Abstract
Mako is a software tool that converts microbiome data and networks into a graph database and visualises query results, thus allowing users without programming knowledge to carry out network-based queries. Mako is accompanied by a database compiled from 60 microbiome studies that is easily extended with the user’s own data. We illustrate mako’s strengths by enumerating association partners linked to propionate production and comparing frequencies of different network motifs across habitat types.
Microbial association network construction and analysis have become well-established tools for the study of microbiomes [1]. While meta-analytical approaches are increasingly adopted in a range of studies [2,3], no framework exists that provides flexible and intuitive methods for the storage of microbiome-derived networks. Network databases and query languages such as neo4J and Cypher implement an alternative data storage and query paradigm that has not been fully exploited for microbiome data yet. Here, we present mako (microbial associations catalog), which fills this gap, thereby enabling rapid and simple construction of network databases from microbiome data and metadata.
For macro-ecology, interaction databases such as Mangal provide a data specification that permits data sharing and reuse in R and Julia [4]. String-DB fulfils a similar role for protein interaction databases [5]. Such databases include validated and observed interactions between species or between proteins. Currently, no such database has been published for microbiome data (although a static database with a web interface appears to be under development by Hu et al., http://www.microbialnet.org/mind_home.html). However, a static network database does not accommodate the flexibility required for microbial associations, since inferred associations change depending on the experimental design used to generate the abundance data and the parameters of the tools used to infer the networks. Additionally, microbial network inference is a rapidly developing field and no standard protocols exist for the inference of these networks. Consequently, the analysis of microbial associations demands a more flexible and local setup that can integrate a wealth of data. To date, this requires a significant amount of data processing and maintenance efforts.
mako provides an interface between standard microbiome formats and Neo4j graph databases via a database schema based on semantic web ontologies (Figure 1). This software package includes a range of methods to interact with Neo4j databases and the pattern-based query language Cypher, requiring only rudimentary computational skills. Additionally, mako includes a curated database derived from 60 separate data sets downloaded from Qiita [6], a platform for hosting microbial studies that facilitates meta-analyses of this scale.
mako features.
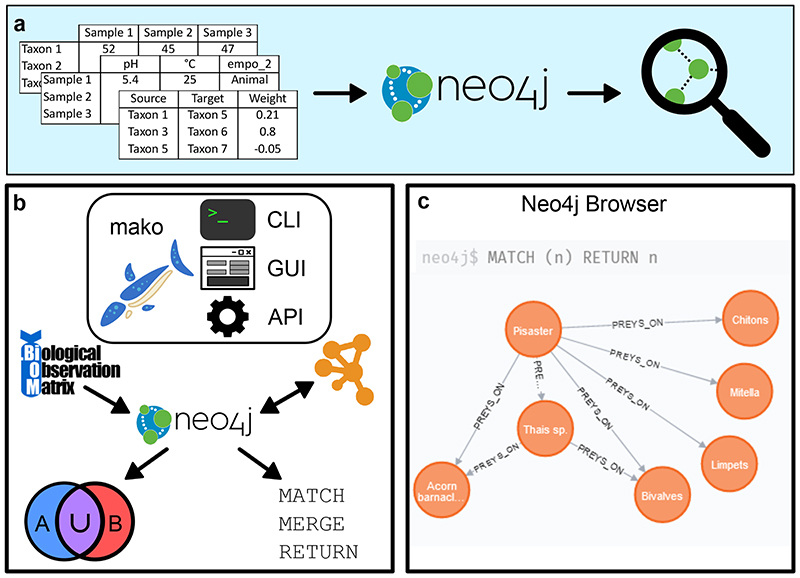
a The mako software supports uploading of tables and other file formats (in particular networks) to a Neo4j database, which can then be used to carry out meta-analyses. b The software includes a command line interface (CLI), graphical user interface (GUI) and application programming interface (API) which can all be used to run the mako functionality. For example, mako can port from BIOM files to a Neo4j database, export to Cytoscape, carry out set operations and several other query-based tasks. c Screenshot of the Neo4j browser, which can be used to run queries and access the database. This screenshot displays a part Paine’s food web including the keystone species Pisaster ochraceus.
Network databases such as Neo4j differ from relational databases in that they do not store data as tables but as networks (graphs) [7]. While association networks only contain links between different microbial taxa or abiotic factors, a graph database containing those association networks can contain different pieces of information in its graph as well. For example, a taxon’s phylum can be stored as a node in a graph database. The taxon’s phylum is then represented as an edge connecting the taxon identifier to the phylum. An edge is therefore a relationship containing two nodes - two pieces of information - and can have multiple meanings, depending on its label. In a graph database, an edge can be an association or an interaction, but it may also be the observation linking a taxon to a sample. Consequently, graph database representations of biological data look more like their conceptual representation. To illustrate, a taxonomic tree can be stored as a tree, a type of graph, rather than a table.
Most standard biological formats use tables of some form and therefore need to be processed. These processes, also referred to as ETL (extract, transform, load) processes, can be combined with semantic web technologies such as OWL (Ontology Web Language) to integrate and structure data from multiple sources [8]. Ontologies, such as the National Cancer Institute (NCI) thesaurus that mako uses for its OWL file, are controlled vocabularies that are machine-and human-readable to improve the accessibility of biological data [9]. Hence, mako includes a conceptual model written in OWL that can be used to check the integrity of the Neo4j database.
To illustrate the speed and ease of use of Cypher, the query language used to access Neo4j databases, we curated 60 BIOM files and generated 60 networks with FlashWeave, constructing a final Neo4j database that contains over 5 million relationships, connecting more than 100.000 nodes. This curated database is provided as a supplementary file with mako. On a Dell Precision laptop with a 3.00 GHz Intel Xeon CPU (sixth generation), an M.2 SSD and 32 GB of RAM, uploading of all BIOM files and networks took approximately 12 minutes. Once this database was initialized, restoring the 1.322 GB database from a database dump took 11 seconds on the same machine.
Pattern-based language like Cypher can be used to identify motifs. Network motifs may indicate the presence of specific dynamic behaviours. For instance, in microbial communities, a negative circuit implementing a rock-paper-scissors game can promote diversity [10]. For association networks, many of these well-characterized motifs are not defined since they consist of directed relationships, while association networks are undirected. Other motifs have not been investigated extensively [11].
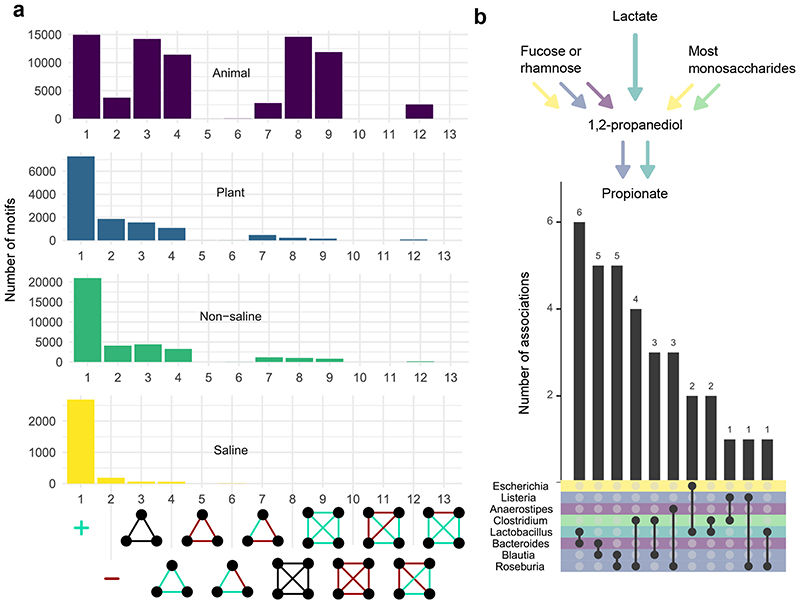
Cypher facilitates the investigation of such motifs in large data sets. To demonstrate this, we wrote Cypher queries to search for motifs that represent either 3-node or 4-node subgraphs (Figure 2a). We used our 60-network database to explore certain combinations of weights in maximally-connected subgraphs across four EMPO (Earth Microbiome Project Ontology) terms [12]. Our results suggest that there are distinct differences between the four EMPO terms. Firstly, there are more associations attributed to non-saline and animal networks, as could be expected given that 33 and 15 of our 60 data sets had the Animal or Non-saline EMPO terms respectively. Secondly, animal-derived networks appear to have higher motif density. These results are supported by previous studies that found differences in edge density across biomes [13, 14]. However, our approach did not allow us to fully explore differences between plant and non-saline microbiomes, since both studies may contain soil samples. The motifs display another striking pattern: internally consistent motifs are overrepresented. Such motifs are balanced graphs, meaning that the product of their edge signs around every cycle is equal to 1. In microbiological terms, balanced graphs suggest that two microbes with a positive association between them are more likely to share negative relationships with other microbes.
Motif identification with Neo4j.
a Overview of motifs in database. The motif frequencies are shown across four EMP ontology terms, which were used to separate 60 data sets into distinct groups. The motifs shown here were identified from 60 FlashWeave association networks. The animal-associated networks differ from the others in the number of densely connected cliques, but only for cliques with mostly positively-weighted edges. For the black motifs, association weights were not taken into consideration, while the other motifs include specific patterns of weights. b Associations between taxa that have previously been linked to propionate synthesis via 1,2-propanediol [15]. Genera are coloured by the steps in the pathway that they have previously been linked to.
In addition to assessing motif structure, taxonomic information can be included in queries. Formation of propionate via 1,2-propanediol can involve different species, depending on the substrate [15]. Here, we looked at three different groups of substrates (sugars, lactate and deoxy sugars fucose and rhamnose), which can be converted to 1,2-propanediol via different genera (Figure 2b). We queried for associations between these and genera that convert 1,2-propanediol to propionate. We found several associations between genera that could theoretically produce propionate together from fucose or rhamnose, but only one association was found that could be linked to lactate degradation. For sugars, only Clostridium and Escherichia could be linked to propionate producers. Overall, the queries suggest that specific substrates could not be linked to associations with a single propionate producer.
To support the use of Neo4j databases for biological data, mako includes several modules. Specifically, the following features are supported:
Imports of biological data. Several types of biological data, commonly used in the analysis of association networks, can be imported via a simple application programming interface (API), command line interface (CLI) or graphical user interface (GUI). These include BIOM files, tab-delimited files, network files and edge lists of custom properties. These data are stored according to a clearly documented database schema, which supports development of custom Cypher queries. The mako software supports batch uploading of node data and uses indices to rapidly speed up database access operations.
Meta-analytical approaches. While the full scope of meta-analytical approaches is too broad to be included in this software package, several methods have been included that are potentially useful. These include options to make “overlay” networks from combinations of networks, such as intersections, unions, or differences. The software also includes methods for merging networks by taxonomic labels, so structure at higher taxonomic levels can be investigated.
Integration with external software, including Cytoscape. The Neo4j database is one large graph, that includes both metadata and network data. Loading this entire database into Cytoscape is not feasible, not only because the number of nodes in Neo4j database can far exceed the number of nodes that can be rendered with Cytoscape, but also because the data model for Cytoscape includes fundamentally different representations of node and edge metadata. The mako software can extract more common network representations from the Neo4j database and import these directly into Cytoscape via an HTTP request, or supply these to other network analysis tools.
Flexible query design. The database schema used by this software has been designed to accommodate a range of biological data. Any data that can be rewritten to an edge list (e.g. property-taxon or property-edge) can be imported into the database. The online manual at at https://ramellose.github.io/mako_docs/ contains a brief introduction to Neo4j database schema design, a detailed explanation of the database schema used by the software and some simple explanations of Cypher queries. Additionally, all scripts and queries used for examples shown in this manuscript have been provided as supplementary files to demonstrate flexibility of the Cypher query language for biological data.
The mako toolbox does not fully exploit the potential of graph databases. For instance, the database schema could be extended to ease standardized queries for different types of data. Especially for large ‘omics data, the increase in speed that could result from additional standardization is likely to be significant. Moreover, the analysis of such linked data in microbiome research is not well established and may require novel analytical methods. In conclusion, while still leaving room for improvement, the mako toolbox makes an important contribution to the adoption of graph databases in microbiome research.
Methods
QIITA-derived networks included with mako
To demonstrate the Neo4j database schema and mako’s functionality, we used 60 files that were downloaded from QIITA. Processed files and derived microbial association networks have been included as part of the database dump.
Processing 60 data sets from QIITA
We downloaded 60 files that could roughly be assigned to the following three categories: human gut microbiome, soil microbiome (including terrestrial plants) and aquatic microbiome. Of the 60 files, 37 had an empo_2 column containing EMPO level 2 annotations. For each of these, the most common empo_2 annotation was used to label the network. The remainder had empo_2 annotations added manually; these were either stored in a differently named column or derived from the study description. After manual curation, 33 data sets had the EMPO 2 label “Animal”, 15 were labelled “Non-saline”, 8 were labelled “Plant” and 4 were labelled “Saline”.
Given the richness of metadata available in QIITA, this data was curated first to speed up database operations. Values that were identical across a data set were filtered. We also chose to remove values that we considered less relevant for future analysis; these included the barcode sequence, the collection timestamp, the run date and the run prefix.
To omit statistical issues with missing values, columns with more than 3 missing values were also filtered. For one specific data set, columns with only “missing”, “not applicable” and “not collected” entries were removed. Finally, we found that columns with mostly numeric values contained characters referring to missing samples; such columns were also removed if they had more than 5 values containing characters.
For a full overview of all downloaded files, we refer to Supplementary Table 1.
Running network inference with FlashWeave
Taxon abundances were first collapsed to the genus level identifier; only taxa that were present in >20% of samples were retained. Taxa that fell below the prevalence threshold were merged into a single synthetic taxon so that the total sum of abundances was not disturbed. The resulting abundance tables were used to infer networks with FlashWeave version 0.18.0, Julia version 1.5.3
[17]. FlashWeave was set to sensitive, non-heterogeneous and to only consider associations with more than 10 observations. Since FlashWeave incorporates a clr transformation, it handles compositional data appropriately without additional pre-processing steps.
Motif analysis with Neo4j
In a Python 3.6.3 environment, we used the mako software package in combination with pandas version 1.1.5 to generate tables of motif counts. The Python script used to query motifs is available via the mako Zenodo repository [18]. Results were further analyzed in R version 4.0.2. Figures were generated with ggplot version 3.3.2, with viridis version 0.5.1 for colour palettes and UpSetR version 1.4.0 for visualizing associations [19].
Creating a database dump with Neo4j
The mako software was used to upload processed BIOM files and network files to a local Neo4j database. Next, the neo4j-admin script was used to create a dump of the complete database. This file, called mako.dump, has also been made available through Zenodo [18]. The file can be used to rapidly restore the complete database.
Database schema
A core component of mako is a database schema constructed from several OWL terms. This database schema also defines constraints for the data and provides guidance on how data needs to be uploaded to become easily accessible. Most functionality of the API uses the schema to construct queries for uploading BIOM files, checking edge weights or finding associations between related species.
To properly integrate with Neo4j’s ability to work with ontology web language (OWL), each node and relationship label is derived from an existing ontology, the NCI Thesaurus [20]. However, since relationships are not well-defined across biological ontologies, constraints for relationships were defined manually with Protégé [9]. For each relationship, domains and ranges (target and source nodes) were specified. While these axioms are not added to Neo4j as actual constraints, mako can run checks to see if domain and range axioms are violated by any nodes or relationships in the database.
The complete database schema represents associations as separate nodes, rather than a relationship between two taxa. This makes it easier to query association properties. This database schema represents a trade-off between both flexibility and speed since the Cypher query planner needs to scan fewer nodes if the labels are provided. Consequently, separate labels are used to indicate pieces of information common to all count tables and microbial association networks. Neo4j offers the opportunity for advanced users to extend the database schema with new labels, so they can write faster queries for specific metadata.
Interfacing with the Neo4j database
The mako software was developed in Python 3.6 and uses the Python API provided by Neo4j [7]. Simply put, the software converts each of the input files (BIOM, tab-delimited files, network files, edge lists) into a list of dictionaries. Each dictionary contains the necessary information, e.g. node names or other properties, that needs to be provided to parameterized queries. Rather than running individual queries, all larger queries are run as batch queries, so that the Cypher query planner can reuse the query plan each time.
Since node labels cannot be added as query parameters, separate queries need to be written for each unique node label. In some cases, e.g. taxonomies, queries are constructed dynamically in Python and then used to construct separate batch queries for each taxonomy node. This permits reuse of query designs across node labels.
Manipulating networks in Neo4j and Python
Some utilities are available for meaningfully interacting with the Neo4j database. For example, mako queries edges that are present in multiple networks and uses this to construct ensemble networks (e.g. intersection, union or difference operations). The software can also find pairs of edges, i.e. edges that have identical taxonomic labels at both ends and merge these. Additionally, mako includes functionality for exporting networks to Cytoscape. While a Cytoscape Neo4j plugin is available, this does not accommodate translations from one database schema to another [21]. The Neo4j database schema used by mako was specifically designed to support meta-analyses of large amounts of information and therefore supports databases with millions of nodes. In contrast, Cytoscape renders networks (partially) and may therefore not be able to represent a full database stored in Neo4j. For this reason, mako includes scripts that extract (specific) networks from the Neo4j database, convert them to a Cytoscape-compatible json file and uses the Python requests HTTP package to transmit this json file to a running instance of Cytoscape.
One potential issue with Cypher is that patterns can be matched in several directions; each time the query planner comes across the pattern, the nodes are returned, even if they were already part of the same pattern but in a different order. Consequently, unlike SQL, some extra parsing of the results can be required. For example, the pattern (a)–(b)–(c)–(a) can be matched to (Node 1)–(Node 2)–(Node 3)–(Node 1), but also to (Node 2)–(Node 3)–(Node 1)–(Node 2). Where applicable, such query parsing is implemented in mako. Example functions to collect unique patterns have also been included in the supplementary script available through Zenodo [18].
Supplementary Material
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Acknowledgements
This project was supported by the KU Leuven under grant number STG/16/006. KF has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme under grant agreement No 801747.
Footnotes
Author Contributions
LR developed the software, carried out the case studies and drafted the manuscript. KF contributed to the design of the software and the case studies, and provided substantial revisions of the the manuscript. Both authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Data availability
The complete Neo4j database used in this manuscript has been used to generate a Neo4j dump file. An archived version of this file has been submitted to Zenodo [18]. Instructions for reconstructing this database are available in mako’s documentation. All BIOM files were downloaded from Qiita [6] and a full overview of these studies can be found in Supplementary Table 1.
Code availability
The latest version of the mako software can be downloaded via
https://github.com/ramellose/mako/. An archived version, including the script used to quantify motifs in the Neo4j database, has been submitted to Zenodo [18]. All mako source code and the included script are available under the Apache License 2.0. An extensive manual for running mako and Neo4j (via Docker) is available via https://ramellose.github.io/mako_docs/. Additionally, an accompanying compute capsule allows a demo of mako and a Neo4j database to be run without requiring any installations [22].
References
- [1].Röttjers L, Faust K. From hairballs to hypotheses–biological insights from microbial networks. FEMS microbiology reviews. 2018;42:761–780. doi: 10.1093/femsre/fuy030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Jackson MA, et al. Detection of stable community structures within gut microbiota co-occurrence networks from different human populations. PeerJ. 2018;6:e4303. doi: 10.7717/peerj.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wang H, et al. Combined use of network inference tools identifies ecologically meaningful bacterial associations in a paddy soil. Soil Biology and Biochemistry. 2017;105:227–235. [Google Scholar]
- [4].Poisot T, et al. mangal–making ecological network analysis simple. Ecography. 2016;39:384–390. [Google Scholar]
- [5].Szklarczyk D, et al. String v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic acids research. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gonzalez A, et al. Qiita: rapid, web-enabled microbiome meta-analysis. Nature methods. 2018;15:796–798. doi: 10.1038/s41592-018-0141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Miller JJ. Graph database applications and concepts with neo4j; Proceedings of the Southern Association for Information Systems Conference; Atlanta, GA, USA. 2013. [Google Scholar]
- [8].Bansal SK. Towards a semantic extract-transform-load (etl) framework for big data integration; 2014 IEEE International Congress on Big Data; 2014. pp. 522–529. [Google Scholar]
- [9].Noy NF, et al. Creating semantic web contents with protege-2000. IEEE intelligent systems. 2001;16:60–71. [Google Scholar]
- [10].Kerr B, Riley MA, Feldman MW, Bohannan BJ. Local dispersal promotes biodiversity in a real-life game of rock–paper–scissors. Nature. 2002;418:171–174. doi: 10.1038/nature00823. [DOI] [PubMed] [Google Scholar]
- [11].Ma ZS, Ye D. Trios—promising in silico biomarkers for differentiating the effect of disease on the human microbiome network. Scientific reports. 2017;7:1–9. doi: 10.1038/s41598-017-12959-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Thompson LR, et al. A communal catalogue reveals earth’s multiscale microbial diversity. Nature. 2017;551:457. doi: 10.1038/nature24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ma B, et al. Earth microbial co-occurrence network reveals interconnection pattern across microbiomes. Microbiome. 2020;8:1–12. doi: 10.1186/s40168-020-00857-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Faust K, et al. Cross-biome comparison of microbial association networks. Frontiers in microbiology. 2015;6:1200. doi: 10.3389/fmicb.2015.01200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Louis P, Flint HJ. Formation of propionate and butyrate by the human colonic microbiota. Environmental microbiology. 2017;19:29–41. doi: 10.1111/1462-2920.13589. [DOI] [PubMed] [Google Scholar]
- [17].Tackmann J, Rodrigues JFM, von Mering C. Rapid inference of direct interactions in large-scale ecological networks from heterogeneous microbial sequencing data. Cell systems. 2019;9:286–296. doi: 10.1016/j.cels.2019.08.002. [DOI] [PubMed] [Google Scholar]
- [18].Röttjers L, Faust K. Fast and flexible analysis of linked microbiome data with mako. Zenodo. 2021 doi: 10.5281/zenodo.4946425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Conway JR, Lex A, Gehlenborg N. Upsetr: an r package for the visualization of intersecting sets and their properties. Bioinformatics. 2017;33:2938–2940. doi: 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sioutos N, et al. Nci thesaurus: a semantic model integrating cancer-related clinical and molecular information. Journal of biomedical informatics. 2007;40:30–43. doi: 10.1016/j.jbi.2006.02.013. [DOI] [PubMed] [Google Scholar]
- [21].Summer G, et al. cyneo4j: connecting neo4j and cytoscape. Bioinformatics. 2015;31:3868–3869. doi: 10.1093/bioinformatics/btv460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Röttjers L, Faust K. Fast and flexible analysis of linked microbiome data with mako. Code Ocean. 2021 doi: 10.24433/CO.0482418.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete Neo4j database used in this manuscript has been used to generate a Neo4j dump file. An archived version of this file has been submitted to Zenodo [18]. Instructions for reconstructing this database are available in mako’s documentation. All BIOM files were downloaded from Qiita [6] and a full overview of these studies can be found in Supplementary Table 1.