/j.immuni.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bottcher JP, et al. NK Cells Stimulate Recruitment of cDC1 into the Tumor Microenvironment Promoting Cancer Immune Control. Cell. 2018;172:1022–1037.:e1014. doi: 10.1016/j.cell.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brewitz A, et al. CD8(+) T Cells Orchestrate pDC-XCR1(+) Dendritic Cell Spatial and Functional Cooperativity to Optimize Priming. Immunity. 2017;46:205–219. doi: 10.1016/j.immuni.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.NI. 2021 [Google Scholar]
- 44.Taylor JS, Raes J. Duplication and divergence: the evolution of new genes and old ideas. Annu Rev Genet. 2004;38:615–643. doi: 10.1146/annurev.genet.38.072902.092831. [DOI] [PubMed] [Google Scholar]
- 45.Rawlins EL, Clark CP, Xue Y, Hogan BL. The Id2+ distal tip lung epithelium contains individual multipotent embryonic progenitor cells. Development. 2009;136:3741–3745. doi: 10.1242/dev.037317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hirota K, et al. Fate mapping of IL-17-producing T cells in inflammatory responses. Nat Immunol. 2011;12:255–263. doi: 10.1038/ni.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luche H, Weber O, Nageswara Rao T, Blum C, Fehling HJ. Faithful activation of an extra-bright red fluorescent protein in “knock-in” Cre-reporter mice ideally suited for lineage tracing studies. Eur J Immunol. 2007;37:43–53. doi: 10.1002/eji.200636745. [DOI] [PubMed] [Google Scholar]
- 48.Choi GB, et al. The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science. 2016;351:933–939. doi: 10.1126/science.aad0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oliphant CJ, et al. MHCII-Mediated Dialog between Group 2 Innate Lymphoid Cells and CD4(+) T Cells Potentiates Type 2 Immunity and Promotes Parasitic Helminth Expulsion. Immunity. 2014;41:283–295. doi: 10.1016/j.immuni.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Intlekofer AM, et al. Anomalous type 17 response to viral infection by CD8+ T cells lacking T-bet and eomesodermin. Science. 2008;321:408–411. doi: 10.1126/science.1159806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eberl G, Littman DR. Thymic origin of intestinal alphabeta T Cells Revealed by Fate Mapping of RORgammat+ Cells. Science. 2004;305:248–251. doi: 10.1126/science.1096472. [DOI] [PubMed] [Google Scholar]
- 52.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol. 2015;109:21 29 21–21 29 29. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng GX, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bourgon R, Gentleman R, Huber W. Independent filtering increases detection power for high-throughput experiments. Proc Natl Acad Sci U S A. 2010;107:9546–9551. doi: 10.1073/pnas.0914005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lun AT, Bach K, Marioni JC. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 2016;17:75. doi: 10.1186/s13059-016-0947-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Street K, et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics. 2018;19:477. doi: 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van de Sande B, et al. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat Protoc. 2020;15:2247–2276. doi: 10.1038/s41596-020-0336-2. [DOI] [PubMed] [Google Scholar]
- 59.Aibar S, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suo S, et al. Revealing the Critical Regulators of Cell Identity in the Mouse Cell Atlas. Cell Rep. 2018;25:1436–1445.:e1433. doi: 10.1016/j.celrep.2018.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ghaem-Maghami M, et al. Intimin-specific immune responses prevent bacterial colonization by the attaching-effacing pathogen Citrobacter rodentium. Infect Immun. 2001;69:5597–5605. doi: 10.1128/iai.69.9.5597-5605.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Withers DR, et al. Transient inhibition of ROR-gammat therapeutically limits intestinal inflammation by reducing TH17 cells and preserving group 3 innate lymphoid cells. Nat Med. 2016;22:319–323. doi: 10.1038/nm.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data availability statement
scRNA-seq data are available in the ArrayExpress database (accession numbers E-MTAB-9795), ATAC-seq data are available in the GEO data base (accession number GSE163149). Further information and requests for resources should be directed to and will be fulfilled by the lead contacts D.R.W. (d.withers@bham.ac.uk) and M.R.H (matthew.hepworth@manchester.ac.uk). Source data are provided with this paper.
scRNA-seq data: “Understanding transcriptional identity in Innate Lymphoid Cells (ILC)” has been assigned ArrayExpress accession E-MTAB-9795.
ATAC-seq data: The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus63 and are accessible through GEO Series accession number GSE163149 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE163149).
scRNA-seq data are available in the ArrayExpress database (accession numbers E-MTAB-9795), ATAC-seq data are available in the GEO data base (accession number GSE163149). Further information and requests for resources should be directed to and will be fulfilled by the lead contacts D.R.W. (d.withers@bham.ac.uk) and M.R.H (matthew.hepworth@manchester.ac.uk). Source data are provided with this paper.
scRNA-seq data: “Understanding transcriptional identity in Innate Lymphoid Cells (ILC)” has been assigned ArrayExpress accession E-MTAB-9795.
ATAC-seq data: The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus63 and are accessible through GEO Series accession number GSE163149 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE163149).
The custom code used did not differ significantly from the intended implementation that we referenced and didn’t contain any new math, the custom R code was all for visualisation was used in this study.