Abstract
Experimental in vitro models that capture pathophysiological characteristics of human tumours are essential for basic and translational cancer biology. Here, we describe a fully synthetic hydrogel extracellular matrix designed to elicit key phenotypic traits of the pancreatic environment in culture. To enable the growth of normal and cancerous pancreatic organoids from genetically engineered murine models and human patients, essential adhesive cues were empirically defined and replicated in the hydrogel scaffold, revealing a functional role of laminin – integrin α3/α6 signalling in establishment and survival of pancreatic organoids. Altered tissue stiffness — a hallmark of pancreatic cancer — was recapitulated in culture by adjusting the hydrogel properties to engage mechano-sensing pathways and alter organoid growth. Pancreatic stromal cells were readily incorporated into the hydrogels and replicated phenotypic traits characteristic of the tumour environment in vivo. This model therefore recapitulates a pathologically remodelled tumour microenvironment for studies of normal and pancreatic cancer cells in vitro.
Introduction
Tumour cells are embedded in a complex environment encompassing conscripted host cells and a pathologically remodelled extracellular matrix (ECM) 1,2. Coerced host cells such as immature myeloid cells and macrophages promote the development of an immune suppressive environment, and cancer-associated fibroblasts (CAFs) deposit and remodel the extracellular matrix, resulting in a rigid and poorly perfused microenvironment 3,4. These features are particularly prominent in pancreatic ductal adenocarcinoma (PDA), where the stromal reaction constitutes, on average, 80% of the total tumour volume 2,5. Normalising the dysfunctional microenvironment improves therapeutic efficiency, where targeting the stiff ECM or the stromal infiltrate improves efficacy of gemcitabine and immune-checkpoint inhibitors 6–9. Models that faithfully replicate the stromal environment are therefore essential to improve the development of therapeutic strategies.
A diverse range of pre-clinical models, including cell lines, genetically-engineered murine models (GEMM) and patient-derived xenografts (PDX) have been instrumental for target discovery and validation. Recently, organoids have gained traction as a complementary model system, enabling the establishment and 3D culture of murine and patient-derived tumour cells at high efficiency 10–12. However, organoid models are typically established in animal-derived hydrogels, most prominently the basement membrane (BM) extract Matrigel, which suffers from batch-to-batch variability and ill-defined composition, disfavours stromal cell propagation, and mostly fails to replicate the pathological ECM of human cancers11,13–15. Consequently, interdependencies between tumour cells and the microenvironment are inadequately modelled in this system. Contemporary synthetic scaffolds, such as the polyethylene glycol (PEG)-based hydrogel scaffolds, offer several advantages to cell- and tissue-derived matrices, including exquisite control over growth conditions 16–19. Although current model systems have been successfully used for primary and induced pluripotent stem cell (iPSC)-derived intestinal organoids 16–19, matrices with tissue-matched adhesion characteristics capable of supporting both epithelial organoids and stromal cells are needed to better replicate the complexity of the tumour ecosystem.
Here, we set out to develop a rationally-designed, synthetic 3D model for pancreatic organoids that simultaneously captures relevant physiological components of the tissue microenvironment. We envisaged an ideal model would replicate essential cell-ECM interactions, mimic tissue stiffness ranges observed across normal and tumour-bearing tissues, support co-culture of epithelial and stromal cells and facilitate growth and development of organoids directly from tissue samples.
Proteomic analysis of the tumour matrisome and functional validation of cellular adhesion requirements were used to develop a relevant PEG hydrogel scaffold. This system successfully replicates the entire stiffness range of normal and tumour-bearing tissue, supports growth of murine and human normal and cancerous organoids, and essential stromal populations, and is compatible with proteomic and single cell analyses.
Results
Defining adhesive requirements of pancreatic cancer cells
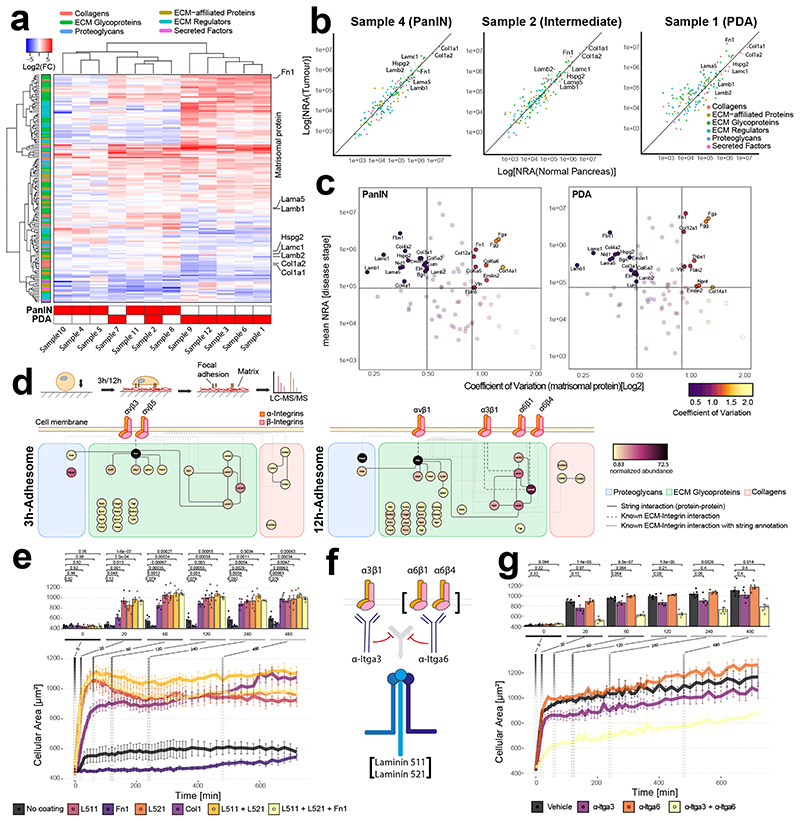
To reconstitute relevant ECM-receptor interactions within a custom-designed PEG environment, we first defined a matrisome reference set for in vivo pancreatic tissues 20. Normal pancreatic tissue (NP), pancreata with Pancreatic Intraepithelial Neoplasia (PanIN), known precursor lesions of PDA, and overt PDA from KrasG12D and KrasG12D; TP53R172H-driven murine models were collected for analysis 21–23 (Supplementary Fig. 1a). We confirmed the high reproducibility of the workflow by analysing 10 normal pancreata by LC-MS/MS (Supplementary Fig. 1a–d) before analysing 12 tumour samples, which resulted in the identification and quantification of 147 matrisomal proteins, 83 of which predominantly exhibit structural functions (termed core matrisome) (Fig. 1a,b and Supplementary Figs. 2-5) 24. Analysis of relative protein abundances by hierarchical clustering segregated samples by their histological disease grade, reflecting the evolving matrisomal remodelling that occurs during disease development 25 (Fig. 1a,b and Supplementary Figs. 2 and 3). Comparison of adhesion-conferring matricellular protein levels with available PDAmatrisome datasets 26 highlighted several proteins, including fibronectin, versican and laminin-332 (L332, α3β3γ2), which were upregulated in both human and murine PDA and correlate with patient outcome (Supplementary Fig. 4). In contrast, the basement membrane proteins laminin 511 (L511, α5β1γ1), laminin 521 (L521, α5β2γ1) and the type 1 and 4 collagens (e.g. Col1a1, Col1a2, Col4a1 and Col4a2) were consistently highly abundant in both normal and disease-bearing pancreas (Supplementary Fig. 3b), with laminin 511 and 521 exhibiting minimal variation across samples (Fig. 1c and Supplementary Fig. 5c). Together, these findings highlight important and potentially complementary roles of individual matrisomal components, including fibronectin, laminins (L322, L511 and L521) and collagens throughout PDA development.
Figure 1. Defining adhesive requirements of pancreatic cancer cells.
a, Hierarchical clustering of matrisomal protein relative abundances (compared to normal pancreas). Histological grade (bottom) and selected matrisomal proteins (right side) are shown. b, Normalised relative abundance (NRA) of matrisomal proteins from 3 representative samples (PanIN: Pancreatic Intraepithelial Neoplasia; Intermediate: mixed sample containing PanIN and PDA; PDA: Pancreatic ductal adenocarcinoma). c, Scatter plot displaying the variance of core matrisome proteins over mean NRA in PanIN (top) or PDA (bottom). Vertical lines represent the 25% quantile and 75% quantile coefficient of variation. Horizontal line indicates median abundance. d, Outline of integrin adhesion complex (IAC) isolation (top) and identified proteins (bottom). Interactions of selected proteins identified in IACs isolated from KPC-1s after 3h (left) or 12h (right) are shown. Proteins are colour-coded according to their relative abundance. e, Time-course of GFP-labelled human Suit-2 PCCs spreading on indicated ECM proteins. f, Schematic outline of blocking antibodies targeting integrins α6β1, α6β4 or α3β1. g, Effect of blocking antibodies on GFP-labelled Suit-2 cells spreading on L511- and L521-coated dishes. e,g, Bar graphs show mean values (n=5), with each data point representing the median cellular area of >100 measured cells for indicated timepoints. Two-sided parametric Welch’s t-test with Benjamini-Hochberg correction; error bars: s.e.m
Integrins constitute a family of cellular ECM receptors that are fundamental to cellular adhesion, and are commonly aberrantly expressed in malignant disease 27–29. To identify which receptors actively engage the ECM to form integrin adhesion complexes (IACs), we seeded pancreatic cancer cells (PCCs) on fibronectin (FN)-coated plates for 3 or 12 hours, and analysed mature IACs by mass spectrometry 30,31 (Fig. 1d and Supplementary Fig. 6). Interestingly, although PCCs were seeded on FN, the isolated IACs changed in composition from the canonical FN-binding integrins αVβ3 and αVβ5 after 3 hours, to laminin-binding integrins α6β1, α6β4, and α3β1, after 12 hours, concurrent with the deposition of laminin 521 and 511 (Fig. 1d and Supplementary Fig. 6). As this indicates PCCs autonomously engineer the ECM to match their specific adhesion dependencies, we analysed the cell-derived matrix (CDM) isolated from PCCs and normal pancreatic ductal epithelial cells (HPDE) by mass spectrometry. This analysis confirmed the deposition of a basement membrane-rich ECM that included both L511 and L521 (Supplementary Fig. 7). Taken together, these data highlight a preference for L511- and L521-mediated tumour cell adhesion through α6- and α3-containing integrins 32.
To test these observations functionally, cell culture dishes were pre-coated with selected ECM ligands that were identified in the proteomic analysis, and the adhesion kinetics of relevant tumour and stromal cells were determined (Supplementary Fig. 8 and Supplementary Videos 1 and 2). Murine and human PCCs preferentially adhered to L511- and L521-precoated dishes, whereas pancreatic fibroblasts adhered to fibronectin-coated dishes (Fig. 1e and Supplementary Figs. 9 and 10). PCCs adhered to collagen-1 in a cell line-dependent manner, but consistently exhibited limited adhesion to fibronectin or non-coated culture dishes (Fig. 1e, Supplementary Figs. 9 and 10 and Supplementary Videos 3–9). Function-blocking antibodies directed against integrin -α3 and -α6 significantly reduced adhesion to L511- and 521-precoated dishes (Fig. 1f-g and Supplementary Fig. 10), confirming the importance of L511/521-integrin α3/6β1/4-mediated PCC adhesion in vitro (Fig. 1g and Supplementary Fig. 10) 33–35.
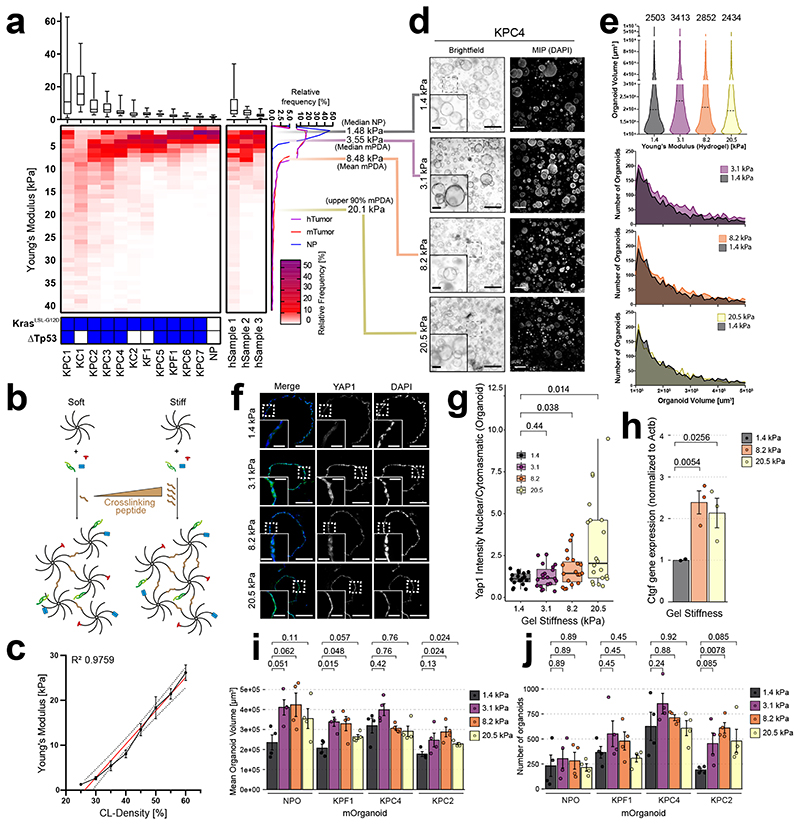
Figure 4. Recapitulating the stiffness range of PDA in PEG hydrogels.
a, Boxplot (top) and heatmap (bottom) displaying stiffness frequency (Young’s modulus) for murine (left, n=11 tumour, n=3 NP) and human (center, n=3) samples. Boxplots represent individual measurements (>800/sample). Bold line represents median, with boxes representing 25th to 75th percentile and whiskers indicate 90% confidence interval. Genotype of murine samples is shown (below). Cumulated relative stiffness frequency (right). Median stiffness for normal (grey) as well as median (purple), mean (orange) and upper 90% border (yellow) of murine cancerous samples. b, Illustration of how crosslinking density after PEG functionalisation control stiffness. c, AFM measurements of PEG CBF-0.5 hydrogels (n=3, error bars, s.e.m). Linear regression (red) and 95% c.i. shown (dashed line). d, Representative brightfield images (left) and maximum intensity projections of DAPI (right) from mPCOs grown at indicated stiffnesses (d4, n=4). Scale bar: 500 μm, inlay: 100 μm (left); 200 μm (right). e, Violin plot of organoid volume (top) and frequency plots of numbers (bottom) from d. Dashed bold lines show median organoid volume. Number of analysed organoids shown above the plot. f, IF of yes-associated protein 1 (YAP1) and DAPI in mPCOs grown at indicated stiffness. IF images are representative of minimum five organoids (n=2). Scale bars: 100 μm, Inlay: 25 μm. g, Nuclear/cytoplasmatic ratio of YAP1 in individual organoids from f. Mean shown as bold line. Two-sided parametric Welch’s t-test used for 1.4, 3.1 or 8.2 kPa and two-sided non-parametric Wilcoxon test for 20.5 kPa with Benjamini-Hochberg correction. Boxes show 25th and 75th percentiles with 50th percentile depicted and whiskers with 1.5 · IQR from hinges. h, Normalised Ctgf expression in mPCOs grown at indicated stiffness (n=3, parametric Welch’s t-test, error bars: s.e.m.). i, j, Mean organoid volume i, and numbers j, of mPCOs grown at indicated stiffness (n=4, two-sided parametric Welch’s t-test with Benjamini-Hochberg correction, error bars: s.e.m.).
Analysis of human PDA by IHC revealed that cytokeratin (CK)-positive pancreatic cancer cells express integrin subunits α6 and β4, whereas β1 and αV are broadly expressed across both tumour and stromal cells. Integrins α2 and α3 appeared mostly intercellular. Lama5 staining was observed basal to CK-positive tumour cells (Supplementary Fig. 11), supporting relevant roles of L511/521-integrin α6β1-, α6β4- and α3β1-mediated PCC adhesion, and collagen- and FN-mediated adhesion via integrins αVβ1 and α2β1, respectively, that may occur in vivo. Thus, we reasoned that inclusion of these adhesion cues would be needed to support epithelial and stromal cells within a synthetic hydrogel scaffold.
Optimising PEG hydrogel composition for pancreatic organoids
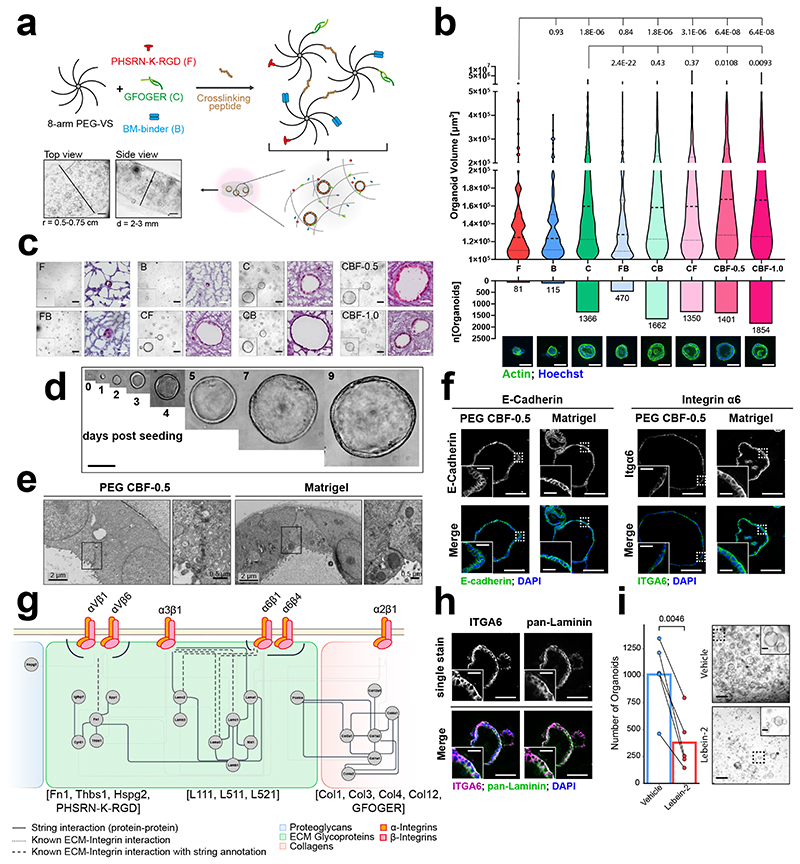
To model relevant cell-ECM interactions for pancreatic epithelial and stromal cells, we utilized an 8-arm poly-ethylene glycol (PEG)-based hydrogel system in which adhesion-linker pre-functionalized vinyl sulfone-activated PEG macromers (f-PEG-VS) are crosslinked via matrix-metalloproteinase (MMP)-sensitive peptides to form the hydrogel scaffold (Fig. 2a) 19,36. To support cell adhesion, we incorporated the fibronectin-mimetic peptide PHSRN-K-RGD (F) and the GFOGER peptide (C), which mimics both fibrillar and network forming collagen, as well as a BM binding peptide to retain cell secreted matricellular proteins more broadly 16,36–40. All adhesion-conferring peptides were fully incorporated into the PEG scaffold with limited effect on the differential elastic gel modulus (Supplementary Fig. 12).
Figure 2. Optimising PEG hydrogel composition for pancreatic organoids.
a, Illustration of 3D PEG hydrogel scaffold crafting and organoid encapsulation. b, Image analysis of murine pancreatic cancer organoids (mPCOs) grown in 3D PEG hydrogels prepared with different adhesion-mimetic peptides as indicated. Violin plots display the volume distribution of individual organoids (top) (d4, n=3, two-tailed Wilcoxon test with Benjamini-Hochberg correction). Dashed bold line indicates median organoid volume. Total organoid number and representative images of analysed mPCOs are shown (bottom). Scale bar = 50 μm. c, Representative brightfield and H&E images of mPCOs in 3D PEG hydrogels using adhesion-mimetic peptides as indicated (d4, n=3). Scale bars: Brightfield: 200 μm; H&E: 50 μm. d, Representative brightfield images of mPCOs developing in 3D PEG CBF-0.5 hydrogels (n=3). Scale bar: 100 μm. e, Representative electron microscopy (EM) images of mPCOs grown as indicated (d4). Scale bars: 2 μm, Inlay: 500 nm. Images are representative of minimum five organoids in each gel. f, Immunofluorescence (IF) images of mPCOs in PEG CBF-0.5 hydrogels or Matrigel. Scale bar: 100 μm, Inlay: 25 μm. Images are representative of minimum 5 organoids in the respective gel (d4, n=2). g, Protein-Protein interaction network of identified integrin-ECM interactions of mPCOs in 3D PEG CBF-0.5 hydrogels highlighting main ECM ligands of identified integrins. h, Dual-IF of pan-Laminin and ITGA6 in mPCOs in 3D PEG CBF-0.5 hydrogels. Images are representative of minimum five organoids (d4, n=2). Scale bar: 100 μm, Inlay: 25 μm. i, Quantification (left) and representative brightfield images (right) of mPCOs in 3D PEG CBF-0.5 hydrogels with vehicle or lebein-2 (d4, n=5, paired parametric two-sided Welch’s t-test). Individual replicates are linked. Scale bar: 500 μm, Inlay: 100 μm. Abbreviations: F – PHSRN-K-RGD, B – BM-binder, C – GFOGER, FB; CB; CF – combinations of indicated peptides, CBF-0.5; CBF-1.0 – combination of indicated peptides with PHSRN-K-RGD at 0.5 or 1 mM.
Established murine pancreatic cancer organoids (mPCOs) were disaggregated into single cells and embedded into PEG-VS hydrogels prepared with different combinations of the adhesion mimetic peptides (Fig. 2b,c). Whilst FN-mimicking and BM-binding peptides in isolation did not support organoid growth, the collagen-mimicking peptide supported expansion of mPCOs (Fig. 2b,c and Supplementary Fig. 13a). This contrasts observations with human colorectal and endometrial carcinoma organoid lines, which were sustained by FN-mimetic peptides 17,19, suggesting that synthetic matrices may need to be engineered to support tissue- and cell type-specific adhesive requirements. Notably, the combination of all three peptide anchors (CBF) significantly enhanced the number and size of organoids compared to GFOGER (C) alone (Fig. 2b,c, Supplementary Fig. 13 and Supplementary Videos 10 and 11), demonstrating a complementary effect of the adhesion peptides on organoid formation and growth. As the matricellular analysis determined the relative in vivo abundance of collagen type I and FN at 2:1 in all pancreatic matrisome samples (Supplementary Fig. 13c), we utilized the CBF-0.5 hydrogel formulation for all subsequent studies.
Using this formulation, organoids matured and developed macroscopically visible spheres throughout the entire gel within 3-5 days of seeding (Fig. 2d, Supplementary Fig. 14a). In addition, organoids exhibited similar growth kinetics (Supplementary Fig. 14c) and established cellular architecture and polarity, comparable to Matrigel when assessed by electron microscopy (EM; Fig. 2e) or immunofluorescence (IF; Fig. 2f and Supplementary Fig. 14b). Finally, organoids from both normal- and tumour-bearing pancreas were supported at similar efficacy, suggesting that cancer cells retain adhesion dependencies that mirror their non-malignant history (Supplementary Fig. 14d).
To determine whether CBF-0.5 hydrogels support the formation of a relevant ECM, we determined the deposition of ECM proteins in the PEG culture systems by LC-MS/MS (Fig. 2g and Supplementary Fig. 15). This resulted in the identification of 104 matrisomal proteins, including laminin-521, laminin-511, type 1 collagen and fibronectin, as well as the FN-binding integrins αVβ1 and αVβ6, the laminin-binding integrins α3β1, α6β1 and α6β4, and the collagen-binding integrin α2β1 (Supplementary Fig. 15). Using a dual IF analysis, we confirmed the basolateral localisation of integrin α6 in close proximity to organoid-derived laminin deposits in the PEG CBF-0.5 gels (Fig. 2h). Importantly, laminin deposits were also identified in PEG hydrogels containing only the GFOGER and PHSRN-K-RGD peptides, indicating the functional importance of the integrin α6/α3-laminin interactions for organoid growth (Supplementary Fig. 16a,b). To functionally verify this interaction, we treated mPCOs grown in PEG CBF-0.5 hydrogels with lebein-2, a disintegrin that selectively blocks the laminin-binding integrins α3β1, α6β1 and α7β1, but does not target the collagen-binding integrins α1β1 and α2β1 41–44. Lebein-2 interfered with adhesion of PCCs onto L511/L521-coated dishes in vitro (Supplemental Fig. 16c,d), and significantly reduced mPCO formation and growth in PEG CBF-0.5 gels (Fig. 2i and Supplementary Fig. 16e-h). Together, these data demonstrate that relevant cell-ECM interactions are established in PEG hydrogels and that laminin-integrin interactions play a functionally important role for the growth of pancreatic cancer organoids. In conclusion, PEG CBF-0.5 hydrogels support the formation of complex cell-ECM interactions to enable the growth and polarization of pancreatic organoids with comparable efficacy to Matrigel.
Human PDO formation in defined PEG matrices
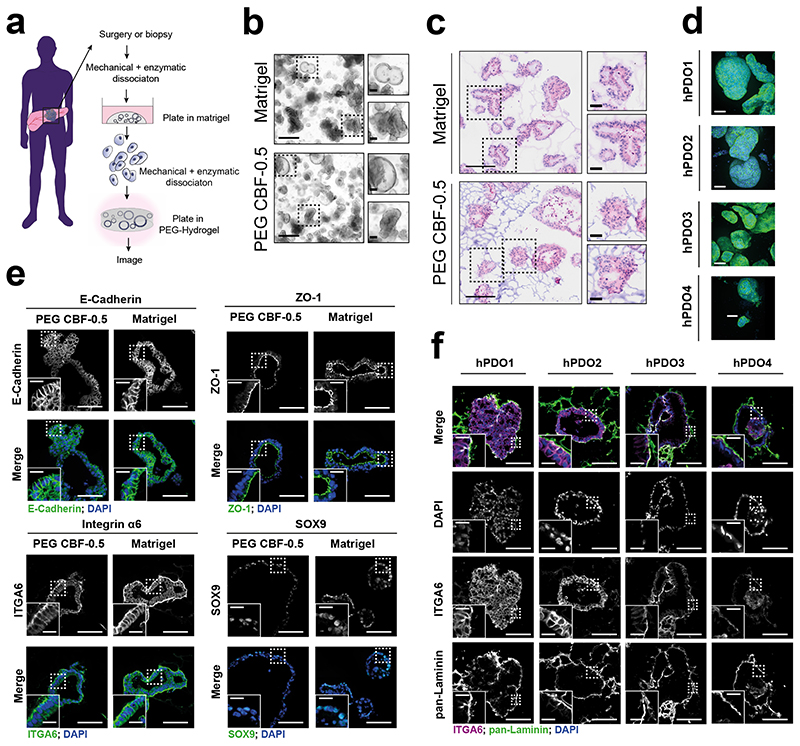
To determine whether the synthetic PEG CBF-0.5 hydrogel scaffold also supports expansion of human patient derived organoids (hPDO), we seeded established hPDOs and tracked their growth and morphology (Fig. 3a). Human PDOs were engrafted as fragments or single cells in PEG CBF-0.5 hydrogels, where they developed into fully mature spheres after 4–12 days of growth and exhibited similar morphology to hPDOs grown in Matrigel (Fig. 3b,c and Supplementary Fig. 17). Consistent with mPCOs, hPDOs exhibited a high degree of morphological heterogeneity in PEG and Matrigel culture conditions (Fig. 3c,d and Supplementary Fig. 17) 45. Immunofluorescence analysis confirmed human organoids were polarised and retained expression of ZO-1, ITGA6, SOX-9 and E-cadherin, (Fig. 3e and Supplementary Fig. 17c-e). All hPDOs express integrin α6 at the cell periphery, close to laminin deposits, indicating that laminin-integrin interactions are conserved in human pancreatic organoids (Fig. 3f). These data demonstrate that PEG CBF-0.5 hydrogels support murine and human pancreatic organoids growth in vitro.
Figure 3. Human PDO formation in defined PEG matrices.
a, Schematic of human pancreatic ductal organoid (hPDO) establishment. b, Representative brightfield images of hPDOs in 3D PEG CBF-0.5 hydrogels or Matrigel (d6). Images are representative of at least 3 independent experiments. Scale bar: 500 μm, Inlay: 100 μm. c, H&E staining of organoids from b. Scale bar: 200 μm, Inlay: 50 μm. d, Maximum intensity projection of hPDOs from four patients stained with Phalloidin and Hoechst to visualize organoid morphology and cellular structures. Images show representative morphology of organoids from each line. Scale bar: 200 μm. e, IF analysis of indicated markers in hPDO2 organoids grown in PEG CBF-0.5 hydrogels or Matrigel (d6, n=2). Scale bar: 100 μm, Inlay: 25 μm. Images are representative of minimum five organoids. f, Dual IF for pan-Laminin and ITGA6 in hPDOs grown in PEG CBF-0.5 hydrogels (d6, n=2). Images are representative of minimum five organoids. e,f, Scale bar: 100 μm, inlay: 25 μm.
Recapitulating the stiffness range of PDA in PEG hydrogels
Deregulated mechano-sensing is commonly observed in solid tumours, where a remodelled ECM promotes malignant traits 46,47. As synthetic hydrogel scaffolds can be configured to recapitulate tissue stiffening 17,36, we set out to determine whether the PEG-hydrogel scaffold reliably replicates the stiffness ranges of normal and tumour-bearing pancreas.
To determine how tissue stiffness changes as normal healthy pancreas transitions into tumour-bearing tissue, we analysed 11 murine and 3 human fresh-frozen pancreatic cancerous samples by atomic force microscopy (AFM) and compared their stiffness profile to that of three murine normal pancreata (Fig. 4a and Supplementary Fig. 18-20). Healthy murine pancreatic tissue exhibited a mean stiffness of 1.48±0.3 kPa, whilst murine tumour-bearing tissue displayed a median stiffness of 3.55 kPa and a mean stiffness of 8.48±2.2 kPa, with 90% of all measurements below ~20.1 kPa, confirming previous analyses 47–49 (Fig. 4a and Supplementary Fig. 19). Similar stiffening was observed for the three human specimens (Fig. 4a and Supplementary Fig. 20). We confirmed these findings using second-harmonic generation (SHG) imaging for linearized collagen fibres, which are known to correlate with matrisomal stiffening 47 (Supplementary Fig. 21).
To model this environment in vitro, we tuned the stiffness of PEG CBF-0.5 hydrogels by increasing the molar ratio of the crosslinking peptide to the number of free vinyl sulfone (VS) arms (hereafter referred to as crosslinking density, Fig. 4b). We profiled the resulting hydrogels using AFM and confirmed the relative increase in stiffening by rheology (Supplementary Fig. 12e), demonstrating that the entire stiffness range from normal pancreas to PDA was recapitulated at high reproducibility (Fig. 4c).
To determine the effects of increased PEG hydrogel stiffening on organoid growth, we seeded murine organoids in hydrogels tuned to replicate the pathological stiffness of PDA (Fig. 4d-e and Supplementary Fig. 22). Organoids formed efficiently in hydrogels across the entire stiffness range, displayed elevated nuclear translocation of yes-associated protein (YAP1) and increased Ctgf levels in stiffened hydrogels, consistent with engagement of mechano-signalling (Fig. 4f-h and Supplementary Fig. 23). To further profile differences in the cellular response to increased hydrogel stiffening, we seeded one mPNO and 3 mPCOs in PEG hydrogels tuned at disease-relevant stiffnesses. While all organoids exhibited a growth advantage when seeded in stiffened gels (Fig. 4i-j and Supplementary Fig. 22) 50, differences existed in the optimal stiffness for growth across the mPCOs, possibly reflecting the stiffness of the tumour from which the organoids were established. These data demonstrate that the PEG CBF-0.5 hydrogels present a suitable and reproducible model to study effects of tissue stiffening.
Modelling a heterocellular tumour microenvironment
To determine whether diverse stromal cell populations from pancreatic tumours are retained in PEG hydrogels, we collected and disaggregated tumours from orthotopically-implanted mPCOs and embedded these in PEG CBF-0.5 hydrogels (Supplementary Fig. 24). Strikingly, stromal cell populations were readily detected by IHC for pan-CK, PDPN and CD45 after eight days of culture, demonstrating the suitability of CBF-0.5 hydrogels for stromal co-cultures.
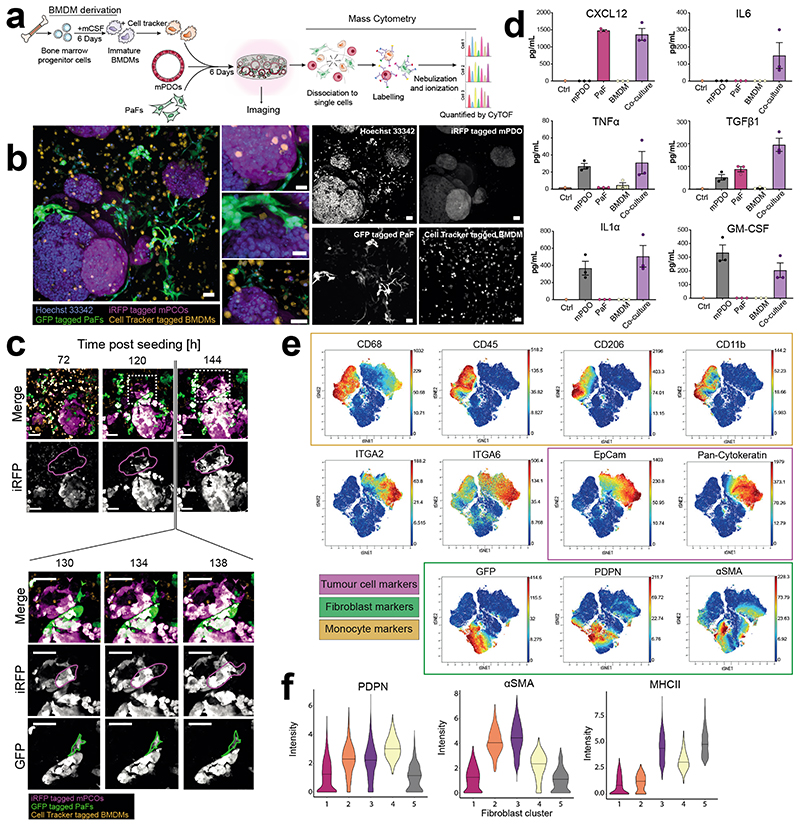
Next, we co-cultured infrared fluorescent protein (iRFP)-labelled mPCOs with green fluorescent protein (GFP)-labelled murine pancreatic fibroblasts (PaFs) and cell-tracker dye-labelled macrophages, derived from macrophage colony stimulating factor 1 (csf-1)-treated murine bone marrow-derived myeloid cells (BMDM), in PEG CBF-0.5 hydrogels (Fig. 5a). Fluorescent image, IHC, and flow analysis after 6 days of culture validated that viable PaFs and macrophages were retained in the PEG CBF-0.5 hydrogels (Fig. 5b, Supplementary Fig. 25). Next, the cellular dynamics within co-cultures was examined by live imaging for 3 days. Dynamic migratory behaviour of both tumour and stromal cells was observed within the PEG CBF-0.5 hydrogels (Supplementary Fig. 26). For example, individual tumour cells in close proximity to fibroblasts exhibited invasive behaviour and migrated away from the organoid structure (Supplementary Fig. 26 and Supplementary Videos 12-15). Moreover, consistent with previous observations, we also observed that fibroblasts exhibited an elongated, mesenchymal morphology; this was especially apparent with fibroblasts proximal to tumour cells 51 (Fig. 5c and Supplementary Fig. 26).
Figure 5. 3D PEG-VS CBF-0.5 gels support stromal co-cultures.
a, Overview of co-culture in PEG-VS CBF-0.5 hydrogels. b, Representative image of co-cultures (d6). Scale bars: 70 μm, images representative of three independent experiments. c, Representative images of co-cultures in 3D PEG-VS CBF-0.5 hydrogels at indicated time after seeding (top) and representative examples of fibroblasts and cancer cells (bottom). Images are representative of at least five individual regions centred around mPCOs. Scale bar: 60 μm. d) ELISA of conditioned medium from mono- and co-cultures grown in PEG-hydrogels for six days (n=3). Error bars: Standard error of mean. e) t-SNE visualisation of mass cytometry analysis of PEG-CBF-0.5 gel co-cultures overlaid with relative quantification of selected markers (d6, n=2). Integrin markers were not used in definition of t-SNE plots. Range of colorimetric scale is indicated for individual markers. f) Violin plots of selected markers for all detected fibroblast clusters. Bold line indicates median intensity of each marker and population.
To further interrogate the interaction of tumour and stromal cells, we determined the production of functionally important soluble signals (Fig. 5d). Several cell-specific signals such as GM-CSF, CXCL12 and IL1α were identified across mono- and co-cultures, whereas other signals (IL6 and TGFβ) increased robustly upon co-culture, demonstrating relevant tumour-stromal interactions (Fig. 5d). To further interrogate cellular phenotypes, we analysed co-cultures at single cell resolution using Cytometry by Time of Flight (CyTOF). Cells were liberated from PEG CBF-0.5 hydrogels after 6 days of co-culture and stained with antibodies directed against known markers of tumour cells, cancer-associated fibroblasts and macrophages followed by analysis (Fig. 5e and Supplementary Fig. 27). Visualisation using t-SNE distinguished cytokeratin- and EpCAM-positive epithelial mPCOs from the CD45pos/CD68pos BMDM-derived macrophages and GFPpos fibroblasts, revealing phenotypic diversity across tumour and stromal cell populations. For example, fibroblasts expressed markers of known subsets, such as αSMApos myCAFs, MHCIIpos apCAFs and MHCIIneg/αSMAneg iCAFs 51,52 (Fig. 5f and Supplementary Fig. 28). This demonstrates that stromal cell populations are readily incorporated with organoids in PEG hydrogels and exhibit signalling, phenotypic and morphological behaviour consistent with in vivo models. Thus, PEG CBF-0.5 hydrogels can be readily utilized for the studies of pancreatic cancer within a functionally relevant stromal context.
Discussion
Contemporary organoid models that replicate physiochemical characteristics and stromal abnormalities of the tumour microenvironment (TME) are needed to improve basic and translational cancer biology.
Here we combined biochemical and functional analysis of cellular adhesion dependencies to inform the design of a synthetic PEG scaffold. Interactions between cell-derived laminin 511, 521 and integrin α6/α3 subunits are recapitulated within a complex cell-ECM niche to support organoid growth. Moreover, PEG hydrogels recapitulate the entire stiffness range of murine and human pancreatic cancers and engage signalling consistent with mechano-sensing. Lastly, stromal cell populations such as pancreatic fibroblasts and bone marrow-derived macrophages 52, were readily co-cultured and fibroblasts express markers and display morphology consistent with adaptation of myCAF, iCAF and apCAF subsets 51, demonstrating that stromal cells acquire relevant phenotypes within these defined growth settings.
This rationally designed and easy to use PEG system provides exquisite growth control in a reproducible and defined manner to support future basic and translational studies of human and murine pancreatic organoids within a relevant microenvironment.
Material & Methods
Mouse models
KPC (KrasLSL-G12D/Wt; Trp53LSL-R172H/Wt; Pdx-1-Cre), KPF (KrasFrt-S-Frt-G12D/Wt; Trp53KO/WT; Pdx-1-Flp), KC (KrasLSL-G12D/Wt; Pdx-1-Cre) and KF (KrasFrt-S-Frt-G12D/+; Pdx-1-Flp) animals have previously been described 21, 23, 54. Animals were bred in-house at CRUK Manchester (CRUK MI) or Beatson Institute (CRUK BI) under pathogen-free conditions. Mice were maintained in purpose-built facility in a 12 hr light/dark cycle at uniform temperature and humidity with continual access to food and water. The genotype of animals was confirmed by Transnetyx (Cordoba, TN, USA). For orthotopic tumour cell implantation studies, 6 weeks old female CD-1 nude animals were obtained from Charles River. All animal experiments were performed under a UK Home Office License and in accordance with the ‘Animal (Scientific Procedures) Act of 1986’ under Project License Number 70/8745 and 70/8375 subject to review by the Animal Welfare and Ethical Review Body of Cancer Research UK Manchester Institute, University of Manchester (CRUK MI) and the University of Glasgow (UoG). Experiments are reported in accordance with Animal Research: Reporting of In Vivo Experiments (ARRIVE) 2.0 guidelines.
Patient Samples
Research samples were obtained from the Manchester Cancer Research Centre (MCRC) Biobank with informed patient consent obtained prior to sample collection. The MCRC Biobank (ethics code: 18/NW/0092) is licensed by the Human Tissue Authority (license number: 30004) and is ethically approved as a research tissue bank by the South Manchester Research Ethics Committee (Ref: 07/H1003/161+5). The role of the MCRC Biobank is to distribute samples. For more information see www.mcrc.manchester.ac.uk/Biobank/Ethics-and-Licensing.
Cell culture of murine and human pancreatic cancer cell lines
Work with mammalian cancer cell lines and organoids was performed under sterile conditions. Cells were grown at 37°C, 5% CO2, humidified air. Murine KPC-1 cells from obtained from Jennifer Morton at CRUK Beatson Institute, UK; iKras cells kindly shared by Dr Ronald DePinho (MD Anderson Cancer Research Centre, TX, USA); KPC-43 cells were a kind gift from Dr Kris Freese (CRUK MI); human Suit-2 cells were obtained from ATCC; human H6c7 immortalized adult pancreatic epithelial cells (HPDE) were obtained from Ming Tsao, University of Toronto, University Health Network Toronto, Canada 55, 56. All cells were confirmed mycoplasma free and cultured in Dulbecco’s Modified Eagle Medium (DMEM; Thermo) supplemented with 10% v/v Fetal Bovine Serum (FBS; Life Technologies) and 1x Hyclone solution (Invitrogen), hereafter referred to as DMEM 10% v/v FBS. HPDE cells were grown in Keratinocyte serum free medium (SFM) supplemented with 0.2 ng/mL EGF (Invitrogen), 30 μg/mL bovine pituitary extract (Invitrogen, #17005042) and 1x HyClone solution. Pancreatic cancer cell lines (PCCs) or HPDEs were allowed to reach a maximum confluence of 80% and split using 0.25% v/v Trypsin/Versene (Invitrogen). iKras cells were cultured in DMEM 10% v/v FBS supplemented with 1 μg/mL Doxycycline (Sigma-Aldrich) at all times.
Establishment and culture of murine normal (mPNO) or cancerous (mPCO) pancreatic organoids (mPOs)
mPOs were isolated from wild-type C57-Black6 pancreata or tumour-bearing KPC mice with histologically verified PDA 10,57. Tissues were minced (<0.5mm in diameter) and enzymatically digested with 5 mg/mL Collagenase-II (ThermoFisher) in advanced DMEM F12 (gibco) supplemented with 1x GlutaMax (gibco), 0.1M HEPES (Thermo) and 50 U of Penicillin-Streptomycin (Thermo), hereafter referred to as AdF Base medium for 45 minutes at 37°C, shaking (700 rpm). Cells were spun, washed and seeded in 20 μL droplets of growth-factor reduced phenol-red- and LDEV-free Matrigel (Corning) in 24 well plates and cultured in AdF base medium supplemented with 1 mM N-acetyl cysteine (Sigma Aldrich), 100 ng/mL Wnt3a (R&D Systems), 100 ng/mL human Noggin (PreproTech), 100 ng/mL FGF-10 (PreproTech), 10 mM Nicotinamide (Sigma Aldrich), 0.5 μM A83-01 (BioTechne), 500 ng/mL R-Spondin 1 (R&D Systems) and 0.01 μM Gastrin (Sigma Aldrich), hereafter termed hPOCM. For mPNOs, EGF (Invitrogen) was added to the hPOCM to a final concentration of 50 ng/mL. Media was changed every 2 days to allow consistent mPCO growth. For passaging, following removal of the growth media, Matrigel domes were depolymerised in ice-cold PBS. mPOs were mechanically dissociated into fragments using a 200 μL pipette tip, and re-plated at a 1:6 split into new 20 μL Matrigel droplets. Frozen organoid stocks were established by mixing dissociated organoid fragments with Recovery Cell Culture Freezing Medium (RCFM, gibco) followed by gentle cryopreservation. Organoids were thawed by rapidly warming cryotubes and following washing mPCOs twice in warm AdF base medium, mPCOs were seeded in 20 μL droplets of Matrigel. Growth media was replaced every other day for the first two days of culture following thawing and mPCOs were passaged at least once before usage for experimental procedures.
Culture and Establishment of human pancreatic organoids (hPDOs)
hPDOs were established from fine-needle biopsy (FNB) or resected pancreatic cancer specimens10. Tumour tissue was minced and enzymatically digested with 5 mg/mL Collagenase-II (ThermoFisher) in AdF Base medium for 45 minutes at 37°C, shaking (700 rpm). Upon removal of red blood cells using red-blood cell lysis buffer (Merck), cells were spun, washed and seeded in 20 μL droplets of Matrigel and grown in hPOCM. For passaging, following removal of the growth media, Matrigel domes were depolymerised in ice-cold PBS. hPDOs were mechanically dissociated into fragments using a 200 μL pipette tip and re-plated at a 1:2 split into new 20 μL Matrigel droplets. Frozen organoid stocks were established by mixing dissociated organoid fragments with RCFM followed by gentle cryopreservation. Organoids were rapidly thawed and washed twice in warm AdF base medium. hPDOs were seeded in 20 μL droplets of Matrigel in enriched hPOCM (enrichment consisted of addition of 50 ng/mL EGF (Invitrogen) and increasing concentrations of Wnt3a [200 ng/mL] and RSpondin 1 [1 μg/mL]). hPDOs were passaged at least once before usage for experimental procedures.
Adhesion Assays
For pancreatic cancer cell (PCC) or pancreatic fibroblast (PaF) adhesion assays, fluorophore-labelled PCCs or PaFs were prepared in single-cell suspension. Cells were then extensively washed in DMEM without FBS and 2000 single cells were seeded in DMEM without FBS onto pre-coated wells of a glass-bottom 96-well plate (CellCarrier ultra, PerkinElmer). For coating, 96-well plates were incubated with ECM ligand overnight at 4°C in 70 μL PBS containing magnesium and calcium (PBS++, ThermoFisher Scientific) followed by another incubation for 30 minutes at RT. Wells were pre-coated with ECM ligands: rat-tail collagen-1 (0.1 mg/mL, Corning, 35429), fibronectin from bovine plasma (5 μg/mL, Sigma Aldrich, F1141), Laminin 10 (5 μg/mL, Biolamina, LN511), Laminin 11 (5 μg/mL, Biolamina, LN521) or combinations of those proteins. For collagen-coating, the collagen-1 solution was carefully neutralized with 1N NaOH (Sigma Aldrich) according to the manufacturer’s instructions in ultrapure water supplemented with 10x PBS. ECM Ligands were then removed and wells were washed twice with PBS. Upon seeding of the cells, the plate was spun at 50 x g for one minute to collect cells at the bottom of the culture plate and imaging started immediately. Live imaging was conducted using the confocal fluorescence-high content screening via the PerkinElmer Opera Phenix (PerkinElmer Waltham, MA), a confocal spinning disk 4 laser (405nm 50mW, 488nm 50mW, 591nm 50mW, 640nm 50mW) fixed light path system, with a range of emission filters (435-550nm, 435-480nm, 500-550nm, 570-630nm, 650-760nm) and environmental control set to 37°C, 5% CO2 throughout the entire experiment. Four Zyla sCMOS cameras, 2160x2160 pixels, 6.5um pixel size (Andor, Belfast, UK) are set up for each dedicated lightpath. A Zeiss W Plan-Apochromat X20 water objective NA 1.0 WD 1.17 mm was used. Prior to the time-course imaging, the vertical position of the z-stack was adjusted and a z-stack of +- 6 μm around the cell-adhesion plane was obtained for each of nine fields of view in each well. Images were obtained every 10 minutes for each well and condition for the first six hours of the experiment followed by imaging every 20 minutes till 12 hours post seeding. For blocking studies on human Suit-2 cells, prior to seeding, 37,500 single cells were pre-incubated for 30 minutes at 37°C, 5% CO2 with blocking antibodies anti-Itga6 (10 μg/mL, Merck Millipore, MAB1378, Clone NKI-GoH3) or anti-Itga3 (10 μg/mL, ThermoFisher, 17-0494042, Clone P1B5) or combinations in 250 μL of DMEM. 15,000 single cells were seeded in 100 μL DMEM for blocking studies. PBS was used as vehicle control. For Lebein-2 adhesion assays, 2,000 murine KPC-1 cells were seeded onto Laminin 511/521 pre-coated surfaces (5 μg/mL each) in the presence of either 10, 20 or 40 μg/mL Lebein-2 or a vehicle control in 100 μL DMEM. 50 mM Hepes, 150 mM NaCl solution (pH 7,4) was used as vehicle control. Following acquisition, cell masks were detected from the fluorophore channel and the cellular area was calculated using the Harmony software suit (Perkin Elmer, V4.8). Minimum 100 cells were analysed for each condition. Representative images of cells from adhesion assays were exported using the Harmony software suit (Perkin Elmer, V4.8) in “highlighted” settings with gamma set to 2.3 to aid the visual interpretation of the images.
PEG, synthetic peptides and hydrogel formulation
8-arm 20 kDa PEG vinyl sulfone (PEG-VS) macromers were purchased from JenKEM Technology (Beijing). All peptides were custom synthesized and purified (>95%) by Aapptec or Genscript. Following peptide were used in this study 16,19: “MMP-CL”, a dithiol crosslinking peptide containing a matrix metalloproteinase (MMP)- and Sortase-sensitive recognition site, (Ac)GCRD-LPRTG-GPQGIWGQ-DRCG(Am); “PHSRN-K-RGD”, a fibronectin (FN) derived adhesion peptide containing the PHSRN synergy site and the RGD motif from the 9th and 10th FN type III repeat respectively in a branched assembly, (Ac)PHSRNGGGK-[GGGERCG(Ac)]-GGRGDSPY(Am)40, labelled with the single letter “F” in the gels; “GFOGER”, a triple-helical collagen-I derived adhesion peptide, (Ac)GGYGGGPG(GPP)5GFOGER(GPP)5GPC(Am)17, labelled with the letter “C” in the gels; “BM-binder”, a peptide with high affinity for BM-derived proteins specifically collagen type IV and laminin, (Ac)GCRE-ISAFLGIPFAEPPMGPRRFLPPEPKKP(Am)40, labelled with the letter “B” in the gels. All peptides were reconstituted in ultrapure water (Thermo) at a concentration of 20 mM for all adhesion mimetic and BM-binding peptides and 50 mM for the MMP-CL crosslinker. The concentration of free thiols was determined using the Ellman’s Reagent (Thermo) according to the manufacturer’s instructions using an L-cysteine standard (Thermo).
Encapsulation of mPCOs as mono or co-cultures with PaFs and BMDMs in PEG-VS hydrogels
Synthetic hydrogels were fabricated as described previously 19. In brief, hydrogels were crafted using Michael-type reaction chemistry inside a 1 mL syringe (BD) modified by removal of the tip. All hydrogels were prepared by reacting 1.5 mg of a 8-arm 20kDa-PEG-VS macromer (2.4 mM PEG-VS (19.7 mM-VS) at 98% substitution) with the corresponding thiol-containing (-SH) adhesion and/or BM-binding peptides, in 1 x PBS/1 M HEPES (pH 7.8, Gibco). For adhesion screening experiments, we prepared a series of peptide-functionalized PEG macromers (fPEG-VS) as follows: for hydrogels C, F, and B, we used 1 mM of GFOGER (C), 1 mM of PHSRN-K-RGDS (F), and 1 mM of BM-binder (B), respectively. For hydrogels CF, CB, and FB, we used a combination of two different peptides (GFOGER, PHSRN-K-RGD, or BM-binder) at 1 mM each, for a final peptide concentration of 2 mM in each gel formulation. The CBF-0.5 hydrogel was made using 1 mM of GFOGER, 1 mM PHSRN-K-RGDS and 0.5 mM BM-binder, Finally, for the CBF-1.0, we combined 1 mM of GFOGER, 1 mM PHSRN-K-RGDS and 1 mM BM-binder. The resulting fPEG-VS solutions were incubated for 30 minutes at 37°C in a humidified incubator to allow complete functionalization of the PEG-VS arms.
Organoids were retrieved from Matrigel, dissociated into single cells by addition of 1x TrypLE Express (gibco) for 10 minutes at 37°C, diluted in AdF base medium and re-suspended in minimal volume of AdF base. GFP-PaF and BMDM cells were collected using Accutase (Thermo) for 5 mins at 37°C and diluted in DMEM v/v 10% FBS followed by re-suspension in a minimal volume of AdF base. The fPEG-VS solution was mixed with a cell-suspension containing 10,000 PDO cells/μL to give a final concentration of 1,000 cells/μL hydrogel for mono-culture experiments. For co-culture experiments PaFs and BMDMs were each included at 50,000 cells/ μL in cell-suspension for a final concentration of 5,000 cells/μL hydrogel generating a 1:5:5 ratio of PDO:Paf:BMDM. The fPEG-VS-cell mixture was polymerised by addition of MMP-CL crosslinker followed by incubation for 45 minutes at 37°C in a humidified incubator. CBF-0.5 hydrogels with different crosslinker density were prepared as follows: 25% (4.95 mM), 30% (5.92 mM), 35% (6.89 mM), 40% (7.88 mM), 45% (8.86 mM), 50% (9.85 mM), 55% (10.83 mM), and 60% (11.82 mM). Polymerised gels were extracted from the modified syringes using a scalpel and dispensed into 24-well plates filled with 1 mL of hPOCM for mono- or m-hPOCM for co-culture experiments. Care was taken to fully submerge the gels in the growth media. Cultures were maintained in a humidified incubator at 37°C, 5.0% CO2 with media changes every 2 days.
Immunofluorescence and Immunohistochemistry of hydrogels
Synthetic hydrogels or Matrigel plugs were embedded in O.C.T. medium (TissueTek) and snap frozen in a dry ice / isopentane (Fisher Scientific, 10407010) mix for 5-10 minutes and prepared as 7 μm thick sections using the cryotome instrument (Thermo Fisher Scientific Cryostar NX70). For immunofluorescence (IF), samples were then fixed for 15 minutes in 4% PFA and washed in PBS followed by blocking with 10% casein solution (Vector labs SP-5020). For E-Cadherin and Sox9, cells were fixed in (1:1) Acetone/Methanol for 15 minutes at 4°C. Primary rabbit anti-ZO1 (1:100, Invitrogen, 61-7300); rat anti-CD49f (1:500, Merck Millipore, MAB1378, clone NKI-GoH3); rabbit anti-E-Cadherin (1:100, Cell Signalling, 3195, Clone 24E10); rabbit anti-Sox9 (1:300, Merck Millipore, AB5535), rabbit anti-Laminin (1:50, abcam, ab11575) and rabbit anti-Yap1 (1:100, Cell Signaling Technology, 4912s) antibodies diluted in PBST were incubated for 1 hour at RT. For anti-Yap1 samples were stained overnight at 4°C. After washing in PBST 3x for 5 minutes each, samples were incubated with Alexa-488 conjugated donkey anti-Rabbit (1:400, Invitrogen, A21206) or Alexa-488 conjugated donkey anti-Rat (1:400, Invitrogen, A21208) secondary for 30 minutes at RT. For dual staining samples were incubated with Alexa-488 conjugated donkey anti-Rabbit (1:400, Invitrogen, A21206) and Alexa-647 conjugated goat anti-Rat (1:400, Invitrogen, A21247) antibodies. Following washing in PBST, cells were incubated for 10minutes at RT with DAPI (1:2000, Thermo Fisher, 62248). Slides were coverslipped using Prolong Gold Antifade (Thermo Fisher P36930) and imaged in confocal fluorescence on the Olympus VS-120 instrument (Olympus) using a 20x objective following pre-scanning using a 4x objective to account for the focus position. Representative organoids were selected and exported from whole scanned slides using the QuPath software58 (QuPath; V: 0.2.3) to Tagged Image Format File (TIF) format. To aid visual interpretation of the data, images were scaled and care was taken to apply the same scaling for comparative samples (Supplementary Data 2 and 3). For quantitation, images were exported without compression or rendering to the TIF format. Quantification of IF staining was conducted on exported ROIs in the open source Fiji software suite (V. 1.52e, NIH)59. For YAP1 quantification, staining intensities were exported using the plot profile function in Fiji from a segmented line (width = 40 px) drawn around the periphery of the organoid of interest without any alteration. DAPI signal was normalized by maximum intensity for each organoid and YAP1 intensities localized in gated nuclear regions (normalized DAPI ≥ 0.25, threshold validated by visual confirmation) were divided by intensities in cytoplasmatic regions for each organoid. At least 20 organoids were quantified from three independent staining. For pan-Laminin deposit quantification, images were converted to 8-bit and a common threshold (10 – 255) was applied which was kept constant between all samples and measurements in Fiji. Organoid area was estimated (Equation 1) where d 1 and d 2 are the organoid’s horizontal or vertical extensions respectively in R (V4.0.0) as measured using Fiji.
| (Equation 1) |
For immunohistochemistry (IHC), hydrogels were embedded in OCT, snap frozen and sectioned at 7 μm. Sections were fixed for 15minutes in 4% PFA at RT and washed in PBS. For podoplanin staining cells were fixed in 1:1 Acetone/Methanol for 15 minutes at 4°C. Endogenous peroxidase was blocked using 3% H2O2 in PBST for 10 minutes at RT followed by 2x PBSt washes for minutes each. Non-specific antibody binding was blocked with 10% casein for 20 minutes at RT. Cells were then stained using primary rabbit anti-F4/80 (1:100, Cell Signalling Technology, 70076), rabbit anti-PanCK (1:100, Abcam, ab9377), rat anti-CD45 (1:100, BD biosciences, 550539, clone 30-F11) or rabbit anti-Podoplanin (1:500, Thermo Fisher, MA5-29742) antibodies diluted in PBST for 1hour at RT. Following extensive washing, cells were stained with HRP conjugated goat anti-Rabbit secondary antibody (RTU, Dako, K4003) or HRP conjugated goat anti-Rat secondary antibody (RTU, Vector Laboratories, MP-7444) for 30 minutes at RT. Liquid DAB+ (Dako, K3468) chromogen was applied for 5 minutes at RT followed by counterstaining in Gills I haematoxylin (Thermo Fisher, 6765006) for 30 seconds. After mounting in Pertex mounting medium (CellPath, SEA-0100-00A), slides were scanned on the Leica SCN400 (Leica Microsystems, Milton Keynes, United Kingdom), using a x20 objective lens under brightfield using the Leica LAS software to control the hardware and export the data to Tagged Image Format File (TIF) format. Raw Leica format files are initially stored onto a Synology RAID system and the exported to a read only central storage solution so to ensure data integrity.
High-content image analysis of hydrogels
Hydrogels were washed in PBS and following fixation for 30 minutes in 4% PFA and permeabilization for 15 minutes in 0.1% Triton X-100 and blocking in 2% BSA, 1% FBS in PBS at RT for 4h-12h, cells were incubated with Alexa 488-conjugated Phalloidin (1:200, Thermo) and Hoechst (1:200, Thermo) in blocking buffer overnight at 4°C with gentle shaking. After extensive washing in PBS, hydrogel plugs were transferred into glass-bottom 24-well imaging plates (CellCarrier; Perkin Elmer) and imaged using confocal fluorescence-high content screening via the Opera Phenix (PerkinElmer Waltham, MA), a confocal spinning disk 4 laser (405nm 50mW, 488nm 50mW, 591nm 50mW, 640nm 50mW), with a range of emission filters (435-550nm, 435-480nm, 500-550nm, 570-630nm, 650-760nm). Four Zyla sCMOS cameras, 2160x2160 pixels, 6.5um pixel size (Andor, Belfast, UK) are set up for each dedicated light-path. A Zeiss EC Plan Neofluar X10 air objective NA 0.3 WD 5.2mm or Zeiss W Plan-Apochromat X20 water objective NA 1.0 WD 1.17mm were used as indicated. Pre-scan directed z-stack acquisition of each individual sphere within the gel plug was used for mono-culture experiments. For quantitative mono and co-culture experiments z-stacks were taken throughout the gel plug and organoids were counted using Harmony 4.8 (PerkinElmer Waltham, MA) software suit in 3D rendered images. Representative images were exported from Harmony 4.8 in the portable network graphic format with scaling equally applied to all conditions.
Lebein-2 isolation and purification
Lebeins were isolated from the venom of Macrovipera lebetina obtusa (L1126, Latoxan, FR) as described previously41, In brief, upon gel filtration of the venom proteins on a Sepharose 6 column (GE Healthcare) in 20 mM sodium phosphate, pH 6.5, 50 mM sodium chloride, and 1 mM EDTA, the relevant fractions were pooled, diluted with 10 mM MES, pH 5.7, and loaded onto a Mono S column (GE Healthcare). Lebein-2 was then eluted in linear sodium chloride gradients at 80 and 140 mM, respectively, and further purified on a C8 reversed-phase column (Nucleosil, Macherey Nagel) in a linear gradient from 0.1% trifluoroacetic acid in water to 80% acetonitrile in 0.08% trifluoroacetic acid/ water. After lyophilization, the lebeins were dissolved in a 50 mM Hepes, 150 mM NaCl solution (pH 7,4) and kept at -80°C until usage. Lebein-2 charges used in this study were validated for effectiveness in adhesion assays.
Comparison of growth between Matrigel and PEG CBF-0.5 hydrogels and Lebein-2 treatment
To evaluate mPDO growth rate, single-cell suspensions of iRFP-labelled mPDOs were generated and encapsulated in PEG CBF-0.5 or Matrigel at 1,000 cells/μL hydrogel as previously described. Growth was assessed by measuring iRFP signal using Odyssey® imaging system (Licor). For Lebein-2 treatment assays, mPCOs were incorporated in PEG CBF-0.5 hydrogels as previously described at a concentration of 1,000 cells per μL hydrogel. Upon gelation, cells grown in hPCOM supplemented with 40 μg/mL Lebein-2 or a vehicle control. Media was half-replenished every day and new Lebein-2 was added to the cells to maintain the inhibition. 50 mM Hepes, 150 mM NaCl solution (pH 7,4) was used as vehicle control.
Reverse transcription - quantitative polymerase-chain reaction (RT-qPCR)
RNA of murine organoids grown in synthetic gels was isolated using the TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. In brief, organoids were washed twice with PBS for 5 minutes at RT shaking and gels were thereafter snap frozen and minced using a mortar and pestle in the presence of liquid nitrogen. Thereafter, 1.5 μg of RNA were reverse transcribed using the RevertAid H Minus Reverse Transcriptase (Thermo Fisher), Random hexamers (Thermo Fisher) and SsoFast EvaGreen Supermix (1725201, Bio-Rad) according to the manufacturers’ instructions. The quantitative polymerase chain reaction (qPCR) was conducted on cDNAs using the QuantStudio real-time PCR system (Thermo Fisher) and analyzed using the QuantStudio Design & Analysis software (applied biosystems by Thermo Fisher Scientific, V1.5.1, 2016). Expression values were obtained by applying the delta-delta Ct method60 and normalized to β-Actin (Actb) in R (V4.0.0). The selected primers are designed to amplify short fragments (99 to 121 bp) and validated to obtain optimal PCR efficiency between 85% and 115% efficiency. The list of primers and their validation parameters are described in Supplemental Methods Table 1.
Atomic force microscopy
Tissue pieces isolated from the diseased portion of the pancreas and spanning the entire width of the tumour for murine samples (d = 1-1.5 cm and 0.5-1 cm for human samples) were embedded in O.C.T. medium and snap-frozen on dry ice submerged in iso-pentane (Sigma Aldrich). 14-μm thick sections were cryosectioned from the frozen tissue blocks stored at -20°C on glass slides (Superfrost Plus, Thermo) until required for AFM. Prior to use, sections were brought up to room temperature and rested for approximately 30 minutes to allow the sections to fully adhere to the glass slide. Following this the sections were washed with copious amounts of ultrapure water to ensure complete removal of OCT. Sections were then air-dried overnight at RT. Each section was then rehydrated in ultrapure water for at least 10 minutes at RT immediately before data acquisition. AFM indentations were acquired using a Bruker Catalyst with a Nanoscope V controller (Bruker UK Ltd., Coventry UK) operating under Nanoscope software (v 4.8). The system was mounted on a Nikon Eclipse Ti-U inverted optical microscope (Nikon Instruments Europe B.V.) equipped with a Hamamatsu ORCA-05G camera. For tissue sample indentations, 5 μm diameter gold colloid spheres mounted on cantilevers with a nominal spring constant of 0.1 N/m were used (CP-qp-CONT-Au-B, sQube, Germany). Cantilevers were calibrated before each experiment using the thermal oscillation method in air. For each tissue sample, force measurements were acquired from nine areas distributed over representative stromal and epithelial areas (as determined from an adjacent, H&E stained section). In each area data was obtained from 100 individual indentations, evenly spaced over a 25 μm2 area, with a ramp size of 4-6 μm and a maximal indentation depth (trigger threshold) of 40 nm (<0.5% of tissue sample depth). For biophysical profiling of hydrogels, gels were prepared 24 hours prior to analysis. 20 μL of gel was pipetted onto pre-cleaned Poly-L-Lysine pre-coated glass slides (Thermo) and allowed to polymerize at 37°C for 40 minutes. Gels were immersed in PBS and stored overnight at 37°C. The gels were washed twice in PBS prior to AFM measurements. For each PEG-gel, ten force maps were obtained consisting of 25 individual indentations evenly acquired over 25 μm2. Data analysis for both the tissue samples and the gels was performed using the Bruker NanoScope Analysis software suite (Version 4.8) after baseline correction. The Hertz model was used to determine the elastic properties of the sample (Equation 2) with F being the force, E being Young’s Modulus, v being the Poisson’s ratio, R being the radius of the indenter and δ being the indentation. The adhesion force was not included and a maximal and minimal force fit boundary of 80% and 20% respectively of the extension curve were used during all calculations. Values exceeding a Young’s modulus of x = μ ± 2 · σ within each force map were removed from the analysis. A Poisson’s ratio of 0.5 was assumed during calculations of the Young’s moduli.
| (Equation 2) |
Integrin adhesion complex isolation
Integrin adhesion complexes (IACs) of PCCs were isolated as previously described30. In brief, six 10 cm diameter dishes for each condition and cell line were coated with 10 μg/mL FN (Sigma-Aldrich; F1141) in PBS containing calcium and magnesium (PBS++, Thermo Fisher Scientific) to stimulate FN-mediated adhesion. PCCs were cultured, passaged and single cells seeded onto each FN-coated 10 cm dish in 5 mL of DMEM 10% (v/v) FBS. Following incubation at 37°C for either 3h or overnight, the media was removed, and cells were washed twice with pre-warmed DMEM-HEPES (Sigma-Aldrich) to remove all non-adherent cells. Integrin-adhesion complexes were then crosslinked by adding a 6 mM dimethyl 3,3’-dithiopropionimidate dihydrochloride (DTBP; Sigma-Aldrich; D2388) in DMEMHEPES solution for 5 min at 37°C. DTBP was removed and quenched with 200 mM Tris-HCl pH 8 (Sigma-Aldrich; Cat.-No.: 252859), for 2 min at RT. Cells were washed twice with ice-cold PBS and lysed in modified radioimmunoprecipitation assay buffer (RIPA: 50 mM Tris·Cl (pH 7.6); 150 mM NaCl; 5 mM disodium EDTA (pH 8); 0.5% w/v SDS; 1% v/v Triton X-100; 1% v/v sodium deoxycholate in ddH2O) buffer for 2 min at 37°C. Cell bodies were removed using a high-pressure water wash. Protein complexes remaining attached to dishes were washed in cold PBS and recovered in 100 μL of adhesion complex recovery solution (125 mM Tris·Cl, pH = 6.8; 1% w/v SDS; 150 mM dithiothreitol (DTT)) by scraping. Protein complexes were then precipitated overnight using four volumes of -20°C acetone at -80°C. Precipitated proteins were centrifuged at 16,000 xg for 20 min at 4°C and protein pellets washed with -20°C acetone (16,000 x g, 20 min, 4°C). Proteins were then allowed to dry at room temperature for about 20 minutes. Precipitated complexes were resuspended in 2x reducing sample buffer (RSB; 50 mM Tris-HCl, pH 6.8, 10% (v/v) glycerol, 4% (w/v) SDS, 0.004% (w/v) bromophenol blue, 15% (v/v) β-mercaptoethanol) and heated to 70 °C for 10 min and then heated for three minutes at 95°C.
In-gel tryptic digest of adhesion complexes and LC-MS/MS
Protein samples were loaded on 4-15% polyacrylamide gels and electrophoretically separated for 3 minutes at 200 volts to allow proteins to fully migrate into the gel. Gels were then stained with Coomassie blue solution (Bio-Rad) for 10 minutes at RT, and protein-bands were cut into 1 mm cubed pieces and transferred to a 96-well PVDF plate (FiltrEx; Corning; Cat.-No.: 3504) with a pore size of 0.2 μm. Gel pieces were then washed with a freshly prepared 25 mM NH4HCO3 (Sigma Aldrich) solution in H2O followed by another wash in 100% (v/v) acetonitrile to dry the pieces. Samples were then washed twice with 100% (v/v) acetonitrile for 5 minutes and gel pieces dried using a vacuum centrifuge for 30 min at RT. Proteins were reduced by incubation in 10 mM dithiothreitol (DTT) diluted in 25 mM NH4HCO3 for 1 h at 56 °C and alkylated in 55 mM iodoacetamide diluted in 25 mM NH4HCO3 for 45 min at RT in the dark. Samples were washed using alternating 25 mM NH4HCO3 and acetonitrile for 5 minutes each and gels dried for 25 min at RT in the vacuum centrifuge. 50 μL of 12 μg trypsin (Pierce) in 25 mM NH4HCO3 was added to each sample and incubated for 15 minutes at 4°C followed by an overnight incubation at 37°C. Upon digestion, peptides were extracted using 100% (v/v) acetonitrile in 0.2% (v/v) formic acid (FA, Thermo Fisher Scientific) for 30 minutes at RT followed by 50% (v/v) acetonitrile in 0.1% (v/v) formic acid into a clean tube. Peptides were dried using a vacuum centrifuge and subsequently resuspended in 5% (v/v) acetonitrile in 0.1% (v/v) formic acid to conduct desalting using 5 mg of POROS R3 beads (Applied Biosystems). Beads were conditioned using 50% (v/v) acetonitrile followed by adding 0.1% (v/v) FA in HPLC-grade water. Peptides were then washed using 0.1% (v/v) FA in HPLC-grade water and eluted in 30% (v/v) acetonitrile in 0.1% (v/v) FA, dried and resuspended in 5% (v/v) acetonitrile in 0.1% (v/v) FA for LC-MS/MS.
Peptides were analysed using an Ultimate 3000 RSLCnano system (Thermo Scientific) coupled to an Orbitrap Elite mass spectrometer (Thermo Fisher Scientific). Peptides were loaded onto a pre-column (200 mm x 180 μm i.d., Waters) and separated on an bridged ethyl hybrid (BEH) C18 column (250 mm x 75 mm i.d., 1.7 μm particle size) (Waters) over a 2h gradient from 8 to 33% (vol/vol) ACN in 0.1% (v/v) FA at a flow rate of 200 nL/min. Liquid chromatography tandem MS analysis was performed using a data-dependent mode to allow selection of peptide fragmentation in an automated manner. The resulting data were searched against the SwissProt database with species set to Mus musculus on an in-house Mascot server (Matrix Science; 2016) in ProteomeDiscoverer (Thermo Fisher Scientific, V. 2.1). Search parameters included peptide modifications for carbamidomethylation (C) as static modification and oxidation (M, P and K) as well as deamination (N, Q) as dynamic modification. A decoy database search was performed to determine the peptide FDR with the Percolator module. A 1% peptide FDR threshold was applied, and peptides were filtered for high peptide confidence, minimum peptide length of 6, and finally peptides without protein reference were removed. All proteins that exhibited a confidence of less than ‘high’ and with less than two uniquely identified peptides were excluded from further analysis.
ECM-enrichment of murine pancreatic tumours
ECM-enrichment was conducted as described previously61. In brief, tumours were snap frozen in liquid nitrogen upon excision from diseased animals histologically verified PDA and stored at -80°C until preparation. For normal pancreata, the organs were excised from 10 healthy C57-Bl/6 animals and immediately flash frozen in liquid nitrogen. Care was taken that access fat was removed from the pancreas prior to preparation. Frozen samples were then weighed, minced using a mortar and pestle in the presence of liquid nitrogen and re-suspended in lysis buffer (buffer C (Merck Millipore) supplemented with 1x protease inhibitor (Merck Millipore)) to a final concentration of 10 mg/100 μL lysis buffer. ECM proteins were enriched from 50 mg of the sample (500 μL buffer homogenate) material using the compartmental protein extraction kit (Merck Millipore) according to the manufacturer’s instructions. In brief, cytosolic, nuclear, membranous and cytoskeletal proteins were extracted and ECM proteins were washed twice in buffer W and three times in PBS before transferring to MS preparation. All buffers were supplemented with 1x protease inhibitor (Merck Millipore) and for buffer N, 3.5 μL Benzonase (ThermoFisher Scientific) were supplemented to digest genomic DNA and RNA. Extractions were conducted at least three times on individual days for all tumour samples unless otherwise stated. For the derivation of a normal pancreas matrisome atlas and to verify the consistency of the method, 10 pancreata were combined, minced, and obtained as homogenate. Individual 10 extractions were performed in batches of 5 on two consecutive days to control for intra- and inter-extraction variability.
S-trap based tryptic digestion of ECM enriched fractions and LC-MS/MS
ECM proteins were prepared for LC-MS/MS using S-Trap midi-columns (Protifi). Samples were pulverized with frozen urea and re-suspended in 8M urea supplemented with 5% w/w SDS, 20 mM HEPES and 50 mM TEAB and harshly sonicated. Samples were then reduced in 5 mM tris(2-carboxyethyl)phosphine (TCEP, Sigma Aldrich) for 2h at 37°C, 1400 rpm (Thermomix) and alkylated using 15 mM MMTS (S-Methyl methanethiosulfonate, Sigma Aldrich) for 30 minutes at RT. Urea was then diluted to 2M by adding 50 mM TEAB and proteins were deglycosylated using PNGaseF (New England Biolabs) for 2h at 37°C, 1400 rpm. Following sonication samples were acidified to a final concentration of 1.2% (v/v) phosphoric acid (Fisher) and dissolved in S-Trap binding buffer (STBB: 90% v/v aqueous methanol, 0.1 M TEAB, pH 7.1) at a volumetric ratio of 1:7 lysate:STBB. The acidified SDS-containing lysate in STBB was then spun into the S-Trap midi spin columns (Protifi) at 4000 x g for 30 sec to trap the proteins until the entire protein-containing lysate was passed through the columns. Trapped proteins were then washed with STBB four times and proteins were digested overnight at 37°C in 50 mM TEAB using sequencing grade trypsin at a ratio of 1:10 in a humidified incubator. Following digestion, peptides were eluted in 500 μL 50 mM TEAB followed by 500 μL 0.2% FA in LC-grade water and 500 μL 50% acetonitrile supplemented with 0.2% FA in LC-grade water at 4000 x g for 60 seconds each. Peptides were then dried down and desalted using Sep-Pak Vac 3cc tC18-cartridges (Waters, WAT036820) according to the manufacturer’s instructions. In brief, following equilibration of the C18-cartridges with 100% ACN supplemented with 0.1% FA and 0.05% TFA in LC-grade water, samples were loaded in 0.05% TFA in LC-grade water and washed with 0.05% TFA in LC-grade water followed by washing with 0.1% FA in LC-grade water. Peptides were then eluted in 30% acetonitrile in LC-grade water and following drying in a speedvac at 60°C for 2h, peptides were re-suspended in 0.05% v/v TFA for LC-MS/MS. 5.55 ng worth peptide in 10 μL of 0.05% TFA were trapped on an PepMap 300 (C18, 300 μm x 5 mm, Thermo Fisher Scientific) column and separated on an Easy spray RSLC C18 columns (75 μm x 500 mm, Thermo Fisher Scientific, ES803) at 220 μL/min using the following gradient profile (minutes:%B); 0:3.0, 45:33, 46:45, 47:80, 49:80, 50:3.0, 70:3.0. The buffers used were: buffer A: 0.1% FA in LC-grade water and B: 100% Acetonitrile. The eluent was directed into an Easy-Spray source (Thermo Fisher Scientific) with temperature set at 60˚C and a source voltage of 1.9 kV. Data was acquired on QExactive HFX (Thermo Fisher Scientific) with precursor scan ranging from 375 to 1200 m/z at 120,000 resolution and automatic gain control (AGC) target of 3e6. The isolation window was set to 1.5 m/z. dd-MS2 scans were conducted at 30,000 resolution and automatic gain control (AGC) target of 1e5 and normalized collision energy (NCE) set to 29%. At least four injections of the identical sample were conducted for each analysed tumour. For normal pancreas samples, at least five injections were conducted.
The resulting data was analysed in Progenesis (v26.45.1656 Nonlinear Dynamics) using a Hi3 approach for each individual replicate and searched against the SwissProt database with species set to Mus musculus on an in-house Mascot server (Matrix Science; 2016) using Mascot Daemon (V 2.6, Matrix Science). Search parameters included peptide modifications for methylthio (C) as static modifications and oxidation (M) as well as deamination (N, Q) as dynamic modifications. Only 2+, 3+, and 4+ charged peptide-ions with a peptide tolerance of 10 ppm were included in the search. A 1% peptide FDR threshold was applied, and peptides were filtered for high peptide confidence, minimum peptide length of 6, and finally peptides without protein reference were removed.
S-trap based tryptic digestion of whole cell lysates from synthetic hydrogels
For LC-MS/MS analysis of cell laden hydrogels, organoids were grown in PEG CBF-0.5 gels for four days from single cell suspension in full organoid growth media as previously described. At the endpoint, gel plugs were removed from the 24 well plates, washed twice with PBS for 5 minutes each and following removal of PBS were gel plugs minced using a mortar and pestle with liquid nitrogen. Fine powder was allowed to thaw to RT and re-suspended in 4% v/v SDS, 50 mM TEAB, 20 mM HEPES (pH 7.8), sonicated and 1 μL Benzonase was per 100 μL lysate to dissociate genomic DNA and RNA for 30 minutes at RT. The lysate was then vigorously vortexed and clarified by centrifuging for 20 minutes at 16,000 x g, RT. Following quantification of protein content using the BCA kit (Thermo Fisher Scientific) according to the manufacturer’s instructions 80 μg of the lysate subjected to S-trap based digestion using S-trap micro spin columns (Protifi). Briefly, proteins were reduced by adding 20 mM DTT (Sigma Aldich) for 10 minutes at 95°C followed by alkylation with 40 mM IAA (Sigma Aldrich) for 30 minutes in the dark at RT. Undissolved matter was then spun out and following acidification of the samples using 12% v/v phosphoric acid which was added to a final concentration of 1.2% v/v was the SDS-containing lysate mixed with S-trap binding buffer (90% v/v Methanol, 0.1 M TEAB, pH 7.1) at a volumetric ratio of 1:7 lysate:STBB. The acidified SDS-containing lysate in STBB was then spun into the S-Trap micro spin columns (Protifi) at 4000 x g for 30 sec to trap the proteins until the entire protein-containing lysate was passed through the columns. Trapped proteins were then washed with STBB four times and proteins were digested overnight at 37°C in 50 mM TEAB using 4 μg of sequencing grade trypsin (mass ratio 1:20 protein:trypsin). Following digestion, peptides were eluted in 50 mM TEAB then 0.2% FA in LC-grade water and finally in 50% acetonitrile, 0.2% FA in LC-grade water. Eluents were combined and peptides were dried down and stored at -80°C until fractionation.
High-pH off-Line fractionation and LC-MS/MS
Peptides were fractionated as previously described27. Briefly, peptides were separated on a Zorbax Extend-C18 column (4.6 x 150 mm, 3.5 μm, Agilent Technologies) at 250 μL/min using the following gradient profile (minutes:%B); 5:0.5, 20:30, 24:40, 26:75, 29:75, 30:0.5, 55:0.5. The buffers used were: buffer A: LC-grade water supplemented with 0.1% v/v NH4OH (pH 10.5) and buffer B: 100% Acetonitrile. The eluent was directed onto 96 round-bottom plates and fractions were collected every 15s. Only fractions in the elution window between 15:50 and 35:00 minutes were used and all of the fractions were concatenated into 24 final fractions with each containing 3.33 μg peptide on average. Following drying in a speedvac at 60°C for 2h, were peptides re-suspended at 100 ng/mL in 0.05% v/v TFA for LC-MS/MS. 100 ng of peptides were trapped on a PepMap 300 (C18, 300 μm x 5 mm, Thermo Fisher Scientific) column and separated on an Easy spray RSLC C18 columns (75 μm x 500 mm, Thermo Fisher Scientific, ES803) at 200 nL/min using the following gradient profile (minutes:%B); 6:1.0, 40:24, 45:45, 46:80, 49:80, 50:1.0, 70:1.0. The buffers used were: buffer A: 0.1% FA in LC-grade water and B: 100% Acetonitrile. The eluent was directed into an Easy-Spray source (Thermo Scientific) with temperature set at 60˚C and a source voltage of 1.9 kV. Data was acquired on QExactive HFX (Thermo Fisher Scientific) with precursor scan ranging from 375 to 1200 m/z at 120,000 resolution and automatic gain control (AGC) target of 3e6. The isolation window was set to 1.5 m/z. dd-MS2 scans were conducted at 30,000 resolution and automatic gain control (AGC) target of 1e5 and normalized collision energy set to 29%.
The resulting data were searched against the SwissProt database with species set to Mus musculus on an in-house Mascot server (Matrix Science; 2016) in Proteome Discoverer (Thermo Fisher Scientific, V. 2.1). Search parameters included peptide modifications for carbamidomethylation (C) as static modification and oxidation (M, P and K) as well as deamination (N, Q) as dynamic modification. A decoy database search was performed to determine the peptide FDR with the Percolator module. A 1% peptide FDR threshold was applied, and peptides were filtered for high peptide confidence, minimum peptide length of 6, and finally peptides without protein reference were removed. Protein grouping was performed by applying strict parsimony principles. All proteins that exhibited a confidence of less than ‘high’ and with less than two uniquely identified peptides were excluded from further analysis.
Electron Microscopy
Samples were extensively washed with PBS and fixed with 4% formaldehyde + 2.5% glutaraldehyde in 0.1M Hepes buffer (pH 7.2). Subsequently samples were post-fixed with 1% osmium tetroxide + 1.5% potassium ferricyanide in 0.1M cacodylate buffer (ph7.2) for 1 hour, then 1% tannic acid in 0.1M cacodylate buffer (ph7.2) for 1 hour and finally in 1% uranyl acetate in water for 1 hour. The samples were then dehydrated in ethanol series infiltrated with TAAB 812 resin and polymerized for 24h at 60C degree. Sections were cut with a Reichert Ultracut ultramicrotome and observed with a FEI Tecnai 12 Biotwin microscope at 100kV accelerating voltage. Images were taken with a Gatan Orius SC1000 CCD camera.
Live imaging of pancreatic cancer organoid co cultures
CBF-0.5 hydrogels containing mPCOs, PaFs and BMDMs 1:5:5 ratio of PDO:Paf:BMDM were prepared as described previously and crafted into 10 μL domes in 24-well Sensoplate (Greiner). Co-culture domes were imaged via the PerkinElmer Opera Phenix microscope. A Zeiss EC Plan Neofluar X10 air objective NA 0.3 WD 5.2mm or Zeiss W Plan-Apochromat X20 water objective NA 1.0 WD 1.17mm were used as indicated. Pre-scan directed z-stack acquisition of individual spheres within the gel was used and co-cultures were imaged every hour over a time-window of 72h starting from day three of culture. Videos and representative still images were generated from maximum intensity projections using Harmony 4.8 software. Representative images were exported from Harmony 4.8 in the portable network graphic format with scaling equally applied to all conditions.
ELISA
Concentrations of GM-CSF (DY415-05), IL6 (DY406-05), IL1α (DY400-05), IL1β (DY401-05), TNFα (DY410-05), CXCL12 (DY453-05) and TGF β (DY1679-05) (all from R & D Systems) in organoid cultures were measured using ELISA DuoSet kits as per manufacturer’s instructions. In brief, CBF-0.5 hydrogels were dissociated using Sortase A pentamutant (SrtA), PBS was removed and 200 μl of 1 mg/ml Srt A was added to the gels for 30 mins at 37°C prior to addition of 18 mM Gly-Gly-Gly (GGG) and 10 mM CaCL2 and a further incubation of 20 mins at 37°C. Following dissociation samples were supplemented with EDTA-free protease inhibitor cocktail set III and frozen at -80°C. Samples were thawed and centrifuged at 14,000 x g for 30 min at 4 °C. Supernatants were mixed in a 1:1 (v/v) ratio with conditioned medium collected from the respective condition, and were used for ELISA assay.
Cell viability analysis
PaFs were encapsulated in either synthetic hydrogel or Matrigel in combination with mPCOs or mPCOs and BMDMs as previously described and cultured for 6 day in hPOCM supplemented with 50 ng/mL EGF and 20 ng/mL M-CSF or reduced media (Minimal media) as indicated. Single cell suspensions were prepared using 0.25% v/v Trypsin/Versene. Following neutralization with DMEM 10% v/v FBS and centrifugation cells were resuspended at 1x106 cells/mL in PBS. Cells were incubated with LIVE/DEAD™ Fixable Blue Dead Cell Stain (1/1000, Thermo) at RT for 30 mins. Following washing in PBS, cells were re-suspended in CSM1 supplemented with Fc-block and subjected to surface-staining with PE conjugated anti-CD45 (1:200, BioLegend, Clone 30-F11) for 45 min on ice. Following staining, cells were washed in PBS, passed through a 70 μm filter, and re-suspended at a concentration of 1 x 106cells/mL and analysed on a LSRFortessa™ X-20 flow cytometer (BD) using 355nm, 488nm, 561nm and 640nm lasers, with 450/50, 515/20, 586/15, 730/45 filter sets respectively. Cell line specific events were selected based on GFP positivity (PaF), iRFP positivity (mPCO) and CD45 positivity (BMDM) and viable cells were selected based on negativity for LIVE/DEAD blue stain.
Sample preparation mass cytometry (MC)
To identify cells in S phase 5-Iodo-2′-deoxyuridine (IdU, Sigma) was added to organoid culture medium at a final concentration of 25 μM and incubated for 30 mins at 37°C. Gels were then dissociated immediately or fixed in 4% paraformaldehyde (PFA) (Thermo Fisher scientific) for 60 min at 37°C followed by 2x wash in PBS (Fisher) and storage at 4°C until required. Both fixed and un-fixed PEG-VS hydrogels were dissolved using SrtA (Supplemental materials and methods). The resultant solution was transferred to a gentleMACS C-Tube (Miltenyi) containing 0.5 mg/ml Dispase II (Sigma), 0.2 mg/ml Collagenase IV (Gibco) and 0.2 mg/ml DNase (Gibco) made up to 5 mL in PBS and processed using the gentleMACS Octo Dissociator (with Heaters) (Miltenyi) at 37°C for 50min using a custom program adapted from C.Tape62. C-tube was spun at 1000g for 1 min to collect all cellular material in the bottom of the tube. Cells were washed in 5 mL of PBS and passed through a 70-μm filter (BD) into polypropylene FACS tubes (BD Falcon) to remove residual cell clusters.
Mass Cytometry analysis of PEG-VS hydrogel co-cultures
Methanol permeabilization: Fixed single-cell suspensions were washed in cell-staining buffer (CSM; Fluidigm) containing 5mM EDTA, hereafter named CSM-1. Cells were then pelleted by centrifugation; this and all subsequent centrifugations were performed at 1000g for 5 min. Supernatant was removed and cell vortexed in void volume before addition of 2 ml of ice-cold 20% methanol for 20 minutes at -20°C. Following incubation cells were washed twice in 3 mL CSM-1 and the cell pellet resuspended in the void volume. 1 μl of Fc block was added for 5 min at RT. 50 μl of the heavy metal tagged antibody mastermix was added in 50 μl CSM and incubated for 45 min at RT (Supplemental Table 2). Following antibody incubation cells were washed twice with CSM-1. The resultant pellet was resuspended in 200 μl PBS and 4% PFA was added under vortex and stored overnight at 4°C.
FOXP3 Fix/Permeabilization: Un-fixed single-cell suspensions were washed in CSM-1. Cells were then pelleted by centrifugation 500 g for 5 min. Supernatant was removed and cell vortexed in void volume before addition of 1μl Fc block (BD Biosciences) for 5 min at RT. Extracellular heavy metal-tagged antibody mastermix (Supplemental Table 2) was added in 50 μl CSM and incubated for 45 mins at RT. Following incubation cells were washed twice with CSM-1. FOXP3 Fix/Perm staining buffer set (Invitrogen) was then used to fix and permeabilise the cells according to the manufacturer’s instructions. In brief, cell pellets were resuspended in 1x Thermo Foxp3 fix buffer and incubated for 30 min at RT. 2 mL of 1xThermo Foxp3 perm buffer was added and cells pelleted. A further 2 mL of 1xThermo Foxp3 perm buffer was added and cells pelleted. The cell pellet was resuspended in the void volume and 1 μl of Fc block added for 5 min at RT. 50 μl of the intracellular heavy metal-tagged antibody mastermix was added in 50 μl CSM and incubated for 45 min at RT. Following antibody incubation cells were washed twice with CSM-1. The resultant pellet was resuspended in 200 μl PBS and 4% PFA was added under vortex and stored overnight at 4°C.
All samples were processed similarly from this point. The following day 1 μl iridium solution (Fluidigm) was added per 3x106 cells followed by incubation for 60 min at RT. The cell suspension was diluted 1:3 in PBS before pelleting. Cells were washed in Maxpar water (Fluidigm) and resuspended in Maxpar water containing 15% (v/v) EQ beads (Fluidigm) to a concentration of cells per ml and filtered twice (70 μm) before measuring on Helios mass cytometer (Fluidigm) (300-500 events per second). Files were normalized against EQ beads, de-barcoded and uploaded to the Cytobank platform (http://www.cytobank.org/). Cell doublets and aggregates were removed based on event length. Live cell events were selected based on cleaved caspase-3 negativity. Cell line specific events were selected based on GFP positivity (PaF) and CD45 positivity (BMDM). Selected populations were exported as FCS files and uploaded to the Cytofkit2 package (version 2.0.1). Cells were clustered using FlowSOM and visualized using UMAP projections and expression overlays, cell data with annotated clusters were exported for further downstream analysis of marker expression using R studio (V4.0.0) (https://www.r-project.org).
Survival Analysis
For comparison of matrisomal protein changes between studies, supplementary data files from Tian et al 201926 were downloaded and Log2FC between PDA and NP samples were used from either the murine or human cohort. For the human cohort, all samples regardless of differentiation grade were used. Pearson or spearman-based rank correlation of the Log2FC(PDA/WT) of matrisomal proteins identified and quantified in this study and Tian et al 2019 was computed using R 4.0.0. For survival analysis, matrisomal proteins with a Log2FC(PDA/WT) ≥ 1 in this study and Tian et al 2019 were used and gene identifiers were translated into entrezgene_ids using the biomaRt R package63. Clinical information and gene-expression data for human pancreatic cancer (TCGA-PAAD) were downloaded from cBioPortal (accessed: 22/04/2020, 64,65) and log2 transformed. The cox-proportional hazard regression p-value and hazard ratio was determined in quantile-categorized patients (top 25% v. bottom 25%) by the matrisomal protein expression for the TCGA dataset (n=179). The survival analysis was done using the survival package (v3.2-3, https://cran.r-project.org/web/packages/survival/index.html) and Kaplan-Meier survival plots were computed using the survminer R-package in R 4.0.0.
Bioinformatics and Statistics
Bioinformatics were conducted using the open-source R software (V4.0.0). Proteins with matrisomal origin were annotated from proteomic datasets using the murine matrisome66. Proteins with basement membrane origin were annotated from matrisome using the gene ontology (GO)-term “basement membrane” (GO:0005604; http://www.informatics.jax.org/go/term/GO:0005604 accessed 2020/10/05 67). Kyoto encyclopaedia of genes and genomes (KEGG)-based pathway- and gene ontology (GO)-term biological process (BP) annotations were obtained from the database for annotation, visualization and integrated discovery (DAVID) functional annotation clustering by assuming the whole genome as statistical background. For GO-biological process (BP)-term enrichment, only terms with at least three members, a FDR ≤ 0.05, Benjamini-Hochberg adjusted p-Value ≤ 0.05 and fold enrichment of ≥ 1.5 were included. For KEGG-pathway annotation, only pathways with at least three members a p-Value ≤ 0.05 and fold enrichment ≥ 1.5 were included. MatriCircos plots were generated using the open-source RCircos R-package68 by applying a custom-made RCircos core-component library containing previously annotated core-matrisome proteins only. Circular visualizations of Yap1 nuclear/cytoplasmatic ratio were implemented using the circlize R package69. Integrin-ECM interactions were obtained from a publicly available manually curated cell-cell interaction database form the Bader lab (https://baderlab.org/CellCellInteractions) and only literature-based interactions with pubmed annotation were included in the analysis. For integrin-ECM interactions, only those interactions nucleating from an integrin-alpha subunit were considered and integrin complexes containing an alpha and beta subunit were subsequently manually assembled. For network analysis and protein-interaction network visualization, the cytoscape software package 70 was utilized by incorporating the String database (STRING CONSORTIUM, V 10.5, 2018, 71 MCODE, and boundaryLayout apps). For String-based network visualization, networks were curated using a very stringent confidence of 0.95 by accepting text-mining, experimental, database, co-expression, neighbourhood, gene-fusion and co-occurrence interaction sources to obtain relevant integrin-ECM interactions. Statistics and visualizations were conducted using GraphPad Prism (Version 8; GraphPad Software Inc.) or the open source R software (V4.0.0). For statistical analysis of high-content imaging data, the non-parametric two-tailed Wilcoxon t-test was utilized with application of Benjamini-Hochberg correction and a FDR threshold of 0.05 in R. For statistical analysis of adhesion assays and PDO growth assessments in stiffened hydrogels, the parametric two-tailed student’s t-test was conducted with application of Benjamini-Hochberg correction and a FDR threshold of 0.05 in R. For statistical analysis of lebein-2 treated organoids, the two-tailed parametric paired student’s t-test was used. For statistical analysis of YAP1 ratio between stiffened hydrogels, the parametric two-sided Student’s t-test was used to compare 1.4, 3.1 and 8.2 kPa conditions whilst the non-parametric Wilcoxon test was utilized for comparison of 1.4 and 20.5 kPa conditions upon removal of outliers and with application of Benjamini-Hochberg correction and a FDR threshold of 0.05 in R. Outliers were identified and removed from each population when exceeding 1.5XIQR and confirmed as outlier by the grubb’s test function from the outliers package in R V.4.0.0. Normality was assessed in R using the Shapiro-Wilk test with a significance level of 0.05.
Supplementary Material
Acknowledgements
This work was supported by a Cancer Research UK Program Grant C13329/A21671 (MH, CJ), Cancer Research UK Institute Awards A19258 (CJ) and A17196 (JPM), Experimental medicine Programme Award (A25236, CJ and JPM), A Rosetrees Trust grant (M286 CJ), and European Research Council Consolidator Award (ERC-2017-COG 772577, CJ), National Science Foundation (grant # CBET-0939511, LGG); National Institutes of Health (Grant # R01EB021908 and Grant # T32GM008334, LGG); Defense Advanced Research Projects Agency (W911NF-12-2-0039, LGG), JAE is financially supported by the Deutsche Forschungsgemeinschaft (DFG grant: SFB1009 project A09). The authors would like to thank Drs David Liu, Adrian Thrasher, Thomas Roberts, Beverly Torok-Storb, Inder Verma, Didier Trono and Prof Tim Somervaille for kindly sharing plasmids, Ming-Sound Tsao (UHN) for HPDE H6c7 cells, Matthew Ball and Edward Mckenzie at Manchester Institute of Biotechnology for Sortase expression and purification, Christopher J Tape at UCL for technical advice, Kenneth Beattie for assistance at FingerPrints Proteomics Facility (University of Dundee), the Cancer Research UK Glasgow Centre (A25142) and the BSU facilities at the Cancer Research UK Beatson Institute and members of Systems Oncology Group at CRUK MI for constructive input.
Footnotes
Author contributions:
CRB, JK, AlB, BYL, JDH, DLS, LGG, MJH, and CJ designed research; CRB, JK, AlB, AnB, JDH, JX, CL, DK, AM, NH, DLS, JB, CC and BYL conducted experiments; CRB, JK, AlB, AnB, and CJ analysed data; AnB, CC, VHG, LS, JAE, BS, XZ, DLS, DK, JA, GA and CH provided technical support; JPM maintained GEMMs and provided murine samples; JPM, LS, LGG, JE, BS provided reagents and cell lines; MAG, JG, LF and DAO help with clinical sample collection; LF provided pathological support; CRB and CJ wrote the paper; CJ and LGG oversaw project. JX contributed to this work while an employee at CRUK MI.
Competing interests:
LGG has patent application pending related to the hydrogel system. The rest of the authors have no competing interests to disclose.
Code Availability
All original R-scripts have been deposited to https://www.zenodo.org/ (https://zenodo.org/record/4664132) and are freely available.
Data Availability
All original source data are freely available: The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE 53 partner repository with the dataset identifiers: NP matrisome atlas (PXD022555 and 10.6019/PXD022555); IAC datasets (PXD022487 and 10.6019/PXD022487); CDM datasets (PXD022509 and 10.6019/PXD022509); 3D PEG CBF-0.5 LC-MS (PXD022520 and 10.6019/PXD022520); Tumour Matrisome LC-MS (PXD022767 and 10.6019/PXD022767). Raw CyTOF data, IF images and AFM force curves as well as source data for all figures (Figures 1-5 and Supplemental Figures 1-28) have been deposited to https://www.zenodo.org/ (https://zenodo.org/record/4664132).
References
- 1.Egeblad M, Nakasone ES, Werb Z. Tumors as Organs: Complex Tissues that Interface with the Entire Organism. Dev Cell. 2010;18:884–901. doi: 10.1016/j.devcel.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feig C, et al. The pancreas cancer microenvironment. Clin Cancer Res. 2012;18:4266–4276. doi: 10.1158/1078-0432.CCR-11-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sahai E, et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer. 2020;6:1–13. doi: 10.1038/s41568-019-0238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeNardo DG, Ruffell B. Macrophages as regulators of tumour immunity and immunotherapy. Nature Reviews Immunology. 2019;7:1–14. doi: 10.1038/s41577-019-0127-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biankin AV, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller BW, et al. Targeting the LOX/hypoxia axis reverses many of the features that make pancreatic cancer deadly: inhibition of LOX abrogates metastasis and enhances drug efficacy. EMBO Molecular Medicine. 2015;7:1063–1076. doi: 10.15252/emmm.201404827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang H, et al. Targeting focal adhesion kinase renders pancreatic cancers responsive to checkpoint immunotherapy. Nat Med. 2016:1–13. doi: 10.1038/nm.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi Y, et al. Targeting LIF-mediated paracrine interaction for pancreatic cancer therapy and monitoring. Nature. 2019;569:1–27. doi: 10.1038/s41586-019-1130-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sherman MH, et al. Vitamin D receptor-mediated stromal reprogramming suppresses pancreatitis and enhances pancreatic cancer therapy. Cell. 2014;159:80–93. doi: 10.1016/j.cell.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boj SF, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tuveson D, Clevers H. Cancer modeling meets human organoid technology. Science. 2019;364:952–955. doi: 10.1126/science.aaw6985. [DOI] [PubMed] [Google Scholar]
- 12.Drost J, Clevers H. Organoids in cancer research. Nat Rev Cancer. 2018;18:407–418. doi: 10.1038/s41568-018-0007-6. [DOI] [PubMed] [Google Scholar]
- 13.Hughes CS, Postovit LM, Lajoie GA. Matrigel: A complex protein mixture required for optimal growth of cell culture. Proteomics. 2010;10:1886–1890. doi: 10.1002/pmic.200900758. [DOI] [PubMed] [Google Scholar]
- 14.Brassard JA, Lutolf MP. Engineering Stem Cell Self-organization to Build Better Organoids. Stem Cell. 2019;24:860–876. doi: 10.1016/j.stem.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 15.Socovich AM, Naba A. The cancer matrisome: From comprehensive characterization to biomarker discovery. Seminars in Cell and Developmental Biology. 2019;89:157–166. doi: 10.1016/j.semcdb.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 16.Cook CD, et al. Local remodeling of synthetic extracellular matrix microenvironments by co-cultured endometrial epithelial and stromal cells enables long-term dynamic physiological function. Integr Biol. 2017;9:271–289. doi: 10.1039/c6ib00245e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gjorevski N, et al. Designer matrices for intestinal stem cell and organoid culture. Nature. 2016;539:560–564. doi: 10.1038/nature20168. [DOI] [PubMed] [Google Scholar]
- 18.Kratochvil MJ, et al. Engineered materials for organoid systems. Nat Rev Mater. 2019;4:606–622. doi: 10.1038/s41578-019-0129-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valdez J, et al. On-demand dissolution of modular, synthetic extracellular matrix reveals local epithelial-stromal communication networks. Biomaterials. 2017;130:90–103. doi: 10.1016/j.biomaterials.2017.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naba A, Clauser KR, Hynes RO. Enrichment of Extracellular Matrix Proteins from Tissues and Digestion into Peptides for Mass Spectrometry Analysis. J Vis Exp. 2015;101:e53057. doi: 10.3791/53057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hingorani SR, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 22.Hruban RH, et al. Pathology of genetically engineered mouse models of pancreatic exocrine cancer: consensus report and recommendations. Cancer Res. 2006;66:95–196. doi: 10.1158/0008-5472.CAN-05-2168. [DOI] [PubMed] [Google Scholar]
- 23.Schönhuber N, et al. A next-generation dual-recombinase system for time- and host-specific targeting of pancreatic cancer. Nat Med. 2014;20:1340–1347. doi: 10.1038/nm.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naba A, et al. The Matrisome: In Silico Definition and In Vivo Characterization by Proteomics of Normal and Tumor Extracellular Matrices. Mol Cell Proteomics. 2012;11:M111.014647-18. doi: 10.1074/mcp.M111.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tian R, et al. Combinatorial proteomic analysis of intercellular signaling applied to the CD28 T-cell costimulatory receptor. Proceedings of the National Academy of Sciences. 2015;112:E1594–603. doi: 10.1073/pnas.1503286112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tian C, et al. Proteomic analyses of ECM during pancreatic ductal adenocarcinoma progression reveal different contributions by tumor and stromal cells. Proc Natl Acad Sci USA. 2019;116:19609–19618. doi: 10.1073/pnas.1908626116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horton ER, et al. Definition of a consensus integrin adhesome and its dynamics during adhesion complex assembly and disassembly. Nat Cell Biol. 2015;17:1577–1587. doi: 10.1038/ncb3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Humphries JD. Integrin ligands at a glance. J Cell Sci. 2006;119:3901–3903. doi: 10.1242/jcs.03098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamidi H, Ivaska J. Every step of the way: integrins in cancer progression and metastasis. Nat Rev Cancer. 2018;18:533–548. doi: 10.1038/s41568-018-0038-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones MC, et al. Isolation of integrin-based adhesion complexes. Curr Protoc Cell Biol. 2015;66:9.8.1–9.8.15. doi: 10.1002/0471143030.cb0908s66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robertson J, et al. Defining the phospho-adhesome through the phosphoproteomic analysis of integrin signalling. Nature Communications. 2015;6 doi: 10.1038/ncomms7265. 6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takizawa M, et al. Mechanistic basis for the recognition of laminin-511 by α6β1 integrin. Sci Adv. 2017;3:e1701497. doi: 10.1126/sciadv.1701497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samarelli AV, et al. Neuroligin 1 induces blood vessel maturation by cooperating with the α6 integrin. Journal of Biological Chemistry. 2014;289:19466–19476. doi: 10.1074/jbc.M113.530972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aumailley M, Timpl R, Sonnenberg A. Antibody to integrin α6 subunit specifically inhibits cell-binding to laminin fragment 8. Experimental Cell Research. 1990;188:55–60. doi: 10.1016/0014-4827(90)90277-h. [DOI] [PubMed] [Google Scholar]
- 35.Lee SP, et al. Sickle cell adhesion to laminin: potential role for the alpha5 chain. Blood. 1998;92:2951–2958. [PubMed] [Google Scholar]
- 36.Hernandez-Gordillo V, et al. Fully synthetic matrices for in vitro culture of primary human intestinal enteroids and endometrial organoids. Biomaterials. 2020;254:120125. doi: 10.1016/j.biomaterials.2020.120125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson G, Moore SW. Identification of a structural site on acetylcholinesterase that promotes neurite outgrowth and binds laminin-1 and collagen IV. Biochemical and Biophysical Research Communications. 2004;319:448–455. doi: 10.1016/j.bbrc.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 38.Brown A, et al. Engineering PEG-based hydrogels to foster efficient endothelial network formation in free-swelling and confined microenvironments. Biomaterials. 2020;243:119921. doi: 10.1016/j.biomaterials.2020.119921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knight CG, et al. The collagen-binding A-domains of integrins alpha(1)beta(1) and alpha(2)beta(1) recognize the same specific amino acid sequence, GFOGER, in native (triple-helical) collagens. J Biol Chem. 2000;275:35–40. doi: 10.1074/jbc.275.1.35. [DOI] [PubMed] [Google Scholar]
- 40.Kuhlman W, Taniguchi I, Griffith LG, Mayes AM. Interplay Between PEO Tether Length and Ligand Spacing Governs Cell Spreading on RGD-Modified PMMA-g-PEO Comb Copolymers. Biomacromolecules. 2007;8:3206–3213. doi: 10.1021/bm070237o. [DOI] [PubMed] [Google Scholar]
- 41.Eble JA, Bruckner P, Mayer U. Vipera lebetinaVenom Contains Two Disintegrins Inhibiting Laminin-binding β 1Integrins. J Biol Chem. 2003;278:26488–26496. doi: 10.1074/jbc.M301860200. [DOI] [PubMed] [Google Scholar]
- 42.Cavaco ACM, et al. The Interaction between Laminin-332 and α3β1 Integrin Determines Differentiation and Maintenance of CAFs, and Supports Invasion of Pancreatic Duct Adenocarcinoma Cells. Cancers. 2019;11:14–20. doi: 10.3390/cancers11010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gasmi A, et al. Amino acid structure and characterization of a heterodimeric disintegrin from Vipera lebetina venom. 2001;1547:51–56. doi: 10.1016/s0167-4838(01)00168-6. [DOI] [PubMed] [Google Scholar]
- 44.Arruda Macêdo JK, Fox JW, de Souza Castro M. Disintegrins from snake venoms and their applications in cancer research and therapy. Curr Protein Pept Sci. 2015;16:532–548. doi: 10.2174/1389203716666150515125002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tiriac H, et al. Organoid Profiling Identifies Common Responders to Chemotherapy in Pancreatic Cancer. Cancer Discov. 2018;8:1112–1129. doi: 10.1158/2159-8290.CD-18-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levental KR, et al. Matrix Crosslinking Forces Tumor Progression by Enhancing Integrin Signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laklai H, et al. Genotype tunes pancreatic ductal adenocarcinoma tissue tension to induce matricellular fibrosis and tumor progression. Nat Med. 2016;22:497–505. doi: 10.1038/nm.4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rice AJ, et al. Matrix stiffness induces epithelial–mesenchymal transition and promotes chemoresistance in pancreatic cancer cells. Oncogenesis. 2019;6:e352. doi: 10.1038/oncsis.2017.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rubiano A, et al. Viscoelastic properties of human pancreatic tumors and in vitro constructs to mimic mechanical properties. Acta Biomaterialia. 2018;67:331–340. doi: 10.1016/j.actbio.2017.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Panciera T, et al. Reprogramming normal cells into tumour precursors requires ECM stiffness and oncogene- mediated changes of cell mechanical properties. Nat Mater. 2020;19:797–806. doi: 10.1038/s41563-020-0615-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Öhlund D, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med. 2017;214:579–596. doi: 10.1084/jem.20162024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Elyada E, et al. Cross-Species Single-Cell Analysis of Pancreatic Ductal Adenocarcinoma Reveals Antigen-Presenting Cancer-Associated Fibroblasts. Cancer Discov. 2019;9:1102–1123. doi: 10.1158/2159-8290.CD-19-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pérez-Riverol Y, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Research. 2018;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu J, et al. Generation of a pancreatic cancer model using a Pdx1-Flp recombinase knock-in allele. PLoS ONE. 2017;12:e0184984-13. doi: 10.1371/journal.pone.0184984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ouyang H, et al. Immortal Human Pancreatic Duct Epithelial Cell Lines with Near Normal Genotype and Phenotype. The American Journal of Pathology. 2010;157:1623–1631. doi: 10.1016/S0002-9440(10)64800-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Furukawa T, et al. Long-term culture and immortalization of epithelial cells from normal adult human pancreatic ducts transfected by the E6E7 gene of human papilloma virus 16. The American Journal of Pathology. 1996;148:1763–1770. [PMC free article] [PubMed] [Google Scholar]
- 57.Huch M, et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 2013;32:2708–2721. doi: 10.1038/emboj.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bankhead P, et al. QuPath: Open source software for digital pathology image analysis. 2018:1–27. doi: 10.1101/099796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 61.Naba A, Clauser KR, Hynes RO. Enrichment of Extracellular Matrix Proteins from Tissues and Digestion into Peptides for Mass Spectrometry Analysis. 2015:e53057. doi: 10.3791/53057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qin X, et al. Cell-type-specific signaling networks in heterocellular organoids. Nat Methods. 2020;17:1–14. doi: 10.1038/s41592-020-0737-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cerami E, et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data: Figure 1. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao J, et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci Signal. 2013;6:pl1-pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Naba A, et al. The extracellular matrix: Tools and insights for the ‘omics’ era. Matrix Biol. 2016;49:10–24. doi: 10.1016/j.matbio.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bult CJ, et al. Mouse Genome Database (MGD) 2019. Nucleic Acids Research. 2019;47:D801–D806. doi: 10.1093/nar/gky1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang H, Meltzer P, Davis S. RCircos: an R package for Circos 2D track plots. BMC Bioinformatics. 2013;14:244–5. doi: 10.1186/1471-2105-14-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gu Z, Gu L, Eils R, Schlesner M, Brors B. circlize Implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 70.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Szklarczyk D, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Research. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All original R-scripts have been deposited to https://www.zenodo.org/ (https://zenodo.org/record/4664132) and are freely available.
All original source data are freely available: The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE 53 partner repository with the dataset identifiers: NP matrisome atlas (PXD022555 and 10.6019/PXD022555); IAC datasets (PXD022487 and 10.6019/PXD022487); CDM datasets (PXD022509 and 10.6019/PXD022509); 3D PEG CBF-0.5 LC-MS (PXD022520 and 10.6019/PXD022520); Tumour Matrisome LC-MS (PXD022767 and 10.6019/PXD022767). Raw CyTOF data, IF images and AFM force curves as well as source data for all figures (Figures 1-5 and Supplemental Figures 1-28) have been deposited to https://www.zenodo.org/ (https://zenodo.org/record/4664132).