Abstract
A substantial fraction of the human genome displays high sequence similarity with at least one other genomic sequence, posing a challenge for the identification of somatic mutations from short-read sequencing data. We here annotate genomic variants in 2,658 cancers from the Pan-Cancer Analysis of Whole Genomes (PCAWG) cohort with links to similar sites across the genome. We train a machine-learning model to use signals distributed over multiple genomic sites to call somatic events in non-unique regions and validate it against linked-read sequencing in an independent dataset. Using this approach, we uncover previously hidden mutations in ~1,700 coding sequences and in thousands of regulatory elements, including in known cancer genes, immunoglobulins, and highly mutated gene families. Mutations in non-unique regions are consistent with mutations in unique regions in terms of mutation burden and substitution profiles. The analysis provides a systematic summary of the mutation events in non-unique regions at a genome-wide scale across multiple human cancers.
Introduction
Catalogs of somatic mutations in cancer promise insights into disease-initiating pathways, underlying evolutionary processes, and the identification of potential therapeutic opportunities. Large-cohort studies, for example The Cancer Genome Atlas (TCGA, https://www.cancer.gov/tcga) and the International Cancer Genome Consortium (ICGC, https://dcc.icgc.org/), have shed light on the complexity of the mutational landscapes in gene-coding regions. The Pan-Cancer Analysis of Whole Genomes (PCAWG) study has further undertaken the analysis of 2,658 whole cancer genomes from the ICGC and TCGA to characterize regions unobserved via exome sequencing studies1. This resource has led to studies of the processes underlying somatic events2, complex structural variation3, driver genes4, and timing5. However, these analyses rely on variants that can be positioned uniquely in the genome and thus the repertoire of variation in non-unique regions still remains unexplored.
Short-read sequencing - the technology used in cancer studies including PCAWG - identifies somatic mutations by comparing fragments of DNA from normal and tumor samples to a reference genome. However, the human genome contains sequences that are present at more than one chromosomal location6,7. At the scale of 100bp, 13% of the genome is affected (9% at 150bp, 8% at 200bp). These regions, which are spread throughout the chromosomes7,8, range in multiplicity from two to several thousand copies and in identity of sequences from vague to perfect matches. This non-uniqueness complicates genetic analyses and creates recurrent blind spots to somatic mutation calling. Irrespective of their amenability to analysis, non-unique regions include genes and regulatory elements that participate in human diseases9, developmental processes10, as well as splicing factors and nuclear RNAs that are recurrently mutated in cancers11,12. Blind spots in these regions thus hinder a systematic understanding of relevant biological processes.
To alleviate the limitations of variant detection from short-read sequencing due to non-unique regions, a technique called thesaurus annotation characterizes mutations in terms of equivalence classes of genetic positions13,14. This approach does not pinpoint the precise location of mutations, but it enables calculation of summary statistics such as mutational load. It also provides sufficient information about somatic events to study mutational signatures15 and to identify affected functional elements. In the present work, we employed this technique to study the PCAWG dataset with the aim to describe the landscape of somatic single-base substitutions. This uncovers a vast set of somatic events across cancer genomes and cancer types.
Results
Thesaurus annotation uncovers a distinct class of mutations
To perform an analysis of somatic mutations that is inclusive to non-unique regions, we set up a pipeline for variant calling on the PCAWG dataset without filtering reads based on mapping quality (Methods). This provided a comprehensive set of candidate positions in all regions of the genome. We then annotated the sites using a procedure that links candidates to possible alternative positions in the genome13. The annotation serves to collect information from reads that may be ambiguously mapped to more than one location in the genome, to avoid identifying the same underlying variant more than once, and to suggest one representative coordinate for each variant (Figure 1a).
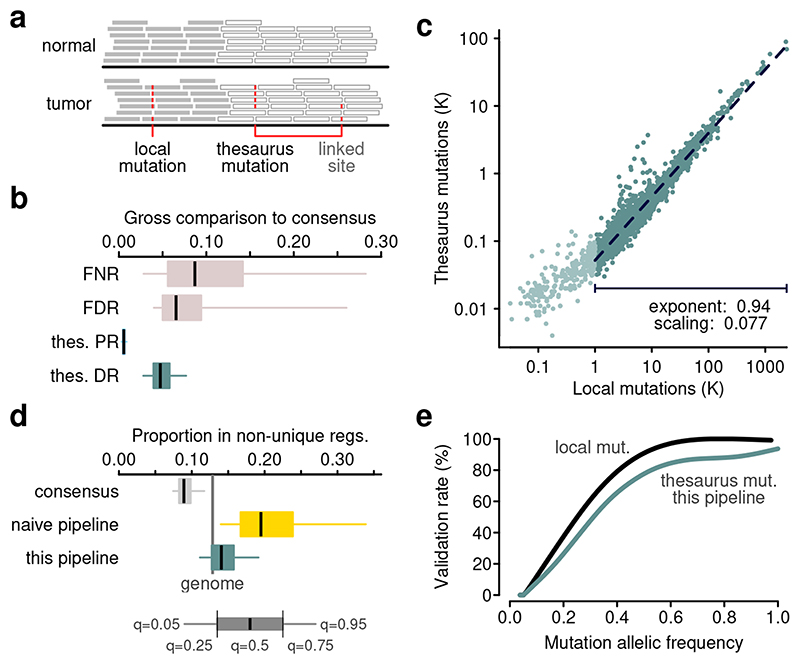
Figure 1. Calling mutations in non-unique regions of the genome.
(a) Schematic of short-read sequencing data with one single-nucleotide substitution in a unique region and one in a non-unique region. Thesaurus annotation links related sites together. (b) Comparison of mutation calls from a mappability agnostic pipeline with the PCAWG mutation set. FDR: false discovery rate; FNR: false negative rate; thes. PR: positive rate among thesaurus mutations; thes. DR: new discovery rate among thesaurus mutations. Box bounds, center line, and whiskers represent 25%-75%, 50%, and 5%-95% quantiles, respectively. (c) Mutation load among simple and thesaurus mutations. A model is fit on a subset of samples with at least 1000 simple mutations. (d) Proportions of mutations that fall in non-unique regions of the genome. Vertical bar shows the fraction of the genome that is non-unique at a resolution of 100bp. The cohort was filtered to exclude samples with fewer than 1000 simple mutations. consensus: PCAWG mutation calls; naive: a mutation set called considering non-unique regions, but without thesaurus annotations. Box bounds, center line, and whiskers represent 25%-75%, 50%, and 5%-95% quantiles, respectively. (e) Validation rate of mutations in an independent cancer sample sequenced with short-read and linked-read technologies.
To search for somatic mutations among the candidates, we utilized curated PCAWG data in two distinct ways. First, we used a panel of 237 genomes from normal tissues to filter out common germline polymorphisms and sequencing artifacts. Second, we trained a machine learning model to classify somatic events. Similar to other machine-driven approaches16,17, our pipeline provided an algorithm with 18 features about candidate sites collected from tumors and matched normal samples. The algorithm then learned a strategy to call mutations from the candidates to match the PCAWG consensus calls1. Crucially, training and testing were performed using only data from unique regions, where the PCAWG call set is expected to be of high quality, and the features provided did not include information about mapping quality. The final classifier achieved a root-mean-square error of 8.9%, with false discovery and false negative rates of 7.6% and 4.3%, respectively (Figure S1). The most important feature for classification was allelic frequency in the matched normal sample (increase in classification error to 37% when omitted), but many other features such as allelic frequency in the tumor and the frequency in the panel of normals contributed as well (Figure S2).
After training the machine-learning model, we processed the entire PCAWG dataset and thereby produced new sets of somatic mutation calls. We split mutations in the new call set into those placed uniquely in the genome, which we describe further as ‘simple’ or ‘local’, and those that can be linked to alternate sites, which we term ‘thesaurus’. Compared to the PCAWG calls, our set of simple mutations showed median false discovery and false negative rates per sample at 7% and 9%, respectively, albeit with variability across the samples in the cohort (Figure 1b). Such discrepancies and variability are not unexpected, as differences among computational pipelines are well-documented18,19. Indeed, modeling revealed that mutation frequency, coverage, and mutation spectrum can explain the largest discrepancies, and that high false discovery and false negative rates are related to internal consistency within the consensus itself (Figure S3). Performance was stable across cancer types (Figure S4).
The set of thesaurus mutations showed little overlap with the PCAWG calls (Figure 1b). This indicates that most of those sites were previously hidden and hence justifies starting our pipeline with a de-novo set of candidate sites. To investigate whether these sites are reasonable additions to the samples’ landscape, we studied total mutation load across samples among the simple and thesaurus calls and found a high correlation (Spearman rho 0.96) that was concordant with direct proportionality (Figure 1c). Other properties such as allele frequency and mutation coverage were also concordant (Figure S5). Moreover, counts of thesaurus mutations correlated only weakly with sequencing coverage (spearman rho 0.16), suggesting the calls were not dominated by noise.
We then studied the position of mutations in relation to annotations of non-unique sequence. As expected, PCAWG calls showed under-representation in regions with low mapping quality (Figure 1d). A simulation of mutation calls that might be obtained using a naive pipeline - considering loci in non-unique regions but without using thesaurus annotation - showed severe over-representation (Figure 1d), providing a justification for common mutation callers to filter out such regions. In contrast, our pipeline produced an intermediate distribution, albeit with a systematic over-representation compared to the proportion of non-unique sequence. This can be due to a residual level of false positives, due to gaps in the sequence of the human reference genome, or due to a propensity for samples to accumulate or tolerate mutations in those regions.
As an orthogonal validation, we performed short-read and linked-read sequencing on one additional cancer sample (Methods). The linked-read protocol uses barcodes to help aligners position reads at their correct coordinates in the reference genome20,21 and thus expands the regions where variation can be assessed by common mutation callers. In the short-read data, our pipeline called 3,074 simple and 189 thesaurus somatic mutations, which we sought to confirm in the linked-read data. Validation rates for simple variants were proportional to the variant allele frequency in the short reads and surpassed 90% at allele frequency of 0.5 (Figure 1e, Figure S6). For calls with thesaurus annotation, the validation rate surpassed 80%; it was on average just 11% lower than for local mutations across allelic frequencies between 0.1 and 1. This suggests that while thesaurus calls may retain more false components than local calls, the large majority of hits from our pipeline represent real events.
Thesaurus mutations are consistent with known processes
Somatic mutations in cancers appear through several biochemical processes, many of which leave distinct patterns in samples’ mutation profiles2,15. To a first approximation, these processes can be presumed to act similarly in unique and non-unique regions and therefore manifest among simple as well as thesaurus-annotated mutations. To test this hypothesis, we stratified mutations by trinucleotide contexts. These profiles, which consist of 96-dimensional vectors, were correlated in most samples (Figure 2a). Representative samples (Figure 2b) suggested that the strength of correlation was influenced by the mutation load. We thus investigated various explanatory variables through linear modeling and found that 85% of the variability in the correlations could be accounted for by mutation load, the entropy of the trinucleotide profiles, and technical features such as depth of coverage (Figure S7).
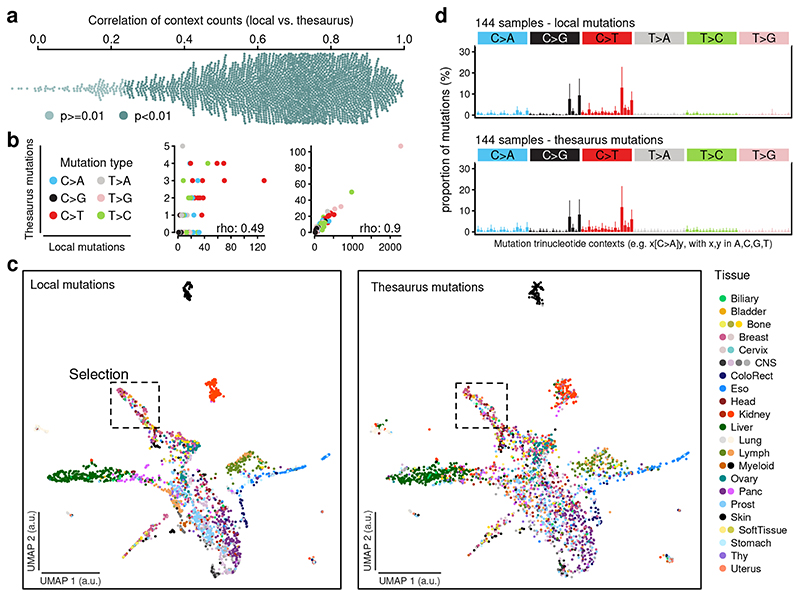
Figure 2. Concordance of simple and thesaurus mutational profiles.
(a) Distribution of spearman correlations, computed for each sample, between 96-dimensional mutational profiles from simple and thesaurus mutations. Color stratification by two-sided p-value. (b) Correlations diagrams between thesaurus and simple mutations for two representative samples. (c) Stereo UMAP embedding of cohort samples based on trinucleotide mutation profiles. The first map is based on simple mutations and a cosine distance. The second view shows a projection of thesaurus profiles onto the same embedding space. Colors indicate histology types and subtypes. (d) Mutation profiles for local and thesaurus mutations for a group of samples selected from the dashed area in (c). Mutation profiles display average profiles for the group. Bars represent 95% quantiles for each substitution type.
To summarize the heterogeneity of the mutation profiles across the cohort, we visualized the similarities between samples in a low-dimensional embedding (Figure 2c). In contrast to analyses that decompose mutation profiles into independent signatures2,15, this technique compares samples in a holistic manner. Profiles based on simple mutations clustered into distinct groups, reproducing known characteristics of the pan-cancer cohort. For example, skin and kidney cancers separated from all others in this visualization, indicating that their mutation profiles are formed by distinct combinations of mutational processes. We then asked to what extent thesaurus mutations capture the same patterns and projected the thesaurus profiles onto the same visualization. Because of the lower overall counts, the resulting patterns were noisier, but nonetheless mirrored the original. This global picture was confirmed by focusing on sets of samples from distinct areas of the embedding. For example, one group of samples exhibited profiles driven by APOBEC activity in both local and thesaurus mutations (Figure 2d). Other sample groups from other parts of the embedding also showed consistent profiles (Figure S7). Correlations of trinucleotide counts in the whole cohort as well as group-level examples together indicate that thesaurus mutations have similar characteristics as simple mutations, both across cancer types and in terms of molecular mutagenic processes.
Thesaurus mutations affect thousands of functional elements
While most mutations in cancer genomes are passengers, some inflict functional effects, for example, by modifying protein structure or altering gene regulation. To create a comprehensive summary of the impact of thesaurus mutations in tumors, we partitioned the genome into non-overlapping regions described by a gene identifier and a functional label (coding sequence, intron, promoter, untranslated, intergenic). These regions are defined through gene annotations, not sequence uniqueness, and thus we found that some carried only simple mutations, others contained only thesaurus mutations, and others harbored both types. In aggregate, thesaurus mutations were associated with thousands of genes, including 1,744 coding sequences (Figure 3a).
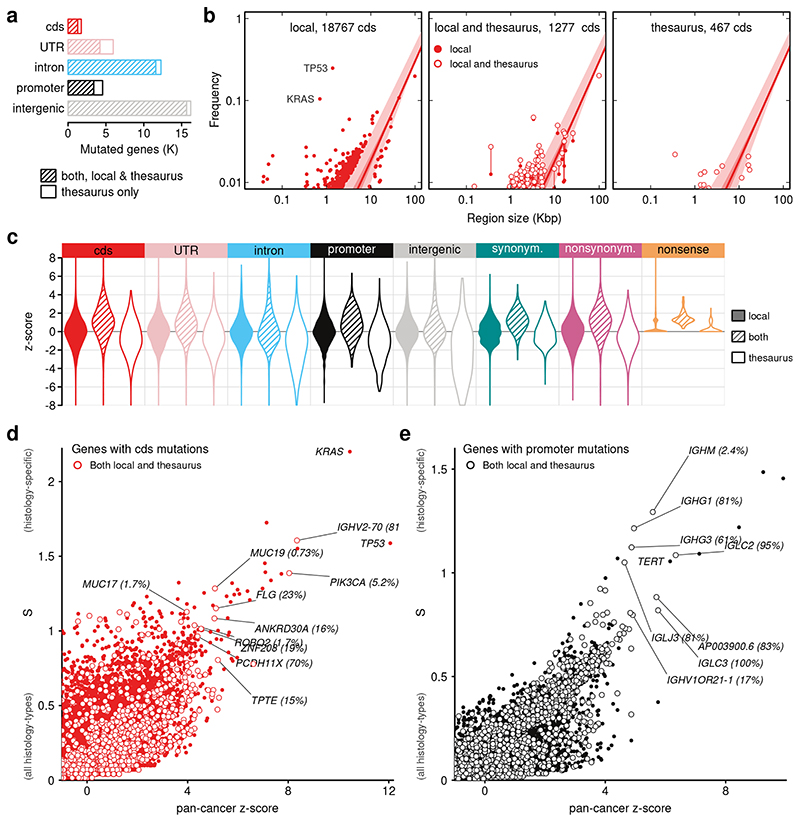
Figure 3. Mutation rates in functional regions.
(a) Counts of functional regions affected by both local and thesaurus mutations, or only by thesaurus mutations. (b) Modeling of cohort frequency in gene coding sequences (cds) as a function of region size using quantile regression. Light and dark-shaded regions indicate 5%-95% and 25%-75% quantile intervals, respectively. Dots display genes outside the range. Subpanels show genes affected only by local mutations, by a mixture of local and thesaurus mutations, or only thesaurus mutations. (c) Summary of quantile modeling on functional regions, and in coding sequences partitioned by mutation effect. (d) Specificity of mutations in coding sequences. Two axes measure over-representation of mutations in the pan-cancer cohort and the entropy across cancer types. Labels for TP53 and KRAS indicate two canonical cancer genes that harbor local mutations, only. Other labels point to genes that contain at least some thesaurus mutations. Percentages show the proportion of mutated samples that carry thesaurus mutations. (e) Analogous to previous panel, showing promoter regions.
Several strategies are used to assess the importance of mutations and identify driver genes22–25. Recurrence in a cohort is a key indicator, but this signal can be confounded by factors such as region size, chromosomal location, proximity between adjacent mutations, sequence composition, and, in the case of coding sequences, effects on protein structure23,26. However, these covariates can themselves be confounded by non-uniqueness in the genome. Here, in order to study mutation patterns across all region types in a way compatible with thesaurus candidates, we performed modeling using only region length as a covariate (Methods). Starting with coding sequences, we fit quantile regression models to describe cohort frequency in genes with unique sequence (Figure 3b). The resultant model captured the expected upward trend, with established driver genes such as TP53 and KRAS as strong outliers. Applying the same model on genes that include non-unique sequences revealed the same trends. Interestingly, the cohort frequencies of many genes shifted across quantile boundaries depending on whether thesaurus mutations were excluded or included.
As the same modeling strategy is also suitable to study mutations outside of coding regions, we carried out a genome-wide analysis and summarized the deviation of individual regions from model trends using z-scores (Figure 3c, Table S1). Distributions of z-scores for elements affected only by local mutations centered around zero by construction. For regions affected by thesaurus mutations, distributions were also centered near zero despite this property not being built into the models. There was a consistent shift toward positive z-scores, but distributions for genes harboring thesaurus mutations exclusively included heavy tails of negative scores, indicating the mutation set may still suffer from false negatives. Indeed, the proportions of thesaurus mutations relative to all mutations in individual genes were consistent, or lower, than the fraction of non-unique sequence in those genes (Figure S8). Scores were consistent when the same modeling was repeated on sub-cohorts and, for coding regions, were correlated with a ranking produced by a specialized scheme accounting for additional covariates (Methods, Figure S9). Overall, the z-scores provide a reasonable, albeit rudimentary, prioritization of hits.
To further refine the prioritization, we computed an entropy-based measure of specificity across cancer types (Methods). We then used the pan-cancer z-scores and specificity measures together to visualize hits in coding sequence (Figure 3d), promoters (defined as regions of at most 2000bp upstream of genes, Figure 3e), and other regions (Figure S10, S11, Table S1). This approach captured expected characteristics, in particular that most genes are neither recurrently mutated nor specific to a cancer-type, and that both dimensions carry outliers. Coding regions in TP53 and KRAS were the top hits for pan-cancer recurrence and specificity, respectively. Genes with thesaurus mutations were found in intermediate regions of the distributions interspersed among other cancer genes. Strikingly, top-ranked thesaurus genes included well-known cancer genes such as PIK3CA, which harbored thesaurus mutations in breast cancers. Another pattern, visible in the analysis of coding sequences, but more pronounced among promoter sequences (Figure 3e), was high recurrence and specificity among immunoglobulin elements of the IGLC, IGHG, IGHJ, and IGHM families. These instances offer leads for more in-depth investigation of thesaurus hits in selected gene families.
Recurrent mutations affect gene families
As some genes with thesaurus mutations already have established links to cancer, we began an exploration of hits by considering the overlap of all thesaurus genes with the cancer gene census27. Our pipeline detected thesaurus mutations in the coding sequences of 35 census genes (Figure 4a) and the untranslated or promoter regions of 29 more (Figure S12). In four of these genes (NUTM2A, NUTM2B, SSX2, SSX4), thesaurus mutations comprised their entire mutational load. This is consistent with these genes being recorded in the census because of translocations and fusions, which are detected by algorithms other than somatic substitution calling. Thesaurus substitutions in these cases provide a complementary set of mutation events. For other genes, the proportion of thesaurus mutations relative to simple mutations was in concordance with the fraction of non-unique sequence within those genes (Figure S12). The proportion of samples affected exclusively by thesaurus mutations ranged from a few percent (e.g. 5% for PIK3CA, 2% for ROBO2) up to a large majority (e.g. 88% for RGPD3).
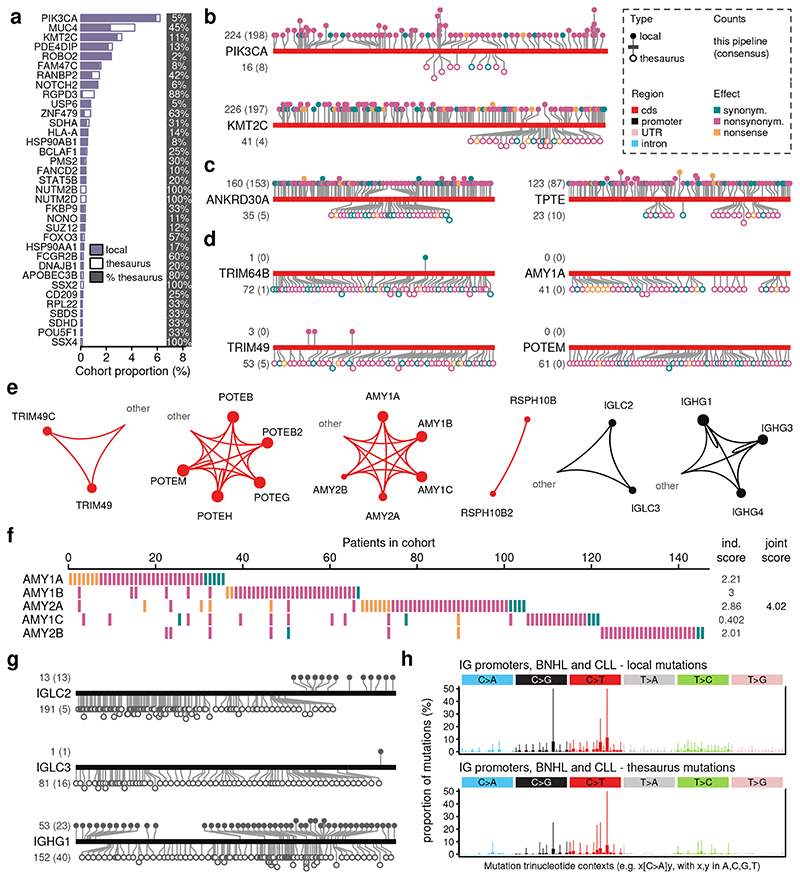
Figure 4. Thesaurus mutations in gene families.
(a) Thesaurus mutations in coding sequences (cds) of the cancer gene census. Percentages on right indicate the proportion of patients that carry exclusively thesaurus mutations. (b) Distribution of mutations along the cds of two known cancer genes. (c) Analogous to (b), genes carrying both local and thesaurus mutations, but not part of cancer census. (d) Analogous to (b) with genes carrying mostly, or exclusively, thesaurus mutations. (e) Dominant linking patterns between selected genes. Genes and links are colored according to region type: cds, untranslated regions (UTR), promoters, and introns. Links from one region type to another, e.g. from cds to UTR, are grouped into a label ‘other’. (f) Cohort summary of mutations in one gene family. (g) Distribution of mutations along sequences upstream (promoters) of selected immunoglobulin (IG) fragments. (h) Mutation profiles in promoter sequences of B-cell non-Hodgkin’s leukemia (BNHL) and chronic lymphocytic leukemia (CLL) consistent with somatic hypermutation by deamination.
To gain more insight into where the mutations lay along the gene structure, we visualized variant-level results along the gene sequences, splitting the results by mutation type and comparing with the PCAWG mutation calls (Figure S13, S14, S15). For PIK3CA and KMT2C, two of the genes with the highest mutation load, our pipeline detected 13% and 15% additional simple substitution events compared to the PCAWG set (Figure 4b). This is broadly consistent with our previous comparisons with the consensus and can in part be attributed to technical differences in the pipelines18. Thesaurus mutations were located in 630bp and 2.63kb segments of their coding regions, respectively. There was a significant enrichment compared to the PCAWG calls (two-sided Fisher tests, PIK3CA, p=4x10-4, KMT2C, p=8x10-23), and, especially for KMT2C, the new mutations filled a noticeable gap in the distribution of simple variants.
Outside of established cancer genes, most genes that harbor thesaurus mutations also contain at least some simple substitutions. Examples with heavy mutation burden include ANKRD30A, an ankyrin-repeat containing gene linked with breast cancer28, and TPTE, a phosphatase linked to the PTEN pathway29 (Figure 4c). Given their high mutation load and interactions with cancer pathways relevant to patient stratification schemes30, thesaurus mutations that fill gaps in their mutation profiles offer direct opportunities to test their translational relevance.
While thesaurus mutations constitute a minority of hits for most genes, they represent the dominant class for others (Figure 4d). Among genes highlighted by our z-score analysis and also by alternative prioritization methods, TRIM49 and TRIM64B, two members of the tripartite motif family of proteins, were prominent with mutations along their entire gene body. This family is involved in innate immunity, autophagy and carcinogenesis31. AMY1B encodes an amylase isoenzyme typically expressed in the salivary gland and the pancreas and is embedded in a region of variable copy-number. It may influence metabolism, but its high mutation burden may also be a corollary of the genomic fragility of its surrounding genomic region32. POTEM is another gene with an ankyrin domain with heavy mutation burden. The gene family has been discussed in studies of expression-based biomarkers33.
Each of the thesaurus mutations in our dataset is annotated by a link to at least one alternative genomic site with a similar surrounding sequence. These links describe ambiguities in assigning the location of the individual somatic events. True to the ubiquity of non-uniqueness across the genome, we found mutations in coding sequences linked to a variety of targets, including untranslated, intronic, promoter, or untranslated regions of other genes. Mutations in PIK3CA, for example, linked to an untranslated region of a pseudogene. Links in other genes often interconnected members from the same family (Figure 4e). These cliques prompted us to consider mutation load across several related genes. Using the amylase gene family as an example, the detected mutations across five genes affected 3.1% of non-hypermutated samples in the cohort. Samples affected by the individual genes were to a large extent non-overlapping (Figure 4f), ruling out the possibility that the mutation set is dominated by double-counting. When we modeled the entire gene family as a single entity in our quantile regression model, the z-score for the family rose from 2.21 for AMY1A alone to 4.04 for the gene group. Analogous patterns, with variability on the degree of effect on the joint scores, occurred in other families (Figure S16-S24).
Among genes highlighted due to mutations in promoter regions were several members of the immunoglobulin (IG) family (Figure 4g, Figure S25, S26). The immunoglobulin locus undergoes hypermutation during B-cell maturation through cytidine deamination34 and genomic rearrangements. Consistent with the role of this process in antibody diversification and immunity, mutations associated to IG gene fragments were enriched in leukemias and lymphomas. Among sequences upstream of all IG gene segments, thesaurus mutations represented 19.7% of all variants in those cancers. Furthermore, the trinucleotide mutation profiles were dominated by C>T substitutions and were consistent with reported sequence hotspot patterns34 (Figure 4h, S15). This suggests that thesaurus annotation reliably detects these non-germline events and can thus inform translational approaches that use immune signatures.
Discussion
The structure of the human genome has been shaped by its evolution, including by duplications and rearrangements. This history leaves a substantial portion of the genome to appear non-unique at the scale of short reads used by high-throughput sequencing studies. As long- and linked-read sequencing protocols become established and widespread, these regions will become accessible for direct analysis20,35,36. However, existing datasets, including efforts to sequence whole genomes of pan-cancer primary tumors1 and metastases37, already offer opportunities to evaluate somatic events in these non-unique regions.
When specific sequences are of interest a priori, targeted approaches can perform re-analysis of subsets of sequencing data. This analytic strategy is effective when there are complex rearrangements as in the case of the HLA locus38, or sequences are present in a large number of copies as in the case of transposons39. However, the human genome also carries areas that are almost exactly duplicated due to recent evolutionary events and are present in fewer than ten copies. Such regions can be studied in a systematic manner through a technique that links specific genomic positions and provides information about clusters based on multi-locus alignments13. Our pipeline collects multi-locus annotations and leverages high-quality mutation calls from PCAWG to train a machine learning model. The resulting calls for somatic substitutions provide a first systematic summary of the mutation events in non-unique regions at a genome-wide scale and across several cancer types.
The landscape of thesaurus-annotated mutations covers over 1700 coding genes, 4500 promoters, and thousands of other functional elements. The mutation burden in these regions is consistent with known properties of cancer types. Trinucleotide substitution patterns that characterize underlying molecular mechanisms are concordant as well. Mutation rates are in line with those in genes with unique sequences. Interestingly, outlier genes with thesaurus mutations are affected to an extent comparable to established cancer genes. These hits provide tantalizing leads toward a more complete picture of mutational processes in cancers.
Our analyses of cohort mutation rates, regional recurrence and hotspots, cancer-type specificity, and co-occurrence are only a first-pass summary of the patterns in these data. Indeed, mutational processes are modulated, directly or indirectly, by a myriad of factors that include nucleotide content, chromatin accessibility, and gene expression23. While methods developed in these areas provide guidance for more refined analyses, they rely on auxiliary data as model covariates. In the context of non-unique regions, these covariates, as they are often acquired through short-read sequencing, are likely to suffer biases related to sequence uniqueness40. A careful examination of those covariates in the non-unique genome is a critical step toward better understanding of the statistical importance of the uncovered mutational landscape. Analyses will also be required to understand the functional impact of these mutations, although preliminary results show that the mutant alleles are expressed in RNA (Figure S27). In an orthogonal direction, our analyses can be extended to other genome assemblies. New assemblies and genome models contain an even higher proportion of sequence that is non-unique (in GRCh38, 19% at the scale of 100bp, 14% at 200bp) and thus more variants require thesaurus annotation (Figure S28). Our approach is directly applicable and can help characterize the mutation landscape in samples aligned with these genome assemblies.
Beyond somatic substitution events, cancer genomes also suffer other types of mutations, including small insertions and deletions and regional copy-number changes4. Such events are fewer in number than SNVs but can have profound functional consequences. Our dataset carries evidence that such events occur in genes with non-unique sequences. Our analysis, however, does not include them because of current limitations in thesaurus annotation. It is thus clear that, despite uncovering thousands of additional somatic events, more still remain hidden. Similar considerations are also relevant for comparative genomics of germline variants and their relation to rare diseases41. The challenges in detecting these events are technical, but can be overcome with careful strategies for variant comparison42, and would help refine views on the genomics of disease.
Online methods
Cancer cohort
Data for cancer samples and matching normal tissue were obtained through the Pan-Cancer Analysis of Whole Genomes (PCAWG) consortium1. The dataset consisted of 2,658 samples from 38 distinct cancer types (22 organ systems) and 47 consortium projects. All alignments were used in their original formats as provided by the consortium, aligned to the hs37d5 reference build of the human genome using bwa-mem43.
A random set of 237 normal-tissue samples reported not to be contaminated by tumor cells were selected to form a panel of normals. These samples originated from all 38 cancer types. The panel thus captures heterogeneity of human populations, although it cannot be treated as a true representative of all genetic variation.
Annotations
All calculations were performed against the hs37d5 genome reference build, including decoy chromosomes. Gene annotations were obtained from GENCODE, release 1944.
Variant calling, thesaurus annotation, and candidate prioritization
An initial set of variants were called from bwa-aligned sequencing data using Bamformatics (v0.2.5) (https://github.com/tkonopka/Bamformatics). Settings were left at their defaults, except for argument --minmapqual 0, which instructs the software to use all primary read alignments irrespective of mapping quality. The software used in this stage is not critical and could be replaced by one of many other tools, as long as capable of processing all reads irrespective of mapping quality.
Variants detected in samples included in the panel of normals were aggregated into a single table. The frequency of each variant in this panel was kept as a proxy for population frequency.
Variants from tumor samples were annotated using GeneticThesaurus (v0.2.1) (https://github.com/tkonopka/GeneticThesaurus). Customized settings included --minmapqual 0, which instructs the software to use all primary aligned reads irrespective of mapping quality, and settings --many 20 --toomany 100, which limit thesaurus links to a smaller number than set by default. The thesaurus annotation process was provided access to alignment data for matched normal samples. Output from this stage included tables linking variants to related sites in the genome, as well as tables associating each variant in the tumor with features such as allelic frequency, coverage, and analogous features from the matched normal. These tables included data based only on the called variant position as well as data pooling information from all thesaurus-linked sites. Variants were automatically assigned filter codes by the software - PASS, which we described as local in this work, ‘thesaurus’ when linked to alternative sites, ‘thesaurusmany’ when potentially linked to more than 100 sites, and ‘thesaurushard’ when in a thesaurus region that could not be processed due to nearby indels. Downstream analyses and modeling were carried out using ‘local’ and ‘thesaurus’ sites only, although sites marked ‘thesaurushard’ were also included in certain visualizations.
Following the variant annotation by the GeneticThesaurus software, variants from each tumor were compared to the panel of normals. Items present at a frequency greater than 1% in normal samples were deemed to consist of common germline variants or sequencing artefacts and were excluded from downstream analysis. Prioritized candidates were further annotated with features from the CIGAR strings of aligned reads using custom scripts.
Machine learning to detect somatic mutations
A set of 300 tumor samples and their matching normal controls were selected at random for machine-learning. The set contained representatives from all the major PCAWG histologies. The set was split into training, testing, and validation sets with 150, 50, and 100 samples, respectively. Separately, PCAWG mutation calls for the same samples were assembled into a truth set. Importantly, the PCAWG mutation calls were filtered using the same panel-of-normals frequency filter used on the candidate sites and then trimmed further to remove items that were not present in the variant candidates. These steps ensured that the candidates and the truth set are consistent, and that all items in the truth set could in principle be obtained from non-missing features in the candidate data. Both the candidate and the truth dataset were restricted to sites with a PASS filter code in the candidate data, i.e. to those sites not linked to any additional locations via thesaurus links. This ensured that the identification of somatic mutations among the candidates could be determined by technical features of the sequencing data and biological aspects of the tumor, and was not confounded by aspects related to mappability.
Machine learning models were trained using xgboost45 - an algorithm based on random forests - with default settings except when stated. To explore the effect of data quantity on classification performance, a series of models were trained based on an increasing number of tumor samples. The samples used in each model were selected with a stochastic procedure that attempted to use distinct samples in replicates. Once the tumor samples were selected, models were trained using all the data from those tumors. After training, all models were evaluated against the entire test set using false-discovery (ratio of false calls among all positive calls) and false-negative rates (ratio of missed calls among all calls in the truth set). After obtaining a satisfactory set of hyperparameters, a final model was trained using the entire set of 150 training samples and re-evaluated on an independent validation set.
Feature importance in the final classifier was assessed using a bootstrapped dropout-loss procedure. This procedure subsamples the testing dataset, permutes values within individual features or certain groups of features, and assesses how the predictions on the adulterated data compare with predictions based on the original data. The downsampling and permutation procedure was repeated 100 times and average dropout-loss values reported.
This model was then applied to call somatic single nucleotide variants on the entire set of candidate sites irrespective of filter code, i.e. on unique and non-uniquely mappable sites. Importantly, for features that can be affected by mappability, the values provided to the classifier were those estimated by the GeneticThesaurus annotation procedure, i.e. averages over all sites linked by the thesaurus. By construction, this procedure ensured that in well-mappable areas, the classifier functioned in the same way as during training. In non-unique regions, the classifier made predictions from data informed by multiple locations in the genome.
Modelling mutation load
Called mutations were classified into two groups - local and thesaurus - based on whether a site was associated with an alternative location via a thesaurus link. The mutation load in each sample was defined as the total number of positions for each type. Cases where mutations were called at more than one site and linked together via a thesaurus annotation were treated as single events; the site with the higher allelic frequency was marked as primary and taken forward for subsequent analysis. The relation between the mutation loads associated with local and thesaurus filter codes was modeled using a power-law equation
where scaling and exponent are free parameters. Under an assumption that thesaurus mutations appear via the same mechanisms as simple mutations, the exponent would be equal to unity and the scaling constant would indicate the relative size of non-unique and unique genomic regions. The model can be reformulated as, log(thesaurus load) ~ scaling + exponent log(simple load). The free parameters were solved using simple regression.
Linked-read sequencing
A sample from a patient with a malignant peripheral nerve sheath tumor was selected for sequencing. The patient provided their written and informed consent to provide samples for this study, which was approved by the National Research Ethics Service (NRES) Committee Yorkshire and The Humber – Leeds East (15/YH/0311).
High molecular weight DNA was extracted from frozen tissue according to the 10x genomics protocol (CG000072). In short, 500 μl of cold nuclei isolation buffer was added (sigma pure prep lysis buffer (NUC201-1KT), 1mM DTT, 10% Triton X-100) to a small piece of frozen tissue in a 1.5 ml tube (and homogenized by moving a pestle up and down 10-20x). After settling, the supernatant was transferred to a new tube and centrifuged for 5 min at 500xg (4˚C). The supernatant was removed without disturbing the pellet and the following was added: 70 μl of cold PBS, 10 μl of proteinase K and 70 μl of Digestion buffer (20mM EDTA, 2nM Tris-HCl, 10mM N-laurylsarcosine sodium salt and water). The pellet was dislodged by tapping the tubes lightly and then left to rotate for 2h at 20˚C. Tween-20 was added to a final concentration of 0.1% and pipette mixed 5 times. An equal amount of 1x SPRIselect reagent (Beckman Coulter B23317) was added followed by rotation for 20 min. The beads were then washed twice with 70% ethanol and resuspended in 50 μl sample elution buffer (Qiagen AE buffer with 0.1% Tween 20). After incubation at 20˚C for 5 min, the beads were put on a shaker at 1400rpm for 3 min (25˚C) to elute the DNA. The samples were quantified using the Qubit. Linked reads libraries were generated using the 10x Chromium, following the manufacturer’s instructions. The library was sequenced using paired-end 150bp reads with an 8bp index on a single lane of an Illumina HiSeq X.
Validation through linked-read sequencing
BCL files were processed and demultiplexed to FASTQ files using bcl2fastq v2.20.0. Reads were mapped to the hs37d5 reference build, de-duplicated and filtered using the LongRanger (v2.2.2) WGS pipeline with the --somatic flag. This pipeline leverages the Chromium molecular barcodes and GATK v4.0.8.1 to call and phase single nucleotide variants, indels, and structural variants. Overall, a total of 1,538,338 GEMs were generated, containing on average 589kb of DNA with an average size of 86kb, each producing a median of 47 linked reads for a final 38x depth of coverage and 32x median depth at mutated sites.
The reported variants were compared to the calls from the machine learning approach on a standard short-read WGS dataset of the same tumor sample. Simple variants called from the short reads were declared validated if they were part of the PASS mutations in the linked reads. Thesaurus calls were declared validated if they were part of PASS mutations in the linked reads, or if their thesaurus-linked sites were part of the linked-read dataset. This approach allows for ambiguity in placing the mutation location based on short-read data, but does not inflate detection rates13.
Mutation trinucleotide profiles
Trinucleotide contexts were extracted for all called mutations in the cohort. These neighborhoods were used to assign each substitution mutation to one of 96 categories as previously described15. Counting the number of mutations of each type produced two profiles - one based on simple mutations in unique genomic regions and one based on mutations with thesaurus links.
For correlation analysis, the simple and thesaurus profiles were treated as 96-dimensional vectors and correlation was evaluated using the Spearman method. Because mutation profiles are degenerate when the overall number of mutations is small and constrained to non-negative counts, statistical significance was estimated by simulation. For a given profile of thesaurus mutations, 10,000 random profiles were generated with an equivalent number of mutations. The Spearman correlation value between the local and thesaurus profiles were compared to the distribution of correlations between the local and simulated profiles. The procedure provided approximations to p-values that were sufficiently precise to determine significance at nominal and multiple-testing adjusted levels.
For visualization of similarities of the mutation profiles, count-based mutation profiles were adjusted using allele-frequency data. This adjustment provided weighting toward well-measured mutation instances and avoided degenerate comparisons based on integer counts. Mutation profiles based on simple mutations were sum-normalized and embedded into a two-dimensional space using UMAP46,47, a dimensional reduction technique, using a euclidean distance metric. Following generation of the embedding based on simple mutations, the resultant model was used to predict the position of allele-frequency-adjusted profiles based on thesaurus mutations.
Modeling of mutation frequency
Mutation frequencies were computed by counting the number of samples (patients) with at least one somatic mutation in regions of interest. In order to avoid counts being inflated by likely passenger mutations, hyper-mutator samples were identified and removed from this calculation and subsequent modeling. Hyper-mutator status was set if a sample contained more than 300 mutations in coding regions, a procedure previously described in other studies of driver mutations23,40. In practice, this criterion omitted 198 samples (7.5% of the cohort) from the mutation frequency analysis.
Mutation frequencies were assessed on non-overlapping genomic regions labeled as coding (cds), intronic (intron), untranslated (UTR), promoter, or intergenic. Each region was associated with a genomic length, a frequency based on simple mutations alone, a frequency based on thesaurus mutations alone, and a frequency based on both simple and thesaurus mutations.
Modeling of the relation between region size and mutation frequency was performed using quantile regression. The model used logarithmically transformed frequency and region size,
With a0, a1, and a2 as free parameters. The quadratic term allows some nonlinearity in the relationship between size and frequency, which is required to allow the growth in frequency to taper for very large regions. Quantile regression with this model was performed at the 50% percentile to describe the primary trend, and at 5%, 25%, 75%, 95% levels to obtain intervals of variability. After fitting the parameters, each genomic region was associated with an expected mutation frequency and an interquartile interval.
Quantile regression based on a linear equation is guaranteed to produce fitted models that preserve ordering of percentiles, e.g. with a model at the 75% quantile always yielding larger values than at 50%. This property is not guaranteed for models with higher-order terms. Predictions from the fitted models were thus adjusted post-hoc. Furthermore, the interquartile interval was forced to correspond to at least 1/N, with N being the number of samples in the modeled cohort. All model predictions were restricted to the unit interval, [0, 1].
Identifying driver genes with dndscv
The dN/dS ratio was computed through maximum likelihood estimates across trinucleotide contexts using dndscv23. After removing hypermutator samples with more than 300 coding mutations, dndscv was ran on the pan-cancer cohort to identify candidate cancer genes. The calculation used the pooled set of thesaurus and simple mutations, but did not correct for epigenetic covariates as these covariates are not annotated for non-uniquely mapping regions.
Cancer type specificity
Specificity of mutations was quantified using information entropy, defined via Shannon’s formula,
with the sum running over cancer types. Probabilities pi for each cancer type were defined as pi = (ci + ɛ)/ ∑j(cj + ɛ) with ci being the count of patients (samples) carrying a mutation in the region and the constant є being a pseudocount regularization. The regularization was set f at 1 for coding sequences and values were adjusted for promoters, UTRs, introns, and intergenic regions proportionally to the median region length. Because entropy is high for quasi-random configurations and low for configurations peaked on one bin, visualizations of specificity were performed using changes in entropy, ΔS. These were defined by subtracting S from the entropy of a hypothetical region with null counts,
The quantity gives mutation patterns with a high number of patients from a single cancer type higher values than cancer-type-agnostic patterns. It also assigns values near zero to all infrequently mutated regions.
Supplementary Material
Supplementary Table 1. Summary of z-scores and histology specificity for all genomic regions. The table contains summary statistics for all genes in the annotation set. Counts, z-scores, and entropy scores are provided based on non-hypermutated samples.
Acknowledgements
This work is supported by The Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001202), the UK Medical Research Council (FC001202) and the Wellcome Trust (FC001202). M.T. is supported as a postdoctoral fellow by the European Union’s Horizon 2020 research and innovation program (Marie Skłodowska-Curie Grant agreement no. 747852-SIOMICS). J.D. is a postdoctoral fellow of the Research Foundation – Flanders (FWO). A.M.F. is a NIHR senior investigator and is supported by the National Institute for Health Research, UCLH Biomedical Research Centre and the CRUK Experimental Cancer Centre. P.V.L. is a Winton Group Leader in recognition of the Winton Charitable Foundation’s support toward the establishment of The Francis Crick Institute. T.K. would like to thank Damian Smedley. This project was enabled through the Crick Scientific Computing STP and through access to the MRC eMedLab Medical Bioinformatics infrastructure, supported by the UK Medical Research Council (grant no. MR/L016311/1). The Bone Cancer Research Trust funded sample biobanking.
Footnotes
Author Contribution
All authors edited and approved the final manuscript. M.T. wrote the first draft of the paper, designed experiments, performed statistical analyses, performed bioinformatics analyses and performed data visualization. J.D. performed bioinformatics analyses of linked-read data. A.V. generated short-read and linked-read data. A.M.F. provided tumor samples and performed pathology assessment. P.V.L. wrote the first draft of the paper, designed experiments, and supervised the study jointly with T.K. T.K. wrote the first draft of the paper, designed experiments, performed statistical analyses, performed bioinformatics analyses, performed data visualization and supervised the study jointly with P.V.L.
Competing Interests Statement
The authors declare no competing interests.
Data availability
The PCAWG dataset is available through the ICGC data portal: https://dcc.icgc.org/pcawg. Somatic mutations called in this study are available at https://www.synapse.org/#!Synapse:syn22297877.
Code Availability
The thesaurus annotation software is available at: sourceforge.net/projects/geneticthesaurus/ and github.com/tkonopka/GeneticThesaurus.
References
- 1.The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature. 2020;578:82–93. doi: 10.1038/s41586-020-1969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexandrov LB, et al. The Repertoire of Mutational Signatures in Human Cancer. Nature. 2020;578:94–101. doi: 10.1038/s41586-020-1943-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y, et al. Patterns of somatic structural variation in human cancer genomes. Nature. 2020;578:112–121. doi: 10.1038/s41586-019-1913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rheinbay E, et al. Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature. 2020;578:102–111. doi: 10.1038/s41586-020-1965-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerstung M, et al. The evolutionary history of 2,658 cancers. Nature. 2020;578:122–128. doi: 10.1038/s41586-019-1907-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karimzadeh M, Ernst C, Kundaje A, Hoffman MM. Umap and Bismap: quantifying genome and methylome mappability. Nucleic Acids Res. 2018;46:e120. doi: 10.1093/nar/gky677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee H, Schatz MC. Genomic dark matter: the reliability of short read mapping illustrated by the genome mappability score. Bioinformatics. 2012;28:2097–2105. doi: 10.1093/bioinformatics/bts330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Treangen TJ, Salzberg SL. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet. 2011;13:36–46. doi: 10.1038/nrg3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mandelker D, et al. Navigating highly homologous genes in a molecular diagnostic setting: a resource for clinical next-generation sequencing. Genetics in Medicine. 2016;18:1282–1289. doi: 10.1038/gim.2016.58. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki IK, et al. Human-Specific NOTCH2NL Genes Expand Cortical Neurogenesis through Delta/Notch Regulation. Cell. 2018;173:1370–1384.:e16. doi: 10.1016/j.cell.2018.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki H, et al. Recurrent noncoding U1 snRNA mutations drive cryptic splicing in SHH medulloblastoma. Nature. 2019;574:707–711. doi: 10.1038/s41586-019-1650-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shuai S, et al. The U1 spliceosomal RNA is recurrently mutated in multiple cancers. Nature. 2019 doi: 10.1038/s41586-019-1651-z. [DOI] [PubMed] [Google Scholar]
- 13.Kerzendorfer C, Konopka T, Nijman SMB. A thesaurus of genetic variation for interrogation of repetitive genomic regions. Nucleic Acids Res. 2015;43:e68. doi: 10.1093/nar/gkv178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konopka T, Nijman SMB. Comparison of genetic variants in matched samples using thesaurus annotation. Bioinformatics. 2016;32:657–663. doi: 10.1093/bioinformatics/btv654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ainscough BJ, et al. A deep learning approach to automate refinement of somatic variant calling from cancer sequencing data. Nat Genet. 2018;50:1735–1743. doi: 10.1038/s41588-018-0257-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anzar I, Sverchkova A, Stratford R, Clancy T. NeoMutate: an ensemble machine learning framework for the prediction of somatic mutations in cancer. BMC Med Genomics. 2019;12:63. doi: 10.1186/s12920-019-0508-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Prieto C, Valencia A, Porta-Pardo E. The consequences of variant calling decisions in secondary analyses of cancer sequencing data. [DOI]
- 19.Ellrott K, et al. Scalable Open Science Approach for Mutation Calling of Tumor Exomes Using Multiple Genomic Pipelines. Cell Syst. 2018;6:271–281.:e7. doi: 10.1016/j.cels.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bishara A, et al. Read clouds uncover variation in complex regions of the human genome. Genome Res. 2015;25:1570–1580. doi: 10.1101/gr.191189.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng GXY, et al. Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat Biotechnol. 2016;34:303–311. doi: 10.1038/nbt.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martincorena I, et al. Universal Patterns of Selection in Cancer and Somatic Tissues. Cell. 2018;173:1823. doi: 10.1016/j.cell.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen H, et al. Comprehensive assessment of computational algorithms in predicting cancer driver mutations. Genome Biol. 2020;21:43. doi: 10.1186/s13059-020-01954-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Araya CL, et al. Identification of significantly mutated regions across cancer types highlights a rich landscape of functional molecular alterations. Nat Genet. 2015;48:117–125. doi: 10.1038/ng.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bailey MH, et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell. 2018;174:1034–1035. doi: 10.1016/j.cell.2018.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tate JG, et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019;47:D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jäger D, et al. Identification of a tissue-specific putative transcription factor in breast tissue by serological screening of a breast cancer library. Cancer Res. 2001;61:2055–2061. [PubMed] [Google Scholar]
- 29.Tapparel C, et al. The TPTE gene family: cellular expression, subcellular localization and alternative splicing. Gene. 2003;323:189–199. doi: 10.1016/j.gene.2003.09.038. [DOI] [PubMed] [Google Scholar]
- 30.Jamaspishvili T, et al. Clinical implications of PTEN loss in prostate cancer. Nat Rev Urol. 2018;15:222–234. doi: 10.1038/nrurol.2018.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hatakeyama S. TRIM Family Proteins: Roles in Autophagy, Immunity, and Carcinogenesis. Trends in Biochemical Sciences. 2017;42:297–311. doi: 10.1016/j.tibs.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 32.Usher CL, et al. Structural forms of the human amylase locus and their relationships to SNPs, haplotypes and obesity. Nat Genet. 2015;47:921–925. doi: 10.1038/ng.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barger CJ, et al. Expression of the POTE gene family in human ovarian cancer. Sci Rep. 2018;8:17136. doi: 10.1038/s41598-018-35567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teng G, Papavasiliou FN. Immunoglobulin somatic hypermutation. Annu Rev Genet. 2007;41:107–120. doi: 10.1146/annurev.genet.41.110306.130340. [DOI] [PubMed] [Google Scholar]
- 35.Amarasinghe SL, et al. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 2020;21:30. doi: 10.1186/s13059-020-1935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marks P, et al. Resolving the full spectrum of human genome variation using Linked-Reads. Genome Res. 2019;29:635–645. doi: 10.1101/gr.234443.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Priestley P, et al. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature. 2019;575:210–216. doi: 10.1038/s41586-019-1689-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGranahan N, et al. Allele-Specific HLA Loss and Immune Escape in Lung Cancer Evolution. Cell. 2017;171:1259–1271.:e11. doi: 10.1016/j.cell.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodriguez-Martin B, et al. Pan-cancer analysis of whole genomes identifies driver rearrangements promoted by LINE-1 retrotransposition. Nat Genet. 2020;52:306–319. doi: 10.1038/s41588-019-0562-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kundaje A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eichler EE. Genetic Variation, Comparative Genomics, and the Diagnosis of Disease. N Engl J Med. 2019;381:64–74. doi: 10.1056/NEJMra1809315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krusche P, et al. Best practices for benchmarking germline small-variant calls in human genomes. Nat Biotechnol. 2019;37:555–560. doi: 10.1038/s41587-019-0054-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frankish A, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47:D766–D773. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Friedman J, Hastie T, Tibshirani R. Additive logistic regression: a statistical view of boosting (With discussion and a rejoinder by the authors) The Annals of Statistics. 2000;28:337–407. [Google Scholar]
- 46.McInnes L, Healy J, Saul N, Großberger L. UMAP: Uniform Manifold Approximation and Projection. Journal of Open Source Software. 2018;3:861. [Google Scholar]
- 47.Becht E, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 2018 doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The PCAWG dataset is available through the ICGC data portal: https://dcc.icgc.org/pcawg. Somatic mutations called in this study are available at https://www.synapse.org/#!Synapse:syn22297877.
The thesaurus annotation software is available at: sourceforge.net/projects/geneticthesaurus/ and github.com/tkonopka/GeneticThesaurus.