Abstract
Precise control of gene expression is fundamental to cell function and development. Although ultimately gene expression relies on DNA-binding transcription factors to guide the activity of the transcription machinery to genes, it has also become clear that chromatin and histone post-translational modification have fundamental roles in gene regulation. Polycomb repressive complexes represent a paradigm of chromatin-based gene regulation in animals. The Polycomb repressive system comprises two central protein complexes, Polycomb repressive complex 1 (PRC1) and PRC2, which are essential for normal gene regulation and development. Our early understanding of Polycomb function relied on studies in simple model organisms, but more recently it has become apparent that this system has expanded and diverged in mammals. Detailed studies are now uncovering the molecular mechanisms that enable mammalian PRC1 and PRC2 to identify their target sites in the genome, communicate through feedback mechanisms to create Polycomb chromatin domains, and control transcription to regulate gene expression. In this Review, we discuss and contextualise the emerging principles that define how this fascinating chromatin-based system regulates gene expression in mammals.
Introduction
The development of multicellular organisms relies on cells and tissues establishing unique gene expression programmes. To achieve this, signalling pathways converge on DNA-binding transcription factors, which guide the binding and activity of RNA polymerase II (Pol II) at gene promoters1. However, through studying these mechanisms it has become apparent that chromatin, and histone post-translational modification, can also profoundly influence transcription and gene expression.
The importance of chromatin modifying complexes in controlling development is exemplified by the Polycomb repressive system. Polycomb genes were initially discovered in Drosophila melanogaster, where they are required for repression of homeotic (Hox) genes and thus for body plan specification2. Polycomb genes were subsequently shown to have mammalian orthologues that have important roles in controlling gene expression throughout development3.
Since these initial discoveries, biochemical analyses have shown that Polycomb proteins typically assemble into one of two large multi-protein complexes that post-translationally modify histones. Polycomb repressive complex 1 (PRC1) mono-ubiquitylates histone H2A at Lys119 (H2AK119ub1)4,5, whereas PRC2 mono-, di- and tri-methylates histone H3 at Lys27 (H3K27me1, H3K27me2 and H3K27me3, respectively)6-9. Importantly, PRC1 and PRC2 tend to spatially converge on the same sites in the genome to form Polycomb chromatin domains, which are uniquely enriched in H2AK119ub1, H3K27me3 and Polycomb complexes10-14. Polycomb chromatin domains are then thought to counteract transcription, though the mechanisms that enable this remain incompletely defined.
Although early genetic and biochemical investigations of Polycomb proteins were focussed on D. melanogaster, detailed studies over the past decade have revealed that the Polycomb system has expanded in mammals and that some key molecular principles that define its function have diverged from D. melanogaster. Based on these discoveries, we have gained a new understanding of how mammalian Polycomb complexes identify their target sites to form Polycomb chromatin domains and ultimately control gene expression. Several Reviews have recently discussed the general features and functions of Polycomb systems across phyla, and in mammalian development and disease2,3,15–18. In this Review we discuss and contextualise our emerging understanding of the molecular principles that enable the functions of mammalian Polycomb complexes in gene regulation. We first introduce the diverse complement of mammalian Polycomb repressive complexes and describe their enzymatic activities. We examine how Polycomb repressive complexes identify their target sites in the genome and communicate with each other to initiate the formation of Polycomb chromatin domains. We then discuss the regulation of Polycomb chromatin domain formation and ultimately how this is integrated with and regulates gene expression. Finally, we conclude by considering future avenues of research that will provide further mechanistic understanding of the principles of Polycomb biology in health and disease.
The wide repertoire of mammalian PRCs
The rapid progress in understanding mammalian Polycomb biology has relied on detailed and systematic biochemical interrogation of Polycomb repressive complexes and their histone modifying activities. This work has shown that Polycomb complexes comprise catalytic cores, which bind auxiliary proteins to create distinct PRC1 and PRC2 assemblies. In this section we introduce the composition, structure and enzymatic activity of the different Polycomb complexes, as a prerequisite for examining how these complexes underpin the functions of mammalian Polycomb complexes in gene regulation.
Polycomb repressive complex 1 and its E3 ubiquitin ligase activity
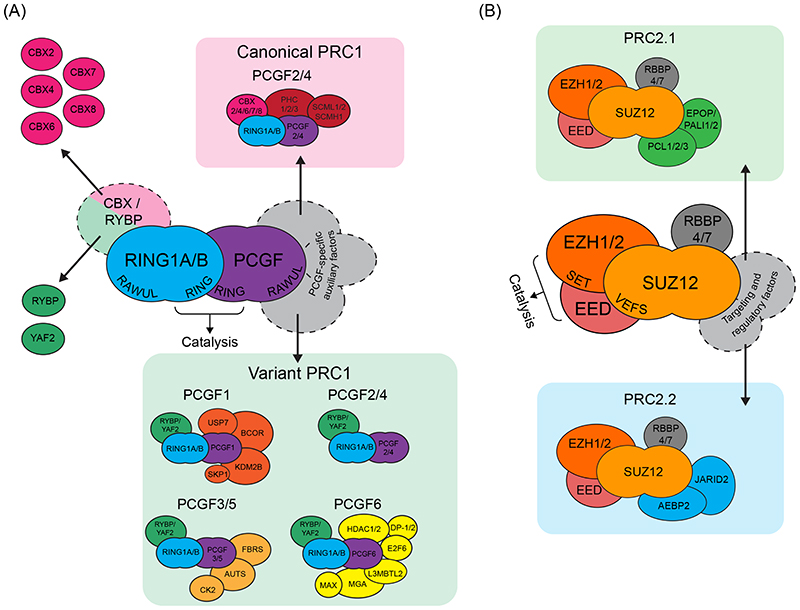
The catalytic core of PRC1 is composed of RING1B or its paralogue RING1A, and one of six Polycomb group ring finger (PCGF) proteins (PCGF1, 2, 3, 4, 5, or 6; Figure 1A)19. RING1 and PCGF proteins have similar domain architecture, including an N-terminal RING domain and C-terminal RING-finger and WD40-associated ubiquitin-like (RAWUL) domain. The RING1 protein and the PCGF protein dimerise through their RING domains, which facilitates their interaction with an E2 conjugating enzyme to enable histone ubiquitylation20-22. Their RAWUL domains bind to a range of auxiliary subunits, which regulate the catalytic activity of PRC1 and target it to specific sites in the genome23,24. Importantly, the PCGF components dictate which auxiliary subunits are incorporated into specific PRC1 complexes. These complexes are generally categorised as either canonical PRC1 (cPRC1) or variant PRC1 (vPRC1; Figure 1A), a nomenclature that reflects the order in which these complexes were initially identifed19,23. cPRC1 complexes, which were the first to be isolated25,26, assemble around either PCGF2 or PCGF4 and include one of five chromodomain-containing paralogues (CBX2, 4, 6, 7 or 8) and a Polyhomeotic (PHC) subunit (PHC1, 2 or 3). By contrast, vPRC1 complexes, which were identified later19,27, can assemble around any of the six PCGF proteins (PCGF1–6) and include RING1 and YY1-binding protein (RYBP), or its paralogue YAF2, and various additional subunits depending on the PCGF component present in the complex.
Figure 1. A diverse repertoire of mammalian Polycomb repressive complexes.
(A) The core of Polycomb repressive complex 1 (PRC1) comprises a RING1A or RING1B protein (RING1A/B) and one of six Polycomb group RING finger (PGCF) proteins (PCGF1–6). RING1 and PCGF proteins dimerise through their RING domains to form the catalytic core of PRC1, while the RING finger and WD40-associated ubiquitin-like (RAWUL) domains of both proteins interact with a range of auxiliary subunits, giving rise to biochemically distinct PRC1 complexes. Canonical PRC1 (cPRC1) complexes (top) assemble around PCGF2 or PCGF4, and include a chromobox protein (CBX2, 4, 6, 7 or 8) and Polyhomeotic (PHC) protein (PHC1, 2 or 3). In some cases, cPRC1 complexes also contain an SCM protein (SCML1 or 2 or SCMH1). By contrast, variant PRC1 (vPRC1) complexes (bottom) can assemble around all six PCGFs and contain RING and YY1 binding protein (RYBP) or YY1-associated factor 2 (YAF2). The identity of the PCGF protein dictates the incorporation of other auxiliary subunits, resulting in a number of distinct vPRC1 complexes.
(B) The catalytic lobe of PRC2 is formed by a SET (Su(var)3-9, Enhancer-of-zeste and Trithorax)-domain containing enhancer of zeste 1 (EZH1) or EZH2 protein, together with embryonic ectoderm development (EED) and the VEFS (VRN2-EMF2-FIS2-SUZ12) domain of suppressor of zeste 12 (SUZ12). The N-terminal part of SUZ12 forms a distinct regulatory lobe that interacts with retinoblastoma-binding protein 4 (RBBP4) or RBBP7 and with other, auxiliary subunits that give rise to distinct PRC2.1 (top) and PRC2.2 (bottom) complexes. PRC2.1 complexes contain a Polycomblike (PCL) subunit (PCL1/2/3) and Elongin BC and Polycomb repressive complex 2-associated protein (EPOP) or PRC2-associated LCOR isoform 1 (PALI1) or PALI2, whereas PRC2.2 complexes contain adipocyte enhancer binding protein 2 (AEBP2) and Jumanji and AT-rich interaction domain containing 2 (JARID2). Protein domains are italicised. AUTS2, autism susceptibility protein 2; BCOR, BCL6 corepressor; CK2, casein kinase 2; DP-1, dimerization partner 1 (also known as transcription factor Dp-1); E2F6, transcription factor E2F6; FBRS, fibrosin; HDAC1, histone deacetylase 1; KDM2B, lysine-specific demethylase 2B; L3MBTL2, lethal(3)malignant brain tumour-like protein 2; MAX, MYC-associated factor X; MGA, MAX gene-associated; SCMH1, sex combs on midleg homolog 1; SCML1, SCM like 1; SKP1, S-phase kinase-associated protein 1; USP7, ubiquitin carboxyl-terminal hydrolase 7.
A key function of PRC1 complexes is to mono-ubiquitylate histone H2A. Given that E3 ubiquitin ligases are notoriously promiscuous, and that histones have a high density of potential Lys acceptor residues, it was initially puzzling how mammalian PRC1 could preferentially target Lys119 of H2A. The solution to this problem was provided by a landmark study detailing the atomic structure of a RING1B–PCGF4 RING domain dimer bound to an E2 ubiquitin conjugating enzyme and the nucleosome28. This structure revealed extensive contacts between PRC1 and the nucleosome acidic patch, which is a binding surface used by many nucleosome-interacting factors29. Furthermore, the E2 enzyme contacts the N-terminus of RING1B and DNA at the nucleosome dyad, thereby orienting itself to transfer ubiquitin to H2AK119 and explaining the substrate specificity of PRC1.
Although all six mammalian PCGF proteins have highly similar RING domains, the ubiquitin ligase activity of the PRC1 complexes they form differ considerably. Notably, vPRC1 complexes are much more active on nucleosome substrates than their cPRC1 counterparts19,30,31. Although these catalytic differences stem in part from the identity of the PCGF component directly influencing the activity of the core complex, incorporation of the vPRC1-specific auxiliary subunit RYBP also dramatically stimulates E3 ligase activity19,30,32. By contrast, incorporation of CBX proteins into cPRC1 complexes has a far less pronounced stimulatory effect and cPRC1 complexes are thus far less active than vPRC1 complexes.
Polycomb repressive complex 2 and its methyltransferase activity
PRC2 complexes form around a tetrameric core complex consisting of EZH2 or its paralogue EZH1, EED, SUZ12, and RBBP4 or RBBP7 (Figure 1B). Biochemical and structural analyses of PRC2 have largely focused on EZH2-containing complexes, which contribute more prominently to H3K27 methylation in most contexts and are essential for early development33. Cryo-EM structures have revealed that PRC2 forms two distinct lobes: a catalytic lobe and a targeting and regulatory lobe, which are bridged by SUZ1234-36. Within the catalytic lobe, EED and the VEFS (VRN2-EMF2-FIS2-SUZ12) domain of SUZ12 interact with EZH1 or EZH2, causing its SET (Su(var)3-9, Enhancer-of-zeste and Trithorax) domain to transition from an autoinhibitory conformation into an active one that supports methyltransferase activity34,37-41. Also important for the catalytic activity of PRC2 is the CXC domain of EZH2, which engages with nucleosomal DNA, and a patch of residues on the surface of EZH2, which interacts with the N-terminal portion of the H3 tail42,43. Together, these interactions position the N-terminal tail of H3 such that Lys27 is adjacent to the active site of PRC2, to enable its methylation.
The targeting and regulatory lobe of PRC2 includes the N-terminal portion of SUZ12, which binds to RBBP4 or RBBP7 and interacts with various additional auxiliary factors in a mutually exclusive manner44, giving rise to PRC2 assemblies with distinct biochemical properties (Figure 1B). PRC2.1 complexes contain a Polycomblike (PCL) protein (PCL1, 2, or 3) and either PRC2-associated LCOR isoform 1 (PALI1), PALI2 or Elongin BC and Polycomb repressive complex 2-associated protein (EPOP)45-47. By contrast, PRC2.2 complexes contain JARID2 and AEBP2. Importantly, PRC2.1-specific and PRC2.2-specific subunits also affect PRC2 methyltransferase activity48. For example, within PRC2.2, AEBP2 contains a patch of basic amino acids that contribute to nucleosome binding and stimulate H3K27 methylation49. Similarly, incorporation of either EPOP or PALI1 into PRC2.1 supports its methyltransferase activity through an undefined mechanism47,50. Interestingly, a protein that is predominantly expressed in germ-cells, EZH inhibitory protein (EZHIP), was recently shown to interact with the core PRC2 complex and inhibit its methyltransferase activity while also blocking interactions with PRC2.1-specific and PRC2.2-specific subunits51-54. This function suggests that context-specific regulators of PRC2 activity have important roles during development.
Target site identification
Despite an intrinsic capacity to engage with nucleosomes, genome-wide studies have shown that Polycomb complexes are enriched at gene promoters and other gene regulatory elements10,11,55-63, indicating that more specific mechanisms must dictate the observed binding patterns. In the following sections, we examine emerging evidence that primary mechanisms of Polycomb complex targeting in mammals rely on a complement of sequence-specific DNA-binding factors, RNA-dependent mechanisms and binding to CpG islands (CGIs).
Target site recognition through sequence-specific DNA-binding factors
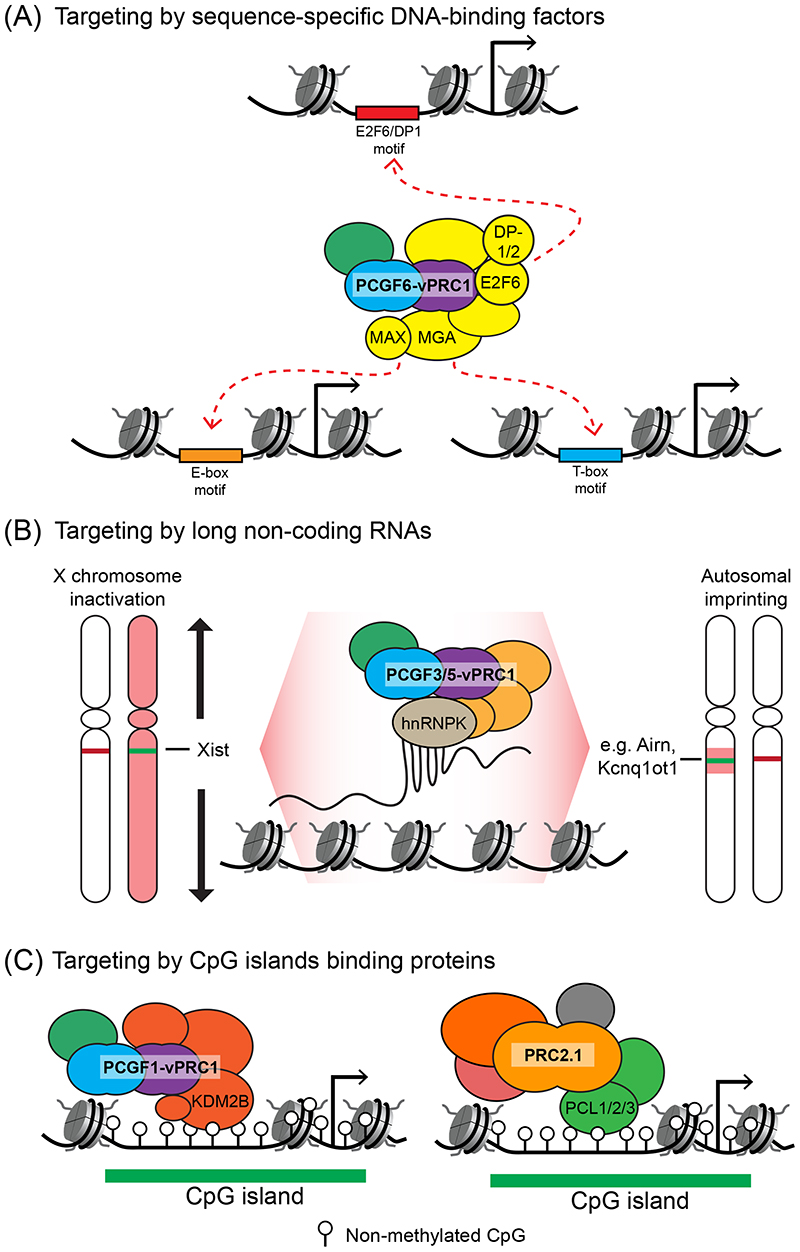
In D. Melanogaster, targeting of Polycomb complexes is thought to rely primarily on sequence-specific DNA-binding factors. However, many of these factors lack clear mammalian orthologues, or if they are present in mammals, do not target Polycomb complexes to chromatin64-66. This difference has brought into question whether these mechanisms are also important for mammalian Polycomb target site identification. In beginning to address this important question, it has recently been shown that the PCGF6-vPRC1 complex, which is absent in D. Melanogaster, stably incorporates auxiliary subunits that function as sequence-specific DNA-binding domains (Figure 2A)19,23,67-69. For example, the subunit MAX gene-associated protein (MGA) contains bHLH and T-box DNA-binding domains70. Studying these domains has revealed that MGA dimerises with MYC-associated factor X (MAX) to bind E-box motifs (5’-CACGTG-3’), and that it can also use its T-box domain to recognise longer T-box motifs (5’-TCACACCT-3’)67,71-73. These domains are important for PCGF6-vPRC1 occupancy at a subset of its target sites, which in somatic cells includes a number of germline-specific genes67,74,75. In addition, E2F6–DP-1 (or DP-2) heterodimers also associate with PCGF6-vPRC1 and provide the complex with distinct target-site specificity in other contexts71,72. Together, these findings demonstrate that PCGF6-vPRC1 utilises sequence-specific DNA-binding activities to identify a subset of its target sites in the genome (Figure 2A), and provide clear and direct evidence for sequence-specific targeting in mammals.
Figure 2. Primary mechanisms of Polycomb target-site identification.
(A) Sequence-specific DNA-binding factors in the Polycomb group RING finger 6 (PCGF6)-containing variant Polycomb repressive complex 1 (vPRC1) complex recognise DNA sequence motifs at target sites. These DNA-binding factors include MAX gene-associated (MGA)–MYC-associated factor X (MAX) and E2F6–dimerization partner 1 or 2 (DP-1/2) dimers, which bind to different DNA sequence motifs and contribute to sequence-specific PCGF6-vPRC1 targeting in different contexts.
(B) Polycomb complexes can identify chromosomal binding sites during X chromosome inactivation through the long-noncoding RNA (lncRNA) X inactivation specific transcript (XIST); in autosomal imprinted regions mono-allelic gene repression is achieved through the lncRNAs Airn and Kcnq1ot1. In these regulatory contexts, the adaptor protein heterogeneous nuclear ribonucleoprotein K (hnRNPK) is thought to interact with the lncRNA and recruit the PCGF3/5-vPRC1 complex. These mono-allelic targeting cases are atypical in that they nucleate the binding of Polycomb complexes at large chromosomal regions, whereas Polycomb complex targeting to the majority of genomic sites is more punctate and associated with gene regulatory elements (see parts A and C).
(C) Both PCGF1-vPRC1 and PRC2.1 are targeted to CpG islands. The lysine-specific demethylase 2B (KDM2B) subunit of PCGF1-vPRC1 contains a zinc finger-CxxC (ZF-CxxC) domain that binds specifically to non-methylated CpG dinucleotides. In PRC2.1, the Polycomb-like 1 (PCL1), PCL2 or PCL3 subunit contains a winged-helix domain that binds non-methylated CpG dinucleotides in certain sequence-contexts.
Other mammalian Polycomb repressive complexes do not stably incorporate proteins with sequence-specific DNA-binding domains. Nevertheless, certain Polycomb complexes can engage in more transient interactions with DNA-binding factors and this engagement appears to be important for target-site recognition in certain contexts. For example, PCGF3-vPRC1 recognises some of its target sites in embryonic stem cells (ESCs) by interacting with the transcription factors upstream stimulatory factor 1 (USF1) and USF272. Other DNA-binding factors including REST, RUNX1 and SNAIL1 have been proposed to contribute to PRC1 or PRC2 targeting in certain cell types76-79. Despite these reports, in most cases direct interactions between DNA-binding factors and Polycomb complexes have not been unequivocally demonstrated, and they often only account for a subset of Polycomb complex binding events in a limited number of developmental contexts. Therefore, it appears unlikely that sequence-specific DNA-binding factors alone can explain how Polycomb complexes recognise their target sites in mammals.
Target site identification through chromatin-associated RNA
As an alternative to DNA-binding transcription factors, it was suggested that targeting of Polycomb complexes could be achieved through long non-coding RNA (lncRNAs) that associate with defined sites in chromatin80. Mammalian X chromosome inactivation (XCI), which is mediated by the lncRNA X inactive specific transcript (XIST), involves Polycomb repressive complexes. During XCI, XIST is expressed from one of the two female X chromosomes and spreads in cis to drive mono-allelic chromosomal gene silencing. Detailed characterization of this process has revealed that the PCGF3/5-vPRC1 complex interacts with the RNA-binding protein heterogeneous nuclear ribonucleoprotein K (hnRNPK), which recognises XIST, leading to PCGF3/5-vPRC1 enrichment on the inactive X chromosome (Figure 2B)81-84. However, this example is somewhat atypical for Polycomb complex targeting, in that XIST leads to enrichment of PCGF3/5-vPRC1 across the entire inactivated X chromosome, whereas on autosomes Polycomb complex enrichment is typically more restricted to gene regulatory elements.
Based on early observations made in XIST studies, numerous other lncRNAs have been proposed to target Polycomb complexes to sites in autosomes. Notably, the lncRNA HOX transcript antisense RNA (HOTAIR) was reported to function as a trans-acting repressor of the HOXD locus by physically interacting with and recruiting PRC285-87. However, recent work has demonstrated that transcription repression by HOTAIR is independent of PRC2, and that PRC2 recruitment to the HOXD locus occurs primarily in response to gene silencing, not through direct targeting by HOTAIR 88. These findings highlight the need for caution when interpreting how lncRNAs influence gene expression and the recruitment of Polycomb complexes80,89,90. However, consistent with a potential role for lncRNAs in targeting the Polycomb complexes to chromatin, the lncRNAs Airn and Kcnq1ot1 promote mono-allelic expression of autosomal genes in mouse trophoblast stem cells through a mechanism that appears to be highly analogous to XCI (Figure 2B)91-93. Like XIST, Airn and Kcnq1ot1 associate with target sites in chromatin and utilise hnRNPK to form megabase-sized transcriptionally repressed genomic domains that are enriched for Polycomb complexes. In light of these findings, it is tempting to speculate that some lncRNAs have co-opted the Polycomb system as a mechanism to achieve mono-allelic gene regulation94.
In addition to lncRNAs, recent studies have suggested that RNA–DNA hybrid structures known as R-loops may also support Polycomb target-site recognition. R-loops were found to occur at a number of Polycomb target genes and their enzymatic resolution perturbed Polycomb complex targeting at some sites95. By contrast, in other contexts, R-loops were reported to negatively affect PRC2 binding and activity96. Although there appears to be a connection between Polycomb complexes and R-loops, the underlying biochemical mechanisms are unknown and the extent to which R-loops mediate Polycomb complex targeting to chromatin remains to be elucidated.
Targeting to CpG islands
Polycomb complexes appear to recognise some of their target sites using sequence-specific DNA-binding factors or through associating with lncRNAs. However, these mechanisms alone cannot explain all Polycomb complex binding observed in mammalian genomes, suggesting that additional targeting mechanisms exist. Early genome-wide studies revealed that Polycomb complexes associate primarily with CGIs61,97. CGIs are short (1–2 kb) regions of CpG-rich DNA that are associated with approximately 70% of mammalian gene promoters98,99. In CGIs, CpG dinucleotides remain free of DNA methylation, whereas elsewhere in the genome they are heavily methylated and function to maintain heterochromatin and repress the expression of parasitic DNA elements100,101. CGIs are absent in non-vertebrate model organisms, including D. melanogaster, which lacks DNA methylation. This led to speculation that CGIs may be used by Polycomb complexes to identify target gene promoters and other regulatory elements in mammals61,102.
Molecular evidence that CGI recognition may be important for Polycomb complex targeting first came from biochemical purifications of mammalian PRC1 complexes, in which the PCGF1-vPRC1 complex was shown to stably integrate lysine-specific demethylase 2B (KDM2B)103,104. KDM2B contains a zinc finger-CxxC domain that specifically binds to non-methylated CpG dinucleotides105,106. Consistent with this specificity, KDM2B can target the PCGF1-vPRC1 complex to CGIs (Figure 2C)107-110. PCL proteins in PRC2.1 complexes were also recently shown to bind to non-methylated CpG-containing DNA through a winged-helix domain that has a preference for certain sequences and is also influenced by the helical properties of DNA (Figure 2C)111,112. Consistent with in vitro evidence that PRC2 may function in some cases as a dimer113, it was recently proposed that a PRC2.1 dimer could stabilise binding to CpG-rich DNA, possibly by enabling simultaneous engagement with multiple non-methylated CpGs114. Although the precise contribution of dimerization to PRC2.1 function in vivo remains to be elucidated, the winged-helix domains of PCL proteins are crucial for PRC2.1 targeting, indicating that CGI binding is also central to target-site identification by PRC2112.
The Polycomb chromatin domain
Once Polycomb complexes engage with target sites, this primary targeting must then be converted into the formation of repressive Polycomb chromatin domains, which can extend up to tens of kilobases and have extremely high levels of H2AK119ub1, H3K27me3, and Polycomb complex occupancy. In the following sections, we describe an intricate series of feedback and communication mechanisms that link the activity of Polycomb complexes on chromatin to enable the formation and maintenance of Polycomb chromatin domains.
H2AK119ub1 is recognised by PRC2.2 and vPRC1
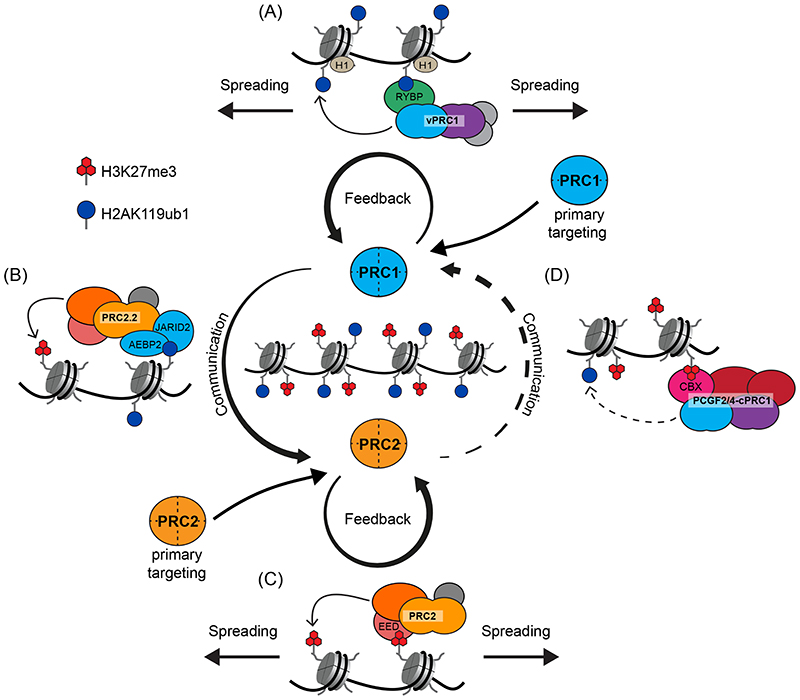
The complex regulation of mammalian gene promoters and the diversity of Polycomb targeting mechanisms have made it challenging to study the formation of Polycomb chromatin domains. To circumvent this difficulty, tethering experiments have been employed, in which specific Polycomb complexes can be artificially nucleated at an ectopic site in the genome and their capacity to communicate with other Polycomb components and to form Polycomb chromatin domains can be examined. Such experiments revealed that tethering of PRC1 was sufficient to drive de novo enrichment of PRC2, accumulation of H3K27me3 and the formation of a Polycomb chromatin domain that spread several kilobases from the primary engagement site32,107,115. Interestingly, this activity was exclusive to vPRC1 complexes and relied on their capacity to ubiquitylate H2AK119 (Ref.30) (Figure 3A). Subsequently, a biochemical link between H2AK119ub1 and PRC2 was narrowed down to the JARID2 subunit of PRC2.2, which can directly bind to H2AK119ub1 through a ubiquitin binding motif116; this interaction also stimulates the methyltransferase activity of PRC2117 (Figure 3B). The structural basis of this interaction was recently elucidated in a cryo-EM model of PRC2.2 bound to an H2AK119ub1-containing mono-nucleosome, which showed JARID2 interacts with mono-ubiquitylated H2A on one face of the nucleosome through its ubiquitin binding motif118. Interestingly, AEBP2 interacts with a second mono-ubiquitylated H2A on the opposite face of the nucleosome through tandem C2H2 zinc fingers. This indicates that auxiliary subunits in PRC2.2 have a specialised role in binding to H2AK119ub1-containing nucleosomes to tri-methylate H3K27, which is consistent with earlier biochemical work116,117. The importance of H2AK119ub1-mediated vPRC1 communication with PRC2 in the formation of Polycomb chromatin domains has also been shown through detailed genetic perturbation studies in mouse ESCs. Disruption of vPRC1 complexes caused major reductions in PRC2 occupancy and H3K27 tri-methylation in Polycomb chromatin domains, and this relied on their E3 ligase activity, consistent with ectopic tethering experiments and recent work in the developing mouse and zebrafish embryo12,30,72,107,115,119-125.
Figure 3. Formation and spreading of Polycomb chromatin domains.
Primary targeting of Polycomb repressive complex 1 (PRC1) and PRC2 is followed by feedback and communication mechanisms that enable the formation and spreading of Polycomb chromatin domains, which are characterised by elevated Polycomb complex occupancy and high levels of mono-ubiquitylated histone H2A Lys119 (H2AK119ub1) and tri-methylated histone H3 Lys27 (H3K27me3).
(A) H2AK119ub1 is recognised by the RING and YY1 binding protein (RYBP) subunit (or by the YY1-associated factor 2 (YAF2) subunit; not shown) of variant PRC1 (vPRC1) complexes. This creates a feedback mechanism, supported by histone H1, that reinforces vPRC1 binding and amplifies H2AK119 ubiquitylation, thereby enabling spreading of vPRC1 and H2AK119ub1 away from the primary vPRC1 targeting site.
(B) PRC1 complexes ubiquitylate H2AK119, which is recognised by the Jumanji and AT-rich interaction domain containing 2 (JARID2) and adipocyte enhancer binding protein 2 (AEBP2) subunits of PRC2.2. This causes elevated PRC2 occupancy and stimulates the tri-methylation of H3K27. Therefore, H2AK119ub1 facilitates communication between PRC1 and PRC2 in Polycomb chromatin domains.
(C) H3K27me3 is recognised by the embryonic ectoderm development (EED) subunit of PRC2, which allosterically activates its methyltransferase activity. H3K27me3 creates a feedback mechanism that reinforces PRC2 binding and amplifies H3K27 tri-methylation, thereby enabling spreading of PRC2 and H3K27me3 away from the primary targeting site.
(D) H3K27me3 is also recognised by the chromobox (CBX) subunit (CBX2, 4, 6, 7 or 8) of canonical PRC1 (cPRC1) complexes. Although cPRC1 complexes are less catalytically active than vPRC1 complexes (as indicated by the dotted arrow), in some contexts their activity may lead to H2AK119 ubiquitylation. Therefore, H3K27me3 can facilitate communication between PRC2 and PRC1 in Polycomb chromatin domains.
In addition to its influence on PRC2.2, H2AK119ub1 also reinforces chromatin binding and modification by vPRC1 through a zinc finger in RYBP, which can directly bind H2AK119ub1 (Figure 3A)32,117,126. This interaction forms the basis of a feedback mechanism that supports spreading of H2AK119ub1 onto neighbouring nucleosomes in a manner that appears to be aided by histone H1 (Ref.32). Together, these data demonstrate that once Polycomb target sites are identified through the primary targeting of vPRC1, feedback mechanisms inherent to vPRC1 complexes and H2AK119ub1 (Figure 3A), coupled with their capacity to communicate with PRC2.2 (Figure 3B), are essential for the formation and spreading of Polycomb chromatin domains.
H3K27me3 stimulates PRC2 activity and is recognised by cPRC1
Although H3K27me3 and the occupancy of PRC2.2 in Polycomb chromatin domains relies on PRC1 and H2AK119ub1, primary targeting modalities also enable PRC2 to identify target sites and catalyse H3K27 tri-methylation. PRC2 can then bind the product of its own catalysis, H3K27me3, through a WD40-repeat domain in EED, causing allosteric activation of its methyltransferase activity39,127,128 (Figure 3C). PRC2 activation creates a feedback mechanism that enables spreading of H3K27me3 along chromatin129. A recently elucidated cryo-EM model of PRC2 bound to a dinucleosome revealed that EED engages with one H3K27me3-containing nucleosome and positions the SET domain of EZH2 to methylate an unmodified H3 tail on an adjacent nucleosome 43. The geometry of this interaction also revealed that a short length of DNA linker between nucleosomes is important for optimal methyltransferase activity, which is consistent with evidence that PRC2 prefers more compact chromatin substrates130.
H3K27me3 is also recognised by a chromodomain in the CBX subunit of cPRC1 complexes, suggesting that PRC2 can communicate with cPRC1 through H3K27me3 (Figure 3D)6,8,131,132. In agreement with this possibility, early functional experiments showed that PRC2 activity was required for cPRC1 occupancy in Polycomb chromatin domains10. However, consistent with the fact that cPRC1 complexes have extremely low E3 ligase activity in vitro 30, PRC2 and cPRC1 contribute only minimally to H2AK119ub1 levels in vivo 12,27,125,133. Furthermore, ectopic tethering of PRC2 or cPRC1 to chromatin in ESCs did not lead to elevated H2AK119ub1 levels, indicating that these complexes are incapable of efficiently driving de novo Polycomb chromatin domain formation in this context107. Therefore, in contrast to vPRC1 complexes, which communicate effectively with PRC2.2 to drive H3K27 tri-methylation, PRC2 appears to work much less efficiently through cPRC1 to reinforce H2AK119 ubiquitylation. Nevertheless, overexpression of the cPRC1 subunit CBX7 was recently shown to support H3K27me3-dependent maintenance of H2AK119ub1 at an artificial target gene promoter134, suggesting that in some circumstances PRC2 may support this process more efficiently. In summary, while primary targeting modalities and recognition of H2AK119ub1 are important for nucleation of PRC2 at target sites, H3K27me3 deposition is necessary to reinforce PRC2 binding, support spreading of PRC2 and H3K27me3, and stabilise cPRC1 occupancy during Polycomb chromatin domain formation.
cPRC1 supports the organization of Polycomb chromatin domains in the nucleus
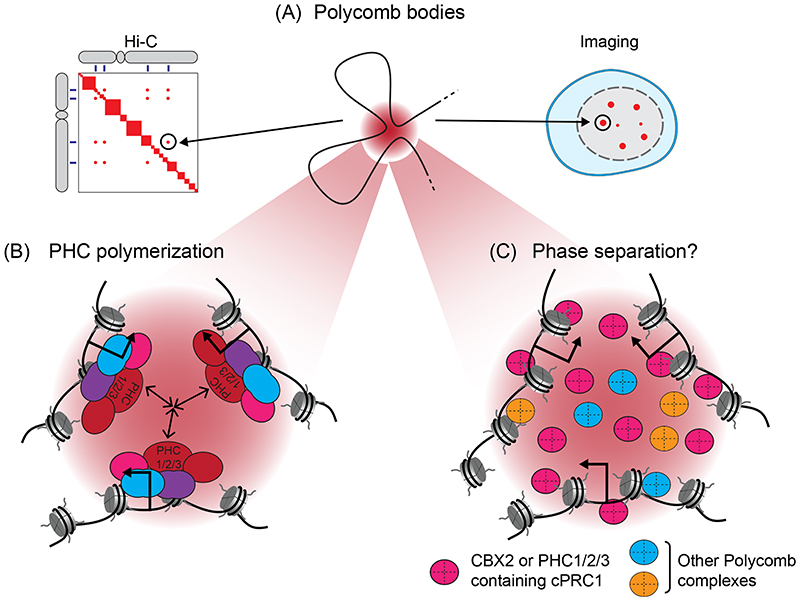
Although cPRC1 complexes contribute minimally to H2AK119 ubiquitylation, they have evolved additional biochemical features that enable them to control the spatial organisation of Polycomb chromatin domains in the nucleus. Imaging and chromatin conformation capture-based approaches have demonstrated that even when separated by up to several megabases across a chromosome, distinct Polycomb chromatin domains can interact in three-dimensional space135-142 (Figure 4A). In imaging experiments, these interactions appear as focal accumulations of Polycomb proteins, which are often referred to as Polycomb bodies143-145. The formation of these interactions relies on PHC proteins in cPRC1 complexes, which at least in vitro can utilise their sterile alpha motif (SAM) to polymerise in a head-to-tail orientation and form long filaments146-150 (Figure 4B). How these filaments support three-dimensional chromatin interactions remains poorly understood, but one could envisage cPRC1 filaments bridging distinct Polycomb chromatin domains. Although Polycomb chromatin domains form prominent interactions in three-dimensional space, they are not static structural entities151. Instead, binding of Polycomb proteins at these sites is highly dynamic144,152-155 and cohesin, which dynamically extrudes chromatin loops, counteracts interactions between Polycomb chromatin domain139,156,157.
Figure 4. Polycomb bodies, long-range interactions, and phase separation.
(A) Distinct Polycomb chromatin domains can interact in three dimensional space, even when separated by very large distances across the genome. In high-throughput chromosome conformation capture (Hi-C), these interactions correspond to regions of high contact frequency (left). In imaging experiments, they correspond to foci of Polycomb complex components, mono-ubiquitylated histone H2A Lys119 (H2AK119ub1) and tri-methylated histone H3 Lys27 (H3K27me3), which are often referred to as Polycomb bodies (right).
(B) Long-range interactions between Polycomb chromatin domains require the Polyhomeotic (PHC) subunit (PHC1, 2 or 3) of canonical Polycomb repressive complex 1 (cPRC1), which can polymerise through its sterile alpha motif (SAM).
(C) Components of cPRC1, including chromobox 2 (CBX2) and the SAM domain of PHC1, 2 or 3, can undergo liquid–liquid phase separation in vitro and form nuclear condensates in vivo, which share similarities with Polycomb bodies. These condensates could possibly augment Polycomb complex activities and may reinforce long-range interactions between Polycomb chromatin domains.
In addition to supporting long-range interactions between Polycomb chromatin domains, components of cPRC1 complexes can undergo liquid–liquid phase separation in vitro and form nuclear condensates in vivo, which appear similar to Polycomb bodies158,159 (Figure 4C). The capacity for liquid–liquid phase separation resides with CBX2, which contains a positively charged disordered region, and with the SAM of PHC proteins through a mechanism that is enhanced by, but not strictly dependent upon, the capacity of PHC to polymerise 158-161. Condensates can enhance cellular processes by sequestering or excluding macromolecules162. In line with this concept, condensates formed by CBX2 or the PHC SAM domain were shown to concentrate nucleosomes and nucleic acids, while also permitting entry of other Polycomb complexes, suggesting that condensates could augment Polycomb activities on chromatin161 or limit the access of other factors to Polycomb chromatin domains. However, quantitative imaging has also shown that concentrations of cPRC1 components inside Polycomb bodies are in some contexts below what is thought to be required to support liquid–liquid phase separation, bringing into question whether Polycomb bodies are formed by this process152,163. Further study of Polycomb protein condensates and their relevance to Polycomb functions is therefore required.
The maintenance of Polycomb chromatin domains following DNA replication
The integrity of Polycomb chromatin domains is challenged by passage of the replication machinery which displaces parental nucleosomes from DNA and necessitates the incorporation of new unmodified histones and the reincorporation of modified parental histones into the replicated daughter strands164,165. Communication and feedback mechanisms inherent to Polycomb complexes32,127 could in theory support copying and epigenetic maintenance of Polycomb chromatin domains, but this would rely on modified parental nucleosomes being deposited in the same location on daughter strands. Elegant experiments examining the kinetics of histone deposition during DNA replication have recently shown that H3K27me3-containing nucleosomes are incorporated into daughter DNA strands almost precisely where they originated from in the parental DNA 166,167, providing a potential conduit for maintaining the identity of Polycomb chromatin domains168,169.
Interestingly, however, a considerable lag exists between DNA replication and the establishment of H3K27me3 on the newly incorporated, unmodified histone H3, despite reports that Polycomb repressive complexes can travel with the replication machinery169-171. Furthermore, H3K27me3 profiles can be re-established de novo following complete erasure of H3K27me3129,172 and work in D. melanogaster has shown that long-term maintenance of H3K27me3 in Polycomb chromatin domains relies on the underlying DNA sequence173,174. Together, these observations have brought into question the extent to which epigenetic maintenance through feedback mechanisms in Polycomb chromatin domains contributes to their re-establishment following DNA replication. It will be important to understand in more detail the kinetics of re-establishment of Polycomb chromatin domains following replication, including whether parental H2AK119ub1 is also faithfully redeposited. Understanding these basic principles will be essential for defining whether epigenetic maintenance is important for the function of Polycomb chromatin domains in actively dividing cells.
Controlling Polycomb chromatin domains
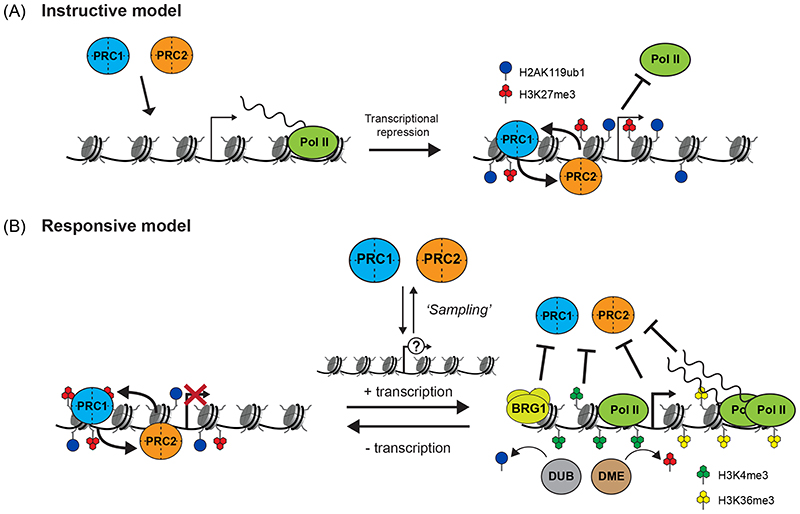
It is often posited that chromatin modifying complexes are recruited to defined target sites in the genome, where they modify histones to drive gene activation or repression175. However, these simple instructive models appear for the most part to be incompatible with our evolving understanding of where and how Polycomb chromatin domains form and function. If instructive processes dictated Polycomb chromatin domain formation (Figure 5A), one would expect that all sites in the genome where primary targeting mechanisms allow Polycomb complexes to engage would acquire repressive Polycomb chromatin domains. However, genome-wide studies have shown that only a small subset of sites where Polycomb complex binding can be detected ultimately go on to form Polycomb chromatin domains that have high levels of H2AK119ub1, H3K27me3 and Polycomb complex occupancy12,133,176,177. For example, the PCGF1-vPRC1 complex is detected at most CGIs in the genome, albeit at low levels12,108-110. Yet, despite the inherent de novo Polycomb chromatin domain-forming activity of PCGF1-vPRC1107, only ~20% of CGIs in a given cell type will acquire a domain108,112, indicating that this process must be highly regulated. In this section we discuss the emerging principles that regulate Polycomb chromatin domain formation and how this is related to transcription.
Figure 5. Models of Polycomb chromatin domain formation and gene regulation.
(A) An instructive model, in which Polycomb complexes are recruited to target sites (left), where they initiate the formation of Polycomb chromatin domains (right) and drive the repression of transcribed genes.
(B) A responsive model, in which Polycomb complexes dynamically ‘sample’ potential target sites for susceptibility to Polycomb chromatin domain formation. In particular, the Polycomb group RING finger 1 (PCGF1)-containing variant Polycomb repressive complex 1 (vPRC1) and PRC2.2 complex, through their capacity to bind CpG islands (not shown), could dynamically engage with approximately 70% of gene promoters. At lowly or untranscribed genes (left), these complexes could potentially sense and respond to the (near) absence of transcription by ubiquitylating histone H2A Lys119 (H2AK119ub1) and tri-methylating histone H3 Lys27 (H3K27me3) to initiate the formation and spreading of Polycomb chromatin domains, which could help counteract low-level or inappropriate transcription and maintain an inactive chromatin state to protect cell identity. However, at expressed genes (right), transcription-associated features including H3K4me3 and H3K36me3, high levels of nascent transcripts, BRG1-mediated chromatin remodelling, and deubiquitylase (DUB) and demethylase (DME) activities, counteract H2AK119 and H3K27 modification, thereby blocking Polycomb chromatin domain formation and limiting Polycomb function at these genes.
Pol II, RNA polymerase II.
Transcription regulates susceptibility to Polycomb chromatin domain formation
A unifying feature of target sites that form Polycomb chromatin domains is extremely low or non-detectable levels of transcription, raising the possibility that transcription itself limits the activity of Polycomb complexes. In agreement with this notion, treating cells with transcription inhibitors can stabilise PRC2 binding and increase H3K27me3 levels at thousands of previously active CGI-associated gene promoters178. When a polyadenylation signal, which promotes transcription termination, was inserted adjacent to an active CGI-associated promoter, this had a similar effect179. These findings are consistent with experiments showing that synthetic CpG-rich non-methylated DNA can recruit Polycomb complexes and lead to the apparent formation of Polycomb chromatin domains, but only when these sites lack binding of transcription activators or are not in close proximity to enhancer elements56,180,181. Furthermore, experiments examining the kinetics of Polycomb chromatin domain formation during cellular-state transitions demonstrated that this primarily occurs following transcription cessation130,182. Together, these observations indicate that despite Polycomb complexes broadly engaging with regulatory elements via targeting modalities such as CGI recognition, transcription potently counteracts the formation of Polycomb chromatin domains.
Nascent RNA regulates the activity of Polycomb repressive complex 2
The realisation that gene activity appears to be incompatible with Polycomb chromatin domain formation has led to a concerted effort to understand the mechanisms that regulate this process. Although some lncRNAs appear to target Polycomb complexes to specific sites, an overwhelming body of evidence now also indicates that PRC2 binds promiscuously to nascent RNA113,183-189. Several studies have reported that nascent RNA competes for PRC2 binding and displaces it from chromatin, while also inhibiting its methyltransferase activity179,190-193, both of which would antagonise the formation of Polycomb chromatin domains at actively transcribed sites. However, other studies have contradicted this view and proposed that interaction with nascent RNA, either directly by PRC2 (Ref.194) or through the pre-mRNA splicing factor RBFOX2 (Ref.195), supports PRC2 occupancy at target sites.
To reconcile these seemingly contradictory effects, an ‘RNA-bridging model’ has been proposed194. In this model, nascent transcripts from very lowly transcribed genes help bring PRC2 into proximity with promoter elements to support chromatin binding and tri-methylation of H3K27. However, at more highly transcribed genes, elevated levels of nascent transcripts would compete with PRC2 for chromatin binding and decrease H3K27 tri-methylation. Although the molecular details of this model remain to be fully tested, it appears that PRC2 is highly attuned to sensing transcript levels and using this information to regulate the formation of Polycomb chromatin domains.
The influence of transcription-associated chromatin features
In addition to producing nascent RNA, transcription also profoundly influences the chromatin environment at transcribed genes. For example, during transcription initiation, histone methyltransferases tri-methylate H3K4 at gene promoters, whereas elongation is associated with H3K36 tri-methylation in gene bodies97,196. Importantly, in vitro studies have demonstrated that the methyltransferase activity of PRC2 is inhibited by the presence of H3K4me3, H3K36me2 or H3K36me3 (Ref.42,197-202), and disruption of H3K4 methyltransferase activity in vivo can lead to enhanced Polycomb chromatin domain formation203. In addition, histone demethylases act as part of transcription co-activator complexes to remove H3K27 methylation204-209, and transcription-associated H3K27, H4K16 and H4K20 acetylation counteracts PRC2 activity210-215. Similarly, the H2AK119ub1-specific deubiquitylases USP16 and 2A-DUB (also known as MYSM1) have been shown to associate with promoters of actively transcribed genes and counteract H2AK119 ubiquitylation216,217.
In addition to histone modification-dependent effects, the chromatin remodelling ATPase BRG1 can actively evict PRC1 from promoters of transcribed target genes218-220. This activity was reported to target RYBP-containing vPRC1 complexes, suggesting that promoter-associated chromatin remodelling activities can displace vPRC1 complexes and limit H2AK119 ubiquitylation221. Together, these findings suggest that although primary targeting activities may enable Polycomb complexes to engage with a wide range of gene promoters, transcription-associated histone modifications and chromatin remodelling activities at regulatory elements can limit the catalysis and feedback mechanisms required for efficient formation of Polycomb chromatin domains.
Target sampling and a responsive model of Polycomb chromatin domain formation
Our growing understanding of Polycomb complex targeting and the mechanisms that control Polycomb chromatin domain formation appear to be consistent with a dynamic sampling mode of action that is responsive to transcription222 (Figure 5B). According to this model, DNA-binding activities associated with Polycomb complexes would provide a mechanism to dynamically engage with, or sample, potential target sites. Central to this process would be PCGF1-vPRC1 and PRC2.1, which can recognise non-methylated DNA in CGI-elements that are associated with most mammalian gene promoters108-112. Sampling could allow Polycomb complexes to constantly interrogate a wide range of potential target sites, with only those that are lowly or not transcribed being susceptible to Polycomb-mediated histone modification, which is necessary to initiate the feedback and communication mechanisms required for efficient formation and spreading of Polycomb chromatin domains.
A responsive mode of Polycomb function based on target site sampling would in theory provide a generic mechanism to form Polycomb chromatin domains at non- or lowly transcribed genes without the need to evolve complex cell-type specific targeting mechanisms. As such, the Polycomb system could function to protect inactive genes against low-level or inappropriate transcription activation signals and help to maintain gene repression in many, if not all, cell types. This model is also consistent with early genetic experiments in fruit flies and mammals, in which Polycomb proteins were observed to maintain, rather than initiate, gene repression3,223-225. Nevertheless, more work is required to test the validity of a sampling-based model for the formation and function of Polycomb chromatin domains in different biological contexts.
Polycomb-mediated gene regulation
After receptive target sites establish Polycomb chromatin domains, these then have an important role in controlling gene expression. In this section we examine the mechanisms of Polycomb chromatin domain-mediated gene repression, discuss atypical cases of gene activation by Polycomb complexes, and consider how CGIs might utilise Polycomb complexes to create bistable chromatin states that shape gene expression during cell differentiation.
Transcription repression by Polycomb chromatin domains
Early models posited that the Polycomb system may repress transcription through cPRC1 binding to H3K27me3 and compacting chromatin to limit access to gene regulatory elements137,149,150,158,159,226,227. Although this mechanism may contribute to Polycomb-mediated gene repression in some contexts, mice deficient for cPRC1 components often have milder phenotypes than mice in which vPRC1 components are perturbed, and cPRC1-deficient ESCs exhibit relatively few gene expression changes12,67,81,107,148,225,228-230. Furthermore, chromatin accessibility does not always change when Polycomb systems are disrupted231,232. Together, these findings indicate that, at least during early development and in ESCs, Polycomb chromatin domain features distinct from H3K27me3 and cPRC1 must contribute centrally to transcription repression.
In line with this possibility, recent studies have pointed towards a central role for H2AK119ub1 and vPRC1 in gene repression. This is evident in ESCs, where disruption of PRC1 resulted in increased expression of thousands of Polycomb target genes, effects which were almost entirely dependent on PCGF1/3/5/6-vPRC1 complexes12. Importantly, disruption of the catalytic activity of PRC1 recapitulated these gene expression defects despite vPRC1 binding being retained at target sites, suggesting that gene repression by PRC1 in this context is mediated by H2AK119 ubiquitylation119,121. H2AK119ub1 having a central role in gene repression is consistent with PRC1 binding to chromatin being highly dynamic and displaying extremely low target site occupancy152,154. Although the precise mechanism(s) by which H2AK119ub1 affects transcription remain to be determined (Figure 6A), it does not appear to rely on PRC2 occupancy233. Furthermore, although H2AK119ub1 reader proteins, including RYBP and the chromatin remodelling factor RSF1, have been linked with transcription repression30,32,126,234, their activities appear insufficient to account for PRC1-mediated gene repression. An alternative possibility is that the addition of a bulky ubiquitin moiety to H2A could more directly counteract the function of the transcription machinery235,236. Consistent with this possibility, acute disruption of PRC1 and loss of H2AK119ub1 caused rapid new binding of Pol II and elevated the frequency of transcription bursts, suggesting PRC1 and H2AK119ub1 affect transcription initiation233, which is in agreement with previous reports that PRC1 might antagonise the assembly or activity of the transcription pre-initiation complex237,238. Other reports have suggested that PRC1 limits transcription by constraining a post-initiation, promoter-proximal poised form of Pol II239,240. More detailed biochemical work is required to define the mechanisms by which H2AK119ub1 influences transcription.
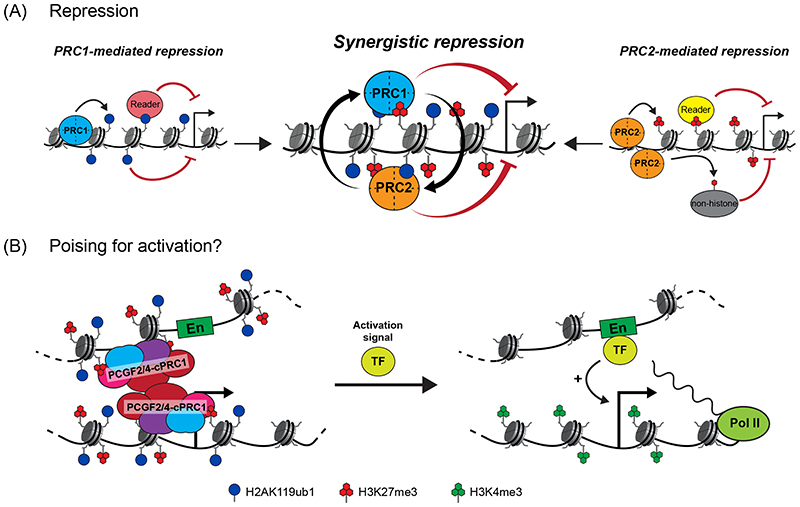
Figure 6. Mechanisms of Polycomb-mediated gene regulation.
(A) Despite the integration of their activities in Polycomb chromatin domains, the mechanisms that enable Polycomb repressive complex 1 (PRC1) and PRC2 to counteract transcription appear to be distinct. PRC1-mediated gene repression is driven by ubiquitylation of histone H2A Lys119 (H2AK119ub1), possibly mediated by the activity of H2AK119ub1-reader proteins or, more directly, by the installation of the bulky ubiquitin moiety into chromatin and thus antagonising some aspect of transcription (left). PRC2-mediated repression appears to involve readers of tri-methylated histone H3 Lys27 (H3K27me3) or methylation of non-histone substrates (right). Importantly, although PRC1 and PRC2 can independently counteract transcription, the communication and feedback between PRC1 and PRC2, which support the formation of Polycomb chromatin domains, appear in some contexts to synergise the repressive activities of PRC1 and PRC2 at target genes (centre), thereby providing a robust barrier against inappropriate gene expression.
(B) In some contexts, Polycomb complexes can activate genes. Canonical PRC1 (cPRC1) mediates the formation of chromatin topologies that can bring poised enhancers (En) and their target promoters into close proximity (left). Once activation signals are received at the enhancer through transcription factor (TF) binding, this could support rapid induction of transcription (right).
PCGF2/4, Polycomb group ring finger 2 or 4; Pol II, RNA polymerase II.
Consistent with the catalytic activity of PRC1 being required for Polycomb-mediated gene repression in ESCs, an essential role for H2AK119ub1 in Polycomb target gene repression was recently demonstrated by examining the early stages of neuronal cell fate restriction241. However, this requirement was less pronounced at later stages of neuronal differentiation, when a shift towards H2AK119ub1-independent gene repression was observed. These findings suggest that H2AK119ub1 could possibly confer a form of repression in stem cells that can be overcome in response to appropriate developmental gene expression cues. By contrast, as cells commit to particular lineages and subsets of genes will no longer be expressed, the Polycomb system may employ additional mechanisms, perhaps involving structural changes to chromatin, that enable long-term and robust maintenance of repression. This possibility has been supported by studies examining the contribution of the catalytic activity of PRC1 to mouse development, which reported that PRC1 uses a combination of H2AK119ub1-dependent and H2AK119ub1-independent mechanisms to control gene expression119,123,133,137,241. However, it has subsequently been shown that PRC1 catalytic mutations used in these studies do not fully eliminate H2AK119 ubiquitylation119,241. Nevertheless, in some cases in D. melanogaster, the repression of Hox genes and other Polycomb target genes can be maintained by PRC1 independently of H2AK119 ubiquitylation242. Clearly more work is required to define the contribution H2AK119ub1-dependent and H2AK119ub1-independent mechanisms to Polycomb-mediated gene repression in different cell types and developmental stages.
PRC2 also has important roles in Polycomb-mediated gene repression, yet its disruption in mammals typically has less severe effects on gene expression and development than removal of PRC110,124,125,243-248. Studies in D. melanogaster have suggested that PRC2-mediated repression relies on H3K27 methylation, as a histone H3 Lys27-substitution mutation that renders this residue refractory to methylation largely phenocopies the gene expression and developmental defects observed when PRC2 is disrupted249. Conversely, a hyperactive form of EZH2, which causes increased levels of H3K27me3, drives inappropriate repression of target genes250. Consistent with these findings, chemical inhibitors that limit the methyltransferase activity of PRC2 and cause a reduction in H3K27 methylation, are associated with derepression of Polycomb target genes in mammals251,252, as is replication-mediated dilution of H3K27me3 (Ref.252). Recently, an evolutionarily-conserved bromo adjacent homology (BAH) domain in BAHD1 and BAHCC1 was shown to specifically recognise H3K27me3 and contribute to gene repression, possibly by recruiting histone deacetylases, which are known to counteract gene expression253-255 (Figure 6A). PRC2 also methylates non-histone substrates, including the transcription factors GATA4 and Elongin A, possibly as an alternative mechanism of counteracting transcription256,257.
PRC1 and PRC2 collaborate in Polycomb chromatin domains to repress transcription
As we understand more about the mechanisms used by Polycomb complexes to counteract transcription, it is becoming increasingly clear that PRC1 and PRC2 have independent gene repression activities, despite being linked by feedback mechanisms in Polycomb chromatin domains258. For example, in mouse ESCs or the developing epidermis, disruption of either PRC1 or PRC2 caused increased expression of a largely overlapping set of target genes231,259,260. However, removal of both PRC1 and PRC2 together caused a more pronounced derepression of target genes, and in the case of the developing epidermis, catastrophic morphological defects. These observations are consistent with the idea that the convergence of Polycomb complexes at target sites provides an opportunity for feedback mechanisms to create more expansive Polycomb chromatin domains in which the independent repressive activities of PRC1 and PRC2 can synergise to create a robust barrier against inappropriate transcription (Figure 6A). These properties may be particularly well suited to protecting gene expression programmes over developmental timescales, where the Polycomb system is known to have important roles10,133,241,243-245,261. The abundance and composition of individual Polycomb repressive complexes can change considerably over development33,176,262-265. This feature may provide an opportunity to regulate the repressive nature of Polycomb chromatin domains in different cellular and developmental contexts, for example by altering the relative contribution of PRC1-dependent and PRC2-dependent repression mechanisms.
Pervasive effects of Polycomb-mediated histone modification on gene expression
Although H2AK119ub1 and H3K27me3 levels are highly elevated in Polycomb chromatin domains, they are also found elsewhere in the genome, and there is mounting evidence that these alternative pools of H2AK119ub1 and H3K27me can affect gene expression in various contexts. The PCGF3/5-vPRC1 complexes deposit a ‘blanket’ of H2AK119ub1 across the genome, with approximately 10% of H2A molecules being modified 12,266. The extent of this pervasive pool of H2AK119ub1 is suppressed by the deubiquitylase BAP1267,268, the absence of which elevates H2AK119ub1 levels and constrains the expression of thousands of genes that are not normally influenced by PRC1 (Ref.269-271). This suggests that pervasive H2AK119 ubiquitylation may function like a rheostat to control the transcriptional potential of the genome.
PRC2 also acts pervasively throughout the genome: approximately 70% of H3K27 residues undergo mono-methylation or di-methylation and up to 15% undergo tri-methylation272-275. Although the mechanisms that control H3K27 methylation outside of Polycomb chromatin domains remain poorly understood, this pervasive activity primarily relies on the core complex and not on PRC2.1-specific and PRC2.2-specific subunits274. Importantly, there is evidence that these alternative pools of H3K27me can broadly antagonise transcription, possibly by counteracting H3K27 acetylation and limiting the activity of enhancers272,276. Therefore, it is clear that Polycomb complexes can also influence gene expression by functioning outside of Polycomb chromatin domains and it has recently been suggested that these activities may be particularly relevant in germ cells and in very early developmental stages-124,125,277,278.
Gene activation by Polycomb complexes
Although Polycomb complexes appear to primarily support transcription repression, there is increasing evidence that in some contexts, Polycomb complexes can potentiate gene expression133,246,279-282. For example, a PCGF5-vPRC1 complex that contains AUTS2 and casein kinase 2 and inefficiently ubiquitylates H2AK119, was shown to potentiate transcription by recruiting the histone acetyltransferase P300 (Ref.279). Alternatively, the H2AK119ub1-specific reader ZRF1 (also known as DNAJC2) was linked to activation of Polycomb-repressed genes during differentiation283. These effects are in agreement with earlier reports of PRC1 supporting gene expression in resting B cells and T cells280.
It has also been suggested in some instances that Polycomb chromatin domains can support gene expression by forming chromatin topologies that aid gene induction57,58,284-286. Specifically, it has been proposed that, by supporting long-range chromatin interactions, cPRC1 can bring into close proximity enhancers that are poised for activation and their target promoters, thereby enabling rapid induction of transcription in response to activation signals during cell differentiation58,284,285 (Figure 6B). This idea is consistent with observations that cPRC1 is associated with enhancer function in breast cancer cells and with activation of genes during cardiac differentiation59,287. The precise mechanisms and extent to which Polycomb complexes potentiate gene expression is an exciting area of active investigation.
Chromatin bistability and transitions in gene expression
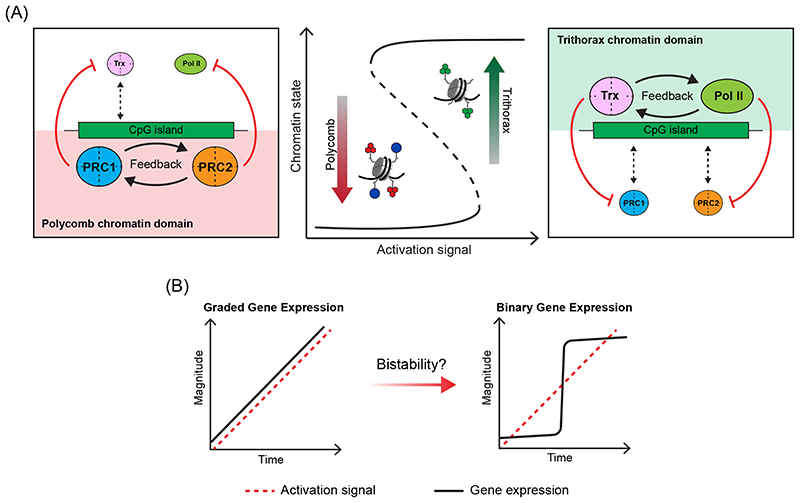
During cellular differentiation, Polycomb target genes can be activated to support new cellular functions288,289. This activation is accompanied by loss of the Polycomb chromatin domain and acquisition of an new type of chromatin domain formed through the function of Trithorax group chromatin modifying complexes, which include myeloid/lymphoid or mixed-lineage leukaemia protein 1 (MLL1; also known as KMT2A)–MLL2 (KMT2D) and SETD1A–SETD1B290. Interestingly, some Trithorax group complexes can also sample CGIs through zinc finger-CxxC DNA binding domains, but in contrast to the Polycomb complexes, they initiate the formation and spreading of transcription-permissive, H3K4me3-containing Trithorax chromatin domains through communication and feedback mechanisms linked to active transcription291-296. Antagonism between the Polycomb and Trithorax complexes helps to ensure that at most gene promoters, either a transcriptionally repressive or a transcriptionally permissive chromatin state predominates.
Based on these feedback mechanisms and antagonism between the Polycomb and Trithorax systems, CGIs have been proposed to support the formation of bistable chromatin states at gene promoters to regulate gene expression99,222,297-300 (Figure 7A). In defined cell types, the capacity to form either Polycomb or Trithorax chromatin domains could provide memory of gene expression programmes and buffer against low-level or spurious transcription activation signals that could be detrimental to cell identity. However, the convergence of these systems on CGIs might also shape gene expression transitions during differentiation. For example, Polycomb chromatin domains maintained by feedback mechanisms at inactive genes could constrain gene activation signals until an appropriate induction threshold is reached301 (Figure 7A). By contrast, following productive initiation of transcription, formation of permissive Trithorax chromatin domains could potentiate transcription while also blocking Polycomb activity. A rapid switch between chromatin states might influence transcription by converting graded gene activation signals into binary gene expression outputs (Figure 7B). In support of this possibility, the Polycomb system has been shown to impart switch-like effects on gene expression in Arabidopsis thaliana 302,303, and there is some evidence that this gene expression switch also occurs in mammals304. We envisage that switch-like transitions in the expression of master regulators of cell fate could help to ensure decisive cell fate decisions, and Polycomb and Trithorax complexes are known to have important roles in regulating these types of genes during mammalian development3. Furthermore, once individual genes acquire defined chromatin states, this could provide hysteresis of their current transcription status to help maintain cell identify in the face of inherently stochastic and pulsatile transcription signals.
Figure 7. CpG islands and chromatin bistability.
(A) A schematic illustrating how chromatin bistablity could form at CpG islands (CGIs). When transcription activation signals are low or absent, communication and feedback between Polycomb repressive complex 1 (PRC1) and PRC2 could drive the formation of repressive Polycomb chromatin domains that antagonise Trithorax (Trx) complexes and RNA polymerase II (Pol II) activity (left). When activation signals are high and persistent, communication and feedback between Trithorax complexes and Pol II could drive the formation of transcription-permissive Trithorax chromatin domains that antagonise PRC1 and PRC2 (right). The capacity of both Polycomb and Trithorax systems to sample CGIs coupled with the feedback and antagonistic mechanisms inherent to the formation of each chromatin state, would provide the opportunity to switch between predominantly Polycomb or predominantly Trithorax chromatin states as gene activation signals increase or decrease. We speculate that this mode of gene regulation could help to shape gene expression transitions (see part B) and also provide a chromatin-encoded hysteresis of the current transcriptional state of the gene in the face of inherently stochastic and pulsatile transcription initiation induced from single gene promoters.
(B) If transitions between gene expression states, for example gene induction during cellular differentiation, scaled linearly with its activation signal, then one would predict graded expression output (left). However, if CGIs help to create bistable chromatin at gene promoters, this could shape binary, switch-like, gene expression transitions in which Polycomb chromatin domains constrain activation signals until appropriate activation thresholds are reached, at which point transcription initiation would precipitate a rapid switch into a Trithorax chromatin state and potentiate transcription. In the context of such a system, one might predict CGIs could help to convert graded gene activation signals into binary switch-like gene expression outputs through chromatin bistability. This could be particularly useful in supporting decisive gene expression transitions during development.
Adapted with permission from REF. 290, Elsevier.
Recent mathematical models incorporating the known activities of Polycomb and Trithorax systems have provided theoretical evidence to support the existence of bistable chromatin states at gene regulatory elements297,298,303. Importantly, interrogation of these models also suggests that bivalent chromatin states, which are characterised by the combined presence of both H3K4me3 and H3K27me3 and previously proposed to define an alternative poised transcriptional state60,305, are unlikely to be stable and may instead correspond to the transition between Polycomb and Trithorax chromatin states. Although many of the underlying biochemical activities of Polycomb and Trithorax systems are consistent with the possibility that bistable chromatin states could be created at their target sites, a major challenge for our research field will be to directly test these models and their relevance to the kinetics of gene expression during cell differentiation.
Conclusion and future perspective
Detailed biochemical and structural characterisation has revealed that the Polycomb system comprises a diverse array of multi-protein complexes that are highly specialised in how they engage with nucleosomes, catalyse histone modifications and function through communication and feedback mechanisms to form Polycomb chromatin domains. Our understanding of the mechanics that underpin these processes has advanced rapidly in recent years, but it still remains poorly understood at the mechanistic level how, once formed, Polycomb chromatin domains influence the transcription machinery to enable gene repression. This represents a central unresolved question in our field. Given the complexity of gene regulatory elements in vivo, we anticipate that addressing this important question will require a reductionist approach leveraging in vitro transcription reactions on reconstituted chromatin templates, and single-molecule approaches that can capture the behaviour of the transcription machinery306,307.
Building on these biochemical underpinnings, the molecular study of how Polycomb systems function in vivo has indicated they are more dynamic than originally anticipated and, somewhat counterintuitively, appear to respond to transcription in forming Polycomb chromatin domains. This is forcing us to rethink how the Polycomb system both influences, and is related to, gene transcription. In this Review, we have discussed some emerging molecular principles that could explain these dynamic behaviours. However, the static and ensemble experimental approaches that our field has relied on until now are largely unsuited to exploring the relevance of these concepts. Instead, this will require live, single-cell, and quantitative measurements. These approaches will be of particular relevance in determining whether the Polycomb system enables chromatin bistablity, how it shapes the kinetics of gene-expression transitions during cell differentiation and development, and how the functions of Polycomb complexes are disrupted in human disease.
Finally, much of our understanding of the mammalian Polycomb system has come from studies in stem cells and in early developmental contexts. Although the ideas and models discussed in this Review primarily draw on this body of work, it is becoming increasingly clear that the mechanisms that guide Polycomb function in specific tissues and at later developmental stages are likely to vary considerably. Therefore, moving forward, a central objective remains to uncover the complement of mechanisms and principles that define mammalian Polycomb-mediated gene regulation in diverse cellular and developmental contexts.
As our field ventures into testing the relevance of new and emerging concepts in Polycomb biology, the coming years will inevitably be a fast-paced and exciting period in the ongoing quest to uncover the molecular principles that define gene regulation by Polycomb repressive complexes.
Acknowledgements
We thank Nadya Fursova, Emilia Dimitrova, Miles Huseyin, Paula Dobrinić and Tom Milne for critical reading of this manuscript. Work in the Klose lab is supported by the Wellcome Trust (209400/Z/17/Z) and the European Research Council (681440).
Footnotes
Author contributions
The authors contributed equally to all aspects of the article.
Competing interests
The authors declare no competing interests.
References
- 1.Haberle V, Stark A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat Rev Mol Cell Biol. 2018;19:621–637. doi: 10.1038/s41580-018-0028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kassis JA, Kennison JA, Tamkun JW. Polycomb and Trithorax Group Genes in Drosophila. Genetics. 2017;206:1699–1725. doi: 10.1534/genetics.115.185116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuettengruber B, Bourbon HM, Di Croce L, Cavalli G. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell. 2017;171:34–57. doi: 10.1016/j.cell.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 4.de Napoles M, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Wang H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 6.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 7.Czermin B, et al. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 8.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muller J, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 10.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 11.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fursova NA, et al. Synergy between Variant PRC1 Complexes Defines Polycomb-Mediated Gene Repression. Mol Cell. 2019;74:1020–1036.:e1028. doi: 10.1016/j.molcel.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tolhuis B, et al. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat Genet. 2006;38:694–699. doi: 10.1038/ng1792. [DOI] [PubMed] [Google Scholar]
- 14.Schwartz YB, et al. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- 15.Piunti A, Shilatifard A. The roles of Polycomb repressive complexes in mammalian development and cancer. Nat Rev Mol Cell Biol. 2021;22:326–345. doi: 10.1038/s41580-021-00341-1. [DOI] [PubMed] [Google Scholar]
- 16.Deevy O, Bracken AP. PRC2 functions in development and congenital disorders. Development. 2019;146:dev181354. doi: 10.1242/dev.181354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laugesen A, Hojfeldt JW, Helin K. Molecular Mechanisms Directing PRC2 Recruitment and H3K27 Methylation. Mol Cell. 2019;74:8–18. doi: 10.1016/j.molcel.2019.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen I, Bar C, Ezhkova E. Activity of PRC1 and Histone H2AK119 Monoubiquitination: Revising Popular Misconceptions. Bioessays. 2020;42:e1900192. doi: 10.1002/bies.201900192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao Z, et al. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell. 2012;45:344–356. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z, et al. Structure of a Bmi-1-Ring1B polycomb group ubiquitin ligase complex. J Biol Chem. 2006;281:20643–20649. doi: 10.1074/jbc.M602461200. [DOI] [PubMed] [Google Scholar]
- 21.Buchwald G, et al. Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 2006;25:2465–2474. doi: 10.1038/sj.emboj.7601144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bentley ML, et al. Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex. EMBO J. 2011;30:3285–3297. doi: 10.1038/emboj.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hauri S, et al. A High-Density Map for Navigating the Human Polycomb Complexome. Cell Rep. 2016;17:583–595. doi: 10.1016/j.celrep.2016.08.096. [DOI] [PubMed] [Google Scholar]
- 24.Wang R, et al. Polycomb group targeting through different binding partners of RING1B C-terminal domain. Structure. 2010;18:966–975. doi: 10.1016/j.str.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levine SS, et al. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol Cell Biol. 2002;22:6070–6078. doi: 10.1128/MCB.22.17.6070-6078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shao Z, et al. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- 27.Tavares L, et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell. 2012;148:664–678. doi: 10.1016/j.cell.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGinty RK, Henrici RC, Tan S. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature. 2014;514:591–596. doi: 10.1038/nature13890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalashnikova AA, Porter-Goff ME, Muthurajan UM, Luger K, Hansen JC. The role of the nucleosome acidic patch in modulating higher order chromatin structure. J R Soc Interface. 2013;10:20121022. doi: 10.1098/rsif.2012.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rose NR, et al. RYBP stimulates PRC1 to shape chromatin-based communication between Polycomb repressive complexes. Elife. 2016;5:e.18591. doi: 10.7554/eLife.18591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taherbhoy AM, Huang OW, Cochran AG. BMI1-RING1B is an autoinhibited RING E3 ubiquitin ligase. Nat Commun. 2015;6:7621. doi: 10.1038/ncomms8621. [DOI] [PubMed] [Google Scholar]
- 32.Zhao J, et al. RYBP/YAF2-PRC1 complexes and histone H1-dependent chromatin compaction mediate propagation of H2AK119ub1 during cell division. Nat Cell Biol. 2020;22:439–452. doi: 10.1038/s41556-020-0484-1. [DOI] [PubMed] [Google Scholar]
- 33.Margueron R, et al. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ciferri C, et al. Molecular architecture of human polycomb repressive complex 2. Elife. 2012;1:e00005. doi: 10.7554/eLife.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kasinath V, et al. Structures of human PRC2 with its cofactors AEBP2 and JARID2. Science. 2018;359:940–944. doi: 10.1126/science.aar5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glancy E, Ciferri C, Bracken AP. Structural basis for PRC2 engagement with chromatin. Curr Opin Struct Biol. 2020;67:135–144. doi: 10.1016/j.sbi.2020.10.017. [DOI] [PubMed] [Google Scholar]
- 37.Brooun A, et al. Polycomb repressive complex 2 structure with inhibitor reveals a mechanism of activation and drug resistance. Nat Commun. 2016;7:11384. doi: 10.1038/ncomms11384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 39.Jiao L, Liu X. Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science. 2015;350:aac4383. doi: 10.1126/science.aac4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Antonysamy S, et al. Structural context of disease-associated mutations and putative mechanism of autoinhibition revealed by X-ray crystallographic analysis of the EZH2-SET domain. PLoS One. 2013;8:e84147. doi: 10.1371/journal.pone.0084147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Justin N, et al. Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat Commun. 2016;7:11316. doi: 10.1038/ncomms11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finogenova K, et al. Structural basis for PRC2 decoding of active histone methylation marks H3K36me2/3. Elife. 2020;9:e.61964. doi: 10.7554/eLife.61964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poepsel S, Kasinath V, Nogales E. Cryo-EM structures of PRC2 simultaneously engaged with two functionally distinct nucleosomes. Nat Struct Mol Biol. 2018;25:154–162. doi: 10.1038/s41594-018-0023-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen S, Jiao L, Shubbar M, Yang X, Liu X. Unique Structural Platforms of Suz12 Dictate Distinct Classes of PRC2 for Chromatin Binding. Mol Cell. 2018;69:840–852.:e845. doi: 10.1016/j.molcel.2018.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beringer M, et al. EPOP Functionally Links Elongin and Polycomb in Pluripotent Stem Cells. Mol Cell. 2016;64:645–658. doi: 10.1016/j.molcel.2016.10.018. [DOI] [PubMed] [Google Scholar]
- 46.Liefke R, Karwacki-Neisius V, Shi Y. EPOP Interacts with Elongin BC and USP7 to Modulate the Chromatin Landscape. Mol Cell. 2016;64:659–672. doi: 10.1016/j.molcel.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Z, et al. PRC2 complexes with JARID2, MTF2, and esPRC2p48 in ES cells to modulate ES cell pluripotency and somatic cell reprogramming. Stem Cells. 2011;29:229–240. doi: 10.1002/stem.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanulli S, et al. Jarid2 Methylation via the PRC2 Complex Regulates H3K27me3 Deposition during Cell Differentiation. Mol Cell. 2015;57:769–783. doi: 10.1016/j.molcel.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee CH, et al. Distinct Stimulatory Mechanisms Regulate the Catalytic Activity of Polycomb Repressive Complex 2. Mol Cell. 2018;70:435–448.:e435. doi: 10.1016/j.molcel.2018.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conway E, et al. A Family of Vertebrate-Specific Polycombs Encoded by the LCOR/LCORL Genes Balance PRC2 Subtype Activities. Mol Cell. 2018;70:408–421.:e408. doi: 10.1016/j.molcel.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 51.Pajtler KW, et al. Molecular heterogeneity and CXorf67 alterations in posterior fossa group A (PFA) ependymomas. Acta Neuropathol. 2018;136:211–226. doi: 10.1007/s00401-018-1877-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Piunti A, et al. CATACOMB: An endogenous inducible gene that antagonizes H3K27 methylation activity of Polycomb repressive complex 2 via an H3K27M-like mechanism. Sci Adv. 2019;5:eaax2887. doi: 10.1126/sciadv.aax2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ragazzini R, et al. EZHIP constrains Polycomb Repressive Complex 2 activity in germ cells. Nat Commun. 2019;10:3858. doi: 10.1038/s41467-019-11800-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jain SU, et al. PFA ependymoma-associated protein EZHIP inhibits PRC2 activity through a H3 K27M-like mechanism. Nat Commun. 2019;10:2146. doi: 10.1038/s41467-019-09981-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cai Y, et al. H3K27me3-rich genomic regions can function as silencers to repress gene expression via chromatin interactions. Nat Commun. 2021;12:719. doi: 10.1038/s41467-021-20940-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mendenhall EM, et al. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet. 2010;6:e1001244. doi: 10.1371/journal.pgen.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ngan CY, et al. Chromatin interaction analyses elucidate the roles of PRC2-bound silencers in mouse development. Nat Genet. 2020;52:264–272. doi: 10.1038/s41588-020-0581-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Loubiere V, Papadopoulos GL, Szabo Q, Martinez AM, Cavalli G. Widespread activation of developmental gene expression characterized by PRC1-dependent chromatin looping. Sci Adv. 2020;6:eaax4001. doi: 10.1126/sciadv.aax4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chan HL, et al. Polycomb complexes associate with enhancers and promote oncogenic transcriptional programs in cancer through multiple mechanisms. Nat Commun. 2018;9:3377. doi: 10.1038/s41467-018-05728-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 61.Ku M, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tanay A, O’Donnell AH, Damelin M, Bestor TH. Hyperconserved CpG domains underlie Polycomb-binding sites. Proc Natl Acad Sci U S A. 2007;104:5521–5526. doi: 10.1073/pnas.0609746104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schlesinger Y, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 64.Bauer M, Trupke J, Ringrose L. The quest for mammalian Polycomb response elements: are we there yet? Chromosoma. 2016;125:471–496. doi: 10.1007/s00412-015-0539-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kassis JA, Brown JL. Polycomb group response elements in Drosophila and vertebrates. Adv Genet. 2013;81:83–118. doi: 10.1016/B978-0-12-407677-8.00003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vella P, Barozzi I, Cuomo A, Bonaldi T, Pasini D. Yin Yang 1 extends the Myc-related transcription factors network in embryonic stem cells. Nucleic Acids Res. 2012;40:3403–3418. doi: 10.1093/nar/gkr1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Endoh M, et al. PCGF6-PRC1 suppresses premature differentiation of mouse embryonic stem cells by regulating germ cell-related genes. Elife. 2017;6 doi: 10.7554/eLife.21064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trojer P, et al. L3MBTL2 protein acts in concert with PcG protein-mediated monoubiquitination of H2A to establish a repressive chromatin structure. Mol Cell. 2011;42:438–450. doi: 10.1016/j.molcel.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- 70.Hurlin PJ, Steingrimsson E, Copeland NG, Jenkins NA, Eisenman RN. Mga, a dual-specificity transcription factor that interacts with Max and contains a T-domain DNA-binding motif. EMBO J. 1999;18:7019–7028. doi: 10.1093/emboj/18.24.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stielow B, Finkernagel F, Stiewe T, Nist A, Suske G. MGA, L3MBTL2 and E2F6 determine genomic binding of the non-canonical Polycomb repressive complex PRC1.6. PLoS Genet. 2018;14:e1007193. doi: 10.1371/journal.pgen.1007193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Scelfo A, et al. Functional Landscape of PCGF Proteins Reveals Both RING1A/B-Dependent- and RING1A/B-Independent-Specific Activities. Mol Cell. 2019;74:1037–1052.:e1037. doi: 10.1016/j.molcel.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huang Y, et al. Combinatorial Control of Recruitment of a Variant PRC1.6 Complex in Embryonic Stem Cells. Cell Rep. 2018;22:3032–3043. doi: 10.1016/j.celrep.2018.02.072. [DOI] [PubMed] [Google Scholar]
- 74.Liu M, et al. The polycomb group protein PCGF6 mediates germline gene silencing by recruiting histone-modifying proteins to target gene promoters. J Biol Chem. 2020;295:9712–9724. doi: 10.1074/jbc.RA119.012121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao W, et al. Essential Role for Polycomb Group Protein Pcgf6 in Embryonic Stem Cell Maintenance and a Noncanonical Polycomb Repressive Complex 1 (PRC1) Integrity. J Biol Chem. 2017;292:2773–2784. doi: 10.1074/jbc.M116.763961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu M, et al. Direct recruitment of polycomb repressive complex 1 to chromatin by core binding transcription factors. Mol Cell. 2012;45:330–343. doi: 10.1016/j.molcel.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ren X, Kerppola TK. REST interacts with Cbx proteins and regulates polycomb repressive complex 1 occupancy at RE1 elements. Mol Cell Biol. 2011;31:2100–2110. doi: 10.1128/MCB.05088-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Herranz N, et al. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol. 2008;28:4772–4781. doi: 10.1128/MCB.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dietrich N, et al. REST-mediated recruitment of polycomb repressor complexes in mammalian cells. PLoS Genet. 2012;8:e1002494. doi: 10.1371/journal.pgen.1002494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Almeida M, Bowness JS, Brockdorff N. The many faces of Polycomb regulation by RNA. Curr Opin Genet Dev. 2020;61:53–61. doi: 10.1016/j.gde.2020.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Almeida M, et al. PCGF3/5-PRC1 initiates Polycomb recruitment in X chromosome inactivation. Science. 2017;356:1081–1084. doi: 10.1126/science.aal2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pintacuda G, et al. hnRNPK Recruits PCGF3/5-PRC1 to the Xist RNA B-Repeat to Establish Polycomb-Mediated Chromosomal Silencing. Mol Cell. 2017;68:955–969.:e910. doi: 10.1016/j.molcel.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chu C, et al. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161:404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zylicz JJ, et al. The Implication of Early Chromatin Changes in X Chromosome Inactivation. Cell. 2019;176:182–197.:e123. doi: 10.1016/j.cell.2018.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Somarowthu S, et al. HOTAIR forms an intricate and modular secondary structure. Mol Cell. 2015;58:353–361. doi: 10.1016/j.molcel.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Portoso M, et al. PRC2 is dispensable for HOTAIR-mediated transcriptional repression. EMBO J. 2017;36:981–994. doi: 10.15252/embj.201695335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brockdorff N. Noncoding RNA and Polycomb recruitment. RNA. 2013;19:429–442. doi: 10.1261/rna.037598.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Davidovich C, Cech TR. The recruitment of chromatin modifiers by long noncoding RNAs: lessons from PRC2. RNA. 2015;21:2007–2022. doi: 10.1261/rna.053918.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pandey RR, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 92.Regha K, et al. Active and repressive chromatin are interspersed without spreading in an imprinted gene cluster in the mammalian genome. Mol Cell. 2007;27:353–366. doi: 10.1016/j.molcel.2007.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schertzer MD, et al. lncRNA-Induced Spread of Polycomb Controlled by Genome Architecture, RNA Abundance, and CpG Island DNA. Mol Cell. 2019;75:523–537.:e510. doi: 10.1016/j.molcel.2019.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen Z, Zhang Y. Maternal H3K27me3-dependent autosomal and X chromosome imprinting. Nat Rev Genet. 2020;21:555–571. doi: 10.1038/s41576-020-0245-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Skourti-Stathaki K, et al. R-Loops Enhance Polycomb Repression at a Subset of Developmental Regulator Genes. Mol Cell. 2019;73:930–945.:e934. doi: 10.1016/j.molcel.2018.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen PB, Chen HV, Acharya D, Rando OJ, Fazzio TG. R loops regulate promoter-proximal chromatin architecture and cellular differentiation. Nat Struct Mol Biol. 2015;22:999–1007. doi: 10.1038/nsmb.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Illingworth RS, Bird AP. CpG islands--’a rough guide’. FEBS Lett. 2009;583:1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 101.Schubeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 102.Garrick D, et al. The role of the polycomb complex in silencing alpha-globin gene expression in nonerythroid cells. Blood. 2008;112:3889–3899. doi: 10.1182/blood-2008-06-161901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gearhart MD, Corcoran CM, Wamstad JA, Bardwell VJ. Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol Cell Biol. 2006;26:6880–6889. doi: 10.1128/MCB.00630-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sanchez C, et al. Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol Cell Proteomics. 2007;6:820–834. doi: 10.1074/mcp.M600275-MCP200. [DOI] [PubMed] [Google Scholar]
- 105.Koyama-Nasu R, David G, Tanese N. The F-box protein Fbl10 is a novel transcriptional repressor of c-Jun. Nat Cell Biol. 2007;9:1074–1080. doi: 10.1038/ncb1628. [DOI] [PubMed] [Google Scholar]
- 106.Voo KS, Carlone DL, Jacobsen BM, Flodin A, Skalnik DG. Cloning of a mammalian transcriptional activator that binds unmethylated CpG motifs and shares a CXXC domain with DNA methyltransferase, human trithorax, and methyl-CpG binding domain protein 1. Mol Cell Biol. 2000;20:2108–2121. doi: 10.1128/mcb.20.6.2108-2121.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Blackledge NP, et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell. 2014;157:1445–1459. doi: 10.1016/j.cell.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Farcas AM, et al. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. Elife. 2012;1:e00205. doi: 10.7554/eLife.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.He J, et al. Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat Cell Biol. 2013;15:373–384. doi: 10.1038/ncb2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wu X, Johansen JV, Helin K. Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol Cell. 2013;49:1134–1146. doi: 10.1016/j.molcel.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 111.Perino M, et al. MTF2 recruits Polycomb Repressive Complex 2 by helical-shape-selective DNA binding. Nat Genet. 2018;50:1002–1010. doi: 10.1038/s41588-018-0134-8. [DOI] [PubMed] [Google Scholar]
- 112.Li H, et al. Polycomb-like proteins link the PRC2 complex to CpG islands. Nature. 2017;549:287–291. doi: 10.1038/nature23881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Davidovich C, Goodrich KJ, Gooding AR, Cech TR. A dimeric state for PRC2. Nucleic Acids Res. 2014;42:9236–9248. doi: 10.1093/nar/gku540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen S, Jiao L, Liu X, Yang X, Liu X. A Dimeric Structural Scaffold for PRC2-PCL Targeting to CpG Island Chromatin. Mol Cell. 2020;77:1265–1278.:e1267. doi: 10.1016/j.molcel.2019.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cooper S, et al. Targeting polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell Rep. 2014;7:1456–1470. doi: 10.1016/j.celrep.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cooper S, et al. Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nat Commun. 2016;7:13661. doi: 10.1038/ncomms13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kalb R, et al. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat Struct Mol Biol. 2014;21:569–571. doi: 10.1038/nsmb.2833. [DOI] [PubMed] [Google Scholar]
- 118.Kasinath V, et al. JARID2 and AEBP2 regulate PRC2 in the presence of H2AK119ub1 and other histone modifications. Science. 2021;371:eabc3393. doi: 10.1126/science.abc3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Blackledge NP, et al. PRC1 Catalytic Activity Is Central to Polycomb System Function. Mol Cell. 2020;77:857–874.:e859. doi: 10.1016/j.molcel.2019.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Healy E, et al. PRC2.1 and PRC2.2 Synergize to Coordinate H3K27 Trimethylation. Mol Cell. 2019;76:437–452.:e436. doi: 10.1016/j.molcel.2019.08.012. [DOI] [PubMed] [Google Scholar]
- 121.Tamburri S, et al. Histone H2AK119 Mono-Ubiquitination Is Essential for Polycomb-Mediated Transcriptional Repression. Mol Cell. 2020;77:840–856.:e845. doi: 10.1016/j.molcel.2019.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hickey GJM, et al. Establishment of Developmental Gene Silencing by Ordered Polycomb Complex Recruitment in Early Zebrafish Embryos. Preprint at bioRxiv. 2021 doi: 10.1101/2021.1103.1116.435592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Illingworth RS, et al. The E3 ubiquitin ligase activity of RING1B is not essential for early mouse development. Genes Dev. 2015;29:1897–1902. doi: 10.1101/gad.268151.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mei H, et al. H2AK119ub1 guides maternal inheritance and zygotic deposition of H3K27me3 in mouse embryos. Nat Genet. 2021;53:539–550. doi: 10.1038/s41588-021-00820-3. [DOI] [PubMed] [Google Scholar]
- 125.Chen Z, Djekidel MN, Zhang Y. Distinct dynamics and functions of H2AK119ub1 and H3K27me3 in mouse preimplantation embryos. Nat Genet. 2021;53:551–563. doi: 10.1038/s41588-021-00821-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Arrigoni R, et al. The Polycomb-associated protein Rybp is a ubiquitin binding protein. FEBS Lett. 2006;580:6233–6241. doi: 10.1016/j.febslet.2006.10.027. [DOI] [PubMed] [Google Scholar]
- 127.Margueron R, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lee CH, et al. Allosteric Activation Dictates PRC2 Activity Independent of Its Recruitment to Chromatin. Mol Cell. 2018;70:422–434.:e426. doi: 10.1016/j.molcel.2018.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Oksuz O, et al. Capturing the Onset of PRC2-Mediated Repressive Domain Formation. Mol Cell. 2018;70:1149–1162.:e1145. doi: 10.1016/j.molcel.2018.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yuan W, et al. Dense chromatin activates Polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science. 2012;337:971–975. doi: 10.1126/science.1225237. [DOI] [PubMed] [Google Scholar]
- 131.Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang L, et al. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell. 2004;14:637–646. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 133.Cohen I, et al. PRC1 Fine-tunes Gene Repression and Activation to Safeguard Skin Development and Stem Cell Specification. Cell Stem Cell. 2018;22:726–739.:e727. doi: 10.1016/j.stem.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Moussa HF, et al. Canonical PRC1 controls sequence-independent propagation of Polycomb-mediated gene silencing. Nat Commun. 2019;10:1931. doi: 10.1038/s41467-019-09628-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Schoenfelder S, et al. Polycomb repressive complex PRC1 spatially constrains the mouse embryonic stem cell genome. Nat Genet. 2015;47:1179–1186. doi: 10.1038/ng.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Vieux-Rochas M, Fabre PJ, Leleu M, Duboule D, Noordermeer D. Clustering of mammalian Hox genes with other H3K27me3 targets within an active nuclear domain. Proc Natl Acad Sci U S A. 2015;112:4672–4677. doi: 10.1073/pnas.1504783112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Eskeland R, et al. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell. 2010;38:452–464. doi: 10.1016/j.molcel.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bonev B, et al. Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell. 2017;171:557–572.:e524. doi: 10.1016/j.cell.2017.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rhodes JDP, et al. Cohesin Disrupts Polycomb-Dependent Chromosome Interactions in Embryonic Stem Cells. Cell Rep. 2020;30:820–835.:e810. doi: 10.1016/j.celrep.2019.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.McLaughlin K, et al. DNA Methylation Directs Polycomb-Dependent 3D Genome Re-organization in Naive Pluripotency. Cell Rep. 2019;29:1974–1985.:e1976. doi: 10.1016/j.celrep.2019.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bantignies F, et al. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell. 2011;144:214–226. doi: 10.1016/j.cell.2010.12.026. [DOI] [PubMed] [Google Scholar]
- 142.Denholtz M, et al. Long-range chromatin contacts in embryonic stem cells reveal a role for pluripotency factors and polycomb proteins in genome organization. Cell Stem Cell. 2013;13:602–616. doi: 10.1016/j.stem.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Saurin AJ, et al. The human polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J Cell Biol. 1998;142:887–898. doi: 10.1083/jcb.142.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ren X, Vincenz C, Kerppola TK. Changes in the distributions and dynamics of polycomb repressive complexes during embryonic stem cell differentiation. Mol Cell Biol. 2008;28:2884–2895. doi: 10.1128/MCB.00949-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Satijn DP, et al. RING1 is associated with the polycomb group protein complex and acts as a transcriptional repressor. Mol Cell Biol. 1997;17:4105–4113. doi: 10.1128/mcb.17.7.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kim CA, Gingery M, Pilpa RM, Bowie JU. The SAM domain of polyhomeotic forms a helical polymer. Nat Struct Biol. 2002;9:453–457. doi: 10.1038/nsb802. [DOI] [PubMed] [Google Scholar]
- 147.Boyle S, et al. A central role for canonical PRC1 in shaping the 3D nuclear landscape. Genes Dev. 2020;34:931–949. doi: 10.1101/gad.336487.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Isono K, et al. SAM domain polymerization links subnuclear clustering of PRC1 to gene silencing. Dev Cell. 2013;26:565–577. doi: 10.1016/j.devcel.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 149.Kundu S, et al. Polycomb Repressive Complex 1 Generates Discrete Compacted Domains that Change during Differentiation. Mol Cell. 2017;65:432–446. doi: 10.1016/j.molcel.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wani AH, et al. Chromatin topology is coupled to Polycomb group protein subnuclear organization. Nat Commun. 2016;7:10291. doi: 10.1038/ncomms10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Belaghzal H, et al. Liquid chromatin Hi-C characterizes compartment-dependent chromatin interaction dynamics. Nat Genet. 2021;53:367–378. doi: 10.1038/s41588-021-00784-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Huseyin MK, Klose RJ. Live-cell single particle tracking of PRC1 reveals a highly dynamic system with low target site occupancy. Nat Commun. 2021;12:887. doi: 10.1038/s41467-021-21130-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Youmans DT, Schmidt JC, Cech TR. Live-cell imaging reveals the dynamics of PRC2 and recruitment to chromatin by SUZ12-associated subunits. Genes Dev. 2018;32:794–805. doi: 10.1101/gad.311936.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhen CY, et al. Live-cell single-molecule tracking reveals co-recognition of H3K27me3 and DNA targets polycomb Cbx7-PRC1 to chromatin. Elife. 2016;5:e.17667. doi: 10.7554/eLife.17667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Vandenbunder B, et al. PRC1 components exhibit different binding kinetics in Polycomb bodies. Biol Cell. 2014;106:111–125. doi: 10.1111/boc.201300077. [DOI] [PubMed] [Google Scholar]
- 156.Cuadrado A, et al. Specific Contributions of Cohesin-SA1 and Cohesin-SA2 to TADs and Polycomb Domains in Embryonic Stem Cells. Cell Rep. 2019;27:3500–3510.:e3504. doi: 10.1016/j.celrep.2019.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kriz AJ, Colognori D, Sunwoo H, Nabet B, Lee JT. Balancing cohesin eviction and retention prevents aberrant chromosomal interactions, Polycomb-mediated repression, and X-inactivation. Mol Cell. 2021;81:1970–1987. doi: 10.1016/j.molcel.2021.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Plys AJ, et al. Phase separation of Polycomb-repressive complex 1 is governed by a charged disordered region of CBX2. Genes Dev. 2019;33:799–813. doi: 10.1101/gad.326488.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tatavosian R, et al. Nuclear condensates of the Polycomb protein chromobox 2 (CBX2) assemble through phase separation. J Biol Chem. 2019;294:1451–1463. doi: 10.1074/jbc.RA118.006620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Eeftens JM, Kapoor M, Brangwynne CP. Epigenetic memory as a time integral over prior history of Polycomb phase separation. Preprint at bioRxiv. 2020 doi: 10.1101/2020.1108.1119.254706. [DOI] [Google Scholar]
- 161.Seif E, et al. Phase separation by the polyhomeotic sterile alpha motif compartmentalizes Polycomb Group proteins and enhances their activity. Nat Commun. 2020;11:5609. doi: 10.1038/s41467-020-19435-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Shin Y, Brangwynne CP. Liquid phase condensation in cell physiology and disease. Science. 2017;357:eaaf4382. doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- 163.Erdel F, et al. Mouse Heterochromatin Adopts Digital Compaction States without Showing Hallmarks of HP1-Driven Liquid-Liquid Phase Separation. Mol Cell. 2020;78:236–249.:e237. doi: 10.1016/j.molcel.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Stewart-Morgan KR, Petryk N, Groth A. Chromatin replication and epigenetic cell memory. Nat Cell Biol. 2020;22:361–371. doi: 10.1038/s41556-020-0487-y. [DOI] [PubMed] [Google Scholar]
- 165.Escobar TM, Loyola A, Reinberg D. Parental nucleosome segregation and the inheritance of cellular identity. Nat Rev Genet. 2021;22:379–392. doi: 10.1038/s41576-020-00312-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Reveron-Gomez N, et al. Accurate Recycling of Parental Histones Reproduces the Histone Modification Landscape during DNA Replication. Mol Cell. 2018;72:239–249.:e235. doi: 10.1016/j.molcel.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Escobar TM, et al. Active and Repressed Chromatin Domains Exhibit Distinct Nucleosome Segregation during DNA Replication. Cell. 2019;179:953–963.:e911. doi: 10.1016/j.cell.2019.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Reinberg D, Vales LD. Chromatin domains rich in inheritance. Science. 2018;361:33–34. doi: 10.1126/science.aat7871. [DOI] [PubMed] [Google Scholar]
- 169.Alabert C, et al. Domain Model Explains Propagation Dynamics and Stability of Histone H3K27 and H3K36 Methylation Landscapes. Cell Rep. 2020;30:1223–1234.:e1228. doi: 10.1016/j.celrep.2019.12.060. [DOI] [PubMed] [Google Scholar]
- 170.Alabert C, et al. Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev. 2015;29:585–590. doi: 10.1101/gad.256354.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hansen KH, et al. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- 172.Hojfeldt JW, et al. Accurate H3K27 methylation can be established de novo by SUZ12-directed PRC2. Nat Struct Mol Biol. 2018;25:225–232. doi: 10.1038/s41594-018-0036-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Laprell F, Finkl K, Muller J. Propagation of Polycomb-repressed chromatin requires sequence-specific recruitment to DNA. Science. 2017;356:85–88. doi: 10.1126/science.aai8266. [DOI] [PubMed] [Google Scholar]
- 174.Coleman RT, Struhl G. Causal role for inheritance of H3K27me3 in maintaining the OFF state of a Drosophila HOX gene. Science. 2017;356:eaai8236. doi: 10.1126/science.aai8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Smith E, Shilatifard A. The chromatin signaling pathway: diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol Cell. 2010;40:689–701. doi: 10.1016/j.molcel.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kloet SL, et al. The dynamic interactome and genomic targets of Polycomb complexes during stem-cell differentiation. Nat Struct Mol Biol. 2016;23:682–690. doi: 10.1038/nsmb.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Morey L, Aloia L, Cozzuto L, Benitah SA, Di Croce L. RYBP and Cbx7 define specific biological functions of polycomb complexes in mouse embryonic stem cells. Cell Rep. 2013;3:60–69. doi: 10.1016/j.celrep.2012.11.026. [DOI] [PubMed] [Google Scholar]
- 178.Riising EM, et al. Gene silencing triggers polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol Cell. 2014;55:347–360. doi: 10.1016/j.molcel.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 179.Kaneko S, Son J, Bonasio R, Shen SS, Reinberg D. Nascent RNA interaction keeps PRC2 activity poised and in check. Genes Dev. 2014;28:1983–1988. doi: 10.1101/gad.247940.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Jermann P, Hoerner L, Burger L, Schubeler D. Short sequences can efficiently recruit histone H3 lysine 27 trimethylation in the absence of enhancer activity and DNA methylation. Proc Natl Acad Sci U S A. 2014;111:E3415-3421. doi: 10.1073/pnas.1400672111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lynch MD, et al. An interspecies analysis reveals a key role for unmethylated CpG dinucleotides in vertebrate Polycomb complex recruitment. EMBO J. 2012;31:317–329. doi: 10.1038/emboj.2011.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hosogane M, Funayama R, Nishida Y, Nagashima T, Nakayama K. Ras-induced changes in H3K27me3 occur after those in transcriptional activity. PLoS Genet. 2013;9:e1003698. doi: 10.1371/journal.pgen.1003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Zhao J, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kaneko S, Son J, Shen SS, Reinberg D, Bonasio R. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Mol Biol. 2013;20:1258–1264. doi: 10.1038/nsmb.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wang X, et al. Targeting of Polycomb Repressive Complex 2 to RNA by Short Repeats of Consecutive Guanines. Mol Cell. 2017;65:1056–1067.:e1055. doi: 10.1016/j.molcel.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 186.Long Y, et al. Conserved RNA-binding specificity of polycomb repressive complex 2 is achieved by dispersed amino acid patches in EZH2. Elife. 2017;6:e.31558. doi: 10.7554/eLife.31558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kanhere A, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol. 2013;20:1250–1257. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Davidovich C, et al. Toward a consensus on the binding specificity and promiscuity of PRC2 for RNA. Mol Cell. 2015;57:552–558. doi: 10.1016/j.molcel.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Beltran M, et al. G-tract RNA removes Polycomb repressive complex 2 from genes. Nat Struct Mol Biol. 2019;26:899–909. doi: 10.1038/s41594-019-0293-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Beltran M, et al. The interaction of PRC2 with RNA or chromatin is mutually antagonistic. Genome Res. 2016;26:896–907. doi: 10.1101/gr.197632.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wang X, et al. Molecular analysis of PRC2 recruitment to DNA in chromatin and its inhibition by RNA. Nat Struct Mol Biol. 2017;24:1028–1038. doi: 10.1038/nsmb.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Zhang Q, et al. RNA exploits an exposed regulatory site to inhibit the enzymatic activity of PRC2. Nat Struct Mol Biol. 2019;26:237–247. doi: 10.1038/s41594-019-0197-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Long Y, et al. RNA is essential for PRC2 chromatin occupancy and function in human pluripotent stem cells. Nat Genet. 2020;52:931–938. doi: 10.1038/s41588-020-0662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wei C, et al. RBFox2 Binds Nascent RNA to Globally Regulate Polycomb Complex 2 Targeting in Mammalian Genomes. Mol Cell. 2016;62:875–889. doi: 10.1016/j.molcel.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 197.Jani KS, et al. Histone H3 tail binds a unique sensing pocket in EZH2 to activate the PRC2 methyltransferase. Proc Natl Acad Sci U S A. 2019;116:8295–8300. doi: 10.1073/pnas.1819029116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Schmitges FW, et al. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell. 2011;42:330–341. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 199.Streubel G, et al. The H3K36me2 Methyltransferase Nsd1 Demarcates PRC2-Mediated H3K27me2 and H3K27me3 Domains in Embryonic Stem Cells. Mol Cell. 2018;70:371–379.:e375. doi: 10.1016/j.molcel.2018.02.027. [DOI] [PubMed] [Google Scholar]
- 200.Voigt P, et al. Asymmetrically modified nucleosomes. Cell. 2012;151:181–193. doi: 10.1016/j.cell.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Yuan W, et al. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem. 2011;286:7983–7989. doi: 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Xu Q, et al. SETD2 regulates the maternal epigenome, genomic imprinting and embryonic development. Nat Genet. 2019;51:844–856. doi: 10.1038/s41588-019-0398-7. [DOI] [PubMed] [Google Scholar]
- 203.Mas G, et al. Promoter bivalency favors an open chromatin architecture in embryonic stem cells. Nat Genet. 2018;50:1452–1462. doi: 10.1038/s41588-018-0218-5. [DOI] [PubMed] [Google Scholar]
- 204.Agger K, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 205.Chen S, et al. The histone H3 Lys 27 demethylase JMJD3 regulates gene expression by impacting transcriptional elongation. Genes Dev. 2012;26:1364–1375. doi: 10.1101/gad.186056.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.De Santa F, et al. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 207.Vernimmen D, et al. Polycomb eviction as a new distant enhancer function. Genes Dev. 2011;25:1583–1588. doi: 10.1101/gad.16985411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Williams K, et al. The histone lysine demethylase JMJD3/KDM6B is recruited to p53 bound promoters and enhancer elements in a p53 dependent manner. PLoS One. 2014;9:e96545. doi: 10.1371/journal.pone.0096545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Lee MG, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 210.Tie F, et al. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 2009;136:3131–3141. doi: 10.1242/dev.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Weaver TM, et al. The EZH2 SANT1 domain is a histone reader providing sensitivity to the modification state of the H4 tail. Sci Rep. 2019;9:987. doi: 10.1038/s41598-018-37699-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Pasini D, et al. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res. 2010;38:4958–4969. doi: 10.1093/nar/gkq244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Reynolds N, et al. NuRD-mediated deacetylation of H3K27 facilitates recruitment of Polycomb Repressive Complex 2 to direct gene repression. EMBO J. 2012;31:593–605. doi: 10.1038/emboj.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Kim TW, et al. Ctbp2 Modulates NuRD-Mediated Deacetylation of H3K27 and Facilitates PRC2-Mediated H3K27me3 in Active Embryonic Stem Cell Genes During Exit from Pluripotency. Stem Cells. 2015;33:2442–2455. doi: 10.1002/stem.2046. [DOI] [PubMed] [Google Scholar]
- 215.Lavarone E, Barbieri CM, Pasini D. Dissecting the role of H3K27 acetylation and methylation in PRC2 mediated control of cellular identity. Nat Commun. 2019;10:1679. doi: 10.1038/s41467-019-09624-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Zhu P, et al. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol Cell. 2007;27:609–621. doi: 10.1016/j.molcel.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Yang W, et al. The histone H2A deubiquitinase Usp16 regulates embryonic stem cell gene expression and lineage commitment. Nat Commun. 2014;5:3818. doi: 10.1038/ncomms4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Ho L, et al. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat Cell Biol. 2011;13:903–913. doi: 10.1038/ncb2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Kadoch C, et al. Dynamics of BAF-Polycomb complex opposition on heterochromatin in normal and oncogenic states. Nat Genet. 2017;49:213–222. doi: 10.1038/ng.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Weber CM, et al. mSWI/SNF promotes polycomb repression both directly and through genome-wide redistribution. Nat Struct Mol Biol. 2021;28:501–511. doi: 10.1038/s41594-021-00604-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Stanton BZ, et al. Smarca4 ATPase mutations disrupt direct eviction of PRC1 from chromatin. Nat Genet. 2017;49:282–288. doi: 10.1038/ng.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Klose RJ, Cooper S, Farcas AM, Blackledge NP, Brockdorff N. Chromatin sampling--an emerging perspective on targeting polycomb repressor proteins. PLoS Genet. 2013;9:e1003717. doi: 10.1371/journal.pgen.1003717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Jones RS, Gelbart WM. Genetic analysis of the enhancer of zeste locus and its role in gene regulation in Drosophila melanogaster. Genetics. 1990;126:185–199. doi: 10.1093/genetics/126.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Struhl G, Akam M. Altered distributions of Ultrabithorax transcripts in extra sex combs mutant embryos of Drosophila. EMBO J. 1985;4:3259–3264. doi: 10.1002/j.1460-2075.1985.tb04075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Akasaka T, et al. Mice doubly deficient for the Polycomb Group genes Mel18 and Bmi1 reveal synergy and requirement for maintenance but not initiation of Hox gene expression. Development. 2001;128:1587–1597. doi: 10.1242/dev.128.9.1587. [DOI] [PubMed] [Google Scholar]
- 226.Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- 227.Grau DJ, et al. Compaction of chromatin by diverse Polycomb group proteins requires localized regions of high charge. Genes Dev. 2011;25:2210–2221. doi: 10.1101/gad.17288211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Lau MS, et al. Mutation of a nucleosome compaction region disrupts Polycomb-mediated axial patterning. Science. 2017;355:1081–1084. doi: 10.1126/science.aah5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Forzati F, et al. CBX7 is a tumor suppressor in mice and humans. J Clin Invest. 2012;122:612–623. doi: 10.1172/JCI58620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Pirity MK, Locker J, Schreiber-Agus N. Rybp/DEDAF is required for early postimplantation and for central nervous system development. Mol Cell Biol. 2005;25:7193–7202. doi: 10.1128/MCB.25.16.7193-7202.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.King HW, Fursova NA, Blackledge NP, Klose RJ. Polycomb repressive complex 1 shapes the nucleosome landscape but not accessibility at target genes. Genome Res. 2018;28:1494–1507. doi: 10.1101/gr.237180.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Hodges HC, et al. Dominant-negative SMARCA4 mutants alter the accessibility landscape of tissue-unrestricted enhancers. Nat Struct Mol Biol. 2018;25:61–72. doi: 10.1038/s41594-017-0007-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Dobrinić P, Szczurek AT, Klose RJ. PRC1 drives Polycomb-mediated gene repression by controlling transcription initiation and burst frequency. Preprint at bioRxiv. 2020 doi: 10.1101/2020.1110.1109.333294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Zhang Z, et al. Role of remodeling and spacing factor 1 in histone H2A ubiquitination-mediated gene silencing. Proc Natl Acad Sci U S A. 2017;114:E7949–E7958. doi: 10.1073/pnas.1711158114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Nakagawa T, et al. Deubiquitylation of histone H2A activates transcriptional initiation via trans-histone cross-talk with H3K4 di- and trimethylation. Genes Dev. 2008;22:37–49. doi: 10.1101/gad.1609708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Aihara H, et al. Histone H2A T120 Phosphorylation Promotes Oncogenic Transformation via Upregulation of Cyclin D1. Mol Cell. 2016;64:176–188. doi: 10.1016/j.molcel.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 237.Lehmann L, et al. Polycomb repressive complex 1 (PRC1) disassembles RNA polymerase II preinitiation complexes. J Biol Chem. 2012;287:35784–35794. doi: 10.1074/jbc.M112.397430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Dellino GI, et al. Polycomb silencing blocks transcription initiation. Mol Cell. 2004;13:887–893. doi: 10.1016/s1097-2765(04)00128-5. [DOI] [PubMed] [Google Scholar]
- 239.Stock JK, et al. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- 240.Brookes E, et al. Polycomb associates genome-wide with a specific RNA polymerase II variant, and regulates metabolic genes in ESCs. Cell Stem Cell. 2012;10:157–170. doi: 10.1016/j.stem.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Tsuboi M, et al. Ubiquitination-Independent Repression of PRC1 Targets during Neuronal Fate Restriction in the Developing Mouse Neocortex. Dev Cell. 2018;47:758–772.:e755. doi: 10.1016/j.devcel.2018.11.018. [DOI] [PubMed] [Google Scholar]
- 242.Pengelly AR, Kalb R, Finkl K, Muller J. Transcriptional repression by PRC1 in the absence of H2A monoubiquitylation. Genes Dev. 2015;29:1487–1492. doi: 10.1101/gad.265439.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Posfai E, et al. Polycomb function during oogenesis is required for mouse embryonic development. Genes Dev. 2012;26:920–932. doi: 10.1101/gad.188094.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Endoh M, et al. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development. 2008;135:1513–1524. doi: 10.1242/dev.014340. [DOI] [PubMed] [Google Scholar]
- 246.Cohen I, et al. PRC1 preserves epidermal tissue integrity independently of PRC2. Genes Dev. 2019;33:55–60. doi: 10.1101/gad.319939.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Chiacchiera F, Rossi A, Jammula S, Zanotti M, Pasini D. PRC2 preserves intestinal progenitors and restricts secretory lineage commitment. EMBO J. 2016;35:2301–2314. doi: 10.15252/embj.201694550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Chiacchiera F, et al. Polycomb Complex PRC1 Preserves Intestinal Stem Cell Identity by Sustaining Wnt/beta-Catenin Transcriptional Activity. Cell Stem Cell. 2016;18:91–103. doi: 10.1016/j.stem.2015.09.019. [DOI] [PubMed] [Google Scholar]
- 249.Pengelly AR, Copur O, Jackle H, Herzig A, Muller J. A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science. 2013;339:698–699. doi: 10.1126/science.1231382. [DOI] [PubMed] [Google Scholar]
- 250.Stepanik VA, Harte PJ. A mutation in the E(Z) methyltransferase that increases trimethylation of histone H3 lysine 27 and causes inappropriate silencing of active Polycomb target genes. Dev Biol. 2012;364:249–258. doi: 10.1016/j.ydbio.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 251.Bradley WD, et al. EZH2 inhibitor efficacy in non-Hodgkin’s lymphoma does not require suppression of H3K27 monomethylation. Chem Biol. 2014;21:1463–1475. doi: 10.1016/j.chembiol.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 252.Jadhav U, et al. Replicational Dilution of H3K27me3 in Mammalian Cells and the Role of Poised Promoters. Mol Cell. 2020;78:141–151.:e145. doi: 10.1016/j.molcel.2020.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Fan H, et al. A conserved BAH module within mammalian BAHD1 connects H3K27me3 to Polycomb gene silencing. Nucleic Acids Res. 2021;49:4441–4455. doi: 10.1093/nar/gkab210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Zhao D, et al. The BAH domain of BAHD1 is a histone H3K27me3 reader. Protein Cell. 2016;7:222–226. doi: 10.1007/s13238-016-0243-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Xu P, et al. FBXO11-mediated proteolysis of BAHD1 relieves PRC2-dependent transcriptional repression in erythropoiesis. Blood. 2021;137:155–167. doi: 10.1182/blood.2020007809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Ardehali MB, et al. Polycomb Repressive Complex 2 Methylates Elongin A to Regulate Transcription. Mol Cell. 2017;68:872–884.:e876. doi: 10.1016/j.molcel.2017.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.He A, et al. PRC2 directly methylates GATA4 and represses its transcriptional activity. Genes Dev. 2012;26:37–42. doi: 10.1101/gad.173930.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Zepeda-Martinez JA, et al. Parallel PRC2/cPRC1 and vPRC1 pathways silence lineage-specific genes and maintain self-renewal in mouse embryonic stem cells. Sci Adv. 2020;6:eaax5692. doi: 10.1126/sciadv.aax5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Cohen I, et al. Polycomb complexes redundantly maintain epidermal stem cell identity during development. Genes Dev. 2021;35:354–366. doi: 10.1101/gad.345363.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Leeb M, et al. Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev. 2010;24:265–276. doi: 10.1101/gad.544410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Inoue A, Jiang L, Lu F, Suzuki T, Zhang Y. Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature. 2017;547:419–424. doi: 10.1038/nature23262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Beguelin W, et al. EZH2 and BCL6 Cooperate to Assemble CBX8-BCOR Complex to Repress Bivalent Promoters, Mediate Germinal Center Formation and Lymphomagenesis. Cancer Cell. 2016;30:197–213. doi: 10.1016/j.ccell.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Klauke K, et al. Polycomb Cbx family members mediate the balance between haematopoietic stem cell self-renewal and differentiation. Nat Cell Biol. 2013;15:353–362. doi: 10.1038/ncb2701. [DOI] [PubMed] [Google Scholar]
- 264.Morey L, et al. Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell. 2012;10:47–62. doi: 10.1016/j.stem.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 265.O’Loghlen A, et al. MicroRNA regulation of Cbx7 mediates a switch of Polycomb orthologs during ESC differentiation. Cell Stem Cell. 2012;10:33–46. doi: 10.1016/j.stem.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Matsui SI, Seon BK, Sandberg AA. Disappearance of a structural chromatin protein A24 in mitosis: implications for molecular basis of chromatin condensation. Proc Natl Acad Sci U S A. 1979;76:6386–6390. doi: 10.1073/pnas.76.12.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Pena-Llopis S, et al. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet. 2012;44:751–759. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Scheuermann JC, et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Campagne A, et al. BAP1 complex promotes transcription by opposing PRC1-mediated H2A ubiquitylation. Nat Commun. 2019;10:348. doi: 10.1038/s41467-018-08255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Conway E, et al. BAP1 enhances Polycomb repression by counteracting widespread H2AK119ub1 deposition and chromatin condensation. Mol Cell. 2021;81:1–16. doi: 10.1016/j.molcel.2021.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Fursova NA, et al. BAP1 constrains pervasive H2AK119ub1 to control the transcriptional potential of the genome. Genes Dev. 2021;35:749–770. doi: 10.1101/gad.347005.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Ferrari KJ, et al. Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol Cell. 2014;53:49–62. doi: 10.1016/j.molcel.2013.10.030. [DOI] [PubMed] [Google Scholar]
- 273.Grzybowski AT, Chen Z, Ruthenburg AJ. Calibrating ChIP-Seq with Nucleosomal Internal Standards to Measure Histone Modification Density Genome Wide. Mol Cell. 2015;58:886–899. doi: 10.1016/j.molcel.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Hojfeldt JW, et al. Non-core Subunits of the PRC2 Complex Are Collectively Required for Its Target-Site Specificity. Mol Cell. 2019;76:423–436.:e423. doi: 10.1016/j.molcel.2019.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Peters AH, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 276.Lee HG, Kahn TG, Simcox A, Schwartz YB, Pirrotta V. Genome-wide activities of Polycomb complexes control pervasive transcription. Genome Res. 2015;25:1170–1181. doi: 10.1101/gr.188920.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Du Z, et al. Polycomb Group Proteins Regulate Chromatin Architecture in Mouse Oocytes and Early Embryos. Mol Cell. 2020;77:825–839.:e827. doi: 10.1016/j.molcel.2019.11.011. [DOI] [PubMed] [Google Scholar]
- 278.Zheng H, et al. Resetting Epigenetic Memory by Reprogramming of Histone Modifications in Mammals. Mol Cell. 2016;63:1066–1079. doi: 10.1016/j.molcel.2016.08.032. [DOI] [PubMed] [Google Scholar]
- 279.Gao Z, et al. An AUTS2-Polycomb complex activates gene expression in the CNS. Nature. 2014;516:349–354. doi: 10.1038/nature13921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Frangini A, et al. The aurora B kinase and the polycomb protein ring1B combine to regulate active promoters in quiescent lymphocytes. Mol Cell. 2013;51:647–661. doi: 10.1016/j.molcel.2013.08.022. [DOI] [PubMed] [Google Scholar]
- 281.Creppe C, Palau A, Malinverni R, Valero V, Buschbeck M. A Cbx8-containing polycomb complex facilitates the transition to gene activation during ES cell differentiation. PLoS Genet. 2014;10:e1004851. doi: 10.1371/journal.pgen.1004851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Maezawa S, et al. Polycomb directs timely activation of germline genes in spermatogenesis. Genes Dev. 2017;31:1693–1703. doi: 10.1101/gad.302000.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Richly H, et al. Transcriptional activation of polycomb-repressed genes by ZRF1. Nature. 2010;468:1124–1128. doi: 10.1038/nature09574. [DOI] [PubMed] [Google Scholar]
- 284.Cruz-Molina S, et al. PRC2 Facilitates the Regulatory Topology Required for Poised Enhancer Function during Pluripotent Stem Cell Differentiation. Cell Stem Cell. 2017;20:689–705.:e689. doi: 10.1016/j.stem.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 285.Kondo T, et al. Polycomb potentiates meis2 activation in midbrain by mediating interaction of the promoter with a tissue-specific enhancer. Dev Cell. 2014;28:94–101. doi: 10.1016/j.devcel.2013.11.021. [DOI] [PubMed] [Google Scholar]
- 286.Pachano T, et al. Orphan CpG islands amplify poised enhancer regulatory activity and determine target gene responsiveness. Nat Genet. 2021:2020.2008.2005.237768. doi: 10.1038/s41588-021-00888-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Morey L, et al. Polycomb Regulates Mesoderm Cell Fate-Specification in Embryonic Stem Cells through Activation and Repression Mechanisms. Cell Stem Cell. 2015;17:300–315. doi: 10.1016/j.stem.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 288.Mohn F, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 289.Xie R, et al. Dynamic chromatin remodeling mediated by polycomb proteins orchestrates pancreatic differentiation of human embryonic stem cells. Cell Stem Cell. 2013;12:224–237. doi: 10.1016/j.stem.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Hughes AL, Kelley JR, Klose RJ. Understanding the interplay between CpG island-associated gene promoters and H3K4 methylation. Biochim Biophys Acta Gene Regul Mech. 2020;1863:194567. doi: 10.1016/j.bbagrm.2020.194567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Ebmeier CC, et al. Human TFIIH Kinase CDK7 Regulates Transcription-Associated Chromatin Modifications. Cell Rep. 2017;20:1173–1186. doi: 10.1016/j.celrep.2017.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Lee JH, Skalnik DG. Wdr82 is a C-terminal domain-binding protein that recruits the Setd1A Histone H3-Lys4 methyltransferase complex to transcription start sites of transcribed human genes. Mol Cell Biol. 2008;28:609–618. doi: 10.1128/MCB.01356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Eberl HC, Spruijt CG, Kelstrup CD, Vermeulen M, Mann M. A map of general and specialized chromatin readers in mouse tissues generated by label-free interaction proteomics. Mol Cell. 2013;49:368–378. doi: 10.1016/j.molcel.2012.10.026. [DOI] [PubMed] [Google Scholar]
- 294.Brown DA, et al. The SET1 Complex Selects Actively Transcribed Target Genes via Multivalent Interaction with CpG Island Chromatin. Cell Rep. 2017;20:2313–2327. doi: 10.1016/j.celrep.2017.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Clouaire T, et al. Cfp1 integrates both CpG content and gene activity for accurate H3K4me3 deposition in embryonic stem cells. Genes Dev. 2012;26:1714–1728. doi: 10.1101/gad.194209.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Vermeulen M, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 297.Reinig J, Ruge F, Howard M, Ringrose L. A theoretical model of Polycomb/Trithorax action unites stable epigenetic memory and dynamic regulation. Nat Commun. 2020;11:4782. doi: 10.1038/s41467-020-18507-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Sneppen K, Ringrose L. Theoretical analysis of Polycomb-Trithorax systems predicts that poised chromatin is bistable and not bivalent. Nat Commun. 2019;10:2133. doi: 10.1038/s41467-019-10130-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Voigt P, Tee WW, Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Binder H, et al. Transcriptional regulation by histone modifications: towards a theory of chromatin re-organization during stem cell differentiation. Phys Biol. 2013;10:026006. doi: 10.1088/1478-3975/10/2/026006. [DOI] [PubMed] [Google Scholar]
- 301.Yakushiji-Kaminatsui N, et al. Variant PRC1 competes with retinoic acid-related signals to repress Meis2 in the mouse distal forelimb bud. Development. 2018;145:dev166348. doi: 10.1242/dev.166348. [DOI] [PubMed] [Google Scholar]
- 302.Angel A, Song J, Dean C, Howard M. A Polycomb-based switch underlying quantitative epigenetic memory. Nature. 2011;476:105–108. doi: 10.1038/nature10241. [DOI] [PubMed] [Google Scholar]
- 303.Berry S, Dean C, Howard M. Slow Chromatin Dynamics Allow Polycomb Target Genes to Filter Fluctuations in Transcription Factor Activity. Cell Syst. 2017;4:445–457.:e448. doi: 10.1016/j.cels.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Kar G, et al. Flipping between Polycomb repressed and active transcriptional states introduces noise in gene expression. Nat Commun. 2017;8:36. doi: 10.1038/s41467-017-00052-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Azuara V, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 306.Rosen GA, et al. Dynamics of RNA polymerase II and elongation factor Spt4/5 recruitment during activator-dependent transcription. Proc Natl Acad Sci U S A. 2020;117:32348–32357. doi: 10.1073/pnas.2011224117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Friedman LJ, Gelles J. Mechanism of transcription initiation at an activator-dependent promoter defined by single-molecule observation. Cell. 2012;148:679–689. doi: 10.1016/j.cell.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]