Abstract
Protein-protein interactions play critical roles in biology, but the structures of many eukaryotic protein complexes are unknown, and there are likely many interactions not yet identified. We take advantage of advances in proteome-wide amino acid coevolution analysis and deep-learning-based structure modeling to systematically identify and build accurate models of core eukaryotic protein complexes within the Saccharomyces cerevisiae proteome. We use a combination of RoseTTAFold and AlphaFold to screen through paired multiple sequence alignments for 8.3 million pairs of yeast proteins, identify 1,505 likely to interact, and build structure models for 106 previously unidentified assemblies and 806 that have not been structurally characterized. These complexes, which have as many as 5 subunits, play roles in almost all key processes in eukaryotic cells and provide broad insights into biological function.
Yeast two hybrid (Y2H), affinity-purification mass spectrometry (APMS), and other high-throughput experimental approaches have identified many pairs of interacting proteins in yeast and other organisms (1–5), but there are discrepancies between sets generated using the different methods and considerable false positive and false negative rates (6–8). Because residues at protein-protein interfaces are expected to coevolve, the likelihood that any two proteins interact can be assessed by identifying and aligning the ortholog sequences of the two proteins in many different species, joining them to create paired multiple sequence alignments (pMSA), and then determining the extent to which changes in the sequences of orthologs for the first protein covary with ortholog sequence changes for the second (9,10). Such amino acid coevolution has been used to guide modeling of complexes for cases in which the structures of the partners are known (11,12), and to systematically identify pairs of interacting proteins in Prokaryotes with accuracy higher than experimental screens (9). Recent deep-learning-based advances in protein structure prediction (13,14) have the potential to increase the power of such approaches as they now enable accurate modeling not only of protein monomer structures but also protein complexes (13).
We set out to combine proteome wide coevolution-guided protein interaction identification with deep learning based protein structure modeling to systematically identify and determine the structures of eukaryotic protein assemblies (Fig. 1A). We faced several challenges in directly applying to eukaryotes the statistical methods we had found effective in identifying coevolving pairs in prokaryotes (8). First, far fewer genome sequences are available for eukaryotes than prokaryotes, and the average number of orthologous sequences (excluding nearly identical copies with > 95% sequence identity) is on the order of 10,000 for bacterial proteins, but 1,000 for eukaryotic proteins. Thus, multiple sequence alignments for pairs of eukaryotic proteins contain fewer diverse sequences, making it more difficult for statistical methods to distinguish true coevolutionary signal from the noise. Second, eukaryotes in general have a larger number of genes, making comprehensive pairwise analysis more computationally intensive, and increasing the background noise. Third, mRNA splicing in eukaryotes further increases the number of protein species, resulting in errors in gene predictions and complicating sequence alignments. Fourth, eukaryotes underwent several rounds of genome duplications in multiple lineages (15), and it can be difficult to distinguish orthologs from paralogs, which is important for detecting coevolutionary signal because the protein interactions of interest are likely to be conserved in orthologs in other species but less so in paralogs.
Figure 1. Evaluation of protein interaction and structure prediction accuracy.
(A) The PPI screen pipeline. (B) Performance (precision at different levels of recall) of different methods in picking out gold standard PPIs from the set of 4.3 million pMSAs (Precision: number of true positives above a cutoff divided by the total number of pairs above this cutoff; recall: number of true positives above cutoff divided by the total number of true positives (gold standard PPIs). Pairs were ranked by the top coevolution score or contact probability between residue pairs. DCA: Direct coupling analysis. RF2t: top contact probability between residues of two proteins by RF 2-track model. RF2t++, optimized RF2t (see methods). RF2t++ predictions better than the cutoff shown in vertical black line (RF2t++L in Fig. 1C) were processed with AF; recall of gold standard PPIs at this cutoff is 29%; and precision is 23%. RF2t++ results with a more stringent cutoff (red vertical line) are also shown in Fig 1C (RF2t++H). (C) AF contact probability ranking of complexes selected by RF2t++ in panel (B); complexes with scores above the horizontal black line were selected for further analysis. (D) Number of high scoring (top contact probability > 0.67) AF predictions in PPI sets from different sources. (E) Distribution of percent of AF predicted inter-protein contacts with predicted error < 8Å found in contact (< 8Å) in closely-related experimental structures.
To mitigate the first three challenges, we chose to predict protein complexes for yeast Saccharomyces cerevisiae as the starting point because there are a large number of fungal genomes (16), the genome is relatively small (6,000 genes total), and there is relatively little mRNA splicing (17). Furthermore, because the interactome of yeast has been extensively studied, there is a “gold standard” set (see supplemental Methods) of known interactions to evaluate the accuracy of predicted interactions and structures.
To distinguish orthologs from paralogs, we started from OrthoDB (18), a hierarchical catalog of orthologs across 1,271 Eukaryote genomes, and supplemented each orthologous group with sequences from 4,325 Eukaryote proteomes we assembled from NCBI (https://www.ncbi.nlm.nih.gov/genome) and JGI (19). Among these, 2,026 are fungal proteomes spanning 14 phyla (47 classes). We compared the sequences for each protein in each of the additional 4,325 proteomes against those of the most closely related species in the OrthoDB database, and used the reciprocal best hit criterion (20) to identify orthologs (fig. S1); these were then added to the corresponding orthologous group. A complication is that each species frequently contains multiple proteins belonging to the same orthologous group, leading to ambiguity in determining which protein should be included in pMSAs. These multiple copies may represent alternatively spliced forms of the same gene, parts of the same gene that were split into multiple pieces due to errors in gene prediction, or recent gene expansions specific to certain lineages. We dealt with these possibilities by keeping only the longest isoform of each gene, merging pieces of the same gene, and selecting the copy with the highest sequence identity to single-copy orthologs in other species. For 4,090 out of ~6,000 yeast proteins, we were able to assign a single-copy yeast protein to orthologs in other species, and we generated pMSAs for all 4,090 * 4,089 / 2 = 8,362,005 pairwise combinations of these proteins (fig. S2). We focused on 4,286,433 pairs with alignments containing over 200 sequences to increase prediction accuracy and less than 1,300 amino acids to accelerate computation (fig. S3).
In a first set of calculations, we found that even with the advantages of S. cerevisiae and improved ortholog identification, the statistical method (Direct Coupling Analysis, DCA) we had used in our previous coevolution-guided protein-protein interaction (PPI) screen in Prokaryotes (9) (the more accurate GREMLIN (11) method is too slow for this) could not effectively distinguish a “gold standard” set of 768 yeast protein pairs known to interact (5) (http://interactome.dfci.harvard.edu/S_cerevisiae/) from the much larger set (768,000 pairs) of primarily non-interacting pairs (Fig. 1B, grey curve, area under the curve: 0.016). Progress required a more accurate and sensitive, but still rapidly computable, method to evaluate protein interactions based on pMSAs.
We explored the application of the deep learning based structure prediction methods, RoseTTAFold (RF) and AlphaFold (AF), to this problem. Even though RF was originally trained on monomeric protein sequences and structures, it can accurately predict the structures of protein complexes given pMSAs with a sufficient number of sequences (13). We found that a lighter-weight (10.7 million parameters) RF two-track model (figs. S4, S5) provided a good tradeoff between compute time and accuracy: the model requires 11 seconds (about 100 times faster than AF) to process a pMSA of 1,000 amino acids on a NVIDIA TITAN RTX graphic processing unit, and it can effectively distinguish gold standard PPIs amongst much larger sets of randomly paired proteins. The very short time required to analyze an individual pMSA made it possible to process all 4.3 million pMSAs. This method considerably outperformed DCA in distinguishing gold standard interactions from random pairs (Fig. 1B, blue curve, area under the curve: 0.219), using the highest predicted contact probability over all pairs of residues in the two proteins as a measure of the propensity for two proteins to interact (fig. S6). Performance was further improved (Fig. 1B, green curve, area under the curve: 0.248) by correcting overestimations of predicted contact probabilities between the C-terminal residues of the first protein and the N-terminal residues of the second protein, and of predicted interactions for a subset of proteins showing hub-like interactions with many other proteins (see Methods and figs. S7, S8). The much better performance of RF than DCA likely stems from the extensive information on protein sequence-structure relationships embedded in the RF deep neural network; DCA by contrast operates solely on protein sequences with no underlying protein structure model.
We next explored whether AF residue-residue contact predictions could further distinguish interacting from non-interacting protein pairs. Like RF, AF was trained on monomeric protein structures, but given the good results with 2-track RF on protein complexes, and the higher accuracy of AF (also a 2-track network followed by a 3D structure module) on monomers, we reasoned that it might similarly have higher accuracy than RF on complexes; to enable modeling of protein complexes using AF, we modified the positional encoding in the AF script (see Methods). AF was too slow to be applied to the entire set of 4.3 million pMSAs (this would require 0.1-1 million GPU hours); instead we applied AF to the 5,495 protein pairs with the highest RF support (indicated by the black vertical line in Fig. 1B). Using the highest AF contact probability over all residue pairs as a measure of interaction strength, we found that the combination of RF followed by AF provided excellent performance (Fig. 1C and figs. S9, S11). Almost all the gold standard pairs were ranked higher than the negative controls, allowing selection of a set of 715 candidate PPIs with an expected precision of 95% at an AF contact probability cutoff of 0.67 (black horizontal line in Fig. 1C); we refer to this RF plus AF procedure as the de novo PPI screen, and the resulting set of predicted interactions, the de novo PPI set, below.
Due to the tradeoff between compute time and accuracy, and the necessity of setting a stringent threshold to avoid large numbers of false positives given the very large number of total pairs, we were concerned that some interacting proteins might not coevolve sufficiently to be identified robustly in our all-vs-all RF screen. Given the excellent performance of AF in distinguishing gold standard interactions amongst the RF filtered pairs, we also applied AF to pMSAs for PPIs reported in the literature, including those identified in high throughput experimental screens. Similarly to our de novo PPI screen procedure, we considered protein pairs with AF contact probability larger than 0.67 to be confident interacting partners. We found that 47% of the gold standard PPIs were confidently predicted, with lower ratios (31% and 24%) for candidate PPIs from the literature (http://interactome.dfci.harvard.edu/S_cerevisiae/download/LC_multiple.txt) (3) or supported by low-throughput experiments according to BIOGRID (21) (Fig. 1D). The ratio of confidently predicted PPIs is even lower for protein pairs identified by Y2H (18%) or APMS (14%) screens (table S1), consistent with the known larger fraction of false positives in large-scale experimental screens (8,22). The fast RF 2-track model used in the de novo screen has comparable or better accuracy than the large-scale experimental screens when assessed in this way: with a high stringency RF cutoff (indicated by the red vertical line in Fig. 1B), the fraction of confidently predicted pairs among PPIs identified by RF is 32%, similar to the accuracy of low-throughput experiments; with a lower stringency cutoff (indicated by the black vertical line in Fig. 1B), this fraction becomes closer to that of the large-scale experimental screens but somewhat fewer true PPIs are missed than with the higher cutoff (Fig. 1D).
In total, we identified 715 likely interacting pairs from the “de novo RF → AF” screen, and 1,251 from the “pooled experimental sets → AF” screen, of which 461 overlap, resulting in a total of 1,505 PPIs (see figs. S11-S13 for interface size and secondary structure distributions for the predicted complex structures). Out of these, 699 have been structurally characterized, 700 have some supporting experimental data from literature and databases, and 106 are not to our knowledge previously described. To evaluate the accuracy of the predicted 3D structure of protein complexes, we used as a benchmark the 699 pairs with experimental structure in the Protein Data Bank (PDB). For 92% of these pairs, at least 50% of confident (predicted aligned error < 8 Å) AF-predicted contacts are present in the experimental structures (Fig. 1E, and fig. S14). The models do miss many contacts observed in the experimental structures however, likely due to lower residue-residue co-evolution (fig. S15).
With these benchmark results providing confidence in the accuracy of the new complex interaction predictions and 3D models of the predicted complexes, we analyzed the structure models for the 806 complexes for which high resolution structural information was not available. We classified these models into groups based on their biological functions, and provide examples of complexes in each functional class in Figs. 2-4. A first set of complexes are involved in maintenance and processing of genetic information: DNA repair, mitosis and meiosis checkpoints, transcription, and translation (Fig. 2). A second set of complexes play roles in protein translocation, transport through the secretory pathway, the cytoskeleton and cell organelles (Fig. 3). A third set of complexes are involved in metabolism (Fig. 4). Examples of protein complexes in which proteins of unknown function are predicted to interact with well characterized ones are shown in Fig. 4: these interactions provide hints about the function of the uncharacterized proteins and could help identify new components of previously characterized assemblies. In cases where three or more proteins were predicted to mutually interact, we generated models of the full assemblies by using as input an sequence alignment for the entire complex (see Methods). Examples of these larger assemblies are shown in Fig. 5; in most cases the pairwise interactions are quite similar to those for the independently built binary complexes, but simultaneous modeling of the full complex has the advantage of allowing conformational changes that could accompany full assembly.
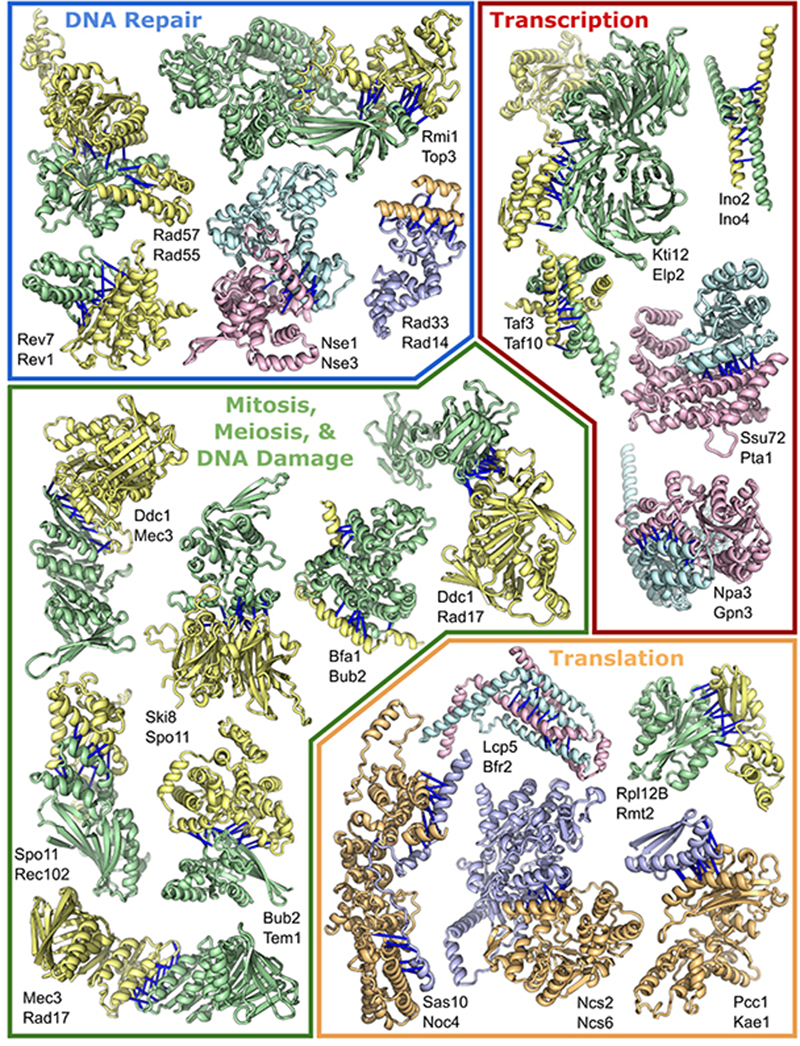
Figure 2. Protein complexes involved in transcription, translation, and DNA repair.
Top predicted residue-residue contacts are indicated with bars. Pair color indicates the method of identification: pairs from the “pooled experimental sets → AF” screen are yellow and green, pairs from the “de novo RF → AF” screen are in blue and light-orange; and pairs present in both datasets are teal and pink. Full names of these proteins are in table S2.
Figure 4. Protein complexes involved in metabolism, GPI anchor biosynthesis or including a protein of unknown function.
Coloring is as in Fig. 2-3. Proteins annotated in the Uniprot database as uncharacterized proteins are denoted with an asterisk. Full names for these proteins are in table S4.
Figure 3. Protein complexes involved in molecule transport, membrane translocation, and mitochondria.
Bars and coloring as in Fig 2. Full names for proteins are in table S3. Membrane spanning regions are annotated on Vtc1-Vtc4 and Sed5-Sft2. Top left: model of Vtc1-Vtc4 complex, with superimposed crystal structure (PDB: 3G3Q, Chain A) of the VTC4 (bright yellow) with phosphate bound (red balls).
Figure 5. Higher order protein complexes.
(A) Top predicted residue-residue contacts for trimers are indicated with bars. Bar color corresponds to the interacting protein pair; protein 1:2 are blue, 1:3 are red, 2:3 are purple. Full names of each protein within the complex are in table S5. (B) Model of Rad55-Rad57-Rad51 and cartoon depiction of placement of this complex in the larger Rad51 filament. Additional information in fig. S18. (C) GARP complex model constructed by predicting structure of central hetero-oligomeric helical bundle, and superimposing models of individual components onto this. 2D class average of GARP complex with minor adaptation (77); reprinted by permission from Springer Nature Customer Service Center GmbH: Springer Nature, Nature Structural and Molecular Biology, CATCHR, HOPS and CORVET tethering complexes share a similar architecture, H-T Chou, D. Dukovski, M.G. Chambers, K.M. Reinisch, and T. Walz, 2016). Alternative GARP models are in fig. S24. (D) Rad33-Rad14 complex model superimposed onto previously determined TFIIH/Rad4-Rad23-Rad33 complex structure (7k04). See fig. S19 for additional details. (E) GPI-T pentamer model highlighting a possible peptide substrate recognition channel adjacent to the catalytic dyad. See fig. S27 for additional details.
It is not possible to analyze the functional implications of all of the new complexes in a single paper. Instead, as an illustration of the insights which can be gained from these, we focus on a few selected examples in the following sections. To enable broader study of the functional implications of the full set of models, we have made them available at https://modelarchive.org/doi/10.5452/ma-bak-cepc and additional information is provided in the supplemental Excel file.
Complexes involved in DNA homologous recombination and repair
The homologous recombination required for accurate chromosome segregation during meiosis is initiated by DNA double-strand breaks made by Spo11 (23). Spo11 is essential for sexual reproduction in most Eukaryotes (24,25), but mechanistic insight has been limited by a deficit of high-resolution structural information. We predict the structures of complexes of Spo11 with its essential partners Ski8 and Rec102 (Fig. 2 and figs. S16, S17). The predicted Spo11–Ski8 structure is supported by crosslinking and mutagenesis data (26,27). Our model resembles a previous model based on the Ski3–Ski8 complex, with Ski8 contacting a sequence in Ski3 that is similar to the sequence QREIF380 in Spo11 (27,28) (fig. S17A), but suggests a more extensive interaction surface than previously appreciated, involving an insertion in Ski8 that is present in Saccharomyces species but not in Schizosaccharomyces pombe and Sordaria macrospora, where Ski8 is also required for meiosis (29,30) (fig. S17B, C). Rec102 was proposed to be a remote homolog of the transducer domain of the Top6B subunit of archaeal topoisomerase VI (31), which couples ATP-dependent dimerization of Top6B subunits to DNA cleavage by Top6A subunits (32). Our predicted Rec102–Spo11 complex resembles the Top6A–Top6B interface: a four-helix bundle consisting of two C-terminal helices from Rec102 and two helices from Spo11 (the first helix of the winged helix domain (WHD) plus a more N-terminally located helix) (fig. S17D). Alanine substitutions in this portion of Rec102 disrupt interaction with Spo11 and block meiotic recombination in vivo (27). The model clarifies the Spo11 portion of this interface, which was not well structured in previous homology models (27,31). Both Rec102 and Top6B have long, helical arms that feed into the Spo11 interface; our model predicts a different angle for this arm and contains a kink that corresponds to a conserved sequence motif EYPMVF192 in Saccharomyces that is missing in both archaeal TopoVI and mammals (fig. S17D, E). Mutations in this region can suppress rec104 conditional alleles (33), suggesting that this part of Rec102 is important for integrating Rec104 function into the Spo11 core complex.
The highly conserved Rad51 protein central to DNA repair carries out key reactions during homologous recombination, and mutations in human paralogs are associated with Fanconi anemia and multiple types of cancer (34). Rad51 paralogs can be positive regulators of Rad51 activity (35); in yeast the Rad51 paralogs Rad55 and Rad57 form a stable homodimer that accelerates assembly of Rad51 filaments on single–stranded DNA (ssDNA) during homologous recombination through a transient interaction with Rad51 (36). The lack of structural data for the Rad55–Rad57 complex and its interface with Rad51 has limited mechanistic understanding of this process. We generated a model of the trimeric Rad55–Rad57–Rad51 complex, which in combination with the known Rad51 filament structure (37), suggests that Rad55–Rad57 binds at the 5’ end of the Rad51 filament where it could promote growth of the Rad51 filament in a directional manner (Fig. 5B and fig. S18).
Nucleotide excision repair (NER) requires a search for lesions in DNA that is mediated by a conserved complex containing Rad4 (XPC), Rad23 (HR23B) and Rad33 (Centrin2) in yeast. The Rad4–Rad23–Rad33 complex is essential for global genome NER and is the major player in initial damage recognition (38). Rad14 (XPA) is recruited at a later stage and activates the helicase Rad3 (XPD) subunit of the general transcription and DNA repair factor IIH complex (TFIIH, consisting of Rad3, Ssl2, Ssl1, Tfb1, Tfb2, Tfb4, and Tfb5) through the release of the TFIIK (CAK) complex following interactions with the TFIIH subunits Tfb5 (p8) and Ssl2 (XPB), and double stranded DNA (39). The structures of Rad14 that are currently available only comprise the extended DNA binding domain and lack the N- and C-terminus, where the latter interacts with Tfb5. We generated a model of the complex between full length Rad14 and Rad33 that resolves much of the current structural ambiguity in this system (Fig. 2 and fig S19B), shedding light on how Rad14 may be recruited to the Rad4–Rad23–Rad33 complex. Placing this model into a cryo EM map comprising XPA (Rad14) and TFIIH bound to DNA (39) suggests how the Rad14 C-terminus, which fits into previously unmodeled density, interacts with TFIIH. The long central helix observed in the Centrin2 (Rad33) structure (40) is kinked about 90° in our Rad33-Rad14 complex model (fig. S19B); both conformations are feasible and are compatible for the interaction with Rad14. In a recent cryo EM structure of the TFIIH/Rad4–Rad23–Rad33 initial recognition complex (41), only the C- terminal part of Rad33 was determined. Superposition of Rad33 in the Rad33-Rad14 complex model onto this structure (Fig. 5D) shows how Rad14 can interact with the Rad4–Rad23–Rad33 recognition complex (38,42) while maintaining the TFIIH interaction, bridging the steps of initial damage recognition and damage verification. Our model suggests that Rad14 and Rad4 can be present at the same time in the repair cascade; crosstalk between these important proteins could modulate downstream events.
Complexes involved in translation and ribosome regulation
Throughout evolution the eukaryotic machinery for protein production has expanded in size and complexity (43), which facilitated the development of sophisticated mechanisms for the regulation of gene expression at the post-transcriptional level (44) and increased integration with the cellular environment (45). The expanded complexity of the eukaryotic translational machinery came at the cost of a highly complex process for ribosome maturation (46). We generate models of complexes which had not been structurally characterized previously that involve components of the translation apparatus (Fig. 2 and fig. S20). Two complexes, Rpl12B–Rmt2 and Rpl7A–Fpr4, involving enzymes that introduce protein modifications such as arginine methylations or proline isomerizations (47), provide insight into mechanisms that expand the chemical diversity of ribosomal proteins at functional sites (48) and possibly regulate translation (49). A complex between components of the U3 ribosome-maturation factor and a protein involved in the regulation of glycerol, Lcp5–Sgd1 (50), could play a role in coupling translation with metabolism. A complex between eIF2B, an auxiliary factor for eIF2 recycling after GTP hydrolysis, and transcriptional factor regulator Dig2 could help couple translation and transcription: the delivery of the first aminoacyl-tRNA (Met-tRNAi Met) is a key event in eukaryotic translation regulation by the GTPase eIF2 (51) and targeting eIF2 via its nucleotide exchanger eIF2B is a basal mechanism of translation regulation. This possible cross-talk between ribosome-maturation pathways and metabolic sensors, and translation initiation regulators such as eIF2 with transcription factors suggests exciting new avenues to further map the highly integrated nature of translation within eukaryotic cells.
Complexes involving ubiquitin and small ubiquitin-like modifier (SUMO) ligases
Reversible covalent modifications of proteins with ubiquitin and SUMO modulate protein-protein interactions, cellular localization, and stability (52). SUMO E3 ligases facilitate SUMO transfer, and Siz1, Siz2, Mms21, and Zip3 are the known SUMO ligases in budding yeast (52). Our model of the Siz2 and Mms21 SUMO ligase complex (fig. S21A) suggests that both E3s could act jointly to modify DNA associated substrates perhaps through the DNA binding SAP domain of Siz2 (53) or involving the Mms21 (Nse2) containing Smc5–6 complex which modulates DNA recombination, replication and repair (54,55). The Smc5–6 complex contains another RING-finger E3 ligase-like subunit, Nse1 (56) that interacts with Nse3 and Nse4. Our model of the yeast Nse1–Nse3–Nse4 complex (fig. S21B) is similar to a structure determined for the Xenopus laevis complex, despite the sequences of the yeast and Xenopus proteins being too distant for similarity to be detectable by BLAST.
SUMO-targeted ubiquitin ligases (STUbLs) are ubiquitin ligases that recognize SUMO-modified proteins. A STUbL consisting of the Slx8 ubiquitin ligase and the associated protein Slx5 functions in proteasome-mediated turnover of several proteins associated with DNA replication, repair and chromosome structure (57–59). Our model of the Slx5-Slx8 complex (fig. S21C) provides insight into how these two proteins may collectively recognize their substrates. In addition, we generated a lower confidence but intriguing model of a previously undescribed complex between Slx8 and Cue3 (Coupling of ubiquitin conjugation to endoplasmic reticulum (ER) degradation protein 3) (fig. S21D), possibly linking ubiquitination of substrates to protein degradation in ER.
Complexes involved in chromosome segregation
The heterodecameric complex DASH/Dam1 (Dam1c) is composed of 10 proteins: Ask1, Dad1, Dad2, Dad3, Dad4, Dam1, Duo1, Hsk3, Spc19, and Spc34 which come together to form a “T” shape, and can further oligomerize into rings (60,61). During mitosis, these heterodecamers strengthen the attachment between kinetochores and microtubules (62) by oligomerizing to form either partial or complete rings around microtubules and further contacting kinetochore components (63–65). Microtubules are required for in-vivo ring formation, but a structure of the Dam1c ring complex from Chaetomium thermophilum was determined in the absence of microtubules using monovalent salts (66). We generated structure models of nine binary complexes (Dad2-Ask1, Dad2-Hsk3, Dad2-Spc1, Dad4-Hsk3, Dam1-Duo1, Duo1-Dad1, Spc19-Dad1, Spc34-Duo1, and Spc34-Spc19) that encompass several members of Dam1c (fig. S22). These complexes are largely consistent with the Dam1c structure, suggesting that the findings from the thermophile structure can likely be extended to S. cerevisiae. We went beyond previous structural data by predicting the structure of a potential inter-decamer interaction between Spc19 and Dad1 involving a flexible loop of Spc19 and the N-terminal region of Dad1, which could be important for ring formation in vivo (66).
Complexes involved in molecule transport and membrane trafficking
The small membrane protein Ksh1 is conserved across eukaryotes, essential for growth, and plays an unknown role in protein secretion (67). We predicted structures of complexes between Ksh1 and two membrane proteins reported to form a complex: Yos1 and Yip1. This complex also includes Yif1 and interacts with Rab GTPases (68) (Fig. 3). These structures suggest Ksh1 is a fourth member of this enigmatic complex essential to the secretory pathway, and explains how Ksh1 can play a role in secretion despite its small size of 72 amino acids.
The vacuolar transporter chaperone (VTC) is a 5-subunit complex that synthesizes polyphosphate to regulate cellular phosphate levels (69). Structures are only known for some soluble portions of this complex, including the catalytic domain of the Vtc4 subunit (70). Our model of the previously non-structurally characterized Vtc1–Vtc4 subcomplex suggests that the cytosolic active site is positioned by the complex to feed the polyphosphate product through a membrane pore into the lumen of the lysosome (Fig. 3).
The ESCRT-III complex is involved in a number of cellular membrane remodeling pathways, including receptor downregulation, membrane repair, and cell division (71,72). Our predicted interface between the Vps2 and Vps24 subunits of the ESCRT-III complex resembles the polymerization interface of a different ESCRT-III subunit Snf7 (73), providing insight into the roles of these previously uncharacterized ESCRT-III subunits, and highlighting the generality of this mode of interaction in ESCRT-III complexes. Notably, previously unpublished mutations (fig. S23) in Vps24 that prevent ESCRT function in multivesicular body sorting are located on the predicted interface between Vps2 and Vps24, supporting our model and the functional importance of the Vps2–Vps24 interaction. Vps55 and Vps68 are conserved membrane proteins that are important for endosomal cargo sorting; our predicted structure (Fig. 2) of their interaction provides clues about the mechanism of their function (74).
The GARP complex is a multisubunit tethering complex (MTC) that mediates docking and fusion of vesicles with the Golgi apparatus (75). Our approach generated models for binary complexes involving the four GARP subunits, and we further modeled the entire complex (fig. S24A). In this model, the four subunits assemble through a four-helix bundle. In each of the three larger subunits, Vps52, Vps53, and Vps54, C-terminal domains comprising “CATCHR” folds emanate from the bundle. This architecture resembles portions of the cryo-EM structure of the Exocyst complex, a distinct MTC that mediates fusion of vesicles at the plasma membrane (76), which possesses two separate four-helix bundles organizing its eight subunits. In our prediction, the “CATCHR” domains appear to be somewhat flexibly linked to the central four-helix bundle, and hence we overlaid the structure predictions for Vps52, Vps53, and Vps54, respectively, onto the central four-helix bundle (Fig. 5C and fig. S24B). The resulting model has a striking resemblance to previously published 2D classes (fig. S24C) from a negative-stain EM analysis of the GARP complex (77). These predictions will facilitate structure-guided experiments to elucidate the mechanism of MTC function.
Golgi-resident protein, Grh1, forms a tethering complex with Uso1 and Bug1 that interacts with the COPII coat protein complex, Sec23/Sec24. The tether is thought to participate in COPII vesicle capture (78,79), but the mechanism remains unclear. The C-terminus of Grh1 contains a predicted intrinsically disordered region (IDR) with a net positively charged cluster and a triple-proline motif (fig. S25A, B). Our model of the Sec23–Grh1 complex contains an interface between the Sec23 gelsolin domain and the PPP motif of Grh1 (80), and an interface between the Grh1 IDR and Sec23 involving a disorder-to-helical transition (fig. S25C). A similar multivalent interface also drives interaction between Sec23 and the COPII coat scaffolding protein, Sec31 (81). Our model suggests that the combinatorial multivalent interaction between Grh1 and Sec23 may compete with the interaction between Sec31 and Sec23 to promote vesicle uncoating; consistent with this model, Grh1 is recruited to GST-Sec23, dependent on the IDR, and competes for Sec31 binding (fig. S25D).
SNARE proteins drive intracellular membrane fusion between transport vesicles and organelles (82). Our predicted complex structure between the SNARE Sed5 and the uncharacterized transmembrane protein Sft2 unexpectedly predicted an interaction between transmembrane domains of the two proteins (Fig. 3). SNARE localization is thought to occur through interactions of cytoplasmic domains with cytoplasmic sorting factors, but this prediction, together with genetic evidence (83), suggests SNARE localization or function may be subject to additional mechanisms via interactions with transmembrane protein regulators. Membrane fusion requires the formation of a 4-helix bundle (called the SNARE complex) between the vesicle SNARE and the target membrane SNAREs (84,85). The bundle is formed by the SNARE motifs, which are 60-70 amino acids with heptad repeats and the ability to form coiled-coil structures. Models of binary complexes of SNARE-motif-containing proteins frequently differ from their classic conformation in the SNARE four-helical bundle (fig. S26A), probably because all four chains are required to form the stable complex (86). Indeed modeling the four SNARE proteins (Ufe1, Use1, Sec20, and Sec22) that are known to mediate the fusion between Golgi-derived retrograde transport vesicles with ER (87), together resulted in a complex that resembles a typical SNARE complex (84) (fig. S26B, C). This example highlights the potential pitfalls of modeling only binary complexes when the functional assembly involves more than two chains.
GPI transamidase complex
Glycosylphosphatidylinositol transamidase (GPI-T) is a pentameric enzyme complex of unknown structure (88–90) which catalyzes the attachment of GPI anchors to the C-terminus of specific substrate proteins, based on recognition of a C-terminal signal peptide (91). GPI-T catalyzes the removal of this signal sequence, replacing it with a new amide bond to an ethanolamine phosphate in the GPI anchor. The five subunits of S. cerevisiae GPI-T are Gpi8 (which contains the catalytic active site), Gpi16, Gaa1, Gpi17, and Gab1 (88,92,93). Our large-scale modeling approach generated models for the following binary complexes: Gpi8-Gpi17, Gab1-Gaa1, Gab1-Gpi17, and Gaa1-Gpi16. We subsequently modeled the full-length, pentameric GPI-T in one shot starting from the sequences of all components (Fig. 5E). Several features of this model are consistent with previous characterization of this enzyme. S. cerevisiae GPI-T can be purified as a core heterotrimer, containing only Gpi8, Gpi16, and Gaa1 (92); our GPI-T model confirms extensive interactions between the soluble domains of these three subunits. This model also recapitulates the disulfide bond between Gpi8 (Cys85) and Gpi16 (Cys202), previously characterized for human GPI-T (94) (the existence of this disulfide bond in yeast GPI-T has been called into question (90)). Gaa1 is essential for binding of the GPI anchor to GPI-T (95) and the hydrophobic Gab1 is also predicted to participate in anchor recognition (88). Our model positions the transmembrane regions of Gaa1 and Gab1 against each other. The catalytic dyad in Gpi8 (Cys199 and His157) faces these transmembrane domains, and abuts against a highly conserved face of Gaa1, proposed to recognize the GPI anchor glycans (96,97). In our model, the positions of these subunits are consistent with binding of the GPI anchor to position the modifying amine in the Gpi8 active site for catalysis. Gpi16 is immediately adjacent to these interactions and is likely to also be involved in anchor recognition. In vivo, GPI-T is expected to be a dimer of pentamers, with dimerization occurring on one face of the caspase-like Gpi8 subunit (92,97,98). This decameric complex was too large for us to model computationally; however the pentameric complex we present here leaves open the dimerization face of Gpi8, consistent with probable dimerization. It also suggests that Gaa1 and Gpi17 would participate in dimerization of this enzyme. The functional role of Gpi17 has been elusive, but our model now suggests Gpi17 together with Gpi8 and Gpi16, forms a recognition channel for the C-terminal GPI-T signal peptide (fig. S27). In humans, mutations in GPI-T subunits are associated with neurodevelopmental disorders (99). Each subunit contributes to different cancer mechanisms, in some cases by perturbing GPI anchoring of specific receptors and in others by separating from GPI-T to alter disparate signal transduction pathways (89). Now, with a structural model in hand, these mechanisms can be examined at a molecular level.
Limitations of the current method
As with any new method, it is important when interpreting the results (our large set of predicted complex structures) to keep in mind the limitations of the approach. First, our study is not comprehensive, so conclusions should not be drawn about absences; in particular we eliminated proteins that arose from recent duplication due to difficulty in identifying orthologs in other organisms, and thus only surveyed 2/3 of the entire yeast proteome. Second, the approach likely misses interactions restricted to a small set of organisms, or which vary rapidly during evolution, due to weaker co-evolutionary signals. Third, the approach likely works less well for transient interactions which generally involve smaller and weaker interfaces which may be under lower selective pressure, in particular those involving intrinsically disordered regions which are poorly represented in the PDB. The majority of known interactions identified by our approach are likely obligate assemblies and involve ordered structural elements. Fourth, interactions between single hydrophobic or amphipathic helices, such as single transmembrane helices or coiled coils, may be overpredicted (in initial studies of human complexes, interactions solely between single-pass transmembrane regions appear to be over represented). Fifth, and perhaps most importantly, for proteins that form high-order obligate protein complexes, binary complex models may be quite inaccurate, as illustrated by the SNARE example.
Conclusion
Our approach extends the range of large scale deep learning based structure modeling from monomeric proteins to protein assemblies. As highlighted by the above examples, following up on the many new complexes presented here should advance understanding of a wide range of eukaryotic cellular processes and provide new targets for therapeutic intervention. The methods can be extended directly to large scale mapping of interactions in the human proteome, but considerably more compute time will be required given the much larger total number of protein pairs, and models may be somewhat less accurate due to weaker co-evolutionary signal for the subset of human proteins unique to higher eukaryotes and for the many closely related paralogs arising from gene duplication. Investigating interactions of individual proteins or subsets of proteins, for example, deorphanization of orphan receptors, should be immediately accessible using our approach provided there are sufficient sequence homologues. Training RF and AF on protein complexes should further improve performance of both methods (100), particularly for protein pairs with fewer homologues and/or weaker and more transient interactions, and reduce the dependence on ortholog identification. Together with the advances in monomeric structure prediction, our results herald a new era of structural biology in which computation plays a fundamental role in both interaction discovery and structure determination.
Methods
As described in detail in the Supplemental Methods, we developed a multistep bioinformatics and deep learning pipeline for identifying pairs of proteins likely to interact and modeling the three dimensional structures of the corresponding protein complexes. The steps of this pipeline are illustrated schematically in Fig. 1A. First, comprehensive orthologous groups of genes were generated and yeast genes were mapped to these groups; second, multiple sequence alignments of orthologous sequences were generated for each pair of yeast proteins; third, contact probability was computed for each protein pair using RoseTTAFold; and fourth, interaction probability was re-evaluated and complex structures were modeled using AlphaFold. The experimental data-guided PPI screening pipeline is very similar except that in the third stage, instead of using RoseTTAFold, we used experimental data primarily derived from large-scale screens to identify PPI candidates.
Supplementary Material
One-sentence summary.
Proteome-wide coevolution and deep-learning methods identify and build accurate models of eukaryotic protein complexes.
Acknowledgements
We thank Eric Horvitz, Nick V. Grishin, Hahnboem Park, and James H. Thomas for helpful discussions, Luki Goldschmidt and Aaron Guillory for computing resource management, and Lance Stewart for logistical support. Additionally, we are grateful to Martin Bard, Trisha N Davis, David G Drubin, Maitreya J Dunham, Scott D Emr, Frederick Hughson, James Hurley, Kenji Murakami, Nobuhiro Nakamura, Eva Nogales, Randy Schekman, Shu-ou Shan, Soyeon Showman, Kaoru Sugasawa, and Sho Suzuki for their correspondence and biological expertise. We thank Stephen Burley, Brinda Vallat, and John Westbrook at the RCSB Protein Data Bank and Torsten Schwede, Gerardo Tauriello, Andrew Waterhouse, and Stefan Bienert at SWISS-MODEL for hosting our model structures at ModelArchive.
Funding
This work was supported by Microsoft (MB, DB, and Azure compute time and expertise), Amgen (DB and IH), Southwestern Medical Foundation (JP and QC), the Washington Research Foundation (MB and QC), Howard Hughes Medical Institute (DB, SB, SK, and generous compute time on Janelia), National Science Foundation (NSF) Cyberinfrastructure for Biological Research (CIBR, Award # DBI 1937533 to DB and IA), CPRIT training grant (RP210041 to JZ), UK Medical Research Council (MRC_UP_1201/10 to EAM.), HHMI Gilliam Fellowship (DJB), the Deutsche Forschungsgemeinschaft (KI-562/11-1 and KI-562/7-1 to CK), NIH/NIGMS (R21AI156595 to SO, R35GM136258 to JCF, R35NS097333 to JR, R35GM118026 and R01CA221858 to ECG), HHMI fellowship of the Damon Runyon Cancer Research Foundation (DRG2273-16 to SB and DRG2389-20 to KL), AIRC investigator and the European Research Council Consolidator (IG23710 and 682190 to DBr), the Defense Threat Reduction Agency (HDTRA1-21-1-0007 to DB). We also thank The National Energy Research Scientific Computing Center (NERSC) for providing computing time (project m3962 at NERSC).
Footnotes
Author contributions:
QC and DB conceived the research; JP and QC prepared the sequence alignments used in the screen; MB implemented the RoseTTAFold pipeline; MB and SO repurposed AlphaFold for complex modeling; JP, JZ, and QC designed the PPI screening procedure; IRH, MB, IA, and QC carried out the screen; IRH, AK, and QC analyzed and presented the results; IRH, AK, QC and DB coordinated the collaborative efforts;TJN, SB, SRB, VGS, XHL, KL, ZZ, DJB, UR, JK, ISF, BS, DB, JR, CK, ECG, SB, SK, EAM, JCF, and TLH provided biological insights on specific examples; QC and DB drafted the manuscript while all other authors contributed to the description of specific examples; all authors discussed the results and commented on the manuscript.
Competing interests: Authors declare that they have no competing interests.
Data and materials availability
Data and materials availability: Structures of highly confident pairs with accompanying pMSAs and metadata are available at ModelArchive: https://modelarchive.org/doi/10.5452/ma-bak-cepc. RoseTTAFold two-track version is available at https://github.com/RosettaCommons/RoseTTAFold or Zenodo (101). AlphaFold was fetched from https://github.com/deepmind/alphafold on July 16th, 2021 (v2.0.0). Code for a GPU implementation of DCA and the modifications to the AlphaFold predictions script are provided in Supplemental Methods.
References
- 1.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins SR, Kemmeren P, Zhao X-C, Greenblatt JF, Spencer F, Holstege FCP, Weissman JS, Krogan NJ. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol Cell Proteomics. 2007;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Reguly T, Breitkreutz A, Boucher L, Breitkreutz B-J, Hon GC, Myers CL, Parsons A, Friesen H, Oughtred R, Tong A, Stark C, et al. Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. J Biol. 2006;5:11. doi: 10.1186/jbiol36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 5.Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, Hao T, et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuchaiev O, Rasajski M, Higham DJ, Przulj N. Geometric de-noising of protein-protein interaction networks. PLoS Comput Biol. 2009;5:e1000454. doi: 10.1371/journal.pcbi.1000454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edwards AM, Kus B, Jansen R, Greenbaum D, Greenblatt J, Gerstein M. Bridging structural biology and genomics: assessing protein interaction data with known complexes. Trends Genet. 2002;18:529–536. doi: 10.1016/s0168-9525(02)02763-4. [DOI] [PubMed] [Google Scholar]
- 8.Mackay JP, Sunde M, Lowry JA, Crossley M, Matthews JM. Protein interactions: is seeing believing? Trends Biochem Sci. 2007;32:530–531. doi: 10.1016/j.tibs.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Cong Q, Anishchenko I, Ovchinnikov S, Baker D. Protein interaction networks revealed by proteome coevolution. Science. 2019;365:185–189. doi: 10.1126/science.aaw6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green AG, Elhabashy H, Brock KP, Maddamsetti R, Kohlbacher O, Marks DS. Large-scale discovery of protein interactions at residue resolution using co-evolution calculated from genomic sequences. Nat Commun. 2021;12:1396. doi: 10.1038/s41467-021-21636-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ovchinnikov S, Kamisetty H, Baker D. Robust and accurate prediction of residue-residue interactions across protein interfaces using evolutionary information. Elife. 2014;3:e02030. doi: 10.7554/eLife.02030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hopf TA, Schärfe CPI, Rodrigues JPGLM, Green AG, Kohlbacher O, Sander C, Bonvin AMJJ, Marks DS. Sequence co-evolution gives 3D contacts and structures of protein complexes. Elife. 2014;3 doi: 10.7554/eLife.03430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baek M, DiMaio F, Anishchenko I, Dauparas J, Ovchinnikov S, Lee GR, Wang J, Cong Q, Kinch LN, Schaeffer RD, Millán C, et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science. 2021;373:871–876. doi: 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer A, Schartl M. Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr Opin Cell Biol. 1999;11:699–704. doi: 10.1016/s0955-0674(99)00039-3. [DOI] [PubMed] [Google Scholar]
- 16.Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R, Riley R, Salamov A, Zhao X, Korzeniewski F, Smirnova T, et al. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014;42:D699-704. doi: 10.1093/nar/gkt1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spingola M, Grate L, Haussler D, Ares M., Jr Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA. 1999;5:221–234. doi: 10.1017/s1355838299981682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zdobnov EM, Kuznetsov D, Tegenfeldt F, Manni M, Berkeley M, Kriventseva EV. OrthoDB in 2020: evolutionary and functional annotations of orthologs. Nucleic Acids Res. 2021;49:D389–D393. doi: 10.1093/nar/gkaa1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clum A, Huntemann M, Bushnell B, Foster B, Foster B, Roux S, Hajek PP, Varghese N, Mukherjee S, Reddy TBK, Daum C, et al. DOE JGI Metagenome Workflow. mSystems. 2021;6 doi: 10.1128/mSystems.00804-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wall DP, Fraser HB, Hirsh AE. Detecting putative orthologs. Bioinformatics. 2003;19:1710–1711. doi: 10.1093/bioinformatics/btg213. [DOI] [PubMed] [Google Scholar]
- 21.Oughtred R, Rust J, Chang C, Breitkreutz B-J, Stark C, Willems A, Boucher L, Leung G, Kolas N, Zhang F, Dolma S, et al. The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021;30:187–200. doi: 10.1002/pro.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang H, Jedynak BM, Bader JS. Where have all the interactions gone? Estimating the coverage of two-hybrid protein interaction maps. PLoS Comput Biol. 2007;3:e214. doi: 10.1371/journal.pcbi.0030214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 24.de Massy B. Initiation of meiotic recombination: how and where? Conservation and specificities among eukaryotes. Annu Rev Genet. 2013;47:563–599. doi: 10.1146/annurev-genet-110711-155423. [DOI] [PubMed] [Google Scholar]
- 25.Murakami H, Keeney S. Regulating the formation of DNA double-strand breaks in meiosis. Genes Dev. 2008;22:286–292. doi: 10.1101/gad.1642308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arora C, Kee K, Maleki S, Keeney S. Antiviral protein Ski8 is a direct partner of Spo11 in meiotic DNA break formation, independent of its cytoplasmic role in RNA metabolism. Mol Cell. 2004;13:549–559. doi: 10.1016/s1097-2765(04)00063-2. [DOI] [PubMed] [Google Scholar]
- 27.Bouuaert CC, Tischfield SE, Pu S, Mimitou EP, Arias-Palomo E, Berger JM, Keeney S. Structural and functional characterization of the Spo11 core complex. Nat Struct Mol Biol. 2021;28:92–102. doi: 10.1038/s41594-020-00534-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Halbach F, Reichelt P, Rode M, Conti E. The yeast ski complex: crystal structure and RNA channeling to the exosome complex. Cell. 2013;154:814–826. doi: 10.1016/j.cell.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 29.Steiner S, Kohli J, Ludin K. Functional interactions among members of the meiotic initiation complex in fission yeast. Curr Genet. 2010;56:237–249. doi: 10.1007/s00294-010-0296-0. [DOI] [PubMed] [Google Scholar]
- 30.Tessé S, Storlazzi A, Kleckner N, Gargano S, Zickler D. Localization and roles of Ski8p protein in Sordaria meiosis and delineation of three mechanistically distinct steps of meiotic homolog juxtaposition. Proc Natl Acad Sci U S A. 2003;100:12865–12870. doi: 10.1073/pnas.2034282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robert T, Nore A, Brun C, Maffre C, Crimi B, Bourbon H-M, de Massy B. The TopoVIB-Like protein family is required for meiotic DNA double-strand break formation. Science. 2016;351:943–949. doi: 10.1126/science.aad5309. [DOI] [PubMed] [Google Scholar]
- 32.Corbett KD, Benedetti P, Berger JM. Holoenzyme assembly and ATP-mediated conformational dynamics of topoisomerase VI. Nat Struct Mol Biol. 2007;14:611–619. doi: 10.1038/nsmb1264. [DOI] [PubMed] [Google Scholar]
- 33.Salem L, Walter N, Malone R. Suppressor analysis of the Saccharomyces cerevisiae gene REC104 reveals a genetic interaction with REC102. Genetics. 1999;151:1261–1272. doi: 10.1093/genetics/151.4.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sullivan MR, Bernstein KA. RAD-ical New Insights into RAD51 Regulation. Genes. 2018;9 doi: 10.3390/genes9120629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Filippo JS, Sung P, Klein H. Mechanism of Eukaryotic Homologous Recombination. Annual Review of Biochemistry. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 36.Roy U, Kwon Y, Marie L, Symington L, Sung P, Lisby M, Greene EC. The Rad51 paralog complex Rad55-Rad57 acts as a molecular chaperone during homologous recombination. Molecular Cell. 2021;81:1043–1057.:e8. doi: 10.1016/j.molcel.2020.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conway AB, Lynch TW, Zhang Y, Fortin GS, Fung CW, Symington LS, Rice PA. Crystal structure of a Rad51 filament. Nat Struct Mol Biol. 2004;11:791–796. doi: 10.1038/nsmb795. [DOI] [PubMed] [Google Scholar]
- 38.Sugasawa K, Akagi J-I, Nishi R, Iwai S, Hanaoka F. Two-step recognition of DNA damage for mammalian nucleotide excision repair: Directional binding of the XPC complex and DNA strand scanning. Mol Cell. 2009;36:642–653. doi: 10.1016/j.molcel.2009.09.035. [DOI] [PubMed] [Google Scholar]
- 39.Kokic G, Chernev A, Tegunov D, Dienemann C, Urlaub H, Cramer P. Structural basis of TFIIH activation for nucleotide excision repair. Nat Commun. 2019;10:2885. doi: 10.1038/s41467-019-10745-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson JR, Ryan ZC, Salisbury JL, Kumar R. The Structure of the Human Centrin 2-Xeroderma Pigmentosum Group C Protein Complex. Journal of Biological Chemistry. 2006;281:18746–18752. doi: 10.1074/jbc.M513667200. [DOI] [PubMed] [Google Scholar]
- 41.van Eeuwen T, Shim Y, Kim HJ, Zhao T, Basu S, Garcia BA, Kaplan CD, Min J-H, Murakami K. Cryo-EM structure of TFIIH/Rad4-Rad23-Rad33 in damaged DNA opening in nucleotide excision repair. Nat Commun. 2021;12:1–17. doi: 10.1038/s41467-021-23684-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Riedl T, Hanaoka F, Egly J-M. The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J. 2003;22:5293–5303. doi: 10.1093/emboj/cdg489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klinge S, Voigts-Hoffmann F, Leibundgut M, Ban N. Atomic structures of the eukaryotic ribosome. Trends Biochem Sci. 2012;37:189–198. doi: 10.1016/j.tibs.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 44.Hinnebusch AG, Ivanov IP, Sonenberg N. Translational control by 5’-untranslated regions of eukaryotic mRNAs. Science. 2016;352:1413–1416. doi: 10.1126/science.aad9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saba JA, Liakath-Ali K, Green R, Watt FM. Translational control of stem cell function. Nat Rev Mol Cell Biol. 2021 doi: 10.1038/s41580-021-00386-2. [DOI] [PubMed] [Google Scholar]
- 46.Klinge S, Woolford JL., Jr Ribosome assembly coming into focus. Nat Rev Mol Cell Biol. 2019;20:116–131. doi: 10.1038/s41580-018-0078-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mulvaney KM, Blomquist C, Acharya N, Li R, Ranaghan MJ, O’Keefe M, Rodriguez DJ, Young MJ, Kesar D, Pal D, Stokes M, et al. Molecular basis for substrate recruitment to the PRMT5 methylosome. Mol Cell. 2021;81:3481–3495.:e7. doi: 10.1016/j.molcel.2021.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watson ZL, Ward FR, Méheust R, Ad O, Schepartz A, Banfield JF, Cate JH. Structure of the bacterial ribosome at 2 Å resolution. Elife. 2020;9 doi: 10.7554/eLife.60482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Małecki JM, Odonohue M-F, Kim Y, Jakobsson ME, Gessa L, Pinto R, Wu J, Davydova E, Moen A, Olsen JV, Thiede B, et al. Human METTL18 is a histidine-specific methyltransferase that targets RPL3 and affects ribosome biogenesis and function. Nucleic Acids Res. 2021;49:3185–3203. doi: 10.1093/nar/gkab088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dragon F, Gallagher JEG, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, Beyer AL, et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kenner LR, Anand AA, Nguyen HC, Myasnikov AG, Klose CJ, McGeever LA, Tsai JC, Miller-Vedam LE, Walter P, Frost A. eIF2B-catalyzed nucleotide exchange and phosphoregulation by the integrated stress response. Science. 2019;364:491–495. doi: 10.1126/science.aaw2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jentsch S, Psakhye I. Control of nuclear activities by substrate-selective and protein-group SUMOylation. Annu Rev Genet. 2013;47:167–186. doi: 10.1146/annurev-genet-111212-133453. [DOI] [PubMed] [Google Scholar]
- 53.Psakhye I, Jentsch S. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell. 2012;151:807–820. doi: 10.1016/j.cell.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 54.Menolfi D, Delamarre A, Lengronne A, Pasero P, Branzei D. Essential Roles of the Smc5/6 Complex in Replication through Natural Pausing Sites and Endogenous DNA Damage Tolerance. Mol Cell. 2015;60:835–846. doi: 10.1016/j.molcel.2015.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Agashe S, Joseph CR, Reyes TAC, Menolfi D, Giannattasio M, Waizenegger A, Szakal B, Branzei D. Smc5/6 functions with Sgs1-Top3-Rmi1 to complete chromosome replication at natural pause sites. Nat Commun. 2021;12:2111. doi: 10.1038/s41467-021-22217-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Piccoli G, Torres-Rosell J, Aragón L. The unnamed complex: what do we know about Smc5-Smc6? Chromosome Res. 2009;17:251–263. doi: 10.1007/s10577-008-9016-8. [DOI] [PubMed] [Google Scholar]
- 57.Psakhye I, Castellucci F, Branzei D. SUMO-Chain-Regulated Proteasomal Degradation Timing Exemplified in DNA Replication Initiation. Mol Cell. 2019;76:632–645.:e6. doi: 10.1016/j.molcel.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Waizenegger A, Urulangodi M, Lehmann CP, Reyes TAC, Saugar I, Tercero JA, Szakal B, Branzei D. Mus81-Mms4 endonuclease is an Esc2-STUbL-Cullin8 mitotic substrate impacting on genome integrity. Nat Commun. 2020;11:5746. doi: 10.1038/s41467-020-19503-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Psakhye I, Branzei D. SMC complexes are guarded by the SUMO protease Ulp2 against SUMO-chain-mediated turnover. Cell Rep. 2021;36:109485. doi: 10.1016/j.celrep.2021.109485. [DOI] [PubMed] [Google Scholar]
- 60.Miranda JJL, De Wulf P, Sorger PK, Harrison SC. The yeast DASH complex forms closed rings on microtubules. Nat Struct Mol Biol. 2005;12:138–143. doi: 10.1038/nsmb896. [DOI] [PubMed] [Google Scholar]
- 61.Westermann S, Avila-Sakar A, Wang H-W, Niederstrasser H, Wong J, Drubin DG, Nogales E, Barnes G. Formation of a dynamic kinetochore-microtubule interface through assembly of the Dam1 ring complex. Mol Cell. 2005;17:277–290. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 62.Asbury CL, Gestaut DR, Powers AF, Franck AD, Davis TN. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc Natl Acad Sci U S A. 2006;103:9873–9878. doi: 10.1073/pnas.0602249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramey VH, Wong A, Fang J, Howes S, Barnes G, Nogales E. Subunit organization in the Dam1 kinetochore complex and its ring around microtubules. Mol Biol Cell. 2011;22:4335–4342. doi: 10.1091/mbc.E11-07-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim JO, Zelter A, Umbreit NT, Bollozos A, Riffle M, Johnson R, MacCoss MJ, Asbury CL, Davis TN. The Ndc80 complex bridges two Dam1 complex rings. Elife. 2017;6 doi: 10.7554/eLife.21069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ng CT, Deng L, Chen C, Lim HH, Shi J, Surana U, Gan L. Electron cryotomography analysis of Dam1C/DASH at the kinetochore-spindle interface in situ. J Cell Biol. 2019;218:455–473. doi: 10.1083/jcb.201809088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jenni S, Harrison SC. Structure of the DASH/Dam1 complex shows its role at the yeast kinetochore-microtubule interface. Science. 2018;360:552–558. doi: 10.1126/science.aar6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wendler F, Gillingham AK, Sinka R, Rosa-Ferreira C, Gordon DE, Franch-Marro X, Peden AA, Vincent J-P, Munro S. A genome-wide RNA interference screen identifies two novel components of the metazoan secretory pathway. EMBO J. 2010;29:304–314. doi: 10.1038/emboj.2009.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heidtman M, Chen CZ, Collins RN, Barlowe C. Yos1p is a novel subunit of the Yip1p-Yif1p complex and is required for transport between the endoplasmic reticulum and the Golgi complex. Mol Biol Cell. 2005;16:1673–1683. doi: 10.1091/mbc.E04-10-0873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Desfougères Y, Gerasimaitė RU, Jessen HJ, Mayer A. Vtc5, a Novel Subunit of the Vacuolar Transporter Chaperone Complex, Regulates Polyphosphate Synthesis and Phosphate Homeostasis in Yeast. J Biol Chem. 2016;291:22262–22275. doi: 10.1074/jbc.M116.746784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hothorn M, Neumann H, Lenherr ED, Wehner M, Rybin V, Hassa PO, Uttenweiler A, Reinhardt M, Schmidt A, Seiler J, Ladurner AG, et al. Catalytic core of a membrane-associated eukaryotic polyphosphate polymerase. Science. 2009;324:513–516. doi: 10.1126/science.1168120. [DOI] [PubMed] [Google Scholar]
- 71.Vietri M, Radulovic M, Stenmark H. The many functions of ESCRTs. Nat Rev Mol Cell Biol. 2020;21:25–42. doi: 10.1038/s41580-019-0177-4. [DOI] [PubMed] [Google Scholar]
- 72.Hurley JH. ESCRTs are everywhere. EMBO J. 2015;34:2398–2407. doi: 10.15252/embj.201592484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tang S, Henne WM, Borbat PP, Buchkovich NJ, Freed JH, Mao Y, Fromme JC, Emr SD. Structural basis for activation, assembly and membrane binding of ESCRT-III Snf7 filaments. Elife. 2015;4 doi: 10.7554/eLife.12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schluter C, Lam KKY, Brumm J, Wu BW, Saunders M, Stevens TH, Bryan J, Conibear E. Global analysis of yeast endosomal transport identifies the vps55/68 sorting complex. Mol Biol Cell. 2008;19:1282–1294. doi: 10.1091/mbc.E07-07-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Siniossoglou S, Pelham HR. An effector of Ypt6p binds the SNARE Tlg1p and mediates selective fusion of vesicles with late Golgi membranes. EMBO J. 2001;20:5991–5998. doi: 10.1093/emboj/20.21.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mei K, Li Y, Wang S, Shao G, Wang J, Ding Y, Luo G, Yue P, Liu J-J, Wang X, Dong M-Q, et al. Cryo-EM structure of the exocyst complex. Nat Struct Mol Biol. 2018;25:139–146. doi: 10.1038/s41594-017-0016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chou H-T, Dukovski D, Chambers MG, Reinisch KM, Walz T. CATCHR, HOPS and CORVET tethering complexes share a similar architecture. Nat Struct Mol Biol. 2016;23:761–763. doi: 10.1038/nsmb.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Behnia R, Barr FA, Flanagan JJ, Barlowe C, Munro S. The yeast orthologue of GRASP65 forms a complex with a coiled-coil protein that contributes to ER to Golgi traffic. J Cell Biol. 2007;176:255–261. doi: 10.1083/jcb.200607151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, Weissman JS, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 80.Ma W, Goldberg J. TANGO1/cTAGE5 receptor as a polyvalent template for assembly of large COPII coats. Proc Natl Acad Sci U S A. 2016;113:10061–10066. doi: 10.1073/pnas.1605916113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stancheva VG, Li X-H, Hutchings J, Gomez-Navarro N, Santhanam B, Babu MM, Zanetti G, Miller EA. Combinatorial multivalent interactions drive cooperative assembly of the COPII coat. J Cell Biol. 2020;219 doi: 10.1083/jcb.202007135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Südhof TC, Rothman JE. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Conchon S, Cao X, Barlowe C, Pelham HR. Got1p and Sft2p: membrane proteins involved in traffic to the Golgi complex. EMBO J. 1999;18:3934–3946. doi: 10.1093/emboj/18.14.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 85.Jahn R, Scheller RH. SNAREs--engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 86.Rizo J. Mechanism of neurotransmitter release coming into focus. Protein Sci. 2018;27:1364–1391. doi: 10.1002/pro.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Burri L, Varlamov O, Doege CA, Hofmann K, Beilharz T, Rothman JE, Söllner TH, Lithgow T. A SNARE required for retrograde transport to the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2003;100:9873–9877. doi: 10.1073/pnas.1734000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hong Y, Ohishi K, Kang JY, Tanaka S, Inoue N, Nishimura J-I, Maeda Y, Kinoshita T. Human PIG-U and yeast Cdc91p are the fifth subunit of GPI transamidase that attaches GPI-anchors to proteins. Mol Biol Cell. 2003;14:1780–1789. doi: 10.1091/mbc.E02-12-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gamage DG, Hendrickson TL. GPI transamidase and GPI anchored proteins: oncogenes and biomarkers for cancer. Crit Rev Biochem Mol Biol. 2013;48:446–464. doi: 10.3109/10409238.2013.831024. [DOI] [PubMed] [Google Scholar]
- 90.Yi L, Bozkurt G, Li Q, Lo S, Menon AK, Wu H. Disulfide Bond Formation and N-Glycosylation Modulate Protein-Protein Interactions in GPI-Transamidase (GPIT) Sci Rep. 2017;8:45912. doi: 10.1038/srep45912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Moran P, Caras IW. A nonfunctional sequence converted to a signal for glycophosphatidylinositol membrane anchor attachment. J Cell Biol. 1991;115:329–336. doi: 10.1083/jcb.115.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fraering P, Imhof I, Meyer U, Strub JM, van Dorsselaer A, Vionnet C, Conzelmann A. The GPI transamidase complex of Saccharomyces cerevisiae contains Gaa1p, Gpi8p, and Gpi16p. Mol Biol Cell. 2001;12:3295–3306. doi: 10.1091/mbc.12.10.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ohishi K, Inoue N, Kinoshita T. PIG-S and PIG-T, essential for GPI anchor attachment to proteins, form a complex with GAA1 and GPI8. EMBO J. 2001;20:4088–4098. doi: 10.1093/emboj/20.15.4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ohishi K, Nagamune K, Maeda Y, Kinoshita T. Two subunits of glycosylphosphatidylinositol transamidase, GPI8 and PIG-T, form a functionally important intermolecular disulfide bridge. J Biol Chem. 2003;278:13959–13967. doi: 10.1074/jbc.M300586200. [DOI] [PubMed] [Google Scholar]
- 95.Vainauskas S, Menon AK. A conserved proline in the last transmembrane segment of Gaa1 is required for glycosylphosphatidylinositol (GPI) recognition by GPI transamidase. J Biol Chem. 2004;279:6540–6545. doi: 10.1074/jbc.M312191200. [DOI] [PubMed] [Google Scholar]
- 96.Meyer U, Benghezal M, Imhof I, Conzelmann A. Active site determination of Gpi8p, a caspase-related enzyme required for glycosylphosphatidylinositol anchor addition to proteins. Biochemistry. 2000;39:3461–3471. doi: 10.1021/bi992186o. [DOI] [PubMed] [Google Scholar]
- 97.Gamage DG, Varma Y, Meitzler JL, Morissette R, Ness TJ, Hendrickson TL. The soluble domains of Gpi8 and Gaa 1 two subunits of glycosylphosphatidylinositol transamidase (GPI-T), assemble into a complex. Arch Biochem Biophys. 2017;633:58–67. doi: 10.1016/j.abb.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 98.Meitzler JL, Gray JJ, Hendrickson TL. Truncation of the caspase-related subunit (Gpi8p) of Saccharomyces cerevisiae GPI transamidase: dimerization revealed. Arch Biochem Biophys. 2007;462:83–93. doi: 10.1016/j.abb.2007.03.035. [DOI] [PubMed] [Google Scholar]
- 99.Nguyen TTM, Murakami Y, Mobilio S, Niceta M, Zampino G, Philippe C, Moutton S, Zaki MS, James KN, Musaev D, Mu W, et al. Bi-allelic Variants in the GPI Transamidase Subunit PIGK Cause a Neurodevelopmental Syndrome with Hypotonia, Cerebellar Atrophy, and Epilepsy. Am J Hum Genet. 2020;106:484–495. doi: 10.1016/j.ajhg.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Evans R, O’Neill M, Pritzel A, Antropova N, Senior A, Green T, Žídek A, Bates R, Blackwell S, Yim J, Ronneberger O, et al. Protein complex prediction with AlphaFold-Multimer. bioRxiv. 2021:2021.10.04.463034 [Google Scholar]
- 101.Baek M, Heo L, Ndem R. neilfleckSCRI, RosettaCommons/RoseTTAFold: RoseTTAFold update: Including the simpler version for PPI screening. 2021 https://zenodo.org/record/5639837.
- 102.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 103.Potter SC, Luciani A, Eddy SR, Park Y, Lopez R, Finn RD. HMMER web server: 2018 update. Nucleic Acids Res. 2018;46:W200–W204. doi: 10.1093/nar/gky448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gabler F, Nam S, Till S, Mirdita M, Steinegger M, Söding J, Lupas AN, Alva V. Protein Sequence Analysis Using the MPI Bioinformatics Toolkit. Current Protocols in Bioinformatics. 2020;72 doi: 10.1002/cpbi.108. [DOI] [PubMed] [Google Scholar]
- 105.Morcos F, Pagnani A, Lunt B, Bertolino A, Marks DS, Sander C, Zecchina R, Onuchic JN, Hwa T, Weigt M. Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc Natl Acad Sci U S A. 2011;108:E1293-301. doi: 10.1073/pnas.1111471108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, Jensen LJ, et al. The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2020;49:D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Käll L, Krogh A, Sonnhammer ELL. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 108.Stark C, Breitkreutz B-J, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535-9. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 110.Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Research. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Banjade S, Tang S, Emr SD. Genetic and Biochemical Analyses of Yeast ESCRT. Methods Mol Biol. 2019;1998:105–116. doi: 10.1007/978-1-4939-9492-2_8. [DOI] [PubMed] [Google Scholar]
- 113.Madrona AY, Wilson DK. Structure of Ski8p, a WD repeat protein involved in mRNA degradation and meiotic recombination. 2004 doi: 10.2210/pdb1sq9/pdb. [DOI] [Google Scholar]
- 114.Nichols MD, De Angelis KA, Keck JL, Berger JM. Structure of the DNA topoisomerase vi a subunit. 1999 doi: 10.2210/pdb1d3y/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Corbett KD, Benedetti P, Berger JM. Crystal structure of the topoisomerase VI holoenzyme from Methanosarcina mazei. 2007 doi: 10.2210/pdb2q2e/pdb. [DOI] [Google Scholar]
- 116.Halbach F, Reichelt P, Rode M, Conti E. Crystal structure of the S cerevisiae Ski2-3-8 complex. 2013 doi: 10.2210/pdb4buj/pdb. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and materials availability: Structures of highly confident pairs with accompanying pMSAs and metadata are available at ModelArchive: https://modelarchive.org/doi/10.5452/ma-bak-cepc. RoseTTAFold two-track version is available at https://github.com/RosettaCommons/RoseTTAFold or Zenodo (101). AlphaFold was fetched from https://github.com/deepmind/alphafold on July 16th, 2021 (v2.0.0). Code for a GPU implementation of DCA and the modifications to the AlphaFold predictions script are provided in Supplemental Methods.