Abstract
Age at first sexual intercourse (AFS) and age at first birth (AFB) have implications for health and evolutionary fitness. In this genome-wide association study (AFS, N=387,338; AFB, N=542,901), we identify 371 SNPs, 11 sex-specific, with a 5-6% polygenic score (PGS) prediction. Heritability of AFB shifted from 9% [CI=4-14] for women born in 1940 to 22% [CI=19-25] in 1965. Signals are driven by the genetics of reproductive biology and externalising behaviour, with key genes related to follicle stimulating hormone (FSHB), implantation (ESR1), infertility, and spermatid differentiation. Our findings suggest that Polycystic Ovarian Syndrome may lead to later AFB, linking with infertility. Late AFB is associated with parental longevity, and reduced incidence of Type 2 Diabetes (T2D) and Cardiovascular disease (CAD). Higher childhood socioeconomic circumstances and those in the highest PGS decile (90%+) experience markedly later reproductive onset. Results are relevant for improving teenage and late-life health, for understanding longevity, and guiding experimentation into mechanisms of infertility.
Subject terms: Genome-wide association studies, Behavioural genetics, Demography, Fertility
The timing of onset of human reproductive behaviour – age at first sexual intercourse (AFS) and age at first birth (AFB) – has implications for reproductive health, adolescent development and evolutionary fitness. Age at first sexual intercourse has occurred increasingly earlier, by the age of 16 for one-third of contemporary UK teenagers.1 Early reproductive onset is associated with teenage pregnancy2 but also adverse health outcomes such as cervical cancer, depression, sexually transmitted diseases2 and substance use disorders.3,4 In contrast to earlier sexual debut, we have witnessed progressively later ages at first birth for women, now reaching an average of 30 years in many modern societies, even later for men (Supplementary Figure 3).5 Late reproductive behaviour is associated with lower fecundity and subfertility6 and infertility traits such as endometriosis and early menopause,7,8 with over 20% of women born after 1970 in many modern countries now remaining childless.9 Earlier ages of sexual debut and later ages at first birth has marked the decoupling of reproduction from sexual behaviour in many contemporary societies, with implications for sexual, reproductive and later-life health (Supplementary Figure 2).
Since reproductive behaviour is shaped by biology and environment, a multidisciplinary approach is required to understand the common genetic aetiology and how it relates to health, reproductive biology, environment and externalising behaviour.10 Since the onset of reproductive behaviour generally occurs in adolescence to early adulthood, it is often linked to externalising behaviour such as self-control and psychiatric (e.g., ADHD) and substance use disorders (e.g., smoking, alcohol use), often moderated by the environment (e.g., childhood socioeconomic conditions).10 Furthermore, it may be that individuals inherit a common genetic liability for a spectrum of interlinked complex traits related to reproduction, health and longevity. There has been limited attention to understanding how these genetic effects are stratified by sex or across different socioeconomic and historical contexts.
In a previous GWAS of AFS (n=125,667)11 and AFB (n=343,072),8 we identified 38 and 10 novel independently-associated single-nucleotide polymorphisms (SNPs), respectively. The current study comprises a markedly expanded sample size for AFS (N=387,338) and AFB (N=542,901), uncovering 371 autosomal or X chromosomal SNPs, some of which are sex-specific, with 99 candidate genes expressed at the protein level in the brain, glands and reproductive organs. The multiple methods applied in this study (Supplementary Figure 1) reveal underlying genetic drivers, common genetic liabilities, heterogeneity by childhood socioeconomic status and historical period and further evidence of the relationship of later reproductive onset with fewer later-life metabolic life diseases and increased longevity.
Results
Changes in reproductive behaviour and heritability over time
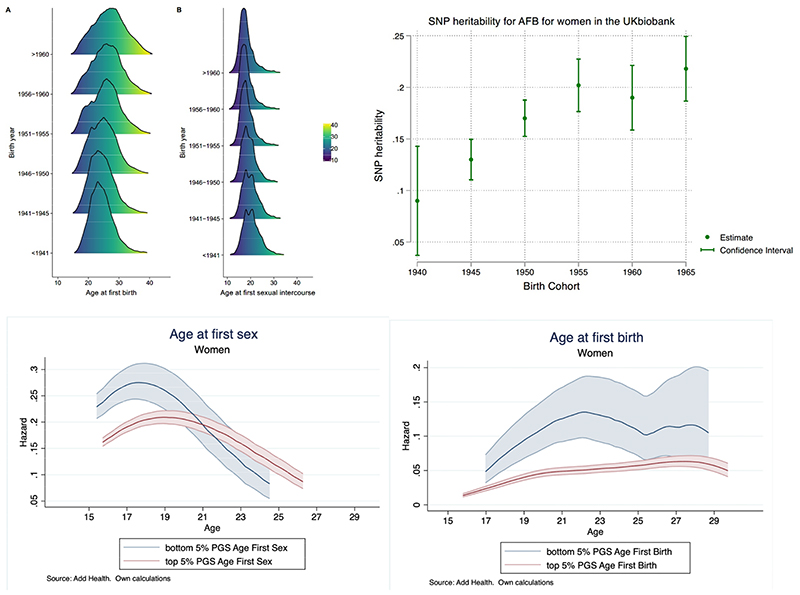
We first examine phenotypic changes in human reproductive behaviour and heritability over time. Descriptive analyses using the UK Biobank illustrate shifts in the mean AFS and AFB, changes in the shape of the distribution by birth cohort, and a bi-modal distribution of AFS in earlier cohorts (Figure 1A, Supplementary Figure 3). Whereas AFB was often in the early 20s for older birth cohorts, this distribution has spread and shifted to older ages over time, with a marked drop in Pearson’s correlation between AFS and AFB from those born <1941 (0.60) to those born >1960 (0.31) (Supplementary Figure 2). Using GREML,12,13 we found a steady increase in SNP-heritability by birth cohort for AFB for women from 9% [CI = 4-14] for those born in 1940, climbing to around 22% [19-25] for the latest cohorts born in 1965. For AFS, heritability ranges between 12 [7-18] for women born around 1940 and 23 [17-28] % for men born around 1940 with a trend for women similar to AFB and a suggestive U-shaped trend for men (Figure 1B for women, Supplementary Figure 1B for men).
Figure 1. Change in age at first sex (AFS) and age at first birth (AFB) over time, heritability by birth cohort and polygenic score (PGS) prediction.
(A) AFB (A) and AFS (B) by birth cohort, UK biobank shows a shift in the ages over time. (B) Increased SNP heritability for women over time by birth cohort, UK Biobank. (C) Nelson-Aalen hazard estimates of AFS by age for women comparing the upper and lower 5% of the PGS for AFS. (D) Nelson-Aalen hazard estimates of AFB by age for women comparing the upper and lower 5% of the PGS for AFB.
Meta-analysis GWAS Human Reproductive Behaviour
We conducted a meta-analysis of GWAS results from 36 cohorts for AFS and AFB in individuals of European ancestry (defined by genetic principal components). We imputed to the 1000 Genomes Project reference panel in a pooled sample and then stratified the analysis by sex (Supplementary Tables 1-8). In total, we discovered 371 associated SNPs. The GWAS of AFS identified 282 (272 pooled of which 4 on the X chromosome; 2 women; 8 men) SNPs at genome-wide significance (p<5 x 10-8, Supplementary Figure 5A-C; Supplementary Table 10). The GWAS of AFB identified 89 (84 pooled of which 4 on the X chromosome; 1 women) independent SNPs at genome-wide significance (p<5 x 10-8, Supplementary Figure 6A-C; Supplementary Table 9). The distribution of genome-wide test statistics for AFS and AFB showed significant inflation (λGC = 1.84 and 1.47, respectively), however LD score regression showed that this could be attributed almost entirely to polygenicity rather than to population substructure (LD intercept AFS 1.07 (SE = 0.01); AFB 1.03 (SE = 0.01, Supplementary Information Section 6). The LD Score intercept test confirmed that only a very small percentage (5.5%) of the observed inflation in the mean χ2 statistic was due to population stratification or other confounders, rather than to a polygenic signal.
Polygenic score prediction
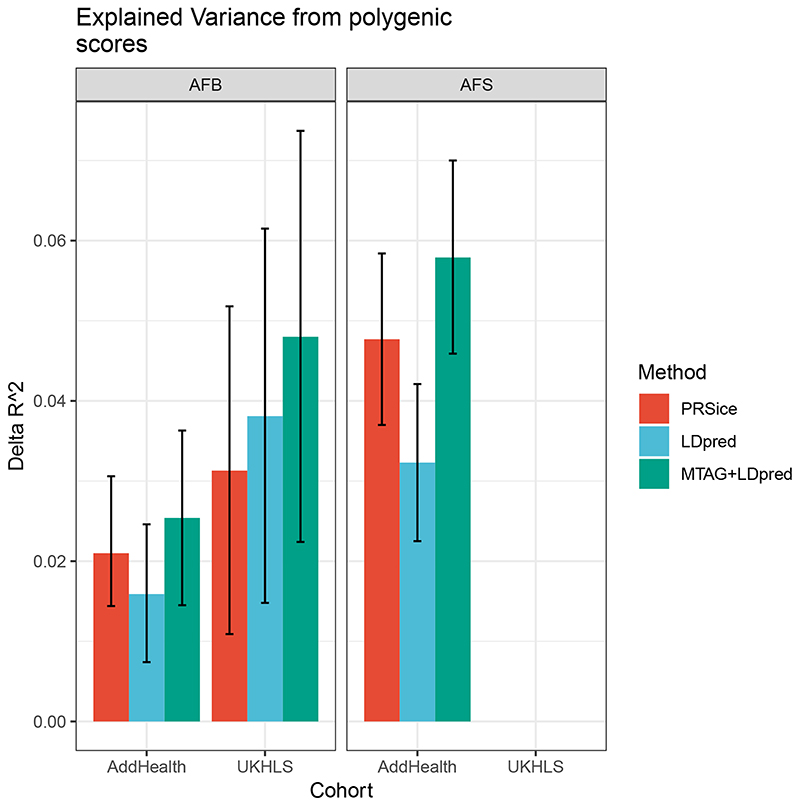
We then calculated polygenic scores (PGSs) using three different specifications (Figure 2, Supplementary Information Section 5). To validate the performance of the PGSs, we performed out-of-sample prediction in Add Health (a survey of adolescence to young adulthood in the US) and UKHLS (a representative survey of adults in the UK) cohorts using ordinary least-squares (OLS) regression models and report the R2 as a measure of goodness-of-fit of the model (Figure 2). PGSs including all SNPs explain up to 5.8% of the variance for AFS and 4.8% for AFB. The difference between the out of sample prediction in the UKHLS and Add Health is related to heterogeneity between the initial GWAS sample, including a large UK older population from the UK Biobank, which is more comparable with the UKHLS population. Add Health participants also have higher levels of right-censoring (i.e., not yet experienced a birth). Previous research has demonstrated that meta-analyses of complex behavioural traits using populations from diverse national contexts and birth cohorts can be influenced by this hidden heritability.13 A 1 standard deviation (SD) change in the AFS/AFB PGS is associated with a 7.3 and 6.3 month delay in AFS and AFB, respectively. We then ran survival models to account for right-censoring, which occurs when an individual does not experience the event of first sex or birth by the time of the interview.14 Using Add Health data, we estimated nonparametric hazard functions and then compared individuals at the top and bottom 5% of the PGS (see Figure 1C AFS, Figure 1D AFB for women, Supplementary Figure 8-9 for men). Those in the top 5% PGS for AFS (i.e., genetic association with later AFS) are less likely to have their sexual debut before age 19. AFS PGSs appear more relevant in explaining women’s AFS in comparison to men. Those in the top 5% PGS for AFB (i.e., genetic association with later AFB) postpone AFB across all ages until approximately age 27, with similar curves for both sexes.
Figure 2. Variance explained from polygenic scores for age at first birth (AFB) and age at first sex (AFS) using different methods in out-of-sample cohorts.
Shows the out-of-sample prediction scores for AFB and AFS performed in two cohorts: The National Longitudinal Study of Adolescence to Adult Health (Add Health) and the UK Household Longitudinal Study (UKLS). The bars represent the delta (Δ) R2, also indicating confidence intervals. R2 measures of the proportion of variance of the phenotype that is explained by the model and Δ R2 shows the change in R2 values when comparing a model without the PGS to one that includes the PGS. The different methods of PRSice, LDPred and MTAG+LDPred are described in the Supplementary Information, Section 5.
We next examined whether these genetic effects were environmentally moderated by childhood socioeconomic status. Disadvantaged socioeconomic status is highly related to early sexual behaviour and teenage pregnancy.15 To explore the impact of environmentally moderated parental genetic effects on our PGSs, we examined PGS prediction across low (0-10%), medium (50-60%) and high (90-100%) PGS percentiles by parents’ education (college versus no college) as a proxy for childhood socioeconomic status. Indeed, those in the highest decile of the PGS (90-100%) for later AFB have a higher AFB, particularly past age 27, which is accentuated for those with highly educated parents (Supplementary Figure 10A). Being in the highest PGS decile for AFS is associated with later sexual intercourse (difference between highest and lowest decile is 2.08 years), especially for those from highest socioeconomic childhood households (2.39 years difference among high SES, compared to 1.62 years for low SES families) (Supplementary Figure 10B).
Genetic correlations
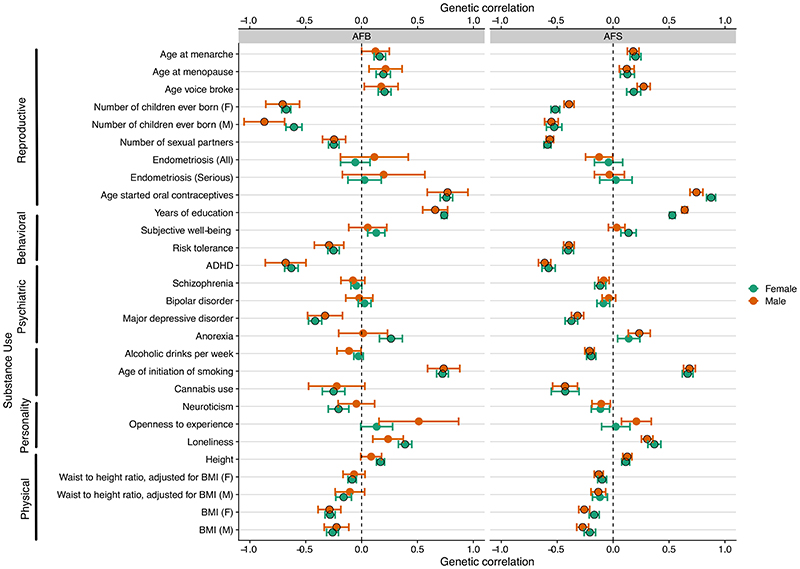
To test the relationships of AFS and AFB with related phenotypes, we calculated genetic correlations using LD score regression16 (Figure 3, Supplementary Figure 11, 13, Supplementary Table 11). Given previous evidence,8 we examined 25 traits by sex from six relevant categories including: reproductive (e.g., age at menarche, number of sexual partners), infertility (e.g., endometriosis and severe endometriosis), behavioural (e.g., years of education, risk tolerance), psychiatric disorders (e.g., ADHD, schizophrenia), substance use (e.g., age of initiation of smoking, cannabis use), personality (e.g., openness to experience) and anthropometric (e.g., BMI, height). Logically, the strongest genetic correlations were observed for reproductive traits, particularly for number of children ever born. We remain cautious regarding the estimates of endometriosis considering the smaller sample size and larger confidence intervals (CIs) of our estimates. In our Mendelian Randomization analysis, discussed shortly, we explore the relationship with infertility in more detail. Behavioural traits also show strong genetic correlations, particularly with AFB and educational attainment in women (0.74, ±0.01), compared to AFS (0.53, ±0.01). There was also a negative genetic correlation between adult risk tolerance and AFS/AFB (AFS ~−0.40; AFB ~−0.25); i.e., those with a low genetic association with risk tolerance corresponds to a genetic association with later reproductive behaviour. Amongst psychiatric traits, often related to externalising behaviour, the strongest correlation was with ADHD (AFS females −0.58, ±0.03, males −0.61, ±0.03; AFB females −0.63, ±0.03; males −0.68, ±0.09) and Major Depressive Disorder (MDD) (AFS females −0.37, ±0.03, males −0.32, ±0.03; AFB females, −0.42, ±0.03; AFB males, −0.33, ±0.08). We also observed strong genetic correlations with age at onset of smoking (AFS ~ 0.68, ±0.03; AFB ~0.74, ±0.03), a trait that provides a unique window into adolescent substance use and externalising behaviour around the same time of early reproductive behaviour. Genetic factors associated with early smoking, early sexual debut and teenage pregnancy are thus – to some extent – shared. As shown in Figure 3, there are few sex differences in these correlations, with the exception of small variations in number of children, anorexia and openness to experience.
Figure 3. Genetic correlations of age at first birth (A) and age at first sex (B) with a selection of related traits.
The horizontal bars represent 95% confidence intervals. If the trait was initially assessed separately for males and females, this is indicated on the left in brackets with F referring to females and M to males. The black circles represent the Bonferonni significant correlations, but the magnitude of the effect (rg, the genetic correlation) is the most informative. Definitions and sources of all traits can be found in the Supplementary Information, Section 7.1 with full results shown in Supplementary Table S11.
Aetiology and causality
To explore aetiology and causality we employed GenomicSEM, Exploratory Factor Analysis and Bi-Directional Mendelian Randomization. To understand the relationships underlying these genetic correlations, we first used GenomicSEM.17 GenomicSEM uses structural equation modelling to decompose the genetic covariance matrix, calculated using multivariate LD score regression, of a set of traits. Parameters are estimated by minimising the difference between the observed genetic covariance matrix and the covariance matrix derived from the model (Supplementary Information Section 8). We fit a series of genetic regression models in which AFB (or AFS) was regressed on both years of education and one other possible mediating trait, such as openness, cognitive performance, ADHD and age of initiation of smoking (Supplementary Figures 12A-B; Supplementary Tables 12A-L). In other words, we wanted to test whether the strong genetic correlation of AFS/AFB with education was the result of another mediating trait such as personality, ADHD or substance use. We found that the genetic correlation of years of education with AFB and AFS was independent of factors like risk tolerance, substance use, and psychiatric disorders. This suggests that the genetic correlation between years of education and AFB is largely a product of a strong bidirectional relationship between these traits, rather than being both downstream of a common identified cause. The exception was age at initiation of smoking – as noted previously, a window into risky adolescent behaviour – which partially mediated the relationship of AFB and AFS with years of education.
Exploratory factor analysis (EFA) was then used to examine whether the genetic signal of the onset of reproductive behaviour originated from two genetically distinguishable subclusters of reproductive biology versus externalising behaviour. Using a two-factor EFA model to fit the genetic covariance matrix AFS and AFB with these two additional traits, we found that the entire model accounted for 47% of the overall variance, with 22% attributed to risk tolerance and 4% to age at menarche. In a more robust analysis we fit a Genomic SEM for AFB in women and regressed several genetic measures of reproductive biology (age at menarche, age at menopause) and a latent factor representing a common genetic tendency for externalising behaviour (age at initiation of smoking, age first used oral contraception, ADHD) (Supplementary Figure 14). These genetic factors predicted 88% of the variance, with the majority of variance significantly predicted by externalising factors (0.90,±0.02), followed by age at menopause (0.20, ±0.04) and age at menarche (0.16,±0.03). We note that selection bias, induced by the fact that AFB can only be measured among individuals with at least one live birth, may have potentially inflated this estimate.
Given the strong genetic correlations between the phenotypes discussed above, we used Mendelian Randomization (MR)-based analyses18 to explore causality and assess the direction of effect between AFB, AFS and years of education19 as well as risk taking (measured in adulthood)4 and age at smoking initiation20 (Supplementary Table 13A; Section 9). For the majority of pairs of phenotypes we found strong evidence of bi-directionality, which was also seen after applying Steiger fitting. The relationship between AFB and years in education appeared to be the explanatory factor that linked AFB to the two risk taking phenotypes. This was not the case, however, for AFS where the analysis suggests that age at initiation of smoking (and the environment and processes that lead to this) are upstream of the start of AFS. In that case, the relationship was significant when assessed as age at smoking to AFS but not the other way round. Of note, associations were much stronger for age at smoking initiation than for risk-taking behaviour assessed in adulthood, suggesting that the timing of this behaviour is key.
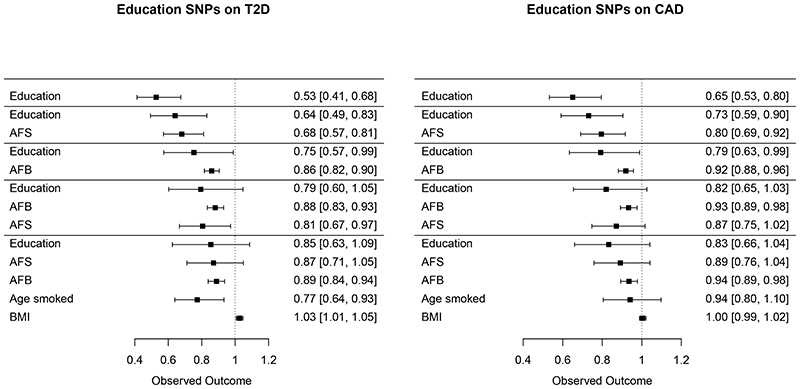
A second set of MR analyses examined whether AFS and AFB PGSs have effects on type 2 diabetes (T2D)21 and coronary artery disease (CAD).22 Acknowledging the substantial overlap between the most significant signals for AFB, AFS and education related phenotypes, we used adjusted models to estimate effects independent of years of education (Figure 4, Supplementary Table 13B, Supplementary Figure 16). Given the extent of the overlap, we were also interested to investigate if AFB or AFS might attenuate any association particularly with education. T2D and CAD were chosen since they are two common major diseases, with broadly defined behavioural risk factors. Findings show that the association with years of education and later life diseases are substantially attenuated by the effects of AFB. We also found a strong association of the BMI weights with the AFS SNPs but note that even when BMI is included the model, the level of attenuation of the educational attainment results by AFB remains striking. This concurs with a large body of research that has established a biological association with the timing of AFB and metabolic diseases including early AFB linked to high blood pressure,23 obesity24 and diabetes.25 Reproductive timing thus appears to capture a latent variable that detects these metabolic effects and is a marker of a broader social trajectory that serves as a more powerful predictor of later life disease than years of education alone. This also suggests that many of the associations with diseases that have been previously ascribed to years of education, may result from this more broadly defined socio-behavioural trajectory captured by AFB.
Figure 4. Mendelian randomisation of years of education on type 2 diabetes (T2D) and coronary artery disease (CAD) adjusted for age at first birth (AFB) and age at first sex (AFS).
Shows the coefficients and confidence intervals (CIs) of Mendelian Randomisation analyses to examine the effects of SNPs associated with years of education on Type 2 Diabetes (T2D) and Coronary Artery Disease (CAD), after adjustment by AFB and AFS and other covariates. The association with years of education and T2D and CAD are substantially attenuated by the effects of AFB. Even when BMI is included in the model, the level of attenuation of educational attainment by AFB remains striking.
Finally, since we were also interested in infertility-related phenotypes, bidirectional MR was performed with AFS and AFB with PCOS (polycystic ovarian syndrome) and given the nature of the disease, on women only.26 Our findings (Supplementary Table 13C) suggest that PCOS leads to later AFB. We find no effect of PCOS on AFS or of either AFS or AFB to PCOS, suggesting that the causal link is infertility-related with PCOS contributing to later AFB.
Cox models of a polygenic score for AFB on longevity
The disposable soma theory of aging hypothesises an evolutionary-trade off between investments in somatic maintenance – such as remaining in education – that in turn reduces resources available for reproduction. To test trade-offs between reproductive behaviour and senescence as argued in the ageing and longevity literature,27 we conducted Cox proportional hazard models analyses to test whether our AFB PGS was associated with (parental) longevity (Supplementary Table 14). We first estimated a baseline Cox proportional hazard model of our AFB PGS on parental longevity and then included the PGS for educational attainment and risk covariates followed by a final model including number of siblings as a proxy for parental fertility. We found that a genetically predicted 1 SD increase in the PGS for AFB is associated with a 2-4% reduction in parental mortality at any age, suggesting that there is likely a trade-off between the timing of reproduction and longevity.
Gene prioritization
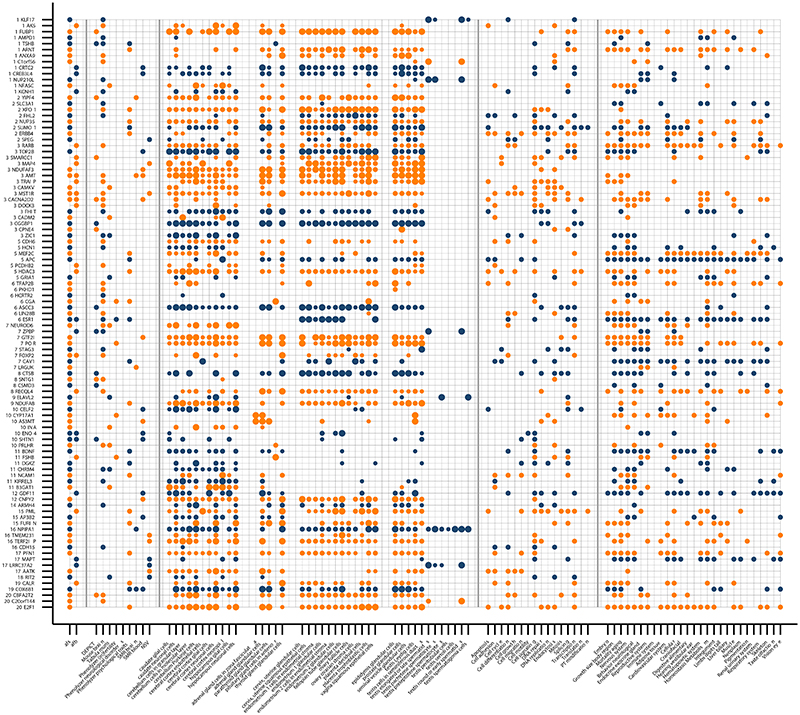
To understand the biology represented by the variants associated with AFS and/or AFB, we performed a gene prioritization analysis that connected variants to genes and prioritized candidate genes based on likely involvement in reproductive biology or psychiatric traits. To this end, we used predicted gene function,28 single-cell RNA sequencing data in mice,29,30 literature mining,31 in silico sequencing,32 and Summary-data based Mendelian Randomization(SMR)33 using eQTL data from brain and blood.34 Integrating results across all approaches resulted in the prioritization of 386 unique genes; 314 genes in 159 loci for AFS and 106 genes in 42 loci for AFB (Supplementary Tables 15A-19C). Of these, 99 were expressed at the protein level in cell types of brain, glands, and/or (fe)male reproductive organs (Figure 5).35 Gene prioritization in sex-specific loci resulted in the prioritization of 11 genes for AFB in women, one gene for AFS in women and 23 genes for AFS in men. Of these, 12 genes at three loci were expressed at the protein level in relevant tissues (Supplementary Figure 17).
Figure 5. Gene prioritization of age at first sex (AFS) and age at first birth (AFB).
Information for 99 genes prioritized in loci identified by GWAS for age at first sex and/or age at first birth that are located within 1 million bp of lead SNPs and are expressed at the protein level in brain, glands and/or reproductive organs. Transitions in colour from blue to orange highlight whether the gene in the next row is still within the same locus or not. Numbers before the genes show the chr. Panels are separated by vertical grey lines. The first panel (left) indicates if the locus was identified as being associated at genome-wide significance with age at first sex (AFS) and/or age at first birth (AFB). The second panel shows which bioinformatic approaches highlighted the gene as a candidate. The third panel shows – from left to right - the cell types in brain, glands, female reproductive organs, and male reproductive organs in which the genes are expressed at a low, moderate or high level (small, medium and large circles), based on data from the Human Protein Atlas. The fourth panel shows gene functions as extracted from Entrez, Uniprot and GeneCards. The fifth panel indicates which phenotypes were observed in mutant mice, as reported by the Mouse Genome Informatics (MGI) database.
Genes that play a role in follicle stimulating hormone (CGA 36), oocyte development (KLF17 37), and implantation and placental growth (ESR1, SUMO1 38, ARNT,39 CAV1,40 E2F1 41) were prioritized for AFS in data from men and women combined, while FSHB 42 and ESR1 were (also) prioritized for AFB. Other genes prioritized in loci identified in the pooled meta-analyses were expressed at the protein level in (developing) sperm – highlighting a role for spermatid differentiation (KLF17 43) for AFS – as well as for sperm morphogenesis and binding between acrosome-reacted sperm and the zona pellucida (ZPBP44) for AFB. The meta-analysis in data from only women yielded genes related to endometriosis (CCR1)45 and spontaneous abortion (CXCR6) for AFB (Supplementary Figure 19).46 Taken together, these results suggest that intrinsic biological processes related to fertility are also related to the onset of sexual behaviour in men and women. Interestingly, NUP210L – prioritized for AFS and highly expressed in developing and mature sperm35 – is normally testis-specific, but was recently shown to be expressed in prefrontal cortex neurons of G allele carriers in rs114697636 (MAF 3%, D’ 0.90 with AFS lead SNP rs113142203), attributed to allele-specific activation through improved binding affinity for testis receptor 2.47 Methylation of, and variants near NUP210L have been associated with psychologic development disorders, intelligence, and mathematical ability,48 illustrating how a testis-specific gene could be linked to brain function in some individuals. We note, however, that the fact that some prioritized proteins are expressed in some relevant tissues does not provide clear evidence supporting a causal role for the prioritized genes.
Several genes prioritized in AFS-associated loci in data from men and women combined have previously been implicated in risk seeking behaviour, sociability and anxiety (GTF2I,49 TOP2B,50 E2F1,51 NCAM1,52 NFASC,53 MEF2C54). In the sex-specific meta-analysis for AFS, a role for externalising behaviour was supported through ERBB4 in women; and through SLC44A1 and NR1H3 in men. ERBB4 has previously been linked to fear, anxiety,55 schizophrenia,56 and polycystic ovary syndrome (PCOS);57 SLC44AI encodes a choline transporter that plays a key role in cerebral inhibition related to substance use and depressive disorders58; and NR1H3 has been implicated in major depressive disorder (MDD).59 These genes provide concrete examples of how genetic variants associated with externalising behaviour may also associate with the initiation of reproductive behaviour.
The gene prioritization results partly mirror and compliment the rigorous post-GWAS in silico association analyses we performed for loci identified for AFS and AFB. However, experimental validation is required before firm conclusions can be drawn about the involvement of, and mechanisms through which prioritized candidate genes influence AFS and AFB. See more information on protein-protein interaction hubs (Supplementary Figure 17), as well as on genes highlighted by literature mining (Supplementary Information Section 11).31
Discussion
In this study, we presented the results of a GWAS of the onset of human reproductive behaviour of age at first sex (AFS) (N=387,338) and age at first birth (AFB) (N=542,901). We identified 371 signals harbouring at least 386 prioritized candidate genes, using 1000G imputed genotype data and an X-Chromosome analysis, which allowed us to detect considerably more signals than ever before (Figure 6). In comparison, a recent GWAS for type 2 diabetes,60 for instance, detected 243 loci. Similar to previous work, we showed that the total SNP heritability accounted for 10-22% of phenotypic variance and varied by birth cohort.13,61 The incremental R2 of our PGSs based on significantly associated SNPs is around 5-6%, similar to direct effect relationships observed for commonly used demographic and social variables (e.g., years of education, age at marriage), classically used to explain the timing of human reproductive behaviour by social scientists. Comparatively, a PGS of 5-6% is in the range observed for other complex traits, like BMI (5.8%)62 and schizophrenia (8.4%).63 The number of signals also opened up opportunities for functional follow-up analyses which suggested a role for spermatid differentiation and oocyte development. The analyses of the correlation and underlying aetiology of these traits revealed a common genetic basis of both AFS and AFB with externalising behaviour and substance use and links to infertility.
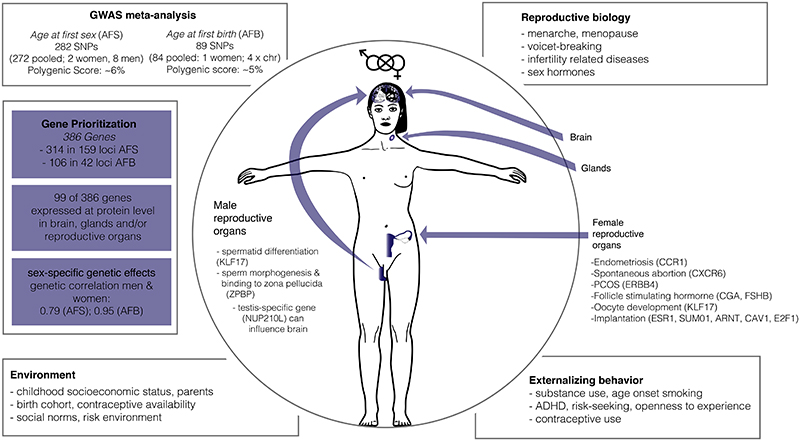
Figure 6. Summary genome-wide association study of timing of onset of reproductive behaviour: age at first sex (AFS) and age at first birth (AFB).
Provides a summary of the GWAS meta-analysis results and highlights that the timing of onset of reproductive behaviour is shaped by the combined effects of reproductive biology and externalizing behaviour but also exogenous environmental factors. Sex-specific genetic correlations are higher for AFB than AFS, with gene prioritization revealing some genes prioritized for men and women as well as sex-specific findings. Of the prioritized genes, 99 were expressed at the protein level in cell types of brain, glands and/or (fe)male reproductive organs. The fact that some prioritised proteins are expressed in some relevant tissues does not provide clear evidence supporting a causal role. Figure created by authors.
Finally, we showed that AFB is an important predictor for late age at onset of disease and longevity, and that it substantially attenuates the effect of years in education. We note that there are some situations where we have a significant Eggers intercept in the MR analysis, including some for the bidirectional data. Here there is some heterogeneity in the data (AFB to education, and AFS to risk), where there are likely important pleotropic effects. However, this does not impinge on our central finding that there is widespread bi-directionality. Since we also find a significant intercept for AFB to CAD, and since in the adjusted model there are not significant effects, we are confident that we are not at risk of a false positive.
Although we opened many new avenues for research, the present GWAS still faces certain limitations. First, the sample sizes for men were appreciably smaller than for women since reproductive and fertility data is routinely collected less often from men. Yet to understand the correlates and causes of infertility in men this information needs to be taken into consideration in future data collection. The paucity of sex differences in the genetic correlations we observe between AFB, AFS, and a variety of related traits, including sex-specific traits such as age at menarche, suggests that the relevant processes overlap between the sexes. Initial within-family analyses showed that our discovery GWAS may actually overestimate causal effects; genotypes associated with later onset of reproductive behaviour genotypes are also associated with parental reproductive genotypes, likely leading to a social environment that affects reproductive and other behaviours. Collection and analysis of family data is clearly a future area of research for reproductive and related complex behaviour. The lack of accessibility of publically available summary statistics from some published research, meant that we were unable to examine the relationship with other traits, particularly with infertility related traits (e.g., larger studies of endometriosis). Future data collection could benefit from focussing on externalising and behavioural disinhibition markers that appear to be highly related to self-control, which has implications for disease prevention and behavioural interventions into lifestyle factors related to obesity, Type 2 diabetes or substance use disorders. A final glaring limitation is our focus on European-ancestry individuals in Western countries. Whilst common in this area of research,64 extension to other ancestries and geographical contexts is required in the future. This is particularly relevant in the context of parental gene-environment interactions, which may be specific to the social and environmental makeup of the sample.
Our detailed correlation, GenomicSEM and MR analyses also provided a deeper understanding of the underlying aetiology of related traits and pleiotropy and the associations between human reproductive behaviour and disease risk. We anticipate that our results will address important interventions in infertility, teenage sexual and mental health, as well as for functional follow-up experiments that will likely yield targets that can be translated in efficient medication to improve fertility (e.g., in IVF) but also for interventions on reproductive health related to earlier sexual debut and teenage pregnancy.
Methods
This article has Supplementary Information with details about data and methods and additional detailed analyses.
Samples
For Age at First Sexual intercourse (AFS), we included 397,338 pooled individuals (n=182,791 males; n=214,547 females) from the UK Biobank. For Age at First Birth (AFB), we included 542,901 individuals (n=124,088 males; n=418,758 females) from 36 studies. We performed a GWAS separately restricted to European ancestry individuals that passed quality control. European ancestry was chosen in this discovery study due to the availability of samples64 and for no biological or substantive reason. We acknowledge that social science research has found large differences in the earlier initiation of AFS and AFB by lower socioeconomic status, which often coincides with societal inequality65,66 and the socially (not biologically) constructed categories of race and ethnicity. Socioeconomic differences are examined in this article, but results are only applicable to European Ancestry groups, with a need for further cross-ancestry discovery research.
The Human Reproductive Behaviour Consortium. This consortium is a collaboration studying the GWAS of human reproductive behaviour including age at first sex and birth, number of children ever born, childlessness and related traits. In some cases we used summary statistics from our first GWAS of AFB and NEB8 on discovery cohorts (see Supp Note Tables S1-S3b).
Phenotypes, genotyping, imputation and meta-analysis
AFS is treated as a continuous measure with individuals considered as eligible if they had given a valid answer and ages lower than 12 excluded (see Supp Note 1.2). Since AFS has a non-normal distribution, a within-sex inverse rank normal transformation is required. AFB is also treated as a continuous measure, assessed for those who have ever given birth to a child. Details about participating cohorts, sample inclusion criteria, genotyping and imputation, models used to test for association, X chromosome analysis, quality control filters and diagnostics, and meta-analysis are in the Supp Note. A sample-size weighted meta-analysis of quality-controlled cohort-level results was performed using the METAL software.67 We performed conditional and joint multiple SNP analyses (COJO) to identify further independent SNPs and sex-specific analyses.
Sex-specific genetic effects
We used LD score bivariate regression68 to estimate the genetic correlation between men and women based on the sex-specific summary statistics from the meta-analysis results. There was a large genetic overlap among the sexes for AFB (0.95) and a somewhat lower overlap for AFS (0.79), suggesting sex-specific effects would be important to examine. In order to determine if there was evidence for sex-specific effects, we compared the allelic effects for these SNPs between men and women and derived a p-value for heterogeneity.69 A multiple testing correction was applied (0.05/242=2 x 10-4) to identify sex-specific associations. We then selected a region of ±1Mb around these lead SNPs to identify the genes that may be represented by these lead SNPs, followed by gene prioritization as we did for the main AFB and AFS analyses.
X chromosome analysis
For AFS, the UK Biobank provided results for between 977,536 and 990,735 variants on the X chromosome after QC (see Table S8). For AFB, 13 cohorts provided information on the X chromosome. Overall, we received 23 files, 13 for women, 8 for men and 2 for the pooled analysis in case there were individuals who were relatives in the data. On average, 275,023 variants survived QC with a minimum of 99,794 in women from WLS to 998,304 for the women in the UK Biobank sample (see TableS7 for full descriptives). Association analyses on the X chromosome were performed using software suggested in the analysis plan (XWAS, SNPtest or BOLT-LMM) using BOLT-LMM for AFS as this was only assessed in the UK Biobank data, for AFB, METAL was used as described above (see sup. note 3.5)
Phenotypic and genotypic historical changes
Descriptive analyses and correlations were undertaken using the UK Biobank data to illustrate phenotypic shifts in the age of AFS and AFB by birth cohort, in addition to changes in the spread of the distribution. Pearson’s correlation coefficients were calculated and correlation graphs illustrate the changing relationship between the two phenotypes over time. Genotypic changes and SNP-heritability by birth cohort were quantified in UK Biobank data using GREML12 as described earlier.13
MTAG: Multi-trait analysis of GWAS
MTAG results70 were calculated using GWA meta-analysis results of the following related phenotypes: AFS, AFB, number of children ever born, childlessness. Using summary statistics from the pooled GWAS of each of the traits, MTAG uses bivariate LD score regression to account for unobserved sample overlap.
Polygenic score prediction
We performed out-of-sample prediction in two cohorts, the National Longitudinal Study of Adolescence to Adult Health (Add Health),71 based in the US and the UK Household Longitudinal Study - Understanding Society (UKHLS).72 We calculated three sets of polygenic risk scores (PGS) with weights based on meta-analysis results excluding the specific cohort from the calculation. First, pruning and thresholding of all SNPs was performed (250kb window; r2=.1) using PRSice73. Second, LDpred PGSs74 with the LD reference were calculated from the same genotyped files, using prior distributions for the causal fraction of SNPs equal to 1 and LDpred weights calculated under the infinitesimal model. Third, MTAG + LDpred PGSs were calculated using the same methodology as in the second PGSs, but this time based on MTAG results70. For both traits, we ran ordinary least-squares (OLS) regression models and report the incremental R2 as a measure of goodness-of-fit of the model. Confidence intervals are based on 1,000 bootstrapped samples.
Population stratification, environment moderated effects
To test whether population stratification biased our results or lead to false positives, we used the LD Score intercept method.75 For each phenotype, we used the “eur_w_ld_chr” files of LD Scores.16 These LD Scores were computed with genotypes from the European-ancestry samples in the 1000 Genomes Project using only HapMap3 SNPs with MAF > 0.01. We then ran survival models to account for right-censoring, which occurs when an individual does not experience the event of first sex or birth by the time of the interview.14 Using Add Health data, we estimated nonparametric hazard functions based on Nelson-Aalen estimates and then compared individuals at the top and bottom 5% of the PGS and plotted the estimated hazards. To further explore the relationship of environmentally moderated parental genetic effects on our PGSs, we examined PGS prediction across low (0-10%), medium (50-60%) and high (90-100%) PGS percentiles by parent’s education status (college versus no college), which serves as a proxy for childhood socioeconomic status.
Genetic correlations
Genetic correlation (rg) values were computed to estimate the genetic correlation between the two traits using all polygenic effects captured by the SNPs and LD-score regression.76 We used summary statistics and the 1000 Genomes reference set, and restricted the analysis to European populations. We also followed the common convention of restricting our analyses to SNPs with MAF >0.01, thus ensuring that all analyses were performed using a set of SNPs that were imputed with reasonable accuracy across all cohorts. The standard errors (SEs) were produced by the LDSC python software package that uses a block jackknife over the SNPs. We estimated the genetic correlation between 28 different traits, pooled by both sexes and then divided by sex. Traits were divided into the six categories of: reproductive, behavioural, psychiatric disorders, substance use disorders, personality and anthropometric.
Genomic SEM and Exploratory Factor Analysis
In an attempt to understand the aetiology of the correlations, we used the R package GenomicSEM to fit genetic multivariable regression models. GenomicSEM17 uses structural equation modelling to decompose the genetic covariance matrix, calculated using multivariate LD score regression, of a set of traits. Formally, structural equation models subsume many statistical methods and are quite flexible. We fit a series of genetic multivariable regression models, in which AFB was regressed on EA (educational attainment) and a trait X, in which we modelled various relevant traits such as openness, cognitive performance and AI (age initiation smoking). We also fit an analogous series of models in which AFS was regressed on EA.
Exploratory factor analysis (EFA) and Genomic SEM by reproductive biology and externalising behaviour. EFA was used to examine whether the genetic signal of the onset of reproductive behaviour originated from two genetically distinguishable sub-clusters of a biological component and an externalising behaviour component. This would suggest distinct causal mechanisms and subtypes of individuals. We tested this by fitting a two factor EFA model to the genetic covariance matrix of AFB, AFS, NEB, and the proxies age at menarche (biological component) and risk tolerance (externalising behaviour). To test this further, we estimated a more robust and additional measures of reproductive biology and externalising behaviour and a sex-specific analysis of AFB for women. We fit a genomic structural equation model (Genomic SEM) where AFB in women is regressed on age at menopause, age at menarche, and a latent factor representing the common genetic tendency to externalising behaviour. The factor is measured by AFS in women, age at initiation of smoking, age first used oral contraception, and ADHD, with the model scaled to unit variance for the latent factor.
Bi-directional Mendelian Randomization
We then tested whether causal pathways linking these phenotypes are potentially bidirectional and whether our phenotypes might offer distinct contributions. We identified 1000 Genomes proxies for our SNPs and used these in multivariate Mendelian Randomisation (MR) models. All data was assumed to be on the forward strand, and as many of these data sets included UK biobank, allele identifier were matched to this study as a reference. First, we modelled the interplay between AFB, AFS and EA (educational attainment)77 as well as risk taking (measured in adulthood)4 and age at smoking initiation (AI).20 In each case IVW78 and MR-EGGER79 methods were performed, with an additional round of IVW performed once a Steiger filter80 had been applied to remove SNPs that appears to show a primary association with the outcome rather than the exposure. Multivariate MR was use to try to dissect causal pathways.81 A second set of MR analyses focused on links to late life diseases, namely type 2 diabetes (T2D)82 and coronary artery disease (CAD)22, using the same methods. Acknowledging the substantial overlap between the most significant signals for AFB, AFS and education related phenotypes, we use multivariate methods to test whether AFS or AFB had independent effects once the well-established links to length of educational attainment, and BMI were controlled for. Finally, as there was a particular interest in infertility related phenotypes, bidirectional MR was performed with AFS and AFB and PCOS (polycystic ovarian syndrome). In this case, given the sex-specific nature of the disease, a specific analysis was also performed on women only.
Cox models of AFB polygenic score on longevity
To test trade-offs between reproductive behaviour and senescence, we conducted additional analyses to test whether our PGS for AFB was predictive of (parental) longevity. We restricted our models to mortality after age 60 to limit the possibility that early mortality affects parental fertility (i.e., collider bias).83 We calculated PGSs for AFB, Educational attainment (EA)84 and risky behaviour4 from the UK Biobank adopting the following procedure. We first split the sample in 10 random groups. We then iteratively estimated genome-wide association results for 9/10th of the sample and used these association results as weights for the calculation of polygenic scores in the remaining 1/10th of the sample. Polygenic scores were calculated using PRSice on a set of independent genotyped SNPs. We then estimated three sets of Cox Proportional hazard models to estimate the effect of the PGS of AFB on maternal and paternal age at death. All models control for the first 10 Genetic Principal Components, sex and year of birth, and are stratified by Local Authority District at birth calculated using the geo-coordinates provided in the UK Biobank due to differences in mortality related to material deprivation.85 We first estimated a baseline model and then included PGSs for EA and risk as covariates, followed by a final model including number of sibling (proxy for parental fertility).
Gene prioritization
We prioritized candidate genes in pooled and sex-specific GWAS-identified loci using predicted gene functions,86 single-cell RNA sequencing data in mice,29,87,88 literature mining,31 in silico sequencing,32 and synthetic Mendelian Randomization89 using eQTL data from brain and blood.90,91
DEPICT and CELLECT for tissue, cell type and gene prioritization were used. First, DEPICT was used to perform pathway analyses, identify enrichment for cell types and tissues, and prioritize candidate genes.86 DEPICT is agnostic to the outcomes analyzed in the GWAS and employs predicted gene functions. For both AFS and AFB, all SNPs with p<1x10-5 in the pooled GWAS meta-analysis were used as input. Based on the results of the tissue enrichment analysis, we used CELLECT88 to identify nervous system cell types that are enriched for expression of genes in loci reaching p<1x10-5 in the GWAS, using RNAseq data from mouse brain.29 A similar approach using tabula muris RNAseq data87 helped prioritize additional central nervous system and pancreatic cell types for AFS. For enriched cell types from mouse brain and tabula muris, the top-10 contributing genes were selected as candidate genes resulting in the prioritization of 296 genes for AFS and 95 for AFB based on mouse brain; and 97 genes for AFS based on tabula muris data.
Phenolyzer to integrate prior knowledge and phenotype information. We used Phenolyzer (v1.1) to prioritize candidate genes by integrating prior knowledge and phenotype information.92 Here we used the regions defined by DEPICT v1.1, reflecting loci reaching P<1x10-5 in first instance. Phenolyzer takes free text input and interprets these as disease names by using a word cloud to identify synonyms. It then queries precompiled databases for the disease names to find and score relevant seed genes. The seed genes are subsequently expanded to include related (predicted) genes based on several types of relationships, e.g., protein-protein interactions, transcriptional regulation and biological pathways. Phenolyzer uses machine-learning techniques on seed genes and predicted gene rankings to produce an integrated score for each gene. We used search terms capturing three broad areas, i.e., (in)fertility, congenital neurological disorders and psychological traits, based on results from pathway, tissue and cell type enrichment analyses.
In silico sequencing. We used in silico sequencing to identify non-synonymous variants with an R2 for LD>0.7 with the lead SNPs in AFS and AFB-associated loci,32 which yielded genes that may drive the GWAS associations through direct effects on protein function.
Summary data-based Mendelian Randomization (SMR) and Heterogeneity in Dependent Instruments (HEIDI).89 We conducted this using eQTL data from brain93 and whole blood.91 This approach provided a list of genes that showed Bonferroni corrected significant evidence (thresholds for blood <3.2x10-6 brain <6.7x10-6) of mediating the association between our phenotypes and GWAS-identified loci based on results from brain and blood.
Integration of findings across all functional approaches. We integrated findings across all approaches and retained genes in loci that reached genome-wide significance, and that were located within 1M bp of a GWAS lead SNP. We next used data from the Human Protein Atlas35 to identify genes amongst 387 genes that are expressed at a low, medium or high protein level in brain, glands, and/or reproductive organs at a ‘supported’ or ‘enhanced’ degree of reliability. For the 97 genes that fulfilled these criteria, we mapped the brain, glandular and reproductive cell types in which they are highly expressed at the protein level;94 used a text-mining approach to extract functions from entries in Entrez, GeneCards and Uniprot; and identified phenotypes in mutant mice from the Mouse Genome Informatics (MGI) database95.
Supplementary Material
Acknowledgements
A detailed list of funding and other acknowledgments for each cohort can be found in Section 14 of the Supplementary Information. This research was conducted using the UK Biobank Resource under application 22276 and 9905. Funding was provided by the ERC to MC Mills SOCIOGENOME (615603) and CHRONO (835079), ESRC/UKRI SOCGEN (ES/N011856/1), Wellcome Trust ISSF, Leverhulme Trust, Leverhulme Centre for Demographic Science. N Barban with ERC GENPOP (865356), FC Tropf, LabEx Ecode, French National Research Agency (ANR) Investissements d’Avenir (ANR-11-LABX-0047). Marcel den Hoed Swedish Heart-Lung Foundation (20170872, 20200781, 20140543, 20170678, 20180706, 20200602), Kjell and Märta Beijer Foundation, Swedish Research Council (2015-03657, 2019-01417). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. This study received Ethical Approval from the Department of Sociology, University of Oxford and relevant Ethics approval was obtained at the local level for the contributing datasets. We thank Evelina T. Akimova and Stine Møllegaard for administrative work in the organization of the cohort information and author list.
Footnotes
Author contributions
MCM and FRD designed and led the study. MCM wrote the paper and Supplementary Information with contributions by authors for respective analyses and comments by all authors. DMB conducted phenotypic changes, phenotype preparation, LD Score and genetic correlations, Genomic SEM and exploratory factor analysis and sex-specific effects. NB conducted GWAS meta-analysis, MTAG, PGS prediction, survival models, and Cox models of longevity. FCT and FRD conducted the cohort QC. FCT conducted GREML cohort heritability analysis and phenotype preparation in UKBB. FRD ran Mendelian Randomization, conducted GWAS analyses, JRBP conducted COJO and X-Chromosome analysis and KKO provided comments and expertise throughout. NvZ conducted DEPICT and Phenolyzer analyses. AV and HS conducted in silico sequencing and SMR analyses. THP conducted cell type enrichment analyses. MdH integrated gene prioritization results and performed downstream analyses, e.g. Human Protein Atlas; Entrez, GeneCards and Uniprot mining; and STRING Protein-Protein interaction analyses. Authors in the Human Reproductive Behaviour Consortium contributed the valuable data, conducted cohort specific GWAS and other analyses, and contributed through the administration, management, and data collection for the participating cohorts. The eQTLGen and BIOS Consortiums provided data for additional analyses. All authors reviewed and approved the final version of the paper and code relies upon the standard packages described above.
Competing interests
The main authors declare no competing interests. The views expressed in this article are those of the author(s) and not necessarily those of the NHS, the NIHR, or the Department of Health. MMcC (Mark McCarthy) has served on advisory panels for Pfizer, NovoNordisk and Zoe Global, has received honoraria from Merck, Pfizer, Novo Nordisk and Eli Lilly, and research funding from Abbvie, Astra Zeneca, Boehringer Ingelheim, Eli Lilly, Janssen, Merck, NovoNordisk, Pfizer, Roche, Sanofi Aventis, Servier, and Takeda. As of June 2019, MMcC is an employee of Genentech, and a holder of Roche stock.
Contributor Information
eQTLGen Consortium:
Mawussé Agbessi, Habibul Ahsan, Isabel Alves, Anand Kumar Andiappan, Wibowo Arindrarto, Philip Awadalla, Alexis Battle, Frank Beutner, Marc Jan Bonder, Dorret I. Boomsma, Mark W. Christiansen, Annique Claringbould, Patrick Deelen, Tõnu Esko, Marie-Julié Fave, Lude Franke, Timothy Frayling, Sina A. Gharib, Greg Gibson, Bastiaan T. Heijmans, Gibran Hemani, Rick Jansen, Mika Kähönen, Anette Kalnapenkis, Silva Kasela, Johannes Kettunen, Yungil Kim, Holger Kirsten, Peter Kovacs, Knut Krohn, Jaanika Kronberg, Viktorija Kukushkina, Zoltan Kutalik, Bernett Lee, Terho Lehtimäki, Markus Loeffler, Urko M. Marigorta, Hailang Mei, Lili Milani, Grant W. Montgomery, Martina Müller-Nurasyid, Matthias Nauck, Michel G. Nivard, Brenda W.J.H. Penninx, Markus Perola, Natalia Pervjakova, Brandon L. Pierce, Joseph Powell, Holger Prokisch, Bruce M. Psaty, Olli T. Raitakari, Samuli Ripatti, Olaf Rotzschke, Sina Rüeger, Ashis Saha, Markus Scholz, Katharina Schramm, Ilkka Seppälä, Eline P. Slagboom, Coen D.A. Stehouwer, Michael Stumvoll, Patrick Sullivan, Peter A.C. ‘t Hoen, Alexander Teumer, Joachim Thiery, Lin Tong, Anke Tönjes, Jenny van Dongen, Maarten van Iterson, Joyce van Meurs, Jan H. Veldink, Joost Verlouw, Peter M. Visscher, Uwe Völker, Urmo Võsa, Harm-Jan Westra, Cisca Wijmenga, Hanieh Yaghootkar, Jian Yang, Biao Zeng, and Futao Zhang
BIOS Consortium (Biobank-based Integrative Omics Study):
Management Team, Bastiaan T. Heijmans, Peter A.C. ‘t Hoen, Joyce van Meurs, Aaron Isaacs, Rick Jansen, Lude Franke, Cohort collection, Dorret I. Boomsma, René Pool, Jenny van Dongen, Jouke Jan Hottenga, Marleen MJ van Greevenbroek, Coen D.A. Stehouwer, Carla J.H. van der Kallen, Casper G. Schalkwijk, Cisca Wijmenga, Lude Franke, Sasha Zhernakova, Ettje F. Tigchelaar, Eline P. Slagboom, Marian Beekman, Joris Deelen, Diana van Heemst, Jan H. Veldink, Leonard H. van den Berg, Cornelia M. van Duijn, Bert A. Hofman, Aaron Isaacs, André G. Uitterlinden66,75, Data Generation, Joyce van Meurs, P. Mila Jhamai, Michael Verbiest, H. Eka D. Suchiman, Marijn Verkerk, Ruud van der Breggen, Jeroen van Rooij, Nico Lakenberg, Data management and computational infrastructure, Hailiang Mei, Maarten van Iterson, Michiel van Galen, Jan Bot, Dasha V. Zhernakova, Rick Jansen, Peter van ‘t Hof, Patrick Deelen, Irene Nooren, Peter A.C. ‘t Hoen, Bastiaan T. Heijmans, Matthijs Moed, Data Analysis Group, Lude Franke, Martijn Vermaat, Dasha V. Zhernakova, René Luijk, Marc Jan Bonder, Maarten van Iterson, Patrick Deelen, Freerk van Dijk, Michiel van Galen, Wibowo Arindrarto, Szymon M. Kielbasa, Morris A. Swertz, Erik. W van Zwet, Rick Jansen, Peter A.C. ‘t Hoen, and Bastiaan T. Heijmans
Human Reproductive Behaviour Consortium:
Evelina T. Akimova, Sven Bergmann, Jason D. Boardman, Dorret I. Boomsma, Marco Brumat, Julie E. Buring, David Cesarini, Daniel I. Chasman, Jorge E. Chavarro, Massimiliano Cocca, Maria Pina Concas, George Davey-Smith, Gail Davies, Ian J. Deary, Tônu Esko, Oscar Franco, Audrey J. Gaskins, Eco J.C. de Geus, Christian Gieger, Giorgia Girotto, Hans Jörgen Grabe, Erica P. Gunderson, Kathleen Mullan Harris, Fernando P. Hartwig, Chunyan He, Diana van Heemst, W. David Hill, Georg Homuth, Bernando Lessa Horta, Jouke Jan Hottenga, Hongyang Huang, Elina Hyppönen, M. Arfan Ikram, Rick Jansen, Magnus Johannesson, Zoha Kamali, Maryam Kavousi, Peter Kraft, Brigitte Kühnel, Claudia Langenberg, Lifelines Cohort Study, Penelope A. Lind, Jian’an Luan, Reedik Mägi, Patrik K.E. Magnusson, Anubha Mahajan, Nicholas G. Martin, Hamdi Mbarek, Mark I. McCarthy, George McMahon, Matthew B. McQueen, Sarah E. Medland, Thomas Meitinger, Andres Metspalu, Evelin Mihailov, Lili Milani, Stacey A. Missmer, Stine Møllegaard, Dennis O. Mook-Kanamori, Anna Morgan, Peter J. van der Most, Renée de Mutsert, Matthias Nauck, Ilja M. Nolte, Raymond Noordam, Brenda W.J.H. Penninx, Annette Peters, Chris Power, Paul Redmond, Janet W. Rich-Edwards, Paul M. Ridker, Cornelius A. Rietveld, Susan M. Ring, Lynda M. Rose, Rico Rueedi, Kári Stefánsson, Doris Stöckl, Konstantin Strauch, Morris A. Swertz, Alexander Teumer, Gudmar Thorleifsson, Unnur Thorsteinsdottir, A. Roy Thurik, Nicholas J. Timpson, Constance Turman, André G. Uitterlinden, Melanie Waldenberger, Nicholas J. Wareham, Gonneke Willemsen, and Jing Hau Zhao
Data availability
Our policy is to make genome-wide summary statistics widely and publically available. Upon publication, summary statistics will be available on the GWAS Catalog website: https://www.ebi.ac.uk/gwas/downloads/summary-statistics The phenotype and genotype data for separate studies used in this GWAS are available upon application to each of the participating cohorts who can be contacted directly to follow their different data access policies. Access to the UK Biobank is available through application with information available at: http://www.ukbiobank.ac.uk).
Code availability
No custom code was used with all analyses and modelling using standard software as described in the Methods section and in detail in the Supplementary Information.
References
- 1.Mercer CH, et al. Changes in sexual attitudes and lifestyles in Britain through the life course and over time: findings from the National Surveys of Sexual Attitudes and Lifestyles (Natsal) Lancet. 2013;382:1781–1794. doi: 10.1016/S0140-6736(13)62035-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lara LAS, Abdo CHN. Age at Time of Initial Sexual Intercourse and Health of Adolescent Girls. J Pediatr Adolesc Gynecol. 2016;29:417–423. doi: 10.1016/j.jpag.2015.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Polimanti R, et al. The Interplay between Risky Sexual Behaviors and Alcohol Dependence: Genome-Wide Association and Neuroimaging Support for LHPP as a Risk Gene. Neuropsychopharmacology. 2017;42:598–605. doi: 10.1038/npp.2016.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karlsson Linnér R, et al. Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat Genet. 2019;51:245–257. doi: 10.1038/s41588-018-0309-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balbo N, Billari FC, Mills M. Fertility in Advanced Societies: A Review of Research. Eur J Popul / Rev Eur Démographie. 2013;29:1–38. doi: 10.1007/s10680-012-9277-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mills MC, et al. Why do people postpone parenthood? Reasons and social policy incentives. Hum Reprod Update. 2011;17:848–860. doi: 10.1093/humupd/dmr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rahmioglu N, et al. Genome-wide enrichment analysis between endometriosis and obesity-related traits reveals novel susceptibility loci. Hum Mol Genet. 2015;24:1185–1199. doi: 10.1093/hmg/ddu516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barban N, et al. Genome-wide analysis identifies 12 loci influencing human reproductive behavior. Nat Genet. 2016;48 doi: 10.1038/ng.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balbo N, Billari FC, Mills MC. Fertility in advanced societies: A review of research. Eur J Popul Eur Démographie. 2013;29:1–38. doi: 10.1007/s10680-012-9277-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin NG, Eaves LJ, Eysenck HJ. Genetical, Environmental and Personality Factors Influencing the Age of First Sexual Intercourse in Twins. J Biosoc Sci. 1977;9:91–97. doi: 10.1017/s0021932000000493. [DOI] [PubMed] [Google Scholar]
- 11.Day FR, et al. Physical and neurobehavioral determinants of reproductive onset and success. Nat Genet. 2016 doi: 10.1038/ng.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tropf FC, et al. Hidden heritability due to heterogeneity across seven populations. Nat Hum Behav. 2017;1:757–765. doi: 10.1038/s41562-017-0195-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mills MC. Introducing survival and event history analysis. Sage Publications; 2011. [Google Scholar]
- 15.Singh S, Darroch JE, Frost JJ. Socioeconomic Disadvantage and Adolescent Women’s Sexual and Reproductive Behavior: The Case of Five Developed Countries. Fam Plann Perspect. 2001;33:251. [PubMed] [Google Scholar]
- 16.Finucane HK, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grotzinger AD, et al. Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nat Hum Behav. 2019;3:513–525. doi: 10.1038/s41562-019-0566-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davey Smith G. What can mendelian randomisation tell us about modifiable behavioural and environmental exposures? BMJ. 2005 doi: 10.1136/bmj.330.7499.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JJ, et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet. 2018;50:1112–1121. doi: 10.1038/s41588-018-0147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu M, et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet. 2019;51:237–244. doi: 10.1038/s41588-018-0307-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarthy M, Mahajan A. Genome-wide association analyses in Type 2 Diabetes: The gift that keeps on giving. Blog. 2018 Available at: http://mccarthy.well.ox.ac.uk/2018/10/gwas-gift-keeps-giving/ [Google Scholar]
- 22.Nelson CP, et al. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet. 2017;49:1385–1391. doi: 10.1038/ng.3913. [DOI] [PubMed] [Google Scholar]
- 23.Lind JM, Hennessy A, Chiu CL. Association Between a Woman’s Age at First Birth and High Blood Pressure. Medicine (Baltimore) 2015;94:e697. doi: 10.1097/MD.0000000000000697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patchen L, Leoutsakos J-M, Astone NM. Early Parturition: Is Young Maternal Age at First Birth Associated with Obesity? J Pediatr Adolesc Gynecol. 2017;30:553–559. doi: 10.1016/j.jpag.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 25.Kim JH, Jung Y, Kim SY, Bae HY. Impact of Age at First Childbirth on Glucose Tolerance Status in Postmenopausal Women: The 2008–2011 Korean National Health and Nutrition Examination Survey. Diabetes Care. 2014;37:671–677. doi: 10.2337/dc13-1784. [DOI] [PubMed] [Google Scholar]
- 26.Day F, et al. Large-scale genome-wide meta-analysis of polycystic ovary syndrome suggests shared genetic architecture for different diagnosis criteria. PLoS Genet. 2018;14:e1007813. doi: 10.1371/journal.pgen.1007813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eisenberg DTA, Hayes MG, Kuzawa CW. Delayed paternal age of reproduction in humans is associated with longer telomeres across two generations of descendants. Proc Natl Acad Sci. 2012;109:10251–10256. doi: 10.1073/pnas.1202092109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pers TH, et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat Commun. 2015;6:5890. doi: 10.1038/ncomms6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeisel A, et al. Molecular Architecture of the Mouse Nervous System. Cell. 2018;174:999–1014.:e22. doi: 10.1016/j.cell.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tabula Muris Consortium et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562:367–372. doi: 10.1038/s41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang H, Robinson PN, Wang K. Phenolyzer: phenotype-based prioritization of candidate genes for human diseases. Nat Methods. 2015;12:841–3. doi: 10.1038/nmeth.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaez A, et al. In Silico Post Genome-Wide Association Studies Analysis of C-Reactive Protein Loci Suggests an Important Role for Interferons. Circ Cardiovasc Genet. 2015;8:487–497. doi: 10.1161/CIRCGENETICS.114.000714. [DOI] [PubMed] [Google Scholar]
- 33.Zhu Z, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48:481–7. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 34.van der Wijst MGP, et al. Single-cell RNA sequencing identifies celltype-specific cis-eQTLs and co-expression QTLs. Nat Genet. 2018;50:493–497. doi: 10.1038/s41588-018-0089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uhlén M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 36.Ellsworth BS, et al. FOXL2 in the pituitary: molecular, genetic, and developmental analysis. Mol Endocrinol. 2006;20:2796–805. doi: 10.1210/me.2005-0303. [DOI] [PubMed] [Google Scholar]
- 37.van Vliet J, et al. Human KLF17 is a new member of the Sp/KLF family of transcription factors. Genomics. 2006;87:474–82. doi: 10.1016/j.ygeno.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 38.Governini L, et al. FOXL2 in human endometrium: hyperexpressed in endometriosis. Reprod Sci. 2014;21:1249–55. doi: 10.1177/1933719114522549. [DOI] [PubMed] [Google Scholar]
- 39.Rico C, et al. HIF1 activity in granulosa cells is required for FSH-regulated Vegfa expression and follicle survival in mice. Biol Reprod. 2014;90:135. doi: 10.1095/biolreprod.113.115634. [DOI] [PubMed] [Google Scholar]
- 40.Dai Z, et al. Caveolin-1 promotes trophoblast cell invasion through the focal adhesion kinase (FAK) signalling pathway during early human placental development. Reprod Fertil Dev. 2019 doi: 10.1071/RD18296. [DOI] [PubMed] [Google Scholar]
- 41.Artini PG, et al. Cumulus cells surrounding oocytes with high developmental competence exhibit down-regulation of phosphoinositol 1,3 kinase/protein kinase B (PI3K/AKT) signalling genes involved in proliferation and survival. Hum Reprod. 2017;32:2474–2484. doi: 10.1093/humrep/dex320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng J, et al. Novel FSHβ mutation in a male patient with isolated FSH deficiency and infertility. Eur J Med Genet. 2017;60:335–339. doi: 10.1016/j.ejmg.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 43.Yan W, Burns KH, Ma L, Matzuk MM. Identification of Zfp393, a germ cell-specific gene encoding a novel zinc finger protein. Mech Dev. 2002;118:233–9. doi: 10.1016/s0925-4773(02)00258-7. [DOI] [PubMed] [Google Scholar]
- 44.Lin Y-N, Roy A, Yan W, Burns KH, Matzuk MM. Loss of zona pellucida binding proteins in the acrosomal matrix disrupts acrosome biogenesis and sperm morphogenesis. Mol Cell Biol. 2007;27:6794–805. doi: 10.1128/MCB.01029-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wieser F, et al. Expression and regulation of CCR1 in peritoneal macrophages from women with and without endometriosis. Fertil Steril. 2005;83:1878–1881. doi: 10.1016/j.fertnstert.2004.12.034. [DOI] [PubMed] [Google Scholar]
- 46.Mei J, et al. CXCL16/CXCR6 interaction Promotes Endometrial Decidualization via the PI3K/AKT Pathway. Reproduction. 2019 doi: 10.1530/REP-18-0417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gusev FE, et al. Epigenetic-genetic chromatin footprinting identifies novel and subject-specific genes active in prefrontal cortex neurons. FASEB J. 2019;33:8161–8173. doi: 10.1096/fj.201802646R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quinn JP, Savage AL, Bubb VJ. Non-coding genetic variation shaping mental health. Curr Opin Psychol. 2019;27:18–24. doi: 10.1016/j.copsyc.2018.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barak B, et al. Neuronal deletion of Gtf2i, associated with Williams syndrome, causes behavioral and myelin alterations rescuable by a remyelinating drug. Nat Neurosci. 2019;22:700–708. doi: 10.1038/s41593-019-0380-9. [DOI] [PubMed] [Google Scholar]
- 50.Li Y, et al. Topoisomerase IIbeta is required for proper retinal development and survival of postmitotic cells. Biol Open. 2014;3:172–84. doi: 10.1242/bio.20146767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Athanasiou MC, et al. The transcription factor E2F-1 in SV40 T antigen-induced cerebellar Purkinje cell degeneration. Mol Cell Neurosci. 1998;12:16–28. doi: 10.1006/mcne.1998.0699. [DOI] [PubMed] [Google Scholar]
- 52.Yang X, et al. The association between NCAM1 levels and behavioral phenotypes in children with autism spectrum disorder. Behav Brain Res. 2019;359:234–238. doi: 10.1016/j.bbr.2018.11.012. [DOI] [PubMed] [Google Scholar]
- 53.Locke AE, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tu S, et al. NitroSynapsin therapy for a mouse MEF2C haploinsufficiency model of human autism. Nat Commun. 2017;8:1488. doi: 10.1038/s41467-017-01563-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shamir A, et al. The Importance of the NRG-1/ErbB4 Pathway for Synaptic Plasticity and Behaviors Associated with Psychiatric Disorders. J Neurosci. 2012;32:2988–2997. doi: 10.1523/JNEUROSCI.1899-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang J-M, et al. erbb4 Deficits in Chandelier Cells of the Medial Prefrontal Cortex Confer Cognitive Dysfunctions: Implications for Schizophrenia. Cereb Cortex. 2019;29:4334–4346. doi: 10.1093/cercor/bhy316. [DOI] [PubMed] [Google Scholar]
- 57.Day FR, et al. Causal mechanisms and balancing selection inferred from genetic associations with polycystic ovary syndrome. Nat Commun. 2015;6:8464. doi: 10.1038/ncomms9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baumgartner HK, et al. Characterization of choline transporters in the human placenta over gestation. Placenta. 2015;36:1362–9. doi: 10.1016/j.placenta.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peng Z, et al. Liver X receptor β in the hippocampus: A potential novel target for the treatment of major depressive disorder? Neuropharmacology. 2018;135:514–528. doi: 10.1016/j.neuropharm.2018.04.014. [DOI] [PubMed] [Google Scholar]
- 60.Mahajan A, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018;50:1505–1513. doi: 10.1038/s41588-018-0241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tropf FC, et al. Human Fertility, Molecular Genetics, and Natural Selection in Modern Societies. PLoS One. 2015;10:e0126821. doi: 10.1371/journal.pone.0126821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Waren EB, et al. Heterogeneity in polygenic scores for common human traits. bioRxiv. 2017 doi: 10.1101/106062. [DOI] [Google Scholar]
- 63.Ripke S, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mills MC, Rahal C. The GWAS Diversity Monitor tracks diversity by disease in real time. Nat Genet. 2020;52:242–243. doi: 10.1038/s41588-020-0580-y. [DOI] [PubMed] [Google Scholar]
- 65.Chen X-K, et al. Teenage pregnancy and adverse birth outcomes: a large population based retrospective cohort study. Int J Epidemiol. 2007;36:368–373. doi: 10.1093/ije/dyl284. [DOI] [PubMed] [Google Scholar]
- 66.Bongaarts J, Mensch BS, Blanc AK. Trends in the age at reproductive transitions in the developing world: The role of education. Popul Stud (NY) 2017;71:139–154. doi: 10.1080/00324728.2017.1291986. [DOI] [PubMed] [Google Scholar]
- 67.Willer CJ, Li Y, Abecasis GR. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Finucane HK, et al. Partionining heritability by functional category using GWAS summary statistics. bioRxiv. 2015 [Google Scholar]
- 69.Altman DG, Bland JM. Interaction revisited: the difference between two estimates. BMJ. 2003;326:219. doi: 10.1136/bmj.326.7382.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Turley P, et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat Genet. 2018;50:229–237. doi: 10.1038/s41588-017-0009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Harris KM, et al. The National Longitudinal Study of Adolescent to Adult Health: Research Design. 2009 Available at: http://www.cpc.unc.edu/projects/addhealth/design.
- 72.Buck N, McFall S. Understanding Society: design overview. Longit Life Course Stud. 2012;3:5–17. [Google Scholar]
- 73.Euesden J, Lewis CM, O’Reilly PF. PRSice: Polygenic Risk Score software. Bioinformatics. 2014;31:btu848-1468. doi: 10.1093/bioinformatics/btu848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vilhjálmsson BJ, et al. Modeling Linkage Disequilibrium Increases Accuracy of Polygenic Risk Scores. Am J Hum Genet. 2015;97:576–592. doi: 10.1016/j.ajhg.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bulik-Sullivan B, et al. An Atlas of Genetic Correlations across Human Diseases and Traits. bioRxiv. 2015;47:1–44. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee JJ, et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet. 2018 doi: 10.1038/s41588-018-0147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37:658–65. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bowden J, Smith GD, Burgess S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015 doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hemani G, Bowden J, Davey Smith G. Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum Mol Genet. 2018;27:R195–R208. doi: 10.1093/hmg/ddy163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Burgess S, Thompson SG. Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol. 2015;181:251–60. doi: 10.1093/aje/kwu283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mahajan A, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018;50:1505–1513. doi: 10.1038/s41588-018-0241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Day FR, Loh P-R, Scott RA, Ong KK, Perry JRB. A Robust Example of Collider Bias in a Genetic Association Study. Am J Hum Genet. 2016;98:392–393. doi: 10.1016/j.ajhg.2015.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee JJ, et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet. 2018;50:1112–1121. doi: 10.1038/s41588-018-0147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Woods LM. Geographical variation in life expectancy at birth in England and Wales is largely explained by deprivation. J Epidemiol Community Heal. 2005;59:115–120. doi: 10.1136/jech.2003.013003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pers TH, et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat Commun. 2015;6:5890. doi: 10.1038/ncomms6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tabula Muris Consortium et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562:367–372. doi: 10.1038/s41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Timshel PN, Thompson JJ, Pers TH. Genetic mapping of etiologic brain cell types for obesity. Elife. 2020;9 doi: 10.7554/eLife.55851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhu Z, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48:481–7. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 90.van der Wijst MGP, et al. Single-cell RNA sequencing identifies celltype-specific cis-eQTLs and co-expression QTLs. Nat Genet. 2018;50:493–497. doi: 10.1038/s41588-018-0089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vosa U, Al E. Unraveling the polygenic architecture of complex traits using blood eQTL metaanalysis. bioRxiv. 2018 doi: 10.1101/447367. [DOI] [Google Scholar]
- 92.Yang H, Robinson PN, Wang K. Phenolyzer: phenotype-based prioritization of candidate genes for human diseases. Nat Methods. 2015;12:841–3. doi: 10.1038/nmeth.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Qi T, et al. Identifying gene targets for brain-related traits using transcriptomic and methylomic data from blood. Nat Commun. 2018;9:2282. doi: 10.1038/s41467-018-04558-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Uhlén M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 95.Bult CJ, et al. Mouse Genome Database (MGD) 2019. Nucleic Acids Res. 2019;47:D801–D806. doi: 10.1093/nar/gky1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Our policy is to make genome-wide summary statistics widely and publically available. Upon publication, summary statistics will be available on the GWAS Catalog website: https://www.ebi.ac.uk/gwas/downloads/summary-statistics The phenotype and genotype data for separate studies used in this GWAS are available upon application to each of the participating cohorts who can be contacted directly to follow their different data access policies. Access to the UK Biobank is available through application with information available at: http://www.ukbiobank.ac.uk).
No custom code was used with all analyses and modelling using standard software as described in the Methods section and in detail in the Supplementary Information.