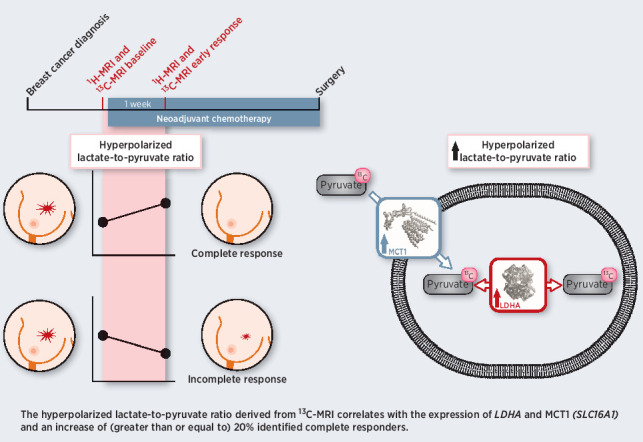
Hyperpolarized carbon-13 MRI allows response assessment in patients with breast cancer after 7–11 days of neoadjuvant chemotherapy and outperformed state-of-the-art and research quantitative proton MRI techniques.
Abstract
Hyperpolarized 13C-MRI is an emerging tool for probing tissue metabolism by measuring 13C-label exchange between intravenously injected hyperpolarized [1–13C]pyruvate and endogenous tissue lactate. Here, we demonstrate that hyperpolarized 13C-MRI can be used to detect early response to neoadjuvant therapy in breast cancer. Seven patients underwent multiparametric 1H-MRI and hyperpolarized 13C-MRI before and 7–11 days after commencing treatment. An increase in the lactate-to-pyruvate ratio of approximately 20% identified three patients who, following 5–6 cycles of treatment, showed pathological complete response. This ratio correlated with gene expression of the pyruvate transporter MCT1 and lactate dehydrogenase A (LDHA), the enzyme catalyzing label exchange between pyruvate and lactate. Analysis of approximately 2,000 breast tumors showed that overexpression of LDHA and the hypoxia marker CAIX was associated with reduced relapse-free and overall survival. Hyperpolarized 13C-MRI represents a promising method for monitoring very early treatment response in breast cancer and has demonstrated prognostic potential.
Significance:
Hyperpolarized carbon-13 MRI allows response assessment in patients with breast cancer after 7–11 days of neoadjuvant chemotherapy and outperformed state-of-the-art and research quantitative proton MRI techniques.
Graphical Abstract
Introduction
Neoadjuvant chemotherapy (NACT) is the standard-of-care treatment for 17%–40% of patients with operable early-stage breast cancer, particularly patients with HER2-positive (HER2+) and triple-negative breast cancer (TNBC; ref. 1). NACT can be used in downstaging of locally advanced breast cancer when breast conservation is considered and for testing novel drugs. Pathological complete response (pCR) at surgery indicates a favorable prognosis and rates of pCR have recently been shown to reach 68%–80% in patients receiving carboplatin or dual HER2 blockade (2). The response to NACT can guide decisions regarding additional adjuvant systemic therapy in nonresponders (3, 4) or de-escalation of therapy if an early response is identified (5).
Early prediction of pCR on imaging can be used to decrease side effects from non-efficacious drugs in nonresponders and could allow these patients to receive alternative regimens or investigational agents. Although early response assessment in breast cancer can be undertaken with multiparametric proton MRI (1H-MRI) using dynamic contrast-enhanced MRI (DCE-MRI) and diffusion-weighted imaging (DWI), a recent meta-analysis demonstrated substantial heterogeneity in sensitivity and specificity between studies (6, 7).
Aggressive breast cancers, such as HER2+ and TNBC, often switch to glycolysis, either as a result of aerobic glycolysis (the Warburg effect) or due to hypoxia, leading to increased intratumoral lactate production (8–10). This switch can be detected by intravenous injection of hyperpolarized [1–13C]pyruvate and monitoring the subsequent exchange of the hyperpolarized 13C-label between pyruvate and lactate using 13C MRI. This experimental clinical imaging tool has been explored in a number of cancer types (11–16). We have demonstrated the feasibility of using hyperpolarized 13C-MRI to assess patients with different subtypes of breast cancer, where higher levels of lactate labeling were observed in higher grade tumors, including TNBCs (11). In the majority of preclinical studies to date, successful treatment response was demonstrated by a decrease in tumor lactate labeling, which can be observed as early as 24 hours after cytotoxic treatment in a range of cancer models, including breast cancer (17). However, in a small number of animal studies the opposite effect occurred, with an increase in 13C-lactate signal following a successful response to therapy (18, 19). We have also recently described the first clinical example of response assessment in human breast cancer following NACT using hyperpolarized 13C-MRI, showing a decreased flux of hyperpolarized 13C-label into lactate after one complete NACT cycle of 3 weeks. DCE-MRI in this patient incorrectly predicted a poor response to therapy (16). Important questions remain about the temporal changes in lactate labeling and how widely applicable this response is across patients and tumor types.
Most studies define early response in breast cancer as that observed after a full cycle or several weeks of NACT (6). We have previously used a similar time point for the first proof-of-principle study in which hyperpolarized 13C-MRI was used to monitor treatment response in breast cancer (16). However, there is an unmet need for very early response assessment in patients with breast cancer, ideally within days or a week of treatment, to allow patients to rapidly change to the most appropriate treatment. Hyperpolarized 13C-MRI is a promising candidate technique for this very early response assessment as metabolic changes in response to treatment have been shown to occur on this timescale (17, 20–22). This prospective clinical study was designed to assess the potential added value of hyperpolarized 13C-MRI for very early response assessment in patients with aggressive breast cancer (TNBC or HER2+) undergoing neoadjuvant treatment in comparison with advanced multiparametric proton MRI techniques. The aim was to determine whether responders could be identified by an early change in lactate 13C-labeling. We have also explored the relationship between lactate labeling in individual patients and the expression, at an RNA level, of those genes that may influence pyruvate metabolism, demonstrating the prognostic significance of this pattern of gene expression in a large group of patients with breast cancer.
Patients and Methods
Patient recruitment
Local research ethics committee approval was obtained for this prospective study [National Research Ethics Service Committee East of England, Cambridge South, Research Ethics Committee number 15/EE/0378; National Institute for Health Research (NIHR) portfolio number 30388]. Written informed consent was obtained from seven patients diagnosed with invasive carcinoma of the breast between 2018 and 2020. Data for the baseline exam of one of these patients were included in a previous publication (11).
pCR was defined as the absence of residual invasive carcinoma in the resected breast specimen at surgery, regardless of the presence of DCIS.
MRI
Multinuclear breast MRI, including 13C-MRI with hyperpolarized [1–13C]pyruvate was performed at baseline before treatment initiation (median number of days between baseline 13C-MRI and treatment initiation was 4). For one of the baseline scans, the 1H-MRI was performed four days before the 13C-MRI for technical reasons. Early follow-up scans for response assessment, including 13C-MRI were performed 7–11 days after the first dose of NACT was administered (median = 7 days).
1H-MRI and postprocessing
All patients underwent proton breast MRI on a clinical 3 T scanner (MR750, GE Healthcare). The inbuilt 1H body coil was used to acquire three-dimensional fast gradient echo scout images and subsequently T1-weighted axial and coronal fast spoiled gradient echo images were used to plan the subsequent 13C-MRI (for specifications see Supplementary Materials). After completion of 13C-MRI, diagnostic quality proton breast MRI was undertaken in the prone position using a dedicated eight-channel phased array receive-only breast coil as described previously (11, 23). Details regarding acquisition, reconstruction, and analysis of DCE-MRI and diffusion weighted MRI are provided in the Supplementary Materials.
13C-MRI and postprocessing
Details regarding pharmacy kit and pyruvate sample preparation and hyperpolarization are provided in the Supplementary Materials.
In five patients, spectral–spatial 13C-imaging was performed (patients 2, 4, 5, 6, and 7 in Supplementary Table S1). A 22.4 ms excitation with flyback gradients was applied (24), alternating between a 15-degree pulse at the pyruvate frequency and a 40-degree pulse at the lactate frequency, each followed by a single-shot spiral readout (40 × 40 points, 20 cm FOV, TR 2 seconds, time resolution 4 seconds).
In one patient (patient 1, Supplementary Table S1), images were acquired using a dynamic coronal-iterative decomposition with echo asymmetry and least-squares estimation (IDEAL) spiral chemical shift imaging sequence (25) and data were processed as described previously (11). Baseline data of this patient were included in a previous publication (11). In another patient (patient 3, Supplementary Table S1), only spectral data at a temporal resolution of 16 seconds were acquired at baseline due to a technical failure; the same scanning approach was repeated at follow up to allow direct comparison. Because of the low temporal resolution in this case, LAC/PYR was calculated on the basis of the summed spectra and this patient was excluded from any summed SNR analyses due to lack of imaging data and from apparent exchange rate constant for pyruvate–lactate exchange (kPL) analyses due to low temporal resolution. Although the 13C-MRI technique varied between patients, it was the same for the baseline and follow-up examination in each patient (no intra-patient variation) to compare measurements between time points for the purpose of response assessment.
Signal-to-noise ratios (SNR) for pyruvate and lactate summed over time and the summed lactate-to-pyruvate ratio (LAC/PYR) were calculated from the sum-of-squares (SOS) image reconstructions (summed SNRPYR, summed SNRLAC, LAC/PYR; Supplementary Table S2). kPL was computed using singular value decomposition (SVD) image reconstructions (kPL; Supplementary Table S2; ref. 26). Further details regarding SOS and SVD reconstructions and analysis of 13C-images are provided in the Supplementary Materials.
Statistical analysis
Statistical analyses were performed using R (version 3.6.3; R Foundation). Relationships between variables were assessed using Pearson's correlation, including the correlation coefficient r. Differences between measurements were compared using a two-sided Student t test (paired if measurements for the same patients at baseline were compared with the follow-up). P values below 0.05 were considered significant. No multiple testing correction was applied: Significant tests should be interpreted as exploratory rather than confirmatory.
RNA sequencing
Biopsy samples were obtained within the Personalized Breast Cancer Program. For biopsy samples of patients included in this study, RNA was extracted from frozen tumor tissue sections obtained using the QIAGEN AllPrep DNA/RNA Mini Kit (catalog no. 80204; QIAGEN; Supplementary Materials). RNA quantification was performed using Qubit RNA BR (Invitrogen/Thermo Fisher Scientific catalog no: Q10211). Assessment of the RNA integrity number was performed using a TapeStation RNA ScreenTape (Agilent Technologies).
RNA sequencing libraries were constructed using the TruSeq Stranded Total RNA Gold library preparation kit (Illumina). The libraries were sequenced as paired-end reads (2 × 75 cycles) on a HiSeq2500 platform to give a mean coverage of 150×. Gene count data were postprocessed included normalization, scaling, and the correction of library preparation effects (Supplementary Materials). In brief, Salmon version 0.14.1 was used to estimate gene expression. The resulting estimated counts were corrected for library size and gene length. Potential library preparation bias was corrected for and further normalization was applied transforming the values to log2 counts for linear modeling. For correlations between RNA expression data and imaging data (LAC/PYR) in our cohort of patients with breast cancer, we only included patients with identical image acquisition (patients 1 and 3 were excluded, Supplementary Table S1).
METABRIC data
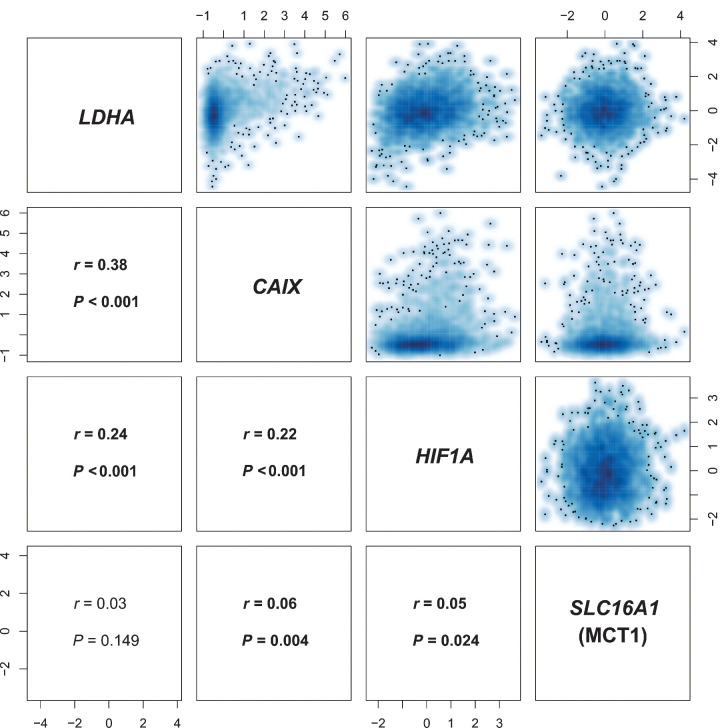
The expression for relevant genes in the METABRIC cohort was normalized, as described previously (27). Log-intensities were standardized using z-scores. Follow-up data and relapse information from METABRIC were curated and processed as described recently (28). The smoothed scatterplots and Pearson correlation coefficients in Fig. 5 were computed between each pair of variables, and the cor.test function in base R was used to test the hypothesis of no correlation. Further details regarding survival analysis are provided in Supplementary Materials.
Figure 5.
Correlation matrix of LDHA, SLC16A1 (MCT1), CAIX, and HIF1A expression in METABRIC. There is a significant correlation between LDHA and SLC16A1 (MCT1) expression (z-scores) with the hypoxia markers CAIX and HIF1A. r, Pearson correlation coefficient.
Data and code availability
Transcriptomic data for those tumors included in our study imaged with hyperpolarized 13C-MRI are deposited at the European Genome-phenome Archive (https://ega-archive.org/datasets/EGAD00001008141). Imaging raw data and MATLAB scripts for data in this article can be obtained from radiology-13c-mri-breast@lists.cam.ac.uk.
Results
Seven patients with a histopathological diagnosis of early-stage breast cancer were enrolled, that is, tumors confined to the breast, with or without locoregional lymph node involvement, but no distant metastatic disease. These included four patients with TNBC (ER and PR negativity defined as Allred score 0 to 3), three of which were invasive cancers of no specific type (IC NST) and one apocrine invasive cancer; and three HER2+ patients with breast cancer (two of which were ER+PR+, and one ER−PR−). All patients underwent standard-of-care NACT, patients with HER2+ breast cancer received dual anti-HER2 therapy in addition to chemotherapy, and two patients with TNBC received additional olaparib, a PARP1/2 inhibitor, as part of a clinical trial (PARTNER Trial, a randomized, phase II/III trial to evaluate the safety and efficacy of the addition of olaparib to platinum-based NACT in patients with breast cancer with TNBC and/or germline BRCA1/2 mutations). After 5–7 cycles of neoadjuvant treatment, three patients demonstrated pCR and four patients non-pCR. Of the three patients with eventual pCR, there was one ER−PR−HER+ patient and two ER−PR−HER2− patients (one with additional olaparib treatment and one without). Further details regarding patient and cancer characteristics and the prescribed treatments are provided in Supplementary Table S1.
Breast MRI was performed at baseline, before treatment initiation (median of 4 days between baseline 13C-MRI and treatment initiation), and as early follow-up for response assessment at 7–11 days after the first dose of NACT was administered (median = 7 days). Examples of one responder and one nonresponder imaged at these time points are shown in Fig. 1. Patient age at baseline was not significantly different between patients with eventual pCR and non-pCR (P > 0.05).
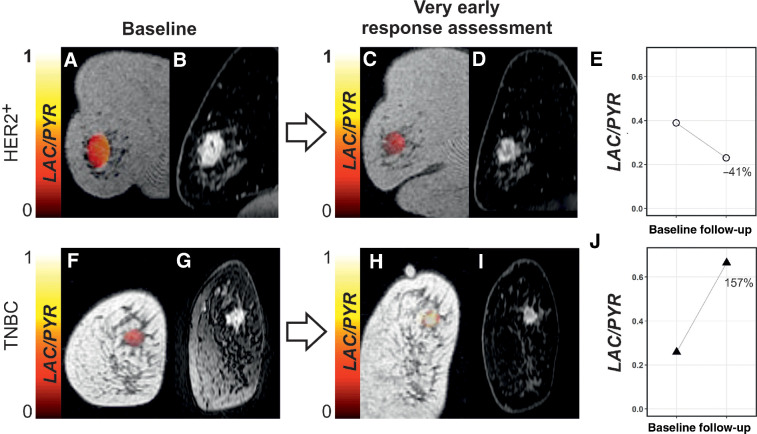
Figure 1.
Changes in LAC/PYR between baseline and very early response assessment in a responder and nonresponder. A, C, F, and H, Coronal T1-weighted 3D spoiled gradient echo (SPGR) images with LAC/PYR map overlaid on the breast tumor. B, D, G, and I, Coronal reformatted DCE images obtained 150 seconds after intravenous injection of a gadolinium-based contrast agent. A patient with HER2+ breast cancer was imaged at baseline (A and B) and for ultra-early response assessment (C and D) following standard-of-care treatment and showed a decrease in LAC/PYR of 41% (E), indicating nonresponse. At surgery, non-pCR with residual invasive cancer was identified. Another patient with TNBC was imaged at baseline (F and G) and for ultra-early response assessment (H and I) following treatment with chemotherapy and a PARP inhibitor and showed an increase in LAC/PYR of 157% (J), indicating response. At surgery, pCR without residual invasive breast cancer was found. HER2+, HER2/neu positive.
13C-MRI analysis
Mean values of 13C-MRI parameters computed for all voxels in each manually drawn region of interest (ROI) were analyzed. The number of voxels included in the 13C-MRI tumor ROIs ranged from 43 to 247 (mean = 102; median = 125). Results of the 13C-MRI and 1H-MRI data for all patients are shown in Supplementary Table S2.
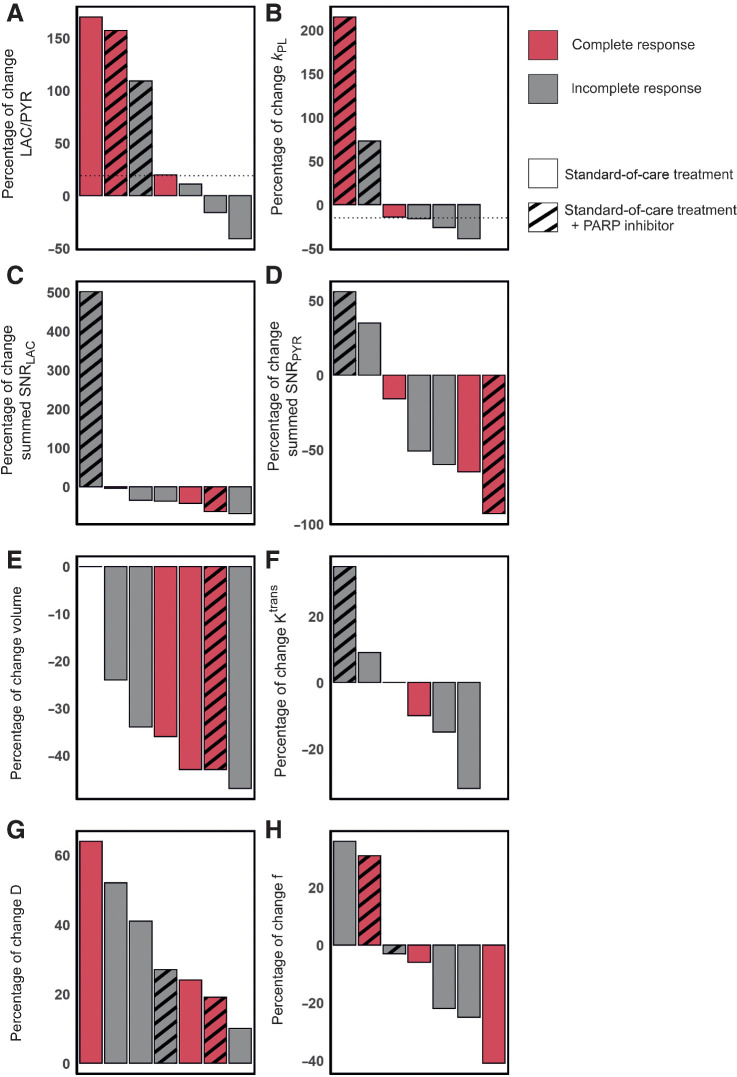
Mean summed SNR of pyruvate (summed SNRPYR) and lactate (summed SNRLAC) decreased between baseline and response assessment (summed SNRPYR at baseline, mean ± SD = 19.7 ± 19.9; versus 16.5 ± 21.9 at response assessment; summed SNRLAC at baseline 7.0 ± 5.6 versus 4.5 ± 3.3 at response assessment) whereas LAC/PYR and kPL increased (LAC/PYR at baseline, mean ± SD = 0.28 ± 0.17 versus 0.34 ± 0.19 at response assessment; kPL at baseline 0.0064 ± 0.0058 vs. 0.0079 ± 0.0078 at response assessment); however, these changes were not significant (P > 0.05). Results for these parameters and those derived from 1H-MRI are shown in Fig. 2 for the entire cohort, and for responders and nonresponders separately in Fig. 3. An increase of ≥20% in LAC/PYR measured using hyperpolarized 13C-MRI 7–11 days after commencing treatment, correctly predicted the three patients with eventual pCR at surgery. All patients with a change in LAC/PYR between baseline and early response assessment that was below a threshold of a 20% increase, demonstrated non-pCR at surgery (Fig. 4A). The only nonresponder with a change in LAC/PYR above this threshold received PARP inhibitor treatment in addition to standard therapy. For the two patients treated with a PARP inhibitor, the responder showed a higher increase in LAC/PYR than the nonresponder. A similar threshold of −15% for kPL separated responders (above threshold) from nonresponders (below threshold), with the patient on PARP inhibition described above being the only nonresponder with an increase above this threshold (Fig. 4B). For the two patients receiving PARP inhibition, the increase in kPL was again larger in the responder than the nonresponder. kPL could not be calculated in one patient due to technical reasons. Changes in LAC/PYR and kPL did not differ significantly between responders and nonresponders in this small cohort (P = 0.165 and P = 0.532, respectively). Neither changes in the summed SNRLAC nor summed SNRPYR could be used to distinguish between responders and nonresponders. Of note, changes in summed SNRPYR may not only reflect biological changes such as perfusion and transport, but may also represent technical differences in polarization and coil sensitivity, and should therefore be interpreted with caution.
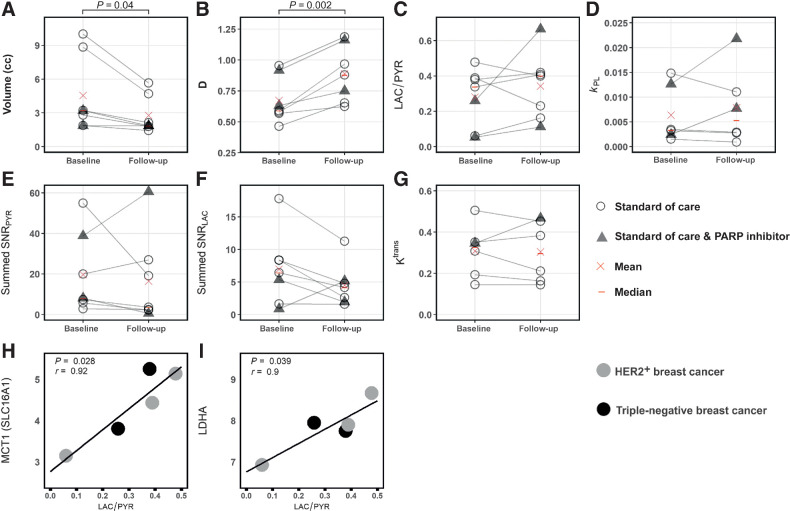
Figure 2.
Parameters obtained from hyperpolarized 13C-MRI and 1H-MRI at baseline and in early follow-up scans. Differences between baseline and follow-up were significant for tumor volume (A) and diffusivity (B) but not for the other parameters (C–G); neither change in volume or diffusivity could distinguish pCR from non-pCR. Correlation of SLC16A1 (MCT1) and LDHA mRNA expression with LAC/PYR was significant (H and I). Only images acquired with identical 13C-MRI acquisition parameters (spectral–spatial excitation) were included in these correlations.
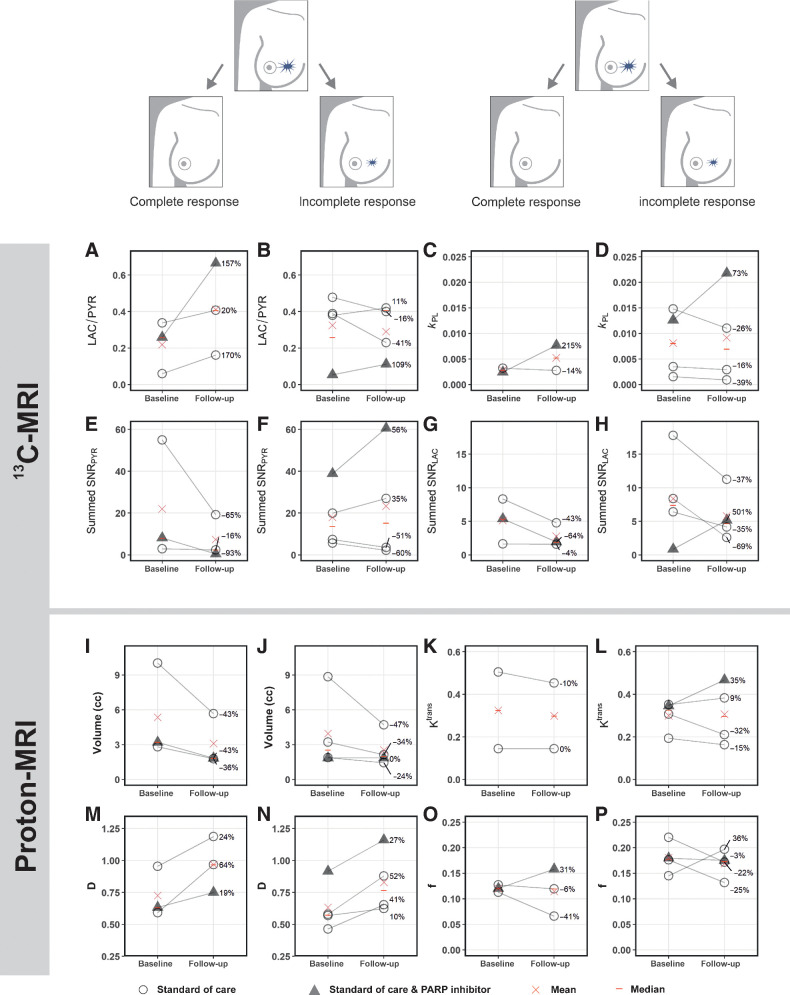
Figure 3.
Changes in hyperpolarized 13C-, but not 1H-MRI–derived metrics, after approximately one week of treatment distinguish responders (pCR) from nonresponders (incomplete response; non-pCR). In the five patients undergoing standard-of-care neoadjuvant treatment, an increase of ≥20% in LAC/PYR was only observed in patients who responded (A), whereas a lower increase or even a decrease in LAC/PYR was observed in nonresponders (B).Both patients treated with a PARP inhibitor in addition showed an increase in LAC/PYR (A and B) and again the increase was highest in the responder (A). Although kPL increased in all patients receiving a PARP inhibitor, but not in the other patients (C and D), neither kPL nor any of the 1H-MRI–based metrics from dynamic contrast-enhanced (DCE) MRI (such as Ktrans) or from intravoxel incoherent motion (IVIM) as part of diffusion-weighted MRI (such as perfusion fraction f and tissue diffusivity D) could distinguish between responders and nonresponders (I–P). None of the parameters differed significantly between baseline and follow-up when evaluated for responders and nonresponders separately (P > 0.05). kPL was not available in one patient due to technical failure (C). Ktrans could not be assessed in one patient due to failed fat saturation (K).
Figure 4.
Changes in hyperpolarized 13C-MRI and 1H-MRI parameters in seven patients with complete and incomplete responses. Results are shown for standard-of-care treatment with and without PARP inhibitor treatment. A, A threshold of +20% change in LAC/PYR distinguished responders from nonresponders on standard-of-care therapy (shown with a dashed horizontal line). One nonresponder receiving PARP inhibitor treatment also showed an increase in LAC/PYR of ≥20%, which may be explained by NAD+ availability (see main text). B, A threshold set at a −15% change in kPL (dashed horizontal line) distinguished responders from nonresponders on standard-of-care therapy. A patient receiving PARP inhibitor treatment in addition, but demonstrating pCR, also showed a change in kPL above this threshold. kPL was not available for one patient due to a technical failure. C–H, There were no thresholds that could be used to distinguish pCR from non-pCR for any of the remaining 1H-MRI or 13C-MRI parameters. Change in Ktrans was not evaluable for one patient where fat saturation failed at baseline.
1H-MRI: volume, DCE, and diffusion-weighted imaging
HER2+ tumors were larger than TNBC at baseline (mean ± SD = 7.4 ± 3.6 mL and 2.4 ± 0.7 mL, respectively) and follow-up (4.2 ± 1.8 mL and 1.7 ± 0.2 mL, respectively), although this difference was not significant (P = 0.140 and P = 0.147, respectively). Tumor volume, as assessed on DCE-MRI, either decreased or remained stable in all tumors. Volume decreased significantly between baseline and response assessment (P = 0.04; Fig. 2A) in the whole cohort, but neither tumor volume at baseline (P > 0.05) nor volume changes could be used to distinguish responders and nonresponders (Fig. 4E).
No significant differences in the following DCE-MRI derived pharmacokinetic parameters were identified between tumors with pCR and non-pCR, or within either group between timepoints (P > 0.05): Ktrans (the volume transfer constant as a measure of capillary permeability to gadolinium-based contrast agent that accumulates in the extravascular extracellular tumor compartment; Figs. 2G and 3K and L); kep (the rate constant reflecting transfer from the extravascular extracellular tumor compartment back into the plasma); ve (volume of the extravascular extracellular space); and iAUC90 (integrated area under the enhancement-time curve 90 seconds after contrast injection). Change in Ktrans was not evaluable for one patient where fat saturation failed at baseline.
The baseline perfusion fraction, f, measured using intravoxel incoherent motion (IVIM) was significantly lower in patients who reached pCR, compared with those with non-pCR (P = 0.026; Fig. 3O and P). The tissue diffusivity (D) increased significantly in all tumors between baseline and early response assessment (P = 0.002; Fig. 2B), whereas changes in f were variable. Importantly, changes in f and D were not significantly different between pCR and non-pCR tumors (P = 0.946 and P = 0.861, respectively) and could not be used to distinguish the two groups (Fig. 4G and H).
In the five patients with an identical 13C-MRI acquisition technique at both scanning timepoints, summed SNRPYR, summed SNRLAC, and LAC/PYR were significantly correlated with volume as assessed on DCE-MRI (r = 0.87, P = 0.001; r = 0.68, P = 0.030; and r = −0.63, P = 0.049, respectively; Supplementary Table S3; Supplementary Fig. S1). Summed SNRPYR was more strongly correlated with volume than summed SNRLAC, resulting in a negative correlation of volume with LAC/PYR. kPL was not significantly correlated with volume (r = 0.42, P = 0.231), but was positively correlated with the perfusion fraction f (r = 0.65, P = 0.044; Supplementary Fig. S1). Correlations between the hyperpolarized 13C-MRI parameters (summed SNRPYR, summed SNRLAC, LAC/PYR and kPL) and the DCE-MRI parameters (Ktrans, kep, ve and iAUC90) were low and non-significant (P > 0.05; Supplementary Table S3), indicating that they may be independent. The percentage changes in kPL and LAC/PYR following treatment did not correlate with changes in volume or diffusion, showing that metabolic alterations were independent of size and cellularity. Percentage changes in ve were significantly correlated with changes in LAC/PYR but not kPL (r = 0.84, P = 0.035 and r = 0.14, P = 0.819, respectively). However, other DCE-MRI parameters such as Ktrans and kep did not correlate with changes in metabolism. Importantly, changes in DCE-MRI, diffusion, and volume could not he used to correctly identify responders.
RNA expression
The hyperpolarized 13C-MRI metrics LAC/PYR and kPL were compared with RNA sequencing of biopsy samples at baseline, in those cases where 13C-MRI was acquired using an identical acquisition technique (n = 5). We have previously demonstrated that LAC/PYR correlates with the expression of the membrane transporter for pyruvate (monocarboxylic acid transporter 1, MCT1) in a cohort of treatment-naïve breast cancers (11). Here, we also observed a significant positive correlation between LAC/PYR at baseline and expression of the solute carrier 16A1 (SLC16A1), the gene encoding MCT1 (r = 0.92, P = 0.028; Fig. 2H), and in addition that LAC/PYR correlates with the expression of lactate dehydrogenase A (LDHA), the gene encoding a subunit of the enzyme LDH that catalyzes the exchange reaction between pyruvate and lactate (r = 0.90, P = 0.039; Fig. 2I). Correlations of SLC16A1 (MCT1) and LDHA gene expression with kPL were not significant (P = 0.984 and P = 0.924, respectively). LAC/PYR and kPL were not significantly correlated with gene expression of carbonic anhydrase 9 (CAIX) and HIF1A (P > 0.05).
Expression of LDHA, SLC16A1 (MCT1), HIF1A, and CAIX and survival in different breast cancer subtypes
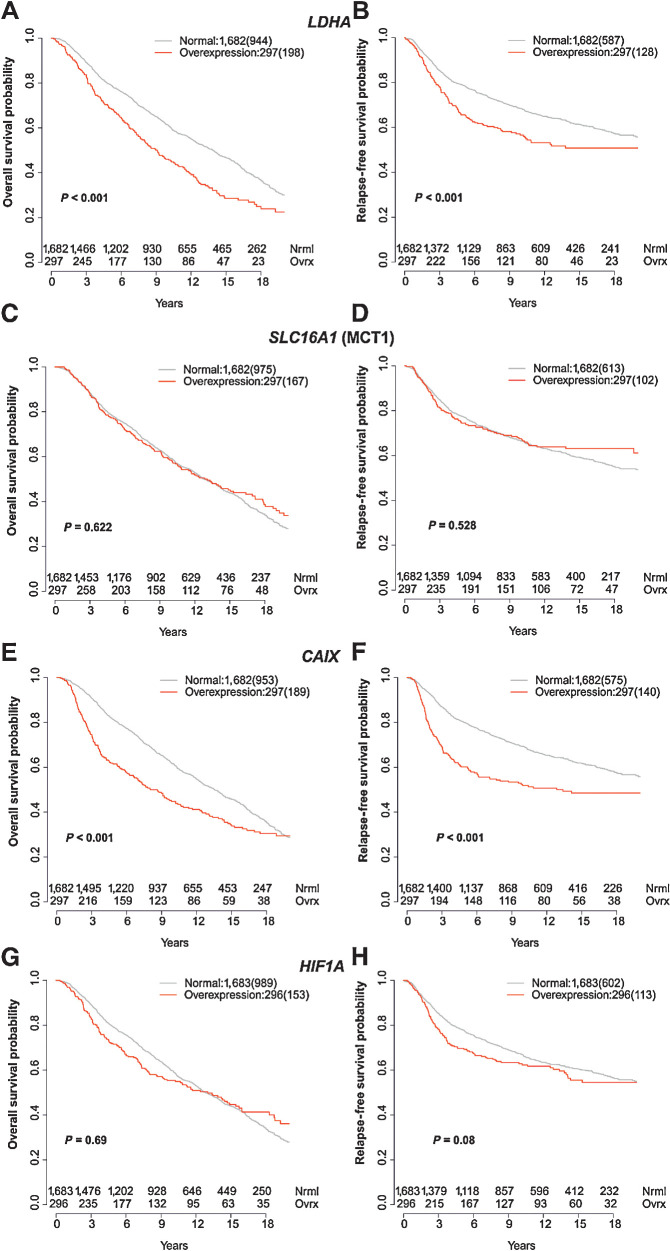
To further explore the significance of these findings, the intercorrelations of LDHA, SLC16A1 (MCT1), HIF1A, and CAIX expressions were analyzed in a larger cohort, to evaluate their association with survival. Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) is a large genomic and transcriptomic dataset acquired from approximately 2,000 breast cancer biopsy samples (27) with its outcome update published in 2019 (28). Although the expression of both, LDHA and SLC16A1 (MCT1), was significantly correlated with expression of HIF1A and CAIX, the expression of LDHA was not significantly correlated with SLC16A1 (MCT1) (Fig. 5). Overexpression of LDHA and the hypoxia marker CAIX [but not SLC16A1 (MCT1) or HIF1A] was also associated with poorer overall survival (OS) and relapse-free survival (Fig. 6).
Figure 6.
Correlation of LDHA, SLC16A1 (MCT1), CAIX, and HIF1A expression with survival in METABRIC. Kaplan–Meier curves for normal expression and overexpression (85th percentile) of LDHA (A and B), SLC16A1 (MCT) (C and D), HIF1A (E and F), and CAIX (G and H). The left column shows overall survival and the right column relapse-free survival. Number of events are shown in brackets.
In addition, we also selected 501 patients from METABRIC whose breast cancer receptor status matched the patients in our imaged cohort (ER−PR−HER2− = 320 patients; ER−PR−HER2+ = 134 patients; ER+PR+HER2+ = 47 patients). Although the expression of LDHA was significantly correlated with expression of HIF1A and CAIX, the expression of SLC16A1 (MCT1) was not significantly correlated with either of the two in this subset of patients (Supplementary Fig. S2I). Overexpression of the hypoxia markers CAIX and HIF1A was associated with poorer OS and overexpression of CAIX was also associated with relapse-free survival (Supplementary Fig. S2A–S2H).
Discussion
Most anticancer therapies induce metabolic alterations that precede the changes in tumor size that are measured routinely on standard-of-care imaging (29, 30). Therefore, noninvasive methods for imaging tumor metabolism offer the possibility for earlier detection of treatment response. Hyperpolarized 13C-MRI measurements using [1–13C]pyruvate as a metabolic probe have shown great promise for early therapy response monitoring in a large number of preclinical cancer models, including breast cancer. We and other groups have demonstrated how lactate labeling varies with tumor grade (11, 13). We have also shown that in breast cancer LAC/PYR correlates with both the expression of MCT1, which is responsible for the cellular uptake of pyruvate, and hypoxia as measured from HIF1α expression (11). To date, there have been two clinical case reports demonstrating a decrease in tumor lactate labeling following treatment in a TNBC and a high-grade prostate cancer after 3 weeks and 6 weeks of standard-of-care therapy, respectively (16, 31). The aim of this study was to evaluate the potential of hyperpolarized 13C-MRI to measure very early response to therapy in breast cancer, 7–11 days after initiation of treatment, and to compare it with 1H-MRI measures of vascular permeability using DCE-MRI and tissue cellularity using IVIM DWI.
The results show that an increase of ≥20% in LAC/PYR measured using hyperpolarized 13C-MRI 7–11 days after commencing standard-of-care treatment correctly predicted patients with eventual pCR at surgery. Changes in no other parameter, either using hyperpolarized 13C-MRI or advanced multiparametric 1H-MRI, could correctly identify patients with pCR. The threshold of 20% for determining response is in line with criteria such as RECIST or Positron Emission Tomography Response Criteria in Solid Tumors (PERCIST), where changes higher than 20% or 30% in size or metabolism, form the basis of determining tumor progression, response, or stable disease (30, 32). These results indicate that the technique holds promise for ultra-early response monitoring of NACT after 7–11 days of standard-of-care treatment, although this requires validation in larger cohorts.
The increase in LAC/PYR following treatment is interesting as the majority of preclinical hyperpolarized 13C-MRI studies have reported decreased lactate labeling following a positive response to therapy (16, 17, 20–22, 31, 33). However, a treatment-induced increase in lactate labeling has been shown in a smaller number of in vitro and in vivo models (18, 19, 34, 35). The accurate prediction of pCR with an increase in lactate labeling, rather than a decrease, raises important questions about the dynamic alterations in metabolism following treatment and the ideal timing for measuring a clinically meaningful metabolic response in future studies. Later timepoints may be dominated by opposing effects such as loss of cellularity, and this has important implications for the timing of treatment response monitoring, which are likely to be tumor- and treatment-specific.
There are a number of potential biological factors contributing to the changes in hyperpolarized 13C-label exchange between pyruvate and lactate following treatment (15). For example, the intracellular pyruvate concentration is determined by both tissue perfusion and membrane transport. Importantly, there were no significant differences in the DCE measures of vascularity (Ktrans, ve, or iAUC90) between baseline and the early treatment timepoint, suggesting that pyruvate delivery via the bloodstream alone cannot account for the changes in lactate labeling. Pyruvate transport is mediated by monocarboxylate transporters (MCT), of which, MCT1 and MCT4 are the most widely expressed in human tissue. MCT1 has a greater affinity for pyruvate and is the main transmembrane transporter for hyperpolarized [1–13C]pyruvate, which has been shown to be rate limiting for hyperpolarized [1–13C]lactate formation in some breast cancer cell lines and can account for treatment-induced changes (19, 36). We have previously demonstrated the importance of SLC16A1 (MCT1) expression for determining lactate labeling in treatment-naïve breast tumors (11), which we have confirmed within this cohort. MCT1 has also been shown to have a role in lactate labeling in prostate cancer (13). Therefore, alterations in SLC16A1 (MCT1) expression or its cellular localization could account for the changes in lactate labeling seen here following treatment. The absence of a significant change in vascular permeability or Ktrans (Fig. 2G) following treatment suggests that vascular delivery of pyruvate is not responsible for the changes in LAC/PYR seen following treatment and further supports a potential role for MCT1 in these changes.
Intracellularly, the enzyme lactate dehydrogenase (LDH) catalyzes the conversion of pyruvate to lactate in the presence of the cofactor NADH. A reduction in LDH expression has been shown to mediate reduced lactate labeling in hyperpolarized 13C-MRI experiments (17, 22, 33), as has a decrease in NADH (20, 21, 37), and a decrease in the intracellular lactate pool (20, 34). Conversely, increasing lactate pool size and LDH expression have been shown to be associated with increasing lactate labeling after treatment (19). Here, we have shown that LDHA expression on an individual patient level, in addition to MCT1, correlates with the LAC/PYR ratio. Taken together with the previous data we have reported, these suggest that changes in LDHA, in addition to MCT1, may explain the after treatment changes we have shown here.
Hypoxia may be the driver for the relationship between LAC/PYR and both MCT1 and LDHA, as well as the changes in LAC/PYR seen after treatment. Tissue lactate is closely related to hypoxia and we have shown previously a correlation between HIF1A expression and LAC/PYR in a varied group of treatment-naïve breast cancers (11). Angiogenesis inhibitors have been shown to increase lactate labeling in a preclinical model of ovarian cancer assessed with hyperpolarized 13C-MRI (18). Antiangiogenesis is also a well-documented effect of taxanes (38) and all patients in our cohort received either weekly doses of paclitaxel or three-weekly docetaxel (Supplementary Table S1). In the absence of significant changes in the measured pharmacokinetic parameters (Ktrans, ve, or iAUC90) on DCE-MRI, any changes in hypoxia are likely to be driven by cellular changes in oxygen demand rather than by changes in the vasculature. Immune cell infiltration has been shown to contribute to the uptake of the radiolabelled glucose analog [18F]fluorodeoxyglucose (18F-FDG) on PET in patients with breast cancer following response to hormonal therapy (39, 40). An increase in tumor-infiltrating immune cells in responders could therefore also increase LAC/PYR more strongly as seen in our study. Future analysis of tumor tissue at this very early response assessment timepoint will further elucidate the mechanisms responsible for the increasing LAC/PYR in responding patients. Very early treatment-induced changes in cellular redox status could also contribute to these findings. For example, the early effects of treatment may generate a reducing environment with increasing NADH-favoring lactate formation, whereas a later oxidized state would increase the relative concentration of NAD+ favoring conversion from lactate-to-pyruvate at later stages of treatment (41). However, tissue redox status is difficult to assess in practice, particularly at multiple timepoints during treatment but future validation of our results with assays quantifying NAD+ (and protein expression of MCT1, LDHA, CAIX, and HIF1A) will be important to fully understand the molecular changes underlying our imaging results.
To validate some of our findings in an external cohort, and to understand the clinical significance of the results, we subsequently analyzed the expression data from almost 2,000 patients in the METABRIC dataset (27, 28). In addition to SLC16A1 (MCT1) and LDHA, CAIX was assessed as a stable membrane-bound isoform of the enzyme carbonic anhydrase that is upregulated in hypoxic conditions and has been shown to be important for pH regulation of tumors (42). LDHA and SLC16A1 (MCT1) expressions were significantly correlated with expression of both hypoxia markers, CAIX and HIF1A, showing the important interrelationship between pyruvate metabolism, lactate formation, and hypoxia. There was a strong correlation between overexpression of both LDHA and CAIX and a statistically significant reduction in relapse-free survival and OS. Therefore, LAC/PYR may provide important prognostic information in addition to detection of early treatment response. It also suggests the importance of combining imaging measures of hypoxia and metabolism with histopathological metrics to more fully phenotype tumors, as well as for prognosis.
Olaparib inhibits PARP1 and 2, two enzymes functioning as DNA damage sensors and facilitators of DNA damage repair mechanisms by using NAD+ as a substrate to PARylate themselves and other target proteins (43, 44). Although up to 90% of cellular NAD+ can be used by PARPs in response to cellular DNA damage, PARP inhibitors compete with NAD+ for the catalytic cages of PARPs and trap PARPs at DNA double-strand breaks to impair PARylation and the DNA damage repair it mediates (44–46). Both patients receiving a PARP inhibitor in addition to standard-of-care treatment (one with eventual pCR and another one with non-pCR) also demonstrated an increase in LAC/PYR greater than 20%, and the increase in LAC/PYR was much higher in the patient with pCR than non-pCR, again pointing toward a greater increase in responders. PARP inhibitors have been shown to increase NAD+ levels in both murine cells and liver tissue (47), and decreasing NAD+ levels have been correlated with decreasing lactate labeling in hyperpolarized 13C-MRI experiments previously (20,21,37). These results suggest that increasing availability of intracellular NAD(H) due to PARP inhibitor treatment may elevate LDH-mediated lactate labeling. PARP inhibitors might, therefore, cause a pharmacodynamic increase in LAC/PYR and kPL regardless of response, and the larger increase in the patient who responded may be accounted for by additional changes in MCT1 and/or LDHA, or an increase in immune cell infiltration. A previous study in the MCF-7 breast cancer cell line has shown that lactate increases with an MEK inhibitor, which was explained by activation of the PI3K and/or AMPK pathways (34). These results suggest that caution is warranted in interpreting results in patients undergoing experimental targeted treatment where the mode of action could influence lactate production.
Pharmacokinetic parameters from DCE-MRI and diffusivity from DWI (including IVIM) have also been explored for early response prediction in breast cancer, with Ktrans typically decreasing and diffusivity increasing in responders compared with nonresponders (48–50). However, in most studies these assessments are made later during NACT than in this study. Our data showed that the perfusion fraction, f, determined using IVIM differs between pCR and non-pCR even at baseline in this cohort. A previous study found significant differences in baseline measurements of f in patients with eventual pCR when compared with those with non-pCR, although f was higher in patients with pCR in their cohort, which contrasts with our results (48). The different composition of the cohorts may account for this difference given the inclusion of mainly ER+ breast cancers previously. As with the findings from hyperpolarized 13C-MRI, these IVIM-derived metrics need validation in larger cohorts. Also, diffusivity D was not found to predict pathological response in our cohort, indicating that there was no significant change in cellularity that could account for the alterations in lactate labeling at this early treatment response timepoint.
We also showed that LAC/PYR is the most reliable metric to distinguish between responders and nonresponders undergoing standard-of-care treatment in our study. Results for kPL were similar, although there was a smaller difference between responders and nonresponders and fewer available data. As the estimation of kPL depends on the SNR in images acquired at single time points, it is less accurate at lower SNR than LAC/PYR, which is calculated from the time-summed images. Our results suggest that LAC/PYR may be the most precise metric for use in the clinical setting to assess a 20% change following treatment.
The limitations of our study include the small size of the dataset. The preliminary findings arising from this work will need to be confirmed and validated in larger cohorts, ideally as part of large multisite studies. However, this was a prospective study and we included patients with the two most common breast cancer subtypes who frequently undergo NACT: TNBC and HER2+. Despite the small group size, we show in both groups undergoing standard-of-care treatment that an increase of LAC/PYR of at least 20% indicates pCR and that in both the pCR and non-pCR groups there was one patient treated with a PARP inhibitor in addition. We have explored the significance of this small dataset by analyzing the very large expression dataset within METABRIC that has shown the potential importance of the technique in demonstrating prognosis.
In conclusion, an increase in the LAC/PYR ratio of ≥20%, measured using hyperpolarized 13C-MRI, has the potential to distinguish between responding and nonresponding patients with breast cancer undergoing standard-of-care neoadjuvant treatment as early as 7–11 days after the start of treatment. LAC/PYR is significantly correlated with both LDHA and SLC16A1 (MCT1) expressions in this cohort. Comparison with a large breast cancer gene expression dataset (METABRIC) showed that LDHA and SLC16A1 (MCT1) expressions are significantly correlated with expression of the hypoxia markers CAIX and HIF1A and that overexpression of the hypoxia markers CAIX and LDHA is significantly associated with shorter OS and relapse-free survival in breast cancer.
Authors' Disclosures
R. Woitek reports grants from AstraZeneca and Cancer Research UK (CRUK), Illumina and CRUK, Austrian Science Fund, and non-financial support from GE Healthcare, and grants from Mark Foundation for Cancer Research, Cancer Research UK Cambridge Center (grant C9685/A25177), Cancer Research UK (CRUK; grants C8742/A18097, C19212/A16628, C19212/A911376, and C197/A16465), CRUK Cambridge Center, the CRUK and Engineering and Physical Sciences Research Council Cancer Imaging Center in Cambridge and Manchester, Addenbrooke's Charitable Trust, and the NIHR Cambridge Biomedical Research Center, and non-financial support from Cambridge Breast Cancer Research Unit Laboratory during the conduct of the study. L. Beer reports grants from AstraZeneca and Cancer Research UK (CRUK), Illumina and CRUK, The Mark Foundation for Cancer Research, Cancer Research UK Cambridge Center, Cancer Research UK, and CRUK Cambridge Center, the CRUK and Engineering and Physical Sciences Research Council Cancer Imaging Center in Cambridge and Manchester, Addenbrooke's Charitable Trust, and the NIHR Cambridge Biomedical Research Center, and non-financial support from Cambridge Breast Cancer Research Unit Laboratory and GE Healthcare during the conduct of the study. G. Baxter reports grants from AstraZeneca and Cancer Research UK (CRUK), Illumina and CRUK, The Mark Foundation for Cancer Research, Cancer Research UK Cambridge Center, Cancer Research UK, and CRUK Cambridge Center, the CRUK and Engineering and Physical Sciences Research Council Cancer Imaging Center in Cambridge and Manchester, Addenbrooke's Charitable Trust, and the NIHR Cambridge Biomedical Research Center, and non-financial support from Cambridge Breast Cancer Research Unit Laboratory and GE Healthcare during the conduct of the study. L. Rundo reports personal fees from Lucida Medical Ltd. outside the submitted work. E. Provenzano reports grants from AstraZeneca and Cancer Research UK (CRUK), Illumina and CRUK, The Mark Foundation for Cancer Research, Cancer Research UK Cambridge Center, CRUK Cambridge Center, the CRUK and Engineering and Physical Sciences Research Council Cancer Imaging Center in Cambridge and Manchester, Addenbrooke's Charitable Trust, and the NIHR Cambridge Biomedical Research Center, non-financial support from Cambridge Breast Cancer Research Unit Laboratory, and non-financial support from GE Healthcare, and personal fees from NIHR Cambridge Biomedical Research Center during the conduct of the study; personal fees and non-financial support from Roche Pharmaceuticals outside the submitted work. J.D. Kaggie reports grants from National Institute of Health Research (NIHR) Cambridge during the conduct of the study; as well as grants from GlaxoSmithKline and grants from EU Horizon 2020 outside the submitted work. A. Frary reports grants from AstraZeneca and Cancer Research UK (CRUK), Illumina and CRUK, The Mark Foundation for Cancer Research, Cancer Research UK Cambridge Center, Cancer Research UK, CRUK Cambridge Center, the CRUK and Engineering and Physical Sciences Research Council Cancer Imaging Center in Cambridge and Manchester, Addenbrooke's Charitable Trust, and the NIHR Cambridge Biomedical Research Center, and non-financial support from Cambridge Breast Cancer Research Unit Laboratory and GE Healthcare during the conduct of the study. J. Kane reports grants from AstraZeneca and Cancer Research UK (CRUK), Illumina and CRUK, The Mark Foundation for Cancer Research, Cancer Research UK Cambridge Center, Cancer Research UK, CRUK Cambridge Center, the CRUK and Engineering and Physical Sciences Research Council Cancer Imaging Center in Cambridge and Manchester, Addenbrooke's Charitable Trust, and the NIHR Cambridge Biomedical Research Center, non-financial support from Cambridge Breast Cancer Research Unit Laboratory and GE Healthcare during the conduct of the study. A.J.V. Benjamin reports grants from AstraZeneca and Cancer Research UK (CRUK), Illumina and CRUK, The Mark Foundation for Cancer Research, Cancer Research UK Cambridge Center, Cancer Research UK, CRUK Cambridge Center, the CRUK and Engineering and Physical Sciences Research Council Cancer Imaging Center in Cambridge and Manchester, Addenbrooke's Charitable Trust, and the NIHR Cambridge Biomedical Research Center, and non-financial support from Cambridge Breast Cancer Research Unit Laboratory and GE Healthcare during the conduct of the study; as well as grants from National Cancer Imaging Translational Accelerator outside the submitted work. A.N. Priest reports personal fees from GE Healthcare outside the submitted work. B. White reports grants from AstraZeneca and Cancer Research UK (CRUK), Illumina and CRUK, The Mark Foundation for Cancer Research, Cancer Research UK Cambridge Center, Cancer Research UK, CRUK Cambridge Center, the CRUK and Engineering and Physical Sciences Research Council Cancer Imaging Center in Cambridge and Manchester, Addenbrooke's Charitable Trust, and the NIHR Cambridge Biomedical Research Center, and non-financial support from Cambridge Breast Cancer Research Unit Laboratory and GE Healthcare during the conduct of the study. B. Carmo reports grants from AstraZeneca and Cancer Research UK (CRUK), Illumina and CRUK, The Mark Foundation for Cancer Research, Cancer Research UK Cambridge Center, Cancer Research UK, CRUK Cambridge Center, the CRUK and Engineering and Physical Sciences Research Council Cancer Imaging Center in Cambridge and Manchester, Addenbrooke's Charitable Trust, and the NIHR Cambridge Biomedical Research Center, as well as grants and non-financial support from Cambridge Breast Cancer Research Unit Laboratory, and non-financial support from GE Healthcare during the conduct of the study. R. Slough reports grants from AstraZeneca and Cancer Research UK (CRUK), Illumina and CRUK, The Mark Foundation for Cancer Research, Cancer Research UK Cambridge Center, Cancer Research UK, CRUK Cambridge Center, the CRUK and Engineering and Physical Sciences Research Council Cancer Imaging Center in Cambridge and Manchester, Addenbrooke's Charitable Trust, and the NIHR Cambridge Biomedical Research Center, and non-financial support from Cambridge Breast Cancer Research Unit Laboratory and GE Healthcare during the conduct of the study. R.F. Schulte reports other support from GE Healthcare during the conduct of the study; and other support from GE Healthcare outside the submitted work. S. Chin reports grants from AstraZeneca and Cancer Research UK (CRUK), Illumina and CRUK, The Mark Foundation for Cancer Research, Cancer Research UK Cambridge Center, Cancer Research UK, CRUK Cambridge Center, the CRUK and Engineering and Physical Sciences Research Council Cancer Imaging Center in Cambridge and Manchester, Addenbrooke's Charitable Trust, and the NIHR Cambridge Biomedical Research Center, and non-financial support from Cambridge Breast Cancer Research Unit Laboratory and GE Healthcare during the conduct of the study. M. Graves reports grants from AstraZeneca and Cancer Research UK (CRUK), Illumina and CRUK, The Mark Foundation for Cancer Research, Cancer Research UK Cambridge Center, Cancer Research UK, CRUK Cambridge Center, the CRUK and Engineering and Physical Sciences Research Council Cancer Imaging Center in Cambridge and Manchester, Addenbrooke's Charitable Trust, and the NIHR Cambridge Biomedical Research Center, non-financial support from Cambridge Breast Cancer Research Unit Laboratory, GE Healthcare, and Rapid Biomedical during the conduct of the study; and personal fees from GE Healthcare and Bayer outside the submitted work. F.J. Gilbert reports grants from AstraZeneca and Cancer Research UK (CRUK), Illumina and CRUK, The Mark Foundation for Cancer Research, Cancer Research UK Cambridge Center, Cancer Research UK, NIHR Cambridge Biomedical Research Center, and non-financial support from GE Healthcare during the conduct of the study; and President European Society of Breast Imaging. J. Abraham reports grants from AstraZeneca and other support from Illumina during the conduct of the study; grants from AstraZeneca and other support from Illumina outside the submitted work. C. Caldas reports grants from AstraZeneca, Cancer Research UK, and Illumina during the conduct of the study; grants from AstraZeneca and Cancer Research UK outside the submitted work; and research grants administered by the University of Cambridge: AstraZeneca, Roche, Servier, Genentech, Varsity Therapeutics; and is a member of external science panel of AstraZeneca iMED and Illumina SAB. K.M. Brindle reports other support from GE Healthcare during the conduct of the study; as well as has a patent for WO2008020764 A1 issued. E. Sala reports grants from AstraZeneca and Cancer Research UK (CRUK), Illumina and CRUK, The Mark Foundation for Cancer Research, Cancer Research UK Cambridge Center, Cancer Research UK, CRUK Cambridge Center, the CRUK and Engineering and Physical Sciences Research Council Cancer Imaging Center in Cambridge and Manchester, Addenbrooke's Charitable Trust, and the NIHR Cambridge Biomedical Research Center, grants and non-financial support from Cambridge Breast Cancer Research Unit Laboratory, and GE Healthcare during the conduct of the study; personal fees from GlaxoSmithKline and other support from Lucida Medical outside the submitted work. F.A. Gallagher reports grants from AstraZeneca/CRUK, Illumina/CRUK, CRUK Cambridge Center, CRUK, NIHR Cambridge Biomedical Research Center, and non-financial support from Cambridge Breast Cancer Research Unit Laboratory, and GE Healthcare during the conduct of the study; as well as grants from GlaxoSmithKline, non-financial support from GE Healthcare, and other support from AstraZeneca outside the submitted work. No disclosures were reported by the other authors.
Supplementary Material
Supplementary Materials (clean)
Acknowledgments
This work was supported by the Mark Foundation for Cancer Research and Cancer Research UK Cambridge Centre (grant C9685/A25177), Cancer Research UK (CRUK; grants C8742/A18097, C19212/A16628, C19212/A27150, and C197/A16465), the Austrian Science Fund (grant J4025-B26), the CRUK Cambridge Center, the CRUK and Engineering and Physical Sciences Research Council Cancer Imaging Center in Cambridge and Manchester, Addenbrooke's Charitable Trust, and the NIHR Cambridge Biomedical Research Center (BRC-1215-20014). The NIHR Cambridge Biomedical Research Center (BRC) is a partnership between Cambridge University Hospitals NHS Foundation Trust and the University of Cambridge, funded by the National Institute for Health Research (NIHR). The views expressed are those of the authors and not necessarily those of the NIHR or the Department of Health and Social Care. The authors also acknowledge funding from the UKRI Medical Research Council (MC UU 00002/16), the Cambridge Experimental Cancer Medicine Center, a Wellcome Trust Strategic Award (095962), and Cambridge University Hospitals National Health Service Foundation Trust. They acknowledge the support of the Cambridge Breast Cancer Research Unit Laboratory in the collection of clinical data and samples and the Cancer Molecular Diagnostics for the extraction of DNA/RNA. The PARTNER Trial (https://clinicaltrials.gov/ct2/show/NCT03150576) was supported by Cancer Research UK and AstraZeneca (CRUK/14/048 and ESR-14-10241). The Personalized Breast Cancer Program is a collaboration between CRUK Cambridge Center and Illumina (C507/A27657). The 3D models of MCT1 and LDH used in the graphical abstract were modified after obtaining them from the National Institutes of Health 3D Print Exchange (https://3dprint.nih.gov/discover/3DPX-013007) and the Biozentrum, University of Basel, Switzerland, The Center for Molecular Life Sciences (https://swissmodel.expasy.org/repository/uniprot/P53985?template=7da5). The authors acknowledge the significant contributions of Ricardo Godinho in preparing the artwork for this article. The authors also acknowledge and are grateful for the involvement of our patients in these research projects. For the purpose of open access, the author has applied a CC BY public copyright license to any Author Accepted Manuscript version arising from this submission.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Contributions
R. Woitek: Conceptualization, resources, data curation, software, formal analysis, investigation, visualization, methodology, writing–original draft. M.A. McLean: Formal analysis, supervision, investigation, methodology, writing–review and editing. S. Ursprung: Data curation, formal analysis, investigation, visualization, methodology, writing–review and editing. O.M. Rueda: Data curation, investigation, visualization, methodology, writing–review and editing. R. Manzano Garcia: Formal analysis, writing–review and editing. M.J. Locke: Resources, project administration, writing–review and editing. L. Beer: Investigation, writing–review and editing. G. Baxter: Formal analysis, writing–review and editing. L. Rundo: Formal analysis, visualization, writing–review and editing. E. Provenzano: Formal analysis, supervision, investigation, writing–review and editing. J.D. Kaggie: Formal analysis, writing–review and editing. A. Patterson: Formal analysis, supervision, investigation, visualization, methodology, writing–review and editing. A. Frary: Investigation, project administration. J. Field-Rayner: Investigation, project administration. V. Papalouka: Investigation, writing–review and editing. J. Kane: Investigation, project administration. A.J.V. Benjamin: Formal analysis, investigation. A.B. Gill: Formal analysis, visualization, methodology, writing–review and editing. A.N. Priest: Formal analysis, writing–review and editing. D.Y. Lewis: Formal analysis, writing–review and editing. R. Russell: Formal analysis, writing–review and editing. A. Grimmer: Resources, investigation, and methodology. B. White: Resources, investigation. B. Latimer-Bowman: Investigation, methodology. I. Patterson: Resources, investigation. A. Schiller: Resources, investigation. B. Carmo: Investigation. R. Slough: Investigation. T. Lanz: Resources, writing–review and editing. J. Wason: Conceptualization, formal analysis, writing–review and editing. R.F. Schulte: Resources, methodology. S. Chin: Formal analysis, supervision, investigation, methodology, writing–review and editing. M.J. Graves: Conceptualization, resources, software, formal analysis, supervision, investigation, methodology, writing–review and editing. F.J. Gilbert: Supervision, investigation, writing–review and editing. J. Abraham: Conceptualization, supervision, funding acquisition, writing–review and editing. C. Caldas: Conceptualization, resources, formal analysis, supervision, funding acquisition, investigation, visualization, methodology, writing–review and editing. K.M. Brindle: Conceptualization, formal analysis, supervision, funding acquisition, investigation, methodology, writing–review and editing. E. Sala: Supervision, funding acquisition, writing–review and editing. F.A. Gallagher: Conceptualization, resources, formal analysis, supervision, funding acquisition, investigation, methodology, project administration, writing–review and editing.
References
- 1. Murphy BL, Day CN, Hoskin TL, Habermann EB, Boughey JC. Neoadjuvant chemotherapy use in breast cancer is greatest in excellent responders: triple-negative and HER2+ subtypes. Ann Surg Oncol 2018;25:2241–8. [DOI] [PubMed] [Google Scholar]
- 2. van Ramshorst MS, van der Voort A, van Werkhoven ED, Mandjes IA, Kemper I, Dezentjé VO, et al. Neoadjuvant chemotherapy with or without anthracyclines in the presence of dual HER2 blockade for HER2-positive breast cancer (TRAIN-2): a multicentre, open-label, randomised, phase 3 trial. Lancet Oncol 2018;19:1630–40. [DOI] [PubMed] [Google Scholar]
- 3. Masuda N, Lee S-J, Ohtani S, Im Y-H, Lee E-S, Yokota I, et al. Adjuvant capecitabine for breast cancer after preoperative chemotherapy. N Engl J Med 2017;376:2147–59. [DOI] [PubMed] [Google Scholar]
- 4. von Minckwitz G, Huang C-S, Mano MS, Loibl S, Mamounas EP, Untch M, et al. Trastuzumab emtansine for residual invasive HER2-positive breast cancer. N Engl J Med 2019;380:617–28. [DOI] [PubMed] [Google Scholar]
- 5. Cortes J, Gebhart G, Ruiz Borrego M, Stradella A, Bermejo B, Escrivá S, et al. Chemotherapy (CT) de-escalation using an FDG-PET/CT (F-PET) and pathological response-adapted strategy in HER2[+] early breast cancer (EBC): PHERGain Trial. J Clin Oncol 2020;38:503. [Google Scholar]
- 6. Cheng Q, Huang J, Liang J, Ma M, Ye K, Shi C, et al. The diagnostic performance of DCE-MRI in evaluating the pathological response to neoadjuvant chemotherapy in breast cancer: a meta-analysis. Front Oncol 2020;10:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Partridge SC, Zhang Z, Newitt DC, Gibbs JE, Chenevert TL, Rosen MA, et al. Diffusion-weighted MRI findings predict pathologic response in neoadjuvant treatment of breast cancer: the ACRIN 6698 multicenter Trial. Radiology 2018;289:618–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Castagnoli L, Iorio E, Dugo M, Koschorke A, Faraci S, Canese R, et al. Intratumor lactate levels reflect HER2 addiction status in HER2-positive breast cancer. J Cell Physiol 2019;234:1768–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gandhi N, Das G. Metabolic reprogramming in breast cancer and its therapeutic implications. Cells 2019;8:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim S, Kim DH, Jung W-H, Koo JS. Metabolic phenotypes in triple-negative breast cancer. Tumor Biol 2013;34:1699–712. [DOI] [PubMed] [Google Scholar]
- 11. Gallagher FA, Woitek R, McLean MA, Gill AB, Manzano Garcia R, Provenzano E, et al. Imaging breast cancer using hyperpolarized carbon-13 MRI. Proc Natl Acad Sci U S A 2020;117:2092–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grist JT, Miller JJ, Zaccagna F, McLean MA, Riemer F, Matys T, et al. Hyperpolarized 13C MRI: a novel approach for probing cerebral metabolism in health and neurological disease. J Cereb Blood Flow Metab 2020;40:1137–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Granlund KL, Tee SS, Vargas HA, Lyashchenko SK, Reznik E, Fine S, et al. Hyperpolarized MRI of human prostate cancer reveals increased lactate with tumor grade driven by monocarboxylate transporter 1. Cell Metab 2020;31:105–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stødkilde-Jørgensen H, Laustsen C, Hansen ESS, Schulte R, Ardenkjaer-Larsen JH, Comment A, et al. Pilot study experiences with hyperpolarized [1-13 C]pyruvate MRI in pancreatic cancer patients. J Magn Reson Imaging 2020;51:961–3. [DOI] [PubMed] [Google Scholar]
- 15. Woitek R, Gallagher FA. The use of hyperpolarised 13C-MRI in clinical body imaging to probe cancer metabolism. Br J Cancer 2021;124:1187–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Woitek R, McLean M, Gill AB, Grist JT, Provenzano E, Patterson AJ, et al. Hyperpolarized 13C-MRI of tumor metabolism demonstrates early metabolic response to neoadjuvant chemotherapy in breast cancer. Radiol Imaging Cancer 2021;2:e200017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hesketh RL, Wang J, Wright AJ, Lewis DY, Denton AE, Grenfell R, et al. Magnetic resonance imaging is more sensitive than PET for detecting treatment-induced cell death-dependent changes in glycolysis. Cancer Res 2019;79:3557–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ravoori MK, Singh SP, Lee J, Bankson JA, Kundra V. In vivo assessment of ovarian tumor response to tyrosine kinase inhibitor pazopanib by using hyperpolarized 13C-pyruvate MR spectroscopy and 18F-FDG PET/CT imaging in a mouse model. Radiology 2017;285:830–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lodi A, Woods SM, Ronen SM. Treatment with the MEK inhibitor U0126 induces decreased hyperpolarized pyruvate to lactate conversion in breast, but not prostate, cancer cells. NMR Biomed 2013;26:299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Day SE, Kettunen MI, Gallagher FA, Hu D-E, Lerche M, Wolber J, et al. Detecting tumor response to treatment using hyperpolarized 13C magnetic resonance imaging and spectroscopy. Nat Med 2007;13:1382–7. [DOI] [PubMed] [Google Scholar]
- 21. Witney TH, Kettunen MI, Hu D, Gallagher FA, Bohndiek SE, Napolitano R, et al. Detecting treatment response in a model of human breast adenocarcinoma using hyperpolarised [1–13C]pyruvate and [1,4–13C2]fumarate. Br J Cancer 2010;103:1400–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ros S, Wright AJ, D'Santos P, en Hu D, Hesketh RL, Lubling Y, et al. Metabolic imaging detects resistance to PI3Kα inhibition mediated by persistent FOXM1 expression in ER+ breast cancer. Cancer Cell 2020;38:516–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bedair R, Graves MJ, Patterson AJ, McLean MA, Manavaki R, Wallace T, et al. Effect of radiofrequency transmit field correction on quantitative dynamic contrast-enhanced MR imaging of the breast at 3.0 T. Radiology 2016;279:368–77. [DOI] [PubMed] [Google Scholar]
- 24. Schulte RF, Sperl JI, Weidl E, Menzel MI, Janich MA, Khegai O, et al. Saturation-recovery metabolic-exchange rate imaging with hyperpolarized [1–13C] pyruvate using spectral-spatial excitation. Magn Reson Med 2013;69:1209–16. [DOI] [PubMed] [Google Scholar]
- 25. Wiesinger F, Weidl E, Menzel MI, Janich MA, Khegai O, Glaser SJ, et al. IDEAL spiral CSI for dynamic metabolic MR imaging of hyperpolarized [1–13C]pyruvate. Magn Reson Med 2012;68:8–16. [DOI] [PubMed] [Google Scholar]
- 26. Rodgers CT, Robson MD. Receive array magnetic resonance spectroscopy: whitened singular value decomposition (WSVD) gives optimal bayesian solution. Magn Reson Med 2010;63:881–91. [DOI] [PubMed] [Google Scholar]
- 27. Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012;486:346–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rueda OM, Sammut SJ, Seoane JA, Chin SF, Caswell-Jin JL, Callari M, et al. Dynamics of breast-cancer relapse reveal late-recurring ER-positive genomic subgroups. Nature 2019;567:399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kurhanewicz J, Vigneron DB, Ardenkjaer-Larsen JH, Bankson JA, Brindle K, Cunningham CH, et al. Hyperpolarized 13C MRI: path to clinical translation in oncology. Neoplasia 2019;21:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Eisenhauer EAA, Therasse P, Bogaerts J, Schwartz LHH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228–47. [DOI] [PubMed] [Google Scholar]
- 31. Aggarwal R, Vigneron DB, Kurhanewicz J. Hyperpolarized 1-[13C]-pyruvate magnetic resonance imaging detects an early metabolic response to androgen ablation therapy in prostate cancer. Eur Urol 2017;72:1028–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wahl RL, Jacene H, Kasamon Y, Lodge MA. From RECIST to PERCIST: evolving considerations for PET response criteria in solid tumors. J Nucl Med 2009;50:122–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dafni H, Larson PEZ, Hu S, Yoshihara HAI, Ward CS, Venkatesh HS, et al. Hyperpolarized 13C spectroscopic imaging informs on hypoxia-inducible factor-1 and Myc activity downstream of platelet-derived growth factor receptor. Cancer Res 2010;70:7400–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lodi A, Woods SM, Ronen SM. MR-detectable metabolic consequences of mitogen-activated protein kinase kinase (MEK) inhibition. NMR Biomed 2014;27:700–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bohndiek SE, Kettunen MI, Hu D, Brindle KM. Hyperpolarized (13)C spectroscopy detects early changes in tumor vasculature and metabolism after VEGF neutralization. Cancer Res 2012;72:854–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Talia H, Galit E, Lucio F, Hadassa D. Kinetics of hyperpolarized 13C1-pyruvate transport and metabolism in living human breast cancer cells. Proc Natl Acad Sci U S A 2009;106:18131–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Witney TH, Kettunen MI, Day SE, Hu D, Neves AA, Gallagher FA, et al. A comparison between radiolabeled fluorodeoxyglucose uptake and hyperpolarized (13)C-labeled pyruvate utilization as methods for detecting tumor response to treatment. Neoplasia 2009;11:574–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bocci G, Di Paolo A, Danesi R. The pharmacological bases of the antiangiogenic activity of paclitaxel. Angiogenesis 2013;16:481–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mamede M, Saga T, Ishimori T, Nakamoto Y, Sato N, Higashi T, et al. Differential uptake of 18F-fluorodeoxyglucose by experimental tumors xenografted into immunocompetent and immunodeficient mice and the effect of immunomodification. Neoplasia 2003;5:179–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mortimer JE, Dehdashti F, Siegel BA, Trinkaus K, Katzenellenbogen JA, Welch MJ. Metabolic flare: indicator of hormone responsiveness in advanced breast cancer. J Clin Oncol 2001;19:2797–803. [DOI] [PubMed] [Google Scholar]
- 41. Broekgaarden M, Bulin A-L, Frederick J, Mai Z, Hasan T. Clinical medicine tracking photodynamic-and chemotherapy-induced redox-state perturbations in 3D culture models of pancreatic cancer: a tool for identifying therapy-induced metabolic changes. J Clin Med 2019;8:1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Serrao EM, Kettunen MI, Rodrigues TB, Dzien P, Wright AJ, Gopinathan A, et al. MRI with hyperpolarised [1–13C]pyruvate detects advanced pancreatic preneoplasia prior to invasive disease in a mouse model. Gut 2016;65:465–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Livraghi L, Garber JE. PARP inhibitors in the management of breast cancer: current data and future prospects. BMC Med 2015;13:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bian C, Zhang C, Luo T, Vyas A, Chen SH, Liu C, et al. NADP+ is an endogenous PARP inhibitor in DNA damage response and tumor suppression. Nat Commun 2019;10:693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer 2010;10:293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu C, Vyas A, Kassab MA, Singh AK, Yu X. The role of poly ADP-ribosylation in the first wave of DNA damage response. Nucleic Acids Res 2017;45:8129–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Almeida GS, Bawn CM, Galler M, Wilson I, Thomas HD, Kyle S, et al. PARP inhibitor rucaparib induces changes in NAD levels in cells and liver tissues as assessed by MRS. NMR Biomed 2017;30:2797–803. [DOI] [PubMed] [Google Scholar]
- 48. Che S, Zhao X, Ou Y, Li J, Wang M, Wu B, et al. Role of the intravoxel incoherent motion diffusion weighted imaging in the pre-treatment prediction and early response monitoring to neoadjuvant chemotherapy in locally advanced breast cancer. Medicine 2016;95:e2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bedair R, Priest AN, Patterson AJ, McLean MA, Graves MJ, Manavaki R, et al. Assessment of early treatment response to neoadjuvant chemotherapy in breast cancer using non–mono-exponential diffusion models: a feasibility study comparing the baseline and mid-treatment MRI examinations. Eur Radiol 2017;27:2726–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tudorica A, Oh KY, Chui SY-C, Roy N, Troxell ML, Naik A, et al. Early prediction and evaluation of breast cancer response to neoadjuvant chemotherapy using quantitative DCE-MRI. Transl Oncol 2016;9:8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials (clean)
Data Availability Statement
Transcriptomic data for those tumors included in our study imaged with hyperpolarized 13C-MRI are deposited at the European Genome-phenome Archive (https://ega-archive.org/datasets/EGAD00001008141). Imaging raw data and MATLAB scripts for data in this article can be obtained from radiology-13c-mri-breast@lists.cam.ac.uk.