Abstract
Malignancy and tumour progression are associated with cancer-cell softening. Yet how the biomechanics of cancer cells affects T-cell mediated cytotoxicity and thus the outcomes of adoptive T-cell immunotherapies is unknown. Here, we show that T-cell-mediated cancer-cell killing is hampered for cortically soft cancer cells, whose plasma membrane is enriched with cholesterol, and that cancer-cell stiffening via cholesterol depletion augments T-cell cytotoxicity and enhances the efficacy of adoptive T-cell therapy against solid tumours in mice. We also show that the enhanced cytotoxicity against stiffened cancer cells is mediated by augmented T-cell forces arising from an increased accumulation of filamentous actin at the immunological synapse, and that cancer-cell stiffening has a negligible influence on T-cell-receptor signalling, on the production of cytolytic proteins such as granzyme B, on the secretion of interferon gamma and tumour necrosis factor alpha, and on Fas-receptor–Fas-ligand interactions. Our findings reveal a mechanical immune checkpoint that could be targeted therapeutically to improve the effectiveness of cancer immunotherapies.
Cancer cells enriched with cholesterol in their plasma membrane impair T-cell mediated cytotoxicity, which can be augmented by stiffening the cancer cells via cholesterol depletion, as shown in mouse models of adoptive T-cell therapy.
Tumours employ certain immune inhibitory pathways, termed immune checkpoints, to evade antitumour immunity and in particular T-cell mediated cytotoxicity. Blockade of ligand–receptor-based immune checkpoints using antibodies, such as anti-cytotoxic T-lymphocyte-associated protein 4 (anti-CTLA-4) and anti-programmed cell death protein 1 (anti-PD-1) antibodies, can reactivate antitumour immunity, and has led to remarkable clinical success in cancer immunotherapy1,2. Identifying new checkpoints and blocking them with therapeutic interventions could potentially benefit patients, especially those who fail to respond to current checkpoint blockade immunotherapies. Despite substantial efforts in seeking new checkpoints on the basis of biochemical signals3,4, potential inhibitory pathways involving biomechanical signals, such as target-cell stiffness, remain largely unexplored. Although tumours are typically stiffer than the paired normal tissues owing to the aberrant production and crosslinking of extracellular matrix proteins5, individual cancer cells are generally softer than their non-malignant counterparts6. Cellular softness is a biomechanical characteristic, which is correlated with the transformation, malignancy and metastasis of cancer cells7,8. The decrease in cancer-cell stiffness has been shown to arise from softening of both the cytoskeletal network9 and the plasma membrane10. T-cells directly interact with the surface of target cells; thus, the mechanical properties of the cell’s cortical structures, including the plasma membrane and the underlying actin cortex11, may impact cancer cell–T-cell interactions. However, the role of cancer-cell stiffness in evading immunosurveillance remains elusive.
T cells do not only sense mechanical environments12–17, but also exert forces at the immunological synapse to potentiate cytotoxicity against target cells18. Cytoskeletal forces19 and effector cytokine production20 are substantially reduced when T cells sense a soft substrate surface or soft target cells. Inspired by these observations, we hypothesized that cancer cells use cellular softness as a mechanical immune checkpoint to develop resistance towards T-cell mediated cytotoxicity by impairing T-cell mechanical forces. Here, we show that cancer cells soften the cortical structure through cholesterol enrichment in the plasma membrane leading to evasion from T-cell mediated cytotoxicity in vitro and in vivo. The cortical stiffness of cancer cells can be augmented by depleting cholesterol in the membrane lipid bilayer using methyl-β-cyclodextrin (MeβCD). Overcoming this mechanical immune checkpoint (target-cell softness / T-cell mechano-sensing axis) by stiffening cancer cells markedly improves the antitumour efficacy of adoptive T-cell transfer (ACT) immunotherapy against solid tumours. We also provide evidence that the enhanced cytotoxicity against stiffened cancer cells is mediated by increased T-cell forces rather than by known cytotoxic pathways based on biochemical signals.
Results
The plasma membrane of cancer cells is enriched with cholesterol
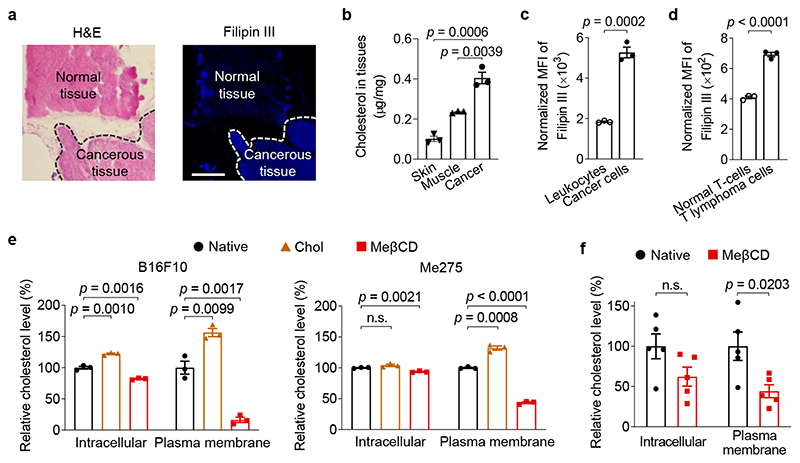
We first explored how cancer cells soften their cortical structure. It has been reported that depleting cholesterol from the plasma membrane in endothelial cells increases cell cortical stiffness21,22. These studies suggest that high membrane cholesterol level correlates with decreased cell stiffness. We therefore measured the cholesterol levels in murine and human tumour tissues as well as cells isolated from those tissues using Filipin III staining, a fluorescent dye that binds specifically to cholesterol23. Interestingly, in the B16F10 murine melanoma model, histological analysis showed that tumour tissues exhibited a substantially higher cholesterol level compared with the paired healthy tissues (Fig. 1a). In line with the results of histological studies, B16F10 tumours showed a 4.0 and 1.7-fold increase of global cholesterol levels compared to adjacent skin and muscle tissues, respectively, as quantified by an Amplex Red cholesterol assay (Fig. 1b). Cholesterol levels were also upregulated in several human tumour biopsies from various cancers including small cell lung cancer, colon cancer, squamous cell lung cancer, and liver cancer (Supplementary Fig. 1), suggesting a possibly common signature of cholesterol dysregulation in diverse cancer types. Further, single cell analyses by flow cytometry revealed that cancer cells in a 4T1 murine breast tumour (defined by tdTomato+) displayed a 2.9-fold greater amount of plasma membrane cholesterol compared to the tumour-infiltrating leukocytes (defined by CD45.2+) (Fig. 1c). In addition, EG7 cancer cells, a murine T lymphoma cell line, enriched 1.7-fold more cholesterol in the plasma membrane than normal murine T-cells (Fig. 1d).
Fig. 1. Cholesterol is enriched in the plasma membrane of cancer cells.
a, B16F10 tumour tissues (indicated with dash lines) and the adjacent normal tissues were stained with hematoxylin and eosin (H&E) and Filipin III (shown in blue colour). Scale bar, 500 μm. b, Cholesterol levels in B16F10 tumour tissues and the adjacent skin and muscle tissues (n = 3). c, d, Membrane cholesterol levels of tumour-infiltrating leukocytes (CD45.2+) and cancer cells (tdTomato+) in 4T1-Fluc-tdTomato tumour (c), and murine T lymphoma cells (EG7-OVA) and normal murine T-cells (d) determined by Filipin III staining (n = 3). The displayed MFI values were normalized by forward scatter area (FSC-A) of corresponding samples. e, f, Relative intracellular and plasma membrane levels of cholesterol (normalized to that of native cells) in murine B16F10 and human Me275 cancer cells treated with water-soluble cholesterol/methyl-β-cyclodextrin complex (Chol) or methyl-β-cyclodextrin (MeβCD) in vitro (e, n = 3) and in vivo (f, B16F10, n = 5). Data are one representative of at least two independent experiments with biological replicates. P values were determined by unpaired Student’s t test. Error bars represent standard error of the mean (SEM). MFI, mean fluorescence intensity; n.s., not significant.
Next, we sought to tune the plasma membrane cholesterol level in order to control cell stiffness. MeβCD, a biocompatible compound widely used as a drug solubilizer in the clinic24, has been reported to scavenge cholesterol through host-guest interaction25. We therefore used MeβCD to extract membrane cholesterol from cancer cells. Upon MeβCD treatment at a concentration of 5 mM for 30 min, the membrane cholesterol level of B16F10 cancer cells dropped markedly to only 16.0% of the native state, whereas the intracellular cholesterol level showed much less alteration (Fig. 1e). Similarly, membrane cholesterol level of cancer cells could be lowered down to 44% of the original level in vivo through a single intratumoural (i.t.) injection of MeβCD (Fig. 1f). In addition, supplementing the plasma membrane with cholesterol using a water-soluble cholesterol/MeβCD complex increased the membrane cholesterol level by 56% (Fig. 1e). Similarly, the plasma membrane cholesterol level was manipulated in various murine cancer cell lines including a murine lymphoma cell line expressing ovalbumin (EG7-OVA) and a murine colon cancer cell line (MC38), and a human melanoma cell line (Me275) (Fig. 1e and Supplementary Fig. 2). Notably, these treatments showed no direct impact on the viability or apoptosis of the cancer cells (Supplementary Fig. 3).
Control of cancer-cell stiffness by manipulating membrane cholesterol levels
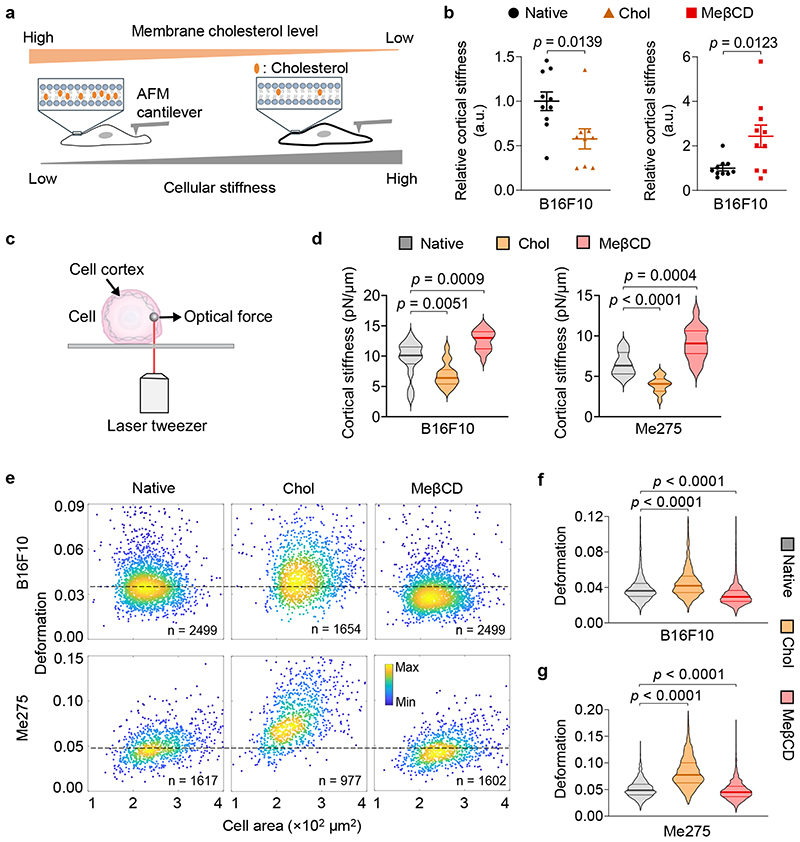
To examine whether membrane cholesterol level indeed influences cell mechanical properties, we directly measured single cell cortical stiffness using an atomic force microscope (AFM)26 (Fig. 2a). We found that cholesterol-supplemented and -depleted B16F10 cancer cells exhibited 40% lower and 2.4-fold higher cortical stiffness, respectively, compared to the untreated cells (Fig. 2b). To confirm the results, we used another well-established technique for the measurement of cell mechanics, optical tweezer27, to probe the cortical stiffness of cancer cells (Fig. 2c). In line with AFM measurements, membrane cholesterol supplementation or depletion could markedly decrease or increase cortical stiffness of both murine B16F10 and human Me275 cancer cells (Fig. 2d). To investigate whether the results of single cell measurements could be extrapolated to cell populations, we employed a recently reported high throughput microfluidic technique called deformability cytometry to measure cellular deformation (Supplementary Fig. 4a-c), which is correlated with cellular stiffness (higher deformation correlates with lower stiffness, and vice versa)28. Murine B16F10 and human Me275 cancer cells showed markedly reduced cellular deformation after MeβCD treatment, suggesting increased cellular stiffness (Fig. 2e-g). In contrast, cholesterol supplementation in B16F10 or Me275 cancer cells increased their cellular deformation (Fig. 2e-g). A similar trend was noted in various cancer cell types (Supplementary Fig. 4d). These results provide evidence that cholesterol enrichment in the plasma membrane contributes to cancer-cell softening, and cholesterol depletion via MeβCD treatment could increase cancer cell stiffness.
Fig. 2. Cancer-cell stiffness can be manipulated via the supplementation or depletion of cholesterol in the cell membrane.
a, Schematic illustration of the correlation between cellular stiffness and membrane cholesterol level. b, Relative cortical stiffness determined by nanoindentation measurements using atomic force microscopy (AFM) for native, Chol- or MeβCD-treated B16F10 cancer cells (n = 9 ~ 10 individual cells). Each data point is the average of at least twenty force curve measurements of a single cancer cell. Native B16F10 cancer cells serve as a standard (100%). Error bars represent SEM. c, Schematic illustration of the optical tweezer setting for cell cortical stiffness measurement. d, Cortical stiffness of native, Chol- or MeβCD-treated murine B16F10 and human Me275 cancer cells measured by the optical tweezer (n = 14 ~ 17 individual cells). e-g, Cellular deformation was measured using deformability cytometry to compare cellular stiffness in a high throughput manner. Shown are representative scatter plots (e; indicated are sample size, outliers not shown) and quantitative deformation of native, Chol- or MeβCD-treated murine B16F10 (f) and human Me275 (g) cancer cells. In all the violin plots (d, f, g), the middle solid line shows median, and lower and upper dash lines show 25th and 75th percentiles, respectively. P values were determined by unpaired Student’s t test. a.u., arbitrary unit; n.s., not significant.
Cancer-cell softness impairs T-cell mediated cytotoxicity
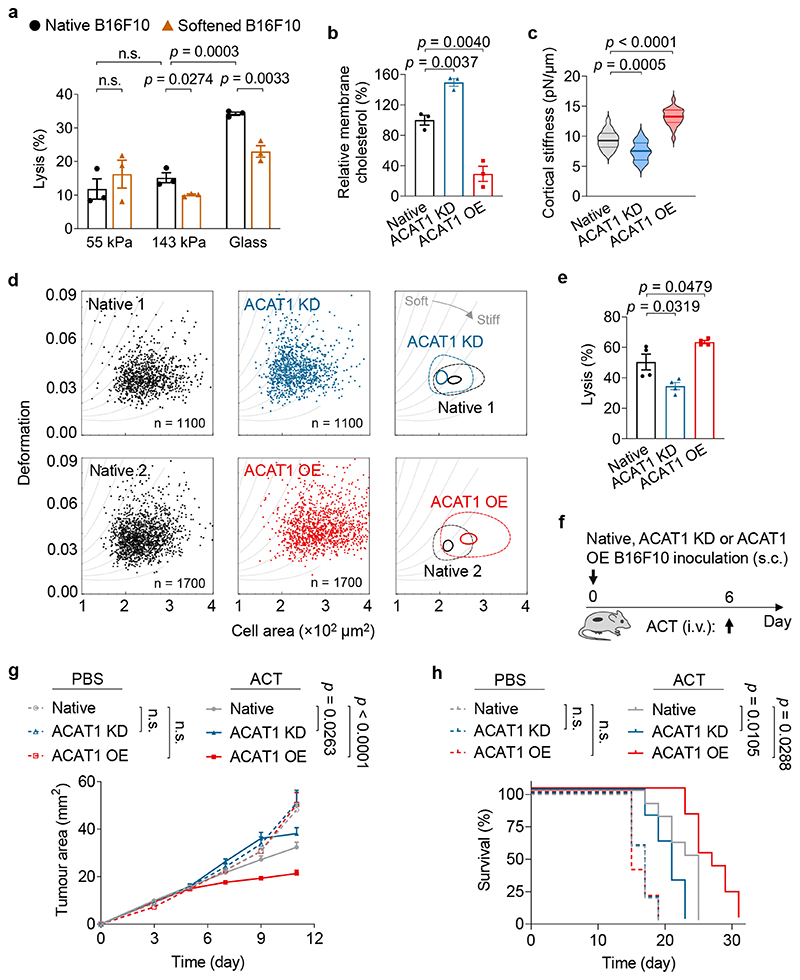
We next investigated whether cellular softness imparted resistance to T-cell mediated cytotoxicity. As culture substrates influence cell mechanics29, we first prepared polyacrylamide (PA) hydrogels of physiologically relevant Young’s modulus as substrates mimicking the tumour mechanical microenvironment (Supplementary Fig. 5b, PA-1 and 2)30. B16F10 cancer cells seeded on hydrogels with varying stiffness were co-cultured with activated Pmel CD8+ T-cells—T-cell receptor (TCR) transgenic T-cells that can recognize the gp100 antigen in B16F10 cancer cells. In accordance with a recent report18, lysis efficiency increased with higher substrate stiffness (Fig. 3a). When supplemented with exogenous cholesterol, B16F10 cancer cells seeded on a hydrogel substrate with stiffness of 143 kPa or a glass substrate (~3 GPa31) survived in higher numbers compared to the native cells suggesting that softened cancer cells were more resistant to T-cell mediated cytotoxicity (Fig. 3a). To validate this observation in vivo, C57BL/6J mice bearing subcutaneous (s.c.) B16F10 tumours were administered with exogenous cholesterol through i.t. injections every other day (2 mg × 8) (Supplementary Fig. 6a). The increased cholesterol level in tumours dampened the anti-tumour efficacy of adoptive transfer of Pmel CD8+ T-cells (Supplementary Fig. 6). Notably, cholesterol administration alone showed no effect on tumour growth or the survival rate of treated mice.
Fig. 3. Cancer-cell softness impairs T-cell mediated cytotoxicity in vitro and in vivo.
a, Lysis percentage of B16F10 cancer cells pre-treated with Chol (softened) or PBS (native) and co-cultured with activated Pmel CD8+ T-cells at an effector:target (E:T) ratio of 10:1 for 5 h (n = 3). b, Relative membrane cholesterol levels of B16F10 cancer cells with ACAT1 knock-down (ACAT1 KD) and ACAT1 overexpression (ACAT1 OE) (n = 3). Native B16F10 cancer cells serve as a standard (100%). c, Cortical stiffness of native, ACAT1 KD, and ACAT1 OE B16F10 cells measured by the optical tweezer (n = 19 ~ 21 individual cells). In the violin plots, the middle solid line shows median, and lower and upper dash lines show 25th and 75th percentiles, respectively. d, Representative scatter plots for native, ACAT1 KD, and ACAT1 OE B16F10 cells by deformability cytometry analysis, and their 50%-density contour plots (the inner contours correspond to 95% event density) with iso-elasticity lines dividing the diagrams into areas of different stiffness. e, Lysis percentage of ACAT1 KD and ACAT1 OE B16F10 cancer cells after 5-h co-culture with Pmel CD8+ T-cells at an E:T ratio of 10:1 (n = 4). Data are one representative of at least two independent experiments with biological replicates (a, b, e). f-h, Mice bearing native, ACAT1 KD, or ACAT1 OE B16F10 tumours were treated with adoptive transfer of Pmel CD8+ T-cells (5 × 106 per mouse), as outlined in the experimental scheme (f) (n = 5 and 10 animals for PBS- and ACT-treated groups, respectively). Shown are tumour growth curves (g) and survival curves (h) of pooled data from two independent experiments with biological replicates. P values were determined by unpaired Student’s t test in (a-c, e), two-way ANOVA in (g), or log-rank test in (h). Error bars represent SEM. ACAT1, acyl-CoA:cholesterol acyltransferase 1; PBS, phosphate-buffered saline; ACT, adoptive cell transfer; s.c., subcutaneous; i.v., intravenous; n.s., not significant.
In order to achieve cancer cell-specific modulation of cholesterol levels in vivo, we first genetically engineered the B16F10 cancer cells by knocking down acyl-CoA:cholesterol acyltransferase 1 (ACAT1) (denoted as ACAT1 KD B16F10) (Supplementary Fig. 7). ACAT1 is an enzyme specific for cholesterol esterification and has been utilized to control membrane cholesterol levels in cells32,33. We found that ACAT1 KD B16F10 cells enriched a 1.5-fold higher level of cholesterol in the plasma membrane as compared to the native B16F10 cells (Fig. 3b). Similar as the cholesterol-supplemented cells, ACAT1 KD B16F10 cells displayed lower stiffness (Fig. 3c, d) as well as increased resistance toward T-cell mediated killing (Fig. 3e) as compared to the native B16F10 cells. Mice inoculated with ACAT1 KD B16F10 cells showed similar tumour progression as the native B16F10 tumour when there was no treatment (Fig. 3f, g). However, ACT of Pmel CD8+ T-cells was less effective in controlling ACAT1 KD B16F10 tumour growth as compared to the native B16F10 tumour (Fig. 3g), leading to a reduced median survival of ACAT1 KD B16F10 tumour-bearing mice (Fig. 3h). Altogether, the results show that cancer cells present decreased cellular stiffness as a mechanically inhibitory pathway to evade T-cell mediated cytotoxicity in vitro and in vivo.
Cancer-cell stiffening enhances the efficacy of ACT therapies
We next investigated whether such mechanical immune checkpoint can be overcome by stiffening cancer cells. We first prepared the ACAT1-overexpressing B16F10 cancer cells (denoted as ACAT1 OE B16F10) (Supplementary Fig. 7), which showed 70% lower membrane cholesterol level than that of native B16F10 cells (Fig. 3b). As measured by both optical tweezer and deformability cytometry, ACAT1 OE B16F10 cells with reduced membrane cholesterol level were stiffer than the native B16F10 cells (Fig. 3c, d). Indeed, ACAT1 OE B16F10 cells displayed increased susceptibility toward T-cell mediated cytotoxicity as compared to the native B16F10 cancer cells (Fig. 3e). To further test this hypothesis in vivo, we inoculated mice with ACAT1 OE B16F10 tumour, which exhibited similar growth rate as the native B16F10 tumour. However, ACT of Pmel CD8+ T-cells better controlled the ACAT1 OE B16F10 tumour compared to the native B16F10 tumour (Fig. 3f, g). Treated mice bearing ACAT1 OE B16F10 tumours also showed prolonged survival (Fig. 3h). In this proof-of-concept study, tumour cells were specifically stiffened leading to enhanced susceptibility toward ACT therapy.
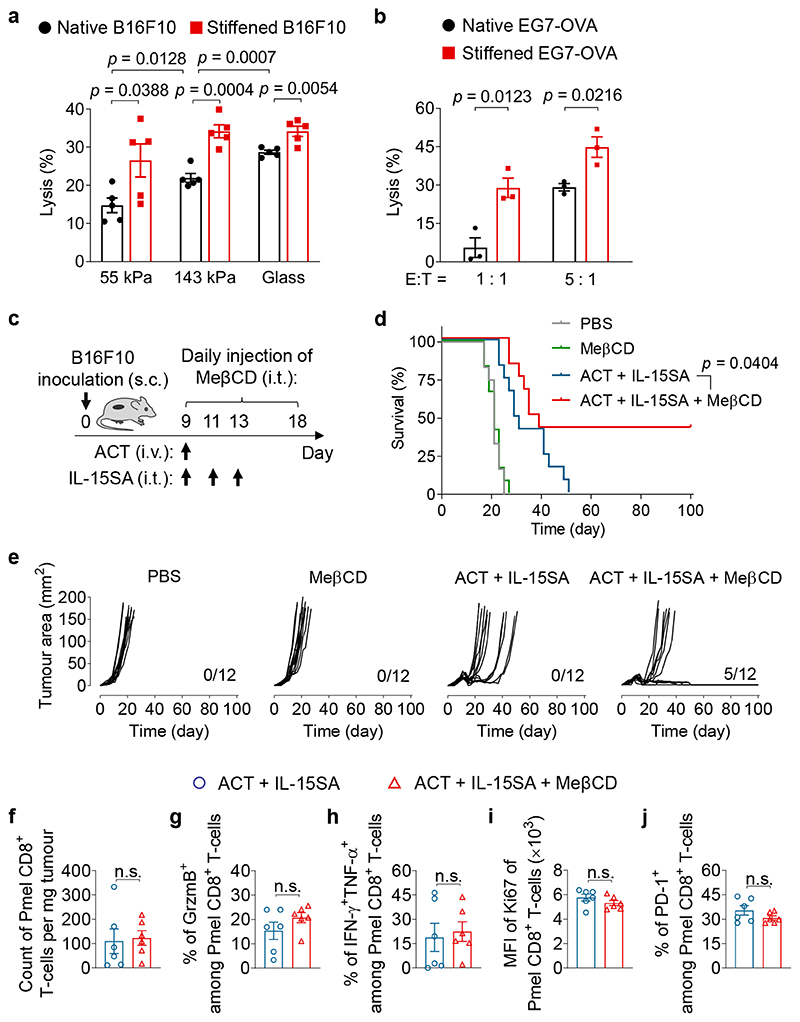
On the basis of these findings, we next sought to develop a potentially applicable therapeutic intervention to stiffen cancer cells for enhanced cancer immunotherapy. We found that B16F10 cancer cells treated with MeβCD maintained a low level of membrane cholesterol up to 5 hours post treatment (Supplementary Fig. 8a). Further, cancer cells that were stiffened by MeβCD pre-treatment displayed considerable increased sensitivity to T-cell mediated killing on all three substrates of different stiffness (55 kPa, 143 kPa, and glass) (Fig. 4a). The enhanced lysis of stiffened cancer cells was most prominent on substrates closely mimicking the physiological mechanical environment in tumours (55 and 143 kPa)30. Once the cholesterol level in the cancer cell membrane recovered to the native state after 12 hours (Supplementary Fig. 8a), the enhancement of killing was abrogated (Supplementary Fig. 8b). To rule out potential effects of a synthetic substrate, EG7-OVA, a suspension cancer cell line, was stiffened with MeβCD and found to exhibit similarly enhanced susceptibility to cytotoxicity mediated by OT-I TCR transgenic CD8+ T-cells that recognize SIINFEKL epitope derived from OVA (Fig. 4b).
Fig. 4. Cancer-cell stiffening by MeβCD enhances the efficacy of ACT immunotherapy.
a, Lysis percentage of B16F10 cancer cells pre-treated with MeβCD (stiffened) or PBS (native) and co-cultured with activated Pmel CD8+ T-cells at an E:T ratio of 10:1 for 5 h (n = 5). b, Lysis percentage of EG7-OVA cancer cells pre-treated with MeβCD (stiffened) or PBS (native) and co-cultured with activated OT-I CD8+ T-cells at indicated E:T ratios for 5 h (n = 3). Data in (a, b) are one representative of at least three independent experiments with biological replicates. c-e, B16F10 tumour-bearing mice were treated with adoptive transfer of Pmel CD8+ T-cells (5 × 106 per mouse) adjuvanted by interleukin-15 super-agonist (IL-15SA, 10 μg per injection) with or without daily MeβCD administration (1 mg per injection) as outlined in the experimental scheme (c). Mice receiving injections of PBS or MeβCD only serve as controls (n = 12 animals per group). Shown are survival curves (d), and individual tumour growth curves (e, indicated are the number of mice with durable responses out of all treated mice) of pooled data of two independent experiments with biological replicates. f-j, Tumour-infiltrating Pmel CD8+ T-cells were analysed by flow cytometry on day 14 (experimental scheme is shown in Supplementary Fig. 12b). Shown are counts (f), frequencies of granzyme B (GrzmB)+ (g), polyfunctional (h), and PD-1+ (j), and Ki67 expression level (i) of tumour-infiltrating Pmel CD8+ T-cells (n = 6 animals per group). Data are one representative of two independent experiments with biological replicates. P values were determined by unpaired Student’s t test in (a, b, f-j) or log-rank test in (d). Error bars represent SEM. PBS, phosphate-buffered saline; ACT, adoptive cell transfer; MFI, mean fluorescence intensity; s.c., subcutaneous; i.v., intravenous; i.t., intratumoural; n.s., not significant.
Next, we extended the stiffening strategy using MeβCD treatment to in vivo studies. C57BL/6J mice bearing s.c. B16F10 tumours were treated with ACT of Pmel CD8+ T-cells adjuvanted with an interleukin-15 super-agonist (IL-15SA, Supplementary Fig. 9) along with or without daily administration of MeβCD (i.t., 1 mg × 10) (Fig. 4c). ACT therapy supported by IL-15SA delayed the tumour growth but failed to achieve durable control of tumour progression in all treated mice. In contrast, the combination therapy of IL-15SA-supported ACT and cancer-cell stiffening intervention using MeβCD led to complete eradication of 5 out of 12 tumours and durable cures in 41.7% of treated mice (Fig. 4d, e). Similarly, ACT plus MeβCD treatment without IL-15SA also induced substantial tumour regression and prolonged the survival of treated mice compared to ACT alone (Supplementary Fig. 10). In addition, the stiffening intervention with MeβCD also effectively reduced the tumour burden in mice bearing s.c. EG7-OVA lymphoma tumours when combined with ACT of OT-I CD8+ T-cells supported by IL-15SA (Supplementary Fig. 11), suggesting that overcoming the mechanical immune checkpoint may be a versatile therapy for different types of cancers. Notably, the stiffening intervention using MeβCD did not cause any side effects, such as body weight drop, splenomegaly, or increased infiltration and activation of CD8+ T-cells in the spleens (Supplementary Fig. 12). MeβCD alone as a monotherapy showed no therapeutic efficacy indicating that the presence of antigen specific cytotoxic T-cells was necessary for enhanced killing of stiffened target cells (Fig. 4d, e).
Biochemical pathways of T-cell mediated cytotoxicity are not affected by cancer-cell stiffening
By comparing IL-15SA-supported ACT therapy with the combination therapy of IL-15SA-supported ACT and MeβCD, we found that the stiffening intervention with MeβCD showed negligible effects on tumour infiltration of adoptively transferred Pmel CD8+ T-cells, as well as their granzyme B production, polyfunctionality, proliferative capacity, or exhaustion phenotypes (Fig. 4f-j; Supplementary Fig. 12b). Further flow-cytometry analyses revealed that MeβCD treatment had almost no impact on the counts and phenotypes of tumour-infiltrating endogenous CD8+ T-cells (Supplementary Fig. 13) or other immune cells including regulatory T-cells, B cells, natural killer cells, macrophages, dendritic cells, and myeloid-derived suppressor cells (Supplementary Fig. 14). To further examine the potential effects of MeβCD on T-cells, we performed an in vitro experiment by co-culturing Pmel CD8+ T-cells and B16F10 cancer cells in the presence of MeβCD. While MeβCD enhanced T-cell mediated killing of cancer cells, it had negligible influence on T-cell proliferation or degranulation (Supplementary Fig. 15). Altogether, these results suggest that the stiffening intervention with MeβCD markedly enhances the efficacy of ACT immunotherapy against solid tumours, an effect that is not owed to the alteration of immune cell infiltration, proliferation, or effector functions in tumours.
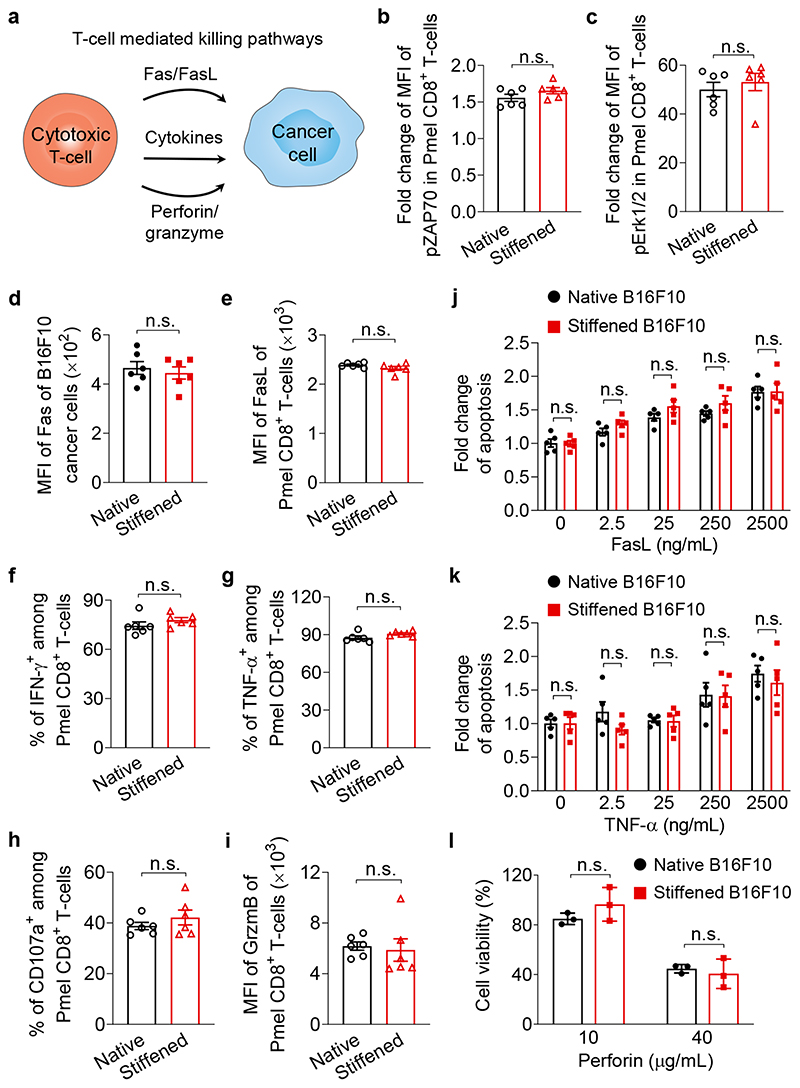
In order to elucidate the mechanism by which the stiffening intervention with MeβCD enhances tumour control by T-cells, we performed mechanistic studies on different pathways that T-cells exploit for cancer-cell killing. Those pathways include the Fas protein (also called CD95)—Fas ligand (FasL) interactions, the secretion of effector cytokines, such as interferon-γ (IFN-γ) and tumour necrosis factor-α (TNF-α), and the granule exocytosis of cytolytic proteins (e.g., perforin and granzymes) (Fig. 5a)34,35. As TCR activation depends on cognate antigen recognition, we first validated that OVA antigen presentation on B16F10-OVA cancer cells, NFAT activation (as indicated by IL-2 production)36 and phosphorylation levels of ZAP70, Erk1/2, and NF-κB (as indicated by p65 phosphorylation)37 as the downstream markers of TCR-signalling were not affected by the MeβCD treatment of cancer cells (Fig. 5b, c and Supplementary Fig. 16a-c). Next, we co-cultured MeβCD-stiffened B16F10 cancer cells with activated Pmel CD8+ T-cells to examine whether MeβCD treatment of cancer cells altered any of the abovementioned biochemical cytotoxic pathways. Proliferation, activation, and exhaustion phenotypes were unchanged in T-cells co-cultured with stiffened B16F10 cancer cells compared to the native cancer cells (Supplementary Fig. 16d-f). Importantly, MeβCD treatment of cancer cells did not influence the expression levels of Fas on B16F10 cancer cells and FasL on T-cells (Fig. 5d, e), or the susceptibility of cancer cells to FasL-induced apoptosis (Fig. 5j). In addition, IFN-γ and TNF-α secretion were unchanged in T-cells co-cultured with native or stiffened B16F10 cancer cells (Fig. 5f, g and Supplementary Fig. 16g, h). Both native and stiffened B16F10 cancer cells exhibited comparable levels of apoptosis when incubated with TNF-α (Fig. 5k). Finally, flow cytometry analyses of degranulation activity and granzyme B production revealed that MeβCD treatment of cancer cells had negligible influence on the granule exocytosis activity of the cytotoxic T-cells in the co-culture (Fig. 5h, i and Supplementary Fig. 16i). Based on these results, we excluded the known biochemical pathways as the major underlying mechanisms of the enhanced T-cell cytotoxicity against stiffened cancer cells.
Fig. 5. Cancer-cell stiffening has negligible influence on biochemical cancer-cell killing pathways mediated by T-cells.
a, Schematic illustration of T-cell mediated killing pathways. b, c, Fold change of MFI of phosphorylated ZAP70 (pZAP70, b) and Erk1/2 (pErk1/2, c) in activated Pmel CD8+ T-cells stimulated by native or MeβCD-treated (stiffened) B16F10 cancer cells at 37 °C for 5 min (n = 6). d, Expression levels of Fas of native and stiffened B16F10 cancer cells (n = 6). e-i, Activated Pmel CD8+ T-cells were co-cultured with native or stiffened B16F10 cancer cells (E:T ratio = 10:1) at 37 °C for 5 h. Shown are expression levels of Fas ligand (FasL) (e) and granzyme B (GrzmB) (i), and frequencies of IFN-γ+ (f), TNF-α+ (g), and CD107a+ (h) of Pmel CD8+ T-cells (n = 6). j, k, Fold change of frequencies of apoptotic native and stiffened B16F10 cancer cells after incubation with FasL (j) or TNF-α (k) at indicated concentrations at 37 °C for 5 h (n = 5). l, Viability of native and stiffened B16F10 cancer cells after incubation with perforin of indicated concentrations at 37 °C for 20 min (n = 3). P values were determined by unpaired Student’s t test. Error bars represent SEM. MFI, mean fluorescence intensity; n.s., not significant. All data are one representative of at least three independent experiments with biological replicates.
Cancer-cell stiffening augments cellular forces and cytotoxicity mediated by T-cells
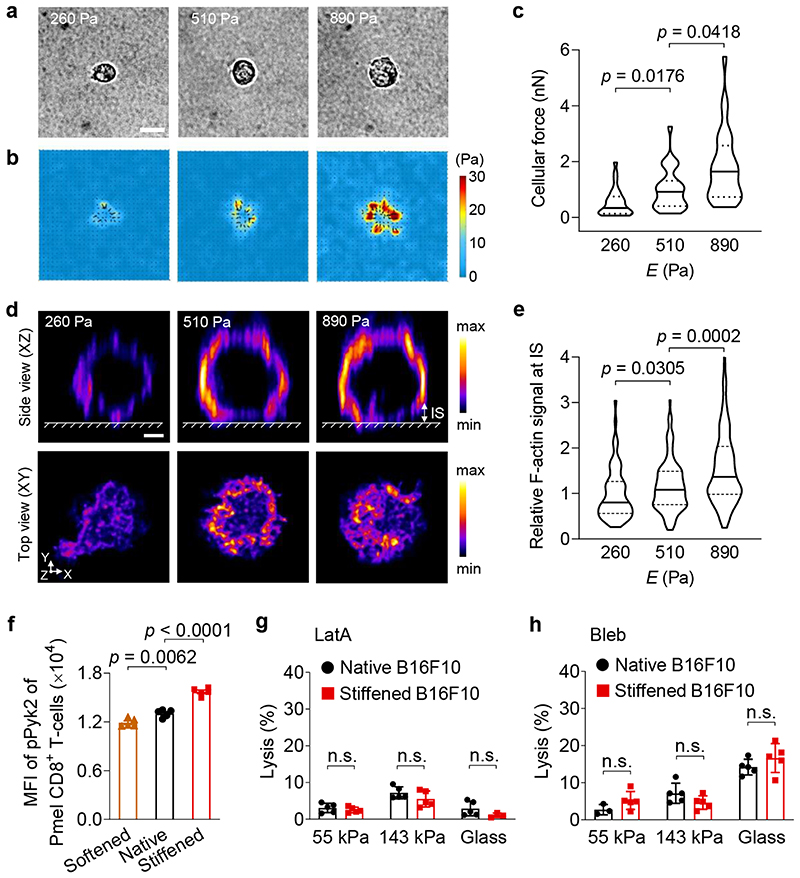
These findings motivated us to investigate whether biomechanical factors contributed to the enhanced T-cell mediated killing. We first incubated MeβCD-stiffened B16F10 cancer cells with perforin, a pore-forming effector protein, in the absence of T-cells, and found that cancer-cell stiffening had no impact on perforin-mediated lysis, suggesting that the enhanced cytotoxicity against stiffened cancer cells is T-cell dependent (Fig. 5l). T-cell forces have been reported to increase membrane tension to promote pore-formation induced by perforin on target cells18. To provide direct evidence that cytotoxic T-cells exert increased forces on stiffer substrates, we used traction force microscopy (TFM)38 to measure forces exerted by primary T-cells on hydrogel substrates of various stiffness. We synthesized PA hydrogels with Young’s modulus from 260 to 890 Pa (Supplementary Fig. 5b, PA-3-5), a range representative of the physiological stiffness of cancer cells39–41. Upon TCR triggering by anti-CD3 and anti-CD28 antibodies coated on the hydrogel surface, T-cell forces were measured by quantifying the displacement of the embedded fluorescent beads in hydrogels (Supplementary Fig. 17a). Pmel CD8+ T-cells exerted markedly higher cellular forces on stiffer substrates (Fig. 6a, b), with average values of 0.5, 1.0 and 1.8 nanonewton per cell on PA hydrogels of 260, 510, and 890 Pa, respectively (Fig. 6c). When the coated antibodies were replaced by anti-CD45 antibody, a non-stimulatory antibody, or T-cells were pre-treated with latrunculin A (LatA), a potent inhibitor of actin polymerization, the cellular forces dropped substantially (Supplementary Fig. 17b). Confocal fluorescence imaging of Pmel CD8+ T-cells on the same PA hydrogels revealed the presence of a filamentous actin (F-actin)-rich peripheral structure across the T-cell/hydrogel interface, which represented the formation of immunological synapse (Fig. 6d). F-actin accumulation in this synaptic interface was notably increased on stiff hydrogel substrates (Fig. 6e). As F-actin polymerization is essential for cellular force generation42, this observation, along with the TFM result, indicates that Pmel CD8+ T-cells generate higher mechanical stress on stiffer surfaces. It has been reported that phosphorylation of proline-rich tyrosine kinase 2 (Pyk2), a member of focal adhesion kinase family, positively correlates with cellular forces exerted by primary T-cells43. To compare the T-cell forces exerted on target cells of various stiffness, we measured the phospho-Pyk2 (pPyk2) levels in Pmel CD8+ T-cells co-cultured with native, softened, or stiffened B16F10 cancer cells. Higher level of pPyk2 was induced in Pmel CD8+ T-cells co-cultured with stiffened B16F10 cancer cells as compared to the native or softened cancer cells (Fig. 6f), suggesting that T-cells indeed exerted higher cellular forces against stiffened cancer cells. Taken together, these results showed that T-cells exert higher forces against a stiffer surface upon TCR triggering.
Fig. 6. Enhanced cytotoxicity against stiffened cancer cells is mediated by T-cell forces.
a-c, Forces exerted by activated Pmel CD8+ T-cells on polyacrylamide (PA) hydrogel substrates of indicated stiffness coated with anti-CD3 and anti-CD28 antibodies were measured using traction force microscopy. Shown are representative bright field images (a) and the corresponding traction stress maps (b), and average total force per cell (c) (n = 29 individual cells). The colour bar indicates the magnitude of stress. Scale bar, 5 μm. d, Representative deconvoluted confocal fluorescence images of F-actin of activated Pmel CD8+ T-cells on PA hydrogel substrates of indicated stiffness coated with anti-CD3 and anti-CD28 antibodies. The upper row shows the side view (XZ plane); the lower row shows the top view (XY plane) of F-actin at the T-cell immunological synapse (IS, defined as the structure between the surface of hydrogel and a height of 2 μm above the surface of the hydrogel). The colour bar indicates the intensity of the F-actin fluorescence signal. Scale bar, 2 μm. e, Relative total fluorescence intensity (normalized by the mean value at 260 Pa) of F-actin at the IS in the images from (d) (n = 66, 119, and 179 individual cells for 260, 510 and 890 Pa, respectively). f, MFI of phosphorylated Pyk2 (pPyk2) in activated Pmel CD8+ T-cells co-cultured with native, Chol-treated (softened) or MeβCD-treated (stiffened) B16F10 cancer cells (n = 5). g, h, Lysis percentage of native and stiffened B16F10 cancer cells co-cultured with activated Pmel CD8+ T-cells (E:T ratio = 10:1), which were pre-treated with latrunculin A (LatA, g) or blebbistatin (Bleb, h) (n = 5). P values were determined by Kruskal-Wallis test in (c, e) or unpaired Student’s t test in (f-h). Error bars represent SEM. In the violin plots (c, e), the middle solid line shows median, and lower and upper dash lines show 25th and 75th percentiles, respectively. MFI, mean fluorescence intensity; n.s., not significant. All data are one representative of at least two independent experiments with biological replicates.
To determine whether enhanced T-cell forces played an important role in augmented cytotoxicity, we pre-treated T-cells with LatA to inhibit actin polymerization and thereby T-cell forces. T-cells pre-treated with LatA exerted greatly reduced cellular forces even 5 hours post the treatment (Supplementary Fig. 17c). As a result, the percentage of cancer cell lysis was substantially reduced on all hydrogel substrates (Fig. 6g, as compared to results shown in Fig. 4a). Importantly, the cytotoxicity enhancement observed following cancer-cell stiffening was completely abrogated on all substrates (Fig. 6g). Similarly, pre-treatment of T-cells with blebbistatin (Bleb), a myosin II inhibitor that inhibits T-cell contractility18, or Mycalolide B (MycaB), an irreversible cytoskeleton inhibitor that covalently binds to globular actin (G-actin) for inhibiting actin polymerization and thus cellular forces44,45 (Supplementary Fig. 17d), led to the complete abrogation of enhanced lysis of stiffened cancer cells (Fig. 6h and Supplementary Fig. 18). Of note, these inhibitors showed no direct effects on T-cell viability or apoptosis at the concentrations used (Supplementary Fig. 19). These results reveal that the augmented killing of stiffened target cells was mediated by cellular forces exerted by T-cells.
Discussion
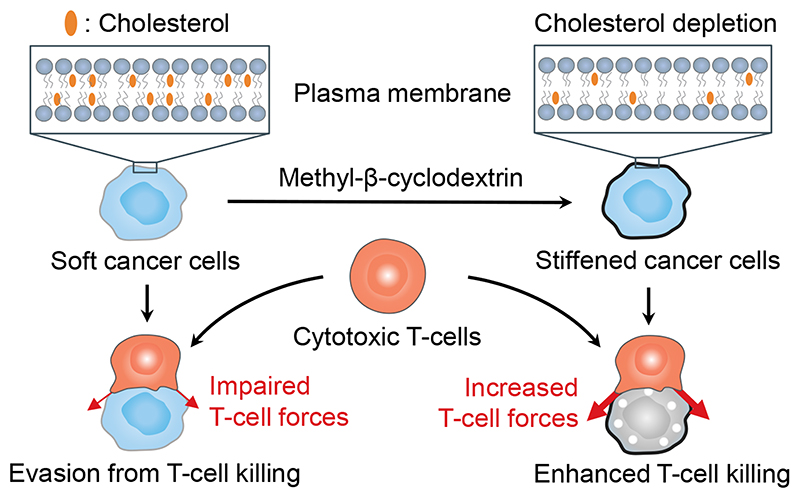
Discovering and targeting new immune checkpoints has the potential to improve patient response rates to cancer immunotherapy. Here we identified cellular softness as an immune checkpoint of biomechanical basis that is employed by cancer cells to impair T-cell forces at the immunological synapse and therefore to evade anti-tumour immunity (Fig. 7). By stiffening cancer cells through depletion of cancer-cell-membrane cholesterol, we show that the mechanical immune checkpoint could be overcome to enhance T-cell forces and cytotoxicity, leading to tumour clearance and durable responses in preclinical mouse tumour models when combined with ACT therapy (Fig. 7).
Fig. 7. Illustration of mechanical immuno-suppression induced by the softness of cancer cells. Stiffening the cancer cells enhances cancer-cell killing by T-cells.
Specifically modulating cell mechanics in vivo is the key to clinical application, but is still challenging. As a proof-of-concept, we used an ex vivo genetic modification approach to regulate the membrane cholesterol levels specifically in cancer cells without perturbing tumour-infiltrating immune cells such as T-cells. Cholesterol in T-cells has been reported to be important in enhancing TCR clustering and thus TCR signalling upon antigen stimulation33. We employed intratumoural injection of MeβCD to transiently deplete cholesterol from plasma membranes. In those experiments, we found that injected MeβCD had negligible influence on the functions of tumour-infiltrating T-cells in vivo likely because the depletion of cholesterol by MeβCD was transient and less potent as compared to genetic modification of T-cells in previous reports33. Future work to target reagents for mechanical modulation, such as MeβCD, specifically to cancer cells using biomaterial-assisted delivery strategies would be necessary to minimize any undesired side effects46.
We found that T-cell forces were critical for enhanced vulnerability of stiffened cancer cells toward T-cell mediated cytotoxicity as inhibition of T-cells forces completely abrogated such effects. In addition, TFM measurement showed that T-cells exerted higher cellular forces against flat substrates with increased stiffness (from 260 to 890 Pa), which mimic the stimulatory surface of target cells. To closely recapitulate the spatial features of target cell surface including the curvature, a recently developed TFM technique based on spherical microparticles may better map the dynamic forces at the T-cell immunological synapse in the future47. Degranulation and cytokine production of CD8+ T-cells is less stiffness-dependent than CD4+ T-cells, particularly on substrates of low stiffness (<8 kPa)20,48. Consistent with previous reports, we found that cancer-cell stiffening via membrane cholesterol depletion had a negligible effect on degranulation or cytokine production of CD8+ T-cells. Recently, overexpression of myocardin-related transcription factors (MRTFs) was shown to increase cancer cell stiffness by inducing rigidification of filamentous actin and promote degranulation and cytokine production in cytotoxic CD8+ T-cells49. These results suggest that different target-cell stiffening methods (membrane cholesterol depletion vs. intracellular cytoskeleton rigidification) may result in different T-cell responses, for which the underlying mechanism is still unknown. Nevertheless, as cellular stiffness is contributed jointly by both cell cortex and cytoskeleton50, modulation of both components is promising to overcome the mechanical immune checkpoint for enhanced cancer immunotherapy.
Despite efforts in searching and investigating immune checkpoints on the basis of biochemical signals, much less is known about how biomechanical cues and interactions could potentially regulate immune responses against diseases such as cancer. The cancer–immunity interactions are multidimensional, involving not only biochemical but also substantial biophysical signals51,52. Our studies provide insight into the multidimensional mechanisms of immune suppression in tumours. The growing knowledge in fundamental mechano-immunology provides the basis for developing new engineering approaches to modulate biomechanical cues for enhanced antitumour immunity53–56. Leveraging cancer-cell mechanics and T-cell forces, as demonstrated in this study, may provide new therapeutic strategies in addition to conventional biochemical modulation. Therapeutically targeting both biochemical and mechanical immune checkpoints could potentially benefit patients with cancer immunotherapies.
Methods
Animals, cell lines, and reagents
All the mouse studies were approved by the Swiss authorities (Canton of Vaud, animal protocol ID 3206 and 3533) and performed in accordance with guidelines from the Center of PhenoGenomics (CPG) in EPFL. Six- to eight-week-old female Thy1.2+ C57BL/6 (C57BL/6J) mice and BALB/cByJ (BALB/c) mice were purchased from Charles River Laboratories (Lyon, France). T-cell receptor (TCR)-transgenic Thy1.1+ pmel-1 (Pmel) mice (B6.Cg-Thy1 a/Cy Tg(TcraTcrb)8Rest/J) and TCR-transgenic OT-I mice (C57BL/6-Tg(TcraTcrb)1100Mjb/J) were purchased from The Jackson Laboratory (Bar Harbor, ME, USA) and maintained in the animal facility in the CPG in EPFL. B16F10 murine melanoma cells and EL4 murine lymphoma cells expressing ovalbumin (EG7-OVA) were originally acquired from the American Type Culture Collection (ATCC; Manassas, VA, USA). MC38 murine colon cancer cells expressing human epidermal growth factor receptor 2 (MC38-HER2) and Me275 human melanoma cells expressing HER2 (Me275-HER2) were kindly provided by Pedro Romero Lab (UNIL, Switzerland). B16F10 murine melanoma cells expressing ovalbumin (B16F10-OVA) and 4T1 murine breast cancer cells expressing luciferase and tdTomato fluorescent protein (4T1-Fluc-tdTomato) were kindly provided by Darrell Irvine Lab (MIT, USA). HEK293T cells and pLKO.1 vector were kindly provided by Didier Trono Lab (EPFL, Switzerland). B16F10, B16F10-OVA, MC38-HER2, and HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with fetal bovine serum (FBS) (10 v/v%, Gibco) and penicillin/streptomycin (1 v/v%, Gibco). EG7-OVA and Me275-HER2 cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco) supplemented with FBS (10 v/v%), HEPES (1 v/v%, Gibco), penicillin/streptomycin (1 v/v%), and β-mercaptoethanol (0.1 v/v%, Gibco). For culturing EG7-OVA cells, G418 (Geneticin) (0.4 mg/mL, Gibco) was supplemented to maintain OVA expression. 4T1-Fluc-tdTomato cells were cultured in Iscove’s Modified Dulbecco’s Medium (IMDM) (Gibco) supplemented with FBS (10 v/v%) and penicillin/streptomycin (1 v/v%).
Filipin III (from Streptomyces filipinensis), methyl-β-cyclodextrin (MeβCD), water-soluble cholesterol/MeβCD complex (Chol), blebbistatin (Bleb), glutaraldehyde solution (25 wt% in H2O), Triton X-100, 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI), propidium iodide (PI) solution (1 mg/mL in H2O), Hoechst 33342, acrylamide, N,N′-methylenebisacrylamide, ammonium persulfate, tetramethylethylenediamine (TEMED), N-sulfosuccinimidyl-6-(4′-azido-2′-nitrophenylamino) hexanoate (Sulfo-SANPAH), deoxyribonuclease I (DNase I, from bovine pancreas), dispase II, hyaluronidase, cholesterol oxidase (from microorganisms), protamine sulfate, bovine serum albumin (BSA), sodium dodecyl sulfate (SDS), calcium chloride dihydrate and polystyrene (PS) bead (carboxylate-modified, 500 nm, orange fluorescence) were purchased from Sigma-Aldrich (St. Louis, MO, USA). 3-aminopropyl-trimethoxysilane (APTMS) was purchased from ACROS Organics (Thermo Fisher Scientific). Methyl-cellulose (4000 cPs) was purchased from Alfa Aesar (Thermo Fisher Scientific). Phalloidin-iFluor™ 488 conjugate was purchased from AAT Bioquest (Sunnyvale, CA, USA). Latrunculin A (LatA) (1 mM in DMSO) was purchased from Calbiochem (Merck, Darmstadt, Germany). Mycalolide B (MycaB) was purchased from Enzo Life Science (Farmingdale, NY, USA). EZ-Link™ NHS-Biotin, collagenase IV, puromycin dihydrochloride, and anti-phospho (Ser536)-NF-κB p65 antibody (clone T.849.2) were purchased from Thermo Fisher Scientific. Streptavidin acrylamide, fluorescent PS bead (carboxylate-modified, 200 nm, red fluorescence), and CellTrace™ CFSE Cell Proliferation Kit were purchased from Invitrogen (Thermo Fisher Scientific). Active Perforin-1 was purchased from Cloud-Clone (Houston, TX, USA). Recombinant human fibronectin fragment (RetroNectin®) was purchased from Takara (Nojihigashi, Shiga, Japan). OVA257-264 (SIINFEKL) and human gp10025-33 (hgp100) peptides were purchased from GenScript (Piscataway, NJ, USA). Recombinant mouse interleulin-2 (IL-2) and interleukin-7 (IL-7), and tumour necrosis factor-α (TNF-α) were purchased from PeproTech (London, UK). Anti-mouse CD3 antibody (clone 17A2), and anti-mouse CD28 antibody (clone 37.51) were purchased from Bioxcell (West Lebanon, NH, USA). Anti-phospho (Tyr402)-Pyk2 antibody (R402) was purchased from EnoGene (New York, NY, USA). Anti-ACAT1 polyclonal antibody was purchased from Cayman Chemical (Ann Arbor, MI, USA). Brefeldin A solution (1000×), Monensin solution (1000×), 7-amino-actinomycin D (7-AAD), and anti-His Tag (J099B12) antibody were purchased from Biolegend (San Diego, CA, USA). Recombinant mouse Fas ligand (FasL) (TNFSF6) was purchased from R&D systems (Minneapolis, MN, USA).
For flow cytometry analyses, fluorescently-labelled anti-phospho (Tyr319/Tyr352)-ZAP70/Syk antibody (n3kobu5), and goat anti-rabbit IgG (H+L) secondary antibody (polyclonal) were purchased from Invitrogen. Fluorescently-labeled Annexin V, and antibodies including anti-mCD4 (RM4-5), anti-mCD8 (YTS156.7.7), anti-mCD3ε (17A2), anti-IFN-γ (XMG1.2), anti-TNF-α (MP6-XT22), anti-IL-2 (JES6-5H4), anti-granzyme B (GB11), anti-CD107a (1D4B), anti-CD69 (H1.2F3), anti-PD-1 (29F.1A12), anti-Ki67 (16A8), anti-Foxp3 (MF-14), anti-NK1.1 (PK136), anti-I-A/I-E (M5/114.15.2), anti-F4/80 (BM8), anti-CD19 (6D5), anti-Gr-1 (RB6-8C5), anti-CD11b (M1/70), anti-CD11c (N418), anti-Siglec-F (S17007L), anti-Thy1.1 (OX-7), anti-CD45.2 (104), anti-phospho (Thr202/Tyr204)-Erk1/2 (4B11B69), anti-FasL (MFL3), anti-H-2Kb-SIINFEKL (25-D1.16), anti-Fas (SA367H8) and anti-PD-L1 (10F.9G2) were purchased from Biolegend.
Histological analyses
Mouse samples including tumour tissues and adjacent tissues (skin and muscle) were harvested from C57BL/6J mice 10 days after subcutaneous inoculation with B16F10 cancer cells (5 × 105). The collected tissues were embedded in O.C.T. compound (Tissue-Tek®, Sakura Finetek, Tokyo, Japan) and frozen with liquid nitrogen for cryosection with Leica CM3050S cryostat (Leica Microsystems, Milton Keynes, UK). Cryosections collected on slides were thawed and hydrated in phosphate-buffered saline (PBS; Gibco) for 15 min at room temperature. The section slides were then stained with Filipin III (100 μg/mL in PBS) for 1 h followed by rinsing with PBS twice. The corresponding adjacent section slides were sent to the Histology Core Facility at EPFL for hematoxylin and eosin (H&E) staining. The slide images were acquired using a confocal microscope (LSM700, Zeiss, Oberkochen, Germany) and processed using ImageJ.
All human biopsy samples were obtained from West China Hospital (Chengdu, China) following the approved protocol (No.120, 2016). Informed consent was obtained from West China Hospital ethic committee. Briefly, the collected cancer samples from human patient were embedded in O.C.T. compound (Tissue-Tek®) and frozen with liquid nitrogen for cryosection with Leica CM3050S cryostat. Filipin III and H&E staining were performed on the section slides for observing the cholesterol level and tissue structure, respectively. The slide images were acquired using an inverted microscope (Eclipse Ti-U, Nikon, Kyoto, Japan) and processed by ImageJ.
Quantification of global cholesterol levels in tumour, skin, and muscle tissues
B16F10 tumour, skin, or muscle tissues adjacent to tumour was harvested separately from tumour-inoculated mice and digested with tissue digestion buffer (0.1 mg/mL DNase I, dispase II and hyaluronidase, and 1 mg/mL collagenase IV in RPMI 1640 medium) on shaker at 37 °C for 1 h. The tissue fluid passing through a cell strainer (70 μm, Fisher Scientific, Pittsburgh, PA, USA) was added with methanol/chloroform (1:2, v/v) for cholesterol extraction and shaked at room temperature for 2 h. Afterward, the organic phase containing cholesterol was collected and the solvent was evaporated under vacuum. Finally, the cholesterol in each sample was quantified using the Amplex Red cholesterol assay kit (Invitrogen) according to the manufacturer’s recommended protocol.
Filipin staining of cholesterol for flow cytometry analyses
Tumour tissue was harvested from BALB/c mice 12 days after subcutaneous inoculation with 4T1-Fluc-tdTomato cancer cells (5 × 105), and digested with tissue digestion buffer on shaker at 37 °C for 45 min. After passing through a cell strainer (70 μm), the red blood cells were lysed with ACK lysing buffer (Gibco) at room temperature for 5 min. The collected cells were then stained with Filipin (10 μg/mL in PBS) at 4 °C for 30 min. After washing with PBS (0.2 w/v% BSA), the cells were resuspended in PI solution (10 μg/mL in PBS) for flow cytometry analyses. Similar Filipin staining was performed with EG7-OVA cancer cells and activated CD8+ T-cells to compare their membrane cholesterol levels.
Modulation of cholesterol levels in plasma membrane of cancer cells
To deplete cholesterol from plasma membrane, B16F10 (or EG7-OVA, MC38-HER2, and Me275-HER2) cancer cells were incubated with DMEM medium supplemented with MeβCD (5 mM) at 37 °C for 30 min, and then washed with PBS twice. To supplement cell plasma membrane with cholesterol, B16F10 (or EG7-OVA, MC38-HER2, and Me275-HER2) cancer cells were treated with Chol (5 mM) in the DMEM medium at 37 °C for 30 min, and then washed with PBS twice33.
Generation of acyl-CoA:cholesterol acyltransferase-1 (ACAT1) knock-down and overexpressing B16F10 cancer cell lines
Lentiviral plasmids containing the ACAT1 knock-down and overexpression constructs were generated by standard molecular cloning methods. To generate ACAT1 knock-down cells, the shRNA target sequence (5’-CCAACCAGAGACTAAACATAT-3’) was cloned into the pLKO.1 vector with the AgeI/EcoRI sites57. To generate ACAT1 overexpressing cells, codon-optimized cDNA encoding ACAT1 (NM_009230.3) was synthesized by Twist Biosciences (South San Francisco, CA, USA) and cloned into the lentiviral expression vector S002 by Gibson assembly cloning.
Lentivirus was produced by transient transfection of HEK293T cells with the S002 or pLKO.1 transgene expression vectors, the pDelta8.9 packaging plasmid, and the VSV-G envelope plasmid. In brief, HEK293T cells were transfected with a mixture of plasmid DNA [VSV-G : pDelta8.9 : S002 (or pLKO.1) with a weight ratio of 1:2:3] assembled in calcium-phosphate particles. After overnight incubation, the medium was replaced with normal culture medium. Supernatants containing viral particles were collected 48 and 72 hours after transfection and filtered through a 0.45-μm filter. Untitrated viral supernatant supplemented with protamine (10 μg/mL) was added to B16F10 cells, and the contact of viral particles with cells was ensured by centrifugation (2000 rpm, 1 h). One day after transduction, stably transduced B16F10 cells were subjected to puromycin selection (0.5 μg/mL) for two weeks.
Quantification of intracellular and plasma membrane cholesterol levels
The total cellular cholesterol level was first quantified using the Amplex Red cholesterol assay kit. In brief, B16F10 (or EG7-OVA, MC38-HER2, and Me275-HER2) cancer cells were fixed with glutaraldehyde (0.1 wt% in PBS) and the total cholesterol was extracted with methanol/chloroform (1:2, v/v) under sonication. After removal of organic solvent under vacuum, the cholesterol level was quantified using the Amplex Red cholesterol assay kit as described above. To quantify the intracellular cholesterol, the fixed cancer cells were treated with cholesterol oxidase (2 U/mL in PBS) to oxidize the plasma membrane cholesterol before extraction. The plasma membrane cholesterol level was calculated by subtracting the intracellular cholesterol level from the total cellular cholesterol level33. The cholesterol levels of cancer cells were measured immediately post the MeβCD- or Chol-treatment. Additionally, a kinetic study was performed to follow the cholesterol levels of B16F10 cancer cells at 0, 1, 3, 5 and 12 h post MeβCD-treatment.
To measure the cellular cholesterol level of cancer cells in B16F10 tumours, tumour tissues were harvested 30 min after a single intratumoural injection of PBS (100 μL) or MeβCD solution (100 μL, 20 mg/mL in PBS) in tumour-bearing mice. Tumour tissues were digested with tissue digestion buffer on shaker at 37 °C for 1 h. Afterward, the cells were collected after passing through a cell strainer (70 μm) and centrifugation (2000 rpm, 5 min). The cells were resuspended in 40 v/v% Percoll (GE Healthcare, Chicago, IL, USA) followed by addition of 55 v/v% Percoll at the bottom. After centrifugation (2000 rpm, 20 min), the concentrated cancer cells were collected at the interface. The intracellular and plasma cholesterol levels were measured using abovementioned procedures.
Viability and apoptosis assays of cancer cells
The viability and apoptosis of MeβCD-treated cancer cells was evaluated using Annexin V and 7-AAD (or DAPI) staining. Briefly, B16F10 (or EG7-OVA, MC38-HER2, and Me275-HER2) cancer cells were incubated with DMEM medium supplied with MeβCD or Chol (5 mM) at 37 °C for 30 min, and washed with PBS twice. The treated cancer cells were then stained with fluorescently-labelled Annexin V and 7-AAD (0.5 μg/mL in PBS, or DAPI, 0.1 μg/mL in PBS) for flow cytometry analysis.
In another assay, B16F10 cancer cells were treated with MeβCD (5 mM in DMEM medium) at 37 °C for 30 min. After washing with PBS twice, the cancer cells were resuspended in DMEM medium (10 v/v% FBS and 10 mM HEPES) followed by the addition of various concentrations of recombinant mouse FasL plus anti-His Tag antibody (5 μg/mL) for multimerization58 or TNF-α in DMEM medium (10 v/v% FBS and 10 mM HEPES) and incubation at 37 °C for 5 h. After washing with PBS (0.2 w/v% BSA), the cancer cells were stained with fluorescently-labelled Annexin V and DAPI for flow cytometry analysis. Similarly, various concentrations of active perforin-1 in PBS were added to cancer cells resuspended in Hank’s Balanced Salt Solution (HBSS, Gibco) supplemented with BSA (0.4 w/v%), HEPES (10 mM), and CaCl2 (5 mM), of which the viability was determined by DAPI staining and flow cytometry analysis.
Measurement of cell cortical stiffness by atomic force microscopy (AFM)
The measurement of cell cortical stiffness using AFM followed a well-established protocol26. In brief, AFM force curves were recorded using a customized Dimension Icon AFM (Bruker, Billerica, MA, USA) mounted on top of an IX81 inverted optical microscope (Olympus, Tokyo, Japan) equipped with a 20× objective and a heating stage for live cell imaging. The samples were mobilized using an XY stage until the cell of interest was placed under the AFM tip as visualized through the optical microscope. Force curves on the cell were recorded at a ~5 μm/s rate in relative trigger mode (15 nm trigger threshold) using a PNP-TR-B cantilever (Nanoworld, Neuchâtel, Switzerland). The cantilever spring constant was 0.08 N/m, measured using the deflection sensitivity (170 nm/V) and a thermal tune. Measurement was performed on single cells at 37 °C before and after treatment with Chol or MeβCD (5 mM, 15 min at 37 °C). Nanoscope Analysis software (Bruker) was used to process the force curves and calculate the sample Young’s Modulus, by doing a fit of the approach curve less than 500 nm indentation (to take into account only the cortical stiffness), assuming a cortex Poisson’s ratio of 0.3.
Measurement of cell cortical stiffness by optical tweezer
The measurement of cell cortical stiffness by optical tweezer was adapted from a well-established method in the literature27. The laser beam (10 W, 1064 nm) was tightly focused through a series of Keplerian beam expanders and a high numerical aperture objective (100×/1.45, oil immersion, Nikon). A high-resolution quadrant detector (PDQ80A, Thorlabs, Newton, NJ, USA) was used for force measurement. To measure the mechanical properties of the cell cortex, polystyrene particles (500 nm in diameter, orange fluorescence) were added to the culture medium and endocytosed by the cells. The particle was then dragged by optical tweezers toward the cell membrane to deform the cell cortex with a speed of 1 μm/s. The displacement of the particle and the resistant force were recorded by the quadrant photodetectors. The stiffness of the cell cortical structure including plasma membrane and cell cortex was defined by the slope of the force-displacement curve. The data collection and post-processing were performed using MATLAB (Mathworks, Natick, MA, USA).
Deformability cytometry (DC)
DC setup was built following a published report28. A 4-inch silicon wafer was selectively etched using photolithography and deep reactive ion etching for the fabrication of the microfluidic device. The height, width, and length of the constriction area were measured using a mechanical profiler (Dektak® XT, Bruker) as 30 μm, 30 μm, and 300 μm, respectively. The chosen geometry ensured that the cells were deformed properly. The projected areas of cells were 90% to 50% of the cross-sectional area of the constriction zone. Devices were casted using a 10:1 mixture of polydimethylsiloxane (PDMS) (Sylgard 184, Dow Corning, Midland, MI, USA) from the microfabricated molds by curing the prepolymer overnight at 65 °C. Glass coverslips (No.1, VWR, Radnor, PA, USA) were cleaned thoroughly by soaking them in acetone, isopropyl alcohol, ethanol and distilled water, and subsequently dried at 65 °C to avoid any possible leakage in the microfluidic device due to the high viscosity of the solutions and high flow rate. The surfaces of the coverslips and PDMS devices were treated with a plasma cleaner (PDC-002-HPCE, Plasma Harrick, Ithaca, NY, USA) at 29 Watt for 45 seconds prior to bonding. They were subsequently compressed using a 1-kg weight overnight at 100 °C to ensure firm and robust attachment.
In a typical DC measurement, cells were flowed through a microfluidic chamber at a rate of 4 μL/min (Me275-HER2) or 5.34 μL/min (B16F10, EG7-OVA, and MC38-HER2). The cells were centred in the microfluidic channel using a sheath flow (Supplementary Fig. 4a). The sheath flow rate was set at 3× the cell flow rates, i.e., 12 μL/min or 16 μL/min, respectively. The flow rates were controlled with a programmable syringe pump (neMESYS 290N, Cetoni, Korbußen, Germany). Prior to the measurement, the cells were treated with MeβCD or Chol as mentioned above or kept in PBS without treatment. The cells were then suspended in a methyl-cellulose solution (0.6 w/v% in HBSS) at a concentration of 2 × 106 cells/mL. The cells were passed through the narrow constriction zone of 30 μm × 30 μm × 300 μm (height × width × length), and visualized using an inverted microscope (Nikon) equipped with a 20× objective and a high-speed camera (VEO640L, Phantom, Wayne, NJ, USA). To ensure that cell deformation was measured at the equilibrium state, a region of interest (ROI) of 128 × 256 pixels was imaged at the end of the channel where the laminar flow was fully developed. The cell imaging was performed using the following camera parameters: exposure time, 1 μs; frame rate, 7000 or 10,000 frame per second (fps). Time-lapse movies containing several thousands of cells were analysed using a custom ImageJ macro (Bioimaging and Optics Platform, EPFL). Briefly, the cell contour was first identified based on greyscale value. Subsequently, a convex hull (Convex Hull, ImageJ) was fitted on the cells to avoid large increase in cell perimeter. The measured projected cellular area and perimeter were used to calculate the deformation (D) defined as:
where A is the projected cell surface area and l is the cell perimeter.
Further post-processing was performed using MATLAB. In particular, cells with irregular shapes or with poor contour identification were eliminated as proposed in published literature59. Briefly, the ratio R of the projected area was calculated using the hull approximation Ahull and the greyscale values Acontour as:
Cells with R ≥ 1.07 were eliminated in the final analysis.
Deformation of cell populations with different cell areas were compared using iso-elasticity lines plotted using Shape-Out (Paul Müller and others, version 2.7.4). In order to input the data in Shape-Out, the above post-processed data in xlsx format were converted into the H5 format using MATLAB.
Fabrication and rheological analysis of polyacrylamide (PA) hydrogel substrates
PA hydrogel substrates were prepared using a protocol adapted from a reported literature60. Briefly, a 96-well glass bottom plate (Falcon, Corning, NY, USA) was treated with a NaOH solution (0.1 M in H2O, 50 μL per well) for 5 min at room temperature (Supplementary Fig. 5a). Upon removal of NaOH solution, APTMS (20 μL per well) was applied for 3 min at room temperature. The well plate was then thoroughly rinsed with de-ionized (DI) water to remove any remaining APTMS followed by the addition of a glutaraldehyde solution (0.5 wt% in H2O, 50 μL per well) and incubation at room temperature for 20 min. The well plate was subsequently rinsed with DI water and dried in air for 30 min. To prepare PA hydrogels of different stiffness (PA-1 and 2 as shown in Supplementary Fig. 5b), we made the PA hydrogel precursor solutions (30 μL per well) with various concentrations of the acrylamide monomer and the bis-acrylamide cross-linker. Ammonium persulfate (0.1 w/v% in final concentration) and TEMED (0.1 v/v% in final concentration) were then added to initiate the polymerization followed by a brief vortexing and incubation at room temperature for 1 h. The hydrogel substrates were then washed with PBS (200 μL per well × 2) to remove any unreacted acrylamide. To facilitate cell attachment, fibronectin was conjugated to the hydrogel surface using the heterobifunctional linker Sulfo-SANPAH. In brief, a Sulfo-SANPAH solution (1 mg/mL in milli-Q H2O, 20 μL per well) was pipetted onto the hydrogel surface followed by UV irradiation (365 nm, 0.8 mW, 20 mA) for 10 min and washing with HEPES buffer (50 mM in PBS) twice. After incubation with fibronectin solution (50 μg/mL in PBS, 50 μL per well) at 4 °C overnight and washing for three times with PBS, the coated PA hydrogels were stored in PBS at 4 °C before use.
DHR3 shear rheometer (TA Instruments, New Castle, DE, USA) with a parallel plate (8 mm in diameter) was used for rheological test. The shear storage modulus G´ was measured using the following parameters: strain, 5%; frequency, 5 rad/s. PA hydrogel sample of a typical thickness of 1.5 mm and a diameter of 14 mm was measured at a constant axial force of 0.5 N and a constant temperature of 37 °C. The tensile elastic modulus E (Young’s modulus) was retrieved using: E = 2 × (1+n) × G´, where n = 0.45 for the Poisson’s ratio of polyacrylamide.
Activation of Pmel and OT-I CD8+ T-cells
Spleens collected from Pmel Thy1.1+ mice were ground through a cell strainer (70 μm) at day 0. Red blood cells were lysed with ACK lysing buffer (2 mL per spleen) at room temperature for 5 min. After washing twice with PBS, splenocytes were cultured in complete RPMI 1640 medium supplemented with FBS (10 v/v%), HEPES (1 v/v%), penicillin/streptomycin (1 v/v%), and β-mercaptoethanol (0.1 v/v%) in the presence of hgp100 peptide (1 μM), recombinant mouse IL-2 (10 ng/mL) and recombinant mouse IL-7 (2 ng/mL) for 3 days. After Ficoll-Pague Plus (GE Healthcare) gradient separation to eliminate dead cells, the activated Pmel CD8+ T-cells (purity > 95 %) were maintained in the medium containing recombinant mouse IL-2 (10 ng/mL) and IL-7 (10 ng/mL) and used between day 4 to 8 post splenocyte collection for in vitro or in vivo studies. Activated OT-I CD8+ T-cells were obtained from the spleens of OT-I mice using similar protocol by replacing hgp100 peptide with SIINFEKL peptide.
In vitro killing assays of cancer cells by T-cells
B16F10 cancer cells were seeded on fibronectin-coated hydrogel substrates and incubated overnight. Afterward, B16F10 cancer cells were pulsed with hgp100 peptide (2 μM in complete RPMI 1640 medium) for 30 min and then treated with MeβCD or Chol (5 mM in DMEM medium) at 37 °C for another 30 min followed by washing with PBS twice. The cancer cells were immediately added with a suspension of activated Pmel CD8+ T-cell in complete RPMI 1640 medium at an effector:target (E:T) ratio of 10:1 and co-cultured at 37 °C for 5 h. In a control sample, a Triton X-100 solution (0.1 wt%) was added to determine the 100 % cell lysis. To quantify target cell death, supernatant from each well was retrieved for a lactate dehydrogenase (LDH) cytotoxicity assay using a CytoTox 96® Non-Radioactive Cytotoxicity Assay kit (Promega, Madison, WI, USA) according to the manufacturer’s recommended protocol. In another experiment, MeβCD-treated B16F10 cancer cells were further cultured in DMEM medium (10 v/v% FBS) at 37 °C for additional 12 h, and pulsed with hgp100 peptide (2 μM in complete RPMI 1640 medium) for 30 min followed by the addition of Pmel CD8+ T-cells for the killing assay. In mechanism experiments, T-cells were pre-treated with various cytoskeleton inhibitors (2 μM LatA for 10 min, 100 μM Bleb for 10 min, or 1 μM MycaB for 15 min) at 37 °C to inhibit T-cell forces before adding to the co-culture.
In the killing assay for EG7-OVA cancer cells, the cancer cells were first pulsed with SIINFEKL (5 μM in complete RPMI 1640 medium) for 30 min and then treated with MeβCD (5 mM in DMEM medium) at 37 °C for another 30 min followed by washing with PBS twice. Next, a suspension of activated OT-I CD8+ T-cell (pre-labelled with 2 μM CFSE at 37 °C for 5 min) in complete RPMI 1640 medium was added at E:T ratios of 1:1 or 5:1. After a co-culture at 37 °C for 5 h, cell death of EG7-OVA cancer cells was quantified by DAPI staining and flow cytometry analysis. Pmel CD8+ T-cell mediated killing of B16F10 (or ACAT1 KD and ACAT1 OE B16F10) cancer cells were analysed at an E:T ratio of 10:1 using a similar killing assay (without MeβCD treatment) by pulsing cancer cells with hgp100 peptide (2 μM).
Production of mouse interleukin-15 super-agonist (IL-15SA)
The engineered IL-15SA construct (gWIZ-mIL-15SA) was a generous gift from Prof. Darrell J. Irvine (MIT, USA). As shown in Supplementary Fig. 9a, the IL-15SA contains a mouse IL-15 fused at the C-terminal of Sushi domain of a mouse IL-15 receptor α (IL-15Rα), which is next fused at the C-terminal with a mouse IgG2 Fc. IL-15SA was expressed by HEK293-E cells (Gibco) in Freestyle medium at the EPFL Protein Production and Structure Core Facility (PTPSP). The supernatant of culture medium containing IL-15SA was harvested by centrifugation after a 7-day culture and was filtered through a filter membrane (0.22 μm) to obtain a clear solution. The IL-15SA was first captured with a HiTrap Protein A affinity chromatography column on an the AKTA pure 25 system (GE Healthcare), and eluted with an elution buffer (0.05 M sodium citrate, 0.3 M sodium chloride, pH = 3.0). The eluted protein was next collected immediately in a neutralization buffer (1 M Tris-HCl, pH = 10.0) followed by concentration with membrane ultrafiltration (molecular weight cut-off 10 kDa) in a Vivaspin (GE Healthcare). The concentrated protein solution was further purified with a Superdex 200 increase size exclusion column (GE Healthcare) at a flow rate of 1.0 mL/min with PBS buffer on the AKTA pure 25 system. The purity and activity of IL-15SA was confirmed with sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (Supplementary Fig. 9b) and T-cell proliferation assay (Supplementary Fig. 9c), respectively. The purified protein was aliquoted and stored at -80 °C before use.
Adoptive T-cell transfer (ACT) therapy in solid tumour models
In an experiment with cholesterol supplementation in tumour (Supplementary Fig. 6), B16F10 melanoma cells (0.5 × 106) in PBS (100 μL) were inoculated subcutaneously in the right flanks of Thy1.2+ C57BL/6J mice at day 0. Recipient mice were randomized before ACT. Activated Pmel Thy1.1+CD8+ T-cells (5 × 106) were intravenously infused via the tail vein into recipient mice at day 7. In another group, post-T-cell transfer mice received the intratumoural injections of Chol every other day (2 mg/dose in 100 μL PBS × 8; day 7 to 21). Tumour area (product of measured orthogonal length and width) and body weight were monitored every 2 days. Mice were euthanized when the body weight loss was higher than 20% of the pre-dosing weight or the tumour area reached 150 mm2.
In the experiments with ACAT1 KD or ACAT1 OE B16F10 tumours, native, ACAT1 KD, or ACAT1 OE B16F10 cells (0.5 × 106) in PBS (100 μL) were inoculated subcutaneously in the right flanks of Thy1.2+ C57BL/6J mice at day 0. Tumour-bearing mice were randomized and treated similarly with PBS or activated Pmel Thy1.1+CD8+ T-cells (5 × 106) through intravenous injection at day 6. Mice were then monitored as described above.
In an experiment for cancer-cell stiffening with MeβCD, B16F10 melanoma cells (0.5 × 106) were inoculated similarly in Thy1.2+ C57BL/6J mice at day 0. At day 9, activated Pmel Thy1.1+CD8+ T-cells (5 × 106) were intravenously infused via the tail vein into recipient mice followed by intratumoural injections of IL-15SA (10 μg/dose in 50 μL PBS) on day 9, 11, and 13, and daily intratumoural injections of MeβCD (1 mg/dose in 50 μL PBS) from day 9 to 18. Mice receiving PBS, MeβCD only, or ACT + IL-15SA served as controls. In another therapeutic experiment (Supplementary Fig. 10), B16F10 tumour-bearing mice received three adoptive transfers of activated Pmel Thy1.1+CD8+ T-cells (5 × 106 per injection) on day 5, 11 and 22 without IL-15SA adjuvant. A similar therapeutic experiment was conducted with Thy1.2+ C57BL/6J mice bearing subcutaneous EG7-OVA lymphoma tumours (0.5 × 106 cancer cells inoculated per mouse at day 0; Supplementary Fig. 11), which received two intravenous infusions of activated OT-I CD8+ T-cells (5 × 106 per injection) on day 11 and 15 followed by intratumoural injections of IL-15SA (5 μg/dose in 50 μL PBS × 5; every other day from day 11 to 19) and MeβCD (1 mg/dose in 50 μL PBS × 10; daily from day 11 to 20). Mice were monitored as described above.
Characterizations of tumour-infiltrating immune cells by flow cytometry analyses
B16F10 melanoma cells (1 × 106) were inoculated subcutaneously in the right flanks of Thy1.2+ C57BL/6J mice at day 0. At day 9, activated Pmel Thy1.1+CD8+ T-cells (5 × 106 per injection) were intravenously infused via the tail vein into tumour-bearing mice followed by intratumoural injections of IL-15SA (10 μg/dose in 50 μL PBS) on day 9 and 11, and daily intratumoural injections of MeβCD (1 mg/dose in 50 μL PBS) from day 9 to 13. Mice receiving PBS, MeβCD only, or ACT + IL-15SA served as controls. Mice were euthanized at day 14 and tumour tissues were collected and ground through a cell strainer (70 μm). Red blood cells were lysed with ACK lysis buffer at room temperature for 5 min. All cells were stained with Aqua live/dead stain (Invitrogen) followed by surface marker staining in buffer (PBS, 0.2 w/v% BSA) with the corresponding fluorescently-labelled antibodies at 4 °C for 20 min. For transcription factor staining, cells were stained for surface markers first followed by fixation and permeabilization with Foxp3/Transcription Factor Staining Buffer Set (eBioscience, San Diego, CA, USA) and addition of fluorescently-labelled antibodies against intracellular transcription factors. For intracellular cytokine staining, cells were first stimulated in complete RPMI 1640 medium containing 1× Cell Activation Cocktail with Brefeldin A (Biolegend) at 37 °C for 4 h. Following surface marker staining, cells were fixed and permeabilized with Cyto-Fast Fix/Perm Buffer Set (Biolegend), and stained with the fluorescently-labelled antibodies against cytokines. Flow cytometry analysis was performed with an Attune NxT flow cytometer (Invitrogen) and data analysis was performed using FlowJo software (Tree Star, BD Biosciences, Franklin Lakes, NJ, USA).
Flow cytometry analyses of cancer cells and T-cells from in vitro co-culture assays
For analysis of B16F10 (or B16F10-OVA) cancer cells only, cells were treated with MeβCD (5 mM in DMEM medium) at 37 °C for 30 min, and then stained with fluorescently labelled anti-H-2Kb-SIINFEKL, anti-Fas, and anti-PD-L1 antibodies for flow cytometry analysis.
In a typical co-culture assay, B16F10 cancer cells were seeded on a 96-well plate and incubated overnight for cell attachment. Afterward, B16F10 cancer cells were pulsed with hgp100 peptide (2 μM in complete RPMI 1640 medium) for 30 min and then treated with MeβCD (5 mM in DMEM medium) for another 30 min followed by washing with PBS twice. Next, a suspension of activated Pmel CD8+ T-cell in complete RPMI medium supplemented with FBS (10 v/v%) was added to each well at an E:T ratio of 10:1. In a similar co-culture assay, B16F10 cancer cells were pulsed with various concentrations of SIINFEKL peptide and then added with a suspension of activated OT-I CD8+ T-cells at an E:T ratio of 10:1. For T-cell function and cytokine secretion analyses, Brefeldin A (5 μg/mL) and Monensin (2 μM) were added to the co-culture. After 5-h co-incubation at 37 °C, T-cells were stained with fluorescently-labelled anti-PD-1, anti-CD69, anti-FasL, anti-CD107a, anti-granzyme B, anti-TNF-α, anti-IFN-γ, and anti-IL-2 antibodies for flow cytometry analysis.
To determine the protein phosphorylation in T-cells, the plate for co-culture was centrifuged at 1500 rpm for 2 min followed by a brief co-incubation for 5 min. To detect phosphorylation of TCR signalling proteins ZAP70, Erk1/2 and NF-κB, T-cells were fixed with a paraldehyde solution (1.5 w/v% in PBS), and then permeabilized by resuspending in ice-cold MeOH at 4 °C for 10 min followed by washing with PBS (0.2 w/v% BSA). Next, T-cells were stained with fluorescently-labelled anti-phospho-ZAP70, anti-phospho-Erk1/2, or anti-phospho-NF-κB p65 antibodies for flow cytometry analysis. The MFI of phospho-ZAP70, Erk1/2, and NF-κB p65 were normalized by the corresponding levels in unstimulated Pmel CD8+ T-cells. To detect phosphorylation of Pyk2, T-cells were fixed with paraldehyde solution (1.5 w/v% in PBS), and then permeabilized by resuspending in Triton X-100 solution (0.1 w/v% in PBS) at 25 °C for 5 min. After washing with PBS (0.2 w/v% BSA), T-cells were stained with anti-phospho-Pyk2 antibody and then fluorescently-labelled secondary antibody for flow cytometry analysis.
In another assay, B16F10 cancer cells were seeded on a 48-well plate and incubated overnight for cell attachment. A suspension of activated Pmel CD8+ T-cell in complete RPMI medium supplemented with FBS (10 v/v%), recombinant mouse IL-2 (10 ng/mL), and MeβCD (0.05 mM) was then added to each well at an E:T ratio of 1:1. After 48-h co-incubation at 37 °C, T-cells and B16F10 cells were collected for flow cytometry analysis.
Preparation of fluorescent bead-embedded PA hydrogel substrates for traction force microscopy (TFM)
To prepare biotinylated anti-CD3 and anti-CD28 antibodies for hydrogel surface coating, a NHS-Biotin (10 mM) solution in DMSO was added to a solution of anti-CD3 or anti-CD28 antibodies (2 mg/mL in PBS) at a final concentration of 300 μM of NHS-Biotin. After 30 min incubation on shaker at room temperature, the biotinylated antibody solution was washed with PBS (500 μL × 5) using an ultra-centrifugal filter with 30-kDa cutoff (Amicon®, Merck) and the final protein concentration was determined with a microvolume UV-Vis spectrophotometer (NanoDrop One, Thermo Fisher Scientific). The biotinylated antibodies were stored at 4 °C before use.
The antibody-coated hydrogel substrates for TFM were prepared according to a protocol adapted from the published literature20. Briefly, NaOH solution (1 mL, 0.1 M in H2O) was added on top of a coverslip (Carl Roth, thickness 0.17 mm, diameter 25 mm) and heated at 70 °C until a film of dried NaOH was formed. APTMS was then added on the coverslip surface and incubated at room temperature for 5 min. After rinsing the coverslips with DI water, glutaraldehyde (0.5 wt% in H2O) was added for 30 min. The amino-silanated coverslip was rinsed with DI water. Next, the hydrogel precursor solution (PA-3, 4, and 5 as shown in Supplementary Fig. 5b) containing fluorescent PS beads (0.6 w/v%, 200 nm), streptavidin acrylamide (0.2 mg/mL), ammonium persulfate (0.1 wt%), and TEMED (0.1 v/v%) was pipetted onto a methyl-silanated glass slide. The amino-silanated coverslip with the treated side facing down was placed on top of the solution to create a sandwich structure. After polymerization for 1 h, the coverslip-hydrogel composite was immersed in PBS for 5 min twice to remove any unpolymerized acrylamide. Afterwards, a PBS solution of biotinylated anti-CD3 (10 μg/mL) and biotinylated anti-CD28 (10 μg/mL) or biotinylated anti-mouse CD45 (20 μg/mL, Clone 30F-11, Biolegend) was added onto the hydrogel surface and incubated at 37 °C for 1 h. Finally, the coverslip-hydrogel composite was immersed in PBS for 5 min to remove free antibodies. The fluorescent bead-embedded, antibody-coated hydrogel substrates were stored in PBS at 4 °C before use.
TFM measurement of T-cell forces
The experiment was performed using an inverted microscope (IX83, Olympus) equipped with a spinning disk confocal scanner (CSU-W1, Visitron, Puchheim, Germany), a 60×/1.42 UPLSAPO objective (Olympus), and a sCMOS camera (Orca Flash 4.0, Hamamatsu, Shizuoka, Japan). The experiment was performed at 37 °C with 5% CO2. The microscope was pre-heated to 37 °C for at least 2 h prior to the experiment. The gels and all reagents were kept at 37 °C during the experiment. At the start of the experiment, activated Pmel T-cells (1 × 105) were added onto the gel in RPMI 1640 medium without phenol red (Gibco) supplemented with FBS (10 v/v%), HEPES (1 v/v%), penicillin/streptomycin (1 v/v%), and β-mercaptoethanol (0.1 v/v%). After a 20-min incubation at 37 °C, unbound or weakly bound cells were removed by aspirating the medium and replacing it with fresh medium. Z-stack imaging of the cells (Brightfield) and the gel (RFP channel) were captured 30 min after the addition of the cells to the gel. Afterwards, SDS (50 μL, 0.5 w/v%) was added to the medium to lyse the cells and the unstressed gel was imaged 5 min later. The same procedure was repeated on hydrogels of various stiffness (PA-3, 4, and 5 as shown in Supplementary Fig. 5b). As a control group, activated Pmel T-cells were treated with LatA (2 μM in PBS) at 37 °C for 10 min before TFM measurement. To measure T-cell forces post pre-treatment with LatA (2 μM at 37 °C for 10 min) or MycaB (1 μM at 37 °C for 15 min), pre-treated T-cells were incubated in complete RPMI medium supplemented with FBS (10 v/v%) at 37 °C for 0 h or 5 h followed by the measurement procedure described above.
The cellular forces were measured using an automated ImageJ macro (Bioimaging and Optics Platform, EPFL). The top layer of the PA gel was automatically found in both stressed and unstressed gels, combined into an image pair, and the drift was corrected using the “Linear stack alignment with SIFT” plugin in ImageJ. The particle displacement and force maps were generated using freely-available particle image velocimetry (PIV) analysis and Fourier transform traction cytometry (FTTC) plugins61. The PIV plugin was run with the cross-correlation iteration option (piv1=128 sw1=128 vs1=64 piv2=64 sw2=64 vs2=32 piv3=32 sw3=32 vs3=16). The FTTC analysis was performed using the following parameters: Poisson ratio, 0.5; Young’s modulus, 210/560/890 Pa. The cell contour was automatically identified from the brightfield Z-stacks. The force field and cell contour were exported and post-processed using MATLAB. Briefly, the stress under the cell was multiplied by the area of final interrogation window (vector spacing vs3) to yield the cellular force (Fcell). The background noise of each gel was calculated by averaging the stress values of the gel away from the cell. The cellular force was corrected (Fcorrected) by subtracting the noise of an area equivalent to the cell area (Fnoise) from Fcell:
F-actin imaging of T-cells on PA hydrogel substrates with varying stiffness
PA hydrogel substrates with varying stiffness were fabricated on glass bottom dishes (35 mm, glass bottom diameter 20 mm, ibidi, Gräfelfing, Germany) and coated with anti-CD3 and anti-CD28 antibodies as described above. Activated Pmel CD8+ T-cell suspension in complete RPMI medium supplemented with FBS (10 v/v%) was added on hydrogels on glass-bottom dishes and centrifuged at low speed (1500 rpm) for 2 min to bring the T-cells into contact with the hydrogel surface. After incubation for 10 min, T-cells were fixed with a formaldehyde solution in PBS (4 w/v%) at room temperature for 20 min followed by washing with PBS twice. T-cells were next permeabilized using Triton X-100 in PBS (0.1 w/v%) at room temperature for 5 min. After washing with PBS twice, T-cells were stained with phalloidin-iFluor™ 488 (1/1000 dilution according to the manufacturer’s recommended protocol) and Hoechst 33342 (5 μg/mL) in PBS (1 wt% BSA) at room temperature in the dark for 30 min. After washing with PBS for three times, Fluoromount-G mounting medium (300 μL, Invitrogen) was added on the hydrogel surface and a coverslip (thickness 0.17 mm, diameter 25 mm) was placed on top to seal the glass bottom. The confocal images were acquired using an inverted microscope (IX83, Olympus) equipped with a spinning disk confocal scanner (CSU-W1, Visitron) and a 100×/1.40 UPLSAPO objective (Olympus). T-cells in Fig. 6d were imaged at high resolution (Z-stack step size, 150 nm) and deconvoluted using Huygens Remote Manager (Scientific Volume Imaging, Hilversum, Netherland). The F-actin fluorescence images in the XZ plane (side view) were obtained by summing 20 slices in the XZ plane in the middle of the cells (Z project plugin). The intensity display settings are identical for each side view image (1500-25000). The F-actin fluorescence images at the immunological synapse (IS, defined as the structure between the surface of hydrogel and a height of 2 μm above the surface of the hydrogel) of T-cell were obtained by overlaying Z-stack slices located within the IS. The intensity display settings are identical for each IS image (0-18000).
The total F-actin fluorescence intensity at the IS of T-cell was measured using a semi-automated ImageJ macro (Bioimaging and Optics Platform, EPFL). Briefly, Z-stack slices located within the IS were summed (Z project plugin). The resulting image was thresholded (Threshold plugin) and holes filled (Fill Holes plugin). Cell contours were automatically identified (Analyze Particles plugin). Further post-processing was performed using MATLAB. The corrected F-actin fluorescence intensity per pixel at the IS was obtained by subtracting the average background value from the average F-actin fluorescence intensity per pixel at the IS. Finally, to calculate the total F-actin fluorescence intensity at the IS, the corrected F-actin fluorescence intensity per pixel was multiplied by the cell area.
Statistical analyses
Data are presented as mean ± standard error of the mean (SEM) unless otherwise noted. Violin plots show frequency distribution curves created by kernel density method in which the middle solid line shows median, and lower and upper dash lines show 25th and 75th percentiles, respectively. Statistical analysis for each experiment is specified in the corresponding figure legend. Statistical analyses were performed using GraphPad Prism 8 software. In all cases, two-tailed test with P values of less than 0.05 were considered significant.
Supplementary Material
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41551-01X-XXXX-X.
Acknowledgements
We thank R. Guiet (EPFL) for assistance on image analysis. We acknowledge the assistance of W. Li (West China Hospital) for histological analyses of human biopsies. We are grateful to EPFL Bioimaging and Optics Platform, Center of PhenoGenomics, Flow Cytometry Core Facility, Protein Production and Structure Core Facility, and Histology Core Facility for technical support. We thank S. M. Leitão and B. Ghadiani (EPFL) for technical assistance on AFM measurement; P. Müller (Max Planck Institute) for assistance on analysing deformability cytometry data; D. J. Irvine (MIT) for providing IL-15SA construct, B16F10-OVA and 4T1-Fluc-tdTomato cell lines; P. Romero (UNIL) for providing MC38-HER2 and Me275-HER2 cell lines; D. Trono (EPFL) for providing HEK293T cells and pLKO.1 vector; E. Amstad (EPFL) for providing access to a rheometer. We are grateful for the discussions with F. Stellacci, M. De Palma, A. Cont, and A. Persat (EPFL). This work was supported in part by the European Research under the ERC grant agreements MechanoIMM (805337) and ROBOCHIP (714609), Swiss National Science Foundation (Project grant 315230_173243), Swiss Cancer Research foundation (No. KFS-4600-08-2018), Kristian Gerhard Jebsen Foundation, Anna Fuller Fund Grant, and École Polytechnique Fédérale de Lausanne (EPFL). A.K. acknowledges funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No. 754354. M.G. was supported by the Chinese Scholarship Council (CSC) (No. 201808320453).
Footnotes
Reporting Summary. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Author contributions
K.L. and L.T. conceived the study and designed the experiments. K.L., A.K., M.K., L.B., Y.H., V.C., M.G., Y.-Q.X., Y.G., M.T.M.H., Y.W., G.Z., M.G., G.E.F., and M.S.S. performed the experiments. K.L., A.K., L.T., M.S.S., M.G., and G.E.F. analysed the data. L.T. supervised the project. K.L., A.K., and L.T. wrote the manuscript. All authors edited the manuscript.
Competing interests
The authors declare no competing interests.
Peer review information Nature Biomedical Engineering thanks Jochen Guck, Bo Huang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Reprints and permissions information is available at www.nature.com/reprints.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Data availability
The data supporting the results in this study are available within the paper and its Supplementary Information. Source data for the figures are provided with this paper.
Code availability
Source code for the custom ImageJ macro for F-actin analysis, cellular-force calculation and deformability-cytometry analysis, and custom MATLAB scripts for optical-tweezer data collection and data post-processing, are available from the corresponding author on reasonable request. Shape-out (version 2.7.4) for deformability cytometry analysis is available at https://github.com/ZELLMECHANIK-DRESDEN/ShapeOut2.
References
- 1.Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science. 2018;359:1350–1355. doi: 10.1126/science.aar4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang H, Mooney DJ. Biomaterial-assisted targeted modulation of immune cells in cancer treatment. Nat Mater. 2018;17:761–772. doi: 10.1038/s41563-018-0147-9. [DOI] [PubMed] [Google Scholar]
- 3.Eil R, et al. Ionic immune suppression within the tumour microenvironment limits T cell effector function. Nature. 2016;537:539–543. doi: 10.1038/nature19364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.André P, et al. Anti-NKG2A mAb is a checkpoint inhibitor that promotes anti-tumor immunity by unleashing both T and NK cells. Cell. 2018;175:1731–1743. doi: 10.1016/j.cell.2018.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levental KR, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cross SE, Jin YS, Rao J, Gimzewski JK. Nanomechanical analysis of cells from cancer patients. Nat Nanotechnol. 2007;2:780–783. doi: 10.1038/nnano.2007.388. [DOI] [PubMed] [Google Scholar]
- 7.Fritsch A, et al. Are biomechanical changes necessary for tumour progression? Nat Phys. 2010;6:730–732. [Google Scholar]
- 8.Swaminathan V, et al. Mechanical Stiffness grades metastatic potential in patient tumor cells and in cancer cell lines. Cancer Res. 2011;71:5075–5080. doi: 10.1158/0008-5472.CAN-11-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alibert C, Goud B, Manneville JB. Are cancer cells really softer than normal cells? Biol Cell. 2017;109:167–189. doi: 10.1111/boc.201600078. [DOI] [PubMed] [Google Scholar]
- 10.Händel C, et al. Cell membrane softening in human breast and cervical cancer cells. New J Phys. 2015;17:083008 [Google Scholar]
- 11.Köster DV, Mayor S. Cortical actin and the plasma membrane: Inextricably intertwined. Curr Opin Cell Biol. 2016;38:81–89. doi: 10.1016/j.ceb.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 12.Mossman KD, Campi G, Groves JT, Dustin ML. Altered TCR signaling from geometrically repatterned immunological synapses. Science. 2005;310:1191–1193. doi: 10.1126/science.1119238. [DOI] [PubMed] [Google Scholar]
- 13.Judokusumo E, Tabdanov E, Kumari S, Dustin ML, Kam LC. Mechanosensing in T lymphocyte activation. Biophys J. 2012;102:L5–L7. doi: 10.1016/j.bpj.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu B, Chen W, Evavold BD, Zhu C. Accumulation of dynamic catch bonds between TCR and agonist peptide-MHC triggers T cell signaling. Cell. 2014;157:357–368. doi: 10.1016/j.cell.2014.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan Y, et al. Mechanogenetics for the remote and noninvasive control of cancer immunotherapy. Proc Natl Acad Sci U S A. 2018;115:992–997. doi: 10.1073/pnas.1714900115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hickey JW, et al. Engineering an artificial T-cell stimulating matrix for immunotherapy. Adv Mater. 2019;31:1807359. doi: 10.1002/adma.201807359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng KP, Majedi FS, Thauland TJ, Butte MJ. Mechanosensing through YAP controls T cell activation and metabolism. J Exp Med. 2020;217:e20200053. doi: 10.1084/jem.20200053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Basu R, et al. Cytotoxic T cells use mechanical force to potentiate target cell killing. Cell. 2016;165:100–110. doi: 10.1016/j.cell.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hui KL, Balagopalan L, Samelson LE, Upadhyaya A. Cytoskeletal forces during signaling activation in Jurkat T-cells. Mol Biol Cell. 2014;26:685–695. doi: 10.1091/mbc.E14-03-0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saitakis M, et al. Different TCR-induced T lymphocyte responses are potentiated by stiffness with variable sensitivity. Elife. 2017;6:e23190. doi: 10.7554/eLife.23190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Byfield FJ, Aranda-Espinoza H, Romanenko VG, Rothblat GH, Levitan I. Cholesterol depletion increases membrane stiffness of aortic endothelial cells. Biophys J. 2004;87:3336–3343. doi: 10.1529/biophysj.104.040634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khatibzadeh N, Gupta S, Farrell B, Brownell WE, Anvari B. Effects of cholesterol on nanomechanical properties of the living cell plasma membrane. Soft Matter. 2012;8:8350–8360. doi: 10.1039/C2SM25263E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Behnke O, Tranum-Jensen J, Van Deurs B. Filipin as a cholesterol probe. II. Filipin-cholesterol interaction in red blood cell membranes. Eur J Cell Biol. 1984;35:200–215. [PubMed] [Google Scholar]
- 24.Jambhekar SS, Breen P. Cyclodextrins in pharmaceutical formulations I: Structure and physicochemical properties, formation of complexes, and types of complex. Drug Discov Today. 2016;21:356–362. doi: 10.1016/j.drudis.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 25.Zidovetzki R, Levitan I. Use of cyclodextrins to manipulate plasma membrane cholesterol content: Evidence, misconceptions and control strategies. Biochim Biophys Acta - Biomembr. 2007;1768:1311–1324. doi: 10.1016/j.bbamem.2007.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vahabikashi A, et al. Probe sensitivity to cortical versus intracellular cytoskeletal network stiffness. Biophys J. 2019;116:518–529. doi: 10.1016/j.bpj.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo M, et al. Cell volume change through water efflux impacts cell stiffness and stem cell fate. Proc Natl Acad Sci U S A. 2017;114:E8618–E8627. doi: 10.1073/pnas.1705179114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otto O, et al. Real-time deformability cytometry: On-the-fly cell mechanical phenotyping. Nat Methods. 2015;12:199–202. doi: 10.1038/nmeth.3281. [DOI] [PubMed] [Google Scholar]
- 29.Azadi S, Tafazzoli-Shadpour M, Soleimani M, Warkiani ME. Modulating cancer cell mechanics and actin cytoskeleton structure by chemical and mechanical stimulations. J Biomed Mater Res Part A. 2019;107A:1569–1581. doi: 10.1002/jbm.a.36670. [DOI] [PubMed] [Google Scholar]
- 30.Lekka M. Discrimination between normal and cancerous cells using AFM. Bionanoscience. 2016;6:65–80. doi: 10.1007/s12668-016-0191-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Butcher DT, Alliston T, Weaver VM. A tense situation: Forcing tumour progression. Nat Rev Cancer. 2009;9:108–122. doi: 10.1038/nrc2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu M, et al. ACAT1 regulates the dynamics of free cholesterols in plasma membrane which leads to the APP-α-processing alteration. Acta Biochim Biophys Sin. 2015;47:951–959. doi: 10.1093/abbs/gmv101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang W, et al. Potentiating the antitumour response of CD8+ T cells by modulating cholesterol metabolism. Nature. 2016;531:651–655. doi: 10.1038/nature17412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barry M, Bleackley RC. Cytotoxic T lymphocytes: All roads lead to death. Nat Rev Immunol. 2002;2:401–409. doi: 10.1038/nri819. [DOI] [PubMed] [Google Scholar]
- 35.Golstein P, Griffiths GM. An early history of T cell-mediated cytotoxicity. Nat Rev Immunol. 2018;18:527–535. doi: 10.1038/s41577-018-0009-3. [DOI] [PubMed] [Google Scholar]
- 36.Chow CW, Rincón M, Davis RJ. Requirement for transcription factor NFAT in interleukin-2 expression. Mol Cell Biol. 1999;19:2300–2307. doi: 10.1128/mcb.19.3.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oeckinghaus A, Ghosh S. The NF-κB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol. 2009;1:a000034. doi: 10.1101/cshperspect.a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Style RW, et al. Traction force microscopy in physics and biology. Soft Matter. 2014;10:4047–4055. doi: 10.1039/c4sm00264d. [DOI] [PubMed] [Google Scholar]
- 39.Weder G, et al. Increased plasticity of the stiffness of melanoma cells correlates with their acquisition of metastatic properties. Nanomed-Nanotechnol Biol Med. 2014;10:141–148. doi: 10.1016/j.nano.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 40.Luo Q, Kuang D, Zhang B, Song G. Cell stiffness determined by atomic force microscopy and its correlation with cell motility. Biochim Biophys Acta - Gen Subj. 2016;1860:1953–1960. doi: 10.1016/j.bbagen.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y, et al. Cell softness prevents cytolytic T-cell killing of tumor-repopulating cells. Cancer Res. 2021;81:476–488. doi: 10.1158/0008-5472.CAN-20-2569. [DOI] [PubMed] [Google Scholar]
- 42.Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J. Actin dynamics, architecture, and mechanics in cell motility. Physiol Rev. 2014;94:235–263. doi: 10.1152/physrev.00018.2013. [DOI] [PubMed] [Google Scholar]
- 43.Bashour KT, et al. CD28 and CD3 have complementary roles in T-cell traction forces. Proc Natl Acad Sci U S A. 2014;111:2241–2246. doi: 10.1073/pnas.1315606111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vaahtomeri K, et al. Locally triggered release of the chemokine CCL21 promotes dendritic cell transmigration across lymphatic endothelia. Cell Rep. 2017;19:902–909. doi: 10.1016/j.celrep.2017.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leithner A, et al. Dendritic cell actin dynamics control contact duration and priming efficiency at the immunological synapse. J Cell Biol. 2021;220:e202006081. doi: 10.1083/jcb.202006081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stack T, et al. Targeted delivery of cell softening micelles to Schlemm’s canal endothelial cells for treatment of glaucoma. Small. 2020;16:2004205. doi: 10.1002/smll.202004205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vorselen D, et al. Microparticle traction force microscopy reveals subcellular force exertion patterns in immune cell–target interactions. Nat Commun. 2020;11:20. doi: 10.1038/s41467-019-13804-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blumenthal D, Chandra V, Avery L, Burkhardt JK. Mouse T cell priming is enhanced by maturation-dependent stiffening of the dendritic cell cortex. Elife. 2020;9:e55995. doi: 10.7554/eLife.55995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tello-Lafoz M, et al. Cytotoxic lymphocytes target characteristic biophysical vulnerabilities in cancer. Immunity. 2021;54:1037–1054.:e7. doi: 10.1016/j.immuni.2021.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Janmey PA, McCulloch CA. Cell mechanics: Integrating cell responses to mechanical stimuli. Annu Rev Biomed Eng. 2007;9:1–34. doi: 10.1146/annurev.bioeng.9.060906.151927. [DOI] [PubMed] [Google Scholar]
- 51.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 52.Chen DS, Mellman I. Oncology meets immunology: The cancer-immunity cycle. Immunity. 2013;39:110. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 53.Huse M. Mechanical forces in the immune system. Nat Rev Immunol. 2017;17:679–690. doi: 10.1038/nri.2017.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu C, Chen W, Lou J, Rittase W, Li K. Mechanosensing through immunoreceptors. Nat Immunol. 2019;20:1269–1278. doi: 10.1038/s41590-019-0491-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jain N, Moeller J, Vogel V. Mechanobiology of macrophages: How physical factors coregulate macrophage plasticity and phagocytosis. Annu Rev Biomed Eng. 2019;21:267–297. doi: 10.1146/annurev-bioeng-062117-121224. [DOI] [PubMed] [Google Scholar]
- 56.Lei K, Kurum A, Tang L. Mechanical immunoengineering of T cells for therapeutic applications. Acc Chem Res. 2020;53:2777–2790. doi: 10.1021/acs.accounts.0c00486. [DOI] [PubMed] [Google Scholar]
- 57.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 58.Rosen K, Shi W, Calabretta B, Filmus J. Cell detachment triggers p38 mitogen-activated protein kinase-dependent overexpression of Fas ligand: A novel mechanism of anoikis of intestinal epithelial cells. J Biol Chem. 2002;277:46123–46130. doi: 10.1074/jbc.M207883200. [DOI] [PubMed] [Google Scholar]
- 59.Hawley TS, Hawley RG. Flow Cytometry Protocols. Humana Press; New York: 2018. [Google Scholar]
- 60.Tse JR, Engler AJ. Preparation of hydrogel substrates with tunable mechanical properties. Curr Protoc Cell Biol. 2010;47:1–16. doi: 10.1002/0471143030.cb1016s47. [DOI] [PubMed] [Google Scholar]
- 61.Tseng Q, et al. Spatial organization of the extracellular matrix regulates cell-cell junction positioning. Proc Natl Acad Sci U S A. 2012;109:1506–1511. doi: 10.1073/pnas.1106377109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the results in this study are available within the paper and its Supplementary Information. Source data for the figures are provided with this paper.
Source code for the custom ImageJ macro for F-actin analysis, cellular-force calculation and deformability-cytometry analysis, and custom MATLAB scripts for optical-tweezer data collection and data post-processing, are available from the corresponding author on reasonable request. Shape-out (version 2.7.4) for deformability cytometry analysis is available at https://github.com/ZELLMECHANIK-DRESDEN/ShapeOut2.