Abstract
The mounting evidence for the involvement of astrocytes in neuronal circuits function and behavior stands in stark contrast to the lack of detailed anatomical description of these cells and the neurons in their domains. To fill this void, we imaged >30,000 astrocytes in hippocampi made transparent by CLARITY, and determined the elaborate structure, distribution, and neuronal content of astrocytic domains. First, we characterized the spatial distribution of >19,000 astrocytes across CA1 lamina, and analyzed the morphology of thousands of reconstructed domains. We then determined the excitatory somatic content of CA1 astrocytes, and measured the distance between inhibitory neuronal somata to the nearest astrocyte soma. We find that on average, there are almost 14 pyramidal neurons per domain in the CA1, increasing toward the pyramidal layer midline, compared to only five excitatory neurons per domain in the amygdala. Finally, we discovered that somatostatin neurons are found in close proximity to astrocytes, compared to parvalbumin and VIP inhibitory neurons. This work provides a comprehensive large-scale quantitative foundation for studying neuron-astrocyte interactions.
Keywords: amygdala, CLARITY, hippocampus, parvalbumin, pyramidal neurons, somatostatin neurons, VIP
1. Introduction
Recent years have seen a surge in development of new methods for neuroscience research, enabling detailed functional investigation of specific cell populations, alongside their elaborate anatomical characterizations. In astrocyte research, manipulations using chemogenetic and optogenetic tools were successfully integrated into the field, resulting in a new understanding of how these cells are an integral part of the circuits controlling emotional and cognitive behavior (Yu et al., 2020). This recent progress in deciphering the functional significance of astrocytes stands in stark contrast to the lack of comprehensive anatomical description of these cells in the hippocampus and many other brain regions, and of the neuron somata in their domains. Compared to the rich anatomical data available describing neurons, no sufficient large-scale characterization of the structure and spatial distribution of astrocytes has been performed.
Two decades ago, astrocytes in the hippocampus were shown to exhibit a unique spatial organization: In contrast to the organization of neuronal dendrites, which are spatially intermingled, astrocytic processes display minimal overlap (Bushong et al., 2002). This finding was later replicated both in the hippocampus (Bushong et al., 2003, 2004; Livet et al., 2007; Ogata & Kosaka, 2002; Wilhelmsson et al., 2006; Xu et al., 2014) and in the cortex (Grosche et al., 2013; Halassa et al., 2007; Lopez-Hidalgo et al., 2016; Oberheim et al., 2008; Wilhelmsson et al., 2006) in different species, using various experimental techniques. The common staining procedures for astrocytes tag only their main processes, representing no more than 15% of their total volume (Bushong et al., 2002), and hence do not expose their true elaborate 3D morphology. To obtain a detailed description of astrocyte discrete domains, early works employed dye filling in CA1 astrocytes (e.g., (Bushong et al., 2002; Ogata & Kosaka, 2002; Wilhelmsson et al., 2006), and later ones induced the expression of fluorophores in astrocytes using transgenic animals (e.g., Halassa et al., 2007; Livet et al., 2007) or viral vectors (Chai et al., 2017; Halassa et al., 2007; Jones et al., 2018). However, most of these studies were performed on thin brain slices, resulting in most astrocytes being truncated due to their large domains, with a diameter up to tens of micrometers, thus radically restricting the number of analyzed cells. Indeed, the number of reconstructed astrocytes in all of the above studies ranged from less than twenty to a few hundreds at most.
Recently developed brain clearing techniques allow large scale, three dimensional, single-cell resolution characterization of multiple cell types simultaneously (Gradinaru et al., 2018; Treweek & Gradinaru, 2016; Ye et al., 2016). Such methods can be adopted to investigate astrocytic domains, and indeed a few studies have already used transgenic mice to show that fluorescent astrocytes can be imaged in the cleared mouse cortex (Chai et al., 2017; Clavreul et al., 2019; Gaire et al., 2018; Lanjakornsiripan et al., 2018; Miller & Rothstein, 2016). However, this was not done in the hippocampus and the amygdala as of yet, two brain areas which play a central role in high cognitive processes such as memory formation or retrieval, and regulation of emotional responses. Thus, a thorough characterization of the morphology and distribution of astrocytic domains in the hippocampus is currently missing.
A crucial open question in the investigation of astrocytic domains is the content of different neuronal cell types within them. The use of sparse labeling, which was often used in past studies, allowed the investigation of the synaptic content of single astrocytes. However, to study the distribution of all neurons among all astrocytes, one must accurately and robustly tag both of these cell types. This approach is suitable for examining the interactions between astrocytes and neuronal somata, which has been mostly ignored. In principle, the average number of any specific neuronal cell type per domain can be fairly accurately calculated by simply dividing the total number of neurons by the total number of astrocytes, even in thin slices. However, such an average estimation relies on the incorrect assumption that astrocytes are similar in shape, volume, and distribution along gray matter lamina, and gives scant information regarding possible unique spatial interactions (e.g., proximity of astrocytes to specific cell types) and domain-specific constrains on neuronal occupancy. Even when full domains of fluorescently-tagged astrocytes were imaged in the past, the common use of sparse astrocytic labeling hindered the accurate evaluation of the neuronal content of their domains, as neurons that are only partially engulfed by one astrocyte may in fact belong to the domain of an adjacent nonfluorescent astrocyte (Chai et al., 2017; Halassa et al., 2007; Octeau et al., 2018). Moreover, previous studies often used general neuronal markers (e.g., NeuN) to tag neurons, thereby preventing cell-type specific distribution analysis. Imaging high penetrance fluorescently tagged cell populations in clear brains does not allow synaptic resolution, but enables a thorough characterization of the spatial interactions between astrocytes and a variety of neuronal cell types somata.
In this work, we employed converging genetic, histological, imaging, and computational tools to determine the elaborate structure, distribution, and neuronal content of hippocampal astrocytic domains. We imaged over 30,000 hippocampal astrocytes, and provide a quantitative characterization of the spatial distribution of over 19,000 astrocytes in CA1 lamina, as well as morphological analysis of thousands of reconstructed astrocytes in the hippocampus (>6700) and hundreds in the amygdala (>890). We offer the first cell-type specific quantitative analysis of the neuronal occupancy of hippocampal and amygdaloid astrocytic domains by pyramidal excitatory neurons. Furthermore, we investigated the proximity of parvalbumin (PV), vasoactive intestinal peptide (VIP), and somatostatin (SST) inhibitory neurons to astrocytes across the CA1 lamina. This article provides a large-scale description of fundamental design principles in the hippocampus, relating the distributions and spatial locations of astrocytes, pyramidal neurons, and inhibitory neurons. Such data defines the range of possibilities for the interdomain effects of astrocytes on neuronal activity, allowing improved interpretation of experimental data, and facilitating the design of future experiments.
2. Meterials and Methods
2.1. Mice
C57BL/6 WT mice, Pv-IRES-Cre (B6.129P2-Pvalbtm1(cre)Arbr/J–stock number 017320; Hippenmeyer et al., 2005), Sst-IRES-Cre (Ssttm2.1[cre]Zjh/J—stock number 013044; Taniguchi et al., 2011), and VIP-IRES-Cre (Viptm1[cre]Zjh/J—stock number 010908; Taniguchi et al., 2011), were used. All mice included in this study were male, hence it does not account for potential gender differences.
Using the Allen Brain Atlas Mouse Connectivity datasets, we found that the different transgenic mouse lines show reasonable penetrance (>94%, >75%, and >92% for SST, PV, and VIP, respectively) and high specificity (>87%, 95%, and >97% for SST, PV, and VIP, respectively) when crossed with Ai14 mice, as indicated by fluorescence in situ hybridization (FISH) co-staining. We next tested the quality of the viral approach used in our study, that is, expression induced by a DIO viral injection into an inhibitory driver mouse line. The DIO infection showed high penetrance (>99% of PV positive cells also expressed enhanced green fluorescent protein (eGFP) and almost complete specificity (>95% of the eGFP expressing cells were also PV positive), indicating that our approach is comparable on both measures.
Mice were group housed on a 12 hr light/dark cycle with ad libitum access to food and water. All mice were maintained under pathogen-free conditions in Tecniplast cages, on Teklad sani-chips (ENVIGO) bedding, at 20–24°C, and fed Teklad 2918SC (ENVIGO) pellets. Experimental protocols were approved by the Hebrew University Animal Care and Use Committee and met the guidelines of the National Institute of Health guide for the Care and Use of Laboratory Animals.
2.2. Stereotactic virus injection
Six to seven weeks old mice were anesthetized with isoflurane, and their head placed in a stereotactic apparatus (Kopf Instruments, USA). The skull was exposed and a small craniotomy was performed. To cover the entire dorsal CA1, mice were bilaterally microinjected using the following coordinates: Anteroposterior, –1.85 mm from Bregma, mediolateral ±1.4 mm, and dorsoventral –1.5 mm. To cover the basolateral amygdala (BLA), mice were bilaterally microinjected using the following coordinates: Anteroposterior, –1.5 mm, mediolateral ±3.2 mm, and dorsoventral –5 mm. All microinjections were carried out using a 10 μl syringe and a 34 gauge metal needle (WPI, Sarasota, FL). The injection volume and flow rate (0.1 μl/min) were controlled by an injection pump (WPI). Following each injection, the needle was left in place for 10 additional minutes to allow for diffusion of the viral vector away from the needle track, and was then slowly withdrawn. The incision was closed using Vetbond tissue adhesive. For postoperative care, mice were subcutaneously injected with Tramadex (5 mg/kg).
2.3. Viral vectors
AAV8-GFAP::eGFP (4.1E12), (UNC vector core). AAV1-GFAP:: TdTomato (5.6E13), AAV5-EF1a-DIO-eYFP (2.11E13), AAV5-CaMKII-H2B-eGFP (2.2E13) (ELSC Vector Core Facility, EVCF). Vectors were injected in a volume of 400 nl/site.
2.4. Clarity
Four to five millimeter thick hippocampal and amygdaloid slices were cleared based on the protocol described by Ye et al. (2016)). Briefly, >10 weeks old mice were transcardially perfused with ice cold phosphate-buffered saline (PBS) followed by 4% paraformaldehyde (PFA), brains were removed and kept in 4% PFA overnight at 4°C. Brains were then transferred to a hydrogel solution (PBS with: 2% acrylamide, bio-rad #161-0140; 0.1% Bisacrylamide, bio-rad #161-0142; 0.25% VA-044 initiator, Wako, 011-19365; 4% PFA) for 2 days. The samples were then degassed with N2 for 45 min and polymerized in 37°C for 3.5 hr. The samples were then washed overnight in 200 mM NaOH-Boric buffer (sigma, #B7901) containing 8% sodium dodecyl sulfate (SDS) (sigma, #L3771), to remove PFA residuals. Samples were then stirred in a clearing solution (100 mM Tris-Boric buffer, bio-lab, #002009239100 with 8% SDS) at 37°C for 3–4 weeks. After the samples became transparent, they were washed with PBST (PBS with 0.2% tritonX100; ChemCruz, #sc-29112A) for 24 hr at 37°C with mild shaking and for another 24 hr with fresh PBST 0.2% at RT. Finally, the samples were incubated in the refractive index matched solution Rapiclear (RI = 1.47; SunJin lab, #RC147002) for 10 hr at 37°C and 2 days at room temperature before imaging.
2.5. Immunohistochemistry
Three weeks postinjection, mice were transcardially perfused with cold PBS followed by 4% PFA in PBS. The brains were extracted, postfixed overnight in 4% PFA at 4°C and cryoprotected in 30% sucrose in PBS. Brains were sectioned to a thickness of 40 μm using a sliding freezing microtome (Leica SM 2010R) and preserved in a cryoprotectant solution (25% glycerol and 30% ethylene glycol in PBS). Free-floating sections were washed in PBS, incubated for 1 hr in blocking solution (1%) bovine serum albumin (BSA) and 0.3% Triton X-100 in PBS) and incubated overnight at 4°C with primary antibodies (see below for a full list of antibodies) in blocking solution. Sections were then washed with PBS and incubated for 2 hr at room temperature with secondary antibodies (see below for a full list of antibodies) in 1% BSA in PBS. Finally, sections were washed in PBS, incubated with 4,6-diamidino-2-phenylindole (DAPI; 1 μg ml−1) and mounted on slides with mounting medium (Fluoromount-G, eBioscience).
Primary antibodies
The following primary antibodies were used: chicken anti-GFAP (Millipore, catalog no. AB5541; diluted 1:500); rabbit anti-Aldh1l1 (Abcam, catalog no. ab87117; diluted 1:500); rabbit anti-S100β (Abcam, catalog no. ab41548; 1:1000); rabbit anti-NeuN (Cell Signaling Technology, catalog no. 12943; diluted 1:400); rabbit anti-vGlut2 (Synaptic Systems, catalog no. 135403; 1:250).
Secondary antibodies
The following secondary antibodies were used, all from Jackson Laboratories: donkey anti-chicken (conjugated to Alexa Fluor 647, catalog no. 703-605-155; diluted 1:500); donkey anti-rabbit (conjugated to Alexa Fluor 647, catalog no. 711-605-152; diluted 1:500).
2.6. Confocal imaging
Confocal images were acquired using an Olympus scanning laser confocal microscope FV1000.
Stained brain slices were imaged using 10× air (UPlanSApo, 0.4 NA) or 40× oil-immersion (UPlanFLN, 1.3 NA) objectives, to obtain 1271 × 1271 μm or 317 × 317 μm images, respectively. Image analysis was performed using either ImageJ (NIH) or Fluoview Viewer v.4.2 (Olympus).
Cleared hippocampi were imaged using a water-immersion 20× objective with 3.4 mm working distance (UMPlanFL N, 0.5 NA), to obtain 635 × 635 μm images. We acquired z-stacks with 3.48 μm intervals between planes. Confocal images were used for somata location identification and somata proximity analyses that did not require full morphological reconstruction.
2.7. 2-Photon imaging
2-Photon imaging was performed using the Neurolabware 2-photon laser scanning microscope (Los Angeles, CA). Excitation light from a Ti:sapphire laser (Chameleon Vision II, Coherent, and then Chameleon Discovery TPC, Coherent) scanned the sample using a 6215 galvometer and a CRS8 resonant mirror (Cambridge Technology). Emitted fluorescence light was detected by GaAsP photo-multiplier tubes (Hamamatsu, H10770-40) after bandpass filtering (Semrock). XYZ motion control was obtained using motorized linear stages, enabled via an electronic rotary encoder (KnobbyII). The Scanbox software, run on MATLAB, was used for microscope control and image acquisition. All images were acquired using a water immersion 16× objective (Nikon, 0.8 NA) with magnification of 2.4, 2.8, or 3.4 to obtain 652 × 483 μm, 601 × 418 μm or 516 × 366 μm fields of view, at 15.5 frames/s. In each sample, we acquired z-stacks with 0.937 μm intervals between planes, centered on the dorsal CA1 pyramidal layer or on the BLA. We scanned each plane 60 times and obtained its mean intensity projection image using FIJI. These images were concatenated into a single volumetric series for subsequent image analysis.
The chosen laser wavelength was optimized for astrocyte fluorescent emission, to enable processes reconstruction in Imaris; Samples with GFP expressing astrocytes were scanned at 920 nm. The co-labeled neuronal somata in these samples were tagged with tdTomato and were successfully detected using the same excitation wavelength. However, in different samples that included tdTomato expressing astrocytes, we used 1040 nm excitation to visualize their processes. This was not sufficient for detecting co-labeled GFP expressing neuronal somata, and hence we conducted a consecutive scan of the same volume using 920 nm excitation.
2.8. Image analysis
Astrocyte processes reconstruction and compact representation
Using a similar approach to a variety of previous studies (Healey et al., 2020; Scofield et al., 2016; Shevelkin et al., 2020; Siemsen et al., 2019; Testen et al., 2019) with mild changes, astrocytes The somata location was extracted to Matlab as 3D points coordinates for further analysis.
Transforming size from CLARITY to nonclarified brains
To estimate the CLARITY induced expansion of the samples, we compared the size of 53 astrocyte somata in slices and in cleared brains. We found CLARITY expansion factor be 1.89 on average, in all dimensions. This factor was used for all subsequent analyses to estimate the real-world distances and volumes.
Calculating the distance to CA1 pyramidal layer
The CA1 pyramidal layer was masked using the manual surface option in Imaris, and then extracted to Matlab and smoothed using a moving mean. Gridded interpolants estimating the top and bottom surfaces of the CA1 pyramidal layer were created from the extracted boundaries, and each point of interest was evaluated accordingly to determine whether it resides below, above, or inside this layer. Next, we found the nearest sampled point on the CA1 pyramidal layer boundary to the point of interest, and created a triangulated surface around it using its four neighboring grid points (above, below, right, and left). Finally, the closest point on the triangulated surface to the point of interest was extracted, as well as the distance between them, using the point2trimesh function.
Because the CA1 pyramidal layer varies in size, we used normalization to present the distances of cells residing within it. First, we calculated the mean size of the CA1 pyramidal layer across all samples. Then, we measured the distances of each cell in this lamina to its boundaries, and calculated the normalized relative cell location as follows:
were segmented by constructing semi-automatic filaments in Imaris 9.1.2 (Bitplane, UK) while maintaining the same parameters for all samples. Using the “autopath algorithm,” starting points diameter was set to 15 μm and seed points diameter was set to 0.85 μm. All of the detected somata were manually confirmed while the threshold for the seed points was automatically selected. Next, all disconnected segments were removed using a maximum gap of 7 μm.
Each astrocytic reconstruction was then extracted from Imaris to Matlab as 3D points coordinates with a connectivity matrix depicting the branching of the astrocyte. Astrocyte somata locations, defined as the filament starting point, were extracted from the processes reconstruction. The length and volume of all processes were extracted from Imaris as well.
Neuronal somata detection
Excitatory neuronal somata were semi-automatically detected using the spots feature in Imaris. All spots diameter was set to 15 μm and the threshold was set to 98% to optimize detection. Post selection, spots were manually verified. Inhibitory somata were manually detected due to their sparseness.
Calculating astrocyte density
To calculate the density of astrocytes in the different hippocampal lamina, we first measured the distance of each astrocyte somata to the pyramidal layer, as described before. Next, we divided the imaging volume into discrete sections based on the pyramidal layer mask (i.e., with parallel boundaries), and counted the number of voxels that resided in each of them to obtain their volumes. Finally, the number of astrocyte somata within a given distance to the pyramidal layer was divided by the corresponding volume.
Determining processes total length
Astrocyte processes total length was determined using the Imaris statistic “Filament Length,” which returns the sum of the lengths of all edges within the filament.
Determining astrocyte volume
Astrocyte volume was determined using the Imaris statistic “Filament volume,” which returns the sum of the volumes of the cones composing a filament.
Calculating the normalized distance between astrocyte center of mass (CoM) to soma
The CoM was defined as the mean of each astrocyte 3D point cloud, and its distance from the soma was measured. The distance was normalized by the astrocyte length.
Calculating astrocyte orientation
To determine the orientation of each astrocyte, we used a similar approach to Lanjakornsiripan et al., 2018, adapted for point cloud representations, instead of objects defined by surfaces. Specifically, we calculated the first principle component based on the 3D point clouds that represent each astrocyte. Next, we found the astrocyte centroid, and its nearest point on the midline surface of the CA1 pyramidal layer (obtained by averaging the bottom and top surfaces), as described above. We then calculated the angle between the normal to the CA1 surface and the principle component, and subtracted it from 90°.
Allocating pyramidal neurons to astrocytic domains
To determine the pyramidal neurons occupancy of each astrocyte, we calculated the minimal distance between each neuron to the end points of all the astrocytes. The neuron was assigned to the astrocyte which had the closest end point relative to the neuron soma. To reduce the probability of including truncated domains in the analysis, we omitted astrocytes with somata that were close to the scanned volume edges (less than 20 μm from the edges) and/or far from the pyramidal layer when applicable (more than 5 μm from the CA1 surfaces). Neurons that were close to the scanned volume edges (less than 5 μm from the edges) and/or were assigned to an omitted astrocyte were also removed from the analysis.
Analyzing somata proximity
In each sample that contained tagged inhibitory neurons, we calculated the distance between each neuron soma to all the astrocyte somata. The minimal distance from each neuron soma to its nearest astrocyte somata in each cubic volume was measured, and used to calculate probability histograms.
2.9. Statistical analysis
Sample number (n) indicates the number of cells or imaging cubes in each experiment and is specified in the figure legends. Results were analyzed by one-way ANOVA, followed by LSD post-hoc tests, paired t-test, independent samples t-test, or a linear regression test, as applicable. All the statistical details of experiments can be found in the results section. Differences in means were considered statistically significant at p <.05. Analyses were performed using the IBM SPSS Statistics software (version 25).
Permutation tests
Permutation tests were conducted to test whether the empirical distributions are significantly different from predicted shuffled distributions. Specifically, we kept the astrocytes in their real locations, and simulated random neural locations. We permuted the neurons on the circumference of a 55.56 μm circle, parallel to the CA1 pyramidal layer midline surface, such that each simulated neuron maintained its distance from this layer. This method enabled us to preserve the cellular spatial distributions relative to CA1, which are unique to each inhibitory cell type (see Figure 4). The simulation was run 1500 times, and the minimal distance between the permuted neuronal somata to astrocyte somata was calculated and binned similarly to the real data. When the simulated distances were smaller than the real minimal distance between the specific inhibitory cell to its nearest astrocyte soma, they were omitted, to avoid nonbiologically plausible structures, that is, cells with overlapping somas. We calculated the mean distributions of the empirical and of the predicted shuffled data, and then compared their expected values to test for significance.
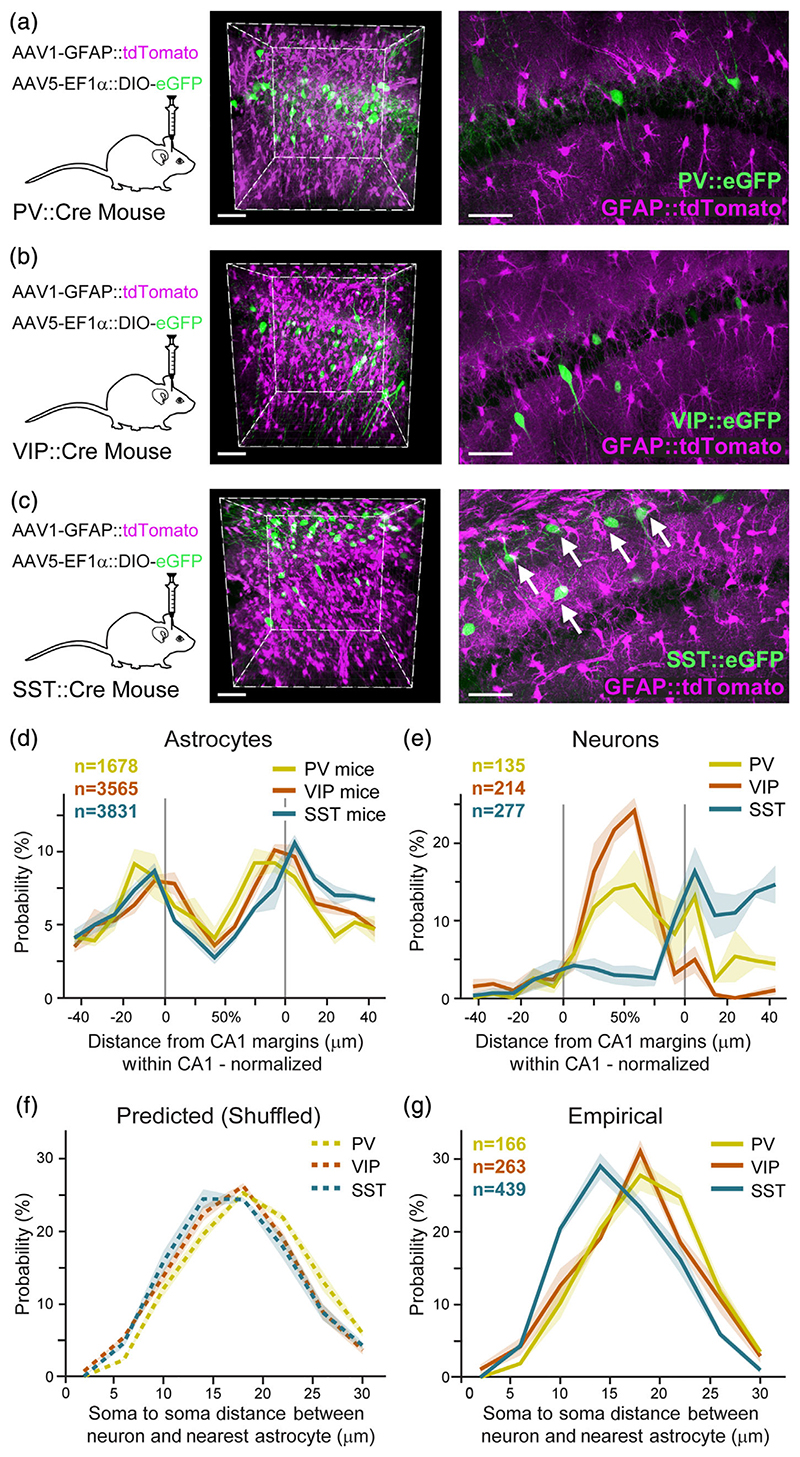
Figure 4. Unique proximity between astrocytes and somatostatin inhibitory neurons.
AAV1-GFAP::tdTomato was injected into CA1 to tag astrocytes (purple), and AAV5-EF1α-DIO-eGFP to tag inhibitory neurons (green) in PV-Cre (n = 3) (a), VIP-Cre (n = 4) (b), or SST-Cre (n = 4) (c) transgenic mice. Co-expression is shown in representative whole 400 × 400 × 450 μm imaging cubes (middle), and one 25 μm-thick optical plane from each cube (right). SST somata were observed in close proximity to astrocytes (white arrows in c right). (d) Astrocyte somata probability distributions in PV (yellow), VIP (orange), and SST (turquoise) mice, as a function of distance from the CA1 pyramidal layer surfaces, showing that astrocytes are highly concentrated on the borders of the pyramidal layer, and show low variance across the different samples. CA1 pyramidal layer borders are marked as in gray. (e) Spatial distribution of PV, VIP, and SST somata, showing their mean density as a function of distance from the CA1 pyramidal layer margins. PV and VIP neurons are concentrated within the pyramidal layer, whereas SST neurons are mostly present in the dendritic layer in stratum oriens, with minor expression in the pyramidal layer. (f) The predicted (permutations-based) distribution of the distances between PV, VIP, and SST somata to their nearest astrocyte soma (g). The empirical probability of SST somata to be in close proximity to the nearest astrocyte soma is significantly larger than that of PV and VIP (p <.01; see Movie S4). Average density for each inhibitory cell type in bold color, with ±SEM shading. Scale bars: 70 μm for cubes, 50 μm for single planes
3. Results
3.1. Large scale automatic detection of astrocytic domains in 3D volumes
Many pioneering studies characterizing astrocytic domains in the hippocampus were limited in the number of fully reconstructed astrocytes, as many of them were truncated (Bushong et al., 2002; Bushong et al., 2003, 2004; Livet et al., 2007; Ogata & Kosaka, 2002; Wilhelmsson et al., 2006; Xu et al., 2014). To rigorously characterize the structural and spatial characteristics of hippocampal astrocytic domains, 2–3 orders of magnitude larger quantities of analyzed cells are required.
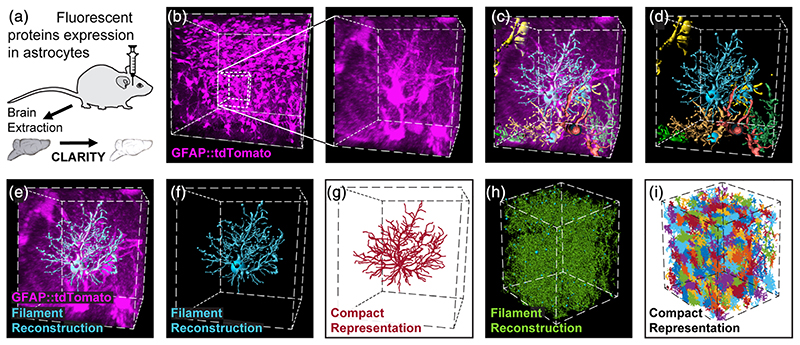
To achieve this demanding goal, we employed viral vectors to fluorescently tag astrocytes densely in WT or transgenic mice. Fluorophores (tdTomato or eGFP) were expressed under the glial fibrillary acidic protein (GFAP) promoter with high penetrance (>96.7%; Figure S1(a)–(d)), and almost complete specificity (>97% Figure S1(a)–(c), (e)). We then rendered thick brain slices (4–5 mm) transparent by CLARITY, and acquired high resolution images of optically-defined cubes (ranging in size from 520 × 370 × 520 μm to 635 × 635 × 1500 μm) from each slice (Figure 1(a), (b)). This analysis was performed for all astrocytes in the entire imaging cube, for 55 cubes sampled from 31 mice, providing quantitative data of over 30,000 astrocytes. Single astrocytic domains were then reconstructed (Figures 1(c)–(f) and S1(f)–(h), Movie S1), and compactly represented for further analysis (Figure 1(g)–(i)). The hippocampal astrocytes occupy visually distinguishable domains, with minimal overlap between their processes.
Figure 1. Automatic detection and reconstruction of astrocytic domains in large 3D volumes.
(a) Wild type or transgenic mice were injected with viral vectors to induce the expression of fluorescent proteins in astrocytes. Following >4 weeks, their brains were extracted, cut into thick hippocampal slices (4–5 mm) and were then made transparent by CLARITY after which they were imaged. (b) Expression of tdTomato (purple) in hippocampal astrocytes is presented for a 450 × 450 × 450 μm cube (left), and a zoomed-in 80 × 80 × 80 μm cube excerpt from it (right). (c, d) Seven representative astrocyte processes reconstructions from this cube are shown (and see Movie S1). Each astrocyte reconstruction, like the one shown in (e, f) was compactly represented (crimson) for further analysis (g). The process of processes reconstruction (green) (h) and compact representation (multicolor) (i) was performed on the entire imaging 500 × 400 × 400 μm cube
3.2. Detailed laminar characterization of astrocyte domains in CA1
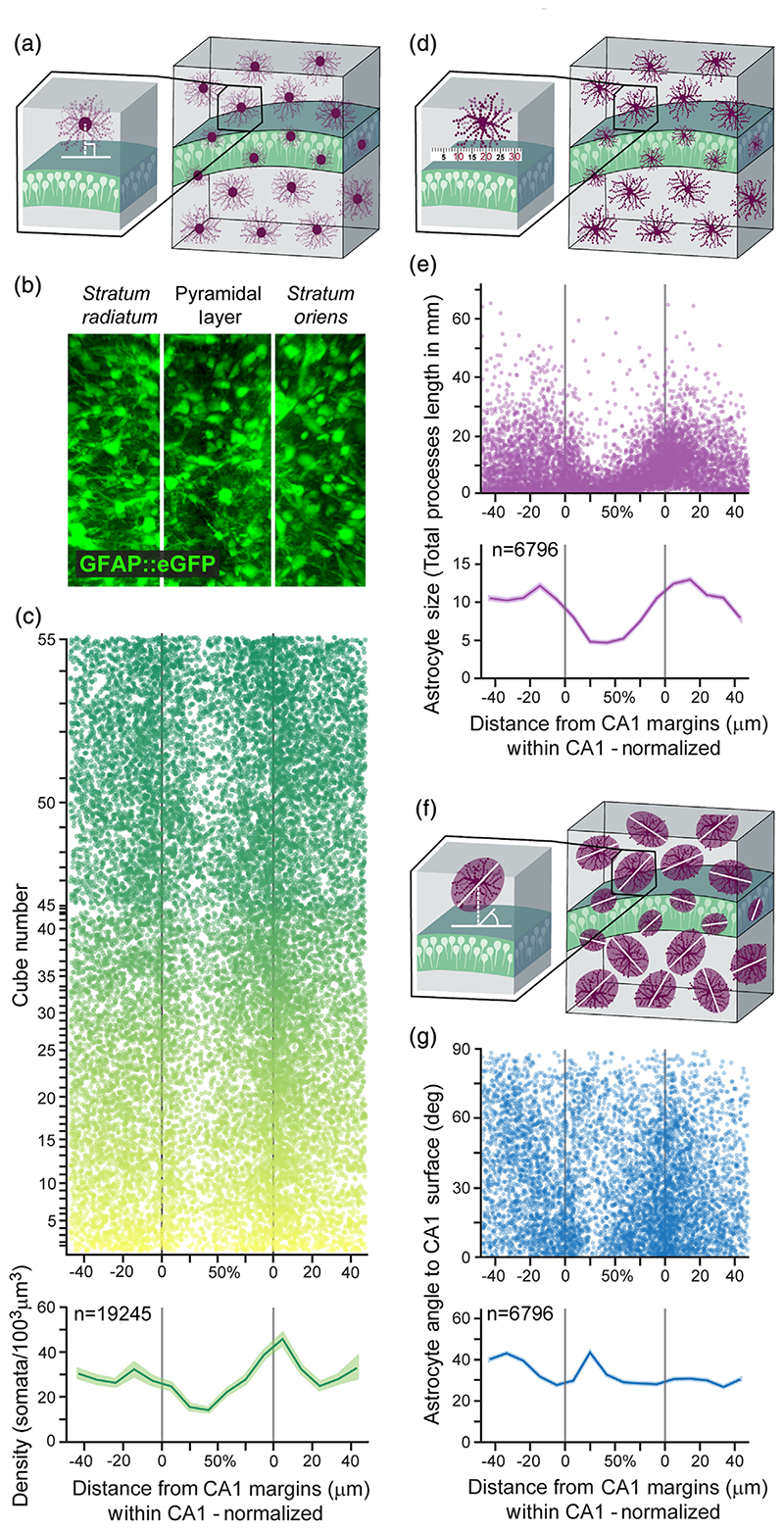
The large number of analyzed astrocytes obtained as detailed above was used to characterize the spatial distribution of astrocytes of different size and orientation along CA1 lamina with unprecedented detail. First, we quantified astrocytic density throughout CA1 lamina (Figure 2(a), (b)) and found that on average, there are 29.2 astrocyte somata per 1003 μm3. Nevertheless, the density of astrocytes is highly variable across the CA1 lamina; In the stratum oriens, the astrocyte density increases dramatically toward the pyramidal layer, reaching 45.9 ± 2.2 somata per 1003 μm3 on average, whereas in the stratum radiatum the density is more uniform, with only a mild increase toward the pyramidal layer. In the pyramidal layer itself astrocytic density dramatically drops toward its middle to a minimum of 14 ± 0.7 somata per 1003 μm3 on average (Figure 2(c)). The pyramidal layer varies in width, and hence we calculated the relative location of cells within it as the ratio between the distance to the lower edge and the total width. Then, we measured the total processes length of the reconstructed astrocytes (Figure 2(d)). As depicted in Figure 2(e), the main finding is that starting ~20 μm from the pyramidal layer on both stratum radiatum and stratum oriens, astrocytic total processes length decreases toward the middle of the pyramidal layer from 12.2 ± 0.1 mm and 13 ± 0.1 mm to 4.7 ± 0.1 mm, respectively. Astrocyte volume and terminal points number showed a similar lamina-dependent pattern (Figure S2(a), (b)). To investigate the dispersion of astrocyte processes relative to their soma, we measured the distance between the CoM and the soma of each astrocyte (Figure S2(c)). Since astrocytes vary in size, we normalized the distance by the astrocyte length. The CoM and somata of astrocytes are closer in the stratum radiatum and stratum oriens than in the pyramidal layer, indicating that their processes are differently spread around the soma. Finally, we calculated the orientation of the same astrocytes relative to the surface of the CA1 pyramidal layer center (Figure 2(f)) and found it to be homogenous, on average, with all angles between 0° and 90° represented throughout hippocampal lamina (Figure 2(g)). This section provides a detailed analysis of the distribution of different astrocyte morphological characteristics that could potentially affect the neuronal content within these domains, which is what we sought to investigate next.
Figure 2. Large scale automatic detection of astrocytic domains and their detailed laminar characterization in CA1.
(a) The distance of each astrocytic soma from the surface of the CA1 pyramidal layer was measured. (b) Representative z-projected cube showing astrocytes (green) in CA1. Lamina borders marked in white. Cube volume: 350 × 300 × 250 μm (c) Astrocyte somata distribution (top; n = 19,245 cells, each represented by a dot, from n = 55 cubes, each displayed in a different semi-transparent color) and average astrocytic density along CA1 lamina (bottom). Density in both stratum radiatum and stratum oriens increases toward the pyramidal layer, and inside this layer density drops toward the middle from both sides. Average density (bottom) presented in bold green, with SEM shading. The distance within the CA1 pyramidal layer is normalized to correct for varying CA1 widths across samples. (d) Astrocyte total processes length was calculated by measuring and summing all the processes. (e) The average total processes length of hippocampal astrocytes (n = 6796 cells, each represented by a semi-transparent dot, from n = 40 cubes) starts to drop at a ~20 μm distance from the pyramidal layer in both stratum radiatum and stratum oriens, and continues to decrease toward the middle. Average total processes length (bottom) presented in bold purple, with SEM shading. (f) Astrocytic orientation was calculated relative to the surface of the CA1 pyramidal layer midline. (g) The orientation of the same astrocytes from panel d (each represented by a semi-transparent dot), relative to the surface of CA1 pyramidal layer. Average orientation (bottom) presented in bold blue, with SEM shading
3.3. Distribution of excitatory pyramidal neurons in astrocytic domains
A large part of the research studying the functional interaction between astrocytes and pyramidal neurons was performed in CA1, and investigations into the role of astrocytes in memory also focused on this region (reviewed in [Santello et al., 2019]). However, little is known about the distribution of excitatory pyramidal neuron somata among the domains of the hippocampal astrocytes around them. Chai et al. (2017) investigated the number of neuronal somata per domain in the stratum radiatum, where very few somata reside, yet to our knowledge, no studies have explored this quantity in the different hippocampal lamina. Previous studies have shown that a large variety of synaptic activity modulations can occur within each domain, in a spatially restricted manner, that is, a single astrocyte selectively affects neurons in its vicinity (Henneberger et al., 2010).
The probable effect of astrocyte processes on neuronal somata in their domains in the pyramidal layer has not been defined as of yet, but pioneering work investigating this interaction in the habenula shows its great potential (Cui et al., 2018).
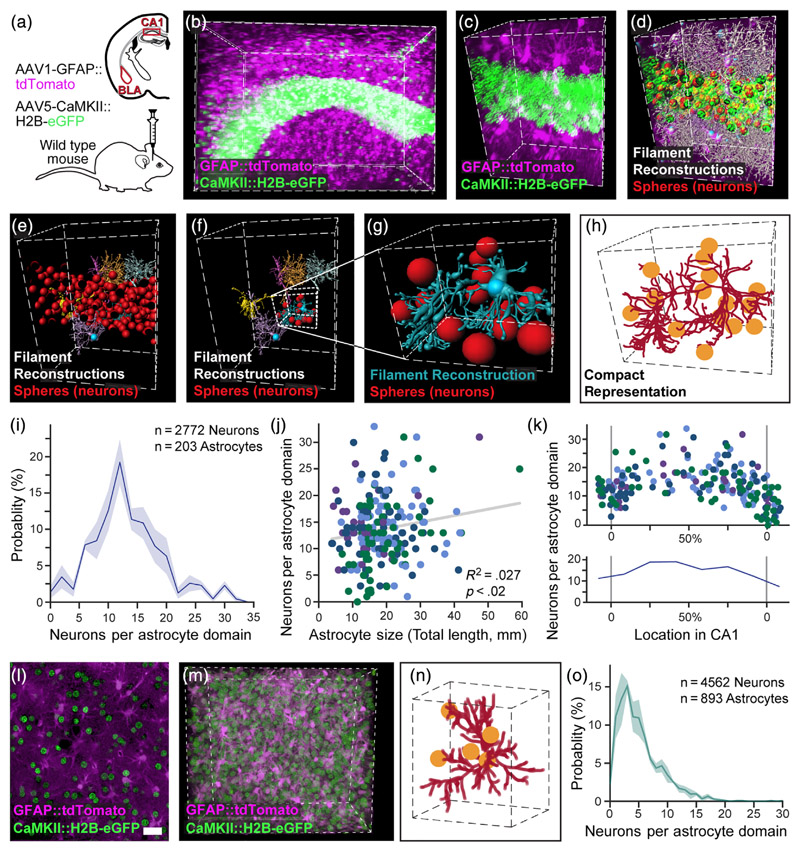
To answer this question, it is crucial to know how many pyramidal neurons are engulfed by each domain and quantify the spatial relations between astrocytes and pyramidal neurons. Therefore, we were required to robustly tag and detect genetically defined excitatory neuronal somata as well as astrocytes in a given volume. To this end, we co-labeled these populations in the hippocampus and the amygdala as follows: Astrocytes were tagged in red using an AAV1-GFAP::tdTomato vector, and pyramidal neurons were labeled in green by a vector inducing the expression of eGFP in their nuclei (AAV5-CaMKII::H2B-eGFP; Figure 3(a)–(c)). This H2B nuclear tagging allows efficient separation of the pyramidal neuron somata, not possible when using a cytoplasm-filling fluorophore (Figure S3(a), (b)). The CaMKII::H2B-eGFP expression in the dorsal CA1 showed high penetrance (>96%) and specificity (>96%; Figure S3(c)–(h)). Astrocytic morphology was reconstructed as detailed above, and each neuronal soma was represented by a sphere (Figure 3(d)–(e)), and associated with a specific astrocytic domain (Figures 3(f)–(h), S3(i); Movie S2 and Stereoscopic Movie 1). While all astrocytes throughout the lamina of the CA1 were reconstructed, the neuronal content was determined only for astrocytes whose soma is in the pyramidal layer, or within 5 μm from its edges since the vast majority of excitatory neurons are located there. The number of pyramidal neurons per domain in these astrocytes ranged from 0 to 33 (with 50% between 10 and 17), averaging 13.8 (Figures 3(i) and S3(i)–(k)). Only a weak positive correlation was found between the total processes length of astrocytic domains and the number of neurons within them (F(l·2oi) = 5.7, p <.02, R2 = .027; Figure 3(j)). However, the number of neurons per domain was more strongly correlated with the location of the astrocytes within the pyramidal layer of CA1 (F(1,201) = 76.6, p <.001, R2 = .28), with increasing numbers of excitatory neurons associated with astrocytic domains toward midline of the layer (Figure 3(k)). Thus, when characterizing the number of pyramidal neurons in CA1 pyramidal layer astrocytic domains, we found it to vary greatly, and be correlated with the location of the astrocyte relative to the pyramidal layer, and to a much lesser degree with the total processes length of the astrocyte.
Figure 3. Automatic detection of excitatory neurons content and its distribution in astrocytic domains.
(a) The hippocampus and amygdala of mice were injected with viral vectors to induce the expression of the red fluorophore tdTomato in astrocytes, and the green fluorophore H2B-eGFP in pyramidal neurons nuclei in the hippocampus or amygdala, and thick slices (4-5 mm) were then rendered transparent by CLARITY. (b) Expression of tdTomato (purple) in hippocampal astrocytes and H2B-eGFP in pyramidal neurons (green) is presented fora 520 × 370 × 520 μm cube, (c) and a zoomed-in 250 × 120 × 150 μm cube. (d) All astrocyte processes reconstructions from this cube are shown (white, and see Movie S2), together with all neuronal somata (red). (e) Six representative filaments (blue, yellow, orange, pink, purple, and gray) of astrocytes crossing the pyramidal layer are shown with all neuronal somata (red). (f) The same six filaments are shown only with the neuronal somata (red) associated with the blue astrocyte. (g) A zoomed-in 80 × 70 × 100 μm cube showing the same reconstructed astrocyte and its associated neurons. (h) A compact representation of the same astrocyte (crimson) and its associated neurons (yellow). (i) The binned distribution (bin size = 2) of pyramidal neurons content of CA1 astrocytes (average = 13.7; n = 2772 neurons in 203 astrocytic domains in 4 cubes). Average presented in bold blue, with SEM shading. (j) A significant, albeit weak, positive correlation is found between the total processes length of the astrocyte and the number of pyramidal neurons in its domain (203 astrocytic domains, each represented by a dot, from n = 4 cubes, each displayed in a different color). (k) The number of pyramidal neurons per astrocytic domain increases toward the middle of the pyramidal layer. Average number (bottom) presented in bold blue, with SEM shading. (l) Expression of tdTomato (purple) in BLA astrocytes and H2B-eGFP in pyramidal neurons (green) is presented for a single slice (m) and a 290 × 250 × 130 μm cube. (see Movie S3). (n) A compact representation of an astrocyte (crimson) and its associated neurons (yellow). Cube volume: 90 × 80 × 60 μm. (o) The distribution of pyramidal neurons content of astrocytes in the BLA (average = 5.2; n = 4562 neurons in 893 astrocytic domains in three cubes). Average presented in bold light green, with SEM shading. Scale bar: 50 μm
Next, we investigated astrocytic domains in the BLA, using a similar approach. Astrocytes were tagged in red, and excitatory neuron somata in green using the viral vectors mentioned above (Figure 3(a), (l), (m)). The labeled astrocytes were reconstructed and the neuronal somata locations were detected and extracted for further analysis as before (Figure 3(n)). The distribution of neurons in the amygdala is extremely different than in the hippocampus; CA1 excitatory neuronal somata are densely packed in the pyramidal layer, whereas the neurons in the BLA are more uniformly spread. Indeed, following assignment of each neuron to its astrocytic domain, we found that the number of pyramidal neuron somata per domain ranged from 0 to 30 (with 50% between 2 and 7) averaging 5.2 (Figure 3(o)), which is significantly smaller than in the pyramidal layer of the CA1 (t(5) = 10.8, p <.001, independent samples t-test). As hippocampal activity is determined not only by pyramidal neurons, but is also continuously modulated by their neighboring inhibitory neurons, we next investigated the spatial interaction between astrocytes and the three most common inhibitory neurons across the CA1 lamina.
3.4. Spatial associations between astrocytes and inhibitory neurons subtypes
After characterizing the excitatory neuronal content of CA1 astrocytic domains, we sought to determine the spatial relations between astrocytes and inhibitory neurons in this region. The importance of astrocytes in GABAergic signaling in the normal and the diseased brain has been repeatedly demonstrated: Astrocytes respond to GABA with intracellular Ca2+ increase and can either mediate inhibition or convert GABAergic inhibition into glutamatergic excitation (Covelo & Araque, 2018; Perea et al., 2016). Furthermore, GABAergic cells depend on astrocytes for normal GABA synthesis (Robel & Sontheimer, 2016). A handful of studies directly investigated the functional interaction between astrocytes and inhibitory cell types, showing specificity in both astrocytic response to- and modulation of- distinct inhibitory populations: On the one hand, astrocytes show distinguishable responses to the activation of different inhibitory celltypes (Deemyad et al., 2018; Mariotti et al., 2018; Matos et al., 2018). For example, cortical astrocytes respond strongly to SST activation, but only weakly to PV activation (Mariotti et al., 2018). On the other hand, astrocytic manipulation can differentially modulate the effects of specific inhibitory neurons and not others (Deemyad et al., 2018; Matos et al., 2018; Perea et al., 2014; Tan et al., 2017). For example, hippocampal astrocytes can detect SST activity and mediate its effects on pyramidal neurons, but they do not mediate the effect of PV cells in the same way (Matos et al., 2018). We found no reports on the interaction between VIP interneurons and astrocytes. While the functional interaction between astrocytes and neighboring inhibitory neurons has been investigated, the only available indirect quantification of spatial relationship between hippocampal astrocytes and neurons reports a sparse occupancy of neurons in the domains of stratum radiatum astrocytes (Chai et al., 2017). The genetic identity of these neurons is not known, but due to their location out of the pyramidal layer they are most likely inhibitory.
To gain insight into the spatial relations between CA1 astrocytes and inhibitory neurons, we co-labeled these populations as follows: Astrocytes were tagged in red, as before, using AAV1-GFAP:: tdTomato. Inhibitory neurons were labeled by injecting a vector encoding a Cre-dependent green fluorophore (AAV5-EF1α-DIO-eGFP) to PV-Cre (Figure 4(a)), or VIP-Cre (Figure 4(b)), or SST-Cre (Figure 4(c)) transgenic mice (Hippenmeyer et al., 2005; Taniguchi et al., 2011). First, we set to quantify the spatial distribution of the somata of astrocytes and inhibitory neurons across CA1 lamina, and the interactions between them: In contrast to the distribution of astrocytic somata, which is highest around the margins of CA1 in all mouse strains (PV-Cre, VIP-Cre, and SST-Cre; Figure 4(d), as in Figure 1(l)), the three inhibitory cell types demonstrated clear differences in spatial distribution (Figures 4(e) and S4(a)). Quantifying these differences, we found that PV neurons were concentrated mainly in the pyramidal layer (65.4% ± 9.3), with sparser expression in the stratum oriens (30.1% ± 8). VIP cells were found almost exclusively in the pyramidal layer (84.1% ± 3.5), especially close to its midline. SST cells presented a very different distribution, and were mostly located in the stratum oriens dendritic layer (66.4% ± 3.1), with low expression in the pyramidal layer (26.5% ± 5.1), and minimal expression in the stratum radiatum (7.2% ± 3.1; Figures 4(e), S4(a)). This spatial distribution is in line with previous reports (Atlas, 2007).
The inhomogeneous distribution of astrocytes along CA1 lamina, together with the differences in the laminar distribution between the three inhibitory populations, may affect the spatial interactions between these cells. Indeed, when the expected distance between the inhibitory neurons somata and their nearest astrocyte soma was calculated for the three populations based on their spatial distribution, a significant difference was observed (distribution mean: 18.2 ± 0.3, 17.1 ± 0.3, 16.8 ± 0.5, for PV, VIP, and SST, respectively; F (2,8) = 4.7, p <.05), and post-hoc tests showed a significant difference between PV cells (p <.05, with a higher expected soma-to-soma distance) and VIP and SST (Figure 4(f)). However, when we quantified the empirical distances between the soma center of each inhibitory cell to its nearest astrocyte soma center we found that SST neurons are located in much closer proximity to their neighboring astrocytes, relative to PV and VIP neurons (Figures 4(c) right, (g) and S4(b), (c); Movie S4). This striking difference was expressed by a significant difference between the average distance of the three populations to their closest astrocyte (distribution mean: 18.5 ± 0.2, 17.5 ± 0.6, 15.9 ± 0.2, for PV, VIP, and SST, respectively; F (2,8) = 8.6, p <.01), and post-hoc tests showed a significant difference between SST neurons and PV or VIP neurons (p <.01 and p <.05, respectively), but not between PV and VIP. We also calculated the minimal distance of excitatory neuron somata to their nearest astrocyte soma (mean: 18.3 ± 0.3, data not shown), comparable to the PV and VIP distributions which are also concentrated in the pyramidal layer. Furthermore, when we compared the empirical distributions to the predicted ones, we found that the SST cells are, on average, significantly closer to their neighboring astrocytes than anticipated (t(3) = 2.9, p <.05, paired t-test). No significant differences were found between empirical and predicted distributions for PV and VIP cells (p >.05). The finding of a unique spatial correlation between astrocytes and SST inhibitory neurons corresponds with the reports of strong functional interaction between these cells.
4. Discussion
To fill the void between the mounting evidence for the functional significance of astrocytes in hippocampal function, and the lack of detailed anatomical characterization of these cells (Yu et al., 2020), we imaged over 30,000 and fully reconstructed thousands of hippocampal astrocytes, to provide a comprehensive quantitative characterization of their spatial distribution, morphology of their processes, excitatory neurons content, and proximity to inhibitory neurons along CA1 lamina. We also report a significant difference in the number of pyramidal neurons per astrocytic domain in the CA1 compared to the BLA.
The investigation of astrocytic morphology to date was hindered mainly by technical barriers (Yu et al., 2020). In a recently published work, imaging of serially sectioned cortex using ChroMS microscopy followed the development of astrocytic clones and the 3D morphology of young and more mature astrocytes in these cortical clones (Clavreul et al., 2019). We found that in the pyramidal layer of the hippocampus astrocytic density dramatically drops toward its middle, where the volume of those astrocytes drops as well. The combination of brain clearing techniques together with tagging of genetically defined of cell types, now give access to large scale 3D imaging of fluorescently labeled cells, and allow simultaneous imaging of more than one cellular population.
Astrocytes have various morphological features and are not uniformly spread in different brain regions, but even within each region substantial variations may exist. Indeed, our results indicate that astrocytes vary across the hippocampal lamina in density, size, and orientation. Moreover, the interface between the pyramidal layer and the stratum oriens differs from the pyramidal-stratum radiatum layer interface. This is unlikely to be an artifact of the viral injection since we did not image or analyze volumes near the injection site, nor see aggregates or empty spaces in the imaged volumes, indicating that the tissue was not damaged. Previous studies have indicated that the deep and superficial neuronal sublayers of the pyramidal layer in CA1 have different characteristics, for example, genetics, morphology, intrinsic electrophysiology, and connectivity, as well as diverse behaviorally relevant functionality (Soltesz & Losonczy, 2018). Perhaps these profound neuronal differences result in significant variability of astrocytes that interact with the different neuronal sublayers.
The number of neuron somata per astrocytic domain also varies greatly across the brain and highly depends on the methodology. For example, in the striatum, the number of neurons was initially estimated to be around 20 (Chai et al., 2017), but using a different methodology this number was then reduced by half (Octeau et al., 2018). In both cases, sparse labeling of astrocytes was used, which can lead to over-estimation of the number of neurons per domain, as a neuron that is mostly engulfed by a nearby, nonfluorescing astrocyte may be erroneously associated with a fluorescent astrocyte. For that reason, our data was generated, by design, from samples with high-penetrance (>96%) expression of fluorophores in the astrocytic population, allowing a more accurate representation of the variance in neuronal content. We found that the number of pyramidal neurons somata in the hippocampus is only mildly correlated with the total processes length of the astrocyte, but is highly correlated to its location, that is, astrocytes whose soma is closer to the pyramidal layer midline have more neurons in their domain. It could be hypothesized that this finding is caused by a bigger part of the surface area of these domains overlapping with the pyramidal layer, allowing more contact of the processes with neuronal somata. Furthermore, we show that astrocytic domain occupancy by excitatory neurons in the amygdala is smaller than in the hippocampus on average.
When quantifying the spatial interaction between astrocytes and inhibitory neurons somata, we found that SST neurons are often “hugged” by astrocytes, that is, they are found in closer proximity to neighboring astrocytes, compared to PV and VIP neurons. This unique interaction does not stem only from the fact that the density of both astrocytic and SST somata is high in the stratum oriens, but rather represents a unique design principle. Interestingly, this spatial proximity goes hand in hand with the reported unique functional interaction between these cells in the hippocampus, where astrocytes mediate the activity of SST (but not PV) cells (Matos et al., 2018). Future studies could investigate the potential mechanisms found in the somata which may underlie this distinctive phenomenon.
The anatomical characterization of astrocytes was so far orthogonal to the investigation of their functional contribution to neuronal activity and behavior. However, the spatial distribution of these cells was shown to follow functional boundaries. For example, astrocyte processes are confined to barrel borders in the somatosensory cortex of mice, and to Cytochrome Oxidase blobs in human V1, and they show layer-specific molecular and morphological phenotype in somatosensory cortex (Eilam et al., 2016; Lanjakornsiripan et al., 2018). Furthermore, long-term potentiation (LTP) blockade in CA1 neurons, induced by calcium clamp in a single astrocyte was shown to be roughly restricted to its domain (Henneberger et al., 2010). We have recently shown that chemogenetic astrocytic activation in CA1 enhances memory and increases the number of active CA1 neurons recruited to support the acquisition of a new memory (Adamsky et al., 2018). We did not study single astrocytes, but such an effect would require within-domain neuronal activity detection, and specific modulation of single neurons or dendrites, but not others, within each domain. The modulatory effects of astrocytes on synapses are well characterized, and the increased number toward the pyramidal layer in the stratum radiatum and even more in the stratum oriens suggest that they may have more effect on the proximal dendrites, than the distal ones. The potential capability of astrocytes to selectively affect neuronal somata within their domains in the pyramidal layer of the hippocampus has not been explored as of yet. Nevertheless, recent work investigating such interaction in the lateral habenula has shown that astrocytes modulate neuronal membrane hyperpolarization and amount of bursting activity via potassium channels found on astrocytic processes that enwrap neuronal somata (Cui et al., 2018). Thus, a functional significance for the local parcellation of other brain regions, such as the hippocampus, by astrocytic domains exerting specific effects on neuronal somatic activity can be easily hypothesized, and needs to be determined in future experiments.
To summarize, in this study we performed a comprehensive quantitative characterization of the spatial distribution and morphology of CA1 astrocytes, in numbers that are orders of magnitudes larger than ever reported in the hippocampus. We then provide the first cell-type specific quantitative analysis of the neuronal somatic occupancy of astrocytic domains by pyramidal neurons, and of their proximity to three inhibitory neurons cell types. These results define the range of neuronal somatic information that each astrocyte in the pyramidal layer is exposed to, and consequently the range of possibilities of within-domain modulation of single neurons. All reconstructed data is available to share with the scientific community. The functional significance of association with a specific astrocytic domain on spontaneous or evoked neuronal activity is yet to be demonstrated, and this article provides the quantitative foundation upon which such future experiments can be designed.
Supplementary Material
Acknowledgments
The authors thank the entire Goshen lab for their support. AD is supported by the Azrieli fellowship and the ELSC graduate students scholarship. This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No 803589), the Israel Science Foundation (ISF grant No. 1815/18), and the Canada-Israel grants (CIHR-ISF, grant No. 2591/18). The authors thank Ami Citri and Adi Kol for the critical reading of the manuscript.
Funding information
H2020 European Research Council, Grant/Award Number: 803589; Israel Science Foundation, Grant/Award Numbers: 1815/18, 2591/18
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- Adamsky A, Kol A, Kreisel T, Doron A, Ozeri-Engelhard N, Melcer T, Refaeli R, Horn H, Regev L, Groysman M, London M, et al. Astrocytic activation generates De novo neuronal potentiation and memory enhancement. Cell. 2018;174(1):59–71.:e14. doi: 10.1016/j.cell.2018.05.002. [DOI] [PubMed] [Google Scholar]
- Atlas AMB. Allen Institute for Brain Science. 2007 Available from: http://mouse.brain-map.org/
- Bushong EA, Martone ME, Ellisman MH. Examination of the relationship between astrocyte morphology and laminar boundaries in the molecular layer of adult dentate gyrus. The Journal of Comparative Neurology. 2003;462(2):241–251. doi: 10.1002/cne.10728. [DOI] [PubMed] [Google Scholar]
- Bushong EA, Martone ME, Ellisman MH. Maturation of astrocyte morphology and the establishment of astrocyte domains during postnatal hippocampal development. International Journal of Developmental Neuroscience. 2004;22(2):73–86. doi: 10.1016/j.ijdevneu.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Bushong EA, Martone ME, Jones YZ, Ellisman MH. Protoplasmic astrocytes in CA1 stratum radiatum occupy separate anatomical domains. The Journal of Neuroscience. 2002;22(1):183–192. doi: 10.1523/JNEUROSCI.22-01-00183.2002. Retrieved from Retrievedfromhttps://www.ncbi.nlm.nih.gov/pubmed/11756501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai H, Diaz-Castro B, Shigetomi E, Monte E, Octeau JC, Yu X, Cohn W, Rajendran PS, Vondriska TM, Whitelegge JP, Coppola G, et al. Neural circuit-specialized astrocytes: Transcriptomic, proteomic, morphological, and functional evidence. Neuron. 2017;95(3):531–549.:e539. doi: 10.1016/j.neuron.2017.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavreul S, Abdeladim L, Hernandez-Garzon E, Niculescu D, Durand J, Ieng SH, Loulier K, Barry R, Bonvento G, Beaurepaire E, Livet J. Cortical astrocytes develop in a plastic manner at both clonal and cellular levels. Nature Communications. 2019;10(1):4884. doi: 10.1038/s41467-019-12791-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covelo A, Araque A. Neuronal activity determines distinct gliotransmitter release from a single astrocyte. eLife. 2018;7(32237) doi: 10.7554/eLife.32237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Yang Y, Ni Z, Dong Y, Cai G, Foncelle A, Ma S, Sang K, Tang S, Li Y, Shen Y, et al. Astroglial Kir4.1 in the lateral habenula drives neuronal bursts in depression. Nature. 2018;554(7692):323–327. doi: 10.1038/nature25752. [DOI] [PubMed] [Google Scholar]
- Deemyad T, Luthi J, Spruston N. Astrocytes integrate and drive action potential firing in inhibitory subnetworks. Nature Communications. 2018;9(1):4336. doi: 10.1038/s41467-018-06338-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilam R, Aharoni R, Arnon R, Malach R. Astrocyte morphology is confined by cortical functional boundaries in mammals ranging from mice to human. Elife. 2016;5:5. doi: 10.7554/eLife.15915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaire J, Lee HC, Ward R, Currlin S, Woolley AJ, Coleman JE, Williams JC, Otto KJ. PrismPlus: A mouse line expressing distinct fluorophores in four different brain cell types. Scientific Reports. 2018;8(1):7182. doi: 10.1038/s41598-018-25208-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradinaru V, Treweek J, Overton K, Deisseroth K. Hydrogeltissue chemistry: Principles and applications. Annual Review of Biophysics. 2018;47:355–376. doi: 10.1146/annurev-biophys-070317-032905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosche A, Grosche J, Tackenberg M, Scheller D, Gerstner G, Gumprecht A, Pannicke T, Hirrlinger PG, Wilhelmsson U, Hüttmann K, Härtig W, et al. Versatile and simple approach to determine astrocyte territories in mouse neocortex and hippocampus. PLoS One. 2013;8(7):e69143. doi: 10.1371/journal.pone.0069143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halassa MM, Fellin T, Takano H, Dong JH, Haydon PG. Synaptic islands defined by the territory of a single astrocyte. The Journal of Neuroscience. 2007;27(24):6473–6477. doi: 10.1523/JNEUROSCI.1419-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healey KL, Kibble S, Hodges S, Reissner KJ, Testen A, Wills TA, Acheson SK, Siemsen BM, McFaddin JA, Scofield MD, Swartzwelder HS. Enduring alterations in hippocampal astrocytesynaptic proximity following adolescent alcohol exposure: Reversal by gabapentin. Neural Regeneration Research. 2020;15(8):1496–1501. doi: 10.4103/1673-5374.274339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneberger C, Papouin T, Oliet SH, Rusakov DA. Long-term potentiation depends on release of D-serine from astrocytes. Nature. 2010;463(7278):232–236. doi: 10.1038/nature08673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippenmeyer S, Vrieseling E, Sigrist M, Portmann T, Laengle C, Ladle DR, Arber S. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biology. 2005;3(5):e159. doi: 10.1371/journal.pbio.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones ME, Paniccia JE, Lebonville CL, Reissner KJ, Lysle DT. Chemogenetic manipulation of dorsal hippocampal astrocytes protects against the development of stress-enhanced fear learning. Neuroscience. 2018;388:45–56. doi: 10.1016/j.neuroscience.2018.07.015. [DOI] [PubMed] [Google Scholar]
- Lanjakornsiripan D, Pior BJ, Kawaguchi D, Furutachi S, Tahara T, Katsuyama Y, Suzuki Y, Fukazawa Y, Gotoh Y. Layer-specific morphological and molecular differences in neocortical astrocytes and their dependence on neuronal layers. Nature Communications. 2018;9(1):1623. doi: 10.1038/s41467-018-03940-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450(7166):56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- Lopez-Hidalgo M, Hoover WB, Schummers J. Spatial organization of astrocytes in ferret visual cortex. The Journal of Comparative Neurology. 2016;524(17):3561–3576. doi: 10.1002/cne.24015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariotti L, Losi G, Lia A, Melone M, Chiavegato A, Gomez-Gonzalo M, Carmignoto G. Interneuron-specific signaling evokes distinctive somatostatin-mediated responses in adult cortical astrocytes. Nature Communications. 2018;9(1):82. doi: 10.1038/s41467-017-02642-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos M, Bosson A, Riebe I, Reynell C, Vallee J, Laplante I, Lacaille JC. Astrocytes detect and upregulate transmission at inhibitory synapses of somatostatin interneurons onto pyramidal cells. Nature Communications. 2018;9(1):4254. doi: 10.1038/s41467-018-06731-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SJ, Rothstein JD. Astroglia in thick tissue with super resolution and cellular reconstruction. PLoS One. 2016;11(8):e0160391. doi: 10.1371/journal.pone.0160391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberheim NA, Tian GF, Han X, Peng W, Takano T, Ransom B, Nedergaard M. Loss of astrocytic domain organization in the epileptic brain. The Journal of Neuroscience. 2008;28(13):3264–3276. doi: 10.1523/JNEUROSCI.4980-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Octeau JC, Chai H, Jiang R, Bonanno SL, Martin KC, Khakh BS. An optical neuron-astrocyte proximity assay at synaptic distance scales. Neuron. 2018;98(1):49–66.:e49. doi: 10.1016/j.neuron.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata K, Kosaka T. Structural and quantitative analysis of astrocytes in the mouse hippocampus. Neuroscience. 2002;113(1):221–233. doi: 10.1016/s0306-4522(02)00041-6. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/12123700. [DOI] [PubMed] [Google Scholar]
- Perea G, Gomez R, Mederos S, Covelo A, Ballesteros JJ, Schlosser L, Araque A. Activity-dependent switch of GABAergic inhibition into glutamatergic excitation in astrocyte-neuron networks. eLife. 2016;5(20362) doi: 10.7554/eLife.20362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea G, Yang A, Boyden ES, Sur M. Optogenetic astrocyte activation modulates response selectivity of visual cortex neurons in vivo. Nature Communications. 2014;5:3262. doi: 10.1038/ncomms4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robel S, Sontheimer H. Glia as drivers of abnormal neuronal activity. Nature Neuroscience. 2016;19(1):28–33. doi: 10.1038/nn.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santello M, Toni N, Volterra A. Astrocyte function from information processing to cognition and cognitive impairment. Nature Neuroscience. 2019;22(2):154–166. doi: 10.1038/s41593-018-0325-8. [DOI] [PubMed] [Google Scholar]
- Scofield MD, Li H, Siemsen BM, Healey KL, Tran PK, Woronoff N, Boger HA, Kalivas PW, Reissner KJ. Cocaine self-administration and extinction leads to reduced glial fibrillary acidic protein expression and morphometric features of astrocytes in the nucleus accumbens core. Biological Psychiatry. 2016;80(3):207–215. doi: 10.1016/j.biopsych.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevelkin AV, Terrillion CE, Hasegawa Y, Mychko OA, Jouroukhin Y, Sawa A, Kamiya A, Pletnikov MV. Astrocyte DISC1 contributes to cognitive function in a brain region-dependent manner. Human Molecular Genetics. 2020;29(17):2936–2950. doi: 10.1093/hmg/ddaa180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemsen BM, Reichel CM, Leong KC, Garcia-Keller C, Gipson CD, Spencer S, McFaddin JA, Hooker KN, Kalivas PW, Scofield MD. Effects of methamphetamine self-administration and extinction on astrocyte structure and function in the nucleus accumbens core. Neuroscience. 2019;406:528–541. doi: 10.1016/j.neuroscience.2019.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltesz I, Losonczy A. CA1 pyramidal cell diversity enabling parallel information processing in the hippocampus. Nature Neuroscience. 2018;21(4):484–493. doi: 10.1038/s41593-018-0118-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z, Liu Y, Xi W, Lou HF, Zhu L, Guo Z, Mei L, Duan S. Glia-derived ATP inversely regulates excitability of pyramidal and CCK-positive neurons. Nature Communications. 2017;8:13772. doi: 10.1038/ncomms13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsani D, Fu Y, Lu J, Lin Y, Miyoshi G, et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71(6):995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testen A, Ali M, Sexton HG, Hodges S, Dubester K, Reissner KJ, Swartzwelder HS, Risher ML. Region-specific differences in morphometric features and synaptic colocalization of astrocytes during development. Neuroscience. 2019;400:98–109. doi: 10.1016/j.neuroscience.2018.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treweek JB, Gradinaru V. Extracting structural and functional features of widely distributed biological circuits with single cell resolution via tissue clearing and delivery vectors. Current Opinion in Biotechnology. 2016;40:193–207. doi: 10.1016/j.copbio.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmsson U, Bushong EA, Price DL, Smarr BL, Phung V, Terada M, Ellisman MH, Pekny M. Redefining the concept of reactive astrocytes as cells that remain within their unique domains upon reaction to injury. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(46):17513–17518. doi: 10.1073/pnas.0602841103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Wang W, Zhou M. Spatial organization of NG2 glial cells and astrocytes in rat hippocampal CA1 region. Hippocampus. 2014;24(4):383–395. doi: 10.1002/hipo.22232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Allen WE, Thompson KR, Tian Q, Hsueh B, Ramakrishnan C, Wang AC, Jennings JH, Adhikari A, Halpern CH, Witten IB, et al. Wiring and molecular features of prefrontal ensembles representing distinct experiences. Cell. 2016;165(7):1776–1788. doi: 10.1016/j.cell.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Nagai J, Khakh BS. Improved tools to study astrocytes. Nature Reviews. Neuroscience. 2020;21:121–138. doi: 10.1038/s41583-020-0264-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.