Abstract
Hepatic activation of protein kinase C isoforms (PKCs) by diacylglycerol (DAG) promotes insulin resistance and contributes to the development of type 2 diabetes (T2D). The closely related protein kinase D (PKD) isoforms act as effectors for DAG and PKC. Here, we showed that PKD3 was the predominant PKD isoform expressed in hepatocytes and was activated by lipid overload. PKD3 suppressed the activity of downstream insulin effectors including the kinase AKT and mechanistic target of rapamycin complex 1 and 2 (mTORC1 and mTORC2). Hepatic deletion of PKD3 in mice improved insulin-induced glucose tolerance. However, increased insulin signaling in the absence of PKD3 promoted lipogenesis mediated by SREBP (sterol regulatory element-binding protein) and consequently increased triglyceride and cholesterol content in the livers of PKD3 deficient mice fed a high-fat diet. Conversely, hepatic-specific overexpression of a constitutively active PKD3 mutant suppressed insulin-induced signaling and caused insulin resistance. Our results indicate that PKD3 provides feedback on hepatic lipid production and suppresses insulin signaling. Therefore, manipulation of PKD3 activity could be used to decrease hepatic lipid content or improve hepatic insulin sensitivity.
Introduction
Hepatocytes are a major target for insulin. On one hand, insulin stimulates hepatic glucose uptake, suppresses de novo glucose production, and therefore lowers systemic glycaemia (1). On the other hand, excessive insulin signaling promotes de novo lipid synthesis and consequently the accumulation of triglycerides (TG) and cholesterol in hepatocytes. This can lead to the development of non-alcoholic fatty liver disease (NAFLD), hepatic insulin resistance, and can eventually lead to the development of non-alcoholic steatohepatitis (NASH) and consequently to liver cirrhosis (2). On the molecular level, insulin stimulates activity and expression of major transcription factors such as sterol regulatory binding proteins (SREBPs) that promote hepatic lipid production (3). Activation of SREBP-dependent transcription requires inputs from various insulin evoked signaling cascades, which include AKT and mechanistic target of rapamycin complex 1 and 2 (mTORC1 and mTORC2) (3, 4).
Obesity-related metabolic overload results in the accumulation of diacylglycerol (DAG) in the liver (5). Protein kinase C (PKC) isoforms mediate DAG-evoked insulin resistance (2, 5). The major PKC isoform expressed in liver, PKCε, promotes insulin resistance by phosphorylating the insulin receptor to inhibit downstream signaling (6, 7). Protein kinase D (PKD) isoforms (PKD1, 2, and 3) are DAG and PKC effectors that integrate multiple nutritional and hormonal inputs (8). However, the impact of PKDs on hepatic metabolism has not been investigated so far. Different PKDs have been implicated in the regulation of muscle differentiation, function of adipose tissue, pathophysiological heart remodeling, immune response, carcinogenesis, blood coagulation, insulin secretion, actin remodeling, trans-Golgi network dynamics, cell proliferation, and migration (8-20).
Here, we showed that PKD3 was the predominant PKD isoform in the liver and was activated in lipid-loaded hepatocytes. Furthermore, we demonstrated that PKD3 suppressed insulin signaling, resulting in impaired AKT phosphorylation as well as activation of mTORC1 and mTORC2. Mice lacking PKD3 in hepatocytes presented improved glucose and insulin tolerance as well as elevated SREBP-dependent lipogenesis, which resulted in increased hepatic TG and cholesterol content. These results indicate that PKD3 attenuates insulin signaling, thereby preventing the development of fatty liver.
Results
Lipid accumulation promotes PKD3 activation in the liver
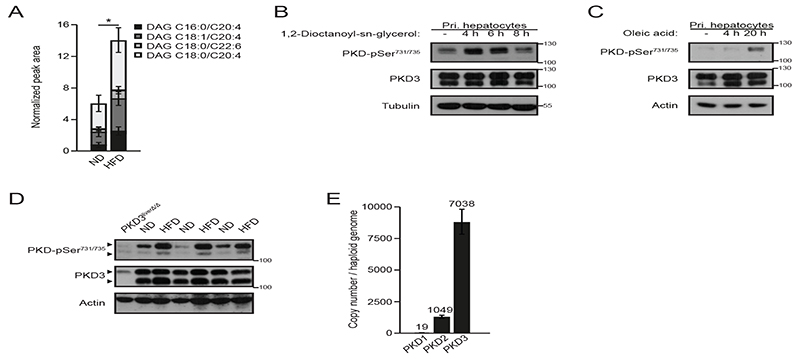
Hepatic DAG accumulation in liver has been postulated to contribute to insulin resistance in obese subjects (5). We found that mice fed a high-fat diet (HFD) accumulated significantly more DAG species compared to normal diet (ND) fed mice in the liver (Fig. 1A). PKD isoforms are activated in response to stimulation of receptors that utilize DAG as a secondary messenger (8). We observed that stimulation of isolated primary hepatocytes with a cell permeable analog of DAG (1,2-dioctanoyl-sn-glycerol) resulted in PKD activation as indicated by the phosphorylation of two serine residues in the activating loop in the kinase domain (Fig. 1B and fig. S1A). Moreover, long-term incubation of hepatocytes with oleic acid, which induces DAG and TG accumulation in cells, also led to PKD activation (Fig. 1C and fig. S1B). Of note, although DAG accumulates during obesity, our data suggests that DAG-evoked signaling might provide negative feedback on the expression of lipogenic genes because stimulation of primary hepatocytes with DAG analog decreased transcription of genes involved in de novo lipid synthesis (fig. S1C). Consistent with the results obtained from cultured hepatocytes, we observed increased PKD activity in livers of mice fed a HFD compared to ND fed animals (Fig. 1D and fig. S1D). Three PKD isoforms (PKD1, 2, and 3) have been identified so far (8). We did not detect PKD1 mRNA in hepatocytes and the expression of the PKD2 isoform was marginal. However, we observed a robust expression of the PKD3 isoform (Fig. 1E). Together, these data indicate that PKD3 could mediate the development of hepatic dysfunction in obese animals.
Fig. 1. PKD3 is the predominant PKD isoform expressed and activated in liver.
A Quantification of the four most abundant DAG classes in livers of normal diet (ND) or high-fat diet (HFD) fed mice determined by LC-MS (n=7 mice/diet).
B, C Western blot analysis of PKD-pSer731/735 in isolated primary hepatocytes stimulated with cell-permeable 1,2-Dioctanoyl-sn-glycerol (DAG) (100 μM) (B) or oleic acid (750 μM) (B) for the indicated time points, respectively (n=3 independent experiments).
D Western blot analysis of the indicated proteins from the livers of ND or HFD fed mice (n=3 mice/diet).
E Absolute quantification of PKD isoform expression in liver by QPCR with in-exon primers and normalizing values to a genomic DNA standard (n=8 biological replicates/group, combined data from three independent experiments).
Data information: In (A, E), data are presented as mean ± SEM. *P>0.05 (unpaired two-tailed Student’s t-test (A) or one way ANOVA with post hoc Tukey’s test (E)).
Hepatic deletion of PKD3 promotes insulin sensitivity
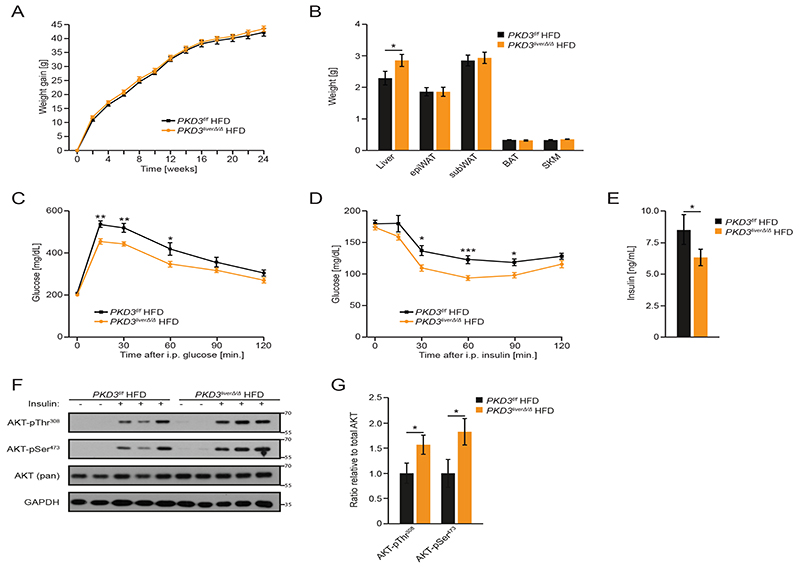
These results prompted us to generate mice deficient for PKD3 specifically in hepatocytes. We crossed PKD3 floxed mice (PKD3f/f) (20) with a mouse strain expressing Cre recombinase under the control of the albumin promoter (B6.Cg-Tg(Alb-cre)21Mgn/J) (21). The resulting pKD3 liverΔ/Δ mice showed specific deletion of PKD3 in liver but not in other organs (fig. S2A and B). pKD3 liverΔ/Δ mice fed a ND or HFD gained similar amounts of body weight as control animals (Fig. 2A and fig. S3A). Notably, mice deficient for PKD3 in hepatocytes presented with increased liver weight compared to control littermates when fed a HFD, whereas the weight of other organs was not affected (Fig. 2B). Similarly, all measured organ weights were not affected by PKD3 deletion in mice fed a ND (fig. S3B). Consistently, energy expenditure, food intake, voluntary movements, and respiratory exchange ratio were not affected by hepatocyte-specific deletion of PKD3 in mice fed HFD or ND (fig. S2C-F, S3C-F). However, mice deficient for PKD3 in hepatocytes were partially protected from HFD-induced glucose intolerance and insulin resistance (Fig. 2C and 2D and fig. S2G, S2H), whereas a ND did not did not alter glucose or insulin sensitivity in these mice (fig. S3G-J). Circulating insulin levels in mice deficient for hepatic PKD3 were significantly reduced (Fig. 2E), which is in line with the improved insulin sensitivity observed in these animals. Accordingly, the activation of the kinase AKT in response to insulin was enhanced in livers of HFD fed pKD3 liverΔ/Δ mice (Fig. 2F, 2G). Together, these results indicate that deletion of PKD3 in hepatocytes partially restores insulin signaling attenuated by HFD feeding.
Fig. 2. PKD3 promotes insulin resistance and glucose intolerance.
A Body weight evolution of control and pKD3 liverΔ/Δ mice fed a HFD for 24 weeks.
B Organ weight of control and pKD3 liverΔ/Δ mice fed a HFD for 24 weeks.
C-E Glucose (2 g/kg BW) (C) and insulin (1.5 U/kg BW) (D) tolerance tests and serum insulin levels (E) in pKD3 liverΔ/Δ and control mice after HFD feeding for 16, 18 and 24 weeks, respectively.
F, G Western blot analysis for the indicated proteins in the livers of mice of the indicated genotypes given insulin (8 U/kg body weight) for 15 min after 24 weeks of HFD feeding and (F) and corresponding densitometric quantification (G) (n=5 mice/group).
Data information: In (A-E), n=8 mice (WT) and n=15 mice (KO) were used and in (A-E, G) data are presented as mean ± SEM. *P>0.05, **P>0.01, ***P>0.001 (unpaired two-tailed Student’s t-test (B, E), one way ANOVA with post hoc Tukey’s test (G), or two way ANOVA with post hoc Tukey’s test (A, C, D)).
Deletion of PKD3 in hepatocytes promotes TG and cholesterol accumulation
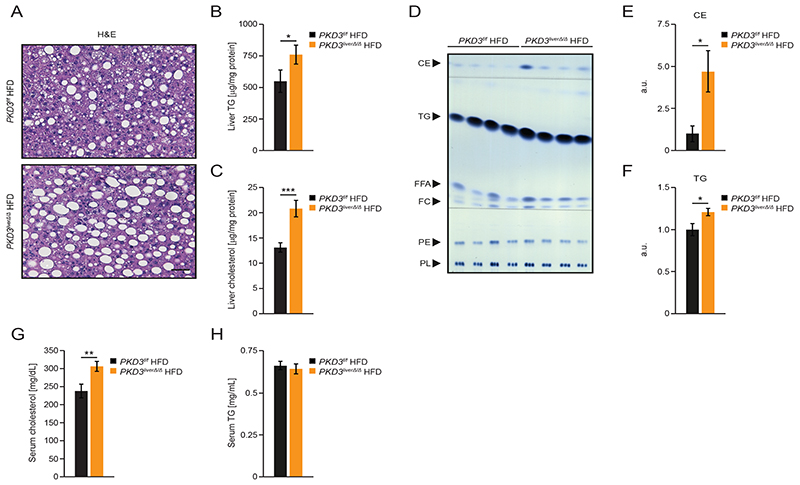
The increased liver weight of pKD3 liverΔ/Δ mice fed a HFD might be caused by enhanced proliferation of hepatocytes, decreased apoptosis, or increased lipid accumulation in the liver. Ki-67 staining, which marks proliferating cells, and cleaved caspase-3, which marks apoptotic cells, showed that PKD3 deficiency did not affect these processes in hepatocytes (fig. S4A and S4B). Similarly, the number of apoptotic (TUNEL-positive) hepatocytes was similar between pKD3 liverΔ/Δ mice and control animals (fig. S4C and S4D). However, H&E staining of liver sections revealed an increased number and enlarged lipid droplets in pKD3 liverΔ/Δ mice fed a HFD as compared to control animals (Fig. 3A), but no changes in the gross liver morphology of pKD3 liverΔ/Δ mice fed a ND (fig. S3K). Accordingly, hepatic TG and total cholesterol content was markedly enhanced in pKD3 liverΔ/Δ mice fed a HFD (Fig. 3B and C3), but not in those fed a ND (fig. S3L and S3M). Thin-layer chromatography (TLC) analysis of neutral lipids isolated from the livers of pKD3 liverΔ/Δ mice fed HFD revealed markedly increased cholesteryl ester and TG content (Fig. 3D-F). Circulating cholesterol levels were also increased in pKD3 liverΔ/Δ mice fed a HFD (Fig. 3G), but not in those fed a ND (fig. S3N and S3O). However, TG levels were not affected by a HFD in mice deficient for PKD3 (Fig. 3H). Increased lipid accumulation characterizes steatosis and NAFLD, which progresses into NASH upon the infiltration of immune cells, and into cirrhosis upon the induction of fibrosis (22). As assessed by immunohistochemistry, infiltration of macrophages (F4/80), T cells (CD3), or B cells (B220) was not altered in the livers from pKD3 liverΔ/Δ animals fed a HFD compared to control mice (fig. S4A). Similarly, collagen type IV and sirius red staining revealed that the extent of fibrosis was similar between HFD fed pKD3 liverΔ/Δ and control animals. Together, these results indicate that hepatic PKD3 suppresses cholesterol and TG accumulation in liver and therefore protects against the development of fatty liver disease.
Fig. 3. Deletion of PKD3 promotes hepatic accumulation of TG and cholesterol.
A Representative microscopy pictures of H&E stained liver sections from control and pKD3 liverΔ/Δ mice fed a HFD for 24 weeks (scale bar = 50 μm; representative of 6 mice/group).
B, C Quantification of TG (B) and cholesterol (C) content in extracted lipid phases normalized to total protein levels in livers from control and pKD3 liverΔ/Δ mice fed a HFD for 24 weeks (n=10 mice/group).
D-F Thin layer chromatography (TLC) separation of extracted hepatic lipids from mice of the indicated genotypes fed a HFD and the corresponding densitometric quantification using standards of known concentrations for cholesteryl ester (CE), triglycerides (TG), free fatty acids (FFA), free cholesterol (FC), phosphatidylethanolamine (PE), and phospholipids (PL) (n=8 mice/group).
G, H Quantification of serum cholesterol (G) and serum TG (H) concentrations in control and pKD3 liverΔ/Δ mice fed a HFD for 20 weeks (n=8 mice/group).
Data information: In (B, C, E-H), data are presented as mean ± SEM. *P>0.05, **P>0.01, ***P>0.001 (unpaired two-tailed Student’s t-test).
PKD3 suppresses de novo lipid synthesis in hepatocytes
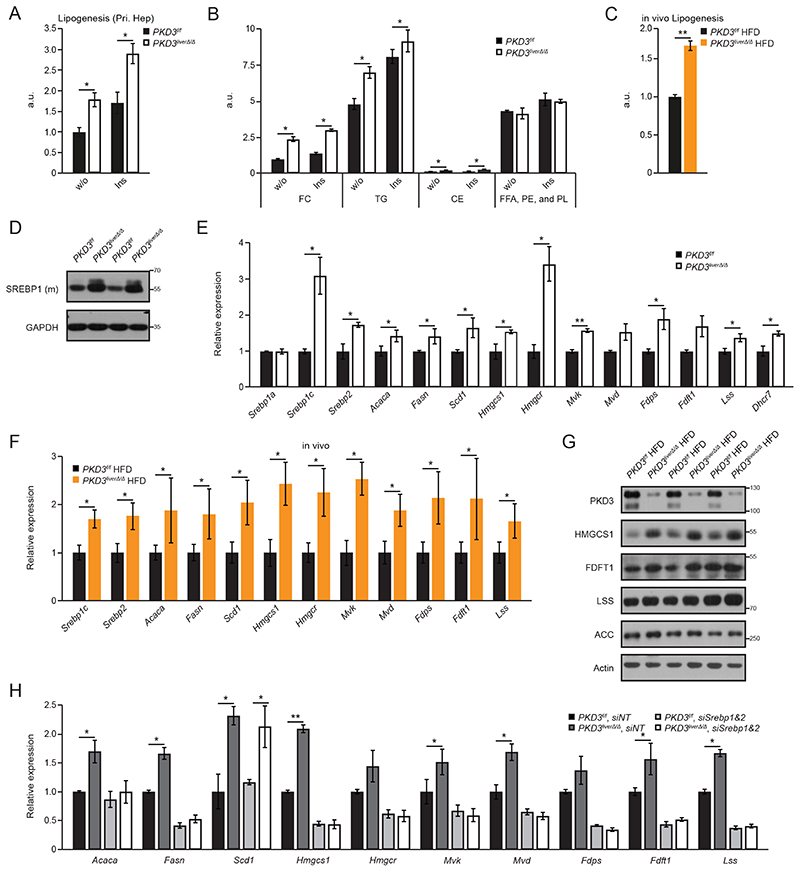
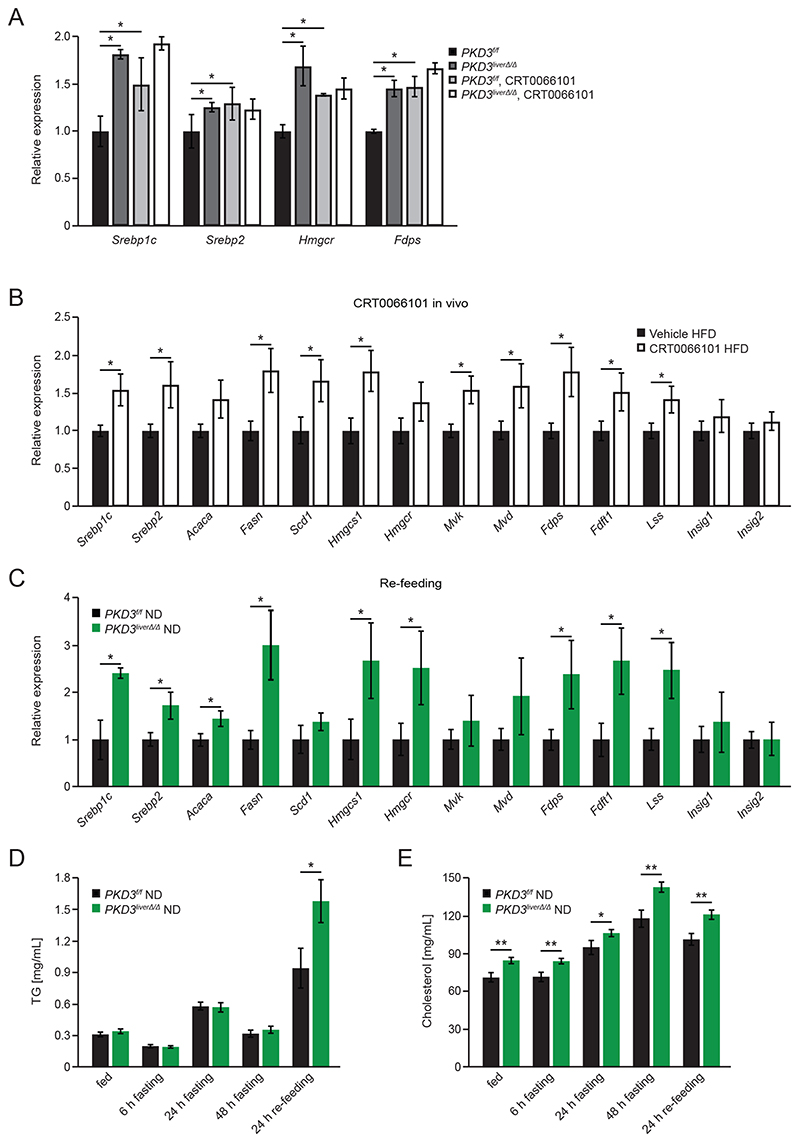
To unravel the cause for increased lipid accumulation in PKD3-deficient livers, we isolated primary hepatocytes and measured the rates of de novo lipogenesis and fatty acid oxidation. In parallel, we also assessed VLDL secretion in pKD3 liverΔ/Δ mice fed a HFD and ND. Overall de novo lipid synthesis was increased in hepatocytes deficient for PKD3 under basal and insulin-stimulated conditions (Fig. 4A), whereas fatty acid oxidation was moderately increased despite higher lipid accumulation in in pKD3 liverΔ/Δ mice (fig. S5A). To determine VLDL secretion, we performed tyloxapol injection (to inhibit lipoprotein lipase which results in blocked peripheral lipid uptake). We did not observe alterations in TG levels between pKD3 liverΔ/Δ and control animals regardless of the diet (fig. S5B, S5C). To further investigate increased de novo lipogenesis, we performed TLC-based separation of the major lipid classes. In the absence of PDK3, we observed increased synthesis rates of cholesterol (both free cholesterol and cholesteryl esters) and TG, whereas the synthesis of other lipids (free fatty acids (FFA), phosphatidylethanolamine (PE), and phospholipids (PL)) was not affected (Fig. 4B). Increased de novo lipogenesis was also observed in vivo in pKD3 liverΔ/Δ mice which synthesized significantly more lipids in liver under lipogenic conditions compared to control mice when fed a HFD (Fig. 4C). Sterol regulatory element-binding proteins (SREBPs) are master regulators of transcription of genes involved in hepatic fatty acids and cholesterol synthesis (3). The level of the cleaved and mature form of SREBP1, which promotes lipogenic gene expression and its own mRNA levels (3), was increased in PKD3-deficient hepatocytes compared to wild type hepatocytes (Fig. 4D and fig. S5D). Consistently, transcriptional analysis revealed increased levels of Srebp1c and Srebp2 as well as their target genes – Acaca (which encodes acetyl-CoA carboxylase alpha), Fasn (which encodes fatty acid synthase), Scd1 (which encodes stearoyl-CoA desaturase), Hmgcs1 (which encodes 3-hydroxyl-3-methylglutaryl-CoA synthase 1), Hmgcr (which encodes 3-hydroxyl-3-methylglutaryl-CoA reductase), Mvk (which encodes mevalonate kinase), Mvd (which encodes mevalonate diphosphate decarboxylase), Fdps (which encodes farnesyl diphosphate synthase), Fdft1 (which encodes farnesyl-diphosphate farnesyltransferase 1), Lss (which encodes lanosterol synthase), and Dhcr7 (which encodes 7-dehydrocholesterol reductase) – in hepatocytes deficient for PKD3 (Fig. 4E). Correspondingly, mRNA and protein levels of factors promoting lipogenesis were also increased in the livers of pKD3 liverΔ/Δ mice compared to control animals when fed a HFD (Fig. 4F, 4G and fig S5E). Conversely, Srebp1c and Srebp2 silencing abolished the significant difference in gene expression of lipogenic genes in PKD3-deficient compared to wild type hepatocytes (Fig. 4H and fig. S5F). Moreover, the PKD-specific inhibitor CRT0066101 increased the mRNA expression of Srebp1c, Srebp2, Hmgcr, and Fdps under insulin-stimulated conditions in primary hepatocytes and brought them to the same level as in PKD3-deficient hepatocytes without or with inhibitor (Fig. 5A). Accordingly, administration of the PKD inhibitor significantly increased the hepatic expression of Srebp1c, Srebp2, and their target genes (Fig. 5B).
Fig. 4. Hepatic PKD3 suppresses de novo lipogenesis in a SREBP-dependent manner.
A, B Basal and insulin-induced total de novo lipogenesis rate (measured under lipogenic conditions: serum deprived, 25 mM glucose, 0.5 mM sodium acetate, and 100 nM insulin) in primary hepatocytes isolated from mice of the indicated genotypes (arbitrary units – a.u.) (A) and quantification of TLC-separated free cholesterol (FC), triglycerides (TG), cholesteryl ester (CE), and other lipids (FFA, PE, and PL) (B) (n=3 biological replicates/group).
C In vivo de novo lipogenesis rate (a.u.) in livers from control and pKD3 liverΔ/Δ mice fed a HFD for 8 weeks, fasted overnight and re-fed for 4 hours before analysis (n=5 mice/group).
D Western blot analysis of mature SREBP1 in primary hepatocytes of the indicated genotypes under the same lipogenic conditions as B (n=3 mice/group; representative of three individual experiments).
E QPCR analysis of Srebp gene and target gene expression in primary hepatocytes of the indicated genotypes under the same lipogenic conditions as B (n=3 mice/group).
F QPCR analysis of Srebp gene and target genes expression in livers from control and pKD3 liverΔ/Δ mice fed for 20 weeks with HFD, subjected to fasting overnight and re-feeding for 4 hours (n=6 mice/group).
G Western blot analysis for indicated proteins in livers from the mice in F (n=3 mice/group).
H QPCR analysis of Srebp gene and target genes expression in primary hepatocytes of the indicated genotypes stimulated with insulin for 4 hours stimulation) that were either transfected with siNonTargeting (siNT) or the combination of slSrebp1 and slSrebp2 as indicated (n=3 biological replicates/condition).
Data information: In (A-C, E-F, and H), data are presented as mean ± SEM. *P>0.05, **P>0.01 (unpaired two-tailed Student’s t-test (C, E, F), or one way ANOVA with post hoc Tukey’s test (A, B, H)).
Fig. 5. The effect of the PKD inhibitor CRT0066101 and re-feeding on lipogenic gene expression and hepatic lipid accumulation.
A QPCR analysis of insulin-stimulated Srebp1c expression in DMSO or CRT0066101 (0.1 μM) treated primary hepatocytes of the indicated genotypes (n=3 biological replicates/condition).
B QPCR analysis of the expression of Srebp and target genes in livers from HFD-fed C57BL/6JRj mice (for 8 weeks) that either received an i.p. injection of vehicle or 10 mg/kg BW CRT0066101 inhibitor for 5 consecutive days. Mice were fasted overnight and re-fed for 4 hours before the livers were excised (n=8 mice/group).
C QPCR analysis of the indicated genes in livers from control and pKD3 liverΔ/Δ mice after overnight fasting and re-feeding for 4 hours (n=4 mice/group; combined data from two individual experiments).
D, E Quantification of serum TG (D) and serum cholesterol (E) concentrations at the indicated time points in control and pKD3 liverΔ/Δ mice fed a ND for 7 weeks that were fasted and re-fed for the indicated time points (n=12 mice/group).
Data information: In (A-E), data are presented as mean ± SEM. *P>0.05, **P>0.01 (unpaired two-tailed Student’s t-test (B, C), one way ANOVA with post hoc Tukey’s test (A), or two way ANOVA with post hoc Tukey’s test (D, E)).
Under physiological conditions, hepatic lipogenesis and the expression of genes promoting this process are induced upon feeding. The expression of Srebp1c, Srebp2, and some of their transcriptional targets were also increased in livers of PKD3-deficient mice subjected to a fasting/re-feeding protocol (Fig. 5C), whereas under fasting conditions, expression of lipogenic genes were not affected by PKD3 deletion (fig. S5G). Moreover, the expression of Insig1 (insulin-inducedgene 1) and Insig2 which encode proteins that prevent the proteolytic cleavage of Srebp did not differ between pKD3 liverΔ/Δ and control mice (Fig. 5C and fig. S5G). PKD3 protein level in liver was increased upon prolonged re-feeding after fasting, but PKD3 abundance and phosphorylation was not affected shortly (within 4 h) after food consumption (fig. S5H-K). Finally, serum TG and cholesterol levels were increased by fasting followed by 24 h of re-feeding in pKD3 liverΔ/Δ mice (Fig. 5D and 5E), indicating that PKD3 limits lipid synthesis evoked by feeding. Together, these data indicate that PKD3 suppresses de novo cholesterol and TG production by attenuating SREBP-dependent transcription of enzymes required for these processes.
PKD3 attenuates the expression of lipogenic genes in an AKT- and mTORC1/2-dependent manner
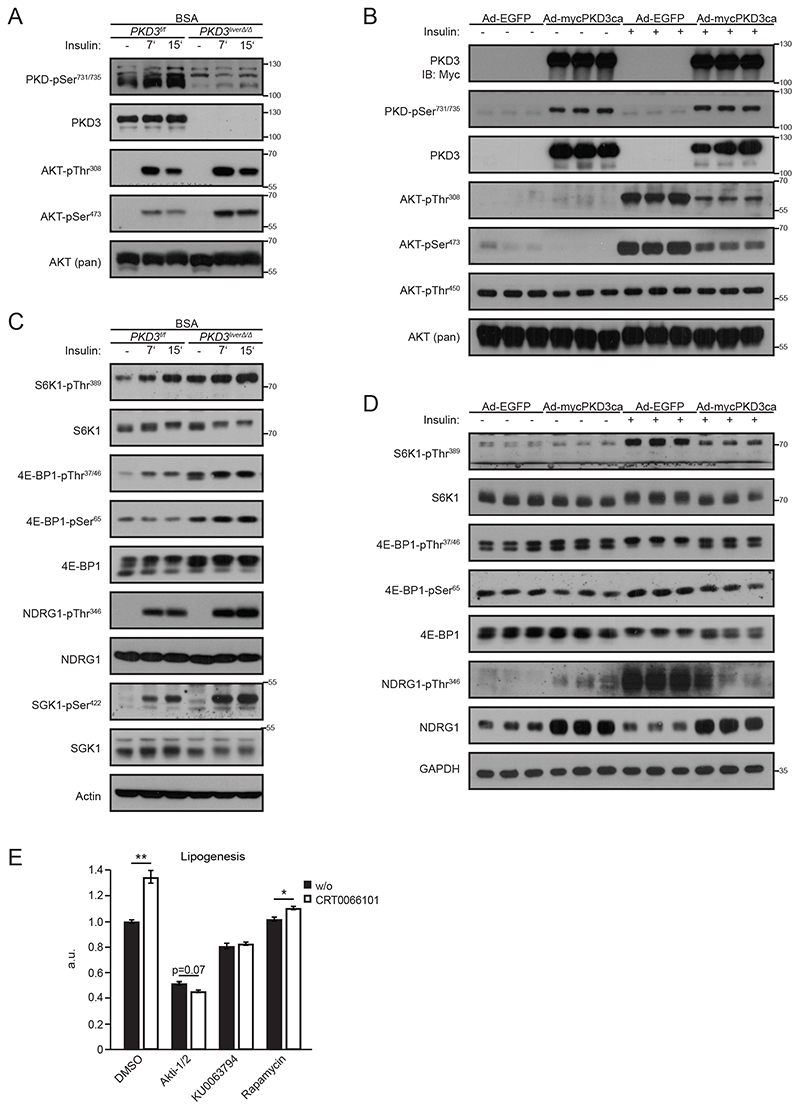
Insulin-stimulated activation of SREBP1c-dependent transcription relies on AKT activation (3, 23-25). We observed increased insulin-stimulated phosphorylation of AKT at Ser473 and Thr308 in hepatocytes deficient for PKD3 compared to control cells (Fig. 6A and fig. S7A). To check if PKD3 activation is sufficient to suppress insulin signaling, we created hepatocytes expressing a constitutively active form of PKD3 (mycPKD3ca), in which the two serine residues in the kinase domain are mutated to glutamic acid (S731E/S735E) (fig. S6C and fig. S8A) (26). This mutation leads to the activation of PKD3 regardless of the upstream signaling events. Consistently with previous results, expression of mycPKD3ca in hepatocytes reduced the phosphorylation of AKT at Ser473 and Thr308 upon insulin stimulation (Fig. 6B and fig. S8B).
Fig. 6. PKD3 suppresses de novo lipogenesis in an AKT and mTORC1/2-dependent manner.
A, C Western blot analysis for the indicated proteins in extracts from control and PKD3-deficient primary hepatocytes stimulated with 100 nM insulin for the indicated time points (n=3 independent experiments).
B, D WB analysis of the indicated proteins in extracts from PKD3-deficient primary hepatocytes transduced with either adenovirus expressing EGFP control (Ad-EGFP) or constitutive active PKD3 (Ad-mycPKD3ca) and stimulated with 100 nM insulin for 15 min (n=3 independent experiments).
E De novo lipogenesis rate in the presence of insulin and under lipogenic conditions in primary hepatocytes treated with DMSO, CRT0066101 (1 μM), Akti-1/2 (10 μM), KU0063794 (0.7 μM), and Rapamycin (0.7 μM) as indicated for 5 hours (n=3 biological replicates/condition).
Data information: In (E), data are presented as mean ± SEM. *P>0.05, **P>0.01 (one way ANOVA with post hoc Tukey’s test).
Insulin-evoked induction of lipogenesis requires the activation of downstream effectors of AKT. This includes activation of mTORC1 and mTORC2, which are required for SREBP1c activation and promote its transcription (4, 27-29). We observed enhanced phosphorylation of mTORC1 target ribosomal protein S6 kinase beta-1 (S6K1) on Thr389 and phosphorylation of 4E-BP1 on Thr37/46 and Ser65 in hepatocytes deficient for PKD3 (Fig. 6C and fig. S7B). Similarly, phosphorylation of downstream targets of mTORC2 such as serum/glucocorticoid-regulated kinase 1 (SGK1) on Ser422 and N-Myc downstream regulated target 1 (NDRG1) on Thr346 were also increased in the absence of PKD3 (Fig. 6C and fig. S7B). Phosphorylation of AKT on Ser473, which is a direct target of mTORC2, was also increased in hepatocytes in the absence of PKD3 (Fig. 6A and fig. S7A). The abundance and/or phosphorylation of other components of mTORC1 and mTORC2 was not affected by deletion of PKD3 (Fig. S6A and fig. S7C). Next, we loaded primary hepatocytes with oleic acid to mimic a high-fat diet in vitro. Under this condition, deletion of PKD3 resulted in increased phosphorylation of AKT on Ser473 and Thr308, 4E-BP1 on Thr37/46 and Ser65, SGK1 on Ser422, and NDRG1 on Thr346 (fig. S6B and fig. S7D, E). Conversely, expression of mycPKD3ca in hepatocytes resulted in decreased phosphorylation of mTORC1 targets (S6K1 on Thr389, 4E-BP1 on Thr37/46 and Ser65) and mTORC2 targets (NDRG1 on Thr346 and AKT on Ser473) (Fig. 6B, 6D and fig. S8B, S8C). However, the abundance of other mTORC1 and mTORC2 components were not affected by expression of mycPKD3ca (Fig. S6D and Fig. S8D, 8E), although the total protein level of NDRG1 was increased in hepatocytes expressing mycPKD3ca (Fig. 6D). Accordingly, the phosphorylation of S6K1 on Thr389, NDRG1 on Thr346, and SGK1 on Ser422 were also increased in the livers of HFD-fed pKD3 liverΔ/Δ mice that received insulin (fig S6E and fig. S8F). Phosphorylation of the insulin receptor substrate at Ser612 was increased in hepatocytes expressing mycPKD3ca (Fig. S6D), which indicates that other signaling pathways might be activated by expression of PKD3. Together, these data indicate that PKD3 suppresses lipogenesis by attenuating AKT-, mTORC1-, and mTORC2-dependent signaling. To support this notion, we measured the lipogenesis rate in hepatocytes treated with the PKD inhibitor CRT0066101 in combination with compounds that block the activity of AKT (Akti-1/2), mTORC1 (rapamycin), and mTORC1/2 (KU0063794) (Fig. 6E). As expected, inhibition of PKD increased the lipogenesis rate, and the addition of the AKT inhibitor resulted in similar inhibition of lipid synthesis in control and CRT0066101 treated hepatocytes (Fig. 6E). Similarly, a mTORC1/2 inhibitor (KU0063794) decreased lipogenesis in control and CRT0066101-treated cells to the same extent (Fig. 6E). However, rapamycin treatment only partially inhibited lipogenesis in CRT0066101 treated cells compared to control cells (Fig. 6E).
The closely related PKD1 isoform promotes lipogenesis in adipocytes by phosphorylating 5′ AMP-activated protein kinase (AMPK) and suppressing its activity towards acetyl-CoA carboxylase 1/2 (ACC1/2) (19). However, our data suggests that PKD3 does not affect AMPK or ACC1/2 activity in hepatocytes (fig. S6D and fig. S8E) but attenuates AKT and mTORC1/2 driven lipogenesis.
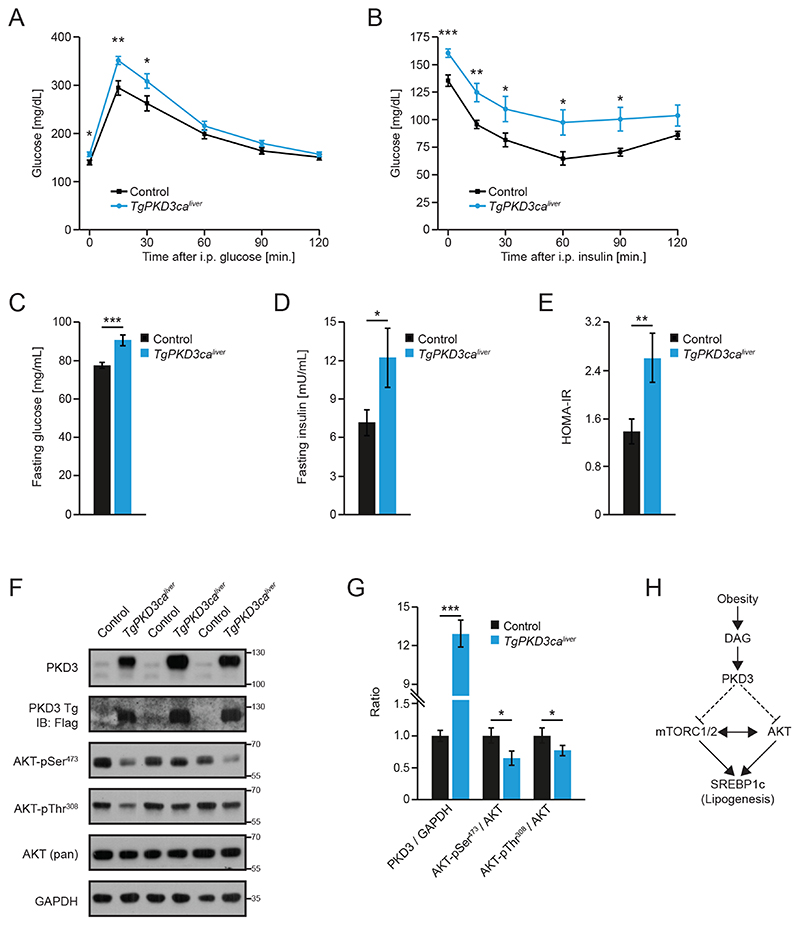
Constitutive activation of PKD3 promotes insulin resistance in liver
To analyze the effect of moderate overexpression of PKD3ca on insulin signaling in vivo, we generated loxP-STOP-loxP-FlagPKD3ca mice and crossed them with mice expressing Cre recombinase under the control of the albumin promoter (21) to restrict expression of PKD3ca to hepatocytes. Liver-specific transgenic PKD3ca mice (TgPKD3caliver) showed impaired glucose tolerance (Fig. 7A and fig. S9A) which was accompanied by reduced insulin sensitivity even when these mice were fed a ND (Fig. 7B and fig. S9B). Accordingly, these observations were also reflected by increased fasting glucose and fasting insulin levels (Fig. 7C and 7D). Overall, TgPKD3caliver mice presented with pronounced insulin resistance as shown by a HOMA-IR index of 2.5 (Fig. 7E). Consistent with the results obtained from isolated hepatocytes, we observed reduced phosphorylation of AKT on Ser473 and Thr308 in livers from TgPKD3calver mice that were re-fed (Fig. 7F and 7G). Expression of lipogenic genes was only marginally reduced, whereas liver triglyceride and cholesterol content were not significantly reduced (P=0.64 and P=0.08, respectively) in TgPKD3caliver mice when fed a ND (fig. S9C-E). Together, these data suggest that activation of PKD3 alone is sufficient to suppress insulin signaling (Fig. 7H).
Fig. 7. Liver-specific expression of a constitutively active PKD3 promotes insulin resistance.
A, B Glucose (2 g/kg BW) (A) and insulin (1 U/kg BW) (B) tolerance test in TgPKD3cahver and control mice fed a ND for 12 and 10 weeks, respectively. (n=11 mice (Control) and n=13 mice (Tg)).
C-E After overnight fast, blood glucose (C) and insulin (D) were measured and used to determine HOMA-IR (E) in mice of the indicated genotypes (n=11 mice (Control) and n=13 mice (Tg)).
F, G Western blot analysis of the indicated proteins from livers of mice of the indicated genotypes that were fasted overnight and refed 4 h before analysis (F) and corresponding densitometric quantification (G) (n=3 mice/group).
H Summary of PKD3 signaling in liver.
Data information: In (A-E and G), data are presented as mean ± SEM. *P>0.05, **P>0.01, ***P>0.001 (unpaired two-tailed Student’s t-test (C-E, G) or two way ANOVA with post hoc Tukey’s test (A, B)).
Discussion
DAG-evoked activation of PKC-dependent signaling is critical for the development of hepatic insulin resistance (2, 5-7). Our study revealed PKD3 as a mediator of DAG-evoked insulin resistance in liver. DAG is an intermediate product of TG synthesis, but it is also utilized by a subclass of G protein coupled receptors as second messenger (5). Our data also demonstrated that PKD3 was activated in livers of obese (HFD fed) mice. Moreover, lipid loading of primary hepatocytes as well as stimulation of cells with insulin resulted in activation of PKD3. However, the precise upstream mechanism promoting PKD3 activation and the relationship of this kinase to the classical PKC-dependent signaling pathway requires further investigation.
Although we showed that PKD3 is one of the signaling molecules aberrantly activated during metabolic overload evoked by high levels of lipids supplementation and its deletion partially ameliorated insulin resistance caused by lipid overload, PKD3 seems to have also a physiological function in liver. We revealed that PKD3 expression was important to provide negative feedback loop on lipogenesis evoked by re-feeding after fasting/starvation, which would be in line with the increase in PKD3 abundance in response to prolonged re-feeding and the increase in PKD activity in response to insulin stimulation.
PKCε suppresses insulin action by directly phosphorylating the insulin receptor (6, 7). Our data suggests that PKD3 also suppressed insulin action, resulting in impaired AKT phosphorylation and reduced mTORC1 and mTORC2 activation in vitro and in vivo. Hepatic deletion of PKD3 improved glucose tolerance in mice, but also resulted in an increased SREBP-dependent lipogenesis and consequently hepatic accumulation of TG and cholesterol. SREBP1c and SREBP2 are critical for the transcriptional activation of lipogenic machinery in hepatocytes (3). However, the phosphorylation of the mTORC2 downstream effector NDRG1 promotes lipogenesis in a PPARγ-dependent manner in adipocytes (30). We found that phosphorylation of NDRG1 was enhanced in the absence of PKD3, suggesting that PKD3 might suppress lipogenesis by utilizing mechanisms complementary to the suppression of SREBP-dependent transcription of lipogenic genes. Similarly, PKD3-dependent suppression of AKT and mTORC1/2 action might influence lipogenesis by inhibiting factors as DNA-dependent protein kinase (DNA-PK) or upstream stimulatory factor (USFs), which also promote the expression of lipogenic enzymes (31). However, we showed that silencing of SREBP isoforms was sufficient to normalize the elevated rates of lipogenesis in PKD3-deficient hepatocytes, which indicates that PKD3 regulates lipogenesis primarily by suppressing SREBP-dependent transcription of lipogenic genes. PKD3 did not affect TG secretion in the form of very low density lipoproteins (VLDL) from hepatocytes. Moreover, deletion of PKD3 seemed to increase FA oxidation in hepatocytes. Increased rates of FA oxidation in hepatocytes would be expected to lead to reduced TG content in liver. However, the lipid accumulation in livers of PKD3-deficient mice, despite increased FA oxidation and unaffected rates of VLDL secretion, indicates that elevated lipogenesis is the primary process responsible for the observed phenotype. Although the closely related PKD1 promotes lipid accumulation in adipocytes in an AMPK-dependent manner (19), PKD3 did not affect the activity of AMPK or its downstream target ACC1/2 in the liver. Additionally, PKD3 seemed to be the predominant PKD isoform that regulates liver metabolism because PKD inhibition did not further stimulate expression of lipogenic genes in the absence of PKD3 and other PKD isoforms were only marginally expressed in hepatocytes. Therefore, our results suggest that different PKD isoforms have a specific set of substrates in different tissues.
Our results indicate also that the expression of the constitutively active form of PKD3 was sufficient to attenuate insulin signaling (AKT and TORC1/2 activity) even in the absence of hyperlipidemia. Mice expressing the constitutively active form of PKD3 in liver showed reduced insulin sensitivity and reduced AKT phosphorylation even when fed normal diet. Cholesterol and triglyceride content was not altered in these livers and the expression of some SREBP target genes was only marginally reduced (fig S9C-E). These results might imply that increased insulin signaling is sufficient to enhance lipogenesis but that reducing insulin signaling does not necessarily block the lipogenic program. Alternatively, the ability of PKD3 to attenuate insulin signaling on lipogenesis might be present only in the livers of animals challenged with high fat diet.
Multiple studies report that hepatic TG and cholesterol levels correlate with the development of insulin resistance (reviewed in (2)). However, we found that deletion of PKD3 promoted glucose utilization and insulin sensitivity while at the same time enhancing lipid accumulation in the liver. In fact, improved glucose utilization and increased lipid deposition are observed in several mouse models in which AKT activity is increased (23, 32). Nevertheless, we did not observe increases in apoptosis, immune cell infiltration or fibrosis in PKD3-deficient livers, which are often associated with the progression of NAFLD into NASH (2). Therefore, PKD3 deletion results in development of metabolically healthy fatty liver.
Finally, our data indicates that the expression of the constitutively active form of PKD3, which preserves its activity in hepatocytes of animals independently of hormonal or metabolic challenges, suppressed insulin signaling and induced insulin resistance and glucose intolerance. The expression of PKD3ca in hepatocytes not only suppressed AKT and mTORC – dependent signaling but also led to phosphorylation of the insulin receptor substrate at Ser612, which indicates that other signaling pathways (for example mitogen-activated protein kinases (33)) might be activated by expression of PKD3. Together, these data indicates that PKD3 expression is not only required to provide negative feedback on insulin signaling under conditions of lipid overload but its activation is also sufficient to partially switch off insulin-dependent signaling.
Material and Methods
Diacylglycerol (DAG) quantification
DAG analysis was performed as described in detail before (19). Briefly, lipids were isolated from liver homogenates by butanol-methanol extraction method. 1,2-Dioctanoyl-sn-glycerol (Enzo Life Sciences) was used as internal standard. Subsequently, lipid extracts were fractionated into lipid classes on a silica matrix column (Phenomenex) and the DAG fraction was analyzed by LC/MS.
Animals
PKD3 floxed mice (PKD3f/f) (20) and loxP-STOP-loxP-(3xFlag)PKD3ca mice, in which the two serines in the PKD3 kinase domain are mutated to glutamic acid (S731E/S735E) (generated by Cyagen with the PiggyBac transgenic method), were mated with mice expressing transgenic Cre recombinase under the control of the albumin promoter/enhancer (The Jackson Laboratory) (21) to generate homozygous liver-specific PKD3-deficient mice (pKD3 liverΔ/Δ) and heterozygous liver-specific transgenic PKD3ca mice (TgPKD3caliver), respectively. All animal studies were approved by the local animal welfare authorities (Regierung von Unterfranken) with the animal protocol number AK55.2-2531.01-124/13 and AK55.2.2-2532-2-741-13. The mice were housed in cages from Tecniplast in a green line IVC-rack system with ad libitum supply of water and normal chow diet (ssniff Spezialdiaeten). In case of HFD (Research Diets, D12331i), mice received the high calorie diet (58% kcal from fat) from the age of 3 weeks for a duration of 24 weeks and body weight measurements were performed weekly. For dissection, mice were sacrificed by cervical dislocation and organs (liver, epiWAT, subWAT, BAT, SKM) were weighted and snap frozen in liquid nitrogen. For fasting/re-feeding experiments, mice were fasted overnight for 16 hours with free access to water and food was restored for 4 hours before livers were removed as described before. For insulin injections, mice received an intraperitoneal (i.p.) dose of 8 U/kg body weight (BW) of insulin (Sigma) and livers were collected after 15 min according to standard procedures. For in vivo experiments using CRT0066101, C57BL/6JRj mice (Janvier Labs) were fed a HFD for 8 weeks and were randomly assigned to two groups that either received an i.p. injection of 10 mg/kg BW of CRT0066101 in 5% DMSO or vehicle for 5 consecutive days.
Glucose and insulin tolerance tests
For glucose tolerance tests, mice were fasted for 4 hours prior to the experiment (starting at 8am) and received an i.p. injection of defined dose of glucose (Carl Roth) as indicated in the figure legends. Glucose concentrations in blood were measured with Accu-chek glucometer (Roche) at 0, 15, 30, 60, 90, and 120 min time points. Insulin tolerance tests were performed similarly except that the mice received a defined dose of insulin (Sigma) as indicated in the figure legends.
Serum metabolite and HOMA-IR analysis
Blood samples were collected after 20 weeks of ND or HFD feeding or after 6 h, 24 h, 48 h of fasting and 24 h of re-feeding for the fasting/re-feeding experiment. Cholesterol (Wako, LabAssay Cholesterol) and TG (Sigma, Triglycerides Kit) concentrations in serum were determined according to the manufacturers’ protocols. Serum insulin levels were quantified by either using a magnetic bead based immunoassay kit (Bio-Rad, Bio-Plex Pro Mouse Insulin) and MAGPIX multiplex reader (Bio-Rad) or by Mouse Insulin ELISA Kit (Crystal Chem) and a SPARK plate reader (Tecan) according to the manufacturers’ guidelines. HOMA-IR was calculated with the following formula: fasting insulin (mU/mL) x fasting glucose (mg/dL) divided by 405, using blood from mice that were fasted overnight.
VLDL secretion
Mice were fasted overnight before they received a retro-orbital injection of 0.5mg/g BW tyloxapol. Blood was collected at 0, 1, 2, 4, and 6 h time points and used for serum TG analysis as described above.
Metabolic measurements
Metabolic parameters such as energy expenditure (EE), food intake, activity, and respiratory exchange ratio (RER) were determined in the Phenomaster (TSE) system as described before (34) after twenty-three weeks on specific diets.
Histological, immunohistochemistry, and immunofluorescent analyses
For histology, liver tissues were fixed in 10% formalin and embedded in paraffin. Hematoxylin-eosin (H&E) or sirius red staining of 2-4 μm liver sections were performed according to standard laboratory procedures. For immunohistochemistry, Ki-67 (H2, 95°C), cleaved caspase-3 (H2), B220 (H2), CD3 (H2, 95°C), F4/80 (E1), and collagen type IV (E1) antibodies were used with EDTA pretreatment (H2) or an enzyme pretreatment kit (E1) (Leica), respectively. For TUNEL staining, fluorescent in situ cell death detection kit (Roche) was utilized according to the manufacturer’s instructions.
Lipid extraction and thin layer chromatography (TLC)
Lipids were extracted according to the protocol of Bligh and Dyer with modifications (35). In brief, 0.5 volumes of liver homogenate (1:100 in PBS) were acidified with 0.3 volumes of 0.2 N HCl. Subsequently, 3 volumes of MeOH/CHCl3 (2:1; v/v), 1 volume of CHCl3, and 1 volume of H2O were added and mixed vigorously stepwise. The phases were separated by centrifugation and the lower phase was transferred to a new tube and evaporated under a stream of nitrogen. The lipids were either resuspended in DMSO/H2O for enzymatic quantification of TG and cholesterol (as described before) or in MeOH/CHCl3 (1:1; v/v) for separation by TLC on a silica gel 60 plate (Merck) in the solvent mixtures of CHCl3/MeOH/20 % acetic acid (65/25/5; v/v/v), hexane/ethyl acetate/acetic acid (59/10/1; v/v/v), and pure hexane. Cholesteryl palmitate (for CE), triolein (for TG), oleic acid (for FFA), cholesterol (for FC), phosphatidylethanolamine (for PE), and phosphatidylcholine (for PL) were used as standards for the TLC. Lipid spots were visualized by dipping the plate in a hydrous solution of 2.5 % 12-molybdophosphoric acid (Alfa Aesar), cerium (IV) sulfate (Sigma), and 6 % H2SO4 and heating at 200 °C until the bands appear. Densitometric analysis was performed using ImageJ and known concentrations of the standards.
Primary hepatocyte isolation and culture
Primary mouse hepatocytes were prepared by the collagen perfusion method. Eight to twelve week old male mice were anaesthetized with ketamine/xylazine and the vena cava was cannulated. The portal vein was cut immediately and the liver was perfused with Earle′s Balanced Salt Solution (EBSS w/o Ca2+/Mg2+ (Thermo Fisher Scientific)) supplemented with 0.5 mM EGTA. Subsequently, the buffer was replaced to Hank’s Balanced Salt Solution with Ca2+/Mg2+ (Biochrom) containing 100 U/mL type I collagenase (Worthington). After sufficient digestion, the liver was excised and the gall bladder was removed. Cells were liberated into Dulbecco’s modified eagle’s medium (DMEM) supplemented with 10 % FBS and 1 % penicillin/streptomycin (P/S), filtered through a 100 μm cell strainer, and centrifuged (50 G, 3 min, 4 °C). Afterwards, Percoll (GE Healthcare) mixed with respective amounts of 10x PBS and culture medium was used to form a gradient that allowed enrichment of the hepatocyte fraction (50 G, 10 min, 4 °C), which was washed 3 times with culture medium (50 G, 3 min, 4 °C). Hepatocytes were then plated on collagen-type I-coated 6 or 12-well plates (BD). After 4-6 hours to allow attachment, the medium was replaced by fasting medium (DMEM supplemented with 0.2 % FFA-free BSA (Sigma) and 1x P/S) and the cells were used the following day unless otherwise noted. For PKD3 phosphorylation studies, primary hepatocytes were either incubated with 1,2-Dioctanoyl-sn-glycerol (Enzo Life Sciences) for 4 h, 6 h, and 8 h or with oleic acid bound to albumin (Sigma) for 4 h and 20 h.
Adenovirus infection
Primary hepatocytes were infected 4-6 h after plating with adenoviruses expressing either EGFP (Ad-EGFP) or a constitutively active form of PKD3 (Ad-mycPKD3ca) at an MOI of 10. Medium was replaced the following morning and cells were used for experiments 36-48 h after infection. Transduction efficiency, which was 100 %, was assessed by analyzing the expression of the EGFP reporter (which was present in all adenoviruses).
siRNA transfections
Primary hepatocytes were transfected 4-6 h after plating with siNonTargeting (60 nM), siSrebp1 (30 nM), or siSrebp2 (30 nM) (Dharmacon) using Lipofectamine RNAiMAX transfection reagent (Invitrogen) according to the manufacturers’ recommendations. Medium was replaced the following morning and cells were used for experiments 36-48 h after infection.
Lipogenesis Assay
De novo lipogenesis assay was performed one day after the isolation of primary hepatocytes. Cells were incubated with a lipogenic medium (DMEM w 25 mM glucose, 0.2 % FFA-free BSA, and 1x P/S) supplemented with 0.5mM sodium acetate and 4 μCi/mL 3H-acetate (Perkin Elmer) at basal or insulin (100 nM) stimulated conditions for 4-6 hours. Cells were then washed twice with PBS and scraped into 0.1 N HCl. Subsequently, the lipids were extracted similarly as described before. 0.8 volumes of homogenate were mixed stepwise with 3 volumes of MeOH/CHCl3 (2:1; v/v), 1 volume of CHCl3, and 1 volume of H2O. The phases were separated by centrifugation (3000 G, 10 min) and the lower phase was transferred to a new tube which was washed once with upper phase MeOH/CHCl3/H2O (15:175:180; v/v/v) (3000 G, 10 min). Finally, the lower phase was transferred to a scintillation tube and evaporated under a stream of nitrogen. The dried lipids were resuspended in 4 mL scintillation liquid before lipid scintillation counting. The lipogenesis rate was calculated as the amount of tritium incorporated into the newly formed lipids (measured in dpm) per total protein (Bradford, Bio-Rad) per hour. Alternatively, the extracted lipids were separated on a TLC plate as described before. The stained lipid spots were scraped and analyzed by lipid scintillation counting. To measure de novo lipogenesis in vivo, mice were fed a HFD for 8 weeks. Following overnight fasting, mice were re-fed with a HFD for 4 hours before they received an i.p. dose of 8 mCi [3H]-water/100g BW. After 2 h, the livers were excised and liver pieces (3 per mouse) were homogenized 1:5 in 0.1 N HCl followed by lipid extraction as described above. For inhibitor treatments, 10 μM Akti-1/2, 0.7 μM KU0063794, and 0.7 μM Rapamycin were added 4-6 hours after plating and 16 hours prior to the experiment, whereas 1 μM CRT0066101 was added one hour before the experiment.
FA oxidation
Hepatocytes were serum-fasted overnight and incubated with DMEM containing 2 μCi/mL 3H-oleic acid (Perkin Elmer) and 0.2 mM oleic acid bound to BSA (Sigma) at basal or insulin (100 nM) stimulated conditions for 3 hours. Subsequently, the supernatant was collected and the cells were washed twice with PBS and scraped in 0.1 N HCl for determining the protein concentration. Then, 1 volume of MeOH/CHCl3 (2:1; v/v) and 1 volume of 2 M KCl/2 M HCl (1:1; v/v) were added to 0.5 volumes of the supernatant and mixed stepwise. The phases were separated by centrifugation (3000 G, 10 min) and the upper phase was transferred to a new tube and the procedure was repeated once more. Finally, the upper phase was transferred to a scintillation tube and 4 mL scintillation liquid were used for scintillation counting. The fatty acid oxidation rate was calculated as the amount of tritium incorporated into 3H2O (measured in dpm) per total protein (Bradford, Bio-Rad) per hour.
Immunoblotting
Western blot was performed according to standard procedures. Briefly, 20 μg of protein lysates were separated by a 10% SDS-polyacrylamide gel electrophoresis and transferred on a polyvinylidenfluorid (PVDF) membrane and then probed overnight with corresponding primary antibodies (table S1). Following incubation with corresponding mouse or rabbit horseradish peroxidase (HRP)-conjugated secondary antibody, proteins were visualized using ECL Reagent in combination with X-ray films (Fuji) or with Amersham Imager 600 (GE Healtcare) for densitometric analysis.
Real-time qPCR analysis
Total RNA was extracted from tissue or cells using Qiazol reagent (Qiagen) according to the manufacturer’s instructions. Reverse transcription of 1 μg of RNA was performed by using First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). Real-time quantitative polymerase chain reaction (QPCR) was performed using PowerUp SYBR Green Master Mix (Thermo Fisher Scientific) on a QuantStudio 5 Real-Time PCR System (Thermo Fisher Scientific). Relative amounts of all mRNAs were calculated using comparative CT method normalized to the reference genes Rpl13a, 36B4, or Hprt1. The primer sequences (sense and antisense) for mouse are listed in table S2. Absolute quantification of PKD isoform copy numbers in liver was performed according to the standard protocol of Applied Biosystems. In brief, the primers were designed to be located within same exons (table S2) and genomic DNA of known concentration was used to create a standard curve reflecting copy numbers.
Statistical analysis
Data are presented as mean values ± standard error of the mean (SEM). Significances were determined by using two-tailed Student’s T-Test for independent groups or by using one-way analysis of variance (ANOVA) followed by the Post hoc Tukey test for multiple comparisons. P-values of 0.05 or lower were considered as statistically significant.
Supplementary Material
Acknowledgments
We thank Olga Sumara for critical comments to our manuscript, Annette Schürmann and Christian Baumeister for introducing us to primary hepatocyte isolation, Jenny Hetzer and Danijela Heide for performing the immunohistochemical stainings, and Katja Aurbach, Isabelle Becker, and Deya Cherpokova for experimental assistance.
Funding
This project was supported by EFSD/Janssen Rising Star Fellowship Programme (2013) from European Foundation for the Study of Diabetes and the Starting Grant (SicMetabol) from the European Research Council (ERC) and internal funds of the Rudolf Virchow Center for Experimental Biomedicine. R. El-Merahbi and G. Sumara were also funded by the Emmy Noether grant from the German Research Foundation (Number Su 820/1-1). M. Heikenwalder was supported by an ERC Consolidator grant (HepatoMetaboPath). A. Schulze was supported by a German Research Foundation grant (FOR2314, SCHU2670/1-1).
Footnotes
Author contributions: A.E.M. and G.S. conceived the study, designed the experimental procedures, and wrote the manuscript. A.E.M. performed the majority of the experimental work and analyzed the data. M.C.L, A.E.L.V., R.M., J.T.V., and M.E. performed parts of the experiments. W.S. participated in experimental design and carried out the mass spectrometry. M.L., T.Z., and U.B. provided the pKD3f/f mice. M.H. and A.S. contributed to the experimental design.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability
The pKD3f/f mice require an MTA by the PKC Research Consult (Cologne, Germany). All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References and Notes
- 1.Fruman DA, et al. The PI3K Pathway in Human Disease. Cell. 2017;170:605–635. doi: 10.1016/j.cell.2017.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Samuel VT, Shulman GI. Nonalcoholic fatty liver disease as a nexus of metabolic and hepatic diseases. Cell metabolism. 2017 doi: 10.1016/j.cmet.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krycer JR, Sharpe LJ, Luu W, Brown AJ. The Akt–SREBP nexus: cell signaling meets lipid metabolism. Trends in Endocrinology & Metabolism. 2010;21:268–276. doi: 10.1016/j.tem.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Ricoult SJ, Manning BD. The multifaceted role of mTORC1 in the control of lipid metabolism. EMBO reports. 2013;14:242–251. doi: 10.1038/embor.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samuel VT, Shulman GI. Mechanisms for insulin resistance: common threads and missing links. Cell. 2012;148:852–871. doi: 10.1016/j.cell.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petersen MC, et al. Insulin receptor Thr 1160 phosphorylation mediates lipid-induced hepatic insulin resistance. The Journal of clinical investigation. 2016;126:4361–4371. doi: 10.1172/JCI86013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samuel VT, et al. Inhibition of protein kinase Cε prevents hepatic insulin resistance in nonalcoholic fatty liver disease. The Journal of clinical investigation. 2007;117:739–745. doi: 10.1172/JCI30400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rozengurt E. Protein kinase D signaling: multiple biological functions in health and disease. Physiology (Bethesda) 2011;26:23–33. doi: 10.1152/physiol.00037.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleger A, et al. Protein kinase D2 is an essential regulator of murine myoblast differentiation. PLoS One. 2011;6:e14599. doi: 10.1371/journal.pone.0014599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steiner TS, Ivison SM, Yao Y, Kifayet A. Protein kinase D1 and D2 are involved in chemokine release induced by toll-like receptors 2, 4, and 5. Cell Immunol. 2010;264:135–142. doi: 10.1016/j.cellimm.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 11.Konopatskaya O, et al. Protein kinase C mediates platelet secretion and thrombus formation through protein kinase D2. Blood. 2011;118:416–424. doi: 10.1182/blood-2010-10-312199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fielitz J, et al. Requirement of protein kinase D1 for pathological cardiac remodeling. Proc Natl Acad Sci U S A. 2008;105:3059–3063. doi: 10.1073/pnas.0712265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim MS, et al. Protein kinase D1 stimulates MEF2 activity in skeletal muscle and enhances muscle performance. Mol Cell Biol. 2008;28:3600–3609. doi: 10.1128/MCB.00189-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li C, et al. Protein kinase D3 is a pivotal activator of pathological cardiac hypertrophy by selectively increasing the expression of hypertrophic transcription factors. J Biol Chem. 2011;286:40782–40791. doi: 10.1074/jbc.M111.263046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matthews SA, et al. Essential role for protein kinase D family kinases in the regulation of class II histone deacetylases in B lymphocytes. Mol Cell Biol. 2006;26:1569–1577. doi: 10.1128/MCB.26.4.1569-1577.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ittner A, et al. Regulation of PTEN activity by p38delta-PKD1 signaling in neutrophils confers inflammatory responses in the lung. J Exp Med. 2012;209:2229–2246. doi: 10.1084/jem.20120677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferdaoussi M, et al. G protein-coupled receptor (GPR)40-dependent potentiation of insulin secretion in mouse islets is mediated by protein kinase D1. Diabetologia. 2012;55:2682–2692. doi: 10.1007/s00125-012-2650-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sumara G, et al. Regulation of PKD by the MAPK p38delta in insulin secretion and glucose homeostasis. Cell. 2009;136:235–248. doi: 10.1016/j.cell.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loffler MC, et al. Protein kinase D1 deletion in adipocytes enhances energy dissipation and protects against adiposity. EMBO J. 2018;37 doi: 10.15252/embj.201899182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang T, Braun U, Leitges M. PKD3 deficiency causes alterations in microtubule dynamics during the cell cycle. Cell Cycle. 2016;15:1844–1854. doi: 10.1080/15384101.2016.1188237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Postic C, et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki A, Diehl AM. Nonalcoholic Steatohepatitis. Annu Rev Med. 2017;68:85–98. doi: 10.1146/annurev-med-051215-031109. [DOI] [PubMed] [Google Scholar]
- 23.Ono H, et al. Hepatic Akt activation induces marked hypoglycemia, hepatomegaly, and hypertriglyceridemia with sterol regulatory element binding protein involvement. Diabetes. 2003;52:2905–2913. doi: 10.2337/diabetes.52.12.2905. [DOI] [PubMed] [Google Scholar]
- 24.Fleischmann M, Iynedjian PB. Regulation of sterol regulatory-element binding protein 1 gene expression in liver: role of insulin and protein kinase B/cAkt. Biochemical Journal. 2000;349:13–17. doi: 10.1042/0264-6021:3490013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leavens KF, Easton RM, Shulman GI, Previs SF, Birnbaum MJ. Akt2 is required for hepatic lipid accumulation in models of insulin resistance. Cell metabolism. 2009;10:405–418. doi: 10.1016/j.cmet.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozgen N, et al. Protein kinase D links Gq-coupled receptors to cAMP response elementbinding protein (CREB)-Ser133 phosphorylation in the heart. J Biol Chem. 2008;283:17009–17019. doi: 10.1074/jbc.M709851200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hagiwara A, et al. Hepatic mTORC2 activates glycolysis and lipogenesis through Akt, glucokinase, and SREBP1c. Cell metabolism. 2012;15:725–738. doi: 10.1016/j.cmet.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 28.Yecies JL, et al. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell metabolism. 2011;14:21–32. doi: 10.1016/j.cmet.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Porstmann T, et al. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell metabolism. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai K, El-Merahbi R, Loeffler M, Mayer AE, Sumara G. Ndrg1 promotes adipocyte differentiation and sustains their function. Sci Rep. 2017;7:7191. doi: 10.1038/s41598-017-07497-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Viscarra J, Kim SJ, Sul HS. Transcriptional regulation of hepatic lipogenesis. Nat Rev Mol Cell Biol. 2015;16:678–689. doi: 10.1038/nrm4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stiles B, et al. Liver-specific deletion of negative regulator Pten results in fatty liver and insulin hypersensitivity. Proceedings of the Natlonal Academy of Sciences. 2004;101:2082–2087. doi: 10.1073/pnas.0308617100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Izawa Y, et al. ERK1/2 activation by angiotensin II inhibits insulin-induced glucose uptake in vascular smooth muscle cells. Exp Cell Res. 2005;308:291–299. doi: 10.1016/j.yexcr.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 34.Trujillo Viera J, El-Merahbi R, Nieswandt B, Stegner D, Sumara G. Phospholipases D1 and D2 Suppress Appetite and Protect against Overweight. PLoS One. 2016;11:e0157607. doi: 10.1371/journal.pone.0157607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The pKD3f/f mice require an MTA by the PKC Research Consult (Cologne, Germany). All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.