Abstract
A prompt public health response to a new epidemic relies on the ability to monitor and predict its evolution in real time as data accumulate. The 2009 A/H1N1 outbreak in the UK revealed pandemic data as noisy, contaminated, potentially biased and originating from multiple sources. This seriously challenges the capacity for real-time monitoring. Here, we assess the feasibility of real-time inference based on such data by constructing an analytic tool combining an age-stratified SEIR transmission model with various observation models describing the data generation mechanisms. As batches of data become available, a sequential Monte Carlo (SMC) algorithm is developed to synthesise multiple imperfect data streams, iterate epidemic inferences and assess model adequacy amidst a rapidly evolving epidemic environment, substantially reducing computation time in comparison to standard MCMC, to ensure timely delivery of real-time epidemic assessments. In application to simulated data designed to mimic the 2009 A/H1N1 epidemic, SMC is shown to have additional benefits in terms of assessing predictive performance and coping with parameter nonidentifiability.
Keywords: Sequential Monte Carlo, resample-move, real-time inference, pandemic influenza, SEIR transmission model
1. Introduction
A pandemic influenza outbreak has the potential to place a significant burden upon healthcare systems. The capacity to monitor and predict its evolution as data progressively accumulate, therefore, is a key component of preparedness strategies for a prompt public health response.
Statistical approaches to real-time monitoring have been used for a number of infectious diseases including: prediction of swine fever cases (Meester et al. (2002)); online estimation of a time-evolving effective reproduction number R(t) for SARS (Wallinga and Teunis (2004), Cauchemez et al. (2006)) and for generic emerging disease (Bettencourt and Ribeiro (2008)); inference of the transmission dynamics of avian influenza in the UK poultry industry (Jewell et al. (2009)); and forecasting of Ebola (Viboud et al. (2018)).
Typically, however, this work relies on the availability of direct data on the number of new cases of an infectious disease over time. In practice, direct data are seldom available, as illustrated by the 2009 outbreak of pandemic A/H1N1pdm influenza in the United Kingdom (UK). More likely, multiple sources of data exist, each indirectly informing the epidemic evolution and each subject to possible sources of bias. These data typically come from routine influenza surveillance systems reporting interactions with healthcare services. They are often: biased towards the more severe cases; subject to the changing healthcare-seeking behaviours of the population; contaminated with cases of people experiencing influenza-like illness; and heavily influenced by governmental policies. These features call for more complex modelling, requiring the synthesis of information from a range of data sources in real time.
In this paper we tackle the problem of online inference and prediction in an influenza pandemic in this more realistic situation. We address this starting from the work of Birrell et al. (2011) who retrospectively reconstructed the A/H1N1 pandemic in a Bayesian framework using multiple data streams collected over the course of the pandemic. In Birrell et al. (2011), posterior distributions of relevant epidemic parameters and related quantities are derived through Markov chain Monte Carlo (MCMC) methods which, if used in real time, pose important computational challenges. MCMC is notoriously inefficient for online inference as it requires repeat browsing of the entire data history as new data accrue. This motivates a more efficient algorithm. Potential alternatives include refinements of MCMC (e.g., Jewell et al. (2009), Banterle et al. (2019)) and Bayesian emulation (e.g., Farah et al. (2014)) where the model is replaced by an easily evaluated approximation readily prepared in advance of the data assimilation process. Here, we explore Sequential Monte Carlo (SMC) methods (Doucet and Johansen (2011)). As batches of data arrive at times t 1,…, tK, SMC techniques allow computationally efficient online inference by combining the posterior distribution πk(·) at time tk, k = 0,…, K with the incoming batch of data to obtain an estimate for πk +1(·).
A further advantage of SMC is that it automatically provides all the posterior predictive distributions necessary to make one-step-ahead probabilistic forecasts of the incoming data. In a pandemic context, monitoring the appropriateness of a model is vital to avoid making public health decisions on the basis of misspecified models. Through formal assessment of the quality of these one step ahead forecasts (Held, Meyer and Bracher (2017)), timely checks of model adequacy and, if necessary, swift adaptations of the model can be made.
Use of SMC in the real-time monitoring of an emerging epidemic is not new. Ong et al. (2010), Dukic, Lopes and Polson (2012), Skvortsov and Ristic (2012), Dureau, Kalogeropoulos and Baguelin (2013), Camacho et al. (2015) and Funk et al. (2018), for instance, provide examples of real-time estimation and prediction for deterministic and stochastic models describing the dynamics of influenza and Ebola epidemics. These models, again, only include a single source of information that has either been preprocessed or is free of any sudden or systematic changes.
In what follows we advance existing literature in three ways: we include a number of data streams, realistically mimicking current data availability in the UK; we consider the situation where a public health intervention introduces a shock to the system, critically disrupting the ability to track the posterior distribution over time; and we demonstrate how the use of SMC can facilitate online assessment of model adequacy.
The paper is organised as follows: in Section 2 the model in Birrell et al. (2011) is reviewed focusing on the data available and the computational limitations of the MCMC algorithm in a real-time context; in Section 3 the idea of SMC is introduced and the algorithm of Gilks and Berzuini (2001) is described; in Section 4 results are presented from the application of Gilks and Berzuini’s SMC algorithm to data simulated to mimic the 2009 outbreak and illustrate the challenges posed by the presence of the informative observations induced by system shocks; in Sections 5 and 6 adjusted SMC approaches that address such challenges are assessed; we conclude with Section 7 in which the ideas explored in the paper are critically reviewed and outstanding issues discussed.
2. A model for pandemic reconstruction
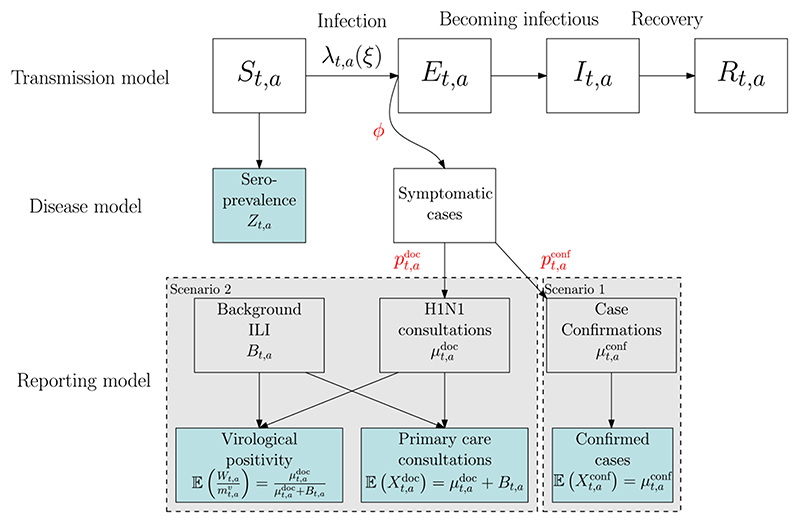
Birrell et al. (2011) estimate the transmission of a novel influenza virus among a fixed population stratified into A age groups (see Figure 1). Disease transmission is approximated by a deterministic age-structured Susceptible (S) Exposed (E) Infectious (I) Recovered (R) model described by a system of differential equations evaluated at discrete times tk= kδt, k = 0,…, K, with δt = 0.5 days. Under this discretisation the number of new infections in interval [tk −1 , tk) is
| (1) |
where Δtk,a ≡ tk,a(ξ) for a vector of transmission parameters ξ and
| (2) |
is the time- and age-varying force of infection, the rate at which susceptible individuals become infected. In (2) R 0(ψ) is the basic reproduction number, the expected number of secondary infections caused by a single primary infection in a fully susceptible population, parameterised in terms of the epidemic growth rate ψ; M tk(m) represent time-varying mixing matrices, parameterised by m, with giving the relative rates of effective contacts between individuals of each pair of age groups (a, b) at time tk, and dL and dI are the mean latent and infectious periods, respectively. The initial conditions of the system are determined by a further parameter ν. Fixing dL = 2 days, the vector of transmission dynamics parameters is ζ= (ψ, ν, dI, m).
Fig. 1.
Schematic diagram showing multiple epidemics surveillance sources linking to an SEIR epidemic model via an observation and reporting model. The shaded blue boxes represent observed data streams.
There is no direct information to estimate ξ as the transmission process is unobserved. Birrell et al. (2011) describe how ξ can be inferred from the combination of different sources linked to the latent transmission through a number of observational models (see Figure 1).
A first source of information is provided by a series of cross-sectional serological survey data Ztk,a on the presence of immunity-conferring antibodies in the general population. Denoting by Na the population size in age group a and the number of blood sera samples tested in time interval [tk −1 , tk), it is assumed that
| (3) |
informing directly the number of susceptibles in age group a at the end of the kth time step. A second source is the time series of virologically confirmed infections (e.g., admission to intensive care) or the number of consultations at general practitioners (GP) for influenza like illness (ILI). Data on consultations are contaminated by a “background” component of individuals attending GP for nonpandemic ILI, strongly influenced by a public’s volatile sensitivity to governmental advice. Both and are assumed to be realisations of negative binomial distributions here expressed in a mean-dispersion (μ, η) parameterisation, such that if X ∼ NegBin(μ, η), then 𝔼(X) = μ, var(X) = μ(η + 1), that is,
| (4) |
and
| (5) |
In (5) the contamination Btk,a is appropriately parameterised in terms of parameters βB (see Section 4) and both and are expressed through a convolution equation, resulting from the process of becoming infected and experiencing a time delay between infection and the relevant healthcare event (see Figure 1). This convolution for is
| (6) |
where the (discretised) delay probability mass function f(·) accounts for both the time from infection to symptoms and the time from symptoms to GP consultation (see Figure 1). Note that where e ∈ {conf, doc} and .
The signal can only be identified by additional virological data from subsamples of size of the primary care consultations. The number of swabs testing positive for the presence of the pandemic strain Wtk,a in each sample is assumed to be distributed:
| (7) |
2.1. Inference
To estimate θ, Birrell et al. (2011) develop a Bayesian approach and use a Markov chain Monte Carlo (MCMC) algorithm to derive the posterior distribution of θ on the basis of 245 days of primary care consultation and swab positivity data, confirmed case and cross-sectional serological data. Their MCMC algorithm is a naively adaptive random walk Metropolis algorithm, requiring 7 × 105 iterations, requiring in excess of 6.3 × 106 evaluations of the transmission model and/or convolutions of the kind in equation (6). MCMC is not easily adapted for parallelised computation, but the likelihood calculations allow for some small-scale parallelisation. The MCMC were thus optimally run on a desktop computer with 8 parallel 3.6 GHz Intel(R) Core(TM) i7-4790 processors, requiring run times of almost four hours. Although this run time might not be prohibitive for real-time inference, this implementation leaves little margin to consider multiple code runs or alternative model formulations. In a future pandemic there will be a greater wealth of data facilitating a greater degree of stratification of the population (Scientific Pandemic Influenza Advisory Committee: Subgroup On Modelling (2011)). With increasing model complexity comes rapidly increasing MCMC run times which can be efficiently addressed through use of SMC methods.
3. An SMC alternative to MCMC
Let Y t denote the vector of all random quantities in (3)–(7), and let y t be the observed values of Y t. Online inference involves the sequential estimation of posterior distributions πk(θ) = p(θ|y 1:k) ∝ π 0(θ)p(y 1:k|θ), k = 1,…, K where π 0(θ) indicates the prior for θ. Estimation of any epidemic feature, for example, the assessment of the current state of the epidemic or prediction of its future course, follows from estimating θ.
Suppose at time tk a set of nk particles with associated weights approximate a sample from the target distribution πk(·). On the arrival of the next batch of data y k +1, πk(·) is used as an importance sampling distribution to sample from πk +1(·). In practice, this involves a reweighting of the particle set. The particles are reweighted according to the importance ratio, πk +1(·)/πk(·), which reduces to the likelihood of the incoming data batch, that is,
| (8) |
Eventually, many particles will carry relatively low weight, leading to sample degeneracy as progressively fewer particles contribute meaningfully to the estimation of πk(·). A measure of this degeneracy is the effective sample size (ESS) (Liu and Chen (1995)),
| (9) |
The ESS is the “required size of an independent sample drawn directly from the target distribution to achieve the same estimating precision attained by the sample contained in the particle set” (Carpenter, Clifford and Fearnhead (1999)), and, as such, values of the ESS that are small in comparison to nk are indicative of an impoverished sample.
This degeneracy can be tackled in different ways. Gordon, Salmond and Smith (1993) introduced a resampling step, removing low weight particles and jittering the remainder. This jittering step was formalised by Gilks and Berzuini (2001) using Metropolis–Hastings (MH) steps to rejuvenate the sample. Fearnhead (2002) and Chopin (2002) provide more general treatises of this SMC method, with Chopin (2002) labelling the algorithm ‘iterated batch importance sampling.’ This was extended by Del Moral, Doucet and Jasra (2006) who unify the static estimation of θ with the filtering problem (estimation of a state vector, x k).
Here, we adapt the resample-move algorithm of Gilks and Berzuini (2001), investigating its real-time efficiency in comparison to successive use of MCMC. The MH steps rejuvenating the sample constitute the computational bottleneck in resample-move as they require a browsing of the whole data history to evaluate the full likelihood, not just the most recent batch. For fast inference the number of such steps should be minimised, without risking Monte Carlo error through sample degeneracy. The resulting algorithm is laid out in full below. It is presumed that it is straightforward to sample from the prior distribution π 0(θ).
3.1. The algorithm
Set k = 0. Draw a sample from the prior distribution, π 0(θ), set the weights .
Set k = k + 1. Observe a new batch of data Y k = y k. Reweigh the particles so that the jth particle has weight, .
Calculate the effective sample size. Set If ESS and return to point (2), else go next.
Resample. Choose nk and sample from the set of particles with corresponding probabilities . Here, we have used residual resampling (Liu and Chen (1998)). Reset .
Move: For each j move from via a MH kernel . If k < K, return to point (2).
End: is a weighed sample from πK(·).
There are a number of algorithmic choices to be made, including tuning any parameters, γ, of the MH kernel and the rejuvenation threshold, ϵL. In a real-time setting it may not be possible to tune an algorithm “on the fly,” so the system has to work “out of the box,” either through prior tuning or through being adaptive (Fearnhead and Taylor (2013)). In what follows we set ϵL = 0.5 (Jasra et al. (2011)), and we focus on the key factors affecting the performance of the algorithm in real time, that is, the MH kernel.
3.1.1. Kernel choice
Correlated random walk
A correlated random walk proposes values in the neighbourhood of the current particle:
| (10) |
where is the sample variance-covariance matrix of the weighted sample . The advantages here are that the parameter γ can be tuned a priori to guarantee a reasonable acceptance rate, or asymptotic results for the optimal scaling of covariance matrices (Roberts and Rosenthal (2001), Sherlock, Fearnhead and Roberts (2010)) could be used. Also, the localised nature of these moves should keep acceptance rates high, leading to quick restoration of the value of the ESS.
Approximate Gibbs’
An independence sampler that proposes (Chopin (2002))
| (11) |
where is the sample mean of the . Here, proposals are drawn from a distribution chosen to approximate the target distribution, only weakly dependent on the current position of the particle. An accept-reject step is still required to correct for this approximation. The quality of the approximation depends on πk −1(·) being well represented by the current particle set, there being sufficient richness in the particle weights after the reweighting step and the target density being sufficiently near-Gaussian. Assuming that the multivariate normal approximation to the target is adequate (and it should be increasingly so as more data are acquired) this type of proposal allows for more rapid exploration of the sample space.
For each type of kernel, both block and componentwise (where individual or subgroups of parameter components are proposed in turn) proposals that use the appropriate conditional distributions derived from (10) and (11) are considered. However, the kernels considered in Step 5 of the resample-move algorithm consist of only a single block proposal or a single proposal for each parameter component.
4. A simulated epidemic
The suitability of the SMC algorithm for real-time epidemic inference is evaluated against the MCMC algorithm used in Birrell et al. (2011) which is taken as a gold standard. Comparisons are made through application to data simulated from the epidemic model in Figure 1. The simulation conditions were chosen so that the resulting epidemic would mimic the timing and dynamics of the 2009 A/H1N1 pandemic in England. This epidemic was characterised by two distinct waves of infection with a first peak induced by an over-summer school holiday and a second peak occurring during the traditional winter flu season.
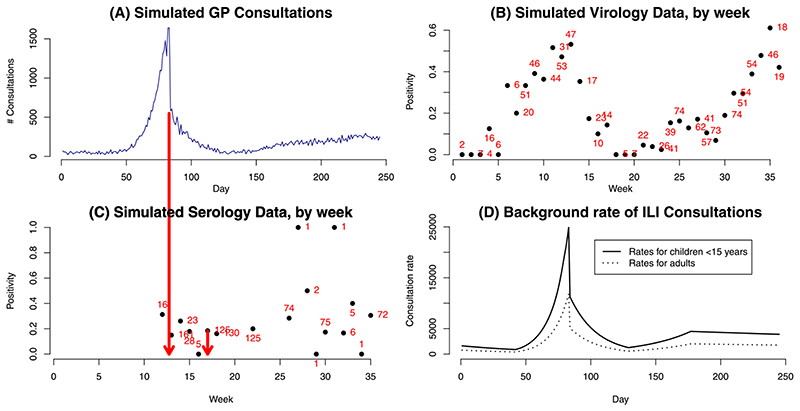
We consider two scenarios. In the first direct information on confirmed cases (e.g., hospitalisation, ICU admissions) is available; in the second we observe the noisy ILI consultations (equation (5)). Alongside either of these data, serological data (equation (3)) are available and, in the second scenario, there are also virological data taken from a subsample of the ILI consultations (see equation (7)). Both scenarios use observations made on 245 consecutive days on a population divided into A = 7 age groups and are characterised by the same underlying epidemic curve, so that the confirmed case and primary care consultation data are subject to similar trends. For both scenarios we introduce a shock at tk= 83 days, similar to the 2009 pandemic, where a public health intervention is assumed to change the way the confirmed cases or consultations occur and are reported. The simulated data for the second scenario are presented in Figure 2(A)–(C) where the timing of the shock is indicated by the red arrow linking (A) and (C). Table A1 in the Supplementary Material (Birrell et al. (2020)) presents the model parameters together with the values used for simulation. Note that the proposed intervention impacts by introducing a changepoint on three groups of parameters: the dispersion in the count data, η, the proportion of infections that appear in the data p ·, and in Scenario 2 the age-specific (i.e., child and adult specific) background consultation rates, Btk,a, which develop over time according to a log-linear spline with a discontinuity at tk= 83. The spline is plotted, by age group, in Figure 2(D) and its parameterisation as a function of the 9-dimensional parameter βB is given in Section A1 of the Supplementary Material (Birrell et al. (2020)).
Fig. 2.
Top row: (A) Number of doctor consultations ; (B) swab positivity data (Wtk,a) with numbers representing the size of the weekly denominator. Bottom row: (C) serological data (Ztk,a); (D) pattern of background consultation rates by age. Arrows between (A) and (C) highlight the timing of some key, informative observations.
Real-time monitoring of the epidemic will begin after an initial outbreak stage, taken here to be the first 50 days. An MCMC implementation of the model is carried out at times tk= 50, 70, 83, 120, 164 and 245 days, and the SMC algorithm is then used to propagate the MCMC-obtained posteriors over the intervals defined by these timepoints. For example, the MCMC-obtained estimate of π 50(θ) will be used as the initial particle set for the SMC algorithm over the interval 50–70 days. This gives an estimate, , for π 70(θ), which is then compared to . The similarity between the two distributions is measured by the Küllback–Leibler (KL) divergence of from the “gold-standard” reference distribution, , calculated using multivariate normal approximations to both distributions.
4.1. Results from a resample-move SMC algorithm
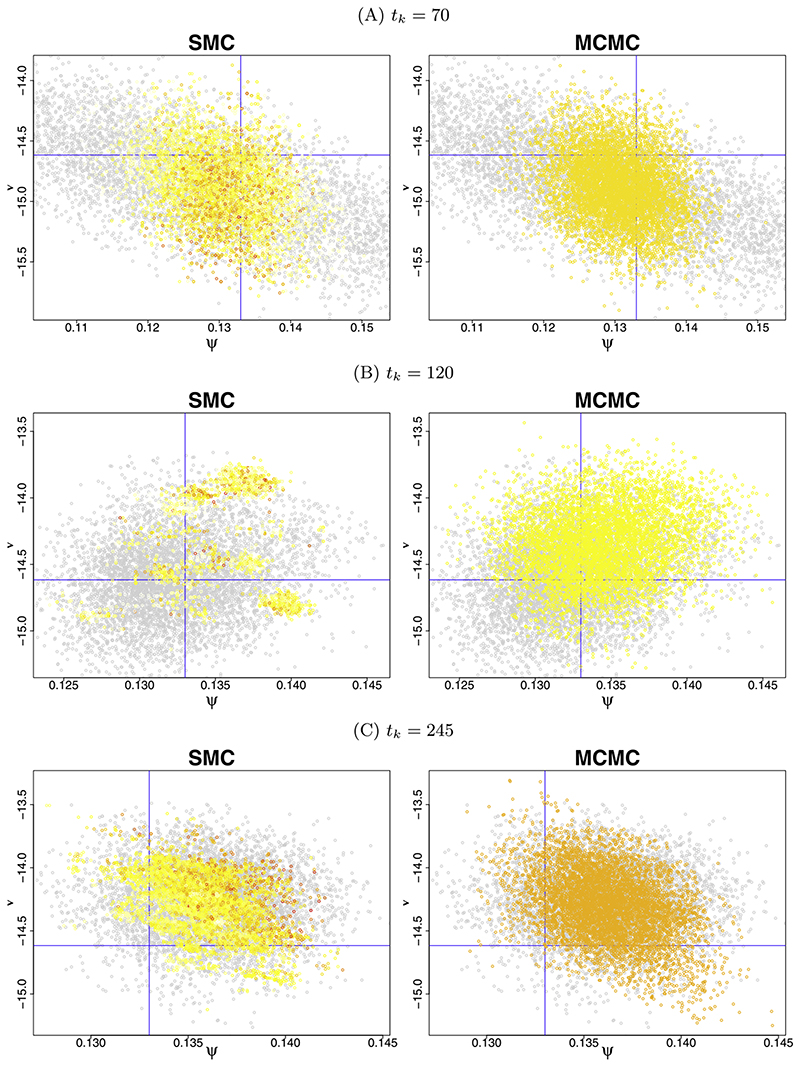
In addition to KL, Table 1 reports Hellinger and Wasserstein divergences for the posterior distributions from Scenario 1, obtained using each of the three different proposal kernels described in Section 3.1.1. The use of the three divergences ensures that inference is not being unduly influenced by the particular characteristics of any single chosen metric. The correlated random walk (10) has the highest KL over the intervals up to 120 days. Beyond 120 days the divergence between distributions πk and πk +1 is small, and the random-walk proposals become progressively more able to bridge the gap. The componentwise approximate Gibbs scheme (11) generally outperforms the block updates. Figure 3 illustrates the performance of the approximate Gibbs componentwise proposal kernel comparing the SMC- and MCMC-obtained scatterplots for the parameter components ψ and ν at tk= 70 (A), tk= 120 (B) and tk= 245 (C). There is close correspondence between the SMC and MCMC obtained distributions at tk= 70 and tk= 245 but substantial departure at tk= 120. This is the only interval for which the block updates perform better (in terms of divergence, Table 1). All of the above findings are consistent irrespective of the metric used. As a result, for ease of presentation we will work with the more familiar KL only from here on. Similar phenomena are observed for Scenario 2 but with magnified KL discrepancies due to the increase in dimensionality (see Table B2, Supplementary Material (Birrell et al. (2020))).
Table 1.
Scenario 1: Küllback–Leibler (KL), Hellinger and Wasserstein statistics and likelihood evaluations per day (“Run Time”) for each resample-move algorithm. Bootstrap standard errors are given in brackets
| Intervals | Proposal method | Correlated random-walk | Componentwise approx. Gibbs | Block approx. Gibbs |
|---|---|---|---|---|
| 0–50 | KL | 2.83 (0.018) | 2.58 (0.011) | 2.61 (0.011) |
| Hellinger | 0.852 (0.0012) | 0.833 (0.0010) | 0.835 (0.00091) | |
| Wasserstein | 19,700 (670) | 12,700 (280) | 12,300 (220) | |
| Run Time | 18,200 | 16,800 | 8000 | |
| 51–70 | KL | 2.00 (0.016) | 0.908 (0.013) | 1.32 (0.018) |
| Hellinger | 0.768 (0.0021) | 0.546 (0.0032) | 0.643 (0.0032) | |
| Wasserstein | 1710 (57) | 112 (2.5) | 230 (3.7) | |
| Run Time | 21,000 | 21,000 | 8000 | |
| 71–83 | KL | 4.44 (0.12) | 0.929 (0.037) | 1.60 (0.037) |
| Hellinger | 0.804 (0.0033) | 0.404 (0.0063) | 0.513 (0.0042) | |
| Wasserstein | 409 (14) | 0.936 (0.065) | 1.35 (0.077) | |
| Run Time | 26,923 | 26,923 | 7692 | |
| 84–120 | KL | 16.3 (0.39) | 6.58 (0.19) | 2.09 (0.085) |
| Hellinger | 0.955 (0.0012) | 0.865 (0.0026) | 0.497 (0.0055) | |
| Wasserstein | 10.5 (0.27) | 8.66 (0.20) | 0.249 (0.0075) | |
| Run Time | 20,811 | 17,027 | 10,000 | |
| 121–164 | KL | 0.106 (0.010) | 0.113 (0.0086) | 0.122 (0.0077) |
| Hellinger | 0.165 (0.0081) | 0.169 (0.0067) | 0.172 (0.0051) | |
| Wasserstein | 0.0342 (0.0045) | 0.0441 (0.0049) | 0.0355 (0.0049) | |
| Run Time | 3182 | 3182 | 4773 | |
| 165–245 | KL | 0.339 (0.013) | 0.471 (0.025) | 1.15 (0.035) |
| Hellinger | 0.274 (0.0047) | 0.296 (0.0065) | 0.424 (0.0046) | |
| Wasserstein | 0.0976 (0.0097) | 0.0406 (0.0044) | 0.109 (0.0046) | |
| Run Time | 8642 | 9506 | 9136 |
Fig. 3.
Comparison of SMC-obtained posteriors and MCMC-obtained posteriors at tk= 70 (A), tk= 120 (B) and tk= 245 (C) days via scatter plots for the parameters ψ and ν. The grey points in both the left and the right panels represent the MCMC-obtained sample at the beginning of the interval, with the overlaid coloured points representing the SMC or MCMC-obtained samples at the end of the interval. In the SMC-obtained samples, the colour of the plotted points represents the weight attached to the particle, with the red particles being those of heaviest weight.
Irrespective of the kernel chosen, it is clear that the basic resample-move SMC algorithm cannot handle the “shock” in the count data occurring at tk= 83, which leads to step changes in some model parameters. The marginal posterior distributions for the new parameter components move rapidly from day 84 as probability density shifts away from uninformative prior distributions. For Scenario 1 the 84–120 day interval is the only one over which the block-update approximate Gibbs method gives the best performance (see KL divergence in Table 1). This arises due to the comparatively low acceptance of the single full block proposals, ensuring that the ESS remains below ϵLnk and leading to further rejuvenations at each following time. This frequent rejuvenation better enables the tracking of the shifting posterior distributions over time (slightly reducing the advantage of this algorithm in terms of computation time, Table 1). Alternatively, componentwise updates lead to a set of nearly unique particles with ESS ≈ nk and fewer subsequent rejuvenations. However, even with the block updates, good correspondence between the SMC- and MCMC-obtained posteriors is not achieved after the shock in Scenario 1 until tk≈ 100, and not at all in Scenario 2.
From these initial results it is clear that a modified algorithmic formulation is needed for computationally efficient inference when target posteriors are highly non-Gaussian and/or are moving fast between successive batches of data as a consequence of highly informative observations.
5. Extending the algorithm-handling informative observations
A key feature of any improved SMC algorithm must be that the ESS retains its interpretation given in Section 3. For example, as the scaling of a random-walk proposal tends to zero (i.e., γ ↓ 0 in equation (10)), acceptance rates will be close to unity, resulting in a set of mostly unique particles and a high value for the ESS. However, in cases where there has been a loss of particle diversity at the resampling stage (because many particles are sampled numerous times) this would give a highly clustered posterior sample, barely distinguishable from the set of resampled particles and definitely not as informative as an independent sample of size nk. Here, the ESS, as calculated from the particle weights, is no longer a reliable guide to the quality of the sample.
We look at three possible improvements to the resample-move algorithm of Section 3 to produce an information-adjusted (IA) SMC algorithm that safeguards the ESS as a good measure of the quality of the sample: we address the timing of rejuvenations; we reconsider the choice of kernels used in the rejuvenations, and we address the problem of choosing the number of iterations we need to run the MCMC sampler before the sample is fully rejuvenated.
5.1. Timing the rejuvenations: A continuous-time formulation
If there is large divergence between consecutive target distributions πk and πk +1, the estimation of intermediate distributions will allow the particle set to move gradually between the two targets (Del Moral, Doucet and Jasra (2006)). These intermediate distributions are generated via tempering (Neal (1996)), introducing gradually the new batch of data into the likelihood at a range of “temperatures,” τ ∈ [0, 1]. These distributions are denoted πk,τ (θ) ∝ πk(θ){p(yk +1|θ) }τ.
We choose to think of data y k +1 arriving uniformly over the (k + 1)th interval and denote to be the weight attached to a particle at an intermediate time tk +τ when the previous rejuvenation took place at time tk +τ0, with τ 0 = 0 corresponding to no prior rejuvenation within the interval (tk, tk +1]. Then, for 0 ≤ τ 0 ≤ τ ≤ 1 and indicator function for an event A denoted 𝟙A,
Therefore, if ESS a further rejuvenation would be proposed at time τ*, such that .
5.2. Choosing kernels-hybrid algorithms
As discussed in Section 4.1, each of the possible MH kernels has its own distinct strengths. These can be exploited by using a combination of kernels. Full block approximate-Gibbs updates are efficient at reducing the clustering that forms around resampled particles. Adding a random walk step would allow the proposal of values outside the space spanned by the principal components of , something of particular necessity if the ESS is very small and is close to singularity.
This motivates a hybridisation of the proposal mechanism, done either by using mixture proposals, for example, a mixture between the approximate Gibbs’ proposals and full block ordinary random walk Metropolis proposals (Kantas, Beskos and Jasra (2014)) or, as will be used in the remainder, by augmenting full block approximate Gibbs updates with componentwise random walk proposals.
5.3. How many MH iterations? Multiple proposals and intraclass correlation
In the MH-step of the algorithm, there are effectively nk parallel MCMC chains. Making proposals until all chains have attained convergence would be an inefficiency. The distribution governing the starting states of these chains forms a biased sample from the target distribution obtained through sampling importance resampling (Chopin (2002)). It then seems a reasonable requirement that we carry out MH steps until the chains have collectively “forgotten” their starting values. This can be monitored through an estimate of an intraclass correlation coefficient (ICC), ρ. First, the particle set is divided into I clusters, each of size di, i = 1,…, I, defined by the parent particle at the resampling stage. For example, if a particular particle is resampled five times, it defines a cluster in the new sample with di= 5. The analysis of variance intraclass correlation coefficient, rA(Donner and Koval (1980), Sokal and Rohlf (1981)), is used to estimate ρ. This estimate is dependent on the mean squared error in a univariate summary statistic, gij= g(θij), calculated for the jth particle in the ith cluster, θ ij both within and between clusters. Here, we choose the “attack rate” of the epidemic, the cumulative number of infections caused by the epidemic:
| (12) |
Details of the calculation of rA are in Section C of the Supplementary Material (Birrell et al. (2020)).
Prior to the MH phase of the algorithm, there is no within-class variation, and rA= 1. However, with each iteration of the chosen MH sampler, ρ will decrease and, in general, so will its estimate rA. We aim to choose a sufficiently small positive threshold, , to be the point beyond which there is no longer any value in carrying out further MH proposals to rejuvenate the sample, as particles spawned from different progenitors become indistinguishable from each other. Ideally, this threshold is as large as is practicably possible to minimise the number of rejuvenations required and, accordingly, we test our algorithms with thresholds . In principle, stopping rules that are based, even indirectly, on the number of accepted proposals can induce bias into the particle-based approximations to the target density. However, here the dependence is sufficiently weak to be of little concern as the stopping time of each chain is dependent on the number of accepted proposals in nk− 1 independent chains as well as itself.
6. Results from IA SMC algorithms
Here, we focus mainly on the intervention-spanning day 83–120 interval. In what follows, a hybrid algorithm is adopted, using combinations of three thresholds for rA with both the continuous and discrete sequential algorithms.
6.1. Scenario 1: Confirmed case and serological data
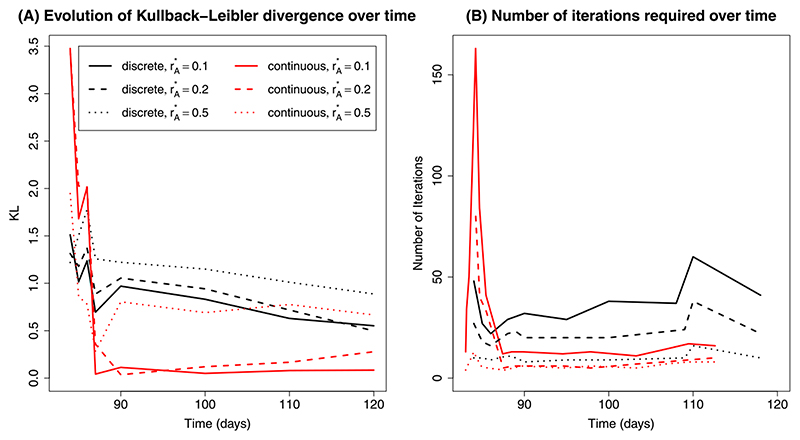
MCMC samples were obtained using data up to and including tk= 84, 85, 86, 87, 90, 100, 110 and 120, with Figure 4 and Table 2 summarising the results. In Figure 4(A) KL discrepancies between and are plotted over time for each combination of algorithm and threshold. To calibrate these KL divergences, a further 40 MCMC chains were obtained at each of these times. The KL divergences between these posterior distributions from the original reference MCMC analysis were then calculated. This formed a distribution of KL values that are typical of MCMC samples from our target distribution. If attains the gold standard, then it should return a KL divergence that could feasibly come from this distribution. Therefore, we generate a “KL target” (see Table 2), the 95% quantile of these sampled KL values and diagnose significant difference in the MCMC and SMC-obtained distributions when their KL divergence is larger than this KL target.
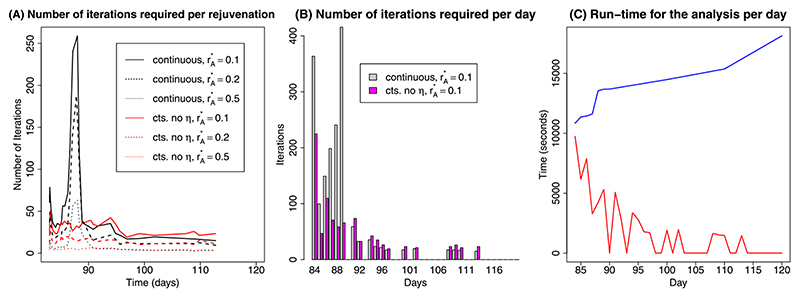
Fig. 4. (A) Kullback–Leibler divergence over time; (B) Number of proposals required at each rejuvenation time by algorithm.
Table 2. Performance in Scenario 1 of the information-adjusted SMC algorithms over the interval 83–120 days (discrete and continuous) by ICC threshold.
| ICC threshold | 0.5 | 0.2 | 0.1 | ICC threshold | 0.5 | 0.2 | 0.1 |
|---|---|---|---|---|---|---|---|
| 84 Days (KL target = 0.732) | 90 Days (KL target = 0.159) | ||||||
| Continuous | 1.95 | 3.46 | 3.48 | Continuous | 0.805 | 0.036 | 0.113 |
| Discrete | 1.22 | 1.31 | 1.51 | Discrete | 1.22 | 1.05 | 0.970 |
| 85 Days (KL target = 0.135) | 100 Days (KL target = 0.135) | ||||||
| Continuous | 0.862 | 2.03 | 1.68 | Continuous | 0.691 | 0.120 | 0.050 |
| Discrete | 1.50 | 1.18 | 1.02 | Discrete | 1.15 | 0.942 | 0.832 |
| 86 Days (KL target = 0.365) | 110 Days (KL target = 0.122) | ||||||
| Continuous | 0.780 | 2.01 | 2.02 | Continuous | 0.776 | 0.167 | 0.080 |
| Discrete | 1.78 | 1.37 | 1.24 | Discrete | 1.01 | 0.719 | 0.630 |
| 87 Days (KL target = 0.276) | 120 Days (KL target 0.119) | ||||||
| Continuous | 0.282 | 0.358 | 0.043 | Continuous | 0.666 | 0.278 | 0.084 |
| Discrete | 1.26 | 0.887 | 0.696 | Discrete | 0.888 | 0.498 | 0.552 |
Performance of the continuous-time algorithm appears strongly linked to the acceptance rate of the block approximate Gibbs’ proposals. This acceptance rate is particularly low (1–2%) prior to tk= 87 when it undergoes a step change to 15–20%. In contrast, the acceptance rates for the discrete-time algorithm are consistently around 5% throughout, as seen from the constant number of iterations required over time (Figure 4(B)). As a result, from day 87 onwards, far fewer proposals are required in total for the continuous-time algorithm, even if the number of rejuvenation times increases.
6.2. Scenario 2: Primary care consulation and serology data
Focusing on the better-performing continuous-time IA algorithm, similar performance to Scenario 1 can be observed (Table 3). The algorithm again suffers from acceptance rates for the approximate-Gibbs’ proposals which, though initially adequate, fall to 0.3% on day 89, illustrated by a peak of over 250 proposals per rejuvenation and over 400 proposals per day in Figures 5(A) and (B), respectively. This low rate is driven by the highly non-Gaussian distribution for the dispersion parameter η 2 which has an unbounded gamma prior and is not well identified from the data. To improve acceptance rates, a “cts. reduced” scheme is devised in which the dispersion parameters are omitted from the block approximate-Gibbs updates and proposed separately. In terms of the resulting KL divergences, there is no significant drop in performance between the continuous and the “cts. reduced” algorithms (Table 3). The “cts. reduced” proposal scheme requires far fewer iterations of the Metropolis–Hastings algorithm over the interval 84–90 days, maintaining acceptance rates of about 10% over this period. On day 90 the “cts. reduced” scheme does give an anomalously high KL value (1.42). Closer inspection found this to be the result of three particles with extremely small values for η. With these three particles removed, the KL divergence falls to 0.086. Over time, as the target distribution converges to a multivariate normal distribution, the number of moves required for both methods equalise and the benefit of the “cts. reduced” proposal scheme vanishes (Figure 5(B)).
Table 3.
Performance in Scenario 2 of the information-adjusted SMC algorithm over the interval 83–120 days in continuous time where the algorithms differ in the inclusion of the η parameters in the block proposals. Parameter βB is omitted from the KL calculations
| ICC threshold | 0.5 | 0.2 | 0.1 | ICC threshold | 0.5 | 0.2 | 0.1 |
|---|---|---|---|---|---|---|---|
| 84 Days (KL target = 6.06) | 90 Days (KL target = 0.120) | ||||||
| Continuous | 2.92 | 2.87 | 2.83 | Continuous | 1.80 | 0.35 | 0.066 |
| Cts. Reduced | 2.97 | 2.85 | 2.86 | Cts. Reduced | 2.10 | 0.093 | 1.42 |
| 85 Days (KL target = 1.90) | 100 Days (KL target = 0.182) | ||||||
| Continuous | 3.05 | 3.00 | 2.98 | Continuous | 0.157 | 0.102 | 0.089 |
| Cts. Reduced | 3.06 | 2.97 | 2.98 | Cts. Reduced | 0.107 | 0.084 | 0.070 |
| 86 Days (KL target = 1.94) | 110 Days (KL target = 0.0936) | ||||||
| Continuous | 3.28 | 3.24 | 3.25 | Continuous | 0.159 | 0.077 | 0.111 |
| Cts. Reduced | 3.27 | 3.22 | 3.26 | Cts. Reduced | 0.197 | 0.037 | 0.035 |
| 87 Days (KL target = 5.44) | 120 Days (KL target = 0.101) | ||||||
| Continuous | 2.54 | 2.45 | 2.42 | Continuous | 0.136 | 0.044 | 0.071 |
| Cts. Reduced | 2.51 | 2.48 | 2.44 | Cts. Reduced | 0.100 | 0.042 | 0.055 |
Fig. 5.
(A) Number of MH-steps required by the continuous-time SMC algorithms per rejuvenation over time; (B) Total number of MH-steps required by the continuous-time SMC algorithms per time interval; (C) The computation time required for model runs on each day using MCMC (blue line) and SMC (red line).
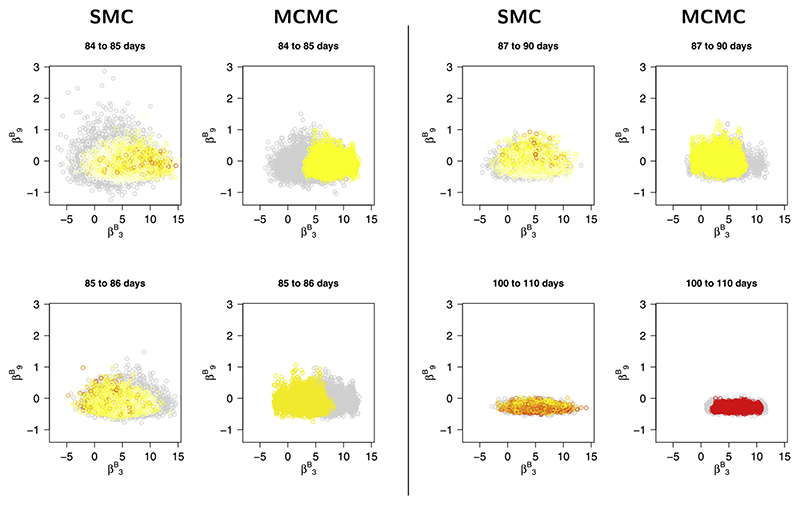
The scatter plots of Figure 6 give a sequence (over time) of marginal posterior distributions for two parameters, , of the background consultation rate model, obtained from the “cts. reduced” SMC scheme and MCMC. These parameters are only weakly identifiable in the immediate period after tk= 83, and a clear discrepancy between the MCMC- and the SMC-obtained posterior scatters emerges. The SMC distributions, being based on many short MCMC chains, cover the full posterior distribution adequately at each tk. The MCMC has difficulty mixing, at tk= 85, 86 in particular, resulting in scatters concentrated in a subregion of the full marginal support.
Fig. 6.
The evolution over time of the marginal joint posterior for two components of the parameter vector βB. Comparison between SMC-obtained and MCMC-obtained posterior distributions. Grey points indicate the distribution at the start of the interval.
Not only does SMC offer an improvement in terms of posterior coverage in the presence of partial identifiability, but its daily implementation is also faster, as shown in Figure 5(C). The run time for SMC decreases almost linearly with increased parallelisation, and so the particles (and hence the parallel MCMC chains) are distributed across 255 Intel(R) Xeon(R) CPU E5-262 2.0 GHz processors on a high-performance computing cluster. This represents modest parallelisation compared to what might be used in a real pandemic. Figure 5 shows that, not only is SMC more computationally efficient on day 84, the day requiring the most MH-updates to rejuvenate the sample, but also the run-times decrease over time, in contrast to the increasing MCMC run times as more data have to be analysed. On days where the sample does not require rejuvenation, run times are negligible.
7. Discussion
This paper addresses the substantive problem of online tracking of an emergent epidemic, assimilating multiple sources of information through the development of an information-adjusted SMC algorithm. When incoming data follow a stable pattern, this process can be automated using standard SMC algorithms, confirming current knowledge (e.g., Dukic, Lopes and Polson (2012), Ong et al. (2010)). However, in the likely presence of interventions or any other event that may provide a system shock, it is necessary to adapt the algorithm appropriately.
Using a simulated epidemic where a public health intervention provides a sudden change to the pattern of case reporting, we have constructed a more robust SMC algorithm by tailoring: (1) the choice of rejuvenation times through tempering; (2) the choice of the MH-kernel by combining local random walk and Gibbs proposals; (3) a stopping rule for the MH steps based on intraclass correlations to minimise the number of iterations within each rejuvenation.
The result is an algorithm that is a hybrid of particle filter and population MCMC (Geyer (1991), Liang and Wong (2001), Jasra, Stephens and Holmes (2007)), is robust to possible shocks, improves over the plain-vanilla MCMC in terms of run times needed to derive accurate inference and can automatically provide all the distributions needed for posterior predictive measures of model adequacy.
7.1. Benefits of SMC
Model run times. From a computational point of view, the SMC algorithm is faster than the plain vanilla MCMC as it is highly parallellisable. However, this may be an unfair comparison as we could have considered more sophisticated MCMC algorithms, as exemplified in an epidemic context by Jewell et al. (2009). The use of differential geometric MCMC (Girolami and Calderhead (2011)), nonreversible MCMC (Bierkens, Fearnhead and Roberts (2016)) or MCMC using parallelisation (Banterle et al. (2019)) could improve run times. However, as MCMC steps are the main computational overhead of the SMC algorithm, any improvements to the MCMC algorithm’s efficiency may also improve the SMC. As target posteriors attain asymptotic normality, it should be progressively easier for SMC to move between distributions over time, as can be seen in Figure 5(C) where the daily running time decreases as data accumulate. For any MCMC algorithm the opposite will be generally true.
Predictive model assessment. A fundamental goal of real-time modelling is to provide online epidemic forecasts with an appropriate quantification of the associated uncertainty. The real-time assessment of the predictive adequacy of a model becomes key and can be carried out through the evaluation of one-step-ahead forecasts based on posterior predictive distributions p(yk +1|y k) (Dawid (1984)). Such assessments can be made informally through, for example, probability integral transform (PIT) histograms (Czado, Gneiting and Held (2009)).
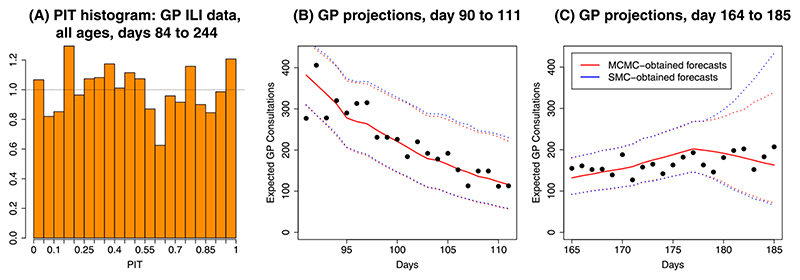
In the example of Section 6.2, Figure 7(A) shows the PIT histogram for one-step-ahead prediction of primary care consultations for all age groups for successive analyses in the range 84–245 days. A good predictive system would give a uniform histogram and, though the histogram here is not entirely uniform, it shows no consistent under or overestimation nor any clear signs of overdispersion.
Fig. 7.
(A) PIT histograms for the one-step-ahead predictions of GP ILI consultation data, calculated over 162 × 7 time and strata combinations. (B) and (C) Comparison of the observed GP data with posterior predictive distributions obtained using the SMC and MCMC algorithms at day 90 and 164, respectively. Solid lines give posterior medians of the distributions, and the dotted lines give 95% credible intervals for the data.
More formally, proper scoring rules (Gneiting and Raftery (2007)) can be used to assess the quality of forecasts, including through formal tests of prediction adequacy (Seillier-Moiseiwitsch and Dawid (1993)). Many different scoring rules exist, but to illustrate a benefit of an SMC algorithm consider the logarithmic score defined for a predictive distribution p(·) and a subsequently realised observation y, to be
Under an SMC scheme for one step ahead forecasts, these are
Weights and are routinely calculated as part of the SMC algorithm in Section 3.1 (equation (8)) whereas additional computation is required if the posterior is derived using MCMC. If the MCMC analyses are not carried out with every new batch of data, then these are not readily available. For further details on the calculation and interpretation of these posterior predictive methods, see Section E of the Supplementary Material (Birrell et al. (2020)).
Figures 7(B) and (C) present longer-term (three week) forecasts for the consultation data obtained via both SMC and MCMC from days 90 and 164 onwards. Whereas in (B) the forecasts are close enough to be identical, there is a divergence in the predictive intervals from tk= 178 onwards, a changepoint in the model for the background ILI rate.
Identifiability
As observed in Section 6.2, the SMC algorithm is better at exploring the full posterior distribution in the presence of parameter nonidentifiability around changepoints. The background ILI rate is modelled using a piecewise log-linear curve (equation (1)) in the Supplementary Material (Birrell et al. (2020)) with linear interpolation giving the value of the curve at intervening points. This results in log-consultation rates in the three days following the change point on day 83 that include the respective sums (neglecting the age effects) μ + α 84, μ + 0.98α 84 + 0.02α 128, μ + 0.96α 84 + 0.04α 128. This makes parameters μ and α 84 only weakly identifiable over this period, inducing convergence problems for MCMC (see Figure 6). Further evidence for this is given in Figure 7(C) by the divergence of the prediction intervals at breakpoint tk= 178 and in Table D3 in the Supplementary Material (Birrell et al. (2020)) where the KL calculations of Table 3 are repeated but with background parameters included. The marked increases in the KL targets from day 90 onwards is a result of significant discrepancy between the MCMC chains. Jasra et al. (2011) claim that, for their example, SMC may well be superior to MCMC, and this is one case where this is certainly true. The population MCMC carried out in the rejuvenation stage achieves good coverage of the sample space, without the individual chains having to do likewise. Reparameterisation may improve the MCMC, but this would also be of benefit to the SMC rejuvenation steps.
Early warning
Changepoints that lead to the lack of identifiability discussed above may coincide with public health interventions. In this paper it is assumed that such times are known, and we have been concerned with the adaptation of inferential procedures to ensure that they can be operated in a semiautomatic fashion at such times.
In general, such changepoints will need to be detected in real time and may be indicative of a change in the underlying epidemic dynamics or in healthcare-seeking behaviours, both of which are of great interest to healthcare managers. A sudden drop in the ESS can raise a flag that the model is no longer suitable and may require modification. Both Whiteley, Johansen and Godsill (2011) and Nemeth, Fearnhead and Mihaylova (2014) discuss automated approaches for the sequential detection of changepoints. However, when considering a complex mechanistic epidemic model, a more fundamental adaptation may be required. Sequential application of MCMC as data arrive over time would not automatically detect this without carrying out a series of exhaustive post hoc diagnostic checks.
7.2. Final considerations
In answer to the question initially posed, we have provided a recipe for online tracking of an emergent epidemic using imperfect data from multiple sources. We have discussed many of the challenges to efficient inference, with particular focus on scenarios where the available information is rapidly evolving and is subject to sudden shocks. Throughout we have inevitably made pragmatic choices and alternative strategies could have been adopted. The choice of the MH-kernels used for rejuvenation is an example. There are many options to tweak the performance of the “vanilla” kernels presented here, including simply scaling the covariance matrix in the approximate-Gibbs moves (West (1993)), treating the composite proposals of Section 5.2 as a single mixture (Kantas, Beskos and Jasra (2014)), using recent developments in kernel SMC methods to design local covariance matrices (Schuster et al. (2017)) and incorporating an adaptive scheme to select an optimal SMC kernel and any tuning parameters (Fearnhead and Taylor (2013)). Equally, we could have adopted multivariate analogues for the intra-class correlation coefficient (e.g., Ahrens (1976), Konishi, Khatri and Rao (1991)) to define a rejuvenation stopping rule; or we could have opted for a particle set expansion by increasing nk as a possible alternative to running long MCMC chains for each particle when new parameters are introduced in the model, for example, through a shock.
We have shown above that the benefits of SMC for online inference extend beyond computational efficiency. It is not claimed, however, that SMC is beneficial when inference is carried out offline, using the full available data. Over the course of any outbreak, the richness of data may grow, interventions may occur and models of increased complexity may be needed. It is therefore important to retain the capacity to fit new models efficiently. Methods such as Hamiltonian MCMC (Girolami and Calderhead (2011)), likelihood-tempered SMC algorithms (Kantas, Beskos and Jasra (2014)), emulation (Farah et al. (2014)), variational (Blei, Kucukelbir and McAuliffe (2017)) and Kalman-filtering approaches (Shaman and Karspeck (2012)) represent potential alternatives to achieve this.
We have focused on an epidemic scenario that has the potential to arise in the UK. Nevertheless, our approach addresses modelling concerns common globally (e.g., Wu et al. (2010), Shubin et al. (2016), te Beest et al. (2015)) and can form a flexible basis for real-time modelling strategies elsewhere. Real-time modelling is, however, more than just a computational problem. It does require the timely availability of relevant data, a sound understanding of any likely biases and effective interaction with experts. In any country only interdisciplinary collaboration between statisticians, epidemiologists and database managers can turn cutting edge methodology into a critical support tool for public health policy.
Supplementary Material
Acknowledgments
The first, fifth and seventh authors were supported in part by the National Institute for Health Research (HTA Project:11/46/03).
The second, third and seventh authors were supported by the UK Medical Research Council (programme codes MC_UU_00002/1, MC_UU_00002/2, and MC_UU_00002/11).
Contributor Information
Paul J. Birrell, Email: paul.birrell@mrc-bsu.cam.ac.uk.
Lorenz Wernisch, Email: lorenz.wernisch@mrc-bsu.cam.ac.uk.
Brian D. M. Tom, Email: brian.tom@mrc-bsu.cam.ac.uk.
Leonhard Held, Email: leonhard.held@uzh.ch.
Gareth O. Roberts, Email: Gareth.O.Roberts@warwick.ac.uk.
Richard G. Pebody, Email: richard.pebody@phe.gov.uk.
Daniela De Angelis, Email: daniela.deangelis@mrc-bsu.cam.ac.uk.
References
- Ahrens H. Multivariate variance-covariance components (MVCC) and generalized intraclass correlation coefficient (GICC) Biom J. 1976;18:527–533. [Google Scholar]
- Banterle M, Grazian C, Lee A, Robert CP. Accelerating Metropolis–Hastings algorithms by Delayed Acceptance. Foundations of Data Science. 2019;1:103–128. [Google Scholar]
- Bettencourt LMA, Ribeiro RM. Real time Bayesian estimation of the epidemic potential of emerging infectious diseases. PLoS ONE. 2008;3:e2185. doi: 10.1371/journal.pone.0002185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierkens J, Fearnhead P, Roberts G. The Zig-Zag process and super-efficient sampling for Bayesian analysis of big data. Ann Stat. 2019;47:1288–1320. [Google Scholar]
- Birrell PJ, Ketsetzis G, Gay NG, Cooper BS, Presanis AM, Harris RJ, Charlett A, Zhang X-S, White P, et al. Bayesian modelling to unmask and predict the influenza A/H1N1pdm dynamics in London. Proc Natn Acad Sci USA. 2011;108:18238–18243. doi: 10.1073/pnas.1103002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birrell PJ, Wernisch L, Tom BDM, Held L, Roberts GO, Pebody RG, De Angelis D. Supplement to “Efficient real-time monitoring of an emerging influenza pandemic: How feasible?”. 2020 doi: 10.1214/19-AOAS1278SUPP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blei DM, Kucukelbir A, Mcauliffe JD. Variational inference: A review for statisticians. J Amer Statist Assoc. 2017;112:859–877.:MR3671776. doi: 10.1080/01621459.2017.1285773. [DOI] [Google Scholar]
- Camacho A, Kucharski A, Aki-Sawyerr Y, White MA, Flasche S, Baguelin M, Pollington T, Carney JR, Glover R, et al. Temporal changes in Ebola transmission in Sierra Leone and implications for control requirements: A real-time modelling study. PLoS Curr. 2015;7 doi: 10.1371/currents.outbreaks.406ae55e83ec0b5193e30856b9235ed2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter J, Clifford P, Fearnhead P. Improved particle filter for nonlinear problems. IEE Proc Radar Sonar Navig. 1999;146:2+. [Google Scholar]
- Cauchemez S, BoëLle PY, Thomas G, Valleron AJ. Estimating in real time the efficacy of measures to control emerging communicable diseases. Am J Epidemiol. 2006;164:591–597. doi: 10.1093/aje/kwj274. [DOI] [PubMed] [Google Scholar]
- Chopin N. A sequential particle filter method for static models. Biometrika. 2002;89:539–551.:MR1929161. doi: 10.1093/biomet/89.3.539. [DOI] [Google Scholar]
- Czado C, Gneiting T, Held L. Predictive model assessment for count data. Biometrics. 2009;65:1254–1261.:MR2756513. doi: 10.1111/j.1541-0420.2009.01191.x. [DOI] [PubMed] [Google Scholar]
- Dawid AP. Statistical theory. The prequential approach. J Roy Statist Soc Ser A. 1984;147:278–292.:MR0763811. doi: 10.2307/2981683. [DOI] [Google Scholar]
- Del Moral P, Doucet A, Jasra A. Sequential Monte Carlo samplers. J R Stat Soc Ser B Stat Methodol. 2006;68:411–436.:MR2278333. doi: 10.1111/j.1467-9868.2006.00553.x. [DOI] [Google Scholar]
- Donner A, Koval JJ. The estimation of intraclass correlation in the analysis of family data. Biometrics. 1980;36:19–25. [PubMed] [Google Scholar]
- Doucet A, Johansen AM. The Oxford Handbook of Nonlinear Filtering. Oxford Univ. Press; Oxford: 2011. A tutorial on particle filtering and smoothing: Fifteen years later; pp. 656–704.MR2884612 [Google Scholar]
- Dukic V, Lopes HF, Polson NG. Tracking epidemics with Google Flu Trends data and a state-space SEIR model. J Amer Statist Assoc. 2012;107:1410–1426.:MR3036404. doi: 10.1080/01621459.2012.713876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dureau J, Kalogeropoulos K, Baguelin M. Capturing the time-varying drivers of an epidemic using stochastic dynamical systems. Biostatistics. 2013;14:541–555. doi: 10.1093/biostatistics/kxs052. [DOI] [PubMed] [Google Scholar]
- Farah M, Birrell P, Conti S, De Angelis D. Bayesian emulation and calibration of a dynamic epidemic model for A/H1N1 influenza. J Amer Statist Assoc. 2014;109:1398–1411.:MR3293599. doi: 10.1080/01621459.2014.934453. [DOI] [Google Scholar]
- Fearnhead P. Markov chain Monte Carlo, sufficient statistics, and particle filters. J Comput Graph Statist. 2002;11:848–862.:MR1951601. doi: 10.1198/106186002321018821. [DOI] [Google Scholar]
- Fearnhead P, Taylor BM. An adaptive sequential Monte Carlo sampler. Bayesian Anal. 2013;8:411–438.:MR3066947. doi: 10.1214/13-BA814. [DOI] [Google Scholar]
- Funk S, Camacho A, Kucharski AJ, Eggo RM, Edmunds WJ. Real-time forecasting of infectious disease dynamics with a stochastic semi-mechanistic model. Epidemics. 2018;22:56–61. doi: 10.1016/j.epidem.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer CJ. Markov chain Monte Carlo maximum likelihood; Computing Science and Statistics: The 23rd Symposium on the Interface; Fairfax Station, VA: Interface Foundation of North America; 1991. pp. 156–163. [Google Scholar]
- Gilks WR, Berzuini C. Following a moving target—Monte Carlo inference for dynamic Bayesian models. J R Stat Soc Ser B Stat Methodol. 2001;63:127–146.:MR1811995. doi: 10.1111/1467-9868.00280. [DOI] [Google Scholar]
- Girolami M, Calderhead B. Riemann manifold Langevin and Hamiltonian Monte Carlo methods. J R Stat Soc Ser B Stat Methodol. 2011;73:123–214.:MR2814492. doi: 10.1111/j.1467-9868.2010.00765.x. [DOI] [Google Scholar]
- Gneiting T, Raftery AE. Strictly proper scoring rules, prediction, and estimation. J Amer Statist Assoc. 2007;102:359–378.:MR2345548. doi: 10.1198/016214506000001437. [DOI] [Google Scholar]
- Gordon NJ, Salmond DJ, Smith AFM. Novel approach to nonlinear/non-Gaussian Bayesian state estimation. IEE Proc-F. 1993;140:107–113. [Google Scholar]
- Held L, Meyer S, Bracher J. Probabilistic forecasting in infectious disease epidemiology: The 13th Armitage lecture. Stat Med. 2017;36:3443–3460.:MR3696502. doi: 10.1002/sim.7363. [DOI] [PubMed] [Google Scholar]
- Jasra A, Stephens DA, Holmes CC. On population-based simulation for static inference. Stat Comput. 2007;17:263–279.:MR2405807. doi: 10.1007/s11222-007-9028-9. [DOI] [Google Scholar]
- Jasra A, Stephens DA, Doucet A, Tsagaris T. Inference for Lévy-driven stochastic volatility models via adaptive sequential Monte Carlo. Scand J Stat. 2011;38:1–22.:MR2760137. doi: 10.1111/j.1467-9469.2010.00723.x. [DOI] [Google Scholar]
- Jewell CP, Kypraios T, Christley RM, Roberts GO. A novel approach to real-time risk prediction for emerging infectious diseases: A case study in Avian Influenza H5N1. Prev vet med. 2009;91:19–28. doi: 10.1016/j.prevetmed.2009.05.019. [DOI] [PubMed] [Google Scholar]
- Kantas N, Beskos A, Jasra A. Sequential Monte Carlo methods for high-dimensional inverse problems: A case study for the Navier-Stokes equations. SIAM/ASA J Uncertain Quantificat. 2014;2:464–489.:MR3283917. doi: 10.1137/130930364. [DOI] [Google Scholar]
- Konishi S, Khatri CG, Rao CR. Inferences on multivariate measures of interclass and intraclass correlations in familial data. J Roy Statist Soc Ser B. 1991;53:649–659.:MR1125722 [Google Scholar]
- Liang F, Wong WH. Real-parameter evolutionary Monte Carlo with applications to Bayesian mixture models. J Amer Statist Assoc. 2001;96:653–666.:MR1946432. doi: 10.1198/016214501753168325. [DOI] [Google Scholar]
- Liu JS, Chen R. Blind deconvolution via sequential imputations. J Amer Statist Assoc. 1995;90:567–576.:MR3363399. doi: 10.1080/01621459.1995.10476549. [DOI] [Google Scholar]
- Liu JS, Chen R. Sequential Monte Carlo methods for dynamic systems. J Amer Statist Assoc. 1998;93:1032–1044.:MR1649198. doi: 10.2307/2669847. [DOI] [Google Scholar]
- Meester R, De Koning J, De Jong MCM, Diekmann O. Modeling and real-time prediction of classical swine fever epidemics. Biometrics. 2002;58:178–184.:MR1891377. doi: 10.1111/j.0006-341X.2002.00178.x. [DOI] [PubMed] [Google Scholar]
- Neal RM. Sampling from multimodal distributions using tempered transitions. Stat Comput. 1996;6:353–366. [Google Scholar]
- Nemeth C, Fearnhead P, Mihaylova L. Sequential Monte Carlo methods for state and parameter estimation in abruptly changing environments. IEEE Trans Signal Process. 2014;62:1245–1255.:MR3168149. doi: 10.1109/TSP.2013.2296278. [DOI] [Google Scholar]
- Ong JBS, Chen MI-C, Cook AR, Chyi H, Lee VJ, Pin RT, Ananth P, Gan L. Real-time epidemic monitoring and forecasting of H1N1-2009 using influenza-like illness from general practice and family doctor clinics in Singapore. PLoS ONE. 2010;5:e10036. doi: 10.1371/journal.pone.0010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts GO, Rosenthal JS. Optimal scaling for various Metropolis-Hastings algorithms. Statist Sci. 2001;16:351–367.:MR1888450. doi: 10.1214/ss/1015346320. [DOI] [Google Scholar]
- Schuster I, Strathmann H, Paige B, Sejdinovic D. In: Machine Learning and Knowledge Discovery in Databases. Ceci M, HollméN J, Todorovski L, Vens C, DžEroski S, editors. Springer; Cham: 2017. Kernel sequential Monte Carlo; pp. 390–409. [Google Scholar]
- Scientific Pandemic Influenza Advisory Committee: Subgroup On Modelling. Modelling Summary. [Accessed 4 February, 2016];SPI-M-O Committee document. 2011 [Google Scholar]
- Seillier-Moiseiwitsch F, Dawid AP. On testing the validity of sequential probability forecasts. J Amer Statist Assoc. 1993;88:355–359.:MR1212496 [Google Scholar]
- Shaman J, Karspeck A. Forecasting seasonal outbreaks of influenza. Proc Natn Acad Sci USA. 2012;109:20425–20430. doi: 10.1073/pnas.1208772109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherlock C, Fearnhead P, Androberts GO. The random walk Metropolis: Linking theory and practice through a case study. Statist Sci. 2010;25:172–190.:MR2789988. doi: 10.1214/10-STS327. [DOI] [Google Scholar]
- Shubin M, Lebedev A, LyytikÄInen O, Auranen K. Revealing the true incidence of pandemic A(H1N1)pdm09 influenza in Finland during the first two seasons—an analysis based on a dynamic transmission model. PLoS Comput Biol. 2016;12:1–3. doi: 10.1371/journal.pcbi.1004803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skvortsov A, Andristic B. Monitoring and prediction of an epidemic outbreak using syndromic observations. Math Biosci. 2012;240:12–19.:MR2974537. doi: 10.1016/j.mbs.2012.05.010. [DOI] [PubMed] [Google Scholar]
- Sokal RR, Rohlf F. Biometry. 2nd. Vol. 668 WH Feeman and Company; New York: 1981. [Google Scholar]
- Te Beest DE, Birrell PJ, Wallinga J, Angelis DD, Van Boven M. Joint modelling of serological and hospitalization data reveals that high levels of pre-existing immunity and school holidays shaped the influenza A pandemic of 2009 in the Netherlands. J R Soc Interface. 2015;12 doi: 10.1098/rsif.2014.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viboud C, Sun K, Gaffey R, Ajelli M, Fumanelli L, Merler S, Zhang Q, Chowell G, Simonsen L, et al. The RAPIDD Ebola forecasting challenge: Synthesis and lessons learnt. Epidemics. 2018;22:13–21. doi: 10.1016/j.epidem.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallinga J, Teunis P. Different epidemic curves for severe acute respiratory syndrome reveal similar impacts of control measures. Am J Epidemiol. 2004;160:509–516. doi: 10.1093/aje/kwh255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M. Mixtures models, Monte Carlo, Bayesian updating and dynamic models. Computer Science and Statistics. 1993;24:325–333. [Google Scholar]
- Whiteley N, Johansen AM, Godsill S. Monte Carlo filtering of piecewise deterministic processes. J Comput Graph Statist. 2011;20:119–139.:MR2816541. doi: 10.1198/jcgs.2009.08052. [DOI] [Google Scholar]
- Wu JT, Cowling BJ, Lau EHY, Ip DKM, Ho LM, Tsang T, Chuang SK, Le-Ung PY, Lo SV, et al. School closure and mitigation of pandemic (H1N1) 2009, Hong Kong. Emerg Infec Dis. 2010;16:538–541. doi: 10.3201/eid1603.091216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.