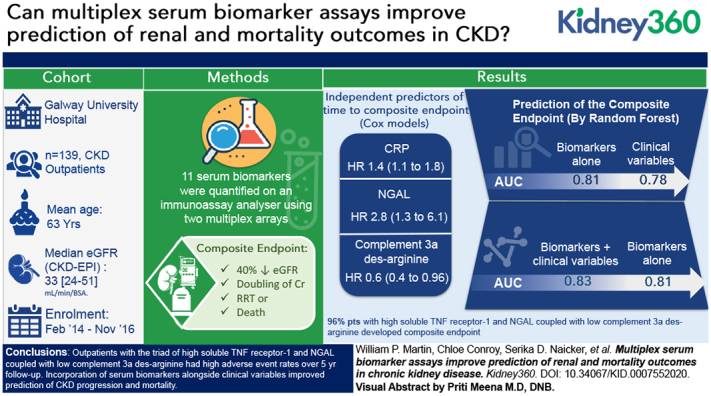
Visual Abstract
Keywords: chronic kidney disease, biomarkers, C3a-desArg, end stage renal disease, machine learning, mortality, multiplex assays, neutrophil gelatinase-associated lipocalin, renal function decline, soluble tumor necrosis factor receptor
Key Points
Incorporation of 11 serum biomarkers alongside clinical variables improved prediction of adverse CKD outcomes over 5-year follow-up.
Patients with the triad of high sTNFR1 and NGAL coupled with low C3a-desArg had particularly high adverse event rates during follow-up.
Biomarkers were quantified on a single, clinical-grade analyzer, with potential for improved translatability to the CKD outpatient setting.
Abstract
Background
We investigated the predictive value of 11 serum biomarkers for renal and mortality end points in people with CKD.
Methods
Adults with CKD (n=139) were enrolled from outpatient clinics between February 2014 and November 2016. Biomarker quantification was performed using two multiplex arrays on a clinical-grade analyzer. Relationships between biomarkers and renal and mortality end points were investigated by random forests and Cox proportional hazards regression.
Results
The cohort was 56% male. The mean age was 63 years and median (IQR) CKD-EPI eGFR was 33 (24–51) ml/min per BSA. A total of 56 (40%) people developed a composite end point defined as ≥40% decline in eGFR, doubling of serum creatinine, RRT, or death over median (IQR) follow-up of 5.4 (4.7–5.7) years. Prediction of the composite end point was better with random forests trained on serum biomarkers compared with clinical variables (area under the curve of 0.81 versus 0.78). The predictive performance of biomarkers was further enhanced when considered alongside clinical variables (area under the curve of 0.83 versus 0.81 for biomarkers alone). Patients (n=27, 19%) with high soluble TNF receptor-1 (≥3 ng/ml) and neutrophil gelatinase-associated lipocalin (≥156 ng/ml), coupled with low complement 3a des-arginine (<2368 ng/ml), almost universally (96%) developed the composite renal and mortality end point. C-reactive protein (adjusted hazard ratio, 1.4; 95% CI, 1.1 to 1.8), neutrophil gelatinase-associated lipocalin (adjusted hazard ratio, 2.8; 95% CI, 1.3 to 6.1) and complement 3a des-arginine (adjusted hazard ratio, 0.6; 95% CI, 0.4 to 0.96) independently predicted time to the composite end point.
Conclusions
Outpatients with the triad of high soluble TNF receptor-1 and neutrophil gelatinase-associated lipocalin coupled with low complement 3a des-arginine had high adverse event rates over 5-year follow-up. Incorporation of serum biomarkers alongside clinical variables improved prediction of CKD progression and mortality. Our findings require confirmation in larger, more diverse patient cohorts.
Introduction
CKD is a growing public health problem, with its prevalence increasing by 29% si nce 1990 to affect 9% of the global population in 2017 (1). Although morbidity and mortality rates from other noncommunicable diseases have declined over the past three decades, no such favorable trends exist for CKD (2). Death due to cardiovascular disease is over-represented among people with CKD, and mortality rates increase as eGFR declines. In a meta-analysis of 21 general population cohorts incorporating >1.2 million participants, eGFR independently predicted mortality risk in an almost linear fashion (3).
Communicating risk of adverse outcomes to patients with CKD is challenging, particularly at earlier, typically asymptomatic, disease stages. Although eGFR and urine albumin-creatinine ratio (uACR) are strongly predictive of adverse outcomes in epidemiologic studies, intraindividual variability weakens their prognostic value in clinical practice (4,5). Measuring multiple circulating biomarkers simultaneously has the potential to uncover subgroups of patients with CKD who have differing risks of progressive renal functional decline and mortality. However, prognostication of adverse CKD outcomes with multiple biomarkers is challenged by the strong intercorrelation between biomarkers from diverse pathways, which may result in marginal improvements in predictive performance when additional biomarkers are studied (6–8). Additionally, many biomarker studies to date have enrolled specific subgroups of patients with CKD, for example those with diabetic kidney disease. Hence, predictive performance of circulating biomarkers across the spectrum of CKD severity and etiology in real-world outpatient nephrology practice is underexplored.
We aimed to evaluate the performance of two multianalyte serum biomarker arrays in patients with CKD. Specifically, we aimed to ascertain the relationships of the 11 biomarkers to each other and to determine their individual and combined predictive value for renal and mortality outcomes. We hypothesized that clusters of patients with differing risks of adverse CKD outcomes could be identified on the basis of serum biomarker profiles. Furthermore, we hypothesized that incorporation of multiple biomarkers into multivariate models would improve prediction of renal and mortality end points over 5-year follow-up, compared with clinical variables alone, in people with a broad range of CKD etiologies and severity attending a tertiary referral nephrology center.
Materials and Methods
Study Cohort
Adults with CKD stages 1–5 were enrolled from nephrology outpatient clinics at Galway University Hospitals between February 2014 and November 2016. As previously described (9), inclusion criteria were as follows: age ≥18 years, diagnosis of CKD, absence of current infection, immunosuppression, cancer, acute cardiovascular event or hematologic condition other than anemia, hemoglobin ≥10 g/dl, not on RRT, and no prior kidney transplant.
Clinical and laboratory data were recorded from enrollment to the end of follow-up on July 15, 2020 in a secure, password-protected, web-based clinical database (Distiller; SlidePath, Dublin, Ireland). Longitudinal measurements of serum creatinine were extracted for each participant using the eMED Renal system (Mediqal H.I., Aston, United Kingdom). A creatininase assay traceable to isotope-dilution mass spectrometry was used to measure creatinine (10). Chronic Kidney Disease Epidemiology Collaboration (CKD-EPI) eGFR was calculated from serum creatinine using standard formulae and expressed as milliliters per minute per body surface area (BSA). Second and subsequent creatinine values on a given day and creatinine values subsequent to RRT initiation were excluded. Individuals with less than three eGFR values were excluded from the dataset. Interim renal outcomes (≥40% decline in CKD-EPI eGFR and doubling of serum creatinine) were defined at their first occurrence after study enrollment compared with baseline study values. RRT was defined as first requirement for dialysis or kidney transplantation after study enrollment. All-cause mortality after study enrollment was recorded. Time to event of renal and mortality end points was recorded for each patient. Duration of study follow-up was calculated as the time from enrollment to date of last study follow-up or death. Duration of renal functional follow-up was calculated as the time from enrollment to date of last eGFR determination. Annual eGFR slopes were calculated by linear regression of eGFR over time for individuals with three or more eGFR determinations over ≥1 year. Median log-transformed uACR was estimated from urinary protein-creatinine ratio (uPCR) using the validated equation of Weaver et al. (11). Four-variable Kidney Failure Risk Equation (KFRE) scores for 2- and 5-year risks of progression to kidney failure were calculated according to regional (non-North American) formulae (12).
The study was approved by the Galway University Hospitals Clinical Research Ethics Committee (reference C.A. 885) and conducted in accordance with the 1964 Declaration of Helsinki. Participants provided written, informed consent.
Blood Sample Collection and Serum Biomarker Determinations
Serum was isolated from peripheral venous blood samples provided at enrollment. Eleven serum biomarkers were quantified using two CKD multiplex arrays on an Evidence Investigator immunoassay analyzer (Randox Teoranta, Donegal, Ireland). Because physiologic serum concentrations vary significantly for the 11 included biomarkers (from pg/mL to μg/mL), low- and high-abundance biomarkers were quantified on separate multiplex arrays to improve assay sensitivity and performance. A seven-analyte array measured the following low-abundance biomarkers: epidermal growth factor (EGF), IL-8, soluble TNF receptor-1 (sTNFR1) and sTNFR2, fatty acid-binding protein-1 (FABP1), D-dimer, and macrophage inflammatory protein-1-α (MIP-1-α). A four-analyte array measured the following high-abundance biomarkers: C-reactive protein, cystatin C, complement 3a with cleaved C-terminal arginine (C3a-desArg), and neutrophil gelatinase-associated lipocalin (NGAL). Additional details on serum isolation, the immunoassay procedure, the rationale behind biomarker selection, and assay performance and validation are provided in Supplemental Appendices 1–3. The interassay coefficient of variation for individual biomarkers across multiplex array plate runs that were used to quantify biomarkers for the study cohort is presented in Supplemental Table 1. The interassay coefficient of variation was 8% for the seven-analyte array, and 10% for the four-analyte array, resulting in an overall interassay coefficient of variation of 9%.
Statistical Analyses
Descriptive and Inferential Statistics, Logistic Regression, and Clustering
Data analysis was performed using the R statistical programming language (R version 4.0.0) in RStudio. A composite renal and mortality end point of ≥40% decline in eGFR, doubling of serum creatinine, RRT, or death was defined and used as the primary outcome for analyses. A renal-specific composite end point of ≥40% decline in eGFR, doubling of serum creatinine, or RRT was used in sensitivity analyses. Biomarkers were transformed using the natural logarithm for analysis. Cohort characteristics were summarized by descriptive statistics. Independent sample t tests, Wilcoxon rank-sum tests, and chi-squared tests assessed for differences in clinical variables and biomarkers among those who did and did not develop the composite renal and mortality end point. P<0.05 was considered statistically significant. Univariate relationships between serum biomarkers and the composite renal and mortality end point were investigated using logistic regression. Odds ratios from logistic regression models are expressed per one unit change in natural logarithm biomarker concentrations. We performed unsupervised clustering of patients on the basis of biomarker concentrations using principal components analysis (13).
Decision Tree and Random Forest Classification Models
A supervised machine-learning approach with binary classification random forests was used to explore the value of all biomarkers considered together in predicting the composite renal and mortality end point. Random forests have several advantages over logistic regression, including seamless handling of regressor collinearity and automatic selection and fitting of nonlinear relationships and statistical interactions. A binary classification decision tree, evaluating classification of the composite renal and mortality end point by serum biomarkers, was generated using default parameters of the R function “rpart” to illustrate the complementary information provided by multiple biomarkers (14,15). Biomarkers were inputted to the decision tree and random forest models as continuous variables. Biomarker thresholds were selected by recursive binary splitting to maximize node purity in each tree (the number of individuals from a single class, who either did or did not develop the composite renal and mortality end point). Binary classification random forests (5000 trees per model), with the composite renal and mortality end point as the response variable, were fit (16). Three model types were created: clinical variables alone (age, sex, hypertension, diabetes, and eGFR), biomarkers alone, and clinical variables plus biomarkers. A leave-one-out cross-validation approach was implemented, which consisted of excluding one individual, in turn, from training the random forest model. Subsequently, the trained random forest model predicted the class of the individual excluded during model training. This process was repeated iteratively for each individual in the dataset, such that a predicted class was assigned to each study participant by each of the three random forest model types. The leave-one-out cross-validation procedure was only performed for random forest models and not for other model types.
Area under the curve (AUC) values were calculated for each of the three model types using the predicted probability of having developed the composite renal and mortality end point and the individual’s actual recorded composite renal and mortality end point status as inputs to the function “roc” in the R package pROC (17). Receiver operating characteristic curves were plotted using the function “ggroc.” Model performance metrics (sensitivity, specificity, positive predictive value, and negative predictive value) were calculated across three probability thresholds (10%, 30%, and 50%) for labeling patients as having developed the composite renal and mortality end point. For example, an individual with a predicted probability of the composite renal and mortality end point by a random forest model of 45% would be labeled as having developed the composite renal and mortality end point by the first two thresholds evaluated, but not the latter. An estimate of variable importance to the random forest models (mean decrease in accuracy) was calculated from mean values of each model iteration during the leave-one-out cross-validation procedure.
Cox Proportional Hazards Regression Models
Multivariable Cox proportional hazards regression models were created to investigate relationships between biomarkers and time to renal and mortality and renal-specific composite end points. The primary Cox model analysis investigated the value of biomarkers when considered in addition to clinical variables in predicting time to the composite renal and mortality end point. For this analysis, two models were constructed: a clinical model adjusting for age, sex, hypertension, diabetes, and eGFR; and a clinical plus biomarker model additionally incorporating all log-transformed biomarker values. Backward elimination of nonsignificant biomarker effects from the clinical plus biomarker model was subsequently performed using stepAIC (18). Biomarkers for which the P-values of their hazard ratios were >0.05 were manually excluded from the Akaike information criterion (AIC)–selected model to create a final parsimonious clinical plus biomarker model. Clinical variables were manually retained in the final model. Hazard ratios from Cox models are expressed per one unit change in natural logarithm biomarker concentrations. Comparisons of Cox model adequacy (clinical model versus clinical plus biomarker model) were assessed using likelihood ratio chi-squared tests.
Cox models were constructed using the R package survival (19). The R package survminer was used to create forest plots and to plot fully adjusted survival curves from the multivariate clinical plus biomarker Cox model for the composite renal and mortality end point according to biomarker tertiles, which were calculated using the function “ntile” from the R package dplyr (20,21). We tested each Cox model for proportionality assumptions using Schoenfeld residuals. AIC values for fitted Cox models were obtained using extractAIC (22).
Sensitivity Analyses
A series of sensitivity analyses was performed to further evaluate the predictive value of serum biomarkers incorporated in the random forest and Cox models outlined above. Random forest models trained on clinical variables, alone, or in combination with biomarkers, were additionally adjusted for baseline uACR. Baseline uACR was included in random forest models in two separate analyses: firstly, random forest models were performed in the subgroup with baseline uACR data available and, secondly, after imputation of missing baseline uACR data using the function “rfImpute” in the R package randomForest (16). Imputation of missing uACR data was only performed for sensitivity analyses involving random forests.
With respect to the final parsimonious clinical plus biomarker Cox model of the composite renal and mortality end point outlined above, the predictive value of biomarkers in this model was separately evaluated in Cox models in the subgroup with CKD stages 3–5/eGFR <60 ml/min per BSA and in the subgroup with baseline uACR data available. The value of these biomarkers for predicting time to a renal-specific composite end point was also evaluated using Cox models, both in the full study cohort and in the subgroup with baseline uACR data available.
Results
Baseline Characteristics and Serum Biomarker Concentrations
Baseline characteristics and serum biomarker concentrations of the study cohort (n=139), stratified by development (n=56) or not (n=83) of the composite renal and mortality end point, are presented in Table 1. The study population had a mean age of 63 years, 56% were male, and median (interquartile range [IQR]) eGFR was 33 (24–51) ml/min per BSA. Of those sampled, study participants had moderate proteinuria with a median (IQR) uACR of 127 (22–479) mg/g and uPCR of 327 (124–814) mg/g. After calculating uACR from uPCR (11), baseline uACR data were available for 113 (81%) individuals, for whom the median (IQR) uACR was 144 (22–532) mg/g. Characteristics of individuals with available and missing baseline uACR data (after conversion of uPCR to uACR where available) are presented in Supplemental Table 2. Those with missing uACR data trended toward being older (68±14 versus 62±17 years, P=0.06) but, otherwise, no significant differences between those with and without baseline uACR data were observed. Over 25% and 80% of the study cohort had diabetes mellitus and hypertension, respectively, and glomerulonephritis (22%) and diabetic kidney disease (17%) were the two most frequently documented CKD etiologies. Median (IQR) 2- and 5-year risks of progression to kidney failure were 1.4% (0.3%–6.5%) and 5.4% (1.2%–23.1%), respectively.
Table 1.
Baseline characteristics and serum biomarker concentrations of the study cohort stratified by development of a composite renal and mortality end point (n=139)
| Characteristic | Data Available, n (%) | Total Cohort, n=139 | Did Not Develop Composite End Point (n=83)a | Developed Composite End Point (n=56)a |
| Clinical parameters | ||||
| Age (yr), (mean±SD) | 139 (100) | 63±17 | 60±17 | 67±15 |
| Male, n (%) | 139 (100) | 78 (56) | 38 (46) | 40 (71) |
| Diabetes mellitus, n (%) | 139 (100) | 35 (25) | 11 (13) | 24 (43) |
| Hypertension, n (%) | 139 (100) | 115 (83) | 64 (77) | 51 (91) |
| Coronary artery disease, n (%) | 139 (100) | 18 (13) | 8 (10) | 10 (18) |
| CKD stage, n (%) | 139 (100) | |||
| Grade 1 | 5 (4) | 5 (6) | 0 (0) | |
| Grade 2 | 17 (12) | 16 (19) | 1 (2) | |
| Grade 3a | 22 (16) | 19 (23) | 3 (5) | |
| Grade 3b | 42 (30) | 25 (30) | 17 (30) | |
| Grade 4 | 46 (33) | 16 (19) | 30 (54) | |
| Grade 5 | 7 (5) | 2 (2) | 5 (9) | |
| CKD etiology, n (%) | 139 (100) | |||
| Diabetes | 23 (17) | 8 (10) | 15 (27) | |
| Hypertension | 12 (9) | 9 (11) | 3 (5) | |
| GN | 30 (22) | 23 (28) | 7 (13) | |
| Congenital | 8 (6) | 7 (8) | 1 (2) | |
| Polycystic kidney disease | 6 (4) | 3 (4) | 3 (5) | |
| Obstructive | 6 (4) | 2 (2) | 4 (7) | |
| Interstitial | 7 (5) | 5 (6) | 2 (4) | |
| Other/unknown | 47 (34) | 26 (31) | 21 (38) | |
| Laboratory data | ||||
| Serum creatinine (mg/dl), mean±SD | 139 (100) | 2.0±1.0 | 1.6±0.6 | 2.6±1.1 |
| CKD-EPI eGFR (ml/min per BSA), median (IQR) | 139 (100) | 33 (24–51) | 43 (31–60) | 26 (18–34) |
| uACR (mg/g), median (IQR) | 58 (42) | 127 (22–479) | 33 (18–144) | 504 (92–1249) |
| uPCR (mg/g), median (IQR) | 76 (55) | 327 (124–814) | 301 (124–637) | 345 (181–1381) |
| Merged uACR (mg/g), median (IQR)b | 113 (81) | 144 (22–532) | 104 (19–269) | 283 (83–993) |
| Hemoglobin (g/dl), mean±SD | 138 (99) | 13.0±1.7 | 13.4±1.9 | 12.3±1.2 |
| KFRE estimates (%), median (IQR) | 113 (81) | |||
| 2 year | 1.4 (0.3–6.5) | 0.6 (0.1–2.4) | 6.4 (1.6–18.7) | |
| 5 year | 5.4 (1.2–23.1) | 2.1 (0.3–9.0) | 22.5 (6.1–55.1) | |
| Multiplex biomarker values, median (IQR) | ||||
| C-reactive protein (ng/ml) | 139 (100) | 3035 (1607–8244) | 2610 (1416–5296) | 4612 (2303–15181) |
| Cystatin C (ng/ml) | 139 (100) | 3715 (2575–5000) | 3241 (2115–4384) | 4984 (3662–5739) |
| C3a-desArg (ng/ml) | 139 (100) | 1762 (1010–2877) | 2240 (1099–3392) | 1467 (947–2178) |
| D-dimer (ng/ml) | 138 (99) | 81 (41–153) | 73 (34–143) | 103 (49–172) |
| EGF (pg/ml) | 138 (99) | 91 (54–126) | 109 (58–134) | 72 (47–104) |
| FABP1 (ng/ml) | 139 (100) | 1.6 (0.7–3.0) | 1.3 (0.6–2.7) | 1.9 (1.2–3.8) |
| IL-8 (pg/ml) | 139 (100) | 3.7 (2.6–8.5) | 3.9 (2.5–8.6) | 3.6 (2.7–7.3) |
| MIP-1-α (pg/ml) | 130 (94) | 4.8 (2.6–10.6) | 3.9 (2.3–6.6) | 7.5 (3.7–15.1) |
| NGAL (ng/ml) | 137 (99) | 167 (116–253) | 137 (104–194) | 213 (173–327) |
| sTNFR1 (ng/ml) | 136 (98) | 3.0 (2.1–4.6) | 2.4 (1.9–3.2) | 4.6 (3.5–6.8) |
| sTNFR2 (ng/ml) | 138 (99) | 1.4 (0.8–2.2) | 1.1 (0.6–1.6) | 2.1 (1.3–2.5) |
Values are given as n (%) for categorical variables, or mean±SD for normally distributed continuous variables, unless otherwise indicated. Median (IQR) values are presented for continuous variables that are not normally distributed. CKD-EPI, Chronic Kidney Disease Epidemiology Collaboration; BSA, body surface area; IQR, interquartile range; uACR, urine albumin-creatinine ratio; uPCR, urine protein-creatinine ratio; KFRE, Kidney Failure Risk Equation (four-variable; non-North America); C3a-desArg, complement 3a with cleaved C-terminal arginine; FABP1, fatty acid-binding protein-1; MIP-1-α, macrophage inflammatory protein-1-α; NGAL, neutrophil gelatinase-associated lipocalin; sTNFR1, soluble TNF receptor-1; sTNFR2, soluble TNF receptor-2.
Composite end point: ≥40% decrease in CKD-EPI eGFR, doubling of serum creatinine, RRT, or mortality.
Merged uACR represents a combination of measured uACR and calculated uACR from uPCR using the validated equation of Weaver et al. (11). uACR values on the natural log scale were exponentiated such that presented values are in absolute units in milligrams per gram.
Compared with individuals who did not develop the composite renal and mortality end point, those who did were older (67±15 versus 60±17 years, P=0.01), more likely to be male (71% versus 46%, P=0.005), had a higher prevalence of diabetes mellitus (43% versus 13%, P<0.001), lower eGFR (26 [IQR, 18–34] versus 43 [31–60] ml/min per BSA, P<0.001), higher uACR (283 [IQR, 83–993] versus 104 [19–269] mg/g, P<0.001), and higher 5-year KFRE scores (22.5% [IQR, 6.1%–55.1%] versus 2.1% [0.3%–9.0%], P<0.001). Concentrations of several serum biomarkers were higher in those who developed the composite renal and mortality end point, including sTNFR1, sTNFR2, NGAL, cystatin C, and—to a lesser extent—C-reactive protein, FABP1, and MIP-1-α. Conversely, concentrations of EGF and C3a-desArg were lower in those who developed the composite renal and mortality end point. Of note, the median (IQR) sTNFR2 concentration (1.4 [0.8–2.2] ng/ml) was lower than that of sTNFR1 (3.0 [2.1–4.6] ng/ml) in the study cohort, which is the converse of what has been observed in several studies (23–25). However, sTNFR1 and sTNFR2 concentrations remained strongly correlated with each other (Pearson r correlation 0.73, P<0.001; Supplemental Figure 1).
Incidence of Renal and Mortality End Points
The median (IQR) duration of study follow-up was 5.4 (4.7–5.7) years. Median (IQR) duration of renal functional follow-up (study enrollment to date of final eGFR determination) was 4.7 (3.5–5.2) years, with participants having a median (IQR) of 22 (12–34) eGFR values (Table 2). The median (IQR) rate of decline in eGFR was −0.9 (−2.3 to 0.5) ml/min per BSA per year. Renal functional decline was greater in those who developed the composite renal and mortality end point compared with those who did not (−2.2 [IQR, −3.9 to −1.4] versus 0 [−1 to 1.6] ml/min per BSA per year, P<0.001). Relevant study end points, including ≥40% decline in CKD-EPI eGFR, doubling of serum creatinine, RRT, and death, occurred in 38 (68%), 14 (25%), 21 (38%), and 15 (27%) patients, respectively. A total of 56 (40%) individuals developed the composite renal and mortality end point, whereas 47 (34%) developed the renal-specific composite end point.
Table 2.
Duration of follow-up and incidence of renal and mortality end points during the study period (n=139)
| Characteristic | Data Available, n (%) | Total Cohort (n=139) | Did Not Develop Composite End Point (n=83)a | Developed Composite End Point (n=56)a |
| Number of eGFR measurements, median (IQR) | 139 (100) | 22 (12–34) | 19 (11–29) | 27 (15–42) |
| Duration of renal functional follow-up (yr), median (IQR)b | 139 (100) | 4.7 (3.5–5.2) | 4.9 (4.1–5.2) | 4.1 (1.8–5.2) |
| Duration of study follow-up (yr), median (IQR)c | 139 (100) | 5.4 (4.7–5.7) | 5.4 (4.8–5.7) | 5.4 (4.4–5.7) |
| Slope of CKD-EPI eGFR (ml/min per BSA per yr), median (IQR)d | 129 (93) | −0.9 (−2.3 to 0.5) | 0 (−1 to 1.6) | −2.2 (−3.9 to −1.4) |
| ≥40% decrease in CKD-EPI eGFR, n (%) | 139 (100) | 38 (27) | 0 (0) | 38 (68) |
| Doubling of serum creatinine, n (%) | 139 (100) | 14 (10) | 0 (0) | 14 (25) |
| Required RRT, n (%) | 139 (100) | 21 (15) | 0 (0) | 21 (38) |
| Death from any cause, n (%) | 139 (100) | 15 (11) | 0 (0) | 15 (27) |
IQR, interquartile range; CKD-EPI, Chronic Kidney Disease Epidemiology Collaboration; BSA, body surface area.
Composite end point: ≥40% decrease in CKD-EPI eGFR, doubling of serum creatinine, RRT, or mortality.
Duration between date of study enrollment and date of final eGFR determination. eGFR values subsequent to RRT initiation were excluded.
Duration between date of study enrollment and date of last study follow-up or date of death.
Slope of CKD-EPI eGFR was calculated only for individuals with three or more eGFR values over at least 1 year.
Univariate Relationships between Serum Biomarkers and the Composite End Point
Figure 1 presents biomarkers with significant associations with the composite renal and mortality end point by univariate logistic regression. EGF (odds ratio [OR], 0.5; 95% CI, 0.3 to 0.9) and C3a-desArg (OR, 0.6; 95% CI, 0.4 to 0.96) were inversely associated with the composite renal and mortality outcome, whereas MIP-1-α (OR, 1.8; 95% CI, 1.2 to 2.8) and C-reactive protein (OR, 1.6; 95% CI, 1.2 to 2.2) were positively associated with it. Furthermore, sTNFR1 (OR, 25.9; 95% CI, 8.6 to 94.5), sTNFR2 (OR, 13.7; 95% CI, 4.5 to 47.4), NGAL (OR, 4.8; 95% CI, 2.4 to 10.6), and cystatin C (OR, 10.4; 95% CI, 4.0 to 31.5) were strongly associated with the composite renal and mortality end point.
Figure 1.
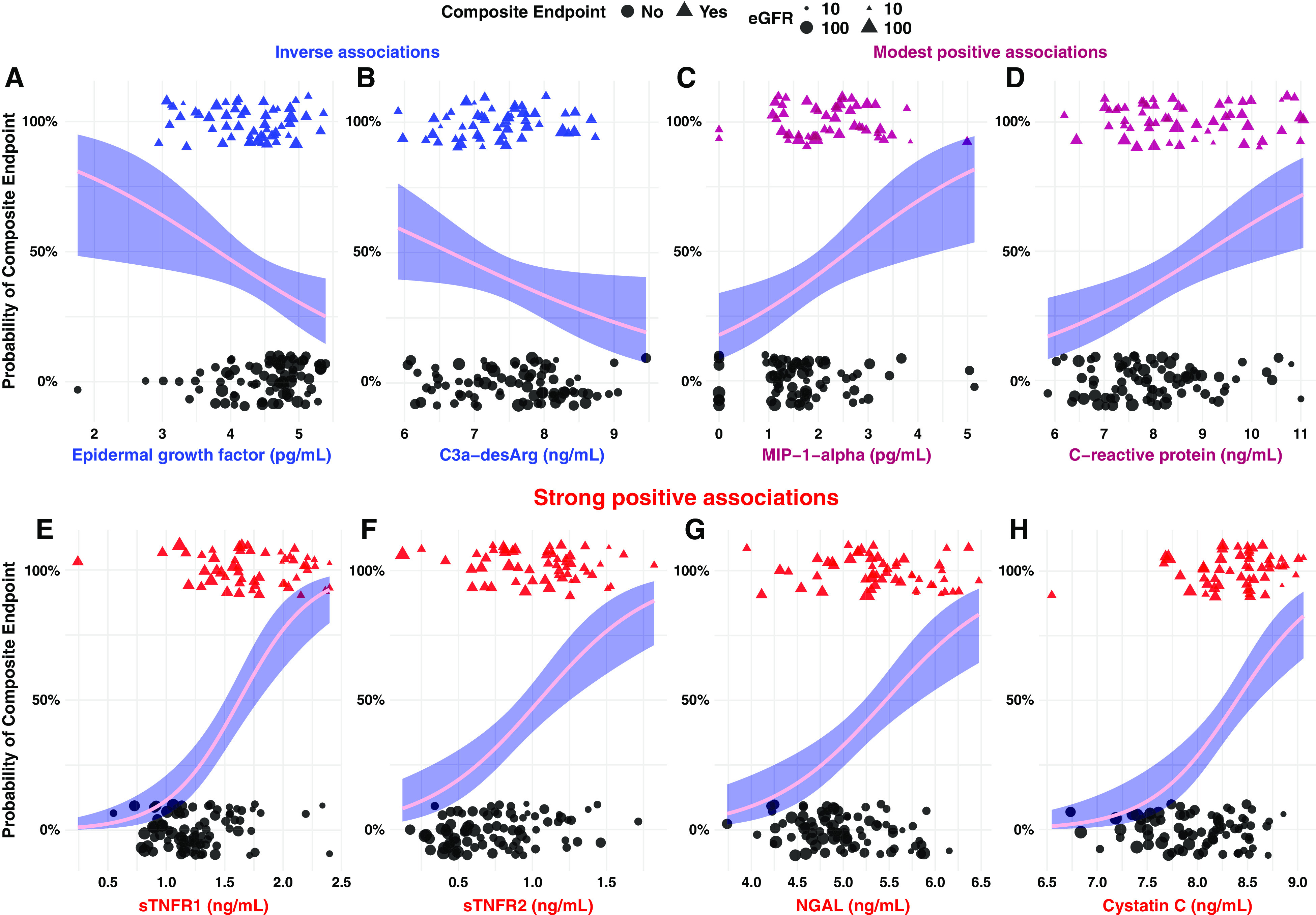
Univariate logistic regression highlights the strength and directionality of relationships between individual serum biomarkers and a composite renal and mortality end point. (A and B) Variables that exhibited a significant inverse relationship with the composite renal and mortality end point, including (A) EGF and (B) complement 3a with cleaved C-terminal arginine (C3a-desArg). (C and D) Biomarkers with a modest positive relationship with the composite renal and mortality end point, including (C) macrophage inflammatory protein-1-α (MIP-1-α) and (D) C-reactive protein. (E–H) Biomarkers that displayed a strong positive relationship with the composite renal and mortality end point, including (E) soluble TNF receptor-1 (sTNFR1), (F) sTNFR2, (G) neutrophil gelatinase-associated lipocalin (NGAL), and (H) cystatin C. Individuals who developed the composite renal and mortality end point are identified as 100% on the y axis and as triangular points. Individuals who did not develop the composite renal and mortality end point are identified as 0% on the y axis and as circular points. Biomarkers that displayed an inverse association with the composite renal and mortality end point are colored in blue, those with a strong positive association are colored in red, and those with modest positive associations are presented in a shade intermediate between both colors. Log transformation of serum biomarkers was performed before modeling. Odds ratios from logistic regression models are expressed per one unit change in natural logarithm biomarker concentrations. The 95% confidence interval is represented by navy shading surrounding the pink regression curve. The size of individual data points is scaled by baseline eGFR to illustrate relationships between serum biomarkers and kidney function at enrollment. Larger points represent higher eGFR; smaller points represent lower eGFR. Composite renal and mortality end point: ≥40% decrease in Chronic Kidney Disease Epidemiology Collaboration eGFR, doubling of serum creatinine, RRT, or mortality.
Clustering of Patients on the Basis of Serum Biomarkers
Unsupervised clustering of patients by principal components analysis identified shifts in biomarkers by CKD stage and by the composite renal and mortality end point (Figure 2, A and B), with the loading plot (Figure 2C) illustrating which biomarkers were important in this regard. Expression of several biomarkers clustered together in a predictable fashion, based on a priori knowledge. sTNFR1 and sTNFR2 clustered together, as did two biomarkers that were inversely associated with the composite renal and mortality end point: EGF and C3a-desArg. Patients separated on the basis of biomarker expression along principal components 1 and 2 by both CKD stage and by development of the composite renal and mortality end point. Biomarkers clustered in the upper left corner (EGF and C3a-desArg) were more strongly expressed in earlier stages of CKD and in those who did not develop the composite renal and mortality end point (Figure 2C, Table 1). Conversely, biomarkers in the far right (NGAL, cystatin C, sTNFR1, and sTNFR2) were more strongly expressed in advanced CKD and in those who developed the composite renal and mortality end point (Figure 2C, Table 1).
Figure 2.
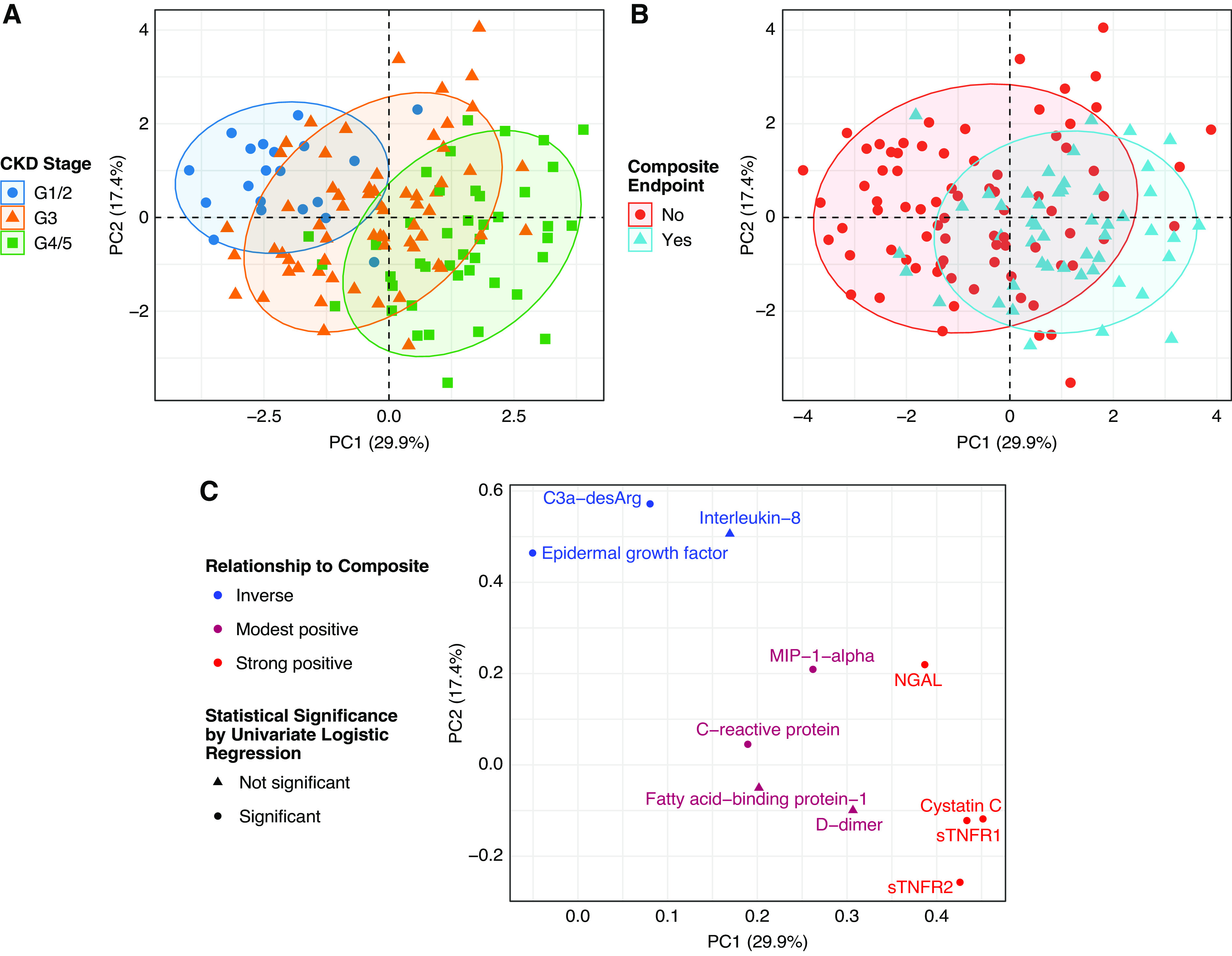
Principal components analysis illustrates relationships between biomarker expression, CKD stage, and a composite renal and mortality end point. (A) Unsupervised clustering of patients by principal components analysis identifies global shifts in biomarkers across categorical CKD stages, grouped as grades 1 (G1) and 2 (eGFR of ≥60 ml/min per body surface area [BSA]), grade 3 (eGFR of 30–59 ml/min per BSA), and grades 4 and 5 (eGFR of <30 ml/min per BSA). (B) Unsupervised clustering of patients by principal components analysis identifies global shifts in biomarker expression profiles between those who did and did not develop a composite renal and mortality end point during follow-up. (C) A loadings plot from principal components analysis reveals the influential biomarkers that drive the shifts in biomarker expression across CKD stages and when stratified by the composite renal and mortality end point. Individuals with advanced CKD who developed the composite renal and mortality end point had higher expression of biomarkers in the right of the plot, including sTNFR1, sTNFR2, NGAL, and cystatin C. Individuals with earlier stages of CKD who did not develop the composite renal and mortality end point had higher expression of protective factors in the upper left corner of the plot, including C3a-desArg and EGF. The color of the points illustrates the directionality of the relationships between biomarkers and the composite renal and mortality end point (blue, inverse; red, strong positive; modest positive relationships are colored in between both). The shape of the points represents statistical significance of the relationship between biomarkers and the composite renal and mortality end point by univariate logistic regression (circle, statistically significant; triangle, not statistically significant). In (A), (B), and (C), the x and y axes represent principal components 1 and 2, respectively. Composite renal and mortality end point: ≥40% decrease in Chronic Kidney Disease Epidemiology Collaboration eGFR, doubling of serum creatinine, RRT, or mortality. PC, principal component.
Prediction of a Renal and Mortality Composite End Point by Serum Biomarkers Using Supervised Machine Learning (Random Forest Classification Models)
C3a-desArg reclassified risk of the composite renal and mortality end point among those with high sTNFR1 and NGAL (Figure 3A). Those with high sTNFR1 (≥3 ng/ml), high NGAL (≥156 ng/ml), but also high C3a-desArg (≥2368 ng/ml) had a 44% risk of the composite renal and mortality end point. Conversely, individuals with high sTNFR1 (≥3 ng/ml), high NGAL (≥156 ng/ml), and low C3a-desArg (<2368 ng/ml), which accounted for 19% of the cohort, almost universally (96%) developed the composite renal and mortality end point.
Figure 3.
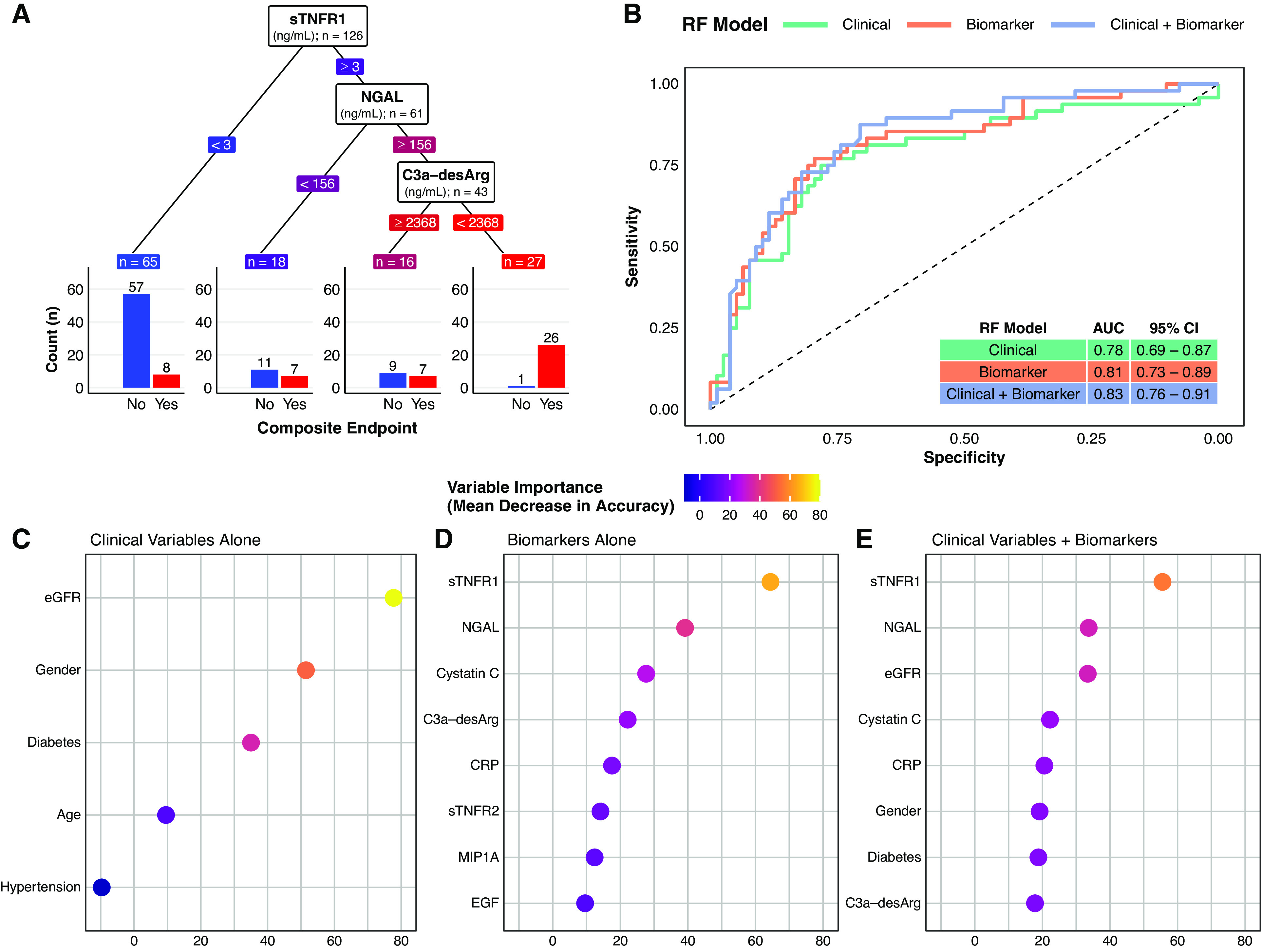
A supervised machine-learning approach (random forest classifier) illustrates the added predictive value of multiple serum biomarkers for a composite renal and mortality end point, both when considered alone and in addition to conventional clinical variables. (A) Decision tree classification of the composite renal and mortality end point by serum biomarkers in the study cohort. The decision tree highlights the predictive value of simultaneously assessing multiple serum biomarkers. In this decision tree, the three biomarkers are ranked by their proximate level of importance to correct classification of the composite renal and mortality end point, from sTNFR1 (highest) to C3a-desArg (lowest). Individuals with low sTNFR1 values (<3 ng/ml) had a relatively low risk of the composite renal and mortality end point (12%). However, not all individuals with high sTNFR1 values had the same risk of the composite renal and mortality end point. Those with high sTNFR1 (≥3 ng/ml) coupled with low NGAL values (<156 ng/ml) had a 39% risk of the composite renal and mortality end point, whereas those with high sTNFR1 (≥3 ng/ml), high NGAL (≥156 ng/ml), but also high C3a-desArg values (≥2368 ng/ml) had a 44% risk of the composite renal and mortality end point. Conversely, individuals with the triad of high sTNFR1 (≥3 ng/ml), high NGAL (≥156 ng/ml), and low C3a-desArg (<2368 ng/ml), which accounted for approximately 20% of the study cohort, almost universally (96%) developed the composite renal and mortality end point during follow-up. Biomarker values in the decision tree are colored in a continuous gradient from left to right from blue (lower risk) to red (higher risk). (B) Receiver operating characteristic curves for three types of random forest (RF) classification models of the composite renal and mortality end point: clinical variables alone (age, sex, hypertension, diabetes, and baseline eGFR; green), serum biomarkers alone (orange), and clinical variables plus serum biomarkers (purple). A leave-one-out cross-validation approach was implemented for the random forest models. The plot illustrates incremental improvements in correct prediction of the composite renal and mortality end point across the three model types. Area under the curve (AUC) values and associated 95% CIs for the three model types are presented in the inset table: 0.78 for clinical variables alone, 0.81 for serum biomarkers alone, and 0.83 for clinical variables plus serum biomarkers. (C–E) Dot plots of variable importance across the three random forest classification models of the composite renal and mortality end point: (C) clinical variables alone (age, sex, hypertension, diabetes, and baseline eGFR), (D) serum biomarkers alone, and (E) clinical variables plus biomarkers. eGFR was the most important clinical variable, whereas sTNFR1 and NGAL were the biomarkers that provided the most predictive value to the models. All five clinical variables are presented (C), whereas the top eight most important variables are presented for the biomarkers alone and clinical variables plus biomarker models (D and E). The dots are colored in a continuous gradient from navy (lower variable importance) to yellow (higher variable importance). Composite renal and mortality end point: ≥40% decrease in Chronic Kidney Disease Epidemiology Collaboration eGFR, doubling of serum creatinine, RRT, or mortality. CRP, C-reactive protein.
Receiver operating characteristic curves and AUC values for classification of the composite renal and mortality end point by random forests trained on clinical variables alone, biomarkers alone, and clinical variables plus biomarkers are presented in Figure 3B. An incremental improvement in predictive performance was observed between models trained on biomarkers compared with those trained on clinical variables (AUC of 0.81 versus 0.78). Predictive performance was further enhanced after inclusion of clinical variables alongside biomarkers to train the models (AUC of 0.83 versus 0.81 for biomarkers alone). Baseline eGFR was the most important predictor of the composite renal and mortality end point in models trained on clinical variables alone, whereas, when incorporated alongside biomarkers, eGFR and cystatin C were ranked as the third and fourth most important variables, respectively (Figure 3, C–E). sTNFR1 and NGAL were the two most important variables to prediction of the composite renal and mortality end point, both when biomarkers were considered alone and alongside clinical variables.
Additional random forest performance metrics stratified by three predicted probability thresholds (10%, 30%, and 50%) for classifying the composite renal and mortality end point are presented in Table 3. At a low (10%) predicted probability threshold, inclusion of serum biomarkers improved sensitivity for classification of the composite end point at the expense of reduced specificity. At the 30% predicted probability threshold, models trained on serum biomarkers again improved sensitivity for classification of the composite end point while also achieving specificity values comparable with models trained on clinical variables alone. At both of these thresholds, the highest sensitivity for classification of the composite end point was observed when serum biomarkers were considered alongside clinical variables. At the 50% predicted probability threshold, favorable trends toward improved sensitivity and specificity were observed in models trained on serum biomarkers, albeit the absolute magnitude of improvement was smaller than at lower predicted probability thresholds.
Table 3.
Random forest model performance metrics for prediction of the composite renal and mortality end point (n=126)
| Parameter | Predicted Probability Threshold >10%a | Predicted Probability Threshold >30%a | Predicted Probability Threshold >50%a | ||||||
| Clinical | Biomarker | Clinical and Biomarker | Clinical | Biomarker | Clinical and Biomarker | Clinical | Biomarker | Clinical and Biomarker | |
| Sensitivity | 0.92 | 0.96 | 0.98 | 0.79 | 0.85 | 0.88 | 0.69 | 0.71 | 0.69 |
| Specificity | 0.36 | 0.24 | 0.21 | 0.69 | 0.65 | 0.71 | 0.81 | 0.83 | 0.82 |
| Positive predictive value | 0.47 | 0.44 | 0.43 | 0.61 | 0.60 | 0.65 | 0.69 | 0.72 | 0.70 |
| Negative predictive value | 0.87 | 0.90 | 0.94 | 0.84 | 0.88 | 0.90 | 0.81 | 0.82 | 0.81 |
Three types of random forest classification models were implemented to evaluate prediction of the composite renal and mortality end point: clinical variables alone (age, sex, hypertension, diabetes, and baseline eGFR), serum biomarkers alone, and clinical variables plus serum biomarkers. A leave-one-out cross-validation approach was implemented, which consisted of excluding one individual from training the random forest model. Subsequently, the trained random forest model predicted the class of the individual excluded during model training. This process was repeated iteratively for each individual in the dataset, such that a predicted class was assigned to each study participant by each of the three random forest model types.
Model performance metrics were calculated across three probability thresholds (10%, 30%, and 50%) for labeling patients as having developed the composite renal and mortality end point. For example, an individual with a predicted probability of the composite renal and mortality end point by a random forest model of 45% would be labeled as having developed the composite renal and mortality end point by the first two thresholds evaluated, but not the latter.
Sensitivity analyses were performed to test the extent to which improved classification of the composite renal and mortality end point by random forest models incorporating serum biomarkers persisted after inclusion of uACR alongside other clinical variables. Although AUC values of random forest models trained on clinical variables alone did improve after incorporation of baseline uACR using two different approaches, the highest AUC values continued to be observed in random forest models trained on both clinical variables plus serum biomarkers (Supplemental Figure 2, A and B).
Prediction of Time to Renal and Mortality and Renal-Specific Composite End Points by Serum Biomarkers (Cox Models)
The AIC value for the Cox model incorporating all clinical and biomarker variables was 382.92, which reduced to 374.83 after stepwise backward elimination of six biomarkers in the following order: EGF, FABP1, cystatin C, IL-8, MIP-1-α, and sTNFR2. Three biomarkers were included in the final clinical plus biomarker model: C-reactive protein, NGAL, and C3a-desArg. Compared with a Cox model incorporating only clinical variables (AIC, 385.31), this parsimonious clinical plus biomarker model (AIC, 375.86) improved prediction of time to the composite renal and mortality end point (P=0.001). C-reactive protein (adjusted hazard ratio [aHR], 1.4; 95% CI, 1.1 to 1.8) and NGAL (aHR, 2.8; 95% CI, 1.3 to 6.1) were positively associated with the composite renal and mortality end point (Figure 4A). C3a-desArg was inversely associated with the composite renal and mortality end point (aHR, 0.6; 95% CI, 0.4 to 0.96). Figure 4, B–D, presents survival without the composite renal and mortality end point in the fully adjusted multivariate clinical plus biomarker Cox model, stratified by tertiles of C-reactive protein (Figure 4B), NGAL (Figure 4C), and C3a-desArg (Figure 4D).
Figure 4.
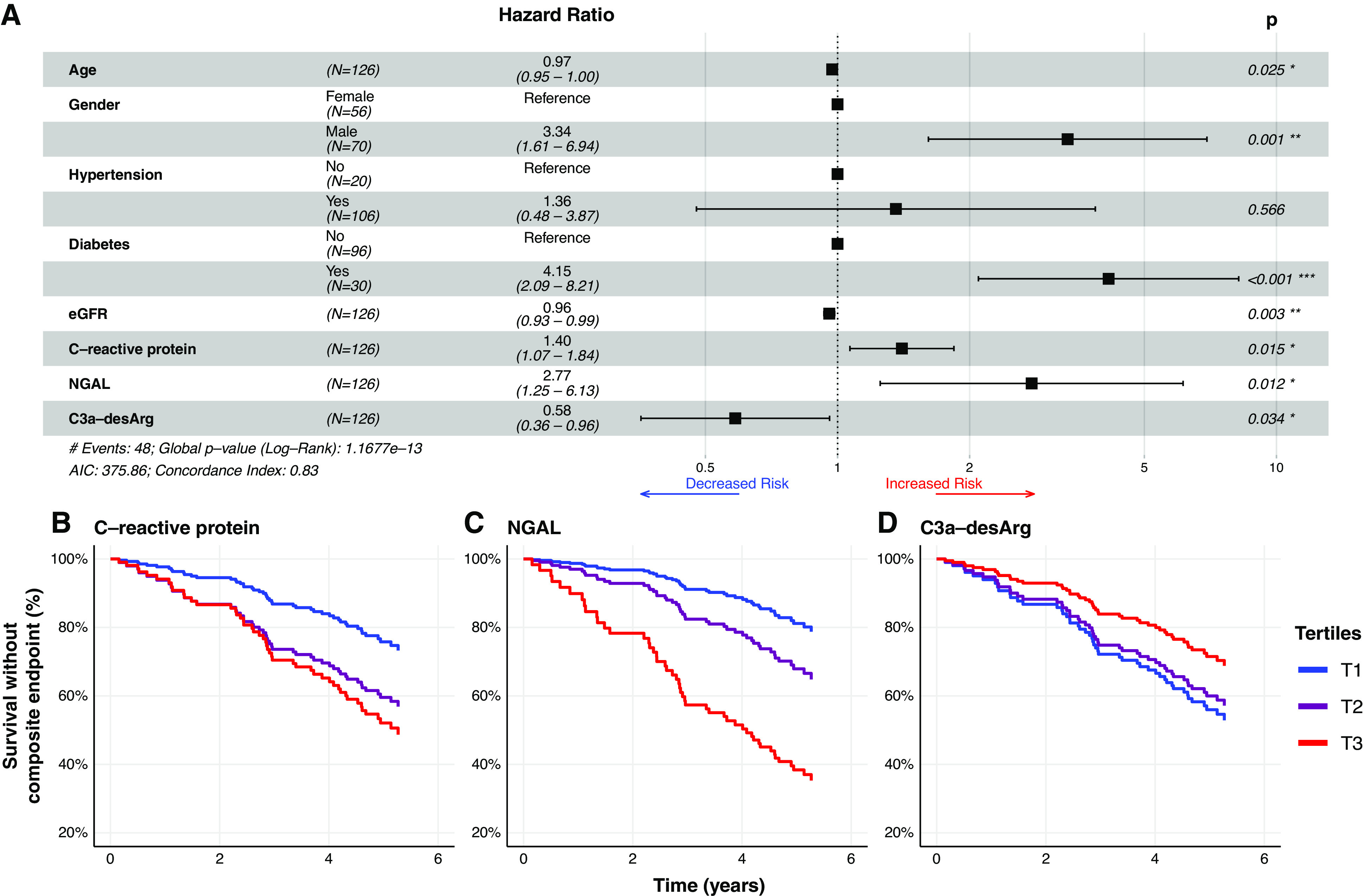
C-reactive protein, NGAL, and C3a-desArg independently predict time to development of a composite renal and mortality end point (Cox proportional hazards regression). (A) Forest plot of parsimonious Cox proportional hazards regression model incorporating clinical variables and serum biomarkers predictive of a composite renal and mortality end point (n=126). C-reactive protein (adjusted hazard ratio [aHR], 1.4; 95% 95% CI, 1.1 to 1.9) and NGAL (aHR, 2.7; 95% CI, 1.3 to 5.8) were positively associated with time to the composite renal and mortality end point. C3a-desArg values were inversely associated with time to the composite renal and mortality end point (aHR, 0.5; 95% CI, 0.3 to 0.9). (B–D) Plots of survival without the composite renal and mortality end point derived from the fully adjusted, multivariate clinical plus biomarker Cox proportional hazards regression model presented in the forest plot in (A), stratified by tertiles of serum (B) C-reactive protein, (C) NGAL, and (D) C3a-desArg. Biomarker values were log transformed for modeling. Hazard ratios from Cox models are expressed per one unit change in natural logarithm biomarker concentrations. Composite renal and mortality end point: ≥40% decrease in Chronic Kidney Disease Epidemiology Collaboration eGFR, doubling of serum creatinine, RRT, or mortality. AIC, Akaike information criterion. T1, tertile 1. *, P<0.05; **, P<0.01; ***, P<0.001
Several sensitivity analyses were performed to further interrogate these findings. In Cox models performed in the subgroup of patients with CKD stages 3–5/eGFR <60 ml/min per BSA, serum biomarkers improved prediction of time to the composite renal and mortality end point (P=0.001). C-reactive protein, NGAL, and C3a-desArg remained independently predictive of time to the composite renal and mortality end point, with hazard ratios similar to those obtained in the full study cohort observed (Supplemental Figure 3A). In Cox models additionally adjusted for uACR, which were performed in the subgroup of patients with baseline uACR available, serum biomarkers improved prediction of time to the composite renal and mortality end point (P=0.001). C-reactive protein (aHR, 1.5; 95% CI, 1.1 to 2.1) and NGAL (aHR, 3.5; 95% CI, 1.5 to 8.2) remained predictive of the composite renal and mortality end point (Supplemental Figure 3B).
Sensitivity analyses were also performed to test the predictive value of the serum biomarkers with respect to a renal-specific composite outcome, defined as ≥40% decline in CKD-EPI eGFR, doubling of serum creatinine, or RRT. Compared with a Cox model incorporating only clinical variables (AIC, 329.01), the clinical plus biomarker model (AIC, 325.00) improved prediction of time to the renal-specific composite end point (P=0.02). NGAL (aHR, 3.0; 95% CI, 1.3 to 6.7) and C3a-desArg (aHR, 0.5; 95% CI, 0.3 to 0.9) independently predicted time to the renal-specific composite end point (Supplemental Figure 4A). In Cox models additionally adjusted for uACR, serum biomarkers considered alongside clinical variables improved prediction of time to the renal-specific composite end point compared with clinical variables alone (P=0.03). NGAL (aHR, 2.7; 95% CI, 1.1 to 6.3) remained predictive of the renal-specific composite end point (Supplemental Figure 4B).
Discussion
This study provides insight into the value of 11 serum biomarkers measured using two multiplex biochip arrays in the identification of outpatients with CKD at high risk of accelerated renal functional decline and mortality. Biomarkers were quantified on a single, clinical-grade platform, which offers a route for improved translatability to the CKD outpatient setting where biomarkers may enhance prognostication afforded by existing clinical risk prediction tools (12,26). Global shifts in serum biomarker profiles, reflecting changes in both protective factors and injury markers, were evident with declining kidney function and in those who subsequently developed a composite renal and mortality end point defined on the basis of CKD progression, need for RRT, or death. We demonstrate, using random forests and multivariate Cox models, that incorporation of biomarkers alongside clinical variables improves prediction of CKD progression and mortality.
We used diverse statistical methods, including univariate logistic regression (for ease of interpretability and visual representation), principal components analysis (to illustrate clustering of patients on the basis of serum biomarker profiles), random forest models (to accurately characterize the predictive value of the biomarkers for adverse outcomes, not least due to seamless handling of regressor collinearity), and Cox models (to incorporate time-to-event data). Replication of the overarching results (improved predictive performance of models incorporating serum biomarkers for adverse CKD outcomes) using a variety of statistical approaches adds to the robustness of these findings.
Annual decline in kidney function was moderate in our study cohort, at −0.9 ml/min per BSA per year, which was significantly lower than a local cohort of patients with type 2 diabetic kidney disease (mean eGFR of 47 ml/min per BSA) attending our hospital for multidisciplinary CKD care, in whom eGFR trajectories ranged from −6 ml/min per BSA per year preintervention to −3 ml/min per BSA per year postintervention (10). However, the latter cohort of patients exclusively had type 2 diabetes for a median duration of 10 years and were enrolled up to a decade before patients in this study (10). The eGFR slope trajectory of this cohort was more similar to, albeit still lower than, a group of patients with type 2 diabetes and CKD (mean eGFR of 42 ml/min per BSA) enrolled in Dublin, Ireland during 2014–2015, in whom annual change in eGFR was −2 ml/min per BSA per year (27). Thus, improvements in CKD management and a lower prevalence of diabetes (25%) in this study cohort may explain the modest rates of renal functional decline observed.
Interestingly, patients with the triad of high sTNFR1 (≥3 ng/ml) and NGAL (≥156 ng/ml) coupled with low C3a-desArg (<2368 ng/ml) almost universally (96%) developed an adverse renal outcome or died during follow-up. This finding requires validation in a larger external cohort, but does suggest this biomarker signature may be useful for enrollment of patients with CKD who are at high risk into prospective studies and may also prove useful as a means of guiding treatment intensification in nephrology practice (28). The discriminant value of sTNFR1 and NGAL for adverse outcomes was improved by C3a-desArg, highlighting that proteins with diverse functions in innate immune responses and inflammation may have complementary prognostic benefit.
To the best of our knowledge, circulating C3a-desArg has not previously been explored as a biomarker of adverse CKD outcomes. C3a-desArg, also known as acylation-stimulating protein, is an adipokine that binds to the C5a receptor-2 to regulate metabolic processes, specifically stimulation of triglyceride accumulation and synthesis in adipocytes and glucose uptake in pancreatic β-cells (29–31). The majority of C3a-desArg is produced by the alternative complement pathway (32). However, adiponectin activates C1q, which generates C3a and C5a fragments via classic pathway activation (33). Increased plasma C3a-desArg has been documented in patients with nephrotic syndrome, and may be implicated in the pathogenesis of the associated dyslipidemia (34,35). In this study, circulating C3a-desArg was inversely associated with adverse CKD outcomes. We hypothesize that decreased C3a-desArg lowers glucose uptake and decreases clearance of triglycerides and fatty acids, thereby promoting glucose intolerance, hyperinsulinemia, and renal glucotoxicity and lipotoxicity (36). Decreased C3a-desArg may partly occur due to obesity-associated hypoadiponectinemia (33,36). By extension, adverse renal outcomes in those with low C3a-desArg in our cohort may be partly accounted for by the loss of the protective effects of adiponectin on glomerular podocytes—if so, this may be reversed by intentional weight loss strategies (37).
Random forests trained on serum biomarkers were superior to models trained on clinical variables in terms of prognosticating CKD progression and death. Predictive performance of the biomarkers was further enhanced when incorporated alongside clinical variables. sTNFR1 and NGAL were the two most important biomarkers to classification by random forests, reaffirming their predictive value, which has been demonstrated across multiple cohorts (23–25,27,38–40). C-reactive protein ranked as the fifth most important variable to random forest classification of the composite renal and mortality end point, both in models trained on serum biomarkers alone and in models trained on both clinical variables and biomarkers, which further endorses its role in prognosticating the risk of renal functional decline in CKD (41,42). Both C-reactive protein and NGAL independently predicted time to the composite renal and mortality end point, which persisted after additional adjustment for baseline albuminuria and in the subgroup of patients with baseline eGFR of <60 ml/min per BSA. Serum NGAL also independently predicted time to a renal-specific composite end point, which persisted after additional adjustment for albuminuria.
The study cohort had a high prevalence of GN, at 22%. Indeed, GN was the most documented CKD etiology in the study cohort. A significant proportion of biomarker research in CKD has focused on the subgroup of patients with diabetic kidney disease, although the Chronic Renal Insufficiency Cohort Study has investigated biomarker prognostication in study populations with CKD who do not exclusively have diabetes (43). Immunosuppressive therapy in people with GN has the potential to modify associations between biomarkers and adverse CKD outcomes (44); however, active immunosuppression was an exclusion criterion for our study cohort. Several of the biomarkers that demonstrated the strongest independent predictive value for adverse CKD outcomes in this study cohort have also demonstrated associations with histopathologic markers of disease severity and risk of renal functional decline in biopsy sample–proven glomerular diseases (such as IgA nephropathy and lupus nephritis), including sTNFR1, NGAL, and C-reactive protein (45–48).
Absolute sTNFR2 concentrations were lower than those of sTNFR1 in the study cohort, which is in contrast to other studies evaluating the prognostic value of both biomarkers in patients with CKD (23–25). At the time of development of the multiplex arrays, limited international reference material was available by which to standardize and harmonize the assays. More generally, assay standardization and harmonization remains a significant challenge in biomarker research (49). Nevertheless, the multiplex arrays used in our study are specific for the 11 target biomarkers contained therein. Indeed, sTNFR1 and sTNFR2 concentrations were strongly correlated with each other, and the two biomarkers clustered together by principal components analysis. Furthermore, sTNFR1 and sTNFR2 were the two biomarkers that were the most strongly associated with development of the composite renal and mortality end point by univariate logistic regression, and both biomarkers ranked as important variables to correct classification of the composite renal and mortality end point by random forest models. Thus, the multiplex arrays are specific for both sTNFR1 and sTNFR2 targets. Although quantification of sTNFR2 by the multiplex arrays appears to provide absolute serum concentrations that are lower than those reported by others using stand-alone immunoassays, this did not affect interpretation of the prognostic value of either sTNFR1 or sTNFR2 in the study cohort.
Our study cohort was relatively small, which limited statistical power, but event rates of renal and mortality outcomes were high and findings were replicated using random forests and Cox models. Ethnicity data were missing, although the majority (>95%) of patients attending nephrology clinics in Ireland are White (10,50). The predictive value of our multiplex arrays requires validation in a larger, more ethnically diverse cohort. Additionally, a head-to-head comparison of the value of the KFRE equation with that of the multiplex biomarker arrays for prediction of kidney failure should be performed in a subsequent, larger-scale study (12). Furthermore, prediction of incident CKD by the multiplex biomarker arrays in populations without established CKD should be evaluated and compared with existing clinical risk prediction tools, such as the equations developed by the CKD Prognosis Consortium (26). Baseline albuminuria data were missing for over half of the cohort. We calculated log-transformed uACR from uPCR using a validated equation (11), providing uACR values for >80% of participants. Individuals with missing uACR data trended to be older, but there were no significant differences between those with available and missing uACR data in terms of CKD stage or etiology, suggesting uACR data was largely missing at random. Furthermore, we performed sensitivity analyses to demonstrate persistence of the predictive value of serum biomarkers in random forest and Cox models, despite additional adjustment for albuminuria.
In summary, simultaneous measurement of 11 serum biomarkers using novel multiplex biochip array technology was technically feasible and robust. Parallel assessment of multiple biomarkers provided complementary prognostic value. In particular, patients with the triad of high sTNFR1, high NGAL, and low C3a-desArg almost universally developed an adverse renal end point or died over 5-year follow-up. Incorporation of serum biomarkers alongside clinical variables improved prediction of CKD progression and mortality. Our findings provide a strong basis for focusing the content of multianalyte biomarker panels as risk prediction tools in the CKD outpatient setting, and for confirmation of their clinical value and cost-effectiveness in larger, more diverse patient cohorts.
Disclosures
P. FitzGerald reports having ownership interest in Randox Laboratories Limited. M.D. Griffin reports receiving honoraria from the American Society of Nephrology, Hebei Medical University (China), and National Institutes of Health; serving on the editorial boards of Frontiers in Immunology, Frontiers in Pharmacology, JASN, Kidney International, and Transplantation; and serving as the section editor of Mayo Clinic Proceedings. T.P. Griffin reports receiving honoraria from Novo Nordisk and Sanofi. J. Lamont reports being named as an inventor on a patent filed on these results. E.M. McCole an d C. Richardson report being employed by Randox Teoranta and do not own any shares. I. McConnell and J. Lamont report being employed by Randox Laboratories Limited and do not own any shares. All remaining authors have nothing to disclose.
Funding
Primary funding for the study derived from the Enterprise Ireland Innovation Partnership grant IP-2013-0248, with cofunding from Randox Teoranta Ltd. (to M.D. Griffin and S.E. Logue). W. P. Martin's and S. Cormican’s contributions were performed within the Irish Clinical Academic Training Programme, supported by the Wellcome Trust, Health Research Board, Health Service Executive of Ireland National Doctors Training and Planning, and the Health and Social Care, Research and Development Division, Northern Ireland (grant number 203930/B/16/Z). S.D. Naicker was supported by a Molecular Medicine Ireland Clinical and Translational Research Scholarship funded by the Government of Ireland Programme for Research in Third Level Institutions, Cycle5 (PRTLI5), and by the College of Medicine, Nursing and Health Sciences, National University of Ireland Galway. T. P. Griffin was supported by a College of Medicine, Nursing and Health Science, National University of Ireland Galway Hardiman Scholarship and an Irish Endocrine Society/Royal College of Physicians of Ireland research bursary. J.P. Ferguson is supported by a Health Research Board Emerging Investigator Award, grant number EIA 2017-017. The work was also supported by European Commission grant 634086 (to W.P. Martin, T.P. Griffin, and M.D. Griffin; via the Horizon 2020 Collaborative Health Project NEPHSTROM), Science Foundation Ireland grants 09/SRC-B1794 (via the REMEDI Strategic Research Cluster; to M.D. Griffin) and 13/RC/2073 (via the CÚRAM Research Centre; to M.D. Griff in), and by the European Regional Development Fund.
Acknowledgments
The authors wish to sincerely thank all of the patients who participated in the study and acknowledge, with gratitude, the doctors, nurses, and phlebotomists at Merlin Park University Hospital, Galway, who contributed to enrollment of study subjects and collection of blood samples.
The authors acknowledge the support of the University College Dublin School of Medicine Summer Student Research Awards program in facilitating C. Conroy’s contribution to the study, including data collection and interpretation.
Author Contributions
C. Conroy, S. Cormican, M. D. Griffin, M. N. Islam, T. P. Griffin, S. E. Logue, W. P. Martin, C. Richardson, E. M. McCole, and S. D. Naicker were responsible for investigation; C. Conroy, S. Cormican, M. D. Griffin, T. P. Griffin, W. P. Martin, S. E. Logue, and S. D. Naicker were responsible for data curation; W. P. Martin and J. P. Ferguson were responsible for formal analysis; P. FitzGerald, M. D. Griffin, J. Lamont, S. E. Logue, I. McConnell, and C. Richardson conceptualized the study; W. P. Martin and M. D. Griffin wrote the original draft; S. Cormican, J. P. Ferguson, M. D. Griffin, T. P. Griffin, S. E. Logue, and W. P. Martin provided supervision; P. FitzGerald, M. D. Griffin, J. Lamont, S. E. Logue, I. McConnell, and C. Richardson were responsible for funding acquisition; M. D. Griffin, J. Lamont, C. Richardson, E. M. McCole, P. FitzGerald, and I. McConnell were responsible for resources; M. D. Griffin, S. E. Logue, E. M. McCole, and C. Richardson were responsible for project administration; and all authors reviewed and edited the manuscript and were responsible for methodology.
Supplemental Material
This article contains the following supplemental material online at http://kidney360.asnjournals.org/lookup/suppl/doi:10.34067/KID.0007552020/-/DCSupplemental.
Blood sample collection and serum isolation. Download Supplemental Appendix 1, PDF file, 359 KB (358.5KB, pdf)
Immunoassay procedure for serum biomarker determinations. Download Supplemental Appendix 2, PDF file, 359 KB (358.5KB, pdf)
Biomarker selection, assay performance, and validation. Download Supplemental Appendix 3, PDF file, 359 KB (358.5KB, pdf)
Inter-assay coefficient of variation of multiplex serum biomarker arrays. Download Supplemental Table 1, PDF file, 359 KB (358.5KB, pdf)
Characteristics of study participants with available and missing baseline uACR data. Download Supplemental Table 2, PDF file, 359 KB (358.5KB, pdf)
Scatterplot of sTNFR1 and sTNFR2 reveals a strong positive correlation between both biomarkers. Download Supplemental Figure 1, PDF file, 359 KB (358.5KB, pdf)
Sensitivity analyses of random forest classification models incorporating additional uACR adjustment illustrate added predictive value of serum biomarkers for a composite renal and mortality endpoint. Download Supplemental Figure 2, PDF file, 359 KB (358.5KB, pdf)
Sensitivity analyses examining the predictive value of serum biomarkers with respect to time to development of a composite renal and mortality endpoint (Cox proportional hazards regression). Download Supplemental Figure 3, PDF file, 359 KB (358.5KB, pdf)
Sensitivity analyses examining the predictive value of serum biomarkers with respect to time to development of a renal-specific composite endpoint (Cox proportional hazards regression). Download Supplemental Figure 4, PDF file, 359 KB (358.5KB, pdf)
References
- 1.GBD Chronic Kidney Disease Collaboration : Global, regional, and national burden of chronic kidney disease, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 395: 709–733, 2020. 10.1016/S0140-6736(20)30045-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jager KJ, Fraser SDS: The ascending rank of chronic kidney disease in the global burden of disease study. Nephrol Dial Transplant 32[suppl_2]: ii121–ii128, 2017. 10.1093/ndt/gfw330 [DOI] [PubMed] [Google Scholar]
- 3.Matsushita K, van der Velde M, Astor BC, Woodward M, Levey AS, de Jong PE, Coresh J, Gansevoort RT; Chronic Kidney Disease Prognosis Consortium: Association of estimated glomerular filtration rate and albuminuria with all-cause and cardiovascular mortality in general population cohorts: A collaborative meta-analysis. Lancet 375: 2073–2081, 2010. 10.1016/S0140-6736(10)60674-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waikar SS, Rebholz CM, Zheng Z, Hurwitz S, Hsu CY, Feldman HI, Xie D, Liu KD, Mifflin TE, Eckfeldt JH, Kimmel PL, Vasan RS, Bonventre JV, Inker LA, Coresh J; Chronic Kidney Disease Biomarkers Consortium Investigators: Biological variability of estimated GFR and albuminuria in CKD. Am J Kidney Dis 72: 538–546, 2018. 10.1053/j.ajkd.2018.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levey AS, Becker C, Inker LA: Glomerular filtration rate and albuminuria for detection and staging of acute and chronic kidney disease in adults: A systematic review. JAMA 313: 837–846, 2015. 10.1001/jama.2015.0602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colhoun HM, Marcovecchio ML: Biomarkers of diabetic kidney disease. Diabetologia 61: 996–1011, 2018. 10.1007/s00125-018-4567-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colombo M, Valo E, McGurnaghan SJ, Sandholm N, Blackbourn LAK, Dalton RN, Dunger D, Groop P-H, McKeigue PM, Forsblom C, Colhoun HM; FinnDiane Study Group and the Scottish Diabetes Research Network (SDRN) Type 1 Bioresource Collaboration: Biomarker panels associated with progression of renal disease in type 1 diabetes. Diabetologia 62: 1616–1627, 2019. 10.1007/s00125-019-4915-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Looker HC, Colombo M, Hess S, Brosnan MJ, Farran B, Dalton RN, Wong MC, Turner C, Palmer CNA, Nogoceke E, Groop L, Salomaa V, Dunger DB, Agakov F, McKeigue PM, Colhoun HM; SUMMIT Investigators: Biomarkers of rapid chronic kidney disease progression in type 2 diabetes. Kidney Int 88: 888–896, 2015. 10.1038/ki.2015.199 [DOI] [PubMed] [Google Scholar]
- 9.Naicker SD, Cormican S, Griffin TP, Maretto S, Martin WP, Ferguson JP, Cotter D, Connaughton EP, Dennedy MC, Griffin MD: Chronic kidney disease severity is associated with selective expansion of a distinctive intermediate monocyte subpopulation. Front Immunol 9: 2845, 2018. 10.3389/fimmu.2018.02845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin WP, Griffin TP, Lappin DW, Griffin DG, Ferguson JP, O’Brien T, Griffin MD: Influence of referral to a combined diabetology and nephrology clinic on renal functional trends and metabolic parameters in adults with diabetic kidney disease. Mayo Clin Proc Innov Qual Outcomes 1: 150–160, 2017. 10.1016/j.mayocpiqo.2017.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weaver RG, James MT, Ravani P, Weaver CGW, Lamb EJ, Tonelli M, Manns BJ, Quinn RR, Jun M, Hemmelgarn BR: Estimating urine albumin-to-creatinine ratio from protein-to-creatinine ratio: Development of equations using same-day measurements. J Am Soc Nephrol 31: 591–601, 2020. 10.1681/ASN.2019060605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tangri N, Grams ME, Levey AS, Coresh J, Appel LJ, Astor BC, Chodick G, Collins AJ, Djurdjev O, Elley CR, Evans M, Garg AX, Hallan SI, Inker LA, Ito S, Jee SH, Kovesdy CP, Kronenberg F, Heerspink HJL, Marks A, Nadkarni GN, Navaneethan SD, Nelson RG, Titze S, Sarnak MJ, Stengel B, Woodward M, Iseki K; CKD Prognosis Consortium: Multinational assessment of accuracy of equations for predicting risk of kidney failure: A meta-analysis. JAMA 315: 164–174, 2016. 10.1001/jama.2015.18202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kassambara A, Mundt F: factoextra: Extract and visualize the results of multivariate data analyses. R package version 1.0.7, 2020. Available at: https://CRAN.R-project.org/package=factoextra. Accessed June 9, 2021
- 14.Therneau T, Atkinson B: rpart: Recursive partitioning and regression trees. R package version 4.1-15, 2019. Available at: https://CRAN.R-project.org/package=rpart Accessed June 9, 2021
- 15.Borkovec M, Madin N: ggparty: “ggplot” visualizations for the “partykit” package. R package version 1.0.0, 2019. Available at: https://CRAN.R-project.org/package=ggparty. Accessed June 9, 2021
- 16.Liaw A, Wiener M: Classification and regression by randomForest. R News 2: 18–22, 2002 [Google Scholar]
- 17.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J-C, Müller M: pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12: 77, 2011. 10.1186/1471-2105-12-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Venables WN, Ripley BD: Modern Applied Statistics with S, 4th Ed., New York, Springer, 2002. 10.1007/978-0-387-21706-2 [DOI] [Google Scholar]
- 19.Therneau TM: A package for survival analysis in R. R package version 3.1-12, 2020. Available at: https://CRAN.R-project.org/package=survival. Accessed June 9, 2021
- 20.Kassambara A, Kosinski M, Biecek P: survminer: Drawing survival curves using “ggplot2.” R package version 0.4.6, 2019. Available at: https://CRAN.R-project.org/package=survminer. Accessed June 9, 2021
- 21.Wickham H, Averick M, Bryan J, Chang W, D’Agostino McGowan L, Francois R, Grolemund G, Hayes A, Henry L, Hester J, Kuhn M, Lin Pedersen T, Miller E, Milton Bache S, Muller K, Ooms J, Robinson D, Paige Seidel D, Spinu V, Takahashi K, Vaughan D, Wilke C, Woo K, Yutani H: Welcome to the Tidyverse. J Open Source Softw 4: 1686, 2019. 10.21105/joss.01686 [DOI] [Google Scholar]
- 22.R Core Team : R: A language and environment for statistical computing. Vienna, Austria, R Foundation for Statistical Computing, 2020. Available at: https://www.R-project.org/. Accessed June 9, 2021 [Google Scholar]
- 23.Niewczas MA, Gohda T, Skupien J, Smiles AM, Walker WH, Rosetti F, Cullere X, Eckfeldt JH, Doria A, Mayadas TN, Warram JH, Krolewski AS: Circulating TNF receptors 1 and 2 predict ESRD in type 2 diabetes. J Am Soc Nephrol 23: 507–515, 2012. 10.1681/ASN.2011060627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coca SG, Nadkarni GN, Huang Y, Moledina DG, Rao V, Zhang J, Ferket B, Crowley ST, Fried LF, Parikh CR: Plasma biomarkers and kidney function decline in early and established diabetic kidney disease. J Am Soc Nephrol 28: 2786–2793, 2017. 10.1681/ASN.2016101101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schrauben SJ, Shou H, Zhang X, Anderson AH, Bonventre JV, Chen J, Coca S, Furth SL, Greenberg JH, Gutierrez OM, Ix JH, Lash JP, Parikh CR, Rebholz CM, Sabbisetti V, Sarnak MJ, Shlipak MG, Waikar SS, Kimmel PL, Vasan RS, Feldman HI, Schelling JR; CKD Biomarkers Consortium and the Chronic Renal Insufficiency Cohort (CRIC) Study Investigators: Association of multiple plasma biomarker concentrations with progression of prevalent diabetic kidney disease: Findings from the Chronic Renal Insufficiency Cohort (CRIC) Study. J Am Soc Nephrol 32: 115–126, 2021. 10.1681/ASN.2020040487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson RG, Grams ME, Ballew SH, Sang Y, Azizi F, Chadban SJ, Chaker L, Dunning SC, Fox C, Hirakawa Y, Iseki K, Ix J, Jafar TH, Köttgen A, Naimark DMJ, Ohkubo T, Prescott GJ, Rebholz CM, Sabanayagam C, Sairenchi T, Schöttker B, Shibagaki Y, Tonelli M, Zhang L, Gansevoort RT, Matsushita K, Woodward M, Coresh J, Shalev V; CKD Prognosis Consortium: Development of risk prediction equations for incident chronic kidney disease. JAMA 322: 2104–2114, 2019. 10.1001/jama.2019.17379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin WP, Tuohy C, Doody A, Jackson S, Canavan RJ, Slattery D, Twomey PJ, McKenna MJ, le Roux CW, Docherty NG: Parallel assessment of albuminuria and plasma sTNFR1 in people with type 2 diabetes and advanced chronic kidney disease provides accurate prognostication of the risks of renal decline and death. Sci Rep 10: 14852, 2020. 10.1038/s41598-020-71684-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamanouchi M, Skupien J, Niewczas MA, Smiles AM, Doria A, Stanton RC, Galecki AT, Duffin KL, Pullen N, Breyer MD, Bonventre JV, Warram JH, Krolewski AS: Improved clinical trial enrollment criterion to identify patients with diabetes at risk of end-stage renal disease. Kidney Int 92: 258–266, 2017. 10.1016/j.kint.2017.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui W, Paglialunga S, Kalant D, Lu H, Roy C, Laplante M, Deshaies Y, Cianflone K: Acylation-stimulating protein/C5L2-neutralizing antibodies alter triglyceride metabolism in vitro and in vivo. Am J Physiol Endocrinol Metab 293: E1482–E1491, 2007. 10.1152/ajpendo.00565.2006 [DOI] [PubMed] [Google Scholar]
- 30.Cui W, Lapointe M, Gauvreau D, Kalant D, Cianflone K: Recombinant C3adesArg/acylation stimulating protein (ASP) is highly bioactive: a critical evaluation of C5L2 binding and 3T3-L1 adipocyte activation. Mol Immunol 46: 3207–3217, 2009. 10.1016/j.molimm.2009.08.013 [DOI] [PubMed] [Google Scholar]
- 31.Kolev M, Kemper C: Keeping it all going–Complement meets metabolism. Front Immunol 8: 1, 2017. 10.3389/fimmu.2017.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paglialunga S, Fisette A, Yan Y, Deshaies Y, Brouillette JF, Pekna M, Cianflone K: Acylation-stimulating protein deficiency and altered adipose tissue in alternative complement pathway knockout mice. Am J Physiol Endocrinol Metab 294: E521–E529, 2008. 10.1152/ajpendo.00590.2007 [DOI] [PubMed] [Google Scholar]
- 33.Peake PW, Shen Y, Walther A, Charlesworth JA: Adiponectin binds C1q and activates the classical pathway of complement. Biochem Biophys Res Commun 367: 560–565, 2008. 10.1016/j.bbrc.2007.12.161 [DOI] [PubMed] [Google Scholar]
- 34.Ozata M, Oktenli C, Gulec M, Ozgurtas T, Bulucu F, Caglar K, Bingol N, Vural A, Ozdemir IC: Increased fasting plasma acylation-stimulating protein concentrations in nephrotic syndrome. J Clin Endocrinol Metab 87: 853–858, 2002. 10.1210/jcem.87.2.8243 [DOI] [PubMed] [Google Scholar]
- 35.Tang JH, Wen Y, Wu F, Zhao XY, Zhang MX, Mi J, Cianflone K: Increased plasma acylation-stimulating protein in pediatric proteinuric renal disease. Pediatr Nephrol 23: 959–964, 2008. 10.1007/s00467-007-0738-1 [DOI] [PubMed] [Google Scholar]
- 36.Escasany E, Izquierdo-Lahuerta A, Medina-Gomez G: Underlying mechanisms of renal lipotoxicity in obesity. Nephron 143: 28–32, 2019. 10.1159/000494694 [DOI] [PubMed] [Google Scholar]
- 37.Martin WP, Docherty NG, Le Roux CW: Impact of bariatric surgery on cardiovascular and renal complications of diabetes: A focus on clinical outcomes and putative mechanisms. Expert Rev Endocrinol Metab 13: 251–262, 2018. 10.1080/17446651.2018.1518130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niewczas MA, Pavkov ME, Skupien J, Smiles A, Md Dom ZI, Wilson JM, Park J, Nair V, Schlafly A, Saulnier PJ, Satake E, Simeone CA, Shah H, Qiu C, Looker HC, Fiorina P, Ware CF, Sun JK, Doria A, Kretzler M, Susztak K, Duffin KL, Nelson RG, Krolewski AS: A signature of circulating inflammatory proteins and development of end-stage renal disease in diabetes. Nat Med 25: 805–813, 2019. 10.1038/s41591-019-0415-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo L, Zhu B, Yuan H, Zhao W: Evaluation of serum neutrophil gelatinase-associated lipocalin in older patients with chronic kidney disease. Aging Med (Milton) 3: 32–39, 2020. 10.1002/agm2.12098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolignano D, Lacquaniti A, Coppolino G, Donato V, Campo S, Fazio MR, Nicocia G, Buemi M: Neutrophil gelatinase-associated lipocalin (NGAL) and progression of chronic kidney disease. Clin J Am Soc Nephrol 4: 337–344, 2009. 10.2215/CJN.03530708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mc Causland FR, Claggett B, Burdmann EA, Eckardt KU, Kewalramani R, Levey AS, McMurray JJ, Parfrey P, Remuzzi G, Singh AK, Solomon SD, Toto RD, Pfeffer MA: C-reactive protein and risk of ESRD: Results from the Trial to Reduce Cardiovascular Events With Aranesp Therapy (TREAT). Am J Kidney Dis 68: 873–881, 2016. 10.1053/j.ajkd.2016.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu EL, Franko MA, Obergfell A, Dekker FW, Gabrielsen A, Jernberg T, Carrero JJ: High-sensitivity C-reactive protein and the risk of chronic kidney disease progression or acute kidney injury in post-myocardial infarction patients. Am Heart J 216: 20–29, 2019. 10.1016/j.ahj.2019.06.019 [DOI] [PubMed] [Google Scholar]
- 43.Hannan M, Ansari S, Meza N, Anderson AH, Srivastava A, Waikar S, Charleston J, Weir MR, Taliercio J, Horwitz E, Saunders MR, Wolfrum K, Feldman HI, Lash JP, Ricardo AC; Chronic Renal Insufficiency Cohort (CRIC) Study Investigators: Risk factors for CKD progression: Overview of findings from the CRIC Study. Clin J Am Soc Nephrol 16: 648–659, 2021. 10.2215/CJN.07830520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kędzierska K, Sindrewicz K, Sporniak-Tutak K, Gołembiewska E, Zair L, Sieńko J, Stańczyk-Dunaj M, Baranowska-Bosiacka I, Ciechanowski K: Does immunosuppressive therapy affect markers of kidney damage? Ann Transplant 21: 137–144, 2016. 10.12659/AOT.895275 [DOI] [PubMed] [Google Scholar]
- 45.Srivastava A, Schmidt IM, Palsson R, Weins A, Bonventre JV, Sabbisetti V, Stillman IE, Rennke HG, Waikar SS: The associations of plasma biomarkers of inflammation with histopathologic lesions, kidney disease progression, and mortality–The Boston Kidney Biopsy Cohort Study. Kidney Int Rep 6: 685–694, 2021. 10.1016/j.ekir.2020.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oh YJ, An JN, Kim CT, Yang SH, Lee H, Kim DK, Joo KW, Paik JH, Kang S-W, Park JT, Lim CS, Kim YS, Lee JP: Circulating tumor necrosis factor α receptors predict the outcomes of human IgA nephropathy: A prospective cohort study. PLoS One 10: e0132826, 2015. 10.1371/journal.pone.0132826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Torres-Salido MT, Cortés-Hernández J, Vidal X, Pedrosa A, Vilardell-Tarrés M, Ordi-Ros J: Neutrophil gelatinase-associated lipocalin as a biomarker for lupus nephritis. Nephrol Dial Transplant 29: 1740–1749, 2014. 10.1093/ndt/gfu062 [DOI] [PubMed] [Google Scholar]
- 48.Kaartinen K, Syrjänen J, Pörsti I, Hurme M, Harmoinen A, Pasternack A, Huhtala H, Mustonen J: Inflammatory markers and the progression of IgA glomerulonephritis. Nephrol Dial Transplant 23: 1285–1290, 2008. 10.1093/ndt/gfm782 [DOI] [PubMed] [Google Scholar]
- 49.Vesper HW, Myers GL, Miller WG: Current practices and challenges in the standardization and harmonization of clinical laboratory tests. Am J Clin Nutr 104[Suppl 3]: 907S–912S, 2016. 10.3945/ajcn.115.110387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin WP, Bauer J, Coleman J, Dellatorre-Teixeira L, Reeve JLV, Twomey PJ, Docherty NG, O’Riordan A, Watson AJ, le Roux CW, Holian J: Obesity is common in chronic kidney disease and associates with greater antihypertensive usage and proteinuria: Evidence from a cross-sectional study in a tertiary nephrology centre. Clin Obes 10: e12402, 2020. 10.1111/cob.12402 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Blood sample collection and serum isolation. Download Supplemental Appendix 1, PDF file, 359 KB (358.5KB, pdf)
Immunoassay procedure for serum biomarker determinations. Download Supplemental Appendix 2, PDF file, 359 KB (358.5KB, pdf)
Biomarker selection, assay performance, and validation. Download Supplemental Appendix 3, PDF file, 359 KB (358.5KB, pdf)
Inter-assay coefficient of variation of multiplex serum biomarker arrays. Download Supplemental Table 1, PDF file, 359 KB (358.5KB, pdf)
Characteristics of study participants with available and missing baseline uACR data. Download Supplemental Table 2, PDF file, 359 KB (358.5KB, pdf)
Scatterplot of sTNFR1 and sTNFR2 reveals a strong positive correlation between both biomarkers. Download Supplemental Figure 1, PDF file, 359 KB (358.5KB, pdf)
Sensitivity analyses of random forest classification models incorporating additional uACR adjustment illustrate added predictive value of serum biomarkers for a composite renal and mortality endpoint. Download Supplemental Figure 2, PDF file, 359 KB (358.5KB, pdf)
Sensitivity analyses examining the predictive value of serum biomarkers with respect to time to development of a composite renal and mortality endpoint (Cox proportional hazards regression). Download Supplemental Figure 3, PDF file, 359 KB (358.5KB, pdf)
Sensitivity analyses examining the predictive value of serum biomarkers with respect to time to development of a renal-specific composite endpoint (Cox proportional hazards regression). Download Supplemental Figure 4, PDF file, 359 KB (358.5KB, pdf)


