Abstract
Highlights
eOPN3 is a mosquito-derived rhodopsin that inhibits neurotransmission in neurons
Activation of eOPN3 activates the Gi/o pathway and reduces Ca2+ channel activity
eOPN3 can suppress neurotransmission in a variety of cell types in vitro and in vivo
Activation of eOPN3 in nigrostriatal dopamine axons modulates locomotor behavior
Summary
Information is carried between brain regions through neurotransmitter release from axonal presynaptic terminals. Understanding the functional roles of defined neuronal projection pathways requires temporally precise manipulation of their activity. However, existing inhibitory optogenetic tools have low efficacy and off-target effects when applied to presynaptic terminals, while chemogenetic tools are difficult to control in space and time. Here, we show that a targeting-enhanced mosquito homolog of the vertebrate encephalopsin (eOPN3) can effectively suppress synaptic transmission through the Gi/o signaling pathway. Brief illumination of presynaptic terminals expressing eOPN3 triggers a lasting suppression of synaptic output that recovers spontaneously within minutes in vitro and in vivo. In freely moving mice, eOPN3-mediated suppression of dopaminergic nigrostriatal afferents induces a reversible ipsiversive rotational bias. We conclude that eOPN3 can be used to selectively suppress neurotransmitter release at presynaptic terminals with high spatiotemporal precision, opening new avenues for functional interrogation of long-range neuronal circuits in vivo.
Introduction
Neurons form local and long-range synaptic connections, through which they interact with neighboring neurons and with distant neuronal circuits, respectively. Long-range neuronal communication is crucial for synchronized activity across the brain and for the transmission of information between brain regions with distinct information processing capabilities. For example, dopaminergic neurons in the substantia nigra project to the dorsal striatum via the nigrostriatal pathway and play a critical role in movement control as part of the basal ganglia circuitry (Alcaro et al., 2007). Manipulating the activity of such long-range projection pathways allows a detailed evaluation of their functional contribution to cognitive and behavioral processes. However, while optogenetics allows robust and temporally precise excitation of long-range projecting axons (Yizhar et al., 2011), silencing such long-range connections with existing optogenetic tools has proven difficult (Wiegert et al., 2017a). We have previously shown that the light-driven chloride pump halorho-dopsin (eNpHR3.0) only partially suppresses neurotransmitter release. The proton-pumping archaerhodopsin (eArch3.0) triggers off-target effects, including an increase in intracellular pH and elevated spontaneous neurotransmission (Mahn et al., 2016), potentially leading to off-target behavioral consequences (Lafferty and Britt, 2020). While halorhodopsin-mediated inhibition has no effect on intra-synaptic pH (Mahn et al., 2016), it does temporarily shift the chloride reversal potential and can lead to GABA-mediated excitation (Raimondo et al., 2012). Furthermore, both halorhodopsin and archaerhodopsin require continuous delivery of high light power to sustain their ion pumping activity (Zhang et al., 2007). Alternative approaches, such as optogenetic induction of synaptic plasticity (Creed et al., 2015; Klavir et al., 2017; Nabavi et al., 2014), or inhibition by disruption of the release machinery (InSynC [Lin et al., 2013]; photo-uncaging of botulinum toxin-B [Liu et al., 2019]), can effectively decrease synaptic transmission but are not as temporally precise as direct optogenetic manipulations.
Chemogenetic tools (Armbruster et al., 2007; Magnus et al., 2011) can effectively suppress presynaptic terminal function upon delivery of the cognate ligands of these engineered receptors (Basu et al., 2016; Stachniak et al., 2014). However, these approaches depend on infusion of the ligand to the location of the targeted presynaptic terminals, and their temporal specificity is fundamentally limited by the binding affinity to and clearance of the ligand. The designer receptor activated by designer drug (DREADD) hM4Di inhibits synaptic transmission (Stachniak et al., 2014) through a mechanism used by native inhibitory GPCRs, presumably through suppression of Ca2+ channel activity (Herlitze et al., 1996) and inhibition of the vesicle release machinery downstream of Ca2+ influx (Gerachshenko et al., 2005; Zhu and Roth, 2014; Zurawski et al., 2019a). We reasoned that a light-activated Gi/o-coupled rhodopsin could potentially trigger the same type of synaptic suppression (Figure 1A). However, while many known vertebrate rhodopsins couple to the Gi/o pathway, these proteins are difficult to utilize as optogenetic tools since they undergo photobleaching after G protein dissociation as part of their natural phototransduction cycle (Bailes et al., 2012) (Figure 1B). Previous studies have revealed that bistable type-II rhodopsins are abundant across vertebrates and invertebrates (Tsukamoto and Terakita, 2010). These photoreceptors form a stable association with both the cis- and transconfiguration of the retinal chromophore (similar to the microbial type-I rhodopsin family including channelrhodopsin) and are therefore often referred to as bistable photopigments (Koyanagi et al., 2004; Terakita, 2005). Importantly, bistable type-II rhodopsins show reduced photobleaching (Bailes et al., 2012) (Figure 1B). We reasoned that members of the bistable type-II rhodopsin family that couple to Gi/o signaling would be suitable candidates for light-mediated silencing of neurotransmitter release from presynaptic terminals.
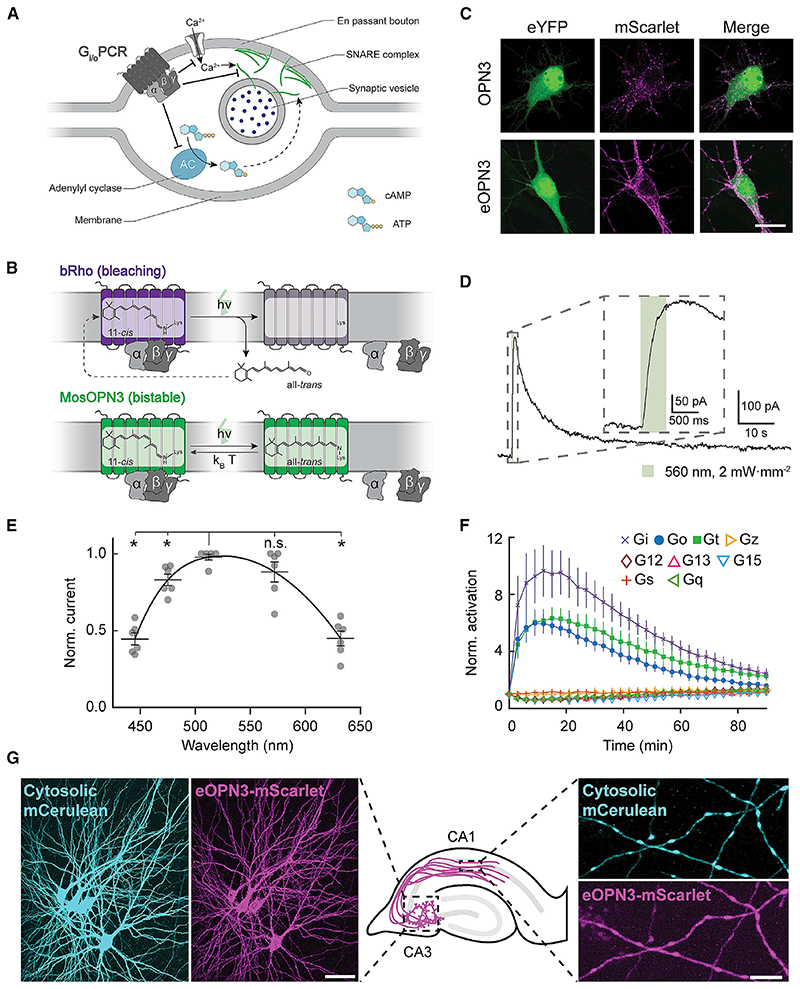
Figure 1. Gi/o-coupled rhodopsins for light-mediated presynaptic inhibition.
(A) Schematic diagram depicting the mechanism through which Gi/o signaling reduces the synaptic vesicle release probability. An activated GPCR leads to inhibition of voltage-gated Ca2+ channels as well as reduced cAMP levels, both leading directly (solid arrow) and indirectly (dashed arrow) to a reduction of Ca2+-dependent vesicle release.
(B) Schematic diagram of distinct retinal binding mechanisms in bleaching (top) and bistable (bottom) rhodopsins. Bleaching rhodopsins release all-trens-retinal following photon absorption (h-v) and need to bind a new 11-c/s-retinal before being ableto enterthe next photocycle. Bistable rhodopsins sustain theircovalent bond with retinal independent of its configuration, removing the influence of 11-c/s-retinal tissue availability. In bistable rhodopsins, all-trans-retinal switches back to 11-c/s-retinal either by absorbing another photon or spontaneously in the darkwith a probability depending on the kinetic energy of the molecule (k B-T). kB = Boltzmann constant; T = thermodynamic temperature; h = Planck constant; n = photon frequency.
(C) Representative confocal images of neurons co-transfected with expression vectors for eYFP and OPN3 or eOPN3. Images show fluorescence in the eYFP channel (left), the mScarlet channel (middle) and the merged images (right). See Figure S2 for all tested rhodopsin variants and quantifications. Scale bar, 15 mm.
(D) Sample whole-cell voltage-clamp recording of a cultured hippocampal neuron co-expressing eOPN3 and GIRK2-1, held at −70 mV. Inset shows an expanded view of the GIRK current onset during the light pulse.
(E) Action spectrum of endogenous GIRK-mediated currents in neurons expressing eOPN3, normalized to peak activation per cell (n = 6, p = 3.45-10− Friedman rank sum test followed by pairwise comparisons using Conover’s test). Peak excitation occurred at 512 nm (p < 4.24-10−3 Holm corrected pairwise comparisons to all other wavelengths except 572 nm).
(F) Light-dependent G protein activation by eOPN3, assayed as in Figure S3. eOPN3 specifically and strongly activated inhibitory G proteins (Gi, Go, Gt) in a lightdependent manner (n = 5). See Figure S3 for complete assay and statistics.
(G) Two-photon maximum-intensity projections of CA3 neurons co-expressing the cytosolic fluorophore mCerulean (cyan) and eOPN3-mScarlet (magenta). Shown are the somatodendritic compartment of neurons electroporated with the two plasmids (left; scale bar, 50 mm) and their axons projecting into stratum radiatum of CA1 (right; scale bar, 5 mm). Plots depict individual data points and average ± SEM.
Here, we tested several bistable rhodopsin variants for use as optogenetic tools, specifically addressing their expression in mammalian neurons and their capacity for Gi/o pathway activation and light-driven inhibition of presynaptic release. While many of these invertebrate opsins failed to express in mammalian neurons, we were able to optimize the expression of a mosquito-derived homolog of the mammalian encephalopsin/panopsin protein (OPN3). The mosquito OPN3 is a bistable photopigment that, upon activation, allows efficient and specific recruitment of the Gi/o signaling cascade (Koyanagi et al., 2013). Using a targeting-enhanced OPN3 (eOPN3) protein, we were able to suppress synaptic release in rodent hippocampal, cortical, thalamic, and mesencephalic neurons. In behaving mice, eOPN3 triggered robust pathway-specific behavioral effects. These findings suggest that eOPN3, and potentially other members of the bistable rhodopsin family, can be utilized as optogenetic tools for potent G protein-mediated modulation of the activity of presynaptic terminals with high spatiotemporal precision.
Results
Expression of naturally occurring and engineered Gi/o-coupled bistable rhodopsins in mammalian neurons
We reasoned that the efficient suppression of presynaptic function by the DREADD hM4Di (Figure 1A; Stachniak et al., 2014) arises from the stable binding of the engineered ligands of these receptors (Sternson and Roth, 2014) and the subsequent, stable Gi/o-mediated signal transduction. We therefore hypothesized that rhodopsins coupling to the Gi/o pathway could serve as potent presynaptic silencing tools provided that persistent activation of such a tool can be achieved with light. While vertebrate visual rhodopsins, which dissociate from their retinal chromophore upon illumination (Figure 1B, bRho), can in principle be used for presynaptic silencing (Li et al., 2005), it remains unclear whether these rhodopsins can provide sufficiently robust activation of the Gi/o pathway at presynaptic terminals to support potent and sustained effects. Recent work has identified several new members of the encephalopsin subfamily of ciliary opsins, which couple to the Gi/o pathway. Encephalopsins exist in a wide range of organisms, including the pufferfish teleost multitissue opsin 3a (PufTMT3a) and the mosquito opsin 3 (OPN3). These rhodopsins are intrinsically bistable, as they retain the covalent bond between the retinal chromophore and the protein moiety (Figure 1B) and display prolonged signal transduction following activation (Koyanagi et al., 2013). We tested several photoreceptors of this family for expression in mammalian neurons.
Generation and characterization of a targeting-enhanced OPN3
We previously showed that addition of an ER export signal (ER) along with a Golgi trafficking signal (ts) to the light-gated chloride channel GtACR2 (eGtACR2) (Mahn et al., 2018) leads to an increase in axonal membrane localization. Applying this modification to OPN3, yielding the enhanced OPN3-ts-mScarlet-ER (eOPN3), led to an increased overall expression and enhanced membrane targeting in cultured hippocampal neurons (Figures 1C and S2A). Green light pulses delivered to neurons co-expressing eOPN3 and G protein-coupled inwardly rectifying potassium (GIRK2-1) channels triggered robust GIRK-mediated currents (Figures 1D and S2B). Activation of GIRK currents was maximal at 512 nm (Figure 1E), consistent with previous characterization of light absorption by OPN3 protein (Koyanagi etal., 2013).
We confirmed that eOPN3 retained its capacity to specifically activate the Gi/o pathway using the GsX assay. Light-activation of GsX-expressing HEK cells yielded selective and strong activation of Gi-, Go-, and Gt-mediated signal transduction, but not of other G proteins (Figures 1F, S2C, and S3B). To rule out undesired consequences of heterologous rhodopsin overexpression, such as impaired cell health or light-independent effects on the physiological activity of expressing neurons, we examined the intrinsic excitability of cultured hippocampal neurons expressing eOPN3-mScarlet. Whole-cell patch-clamp recordings revealed no significant difference in intrinsic properties between neurons expressing eOPN3-mScarlet and neighboring, non-expressing neurons from the same neuronal culture (Figure S4). We therefore conclude that expression of eOPN3 is well tolerated in mammalian neurons and does not result in significant light-independent physiological changes in neuronal excitability.
Next, we tested eOPN3 in pyramidal neurons of organotypic hippocampal slice cultures, a preparation that preserves the anatomical and functional connectivity between neurons in the CA3 and CA1 regions. Light delivery directly to the somatodendritic region of cells co-expressing eOPN3-mScarlet with cytoplasmic mCerulean (Figure 1G) triggered long-lasting photocurrents reversing at –105.1 ± 0.9 mV (Figure S5A), close to the calculated K+ reversal potential of –102.5 mV, indicating activation of endogenous GIRK channels. This eOPN3-dependent K+-conductance led to a lower input resistance (Figure S5B), a decrease in electrically evoked action potential firing (Figure S5C), a slight hyperpolarization of the resting membrane potential (Figure S5D), and an increased rheobase (Figure S5E).
Activation of eOPN3 leads to suppression of neurotransmitter release
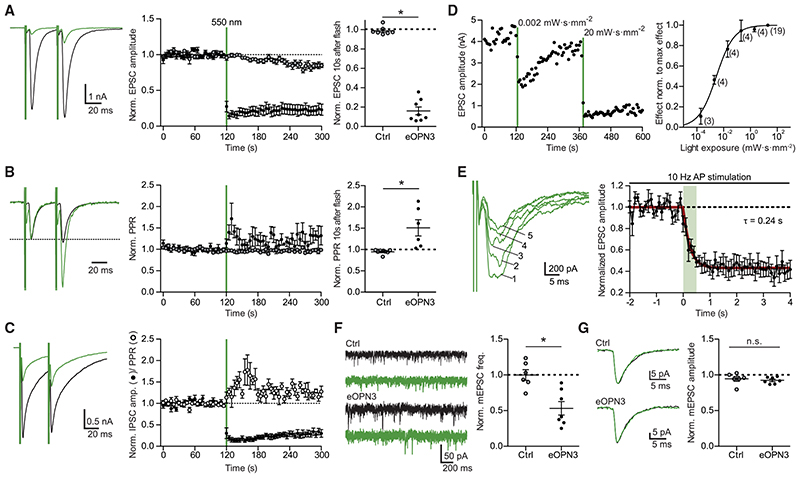
Our findings demonstrated that eOPN3 reliably couples to the Gi/o-signaling pathway and evokes GIRK-mediated currents. Axons and boutons of mCerulean-expressing CA3 pyramidal neurons in the stratum radiatum in CA1 of hippocampal slice cultures (Figure 1G) showed expression of eOPN3-mScarlet, indicating that the rhodopsin is present at presynaptic terminals as well. We therefore used the autaptic neuron preparation (Bekkers and Stevens, 1991) to ask whether activation of eOPN3 triggers changes in neurotransmission via G-protein activation, similar to the DREADD hM4Di (Figure S6). Light delivery to eOPN3-expressing autaptic neurons resulted in a robust and long-lasting decrease of excitatory postsynaptic currents (EPSCs; Figure 2A) and led to an increase in the paired-pulse ratio (Figure 2B), consistent with a decrease in release probability (Dobrunz et al., 1997). Light-triggered suppression of release was also found in autaptic hippocampal interneurons and was similarly accompanied by an increase in the paired-pulse ratio of the inhibitory postsynaptic currents (Figure 2C). To determine the light sensitivity of eOPN3, we varied the light exposure between 0.2 μW·s·mm−2 and 20 mW·s·mm−2 (Figure 2D). The half-maximal effect size was reached at 2.90 mW·s·mm−2, meaning that 1 s continuous illumination at 2.9 mW·mm−2 was sufficient to reach half maximal inhibition of synaptic vesicle release. The onset of eOPN3-mediated suppression of release was rapid, with a time constant (ton) of 0.24 s, and saturated after 1 s (Figure 2E). Furthermore, activation of eOPN3 significantly decreased the frequency of AP-independent miniature EPSCs (Figure 2F), but not their amplitude (Figure 2G). Together, these results are consistent with a presynaptic action of this photoreceptor on neurotransmission.
Figure 2. Light-induced inhibition of neurotransmitter release in autaptic hippocampal neurons expressing eOPN3.
(A) Typical autaptic EPSCs evoked by a pair of 1 ms depolarizing current injections (40 ms inter-stimulus interval, injected currents clipped for presentation) before (black) and after (green) illumination with 550 nm light (40 mW-mm−2, unless otherwise indicated). Traces are averages of 6 sweeps. A 500 ms light pulse caused sustained suppression of EPSCs in eOPN3-expressing neurons. EPSCs decreased to 16 ± 4% of baseline (n = 8), while EPSCs in control neurons were not affected by illumination (open circles, n = 7, p = 3·10−4 two-tailed Mann-Whitney test).
(B) Traces from (A) scaled to the amplitude of the first EPSC (dashed line). Illumination increased the paired-pulse ratio (EPSC2/EPSC1) in the eOPN3-positive neurons (n = 6) compared to controls (p = 1.2-10−3 unpaired, two-tailed Student’s t test).
(C) Amplitudes and PPR of evoked autaptic IPSCs in GABAergic neurons, compared to the pre-light baseline (IPSCs: n = 7; PPR: n = 5).
(D) Quantification of light exposure required for half maximal synaptic inhibition. Normalized effect size was fit as a sigmoidal dose-response curve (n is reported next to the measurement points, EC50 = 2.895 mW-s-mm−2).
(E) Time-course of the eOPN3 activation on EPSC amplitudes evoked by APs triggered at 10 Hz. Traces show five consecutive EPSCs of the train following the onset of a single 500 ms light pulse. EPSCs decreased with a time constant ton of 240 ms (n = 6).
(F) Representative traces of mEPSCs (left) and quantification (right). eOPN3 activation decreased mEPSC frequency to 53 ± 9% compared to baseline (n = 7), significantly different from controls (n = 6, p = 3·10−3, two-tailed Mann-Whitney test).
(G) Quantal EPSC amplitude in eOPN3-expressing and control neurons after illumination (p = 0.3 unpaired, two-tailed Student’s t test). Plots show individual data points and average (black) ± SEM.
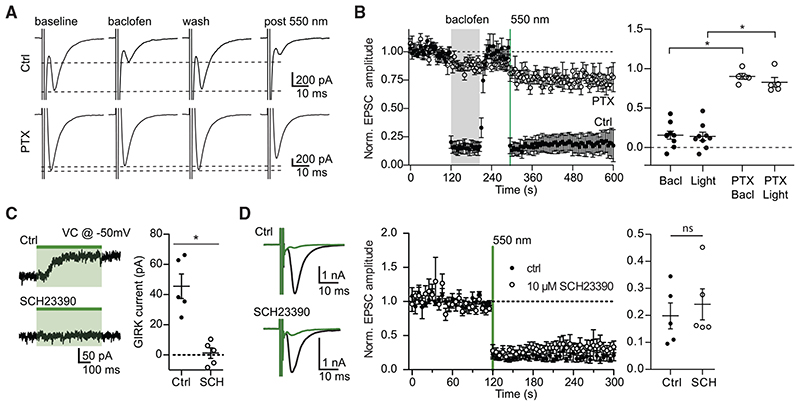
The effect of eOPN3 activation on synaptic transmission was similar to the effect of the GABAB agonist baclofen, a potent modulator of neurotransmitter release (Figures 3A and 3B; Rost et al., 2011; Scanziani et al., 1992), indicating that they both act through the Gi/o signaling pathway. Accordingly, preincubating the neurons with the Gα i/o subunit blocker pertussis toxin (PTX) blocked both the eOPN3- and the baclofen-mediated effects (Figures 3A and 3B), indicating that eOPN3 acts through the PTX-sensitive Gi/o protein signaling cascade. To examine whether the effects on synaptic transmission are dependent on GIRK channel activation, we applied SCH23390, which blocks GIRK channel currents (Kuzhikandathil and Oxford, 2002). Bath application of SCH23390 abolished the outward currents evoked by green light at the somatic compartment (Figure 3C) but had no detectable impact on the light-activated suppression of synaptic release in the same neurons (Figure 3D). These results suggest that the synaptic effects of eOPN3 are not mediated by blocking the propagation of APs, but rather by direct G protein-mediated effects at the presynaptic compartment (Wu and Saggau, 1994; Zurawski et al., 2019b).
Figure 3. The effect of eOPN3 on neurotransmitter release is sensitive to pharmacological inhibition of Gi/o-protein signaling but is not affected by a GIRK channel blocker.
(A) Action potential-evoked EPSCs in control neurons (upper row) were suppressed both by the GABABR agonist baclofen (30 μM) and by subsequent activation of eOPN3 with 550 nm light (500 ms, 40 mW·mm 2). In pertussis toxin (PTX)-treated neurons (20-26 h pre-treatment, 0.5 μg-mL ‘, bottom row), both baclofen and eOPN3 largely failed to suppress release.
(B) Averaged time-course of EPSCs recorded in neurons treated with PTX (open circles; n = 5) and neurons not treated with PTX (filled circles; n = 9; p = 3·10 4 Kruskal-Wallis test followed by Dunn’s multiple comparison tests: p < 0.05 for Bacl versus PTX Bacl, Light versus PTX Bacl and Light versus PTX Light).
(C) Illumination of eOPN3-expressing neurons evokes robust outward currents (45.5 ± 8.1 pA, n = 5), which are abolished in the presence of the GIRK channel blocker SCH23390 (10 μM, 1.2 ± 3.5 pA; n = 5; p = 1·10 3 unpaired, two-tailed Student’s t test).
(D) The extent and time-course of EPSC suppression by eOPN3 activation is not affected by the GIRK channel blocker SCH23390 (filled circles: ctrl recordings, n = 5; open circles: SCH23390, n = 5; p = 0.59 unpaired, two-tailed Student’s t test). Plots show individual data points and average ± SEM.
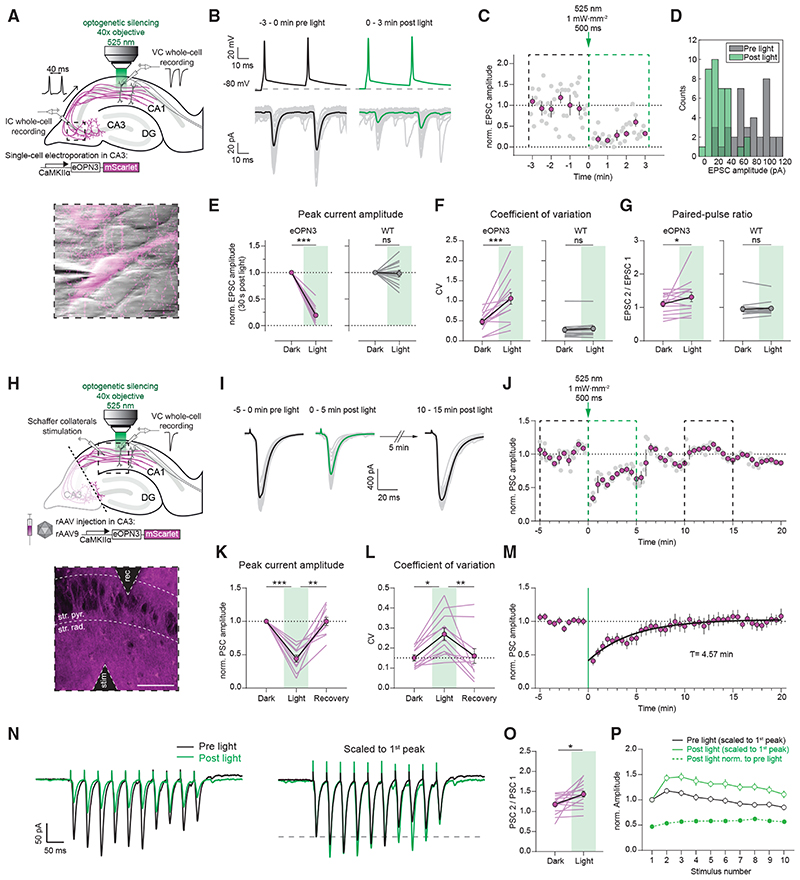
We next tested whether presynaptically expressed eOPN3 can be used to inhibit synaptic transmission in organotypic slices, where axon terminals can be locally illuminated independently of the neuronal soma (Figure 4A). In whole-cell recordings from pairs of CA3 and CA1 neurons, local illumination of the axonal terminals in CA1 induced a potent, long-lasting, and reversible reduction of the evoked EPSC amplitude (Figures 4B–4E and S7). Light application in CA1 neither induced AP failure nor GIRK-mediated hyperpolarization in the recorded presynaptic neurons (Figure S7), suggesting that activation of eOPN3 in the axonal compartment does not reduce somatic excitability. In accordance with a reduction in evoked release and thus a direct effect of eOPN3 on neurotransmitter release, we found that both the coefficient of variation (CV, Figure 4F) and the paired-pulse ratio (PPR, Figure 4G) increased following illumination in nearly all recorded pairs. The time until 50% EPSC recovery was 6.58 ± 1.78 min (Figures S7C-S7F). Synaptic transmission in non-expressing CA3-CA1 control pairs was unaffected by light stimulation (Figures 4E-4G). We therefore conclude that eOPN3 robustly activates the Gi/o pathway in neurons, leading to efficient suppression of presynaptic vesicle release that recovers spontaneously within minutes.
Figure 4. eOPN3 activation induces long-lasting, reversible inhibition of synaptic transmission at Schaffer collateral synapses.
(A) Schematic diagram of experimental setup for whole-cell paired-recordings in organotypic hippocampal slices (see STAR Methods for details). Inset: IR-scanning gradient contrast image overlaid with the fluorescence image of patch-clamped, eOPN3 expressing CA3 neuron. Scale bar, 20 mm.
(B) Top: representative voltage traces of electrically induced APs from an eOPN3 expressing CA3 neuron, before and after light delivery to the CA1 region (dashed line shows the resting membrane potential at the beginning of the experiment. Note that APs were still reliably evoked after light stimulation). Bottom: corresponding current traces from a postsynaptic CA1 neuron in response to the paired-pulse stimulation, before and after light delivery (gray: single trials, black and green: averaged trials).
(C) Time course of the normalized EPSCs peak amplitudes from the example shown in (B) (gray circles: single trials, magenta: means of 30 s time bins ± SEM).
(D) Histogram count of peak current amplitudes of the example shown in (B).
(E) Normalized EPSC amplitudes in the eOPN3 group (left) and wild-type (WT) control group (right) (eOPN3: 0.19 ± 0.04, n = 14 pairs from 14 slices, p = 1.10−4, Wilcoxon test; WT: 0.98 ± 0.06, n = 13 pairs from 13 slices, p = 0.5, Wilcoxon test).
(F) Coefficient of variation of EPSCs in the dark and after light application for the eOPN3 (left) and control group (right) (eOPN3 dark: 0.48 ± 0.06, eOPN3 light: 1.06 ± 0.15, n = 14 pairs from 14 slices, p = 4·10−4, paired t test; WT dark: 0.27 ± 0.06, WT light: 0.31 ± 0.06, n = 13 pairs from 13 slices, p = 0.11, Wilcoxon test).
(G) Paired-pulse ratio change in the dark compared to after light application for the eOPN3 (left) and control group (right) (eOPN3 dark: 1.11 ± 0.08, eOPN3 light: 1.32 ± 0.14, n = 14 pairs from 14 slices, p = 0.02, Wilcoxon test; WT dark: 0.95 ± 0.07, WT light: 0.97 ± 0.06, n = 13 pairs from 13 slices, p = 0.59, Wilcoxon test). Circles in (E-G): mean ± SEM.
(H) Schematic diagram of experimental setup for field stimulation (see STAR Methods for details). Inset: two-photon single-plane image of the CA1 region with the stimulating and recording electrodes. eOPN3-expressing axons (magenta) surround CA1 pyramidal neurons (dark shadows). Scale bar, 50 γm.
(I) Representative voltage traces (PSCs) before, immediately and 10 min after light (gray: single trials, black and green: average trials).
(J) Time course of the normalized PSC peak amplitudes from the example shown in (I). Dashed boxes indicate the time periods shown in (I) (gray circles: single trials, magenta: 30 s time bins ± SEM).
(K) Quantification of eOPN3 effect on PSC peak amplitudes (“Dark”: 5 min period before light; “Light”: maximal eOPN3 effect during first 30 s post light, 0.44 ± 0.05, p < 1·10−4; “Recovery”: 10−15 min period after light, 0.99 ± 0.06, p = 1.9·10−3; n = 11 slices, Friedman test with Dunn’s multiple comparison test).
(L) Quantification of the effect of eOPN3 activation on the coefficient of variation. “Light” refers to the 5 min post light application matching the duration of the two other conditions (“Dark”: 0.15 ± 0.02; “Light”: 0.27 ± 0.03, p = 0.02; “Recovery”: 0.16 ± 0.04, p = 8·5-10−3, n = 11 slices, Friedman test with Dunn’s multiple comparison test).
(M) Summary of all field stimulation experiments. Mono-exponential fit is shown in black.
(N) Left: representative voltage traces in response to a 10-pulse stimulus train (25 Hz). Traces are averages of5 sweeps each. Right: same traces as on the left, each scaled to its 1st PSC peak amplitude.
(O) Quantification of the PPR (PSC 2 / PSC 1 of the train), showing increased facilitation (Dark: 1.18 ± 0.05, Light: 1.43 ± 0.07, p = 0.01, n = 16 slices, Paired t test).
(P) Summary of all train stimulation experiments. Circles in (K-P): mean ± SEM.
To predict the effects of eOPN3-mediated inhibition in vivo, we virally transduced CA3 pyramidal cells in organotypic hippocampal slice cultures, emulating the most commonly used method for gene transfer in vivo (Figures 4H-4M). To avoid both recurrent polysynaptic activity of the CA3 network and contribution of somatic eOPN3 activation, CA3 axons were dissected from their somata at the boundary of CA3 to CA1 (Figure 4H). The PSC amplitude evoked by electrical stimulation of isolated Schaffer collateral axons was attenuated by 56 ± 5% following a single 500 ms light pulse to the terminal field in the CA1 (Figures 4I-4L) and recovered to baseline levels with a time constant of 4.57 min (95% CI: 4.19 to 4.97; R2: 0.90; Figure 4M). As before, the CV of synaptic responses increased in the 5 min following light stimulation, and eventually returned to baseline values. The lower efficacy of PSC amplitude reduction recorded in this experimental setup (Figure 4K) compared with the efficacy observed in paired recordings (81 ± 4%, Figure 4E) is likely due to the contribution of non-expressing axons to the PSCs evoked by field stimulation.
GPCRs may act at presynaptic terminals as canonical or non-canonical modulators of synaptic transmission (Zurawski et al., 2019a). It has been reported that canonical GPCR-mediated pre-synaptic inhibition decreases neurotransmission by altering the probability of vesicle release and changing the short-term plasticity profile of modulated synapses (Chalifoux and Carter, 2011), leading in some cases to suppression of initial release but facilitation of subsequent responses. To better characterize the efficacy of eOPN3-mediated synaptic inhibition during higher firing rates, we applied trains of 10 stimulations at 25 Hz (Figures 4N-4P). Postsynaptic responses in the dark showed facilitation for the initial pulses while displaying depression toward the end of the train. In accordance with our previous single-pulse field stimulation results, light activation of eOPN3 inhibited the first pulse by an almost identical amount (single pulse stimulation: 44 ± 5% versus train stimulation: 47 ± 5% of initial strength). Consistent with our paired recording data, eOPN3 increased the PPR of the initial two pulses (PSC2 / PSC1) and maintained facilitation throughout the train. Nonetheless, light activation of eOPN3 robustly suppressed the entire sequence of PSCs in the stimulus train, albeit to a slightly lower degree for all the consecutive pulses relative to the initial one (suppression of the 10th pulse was 43 ± 2% of the initial strength).
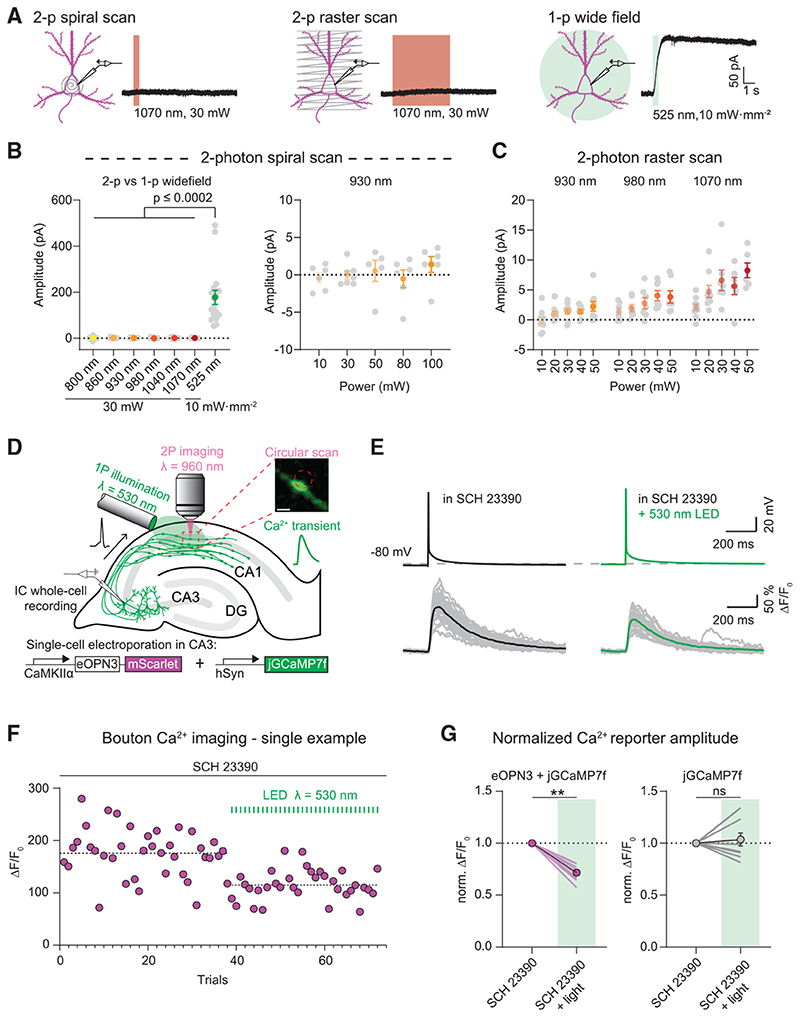
Integration of eOPN3-based manipulation with two-photon Ca2+ imaging
To assess whether eOPN3 can be combined with two-photon imaging, we tested eOPN3 activation by two-photon absorption. In CA3 pyramidal cells of organotypic hippocampal cultures expressing eOPN3 and GIRK2-1, we compared green light-evoked GIRK channel currents to fast spiral scanning on the soma or slow raster scanning across the somatodendritic compartment with a femtosecond-pulsed infrared laser at wavelengths ranging from 800 to 1070 nm and at intensities ranging from 10 to 100 mW (Figures 5A-5C). Spiral scans did not evoke any detectable photocurrents (Figure 5B). Only slow raster scans at wavelengths above 980 nm and intensities above 30 mW resulted in very small photocurrents of less than 10 pA on average (Figure 5C). In contrast, green-light activation of eOPN3 in the same cells evoked more than 20-fold larger photocurrents (Figure 5B). Thus, eOPN3 can be combined with two-photon imaging of blue-shifted sensors with minimal cross-activation.
Figure 5. eOPN3 two-photon activation properties and modulation of presynaptic voltage-gated Ca2+ channels.
(A) Two-photon (left, middle) versus single-photon (right) activation of eOPN3 in CA3 pyramidal neurons in organotypic hippocampal slice cultures expressing eOPN3-mScarlet and GIRK2-1. Somatic 500 Hz spiral scans (2 ms/spiral, 250 cycles, 500 ms total duration) or raster scans (FOV = 106*106 μm, 512x512 pixels, 1.8 ms/line, 5 frames, 4.6 s total duration) at 1.09 Hz over the somatodendritic compartment were used for two-photon activation characterization. Example voltage-clamp traces show photocurrents obtained by the different stimulation modalities in the same cell.
(B) Quantification of the photocurrents elicited by two-photon versus single-photon illumination. Left: GIRK-mediated currents in eOPN3 expressing neurons stimulated with two-photon spiral scanning at wavelengths from 800 nm to 1070 nm at 30 mW, or with full-field 525 nm light (Kruskal-Wallis test, Dunn’s multiple comparisons test). Right: Increasing laser intensity during spiral scans at930 nm did not result in significant photocurrent.
(C) Slower and longer raster scanning over a larger field of view resulted in minimal outward currents and was wavelength and laser-intensity dependent (Linear regression indicated positive slopes. Bonferroni-Holm corrected p values: wavelength: p = 6.1·10−4; laser power: 930 nm: p = 0.01; 980 nm: p = 7.2·10−3; 1070 nm: p = 1.2·10−3).
(D) Schematic diagram of presynaptic Ca2+ imaging experiments (see STAR Methods for details). Inset shows a single-plane jGCaMP7f image of an en passant bouton and the circular imaginglaser scanning path (red dashed circle, scale bar, 1 μm). A fiber-coupled LED was used to locally activate eOPN3 in CA1 the presence of the GIRK channel blocker SCH 23390.
(E) Top: representative voltage traces of electrically evoked APs in a transfected CA3 pyramidal neuron in the dark and after a green light pulse (dashed line shows the resting membrane potential at the beginning of the experiment). Bottom: corresponding Ca2+ responses from a presynaptic bouton. Single trials are shown in gray; black and green traces represent the averaged responses before and after light, respectively.
(F) Peak jGCaMP7f transients in the dark and after green light pulses in a single experiment, indicating a light-dependent decrease in presynaptic Ca2+ influx. Dashed lines show the average for the two conditions.
(G) Quantification of normalized eOPN3-jGCaMP7f transients (left) (SCH 23390 + light = 0.72 ± 0.026, p = 2.10_3, Wilcoxon-test, n = 10 slices) and jGCaMP7f alone (right) (SCH 23390 + light = 1.04 ± 0.06, p = 0.89, paired t test, n = 10 slices). Plots show individual data points (lines), and average (circles) ± SEM.
Based on this characterization, we used two-photon imaging to determine whether eOPN3 alters Ca2+ influx through presynaptic voltage-gated Ca2+ channels, as shown for different neuromodulators (Wu and Saggau, 1994; Ikeda, 1996; Herlitze et al., 1996; Chalifoux and Carter, 2011; Burke etal., 2018). Gi-coupled GPCRs can suppress neurotransmitter release via Gβγ-mediated inhibition of voltage-gated Ca2+ channels (Herlitze et al., 1996; Kajikawa etal., 2001), possibly by delaying the time of first opening or by shifting the voltage-dependency of channel activation (Bean, 1989). We therefore tested whether eOPN3 activation in presynaptic terminals reduces AP-evoked Ca2+ influx. We evoked single APs in CA3 cells co-expressing eOPN3 and jGCaMP7f (Dana et al., 2019) while imaging the corresponding presynaptic Ca2+ transients in CA3 cell axonal boutons in CA1 stratum radiatum (Figures 5D and 5E). The GIRK channel blocker SCH23390 was added to exclude potentially confounding GIRK channel-mediated hyperpolarization effects. Green light pulses locally applied to the CA1 region before each trial significantly reduced presynaptic Ca2+ influx in a GIRK-independent manner (Figures 5F-5G), indicating that eOPN3 acts directly at voltagedependent Ca2+ channels at presynaptic terminals similar to native Gi-coupled receptors.
In vivo characterization of eOPN3-mediated terminal inhibition
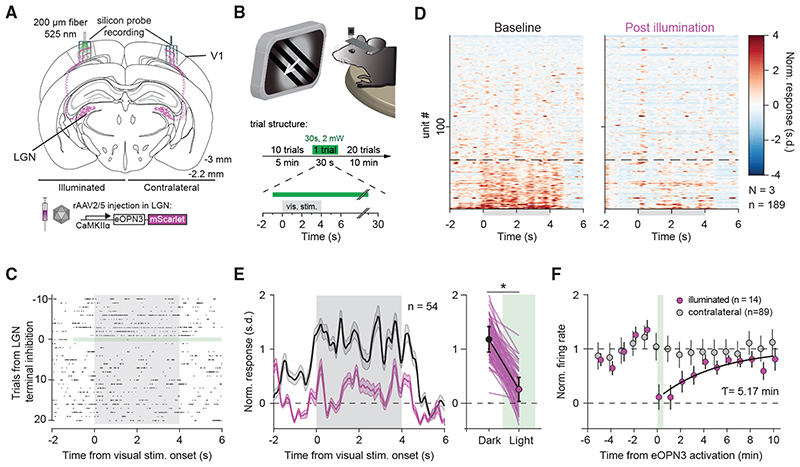
Next, we examined the efficacy and kinetics of eOPN3-mediated presynaptic silencing using in vivo electrophysiology. We chose to modulate the visual thalamocortical pathway, since the visual responses of V1 neurons depend on input from the lateral geniculate nucleus of the thalamus (LGN), which constitutes the main feed-forward projection from the retina to V1 (Niell and Stryker, 2008; Froudarakis etal., 2019). Using multi-shank silicon probes, we recorded bilaterally from V1 in mice expressing eOPN3 in the LGN (Figure 6A). Visual stimulation (4 s compound visual stimulus every 30 s) led to reliable evoked responses in V1 (Figures 6C and 6D left). A subset of units showed an increase in their average firing rates during visual stimulus presentation (Figure 6D). After 10 trials of visual stimulus presentation, we activated eOPN3 in LGN terminals unilaterally by 30 s continuous illumination (2 mW at the fiber tip) directed at V1. eOPN3 activation resulted in a reduced impact of visual stimulation on evoked network activity in V1 (Figures 6C and 6D), with responsive units reducing their response amplitude (Figure 6E). In units that showed a strong suppression of visually evoked responses (more than 50% during eOPN3 activation; 14 of 54 units), the average response amplitude recovered with a time constant of 5.17 min (95% CI: 1.12 to 7.20 min; R2: 0.82; Figure 6F). By contrast, units recorded simultaneously at the contralateral (non-illuminated) side did not show a change in their visual stimulus presentation response after eOPN3 activation on the ipsilateral hemisphere (Figure 6F), demonstrating the spatial specificity of the manipulation.
Figure 6. eOPN3 mediated suppression of thalamocortical inputs in awake head-fixed mice.
(A) Schematic diagram of the investigated circuit. Lateral geniculate nucleus (LGN) neurons were bilaterally transduced with eOPN3. Acute silicon probe recordings were performed bilaterally in primary visual cortex (V1) before and after unilateral illumination of LGN terminals in V1.
(B) During recordings, head-fixed mice were presented with a compound drifting grating stimulus (4 s duration) every 30 s for 21 trials (top). Ten baseline trials were followed by a single trial paired with 30 s of light delivery (525 nm at ~2 mW from a 200 mm, 0.5 NA optical fiber) to V1, and 20 post-light trials.
(C) Raster plot of a representative V1 unit with reduced firing rate induced by eOPN3 activation.
(D) Heat plot of the population response to visual stimulus presentation of all recorded units (189 units from 3 mice) on the hemisphere of eOPN3 activation before (left) and after (right) eOPN3 activation. Units were sorted by their response magnitude to visual stimulus presentation during baseline condition. Units below the dashed line (n = 54) show a positive average response during the 4 s visual stimulus presentation.
(E) Left: Average peristimulus time histogram of the visual stimulus responsive units (below dashed line in D). Each unit’s activity was normalized to the average firing rate in the 15 s prior to stimulus presentation during the two trials before eOPN3 activation. Right: Quantification of the average response during 4 s visual stimulus presentation in the two trials before (Dark) and first two trials after eOPN3 activation onset (Light). Dark: 1.17 ± 0.23, Light: 0.25 ± 0.22, p < 1 o 10_3, Wilcoxon test, n = 54 units. Plot shows individual units (lines), and population average (circles) ± SEM.
(F) Kinetics of the recovery of visual stimulus response amplitude for units that showed a reduction >50% in their visual stimulus response (magenta), fitted with a mono-exponential function (black line). Units recorded simultaneously from the contralateral hemisphere (gray) did not change their response following ipsilateral eOPN3 activation. During the baseline and post light period, the plot shows the averages of two consecutive trials (circles) ± SEM.
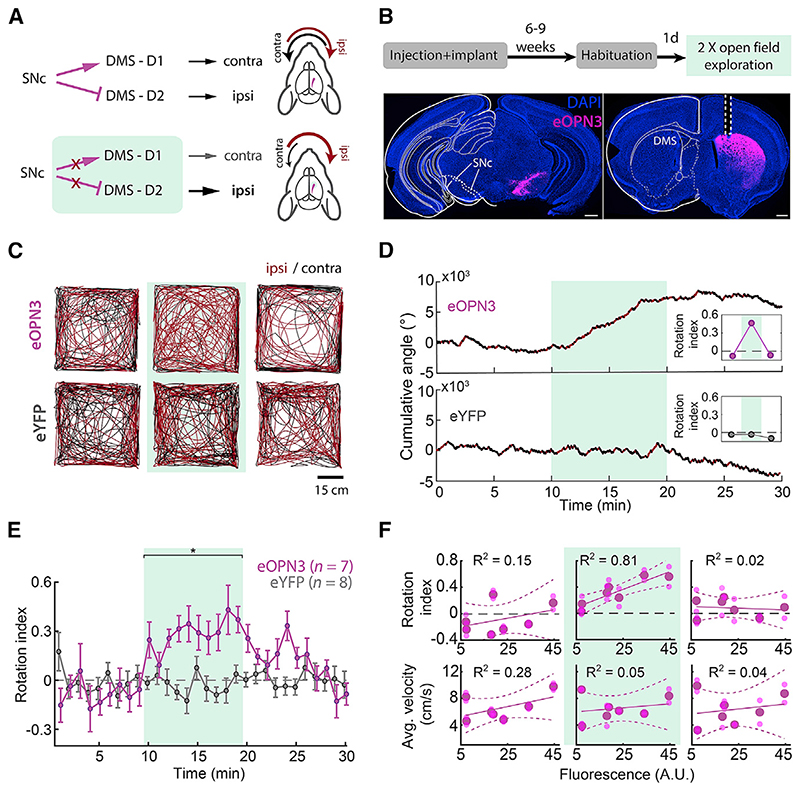
To examine the efficacy and kinetics of eOPN3-mediated presynaptic silencing in vivo on the behavioral level, we used eOPN3 to inhibit dopaminergic (DA) input to the dorsomedial striatum (DMS) of mice during free locomotion. Previous work has demonstrated the important role of nigrostriatal DA projections in the control of animal locomotion (Alcaro et al., 2007; Kravitz et al., 2010; Grealish et al., 2010; Tecuapetla et al., 2014; Barter et al., 2015; Borgkvist et al., 2015; da Silva et al., 2018). Briefly, striatal D1-expressing medium spiny neurons (D1-MSNs) facilitate motion upon selective, bilateral activation and induce a contralateral rotation upon unilateral stimulation. Conversely, D2-expressing MSNs (D2-MSNs) decrease motion and, upon unilateral stimulation, induce ipsilateral rotation. While D1 and D2 neurons drive motion in opposite directions, their common substantia nigra pars compacta (SNc) dopaminergic input stimulate D1-MSNs while inhibiting D2-MSNs. Overall, these studies suggest that unilateral inhibition of SNc DA projections would introduce an ipsiversive bias in free locomotion (Figure 7A). We thus expressed an eOPN3- or an eYFP-expressing control vector unilaterally in SNc DA neurons and implanted an optical fiber above the ipsilateral DMS to allow illumination of nigrostriatal DA projections (Figure 7B). Activation of eOPN3 in DA terminals (500 ms light pulses at 0.1 Hz, 540 nm, 10 mW at the fiber tip) triggered an ipsiversive bias in locomotion (Figures 7C and 7D). The rotational preference was not observed during the baseline period, became evident within the first minute following light onset, and recovered within <10 min of the last light pulse (Figure 7E), in line with the recovery kinetics of eOPN3 observed in our experiments in vitro and in vivo (Figures 4M, 6F, and S7C-S7F). Control eYFP-expressing mice did not show such side bias or light-induced equivalent dynamics (Figures 7C-7E). Apart from their strong side preference, eOPN3 mice did not differ from control mice in distance traveled (p = 0.54, Kruskal-Wallis test), center entries (p = 0.99, Kruskal-Wallis test), or time in center (p = 0.69, Kruskal-Wallis test). The magnitude of the observed behavioral effect of eOPN3 activation, quantified as the rotation index (Figure 7D, insets; see STAR Methods), was positively correlated with expression levels across individual mice (p = 6.1·10−3, R2 = 0.81) during the light activation period, but not before light delivery or after its termination (Figure 7F). No significant correlation was found with the average velocity before, during, or after eOPN3 activation (Figure 7F). Finally, one week after the initial test, we repeated the test using the same parameters. We found a high correlation in the light evoked rotational bias between the first and second trial in each mouse (Pearson’s correlation coefficient: 0.8147; p = 0.0256). Taken together, our results demonstrate that eOPN3 can be used for synaptic terminal inhibition in behaving animals, with high light-sensitivity, precisely timed onset, and behaviorally relevant recovery time.
Figure 7. eOPN3-mediated suppression of dopaminergic projections from the substantia nigra to the dorsomedial striatum leads to ipsiversive bias during free locomotion.
(A) Schematic diagram of the experimental setup and hypothesis. Unilateral expression of eOPN3 in SNc dopaminergic neurons and light-mediated suppression of their striatal projections would induce an ipsiversive side bias during free locomotion.
(B) Top: experimental timeline. Bottom: Representative images of neurons expressing eOPN3-mScarlet in the SNc (left) and their striatal projections (right) in DAPI-stained brain sections. Scale bars, 500 mm.
(C) Locomotion trajectories of representative eOPN3 (top) and eYFP (bottom) mice, over successive 10-min periods: (left to right) before, during and after light delivery (540 nm, 500 ms pulses at 0.1 Hz, 10 mW from the fiber tip), together covering continuous 30 min sessions. Red and black color code trajectory segments where the mice showed ipsilateral or contralateral angle gain, respectively.
(D) Representative cumulative angle traces of individual eOPN3-expressing (top) and eYFP-expressing (bottom) mice, over 30 min of free locomotion in an open field arena. Red and black colors depict ipsilateral or contralateral segments, respectively. Green shaded region marks the light delivery period.
(E) The rotation index (mean ± SEM), calculated as the difference between cumulative ipsilateral and contralateral rotations, divided by their sum, over
1-min bins for eOPN3-expressing mice (magenta, n = 7) and eYFP controls (gray, n = 8). Green shaded region marks the light delivery period, where eOPN3 demonstrate significant ipsiversive bias (p = 1.3·10 3 Kruskal-Wallis test followed by Bonferroni-Holm corrected pairwise comparisons using Wilcoxon rank sum tests. Baseline: ctrl versus eOPN3 p = 1; light: ctrl versus eOPN3 p = 1.9·10 3; post light: ctrl versus eOPN3 p = 0.09).
(F) Top: rotation index, calculated for individual mice before (left), during (middle), and after (right) light-induced activation of eOPN3, plotted against eOPN3 expression levels measured at the DMS projections (symbols). Solid and dashed lines are linear regression fit with 95% confidence intervals, respectively. Bottom: average velocity of individual mice, plotted against expression levels in the same manner shown above. R2 values are indicated separately for each plot.
Discussion
Optogenetic silencing is a powerful tool for functionally dissecting neuronal circuits and understanding the contribution of defined neuronal populations to behavioral processes. However, silencing of long-range axonal projections has posed a formidable challenge. Our results demonstrate that a mosquito homolog of encephalopsin (OPN3) can selectively recruit Gi/o signaling in mammalian neurons. Optimization of this rhodopsin (yielding eOPN3) led to enhanced membrane targeting and improved expression in long-range axons. Activation of eOPN3 in four different neuronal preparations (autaptic hippocampal neurons, organotypic hippocampal slices, thalamocortical afferents, and nigrostriatal DA fibers) led to effects that are consistent with robust suppression of neurotransmitter release. In autaptic neurons, eOPN3 activation led to an inhibitory effect that was similar in its magnitude to the effect of activating endogenous GABAB receptors and was blocked by pertussis toxin, consistent with Gi/o-mediated inhibition. One potential caveat to the use of Gi/ o-mediated inhibition for the manipulation of neuronal and synaptic activity is that the biochemical signaling pathways and the effector proteins might differ among cell types and subcellular compartments. Furthermore, Gi/o-mediated inhibition is known to be activity-dependent to some extent (Brenowitz et al., 1998), and its efficacy might be dependent on the initial firing patterns and short-term synaptic plasticity features of the targeted neurons. We therefore recommend that eOPN3 effects are rigorously characterized using electrophysiology before this tool is applied in a behavioral setting.
Although we detected eOPN3-mediated GIRK currents, the effect of eOPN3 activation on the intrinsic excitability of expressing neurons was relatively weak. This suggests that activation of eOPN3 in the somatodendritic compartment induces a less efficient inhibition of neuronal spiking compared to other K+ channel-mediated optogenetic silencing approaches (Bernal Sierra et al., 2018; Beck et al., 2018). In contrast, silencing of synaptic transmission with eOPN3 was highly efficient and independent of GIRK channel activity, suggesting that eOPN3-mediated synaptic inhibition occurs through direct activity on the highly conserved presynaptic release apparatus and on Ca2+ channel function (Dittman and Regehr, 1996; Kajikawa et al., 2001; Sakaba and Neher, 2003; Zurawski et al., 2019b). This is consistent with our observation of GIRK-channel-independent suppression of spike-evoked Ca2+ transients after eOPN3 activation. Thus, if locally activated at synaptic terminals, eOPN3 is a robust and broadly applicable optogenetic tool for inhibition of synaptic neurotransmission, similar to the DREADD receptor hM4Di, which has been successfully used for presynaptic silencing in a variety of neuronal cell types and systems (Stachniak et al., 2014; Evans et al., 2018; Malvaez et al., 2019).
The effects of GPCRs on presynaptic neurotransmitter release have been partially attributed to G-protein modulation of presynaptic Ca2+influx (Herlitze et al., 1996). Meanwhile, non-canonical presynaptic GPCR modulators have been shown to decrease the vesicle release probability without a concomitant change in short term plasticity, through Ca2+-dependent and independent mechanisms (Hamid et al., 2014; Burke et al., 2018). Our paired-pulse facilitation results suggest that eOPN3 acts as a canonical presynaptic GPCR modulator, suppressing the initial synaptic response more strongly than it does the consecutive pulses (Figures 4N-4P). This could be due to presynaptic Ca2+ accumulation (Jackman and Regehr, 2017) and a depolarization-triggered relief of the G-protein interaction with voltage-gated Ca2+ channels (Currie, 2010). Thus, eOPN3 activation biases short-term synaptic plasticity toward short-term facilitation.
We have previously shown that current approaches utilizing ion pumps for vesicle release inhibition are not suitable for suppressing presynaptic release for extended time periods (Mahn et al., 2016; Wiegert et al., 2017a; Lafferty and Britt, 2020). Although bistable rhodopsins such as eOPN3 cannot replace ion-pumping type-I rhodopsins in the sub-second range, eOPN3 can be used for experiments that require modulation in the range of minutes to hours. Foreven longer inhibition periods, tools such as the photoactivatable botulinum neurotoxin are likely also suitable (Liu et al., 2019). Silencing synaptic transmission using hM4Di with local agonist infusion at the terminal field (Stachniak et al., 2014) should in principle allow for similar efficiency compared to eOPN3. However, eOPN3 has the advantage of more precise temporal control and reduced problems with agonist microinfusion such as potential off-site effects due to leakage to the cerebrospinal fluid. The time course of recovery after eOPN3 activation that we observed in vitro (Figures 4M and S7C-S7F) and in vivo (Figures 6F and 7E) is consistent across the four preparations and three cell types used. However, we would like to emphasize that the exact time constants will depend on cell type and expression level and should ideally be determined experimentally in every preparation.
Our in vitro experiments showed that eOPN3 is highly light sensitive (Figure 2D), likely due to its recovery kinetics. By relaxing the limitations imposed by tissue heating in vivo, eOPN3 allows for optical access to large brain volumes, a major constraint of type-I rhodopsins such as NpHR and Arch (Stujenske et al., 2015; Owen et al., 2019). In our single-photon excitation experiments, we used light exposures above 0.5 mW·s·mm−2, leading to complete eOPN3 activation. This approach was aimed at achieving the maximal effect, making the effect of light exposures comparable as long as they are beyond saturation while not leading to tissue heating. However, for experiments where subsets of postsynaptic targets need to be specifically inhibited, light exposure should be minimized to prevent inadvertent eOPN3 activation in neighboring areas. Furthermore, the high light sensitivity of eOPN3 necessitates working in light shielded conditions when using in vitro preparations ortransparent organisms. For behavioral experiments, we used single light pulses spaced at 0.1 Hz. The exact irradiance and duty cycle in such experiments should be calibrated based on the volume of the targeted terminal field and the distance from other projections and somata that should remain unaffected.
We also show that eOPN3 has a small two-photon absorption cross section at the typical wavelength ranges used for two-photon Ca2+ indicator imaging (Figure 5B). Even continuous raster scanning on the soma and proximal dendrites of neurons expressing eOPN3 and GIRK2-1 only led to a mild somatic hyperpolarization, indicating that eOPN3 is not effectively activated. A potential use case would be to image the activity of a local network before and during inhibition of a given afferent via eOPN3 activation. Here, one potential concern is that the slow recovery kinetics of eOPN3 might lead to an accumulation of Gi/o signaling overtime, even with the low two-photon absorption properties of eOPN3. This certainly warrants careful controls, but we do not expect this to represent a major constraint in classical raster scanning two-photon imaging. Typical experiments in which network activity is continuously imaged typically involve a larger field of view (1x1 mm versus 106x106 μm used here). This effectively reduces the irradiance per illuminated presynaptic terminal. Second, whatever activation of eOPN3 molecules does take place, it will be limited to the imaging plane, meaning that out-of-focus eOPN3 molecules will not be affected. In contrast, combination of eOPN3-mediated inhibition with scanless two-photon approaches, such as temporal focusing or holographic imaging, might lead to an increased crosstalk. Although we did not observe such an effect in our experiments, one should also take into account that eOPN3 can potentially be activated by the emission light of the imaged indicator. In both types of experiments, the imaging parameters should be optimized to minimize such cross-activation.
To the best of our knowledge, this study along with the adjoining manuscript from the Bruchas and Gereau labs using the lamprey parapinopsin (PPO; Copits et al., 2021) are the first to describe an optogenetic application of bistable nonvisual rhodopsins for efficient light-gated silencing of synaptic transmission. The unique spectral features of eOPN3 and PPO, particularly in their two-photon cross sections, will potentially allow them to be utilized in concert for dual-channel optogenetic control of intracellular signaling. These two rhodopsins are part of a widespread family of non-visual rhodopsins, some of which have been shown to similarly couple to Gi/o signaling when expressed heterologously (Koyanagi and Terakita, 2014). Thus, additional members of this rhodopsin family could potentially serve as effective tools for controlling the activity of presynaptic terminals and might be further engineered for spectral tuning or G-protein coupling specificity. Further work is needed to examine the functional properties of these little-explored photoreceptors and adapt them for optogenetic applications. Nevertheless, eOPN3-mediated silencing of transmitter release constitutes a much-needed experimental approach for light-triggered suppression of neuronal communication in the target area of long-range projections, and we expect its application will facilitate research in a variety of neurobiological studies.
Star Methods
Table 1. Key Resources Table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| rAAV2/1&2.CamKIIa(0.4).OPN3-mScarlet | This paper | N/A |
| rAAV2/1&2.CamKIIa.eYFP.WPRE | This paper | N/A |
| rAAV2/1&2.CamKIIa(0.4).eOPN3-mScarlet | This paper | http://www.addgene.org/125712/ |
| rAAV2/1&2.hSyn.SIO-eOPN3-mScarlet | This paper | http://www.addgene.org/125713/ |
| Chemicals, peptides, and recombinant proteins | ||
| (R)-baclofen | Tocris | Cat#0796 |
| Clozapine-N-Oxide | Enzo Life Science | Cat#-BML-NS105 |
| CPPene | Tocris | Cat#1265 |
| Gabazine | Tocris | Cat#1262 |
| NBQX | Tocris | Cat#1044 |
| Pertussis toxin | Sigma-Aldrich | Cat#516560 |
| Picrotoxin | Tocris | Cat#1128 |
| SCH23390 | Tocris | Cat#0925 |
| Critical commercial assays | ||
| GloSensor cAMP Assay | Promega | Cat#E1171 |
| Experimental models: Cell lines | ||
| HEK293T | Sigma-Aldrich | Cat#12022001 RRID:CVCL_0063 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6JRccHsd | Envig | Cat#043 |
| Mouse: C57BL/6NHsd | Envig | Cat#044 |
| Mouse: DAT-IRES-Cre | The Jackson Laboratory | Strain #006660 |
| Rattus norvegicus: Sprague-Dawley | Envig | Cat#002 |
| Rattus norvegicus: Wistar | Charles River, bred in the animal facility, UKE Hamburg | Cat#003 |
| Recombinant DNA | ||
| pAAV-CaM KIIa(0.4)-OPN3-mScarlet | This Paper | N/A |
| pAAV-CaM KIIa(0.4)-PufTMT3a-mScarlet | This Paper | N/A |
| pAAV-CaMKIIa(0.4)-OPN3-M4-mScarlet | This Paper | N/A |
| pAAV-CaM KIIa(0.4)-PufTMT3a-M4-mScarlet | This Paper | N/A |
| pAAV-CamKIIa-eYFP | Karl Deisseroth | RRID:Addgene_105622; www.addgene.org/105622 |
| pcDNA3.1-GIRK2-1 | Eitan Reuveny | GenBank: NM_001025584.2 |
| pcDNA3.1-mCerulean | Dave Piston; Rizzo et al.,2004 | RRID:Addgene_15214; www.addgene.org/15214/ |
| pAAV-CaMKIIa(0.4)-eOPN3-mScarlet | This Paper | RRID:Addgene_125712; www.addgene.org/125712/ |
| pAAV-hSyn-SIO-eOPN3-mScarlet | This Paper | RRID:Addgene_125713; www.addgene.org/125713/ |
| Software and algorithms | ||
| Fiji | Schindelin et al., 2012 | RRID:SCR_002285; http://imagej.net/Fiji |
| MATLAB 2018b | Mathworks | RRID:SCR_001622; www.mathworks.com |
| Prism 8.2.1 | Graphpad | RRID:SCR_002798; https://www.graphpad.com |
| RStudio Desktop | RStudi | RRID:SCR_000432; https://www.rstudio.com |
| Ephus | Suter et al., 2010 | https://doi.org/10.3389/fncir.2010.00100 |
| WaveSurfer | Janelia | https://wavesurfer.janelia.org |
| ScanImage | Vidrio Technologies | RRID:SCR_014307; v2017b http://www.scanimage.org/ |
| EthoVision XT 11.5 | Noldus | RRID:SCR_000441; https://www.noldus.com/ethovision-xt |
| DeepLabCut | Mathis et al., 2018 | www.mackenziemathislab.org/deeplabcut |
Resource Availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ofer Yizhar (ofer.yizhar@weizmann.ac.il).
Experimental Model and Subject Details
Animals
Animal experiments were carried out according to the guidelines stated in directive 2010/63/EU of the European Parliament on the protection of animals used for scientific purposes. Animal experiments at the Weizmann Institute were approved by the Weizmann Institute Institutional Animal Care and Use Committee (IACUC); experiments in Berlin were approved by local authorities in Berlin and the animal welfare committee of the Charité – Universitätsmedizin Berlin, Germany. Experiments in Hamburg were done in accordance with the guidelines of local authorities and Directive 2010/63/EU. Experiments in Basel were done in accordance with institutional guidelines at the Friedrich Miescher Institute for Biomedical Research and were approved by the Veterinary Department of the Canton of Basel-Stadt. For in vivo electrophysiological recordings male mice (C57BL/6JRccHsd; Envigo, Cat#043) at 8-9 weeks old were used. Mean weight at the day of surgery was 23.8 g. Experimental mice were individually housed. All mice were assigned to the same experimental group. For in vivo behavioral experiments male and female mice (DAT-IRES-Cre; The Jackson Laboratory, Strain #006660) were used. Mice were housed in single gender groups, 2-4 littermates/cage. Littermates from single cages underwent surgery on the same day and were assigned to the eOPN3 or control group such that cages always included mixed groups. The control group included 8 mice (3 malesand 5 females). Age at day of surgery was 9-14 weeks (mean = 12 weeks). Mean weight at the day of surgery was 19.6 g for females and 24.6 g for males. The eOPN3 group included 7 mice (3 males and 4 females). Age at day of surgery was 9-14 weeks (mean = 11.9 weeks). Mean weight at the day of surgery was 19.2 g for females and 24.75 g for males. The room temperature was set at 22°C (±2°C) and room humidity was set at 55% (±10%). Mice were kept in a 12-h light/dark cycle with access to food and water ad libitum. Mice were checked daily by animal caretakers.
Cell lines
HEK293T cells (RRID:CVCL_0063) were incubated at 37°C (5% CO2) in DMEM containing 4500 mg/L glucose, L-glutamine, (Sigma) with penicillin (100 U/mL), streptomycin (100 mg/mL), and 10% FBS. The cell line is authenticated by the European collection of authenticated cell cultures. Sex of these cells is female, and the cell line is derived from fetal human tissue.
Primary cell cultures
Primary cultured hippocampal neurons were prepared from post-natal day 0 Sprague-Dawley rat pups (Envigo, Cat#002) of either sex.
Autaptic cultures of primary hippocampal neurons on glia cell micro-islands were prepared from newborn mice (C57BL/6NHsd; Envigo, Cat#044) of either sex.
Organotypic hippocampal slices were prepared from post-natal day 5-7 Wistar rats (Charles River Cat#003 bred in the animal facility, UKE Hamburg) of either sex.
Method Details
Molecular cloning of bistable rhodopsin constructs
The genes encoding mScarlet (Bindels et al., 2017), OPN3, PufTMT3a, OPN3-M4 and PufTMT3a-M4 were synthesized using the Twist gene synthesis service (Twist Bioscience, USA). The Rho1D4 sequence (TETSQVAPA) was added at the C terminus of all rhodopsins. All genes were subcloned into pAAV vectors under the CamKIIa promoter and in-frame with mScarlet at the C terminus. The eOPN3 plasmid was generated by adding the Kir2.1 membrane trafficking signal (KSRITSEGEYIPLDQIDINV) between the OPN3 and the mScarlet coding sequences and the Kir2.1 ER export signal (FCYENEV) following the C terminus of mScarlet. eOPN3 constructs and viruses are available from Addgene: https://www.addgene.org/Ofer_Yizhar/
Production of recombinant AAV vectors
HEK293T cells were seeded at 25%-35% confluence. The cells were transfected 24 h later with plasmids encoding AAV rep, cap of AAV1 and AAV2 and a vector plasmid for the rAAV cassette expressing the relevant DNA using the PEI method (Grimm et al., 2003). Cells and medium were harvested 72 h after transfection, pelleted by centrifugation (300 g), resuspended in lysis solution ([mM]: 150 NaCl, 50 Tris-HCl; pH 8.5 with NaOH) and lysed by three freeze-thaw cycles. The crude lysate was treated with 250 U benzonase (Sigma) per 1 mL of lysate at 37°C for 1.5 h to degrade genomic and unpackaged AAV DNA before centrifugation at 3,000 g for 15 min to pellet cell debris. The virus particles in the supernatant (crude virus) were purified using heparin-agarose columns, eluted with soluble heparin, washed with phosphate buffered saline (PBS) and concentrated by Amicon columns. Viral suspension was ali-quoted and stored at −80°C. Viral titers were measured using real-time PCR. In experiments that compared between different constructs, viral titers were matched by dilution to the lowest concentration. AAV vectors used for neuronal culture transduction were added 4 days after cell seeding. Recordings were carried out between 4-20 days after viral transduction. The following viral vectors were used in this study:
AAV2/1&2.CamKIIa(0.4).OPN3-mScarlet, AAV2/1&2.CamKIIa(0.4).eOPN3-mScarlet, AAV2/5.CamKIIa(0.4).eOPN3-mScarlet, AAV2/ 9.CamKIIa(0.4).eOPN3-mScarlet AAV2/1&2.CamKIIa.eYFP.WPRE, AAV2/1&2.hSyn.SIO-eOPN3-mScarletAAV2/1&2.EF1a.DIO.eYFP. WPRE.
Primary hippocampal neuron culture
Primary cultured hippocampal neurons were prepared from male and female P0 Sprague-Dawley rat pups (Envigo). CA1 and CA3 were isolated, digested with 0.4 mg ml−1 papain (Worthington), and plated into a 24-well plate at a density of 65,000 cells per well, onto glass coverslips pre-coated with 1:30 Matrigel (Corning). Cultured neurons were maintained in a 5% CO2 humidified incubator in Neurobasal-A medium (Invitrogen) containing 1.25% fetal bovine serum (FBS, Biological Industries), 4% B-27 supplement (GIBCO), and 2 mM Glutamax (GIBCO). To inhibit glial overgrowth, 200 μM fluorodeoxyuridine (Sigma) was added after 4 days of in vitro culture (DIV).
Neurons were transfected using the Ca2+ phosphate method (Graham and van der Eb, 1973). Briefly, the medium of primary hippocampal neurons cultured in a 24 well plate was collected and replaced with 400 μl serum-free modified eagle medium (MEM, Thermo Fisher Scientific). 30 μl transfection mix (2 μg plasmid DNA and 250 μM CaCl2 in HBS at pH 7.05) were added per well. After 1 h incubation the cells were washed 2 times with MEM and the medium was changed back to the collected original medium. Cultured neurons were used between 14 − 17 DIV for experiments. The following plasmids were used in this study: pAAV-CamKIIa(0.4)-OPN3-mScarlet, pAAV-CamKIIa(0.4)-eOPN3-mScarlet, pAAV-CamKIIa(0.4)-PufTMT3a-mScarlet, pAAV-CamKIIa(0.4)-OPN3-M4-mScarlet, pAAV-CamKIIa (0.4)PufTMT3a-M4-mScarlet, pAAV-CamKIIa(0.4)-eYFP. The pcDNA3.1-GIRK2-1 plasmid was a gift from Eitan Reuveny.
Autaptic cultures of primary hippocampal neurons on glia cell micro-islands were prepared from newborn mice (C57BL/6NHsd; Envigo, Cat#044) of either sex as previously described (Rost et al., 2010). Briefly, 300 μm diameter spots of growth permissive substrate consisting of 0.7 mg ml−1 collagen and 0.1 mg ml−1 poly-D-lysine was applied with a custom-made stamp on coverslips coated with a thin film of agarose. Astrocytes were seeded onto the glass coverslips and were allowed to proliferate in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% fetal calf serum and 0.2% penicillin/streptomycin (Invitrogen) for one more week to form glia micro-islands. After changing the medium to Neurobasal-A supplemented with 2% B27 and 0.2% penicillin/streptomycin, hippocampal neurons prepared from P0 mice were added at a density of 370 cells cm−2. Neurons were infected with AAVs at DIV 1-3 and recorded between DIV 14 and DIV 21.
Confocal imaging and quantification
Primary cultured hippocampal neurons were transfected at 5 DIV with plasmids encoding a rhodopsin protein (mScarlet, OPN3, PufTMT3a, OPN3-M4, PufTMT3a-M4, eOPN3) along with pAAV-CamKIIa-eYFP. Four days after transfection, cells were fixed and permeabilized, washed 4 times with PBS and stained for 3 min with DAPI (5 mg/mL solution diluted 1:30,000 prior to staining). Coverslips were then mounted using PVA-DABCO (Sigma) and allowed to dry. Images of mScarlet and EYFP fluorescence were acquired using a Zeiss LSM 700 confocal microscope with a 20X magnification objective. Fluorescence was quantified using ImageJ (Schindelin et al., 2012) by marking a region containing the somatic cytoplasm using the EYFP fluorescence and then measuring the average pixel intensity in the red imaging channel.
Histology, imaging, and quantification
Mice were deeply anesthetized using pentobarbital (130 mg per kg, intraperitoneally) and then transcardially perfused with ice-cold PBS (pH 7.4,10 ml) followed by 4% paraformaldehyde (PFA, 10 ml) solution. Heads were removed and post-fixed overnight at 4 °C in 4% PFA. Then, brains were extracted and transferred to 30% sucrose solution for at least 24 h. Coronal sections (40 μm) were acquired using a microtome (Leica Microsystems) and stained with a nucleic acid dye (4,6-diamidino-2-phenylindole (DAPI), 1:10,000). Slices were then mounted on gelatin-coated slides, dehydrated, and embedded in DABCO mounting medium (Sigma). Slices were imaged using aVS120 microscope (Olympus), at 10x magnification with two channels: 1) DAPI, to identify brain structures, the corresponding anterior-posterior coordinates and sites of lesions created by the optic fiber. 2) Either Cy3 (mScarlet - eOPN3 mice) or FITC (eYFP - control mice), to measure expression levels in cells and projections. The resulting images were then analyzed using ImageJ to measure the fluorescence of DAPI and additional fluorophores within specific target regions. For each slice, a rectangle outlining the target site was defined and copied to the contralateral (non-expressing) hemisphere. Mean fluorescence values were measured separately for each channel and compared between hemispheres, demonstrating differences in fluorophore expression but not in DAPI staining. Imaging acquisition parameters and the ensuing analysis pipeline were kept constant across mice to allow comparison between the eOPN3 and the control groups.
Cell culture and live-cell cAMP assay
Optical activation and G protein coupling of mosOPN3-mScarlett and chimeric GPCR constructs was tested in HEK293T cells using a live cell assay (Ballister et al., 2018). Briefly, GPCR constructs were subcloned into pcDNA3.1 (ThermoFisher). HEK293T cells were incubated at 37°C (5% CO2) in DMEM containing 4500 mg/L glucose, L-glutamine (Sigma Aldrich) with penicillin (100 U/mL), streptomycin (100 mg/mL), and 10% FBS. For transfection, cells were seeded into solid white 96-well plates (Greiner) coated with poly-L-Lysine (Sigma Aldrich) and transfected with Lipofectamine 2000 (ThermoFisher) together with individual G protein chimera (GsX) and Glo22F (Promega). Cells were incubated for 24 h at 37°C, 5% CO2 and, subsequently, in L-15 media (without phenol-red, with L-glutamine, 1% FBS, penicillin, streptomycin (100 mg/mL)) and 9-cis retinal (10 mM) and beetle luciferin (2 mM in 10 mM HEPES pH 6.9) for 1 h at RT. Cells were kept in the dark throughout the entire time. Baseline luminescence was measured 3 times and opto-GPCR activation was then induced by illuminating cells for 1 s with an LED plate (530 nm, 5.5 μW-mm−2, Phlox Corp.) Changes in cAMP levels were measured over time using GloSensor luminescence. For the assay quantification each technical repeat was normalized to its pre-light baseline.
Slice culture preparation and transgene delivery
Organotypic hippocampal slices were prepared from Wistar rats at postnatal day 5-7 as described (Gee et al., 2017). Briefly, dissected hippocampi were cut into 400 μm slices with a tissue chopper and placed on a porous membrane (Millicell CM, Millipore). Cultures were maintained at 37°C, 5% CO2 in a medium containing 80% MEM (Sigma M7278), 20% heat-inactivated horse serum (Sigma H1138) supplemented with 1 mM L-glutamine, 0.00125% ascorbic acid, 0.01 mg/mL insulin, 1.44 mM CaCl2, 2 mM MgSO4 and 13 mM D-glucose. No antibiotics were added to the culture medium.
For transgene delivery in organotypic slices, individual CA3 pyramidal cells were transfected by single-cell electroporation between DIV 15-20 as previously described (Wiegert et al., 2017b). The plasmids pAAV-CKIIa(0.4)-eOPN3-mScarlet, pCI-hSyn-mCerulean, CAG-GIRK2-1 and pGP-AAV-hSyn-jGCaMP7f-WPRE were all diluted to 50 ng/μl in K-gluconate-based solution consisting of (in mM): 135 K-gluconate, 10 HEPES, 0.2 EGTA, 4 Na2-ATP, 0.4 Na-GTP, 4 MgCl2, 3 ascorbate, 10 Na2- phosphocreatine, pH 7.2, 295 mOsm/kg. An Axoporator 800A (Molecular Devices) was used to deliver 25 hyperpolarizing pulses (−12 V, 0.5 ms) at 50 Hz. During electroporation slices were maintained in pre-warmed (37°C) HEPES-buffered solution in (mM): 145 NaCl, 10 HEPES, 25 D-glucose, 2.5 KCl, 1 MgCl2 and 2 CaCl2 (pH 7.4, sterile filtered).
For targeted viral vector-based transduction of organotypic hippocampal slice cultures (Wiegert et al., 2017c), adeno-associated viral particles encoding AAV2/9.CamKIIa(0.4).eOPN3-mScarlet were pressure injected (20 PSI/2-2.5 bar, 50 ms duration) using a Picospritzer III (Parker) under visual control (oblique illumination) into CA3 stratum pyramidale between DIV 2-5. Slice cultures were then maintained in the incubator for 2-3 weeks allowing for virus payload expression.
Electrophysiology in cultured neurons
Whole-cell patch clamp recordings in dissociated cultures were performed under visual control using differential interference contrast infrared (DIC-IR) illumination on an Olympus IX-71 microscope equipped with a monochrome scientific CMOS camera (Andor Neo). Borosilicate glass pipettes (Sutter Instrument BF100-58-10) with resistances ranging from 3−7 MΩ were pulled using a laser micropipette puller (Sutter Instrument Model P-2000). For hippocampal neuron cultures, electrophysiological recordings from neurons were obtained in Tyrode’s medium ([mM] 150 NaCl, 4 KCl, 2 MgCl2, 2 CaCl2, 10 D-glucose, 10 HEPES; 320 mOsm; pH adjusted to 7.35 with NaOH). The recording chamber was perfused at 0.5 mL min−1 and maintained at 29°C or 23°C (Figure S4A). Pipettes were filled using a potassium gluconate-based intracellular solution ([mM] 135 K-gluconate, 4 KCl, 2 NaCl, 10 HEPES, 4 EGTA, 4 MgATP, 0.3 NaGTP; 280 mOsm kg−1; pH adjusted to 7.3 with KOH). Whole-cell voltage clamp recordings were performed using a MultiClamp 700B amplifier, filtered at 8 kHz and digitized at 20 kHz using a Digidata 1440A digitizer (Molecular Devices). Light was delivered using a Lumencor SpecraX light engine, using band-pass filters at 445/20, 475/28, 512/25, 572/35 and 632/22 nm (peak wavelength/bandwidth). Photon flux was calibrated to be similar for all five wavelengths at the sample plane to allow comparison of activation efficiency. Remaining photon flux differences were less than 6%.
Whole-cell recordings in autaptic neurons were performed on an Olympus IX73 microscope using a Multiclamp 700B amplifier (Molecular Devices) under control of Clampex 10 (Molecular Devices). Data was acquired at 10 kHz and filtered at 3 kHz. Extracellular solution contained (in mM): 140 NaCl, 2.4 KCl, 10 HEPES, 10 glucose, 2 CaCl2, and 4 MgCl2 (pH adjusted to 7.3 with NaOH, 300 mOsm). Internal solution contained the following (in mM): 136 KCl, 17.8 HEPES, 1 EGTA, 0.6 MgCl2, 4 MgATP, 0.3 Na2GTP, 12 Na2 phosphocreatine, 50 U ml−1 phosphocreatine kinase (300 mOsm); pH adjusted to 7.3 with KOH. Fluorescence light from a TTL-controlled LED system (pE4000, CoolLED) was filtered using single band-pass filters (AHF F66-415), coupled into the back port of the microscope by a liquid light guide, and delivered through an Olympus UPLSAPO 20x, 0.75 NA objective. Membrane potential was set to −70 mV, and series resistance and capacitance were compensated by 70%. To obtain strong GIRK currents, cells were voltage clamp briefly to −50 mV for the light flash only, while EPSCs were recorded at −70 mV. Synaptic transmitter release was elicited by 1 ms depolarization to 0 mV, causing an unclamped AP in the axon. To estimate the onset time course of the eOPN3-mediated effect on synaptic release, trains of APs were evoked at 10 Hz. Light was applied after 200 such APs, when EPSC amplitudes reached a steady state. Baclofen and SCH23390 were applied via a rapid perfusion system (Rost et al., 2010). Pertussis toxin was applied to the cultures 24 h before the recordings, at a concentration of 0.5 μg ml−1. Cells were excluded from the analysis of the paired-pulse ratio if eOPN3 activation completely abolished the first EPSC, and mEPSCs were not analyzed when noise-events detected by an inverted template occurred at > 1 Hz, as previously described (Rost et al., 2015).
Slice culture electrophysiology and two-photon microscopy
To characterize the effects of eOPN3-activation on neuronal cell parameters, targeted whole-cell recordings of transfected CA3 pyramidal neurons were performed at room temperature (21-23°C), between 1-2 weeks after electroporation or viral transduction, undervisual guidance using a BX51WI microscope (Olympus) and a Multiclamp 700B amplifier (Molecular Devices) controlled by either Ephus (Suter et al., 2010) or WaveSurfer software (https://www.janelia.org/open-science/wavesurfer), both written in MATLAB. Patch pipettes with a tip resistance of 3-4 MΩ were filled with (in mM): 135 K-gluconate, 4 MgCl2, 4 Na2-ATP, 0.4 Na-GTP, 10 Na2-phosphocreatine, 3 ascorbate, 0.2 EGTA, and 10 HEPES (pH 7.2). Artificial cerebrospinal fluid (ACSF) consisted of (in mM): 135 NaCl, 2.5 KCl, 4 CaCl2, 4 MgCl2, 10 Na-HEPES, 12.5 D-glucose, 1.25 NaH2PO4 (pH 7.4). To block synaptic transmission, 10 μM CPPene, 10 μM NBQX, and 100 μM picrotoxin (Tocris, Bristol, UK) were added to the recording solution. Measurements were corrected for a liquid junction potential of —14 mV.
In dual patch-clamp experiments (Figure 4), we recorded from pairs of synaptically connected CA3 pyramidal cells expressing eOPN3 and non-expressing CA1 pyramidal cells. CA3 pyramidal neurons were stimulated in current clamp to elicit 2 action potentials (40 ms Inter Stimulus Interval, 0.2 Hz) by brief somatic current injection (2-3 ms, 3 - 4 nA) in the absence of synaptic blockers while recording EPSCs by holding the CA1 cell at −60 mV in voltage clamp mode. A brief light pulse (500 ms, 525 nm, 1 mW-mm−2) through the objective (illuminated area = 0.322 mm2) in CA1 was used to activate eOPN3 locally at axon terminals innervating the postsynaptic CA1 pyramidal cell. For extracellular stimulation, afferent Schaffer collateral axons were stimulated (0.2 ms, 20-70 μA every 10 s) with a monopolar glass electrode connected to a stimulus isolator (IS4 stimulator, Scientific Devices). Fortrain stimulation, 10 pulses were delivered every 40 ms. Access resistance of the recorded non-transfected CA1 neuron was continuously monitored and recordings above 20 MΩ and/or with adrift >30% were discarded. A 16-channel pE-4000 LED light engine (CoolLED, Andover, UK) was used for epifluorescence excitation and light activation of eOPN3 (500 ms, 525 nm, 1 mW mm−2). Light intensity was measured in the object plane with a 1918 R power meter equipped with a calibrated 818 ST2 UV/D detector (Newport, Irvine CA) and divided by the illuminated field of the Olympus LUMPLFLN 60XW objective (0.134 mm2) or of the Olympus LUMPLFLN 40XW objective (0.322 mm2). All the electrophysiological synaptic measurements in organotypic hippocampal slice cultures were performed at 33 ± 1°C.
For the eOPN3 two-photon stimulation experiments (Figure 5), a custom-built two-photon imaging setup was used based on an Olympus BX51WI microscope controlled by ScanImage 2017b (Vidrio Technologies). Electrophysiological recordings were acquired using a Multiclamp 700B amplifier controlled by the WaveSurfer software written in MATLAB (https://www.janelia.org/open-science/wavesurfer). A tunable, pulsed Ti:Sapphire laser (MaiTai DeepSee, Spectra Physics) controlled by an electro-optic modulator (350-80, Conoptics) tuned to 1040 nm was used to excite the mScarlet-labeled eOPN3. Red fluorescence was detected through the objective (LUMPLFLN 60XW, 60x, 1.0 NA, Olympus) and through the oil immersion condenser (numerical aperture 1.4, Olympus) by photomultiplier tubes (H7422P-40SEL, Hamamatsu). 560 DXCRdichroic mirrorsand 525/50and 607/70 emission filters (ChromaTechnology) were used to separate green and red fluorescence. Excitation light was blocked by short-pass filters (ET700SP-2P, Chroma). In addition, the forward-scattered IR laser light was collected through the condenser, spatially filtered by a Dodt contrast tube (Luigs&-Neumann) attached to the trans-illumination port of the microscope and detected with a photodiode connected to a detection channel of the laser scanning microscope. This generated an IR-scanning gradient contrast image (IR-SGC) synchronized with the fluorescence images(Wimmer et al., 2004). This approach was used for targeted patch-clamp recordings avoiding prior activation of the ultrasensitive eOPN3 with epifluorescence illumination. The two-photon laser scanning pattern used for stimulation was either a spiral scan with a repetition rate of 500 Hz above the soma (2 ms/spiral, 250 cycles, 500 ms total duration) or standard raster scans at 1.09 Hz overthe somatodendritic compartment (FOV =106x106 μm, 512x512 pixels, 1.8 ms/line, 5 frames, 4.6 s total duration). The laser wavelengths used for stimulation were 800 nm, 860 nm, 930 nm, 980 nm and 1040 nm, all at 30 mW, measured at the back focal aperture of the objective. Wide field illumination at 525 nm (10 mW/mm2) was done with a 16 channel pE-4000 LED light engine (CoolLED, Andover, UK) for 500 ms. An additional set of experiments was performed on a second custom-modified two-photon imaging setup (DF-Scope, Sutter) based on an Olympus BX51WI microscope controlled by ScanImage 2017b (Vidrio Technologies) and equipped with an Ytterbium-doped 1070-nm pulsed fiber laser (Fidelity-2, Coherent) for far infrared stimulation. Electrophysiological recordings were performed using a Double IPA integrated patch amplifier controlled with SutterPatch software (Sutter Instrument).
The same microscope was used to acquire images of eOPN3-expressing CA3 cells co-transfected with the cyan cell-filler fluorophore mCerulean (Rizzo etal., 2004) and their projecting axons in stratum radiatum of CA1 (Figure 1). The 1070-nm laser was used to excite fluorescence of mScarlet-labeled eOPN3. mCerulean was excited by a pulsed Ti:Sa laser (Vision-S, Coherent) tuned to 810 nm. Laser power was controlled by electro-optic modulators (350-80, Conoptics). Red and cyan fluorescence were detected through the objective (Olympus LUMPLFLN 60XW, 1.0 NA, or Leica HC FLUOTAR L25x/0.95 W VISIR) and through the oil immersion condenser (numerical aperture 1.4, Olympus) by GaAsP photomultiplier tubes (Hamamatsu, H11706-40). Dichroic mirrors (560 DXCR, Chroma Technology) and emission filters (ET525/70 m-2P, ET605/70 m-2P, Chroma Technology) were used to separate cyan and red fluorescence. Excitation light was blocked by short-pass flters (ET700SP-2P, Chroma Technology). All electrophysiology recordings were analyzed using custom written scripts in MATLAB except for recordings acquired with the Double IPA integrated patch amplifier, which were analyzed with the SutterPatch software.
For presynaptic Ca2+ imaging experiments (Figure 5), single action potentials were triggered via a patch pipette in a CA3 pyramidal neuron co-expressing eOPN3 and jGCaMP7f or jGCaMP7f alone as control while evoked Ca2+ influx at distal presynaptic terminals in stratum radiatum of CA1 was monitored by two-photon microscopy. A custom-modified version of ScanImage 3.8 (Pologruto et al., 2003) was used to allow user-defined arbitrary line scans. jGCaMP7f was excited at 960 nm. Similar to the two-photon stimulation experiments, targeted patch-clamp recordings were achieved using IR-scanning gradient contrast image (IR-SGC) synchronized with the fluorescence images. Action potentials were triggered by brief somatic current injection (2-3 ms, 3-4 nA) in the absence of synaptic blockers while monitoring fluorescent transients at single Schaffer collateral terminals in CA1 (70-80 trials on average at 0.1 Hz). User-defined circular scans at 500 Hz across the bouton were used to repeatedly sample the fluorescent changes. During each trial (3 s), laser exposure was restricted to the periods of expected Ca2+ response (~1.3 s) to minimize bleaching. To activate eOPN3 selectively at the terminals, we used a fiber-coupled LED (400 μm fiber, NA 0.39, M118L02, ThorLabs) to deliver 500 ms green light pulses (l = 530 nm, 83 μW at the fiber tip) 1 s prior to the onset of electrical stimulation. During the LED pulses, upper and lower PMTs were protected by TTL triggered shutters (NS45B, Uniblitz). GIRK channels were blocked by SCH 23390 (10 μM, Tocris, Bristol, UK) throughout the entire experiment to exclude hyperpolarization-mediated effects on action potential propagation and presynaptic Ca2+ influx.
The photon shot-noise subtracted relative change in jGCaMP7f fluorescence (ΔF/F0) was measured by using a template-based fitting algorithm. The characteristic fluorescence time constant was extracted for every bouton by fitting a double exponential function (trise, tdecay) to the average jGCaMP7f signal. To estimate the Ca2+ transient amplitude for every trial, we fitted the bouton-specific template to every response, amplitude being the only free parameter. Response amplitude was defined as the value of the fit function at its maximum.
In vivo electrophysiological recordings
8-9 weeks old male C75/Bl6 mice were pressure injected (Picospritzer III; Parker) bilaterally into LGN (AP: - 2.2 mm; ML: +/- 2.3 mm; DV: -3.1 mm) at 50 nL/min with 200 nL adeno-associated viral particles encoding eOPN3 (AAV2/5.CKIIa(0.4).eOPN3-mScarlet) diluted to 2.5 x 1012 viral genomes per ml using a pulled glass capillary. Following 5-6 weeks of recovery, mice underwent 3-4 head fixation habituation sessions starting with 15 min and gradually increasing to 25 min. 7-12 weeks after virus injection, craniotomies were performed bilaterally to provide access to V1 spanning from −2.3 mm to −4.7 mm in the anterior posterior direction and 2 mm at its widest part (at AP: −3.8 mm) from ±1.3 mm to ± 3.3 mm along the medio-lateral axis. Craniotomies were covered with Kwik-Cast (WPI Inc) to protect the brain surface from mechanical impact, dehydration, and light exposure between the silicon probe recording sessions.
For the electrophysiological recordings, two 4-shank, 128 channel silicon microprobes (128DN; 4 shanks, 150 μm shank spacing, 25 μm channel spacing, 100 μm2 electrode area, 7 mm x 65 μm x 23 μm shank dimensions) (Yang et al., 2020) (kindly provided by Dr.S. Masmanidis, UCLA) were inserted bilaterally in the V1 at a depth of approximately 1 mm, with an insertion speed of 100 μm/min. Before each recording session, silicon probe recording sites were electroplated in a PEDOT solution to an impedance of ~100 kOhm. Each silicon probe was connected to an RHD2000 chip-based 128 channel amplifier board (Intan Technologies). Broadband (0.1 Hz-7.5 kHz) signals were acquired at 30 kHz. Signals were digitized at 16 bit and transmitted to an OpenEphys recording controller (OEPS).
Raw data were processed to detect spikes and extract single-unit activity. Briefly, the wide-band signals were band-pass fltered (0.6 kHz-6 kHz), spatially whitened across channels and thresholded for isolation of putative spikes. Clustering was performed using template matching implemented in Kilosort2 (Pachitariu et al., 2016) and computed cluster metrics were used to pre-select units for later manual curation using custom-written software.
For the optogenetic inhibition of LGN axons, the silicon probe inserted in one of the two craniotomies was coupled with a 200 μm 0.5 NA optic fiber (Thorlabs, FP200URT), placed between the two middle shanksand at ~300 μm above the top-most channel of the silicon probe, thus the optic fiber remained just outside the surface of the cortex during the recordings. This fiber was coupled with a 525 nm LED (PlexBright, Plexon), controlled using a Cyclops 3.6 LED driver and a custom Teensy3.2-based stimulation system, calibrated to deliver ~2 mW of light at the tip of the fiber.
Following a long baseline period, the paradigm used to investigate the effect of eOPN3 on the synaptic vesicle release in vivo consisted of 31 presentations of a visual stimulus every 30 s. The 10 first trials were used to establish the baseline of the visual response and the 11th trial was coupled with optogenetic stimulation, starting 1 s before the visual stimulation and lasting for a total of 30 s. Each visual stimulus presentation trial consisted of 8 repeats of a 500 ms visual drifting grating presentations in the cardinal and intercardinal directions. The stimuli were presented on a 23.5” monitor placed 20 cm centrally in front of the mouse, so that the monitor was visible to both eyes. The stimulus presentation was controlled using a custom-written Python program and utilized PsychoPy3.0. For the accurate detection of the stimulus onset to allow for alignment with electrophysiological data, a photodetector was mounted in one corner of the monitor. The mouse was gradually habituated to head-fixation over multiple sessions and was running freely on a horizontal wheel. Each mouse was recorded for 1 or 2 identical sessions on different days and data were pooled for the subsequent analyses. Recording sessions in which no units showed visual stimulus-evoked activity were excluded from the analysis.
For visual stimulus response characterization, the spike rates were calculated in 50 ms bins. Each unit’s activity was normalized to the average firing rate in the 15 s prior to stimulus presentation during the baseline period. The baseline period in Figure 6D was defined as the activity during the two trials before eOPN3 activation. For clarity, the peristimulus time histograms shown in Figure 6E were low pass filtered using a Gaussian function (window: 250 ms, σ = 100 ms). The recovery time constant shown in Figure 6F was calculated by fitting the post eOPN3 activation visual stimulus response to f(t) = 1-a·exp(-t/tau), with the effect size (a) and recovery time constant (tau) as free parameters.
In vivo optogenetic silencing of the nigrostriatal pathway
AAV vectors encoding a Cre-dependent eOPN3-mScarlet transgene (AAV2/1&2.hSyn.SIO-eOPN3-mScarlet; 6 x 1012 viral genomes / ml) or eYFP (AAV2/1&2.EF1a.DIO.eYFP; 2 x 1013 viral genomes / ml) were unilaterally injected into the substantia nigra (AP: - 3.5 mm, ML: + or -1.4 mm DV: - 4.25 mm; 500 nL per mouse) of DAT-Cre transgenic mice. Optical fibers (200 μm diameter, NA 0.5) were unilaterally implanted above the ipsilateral dorsomedial striatum (AP: + 0.6 mm, ML: + or - 1.5 mm DV: - 2.1 mm). Left and right implanted mice were counterbalanced among the eOPN3 and control groups. Mice were allowed to recover for 6-9 weeks to allow for viral expression. Following recovery, mice underwent a single 10-min habituation session, to habituate to handling, patch cord attachment and the open field arena. In experimental sessions, we attached individual mice to a patch cord and video recorded their free locomotion continuously in the open field under near-infrared illumination.
To measure eOPN3 induced bias in locomotion, we video recorded the free locomotion of single mice in an open field arena (50x 50x50 cm) continuously over 30 min. After a 10-min baseline no-light period, we delivered 500 ms light pulses (540 nm, 10 mW at the fiber tip), at 0.1 Hzfor 10 min, followed by an additional 10-min no-light period. Offline video processing and mouse tracking was done using DeepLabCut (DLC; (Mathis et al., 2018)). Briefly, we trained DLC to detect 6 features on the mouse body (nose, head center, left and right ears, center of mass, tail) and 3 bottom corners of the arena. X-Y coordinates of each feature were then further processed to complete missing or noisy values (high amplitude and short duration changes in X or Y dynamics) using linear interpolation (interpl) of data from neighboring frames. This was followed by a low pass filtering of the signals (malowess, with 50 points span and of linear order). Finally, a pixel to cm conversion was done based on the video-detected arena features and its physical measurements. A linear fit to the nose, head, center and tail features defined the mouse angle with respect to the south arena wall at each frame. Following its dynamics over the session, we identified direction shifts as a direction change in angle that exceeds 20° and 1 s. To achieve a comparable measurement between right- and left- hemisphere injected mice, we measured motion in the ipsilateral direction as positive and contralateral motion as negative from the cumulative track of angle. The net angle gain was calculated as the sum of ipsilateral and contralateral angle gained over each time bin (1- or 10-min bins as indicated). For each time bin we then calculated a rotation index, based on angle gains, as follows:
For each mouse, rotation index scores were calculated for two complete sessions on different days. Individual scores were plotted for each mouse against the expression levels measured in that mouse (see section: Histology, imaging, and quantification). Results were then averaged across individual sessions, and used for all statistical comparisons, and linear regressions analysis. Mouse positions and velocities were measured by the “center of mass” feature.
Quantification and Statistical Analysis
Mean was used as center measure and standard error of the mean (SEM) as dispersion measure throughout the manuscript. The data was tested for violations of assumptions of parametric tests (Gaussian distribution of the residuals was assessed using the Kolmogorov-Smirnov test; Equality of variances was assessed using the Levene’s test), and non-parametric tests were utilized where assumptions were violated. The statistical details for the specific experiments, including the statistical tests used, exact value of n, what n represents (e.g., number of animals, number of brain slices, number of cells, or number of trials), can be found in the figures, figure legends or Results text. Significance was determined at a level of 0.05 using the statistical test as reported in the figure legend or Results. P values were corrected for multiple comparison as reported in the figure legends or Results. For fitting results, confidence intervals are reported. No statistical tests were run to predetermine sample size, but sample sizes were similar to those commonly used in the field. Blinding and randomization were performed only in the behavioral experiments (Figure 7); in other experiments, automated analysis was used whenever possible. For autaptic neuron recordings (Figure 2), cells were excluded from the analysis of the paired-pulse ratio if eOPN3 activation completely abolished the first EPSC, and mEPSCs were not analyzed when noise-events detected by an inverted template occurred at > 1 Hz, as previously described (Rost et al., 2015). For organotypic slice culture recordings the access resistance of the recorded non-transfected CA1 neuron was continuously monitored and recordings with access resistance above 20 MΩ and/or with a drift > 30% were discarded. For in vivo electrophysiology (Figure 6), recording sessions in which no units showed visual stimulus-evoked activity were excluded from the analysis. Statistical analysis was performed using MATLAB (Mathworks), RStudio Desktop (RStudio), and Prism (Graphpad).
Supplementary Material
Acknowledgments
We thank the members of the Yizhar, Wiegert, Soba, and Schmitz labs for ideas, criticism, and discussions throughout this project. We thank Bryan Copits, Michael Bruchas, and Robert Gereau for insightful comments on the manuscript. We would also like to thank Thomas Oertner for generous sharing of equipment and Eitan Reuveny for the GIRK expression plasmids. This work was supported by funding from the European Research Commission (ERC CoG PrefrontalMap 819496 to O.Y., ERC StG LIFE synapses 714762 to J.S.W., and ERC BrainPlay to D.S.), the Israel Science Foundation (COEX 3131/20), the Adelis Brain Research Award, the Ilse Katz Institute for Material Sciences and Magnetic Resonance Research (to O.Y.), EMBO (ALTF 352-2019 to M.M., ALTF 914-2018 to N.K., and ALTF 378-2019 to J.W.), the Achar Research Fellow Chair in Electrophysiology (to J.D.), the Minerva Foundation (to J.W.), and the German Research Foundation (DFG FOR2419 and SFB936 to J.S.W., DFG EXC-2049, SFB 958, SFB 1315, and SFB 1665 to D.S., and DFG SPP 1926 jointly to O.Y., P.S., B.R.R., and J.S.W.).
Footnotes
Author Contributions
Contributions of the authors, according to CRediT, are as follows: conceptualization (M.M., O.Y.), formal analysis (M.M., I.S.-S., P.P., M.P., E.B., N.K., J.D., J.W., B.R.R.), investigation (M.M., I.S.-S., P.P., M.P., E.B., N.K., F.B., S.P., A.G., J.D., J.W., R.L., A. Litvin, F.Z., B.R.R., O.Y.), methodology (M.M., I. S.-S., P.P., M.P., E.B., N.K., R.L., F.Z., K.S., J.S.W.), resources (R.L., P.S., D.S., A. Luthi, B.R.R., J.S.W., O.Y.), supervision (P.S., D.S., A. Lüthi, B.R.R., J. S.W., O.Y.), writing the original draft (M.M., I.S.-S., M.P., B.R.R., J.S.W., O.Y.), review and editing (M.M., I.S.-S., P.P., M.P., N.K., B.R.R., J.W., D.S., A. Lüthi, J.S.W., O.Y.), funding acquisition (P.S., J.S.W., O.Y.), overall project administration (O.Y.).
Declaration Of Interests O.Y. and M.M. have disclosed these findings to Yeda, the Weizmann Institute Technology Transfer Arm, which is filing a patent application on these developments. The constructs and viral vectors remain freely available from the authors and through Addgene.
Supporting Citations The following references appear in the supplemental information: Edgar et al. (2004); Lesage et al. (1994).
Materials availability
Plasmids and viral vectors for expression of eOPN3 are available from Addgene (https://www.addgene.org/Ofer_Yizhar/).
Data and code availability
The datasets and the code that support the findings of this study are available from the lead contact upon reasonable request.
References
- Alcaro A, Huber R, Panksepp J. Behavioral functions of the mesolimbic dopaminergic system: an affective neuroethological perspective. Brain Res Brain Res Rev. 2007;56:283–321. doi: 10.1016/j.brainresrev.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster BN, Li X, Pausch MH, Herlitze S, Roth BL. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc Natl Acad Sci USA. 2007;704:5163–5168. doi: 10.1073/pnas.0700293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailes HJ, Zhuang L-Y, Lucas RJ. Reproducible and sustained regulation of Gas signalling using a metazoan opsin as an optogenetic tool. PLoS ONE. 2012;7:e30774. doi: 10.1371/journal.pone.0030774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballister ER, Rodgers J, Martial F, Lucas RJ. A live cell assay of GPCR coupling allows identification of optogenetic tools for controlling Go and Gi signaling. BMC Biol. 2018;76:10. doi: 10.1186/s12915-017-0475-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barter JW, Li S, Lu D, Bartholomew RA, Rossi MA, Shoemaker CT, >Salas-Meza D, Gaidis E, Yin HH. Beyond reward prediction errors: the role of dopamine in movement kinematics. Front Integr Nuerosci. 2015;9:39. doi: 10.3389/fnint.2015.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu J, Zaremba JD, Cheung SK, Hitti FL, Zemelman BV, Losonczy A, Siegelbaum SA. Gating of hippocampal activity, plasticity, and memory by entorhinal cortex long-range inhibition. Science. 2016;357:aaa5694. doi: 10.1126/science.aaa5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean BP. Neurotransmitter inhibition of neuronal calcium currents by changes in channel voltage dependence. Nature. 1989;340:153–156. doi: 10.1038/340153a0. [DOI] [PubMed] [Google Scholar]
- Beck S, Yu-Strzelczyk J, Pauls D, Constantin OM, Gee CE, Ehmann N, Kittel RJ, Nagel G, Gao S. Synthetic Light-Activated ion channels for optogenetic activation and inhibition. Front Neurosci. 2018;72:643. doi: 10.3389/fnins.2018.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekkers JM, Stevens CF. Excitatory and inhibitory autaptic currents in isolated hippocampal neurons maintained in cell culture. Proc Natl Acad Sci USA. 1991;88:7834–7838. doi: 10.1073/pnas.88.17.7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal Sierra YA, Rost BR, Pofahl M, Fernandes AM, Kopton RA, Moser S, Holtkamp D, Masala N, Beed P, Tukker JJ, et al. Potassium channel-based optogenetic silencing. Nat Commun. 2018;9:4611. doi: 10.1038/s41467-018-07038-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindels DS, Haarbosch L, van Weeren L, Postma M, Wiese KE, Mastop M, Aumonier S, Gotthard G, Royant A, Hink MA, Gadella TWJ., Jr mScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nat Methods. 2017;74:53–56. doi: 10.1038/nmeth.4074. [DOI] [PubMed] [Google Scholar]
- Borgkvist A, Avegno EM, Wong MY, Kheirbek MA, Sonders MS, Hen R, Sulzer D. Loss of Striatonigral GABAergic Presynaptic Inhibition Enables Motor Sensitization in Parkinsonian Mice. Neuron. 2015;87:976–988. doi: 10.1016/j.neuron.2015.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenowitz S, David J, Trussell L. Enhancement of synaptic efficacy by presynaptic GABA(B) receptors. Neuron. 1998;20:135–141. doi: 10.1016/s0896-6273(00)80441-9. [DOI] [PubMed] [Google Scholar]
- Burke KJ, Jr, Keeshen CM, Bender KJ. Two Forms of Synaptic Depression Produced by Differential Neuromodulation of Presynaptic Calcium Channels. Neuron. 2018;99:969–984.:e7. doi: 10.1016/j.neuron.2018.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalifoux JR, Carter AG. GABAB receptor modulation of voltage-sensitive calcium channels in spines and dendrites. J Neurosci. 2011;37:4221–4232. doi: 10.1523/JNEUROSCI.4561-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copits BA, Gowrishankar R, O’Neill PR, Li J-N, Girven KS, Yoo JJ, Meshik X, Parker KE, Spangler SM, Elerding AJ, et al. A photoswitchable GPCR-based opsin for presynaptic inhibition. Neuron. 2021;709 doi: 10.1016/j.neuron.2021.04.026. Published online May 11,2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creed M, Pascoli VJ, Lüscher C. Addiction therapy.Refining deep brain stimulation to emulate optogenetic treatment of synaptic pathology. Science. 2015;347:659–664. doi: 10.1126/science.1260776. [DOI] [PubMed] [Google Scholar]
- Currie KPM. G protein modulation of CaV2 voltage-gated calcium channels. Channels (Austin) 2010;4:497–509. doi: 10.4161/chan.4.6.12871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva JA, Tecuapetla F, Paixão V, Costa RM. Dopamine neuron activity before action initiation gates and invigorates future movements. Nature. 2018;554:244–248. doi: 10.1038/nature25457. [DOI] [PubMed] [Google Scholar]
- Dana H, Sun Y, Mohar B, Hulse BK, Kerlin AM, Hasseman JP, Tsegaye G, Tsang A, Wong A, Patel R, et al. High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nat Methods. 2019;76:649–657. doi: 10.1038/s41592-019-0435-6. [DOI] [PubMed] [Google Scholar]
- Dittman JS, Regehr WG. Contributions of calcium-dependent and calcium-independent mechanisms to presynaptic inhibition at a cerebellar synapse. J Neurosci. 1996;76:1623–1633. doi: 10.1523/JNEUROSCI.16-05-01623.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrunz LE, Huang EP, Stevens CF. Very short-term plasticity in hippocampal synapses. Proc Natl Acad Sci USA. 1997;94:14843–14847. doi: 10.1073/pnas.94.26.14843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DA, Stempel AV, Vale R, Ruehle S, Lefler Y, Branco T. A synaptic threshold mechanism for computing escape decisions. Nature. 2018;558:590–594. doi: 10.1038/s41586-018-0244-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froudarakis E, Fahey PG, Reimer J, Smirnakis SM, Tehovnik EJ, Tolias AS. The Visual Cortex in Context. Annu Rev Vis Sci. 2019;5:317–339. doi: 10.1146/annurev-vision-091517-034407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee CE, Ohmert I, Wiegert JS, Oertner TG. Preparation of Slice Cultures from Rodent Hippocampus. Cold Spring Harb Protoc. 2017;2077 doi: 10.1101/pdb.prot094888. [DOI] [PubMed] [Google Scholar]
- Gerachshenko T, Blackmer T, Yoon E-J, Bartleson C, Hamm HE, Alford S. Gbetagamma acts at the C terminus of SNAP-25 to mediate presynaptic inhibition. Nat Neurosci. 2005;8:597–605. doi: 10.1038/nn1439. [DOI] [PubMed] [Google Scholar]
- Graham FL, van der Eb AJ. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Grealish S, Mattsson B, Draxler P, Björklund A. Characterisation of behavioural and neurodegenerative changes induced by intranigral 6-hydroxydopamine lesions in a mouse model of Parkinson’s disease. Eur J Neurosci. 2010;37:2266–2278. doi: 10.1111/j.1460-9568.2010.07265.x. [DOI] [PubMed] [Google Scholar]
- Grimm D, Kay MA, Kleinschmidt JA. Helper virus-free, optically controllable, and two-plasmid-based production of adeno-associated virus vectors of serotypes 1 to 6. MolTher. 2003;7:839–850. doi: 10.1016/s1525-0016(03)00095-9. [DOI] [PubMed] [Google Scholar]
- Hamid E, Church E, Wells CA, Zurawski Z, Hamm HE, Alford S. Modulation of neurotransmission by GPCRs is dependent upon the microarchitecture of the primed vesicle complex. J Neurosci. 2014;34:260–274. doi: 10.1523/JNEUROSCI.3633-12.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlitze S, Garcia DE, Mackie K, Hille B, Scheuer T, Catterall WA. Modulation of Ca2+ channels by G-protein beta gamma subunits. Nature. 1996;380:258–262. doi: 10.1038/380258a0. [DOI] [PubMed] [Google Scholar]
- Ikeda SR. Voltage-dependent modulation of N-type calcium channels by G-protein beta gamma subunits. Nature. 1996;380:255–258. doi: 10.1038/380255a0. [DOI] [PubMed] [Google Scholar]
- Jackman SL, Regehr WG. The mechanisms and functions of synaptic facilitation. Neuron. 2017;94:447–464. doi: 10.1016/j.neuron.2017.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajikawa Y, Saitoh N, Takahashi T. GTP-binding protein β γ subunits mediate presynaptic calcium current inhibition by GABA(B) receptor. Proc NatlAcad Sci USA. 2001;98:8054–8058. doi: 10.1073/pnas.141031298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klavir O, Prigge M, Sarel A, Paz R, Yizhar O. Manipulating fear associations via optogenetic modulation of amygdala inputs to prefrontal cortex. Nat Neurosci. 2017;20:836–844. doi: 10.1038/nn.4523. [DOI] [PubMed] [Google Scholar]
- Koyanagi M, Terakita A. Diversity of animal opsin-based pigments and their optogenetic potential. Biochim BiophysActa. 2014;1837:710–716. doi: 10.1016/j.bbabio.2013.09.003. [DOI] [PubMed] [Google Scholar]
- Koyanagi M, Kawano E, Kinugawa Y, Oishi T, Shichida Y, Tamotsu S, Terakita A. Bistable UV pigment in the lamprey pineal. Proc Natl Acad Sci USA. 2004;101:6687–6691. doi: 10.1073/pnas.0400819101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyanagi M, Takada E, Nagata T, Tsukamoto H, Terakita A. Homologs of vertebrate Opn3 potentially serve as a light sensor in nonphotor-eceptive tissue. Proc Natl Acad Sci U S A. 2013;110:4998–5003. doi: 10.1073/pnas.1219416110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravitz AV, Freeze BS, Parker PR, Kay K, Thwin MT, Deisseroth K, Kreitzer AC. Regulation of parkinsonian motor behaviours by optogenetic control of basal ganglia circuitry. Nature. 2010;466:622–626. doi: 10.1038/nature09159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzhikandathil EV, Oxford GS. Classic D1 dopamine receptor antagonist R-(+)-7-chloro-8-hydroxy-3-methyl-1-phenyl-2,3,4,5-tetrahydro-1H-3-benzazepine hydrochloride (SCH23390) directly inhibits G proteincoupled inwardly rectifying potassium channels. Mol Pharmacol. 2002;62:119–126. doi: 10.1124/mol.62.1.119. [DOI] [PubMed] [Google Scholar]
- Lafferty CK, Britt JP. Off-Target Influences of Arch-Mediated Axon Terminal Inhibition on Network Activity and Behavior. Front Neural Circuits. 2020;14:10. doi: 10.3389/fncir.2020.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage F, Duprat F, Fink M, Guillemare E, Coppola T, Lazdunski M, Hugnot JP. Cloning provides evidence for a family of inward rectifier and G-protein coupled K+ channels in the brain. FEBS Lett. 1994;353:37–42. doi: 10.1016/0014-5793(94)01007-2. [DOI] [PubMed] [Google Scholar]
- Li X, Gutierrez DV, Hanson MG, Han J, Mark MD, Chiel H, Hegemann P, Landmesser LT, Herlitze S. Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc Natl Acad Sci USA. 2005;102:17816–17821. doi: 10.1073/pnas.0509030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JY, Sann SB, Zhou K, Nabavi S, Proulx CD, Malinow R, Jin Y, Tsien RY. Optogenetic inhibition of synaptic release with chromophore-assisted light inactivation (CALI) Neuron. 2013;79:241–253. doi: 10.1016/j.neuron.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Sinnen BL, Boxer EE, Schneider MW, Grybko MJ, Buchta WC, Gibson ES, Wysoczynski CL, Ford CP, Gottschalk A, et al. A Photoactivatable Botulinum Neurotoxin for Inducible Control of Neurotransmission. Neuron. 2019;101:863–875. doi: 10.1016/j.neuron.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnus CJ, Lee PH, Atasoy D, Su HH, Looger LL, Sternson SM. Chemical and genetic engineering of selective ion channel-ligand interactions. Science. 2011;333:1292–1296. doi: 10.1126/science.1206606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahn M, Prigge M, Ron S, Levy R, Yizhar O. Biophysical constraints of optogenetic inhibition at presynaptic terminals. Nat Neurosci. 2016;19:554–556. doi: 10.1038/nn.4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahn M, Gibor L, Patil P, Cohen-Kashi Malina K, Oring S, Printz Y, Levy R, Lampl I, Yizhar O. High-efficiency optogenetic silencing with soma-targeted anion-conducting channelrhodopsins. Nat Commun. 2018;9:4125. doi: 10.1038/s41467-018-06511-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvaez M, Shieh C, Murphy MD, Greenfeld VY, Wassum KM. Distinct cortical-amygdala projections drive reward value encoding and retrieval. Nat Neurosci. 2019;22:762–769. doi: 10.1038/s41593-019-0374-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis A, Mamidanna P, Cury KM, Abe T, Murthy VN, Mathis MW, Bethge M. DeepLabCut: markerless pose estimation of user- defned body parts with deep learning. Nat Neurosci. 2018;21:1281–1289. doi: 10.1038/s41593-018-0209-y. [DOI] [PubMed] [Google Scholar]
- Nabavi S, Fox R, Proulx CD, Lin JY, Tsien RU, Malinow R. Engineering a memory with LTD and LTP. Nature. 2014;511:348–352. doi: 10.1038/nature13294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niell CM, Stryker MP. Highly selective receptive fields in mouse visual cortex. J Neurosci. 2008;28:7520–7536. doi: 10.1523/JNEUROSCI.0623-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen SF, Liu MH, Kreitzer AC. Thermal constraints on in vivo optogenetic manipulations. Nat Neurosci. 2019;22:1061–1065. doi: 10.1038/s41593-019-0422-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachitariu M, Steinmetz N, Kadir S, Carandini M, Harris KD. Kilosort: realtime spike-sorting for extracellular electrophysiology with hundreds of channels. bioRxiv. 2016 doi: 10.1101/061481. Published online June 30,2016. [DOI] [Google Scholar]
- Pologruto TA, Sabatini BL, Svoboda K. ScanImage: Flexible software for operating laser scanning microscopes. BioMed Eng Online. 2003;2 doi: 10.1186/1475-925X-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimondo JV, Kay L, Ellender TJ, Akerman CJ. Optogenetic silencing strategies differ in their effects on inhibitory synaptic transmission. Nat Neurosci. 2012;15:1102–1104. doi: 10.1038/nn.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo MA, Springer GH, Granada B, Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- Rost BR, Breustedt J, Schoenherr A, Grosse G, Ahnert-Hilger G, Schmitz D. Autaptic cultures of single hippocampal granule cells of mice and rats. Eur J Neurosci. 2010;32:939–947. doi: 10.1111/j.1460-9568.2010.07387.x. [DOI] [PubMed] [Google Scholar]
- Rost BR, Nicholson P, Ahnert-Hilger G, Rummel A, Rosenmund C, Breustedt J, Schmitz D. Activation of metabotropic GABA receptors increases the energy barrier for vesicle fusion. J Cell Sci. 2011;124:3066–3073. doi: 10.1242/jcs.074963. [DOI] [PubMed] [Google Scholar]
- Rost BR, Schneider F, Grauel MK, Wizny C, Bentz C, Blessing A, Rosenmund T, Jentsch TJ, Schmitz D, Hegemann P, Rosenmund C. Optogenetic acidifcation of synaptic vesicles and lysosomes. Nat Neurosci. 2015;18:1845–1852. doi: 10.1038/nn.4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaba T, Neher E. Direct modulation of synaptic vesicle priming by GABA(B) receptor activation at a glutamatergic synapse. Nature. 2003;424:775–778. doi: 10.1038/nature01859. [DOI] [PubMed] [Google Scholar]
- Scanziani M, Capogna M, Gähwiler BH, Thompson SM. Presynaptic inhibition of miniature excitatory synaptic currents by baclofen and adenosine in the hippocampus. Neuron. 1992;9:919–927. doi: 10.1016/0896-6273(92)90244-8. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B. Fiji: an opensource platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachniak TJ, Ghosh A, Sternson SM. Chemogenetic synaptic silencing of neural circuits localizes a hypothalamus/midbrain pathway for feeding behavior. Neuron. 2014;82:797–808. doi: 10.1016/j.neuron.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternson SM, Roth BL. Chemogenetic tools to interrogate brain functions. Annu RevNeurosci. 2014;37:387–407. doi: 10.1146/annurev-neuro-071013-014048. [DOI] [PubMed] [Google Scholar]
- Stujenske JM, Spellman T, Gordon JA. Modeling the Spatiotemporal Dynamics of Light and Heat Propagation for In Vivo Optogenetics. Cell Rep. 2015;12:525–534. doi: 10.1016/j.celrep.2015.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter BA, O’Connor T, Iyer V, Petreanu LT, Hooks BM, Kiritani T, Svoboda K, Shepherd GM. Ephus: multipurpose data acquisition software for neuroscience experiments. Front Neural Circuits. 2010;4:100. doi: 10.3389/fncir.2010.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tecuapetla F, Matias S, Dugue GP, Mainen ZF, Costa RM. Balanced activity in basal ganglia projection pathways is critical for contraver-sive movements. Nat Commun. 2014;5:4315. doi: 10.1038/ncomms5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terakita A. The opsins. Genome Biol. 2005;6:213. doi: 10.1186/gb-2005-6-3-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto H, Terakita A. Diversity and functional properties of bistable pigments. Photochem Photobiol Sci. 2010;9:1435–1443. doi: 10.1039/c0pp00168f. [DOI] [PubMed] [Google Scholar]
- Wiegert JS, Mahn M, Prigge M, Printz Y, Yizhar O. Silencing Neurons: Tools, Applications, and Experimental Constraints. Neuron. 2017a;95:504–529. doi: 10.1016/j.neuron.2017.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegert JS, Gee CE, Oertner TG. Single-Cell Electroporation of Neurons. Cold Spring Harb Protoc. 2017b;2017 doi: 10.1101/pdb.prot094904. [DOI] [PubMed] [Google Scholar]
- Wiegert JS, Gee CE, Oertner TG. Viral Vector-Based Transduction of Slice Cultures. Cold Spring Harb Protoc. 2017c;2017 doi: 10.1101/pdb.prot094896. [DOI] [PubMed] [Google Scholar]
- Wimmer VC, Nevian T, Kuner T. Targetedin vivoexpression of proteins in the calyx of Held. Pflugers Arch. 2004;449:319–333. doi: 10.1007/s00424-004-1327-9. [DOI] [PubMed] [Google Scholar]
- Wu LG, Saggau P. Adenosine inhibits evoked synaptic transmission primarily by reducing presynaptic calcium influx in area CA1 of hippocampus. Neuron. 1994;12:1139–1148. doi: 10.1016/0896-6273(94)90321-2. [DOI] [PubMed] [Google Scholar]
- Yang L, Lee K, Villagracia J, Masmanidis SC. Open source silicon microprobes for high throughput neural recording. J Neural Eng. 2020;17:016036. doi: 10.1088/1741-2552/ab581a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yizhar O, Fenno LE, Davidson TJ, Mogri M, Deisseroth K. Optogenetics in neural systems. Neuron. 2011;71:9–34. doi: 10.1016/j.neuron.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Zhang F, Wang LP, Brauner M, Liewald JF, Kay K, Watzke N, Wood PG, Bamberg E, Nagel G, Gottschalk A, Deisseroth K. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- Zhu H, Roth BL. Silencing synapses with DREADDs. Neuron. 2014;82:723–725. doi: 10.1016/j.neuron.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski Z, Thompson Gray AD, Brady LJ, Page B, Church E, Harris NA, Dohn MR, Yim YY, Hyde K, Mortlock DP, et al. Disabling the Gβγ-SNARE interaction disrupts GPCR-mediated presynaptic inhibition, leading to physiological and behavioral phenotypes. Science. 2019a;12:eaat8595. doi: 10.1126/scisignal.aat8595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski Z, Yim YY, Alford S, Hamm HE. The expanding roles and mechanisms of G protein-mediated presynaptic inhibition. J Biol Chem. 2019b;294:1661–1670. doi: 10.1074/jbc.TM118.004163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Plasmids and viral vectors for expression of eOPN3 are available from Addgene (https://www.addgene.org/Ofer_Yizhar/).
The datasets and the code that support the findings of this study are available from the lead contact upon reasonable request.