Abstract
The efficacy of adoptive cell therapy for solid tumours is hampered by the poor accumulation of the transferred T cells in tumour tissue. Here, we show that the forced expression of the C-X-C chemokine receptor type 6 (CXCR6, whose ligand is highly expressed by human and murine pancreatic cancer cells and by tumour-infiltrating immune cells) in antigen-specific T cells enhanced the recognition and lysis of pancreatic cancer cells and the efficacy of adoptive cell therapy for pancreatic cancer. In mice with subcutaneous pancreatic tumours treated with T cells with either a transgenic T-cell receptor or a murine chimeric antigen receptor targeting the tumour-associated antigen epithelial cell-adhesion molecule, and in mice with orthotopic pancreatic tumours or patient-derived xenografts treated with T cells expressing a chimeric antigen receptor targeting mesothelin, the T cells exhibited enhanced intratumoral accumulation, exerted sustained antitumoral activity and prolonged animal survival only when co-expressing CXCR6. Arming tumour-specific T cells with tumour-specific chemokine receptors may represent a promising strategy for the realization of adoptive cell therapy for solid tumours.
Adoptive T cell therapy (ACT) harnesses the power of tumour-specific T cells for cancer treatment1,2. These T cells can either be directly isolated from cancer patients or obtained by genetic engineering with a tumour antigen-specific T cell receptor (TCR) or chimeric antigen receptor (CAR)1. CAR-engineered T cells have shown promising outcomes in treating hematologic malignancies which led to FDA-approval of anti-CD19-CAR T cells in 2017, representing the first approved T cell therapy3. However, for patients suffering from solid tumours, the therapeutic potential of CAR T cells is still far from being realized4,5. Although anti-tumour effects of CAR T cell against solid tumours have been demonstrated in preclinical models, their efficiency in clinical trials has been limited6–8. Major reasons for this have been extensively reviewed elsewhere and include limited accumulation of T cells resulting from inefficient trafficking and poor local persistence in tumour tissue9–12. So far, efforts to enable ACT in solid tumours have mostly focused on identifying optimal antigens and CAR structures to promote specificity and targeting13. With the identification of immune checkpoint blocking antibodies, efforts have been centred on counteracting the tumour-induced immune suppression by combination therapies. Additional T cell engineering and combination therapies typically demonstrates strong in vitro activity but so far have fallen short of translating into objective clinical response in solid tumours14. We and others have argued that these approaches will only be successful if the modified T cells are able to act in the right place at the right time13,15. If T cells cannot enter or access the cancer site, it is unlikely that any observed in vitro activity against cancer cells would translate into treatment benefits. However, there is only limited work on strategies to specifically improve T cell recruitment to cancer tissues, even though this might be the most critical requirement for ACT efficacy in solid tumours.
Chemokines and their receptors are crucial for the migration and homing of lymphocytes and play a critical role in the development and haemostasis of the immune system. Vital to lymphocyte homing is a multi-step process of rolling and adhesion which results in lymphocyte extravasation and infiltration of healthy and inflamed or diseased tissue16. Besides lymphocytes, many other cell types express chemokines and chemokine receptors which are associated with various biological functions. Tumour tissues utilize mechanisms such as chemokine induction, integrin regulation and activation as well as enhanced tissue permeability both for recruiting accessory immune cells and for their own migration. Similarly, tumours downregulate chemokines that attract cytotoxic cell populations such as CD8+ T cells and Th1 cells to evade the immune system17. Instead, chemokine gradients in tumours attract immune suppressive myeloid or regulatory T cells supporting tumour progression9,17. Previous studies reported that these characteristics of solid tumours can be utilized to enhance trafficking of therapeutic T cells by using chemokine receptors18. However, so far only a few chemokine receptors have been studied in this context, including CCR2, CCR4, CXCR2, CXCR3, CXCR4 and CX3CR1. These studies have demonstrated an enhanced migration of T cells, but only limited additional efficacy as compared to regular tumour targeting approaches15,19–26. Currently, CXCR2 and CCR4 are the only chemokine receptor to have entered clinical trials (NCT 01740557 CXCR2-transduced autologous tumour-infiltrating T cells (TIL), NCT 03602157 combining CD30-specific CAR T cells with CCR4) in 2015 and 2018 but so far, no outcome has been reported.
Within the complex chemokine network, CXCL16 has several unique characteristics: 1) it interacts with only one known cognate receptor, CXCR6; 2) it exists both in a transmembrane form mediating adhesion and a soluble form that acts as a chemoattractant1,27; 3) it is expressed by a variety of cancer cells28; and 4) in pancreatic ductal adenocarcinoma (PDAC), CXCL16 has been reported to contribute to disease progression and patient prognosis29. PDAC is particularly difficult to treat with ACT because of a pronounced desmoplastic reaction together with poor vascularization which limit immune cell infiltration30,31. We hypothesized that the CXCL16-CXCR6 axis is an attractive candidate for enhancing ACT in PDAC because of its dual functionality and the specific interaction of receptor and chemokine. By screening two murine pancreatic cancer models, we identified the chemokine ligand CXCL16 to be highly expressed by pancreatic cancer and myeloid stromal cells, while its receptor CXCR6 was mostly absent from cytotoxic T cells. Transduction of CXCR6 into primary T cells enabled their migration towards CXCL16 both in vitro and in vivo. In addition, CXCR6 on antigen-specific T cells promoted their adhesion to cancer cells, thereby enhancing tumour cell recognition and killing. We demonstrate that co-transducing CXCR6 with a tumour-specific TCR or CAR strongly enhances activity of ACT in subcutaneous cancer models by facilitating T cell influx at the tumour site. High expression of CXCL16 was confirmed in primary human pancreatic cancer tissue, providing a rationale for testing the strategy in human diseases. Arming T cells with both CXCR6 and a mesothelin (MSLN)-specific CAR enabled T cell migration towards CXCL16-producing human pancreatic cancer cells with subsequent tumour cell lysis. Engineered T cells were specifically recruited into pancreatic cancer patient-derived organoids (PDO) and mediated complete tumour rejection in subcutaneous and orthotopic pancreatic cancer xenograft model as well as anti-tumour response in a patient-derived xenograft model (PDX). Improved infiltration of CXCR6-expressing T cells into ovarian cancer resection specimens confirmed the applicability to other solid tumour entities and potential for infiltration into patient tumour tissues. Our study therefore provides a tool enabling the selective recruitment of genetically engineered T cells to cancer tissues expressing CXCL16.
Results
CXCL16 is expressed in murine pancreatic cancer and its receptor CXCR6 is absent from cytotoxic T cells
To identify suitable targets for T cell recruitment to tumours, we analysed RNA expression levels of C-X-C motif chemokines in two syngeneic murine pancreatic cancer models, Panc02 and T110299, which both express the model antigen ovalbumin (OVA) (figure 1a and supplementary figure 1a). CXCL16 was one of the major ligands identified in both models. Its receptor, CXCR6, has been reported to be expressed in a minority (<5 %) of circulating T cells under physiological conditions, but can be up-regulated upon activation32,33. Thus, the CXCL16 – CXCR6 axis was selected for further investigation. We confirmed high expression of the CXCL16 protein in Panc02-OVA and T110299-OVA tumours (figure 1b and supplementary figure 1b). In both tumour models, CXCL16 was most abundantly expressed in the tumour tissue, followed by expression in kidney, lung and lymph nodes. Analyses of plasma revealed a higher concentration of CXCL16 in Panc02-OVA tumour-bearing mice (supplementary figure 1c) and a positive correlation between tumour size and plasma CXCL16 levels (supplementary figure 1d). CXCL16 is produced and secreted by the tumour cells themselves, as spontaneous and inducible CXCL16 secretion was found for both cell lines (figure 1c and supplementary figure 1e). Both, Panc02-OVA and T112099-OVA, showed a higher level of secreted CXCL16 compared to the membranous form, which was further increased after stimulation with IFN-γ (supplementary figure 1f and 1g). Next, CRISPR-Cas9 was used to knock out CXCL16 in Panc02-OVA tumour cells (supplementary figure 1h). In explanted CXCL16-/- Panc02-OVA tumour tissue, the expression of the chemokine was strongly reduced, but not abolished, which indicated additional non-tumour cell sources of CXCL16 within the tumour tissue (figure 1d). Subsequent analysis identified CD11c+ myeloid cells as an additional intratumoral CXCL16 source (figure 1e). This observation was highly relevant for our study, since CXCL16 expressed by infiltrating myeloid cells complements the chemokine gradient produced by tumour cells. Together, these results highlight the CXCR6 – CXCL16 axis as a valuable candidate for targeting ACT.
Fig. 1. CXCL16 is expressed in murine pancreatic tumours and affects CXCR6-engineered T cells.
(a) C-X-C chemokine expression profile of Panc02-OVA tumours quantified by qPCR (n = 6). (b) ELISA evaluating murine CXCL16 protein concentrations in different organs of Panc02-OVA tumour-bearing mice (n = 12). (c) Expression level of murine CXCL16 by Panc02-OVA tumour cells after stimulation with 20 ng/ml IFN-γ, 20 ng/ml TNF-α or a combination of both, quantified using ELISA. (d) CXCL16 levels of CXCL16-knockout Panc02-OVA (n = 10) and CRISPR control Panc02-OVA tumours (n = 15) determined using CXCL16 ELISA. (e) CD11c- and CD11c+ cells were isolated from Panc02-OVA tumour tissue by FACS sorting, and qPCR was used to analyse CXCL16 expression levels of both populations (n = 10 mice). (f) Trans-well migration of GFP- and CXCR6-transduced T cells towards descending concentrations of recombinant murine CXCL16. After 3 h the number of migrated T cells was quantified by flow cytometry. (g) Target cell lysis of CXCL16-overexpressing Panc02-OVA by CXCR6- and GFP-transduced OT-1 T cells following migration through a permeable membrane (suppl. fig.2h). After a migration period of 3 h, migrated T cells and tumour cells were co-cultured for further 1.5 h. (h) ELISA revealing time-dependent activation levels of CXCR6- and GFP-transduced OT-1 T cells upon co-culture with Panc02-OVA tumour cells. E:T ratio 5:1. (i) Panc02-OVA tumour cells were co-incubated with GFP- and CXCR6-transduced OT-1 T cells, and lysis of tumour cells was measured after 6.5 h. (j) Adherence of GFP- or CXCR6-transduced T cells to a CXCL16-coated (9 pmol) or control BSA-coated (9 pmol) surface. As an additional control, T cells were pre-incubated with soluble recombinant CXCL16 (2 μg/ml). (k) Membrane expression of CXCR6 upon stimulation with 200 ng recombinant CXCL16 or CCL1 (arrow) indicating intracellular trafficking and receptor recycling.
In vitro experiments (c, d, f, g, h, I, j) show mean values ± SEM of at least two biological replicates and are representative of three independent experiments (n = 3). p-values are based on two-sided unpaired t-test. Data shown in k are representative for two independent experiments (n = 2). Ex vivo experiments shown are representative of n = 2 (a, d) or n = 3 (b, e). Data shown in e are comprised of two independent experiments (n = 2). Analyses of differences between groups were performed using unpaired Mann-Whitney test.
Transduction of CXCR6 into primary murine TCR-transgenic T cells enables cell migration, adhesion and enhanced recognition of CXCL16 producing pancreatic cancer cells
Based on CXCL16 and CXCR6 expression analyses, we hypothesized that arming antigen-specific cytotoxic T cells with CXCR6 might improve T cell homing into CXCL16-producing tumours and, thus, therapeutic efficacy of ACT. First, we studied the endogenous expression of CXCR6 in murine splenocytes and found the highest expression in TEM cells while <10% of Teff cells expressed CXCR6 (supplementary figure 2a). Splenocytes were activated and either transduced with CXCR6 or GFP as a control resulting in transduction efficiencies of approximately 40-45% (supplementary figure 2b). Transgenic CXCR6 expression was stable and both in vitro and in vivo TCR-activation only led to a minor upregulation of CXCR6 in mCherry control or CXCR6-transduced T cells (supplementary figure 2c and 2d). In a trans-well migration assay, CXCR6-transduced OT-1 T cells specifically acquired the capacity to migrate towards CXCL16 in a dose-dependent manner, whereas control-transduced OT-1 T cells failed to do so (figure 1f). Similar results were obtained when migration was directed towards supernatants of the pancreatic cancer cells Panc02-OVA-CXCL16 and T110299-OVA or the lymphoma cell line E.G7-OVA-CXCL16 (supplementary figures 2e-g). Addition of a neutralizing antibody confirmed the CXCL16-dependence of the migratory effect (supplementary figure 2f). Combining migration and cytotoxicity assays, we found that CXCR6 increased the migration of OT-1 T cells towards CXCL16-expressing tumour cells (supplementary figure 2h), resulting in enhanced target cell lysis, as compared to control-transduced OT-1 T cells (figure 1g). In co-cultures of either Panc02-OVA or T110299-OVA with transduced OT-1 T cells, we found that CXCR6 transduction enhanced and accelerated tumour cell recognition and T cell activation (figure 1h and supplementary figure 2i). This improved recognition resulted in increased target cell lysis by CXCR6-transduced OT-1 T cells compared to control-transduced OT-1 T cells (figure 1i and supplementary figure 2j). As CXCL16 exists as a transmembrane form before shedding and because CXCR6 mediates adhesion to the transmembrane form, we hypothesized that the observed improved cytotoxic effect might be due to enhanced T cell adhesion to the tumour cell. We thus analysed adhesion of CXCR6-transduced T cells to plate-bound CXCL16 in comparison to control-transduced T cells and found an enhanced adhesion of CXCR6-transduced T cells. This effect was specific as pre-incubation with recombinant CXCL16 abolished the adhesive effect (figure 1j). Comparative analysis revealed that the adhesive effect mediated through the CXCR6-CXCL16 axis is superior to the one mediated by the anti-EpCAM-CAR axis. Co-expression of both receptors, CXCR6 and anti-EpCAM-CAR, resulted in a cumulative adhesive effect and improved adhesion to a CXCL16+ EpCAM+ double coated surface (supplementary figure 2k and 2l). We validated this effect by using confocal microscopy. Following co-culture of CXCR6 or control-transduced OT-1 T cells with either Panc02-OVA or T110299-OVA cells, we found a preferential adhesion of CXCR6-transduced T cells to tumour cells (supplementary figure 2m). Again, specificity was confirmed by addition of an anti-CXCL16 neutralizing antibody, which abrogated the differences in adhesion. The phenotype and proliferation of CXCR6-transduced T cells was not affected by in vitro stimulation with recombinant CXCL16 (supplementary figure 8b-e).
The internalization and intracellular trafficking of chemokine receptors upon ligand binding are of major importance for the subsequent cellular response. Therefore, we investigated the dynamics of the receptor after ligand binding to rule out desensitization of the receptor upon engagement. In the presence of an oversaturated concentration of recombinant CXCL16, CXCR6 became rapidly internalized and was thereafter recycled to the cell surface to be re-exposed to the recombinant ligand (figure 1k and supplementary figure 2n). This effect was specific for CXCL16, since the presence of an irrelevant chemokine (CCL1) did not affect the expression of CXCR6 on the cell surface. These results indicate that ligand engagement does not result in durable downregulation of CXCR6 expression as occurs for some other receptor interactions, which might prevent sustained attraction and adhesion of transduced T cells to CXCL16.
Transduction of CXCR6 in murine T cells enhances TCR and CAR efficacy in subcutaneous murine tumour models
To next decipher the in vivo relevance of the above-described findings and the functional advantages of CXCR6-transduced T cells, we treated mice bearing established Panc02-OVA tumours with CXCR6-transduced OT-1 T cells or with control-transduced OT-1 T cells. Mice treated with CXCR6-transduced OT-1 T cells showed a significantly prolonged tumour control with a complete tumour rejection in 2 out of 5 mice (figure 2a and supplementary figure 3a). In contrast, all mice treated with control-transduced OT-1 T cells reached the pre-defined abort criteria due to tumour burden. This effect was mediated by tumour-derived CXCL16, since the therapeutic benefit of CXCR6-transduced OT-1 T cells was lost in mice implanted with CXCL16-/- Panc02-OVA tumours (figure 2b), whereas treatment experiments with CXCL16-expressing CRISPR-control Panc02-OVA tumour cells confirmed the enhanced anti-tumour effect (figure 2c).
Fig. 2. CXCR6-transduced T cells induce tumour regression.
(a) Tumour growth curves of Panc02-OVA-bearing mice with adoptive transfer of 107 GFP- or CXCR6-transduced OT-1 T cells (n= 5 mice per group). T cells were transferred when tumours were palpable (day 5). 2 out of 5 mice treated with CXCR6-transduced T cells showed complete response (CR). (b) C57BL/6 mice inoculated with CXCL16-knockout Panc02-OVA (clone 55) or (c) CRISPR control Panc02-OVA (clone 50) were treated with a single i.v. injection of 107 GFP- or CXCR6-transduced OT-1 T cells (n = 5-12 mice per group). (d) Tumour growth of subcutaneous E.G7-OVA-CXCL16 tumours following treatment with a single injection of 107 mCherry- or CXCR6-transduced OT-1 T cells (n = 4-5 mice per group). (e) Tumour growth of subcutaneous Panc02-OVA-pCAM tumour with adoptive transfer of 107 T cells transduced with either anti-EpCAM-CAR or anti-EpCAM-CAR-CXCR6 (n = 5 mice per group). (f) Tumour growth of subcutaneous Panc02-OVA-EpCAM tumours with adoptive transfer of 107 T cells transduced with anti-EpCAM-CAR, anti-EpCAM-CAR-CXCR3, anti-EpCAM-CAR-CXCR6 or anti-EpCAM-CAR-CCR4 (n = 10-14 mice per group).
Experiments shown are representative of two (b, c, d, e, f) or three independent (a) experiments. Analyses of differences between groups were performed using two-way ANOVA with correction for multiple testing by the Bonferroni method.
The therapeutic efficacy was validated in a second tumour model, the E.G7-OVA-CXCL16 lymphoma model. Here, CXCR6-transduced OT-1 T cells mediated complete tumour rejection in 4 out of 5 mice, and significantly prolonged overall survival (figure 2d and supplementary figure 3b). We next combined transgenic CXCR6 expression in T cells with an anti-EpCAM- CAR for the treatment of Panc02-OVA-EpCAM tumours. While T cells transduced exclusively with the anti-EpCAM-CAR failed to mediate tumour rejection, the combination with CXCR6 mediated prolonged tumour control and tumour rejection in 4 out of 5 mice (figure 2e and supplementary figure 3c).
As different chemokine receptors were shown to improve lymphocyte trafficking in cancer, we next performed a comparative analysis in order to quantify the functional effect of CXCR6 co-expression. We compared the therapeutic activity of anti-EpCAM-CAR-CXCR6 co-transduced T cells with anti-EpCAM-CAR-CXCR3 and anti-Epcam-CAR-CCR4 co-expressing T cells. Both chemokine receptors, CXCR3 and CCR4, were shown to impact tumour-homing of lymphocytes and our data and another study15 revealed the expression of CXCR3 ligands (CXCL4, CXCL9, CXCL10 and CXCL11; figure 1a) and CCR4 ligands (CCL17 and CCL22) in Panc02 tumours15,26. To assure comparability, similar transduction efficiencies for all constructs were ascertained through flow cytometry prior to administration (supplementary figure 3d). In accordance with our previous data, the combination with CXCR6 mediated prolonged tumour control and led to tumour rejection in 4 out of 12 mice. The therapeutic effect mediated by CXCR6 co-expressing CAR T cells was superior to the anti-tumour response of CXCR3 or CCR4 co-expressing CAR T cells, that resulted in tumour rejection in 2 out of 12 mice or no complete response (figure 2f and supplementary figure 3e). After tumour clearance all mice stayed tumour-free till the end of the observation period (100 days). Together these results demonstrate the potency and superiority of CXCR6 to enhance adoptive cell therapy in solid tumour models.
CXCR6 recruits T cells to tumour tissue in vivo
To analyse the underlying mechanisms of CXCR6-transduced T cells in vivo, we performed tracking experiments in tumour bearing mice. In contrast to control-transduced OT-1 T cells, we found a strong accumulation of CXCR6-transduced OT-1 T cells in Panc02-OVA tumours and only a marginal infiltration of other organs including kidney which showed the highest CXCL16 level of all healthy tissues (figure 3a and supplementary figure 4a). We, however, noted an accumulation of CXCR6-expressing T cells in lung tissue and in a lesser extent in Peyer plaques (supplementary figure 4b and 4c). The enhanced tumour homing of CXCR6-expressing T cells was dependent on CXCL16-producing tumour cells, since CXCL16-/- Panc02-OVA tumours showed no significantly enhanced accumulation of CXCR6-transduced OT-1 T cells compared to other tissues indicating that tumour-infiltrating myeloid cells as single source for CXCL16 are not sufficient to substantially improve tumour infiltration (supplementary figure 4d). Next, we studied the contribution of CAR- and CXCR6-signaling for tumour accumulation. As a reference we used T cells expressing a synthetic antigen receptor (SAR) which consists of identical T cell activation domains as the CAR molecule but requires the presence of a bispecific antibody for T cell activation34. After normalization to control SAR T cells, we observed a higher accumulation of CXCR6-expressing T cells compared to anti-EpCAM-CAR T cells in tumour tissue confirming the superiority of CXCR6 for T cell recruitment (supplementary figure 4e). To further characterize the adoptively transferred T cells after in vivo activation, we examined the expression of activation markers, other potentially relevant chemokine receptors, effector molecules and adhesion receptors on control-transduced and CXCR6-transduced tumour-infiltrating OT-1 T cells (supplementary figure 4f). We found a higher expression of the activation markers 4-1BB and IFN-γ in CXCR6-transduced OT-1 isolated from tumour tissue in comparison to control-transduced T cells. Furthermore, CXCR6-transduced OT-1 T cells showed a stronger expression of the chemokine receptors CCR2, CCR5 and CX3CR1 compared to control-transduced T cells. Interestingly, CXCR6-transduced T cells also showed an elevated expression of the adhesion molecule VLA-4 highlighting a potential role of CXCR6 in integrin-mediated adhesion and transendothelial migration.
Fig. 3. CXCR6-transduced T cells are recruited into tumour tissue.
(a) Flow cytometry analysis evaluating the number of OT-1 T cells in Panc02-OVA bearing mice after treatment with CD45.1+ GFP- and CD90.1+ CXCR6-transduced OT-1 T cells in a ratio of 1:1 (n = 5 mice). (b, c) Ex vivo quantification of tumour-infiltrating CXCR6- or GFP-transduced OT-1 T cells in Panc02-OVA tumours by two-photon microscopy (n = 5 mice per group). (d) Flow cytometry analysis quantifying homing of mCherry- and CXCR6-transduced T cells into Panc02 tumours (n = 3 mice per group). (e, f) Before flow cytometry analysis, tumour infiltration and T cell velocity was investigated using DSFC and intravital imaging (n = 4 mice per group). (g) Representative coronal and axial granzyme B PET image taken from Panc02 (left shoulder; white circle) and Panc02-OVA (right shoulder, green circle) tumour-bearing mice treated with either CXCR6-transduced or GFP-transduced (mock) OT-1 T cells. (h) In order to assess granzyme B levels, tracer accumulation in tumour in relation to heart (background radioactivity) was measured (n = 4 mice per group).
Experiments shown are representative of two (d, e, f) or three independent (a, b, c) experiments with n = 3-5 mice per group. Data shown in h are comprised of two independent experiments with n = 4 mice per group. Analyses of differences between groups were performed using unpaired Mann-Whitney test or two-way ANOVA with correction for multiple testing by the Bonferroni method.
We next mapped the distribution of CXCR6-transduced OT-1 T cells compared to control-transduced OT-1 T cells in Panc02-OVA tumours using two-photon microscopy. Here, the improved tumoral accumulation of CXCR6-transduced T cells compared to control-transduced T cells was confirmed (figure 3b and 3c). Intravital live cell tracking experiments confirmed the accumulation of CXCR6-transduced T cell infiltration at the tumour site (figure 3d and 3e). In addition, we found a greater mobility of CXCR6-transduced T cells within the tumour tissue as compared to control-transduced T cells (figure 3f). To further probe the specific accumulation of CXCR6-transduced OT-1 T cells in ovalbumin-expressing tumour tissue, we injected mice with Panc02 cells on the left shoulder and Panc02-OVA cells on the right shoulder. Tumour-bearing mice were treated with CXCR6- or GFP-transduced OT-1 T cells and trafficking and activation of the T cells was monitored by granzyme B PET imaging. After ACT, specific activation of OT-1 T cells in ovalbumin-positive tumours was observed. Mice treated with CXCR6-transduced OT-1 T cells had a higher tracer accumulation in the tumour than mice treated with control-transduced OT-1 T cells indicating an improved tumour-homing and consequently anti-tumour activity of CXCR6-transduced OT-1 T cells (figure 3g and 3h). Model-antigen negative tumours and other tissues (liver, lung and bone marrow) showed no evidence of tracer accumulation, confirming the specificity of the presented study (supplementary figure 4g). Importantly, we did not observe an activation of T cells in the lung, although CXCR6-expressing T cells get trapped in the tissue after i.v. administration. In summary, we found an increased number and activation of CXCR6-transduced T cells in tumour tissue using flow cytometry, confocal and intravital microscopy and granzyme B PET scan indicating the improved tumour homing and consequently enhanced anti-tumour activity.
CXCL16 is expressed by human pancreatic cancer cells and recruits CXCR6-transduced anti-mesothelin CAR T cells for enhanced therapeutic activity of T cells in vitro and in vivo
To translate our findings from murine models into the human system, we first investigated the expression of CXCR6 compared to other chemokine receptors, which have been reported to enhance T cell trafficking. We found CXCR6 to be expressed in <1.5% of CD4+ and <6.5% of CD8+ T cells, whereas no difference between healthy donor and PDAC patient PBMC was detected (supplementary figure 5a and 5b). In addition, we used formalin-fixed paraffin-embedded (FFPE) PDAC specimens to examine the expression of CXCR6 on tumour cells and tumour-infiltrating immune cells and found a limited expression on tumour cells and a heterogeneous expression on immune cells (supplementary figure 5c and 5d). A TCGA database analysis demonstrated an expression of CXCR6 in various tumour entities with some showing higher CXCR6 levels in tumours compared to matched normal tissues (supplementary figures 5e). Further analyses are required to discriminate between CXCR6-positive tumour cells and CXCR6-positive immune cells in these tumours.
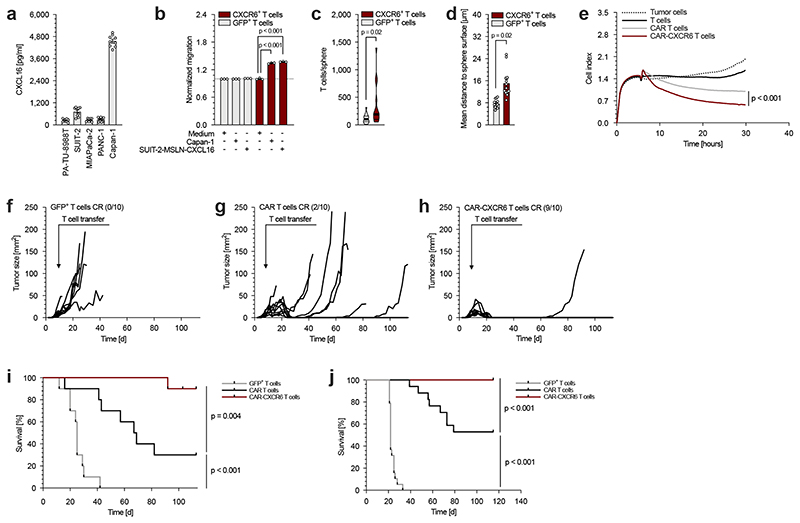
Next, we analysed a panel of pancreatic cancer cell lines for the secretion of CXCL16. All cell lines expressed and secreted CXCL16 at varying levels with the highest secretion by Capan-1 cells (figure 4a), demonstrating their principal amenability to the above strategy. We therefore transduced primary human T cells with human CXCR6 or GFP as a control and tested their ability to migrate towards a gradient of recombinant human CXCL16. CXCR6, but not control-transduced T cells specifically migrated towards the CXCL16 gradient (supplementary figure 5f). Similarly, supernatants of CXCL16-producing SUIT-2 or Capan-1 tumour cells specifically attracted CXCR6-transduced, but not control-transduced T cells (figure 4b). To analyse the ability of CXCR6-transduced T cells to infiltrate tissue, we took advantage of the 3D sphere-forming ability of the cell lines Capan-1 and HEK-CXCL16. In this system, CXCR6-transduced T cells showed enhanced infiltrating abilities and penetrated deeper into tumour spheres compared to control-transduced T cells (figure 4c and 4d and supplementary figure 4g). These findings are consistent with our tracking, two-photon and intravital microscopy data that showed an enhanced accumulation of CXCR6-transduced murine T cells in tumour tissue.
Fig. 4. CXCL16 expressed by human pancreatic cancer cells enhances cytotoxic activity of engineered T cells.
(a) Secretion of CXCL16 by human pancreatic cancer cells was measured by ELISA. (b) Migration capability of GFP- and CXCR6-transduced human T cells toward Capan-1 and MSLN-CXCL16-overexpressing SUIT-2 supernatants. The number of migrated cells was normalized to the medium control condition. (c, d) Number of sphere-penetrating GFP- and CXCR6-transduced human T cells and infiltration depth into spheres formed by HEK overexpressing human CXCL16. (e) In a combined migration-cytotoxicity assay anti-MSLN-CAR and anti-MSLN-CAR-CXCR6-transduced human T cells are compared with regard of specific migratory and cytotoxic efficiency. T cells migrated towards MSLN-CXCL16-overexpressing SUIT-2 tumour cells (Suppl. fig. 6e) followed by CAR-mediated cytotoxicity presented by real-time target cell lysis. (f - i) In a subcutaneous xenograft model, MSLN-CXCL16-overexpressing SUIT-2 tumour bearing mice were treated with GFP-transduced (f), anti-MSLN-CAR-transduced (g) or anti-MSLN-CAR-CXCR6 co-transduced T cells (h). Tumour growth and survival was measured over 110 days (n = 10 mice per group). One mouse of the anti-MSLN-CAR-CXCR6 treated group was sacrificed on day 103 post tumour injection due to (unclear) neck swelling, presumably unrelated to subcutaneous tumour injection and was therefore censored at the timepoint. (i). (j) For orthotopic treatment experiments, SUIT-2-MSLN-CXCL16 tumour cells were implanted into the pancreas. Mice were treated with a single i.v. injection of either non-transduced human T cells, anti-MSLN-CAR- or anti-MSLN-CAR-CXCR6-transdcued T cells. Tumour growth and survival was monitored over 115 days (n = 17-20 mice per group).
In vitro experiments (b, e) show mean values ± SEM of at least two biological replicates and are representative of three independent experiments (n = 3). Data shown in a, c and d are comprised of three independent experiments (n = 3) each with three biological replicates. In vivo experiments (f – j) are summarized from two independent experiments. p-values are based on two-sided unpaired t-test or two-way ANOVA with correction for multiple testing by the Bonferroni method. Comparison of survival rates was performed with the Log-rank (Mantel-Cox) test.
To test the therapeutic potential in human T cells, we co-transduced T cells with CXCR6 and with a mesothelin-specific CAR. Activation and CAR transduction did not affect endogenous CXCR6 expression levels and both constructs, CAR and CAR-CXCR6, were stably expressed in primary human T cells (supplementary figure 6a-d). Co-expression of CXCR6 enhanced the migration of anti-MSLN-CAR T cells towards recombinant human CXCL16 gradients (supplementary figure 6e), although it did not improve their activation or lytic potential towards MSLN-CXCL16-expressing SUIT-2 tumour cells (supplementary figure 6f and 6g). However, when the lytic potential was analysed together with the enhanced migration (by combining migration and lysis in one assay), anti-MSLN-CAR-CXCR6 co-transduced T cells both migrated towards MSLN-CXCL16-expressing SUIT-2 tumour cells (supplementary figure 6h) and also mediated enhanced lysis compared to anti-MSLN-CAR-transduced T cells (figure 4e). To further characterize CAR-transduced and CAR-CXCR6 co-transduced T cells, we studied the Ca2+ influx in those cells after interaction with CXCL16-expressing tumour cells. CAR-CXCR6 co-expressing T cells showed an increased initial Ca2+ influx compared to CAR T cells whereas the Ca2+ level at the plateau was not affected, confirming signal transduction through transgenic CXCR6 and involvement in T cell activation kinetics previously observed in murine cells (figure 1h and supplementary figures 2i).
Next, we assessed the functional relevance of these findings by inducing subcutaneous MSLN-CXCL16-overexpressing SUIT-2 tumours in immune compromised NSG mice. After the tumours were established, mice were treated once with either control-transduced, anti-MSLN-CAR-transduced or anti-MSLN-CAR-CXCR6 co-transduced T cells. All tumour-bearing mice treated with control-transduced T cells reached the pre-defined abort criteria due to tumour burden within 42 days (figure 4f and 4i). In the anti-MSLN-CAR-transduced T cell group, 8 out of 10 mice relapsed and 7 died due to the tumour burden (figure 4g and 4i). In contrast, 9 out of 10 mice treated with anti-MSLN-CAR-CXCR6 co-transduced T cells fully rejected the tumour and remained tumour-free throughout the 100 days observation period (figure 4h). As a consequence, the survival of mice treated with anti-MSLN-CAR-CXCR6 co-transduced T cells was significantly prolonged, indicative of the transformative potential of our strategy (figure 4i). To validate the enhanced tumour-homing of anti-MSLN-CAR-CXCR6 co-transduced T cells compared to anti-MSLN-CAR-transduced T cells, we quantified the number of tumour-infiltrating CAR T cells after ACT. We found higher numbers of anti-MSLN-CAR-CXCR6 co-expressing CD4+ and CD8+ T cells in the tumours (supplementary figure 6j and 6k), demonstrating the effectiveness of CXCR6 to improve homing to solid tumours and confirming our finding in the syngenic models.
To substantiate the clinical relevance of the current study, we used an orthotopic pancreatic cancer xenograft mouse model. Five days following implantation of MSLN-CXCL16-expressing SUIT-2 tumour cells into the pancreas, mice were treated with i.v. injection of anti-MSLN-CAR-transduced, anti-MSLN-CAR-CXCR6 co-transduced or non-transduced human T cells and the survival of the mice was monitored. Animals were sacrificed at signs of disease such as weight loss, behavioural or physiological changes and survival data were plotted in a Kaplan-Meier survival curve (figure 4j). All mice treated with non-transduced T cells had to be sacrificed within 33 days after tumour implantation. Treatment with anti-MSLN-CAR-transduced T cells led to improved survival and tumour remission in 9 out of 17 mice. All 20 mice treated with anti-MSLN-CAR-CXCR6 co-transduced T cells showed tumour rejection and long-term remission.
CXCL16 is expressed by tumour cells and infiltrating immune cells in primary pancreatic cancer tissue and mediates enhanced attraction of CXCR6-transduced T cells into pancreatic cancer patient-derived organoids (PDO) and xenografts (PDX)
Assessing CXCL16 gene expression levels in pancreatic cancer cells, as compared to healthy pancreatic tissue, we found a specific up-regulation of CXCL16 in the patient cohort (n = 36 patients with PDAC compared to n = 12 healthy controls) (figure 5a). These results were corroborated by a TCGA database analysis (n = 178 patients with PDAC compared to n = 165 healthy controls) further highlighting the tumour-associated expression of CXCL16 (figure 5b). In an additional cohort of pancreatic cancer patients (n = 399), immunohistochemical analysis revealed CXCL16 expression in 66.9% of analysed tumours. Both, CXCL16-positive tumour cells as well as CXCL16-positive immune cells were detected in the tumour tissue (figure 5c and 5d and supplementary figure 7a). It should be noted that only a considerably low number of CXCL16-positive tumour cells were detected in IHC staining, which most likely is due to the low sensitivity of the anti-CXCL16 antibody, since SUIT-2 overexpressing CXCL16 also showed a weak staining (supplementary figure 8a). For this reason and in order to further characterize tumour-infiltrating CXCL16-positive immune cells, we analysed previously published single cell RNA (scRNA) sequencing data. These data confirmed hat, besides malignant ductal cells, macrophages are a main source of intratumoral CXCL16 (figure 5e and supplementary figure 7b), which is in line with our observation of CD11c+ myeloid cells in CXCL16-/- Panc02-OVA tumour tissue (figure 1e). Importantly, healthy ductal and acinar cells of the pancreas showed no or low CXCL16 expression suggesting an intratumoral rather than peritumoral accumulation of CXCR6-expressing T cells. To assess the potential of misguidance, we analysed additional scRNA seq data sets of healthy human tissues that we had previously found to express CXCL16 (figure 1b and supplementary figure 1b). This analysis revealed that myeloid-derived cells, especially monocytes, macrophages and dendritic cells, are major sources of CXCL16 in lung, lymph node and kidney (supplementary figure 7c).
Fig. 5. CXCL16 is expressed by PDAC specimens and attracts CXCR6 transduced T cells.
(a) Gene expression analysis (mRNA) of pancreatic cancer specimens in comparison to healthy pancreatic tissue (n = 36 PDAC patients and n = 12 healthy controls) using NanoString nCounter® System. (b) TCGA data analysis comparing CXCL16 expression (mRNA) by PDAC and healthy tissue (n = 178 PDAC patients and n = 165 healthy controls). (c) Representative images of PDAC specimens stained for CXCL16. (d) Quantification of CXCL16 expression by tumour cells and tumour-infiltrating immune cells validated by immunohistochemical staining of CXCL16 in PDAC specimens (n = 399 with three biopsies per patient). (e) Single cell RNA (scRNA) sequencing analysis of PDAC and healthy pancreas tissue comparing CXCL16 expression levels. (f) CXCL16 levels in plasma of PDAC patients and healthy specimens quantified by ELISA (n = 10 PDAC patients and n = 11 healthy specimens). (g) Activation level of human T cells (quantified by IFN-γ concentrations) following co-culture with pancreatic cancer PDO (summarized data from independent co-cultures with 3 different PDO specimens: B34, B54 and B61; n = 3). (h) Representative images showing confocal analysis of pancreatic cancer PDO (specimen B61) infiltrated by GFP- or CXCR6-transduced T cells. (i) Quantification of GFP- and CXCR6-transduced T cells penetrating into PDO (summarized from specimen B34 and B48; n = 2) in the absence or presence of an anti-CXCL16 neutralization antibody. (j) For PDX experiments, PDO (MGH1247) were heterotopically implanted in NCG mice and treated with non-transduced, anti-MSLN-CAR or anti-MSLN-CAR-CXCR6-transduced T cells. Tumour growth was monitored for 35 days post ACT (n = 5 mice per group). (k) PDX tumour weight after treatment with either non-transduced, anti-MSLN-CAR or anti-MSLN-CAR-CXCR6-transduced T cells. (l) Quantification of GFP- and CXCR6-transduced T cells after penetration into surgical ovarian cancer (OC) specimens of seven patients.
Analyses of differences between groups in a, b and f were performed using unpaired Mann-Whitney test. Data shown in g are comprised of three independent experiments (n = 3). Data shown in i are comprised of two independent experiments with mean values ± SEM of at least 10 organoids per condition (i; n = 2). p-values are based on two-sided unpaired t-test. Data shown in j, k and l resulted from one single experiment (n = 1). Differences in tumour growth were analysed by using two-way ANOVA with correction for multiple testing by the Bonferroni method and differences in tumour weight were analysed by using unpaired Mann-Whitney test. Data shown in l are mean values ± SEM of seven OC specimen co-cultures with the same T cell donor. p-vales in k are based Wilcoxon signed-rank test.
As shown in figure 5a and 5d, there is substantial interpatient heterogeneity in CXCL16 expression, therefore the quantification of CXCL16 in plasma and tumour tissue is crucial to predict the therapeutic benefit of CXCR6-engineered tumour-specific T cells. By using ELISA, we found elevated CXCL16 plasma levels in PDAC patients in comparison to healthy donors. This makes CXCL16 a convenient companion biomarker and represents an important addition to IHC analysis (figures 5f).
In a further analysis, we demonstrated that CXCL16 is produced in various concentrations by pancreatic cancer PDO (supplementary figure 7d). Co-culture experiments of PDO and anti-MSLN-CAR-transduced or anti-MSLN-CAR-CXCR6 co-transduced T cells resulted in effective T cell activation for all patients tested (with n = 3 PDAC patients) (figure 5g). When we examined the migratory capacity of CXCR6-transduced T cells towards pancreatic cancer PDO, we found an efficient penetration of CXCR6-expressing T cells into these organoids compared to control-transduced T cells (figure 5h and supplementary figure 7e). Specificity of migration into PDO was demonstrated through addition of a neutralizing antibody. In the presence of CXCL16-neutralizing antibodies, the superior migratory potential of CXCR6-transduced T cells was abolished, and the number of penetrating T cells was comparable to control-transduced T cells (figure 5i). Finally, to demonstrate the transferability of the concept to a more clinically relevant setting, we heterotopically implanted patient-derived xenograft (PDX) tumours that express CXCL16 and mesothelin into NCG mice and transferred anti-MSLN-CAR-transduced or anti-MSLN-CAR-CXCR6 co-transduced T cells, when the tumour volume reached >60 mm3 (supplementary figure 7f and 7g). Compared to anti-MSLN-CAR T cells, treatment with anti-MSLN-CAR-CXCR6 co-transduced T cells resulted in reduced tumour growth and enhanced tumour control (figure 5j). Consequently, when terminating the experiment, mice treated with anti-MSLN-CAR-CXCR6 co-transduced T cell had substantially smaller tumours than those treated with anti-MSLN-CAR-transduced T cells (figure 5k). Together these data indicate the translational potential of CXCR6 as an enhancer of CAR T cell activity in pancreatic cancer.
Moreover, we argue that the described strategy may have a broad applicability, since a TCGA database analysis revealed expression of CXCL16 in various tumour entities such as ovarian cancer (supplementary figure 7h). To prove that the strategy is amenable to multiple disease and to provide direct evidence for penetration capabilities into patient tissue, we took advantage of a method allowing the use of tissue explants. CXCR6-transduced and as a control GFP-transduced primary human T cells were co-cultivated with unprocessed surgical resection specimens of ovarian cancer patients. Quantification of tissue-infiltrating T cells was performed on whole slide sections and showed a strong increase of CXCR6-transduced T cells compared to control-transduced T cells (figure 5l and supplementary figure 7i). Importantly, this finding demonstrates the broad applicability of CXCR6 to enhance recruitment of T cells into primary patient tissue.
Discussion
Together our results demonstrate that the addition of the chemokine receptor CXCR6 to both TCR- and CAR-based cell therapies increases antitumour efficacy in murine and human models of pancreatic cancer as well as in pancreatic cancer patient-derived organoids and xenograft models. Additionally, CXCR6-equipped T cells are able to infiltrate primary patient tissue. This therapeutic benefit is mainly driven by the enhanced access of T cells to the tumour tissue and appears to be promoted by the adhesive effect of the CXCL16-CXCR6 axis. The chemokine receptor CXCR6 might thus be a universal tool to enable ACT in CXCL16-expressing cancers.
Trafficking of T cells to the tumour tissue is one of the most critical requirements for ACT efficacy in solid tumours18. Approaches that have been pursued to enhance T cell infiltration upon ACT include total body irradiation35, the administration of bispecific antibodies36 or anti-angiogenic therapies37. While these methods increase the efficacy of ACT to some degree, none of them markedly and – more importantly - specifically enhance T cell migration and trafficking to tumour tissue. To date, only few chemokine receptors, including CCR2, CCR4, CXCR2, CXCR3, CXCR4 and CX3CR1, have been studied to enhance T cell trafficking and thus ACT in solid tumours15,19–26. For pancreatic cancer, we previously reported an improved anti-tumour efficiency of CCR4 co-expressing T cells and more recently the combination of radiation therapy and CAR-CXCR1 or CAR-CXCR2 co-expressing T cells has been reported to improve ACT in a preclinical PDAC model15,38. In this study, local ionizing radiation was used as a pre-treatment to enhance the tumoral chemokine production whereas our approach utilizes the physiologic CXCL16 gradient.
Low infiltration of PDAC tissue by lymphocytes is attributed to a profoundly desmoplastic stroma with a large proportion of extracellular matrix (ECM) and a high number of immune suppressive fibroblasts39,40. In context with our approach, the ECM could potentially lead to a contact guidance-dependent inactivation of the chemokine-induced migration as reported by other groups41. Although we utilized various in vitro and in vivo models, the lack of models with high levels of desmoplasia is a limitation of our study. Nevertheless, we were able to confirm our findings in a PDX model and minimally processed tissue explants which most closely reflect actual tumour structures. Future studies may combine CAR-CXCR6 co-transduced T cells microenvironment-targeting agents, such as all-trans retinoic acid (ATRA)42, Nab-paclitaxel43 or other innovative cell-based approaches (e.g. anti-FAP CAR T cells44) to address this issue.
Among chemokines, CXCL16 is one of the few that exists both in a secreted and transmembrane form45. Accordingly, it not only functions as a chemoattractant but also mediates cell-cell adhesion46. These properties identify CXCL16 as an attractive mediator for enhancing ACT, as it promotes two important functions in the efficacy of T cell therapies – increased recruitment as well as strengthened cell-cell interactions between tumour cells and cytotoxic T cells. Interestingly, transmembrane CXCL16 has been described to be expressed on activated endothelial cells of the vasculature and the CXCL16-CXCR6 axis seems to be involved in adhesion of PBMCs to endothelium and their recruitment into tissues11,47. Transgenic expression of CXCR6 in T cell may therefore result in similar effects. This hypothesis is supported by the up-regulation of VLA-4 in CXCR6-expressing cells, which is a key integrin involved in transendothelial migration of lymphocytes48, indicating that the CXCL16-CXCR6 axis is not only important for the adhesion of lymphocytes to endothelium but also for their transendothelial migration.
Under steady state conditions, CXCR6 is mostly absent from peripheral CD8+ T cells but is expressed on T cells in peripheral tissues, in certain pathologies or upon exposure to defined stimuli49. In cancer, however, CXCR6 has been shown to not only facilitate infiltration of suppressive immature myeloid cells and regulatory T cells (Treg) but also to promote migration of the cancer cells themselves50–52. In pancreatic cancer, previous reports suggested that the CXCR6 - CXCL16 axis is important for tumour progression29,53. These important functions in immune suppression and cancer biology reduce the likelihood that the cancer tissue might lose CXCL16 expression upon therapy, a vital requisite for our approach. Using scRNA sequencing analysis, we found CXCL16-expressing tumour-infiltrating macrophages besides malignant ductal cells as a source of CXCL16 which further supports the aforementioned hypothesis. In addition, CAFs have been reported to secrete chemokines which could be an alternative intratumoral source of CXCL16, although we could not verify this observation in our scRNAseq analysis which might be due to technical reasons54. Importantly, our scRNA sequencing analysis revealed no or only low CXCL16 expression in healthy ductal cells, an essential observation supporting the idea of enhanced intratumoral accumulation of CXCR6-expressing T cells. Previous studies, however, found a strong CXCL16 positivity of inflamed peritumoral tissue in PDAC and chronic pancreatitis specimens29. This observation could potentially lead to a misguidance of CXCR6-expressing T cells which has not been observed in our studies.
It is important to note that CXCL16 is expressed by a number of other solid cancer entities including ovarian, lung and breast cancer52,55. Our TCGA data base analysis confirms up-regulation of CXCL16 in several solid tumour indications, pointing towards additional entities accessible for ACT using CXCR6-expressing tumour antigen-specific T cells. The effective infiltration of CXCR6-expressing T cells into ovarian cancer resection specimens, emphasizes the pan-cancer translation of CXCR6 expression to improve T cell trafficking.
We could recapitulate a physiological function of CXCR6 through transduction: overexpression of CXCR6 in T cells enhanced adhesion to CXCL16-expressing tumour cells and thereby recognition and lysis. Importantly, the adhesive effect mediated by the CXCL16-CXCR6 axis was superior to the CAR-mediated adhesion and the expression of both receptors led to an additive effect. Under therapeutic settings, enhanced adhesion might facilitate the recognition of tumour cells, especially of cells expressing lower levels of the CAR-target antigen. Avidity to the target cell might then enhance activation and lysis capabilities of CAR T cells, as seen for polyvalent antibodies56. At the same time, the enhanced velocity of CXCR6-expressing T cells without tumour-specificity that we observed might compensate for unwanted and perturbing immobilization. These points, however, require further investigation.
TCR- and CAR-based strategies are the most advanced ACT modalities, with anti-CD19 CAR T cells being the first approved T cell therapy for cancer1. A promising target for solid tumours is mesothelin (MSLN) and the potential of anti-MSLN CAR T cells for the treatment of multiple solid cancers is currently investigated in several preclinical and clinical studies57. The anti-MSLN-CAR used in the present study is based on a clinical CAR candidate: the anti-MSLN-CAR with an SS1-antibody backbone is currently investigated in clinical trials for the treatment of pancreatic ductal adenocarcinoma (including NCT01583686 or NCT01355965). In the latter study, anti-MSLN-CAR T cell treatment resulted in stable disease in three out of six patients and in a partial response for one patient58. Co-transduction of CXCR6 and anti-MSLN-CAR into T cells may overcome ACT limitations observed for PDAC by improving tumour homing of CAR T cells, a prerequisite for anti-tumour efficacy. Furthermore, generation of TIL can be achieved in a variety of cancer entities, including pancreatic and ovarian cancer59,60. As demonstrated for other chemokine receptors, we hypothesize, that genetic modification of TIL to express CXCR6 may be clinically relevant to optimize TIL trafficking in these malignant diseases. Currently, the phase I/II trial study of CXCR2-modified TIL for treating metastatic melanoma patient is exploring this question (NCT01740557).
TCGA and scRNA data analysis revealed expression of CXCL16 in healthy tissue, especially in testis, kidney and lung, indicating the possibility of misguidance of CXCR6-modified T cells to healthy tissues. Additionally, CXCR6 is involved in lymphocyte migration into inflamed tissues in, for example, arthritis or inflamed liver30,61. Accordingly, we found a higher number of CXCR6-expressing T cells in lung tissue after i.v. administration, although we did not observe an accumulation of CXCR6-expressing T cells in kidneys, which showed the highest CXCL16 levels of healthy tissues. It is therefore crucial to combine CXCR6 with a highly tumour-specific TCR or CAR to ensure activation of transduced T cells exclusively in the tumour tissue and to minimize the risk of off-target accumulation, which would dampen the therapeutic response. Along these lines, analysing T cell activation by PET imaging, we found selective antigen-dependent activation of T cells only at the site of antigen-positivity and no T cell activation in non-tumour tissue, including lung, or antigen-negative tumour sites. These findings are a strong argument for the safety of CXCR6-coexpression, since T cell activation and possible toxicity is mainly regulated by the expression of the CAR or TCR. At the same time, this highlights the dependence of our approach on a suitable immune target, which together with limited trafficking and immune suppression is a major challenge for ACT of solid tumours. In fact, several clinical trials with CAR T cell therapy in pancreatic cancer were recently completed and are currently ongoing, as summarized in ref. 62.
In summary, our study provides a rationale for the further development and testing of CXCR6 as a universal receptor promoting migration and cell-cell interactions for the T-cell-based treatment of pancreatic cancer and other solid tumours refractory to T-cell treatments.
Methods
Cell lines
The ovalbumin overexpressing murine pancreatic cancer cell line Panc02-OVA, a chemically induced pancreatic cancer cell line, and the murine lymphoma cell line E.G7-OVA have previously been described63,64. Panc02-OVA-CXCL16 and E.G7-OVA-CXCL16 were generated by transduction with pMXs vector containing the full length murine CXCL16 sequence (UNIPROT entry Q8BSU2). To generate the Panc02-OVA-EpCAM cell line overexpressing the murine epithelial cell adhesion molecule EpCAM, Panc02-OVA tumour cells were stably transduced with the pMXs vector containing the full murine EpCAM sequence (UNIPROT entry Q99JW5). The ovalbumin-overexpressing murine cell line T110299-OVA, a cell line derived from a primary tumour of Kras- and p53-mutant KPC mice, has previously been described (obtained from Prof Siveke, Essen, Germany). For multi-photon intra-vital microscopy, Panc02 tumour cells were transduced with pMP71 containing a fusion of histone H2B to cerulean fluorescent protein. For the generation of CXCL16-knockout Panc02-OVA, the CRISPR/Cas9 system was used targeting exon 2 (gRNA sequence 5’ – 3’ ACTTCCAGCGACACTGCCCTGG) of the murine CXCL16 gene. Efficient gene knockout of single cell clones was validated by genome sequencing and CXCL16 ELISA after stimulation with IFN-γ. As a CRISPR control single cell clones with an insufficient CXCL16 gene knockout were used. The human pancreatic cancer cell lines SUIT-2 was obtained from K. Lauber, Munich, Germany. PA-TU-8988T (DSMZ: ACC 162), MIA PaCa-2 (ATCC: CRL-1420), PANC-1 (ATCC: CRL-1469), Capan-1 (ATCC: HTB-79), Flp-In™ 293 (Thermo Fisher, USA) and the human lung cancer cell line H3122 (ATCC: CRM-CLL-119) were purchased. SUIT-2-MSLN were generated by transduction of SUIT-2 with pMXs containing full length human MSLN (UNIPROT entry Q13421). Flp-In™ 293-CXCL16 (HEK-CXCL16) and SUIT-2-MSLN-CXCL16 were generated by transduction with pMXs containing full length human CXCL16 (UNIPROT entry Q9H2A7). In case of SUIT-2-MSLN-CXCL16, single cell clones were generated and one MSLN- and CXCL16-positive clone was used for further experiments. The Platinum-A and Platinum-E packaging cell lines were purchased from Cell Biolabs Inc. (Hoelzel Diagnostika, Cologne, Germany). 293Vec-Galv, 293Vec-Eco and 293Vec-RD114 were a kind gift of Manuel Caruso, Québec, Canada and have been previously described65. For virus production, retroviral pMP71 vectors carrying the sequence of the relevant transgene were stably introduced in packaging cell lines. Single cell clones were generated and indirectly screened for highest level of virus production by determining transduction efficiency of primary T cells. This method was used to generate the producer cell lines 293Vec-RD114-GFP, 293Vec-RD114-CXCR6, 293Vec-RD114-anti-MSLN-CAR-CXCR6, 293Vec-RD114-anti-MSLN-CAR, 293Vec-Eco-GFP, 293Vec-Eco-mCherry, 293Vec-Eco-CXCR6, 293Vec-Eco-anti-EpCAM-CAR, 293Vec-Eco-anti-EpCAM-CAR-CXCR6, 293Vec-Eco-anti-EpCAM-CAR-CCR4 and 293Vec-Eco-anti-EpCAM-CAR-CXCR3. All cells, with the exception of E.G7-OVA and E.G7-OVA-CXCL16, were cultured in DMEM with 10% fetal bovine serum (FBS, Life Technologies, USA), 1% penicillin and streptomycin (PS) and 1% L-glutamine (all from PAA, Germany). 10 μg/ml puromycin and 1 μg/ml blasticidin (Sigma, Germany) selection antibiotics were added to the Plat-A or Plat-E medium. Producer cell lines were cultured in DMEM with 10% FBS, 1% PS and 2% L-glutamine. E.G7 (derivate of EL4) and E.G7- OVA-CXCL16 as well as primary murine T cells were cultured in RPMI 1640 (Lonza, Basel, Switzerland) containing 10% FBS, 1% PS, 1% L glutamine, 1% sodium pyruvate and 1 mM HEPES (T cell medium TCM). 50 μM β-mercaptoethanol and 1 mg/ml IL-15 were added to murine TCM immediately when culturing primary murine T cells. Primary human T cells were cultured in VLE-RPMI 1640 (Biochrom, Germany) containing 2.5% human serum, 1% PS, 1% L-glutamine, 1% NEAA, 1% sodium pyruvate (human TCM). 50 μM β-mercaptoethanol, 1 μg/ml IL-2 and 100 μg/ml IL-15 were added to human TCM immediately when culturing the T cells.
All cell lines used in experiments were regularly checked for contamination with Mycoplasma using the MycoAlert Mycoplasma Detection Kit (Lonza). Authentication of human cell lines by STR DNA profiling analysis was conducted in house.
Animal experiments
Wild type C57Bl/6RJ mice were purchased from Janvier (St Bertevin, France) or Charles River (Sulzfeld, Germany). C57Bl/6RJ mice transgenic for an ovalbumin-specific T cell receptor (OT-1) were purchased from The Jackson Laboratory, USA (stock number 003831). OT-1 mice were crossed with CD45.1 or CD90.1 congenic marker mice (obtained from The Jackson Laboratory, stock number 002014 or as a kind gift from R. Obst, Munich, Germany). NSG mice (NOD.Cg-Prkdcscid Il2rgtm1WjI/SzJ; stock number 005557) were purchased from Charles River (Sulzfeld, Germany) or bred within the local animal facility (Zentrale Versuchstierhaltung Innenstadt). Animal experiments were approved by the local regulatory agency (Regierung von Oberbayern) or the MGH Institutional Animal Care and Use Committee (IACUC) and adhere to the NIH guide for the care and use of laboratory animals.
Tumours were induced by subcutaneous injection of 2 x 106 Panc02-OVA, Panc02-OVA-EpCAM, Panc02-OVA-CXCL16-/- or Panc02-OVA-CRISPR control, 0.5 x 106 E.G7- OVA-CXCL16 or 4 x 106 T110299-OVA. For treatment experiments mice were i. v. injected with 107 T cells when tumour was palpable. For tracking experiments, mice were i. v. injected with 107 T cells, but here equal numbers of CD90.1+ CXCR6-transduced and CD45.1+ GFP-transduced OT-1 T cells (1:1 ratio) or CXCR6-transduced and mCherry-transduced OT-1 T cells (1:1 ratio) were co-injected in one mouse. Alternatively, equal numbers (1:1 ratio) of SAR-transduced T cells to CXCR6- or CAR-transduced T cells were co-injected i.v. into mice bearing Panc02-OVA-EpCAM tumours. To investigate production of secretory proteins in in vivo activated T cells, mice were i.p. injected with 250 μg brefeldin A (Sigma Aldrich) five hours prior to euthanasia. For multi-photon intravital microscopy, H2A-Cerulean fluorescent protein expressing Panc02 tumour cells were implanted in the back of mice after removal of hair. Engrafted tumours were framed within a dorsal skin-fold chamber, implanted by means of an aseptical surgical procedure under anaesthesia. In order to compare tumour homing, CXCR6-GFP-transduced T cells were co-injected with mCherry control-transduced T cells. For the identification of intravascular or intratumoral localization of traced T cells, blood vessels were stained by injecting mice intravenously with 10 μl Qdot 655 prior to imaging. Mice were monitored daily for tumour growth as well as for pain and local or systemic inflammatory signs. For Granzyme B PET imaging, C57Bl/6 mice were depleted of endogenous CD8+ T cells using a depleting antibody (YTS169.4) prior to implantation of 2 x 106 Panc02 cells on the left shoulder and the same number of Panc02-OVA cells on the right shoulder. Seven days following tumour implantation, separate groups of mice were injected intravenously with 100 μl of either CXCR6- transduced OT-1 T cells or GFP-transduced OT-1 T cells diluted at 108 cells/ml in PBS.
For the xenograft tumour model, 106 SUIT-2-MSLN-CXCL16 tumour cells were injected subcutaneously into NSG mice. When the tumour was established (9 days post tumour injection), mice were treated by i. v. injection of 107 T cells. Orthotopic tumours were implanted as described before66. Briefly, NSG mice were anaesthetized and a small surgical incision was made to mobilize the pancreas. Following injection of 5 x 105 SUIT-2-MSLN-CXCL16 in 25 μl PBS, the pancreas was re-located, and the incision was closed. Five days after tumour implantation, the mice were treated by i.v. injection of 107 T cells. For tracking experiments, 106 SUIT-2-MSLN-CXCL16 were injected subcutaneously into NSG mice. When the tumour size was >10 mm2, 6 x 106 anti-MSLN-CAR-CXCR6 or anti-MSLN-CAR transduced T cells were injected into the tail vein. 5-7 days after ACT, mice were sacrificed and the number of tumour-infiltrating CAR-positive T cells was quantified by flow cytometry.
For PDX studies, patient-derived xenograft tumours of MGH1247 were employed. MGH1247 contains mutations in genes frequently altered in PDAC, including those that alter KRAS (G12D) and TP53 (Y181C). Expression of CXCL16 and MSLN was confirmed by RNA sequencing and MSLN expression was further validated using IHC. Fifth passage tumours were harvested and approximately 40 mg of Matrigel-coated tumour pieces were implanted heterotopically into NOD-Prkdcem26Cd52Il2rgem26Cd22/NjuCrl (NCG) mice. After 22 days of tumour growth, mice were randomly distributed into three experimental arms: non-transduced T cells (n = 5), anti-MSLN-CAR transduced T cells (n = 5), and anti-MSLN-CAR-CXCR6 co-transduced T cells (n = 5). Mice were injected with 107 T cells resuspended in 100 μl PBS through the tail vein. Equivalent viability and transduction efficiency between T cell populations was determined prior to injections.
All studies are conducted randomized, blinded and with adequate controls. In accordance with the animal experiment application, tumour growth and health status of mice were monitored every other day.
Generation of fusion constructs and chimeric antigen receptors
All constructs were generated by overlap extension PCR and recombinant expression cloning into the retroviral pMP71 vector. CXCR6-GFP consists of the full length murine CXCR6 (UNIPROT entry Q9EQ16 amino acids 1-351) fused to GFP via a self-cleaving 2A sequence, hereinafter referred to as CXCR6. The human CXCR6-GFP consists of the full length human CXCR6 (UNIPROT entry O00574 amino acids 1-342) fused to GFP via a self-cleaving 2A sequence. The anti-EpCAM-CAR construct consists of a single-chain variable fragment that recognizes the murine EpCAM antigen (clone G8.8), fused to the transmembrane and signalling domains of the murine T cell co-stimulatory receptor CD28 (UNIPROT entry P31041 AA 151-218) and the cytoplasmic signalling domain of the murine zeta chain of the TCR/CD3 complex (UNIPROT entry P24161 AA 52-164). The anti-EpCAM-CAR-CXCR6 consists of the anti-EpCAM-CAR fused to full length murine CXCR6 via a self-cleaving 2A sequence. The anti-MSLN-CAR construct consists of a single chain variable fragment that recognizes human mesothelin (clone SS1), fused to an extracellular CD8a hinge domain as well as the transmembrane and intracellular signalling domains of the T cell co-stimulatory receptor CD28 (UNIPROT entry P10747 AA 153-220) and the cytoplasmatic signalling domain of the zeta chain of the human TCR/CD3 complex (UNIPROT entry P20963 AA 52-164). The anti-MSLN-CAR-CXCR6 construct consists of the anti-MSLN-CAR fused via a self-cleaving 2A peptide to the full length human CXCR6.
Murine T cell transduction
The transduction of primary murine OVA-specific T cells (OT-1 T cells) was conducted following the previously described protocol25. In brief, the ecotrophic packaging cell line Platinum E (Cell Biolabs) was transfected with 18 μg of the retroviral vector plasmid pMP71 (kindly provided by C. Baum, Hannover) using calcium phosphate precipitation. After 48 h and 72 h, the virus-containing supernatant was harvested and used to transduce murine T cells. If working with 293Vec-Eco producer cell lines, 1.2 x 106 cells were seeded into a 6-well plate and virus-containing supernatants were used for transduction on two consecutive days. In parallel, primary murine T cells were activated with anti-CD28 and anti-CD3 antibodies in murine TCM (eBioscience, Frankfurt, Germany, clones 145-2C11 and 37.51) supplemented with IL-2 (Peprotech, Hamburg, Germany) for 24 h. During the transduction process T cells were stimulated with Dynabeads® Mouse T-Activator CD3/CD28 (Life technologies, Darmstadt, Germany). Transduced murine T cells were cultured with murine TCM supplemented with human IL-15 (Peprotech, Hamburg, Germany) and β-mercaptoethanol. T cells were checked for transgene expression by FACS analysis and re-cultured in TCM supplemented with IL-15 (Peprotech, Hamburg, Germany) and β-mercaptoethanol and maintained at a concentration of 106 cells/ml every second day. For all functional assays, GFP or mCherry control-transduced T cells were used to exclude secondary effects of the genetic modification. Whenever necessary, transduction efficiencies of CAR-transduced and CAR-CXCR6-co-transduced T cells were adjusted to similar levels to avoid any bias.
Human T cell transduction
The retroviral vector pMP71 was used for transfection of the amphotrophic packaging cell line Platinum A. Transfection and virus production using 293Vec-RD114 producer cell lines were performed as described above. Human PBMCs were enriched using Ficoll density gradient separation. CD3+ T cells were isolated by MACS® Technology (CD3 MicroBeads, Miltenyi, Biotec, Germany) and activated on anti-CD3 and anti-CD28 coated wells (eBioScience, Frankfurt, Germany, clones HIT3a and CD28.2) in hTCM supplemented with IL-2 (Peprotech, Hamburg, Germany) and Dynabeads® Human T-Activator CD3/CD28 (Life technologies, Darmstadt, Germany). After two days, retrovirus was coated onto 24-well culture plates coated with 12.5 μg/ml RetroNectin (TaKaRa Biotech, Japan). 106 activated human T cells in hTCM supplemented with IL-2 (Peprotech, Hamburg, Germany), IL-15 (Peprotech, Hamburg, Germany) and β-mercaptoethanol were seeded onto virus-coated wells. The following day, a second transduction was performed using the same protocol. T-cells were checked for their transduction efficiency using FACS analysis and re-cultured in hTCM supplemented with IL-2 (Peprotech, Hamburg, Germany), IL-15 (Peprotech, Hamburg, Germany) and β-mercaptoethanol and maintained at a concentration of 106 cells/ml every second day. To avoid any bias, CAR- and CAR-CXCR6-transduction efficiencies were titrated whenever required.
Ex vivo chemokine assay of tissue lysates and plasma
To determine the expression of CXCL16 in lung, spleen, kidney, liver, tumour and lymph nodes of wild type mice, organs were homogenized and resuspended in lysis buffer (BioRad Laboratories, CA, USA). Following centrifugation, protein concentrations were determined by Bradford assay (BioRad Laboratories, CA, USA). All samples were diluted to a protein concentration of 50 mg/ml and CXCL16 concentrations were analysed by ELISA (R&D systems). Absorbance was measured with Mithras LB 940 Multimode Microplate reader (Software MicroWin 2000). Final CXCL16 concentrations were calculated as picogram cytokine per milligram protein in respective lysates. Plasma was analysed without further dilution and CXCL16 levels were calculated as picogram per ml plasma.
In vitro chemokine assay of tumour cell supernatant
To analyse spontaneous and inducible CXCL16 secretion by tumour cells 104 Panc02-OVA or 2 x 104 T110299-OVA tumour cells were seeded into a 96-well flat bottom plate and stimulated with 20 ng/ml IFN-γ (Peprotech), 20 ng/ml TNF-γ (Peprotech) or a combination of both for 48 h. To analyse intracellular and transmembrane CXCL16 concentration 2 x 105 Panc02-OVA or 2 x 105 T110299-OVA were plated in a 6-well plate and stimulated with or without 20 ng/ml IFN-γ. After 72 h supernatants were harvested, cells were washed once with PBS and cells were lysed using RIPA Lysis Buffer system (Santa Cruz Biotechnology). CXCL16 concentrations in supernatants and lysates were determined using ELISA (R&D systems). Absorbance was measured with Mithras LB 940 Multimode Microplate reader (Software MicroWin 2000). To quantify CXCL16 secretion by human pancreatic tumour cell lines, 2 x 105 cells were seeded in a 6-well plate and supernatants were analysed after 72 h by ELISA (R&D systems).
T cell stimulation assay
104 Panc02-OVA or T110299-OVA target cells were co-incubated with 5 x 104 OT-1 T cells in a 96-well flat bottom plate (Corning, Kaiserslautern, Germany) for up to 48 h (4, 8, 12, 24h). Analogous, 2 x 104 SUIT-2-MSLN-CXCL16 tumour cells were seeded into 96-well flat bottom plates 24 h prior to addition of T cells in a 10:1 effector to target ratio. Following incubation, supernatants were collected and IFN-γ levels were quantified by ELISA (BD bioscience, USA).
Migration assay
Murine and human T cell migration was investigated by trans-well migration assays (Corning). 106 transduced T cells were placed into the upper chamber of a trans-well plate with a 3 μm pore filter. The lower chamber contained different concentrations of recombinant murine or human CXCL16 (Peprotech) or tumour cell supernatant. To generate tumour cell supernatant, 105 T110299, 2 x 105 Panc02-OVA-CXCL16 or 5 x 105 E.G7-OVA-CXCL16 tumour cells were seeded into 6-well plates and when indicated stimulated with 20 ng/ml IFN-γ and 20 ng/ml TNF-γ and after 48 h supernatants were harvested and used for migration assay. SUIT-2-MSLN-CXCL16 supernatants were generated by seeding 2 x 105 cells in a 6-well plate and incubation for 72 h. To antagonize CXCL16-mediated migration 4 μg/ml neutralizing antibody (anti-mouse CXCL16, clone Q8BSU2, R&D systems) was added to the lower chamber. The numbers of migrated cells in the lower chamber were quantified by FACS analysis after an incubation at 37°C for 3 – 4 hours. When indicated count bright absolute counting beads (Life Technologies) were used for quantification. For flow cytometry analysis, migrated cells were stained with anti-mouse CD8a (Pacific Blue, clone 53-6.7, Biolegend) or anti-human CD8a (APC, clone SK1, Biolegend) and anti-c-myc (FITC, clone SH1-26E7.1.6, Miltenyi Biotec).
Migration cytotoxicity assay
105 Panc02-OVA-CXCL16 tumour cells were seeded in the lower chamber of a Polylysin coated (10 μg/well) trans-well migration plate (Corning) and cultured at 37°C. After 24 h, 0.5 or 1 x 106 T cells were added to the upper chamber and incubated for 3 h. Following incubation, the upper chamber with remaining T cells was removed and migrated T cells in the lower chamber were further incubated with tumour cells (1.5 h). Target cell lysis was quantified using the CytoTox 96® Non-Radioactive Cytotoxicity Assay (Promega).
Migration cytotoxicity assay using iCELLigence
2.5 – 5 x 104 SUIT-2-MSLN-hCXCL16 were seeded in the lower chamber of a trans-well migration plate (Corning) and incubated at 37°C. 24 h later, 5 x 105 T cells were added to the upper chamber of the trans-well plate. After 4 h incubation at 37°C a fraction of the migrated T cells was transferred to iCELLigence 8-well E-plates (OLS OMNI Life Science, Bremen). SUIT-2-MSLN had previously been seeded in the iCELLigence E-plates and had reached a cell index of approximately 1 before addition of migrated T cells. Killing of tumour cells was analysed for up to 60 h by measuring the Cell index in real-time using the iCELLigence device (ACEA Bioscience, USA). Migration of T cells was determined by flow cytometry as described above.
Cytotoxicity assay
3 x 104 Panc02-OVA or T110299-OVA target cells were co-incubated with 3 x 105 or 1.5 x 105 T cells in a 96-well flat bottom plate (Corning, Kaiserslautern, Germany) for up to 36 h. 2.5 x 104 E.G7-OVA target cells were co-incubated with 2.5 x 105 T cells in a 96-well flat bottom plate for 18 h. Target cell lysis was quantified using the CytoTox 96® Non-Radioactive Cytotoxicity Assay (Promega
Cytotoxicity assay using iCELLigence
SUIT-2-MSLN or SUIT-2-MSLN-hCXCL16 were seeded into iCELLigence 8-well E-plates (OLS OMNI Life Science, Bremen) and left to grow until they reached a cell index of approximately 1. T cells were added in varying effector to target ratios to the tumour cells and tumour cell death was analysed by measuring the cell index in real-time for up to 50 h using the iCELLigence device (ACEA Bioscience, USA).
Adhesion assay
T cells were either stained with Calcein (Life Technologies, Carlsbad, CA, USA) or CSFE (Cat. Number C34554, ThermoScientific, Darmstadt) and pre-incubated with or without 9 pmol recombinant mouse CXCL16 (Cat. Number 250-28, Peprotech, London, UK). Nickel-coated 96-well plates (Cat. Number 15442, ThermoScientific, Darmstadt) were coated with 9 pmol His-tagged CXCL16 (Cat. Number 50142-M08H, SinoBiological, Peking, China), 9 pmol His-tagged EpCAM (Cat. Number 50591-M08H, SinoBiological, Peking, China) or 9 pmol BSA. The pre-stimulated T cells were transferred to the CXCL16- or BSA-coated Nickel plate. After 25 minutes incubation and a washing step, adherent cells were lysed using RIPA buffer. Calcein or CSFE was detected with the Mithras LB 940 Multimode Microplate Reader (Berthold Technologies, Bad Wildbad), where the fluorescent signal intensity is proportional to the quantity of adherent cells.
Flow cytometry analysis
To isolate CD11c+ myeloid cells from tumour tissue, tumours were mechanically disrupted, incubated with 1mg/ml collagenase (Sigma Aldrich, Germany) and 0.05 mg/ml DNase (Sigma Aldrich, Germany) and passed through a cell strainer. Single cell suspensions were layered on a density gradient of 44 % Percoll (Biochrome, Berlin, Germany) and 67 % Percoll followed by centrifugation at 800 g for 30 min. Lymphocytes obtained from the interphase were washed with PBS and stained using anti-mouse CD11c (APC, clone N418, Biolegend). CD11c+ and CD11c- cells were separated by BD FACS Aria II (BD bioscience, Germany). For tracking experiments, lymphoid cells were isolated from tumours, spleens, kidney, lung, Peyer plaques, ipsilateral lymph nodes (LNi) and contralateral lymph nodes (LNk). Tumours, lungs and kidneys were mechanically disrupted, incubated with 1mg/ml collagenase (Sigma Aldrich, Germany) and 0.05 mg/ml DNase (Sigma Aldrich, Germany) and passed through a cell strainer. For tumours > 25mm2 and for kidneys, TILs and other mononuclear cells were enriched using Percoll density gradient centrifugation. Spleens, Peyer plaques and lymph nodes were smashed through a cell strainer. Red blood cell lysis was carried out for splenic samples. For tracking experiments, single cell suspensions of all organs were stained for anti-mouse CD3 (PE/Cy7, clone 17A2, Biolegend), anti-mouse CD8a (PerCP, clone 53-6.7, Biolegend), anti-mouse CD90.1 (Pacific Blue, clone OX-7, Biolegend) and anti-mouse CD45.1 (APC/Cy7, clone A20, Biolegend). CD90.1+ CXCR6+ and control transduced CD45.1+ GFP+ T cells were identified. Antibodies used to the characterization of in vivo activated OT-1 T cells are specified in supplementary table 2. After staining, samples were resuspended in PBS containing count bright absolute counting beads (Life Technologies) and analysed by FACS Canto II and FACS Fortessa (BD bioscience, Germany).
CAR expression on human T cells was determined by staining with anti-c-myc (FITC, clone SH1-26E7.1.6, Miltenyi Biotec) or the corresponding isotype control (mouse IgG1κ-FITC, Miltenyi Biotec). Human non-transduced T cells were stained with anti-human CD3 (FITC, clone UCHT1, Biolegend), if needed. Human CXCR6 expression was determined by staining with anti-human CXCR6 antibody (APC, clone K041E5, Biolegend) or the respective isotype control (APC mouse IgG2aκ, Biolegend). To analyse the expression of chemokine receptors, T cells were stained with Fixable Viability Dye (eBioScience, eFluor780) and anti-human CD3 (FITC, clone HIT3a, Biolegend), anti-human CD4 (Pacific Blue, clone OKT4, Biolegend), anti-human CD8 (PE, clone SK1, Biolegend), anti-human CXCR6 (APC, clone K041E5, Biolegend), anti-human CXCR3 (PE/Cy7, clone G025H7, Biolegend), anti-human CCR4 (PerCP/Cy5.5, clone L291H4, Biolegend) and anti-human CCR2 (Brilliant Violet 605, clone K036C2, Biolegend) or the respective isotype controls (FITC mouse IgG2aκ, Pacific Blue mouse IgG2bκ, PE mouse IgG1κ, APC mouse IgG2aκ, PE/Cy7 mouse IgG1κ, PerCP/Cy5.5 mouse IgG1κ, Brilliant Violet 605 mouse IgG2aκ, Biolegend). For xenograft tracking experiments, single cell suspensions of tumour and spleen were stained with Fixable Viability Dye (eBioScience, eFluor780) and anti-myc (FITC, clone SH1-26E7.1.6, Miltenyi Biotec), anti-human CD45 (PE/Cy7, clone 2D1, Biolegend), anti-human CD4 (AlexaFluor700, OKT4, Biolegend) and anti-CD8a (PE, clone HIT8a, Biolegend).
RNA isolation and quantitative real-time PCR
Total RNA was extracted from frozen organs or cells using pegGOLD TriFast™ (Peqlab, Germany) according to the manufacturer’s instructions. 2 μg of total RNA was used as a template for cDNA synthesis with the Superscript II kit (Life Technologies). Primers were design with help of the Roche Universal ProbeLibrary Assay Design Centre using NCBI GenBank sequences (for primer sequences see supplementary table 1). After the initial screen of different CXC chemokines, 5’ Primer TGA ACT AGT GGA CTG CTT TGA GC and 3’ Primer GCA AAT GTT TTT GGT GGT GA combined with probe #103 were used for CXCL16 quantification. The LightCycler 480 system (Roche Diagnostics) was used to perform quantitative real-time PCR. Relative gene expression levels are shown as the expression level of the gene of interest in relation to the expression level of hypoxanthine phosphoribosyl-transferase (HPRT).
Intracellular calcium measurement
CAR T cells were incubated for 30 min at 37°C with 1 μM fura-2 AM (Life Technologies). T cells were then washed in HBSS and added to the tumour cell layer cultured in ibidi μ-slides. Images were acquired every 10 seconds at 350 and 380 nm. Emissions at 510 nm were used for the analysis of Ca2+ responses with the use of the Fiji Trackmate plugin. Ca2+ values were represented as a ratio: fluorescence intensity at 350 nm/fluorescence intensity at 380 nm. CAR T cells were considered responsive when the amplitude of their responses reached at least twice that of the background. When Ca2+ traces were averaged, the rising phases of the traces were synchronized.
Confocal microscopy assay
To monitor the trafficking of CXCR6 after interaction with CXCL16, 5 x 103 CXCR6-GFP transduced T cells were stimulated with 10 ng/ml recombinant CXCL16 (Peprotech, Hamburg, Germany). For visualization of the receptor, in this experiment T cells were expressing CXCR6 fused to GFP via a non-cleavable 2A sequence. Receptor trafficking was imaged over a period of 1 h and membrane expression of CXCR6 was quantified by blinded validation of at least 75 representative cells per time point. Live fluorescent microscopy was conducted with a Leica SP5 AOBS confocal microscope.
To analyse the adhesion of CXCR6-transduced T cells to tumours cells, T cells were enriched by MACS sort one day before the co-culture. One day after enrichment, 5 x 103 Panc02-OVA or T110299-OVA tumour cells were co-incubated with 5 x 104 enriched CXCR6+ T cells, non-transduced T cells or a mixture of both (1:1), which were previously labelled with PKH-67 and PKH-26 (Sigma, Germany) according to the manufacturer’s instruction. To neutralize CXCL16-mediated effect, 4 μg/ml anti-CXCL16 antibody was added. Following an incubation period of 6 h, cell clusters were gently transferred to a glass-bottom dish and analysed by confocal microscopy. The amount of CXCR6+ and non-transduced T cells per cluster was quantified by blinded counting of at least 20 representative clusters for each condition.
Ex vivo imaging of tumour-infiltrating T cells
On day five after T cell transfer, the amount of CXCR6-GFP and GFP transduced T cells in tumour tissue was determined by 2-photon laser scanning microscopy (TPLSM). To distinguish intravascular and intratumoral T cells, blood vessels were stained by injecting mice with 3 μg anti-mouse CD31 antibody (eFluor450, clone 390, eBioScience) 30 min. before euthanasia. Imaging of tumour-infiltrating T cells was performed using a resonant scanning Leica SP5IIMP system equipped with a Spectra Physics MaiTai DeepSee Ti:Sa pulsed laser turned to 890 nm and a 20X NA 1.00 objective (Leica). Images with 1.5 to 2.0 μm spacing were acquired and processed using the Leica LAS X 3.1 software. The number of tumour-infiltrating T cells was quantified by counting of at least six representative areas per tumour.
Multi-photon intravital microscopy
Mice were anesthetized and imaged every other day. Multiphoton excitation was done with a MaiTai Ti:sapphire laser (Spectra-Physics) tuned to 950 nm to excite all fluorescent probes used. Sections with 4 to 5 μm z spacing were acquired on an Ultima multiphoton microscope (Prairie Technologies) every 60 sec, as described 10. Emitted fluorescence was detected through 460/50, 525/50, 595/50, and 660/40 band-pass filters and non-descanned detectors to generate 4-color images. Quantification was performed with the Imaris software (Bitplane).
Granzyme B PET imaging
PET images were acquired 1 and 3 days after T cell injection using the previously established methods 67,68. On the day of imaging, 68Ga-NOTA-GZP was prepared and mice were injected intravenously with 100 μl of radiolabelled peptide and subjected to PET/CT scan after one hour. All scans were completed on a rodent Triumph PET/CT (GE Healthcare) and PET images were obtained for 15 min, which was followed by CT imaging. All images were reconstructed using 3D-MLEM (4 iterations with 20 subsets). The mean regions of interest were drawn around the tumour and heart using anatomic guidance with VivoQuant software (InviCRO) and standard uptake value (SUV) was calculated for each tumour and heart to generate target-to-background (tumour:blood) ratios. An accumulation in kidneys was found to be due to renal extraction and excretion of the radiotracer.
Tumour spheroids and microscopic imaging
Seeding 2 x 103 or 250 tumour cells into agarose-coated 96-well plate generated Capan-1 and HEK-CXCL16 spheroids. On day 7, spheroids were co-incubated with 1.5 x 104 CXCR6-GFP- or GFP-transduced human T cells for 18 h. Non-invaded T cells were removed prior to fixation with 4 % paraformaldehyde. Samples were imaged using a selective plane illumination microscope and invaded T cells were quantified using Fiji software as described previously69,70.
Quantification of CXCL16- and CXCR6-positive cells in tissue microarray of pancreatic cancers
Tumour containing formalin-fixed paraffin-embedded (FFPE) tissue blocks of 399 patients which underwent curative intend resection for pancreatic ductal adenocarcinoma (Whipple / modified Whipple procedure, pancreatic tail resections or total pancreatectomy) between 2001 and 2015 were retrieved from the archives of the institute of pathology. This retrospective study using archival patient material was approved by the ethics committee of the faculty of medicine (approval number 20-081 to SO). A tissue microarray (TMA) comprising three tissue cores of one mm in diameter of different representative and vital tumour regions was constructed using a semi-automatic tissue arrayer (Beecher Instruments, Sun Prairie, WI, USA). For immunohistochemical detection of CXCL16 and CXCR6, 4 μm thick TMA sections were dewaxed and incubated with primary antibodies after heat mediated antigen retrieval (rabbit polyclonal anti-CXCL16 antibody, dilution 1:50, HPA066315, Atlas antibodies, Stockholm, Sweden; rabbit polyclonal anti-CXCR6 antibody, dilution 1:100, PA5-27171, Thermo Fisher Scientific, Waltham, MA, USA). Nuclei were counterstained using haematoxylin and the signal was detected using diaminobenzidine (DAB+, Agilent Technologies, Santa Clara, CA, USA) or alkaline phosphatase red (Permanent AP Red, Zytomed Systems, Bargteheide, Germany) after secondary antibody incubation. In each tumour core, the expression of both antigens was semi-quantitatively evaluated in the mononuclear cell infiltrate and the carcinoma epithelia using Zeiss Axiovert 200M microscope with Zen2012 software. CXCR6 expression in FFPE tissue blocks of five patients (10-17 fields per vision) was graded as absent (negative), weak, moderate or strong.
NanoString gene expression analysis
The retrospective analysis of gene expression using FFPE material from curative intent resections of PDAC patients (n = 36) and healthy controls (n = 12) was approved by the local ethics committee (project number: 629-16 “Immune Cell Profiling to develop immunotherapeutic strategies in Pancreatic Cancer”). Briefly, vital tumour tissue or normal pancreatic parenchyma was identified on HE stained sections by a board-certified pathologist (SO) and tumour RNA was extracted from consecutive 10 μm thick sections using Qiagen RNeasy extraction kits (Qiagen, Hilden Germany). Gene expression was examined using nCounter® PanCancer Immune Profiling Panel (NanoString Technologies, Seattle, WA, USA) and nSolver™ Software.
Generation of patient derived organoids (PDO) and co-culture with T cells
PDO were established from pancreatic cancer patients according to the protocols described previously71–73 with Ethical Committee Agreement Project-Number: 207/15 and 1946/07 (generation of Organoid-Bank). In order to get single cell suspensions, PDO were mechanically broken and enzymatically dissociated with dispase and trypsin for 30 min. 1.5 to 2.5 x 104 single cells were resuspended in 100 μl PDO medium and plated in duplicate in the 96-well plate. Cells were incubated 48 h at 37°C. Then, T cells were resuspended in medium and added to the target cells in a 1:1 effector to target ratio. Following incubation for 24 h, cells were spun down, supernatant was collected and IFN-γ levels were analysed with ELISA. Experiments were conducted on three individual PDO (B34, B54 and B61).
For confocal microscopy, partially digested PDO were resuspended in 50 μl Matrigel (Corning) and plated in duplicates as droplets in the wells of chamber slides (Thermo Fisher Scientific). Chamber slides were incubated at 37°C for 30 min to solidify the Matrigel. Droplets were overlaid with 5 x 105 CXCR6-GFP or GFP-transduced human T cells per well resuspended in 500 μl PDO medium. To antagonize CXCL16-mediated infiltration 5 μg/ml neutralizing antibody (anti-human CXCL16, clone 256213, R&D systems) was added. After 72 h, medium was aspirated, and droplets were gently washed twice with PBS. After fixation with 4% paraformaldehyde in PBS for 20 min. at room temperature, cells were permeabilized and stained with Phalloidin Alexa Fluor 594 (dilution 1:40, Thermo Fisher Scientific) and DAPI (dilution 1:14,000, Invitrogen) according to published protocol74. SlowFade™ Diamond Antifade Mountant (Thermo Fisher Scientific) was used to prepare slides. Slides were analysed with Leica TCS SP5 confocal laser scanning microscope. Quantification of GFP-positive T cells was performed with Imaris 7.6.5 software. Experiments without neutralization antibody (figure 5h and supplementary figure 7e) were conducted on four PDO (B34, B54, B61 and B79). For neutralization experiments two pancreatic cancer PDO (B34 and B48) were co-cultured with T cells (figure 5i).
Migration testing in organotypic functional tumour explant models
As described previously75, resected specimens were used for culture treatment. Briefly, the resected specimen rapidly was transferred into the lab in a sterile container, cut (size approximately 5 x 3 x 1 mm) and treated with respectively labelled and genetically modified cells. After culturing, the tissue was harvested and sectioned, subsequently being stained for human CD3epsilon (1:100 dilution, clone PS1, Novocastra, UK), CD8 (1:100 dilution, clone 4B11, Novocastra, UK) and for migrated cells a monoclonal anti-GFP antibody (clone FM264G, Biolegend) was used. Quantification was performed on whole slide sections as described previously76. All material was obtained after approval by the medical ethics committee of the University of Heidelberg (S-069), written consent was obtained from all patients prior to analysis.
TCGA data analysis
With help of the bioinformatics tool UCSC Xena, TCGA (The Cancer Genome Atlas) RNA sequencing datasets were analysed in comparison to GTex Portal (Genotype-tissue Expression) healthy tissue reference datasets concerning the expression of CXCL16 and CXCR677. The healthy tissue references EcGj (esophageal mucosa and gastroesophageal junction), brain (brain cortex, cerebellum, hippocampus, substantia nigra, anterior cingulate cortex [Ba 24], cerebellar hemisphere [basal ganglia], nucleus accumbens [basal ganglia], putamen [basal ganglia], hypothalamus, amygdala), skin (non-sun and sun-exposed skin) and CoSi (colon and sigmoid) have been summarized from datasets as indicated.
Single cell RNA (scRNA) sequencing data analysis
To quantitatively compare CXCL16 expression in public single cell RNA-Seq datasets, comparable pre-processing was carried out for each dataset separately. All pre-processing and analysis steps were run using the python-based Scanpy toolkit78. For the datasets of Travaglini, Madissoon, Reyfman, Peng and Baron27,79–82, batch balanced k nearest neighbours (BBKNN) were calculated to account for batches along the respective samples12. Pre-processing of droplet-based single cell RNA-Seq data involved basic quality control (removing low quality cells and lowly expressed genes), cell count normalization using R-based scran83, selecting highly variable genes based on normalized dispersion as described in ref. 84, and visualizing the cells in a two-dimensional Uniform Manifold Approximation and Projection (UMAP) embedding85. For cell type identification, we used barcode annotations provided by the authors of the respective study. For the lung datasets of Travaglini, Madissoon and Reyfman, cell annotations were obtained from a recent preprint integrating single cell RNA-Seq datasets86. All analyses from UMI count matrices were run with python 3 with the Scanpy API v.1.4.6 and anndata v.0.7.1. All figures were plotted with matplotlib and seaborn.
Statistics
The FACS data was analysed with FlowJo V9.2 or V10.3 software. Statistical analyses were performed by using GraphPad Prism software 9.0. For the comparison of experimental conditions unpaired two-tailed Student’s t test, Mann-Whitney test or Wilcoxon signed-rank test were used as indicated. For in vivo experiments, two-way ANOVA with correction for multiple testing by the Bonferroni method was used to analyse differences between the groups. Log-rank (Mantel-Cox) test was performed to determine significance of survival curve differences. p-values <0.05 were considered to be significant. Data are shown as mean values ± SEM of a minimum of two biological replicates or independent experiments, as indicated.
Supplementary Material
Supplementary information is available for this paper at https://doi.org/10.1038/s41551-01X-XXXX-X.
Acknowledgements
This study was supported by grants from the Wilhelm Sander Stiftung (grant number 2014.018.1 to SE and SK), the international doctoral program “i-Target: Immunotargeting of cancer” funded by the Elite Network of Bavaria (to SK and SE), the Melanoma Research Alliance (grant number N269626 to SE and 409510 to SK), the Marie-Sklodowska-Curie “Training Network for the Immunotherapy of Cancer (IMMUTRAIN)” funded by the H2020 program of the European Union (to SE and SK), the Marie-Sklodowska-Curie Program Training Network for Optimizing Adoptive T Cell Therapy of Cancer funded by the H2020 Program of the European Union (Grant 955575, to S.K.), the Else Kröner-Fresenius-Stiftung (to SK), the German Cancer Aid (to SK), the Ernst-Jung-Stiftung (to SK) by LMU Munich’s Institutional Strategy LMUexcellent within the framework of the German Excellence Initiative (to SE and SK), by the Bundesministerium für Bildung und Forschung (to SE and SK), by the European Research Council Starting Grant (grant number 756017 to SK), the DFG (to SK), the Fritz-Bender-Foundation (to SK), the José-Carreras Foundation (to SK) and the Hector Foundation (to SK). RM is supported by the DFG INST409/97-1 FUGG, SFB1123/Z1, and ERA-CVD (AtheroInside), DFG SFB1321 to MS. AGA Moti L. @ Kamla Rustgi International Travel Award to ZD. MR was supported by the German Cancer Aid Foundation (Max Eder Program, Deutsche Krebshilfe 111273, MR) and the German Research Foundation (Deutsche Forschungsgemeinschaft, SFB1321 Modeling and Targeting Pancreatic Cancer and RE 3723/4-1). ED was supported by a grant from INSERM (HTE: chemotaxis in cancer). MT is funded by the Volkswagen Foundation (project OntoTime). CM has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (Grant agreement No. 866411). MSch was supported by Deutsche Forschungsgemeinschaft, SFB1321 Modeling and Targeting Pancreatic Cancer (Project Number 329628492). The authors thank Life Science Editors for editorial assistance and acknowledge the iFlow Core Facility of the university hospital Munich for assistance with the generation of flow cytometry data. Image processing using the Imaris 7.6.5 software was performed at the core facility for bioimaging of the Biomedical Center of the Ludwig-Maximilians-Universität München.
Footnotes
Reporting Summary. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Author contributions
SL, VB, SS, JO, BLC, ZD, FR, KD, JL, CHK, CoH, MK, BML, SG, MR, AN, AG, StK, NT, PM, CH, MRB, DD, AO, RG, MS, SJ, ÖU, LV, MT, TT, TH, TB, DH, RTAM, KPJ, MJ, DL, SRu, MDP, JNP, MR, SO, CM, ET, ED, MH, AR, SRo, PD, LMK and MSch performed or assisted with experiments, analysed data and supported the project. SK and SE supervised the project and did the funding acquisition. SK, SL, VB, SS, JO, BLC, MS, AL, NH, MR and TRM designed the experiments. SK and SL wrote the manuscript. All authors critically read and approved the final manuscript.
Competing interests
Parts of this work have been performed for the doctoral theses of SL, VB, SS, KD and JL at the Ludwig-Maximilians-Universität München. MR, SG, SE and SK are inventors on a patent application related to this work filed by the Ludwig-Maximilians-Universität München. SE and SK received research support from TCR2 Inc and Arcus Biosciences for work on T cell therapies unrelated to the present manuscript. The remaining authors declare no competing interests.
Additional information
Peer review information Nature Biomedical Engineering thanks Eduard Ryschich, Prasad Adusumilli and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Reprints and permissions information is available at www.nature.com/reprints.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Data availability
The main data supporting the results in this study are available within the paper and its Supplementary Information. The raw and analysed datasets generated during the study are too large to be publicly shared, yet they are available for research purposes from the corresponding authors on reasonable request. They also contain personal and patient data and are available for research purposes pending completion of adequate paper work ensuring personal data protection and ethical approval. RNA sequencing data in this study have been published previously and are accessible through NCBI GEO: GSE84133, GSE122960, NCBI PRJEB31843, GSA: CRA001160, EGAS00001004344.
References
- 1.Rosenberg SA, Restifo NP. Adoptive cell transfer as personalized immunotherapy for human cancer. Science. 2015;348:62–68. doi: 10.1126/science.aaa4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kobold S, et al. Immunotherapy in tumors. Deutsches Arzteblatt international. 2015;112:809–815. doi: 10.3238/arztebl.2015.0809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sheridan C. First approval in sight for Novartis’ CAR-T therapy after panel vote. Nat Biotechnol. 2017;35:691–693. doi: 10.1038/nbt0817-691. [DOI] [PubMed] [Google Scholar]
- 4.Ahmed N, et al. Human epidermal growth factor receptor 2 (HER2) - specific chimeric antigen recpetor-modified T cells for the immunotherapy of HER2-positive sarcoma. J Clin Oncol. 2015;33:1688–1696. doi: 10.1200/JCO.2014.58.0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adusumilli PS, et al. Regional delivery of mesothelin-targeted CAR T cell therapy generates potent and long-lasting CD4-dependent tumor immunity. Sci Transl Med. 2014;6:261–151. doi: 10.1126/scitranslmed.3010162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown CE, et al. Regression of glioblastoma after chimeric antigen receptor T cell therapy. N Engl J Med. 2016;375:2561–2569. doi: 10.1056/NEJMoa1610497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Rourke DM, et al. A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Sci Transl Med. 2017;9:eaaa0984. doi: 10.1126/scitranslmed.aaa0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tchou J, et al. Safety and efficacy of intratumoral injections of chimeric antigen receptor (CAR) T cells in metastatic breast cancer. Cancer Immunol Res. 2017;5:1152–1161. doi: 10.1158/2326-6066.CIR-17-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akbay EA, et al. Interleukin-17A promotes lung tumor progression through neutrophil attraction to tumor sites and mediating resistance to PD-1 blockade. J Thorac Oncol. 2017;12:1268–1279. doi: 10.1016/j.jtho.2017.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bauer CA, et al. Dynamic Treg interactions with intratumoral APCs promote local CTL dysfunction. J Clin Invest. 2014;124:2425–2440. doi: 10.1172/JCI66375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linke B, et al. CXCL16/CXCR6-mediated adhesion of human peripheral blood mononuclear cells to inflamed endothelium. Cytokine. 2019;122:154081. doi: 10.1016/j.cyto.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Polański K, et al. BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics. 2020;36:964–965. doi: 10.1093/bioinformatics/btz625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tokarew N, Ogonek J, Endres S, von Bergwelt-Baildon M, Kobold S. Teaching an old dog new tricks: next-generation CAR T cells. Br J Cancer. 2019 doi: 10.1038/s41416-018-0325-1. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grosser R, Cherkassky L, Chintala N, Adusumilli PS. Combination Immunotherapy with CAR T Cells and Checkpoint Blockade for the Treatment of Solid Tumors. Cancer cell. 2019;36:471–482. doi: 10.1016/j.ccell.2019.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rapp M, et al. C-C chemokine receptor type-4 transduction of T cells enhances interaction with dendritic cells, tumor infiltration and therapeutic efficacy of adoptive T cell transfer. Oncoimmunology. 2016;5:e1105428. doi: 10.1080/2162402X.2015.1105428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hughes CE, Nibbs RJB. A guide to chemokines and their receptors. FEBS J. 2018;285:2944–2971. doi: 10.1111/febs.14466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Curiel TJ, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med. 2004;10:942–949. doi: 10.1038/nm1093. [DOI] [PubMed] [Google Scholar]
- 18.Lim WA, June CH. The principles of engineering immune cells to treat cancer. Cell. 2017;168:724–740. doi: 10.1016/j.cell.2017.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garetto S, et al. Tailored chemokine receptor modification improves homing of adoptive therapy T cells in a spontaneous tumor model. Oncotarget. 2016;7:43010–43026. doi: 10.18632/oncotarget.9280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siddiqui I, Erreni M, van Brakel M, Debets R, Allavena P. Enhanced recruitment of genetically modified CX3CR1-positive human T cells into Fractalkine/CX3CL1 expressing tumors: importance of the chemokine gradient. J Immunother Cancer. 2016;4:21. doi: 10.1186/s40425-016-0125-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muller N, et al. Engineering NK cells modified with an EGFRvIII-specific chimeric antigen receptor to overexpress CXCR4 improves immunotherapy of CXCL12/SDF-1alpha-secreting glioblastoma. J Immunother. 2015;38:197–210. doi: 10.1097/CJI.0000000000000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moon EK, et al. Expression of a functional CCR2 receptor enhances tumor localization and tumor eradication by retargeted human T cells expressing a mesothelin-specific chimeric antibody receptor. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:4719–4730. doi: 10.1158/1078-0432.CCR-11-0351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng W, et al. Transduction of tumor-specific T cells with CXCR2 chemokine receptor improves migration to tumor and antitumor immune responses. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16:5458–5468. doi: 10.1158/1078-0432.CCR-10-0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimaoka T, et al. Cell surface-anchored SR-PSOX/CXC chemokine ligand 16 mediates firm adhesion of CXC chemokine receptor 6-expressing cells. J Leukoc Biol. 2004;75:267–274. doi: 10.1189/jlb.1003465. [DOI] [PubMed] [Google Scholar]
- 25.Kobold S, et al. Impact of a new fusion receptor on PD-1-mediated immunosuppression in adoptive T cell therapy. J Natl Cancer Inst. 2015;107:146. doi: 10.1093/jnci/djv146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li K, et al. Impact of chemokine receptor CXCR3 on tumor-infiltrating lymphocyte recruitment associated with favorable prognosis in advanced gastric cancer. Int J Clin Exp Pathol. 2015;8:14725–14732. [PMC free article] [PubMed] [Google Scholar]
- 27.Madissoon E, et al. scRNA-seq assessment of the human lung, spleen, and esophagus tissue stability after cold preservation. Genome Biol. 2019;21:1. doi: 10.1186/s13059-019-1906-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng L, Chen N, Li Y, Zheng H, Lei Q. CXCR6/CXCL16 functions as a regulator in metastasis and progression of cancer. Biochim Biophys Acta. 2010;1806:42–49. doi: 10.1016/j.bbcan.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 29.Wente MN, et al. Expression and potential function of the CXC chemokine CXCL16 in pancreatic ductal adenocarcinoma. International journal of oncology. 2008;33:297–308. [PubMed] [Google Scholar]
- 30.Heydtmann M, et al. CXC chemokine ligand 16 promotes integrin-mediated adhesion of liver-infiltrating lymphocytes to cholangiocytes and hepatocytes within the inflamed human liver. J Immunol. 2005;174:1055–1062. doi: 10.4049/jimmunol.174.2.1055. [DOI] [PubMed] [Google Scholar]
- 31.Rataj F, et al. PD1-CD28 fusion protein enables CD4+ T cell help for adoptive T cell therapy in models of pancreatic cancer and non-hodgkin lymphoma. Front Immunol. 2018;9 doi: 10.3389/fimmu.2018.01955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato T, et al. Role for CXCR6 in recruitment of activated CD8+ lymphocytes to inflamed liver. J Immunol. 2005;174:277–283. doi: 10.4049/jimmunol.174.1.277. [DOI] [PubMed] [Google Scholar]
- 33.Unutmaz D, et al. The primate lentiviral receptor Bonzo/STRL33 is coordinately regulated with CCR5 and its expression pattern Is conserved between human and mouse. J Immunol. 2000;165:3284–3292. doi: 10.4049/jimmunol.165.6.3284. [DOI] [PubMed] [Google Scholar]
- 34.Karches CH, et al. Bispecific Antibodies Enable Synthetic Agonistic Receptor-Transduced T Cells for Tumor Immunotherapy. Clin Cancer Res. 2019;25:5890–5900. doi: 10.1158/1078-0432.Ccr-18-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paulos CM, et al. Microbial translocation augments the function of adoptively transferred self/tumor-specific CD8+ T cells via TLR4 signaling. J Clin Invest. 2007;117:2197–2204. doi: 10.1172/JCI32205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kobold S, et al. Selective bispecific T cell recruiting antibody and antitumor activity of adoptive T cell transfer. J Natl Cancer Inst. 2015;107:364. doi: 10.1093/jnci/dju364. [DOI] [PubMed] [Google Scholar]
- 37.Chinnasamy D, et al. Local delivery of interleukin-12 using T cells targeting VEGF receptor-2 eradicates multiple vascularized tumors in mice. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:1672–1683. doi: 10.1158/1078-0432.CCR-11-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin L, et al. CXCR1- or CXCR2-modified CAR T cells co-opt IL-8 for maximal antitumor efficacy in solid tumors. Nat Commun. 2019;10:4016. doi: 10.1038/s41467-019-11869-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bailey P, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 40.Schizas D, et al. Immunotherapy for pancreatic cancer: A 2020 update. Cancer Treat Rev. 2020;86:102016. doi: 10.1016/j.ctrv.2020.102016. [DOI] [PubMed] [Google Scholar]
- 41.Hartmann N, et al. Prevailing Role of Contact Guidance in Intrastromal T-cell Trapping in Human Pancreatic Cancer. Clinical Cancer Research. 2014;20:3422. doi: 10.1158/1078-0432.CCR-13-2972. [DOI] [PubMed] [Google Scholar]
- 42.Kocher HM, et al. Phase I clinical trial repurposing all-trans retinoic acid as a stromal targeting agent for pancreatic cancer. Nat Commun. 2020;11:4841. doi: 10.1038/s41467-020-18636-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alvarez R, et al. Stromal disrupting effects of nab-paclitaxel in pancreatic cancer. British Journal of Cancer. 2013;109:926–933. doi: 10.1038/bjc.2013.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lo A, et al. Tumor-Promoting Desmoplasia Is Disrupted by Depleting FAP-Expressing Stromal Cells. Cancer Research. 2015;75:2800. doi: 10.1158/0008-5472.CAN-14-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matloubian M, David A, Engel S, Ryan JE, Cyster JG. A transmembrane CXC chemokine is a ligand for HIV-coreceptor Bonzo. Nat Immunol. 2000;1:298–304. doi: 10.1038/79738. [DOI] [PubMed] [Google Scholar]
- 46.Linke B, et al. CXCL16/CXCR6-mediated adhesion of human peripheral blood mononuclear cells to inflamed endothelium. Cytokine. 2017 doi: 10.1016/j.cyto.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 47.Collado A, et al. Functional role of endothelial CXCL16/CXCR6-platelet-leucocyte axis in angiotensin II-associated metabolic disorders. Cardiovasc Res. 2018;114:1764–1775. doi: 10.1093/cvr/cvy135. [DOI] [PubMed] [Google Scholar]
- 48.Sackstein R, Schatton T, Barthel SR. T-lymphocyte homing: an underappreciated yet critical hurdle for successful cancer immunotherapy. Laboratory Investigation. 2017;97:669–697. doi: 10.1038/labinvest.2017.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agostini C, et al. Role for CXCR6 and its ligand CXCL16 in the pathogenesis of T-cell alveolitis in sarcoidosis. Am J Respir Crit Care Med. 2005;172:1290–1298. doi: 10.1164/rccm.200501-142OC. [DOI] [PubMed] [Google Scholar]
- 50.Oldham KA, et al. T lymphocyte recruitment into renal cell carcinoma tissue: a role for chemokine receptors CXCR3, CXCR6 CCR5, and CCR6. Eur Urol. 2012;61:385–394. doi: 10.1016/j.eururo.2011.10.035. [DOI] [PubMed] [Google Scholar]
- 51.La Porta CA. CXCR6: the role of environment in tumor progression. Challenges for therapy. Stem Cell Rev. 2012;8:1282–1285. doi: 10.1007/s12015-012-9383-6. [DOI] [PubMed] [Google Scholar]
- 52.Allaoui R, et al. Cancer-associated fibroblast-secreted CXCL16 attracts monocytes to promote stroma activation in triple-negative breast cancers. Nat Commun. 2016;7:13050. doi: 10.1038/ncomms13050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chalabi-Dchar M, et al. Loss of somatostatin receptor subtype 2 promotes growth of KRAS-induced pancreatic tumors in mice by activating PI3K signaling and overexpression of CXCL16. Gastroenterology. 2015;148:1452–1465. doi: 10.1053/j.gastro.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 54.Elyada E, et al. Cross-Species Single-Cell Analysis of Pancreatic Ductal Adenocarcinoma Reveals Antigen-Presenting Cancer-Associated Fibroblasts. Cancer Discovery. 2019;9:1102. doi: 10.1158/2159-8290.CD-19-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu W, Liu Y, Zhou W, Si L, Ren L. CXCL16 and CXCR6 are coexpressed in human lung cancer in vivo and mediate the invasion of lung cancer cell lines in vitro. PloS one. 2014;9:e99056. doi: 10.1371/journal.pone.0099056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Slaga D, et al. Avidity-based binding to HER2 results in selective killing of HER2-overexpressing cells by anti-HER2/CD3. Sci Transl Med. 2018;10:eaat5775. doi: 10.1126/scitranslmed.aat5775. [DOI] [PubMed] [Google Scholar]
- 57.Morello A, Sadelain M, Adusumilli PS. Mesothelin-Targeted CARs: Driving T Cells to Solid Tumors. Cancer Discovery. 2016;6:133. doi: 10.1158/2159-8290.CD-15-0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beatty GL, et al. Activity of mesothelin-specific chimeric antigen receptor T cells against pancreatic carcinoma metastases in a phase 1 trial. Gastroenterology. 2018;155:29–32. doi: 10.1053/j.gastro.2018.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fujita K, et al. Prolonged disease-free period in patients with advanced epithelial ovarian cancer after adoptive transfer of tumor-infiltrating lymphocytes. Clinical Cancer Research. 1995;1:501. [PubMed] [Google Scholar]
- 60.Hall M, et al. Expansion of tumor-infiltrating lymphocytes (TIL) from human pancreatic tumors. Journal for immunotherapy of cancer. 2016;4:61–61. doi: 10.1186/s40425-016-0164-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nanki T, et al. Pathogenic role of the CXCL16–CXCR6 pathway in rheumatoid arthritis. Arthritis Rheum. 2005;52:3004–3014. doi: 10.1002/art.21301. [DOI] [PubMed] [Google Scholar]
- 62.Akce M, Zaidi MY, Waller EK, El-Rayes BF, Lesinski GB. The Potential of CAR T Cell Therapy in Pancreatic Cancer. Front Immunol. 2018;9:2166–2166. doi: 10.3389/fimmu.2018.02166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jacobs C, et al. An ISCOM vaccine combined with a TLR9 agonist breaks immune evasion mediated by regulatory T cells in an orthotopic model of pancreatic carcinoma. Int J Cancer. 2011;128:897–907. doi: 10.1002/ijc.25399. [DOI] [PubMed] [Google Scholar]
- 64.Anz D, et al. Suppression of intratumoral CCL22 by type I interferon inhibits migration of regulatory T cells and blocks cancer progression. Cancer Res. 2015;75:4483–4493. doi: 10.1158/0008-5472.CAN-14-3499. [DOI] [PubMed] [Google Scholar]
- 65.Ghani K, et al. Efficient human hematopoietic cell transduction using RD114- and GALV-pseudotyped retroviral vectors produced in suspension and serum-free media. Hum Gene Ther. 2009;20:966–974. doi: 10.1089/hum.2009.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Metzger P, et al. Immunostimulatory RNA leads to functional reprogramming of myeloid-derived suppressor cells in pancreatic cancer. Journal for ImmunoTherapy of Cancer. 2019;7:288. doi: 10.1186/s40425-019-0778-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Larimer BM, et al. Granzyme B PET Imaging as a Predictive Biomarker of Immunotherapy Response. Cancer Research. 2017;77:2318–2327. doi: 10.1158/0008-5472.can-16-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Larimer BM, et al. The Effectiveness of Checkpoint Inhibitor Combinations and Administration Timing Can Be Measured by Granzyme B PET Imaging. Clin Cancer Res. 2019;25:1196–1205. doi: 10.1158/1078-0432.CCR-18-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rühland S, et al. Quantification of in vitro mesenchymal stem cell invasion into tumor spheroids using selective plane illumination microscopy. J Biomed Opt. 2015;20:3. doi: 10.1117/1.JBO.20.4.040501. [DOI] [PubMed] [Google Scholar]
- 70.Schmohl KA, et al. Thyroid hormones and tetrac: new regulators of tumour stroma formation via integrin αvβ3. Endocr Relat Cancer. 2015;22:941–952. doi: 10.1530/erc-15-0245. [DOI] [PubMed] [Google Scholar]
- 71.Renz BW, et al. Adrenergic-neurotrophin feedforward loop promotes pancreatic cancer. Cancer Cell. 2018;33:75–90.:e77. doi: 10.1016/j.ccell.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Renz BW, et al. Cholinergic signaling via muscarinic receptors directly and indirectly suppresses pancreatic tumorigenesis and cancer stemness. Cancer Discov. 2018;8:1458–1473. doi: 10.1158/2159-8290.CD-18-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ruess DA, et al. Mutant KRAS-driven cancers depend on PTPN11/SHP2 phosphatase. Nat Med. 2018;24:954–960. doi: 10.1038/s41591-018-0024-8. [DOI] [PubMed] [Google Scholar]
- 74.Reichert M, et al. Isolation, culture and genetic manipulation of mouse pancreatic ductal cells. Nat Protoc. 2013;8:1354–1365. doi: 10.1038/nprot.2013.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Halama N, et al. Tumoral Immune Cell Exploitation in Colorectal Cancer Metastases Can Be Targeted Effectively by Anti-CCR5 Therapy in Cancer Patients. Cancer Cell. 2016;29:587–601. doi: 10.1016/j.ccell.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 76.Halama N, et al. Localization and Density of Immune Cells in the Invasive Margin of Human Colorectal Cancer Liver Metastases Are Prognostic for Response to Chemotherapy. Cancer Research. 2011;71:5670. doi: 10.1158/0008-5472.CAN-11-0268. [DOI] [PubMed] [Google Scholar]
- 77.Goldman M, et al. The UCSC Xena Platform for cancer genomics data visualization and interpretation. bioRxiv. 2018 [Google Scholar]
- 78.Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15–15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Baron M, et al. A Single-Cell Transcriptomic Map of the Human and Mouse Pancreas Reveals Inter- and Intra-cell Population Structure. Cell Syst. 2016;3:346–360.:e344. doi: 10.1016/j.cels.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peng J, et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019;29:725–738. doi: 10.1038/s41422-019-0195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reyfman PA, et al. Single-Cell Transcriptomic Analysis of Human Lung Provides Insights into the Pathobiology of Pulmonary Fibrosis. Am J Respir Crit Care Med. 2019;199:1517–1536. doi: 10.1164/rccm.201712-2410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Travaglini KJ, et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature. 2020;587:619–625. doi: 10.1038/s41586-020-2922-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lun ATL, Bach K, Marioni JC. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 2016;17:75. doi: 10.1186/s13059-016-0947-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zheng GXY, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049–14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McInnes L, Healy J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. ArXiv. 2018 abs/1802.03426. [Google Scholar]
- 86.Muus C, et al. Integrated analyses of single-cell atlases reveal age, gender, and smoking status associations with cell type-specific expression of mediators of SARS-CoV-2 viral entry and highlights inflammatory programs in putative target cells. bioRxiv. 2020:2020.2004.2019.049254. doi: 10.1101/2020.04.19.049254. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The main data supporting the results in this study are available within the paper and its Supplementary Information. The raw and analysed datasets generated during the study are too large to be publicly shared, yet they are available for research purposes from the corresponding authors on reasonable request. They also contain personal and patient data and are available for research purposes pending completion of adequate paper work ensuring personal data protection and ethical approval. RNA sequencing data in this study have been published previously and are accessible through NCBI GEO: GSE84133, GSE122960, NCBI PRJEB31843, GSA: CRA001160, EGAS00001004344.